
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 29 May 2020
Sec. Food Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00905
This article is part of the Research TopicInsights of Fermented Foods and Beverages: Microbiology and Health-Promoting BenefitsView all 19 articles
Filamentous fungi are important organisms in traditionally prepared amylase and alcohol-producing dry starters in India. We collected 40 diverse types of amylase and alcohol-producing starters from eight states in North East India viz. marcha, thiat, humao, hamei, chowan, phut, dawdim, and khekhrii. The average fungal population was 4.9 × 105 cfu/g with an average of pH 5.3 and 10.7%, respectively. In the present study, 131 fungal isolates were isolated and characterized based on macroscopic and microscopic characteristics and were grouped into 44 representative fungal strains. Based on results of morphological characteristics and ITS gene sequencing, 44 fungal strains were grouped into three phyla represented by Ascomycota (48%), Mucoromycota (38%), and Basidiomycota (14%). Taxonomical keys to species level was illustrated on the basis of morphological characteristics and ITS gene sequencing, aligned to the fungal database of NCBI GenBank, which showed seven genera with 16 species represented by Mucor circinelloides (20%), Aspergillus sydowii (11%), Penicillium chrysogenum (11%), Bjerkandera adusta (11%), Penicillium citrinum (7%), Rhizopus oryzae (7%), Aspergillus niger (5%), Aspergillus flavus (5%), Mucor indicus (5%) Rhizopus microsporus (5%), Rhizopus delemar (2%), Aspergillus versicolor (2%), Penicillium oxalicum (2%), Penicillium polonicum (2%), Trametes hirsuta (2%), and Cladosporium parahalotolerans (2%). The highest Shannon diversity index H was recorded in marcha of Sikkim (H: 1.74) and the lowest in hamei of Manipur (H: 0.69). Fungal species present in these amylolytic starters are morphologically, ecologically and phylogenetically diverse and showed high diversity within the community.
Drinking alcoholic beverages has a cultural connotation in India from the Indus Valley Civilization dating back to 8,000 years (Sarkar et al., 2016), mostly through fermentation (Singh et al., 2010) and distillation (Achaya, 1991). Traditionally malting, brewing (such as beer), and vinification (fermentation of grapes into wine) processes are unknown in Indian food culture. Instead, traditional alcoholic beverages are prepared either by natural fermentation of plants or cereals, or by using traditionally prepared dry starters in India (Tamang, 2010). Some ethnic people in India traditionally prepare amylase and alcohol-producing starters to ferment alcoholic beverages for home consumption, which are known by different names in different languages spoken locally in regions such as marcha in Sikkim and Darjeeling hills, thiat in Meghalaya, humao in Assam, hamei in Manipur, chowan in Tripura, phut in Arunachal Pradesh, dawdim in Mizoramand khekhrii in Nagaland (Anupma et al., 2018), dhehli, balam, maler, treh, and bakhar of Himachal Pradesh and Uttarakhand (Thakur et al., 2015), and ranu dabai/goti of West Bengal, Odisha and Jharkhand (Ghosh et al., 2015). Traditional methods of the preparation of Indian starters are almost the same with some differences in use of starch-rich substrates such as rice or wheat or barley, and wrapping materials either in fern fronds or dry paddy-straw, or in fresh leaves of locally available wild plants (Shrivastava et al., 2012; Tamang et al., 2016). Soaked, dewatered, and ground cereal (rice/wheat/barley) flours are mixed with some wild plants, with a few spices such as sun-dried chilies or garlics and supplemented with 1–2% of previously prepared dry starters in powder forms (“back-slopping method” for sub-culturing the microbiota) to make thick doughs with addition of water. Thoroughly mixed dough mixtures are made into round or flat cakes of varying shapes and sizes, placed on fresh ferns or other plant leaves/dry paddy straws and allowed to ferment under semi-anaerobic conditions for 2–3 days at room temperature inside the room. After desirable fermentation, fermented doughs are then sun dried for 2–3 days to obtain dry starters which are exclusively used to ferment cereals into mild/strong alcoholic beverages (Tamang, 2010; Anupma et al., 2018). However, khekhrii, a dry starter from Nagaland in India is prepared by naturally fermenting sprouted-rice grains which are then dried in the sun to obtain dry starter granules to prepare an alcoholic beverage locally called zutho. Indian amylase and alcohol-producing starters are similar to starters from Asian countries such as daqu or chiu from China (Zheng et al., 2012), benh from Vietnam (Dung et al., 2007), nuruk from Korea (Jung et al., 2012), ragi from Indonesia (Roslan et al., 2018), bubod from the Philippines (Fronteras and Bullo, 2017), loogpang from Thailand (Daroonpunt et al., 2016) and dombea or medombae from Cambodia (Ly et al., 2018).
Several species of filamentous molds (Hesseltine et al., 1988; Yang et al., 2011; Lv et al., 2012a; Chen et al., 2014; Das et al., 2017); yeasts (Hesseltine and Kurtzman, 1990; Tamang and Sarkar, 1995; Tsuyoshi et al., 2005; Jeyaram et al., 2008, 2011; Thanh et al., 2008; Fronteras and Bullo, 2017; Sha et al., 2017, 2018, 2019), and bacteria (Hesseltine and Ray, 1988; Tamang et al., 2007; Sha et al., 2017; Roslan et al., 2018) are found to coexist in traditionally prepared dry starters as “micro-resources” which have been sub-cultured to preserve essential microbiota for alcohol production by Asian people for centuries (Tamang et al., 2020). Filamentous fungi present in traditional starters from Asia have several functionalities such as saccharification (Lee and Lee, 2002; Thapa and Tamang, 2004), liquefaction (Suesse et al., 2016), and ethanol production (Dung et al., 2007; Chen et al., 2014) to produce different types of low-alcoholic beverages and high-alcoholic distilled liquor. Filamentous molds are also responsible for the quality of alcoholic beverages including nutritional values and organoleptic properties such as flavor, taste, and color (Zhang et al., 2015; Tamang et al., 2016). Taxonomical identification of filamentous molds isolated from traditionally prepared dry starters from India have not been reported yet except from marcha (Tamang et al., 1988; Sha et al., 2017, 2019), thiat (Sha et al., 2017, 2019), amou, and perok-kushi (Das et al., 2017). Mucor circinelloides, Rhizopus chinensis, and Rhizopus stolonifer were reported earlier from marcha samples collected from Nepal, Darjeeling, and Sikkim (Tamang et al., 1988; Tamang and Sarkar, 1995; Thapa and Tamang, 2006; Sha et al., 2017, 2018), Amylomyces rouxii and Rhizopus oryzae from samples of amou, and perok-kushi, traditional starters of Assam (Das et al., 2017). Sha et al. (2017) reported fungal Phylum Ascomycota (98.6%) followed by Mucoromycota (1.4%), while in marcha samples only Phylum Ascomycota by high-through sequencing was reported. The present study aimed to identify the filamentous molds isolated from eight different types of traditionally prepared starters from North East India, viz. marcha, thiat, humao, hamei, chowan, phut, dawdim, and khekhriii, to species level by morphological and molecular identifications, and to profile their diversity within the fungal community.
A total of 40 samples of traditionally prepared dry starters viz marcha from Sikkim, thiat from Meghalaya, humao from Assam, hamei from Manipur, chowan from Tripura, phut from Arunachal Pradesh, dawdim from Mizoram, and khekhrii from Nagaland (Table 1) were collected directly from local markets and the homes of local producers in North East India (Figure 1) in pre-sterile containers. Dry starter samples were transported to the laboratory and stored in desiccators at room temperature as traditionally prepared dry starters have a shelf life of more than 1 year (Sha et al., 2018).
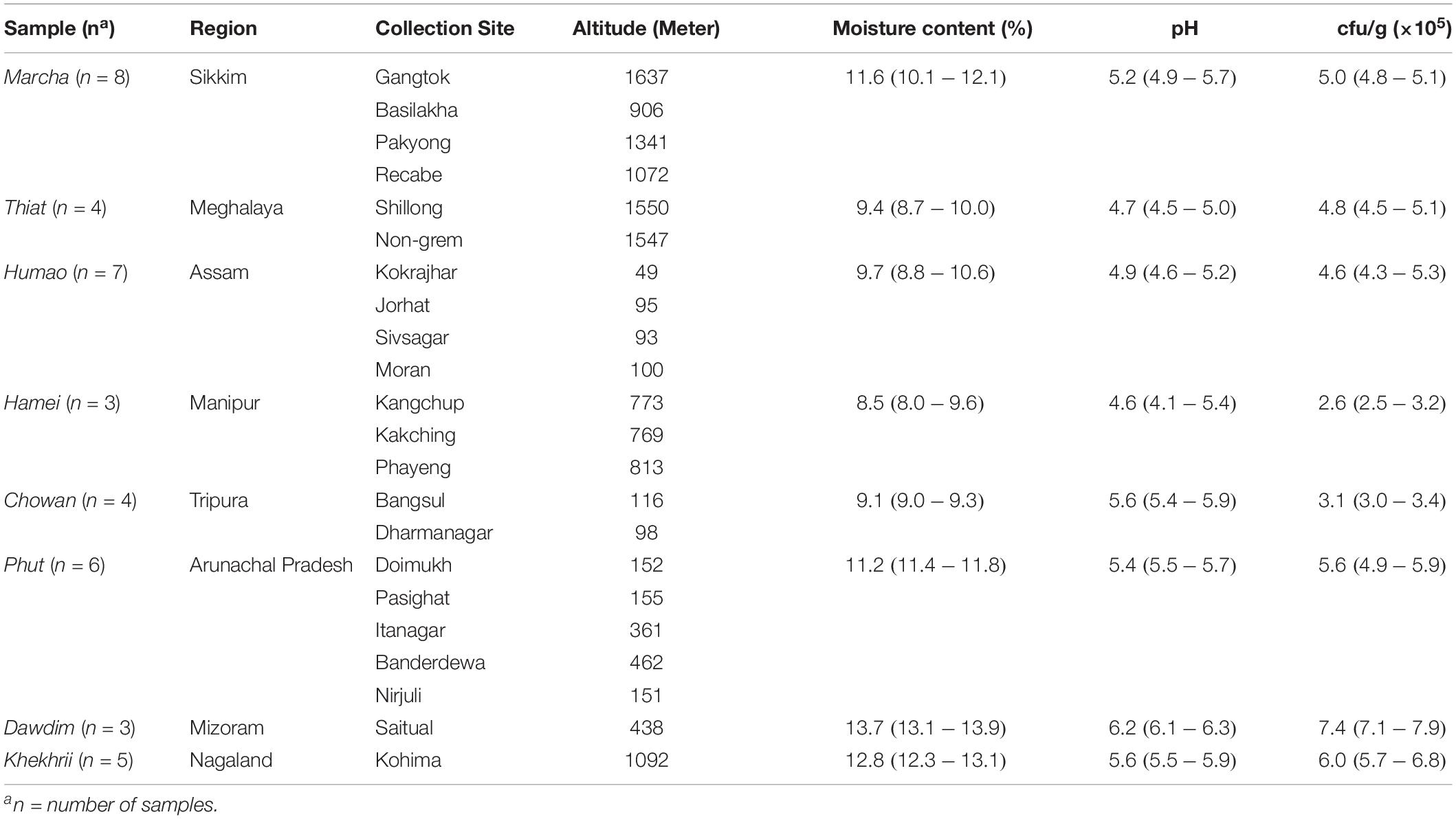
Table 1. Geographical locations, pH, moisture content, and fungal populations of dry starters from North East India.
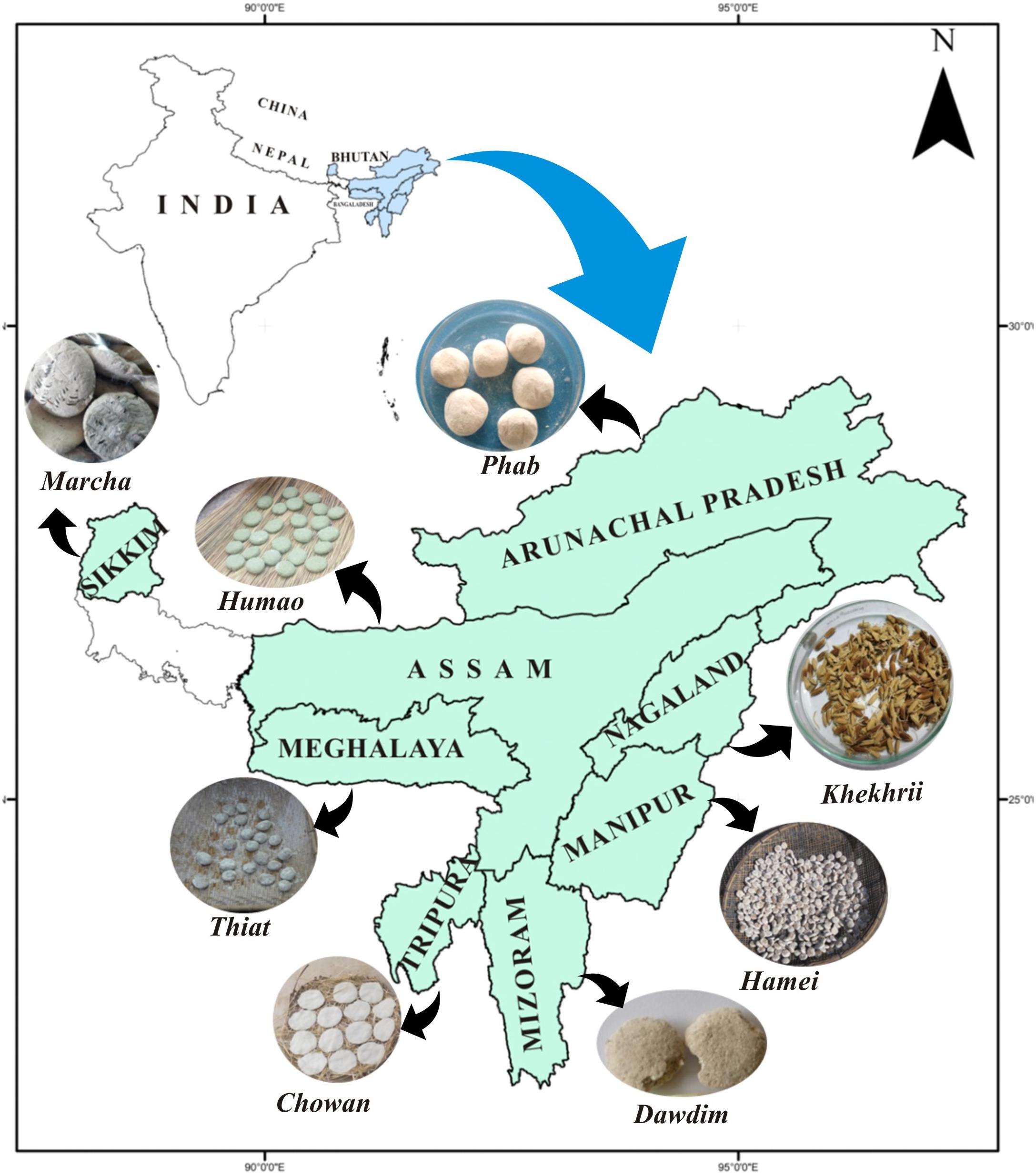
Figure 1. Location map of North East India showing collection sites of traditionally prepared dry starters.
The pH of homogenized samples was recorded by digital pH-meter (Orion 910003, Thermo Fisher Scientific, United States). The moisture content of the samples was estimated by a moisture analyzer (OHAUS/MB-45, United States).
Each dry sample starter was taken from the desiccator, then crushed coarsely by sterile spatula and 10 g of the crushed powered sample was homogenized with 90 mL of 0.85% physiological saline in a stomacher lab blender 40 (Seward, United Kingdom) for 2 min to obtain serial dilutions. One milliliter of each diluted sample (10–4, 10–5, 10–6, and 10–7) was poured onto malt extract agar (M137, HiMedia, Mumbai, India) and potato dextrose agar (M096, HiMedia, Mumbai, India) with an addition of antibiotics (1% streptomycin) to suppress the growth of bacteria, and plates were then incubated under 28°C and observed for the appearance of colonies for up to 1 week. The colonies that appeared on plates were counted as a colony forming unit (cfu/g) on the dry weight of starters. Colonies were selected on the basis of macroscopic and microscopic characteristics. Selected filamentous molds were sub-cultured on new plates and purified and stored on slants at 4°C for further studies.
For each isolate, one- or three-point inoculations on petri plates containing ~25 mL of media were applied. Fungal morphology was studied macroscopically by observing the colony features (surface color, reverse side color, shape, and diameter), and microscopically by observation of fruiting bodies using a stereomicroscope, and the vegetative and asexual stages were observed by a DE/Axio Imager A1 microscope (Carl Zeiss, Oberkochen, Germany) after staining freshly grown mycelia stained with cotton blue in MEA plates (Gaddeyya et al., 2012). Filamentous molds were identified on the basis of morphological features using the taxonomical keys described by Samson et al. (2004) and Pitt and Hocking (2009).
The genomic DNA was extracted from mold cultures following the methods of Umesha et al. (2016). Mycelial mass from the culture plate was scraped out by a sterile surgical blade and ground in a sterile mortar and pestle using 500 μL extraction buffer [100 mM Tris–HCl (pH 8.0), 20 mM EDTA (pH 8.0), 1.4 M NaCl, 2% CTAB, and 0.2% 2 mercaptoethanol]. The mixture was transferred to a fresh 1.5 mL tube with addition of 4-μL RNase, vortexed and incubated for 60 min at 37°C, and kept in a water bath for 60 min at 55°C. 500 μL phenol: chloroform: isoamyl alcohol (25:24:1) was added to the solution, mixed thoroughly for 5 min, and then centrifuged at 14,000 rpm for 10 min. The aqueous clear phase was recovered and mixed with chloroform: isoamyl alcohol (24:1), centrifuged at 12,000 rpm for 5 min, and the aqueous phase was recovered, adding 0.8 volume of cold 7.5 M ammonium acetate and 0.54 volume of ice-cold isopropanol, and finally mixed well and stored overnight for precipitation of DNA in a deep freezer. The solution was centrifuged at 14,000 rpm for 3 min and precipitated with absolute ethanol to recover DNA. The DNA was then rinsed twice with 1 mL of 70% ethanol and resuspended in 100 μL of 1X TE [200 mM Tris–HCl (pH 8.0), 20 mM EDTA (pH 8.0)] buffer for further use and stored at −20°C. The quality of DNA was checked on agarose gel and the concentration was measured using a nanodrop spectrometer (ND-1000 spectrometer, NanoDrop Technologies, Willington, United States) (Kumbhare et al., 2015).
Polymerase chain reactions (PCR) of the internal transcribed spacer (ITS) region of filamentous molds was amplified using the primer ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (Adekoya et al., 2017). PCR reactions were performed in 25 μL of PCR pre-master mix solution (Promega, United States). The amplification steps were followed: initial denaturation at 94°C for 5 min followed by 35 cycles consisting of 94°C for 1 min, 54°C for 1 min, and 72°C for 1 min, respectively; and a final extension at 72°C for 10 min in a Thermal Cycler (Applied biosystems-2720, United States). The PCR products were verified by electrophoresis on 1.0% agarose gel containing 0.7 mg/mL of ethidium bromide and visualized under UV light (Gel doc 1000, Bio-Rad, 97-0186-02, United States). Approximate size of amplicons was determined using standard molecular markers (Himedia-100 bp DNA ladder, Mumbai, India).
The amplified PCR products were purified using PEG (polyethylene glycol)-NaCl (sodium chloride) and precipitation solution (20% w/v of PEG, 2.5 M NaCl) with the addition of 0.6 volumes of 20% PEG-NaCl to the final volume of the PCR products (Schmitz and Riesner, 2006). The mixture was centrifuged at 12,000 rpm for 30 min, incubated at 37°C for 30 min, the aqueous solution was discarded, and the pellet was washed twice with 1 mL ice cold 70% freshly prepared ethanol (70%). The collected pellet was then air dried prior to elution in 20 μL of nuclease-free water, and finally, the purified product was loaded in 1% agarose gel.
PCR-amplified products had been sequenced in a forward and reverse direction using ITS1 primer (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 primer (5′-TCCTCCGCTTATTGATATGC-3′), respectively, as per the method described by Martin and Rygiewicz (2005). The PCR reaction was carried out in 50 μL reaction volume containing 2.0 mM MgCl2, 0.2 μM each primer, 0.2 mM dNTP, 0.5 mg [mL]–1 bovine serum albumin (BSA) and 0.04 U [μL]–1 tTaq DNA polymerase on a thermal cycler equipped with a heated lid. The thermal program included initial denaturation, enzyme activation at 95°C (6–10 min) followed by 35 cycles to complete the step [95°C (1 min), 40°C (2 min) and 72°C (1 min)] and one cycle at 72°C (10 min). The amplified products were sequenced by an automated DNA Analyzer (ABI 3730XL Capillary Sequencers, Applied Biosystems, Foster City, CA, United States). These high-quality, double-stranded sequence data were analyzed with the help of the BLASTn program and multiple sequence alignment.
The qualities of the raw sequences were checked by Sequence Scanner version 1.0 (Applied Biosystems, Foster City, CA, United States) and were edited using software ChromasPro version 1.34. Sequences were compared with sequence entries in the GenBank of NCBI (National Center for Biotechnology Information)1 using the Basic Local Alignment Search Tool for nucleotides (BLASTn) on the NCBI website (Pinto et al., 2012). For phylogenetic analysis, the available sequence of similar related organisms was retrieved in FASTA format and aligned using the clustal-W. Sequence alignment and a phylogenetic tree were constructed using MEGA7.0 software by Neighbor-Joining methods using 1000-bootstrap replicates (Lutzoni et al., 2004).
Percentages of frequency and relative density of fungal species in samples were determined as per the method described by Doi et al. (2018). Frequency (%) was calculated by the equation:
Relative Density (%) was calculated by the equation:
Diversity indexes of filamentous molds in samples were calculated by species richness (R), Shannon’s diversity index (H), and species evenness (E) (Panda et al., 2010) using PAST (Paleontological STatistics) software version 3.26 (Hammer et al., 2001).
The sequences obtained in this study were deposited at the GenBank-NCBI database under accession numbers: MK396469–MK396484, MK396486–MK396500, MK778442–MK778449, and MK796041–MK796045.
The microbial load of filamentous molds in 40 samples of traditionally prepared dry starters collected from different regions of North East India were 2.5 to 7.9 × 105 cfu/g (Table 1). The pH and moisture contents of all samples analyzed were pH 4.1–6.3 and 8.0–13.9%, respectively (Table 1).
We isolated 131 total fungal isolates from 40 different samples of traditionally prepared dry starters (marcha, thiat, humao, hamei, chowan, phut, dawdim, and khekhrii) collected from eight states of North East India (Table 1). Based on the morphological characteristics (such as color, texture, size, and appearance of colony), microscopic characteristics (sporangia, sporangiospores, chlamydospores, conidia, conidiophore, and rhizoid structure), 44 representative fungal isolates were grouped (seven isolates from marcha, five from thiat, six from humao, two from hamei, five from chowan, six from phut, six from dawdim, and seven from khekhrii). Mucor, Rhizopus, Aspergillus, Penicillium, and Cladosporium and a few unidentified basidiomycetes fungi were tentatively identified on the basis of detailed morphological characters using the taxonomical keys described by Samson et al. (2004) and Pitt and Hocking (2009) (Supplementary Table S1).
Genomic DNA of each isolate of 44 representative fungal strains was extracted and PCR products were prepared for identification by ITS gene sequencing. DNA sequences of fungal isolates were assigned by comparison with those available in the GenBank of NCBI database using the ITS gene sequence (ITS1 and ITS4) based on the Basic Local Alignment Search Tool (BLAST) 2.0 program (Raja et al., 2017). The phylogenetic trees of nucleotide sequences of the 44 fungal isolates from the samples were constructed using the Neighbor-joining method with 1000 replicates bootstrap values (Figure 2). ITS gene sequencing results showed three fungal phyla represented by Ascomycota (48%), Mucoromycota (38%), and Basidiomycota (14%) (Figure 3). Distribution percentage of the phyla in the starter showed the highest percentage of Ascomycota (86%) in khekhrii, Mucoromycota (60%) in dawdim, and Basidiomycota (20%) in chowan, dawdim, and thiat, respectively. Phyla Ascomycota and Mucoromycota were present in all starters, whereas Basidiomycota was present only in marcha, thiat, chowan, and dawdim.

Figure 2. Molecular phylogenetic analysis of 44 filamentous fungal isolates from starters from North East India using the Neighbor-Joining method in MEGA7 software. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branch. The tree is drawn to scale with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method and are in the units of the number of base substitutions per site. The phylogenetic tree branches are collapsed at 50%.
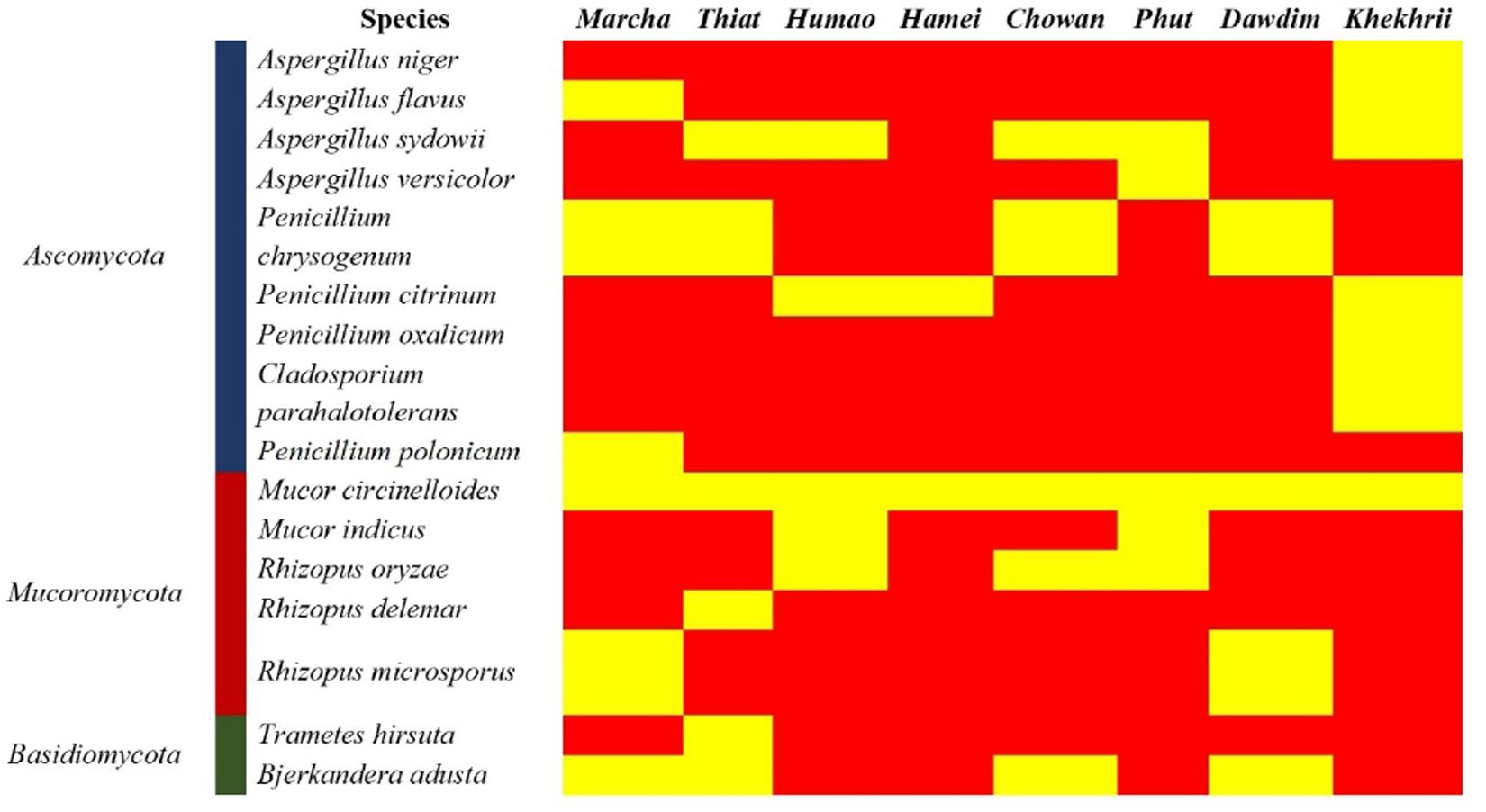
Figure 3. Heatmap showing the consensus species diversity resulted by ITS-region gene sequencing of filamentous fungal isolates. We used presence/absence value for fungal species to generate the heatmap, where the yellow color indicates the presence and red indicates absence.
Based on results of morphological characteristics and ITS gene sequencing, 44 representative strains of filamentous molds isolated from traditionally prepared dry starters from India were grouped into seven genera with 16 species, which were represented by Mucor circinelloides (20%), Aspergillus sydowii (11%), Penicillium chrysogenum (11%), Bjerkandera adusta (11%), Penicillium citrinum (7%), Rhizopus oryzae (7%), Aspergillus niger (5%), Aspergillus flavus (5%), Mucor indicus (5%) Rhizopus microsporus (5%), Rhizopus delemera (2%), Aspergillus versicolor (2%), Penicillium oxalicum (2%), Penicillium polonicum (2%), Trametes hirsuta (2%), and Cladosporium parahalotolerans (2%) (Table 2 and Figure 4). Interestingly we detected few basidiomycetes fungi represented by Bjerkandera adusta and Trametes hirsuta in marcha, thiat, chowan and dawdim samples. Colony morphology and microscopic images of 16 species of seven genera of filamentous molds isolated from dry starters from India were illustrated for fungal taxonomy (Figure 5).
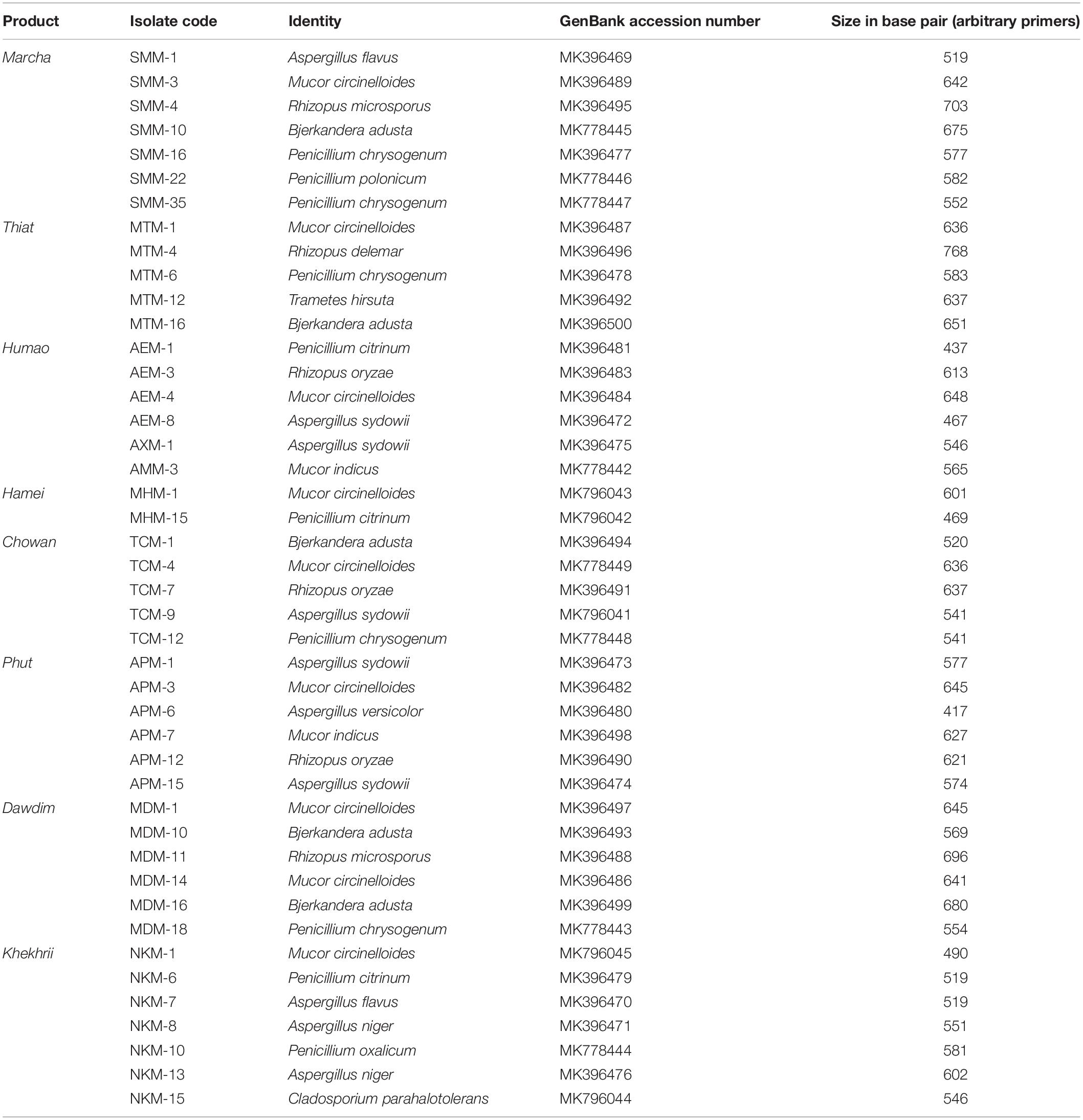
Table 2. Molecular identification of filamentous molds isolated from starters from North East India by ITS gene sequence (ITS1 and ITS4) based on BLAST.
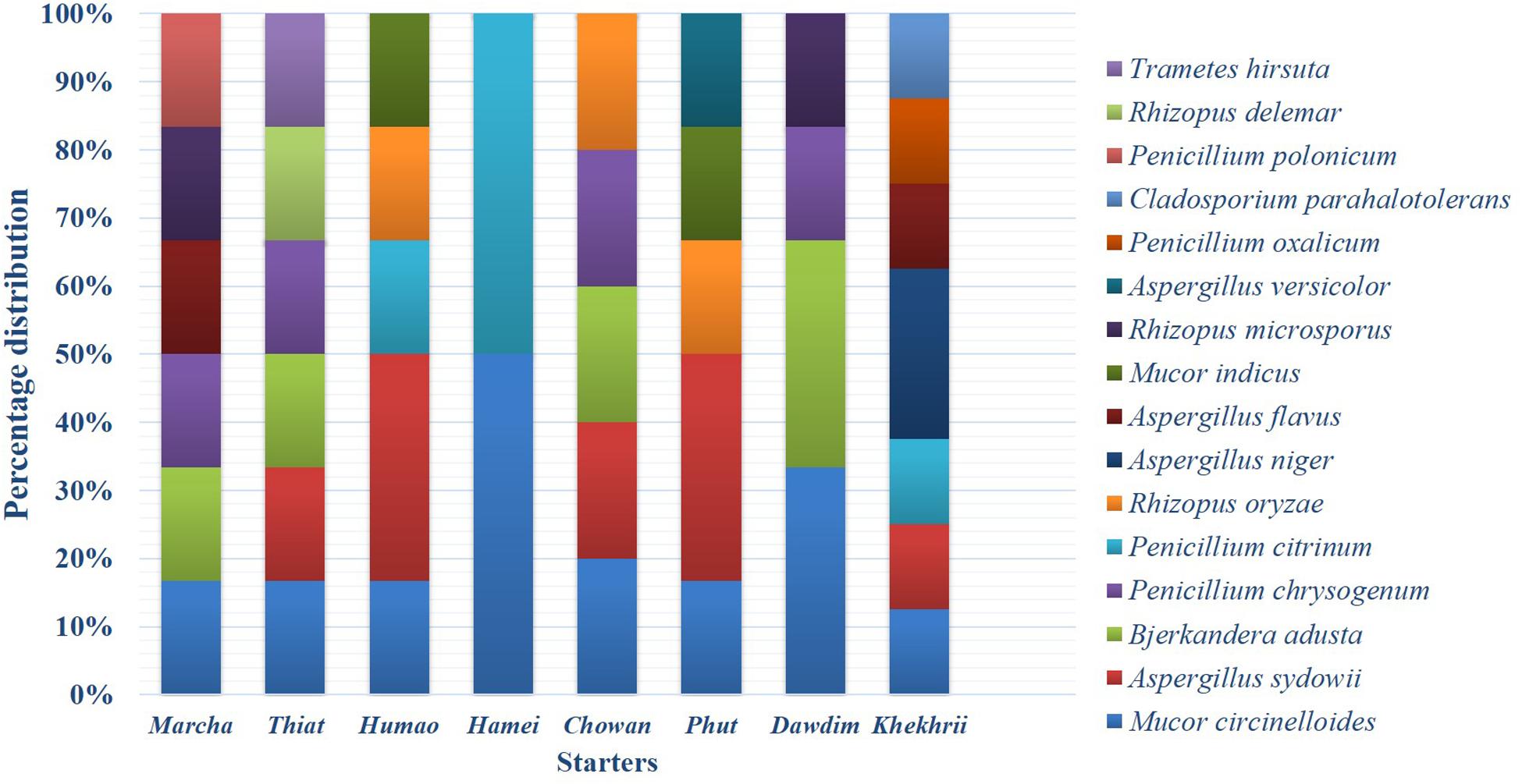
Figure 4. Abundance distribution of the filamentous fungi isolated from dry starters from North East India.
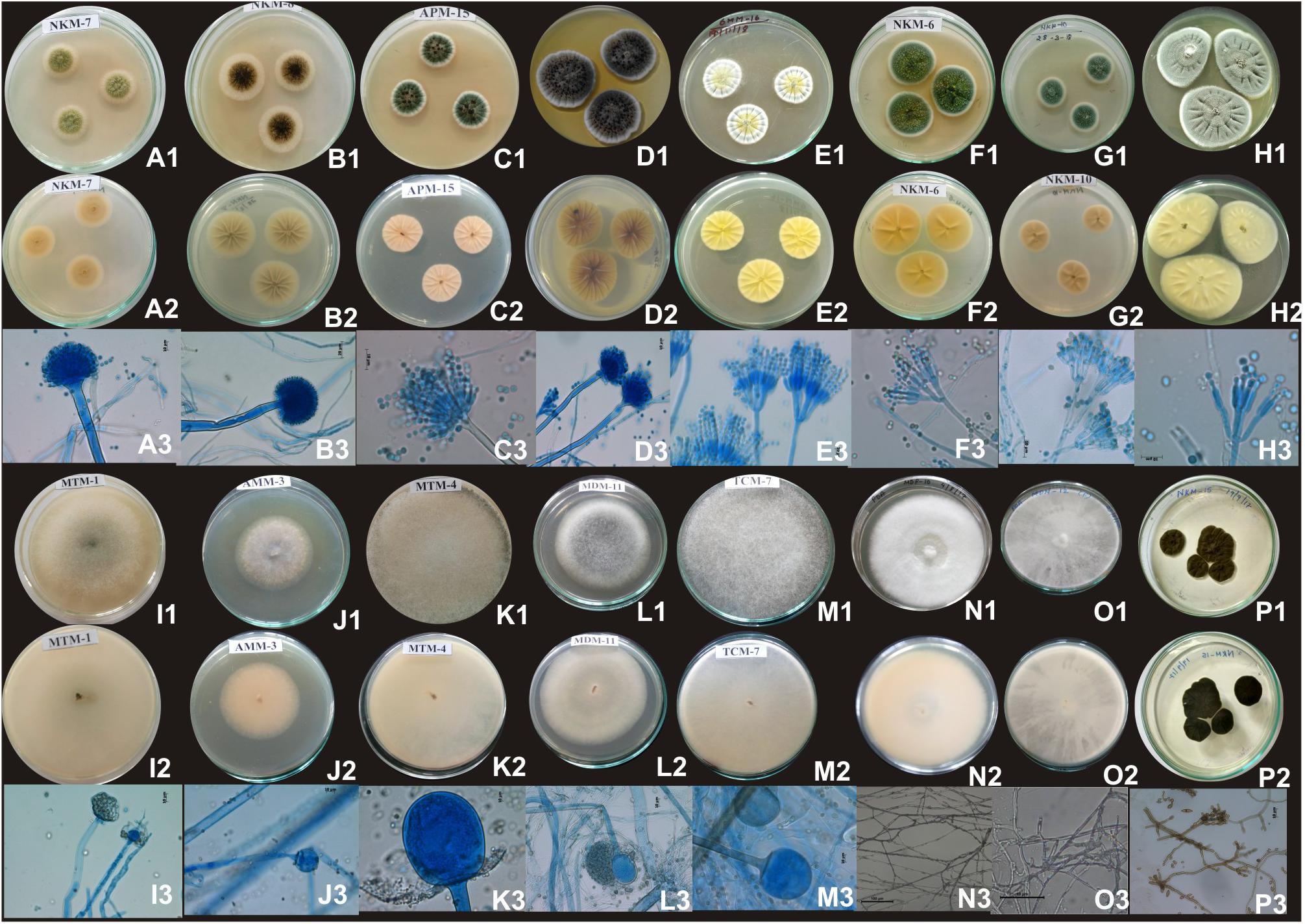
Figure 5. Images of colony morphology and microscopic features of filamentous molds that grew on MEA media: Aspergillus flavus colonies top (A1), reverse (A2), Conidiophores (A3); Aspergillus niger colonies top (B1), reverse (B2), mature conidia globose conidial head contain conidia (B3); Aspergillus sydowii colonies top (C1), reverse (C2), mature conidiophore with vesicle bearing conidiogenous metulae and phailides (biserate) (C3); Aspergillus versicolor colonies top (D1), reverse (D2), conidial heads supported vesicles with which are biseriate with metulae about the same size of philiades (D3); Penicillium chrysogenum colonies top (E1), reverse (E2), smooth-walled conidiophores stipes (150–280 μm) and biverticillate (E3); Penicillium citrinum colonies top (F1), reverse (F2), conidiophores stipes (150–280 μm) and biverticillate, phialides ampuliform (flask-shaped) (F3); Penicillium oxalicum colonies top (G1), reverse (G2), mature conidiophores monoverticillate, or biverticillate and asymmetrical, phialides were cylindrical; Penicillium polonicum colonies top (H1), reverse (H2), conidiophore were terverticillate, phialides (H); Mucor circinelloides colonies top (I1), reverse (I2), mature sporangiosphores contain sporangiospores (I3); Mucor indicus colonies top (J1), reverse (J2), mature sporangiosphores contain sporangiospores (J3); Rhizopus delemar colonies top (K1), reverse (K2), globose sporangium (K3); Rhizopus oryzae colonies top (L1), reverse (L2), sporangiophores were usually straight, mostly 10∼20 μm (L3); Rhizopus microsporus colonies top (M1), reverse (M2), sporangia globose, smooth and released spore (M3); Trametes hirsuta colonies top (N1), reverse (N2), hyphal structure (N3); Bjerkandera adusta colonies top (O1), reverse (O2), dichotomously branched hyphae (O3); Cladosporium parahalotolerans colonies top (P1), reverse (P2), conidiophores and conidial chain (P3).
Frequency and density of fungal species in samples showed that Aspergillus niger was colonized with khekhrii; a species from the Mucor circinelloides complex was observed with a high dominance in samples, whereas Trametes hirsuta was less diversified and observed only in thiat samples (Table 3).
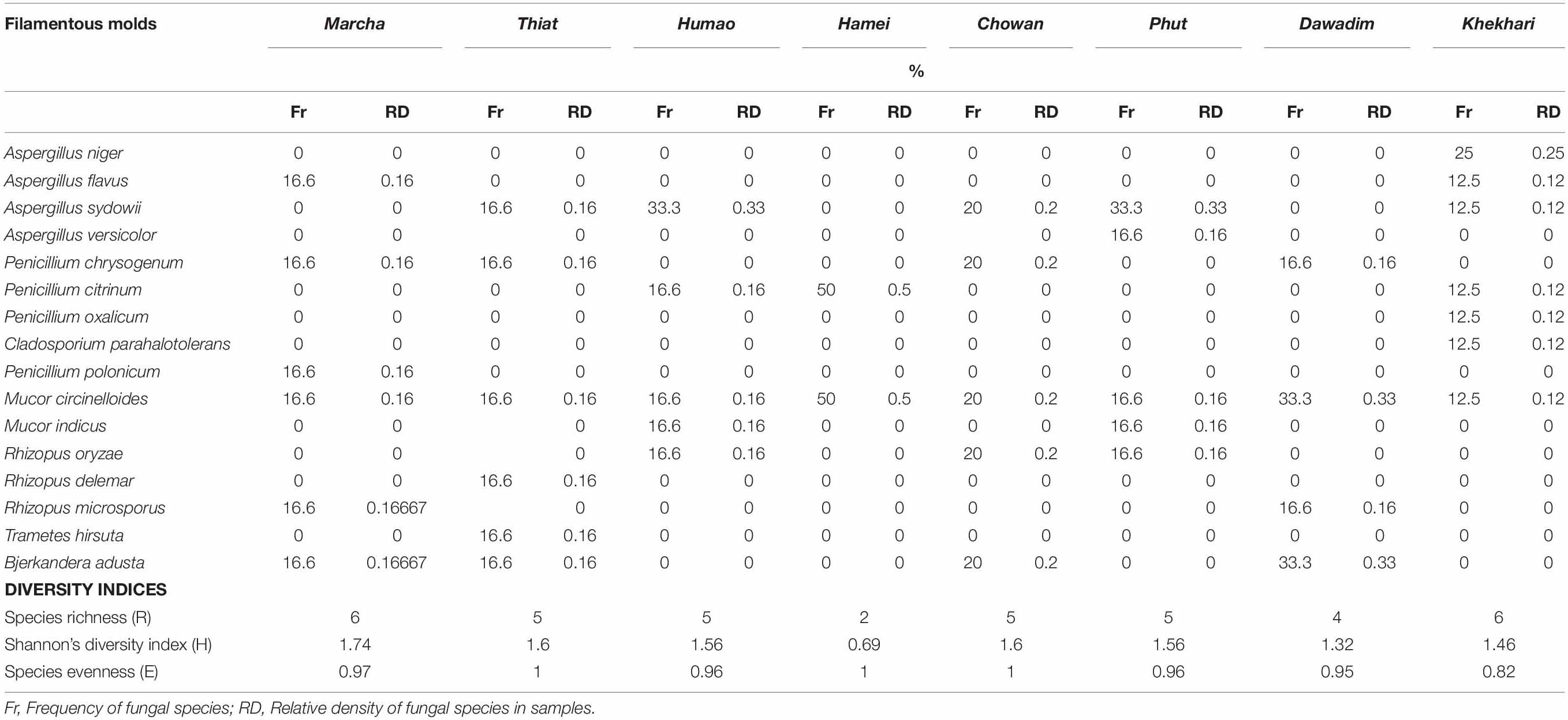
Table 3. Frequency, density, and diversity indices of filamentous molds observed in dry starters from North East India.
Diversity indexes of filamentous molds of dry starters were characterized by species richness (R), Shannon’s diversity index (H), and species evenness (E) (Table 3). The Shannon diversity index H was recorded as the highest in marcha from Sikkim (H: 1.74) and the lowest in hamei from Manipur (H: 0.69). Species Evenness (E) values were 0.97 in marcha followed by humao from Assam and phut from Arunachal Pradesh. The Species Richness (R), values were recorded highest in marcha and khekhrii samples (Table 3).
Drinking of cereal-based mild to strong alcoholic beverages produced by traditionally prepared amylase and alcohol-producing starters has been a traditional food culture of the ethnic people from the North East states of India for centuries. Traditionally prepared dry starters have consortia of co-existed microbiota containing filamentous molds, yeasts, and bacteria and are crudely sub-cultured through a “back-slopping” process by traditional starter-makers (Hesseltine et al., 1988; Tamang and Sarkar, 1995; Tamang et al., 2007; Sha et al., 2018, 2019), for alcohol production by the Indian people. The pH of traditionally prepared dry starters from India were slightly acidic in nature, perhaps due to accumulation of metabolic organic acids (Ma et al., 2019). Moreover, low pH is favorable for the growth of mycelial fungi (Abubakar et al., 2013). Low content of moisture in starter cultures is due to the sun-drying process during the traditional method of preparation practiced by the ethnic people of India, which may increase the shelf life of the starter for a year or more at room temperature (Tsuyoshi et al., 2005; Tamang, 2010).
Some traditionally prepared starters from North East India have been microbiologically analyzed in earlier works and several species of yeasts (Tsuyoshi et al., 2005; Jeyaram et al., 2008, 2011; Sha et al., 2017, 2018, 2019) and bacteria (Tamang et al., 2007; Pradhan and Tamang, 2019) were reported. However, detailed taxonomical studies of filamentous molds isolated from traditionally prepared dry starters from North East India have not been reported yet, except for marcha (Tamang et al., 1988; Tamang and Sarkar, 1995; Sha et al., 2017, 2019), thiat (Sha et al., 2017, 2019), amou, perok-kushi (Das et al., 2017). Hence, we studied the taxonomy and diversity of filamentous fungi associated with traditionally prepared dry starter cultures from North East India viz., marcha from Sikkim, thiat from Meghalaya, humao from Assam, hamei from Manipur, chowan from Tripura, phut from Arunachal Pradesh, dawdim from Mizoram, and khekhrii from Nagaland based on morphological characters and molecular identifications. The average fungal population in traditionally prepared dry starters from North East India was 105 cfu/g, which was in accordance with earlier reports on fungal populations in marcha of Sikkim, and the Darjeeling hills in India (Tamang et al., 1988; Tamang and Sarkar, 1995). No such data on fungal population in other starters of India are available except for marcha. In the present study, we first isolated and characterized 131 fungal isolates from 40 different starters from North East India based on macroscopic and microscopic characteristics and grouped them into 44 representative fungal strains. Morphological examination and identification of fungi are useful for identification up to the family or genus level (Alsohaili and Bani-Hasan, 2018). However, morphological-based identification is not adequate to identify the fungi up to species level (Lutzoni et al., 2004). The sequence-based identification tool is widely applied to confirm the exact identify of the fungal species (Romanelli et al., 2010; Xu, 2016).
We applied polymerase chain reactions (PCR) of the internal transcribed spacer (ITS) region of 44 strains of filamentous fungi isolated from starters from North East India using the primers ITS1 and ITS4 and grouped into three phyla represented by Ascomycota (48%), Mucoromycota (38%), and Basidiomycota (14%). A similar type of phylum distribution was also reported earlier in a nuruk dry starter from Korea (Carroll et al., 2017) and daqu from China (Shoubao et al., 2019). Seven genera with 16 species of filamentous fungi, isolated from Indian amylase and alcohol-producing starters, were identified based on the morphological and microscopic characteristics, and molecular identification which were represented by Aspergillus flavus, A. niger, A. sydowii, A. versicolor, Bjerkandera adusta, Cladosporium parahalotolerans, Mucor circinelloides, M. indicus, Penicillium chrysogenum, P. citrinum, P. oxalicum, P. polonicum, Rhizopus delemar, R. microsporus, R. oryzae, and Trametes hirsuta. Illustration of taxonomical keys based on morphological and molecular identification is more accurate and reliable in fungal taxonomy (Xing et al., 2018). Our earlier findings of Rhizopus oryzae and species from the Mucor circinelloides complex in traditionally prepared starters of North East India by PCR-DGGE method (Sha et al., 2018) supported the present study. Hesseltine and Kurtzman (1990) reported species from the M. circinelloides complex in bubod from the Philippines. Species from the M. circinelloides complex, M. indicus, Rhizopus oryzae, and R. microsporus were reported in benh men from Vietnam (Dung et al., 2007; Thanh et al., 2008). In marcha and khekhrii we detected Aspergillus flavus, which was also reported in mana, an amylolytic starter from Nepal (Nikkuni et al., 1996).
Aspergillus belonging to order Eurotials is a phenotypically polythetic genus and is widely distributed in the environment (Tsang et al., 2018). Samson et al. (2014) proposed phylogenic identification of Aspergillus with ITS sequence data, and calmodulin as a secondary identification marker, according to the decision of the International Commission of Penicillium and Aspergillus2. Application of ITS with β-tubulin sequences for identification of Aspergillus species has also been reported by Zulkifli and Zakaria (2017). However, in this study we have applied both ITS sequence and morphological characteristics, such as the conidiophore with straight ending in a large vesicle from where primary and secondary sterigmata arise bearing conidia in chains, for identification of species of Aspergillus. Aspergillus niger and A. flavus cannot be distinguished only by their ITS sequences, the morphological characters are also essential in species identification (Zulkifli and Zakaria, 2017). We identified genus Aspergillus with four species in dry starter samples from India which included A. niger, A. flavus, A. sydowii, and A. versicolor. Among Aspergillus A. flavus, A. niger and A. sydowii were most prevalent in food samples due to their sporulating ability in the environment (Adekoya et al., 2017). Aspergillus is a dominant fungal genus in daqu from China (Ji et al., 2018), and may contribute to the saccharification process (Wang et al., 2019). We detected two strains of Aspergillus flavus in a marcha sample from Sikkim (Aspergillus flavus SMM-1) and in a khekhrii sample from Nagaland (A. flavus NKM-7). Though the distribution percentage was only 5%, the presence of A. flavus in samples of marcha and khekhrii is alarming. A. flavus is a saprotrophic with cosmopolitan distribution (Ramírez-Camejo et al., 2012), which produces aflatoxin (Saori and Keller, 2011; Priyanka et al., 2012; Mudili et al., 2014). Probable sources of A. flavus in starters may be from contaminated rice grains (Lai et al., 2015) since rice is the main base substrates for the preparation of starters for the production of alcohol. Moreover starter-makers commonly use low-quality, old-stocked and discarded rice grains for preparation of starters. However due to co-existence of other species of filamentous molds, yeasts and lactic acid bacteria in traditionally prepared starters may antagonize against A. flavus in marcha and khekhrii, which may reduce aflatoxin production in the sample (Karlovsky et al., 2016; Adebo et al., 2019). Lactic acid bacteria isolated from marcha showed an antagonistic property (Tamang et al., 2007), similarly, some bacteria have antifungal activity against aflatoxin-producing A. flavus (Shakeel et al., 2018). Rhizopus spp. from tempeh, a fermented soybean food from Indonesia, were reported for detoxification of alfatoxins (Nakazato et al., 1990). A. sydowii present in samples humao, phut and chowan, is an industrially important filamentous mold, which produces monosaccharides and indole alkaloids (Zhou et al., 2018). None of the amylolytic starters of North East India showed the presence of A. versicolor except in phut samples from Arunachal Pradesh. A. versicolor is a slow-growing filamentous fungus commonly found in/on damp indoor environments (Samson et al., 2004), foods, and feeds (Jurjevic et al., 2012), and produces toxic metabolites (Piontek et al., 2016). Contamination of A. versicolor in phut samples might be from the damp room where preparation of phut is often practiced by starter-producers in Arunachal Pradesh.
Mucor circinelloides was found to be the most dominant fungus in dry starter cultures from North East India. M. circinelloides has a sub-globose sporangiospore with a sympodial branching pattern. Using the ITS sequencing tool, it is difficult to distinguish among the different species of the Mucor circinelloides complex (MCC) which include M. circinelloides, M. griseocyanus, M. janssenii, M. lusitanicus, M. ramosissimus, M. variicolumellatus, and M. velutinosus (Wagner et al., 2019). We therefore used species from the Mucor circinelloides complex. Mucor circinelloides contributes in saccharification and liquefaction of cereal during fermentation of kodo ko jaanr, an alcoholic product of Sikkim fermented by starter marcha (Thapa and Tamang, 2004; Tamang and Thapa, 2006). M. circinelloides is an oleaginous fungus (Qiao et al., 2018) which produces lipids (Wei et al., 2013), cellulose degrading enzymes (Huang et al., 2014), and has several functional properties including antioxidants (Hameed et al., 2017). Phylum Mucoromycota does not produce mycotoxins, however, some species that belong to this M. circincelloides forma circinelloides group has been described to be putatively responsible for human illnesses after consumption of mold-contaminated yogurt (Lee et al., 2014) although its involvement was not clearly proven. M. circinelloides was also reported earlier in marcha samples (Tamang et al., 1988; Tamang and Sarkar, 1995). M. indicus, isolated from humao from Assam and phut from Arunachal Pradesh, is a dimorphic and ethanolic fungus which is able to produce ethanol from glucose, mannose, fructose and galactose (Karimi and Zamani, 2013) and oil, protein, and glucosamine (Sharifyazd and Karimi, 2017).
Phylogenetic and phylogenomic approaches show that genus Rhizopus has three major clades viz. R. microsporus with its sister taxon R. stolonifer, R. arrhizus, and R. delemar (Gryganskyi et al., 2018). Rhizopus oryzae, commonly inhabits soils, animal excrement, and rotting vegetables (Ghosh and Ray, 2011), and is very similar to Rhizopus stolonifer, except for its smaller sporangia with air-dispersed sporangiospores (Pitt and Hocking, 2009). R. oryzae and R. microsporus are detected in yao qu from China and banh men from Vietnam, which are strong amylase producers (Dung et al., 2007; Thanh et al., 2008; Lv et al., 2012b). R. oryzae is considered as a GRAS filamentous fungus (Londoño-Hernández et al., 2017), which is commonly used for production of some Asian fermented foods (Tamang et al., 2016). Rhizopus microsporus is the major fungus in tempe, a fermented soybean food from Indonesia (Hartanti et al., 2015). R. delemar was found in the thiat sample only, which naturally accumulates fumaric acid with a fruity taste (Odoni et al., 2017), and it probably imparts taste and flavor in kiad, an alcoholic product fermented by the starter thiat. R. delemar has also been reported in xajpitha, starter from Assam in India (Bora et al., 2016). Presence of Rhizopus spp. in starters from North East India may contribute functionalities in end products during acholic fermentation.
Penicillium chrysogenum was found in only four types of starters viz. marcha (Sikkim), thiat (Meghalaya), chowan (Tripura), and dowdim (Mizoram). The probable entry of P. chrysogenum during traditional preparation may be from damp and moist rooms where preparation for such starters is usually done, since P. chrysogenum is also found in damp buildings (Andersen et al., 2011). Due to the ability of P. chrysogenum to produce antibiotics, mostly penicillin (Bajaj et al., 2014), its presence in starters may have an antagonist property in the end product. P. citrinum was recovered in samples of humao, hamei and khekhrii, probably from indoor environments (Samson et al., 2004). P. oxalicum was found in samples of khekhrii (Nagaland) and P. polonicum in marcha samples. P. oxalicum produces various enzymes and natural products (Li et al., 2016). P. polonicum has also been reported in fermented black table olives (Bavaro et al., 2017).
It is interesting to note that we detected Basidiomycetous fungi represented by Bjerkandera adusta in samples of marcha, thiat, dawdim, and chowan, and also Trametes hirsuta in thiat samples. Bjerkandera adusta and Trametes hirsuta are wood decaying white-rot fungi (Rosales et al., 2005; Horisawa et al., 2019). B. adusta grows on a natural cellulosic substrate, imparts a refreshing aroma (Zhang et al., 2015), contributes to saccharification (Quiroz-Castañeda et al., 2009), and produces ethanol (Horisawa et al., 2019). Trametes hirsuta is lignin-degrading fungus due its ability to synthesize laccase (Cilerdzic et al., 2011). Traditional methods of preparation of these amylolytic starter cultures require locally grown wild herbs and spices used as ingredients by local starter-makers (Anupma et al., 2018). We assume that during collection of wild herbs from forest grounds, people might have collected whole wild plants in situ, where wood-rooting fungi have been reported in forests of North East India (Chuzho et al., 2017). There is no practice of filtering and cleaning of collected wild plants during starter preparation, hence chances for contamination of these basidiomycetous fungi may be possible during preparation. B. adusta and T. hirsuta were not reported earlier in any starter culture or in any fermented food.
Cladosporium parahalotolerance was found only in samples of khekhrii. C. parahalotolerance mostly occurred in plant debris, foods, and indoors (Bensch et al., 2012). Source of Cladosporium in khekhrii might be from wild herbs used as ingredients during traditional preparation of khekhrii in Nagaland. Species of Bjerkandera, Trametes, and Cladosporium have not been reported in any fermented foods elsewhere.
Diversity indexes determine the phylogenetic relations within different fungal species in a community (Fernandes et al., 2015). We calculated diversity indexes of fungal community present in starters of North East India by Shannon’s diversity index (H), species evenness (E), and species richness (R). Shannon diversity index H for evaluating fungal diversity was recorded highest in marcha samples collected from Sikkim (H: 1.74) and lowest in hamei samples of Manipur (H: 0.69) indicating higher fungal diversity in marcha samples of Sikkim as compared to starters of other states. The diversity index, which considers both the number of species as well as relative abundance of each species for evaluating diversity (Lucas et al., 2017), showed the highest value for marcha of Sikkim. Species richness is the number of different species represented in an ecological community, where it reflects the abundances of species or their distributions (Unterseher et al., 2008). Species Richness (R) values in samples of marcha and khekhrii were recorded as the highest showing more diversity in species level of filamentous molds. Species evenness refers to how equal the community is numerically, ranging from 0 to 1 (Savary et al., 2018) signifying that the value 1.0 in thiat, hamei, and chowan have a complete evenness in comparison to other starters. Hence diversity index of filamentous fungal community present in dry starters of North East India showed high diversity within the community. It was observed that there was variation in fungal species distribution in each type of amylolytic starters in North East India which determines the quality of the acholic product, preferred by the local consumers. This might be due to varied geographical regions, environmental conditions, and different plant species used in the preparation methods of amylolytic starters. It therefore shows that fungal diversity, present in amylase and alcohol-producing starters, traditionally prepared by ethnic Indian people using their indigenous knowledge of “back-slopping,” are morphologically, ecologically, and phylogenetically diverse. Our findings on fungal diversity in amylolytic starters from North East India may supplement the microbial diversity in eco-systems of North East India, which is one of the biodiversity hot spots of the world.
Traditionally prepared amylolytic starters are consortia of filamentous fungi, yeasts, and bacteria which are traditionally sub-cultured and preserved using traditional methods of “back-slopping” by the ethnic people of North East India for production of alcoholic beverages. Yeasts and bacteria present in these starters have already been reported in earlier studies. However, no information on fungal communities and their diversity in Indian amylolytic starters is available. We therefore identified the filamentous molds isolated from marcha, thiat, humao, hamei, chowan, phut, dawdim, and khekhrii based on morphological and sequence-based identifications. We identified seven genera with 16 species represented by Aspergillus flavus, Aspergillus niger, Aspergillus sydowii, Aspergillus versicolor, Bjerkandera adusta, Cladosporium parahalotolerans, Mucor circinelloides, Mucor indicus, Penicillium chrysogenum, Penicillium citrinum, Penicillium oxalicum, Penicillium polonicum, Rhizopus delemar, Rhizopus microsporus, Rhizopus oryzae, and Trametes hirsuta. Fungal species present in these traditionally prepared dry starters are morphologically, ecologically, and phylogenetically diverse and showed high diversity within the community.
The sequences of the internal transcribed spacers (ITS) region obtained in this study were deposited at the GenBank-NCBI database 6S rRNA sequencing were deposited at GenBank-NCBI numbers: MK396469-MK396484, MK396486-MK396500, MK778442-MK778449, MK796041-MK796045.
AA performed the experiments. JT supervised the experiments and finalized the manuscript.
We are grateful to Department of Biotechnology (DBT), Government of India for financial support. AA is grateful to DBT for the award of the Studentship in DBT-funded Bioinformatics Centre of Sikkim University sanctioned to JT.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00905/full#supplementary-material
Abubakar, A., Suberu, H. A., Bello, I. M., Abdulkadir, R., Daudu, O. A., and Lateef, A. A. (2013). Effect of pH on mycelial growth and sporulation of Aspergillus parasiticus. J. Plant Sci. 1, 64–67. doi: 10.11648/j.jps.20130104.13
Achaya, K. T. (1991). Alcoholic fermentation and its products in ancient India. Indian J. History Sci. 26, 123–129.
Adebo, O. A., Kayitesi, E., and Njobeh, P. B. (2019). Reduction of mycotoxins during fermentation of whole grain sorghum to whole grain ting (a Southern African Food). Toxins 11:180. doi: 10.3390/toxins11030180
Adekoya, I., Obadina, A., Phoku, J., Nwinyi, O., and Njobeh, P. (2017). Contamination of fermented foods in Nigeria with fungi. LWT-Food Sci. Technol. 86, 76–84.
Alsohaili, S. A., and Bani-Hasan, B. A. (2018). Morphological and molecular identification of fungi isolated from different environmental sources in the Northern Eastern desert of Jordan. Jordan J. Biol. Sci. 11, 329–337.
Andersen, B., Frisvad, J. C., Søndergaard, I., Rasmussen, I. S., and Larsen, L. S. (2011). Associations between fungal species and water-damaged building materials. Appl. Environ. Microbiol. 77, 4180–4188. doi: 10.1128/AEM.02513-10
Anupma, A., Pradhan, P., Sha, S. P., and Tamang, J. P. (2018). Traditional skill of ethnic people of the Eastern Himalayas and North East India in preserving microbiota as dry amylolytic starters. Indian J. Trad. Know. 17, 184–190.
Bajaj, I., Veiga, T., van Dissel, D., Pronk, J. T., and Daran, J. M. (2014). Functional characterization of a Penicillium chrysogenum mutanase gene induced upon co-cultivation with Bacillus subtilis. BMC Microbiol. 14:114. doi: 10.1186/1471-2180-14-114
Bavaro, S. L., Susca, A., Frisvad, J. C., Tufariello, M., Chytiri, A., Perrone, G., et al. (2017). Isolation, characterization, and selection of molds associated to fermented black table olives. Front. Microbiol. 8:1356. doi: 10.3389/fmicb.2017.01356
Bensch, K., Braun, U., Groenewald, A., and Crous, P. W. (2012). The genus Cladosporium. Stud. Mycol. 72, 1–401. doi: 10.3114/sim0003
Bora, S. S., Keot, J., Das, S., Sarma, K., and Barooah, M. (2016). Metagenomics analysis of microbial communities associated with a traditional rice wine starter culture (Xaj-pitha) of Assam. India. 3 Biotech. 6:153. doi: 10.1007/s13205-016-0471-1
Carroll, E., Trinh, T. N., Son, H., Lee, Y. W., and Seo, J. A. (2017). Comprehensive analysis of fungal diversity and enzyme activity in nuruk, a Korean fermenting starter, for acquiring useful fungi. J. Microbiol. 55:357. doi: 10.1007/s12275-017-7114-z
Chen, B., Wu, Q., and Xu, Y. (2014). Filamentous fungal diversity and community structure associated with the solid-state fermentation of Chinese Maotai-flavor liquor. Int. J. Food Microbiol. 179, 80–84. doi: 10.1016/j.ijfoodmicro.2014.03.011
Chuzho, K., Dkhar, M. S., and Lyngdoh, A. (2017). Wood-rotting fungi in two forest stands of Kohima, North-East India: a preliminary report. Cur. Res. Environ. Appl. Mycol. 7, 1–7.
Cilerdzic, J., Stajic, M., Vukojevic, J., Duletic-Lausevic, S., and Knezevic, A. (2011). Potential of Trametes hirsuta to produce ligninolytic enzymes during degradation of agricultural residues. Bioresearch 6, 2885–2895.
Daroonpunt, R., Tanasupawat, S., and Keeratipibul, S. (2016). Characterization and amylolytic activity of yeast and mold strains from Thai sweet rice. Malaysian J Microbiol. 12, 121–131.
Das, A. J., Miyaji, T., and Deka, S. C. (2017). Amylolytic fungi in starter cakes for rice beer production. J. Gen. Appl. Microbiol. 63, 236–245. doi: 10.2323/jgam.2016.11.004
Doi, S. A., Pinto, A. B., Canali, M. C., Polezel, D. R., Chinellato, R. A. M., and de Oliveira, A. J. F. C. (2018). Density and diversity of filamentous fungi in the water and sediment of Araçá bay in São Sebastião, São Paulo, Brazil. Biota Neotrop. Campinas 18:e20170416. doi: 10.1590/1676-0611-bn-2017-0416
Dung, N. T. P., Rombouts, F. M., and Nout, M. J. R. (2007). Characteristics of some traditional Vietnamese starch-based rice wine fermentation starters (men). LWT-Food Sci. Technol. 40, 130–135.
Fernandes, E. G., Pereira, O. L., da Silva, C. C., Bento, C. B. P., and de Queiroz, M. V. (2015). Diversity of endophytic fungi in Glycine max. Microbiol. Res. 181, 84–92. doi: 10.1016/j.micres.2015.05.010
Fronteras, J. P., and Bullo, L. L. R. (2017). Raw starch-digesting amylase from Saccharomycopsis fibuligera 2074 isolated from bubod starter. Philippine J. Sci. 146, 27–35.
Gaddeyya, G., Niharika, P. S., Bharathi, P., and Kumar, P. R. (2012). Isolation and identification of soil mycoflora in different crop fields at Salur Mandal. Adv. Appl. Sci. 3, 2020–2026.
Ghosh, B., and Ray, R. R. (2011). Current commercial perspective of Rhizopus oryzae: a review. J. Appl. Sci. 11, 2470–2486.
Ghosh, K., Ray, M., Adak, A., Dey, P., Halder, S. K., Das, A., et al. (2015). Microbial, saccharifying and antioxidant properties of an Indian rice based fermented beverage. Food Chem. 168, 196–202. doi: 10.1016/j.foodchem.2014.07.042
Gryganskyi, A. P., Golan, J., Dolatabadi, S., Mondo, S., Robb, S., Idnurm, A., et al. (2018). Phylogenetic and phylogenomic definition of Rhizopus species. G3 8, 2007–2018. doi: 10.1534/g3.118.200235
Hameed, A., Hussain, S. A., Yang, J., Ijaz, M. U., Liu, Q., Suleria, H. A. R., et al. (2017). Antioxidants potential of the filamentous fungi (Mucor circinelloides). Nutrients 9:1101. doi: 10.3390/nu9101101
Hammer, Ø, Harper, D. A. T., and Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron 4:9.
Hartanti, A. T., Rahayu, G., and Hidayat, I. (2015). Rhizopus species from fresh tempeh collected from several regions in Indonesia. HAYATI J. Bios. 22, 136–142.
Hesseltine, C. W., and Kurtzman, C. P. (1990). Yeasts in amylolytic food starters. Anal. Inst. Biol. Univ. Nac. Autón. México. Ser. Bot. 1–7. doi: 10.1016/B978-0-444-52149-1.00187-7
Hesseltine, C. W., and Ray, M. L. (1988). Lactic acid bacteria in murcha and ragi. J. Appl. Microbiol. 64, 395–401.
Hesseltine, C. W., Rogers, R., and Winarno, F. G. (1988). Microbiological studies on amylolytic Oriental fermentation starters. Mycopathology 101, 141–155.
Horisawa, S., Inoue, A., and Yamanaka, Y. (2019). Direct ethanol production from lignocellulosic materials by mixed culture of wood rot fungi Schizophyllum commune. Bjerkandera adusta, and Fomitopsis palustris. Ferment 5:21. doi: 10.3390/fermentation5010021
Huang, Y., Busk, P. K., Grell, M. N., Zhao, H., and Lange, L. (2014). Identification of a β-glucosidase from the Mucor circinelloides genome by peptide pattern recognition. Enzyme Micro. Technol. 67, 47–52. doi: 10.1016/j.enzmictec.2014.09.002
Jeyaram, K., Singh, W., Capece, A., and Romano, P. (2008). Molecular identification of yeast species associated with “Hamei” — A traditional starter used for rice wine production in Manipur. India. Int. J. Food Microbiol. 124, 115–125. doi: 10.1016/j.ijfoodmicro.2008.02.029
Jeyaram, K., Tamang, J. P., Capece, A., and Romano, P. (2011). Geographical markers for Saccharomyces cerevisiae strains with similar technological origins domesticated for rice-based ethnic fermented beverages production in North East India. Antonie. Van Leeuwen. 100, 569–578. doi: 10.1007/s10482-011-9612-z
Ji, Z., Jin, J., Yu, G., Mou, R., Mao, J., Liu, S., et al. (2018). Characteristic of filamentous fungal diversity and dynamics associated with wheat Qu and the traditional fermentation of Chinese rice wine. Int. J. Food Sci. Technol. 53, 1611–1621.
Jung, M. J., Nam, Y. D., Roh, S. W., and Bae, J. W. (2012). Unexpected convergence of fungal and bacterial communities during fermentation of traditional Korean alcoholic beverages inoculated with various natural starters. Food Microbiol. 30, 112–123. doi: 10.1016/j.fm.2011.09.008
Jurjevic, Z., Peterson, S. W., and Horn, B. W. (2012). Aspergillus section Versicolores: nine new species and multilocus DNA sequence based phylogeny. IMA Fungus 3, 59–79. doi: 10.5598/imafungus.2012.03.01.07
Karimi, K., and Zamani, A. (2013). Mucor indicus: biology and industrial application perspectives: a review. Biotechnol. Adv. 31, 466–481. doi: 10.1016/j.biotechadv.2013.01.009
Karlovsky, P., Suman, M., Berthiller, F., De, Meester J, Eisenbrand, G., Perrin, I., et al. (2016). Impact of food processing and detoxification treatments on mycotoxin contamination. Mycotoxin Res. 32, 179–205. doi: 10.1007/s12550-016-0257-7
Kumbhare, S. V., Dhotre, D. P., Dhar, S. K., Jani, K., Apte, D. A., Shouche, Y. S., et al. (2015). Insights into diversity and imputed metabolic potential of bacterial communities in the continental shelf of Agatti Island. PLoS One 10:e129864. doi: 10.1371/journal.pone.0129864
Lai, X., Zhang, H., Liu, R., and Liu, C. (2015). Potential for aflatoxin B1 and B2 production by Aspergillus flavus strains isolated from rice samples. Saudi J. Biol. Sci. 22, 176–180. doi: 10.1016/j.sjbs.2014.09.013
Lee, C. H., and Lee, S. S. (2002). Cereal fermentation by fungi. Appl. Mycol. Biotechnol. 2, 151–170.
Lee, S. C., Billmyre, R. B., Li, A., Carson, S., Sykes, S. M., Huh, E. Y., et al. (2014). Analysis of a food-borne fungal pathogen outbreak: virulence and genome of a Mucor circinelloides isolate from yogurt. mBio 5:e01390-14. doi: 10.1128/mBio.01390-14
Li, Y., Zheng, X., Zhang, X., Bao, L., Zhu, Y., Qu, Y., et al. (2016). The different roles of Penicillium oxalicum LaeA in the production of extracellular cellulase and β-xylosidase. Front. Microbiol. 7:2091. doi: 10.3389/fmicb.2016.02091
Londoño-Hernández, L., Ramírez-Toro, C., Ruiz, H. A., Ascacio-Valdés, J. A., Aguilar-Gonzalez, M. A., Rodríguez-Herrera, R., et al. (2017). Rhizopus oryzae – ancient microbial resource with importance in modern food industry. Int. J. Food Microbiol. 257, 110–127. doi: 10.1016/j.ijfoodmicro.2017.06.012
Lucas, R., Groeneveld, J., Harms, H., Johst, K., Frank, K., and Kleinsteuber, S. (2017). A critical evaluation of ecological indices for the comparative analysis of microbial communities based on molecular datasets. FEMS Microbiol. Ecol. 93:1. doi: 10.1093/femsec/fiw209
Lutzoni, F., Kauff, F., Cox, C. J., McLaughlin, D., Celio, G., Dentinger, B., et al. (2004). Assembling the fungal tree of life: progress, classification, and evolution of subcellular traits. American J. Bot. 91, 1446–1480. doi: 10.3732/ajb.91.10.1446
Lv, X. C., Huang, Z. Q., Zhang, W., Rao, P. F., and Ni, L. (2012a). Identification and characterization of filamentous fungi isolated from fermentation starters for Hong Qu glutinous rice wine brewing. J. Gen. Appl. Microbiol. 58, 33–42. doi: 10.2323/jgam.58.33
Lv, X. C., Weng, X., Zhang, W., Rao, P. F., and Ni, L. (2012b). Microbial diversity of traditional fermentation starters for Hong Qu glutinous rice wine as determined by PCR-mediated DGGE. Food Control 28, 426–434.
Ly, S., Mith, H., Tarayre, C., Taminiau, B., Daube, G., Fauconnier, M. L., et al. (2018). Impact of microbial composition of Cambodian traditional dried starters (Dombea) on flavor compounds of rice wine: combining amplicon sequencing with HP-SPME-GCMS. Front. Microbiol. 9:894. doi: 10.3389/fmicb.2018.00894
Ma, R., Sui, L., Zhang, J., Hu, J., and Liu, P. (2019). Polyphasic characterization of yeasts and lactic acid bacteria metabolic contribution in semi-solid fermentation of Chinese baijiu (traditional fermented alcoholic drink): towards the design of a tailored starter culture. Microorganisms 7:147. doi: 10.3390/microorganisms705014
Martin, K. J., and Rygiewicz, P. T. (2005). Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 5:28. doi: 10.1186/1471-2180-5-28
Mudili, V., Siddaih, C. N., Nagesh, M., Garapati, P., Naveen Kumar, K., Murali, H. S., et al. (2014). Mould incidence and mycotoxin contamination in freshly harvested maize kernels originated from India. J. Sci. Food Agri. 94, 2674–2683. doi: 10.1002/jsfa.6608
Nakazato, M., Morozumi, S., Saito, K., Fujinuma, K., Nishima, T., and Kasai, N. (1990). Interconversion of aflatoxin B1 and aflatoxicol by several fungi. Appl. Environ. Microbiol. 56, 1465–1470.
Nikkuni, S., Karki, T. B., Terao, T., and Suzuki, C. (1996). Microflora of mana, a Nepalese rice koji. J. Ferment.Bioengin. 81, 168–170. doi: 10.1016/0922-338X(96)87597-87590
Odoni, D. I., Tamayo-Ramos, J. A., Sloothaak, J., van Heck, R. G. A., Martins Dos Santos, V. A. P., de Graaff, L. H., et al. (2017). Comparative proteomics of Rhizopus delemar ATCC 20344 unravels the role of amino acid catabolism in fumarate accumulation. Peer J. 5:e3133. doi: 10.7717/peerj.3133
Panda, T., Pani, P. K., Mishra, N., and Mohanty, R. B. (2010). A comparative account of the diversity and distribution of fungi in tropical forest soils and sand dunes of Orissa. India. J. Biodiv. 1, 27–41.
Pinto, F. C. J., Lima, D. B. D., Agustini, B. C., Dallagassa, C. B., Shimabukuro, M. F., Chimelli, M., et al. (2012). Morphological and molecular identification of filamentous fungi isolated from cosmetic powders. Brazilian Arch. Biol. Technol. 55, 897–901.
Piontek, M., Łuszczyñska, K., and Lechów, H. (2016). Occurrence of the toxin-producing Aspergillus versicolor Tiraboschi in residential buildings. Int. J. Environ. Res. Public Health 13, e862. doi: 10.3390/ijerph13090862
Pitt, J. I, and Hocking, A. D. (2009). Fungi and Food Spoilage, 3rd Edn. New York, NY: Springer Dordrecht.
Pradhan, P., and Tamang, J. P. (2019). Phenotypic and genotypic identification of bacteria isolated from traditionally prepared dry starters of the Eastern Himalayas. Front. Microbiol. 10:2526. doi: 10.3389/fmicb.2019.02526
Priyanka, S. R., Ramana, M. V., Balakrishna, K., Murali, H. S., and Batra, H. V. (2012). A novel non radioactive PCR-DNA probe for the detection of aflatoxin producing Aspergillus species from major food crops grown in India. Adv. Microbiol. 2, 577–586. doi: 10.4236/aim.2012.24075
Qiao, W., Tao, J., Luo, Y., Tang, T., Miao, J., and Yang, Q. (2018). Microbial oil production from solid-state fermentation by a newly isolated oleaginous fungus, Mucor circinelloides Q531 from mulberry branches. R. Soc. Open Sci. 5:180551. doi: 10.1098/rsos.180551
Quiroz-Castañeda, R. E., Balcázar-López, E., Dantán-González, E., Martinez, A., Folch-Mallol, J., and Martínez Anaya, C. (2009). Characterization of cellulolytic activities of Bjerkandera adusta and Pycnoporus sanguineus on solid wheat straw medium. Electr. J. Biotechnol. 12, 5–6. doi: 10.2225/vol12-issue4-fulltext-3
Raja, H. A., Miller, A. N., Pearce, C. J., and Oberlies, N. H. (2017). Fungal identification using molecular tools: a primer for the natural products research community. J. Nat. Prod. 80, 756–770. doi: 10.1021/acs.jnatprod.6b01085
Ramírez-Camejo, L. A., Zuluaga-Montero, A., Lázaro-Escudero, M. A., Hernández-Kendall, V. N., and Bayman, P. (2012). Phylogeography of the cosmopolitan fungus Aspergillus flavus: is everything everywhere?”. Fungal Biol. 116, 452–463. doi: 10.1016/j.funbio.2012.01.006
Romanelli, A. M., Sutton, D. A., Thompson, E. H., Rinaldi, M. G., and Wickes, B. L. (2010). Sequence-based identification of filamentous basidiomycetous fungi from clinical specimens: a cautionary note. J. Clin. Microbiol. 48, 741–752. doi: 10.1128/JCM.01948-09
Rosales, E., Couto, S. R., and Sanromán, M. A. (2005). Reutilisation of food processing wastes for production of relevant metabolites: application to laccase production by Trametes hirsuta. J. Food Eng. 66, 419–423.
Roslan, R., Rehan, M. M., Kamarudin, K. R., Noor, H. M., Huda-Faujan, N., and Radzi, S. M. (2018). Isolation and identification of amylolytic bacteria from Ragi. Malaysian Appl. Biol. 47, 83–88.
Samson, R. A., Hoekstra, E. S., and Frisvad, J. C. (2004). Introduction to Food and Airborne Fungi, 7th Edn. Utrecht: Central Bureau Voor Schimmer Cultures (CBS).
Samson, R. A., Visagie, C. M., Houbraken, J., Hong, S. B., Hubka, V., Klaassen, C. H. W., et al. (2014). Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 78, 141–173. doi: 10.1016/j.simyco.2014.07.004
Sarkar, A., Mukherjee, A., Bera, M. K., Das, A., Juyal, N., Morthekai, R., et al. (2016). Oxygen isotope in archaeological bioapatite from India: implications to climate change and decline of Bronze Age Harappan civilization. Sci. Rep. 6:26555. doi: 10.1038/srep26555
Savary, R., Villard, L., and Sanders, I. R. (2018). Within-species phylogenetic relatedness of a common mycorrhizal fungus affects evenness in plant communities through effects on dominant species. PLoS One 13:e0198537. doi: 10.1371/journal.pone.0198537
Schmitz, A., and Riesner, D. (2006). Purification of nucleic acids by selective precipitation with polyethylene glycol 6000. Anal. Biochem. 354, 311–313. doi: 10.1016/j.ab.2006.03.014
Sha, S. P., Jani, K., Sharma, A., Anupma, A., Pradhan, P., Shouche, Y., et al. (2017). Analysis of bacterial and fungal communities in Marcha and Thiat, traditionally prepared amylolytic starters of India. Sci. Rep. 7:10967. doi: 10.1038/s41598-017-11609-y
Sha, S. P., Suryavanshi, M. S., and Tamang, J. P. (2019). Mycobiome diversity in traditionally prepared starters for alcoholic beverages in India by high-throughput sequencing method. Front. Microbiol. 10:348. doi: 10.3389/fmicb.2019.003482237
Sha, S. P., Suryavanshi, M. V., Jani, K., Sharma, A., Shouche, Y. S., and Tamang, J. P. (2018). Diversity of yeasts and molds by culture-dependent and culture-independent methods for mycobiome surveillance of traditionally prepared dried starters for the production. Front. Microbiol. 9:2237. doi: 10.3389/fmicb.2018.02237
Shakeel, Q., Lyu, A., Zhang, J., Wu, M., Li, G., Hsiang, T., et al. (2018). Biocontrol of Aspergillus flavus on peanut kernels using Streptomyces yanglinensis 3-10. Front. Microbiol. 9:1049. doi: 10.3389/fmicb.2018.01049
Sharifyazd, S., and Karimi, K. (2017). Effects of fermentation conditions on valuable products of ethanolic fungus Mucor indicus. Electr. J. Biotechnol. 30, 77–82.
Shoubao, Y., Xiangsong, C., and Jiaquan, G. (2019). Bacterial and fungal diversity in the traditional Chinese strong flavour liquor Daqu. J. Inst. Brew. 125, 443–452. doi: 10.1002/jib.574
Shrivastava, K., Greeshma, A. G., and Srivastava, B. (2012). Biotechnology in tradition – a process technology of alcoholic beverages practiced by different tribes of Arunachal Pradesh. North East India. Indian J. Trad. Knowl. 11, 81–89.
Singh, N. L., Ramprasad Mishra, P. K., Shukla, S. K., Kumar, J., and Singh, R. (2010). Alcoholic fermentation techniques in early Indian tradition. Indian J. History Sci. 45, 163–173.
Suesse, A. R., Norton, G. A., and van Leeuwen, J. (2016). Pilot-scale continuous-flow hydrothermal liquefaction of filamentous fungi. Energy Fuels 30, 7379–7386. doi: 10.1021/acs.energyfuels.6b01229
Tamang, J. P. (2010). Himalayan Fermented Foods: Microbiology, Nutrition, and Ethnic Values. New York, NY: CRC Press.
Tamang, J. P., Cotter, P., Endo, A., Han, N. S., Kort, R., Liu, S. Q., et al. (2020). Fermented foods in a global age: east meets west. Comprehen. Rev. Food Sci. Food Saf. 19, 184–217. doi: 10.1111/1541-4337.12520
Tamang, J. P., Dewan, S., Tamang, B., Rai, A., Schillinger, U., and Holzapfel, W. H. (2007). Lactic acid bacteria in hamei and marcha of North East India. Indian J. Microbiol. 47, 119–125. doi: 10.1007/s12088-007-0024-8
Tamang, J. P., and Sarkar, P. K. (1995). Microflora of murcha: an amylolytic fermentation starter. Microbios. 81, 115–122.
Tamang, J. P., Sarkar, P. K., and Hesseltine, C. W. (1988). Traditional fermented foods and beverages of Darjeeling and Sikkim - a review. J. Sci. Food Agri. 44, 375–385.
Tamang, J. P., and Thapa, S. (2006). Fermentation dynamics during production of bhaati jaanr, a traditional fermented rice beverage of the Eastern Himalayas. Food Biotechnol. 20, 251–261.
Tamang, J. P., Watanabe, K., and Holzapfel, W. H. (2016). Diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 7:377. doi: 10.3389/fmicb.2016.00377
Thakur, N., Saris, P. E., and Bhalla, T. C. (2015). Microorganisms associated with amylolytic starters and traditional fermented alcoholic beverages of North Western Himalayas in India. Food Biosci. 11, 92–96.
Thanh, V. N., Mai, L. T., and Tuan, D. A. (2008). Microbial diversity of traditional Vietnamese alcohol fermentation starters (banh men) as determined by PCR-mediated DGGE. Int. J. Food Microbiol. 128, 268–273. doi: 10.1016/j.ijfoodmicro.2008.08.020
Thapa, S., and Tamang, J. P. (2004). Product characterization of kodo ko jaanr: fermented finger millet beverage of the Himalayas. Food Microbiol. 21, 617–622.
Thapa, S., and Tamang, J. P. (2006). Microbiological and physio-chemical changes during fermentation of kodo ko jaanr, a traditional alcoholic beverage of the Darjeeling hills and Sikkim. Indian J. Microbiol. 46, 333–341.
Tsang, C. C., Tang, J. Y., Lau, S. K., and Woo, P. C. (2018). Taxonomy and evolution of Aspergillus, Penicillium and Talaromyces in the omics era–Past, present and future. Computer Struct. Biotech. 16, 197–210. doi: 10.1016/j.csbj.2018.05.003
Tsuyoshi, N., Fudou, R., Yamanaka, S., Kozaki, M., Tamang, N., Thapa, S., et al. (2005). Identification of yeast strains isolated from marcha in Sikkim, a microbial starter for amylolytic fermentation. Int. J. Food Microbiol. 99, 135–146. doi: 10.1016/j.ijfoodmicro.2004.08.011
Umesha, S., Manukumar, H. M., and Raghava, S. (2016). A rapid method for isolation of genomic DNA from food-borne fungal pathogens. 6, 123. doi: 10.1007/s13205-016-0436-4
Unterseher, M., Schnittler, M., Dormann, C., and Sickert, A. (2008). Application of species richness estimators for the assessment of fungal diversity. FEMS Microbiol. Lett. 282, 205–213. doi: 10.1111/j.1574-6968.2008.01128.x
Wagner, L., Stielow, J. B., de Hoog, S., Bensch, K., Schwartze, V., Voigt, K., et al. (2019). A new species concept for the clinically relevant Mucor circinelloides complex. Personia 44, 67–97.
Wang, J., Chio, C., Chen, X., Su, E., Cao, F., Jin, Y., et al. (2019). Efficient saccharification of agave biomass using Aspergillus niger produced low-cost enzyme cocktail with hyperactive pectinase activity. Biores. Technol. 272, 26–33. doi: 10.1016/j.biortech.2018.09.069
Wei, H., Wang, W., Yarbrough, J. M., Baker, J. O., Laurens, L., and Van Wychen, S. (2013). Genomic, proteomic, and biochemical analyses of oleaginous Mucor circinelloides: evaluating its capability in utilizing cellulolytic substrates for lipid production. PLoS One 8:e71068. doi: 10.1371/journal.pone.0071068
Xing, J. H., Sun, Y. F., Han, Y. L., Cui, B. K., and Dai, Y. C. (2018). Morphological and molecular identification of two new Ganoderma species on Casuarina equisetifolia from China. Mycol. Keys 34, 93–108. doi: 10.3897/mycokeys.34.22593
Yang, S., Lee, J., Kwak, J., Kim, K., Seo, M., and Lee, Y. W. (2011). Fungi associated with the traditional starter cultures used for rice wine in Korea. J. Korean Soc. Appl. Biol. Chem. 54, 933–943.
Zhang, Y., Fraatz, M. A., Müller, J., Schmitz, H. J., Birk, F., Schrenk, D., et al. (2015). Aroma characterization and safety assessment of a beverage fermented by Trametes versicolor. J. Agric. Food Chem. 63, 6915–6921. doi: 10.1021/acs.jafc.5b02167
Zheng, X. W., Yan, Z., Han, B. Z., Zwietering, M. H., Samson, R. A., Boekhout, T., et al. (2012). Complex microbiota of a Chinese “Fen” liquor fermentation starter (Fen-Daqu), revealed by culture-dependent and culture-independent methods. Food Microbiol. 31, 293–300. doi: 10.1016/j.fm.2012.03.008
Zhou, B., Ma, C., Wang, H., and Xia, T. (2018). Biodegradation of caffeine by whole cells of tea-derived fungi Aspergillus sydowii, Aspergillus niger and optimization for caffeine degradation. BMC Microbial. 18:53. doi: 10.1186/s12866-018-1194-8
Keywords: filamentous molds, amylolytic starter, India, Mucor, Rhizopus, Aspergillus, Penicillium
Citation: Anupma A and Tamang JP (2020) Diversity of Filamentous Fungi Isolated From Some Amylase and Alcohol-Producing Starters of India. Front. Microbiol. 11:905. doi: 10.3389/fmicb.2020.00905
Received: 01 December 2019; Accepted: 16 April 2020;
Published: 29 May 2020.
Edited by:
Abd El-Latif Hesham, Assiut University, EgyptReviewed by:
Venkataramana M., Independent Researcher, Hyderabad, IndiaCopyright © 2020 Anupma and Tamang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jyoti Prakash Tamang, anlvdGlfdGFtYW5nQGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.