- 1National and Regional Joint Engineering Laboratory for Medicament of Zoonoses Prevention and Control, Guangdong Laboratory for Lingnan Modern Agriculture, Laboratory of Veterinary Vaccine Innovation of the Ministry of Agriculture, Key Laboratory of Zoonosis Prevention and Control of Guangdong Province, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
- 2Shanghai Municipal Center for Disease Control and Prevention, Shanghai, China
In this study, we investigated the pattern of antimicrobial resistance in Salmonella enterica serotype Enteritidis isolates in Shanghai, China from 2005 to 2014. We found the first isolates with resistance to the fourth-generation cephalosporin cefepime starting in 2010. Furthermore, we analyzed the epidemic characteristics and mechanisms of underlying cefepime resistance in S. Enteritidis isolates found from 2010. In total, 38 of 2,914 (1.30%) isolates were identified as cefepime-resistant S. Enteritidis (CRSE) isolates by Kirby–Bauer disk diffusion. Two isolates were from animal derived food sources; 36 isolates were from fecal samples of human patients with salmonellosis. Antimicrobial susceptibility testing using the agar dilution method revealed that all CRSE isolates showed additional resistances at least to ceftazidime, cefotaxime, and ampicillin. Additionally, pulsed-field gel electrophoresis (PFGE) profiles indicated that 89.47% of CRSE isolates also displayed similar PFGE patterns. Five types of β-lactamase genes, blaCTX–M (100.00%, 38/38), blaSHV (65.79%, 25/38), blaTEM (52.63%, 20/38), blaACC (18.42%, 7/38), and blaPSE (5.26%, 2/38) were detected by PCR and sequencing. Among blaCTX–M genes, blaCTX–M–55 was the dominant type (84.21%, 32/38). Conjugation and transformation experiments along with plasmid replicon typing revealed that blaCTX–M–55 was located on plasmids of various replicon types with sizes ranging from 76.8 to 138.9 kb. Plasmid sequence analysis also showed that the blaCTX–M–55 gene was mobilized mainly by the ISEcp1-blaCTX–M–55-ORF477 transposition unit and had its own ISEcp1-based promoter, which accelerated the expression and transmission of blaCTX–M–55. Analysis of whole genome sequences (Illumina) of one selected transformant SH12G706-C showed high similarity of the blaCTX–M–55 carrying plasmid with the IncI1 plasmid backbone p628-CTX-M of Klebsiella pneumoniae detected in 2010 in China. The present study demonstrated that the blaCTX–M–55 gene mobilized by ISEcp1- blaCTX–M–55-ORF477 was the main feature shared by CRSE isolates and seems to play an important role for transmission of cefepime resistance. The number of CRSE isolates is rising annually, and the strong dissemination ability of ISEcp1-blaCTX–M–55-ORF477-harboring plasmids among different species represents an important threat to the therapeutic effectiveness of cefepime.
Introduction
Salmonella is one of the most common foodborne pathogens worldwide (Tabu et al., 2012). Gastroenteritis caused in both human and animals by non-typhoid Salmonella (NTS) has become a global public health concern. To date, more than 2,500 Salmonella serotypes have been identified (Yang et al., 2010), and S. Enteritidis is the predominant serotype in many countries (Galanis et al., 2006; Matheson et al., 2010; Zhang et al., 2014a). According to a report by Chinese Centers for Disease Control (CDC), around 792 people died as a result of S. Enteritidis infection annually (Ren et al., 2016).
In general, compared to other NTS serotypes, such as S. Typhimurium or S. Derby, S. Enteritidis is usually self-limiting and shows susceptibility to most clinical first-line medicines (Ke et al., 2014). But antibiotics are needed for complicated cases and are used if the infection spreads or is highly likely to spread from the intestines to the bloodstream and other organs. Third generation cephalosporins have excellent curative effects, but with the abuse and inappropriate use of these antibiotics, an increasing number of S. Enteritidis strains are nowadays resistant, mainly caused by production of extended-spectrum beta-lactamases (ESBLs) (Tenover, 2006). This phenomenon represents an ever-increasing threat to public health and a challenge that must be met by the development of alternative therapeutic strategies. Cefepime, which is the first fourth generation cephalosporin to be approved for use in China, has been reported to show low toxicity and high activity against Enterobacteriaceae that resistant to third-generation cephalosporins (Chapman and Perry, 2003; Yahav et al., 2007). There are some reports stated that cefepime should be used with caution for serious ESBL infections based on its failure on some clinical experiments (Labombardi et al., 2006; Wang et al., 2016; Tamma and Rodriguez-Bano, 2017). But it can be considered for treatment if the MIC ≤ 1 μg/mL and meanwhile taken the patient’s health conditions into consideration, such as the severity of infection, resistance to antibiotics (Patel et al., 2019).
Despite the important role of cefepime in treating ESBL-producing S. Enteritidis infections, the epidemiological data in this field are scarce. Therefore, in this study, we conducted a systematic characterization of the epidemiology of S. Enteritidis infections in Shanghai, China from 2005 to 2014 to clarify the mechanisms underlying the development of cefepime-resistant S. Enteritidis.
Materials and Methods
Bacterial Strains
During the period from 2005 to 2014, a total of 2,914 S. Enteritidis isolates were collected by the Shanghai CDC from human diarrhea samples obtained from 134 hospitals and animal-derived food obtained from 123 retail markets. The Salmonella isolates were serotyped according to the White Kauffmann Le Minor scheme (Issenhuth-Jeanjean et al., 2014) or National Food Safety Standard-Food microbiological examination: Salmonella (GB 4789.4–2016) by slide agglutination, using specific O and H antisera (S&A Reagents Lab Ltd., Bangkok, Thailand). All isolates were then transported to the Key Laboratory of Zoonosis Prevention and Control of Guangdong Province in South China Agriculture University by a medium that can keep bacteria alive at room temperature for 12 months (Kemajia, Shanghai, China). The Kirby–Bauer disk diffusion method was used to select the cefepime-resistant S. Enteritidis (CRSE) isolates and characterize their antimicrobial susceptibility profiles. This test was performed on Mueller–Hinton agar with seventeen impregnated disks (Oxoid, United Kingdom) as follows: amoxicillin-clavulanic acid 30 μg (AMC), ampicillin 10 μg (AMP), cefotaxime 30 μg (CTX), ceftazidime 30 μg (CAZ), cefepime 5 μg (FEP), imipenem 10 μg (IPM), amikacin 30 μg (AMK), gentamicin 10 μg (GEN), streptomycin 10 μg (STR), sulfisoxazole 300 μg (SIZ), trimethoprim-sulfamethoxazole 23.75/1.2 μg (SXT), polymyxin B 300 IU (PMB), chloramphenicol 30 μg (CHL), tetracycline 30 μg (TET), nalidixic acid 30 μg (NAL), ciprofloxacin 5 μg (CIP), and ofloxacin 5 μg (OFX). The minimum inhibitory concentrations (MICs) of the selected cefepime-resistant isolates and their transconjugants to three beta-lactam antibiotics (cefotaxime, ceftazidime and cefepime) were determined by agar dilution in accordance with the standards and guidelines described by the Clinical and Laboratory Standards Institute (CLSI). E. coli ATCC 25922 was used as the quality control and the results were interpreted according to the CLSI standards and guidelines 2017.
Detection of Beta-Lactamase Genes
Salmonella isolates showing resistance to cefepime were further subjected to screening for the presence of beta-lactamase genes (blaCTX–M, blaTEM, blaSHV, blaACC, blaOXA, blaPSE, blaVEB, blaPER, and blaGES) by PCR assays (Kiratisin et al., 2008; Usha et al., 2008). All blaCTX–M positive PCR products were sequenced and aligned using GenBank online BLAST software. The primers used in this study are shown in Supplementary Table S1.
Pulsed-Field Gel Electrophoresis (PFGE)
PFGE subtyping of 38 CRSE was performed using XbaI-digestion of genomic DNA according to the protocol recommended by the CDC (Ribot et al., 2006). The PFGE patterns were then compared using BioNumerics software (Version 5.1; Applied-Maths, Sint-Martens-Latem, Belgium).
Conjugation and Transformation Experiments
Conjugation experiments were conducted in Luria-Bertani broth, using CRSE as donor strains and the streptomycin-resistant E. coli strain C600 as the recipient. Cultures of donor and recipient cells in the logarithmic phase (1 and 4 mL, respectively) were mixed and incubated overnight at 37°C without shaking. If conjugation failed the mixture was exposed to a brief pulse of a high-voltage electric field (1.8 kv). 1 mL LB broth was added to the mixture after electric shock to recover the bacteria. E. coli C600 transconjugants/transformants with ESBL genes on plasmids provided by S. Enteritidis were identified on the basis of the ability to generate blue bacterial colonies on CHROMagarTM containing ceftazidime (2 μg/mL) plus streptomycin (3,000 μg/mL, Sigma–Aldrich Co., St Louis, MO, United States). To further verify the transconjugants/transformants, MICs of cefotaxime, ceftazidime, cefepime and the presence of blaCTX–M were determined as described above.
Plasmid Characterization
The incompatibility groups of plasmids harbored by transconjugants/transformants were determined by PCR-based replicon typing using previously reported primers (Carattoli et al., 2005). The sizes of bacterial plasmids carrying blaCTX–M gene were determined by PFGE of S1 nuclease (TaKaRa Biotechnology, Dalian, China)-digested whole genomic DNA; the location of the blaCTX–M gene was determined by Southern blot hybridization using probes for the specific detection of the DIG-labeled blaCTX–M fragment according to the manufacturer’s instructions (DIG High Prime DNA Labeling and Detection Starter Kit I, Roche Applied Science, Mannheim, Germany) (Yang et al., 2012). The genetic context of blaCTX–M was determined by PCR using previously described primers and conditions (Pan et al., 2013).
Sequencing and Analysis of a blaCTX–M–55-Carrying Plasmid
The plasmid that transferred from the blaCTX–M–55 positive S. Enteritidis isolate SH12G706 to E. coli C600 was extracted using the Omega Plasmid BAC/PAC DNA Kit D2156 (Omega, Bio-Tek) and then sent to GENEWIZ (Guangzhou, China) for whole genome de novo sequencing by using Illumina HiSeq sequencing technology. The contigs was assembled using Velvet (version 1.2.10), SSPACE (version 3.0), and GapFiller (version 1–10) software and the annotation of genes was conducted using Non-Redundant Protein, gene ontology and Kyoto encyclopedia of genes and genomes databases. The nucleotide and amino acid sequences were analyzed and compared through BLAST queries against the GenBank database. The datasets generated for this study can be found in the NCBI-Sequence Read Archive (SRA): SRR9734360.
Results
Prevalence of CRSE
In total, 2,914 S. Enteritidis isolates were obtained, of which, 222 isolates were collected in 2010, 679 isolates were collected in 2011, 547 isolates were collected in 2012, 629 isolates were collected in 2013 and 462 isolates were collected in 2014. In total, 2,566 isolates were human-derived, while 348 isolates were from animal-derived food, see detailed information in Table 1. S. Enteritidis could be detected all year-round, but mainly in summer and autumn. Furthermore, adults (men and women) and children were more often to be infected than infants and the elderly. CRSE first appeared in a clinical diarrhea sample in 2010 and more recently, in foodborne source in 2013. Subsequently, 38 CRSE isolates (1.3%; 38/2914) were obtained with increasing frequency year by year; 36 CRSE were from human patients (17 hospitals) and 2 from animal-derived food (chicken wings and duck breast).

Table 1. The prevalence of Salmonella Enteritidis isolates showing resistance to cefepime in Shanghai, China from 2005 to 2014.
Antimicrobial Susceptibility Profiles
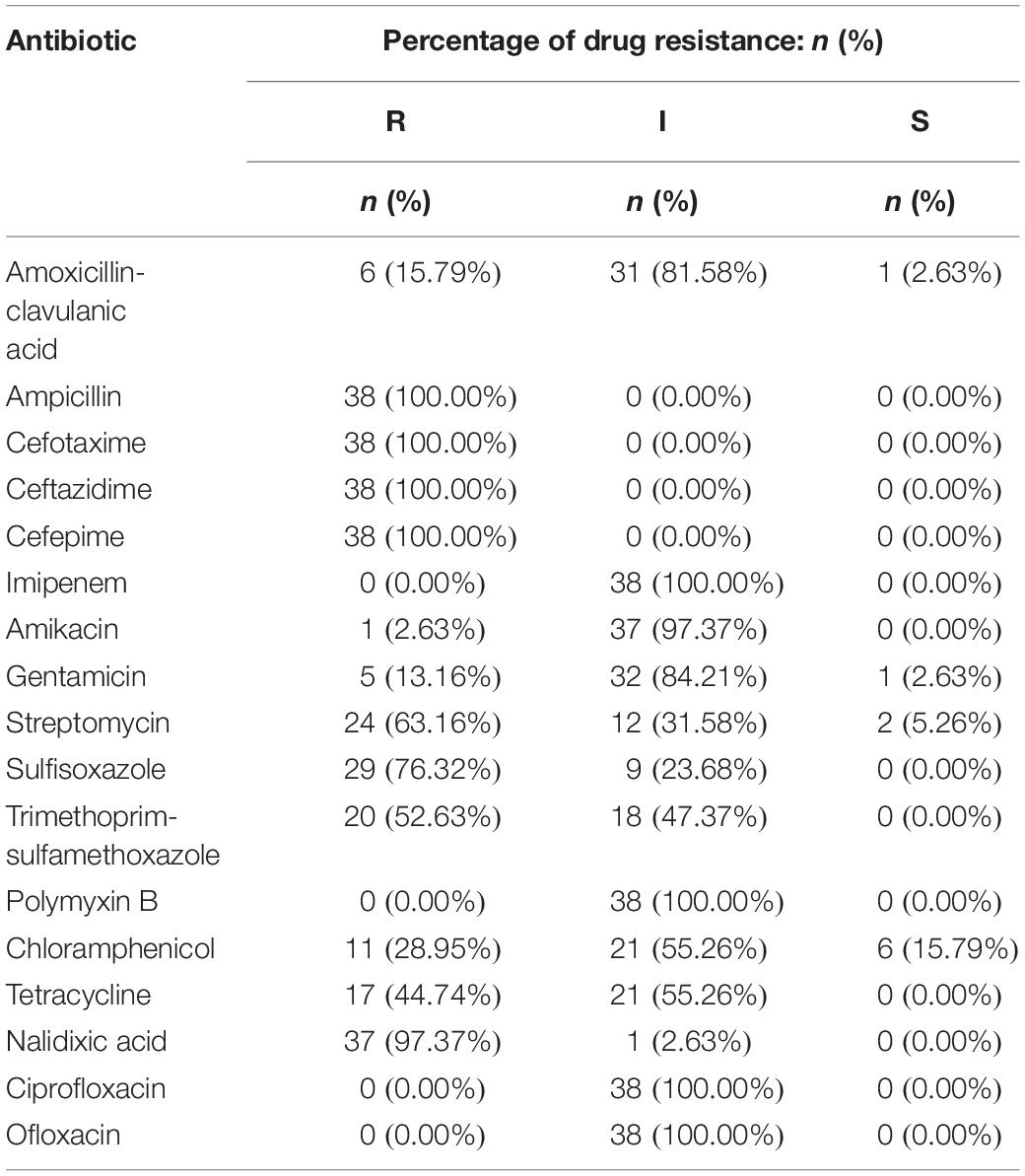
The 38 isolates with cefepime resistance were simultaneously resistant to ceftazidime (100%), cefotaxime (100%) and ampicillin (100%) and also showed high resistance to nalidixic acid (97.37%), sulfafurazole (76.32%), streptomycin (63.16%) and trimethoprim-sulfamethoxazole (52.63%), followed by tetracycline (44.74%) and chloramphenicol (28.95%). However, the CRSE isolates were more susceptible to amoxicillin-clavulanic acid (15.79%), gentamicin (13.16%), amikacin (13.16%). They all showed intermediate susceptibility to ciprofloxacin, ofloxacin, polymyxin B and imipenem (Table 2 and Supplementary Table S2).
Isolates resistant to cefepime were 100% multidrug-resistant (MDR), with resistance to a minimum of five antibiotics and a maximum of 10 antibiotics and 6 isolates were resistant to ten different antibiotics. The spectrum of resistance types reached 19 with the following three combinations detected in higher proportions, “AMP-CTX-CAZ-FEP-STR-SIZ-SXT-CHL-TET-NAL” (13.16%, 5/38), “AMP-CTX-CAZ-FEP-STR-SIZ-TET-NAL” (13.16%, 5/38), and “AMP-CTX-CAZ-FEP-GEN”(13.16%, 5/38) (Supplementary Table S3).
Prevalence of Beta-Lactamase Genes
The beta-lactamase-encoding genes blaCTX–M (100.00%), blaSHV (65.79%), blaTEM (52.63%), blaACC (5.26%) and blaPSE (5.26%) were identified. Coexistence of two or more genes in a single CRSE was very common. Sequencing of the blaCTX–M genes revealed presence of various types: blaCTX–M–55 (84.21%, 32/38), blaCTX–M–64 (5.26%), blaCTX–M–123 (5.26%), blaCTX–M–3 (2.63%), and blaCTX–M–15 (2.63%).
Pulse Field Gel Electrophoresis (PFGE)
A total of 18 different PFGE patterns were obtained from the 38 CRSE isolates (Figure 1). The similarity ranged from 65.44 to 100%. PFGE genotypes of >85% similarity were grouped into 2 clusters and another two individual isolates. The PFGE genotype of one foodborne isolate with CTX-M-55 (SH14SF008) was identical to four CTX-M-55 producing isolates from clinical samples (SH13G1882, SH14G065, SH14G169, SH14G548). However, several isolates from different years and hospitals also shared the same PFGE genotype (Figure 1).
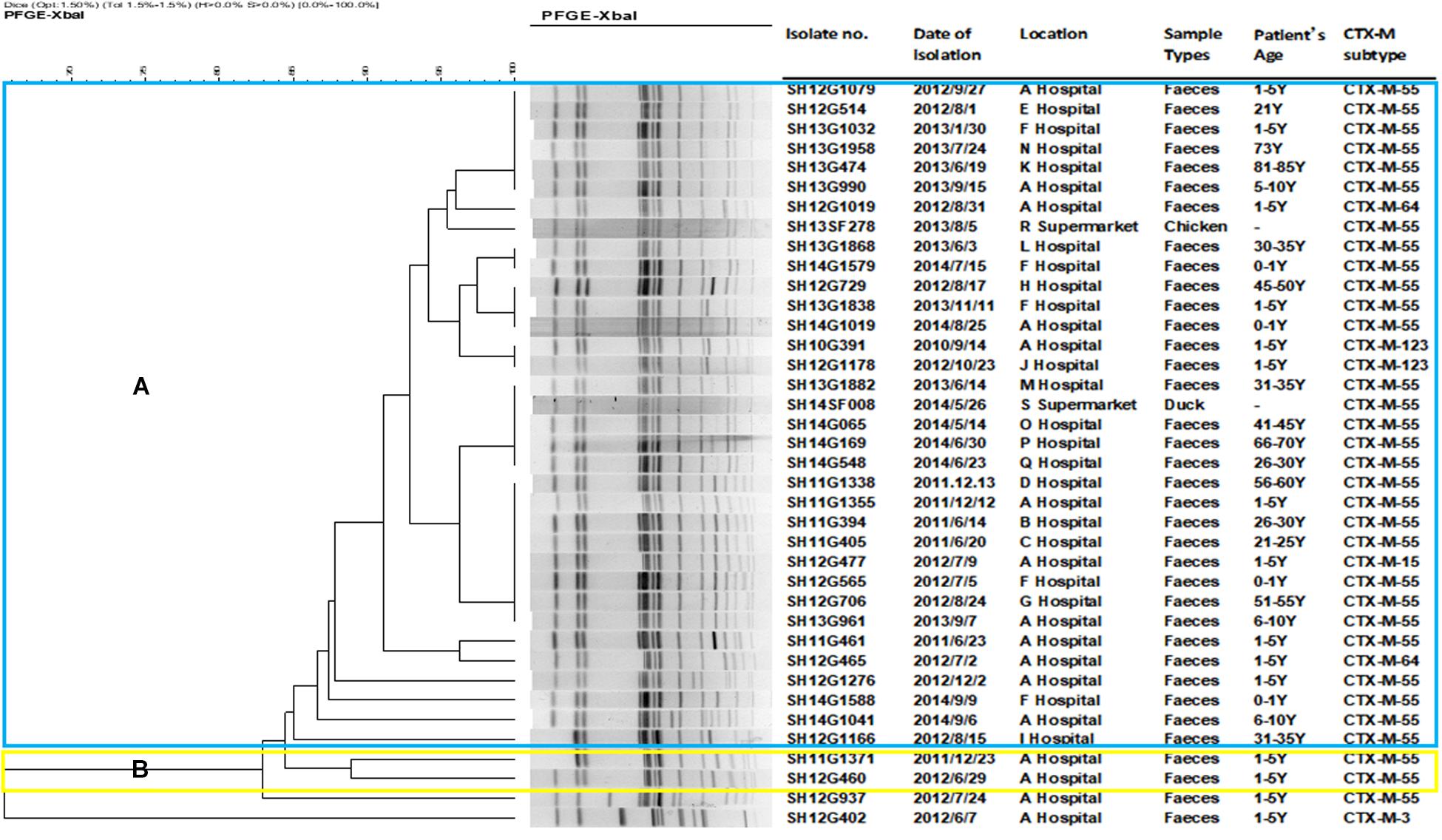
Figure 1. Dendrogram of pulsed-field gel electrophoresis (PFGE) profiles of 38 cefepime-resistant Salmonella Enteritidis isolates. The blue color represents cluster (A), which includes 34 isolates; the yellow color represents cluster (B), which including 2 isolates; another 2 isolates were not assigned to clusters (A,B).
Plasmid Characterization
In total, 35 transconjugants were successfully obtained from 38 CRSE isolates through the conjugation experiments; the transfer efficiency was 92.11% (35/38). Transfer of the blaCTX–M gene alone was detected in 33 of 35 transconjugants; two transconjugants harbored combinations (blaCTX–M–3 + blaTEM; blaCTX–M–55 + blaSHV) (Supplementary Figures S1–S3). blaCTX–M–55 gene could be found on 76.8 kb IncI1 (n = 9), 76.8 kb IncFrepB (n = 5), 104.5 kb IncFrepB (n = 1), 138.9 kb IncFrepB (n = 1), 138.9 kb IncFrepB/N (n = 1), 104.5 kb IncFrepB/FIIA (n = 1), 76.8 kb IncFIIA/FIIB/FrepB (n = 3), 104.5 kb IncFIIA/FIIB/FrepB (n = 4), 104.5∼138.9 kb IncFIIA/FIIB/FrepB (n = 1), 138.9 kb IncFIIA/FIIB/FrepB (n = 1) and >138.9 kb IncN/FIIB/FrepB (n = 1) plasmids; while blaCTX–M–3 and blaTEM could be observed on 76.8 kb IncL/M plasmid (n = 1); blaCTX–M–55 and blaSHV could be observed on 104.5 kb IncFrepB/FIIA plasmid (n = 1). Comparing to the recipient strain E. coli C600, horizontal transfer of blaCTX–M genes resulted in an increase of MICs for cefotaxime (from 128 to 2,048 mg/L), ceftazidime (from 4 to 64 mg/L) and cefepime (from 16 to 64 mg/L) (Table 3). The simultaneous presence of two or three plasmid replicons in one blaCTX–M–55-carrying plasmid was observed in this study. Southern blot hybridization suggested that the size of the blaCTX–M gene carrying plasmids varied between 76.8 and 138.9 kb (Supplementary Figures S1–S3). In genetic context detection tests, the insertion sequence ISEcp1 or IS903 appeared frequently upstream and downstream of blaCTX–M, functioning as a mobile element in the process of horizontal transmission (Yi et al., 2010). Totally, 45.7% (16/35) transconjugants harbored “ISEcp1-F-blaCTX–M–55-ORF477” element, which had been reported in Shigella sonnei strain before (Qu et al., 2014). The characteristics of the transconjugant plasmids are shown in Table 3.
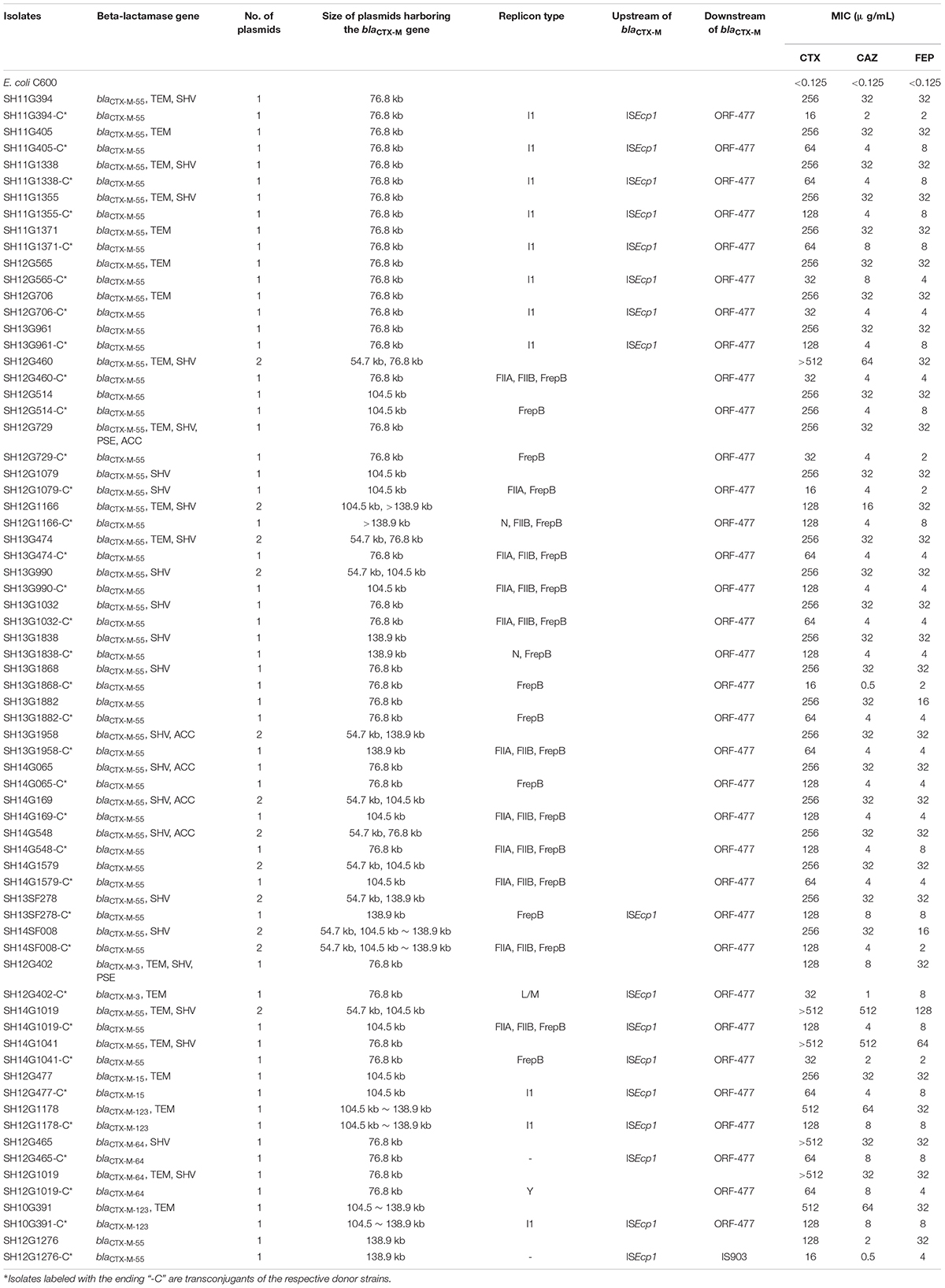
Table 3. Characteristics of cefepime-resistant Salmonella Enteritidis donor strains and transconjugants.
Sequence Analysis of a blaCTX–M–55 Carrying Plasmid
Plasmid sequencing and assembly revealed that the plasmid transferred from the blaCTX–M–55 positive S. Enteritidis isolate SH12G706 to E. coli C600 (designated 44C) was 85,897 bp in length with an average G+C content of 49.67% and harbored a total of 95 ORFs annotated to genes associated with plasmid replication (repA), IncI1 conjugation transfer (traA-Y, trbA-C, pilA-X), plasmid SOS inhibition (psiA, psiB), intracellular multiplication (icmB, icmO, icmP, dotL, dotO), plasmid segregation (parA, parM, soj) and plasmid stability (stbA, stbB) (Figure 2). When compared with a range of plasmids that harbored blaCTX–M–55 gene, the flanking region of blaCTX–M–55 on plasmid 44C showed high similarity with plasmid p628-CTX-M (KP987217, K. pneumoniae, china, 2010) that had a 2,980 bp ISEcp1-related element that functioned as a mobile unit (Figure 3).” Both plasmids showed the linear structure “repZ-yafA-yafB -tnpA-blaCTX–M–55-ΔyagA.”
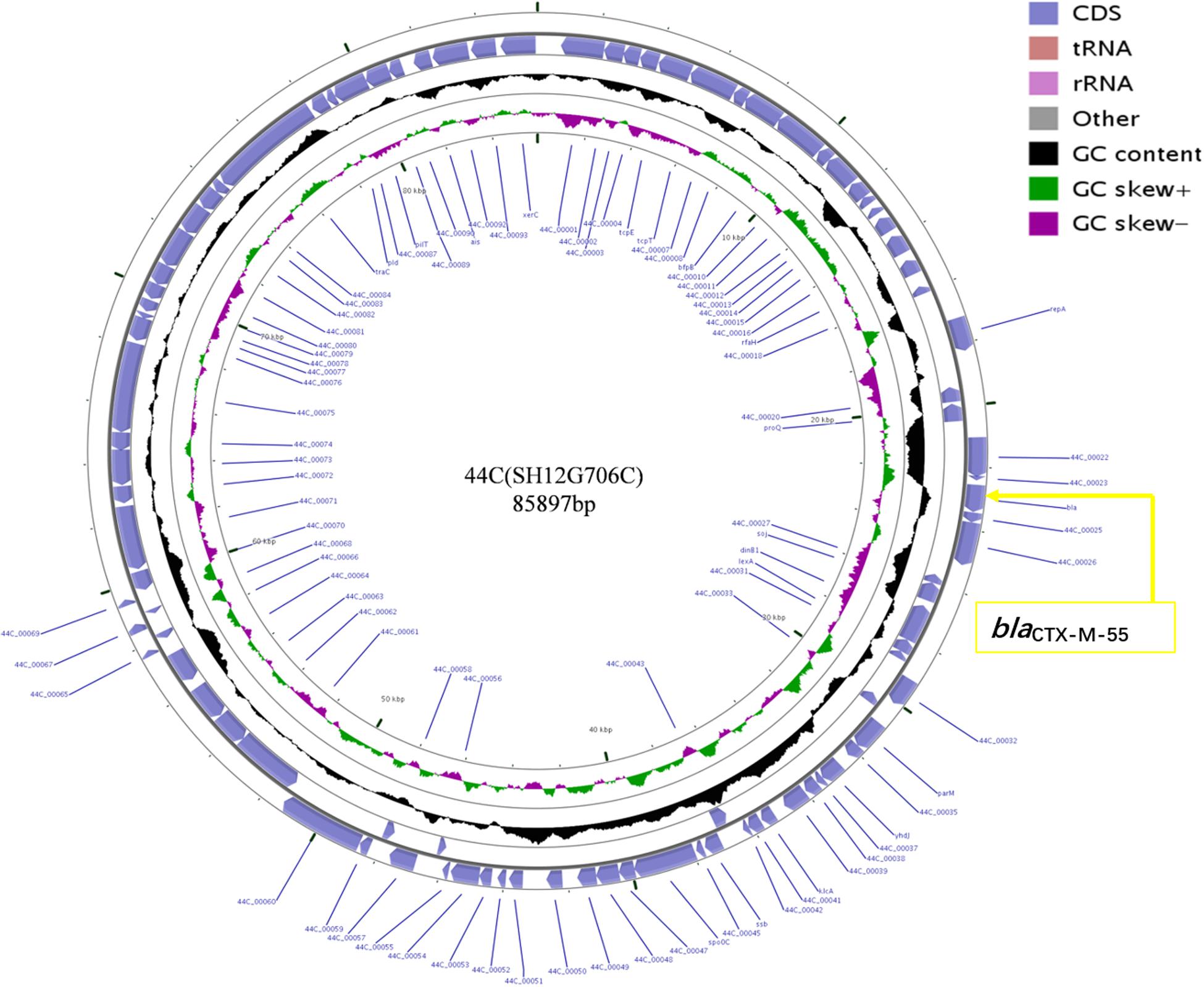
Figure 2. Circular map of plasmid 44C isolated from a transconjugant (donor strain S. Enteritidis no. SH12G706). The blue arrows represent open reading frames and their direction of transcription. The yellow box represents the location of gene blaCTX–M–55.
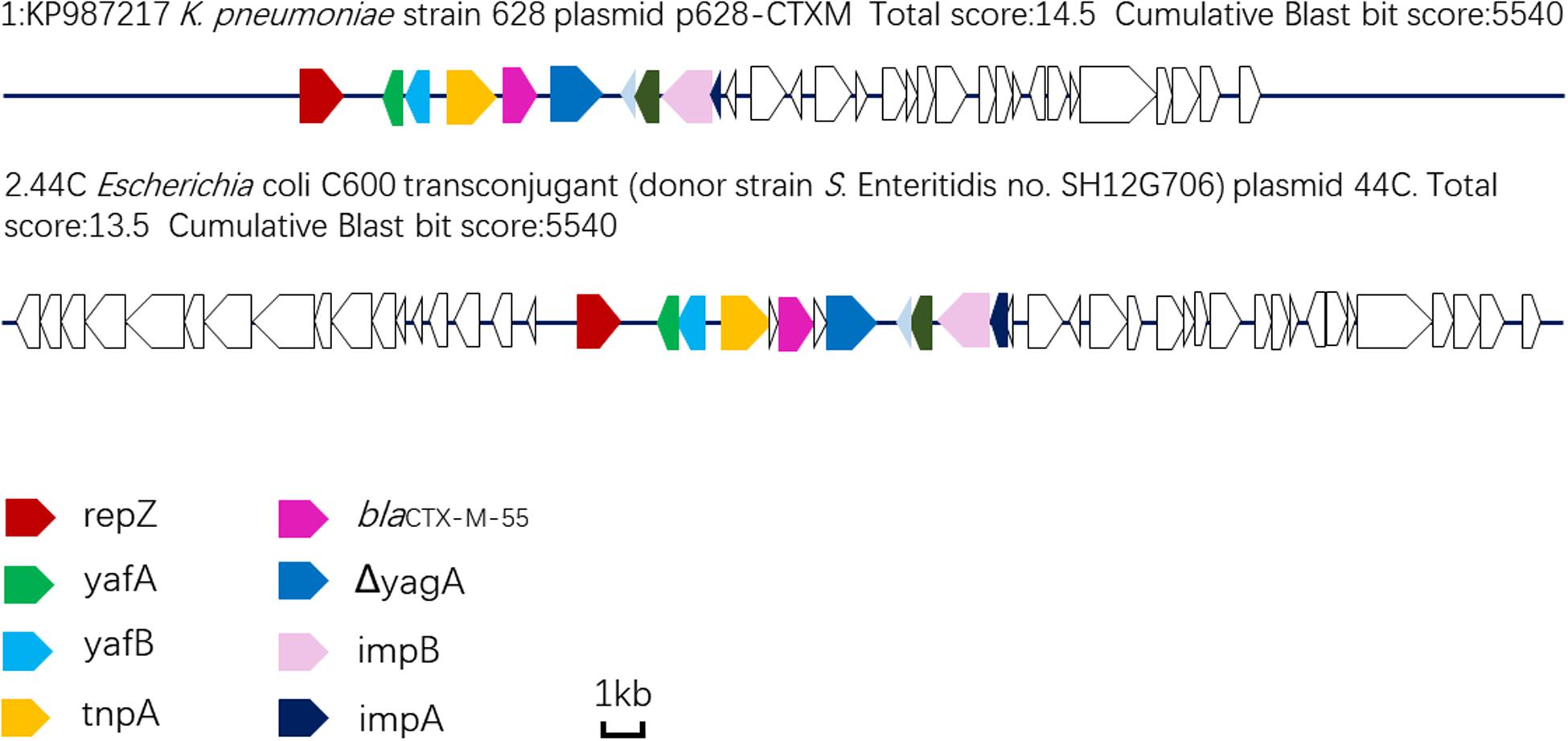
Figure 3. Linear comparison of DNA sequences of plasmid 44C isolated from a transconjugant (donor strain S. Enteritidis no. SH12G706) and plasmid p628-CTX-M from a K. pneumoniae isolate china, 2010.
Discussion
In the investigation period from 2005 to 2014 in Shanghai, China, S. Enteritidis strains showed only a slight variation in resistance to commonly used drugs, including third generation cephalosporins (Supplementary Table S4). This finding is in contrast to previous reports that other Salmonella serotypes (e.g., Typhimurium), exhibited an increasing tendency of resistance to at least 10 types of antibiotics during the same period (Ni et al., 2013). However, it is noteworthy that CRSE strains emerged in 2010 and their frequency is rising annually.
In recent years, cefepime ranked the leading anti-infective medicine in many hospitals in China. In order to prevent the spread of antibiotic resistance, cefepime is rarely used for animals or in breeding industry. However, our study shows that cefepime-resistant S. Enteritidis has also been isolated from animal-derived foodborne sources since 2013.
In this study, we analyzed 38 CRSE isolates, all of which exhibited MDR and the frequency of isolates resistant to more than five antibiotics was very high (100%). The emergence of severe drug-resistant status in China is largely related to the abuse of antibiotics as feed additives in veterinary clinical practice and the transfer of movable components carrying drug-resistant genes between isolates, which has become a common problem in both human and veterinary clinical practice (Rahmani et al., 2013). However, a relatively low resistance to amoxicillin-clavulanic acid (15.79%), gentamicin (13.16%), and amikacin (13.16%) was detected in the 38 cefepime-resistant S. Enteritidis isolates. They all showed intermediate susceptibility to ciprofloxacin, ofloxacin, polymyxin B and imipenem; therefore, these drugs should be used with caution. The fact also emphasizes the importance of standardization of clinical drug application and to prolong the effectiveness of new drugs. In addition, although the isolates were sensitive to aminoglycosides and fluoroquinolones, these agents should be used with caution due to the ototoxicity and adverse effects of cartilage on children.
In this study, nine kinds of β-lactamase resistance genes were detected, among them, subtype blaCTX–M–55 was predominant. Based on amino acid sequence differences, CTX-M-type ESBLs can be divided into more than five groups and hundreds of variants. CTX-M-55 belongs to the CTX-M-1 group, which was first detected in E. coli in Thailand in 2005 (Kiratisin et al., 2007) and first detected in Salmonella in 2011 (Yu et al., 2011). CTX-M-55 has only one amino acid site difference (valine to alanine) compared with CTX-M-15 (Kiratisin et al., 2007), and exhibits high hydrolytic activity to cefotaxime and ceftazidime (Dallenne et al., 2010). The prevalence of CTX-M-55-producing bacteria in China has grown significantly in recent years in both animal and human populations (Bevan et al., 2017). Data showed that CTX-M-55 is widely distributed in foodborne E. coli in Asia regions (Zhang et al., 2014b), which was the second most common CTX-M genotype (26.1%, 29/111) following CTX-M-14 (Zheng et al., 2012). Regarding hospital-associated Enterobacteriaceae infections, CTX-M-55 was found to be more common than CTX-M-15 in China (Xia et al., 2014). Also, CTX-M-55 was the most prevalent ESBL type observed in Salmonella isolates from livestock animals in China (Zhang et al., 2019). It is known that CTX-M-55 exists in bacteria that show resistance to second or third generation cephalosporins, and in this study, we found that CTX-M-55-producing S. Enteritidis was also with resistance to fourth-generation cephalosporin, cefepime. Therefore, CTX-M-55 should be considered as an important surveillance target around the world.
The blaCTX–M genes of 35 of the 38 isolates were transferred to the recipient by conjugation. Under the influence of blaCTX–M, the transconjugants showed resistance to cephalosporins but the MICs varied. They did not reach the same MIC values as the donor bacterium, a difference that may be related to the complex mechanisms of antibiotic resistance such as efflux pumps and penicillin-binding protein in the donor bacterium.
In this study, transconjugants carrying blaCTX–M shared different types of replicons, mainly IncI1 and InFrepB, with sizes ranging from 76.8 to 138.9 kb. The genetic context of blaCTX–M was mainly “ISEcp1-F-blaCTX–M–55-ORF477.” ISEcp1, like IS26 and ISCR1, is a member of the IS1380 family. This component shares a similar structure with a promoter, which positively regulates the high-level expression of blaCTX–M and facilitates its horizontal transmission among various species (Dhanji et al., 2011; Singh et al., 2018).
PFGE analysis showed that most CRSE displayed similar PFGE patterns (>82.90%). Notably, the pattern of one foodborne isolate (SH14SF008) was identical to that of another four isolates collected from clinical diarrhea human samples and all had the same blaCTX–M–55 gene, which suggested that we should pay attention to food consumption. Furthermore, S. Enteritidis isolates from different years showed 100% homology, indicating possible long-term outbreaks caused by clonal transfer of S. Enteritidis strains within the hospital or recurring introduction and spread of Salmonella in the hospital by colonized humans or contaminated food (Blahova et al., 1998). CRSE long term presence in animal breeding and especially fattening lots has been described (Ramchandani et al., 2005; Parveen et al., 2007). On the other hand, the limited discriminatory power of PFGE for several Salmonella serovars is known; this might falsely indicate clonal transfer. Therefore, WGS-based phylogenetic analyses are needed in future to assess the genetic relationship of the CRSE isolates in more detail.
WGS analysis of the plasmid 44C, which was from CRSE isolate of a diarrhea patient, showed that it had genes associated with plasmid replication, conjugation transfer, inhibition, intracellular multiplication, segregation and stability, functional genes that confer the ability to transfer blaCTX–M–55 and integrate into the genome of other bacteria. Moreover, compared with other blaCTX–M–55-harboring plasmids, we found that plasmid 44C showed high sequence similarity with plasmid p628-CTX-M isolated from a K. pneumoniae strain in October 2010 (Wang et al., 2015). In particular, both plasmids shared an ISEcp1-related component in the genetic context around blaCTX–M–55, indicating that such structures may transfer from K. pneumoniae to Salmonella. Thus, it is crucial to implement precautionary measures to prevent the spread of bacteria with these mobile resistance genes in both clinical settings and breeding industry by strengthening the detection and analysis of this mobile genetic element.
Conclusion
In conclusion, increase of cefepime resistance in S. Enteritidis is a serious public health concern. Our data demonstrate that the blaCTX–M–55 gene mobilized by the ISEcp1-blaCTX–M–55-ORF477 transposition unit is the main feature shared by CRSE and plays an important role in the transmission among different species.
Data Availability Statement
The data sets generated for this study can be found in the NCBI Sequence Read Archive (SRA) under accession number SRR9734360 (https://www.ncbi.nlm.nih.gov/search/all/?term=SRR9734360).
Ethics Statements
Ethical approval for this study was provided by Shanghai Municipal Center for Disease Control and Prevention (Shanghai, China). This study is a retrospective study and individual patient identification is not accessed and informed consent is not required.
Author Contributions
YF, JZ, and ML conceived and designed the experiment. YF and YM carried out the experiment. YF, LZ, ZX, YM, YW, ZC, and JB contributed to sample preparation and collected the data. YF, LZ, ZX, and YM performed the analysis. YF wrote the manuscript. JZ and ML revised the manuscript. XX provided the samples. ML and JZ fund the project. All authors discussed the results and commented on the manuscript.
Funding
This work was supported by the National Key R&D Program of China (2017YFC1600101, 2018YFD0500500); National Natural Science Foundation of China (31972762); Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2018); Pearl River S&T Nova Program of Guangzhou (201806010183); Province Science and Technology of Guangdong Research Project (2017A020208055); Walmart Foundation (SA1703162); Guangdong Key S&T Program (Grant no. 2019B020217002) from Department of Science and Technology of Guangdong Province; National Broiler Industry Technology System Project (cARS-41-G16).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00910/full#supplementary-material
References
Bevan, E. R., Jones, A. M., and Hawkey, P. M. (2017). Global epidemiology of CTX-M beta-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Blahova, J., Lesicka-Hupkova, M., Kralikova, K., Krcmery, V. Sr., and Krcmeryova, T. (1998). Further occurrence of extended-spectrum beta-lactamase-producing Salmonella enteritidis. J. Chemother. 10, 291–294. doi: 10.1179/joc.1998.10.4.291
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Chapman, T. M., and Perry, C. M. (2003). Cefepime: a review of its use in the management of hospitalized patients with pneumonia. Am. J. Respir. Med. 2, 75–107. doi: 10.1007/bf03256641
Dallenne, C., Da Costa, A., Decre, D., Favier, C., and Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495. doi: 10.1093/jac/dkp498
Dhanji, H., Doumith, M., Hope, R., Livermore, D. M., and Woodford, N. (2011). ISEcp1-mediated transposition of linked blaCTX-M-3 and blaTEM-1b from the IncI1 plasmid pEK204 found in clinical isolates of Escherichia coli from Belfast. U. K. J. Antimicrob. Chemother. 66, 2263–2265. doi: 10.1093/jac/dkr310
Galanis, E., Lo Fo Wong, D. M., Patrick, M. E., Binsztein, N., Cieslik, A., Chalermchikit, T., et al. (2006). Web-based surveillance and global Salmonella distribution, 2000-2002. Emerg. Infect. Dis. 12, 381–388. doi: 10.3201/eid1205.050854
Issenhuth-Jeanjean, S., Roggentin, P., Mikoleit, M., Guibourdenche, M., de Pinna, E., Nair, S., et al. (2014). Supplement 2008-2010 (no. 48) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 165, 526–530. doi: 10.1016/j.resmic.2014.07.004
Ke, B., Sun, J., He, D., Li, X., Liang, Z., and Ke, C. W. (2014). Serovar distribution, antimicrobial resistance profiles, and PFGE typing of Salmonella enterica strains isolated from 2007-2012 in Guangdong, China. BMC Infect. Dis. 14:338. doi: 10.1186/1471-2334-14-338
Kiratisin, P., Apisarnthanarak, A., Laesripa, C., and Saifon, P. (2008). Molecular characterization and epidemiology of extended-spectrum-beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates causing health care-associated infection in Thailand, where the CTX-M family is endemic. Antimicrob. Agents Chemother. 52, 2818–2824. doi: 10.1128/AAC.00171-08
Kiratisin, P., Apisarnthanarak, A., Saifon, P., Laesripa, C., Kitphati, R., and Mundy, L. M. (2007). The emergence of a novel ceftazidime-resistant CTX-M extended-spectrum beta-lactamase, CTX-M-55, in both community-onset and hospital-acquired infections in Thailand. Diagn. Microbiol. Infect. Dis. 58, 349–355. doi: 10.1016/j.diagmicrobio.2007.02.005
Labombardi, V. J., Rojtman, A., and Tran, K. (2006). Use of cefepime for the treatment of infections caused by extended spectrum beta-lactamase-producing Klebsiella pneumoniae and Escherichia coli. Diagn. Microbiol. Infect. Dis. 56, 313–315. doi: 10.1016/j.diagmicrobio.2006.03.019
Matheson, N., Kingsley, R. A., Sturgess, K., Aliyu, S. H., Wain, J., Dougan, G., et al. (2010). Ten years experience of Salmonella infections in Cambridge, UK. J. Infect. 60, 21–25. doi: 10.1016/j.jinf.2009.09.016
Ni, J., Xu, H., Hu, X., Zhong, H.-M., Zhang, J., Jin, H.-M., et al. (2013). Laboratory network based surveillance of drug resistance of Salmonella enteritidis in Shanghai, 2006-2012. Res. Microbiol. 28, 369–375.
Pan, Y. S., Liu, J. H., Hu, H., Zhao, J. F., Yuan, L., Wu, H., et al. (2013). Novel arrangement of the blaCTX-M-55 gene in an Escherichia coli isolate coproducing 16S rRNA methylase. J. Basic Microbiol. 53, 928–933. doi: 10.1002/jobm.201200318
Parveen, S., Taabodi, M., Schwarz, J. G., Oscar, T. P., Harter-Dennis, J., and White, D. G. (2007). Prevalence and antimicrobial resistance of Salmonella recovered from processed poultry. J. Food Prot. 70, 2466–2472. doi: 10.4315/0362-028x-70.11.2466
Patel, H. B., Lusk, K. A., and Cota, J. M. (2019). The role of cefepime in the treatment of extended-spectrum beta-lactamase infections. J. Pharm. Pract. 32, 458–463. doi: 10.1177/0897190017743134
Qu, F., Ying, Z., Zhang, C., Chen, Z., Chen, S., Cui, E., et al. (2014). Plasmid-encoding extended-spectrum beta-lactamase CTX-M-55 in a clinical Shigella sonnei strain, China. Future Microbiol. 9, 1143–1150. doi: 10.2217/fmb.14.53
Rahmani, M., Peighambari, S. M., Svendsen, C. A., Cavaco, L. M., Agerso, Y., and Hendriksen, R. S. (2013). Molecular clonality and antimicrobial resistance in Salmonella enterica serovars enteritidis and infantis from broilers in three Northern regions of Iran. BMC Vet. Res. 9:66. doi: 10.1186/1746-6148-9-66
Ramchandani, M., Manges, A. R., DebRoy, C., Smith, S. P., Johnson, J. R., and Riley, L. W. (2005). Possible animal origin of human-associated, multidrug-resistant, uropathogenic Escherichia coli. Clin. Infect. Dis. 40, 251–257. doi: 10.1086/426819
Ren, X., Li, M., Xu, C., Cui, K., Feng, Z., Fu, Y., et al. (2016). Prevalence and molecular characterization of Salmonella enterica isolates throughout an integrated broiler supply chain in China. Epidemiol. Infect. 144, 2989–2999. doi: 10.1017/S0950268816001515
Ribot, E. M., Fair, M. A., Gautom, R., Cameron, D. N., Hunter, S. B., Swaminathan, B., et al. (2006). Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3, 59–67. doi: 10.1089/fpd.2006.3.59
Singh, N. S., Singhal, N., and Virdi, J. S. (2018). Genetic environment of blaTEM-1, blaCTX-M-15, blaCMY-42 and characterization of integrons of Escherichia coli isolated from an indian urban aquatic environment. Front. Microbiol. 9:382. doi: 10.3389/fmicb.2018.00382
Tabu, C., Breiman, R. F., Ochieng, B., Aura, B., Cosmas, L., Audi, A., et al. (2012). Differing burden and epidemiology of non-Typhi Salmonella bacteremia in rural and urban Kenya, 2006-2009. PLoS One 7:e31237. doi: 10.1371/journal.pone.0031237
Tamma, P. D., and Rodriguez-Bano, J. (2017). The use of noncarbapenem beta-lactams for the treatment of extended-spectrum beta-lactamase infections. Clin. Infect. Dis. 64, 972–980. doi: 10.1093/cid/cix034
Tenover, F. C. (2006). Mechanisms of antimicrobial resistance in bacteria. Am. J. Med. 119(6 Suppl. 1), S3–S10. doi: 10.1016/j.amjmed.2006.03.011
Usha, G., Chunderika, M., Prashini, M., Willem, S. A., and Yusuf, E. S. (2008). Characterization of extended-spectrum beta-lactamases in Salmonella spp. at a tertiary hospital in Durban, South Africa. Diagn. Microbiol. Infect. Dis. 62, 86–91. doi: 10.1016/j.diagmicrobio.2008.04.014
Wang, L., Fang, H., Feng, J., Yin, Z., Xie, X., Zhu, X., et al. (2015). Complete sequences of KPC-2-encoding plasmid p628-KPC and CTX-M-55-encoding p628-CTXM coexisted in Klebsiella pneumoniae. Front. Microbiol. 6:838. doi: 10.3389/fmicb.2015.00838
Wang, R., Cosgrove, S. E., Tschudin-Sutter, S., Han, J. H., Turnbull, A. E., Hsu, A. J., et al. (2016). Cefepime therapy for cefepime-susceptible extended-spectrum beta-lactamase-producing Enterobacteriaceae Bacteremia. Open Forum Infect. Dis. 3:ofw132. doi: 10.1093/ofid/ofw132
Xia, S., Fan, X., Huang, Z., Xia, L., Xiao, M., Chen, R., et al. (2014). Dominance of CTX-M-type extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from patients with community-onset and hospital-onset infection in China. PLoS One 9:e100707. doi: 10.1371/journal.pone.0100707
Yahav, D., Paul, M., Fraser, A., Sarid, N., and Leibovici, L. (2007). Efficacy and safety of cefepime: a systematic review and meta-analysis. Lancet Infect. Dis. 7, 338–348. doi: 10.1016/S1473-3099(07)70109-3
Yang, B., Qu, D., Zhang, X., Shen, J., Cui, S., Shi, Y., et al. (2010). Prevalence and characterization of Salmonella serovars in retail meats of marketplace in Shaanxi, China. Int. J. Food Microbiol. 141, 63–72. doi: 10.1016/j.ijfoodmicro.2010.04.015
Yang, J., Chen, Y., Jia, X., Luo, Y., Song, Q., Zhao, W., et al. (2012). Dissemination and characterization of NDM-1-producing Acinetobacter pittii in an intensive care unit in China. Clin. Microbiol. Infect. 18, E506–E513. doi: 10.1111/1469-0691.12035
Yi, H., Xi, Y., Liu, J., Wang, J., Wu, J., Xu, T., et al. (2010). Sequence analysis of pKF3-70 in Klebsiella pneumoniae: probable origin from R100-like plasmid of Escherichia coli. PLoS One 5:e8601. doi: 10.1371/journal.pone.0008601
Yu, F., Chen, Q., Yu, X., Li, Q., Ding, B., Yang, L., et al. (2011). High prevalence of extended-spectrum beta lactamases among Salmonella enterica Typhimurium isolates from pediatric patients with diarrhea in China. PLoS One 6:e16801. doi: 10.1371/journal.pone.0016801
Zhang, C. Z., Ding, X. M., Lin, X. L., Sun, R. Y., Lu, Y. W., Cai, R. M., et al. (2019). The emergence of chromosomally located bla CTX-M-55 in Salmonella from foodborne animals in China. Front. Microbiol. 10:1268. doi: 10.3389/fmicb.2019.01268
Zhang, J., Jin, H., Hu, J., Yuan, Z., Shi, W., Ran, L., et al. (2014a). Serovars and antimicrobial resistance of non-typhoidal Salmonella from human patients in Shanghai, China, 2006-2010. Epidemiol. Infect. 142, 826–832. doi: 10.1017/S0950268813001659
Zhang, J., Zheng, B., Zhao, L., Wei, Z., Ji, J., Li, L., et al. (2014b). Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect. Dis. 14:659. doi: 10.1186/s12879-014-0659-0
Keywords: antimicrobial susceptibility, cefepime-resistant Salmonella Enteritidis, extended-spectrum β-lactamase genes, pulsed-field gel electrophoresis, transconjugants
Citation: Fu Y, Xu X, Zhang L, Xiong Z, Ma Y, Wei Y, Chen Z, Bai J, Liao M and Zhang J (2020) Fourth Generation Cephalosporin Resistance Among Salmonella enterica Serovar Enteritidis Isolates in Shanghai, China Conferred by blaCTX–M–55 Harboring Plasmids. Front. Microbiol. 11:910. doi: 10.3389/fmicb.2020.00910
Received: 06 September 2019; Accepted: 17 April 2020;
Published: 15 May 2020.
Edited by:
Kristina Kadlec, Independent Researcher, Wunstorf, GermanyReviewed by:
Yvonne Pfeifer, Robert Koch Institute, GermanyJessica C. Chen, Centers for Disease Control and Prevention, United States
Jason Patrick Folster, Centers for Disease Control and Prevention (CDC), United States
Copyright © 2020 Fu, Xu, Zhang, Xiong, Ma, Wei, Chen, Bai, Liao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Liao, bWxpYW9Ac2NhdS5lZHUuY24=; Jianmin Zhang, anVuZmVuZy12QDE2My5jb20=; am16aGFuZ0BzY2F1LmVkdS5jbg==
 Ying Fu
Ying Fu Xuebin Xu2
Xuebin Xu2 Lina Zhang
Lina Zhang Ming Liao
Ming Liao Jianmin Zhang
Jianmin Zhang