- 1The First Clinical Medical College, Lanzhou University, Lanzhou, China
- 2Department of General Surgery, The First Hospital of Lanzhou University, Lanzhou, China
- 3Key Laboratory of Biological Therapy and Regenerative Medicine Transformation Gansu Province, Lanzhou, China
- 4The Department of Infectious Diseases, The First Hospital of Lanzhou University, Lanzhou, China
The coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has spread rapidly worldwide, seriously endangering human health. In addition to the typical symptoms of pulmonary infection, patients with COVID-19 have been reported to have gastrointestinal symptoms and/or intestinal flora dysbiosis. It is known that a healthy intestinal flora is closely related to the maintenance of pulmonary and systemic health by regulating the host immune homeostasis. Role of the “gut-lung axis” has also been well-articulated. This review provides a novel suggestion that intestinal flora may be one of the mediators of the gastrointestinal responses and abnormal immune responses in hosts caused by SARS-CoV-2; improving the composition of intestinal flora and the proportion of its metabolites through probiotics, and personalized diet could be a potential strategy to prevent and treat COVID-19. More clinical and evidence-based medical trials may be initiated to determine the strategy.
Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was first reported in December 2019 (Wu et al., 2020). Since then, the disease has spread rapidly worldwide and has been declared a global pandemic by the World Health Organization. As of 25 May 2020, there were 5,307,298 confirmed cases, including 342,070 deaths (WHO, 2020; Zhu et al., 2020). Considering its strong infectivity, poor prognosis, and lack of effective/targeted drugs, potential prevention and treatment strategies for COVID-19 need to be urgently developed. The damage to host immune defense and the “cytokine storm,” an excessive production of inflammatory cytokines, are believed to be the critical causes of deteriorated health and even death of patients with COVID-19 (Zumla et al., 2016; Pedersen and Ho, 2020; Ye et al., 2020). Given the crucial role played by intestinal flora and its metabolites in regulating immune and inflammatory response of the host, the prospect of modulating intestinal flora for preventing and treating COVID-19 and related illnesses (e.g., viral and/or bacterial pneumonia, acute respiratory infections, or influenza) has attracted considerable attention from the scientific community (Belkaid and Harrison, 2017; Dang and Marsland, 2019; Xu et al., 2020). In order to develop a potential strategy for COVID-19 prevention and treatment by targeting the intestinal flora, we focused mainly on the effects of SARS-CoV-2 on the host intestinal microecology, as well as the possible mechanisms through which intestinal flora regulate immune and inflammatory responses in patients with COVID-19 and related diseases. Particularly, the role of “gut-lung axis” and some indirect evidence for the effect of intestinal flora on the prevention and treatment of COVID-19 have been highlighted here.
Clinical Manifestations and the Possible Mechanism of Intestinal Microecology Disorders In Patients With COVID-19
The main clinical manifestations of COVID-19 are fever, cough, and acute respiratory distress syndrome. However, gastrointestinal symptoms, such as diarrhea, nausea and vomiting, abdominal pain, and loss of appetite have been reported in an increasing number of COVID-19 patients (Cholankeril et al., 2020; Goyal et al., 2020; Guan et al., 2020; Lin et al., 2020). Interestingly, the COVID-19 patients with gastrointestinal symptoms had more severe disease and these symptoms could be used to predict the development of severe respiratory disorders (Gou et al., 2020; Wan et al., 2020). It is noteworthy that some COVID-19 patients also showed microbial dysbiosis with decreased levels of Lactobacillus and Bifidobacterium (Xu et al., 2020); the abundance of Clostridium hathewayi, Clostridium ramosum, and Coprobacillus was positively correlated while that of Faecalibacterium prausnitzii was inversely correlated with the severity of the disease (Zuo et al., 2020).
SARS-CoV-2 can identify and invade human cells through the interaction of spike proteins with human angiotensin-converting enzyme 2 (ACE2) (Wu et al., 2020). ACE2 is expressed not only in the lung tissue but also on esophageal and intestinal epithelium; this is the basis of SARS-CoV-2 attacking the digestive tract of the host and leading to intestinal flora dysbiosis and gastrointestinal symptoms (Guan et al., 2020; Holshue et al., 2020; Li M. Y. et al., 2020). Moreover, some studies have reported that SARS-CoV-2 and its nucleic acid were isolated from stool samples of patients with diarrhea (Lamers et al., 2020; Zhou et al., 2020; Zou et al., 2020). These evidences suggest that SARS-CoV-2 may be harbored in the digestive tract of patients and transmitted via the fecal-oral route, affecting the health of the gastrointestinal tract and intestinal flora.
ACE2 is a negative regulator of renin-angiotensin system and is critical for maintaining the homeostasis of blood pressure and the balance of salts and fluid; and ACE2 has local regulatory effects in the pathological changes in several organs, including the heart, kidneys, and lungs (Patel et al., 2017). The association between intestinal flora and ACE2 has also been reported previously: deficiency of ACE2 caused critical impairment of local tryptophan homeostasis in a mouse model, which could alter the intestinal microbiome and susceptibility to inflammation (Hashimoto et al., 2012). ACE2 can also regulate the absorption of nutrients by binding with amino acid transporters on intestinal epithelial cells, which suggests that SARS-CoV-2 might compete with protein nutrients and interfere in their absorption through ACE2 on the intestinal epithelium (Singer et al., 2012; Vuille-Dit-Bille et al., 2015; Javed and Broer, 2019). Cole-Jeffrey et al. indicated that the protective actions of ACE2 against cardiopulmonary disorders could be mediated by its actions on the gastrointestinal tract and intestinal flora (Cole-Jeffrey et al., 2015). A recent study also reported that some specific intestinal microorganisms that can downregulate ACE2 expression in murine gut, such as Bacteroides thetaiotaomicron, Bacteroides dorei, and Bacteroides massiliensis, correlated inversely with the SARS-CoV-2 load in patient's fecal samples (Zuo et al., 2020). It is known that a healthy intestinal flora plays a vital role in maintaining immune homeostasis and gastrointestinal tract health of the host (Lynch and Pedersen, 2016; Shi et al., 2017). It could be speculated that gastrointestinal symptoms and the changes in immune homeostasis induced by SARS-CoV-2 might be mediated, in part, by the intestinal flora. There could be a potential strategy to fight SARS-CoV-2 infection by targeting intestinal flora.
Intestinal Flora and the GUT-LUNG Axis
Intestinal flora widely affects host health and is highly correlated with a variety of illnesses, including metabolic, digestive system, and even the respiratory diseases. As shown by 16S rRNA and metagenomics sequencing, the human intestinal flora contains more than 1,000 different microbial species, including bacteria, fungi, and viruses (Grice and Segre, 2012; Sender et al., 2016). On average, each host contains about 160 dominant bacterial species, depending on genetics, environmental factors, and dietary habits (Eckburg et al., 2005; Voreades et al., 2014; Zhernakova et al., 2016). The human intestinal microbiome is a highly dynamic microecosystem and interacts with the immune system. Immune cells induced by a variety of antigens can move between the gut and the lungs through the lymphatic system and/or blood, resulting in the regulation of immune response of both organs. The cross-talk between intestinal and pulmonary tissues mediated by the microbiome and immune cells is called the “gut–lung axis” (Mcghee and Fujihashi, 2012; Date et al., 2017; Ipci et al., 2017).
Many studies have reported that disorders of the intestinal flora were related to lung diseases and respiratory tract infections (Hand et al., 2016; Belkaid and Harrison, 2017; Selber-Hnatiw et al., 2017; Gong et al., 2018; Schirmer et al., 2018). In the mouse model, the depletion of sensitive intestinal bacteria (e.g., Bifidobacteria) after neomycin administration increased the susceptibility of the mice to influenza virus infection and pulmonary allergic inflammation (Dharmage et al., 2015; Metsala et al., 2015; Pang et al., 2018); and a recent study reported that the gut population of endogenous Bifidobacterium increased to enhance the hosts' resistance to influenza when a lethal influenza infection occurred (Zhang et al., 2020). Bradley et al. showed that the abundance of segmented filamentous bacteria could stimulate the migration of Th17 cells to the lung, augmenting the autoimmune response and aggravating pulmonary lesions (Bradley et al., 2017). Moreover, the intestinal flora and its metabolites, such as short-chain fatty acids (SCFAs) and lipopolysaccharides (LPS), are parts of the intestinal mucosal immune barrier and maintain their normal functions during respiratory tract infections (Leblanc et al., 2017; Sittipo et al., 2019; Visconti et al., 2019). The mucosal immune barrier provides protection against thousands of microorganisms and environmental antigens, and is closely related to the systemic and pulmonary immune function of the host (Abt et al., 2012; Abrahamsson et al., 2014; Donaldson et al., 2016). If the intestinal mucosal immune barrier is disrupted, invading organisms are able to enter the blood or lungs and this could result in septicaemia and acute respiratory distress syndrome (Dickson et al., 2016).
Interestingly, Changes in the pulmonary microenvironment (e.g., by influenza virus or SARS-CoV-2 infection) can also alter the structure and function of intestinal flora (Budden et al., 2017; Dang and Marsland, 2019). In the mouse model, influenza virus infection of the respiratory tract increased the number of Enterobacteria in the intestinal flora while decreasing the number of Lactobacillus and Lactococcus (Looft and Allen, 2012; Tirone et al., 2019). Similarly, LPS injection in mice lungs resulted in an imbalance of pulmonary microbiota that was accompanied by an intestinal microbiota disbalance, which was caused by bacteria that entered the blood and intestinal mucosa coming from the lung tissue (Sze et al., 2014; Hanada et al., 2018). As mentioned above, many COVID-19 patients have also been reported to present apparent microbial dysbiosis; and intestinal flora alterations were associated with COVID-19 susceptibility and severity (Xu et al., 2020; Zuo et al., 2020).
To summarize, interaction between the intestinal flora and lungs and ways to promote optimum lung health should be further investigated. Based on the current knowledge regarding the “gut–lung axis,” it can be hypothesized that SARS-CoV-2 not only directly invade human intestinal epithelium cells by being transmitted via the fecal-oral route but also indirectly affect the intestine and intestinal flora along the gut-lung axis, and lung lesions caused by SARS-CoV-2 could potentially be prevented and treated by targeting the intestinal flora (Figure 1A).
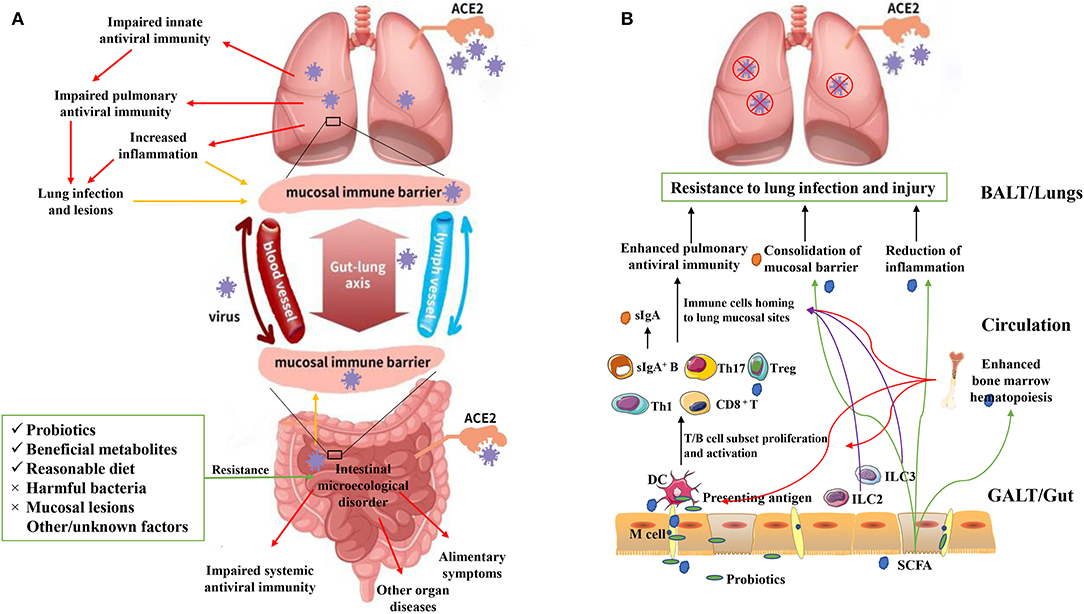
Figure 1. (A) Adverse effects of SARS-CoV-2 on the human lungs and intestine. SARS-CoV-2 can be transmitted through the respiratory or digestive tract, directly infecting the host by binding with the ACE2 receptor of pulmonary epithelial or intestinal epithelial cells. This leads to lung and/or intestinal tissue damage and a systemic immune response. However, after initially infecting the lungs, SARS-CoV-2 can break through the mucosal immune barrier and indirectly affect the intestine along the “gut-lung axis”, and vice versa. Intestinal tissue damage, an excessive inflammatory response, and a dysfunctional immune response can lead to intestinal microecological disorder. However, some interventions (e.g., probiotics, beneficial metabolites, and eliminating harmful bacteria) can provide resistance against these adverse effects. The “gut-lung axis” refers to the cross-talk between these two mucosal parts of the human body, which may take place via blood and lymphatic circulation. The yellow arrows represent “increased barrier dysfunction.” The red arrows represent “adverse effects of SARS-CoV-2 infection.” (B) Possible model for using probiotics and beneficial metabolites (e.g., short chain fatty acids; SCFAs) against lung infection and injury. Probiotics and metabolites such as SCFAs can be taken up by M cells and presented to T cells as antigens via dendritic cells, leading to T/B cell proliferation and activation. Guided by immune mediators, immune cells are then localized at the lung infection site, enhancing antiviral immunity, and providing protection to the lungs. The intestinal innate lymphoid cells (ILC2 and ILC3) can migrate to the lungs to enhance antiviral immunity via lymphatic and blood circulations (purple arrows). Surface IgA can be produced and transported from gut-associated lymphoid tissues to the surface of the pulmonary mucosa, which can prevent virus adhesion and consolidate the mucosal barrier. SCFAs produced by intestinal flora can be transported to the lungs through the blood, where they can play an anti-inflammatory role and consolidate the lung mucosal barrier (green arrows). SCFAs can also be transported to the bone marrow and enhance its hematopoietic function, further promoting the proliferation and activation of dendritic cells and other immune cells. Overall, these phenomena can enhance the antiviral immunity of the host (red arrows).
Possible Mechanism Of the Intestinal Flora Regulation of Immune and Inflammatory Responses of the Host
Intestinal flora is supposed to significantly regulate the development and function of the innate and adaptive immune system, tune the immune cells for pro- and anti-inflammatory responses, and maintain immune homeostasis thereby affecting the host susceptibility to various diseases. In case of a pathogenic SARS-CoV-2 infection, a healthy intestinal flora could be essential in maintaining an optimal immune system to prevent excessive inflammatory responses that eventually become detrimental to lungs and vital organ systems. The intestinal flora might regulate host's immune and inflammatory responses along the gut-lung axis, by means of microbial metabolites and the mucosal immune system (Figure 1B).
Microbial Metabolites
In many microbial metabolites, SCFAs, including butyric acid, acetic acid, and propionic acid, are the most critical metabolites of the intestinal flora. They are extremely important in regulating systemic and pulmonary immune and inflammatory responses (Budden et al., 2017; Goncalves et al., 2018). The most direct function of SCFAs is to reduce the intestinal pH and increase mucin production, which reduces the growth and adhesion of pathogenic microorganisms and improves epithelial integrity, further enhancing the systemic immunity of the host (Fukuda et al., 2011; Jung et al., 2015). SCFAs exert biological effects mainly by inhibiting histone deacetylase (HDAC) and activating G protein–coupled receptors (GPCRs) (Tan et al., 2014; Husted et al., 2017; Li et al., 2018). More specifically, SCFAs can increase the number and function of T regulatory (Treg) cells, T helper (Th) 1 cells, and Th17 effector cells through HDAC inhibition, thus impairing excessive inflammation and immune response in airway diseases along the gut-lung axis (Meijer et al., 2010; Furusawa et al., 2013; Hull et al., 2016; Li et al., 2018). Many studies have shown that GPCRs, especially GPR43, GPR41, and GPR109A, play important roles in the regulation of metabolism, inflammation, and immunity (Den Besten et al., 2013; Kim et al., 2013; Husted et al., 2017; Sun et al., 2017). SCFAs, especially butyrate, have a wide range of anti-inflammatory functions, which are mediated via activation of GPR43 and subsequent activation of β-arrestin 2 by inhibition of the NF-κB pathway (Meijer et al., 2010; Furusawa et al., 2013; Li et al., 2018). SCFAs can also regulate Ly6c(–) patrolling monocyte haematopoiesis and enhance the function of CD8+ T cells to confer protection against influenza virus infection through GPR41 activation (Trompette et al., 2018). Butyrate has been reported to induce the differentiation of Treg cells and IL-10/18-producing T cells through GPR109A activation (Singh et al., 2014).
Recent studies have shown that SCFAs represent a link between the bone marrow, gut, and airways (Dang and Marsland, 2019). These molecules have been detected in very small quantities in the lungs, indicating that the lung microbiome does not produce them in large quantities and the circulating SCFAs do not accumulate in the lung tissue. Thus, SCFAs may have a negligible role in the respiratory tract. However, the metabolized intestinal SCFAs can enter the peripheral blood circulation and bone marrow and affect the development of immune cells, which could then be recruited to the lungs and promote lung homeostasis and immunity (Trompette et al., 2014, 2018; Kopf et al., 2015). SCFAs can also promote the generation of progenitors of macrophages and dendritic cells (DCs) in the bone marrow; phagocytic DCs compose the majority of cells that enter the lungs, thus enhancing the function of the T cell subset and triggering a protective mechanism against allergic airway inflammation and respiratory tract infection (Liu et al., 2009; Trompette et al., 2014; Kopf et al., 2015).
In addition to SCFAs, many metabolites of the symbiotic intestinal flora have been reported to be related to host immunity (Rooks and Garrett, 2016). Tryptophan can be used as an energy source by Lactobacillus to produce ligands for an aryl hydrocarbon receptor; this receptor is essential not only for the organogenesis of intestinal lymphoid follicles but also for maintaining the homeostasis of the epithelial barrier and intraepithelial lymphocytes (Kiss et al., 2011; Lee et al., 2011; Wynn et al., 2013; Gao et al., 2018). Retinoic acid plays an important role in maintaining intestinal immune homeostasis as it promotes IgA production by B cells and Treg cells development through transforming growth factor β (Kang et al., 2007; Sun et al., 2007; Levy et al., 2016). Niacin has been reported to promote anti-inflammatory properties of colonic macrophages and DCs through GPR109A signaling and to enable them to induce Treg cells and IL-10-producing T cells (Singh et al., 2014). LPS can enhance the mucosal immune response and provide improved resistance against infection by respiratory influenza A virus (Ichinohe et al., 2011). Lactate and pyruvate produced by intestinal bacteria can enhance the immune response by inducing the dendrite protrusion of small intestinal mononuclear cells that express CX3CR1+ via GPR31 signaling (Morita et al., 2019). Desaminotyrosine produced by Clostridium orbiscindens has been reported to provide protection against the influenza virus via type I interferon (IFN-1) (Steed et al., 2017).
Common Mucosal Immune System
The common mucosal immune system (MIS) is an important part of the systemic immunity and forms the first line of defense against infections. It is mainly comprised of mucosa-associated lymphoid tissue, such as gut-associated lymphoid tissues (GALT) and bronchial-associated lymphoid tissue (Mcghee and Fujihashi, 2012). The GALT consist of Peyer's patches (PP), mesenteric lymph nodes (MLN), and numerous lymphocytes scattered in the lamina propria (LP) and intestinal epithelium. The intestinal epithelium also has widely distributed microfold (M) cells (Bekiaris et al., 2014; Brugman et al., 2015). M cells take up antigens from the intestinal mucosa and present them to T cells through DCs, which leads to the proliferation and activation of the T cell subset (Cesta, 2006; Qi et al., 2006). Moreover, the GALT are rich in surface IgA (sIgA) (Mcghee and Fujihashi, 2012). When pathogenic bacteria come in contact with sIgA, they are eliminated, whereas non-pathogenic and beneficial bacteria are not disturbed and remain on the mucosal surface (Bunker et al., 2017; Bunker and Bendelac, 2018). Interestingly, these immune cells and immune factors can be transferred from the GALT to the bronchial-associated lymphoid tissue through blood and lymph vessels (Qi et al., 2006; Samuelson et al., 2015), providing and enhancing resistance to respiratory infections. In a mouse model, activated intestinal group 2 innate lymphoid cells (ILC2s) were found in lungs injected with IL-25, and lungs with pneumonia were reported to have intestinal ILC3s, which could help the body in resisting infection (Huang et al., 2018). In addition, a healthy intestinal flora also plays an important role in regulating Toll-like receptor 7 (a kind of pattern recognition receptor) signal transduction after respiratory influenza virus infection, which has been found to alleviate MIS damage caused by antibiotic treatment in mice (Wu et al., 2013).
Altogether, understanding the possible mechanism of intestinal flora regulating host immune and inflammatory response may provide another preliminary theoretical basis for the prevention and treatment of COVID-19 by targeting intestinal flora. More trials need to be initiated to further study the role played by intestinal microorganisms in regulating the immunity of COVID-19 patients.
Intestinal Flora-Mediated Enhancement of Antiviral Immunity
Improving the intestinal microecology (e.g., by taking probiotics and beneficial metabolites) (Table 1) may maintain an optimal immune system and prevent an array of excessive inflammation reactions, while also preventing secondary bacterial infections (Levy et al., 2016; Hanada et al., 2018; Descamps et al., 2019). A high-fiber diet was reported to change the proportion of Firmicutes and Bacteroidetes that could increase the level of SCFAs in the intestine and blood, which in turn reduced the lung damage caused by a respiratory syncytial virus infection. In the mouse model, the same effect was also observed by supplementing acetic acid in drinking water (Meijer et al., 2010; Den Besten et al., 2013; Trompette et al., 2014). The probiotic bacteria of the genus Lactobacillus were shown to stimulate respiratory immune responses in mice by increasing inflammatory signals, thereby enhancing the host's defense against respiratory infections (Salva et al., 2010; Yoda et al., 2012). Lactobacillus casei enhanced the phagocytic and killing activity of alveolar macrophages and increased IgA, IFN-γ, and TNF-α expression, assisting the host in the fight against influenza virus (Hori et al., 2001). Interestingly, Bifidobacterium, Lactobacillus paracasei, and Lactobacillus rhamnosus also presented an effect in preventing respiratory infections (e.g., H1N1, H5N1, and H3N2) by enhancing the vaccine response (Lei et al., 2010; Samuelson et al., 2015). Recent studies have shown that traditional Chinese medicine may have beneficial effects on the recovery of patients with COVID-19, possibly by enhancing the intestinal microecological balance to improve immunity (Xu et al., 2017, 2020; Wang et al., 2019). Notably, a pre-published study (as yet, not peer-reviewed) suggested that the risk of developing severe COVID-19 among patients with vitamin D deficiency was significantly higher than in patients with normal vitamin D levels; moreover, vitamin D may reduce COVID-19 severity by suppressing the cytokine storm displayed by COVID-19 patients (Daneshkhah et al., 2020). However, whether this difference is mediated by intestinal flora remains to be further studied. Nonetheless, what we already know is that Vitamins A and D can enhance the intestinal barrier function and mucosal immune response by maintaining the normal function of ILC3s and T cells (Cantorna et al., 2019). Overall, it is apparent that probiotics and diet-mediated modulation of intestinal flora can influence immunity. The administration of personalized probiotics and diet may be thoughtfully considered for COVID-19 patients to accelerate recovery and improve prognosis.
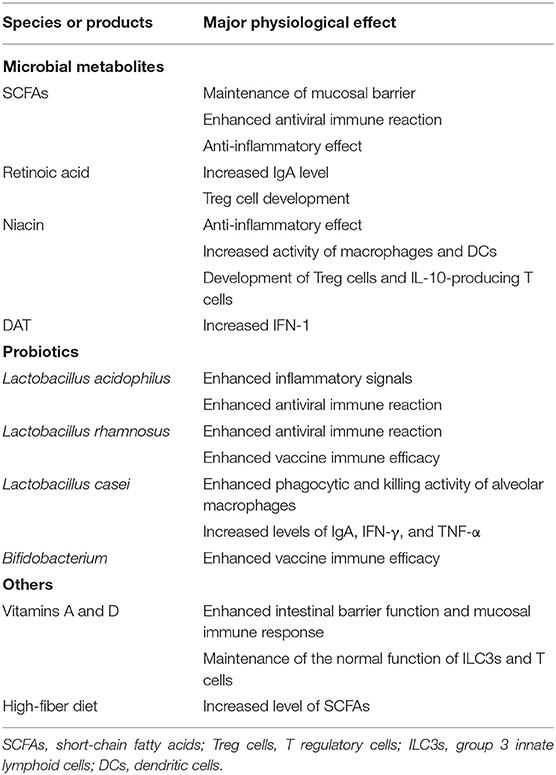
Table 1. Changes in the immune and inflammatory response upon administration of different probiotics and products for pulmonary infectious disease treatment.
Conclusion and Prospects
The COVID-19 pandemic presents a significant social and economic burden worldwide, and studies on the disease, unfortunately, remain at a very preliminary stage. This review provides a novel suggestion that the intestinal flora may partially mediate the effects of SARS-CoV-2 on the both local gastrointestinal response and systemic immune response of the host, and thus be a target for COVID-19 prevention and treatment. This is consistent with emerging data, which suggest that the gut microbiota plays a key role in predicting the blood proteomic biomarkers that determine the abnormal inflammatory state of individuals with severe COVID-19 (Gou et al., 2020). Moreover, considering the fact that elderly people have less diverse intestinal flora in which beneficial microorganisms (e.g., Bifidobacterium) lose ground (Nagpal et al., 2018), this is in line with the observation that older individuals are more susceptible to SARS-CoV-2 and more severe COVID-19 (Goyal et al., 2020; Lake, 2020). The structure and function of the intestinal flora could be a potential biological mechanism behind the diverse susceptibility of different groups of people to SARS-CoV-2. However, the specific mechanisms through which the SARS-CoV-2 infection influences the intestinal microbial community among patients of different ages, ethnic groups, and geographical locations remain to be seen.
The prevention and treatment strategies for SARS-CoV-2 infection considering gastroenterology and intestinal microbiota have received great attention (Gou et al., 2020; Li L. Y. et al., 2020). In February 2020, China's National Health Commission and National Administration of Traditional Chinese Medicine suggested the use of probiotics in patients with severe COVID-19, which has shown good efficacy (National Health Committee of the People's Republic of China, 2020). As shown by existing indirect evidences, there is a potential strategy to prevent and treat COVID-19 through the improvement of intestinal flora composition and of its metabolites. This could be performed using probiotics, personalized diet, and traditional Chinese medicine to balance the immune function and suppress “cytokine storm.” Some specific intestinal microorganisms that can downregulate intestinal ACE2 expression has also been considered as the potential target to fight against SARS-CoV-2 (Zuo et al., 2020). These insights will add new dimensions to understanding SARS-CoV-2 and COVID-19, and can also be helpful for designing a more reasonable and personalized treatment plan for patients, which would be of great significance for assigning medical resources.
It is worth noting that the rationale for using probiotics in COVID-19 is derived from indirect evidence. Different kinds of probiotics and/or different doses of probiotics usually have different biological effects on the host. To further understand the specific mechanism and elucidate the benefits of personalized functional food including probiotics and metabolite products in the prevention and treatment of COVID-19, more clinical trials and evidence-based medical data are needed.
Author Contributions
All authors contributed to the critical analysis of the collected data and writing the manuscript, and all authors approved the final manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (31960236 and 31770536) and the Talent Innovation and Entrepreneurship Project of Lanzhou City (2019-RC-34).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Mrs. Hong-Yu Liu and Editage (www.editage.cn) for English language editing and Prof. Hong-Mei Chen for providing critical revisions to the manuscript.
References
Abrahamsson, T. R., Jakobsson, H. E., Andersson, A. F., Bjorksten, B., Engstrand, L., and Jenmalm, M. C. (2014). Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Allergy 44, 842–850. doi: 10.1111/cea.12253
Abt, M. C., Osborne, L. C., Monticelli, L. A., Doering, T. A., Alenghat, T., Sonnenberg, G. F., et al. (2012). Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170. doi: 10.1016/j.immuni.2012.04.011
Bekiaris, V., Persson, E. K., and Agace, W. W. (2014). Intestinal dendritic cells in the regulation of mucosal immunity. Immunol. Rev. 260, 86–101. doi: 10.1111/imr.12194
Belkaid, Y., and Harrison, O. J. (2017). Homeostatic immunity and the microbiota. Immunity 46, 562–576. doi: 10.1016/j.immuni.2017.04.008
Bradley, C. P., Teng, F., Felix, K. M., Sano, T., Naskar, D., Block, K. E., et al. (2017). Segmented filamentous bacteria provoke lung autoimmunity by inducing gut-lung axis Th17 cells expressing dual TCRs. Cell Host Microbe. 22, 697–704 e694. doi: 10.1016/j.chom.2017.10.007
Brugman, S., Perdijk, O., Van Neerven, R. J., and Savelkoul, H. F. (2015). Mucosal immune development in early life: setting the stage. Arch. Immunol. Ther. Exp. 63, 251–268. doi: 10.1007/s00005-015-0329-y
Budden, K. F., Gellatly, S. L., Wood, D. L., Cooper, M. A., Morrison, M., Hugenholtz, P., et al. (2017). Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 15, 55–63. doi: 10.1038/nrmicro.2016.142
Bunker, J. J., and Bendelac, A. (2018). IgA responses to microbiota. Immunity 49, 211–224. doi: 10.1016/j.immuni.2018.08.011
Bunker, J. J., Erickson, S. A., Flynn, T. M., Henry, C., Koval, J. C., Meisel, M., et al. (2017). Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 358:eaan6619. doi: 10.1126/science.aan6619
Cantorna, M. T., Snyder, L., and Arora, J. (2019). Vitamin A and Vitamin D regulate the microbial complexity, barrier function, and the mucosal immune responses to ensure intestinal homeostasis. Crit. Rev. Biochem. Mol. Biol. 54, 184–192. doi: 10.1080/10409238.2019.1611734
Cesta, M. F. (2006). Normal structure, function, and histology of mucosa-associated lymphoid tissue. Toxicol. Pathol. 34, 599–608. doi: 10.1080/01926230600865531
Cholankeril, G., Podboy, A., Aivaliotis, V. I., Tarlow, B., Pham, E. A., Spencer, S., et al. (2020). High prevalence of concurrent gastrointestinal manifestations in patients with SARS-CoV-2: early experience from California. Gastroenterology S0016-5085(20)30471-6. doi: 10.1053/j.gastro.2020.04.008
Cole-Jeffrey, C. T., Liu, M., Katovich, M. J., Raizada, M. K., and Shenoy, V. (2015). ACE2 and microbiota: emerging targets for cardiopulmonary disease therapy. J. Cardiovasc. Pharmacol. 66, 540–550. doi: 10.1097/FJC.0000000000000307
Daneshkhah, A., Agrawal, V., Eshein, A., Subramanian, H., Roy, H. K., and Backman, V. (2020). The possible role of Vitamin D in suppressing cytokine storm and associated mortality in COVID-19 Patients. medRXiv 2020.2004.2008.20058578. doi: 10.1101/2020.04.08.20058578
Dang, A. T., and Marsland, B. J. (2019). Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 12, 843–850. doi: 10.1038/s41385-019-0160-6
Date, Y., Ebisawa, M., Fukuda, S., Shima, H., Obata, Y., Takahashi, D., et al. (2017). NALT M cells are important for immune induction for the common mucosal immune system. Int. Immunol. 29, 471–478. doi: 10.1093/intimm/dxx064
Den Besten, G., Van Eunen, K., Groen, A. K., Venema, K., Reijngoud, D. J., and Bakker, B. M. (2013). The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54, 2325–2340. doi: 10.1194/jlr.R036012
Descamps, H. C., Herrmann, B., Wiredu, D., and Thaiss, C. A. (2019). The path toward using microbial metabolites as therapies. EBioMedicine 44, 747–754. doi: 10.1016/j.ebiom.2019.05.063
Dharmage, S. C., Lodge, C. J., Lowe, A. J., and Allen, K. J. (2015). Antibiotics and risk of asthma: a debate that is set to continue. Clin. Exp. Allergy 45, 6–8. doi: 10.1111/cea.12424
Dickson, R. P., Singer, B. H., Newstead, M. W., Falkowski, N. R., Erb-Downward, J. R., Standiford, T. J., et al. (2016). Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol. 1:16113. doi: 10.1038/nmicrobiol.2016.113
Donaldson, G. P., Lee, S. M., and Mazmanian, S. K. (2016). Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32. doi: 10.1038/nrmicro3552
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi: 10.1126/science.1110591
Fukuda, S., Toh, H., Hase, K., Oshima, K., Nakanishi, Y., Yoshimura, K., et al. (2011). Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547. doi: 10.1038/nature09646
Furusawa, Y., Obata, Y., Fukuda, S., Endo, T. A., Nakato, G., Takahashi, D., et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450. doi: 10.1038/nature12721
Gao, J., Xu, K., Liu, H., Liu, G., Bai, M., Peng, C., et al. (2018). Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell. Infect. Microbiol. 8:13. doi: 10.3389/fcimb.2018.00013
Goncalves, P., Araujo, J. R., and Di Santo, J. P. (2018). A cross-talk between microbiota-derived short-chain fatty acids and the host mucosal immune system regulates intestinal homeostasis and inflammatory bowel disease. Inflamm. Bowel Dis. 24, 558–572. doi: 10.1093/ibd/izx029
Gong, S., Lan, T., Zeng, L., Luo, H., Yang, X., Li, N., et al. (2018). Gut microbiota mediates diurnal variation of acetaminophen induced acute liver injury in mice. J. Hepatol. 69, 51–59. doi: 10.1016/j.jhep.2018.02.024
Gou, W., Fu, Y., Yue, L., Chen, G.-D., Cai, X., Shuai, M., et al. (2020). Gut microbiota may underlie the predisposition of healthy individuals to COVID-19. medRXiv. 2020.2004.2022.20076091. doi: 10.1101/2020.04.22.20076091
Goyal, P., Choi, J. J., Pinheiro, L. C., Schenck, E. J., Chen, R., Jabri, A., et al. (2020). Clinical characteristics of Covid-19 in New York city. N Engl J Med. NEJMc2010419. doi: 10.1056/NEJMc2010419
Grice, E. A., and Segre, J. A. (2012). The human microbiome: our second genome. Annu. Rev. Genomics Hum. Genet. 13, 151–170. doi: 10.1146/annurev-genom-090711-163814
Guan, W. J., Ni, Z. Y., Hu, Y., Liang, W. H., Ou, C. Q., He, J. X., et al. (2020). Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 382, 1708–1720. doi: 10.1056/NEJMoa2002032
Hanada, S., Pirzadeh, M., Carver, K. Y., and Deng, J. C. (2018). Respiratory viral infection-induced microbiome alterations and secondary bacterial pneumonia. Front. Immunol. 9:2640. doi: 10.3389/fimmu.2018.02640
Hand, T. W., Vujkovic-Cvijin, I., Ridaura, V. K., and Belkaid, Y. (2016). Linking the microbiota, chronic disease, and the immune system. Trends Endocrinol. Metab. 27, 831–843. doi: 10.1016/j.tem.2016.08.003
Hashimoto, T., Perlot, T., Rehman, A., Trichereau, J., Ishiguro, H., Paolino, M., et al. (2012). ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 487, 477–481. doi: 10.1038/nature11228
Holshue, M. L., Debolt, C., Lindquist, S., Lofy, K. H., Wiesman, J., Bruce, H., et al. (2020). First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 382, 929–936. doi: 10.1056/NEJMoa2001191
Hori, T., Kiyoshima, J., Shida, K., and Yasui, H. (2001). Effect of intranasal administration of Lactobacillus casei Shirota on influenza virus infection of upper respiratory tract in mice. Clin. Diagn. Lab. Immunol. 8, 593–597. doi: 10.1128/CDLI.8.3.593-597.2001
Huang, Y., Mao, K., Chen, X., Sun, M. A., Kawabe, T., Li, W., et al. (2018). S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science 359, 114–119. doi: 10.1126/science.aam5809
Hull, E. E., Montgomery, M. R., and Leyva, K. J. (2016). HDAC inhibitors as epigenetic regulators of the immune system: impacts on cancer therapy and inflammatory diseases. Biomed Res. Int. 2016:8797206. doi: 10.1155/2016/8797206
Husted, A. S., Trauelsen, M., Rudenko, O., Hjorth, S. A., and Schwartz, T. W. (2017). GPCR-mediated signaling of metabolites. Cell Metab. 25, 777–796. doi: 10.1016/j.cmet.2017.03.008
Ichinohe, T., Pang, I. K., Kumamoto, Y., Peaper, D. R., Ho, J. H., Murray, T. S., et al. (2011). Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U.S.A. 108, 5354–5359. doi: 10.1073/pnas.1019378108
Ipci, K., Altintoprak, N., Muluk, N. B., Senturk, M., and Cingi, C. (2017). The possible mechanisms of the human microbiome in allergic diseases. Eur. Arch. Otorhinolaryngol. 274, 617–626. doi: 10.1007/s00405-016-4058-6
Javed, K., and Broer, S. (2019). Mice lacking the intestinal and renal neutral amino acid transporter SLC6A19 demonstrate the relationship between dietary protein intake and amino acid malabsorption. Nutrients 11:2024. doi: 10.3390/nu11092024
Jung, T. H., Park, J. H., Jeon, W. M., and Han, K. S. (2015). Butyrate modulates bacterial adherence on LS174T human colorectal cells by stimulating mucin secretion and MAPK signaling pathway. Nutr. Res. Pract. 9, 343–349. doi: 10.4162/nrp.2015.9.4.343
Kang, S. G., Lim, H. W., Andrisani, O. M., Broxmeyer, H. E., and Kim, C. H. (2007). Vitamin A metabolites induce gut-homing FoxP3+ regulatory T cells. J. Immunol. 179, 3724–3733. doi: 10.4049/jimmunol.179.6.3724
Kim, M. H., Kang, S. G., Park, J. H., Yanagisawa, M., and Kim, C. H. (2013). Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 145, 396–406 e391–e310. doi: 10.1053/j.gastro.2013.04.056
Kiss, E. A., Vonarbourg, C., Kopfmann, S., Hobeika, E., Finke, D., Esser, C., et al. (2011). Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 334, 1561–1565. doi: 10.1126/science.1214914
Kopf, M., Schneider, C., and Nobs, S. P. (2015). The development and function of lung-resident macrophages and dendritic cells. Nat. Immunol. 16, 36–44. doi: 10.1038/ni.3052
Lake, M. A. (2020). What we know so far: COVID-19 current clinical knowledge and research. Clin Med. 20, 124–127. doi: 10.7861/clinmed.2019-coron
Lamers, M. M., Beumer, J., Van Der Vaart, J., Knoops, K., Puschhof, J., Breugem, T. I., et al. (2020). SARS-CoV-2 productively infects human gut enterocytes. Science. doi: 10.1126/science.abc1669. [Epub ahead of print].
Leblanc, J. G., Chain, F., Martin, R., Bermudez-Humaran, L. G., Courau, S., and Langella, P. (2017). Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb. Cell Fact. 16:79. doi: 10.1186/s12934-017-0691-z
Lee, J. S., Cella, M., Mcdonald, K. G., Garlanda, C., Kennedy, G. D., Nukaya, M., et al. (2011). AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 13, 144–151. doi: 10.1038/ni.2187
Lei, H., Xu, Y., Chen, J., Wei, X., and Lam, D. M. (2010). Immunoprotection against influenza H5N1 virus by oral administration of enteric-coated recombinant Lactococcus lactis mini-capsules. Virology 407, 319–324. doi: 10.1016/j.virol.2010.08.007
Levy, M., Thaiss, C. A., and Elinav, E. (2016). Metabolites: messengers between the microbiota and the immune system. Genes Dev. 30, 1589–1597. doi: 10.1101/gad.284091.116
Li, L. Y., Wu, W., Chen, S., Gu, J. W., Li, X. L., Song, H. J., et al. (2020). Digestive system involvement of novel coronavirus infection: prevention and control infection from a gastroenterology perspective. J Dig Dis. 21, 199–204. doi: 10.1111/1751-2980.12862
Li, M., Van Esch, B., Wagenaar, G. T. M., Garssen, J., Folkerts, G., Henricks, P., et al. (2018). Pro- and anti-inflammatory effects of short chain fatty acids on immune and endothelial cells. Eur. J. Pharmacol. 831, 52–59. doi: 10.1016/j.ejphar.2018.05.003
Li, M. Y., Li, L., Zhang, Y., and Wang, X. S. (2020). Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty 9:45. doi: 10.1186/s40249-020-00662-x
Lin, L., Jiang, X., Zhang, Z., Huang, S., Zhang, Z., Fang, Z., et al. (2020). Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 69, 997–1001. doi: 10.1136/gutjnl-2020-321013
Liu, K., Victora, G. D., Schwickert, T. A., Guermonprez, P., Meredith, M. M., Yao, K., et al. (2009). In vivo analysis of dendritic cell development and homeostasis. Science 324, 392–397. doi: 10.1126/science.1170540
Looft, T., and Allen, H. K. (2012). Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes 3, 463–467. doi: 10.4161/gmic.21288
Lynch, S. V., and Pedersen, O. (2016). The human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379. doi: 10.1056/NEJMra1600266
Mcghee, J. R., and Fujihashi, K. (2012). Inside the mucosal immune system. PLoS Biol. 10:e1001397. doi: 10.1371/journal.pbio.1001397
Meijer, K., De Vos, P., and Priebe, M. G. (2010). Butyrate and other short-chain fatty acids as modulators of immunity: what relevance for health? Curr. Opin. Clin. Nutr. Metab. Care 13, 715–721. doi: 10.1097/MCO.0b013e32833eebe5
Metsala, J., Lundqvist, A., Virta, L. J., Kaila, M., Gissler, M., and Virtanen, S. M. (2015). Prenatal and post-natal exposure to antibiotics and risk of asthma in childhood. Clin. Exp. Allergy 45, 137–145. doi: 10.1111/cea.12356
Morita, N., Umemoto, E., Fujita, S., Hayashi, A., Kikuta, J., Kimura, I., et al. (2019). GPR31-dependent dendrite protrusion of intestinal CX3CR1(+) cells by bacterial metabolites. Nature 566, 110–114. doi: 10.1038/s41586-019-0884-1
Nagpal, R., Mainali, R., Ahmadi, S., Wang, S., Singh, R., Kavanagh, K., et al. (2018). Gut microbiome and aging: physiological and mechanistic insights. Nutr Healthy Aging 4, 267–285. doi: 10.3233/NHA-170030
National Health Committee of the People's Republic of China N.a.O.T.C.M. (2020). Diagnostic and Therapeutic Guidance for 2019 Novel Coronavirus Disease (version 5). Available online at: http://www.nhc.gov.cn/yzygj/s7653p/202002/d4b895337e19445f8d728fcaf1e3e13a/files/ab6bec7f93e64e7f998d802991203cd6.pdf (accessed February 8, 2020).
Pang, P., Yu, B., Shi, Y., Deng, L., Xu, H., Wu, S., et al. (2018). Alteration of intestinal flora stimulates pulmonary microRNAs to interfere with host antiviral immunity in influenza. Molecules 23:3151. doi: 10.3390/molecules23123151
Patel, S., Rauf, A., Khan, H., and Abu-Izneid, T. (2017). Renin-angiotensin-aldosterone (RAAS): The ubiquitous system for homeostasis and pathologies. Biomed. Pharmacother. 94, 317–325. doi: 10.1016/j.biopha.2017.07.091
Pedersen, S. F., and Ho, Y. C. (2020). SARS-CoV-2: a storm is raging. J. Clin. Invest. 130, 2202–2205. doi: 10.1172/JCI137647
Qi, H., Egen, J. G., Huang, A. Y., and Germain, R. N. (2006). Extrafollicular activation of lymph node B cells by antigen-bearing dendritic cells. Science 312, 1672–1676. doi: 10.1126/science.1125703
Rooks, M. G., and Garrett, W. S. (2016). Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352. doi: 10.1038/nri.2016.42
Salva, S., Villena, J., and Alvarez, S. (2010). Immunomodulatory activity of Lactobacillus rhamnosus strains isolated from goat milk: impact on intestinal and respiratory infections. Int. J. Food Microbiol. 141, 82–89. doi: 10.1016/j.ijfoodmicro.2010.03.013
Samuelson, D. R., Welsh, D. A., and Shellito, J. E. (2015). Regulation of lung immunity and host defense by the intestinal microbiota. Front. Microbiol. 6:1085. doi: 10.3389/fmicb.2015.01085
Schirmer, M., Franzosa, E. A., Lloyd-Price, J., Mciver, L. J., Schwager, R., Poon, T. W., et al. (2018). Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat Microbiol 3, 337–346. doi: 10.1038/s41564-017-0089-z
Selber-Hnatiw, S., Rukundo, B., Ahmadi, M., Akoubi, H., Al-Bizri, H., Aliu, A. F., et al. (2017). Human gut microbiota: toward an ecology of disease. Front. Microbiol. 8:1265. doi: 10.3389/fmicb.2017.01265
Sender, R., Fuchs, S., and Milo, R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14:e1002533. doi: 10.1371/journal.pbio.1002533
Shi, N., Li, N., Duan, X., and Niu, H. (2017). Interaction between the gut microbiome and mucosal immune system. Mil. Med. Res. 4:14. doi: 10.1186/s40779-017-0122-9
Singer, D., Camargo, S. M., Ramadan, T., Schafer, M., Mariotta, L., Herzog, B., et al. (2012). Defective intestinal amino acid absorption in Ace2 null mice. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G686–695. doi: 10.1152/ajpgi.00140.2012
Singh, N., Gurav, A., Sivaprakasam, S., Brady, E., Padia, R., Shi, H., et al. (2014). Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139. doi: 10.1016/j.immuni.2013.12.007
Sittipo, P., Shim, J. W., and Lee, Y. K. (2019). Microbial metabolites determine host health and the status of some diseases. Int. J. Mol. Sci. 20:5296. doi: 10.3390/ijms20215296
Steed, A. L., Christophi, G. P., Kaiko, G. E., Sun, L., Goodwin, V. M., Jain, U., et al. (2017). The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science 357, 498–502. doi: 10.1126/science.aam5336
Sun, C. M., Hall, J. A., Blank, R. B., Bouladoux, N., Oukka, M., Mora, J. R., et al. (2007). Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 204, 1775–1785. doi: 10.1084/jem.20070602
Sun, M., Wu, W., Liu, Z., and Cong, Y. (2017). Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J. Gastroenterol. 52, 1–8. doi: 10.1007/s00535-016-1242-9
Sze, M. A., Tsuruta, M., Yang, S. W., Oh, Y., Man, S. F., Hogg, J. C., et al. (2014). Changes in the bacterial microbiota in gut, blood, and lungs following acute LPS instillation into mice lungs. PLoS ONE 9:e111228. doi: 10.1371/journal.pone.0111228
Tan, J., Mckenzie, C., Potamitis, M., Thorburn, A. N., Mackay, C. R., and Macia, L. (2014). The role of short-chain fatty acids in health and disease. Adv. Immunol. 121, 91–119. doi: 10.1016/B978-0-12-800100-4.00003-9
Tirone, C., Pezza, L., Paladini, A., Tana, M., Aurilia, C., Lio, A., et al. (2019). Gut and lung microbiota in preterm infants: immunological modulation and implication in neonatal outcomes. Front. Immunol. 10:2910. doi: 10.3389/fimmu.2019.02910
Trompette, A., Gollwitzer, E. S., Pattaroni, C., Lopez-Mejia, I. C., Riva, E., Pernot, J., et al. (2018). Dietary fiber confers protection against flu by shaping Ly6c(-) patrolling monocyte hematopoiesis and CD8(+) T Cell Metabolism. Immunity 48, 992–1005 e1008. doi: 10.1016/j.immuni.2018.04.022
Trompette, A., Gollwitzer, E. S., Yadava, K., Sichelstiel, A. K., Sprenger, N., Ngom-Bru, C., et al. (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166. doi: 10.1038/nm.3444
Visconti, A., Le Roy, C. I., Rosa, F., Rossi, N., Martin, T. C., Mohney, R. P., et al. (2019). Interplay between the human gut microbiome and host metabolism. Nat. Commun. 10:4505. doi: 10.1038/s41467-019-12476-z
Voreades, N., Kozil, A., and Weir, T. L. (2014). Diet and the development of the human intestinal microbiome. Front. Microbiol. 5:494. doi: 10.3389/fmicb.2014.00494
Vuille-Dit-Bille, R. N., Camargo, S. M., Emmenegger, L., Sasse, T., Kummer, E., Jando, J., et al. (2015). Human intestine luminal ACE2 and amino acid transporter expression increased by ACE-inhibitors. Amino Acids 47, 693–705. doi: 10.1007/s00726-014-1889-6
Wan, Y., Li, J., Shen, L., Zou, Y., Hou, L., Zhu, L., et al. (2020). Enteric involvement in hospitalised patients with COVID-19 outside Wuhan. Lancet Gastroenterol Hepatol. 5, 534–535 doi: 10.1016/S2468-1253(20)30118-7
Wang, Y., Qin, S., Jia, J., Huang, L., Li, F., Jin, F., et al. (2019). Intestinal microbiota-associated metabolites: crucial factors in the effectiveness of herbal medicines and diet therapies. Front. Physiol. 10:1343. doi: 10.3389/fphys.2019.01343
WHO (2020). Coronavirus Disease (COVID-19) Outbreak Situation. Available online at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed May 25, 2020).
Wu, F., Zhao, S., Yu, B., Chen, Y. M., Wang, W., Song, Z. G., et al. (2020). A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269. doi: 10.1038/s41586-020-2202-3
Wu, S., Jiang, Z. Y., Sun, Y. F., Yu, B., Chen, J., Dai, C. Q., et al. (2013). Microbiota regulates the TLR7 signaling pathway against respiratory tract influenza A virus infection. Curr. Microbiol. 67, 414–422. doi: 10.1007/s00284-013-0380-z
Wynn, T. A., Chawla, A., and Pollard, J. W. (2013). Macrophage biology in development, homeostasis and disease. Nature 496, 445–455. doi: 10.1038/nature12034
Xu, J., Chen, H. B., and Li, S. L. (2017). Understanding the molecular mechanisms of the interplay between herbal medicines and gut microbiota. Med. Res. Rev. 37, 1140–1185. doi: 10.1002/med.21431
Xu, K., Cai, H., Shen, Y., Ni, Q., Chen, Y., Hu, S., et al. (2020). [Management of corona virus disease-19 (COVID-19): the Zhejiang experience]. Zhejiang Da Xue Xue Bao Yi Xue Ban 49, 147–157.
Ye, Q., Wang, B., and Mao, J. (2020). The pathogenesis and treatment of the ‘Cytokine Storm' in COVID-19. J Infect. 80, 607–613. doi: 10.1016/j.jinf.2020.03.037
Yoda, K., He, F., Miyazawa, K., Kawase, M., Kubota, A., and Hiramatsu, M. (2012). Orally administered heat-killed Lactobacillus gasseri TMC0356 alters respiratory immune responses and intestinal microbiota of diet-induced obese mice. J. Appl. Microbiol. 113, 155–162. doi: 10.1111/j.1365-2672.2012.05316.x
Zhang, Q., Hu, J., Feng, J. W., Hu, X. T., Wang, T., Gong, W. X., et al. (2020). Influenza infection elicits an expansion of gut population of endogenous Bifidobacterium animalis which protects mice against infection. Genome Biol. 21:99. doi: 10.1186/s13059-020-02007-1
Zhernakova, A., Kurilshikov, A., Bonder, M. J., Tigchelaar, E. F., Schirmer, M., Vatanen, T., et al. (2016). Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569. doi: 10.1126/science.aad3369
Zhou, P., Yang, X. L., Wang, X. G., Hu, B., Zhang, L., Zhang, W., et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579, 270–273. doi: 10.1038/s41586-020-2012-7
Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., et al. (2020). A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 382, 727–733. doi: 10.1056/NEJMoa2001017
Zou, X., Chen, C., Zou, J., Han, P., Hao, J., and Han, Z. (2020). The single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to Wuhan 2019-nCoV infection. Front. Med. 14, 185–192. doi: 10.1007/s11684-020-0754-0
Zumla, A., Chan, J. F., Azhar, E. I., Hui, D. S., and Yuen, K. Y. (2016). Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 15, 327–347. doi: 10.1038/nrd.2015.37
Keywords: COVID-19, SARS-CoV-2, intestinal flora, gut-lung axis, immunity, SCFAs
Citation: He L-H, Ren L-F, Li J-F, Wu Y-N, Li X and Zhang L (2020) Intestinal Flora as a Potential Strategy to Fight SARS-CoV-2 Infection. Front. Microbiol. 11:1388. doi: 10.3389/fmicb.2020.01388
Received: 06 March 2020; Accepted: 29 May 2020;
Published: 09 June 2020.
Edited by:
Daniel Marc, Institut National de Recherche pour l'agriculture, l'alimentation et l'environnement (INRAE), FranceReviewed by:
Zhongming Ge, Massachusetts Institute of Technology, United StatesNoton Kumar Dutta, Johns Hopkins University, United States
Copyright © 2020 He, Ren, Li, Wu, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xun Li, bHhkcjIxQDEyNi5jb20=; Lei Zhang, ZHJ6aGFuZ2xlaUBsenUuZWR1LmNu
†These authors have contributed equally to this work
 Li-Hong He
Li-Hong He Long-Fei Ren1,2,3†
Long-Fei Ren1,2,3†