- 1Nuclear Agriculture and Biotechnology Division, Bhabha Atomic Research Centre, Mumbai, India
- 2Homi Bhabha National Institute, Mumbai, India
- 3Radiation Biology & Health Sciences Division, Bhabha Atomic Research Centre, Mumbai, India
Trichoderma virens is a commercial biofungicide used in agriculture. We have earlier isolated a mutant of T. virens using gamma ray-induced mutagenesis. This mutant, designated as M7, is defective in morphogenesis, secondary metabolism, and mycoparasitism. The mutant does not produce conidia, and the colony is hydrophilic. M7 cannot utilize cellulose and chitin as a sole carbon source and is unable to parasitize the plant pathogens Rhizoctonia solani and Pythium aphanidermatum in confrontation assay. Several volatile (germacrenes, beta-caryophyllene, alloaromadendrene, gamma-muurolene) and non-volatile (viridin, viridiol, gliovirin, heptelidic acid) metabolites are not detected in M7. In transcriptome analysis, many genes related to secondary metabolism, carbohydrate metabolism, hydrophobicity, and transportation, among others, were found to be downregulated in the mutant. Using whole genome sequencing, we identified five deletions in the mutant genome, totaling about 250 kb (encompassing 71 predicted ORFs), which was confirmed by PCR. This study provides novel insight into genetics of morphogenesis, secondary metabolism, and mycoparasitism and eventually could lead to the identification of novel regulators of beneficial traits in plant beneficial fungi Trichoderma spp. We also suggest that this mutant can be developed as a microbial cell factory for the production of secondary metabolites and proteins.
Introduction
Trichoderma virens is an agriculturally important fungus used for plant growth promotion and biocontrol of pathogens in commercial settings (Mukherjee et al., 2013b)1,2. Two strains of this fungus exist in nature: the “P” strains and the “Q” strains (Howell et al., 1993). Two isolates of T. virens have been widely used for genetic and biocontrol studies: Gv29-8, a “Q” strain, and IMI304061, a “P” strain (Mukhopadhyay et al., 1992; Djonović et al., 2006a, b, 2007a,b; Mukherjee et al., 2006, 2007; Bansal et al., 2018). T. virens is a mycoparasite on many plant pathogenic fungi and also produces several secondary metabolites, both volatile and non-volatile. Among these are gliotoxin (produced by “Q” strains), gliovirin (produced by “P” strains), viridin, viridiol, and heptelidic (koningic) acid, and several volatile sesquiterpenes, including beta-caryophyllene and germacrenes (Mukherjee et al., 2006; Crutcher et al., 2013; Zeilinger et al., 2016; Pachauri et al., 2019b). T. virens is also reported to induce systemic defense responses in plants (Djonović et al., 2007a; Gaderer et al., 2015; Mukherjee et al., 2018).
Whole genome of Gv29-8 and IMI304061 have been sequenced and analyzed, resulting in the identification of biosynthesis gene clusters for many unknown metabolites as well as for gliotoxin, gliovirin, viridin/viridiol, siderophores, and volatile sesquiterpenes (Kubicek et al., 2011; Crutcher et al., 2013; Sherkhane et al., 2017; Bansal et al., 2018; Mukherjee et al., 2018). Many hydrolytic enzymes and signal transduction proteins have been implicated to be involved in mycoparasitism of Trichoderma spp. on plant pathogenic fungi (Haran et al., 1996; Carsolio et al., 1999; Mendoza-Mendoza et al., 2003; Mukherjee et al., 2003; Druzhinina et al., 2011). A few elicitor proteins/peptides have been identified to be involved in triggering of plant defense (Djonović et al., 2006a; Viterbo et al., 2007; Mendoza-Mendoza et al., 2018). T. virens has also been studied for genomic presence and biological role of hydrophobins (Przylucka et al., 2006, 2017; Kubicek et al., 2008; Guzmán-Guzmán et al., 2017; Mendoza-Mendoza et al., 2018). Most of these studies involved bioinformatically identifying candidate genes, obtaining knockout mutants, and comparative phenotyping. Classical mutagenesis followed by complementation had earlier led to the discovery of several novel genes in many fungi (Mattern et al., 1992; Casselton and Zolan, 2002). In recent years, the advancements in whole genome sequencing tools have allowed for identification of mutations in classically induced mutants of fungi (McCluskey et al., 2011). The Trichoderma reesei strain RutC30 that is the major source of industrial cellulases was originally obtained by repeated chemical and physical mutagenesis of the wild type T. reesei strain QM6a. Using whole genome sequence analysis, a large deletion of 100 kb, 15 small deletions or insertions, and 223 single nucleotide variants were identified in RutC30 (Le Crom et al., 2009). We earlier obtained a mutant of T. virens, induced by gamma ray that does not produce conidia or detectable amounts of non-volatile secondary metabolites but has normal vegetative growth and produces excess chlamydospores (Mukherjee et al., 2006). Using SSH and this mutant, we identified a few genes involved in secondary metabolism and conidiation (Mukherjee et al., 2006; Crutcher et al., 2013; Bansal et al., 2019). Here, we describe several novel phenotypes associated with T. virens mutant M7, expand the repertoire of genes that are downregulated in this mutant by RNAseq and, using whole genome sequencing, attempt to identify the alterations in the genome composition of the mutant.
Materials and Methods
Fungal Strains and Growth Conditions
Trichoderma virens (IMI 304061), Pythium aphanidermatum, and Rhizactonia solani (ITCC 4110) were used from our previous studies (Mukherjee et al., 2007). The non-conidiating mutant M7 of T. virens was obtained from our previous study (Mukherjee et al., 2006). The fungal cultures were grown in potato dextrose medium at 28°C and maintained at −80°C for long-term storage.
Test of Hydrophobicity
Wild-type T. virens and M7 mutant were grown for 5 days on PDA at 28°C, and hydrophobicity was tested by adding 0.5% aqueous aniline blue dye or water and recording the disappearance of the drop after 1 h.
Assays for Mycoparasitism
The mycoparasitic ability of wild-type and mutant M7 was determined using confrontation assay by pairing the Trichoderma and the test pathogens simultaneously on the same plate, placed opposite each other. Because T. virens wild-type takes 4–5 days to completely overgrow these test pathogens (Mukherjee et al., 2007), ability of wild-type or mutant to overgrow and lyse the mycelia of P. aphanidermatum and R. solani was recorded after 5 days of co-inoculation. P. aphanidermatum and R. solani cell walls are primarily composed of cellulose and chitin, respectively, and hence, we assessed the ability of wild-type and M7 to utilize chitin and cellulose as a carbon source by growing them in Vogel’s minimal medium containing chitin or cellulose as the sole carbon source in liquid shake culture at 28°C and 150 rpm. For initiation of growth, 0.1% sucrose was added to the medium. All the experiments were conducted in four replicates.
Identification of Secondary Metabolites Produced by Wild-Type and M7
The non-volatile compounds were detected in wild-type T. virens and M7 using thin layer chromatography as described earlier (Mukherjee and Kenerley, 2010). WT and M7 cultures were grown for 3 days in potato dextrose broth and extracted with equal volumes of ethyl acetate. The extract was dried and reconstituted in 0.1 volume methanol, and 100 μl was spotted on precoated TLC plate. TLC plate was developed in acetone:chloroform:formic acid (28:70:2) and visualized under short-wavelength UV (254 nm). The spots were eluted with 100% methanol and subjected to LC-MS/MS analysis as described earlier (Bansal et al., 2018). For confirmation, LC-MS/MS analysis of standard viridin and viridiol (a kind gift from the late Dr. C. R. Howell) and heptelidic acid (Cayman Chemical Company, United States) were also analyzed and matched with the sample.
The head space solid-phase microextraction (HS-SPME) and gas chromatography–mass spectrometry (GC–MS) technique was used for volatile compound detection in wild-type T. virens and M7. The instrument used for analysis was GC–MS 2010 Plus (Shimadzu, Kyoto, Japan). The GC–MS instrument was equipped with an injection port having SPME glass liner (Supelco) and RTX-5 column (5% diphenyl dimethyl polysiloxane, 10 m × 0.1 mm I.D.; Restek Corporation, Bellefonte, PA, United States). The injection port was maintained at 270°C with no solvent cut. Helium was used as a carrier gas. GC column temperature was programmed as follows: 40°C for 5 min and then increased to 200°C at 4°C min–1, held for 5 min and then increased to 280°C at 10°C min–1 with a final hold of 10 min. MS parameters were ionization voltage 70 eV, electron multiplier voltage 1 kV, and scan mode from m/z 35 to 350. Identification of the peaks was done by comparing their mass fragmentation pattern, Kovats retention indices and from the data available in the spectral (Wiley/NIST) libraries of the instrument. The experiments were conducted in three replicates and repeated once.
Transcriptome Analysis
Wild-type and M7 were grown on PDA plates overlaid with dialysis membrane (MWCO 12 kDa) opposite of R. solani for 2 days. For control, wild-type T. virens and M7 were confronted with self. Mycelial mat from the zone of contact was harvested with a sterile spatula and frozen in liquid nitrogen and stored at −80°C until further use. Cultures were grown in four replicates, and samples were pooled before RNA extraction. RNA was extracted with TriReagent, and transcriptome sequencing was performed on Illumina HiSeq 2500 platform at M/S Scigenom, Cochin, Kerala, India, as discussed earlier (Mukherjee et al., 2019). A paired-end library was prepared and sequenced, and Trinity software was used for assembly of the cleaned reads produced by standard data preprocessing methods, including adaptor trimming, quality filtering, and end trimming. Gene expression estimation was performed by aligning the trimmed reads to the assembled transcriptome using Bowtie2 program. DESeq program was employed for differential gene expression analysis, and an in-house pipeline CANoPI (Contig Annotator Pipeline) was used for transcriptome annotation. FPKM values were considered for fold-change calculation, and log2 fold change ≥ 2 was considered for comparison of gene expression.
Whole Genome Sequencing and Analysis
Total genomic DNA was extracted, and whole genome sequencing of the M7 mutant was performed using Illumina platform at M/S Xcelris Genomics, Ahmedabad, India. Illumina TruSeq Nano DNA HT Library Preparation Kit was used for paired end sequencing library preparation. The generated libraries were sequenced on Illumina Nextseq 500 using 2 × 150 bp chemistry. T. virens wild-type sequence (LQCH00000000) was used as a reference genome for M7 genome mapping using a Burrows-Wheeler Aligner (BWA) (v. 0.7.5a) program with optimized mapping parameters. The genome coverage, gene prediction, and total number of single nucleotide polymorphisms (SNPs) and insertion/deletion polymorphisms (INDELs) were analyzed and compared with the reference genome. Absence of the genes in the deleted regions was further confirmed by PCR amplification using gene specific primers (Supplementary Table S1) of representative genes using the following conditions: denaturation at 95°C for 5 min, annealing at 57°C for 45 s, extension at 72°C for 1 min, and 30 cycles.
Results
M7 Mutant Is Deficient in Hydrophobicity in Addition to Conidiation
As reported earlier (Mukherjee et al., 2006), the M7 mutant does not conidiate while wild-type T. virens produces green pigmented conidia (Supplementary Figure S1). The deficiency in conidiation is associated with loss of hydrophobicity. Aqueous aniline blue dye or water drops stay on the wild-type colony for several hours but diffuse through M7 colony immediately after application (Figure 1).
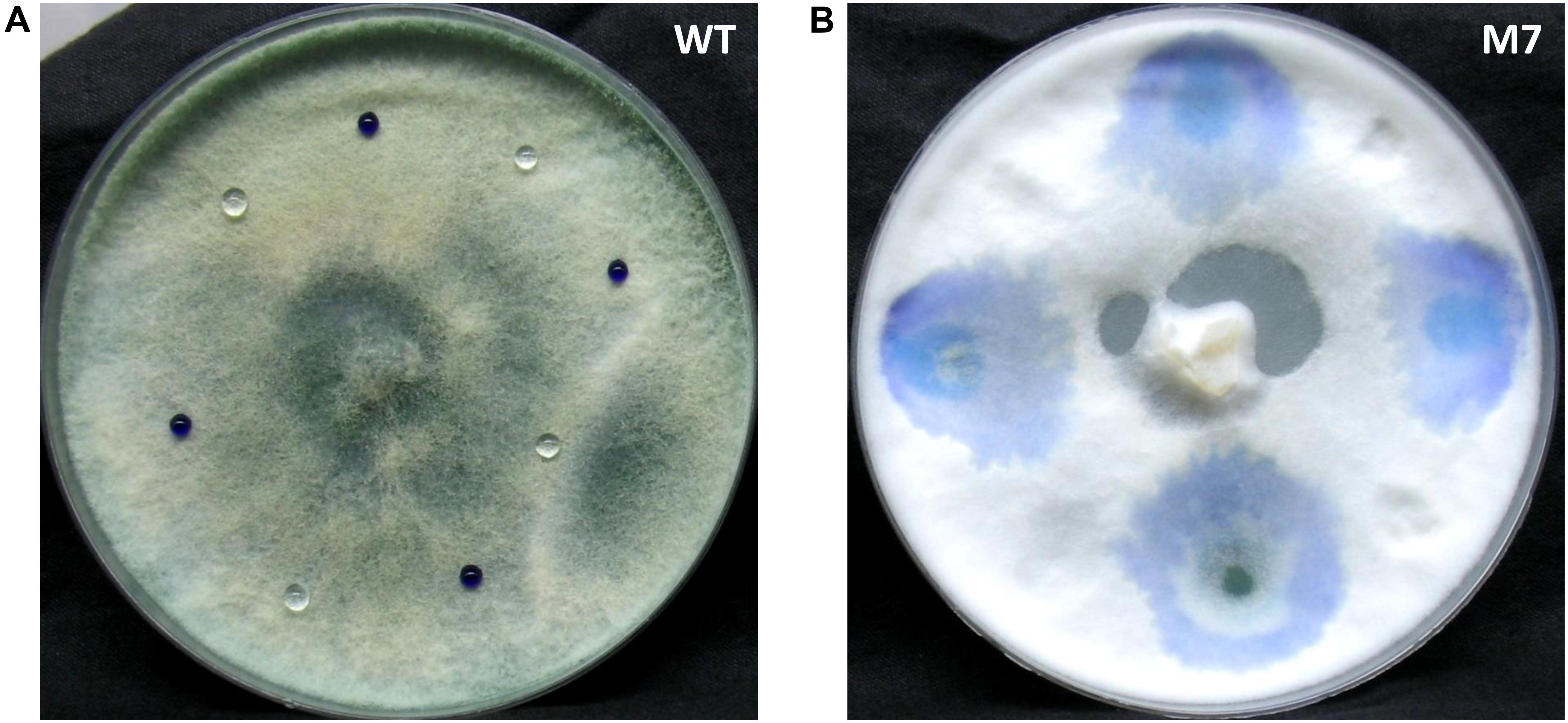
Figure 1. Culture of Trichoderma virens wild-type and M7 with aniline blue dye and water placed on it. (A) Wild-type. (B) M7.
M7 Is Defective in Mycoparasitism
A confrontation assay demonstrated that M7 is not able to overgrow the plant pathogens R. solani and P. aphanidermatum while wild-type T. virens completely overgrew the colonies in 5 days (Figure 2). Although wild-type showed mycoparasitic coiling on R. solani and P. aphanidermatum, M7 failed to do so (Supplementary Figure S2). Moreover, M7 could not utilize chitin and cellulose as a carbon source even when the medium was supplemented with 0.01% sucrose (Supplementary Figure S3).
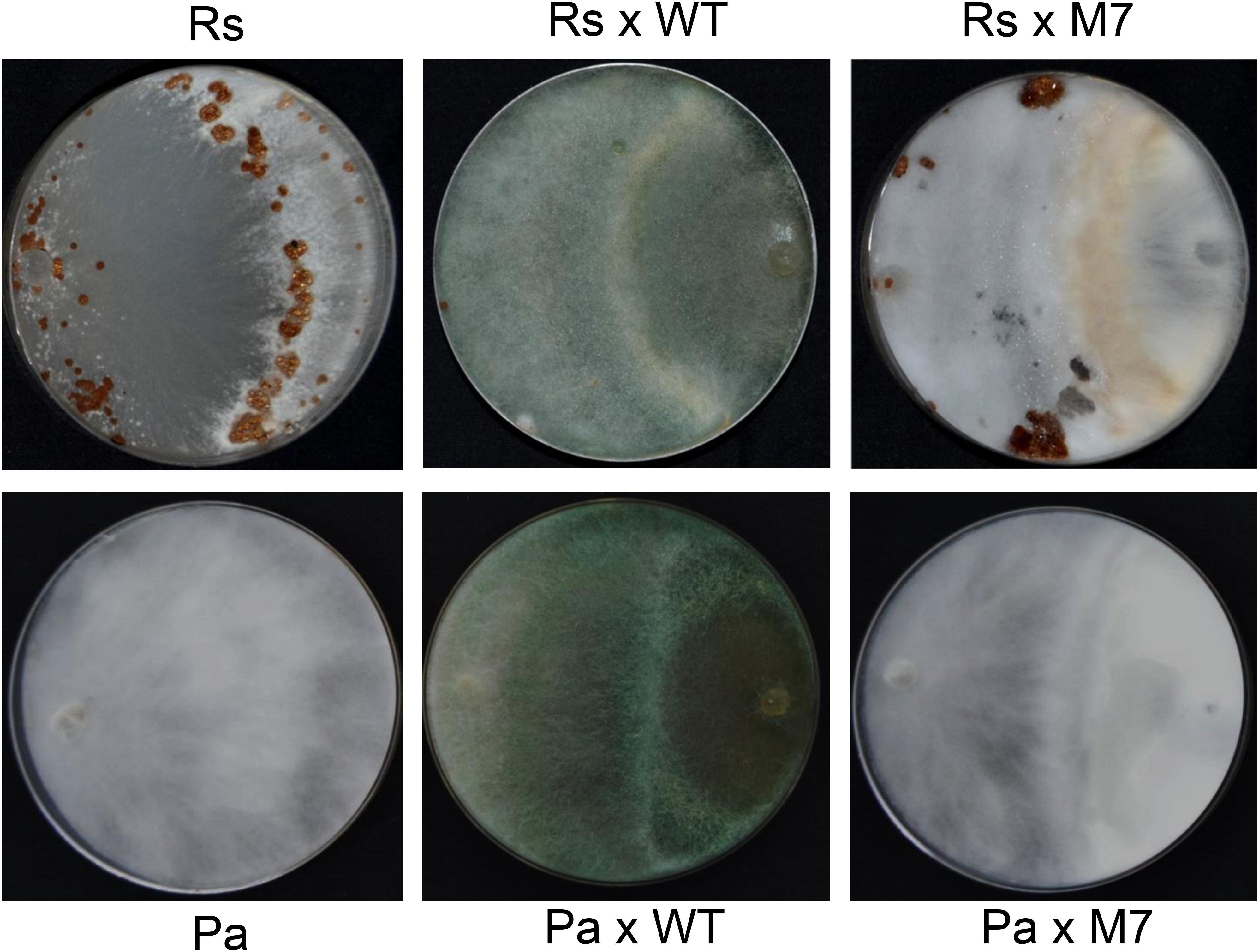
Figure 2. Confrontation assay demonstrating that M7 is non-mycoparasitic in nature as it cannot overgrow and lyse Rhizoctonia solani (Rs) and Pythium aphanidermatum (Pa) after 5 days of co-inoculation, and wild-type (WT) T. virens is able to overgrow the test pathogens.
M7 Is Downregulated in the Biosynthesis of Secondary Metabolites
The TLC profile of wild-type and M7 culture filtrate shows that none of the non-volatile metabolites, including viridin, viridiol, and heptelidic acid, are detected in M7 (Figure 3A). Similarly, GC–MS analysis of head-space gas shows that M7 is downregulated in biosynthesis of volatile metabolites that are present in wild-type T. virens; only 13 of 73 metabolites being detected in M7 mutant (Figure 3B and Supplementary Table S2).
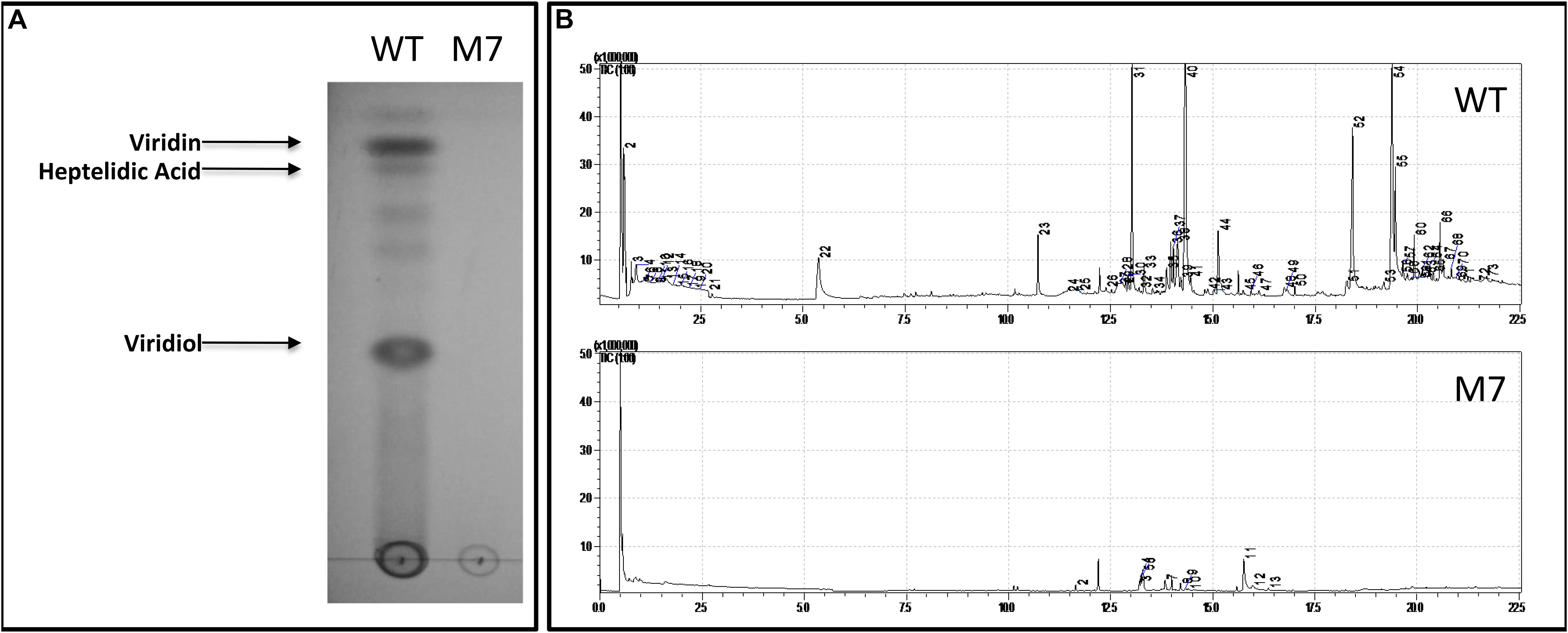
Figure 3. Secondary metabolite profile of T. virens wild-type (WT) and M7. (A) Thin layer chromatography analysis of WT and M7 filtrate. (B) GC–MS analysis of WT and M7 head-space gas. The peak numbers correspond to the volatile metabolites detected as described in Supplementary Table S2.
Several Genes for Secondary Metabolism, Hydrolytic Enzymes, and Hydrophobicity Are Downregulated in M7
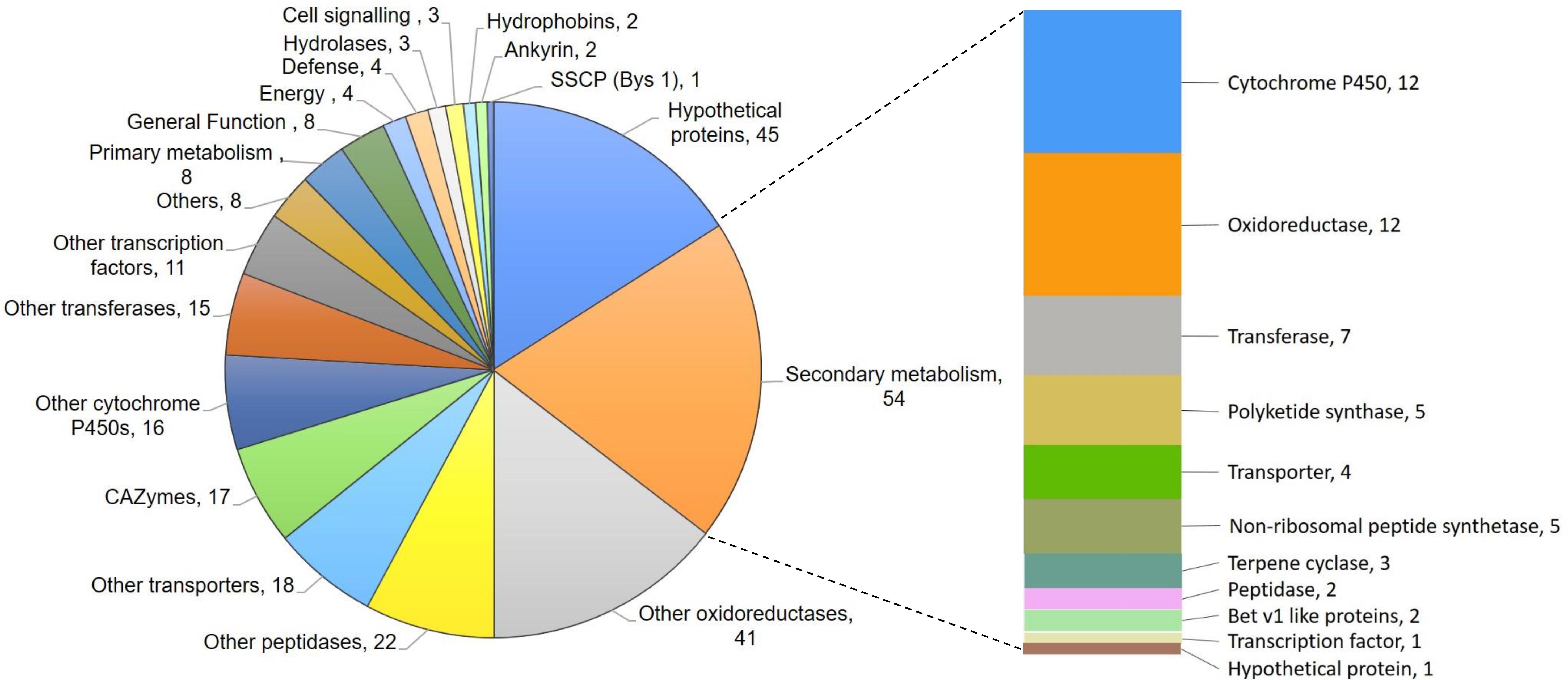
De novo transcriptome analysis of wild-type and M7, while interacting with self or R. solani (Supplementary Figure S4), indicated that as many as 463 genes are downregulated in M7 (Supplementary Tables S3, S4, Supplementary Figures S5, S6, and Figures 4, 5). In self-confrontation, 294 genes were downregulated in M7 compared to wild-type (Supplementary Table S3). Among these are genes for secondary metabolism (including genes coding for NRPSs, PKSs, and terpene cyclases, modifying enzymes, transporters, and transcription factors), other cytochrome P450s (not associated with secondary metabolism gene clusters), carbohydrate-active enzymes (CAZymes), peptidases, and hydrophobins (Figure 4). In confrontation with the plant pathogen R. solani, a larger set of genes (375) were downregulated in M7, 206 genes being common under both the conditions (Supplementary Table S4 and Supplementary Figure S7). A large number of genes involved in secondary metabolism (69), carbohydrate utilization (31), and transporters (30) are downregulated in M7 in confrontation with R. solani (Figure 5). This is in agreement with the fact that M7 does not produce detectable amount of secondary metabolites and also does not parasitize the plant pathogens P. aphanidermatum and R. solani. Six of the genes (protein IDs. TRIVIDRAFT_49849, TRIVIDRAFT_74289, TRIVIDRAFT_74291, TRIVIDRAFT_56195, TRIVIDRAFT_23 0740, TRIVIDRAFT_53375) that are downregulated in M7 were identified in our earlier study as under-expressed in M7 in RT-PCR analysis (Mukherjee et al., 2006).
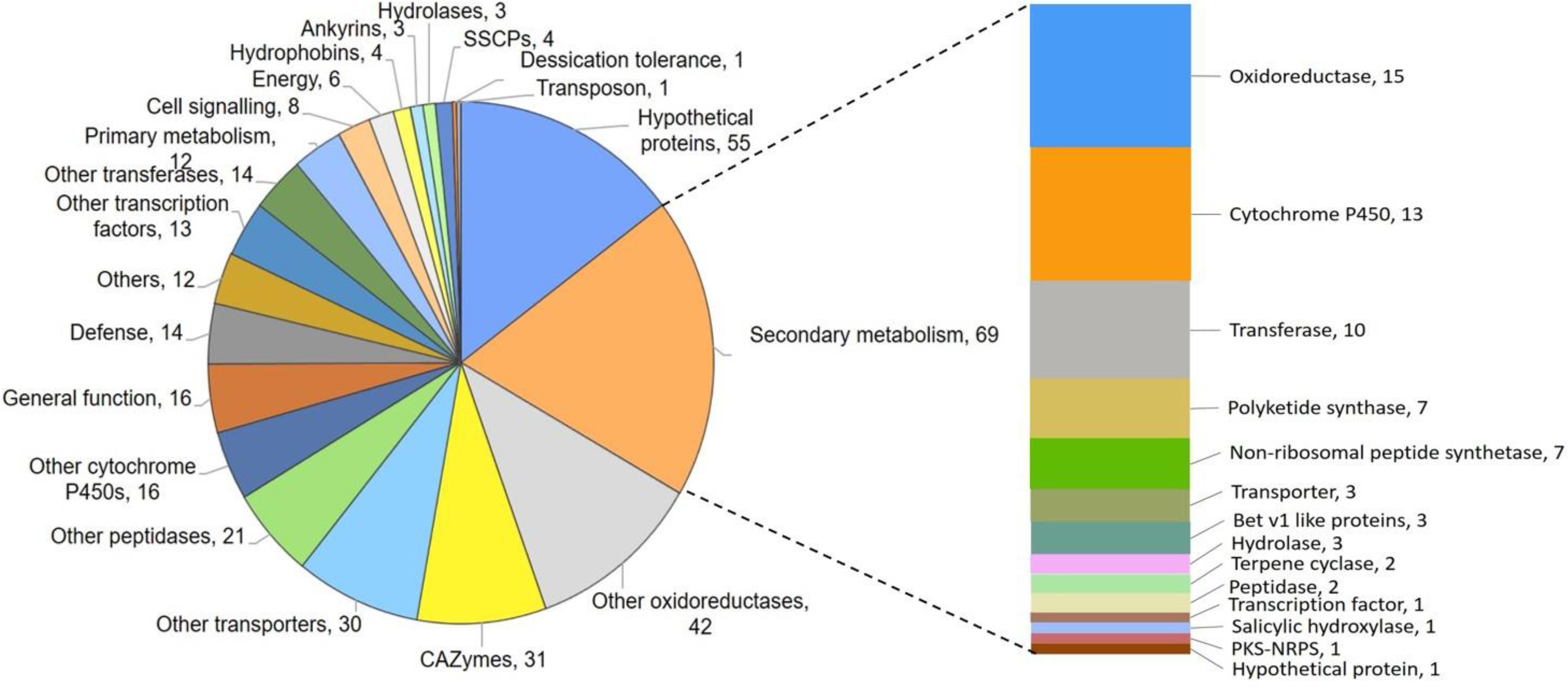
Figure 5. Pie chart representing genes downregulated in M7 in confrontation with Rhizoctonia solani.
M7 Genome Has Large Deletions
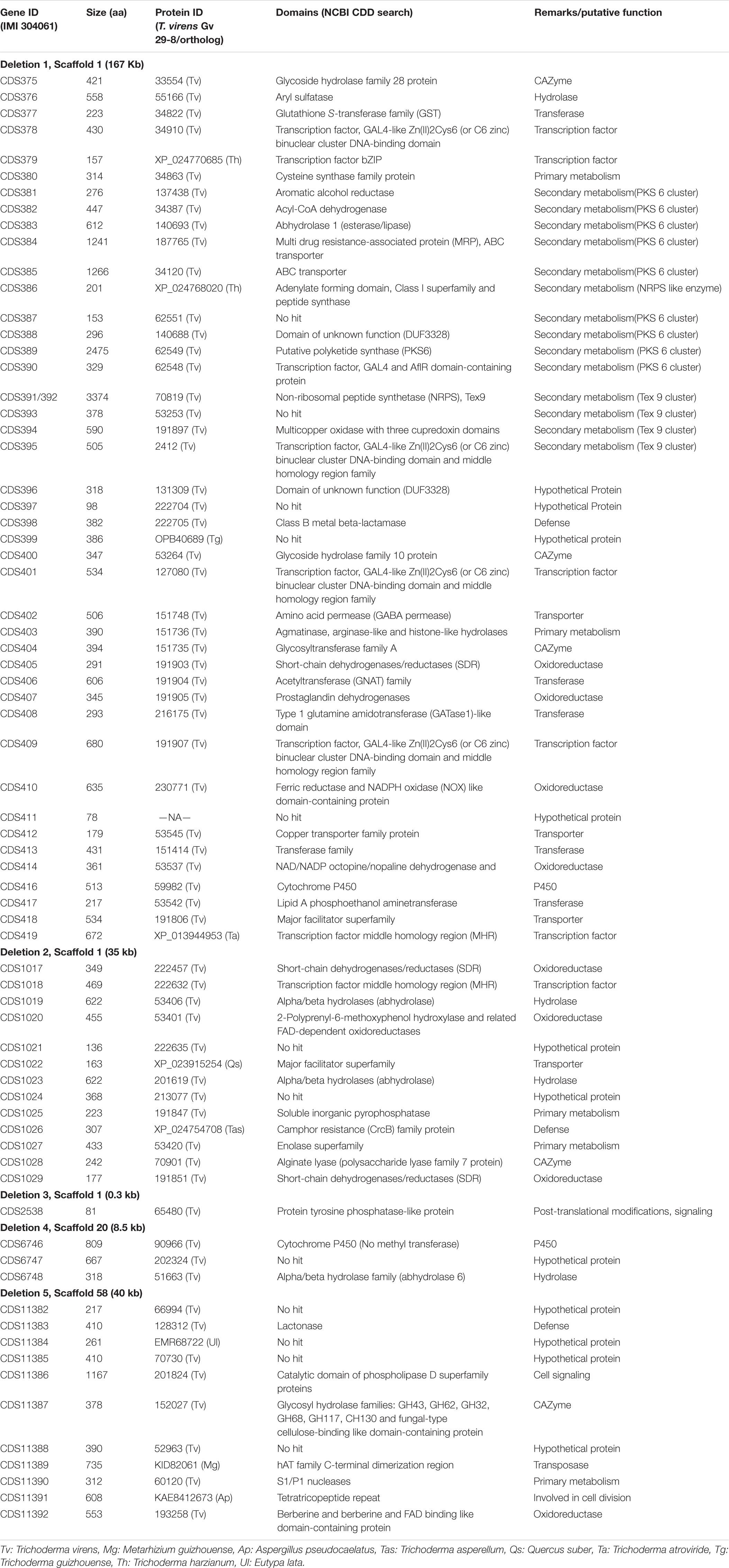
Whole genome sequencing of the M7 mutant was performed using the Illumina platform to generate 2.1 GB data. The paired end sequencing library was of mean size 624 bp. The generated library was sequenced and mapped to T. virens wild-type assembly as a reference genome. A genome alignment revealed five deletions across three scaffolds (scaffold 1, 20, and 58) totaling about 250 Kb, comprising 71 genes (Table 1). Among the genes that are absent in the M7 genome are two large secondary metabolism gene clusters (PKS6 and Tex9 clusters), eight transcription factors (two associated with secondary metabolism gene clusters), five genes for carbohydrate metabolism, two genes related to cell signaling and as many as 11 oxidoreductases (Table 1 and Supplementary Figure S8). To further confirm these deletions, PCR of representative genes was performed using wild-type and M7 genomic DNA as template. No amplification of these genes was obtained in M7, confirming the gene deletions (Figure 6). In addition, 118 SNPs and 57 small INDELs were also detected in M7 (Supplementary Table S5).
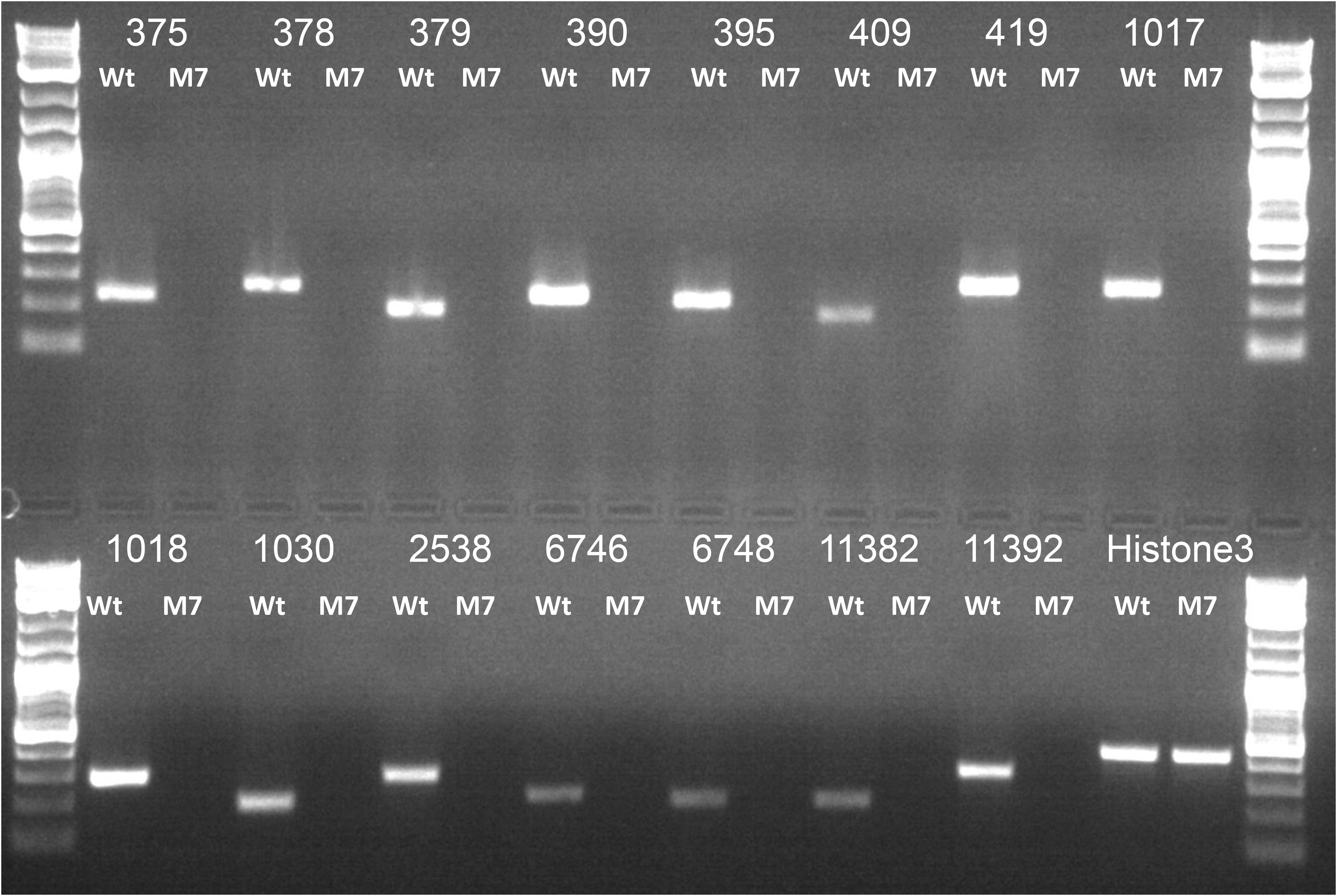
Figure 6. PCR analysis of representative genes that are deleted in M7, using genomic DNA as template. Histone 3 was used as a positive control. The number corresponds to gene IDs as per Table 1. Ladder: Quick-Load Purple 1 kb Plus DNA Ladder (NEB).
Discussion
Trichoderma spp. are commercially important filamentous fungi and are used extensively in agriculture (Lorito et al., 2010; Mukherjee et al., 2013a). We had earlier isolated a non-conidiating mutant of the biocontrol agent T. virens that is also downregulated in secondary metabolite biosynthesis (Mukherjee et al., 2006). In the present study, we provide evidence that the mutant also lacks hydrophobicity. Hydrophobicity is imparted in fungi by small secreted cysteine-rich proteins known as hydrophobins. One hydrophobin (Trividraft_49849) was reported earlier to be downregulated in M7 (Mukherjee et al., 2006). In the transcriptome data, we noted one additional hydrophobin to be downregulated in the mutant. Hydrophobins are associated with conidiation (Wessels, 1996; Bayry et al., 2012) and loss of hydrophobicity in this non-conidiating mutant is not unexpected. Trichoderma hydrophobins also play important roles in attachment to root (Viterbo and Chet, 2006), elicitation of plant defense (Zhang et al., 2019), and tolerance to biotic and abiotic stresses (Przylucka et al., 2017). Hydrophobins are also involved in mycoparasitic interactions in Trichoderma (Guzmán-Guzmán et al., 2017). We have noticed two additional hydrophobins to be downregulated in the mutant in interaction with R. solani (Supplementary Table S4). These hydrophobins might, thus, be involved in mycoparasitism of T. virens on R. solani.
Trichoderma virens is an aggressive mycoparasite on many plant pathogenic fungi (Druzhinina et al., 2011). The mutant, however, has lost this property (Figure 2 and Supplementary Figure S2), which can be explained by the lack of upregulation of a large number of CAZymes known to be involved in parasitism on other fungi (Haran et al., 1996; Carsolio et al., 1999; Djonović et al., 2006b, 2007b). CAZymes of glycoside hydrolase 2 (GH-2), GH-3, GH-18, GH-30, GH-47, GH-55, GH-75, and GH-92 families were downregulated in M7. Furthermore, the downregulation of CAZymes of GH-16, GH-20, GH-78, and GH-79 families only during confrontation with R. solani suggests the role of these CAZymes in mycoparasitism. Atanasova et al. (2013a) reported upregulation of many gene families, such as those involved in metabolism, transporters, signal transduction, transcriptional regulators, defense, etc., in T. virens during confrontation with R. solani. Similarly, in our study, we have detected downregulation of different groups of genes involved in secondary metabolism (69), genes for oxidoreductases (42), CAZymes (31), peptidases (21), transporters (30), transcription factors (13), transferases (14), and defense-related genes (14) in T. virens during confrontation with R. solani. Approximately twice the number of genes for transporters (30) and carbohydrate utilization groups (31) were downregulated in M7 during confrontation with R. solani as compared to confrontation with self, where only 18 genes for transporter and 17 for carbohydrate utilization were downregulated (Figure 4). These results indicate that the mutant, which cannot parasitize the host fungus R. solani, fails to respond to its presence. The role of most of these genes in mycoparasitism is not known, and further studies on the role of these genes could throw novel insights into the phenomenon of fungus–fungus interactions.
Trichoderma spp. produce a large number of useful secondary metabolites with diverse biological activities (Zeilinger et al., 2016; Pachauri et al., 2019b). A large number of genes for secondary metabolism (69) were downregulated in M7 during confrontation with R. solani. Viridin is one of the oldest known metabolites produced by T. virens possessing anti-cancer and anti-fungal activities, and viridiol is a phytotoxic agent produced by irreversible enzymatic conversion of viridin (Jones and Hancock, 1987; Howell et al., 1993). Gliovirin is a ‘P’ strain-specific non-ribosomal peptide with anti-oomycete activities (Howell and Stipanovic, 1983). Non-volatile compounds, such as viridin, viridiol, and heptelidic acid, were detected by TLC followed by LC-MS/MS analysis, and gliovirin was detected by LC-MS in T. virens wild-type (data not presented); the mutant does not produce these metabolites. In addition, M7 is downregulated in biosynthesis of volatile organic compounds, including volatile sesquiterpenes, such as germacrenes, caryphyllene, alloaromadendrene, and gamma-muurolene, all having important biological activities (Figure 3 and Supplementary Table S2). Volatile compounds produced by Trichoderma spp. promote plant growth and have antimicrobial effects against plant pathogens (Vinale et al., 2008; Hung et al., 2013). In transcriptome analysis, many secondary metabolism-related genes and gene clusters were downregulated in M7 (Supplementary Tables S3, S4). The viridin biosynthesis gene cluster comprises of 21 genes (Bansal et al., 2018), out of which, 16 genes were detected to be downregulated in M7. Similarly, the gliovirin biosynthetic gene cluster has 22 genes (Sherkhane et al., 2017), of which 16 genes were detected to be downregulated in M7. All the eight genes from the “vir” cluster, which are associated with volatile sesquiterpene compound production (Crutcher et al., 2013; Pachauri et al., 2019a), were downregulated in M7. In addition, eight NRPSs, one NRPS/PKS, seven PKSs, and three terpene cyclase genes were downregulated in M7. The NRPS genes that are downregulated in transcriptome analysis of M7 are Tex1, Tex2, Tex5, Tex7, Tex21, Glv21, NRPS (Trividraft 50383), NRPS-like (Trividraft_222800) (NRPSs), and Tex14 (NRPS/PKS). In addition, certain genes belonging to the Tex6, Tex8, Tex10 (NRPSs), and Tex12 (NRPS/PKS) clusters were also downregulated in M7. Tex1 and Tex2 code for peptaibol synthetase (Wiest et al., 2002; Mukherjee et al., 2011). Tex10 is involved in the biosynthesis of intracellular siderophore ferricrocin while Tex21 is responsible for the biosynthesis of extracellular siderophores (Mukherjee et al., 2018). The metabolites produced by Tex5 and Tex7-NRPS genes and Tex12 and Tex14-NRPS/PKS hybrid genes are not known. Of these NRPS genes, Tex5, Tex14, and Tex21 are downregulated in M7 during confrontation with R. solani (Supplementary Table S4). Seven PKS genes, namely PKS4, PKS8, PKS14, and PKS17, one un-annotated PKS (Trividraft_53518), and two orthologs of Arthroderma benhamiae (Acc. Nos. DAA76265 and EGE05473) PKSs were downregulated in M7 during confrontation with R. solani. The biosynthetic products of none of these PKS genes are known. In addition, some members of PKS3 and PKS6 gene clusters were also found to be downregulated in M7. PKS4 is an ortholog of pigment-forming PKS from T. reesei responsible for green conidial pigment biosynthesis and is involved in mechanical stability and stress tolerance (Atanasova et al., 2013a). Because M7 does not produce conidia, it is not surprising that the pigment PKS4 gene is downregulated in M7. In addition to PKS4, another gene (con6) that is associated with conidiation in Neurospora crassa (White and Yanofsky, 1993) was also downregulated in M7.
Having noted that the mutant M7 has a deficiency in conidiation, hydrophobicity, secondary metabolism, and mycoparasitism (all traits that dictate the success of Trichoderma spp. as plant beneficial fungi), which was apparent from the phenotyping and transcriptome data analysis, we did a whole genome sequence analysis of the mutant vis-à-vis the wild-type strain. It was interesting to note that the M7 genome has five deletions comprising as many as 71 genes. Because the mutant was generated using gamma ray-induced mutagenesis, deletions may be expected. Two large secondary metabolism-related gene clusters (PKS6 and Tex9) are deleted in the mutant. Four genes for glycoside hydrolase (GH) are also deleted, and so are several oxidoreductases. Because the data set of genes that are under-expressed in M7 is much larger that the number of genes that are deleted in the genome, it is likely that one or a few of these genes that are deleted or affected due to other mutations (Table 1 and Supplementary Table S5) could have a pleiotropic phenotype regulating a big set of genes. Our findings identify possible candidates for future research leading to identification of master regulator gene(s) in Trichoderma, a biotechnologically important fungal genus. Also, because the mutant is under-regulated in secondary metabolism and a large number of carbohydrate active enzymes, it could be possible to develop this mutant as a microbial cell factory for production of secondary metabolites and proteins.
We have recently reported the isolation and characterization of a mutant (G2) of T. virens that is upregulated in secondary metabolite biosynthesis (Mukherjee et al., 2019). A total of 140 genes were found to be upregulated in G2 over wild-type when grown on PDA medium. Of these, as many as 45 genes are downregulated in M7, including genes for CAZymes (13), secondary metabolism (5), Cytochrome P450 (3), SSCPs (2), oxidoreductase (3), peptidases (3), metabolism (5), hydrophobins (3), hypothetical proteins (5), transporter (1), and others (2). G2 and M7, thus, seem to be two contrasting “gene-regulation” mutants of T. virens. Although G2 has been developed as an improved biocontrol formulation (Mukherjee et al., 2019), M7 can serve as an excellent genetic tool for understanding Trichoderma biology.
Conclusion
We have identified a mutant of the plant beneficial fungus T. virens that is downregulated in conidiation, hydrophobicity, secondary metabolism, carbohydrate metabolism, and mycoparasitism. These traits are important for development of formulations, enzyme production, plant interactions, and biocontrol applications. By transcriptome analysis, we have also identified novel candidate genes for future research. We have also been able to identify 71 ORFs that are deleted in the mutant, which could be further studied for a role as novel regulator(s) of morphogenesis, secondary metabolism, and biocontrol properties in these plant beneficial fungal bioagents.
Data Availability Statement
The datasets generated for this study can be found in the attached Supplementary Material. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JABENJ000000000. The version described in this manuscript is version JABENJ010000000.
Author Contributions
SP and PS performed the laboratory experiments. PM and VK conceptualized, designed, and coordinated the studies. SP and PM analyzed the data. SP, PM, and VK wrote the manuscript.
Funding
SP was supported by a fellowship from the Department of Atomic Energy, Government of India.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
PM thanks Dr. P. Suprasanna, Head, NA&BT Division and Dr. V. P. Venugopalan, Associate Director, Bioscience Group, BARC, for encouragement and support. We would like to acknowledge Dr. Sumit Gupta, Food Technology Division, BARC for helping us in GC–MS analysis and Dr. Kaushik Banerjee, NRC-Grapes, Pune, for LC-MS/MS analysis of the metabolites.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01030/full#supplementary-material
Footnotes
- ^ https://www.certisusa.com/pest_management_products/biofungicides/soilgard_12g_microbial_fungicide
- ^ http://www.acceleronsas.com/products/Pages/QuickRoots-Corn.aspx
References
Atanasova, L., Crom, S. L., Gruber, S., Coulpier, F., Seidl-Seiboth, V., Kubicek, C. P., et al. (2013a). Comparative transcriptomics reveals different strategies of Trichoderma mycoparasitism. BMC Genomics 14:121. doi: 10.1186/1471-2164-14-121
Atanasova, L., Knox, B. P., Kubicek, C. P., Druzhinina, I. S., and Baker, S. E. (2013b). The polyketide synthase gene pks4 of Trichoderma reesei provides pigmentation and stress resistance. Eukaryot. Cell 12, 1499–1508. doi: 10.1128/EC.00103-13
Bansal, R., Mukherjee, M., Horwitz, B. A., and Mukherjee, P. K. (2019). Regulation of conidiation and antagonistic properties of the soil-borne plant beneficial fungus Trichoderma virens by a novel proline-, glycine-, tyrosine-rich protein and a GPI-anchored cell wall protein. Curr. Genet. 65, 953–964. doi: 10.1007/s00294-019-00948-0
Bansal, R., Sherkhane, P. D., Oulkar, D., Khan, Z., Banerjee, K., and Mukherjee, P. K. (2018). The viridin biosynthesis gene cluster of Trichoderma virens and its conservancy in the bat white-nose fungus Pseudogymnoascus destructans. Chem. Select. 3, 1289–1293. doi: 10.1002/slct.201703035
Bayry, J., Aimanianda, V., Guijarro, J. I., Sunde, M., and Latgé, J. P. (2012). Hydrophobins-unique fungal proteins. PLoS Pathog. 8:e1002700. doi: 10.1371/journal.ppat.1002700
Carsolio, C., Benhamou, N., Haran, S., Cortés, C., Gutierrez, A., Chet, I., et al. (1999). Role of the Trichoderma harzianum endochitinase gene, ech42, in mycoparasitism. Appl. Environ. Microbiol. 65, 929–935. doi: 10.1128/aem.65.3.929-935.1999
Casselton, L., and Zolan, M. (2002). The art and design of genetic screens: Filamentous fungi. Nat. Rev. Genet. 3, 683–697. doi: 10.1038/nrg889
Crutcher, F. K., Parich, A., Schuhmacher, R., Mukherjee, P. K., Zeilinger, S., and Kenerley, C. M. (2013). A putative terpene cyclase, vir4, is responsible for the biosynthesis of volatile terpene compounds in the biocontrol fungus Trichoderma virens. Fungal Genet. Biol. 56, 67–77. doi: 10.1016/j.fgb.2013.05.003
Djonović, S., Pozo, M. J., Dangott, L. J., Howell, C. R., and Kenerley, C. M. (2006a). Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol. Plant Microbe Interact. 19, 838–853. doi: 10.1094/mpmi-19-0838
Djonović, S., Pozo, M. J., and Kenerley, C. M. (2006b). Tvbgn3, a β-1,6-glucanase from the biocontrol fungus Trichoderma virens, is involved in mycoparasitism and control of Pythium ultimum. Appl. Environ. Microbiol. 72, 7661–7670. doi: 10.1128/aem.01607-06
Djonović, S., Vargas, W., Kolomiets, M., Horndeski, M., Wiest, A., and Kenerley, C. (2007a). A Proteinaceous Elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in Maize. Plant Physiol. 145, 875–889. doi: 10.1104/pp.107.103689
Djonović, S., Vittone, G., Mendoza-Herrera, A., and Kenerley, C. M. (2007b). Enhanced biocontrol activity of Trichoderma virens transformants constitutively coexpressing β-1,3- and β-1,6-glucanase genes. Mol. Plant Pathol. 8, 469–480. doi: 10.1111/j.1364-3703.2007.00407.x
Druzhinina, I. S., Seidl-Seiboth, V., Herrera-Estrella, A., Horwitz, B. A., Kenerley, C. M., Monte, E., et al. (2011). Trichoderma: the genomics of opportunistic success. Nat. Rev. Microbiol. 9, 749–759. doi: 10.1038/nrmicro2637
Gaderer, R., Lamdan, N. L., Frischmann, A., Sulyok, M., Krska, R., Horwitz, B. A., et al. (2015). Sm2, a paralog of the Trichoderma cerato-platanin elicitor Sm1, is also highly important for plant protection conferred by the fungal-root interaction of Trichoderma with maize. BMC Microbiol. 15:2. doi: 10.1186/s12866-014-0333-0
Guzmán-Guzmán, P., Alemán-Duarte, M. I., Delaye, L., Herrera-Estrella, A., and Olmedo-Monfil, V. (2017). Identification of effector-like proteins in Trichoderma spp. and role of a hydrophobin in the plant-fungus interaction and mycoparasitism. BMC Genet. 18:16. doi: 10.1186/s12863-017-0481-y
Haran, S., Schickler, H., and Chet, I. (1996). Molecular mechanisms of lytic enzymes involved in the biocontrol activity of Trichoderma harzianum. Microbiology 142, 2321–2331. doi: 10.1099/00221287-142-9-2321
Howell, C. R., and Stipanovic, R. D. (1983). Gliovirin, a new antibiotic from Gliocladium virens, and its role in the biological control of Pythium ultimum. Can. J. Microbiol. 29, 321–324. doi: 10.1139/m83-053
Howell, C. R., Stipanovic, R. D., and Lumsden, R. D. (1993). Antibiotic production by strains of Gliocladium virens and its relation to the biocontrol of cotton seedling diseases. Biocontrol Sci. Techn. 3, 435–441. doi: 10.1080/09583159309355298
Hung, R., Lee, S., and Bennett, J. W. (2013). Arabidopsis thaliana as a model system for testing the effect of Trichoderma volatile organic compounds. Fungal Ecol. 6, 19–26. doi: 10.1016/j.funeco.2012.09.005
Jones, R. W., and Hancock, J. G. (1987). Conversion of viridin to viridiol by viridin-producing fungi. Can. J. Microbiol. 33, 963–966. doi: 10.1139/m87-169
Kubicek, C. P., Baker, S., Gamauf, C., Kenerley, C. M., and Druzhinina, I. S. (2008). Purifying selection and birth-and-death evolution in the class II hydrophobin gene families of the ascomycete Trichoderma/Hypocrea. BMC Evol. Biol. 8:4. doi: 10.1186/1471-2148-8-4
Kubicek, C. P., Herrera-Estrella, A., Seidl-Seiboth, V., Martinez, D. A., Druzhinina, I. S., Thon, M., et al. (2011). Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 12:R40. doi: 10.1186/gb-2011-12-4-r40
Le Crom, S., Schackwitz, W., Pennacchio, L., Magnuson, J. K., Culley, D. E., Collett, J. R., et al. (2009). Tracking the roots of cellulase hyperproduction by the fungus Trichoderma reesei using massively parallel DNA sequencing. PNAS 106, 16151–16156. doi: 10.1073/pnas.0905848106
Lorito, M., Woo, S. L., Harman, G. E., and Monte, E. (2010). Translational research on Trichoderma: from ’Omics to the Field. Annu. Rev. Phytopathol. 48, 395–417. doi: 10.1146/annurev-phyto-073009-114314
Mattern, I. E., van Noort, J. M., van den Berg, P., Archer, D. B., Roberts, I. N., and van den Hondel, C. A. M. J. J. (1992). Isolation and characterization of mutants of Aspergillus niger deficient in extracellular proteases. Mol. Gen. Genet. 234, 332–336. doi: 10.1007/bf00283855
McCluskey, K., Wiest, A. E., Grigoriev, I. V., Lipzen, A., Martin, J., Schackwitz, W., et al. (2011). Rediscovery by whole genome sequencing: classical mutations and genome polymorphisms in Neurospora crassa. G3 1, 303–316. doi: 10.1534/g3.111.000307
Mendoza-Mendoza, A., Pozo, M. J., Grzegorski, D., Martínez, P., García, J. M., Olmedo-Monfil, V., et al. (2003). Enhanced biocontrol activity of Trichoderma through inactivation of a mitogen-activated protein kinase. PNAS 100, 15965–15970. doi: 10.1073/pnas.2136716100
Mendoza-Mendoza, A., Zaid, R., Lawry, R., Hermosa, R., Monte, E., Horwitz, B. A., et al. (2018). Molecular dialogues between Trichoderma and roots: role of the fungal secretome. Fungal Biol. Rev. 32, 62–85. doi: 10.1016/j.fbr.2017.12.001
Mukherjee, M., Horwitz, B. A., Sherkhane, P. D., Hadar, R., and Mukherjee, P. K. (2006). A secondary metabolite biosynthesis cluster in Trichoderma virens: evidence from analysis of genes underexpressed in a mutant defective in morphogenesis and antibiotic production. Curr. Genet. 50, 193–202. doi: 10.1007/s00294-006-0075-0
Mukherjee, P. K., and Kenerley, C. M. (2010). Regulation of morphogenesis and biocontrol properties in Trichoderma virens by a VELVET protein, Vel1. Appl. Environ. Microbiol. 76, 2345–2352. doi: 10.1128/AEM.02391-09
Mukherjee, M., Mukherjee, P. K., and Kale, S. P. (2007). cAMP signalling is involved in growth, germination, mycoparasitism and secondary metabolism in Trichoderma virens. Microbiology 153, 1734–1742. doi: 10.1099/mic.0.2007/005702-0
Mukherjee, P. K., Horwitz, B. A., Herrera-Estrella, A., Schmoll, M., and Kenerley, C. M. (2013a). Trichoderma research in the genome era. Annu. Rev. Phytopathol. 51, 105–129.
Mukherjee, P. K., Horwitz, B. A., Singh, U. S., Mukherjee, M., and Schmoll, M. (eds) (2013b). Trichoderma Biology and Applications. Wallingford: CABI.
Mukherjee, P. K., Hurley, J. F., Taylor, J. T., Puckhaber, L., Lehner, S., Druzhinina, I., et al. (2018). Ferricrocin, the intracellular siderophore of Trichoderma virens, is involved in growth, conidiation, gliotoxin biosynthesis and induction of systemic resistance in maize. Biochem. Biophys. Res. Commun. 505, 606–611. doi: 10.1016/j.bbrc.2018.09.170
Mukherjee, P. K., Latha, J., Hadar, R., and Horwitz, B. A. (2003). TmkA, a mitogen-activated protein kinase of Trichoderma virens, is involved in biocontrol properties and repression of conidiation in the dark. Eukaryot. Cell. 2, 446–455. doi: 10.1128/ec.2.3.446-455.2003
Mukherjee, P. K., Mehetre, S. T., Sherkhane, P. D., Muthukathan, G., Ghosh, A., Kotasthane, A. S., et al. (2019). A novel seed-dressing formulation based on an improved mutant strain of Trichoderma virens, and its field evaluation. Front. Microbiol. 10:1910. doi: 10.3389/fmicb.2019.01910
Mukherjee, P. K., Wiest, A., Ruiz, N., Keightley, A., Moran-Diez, M. E., McCluskey, K., et al. (2011). Two classes of new peptaibols are synthesized by a single non-ribosomal peptide synthetase of Trichoderma virens. J. Biol. Chem. 286, 4544–4554. doi: 10.1074/jbc.M110.159723
Mukhopadhyay, A. N., Shrestha, S. M., and Mukherjee, P. K. (1992). Biological seed treatment for control of soil-borne plant pathogens. FAO Plant Prot. Bull. 40, 21–30.
Pachauri, S., Chatterjee, S., Kumar, V., and Mukherjee, P. K. (2019a). A dedicated glyceraldehyde-3-phosphate dehydrogenase is involved in the biosynthesis of volatile sesquiterpenes in Trichoderma virens—evidence for the role of a fungal GAPDH in secondary metabolism. Curr Genet. 65, 243–252. doi: 10.1007/s00294-018-0868-y
Pachauri, S., Sherkhane, P., and Mukherjee, P. K. (2019b). “Secondary metabolism in trichoderma: chemo- and geno-diversity” in Microbial Diversity in Ecosystem Sustainability and Biotechnological Applications, eds T. Satyanarayana, S. K. Das, and B. N. Johri (Berlin: Springer Nature)Google Scholar
Przylucka, A., Akcapinar, G. B., Chenthamara, K., Cai, F., Grujic, M., Karpenko, J., et al. (2006). Multiple roles and effects of a novel Trichoderma hydrophobin. Mol. Plant Pathol. 7, 249–258. doi: 10.1094/MPMI-07-14-0194-R
Przylucka, A., Akcapinar, G. B., Chenthamara, K., Cai, F., Grujic, M., Karpenko, J., et al. (2017). HFB7 – A novel orphan hydrophobin of the Harzianum and Virens clades of Trichoderma, is involved in response to biotic and abiotic stresses. Fungal Genet. Biol. 102, 63–76. doi: 10.1016/j.fgb.2017.01.002
Sherkhane, P. D., Bansal, R., Banerjee, K., Chatterjee, S., Oulkar, D., Jain, P., et al. (2017). Genomics-driven discovery of the gliovirin biosynthesis gene cluster in the plant beneficial fungus Trichoderma virens. ChemistrySelect 2, 3347–3352. doi: 10.1002/slct.201700262
Vinale, F., Sivasithamparam, K., Ghisalberti, E. L., Marra, R., Woo, S. L., and Lorito, M. (2008). Trichoderma–plant–pathogen interactions. Soil Biol. Biochem. 40, 1–10.
Viterbo, A., and Chet, I. (2006). TasHyd1, a new hydrophobin gene from the biocontrol agent Trichoderma asperellum, is involved in plant root colonization. Mol. Plant Pathol. 7, 249–258. doi: 10.1111/j.1364-3703.2006.00335.x
Viterbo, A., Wiest, A., Brotman, Y., Chet, I., and Kenerley, C. (2007). The 18mer peptaibols from Trichoderma virens elicit plant defence responses. Mol Plant Pathol. 8, 737–746. doi: 10.1111/j.1364-3703.2007.00430.x
Wessels, J. G. H. (1996). Fungal hydrophobins: proteins that function at an interface. Trends Plant Sci. 1, 9–15. doi: 10.1016/s1360-1385(96)80017-3
White, B. T., and Yanofsky, C. (1993). Structural characterization and expression analysis of the Neurospora conidiation gene con-6. Dev. Biol. 160, 254–264. doi: 10.1006/dbio.1993.1303
Wiest, A., Grzegorski, D., Xu, B. W., Goulard, C., Rebuffat, S., Ebbole, D. J., et al. (2002). Identification of peptaibols from Trichoderma virens and cloning of a peptaibol synthetase. J. Biol. Chem. 277, 20862–20868. doi: 10.1074/jbc.m201654200
Zeilinger, S., Gruber, S., Bansal, R., and Mukherjee, P. K. (2016). Secondary metabolism in Trichoderma - Chemistry meets genomics. Fungal Biol. Rev. 30, 74–90. doi: 10.1016/j.fbr.2016.05.001
Keywords: Trichoderma virens, mutant, transcriptome, NGS, secondary metabolism
Citation: Pachauri S, Sherkhane PD, Kumar V and Mukherjee PK (2020) Whole Genome Sequencing Reveals Major Deletions in the Genome of M7, a Gamma Ray-Induced Mutant of Trichoderma virens That Is Repressed in Conidiation, Secondary Metabolism, and Mycoparasitism. Front. Microbiol. 11:1030. doi: 10.3389/fmicb.2020.01030
Received: 03 February 2020; Accepted: 27 April 2020;
Published: 12 June 2020.
Edited by:
Francesco Vinale, University of Naples Federico II, ItalyReviewed by:
Birinchi Kumar Sarma, Banaras Hindu University, IndiaEliane Ferreira Noronha, University of Brasilia, Brazil
Vishukumar Aimanianda, Institut Pasteur, France
Copyright © 2020 Pachauri, Sherkhane, Kumar and Mukherjee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prasun K. Mukherjee, cHJhc3VubUBiYXJjLmdvdi5pbg==; cHJhc3VubXVraGVyamVlMUBnbWFpbC5jb20=
 Shikha Pachauri
Shikha Pachauri Pramod D. Sherkhane
Pramod D. Sherkhane Vinay Kumar2,3
Vinay Kumar2,3 Prasun K. Mukherjee
Prasun K. Mukherjee