
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 May 2020
Sec. Microbiotechnology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00924
This article is part of the Research Topic Trace Elements in the Environment: Biogeochemical Cycles and Bioremediation View all 6 articles
Salix integra Thunb., a fast-growing woody plant species, has been used for phytoremediation in recent years. However, little knowledge is available regarding indigenous soil microbial communities associated with the S. integra phytoextraction process. In this study, we used an Illumina MiSeq platform to explore the indigenous microbial composition after planting S. integra at different lead (Pb) contamination levels: no Pb, low Pb treatment (Pb 500 mg kg–1), and high Pb treatment (Pb 1500 mg kg–1). At the same time, the soil properties and their relationship with the bacterial communities were analyzed. The results showed that Pb concentration was highest in the root reaching at 3159.92 ± 138.98 mg kg–1 under the high Pb treatment. Planting S. integra decreased the total Pb concentration by 84.61 and 29.24 mg kg–1, and increased the acid-soluble Pb proportion by 1.0 and 0.75% in the rhizosphere and bulk soil under the low Pb treatment compared with unplanted soil, respectively. However, it occurred only in the rhizosphere soil under the high Pb treatment. The bacterial community structure and microbial metabolism were related to Pb contamination levels and planting of S. integra, while the bacterial diversity was only affected by Pb contamination levels. The dominant microbial species were similar, but their relative abundance shifted in different treatments. Most of the specific bacterial assemblages whose relative abundances were promoted by root activity and/or Pb contamination were suitable for use in plant-microbial combination remediation, especially many genera coming from Proteobacteria. Redundancy analysis (RDA) showed available nitrogen and pH having a significant effect on the bacteria relating to phytoremediation. The results indicated that indigenous bacteria have great potential in the application of combined S. integra-microbe remediation of lead-contaminated soil by adjusting soil properties.
Lead (Pb) is a widely distributed toxic element in soil with no biological function (Kushwaha et al., 2018). It is produced by mining and smelting activities, burning of leaded gasoline, as well as disposal of sewage sludge, batteries, and other lead-containing products (Huang et al., 2006). The accumulation of Pb causes serious harm to human, particularly on children. Many techniques to reduce its effects on ecosystems and humans have been tried, including physical, chemical, and biological processes, while most of them are high cost and also lead to secondary pollution (Ma et al., 2016). Combined microbial and plant remediation has become an popular way for its low cost, no secondary contamination, and being superior to single phytoremediation and microbiological methods (Khalid et al., 2017). Hence, plant-microbial remediation has become a research hotspot.
Although bacterially-assisted phytoremediation may seem pretty good at first for remediation of heavy metal contaminated soils, previous research has been performed only in pot culture and focused on the function of a single microbial inoculum (Hou et al., 2018; Zhang et al., 2018; Ramirez et al., 2019; Ju et al., 2020). They overlooked the complexity of the plant-soil environment in which abundant indigenous microbes may surpass or suppress such inoculated microbes (Wood et al., 2016; Kong et al., 2019; Xiao et al., 2020), and therefore the practical application of bacterially-assisted phytoremediation has generally been restricted (Xuliang et al., 2007; Kong et al., 2019; Xiao et al., 2020). However, mounting research has proved the importance of indigenous rhizosphere microbes in the process of phytoremediation (Thijs et al., 2016; Egamberdieva et al., 2017; Hou et al., 2019). A unique microbiome harboring in the rhizosphere of hyperaccumulator Sedum alfredii made a great contribution to the accumulation of trace metals (Hou et al., 2017a). Plants can accumulate more trace metals in natural soil than in γ-irradiated soil due to the differences in the rhizosphere microbial communities (Muehe et al., 2015; Hou et al., 2017b). Bacterial inoculation enhanced phytoremediation through interactions with indigenous rhizosphere bacteria (Hou et al., 2019; Joe et al., 2019). The study of the indigenous microbial community is, therefore, very important for phytoremediation, especially in the rhizosphere microenvironment.
Rhizosphere microbes are affected by soil physicochemical properties and heavy metal concentrations (Philippot et al., 2013; Guo et al., 2017; Lin et al., 2019; Cao et al., 2020). Soil organic matter, nitrogen, and pH can greatly influence the soil microbial structure and heavy metal content by affecting the bioavailability of heavy metals in soil (Lin et al., 2019; Wang et al., 2019a; Zhen et al., 2019; Cao et al., 2020). The soil N: P ratios and available phosphorus affected bacterial biodiversity and community composition in a reddish paddy soil (Huang et al., 2019). Soil Na+ significantly changes dominant microbes in Cd-contaminated soil (Wang et al., 2019b). Adjusting rhizosphere microbial community structure could, therefore be used to promote trace metal absorption by plants.
Salix integra Thunb., is a fast-growing woody species, with a large biomass and deep root system (Kuzovkina and Volk, 2009; Niu et al., 2019). It is a well-known Pb/Zn/Cd co-accumulator native to China (Liu et al., 2011). Previous studies have evaluated the ability of S. integra to absorb heavy metals such as Pb, Cu, Cd, and Zn (Yang et al., 2014; Shi et al., 2017; Cao et al., 2018, 2020; Yang et al., 2018). However, little is known regarding the indigenous rhizosphere microorganism during the phytoremediation by S. integra., which hinders the further application of microbial remediation with S. integra. This present study, therefore, investigates the characteristics of the indigenous bacterial community after phytoremediation by S. integra under different Pb levels using 16S rRNA gene amplicon sequencing as well as the interaction impacts of Pb contamination and root activity on the structure of soil bacterial communities. We aimed to (i) evaluate the effect of planting S. integra and Pb exposure on the indigenous bacterial community from different parts of the experiment (rhizosphere, bulk, and unplanted) in response to different Pb contamination levels, (ii) the effect of planting S. integra on Pb bioavailability from different parts of the experiment, and (iii) explore the key bacterial taxa involved in the phytoremediation process of Pb-contaminated soil, and their relationship with the soil properties.
The experiment was conducted in Baoding experimental station of Hebei Agricultural University, Baoding City, Hebei Province, China, which has a temperate climate (38°45′21″ N, 115°24′37″ E) with mean annual temperature ∼13.0°C and annual precipitation about 532 mm. The soil is a typical meadow cinnamon soil in which the average concentration of Pb is 14.15 mg kg–1 lower than the soil background value (21.5 mg kg–1 in Hebei province). S. integra is a shrub of the family Salicaceae, and 1-year-old cuttings were selected for use with similar growth and vigor in this study.
The experiment was conducted in three square plots, 4 × 4 m and 0.6 m deep. This method was closer to the natural growth of the plant than pot experiments. Three Pb treatment regimens were conducted on each of the plots: 0 mg kg–1 (CK), low Pb treatment of 500 mg kg–1 (LT), and high Pb treatment of 1,500 mg kg–1 (HT). These correspond to the levels of Pb pollution found in urban soils and lead mines in China (Wu et al., 2003; Qin et al., 2019). Sheets of a perfluorinated ethylene-propylene copolymer plastic were put on the bottom and sides of the three plots, to prevent seepage. In each treatment, we planted S. integra and set out the unplanted area at the same time. They were separated by a PVC sheet in each plot. The layout of the study site is shown in Figure 1.
Pb was added into the soil in the form of Pb(NO3)2 solution in February 2017. The water volume was calculated by the maximum water-holding capacity, and the weight of Pb(NO3)2 was calculated by dry soil weight and treat concentration. After the powder of Pb(NO3)2 dissolving completely, it was sprayed and mixed with soil. The soil was plowed and mixed every 10 days, and were aged and equilibrated for 3 months under natural conditions (the soil moisture being at ∼75% of the maximum water-holding capacity), then the Pb concentration was measured. One-year-old cuttings were grown in May 2017. Eight cuttings were grown in each treatment. The awning with a transparent plastic sheet was constructed above the experiment area to avoid contaminating other soil when it rained. The control and treatments were all take the same agronomic management measures in the study period.
The initial soil properties were as follows: “organic carbon 2,780 mg kg–1, total nitrogen 240 mg kg–1, available nitrogen 22.28 mg kg–1, total phosphorus 510 mg kg–1, available phosphorus 10.54 mg kg–1, total potassium 8,080 mg kg–1, available potassium 278.4 mg kg–1, cation exchange capacity 8.46 cmol kg–1, and pH 8.14” (Niu et al., 2019). The Pb concentrations were as follows: total Pb concentrations were 14.21 mg kg–1, 510.28 mg kg–1, 1513.45 mg kg–1 and the acid-soluble Pb concentrations were 0.42 mg kg–1, 15.32 mg kg–1, 81.52 mg kg–1 in CK, LT and HT, respectively.
After 17 months growth (in September 2018), samples of the plants, rhizospheric soil, bulk soil, and unplanted soil were collected in three replicates under each treatment. It is important to note that the aerial parts of the plant were ever reaped in November 2017. The plants were completely uprooted and shaken gently. After that, the soil adhering to the root was collected by brush as the rhizosphere soil sample. The soil from five plant roots was pooled as a mixed sample of rhizospheric soil. The bulk soil and unplanted soil were all obtained by soil-drilling at a depth of 20–30 cm, and five random spots were mixed as one replicate. Approximately 3 g soil from each replication was stored at −80°C until DNA extraction. Residual soils were air dried to measure soil physicochemical properties and Pb content. When sampling, all tools were sterilized in different treatments.
At harvest, roots, trunk, branch, and leaves were harvested separately, then rinsed thoroughly with water, and finally rinsed with deionized water. The samples were through drying at 65°C, weighing, grinding, and passing through a 60 mesh sieve to measure Pb content.
After the soil drying and passing through a 10 mesh sieve, the soil properties were measured, such as available nitrogen (AN), available phosphorus (AP), available potassium (AK), pH, and cation exchange capacity (CEC), following the methods of Niu et al. (2019).
0.5 g of the powdered root, branch, leaf or soil was digested in a mixture of HNO3 and HClO4 (10:1, v/v, 10 mL) by microwave digestion (Sined, MDS-6, Shanghai, China). The mixture was digested at 180°C for about 3.5 h, until the solution become clarification. It was diluted to 50 mL, and filtered by 0.45 μm membrane. Total Pb in the extractant was measured by atomic absorption spectrophotometry (AA-680, SHIMADZU, Kyoto, Japan).
Partitioning of Pb was measured by the sequential extraction procedure of Rauret et al. (1999), including the “acid-soluble,” “reducible,” “oxidizable,” and “residual” fractions, using 0.5 g soil. The extractions were as follows: acid-soluble extraction using acetic acid (0.11 mol L–1, 16 h), reducible extraction using hydroxylamine hydrochloride (0.5 mol L–1, pH = 1.5, 16 h), oxidizable extraction using H2O2 (8.8 mol L–1, 2 × 1 h, 85°C) and then following extraction using ammonium acetate 1.0 mol L–1. The digestion of residual fraction was through adding three acids (4 mL HCl, 2 mL HNO3, and 2 mL HF). The different forms of Pb were measured by atomic absorption spectrophotometry.
It is important to note that we measured only rhizosphere and bulk soil samples in CK and the low Pb treatment (LT), but measured rhizosphere, bulk, and unplanted soils in the high Pb treatment (HT). The PowerSoil DNA isolation kit (MOBIO, San Diego, United States) was used to extract soil genomic DNA from 0.5 g soil following the manufacturer’s instructions, and a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, United States) was used to measure DNA concentration. The V3-V4 regions of the bacterial 16S rRNA genes were amplified using the primer set DBV34F (5′-GTACTCCTACGGGAGGCAGCA-3′) and DBV34R (5′-GTGGACTACHVGG GTWTCTAAT-3′). All PCR amplifications were conducted in triplicate with a total reaction volume of 25 μL containing 1 μL (5 μmol L−1) of each forward/reverse primer, 30 ng genomic DNA, BSA 3 μL (2 ng μL–1), 2 × Taq Master Mix 13.5 μL (Allwegene Biotech Co., Ltd., Beijing, China). The PCR was carried out under the conditions: initial denaturation: 94°C, 5 min; 25 cycles (denaturation: 94°C, 30 s; annealing: 50°C, 30 s; and extension: 72°C, 60 s); with a final extension: 72°C, 7 min. PCR amplicons were purified using a Purification Kit (Qiagen, Shenzhen, China), and the concentration of purification was measured by a NanoDrop 2000 spectrophotometer. Equivalent PCR products from all repeats were combined into a mixed sample, and sequenced using a MiSeqPE300 platform (Illumina, United States; Allwegene Biotech Co., Ltd., Beijing, China).
QIIME v1.7.0 was used to de-multiplex, quality filter, and analyze raw Illumina fastq files (Caporaso et al., 2012). Reads were filtered with less than 200 bp long or 25 of the average quality score. Chimeric sequences were removed using Chimera and filtered by USEARCH (Edgar et al., 2011), and singletons were also removed. Operational taxonomic units (OTU) were divided by UCLUST according to 97% similarity threshold (Edgar, 2013). Phylogenetic classification of the reads was identified according to the Ribosomal Database Project (RDP), following 80% threshold (Wang et al., 2007). The classification of representative OTUs were according to similarity to the SILVA (release_132) database. OTUs belonging to plants were removed. In order to decrease the effects induced by sequencing depth, all samples were rarefied to the same sequence depth. Shannon, Chao 1, and Observed species were used to estimate the alpha microbial biodiversity (Pagaling et al., 2014).
Each data point represented the results of three replications performed as average value ± standard error. The effects of Pb contamination levels and planting S. integra on different treatments were tested by a two-way ANOVA. The Shapiro-Wilk test was used to check the analyzed features’ normality. The significant differences were checked by the least significant difference (LSD) in the parameter test or the Kruskal-Wallis test in the non-parameter according to the distribution of the estimated parameters. In all analyses, p-value < 0.05 meant significant difference. The package SPSS 19.0 (IBM, America) was used to analyze the data. Redundancy analysis (RDA) was conducted to test the relationship of soil properties and bacterial communities performed by the software CANOCO (Windows version 4.5) (Biometris-Plant research international, Wageningen, the Netherlands). Non-metric multidimensional scaling (NMDS) was used to visualize differences based on community composition by the metaMDS function of the vegan package in R package. The microbial metabolism heat map and cluster analyses were conducted, based on relative abundance and Bray-Curtis distances, with the agnes function of the cluster package in R package.
Plants were exposed to the Pb-polluted and unpolluted soil for 17 months and appeared significant difference in biomass (Supplementary Table S1). On the whole, the growth of S. integra was not inhibited in the low Pb treatment (Pb500). The plant biomass in CK (Pb 0) and the low Pb treatment had no significant difference, but both were higher than in the high Pb treatment (Pb1500). The total amount of Pb uptake by S. integra was highest in the high Pb treatment, followed by the low Pb treatment. Pb concentration was highest in the root, reaching 1221.36 ± 111.36 mg kg–1 in the low Pb treatment and 3159.92 ± 138.98 mg kg–1 in the high Pb treatment, followed by the trunk and leaf, and lowest in the branch whether in CK, low Pb treatment, or high Pb treatment. It may be contributed to the lower transfer factor from the root part to the aerial part. The Pb concentrations in S. integra planted in Pb-treated soils were significantly greater than that planted in the control soils. Heavy metal accumulation in the plant was related to heavy metal concentration in soil. The Pb accumulation of S. integra grown in soil containing 1,500 mg kg–1 Pb was 1.98 times as much as that grown in soil containing 500 mg kg–1 Pb. The results got here clearly show that S. integra has great potential to tolerate high Pb concentrations.
The contents of available potassium and available nitrogen were affected by the interaction of Pb contamination level with soils from different parts of the experiment (rhizosphere, bulk, and unplanted), while available phosphorus and CEC were only affected by Pb contamination or by differences in the soil samples (Supplementary Figures S1A–E). Overall, Pb contamination had a negative effect on CEC, available potassium, and available phosphorus, and had a positive effect on available nitrogen. However, root activity promoted them all, except CEC.
The total Pb concentration after the phytoremediation decreased by 84.61 mg kg–1 in rhizosphere and 29.24 mg kg–1 in bulk soil in the low Pb treatment (LT), but it occurred only in the rhizosphere soil in the high Pb treatment (HT) (Figure 2A). However, there was no significant difference between bulk and unplanted soil in the high Pb treatment. Four different fractions of Pb from soil samples were extracted and analyzed (Figures 2B–E). Phytoremediation decreased the acid-soluble Pb concentration of planted soil in the high Pb treatment, but showed the opposite effect on the low Pb treatment. It increased the concentration of oxidizable Pb and decreased the concentration of residual Pb in planted soil compared with unplanted soil. The concentration of acid-soluble Pb, oxidizable Pb, and residual Pb were all higher in bulk than in rhizosphere soil, except residual Pb in the high Pb treatment. The concentration of reducible Pb showed no significant difference between planted soil and unplanted soil in the low Pb treatment. However, it was higher in the bulk and lower in rhizosphere soil compared with unplanted soil in the high Pb treatment, respectively.
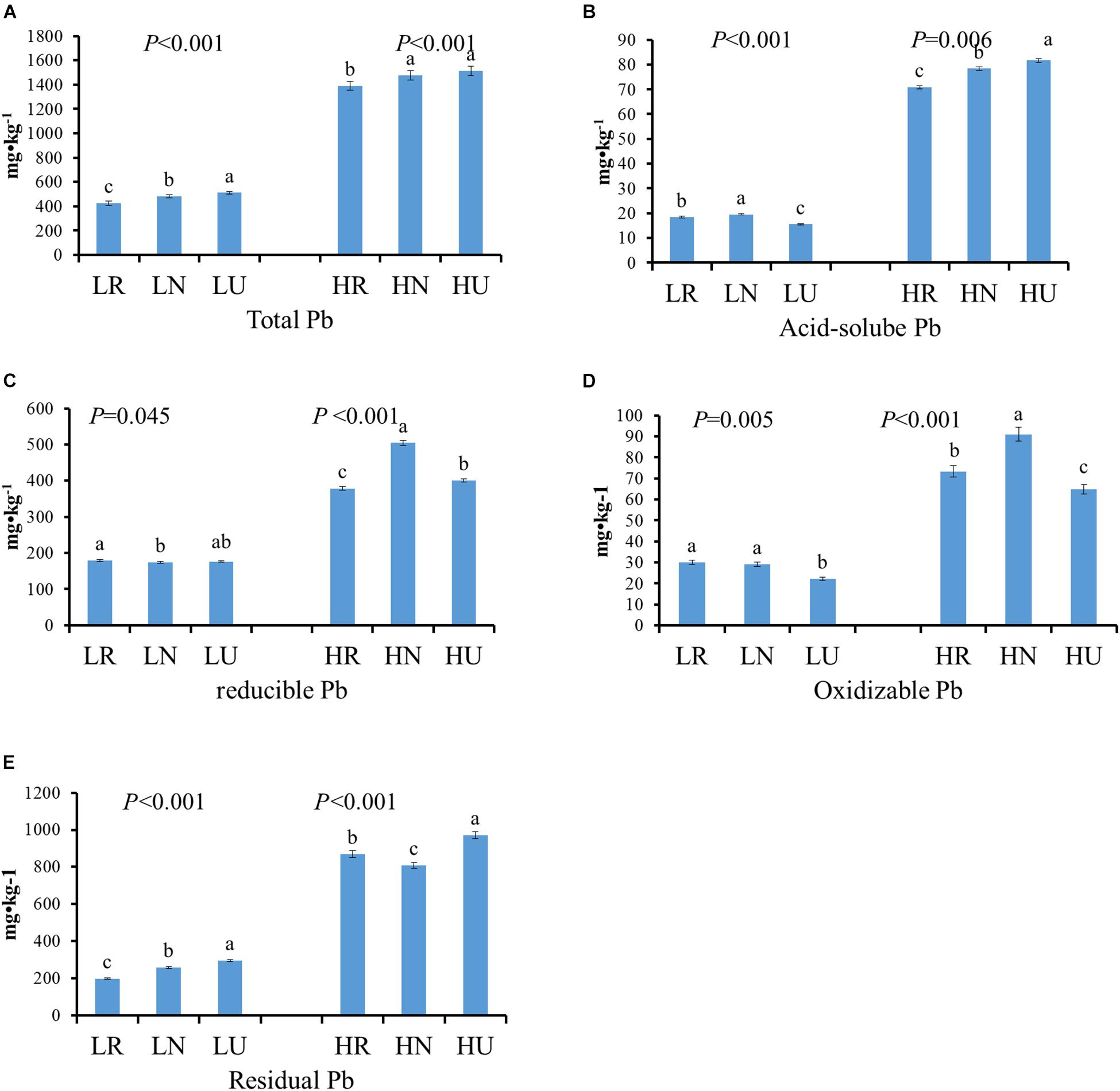
Figure 2. Different fractions Pb content in rhizosphere, bulk, and unplanted soil after phytoremediation. (A) Total Pb. (B) Acid-soluble Pb. (C) Reducible Pb. (D) Oxidizable Pb. (E) Residual Pb. LR, rhizosphere soil in the low Pb treatment; LN, bulk soil in the low Pb treatment; LU, unplanted soil in the low Pb treatment; HR, rhizosphere soil in the high Pb treatment; HN, bulk soil in the high Pb treatment; HU, unplanted soil in the high Pb treatment. Different letters indicate that the values differ significantly at p < 0.05.
The ratio of Pb under four fractionations was similar in different samples (Figure 3). Quite a large amount of residual and reducible Pb was found in the soil, but very little oxidizable and acid-soluble Pb. Planting S. integra promoted the proportion of bioavailable fractions in the low Pb treatment, but not in the high Pb treatment. Compared with the unplanted soil, the acid-soluble Pb proportion in the rhizosphere and bulk soil was increased by 1.0 and 0.75%, respectively, and the residual Pb proportion was decreased in the low Pb treatment. Furthermore, the acid-soluble Pb proportion of the rhizosphere soil was highest in the low Pb treatment, but decreased in the high Pb treatment when compared with the unplanted soil. The acid-soluble Pb proportion was higher in the low Pb treatment than in the high Pb treatment, whether in the rhizosphere or bulk soil, while the residual Pb proportion showed the opposite trend. The reducible and oxidizable Pb proportion showed a similar trend, and they were all promoted by phytoremediation in the rhizosphere and bulk soil when compared with unplanted soil.
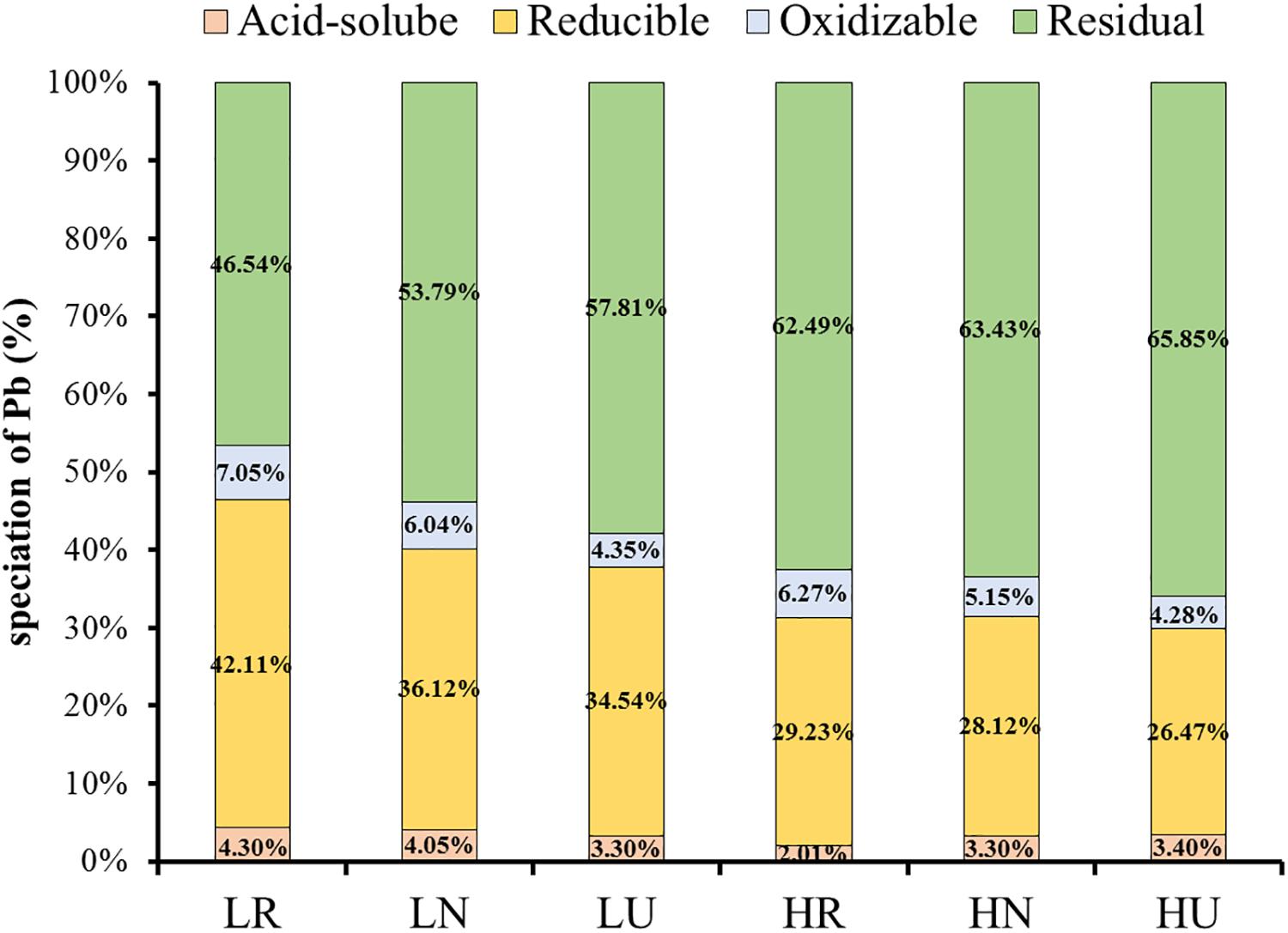
Figure 3. Effect of S. integra phytoremediation on speciation of Pb (%) in the low and high Pb treatments. LR, rhizosphere soil in the low Pb treatment; LN, bulk soil in the low Pb treatment; LU, unplanted soil in the low Pb treatment; HR, rhizosphere soil in the high Pb treatment; HN, bulk soil in the high Pb treatment; HU, unplanted soil in the high Pb treatment. Different letters indicate that the values differ significantly at p < 0.05.
The high-quality reads generated by 16S rRNA gene amplicon sequencing were classified according to a 97% sequence identity threshold, which produced 5,505 bacterial operational taxonomic units (OTUs). A total of 5,505 different OTUs belonged to 660 genera, 396 families, 257 orders, 117 classes, and 41 phyla. Of these OTUs, only 0.1% belonged to archaea. The coverage rang were between 94.45 and 95.57%, showing that most of the bacterial taxa were detected in the soil samples (Table 1).
The observed species, Chao 1, and Shannon index were used to compare alpha-diversity of the bacterial community among different samples, and they were only significantly influenced by Pb contamination levels (Supplementary Figure S2A). Chao 1 appeared decreasing trend with increase of Pb levels regardless of sampling from different parts of the experiment. However, observed species and Shannon’s index were only decreased in the high Pb treatment, and showed no significant difference between the CK and low Pb treatments. The shared genera of bacterial communities were performed by Venn diagrams among samples (Supplementary Figure S2B). The vast majority of the genera were common to all samples, which was likely caused by the same soil source and similar experimental conditions.
The non-metric multidimensional scaling (NMDS) analysis revealed a clear differentiation of bacterial community structure (Figure 4A). The bacterial community composition of rhizosphere, bulk, and unplanted soil samples from different Pb contaminations formed distinct clusters in the NMDS ordination plots, implying that the bacterial community structure was affected by the root activity and Pb contamination. To provide a functional understanding of the microbial community in the different samples, the metabolic functions were predicted by the software Picrust and the KEGG database (Langille et al., 2013). From the metabolism heat map (Figure 4B), we can see that microbial metabolism has significant differences in different samples, especially amino acid metabolism and carbohydrate metabolism. They were also clustered in relation to the Pb contamination levels and root activity. The microbial metabolism of the low Pb treatment was more similar to CK.
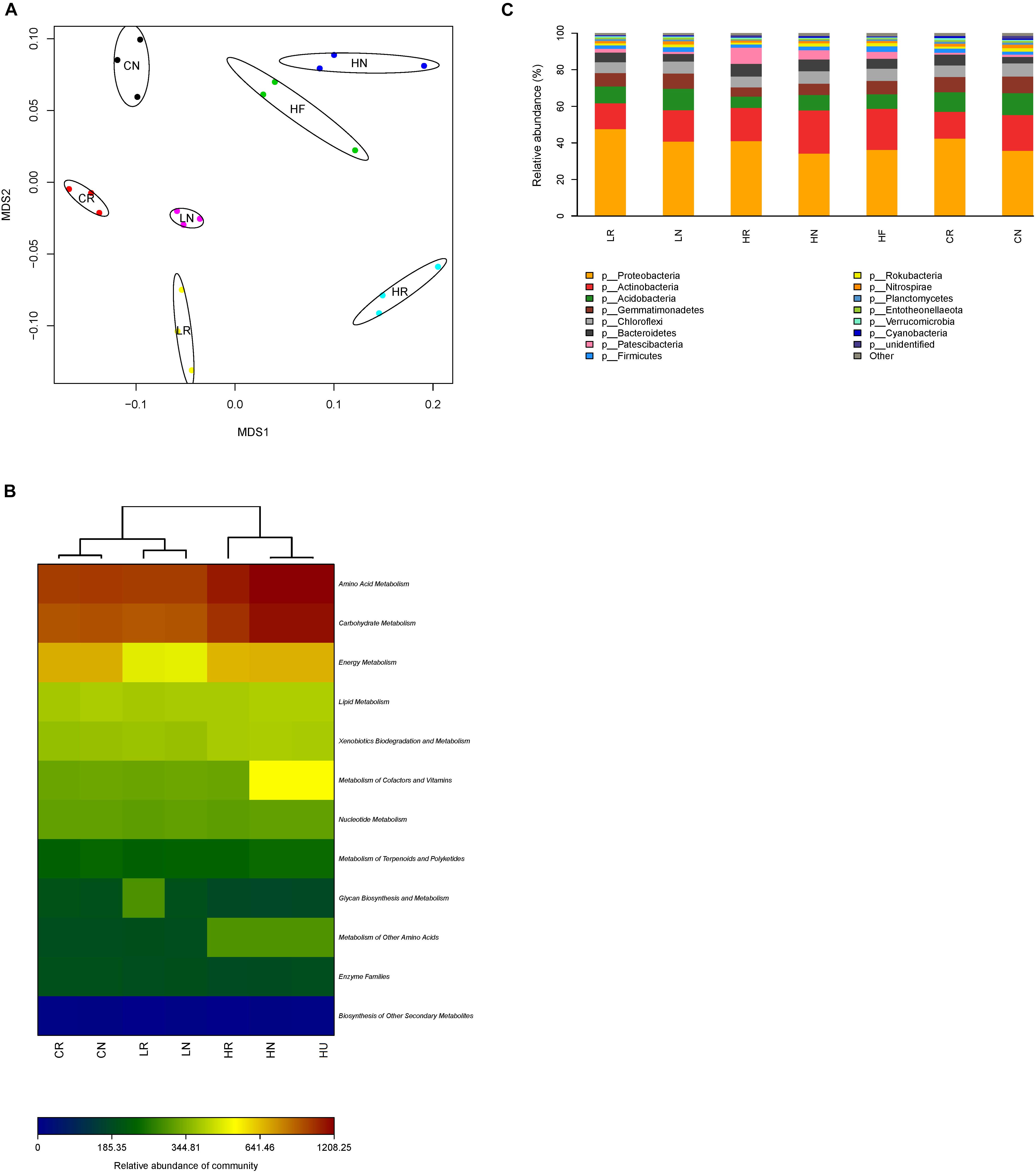
Figure 4. Bacterial community structure. (A) Non-metric multidimensional scaling (NMDS) ordination plots derived from Unifrac distance matrix, (B) microbial metabolism heat map, and (C) the relative abundance of bacteria in rhizosphere, bulk, and unplanted soil with different treatments at phylum level. CR, rhizosphere soil in CK; CN, bulk soil in CK; LR, rhizosphere soil in the low Pb treatment; LN, bulk soil in the low Pb treatment; HR, rhizosphere soil in the high Pb treatment; HN, bulk soil in the high Pb treatment; HU, unplanted soil in the high Pb treatment.
The sequencing reads obtained from all samples classified at the phylum level were attached to 14 bacterial phyla (Figure 4C). The dominant phyla with relative abundance more than 1% of the overall community in each sample were similar, while the relative abundance shifted. The relative abundance of Proteobacteria was highest, and was increased by root activity which was higher in rhizosphere soil, and promoted in the low Pb treatment compared with CK (Table 2). However, Actinobacteria showed the opposite trend, being higher in bulk soil, and improved in the high Pb treatment compared with CK. The interaction of soils from different parts of the experiment and Pb contamination had a significant effect on Acidobacteria and Patescibacteria (Table 2). Overall, Acidobacteria was decreased in the high Pb treatment regardless of whether in rhizosphere soil, bulk soil, or unplanted soil, but not in the low Pb treatment. Patescibacteria was increased only in the high Pb treatment and higher in the bulk soil. Both the relative abundance of Gemmatimonadetes and Chloroflexi were higher in the bulk soil, while only Gemmatimonadetes was decreased by Pb contamination. Bacteroidetes was higher in the bulk soil and increased in the high Pb content (Table 2).
The metastasis were used to analyze the specific bacterial assemblages whose relative abundance showed significant changes between rhizosphere and bulk soil in different treatments (Table 3). There were 88, 62, and 91 specific bacterial assemblages at a general level in CK, the low Pb treatment, and the high Pb treatment, respectively. In order to measure the effect of Pb contamination and root activity on the relative abundance of specific bacterial assemblages, two-way ANOVA was used to test 17 general bacteria that emerged in the top 10 microbes of relative abundance in each sample. From Tables 3A,B, we can see that Lysobacter, Pseudomonas, Allorhizobium, Steroidobacter, Skermanella, and Acidovorax were affected by the interaction of Pb contamination and root activity. The others were affected by Pb contamination and/or root activity. Overall, the relative abundances of partial bacterial genera were increased by Pb contamination and root activity; examples were Lysobacter, Pseudomonas, Allorhizobium, Acidovorax, and Ensifer, especially Pseudomonas and Ensifer which increased to more than 1%, while others were decreased by Pb contamination, such as Steroidobacter and Skermanella. Pb contamination had no significant effect on the relative abundance of Nitrospira, Ramlibacter, Microvirga, Ellin6067, and MND1 in the low Pb treatment, but had a positive effect in the high Pb treatment. In addition, root activity also had a positive effect on the relative abundance of Ramlibacter, Microvirga, and MM2. The relative abundances of uncultured_bacterium and uncultured were higher in all the treatments, while they were uncultured and not promoted by the root activity.
The relationships of soil properties and specific bacterial assemblages were measured by redundancy analysis (RDA) (Figure 5). Eigenvalues of RDA showed that axes 1 and 2 explained 65.7 and 22.1% of the variance of the microbial quantities data, respectively. Available nitrogen (AN) and pH were significant (F = 5.315, p = 0.008 and F = 3.755, p = 0.048, respectively) in explaining the relative abundance of specific bacterial assemblages, and could account for up to 14.9 and 12.6%, respectively. The contributions of other factors to the observed variation were as follows: residual Pb 12.5%, acid-soluble Pb 12.4%, total soluble Pb 12.2%, reducible Pb 11.7%, oxidizable Pb 11.8%, available potassium 10.1%, CEC 6.5%, and available phosphorus 9.8%.
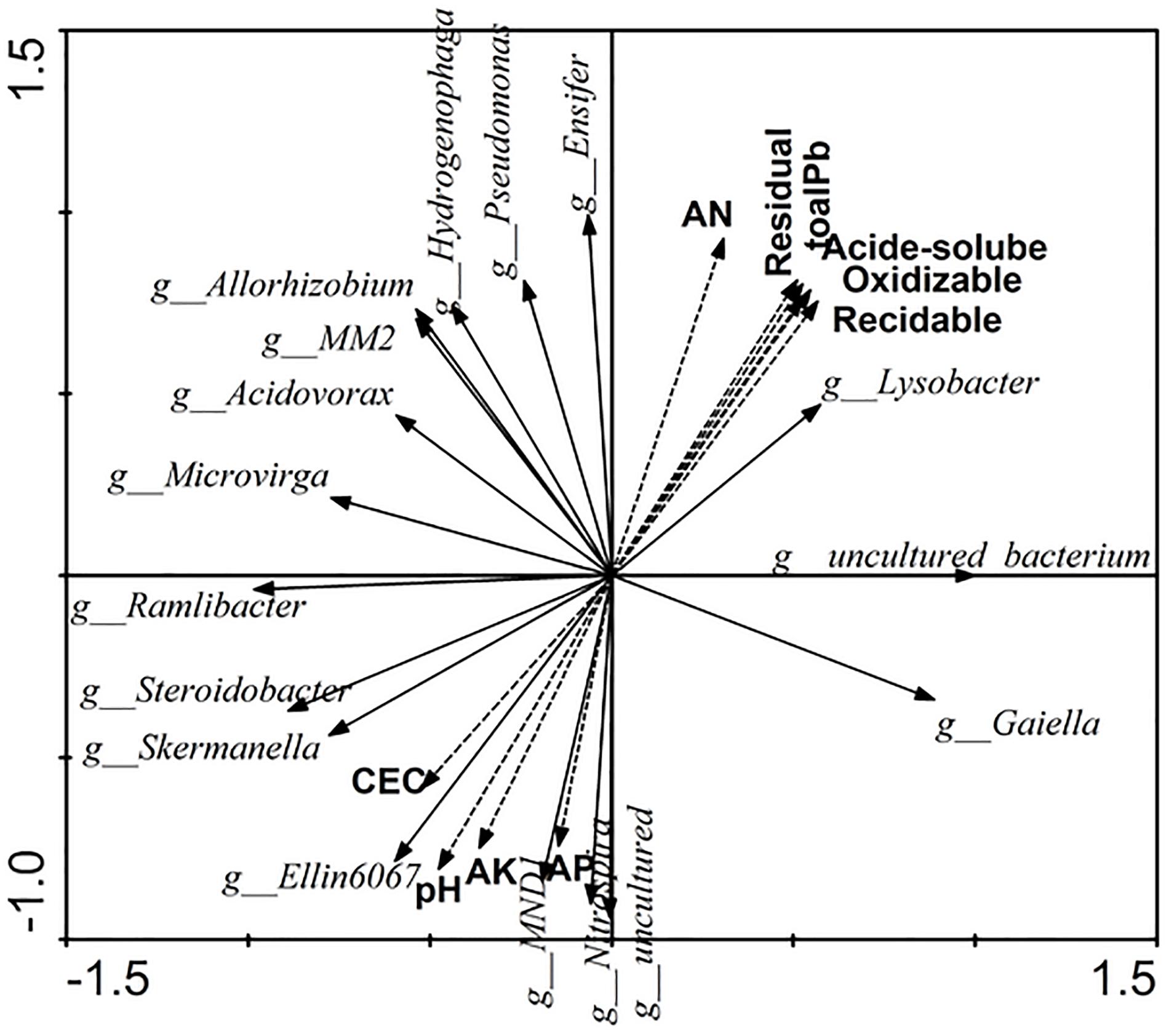
Figure 5. The relationships of soil properties and significance change bacterial of the relative abundance by Redundancy analysis. AN, available nitrogen; AK, available potassium; AP, available phosphorus.
From Figure 5, we can see that Lysobacter, Pseudomonas, Allorhizobium, Acidovorax, and Ensifer, which were promoted by Pb and root activity, had a strong positive correlation with the available nitrogen and Pb content, and had a negative correlation with CEC, pH, available potassium, and available phosphorus. Furthermore, uncultured bacterium, which was the dominant genus, also had a strong correlation with the available nitrogen and Pb content. Lysobacter, Pseudomonas, Allorhizobium, Acidovorax, Ensifer, Ramlibacter, Microvirga, and MM2 had stronger correlations with each other.
Salix integra has been reported to be potentially valuable for phytoremediation of Pb contaminated soils (Shi et al., 2017; Cao et al., 2018, 2019), which is supported by the results of our study (Supplementary Table S1 and Figure 2). However, the potential of S. integra for heavy metal extraction relies not only on its fast-growth, large biomass and deeper, more integrated root systems and powerful tolerance to Pb, but also on the underground processes that promote root uptake of the heavy metal. A high-throughput sequencing approach was therefore used to measure rhizospheric bacterial community that may influence the accumulation of heavy metal in S. integra.
Planting S. integra decreased the total Pb concentration both in rhizosphere and bulk soil in the low Pb treatment, which could be explained by the effects of root activity not being limited to the root surface, though the effect declined when the distances increase (Yang et al., 2017). Furthermore, our previous study also showed that microbial quantity and metabolism were promoted both in the rhizosphere and bulk soil in the low Pb treatment (Niu et al., 2019). However, it only occurred in the rhizosphere soil under high Pb treatment. The efficiency of phytoextraction relies greatly on the bioavailability of heavy metals. The concentration and proportion of acid-soluble Pb with the highest bioavailability were not decreased for S. integra uptake but increased in planted soil when compared with unplanted soil in the low Pb treatment, which may be attributed to higher transformation rate than uptake for the acid-soluble fraction. However, the opposite trend appeared in high Pb treatment. The important components of root exudates, such as organic and amino acids can promote the transformation of acid-soluble Pb from other species of the heavy metals in the low Pb treatment (Luo et al., 2017, 2019, 2020; Liu et al., 2018; Moshiri et al., 2019). However, the plant root may exude carboxyl groups and phenols to inhibit the transformation of acid-soluble Pb due to its high toxicity, and thus protect themselves in the high Pb treatment (Liu et al., 2017; Moshiri et al., 2019).
The soil microbes alter their structure to acclimatize different levels of heavy metal pollution and root environments, which is consistent with the previous studies (Edwards et al., 2015; Li et al., 2017; Liu et al., 2018; Lin et al., 2019; Cao et al., 2020). The shift of microbial composition led to metabolic function variation (Tipayno et al., 2018; Liu et al., 2020), and further caused Pb speciation differences in different treatments. It could explain the difference of acid-soluble Pb concentration between the low and high Pb treatments. Heavy metal pollution inhibits the metal-sensitive species, and stimulates metal-resistant species, microbial diversity therefore affects (Xie et al., 2016; Cao et al., 2020). Bacterial diversity was not affected by root activity, contrary to the rhizosphere bacterial community associated with Sedum alfredii (Hou et al., 2018), and that may result from the differences of plant root and soil environment.
Dominant bacterial species were similar among different treatments, which may be related to the same soil environment (Fan et al., 2016), and their relative abundances shifted that contributed to the selection of root activity and Pb contamination. Most of the specific bacterial assemblages between rhizosphere and bulk soil were promoted by root activity and/or Pb contamination, and are suitable for use in plant-microbial combination remediation, for example Proteobacteria, Actinobacteria, Patescibacteria, Bacteroidetes, and Gemmatimonadetes. The relative abundance of Proteobacteria was the highest of any treatment (more than 35%), which were also reported to be the dominant phyla in soils contaminated by polycyclic aromatic hydrocarbons, Cr, Cu, Pb, and Zn, always exceeding 50% in Pb contaminated soils (Bourceret et al., 2016; Fan et al., 2016; Tipayno et al., 2018; Lin et al., 2019). We, therefore, infer that it plays an important role in phytoremediation by S. integra. Actinobacteria are also increased after phytoremediation by Sedum in Cd-contaminated soil (Hou et al., 2018). Many of them can produce IAA to stimulate plant growth, as well as siderophores to form stable complexes with heavy metals (Rajkumar et al., 2010). Patescibacteria and Bacteroidetes have been reported as dominant phyla in heavy metal-polluted soils (Ellis et al., 2003; Idris et al., 2004), although their relationship with root activity was small in our study. Gemmatimonadetes was decreased by Pb contamination, but Hou et al. (2018) found it was positively associated with Cd levels. These conflicting results may be caused by the difference in the metal levels and soil physicochemical properties and the history of contamination in the different studies (Fan et al., 2016).
At a general level, partial bacterial genera, such as Lysobacter, Pseudomonas, Allorhizobium, Acidovorax, and Ensifer were increased by Pb contamination and root activity. Among them, Pseudomonas and Ensifer which increased to more than 1% played key roles during phytoremediation by S. integra. Moreover, Pseudomonas strains have been developed as bioinoculants for phytoremediation, bioremediation of metals, and methylmercury degradation (Boeris et al., 2016; Cabral et al., 2016; Sun et al., 2017). Uncultured bacteria with higher abundance in all the treatments were promoted only by Pb contamination, but their functions cannot be ignored. From all of these observations, we can infer that indigenous soil microbial communities played very important roles in promoting heavy metals absorption by S. integra.
In this study, the specific bacterial assemblages were promoted by available nitrogen and decreased by high pH, which is consistent with previous studies (Guo et al., 2019; Huang et al., 2019; Wang et al., 2019a; Zhen et al., 2019). Available nitrogen and available phosphorus are a significant energy source for microbes and play an important role in regulating their adaptation to different levels of heavy metal contamination (Guo et al., 2017; Lin et al., 2019). We also found that the specific bacterial assemblages, such as Pseudomonas, Allorhizobium, Acidovorax, Ensifer, Ramlibacter, Microvirga, and MM2, all had stronger correlations, which implied that plant-microbe combined remediation cannot rely on only a kind of microbe. We could, therefore, shift microbial composition by adjusting soil properties, for example adding available nitrogen, so promoting S. integra absorption of Pb.
During phytoremediation, the relative abundance of dominant microbial species shifted. The bacteria were promoted by root activity and/or Pb contamination, especially many genera from Proteobacteria, which are suitable for using in plant-microbial combination remediation. Indigenous bacteria have great potential, therefore, in the application of combined S. integra-microorganism remediation of lead-contaminated soil by adjusting soil properties, for example available nitrogen and pH. Further study on promoting plant-indigenous bacteria combined remediation through by adjusting soil properties and its practical application will be conducted.
The datasets generated for this study can be found in https://www.ncbi.nlm.nih.gov/sra/PRJNA588741.
XN, JZ, and DH conceived, designed, and directed the project, contributed to the interpretation of the results, and designed the figures. XN and JZ worked out almost all of the technical details, contributed to sample preparation, carried out the experiments, and performed the numerical calculations. XW and SD participated in the acquisition of the data. XS, YZ, and JY partially analyzed the data. All authors provided critical feedback, helped to shape the research, analysis, and manuscript; all discussed the results and contributed to the final version of the manuscript.
This research was funded by Youth project of Hebei Provincial Ministry of Education, Grant No. QN2016131. It was also partly supported by National Natural Science Foundation of China (Nos. 31971651 and 31600486), Natural Science Foundation of Hebei Province (Grant No. C2017204041), Youth top-notch talent project of Hebei Provincial Ministry of Education (Grant No. BJ2017053).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00924/full#supplementary-material
Boeris, P. S., Agustin, M. D., Acevedo, D. F., and Lucchesi, G. I. (2016). Biosorption of aluminum through the use of non-viable biomass of Pseudomonas putida. J. Biotechnol. 236, 57–63. doi: 10.1016/j.jbiotec.2016.07.026
Bourceret, A., Cebron, A., Tisserant, E., Poupin, P., Bauda, P., Beguiristain, T., et al. (2016). The bacterial and fungal diversity of an aged PAH- and heavy metal-contaminated soil is affected by plant cover and edaphic parameters. Microb. Ecol. 71, 711–724. doi: 10.1007/s00248-015-0682-8
Cabral, L., Yu, R. Q., Crane, S., Giovanella, P., Barkay, T., and Camargo, F. A. O. (2016). Methylmercury degradation by Pseudomonas putida V1. Ecotoxicol. Environ. Safety 130, 37–42. doi: 10.1016/j.ecoenv.2016.03.036
Cao, Y. N., Ma, C. X., Chen, H. J., Chen, G. C., White, J. C., and Xing, B. X. (2020). Copper stress in flooded soil: Impact on enzyme activities, microbial community composition and diversity in the rhizosphere of Salix integra. Sci. Total Environ. 704:135350. doi: 10.1016/j.scitotenv.2019.135350
Cao, Y. N., Ma, C. X., Zhang, J. F., Wang, S. F., White, J. C., Chen, G. C., et al. (2019). Accumulation and spatial distribution of copper and nutrients in willow as affected by soil flooding: A synchrotron-based X-ray fluorescence study. Environ. Pollut. 246, 980–989. doi: 10.1016/j.envpol.2018.12.025
Cao, Y. N., Zhang, Y., Ma, C. X., Li, H. M., Zhang, J. F., and Chen, G. C. (2018). Growth, physiological responses, and copper accumulation in seven willow species exposed to Cu-a hydroponic experiment. Environ. Sci. Pollut. Res. 25, 19875–19886. doi: 10.1007/s11356-018-2106-z
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. Isme J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/Nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Edwards, J., Johnson, C., Santos-Medellin, C., Lurie, E., Podishetty, N. K., Bhatnagar, S., et al. (2015). Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. U.S.A. 112, E911–E920. doi: 10.1073/pnas.1414592112
Egamberdieva, D., Wirth, S., Li, L., Abd-Allah, E. F., and Lindstrom, K. (2017). Microbial cooperation in the rhizosphere improves liquorice growth under salt stress. Bioengineered 8, 433–438. doi: 10.1080/21655979.2016.1250983
Ellis, R. J., Morgan, P., Weightman, A. J., and Fry, J. C. (2003). Cultivation-dependent and -independent approaches for determining bacterial diversity in fleavy-metal-contaminated soil. Appl. Environ. Microbiol. 69, 3223–3230. doi: 10.1128/Aem.69.6.3223-3230.2003
Fan, M. C., Lin, Y. B., Huo, H. B., Liu, Y., Zhao, L., Wang, E. T., et al. (2016). Microbial communities in riparian soils of a settling pond for mine drainage treatment. Water Res. 96, 198–207. doi: 10.1016/j.watres.2016.03.061
Guo, H. H., Nasir, M., Lv, J. L., Dai, Y. C., and Gao, J. K. (2017). Understanding the variation of microbial community in heavy metals contaminated soil using high throughput sequencing. Ecotoxicol. Environ. Safety 144, 300–306. doi: 10.1016/j.ecoenv.2017.06.048
Guo, Q. X., Yan, L. J., Korpelainen, H., Niinemets, U., and Li, C. Y. (2019). Plant-plant interactions and N fertilization shape soil bacterial and fungal communities. Soil Biol. Biochem. 128, 127–138. doi: 10.1016/j.soilbio.2018.10.018
Hou, D. D., Lin, Z., Wang, R. Z., Ge, J., Wei, S., Xie, R. H., et al. (2018). Cadmium exposure-Sedum alfredii planting interactions shape the Bacterial Community in the hyperaccumulator Plant Rhizosphere. Appl. Environ. Microbiol. 84, e2797–e2717. doi: 10.1128/AEM.02797-17
Hou, D. D., Wang, K., Liu, T., Wang, H. X., Lin, Z., Qian, J., et al. (2017a). Unique rhizosphere micro-characteristics facilitate phytoextraction of multiple metals in soil by the hyperaccumulating plant Sedum alfredii. Environ. Sci. Technol. 51, 5675–5684. doi: 10.1021/acs.est.6b06531
Hou, J. Y., Liu, W. X., Wu, L. H., Hu, P. J., Ma, T. T., Luo, Y. M., et al. (2017b). Modulation of the efficiency of trace metal phytoremediation by Sedum plumbizincicola by microbial community structure and function. Plant Soil. 421, 285–299. doi: 10.1007/s11104-017-3466-8
Hou, J. Y., Liu, W. X., Wu, L. H., Ge, Y. Y., Hu, P. J., Li, Z., et al. (2019). Rhodococcus sp NSX2 modulates the phytoremediation efficiency of a trace metal-contaminated soil by reshaping the rhizosphere microbiome. Appl. Soil Ecol. 133, 62–69. doi: 10.1016/j.apsoil.2018.09
Huang, D. L., Zeng, G. M., Jiang, X. Y., Feng, C. L., Yu, H. Y., Huang, G. H., et al. (2006). Bioremediation of Pb-contaminated soil by incubating with Phanerochaete chrysosporium and straw. J. Hazard. Mater. 134, 268–276. doi: 10.1016/j.jhazmat.2005.11.021
Huang, Q., Wang, J. L., Wang, C., and Wang, Q. (2019). The 19-years inorganic fertilization increased bacterial diversity and altered bacterial community composition and potential functions in a paddy soil. Appl. Soil Ecol. 144, 60–67. doi: 10.1016/j.apsoil.2019.07.009
Idris, R., Trifonova, R., Puschenreiter, M., Wenzel, W. W., and Sessitsch, A. (2004). Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense. Appl. Environ. Microbiol. 70, 2667–2677. doi: 10.1128/Aem.70.5.2667-2677.2004
Joe, M. M., Gomathi, R., Benson, A., Shalini, D., Rengasamy, P., Henry, A. J., et al. (2019). Simultaneous application of biosurfactant and bioaugmentation with rhamnolipid-producing shewanella for enhanced bioremediation of oil-polluted soil. Appl. Sci. Basel. 9:3773. doi: 10.3390/app9183773
Ju, W. L., Jin, X. L., Shen, G. T., Zhao, W., Duan, C. J., et al. (2020). Rhizobacteria inoculation benefits nutrient availability for phytostabilization in copper contaminated soil: drivers from bacterial community structures in rhizosphere. Appl. Soil Ecol. 150:103450. doi: 10.1016/j.apsoil.2019.103450
Khalid, S., Shahid, M., Niazi, N. K., Murtaza, B., Bibi, I., and Dumat, C. (2017). A comparison of technologies for remediation of heavy metal contaminated soils. J. Geochem. Exp. 182, 247–268. doi: 10.1016/j.gexplo.2016.11.021
Kong, Z. Y., Wu, Z. J., Glick, B. R., He, S. Y., Huang, C., and Wu, L. (2019). Co-occurrence patterns of microbial communities affected by inoculants ofplant growth-promoting bacteria during phytoremediation of heavy metalcontaminated soils. Ecotoxicol. Environ. Safety. 183:109504. doi: 10.1016/j.ecoenv.2019.109504
Kushwaha, A., Hans, N., Kumar, S., and Rani, R. (2018). A critical review on speciation, mobilization and toxicity of lead in soil microbe-plant system and bioremediation strategies. Ecotoxicol. Environ. Safety 147, 1035–1045. doi: 10.1016/j.ecoenv.2017.09.049
Kuzovkina, Y. A., and Volk, T. A. (2009). The characterization of willow (Salix L.) varieties for use in ecological engineering applications: Co-ordination of structure, function and autecology. Ecol. Eng. 35, 1178–1189. doi: 10.1016/j.ecoleng.2009.03.010
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Li, X. Q., Meng, D. L., Li, J., Yin, H. Q., Liu, H. W., Liu, X. D., et al. (2017). Response of soil microbial communities and microbial interactions to long-term heavy metal contamination. Environ. Pollut. 231, 908–917. doi: 10.1016/j.envpol.2017.08.057
Lin, Y. B., Ye, Y. M., Hu, Y. M., and Shi, H. K. (2019). The variation in microbial community structure under different heavy metal contamination levels in paddy soils. Ecotoxicol. Environ. Safety 180, 557–564. doi: 10.1016/j.ecoenv.2019.05.057
Liu, C. J., Lin, H., Dong, Y. B., Li, B., and Liu, Y. (2018). Investigation on microbial community in remediation of lead-contaminated soil by Trifolium repens L. Ecotoxicol. Environ. Safety 165, 52–60. doi: 10.1016/j.ecoenv.2018.08.054
Liu, H. K., Xie, Y. L., Li, J. J., Zeng, G. Q., Li, H., Xu, F., et al. (2020). Effect of Serratia sp. 10 combined with organic materials on cadmium migration in soil-vetiveria zizanioides L. system and bacterial community in contaminated soil. Chemosphere 242:125164. doi: 10.1016/j.chemosphere.2019.125164
Liu, X., Fu, J. W., Da Silva, E., Shi, X. X., Cao, Y., Rathinasabapathi, B., et al. (2017). Microbial siderophores and root exudates enhanced goethite dissolution and Fe/As uptake by As-hyperaccumulator Pteris vittata. Environ. Pollut. 223, 230–237. doi: 10.1016/j.envpol.2017.01.016
Liu, Y. Q., Chen, G. C., Zhang, J. F., Shi, X., and Wang, R. M. (2011). Uptake of cadmium from hydroponic solutions by willows (Salix spp.) seedlings. Afr. J. Biotechnol. 10, 16209–16218. doi: 10.5897/Ajb11.2027
Luo, Q., Wang, S. Y., Sun, L. N., and Wang, H. (2017). Metabolic profiling of root exudates from two ecotypes of Sedum alfredii treated with Pb based on GC-MS. Sci. Rep. 7:39878. ARTN 39878 doi: 10.1038/srep39878
Luo, Y. F., Wu, X. Y., Sun, H., and Wu, Y. G. (2020). Root-induced changes in aggregation characteristics and potentially toxic elements (PTEs) speciation in a revegetated artificial zinc smelting waste slag site. Chemosphere 243, 125414. doi: 10.1016/j.chemosphere.2019.125414
Luo, Y. F., Wu, Y. G., Qiu, J., Wang, H., and Yang, L. (2019). Suitability of four woody plant species for the phytostabilization of a zinc smelting slag site after 5years of assisted revegetation. J Soils Sedim. 19, 702–715. doi: 10.1007/s11368-018-2082-4
Ma, Y., Rajkumar, M., Zhang, C., and Freitas, H. (2016). Beneficial role of bacterial endophytes in heavy metal phytoremediation. J. Environ. Manag. 174, 14–25. doi: 10.1016/j.jenvman.2016.02.047
Moshiri, F., Ebrahimi, H., Ardakani, M. R., Rejalia, F., and Mousavia, S. M. (2019). Biogeochemical distribution of Pb and Zn forms in two calcareous soils affected by mycorrhizal symbiosis and Alfalfa rhizosphere. Ecotoxicol. Environ. Safety 179, 241–248. doi: 10.1016/j.ecoenv.2019.04.055
Muehe, E. M., Weigold, P., Adaktylou, I. J., Planer-Friedrich, B., Kraemer, U., Kappler, A., et al. (2015). Rhizosphere microbial community composition affects Cadmium and Zinc uptake by the metal-hyperaccumulating plant Arabidopsis halleri. Appl. Environ. Microbiol. 81, 2173–2181. doi: 10.1128/Aem.03359-14
Niu, X. Y., Zhou, Y., Zhou, J., Wang, X. N., Gao, Z. T., and Huang, D. Z. (2019). The Effects of different Lead pollution levels on soil microbial quantities and metabolic function with/without Salix integra Thunb. Planting. Forests 10:77. doi: 10.3390/f10020077
Pagaling, E., Yang, K., and Yan, T. (2014). Pyrosequencing reveals correlations between extremely acidophilic bacterial communities with hydrogen sulphide concentrations, pH and inert polymer coatings at concrete sewer crown surfaces. J. Appl. Microbiol. 117, 50–64. doi: 10.1111/jam.12491
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Qin, Y., Zhu, X., Su, Q., Lv, W., and Wang, B. (2019). Study on contents and distributions of heavy metals in soils of a typical Lead-Zinc. Bull. Mineral. Petrol. Geochem. [Preprint]. Available online at: http://kns.cnki.net/kcms/detail/52.1102.p.20190823.1230.003.html (accessed Augest 26, 2019).
Rajkumar, M., Ae, N., Prasad, M. N. V., and Freitas, H. (2010). Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 28, 142–149. doi: 10.1038/s41467-018-05188-3
Ramirez, V., Baez, A., Lopez, P., Bustillos, R., Villalobos, M. A., Carreno, R., et al. (2019). Chromium Hyper-Tolerant Bacillus sp. MH778713 assists phytoremediation of heavy metals by mesquite trees (Prosopis laevigata). Front. Microbiol. 10:1833. doi: 10.3389/fmicb.2019.01833
Rauret, G., Lopez-Sanchez, J. F., Sahuquillo, A., Rubio, R., Davidson, C., Ure, A., et al. (1999). Improvement of the BCR three step sequential extraction procedure prior to the certification of new sediment and soil reference materials. J. Environ. Monitor. 1, 57–61. doi: 10.1039/a807854h
Shi, X., Wang, S. F., Sun, H. J., Chen, Y. T., Wang, D. X., Pan, H. W., et al. (2017). Comparative of Quercus spp. and Salix spp. for phytoremediation of Pb/Zn mine tailings. Environ. Sci. Pollut. Res. 24, 3400–3411. doi: 10.1007/s11356-016-7979-0
Sun, D. L., Zhuo, T., Hu, X., Fan, X. J., and Zou, H. S. (2017). Identification of a Pseudomonas putida as biocontrol agent for tomato bacterial wilt disease. Biol. Control 114, 45–50. doi: 10.1016/j.biocontrol.2017.07.015
Thijs, S., Sillen, W., Rineau, F., Weyens, N., and Vangronsveld, J. (2016). Towards an enhanced understanding of plant-microbiome interactions to improve phytoremediation: engineering the metaorganism. Front. Microbiol. 7:341. doi: 10.3389/fmicb.2016.00341
Tipayno, S. C., Truu, J., Samaddar, S., Truu, M., Preem, J. K., Oopkaup, K., et al. (2018). The bacterial community structure and functional profile in the heavy metal contaminated paddy soils, surrounding a nonferrous smelter in South Korea. Ecol. Evol. 8, 6157–6168. doi: 10.1002/ece3.4170
Wang, C. Y., Wu, B. D., Jiang, K., Wei, M., and Wang, S. (2019a). Effects of different concentrations and types of Cu and Pb on soil N-fixing bacterial communities in the wheat rhizosphere. Appl. Soil Ecol. 144, 51–59. doi: 10.1016/j.apsoil.2019.07.008
Wang, M., Chen, S. B., Chen, L., and Wang, D. (2019b). Saline stress modifies the effect of cadmium toxicity on soil archaeal communities. Ecotoxicol. Environ. Safety 182:109431. doi: 10.1016/j.ecoenv.2019.109431
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/Aem.00062-07
Wood, J. L., Zhang, C., Mathews, E. R., Tang, C., and Franks, A. E. (2016). Microbial community dynamics in the rhizosphere of a cadmium hyper-accumulator. Sci. Rep. 6:36067. doi: 10.1038/srep36067
Wu, X., Li, L., Pan, G., Ju, Y., and Jiang, H. (2003). Soil pollution of Cu, Zn, Pb and Cd in different city zones of Nanjing. Environ. Sci. 24, 105–109. doi: 10.13227/j.hjkx.2003.03.021
Xiao, Y., Zhao, Z. J., Chen, L., and Li, Y. (2020). Arbuscular mycorrhizal fungi and organic manure have synergistic effects on Trifolium repens in Cd-contaminated sterilized soil but not in natural soil. Appl. Soil Ecol. 149:103485. doi: 10.1016/j.apsoil.2019.103485
Xie, Y., Fan, J. B., Zhu, W. X., Amombo, E., Lou, Y. H., Chen, L., et al. (2016). Effect of heavy metals pollution on soil microbial diversity and Bermudagrass genetic variation. Front. Plant Sci. 7:755. doi: 10.3389/fpls.2016.00755
Xuliang, Z., Jian, C., Hojae, S., and Zhihui, B. (2007). New advances in plant growth-promoting rhizobacteria for bioremediation. Environ. Int. 33, 406–413. doi: 10.1016/j.envint.2006.12.005
Yang, W. D., Wang, Y. Y., Zhao, F. L., Ding, Z. L., Zhang, X. C., Zhu, Z. Q., et al. (2014). Variation in copper and zinc tolerance and accumulation in 12 willow clones: implications for phytoextraction. J. Zhej. Univ. Sci. B 15, 788–800. doi: 10.1631/jzus.B1400029
Yang, W. D., Zhao, F. L., Ding, Z. L., Wang, Y. Y., Zhang, X. C., Zhu, Z. Q., et al. (2018). Variation of tolerance and accumulation to excess iron in 24 willow clones: implications for phytoextraction. Int. J. Phytor. 20, 1284–1291. doi: 10.1080/15226514.2014.922927
Yang, W. H., Zhang, T. X., Lin, S., and Ni, W. Z. (2017). Distance-dependent varieties of microbial community structure and metabolic functions in the rhizosphere of Sedum alfredii Hance during phytoextraction of a cadmium-contaminated soil. Environ. Sci. Pollut. Res. 24, 14234–14248. doi: 10.1007/s11356-017-9007-4
Zhang, X., Li, X. X., Yang, H. H., and Cui, Z. J. (2018). Biochemical mechanism of phytoremediation process of lead and cadmium pollution with Mucor circinelloides and Trichoderma asperellum. Ecotoxicol. Environ. Safety 157, 21–28. doi: 10.1016/j.ecoenv.201803.047
Zhen, Z., Wang, S. B., Luo, S. W., Ren, L., Liang, Y. Q., Yang, R. C., et al. (2019). Significant impacts of both total amount and availability of heavy metals on the functions and assembly of soil microbial communities in different Land use patterns. Front. Microbiol. 10:02293. doi: 10.3389/fmicb.2019.02293
Keywords: bacterial community, soil properties, Pb contamination, plant-microbe combined remediation, redundancy analysis
Citation: Niu X, Zhou J, Wang X, Su X, Du S, Zhu Y, Yang J and Huang D (2020) Indigenous Bacteria Have High Potential for Promoting Salix integra Thunb. Remediation of Lead-Contaminated Soil by Adjusting Soil Properties. Front. Microbiol. 11:924. doi: 10.3389/fmicb.2020.00924
Received: 17 November 2019; Accepted: 20 April 2020;
Published: 19 May 2020.
Edited by:
Prayad Pokethitiyook, Mahidol University, ThailandReviewed by:
Santosh Kr. Karn, Sardar Bhagwan Singh Post Graduate Institute of Biomedical Science & Research, IndiaCopyright © 2020 Niu, Zhou, Wang, Su, Du, Zhu, Yang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dazhuang Huang, eWx6aGpAaGViYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.