- 1Department of Biophysics, University of Delhi, New Delhi, India
- 2Regional Centre for Biotechnology, NCR-Biotech Science Cluster, Faridabad, India
Nascent polypeptide-associated complex (NAC) is a ribosome-associated molecular chaperone which is present only in archaea and eukaryotes. The primary function of NAC is to shield the newly synthesized polypeptide chains from inappropriate interactions with the cytosolic factors. Besides that, NAC has been implicated in diverse biological functions, which suggest that it might be a multifunctional protein. An elaborate study on NAC can provide useful information on protein folding in extreme conditions in which many archaea grow. Thus, in the present study, we have studied the biophysical and the biochemical characteristics of NAC of Picrophilus torridus, an extreme thermoacidophilic archaeon. The study of protein–protein interactions and binding partners of a protein provides useful insights into the new/unreported roles of a protein. Thus, in this study, we have identified the binding partners of NAC in P. torridus. The NAC protein of P. torridus was cloned, expressed, and purified, and its binding partners were isolated by a pull down assay followed by identification with liquid chromatography–mass spectrometry. To the best of our knowledge, this is the first report on the biophysical and the biochemical characterization of NAC from P. toridus and the identification of its interacting partners.
Introduction
Nascent polypeptide-associated complex (NAC) is a ribosome-associated molecular chaperone which is present near the peptide exit site of the translating ribosomes (Wiedmann et al., 1994). The major function associated with the NAC complex is thought to be to shield the newly synthesized polypeptide chains from inappropriate interactions with the cytosolic factors. Since most of the translating ribosomes likely associated with NAC, it was considered an abundant cellular protein, expressed in equimolar quantities relative to ribosomes (Raue et al., 2007; Hoffmann et al., 2010). Ribosome-associated chaperones vary across the three domains of life. While the prokaryotes employ the bacterial trigger factors in preventing the inappropriate folding of newly synthesized polypeptide chains (Rospert et al., 2002; Kaiser et al., 2006) in eukaryotes and archaea, this function is performed by NAC (Koplin et al., 2010; del Alamo et al., 2011). In eukaryotes, NAC is reported to be a heterodimeric protein composed of alpha- and beta-subunits, while in archaea, it was reportedly a homodimer composed of only alpha-subunits (Spreter et al., 2005).
Several recent studies indicate that, besides its primary function as a molecular chaperone, NAC is involved in many other biological functions. It has been reported to be involved in the translation and the subcellular targeting of nascent polypeptides (Wickner, 1995; Powers and Walter, 1996) and in the prevention of mistargeting of ribosome nascent chain complex (Arsenovic et al., 2012; Gamerdinger et al., 2015). Besides that, NAC has been functionally implicated in the folding of the polypeptide chain, in ribosome biogenesis, in modulating protein secretion, in regulating apoptosis, as a transcription factor, etc. (Creagh et al., 2009; Kogan and Gvozdev, 2014). Despite being implicated in diverse cellular functions, its in vivo role and knowledge regarding its mechanism of action are still fragmentary. An elaborate study on NAC is expected to provide further insights into protein folding in extreme conditions in which many archaea grow. Thus, in the present study, we have performed biophysical and biochemical characterization of NAC of Picrophilus torridus to understand its role, if any, in the thermoacidophilic adaptation of this archaeon. P. torridus is an interesting organism which has some unique characteristics like small genome size (1.55 Mbp), adaptability to thrive in extremely low pH (0–1) and moderately high temperatures (50–60°C), and with an intracellular pH of about 4.0 (Schleper et al., 1995).
The study of protein–protein interactions (PPIs) adds crucially to our understanding of protein function(s) and helps in the characterization of the various pathways in which cellular proteins might be involved. The identification of the binding partners of a protein provides useful insights into the new/unreported roles of a protein. Also, an analysis of the interaction network aids in identifying the mechanism(s) underlying various related biological processes. Thus, in this study we have identified the binding partners of NAC in P. torridus. The NAC protein of P. torridus was cloned, expressed, and purified, and its binding partners were isolated by a pull down assay followed by identification using mass spectrometry. To the best of our knowledge, this is the first report on the biophysical and the biochemical characterization of NAC of P. torridus and the identification of its interacting partners.
Materials and Methods
Cloning and Expression of P. torridus Nascent Polypeptide-Associated Chaperone
The gene encoding PtNAC (327 bases) (accession no. AE017261.1) was synthesized at a commercial facility (Genscript, United States) in pUC57 plasmid with flanking NheI and SalI restriction sites. The plasmid with the insert was digested with restriction enzymes NheI and SalI (30-μl reaction mixture: 3.0 μl 1X buffer, 25 μl plasmid DNA with insert, 1.0 μl NheI, and 1.0 μl SalI) to obtain the free insert. The insert obtained was purified from the gel using a gel extraction kit and ligated with similarly digested pET28a(+) vector in 10 μl reaction mixture [ligation reaction in 10 μl reaction mixture: 1.0 μl ligase buffer, 1.0 μl insert, 7.5 μl digested pET28a(+) plasmid, and 0.5 μl ligase enzyme]. Escherichia coli DH5α cells were transformed with the pET28a(+)-PtNAC construct. Colony PCR and double-restriction digestion with enzymes NheI and SalI were used for the screening of the transformants.
The pET28a(+)-PtNAC construct was isolated from the transformed E. coli DH5α cells and further transformed in the expression vector E. coli BL21 (DE3). The transformants were grown overnight in 50 ml Luria–Bertani broth, containing 50 μg ml–1 kanamycin, at 37°C and 200 rpm and inoculated (2%, v/v) again in 1 L of the same medium. The culture was kept for incubation at 37°C and 200 rpm until A600 of 0.6 was achieved when the protein expression was induced by adding 1 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG). The induced cells were harvested after 4 h by centrifuging at 8,000 × g, resuspended in 70 ml of lysis buffer (1X phosphate-buffered saline, PBS; pH 7.2), and sonicated (Sonics, VibraTM cell, Connecticut, United States) intermittently for 30 min to release the intracellular protein. The cell debris was removed by centrifugation at 10,000 × g (30 min at 4°C). Column chromatography was performed on an AKTA Prime Plus (GE Healthcare) system. The clear supernatant was filtered through a 0.22-μm membrane (mdi: SY25KG-S) and applied to 10 ml Co2+-NTA beads (G Biosciences) packed in a XK16 column (Pharmacia Biotech) pre-equilibrated with 1X PBS, pH 7.2. The column was washed with five column volumes of equilibration buffer and then with wash buffer W1 (1X PBS with 20 mM imidazole), and the recombinant PtNAC was eluted using 1X PBS containing 200 mM imidazole. To remove imidazole, the eluted PtNAC protein was dialyzed in 1X PBS buffer (pH 7.2) overnight. The purity and the identity of the expressed protein was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and matrix-assisted laser desorption/ionization (MALDI-TOF) analysis, respectively.
Chaperone Assay
The chaperone activity of PtNAC was evaluated by its capability to prevent the thermal aggregation of the substrate protein, bovine carbonic anhydrase (BCA II) (Rajaraman et al., 1996; Tomar et al., 2013). A cuvette with the sample (2 ml) was inserted in the pre-heated sample chamber (65°C) of a spectrofluorimeter containing a Peltier-controlled stirred cell, and scattering was monitored with time. The excitation and emission monochromators were set at 400 nm. BCA II (0.75 μM) heated at 65°C in 50 mM Tris-HCl buffer (pH 7.5) was used as the substrate control. Keeping the BCA II concentration (0.75 μM) constant, PtNAC was added in increasing concentration ratios, i.e., 1:0.5, 1:1, 1:2, and 1:4. For this experiment, purified NAC was dialyzed overnight in 50 mM Tris-HCl buffer (pH 7.5) and then used for estimating the chaperone activity.
Biophysical Characterization of PtNAC Using Circular Dichroism Spectroscopy
Circular dichroism (CD) experiments were conducted in a Jasco J-815 spectropolarimeter with a Peltier-type temperature controller (Jasco CDF-426 S/15). The far-UV CD spectra of PtNAC were recorded in the wavelength range 260–190 nm using a quartz cuvette with 0.1 cm path length. The data points were collected using step resolution 0.1 nm, time constant of 2 s, and scan speed of 100 nm/min, with a spectral band width of 2.0 nm. Three accumulations per sample were recorded. The software BestSel was used for predicting the percentage of α-helices and β-sheets1.
Effect of pH and Temperature
The effect of pH on the secondary structure of PtNAC was studied by incubating NAC protein (0.5 mg/ml) in a buffer of different pH, ranging from 2 to 10 (20 mM glycine HCl, pH 2.0 and 3.0; 20 mM acetate buffer, pH 4.0 and 5.0; 20 mM phosphate buffer, pH 7.0 and 8.0; and 20 mM glycine NaOH buffer, pH 9.0 and 10). The effect of temperature on the secondary structure of PtNAC was studied by incubating the protein (0.5 mg ml–1) for 2 min at different temperatures (20–80°C), after which changes in the structural conformation were recorded. For this, the protein was dialyzed overnight in 20 mM Tris HCl buffer (pH 7.5).
Effect of Denaturants on the Secondary Structure of NAC
A fixed volume of PtNAC (0.5 mg ml–1) was incubated for 3 h with different concentrations of denaturant, guanidine hydrochloride (Gdn-HCl) (1–6 M), and urea (1–6 M), and changes in the structural conformation of PtNAC were recorded.
Conditions of P. torridus Culture and Preparation of Cell Lysate
P. torridus (DSM 9790) was purchased from DSMZ-German Collection of Microorganisms and Cell Cultures, GmbH Leibniz Institute, Germany. Archaea were grown in 500 ml of culture medium (Arora et al., 2014) at 55°C in a shaking incubator (100 rpm). The archaeal cells were harvested in the late log growth phase by centrifuging at 10,000 × g for 10 min at 4°C and washed with 10 mM phosphate-buffered saline (pH 7.4). The cells were resuspended in cell lysis buffer (50 mM Tris–HCl, 10 mM MgCl2, 1 mM EDTA; pH 7.4) and lysed by intermittent sonication on ice for 10 min. The cell lysate was cleared by centrifuging at 14,000 × g for 20 min (4°C).
Isolation of Interacting Proteins of PtNAC by Pull Down Assay
His6-PtNAC was mixed with Co2+-NTA-Agarose beads (the beads were washed and equilibrated with 1X PBS, pH 7.2) and incubated at 4°C for 1 h to allow it to bind with the beads. These beads were then transferred into a glass column and washed with five column volumes of the equilibration buffer, followed by washing with 1X PBS containing 50 mM imidazole, and the samples were collected to remove non-specific proteins. The beads were then washed extensively with 1X PBS to remove imidazole. Following this, 10 ml of P. torridus cell lysate was added to the column, which was incubated overnight at 4°C on a rocker, allowing the NAC interacting proteins to bind with the PtNAC protein associated with the beads. On the next day, the column was washed with 1X PBS buffer to remove non-interacting proteins, and the elution of the bound proteins was done in 1X PBS containing 200 mM imidazole. The samples were subjected to SDS-PAGE analysis. The interacting proteins were identified using liquid chromatography–mass spectrometry (LC–MS).
Sample Processing for LC–MS Analysis
The methods for sample preparation for LC–MS analysis have been described previously (Kumar et al., 2018). Briefly, the proteins in the eluate were incubated with 10% trichloroacetic acid overnight at 4°C. The resulting protein precipitate was washed with 2% sodium acetate in ethanol, air-dried, and resuspended in 200 μl of 8 M urea buffer (UB), loaded in a 3-kDa filter unit (Amicon-Millipore), and centrifuged at 14,000 × g for 15 min. Then, 100 μl of 0.05 M iodoacetamide prepared in UB was added, mixed at 600 rpm for 1 min, again kept for incubation for 20 min, followed by centrifugation at 14,000 × g for 10 min, and followed by twice washing with 100 μl of UB and centrifugation at 14,000 × g for 15 min. Then, two washings were done with 100 μl of 0.05 M ammonium bicarbonate (ABC), and centrifugation was done at 14,000 × g for 10 min. After this, 40 μl of ABC and trypsin (Promega V511A) solution (enzyme/protein ratio, 1:100) was added, mixed at 600 rpm for 1 min, and kept for 16–18 h of incubation in a water bath at 37°C. The digested peptides were eluted at 14,000 × g for 10 min, followed by recovering the remaining uneluted peptides with another 20–30 μl of ABC. The total eluted sample was acidified with 0.1% formic acid and concentrated by speed vac to attain a final volume of 10 μl. LC–MS/MS analysis was performed in a AB SCIEX Triple TOF 5600. The MASCOT and PARAGON search engines were used for peptide identification. Proteins were selected for further study based on a 5% false-discovery rate cutoff and a minimum of two peptides per protein.
Results
Cloning, Expression, and Purification of NAC
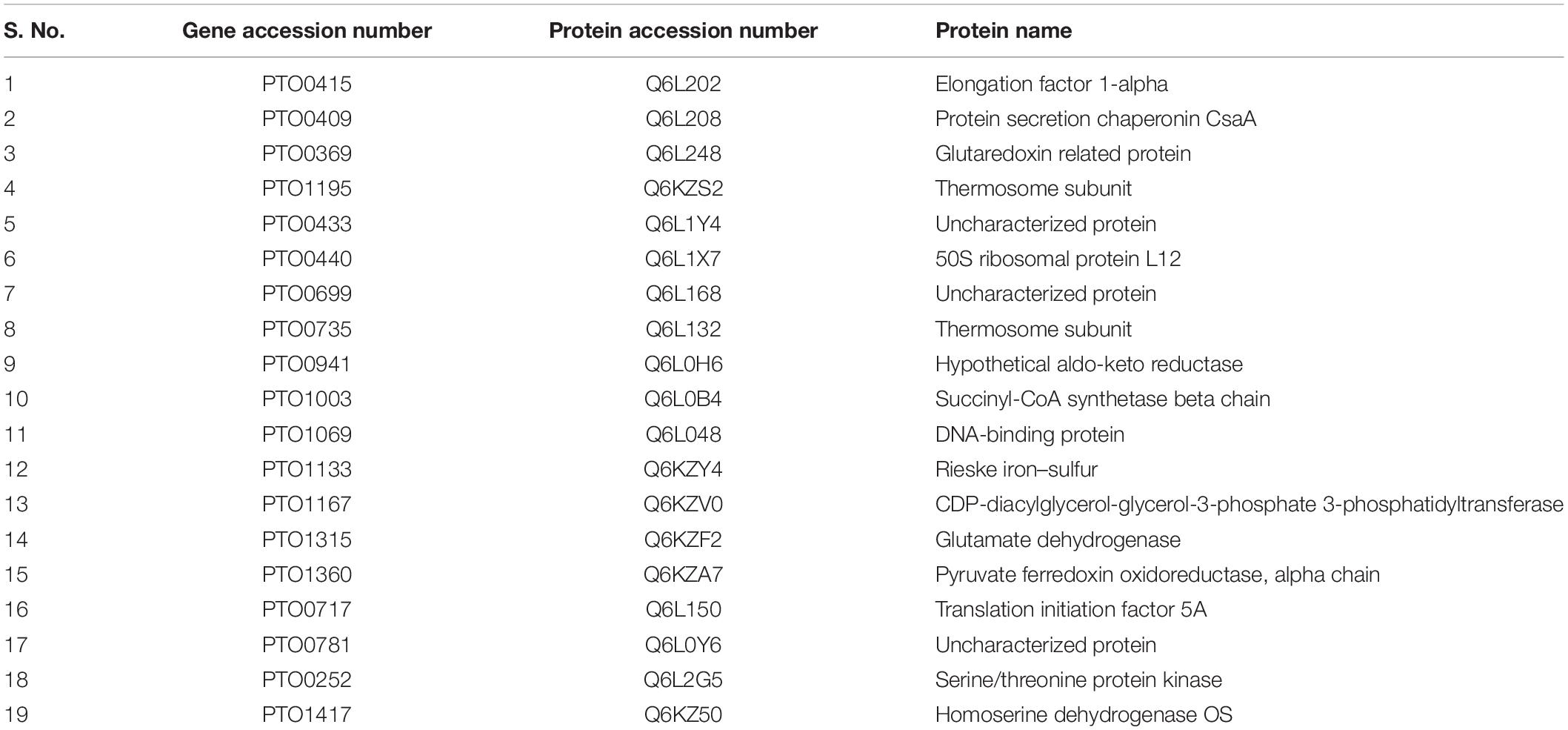
The positive clones encoding PtNAC were confirmed by double-digestion method (Figure 1A). The protein was found to be overexpressed in E. coli BL21 (DE3) (Figure 1B). The protein was purified using Co2+-NTA His6-affinity chromatography, followed by size exclusion chromatography on a HiPrepTM S-200 HR column (GE Healthcare). The molecular mass of the recombinant NAC protein after electrophoresis on 15% SDS-PAGE was observed to be ∼14 kDa (Figure 1C). The MALDI-TOF analysis further confirmed that the recombinant, overexpressed protein was the NAC protein of P. torridus. The MALDI-TOF analysis revealed two peaks corresponding to the molecular weights of 14.5 and 29.0 kDa, suggesting that the PtNAC protein purified by affinity chromatography was a mixture of dimer and monomer population (Supplementary Figure S1). However, only a single peak was observed in the elution on the gel filtration chromatography, which corresponds to the dimer population when compared to the protein standards run on this column in our laboratory (Supplementary Figure S2).
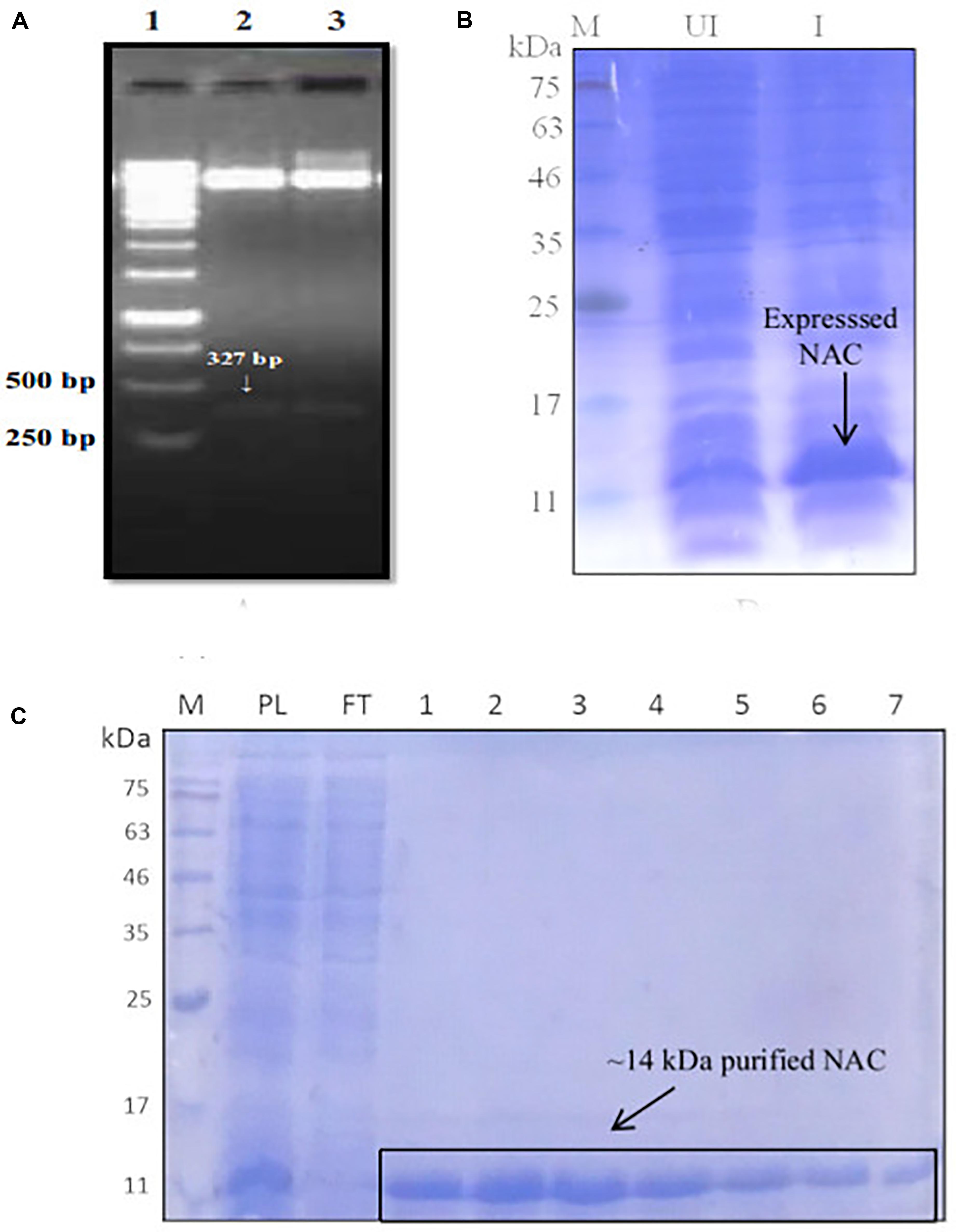
Figure 1. (A) Confirmation of positive clones of PtNAC by double-digestion method. Lanes: 1—marker (Thermo Scientific O’ GeneRuler 1-kb DNA ladder); 2, 3—double-digested pET28a(+) with the fallout. (B) Expression of PtNAC in E. coli BL21(DE3). Lanes: 1—SDS-PAGE marker, 2—uninduced: total cellular extract from E. coli BL21(DE3) before isopropyl β-D-1-thiogalactopyranoside (IPTG) induction, and 3—induced: total cellular extract after IPTG induction shown by arrow. (C) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of recombinant NAC purified in pET28a(+) systems (samples were resolved on 15% polyacrylamide gel and stained with Coomassie Brilliant Blue R-250). Lanes: M—SDS protein marker, PL—preload, FT—flow through, 1–7 purified recombinant nascent polypeptide-associated complex eluents obtained from Co2+-NTA metal affinity column using imidazole (200 mM). BlueRAY prestained protein marker was used as the SDS-PAGE marker.
Chaperone Activity
There was a slight reduction in the thermal aggregation of BCAII in the presence of an equimolar concentration of NAC (BCAII/NAC, 1:1) compared to the sample containing BCAII only, suggesting that the PtNAC protein might have a chaperone-like activity in vitro (Figure 2). The higher concentrations of NAC were accompanied by a greater aggregation (BCAII/NAC, 1:2 and 1:4). However, the effect of the chaperone-like activity on cell viability under in vivo stress conditions remains to be studied.
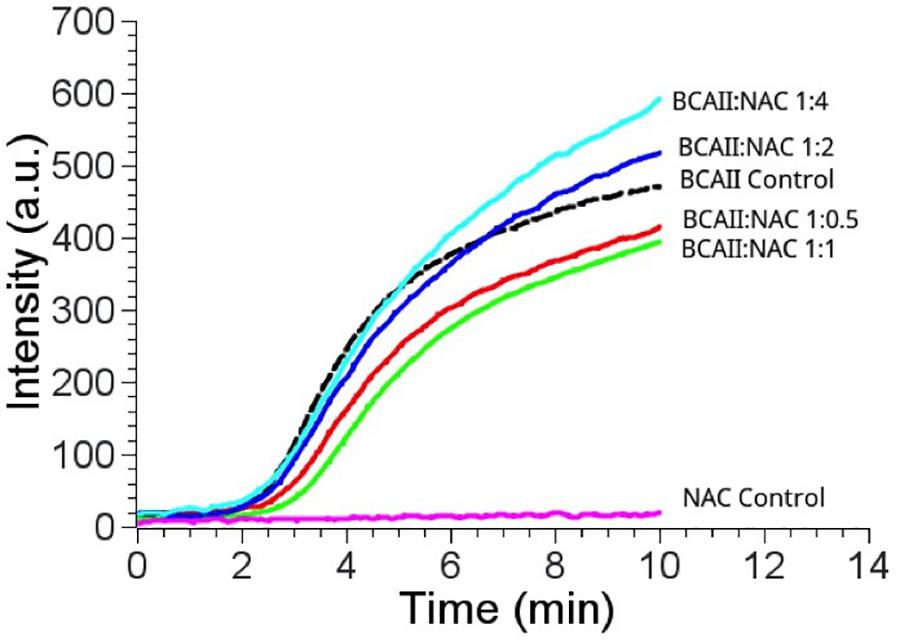
Figure 2. Chaperone activity of PtNAC/BCAII control is substrate control (black); BCAII/NAC in different ratios; 1:0.5 (red), 1:1 (green), 1:2 (blue), and 1:4 (cyan). The best prevention of aggregation of BCAII in the presence of NAC was observed at equimolar BCAII/NAC concentration.
Effect of Temperature and pH on PtNAC Structure
The CD spectrum showed the predominance of β-sheets in the secondary structure of PtNAC at pH 7.5 and 20°C (Figure 3A). The changes in the secondary structure of PtNAC were studied at different temperatures (20–80°C), and it was observed that there was no significant change in the secondary structure with the increase in temperature up to 80°C, indicating that PtNAC was a thermostable protein (Figure 3B). The secondary structure of PtNAC was stable over a broad range of pH between 2 and 10 (Figure 3C).
Figure 3. Changes in the secondary structure of PtNAC investigated using circular dichroism spectroscopy: (A) secondary structure estimation at pH 7.5 and 20°C, (B) effect of different temperatures ranging from 20 to 80°C, and (C) effect of different pH values ranging from 2 to 10.
CD Spectra of the Native and Denatured PtNAC
Guanidine hydrochloride (Gdn-HCl), a strong chaotrophic agent, was used to determine the effect of denaturants on the secondary structure of PtNAC. BestSel analysis predicted the presence of 21.9% ɑ-helices and 28.7% antiparallel β-sheet in the native protein (Supplementary Figure S3). However, in the presence of 6 M Gdn-HCl, the ɑ-helices content decreased significantly to 12.2%, while the antiparallel β-sheet content showed a smaller decrease to 24.9%. A progressive shift in the spectral wavelength and a decrease in negative ellipticity were observed with an increase in the concentration of Gdn-HCl up to 6 M (Figure 4, the change in secondary structure at 222 nm is shown in the inset). However, when PtNAC was incubated with increasing concentrations of urea, the decrease in negative ellipticity and the shift in spectral wavelength were not very significant (Figure 5, the change in secondary structure at 222 nm is shown in the inset).
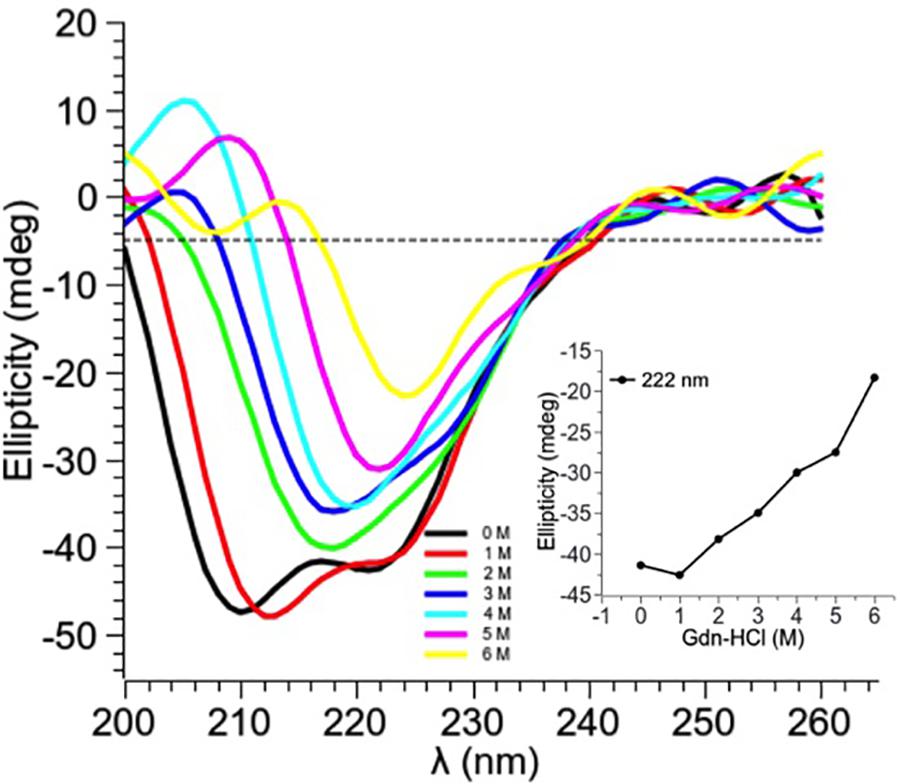
Figure 4. Effect of increasing concentrations of Gdn-HCl (1–6 M) on PtNAC: Inset: ellipticity at 222 nm versus Gdn-HCl concentration. On increasing Gdn-HCl concentration from 1 to 2 M, a gradual change in the secondary structure was observed. A decrease in the negative ellipticity and a right shift in the spectral wavelength were observed with the progressive increase in Gdn-HCl concentration up to 6 M.
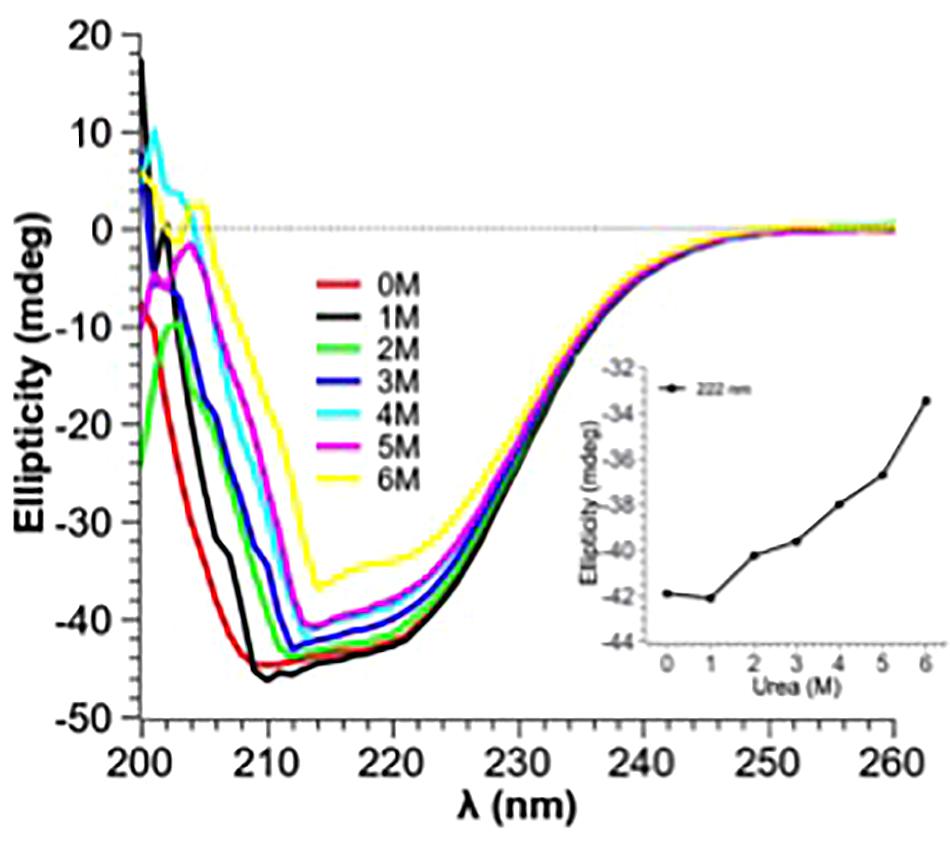
Figure 5. Effect of increasing concentrations of urea (1–6 M) on PtNAC: Inset: ellipticity at 222 nm versus urea concentration. A gradual change in the secondary structure was observed when the concentration of urea was increased from 1 to 6 M.
PtNAC-Binding Proteins
Protein interactions play an important role in cellular organization, and information about the functional partners of a protein can help in understanding its role in biological processes. Thus, an attempt was made to identify the interacting protein partners of PtNAC. PtNAC-binding proteins were isolated by pull down assay, separated on an SDS-PAGE (Figure 6) and identified by LC–MS. The LC–MS analysis identified 19 proteins as the binding partners of PtNAC (Table 1). These were elongation factor 1-alpha (Q6L202), protein secretion chaperonin CsaA (Q6L208), glutaredoxin (Q6L248)-related protein, thermosome subunit (Q6KZS2), 50S ribosomal protein L12 (Q6L1X7), thermosome subunit (Q6L132), hypothetical aldo-keto reductase (Q6L0H6), succinyl-CoA synthetase beta chain (Q6L0B4), DNA-binding protein (Q6L048), Rieske iron–sulfur protein (Q6KZY4), diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase (Q6KZV0), glutamate dehydrogenase (Q6KZF2), pyruvate ferredoxin oxidoreductase, alpha chain (Q6KZA7), translation initiation factor 5A (Q6L150), serine/threonine protein kinase (Q6L2G5), homoserine dehydrogenase (Q6KZ50), and uncharacterized proteins (Q6L1Y4, Q6L168, and Q6L0Y6).
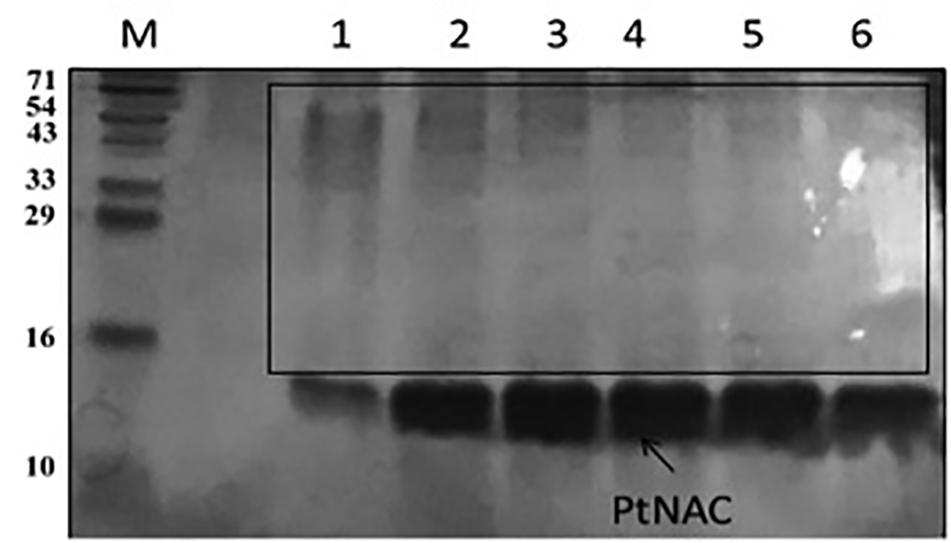
Figure 6. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis protein elution profile of PtNAC with 200 mM imidazole after binding with P. torridus cell lysate: lane M—three-color prestained protein ladder from PUReGeNe; lanes 1–6 show the eluents with interacting proteins (boxed).
Discussion
The present study aimed to unravel the biophysical and the biochemical characteristics of PtNAC and discern its binding partners. The homologs of PtNAC have been reported in all archaeal species, except Nanarchaeum equitans, Sulfolobus solfataricus P2, Sulfolobus islandicus REY15A, and Sulfolobus islandicus M.14.25 (Rani et al., 2016). To date, the crystal structure has been reported for NAC from only one archaeal species Methanothermobacter marburgensis, where it was found to be a homodimer of alpha-subunits (Spreter et al., 2005). The results of the CD spectroscopy of PtNAC indicated that ß-sheet is the predominant form of secondary structure present in PtNAC, similar to that reported in the crystal structure of NAC of M. marburgensis (Spreter et al., 2005).
Earlier studies had proposed that the NAC protein has a chaperone-like activity as it binds to ribosomes and interacts with nascent polypeptides (Bukau et al., 2000; Hartl and Hayer-Hartl, 2002; Wegrzyn and Deuerling, 2005). Since PtNAC mildly prevented the aggregation of heat-denatured BCAII, it might also have a chaperone-like activity. However, the effect of a chaperone-like activity on cell viability under in vivo stress conditions remains to be studied. The CD spectroscopic studies also revealed that the secondary structure of PtNAC was stable over a wide range of temperature and pH, which fortifies its role in aiding the adaptability of this thermoacidophilic archaea in extreme growth conditions, possibly by contributing to the prevention of aggregation and helping in the proper folding of other cellular proteins. The denaturants Gdn-HCl and urea showed different effects on the secondary structure conformations of PtNAC. This might be attributed to the difference in the nature of both denaturants. Gdn-HCl is a charged denaturant, while urea is neutral by nature. Gdn-HCl, due to its charged nature, hides the favorable electrostatic interactions that might stabilize the native state of protein, affecting the protein structure, while urea specifically binds to the amide units and therefore denatures proteins by minimizing the hydrophobic effect.
To understand the participation of NAC in metabolic processes and pathways, the present study was designed to isolate the binding proteins of P. torridus NAC by affinity separation followed by LC–MS analysis. The results of the pull down assay indicated that the NAC of P. torridus interacted with a diverse set of proteins, suggesting that it might be a multifaceted protein. The Kyoto Encyclopedia of Genes and Genomes-based metabolic pathway analysis of the NAC-binding proteins revealed that many interacting protein partners of NAC were multi-functional. A functional categorization of these proteins revealed that two of them were associated with amino acid metabolism (Q6KZ50 and Q6KZF2), two with carbohydrate metabolism (Q6L0B4 and Q6KZA7), three with energy metabolism (Q6L0B4, Q6KZA7, and Q6KZF2), four with global and overview maps (Q6L0B4, Q6KZA7, Q6KZF2, and Q6KZ50), one with metabolism of other amino acids (Q6KZF2), and one with translation (Q6L1X7) (Table 2). Interestingly, four of the interacting protein partners of PtNAC were archaeal chaperones. Two of them (Q6L132 and Q6KZS2) were subunits of thermosome that belong to the family of Group II archaeal chaperonins, one was CsaA (Q6L208) and the other was a glutaredoxin-related protein (Q6L248), a member of the thioredoxin superfamily of archaeal chaperones (Table 2).
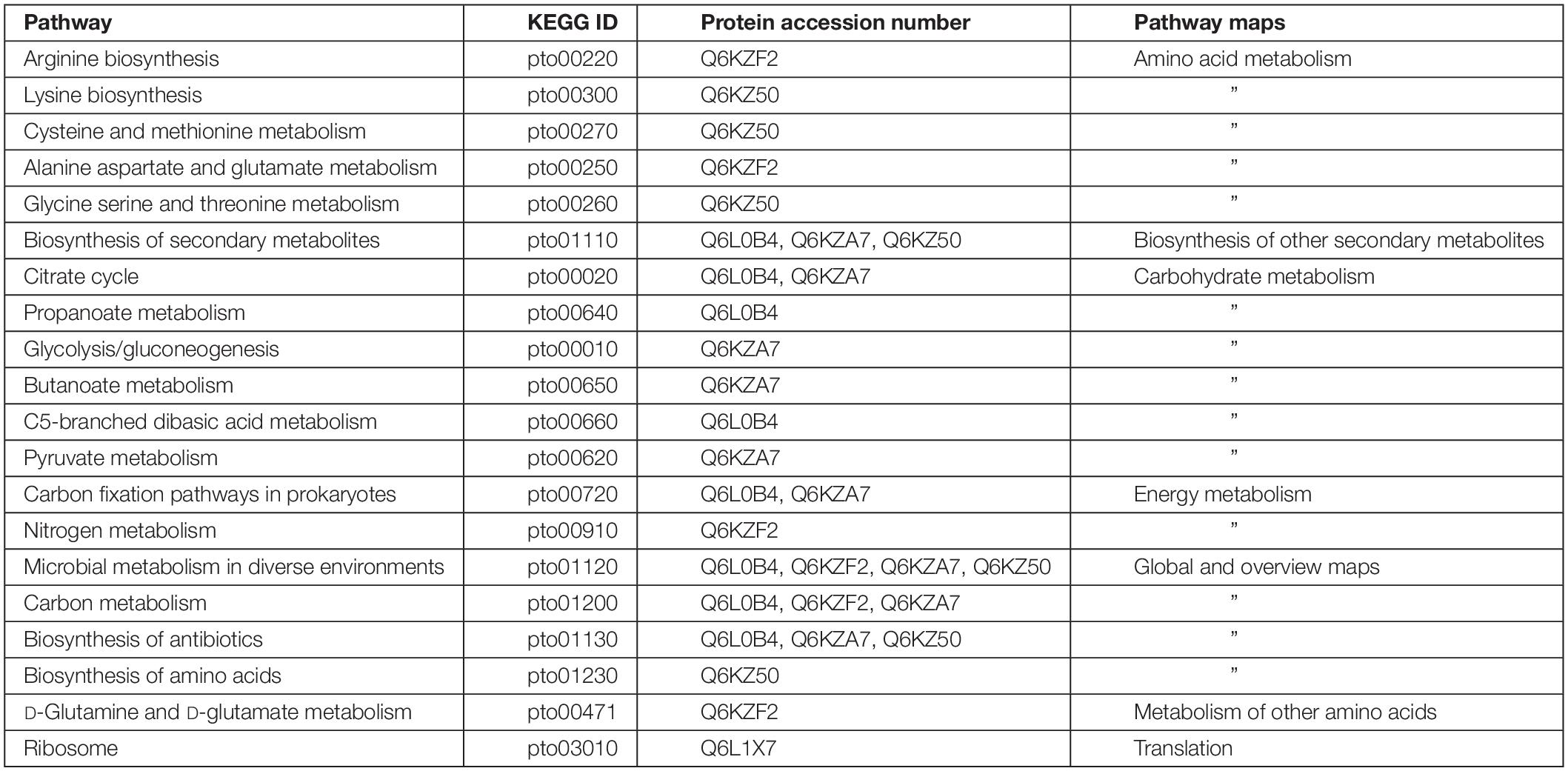
Table 2. Annotation of Kyoto Encyclopedia of Genes and Genomes pathways of the interacting protein partners of P. torridus NAC.
The NAC-binding partner Q6L202 was identified as elongation factor 1-alpha. Elongation factor 1-alpha promotes the GTP-dependent binding of aminoacyl-tRNA to the A-site of ribosomes during the biosynthesis of proteins. The NAC-binding protein Q6L208 was recognized as CsaA. CsaA is one of the few chaperones which are present in bacteria and archaea but is absent in eukaryotes. CsaA prevents the aggregation of unfolded proteins and helps in the translocation of proteins across the cytoplasmic membrane (Sharma et al., 2018). The binding partner Q6L248 was identified as a glutaredoxin-related protein. Glutaredoxin is a member of the thioredoxin superfamily of proteins which is crucial for maintaining a reduced intracellular redox state. It also helps in protein folding and has been shown to have a chaperone-like activity (Berndt et al., 2008; Rani et al., 2016).
The NAC-binding proteins Q6L132 and Q6KZS2 were identified as thermosome subunits. Thermosomes belong to the family of Group II chaperonins which are ubiquitously present in archaea and impart extreme thermal stability (Phipps et al., 1991; Bigotti and Clarke, 2008). The Group II chaperonins comprise up to 40% of the total cellular protein and are abundantly produced by the cells exposed to heat shock (Trent et al., 1991). The binding partner Q6L1X7 was identified as 50S ribosomal protein L12. The 50S ribosomal protein L12 binds to the 23S rRNA and is an important constituent of the secondary structure of the ribosome. The NAC-binding protein Q6L0H6 was identified as a hypothetical aldo-keto reductase. Aldo-keto reductases are a superfamily of enzymes which are involved in the reduction of aldehydes and ketones. The binding protein Q6L0B4 was identified as the beta chain of succinyl-CoA synthase. It is a mitochondrial matrix enzyme composed of two subunits, alpha and beta. Succinyl-CoA synthase is an enzyme in the Krebs cycle that converts succinyl-CoA to succinate and free coenzyme A and converts ADP or GDP to ATP or GTP, respectively (Johnson et al., 1998).
The PtNAC-binding partner Q6KZY4 was identified as Rieske iron–sulfur protein. Proteins containing Rieske-type [2Fe-2S] clusters are associated with essential functions in all the three domains of life. The Rieske proteins occur as subunits in the cytochrome bc1 and cytochrome b6f complexes of prokaryotes and eukaryotes or form components of archaeal electron transport systems (Schmidt and Shaw, 2005). The families encoding for Rieske iron–sulfur proteins are more common in bacteria and archaea than in eukaryotes. Recent studies suggest that Rieske proteins are functionally versatile like other redox proteins, and the use of multiple Rieske proteins in electron transfer reactions helps in microbial adaptation to changing environmental conditions (Schneider and Schmidt, 2005). The binding protein Q6KZF2 was identified as glutamate dehydrogenase. Glutamate dehydrogenase is one of the most widely studied enzymes for understanding the thermostability of hyperthermophiles (Vetriani et al., 1998; Vieille and Zeikus, 2001). The binding protein Q6KZA7 was identified as pyruvate ferredoxin oxidoreductase alpha chain. In enzymology, a pyruvate ferredoxin oxidoreductase/pyruvate synthase is an enzyme that catalyzes the interconversion of pyruvate and acetyl-CoA.
The PtNAC-binding protein Q6KZ50 was identified as reversed homoserine dehydrogenase. Homoserine dehydrogenase is a key enzyme in the aspartate pathway involved in the NAD (P)-dependent reduction of aspartate beta-semialdehyde into homoserine. Homoserine is an intermediate in the biosynthesis of three amino acids—threonine, methionine, and isoleucine—in plants and microorganisms. The binding protein Q6L2G5 was identified as a serine/threonine protein kinase. Serine/threonine protein kinases are involved in protein phosphorylation, one of the most important post-translational modifications that regulate almost every cellular process, including signal transduction. The binding protein Q6L150 was identified as translation initiation factor 5A. The translation initiation factor 5A promotes the formation of the first peptide bond at the onset of protein synthesis. The binding protein Q6KZV0 was identified as CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase. The CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase participates in the metabolism of glycerophospholipids/phosphoglycerides. Glycerophospholipids are present abundantly in the cell membranes where they serve as an anchor for proteins in cell membranes and also participate in cell signaling.
When String database version 10.5 (Szklarczyk et al., 2017) was used to discern the interacting protein partners of PtNAC, it was observed that, of the 19 interacting protein partners predicted by the STRING database, only one protein, 50S ribosomal protein L12 (rpl12), was present in the experimental interactome (Figure 7A). This is, however, not surprising because to date no experimental interactome studies have been conducted for the NAC proteins of archaea. Also, because NAC proteins are absent in prokaryotes, the NAC interaction networks in STRING might have been built on the basis of information available about eukaryotic NAC proteins. The STRING database suggests the maximum interacting partners of PtNAC to be ribosomal proteins, as predicted by other studies as well (Pech et al., 2010); however, only a single ribosomal protein was identified in our study. It is possible that the inclusion of EDTA in the binding buffers could have possibly led to the dissociation of ribosomes and affected their binding to PtNAC. Since the binding of most proteins in vivo is transient, the interacting partners identified in the study are also expected to be sensitive to conditions such as the buffers used for facilitating binding and subsequent separation from the column. More elaborate studies (e.g., similar pull down studies at different pH and buffer concentrations) may be required to make the predictions more robust. However, the final validation of any such predicted interaction lies in seeking the effect of such interactions on the physiology of the organism, such as growth and reproduction. However, this preliminary study does provide an opportunity to investigate the role of NAC protein in pathways or cellular functions hitherto unassigned to this protein. We therefore made an attempt to find if the interacting partners predicted in this study can be predicted to be part of a common pathway or have been shown to interact with each other in any previous studies.
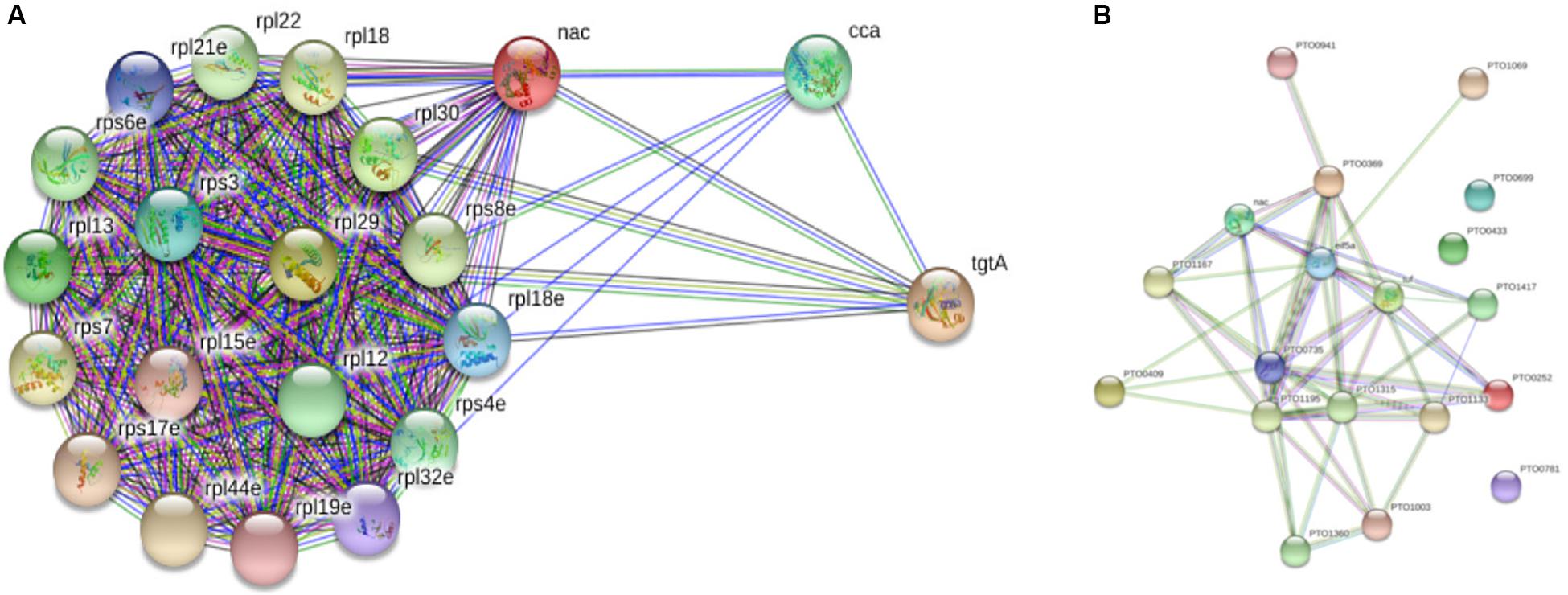
Figure 7. STRING database version 10.5-based protein–protein interaction analysis of PtNAC: (A) STRING-predicted protein binding partners of PtNAC. (B) Network of PtNAC and experimentally determined PtNAC-binding proteins created using STRING.
Next, a network analysis of PtNAC and its interacting protein partners experimentally identified in our study was performed using String database version 10.5 (Figure 7B). Interestingly, the STRING analysis revealed a strong interaction network among the experimentally identified PtNAC-binding proteins. Sixteen of the 19 experimentally identified proteins showed mutual connections with each other on the PPI network map. The three PtNAC interacting proteins which did not integrate in the interaction network were uncharacterized proteins (protein accession/gene accession—Q6L1Y4/PTO0433, Q6L168/PTO0699, and Q6L0Y6/PTO0781). Thus, the results of the STRING analysis corroborated our experimental findings and suggested novel functional possibilities for the NAC protein.
Conclusion
The present work is the first study of the interactome analysis of NAC of any archaeal species and, to the best of our knowledge, the first report on the biophysical and the biochemical characterization of PtNAC. Although this is a preliminary study, our results do provide an opportunity to investigate the role of NAC in pathways or cellular functions hitherto unassigned to this protein.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
MG conceptualized the study. NS, AS, SK, RR, and NS worked on the methodology. AG, MK, and NS were responsible for the software. NS, AS, SK, RR, and NS validated the study. NS contributed to the formal analysis. MG helped with the resources. NS and AS prepared the original draft. NS, AS, and MG reviewed and edited the manuscript.
Funding
This research was funded by the Indian Council of Medical Research (ICMR) (BIC/12(14)/2015).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
AS is thankful to the Department of Biotechnology, Government of India, for the research grant (BT/Bio-CARe/01/9935/2013-14, dated 30.09.2014) received under BioCARe Scheme that supported this work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00915/full#supplementary-material
Footnotes
References
Arora, J., Goswami, K., and Saha, S. (2014). Characterization of the replication initiator Orc1/Cdc6 from the archaeon Picrophilus torridus. J. Bacteriol. 196, 276–286. doi: 10.1128/JB.01020-13
Arsenovic, P. T., Maldonado, A. T., Colleluori, V. D., and Bloss, T. A. (2012). Depletion of the C. elegans NAC engages the unfolded protein response, resulting in increased chaperone expression and apoptosis. PLoS One 7:e44038. doi: 10.1371/journal.pone.0044038
Berndt, C., Lillig, C. H., and Holmgren, A. (2008). Thioredoxins and glutaredoxins as facilitators of protein folding. Biochim. Biophys. Acta 1783, 641–650. doi: 10.1016/j.bbamcr.2008.02.003
Bigotti, M. G., and Clarke, A. R. (2008). Chaperonins: the hunt for the Group II mechanism. Arch. Biochem. Biophys. 474, 331–339. doi: 10.1016/j.abb.2008.03.015
Bukau, B., Deuerling, E., Pfund, C., and Craig, E. A. (2000). Getting newly synthesized proteins into shape. Cell 101, 119–122.
Creagh, E. M., Brumatti, G., Sheridan, C., Duriez, P. J., Taylor, R. C., Cullen, S. P., et al. (2009). Bicaudal is a conserved substrate for Drosophila and mammalian caspases and is essential for cell survival. PLoS One 4:e5055. doi: 10.1371/journal.pone.0005055
del Alamo, M., Hogan, D. J., Pechmann, S., Albanese, V., Brown, P. O., and Frydman, J. (2011). Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biol. 9:e1001100. doi: 10.1371/journal.pbio.1001100
Gamerdinger, M., Hanebuth, M. A., Frickey, T., and Deuerling, E. (2015). The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science 348, 201–207. doi: 10.1126/science.aaa5335
Hartl, F. U., and Hayer-Hartl, M. (2002). Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 95, 1852–1858.
Hoffmann, A., Bukau, B., and Kramer, G. (2010). Structure and function of the molecular chaperone Trigger Factor. Biochim. Biophys. Acta 1803, 650–661. doi: 10.1016/j.bbamcr.2010.01.017
Johnson, J. D., Mehus, J. G., Tews, K., Milavetz, B. L., and Lambeth, D. O. (1998). Genetic evidence for the expression of ATP- and GTP-specific succinyl-CoA synthetases in multicellular eucaryotes. J. Biol. Chem. 273, 27580–27586.
Kaiser, C. M., Chang, H. C., Agashe, V. R., Lakshmipathy, S. K., Etchells, S. A., Hayer-Hart, M., et al. (2006). Real-time observation of trigger factor function on translating ribosomes. Nature 444, 455–460.
Kogan, G. L., and Gvozdev, V. A. (2014). Multifunctional protein complex NAC (nascent polypeptide associated complex). Mol. Biol. 48, 223–231.
Koplin, A., Preissler, S., Ilina, Y., Koch, M., Scior, A., Erhardt, M., et al. (2010). Dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol. 189, 57–68. doi: 10.1083/jcb.200910074
Kumar, R., Singh, N., Abdin, M. Z., Patel, A. H., and Medigeshi, G. R. (2018). Dengue virus capsid interacts with DDX3X-a potential mechanism for suppression of antiviral functions in dengue infection. Front. Cell Infect. Microbiol. 7:542. doi: 10.3389/fcimb.2017.00542
Pech, M., Spreter, T., Beckmann, R., and Beatrix, B. (2010). Dual binding mode of the nascent polypeptide-associated complex reveals a novel universal adapter site on the ribosome. J. Biol. Chem. 285, 19679–19687. doi: 10.1074/jbc.M109.092536
Phipps, B. M., Hoffmann, A., Stetter, K. O., and Baumeister, W. A. (1991). Novel ATPase complex selectively accumulated upon heat shock is a major cellular component of thermophilic archaebacteria. EMBO J. 10, 1711–1722.
Powers, T., and Walter, P. (1996). The nascent polypeptide-associated complex modulates interactions between the signal recognition particle and the ribosome. Curr. Biol. 6, 331–338.
Rajaraman, K., Raman, B., and Rao, C. M. (1996). Molten-globule state of carbonic anhydrase binds to the chaperone-like alpha-crystallin. J. Biol. Chem. 271, 27595–27600.
Rani, S., Srivastava, A., Kumar, M., Goel, M., and Lund, P. (2016). CrAgDb-a database of annotated chaperone repertoire in archaeal genomes. FEMS Microbiol. Lett. 364:fnw030. doi: 10.1093/femsle/fnx030
Raue, U., Oellerer, S., and Rospert, S. (2007). Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J. Biol. Chem. 282, 7809–7816.
Rospert, S., Dubaquié, Y., and Gautschi, M. (2002). Nascent-polypeptide-associated complex. Cell Mol. Life Sci. 59, 1632–1639.
Schleper, C., Puehler, G., Holz, I., Gambacorta, A., Janekovic, D., Santarius, U., et al. (1995). Picrophilus gen. nov., fam. nov.: a novel aerobic, heterotrophic, thermoacidophilic genus and family comprising archaea capable of growth around pH 0. J. Bacteriol. 177, 7050–7059.
Schmidt, C. L., and Shaw, L. A. (2005). A comprehensive phylogenetic analysis of Rieske and Rieske-type iron-sulfur proteins. J. Bioenerg. Biomembr. 33, 9–26.
Schneider, D., and Schmidt, C. L. (2005). Multiple Rieske proteins in prokaryotes: where and why? Biochim. Biophys. Acta 1710, 1–12.
Sharma, A., Rani, S., and Goel, M. (2018). Navigating the structure-function-evolutionary relationship of CsaA chaperone in archaea. Crit. Rev. Microbiol. 44, 274–289. doi: 10.1080/1040841X.2017.1357535
Spreter, T., Pech, M., and Beatrix, B. (2005). The crystal structure of archaeal nascent polypeptide-associated complex (NAC) reveals a unique fold and the presence of a ubiquitin-associated domain. J. Biol. Chem. 280, 15849– 15854.
Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., et al. (2017). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. doi: 10.1093/nar/gkw937
Tomar, R., Garg, D., Mishra, R., Thakur, A., and Kundu, B. (2013). N-terminal domain of Pyrococcus furiosus L-asparaginase functions as a non-specific, stable, molecular chaperone. FEBS J. 280, 2688–2699. doi: 10.1111/febs.12271
Trent, J. D., Nimmesgern, E., Wall, J. S., Hartl, F. U., and Horwich, A. L. (1991). A molecular chaperone from a thermophilic archaebacterium is related to the eukaryotic protein t-complex polypeptide-1. Nature 354, 490–493.
Vetriani, C., Maeder, D. L., Tolliday, N., Yip, K. S., Stillman, T. J., Britton, K. L., et al. (1998). Protein thermostability above 100 degrees C: a key role for ionic interactions. Proc. Natl. Acad. Sci. U.S.A. 95, 12300–12305.
Vieille, C., and Zeikus, G. J. (2001). Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65, 1–43.
Wegrzyn, R. D., and Deuerling, E. (2005). Molecular guardians for newborn proteins: ribosome-associated chaperones and their role in protein folding. Cell Mol. Life Sci. 62, 2727–2738.
Wickner, W. (1995). The nascent-polypeptide-associated complex: having a “NAC” for fidelity in translocation. Proc. Natl. Acad. Sci. U.S.A. 92, 9433–9434.
Keywords: chaperone, circular dichroism, liquid chromatography–mass spectrometry, STRING, interactome
Citation: Singhal N, Sharma A, Kumari S, Garg A, Rai R, Singh N, Kumar M and Goel M (2020) Biophysical and Biochemical Characterization of Nascent Polypeptide-Associated Complex of Picrophilus torridus and Elucidation of Its Interacting Partners. Front. Microbiol. 11:915. doi: 10.3389/fmicb.2020.00915
Received: 20 December 2019; Accepted: 17 April 2020;
Published: 26 May 2020.
Edited by:
Jerry Eichler, Ben-Gurion University of the Negev, IsraelReviewed by:
Bruno Franzetti, GDR3635 Biodiversité, Origine, Processus Cellulaires Fondamentaux, Biotechnologies (Archaea), FranceMasafumi Yohda, Tokyo University of Agriculture and Technology, Japan
Copyright © 2020 Singhal, Sharma, Kumari, Garg, Rai, Singh, Kumar and Goel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manisha Goel, bWFuaXNoYWdvZWxAc291dGguZHUuYWMuaW4=
†These authors have contributed equally to this work
 Neelja Singhal
Neelja Singhal Archana Sharma1†
Archana Sharma1† Manish Kumar
Manish Kumar Manisha Goel
Manisha Goel