
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 09 June 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.01089
This article is part of the Research TopicRegulation of Antibiotic Production in ActinomycetesView all 15 articles
 Yuya Misaki1,2†
Yuya Misaki1,2† Shouji Yamamoto2†‡
Shouji Yamamoto2†‡ Toshihiro Suzuki2‡
Toshihiro Suzuki2‡ Miyuki Iwakuni2
Miyuki Iwakuni2 Hiroaki Sasaki2
Hiroaki Sasaki2 Yuzuru Takahashi2
Yuzuru Takahashi2 Kuninobu Inada3
Kuninobu Inada3 Haruyasu Kinashi2
Haruyasu Kinashi2 Kenji Arakawa1,2*
Kenji Arakawa1,2*Streptomyces rochei 7434AN4, a producer of lankacidin (LC) and lankamycin (LM), carries many regulatory genes including a biosynthesis gene for signaling molecules SRBs (srrX), an SRB receptor gene (srrA), and a SARP (Streptomyces antibiotic regulatory protein) family activator gene (srrY). Our previous study revealed that the main regulatory cascade goes from srrX through srrA to srrY, leading to LC production, whereas srrY further regulates a second SARP gene srrZ to synthesize LM. In this study we extensively investigated the function of srrB, a pseudo-receptor gene, by analyzing antibiotic production and transcription. Metabolite analysis showed that the srrB mutation increased both LC and LM production over four-folds. Transcription, gel shift, and DNase I footprinting experiments revealed that srrB and srrY are expressed under the SRB/SrrA regulatory system, and at the later stage, SrrB represses srrY expression by binding to the promoter region of srrY. These findings confirmed that SrrB acts as a negative regulator of the activator gene srrY to control LC and LM production at the later stage of fermentation in S. rochei.
Secondary metabolites production is strictly controlled by small diffusible signaling molecules that constitute signaling-molecule/receptor regulatory systems in Streptomyces species (Bibb, 2005; Takano, 2006; Horinouchi and Beppu, 2007; Martín and Liras, 2019). The most-studied signaling-molecule/receptor system is A-factor/ArpA in Streptomyces griseus for streptomycin and grixazone production (Ohnishi et al., 1999, 2005). In the absence of A-factor, ArpA protein specifically binds to the promoter region of the target activator gene adpA and represses its transcription. When A-factor reaches a critical concentration, A-factor/ArpA complex dissociates from the promoter region of adpA, leading to the onset of adpA transcription. Then, the gene product of adpA binds to its targets (AdpA-regulons) to activate streptomycin and grixazone production and morphological differentiation (Ohnishi et al., 1999, 2005). Gene sets involved in the signaling-molecule-dependent regulatory pathways for secondary metabolite production are listed in Table 1 (Arakawa, 2018; Xu and Yang, 2019); e.g., streptomycin and grixazone production in S. griseus (Hara and Beppu, 1982; Onaka et al., 1995; Ohnishi et al., 1999, 2005), lankamycin (LM) and lankacidin (LC) (Figure 1) in Streptomyces rochei (Arakawa et al., 2007, 2012; Yamamoto et al., 2008; Suzuki et al., 2010), tylosin in Streptomyces fradiae (Bate et al., 1999, 2002; Stratigopoulos and Cundliffe, 2002), coelimycin P-1 in Streptomyces coelicolor (Takano et al., 2001, 2005; Hsiao et al., 2009; Gottelt et al., 2010; Li et al., 2015), actinorhodin and undecylprodigiosin in Streptomyces coelicolor (Xu et al., 2010; Wang et al., 2011), virginiamycin in Streptomyces virginiae (Kondo et al., 1989; Kinoshita et al., 1997; Kawauchi et al., 2000), jadomycin in Streptomyces venezuelae (Yang et al., 1995; Wang and Vining, 2003; Wang et al., 2009; Xu et al., 2010; Zou et al., 2014), kinamycin in Streptomyces ambofaciens (Aigle et al., 2005; Bunet et al., 2008, 2011), and avermectin in Streptomyces avermitilis (Kitani et al., 2011; Wang J. B., et al., 2014; Zhu et al., 2016).
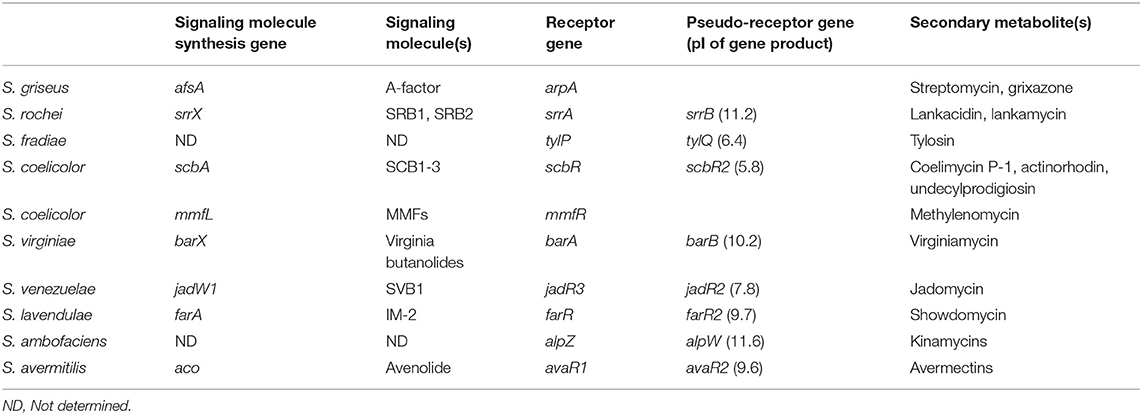
Table 1. Gene sets of signaling molecule(s)/receptor/pseudo-receptor for secondary metabolite production in Streptomyces species.
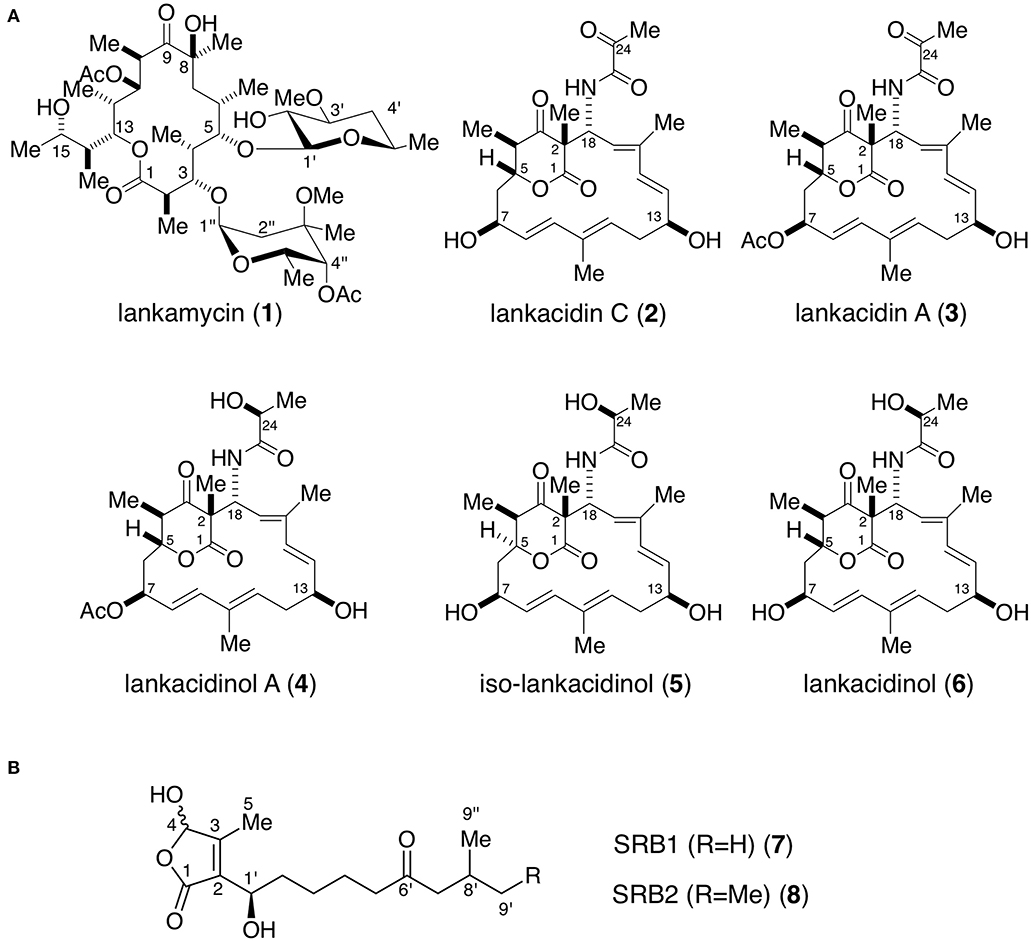
Figure 1. Chemical structures of antibiotics (A) and signaling molecules (B) produced in Streptomyces rochei. (A) Antibiotics lankamycin (1), lankacidin C (2), lankacidin A (3), lankacidinol A (4), iso-lankacidinol (5), and lankacidinol (6). (B) Signaling molecules SRB1 (7) and SRB2 (8). Me, methyl; Ac, acetyl.
Streptomyces rochei 7434AN4 produces two structurally unrelated polyketide antibiotics, LM and LC (Figure 1A) and carries three linear plasmids pSLA2-L, -M, and -S (Kinashi et al., 1994). Together with the biosynthetic genes for LM and LC, many regulatory genes including a biosynthetic gene for signaling molecules SRBs (Figure 1B) (srrX), six tetR-type repressor genes (srrA, srrB, srrC, srrD, srrE, and srrF), and three SARP (Streptomyces antibiotic regulatory protein) family activator genes (srrY, srrZ, and srrW) are located on giant linear plasmid pSLA2-L (210,614 bp) (Mochizuki et al., 2003). Our group revealed that SRBs/SrrA complex dissociates from the promoter region of srrY, leading to the activation of LC production (Yamamoto et al., 2008), whereas the gene product of srrY further activates a second activator gene srrZ to produce LM (Suzuki et al., 2010). In addition, mutation of an additional receptor gene srrB greatly increased the production of both LC and LM (Arakawa et al., 2007). This finding suggested that srrB negatively regulates LC and LM production, however, its functional role has not been clarified.
In this study, we extensively investigated the function of srrB by analyzing antibiotic production and transcription, the results of which indicated that SrrB acts as a negative regulator by binding to the promoter region of the activator gene srrY to control LC and LM production at the later stage of fermentation in S. rochei.
S. rochei strain 51252 carrying only the linear plasmid pSLA2-L was used as the parent strain (Kinashi et al., 1994). All strains, plasmids, and oligonucleotides used in this study are listed in Table 2. Streptomyces strains were grown in YM medium (0.4% yeast extract, 1.0% malt extract, 0.4% D-glucose, pH 7.3) for antibiotic production and RNA isolation. Escherichia coli strains were grown in Luria-Bertani (LB) medium supplemented with ampicillin (100 μg/ml), apramycin (50 μg/ml), and/or chloramphenicol (25 μg/ml) when necessary. For protoplasts preparation, Streptomyces strains were grown in YEME medium (Kieser et al., 2000). Protoplasts were regenerated on R1M plates (Zhang et al., 1997). DNA manipulations for E. coli (Sambrook et al., 1989) and Streptomyces (Kieser et al., 2000) were performed according to the standard protocols. PCR amplification was done on a 2720 Thermal Cycler (Thermo Fisher Scientific, Waltham, MA, USA) with KOD-Plus- DNA polymerase (Toyobo, Osaka, Japan).
The target plasmid pKAR3014 that carries in-frame deletion of srrA in E. coli-Streptomyces shuttle vector pRES18 (Ishikawa et al., 1996) was constructed as described previously (Arakawa et al., 2007). Targeted mutagenesis was performed as follows. Plasmid pKAR3014 was transformed into protoplasts of S. rochei strain KA07 (srrB mutant), and thiostrepton-resistant strains were obtained. Among these transformants, single-crossover strains were selected by Southern hybridization. Some single-crossover colonies were continuously grown in YEME liquid medium to facilitate a second crossover. Finally, thiostrepton sensitive strains were selected as double crossover strains, to obtain a strain TS03 (srrAB mutant). Gene disruption was checked by Southern hybridization analysis using DIG DNA Labeling and Detection Kit (Roche Diagnostics GmbH, Mannheim, Germany).
The target plasmid pKAR3055 that carries in-frame deletion of srrY in pRES18 was constructed as described previously (Yamamoto et al., 2008). This plasmid was transformed into protoplasts of S. rochei strain KA07, and an srrB-srrY double mutant KA64 was constructed in a similar manipulation as above mentioned.
The srrB gene was amplified using cosmid A8 (Mochizuki et al., 2003) and primers, srrB-8600f1 and srrB-8600r1. The resulting PCR product was digested with NdeI and XbaI and cloned into pIJ8600, an E. coli-Streptomyces shuttle vector carrying a tipA promoter (Sun et al., 1999), to obtain pKAR3065.
This plasmid was introduced into strain 51252, and transformants were cultured for 24 h at 28°C in YM liquid medium with 10 μg/ml apramycin. Thiostrepton (10 μg/ml as final concentration) was added at 24 h to induce srrB expression. After cultivation for additional 24 h, the broth filtrate was extracted twice with equal volume of ethyl acetate. The combined organic phase was dried with Na2SO4, and concentrated in vacuo to obtain crude extracts.
The srrB-coding sequence was PCR amplified using the template cosmid A8 (Mochizuki et al., 2003) and primers, KAR7903OE and KAR7902OE. The amplified fragment was digested with BglII and EcoRI and cloned into pET32b(+), a (His)6-tagged expression vector, to obtain pKAR3036.
The 48-h cultures of S. rochei strains were harvested, and the supernatant was extracted twice with equal volume of ethyl acetate. The crude extracts were purified by Sephadex LH-20 chromatography (1 × 40 cm, GE Healthcare, Chicago, IL) with methanol. Then the fractions containing antibiotics were purified by silica gel chromatography with chloroform-methanol (80:1–10:1, v/v). NMR spectra were recorded on an ECA-500 spectrometer (JEOL, Tokyo, Japan) equipped with a field gradient accessory. Chloroform-d and methanol-d4 were used as solvents. Chemical shifts were recorded in δ value based on the solvent signals (δC = 77.0 in CDCl3, δC = 49.0 in CD3OD, and δH = 3.30 in residual CH3OH) or an internal standard tetramethylsilane (δH = 0). High resolution ESI-MS spectra were measured by a LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific). The 1H- and 13C-NMR assignments for lankamycin (1), lankacidin C (2), lankacidin A (3), lankacidinol A (4), iso-lankacidinol (5), and lankacidinol (6) have already been reported (Suzuki et al., 2010; Arakawa et al., 2011; Yamamoto et al., 2018).
Two strains KA61 (ΔsrrY) and KA64 (ΔsrrYΔsrrB) were cultured at 28°C for 30 h, and the supernatant (60 ml) was acidified to pH 3 and extracted with equal volume of ethyl acetate twice. The combined organic phase was concentrated in vacuo. Appropriately diluted culture extract (100 μl) was added to the fresh culture (5 ml) of strain KA20, an srrX-deficient strain, and cultured at 28°C for 36 h to restore LM and LC production.
S. rochei strains were grown in YM liquid medium and harvested at various time periods at 12-36 h. Cells were used for measurement of dry cell weight (dcw) and isolation of total RNA, while the culture supernatant was for measurement of antibiotic production.
Cultures were collected at various time periods and centrifuged at 5,000 rpm for 10 min. The resulting pellet was washed twice with 10.3% sucrose, and then placed in a 60°C dry oven until the weight reaches to a constant value.
S. rochei strains were cultured at 28°C in YM liquid medium for various time periods. Total RNAs was extracted from cells with a TRI reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Trace amounts of DNA were removed with RNase-free DNase I (Takara, Kyoto, Japan). The concentration of purified RNA was determined by UV absorbance at 260 nm using Ultrospec 3300 pro spectrometer (GE Healthcare). The cDNAs were synthesized using Transcriptor Reverse Transcriptase (Roche Diagnostics). Each reaction mixture contained 1 μg of total RNA and 0.08 A260 units random primer. Each mixture was sequentially treated at 85°C for 5 min, at 25°C for 10 min, and 55°C for 45 min for the cDNA synthesis. The 16S rRNA was used as an internal standard (Lane, 1991;Turner et al., 1999).
Transcriptional start site (TSS) of srrB was determined using 5′RACE System, Version 2.0 (Invitrogen, Carlsbad, CA, USA). Total RNA was prepared from a 24-h culture sample of parent strain. One microgram of total RNA was converted to the cDNA using specific primer srrB-GSP1, and the resultant was treated with ribonuclease and purified through spin column to afford cDNA. A homopolymeric tail was then added to the 3'-end of cDNA using terminal deoxynucleotidyl transferase and dCTP. PCR was performed with poly C tailed cDNA as a template using abridged anchor primer and inner specific primer RT79-R2. TSS was determined from nucleotide sequence of amplified PCR product using ABI PRISM 310 Genetic Analyzer (Life Technologies, Carlsbad, CA, USA).
E. coli BL21(DE3)pLysS was used as hosts for plasmid pKAR3036. Cells were grown in LB liquid medium supplemented with 100 μg/ml ampicillin and 25 μg/ml chloramphenicol at 37°C to OD600 = 0.6 and then were induced with 1 mM isopropyl β-thiogalactopyranoside (IPTG). Cultivation was continued for 12 h at 16°C, and then cells were harvested and disrupted by SONIFER 250 ultrasonic homogenizer (Branson Ultrasonics Corporation, Danbury, CT, USA). The (His)6-fusion protein was purified by Ni2+-nitrotriacetic acid agarose (Qiagen GmbH, Hilden, Germany) according to the manufacture's protocol. After dialysis with PBS buffer (137 mM NaCl, 8.1mM Na2HPO4·12H2O, 2.68 mM KCl, 1.47 mM KH2PO4), the (His)6-tagged SrrB protein was treated with enterokinase (Novagen, Madison, WI, USA), and the (His)6-tag peptide upstream of the N-terminal SrrB was removed by Enterokinase Cleavage Capture Kit (Novagen) according to the manufacture's protocol. The protein was analyzed by SDS-PAGE with 15% polyacrylamide gel. The protein concentration was determined according to the methods of Bradford using Bio-Rad protein assay (Bio-Rad, Hercules, CA, USA) with bovine serum albumin as a standard.
The srrB probes for gel shift assay were prepared as follows. For preparation of probe B1, a 564-bp DNA fragment containing the upstream region of srrB was amplified using pKAR3004 as a template and primers SRRBf3 and SRRBr3 (positions −81 to +483 from TSS of srrB; nt 140,677-141,240 of pSLA2-L). For preparation of probe B2, a 386-bp DNA fragment containing the internal region of srrB was amplified using pKAR3004 as a template and primers SRRBf1E and SRRBr4 (positions +574 to +959 from TSS of srrB; nt 140,201-140,586 of pSLA2-L).
Probe B1 was then 3′ -end labeled with [γ-32P]ATP (GE Healthcare) and T4 polynucleotide kinase (Toyobo). The reaction mixture contained the binding buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM dithiothreitol, 0.1 mg of bovine serum albumin and 5% glycerol), 0.5 nM labeled DNA and 2 μM SrrA protein. SrrA protein was prepared as reported previously (Yamamoto et al., 2008). When necessary, synthetic SRB1 [(1–′R)-isomer; Figure 1B] (Arakawa et al., 2012) was added to the reaction mixture. For competition experiment, unlabeled probes B1 and B2 were used at a final concentration of 200 nM. The reaction mixture was incubated at 26°C for 30 min, and subjected to electrophoresis at room temperature on a native 4.5% polyacrylamide gel in 0.5 × TBE buffer (46 mM Tris base, 46 mM boric acid, 1 mM EDTA). The 32P-labeled DNAs were detected by autoradiography.
Preparation of srrY probes for gel shift assay was described previously (Yamamoto et al., 2008). To analyze the effect of SRB on the binding of SrrA and SrrB, various concentration of synthetic SRB1 [(1′R)-isomer; Figure 1B] (Arakawa et al., 2012) was added to the reaction mixture. In order to evaluate the effect of endogenous metabolites in S. rochei and other antibiotics on the binding of SrrB, the following compounds (1 mM) were separately added to the reaction mixture; LC, LM, chlorotetracycline, kanamycin, and ampicillin.
The method used for DNase I footprinting analysis for the upstream region of srrY was described previously (Yamamoto et al., 2008). For the upstream region of srrB, the primer SRRBf3 was 5′-end labeled using [γ-32P]ATP (GE Healthcare) and T4 polynucleotide kinase (Toyobo), and then PCR reaction was performed with unlabeled primer SRRBr3 and pKAR3004 as a template to afford a 564-bp product containing the upstream region of srrB (positions −81 to +483 from TSS of srrB; nt 140,677-141,240 of pSLA2-L). Binding reaction mixture (50 μl) contained 10 nM labeled DNA, 20 mM Tris-HCl (pH8.0), 1 mM MgCl2, 100 mM NaCl, 1 mM dithiothreitol, 0.1 mg/ml BSA, 5% glycerol, and various concentrations of SrrA. The binding reaction mixture was incubated for 30 min at 25°C, and then a mixture was treated with DNase I (Roche Diagnostics) solution [1 ng in 50 μl of 5 mM MgCl2 and 5 mM CaCl2] for 2 minutes at room temperature. The reaction was terminated by 100 μl of phenol-chloroform. The aqueous fraction containing DNAs was precipitated by ethanol and separated on a 5% polyacrylamide gel containing 6 M urea. The labeled DNAs were detected by autoradiography. Sequencing ladders were generated by Maxam-Gilbert sequencing of the labeled DNA used for binding reaction.
Alignment of Amino acid sequences of the pseudo-receptors including SrrB was performed by BioEdit version 7.2.5 software (https://bioedit.software.informer.com/) (Hall, 1999) (Figure S1A). Phylogenetic tree was constructed by the neighbor-joining algorithm of MEGA X version 10.1.5 software (Kumar et al., 2018) (Figure S1B).
The gene product of srrB belongs to the TetR-type transcriptional regulator family proteins, which contains a helix-turn-helix DNA binding motif at the N-terminal region (Figure S1). We previously reported overproduction of LM and LC in the srrB mutant KA07 based on TLC bioautography (Arakawa et al., 2007). In this study, we performed comparative metabolite analysis of the srrB mutant and its parent strain 51252. As shown in Figures 2A,B, the srrB mutant KA07 accumulated larger amount of compounds 1-6 compared with the parent strain 51252. Namely, KA07 produced 6-folds of lankamycin (1) (Figure 2A) and 9.9, 25, 4.2, and 5.7-folds of lankacidin C (2), lankacidinol A (4), iso-lankacidinol (5), and lankacidinol (6), respectively (Figure 2B). To investigate the effect of SrrA on antibiotic production, we further analyzed two mutants, an srrA mutant KA12 and an srrA-srrB double mutant TS03 (Figure S2). KA12 produced about 40% of metabolites when compared with the parent, while TS03 overproduced metabolites 1-6 at the same level with the srrB mutant KA07 (Figures 2A,B). These results confirmed the following two aspects; srrA mutation causes a slight decrease of the metabolic titer, whereas SrrB acts as a negative regulator for lankacidin and lankamycin production in S. rochei.
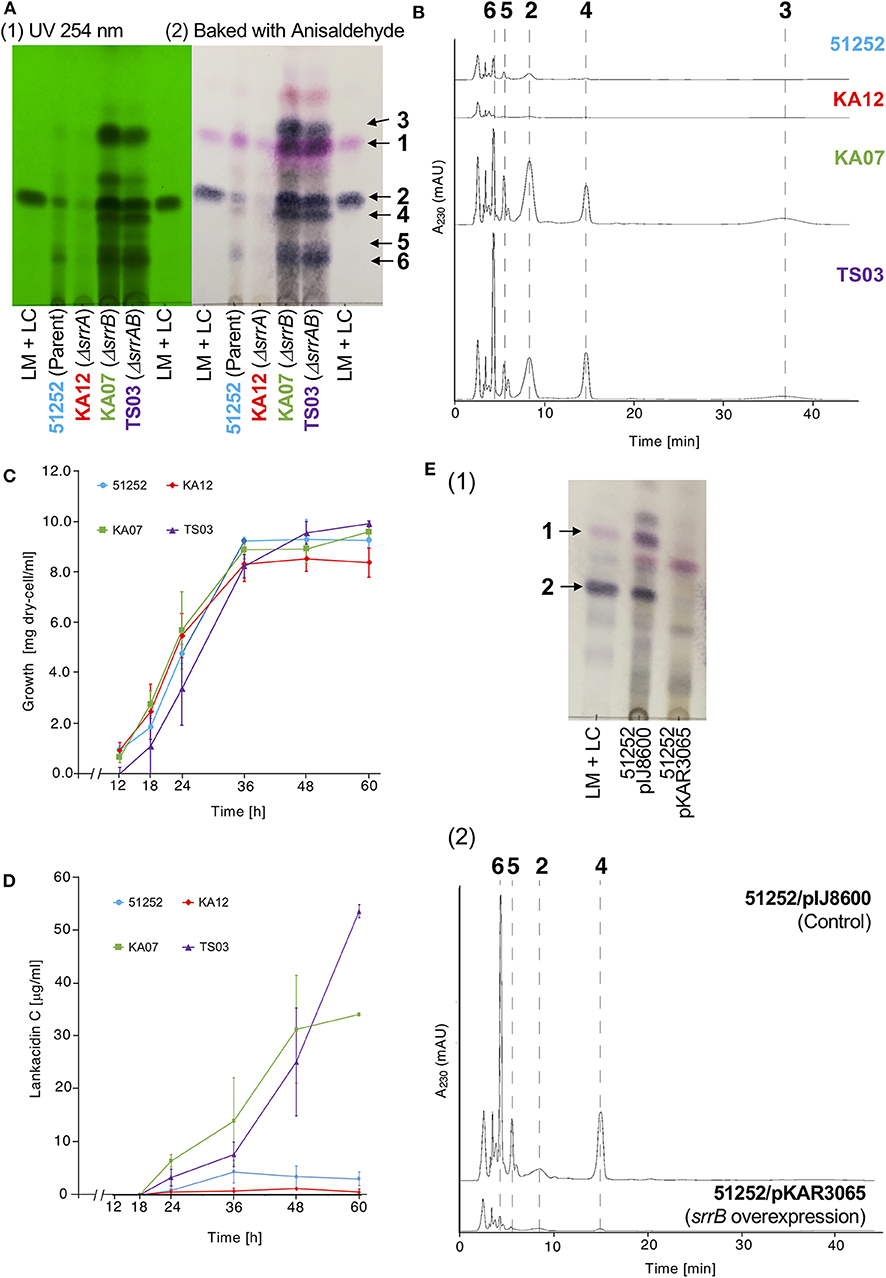
Figure 2. Metabolite profiles and cell growth of four S. rochei strains; 51252 (parent), KA12 (ΔsrrA), KA07 (ΔsrrB), and TS03 (ΔsrrA-srrB). (A) TLC analysis of the crude extract of S. rochei strains. All strains were grown at 28°C for 48 h. The left panel represents the TLC plate under ultraviolet irradiation (254 nm). The right panel represents the TLC plate after baking with anisaldehyde-H2SO4. TLC plates were developed with chloroform-methanol = 15:1 (v/v). (B) HPLC analysis of metabolites produced by S. rochei strains. The crude extracts were applied on a COSMOSIL Cholester column (4.6 × 250 mm, Nacalai Tesque) and eluted with a mixture of acetonitrile-10 mM sodium phosphate buffer (pH 8.2) (3:7, v/v) at a flow rate of 1.0 ml/min. (C) Time-course growth of S. rochei strains. Symbols represent each dry cell weight (DCW; g/l); strain 51252, blue circles and line; strain KA12, red diamonds and line; strain KA07, green squares and line; TS03, purple triangles and line. Results are representative of at least three independent experiments. (D) Time-course production of 2 in S. rochei strains. Symbols represent each production yield of 2 (μg/ml); strain 51252, blue circles and line; strain KA12, red diamonds and line; strain KA07, green squares and line; TS03, purple triangles and line. Results are representative of at least three independent experiments. (E) Effect of srrB overexpression on antibiotic production. Thiostrepton (10 μg/ml) was added to the 24-h culture of S. rochei 51252 recombinants harboring either pIJ8600 (control) or pKAR3065 (intact srrB), and then the cultures were further incubated at 28°C for 24 h. (1) TLC analysis of crude extracts. Crude extracts were separated by TLC [eluent; chloroform-methanol = 15:1 (v/v)] and detected by baking with anisaldehyde-H2SO4. Lane 1, LM and LC standard; lane 2, recombinant harboring pIJ8600 (control); lane 3, recombinant harboring pKAR3065 (intact srrB). (2) HPLC analysis of crude extracts. The crude extracts were applied on a COSMOSIL Cholester column (4.6 x 250 mm, Nacalai Tesque) and eluted with a mixture of acetonitrile-10 mM sodium phosphate buffer (pH 8.2) (3:7, v/v) at a flow rate of 1.0 ml/min.
To determine the role of srrB in the regulation of lankacidin and lankamycin production, we further performed time-course analysis of metabolite profile, growth curve, and transcription in the parent and three mutants (KA07, KA12, and TS03) at various time periods. As shown in Figure 2C, all strains grew in a similar proportion, indicating that overproduction in KA07 and TS03 was due to srrB mutation but not to cell growth difference. The time-course of antibiotic production was analyzed by the titer of 2, a major product among lankacidin derivatives (2-6; Figure 1A). As shown in Figure 2D, 2 was detected after 18 h, and its titer at 48-h growth in the srrB deficient strains, KA07 and TS03, were 9.0- and 7.2-times of 51252, respectively, which agrees with the overproduction profiles in KA07 and TS03 in Figures 2A,B. To confirm the negative regulatory property of SrrB in vivo, overexpression of SrrB in S. rochei was carried out. The intact srrB gene was introduced into plasmid pIJ8600, an E. coli-Streptomyces shuttle plasmid with a thiostrepton-inducible tipA promoter, to give pKAR3065. We tested antibiotic production in the S. rochei 51252 recombinants containing either the empty vector pIJ8600 or the srrB overexpression plasmid (pKAR3065). Compared with the control recombinant S. rochei 51252/pIJ8600, the S. rochei 51252/pKAR3065 recombinant significantly reduced antibiotic production (Figure 2E). These results clearly indicated that SrrB acts as a negative regulator for antibiotic production in S. rochei.
In our preliminary experiment, gel shift assay indicated that SrrA and SrrB could bind to the upstream region of srrX, a gene responsible for SRB biosynthesis. This finding suggested that the signaling molecule receptor/pseudo-repressor repress the transcription of srrX. Two strains were used to evaluate the comparative yield of SRB; an srrY single mutant KA61 and an srrY-srrB double mutant KA64 (Figure S3), both of which are unable to produce LC or LM due to a mutation on the major activator srrY. The yields of SRBs were evaluated by a help of bioassay using an srrX-deficient mutant as described previously (Arakawa et al., 2012). One-eighth of the crude extract of KA64 contained an equivalent amount of SRBs to that of KA61 (Figure S4), suggesting that srrB negatively controls the titer of SRBs.
To analyze the role of srrB in the SRB/SrrA regulatory system in S. rochei, we performed comparative transcriptional analysis of the selected regulatory genes in the parent and three mutants (srrA, srrB, and srrA-srrB). Transcription of srrY in the parent appeared around 18 h and diminished after 32 h (Figure 3A), while that in the srrB mutant KA07 continued until the later stage (Figure 3C). On the other hand, srrB transcription in the parent appeared around 16 h and prolonged until the late stage of fermentation (Figure 3A). In the srrA mutant KA12, transcription of both srrY and srrB appeared at 12 h or earlier (Figure 3B), whereas srrY transcription in the srrA-srrB mutant TS03 was detected through all time periods (12–36 h) (Figure 3D). These findings together with our previous result (Yamamoto et al., 2008) showed that SRB/srrA regulatory system controls transcription of both srrY and srrB, and SrrB represses srrY transcription at the later stage.
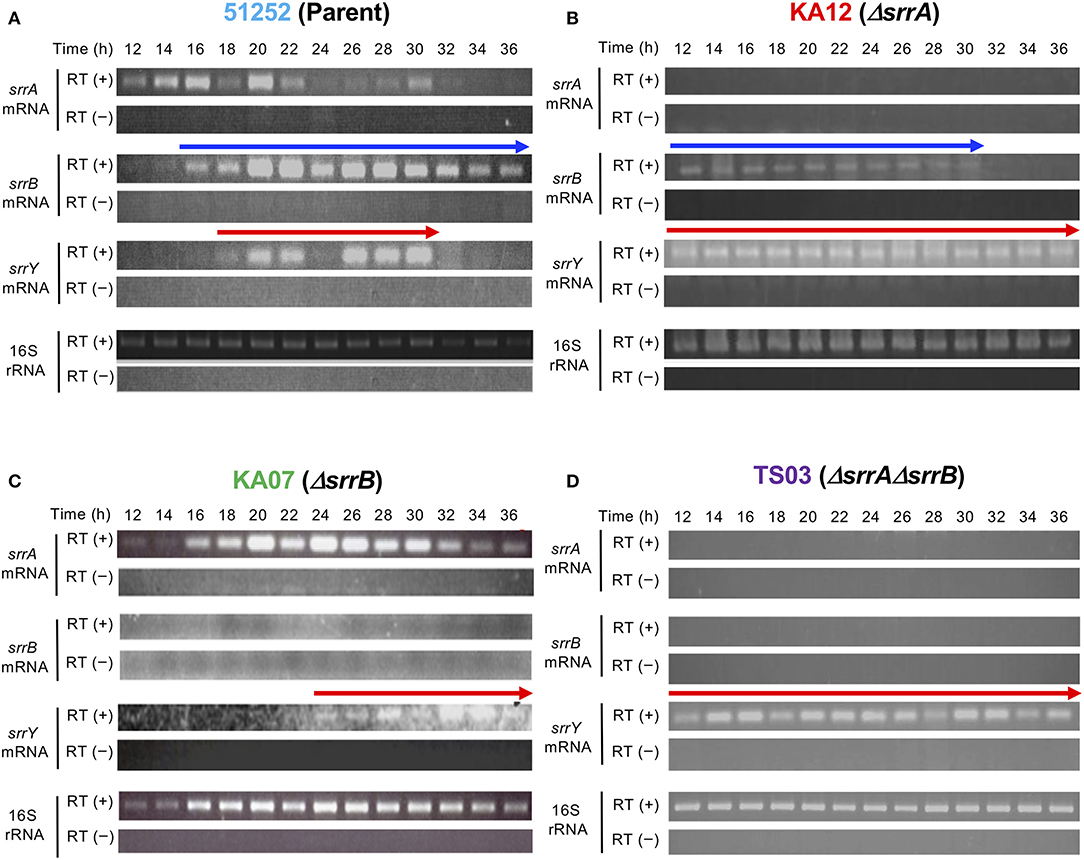
Figure 3. Time-course RT-PCR analysis of strains 51252 (A), KA12 (B), KA07 (C), and TS03 (D). Three upper panels represent srrA mRNA, srrB mRNA, and srrY mRNA. The lowest panels represents 16S rRNA gene as a control. RT (+) indicates the treatment of total RNA with Transcriptor Reverse Transcriptase, while RT (–) dues no treatment of transcriptase. Red arrow indicates transcription of srrY, while blue arrow does transcription of srrB.
A transcriptional start point (TSS) of srrB was determined be 401-bp upstream of its translational start codon by 5′-RACE (Figure 4 and Figure S5). To determine whether SrrA binds to the upstream promoter region of srrB (srrBp), gel shift assay was performed using 32P-labeled probe B1 (nt 140,677-141,240 of pSLA2-L) that contained the upstream region of srrB (Figure 5A). SrrA protein was overexpressed in E. coli as described previously (Yamamoto et al., 2008). A band shift of probe B1 was observed in the presence of SrrA protein in a concentration-dependent manner (Figure 5B). Competition experiments using unlabeled probes B1 and B2 (nt 140,201-140,585 of pSLA2-L) (Figure 5C) were performed to determine the specific binding of SrrA to the region of probe B1. A band shift disappeared in the large excess of unlabeled probe B1, whereas probe B2 gave no effect on band shift (Figure 5C). Addition of SRB led to dissociation of SrrA from probe B1 (Figure 5C), indicating that the srrB transcription is controlled by SRB/SrrA regulatory system.
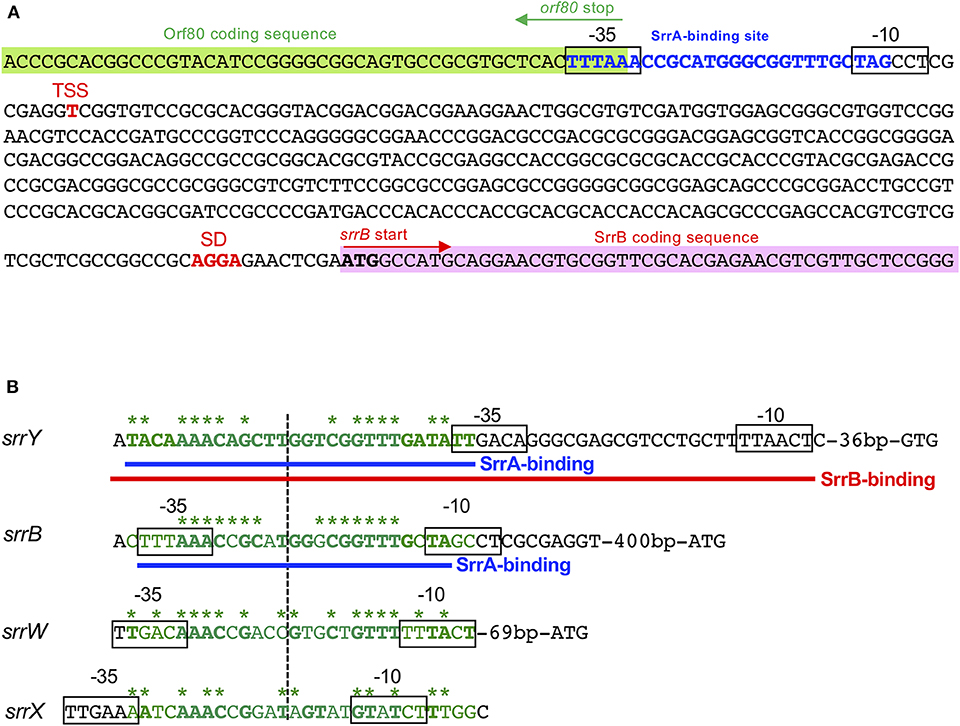
Figure 4. SrrA/SrrB binding sequences. (A) Characterization of the upstream region of srrB. Putative srrB promoter regions (−35 and −10) are boxed. Shine-Dalgarno (SD) sequence and transcriptional start site (TSS) are shown as red boldface letters. SrrA-binding sequence is shown as blue boldface letters. Pink and green highlights indicate SrrB and Orf80 coding sequences, respectively. (B) Comparison of the binding sequences for SrrA and SrrB. The confirmed SrrA- and SrrB-binding sequences are shown as blue and red underlines, respectively. Possible SrrA-binding sites at upstream of srrW and srrX are deduced from sequence data. For comparison of consensus sequence, SrrA-binding sites at the upstream of srrY (SrrA-srrY) are shown as green. Bases identical with SrrA-srrY are shown in boldface letters. Complementary bases are indicated as asterisks. The center of palindromes is shown as a vertical dashed line.
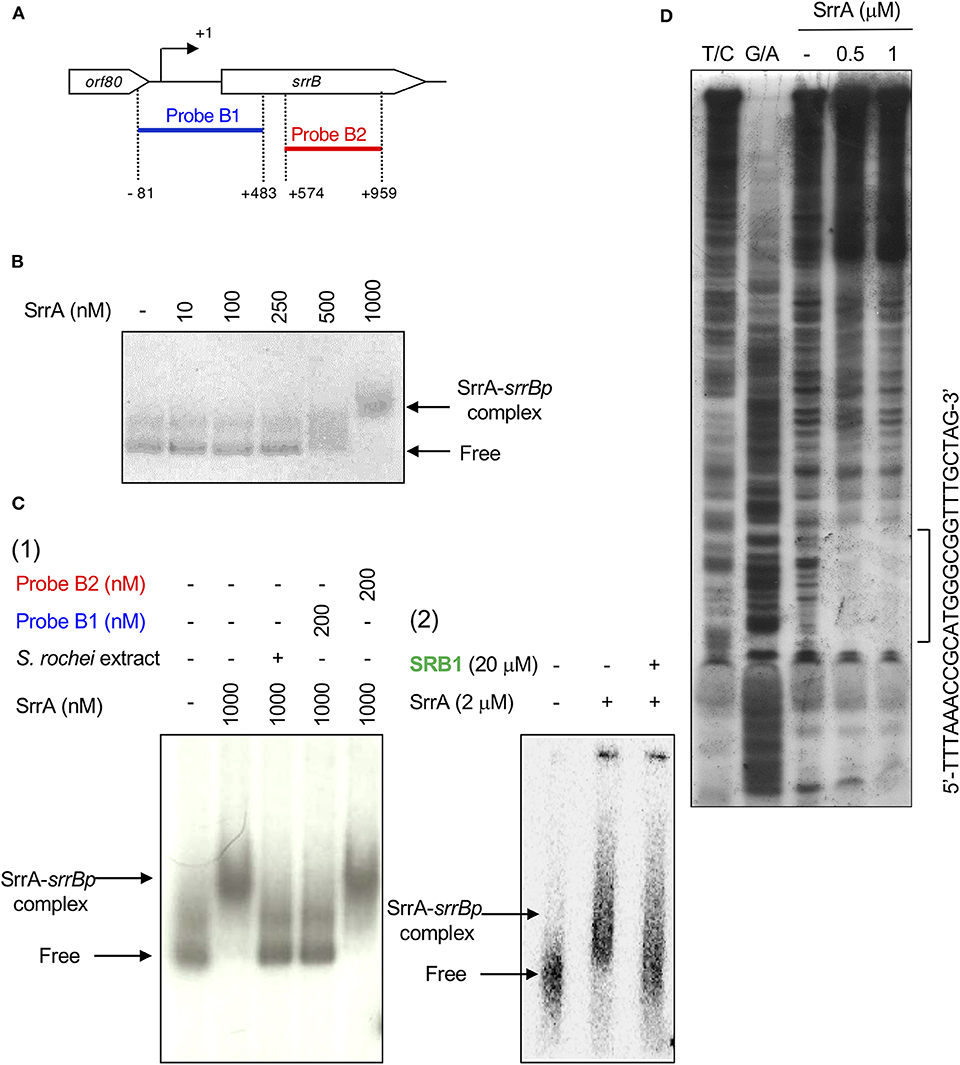
Figure 5. Gel shift assay and DNase I footprinting analysis of SrrA-binding in the upstream region of srrB. (A) Location of the two probes B1 and B2. The TSS of srrB is numbered +1. (B) The concentration-dependent binding of SrrA to the upstream promoter region of srrB (srrBp). The 32P-labeled probe B1 (1 nM) was mixed with various concentration of SrrA (0–1,000 nM). (C) (1) Competition experiments to investigate specific binding of SrrA to the upstream region of srrB. Each reaction mixture contained 0.5 nM 32P-labeled probe B1 (lane 1) and 1,000 nM SrrA (lane 2). Then, S. rochei culture extract (lane 3), 200 nM unlabeled probe B1 (lane 4) or unlabeled probe B2 (lane 5) was added. (2) Effect of endogenous SRB to investigate specific binding of SrrA to the upstream region of srrB. Each reaction mixture contained 0.5 nM 32P-labeled probe B1 (lane 1) and 2,000 nM SrrA (lane 2). Then, 20 μM synthetic SRB1 [(1′R)-isomer; Figure 1B] (Arakawa et al., 2012) (lane 3) was added. (D) DNase I footprinting analysis of SrrA-binding site on the upstream of srrB. Probe B1 was end labeled on the non-template strand. Each reaction mixture contained 2 nM labeled DNA and SrrA (0.5 and 1 μM). Sequencing ladders were generated by Maxam-Gilbert sequencing of the labeled probe B1. Capital letters at right side indicate SrrA-binding sequences.
DNase I footprinting experiment was performed to identify the SrrA binding sequence in the upstream of srrB. Positions −36 to −11 of the non-template strand was protected by SrrA (Figure 5D). The protected region overlapped with a possible srrB promoter containing a palindromic sequence (asterisks in Figure 4B), whose sequence well-matched with the SrrA-binding sequence of srrY (Yamamoto et al., 2008). Taking account of transcriptional analysis above mentioned, SrrA binds to the upstream regions of both srrB and srrY to repress their transcription at the early growth phase (~16 h).
To analyze whether SrrB binds to the promoter region of srrY (srrYp), gel shift assay was performed using probe Y1 (positions −452 to +100 from TSS of srrY) (Figure 6A) containing the promoter region of srrY, which was constructed previously (Yamamoto et al., 2008). SrrB protein was overexpressed in the E. coli BL21(DE3)pLysS/pKAR3036 recombinant with IPTG induction and purified by Ni-NTA agarose (Figure S6). The addition of SrrB protein gave a shifted band of probe Y1 in a concentration-dependent manner (Figure 6B). Competitive experiments (Figure 6C) revealed that SrrB specifically binds to probe Y1, not to probe Y2 (positions +101 to +333 from TSS of srrY). The pseudoreceptors hitherto studied are insensitive to endogenous signaling molecules and interact with endogenous antibiotics (Martín and Liras, 2019; Xu and Yang, 2019) (details are described in Discussion Section). We tested the effects of various signaling-molecule/antibiotics on the binding of SrrB to srrYp through gel shift analysis by using endogenous signaling molecule SRB, endogenous antibiotics (LC and LM), and other exogenous antibiotics (Figures 6D,E). Dissociation of SrrB from srrYp could be caused by SRB, however, a higher concentration of 1 mM was required (500-fold excess against SrrB protein). The sensitivity of SrrB against SRB was 50-fold lower compared with the signaling molecule receptor SrrA (Figure 6D). Dissociation of SrrB from srrYp was not caused by endogenous antibiotics LC and LM in S. rochei and neither by exogenous antibiotics (chlorotetracycline, kanamycin, ampicillin) at even 1 mM concentration (500-fold excess against SrrB protein) (Figure 6E).
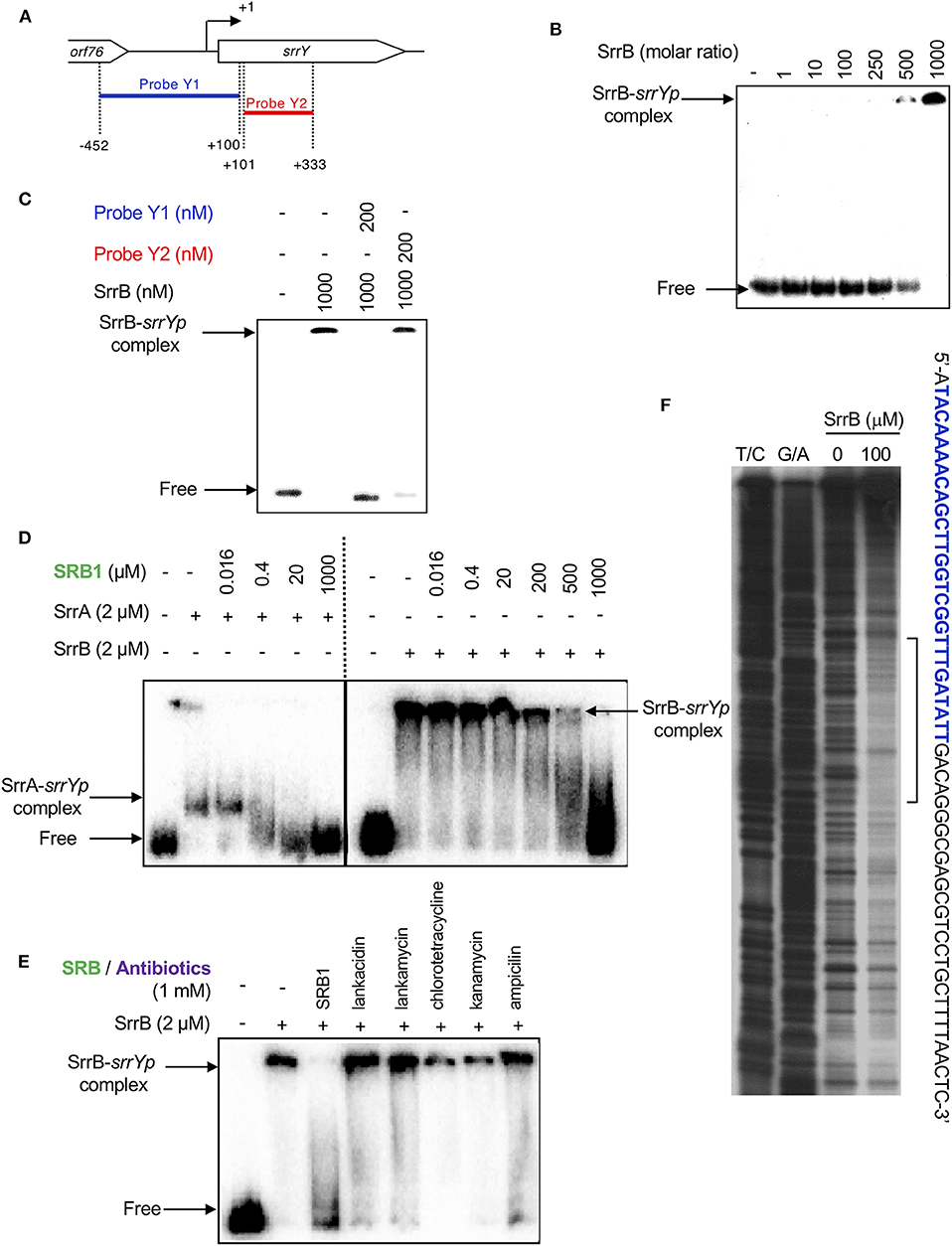
Figure 6. Gel shift assay and DNase I footprinting analysis of SrrB-binding in the upstream region of srrY. (A) Location of the two probes, probe Y1 and Y2. The description of probe preparation was described previously (Yamamoto et al., 2008). (B) The concentration-dependent binding of SrrB to the upstream promoter region of srrY (srrYp). Labeled probe Y1 (1 nM) was mixed with various concentration of SrrB (0–1,000 nM). (C) Competition experiments to investigate specific binding of SrrB to the upstream region of srrY. Each reaction mixture contained 1 nM 32P-labeled probe Y1 and 1,000 nM SrrB. Then, 200 nM unlabeled probe Y1 (lane 3) or unlabeled probe Y2 (lane 4) was added. (D) Effect of SRB1 on the binding of SrrA (Right panel) and SrrB (left panel). Each reaction mixture contained 0.5 nM probe Y1 and 2,000 nM recombinant protein. To the reaction mixture, various concentration of synthetic SRB1 [(1′R)-isomer; Figure 1B) (Arakawa et al., 2012) was added. (E) Effect of endogenous metabolites in S. rochei and other antibiotics on the binding of SrrB. To the same reaction mixture described for panel D, various compounds including SRB1, LC, LM, chlorotetracycline, kanamycin, and ampicillin (each 1 mM) were separately added. (F) DNase I footprint analysis of SrrB-binding site on the upstream of srrY. Probe Y1 was end labeled on the non-template strand. Each reaction mixture contained 2 nM labeled DNA and SrrB (100 nM). Sequencing ladders were generated by Maxam-Gilbert sequencing of the labeled probe Y1. Capital letters at right side indicate SrrB-binding sequences, among which blue letters indicate SrrA-binding sequences.
We further performed DNase I footprinting experiment to identify the SrrB binding sequence in the upstream of srrY. As shown in Figure 6F, positions −61 to −4 of non-template strand were protected by SrrB. Although SrrB covers larger upstream region (58 bp) than SrrA does (28 bp; blue letters in Figure 6F), both SrrA and SrrB could bind to the promoter region of srrY (Figure 4B).
In this study, we revealed that srrB acts as a negative regulator by binding to the promoter region of the target gene srrY to repress LC and LM production in S. rochei. Expression of srrB is controlled by SRB/SrrA regulatory system.
TetR-type receptors have a conserved DNA-binding helix-turn-helix motif in the N-terminus and a ligand-binding pocket in the C-terminus (Yu et al., 2010). Particularly, the signaling molecule receptors and the pseudo-receptors constitute one of a major class of TetR-type regulators (Figure S1). The signaling molecule receptors have a helix-turn-helix DNA-binding motif in the N-terminus and a ligand-binding Trp residue in the C-terminus. It is noteworthy that the signaling molecule synthase and its cognate receptor gene pairs usually locate adjacently on the genome (Biarnes-Carrera et al., 2015), which allows us to predict signaling molecule/receptor systems in Streptomyces species (Niu et al., 2016). On the other hand, the pseudo-receptors also have a conserved DNA-binding motif like the signaling molecule receptors (Figure S1A), however, their location has no relationship with the signaling molecule synthase genes. Many of them act as negative regulators for antibiotic production; for example, TylQ for tylosin production in Streptomyces fradiae (Stratigopoulos and Cundliffe, 2002), BarB for virginiamycin in Streptomyces virginiae (Matsuno et al., 2004), ScbR2 for coelimycin P-1 in Streptomyces coelicolor (Gottelt et al., 2010), AlpW for orange pigment kinamycin in Streptomyces ambofaciens (Bunet et al., 2008), and AvaR2 for avermectin in Streptomyces avermitilis (Zhu et al., 2016) (Table 1).
In general, the pseudo-receptors are insensitive to endogenous signaling molecules. BarB has no binding affinity to virginia butanolides in S. virginiae (Matsuno et al., 2004). Surprisingly, ScbR2 from S. coelicolor does not bind to the signaling molecules SCB1-3 but binds to two endogenous antibiotics, actinorhodin and undecylprodigiosin (Xu et al., 2010), and exogenous antibiotic jadomycin (Wang W., 2014). In S. venezuelae, JadR2 binds to endogenous jadomycin and chloramphenicol as ligands, (Xu et al., 2010). Thus, ScbR2 and JadR2 bind to multiple antibiotics, and coordinate their biosynthesis (Xu et al., 2010; Zou et al., 2014). In S. avermitilis; AvaR2 binds to the endogenous signaling molecule avenolide, but not to oligomycin and avermectin (Zhu et al., 2016). Its mutational analysis revealed that AvaR2 plays a negative regulatory role in avermectin production and cell growth (Zhu et al., 2016). In S. rochei, SrrB-srrYp complex was disrupted by endogenous signaling molecule SRB at 1 mM concentration, although its minimum dissociation concentration for SrrB was 50-fold higher than that for SrrA, the SRB receptor. SrrB showed no binding activity to endogenous polyketide antibiotics LM or LC in S. rochei and neither to exogenous antibiotics including aromatic polyketide chlorotetracycline, aminoglycoside antibiotic kanamycin, and β-lactam antibiotic ampicillin even at 1 mM concentration (500-fold excess against SrrB). Thus, functions of the pseudo-receptors are variable in Streptomyces species.
The possible regulatory pathway in S. rochei is shown in Figure 7. At the early growth phase, SrrA represses transcription of both srrY and srrB (panel A). When SRB reaches a critical concentration, SrrA-SRB complex dissociates from both promoter regions to induce expression of srrY and srrB (panel B). Then SrrB represses srrY transcription at the later stage fermentation (panel C), suggesting a transient expression of srrY by two receptor proteins SrrA and SrrB in S. rochei. A similar regulatory pathway was proposed for kinamycin production in S. ambofaciens (Bunet et al., 2008) although its ligand has not yet been identified. In the early stage of growth, the signaling molecule receptor AlpZ represses both transcription of alpV (an SARP-type activator gene) and alpW (a pseudo-receptor gene). When an unidentified ligand interacts with AlpZ, this protein dissociates from the promoter regions in both alpV and alpW, leading to kinamycin production. At the later stage of fermentation, AlpW represses alpV transcription again to cease kinamycin production. Another interesting features in the S. rochei regulatory pathway is the presence of srrY-srrC cistron (Figure 7). The srrC mutant showed no sporulation, suggesting that srrC acts as a positive regulator for morphological differentiation (Arakawa et al., 2007). As shown in Figure 7, srrB negatively regulates the transcription of both srrY and srrC, which leads to transient controls for antibiotic production and morphological differentiation, respectively.
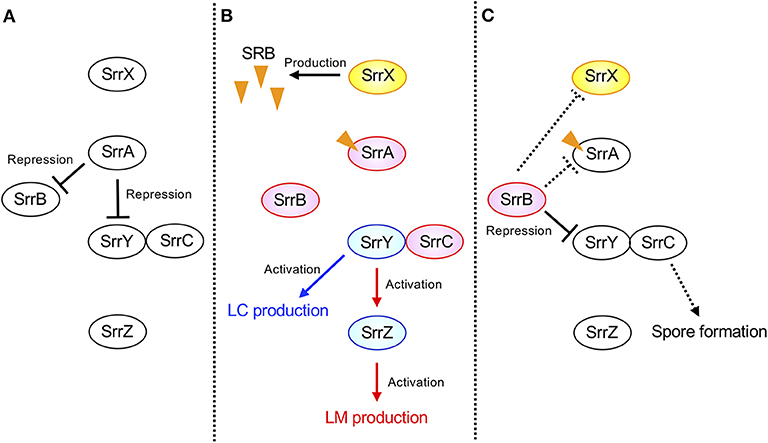
Figure 7. Model of transient srrY expression for LM and LC production in S. rochei. (A) SrrA represses both srrY and srrB expression in the early stage of growth. (B) Dissociation of SrrA from the promoter region of both srrY and srrB by SRB in middle stage of growth. (C) Repression of srrY by SrrB in the late stage of growth. Solid lines indicate the confirmed regulatory pathway hitherto. Additional dashed lines were suggested by unpublished results. Orange triangles indicate the signaling molecules SRBs.
The srrB mutation increased the titers of antibiotics as well as SRBs. This result well agreed with our preliminary gel shift assay that both SrrA and SrrB bind to the upstream region of SRB biosynthesis gene srrX (data not shown). Large excess of SRBs has no effect on antibiotic overexpression in S. rochei (Arakawa et al., 2012), hence, exact mechanism of the srrX repression by SrrB at the later stage remains to be clarified.
Manipulation of regulatory genes often causes activation of “silent” secondary metabolites (Olano et al., 2008; Zerikly and Challis, 2009; Rutledge and Challis, 2015; Arakawa, 2018). In S. ambofaciens, a mutant of the pseudo-receptor gene alpW accumulated kinamycin (Bunet et al., 2011). A mutant of the repressor gene ksbC accumulated β-carboline compound kitasetaline in Kitasatospora setae (Aroonsri et al., 2012). To our surprise, azoxyalkene compound KA57-A accumulated in a triple knockout mutant of S. rochei that have mutations on two biosynthetic gene clusters for LC, LM, and srrB (Kunitake et al., 2015). The genome sequence of the S. rochei chromosome has been determined to be 8.36 Mb in size, and at least 35 secondary metabolites gene clusters are coded on the chromosome (Nindita et al., 2019). Comprehensive mutational analysis on various regulatory genes may lead to activate silent secondary metabolite gene clusters in S. rochei, which is in progress in our laboratory. In conclusion, we have extensively characterized the role of the pseudo-receptor SrrB for antibiotic production in S. rochei. Further understanding and manipulation of the regulatory system in Streptomyces will lead to a natural product discovery with notable biological activities.
The datasets generated for this study are available on request to the corresponding author.
YM, SY, TS, HK, and KA designed the experiments. YM, SY, TS, MI, HS, YT, KI, and KA performed the experiments. YM, SY, TS, MI, HS, and KA analyzed the data. YM, SY, TS, HK, and KA wrote the manuscript with input from all of the authors. All authors approved the final version of the manuscript.
This work was supported by Grants-in-Aid for Scientific Research on Innovative Areas (23108515, 25108718, 17H05446, and 19H04659 to KA) from Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT), Grants-in-Aid for Scientific Research (B) (16H04917 to KA) from the Japan Society for the Promotion of Science (JSPS), and JSPS A3 Foresight Program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Dr. D. Kajiya and Mrs. T. Amimoto (N-BARD, Hiroshima University) for measurement of the high resolution mass spectra.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01089/full#supplementary-material
Aigle, B., Pang, X., Decaris, B., and Leblond, P. (2005). Involvement of AlpV, a new member of the Streptomyces antibiotic regulatory protein family, in regulation of the duplicated type II polyketide synthase alp gene cluster in Streptomyces ambofaciens. J. Bacteriol. 187, 2491–2500. doi: 10.1128/JB.187.7.2491-2500.2005
Arakawa, K. (2018). Manipulation of metabolic pathways controlled by signaling molecules, inducers of antibiotic production, for genome mining in Streptomyces spp. Antonie. Leeuwen. 111, 743–751. doi: 10.1007/s10482-018-1052-6
Arakawa, K., Cao, Z., Suzuki, N., and Kinashi, H. (2011). Isolation, structural elucidation, and biosynthesis of 15-norlankamycin derivatives produced by a type-II thioesterase disruptant of Streptomyces rochei. Tetrahedron 67, 5199–5205 doi: 10.1016/j.tet.2011.05.047
Arakawa, K., Mochizuki, S., Yamada, K., Noma, T., and Kinashi, H. (2007). γ-Butyrolactone autoregulator-receptor system involved in lankacidin and lankamycin production and morphological differentiation in Streptomyces rochei. Microbiology 153, 1817–1827. doi: 10.1099/mic.0.2006/002170-0
Arakawa, K., Tsuda, N., Taniguchi, A., and Kinashi, H. (2012). The butenolide signaling molecules SRB1 and SRB2 induce lankacidin and lankamycin production in Streptomyces rochei. ChemBioChem 13, 1447–1457. doi: 10.1002/cbic.201200149
Aroonsri, A., Kitani, S., Hashimoto, J., Kosone, I., Izumikawa, M., Komatsu, M., et al. (2012). Pleiotropic control of secondary metabolism and morphological development by KsbC, a butyrolactone autoregulator receptor homologue in Kitasatospora setae. Appl. Environ. Microbiol. 78, 8015–8024. doi: 10.1128/AEM.02355-12
Bate, N., Butler, A. R., Gandecha, A. R., and Cundliffe, E. (1999). Multiple regulatory genes in the tylosin biosynthetic cluster of Streptomyces fradiae. Chem. Biol. 6, 617–624. doi: 10.1016/S1074-5521(99)80113-6
Bate, N., Stratigopoulos, G., and Cundliffe, E. (2002). Differential roles of two SARP-encoding regulatory genes during tylosin biosynthesis. Mol. Microbiol. 43, 449–458. doi: 10.1046/j.1365-2958.2002.02756.x
Biarnes-Carrera, M., Breitling, R., and Takano, E. (2015). Butyrolactone signaling circuits for synthetic biology. Curr. Opin. Chem. Biol. 28, 91–98. doi: 10.1016/j.cbpa.2015.06.024
Bibb, M. J. (2005). Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8, 208–215. doi: 10.1016/j.mib.2005.02.016
Bunet, R., Mendes, M. V., Rouhier, N., Pang, X., Hotel, L., Leblond, P., et al. (2008). Regulation of the synthesis of the angucyclinone antibiotic alpomycin in Streptomyces ambofaciens by the autoregulator receptor AlpZ and its specific ligand. J. Bacteriol. 190, 3293–3305. doi: 10.1128/JB.01989-07
Bunet, R., Song, L., Mendes, M. V., Corre, C., Hotel, L., Rouhier, N., et al. (2011). Characterization and manipulation of the pathway-specific late regulator AlpW reveals Streptomyces ambofaciens as a new producer of kinamycins. J. Bacteriol. 193, 1142–1153. doi: 10.1128/JB.01269-10
Gottelt, M., Kol, S., Gomez-Escribano, J. P., Bibb, M., and Takano, E. (2010). Deletion of a regulatory gene within the cpk gene cluster reveals novel antibacterial activity in Streptomyces coelicolor A3(2). Microbiology 156, 2343–2353. doi: 10.1099/mic.0.038281-0
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41, 95–98.
Hara, O., and Beppu, T. (1982). Mutants blocked in streptomycin production in streptomyces griseus - The role of A-factor. J. Antibiot. 35, 349–358. doi: 10.7164/antibiotics.35.349
Horinouchi, S., and Beppu, T. (2007). Hormonal control by A-factor of morphological development and secondary metabolism in streptomyces. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 83, 277–295. doi: 10.2183/pjab.83.277
Hsiao, N. H., Nakayama, S., Merlo, M. E., de Vries, M., Bunet, R., Kitani, S., et al. (2009). Analysis of two additional signaling molecules in streptomyces coelicolor and the development of a butyrolactone-specific reporter system. Chem. Biol. 16, 951–960. doi: 10.1016/j.chembiol.2009.08.010
Ishikawa, J., Niino, Y., and Hotta, K. (1996). Construction of pRES18 and pRES19, streptomyces-escherichia coli shuttle vectors carrying multiple cloning sites. FEMS Microbiol. Lett. 145, 113–116. doi: 10.1016/0378-1097(96)00397-7
Kawauchi, R., Akashi, T., Kamitani, Y., Sy, A., Wangchaisoonthorn, U., Nihira, T., et al. (2000). Identification of an AfsA homologue (BarX) from streptomyces virginiae as a pleiotropic regulator controlling autoregulator biosynthesis, virginiamycin biosynthesis and virginiamycin M1 resistance. Mol. Microbiol. 36, 302–313. doi: 10.1046/j.1365-2958.2000.01819.x
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000). Practical Streptomyces Genetic: A Laboratory Manual. Norwich: John Innes Foundation.
Kinashi, H., Mori, E., Hatani, A., and Nimi, O. (1994). Isolation and characterization of large linear plasmids from lankacidin-producing Streptomyces species. J. Antibiot. 47, 1447–1455. doi: 10.7164/antibiotics.47.1447
Kinoshita, H., Ipposhi, H., Okamoto, S., Nakano, H., Nihira, T., and Yamada, Y. (1997). Butyrolactone autoregulator receptor protein (BarA) as a transcriptional regulator in Streptomyces virginiae. J. Bacteriol. 179, 6986–6993. doi: 10.1128/JB.179.22.6986-6993.1997
Kitani, S., Miyamoto, K. T., Takamatsu, S., Herawati, E., Iguchi, H., Nishitomi, K., et al. (2011). Avenolide, a Streptomyces hormone controlling antibiotic production in streptomyces avermitilis. Proc. Natl. Acad. Sci. U.S.A. 108, 16410–16415. doi: 10.1073/pnas.1113908108
Kondo, K., Higuchi, Y., Sakuda, S., Nihira, T., and Yamada, Y. (1989). New virginiae butanolides from Streptomyces virginiae. J. Antibiot. 42, 1873–1876. doi: 10.7164/antibiotics.42.1873
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kunitake, H., Hiramatsu, T., Kinashi, H., and Arakawa, K. (2015). Isolation and biosynthesis of an azoxyalkene compound produced by a multiple gene disruptant of streptomyces rochei. ChemBioChem 16, 2237–2243. doi: 10.1002/cbic.201500393
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (New York, NY: John Wiley and Sons, 115–175.
Li, X., Wang, J., Li, S., Ji, J., Wang, W., and Yang, K. (2015). ScbR- and ScbR2-mediated signal transduction networks coordinate complex physiological responses in Streptomyces coelicolor. Sci. Rep. 5:14831. doi: 10.1038/srep14831
Martín, J. F., and Liras, P. (2019). Harnessing microbiota interactions to produce bioactive metabolites: communication signals and receptor proteins. Curr, Opin. Pharmacol. 48, 8–16. doi: 10.1016/j.coph.2019.02.014
Matsuno, K., Yamada, Y., Lee, C. K., and Nihira, T. (2004). Identification by gene deletion analysis of barB as a negative regulator controlling an early process of virginiamycin biosynthesis in Streptomyces virginiae. Arch. Microbiol. 181, 52–59. doi: 10.1007/s00203-003-0625-5
Mochizuki, S., Hiratsu, K., Suwa, M., Ishii, T., Sugino, F., Yamada, K., et al. (2003). The large linear plasmid pSLA2-L of Streptomyces rochei has an unusually condensed gene organization for secondary metabolism. Mol. Microbiol. 48, 1501–1510. doi: 10.1046/j.1365-2958.2003.03523.x
Nindita, Y., Cao, Z., Fauzi, A. A., Teshima, A., Misaki, Y., Muslimin, R., et al. (2019). The genome sequence of Streptomyces rochei 7434AN4, which carries a linear chromosome and three characteristic linear plasmids. Sci. Rep. 9:10973. doi: 10.1038/s41598-019-47406-y
Niu, G., Chater, K. F., Tian, Y., Zhang, J., and Tan, H. (2016). Specialized metabolites regulating antibiotic biosynthesis in streptomyces spp. FEMS Microbiol. Rev. 40, 554–573. doi: 10.1093/femsre/fuw012
Ohnishi, Y., Kameyama, S., Onaka, H., and Horinouchi, S. (1999). The A-factor regulatory cascade leading to streptomycin production in streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34, 102–111. doi: 10.1046/j.1365-2958.1999.01579.x
Ohnishi, Y., Yamazaki, H., Kato, J. Y., Tomono, A., and Horinouchi, S. (2005). AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 69, 431–439. doi: 10.1271/bbb.69.431
Olano, C., Lomb,ó, F., Méndez, C., and Salas, J. A. (2008). Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab. Eng. 10, 281–292. doi: 10.1016/j.ymben.2008.07.001
Onaka, H., Ando, N., Nihira, T., Yamada, Y., Beppu, T., and Horinouchi, S. (1995). Cloning and characterization of the A-factor receptor gene from Streptomyces griseus. J. Bacteriol. 177, 6083–6092. doi: 10.1128/JB.177.21.6083-6092.1995
Rutledge, P. J., and Challis, G. L. (2015). Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 13, 509–523. doi: 10.1038/nrmicro3496
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Stratigopoulos, G., and Cundliffe, E. (2002). Expression analysis of the tylosin-biosynthetic gene cluster: pivotal regulatory role of the tylQ product. Chem. Biol. 9, 71–78. doi: 10.1016/S1074-5521(01)00095-3
Sun, J., Kelemen, G. H., Fernandez-Abalos, J. M., and Bibb, M. J. (1999). Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145, 2221–2227. doi: 10.1099/00221287-145-9-2221
Suzuki, T., Mochizuki, S., Yamamoto, S., Arakawa, K., and Kinashi, H. (2010). Regulation of lankamycin biosynthesis in streptomyces rochei by two SARP genes, srrY and srrZ. Biosci. Biotechnol. Biochem. 74, 819–827. doi: 10.1271/bbb.90927
Takano, E. (2006). γ-Butyrolactones: Streptomyces signaling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol. 9, 287–294. doi: 10.1016/j.mib.2006.04.003
Takano, E., Chakaraburtty, R., Nihira, T., Yamada, Y., and Bibb, M. J. (2001). A complex role for the γ-butyrolactone SCB1 in regulating antibiotic production in streptomyces coelicolor A3(2). Mol. Microbiol. 41, 1015–1028. doi: 10.1046/j.1365-2958.2001.02562.x
Takano, E., Kinoshita, H., Mersinias, V., Bucca, G., Hotchkiss, G., Nihira, T., et al. (2005). A bacterial hormone (the SCB1) directly controls the expression of a pathway-specific regulatory gene in the cryptic type I polyketide biosynthetic gene cluster of Streptomyces coelicolor. Mol. Microbiol. 56, 465–479. doi: 10.1111/j.1365-2958.2005.04543.x
Turner, S., Pryer, K. M., Miao, V. P., and Palmer, J. D. (1999). Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol. 46, 327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x
Wang, J., Wang, W., Wang, L., Zhang, G., Fan, K., Tan, H., et al. (2011). A novel role of 'pseudo' γ-butyrolactone receptors in controlling γ-butyrolactone biosynthesis in streptomyces. Mol. Microbiol. 82, 236–250. doi: 10.1111/j.1365-2958.2011.07811.x
Wang, J. B., Zhang, F., Pu, J. Y., Zhao, J., Zhao, Q. F., and Tang, G. L. (2014). Characterization of AvaR1, an autoregulator receptor that negatively controls avermectins production in a high avermectin-producing strain. Biotechnol. Lett. 36, 813–819. doi: 10.1007/s10529-013-1416-y
Wang, L., Tian, X., Wang, J., Yang, H., Fan, K., Xu, G., et al. (2009). Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc. Natl. Acad. Sci. U.S.A. 106, 8617–8622. doi: 10.1073/pnas.0900592106
Wang, L., and Vining, L. C. (2003). Control of growth, secondary metabolism and sporulation in Streptomyces venezuelae ISP5230 by jadW1, a member of the afsA family of γ-butyrolactone regulatory genes. Microbiology 149, 1991–2004. doi: 10.1099/mic.0.26209-0
Wang, W., Ji, J., Li, X., Wang, J., Li, S., Pan, G., et al. (2014). Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. Proc. Natl. Acad. Sci. U.S.A. 111, 5688-5693. doi: 10.1073/pnas.1324253111
Xu, G., Wang, J., Wang, L., Tian, X., Yang, H., Fan, K., et al. (2010). “Pseudo” γ-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J. Biol. Chem. 285, 27440–27448. doi: 10.1074/jbc.M110.143081
Xu, G., and Yang, S. (2019). Regulatory and evolutionary roles of pseudo γ-butyrolactone receptors in antibiotic biosynthesis and resistance. Appl. Microbiol. Biotechnol. 103, 9373–9378. doi: 10.1007/s00253-019-10219-0
Yamamoto, S., He, Y., Arakawa, K., and Kinashi, H. (2008). γ-Butyrolactone-dependent expression of the SARP gene srrY plays a central role in the regulatory cascade leading to lankacidin and lankamycin production in Streptomyces rochei. J. Bacteriol. 190, 1308–1316. doi: 10.1128/JB.01383-07
Yamauchi, Y., Nindita, Y., Hara, K., Umeshiro, A., Yabuuchi, Y., Suzuki, T., et al. (2018). Quinoprotein dehydrogenase functions at the final oxidation step of lankacidin biosynthesis in Streptomyces rochei 7434AN4. J. Biosci. Bioeng. 126, 145–152. doi: 10.1016/j.jbiosc.2018.03.006
Yang, K., Han, L., and Vining, L. C. (1995). Regulation of jadomycin B production in Streptomyces venezuelae ISP5230: involvement of a repressor gene, jadR2. J. Bacteriol. 177, 6111–6117. doi: 10.1128/JB.177.21.6111-6117.1995
Yu, Z., Reichheld, S. E., Savchenko, A., Parkinson, J., and Davidson, A. R. (2010). A comprehensive analysis of structural and sequence conservation in the TetR family transcriptional regulators. J. Mol. Biol. 400, 847–864. doi: 10.1016/j.jmb.2010.05.062
Zerikly, M., and Challis, G. L. (2009). Strategies for the discovery of new natural products by genome mining. ChemBioChem 10, 625–633. doi: 10.1002/cbic.200800389
Zhang, H., Shinkawa, H., Ishikawa, J., Kinashi, H., and Nimi, O. (1997). Improvement of transformation system in Streptomyces using a modified regeneration medium. J. Ferment. Bioeng. 83, 217–221. doi: 10.1016/S0922-338X(97)80982-8
Zhu, J., Sun, D., Liu, W., Chen, Z., Li, J., and Wen, Y. (2016). AvaR2, a pseudo γ-butyrolactone receptor homologue from Streptomyces avermitilis, is a pleiotropic repressor of avermectin and avenolide biosynthesis and cell growth. Mol. Microbiol. 102, 562–578. doi: 10.1111/mmi.13479
Keywords: Streptomyces, regulatory cascade, pseudo-receptor, antibiotic, biosynthesis
Citation: Misaki Y, Yamamoto S, Suzuki T, Iwakuni M, Sasaki H, Takahashi Y, Inada K, Kinashi H and Arakawa K (2020) SrrB, a Pseudo-Receptor Protein, Acts as a Negative Regulator for Lankacidin and Lankamycin Production in Streptomyces rochei. Front. Microbiol. 11:1089. doi: 10.3389/fmicb.2020.01089
Received: 17 December 2019; Accepted: 30 April 2020;
Published: 09 June 2020.
Edited by:
Evi Stegmann, University of Tübingen, GermanyReviewed by:
Yihua Chen, Institute of Microbiology (CAS), ChinaCopyright © 2020 Misaki, Yamamoto, Suzuki, Iwakuni, Sasaki, Takahashi, Inada, Kinashi and Arakawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kenji Arakawa, a2FyYWthd2FAaGlyb3NoaW1hLXUuYWMuanA=
†These authors have contributed equally to this work
†Present address: Shouji Yamamoto, Department of Bacteriology I, National Institute of Infectious Diseases, Tokyo, Japan
Toshihiro Suzuki, Department of Fermentation Science, Faculty of Applied Biosciences, Tokyo University of Agriculture, Tokyo, Japan
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.