- 1Department of Population Health, University of Georgia, Athens, GA, United States
- 2Department of Biomedical Sciences and Pathobiology, Virginia Polytechnic Institute and State University, Blacksburg, VA, United States
- 3Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration, Laurel, MD, United States
- 4Department of Environmental Health Science, University of Georgia, Athens, GA, United States
- 5Department of Animal and Poultry Sciences, Virginia Polytechnic Institute and State University, Blacksburg, VA, United States
Salmonella enterica cause significant illnesses worldwide. There has been a marked increase in resistance to fluoroquinolones and β-lactams/cephalosporins, antibiotics commonly used to treat salmonellosis. However, S. enterica serovars vary in their resistance to these and other antibiotics. The systemic virulence of some Salmonella serovars is due to a low copy number, IncF plasmid (65–100 kb) that contains the ADP-ribosylating toxin, SpvB. This virulence plasmid is present in only nine Salmonella serovars. It is possible that the spvB-virulence plasmid excludes other plasmids and may explain why antibiotic resistance is slow to develop in certain Salmonella serovars such as S. Enteritidis. The distribution of plasmid entry exclusion genes traS/traT and traY/excA are variable in Salmonella IncF and IncI plasmids, respectively and may account for differences in emergent antimicrobial resistance for some Salmonella serovars. The goal of this study is to determine the contribution of the Salmonella spvB-virulence plasmid in F-plasmid exclusion. From conjugation experiments, S. Typhimurium exhibited lower conjugation frequency with incFI and incFII plasmids when the spvB-virulence plasmid is present. Furthermore, introduction of cloned incFI traS into a “plasmidless” S. Typhimurium LT2 strain and Escherichia coli DH5α excluded incFI plasmid. However, deletion of the virulence plasmid traS did not affect plasmid exclusion significantly compared to a spvB control deletion. In addition, differences in F plasmid conjugation in natural Salmonella isolates did not correlate with IncF or SpvB-virulence plasmid genotype. There appear to be other plasmid or chromosomal genes at play in plasmid exclusion that may be responsible for the slow development of antibiotic resistance in certain serovars.
Introduction
Salmonella enterica is responsible for 78 million illnesses and 59 thousand deaths per year, worldwide (Havelaar et al., 2015). Non-typhoid Salmonella are primarily transmitted via fecal contamination of meat, eggs, dairy products, fruits, nuts, and vegetables (De Buyser et al., 2001; Brar and Danyluk, 2018; De Cesare, 2018; Li et al., 2018; Omer et al., 2018). In the United States, S. enterica causes 1 million illnesses each year, resulting in 19 thousand hospitalizations (Scallan et al., 2011). Although most Salmonella infections are largely treatable with antibiotics, a disturbing trend is the rise of multidrug resistant (MDR) Salmonella (Wasyl et al., 2015; Iwamoto et al., 2017; Tyson et al., 2017; Duong et al., 2018), especially to fluoroquinolones, β-lactams, and cephalosporins; antibiotics frequently used to treat these infections (Collard et al., 2007; Tribble, 2017; Duong et al., 2018). According to the National Antimicrobial Resistance Monitoring System (NARMS) 2015 Annual Report, 12.4% of S. enterica isolates tested were resistant to three or more classes of antibiotics [Centers for Disease Control and Prevention (CDC), 2016]. However, Salmonella serovars vary substantially in their susceptibility to antimicrobials tested in the NARMS panel. For example, 77.7% of S. Enteritidis isolates are pan-susceptible compared to just 28.3% of S. Typhi isolates [Centers for Disease Control and Prevention (CDC), 2016].
Mobile genetic elements, such as plasmids and transposons, are responsible for the transmission and subsequent dissemination of antimicrobial resistance (Partridge et al., 2018). However, a few Salmonella serovars such as Enteritidis are slow to develop antimicrobial resistance despite their presence in environments rich in antimicrobial resistance genes (Liljebjelke et al., 2017). The question that arises is why are some Salmonella serovars slow to develop antibiotic resistance compared to others?
Plasmids are often important vehicles for disseminating antibiotic resistance, however some Salmonella serovars are slower in developing antibiotic resistance compared to others. A significant genetic barrier to plasmid transmission and therefore development of antimicrobial resistance may be the resident spvB-virulence plasmid. Like virulence plasmids in E. coli pathovars (Johnson and Nolan, 2009), the Salmonella virulence plasmid belongs to IncF incompatibility group, and specifically contains the FIC and FII replicons present in F and R100 plasmids, respectively (Villa et al., 2010). Similarly, newer β-lactam/cephalosporin and quinolone resistance genes reside on IncF plasmids. Those same resistance genes in Salmonella, however, reside on plasmid incompatibility groups other than IncF (Carattoli, 2009).
Some Salmonella serovars contain a large molecular weight plasmid that enables it to proliferate in the reticuloendothelial system and cause systemic infection (Gulig and Curtiss, 1987). The key virulence factor SpvB is an ADP-ribosylating toxin that interferes with phagocyte function in the host (Lesnick et al., 2001). This plasmid is present in the non-typhoid Salmonella serotypes Abortusovis, Choleraesuis, Derby, Dublin, Enteritidis, Gallinarum/Pullorum, Paratyphi C, and Typhimurium (Boyd and Hartl, 1998; Feng et al., 2012). While the spv operon provides a small, but crucial growth advantage in bovine and human monocyte-derived macrophages (Libby et al., 2000), it does not appear to be essential to Salmonella's ability to cause gastroenteritis (Horiuchi et al., 1991). Other virulence loci present on the virulence plasmid are: the pef genes (plasmid-encoded fimbriae) which mediate adhesion to the small intestine and contributes to fluid accumulation in infant mouse model (Baumler et al., 1996); srgA, SdiA-regulated genes and putative disulphide bond oxidoreductase (Feng et al., 2012); mig-5, a macrophage-inducible gene and putative carbonic anhydrase (Feng et al., 2012); and rck, which encodes a 17-KD outer membrane protein that confers complement resistance (Vandenbosch et al., 1989).
The spvB-virulence plasmid is a conjugative, IncF plasmid (Ahmer et al., 1999; Villa et al., 2010). The F-plasmid, or “Fertility” (F) factor, was the earliest of self-transmissible plasmids studied, and it has served as a model for understanding plasmid replication, partitioning, maintenance, and transfer. This plasmid contains 40 genes clustered together in a 33.3 kb transfer region (tra) that mediates physical transfer of plasmids between bacterial cells. The F plasmid transfer begins when the F pilus makes contact with one or more recipient cells, which leads to the formation of a mating pair aggregate (Frost et al., 1994). The F pilus retracts to bring donor and recipient adhesion sites into contact to form a transfer pore. The OmpA protein along with TraG and TraN stabilize the pore, which leads to the stabilization of the mating pair aggregates (Klimke and Frost, 1998; Anthony et al., 1999). Over time, the mating pair stabilize and they will not disassociate, even with the addition of sodium dodecyl sulfate (SDS) (Achtman et al., 1978). There are two main genes reported to be involved in mating pair stabilization: traN in the outer-membrane and traG in the inner-membrane (Manning et al., 1981; Klimke and Frost, 1998).
Plasmids are similar to bacteriophages in that they are “selfish DNA” that contain attributes that favors their spread and retention within their bacterial host, while excluding similar competing DNA molecules. The distribution of plasmid incompatibility groups, within a bacterial population or microbial community, affects plasmid transmission. Competition for initiation of DNA replication or partitioning between the dividing daughter cells affects the plasmid's persistence within a community of like plasmids (Novick, 1987; Bouet et al., 2007). Plasmids with similar replicons and partitioning apparatus are incompatible. This incompatibility is often tied to exclusion, mechanisms that limit plasmid transfer by disrupting mating aggregates (surface exclusion) or inhibiting DNA transfer in the presence of mating aggregates (entry exclusion) (Garcillan-Barcia and de la Cruz, 2008).
Like lysogenic bacteriophages, plasmids have several mechanisms for preventing “super infection” of its bacterial host with similar plasmids. Plasmid incompatibility, the inability of similar plasmids to coexist in the same bacterial cell, involves either interference with initiation of plasmid replication (Novick, 1987) or partitioning of competing plasmids between daughter cells (Bouet et al., 2007). Another mechanism, exclusion involves interference with plasmid transfer of those belonging to the same incompatibility group. For plasmids belonging to IncF incompatibility group, traS and traT are primarily responsible for this exclusion (Garcillan-Barcia and de la Cruz, 2008). TraT is an outer membrane lipoprotein that blocks conjugation by preventing the formation of stable mating aggregates (Sukupolvi and O'Connor, 1990). TraS, localized in the inner membrane, functions to inhibit DNA transfer even after the formation of stable mating aggregates by recognizing its cognate TraG in the donor cell (Achtman et al., 1979; Firth and Skurray, 1992).
Genomic comparisons of spvB-virulence plasmids in S. Choleraesuis, S. Dublin, and S. Enteritidis, using S. Typhimurium LT2 pSLT as the prototype, self-transmissible virulence plasmid, revealed large deletions within the tra operon, including traS responsible for entry exclusion (Yu et al., 2006; Hong et al., 2008). The S. Choleraesuis, S. Dublin and S. Enteritidis virulence plasmids are neither mobilizable nor self-transmissible due to the absence of oriT and most tra genes (Yu et al., 2006; Hong et al., 2008). As more Salmonella virulence plasmids have been sequenced, there is significant differences with regards to tra gene(s) deletion(s); where most deletions include both entry exclusion genes traS and traT or traS alone (Figure 1A).

Figure 1. Organization of conjugation genes associated with IncF (A) and IncI (B) plasmids in Salmonella isolated from non-domestic animals and water. R64 (IncI) and Salmonella Typhimurium LT2 spvB-virulence plasmid (pSLT; IncF) served as templates for comparing gene organization of Salmonella plasmids. Different colors were used to depict the different classes of conjugation genes. Gene (s) or contiguous plasmid segment missing in Salmonella isolates are denoted as Δ or Δ with dotted line. A solid line was used to connect pil with tra operon in R64, IncI plasmid. Blocks above plasmid backbone for R64 or pSLT denotes gene(s) only present in other Salmonella plasmids. The exclusion genes traS, traT and traY, excA are bracketed in red.
The SpvB virulence plasmid gene, traS is an important genetic barrier to development of plasmid-mediated antimicrobial resistance. If this is the case, natural Salmonella isolates possessing the SpvB virulence plasmid are less likely to acquire other large molecular weight plasmids including those that mediate plasmid-mediated antibiotic resistance. The purpose of this study is to determine whether the Salmonella SpvB-virulence plasmid, and specifically traS, can actively exclude other incF plasmids from entering the cell.
Materials and Methods
Bacterial Strains and Plasmids
Table 1 describes bacterial strains and plasmids used in this study. Escherichia coli strains XK1200 and MC4100 served as bacterial hosts for IncFI and IncFII plasmids pOX38-km and R100, respectively. Bacterial cultures were grown overnight at 37°C in Luria-Bertani (LB) medium with appropriate antibiotic at the following concentrations: tetracycline (10 μg/ml), kanamycin (30 μg/ml), nalidixic acid (64 μg/ml), rifampicin (64 μg/ml), and chloramphenicol (64 μg/ml). Salmonella Typhimurium pSLT− strain was constructed by transducing pStLT203 Ω parA::Km (Tinge and Curtiss, 1990) into S. Typhimurium strain LT2 using P22 HT int (Provence and Curtiss, 1994). A kanamycin-resistant, S. Typhimurium transductant was passaged in LB without antibiotics and subsequently screened for sensitivity to kanamycin (Tinge and Curtiss, 1990). Loss of pSLT virulence plasmid was confirmed by polymerase chain reaction (PCR) analysis (Swamy et al., 1996). Defined deletions in traS or spvB were introduced into S. Typhimurium virulence plasmid using λ red recombineering approach described by Datsenko and Wanner (2000). Table 2 describes PCR primers and conditions used to construct λ red knockouts. Escherichia coli and S. Typhimurium LT2 strains were transformed using electroporation protocol described by Dower et al. (1988).
Mating Assay
Conjugations were performed as follows. Bacteria grew as standing overnight cultures at 37°C in Luria-Bertani (LB) broth. The mating mix consisted of overnight cultures of donor strain (5 μl) and recipient strain (50 μl), in 5 ml of 10 mM MgSO4. Cells were collected on a 0.45 μm pore size cellulose filter membrane (Millipore Sigma; St. Louis, MO), and the filter was aseptically placed on Luria-Bertani (LB) plate containing 0.2% glucose and 10 mM MgSO4, cell side up. After overnight incubation at 37°C, a cell suspension was made by aseptically placing the filter in 5 ml of 10 mM MgSO4 and vortexing. The cell suspension was diluted 10-fold and plated on LB plates containing the appropriate antibiotic for selecting recipients or transconjugants. The conjugation frequency was determined from the number of transconjugants divided by recipients; averaging the results of duplicate matings, for three separate trials.
PCR
Web-based software analysis program, Primer3Plus (https://primer3plus.com/cgi-bin/dev/primer3plus.cgi) was used to design primers, targeting Salmonella Typhimurium LT2 virulence plasmid genes spvB and traS genes. See Table 2 for description of primer sequences, PCR conditions, and expected size for PCR amplicons. The University of Georgia Molecular Instrumentation Laboratory synthesized the PCR primers. Genomic DNA was prepared as described by Sambrook et al. (1989). The PCR reaction mix contained 2 mM MgCl2, 0.1 mM primer, 0.2 mM nucleotide and 0.5-unit Taq DNA polymerase (Roche Molecular Biochemicals; Indianapolis, IN). PCR screens were performed using the Rapidcycler hot-air thermocycler (Idaho Technology; Salt Lake City, UT) with denaturation set at 93°C for 1 min; annealing as described in Table 2 for each primer set for 1 s, and primer extension at 72°C for 15 s for 30 cycles. Probes, for DNA: DNA hybridization, were prepared by PCR, substituting standard nucleotides with digoxigenin-labeled nucleotides (Roche Molecular Biochemicals) in the PCR reaction mix.
Plasmid Extraction
Bacterial isolates were streaked from frozen glycerol stocks onto tryptic soy agar (Fisher Scientific) and plates were incubated overnight at 37°C. A bacterial suspension was made by inoculating Superbroth (Provence and Curtiss, 1994) (6 ml) with a single colony. Bacteria grew overnight at 37°C with aeration (235 rpm). Plasmid DNA was extracted from overnight cultures using the FosmidMAX DNA Purification Kit (Epicenter; Grand Island, NY). DNA samples were stored at −20°C. Gel electrophoresis separated plasmid DNA on a 0.5% agarose gel; at 44 V in E buffer (40 mM Tris-acetate, 2 mM sodium EDTA) for 16.5 h (88). Gels were stained with 1 X Sybr Gold (Invitrogen) in 1X TAE (40 mM Tris-acetate, 1 mM EDTA; pH 8.0) buffer while shaking for 30 min at 40 rpm. Gel images, illuminated on UV transilluminator, were captured with Molecular Imager Gel Doc XR System digital camera (BioRad; Hercules, CA).
DNA: DNA Hybridization
Agarose gels were stored at 4°C before DNA transfer to nylon membranes. Gels were treated with HCl, followed with NaOH treatment (Sambrook et al., 1989), before the single stranded DNA was transferred to a nylon membrane using BioRad vacuum blotter. Single stranded DNA was UV-cross linked onto the nylon membranes. Membranes were covered in aluminum foil and stored at −80°C. DNA:DNA hybridization was performed as described by Sambrook et al. (1989) with hybridization and washes performed at 68°C. Bound probe was visualized, on nylon membranes, with anti-digoxigenin alkaline phosphatase conjugate and the nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate substrate as described by the manufacturer (Roche Molecular Biochemicals). The DNA probe generated using FI traS primers is specific for F-plasmid and its F-plasmid derivative pOX38. There is only 47% identity at the nucleotide level between the traS of pSLT and F-plasmid.
Sequencing of Salmonella Genomes
Salmonella enterica strains were sub-cultured from frozen stocks onto Tryptic Soy Agar (TSA) (Fisher Scientific) plates amended with 5% sheep blood and incubated overnight at 37°C. Single isolated colonies were inoculated into Tryptic Soy Broth (TSB) (Fisher Scientific) and incubated overnight at 37°C, with shaking. Cell pellets were harvested by centrifugation at 6,010 × g for 5 min, and genomic DNA was extracted with the QIAcube automated sample preparation platform, using the QIAamp DNA mini protocol (Qiagen, Valencia, CA, USA).
Extracted genomic DNA was quantified using a Qubit 2.0 Fluorometer (Invitrogen- ThermoFisher, Waltham, MA, USA). Genomic DNA was diluted with nuclease-free water and sequencing libraries were prepared using the Nextera XT DNA Library Prep kit (Illumina, San Diego, CA, USA). Whole genome sequencing was performed on the MiSeq benchtop sequencer (Illumina, San Diego, CA, USA), utilizing 2 × 250 bp paired-end V2 chemistry. Raw sequence reads were deposited in the Sequence Read Archive (SRA) at NCBI. Fastq datasets were trimmed for quality, ambiguities (n = 0), and length (l > 150 bp), and then de novo assembled with CLC Genomics Workbench version 9.0 (CLC bio, Aarhus, Denmark). The draft genome sequence assemblies were annotated using Rapid Annotation using Subsystem Technology (RAST) (Aziz et al., 2008). The Salmonella genomes were searched for contigs containing genes annotated as “tra,” including traI. Plasmid incompatibility group was identified based on homology to the conjugative relaxase TraI of published, reference plasmid genomes for the F plasmid (IncF) (NCBI GenBank AP001918.1), R100 (IncF) (NCBI GenBank NC_002134.1), and R64 (IncI) (NCBI GenBank AP005147) (Fernandez-Lopez et al., 2017). Plasmid genome comparisons were limited to those belonging either to IncF or IncI, as these are the most studied conjugative plasmids, especially in terms of understanding entry exclusion (Furuya and Komano, 1994; Garcillan-Barcia and de la Cruz, 2008).
Statistical Analysis
Chi-squared; and paired and unpaired Student tests were used to determine whether differences observed were significant.
Results
Distribution of Conjugative Plasmids in Salmonella Isolated From Non-domestic Animals and Water
Whole genome sequencing was performed on 161 Salmonella isolated from various animal species (reptiles, opossum, racoon, songbirds) and water in order to identify major conjugative plasmids. These isolates were chosen from sources that are not likely to be exposed to antibiotics; and their resident plasmids are more likely to reflect their natural state prior to antibiotic selection pressure. Approximately half of Salmonella isolates (n =161) possessed IncI (34.2%) or IncF plasmids (18.0%) (Figure 1B, Table 3). None of these plasmids possessed genes associated with antimicrobial or heavy metal resistance. Salmonella IncI and incF plasmids possessed plasmid exclusion genes traY/excA and traS,T, respectively. However, of the isolates that possessed one of these two plasmid types, only 10.9 and 13.8% had both plasmid exclusion genes traY/excA or traS,T, respectively (Table 3). Of Salmonella IncF plasmids (n = 29) identified, five possessed spvB (17.9%); the signature gene of Salmonella virulence plasmids. These spvB virulence plasmids, as well as the other IncF plasmids, were variable in the distribution of plasmid exclusion gene traS (Table 3).
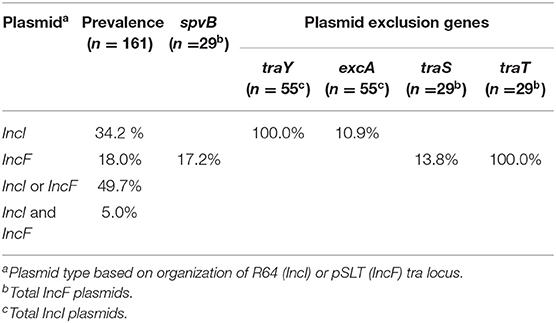
Table 3. Distribution of conjugative IncI and IncF plasmids in Salmonella isolated from non-domestic animals and water.
Plasmid Profile and Prevalence of spvB-virulence Plasmids in S. enterica Serovars S. Dublin, S. Enteritidis, S. Kentucky, and S. Typhimurium Isolated From Domestic Animals
Single and multiple, large molecular weight plasmids (>55 kb) were identified in the S. enterica serovars screened (Figures 2, 3; and summarized in Table 4). The spvB-virulence plasmid was present in Salmonella serovars S. Dublin, S. Enteritidis, and S. Typhimurium, as one of these large sized plasmids (Figure 2). However, none of the S. Dublin spvB-virulence positive isolates contained the entry exclusion gene, traS (Table 4). Ninety-three percent of S. Kentucky isolates contained one or more, large molecular weight plasmids (Table 4), but none were identified as the spvB-virulence plasmid (Figure 3). The prevalence of other large sized plasmids (>55 kb), among spvB-virulence positive isolates varied from 7.7 to 62% among S. enterica serovars screened (Table 4). There was a statistically significant difference in the distribution of these plasmids among S. serovars screened relative to the prevalence of spvB-virulence plasmid and specifically the plasmid's resident traS in these same isolates (Table 4; Chi-Squared test, p < 0.05).
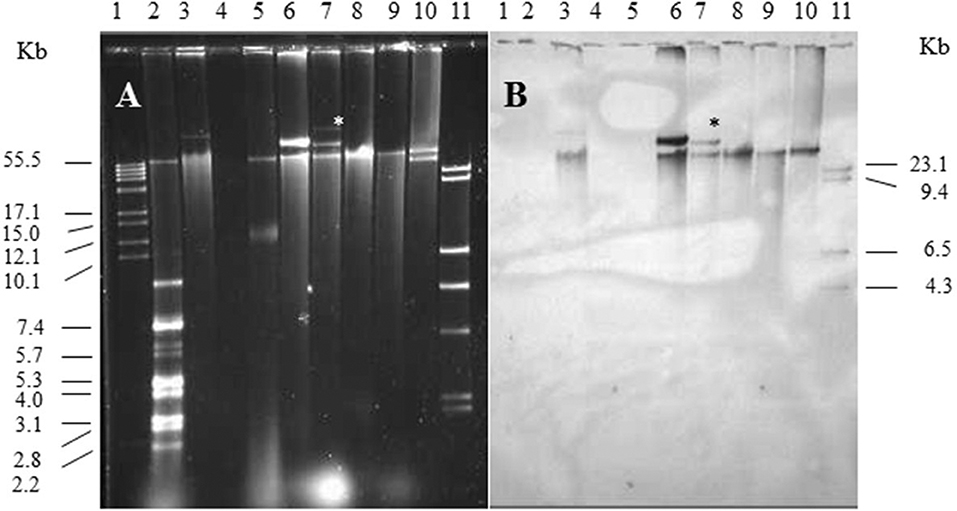
Figure 2. Plasmid profile and identification of spvB-virulence plasmid in S. Dublin and S. Enteritidis isolates. Gel electrophoresis of Salmonella plasmids (A), DNA transfer, and hybridization with spvC DNA probe (B). Lane 1: supercoiled plasmid, VI molecular weight standards (Roche); lane 2: V517 plasmid, molecular weight standards (Macrina et al., 1978); lane 3: S. Typhimurium LT2 (90 kb spvB-virulence plasmid control); lane 4: S. Typhimurium LT2 pSLT− (spvB-virulence plasmid negative control); lane 5: E. coli XK1200 with pOX38-Km; lanes 6–8: S. Dublin isolates 564, 2078, and 2098; lanes 9,10: S. Enteritidis isolates 415 and 98; and lane 11: digoxigenin-labeled l Hind III molecular weight standards (Roche). *Faint, large MW plasmid that did not hybridize with spvC DNA probe.
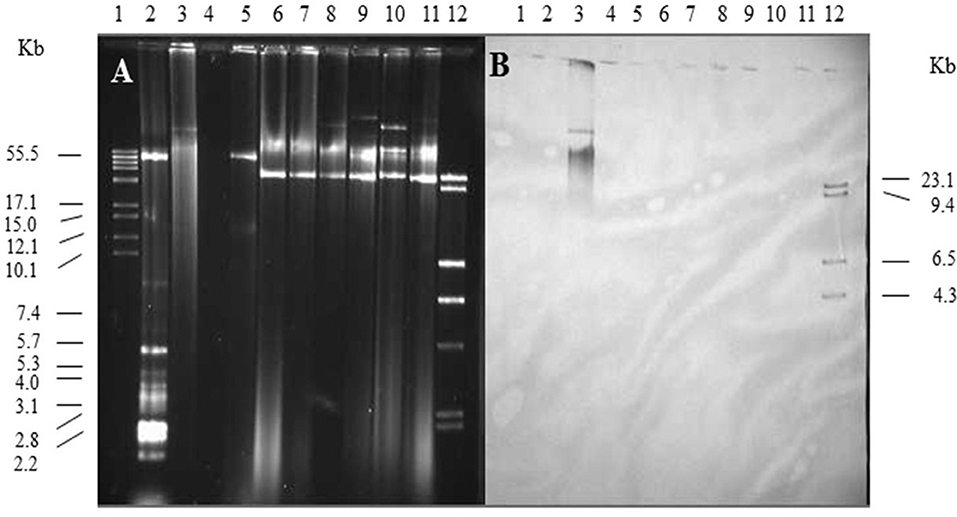
Figure 3. Plasmid profile and identification of spvB-virulence plasmid in S. Kentucky isolates. Gel electrophoresis of Salmonella plasmids (A), DNA transfer, and hybridization with spvC DNA probe (B). Lane 1: supercoiled plasmid, VI molecular weight standards (Roche); lane 2: V517 plasmid, molecular weight standards (Macrina et al., 1978); lane 3: S. Typhimurium LT2 (90 kb spvB-virulence plasmid control); lane 4: S. Typhimurium LT2 pSLT− (spvB-virulence plasmid negative control); lane 5: E. coli XK1200 with pOX38-Km; lanes 6–11: S. Kentucky isolates 102, 116, 117, 112, 118, and 105; and lane 12: digoxigenin-labeled l Hind III molecular weight standards (Roche).
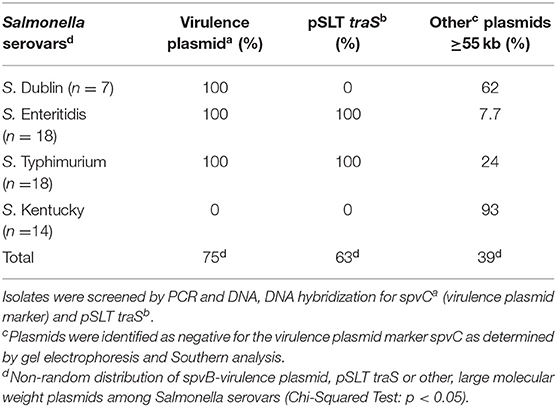
Table 4. Plasmid composition and prevalence of spvB-virulence plasmids and pSLT traS in S. enterica isolates from domestic animals.
Contribution of the spvB-virulence Plasmid and Specifically Its Resident traS to Exclusion of IncF Plasmids in S. Typhimurium
The contribution of the recipient strain's genetic background in IncF plasmid transfer, specifically focusing on spvB-virulence plasmid and the plasmid's resident traS was examined. Several S. Typhimurium LT2 strains were created that were negative for spvB-virulence plasmid (pSLT−), or contained targeted deletions in traS or another plasmid gene, unrelated to plasmid transfer exclusion (spvB). In addition to these strain constructs, S. Typhimurium LT2 pSLT− and E. coli DH5α strains with the cloned FI traS (pRS31) (Collard et al., 2007) served as FI plasmid exclusion controls. Conjugation frequency was calculated as transconjugants per recipient (Cottell et al., 2012; Anjum et al., 2016) rather commonly used transconjugants per donor (Klimke and Frost, 1998; Ahmer et al., 1999). The latter calculation describes the plasmid's properties, in a given host, while in this circumstance, the former is preferred in describing plasmid transfer in relation to the recipient strains used in this study. The spvB-virulence plasmid significantly reduced FI and FII plasmid transfer 10 to 100-fold into S. Typhimurium LT2 recipient strain background (Table 5; S. Typhimurium LT2 wild-type strain vs. LT2 pSLT−, p < 0.05). Introduction of pRS31 (FI traS+) into S. Typhimurium LT2 pSLT− strain restored plasmid exclusion of both F plasmids (Table 5; p < 0.05), but exclusion was most pronounced for the FI plasmid pOX38-km in S. Typhimurium LT2 strain [Table 5; Exclusion Index (EI) 22,741.12 vs. 83.78]. Similarly, pRS31 with FI traS+ was able to exclude F1 plasmid pOX38-km from E. coli DH5α and exhibited plasmid specificity in its exclusion of FI vs. FII plasmids (Table 5; Collard et al., 2007). If spvB-virulence plasmid's traS is responsible for F plasmid exclusion in S. Typhimurium LT2, then deletion of this gene is expected to significantly increase plasmid transfer compared to wild-type or S. Typhimurium LT2 strain with a deletion in another, unrelated plasmid gene. The traS deletion did not significantly alter plasmid transfer frequency compared to either the wild type or spvB deletion strain for F plasmids pOX38-km (FI) or R100 (FII) (Table 5).
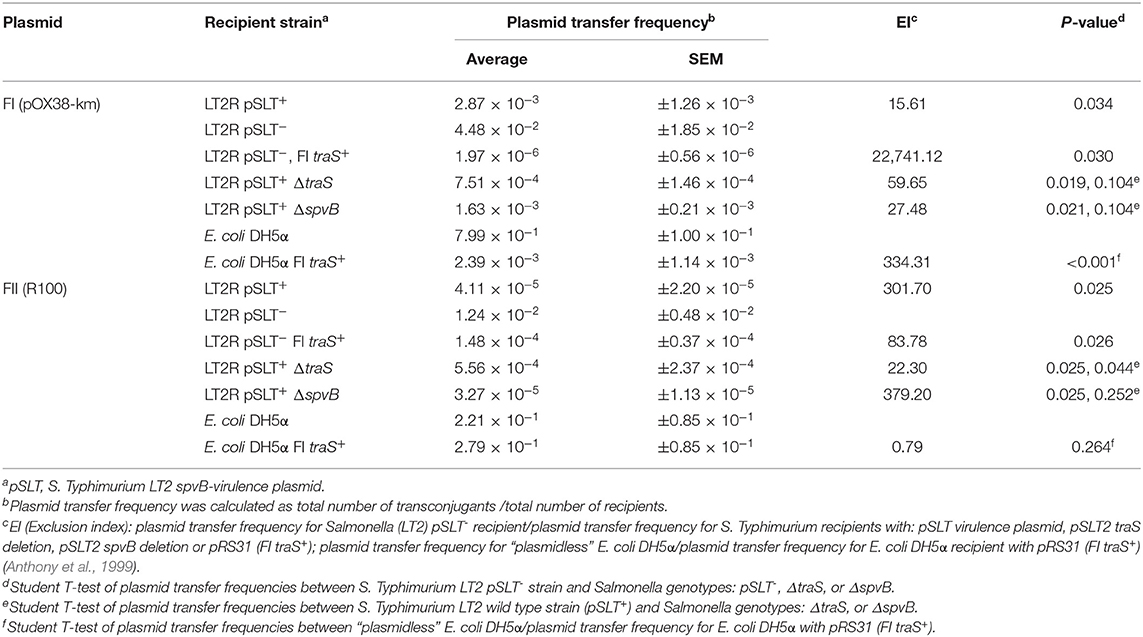
Table 5. The contribution of the Salmonella spvB-virulence plasmid and the resident plasmid traS on exclusion of IncF plasmids.
Contribution of the spvB-virulence Plasmid and Specifically Its Resident FII traS to Exclusion of IncF Plasmids in Natural Salmonella Isolates
The ability of spvB-virulence plasmid to exclude F plasmids was examined in natural Salmonella isolates. Critical to this study was the identification of a natural S. Typhimurium clone (Hudson et al., 2000; Hernandez et al., 2012), with and without the spvB-virulence plasmid, and several Salmonella isolates with the virulence plasmid, minus traS (Table 1). The natural isolates exhibited 20 to 700-fold, lower conjugation frequency for FI plasmid compared to the spvB-virulence plasmid positive, S. Typhimurium LT2 control (Table 6). Presence or absence of the virulence plasmid or the plasmid's traS in natural Salmonella isolates did not correlate with changes in conjugation frequency for either F plasmids (Table 6). The virulence plasmid alone does not appear to be a significant barrier to F-plasmid exclusion in these isolates.
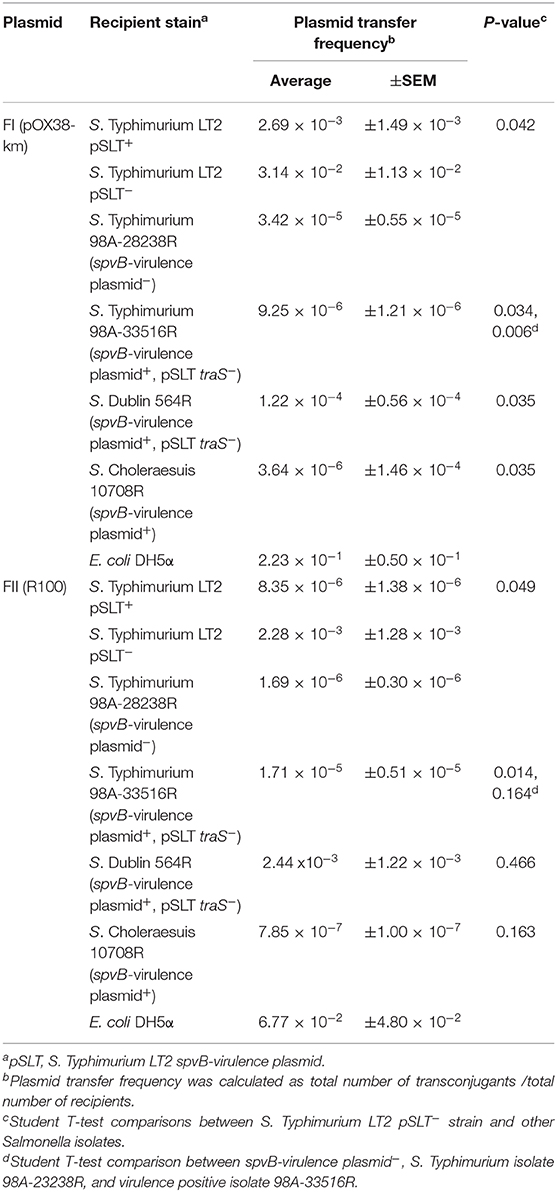
Table 6. The contribution of the Salmonella spvB-virulence plasmid on exclusion of IncF plasmids in natural S. enterica isolates.
F-plasmid Exists Autonomously in S. Typhimurium LT2 Containing IncF, spvB-virulence Plasmid
The spvB-virulence plasmid is comprised of the FIB and FII replicons present in F and R100 plasmids, respectively (Villa et al., 2010). These plasmid replicons play an important role in plasmid incompatibility for IncF group of plasmids (Novick, 1987). Therefore, plasmid incompatibility is expected to affect the persistence of the resident plasmid in wild-type S. Typhimurium LT2 (pSLT+) transconjugants. With continued antibiotic selection pressure on pOX38 and plasmid incompatibility, one expectation is the loss of SpvB-virulence plasmid while another possible outcome is the recombination between the two plasmids. Plasmid recombination was expected to result in change in size of either plasmid and pSLT and F-plasmid specific probes binding to the same-size DNA band(s). On the other hand, if the pSLT and F-plasmid specific probes bound to distinctly different size DNA bands, similar in size to the plasmid controls, then these plasmids existed as separate entities in Salmonella (pSLT+) transconjugants
Kanamycin-resistant, S. Typhimurium LT2 (pSLT+) transconjugants were positive for the SpvB-virulence plasmid. The pSLT and pOX38 controls produced two distinct, DNA bands that most likely represent their relaxed and supercoiled states (Figure 4, lanes 3 and 5, respectively). Salmonella Typhimurium LT2 pSLT+ transconjugants had similar size DNA bands, recognized by pSLT-probe as the virulence plasmid control (Figures 4A,B, lanes 6–8 vs. lane 3). However, one of the two DNA bands recognized by F-plasmid specific probe was absent in S. Typhimurium pSLT+ transconjugants (Figure 4B, lane 5 vs. lanes 6–8).
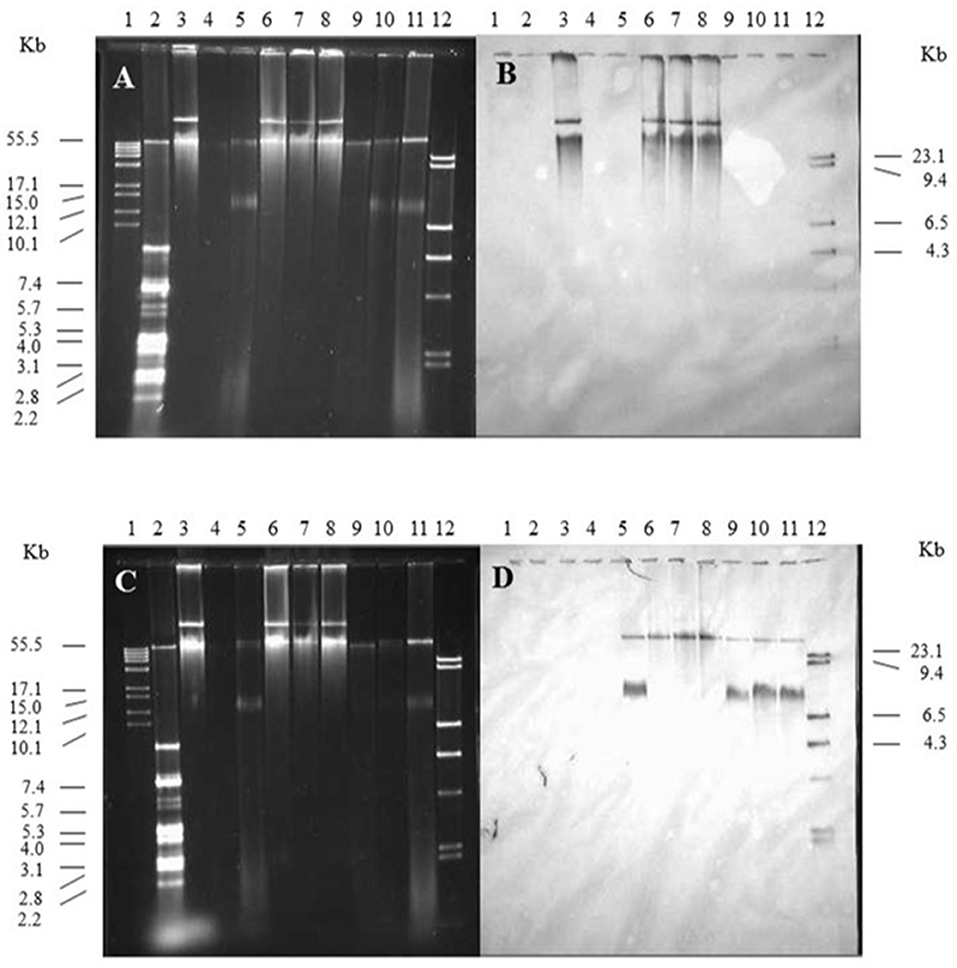
Figure 4. The contribution of the spvB-virulence plasmid on localization of F-plasmid in S. Typhimurium transconjugants. Gel electrophoresis of Salmonella plasmids (A,C), DNA transfer, and hybridization with spvC (B) or FI traS (D) DNA probes. Lane 1: supercoiled plasmid, VI molecular weight standards (Roche); lane 2: V517 plasmid, molecular weight standards (Macrina et al., 1978); lane 3: S. Typhimurium LT2 (90 kb spvB-virulence plasmid control); lane 4: S. Typhimurium LT2 pSLT− (spvB-virulence plasmid negative control); lane 5: E. coli XK1200 with pOX38-Km; lanes 6–8: S. Typhimurium LT2R transconjugants (pOX38-Km); lanes 9–11: S. Typhimurium LT2 pSLT− transconjugants (pOX38-Km); and lane 12: digoxigenin-labeled l Hind III molecular weight standards (Roche).
Discussion
traS and traT are responsible for F-plasmid exclusion. In genomic comparisons of the virulence plasmid of Salmonella serovars, Choleraesuis, Dublin, Enteritidis, and Kentucky, the most notable genetic difference linking plasmid exclusion to the slow development of plasmid-mediated antibiotic resistance in Salmonella serovars was traS. The distribution of the spvB-virulence plasmid and specifically traS among Salmonella serovars adversely affects the prevalence of other large molecular weight plasmids. The virulence plasmid significantly excluded F plasmid transfer to S. Typhimurium LT2 strain. Introduction of pRS31 containing FI traS into a virulence plasmid-deficient S. Typhimurium LT2 restored exclusion, with plasmid specificity exhibited by traS (Audette et al., 2007). However, the plasmid exclusion was not linked to the plasmid's resident traS as its deletion did not significantly decrease exclusion compared to plasmid gene deletion control, ΔspvB. One possibility is the overnight mating period, even at a 1:10 donor to recipient ratio, favored multiple plasmid transfer events (Simonsen et al., 1990; Anthony et al., 1994), which overwhelmed entry exclusion by traS or traT and therefore masked the impact traS deletion had on plasmid exclusion. While traS was cloned into a low-copy number plasmid, its expression is under plac promoter and in Salmonella, without the plac repressor lacI, this gene is expected to be constitutively expressed. Therefore, overexpression of TraS would explain plasmid exclusion, even under conditions conducive to multiple, repeated plasmid transfers in the recipient population, as transconjugants become new plasmid donors.
It is also possible that traT plays a more significant role in plasmid exclusion. While traT exhibits greater conservation in its amino acid sequence compared to traS of F and R100 plasmids (Harrison et al., 1992), like traS (Garcillan-Barcia and de la Cruz, 2008), traT exhibits plasmid specificity in its exclusion (Harrison et al., 1992). There is significant sequence divergence in F plasmid replicons and tra genes, including traS and traT, where F plasmid evolution in Klebsiella, Salmonella, and Yersinia mirrors the divergence of these genera (Villa et al., 2010). This in part explains how F plasmid pOX38-km can exist as an autonomous replicon in Salmonella host with the FII/FIC virulence plasmid pSLT but does not explain how this plasmid can exclude F plasmids from entry into the Salmonella cell. Maybe the pSLT TraT shares some amino acid sequence or motif with both FI and FII TraT or there is some other plasmid gene(s) responsible for F-plasmid exclusion.
While plasmid exclusion, linked to the virulence plasmid in S. Typhimurium laboratory strain LT2 was observed, similar plasmid exclusion was not observed for natural Salmonella isolates, that varied in their spvB-virulence plasmid or traS genotype. Despite conditions that favor multiple plasmid transfer events (Simonsen et al., 1990; Anthony et al., 1994), conjugation frequencies were significantly lower in natural Salmonella isolates compared to S. Typhimurium LT2 strains. As these isolates were not screened for traT, it is possible that while negative for traS, traT is sufficient for excluding IncF plasmids in these isolates. Another possibility, is that these isolates contain other IncF plasmids (Villa et al., 2010) not recognized by our traS probes. No difference in plasmid exclusion for two genetically related S. Typhimurium isolates ± spvB virulence plasmid was observed. In fact, the spvB-virulence plasmid, negative S. Typhimurium isolate, which one would expect to be more receptive to plasmid acquisition, acquired either IncF plasmids at a much lower frequency compared to LT2, pSLT− control (10−5 to 10−6 vs. 10−2 to 10−3). Another plasmid exclusion mechanism may be involved in some Salmonella serovars or strains. One possible candidate for plasmid exclusion is clustered regularly interspaced short palindromic repeat (CRISPR) system that functions to exclude foreign genetic elements from entering the cell by forming a perfect sequence match between the spacer in CRISPR and the spacer located in invading DNA (Marraffini and Sontheimer, 2008; Shariat et al., 2015). Unraveling the contribution of these candidate genes (traT, CRISPR-cas) to plasmid exclusion will require creating S. Typhimurium strains with single or multiple deletions in targeted genes, and comparative genomics of multi-drug resistant and pan-susceptible Salmonella isolates.
Conclusion
Antibiotics have been a great panacea in reducing morbidity and mortality attributed to bacterial infections. Unfortunately, resistance to these “wonder” drugs often quickly follows their introduction (Abraham and Chain, 1988). Usage of antibiotics in agriculture has long been a contentious issue; with fears that antimicrobial resistance has spilled over into human pathogens through the food chain 1975. However, there are several circumstances where there is a disconnect between antibiotic usage and resistance in bacteria colonizing food animals (Idris et al., 2006; Simjee et al., 2007; Smith et al., 2007; Liljebjelke et al., 2017). There is also a disparity in the antimicrobial susceptibility of microbes that inhabit the same environment which may be high in antibiotic resistance gene load (Nandi et al., 2004), and encounter the same selection pressures (e.g., antibiotic usage) (Simjee et al., 2007). This disparity in antimicrobial susceptibility also occurs within the same species, as is the case for S. enterica (Liljebjelke et al., 2017). It appears that several genetic factors are at play that affects the speed at which antibiotic resistance develops and spreads within a bacterial population. A systems-based approach will provide a better understanding of how and when antimicrobial resistance emerges in zoonotic pathogens like Salmonella.
Data Availability Statement
The datasets generated for this study can be found in the NCBI BioProject PRJNA186035.
Author Contributions
JM, ML, and EL contributed to the conception and design of this study. MO, YC, and CG were responsible for the acquisition of data analyzed in this study. JM and CG were involved in the analysis and interpretation associated with this work. All authors were involved in manuscript writing, revisions, and final approval of this manuscript.
Funding
This work was supported by grants from the United States Department of Agriculture (2017-05586; VA-160130).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work, presented here, was part of MO M.S. thesis at the University of Georgia (Oluwadare, 2013). We wish to thank Susan Sanchez for providing archived S. Dublin isolates and Laura Frost for providing plasmids pOX38-Km, pRS31, and R100-1.
Abbreviations
Ap, ampicillin; Cm, chloramphenicol; Fa, fusaric acid; Km, kanamycin; LB, Luria-Bertani; Nal, nalidixic; PCR, polymerase chain reaction; Rif, rifampicin; Sp, spectinomycin; Sm, streptomycin; Su, sulfonamide; TAE, Tris acetate EDTA buffer; Tc, tetracycline; TSA, tryptic soy agar; TSB, tryptic soy broth; UV, ultra violet.
References
Abraham, P., and Chain, E. (1988). An enzyme from bacteria able to destroy penicillin. 1940. Rev. Infect. Dis. 10, 677–678.
Achtman, M., Manning, P. A., Edelbluth, C., and Herrlich, P. (1979). Export without proteolytic processing of inner and outer membrane proteins encoded by F sex factor tra cistrons in Escherichia coli minicells. Proc. Natl. Acad. Sci U.S.A. 76, 4837–4841. doi: 10.1073/pnas.76.10.4837
Achtman, M., Morelli, G., and Schwuchow, S. (1978). Cell-cell interactions in conjugating Escherichia coli: role of F pili and fate of mating aggregates. J. Bacteriol. 135, 1053–1061. doi: 10.1128/JB.135.3.1053-1061.1978
Ahmer, M., Tran, M., and Heffron, F. (1999). The virulence plasmid of Salmonella Typhimurium is self-transmissible. J. Bacteriol. 181, 1364–1368. doi: 10.1128/JB.181.4.1364-1368.1999
Anjum, M. F., Duggett, N. A., AbuOun, M., Randall, L., Nunez-Garcia, J., Ellis, R. J., et al. (2016). Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in great britain. J. Antimicrob. Chemother. 71, 2306–2313. doi: 10.1093/jac/dkw149
Anthony, K. G., Kathir, P., Moore, D., Ippen-Ihler, K., and Frost, L. S. (1996). Analysis of the traLEKBP sequence and the TraP protein from three F-like plasmids: F, R100-1 and ColB2. J. Bacteriol. 178, 3194–3200. doi: 10.1128/JB.178.11.3194-3200.1996
Anthony, K. G., Klimke, W. A., Manchak, J., and Frost, L. S. (1999). Comparison of proteins involved in pilus synthesis and mating pair stabilization from the related plasmids F and R100-1: insights into the mechanism of conjugation. J. Bacteriol. 181, 5149–5159. doi: 10.1128/JB.181.17.5149-5159.1999
Anthony, K. G., Sherburne, C., Sherburne, R., and Frost, L. S. (1994). The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol. Microbiol. 13, 939–953. doi: 10.1111/j.1365-2958.1994.tb00486.x
Audette, G. F., Manchak, J., Beatty, P., Klimke, W. A., and Frost, L. S. (2007). Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology 153(Pt 2), 442–451. doi: 10.1099/mic.0.2006/001917-0
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Baumler, J., Tsolis, R. M., Bowe, F. A., Kusters, J. G., Hoffmann, S., and Heffron, F. (1996). The pef fimbrial operon of Salmonella Typhimurium mediates adhesion to murine small intestine and is necessary for fluid accumulation in the infant mouse. Infect. Immun. 64, 61–68. doi: 10.1128/IAI.64.1.61-68.1996
Bouet, J. Y., Nordstrom, K., and Lane, D. (2007). Plasmid partition and incompatibility–the focus shifts. Mol. Microbiol. 65, 1405–1414. doi: 10.1111/j.1365-2958.2007.05882.x
Boyd, E. F., and Hartl, D. L. (1998). Salmonella virulence plasmid. Modular acquisition of the spv virulence region by an F-plasmid in Salmonella enterica subspecies I and insertion into the chromosome of subspecies II, IIIa, IV and VII isolates. Genetics 149, 1183–1190.
Brar, P. K., and Danyluk, M. D. (2018). Nuts and grains: microbiology and preharvest contamination risks. Microbiol. Spectr. 6, 1–12. doi: 10.1128/microbiolspec.PFS-0023-2018
Carattoli, A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238. doi: 10.1128/AAC.01707-08
Centers for Disease Control and Prevention (CDC) (2016). National Antimicrobial Resistance Monitoring System 2015 Human Isolates Surveillance Report. Atlanta, GA.
Cherepanov, P. P., and Wackernagel, W. (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14. doi: 10.1016/0378-1119(95)00193-A
Collard, J. M., Place, S., Denis, O., Rodriguez-Villalobos, H., Vrints, M., Weill, F. X., et al. (2007). Travel-acquired salmonellosis due to Salmonella Kentucky resistant to ciprofloxacin, ceftriaxone and co-trimoxazole and associated with treatment failure. J. Antimicrob. Chemother. 60, 190–192. doi: 10.1093/jac/dkm114
Cottell, J. L., Webber, M. A., and Piddock, L. J. (2012). Persistence of transferable extended-spectrum-beta-lactamase resistance in the absence of antibiotic pressure. Antimicrob Agents Chemother 56, 4703–4706. doi: 10.1128/AAC.00848-12
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
De Buyser, M. L., Dufour, B., Maire, M., and Lafarge, V. (2001). Implication of milk and milk products in food-borne diseases in France and in different industrialised countries. Int. J. Food Microbiol. 67, 1–17. doi: 10.1016/S0168-1605(01)00443-3
De Cesare, A. (2018). Salmonella in foods: a reemerging problem. Adv. Food Nutr. Res. 86, 137–179. doi: 10.1016/bs.afnr.2018.02.007
Dower, W. J., Miller, J. F., and Ragsdale, C. W. (1988). High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16, 6127–6145. doi: 10.1093/nar/16.13.6127
Duong, V. T., Tuyen, H. T., Van Minh, P., Campbell, J. I., Phuc, H. L., Nhu, T. D. H., et al. (2018). No clinical benefit of empirical antimicrobial therapy for pediatric diarrhea in a high-usage, high-resistance setting. Clin. Infect. Dis. 66, 504–511. doi: 10.1093/cid/cix844
Feng, Y., Liu, J., Li, Y. G., Cao, F. L., Johnston, R. N., Zhou, J., et al. (2012). Inheritance of the Salmonella virulence plasmids: mostly vertical and rarely horizontal. Infect. Genet. Evol. 12, 1058–1063. doi: 10.1016/j.meegid.2012.03.004
Fernandez-Lopez, R., Redondo, S., Garcillan-Barcia, M. P., and de la Cruz, F. (2017). Towards a taxonomy of conjugative plasmids. Curr. Opin. Microbiol. 38, 106–113. doi: 10.1016/j.mib.2017.05.005
Firth, N., and Skurray, R. (1992). Characterization of the F plasmid bifunctional conjugation gene, traG. Mol. Gen. Genet. 232, 145–153. doi: 10.1007/BF00299147
Frost, L. S., Ippen-Ihler, K., and Skurray, R. A. (1994). Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58, 162–210. doi: 10.1128/MMBR.58.2.162-210.1994
Furuya, N., and Komano, T. (1994). Surface exclusion gene of IncI1 plasmid R64: nucleotide sequence and analysis of deletion mutants. Plasmid 32, 80–84. doi: 10.1006/plas.1994.1047
Garcillan-Barcia, M. P., and de la Cruz, F. (2008). Why is entry exclusion an essential feature of conjugative plasmids? Plasmid 60, 1–18. doi: 10.1016/j.plasmid.2008.03.002
Gulig, P. A., and Curtiss, R. III. (1987). Plasmid-associated virulence of Salmonella Typhimurium. Infect. Immun. 55, 2891–2901. doi: 10.1128/IAI.55.12.2891-2901.1987
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/S0022-2836(83)80284-8
Harrison, J. L., Taylor, I. M., Platt, K., and O'Connor, C. D. (1992). Surface exclusion specificity of the TraT lipoprotein is determined by single alterations in a five-amino-acid region of the protein. Mol. Microbiol. 6, 2825–2832. doi: 10.1111/j.1365-2958.1992.tb01462.x
Havelaar, H., Kirk, M. D., Torgerson, P. R., Gibb, H. J., Hald, T., Lake, R. J., et al. (2015). World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS. Med. 12:e1001923. doi: 10.1371/journal.pmed.1001923
Hernandez, S. M., Keel, K., Sanchez, S., Trees, E., Gerner-Smidt, P., Adams, J. K., et al. (2012). Epidemiology of a Salmonella enterica subsp. enterica serovar Typhimurium strain associated with a songbird outbreak. Appl. Environ. Microbiol. 78, 7290–7298. doi: 10.1128/AEM.01408-12
Hong, S. F., Chiu, C. H., Chu, C., Feng, Y., and Ou, J. T. (2008). Complete nucleotide sequence of a virulence plasmid of Salmonella enterica serovar Dublin and its phylogenetic relationship to the virulence plasmids of serovars Choleraesuis, Enteritidis and Typhimurium. FEMS Microbiol. Lett. 282, 39–43. doi: 10.1111/j.1574-6968.2008.01096.x
Horiuchi, S., Goto, N., Inagaki, Y., and Nakaya, R. (1991). The 106-kilobase plasmid of Salmonella braenderup and the 100-kilobase plasmid of Salmonella Typhimurium are not necessary for the pathogenicity in experimental models. Microbiol. Immunol. 35, 187–198. doi: 10.1111/j.1348-0421.1991.tb01547.x
Hudson, C. R., Quist, C., Lee, M. D., Keyes, K., Dodson, S. V., Morales, C., et al. (2000). Genetic relatedness of Salmonella isolates from nondomestic birds in Southeastern United States. J. Clin. Microbiol. 38, 1860–1865. doi: 10.1128/JCM.38.5.1860-1865.2000
Idris, U., Lu, J., Maier, M., Sanchez, S., Hofacre, C. L., Harmon, B. G., et al. (2006). Dissemination of fluoroquinolone-resistant Campylobacter spp. within an integrated commercial poultry production system. Appl. Environ. Microbiol. 72, 3441–3447. doi: 10.1128/AEM.72.5.3441-3447.2006
Iwamoto, M., Reynolds, J., Karp, B. E., Tate, H., Fedorka-Cray, P. J., Plumblee, J. R., et al. (2017). Ceftriaxone-resistant nontyphoidal salmonella from humans, retail meats, and food animals in the United States, 1996-2013. Foodborne Pathog. Dis. 14, 74–83. doi: 10.1089/fpd.2016.2180
Johnson, T. J., and Nolan, L. K. (2009). Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 73, 750–774. doi: 10.1128/MMBR.00015-09
Klimke, W. A., and Frost, L. S. (1998). Genetic analysis of the role of the transfer gene, traN, of the F and R100-1 plasmids in mating pair stabilization during conjugation. J. Bacteriol. 180, 4036–4043. doi: 10.1128/JB.180.16.4036-4043.1998
Lesnick, M. L., Reiner, N. E., Fierer, J., and Guiney, D. G. (2001). The Salmonella spvB virulence gene encodes an enzyme that ADP-ribosylates actin and destabilizes the cytoskeleton of eukaryotic cells. Mol. Microbiol. 39, 1464–1470. doi: 10.1046/j.1365-2958.2001.02360.x
Li, M., Baker, C. A., Danyluk, M. D., Belanger, P., Boelaert, F., Cressey, P., et al. (2018). Identification of biological hazards in produce consumed in industrialized countries: a review. J. Food Prot. 81, 1171–1186. doi: 10.4315/0362-028X.JFP-17-465
Libby, S. J., Lesnick, M., Hasegawa, P., Weidenhammer, E., and Guiney, D. G. (2000). The Salmonella virulence plasmid spv genes are required for cytopathology in human monocyte-derived macrophages. Cell Microbiol. 2, 49–58. doi: 10.1046/j.1462-5822.2000.00030.x
Liljebjelke, K. A., Hofacre, C. L., White, D. G., Ayers, S., Lee, M. D., and Maurer, J. J. (2017). Diversity of antimicrobial resistance phenotypes in Salmonella isolated from commercial poultry farms. Front. Vet. Sci. 4:96. doi: 10.3389/fvets.2017.00096
Macrina, L., Kopecko, D. J., Jones, K. R., Ayers, D. J., and McCowen, S. M (1978). A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid 1, 417–420. doi: 10.1016/0147-619X(78)90056-2
Manning, P. A., Morelli, G., and Achtman, M. (1981). traG protein of the F sex factor of Escherichia coli K-12 and its role in conjugation. Proc. Natl. Acad. Sci. U.S.A. 78, 7487–7491. doi: 10.1073/pnas.78.12.7487
Marraffini, L. A., and Sontheimer, E. J. (2008). CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845. doi: 10.1126/science.1165771
Nandi, S., Maurer, J. J., Hofacre, C., and Summers, A. O. (2004). Gram-positive bacteria are a major reservoir of Class 1 antibiotic resistance integrons in poultry litter. Proc. Natl. Acad. Sci. U.S.A. 101, 7118–7122. doi: 10.1073/pnas.0306466101
Novick, R. P. (1987). Plasmid incompatibility. Microbiol. Rev. 51, 381–395. doi: 10.1128/MMBR.51.4.381-395.1987
Oluwadare, M. (2013). Salmonella Are Less Likely to Develop Plasmid Mediated Drug Resistance Due to Its Resident F-Like Plasmid And Specifically the Entry Exclusion Gene, traS, 1–63. Available online at: http://purl.galileo.usg.edu/uga_etd/oluwadare_mopelola_o_201305_ms (accessed February 06, 2020).
Omer, M. K., Alvarez-Ordonez, A., Prieto, M., Skjerve, E., Asehun, T., and Alvseike, O. A. (2018). A systematic review of bacterial foodborne outbreaks related to red meat and meat products. Foodborne Pathog. Dis. 15, 598–611. doi: 10.1089/fpd.2017.2393
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088-17. doi: 10.1128/CMR.00088-17
Provence, L., and Curtiss, R. III. (1994). Gene Transfer in Gram-Negative Bacteria. Washington, DC: ASM Press.
Sambrook, J. E., Fritsch, F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Sanderson, K. E., and Roth, J. R. (1988). Linkage map of Salmonella Typhimurium, edition VII. Microbiol. Rev. 52, 485–532. doi: 10.1128/MMBR.52.4.485-532.1988
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Shariat, N., Timme, R. E., Pettengill, J. B., Barrangou, R., and Dudley, E. G. (2015). Characterization and evolution of Salmonella CRISPR-Cas systems. Microbiology 161, 374–386. doi: 10.1099/mic.0.000005
Simjee, S., McDermott, P. F., White, D. G., Hofacre, C., Berghaus, R. D., Carter, P. J., et al. (2007). Antimicrobial susceptibility and distribution of antimicrobial-resistance genes among Enterococcus and coagulase-negative Staphylococcus isolates recovered from poultry litter. Avian Dis. 51, 884–892. doi: 10.1637/7973-032607-REGR.1
Simonsen, L., Gordon, D. M., Stewart, F. M., and Levin, B. R. (1990). Estimating the rate of plasmid transfer: an end-point method. J. Gen. Microbiol. 136, 2319–2325. doi: 10.1099/00221287-136-11-2319
Smith, J. L., Drum, D. J., Dai, Y., Kim, J. M., Sanchez, S., Maurer, J. J., et al. (2007). Impact of antimicrobial usage on antimicrobial resistance in commensal Escherichia coli strains colonizing broiler chickens. Appl. Environ. Microbiol. 73, 1404–14014. doi: 10.1128/AEM.01193-06
Sukupolvi, S., and O'Connor, C. D. (1990). TraT lipoprotein, a plasmid-specified mediator of interactions between gram-negative bacteria and their environment. Microbiol. Rev. 54, 331–341. doi: 10.1128/MMBR.54.4.331-341.1990
Swamy, S. C., Barnhart, H. M., Lee, M. D., and Dreesen, D. W. (1996). Virulence determinants invA and spvC in salmonellae isolated from poultry products, wastewater, and human sources. Appl. Environ. Microbiol. 62, 3768–3771. doi: 10.1128/AEM.62.10.3768-3771.1996
Tinge, S. A., and Curtiss, R. III. (1990). Conservation of Salmonella Typhimurium virulence plasmid maintenance regions among Salmonella serovars as a basis for plasmid curing. Infect. Immun. 58, 3084–3092. doi: 10.1128/IAI.58.9.3084-3092.1990
Tribble, D. R. (2017). Resistant pathogens as causes of traveller's diarrhea globally and impact(s) on treatment failure and recommendations. J. Travel. Med. 24(Suppl. 1), S6–S12. doi: 10.1093/jtm/taw090
Tyson, G. H., Tate, H. P., Zhao, S., Li, C., Dessai, U., Simmons, M., and McDermott, P. F (2017). Identification of plasmid-mediated quinolone resistance in salmonella isolated from Swine Ceca and retail pork chops in the United States. Antimicrob. Agents Chemother. 61:e01318-17. doi: 10.1128/AAC.01318-17
Vandenbosch, J. L., Rabert, D. K., Kurlandsky, D. R., and Jones, G. W. (1989). Sequence analysis of rsk, a portion of the 95-kilobase plasmid of Salmonella Typhimurium associated with resistance to the bactericidal activity of serum. Infect. Immun. 57, 850–857. doi: 10.1128/IAI.57.3.850-857.1989
Villa, L., Garcia-Fernandez, A., Fortini, D., and Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Wasyl, D., Kern-Zdanowicz, I., Domanska-Blicharz, K., Zajac, M., and Hoszowski, A. (2015). High-level fluoroquinolone resistant Salmonella enterica serovar Kentucky ST198 epidemic clone with IncA/C conjugative plasmid carrying bla(CTX-M-25) gene. Vet. Microbiol. 175, 85–91. doi: 10.1016/j.vetmic.2014.10.014
Keywords: Salmonella, virulence plasmid, antibiotic resistance, entry exclusion, conjugation
Citation: Oluwadare M, Lee MD, Grim CJ, Lipp EK, Cheng Y and Maurer JJ (2020) The Role of the Salmonella spvB IncF Plasmid and Its Resident Entry Exclusion Gene traS on Plasmid Exclusion. Front. Microbiol. 11:949. doi: 10.3389/fmicb.2020.00949
Received: 27 February 2020; Accepted: 21 April 2020;
Published: 15 May 2020.
Edited by:
Clay Fuqua, Indiana University Bloomington, United StatesReviewed by:
Brian Ahmer, The Ohio State University, United StatesMark Glover, University of Alberta, Canada
Copyright © 2020 Oluwadare, Lee, Grim, Lipp, Cheng and Maurer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John J. Maurer, amptYXVyZXJAdnQuZWR1
†Present address: Margie D. Lee, Department of Biomedical Sciences and Pathobiology, Virginia Polytechnic Institute and State University, Blacksburg, VA, United States
John J. Maurer, Department of Animal and Poultry Sciences, Virginia Polytechnic Institute and State University, Blacksburg, VA, United States
 Mopelola Oluwadare1
Mopelola Oluwadare1 Margie D. Lee
Margie D. Lee Christopher J. Grim
Christopher J. Grim Erin K. Lipp
Erin K. Lipp John J. Maurer
John J. Maurer