- 1Department of Pharmacy, College of Pharmacy, Hanyang University, Ansan, South Korea
- 2Institute of Pharmacological Research, Hanyang University, Ansan, South Korea
- 3Microbiome Laboratory, Rajiv Gandhi Centre for Biotechnology, Thiruvananthapuram, India
Vibrio cholerae O1 serogroup strains have been classified into classical and El Tor biotypes. Cholera, a life-threatening diarrheal disease, can be caused by either biotype through the cholera toxin (CT) that they produce. To increase our knowledge of the pathogenicity of bacteria, we must understand the toxigenicity of bacteria. CT production by classical biotype strains in simple single-phase cell cultures has been established; however, special culture media and growth conditions that are not appropriate for mass production of CT are required to facilitate CT production in El Tor biotype strains. In this report, we produced CT in El Tor biotype strains using simple media and single-phase culture conditions. A single point mutation in ToxT, a transcriptional activator of toxin co-regulated pilus (TCP) and CT, enabled the El Tor biotype strains to produce CT in similar quantities as classical biotype strains in single-phase laboratory culture conditions. CT production capacity varied between El Tor biotype strains. Wave 2 and 3 atypical El Tor strains tended to produce more CT than prototype Wave 1 strains. Wave 2 and 3 strains lack neutral fermentation; however, the capacity for neutral fermentation was not associated with significant differences in CT production by El Tor biotype strains. The Wave 3 strain that caused the 2010 cholera outbreak in Haiti produced CT only when neutral fermentation was abolished. The disparity in CT production between the seventh cholera pandemic strains highlight the differences in virulence between strains and the cause of population changes in V. cholerae.
Introduction
Serogroup O1 and O139 Vibrio cholerae strains cause a fatal diarrheal disease, cholera (Kaper et al., 1995). The symptoms of cholera are induced primarily by the cholera toxin (CT) that is produced by V. cholerae bacteria. The CT gene ctxAB is carried by a filamentous phage, CTXΦ, which can be integrated into both chromosomes in V. cholerae (Waldor and Mekalanos, 1996; Kim et al., 2017). Two biotypes of V. cholerae O1 serogroup strains—classical and El Tor—have been recognized as the causative agents of the first to sixth and the seventh cholera pandemics, respectively (Safa et al., 2010; Kim et al., 2015).
Classical biotype strains produce classical biotype-specific CT, whereas El Tor biotype strains synthesize El Tor biotype-specific CT (Raychoudhuri et al., 2009). The 2 toxins differ by 2 amino acids at positions 39 and 68 in the binding subunit (CTB) of the toxin, but the amino acid sequences are identical in the active subunit (CTA). Recently, atypical El Tor strains that produce classical biotype CT have replaced prototype El Tor strains globally (Kim et al., 2015).
Cholera is caused by the CT that is produced by and secreted from the bacteria in the intestinal environment of human hosts; however, inducing CT production in vitro is challenging. Laboratory conditions for stimulating CT production in classical biotype strains have been established, whereas efforts to establish CT production in El Tor biotype strains in vitro have demonstrated limited success (Ghosh-Banerjee et al., 2010). Culture in media that contain bicarbonate stimulates CT production in El Tor biotype strains (Iwanaga and Yamamoto, 1985; Abuaita and Withey, 2009). An unusual bi-phasic growth condition—described as AKI conditions—has been created for El Tor biotype strains, in which the bacterial culture is kept under static conditions without shaking for 4 h, followed by vigorous shaking for 16 h in AKI media (Iwanaga et al., 1986). However, monitoring CT production and especially, mass production of CT by El Tor biotype strains remains difficult when using these methods.
Recently, we constructed El Tor biotype strains that can be transduced by CTXΦ by inducing the expression of CTX phage receptor, toxin co-regulated pilus (TCP) (Kim et al., 2017). These El Tor strains were constructed by introducing a point mutation in the ToxT transcription factor, which regulates the expression of TCP and CT (Childers et al., 2011). In V. cholerae strains, ToxT typically contains a tyrosine at position 139 (139Y) and does not induce CT and TCP under conventional laboratory conditions; however, CT and TCP expression in El Tor biotype strains has been affected under laboratory conditions when the Tyr-139 is replaced by phenylalanine (139F) (Kim et al., 2017). CT production in El Tor strains that contain the toxT-139F allele was briefly demonstrated under agglutinating conditions (culture at 30°C in media that was adjusted to pH 6.5 with 50 mM Tris buffer), which had been established for classical biotype strains (Davis et al., 1999). Although CT production by classical biotype strains has been optimized under agglutinating conditions, we expected El Tor biotype strains to have different optimal conditions for CT production.
In this study, we determined the optimal conditions for CT production by El Tor biotype strains that contain the toxT-139F allele by varying the pH of the culture medium and growth temperature. CT production in El Tor strains varied, depending on the strain: CT production in prototype (Wave 1) El Tor biotype strains was induced by the toxT-139F allele but was less than 50% of that in classical biotype strains; atypical El Tor strains (Wave 2 and 3) synthesized more CT than Wave 1 strains, with several Wave 2 strains producing 200% more toxin than classical biotype strains under agglutinating conditions. Atypical El Tor strains produced CT at 30°C and 37°C, whereas 30°C was more favorable for toxin production in classical and Wave 1 El Tor biotype strains.
We also constructed a derivative of the classical biotype strain that contained the toxT-139F allele to confirm that this allele also enhances toxin production in classical biotype strains. When compared with CT production by classical biotype strains under agglutinating condition, the engineered classical biotype strain generated 400% more CT in regular LB media and under agglutinating conditions.
Although the CT production condition that we have described in this report might not reflect the physiological conditions in the human host directly, our results provide insights into the pathogenicity of V. cholerae.
Materials and Methods
Bacterial Strains and Bacterial Culture
The bacterial strains used in this study are listed in Table 1.
toxT Allele Exchange
Two 843 bp DNA fragments, encompassing the 50 nucleotides upstream of the translation start codon to nucleotide 793 of toxT, were amplified by PCR using primers the toxT-XbaIF: CCG GCC TCT AGA TAC GTG GAT GGC TCT CTG CG and toxT-SacIR: CCG GCC GAG CTC CAC TTG GTG CTA CAT TCA from strains MG116025 and N16961, respectively. The fragments were inserted into a suicide plasmid, pCVD442. The SNP at nucleotide 416 (A416 in N16961 and T416 in MG116025) lies in the center of these fragments. The toxT-139F allele of MG116025 was replaced by the toxT-139Y allele of N16961 by allelic exchange, and similarly, the toxT-139Y alleles of other strains were replaced with the toxT-139F allele of MG116025 (Donnenberg and Kaper, 1991).
VC1589 Allele Exchange
Using the VC1589-XbaIF (CGG TCT AGA CGG CGG GCG TCA ACT CAA CG) and VC1589-SacIR (CGG GAG CTC TTG GGC GAT AAG TTG TGC TC) primer set, a 1,261-bp fragment that encompassed the functional VC1589 allele from the N16961 Wave 1 El Tor biotype strain and a 1,260-bp fragment that contained the nonfunctional VC1589 allele from the B33 Wave 2 atypical El Tor strain were amplified by PCR and subcloned into the suicide plasmid pCVD442 to generate pCVD442-VC1589-T7 and pCVD442-VC1589-T6, respectively. pCVD442-VC1589-T7 was conjugally transferred to Wave 2 and 3 atypical El Tor strains to replace their nonfunctional VC1589 with a functional VC1589 allele. Similarly, functional VC1589 in Wave 1 El Tor biotype strains was replaced by allelic exchange using pCVD442-VC1589-T6. The replacement of the VC1589 allele in each strain was confirmed by sequencing.
Measurement of Cholera Toxin Production
The GM1 enzyme-linked immunosorbent assay was used to determine the CT production in culture supernatant (Lee et al., 2012). Purified whole CT (Cayman Chemical, Ann Arbor, MI, United States) was used to provide a standard curve. Rabbit polyclonal anti-CTB (GeneTex, Irvine, CA, United States) and anti-rabbit IgG (GeneTex) were used for detection. The optimal laboratory conditions for CT production from classical biotype strains were as follows: culture at 30°C, LB pH 6.5 buffered with 50 mM Tris–which has been described as “agglutinating condition” (Waldor and Mekalanos, 1996). CT production (amount of CT/1 OD600) in the classical biotype strain O395 under such conditions was considered the reference CT production value. CT production from El Tor strains was measured in culture media after 16 h of single-phase shake culture at 30 and 37°C in LB, PBS-buffered LB, AKI media (0.5% NaCl, 0.3% NaHCO3, 0.4% yeast extract, and 1.5% Bacto-Peptone), and LB buffered with 50 mM Tris pH 6.5. CT production (amount of CT/1 OD600) values of mean ± standard deviation in El Tor strains were obtained from three independent experiments and were compared with the reference value. Expression of CT in V. cholerae strains was additionally confirmed by immunoblot analysis using anti-CT (Supplementary Figure S1).
Results
Cholera Toxin Production in Wave 1 El Tor Biotype Strains
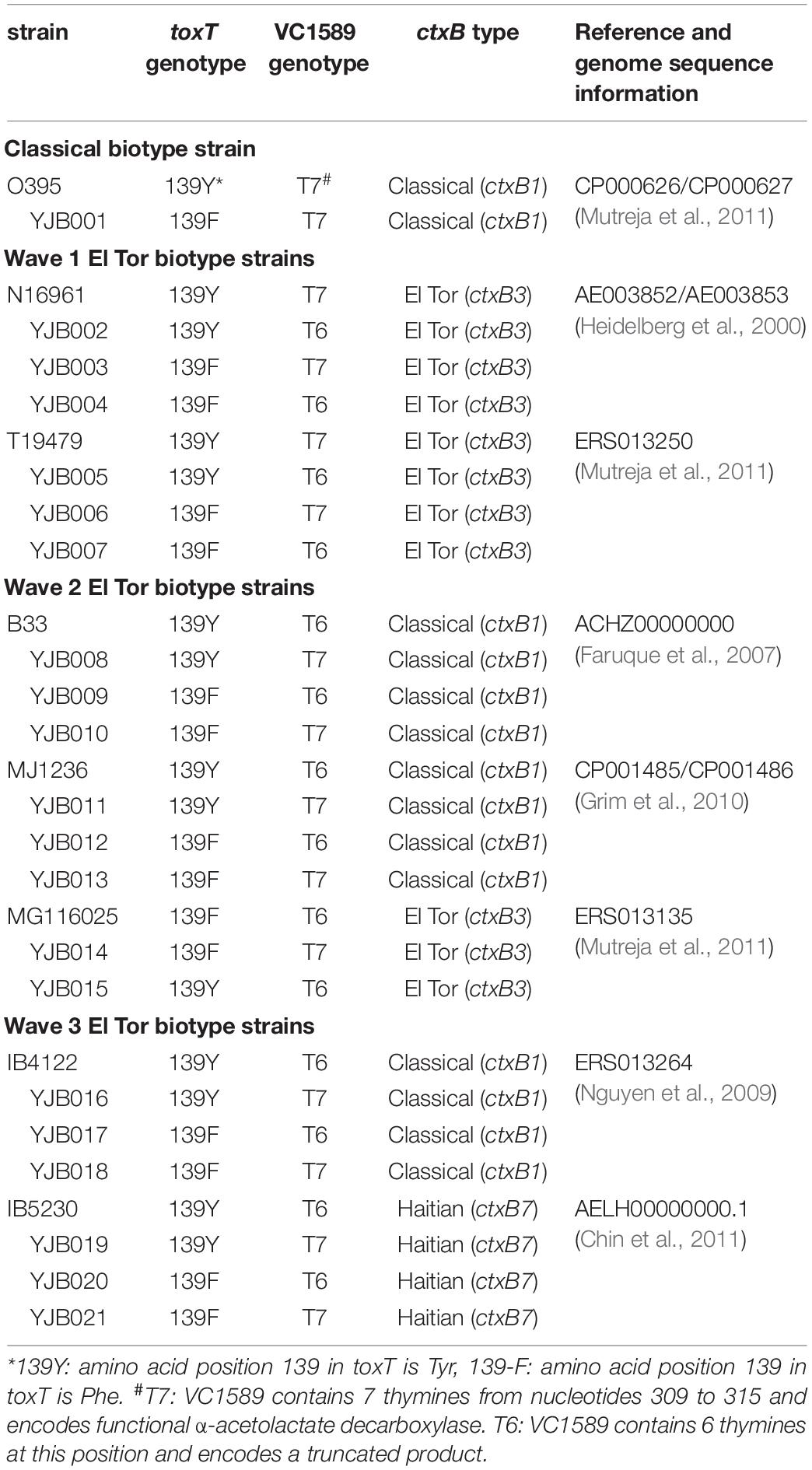
Cholera toxin production by various El Tor biotype strains was compared with that of O395 under agglutinating conditions (reference CT production). V. cholerae strains usually contain the toxT-139Y allele (1 strain, MG116025, reportedly contains the toxT-139F allele), and no CT is produced in El Tor strains under single-phase laboratory shake culture conditions (Kim et al., 2017). We confirmed that regardless of the culture medium, no measurable CT was produced in El Tor strains that carry the toxT-139Y allele, except for 1 Wave 3 strain, described below. On replacing the toxT-139Y allele with toxT-139F in a Wave 1 El Tor biotype strains—T19479-toxT-139F, cultured under agglutinating conditions—CT production was detected (Figure 1), albeit at levels that were less than 40% versus the classical biotype strain, and no CT was produced at 37°C. The function of the neutral fermentation gene (VC1589) did not influence CT production in this strain (Figure 1).
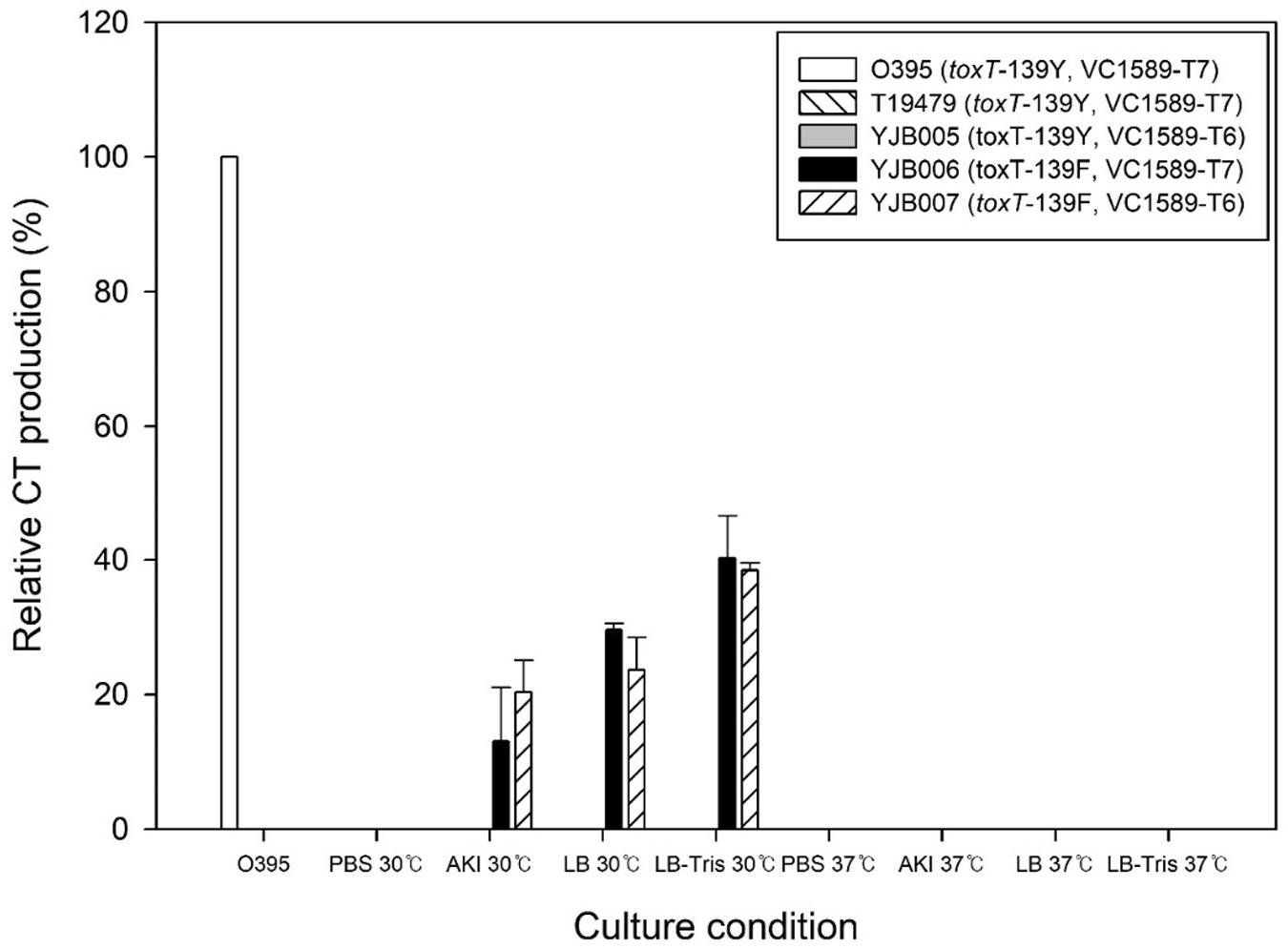
Figure 1. CT production in the Wave 1 strain T19479 and its derivatives. Wave 1 strains primarily contain toxT-139Y and functional VC1589 (VC1589-T7) alleles. Derivatives of T19479 (YJB006 and YJB007), which contain the toxT-139F allele, produced CT in single phase cultures at 30°C in amounts equivalent to approximately 20–40% of the reference value (CT production from classical biotype strain O395 in 30°C in Tris–buffered LB). However, these strains did not produce CT at 37°C. The following media were used: PBS; PBS-buffered LB, AKI; AKI media, LB-Tris; LB buffered with 50 mM Tris pH 6.5.
The other Wave 1 El Tor biotype strain, N16961, which produces CT in amounts that are comparable with classical biotype strains under AKI conditions, did not generate CT in any medium under single-phase culture conditions, despite the toxT-139Y allele being replaced by toxT-139F (Supplementary Figure S2). Based on these results, we determined that Wave 1 El Tor biotype strains can be manipulated to produce CT in single-phase culture.
Cholera Toxin Production in Wave 2 Strains
Three Wave 2 El Tor strains that harbored the toxT-139Y allele—B33, MJ1236, and MG116025—were tested with regard to their CT production. None of these strains generated CT under conditions tested in this study (Figure 2).
Figure 2. CT production in Wave 2 strains and their derivatives. (A) Strain B33 and its derivatives. B33 and its derivative, YJB008, which contains the toxT-Y allele, did not produce CT, whereas the B33 derivatives YJB009 and YJB010 that contained the toxT-139F allele produced CT at up to 200% of the reference value. (B) CT production in the MJ1236 strain and its derivatives was similar to B33 and its derivatives. (C) CT production in the MG116025 strain and its derivatives. MG116025 contained the toxT-139F allele and produced CT under laboratory conditions. No detectable CT was produced in a derivative of MG116025 (YJB015) that contained the toxT-139Y allele. CT produced in MG116025 is the El Tor type (ctxB3), whereas CT produced from two other Wave 2 strains is the classical type (ctxB1). A notable difference in CT production in the Wave 2 strains B33 and MJ1236 compared with Wave 1 strains is that the former produced CT at 37 and 30°C.
Derivatives of B33 That Contain the toxT-139F Allele
B33 (toxT-139Y, VC1589-T6) has a tandem repeat of CTX-2 on chromosome 2. YJB009 (toxT-139F, VC1589-T6) and YJB010 (toxT-139F, VC1589-T7) generally produced as much CT as classical biotype strains in the media that were tested. In cultures in AKI media at 37°C, CT production was approximately twice that of the O395 classical biotype strain. In the same medium, slightly more CT was synthesized at 37°C than 30°C. The function of the V1589 gene (nonfunctional VC1589 in YJB009 and functional VC1589 in YJB010) did not affect CT production significantly (Figure 2A). The CT that was produced from these strains was of the classical type, because the CTX-2 in them contains the classical type ctxB allele (ctxB1).
Derivatives of MJ1236 That Contain the toxT-139F Allele
MJ1236 is closely related to B33, although these strains were isolated in Bangladesh and Mozambique, respectively. They contain the same CTX phage array (a tandem repeat of CTX-2 on chromosome 2). When cultured at 30°C, CT production by YJB012 (toxT-139F, VC1589-T6) and YJB013 (toxT-139F, VC1589-T7), which contained toxT-139F allele, was similar to the reference CT production, regardless of culture medium (Figure 2B). The function of VC1589 (nonfunctional VC1589 in YJB012 and functional VC1589 in YJB013) did not influence CT production significantly in these strains. At 37°C in AKI media, CT production by YJB012 was approximately two-fold that of the reference strain under agglutinating conditions.
MG116025
The original MG116025 strain contains the toxT-139F allele. We constructed a derivative of MG116025, YJB015, that contained toxT-139Y and VC1589-T6 to confirm that native MG116025 produces CT but the strain contained toxT-139Y allele does not (Figure 2C). Another derivative of MG116025 that contained toxT-139Y and functional VC1589 did not produce CT either (data not shown). Although MG116025 is included among Wave 2 atypical El Tor strains, the CT that it produces is the El Tor-type toxin (ctxB3). Approximately equal amounts of CT were produced from MG116025 and the reference O395 at 30°C, whereas CT production at 37°C was negligible (Figure 2C).
In summary, CT production varies between Wave 2 El Tor strains, and approximately twice as much CT can be produced from a derivative of B33 that contains the toxT-139F allele versus the reference classical biotype strain. Although MG116025 is a Wave 2 strain, its pattern of CT production was similar to that of Wave 1 strains, because CT was produced at 30°C. The function of VC1589, and thus the capacity for neutral fermentation, did not affect CT production in Wave 2 strains.
Cholera Toxin Production in Wave 3 Strains
Two Wave 3 atypical El Tor strains were examined. IB4122 was isolated from a cholera outbreak in Northern Vietnam in 2009, and IB5230 was isolated from the 2010 cholera outbreak in Haiti (Nguyen et al., 2009; Chin et al., 2011). Both strains originally contained the toxT-139Y allele.
Derivatives of IB4122 That Contain the toxT-139F Allele
The CT that was produced by this strain was the classical type toxin (ctxB1). Approximately equal amounts of CT, with the reference strain, were produced by YJB017 (toxT-139F, VC1589-T6) and YJB018 (toxT-139F, VC1589-T7), which contained the toxT-139F allele, at 37°C in AKI media; slightly more CT was generated at 37°C than at 30°C (Figure 3A).
Figure 3. CT production in Wave 3 El Tor strains and their derivatives. (A) IB4122 and one of its derivatives (YJB016) that contained the toxT-139Y allele produced no CT. The IB4122 derivatives YJB017 and YJB018 that contained the toxT-139F allele produced CT under laboratory conditions. The optimal conditions for CT production were cultured at 37°C in AKI media. The function of the VC1589 was not associated with significant differences in CT production in YJB017 or YJB018. (B) CT production in IB230 is unique compared with other El Tor strains. IB5230 that contained the toxT-139Y allele and nonfunctional VC1589 produced CT at 37°C in PBS-buffered LB and AKI media, whereas other El Tor biotype strains that contained toxT-139Y did not produce CT. Moreover, YJB020, a derivative of IB5230 that contained the toxT-139F allele and nonfunctional VC1589, produced CT, and YJB021 that contained toxT-139F and functional VC1589 did not produce CT.
IB5230 and Derivatives of IB5230 That Contain the toxT-139F Allele
IB5230 is considered a hypervirulent strain due to its elevated CT production and colonization of the intestine (Satchell et al., 2016; Ghosh et al., 2019). The CT that was produced by this strain is ctxB7, which differs from the classical type ctxB (ctxB1) at amino acid residue 20. However, the secreted form of CT from this strain is the same as classical type CT, because the first 21 amino acids are removed during secretion (Sanchez and Holmgren, 2008).
Notably, although IB5230 harbors the toxT-139Y allele, CT production in PBS-buffered LB or AKI media at 37°C was similar to that by the reference strain (Figure 3B). The function of VC1589 did not affect the production of CT under these culture conditions as similar amount of CT was produced from strain YJB19 (toxT-139Y, and VC1589-T7), implying that IB5230 can be applied directly to the production of CT without alteration in toxT.
Another unique characteristic of IB5230 is that the function of VC1589, and hence the capacity for neutral fermentation, interfered with CT production when the toxT-139Y allele was replaced with toxT-139F. A derivative of IB5230, YJB20, containing the toxT-139F allele and nonfunctional VC1589, produced as much CT as the reference strain, similar to other strains that contain toxT-139F; however, YJB021, which harbors the toxT-139F allele and functional VC1589, did not produce CT under any culture condition (Figure 3B). Although the function of VC1589 affects the fermentation switch in media that has been supplemented with additional glucose, it appears that the operative metabolic pathway also influences toxin production in V. cholerae. More detailed studies of CT production in IB5230 and its derivatives might identify additional important characteristics that underlie the hypervirulent nature of this strain.
Enhanced Cholera Toxin Production in a V. cholerae Classical Biotype Strain
A derivative of O395, YJB001, that contained the toxT-139F allele produced 400% more CT than O395 that harbored the toxT-139Y allele when cultured in regular LB media at 30°C and under agglutinating conditions (Figure 4). More CT was produced at 30°C than 37°C. In conclusion, the toxT-139F allele induces classical V. cholerae strains to produce more CT under laboratory conditions.
Figure 4. CT production in a classical biotype strain O395 and its derivative that contains the toxT-139F allele. YJB001, a derivative of O395 that contained the toxT-139F allele, produced up to 200% CT in PBS-buffered LB and AKI media and up to 400% CT in LB and Tris–buffered LB at 30°C relative to the reference. O395 did not produce detectable amounts of CT at 37°C, but YJB001 produced CT in amounts comparable with the reference at 37°C.
Discussion
The toxigenesis of V. cholerae has received much attention with regard to understanding the virulence of bacteria (Clemens et al., 2017). A protocol for generating CT under laboratory conditions has been developed for classical biotype strains—described as “agglutinating conditions” (DiRita et al., 1996). Several special culture methods have been developed to promote CT production in El Tor biotype strains: the use of a bi-phasic culture method for CT production under the AKI condition (Sanchez et al., 2004); increases in the surface area-to media-volume ratio to stimulate CT production under laboratory conditions (Iwanaga et al., 1986); and a shallow static culture method for CT production (Cobaxin et al., 2014).
However, these culture methods are not appropriate for monitoring toxigenesis in bacteria or large-scale CT production (Cobaxin et al., 2014). In the present study, we developed laboratory culture conditions under which El Tor biotype V. cholerae strains produced CT in a simple single-phase culture.
The expression of CT and TCP is stimulated by ToxT, an AraC/XylS-type transcriptional regulator (Yu and DiRita, 2002; Taylor et al., 2004). toxT expression is tightly regulated by ToxR/ToxS and TcpP/TcpH, which in turn are controlled by various regulatory proteins and stimuli in the intestinal environment (Sanchez and Holmgren, 2008). We identified a V. cholerae Wave 2 atypical El Tor strains, MG116025, that contains a point mutation in toxT (Phe at amino acid position 139, instead of Tyr in most other strains) (Kim et al., 2014, 2017). Amino acid position 139 of ToxT belongs to the N-terminal domain (amino acids 1–160), which has been implicated in regulation of binding to the cognate DNA sites of ToxT (Thomson and Withey, 2014; Thomson et al., 2015). The expression of TCP and CT is higher in this strain, and we anticipated the toxT-139F allele to be useful for examining and producing CT in V. cholerae strains (Kim et al., 2017). As expected, CT production in El Tor strains was stimulated by the same point mutation in toxT, but the toxin production varies by strain. These results also imply that the expression of TCP and CT is constitutively stimulated by pre-existing ToxT but not by newly synthesized ToxT.
We determined the optimum single-phase culture methods for CT production in El Tor strains. A representative Wave 1 El Tor biotype strain, N16961, produces CT under specific culture conditions (Lee et al., 2012; Satchell et al., 2016). However, this strain failed to produce CT under regulation of the toxT-139F allele. When toxT-139F was introduced, another Wave 1 strain, T19479, generated approximately 40% of the CT that was produced by the classical biotype strain O395 under agglutinating conditions. CT production varies between atypical El Tor strains. Atypical El Tor strains tend to produce more CT than prototype El Tor biotype strains under regulation of the toxT-139F allele. A notable characteristic of classical and Wave 1 El Tor strains is that they produce more CT at 30°C than at 37°C, whereas Wave 2 and 3 atypical El Tor strains synthesize similar amounts of CT at both temperatures. This might be an important factor for population changes or the Waves in El Tor biotype strains within the seventh cholera pandemic.
Recently, we found that Wave 2 and 3 atypical El Tor strains lack neutral fermentation due to the loss of function of the acetolactate decarboxylase that is encoded by VC1589 (Lee et al., 2020). This change in phenotype did not affect CT production in most El Tor strains, except for the hypervirulent 2010 Haiti cholera outbreak strain that produced CT only when neutral fermentation was disrupted (Chin et al., 2011). However, acidic fermentation occurred when glucose was sufficiently supplied in the media, and the bacteria did not experience much difference in growth without fortified glucose, regardless of whether the VC1589 was functional. More detailed follow-up studies are required to determine the link between the fermentation pathways and toxigenesis in this strain, especially because it is considered a hypervirulent V. cholerae strain.
We also found that CT production increased approximately four-fold under agglutinating conditions in classical biotype strains when the toxT-139Y allele was replaced by toxT-139F. The optimal culture conditions for CT production by classical biotype strains that contained the toxT-139F allele were regular LB media at 30°C and agglutinating conditions.
Cholera toxin has tremendous potential as a mucosal vaccine adjuvant (Terrinoni et al., 2019). Efforts to develop mutated CT to avoid enterotoxicity when used as a vaccine adjuvant are ongoing. The method that we have established to mass-produce CT might support the development of a potent CT-based vaccine adjuvant (Lebens et al., 2016).
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
EK and DK designed the research. YB, DL, EK, JL, and YY performed the research. GN contributed the bacterial strains. YB, DL, EK, JL, and YY analyzed the data and created figures. EK and DK wrote the manuscript. All authors reviewed the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants NRF-2018R1A2A2A05018341 and NRF-2015M3C9A2054024 from the National Research Foundation (NRF) of Korea.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00825/full#supplementary-material
FIGURE S1 | Immunoblot analysis of cholera toxin production in V. cholerae strains. As a representative, Western blot analysis of CT in strains MG116025 and one of its derivatives YJB014 is shown in this figure. Bacterial culture that contains approximately 5 × 107 cells were loaded onto each lane. Lanes M: Protein molecular weight marker, Lane 1: O395 reference, Lanes 2, 3: MG116025 cultured in PBS-buffered LB at 30°C, Lanes 4, 5: MG116025 cultured in PBS-buffered LB at 37°C, Lanes 6, 7: YJB014 cultured in LB at 30°C, Lanes 8, 9: YJB014 cultured in LB at 37°C. White and black arrows indicate CTA and CTB, respectively.
FIGURE S2 | CT production in Wave 1 strains N16961 and its derivatives. N16961 has been shown to produce CT under the AKI conditions: however, no detectable CT was produced from N16961 or its derivatives that contained toxT-139F allele.
References
Abuaita, B. H., and Withey, J. H. (2009). Bicarbonate induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect. Immun. 77, 4111–4120. doi: 10.1128/IAI.00409-09
Childers, B. M., Cao, X., Weber, G. G., Demeler, B., Hart, P. J., and Klose, K. E. (2011). N-terminal residues of the Vibrio cholerae virulence regulatory protein ToxT involved in dimerization and modulation by fatty acids. J. Biol. Chem. 286, 28644–28655. doi: 10.1074/jbc.M111.258780
Chin, C. S., Sorenson, J., Harris, J. B., Robins, W. P., Charles, R. C., Jean-Charles, R. R., et al. (2011). The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 364, 33–42. doi: 10.1056/NEJMoa1012928
Clemens, J. D., Nair, G. B., Ahmed, T., Qadri, F., and Holmgren, J. (2017). Cholera. Lancet 390, 1539–1549. doi: 10.1016/S0140-6736(17)30559-7
Cobaxin, M., Martinez, H., Ayala, G., Holmgren, J., Sjoling, A., and Sanchez, J. (2014). Cholera toxin expression by El Tor Vibrio cholerae in shallow culture growth conditions. Microb. Pathog. 66, 5–13. doi: 10.1016/j.micpath.2013.11.002
Davis, B. M., Kimsey, H. H., Chang, W., and Waldor, M. K. (1999). The Vibrio cholerae O139 Calcutta bacteriophage CTXΦ is infectious and encodes a novel repressor. J. Bacteriol. 181, 6779–6787.
DiRita, V. J., Neely, M., Taylor, R. K., and Bruss, P. M. (1996). Differential expression of the ToxR regulon in classical and E1 Tor biotypes of Vibrio cholerae is due to biotype-specific control over toxT expression. Proc. Natl. Acad. Sci. U.S.A. 93, 7991–7995. doi: 10.1073/pnas.93.15.7991
Donnenberg, M. S., and Kaper, J. B. (1991). Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59, 4310–4317.
Faruque, S. M., Tam, V. C., Chowdhury, N., Diraphat, P., Dziejman, M., Heidelberg, J. F., et al. (2007). Genomic analysis of the Mozambique strain of Vibrio cholerae O1 reveals the origin of El Tor strains carrying classical CTX prophage. Proc. Natl. Acad. Sci. U.S.A. 104, 5151–5156. doi: 10.1073/pnas.0700365104
Ghosh-Banerjee, J., Senoh, M., Takahashi, T., Hamabata, T., Barman, S., Koley, H., et al. (2010). Cholera toxin production by the El Tor variant of Vibrio cholerae O1 compared to prototype El Tor and classical biotypes. J. Clin. Microbiol. 48, 4283–4286. doi: 10.1128/JCM.00799-10
Ghosh, P., Sinha, R., Samanta, P., Saha, D. R., Koley, H., Dutta, S., et al. (2019). Haitian variant Vibrio cholerae O1 strains manifest higher virulence in animal models. Front. Microbiol. 10:111. doi: 10.3389/fmicb.2019.00111
Grim, C. J., Hasan, N. A., Taviani, E., Haley, B., Chun, J., Brettin, T. S., et al. (2010). Genome sequence of hybrid Vibrio cholerae O1 MJ-1236, B-33, and CIRS101 and comparative genomics with V. cholerae. J. Bacteriol. 192, 3524–3533. doi: 10.1128/JB.00040-10
Heidelberg, J. F., Eisen, J. A., Nelson, W. C., Clayton, R. A., Gwinn, M. L., Dodson, R. J., et al. (2000). DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483. doi: 10.1038/35020000
Iwanaga, M., and Yamamoto, K. (1985). New medium for the production of cholera toxin by Vibrio cholerae O1 biotype El Tor. J. Clin. Microbiol. 22, 405–408.
Iwanaga, M., Yamamoto, K., Higa, N., Ichinose, Y., Nakasone, N., and Tanabe, M. (1986). Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol. Immunol. 30, 1075–1083. doi: 10.1111/j.1348-0421.1986.tb03037.x
Kim, E. J., Lee, C. H., Nair, G. B., and Kim, D. W. (2015). Whole-genome sequence comparisons reveal the evolution of Vibrio cholerae O1. Trends Microbiol. 23, 479–489. doi: 10.1016/j.tim.2015.03.010
Kim, E. J., Lee, D., Moon, S. H., Lee, C. H., Kim, S. J., Lee, J. H., et al. (2014). Molecular insights into the evolutionary pathway of Vibrio cholerae O1 atypical El Tor variants. PLoS Pathog. 10:e1004384. doi: 10.1371/journal.ppat.1004384
Kim, E. J., Yu, H. J., Lee, J. H., Kim, J. O., Han, S. H., Yun, C. H., et al. (2017). Replication of Vibrio cholerae classical CTX phage. Proc. Natl. Acad. Sci. U.S.A. 114, 2343–2348. doi: 10.1073/pnas.1701335114
Lebens, M., Terrinoni, M., Karlsson, S. L., Larena, M., Gustafsson-Hedberg, T., Kallgard, S., et al. (2016). Construction and preclinical evaluation of mmCT, a novel mutant cholera toxin adjuvant that can be efficiently produced in genetically manipulated Vibrio cholerae. Vaccine 34, 2121–2128. doi: 10.1016/j.vaccine.2016.03.002
Lee, D., Kim, E. J., Baek, Y., Lee, J., Yoon, Y., Nair, G. B., et al. (2020). Alterations in glucose metabolism in Vibrio cholerae serogroup O1 El Tor biotype strains. Sci. Rep. 10:308. doi: 10.1038/s41598-019-57093-4
Lee, K. M., Park, Y., Bari, W., Yoon, M. Y., Go, J., Kim, S. C., et al. (2012). Activation of cholera toxin production by anaerobic respiration of Trimethylamine N-oxide in Vibrio cholerae. J. Biol. Chem. 287, 39742–39752. doi: 10.1074/jbc.M112.394932
Mutreja, A., Kim, D. W., Thomson, N. R., Connor, T. R., Lee, J. H., Kariuki, S., et al. (2011). Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477, 462–465. doi: 10.1038/nature10392
Nguyen, B. M., Lee, J. H., Cuong, N. T., Choi, S. Y., Hien, N. T., Anh, D. D., et al. (2009). Cholera outbreaks caused by an altered Vibrio cholerae O1 El Tor biotype strain producing classical cholera toxin B in Vietnam in 2007 to 2008. J. Clin. Microbiol. 47, 1568–1571. doi: 10.1128/jcm.02040-08
Raychoudhuri, A., Patra, T., Ghosh, K., Ramamurthy, T., Nandy, R. K., Takeda, Y., et al. (2009). Classical ctxB in Vibrio cholerae O1, Kolkata, India. Emerg. Infect. Dis. 15, 131–132. doi: 10.3201/eid1501.080543
Safa, A., Nair, G. B., and Kong, R. Y. (2010). Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 18, 46–54. doi: 10.1016/j.tim.2009.10.003
Sanchez, J., and Holmgren, J. (2008). Cholera toxin structure, gene regulation and pathophysiological and immunological aspects. Cell. Mol. Life Sci. 65, 1347–1360. doi: 10.1007/s00018-008-7496-5
Sanchez, J., Medina, G., Buhse, T., Holmgren, J., and Soberon-Chavez, G. (2004). Expression of cholera toxin under non-AKI conditions in Vibrio cholerae El Tor induced by increasing the exposed surface of cultures. J. Bacteriol. 186, 1355–1361. doi: 10.1128/jb.186.5.1355-1361.2004
Satchell, K. J., Jones, C. J., Wong, J., Queen, J., Agarwal, S., and Yildiz, F. H. (2016). Phenotypic analysis reveals that the 2010 Haiti cholera epidemic is linked to a hypervirulent strain. Infect. Immun. 84, 2473–2481. doi: 10.1128/IAI.00189-16
Taylor, R. K., Kirn, T. J., Meeks, M. D., Wade, T. K., and Wade, W. F. (2004). A Vibrio cholerae classical TcpA amino acid sequence induces protective antibody that binds an area hypothesized to be important for toxin-coregulated pilus structure. Infect. Immun. 72, 6050–6060. doi: 10.1128/IAI.72.10.6050-6060
Terrinoni, M., Holmgren, J., Lebens, M., and Larena, M. (2019). Proteomic analysis of cholera toxin adjuvant-stimulated human monocytes identifies Thrombospondin-1 and Integrin-beta1 as strongly upregulated molecules involved in adjuvant activity. Sci. Rep. 9:2812. doi: 10.1038/s41598-019-38726-0
Thomson, J. J., Plecha, S. C., and Withey, J. H. (2015). A small unstructured region in Vibrio cholerae ToxT mediates the response to positive and negative effectors and ToxT proteolysis. J. Bacteriol. 197, 654–668. doi: 10.1128/jb.02068-14
Thomson, J. J., and Withey, J. H. (2014). Bicarbonate increases binding affinity of Vibrio cholerae ToxT to virulence gene promoters. J. Bacteriol. 196, 3872–3880. doi: 10.1128/jb.01824-14
Waldor, M. K., and Mekalanos, J. J. (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272, 1910–1914. doi: 10.1126/science.272.5270.1910
Keywords: Vibrio cholerae, cholera, biotype, cholera toxin, El Tor biotype
Citation: Baek Y, Lee D, Lee J, Yoon Y, Nair GB, Kim DW and Kim EJ (2020) Cholera Toxin Production in Vibrio cholerae O1 El Tor Biotype Strains in Single-Phase Culture. Front. Microbiol. 11:825. doi: 10.3389/fmicb.2020.00825
Received: 14 November 2019; Accepted: 07 April 2020;
Published: 05 May 2020.
Edited by:
Mattias Collin, Lund University, SwedenReviewed by:
Jeffrey H. Withey, Wayne State University, United StatesNityananda Chowdhury, Medical University of South Carolina, United States
Copyright © 2020 Baek, Lee, Lee, Yoon, Nair, Kim and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong Wook Kim, ZG9uZ3dvb2tAaGFueWFuZy5hYy5rcg==; Eun Jin Kim, ZWpraW0wODE2QGhhbnlhbmcuYWMua3I=
†These authors have contributed equally to this work
 Yeongjun Baek1,2†
Yeongjun Baek1,2† Dong Wook Kim
Dong Wook Kim