
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 07 May 2020
Sec. Fungi and Their Interactions
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00838
Eukaryotic species are reproductively isolated by sterility barriers that prevent interspecies fertilization (prezygotic sterility barrier) or the fertilization results in infertile offspring (postzygotic sterility barrier). The Saccharomyces species are isolated by postzygotic sterility barriers. Their allodiploid hybrids form no viable gametes (ascospores) and the viable ascospores of the allotetraploids cannot fertilize (conjugate). Our previous work revealed that this mechanism of reproductive isolation differs from those operating in plants and animals and we designated it double sterility barrier (the failure of homeologous chromosomes to pair and the repression of mating by mating-type heterozygosity). Other studies implicated nucleo-mitochondrial incompatibilities in the sterility of the Saccharomyces hybrids, a mechanism assumed to play a central role in the reproductive isolation of animal species. In this project the mitochondrial genomes of 50 cevarum (S. cerevisiae × S. uvarum) hybrids were analyzed. 62% had S. cerevisiae mitotypes, 4% had S. uvarum mitotypes, and 34% had recombinant mitotypes. All but one hybrid formed viable spores indicating that they had genomes larger than allodiploid. Most of these spores were sterile (no sporulation in the clone of vegetative descendants; a feature characteristic of allodiploids). But regardless of their mitotypes, most hybrids could also form fertile alloaneuploid spore clones at low frequencies upon the loss of the MAT-carrying chromosome of the S. uvarum subgenome during meiosis. Hence, the cevarum alloploid nuclear genome is compatible with both parental mitochondrial genomes as well as with their recombinants, and the sterility of the hybrids is maintained by the double sterility barrier (determined in the nuclear genome) rather than by nucleo-mitochondrial incompatibilities. During allotetraploid sporulation both the nuclear and the mitochondrial genomes of the hybrids could segregate but no correlation was observed between the sterility or the fertility of the spore clones and their mitotypes. Nucleo-mitochondrial incompatibility was manifested as respiration deficiency in certain meiotic segregants. As respiration is required for meiosis-sporulation but not for fertilization (conjugation), these segregants were deficient only in sporulation. Thus, the nucleo-mitochondrial incompatibility affects the sexual processes only indirectly through the inactivation of respiration and causes only partial sterility in certain segregant spore clones.
The biological species are defined as groups of interbreeding natural populations that are reproductively isolated from other such groups (Mayr, 1942). Because of the reproductive isolation, interspecies hybridisation is a rare event in most groups of eukaryotes and usually results in infertile offspring. Under this concept, two interbreeding taxa (populations) are conspecific, if they produce fertile hybrids and allospecific, if they fail to produce zygotes (prezygotic sterility barrier) or their hybrids are infertile (postzygotic sterility barrier). Interspecies hybrids occur quite commonly in closely related, vegetatively propagating groups of species such as certain yeasts and fungi (for clonal propagation, gametes are not needed) or tolerate high ploidy (e.g., certain groups of higher plants; allopolyploid plants can produce viable diploid gametes). It is assumed that most extant plant species have emerged via interspecies hybridisation (Soltis et al., 2009).
The budding yeast genus Saccharomyces evolved from an ancient whole genome duplication event (Marcet-Houben and Gabaldon, 2015; Wolfe, 2015) is an excellent model system for the investigation of interspecies hybridisation. It accommodates seven “natural” species and two highly heterogeneous “hybrid” species of diverse chimeric genomes (e.g., Naseeb et al., 2017 and references therein). The species of the genus are reproductively isolated by a postzygotic double sterility barrier (Pfliegler et al., 2012). Under laboratory conditions any of the Saccharomyces species can form hybrids with any other species of the genus (e.g., Banno and Kaneko, 1989; Hawthorne and Philippsen, 1994; Naumov, 1996; Marinoni et al., 1999; Lorenz et al., 2002; Antunovics et al., 2005; Solieri et al., 2005; Rainieri et al., 2008; Bellon et al., 2011, 2013; Garcia Sanchez et al., 2012; Pérez-Través et al., 2014; Lopandic et al., 2016; Karanyicz et al., 2017; Nikulin et al., 2018; Peris et al., 2019; Sipiczki, 2019; Lairón-Peris et al., 2020). The interspecies hybrids are viable but sterile because cannot produce functional gametes (ascospores). The allodiploid two-species hybrids having single sets of parental chromosomes either do not produce ascospores or if they do so, the spores are very rarely viable (e.g., Hawthorne and Philippsen, 1994; Kishimoto, 1994; Hunter et al., 1996; Zambonelli et al., 1997; Marinoni et al., 1999; Liti et al., 2006; Kunicka-Styczyńska and Rajkowska, 2011; Pfliegler et al., 2012).
According to the most widely accepted model, the sporulation deficiency of the Saccharomyces allodiploid hybrids is due to the inability of the chromosomes of the subgenomes to pair correctly in prophase-I of meiosis. The failure of the allosyndetic (homeologous) chromosomes to pair is the first part of the double sterility barrier (for a review, see Sipiczki, 2018). It can be overcome by genome duplication. Much like in the case of the hybrids of many plant species, the duplication of the Saccharomyces hybrid genome “normalizes” meiosis because each chromosome will have a homologous (autosyndetic) partner to pair with. The allotetraploid meiosis produces viable allodiploid spores which, however, are sterile as the allodiploid hybrids. The sterility of the cells produced by these spores (F1 spore clones) is the second part of the double sterility barrier and is due to MAT heterozygosity (Pfliegler et al., 2012) that suppresses the activity of the genes required for mating (fertilization) (Herskowitz, 1988). Thus, genome duplication does not turn the interspecies hybrid fertile. It only provides a possibility for genomic changes that result in fertile alloaneuploid and chimeric derivatives by a process called GARMe (Genome Autoreduction in Meiosis) (Karanyicz et al., 2017). The key event that launches GARMe is the loss of the MATa/MATalpha heterozygosity that unblocks the processes which are required for the cells to function as gametes (an important difference from the allodiploid products of the plant allotetraploid meiosis). The MAT heterozygosity is lost when the meiotic division machinery fails to transfer copies of the MAT-carrying chromosomes of both subgenomes to a spore. The resulting alloaneuploid spore nullisomic for one parental MAT-carrying chromosome forms a clone of fertile vegetative cells with reactivated mating-type switching and mating programs. These cells can mate with each other within the clone and the resulting “intraclonal hybrids” form fertile F2 spore clones. The intraclonal mating and sporulation of the F2 spore clone then will give rise to additional fertile filial generations. Meanwhile additional chromosomes can be lost and recombination can also take place. Thus, GARMe produces diverse chimeric genomes similar to those found in “natural hybrids” of wine yeasts (for a review, see Sipiczki, 2018).
Less is known about the fate of the mitochondrial genome during the process of interspecies hybridisation. Most of the de-novo hybridisation studies found the synthetic hybrids homoplasmic. The hybrid cells received mtDNA from only one of the parental species and the donor was usually the same in all hybrids produced by the same combination of parental strains (e.g., Marinoni et al., 1999; Pulvirenti et al., 2000; De Vero et al., 2003; Antunovics et al., 2005; Lee et al., 2008; Rainieri et al., 2008; Solieri et al., 2008; Garcia Sanchez et al., 2012; Albertin et al., 2013; Karanyicz et al., 2017; Li and Fay, 2017; Origone et al., 2018; Verspohl et al., 2018). For example, in Saccharomyces cerevisiae x S. uvarum hybridisation, the “cevarum” hybrids had S. cerevisiae mitotypes (e.g., Antunovics et al., 2005), whereas in the S. kudriavzevii x S. uvarum combination the resulting “kudvarum” hybrids had S. uvarum mtDNA (Karanyicz et al., 2017). In the synthetic interspecies hybrids an interesting correlation was noticed between the origin of the mitochondrial genome and the higher stability of one of the nuclear subgenomes in GARMe. In cevarum hybrids having S. cerevisiae mitotypes, the S. cerevisiae subgenome was more stable (Antunovics et al., 2005), in kudvarum hybrids having S. uvarum mitotypes, the S. uvarum subgenome was more stable (Karanyicz et al., 2017). Consistent with these findings, a recent study involving six species also found that in the instable hybrids the mitochondrial genome donor tended to retain more nuclear chromosomes (Peris et al., 2019). Similar trends can also be observed in the nature. Recent studies analyzed the mitochondrial genomes of large numbers of chimeric (hybrid) brewing yeasts and also found correlation between mitotypes and the structure of the nuclear genomes, but did not address the impact of the mitotypes on the sexual activities (reproductive barrier) of the strains (Langdon et al., 2019). These observations hint at the possibility that the mitotype of the hybrid may affect the direction of the postzygotic evolution of the hybrid nuclear genome perhaps by impairing the fitness and viability of spores inheriting allospecific (mismatching) combinations of alleles of certain nuclear and mitochondrial genes. This notion fits well with the incompatibilities discovered between certain chromosomal and mitochondrial genes of S. cerevisiae and S. uvarum. For example in a cevarum hybrid having S. cerevisiae mitotype, the lack of the S. cerevisiae AEP2 gene in the nucleus was found to cause respiration deficiency because its S. uvarum ortholog is incompatible with the OLI1/ATP9 gene of the S. cerevisiae mitochondrion (Lee et al., 2008). The S. uvarum Aep2 protein cannot regulate the translation of the S. cerevisiae ATP9 mRNA. Similar incompatibility was detected between the S. uvarum nuclear gene CCM1 and the S. cerevisiae mitochondrion (Jhuang et al., 2017). The Ccm1 protein is required for the maturation of the COX1 mRNA in the mitochondrion but its S. uvarum version has reduced affinity for the S. cerevisiae mRNA. Nucleo-cytoplasmic incompatibility in the opposite direction was also found in cells with incomplete sets of parental chromosomes. The loss of either of the S. uvarum nuclear genes MRS1 and AIM22 in a hybrid having S. uvarum mitotype caused respiration deficiency because their S. cerevisiae orthologs cannot serve the S. uvarum mitochondrion (Chou et al., 2010). The S. cerevisiae Mrs1 protein cannot splice correctly the S. uvarum COX1 transcript. AIM22 codes for an enzyme that lipoylates several mitochondrial enzymes. All these incompatibilities prevent sporulation by causing respiration deficiency. Respiration-deficient (petite) cells cannot sporulate (e.g., Kuenzi et al., 1974) because respiration is necessary for the expression of IME1, a key gene in the regulation of meiosis and sporulation (Treinin and Simchen, 1993). The incompatibility of the S. uvarum mitochondrion with the S. cerevisiae nucleus was reinforced by the respiration deficiency of S. cerevisiae rho strains supplemented with S. uvarum mitochondria (Osuský et al., 1997; Špírek et al., 2000, 2015). However, when examined, the incompatible nuclear genes (alleles) were found recessive. Thus, it is unlikely that these incompatibilities play a significant role in the sterility of the alloploid hybrids because they have also the dominant compatible alleles.
In the present study, we attempt to find correlation between the formation of fertile spores (production of functional gametes) and mitotypes in hybrids of the species S. cerevisiae and S. uvarum, whose nuclear and mitochondrial genomes are most distantly related in the Saccharomyces genus (Kawahara and Imanishi, 2007; Ruan et al., 2017). We selected these species for the experiments because the pioneering studies proposing the involvement of the nucleo-cytoplasmic incompatibility in hybrid sterility (Lee et al., 2008; Chou et al., 2010; Jhuang et al., 2017) were performed with these species and numerous other studies also used these species to investigate the relationship of the nuclear and the mitochondrial genomes (e.g., Osuský et al., 1997; Albertin et al., 2013; Špírek et al., 2015; Verspohl et al., 2018; Li et al., 2019). Besides, we have previously used this species pair to investigate the involvement of the nuclear genome and its changes in the maintenance and break-down of the sterility barrier (Antunovics et al., 2005; Pfliegler et al., 2012).
For the investigation of the mitochondrial genomes in cells of hybrid nuclear genomes, we isolated a large number of cevarum hybrids produced by mass-mating of a S. cerevisiae strain and a S. uvarum strain marker with complementary auxotrophies. We find high mtDNA diversity in the prototrophic hybrids despite the fact that all hybrids were produced by the same pair of parental strains. Consistent with previous observations made on smaller numbers of hybrids, the majority of the cevarum hybrids inherited the mitochondrial genome from S. cerevisiae. Only two hybrids had S. uvarum mitochondria but a surprisingly high number of hybrids had recombinant mtDNAs. In spite of the differences in the structure of their mitochondrial genomes, all but one hybrid formed viable but mostly sterile spores and most of them could also produce fertile spores at low frequencies when losing the MAT-carrying chromosome of the S. uvarum subgenome. During meiosis of the fertile F1 spore clones, the recombinant mitochondrial genomes frequently underwent additional structural modifications resulting in respiration deficiency and concomitant sporulation deficiency in certain F2 spores. The results indicate that the cevarum alloploid nuclear genome is compatible with both parental mitochondrial genomes as well as with their recombinants. Hence, the mitochondrial genome is not directly involved in hybrid sterility but can impair the competitiveness of certain segregants by compromising their respiratory capabilities.
The yeast strains used for hybridisation were 10–170 Saccharomyces cerevisiae MATa leu2 mel– and the homothallic 10–522 S. uvarum ura3 mel+ ts (Antunovics et al., 2005). The strains were maintained at −70 °C and samples of the frozen material were defrosted and propagated on YEA (yeast-extract-agar: 0.5% yeast extract, 1% glucose, 2% agar) or in YEL broth (YEA without agar) to obtain cultures for the experiments. YNBA (yeast-nitrogen-base-agar: 6.7% Bacto Yeast Nitrogen Base, 2% agar) plates supplemented with 2% melibiose were used for sugar assimilation tests. Sporulation was examined on acetate SPA (sporulation agar: 1% K-acetate, 0.1% yeast extract, 0.05% glucose, 2% agar). Hybrids were selected on SMA [synthetic minimal agar medium: 1% glucose, 2% agar, 0.5% (NH4)2SO4, 0.01% KH2PO4, 0.005% MgSO4 and vitamins]. Tests for the presence of auxotrophy markers were performed on SMA supplemented with leucine or uracil or both. The composition of the media was described by Sipiczki and Ferenczy (1977) and Van der Walt and Yarrow (1984).
To obtain prototrophic interspecies hybrids the auxotrophic strains were hybridized in two ways. (1) By mass-mating in intersections of line-shaped cultures replica-plated on SMA plates as described by Sipiczki et al. (2018). The strains to be hybridized were streaked on YEA plates (two strains on one plate). After 3 days of incubation the line-shaped cultures were replica-plated onto fresh plates perpendicularly to each other to produce grids of prints in which the parental strains intersected each other. After 5 days of incubation at 20°C, the “grids” were replica-plated onto SMA plates on which the parental strains could not grow. If hybridisation took place, prototrophic colonies appeared at the intersections. Individual colonies were isolated from the intersections, suspended in sterile water and spread onto fresh SMA plates to obtain pure hybrid cultures. (2) By mass-mating of mixed populations of cells and spores of cultures of the auxotrophic strains in YEL using the slightly modified method described in Antunovics et al. (2005). Cultures of the hybridisation partners, grown on the sporulation medium SPA at 20°C for 3 days, were suspended in YEL. After 1.5 h of incubation at room temperature, samples were spread onto SMA plates. The plates were incubated at 30°C for 5 days. The colonies produced were isolated as putative hybrids. The isolates were kept at −70°C to prevent spontaneous segregation.
For random spore analysis samples of the cultures of the isolates grown on plates of sporulation medium for 5 days at room temperature were suspended in Zymolyase-T20 (0.05 mg ml−1) solution to dissolve the vegetative cells and lyse the walls of the asci. After incubation at 37°C for 1 h, the suspension was sonicated and aliquots were then spread on YEA plates where the viable spores formed colonies. For tetrad analysis samples of the sporulating cultures were suspended in the Zymolyase-T20 solution and incubated at 37°C for 15 min. Aliquots were then streaked on YEA plates, and four-spored asci were pulled from the streaks by micromanipulation. The asci were dissected and their spores were separated from each other on YEA plates to let them form individual colonies. The colonies produced by the spores (spore clones) were tested for the presence of the auxotrophic markers of the parental strains by replica-plating on SMA plates and SMA plates supplemented with the corresponding compounds.
Sporulation was examined microscopically in cultures smeared on SPA plates and inoculated at 25°C for 5 days. The ability to grow at 37°C or on melibiose and glycerol as carbon sources was examined by drop tests on agar plates. Growth at 37°C was tested on YEA plates. Utilization of glycerol and melibiose was examined on YEA containing 3% glycerol instead of glucose and YNB (yeast nitrogen base) supplemented with 2% melibiose, respectively. 0.2 OD suspensions were prepared from overnight cultures of the strains to be tested and 15-μl samples of the suspensions were dropped on the plates. Results were read after 6 days.
Karyotype analysis was performed as described by Antunovics et al. (2005). Chromosomal DNA was prepared in agarose plugs as described by Nguyen and Gaillardin (1997). Plugs were washed in TE and inserted into wells of 1.1% agarose (Chromosomal grade, Bio-Rad) gel prepared in 0.5 × TBE buffer. The chromosomes were separated by electrophoresis in 0.5 × TBE with a CHEF-Mapper apparatus (Bio-Rad). The running parameters were: 200V, linear ramping from 40 to 120 s for 24 h at 14°C. The chromosomal bands were visualized by staining with ethidium-bromide and destaining in sterile water.
Mitochondrial DNA was prepared from exponential-phase YEL cultures with the method described by Nguyen et al. (2000) and digested with HaeIII and MboI. The fragments were separated by electrophoresis in 0.7% agarose, 0.5 × TBE.
Segments of the mitochondrial loci ATP6, COX2, and COX3 were amplified with primers described by Albertin et al. (2013). The ATP6 and COX3 primers allow differentiating the S. uvarum alleles from the S. cerevisiae alleles by the PCR product length (the S. cerevisiae fragments are larger). The COX2 primers amplified fragments of identical length from both species, but they could be differentiated by digestion with the restriction endonuclease SfcI. The PCR reactions were carried out with extracted mtDNA as described previously for genomic DNA (Albertin et al., 2013). The introns of the COX1 gene were amplified with the primers 3L, 3R, 4L, 5R as described by Lopez et al. (2003). PCR fragment sizes were compared by electrophoresis in 1% agarose (w/v), 1 × TBE.
With two methods of mass-mating of auxotrophic parental strains, 10 (method 1) and 40 (method 2) prototrophic colonies were produced (Figure 1). All proved to have alloploid karyotypes consisting of complete parental sets of chromosomes (example is shown in Figure 2). All grew at 37°C (a trait inherited from the S. cerevisiae parent) and assimilated melibiose (a property of the S. uvarum parent). In previous studies both traits were found dominant in interspecies hybrids (e.g., Kishimoto, 1994; Zambonelli et al., 1997; Antunovics et al., 2005; Pfliegler et al., 2012). Thus, the prototrophic colonies were interspecies hybrids.
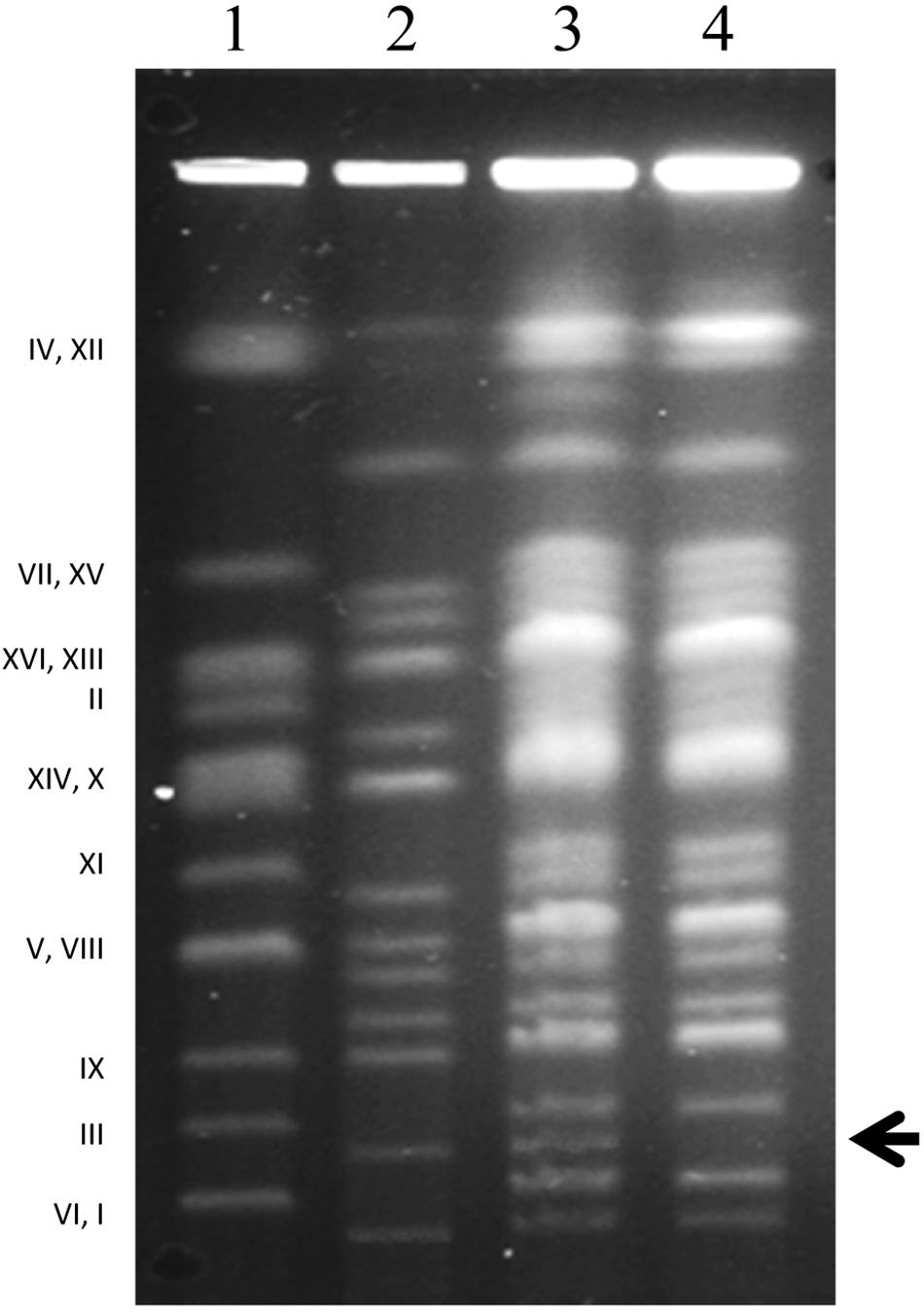
Figure 2. Karyotyping. Chromosomal patterns of S. cerevisiae 10–170 (1), S. uvarum 10-522 (2), hybrid A4 (3) and the fertile F1 spore clone A4.5a of the hybrid A4 (4). Note the missing band in lane 4 (arrowhead), corresponding to the S. uvarum chromosome III (2). Designation (numbering) of S. cerevisiae chromosomes is shown on the left side.
All but one hybrid sporulated (Table 1). The non-sporulating hybrid (A27; category II in Table 1) was most probably allodiploid having single parental sets of chromosomes (haploid subgenomes). Saccharomyces cells having single sets of two different species are unable to produce viable spores, due to the failure of the allosyndetic (homeologous) chromosomes of the subgenomes to pair in meiosis-I (the first sterility barrier; Pfliegler et al., 2012).
The rest of the isolates produced viable spores that germinated and formed colonies (F1 spore clones), albeit with diverse efficiencies. From the viability of the spores we inferred that meiosis took place in their cells. Numerous previous studies demonstrated that alloploid hybrids having genomes higher than diploid can form viable spores (e.g., Banno and Kaneko, 1989; Greig et al., 2002; Pfliegler et al., 2012; Karanyicz et al., 2017; García-Ríos et al., 2019). As meiosis necessitates efficient chromosome pairing, each chromosome in these isolates must have had an autosyndetic (homologous) partner to pair with. Thus, these hybrids were allopolyploids (e.g., most probably allotetraploids or allotriploids). Allotetraploids have two sets of chromosomes in both subgenomes, hence each chromosome has an autosyndetic partner for pairing. Allotriploids can also produce viable spores because their cells have two sets of the chromosomes of one of the parents which can pair within their subgenome. As meiosis produces four ascospores, the spores of a tetraploid yeast cell are diploid (Leupold, 1956); in the case of allotetraploid meiosis the spores are allodiploid. The spores of the allotriploid cells can have haploid sets of chromosomes from the diploid subgenome and randomly distributed chromosomes of the haploid subgenome (alloaneuploid spores). To examine the genomes of the viable spores, we karyotyped certain spore-derived colonies (spore clones) (Figure 1). Almost all had complete sets of chromosomal bands of both parents in their karyotypes, demonstrating that they had at least allodiploid genomes, and the genomes of the hybrids were tetraploid rather than triploid. The exceptions lacked the band corresponding to Chr III of S. uvarum (Figure 2). The different efficiencies of the isolates to produce viable spores, however, hint at the possibility that certain hybrids had allotriploid genomes or their cultures were heterogeneous in ploidy, containing also cells with ploidies different from allotetraploidy. We considered the asci containing four viable spores (complete tetrads) products of allotetraploid cells.
The F1 spore clones that possessed all types of chromosomes of both parents were prototrophic, most probably because they were heterozygous for both recessive auxotrophic markers (leu1– and ura3–) used for hybrid selection. The majority of the hybrids produced only prototrophic spores (categories I and III in Table 1) which corroborates the notion that the allotetraploid meiosis produces allodiploid spores (heterozygous for the markers). However, 17 hybrids segregated. 14 hybrids formed less than 1% (categories IV and V in Table 1 and one hybrid in category I) and 4 hybrids produced more than 30 % auxotrophic spores in random spore analysis (category VI, Table 1). The loss of the wild-type alleles of these markers implies that meiosis failed to transmit copies of all chromosomes into all spores. The auxotrophic spores did not receive copies of the parental chromosomes carrying the wild-type alleles of the markers. Remarkably, most segregating hybrids lost the LEU1 allele of S. uvarum (categories IV and VI, Table 1). Only 3 hybrids produced both leu– and ura– segregants (category V, Table 1).
To verify the association of the loss of marker heterozygosity with the loss of a chromosome, we subjected certain F1 spore clones to karyotyping (Figure 1). The leu– clones lacked the band corresponding to Chr. III of S. uvarum (designated Chr. 2 in Nguyen et al., 2000) (Figure 2), whereas the prototrophs had complete alloploid karyotypes. As Chr. III carries also the MAT locus, we supposed that the loss of the wild-type LEU1 allele was entailed with the simultaneous loss of MAT heterozygosity. In a previous study we noticed that the loss of MAT heterozygosity reactivated mating-type switching and the mating program repressed in the MATa/MATalpha hybrid cells (Pfliegler et al., 2012). Because of these changes, the vegetative descendants of the aneuploid spore (nullisomic for one parental Chr. III) could switch their mating type and conjugate with each other to double their genomes (resulting in incomplete allotetraploids nullisomic for one parental Chr. III). In that study, the outcomes of the process were cells with restored fertility that formed viable F2 spores. To find out if the loss of MAT heterozygosity by the loss of the S. uvarum chromosome Chr. III had a similar effect in the hybrids produced in this study, we isolated F2 spores from certain asci of the F1 leu– spore clones of the four unstable hybrids (Figure 1). The isolated spores were viable and formed colonies (F2 spore clones) (Table 2), which clearly demonstrated that the loss of this chromosome broke down the sterility barrier.
As all karyotyped hybrids had complete sets of parental chromosomes, we asked whether they also had the mitochondrial genomes of both parents. For gross comparison of the structures of the mitochondrial genomes, mtDNA was isolated from the cells and digested with restriction endonucleases (Figure 1). After separation of the fragments by gel electrophoresis, we compared the banding patterns of the hybrids with each other and with those of the parental strains (Figure 3 and Table 3). The banding patterns of the parental strains were clearly different; 30 hybrids had S. cerevisiae-type patterns whereas only 2 hybrids had S. uvarum-type mtDNA patterns. 18 hybrids differed from both parents. One of these hybrids (a3) turned out to be a mixed clone consisting of a subpopulation showing the S. cerevisiae pattern and a subpopulation having a non-parental pattern. The latter subclone and the rest of the group had various combinations of bands corresponding in size to certain parental bands. None of these patterns had all bands of both parents and six patterns contained extra bands not seen in the parents. Altogether eight non-parental band combinations (R1–R8 in Table 3) could be distinguished.
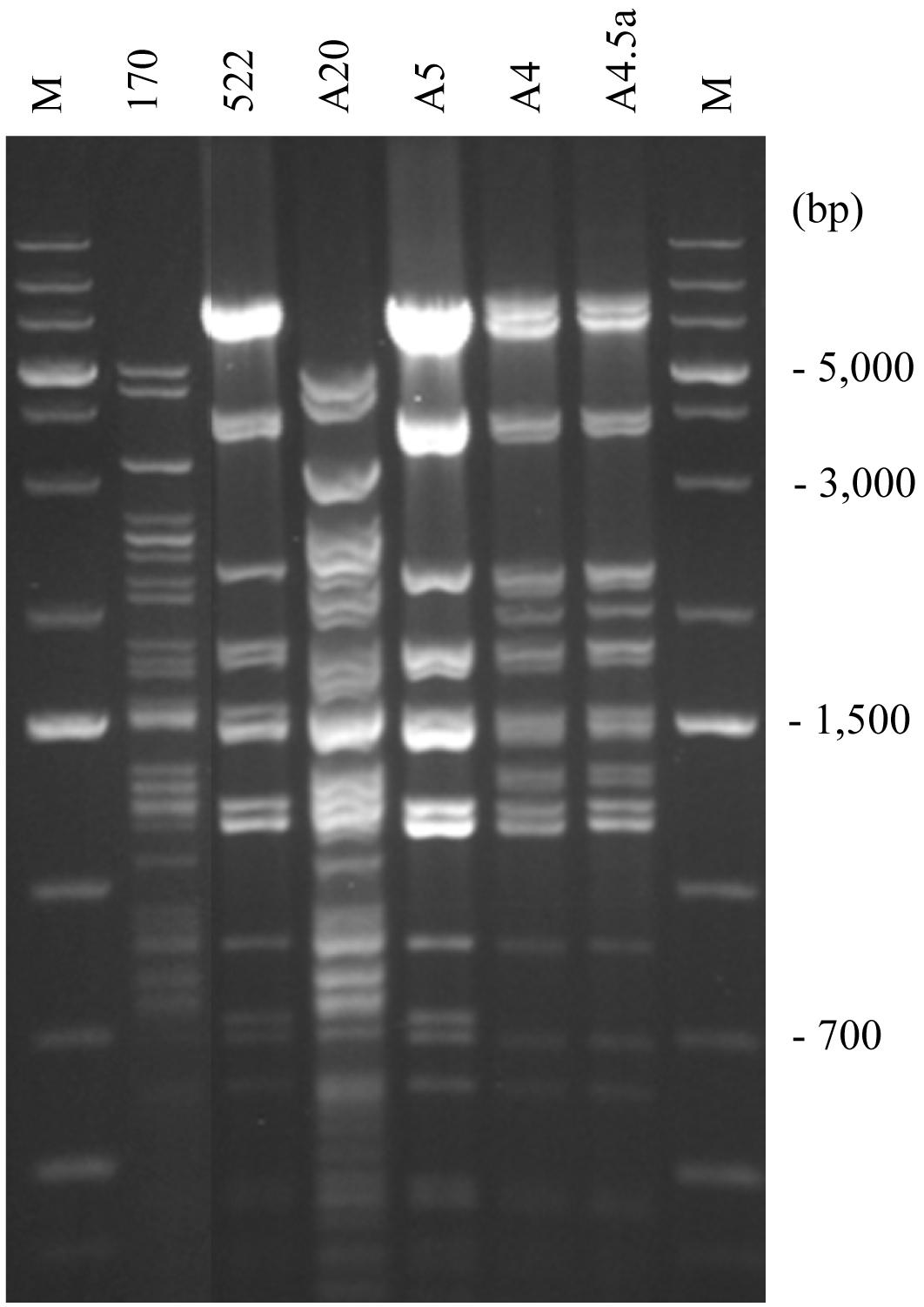
Figure 3. RFLP analysis of mitochondrial genomes. S. cerevisiae 10-170 (170); S. uvarum 10-522 (522); S. cerevisiae mitotype in the hybrid A20 (A20); S. uvarum mitotype in the hybrid A5 (A5); R7-type recombinant mitotype in the hybrid A4 (A4); one of the F1 spore clones of the hybrid A4 (A4.5a); M, size ladder.
Non-parental RFLP patterns can be generated either by rearrangement in one or the other parental mtDNA or by their recombination at conjugation or thereafter, during the vegetative propagation of the hybrid cells. As no drastic genome-size changes were noticed, the more probable mode of interaction was the exchange of segments between the parental mitochondrial genomes via recombination resulting in mosaic (chimeric) genome structures. To examine this possibility, we set out to determine the origin of the loci ATP6, COX1, COX2, and COX3 (Figure 1). These markers were used in previous studies for monitoring the transmission and recombination of mtDNA in yeast hybridisation experiments (e.g., Rainieri et al., 2008; Solieri et al., 2008; Garcia Sanchez et al., 2012; Albertin et al., 2013; Špírek et al., 2015; Verspohl et al., 2018). By amplification with locus-specific primers and restriction analysis of the amplicons, the S. cerevisiae and S. uvarum ATP6, COX2, and COX3 genes can be distinguished (Albertin et al., 2013). All hybrids showing non-parental RFLP patterns had non-recombinant, parental-type ATP6 and COX3 genes (Table 3). Equal numbers of hybrids had S. cerevisiae-type and S. uvarum-type ATP6 genes. All but one of these hybrids had the COX3 gene from S. uvarum. Fourteen hybrids had the S. uvarum version of COX2, three hybrids had recombinant COX2 and only one hybrid had this gene from S. cerevisiae. The origin of the COX1 genes was examined with two pairs of primers. In the four-primer amplification, the parental strains differed in the number and size of the bands. Two hybrids had recombinant COX1 banding patterns; the rest of the hybrids had S. cerevisiae patterns (Figure 4). The analysis of the four loci reinforced that the mitochondrial genomes of non-parental RFLP patterns were recombinants consisting of mosaics of the parental genomes. Recombination took place both in the intergenic regions and within genes (four examples of chimeric genomes are shown in Figure 5).
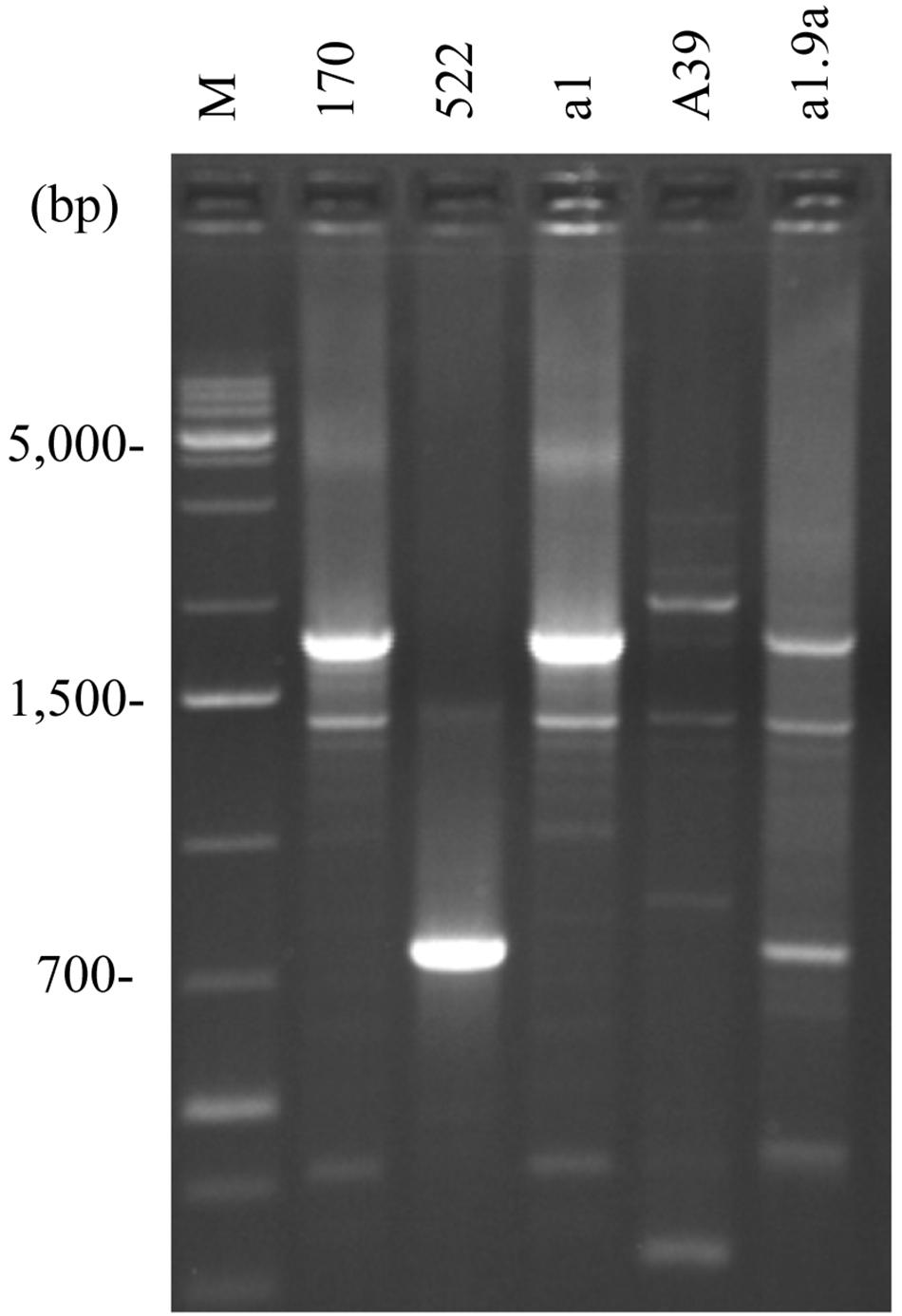
Figure 4. Intron diversity in COX1. S. cerevisiae 10–170 (170); S. uvarum 10–522 (522); hybrid a1 with S. cerevisiae pattern (a1); hybrid A39 with Ri1-type recombinant pattern (A39); F1 spore clone of the hybrid a1 showing Ri3-type recombinant pattern (a1.9a); M, size ladder.

Figure 5. Structure of mitochondrial genomes. S. cerevisiae 170 (S. c.), S. uvarum 522 (S. u.) and four hybrid strains that have recombinant mitochondrial genomes. Markers found are bold.
The hybrid defective in sporulation (A27) had a unique chimeric mtDNA, but as long as more non-sporulating hybrids are not analyzed, it cannot be inferred that this mitochondrial genome structure causes the sporulation deficiency. Viable spores were produced in hybrids with either type of parental mitochondrial genomes and with their various chimeric versions (Table 3). No correlation was found between the mitochondrial genomes and the instability of the nuclear genomes either. The four hybrids of category VI that formed auxotrophic spores at high frequencies had three types of mitochondrial genomes, two of which were parental (Table 2). The three hybrids, which produced both leu– and ura– spores (category V), had S. cerevisiae-type mitochondria, but 37 hybrids belonging to other categories also had this parental mitotype.
Saccharomyces cells gain energy from glucose by glycolysis leading to pyruvate, which is then converted either to ethanol, lactic acid (fermentation) or CO2 (respiration). The latter process requires oxygen and active mitochondria. Cells having functional mitochondria can grow on non-fermentable carbon sources such as glycerol if sufficient amount of oxygen is available. Strains unable to grow on glycerol are considered respiration deficient (Nagai et al., 1961). As the hybrids had diverse mitochondrial genome structures, we tested them for growth on glycerol (Table 3 and Supplementary Figure 1). Both parents and all but one hybrid grew on glycerol as the only carbon source at 25°C. The exception was the non-sporulating hybrid A27. At 37°C, a temperature restrictive for S. uvarum, the S. uvarum parent and certain hybrids did not grow on glycerol or their growth was retarded, indicating that their respiration was temperature sensitive. 37°C inhibited very severely the growth of the III-type hybrids A35, A37, A39, A40, the IV-type hybrids 21, 24 and the VI-type hybrid A4. The III-type hybrid A33 was also retarded in growth at 37°C but less drastically than the previous hybrids. Remarkably, the respiration of the other hybrids of these categories was not sensitive to 37°C. A common feature of all hybrids showing temperature-sensitive respiration was the lack of the S. cerevisiae alleles of ATP6, COX2, and COX3. It is pertinent to mention here that the proteins encoded by these genes in the S. cerevisiae and S. uvarum type strains are less similar than the Cox1 proteins. When comparing the amino acid sequences (EMBOSS Matcher1), we found 93.8, 96.8, and 97.8% interspecies sequence identity for Atp6 (accession numbers: AOT85185.1 and AOT85145.1), Cox2 (accession numbers: AOT85175.1 and AOT85135.1) and Cox3 (accession numbers: AOT85177.1 and AOT85137.1), respectively, whereas the Cox1 proteins were 98.5% identical (accession numbers: AOT85178.1 and AOT85141.1) in sequence. The higher similarity between the Cox1 proteins suggests that the Cox1 orthologs have better interchangeable compatibility for complex IV.
To investigate the stability of the mitochondrial genomes at meiotic division found in previous studies to be prone to destabilize the allotetraploid nuclear genome (for a review, see Sipiczki, 2018), spores were isolated from the asci of certain hybrids (Figure 1). When tested for auxotrophy, the spores of the complete tetrads (four viable spores in the ascus) usually formed prototrophic F1 spore clones or prototrophic and leu– spore clones mostly in 1:1 ratio. The mitochondrial genomes of the clones were then examined with whole-genome RFLP analysis (Figure 6) and the amplification of the genes ATP6, COX1, COX2, and COX3 (examples are shown in Table 4). The hybrids having parental-type mitochondrial genomes usually transmitted their mtDNA unchanged into the ascospores. A5 was an exception because its S. uvarum-type mitochondrial genome underwent changes in certain spores. In contrast, the hybrids which had recombinant mitochondrial genomes produced F1 spore clones with additional modifications in the structure of the mtDNA at higher frequency. In the whole-genome RFLP patterns of their mitochondria, bands were missing or replaced by new bands. These changes were occasionally associated with the loss of one or the other of the genes monitored by PCR-amplification and/or with rearrangements in the intron structure of COX1. However, no correlation was observed between the segregation of the nuclear genome and the changes in the mitochondrial genomes. The loss of the S. uvarum LEU1 allele (by the loss of the S. uvarum chromosome III) did not affect the stability of the mitochondrial genome. As none of the hybrids were heterozygous for any of the examined loci, the changes of their mitotypes during meiosis-sporulation can be attributed to intragenomic rearrangements rather than to recombination between parental mtDNAs.
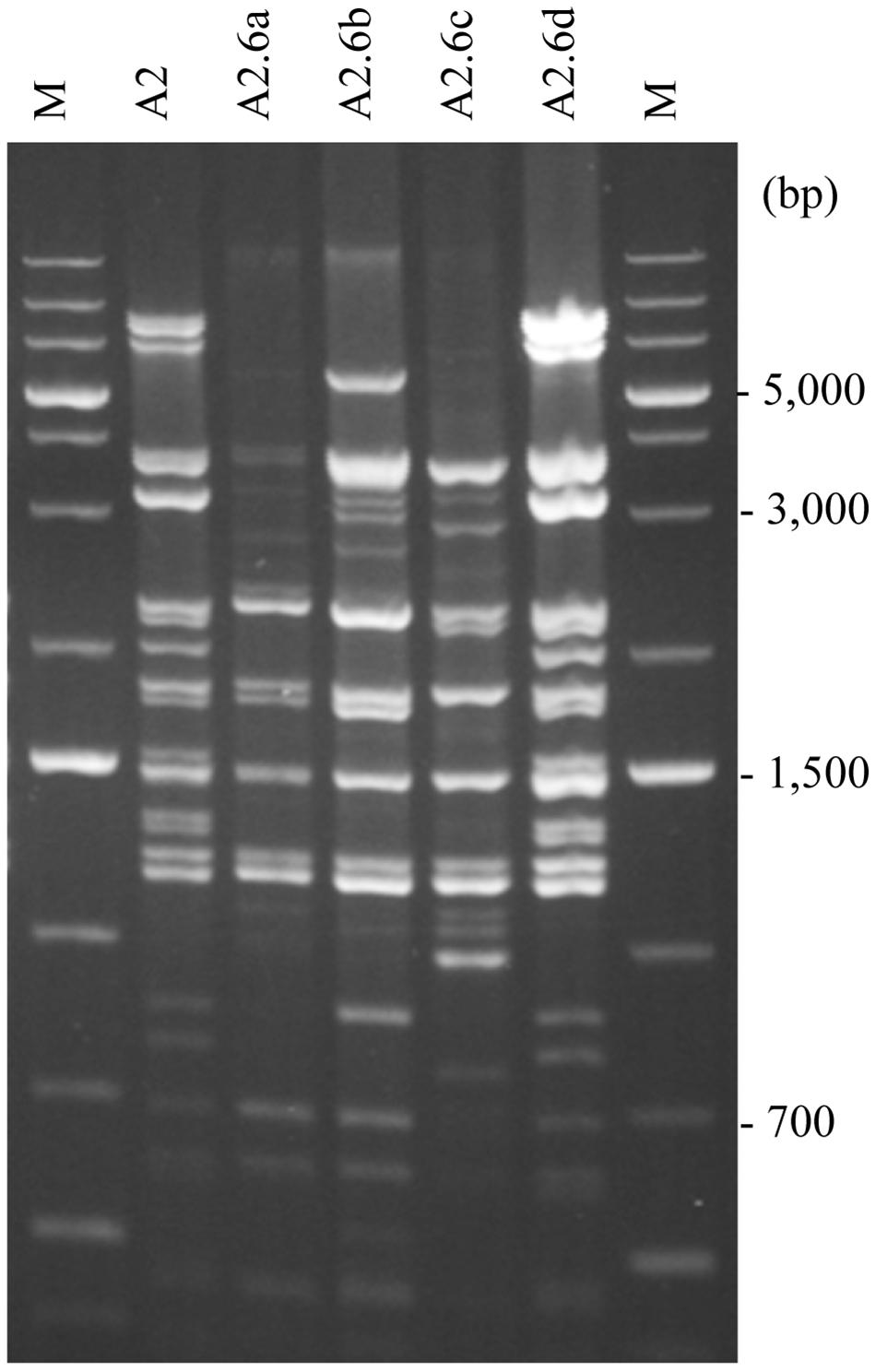
Figure 6. Meiotic segregation of mitotypes. RFLP patterns of the hybrid A2 and its F1 spore clones isolated from a complete tetrad. M, size ladder.
By two methods based on natural mating of mutants with complementary auxotrophy markers, 50 prototrophic cevarum (S. cerevisiae x S. uvarum) hybrids were isolated. One hybrid did not form spores in spite of having complete sets of parental chromosomes. Its sporulation deficiency can be attributed to the first part of the postzygotic double sterility barrier. This barrier operates in allodiploids and is attributable to the inability of the allosyndetic (homeologous) chromosomes of the subgenomes to pair during prophase-I of meiosis (Pfliegler et al., 2012).
The rest of the hybrids produced viable spores, indicating that the first sterility barrier was ineffective in their cells. The spores germinated and developed clones of vegetatively propagating cells (F1 spore clones). Previous studies revealed that allopolyploid (e.g. allotetraploid) Saccharomyces hybrids produce viable ascospores (Banno and Kaneko, 1989; Greig et al., 2002; Sebastiani et al., 2002; Antunovics et al., 2005; Pfliegler et al., 2012) because chromosome pairing becomes can take place within the duplicated subgenomes (Karanyicz et al., 2017). Since the subgenomes of the allotetraploids are autodiploid, the process is called autodiploidisation of the allotetraploid meiosis. The phenomenon was first observed in allotetraploid plant hybrids (for a review, see Ramsey and Schemske, 2002) and recently described also in Saccharomyces interspecies hybrids (Karanyicz et al., 2017). From the viability of their spores we inferred, that the sporulation-proficient hybrids produced in this study had allotetraploid genomes. However, the production of viable spores does not imply that genome duplication overrides the sterility barrier. In a previous study (Pfliegler et al., 2012) we found that the cells of the F1 spore clones were sterile. Being allodiploid, their cells could neither conjugate nor sporulate due to the repression of the genes of the mating (fertilization) process by MATa/MATalpha heterozygosity and the inability of the allosyndetic chromosomes to pair in prophase-I (the second sterility barrier). Consistent with our earlier observations, the F1 spore clones of most sporulation-proficient hybrids of this study failed to produce F2 spore clones. Hence, these hybrids were also sterile.
As the inability of the F1 spore clones to mate is the consequence of the interaction between MATa and MATalpha, the loss of MAT heterozygosity by malsegregation of the MAT-carrying chromosomes of one of the partner genomes during tetraploid meiosis can abolish the second sterility barrier (Pfliegler et al., 2012). Being diplontic organisms, the Saccharomyces strains have a meiotic apparatus optimal for handling diploid sets of chromosomes. When the ploidy increases, the fidelity of meiosis decreases (Loidl, 1995). If the allotetraploid meiotic cell fails to transfer copies of both parental chromosomes carrying the MAT locus into each spore, alloaneuploid spores are formed which have only one MAT allele. The vegetative descendants of these spores can switch their mating types and can mate with other cells of the clone. In our hybrids such alloaneuploids could be identified as leu– spores because the LEU1 gene is on the MAT-carrying chromosome in both parental species, and the S. uvarum subgenome had the wild-type allele of the gene. These spore clones lacked the corresponding S. uvarum chromosomal band in their karyotypes. 34% of the hybrids formed leu– spores at low frequency, but four of them produced more than 30%. These results are consistent with those of previous works which showed that the synthetic polyploid cevarum hybrids occasionally produce fertile spores by losing the MAT-carrying chromosome of the S. uvarum subgenome (Antunovics et al., 2005; Pfliegler et al., 2012).
Consistent with previous reports (e.g., Antunovics et al., 2005; Solieri et al., 2008; Albertin et al., 2013; Karanyicz et al., 2017; Verspohl et al., 2018), all but one hybrid constructed in this study were homoplasmic. 64% of the hybrids had one or the other clearly distinguishable parental-type mtDNA RFLP pattern. The rest had non-parental RFLP patterns and various combinations of the parental orthologs of genes used as markers in the analysis. None of these hybrids had complete parental sets of RFLP bands and both parental versions of the marker genes. The only exception, hybrid a3 formed a mixed culture consisting of cells having S. cerevisiae mtDNA and cells having recombinant mtDNA.
As the cells of different Saccharomyces species conjugate in much the same way as the S. cerevisiae cells, it can be assumed that the transmission of parental mitochondria into the zygotes also takes place similarly in the intra- and the interspecies conjugations. When S. cerevisiae cells conjugate, the zygote contains the mitochondria of both parental cells (reviewed in Birky, 1978). If their mitochondria are different, the zygote is heteroplasmic. After karyogamy, the zygote produces a bud that receives the fused nucleus and copies of both types of mitochondrial genomes. When the bud divides, its vegetative descendants inherit both mitochondrial genomes (biparental inheritance). However, the heteroplasmic state is only transient. After a few rounds of division, the clone becomes homoplasmic (Dujon et al., 1974; Birky et al., 1978a). The results of a recent study of Albertin et al. (2013) suggests that gradual transition from heteroplasmy to homoplasmy takes place also in the interspecies hybrids. They found two types of mitochondria in certain cevarum hybrids after 20 rounds of vegetative divisions but only one type after 80 generations. But they did not examine the possibility that the culture at the 20th generation could have been a mixture of different homoplasmic subpopulations, from which one overgrew the other subpopulations by the 80th generation. Our hybrid a3 indicates that the transition from heteroplasmy to homoplasmy can take place in more than one way within a hybrid culture, converting it in a mixture of different homoplasmic subpopulations.
The mechanism of the heteroplasmy-to-homoplasmy transition is not fully understood but appears to be influenced in S. cerevisiae intraspecies hybrids by several factors such as the position of the bud arising from the zygote (Strausberg and Perlman, 1978; Zinn et al., 1987), difference in the number of mitochondria in the conjugating cells, different copy numbers of the parental mtDNA molecules (Fangman et al., 1989), different density and activity of replication origins in the parental mtDNAs (de Zamaroczy et al., 1981), different level of membrane polarization (due to decreased respiratory chain activity) and the degradation of the less polarized mitochondria by autophagy (Karavaeva et al., 2017). The process can start already in the zygote because heteroplasmic yeast zygotes have upregulated levels of mitochondrial degradation by autophagy (Karavaeva et al., 2017). The model based on the observation that mtDNA replicates in a rolling-circle model on randomly selected mtDNA molecules into the buds can explain the establishment of homoplasmy, but does not explain how the hybrid clone becomes uniformly homoplasmic (Ling and Shibata, 2004).
Similar mechanisms can be assumed to operate in the cevarum hybrids as well. Nevertheless, from the high proportion of hybrids with recombinant mitotypes we infer that both parental mitochondria were present in the cevarum zygotes which then fused to allow their DNAs to recombine. Further we suppose that the subpopulations having recombinant mitochondrial genomes and being more fit in the culturing conditions outcompeted the subpopulations which had parental or less favorable recombinant mitochondrial genomes. The structural diversity of the recombinant mitochondrial genomes can be ascribed to recombination events at random positions. The intensifying research into yeast mitochondrial biology (e.g., Wu et al., 2015; Higuchi-Sanabria et al., 2016; Pernice et al., 2016, 2018; Chakraborty et al., 2017; Guaragnella et al., 2018; Prevost et al., 2018; Ling et al., 2019; Dhar et al., 2019; Peris et al., 2019) will surely provide novel tools for the deciphering of the complexity of the mechanisms underlying the transmission of the mitochondrial genomes into the hybrid bud formed on the zygote and their subsequent evolution.
Sixty six percent of the hybrids had parental mitotypes, either from S. cerevisiae or from S. uvarum, demonstrating that neither parental mitochondrial genome is incompatible with the alloploid nuclear genome. Although both types of hybrids were viable and propagated equally well on glucose-containing media, only two of them had S. uvarum mitotypes. Previous studies including our own work also observed preferential transmission of S. cerevisiae mitochondria to cevarum hybrids (e.g., Marinoni et al., 1999; Delneri et al., 2003; Antunovics et al., 2005; Lee et al., 2008; Solieri et al., 2008; Albertin et al., 2013; Li and Fay, 2017; Origone et al., 2018). Marinoni et al. (1999) found that cevarum hybrids had S. uvarum mitochondria only when the S. cerevisiae parent was petite (did not contain functional mitochondrial genome). Other authors (Solieri et al., 2008; Verspohl et al., 2018) found that the hybrids obtained from mating of the same parental strains always had the same parental mitotype but hybrids of other strains of the species could have mitochondria from the other species. In our hands, the hybrids produced by the same pair of parental strains were heterogeneous. 62% had S. cerevisiae mitotypes, 4% had S. uvarum mitotypes, and 34% had diverse recombinant mitotypes. The observed diversity indicates that neither parent can exclusively determine the hybrid mitotype, although the S. cerevisiae parent is more successful in transferring its mitochondrial genome.
The mating of yeast cells is an isogamic process, resulting in a heteroplasmic zygote containing all mitochondria of the conjugating cells (see above). Why is then the S. cerevisiae mitochondrial genome preferentially transmitted to the hybrid cells? Since the underlying mechanism is not known, we propose two alternative models. One possibility is that certain components of the mechanism controlling the replication of mtDNA promote the transmission of copies of the S. cerevisiae mtDNA into the first bud formed on the zygote. Its preferential transmission could be due to its faster replication. Cardazzo et al. (1998) observed greater mtDNA replication capacity in the S. cerevisiae mitochondrial genome than in the S. uvarum mitochondrial genome. The difference was attributed to the different numbers of replication origins in the two species. The S. cerevisiae mitochondrial genome was estimated to have 7 or 8 replication origins whereas the S. bayanus (uvarum) mitochondrial genome contained only 4 origins (de Zamaroczy et al., 1981; Cardazzo et al., 1998; Piskur et al., 1998). The alternative possibility is that the S. cerevisiae mtDNA is not replicated faster but makes the hybrid cells somewhat more competitive compared to the hybrid cells possessing S. uvarum mtDNA or recombinant mitochondrial genomes. In this model we assume that the bud of the zygote produces heteroplasmic hybrid cells, but these cells then segregate (much like the intraspecies S. cerevisiae hybrids; Birky et al., 1978b). Their culture becomes a mixture of homoplasmic subpopulations having parental and recombinant mitotypes. Those having S. cerevisiae mitotypes may have a better chance to overgrow the other subpopulations on the medium used for hybrid selection. We found two types of mtDNA in the culture of only one hybrid characterized in this study.
One third of the hybrids had non-parental (recombinant) mitotypes. This recombination rate is much higher than those detected previously in cevarum hybrids. da Silva et al. (2015) found no recombinant mitochondria in 28 cevarum hybrids. Albertin et al. (2013) detected recombinant mtDNA in 10% of their cevarum hybrids. Verspohl et al. (2018) found recombinant mitotypes in three out of 83 hybrids. The different results can be attributed to different strains used for hybridisation or to different detection methods. Because of the lower resolution genotyping approaches in the previous studies many recombination events might have gone undetected. Verspohl et al. (2018) included only three genes (COX2, COX3, and ATP6) in the analysis. When we consider only those genes, 5 of our hybrids can be categorized as having S. uvarum mitochondrial genomes. In our analysis we combined whole-genome RFLP with the comparison of the amplified segments of four protein-encoding genes. In S. cerevisiae, mtDNA encodes eight proteins, such as subunits of the respiratory complexes III (Cytb), IV (Cox1, Cox2, and Cox3) and V (Atp6, Atp8, and Atp9) (Rps3/Var1) (Foury et al., 1998). We used 4 of these genes for the detection of recombination. With the combined approach we found mitotypes different from both parental mitotypes at a frequency (34%) comparable to the recombination frequencies found in S. cerevisiae intraspecies crosses. Dujon et al. (1974) described a recombination rate in crosses of S. cerevisiae strains of up to 25%. Recently Wolters et al. (2018) observed that approximately 40% of the mated S. cerevisiae cells contained recombinant mtDNAs.
The astonishingly high proportion of cevarum hybrids with recombinant mitotypes implies that the S. cerevisiae mitochondria can fuse easily with the S. uvarum mitochondria during conjugation to let their DNAs physically interact and then the fused mitochondria divide (fission) to allow the separation of the recombinant (chimeric) genomes from the parental genomes. It has been shown that in vegetatively propagating cells, the mitochondria undergo constant fusion and fission events (Nunnari et al., 1997; Westermann, 2010; Murley et al., 2013; Higuchi-Sanabria et al., 2016). Analogous processes may take place in the cevarum zygotes.
Given that the mitochondrial genomes of the two species differ in size and gene order (e.g., Cardazzo et al., 1998; Ruan et al., 2017), the observed rate is surprisingly high. The mtDNA of the S. uvarum type strain is smaller by nearly one third than the mitochondrial genome of the S. cerevisiae neotype strain (Sulo et al., 2017) and certain segments have different and even inverted positions in the two species (Ruan et al., 2017; Sulo et al., 2017). It can be assumed that because of the synteny differences, multiple recombination events are required for the formation of complete chimeric marker sets (co-segregation of markers inherited from different parents). Interestingly, recombination took place mainly outside of the markers, which might be attributed to the preferential location of recombination hotspots in non-protein-coding regions of the genomes. In S. cerevisiae, recombination occurs mainly in the intergenic GC-rich regions (e.g., Dieckmann and Gandy, 1987; Fritsch et al., 2014). Another interesting feature of the recombinant mitochondrial genomes is the high representation of the S. uvarum alleles of COX2 and COX3. Their high occurrence implies that either recombination does not occur randomly between the mitochondrial genomes of the partner species or the observed allele (ortholog) combinations fit the alloploid nuclear genome better than other combinations.
Viable spores were produced in all but one hybrid regardless of their mitotypes. Thus, the cevarum nuclear genome can accommodate either of the parental mitochondrial genomes and also their recombinant versions with no severe impact on meiosis and sporulation. Nonetheless, the viability of the spores of these hybrids does not contradict the observations that certain nuclear genes of one species are incompatible with the mitochondria of the other species and their incompatibility causes sporulation deficiency (Lee et al., 2008; Chou et al., 2010; Špírek et al., 2015; Jhuang et al., 2017). Those incompatibilities were detected in segregants of sporulation-proficient hybrids and were usually recessive. In the alloploid nuclei of the cevarum hybrids the dominant compatible alleles are also present for both the S. cerevisiae and the S. uvarum mitochondria. The nucleo-mitochondrial conflict therefore cannot play a role in hybrid sterility.
Due to the second sterility barrier, the overwhelming majority of the spores were sterile allodiploids but one third of the hybrids produced also at least a few aneuploid spores that lost the MAT heterozygosity responsible for the sterility of the allodiploid spores. The break-down of the sterility barrier did not correlate with any mitotype; leu– segregants nullisomic for the MAT-carrying chromosome of S. uvarum were produced independently of the mitotypes of the hybrids. This finding indicates that the restoration of fertility by malsegregation in tetraploid meiosis is not affected by the mitotype.
By (taxonomic) definition, S. cerevisiae tolerates high temperatures, whereas S. uvarum (formerly also known as S. bayanus var. uvarum) is inhibited by temperatures above 34°C (Vaughan-Martini and Martini, 1993) but numerous studies described strains of the latter species that could grow also at high temperatures (for a review, see Sipiczki, 2002 and references therein). All hybrids generated in this study grew well on glucose-containing medium at 37°C regardless of their mitotypes. Thus, the mitotype is not involved in the determination of temperature tolerance (of the hybrid) or it may play only a minor role. This finding is consistent with the observation that cevarum hybrids with no functional mitochondria can grow at this temperature (Li et al., 2019) and with the findings suggesting that the main genes determining temperature tolerance are not in the mitochondrial but in the nuclear genome (Serra et al., 2005).
However, this conclusion does not hold true for media containing glycerol as the energy and carbon source. In a recent study, cevarum hybrids with S. uvarum mitotype exhibited reduced growth intensity on a glucose medium and almost no growth on the glycerol medium at 37°C (Li et al., 2019). Unexpectedly, our experiments could not confirm this correlation. Our hybrids showing S. uvarum-type RFLP grew well on both media at 37°C. In contrast, the hybrids that did not grow on glycerol at 37°C had recombinant mitochondria lacking the S. cerevisiae orthologs of the ATP6, COX3, and COX2 genes. But they grew on glycerol at 25°C, indicating that they were not petite. This correlation is consistent with the temperature sensitive glycerol utilization (respiratory growth) recently observed upon the replacement of the S. cerevisiae genes COX1 and ATP6 with their S. uvarum counterparts in the S. cerevisiae mitochondrial genome (Li et al., 2019). The replacement of COX2 and COX3 only reduced the efficiency. Interestingly, the only non-sporulating hybrid found in our study was defective in glycerol utilization at 25°C as well. It had recombinant mtDNA. Albertin et al. (2013) found two hybrids with recombinant mitotypes in a collection of 20 cevarum hybrids. One of them was unable to respire. It is pertinent to note here that a recent study demonstrated that certain allele combinations of mitochondrial genes can impair the growth on glycerol at high temperature even in S. cerevisiae intraspecies hybrids (Wolters et al., 2018).
Because of the autodiploidisation of their meiosis, the allotetraploid cells produce viable but sterile allodiploid F1 spores. As the process is error-prone, fertile alloaneuploid spores are occasionally also formed (see above). The alloaneuploid spore monosomic for the MAT-carrying chromosome produces fertile vegetative cells that are prone to lose additional chromosomes from one or the other subgenome during subsequent meiotic divisions (GARMe) (Karanyicz et al., 2017). If the recurrent loss of chromosomes is accompanied with recombination between allosyndetic (homeologous) chromosomes, GARMe results in chimeric nuclear genomes. In this study we find that the mitochondrial genomes of the cevarum hybrids can also undergo changes during meiosis. The F1 spore clones produced by hybrids of recombinant (chimeric) mitochondrial genomes frequently had whole-genome RFLP patterns different from those of the hybrids. Assuming homoplasmy in these hybrids, we inferred that the pattern changes could be attributed to intramolecular rearrangements and ectopic recombination. The structural alterations of the mtDNA structure were associated with the loss of the ability to utilize glycerol as the only carbon source (petite phenotype) and sporulation deficiency in almost all mitochondrial segregants. Thus, the recombination of the parental mitochondrial genomes during hybridisation generates mtDNA molecules with inherent instability associated with potential fatal consequences on mitochondrial functions. Cevarum hybrids producing F1 spore clones unable to grow on glycerol were previously described by Sebastiani et al. (2002), but the background of the segregation was not investigated.
The postzygotic reproductive isolation of the Saccharomyces species seems to be the result of the combined and synergistic effects of several factors, such as (1) the sequence and structural differences between the homeologous (allosyndetic) chromosomes (impairing chromosome pairing in the prophase of meiosis-I), (2) mating-type heterozygosity (repressing the mating process) and (3) functional incompatibility of certain nuclear and mitochondrial genes (preventing sporulation in certain segregant spore clones by causing respiration deficiency). None of these mechanisms appears sufficient to ensure total reproductive isolation of the species because each can be overcome: (1) genome duplication overrides the chromosomal pairing problem (by allowing autodiploidisation of the allopolyploid meiosis; Karanyicz et al., 2017), (2) loss of MAT heterozygosity (by malsegregation of MAT-carrying chromosomes; Pfliegler et al., 2012) abrogates the suppression of conjugation, and (3) rearrangements (chimerisation) in the hybrid nuclear genome accompanied with recombination of the parental mitochondrial genomes creates segregants avoid of incompatible combinations of nuclear and mitochondrial genes (Lee et al., 2008; Chou et al., 2010) and results of this study.
When concerning the involvement of the mitochondrial genomes in reproductive isolation of species, one has to bear in mind that the nucleo-cytoplasmic incompatibilities do not cause sporulation deficiency directly in the hybrids but only in their segregants (e.g., spore clones) that lose the heterozygosity of the nuclear genes by inheriting only the alleles (orthologs) incompatible with the mitochondrion (Lee et al., 2008; Chou et al., 2010). As only a few incompatible combinations are known (Lee et al., 2008; Chou et al., 2010; Špírek et al., 2015; Jhuang et al., 2017), considerable proportions of the spores can be fertile. These represent escape routes from the sterility caused by nucleo-mitochondrial incompatibility. Besides, even the spores inheriting incompatible gene combinations are only partially sterile, because respiration-deficiency, which prevents sporulation, does not represses mating (Clark-Walker and Miklos, 1975). It has also to be taken into consideration that incompatible alleles occur also in conspecific strains (e.g., Codón et al., 1995) and some interspecies incompatibilities appear to be strain-specific only (Sulo et al., 2003). Figure 7 shows a model of the postzygotic evolution of the nuclear and mitochondrial genomes of cevarum hybrids producing both sterile and fertile segregants due to the break-down of the second sterility barrier. The replacement of mitochondrial genes in certain Saccharomyces wine strains with orthologs from a different species, promiscuously called horizontal gene transfer (HTG) and/or introgression (without any clues on the transfer mechanism) (Peris et al., 2017) can be attributed to processes similar to those depicted in Figure 7. If nucleo-mitochondrial incompatibility made all spores sterile, it would block gene flow between populations of different species (as hypothesized by Chou et al., 2010), but then no Saccharomyces strains with chimeric genomes could have been produced in the nature and no fertile filial spore clones described in studies on synthetic hybrids could be obtained after the loss of MAT heterozygosity (reviewed in Sipiczki, 2018). The nucleo-mitochondrial incompatibility seems to play a minor indirect role in the biological isolation of the Saccharomyces species by rendering the cells of certain spore clones (but not the hybrid cells) deficient in respiration. The non-respiring cells propagate more slowly than the respiring cells.
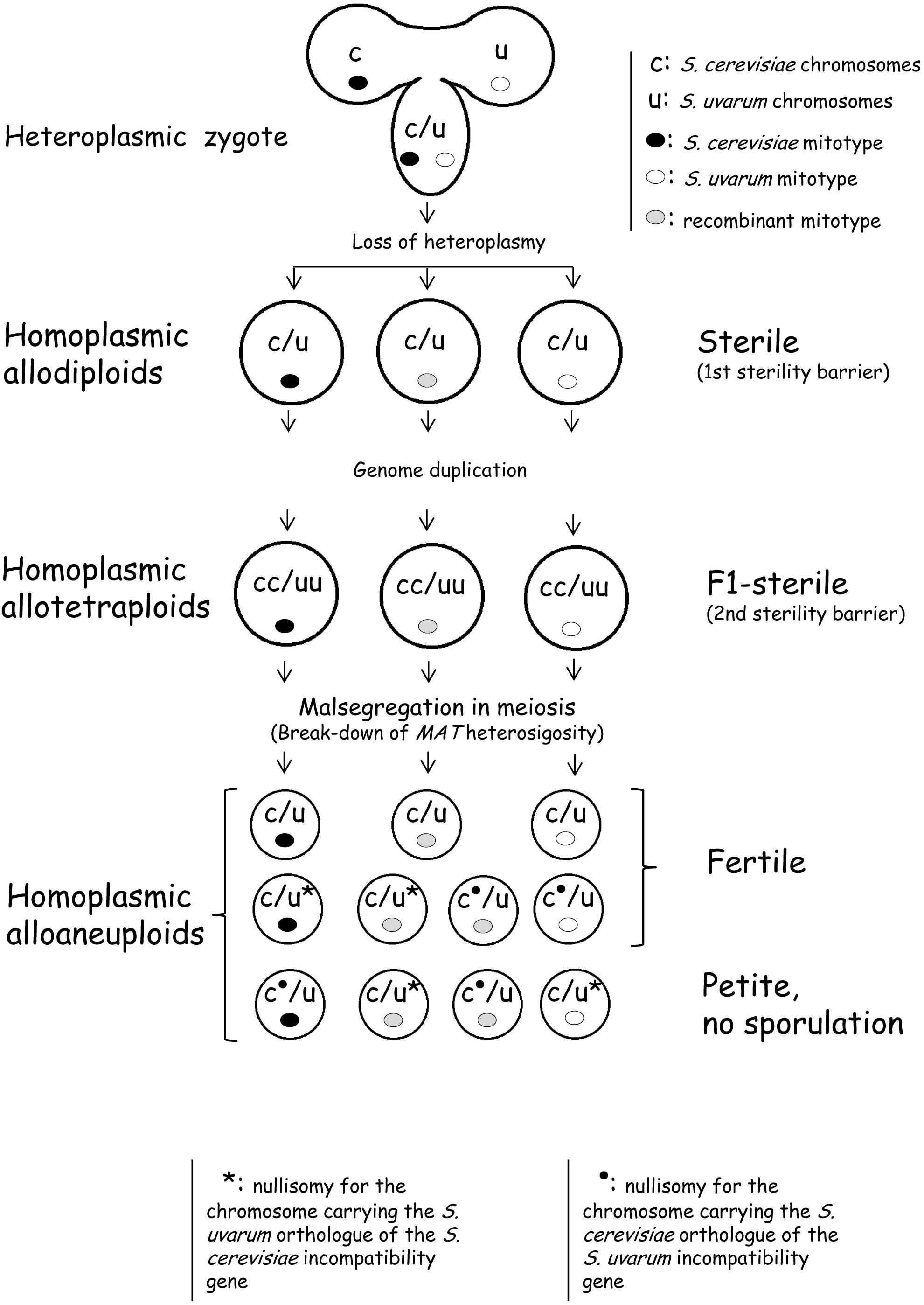
Figure 7. A model depicting the postzygotic evolution of the nuclear and mitochondrial genomes of sterile cevarum hybrids in which the sterility barrier breaks down and both fertile and sterile segregants are formed.
Neither postzygotic reproductive isolation nor mitonuclear incompatibilities are specific to Saccharomyces; both phenomena are known in other groups of fungi as well (e.g., Olson and Stenlid, 2001; Le Gac et al., 2007; De Vienne et al., 2009; Turner et al., 2011; Giordano et al., 2018; Verma et al., 2018) and the MAT loci were also implicated in the isolation of other species (e.g., Ortiz-Merino et al., 2017; Watanabe et al., 2017; Braun-Galleani et al., 2018; Bizzarri et al., 2019). However, the current knowledge on the role and the effect of mitonuclear interactions if fungal hybrids is still limited and mainly focused on virulence (e.g., Olson and Stenlid, 2001; Verma et al., 2018; Giordano et al., 2018).
The results indicate that in the petite-positive genus Saccharomyces, in contrast to metazoans, the nucleo-mitochondrial incompatibility is only marginally involved in the reproductive barrier. The nuclear genome of the cevarum (S. cerevisiae × S. uvarum) hybrid is compatible with both parental mitochondrial genomes and their recombinants. Our previous studies (e.g., Pfliegler et al., 2012) demonstrated that the allopolyploid (allotetraploid) cells produce viable, mostly sterile alloploid (allodiploid) ascospores, but the loss (malsegregation) of the MAT-carrying chromosome of one of the subgenomes during meiosis (loss of MAT-heterozygosity) results in (alloaneuploid) ascospores that can act as gametes because MAT homozygosity reactivates the mating (fertilization) process. The results of this study show that neither the sterility nor the fertility of the ascospores (gametes) can be associated with parental or recombinant mitochondrial genomes. Nucleo-mitochodrial incompatibility was observed only in certain recombinant ascospores as respiration deficiency. As respiration is not essential for life in Saccharomyces, these gametes produce vegetative clones of cells which can fertilize (conjugate with) each other or the cells of other spore clones of different mating type. From these results we concluded that the sterility barrier separating the Saccharomyces species is due to the failure of allosyndetic (homeologous) chromosomes to pair and to mating-type (MAT) heterozygosity (described in detail in our previous works and reviewed in Sipiczki, 2018), rather than to nucleo-mitochondrial incompatibilities. The postzygotic evolution of the mitochondrial genomes including interspecies recombination may have evolutionary relevance and the analysis of an even larger number of hybrids could contribute to the better understanding of the evolutionary processes that have shaped the yeast mitochondrial genomes (Freel et al., 2015; Wolters et al., 2018).
All datasets generated for this study are included in the article/Supplementary Material.
MS and ZA designed the experiments. AS and EK performed the experiments. MS provided funding for this research, wrote the manuscript, and performed the data analysis. AS, EK, and ZA participated in manuscript revision.
This study was funded by the grant K-124417 provided by the National Research, Development and Innovation Office of Hungary.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Anita Kovacs-Bordan for skilful technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00838/full#supplementary-material
Albertin, W., da Silva, T., Rigoulet, M., Salin, B., Masneuf-Pomarede, I., de Vienne, D., et al. (2013). The mitochondrial genome impacts respiration but not fermentation in interspecific Saccharomyces hybrids. PLoS One 8:e75121. doi: 10.1371/journal.pone.0075121
Antunovics, Z., Nguyen, H. V., Gaillardin, C., and Sipiczki, M. (2005). Gradual genome stabilisation by progressive reduction of the Saccharomyces uvarum genome in an interspecific hybrid with Saccharomyces cerevisiae. FEMS Yeast Res. 5, 1141–1150. doi: 10.1016/j.femsyr.2005.04.008
Banno, I., and Kaneko, Y. (1989). A genetic analysis of taxonomic relation between Saccharomyces cerevisiae and Saccharomyces bayanus. Yeast 5, S373–S377.
Bellon, J. R., Eglinton, J. M., Siebert, T. E., Pollnitz, A. P., Rose, L., de Barros Lopes, M., et al. (2011). Newly generated interspecific wine yeast hybrids introduce flavor and aroma diversity to wines. Appl. Microbiol. Biotechnol. 91, 603–612. doi: 10.1007/s00253-011-3294-3
Bellon, J. R., Schmid, F., Capone, D. L., Dunn, B. L., and Chambers, P. J. (2013). Introducing a new breed of wine yeast: interspecific hybridization between a commercial Saccharomyces cerevisiae wine yeast and Saccharomyces mikatae. PLoS One 8:e62053. doi: 10.1371/journal.pone.0062053
Birky, C. W. (1978). Transmission genetics of mitochondria and chloroplasts. Ann. Rev. Genet. 12, 471–512.
Birky, C. W., Demko, C. A., Perlman, P. S., and Strausberg, R. (1978a). Uniparental inheritance of mitochondrial genes in yeast: dependence on input bias of mitochondrial DNA and preliminary investigations of the mechanism. Genetics 89, 615–651.
Birky, C. W., Strausberg, R. L., Forster, J. L., and Perlman, P. S. (1978b). Vegetative segregation of mitochondria in yeast: estimating parameters using a random model. Mol. Gen. Genet. 158, 251–261.
Bizzarri, M., Cassanelli, S., Bartolini, L., Pryszcz, L. P., Dušková, M., Sychrová, H., et al. (2019). Interplay of chimeric mating-type loci impairs fertility rescue and accounts for intra-strain variability in Zygosaccharomyces rouxii interspecies hybrid ATCC42981. Front. Genet. 10:137. doi: 10.3389/fgene.2019.00137
Braun-Galleani, S., Ortiz-Merino, R. A., Wu, Q., Xu, Y., and Wolfe, K. H. (2018). Zygosaccharomyces pseudobailii, another yeast interspecies hybrid that regained fertility by damaging one of its MAT loci. FEMS Yeast Res. 18, foy079. doi: 10.1093/femsyr/foy079
Cardazzo, B., Minuzzo, S., Sartori, G., Grapputo, A., and Carignani, G. (1998). Evolution of mitochondrial DNA in yeast: gene order and structural organization of the mitochondrial genome of Saccharomyces uvarum. Curr. Genet. 33, 52–59. doi: 10.1007/s002940050308
Chakraborty, A., Lyonnais, S., Battistini, F., Hospital, A., Medici, G., Prohens, R., et al. (2017). DNA structure directs positioning of he mitochondrial genome packaging protein Abf2p. Nucleic Acids Res. 45, 951–967. doi: 10.1093/nar/gkw1147
Chou, J. Y., Hung, Y. S., Lin, K. H., Lee, H. Y., and Leu, J. Y. (2010). Multiple molecular mechanisms cause reproductive isolation between three yeast species. PLoS Biol. 8:e1000432. doi: 10.1371/journal.pbio.1000432
Clark-Walker, G. D., and Miklos, G. L. (1975). Complementation in cytoplasmic petite mutants of yeast to form respiratory competent cells. Proc. Natl. Acad. Sci. U.S.A. 72, 372–375. doi: 10.1073/pnas.72.1.372
Codón, A. C., Gasent-Ramírez, J. M., and Benítez, T. (1995). Factors which affect the frequency of sporulation and tetrad formation in Saccharomyces cerevisiae baker’s yeasts. Appl. Environ. Microbiol. 61, 630–638.
da Silva, T., Albertin, W., Dillmann, C., Bely, M., la Guerche, S., Giraud, C., et al. (2015). Hybridization within Saccharomyces genus results in homoeostasis and phenotypic novelty in winemaking conditions. PLoS One 10:e0123834. doi: 10.1371/journal.pone.0123834
De Vero, L., Pulvirenti, A., Gullo, M., Bonatti, P. M., and Giudici, P. (2003). Sorting of mitochondrial DNA and proteins in the progeny of Saccharomyces interspecific hybrids. Ann. Microbiol. 53, 219–231.
De Vienne, D. M., Refrégier, G., Hood, M. E., Guigue, A., Devier, B., Vercken, E., et al. (2009). Hybrid sterility and inviability in the parasitic fungal species complex Microbotryum. J. Evol. Biol. 22, 683–698. doi: 10.1111/j.1420-9101.2009.01702.x
de Zamaroczy, M., Marotta, R., Faugeron-Fonty, G., Goursot, R., Mangin, M., Baldacci, G., et al. (1981). The origins of replication of the yeast mitochondrial genome and the phenomenon of suppressivity. Nature 292, 75–78. doi: 10.1038/292075a0
Delneri, D., Colson, I., Grammenoudi, S., Roberts, I. N., Louis, E. J., and Oliver, S. G. (2003). Engineering evolution to study speciation in yeasts. Nature 422, 68–72. doi: 10.1038/nature01418
Dhar, R., Missarova, A. M., Lehner, B., and Carey, L. B. (2019). Single cell functional genomics reveals the importance of mitochondria in cell-to-cell phenotypic variation. elife 14:8. doi: 10.7554/eLife.38904
Dieckmann, C. L., and Gandy, B. (1987). Preferential recombination between GC clusters in yeast mitochondrial DNA. EMBO J. 6, 4197–4203.
Dujon, B., Slonimski, P. P., and Weill, L. (1974). Mitochondrial genetics. IX. A model for recombination and segregation of mitochondrial genomes in Saccharomyces cerevisiae. Genetics 78, 415–437.
Fangman, W. L., Henly, J. W., Churchill, G., and Brewer, B. J. (1989). Stable maintenance of a 35-base-pair yeast mitochondrial genome. Mol. Cell. Biol. 9, 1917–1921. doi: 10.1128/mcb.9.5.1917
Foury, F., Roganti, T., Lecrenier, N., and Purnelle, B. (1998). The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440, 325–331.
Freel, K. C., Friedrich, A., and Schacherer, J. (2015). Mitochondrial genome evolution in yeasts: an all-encompassing view. FEMS Yeast Res. 15:fov023. doi: 10.1093/femsyr/fov023
Fritsch, E. S., Chabbert, C. D., Klaus, B., and Steinmetz, L. M. (2014). A genome-wide map of mitochondrial DNA recombination in yeast. Genetics 198, 755–771. doi: 10.1534/genetics.114.166637
Garcia Sanchez, R., Solodovnikova, N., and Wendland, J. (2012). Breeding of lager yeast with Saccharomyces cerevisiae improves stress resistance and fermentation performance. Yeast 29, 343–355. doi: 10.1002/yea.2914
García-Ríos, E., Guillén, A., de la Cerda, R., Pérez-Través, L., Querol, A., and Guillamón, J. M. (2019). Improving the cryotolerance of wine yeast by interspecific hybridization in the genus Saccharomyces. Front. Microbiol. 9:3232. doi: 10.3389/fmicb.2018.03232
Giordano, L., Sillo, F., Garbelotto, M., and Gonthier, P. (2018). Mitonuclear interactions may contribute to fitness of fungal hybrids. Sci. Rep. 8:1706. doi: 10.1038/s41598-018-19922-w
Greig, D., Borts, R. H., Louis, E. J., and Travisano, M. (2002). Epistasis and hybrid sterility in Saccharomyces. Proc. R. Soc. Lond. B. 269, 1167–1171. doi: 10.1098/rspb.2002.1989
Guaragnella, N., Coyne, L. P., Chen, X. J., and Giannattasio, S. (2018). Mitochondria-cytosol-nucleus crosstalk: learning from Saccharomyces cerevisiae. FEMS Yeast Res. 18:foy088. doi: 10.1093/femsyr/foy088
Hawthorne, D., and Philippsen, P. (1994). Genetic and molecular analysis of hybrids in the genus Saccharomyces involving S. cerevisiae. S. uvarum and a new species, S. douglasii. Yeast 10, 1285–1296. doi: 10.1002/yea.320101005
Herskowitz, I. (1988). Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol. Rev. 52, 536–553.
Higuchi-Sanabria, R., Charalel, J. K., Viana, M. P., Garcia, E. J., Sing, C. N., Koenigsberg, A., et al. (2016). Mitochondrial anchorage and fusion contribute to mitochondrial inheritance and quality control in the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 27, 776–787. doi: 10.1091/mbc.E15-07-0455
Hunter, N., Chambers, S. R., Louis, E. J., and Borts, R. H. (1996). The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 15, 1726–1733.
Jhuang, H.-Y., Lee, H.-Y., and Leu, J.-Y. (2017). Mitochondrial–nuclear co-evolution leads to hybrid incompatibility through pentatricopeptide repeat proteins. EMBO Rep. 18, 87–101. doi: 10.15252/embr.201643311
Karanyicz, E., Antunovics, Z., Kallai, Z., and Sipiczki, M. (2017). Non-introgressive genome chimerisation by malsegregation in autodiploidised allotetraploids during meiosis of Saccharomyces kudriavzevii x Saccharomyces uvarum hybrids. Appl. Microbiol. Biotechnol. 101, 4617–4633. doi: 10.1007/s00253-017-8274-9
Karavaeva, I. E., Golyshev, S. A., Smirnova, E. A., Sokolov, S. S., Severin, F. F., and Knorre, D. A. (2017). Mitochondrial depolarization in yeast zygotes inhibits clonal expansion of selfish mtDNA. J. Cell. Sci. 130, 1274–1284. doi: 10.1242/jcs.197269
Kawahara, Y., and Imanishi, T. (2007). A genome-wide survey of changes in protein evolutionary rates across four closely related species of Saccharomyces sensu stricto group. BMC Evol. Biol. 7:9. doi: 10.1186/1471-2148-7-9
Kishimoto, M. (1994). Fermentation characteristics of hybrids between the cryophilic wine yeast Saccharomyces bayanus and the mesophilic wine yeast Saccharomyces cerevisiae. J. Ferment. Bioeng. 77, 432–435.
Kuenzi, M. T., Tingle, M. A., and Halvorson, H. O. (1974). Sporulation of Saccharomyces cerevisiae in the absence of a functional mitochondrial genome. J. Bacteriol. 117, 80–88.
Kunicka-Styczyńska, A., and Rajkowska, K. (2011). Physiological and genetic stability of hybrids of industrial wine yeasts Saccharomyces sensu stricto complex. J. Appl. Microbiol. 110, 1538–1549. doi: 10.1111/j.1365-2672.2011.05009.x
Lairón-Peris, M., Pérez-Través, L., Muñiz-Calvo, S., Guillamón, J. M., Heras, J. M., Barrio, E., et al. (2020). Differential contribution of the parental genomes to a S. cerevisiae S. uvarum hybrid, inferred by phenomic, genomic, and transcriptomic analyses, at different industrial stress conditions. Front. Bioeng. Biotechnol. 8:129. doi: 10.3389/fbioe.2020.00129
Langdon, Q. K., Peris, D., Baker, E. P., Opulente, D. A., Nguyen, H. V., Bond, U., et al. (2019). Fermentation innovation through complex hybridization of wild and domesticated yeasts. Nat. Ecol. Evol. 3, 1576–1586. doi: 10.1038/s41559-019-0998-8
Le Gac, M., Hood, M. E., and Giraud, T. (2007). Evolution of reproductive isolation within a parasitic fungal species complex. Evolution 61, 1781–1787. doi: 10.1111/j.1558-5646.2007.00144.x
Lee, H. Y., Chou, J. Y., Cheong, L., Chang, N. H., Yang, S. Y., and Leu, J. Y. (2008). Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135, 1065–1073. doi: 10.1016/j.cell.2008.10.047
Li, X. C., and Fay, J. C. (2017). Cis-regulatory divergence in gene expression between two thermally divergent yeast species. Genome Biol. Evol. 9, 1120–1129. doi: 10.1093/gbe/evx072
Li, X. C., Peris, D., Hittinger, C. T., Sia, E. A., and Fay, J. C. (2019). Mitochondria-encoded genes contribute to evolution of heat and cold tolerance in yeast. Sci. Adv. 5:eaav1848. doi: 10.1126/sciadv.aav1848
Ling, F., Bradshaw, E., and Yoshida, M. (2019). Prevention of mitochondrial genomic instability in yeast by the mitochondrial recombinase Mhr1. Sci. Rep. 9:5433. doi: 10.1038/s41598-019-41699-9
Ling, F., and Shibata, T. (2004). Mhr1p-dependent concatemeric mitochondrial DNA formation for generating yeast mitochondrial homoplasmic cells. Mol. Biol. Cell. 15, 310–322.
Liti, G., Barton, D. B., and Louis, E. J. (2006). Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics 174, 839–850.
Loidl, J. (1995). Meiotic chromosome pairing in triploid and tetraploid Saccharomyces cerevisiae. Genetics 139, 1511–1520.
Lopandic, K., Pfliegler, W., Tiefenbrunner, W., Gangl, H., Sipiczki, M., and Sterflinger, K. (2016). Genotypic and phenotypic evolution of yeast interspecies hybrids during high-sugar fermentation. Appl. Microbiol. Biotechnol. 100, 6331–6343. doi: 10.1007/s00253-016-7481-0
Lopez, V., Fernández-Espinar, M. T., Barrio, E., Ramón, D., and Querol, A. (2003). A new PCR-based method for monitoring inoculated wine fermentations. Int. J. Food Microbiol. 81, 63–71.
Lorenz, A., Fuchs, J., Trelles-Sticken, E., Scherthan, H., and Loidl, J. (2002). Spatial organisation and behaviour of the parental chromosome sets in the nuclei of Saccharomyces cerevisiae x S. paradoxus hybrids. J. Cell. Sci. 115, 3829–3835.
Marcet-Houben, M., and Gabaldon, T. (2015). Beyond the whole-genome duplication: phylogenetic evidence for an ancient interspecies hybridization in the baker’s yeast Lineage. PLoS Biol. 13:e1002220. doi: 10.1371/journal.pbio.1002220
Marinoni, G., Manuel, M., Petersen, R. F., Hvidtfeldt, J., Sulo, P., and Piskur, J. (1999). Horizontal transfer of genetic material among Saccharomyces yeasts. J. Bacteriol. 181, 6488–6496.
Murley, A., Lackner, L. L., Osman, C., West, M., Voeltz, G. K., Walter, P., et al. (2013). ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife 2:e00422. doi: 10.7554/eLife.00422
Nagai, S., Yanagishima, N., and Nagai, H. (1961). Advances in the study of respiration-deficient (RD) mutation in yeast and other microorganisms. Bacteriol. Rev. 25, 404–426.
Naseeb, S., James, S. A., Alsammar, H., Michaels, C. J., Gini, B., Nueno-Palop, C., et al. (2017). Saccharomyces jurei sp. nov., isolation and genetic identification of a novel yeast species from Quercus robur. Int. J. System. Evol. Microbiol. 67, 2046–2052. doi: 10.1099/ijsem.0.002013
Naumov, G. I. (1996). Genetic identification of biological species in the Saccharomyces sensu stricto complex. J. Ind. Microbiol. 17, 295–302. doi: 10.1007/BF01574704
Nguyen, H. V., and Gaillardin, C. (1997). Two subgroups within the Saccharomyces bayanus species evidenced by PCR amplification and restriction fragment length polymorphism of the nontranscribed spacer 2 in the ribosomal DNA unit. Syst. Appl. Micobiol. 20, 286–294.
Nguyen, H. V., Lepingle, A., and Gaillardin, C. A. (2000). Molecular typing demonstrates homogeneity of Saccharomyces uvarum strains and reveals the existence of hybrids between S. uvarum and S. cerevisiae, including the S. bayanus type strain CBS 380. Syst. Appl. Microbiol. 23, 71–85.
Nikulin, J., Krogerus, K., and Gibson, B. (2018). Alternative Saccharomyces interspecies hybrid combinations and their potential for low-temperature wort fermentation. Yeast 35, 113–127. doi: 10.1002/yea.3246
Nunnari, J., Marshall, W. F., Straight, A., Murray, A., Sedat, J. W., and Walter, P. (1997). Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol Cell 8, 1233–1242.
Olson, A., and Stenlid, J. (2001). Plant pathogens: mitochondrial control of fungal hybrid virulence. Nature 411, 438–438.
Origone, A. C., Rodríguez, M. E., Oteiza, J. M., Querol, A., and Lopes, C. A. (2018). Saccharomyces cerevisiae × Saccharomyces uvarum hybrids generated under different conditions share similar winemaking features. Yeast 35, 157–171. doi: 10.1002/yea.3295
Ortiz-Merino, R. A., Kuanyshev, N., Braun-Galleani, S., Byrne, K. P., Porro, D., Branduardi, P., et al. (2017). Evolutionary restoration of fertility in an interspecies hybrid yeast, by whole-genome duplication after a failed mating-type switch. PLoS Biol. 15:e2002128. doi: 10.1371/journal.pbio.2002128
Osuský, M., Kissová, J., and Kováè, L. (1997). Interspecies transplacement of mitochondria in yeasts. Curr. Genet. 32, 24–26.
Pérez-Través, L., Lopes, C. A., Barrio, E., and Querol, A. (2014). Stabilization process in Saccharomyces intra and interspecific hybrids in fermentative conditions. Int. Microbiol. 17, 213–224. doi: 10.2436/20.1501.01.224
Peris, D., Arias, A., Orliæ, S., Belloch, C., Pérez-Través, L., Querol, A., et al. (2017). Mitochondrial introgression suggests extensive ancestral hybridization events among Saccharomyces species. Mol. Phylogenet. Evol. 108, 49–60. doi: 10.1016/j.ympev.2017.02.008
Peris, D., Moriarty, R. V., Alexander, W. G., Wrobel, R. L., and Hittinger, C. T. (2019). Allododecaploid yeasts: synthetic hybrids of six species. BioRxiv [Preprint] doi: 10.1101/597633
Pernice, W. M., Swayne, T. C., Boldogh, I. R., and Pon, L. A. (2018). Mitochondrial tethers and their impact on lifespan in budding yeast. Front. Cell Dev. Biol. 5:120. doi: 10.3389/fcell.2017.00120
Pernice, W. M., Vevea, J. D., and Pon, L. A. (2016). A role for Mfb1p in region-specific anchorage of high-functioning mitochondria and lifespan in Saccharomyces cerevisiae. Nat. Commun. 7:10595. doi: 10.1038/ncomms10595
Pfliegler, W. P., Antunovics, Z., and Sipiczki, M. (2012). Double sterility barrier between Saccharomyces species and its breakdown in allopolyploid hybrids by chromosome loss. FEMS Yeast Res. 12, 703–718. doi: 10.1111/j.1567-1364.2012.00820.x
Piskur, J., Smole, S., Groth, C., Petersen, R. F., and Pedersen, M. B. (1998). Structure and genetic stability of mitochondrial genomes vary among yeasts of the genus Saccharomyces. Int. J. Syst. Bacteriol. 48, 1015–1024.
Prevost, C. T., Peris, N., Seger, C., Pedeville, D. R., Wershing, K., Sia, E. A., et al. (2018). The influence of mitochondrial dynamics on mitochondrial genome stability. Curr. Genet. 64, 199–214. doi: 10.1007/s00294-017-0717-4
Pulvirenti, A., Caggia, C., Restuccia, C., Giudici, P., and Zambonelli, C. (2000). Inheritance of mitochondrial DNA in interspecific Saccharomyces hybrids. Ann. Microbiol. 50, 61–64.
Rainieri, S., Kodama, Y., Nakao, Y., Pulvirenti, A., and Giudici, P. (2008). The inheritance of mtDNA in lager brewing strains. FEMS Yeast Res. 8, 586–596. doi: 10.1111/j.1567-1364.2008.00363.x
Ramsey, J., and Schemske, D. W. (2002). Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 33, 589–639.
Ruan, J., Cheng, J., Zhang, T., and Jiang, H. (2017). Mitochondrial genome evolution in the Saccharomyces sensu stricto complex. PLoS One 12:e0183035. doi: 10.1371/journal.pone.0183035
Sebastiani, F., Barberio, C., Casalone, E., Cavalieri, D., and Polsinelli, M. (2002). Crosses between Saccharomyces cerevisiae and Saccharomyces bayanus generate fertile hybrids. Res. Microbiol. 153, 53–58.
Serra, A., Strehaiano, P., and Taillandier, P. (2005). Influence of temperature and pH on Saccharomyces bayanus var. uvarum growth: impact of a wine yeast interspecific hybridization on these parameters. Int. J. Food Microbiol. 104, 257–265.
Sipiczki, M. (2002). “Taxonomic and physiological diversity of Saccharomyces bayanus,” in Biodiversity and Biotechnology of Wine Yeasts, ed. M. Ciani (Kerala: Research Signpost), 53–69.
Sipiczki, M. (2018). Interspecies hybridisation and genome chimerisation in Saccharomyces: Combining of gene pools of species and its biotechnological perspectives. Front. Microbiol. 9:3071. doi: 10.3389/fmicb.2018.03071
Sipiczki, M. (2019). Yeast two- and three-species hybrids and high-sugar fermentation. Microbial Biotechnol. 2019:5. doi: 10.1111/1751-7915.13390
Sipiczki, M., and Ferenczy, L. (1977). Protoplast fusion of Schizosaccharomyces pombe auxotrophic mutants of identical mating mating-type. Mol. Gen. Genet. 151, 77–81.
Sipiczki, M., Horvath, E., and Pfliegler, W. P. (2018). Birth-and-death evolution and reticulation of ITS segments of Metschnikowia andauensis and Metschnikowia fructicola rDNA repeats. Front. Microbiol. 9:1193. doi: 10.3389/fmicb.2018.01193
Solieri, L., Antúnez, O., Pérez-Ortín, J. E., Barrio, E., and Giudici, P. (2008). Mitochondrial inheritance and fermentative: oxidative balance in hybrids between Saccharomyces cerevisiae and Saccharomyces uvarum. Yeast 25, 485–500. doi: 10.1002/yea.1600
Solieri, L., Gullo, M., De Vero, L., Antúnez, O., Pérez-Ortín, J. E., and Giudici, P. (2005). Homogeneity of interspecific hybrids between Saccharomyces cerevisiae and Saccharomyces uvarum by phenotypic and transcriptional analysis. Int. J. Biotechnol. Biochem. 1, 11–21.
Soltis, D. E., Albert, V. A., Leebens-Mack, J., Bell, C. D., Paterson, A. H., Zheng, C., et al. (2009). Polyploidy and angiosperm diversification. Am. J. Bot. 96, 336–348. doi: 10.3732/ajb.0800079
Špírek, M., Horvath, A., Piskur, J., and Sulo, P. (2000). Functional co-operation between the nuclei of Saccharomyces cerevisiae and mitochondria from other yeast species. Curr. Genet. 38, 202–207.
Špírek, M., Poláková, S., Jatzová, K., and Sulo, P. (2015). Post-zygotic sterility and cytonuclear compatibility limits in S. cerevisiae xenomitochondrial cybrids. Front. Genet. 5:454. doi: 10.3389/fgene.2014.00454
Strausberg, R. L., and Perlman, P. S. (1978). The effect of zygotic bud position on the transmission of mitochondrial genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 163, 131–144.
Sulo, P., Spírek, M., Soltésová, A., Marinoni, G., and Piskur, J. (2003). The efficiency of functional mitochondrial replacement in Saccharomyces species has directional character. FEMS Yeast Res. 4, 97–104.
Sulo, P., Szabóová, D., Bielik, P., Poláková, S., Šoltys, K., Jatzová, K., et al. (2017). The evolutionary history of Saccharomyces species inferred from completed mitochondrial genomes and revision in the ‘yeast mitochondrial genetic code’. DNA Res. 24, 571–583.
Treinin, M., and Simchen, G. (1993). Mitochondrial activity is required for the expression of IME1, a regulator of meiosis in yeast. Curr. Genet. 23, 223–227.
Turner, E., Jacobson, D. J., and Taylor, J. W. (2011). Genetic architecture of a reinforced, postmating, reproductive isolation barrier between Neurospora species indicates evolution via natural selection. PLoS Genet. 7:e1002204. doi: 10.1371/journal.pgen.1002204
Van der Walt, J. P., and Yarrow, D. (1984). “Methods for the isolation, maintenance, classification and identification of yeasts,” in The Yeasts: A Taxonomic Study, ed. N. J. W. Kreger-van Rij (Amsterdam: Elsevier), 45–104.
Vaughan-Martini, A., and Martini, A. (1993). A taxonomic key to the genus Saccharomyces. Syst. Appl. Microbiol. 16, 113–119.
Verma, S., Shakya, V. P. S., and Idnurm, A. (2018). Exploring and exploiting the connection between mitochondria and the virulence of human pathogenic fungi. Virulence 9, 426–446. doi: 10.1080/21505594.2017.1414133
Verspohl, A., Pignedoli, S., and Giudici, G. (2018). The inheritance of mitochondrial DNA in interspecific Saccharomyces hybrids and their properties in winemaking. Yeast 35, 173–187. doi: 10.1002/yea.3288
Watanabe, J., Uehara, K., Mogi, Y., and Tsukioka, Y. (2017). Mechanism for restoration of fertility in hybrid Zygosaccharomyces rouxii generated by interspecies hybridization. Appl. Environ. Microbiol. 83:e01187-17. doi: 10.1128/AEM.01187-17
Westermann, B. (2010). Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11, 872–884. doi: 10.1038/nrm3013
Wolfe, K. H. (2015). Origin of the yeast whole-genome duplication. PLoS Biol. 13:e1002221. doi: 10.1371/journal.pbio.1002221
Wolters, J. F., Charron, G., Gaspary, A., Landry, C. R., Fiumera, A. C., and Fiumera, H. L. (2018). Mitochondrial recombination reveals mito-mito epistasis in yeast. Genetics 209, 307–319. doi: 10.1534/genetics.117.300660
Wu, B., Buljic, A., and Hao, W. (2015). Extensive horizontal transfer and homologous recombination generate highly chimeric mitochondrial genomes in yeast. Mol. Biol. Evol. 32, 2559–2570. doi: 10.1093/molbev/msv127
Zambonelli, C., Passarelli, P., Rainieri, S., Bertolini, L., Giudici, P., and Castellari, L. (1997). Technological properties and temperature response of interspecific Saccharomyces hybrids. J. Sci. Food Agric. 74, 7–12.
Keywords: Saccharomyces, hybrid, sterile, mitotype, fermentation, respiration, alloploid, meiosis
Citation: Szabó A, Antunovics Z, Karanyicz E and Sipiczki M(2020) Diversity and Postzygotic Evolution of the Mitochondrial Genome in Hybrids of Saccharomyces Species Isolated by Double Sterility Barrier. Front. Microbiol. 11:838. doi: 10.3389/fmicb.2020.00838
Received: 17 February 2020; Accepted: 07 April 2020;
Published: 07 May 2020.
Edited by:
Michael H. Perlin, University of Louisville, United StatesReviewed by:
Estéfani García Ríos, Consejo Superior de Investigaciones Científicas (CSIC), SpainCopyright © 2020 Szabó, Antunovics, Karanyicz and Sipiczki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthias Sipiczki, Z2VjZWxhQHBvc2suc2s=; bGlwb3Z5QGdteC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.