- Département de Biochimie, Université de Lausanne, Lausanne, Switzerland
Cells face major changes in demand for and supply of inorganic phosphate (Pi). Pi is often a limiting nutrient in the environment, particularly for plants and microorganisms. At the same time, the need for phosphate varies, establishing conflicts of goals. Cells experience strong peaks of Pi demand, e.g., during the S-phase, when DNA, a highly abundant and phosphate-rich compound, is duplicated. While cells must satisfy these Pi demands, they must safeguard themselves against an excess of Pi in the cytosol. This is necessary because Pi is a product of all nucleotide-hydrolyzing reactions. An accumulation of Pi shifts the equilibria of these reactions and reduces the free energy that they can provide to drive endergonic metabolic reactions. Thus, while Pi starvation may simply retard growth and division, an elevated cytosolic Pi concentration is potentially dangerous for cells because it might stall metabolism. Accordingly, the consequences of perturbed cellular Pi homeostasis are severe. In eukaryotes, they range from lethality in microorganisms such as yeast (Sethuraman et al., 2001; Hürlimann, 2009), severe growth retardation and dwarfism in plants (Puga et al., 2014; Liu et al., 2015; Wild et al., 2016) to neurodegeneration or renal Fanconi syndrome in humans (Legati et al., 2015; Ansermet et al., 2017). Intracellular Pi homeostasis is thus not only a fundamental topic of cell biology but also of growing interest for medicine and agriculture.
Phosphate Control Is a Challenge for Cells
Cells should coordinate their systems for the uptake, export, and storage of Pi in order to strike a delicate balance between the biosynthetic requirements for Pi and the risks of an excessive cytoplasmic Pi concentration. To achieve this, they may use signaling networks that sense extra- and intracellular Pi and a buffering system for cytosolic Pi. In this review, we do not provide a global overview of phosphate homeostasis in all model organisms studied in this respect because this adds significant complexity and detail, resulting for example from multicellularity, tissue differentiation, or the complexity of their life cycles. We will rather focus on the yeast Saccharomyces cerevisiae because it is a unicellular model and the eukaryotic model system in which relevant pathways and mechanisms have been best characterized. Focusing on a single well-characterized and simple model provides the best basis for our effort to develop a conceptual framework for intracellular phosphate homeostasis, which may provide leads to dissect this crucial homeostatic system also in other eukaryotic organisms (Table 1). We therefore add information from other models only where it is necessary to provide information that is not available for S. cerevisiae.
Strategies for Phosphate Homeostasis
Multicellular organisms can regulate Pi concentration at the organismal level. Humans, for example, maintain Pi concentration in circulating body fluids through filtration at the level of the kidneys and controlled reabsorption (Biber et al., 2013; Sabbagh, 2013). They can access the apatite in bones as a huge Pi reserve. These well-studied mechanisms provide the individual cells with a relatively constant environment of extracellular Pi, alleviating the need for complex Pi-foraging programs at the cellular level. Nevertheless, human tissues widely express a regulated Pi exporter in the plasma membrane (XPR1) (Giovannini et al., 2013), which suggests that they might also maintain a safeguard against peaks of cytosolic Pi. Dysregulation of XPR1 leads to neurodegeneration and brain calcification, suggesting that XPR1 might dampen the significant changes of Pi concentration in brain cells that can occur depending on its metabolic state (McIlwain et al., 1951). Interestingly, XPR1 is even important for Pi homeostasis at the organismal level because it is implicated in the reabsorption of Pi across the renal tubules of the kidney (Ansermet et al., 2017).
Whereas Pi regulation at the organismal level has been intensely studied, particularly in humans (Biber et al., 2013; Sabbagh, 2013), it is poorly understood how cytosolic Pi concentration is measured and regulated at the level of individual cells. Unicellular organisms can experience rapid changes in Pi availability. They use multiple systems to maintain Pi homeostasis, which allow the cell to mount a graded response that is tuned to the degree of Pi availability and consumption (Bostian et al., 1983). Cells respond to Pi scarcity with various foraging strategies (Conrad et al., 2014; Puga et al., 2017), in which they try to liberate Pi from a variety of extracellular substrates. They can express Pi importers of low and high affinity, which allow them to acquire Pi under a wide range of external concentrations. They maintain important Pi stores in acidocalcisome-like organelles, which, in case of sudden Pi starvation, can guarantee them sufficient reserves to finish the next cell cycle and make an ordered transition into a robust quiescent state. Finally, cells can also “recycle” and liberate Pi from internal sources, such as nucleotides and phospholipids. While such recycling appears senseless for a growing cell—because it will need those compounds to grow—it may become critical for cells that have already arrested growth. They may choose to reallocate their internal Pi pool in order to perform new biosyntheses that are critical to survive in the non-dividing, starved state. Since Pi scarcity retards growth but as such does not seem to be lethal, mounting starvation responses can rely on (relatively slow) transcriptional Pi starvation programs. But cells may also experience a sudden excess of Pi and may need highly reactive mechanisms to protect themselves against the potentially lethal consequences. In this situation, posttranslational signaling becomes important, allowing to rapidly and simultaneously regulate multiple systems for import, export, and storage of Pi and to thus dampen cytosolic Pi peaks.
Acidocalcisomes: a Conserved Organelle With a Role in Phosphate Buffering?
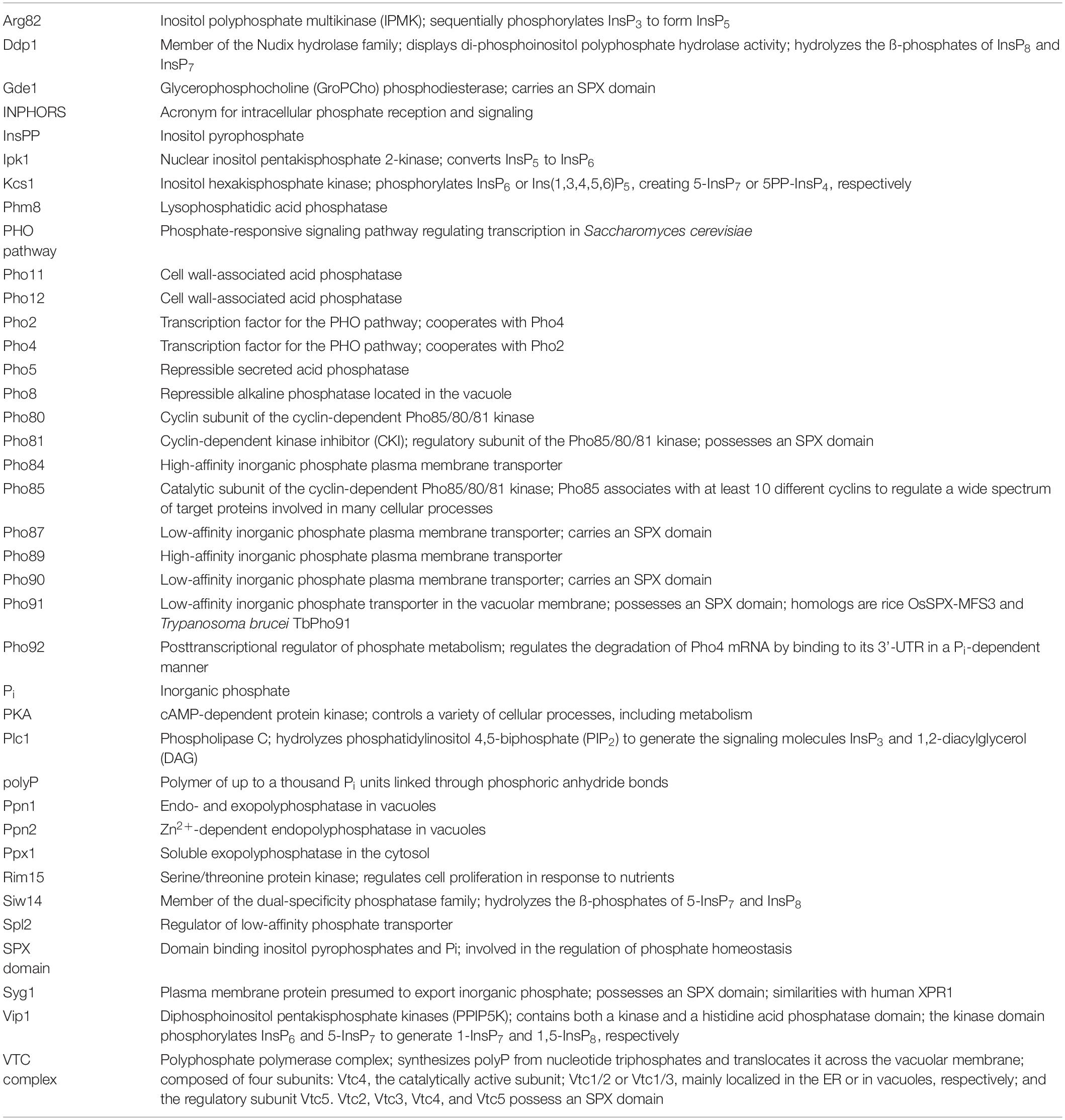
Acidocalcisomes are membrane-bound organelles that are conserved from bacteria to mammals (Docampo and Huang, 2016), but their functions are poorly understood. Yeast cells have an acidocalcisome-like compartment, the vacuole, which carries transporters for all compounds that are typically concentrated in acidocalcisomes (Figure 1). Typical acidocalcisome features comprise high concentrations of divalent cations, an acidic pH, and several hundred millimolars of the basic amino acids arginine as lysine. Pi is accumulated to similarly high concentrations in the form of polyphosphate (polyP), a polymer of up to a thousand Pi units linked through phosphoric anhydride bonds. PolyP is stored in the acidocalcisome lumen, where polyphosphatases are also located. It is assumed that these enzymes can hydrolyze polyP, which may allow re-export of the liberated Pi into the cytosol (Gerasimaite and Mayer, 2016, 2017). Thereby, acidocalcisome-like organelles might be important buffering devices for cytosolic Pi. In line with this, cells lacking polyP show an accelerated activation of the transcriptional phosphate starvation response on low-Pi media (Neef and Kladde, 2003; Thomas and O’Shea, 2005). They also show delays in the S-phase and slower dNTP synthesis, probably because a rapid duplication of nucleic acids and phospholipids generates a Pi requirement that transiently exceeds the uptake capacity of the cell and necessitates the engagement of internal Pi reserves (Neef and Kladde, 2003; Bru et al., 2016). A major open question is whether and how acidocalcisome-like vacuoles behave in these situations, i.e., how the turnover and release of Pi back into the cytosol are triggered, such that futile cycles of polyP synthesis and hydrolysis are avoided. This is a pre-condition for acidocalcisome-like vacuoles to constitute an efficient regulated Pi buffer.
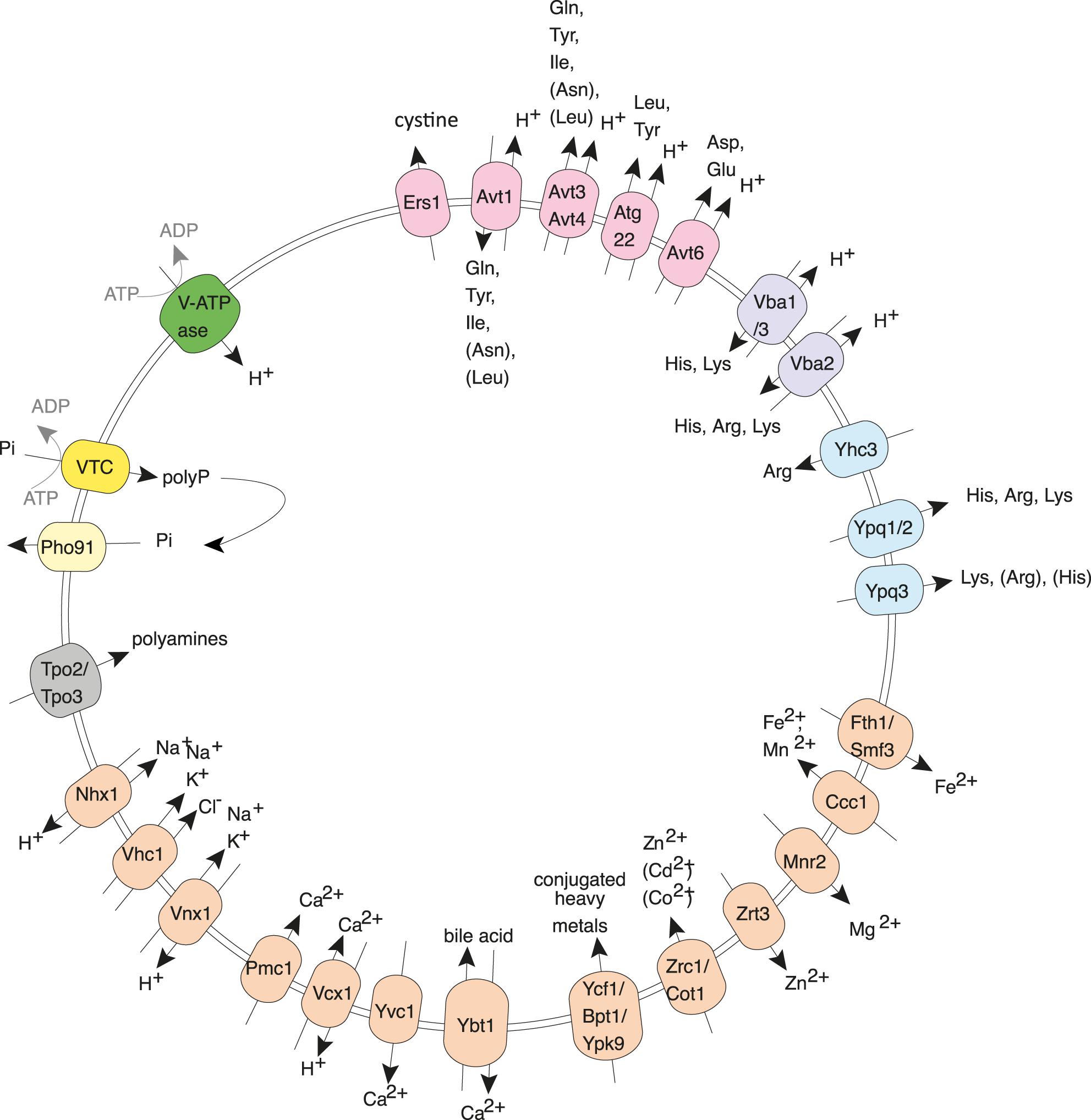
Figure 1. Yeast vacuoles as an example of an acidocalcisome-like organelle. The polyphosphate polymerase VTC transfers inorganic phosphate (Pi) into vacuoles by converting it into polyphosphate (polyP) and translocating it across the membrane. Here, the lumenal polyphosphatases can convert polyP back into Pi, which can be re-exported into the cytosol through Pho91. Apart from polyP, yeast vacuoles actively accumulate many other compounds that are typical of acidocalcisomes, such as Ca2+, Mn2+, Zn2+, and basic amino acids. Examples of the involved transporters are shown in the illustration.
While acidocalcisomes store high concentrations of phosphate and are, hence, prime candidates for a Pi-buffering system, it must be kept in mind that other organelles also use and/or liberate Pi as part of their metabolic functions. For example, the endoplasmic reticulum (ER) lumen contains many chaperones that hydrolyze ATP (Depaoli et al., 2019). The Golgi liberates Pi as a by-product of the glycosylation reactions in this compartment, and it is likely that this Pi is recycled back to the cytosol through a dedicated channel, Erd1 (Snyder et al., 2017). Likewise, the mitochondria permanently import large quantities of Pi in order to regenerate ATP from ADP (Palmieri and Monné, 2016). Phosphate homeostasis in the cytosol will be influenced by these different processes, but their impact has yet remained essentially unaddressed.
Basic Setup for Pi Regulation in Yeast
Pi homeostasis can be achieved by controlling a variety of processes in a synergistic manner, such as the import and export of Pi, its intracellular storage and re-mobilization, active foraging for Pi in the environment, and Pi recycling within the cell. In yeast, many components contributing to these processes are known. At least 25 out of the approximately 6,000 genes in yeast are directly implicated in Pi homeostasis, illustrating the crucial importance of this parameter for the cells. They comprise secreted acid phosphatases to liberate Pi from a variety of substrates in the environment (Oshima, 1997): Pi importers of high (Pho84 and Pho89) and low (Pho87 and Pho90) affinity and a putative Pi exporter in the plasma membrane (Syg1; Figure 2) (Persson et al., 2003). Cells can also recycle Pi from internal sources, e.g., by liberating it from nucleotides or phospholipids (Phm8 and Gde1) (Patton-Vogt, 2007; Xu et al., 2013). An ATP-driven protein complex (VTC complex) exists for storing and concentrating Pi as an osmotically inactive polyphosphate and depositing it inside the vacuole, from where it can be re-mobilized by polyphosphatases (Ppn1 and Ppn2) (Sethuraman et al., 2001; Gerasimaite and Mayer, 2016, 2017). The vacuolar membrane carries a Pi transporter (Pho91) (Hürlimann et al., 2009), which might participate in the re-mobilization of polyP and export Pi resulting from polyP hydrolysis into the cytoplasm.
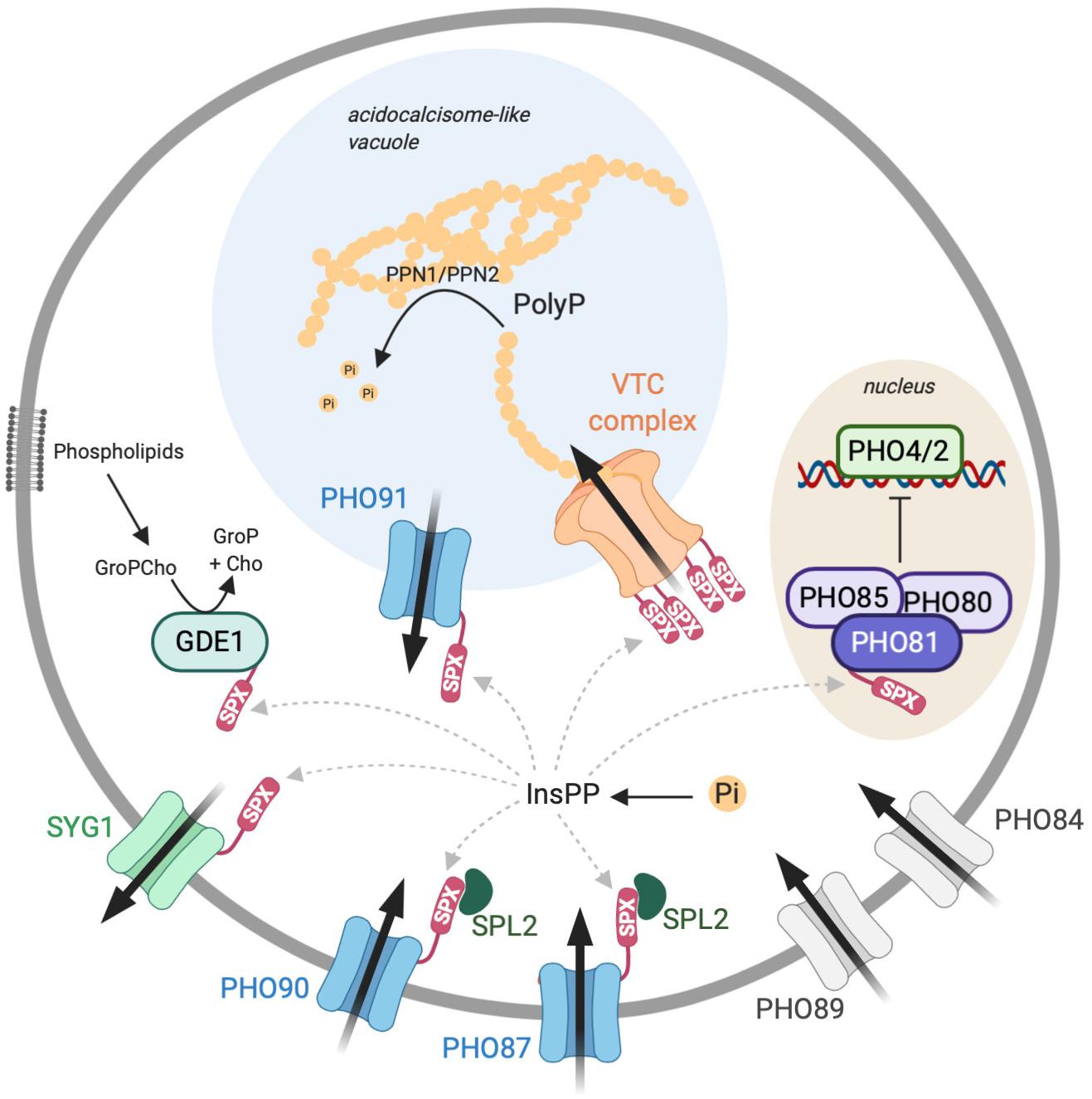
Figure 2. The INPHORS (intracellular phosphate reception and signaling) pathway for inorganic phosphate (Pi) homeostasis in the cytosol, based on known components from Saccharomyces cerevisiae. Pi can be imported into the cytosol by high-affinity (Pho89/84) and low-affinity importers (Pho87/90) and by a Pi-permeable vacuolar channel (Pho91). It can be exported from the cytosol by secretion into the extracellular space (Syg1) or by conversion into polyphosphate (polyP) and transferred into the acidocalcisome-like vacuole (VTC). Inside this organelle, polyphosphatases can reconvert polyP into Pi to make it available for export into the cytosol. A Pi-regulated kinase (Pho80/85/81) phosphorylates transcription factors (Pho4/2) for the transcriptional phosphate starvation response. A sensor for cytosolic Pi might regulate the levels of inositol pyrophosphates (InsPPs; e.g., 5-InsP7), which then act on SPX domains. These should, when bound to InsPPs, inactivate proteins importing Pi and activate proteins exporting Pi from the cytosol.
Many genes involved in Pi homeostasis are expressed through a transcriptional control mechanism, the phosphate-responsive signal transduction (PHO) pathway (Yoshida et al., 1989b). The PHO pathway is regulated through a Pi-responsive kinase (Pho80/Pho85/Pho81), which phosphorylates the transcription factor Pho4, thereby keeping it inactive. Pi starvation favors the dephosphorylation of Pho4 and allows it to activate a wide spectrum of genes dedicated not only to Pi import and storage but also to foraging for extracellular Pi and the recycling of intracellular phosphate (Carroll and O’Shea, 2002). The response is graded: Upon moderate phosphate starvation, high-affinity Pi importers (Pho84) and the polyP polymerase VTC are induced, whereas proteins for Pi foraging are upregulated only upon further Pi limitation (Vardi et al., 2014). This includes phosphatases that are secreted from the cells in order to recover Pi from phosphorylated substrates in their environment (Oshima, 1997). In addition, phosphate starvation leads to the enhanced production of proteins for Pi recycling, which allows the cell to recover Pi from internal molecules (Ogawa et al., 2000). Examples are the glycerophosphodiesterase Gde1 (Fisher et al., 2005; Patton-Vogt, 2007), which can remove Pi from intermediates of lipid metabolism, or Phm8, which dephosphorylates nucleoside monophosphates (Xu et al., 2013). For all of these systems, it has remained enigmatic how Pi availability is measured in order to regulate them.
Yeast cells thus use multiple systems and mechanisms to control cytosolic Pi. We can expect that this results in a high degree of redundancy, which can be taken as an indication that Pi homeostasis is of critical importance for the cells. For the exploration of Pi homeostasis, this comes as a blessing and a curse at the same time: On the one hand, mutants in a single compound of this complex system will usually be viable and amenable to investigation, but, on the other hand, redundancy renders it more difficult to generate clear phenotypes that are necessary to analyze its function.
Cells should benefit from information about the concentrations of free Pi in their environment, within the cytoplasm, and within the subcellular compartments. How cells perceive or measure these important parameters is not well understood at this point. Available evidence suggests a transceptor for extracellular Pi. A transceptor is a transporter that senses an external substrate independently of its transport activity. Pho84 was (together with the low-affinity transporter Pho87) proposed to sense extracellular Pi in this manner and to regulate intracellular responses, such as the protein kinase A (PKA) pathway (Giots et al., 2003; Mouillon and Persson, 2005, 2006; Popova et al., 2010). The transceptor does not appear to address all Pi-regulated events because a major Pi-dependent response, the transcriptional starvation response (PHO pathway), reacts to intracellular rather than extracellular Pi, arguing against a general role of Pho84 or Pho87 in Pi sensing (Wykoff and O’Shea, 2001; Auesukaree et al., 2004; Desfougères et al., 2016). We thus face the possibility that cells may use at least two different signaling mechanisms that distinguish between cytosolic and extracellular Pi. Such a differentiation might be useful because the PKA pathway is particularly important when cells exit a growth arrest resulting from nutrient limitation (Conrad et al., 2014). In this situation—of a non-dividing cell—cytosolic Pi is not a useful readout because there is no Pi consumption and, hence, the cytosolic concentration may easily be sufficient. The decision to reenter the cell cycle can more reliably be instructed by information about the external Pi supply, which will be necessary to support the next S-phase. Hence, the interest of a transceptor. In contrast, the measurement of cytosolic Pi is highly relevant in an actively growing and dividing cell because, here, the duplication of all cellular components requires the uptake of enormous amounts of Pi. In this situation, the exhaustion of an existing external Pi resource usually occurs gradually, calling for the activation of additional Pi foraging, which is one of the main purposes of the phosphate starvation response. However, the consumption of cytosolic Pi changes drastically in the different phases of the cell cycle, being by far the highest in the S-phase. A dividing cell will therefore have to closely monitor and regulate its cytosolic Pi in order to avoid large changes in this critical parameter. The sensing mechanism for intracellular Pi should hence address multiple proteins for the mobilization, transport, and storage of Pi in order to stabilize the Pi concentration in the cytosol in the face of a grossly fluctuating demand.
INPHORS: a Conserved Phosphate Signaling Pathway for Intracellular Pi
A yeast cell thus uses multiple and potentially redundant systems to supply Pi to the cytoplasm or withdraw it from there. A key question is how the activities of these systems are coordinated. An important hint in this respect comes from the sequences of these proteins since many of them carry an SPX (Syg1/Pho81/XPR1) domain. The fact that all of the 10 yeast proteins that contain an SPX domain are involved in Pi homeostasis strongly suggests a role of this domain in coordinating Pi signaling (Secco et al., 2012a, b). SPX appears to act in conjunction with inositol pyrophosphates (InsPPs), molecules which are also critical for Pi homeostasis (Auesukaree et al., 2005; Wild et al., 2016).
InsPPs are highly phosphorylated inositol species with at least one pyrophosphate moiety. Their abundance increases in correlation with the availability of Pi in the media (and hence presumably in response to the resulting changes in cytosolic Pi) (Lee et al., 2007; Lonetti et al., 2011; Wild et al., 2016; Gu et al., 2017). They bind to the SPX domains, through which they modulate the activity of the target proteins associated with these domains, such as polyP polymerases, Pi transporters, or Pi-dependent plant transcription factors (Wild et al., 2016; Puga et al., 2017). A variety of InsPP isomers exist in cells (Figure 3), but it is unclear whether different isomers assume different signaling functions, and might represent an inositol pyrophosphate “code” (Azevedo and Saiardi, 2017; Gerasimaite et al., 2017; Shears, 2017). Available data do not provide a consistent picture. Whereas studies on the transcriptional phosphate starvation response (PHO pathway) concluded that phosphate starvation is signaled through increasing the 1-InsP7 concentration (Lee et al., 2007, 2008), later studies on the VTC complex indicated that phosphate starvation is signaled by a decline of 5-InsP7 (Lonetti et al., 2011; Wild et al., 2016; Gerasimaite et al., 2017). In contrast, studies on the mammalian Pi exporter XPR1 provided strong evidence for its exclusive regulation through 1,5-InsP8 (Li et al., 2020). Further work will be necessary to clarify whether the transcriptional phosphate starvation response and the transport and storage of Pi through SPX-containing proteins are indeed regulated by different inositol pyrophosphate isomers or whether these isomers might serve as signals to integrate different physiological parameters with Pi signaling (Azevedo and Saiardi, 2017; Shears, 2017).
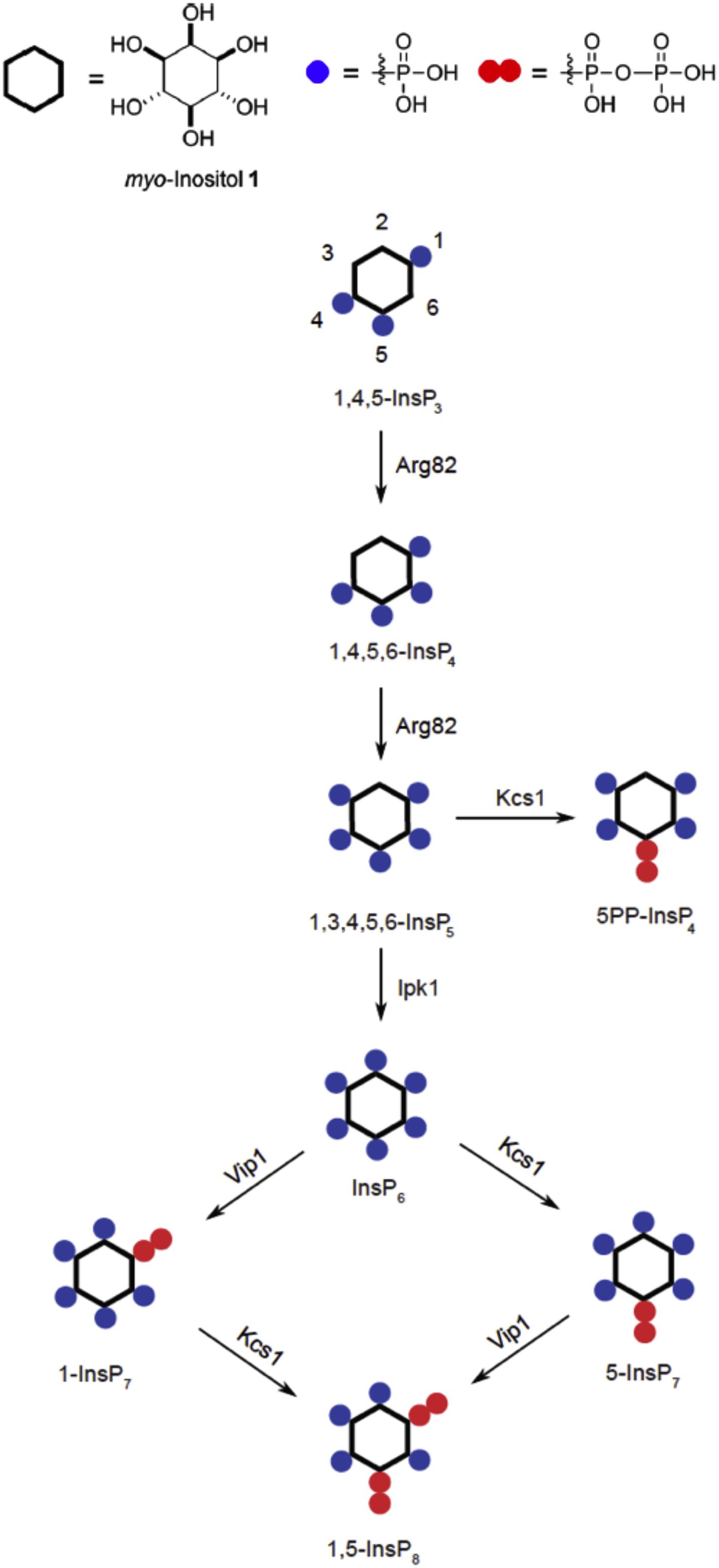
Figure 3. Synthesis of inositol pyrophosphates (InsPPs) in yeast cells, starting from InsP3. Responsible enzymes are indicated for each step. Blue dots indicate a phosphoryl group and pairs of red dots a pyrophosphoryl group on the inositol ring.
SPX domains and InsPPs were found in many eukaryotes, and mutations affecting them give corresponding phenotypes (Secco et al., 2012a; Wilson et al., 2013; Shears, 2015; Wild et al., 2016; Puga et al., 2017). Since, in addition, SPX domains are frequently associated with proteins that mediate the uptake, export, storage, or foraging for Pi, we can postulate an evolutionarily conserved signaling pathway in which cytosolic Pi is measured, translated into a corresponding change of InsPPs, and thereby communicated to a multitude of SPX-containing proteins (Figure 2). We term this pathway INPHORS (intracellular phosphate reception and signaling). We postulate that InsPPs address these proteins in a coordinated fashion in order to maintain cytosolic Pi in a suitable range. Then, we must expect that SPX domains bound to inositol pyrophosphates can have either activating or inactivating effects, depending on the proteins that they associate with. Since the levels of inositol pyrophosphates increase with the level of available Pi, proteins exporting Pi, either across the plasma membrane or into intracellular storage places such as the acidocalcisome-like vacuole, should be activated by inositol pyrophosphate-bound SPX. Likewise, we should expect that the activation of the transcriptional phosphate starvation response, the PHO pathway, should be repressed by InsPPs. This scheme is partially consistent with existing data because the SPX-containing polyP polymerase VTC is activated through the interaction between its SPX domains and the inositol pyrophosphate 5-InsP7 (Auesukaree et al., 2005; Lonetti et al., 2011; Wild et al., 2016; Gerasimaite et al., 2017) and because the low-affinity Pi importers Pho87 and Pho90 are inhibited by their respective SPX domains (Hürlimann et al., 2009). In contrast, conflicting data exist on other SPX proteins. The SPX-containing kinase Pho85/80/81, which represses the PHO pathway, was reported to be silenced by the inositol pyrophosphate 1-InsP7 (Lee et al., 2007). Furthermore, patch-clamp experiments suggested that the vacuolar Pi exporter Pho91 is activated by 5-InsP7 (Potapenko et al., 2018), although we would expect the inverse. It has also been suggested that inorganic pyrophosphate (PPi) can stimulate Pho91 (Potapenko et al., 2019). An argument against this hypothesis is that stimulation was observed with a high concentration of this compound, which is toxic in yeast and maintained at very low cytosolic levels by the essential enzyme Ipp1 (Serrano-Bueno et al., 2013). Further analyses will be needed in order to elucidate the reasons for the inconsistencies mentioned above and/or to allow us to revise our working hypothesis.
The Pi Transporters of Yeast
Yeast cells can take up Pi through five transporters (Pho84, Pho87, Pho89, Pho90, and Pho91). The deletion of all five transporters together is lethal, but the quintuple mutant can be rescued by the overexpression of any one of these five importers (Wykoff and O’Shea, 2001) or by the overexpression of GIT1, a glycerophosphoinositol permease that also transports Pi and glycerol-3-phosphate (Wykoff and O’Shea, 2001; Popova et al., 2010). A further transporter in the plasma membrane, Syg1, is presumed to export Pi due to its homology to the mammalian Pi exporter XPR1 (Giovannini et al., 2013).
High-Affinity Pi Transporters: Pho84 and Pho89
Pho84 and Pho89 are high-affinity transporters since they permit the uptake of Pi from low-Pi media (<0.2 mM) with a Km of 8–40 μM (Bun-ya et al., 1991; Wykoff and O’Shea, 2001; Auesukaree et al., 2003; Levy et al., 2011). While Pho84 is a H+ symporter with a pH optimum in the acidic range (Bun-ya et al., 1991; Pérez-Sampietro et al., 2016), Pho89 is active at alkaline pH and driven by Na+ (Martinez and Persson, 1998; Zvyagilskaya et al., 2008; Serra-Cardona et al., 2014). In addition to Pi, Pho84 serves also for the low-affinity co-transport of manganese ions (Jensen et al., 2003) and for the import of selenite (Lazard et al., 2010). Pho84 is highly expressed at intermediate concentrations of Pi (<0.5 mM), but becomes transferred to the vacuole for degradation in the absence of Pi or at high excess of Pi (Petersson et al., 1999; Pratt et al., 2004; Lundh et al., 2009). Pho84 allows the cells to exploit very low Pi concentrations, and their growth arrests only below a threshold of 5 μM. In standard Pi-rich media, Pho84 is required for the maintenance of normal polyP levels (Hürlimann et al., 2007) and for repressing transcription through the PHO pathway (Wykoff et al., 2007). However, constitutive activation of the PHO pathway in cells lacking Pho84 does not reflect a major contribution of this high-affinity transporter to Pi uptake under Pi-replete conditions. It is rather the consequence of the downregulation of the low-affinity transporters, which results from the deletion of PHO84 due to feedback regulation (Wykoff et al., 2007; Vardi et al., 2013). This feedback regulation creates a bistable, Spl2-dependent switch, which dedicates cells either to the Pi-starved regulatory state (downregulating the low-affinity Pi transporters and inducing the PHO pathway) or to the Pi-replete state (stabilizing the low-affinity transporters and repressing the PHO pathway). 14-3-3 proteins (Bmh1 and Bmh2, which also interact with the inositol hexakisphosphate kinase Kcs1) were proposed to influence this commitment (Teunissen et al., 2017), but the nature of their influence has not yet been resolved. The decision between the two states may depend on small differences in the cytosolic Pi concentration, i.e., on the degree to which cells experience Pi limitation, or on stochastic fluctuations. The bistable switch allows a fraction of the cells in a population to stably maintain their commitment to the phosphate starvation response for multiple generations—even after phosphate has been replenished. This can be seen as a potential advantage because these cells would be best prepared for a sudden drop in Pi availability and facilitate the survival of the population under such conditions.
As mentioned above, Pho84 has also been proposed to act as a Pi transceptor, which could provide an alternative explanation how it represses the PHO pathway under phosphate-replete conditions. Obtaining clear evidence for an activity as a transceptor is difficult because it requires maintaining signaling while suppressing the transporter function entirely. In the case of Pho84, one caveat is that the substrates that were used to induce putative transceptor signaling in the absence of transport, such as glycerol-3-phosphate, might be hydrolyzed extracellularly by yeast and the released Pi taken up by Pho84 (Popova et al., 2010). Also, transport-deficient point mutants retain low-capacity transport, which, in a Pi-starved, non-dividing cell, may nevertheless suffice to satisfy the needs. Finally, the complex feedback regulation mentioned above complicates the interpretation of the experiments that suggested Pho84 as a transceptor (Giots et al., 2003; Mouillon and Persson, 2005; Popova et al., 2010; Samyn et al., 2012).
SPX-Containing Low-Affinity Pi Transporters: Pho87 and Pho90
Pho87 and Pho90 are two low-affinity plasma membrane transporters which mediate the uptake of Pi from the environment with Km values of 200–800 μM (Tamai et al., 1985; Wykoff and O’Shea, 2001; Auesukaree et al., 2003; Giots et al., 2003; Pinson et al., 2004; Hürlimann et al., 2007, 2009). They belong to the divalent anion:Na+ symporter (DASS) family. Both Pho87 and Pho90 harbor an SPX domain at their N-terminus. Removal of this SPX domain leads to phenotypes suggesting an enhanced, unrestricted flux of Pi across these transporters: up to an eightfold increased Pi uptake activity, higher total phosphate, and higher polyP content. Cells expressing Pho90 without an SPX domain also become sensitive to high Pi concentrations in the media, and they show an enhanced leakage of cellular Pi on low-Pi media (Hürlimann et al., 2009). Together, these observations indicate that the SPX domain has a restrictive function on Pho87 and Pho90, which is necessary to close these transporters at excessively high and low concentrations of Pi.
The transcription of the PHO87 and PHO90 genes is independent of Pi availability, but the transporters are targeted to the vacuole and degraded in response to Pi limitation (Auesukaree et al., 2003; Wykoff et al., 2007; Hürlimann et al., 2009; Ghillebert et al., 2011). This targeting requires their SPX domain and, for Pho87, also Spl2. But why do cells have low-affinity transporters in addition to a high-affinity system? When the low-affinity importers are ablated, the cells compensate by expressing more Pho84, and this largely rescues their Pi uptake activity. While this demonstrates that a high-affinity transporter can substitute for the low-affinity system, even under high-Pi conditions (Wykoff and O’Shea, 2001; Pinson et al., 2004; Ghillebert et al., 2011), it brings up the question what benefit a cell draws from expressing low-affinity transporters when it has high-affinity transporters available. An advantage becomes apparent in situations where Pi gradually becomes limiting, for example in a culture that is exhausting the available Pi as it grows. At an intermediate Pi concentration, both high- and low-affinity transporters are expressed. The decrease in Pi availability can be detected earlier and over a much larger range of external Pi concentrations when the cells use low-affinity transporters as long as Pi is abundant and begin to express high-affinity transporters only once Pi becomes scarcer. In an environment of gradually depleting Pi, this provides for a longer transition phase from Pi-replete conditions to full Pi starvation, giving the cells much more time to adapt and prepare for Pi starvation (Levy et al., 2011). This leads to enhanced recovery from growth arrest once Pi becomes available again.
SPX-Controlled Vacuolar Pi Transporter: Pho91
Pho91 is a low-affinity Pi transporter. Like the low-affinity Pi transporters Pho90 and Pho87, it belongs to the DASS family and its expression is independent of Pi supply (Auesukaree et al., 2003). However, a green fluorescent protein (GFP) fusion of Pho91 is localized to vacuoles (Hürlimann et al., 2007). When overexpressed in a mutant lacking all other Pi importers, Pho91 can nevertheless support the growth and Pi uptake of cells with a Km of around 200 μM (Wykoff and O’Shea, 2001). Initially, this was taken as an indication that it operates at the plasma membrane. However, yeast cells can take up nutrients also by other routes—as shown for magnesium, which can be acquired by endocytic transfer to the vacuole lumen and subsequent transport across the vacuolar membrane into the cytosol (Klompmaker et al., 2017). Such an uptake route via vacuoles might also allow Pi acquisition through Pho91. In further support of a function at the vacuolar membrane, the ablation of Pho91 leads to an overaccumulation of phosphate in vacuoles, whereas its overexpression depletes this vacuolar pool. This led to the proposal that Pho91 transfers Pi from vacuoles to the cytosol (Hürlimann et al., 2007).
This view could be confirmed by electrophysiological analysis of Pho91 and of its homologs from rice OsSPX-MFS3 and Trypanosoma brucei TbPho91 (Wang et al., 2015; Potapenko et al., 2018), where a Na+-dependent transport of Pi into the cytosol with Km values of 1–10 mM could be demonstrated. The transport activity is highly pH-dependent, as demonstrated for OsSPX-MFS3 (Wang et al., 2015). Whereas net charge transport occurs at neutral pH, the protein does not mediate significant Pi transport under these conditions. Pi is only transported in the presence of a pH gradient, when the extra-cytosolic face of the protein is exposed to acidic pH. This pH dependency corresponds to the natural condition under which these Pho91-like transporters operate because the lumen of the vacuoles in which they reside is much more acidic than the cytosol. In the absence of a pH gradient, OsSPX-MFS3 mediates Pi efflux from the cytosol (Wang et al., 2015). For TbPho91 and Pho91, it was shown that charge transport depends on their SPX domain and on 5-InsP7. Other inositol polyphosphates, such as 1-InsP7 or InsP6, do not activate the channel (Potapenko et al., 2018). However, these experiments have been conducted in the absence of a proton gradient across the membrane. Thus, while they demonstrate a regulatory impact of SPX and 5-InsP7 on the transporter, their effects on Pi transport under a pH gradient remain to be determined.
Overall, these observations suggest that Pho91 transports Pi from the vacuole into the cytosol (Hürlimann et al., 2007), which might be necessary to allow the degradation of vacuolar polyP. It might then be a critical element of the system to buffer cytosolic Pi via polyP (Neef and Kladde, 2003; Thomas and O’Shea, 2005). However, direct evidence for an impact of Pho91 on cytosolic Pi and a coherent model for the functioning of a vacuolar, polyP-based Pi buffer is still missing.
The SPX-Containing Putative Pi Exporter Syg1
Syg1 was identified as a genetic suppressor of defects in pheromone signaling and predicted to be on the plasma membrane (Spain et al., 1995). Syg1, the mammalian transporter XPR1, and the related PHO1 from Arabidopsis share around 30% of sequence identity and some common features, such as an N-terminal SPX and a C-terminal EXS (for ERD1/XPR1/SYG1) domain. The function of the EXS domain is still unknown. Although the transport activity for Syg1 itself has not yet been shown, its human homolog XPR1 does facilitate Pi export across the plasma membrane (Giovannini et al., 2013; Wilson et al., 2019; Li et al., 2020). Export depends on its SPX domain and on 1,5-InsP8. Thus, Syg1 likely acts as a Pi exporter in yeast.
Polyphosphate Metabolism and Phosphate Homeostasis
Inorganic polyphosphate is a polymer of dozens to hundreds of orthophosphate (Pi) linked by energy-rich phosphoric anhydride bonds. PolyP efficiently chelates ions such as Ca2+, Mg2+, and Mn2+. Yeast cells can accumulate up to a quarter of their dry weight in the form of polyphosphate (Langen and Liss, 1958), located mostly in their acidocalcisome-like vacuole (Saito et al., 2005). Minor amounts have also been associated with other organelles, such as mitochondria, the ER, or the periplasmic space (Pestov et al., 2004; Lichko L. et al., 2006; Kulakovskaya et al., 2010). The sequestered polyP is not an immobile aggregate. Instead, it appears to be highly mobilizable (Wiame, 1947; Urech et al., 1978). PolyP is necessary to rapidly buffer changes in the cytosolic phosphate levels that can result from sudden changes in Pi availability or consumption (Neef and Kladde, 2003; Thomas and O’Shea, 2005). The polyP in the acidocalcisomes of other organisms can also be mobilized, for example upon osmotic challenges or changes in nutrient supply (Docampo and Huang, 2016). The accumulation of polyP has a strong impact on cytosolic Pi homeostasis and should hence be carefully controlled by the cell (Desfougères et al., 2016). However, it remains a major unsolved question how the synthesis and degradation of polyP are coordinated to achieve this goal. While we have some insights into the synthesis of polyP, it remains an enigma how the cells can store polyP in a compartment that contains considerable polyphosphatase activities and how the conversion of polyP back into cytosolic Pi is regulated (Gerasimaite and Mayer, 2016).
PolyP Synthesis by the VTC Complex
The VTC proteins form complexes which all contain Vtc1, a 14-kDa integral membrane protein that spans the membrane three times, and Vtc4, the catalytically active subunit that synthesizes polyP from nucleotide triphosphates (Müller et al., 2002, 2003; Hothorn et al., 2009). Catalytic activity requires Mn2+, which is located in the active site. All VTC proteins have a transmembrane region similar to Vtc1. In contrast to Vtc1, the other VTC proteins contain an additional SPX domain at the N-terminus and a hydrophilic central domain, both facing the cytosolic side of the membrane (Müller et al., 2003). The central domain accommodates the catalytic center in Vtc4, whereas it is assumed to have only regulatory, non-catalytic function in Vtc2, Vtc3, and Vtc5 (Hothorn et al., 2009; Desfougères et al., 2016). VTC complexes exist in different isoforms, which contain Vtc1 and Vtc4, plus either Vtc2 or Vtc3. Vtc5 can associate with VTC to increase its activity, but VTC functions at a lower basal activity without it (Desfougères et al., 2016). Vtc1/2/4 is mainly localized in the ER, whereas Vtc1/3/4 concentrates on the vacuoles, when the cells are cultivated in Pi-replete media. Under Pi limitation, both complexes localize to the vacuoles (Hothorn et al., 2009).
VTC acts not only as a polyP polymerase but at the same time as a polyP translocase. It couples the synthesis of polyP at the cytosolic face of the membrane with its translocation into the lumen of the organelle (Gerasimaite et al., 2014). Continued synthesis of polyP by VTC requires the electrochemical gradient across the vacuolar membrane, which is established by V-ATPase. This gradient is assumed to provide the driving force translocating the negatively charged polyP chain (Wurst et al., 1995; Gerasimaite et al., 2014).
The activity of VTC can be assayed on isolated yeast vacuoles (Gerasimaite et al., 2014). This in vitro system allowed the discovery of the regulation of SPX domains through inositol pyrophosphates (Wild et al., 2016). While SPX domains bind various inositol polyphosphates and pyrophosphates with Kd values in the low micromolar or even sub-micromolar range (Wild et al., 2016), these different isomers show strikingly different agonist properties on VTC. Only InsPPs stimulate the enzyme at low micromolar concentrations, with 5-InsP7 being the isomer that appears to regulate VTC in vivo (Gerasimaite et al., 2017). Since InsP7 increases when cells are in Pi-replete conditions and decreases under Pi starvation (Lonetti et al., 2011; Wild et al., 2016), VTC should synthesize polyP when cytosolic Pi is sufficiently high, but it should be switched off when this parameter is too low. This reflects the in vivo situation because cells accumulate polyP stocks when sufficient Pi is still available and they deplete this stock under Pi starvation or upon a transient excessive consumption of Pi during the S-phase (Langen and Liss, 1960; Bru et al., 2016). That VTC must be carefully controlled by the cells is also suggested by the fact that it has a major impact on cytosolic Pi. Inappropriate overactivation of VTC can lead to Pi depletion from the cytosol and activate the PHO starvation pathway even on Pi-rich media, whereas its silencing can suppress the PHO pathway on media with limiting Pi (Desfougères et al., 2016). PolyP storage is a major function of yeast vacuoles, which is probably the reason why polyP production also activates the vacuolar membrane fusion machinery. The resulting fusion of several vacuoles together increases the volume of the compartment, thereby accommodating the need for increasing storage space in a rapid and efficient manner (Müller et al., 2002; Desfougères et al., 2016).
Polyphosphatases
The Exopolyphosphatase Ppx1
Ppx1 is a member of the DHH phosphoesterase superfamily, together with h-prune, a mammalian exopolyphosphatase (Tammenkoski et al., 2008). Ppx1 is a soluble enzyme which hydrolyzes polyP to release Pi and PPi (Wurst and Kornberg, 1994). Hydrolysis is processive, i.e., the enzyme does not leave the polyP chain after having hydrolyzed its terminal Pi residue. Ppx1 hydrolyzes polyP chains as short as three Pi residues, but cannot degrade PPi and ATP. The enzyme is metal-dependent, with a preference for Mg2+, and active from pH 5.5 to 9 (Tammenkoski et al., 2007).
Ppx1 is expressed irrespective of Pi availability (Ogawa et al., 2000). Initial studies ascribed it to the cytosol, but exopolyphosphatase activities in the plasma membrane and mitochondrial fractions were also related to Ppx1 (Lichko et al., 2003; Lichko L.P. et al., 2006). High-throughput localization studies through GFP fusion proteins suggested localization in the cytosol, but also some enrichment in the nucleus (Huh et al., 2003). In any case, Ppx1 appears to be localized outside the vacuoles, where the major polyP stores are kept. In line with this, its deletion affects neither the chain length nor the abundance of polyP in the cell (Lonetti et al., 2011). It was hence proposed that Ppx1 might counteract an the accumulation of polyP in the cytosol, which is toxic for cells (Gerasimaite et al., 2014). A further attractive possibility is that Ppx1 might trim polyP from proteins that are covalently modified with this polymer (Azevedo et al., 2015, 2018, 2019).
The Vacuolar Endopolyphosphatases Ppn1 and Ppn2
In its active, homo-tetrameric state, Ppn1 is a non-processive endopolyphosphatase which preferentially hydrolyzes long polyP in the midst of the chain rather than at its terminal phosphate residues. Depending on the conditions, however, it was also reported to have exopolyphosphatase activity in vitro (Kumble and Kornberg, 1996; Andreeva et al., 2015). Ppn1 activity requires Mn2+ or Mg2+. The enzyme can hydrolyze polyP down to Pi and tripolyphosphate, whereas pyrophosphate and ATP are potent inhibitors (Kumble and Kornberg, 1996). PPN1 gene expression is induced under Pi limitation, i.e., when vacuolar polyP pools are consumed (Kumble and Kornberg, 1996). PPN1 knockouts retain polyP in similar amounts as wild types, but their polyP is of higher chain length.
Ppn2 is a member of the phospoprotein phosphatase (PPP) superfamily of metallophosphatases that resides in the vacuolar lumen. The enzyme depends on Zn2+ and exclusively shows endopolyphosphatase activity (Gerasimaite and Mayer, 2017). Ppn1 and Ppn2 together constitute the major part of the vacuolar polyphosphatase activity. They are necessary to mobilize polyP stores under Pi starvation. In their absence, the cells accumulate polyP of excessively high chain length and they virtually show no short-chain polyP anymore.
Phosphate “Recycling”
When Pi becomes limiting, the cells use the PHO pathway to induce first the high-affinity transporter Pho84. They secrete phosphatases to recover Pi from external sources and induce VTC expression to maximize their polyP stores (Ogawa et al., 2000; Springer et al., 2003; Thomas and O’Shea, 2005; Wykoff et al., 2007; Vardi et al., 2014). When the cells really starve for Pi, the polyP stores are hydrolyzed (Langen and Liss, 1958; Sethuraman et al., 2001; Neef and Kladde, 2003; Thomas and O’Shea, 2005; Gerasimaite and Mayer, 2017). Using this strategy might be helpful to allow redifferentiation and ordered transition into the quiescent state (the G0 phase of the cell cycle), in which cells arrest growth and become more resistant to stresses (De Virgilio, 2011). This important transition often coincides with the induction of autophagy, which transfers large amounts of cytosolic material, including RNA and organelles, into vacuoles for degradation (De Virgilio, 2011). Their hydrolysis provides the cells with a source of degradable nucleotides and phospholipids. Profound Pi starvation induces the expression of enzymes that release Pi from these internal molecules. It appears likely that this represents a “last resort” because it makes little sense for a cell to deplete its pools of nucleotides or phospholipids, which are indispensable for the next cell division, unless it is the only means to survive. In line with this, cells in which these recycling pathways have been ablated show poor long-term survival on Pi-free media (Xu et al., 2013).
Gde1 is a glycerophosphocholine phosphodiesterase which hydrolyzes glycerophosphocholine to glycerol-3-phosphate and choline (Fisher et al., 2005). Glycerol-3-phosphate can either be channeled into the synthesis of new phospholipids, into glycolysis, or it can be hydrolyzed by the glycerol-3-phosphatases Gpp1 and Gpp2 (Patton-Vogt, 2007). All these pathways effectively lead to the recycling of internal Pi. They are of sufficiently high capacity to allow the cells to grow on glycerophosphocholine as the sole source of phosphate. That the recycling activity of Gde1 may be relevant to the maintenance of cytosolic Pi homeostasis is suggested by the fact that GDE1 gene expression is regulated by the PHO pathway and strongly induced in low-Pi conditions (Ogawa et al., 2000; Almaguer et al., 2004). Furthermore, Gde1 carries an N-terminal SPX domain, which is expected to subject the enzyme to regulation by the INPHORS pathway. However, the regulatory role of this SPX domain has not yet been experimentally confirmed. Interestingly, Gde1 shares its glycerophosphodiesterase domain with Pho81, a key regulator of the PHO pathway. Whether the glycerophosphodiesterase domain of Pho81 is catalytically active is also unknown.
Phm8 has originally been identified as a lysophosphatidic acid phosphatase (Reddy et al., 2008). Its expression is strongly induced by Pi starvation (Ogawa et al., 2000). Under Pi starvation, yeast cells show a significant reduction in lysophosphatidic acid, and this reduction has been ascribed to Phm8 (Reddy et al., 2008). The enzyme might thus participate in the recycling of phosphate from degraded phospholipids. Phm8 has also been identified as a nucleotide monophosphate phosphatase (Xu et al., 2013). The enzyme allows liberating Pi from a wide variety of nucleoside monophosphates. In its absence, cells cannot survive Pi starvation for prolonged periods of time, underscoring the relevance of its Pi recycling activities in this situation.
The repressible “alkaline” phosphatase Pho8 (Kaneko et al., 1982) shows maximal activity on artificial chromogenic substrates, such as p-nitrophenyl phosphate, at alkaline pH. However, the enzyme is membrane-anchored and resides within the vacuole, which is an acidic compartment with a pH ranging from 5 to 6. At this pH, Pho8 exhibits maximal activity toward phosphoserine and phosphothreonine, and it has therefore been proposed that it may serve to dephosphorylate peptides (Donella-Deana et al., 1993), which are imported into vacuoles through autophagy. The expression of the enzyme is induced upon Pi starvation through the PHO pathway (Kaneko et al., 1985). Pho8 can also dephosphorylate fructose-2,6-bisphosphate, which is generated in the cytosol, but the physiological relevance of this activity has remained unexplored, and it remains unclear how fructose-2,6-bisphosphate could be translocated into vacuoles, where Pho8 is located (Plankert et al., 1991). Pho8 may also participate in the recuperation of Pi from NAD+. Upon Pi starvation, part of the nicotinamide nucleotide pool may be converted into nicotinamide riboside, liberating Pi (Lu and Lin, 2011). This conversion requires Pho8 and the vacuolar nucleoside transporter Fun26. Yeast cells with a constitutively activated PHO pathway show increased production and secretion of nicotinamide riboside.
Elements of the INPHORS Pathway
The PIPP5 Kinase Vip1
Vip1 belongs to a conserved family of diphosphoinositol pentakisphosphate kinases (PPIP5Ks) (Randall et al., 2019). These enzymes contain both a kinase and a histidine acid phosphatase domain, which compete with each other (Mulugu et al., 2007). Their enzymatic and structural properties have mainly been uncovered through studies of the mammalian enzymes (Wang et al., 2012; Weaver et al., 2013; Gu et al., 2017; Nair et al., 2018; Randall et al., 2019), but the essential features could be confirmed for the plant and yeast members of the family (Pöhlmann et al., 2014; Dong et al., 2019; Zhu et al., 2019). The kinase domain phosphorylates InsP6 and 5-InsP7 to generate 1-InsP7 and 1,5-InsP8, respectively, whereby 5-InsP7 appears to be the preferred substrate over InsP6 (Wang et al., 2012; Weaver et al., 2013). The simultaneous presence of competing kinase and phosphatase domains, which, in addition, appear to be coordinated by allosteric effects (Yousaf et al., 2018), has important consequences. It can translate small changes in the concentration of the substrates into much larger changes of product concentration, i.e., it can amplify the response in a signaling cascade. Under suitable conditions (for PIPP5Ks, when only the phosphatase but not the kinase domain is substrate-limited), it can also make the net kinase activity quite insensitive to changes in substrate concentrations (Gu et al., 2017). This property would allow the cell to modulate 5-InsP7 without an immediate impact on the level of 1,5-InsP8, providing an important prerequisite to use these inositol pyrophosphates to communicate different cellular parameters, which might then be integrated by proteins that can “read” several different inositol pyrophosphates, such as SPX domains.
Importantly, the phosphatase activity of PIPP5Ks is inhibitable by Pi in the low millimolar range—a concentration range that is commonly found in the cytosol—and their kinase activity is boosted by an increasing ATP concentration (Gu et al., 2017; Zhu et al., 2019). Since cellular ATP concentration diminishes with cellular Pi availability (Boer et al., 2003), Pi limitation in the cytosol should tip the equilibrium between the kinase and phosphatase activities in favor of dephosphorylation, decreasing the pools of 1,5-InsP8 and 1-InsP7. PIPP5Ks might thus provide an important link between the Pi concentration and the corresponding changes in the inositol pyrophosphate levels. However, whether PIPP5Ks represent the critical Pi sensor is not yet clear. Also, the physiological implications of the sensitivity of PIPP5K to phosphoinositides, such as PI(4,5)P2 (Nair et al., 2018), are not yet understood.
The InsP6 Kinase Kcs1
Kcs1 is a member of the inositol hexakisphosphate kinase family and phosphorylates the phosphate on carbon 5 of InsP6 or Ins(1,3,4,5,6)P5, creating 5-InsP7 or 5PP-InsP4, respectively (Saiardi et al., 1999, 2000; Figure 3). The physiological relevance of 5PP-InsP4 is uncertain because it accumulates to appreciable levels only in a mutant lacking IPK1, which cannot convert Ins(1,3,4,5,6)P5 into InsP6 and, hence, does not offer InsP6, which is the far more abundant substrate of Kcs1 in a wild-type cell, where it probably outcompetes Ins(1,3,4,5,6)P5. Kcs1 shares a PxxxDxKxG motif in its catalytic site with other members of the family. The ablation of Kcs1 activity leads to the constitutive activation of the PHO transcription pathway even on high-Pi media, to an overaccumulation of Pi and ATP, and to a complete absence of polyP synthesis (Auesukaree et al., 2005; Lonetti et al., 2011; Szijgyarto et al., 2011; Wild et al., 2016). In contrast, KCS1 overexpression represses the PHO pathway even under Pi limitation, where it would normally be maximally active (Auesukaree et al., 2005). The inactivation of this enzyme thus produces the effects that we would expect if its product 5-InsP7 was a negative regulator of the phosphate starvation response. Erroneous activation of the phosphate starvation program in a kcs1Δ cell under high-Pi conditions should then lead to an overaccumulation of Pi. Although the synthesis of polyP normally occurs under high-Pi conditions (Langen and Liss, 1958), this requires the activation of the VTC complex by 5-InsP7, which is not present in kcs1Δ cells (Lonetti et al., 2011; Wild et al., 2016). Therefore, kcs1Δ mutants cannot accumulate polyP.
Kcs1 has a Km for InsP6 of 0.6 μM and an exceptionally high Km for ATP of around 1 mM (Saiardi et al., 1999). By consequence, its activity should be sensitive to the fluctuations of cytosolic ATP concentrations that normally occur in living yeast cells (Saiardi et al., 1999). This may be one reason why mutants in nucleotide metabolism that show constitutively lower cellular ATP levels—and thereby probably lower production of 5-InsP7 and 1,5-InsP8—lead to a constitutive activation of the PHO pathway (Choi et al., 2017). The cellular ATP concentration in wild-type cells declines as a result of Pi limitation (Boer et al., 2010). This opens the possibility that Pi limitation might be translated into a reduction of 5-InsP7 and 1,5-InsP8 through a reduced ATP production.
The Inositol Pyrophosphatases Ddp1 and Siw14
Inositol pyrophosphates can be turned over by two dedicated phosphatases, which selectively hydrolyze the phosphoric anhydride bonds in these molecules. They have considerable influence on the steady-state levels of inositol pyrophosphates, but it is unknown whether their activity is constitutive or regulated.
Ddp1 belongs to the Nudix hydrolase family. In cells lacking the DDP1 gene, the abundance of InsP7 increases up to sixfold (Lonetti et al., 2011; Steidle et al., 2016). Ddp1 displays di-phosphoinositol polyphosphate hydrolase activity (Safrany et al., 1999). It also exhibits di-adenosine polyphosphate hydrolase activity (Cartwright and McLennan, 1999; Safrany et al., 1999), which is of unclear physiological significance in yeast. Ddp1 can hydrolyze polyP (Lonetti et al., 2011). However, the enzyme is localizing to the cytosol and the nucleus, i.e., out of reach of the major polyP reserves, which are localized in the vacuole (Saito et al., 2005). This makes it unlikely that Ddp1 influences Pi homeostasis through polyP turnover. In line with this, its deletion has no significant impact on the polyP pool (Lonetti et al., 2011). Cells lacking Ddp1 show a 20% reduction of polyP rather than an increase. This reduction might be a secondary consequence of the deregulation of the INPHORS pathway and of an altered cellular Pi homeostasis due to the influence of Ddp1 on inositol pyrophosphates. The polyphosphatase activity of Ddp1 might, however, serve to modify the polyphosphorylation of proteins in the nucleus (Azevedo et al., 2015).
Siw14 belongs to the atypical dual-specificity phosphatase family and hydrolyzes the ß-phosphate of 5-InsP7 with high specificity (Steidle et al., 2016; Wang et al., 2018). Mutants lacking SIW14 display a sixfold increase in InsP7 content. This effect is synergistic with the simultaneous deletion of DDP1 and the double mutants display a 20-fold increase in InsP7 (Steidle et al., 2016). Overexpression of SIW14 depletes the InsP7 pool.
Inositol Pyrophosphate Receptors: SPX Domains
SPX domains are found in all eukaryotic kingdoms and share common sequence features, most notably clusters of positively charged amino acids. These conserved arginine and lysine residues cluster in a surface patch which forms a high-affinity (sub-micromolar) binding site for inositol polyphosphates and pyrophosphates. Purified SPX domains show a relatively low discrimination in binding different inositol polyphosphates or pyrophosphate isomers, and their affinity decreases in parallel to the net charge of the compound (InsP8 > InsP7 > InsP6). In marked contrast to the low selectivity in binding, the agonist properties of different isomers vary considerably. The VTC complex, for example, is strongly activated by 1,5-InsP8, but very poorly by 5-PPP-InsP5, which carries the same number of phosphate groups (Gerasimaite et al., 2017). Likewise, 5-InsP7 activates VTC, whereas InsP6 has virtually no effect. Thus, despite the extremely high charge density of the inositol polyphosphates and their similar binding affinities, the SPX domain must decode precise structural features of the ligands and translate them into different degrees of activation.
Neither the binding affinity nor the EC50 values of inositol pyrophosphate isomers seem to faithfully reflect their physiological relevance for controlling an SPX domain. This could be illustrated with VTC, which has an EC50 for 5-InsP7 that is similar to that of 1-InsP7 and is 20-fold higher than that of 1,5-InsP8. Nevertheless, genetic ablation of the enzyme synthesizing 1-InsP7 and 1,5-InsP8 (Vip1) has no significant impact on polyP synthesis in vivo. The ablation of 5-InsP7 synthesis (Kcs1), in contrast, eliminates polyP synthesis, strongly suggesting this isomer as the relevant controller of the SPX domains of VTC in vivo. Whether 5-InsP7 is the principal regulator also for the other SPX-controlled proteins in the cell is an important issue that remains to be explored. The mammalian Pi exporter XPR1 provides an important example in this respect because this SPX-controlled transporter is opened by 1,5-InsP8 rather than by 1-InsP7 or 5-InsP7 (Li et al., 2020).
Spl2
Spl2 interacts with the SPX domains of Pho90 and Pho87 and restrains the flux of Pi through Pho90 (Wykoff et al., 2007; Hürlimann et al., 2009). It is transcribed at a basal constitutive level, but it becomes further induced through the PHO pathway upon Pi limitation (Ogawa et al., 1995; Flick and Thorner, 1998). Together with the PHO81 gene, SPL2 was originally identified as a multicopy suppressor of the growth defect resulting from the ablation of the phospholipase C Plc1 (Flick and Thorner, 1998). Plc1 hydrolyzes phosphatidylinositol 4,5-biphosphate into 1,2-diacylglycerol and inositol 1,4,5-triphosphate, which is the precursor for the synthesis of inositol pyrophosphates (Saiardi et al., 2000). The genetic interactions of Spl2, Pho81, and Plc1 can thus be rationalized based on the premise that low cytosolic Pi is signaled by low levels of inositol pyrophosphates. The absence of Plc1 activity impairs the synthesis of inositol pyrophosphates and induces the phosphate starvation response (Auesukaree et al., 2005). When such cells are cultivated on Pi-rich standard yeast media, one should expect that they behave incorrectly, i.e., they are maximizing their efforts for Pi uptake although this nutrient is abundant. A resulting excessive Pi concentration in the cytosol is the likely reason for their growth defect. This inappropriate regulation can be mitigated by the overexpression of Spl2, which limits Pi transporter activity (Hürlimann et al., 2009), or by the overexpression of Pho81 (Creasy et al., 1993; Ogawa et al., 1995), which inactivates the transcriptional phosphate starvation response. This sets cells back into the high-Pi mode, which then corresponds again to the Pi-rich conditions of standard media, which had been used for these experiments (Flick and Thorner, 1998).
Upon Pi starvation, the low-affinity phosphate transporters Pho87 and Pho90 are endocytosed and targeted to the vacuole. For Pho87 and Pho90, this process depends on their SPX domains. Although the SPX domains of both transporters interact with Spl2, Spl2 is only necessary for the vacuolar targeting of Pho87, which occurs rapidly upon phosphate limitation. The degradation of Pho90, which occurs only after prolonged Pi limitation, is independent of Spl2 (Ghillebert et al., 2011; Pérez-Sampietro et al., 2016). Thus, Spl2 may allow the cells to differentially degrade their low-affinity transporters as a function of declining Pi availability.
The deletion of SPL2 does not produce a striking growth phenotype on Pi-rich standard yeast media (Flick and Thorner, 1998), but it results in an exacerbated sensitivity to selenite, probably due to the resulting hyperactivation of the low-affinity transporter Pho90, which is involved in selenite toxicity (Lazard et al., 2010).
The PHO Pathway: a Transcriptional Phosphate Starvation Response
The PHO pathway is a transcriptional response pathway that regulates the expression of a wide variety of genes (Ogawa et al., 2000), many of which have been proven beneficial or essential for growth and survival under Pi limitation. The PHO pathway has been studied extensively, providing us with detailed insights into its regulation and with numerous tools to manipulate it. A key element is the nuclear Pho80/85/81 kinase, which is inactivated through its Pho81 regulatory subunit when Pi becomes limiting (Schneider et al., 1994). The kinase subunit Pho85 phosphorylates the transcription factor Pho4 and thereby shifts its localization toward the cytosol (Kaffman et al., 1994; O’Neill et al., 1996). Pho4 localization is hence defined through an equilibrium of Pho85-controlled import and export across the nuclear membrane. The removal of Pho4 from the nucleus under Pi-replete conditions represses the expressions of genes controlled by the PHO pathway. In the following, we describe the key regulatory factors, but we will discuss only the most important PHO-regulated target genes.
The PHO Regulon: Phosphate-Responsive Genes (the PHO Genes)
The transcription factors Pho4 and Pho2 co-regulate the expressions of dozens of genes (Bun-ya et al., 1991; Oshima, 1997; Ogawa et al., 2000). Among these genes are the Pi transporters Pho84 (Bun-ya et al., 1991), Pho89 (Martinez and Persson, 1998), and the ER protein Pho86, which is involved in the targeting and exit of Pho84 from the ER (Yompakdee et al., 1996). PHO genes also encode several phosphatases (Persson et al., 2003): the repressible secreted acid phosphatase Pho5 and the cell wall-associated acid phosphatases Pho11 and Pho12 (Toh-e and Kakimoto, 1975; Arima et al., 1983; Bostian et al., 1983), which liberate Pi from phosphoester substrates; the vacuolar “alkaline” phosphatase Pho8 (Kaneko et al., 1985); and the glycerophosphocholine (GroPCho) phosphodiesterase Gde1 (Ogawa et al., 2000). All genes for the synthesis and degradation of vacuolar polyphosphate (VTC1 through VTC5, PPN1, and PPN2) are also controlled by the PHO pathway (Ogawa et al., 2000).
The genes of some regulators of the PHO pathway are themselves transcribed under the control of this pathway, such as PHO81 and SPL2 (Wykoff et al., 2007; Hürlimann et al., 2009; Ghillebert et al., 2011). As described above, this generates the possibility of positive and negative feedback loops, which can lead to heterogeneity in a yeast population, e.g., with a fraction of cells remaining stably committed to the Pi starvation program even under Pi-replete conditions (Wykoff et al., 2007; Vardi et al., 2013, 2014; Estill et al., 2015).
Besides the genes mentioned above, the PHO pathway regulates numerous other genes, among them many open reading frames of unknown function, for which the relationship to Pi homeostasis has not yet been understood (Ogawa et al., 2000). A similar complexity of Pi-regulated transcription has been found in other fungi, e.g., in Cryptococcus neoformans and Schizosaccaromyces pombe, where the number of PHO genes has been determined to be in the range of 130–160 (Henry et al., 2011; Carter-O’Connell et al., 2012; Lev and Djordjevic, 2018). They suggest links of the PHO signaling pathway to cellular transport, carbohydrate and lipid metabolism, and the responses to stress and to chemicals.
Regulation of PHO Genes During Pi Limitation and Serious Pi Starvation
The transition of S. cerevisiae into phosphate starvation is a complex process in which the cells show a graded adaptation to the declining Pi availability and temporally separate waves of gene induction. When Pi gradually becomes limiting, the activity of Pho85 kinase declines, leading first to a partial dephosphorylation of the Pho4 transcription factor. The partially dephosphorylated Pho4 induces only part of the PHO genes, such as the high-affinity transporter Pho84. Others, such as the secreted acid phosphatase Pho5, follow only upon more profound Pi starvation, leading to the inactivation of Pho85 kinase and the full dephosphorylation of Pho4 (O’Neill et al., 1996; Springer et al., 2003; Thomas and O’Shea, 2005). This establishes a graded response, which first maximizes phosphate acquisition and, upon more serious starvation, activates the recycling of Pi from internal resources.
The activation of the PHO pathway is also influenced by a metabolic intermediate of purine biosynthesis, 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) (Pinson et al., 2009). AICAR binds Pho2 and Pho4 in vitro and promotes their interaction in vivo, suggesting a direct effect on these transcription factors. Since AICAR also transcriptionally regulates purine biosynthesis, it may coordinate phosphate acquisition with nucleotide synthesis. Nucleotide synthesis is a major sink for Pi, and such coordination might be useful for this reason. Importantly, the activation of the PHO pathway through AICAR does not coincide with a major accumulation of Pho4 in the nucleus, in striking contrast to the induction through Pho85/80/81. AICAR thus represents an alternative pathway to activate the transcription of PHO pathway genes.
Detailed analyses of the temporal pattern of PHO gene activation revealed further features that stabilize the transcriptional response and keep the cells from erroneously self-inactivating the PHO pathway (Vardi et al., 2014). This is relevant under moderate Pi limitation, where such self-inactivation might occur, because the measures by which the cells seek to improve Pi acquisition may completely rectify the initial decline in cytosolic Pi that triggered the starvation response, thereby leading to an oscillation of the PHO pathway activation and inactivation. This cannot happen upon complete depletion of Pi from the media. A decisive difference is thus whether the cells experience only moderate Pi limitation or profound Pi starvation. Upon moderate limitation of Pi, a first wave of transcription induces a subset of genes: for the high-affinity Pi importer Pho84; Pho86, which is involved in its biogenesis; the secreted acid phosphatases Pho12 and Pho11 (Vardi et al., 2014); the genes for the VTC complex, which depletes cytosolic Pi and transfers it as polyP into vacuoles; Spl2, which shuts down the low-affinity Pi transporters; and Pho81, which reduces Pho85 kinase activity and promotes Pho4-dependent transcription. This wave of gene induction enhances global Pi absorption and storage by the cells and, at the same time, establishes a positive feedback loop that stabilizes the induction of Pho4-dependent gene expression (Wykoff et al., 2007; Vardi et al., 2013, 2014). This feedback loop maintains the activation of the PHO pathway for several generations, even after the cells have been transferred back into high-Pi media.
Cells experiencing profound Pi starvation (no Pi in the medium) reveal other features. They induce the same set of immediately activated genes as cells in intermediate Pi. However, due to the absence of Pi in the media, the induction of these genes cannot suffice to stabilize their cytosolic Pi. The more pronounced and stable decline in cytosolic Pi is believed to trigger a second wave of gene inductions 2 h after the first wave (Vardi et al., 2014). This second wave additionally activates genes for the intracellular recuperation of Pi from nucleotides, lipids, and other substrates (Phm8 and Pho8); the high-affinity Pi importer Pho89; and the secreted phosphatase Pho5. This second wave of transcription coincides with the induction of the environmental stress response and a decline in the growth rate. The partial dephosphorylation of the transcription factor Pho4 may be the trigger for the first transcriptional wave (Springer et al., 2003; Vardi et al., 2014), whereas full dephosphorylation of Pho4 might trigger the second wave of induction. Furthermore, the promoter regions of the genes induced in these two waves show characteristic differences that may support this differential activation through their impact on the chromatin structure (Lam et al., 2008), which is also influenced by inositol polyphosphates (Steger et al., 2003).
Upon serious Pi starvation, the cells finally arrest their cell cycle after two further divisions, and they enter into the quiescent G0 phase of the cell cycle. Pho85/80/81 kinase influences the entry and exit into quiescence in at least two ways: It phosphorylates and thereby inhibits the Rim15 kinase, which is a key factor for G0 entry (Wanke et al., 2005). Under Pi-replete conditions, Pho85/80/81 is active and phosphorylates Rim15, which then concentrates in the cytosol, where it is sequestered from its nuclear targets that initiate the G0 program. The residue that becomes phosphorylated on Rim15 is the same as the one phosphorylated through the TOR pathway, suggesting that Pi availability is integrated with information about the availability of other nutrients, such as amino acids, at the level of Rim15. Pho85/80/81 also phosphorylates and thereby destabilizes the cyclin Cln3, which is necessary for the exit from G0 (Menoyo et al., 2013). Both activities may synergize to favor an entry into G0 when Pi and, hence, Pho85/80/81 activities are low.
Elements of the PHO Pathway
Pho85, Pho80, and the Regulation of Transcription Through Pho4 and Pho2
Pho4 is a transcription factor which is expressed independently of the Pi concentration in the medium (Yoshida et al., 1989a). It carries a transactivation domain and a basic helix–loop–helix motif. Pho2 is a transcriptional activator bearing a homeobox (Bürglin, 1988). It interacts with Pho4 (controlling the PHO pathway), but also with a variety of transcription factors, such as Swi5 (controlling mating type switching) and Bas1 (controlling purine and histidine biosynthesis) (Bhoite et al., 2002). Its interaction with Pho4 increases the affinity for Pho4 binding sites in the promoter region of the PHO genes and allows them to recruit the general transcription machinery (Shao et al., 1996; Magbanua et al., 1997a, b).
Pho85 is a cyclin-dependent kinase subunit that associates with at least 10 different cyclins to regulate a wide spectrum of target proteins involved in many cellular processes, such as cell cycle control, storage and metabolism of carbohydrates, amino acid metabolism, and calcium signaling (Toh-E et al., 1988; Toh-E and Nishizawa, 2001; Huang et al., 2007). The cyclins confer target specificity to Pho85. Virtually all Pho85 effects that are directly relevant to phosphate homeostasis are mediated through its association with the cyclin Pho80. Pho80, and by consequence also the Pho85/80 kinase, are localized in the nucleus. This localization is independent of Pi availability (Hirst et al., 1994; Schneider et al., 1994). Pho85/80 is constitutively associated with Pho81, which inhibits its kinase activity under Pi limitation. Nuclear Pho85/80/81 phosphorylates the transcription factor Pho4 at five sites (O’Neill et al., 1996; Komeili and O’Shea, 1999). One of these phosphorylations blocks the association of Pho4 with the transcription factor Pho2; others promote the interaction of Pho4 with the nuclear export receptor Msn5 (Kaffman et al., 1998a) and impair its interaction with the nuclear import receptor Pse1 (Kaffman et al., 1998b). By means of these phosphorylations, Pho85/80 hinders the transcription of PHO genes in two ways: by impairing the interaction of Pho4 with the second necessary transcription factor Pho2 and by favoring the export of Pho4 into the cytosol.
Pho81
Pho81 is a cyclin-dependent kinase inhibitor (CKI) and the major regulator of Pho85/80 kinase and the PHO pathway (Schneider et al., 1994). Pho81 binds the Pho80 subunit of the Pho85/80 complex independently of Pi availability, but it inhibits the kinase only in low-Pi conditions (Schneider et al., 1994; Ogawa et al., 1995; O’Neill et al., 1996; Huang et al., 2001). Pho81 can also regulate the Pho85/Pcl7 cyclin-CDK complex in a Pi-dependent manner, which controls glycogen metabolism (Lee et al., 2000). PHO81 gene expression is induced by Pho4 upon Pi limitation (Bun-ya et al., 1991; Creasy et al., 1993, 1996; Ogawa et al., 2000), and Pho81 itself is also a substrate for Pho85/80 kinase (Knight et al., 2004; Waters et al., 2004). This creates a possibility for feedback regulation.
Pho81 is a large protein of almost 1,200 amino acids, which can be dissected into three functional domains (Ogawa et al., 1995). An N-terminal SPX domain and a C-terminal part, both of which influence the degree to which Pi starvation can induce the PHO pathway. The central part of the protein contains six ankyrin repeats. Part of these repeats and an adjacent region form a 141-amino acid piece that is sufficient to maintain some Pi-dependent regulation of the PHO pathway in vivo. The resulting inducibility is limited, however, because this central domain yields only a fivefold induction of the PHO5 gene on Pi-free media, and this only upon overexpression, whereas full-length Pho81, expressed at normal levels from its native promoter, yields an almost 200-fold induction (Ogawa et al., 1995). This central region was later trimmed further to remove the remaining ankyrin repeats, resulting in an 80-amino acid region, termed “minimum domain.” Also, the minimum domain allows only a less than 10-fold induction of PHO5 when overexpressed (Huang et al., 2001). Nevertheless, the minimum domain was ascribed a key function in the regulation of the PHO pathway through Pho81 (Lee et al., 2007, 2008). That the other parts of Pho81 may as well play a crucial role in regulation is suggested by the observation that the minimum domain is not sufficient to allow Pho85/80 to regulate the stress response pathway in a Pi-dependent manner (Swinnen et al., 2005), which full-length Pho81 does. Furthermore, several mutations leading to the constitutive activation of the PHO pathway lie in the N-terminal part of Pho81 (Creasy et al., 1993; Ogawa et al., 1995), underlining the physiological relevance of this region.
It has been reported that Pho81 requires the PIPP5K Vip1 and its product 1-InsP7 to inhibit Pho85/80 kinase and that InsP7 shows a corresponding increase under Pi starvation (Lee et al., 2007). Although this model has been widely accepted, several observations cannot easily be reconciled with it: Firstly, the InsP7 concentration actually decreases with decreasing Pi availability and Vip1 makes only a minor contribution to the InsP7 pool in rich media (Lonetti et al., 2011; Steidle et al., 2016; Wild et al., 2016). Secondly, the inactivation of the enzymes Plc1, Ipk1, and Arg82, which ablates the synthesis of 1-InsP7 and its precursor InsP6, inhibits Pho85/80 (Auesukaree et al., 2005) instead of showing the activation that one should expect if InsP7 inhibited the enzyme. Thirdly, vip1Δ mutants can also activate the PHO pathway, though with a moderate delay relative to wild-type cells (Choi et al., 2017). Thus, it is currently unclear how the PHO pathway is regulated through inositol pyrophosphates and the availability of Pi.
Control of the PHO Pathway Through Other Parameters Than Pho4 Activation
Besides phosphate limitation, several other conditions can activate PHO genes. Potassium starvation upregulates the PHO genes PHO84, PHO5, and SPL2 using the PHO signaling pathway (Anemaet and van Heusden, 2014; Canadell et al., 2015). Also, growth on acidic or alkaline pH, or the activation of calcineurin, can trigger the PHO pathway (Causton et al., 2001; Serrano et al., 2002; Ruiz et al., 2003; Luan and Li, 2004). The cell cycle activates PHO genes transiently and in a cyclic manner through Pho4 (Spellman et al., 1998), but also through the transcription factor Fkh2 (Pondugula et al., 2009; Korber and Barbaric, 2014; Bru et al., 2016).
While some activation of the PHO pathway may be related to secondary effects of the tested conditions on cytosolic Pi concentration, it is clear that Pho4/Pho2 are not the only transcription factors inducing PHO gene expression. These genes are embedded into a complex network of transcriptional control, which engages many other players, including the SAGA complex (Nishimura et al., 1999), the SWI-SNF complex (Gregory et al., 1999), the nucleosome spacing factor Ino80 (Barbaric et al., 2007), the arginine methyl transferase Hmt1 (Chia et al., 2018), or the transcriptional activator Crz1 (Serra-Cardona et al., 2014). These factors can either directly activate transcription or they may regulate promoter activity through the chromatin structure (Steger et al., 2003; Lam et al., 2008; Korber and Barbaric, 2014).
Yeast cells also perform pervasive transcription, which produces non-coding RNAs (Xu et al., 2009). Antisense transcripts for PHO5 and PHO84 mediate an exosome-dependent reduction in the abundance of messenger RNA (mRNA) from these two genes (Camblong et al., 2007) and affect chromatin remodeling during the activation of PHO5 transcription (Uhler et al., 2007). On the PHO84 promoter, it recruits the histone deacetylase Hda1, which downregulates transcription (Camblong et al., 2007). At the KCS1 locus, antisense and intragenic transcripts are produced, and their production depends on the transcription factor Pho4 and on Pi starvation (Nishizawa et al., 2008). These transcripts lead to the synthesis of truncated Kcs1, which is presumed to show lower activity.
Phosphate-dependent regulation of gene expression appears to act also on steps subsequent to the initiation of transcription. For example, Pho92 regulates the degradation of Pho4 mRNA by binding to its 3′-UTR in a Pi-dependent manner (Kang et al., 2014). The nonsense-mediated decay pathway is probably involved in this mRNA degradation reaction through Pop2 and the Ccr4-NOT complex.
Outlook
Pi homeostasis is an essential and complex aspect of metabolism. Even a simple unicellular organism such as yeast dedicates a significant percentage of its only 6,000 genes on it, underlining its fundamental importance. Many components involved in the Pi homeostasis in yeast are conserved in other organisms, suggesting that there are common approaches to the problem. Others appear functionally conserved, but realized in a quite organism-specific manner. For example, Pi-dependent transcriptional control is a widespread phenomenon which is conserved in that it employs SPX domains and InsPPs, and we expect that the way in which SPX domains interact with their target proteins may reveal common features. However, in other organisms, the mediators transmitting this control to the transcription factors can differ from the PHO pathway components acting in yeast. Work in a variety of different model organisms will thus contribute essential information necessary to grasp the full spectrum of strategies underlying Pi homeostasis.
Yeast cells are a prime model to study phosphate homeostasis in a eukaryotic cell system because many of the proteins immediately implicated in stabilizing cytosolic Pi are known and important elements of the signaling cascades regulating their expression and activity have begun to emerge. A major challenge lies in the high degree of redundancy that cells use in order to regulate their internal phosphate store. In order to study the regulatory mechanisms behind Pi homeostasis, it will therefore be necessary to establish reduced and simplified systems, both in vivo and in vitro, which will allow isolating individual aspects of the complex regulatory network and exploring them without too much interference by redundant mechanisms. The potent and convenient methods for manipulating the yeast genome provide the necessary tools to tackle this task.
Author Contributions
Both authors wrote all parts of the review together.
Funding
This work was supported by grants from the SNSF (CRSII5_170925) and the ERC (788442) to AM.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Geun-Don Kim, Andrea Schmidt, Valentin Chabert, and Juan Francisco Bada for their comments on our manuscript.
References
Almaguer, C., Cheng, W., Nolder, C., and Patton-Vogt, J. (2004). Glycerophosphoinositol, a novel phosphate source whose transport is regulated by multiple factors in Saccharomyces cerevisiae. J. Biol. Chem. 279, 31937–31942. doi: 10.1074/jbc.M403648200
Andreeva, N., Trilisenko, L., Eldarov, M., and Kulakovskaya, T. (2015). Polyphosphatase PPN1 of Saccharomyces cerevisiae: switching of exopolyphosphatase and endopolyphosphatase activities. PLoS One 10:e0119594. doi: 10.1371/journal.pone.0119594
Anemaet, I. G., and van Heusden, G. P. H. (2014). Transcriptional response of Saccharomyces cerevisiae to potassium starvation. BMC Genomics 15:1040. doi: 10.1186/1471-2164-15-1040
Ansermet, C., Moor, M. B., Centeno, G., Auberson, M., Hu, D. Z., Baron, R., et al. (2017). Renal fanconi syndrome and hypophosphatemic rickets in the absence of xenotropic and polytropic retroviral receptor in the nephron. J. Am. Soc. Nephrol. 28, 1073–1078. doi: 10.1681/ASN.2016070726
Arima, K., Oshima, T., Kubota, I., Nakamura, N., Mizunaga, T., and Toh-e, A. (1983). The nucleotide sequence of the yeast PHO5 gene: a putative precursor of repressible acid phosphatase contains a signal peptide. Nucleic Acids Res. 11, 1657–1672. doi: 10.1093/nar/11.6.1657
Auesukaree, C., Homma, T., Kaneko, Y., and Harashima, S. (2003). Transcriptional regulation of phosphate-responsive genes in low-affinity phosphate-transporter-defective mutants in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 306, 843–850. doi: 10.1016/s0006-291x(03)01068-4
Auesukaree, C., Homma, T., Tochio, H., Shirakawa, M., Kaneko, Y., and Harashima, S. (2004). Intracellular phosphate serves as a signal for the regulation of the PHO pathway in Saccharomyces cerevisiae. J. Biol. Chem. 279, 17289–17294. doi: 10.1074/jbc.M312202200
Auesukaree, C., Tochio, H., Shirakawa, M., Kaneko, Y., and Harashima, S. (2005). Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. J. Biol. Chem. 280, 25127–25133. doi: 10.1074/jbc.M414579200
Azevedo, C., Desfougères, Y., Jiramongkol, Y., Partington, H., Trakansuebkul, S., Singh, J., et al. (2019). Development of a yeast model to study the contribution of vacuolar polyphosphate metabolism to lysine polyphosphorylation. J. Biol. Chem. 295, 1439–1451. doi: 10.1074/jbc.RA119.011680
Azevedo, C., Livermore, T., and Saiardi, A. (2015). Protein polyphosphorylation of lysine residues by inorganic polyphosphate. Mol. Cell 58, 71–82. doi: 10.1016/j.molcel.2015.02.010
Azevedo, C., and Saiardi, A. (2017). Eukaryotic phosphate homeostasis: the inositol pyrophosphate perspective. Trends Biochem. Sci. 42, 219–231. doi: 10.1016/j.tibs.2016.10.008
Azevedo, C., Singh, J., Steck, N., Hofer, A., Ruiz, F. A., Singh, T., et al. (2018). Screening a protein array with synthetic biotinylated inorganic polyphosphate to define the human polyP-ome. ACS Chem. Biol. 13, 1958–1963. doi: 10.1021/acschembio.8b00357
Barbaric, S., Luckenbach, T., Schmid, A., Blaschke, D., Hörz, W., and Korber, P. (2007). Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J. Biol. Chem. 282, 27610–27621. doi: 10.1074/jbc.M700623200
Bhoite, L. T., Allen, J. M., Garcia, E., Thomas, L. R., Gregory, I. D., Voth, W. P., et al. (2002). Mutations in the pho2 (bas2) transcription factor that differentially affect activation with its partner proteins bas1, pho4, and swi5. J. Biol. Chem. 277, 37612–37618. doi: 10.1074/jbc.M206125200
Biber, J., Hernando, N., and Forster, I. (2013). Phosphate transporters and their function. Annu. Rev. Physiol. 75, 535–550. doi: 10.1146/annurev-physiol-030212-183748
Boer, V. M., Crutchfield, C. A., Bradley, P. H., Botstein, D., and Rabinowitz, J. D. (2010). Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations. Mol. Biol. Cell 21, 198–211. doi: 10.1091/mbc.e09-07-0597
Boer, V. M., de Winde, J. H., Pronk, J. T., and Piper, M. D. W. (2003). The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 278, 3265–3274. doi: 10.1074/jbc.M209759200
Bostian, K. A., Lemire, J. M., and Halvorson, H. O. (1983). Physiological control of repressible acid phosphatase gene transcripts in Saccharomyces cerevisiae. Mol. Cell Biol. 3, 839–853. doi: 10.1128/mcb.3.5.839
Bru, S., Martínez-Laínez, J. M., Hernández-Ortega, S., Quandt, E., Torres-Torronteras, J., Martí, R., et al. (2016). Polyphosphate is involved in cell cycle progression and genomic stability in Saccharomyces cerevisiae. Mol. Microbiol. 101, 367–380. doi: 10.1111/mmi.13396
Bun-ya, M., Nishimura, M., Harashima, S., and Oshima, Y. (1991). The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol. Cell Biol. 11, 3229–3238. doi: 10.1128/mcb.11.6.3229
Bürglin, T. R. (1988). The yeast regulatory gene PHO2 encodes a homeo box. Cell 53, 339–340. doi: 10.1016/0092-8674(88)90153-5
Camblong, J., Iglesias, N., Fickentscher, C., Dieppois, G., and Stutz, F. (2007). Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 131, 706–717. doi: 10.1016/j.cell.2007.09.014
Canadell, D., González, A., Casado, C., and Ariño, J. (2015). Functional interactions between potassium and phosphate homeostasis in Saccharomyces cerevisiae. Mol. Microbiol. 95, 555–572. doi: 10.1111/mmi.12886
Carroll, A. S., and O’Shea, E. K. (2002). Pho85 and signaling environmental conditions. Trends Biochem. Sci. 27, 87–93. doi: 10.1016/s0968-0004(01)02040-0
Carter-O’Connell, I., Peel, M. T., Wykoff, D. D., and O’Shea, E. K. (2012). Genome-wide characterization of the phosphate starvation response in Schizosaccharomyces pombe. BMC Genomics 13:697. doi: 10.1186/1471-2164-13-697
Cartwright, J. L., and McLennan, A. G. (1999). The Saccharomyces cerevisiae YOR163w gene encodes a diadenosine 5′, 5″′-P1,P6-Hexaphosphate (Ap6A) hydrolase member of the MutT motif (Nudix hydrolase) family. J. Biol. Chem. 274, 8604–8610. doi: 10.1074/jbc.274.13.8604
Causton, H. C., Ren, B., Koh, S. S., Harbison, C. T., Kanin, E., Jennings, E. G., et al. (2001). Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12, 323–337. doi: 10.1091/mbc.12.2.323
Chia, S. Z., Lai, Y.-W., Yagoub, D., Lev, S., Hamey, J. J., Pang, C. N. I., et al. (2018). Knockout of the Hmt1p arginine methyltransferase in Saccharomyces cerevisiae leads to the dysregulation of phosphate-associated genes and processes. Mol. Cell. Proteomics 17, 2462–2479. doi: 10.1074/mcp.RA117.000214
Choi, J., Rajagopal, A., Xu, Y.-F., Rabinowitz, J. D., and O’Shea, E. K. (2017). A systematic genetic screen for genes involved in sensing inorganic phosphate availability in Saccharomyces cerevisiae. PLoS One 12:e0176085. doi: 10.1371/journal.pone.0176085
Conrad, M., Schothorst, J., Kankipati, H. N., Van Zeebroeck, G., Rubio-Texeira, M., and Thevelein, J. M. (2014). Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 38, 254–299. doi: 10.1111/1574-6976.12065
Creasy, C. L., Madden, S. L., and Bergman, L. W. (1993). Molecular analysis of the PHO81 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 21, 1975–1982. doi: 10.1093/nar/21.8.1975
Creasy, C. L., Shao, D., and Begman, L. W. (1996). Negative transcriptional regulation of PH081 expression in Saccharomyces cerevisiae. Gene 168, 23–29. doi: 10.1016/0378-1119(95)00737-7
De Virgilio, C. (2011). The essence of yeast quiescence. FEMS Microbiol. Rev. 36, 306–339. doi: 10.1111/j.1574-6976.2011.00287.x
Depaoli, M. R., Hay, J. C., Graier, W. F., and Malli, R. (2019). The enigmatic ATP supply of the endoplasmic reticulum. Biol. Rev. Camb. Philos. Soc. 94, 610–628. doi: 10.1111/brv.12469
Desfougères, Y., Gerasimaitë, R. U., Jessen, H. J., and Mayer, A. (2016). Vtc5, a novel subunit of the vacuolar transporter chaperone complex, regulates polyphosphate synthesis and phosphate homeostasis in yeast. J. Biol. Chem. 291, 22262–22275. doi: 10.1074/jbc.M116.746784
Docampo, R., and Huang, G. (2016). Acidocalcisomes of eukaryotes. Curr. Opin. Cell Biol. 41, 66–72. doi: 10.1016/j.ceb.2016.04.007
Donella-Deana, A., Ostojić, S., Pinna, L. A., and Barbarić, S. (1993). Specific dephosphorylation of phosphopeptides by the yeast alkaline phosphatase encoded by PHO8 gene. Biochim. Biophys. Acta 1177, 221–228. doi: 10.1016/0167-4889(93)90044-p
Dong, J., Ma, G., Sui, L., Wei, M., Satheesh, V., Zhang, R., et al. (2019). Inositol pyrophosphate InsP8 acts as an intracellular phosphate signal in Arabidopsis. Mol. Plant 12, 1463–1473. doi: 10.1016/j.molp.2019.08.002
Estill, M., Kerwin-Iosue, C. L., and Wykoff, D. D. (2015). Dissection of the PHO pathway in Schizosaccharomyces pombe using epistasis and the alternate repressor adenine. Curr. Genet. 61, 175–183. doi: 10.1007/s00294-014-0466-6
Fisher, E., Almaguer, C., Holic, R., Griac, P., and Patton-Vogt, J. (2005). Glycerophosphocholine-dependent growth requires Gde1p (YPL110c) and Git1p in Saccharomyces cerevisiae. J. Biol. Chem. 280, 36110–36117. doi: 10.1074/jbc.M507051200
Flick, J. S., and Thorner, J. (1998). An essential function of a phosphoinositide-specific phospholipase C is relieved by inhibition of a cyclin-dependent protein kinase in the yeast Saccharomyces cerevisiae. Genetics 148, 33–47.
Gerasimaite, R., and Mayer, A. (2016). Enzymes of yeast polyphosphate metabolism: structure, enzymology and biological roles. Biochem. Soc. Trans. 44, 234–239. doi: 10.1042/BST20150213
Gerasimaite, R., and Mayer, A. (2017). Ppn2, a novel Zn(2+)-dependent polyphosphatase in the acidocalcisome-like yeast vacuole. J. Cell Sci. 130, 1625–1636. doi: 10.1242/jcs.201061
Gerasimaite, R., Pavlovic, I., Capolicchio, S., Hofer, A., Schmidt, A., Jessen, H. J., et al. (2017). Inositol pyrophosphate specificity of the SPX-dependent polyphosphate polymerase VTC. ACS Chem. Biol. 12, 648–653. doi: 10.1021/acschembio.7b00026
Gerasimaite, R., Sharma, S., Desfougères, Y., Schmidt, A., and Mayer, A. (2014). Coupled synthesis and translocation restrains polyphosphate to acidocalcisome-like vacuoles and prevents its toxicity. J. Cell Sci. 127, 5093–5104. doi: 10.1242/jcs.159772
Ghillebert, R., Swinnen, E., De Snijder, P., Smets, B., and Winderickx, J. (2011). Differential roles for the low-affinity phosphate transporters Pho87 and Pho90 in Saccharomyces cerevisiae. Biochem. J. 434, 243–251. doi: 10.1042/BJ20101118
Giots, F., Donaton, M., and Thevelein, J. M. (2003). Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 47, 1163–1181. doi: 10.1046/j.1365-2958.2003.03365.x
Giovannini, D., Touhami, J., Charnet, P., Sitbon, M., and Battini, J.-L. (2013). Inorganic phosphate export by the retrovirus receptor XPR1 in metazoans. Cell Rep. 3, 1866–1873. doi: 10.1016/j.celrep.2013.05.035
Gregory, P. D., Schmid, A., Zavari, M., Münsterkötter, M., and Hörz, W. (1999). Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 18, 6407–6414. doi: 10.1093/emboj/18.22.6407
Gu, C., Nguyen, H.-N., Hofer, A., Jessen, H. J., Dai, X., Wang, H., et al. (2017). The significance of the bifunctional kinase/phosphatase activities of PPIP5Ks for coupling inositol pyrophosphate cell-signaling to cellular phosphate homeostasis. J. Biol. Chem. 292, 4544–4555. doi: 10.1074/jbc.M116.765743
Henry, T. C., Power, J. E., Kerwin, C. L., Mohammed, A., Weissman, J. S., Cameron, D. M., et al. (2011). Systematic screen of Schizosaccharomyces pombe deletion collection uncovers parallel evolution of the phosphate signal transduction pathway in yeasts. Eukaryot. Cell 10, 198–206. doi: 10.1128/ec.00216-10
Hirst, K., Fisher, F., McAndrew, P. C., and Goding, C. R. (1994). The transcription factor, the Cdk, its cyclin and their regulator: directing the transcriptional response to a nutritional signal. EMBO J. 13, 5410–5420. doi: 10.1002/j.1460-2075.1994.tb06876.x
Hothorn, M., Neumann, H., Lenherr, E. D., Wehner, M., Rybin, V., Hassa, P. O., et al. (2009). Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science 324, 513–516. doi: 10.1126/science.1168120
Huang, D., Friesen, H., and Andrews, B. (2007). Pho85, a multifunctional cyclin-dependent protein kinase in budding yeast. Mol. Microbiol. 66, 303–314. doi: 10.1111/j.1365-2958.2007.05914.x
Huang, S., Jeffery, D. A., Anthony, M. D., and O’Shea, E. K. (2001). Functional analysis of the cyclin-dependent kinase inhibitor Pho81 identifies a novel inhibitory domain. Mol. Cell Biol. 21, 6695–6705. doi: 10.1128/mcb.21.19.6695-6705.2001
Huh, W.-K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., et al. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691. doi: 10.1038/nature02026
Hürlimann, H. C. (2009). Investigations on the Physiology and Molecular Basis of Polyphosphate Hyper- and Hypo-Accumulation in Saccharomyces cerevisiae. Zurich: ETH.
Hürlimann, H. C., Pinson, B., Stadler-Waibel, M., Zeeman, S. C., and Freimoser, F. M. (2009). The SPX domain of the yeast low-affinity phosphate transporter Pho90 regulates transport activity. EMBO Rep. 10, 1003–1008. doi: 10.1038/embor.2009.105
Hürlimann, H. C., Stadler-Waibel, M., Werner, T. P., and Freimoser, F. M. (2007). Pho91 is a vacuolar phosphate transporter that regulates phosphate and polyphosphate metabolism in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 4438–4445. doi: 10.1091/mbc.e07-05-0457
Jensen, L. T., Ajua-Alemanji, M., and Culotta, V. C. (2003). The Saccharomyces cerevisiae high affinity phosphate transporter encoded by PHO84 also functions in manganese homeostasis. J. Biol. Chem. 278, 42036–42040. doi: 10.1074/jbc.M307413200
Kaffman, A., Herskowitz, I., Tjian, R., and O’Shea, E. K. (1994). Phosphorylation of the transcription factor-Pho4 by a cyclin-Cdk complex, Pho80-Pho85. Science 263, 1153–1156. doi: 10.1126/science.8108735
Kaffman, A., Rank, N. M., O’Neill, E. M., Huang, L. S., and O’Shea, E. K. (1998a). The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396, 482–486. doi: 10.1038/24898
Kaffman, A., Rank, N. M., and O’Shea, E. K. (1998b). Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 12, 2673–2683. doi: 10.1101/gad.12.17.2673
Kaneko, Y., Tamai, Y., Toh-e, A., and Oshima, Y. (1985). Transcriptional and post-transcriptional control of PHO8 expression by PHO regulatory genes in Saccharomyces cerevisiae. Mol. Cell Biol. 5, 248–252. doi: 10.1128/mcb.5.1.248
Kaneko, Y., Toh-e, A., and Oshima, Y. (1982). Identification of the genetic locus for the structural gene and a new regulatory gene for the synthesis of repressible alkaline phosphatase in Saccharomyces cerevisiae. Mol. Cell Biol. 2, 127–137. doi: 10.1128/mcb.2.2.127
Kang, H.-J., Jeong, S.-J., Kim, K.-N., Baek, I.-J., Chang, M., Kang, C.-M., et al. (2014). A novel protein, Pho92, has a conserved YTH domain and regulates phosphate metabolism by decreasing the mRNA stability of PHO4 in Saccharomyces cerevisiae. Biochem. J. 457, 391–400. doi: 10.1042/BJ20130862
Klompmaker, S. H., Kohl, K., Fasel, N., and Mayer, A. (2017). Magnesium uptake by connecting fluid-phase endocytosis to an intracellular inorganic cation filter. Nat. Commun. 8:1879.
Knight, J. P., Daly, T. M., and Bergman, L. W. (2004). Regulation by phosphorylation of Pho81p, a cyclin-dependent kinase inhibitor in Saccharomyces cerevisiae. Curr. Genet. 46, 10–19. doi: 10.1007/s00294-004-0502-z
Komeili, A., and O’Shea, E. K. (1999). Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284, 977–980. doi: 10.1126/science.284.5416.977
Korber, P., and Barbaric, S. (2014). The yeast PHO5 promoter: from single locus to systems biology of a paradigm for gene regulation through chromatin. Nucleic Acids Res. 42, 10888–10902. doi: 10.1093/nar/gku784
Kulakovskaya, T. V., Lichko, L. P., Vagabov, V. M., and Kulaev, I. S. (2010). Inorganic polyphosphates in mitochondria. Biochem. Mosc. 75, 825–831.
Kumble, K. D., and Kornberg, A. (1996). Endopolyphosphatases for long chain inorganic polyphosphate in yeast and mammals. J. Biol. Chem. 271, 27146–27151. doi: 10.1074/jbc.271.43.27146
Lam, F. H., Steger, D. J., and O’Shea, E. K. (2008). Chromatin decouples promoter threshold from dynamic range. Nature 453, 246–250. doi: 10.1038/nature06867
Langen, P., and Liss, E. (1958). [Formation and conversion of yeast polyphosphates]. Biochem. Z. 330, 455–466.
Langen, P., and Liss, E. (1960). [Differentiation of orthophosphates of yeast cells]. Biochem. Z. 332, 403–406.
Lazard, M., Blanquet, S., Fisicaro, P., Labarraque, G., and Plateau, P. (2010). Uptake of selenite by Saccharomyces cerevisiae involves the high and low affinity orthophosphate transporters. J. Biol. Chem. 285, 32029–32037. doi: 10.1074/jbc.M110.139865
Lee, M., O’Regan, S., Moreau, J. L., Johnson, A. L., Johnston, L. H., and Goding, C. R. (2000). Regulation of the Pcl7-Pho85 cyclin-Cdk complex by Pho81. Mol. Microbiol. 38, 411–422. doi: 10.1046/j.1365-2958.2000.02140.x
Lee, Y.-S., Huang, K., Quiocho, F. A., and O’Shea, E. K. (2008). Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 4, 25–32. doi: 10.1038/nchembio.2007.52
Lee, Y. S., Mulugu, S., York, J. D., and O’Shea, E. K. (2007). Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316, 109–112. doi: 10.1126/science.1139080
Legati, A., Giovannini, D., Nicolas, G., López-Sánchez, U., Quintáns, B., Oliveira, J. R. M., et al. (2015). Mutations in XPR1 cause primary familial brain calcification associated with altered phosphate export. Nat. Genet. 47, 579–581. doi: 10.1038/ng.3289
Lev, S., and Djordjevic, J. T. (2018). Why is a functional PHO pathway required by fungal pathogens to disseminate within a phosphate-rich host: a paradox explained by alkaline pH-simulated nutrient deprivation and expanded PHO pathway function. PLoS Pathog. 14:e1007021. doi: 10.1371/journal.ppat.1007021
Levy, S., Kafri, M., Carmi, M., and Barkai, N. (2011). The competitive advantage of a dual-transporter system. Science 334, 1408–1412. doi: 10.1126/science.1207154
Li, X., Gu, C., Hostachy, S., Sahu, S., Wittwer, C., Jessen, H. J., et al. (2020). Control of XPR1-dependent cellular phosphate efflux by InsP8 is an exemplar for functionally-exclusive inositol pyrophosphate signaling. Proc. Natl. Acad. Sci. U.S.A. 14:201908830. doi: 10.1073/pnas.1908830117
Lichko, L., Kulakovskaya, T., Pestov, N., and Kulaev, I. (2006). Inorganic polyphosphates and exopolyphosphatases in cell compartments of the yeast Saccharomyces cerevisiae under inactivation of PPX1 and PPN1 genes. Biosci. Rep. 26, 45–54. doi: 10.1007/s10540-006-9003-2
Lichko, L. P., Kulakovskaya, T. V., and Kulaev, I. S. (2006). Inorganic polyphosphate and exopolyphosphatase in the nuclei of Saccharomyces cerevisiae: dependence on the growth phase and inactivation of the PPX1 and PPN1 genes. Yeast 23, 735–740. doi: 10.1002/yea.1391
Lichko, L. P., Pestov, N. A., Kulakovskaya, T. V., and Kulaev, I. S. (2003). Effect of PPX1 inactivation on the exopolyphosphatase spectra in cytosol and mitochondria of the yeast Saccharomyces cerevisiae. Biochem. Mosc. 68, 740–746.
Liu, J., Yang, L., Luan, M., Wang, Y., Zhang, C., Zhang, B., et al. (2015). A vacuolar phosphate transporter essential for phosphate homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 112, E6571–E6578. doi: 10.1073/pnas.1514598112
Lonetti, A., Szijgyarto, Z., Bosh, D., Loss, O., Azevedo, C., and Saiardi, A. (2011). Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J. Biol. Chem. 286, 31966–31974. doi: 10.1074/jbc.M111.266320
Lu, S.-P., and Lin, S.-J. (2011). Phosphate-responsive signaling pathway is a novel component of NAD+ metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 286, 14271–14281. doi: 10.1074/jbc.M110.217885
Luan, Y., and Li, H. (2004). Model-based methods for identifying periodically expressed genes based on time course microarray gene expression data. Bioinformatics 20, 332–339. doi: 10.1093/bioinformatics/btg413
Lundh, F., Mouillon, J.-M., Samyn, D., Stadler, K., Popova, Y., Lagerstedt, J. O., et al. (2009). Molecular mechanisms controlling phosphate-induced downregulation of the yeast Pho84 phosphate transporter. Biochemistry 48, 4497–4505. doi: 10.1021/bi9001198
Magbanua, J. P., Fujisawa, K., Ogawa, N., and Oshima, Y. (1997a). The homeodomain protein Pho2p binds at an A/T-rich segment flanking the binding site of the basic-helix-loop-helix protein Pho4p in the yeast PHO promoters. Yeast 13, 1299–1308. doi: 10.1002/(sici)1097-0061(199711)13:14<1299::aid-yea178>3.0.co;2-a
Magbanua, J. P., Ogawa, N., Harashima, S., and Oshima, Y. (1997b). The transcriptional activators of the PHO regulon, Pho4p and Pho2p, interact directly with each other and with components of the basal transcription machinery in Saccharomyces cerevisiae. J. Biochem. 121, 1182–1189. doi: 10.1093/oxfordjournals.jbchem.a021713
Martinez, P., and Persson, B. L. (1998). Identification, cloning and characterization of a derepressible Na+-coupled phosphate transporter in Saccharomyces cerevisiae. Mol. Gen. Genet. 258, 628–638. doi: 10.1007/s004380050776
McIlwain, H., Buchel, L., and Cheshire, J. D. (1951). The inorganic phosphate and phosphocreatine of brain especially during metabolism in vitro. Biochem. J. 48, 12–20. doi: 10.1042/bj0480012
Menoyo, S., Ricco, N., Bru, S., Hernández-Ortega, S., Escoté, X., Aldea, M., et al. (2013). Phosphate-activated cyclin-dependent kinase stabilizes G1 cyclin to trigger cell cycle entry. Mol. Cell Biol. 33, 1273–1284. doi: 10.1128/mcb.01556-12
Mouillon, J.-M., and Persson, B. L. (2005). Inhibition of the protein kinase A alters the degradation of the high-affinity phosphate transporter Pho84 in Saccharomyces cerevisiae. Curr. Genet. 48, 226–234. doi: 10.1007/s00294-005-0019-0
Mouillon, J.-M., and Persson, B. L. (2006). New aspects on phosphate sensing and signalling in Saccharomyces cerevisiae. FEMS Yeast Res. 6, 171–176. doi: 10.1111/j.1567-1364.2006.00036.x
Müller, O., Bayer, M. J., Peters, C., Andersen, J. S., Mann, M., and Mayer, A. (2002). The Vtc proteins in vacuole fusion: coupling NSF activity to V(0) trans-complex formation. EMBO J. 21, 259–269. doi: 10.1093/emboj/21.3.259
Müller, O., Neumann, H., Bayer, M. J., and Mayer, A. (2003). Role of the Vtc proteins in V-ATPase stability and membrane trafficking. J. Cell Sci. 116, 1107–1115. doi: 10.1242/jcs.00328
Mulugu, S., Bai, W., Fridy, P. C., Bastidas, R. J., Otto, J. C., Dollins, D. E., et al. (2007). A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316, 106–109. doi: 10.1126/science.1139099
Nair, V. S., Gu, C., Janoshazi, A. K., Jessen, H. J., Wang, H., and Shears, S. B. (2018). Inositol pyrophosphate synthesis by diphosphoinositol pentakisphosphate kinase-1 is regulated by phosphatidylinositol(4,5)bisphosphate. Biosci. Rep. 38:BSR20171549. doi: 10.1042/BSR20171549
Neef, D. W., and Kladde, M. P. (2003). Polyphosphate loss promotes SNF/SWI- and Gcn5-dependent mitotic induction of PHO5. Mol. Cell Biol. 23, 3788–3797. doi: 10.1128/mcb.23.11.3788-3797.2003
Nishimura, K., Yasumura, K., Igarashi, K., Harashima, S., and Kakinuma, Y. (1999). Transcription of some PHO genes in Saccharomyces cerevisiae is regulated by spt7p. Yeast 15, 1711–1717. doi: 10.1002/(sici)1097-0061(199912)15:16<1711::aid-yea497>3.0.co;2-8
Nishizawa, M., Komai, T., Katou, Y., Shirahige, K., Ito, T., and Toh-E, A. (2008). Nutrient-regulated antisense and intragenic RNAs modulate a signal transduction pathway in yeast. PLoS Biol. 6:e326. doi: 10.1371/journal.pbio.0060326
Ogawa, N., DeRisi, J., and Brown, P. O. (2000). New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell 11, 4309–4321. doi: 10.1091/mbc.11.12.4309
Ogawa, N., Noguchi, K., Sawai, H., Yamashita, Y., Yompakdee, C., and Oshima, Y. (1995). Functional domains of Pho81p, an inhibitor of Pho85p protein kinase, in the transduction pathway of Pi signals in Saccharomyces cerevisiae. Mol. Cell Biol. 15, 997–1004. doi: 10.1128/mcb.15.2.997
O’Neill, E. M., Kaffman, A., Jolly, E. R., and O’Shea, E. K. (1996). Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271, 209–212. doi: 10.1126/science.271.5246.209
Oshima, Y. (1997). The phosphatase system in Saccharomyces cerevisiae. Genes Genet. Syst. 72, 323–334. doi: 10.1266/ggs.72.323
Palmieri, F., and Monné, M. (2016). Discoveries, metabolic roles and diseases of mitochondrial carriers: a review. Biochim. Biophys. Acta 1863, 2362–2378. doi: 10.1016/j.bbamcr.2016.03.007
Patton-Vogt, J. (2007). Transport and metabolism of glycerophosphodiesters produced through phospholipid deacylation. Biochim. Biophys. Acta 1771, 337–342. doi: 10.1016/j.bbalip.2006.04.013
Pérez-Sampietro, M., Serra-Cardona, A., Canadell, D., Casas, C., Ariño, J., and Herrero, E. (2016). The yeast Aft2 transcription factor determines selenite toxicity by controlling the low affinity phosphate transport system. Sci. Rep. 6:32836. doi: 10.1038/srep32836
Persson, B. L., Lagerstedt, J. O., Pratt, J. R., Pattison-Granberg, J., Lundh, K., Shokrollahzadeh, S., et al. (2003). Regulation of phosphate acquisition in Saccharomyces cerevisiae. Curr. Genet. 43, 225–244. doi: 10.1007/s00294-003-0400-9
Pestov, N. A., Kulakovskaya, T. V., and Kulaev, I. S. (2004). Inorganic polyphosphate in mitochondria of Saccharomyces cerevisiae at phosphate limitation and phosphate excess. FEMS Yeast Res. 4, 643–648. doi: 10.1016/j.femsyr.2003.12.008
Petersson, J., Pattison, J., Kruckeberg, A. L., Berden, J. A., and Persson, B. L. (1999). Intracellular localization of an active green fluorescent protein-tagged Pho84 phosphate permease in Saccharomyces cerevisiae. FEBS Lett. 462, 37–42. doi: 10.1016/s0014-5793(99)01471-4
Pinson, B., Merle, M., Franconi, J.-M., and Daignan-Fornier, B. (2004). Low affinity orthophosphate carriers regulate PHO gene expression independently of internal orthophosphate concentration in Saccharomyces cerevisiae. J. Biol. Chem. 279, 35273–35280. doi: 10.1074/jbc.M405398200
Pinson, B., Vaur, S., Sagot, I., Coulpier, F., Lemoine, S., and Daignan-Fornier, B. (2009). Metabolic intermediates selectively stimulate transcription factor interaction and modulate phosphate and purine pathways. Genes Dev. 23, 1399–1407. doi: 10.1101/gad.521809
Plankert, U., Purwin, C., and Holzer, H. (1991). Yeast fructose-2,6-bisphosphate 6-phosphatase is encoded by PHO8, the gene for nonspecific repressible alkaline phosphatase. Eur. J. Biochem. 196, 191–196. doi: 10.1111/j.1432-1033.1991.tb15803.x
Pöhlmann, J., Risse, C., Seidel, C., Pohlmann, T., Jakopec, V., Walla, E., et al. (2014). The Vip1 inositol polyphosphate kinase family regulates polarized growth and modulates the microtubule cytoskeleton in fungi. PLoS Genet. 10:e1004586. doi: 10.1371/journal.pgen.1004586
Pondugula, S., Neef, D. W., Voth, W. P., Darst, R. P., Dhasarathy, A., Reynolds, M. M., et al. (2009). Coupling phosphate homeostasis to cell cycle-specific transcription: mitotic activation of Saccharomyces cerevisiae PHO5 by Mcm1 and forkhead proteins. Mol. Cell Biol. 29, 4891–4905. doi: 10.1128/mcb.00222-09
Popova, Y., Thayumanavan, P., Lonati, E., Agrochão, M., and Thevelein, J. M. (2010). Transport and signaling through the phosphate-binding site of the yeast Pho84 phosphate transceptor. Proc. Natl. Acad. Sci. U.S.A. 107, 2890–2895. doi: 10.1073/pnas.0906546107
Potapenko, E., Cordeiro, C. D., Huang, G., and Docampo, R. (2019). Pyrophosphate stimulates the phosphate-sodium symporter of Trypanosoma brucei Acidocalcisomes and Saccharomyces cerevisiae vacuoles. mSphere 4:e00045-19.
Potapenko, E., Cordeiro, C. D., Huang, G., Storey, M., Wittwer, C., Dutta, A. K., et al. (2018). 5-diphosphoinositol pentakisphosphate (5-IP7) regulates phosphate release from Acidocalcisomes and yeast vacuoles. J. Biol. Chem. 293, 19101–19112. doi: 10.1074/jbc.RA118.005884
Pratt, J. R., Mouillon, J.-M., Lagerstedt, J. O., Pattison-Granberg, J., Lundh, K. I., and Persson, B. L. (2004). Effects of methylphosphonate, a phosphate analogue, on the expression and degradation of the high-affinity phosphate transporter Pho84, in Saccharomyces cerevisiae. Biochemistry 43, 14444–14453. doi: 10.1021/bi049327t
Puga, M. I., Mateos, I., Charukesi, R., Wang, Z., Franco-Zorrilla, J. M., de Lorenzo, L., et al. (2014). SPX1 is a phosphate-dependent inhibitor of PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 14947–14952. doi: 10.1073/pnas.1404654111
Puga, M. I., Rojas-Triana, M., de Lorenzo, L., Leyva, A., Rubio, V., and Paz-Ares, J. (2017). Novel signals in the regulation of Pi starvation responses in plants: facts and promises. Curr. Opin. Plant Biol. 39, 40–49. doi: 10.1016/j.pbi.2017.05.007
Randall, T. A., Gu, C., Li, X., Wang, H., and Shears, S. B. (2019). A two-way switch for inositol pyrophosphate signaling: evolutionary history and biological significance of a unique, bifunctional kinase/phosphatase. Adv. Biol. Regul. 75:100674. doi: 10.1016/j.jbior.2019.100674
Reddy, V. S., Singh, A. K., and Rajasekharan, R. (2008). The Saccharomyces cerevisiae PHM8 gene encodes a soluble magnesium-dependent lysophosphatidic acid phosphatase. J. Biol. Chem. 283, 8846–8854. doi: 10.1074/jbc.M706752200
Ruiz, A., Yenush, L., and Ariño, J. (2003). Regulation of ENA1 Na(+)-ATPase gene expression by the Ppz1 protein phosphatase is mediated by the calcineurin pathway. Eukaryot. Cell 2, 937–948. doi: 10.1128/ec.2.5.937-948.2003
Sabbagh, Y. (2013). Phosphate as a sensor and signaling molecule. Clin. Nephrol. 79, 57–65. doi: 10.5414/CN107322
Safrany, S. T., Ingram, S. W., Cartwright, J. L., Falck, J. R., McLennan, A. G., Barnes, L. D., et al. (1999). The diadenosine hexaphosphate hydrolases from Schizosaccharomyces pombe and Saccharomyces cerevisiae are homologues of the human diphosphoinositol polyphosphate phosphohydrolase. Overlapping substrate specificities in a MutT-type protein. J. Biol. Chem. 274, 21735–21740. doi: 10.1074/jbc.274.31.21735
Saiardi, A., Caffrey, J. J., Snyder, S. H., and Shears, S. B. (2000). The inositol hexakisphosphate kinase family. Catalytic flexibility and function in yeast vacuole biogenesis. J. Biol. Chem. 275, 24686–24692. doi: 10.1074/jbc.m002750200
Saiardi, A., Erdjument-Bromage, H., Snowman, A. M., Tempst, P., and Snyder, S. H. (1999). Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 9, 1323–1326. doi: 10.1016/s0960-9822(00)80055-x
Saito, K., Ohtomo, R., Kuga-Uetake, Y., Aono, T., and Saito, M. (2005). Direct labeling of polyphosphate at the ultrastructural level in Saccharomyces cerevisiae by using the affinity of the polyphosphate binding domain of Escherichia coli exopolyphosphatase. Appl. Environ. Microbiol. 71, 5692–5701. doi: 10.1128/AEM.71.10.5692-5701.2005
Samyn, D. R., Ruiz-Pávon, L., Andersson, M. R., Popova, Y., Thevelein, J. M., and Persson, B. L. (2012). Mutational analysis of putative phosphate- and proton-binding sites in the Saccharomyces cerevisiae Pho84 phosphate:H(+) transceptor and its effect on signalling to the PKA and PHO pathways. Biochem. J. 445, 413–422. doi: 10.1042/BJ20112086
Schneider, K. R., Smith, R. L., and O’Shea, E. K. (1994). Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science 266, 122–126. doi: 10.1126/science.7939631
Secco, D., Wang, C., Arpat, B. A., Wang, Z., Poirier, Y., Tyerman, S. D., et al. (2012a). The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 193, 842–851. doi: 10.1111/j.1469-8137.2011.04002.x
Secco, D., Wang, C., Shou, H., and Whelan, J. (2012b). Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett. 586, 289–295. doi: 10.1016/j.febslet.2012.01.036
Serra-Cardona, A., Petrezselyova, S., Canadell, D., Ramos, J., and Ariño, J. (2014). Coregulated expression of the Na+/phosphate Pho89 transporter and Ena1 Na+-ATPase allows their functional coupling under high-pH stress. Mol. Cell Biol. 34, 4420–4435. doi: 10.1128/mcb.01089-14
Serrano, R., Ruiz, A., Bernal, D., Chambers, J. R., and Ariño, J. (2002). The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol. Microbiol. 46, 1319–1333. doi: 10.1046/j.1365-2958.2002.03246.x
Serrano-Bueno, G., Hernández, A., López-Lluch, G., Pérez-Castiñeira, J. R., Navas, P., and Serrano, A. (2013). Inorganic pyrophosphatase defects lead to cell cycle arrest and autophagic cell death through NAD+ depletion in fermenting yeast. J. Biol. Chem. 288, 13082–13092. doi: 10.1074/jbc.M112.439349
Sethuraman, A., Rao, N. N., and Kornberg, A. (2001). The endopolyphosphatase gene: essential in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 98, 8542–8547. doi: 10.1073/pnas.151269398
Shao, D., Creasy, C. L., and Bergman, L. W. (1996). Interaction of Saccharomyces cerevisiae Pho2 with Pho4 increases the accessibility of the activation domain of Pho4. Mol. Gen. Genet. 251, 358–364. doi: 10.1007/BF02172527
Shears, S. B. (2015). Inositol pyrophosphates: why so many phosphates? Adv. Biol. Regul. 57, 203–216. doi: 10.1016/j.jbior.2014.09.015
Shears, S. B. (2017). Intimate connections: inositol pyrophosphates at the interface of metabolic regulation and cell signaling. J. Cell Physiol. 233, 1897–1912. doi: 10.1002/jcp.26017
Snyder, N. A., Stefan, C. P., Soroudi, C. T., Kim, A., Evangelista, C., and Cunningham, K. W. (2017). H(+) and Pi byproducts of glycosylation affect Ca(2+) homeostasis and are retrieved from the golgi complex by homologs of TMEM165 and XPR1. G3 7, 3913–3924. doi: 10.1534/g3.117.300339
Spain, B. H., Koo, D., Ramakrishnan, M., Dzudzor, B., and Colicelli, J. (1995). Truncated forms of a novel yeast protein suppress the lethality of a G protein alpha subunit deficiency by interacting with the beta subunit. J. Biol. Chem. 270, 25435–25444. doi: 10.1074/jbc.270.43.25435
Spellman, P. T., Sherlock, G., Zhang, M. Q., Iyer, V. R., Anders, K., Eisen, M. B., et al. (1998). Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9, 3273–3297. doi: 10.1091/mbc.9.12.3273
Springer, M., Wykoff, D. D., Miller, N., and O’Shea, E. K. (2003). Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol. 1:E28. doi: 10.1371/journal.pbio.0000028
Steger, D. J., Haswell, E. S., Miller, A. L., Wente, S. R., and O’Shea, E. K. (2003). Regulation of chromatin remodeling by inositol polyphosphates. Science 299, 114–116. doi: 10.1126/science.1078062
Steidle, E. A., Chong, L. S., Wu, M., Crooke, E., Fiedler, D., Resnick, A. C., et al. (2016). A novel inositol pyrophosphate phosphatase in Saccharomyces cerevisiae: Siw14 PROTEIN SELECTIVELY CLEAVES THE β-PHOSPHATE FROM 5-DIPHOSPHOINOSITOL PENTAKISPHOSPHATE (5PP-IP5). J. Biol. Chem. 291, 6772–6783. doi: 10.1074/jbc.M116.714907
Swinnen, E., Rosseels, J., and Winderickx, J. (2005). The minimum domain of Pho81 is not sufficient to control the Pho85-Rim15 effector branch involved in phosphate starvation-induced stress responses. Curr. Genet. 48, 18–33. doi: 10.1007/s00294-005-0583-3
Szijgyarto, Z., Garedew, A., Azevedo, C., and Saiardi, A. (2011). Influence of inositol pyrophosphates on cellular energy dynamics. Science 334, 802–805. doi: 10.1126/science.1211908
Tamai, Y., Toh-e, A., and Oshima, Y. (1985). Regulation of inorganic phosphate transport systems in Saccharomyces cerevisiae. J. Bacteriol. 164, 964–968.
Tammenkoski, M., Koivula, K., Cusanelli, E., Zollo, M., Steegborn, C., Baykov, A. A., et al. (2008). Human metastasis regulator protein H-prune is a short-chain exopolyphosphatase. Biochemistry 47, 9707–9713. doi: 10.1021/bi8010847
Tammenkoski, M., Moiseev, V. M., Lahti, M., Ugochukwu, E., Brondijk, T. H. C., White, S. A., et al. (2007). Kinetic and mutational analyses of the major cytosolic exopolyphosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 282, 9302–9311. doi: 10.1074/jbc.M609423200
Teunissen, J. H. M., Crooijmans, M. E., Teunisse, P. P. P., and van Heusden, G. P. H. (2017). Lack of 14-3-3 proteins in Saccharomyces cerevisiae results in cell-to-cell heterogeneity in the expression of Pho4-regulated genes SPL2 and PHO84. BMC Genomics 18:701. doi: 10.1186/s12864-017-4105-8
Thomas, M. R., and O’Shea, E. K. (2005). An intracellular phosphate buffer filters transient fluctuations in extracellular phosphate levels. Proc. Natl. Acad. Sci. U.S.A. 102, 9565–9570. doi: 10.1073/pnas.0501122102
Toh-e, A., and Kakimoto, S. (1975). Genes coding for the structure of the acid phosphatases in Saccharomyces cerevisiae. Mol. Gen. Genet. 143, 65–70. doi: 10.1007/bf00269421
Toh-E, A., and Nishizawa, M. (2001). Structure and function of cyclin-dependent Pho85 kinase of Saccharomyces cerevisiae. J. Gen. Appl. Microbiol. 47, 107–117. doi: 10.2323/jgam.47.107
Toh-E, A., Tanaka, K., Uesono, Y., and Wickner, R. B. (1988). PHO85, a negative regulator of the PHO system, is a homolog of the protein kinase gene, CDC28, of Saccharomyces cerevisiae. Mol. Gen. Genet. 214, 162–164. doi: 10.1007/BF00340196
Uhler, J. P., Hertel, C., and Svejstrup, J. Q. (2007). A role for noncoding transcription in activation of the yeast PHO5 gene. Proc. Natl. Acad. Sci. U.S.A. 104, 8011–8016. doi: 10.1073/pnas.0702431104
Urech, K., Dürr, M., Boller, T., Wiemken, A., and Schwencke, J. (1978). Localization of polyphosphate in vacuoles of Saccharomyces Cerevisiae. Arch. Microbiol. 116, 275–278. doi: 10.1007/bf00417851
Vardi, N., Levy, S., Assaf, M., Carmi, M., and Barkai, N. (2013). Budding yeast escape commitment to the phosphate starvation program using gene expression noise. Curr. Biol. 23, 2051–2057. doi: 10.1016/j.cub.2013.08.043
Vardi, N., Levy, S., Gurvich, Y., Polacheck, T., Carmi, M., Jaitin, D., et al. (2014). Sequential feedback induction stabilizes the phosphate starvation response in budding yeast. Cell Rep. 9, 1122–1134. doi: 10.1016/j.celrep.2014.10.002
Wang, C., Yue, W., Ying, Y., Wang, S., Secco, D., Liu, Y., et al. (2015). Rice SPX-major facility superfamily3, a vacuolar phosphate efflux transporter, is involved in maintaining phosphate homeostasis in rice. Plant Physiol. 169, 2822–2831. doi: 10.1104/pp.15.01005
Wang, H., Falck, J. R., Hall, T. M. T., and Shears, S. B. (2012). Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat. Chem. Biol. 8, 111–116. doi: 10.1038/nchembio.733
Wang, H., Gu, C., Rolfes, R. J., Jessen, H. J., and Shears, S. B. (2018). Structural and biochemical characterization of Siw14: a protein-tyrosine phosphatase fold that metabolizes inositol pyrophosphates. J. Biol. Chem. 293, 6905–6914. doi: 10.1074/jbc.RA117.001670
Wanke, V., Pedruzzi, I., Cameroni, E., Dubouloz, F., and De Virgilio, C. (2005). Regulation of G0 entry by the Pho80-Pho85 cyclin-CDK complex. EMBO J. 24, 4271–4278. doi: 10.1038/sj.emboj.7600889
Waters, N. C., Knight, J. P., Creasy, C. L., and Bergman, L. W. (2004). The yeast Pho80-Pho85 cyclin-CDK complex has multiple substrates. Curr. Genet. 46, 1–9.
Weaver, J. D., Wang, H., and Shears, S. B. (2013). The kinetic properties of a human PPIP5K reveal that its kinase activities are protected against the consequences of a deteriorating cellular bioenergetic environment. Biosci. Rep. 33:e00022. doi: 10.1042/BSR20120115
Wiame, J. M. (1947). Étude d’une substance polyphosphorée, basophile et métachromatique chez les levures. Biochim. Biophys. Acta 1, 234–255. doi: 10.1016/0006-3002(47)90137-6
Wild, R., Gerasimaite, R., Jung, J.-Y., Truffault, V., Pavlovic, I., Schmidt, A., et al. (2016). Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352, 986–990. doi: 10.1126/science.aad9858
Wilson, M. S., Jessen, H. J., and Saiardi, A. (2019). The inositol hexakisphosphate kinases IP6K1 and -2 regulate human cellular phosphate homeostasis, including XPR1-mediated phosphate export. J. Biol. Chem. 294, 11597–11608. doi: 10.1074/jbc.RA119.007848
Wilson, M. S. C., Livermore, T. M., and Saiardi, A. (2013). Inositol pyrophosphates: between signalling and metabolism. Biochem. J. 452, 369–379. doi: 10.1042/BJ20130118
Wurst, H., and Kornberg, A. (1994). A soluble exopolyphosphatase of Saccharomyces cerevisiae. Purif. charact. 269, 10996–11001.
Wurst, H., Shiba, T., and Kornberg, A. (1995). The gene for a major exopolyphosphatase of Saccharomyces cerevisiae. J. Bacteriol. 177, 898–906. doi: 10.1128/jb.177.4.898-906.1995
Wykoff, D. D., and O’Shea, E. K. (2001). Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159, 1491–1499.
Wykoff, D. D., Rizvi, A. H., Raser, J. M., Margolin, B., and O’Shea, E. K. (2007). Positive feedback regulates switching of phosphate transporters in S. cerevisiae. Mol. Cell 27, 1005–1013. doi: 10.1016/j.molcel.2007.07.022
Xu, Y.-F., Létisse, F., Absalan, F., Lu, W., Kuznetsova, E., Brown, G., et al. (2013). Nucleotide degradation and ribose salvage in yeast. Mol. Syst. Biol. 9:665. doi: 10.1038/msb.2013.21
Xu, Z., Wei, W., Gagneur, J., Perocchi, F., Clauder-Münster, S., Camblong, J., et al. (2009). Bidirectional promoters generate pervasive transcription in yeast. Nature 457, 1033–1037. doi: 10.1038/nature07728
Yompakdee, C., Bun-ya, M., Shikata, K., Ogawa, N., Harashima, S., and Oshima, Y. (1996). A putative new membrane protein, Pho86p, in the inorganic phosphate uptake system of Saccharomyces cerevisiae. Gene 171, 41–47. doi: 10.1016/0378-1119(96)00079-0
Yoshida, K., Kuromitsu, Z., Ogawa, N., and Oshima, Y. (1989a). Mode of expression of the positive regulatory genes PHO2 and PHO4 of the phosphatase regulon in Saccharomyces cerevisiae. Mol. Gen. Genet. 217, 31–39. doi: 10.1007/bf00330939
Yoshida, K., Ogawa, N., and Oshima, Y. (1989b). Function of the PHO regulatory genes for repressible acid phosphatase synthesis in Saccharomyces cerevisiae. Mol. Gen. Genet. 217, 40–46. doi: 10.1007/bf00330940
Yousaf, R., Gu, C., Ahmed, Z. M., Khan, S. N., Friedman, T. B., Riazuddin, S., et al. (2018). Mutations in diphosphoinositol-pentakisphosphate kinase PPIP5K2 are associated with hearing loss in human and mouse. PLoS Genet. 14:e1007297. doi: 10.1371/journal.pgen.1007297
Zhu, J., Lau, K., Puschmann, R., Harmel, R. K., Zhang, Y., Pries, V., et al. (2019). Two bifunctional inositol pyrophosphate kinases/phosphatases control plant phosphate homeostasis. eLife 8:e43582. doi: 10.7554/eLife.43582
Keywords: nutrient signaling, phosphate, acidocalcisome, SPX, polyphosphate, inositol pyrophosphate, Tor, PKA
Citation: Austin S and Mayer A (2020) Phosphate Homeostasis − A Vital Metabolic Equilibrium Maintained Through the INPHORS Signaling Pathway. Front. Microbiol. 11:1367. doi: 10.3389/fmicb.2020.01367
Received: 23 March 2020; Accepted: 27 May 2020;
Published: 14 July 2020.
Edited by:
Jose Roman Perez-Castineira, Universidad de Sevilla, SpainReviewed by:
Silvia N. J. Moreno, University of Georgia, United StatesDennis D. Wykoff, Villanova University, United States
Copyright © 2020 Austin and Mayer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andreas Mayer, YW5kcmVhcy5tYXllckB1bmlsLmNo
 Sisley Austin
Sisley Austin Andreas Mayer
Andreas Mayer