- 1Shanghai Engineering Research Center of Aquatic Product Processing & Preservation, Shanghai Ocean University, Shanghai, China
- 2National Experimental Teaching Demonstration Center for Food Science and Engineering, Shanghai Ocean University, Shanghai, China
- 3College of Food Science and Technology, Shanghai Ocean University, Shanghai, China
Food spoilage by certain species of bacteria is reported to be regulated by quorum sensing (QS). Acinetobacter johnsonii and Pseudomonas fluorescens, the major specific spoilage organisms, are found to be limited in their QS and co-culture interactions. The aim of this study was to determine how QS-regulated proteins affect the spoilage potential of co-cultured A. johnsonii and P. fluorescens obtained from spoiled bigeye tuna (Thunnus obesus) using a proteomics approach. The A. johnsonii, P. fluorescens, and their co-culture tested the N-acyl-homoserine lactone (AHL) activities using reporter Chromobacterium violaceum CV026 and LC-MS/MS in qualitative and quantitative approaches, respectively. These latter showed that, of the 470 proteins and 444 proteins in A. johnsonii (A) and P. fluorescens (P), respectively, 80 were significantly up-regulated and 97 were significantly down-regulated in A vs. AP, whereas 90 were up-regulated and 65 were down-regulated in P vs. AP. The differentially expressed proteins included the AI-2E family transporter OS, 50S ribosomal protein L3, thioredoxin reductase OS, cysteine synthase CysM OS, DNA-binding response regulator, and amino acid ABC transporter ATPase OS. The cellular process (GO:0009987), metabolic process (GO:0008152), and single-organism process (GO:0044699) were classified into the gene ontology (GO) term. In addition, energy production and conversion, amino acid transport and metabolism, translation, ribosomal structure and biogenesis, post-translational modification, protein turnover, and chaperones were distributed into the clusters of orthologous groups of proteins (COG) terms. The KEGG pathways revealed that 84 and 77 differentially expressed proteins were divided into 20 KEGG pathways in A vs. AP and P vs. AP, respectively, and amino acid metabolism, carbohydrate metabolism, energy metabolism, and translation were significantly enriched. Proteins that correlated with the spoilage-related metabolic pathways, including thioredoxin reductase OS, cysteine synthase OS, and pyridoxal phosphate-dependent enzyme family protein OS, were identified. AI-2E family transporter OS and LuxR family transcriptional regulator OS were identified that related to the QS system. These findings provide a differential proteomic profile of co-culture in A. johnsonii and P. fluorescens, and have potential applications in QS and the regulation of spoilage potential.
Introduction
Bigeye tuna (Thunnus obesus) is a highly sought after fish species used to prepare sashimi in many countries around the world (Wang and Xie, 2019). However, bigeye tuna is easily spoiled by specific spoilage organisms during refrigerated storage, which leads to a reduced shelf life (Sun et al., 2013; Silbande et al., 2016). Currently, more attention has been paid to convenient methods of refrigeration for storing bigeye tuna than to how spoilage bacteria in refrigerated tuna develop through their interactions with each other (Wang et al., 2017). The microbial spoilage of aquatic products is correlated mainly with Gram-negative bacteria, including Acinetobacter spp., Shewanella spp., Pseudomonas spp., Aeromonas spp., lactic acid bacteria, and the Enterobacteriaceae family, when stored under different storage conditions. The main species of bacteria leading to the spoilage of aquatic products during cold storage are Acinetobacter johnsonii and Pseudomonas fluorescens, which are commonly referred to as specific spoilage organisms (SSOs) (Jia et al., 2018; Pang and Yuk, 2019; Zhu et al., 2019). It is a significant SSOs due to its ability to produce volatile sulfides, amines, trimethylamine extracellular enzymes, trimethylamines, organic acids and some spoilage metabolites.
Quorum sensing (QS) is the mechanism by which cell population-dependent signaling and interactions are recognized by bacteria in order to modulate their collective behaviors, including spoilage activity, enzyme secretion, bioluminescence, biofilm formation, virulence, and several signal molecules that mediate this mechanism have now been reported (Natrah et al., 2012; Zhu S. et al., 2015). There are various QS molecules, such as CAI-1 (cholera autoinducer 1), DKPs (Diketopiperazines), HAQs (4-hydroxy-2-alkylquinolines), DSFs (Diffusible Signal Factors), AI-2 (Autoinducer-2) and indole, etc (Monnet and Gardan, 2015; Papenfort and Bassler, 2016). QS-mediated communication is based on the prevailing interspecies communication in both Gram-negative and Gram-positive bacteria occurring via autoinducer-2 (AI-2) and auto-inducing peptides (AIPs) (Stephens et al., 2019). P fluorescens is a Gram-negative bacteria that has been reported to use N-acyl-homoserine lactone (AHL) signals to monitor its local population through the exchange of extracellular signal molecules (Li et al., 2018). SSOs utilize QS communication of circuits to regulate a diverse array of physiological activities microbial, including eavesdropping, biofilm genesis, and bioluminescence. Recently, several studies have found that QS signal molecules were N-butyryl-DL-homoserine lactone (C4-HSL), N-hexanoyl-DL-homoserine lactone (C6-HSL), octanoyl-L-homoserine lactone (C8-HSL), decanoyl-homoserine lactone (C10-HSL), and N-dodecanoyl-L-homoserine lactone (C12-HSL), which significantly reduced the protease activities and spoilage potential of SSOs. Moreover, QS systems can govern bacterial behavior in food spoilage ecology (Whiteley et al., 2017). Bacterial growth, protease production, spoilage potential, electrochemical tests, and spoilage protein expression were significantly enriched through AHL signal regulation (Zhu et al., 2017). However, few studies have focused on how two species of SSOs regulate their protein function in relation to spoilage potential.
Recently, proteomic analysis has become a beneficial and quantifiable technique for providing relative measurements of proteins from different samples by nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS/MS) without any isotope labeling. In a previous study on QS signaling, an absolute quantitative proteomic experiment was designed to investigate how Cyclo (L-phenylalanine-L-proline) effects protein expression in Staphylococcus aureus. Cyclo (L-phenylalanine-L-proline) has focused mainly on LuxR-mediated QS systems in bacteria, while the mechanism of extracellular QS signal molecule remained known (Ai et al., 2019). Wang et al. (2019) identified 1103 acetylated proteins and 2929 acetylation sites in Shewanella baltica from aquatic products to evaluate spoilage activity by LC-MS/MS analysis. A label-free quantitative proteomic approach has been applied to analyze the differential protein expression of Enterococcus faecalis SK460 to determine potential factors related to enterococcal biofilm formation. The results of this study showed that the related protein in the relevance of LuxS QS and pheromone in the biofilm development of E. faecalis was due to the Fsr being lacking in QS (Suryaletha et al., 2019). Based on proteomic analysis, a total of 338 vesicular proteins of Pseudomonas aeruginosa were identified with high confidence by LC-MS/MS analysis. This proteome profile provides a basis for future studies to illustrate the pathological functions of outer membrane vesicles from P. aeruginosa (Choi et al., 2011). However, there are very few studies demonstrating the fact that two SSOs have a possible role in QS for controlling expression of their target proteins and regulating bacterial behavior.
The aim of this study was to examine AHL production and AI-2 activity in A. johnsonii, P. fluorescens, and their co-culture, and to examine AHL-based QS systems regulating biofilm formation, protease activity, spoilage potential, and key proteins. Proteomic analysis of A. johnsonii, P. fluorescens, and their co-culture was used to elucidate the potential role of QS systems and their expression of proteins, interspecies communication, and metabolic pathways, which might be useful for developing effective methods for detecting spoilage capability.
Materials and Methods
Preparation of Bacterial Strain Cultures
Two kinds of SSOs (A. johnsonii and P. fluorescens) were originally isolated from spoiled bigeye tuna muscle (Zhejiang Fenghui Ocean Fishing Co., Ltd., China) by 16s rRNA gene sequences and VITEK® 2 CompactA system (bioMérieux, France). Stock cultures containing 25% glycerine were stored at −80°C. Before use, A. johnsonii and P. fluorescens were pre-cultured individually over two successive periods of 18 h in a brain-heart infusion broth at 30°C and then cultured in tryptic soya broth until the maximal concentration (108 CFU/mL) was attained. The bioreactors were inoculated with single strains overnight culture at a 1% (v/v) inoculation level. The co-culture consisted of a mixture of equal amounts (v/v) of A. johnsonii and P. fluorescens. All bacterial strains were cultured overnight at 30°C, then centrifuged at 10,000 g for 10 min and collected in a 15 mL centrifuge tube to remove the medium. The precipitate was finally collected and washed three times with pre-cooled 10 mL 1 × PBS solution. The precipitate obtained was transferred to a new 2 mL centrifuge tube and immediately stored at −80°C. Chromobacterium violaceum CV026 and Vibrio harveyi BB170 were kindly donated by Dr. Mi (Bohai University, Jinzhou, China).
Preparation of Reagents
C4-HSL, C6-HSL, and C8-HSL with purities of over 96% were purchased from Sigma-Aldrich (St.Louis, MO, United States). Other reagents were analytical grade and were purchased from Sangon Biotech Co., Ltd (Shanghai, China) and Aladdin Industrial Corporation (Shanghai, China).
Detection of AHLs and AI-2 Activity
Detection of AHLs by Cross-Feeding Plate Assay
Detection of AHLs was carried out by the cross-feeding plate assay reported by Chu et al. (2011). Test strains (A. johnsonii, P. fluorescens and co-culture) were cultured with shaking in TSB at 30°C for 24 h. CV026 was cultured with shaking in Luria-Bertani (LB) broth supplemented with kanamycin (20 μg/mL) as the reporter strain at 30°C for 24 h. All strains were grown to approximately 108 CFU/mL (OD600≈0.8). The test strain and CV026 were struck in parallel on the LB nutrient agar plate. The plate was dried with sterile air and then incubated at 28°C for 24 h. The reporter strain was used as the negative control.
Detection of AI-2 Activity
Vibrio harveyi BB170 was inoculated into the Autoinducer Bioassay (AB) liquid medium (Bodor et al., 2008) and cultured overnight with consistent shaking in AB liquid medium adjusted to 108 CFU/mL (OD600≈0.8). The fresh AB medium and the test bacterial solution were mixed and diluted at a ratio of 1:5000. A volume of 20 μL of culture supernatant and 180 μL of V.harveyi BB170 dilution were mixed at 100 r/min for 4h at 30°C in a 96-well black microtiter plate. Finally, a light unit at a wavelength of 460 nm was measured every 1d with a multiplate reader (Synergy 2, BioTek, Winooski, VT, United States). The cell-free culture of V. harveyi BB170 supernatant was defined as a positive control. Sterile AB medium served as a blank control. This test was repeated three times.
Extraction and Detection of AHLs by HPLC-MS
To acquire the supernatant, the concentrated bacteria of the test bacterial cultures during the logarithmic phase of growth was centrifuged at 10,000 r/min for 10 min at 4°C. A volume of 50mL of the supernatant was extracted three times into an equal volume of acidified ethyl acetate (0.5% acetic acid). The AHLs were dissolved in an appropriate volume of methanol and stored at −20°C for further experiments.
The solution of AHLs was drained of solvent, and then 1mL of the extracted solution (methanol:water = 80:20) was added to an ultrasound steel ball grinding machine (6 min, 50 Hz) at −20°C for 30 min and centrifuged at 13000 g for 15 min at 4°C to obtain 200 μL of supernatant.
Quantitative experiments involving the AHLs were performed on a Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific, Waltham, MA, United States) coupled to an Easy-nLC 1200 nano-flow UHPLC. The chromatographic parameters were as follows: column: BEH C18 (100 × 2.1 mm i.d., 1.7 μm; Waters Corporation, Milford, MA, United States), flow rate: 0.4 mL/min, sample injection volume: 5 μL, gradient mobile phase: water with 0.1% formic acid as solvent A and 0.1% formic acid and methanol as solvent B, column temperature: 40°C, and ion mode: positive ion mode (ESI+). The gradient mobile phase program applied was as follows, t = 0 min, 90% A; t = 4 min, 5% A; t = 9 min, 5% A; t = 7.1 min, 90% A; t = 10 min, 90% A. The conditions of MS/MS included the ion source temperature: 320°C, spray voltage: 3.5kV, sheath gas flow rate: 50 Arb and MS/MS collision energy: 70–1050 V.
Protease Activity Assay
The protease activity of the culture supernatant was determined according to the method described by Zhu J. et al. (2015), with a slight modification. A volume of 20 μL of the culture supernatant was added to 2.5 μL of the protease substrate solution. Incubation buffer was added to adjust the final assay volume to 50 μL. The tube was sealed and incubated at 37°C for 0.5 h. Following incubation, 50 μL of precipitation agent were added, and the contents were mixed and incubated again at 37°C for 10 min. The tubes were then centrifuged at 12,000 g for 5min. All experiments were conducted in triplicate.
Biofilm Formation Assay
Biofilm formation was investigated using an alteration of the method reported by Li et al. (2018). Biofilm formation was measured by crystal violet assay. Briefly, the above 96-well polystyrene microtiter plates inoculated A. johnsonii, P. fluorescens and co-culture. Following incubation, the biofilm in the plates in each well was carefully washed thrice with sterile phosphate buffered saline (PBS, pH.0) to remove unattached cells. After drying, the wells were added 0.2% (w/v) crystal violet to stain 15 min. The wells were then washed and dried as described above, then the dye attached to the biofilm was re-solubilized with 95% ethanol for 5 min. A volume of 200 μL of the sample solution was measured at 595 nm using a microplate reader.
Proteome Analysis
Protein Extraction and Digestion
The bacterial cells were mixed with lysis solution (8 M urea, protease inhibitor cocktail), and incubated in ice-bath for 30 min, vortex-oscillated for 5 s every 5 min. The mixture was centrifuged at 12,000 g, 4°C for 30 min to obtain supernatant. Protein concentrations were measured using a BCA Assay Kit (Thermo Fisher Scientific, United States). Each sample tube contained 150 μg of protein. The sample solution was added to tris(2-carboxyethyl)phosphine (TCEP) to reach a final concentration of 10 mM and incubated at 37°C for 60 min. Then, an appropriate quantity of iodoacetamide (IAM) was added to achieve a final concentration of 40mM and incubated for 40min in the dark. Finally, 100 mM triethylammonium bicarbonate (TEAB) buffer was added to dilute the concentration of the solution. To each sample tube was then added trypsin solution in the ratio 1:50, and the tubes were thenincubated at 37°C overnight.
Mass Spectrometry Analysis and Protein Identification
A total of nine samples from three groups (A, P, AP groups) were analyzed on a Q Exactive HF-X mass spectrometer coupled to an Easy-nLC 1200 nano-flow UHPLC. Three biological replicates were used per sample. Each peptide sample (0.5 μgμL) was injected for nano-LC-MS/MS analysis. Each sample was loaded onto the C18 reversed phase column (75 μm × 25 cm, Thermo Fisher Scientific, United States) having two solvent systems (buffer A: 2% acetonitrile and 0.1% formic acid; buffer B: 80% acetonitrile and 0.1% formic acid) for 160 min at a flow rate of 300 nL/min. The full scan MS spectra ranged from 350 to 1300m/z and were acquired with a mass resolution of 70 K. Three biological replicates were used per sample.
MS/MS spectra were screened by Proteome DiscovererTM 2.2 software (Thermo Fisher Scientific, United States) against the uniprot-Acinetobacter johnsonii-taxonomy_40214-20190813, uniprot-Pseudomonas fluorescens-taxonomy_294-20190813 and the decoy database with following parameters. The highest score for a given peptide mass (best match to that predicted in the database) was used to identify parent proteins. The parameters for protein searching were set as follows: tryptic digestion with up to two missed cleavages, carbamidomethylation of cysteines as a fixed modification, and oxidation of methionines and protein N-terminal acetylation as variable modifications. A 1% false discovery rate (FDR) was used to identify peptide spectral matches based on q-Values.
Statistical Analysis
All statistical values are expressed as the mean ± standard deviation. The biofilm formation, AHLs, AI-2 expression, and protease activity were analyzed by one-way analysis using SPSS 22.0 software (SPSS Inc., Chicago, IL, United States). The differential expression proteins were assigned to functional analysis according to the gene ontology (GO), clusters of orthologous groups of proteins (COG), and KEGG pathway 3 databases.
Results and Discussion
Identification and Detection of AHLs in A. johnsonii, P. fluorescens, and Co-culture
The reporter strain of the cross-feeding plate method is the fastest and most direct method to qualitatively detect AHLs in bacteria. CV026 is one of the commonly used reporter strains. CV026 does not produce AHLs by itself; however, CV026 is able to sense some of the AHLs of CivR protein (Yu et al., 2019). CV026 is fully capable of producing violacein in response to its cognate signal molecule. As shown in Figure 1A, P. fluorescens and the co-culture were capable of producing AHLs induced CV026 color reaction, while A. johnsonii could have low capable of producing AHLs not induce CV026 color reaction. The AHL signals for A. johnsonii, P. fluorescens, and their co-culture are shown in Figure 2 and Supplementary Figure S2. The three peaks with retention times of 3.26, 4.29, and 4.84 min were identified as C4-HSL, C6-HSL, and C8-HSL, respectively. The AHL (C4-HSL, C6-HSL, and C8-HSL) concentrations of the co-culture samples were significantly higher than those of A. johnsonii and P. fluorescens, indicating that the co-culture had pronounced AHL activities. The cooperative behaviors of the A. johnsonii and P. fluorescens cultures in response to AHLs activities might be to accelerate aquatic products spoilage. These trends are in agreement with the results presented in Table 1. The HPLC-MS chromatograms of the AHLs of the co-culture in C4-HSL, C6-HSL, and C8-HSL contained 3.145, 7.359, and 1.560 ng/ml, respectively (Table 1). Though several studies have reported that AHL signal molecules were the strongest autoinducers in Gram-negative bacteria from aquatic products (Zhu J. et al., 2015), reports of relative AHL production in A. johnsonii and its co-culture are limited.
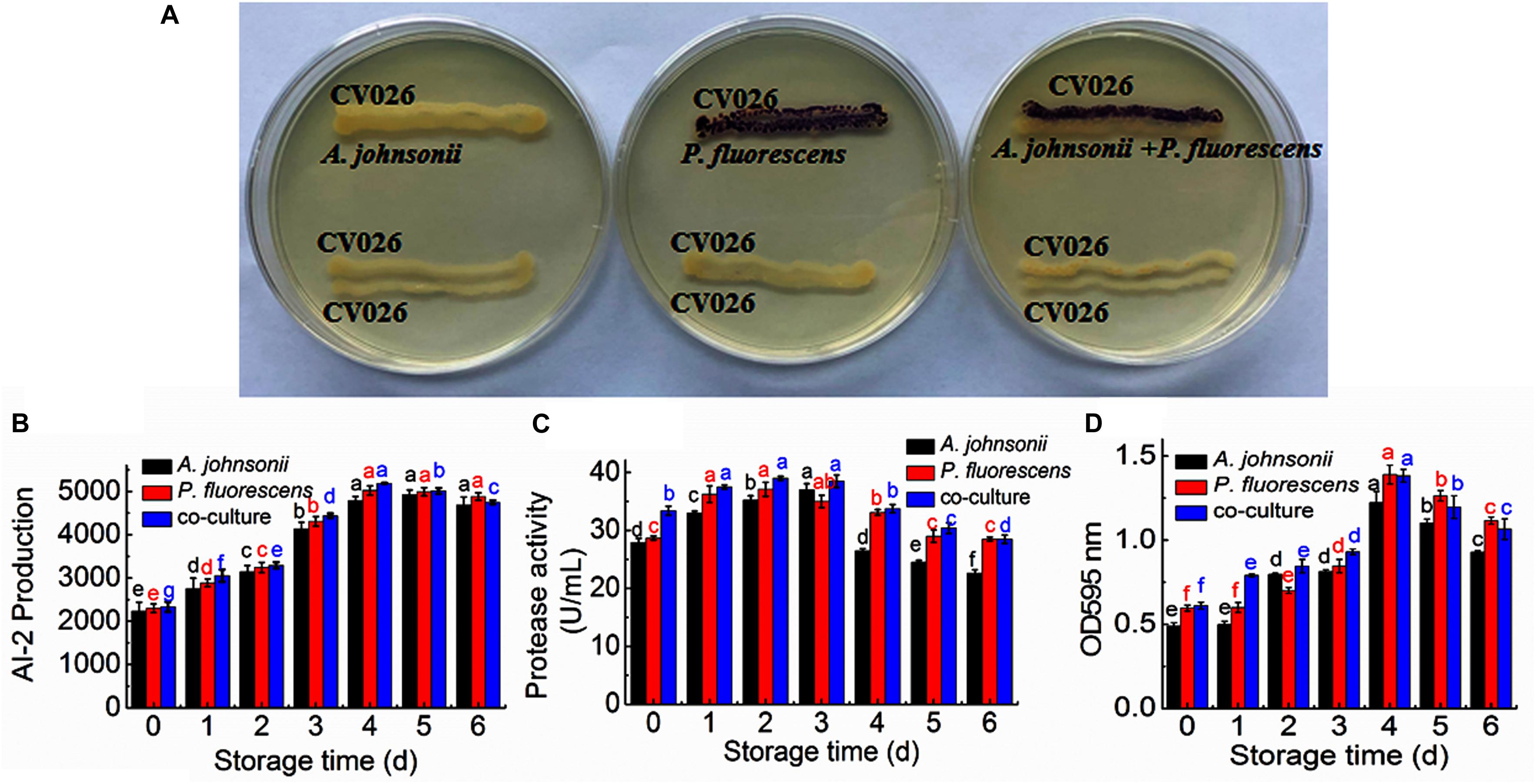
Figure 1. (A) Detection of AHL production in A. johnsonii, P. fluorescens, and their co-culture; changes in: (B) AI-2 production; (C) protease activity; and (D) biofilm production in A. johnsonii, P. fluorescens, and their co-culture after 6 days.
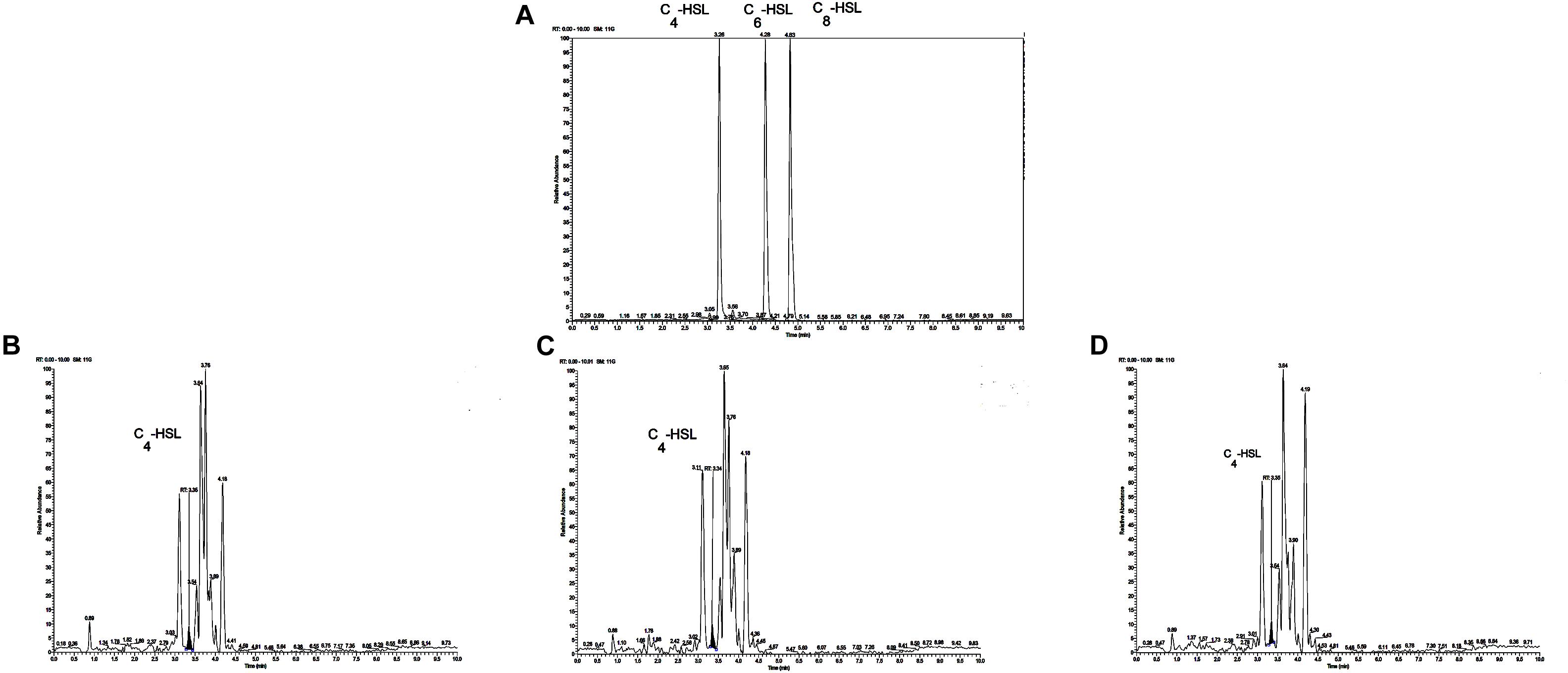
Figure 2. (A) Chromatograms of three kinds of AHL mixed standards. Chromatograms of C4-HSL produced by: (B) A. johnsonii; (C) P. fluorescens; and (D) their co-culture.
Detection of AI-2 Activity in A. johnsonii, P. fluorescens, and Co-culture
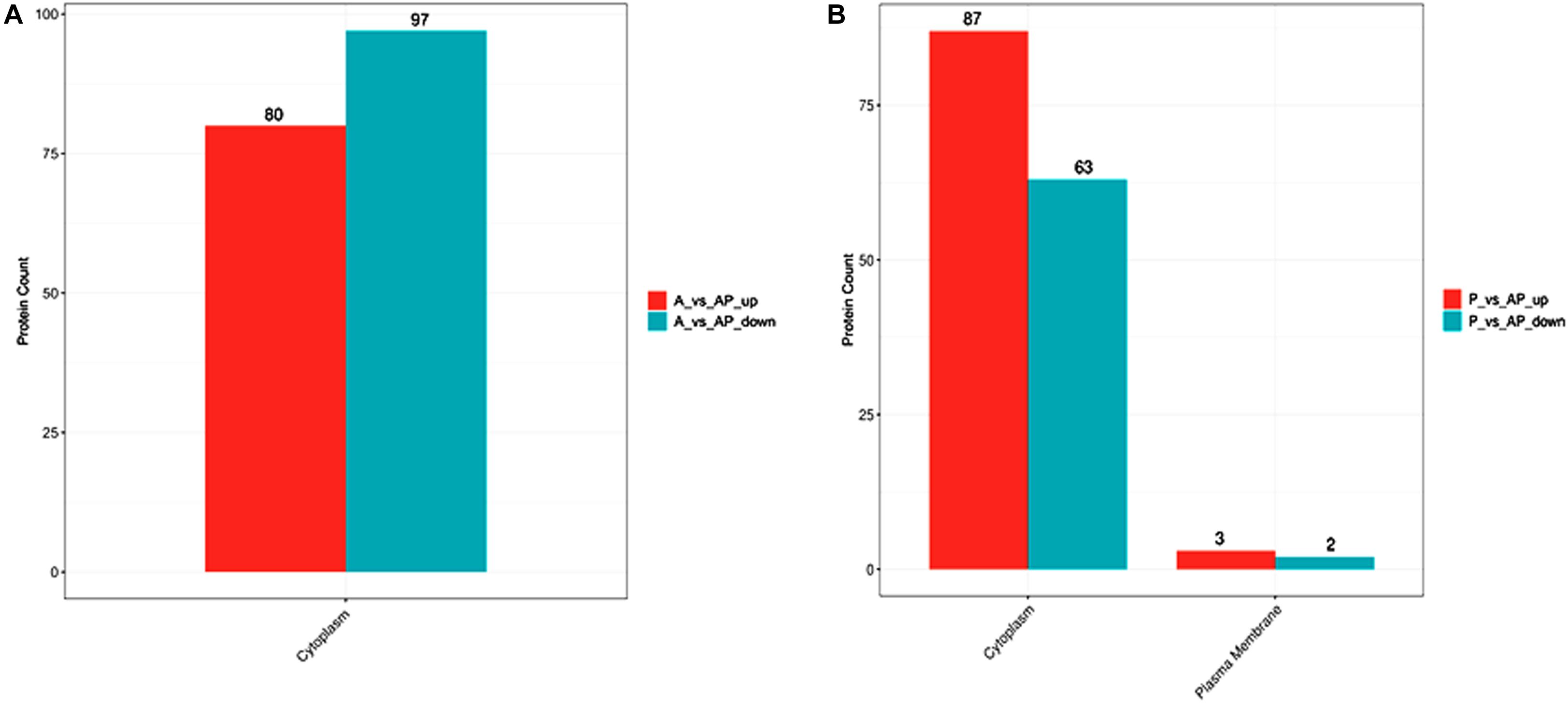
The activities of AI-2 signal molecules of A. johnsonii, P. fluorescens, and their co-culture were detected (Figure 1B). There were significant differences in the AI-2 activity of A. johnsonii, P. fluorescens, and their co-culture at different incubation times (p < 0.05). With increased culture time, the AI-2 activity first increased significantly from 0 to 4 days, then decreased, which was caused by increased bacterial growth density of bacteria, and bacteria in logarithmic phase. At the end of culture time, bacteria were in stationary phase and decline phase. Therefore, AI-2 activity decreased. It indicated that the changes in AI-2 activity were related to the secretion of the spoilage bacteria and environmental changes. Similar studies have shown that the AI-2 of QS is a global regulatory factor in aquatic products and influences the potential for spoilage (Peng et al., 2018; Li S. et al., 2019).
Protease Activity of A. johnsonii, P. fluorescens, and Co-culture
Protease activity plays an essential role in food SSOs and is regulated by QS (Moradi et al., 2019; Li T. et al., 2020). Moreover, protease activity decomposes aquatic food proteins into small peptides and amino acids that are metabolized into volatile nitrogenous end products (Li J. et al., 2019). In this study, protease activity exhibited no significant effect after 3 days. The protease activity of A. johnsonii, P. fluorescens, and their co-culture first increased and then decreased (p < 0.05) during different culture periods (Figure 1C). The previous study reported that SSOs growth consumed low-molecular-weight compounds, and then protein was degraded by protease which caused SSOs growth and protease activity increase (Moradi et al., 2019). At the end of culture time, molecular-weight compounds were almost depleted. Therefore, protease activity decreased. The protease activities of the co-cultured samples were also higher than those of the single bacteria groups, in accord with the results reported in the literature (Li J. et al., 2019). This result suggests that protease activity as a key spoilage characteristic of co-cultured samples was regulated significantly, and at least partially, by an AHL-based QS system.
Biofilm Formation by A. johnsonii, P. fluorescens, and Co-culture
Not only can biofilms influence food spoilage, resulting in reduced shelf life, but they can also adhere to bacteria and colonize surfaces (Zhou et al., 2019). Previous studies have provided some evidence to verify the correlation between biofilm formation and the SSOs of QS systems, by which exogenous bacteria can affect the formation of biofilms (Cui et al., 2020). Figure 1D shows that A. johnsonii, P. fluorescens, and their co-culture had the ability to form biofilms, which were greater in the co-cultured samples than those of the single bacteria. After 4d of culture, biofilm production reached maximum levels of 1.22, 1.38, and 1.39 by A. johnsonii, P. fluorescens, and their co-culture, respectively. Biofilm production increased when the incubation period was extended to 4d, but slowly decreased after 5d. Among the three groups, the presence of the co-cultured samples resulted in a significant increase in biofilm formation. This revealed that the cooperative behaviors of the A. johnsonii and P. fluorescens cultures in response to various signaling molecules helped to accelerate biofilm production.
Proteome Analysis of A. johnsonii, P. fluorescens, and Co-culture
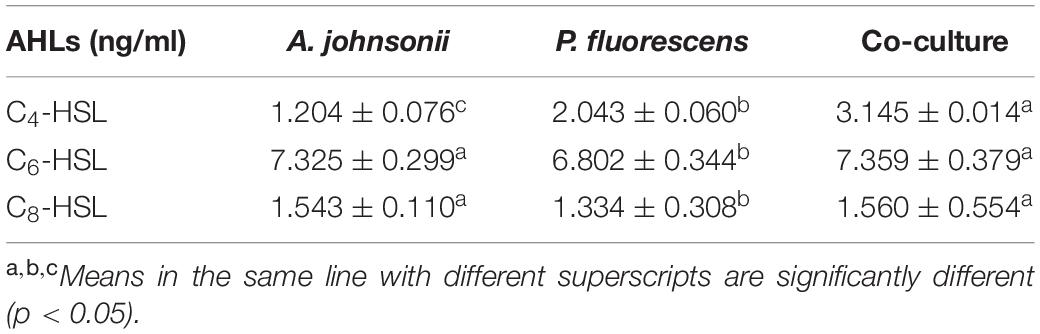
Acinetobacter johnsonii, P. fluorescens, and their co-cultured samples were prepared using the label-free technique, which showed that a total of 1,176,121 spectra were identified in A. johnsonii and P. fluorescens by Proteome DiscovererTM 2.2 software with a peptide FDR ≤ 0.01. As the fold change was >1.2 or <0.83, a p-value of <0.05 was used as the threshold to define the significance of the difference in protein expression. This enabled the quantification of 470 proteins and 444 proteins in A. johnsonii and P. fluorescens, respectively. As shown in Figure 3A, there were 80 significant up-regulated proteins and 97 down-regulated proteins in the A group, compared with the AP group. In addition, 90 up-regulated and 65 down-regulated proteins were in the P group, compared with the AP group (Figure 3B). The differences in protein expression are given in Tables 2, 3. Among the remarkably up-regulated proteins, the AI-2E family transporter OS, such as those of A0A2S8XHJ7, A0A166PHR0, A0A423M0X2, A0A2N1DUG2, and A0A165YHM6, showed notable up-regulation, which mediated the regulation of QS system expression (Quintieri et al., 2019; Saipriya et al., 2020). The result of AI-2 protein expression obtained above was similar to those shown in Figures 1B, 2. Moreover, some ribosomal proteins were significantly up-regulated, of which 50S ribosomal protein L3 OS, 50S ribosomal protein L25 OS, 30S ribosomal protein S17 OS, 50S ribosomal protein L23 OS, 30S ribosomal protein S2 OS, 50S ribosomal protein L29 OS, 50S ribosomal protein L10 OS, 50S ribosomal protein L4 OS, 30S ribosomal protein S10 OS, 30S ribosomal protein S4 OS, 50S ribosomal protein L9 OS, 50S ribosomal protein L15 OS, 50S ribosomal protein L11 OS, 50S ribosomal protein L6 OS, 50S ribosomal protein L7/L12 OS, 50S ribosomal protein L2 OS, and 50S ribosomal protein L1 OS were expressed at much higher levels, and their fold changes were all greater than 139.12. A similar result was tested for the progression of the ribosome through a regulatory open reading frame (ORF), which controlled the protein synthesis expression of many genes in P. fluorescens ITEM 17298 and influenced the expression of the downstream gene (Vazquez-Laslop et al., 2011; Gupta et al., 2013; Quintieri et al., 2019). The results showed that a majority of up-regulated proteins could accelerate protein expression and biological activity. There were some down-regulated proteins, such as thioredoxin reductase OS (A0A2K9M4Z2), cysteine synthase CysM OS (A0A506RJG5), DNA-binding response regulator (A0A423MKX5), amino acid ABC transporter ATPase OS (A0A0F4T6P4), that were expressed at a higher level in A vs. AP and P vs. AP. In the present study, bacteria played a critical role in transporting some molecules, including sugars, amino acids, vitamins, peptides, polysaccharides, lipids, thioredoxin, and ABC transporters (Rees et al., 2009; Zhong et al., 2019). Interestingly, more down-regulated proteins of the AI-2 family transporter OS were obtained in A vs. AP than in P vs. AP, which demonstrated more down-regulated proteins affecting AI-2 expression in A. johnsonii. Further investigations of AI-2 activity in A. johnsonii revealed it to be at a lower level than in the co-culture, suggesting that the latter might more easily contribute to AI-2 protein expression.
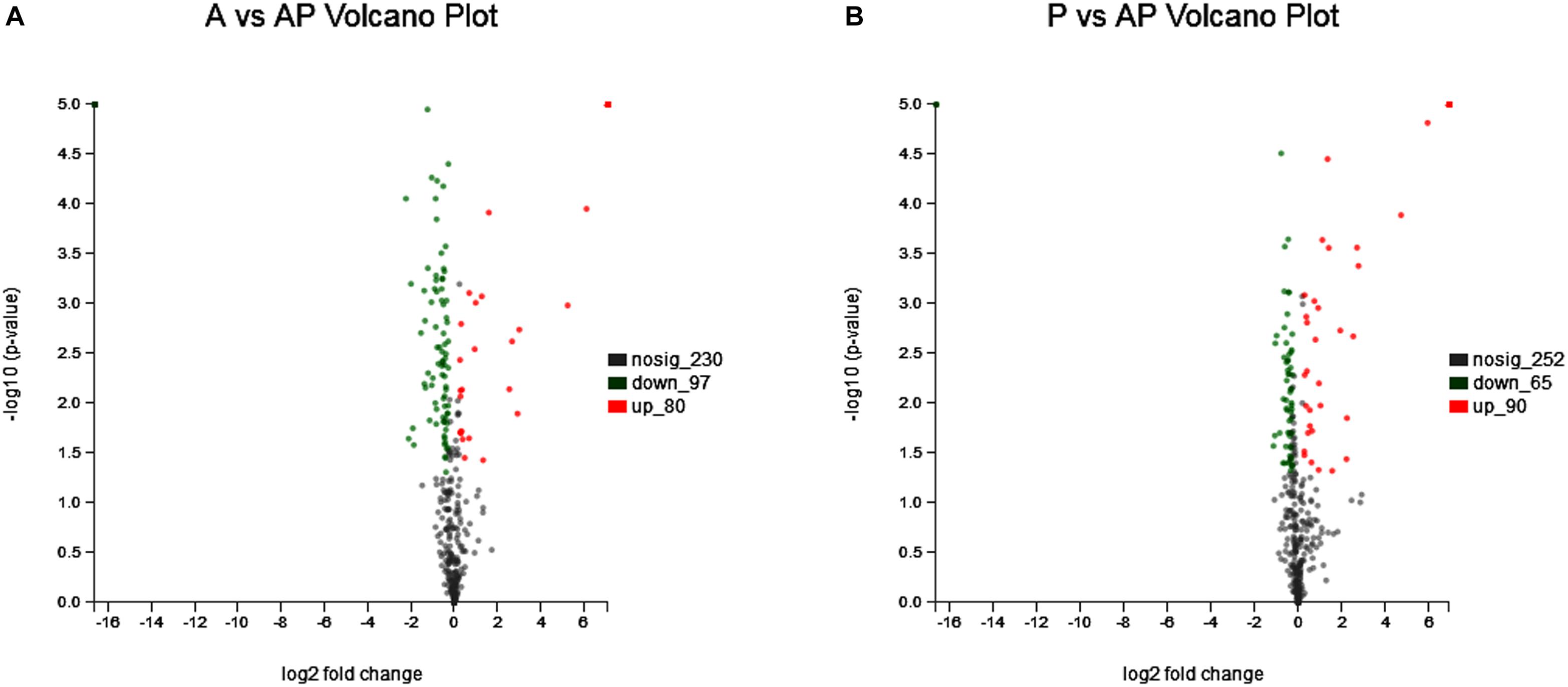
Figure 3. Volcanic map of all identified proteins: (A) volcanic map of all identified proteins in A vs. AP; (B) volcanic map of all identified proteins in P vs. AP. Red points: up-regulated proteins (fold change > 1.2, p < 0.05); green points: down-regulated proteins (fold change > 0.83, p < 0.05); black points: unchanged proteins.
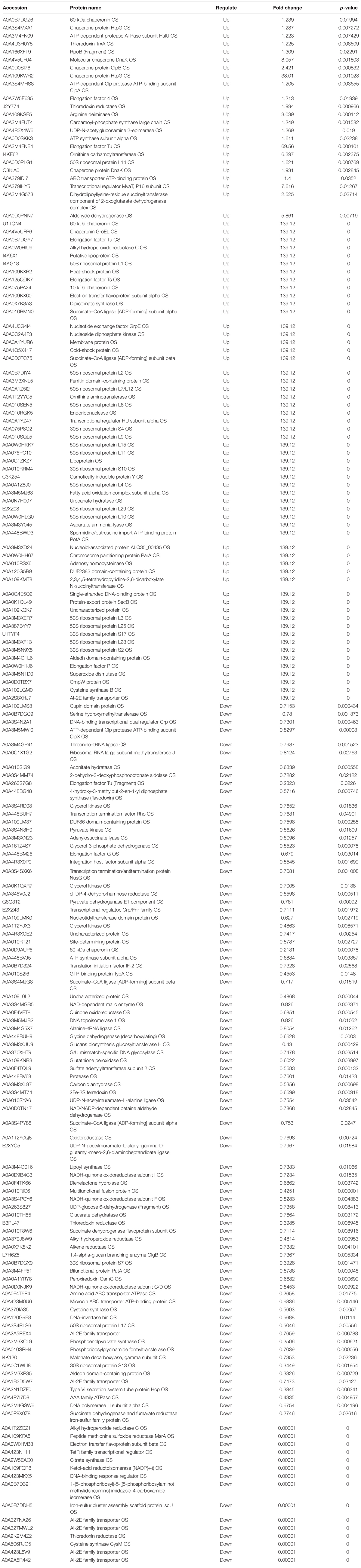
Table 2. The proteins differentially expression and most abundant proteins uniquely identified in A vs AP.
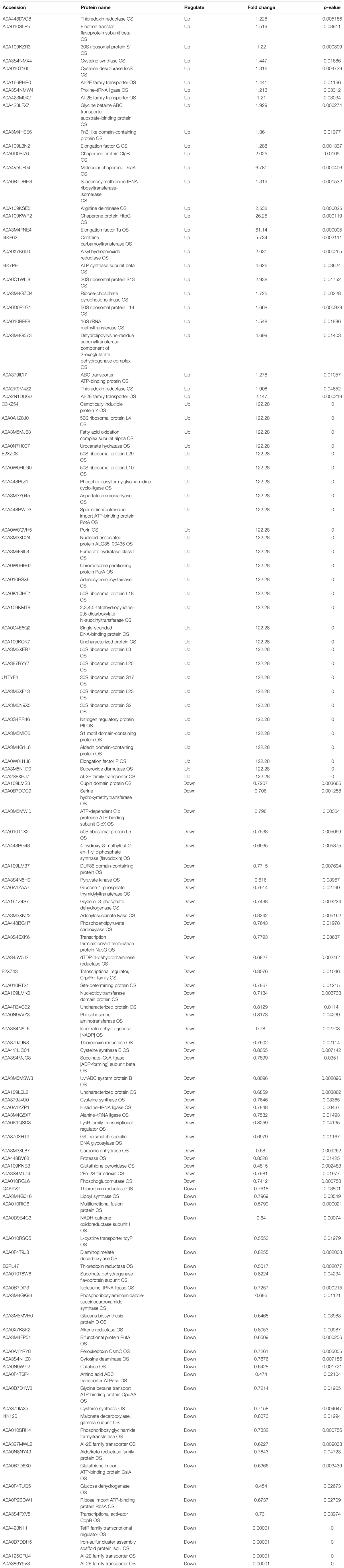
Table 3. The proteins differentially expression and most abundant proteins uniquely identified in P vs. AP.
Bioinformatics Functional Analysis of A. johnsonii, P. fluorescens, and Co-culture
GO and COG Enrichment Analysis of A. johnsonii, P. fluorescens, and Co-culture
A GO classification and COG enrichment of the 177 and 155 differentially expressed proteins were performed in A vs. AP and P vs. AP, respectively (Figures 4A,B). There were three main categories of cellular components, biological processes, and molecular functions in the GO classification. The cellular process (GO:0009987), the metabolic process (GO:0008152), and the single-organism process (GO:0044699) were the three main distributed terms in the biological processes. The 85 and 79 differentially expressed proteins were annotated as belonging to the cell in A vs. AP and P vs. AP, respectively. In addition, A vs. AP (80 out of 177) and P vs. AP (76 out of 155) of the differentially expressed proteins were localized in the cell part. This suggested that A. johnsonii, P. fluorescens, and their co-culture played an essential role in the transmembrane transport function and intracellular and extracellular substance migration, thereby promoting nutrient absorption and excretion of metabolic products. In the molecular functions category, the proteins were related to catalytic activity and binding.
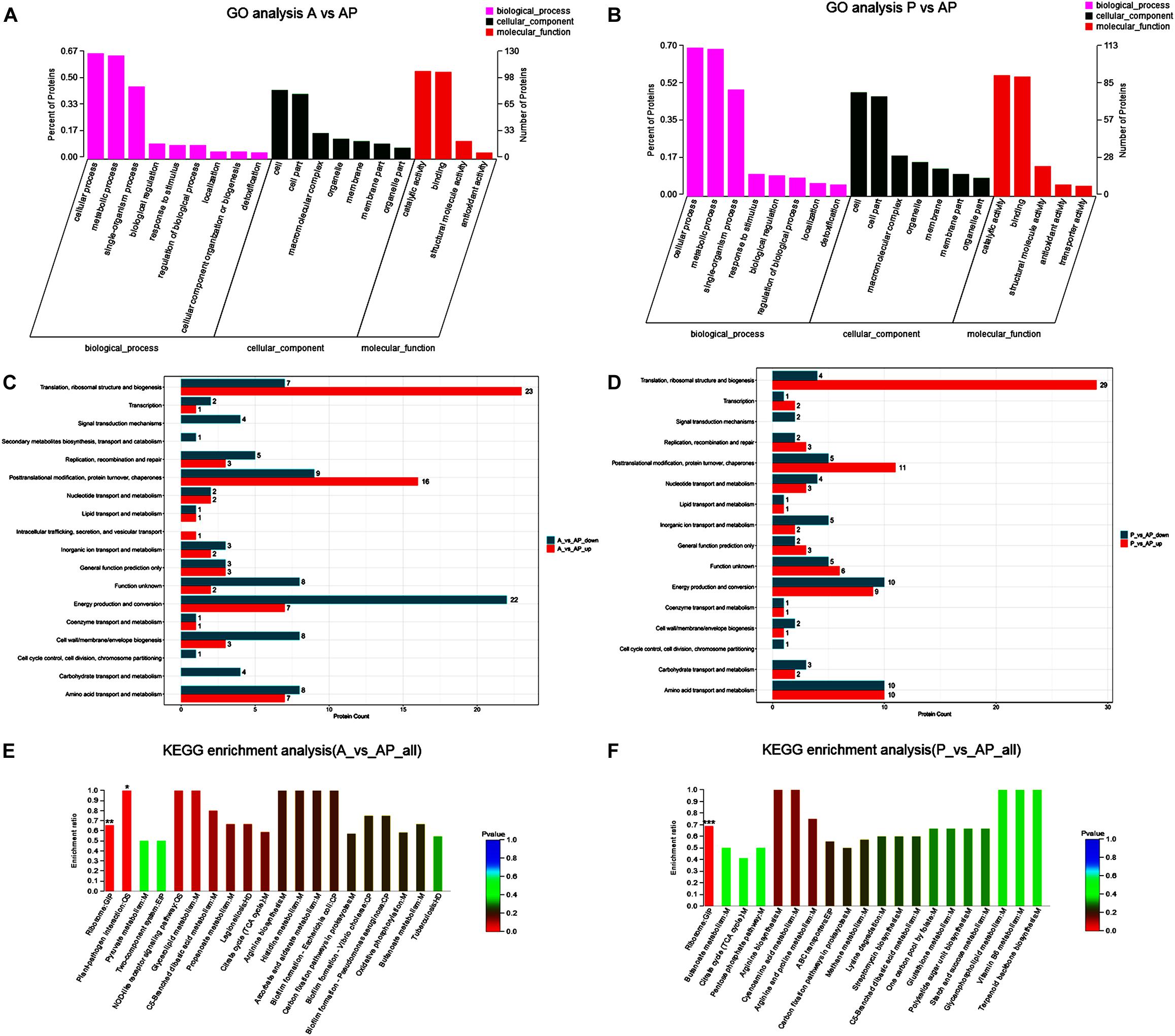
Figure 4. Gene ontology terms of the differentially expressed proteins in: (A) A vs. AP; (B) P vs. AP. COG terms of the differentially expressed proteins in: (C) A vs. AP; (D): P vs. AP. KEGG pathway analysis of the differentially expressed proteins in: (E) A vs. AP; (F) P vs. AP. The red bars represented the up-regulated proteins, and the blue bars represent the down-regulated proteins in the KEGG pathway analysis of A. johnsonii, P. fluorescens, and their co-culture. Candida albicans glycolysis/gluconeogenesis pathways (A) and peroxisomal assembly and fatty acid oxidation pathways.
Figures 4C,D displays the COG enrichment analysis. A total of 18 categories were classified, in which the top 6 COG terms were (i) energy production and conversion, (ii) amino acid transport and metabolism, (iii) translation, ribosomal structure, and biogenesis, (iv) post-translational modification, (v) protein turnover, and (vi) chaperones. A total of 22 down-regulated proteins were involved in energy production and conversion in A vs. AP, while 23 up-regulated proteins were involved in translation, ribosomal structure, and biogenesis in A vs. AP. Interestingly, translation, ribosomal structure, and biogenesis were significantly up-regulated in P vs. AP, and amino acid transport and metabolism and energy production and conversion were significantly down-regulated in P vs. AP, suggesting that translation, ribosomal structure, and energy production and conversion were valuable targets for co-culturing, and thus deserve further investigation. It has been suggested that under co-culture conditions, energy production is capable of involving translation of the bacteria, resulting in the activation of the ribosomal structure (Gupta et al., 2013). These proteins might also participate in nucleotide catabolism, allowing bacteria to use deoxynucleotides as energy sources. The results of GO analysis and COG enrichment provide a significant view of the proteins differentially expressed in A. johnsonii, P. fluorescens, and their co-culture that can elevate protein functions.
Pathway Enrichment Analysis of A. johnsonii, P. fluorescens, and Co-culture
Pathway enrichment analysis of the KEGG database was conducted on the differentially expressed proteins to reveal the metabolic and signal transduction pathways of A. johnsonii, P. fluorescens, and their co-culture (Figures 4E,F). In our study, taken together, the up/down-regulated differentially expressed proteins of A. johnsonii, P. fluorescens, and their co-culture involved in amino acid metabolism, carbohydrate and energy metabolism, and nucleotide metabolism enabled predictions of the change in the culture of the A. johnsonii, P. fluorescens samples in the co-culture, indicating that protein changes and carbohydrate transformation contributed to bacteria co-culturing. A total of 84 and 77 differentially expressed proteins in A vs. AP and P vs. AP, respectively, were divided into 20 KEGG pathways, and a majority of the metabolic pathways included genetic information processing, environmental information processing, cellular processes, organismal systems, and human diseases. The KEGG pathways of amino acid metabolism, carbohydrate metabolism, energy metabolism, and translation were significantly enriched. A similar result for carbohydrate metabolism and energy metabolism of Vibrio parahaemolyticus revealed that carbohydrate metabolism is the key factor, indicating that carbohydrate can either be converted into glucan and fructose through a glycosyltransferase reaction or transported by sugar transport systems and subsequently metabolized through glycolysis (Zhong et al., 2019).
The main significant items relevant to the regulation of biofilm formation of A vs. AP and P vs. AP included the map 02026, map 05111, and map 02025 pathways (Supplementary Tables S1, S2). Analysis of the proteins of A. johnsonii, P. fluorescens, and their co-culture indicated that all of the pathways were present. In addition, the pathway of map 02024 was that which regulated QS, which is a crucial feature affecting the regulation of biofilm formation by AHLs, bacterial growth, protease activity, and the spoilage potential of bacteria (Jie et al., 2018).
Furthermore, as shown in Supplementary Tables S1, S2, ribosome was the most significantly enriched pathway, which indicated that protein expression was substantially promoted to achieve a large demand for bacterial growth (Li J. et al., 2020). Ribosome was the most significant pathway, with 43 differentially expressed proteins, of which 22 were up-regulated proteins and 21 were down-regulated proteins. In addition, abundant proteins were associated with the tricarboxylic acid (TCA) cycle and oxidative phosphorylation in response to co-culture conditions (Supplementary Figure S1). The TCA cycle pathway has 7 up-regulated proteins and 9 down-regulated proteins. The most abundant protein detected among the 6 up-regulated and 11 down-regulated proteins was associated with oxidative phosphorylation. These pathways were essential for bacterial growth and cell interactions, which have potential for enhancing bacterial spoilage and QS regulation (Remenant et al., 2015; Otwell et al., 2018).
Subcellular Localization Prediction of A. johnsonii, P. fluorescens, and Co-culture
The subcellular localization prediction of proteins has attracted considerable attention in protein functional annotation. There were two subcellular localization predictions, including for cytoplasm and plasma membranes. Figure 5 shows that subcellular localization in P vs. AP was located mainly in the cytoplasm and the plasma membrane, but mainly in the cytoplasm in A vs. AP. Previous studies have focused mainly on single bacteria producing a few metabolites that migrate from the inner membrane to cell-extracellular membranes during culture (Ellepola et al., 2019), resulting in cell communications. However, few studies have reported explanations for two bacterial subcellular localization predictions. As shown in Figure 5, expression of cytoplasmic proteins was also regulated in the co-culture.
Quorum Sensing and Spoilage-Related Proteins
The QS mechanism is the means by which cell population communication can regulate specific proteins to express the physiological characteristics of microorganisms (Li S. et al., 2019). As given in Table 4, the QS system was related to the protein of the AI-2E family transporter OS and the LuxR family transcriptional regulator OS. The LuxR family transcriptional regulator was found in A vs. AP and P vs. AP. The LuxR protein (AHL receptor) commonly consists of 200–260 amino acids blinding with the key protein, resulting in the expression of the key protein. Five AI-2E family transporter up-regulated proteins and 11 AI-2E family transporter up-regulated proteins were found in A vs. AP and P vs. AP, respectively. This suggests that the AI-2E family transporter played a vital role in regulating QS. Previous studies have shown that P. fluorescens can produce AI-2 proteins and AHLs (Sharma et al., 2006), which is consistent with the results described in Section “Protease Activity of A. johnsonii, P. fluorescens, and Co-culture.” Moreover, there were too many spoilage-related proteins, including thioredoxin reductase OS (6 down-regulated proteins, 5 up-regulated proteins), cysteine synthase OS (7 down-regulated proteins, 1 up-regulated proteins), and pyridoxal phosphate-dependent enzyme family protein OS. Notably, the spoilage-related proteins in A. johnsonii, P. fluorescens, and their co-culture were similar to those of Shewanella baltica and P. fluorescens ITEM 17298 (Quintieri et al., 2019).

Table 4. Quorum sensing system proteins and spoilage related proteins of A. johnsonii, P. fluorescens and co-culture.
Conclusion
This study was carried out in order to explore cultures of A. johnsonii and P. fluorescens and compare them with their co-cultured state for QS and spoilage potential by means of their proteomic profiles. The results show that the products of AHL production (C4-HSL, C6-HSL, C8-HSL), biofilm production, protease activity, and spoilage potential were at a higher level in the co-culture than those of A. johnsonii and P. fluorescens single cultures alone. The proteomic results revealed that there were differences in the proteins involved in the metabolism of amino acids, carbohydrates, energy, and translation. The differentially expressed proteins that were spoilage-related included thioredoxin reductase OS, cysteine synthase OS, and pyridoxal phosphate-dependent enzyme family protein OS, as well as specific QS system proteins of the AI-2E family transporter OS and the LuxR family transcriptional regulator OS, which could be used as biomarkers in A. johnsonii, P. fluorescens, and their co-cultured state. These results may provide an understanding of how the QS and spoilage mechanisms of A. johnsonii, P. fluorescens, and their co-cultures can be regulated in future.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD018646) and the iProX (https://www.iprox.org/page/project.html?id=IPX0002129000).
Author Contributions
X-YW analyzed the data, wrote the manuscript, and performed the experiments. JX made suggestions for revision and guided the experiments.
Funding
This research was financially supported by the National Natural Science Foundation of China (Grant Nos. 31972142 and 31571914), the China Agriculture Research System (CARS-47), and was also supported by the Shanghai Municipal Science and Technology Project to enhance the capabilities of the platform (Grant No. 19DZ2284000).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge Jun Yan for his helpful comments and suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00940/full#supplementary-material
FIGURE S1 | (A) KEGG pathway map of ribosome; (B) TCA cycle; and (C) oxidative phosphorylation. Proteins in the blue block belong to the experimental species. Red frames indicate up-regulated proteins, and green frames indicate down-regulated proteins.
FIGURE S2 | Chromatograms of C6-HSL produced by: (A) A. johnsonii; (B) P. fluorescens; and (C) their co-culture. Chromatograms of C8-HSL produced by: (D) A. johnsonii; (E) P. fluorescens; and (F) their co-culture.
TABLE S1 | KEGG pathway of the A group compared with the AP group.
TABLE S2 | KEGG pathway of the P group compared with the AP group.
References
Ai, D., Zhang, W., Yun, J., and Cao, Y. (2019). Analysis of the influence of cyclo (L-phenylalanine-L-proline) on the proteome of Staphylococcus aureus using iTRAQ. Ann. Microbiol. 69, 1247–1257. doi: 10.1007/s13213-019-01508-0
Bodor, A., Elxnat, B., Thiel, V., Schulz, S., and Wagner-Döbler, I. (2008). Potential for luxS related signalling in marine bacteria and production of autoinducer-2 in the genus Shewanella. BMC Microbiol. 8:13. doi: 10.1186/1471-2180-8-13
Choi, D. S., Kim, D. K., Choi, S. J., Lee, J., Choi, J. P., Rho, S., et al. (2011). Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics 11, 3424–3429. doi: 10.1002/pmic.201000212
Chu, W., Vattem, D. A., Maitin, V., Barnes, M. B., and McLean, R. J. (2011). Bioassays of quorum sensing compounds using Agrobacterium tumefaciens and Chromobacterium violaceum. Methods Mol. Biol. 692, 3–19. doi: 10.1007/978-1-60761-971-0_1
Cui, T., Bai, F., Sun, M., Lv, X., Li, X., Zhang, D., et al. (2020). Lactobacillus crustorum ZHG 2-1 as novel quorum-quenching bacteria reducing virulence factors and biofilms formation of Pseudomonas aeruginosa. LWT 117:108696. doi: 10.1016/j.lwt.2019.108696
Ellepola, K., Truong, T., Liu, Y., Lin, Q., Lim, T. K., Lee, Y. M., et al. (2019). Multi-omics analyses reveal synergistic carbohydrate metabolism in Streptococcus mutans-Candida albicans mixed-species biofilms. Infect. Immun. 87:e00339-19. doi: 10.1128/IAI.00339-19
Gupta, P., Kannan, K., Mankin, A. S., and Vazquez-Laslop, N. (2013). Regulation of gene expression by macrolide-induced ribosomal frameshifting. Mol. Cell. 52, 629–642. doi: 10.1016/j.molcel.2013.10.013
Jia, S., Huang, Z., Lei, Y., Zhang, L., Li, Y., and Luo, Y. (2018). Application of Illumina-MiSeq high throughput sequencing and culture-dependent techniques for the identification of microbiota of silver carp (Hypophthalmichthys molitrix) treated by tea polyphenols. Food Microbiol. 76, 52–61. doi: 10.1016/j.fm.2018.04.010
Jie, J., Yu, H., Han, Y., Liu, Z., and Zeng, M. (2018). Acyl-homoserine-lactones receptor LuxR of Shewanella baltica involved in the development of microbiota and spoilage of refrigerated shrimp. J. Food Sci. Technol. 55, 2795–2800. doi: 10.1007/s13197-018-3172-4
Li, J., Yu, H., Yang, X., Dong, R., Liu, Z., and Zeng, M. (2019). Complete genome sequence provides insights into the quorum sensing-related spoilage potential of Shewanella baltica 128 isolated from spoiled shrimp. Genomics 112, 736–748. doi: 10.1016/j.ygeno.2019.05.010
Li, J., Zhang, X., Ashokkumar, M., Liu, D., and Ding, T. (2020). Molecular regulatory mechanisms of Escherichia coli O157:H7 in response to ultrasonic stress revealed by proteomic analysis. Ultrason Sonochem. 61:104835. doi: 10.1016/j.ultsonch.2019.104835
Li, S., Tang, S., He, Q., Hu, J., and Zheng, J. (2019). Changes in proteolysis in fermented milk produced by Streptococcus thermophilus in co-culture with Lactobacillus plantarum or Bifidobacterium animalis subsp. lactis during refrigerated storage. Molecules 24:3699. doi: 10.3390/molecules24203699
Li, T., Sun, X., Chen, H., He, B., Mei, Y., Wang, D., et al. (2020). Methyl anthranilate: a novel quorum sensing inhibitor and anti-biofilm agent against Aeromonas sobria. Food Microbiol. 86:103356. doi: 10.1016/j.fm.2019.103356
Li, T., Yang, B., Li, X., Li, J., Zhao, G., and Kan, J. (2018). Quorum sensing system and influence on food spoilage in Pseudomonas fluorescens from turbot. J. Food Sci. Technol. 55, 3016–3025. doi: 10.1007/s13197-018-3222-y
Monnet, V., and Gardan, R. (2015). Quorum-sensing regulators in Gram-positive bacteria:‘cherchez le peptide’. Mol. Microbiol. 97, 181–184. doi: 10.1111/mmi.13060
Moradi, M., Sun, Z., Song, Z., and Hu, H. (2019). Effect of proteases secreted from a marine isolated bacterium Bacillus vietnamensis on the corrosion behaviour of different alloys. Bioelectrochemistry 126, 64–71. doi: 10.1016/j.bioelechem.2018.08.003
Natrah, F. M., Alam, M. I., Pawar, S., Harzevili, A. S., Nevejan, N., Boon, N., et al. (2012). The impact of quorum sensing on the virulence of Aeromonas hydrophila and Aeromonas salmonicida towards burbot (Lota lota L.) larvae. Vet. Microbiol. 159, 77–82. doi: 10.1016/j.vetmic.2012.03.014
Otwell, A. E., Callister, S. J., Sherwood, R. W., Zhang, S., Goldman, A. R., Smith, R. D., et al. (2018). Physiological and proteomic analyses of Fe(III)-reducing co-cultures of Desulfotomaculum reducens MI-1 and Geobacter sulfurreducens PCA. Geobiology 16, 522–539. doi: 10.1111/gbi.12295
Pang, X., and Yuk, H. G. (2019). Effects of the colonization sequence of Listeria monocytogenes and Pseudomonas fluorescens on survival of biofilm cells under food-related stresses and transfer to salmon. Food Microbiol. 82, 142–150. doi: 10.1016/j.fm.2019.02.002
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14:576. doi: 10.1038/nrmicro.2016.89
Peng, L.-Y., Yuan, M., Cui, Z.-Q., Wu, Z.-M., Yu, Z.-J., Song, K., et al. (2018). Rutin inhibits quorum sensing, biofilm formation and virulence genes in avian pathogenic Escherichia coli. Microb. Pathog. 119, 54–59. doi: 10.1016/j.micpath.2018.04.007
Quintieri, L., Zuhlke, D., Fanelli, F., Caputo, L., Liuzzi, V. C., Logrieco, A. F., et al. (2019). Proteomic analysis of the food spoiler Pseudomonas fluorescens ITEM 17298 reveals the antibiofilm activity of the pepsin-digested bovine lactoferrin. Food Microbiol. 82, 177–193. doi: 10.1016/j.fm.2019.02.003
Rees, D. C., Johnson, E., and Lewinson, O. (2009). ABC transporters: the power to change. Nat. Rev. Mol. Cell Biol. 10, 218–227. doi: 10.1038/nrm2646
Remenant, B., Jaffres, E., Dousset, X., Pilet, M. F., and Zagorec, M. (2015). Bacterial spoilers of food: behavior, fitness and functional properties. Food Microbiol. 45(Pt A), 45–53. doi: 10.1016/j.fm.2014.03.009
Saipriya, K., Swathi, C. H., Ratnakar, K. S., and Sritharan, V. (2020). Quorum-sensing system in Acinetobacter baumannii: a potential target for new drug development. J. Appl. Microbiol. 128, 15–27. doi: 10.1111/jam.14330
Sharma, S., Sundaram, C. S., Luthra, P. M., Singh, Y., Sirdeshmukh, R., and Gade, W. N. (2006). Role of proteins in resistance mechanism of Pseudomonas fluorescens against heavy metal induced stress with proteomics approach. J. Biotechnol. 126, 374–382. doi: 10.1016/j.jbiotec.2006.04.032
Silbande, A., Adenet, S., Smith-Ravin, J., Joffraud, J.-J., Rochefort, K., and Leroi, F. (2016). Quality assessment of ice-stored tropical yellowfin tuna (Thunnus albacares) and influence of vacuum and modified atmosphere packaging. Food Microbiol. 60, 62–72. doi: 10.1016/j.fm.2016.06.016
Stephens, K., Pozo, M., Tsao, C. Y., Hauk, P., and Bentley, W. E. (2019). Bacterial co-culture with cell signaling translator and growth controller modules for autonomously regulated culture composition. Nat. Commun. 10:4129. doi: 10.1038/s41467-019-12027-6
Sun, J., Wang, Q. J., Huang, J., Hou, Y. D., Chen, Y. F., and Su, X. R. (2013). Influence of heating temperature on the development of volatile compounds in bigeye tuna meat (Thunnus obesus) as assessed by E-nose and SPME-GC/MS. Int. Food Res. J. 20, 3077–3083.
Suryaletha, K., Narendrakumar, L., John, J., Radhakrishnan, M. P., George, S., and Thomas, S. (2019). Decoding the proteomic changes involved in the biofilm formation of Enterococcus faecalis SK460 to elucidate potential biofilm determinants. BMC Microbiol. 19:146. doi: 10.1186/s12866-019-1527-2
Vazquez-Laslop, N., Klepacki, D., Mulhearn, D. C., Ramu, H., Krasnykh, O., Franzblau, S., et al. (2011). Role of antibiotic ligand in nascent peptide-dependent ribosome stalling. Proc. Natl. Acad. Sci. U.S.A. 108, 10496–10501. doi: 10.1073/pnas.1103474108
Wang, X., Geng, L., Xie, J., and Qian, Y.-F. (2017). Relationship between water migration and quality changes of yellowfin tuna (Thunnus albacares) during storage at 0°C and 4°C by LF-NMR. J. Aquat. Food Product Technol. 27, 35–47. doi: 10.1080/10498850.2017.1400630
Wang, X.-Y., and Xie, J. (2019). Evaluation of water dynamics and protein changes in bigeye tuna (Thunnus obesus) during cold storage. LWT 108, 289–296. doi: 10.1016/j.lwt.2019.03.076
Wang, Y., Wang, F., Bao, X., and Fu, L. (2019). Systematic analysis of lysine acetylome reveals potential functions of lysine acetylation in Shewanella baltica, the specific spoilage organism of aquatic products. J. Proteomics 205:103419. doi: 10.1016/j.jprot.2019.103419
Whiteley, M., Diggle, S. P., and Greenberg, E. P. (2017). Progress in and promise of bacterial quorum sensing research. Nature 551, 313–320. doi: 10.1038/nature24624
Yu, H., Li, J., Han, Y., Shi, G., Liu, Z., and Zeng, M. (2019). AHLs-produced bacteria in refrigerated shrimp enhanced the growth and spoilage ability of Shewanella baltica. J. Food Sci. Tech. 56, 114–121. doi: 10.1007/s13197-018-3464-8
Zhong, Q., Wang, B., Wang, J., Liu, Y., Fang, X., and Liao, Z. (2019). Global proteomic analysis of the resuscitation state of Vibrio parahaemolyticus compared with the normal and viable but non-culturable state. Front. Microbiol. 10:1045. doi: 10.3389/fmicb.2019.01045
Zhou, S., Yu, Z., and Chu, W. (2019). Effect of quorum-quenching bacterium Bacillus sp. QSI-1 on protein profiles and extracellular enzymatic activities of Aeromonas hydrophila YJ-1. BMC Microbiol. 19:135. doi: 10.1186/s12866-019-1515-6
Zhu, J., Huang, X., Zhang, F., Feng, L., and Li, J. (2015). Inhibition of quorum sensing, biofilm, and spoilage potential in Shewanella baltica by green tea polyphenols. J. Microbiol. 53, 829–836. doi: 10.1007/s12275-015-5123-3
Zhu, J., Yan, Y., Wang, Y., and Qu, D. (2019). Competitive interaction on dual-species biofilm formation by spoilage bacteria, Shewanella baltica and Pseudomonas fluorescens. J. Appl. Microbiol. 126, 1175–1186. doi: 10.1111/jam.14187
Zhu, S., Wu, H., Zeng, M., Liu, Z., and Wang, Y. (2015). The involvement of bacterial quorum sensing in the spoilage of refrigerated Litopenaeus vannamei. Int. J. Food Microbiol. 192, 26–33. doi: 10.1016/j.ijfoodmicro.2014.09.029
Keywords: quorum sensing, autoinducers (AHLs), AI-2, proteome, Acinetobacter johnsonii, Pseudomonas fluorescens, spoilage
Citation: Wang X-Y and Xie J (2020) Quorum Sensing System-Regulated Proteins Affect the Spoilage Potential of Co-cultured Acinetobacter johnsonii and Pseudomonas fluorescens From Spoiled Bigeye Tuna (Thunnus obesus) as Determined by Proteomic Analysis. Front. Microbiol. 11:940. doi: 10.3389/fmicb.2020.00940
Received: 29 December 2019; Accepted: 20 April 2020;
Published: 14 May 2020.
Edited by:
Giovanna Suzzi, University of Teramo, ItalyReviewed by:
Laura Quintieri, Institute of Sciences of Food Production (CNR), ItalyJianjun Qiao, Tianjin University, China
Copyright © 2020 Wang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Xie, anhpZUBzaG91LmVkdS5jbg==
 Xin-Yun Wang
Xin-Yun Wang Jing Xie
Jing Xie