
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 26 June 2020
Sec. Fungi and Their Interactions
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01392
 Xin Liu1,2,3†
Xin Liu1,2,3† Yichen Jiang1,4†
Yichen Jiang1,4† Dan He1
Dan He1 Xin Fang1,3
Xin Fang1,3 Jianhong Xu1,3
Jianhong Xu1,3 Yin-Won Lee1,5
Yin-Won Lee1,5 Nancy P. Keller2,6*
Nancy P. Keller2,6* Jianrong Shi1,3*
Jianrong Shi1,3*All organisms must secure essential trace elements (e.g., Cu) for survival and reproduction. However, excess trace element accumulation in cells is highly toxic. The maintenance of copper (Cu) homeostasis has been extensively studied in mammals, bacteria, and yeast but not in plant pathogens. In this study, we investigated the molecular mechanisms of copper tolerance in Fusarium graminearum, the important wheat head scab fungus. RNA-seq revealed induced expression of the P-type ATPase transporter FgCrpA and metallothionein (MT) FgCrdA after excess Cu treatment. Deletion of FgCrpA but not FgCrdA resulted in reduced tolerance to Cu toxicity. The “Cu fist” transcription factor FgAceA was involved in Cu detoxification through activation of FgCrpA. △FgAceA was more sensitive to copper toxicity than △FgCrpA and overexpression of FgCrpA restored copper tolerance in △FgAceA. FgAceA negatively regulated aurofusarin production and its biosynthetic gene expression. △FgCrpA and △FgAceA were reduced in virulence in flowering wheat heads and synthesized decreased amounts of the mycotoxin deoxynivalenol when challenged with excess Cu. Taken together, these results suggest that mediation of Cu tolerance in F. graminearum mainly relies on the Cu efflux pump and that FgAceA governs Cu detoxification through activation of FgCrpA.
Copper (Cu) is an essential trace element that cycles between Cu1+ (reduced) and Cu2+ (oxidized) states in virtually all organisms (Dalecki et al., 2017; Gerwien et al., 2018). By coordinating with proteins and serving as a redox cofactor, Cu confers changes in protein structure, catalytic activity and protein-protein interaction, thus controlling a variety of cellular biochemical processes (Kim et al., 2008). However, these same properties can be highly detrimental to microorganisms when Cu is present in excess. Free Cu can lead to oxidative stress by reacting with reactive oxygen species (ROS) to generate hydroxyl radicals that damage many biomolecules, including DNA, membrane lipids and proteins (Kim et al., 2008). Furthermore, Cu can disrupt the active sites of metallothioneins (MTs), such as iron-sulfur enzymes, and displace other metals from their cognate enzymes, leading to their inactivation (Besold et al., 2016).
Given its toxic properties to microorganisms, Cu-containing compounds have been historically used as antimicrobials both in healthcare and agriculture (Dollwet and Sorenson, 1985; Grass et al., 2011; Festa et al., 2014; Michels et al., 2015). There is an emerging concept in innate immunity that animal hosts intentionally exploit copper toxicity as a weapon to combat invading microbes. For example, macrophages can attack invading microbes with high Cu levels, and Cu is elevated at sites in the lung and serum during infection (Besold et al., 2016).
In the host-microbe battle, on the other hand, the microbial intruder is equipped with a battery of Cu detoxification defenses that promotes survival and colonization in the host, including Cu binding MTs and Cu exporting P-type ATPases to defend against toxic levels of Cu. MTs are cysteine (Cys)-rich low molecular weight polypeptides found in organisms from prokaryotes to mammals (Ding et al., 2014). MTs are utilized in response to high Cu levels and detoxify Cu through binding the Cys of Cu via Cys-thiolate bonds (Palacios et al., 2011; Babula et al., 2012). P-type ATPases are heavy metal translocators conserved in all biological kingdoms that regulate intercellular Cu levels (Odermatt et al., 1993; Solioz and Odermatt, 1995; Ladomersky and Petris, 2015; Antsotegi-Uskola et al., 2017; Wiemann et al., 2017).
In the model yeast Saccharomyces cerevisiae, the metallo-regulation of the Cu-binding transcription factor (TF) Ace1 functions in Cu-replete cells and regulates the expression of Cu detoxification-related genes, including those encoding MTs (Cup1 and Crs5) and Cu/Zn super oxide dismutase, SOD (SOD1) under excess Cu conditions (Keller et al., 2005; Cyert and Philpott, 2013). In the human fungal pathogen Cryptococcus neoformans, CMT1 and CMT2 genes encode cysteine-rich Cu-binding MTs, whose expression levels are dramatically upregulated in response to elevated Cu levels, play essential roles in Cu detoxification (Ding et al., 2011) and are critical for fungal virulence (Ding et al., 2013). While MTs have been proven to be the major players in Cu detoxification in S. cerevisiae and C. neoformans, P-type ATPases that pump Cu extracellularly through the transmembrane channel are utilized for Cu export in Candida albicans and Aspergillus spp. In the pathogenic yeast C. albicans, CRP1, which encodes a plasma membrane-localized Cu exporter P-type ATPase (Crp1), is transcriptionally upregulated by high Cu conditions in an Ace1-dependent manner, and deletion of CRP1 resulted in a Cu-sensitive growth phenotype as well as massive intracellular Cu accumulation (Weissman et al., 2000). Recent studies have shown that A. fumigatus, A. nidulans, and A. flavus share a similar Cu detoxification machinery; under excess Cu conditions, the Cu-binding TF AceA induces the expression of the P-type ATPase CrpA for Cu detoxification (Antsotegi-Uskola et al., 2017; Wiemann et al., 2017; Cai et al., 2018; Yang et al., 2018). Both AceA and CrpA are virulence determinants in the human pathogen A. fumigatus (Wiemann et al., 2017). Recently, Yang et al. illustrated that in the crossover pathogen A. flavus, deletion of P-type ATPases (A. flavus contains two, CrpA and CrpB) and the TF AceA resulted in significantly reduced virulence in mice but had no impact on virulence in maize seeds (Yang et al., 2018).
Fusarium graminearum species complex is the major causal agent of Fusarium head blight (FHB) or scab of wheat and barley, which is among the most important diseases of cereal crops worldwide (Bai and Shaner, 2004; Goswami and Kistler, 2004). Epidemics of FHB not only cause huge yield losses but also contaminate grains with harmful trichothecene mycotoxins, threatening the health of both humans and animals (Lombaert et al., 2003; Rasmussen et al., 2003; Tutelyan, 2004; Isebaert et al., 2005; Ji et al., 2014). Despite in-depth studies of copper homeostasis in human pathogens, information on the Cu detoxification machinery and the roles of this machinery in the virulence of plant pathogens is still limited, and thus we examined the possible existence and importance of Cu-exporting mechanisms in the wheat pathogen F. graminearum.
The objectives of this study were to identify and characterize the major factors determining copper detoxification and their possible regulatory mechanisms in F. graminearum. Our results indicate that copper detoxification in the wheat pathogen F. graminearum relies mainly on Cu export similar to that in C. albicans and Aspergillus spp.
Fusarium graminearum strain PH-1 (NRRL 31084) (Cuomo et al., 2007) was used as the parental wild-type. To assess the mycelial growth, the wild-type strain PH-1 and its corresponding mutants were grown on solidified FMM plates (Fusarium minimal medium) (Leslie and Summerell, 2007) amended under different Cu and/or menadione concentrations and incubated at 25°C. Fungal biomass was compared by collecting mycelia from 3-day-old liquid FMM culture incubated at 25°C in a shaker (180 rpm). For conidiation, five 5 mm mycelial plugs of the wild type strain PH-1 and its mutants taken from the edge of a 3-day-old colony were inoculated in a 150 ml triangular flask containing 50 ml of CMC (carboxymethyl cellulose) medium and incubated at 25°C, 180 rpm for 3 days in a shaker with light (Iida et al., 2008; Chen et al., 2018). The number of conidia in CMC medium was counted using a hemacytometer. Conidial germination rates were compared by re-suspending conidia in 2% sucrose solutions amended with or without 20 μM Cu at 25°C for 4 or 6 h. Each experiment was carried out in triplicate and repeated for three times.
Using double-joint PCR strategy (Yu et al., 2004), deletion cassettes for each targeted gene were generated and transformed into the wild type PH-1 by using polyethylene glycol (PEG)-mediated protoplast transformation method (Proctor et al., 1995b). PCR primers used in this study were synthesized by Sangon Biotech (Shanghai, China) and are listed in Supplementary Table S1. Genes were deleted by creating constructs where 5′ and 3′ flanking regions were fused to the hygromycin gene HPH obtained from pBlueScript-hph or the G418 sulfate gene NEO from pBlueScript-neo. Solidified FMM supplemented with hygromycin (100 mg/L) or G418 sulfate (100 mg/L) was used to select transformants. PCR screened-positive transformants were single spore isolated and stored in 15% glycerol at −80°C for further experiments. To construct FgAceAc or FgCrpAc, the full-length DNA of FgAceA or FgCrpA, including their promoter region (about 1.0 kb), was cloned into the pYF11 plasmid (Bruno et al., 2004), and then the recombinant plasmid pYF11-FgAceA or pYF11-FgCrpA was transformed into its corresponding deletion mutant. Southern blot hybridization analysis of the mutants and its wild-type strain was performed using a detection starter kit II according to the manufacturer’s instructions (Roche Diagnostics, Mannheim, Germany).
Predicted protein sequences released in FungiDB1 were aligned with Clustal W (Thompson et al., 1994) and a phylogenetic tree was constructed with MEGA 5.10 software using a neighbor-joining method (Kumar et al., 2008).
Conidia (105) of F. graminearum wild-type strain PH-1 and its corresponding mutants were inoculated into 50 ml liquid FMM in triplicates and cultured at 25°C, 180 rpm for 3 days in a shaker. Mycelia was collected by filtering through sterile miracloth, washed with sterile water and then one half mycelia was grown under the same conditions with the other half shifted to a Cu treatment for several time points. Mycelia was then collected, washed with sterile water, frozen with liquid nitrogen and lyophilized for 24 h. Total RNA of F. graminearum strains was extracted using TaKaRa RNAiso Plus (TaKaRa, Dalian, China) and cDNA was reverse transcripted using TaKaRa PrimeScriptTM RT Master Mix (TaKaRa, Dalian, China). For quantitative Real-Time PCR (qRT-PCR) assays, expression of genes was determined with primer pairs listed in Supplementary Table S1. qRT-PCR amplifications were performed in a LightCycler® 96 (Roche Molecular Systems, Inc.) using the TaKaRa SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) (TaKaRa, Dalian, China).
For RNA-seq analysis, 105 conidia of the wild-type strain PH-1 was inoculated into 50 ml liquid FMM without Cu and cultured at 25°C with shaking (180 rpm) for 3 days with a subsequent shift to Cu-treated condition (100 μM) for 1 h with triplicates as described above. RNA samples were prepared to perform digital transcriptome analysis by the RNA-seq approach (Shanghai Majorbio Bio-pharm Technology Co., Ltd.). The data were analyzed on the free online platform of Majorbio I-Sanger Cloud Platform2.
Conidia (105) of F. graminearum wild-type strain PH-1 and corresponding mutants were inoculated into 50 ml liquid FMM in four replicates and cultured at 25°C, 180 rpm for 3 days in a shaker. Subsequently, Cu was added to one half of the culture at a final concentration of 100 μM and fungi further cultured for 12 h. Mycelia were then collected, washed by sterile water, and lyophilized. Quantification of Cu from F. graminearum strains was carried out according to the method previously described by Shanghai Microspectrum Chemical Technology Service Co., Ltd. (Yang et al., 2018).
Conidia of F. graminearum strains were harvested from 4-day-old CMC cultures, re-suspended in 0.01% (vol/vol) Tween 20 and adjusted to 1 × 105 conidia/ml. Virulence tests were performed using a single floret injection method as previously described (Wu et al., 2005). Briefly, 10 μl of conidial suspension (1 × 105/ml) was injected into a single floret in the central spikelet of single flowering wheat heads of susceptible cultivar Zhenmai 10 at early anthesis with or without 3 g/L Cu fungicide (Nordox Cuprousoxide) spraying. Ten spikes were used for each strain. Infected spikelets in each inoculated wheat head were recorded and photographed 15 days after inoculation. The experiments were repeated three times.
Conidia (105) of F. graminearum wild-type strain PH-1 and corresponding mutants were inoculated into 30 ml trichothecene biosynthesis inducing (TBI) liquid with or without Cu in three replicates and cultured at 28°C for 7 days. The filtrate and fungal mass were then collected separately, frozen with liquid nitrogen and lyophilized for 24 h. The filtrate was re-dissolved in methanol and fungal mass was measured. The amount of DON in each sample was determined using a high-performance liquid chromatography-mass spectrometer/mass spectrometer (HPLC-MS/MS) system (Shimadzu 30A LC system coupled to a Triple Quad 6500 plus, Sciex, United States). Mass spectrometric parameters were according to a previously described method (Dong et al., 2016). The experiment was repeated two times.
All data were presented as the means ± standard deviation (SD). Statistical significance analysis was performed between the wild type parent PH-1 and the deletion mutants with group t-tests with software package SPSS (version 13 for Windows, 2004). A p-value less than 0.05 was labeled as statistically significant.
To gain a genomic perspective of the Cu tolerance mechanism in F. graminearum, we treated the wild-type strain PH-1 with or without 100 μM CuSO4 for 1 h. Subsequently, RNA was extracted and prepared for RNA sequencing (RNA-Seq). All RNA-Seq raw data were deposited at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA)3 under bioproject accession number PRJNA601796.
Overall, by comparing RNA-Seq data, we found that genes encoding putative Cu transporters were significantly differentially regulated following 100 μM CuSO4 treatment. In F. graminearum, six genes encoding putative Cu-importing transporters, including the S. cerevisiae high-affinity Cu transporter Ctr1p ortholog FGRAMPH1_01G09843, the S. cerevisiae high-affinity Cu transporter Ctr2p homologs FGRAMPH1_01G19393 and FGRAMPH1_01G23855, the A. fumigatus Cu transporter CtrA1 orthologs FGRAMPH1_01G10595 and FGRAMPH1_01G13479, and the S. cerevisiae intracellular Cu ATPase Ccc2p ortholog FGRAMPH1_01G09315, were significantly downregulated (Supplementary Table S2). Furthermore, the C. albicans Cu-exporting ATPase Crp1 ortholog FgCrpA (FGRAMPH1_01G10037) and the C. albicans MT Crd2 ortholog FgCrdA (FGRAMPH1_01G09281) were significantly upregulated (Supplementary Table S2). Based on the observations from the RNA-Seq data, we next characterized the possible roles of the F. graminearum Crp1 ortholog (FgCrpA) and the Crd2 ortholog (FgCrdA) in Cu detoxification.
Previous studies have shown that the “Cu fist” DNA binding domain protein AceA was crucial in mediating Cu tolerance in Aspergillus spp. by activating the Cu-exporting transporters. The S. cerevisiae Ace1/YGL166W ortholog FgAceA (FGRAMPH1_01G09843) was identified using BlastP. Phylogenetic analysis showed that the Cu-exporting ATPase FgCrpA, the MT FgCrdA, and the “Cu fist” DNA binding domain protein FgAceA shared high level of sequence identities with their orthologs in yeasts and Aspergillus spp. (Supplementary Figures S1A–C).
Quantitative real-time PCR (qRT-PCR) results showed that the expression of FgCrpA and FgCrdA in F. graminearum was induced rapidly by exposure to 100 μM Cu. FgCrpA reached its maximal expression level after exposure to Cu treatment at 15 min, followed by a decrease in expression (Figure 1A). FgCrdA was also induced by Cu treatment, and its expression displayed a similar pattern as FgCrpA; the maximal expression level was observed at 10 min, followed by a decrease (Figure 1A).
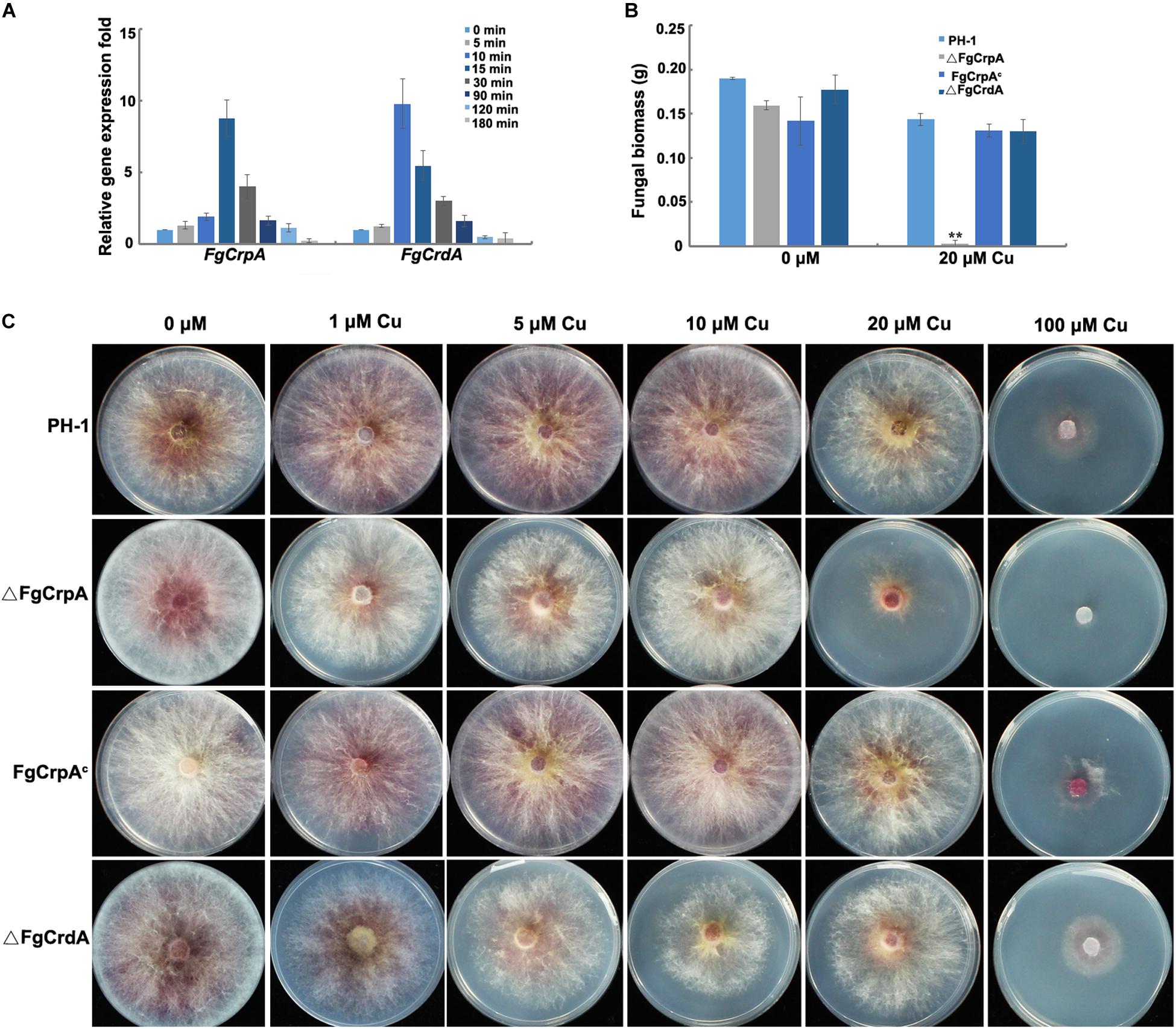
Figure 1. FgCrpA and FgCrdA were induced by Cu treatment and their roles in Cu detoxification. (A) Expression of FgCrpA and FgCrdA in F. graminearum after Cu treatment analyzed by quantitative Real-time PCR assays. The wild-type strain PH-1 was grown in liquid FMM (Cu free) for 3 days at 25°C and then collected, washed using sterile water and transferred to liquid FMM supplemented with CuSO4 at 100 μM for different time courses. (B) Fungal growth was measured by determining the dry weight of the wild-type strain and mutants grown in 50 ml liquid FMM amended with or without CuSO4 at 20 μM in a shaker at 180 rpm at 25°C for 3 days. Bars indicated means and error bars denote standard deviation from three repeated experiments. A t-test was performed to determine significant differences, **p < 0.01. (C) Phenotypic characterization of △FgCrpA, FgCrpAc, and △FgCrdA under Cu stress, equal numbers of conidia (2 × 103) were spot inoculated onto solidified FMM amended with CuSO4 at different concentrations indicated in the figure and cultured at 25°C for 3days.
To determine the roles of FgCrpA and FgCrdA in Cu tolerance in F. graminearum, we generated the deletion mutants, △FgCrpA and △FgCrdA, which were further confirmed by diagnostic PCR and Southern blot (Supplementary Figures S2, S3). Deletion of FgCrpA resulted in reduced tolerance to Cu concentrations of 20 μM or higher (Figures 1B,C). However, no obvious difference in Cu tolerance was noticed between △FgCrdA and the wild-type PH-1 (Figures 1B,C). Further, there were no additive Cu sensitivity effect observed in the △FgCrdA△FgCrpA double mutant as compared with △FgCrpA (data not shown). Transformation of the full-length FgCrpA with its promoter region into △FgCrpA rescued tolerance to Cu (Figures 1B,C).
The “Cu fist” DNA binding domain protein AceA has been identified in several Aspergillus spp. and proven to regulate the response to excess Cu by activating the Cu-exporting transporters CrpA (and CrpB in A. flavus). In contrast, in Aspergillus the MT, CrdA, is not activated by AceA (Wiemann et al., 2017; Yang et al., 2018). To explore the roles of the F. graminearum AceA ortholog FgAceA in mediating Cu detoxification, an FgAceA full-length deletion strain, ΔFgAceA, was generated and confirmed by diagnostic PCR and Southern blot (Supplementary Figure S4). Deletion of FgAceA resulted in even more Cu sensitivity compared to ΔFgCrpA. ΔFgAceA barely grew on FMM supplemented with 5 μM Cu. The double mutant ΔFgCrpAΔFgAceA was more sensitive than ΔFgAceA when the FMM was treated with 20 μM Cu (Figures 2A,B). Complementation with the full-length FgAceA driven by its native promoter restored Cu tolerance to that of wild type (Figures 2A,B).
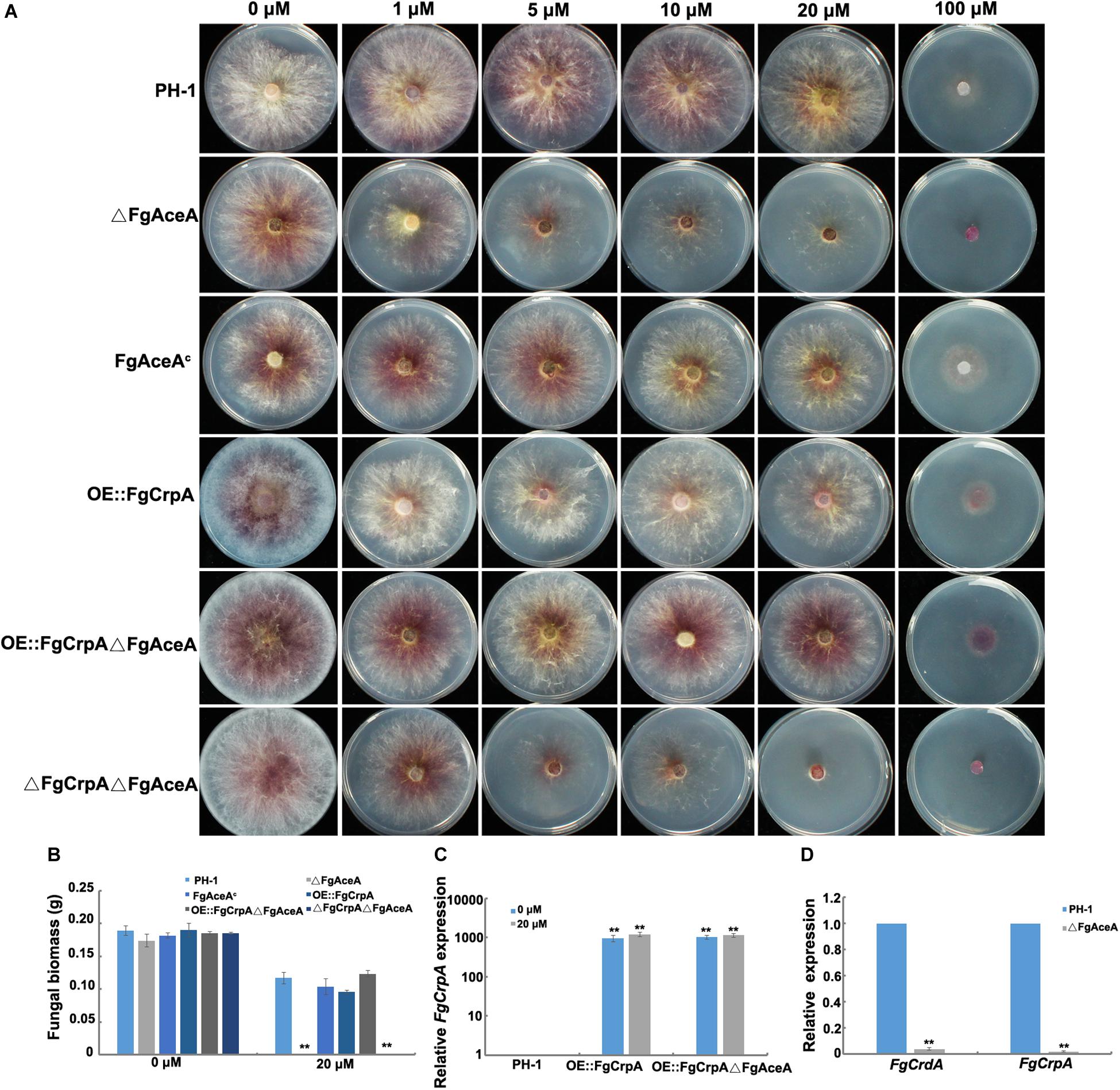
Figure 2. Phenotypic analysis of △FgAceA and its regulation on FgCrpA and FgCrdA. (A) Phenotypic characterization of the wild type strain PH-1 and mutants under Cu stress, equal numbers of conidia (2 × 103) were spot inoculated onto solidified FMM amended with CuSO4 at different concentrations indicated in the figure and cultured at 25°C for 3 days. (B) Fungal growth was measured by determining the dry weight of the wild type strain PH-1, △FgAceA, FgAceAc, OE:FgCrpA, OE:FgCrpA△FgAceA, and △FgCrpA△FgAceA grown in liquid FMM amended with or without CuSO4 at 20 μM in a shaker at 180 rpm at 25°C for 3 days. A t-test was performed to determine significant differences, **p < 0.01. (C) Relative expression of FgCrpA in OE:FgCrpA or OE:FgCrpA△FgAceA analyzed by qRT-PCR assays. (D) Relative expression of FgCrpA and FgCrdA in △FgAceA analyzed by qRT-PCR assays.
To investigate the regulatory mechanism of FgAceA in F. graminearum Cu tolerance, we generated an overexpression strain of FgCrpA (OE:FgCrpA) by replacing its native promoter with the constitutive promoter gpdA from A. nidulans in both the wild-type strain PH-1 and ΔFgAceA. OE:FgCrpA strains were screened by diagnostic PCR and further confirmed by Southern blot (Supplementary Figure S5) and qRT-PCR. The transcriptional level of FgCrpA was significantly upregulated with or without Cu treatment (Figure 2C). OE:FgCrpA exhibited a restored Cu tolerance in the ΔFgAceA background (Figures 2A,B), however, OE::FgCrpA in the wild-type strain PH-1 did not exhibit an increased Cu tolerance (Figure 2A). Deletion of FgAceA ameliorated the induced FgCrpA and FgCrdA expression after excess Cu exposure (Figure 2D).
F. graminearum produced atypical conidia with shorter sizes and fewer septa when cultured under excess Cu conditions (Figures 3A–C). Conidial formation and germination were significantly reduced in ΔFgCrpA (p < 0.05), ΔFgAceA (p < 0.01) and ΔFgCrpAΔFgAceA (p < 0.01) under 100 μM Cu treatment (Figures 3D,E). The double deletion mutant ΔFgCrpAΔFgAceA was more sensitive to Cu as reflected by greater impairment of both conidial formation and germination compared with both single deletion mutants (Figures 3D,E). Noticeably, deletion of FgAceA caused reduced conidial germination even without excess Cu challenge at 4 h but was restored to wild-type level when the germination time was extended to 6 h (Figure 3E).
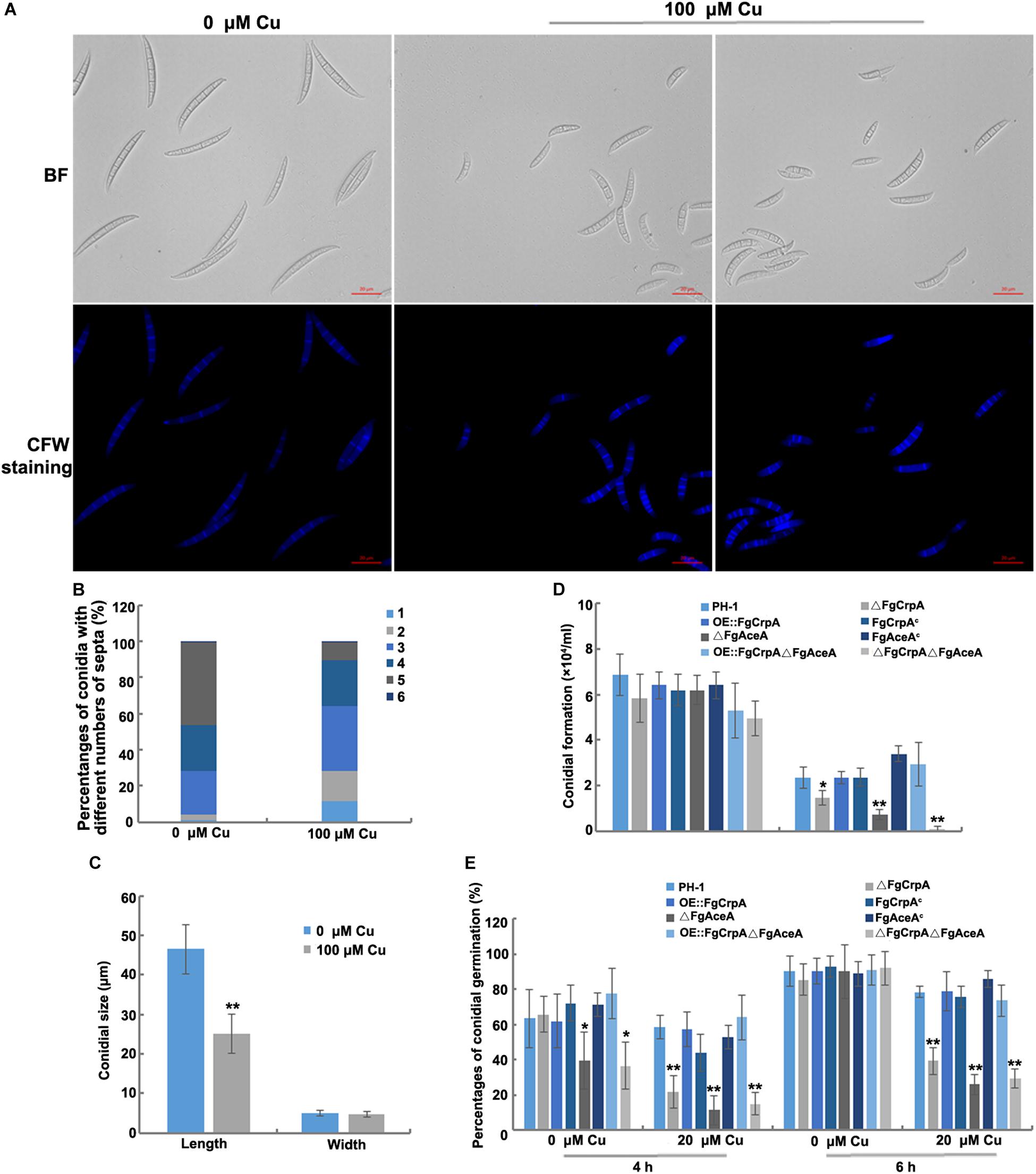
Figure 3. Excess Cu treatment caused defects in conidial morphogenesis. Differences in F. graminearum conidial morphology (A), numbers of septa (B), and conidial size (C) between conidia treated and untreated with Cu at 100 μM. (D) Conidial formation was measured by inoculating five mycelial plugs taken from wild-type strain PH-1 and its mutants in 50 ml liquid CMC amended with or without CuSO4 at 20 μM in a shaker with light at 180 rpm at 25°C for 3 days. (E) Conidial germination was also compared between wild-type strain PH-1 and its mutants by re-suspending the conidia in 10 ml liquid FMM amended with or without CuSO4 at 20 μM in a shaker with light at 180 rpm at 25°C for 4 h and 6 h. A t-test was performed to determine significant differences, *p < 0.05, **p < 0.01.
One immediately observed phenotype of ΔFgAceA grown in high Cu medium was an intense red pigmentation (Figure 4A). We hypothesized that this pigment was the known secondary metabolite aurofusarin and confirmed this likelihood by the dense red color produced. qRT-PCR results which showed that in liquid FMM with or without Cu, the expression of six genes (PKS12, GIP1, GIP2, AURJ, AURF, and AURO) involved in aurofusarin biosynthesis increased dramatically in ΔFgAceA, and trace amounts of Cu stimulated the expression of these six genes (Figure 4B).
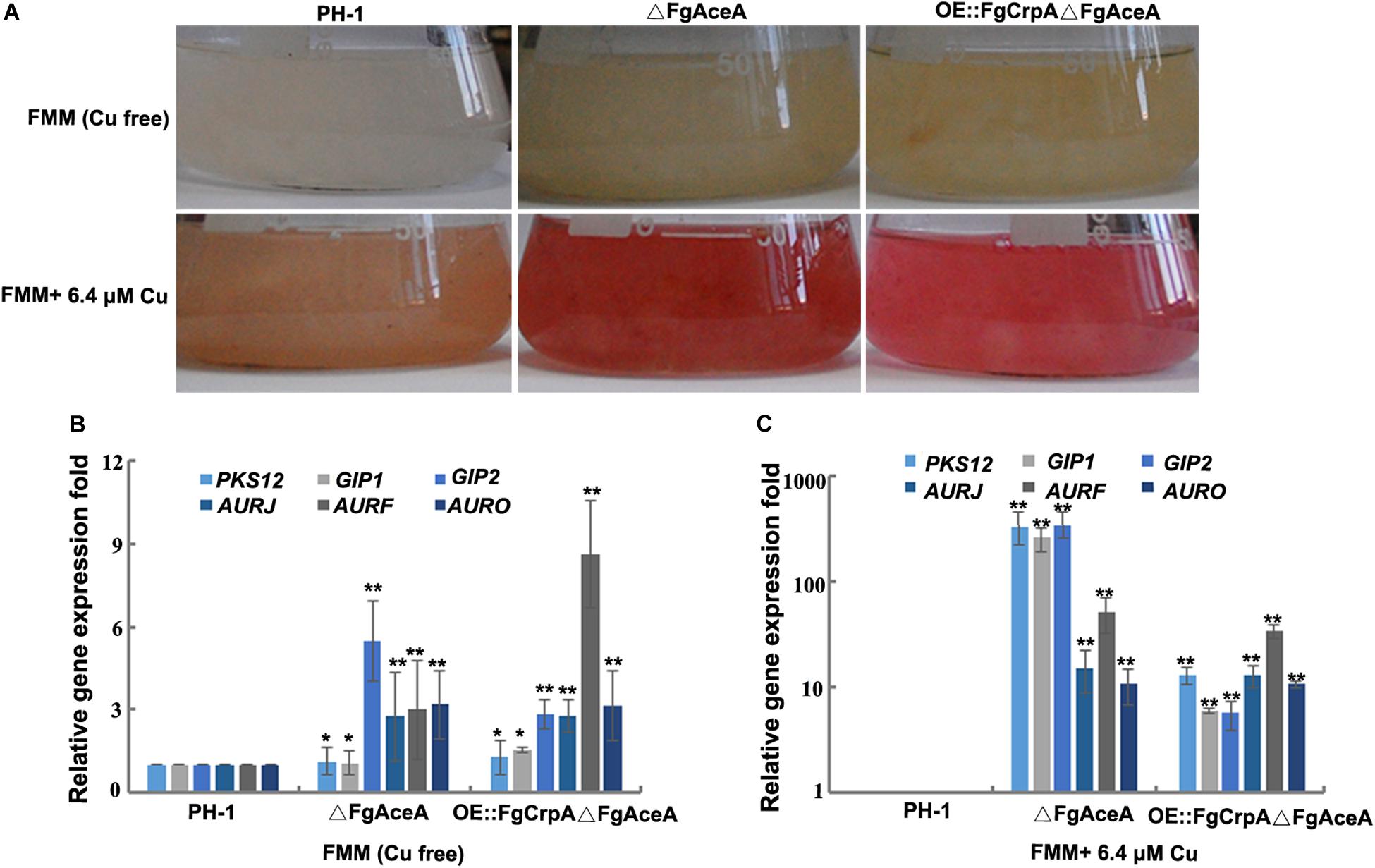
Figure 4. Cu homeostasis affects aurofusarin accumulation in F. graminearum. (A) F. graminearum wild-type strain PH-1, △FgAceA, or OE:FgCrpA△FgAceA was cultured in 50 ml liquid FMM with or without 5 μM (trace element level) Cu in a shaker with light at 180 rpm at 25°C for 4 days. Relative expression of six genes involved in aurofusarin biosynthesis (PKS12, GIP1, GIP2, AURJ, AURF, and AURO) in △FgAceA or OE:FgCrpA△FgAceA when grown in 50 ml liquid FMM with (B) or without 5 μM (trace element level) (C) Cu in a shaker with light at 180 rpm at 25°C for 4 days. A t-test was performed to determine significant differences, *p < 0.05, **p < 0.01.
Previous studies have shown that excess Cu could generate reactive oxygen species such as hydroxyl radicals (Ding et al., 2013, 2014). To test whether Cu detoxification was involved in mediating reactive oxygen intermediates (ROI) stress in F. graminearum, wild type strain PH-1, ΔFgCrpA, ΔFgAceA and their complementary strains were challenged with excess Cu and menadione. Results showed that 30 μM menadione and increasing Cu had inhibitory effects on all the F. graminearum strains tested. Reduced Cu tolerance caused by deletion of FgAceA or FgCrpA aggravates ROS sensitivity as demonstrated by the reduced growth of ΔFgCrpA and ΔFgAceA when grown on Cu containing FMM media supplemented with 30 μM menadione (Figure 5). Moreover, the reductant L-glutathione (GSH) could relieved the severe phenotype of all strains (Figure 5), suggesting that compromises in the Cu detoxification pathway aggravates ROI toxicity.
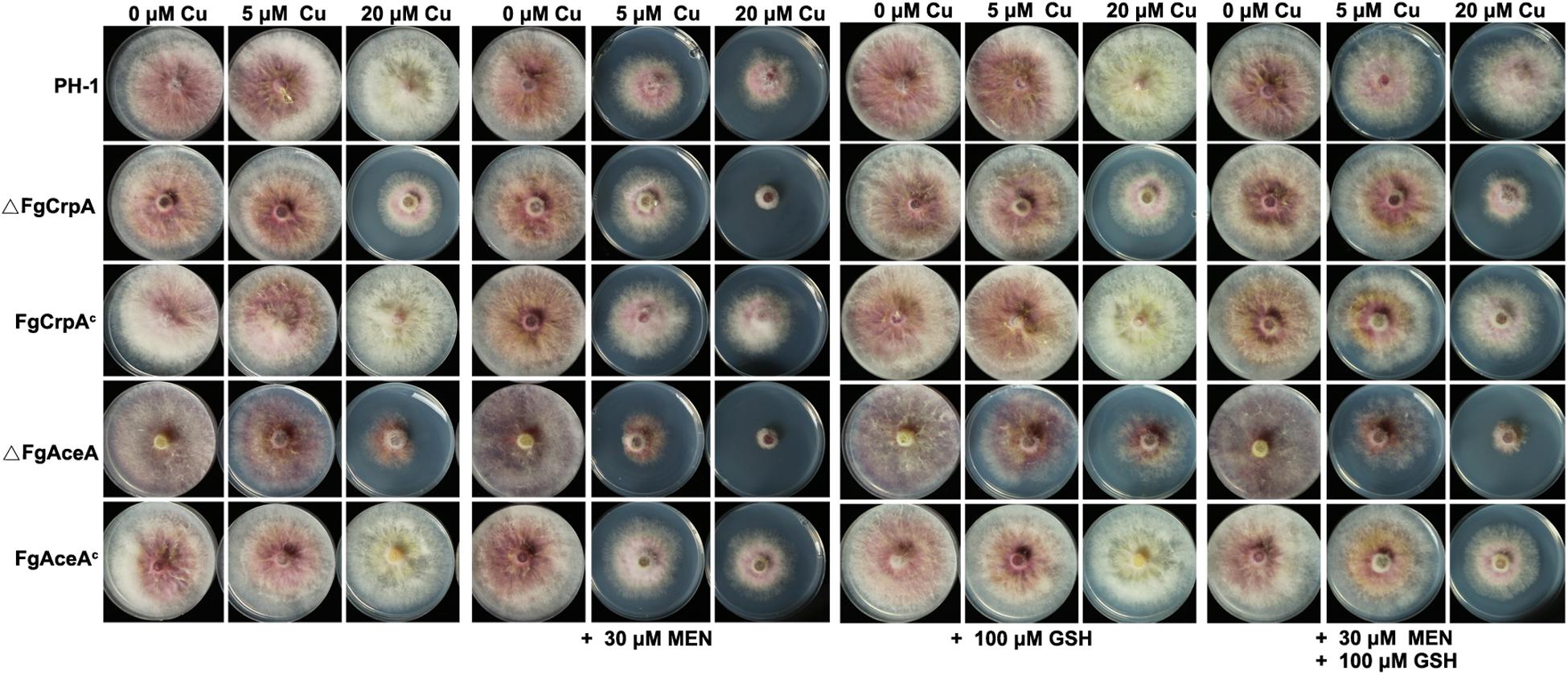
Figure 5. Reduced Cu tolerance caused by FgAceA or FgCrpA deletion aggravates ROS sensitivity in F. graminearum. Growth of F. graminearum strains on Cu-containing FMM media added with supplements (menadione, MEN and/or L-glutathione, GSH) at various concentrations as indicated at 25°C for 4 days.
Pathogenicity assay results showed that the ability to invade untreated flowering wheat heads was not impaired by the single or double deletion of FgAceA and FgCrpA, as shown in Figure 6A (upper panel). However, when inoculated onto Cu fungicide-treated flowering wheat heads, ΔFgCrpA, ΔFgAceA, and ΔFgCrpAΔFgAceA could not successfully colonize the inoculated spikelet, thus completely losing their aggressiveness (Figure 6A, lower panel). Reintroduction of FgCrpA or FgAceA to their respective deletion mutants and overexpression of FgCrpA in ΔFgAceA rescued their lost virulence on Cu fungicide-treated flowering wheat heads (Figure 6A, lower panel). Deletion of FgCrdA did not result in visible changes in virulence in F. graminearum regardless of Cu treatment (Figure 6A).
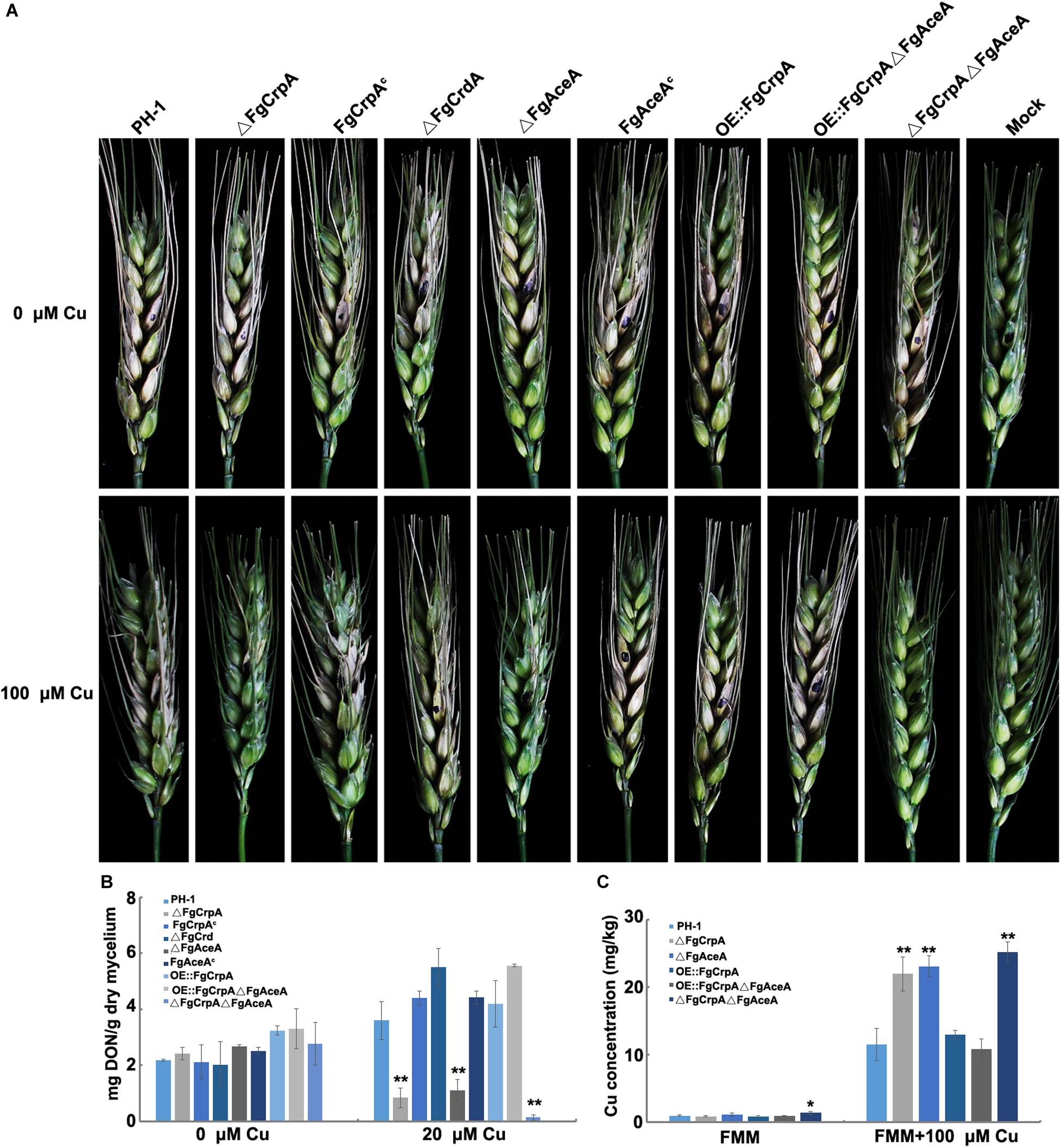
Figure 6. Comparisons of virulence, DON accumulation and Cu concentration between wild-type strain PH-1 and its mutants. (A) Difference in virulence of F. graminearum strains inoculated on Cu treated and untreated flowering wheat heads. (B) Difference in the ability of DON accumulation virulence of F. graminearum strains inoculated in 30 ml liquid TBI (inoculum: 105 spores) with or without 20 μM Cu and cultured at 28°C for 7 days. (C) Difference in Cu concentration in mycelium collected from the wild-type strain PH-1 and its mutants treated or untreated with 100 μM Cu for 12 h. A t-test was performed to determine significant differences, *p < 0.05, **p < 0.01.
The toxic secondary metabolite deoxynivalenol (DON) produced by F. graminearum is a known virulence factor during plant infection (Proctor et al., 1995a; Ilgen et al., 2008). The reduced Cu tolerance caused by disruption of FgAceA and/or FgCrpA also affected DON accumulation. As shown in Figure 6B, ΔFgCrpA, ΔFgAceA, and ΔFgCrpAΔFgAceA produced significantly (p < 0.01) lower amounts of DON under 20 μM Cu treatment than did the wild-type strain PH-1. Reintroduction of FgCrpA or FgAceA to their respective deletion mutants and overexpression of FgCrpA in ΔFgAceA rescued DON biosynthesis. Deletion of FgCrdA did not result in changes in DON biosynthesis in F. graminearum. When treated with 20 μM Cu, the F. graminearum wild-type strain PH-1 accumulated more DON than the untreated TBI liquid (Figure 6B).
Next, we determined the Cu concentration in the mycelium and compared the differences between the mutants and their wild-type strain PH-1. The results showed that when cultured in FMM liquid (trace level of Cu), only ΔFgCrpAΔFgAceA showed significantly (p < 0.05) increased cellular accumulation of Cu compared to the wild-type strain PH-1 (Figure 6C). However, when cultured in excess Cu (FMM liquid amended with 100 μM), the Cu concentration in ΔFgCrpA, ΔFgAceA, and ΔFgCrpAΔFgAceA increased significantly (p < 0.01) compared with the wild-type strain PH-1 or the complementary strain. Additionally, the overexpression of FgCrpA in ΔFgAceA rescued the Cu export deficiency of the mutant, and the Cu level decreased to that of the wild-type (Figure 6C).
Accumulating evidence suggests that during infection, invading fungal pathogens will be confronted with elevated levels of essential trace nutrients harnessed by the host, collectively known as “nutritional immunity” (Ballou and Wilson, 2016). Moreover, fungal pathogens have also evolved an accurate mechanism to balance the fine lines between the essentiality and toxicity of these essential trace nutrients to survive and colonize within the infected host. For example, all living organisms maintain Cu homeostasis by regulating the balance between Cu uptake, utilization and detoxification (Ding et al., 2014; Ballou and Wilson, 2016). In this study, we characterized the copper tolerance determinants in the important wheat head blight fungus F. graminearum. Overall, our study showed that F. graminearum Cu resistance is mainly mediated by the ATP-based export system and not by a Cu MT.
The P-type ATPase FgCrpA and the MT FgCrdA were both found to be significantly upregulated when treated with excess Cu in RNA-Seq analysis and further verified by qRT-PCR. This was consistent with observation in Aspergillus spp. Cu-induced CrpA and CrpB expression was confirmed in A. flavus by using Northern Blot analysis (Yang et al., 2018). In A. nidulans, Cu-induced CrpA and CrdA protein expression was confirmed by Western blot (Antsotegi-Uskola et al., 2017). As Crp and Crd shared conserved expression pattern and functions in response to Cu toxicity, we expected protein expression of FgCrpA and FgCrdA to be induced in F. graminearum. The results of gene disruption assays indicated crucial roles of FgCrpA but not FgCrdA in mediating Cu resistance in F. graminearum, indicating that Cu efflux is the predominant mechanism of Cu detoxification rather than MT-mediated Cu buffering. Together with previous studies carried out in other fungi, including C. albicans and Aspergillus spp. (A. fumigatus, A. flavus, A. nidulans), these results indicated that ATP-based efflux, as the principal Cu detoxification mechanism, is not unique and likely a well-conserved feature among filamentous fungal species (Antsotegi-Uskola et al., 2017; Wiemann et al., 2017; Cai et al., 2018; Yang et al., 2018).
Apart from Cu efflux, MT-related buffering also plays critical roles in Cu detoxification for some fungi. MTs function as Cu storage proteins by chelating excess Cu in yeast species. In S. cerevisiae, cells are protected from excess Cu by activating the MT-encoding genes cup1 and crs5, and surplus Cu is then coordinated (Ecker et al., 1986; Culotta et al., 1994; Palacios et al., 2011; Thiele, 2015). In the human fungal pathogen Cryptococcus neoformans, the Cu metallo-regulatory TF Cuf1 activates the Cu-buffering MT-encoding genes Mt1 and Mt2 for Cu detoxification (Ding et al., 2013; Garcia-Santamarina et al., 2017). In the soil organism F. oxysporum, the Cu metallothionein Mt1 has been shown to be involved in mediating resistance to metal toxicity and virulence (Lorenzo-Gutiérrez et al., 2019). Despite the important roles of MTs in Cu detoxification in yeast species and F. oxysporum, their homologs in other filamentous fungi, including A. flavus, A. fumigatus, and A. nidulans, have been reported to be not particularly important. Deletion of CrdA caused no significant differences in Cu tolerance in these Aspergillus spp. In F. graminearum, although FgCrdA has been identified to respond to excess Cu treatment, its disruption did not alter Cu tolerance in △FgCrdA, the exact role of FgCrdA in Cu detoxification remains to be explored. Alternatively, Cu binding secondary metabolites – possibly filling a MT-like function – are hypothesized to be involved in Cu homeostasis in filamentous fungi (Raffa et al., 2019).
Cu homeostasis is tightly regulated in F. graminearum by the Cu-responsive TF FgAceA. We showed that deletion of FgAceA resulted in reduced Cu tolerance and ceased the induction of FgCrpA expression by excess Cu loading, suggesting that FgCrpA is transcriptionally expressed in an FgAceA-dependent manner under excess Cu stress. Overexpression of FgCrpA in △FgAceA could compensate for the reduced Cu tolerance, indicating that FgAceA mediates Cu homeostasis by regulating the expression of the copper exporting P-type ATPase FgCrpA. In S. cerevisiae, Ace1 functions by recognizing and binding to the Cu-dependent responsive element ACE in the promoter regions of its target MT-encoding genes cup1 and crs5 and the superoxide dismutase gene sod1. ACE contains the core sequence 5′-HTHXXGCTGD-3′ (D = A, G, or T; H = A, C or T; and X = any residue). The promoter region of FgCrpA contains one core ACE sequence (TTATGCTGT), indicating a potential FgAceA binding domain.
Cu could not be properly transported out of the cell and was accumulated in mutants lacking FgAceA or FgCrpA (Figure 6C); thus, these mutants displayed compromised phenotypes when challenged with excess Cu, as expected. Conidial formation and germination as well as DON biosynthesis and the ability to colonize flowering wheat heads were impaired in △FgAceA and △FgCrpA when treated with high levels of Cu. We found that FgAceA negatively regulates aurofusarin biosynthesis. In both liquid FMM medium with and without trace amounts of Cu, the accumulation of the red pigment aurofusarin increased significantly in △FgAceA compared with its wild-type parent. Six genes involved in aurofusarin biosynthesis were significantly upregulated in △FgAceA and OE:FgCrpA△FgAceA (Figures 4B,C). Overexpression of FgCrpA in △FgAceA did not restore the overproduction of aurofusarin, suggesting that FgAceA has additional roles in negatively regulating secondary metabolism and bypassing the mediation of Cu tolerance.
FgAceA and/or FgCrpA deletion mutants could not colonize on Cu fungicide-treated flowering wheat heads. Several reasons could account for the virulence loss of the mutants. First, it could not be separated from the defects in conidial germination and reduced aerial hyphae under excess Cu treatment. Second, ROS such as hydroxyl radicals generated with excess Cu or plant oxidative burst could also be responsible for the unsuccessful colonization of the mutants. Additionally, DON has been identified as a crucial virulence factor and plays an important role in aggressiveness of the fungus among the spikelets of the infected wheat head, the decreased DON production in the mutants challenged with excess Cu may also account for the loss of full virulence.
In summary, we elucidated the copper tolerance mechanism in the important wheat head blight fungus F. graminearum, which is mainly mediated by the ATP-based extrusion system. The P-type ATPase FgCrpA is transcriptionally regulated by the Cu-fist TF FgAceA, and both are crucial for Cu tolerance.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.
XL and YJ carried out the experimentation of this work. JS and NK conceived the experiments and wrote the manuscript. DH, XF, and JX helped create strains for this research. Y-WL helped revise this manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key R&D Program of China (2018YFE0206000), National Natural Science Fund of China (31772118, 31701748, and 31601594), Natural Science Fund of Jiangsu Province (BK20181246), Jiangsu Agriculture Science and Technology Innovation Fund (CX(19)3004), Special Fund for Risk Assessment of China (GJFP2019002), and Shanghai Agriculture Applied Technology Development Program (2019-02-08-00-12-F01148).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the native English-speaking scientists of American Journal Experts (Durham, NC, United States) for editing our manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01392/full#supplementary-material
FIGURE S1 | Bioinformatic analysis of putative Cu detoxification determinants.
FIGURE S2 | Diagram for generation and verification for △FgAceA by Southern blotting analysis.
FIGURE S3 | Diagram for generation and verification for ΔFgCrpA by Southern blotting analysis.
FIGURE S4 | Diagram for generation and verification for △FgCrdA by Southern blotting analysis.
FIGURE S5 | Diagram for generation and verification for OE:FgCrpA by Southern blotting analysis.
TABLE S1 | PCR primers used in this study.
TABLE S2 | Expression of genes encoding putative copper-binding proteins revealed by RNA-seq.
Antsotegi-Uskola, M., Markina-Iñarrairaegui, A., and Ugalde, U. (2017). Copper resistance in Aspergillus nidulans relies on the P-type ATPase CrpA, regulated by the transcription factor AceA. Front. Microbiol. 8:912. doi: 10.3389/fmicb.2017.00912
Babula, P., Masarik, M., Adam, V., Eckschlager, T., Stiborova, M., Trnkova, L., et al. (2012). Mammalian metallothioneins: properties and functions. Metallomics 4, 739–750. doi: 10.1039/c2mt20081c
Bai, G., and Shaner, G. (2004). Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 42, 135–161. doi: 10.1146/annurev.phyto.42.040803.140340
Ballou, E. R., and Wilson, D. (2016). The roles of zinc and copper sensing in fungal pathogenesis. Curr. Opin. Microbiol. 32, 128–134. doi: 10.1016/j.mib.2016.05.013
Besold, A. N., Culbertson, E. M., and Culotta, V. C. (2016). The Yin and Yang of copper during infection. J. Biol. Inorg. Chem. 21, 137–144. doi: 10.1007/s00775-016-1335-1
Bruno, K. S., Tenjo, F., Li, L., Hamer, J. E., and Xu, J. R. (2004). Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot. Cell 3, 1525–1532. doi: 10.1128/ec.3.6.1525-1532.2004
Cai, Z., Du, W., Zhang, Z., Guan, L., Zeng, Q., Chai, Y., et al. (2018). The Aspergillus fumigatus transcription factor AceA is involved not only in Cu but also in Zn detoxification through regulating transporters CrpA and ZrcA. Cell. Microbiol. 20:e12864. doi: 10.1111/cmi.12864
Chen, L., Tong, Q., Zhang, C., and Ding, K. (2018). The transcription factor FgCrz1A is essential for fungal development, virulence, deoxynivalenol biosynthesis and stress responses in Fusarium graminearum. Curr. Genet. 65, 1–14. doi: 10.1007/s00294-018-0853-5
Culotta, V. C., Howard, W. R., and Liu, X. F. (1994). CRS5 encodes a metallothionein-like protein in Saccharomyces cerevisiae. J. Biol. Chem. 269, 25295–25302.
Cuomo, C. A., Güldener, U., Xu, J. R., Trail, F., Turgeon, B. G., Di Pietro, A., et al. (2007). The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317, 1400–1402. doi: 10.1126/science.1143708
Cyert, M. S., and Philpott, C. C. (2013). Regulation of cation balance in Saccharomyces cerevisiae. Genetics 193, 677–713. doi: 10.1534/genetics.112.147207
Dalecki, A. G., Crawford, C. L., and Wolschendorf, F. (2017). Copper and antibiotics: discovery, modes of action, and opportunities for medicinal applications. Adv. Microb. Physiol. 70, 193–260. doi: 10.1016/bs.ampbs.2017.01.007
Ding, C., Festa, R. A., Chen, Y. L., Espart, A., Palacios, Ò, Espín, J., et al. (2013). Cryptococcus neoformans copper detoxification machinery is critical for fungal virulence. Cell Host Microbe 13, 265–276. doi: 10.1016/j.chom.2013.02.002
Ding, C., Festa, R. A., Sun, T. S., and Wang, Z. Y. (2014). Iron and copper as virulence modulators in human fungal pathogens. Mol. Microbiol. 93, 10–23. doi: 10.1111/mmi.12653
Ding, C., Yin, J., Tovar, E. M. M., Fitzpatrick, D. A., Higgins, D. G., and Thiele, D. J. (2011). The copper regulon of the human fungal pathogen Cryptococcus neoformans H99. Mol. Microbiol. 81, 1560–1576. doi: 10.1111/j.1365-2958.2011.07794.x
Dollwet, H. H. A., and Sorenson, J. R. J. (1985). Historic uses of copper compounds in medicine. J Trace Elem. Med. Bio. 2, 80–87. doi: 10.1093/eurpub/11.2.238
Dong, F., Qiu, J., Xu, J., Yu, M., Wang, S., Sun, Y., et al. (2016). Effect of environmental factors on Fusarium population and associated trichothecenes in wheat grain grown in Jiangsu province, China. Int. J. Food Microbiol. 230, 58–63. doi: 10.1016/j.ijfoodmicro.2016.04.020
Ecker, D. J., Butt, T. R., Sternberg, E. J., Neeper, M. P., Debouck, C., Gorman, J. A., et al. (1986). Yeast metallothionein function in metal ion detoxification. J. Biol. Chem. 261, 16895–16900.
Festa, R. A., Helsel, M. E., Franz, K. J., and Thiele, D. J. (2014). Exploiting innate immune cell activation of a copper-dependent antimicrobial agent during infection. Chem. Biol. 21, 977–987. doi: 10.1016/j.chembiol.2014.06.009
Garcia-Santamarina, S., Uzarska, M. A., Festa, R. A., Lill, R., and Thiele, D. J. (2017). Cryptococcus neoformans iron-sulfur protein biogenesis machinery is a novel layer of protection against Cu Stress. MBio 8, 1–18. doi: 10.1128/mbio.01742-17
Gerwien, F., Skrahina, V., Kasper, L., Hube, B., and Brunke, S. (2018). Metals in fungal virulence. FEMS Microbiol. Rev. 42, 1–21. doi: 10.1093/femsre/fux050
Goswami, R. S., and Kistler, H. C. (2004). Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. doi: 10.1111/J.1364-3703.2004.00252.X
Grass, G., Rensing, C., and Solioz, M. (2011). Metallic copper as an antimicrobial surface. Appl. Environ. Microbiol. 77, 1541–1547. doi: 10.1128/AEM.02766-10
Iida, Y., Kurata, T., Harimoto, Y., and Tsuge, T. (2008). Nitrite reductase gene upregulated during conidiation is involved in macroconidium formation in Fusarium oxysporum. Phytopathology 98, 1099–1106. doi: 10.1094/PHYTO-98-10-1099
Ilgen, P., Maier, F., and Schäfer, W. (2008). Trichothecenes and lipases are host-induced and secreted virulence factors of Fusarium graminearum. Cereal Res. Commun. 36, 421–428. doi: 10.1556/CRC.36.2008.Suppl.B.35
Isebaert, S., Haesaert, G., Devreese, R., Maene, P., Fremaut, F., and Vlaemynck, G. (2005). Fusarium spp. and Fusarium mycotoxins in maize: a problem for Flanders? Commun. Agric. Appl. Biol. Sci. 70, 129–136.
Ji, F., Xu, J., Liu, X., Yin, X., and Shi, J. (2014). Natural occurrence of deoxynivalenol and zearalenone in wheat from Jiangsu province, China. Food Chem. 157, 393–397. doi: 10.1016/j.foodchem.2014.02.058
Keller, G., Bird, A., and Winge, D. R. (2005). Independent metalloregulation of Ace1 and Mac1 in Saccharomyces cerevisiae. Eukaryot. Cell. 4, 1863–1871. doi: 10.1128/EC.4.11.1863-1871.2005
Kim, B. E., Nevitt, T., and Thiele, D. J. (2008). Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 4, 176–185. doi: 10.1038/nchembio.72
Kumar, S., Nei, M., Dudley, J., and Tamura, K. (2008). MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9, 299–306. doi: 10.1093/bib/bbn017
Ladomersky, E., and Petris, M. J. (2015). Copper tolerance and virulence in bacteria. Metallomics 7, 957–964. doi: 10.1039/c4mt00327f
Leslie, J. F., and Summerell, B. A. (2007). The Fusarium Laboratory Manual. Oxford: Blackwell Publishing. doi: 10.1002/9780470278376
Lombaert, G. A., Pellaers, P., Roscoe, V., Mankotia, M., Neil, R., and Scott, P. M. (2003). Mycotoxins in infant cereal foods from the Canadian retail market. Food Addit. Contam. 20, 494–504. doi: 10.1080/0265203031000094645
Lorenzo-Gutiérrez, D., Gómez-Gil, L., Guarro, J., Roncero, M. I. G., Fernández-Bravo, A., Capilla, J., et al. (2019). Role of the Fusarium oxysporum metallothionein Mt1 in resistance to metal toxicity and virulence. Metallomics 11, 1230–1240. doi: 10.1039/c9mt00081j
Michels, H. T., Keevil, C. W., Salgado, C. D., and Schmidt, M. G. (2015). From laboratory research to a clinical trial: copper alloy surfaces kill bacteria and reduce hospital-acquired infections. Heal. Environ. Res. Des. J. 9, 64–79. doi: 10.1177/1937586715592650
Odermatt, A., Suter, H., Krapf, R., and Solioz, M. (1993). Primary structure of two P-type ATPases involved in copper homeostasis in Enterococcus hirae. J. Biol. Chem. 268, 12775–12779. doi: 10.1016/j.brainres.2016.08.031
Palacios, Ò, Atrian, S., and Capdevila, M. (2011). Zn-and Cu-thioneins: a functional classification for metallothioneins? J. Biol. Inorg. Chem. 16, 991–1009. doi: 10.1007/s00775-011-0827-2
Proctor, R. H., Hohn, T. M., and McCormick, S. P. (1995a). Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol. Plant Microbe Interact. 8, 593–601. doi: 10.1094/MPMI-8-0593
Proctor, R. H., Hohn, T. M., McCormick, S. P., and Desjardins, A. E. (1995b). Tri6 encodes an unusual zinc finger protein involved in regulation of trichothecene biosynthesis in Fusarium sporotrichioides. Appl. Environ. Microbiol. 61, 1923–1930. doi: 10.1128/aem.61.5.1923-1930.1995
Raffa, N., Osherov, N., and Keller, N. P. (2019). Copper utilization, regulation, and acquisition by Aspergillus fumigatus. Int. J. Mol. Sci. 20:1980. doi: 10.3390/ijms20081980
Rasmussen, P. H., Ghorbani, F., and Berg, T. (2003). Deoxynivalenol and other Fusarium toxins in wheat and rye flours on the Danish market. Food Addit. Contam. 20, 396–404. doi: 10.1080/0265203031000082495
Solioz, M., and Odermatt, A. (1995). Copper and silver transport by CopB-ATPase in membrane vesicles of Enterococcus hirae. J. Biol. Chem. 270, 9217–9221. doi: 10.1074/jbc.270.16.9217
Thiele, D. J. (2015). ACE1 regulates expression of the Saccharomyces cerevisiae metallothionein gene. Mol. Cell. Biol. 8, 2745–2752. doi: 10.1128/mcb.8.7.2745
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tutelyan, V. A. (2004). Deoxynivalenol in cereals in Russia. Toxicol. Lett. 153, 173–179. doi: 10.1016/j.toxlet.2004.04.042
Weissman, Z., Berdicevsky, I., Cavari, B.-Z., and Kornitzer, D. (2000). The high copper tolerance of Candida albicans is mediated by a P-type ATPase. Proc Natl Acad Sci U.S.A. 97, 3520–3525. doi: 10.1073/pnas.97.7.3520
Wiemann, P., Perevitsky, A., Huttenlocher, A., Osherov, N., and Keller, N. P. (2017). Aspergillus fumigatus copper export machinery and reactive oxygen intermediate defense counter host copper-mediated oxidative antimicrobial offense. Cell Rep. 19, 1008–1021. doi: 10.1016/j.celrep.2017.04.019
Wu, A. B., Li, H. P., Zhao, C. S., and Liao, Y. C. (2005). Comparative pathogenicity of Fusarium graminearum isolates from China revealed by wheat coleoptile and floret inoculations. Mycopathologia 160, 75–83. doi: 10.1007/s11046-005-1153-4
Yang, K., Shadkchan, Y., Tannous, J., Landero Figueroa, J. A., Wiemann, P., Osherov, N., et al. (2018). Contribution of atpase copper transporters in animal but not plant virulence of the crossover pathogen Aspergillus flavus. Virulence 9, 1273–1286. doi: 10.1080/21505594.2018.1496774
Keywords: Fusarium graminearum, copper tolerance, FgAceA, copper transporters, metallothionein, virulence
Citation: Liu X, Jiang Y, He D, Fang X, Xu J, Lee Y-W, Keller NP and Shi J (2020) Copper Tolerance Mediated by FgAceA and FgCrpA in Fusarium graminearum. Front. Microbiol. 11:1392. doi: 10.3389/fmicb.2020.01392
Received: 03 April 2020; Accepted: 29 May 2020;
Published: 26 June 2020.
Edited by:
Daisuke Hagiwara, University of Tsukuba, JapanCopyright © 2020 Liu, Jiang, He, Fang, Xu, Lee, Keller and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianrong Shi, c2hpamlAamFhcy5hYy5jbg==; Nancy P. Keller, bnBrZWxsZXJAd2lzYy5lZHU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.