- 1UMR Transfrontalière BioEcoAgro N° 1158, Université de Lille, INRAE, Université de Liège, UPJV, YNCREA, Université d’Artois, Universite du Littoral Côte d’Opale, ICV – Institut Charles Viollette, Lille, France
- 2Laboratoire de Microbiologie Appliquée, Faculté des Sciences de la Nature et de la Vie, Université de Bejaia, Béjaïa, Algeria
- 3ANSES, Laboratoire de Ploufragan-Plouzané-Niort, Unité Mycoplasmologie Bactériologie Antibiorésistance, Ploufragan, France
- 4Université Bretagne Loire, Rennes, France
- 5Université de Lille, CNRS, Centrale Lille, ISEN, Univ. Valenciennes, UMR 8520 – IEMN, Lille, France
Lactobacillus paracasei CNCM I-5369 isolated from a traditional Algerian dairy product produces extracellular inhibitory substances, namely, bacteriocins, which are active against a panel of pathogenic Escherichia coli strains. This activity was observed only at a narrow pH 4.5–5, and resulted to be heat stable and sensitive to the action of proteolytic enzymes, which indicate a proteinaceous nature. This new strain has a genome of 2,752,975 bp, with a 46.6% G + C ratio and contains at least 2664 coding sequences. The Bagel software analysis identified five open reading frames (ORFs) that are translated to new class II bacteriocin. Each ORF was cloned in frame with a His-tag tail and expressed in E. coli BL21 (DE3) (pLysS) strain. Of note, each fusion protein carrying any of these ORFs at the C- or N-terminal position resulted to be active against E. coli 184 strain used as target organism. This manuscript reports the first multi-bacteriocinogenic strain producing five new class II bacteriocins with activity against Gram-negative bacilli (GNB), namely, E. coli. Heterologous expression and activity of each new class II bacteriocin were demonstrated.
Introduction
Bacteriocins are a family of ribosomally synthesized antimicrobial peptides (AMP) of low molecular sizes and proteinaceous nature. They are produced by Gram-negative bacteria (GNB) and Gram-positive bacteria (GPB) (Drider and Rebuffat, 2011). Bacteriocins from GPB are abundantly and diversely produced by lactic acid bacteria (LAB). They can be of narrow spectra, acting therefore only on members of the same species, or of broad spectra targeting other species and genera (Cotter et al., 2013). Bacteriocin classification is evolving with accumulated new knowledge, and there is not a single classification scheme that is universally accepted. Alvarez-Sieiro et al. (2016) classified bacteriocins into three classes. Briefly, class I contains small post-translationally modified peptides, designated RiPPs and are less than 10 kDa; class II contains unmodified bacteriocins smaller than 10 kDa; and class III contains unmodified bacteriocins larger than 10 kDa and endowed with bacteriolytic or non-lytic mechanisms (Alvarez-Sieiro et al., 2016). Unlike non-ribosomally synthesized peptides (NRPS), LAB-bacteriocins are synthesized and secreted during bacterial metabolism (Heng et al., 2007), following a scheme of primary metabolites (Lu et al., 2005). Moreover, LAB-bacteriocins were assumed to have limited toxicity for eukaryotic cells (Belguesmia et al., 2011; Dicks et al., 2018). LAB-bacteriocins have been used in the food sector to preserve foodstuffs from different bacterial contaminations attributable to spoilage and pathogens microorganisms. The last two decades have seen a stream of reports associating LAB-bacteriocins with a variety of multifaceted activities (Drider et al., 2016; Chikindas et al., 2018; Todorov et al., 2019). Bacteriocins represent an important alternative to traditional antibiotics in the face of antimicrobial resistance. Of note, LAB-bacteriocins can be active at nanomolar concentrations, unlike traditional antibiotics. They have two recognized modes of action (MoA). The first is associated with their capability to interact with the charged bacterial cell membrane, leading to ATP depletion, ionic imbalance, and membrane potential disruption (Naghmouchi et al., 2007). This MoA, known as a pore-forming mechanism, is mediated by positive charges of LAB-bacteriocins and the negatively charged bacterial membrane through electrostatic interactions (Egan et al., 2016; Vieco-Saiz et al., 2019). The second MoA is based on the use of a specific receptor located on the cell membrane. Nisin, which is a class I bacteriocin or lantibiotic, can target the lipid-anchored precursor of peptidoglycan (lipid II), which is a key component of peptidoglycan biosynthesis. This mechanism causes pore formation in the bacterial cell membrane, and consequently cell death (Bierbaum and Sahl, 2009; Egan et al., 2016; Vieco-Saiz et al., 2019). LAB-bacteriocin–lipid II interactions result in a loosened peptidoglycan meshwork, and ultimately in the leakage of vital cytosolic components by the formation of pores. Importantly, lipid II is also a target of the glycopeptide antibiotic vancomycin. Lipid II is not the unique receptor of LAB-bacteriocins; other ones have been described in the literature (Cotter et al., 2013; Cotter, 2014). Gram-negative pathogens have proven to be recalcitrant to LAB-bacteriocins by limiting access to the cell membrane. The discovery of LAB-bacteriocins with antimicrobial activity against GNB is of paramount interest for academic and applied research. Class II bacteriocins reported here constitute the first models to be simultaneously produced by a single LAB strain, and their activity against Escherichia coli carrying mcr-1 gene opens a new window for further study.
In this paper, we report a multibacteriocinogenic strain, Lactobacillus paracasei CNCM I-5369, capable of producing five new class II bacteriocins with anti-E. coli activity. Each of these new class II bacteriocins was successfully cloned and heterologously expressed.
Materials and Methods
Bacterial Strains and Culture Conditions
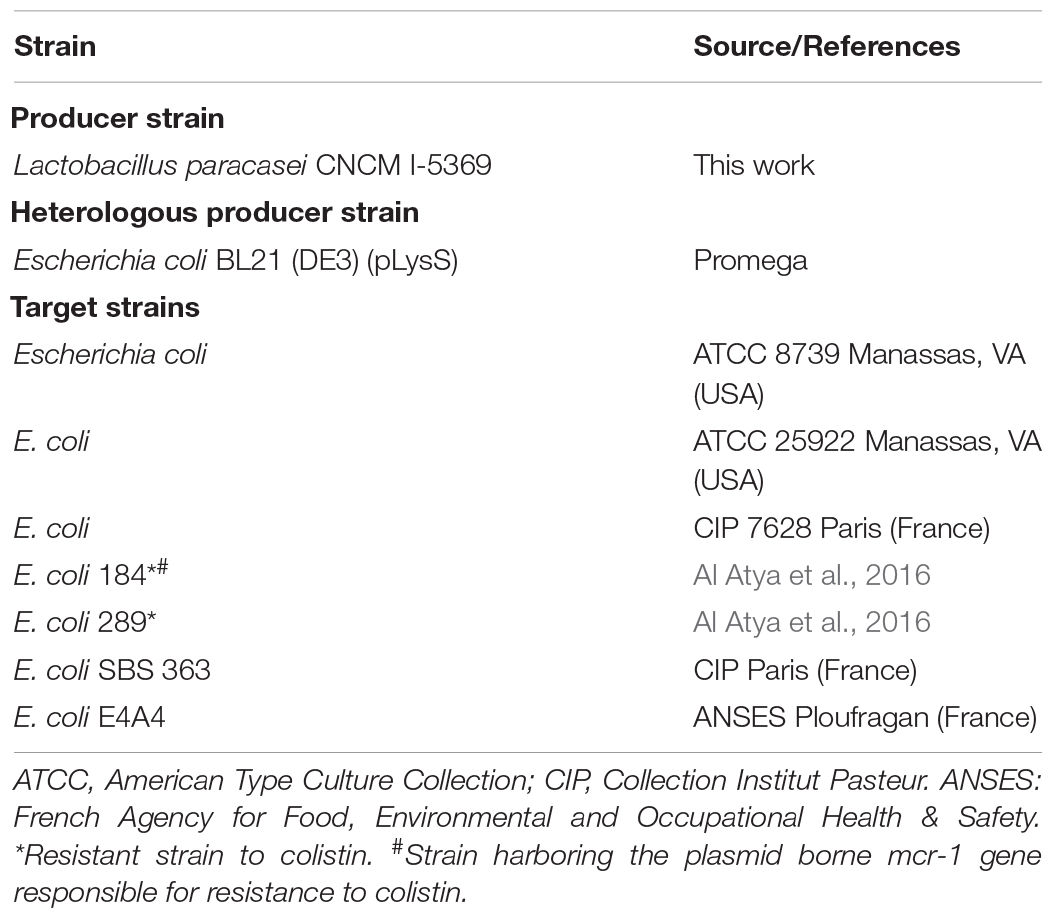
Bacteria used in this work are listed in Table 1. All target E. coli strains were propagated, without agitation, in Brain Heart Infusion (BHI) media at 37°C for 12–18 h before use. Lb. paracasei CNCM I-5369 was grown at 37°C for 18–24 h, without agitation, in MRS medium (de Man, Rogosa and Sharpe) (De Man et al., 1960).
Bacteria Identification
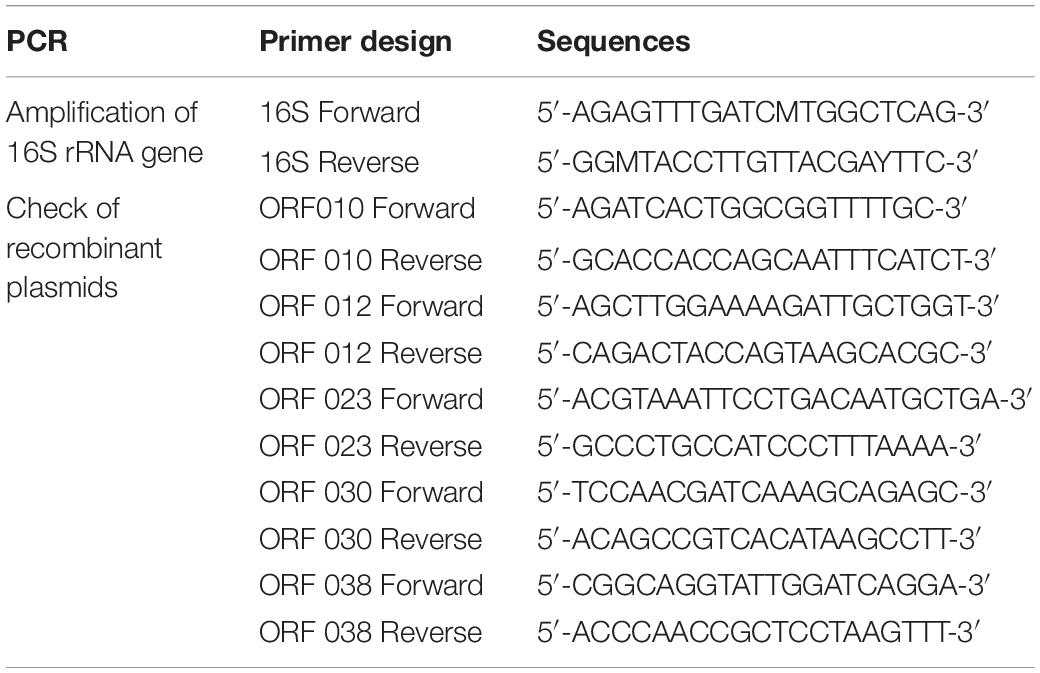
The bacteriocinogenic strain used in this work was first identified as Lb. paracasei with classical methods of bacteriology based on different criteria such as the Gram staining, catalase activity, and sugar assimilation profile. This identification was confirmed with the MALDI-TOF Mass Spectrometry and 16S rRNA gene sequencing. The mass spectrometry (MS) profile was done as recently reported by Zidour et al. (2017). Genes encoding 16S rRNA were PCR-amplified from the total DNA extracted with the Wizard® Genomic DNA Purification Kit (Promega, Charbonnières-les-Bains, France), and primers formerly designed by Drago et al. (2011) (Table 2).
The 16S rRNA codifying gene was amplified with the following PCR program: 94°C/5 min, 29 cycles at 94°C/1 min, 55°C/1 min, and 72°C/1 min and finally 72°C/5 min. PCR products were purified with a PCR purification kit (Qiagen, Courtaboeuf, France), and sequenced by Eurofins Genomics (Munich, Germany). The obtained 16S rDNA nucleotide sequence was blasted with the BLASTn online software1.
Production and Purification of Antibacterial Molecule(s) Produced by Lb. paracasei CNCM I-5369 Strain
The kinetics of production of inhibitory compounds was followed for 72 h. Thus, 100 ml of a sterile MRS medium was inoculated with 1 ml of an overnight culture of Lb. paracasei CNCM I-5369 grown at 37°C in MRS medium. The OD600nm was measured after 2, 4, 6, 8, 10, 26, 48, and 72 h of growth at 37°C. Samples of 1 ml each were taken and centrifuged (8000 g, 4°C, 10 min), and the resulting cell-free supernatant (CFS) was assessed for its antibacterial activity as previously reported (Batdorj et al., 2006). Briefly, each sample of CFS was serially diluted in the MRS medium using the following dilution ratios: 1/2, 1/4, 1/8, 1/16, 1/32, and 1/64. Then 10 μl of each diluted CFS was deposited on 1% (w/v) BHI medium agar inoculated with the indicator strain, E. coli ATCC 8739, or E. coli 184 carrying mcr-1 gene. The plates were incubated at 4°C for 1 h and then at 37°C for 18 h. The antibacterial activity was expressed in arbitrary units/ml (AU/ml), which correspond to the reciprocal of the highest dilution (2n), resulting in the inhibition of the indicator strain. The total activity expressed in AU/ml corresponds to 2n × 100 μl/volume deposited (μl) (Batdorj et al., 2006). The concentration of proteins present in each CFS was determined with the BCA (bicinchoninic acid) Assay protein kit (Sigma-Aldrich, St Louis, MO, USA), as recommended by the supplier.
To purify the active compound(s), a culture of Lb. paracasei CNCM I-5369, grown in MRS medium for 24 h to 30 h at 37°C, was centrifuged (8000 g, 4°C, 10 min), and 40 ml of the CFS was loaded onto a reversed-phase C18 (Agilent, Santa Clara, CA, USA) cartridge. A washing step was done with 40 ml of 10% (v/v) acetonitrile, followed-up by an elution with 40 ml of 20% (v/v) acetonitrile solution. The active fraction of ca. 40 ml was dried using SpeedVac and resuspended in 4 ml of ultrapure water. The active fraction, designed E20, was stored at 4°C until use.
Spectrum of Activity of E20 Fraction
The E20 fraction was tested against E. coli strains listed in Table 1. The antimicrobial activity was done using the agar diffusion test. Plates were flooded with a target bacterial suspension of ∼107 CFU/ml and dried at room temperature (20°C) for 15 min. Then, wells of ∼6 mm diameter and 4 mm depth were made in 1% (w/v) BHI soft agar. Next, 50 μl of E20 fraction of a total protein concentration of 16 mg/ml (pH 4.5 or pH 7.0) was introduced in these wells. Notably, aliquots of 50 μl of MRS medium adjusted to the same pH were deposited and used as controls. Plates were pre-incubated for 2 h at 4°C to stop the growth of the target strain and allow inhibitory compounds present in the CFS to diffuse through agar. Afterward, plates were incubated for 24 h at 37°C, and the antimicrobial activity was evaluated by measuring the diameter of the inhibition zones formed around wells.
Nature of Antimicrobial Activity of E20 Fraction
The E20 fraction was treated with different proteases, such as proteinase K, trypsin, papain, pepsin, and α-chemotrypsin, and also by other enzymes such as α-amylase and lipase (Sigma Aldrich) at a final concentration of 2 mg/ml. These treatments aimed at identifying the nature of the substance(s) responsible for activity. Briefly, enzymes were prepared in an adequate buffer at a concentration of 20 mg/ml. Then, 100 μl of each solution was added to 1 ml of E20 fraction and incubated for 2 h at 37°C. The residual antimicrobial activity was determined against E. coli ATCC 8739 used as a target strain (Batdorj et al., 2006).
The stability of this antimicrobial activity was tested at different temperatures and pH ranges. The E20 fraction was treated for 5 min at temperatures varying from 60 to 100°C, and then for 10–20 min at a temperature of 120°C.
For the pH stability, the E20 fraction was adjusted with 1 M of sterile NaOH or HCl to pH values ranging from 2 to 10. The E20 fraction was then left at room temperature for 2 h. Afterward, the residual activity was determined against E. coli ATCC 8739 as previously reported (Batdorj et al., 2006).
In silico Analysis of the Genome of Lb. paracasei CNCM I-5369 Strain
The genome of Lb. paracasei CNCM I-5369 was sequenced using the Illumina MiSeq and HiSeq 2500 technology platforms and 2 × 250 bp paired-end reads (University of Liège, Belgium). The functional annotation of predicted genes was done with the RAST online server2, which predicted open reading frames (ORFs), and appropriate annotation according to the free SEED database (Overbeek et al., 2014).
To identify putative bacteriocins, the genome of Lb. paracasei CNCM I-5369 was analyzed with the Bagel 3 online software3, and the amino acid sequence translation was done with the Jpred software4.
Heterologous Expression of Each ORF and Assessment of Its Antibacterial Activity
Each ORF coding for a putative bacteriocin was cloned under the control of the inducible T7 promoter, with the e-Zyvec technology5. Each ORF was cloned either upstream (N-terminal position) or downstream (C-terminal position) of the His-Tag (6 His) tail. This strategy enabled us to obtain 10 recombinant plasmids. Each recombinant plasmid was then introduced in E. coli BL21 (DE3)(plysS) competent cells. The transformed E. coli cells were regenerated in the SOC (Super Optimal broth with Catabolic repression) medium for 1 h at 37°C with shaking, at 160 rpm, and then selected on the Luria-Bertani (LB) agar medium supplemented with ampicillin (100 μg/ml) + chloramphenicol (30 μg/ml) (Sigma Aldrich). After overnight incubation at 37°C, the colonies were checked for plasmid carriage, targeting the appropriate ORF. This was done with PCR, using primers listed in Table 2, and the following PCR program: 94°C/3 min, 30 cycles at 94°C/1 min, 60°C/30 s, and 72°C/45 s, and finally 72°C/10 min.
For the heterologous expression assays, overnight cultures of E. coli strain BL21 (DE3) (pLysS), harboring recombinant plasmids were diluted to 1% (v/v) in LB medium + ampicillin (100 μg/ml) and chloramphenicol (30 μg/ml), and then grew aerobically at 37°C, until they had reached an OD600nm of 0.8. The gene expression was induced by adding the isopropyl-β-d-thiogalactopyranoside (IPTG) at 1 mM (Sigma-Aldrich). Bacteria were let to grow for five additional hours. Samples were harvested by centrifugation (8000 g, 10 min, 4°C), and the pellets were washed in a phosphate buffer solution (PBS) (pH 7.4), and resuspended in 10 mM imidazole (Sigma-Aldrich) and PBS buffer (pH 7.9). Then, they were sonicated five times, with a cycle of 2 min each, and cells were then lysed. The separation of the cytoplasmic soluble fraction (CSF) from the cytoplasmic insoluble fraction (CIF) and cell debris was done by centrifugation (14,000 g, 15 min, 4°C). The CSF was filtered (0.45-μm-pore-size filter) and loaded onto a 1-ml nickel His-Trap chelating column (Thermo Fisher Scientific, Waltham, MA, USA). The column was washed with 30 mM imidazole and PBS (pH 7.9). Peptides encoded by cloned ORFs with His-Tag were eluted with 2 ml of 250 mM imidazole and PBS (pH 7.9). After a desalting step with PD miditrap columns (GE Healthcare Life Science, Pollard, United Kingdom). The obtained solution was adjusted to pH 4.5 with the acetic acid, and its activity was evaluated against E. coli ATCC 8739 and E. coli 184 colistin-resistant strain.
Results
The bacteriocin-producing strain described here was initially identified as a Lactobacillus species. The MALDI-TOF-MS and 16 rRNA sequencing enabled its identification as Lb. paracasei with a high degree of confidence. According to the NCBI database, the strains related were Lb. paracasei M0116 with 99.79% of identity (accession number EU780145.1), Lb. paracasei SK04B2 (accession number KJ764645.1), and Lb. paracasei Y132 (accession number MK774551.1) with 99.72% identity.
Kinetics of Production of Inhibitory Compound(s)
The antimicrobial activity attributable to inhibitory compounds present in the CFS started to be detected after 10 h of culture on MRS medium at 37°C. This activity was estimated to 100 AU/ml and increased as seen in Figure 1. The highest activity was obtained at the end of the lag phase, with a total activity of 400 AU/ml (Figure 1). Then, it remained stable until the end of the experiment.
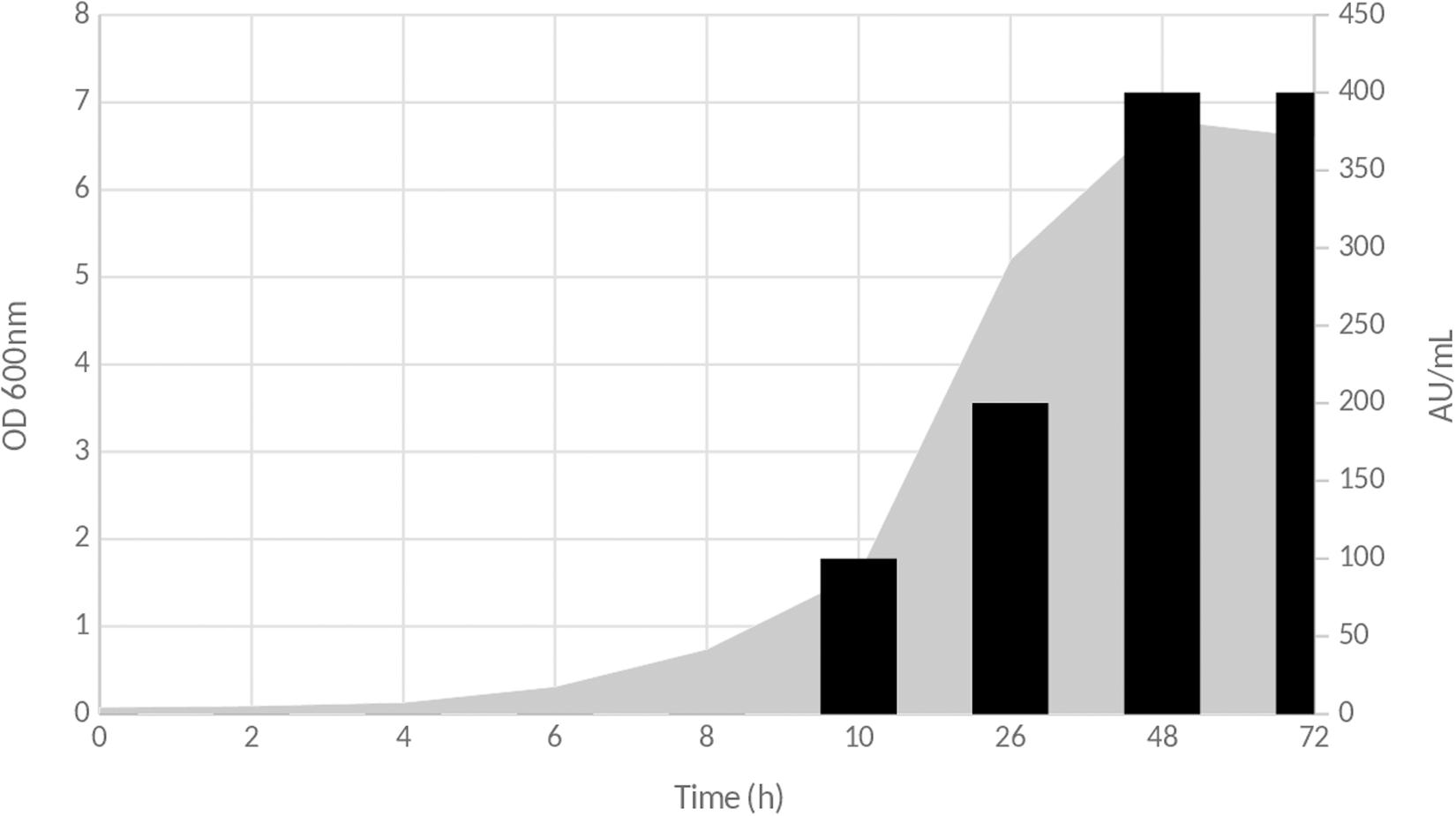
Figure 1. Time course production of inhibitory compound(s) during the growth of Lb. paracasei CNCM I-5369 on MRS medium. Gray zone corresponds to OD600nm evolution, and black bars represent antimicrobial activity in arbitrary units per milliliter (AU/ml).
The E20 Fraction Contains Putative Bacteriocin(s) With Activity Against E. coli
After each step of the purification procedure, several parameters including the antibacterial activity (AU/ml), total activity (UA), and specific activity (AU/mg) were determined. As reported in Table 3, the E20 fraction has a total activity of 8,000 AU.
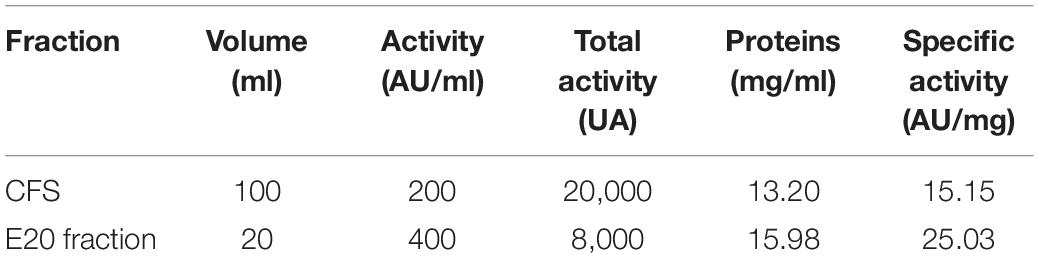
Table 3. Antibacterial activities of the cell-free supernatant (CFS) and the semi-purified E20 fraction.
The E20 fraction was adjusted to pH 4.5 and tested against different E. coli strains. Subsequently, all these target strains appeared to be, in a strain dependent-manner, sensitive to the action of E20 fraction. Notably, control assays were done at a same pH to discard any false-positive reaction attributable to the acidity of the medium (data not shown). Therefore, strong inhibition activities were observed on E. coli target strains based on the radii of inhibition zones formed around wells (Table 4).
Notably, the activity of E20 fraction was lost following its treatment with proteolytic enzymes such as papain or proteinase K, but not with α-chymotrypsin or pepsin. These results indicate a proteinaceous nature of these antimicrobial substances. Notably, activity of E20 fraction remained stable for 20 min at 100°C but not for a longer heating period of incubation. Indeed, this antimicrobial activity decreased significantly after 30 min, at 100°C, and was completely abolished after 60 min, at the same temperature. Taken together, these results indicate that the E20 fraction might contain at least one active bacteriocin.
Additionally, the activity of E20 fraction was highly stable during storage at 4 to 8°C, with a very limited loss of its activity after 12 weeks. Importantly, the activity of E20 fraction was tightly pH-dependent. Indeed, at pH values ranging from 2 to 5, the E20 fraction showed a strong antimicrobial activity and then started to decrease drastically until disappearing at a pH of 6 or higher.
Identification of Bacteriocin-Coding ORFs
Software such as Bagel or AntiSmash are routinely used to identify genes coding for bacteriocins (van Heel et al., 2013; Weber et al., 2015; Collins et al., 2018). They have algorithms with a capability to recognize specific amino acids on the bacteriocin sequence, and the Bagel software is currently the most suitable one for this application (Oliveira et al., 2017; Collins et al., 2018). The genome of Lb. paracasei CNCM I-5369 contains 2,752,975 bp, with a 46.6% G + C ratio, and 2,664 coding sequences. The in silico analysis of this genome with the Bagel 3 tool6 enabled us to identify five ORFs (Figure 2) and reliably predict the likelihood that these sequences are translated to produce new class II bacteriocins with molecular sizes varying from 3.199 to 12.252 Da (Table 5). The first bacteriocin (ORF010) has a predicted molecular size of 3.199 Da and a pI of 5.17. Those of the ORF012 and ORF023 were 6.300 and 6.582 Da, with pI values of 4.86 and 8.25, respectively. The fourth and fifth ones (ORF030 and ORF038) have the largest molecular sizes with 10.395 and 12.252 Da, and pI of 8.62 and 6, respectively.
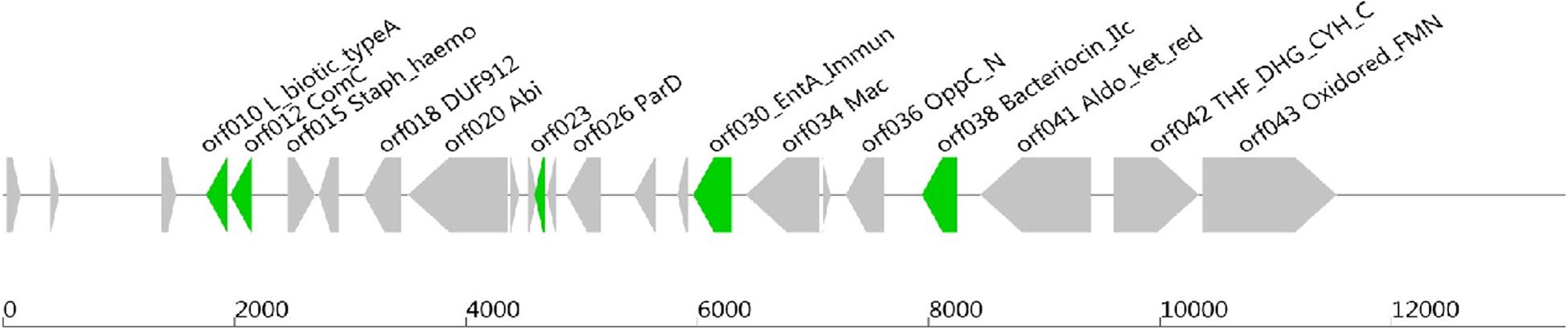
Figure 2. Open reading frames coding (ORFs), for presumed new class II bacteriocins, located on the chromosome of Lb. paracasei CNCM I-5369.
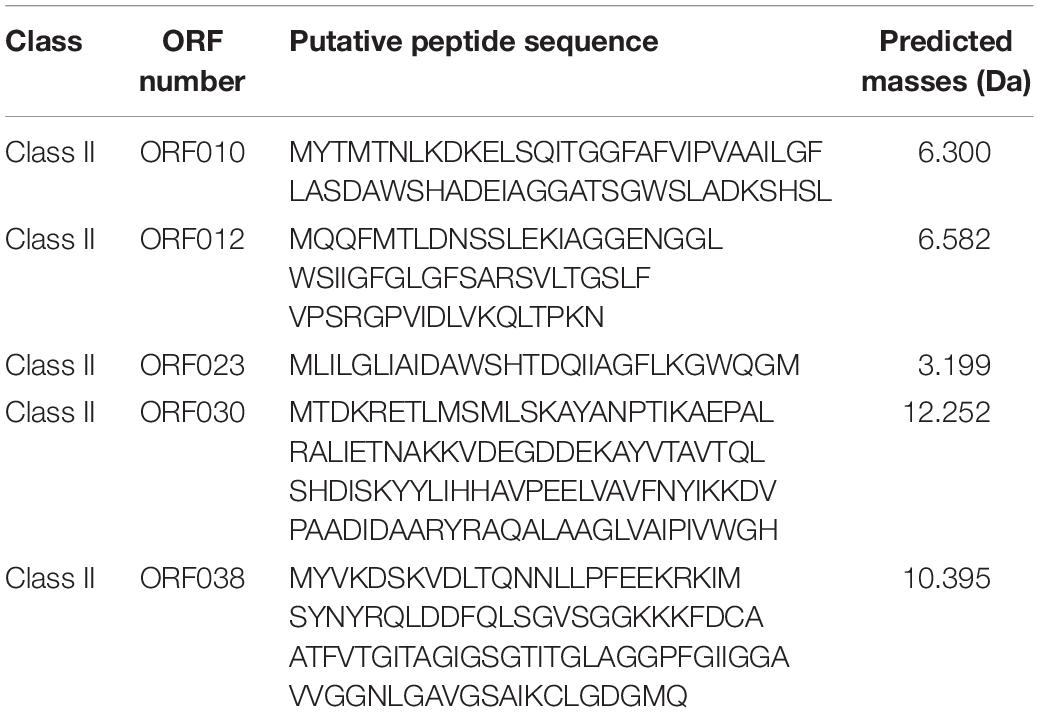
Table 5. Amino acid sequence of each putative new class II bacteriocins produced by Lb. paracasei CNCM I-5369.
Amino acid sequences identified with Bagel 3 and Blastp online software1 revealed some similarities between the bacteriocin predicted from ORF010 and enterocin X or Lactococcin-like family Lantibiotic (Hu et al., 2010). Of note, these new bacteriocins have only 24 common amino acids with the β-chain of Enterocin-X (Figure 3). The ORF012 may be translated into a bacteriocin similar to the ComC/BlpC leader-containing pheromone/bacteriocin family (Wang et al., 2018), while ORF023 matches with bacteriocins of the lactobin A/cerein 7B family (Escamilla-Martínez et al., 2017). The bacteriocin predicted from the ORF030 is 33% similar with carnobacteriocin B2 and a class IIa immunity protein (Quadri et al., 1995). Finally, the sequence predicted from ORF038 is 36% similar to that of thermophilin A, a bacteriocin with characteristics of class IIc (Marciset et al., 1997).

Figure 3. Sequence alignments of putative new bacteriocins translated from ORFs located and identified using the Bagel 3 software and the NCBI alignment tool (black zone = consensus domain of Ent_A Immun Superfamily; gray zone = conserved domain of class IIc bacteriocin).
Heterologous Expression and Activity of Each New Bacteriocin
Data depicted in Figure 4 and Table 6 report the activity of each new bacteriocin following its heterologous expression in E. coli BL21 (DE3) (pLysS). Remarkably, this activity was found for each ORF cloned in frame with a His-tag at the N- or C-terminal position. These activities obtained in a heterologous system were slightly different from those attributed to the native system, measured in the CFS or in the E20 fraction (Figure 4 and Table 3). Antimicrobial activities comprised between 200 and 800 AU/ml, and the upmost one was registered for ORF038, when its corresponding gene was cloned at the C-terminal position (His-tag at the N-terminal position). This activity was two to four times higher than those obtained from other constructions (Table 6).
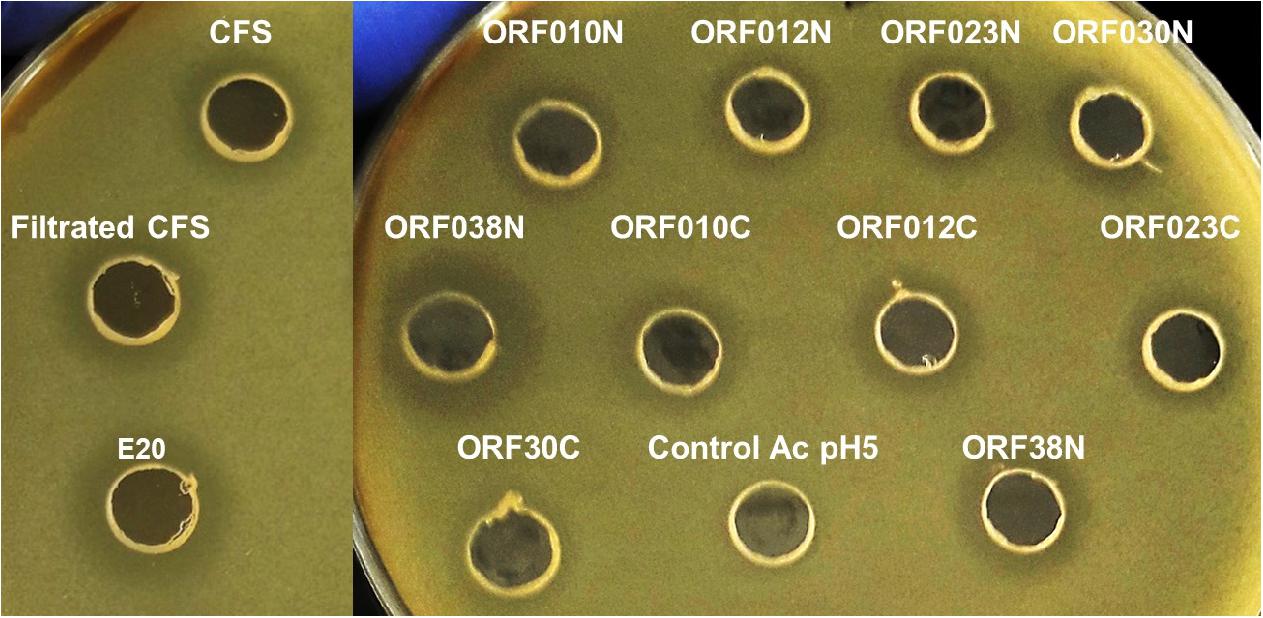
Figure 4. Agar diffusion test on E. coli 184 (mcr-1+) strain of (Control Ac pH5) PBS adjusted to pH 5 with acetic acid (negative-control), CFS (cell free culture supernatant); filtrated CFS (0.2 μM); E20 semi-purified fraction; N-terminal Histidine-tagged recombinant peptides (ORF010N, ORF012N, ORF023N, ORF030N, and ORF038N) and C-terminal Histidine-tagged recombinant peptides (ORF010C, ORF012C, ORF023C, ORF030C, and ORF038C).
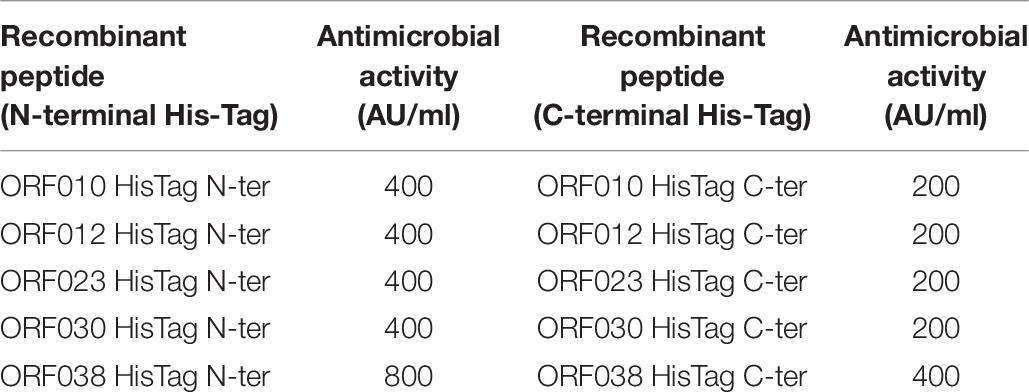
Table 6. Antimicrobial activity of new class II bacteriocins as fusion-proteins against E. coli 184.
Discussion
Bacteriocins are presently of common use in the food industry, in spite of their great potential for animal feeds, organic fertilizers, environmental protection, microbiota regulators, and personal care products. Nisin is the only LAB-bacteriocin to be approved by the FDA as a food preservative (E234) under the Annex of the EC regulation 1333/2008. Nevertheless, during the last decade, ca. 37% of published research on bacteriocins has focused on their potential as therapeutics (López-Cuellar et al., 2016; Mathur et al., 2017; Meade et al., 2020). LAB-bacteriocins differ in many aspects from traditional antibiotics (Cleveland et al., 2001; Perez et al., 2014), and their gene-encoded nature renders them genetically amendable and their activity improvable (Field et al., 2015).
By 2050, the expected number of deaths from antimicrobial resistance will increase to 10 million deaths per year globally (O’Neill, 2016), indicating that action to avert this crisis is needed. Major objectives should include new antibiotic discovery to rekindle the pipeline and the pursuit of antibiotic alternatives such as antimicrobial peptides. LAB-bacteriocins, which warrant serious consideration, are most often active against genetically related strains, and seldom against phylogenetically distant target strains such as GNB. LAB-bacteriocins having activity against GNB activity are of academic interest and will open new opportunities for medical applications, mainly to treat GNB-associated infections. The first anti-GNB LAB-bacteriocin was reported by Line et al. (2008) who reported the in vitro effectiveness of Enterocin E-760 against Campylobacter jejuni, a leading food-borne and human pathogen. Subsequently, Svetoch et al. (2011) reported the activity of a class IIa bacteriocin, named L-1077, against different Salmonella Typhimurium, Salmonella Enteritidis, and E. coli. Then, Messaoudi et al. (2012) reported the activity of the bacteriocin SMXD51 against C. jejuni. All these examples showed that activity of LAB-bacteriocins is not restricted to GPB, but can be active against GNB.
In this work, we report a Lb. paracasei CNCM I-5369 strain, capable of producing five new class II bacteriocins. The genome sequencing and analysis enabled us to locate at least five different ORFs coding for presumed new class II bacteriocins. Attempts to identify these new bacteriocins were first undertaken by the mass spectrometry approach on the CFS prepared from Lb. paracasei CNCM I-5369 culture and then on the semi-purified E20 fraction. Because of a very limited amount of pure peptides obtained at the end of the purification procedure, and because of their possible post-translational modifications, we decided to identify these new bacteriocins with a bioinformatics approach. Bacteriocins translated from ORF010 and ORF023 were less than 10 kDa and exhibited similarity with the β-chain of enterocin X, a class IIb bacteriocin (Hu et al., 2010). Interestingly, the GxxxG motif present in the amino acid sequence of bacteriocins encoded by ORF010 and ORF023 have been shown to play a role in the sec-independent export machinery of enterocin X (Nissen-Meyer et al., 2010; Escamilla-Martínez et al., 2017). Of note, ORF010 and ORF012 were separated only by 28 nucleotides, suggesting that their bacteriocins can be affiliated to class IIb. The BLASTP 2.9.0 software (Altschul et al., 1997) analysis done on sequences translated from ORF010 and ORF012 revealed large amino acid differences between them. Moreover, the predicted bacteriocin from ORF012 is highly similar to bacteriocins from the ComC/BlpC family (Hu et al., 2010). The bacteriocin predicted from ORF030 contains amino acid sequences partly similar to immunity proteins of Enterocin A, a class IIa bacteriocin organized in antiparallel form, with four alpha-helical globular bundles and a flexible fifth divergent C-terminal helical hairpin (Fimland et al., 2002). Finally, the bacteriocin predicted from ORF038 was associated with the thermophilin A bacteriocin and contains amino acids typical for class IIc bacteriocins (Marciset et al., 1997). Moreover, a clear ribosome binding site (RBS) was identified and located upstream of each located ORF (data not shown). Of note, the CFS and semi-purified E20 fraction prepared from Lb. paracasei CNCM I-5369 inhibited E. coli 184, a strain from swine origin carrying on a plasmid, the gene mcr-1 gene, which is responsible for resistance to colistin. The discovery of plasmid-borne mcr-1 gene was reported for the first time by Liu et al. (2016), and its rapid dissemination worldwide has consequently limited colistin as a drug of last resort for treatments of infections associated with multidrug-resistant GNB (Teo et al., 2016). Similarly, CFS and E20 fraction inhibited the growth of E. coli 289 strain from swine origin, which is as well resistant to colistin, but by a mechanism other than mcr-1 gene. Of note, the activity obtained against the short-chain LPS E. coli SBS363 and the sensitive E. coli E4A4 strain were, surprisingly, in the same range as those obtained for other E. coli strains tested here.
As effective antimicrobials, LAB-bacteriocins may replace or prolong the effectiveness of antibiotics such as colistin (Naghmouchi et al., 2013). Here, we report a multibacteriocinogenic strain, Lb. paracasei CNCM I-5369, capable of inhibiting different E. coli strains including those resistant to colistin, through production of new class II bacteriocins. Noteworthy, each of these peptides was heterologously produced and its activity was confirmed on E. coli 184 strain. The activity obtained with E20 fraction may result from a synergistic interaction between these five new bacteriocins, in acidic pH, as it has been previously suggested for other bacteriocins (Yang and Ray, 1994; Houlihan et al., 2004).
In summary, we report a multibacteriocinogenic strain, namely, Lb. paracasei CNCM I-5369, with activity against E. coli. Analysis of the whole-genome sequences enabled us to identify ORFs presumed to code for five new class II bacteriocins. Each ORF was heterologously expressed in E. coli BL21 (DE3) (pLysS), and its activity was determined against E. coli 184 (mcr-1+). Independently of its C or N orientation in the fusion protein, each ORF was shown to be active against the aforementioned target strain. Further experiments aimed at understanding the regulation and expression of each ORF in the natural host constitute our next focus.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given their approval to the final version of the manuscript.
Funding
This work was supported by ANR Sincolistin-15-CE21-0015, and partly by ALIBIOTECH (2016–2021) from la Région des Hauts-de-France. Part of this work is included in the EP19178926.2.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Dr. Bruce Seal from the Oregon State University Cascades (USA) and Prof. Brian Oaklen from Western University (USA) for their critical reading of the manuscript. The authors are indebted to Dr. Yohan Faivre (e-Zyvec SAS, Loos) for the conception of the expression vectors.
Footnotes
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://rast.nmpdr.org/
- ^ http://bagel4.molgenrug.nl
- ^ www.compbio.dundee.ac.uk/jpred/
- ^ https://www.e-zyvec.com/
- ^ http://bagel.molgenrug.nl//
References
Al Atya, A. K., Belguesmia, Y., Chataigne, G., Ravallec, R., Vachée, A., Szunerits, S., et al. (2016). Anti-MRSA activities of enterocins DD28 and DD93 and evidences on their role in the inhibition of biofilm formation. Front. Microbiol. 7:817. doi: 10.3389/fmicb.2016.00817
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Alvarez-Sieiro, P., Montalbán-López, M., Mu, D., and Kuipers, O. P. (2016). Bacteriocins of lactic acid bacteria: extending the family. Appl. Microbiol. Biotechnol. 100, 2939–2951. doi: 10.1007/s00253-016-7343-9
Batdorj, B., Dalgalarrondo, M., Choiset, Y., Pedroche, J., Métro, F., Prévost, H., et al. (2006). Purification and characterization of two bacteriocins produced by lactic acid bacteria isolated from Mongolian airag. J. Appl. Microbiol. 101, 837–848. doi: 10.1111/j.1365-2672.2006.02966.x
Belguesmia, Y., Madi, A., Sperandio, D., Merieau, A., Feuilloley, M., Prévost, H., et al. (2011). Growing insights into the safety of bacteriocins: the case of enterocin S37. Res. Microbiol. 162, 159–163. doi: 10.1016/j.resmic.2010.09.019
Bierbaum, G., and Sahl, H. G. (2009). Lantibiotics: mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotech. 10, 2–18. doi: 10.2174/138920109787048616
Chikindas, M. L., Weeks, R., Drider, D., Chistyakov, V. A., and Dicks, L. M. (2018). Functions and emerging applications of bacteriocins. Curr. Opin. Biotechnol. 49, 23–28. doi: 10.1016/j.copbio.2017.07.011
Cleveland, J., Montville, T. J., Nes, I. F., and Chikindas, M. L. (2001). Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71, 1–20. doi: 10.1016/s0168-1605(01)00560-8
Collins, F. W. J., Mesa-Pereira, B., O’Connor, P. M., Rea, M. C., Hill, C., and Ross, R. P. (2018). Reincarnation of Bacteriocins From the Lactobacillus Pangenomic Graveyard. Front. Microbiol 9:1298. doi: 10.3389/fmicb.2018.01298
Cotter, P. D. (2014). An ‘Upp’-turn in bacteriocin receptor identification. Mol. Microbiol. 92, 1159–1163. doi: 10.1111/mmi.12645
Cotter, P. D., Ross, R. P., and Hill, C. (2013). Bacteriocins - a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95–105. doi: 10.1038/nrmicro2937
De Man, J. C., Rogosa, M., and Sharpe, M. E. (1960). A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23, 130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x
Dicks, L. M. T., Dreyer, L., Smith, C., and van Staden, A. D. (2018). A review: the fate of bacteriocins in the human gastro-intestinal tract: do they cross the gut–blood barrier? Front. Microbiol. 9:2297. doi: 10.3389/fmicb.2018.02297
Drago, L., Mattina, R., Nicola, L., Rodighiero, V., and De Vecchi, E. (2011). Macrolide resistance and in vitro selection of resistance to antibiotics in lactobacillus isolates. J. Microbiol. 49, 651–656. doi: 10.1007/s12275-011-0470-1
Drider, D., Bendali, F., Naghmouchi, K., and Chikindas, M. L. (2016). Bacteriocins: not only antibacterial agents. Probiotics Antimicrob. Proteins. 8, 177–182. doi: 10.1007/s12602-016-9223-0
Drider, D., and Rebuffat, S. (2011). Prokaryotic Antimicrobial Peptides: From Genes to Applications. Berlin: Springer. Science Business Media, 451.
Egan, K., Field, D., Rea, M. C., Ross, R. P., Hill, C., and Cotter, P. D. (2016). Bacteriocins: novel solutions to age old spore-related problems? Front. Microbiol. 7:461. doi: 10.3389/fmicb.2016.00461
Escamilla-Martínez, E. E., Cisneros, Y. M. Á, Fernández, F. J., Quirasco-Baruch, M., and Ponce-Alquicira, E. (2017). Identification of structural and immunity genes of a class IIb bacteriocin encoded in the enterocin A Operon of Enterococcus faecium Strain MXVK29. J. Food Prot. 9, 1851–1856. doi: 10.4315/0362-028x.jfp-17-039
Field, D., Cotter, P. D., Ross, R. P., and Colin, H. (2015). Bioengineering of the model lantibiotic nisin. Bioengineered 6, 187–192. doi: 10.1080/21655979.2015.1049781
Fimland, G., Eijsink, V. G., and Nissen-Meyer, J. (2002). Comparative studies of immunity proteins of pediocin-like bacteriocins. Microbiology 148, 3661–3670. doi: 10.1099/00221287-148-11-3661
Heng, N. C. K., Wescombe, P. A., Burton, J. P., Jack, R. W., and Tagg, J. R. (2007). “The diversity of bacteriocins in gram-positive bacteria,” in Bacteriocins, eds M. A. Riley and M. A. Chavan (Berlin: Springer), 45–92. doi: 10.1007/978-3-540-36604-1_4
Houlihan, A. J., Mantovani, H. C., and Russell, J. B. (2004). Effect of pH on the activity of bovicin HC5, a bacteriocin from Streptococcus bovis HC5. FEMS Microbiol. Lett. 231, 27–32. doi: 10.1016/s0378-1097(03)00922-4
Hu, C. B., Malaphan, W., Zendo, T., Nakayama, J., and Sonomoto, K. (2010). Enterocin X, a novel two-peptide bacteriocin from Enterococcus faecium KU-B5, has an antibacterial spectrum entirely different from those of its component peptides. Appl. Env. Microbiol. 76, 4542–4545. doi: 10.1128/aem.02264-09
Line, J. E., Svetoch, E. A., Eruslanov, B. V., Perelygin, V. V., Mitsevich, E. V., Mitsevich, I. P., et al. (2008). Isolation and purification of enterocin E-760 with broad antimicrobial activity against Gram-positive and Gram-negative bacteria. Antimicrob. Agents Chemother. 52, 1094–1100. doi: 10.1128/aac.01569-06
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
López-Cuellar, M. D. R., Rodríguez-Hernández, A. I., and Chavarría-Hernández, N. (2016). LAB bacteriocin applications in the last decade. Biotech. Biotechnol. Equipment. 30, 1039–1050. doi: 10.1080/13102818.2016.1232605
Lu, W., Zhang, X., and Cong, W. (2005). Modelling the production of nisin by Lactococcus lactis in fed-batch culture. Appl. Microbiol. Biotechnol. 68, 322–326. doi: 10.1007/s00253-005-1892-7
Marciset, O., Jeronimus-Stratingh, M. C., Mollet, B., and Poolman, B. (1997). Thermophilin 13, a nontypical antilisterial poration complex bacteriocin, that functions without a receptor. J. Biol. Chem. 272, 14277–14284. doi: 10.1074/jbc.272.22.14277
Mathur, H., Field, D., Rea, M. C., Cotter, P. D., Hill, C., and Ross, R. P. (2017). Bacteriocin-antimicrobial synergy: a medical and food perspective. Front. Microbiol. 8:1205. doi: 10.3389/fmicb.2017.01205
Meade, E., Slattery, M. A., and Garvey, M. (2020). Bacteriocins, potent antimicrobial peptides and the fight against multi drug resistant species: resistance is futile? Antibiotics 9:E32. doi: 10.3390/antibiotics9010032
Messaoudi, S., Kergourlay, G., Dalgalarrondo, M., Choiset, Y., Ferchichi, M., Prévost, H., et al. (2012). Purification and characterization of a new bacteriocin active against Campylobacter produced by Lactobacillus salivarius SMXD51. Food Microbiol. 32, 129–134. doi: 10.1016/j.fm.2012.05.002
Naghmouchi, K., Baah, J., Hober, D., Jouy, E., Rubrecht, C., Sané, F., et al. (2013). Synergistic effect between colistin and bacteriocins in controlling Gram-negative pathogens and their potential to reduce antibiotic toxicity in mammalian epithelial cells. Antimicrob. Agents Chemother. 57, 2719–2725. doi: 10.1128/aac.02328-12
Naghmouchi, K., Drider, D., and Fliss, I. (2007). Action of divergicin M35, a class IIa bacteriocin, on Liposomes and Listeria. J. Appl. Microbiol. 102, 1508–1517. doi: 10.1111/j.1365-2672.2006.03206.x
Nissen-Meyer, J., Oppegård, C., Rogne, P., Haugen, H. S., and Kristiansen, P. E. (2010). Structure and mode-of-action of the two-peptide (class-IIb) bacteriocins. Probiotics Antimicrob. Proteins 2, 52–60. doi: 10.1007/s12602-009-9021-z
Oliveira, L. C., Silveira, A. M. M., Monteiro, A. S., Dos Santos, V. L., Nicoli, J. R., Azevedo, V. A. C., et al. (2017). In silico prediction, in vitro antibacterial spectrum, and physicochemical properties of a putative bacteriocin produced by lactobacillus rhamnosus Strain L156.4. Front. Microbiol. 8:876. doi: 10.3389/fmicb.2017.00876
O’Neill, J. (2016). Tackling Drug-Resistant Infections Globally: Final Report and Recommendationsthe Review on Antimicrobial Resistance. Available at; https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (accessed October 25, 2019).
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214.
Perez, R. H., Zendo, T., and Sonomoto, K. (2014). Novel bacteriocins from lactic acid bacteria (LAB): various structures and applications. Microb. Cell Fact. 29(13 Suppl. 1):S3.
Quadri, L. E., Sailer, M., Terebiznik, M. R., Roy, K. L., Vederas, J. C., and Stiles, M. E. (1995). Characterization of the protein conferring immunity to the antimicrobial peptide carnobacteriocin B2 and expression of carnobacteriocins B2 and BM1. J. Bact. 177, 1144–1151. doi: 10.1128/jb.177.5.1144-1151.1995
Svetoch, E. A., Eruslanov, B. V., Levchuk, V. P., Perelygin, V. V., Mitsevich, E. V., Mitsevich, I. P., et al. (2011). Isolation of Lactobacillus salivarius 1077 (NRRL B-50053) and characterization of its bacteriocin, including the antimicrobial activity spectrum. Appl. Env. Microbiol. 77, 2749–2754. doi: 10.1128/aem.02481-10
Teo, J. W., Chew, K. L., and Lin, R. T. (2016). Transmissible colistin resistance encoded by mcr-1 detected in clinical Enterobacteriaceae isolates in Singapore. Emerg. Microb. Infect. 5, 1–12.
Todorov, S. D., de Melo Franco, B. D. G., and Tagg, J. R. (2019). Bacteriocins of Gram-positive bacteria having activity spectra extending beyond closely-related species. Benef. Microbes 10, 315–328. doi: 10.3920/bm2018.0126
van Heel, A. J., de Jong, A., Montalbán-López, M., Kok, J., and Kuipers, O. P. (2013). BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal post-translationally modified peptides. Nucleic Acids Res. 41, W448–W453. doi: 10.1093/nar/gkt391
Vieco-Saiz, N., Belguesmia, Y., Raspoet, R., Auclair, E., Gancel, F., Kempf, I., et al. (2019). Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front. Microbiol. 10:57. doi: 10.3389/fmicb.2019.00057
Wang, C. Y., Patel, N., Wholey, W. Y., and Dawid, S. (2018). ABC transporter content diversity in Streptococcus pneumoniae impacts competence regulation and bacteriocin production. Proc. Nat. Acad. Sci. U.S.A. 115, E5776–E5785.
Weber, T., Blin, K., Duddela, S., Krug, D., Kim, H. U., Bruccoleri, R., et al. (2015). antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243.
Yang, R., and Ray, B. (1994). Factors influencing production of bacteriocins by lactic acid bacteria. Food Microbiol. 11, 281–291.
Zidour, M., Chevalier, M., Belguesmia, Y., Cudennec, B., Grard, T., Drider, D., et al. (2017). Isolation and characterization of bacteria colonizing acartia tonsa copepod eggs and displaying antagonist effects against vibrio anguillarum, vibrio alginolyticus and other pathogenic strains. Front. Microbiol. 6:1919. doi: 10.3389/fmicb.2017.01919
Keywords: multibacteriocinogenic strain, Lactobacillus paracasei, new class II bacteriocins, Escherichia coli, antimicrobials
Citation: Belguesmia Y, Bendjeddou K, Kempf I, Boukherroub R and Drider D (2020) Heterologous Biosynthesis of Five New Class II Bacteriocins From Lactobacillus paracasei CNCM I-5369 With Antagonistic Activity Against Pathogenic Escherichia coli Strains. Front. Microbiol. 11:1198. doi: 10.3389/fmicb.2020.01198
Received: 28 October 2019; Accepted: 12 May 2020;
Published: 19 June 2020.
Edited by:
Octavio Luiz Franco, Catholic University of Brasilia (UCB), BrazilCopyright © 2020 Belguesmia, Bendjeddou, Kempf, Boukherroub and Drider. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanath Belguesmia, eWFuYXRoLmJlbGd1ZXNtaWFAdW5pdi1saWxsZS5mcg==; Djamel Drider, ZGphbWVsLmRyaWRlckB1bml2LWxpbGxlLmZy; ZGphbWVsLmRyaWRlckB1bml2LWxpbGxlMS5mcg==
 Yanath Belguesmia1*
Yanath Belguesmia1* Kamel Bendjeddou
Kamel Bendjeddou Isabelle Kempf
Isabelle Kempf Djamel Drider
Djamel Drider