
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 May 2020
Sec. Extreme Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00824
This article is part of the Research Topic Polar and Alpine Microbiological and Biogeochemical Processes in the Warming World View all 13 articles
 Noor Hassan1,2
Noor Hassan1,2 Alexandre M. Anesio3
Alexandre M. Anesio3 Muhammad Rafiq1,2,4
Muhammad Rafiq1,2,4 Jens Holtvoeth5
Jens Holtvoeth5 Ian Bull5
Ian Bull5 Abdul Haleem1
Abdul Haleem1 Aamer Ali Shah1
Aamer Ali Shah1 Fariha Hasan1*
Fariha Hasan1*Bacteria inhabiting non-polar glaciers are exposed to large variations in temperature, which significantly affects the fluidity of bacterial cell membranes. In order to maintain normal functions of the cell membranes, psychrophilic bacteria adapt by changing the composition of cell membrane fatty acids. However, information on the exact pattern of cell membrane adaptability in non-polar low-temperature habitats is scarce. In the present study, 42 bacterial strains were isolated from the Ghulmet, Ghulkin, and Hopar glaciers of the Hunza Valley in the Karakoram Mountain Range, Pakistan and their cell membrane fatty acid distributions studied, using gas chromatography/mass spectrometry (GC-MS) for the analysis of fatty acid methyl esters (FAMEs) liberated by acid-catalyzed methanolysis. Furthermore, Gram-negative and Gram-positive groups were grown under different temperature settings (5, 15, 25, and 35°C) in order to determine the effect of temperature on cell membrane (CM) fatty acid distribution. The analyses identified the major groups of cell membrane fatty acids (FA) as straight-chain monounsaturated fatty acids (n-MUFAs) and branched fatty acids (br-FAs), accounting for more than 70% of the fatty acids analyzed. The distribution of br-FAs and n-FAs in bacterial cell membranes was significantly affected by temperature, with the level of br-FAs decreasing relative to n-FAs with increasing temperature. Notably, the production of polyunsaturated fatty acids (PUFAs) was only seen at lower temperatures. This study contributes to understanding, for the first time, the role of br-FAs in the maintenance of cell membrane fluidity of bacteria inhabiting non-polar habitats.
By far, the largest part of the biosphere on Earth, which includes the oceans, is exposed to temperatures below 5°C either perennially or seasonally (Margesin et al., 2007; Margesin and Miteva, 2011; Hoshino and Matsumoto, 2012). Arctic and Antarctic habitats, ranging from high mountains to the deep ocean, experience subzero temperatures as they are deprived of direct sunlight. Among these cold habitats the deep sea represents the major component (almost 71% of the Earth is covered by ocean), with 90% of the volume of the ocean having a temperature of ∼5°C. Other large cold habitats include snow, permafrost, sea ice and glacial habitats (35, 24, 13, and 10% coverage of the Earth surface, respectively) in addition to various other cold habitats such as lakes, cold soils (especially surface soils), cold deserts, and caves (Singh et al., 2006; Margesin and Miteva, 2011).
Bacteria inhabiting cold environments face prolonged frigid temperatures and daily freeze-thaw cycles (Montiel, 2000). They have to cope with special challenges to thrive under subzero temperatures, such as slow chemical reaction rates and limited enzyme activity, denaturation of proteins, increased water viscosity, decreased cell membrane fluidity (Russell, 1990; Crowe et al., 1992; Hassan et al., 2016) and limited water availability as a solvent for biochemical reactions (Wynn-Williams, 1990). However, bacteria have remarkable adaptive abilities that enable them to survive harsh and highly variable environmental conditions, including changes in pH and temperature (Ganzert et al., 2011; Bajerski and Wagner, 2013). These adaptation mechanisms include the expression of heat/cold shock proteins, production of protective compatible solutes or an altered metabolism (Georlette et al., 2004). Another strategy employed by bacteria is the modification of the cell membrane structure as it is involved in important metabolic processes and is a vital interface in the electron transport chain (Denich et al., 2003).
Homeoviscous adaptation, i.e., the alteration of cell membrane composition to maintain cell membrane fluidity in response to environmental changes, has long been observed in bacterial communities (Sinensky, 1974) and in bacteria involved in the direct synthesis of fatty acids, with diverse melting points, to maintain the required physical properties needed of membrane lipids (Mansilla et al., 2004). An increase in the amount of low-melting-point fatty acids (such as monounsaturated fatty acids, polyunsaturated fatty acids and branched-chain fatty acids relative to their saturated straight analogs) provides adequate membrane fluidity in almost all organisms (Fulco, 1983; Hazel and Williams, 1990; Suutari and Laakso, 1994). Low temperature and high pressure play a significant role in probing the integrity of membranes, with evidence even for irreversible change, i.e., from a fluid to a rigid state (Cossins and MacDonald, 1984; Hazel and Williams, 1990). Bacterial adaptation to low temperature and high pressure typically involves an increased proportion of branched chain fatty acids (BCFA) and unsaturated fatty acids (UFA) into membrane phospholipids (DeLong and Yayanos, 1986; Wirsen et al., 1986; Nichols et al., 1997). However, to the best of our knowledge, these results have only been obtained by studying Gram-negative bacteria residing in deep sea or a very few Antarctic habitats. The current study aims to characterize the cell membrane fatty acid profiles of Gram-positive and Gram-negative psychrotolerant bacteria isolated from non-polar glaciers and determine the role of saturated and unsaturated, straight and BCFAs in bacterial adaptation to low temperatures.
Samples (ice, melted water, and sediment) were collected from three different glaciers, Ghulkin (36.42791 N, 74.80659 E), Ghulmet (36°12.474 N, 74°29.035 E) and Hopar (36.2108228 N, 74.7724664 E) of the Hunza Valley, Karakoram Mountains Range, Pakistan using Nasco Whirl–Pak bags and bottles (Fisher Scientific). A total of 42 different bacterial strains were isolated using R2A and Nutrient agar at 5, 15, and 35°C as incubation temperatures. Culture of each bacteria isolate was then preserved in 35% glycerol and stored at −20°C prior to further analysis.
The genomic DNA of all the bacteria isolates was extracted using the Invitrogen PureLink Microbiome DNA Kit (Invitrogen) following instructions given by manufacturer. After that 16S rRNA gene for all of the bacterial isolates was amplified using primers 27F and 1492R. PCR conditions used for 16S rRNA amplification were adjusted as preliminary denaturation at 94°C for 5 min, then 40 cycles of 94, 56, and 72°C each for 30 s and a final step of extension at 72°C for 8 min. PCR products were then purified using QIAquick PCR Purification Kit (QIAGEN) and sequenced from MRC PPU DNA sequencing and services, University of Dundee, United Kingdom. Subsequently, obtained sequences were trimmed via BioEdit software and submitted to GenBank for getting accession numbers (Tables 1, 2).
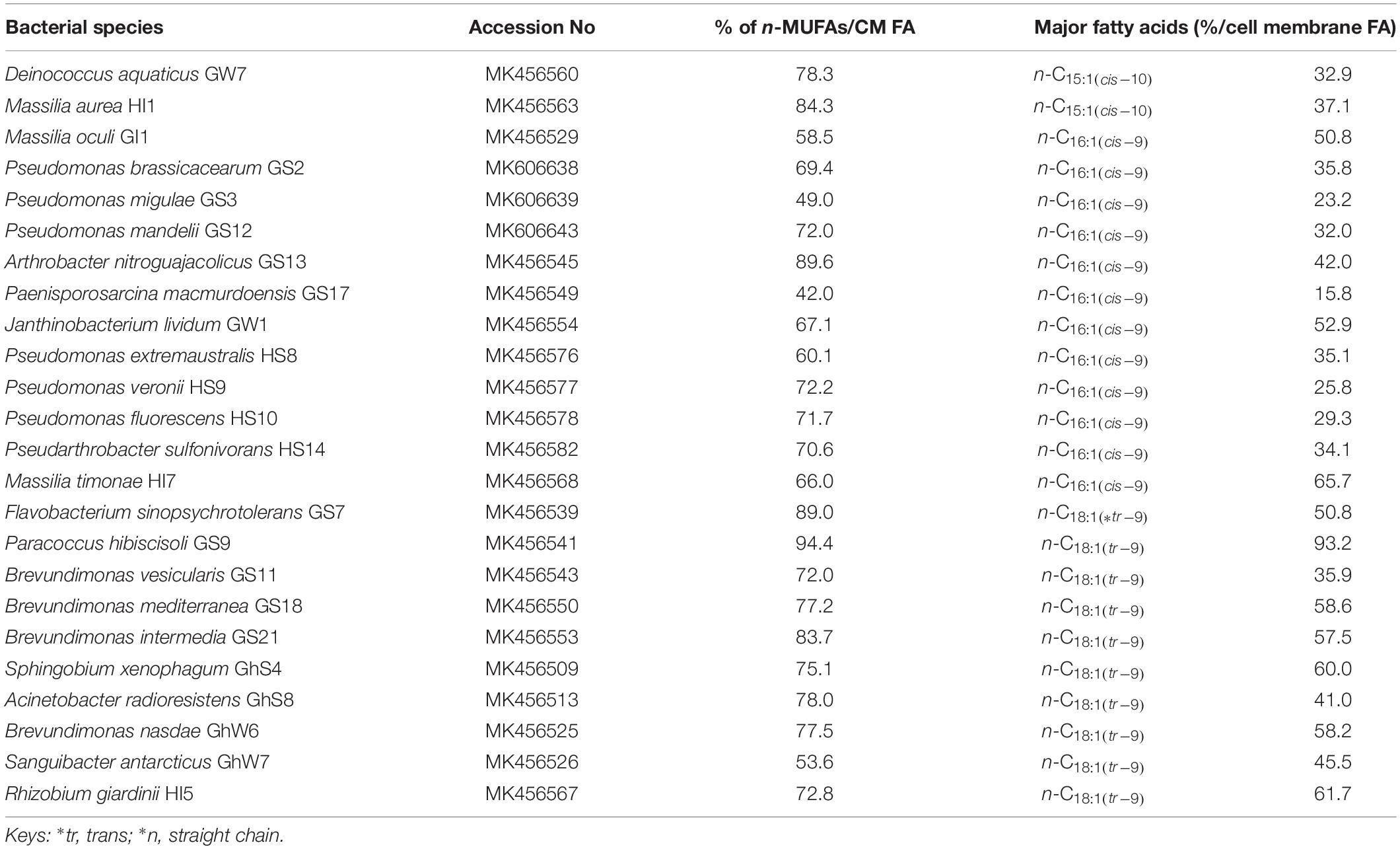
Table 1. List of bacteria species produced straight chain monounsaturated fatty acids (∗n-MUFAs) as major group of cell membrane (CM) fatty acids (FA).
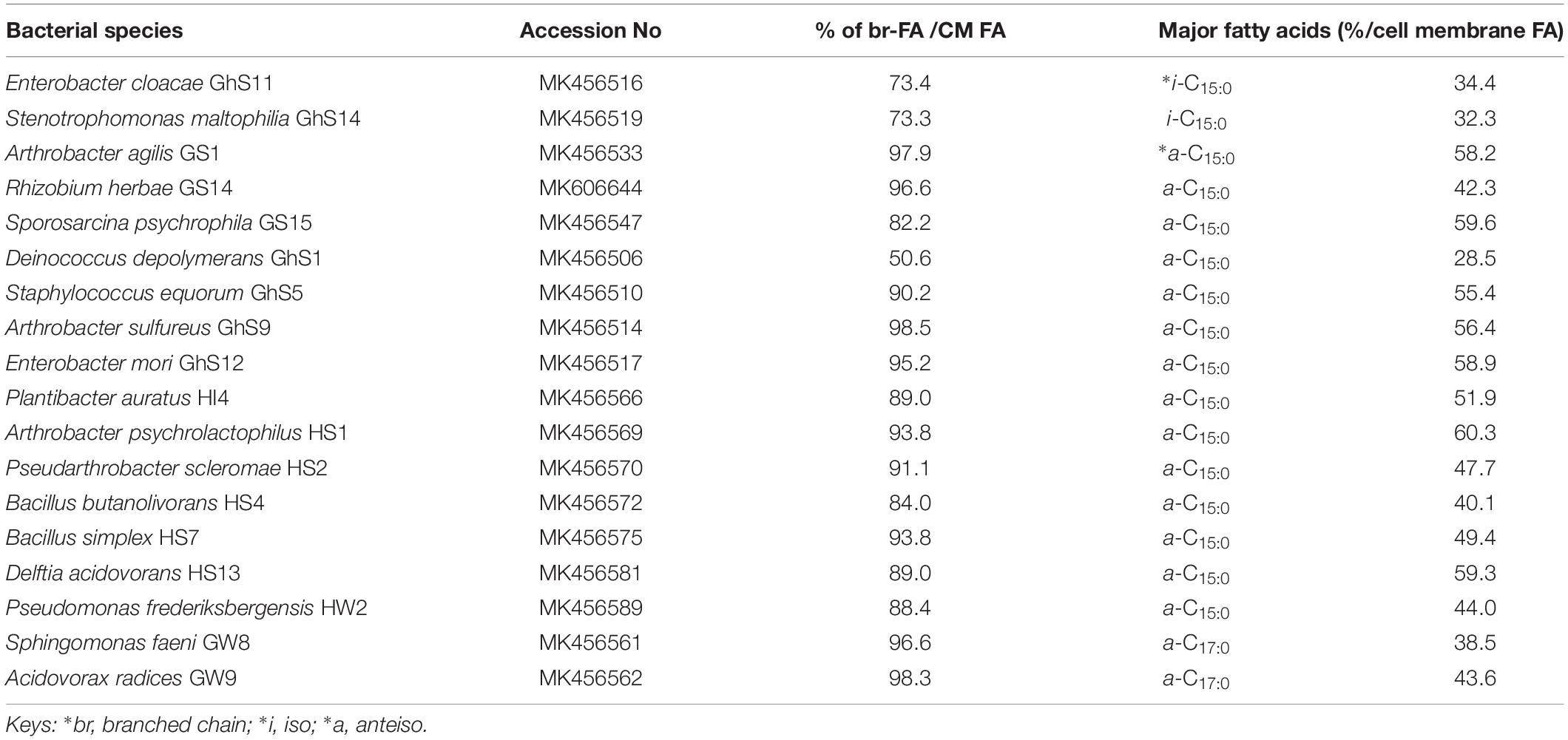
Table 2. List of bacteria species produced branched chain fatty acids (∗br-FAs) as major group of cell membrane (CM) fatty acids (FA).
The experiments for the analysis of cell membrane fatty acids of all bacteria isolates were performed in Nutrient Agar (NA) [g L–1; D(+)-glucose 1, peptone 15, sodium chloride 6, yeast extract 3] and Reasoner’s 2A (R2A) (gL–1; casein acid hydrolysate 0.5, dextrose 0.5, dipotassium phosphate 0.3, magnesium sulfate 0.024, protease peptone 0.5, sodium pyruvate 0.3, starch soluble 0.5, yeast extract 0.5). The growth of isolates was conducted in 250 mL Erlenmeyer flasks with 50 mL production media at 15°C for 7 days. The biomass collection of bacteria isolates was achieved by centrifuging the cultures in 50 mL falcon tubes at 4,500 g at 10°C for 30 min. The resulting cell pellets were stored in 2 mL microcentrifuge tubes and freeze-dried prior to subsequent use.
Fatty acid methyl ester from all bacterial cell cultures were extracted using the protocol described by El Razak et al. (2014). Two mL of 5% methanolic HCl was mixed with 0.1 g of each dried bacterial cell culture and heated at 70°C in a water bath for 120 min in sealed glass tubes. The sealed glass tubes were allowed to cool at 23°C for 30 min. Then, 1 mL Milli-Q® water was added to the tubes and vortexed, followed by 2 mL hexane and robust vortexing of the vial in order to extract the liberated fatty acid methyl esters (FAMEs). After phase separation, the solvent layer was transferred into a clean vial and the hexane removed under a gentle stream of nitrogen.
A total of 10 bacterial species, representing 10 different bacterial genera belonging to both Gram-negative and Gram-positive groups, were selected in order to determine the variation or similarity in composition of bacterial cell membrane fatty acids growing under different temperature regimes. The selection criteria of bacteria isolates was based on their ability to grow over a wide range of temperatures and their first time study on selected temperatures. Briefly, bacteria isolates were grown at 5, 15, 25, and 35°C using NB medium for 7 days. The bacterial biomass was obtained by centrifuging 40 mL falcon tubes containing bacterial cultures at 4,500 g at 10°C for 30 min. The pelleted bacterial cells were then subjected to FAME extraction as above.
Fatty acid methyl esters derived from the bacteria isolates were analyzed by GC-MS. The GC-MS comprised a Thermo Scientific Trace 1300 gas chromatograph coupled via a heated transfer line to a Thermo Scientific ISQ LT single quadrupole mass spectrometer. The GC was fitted with a programmable temperature vaporising (PTV) injector and an Agilent-HP 1 capillary column coated with a 100% dimethylpolysiloxane stationary phase (50 m × 0.32 mm internal diameter × 0.17 mm film thickness). The carrier gas was helium at a flow rate of 2 mL min–1. The PTV was programmed to heat up to 300°C at 14°C sec–1, transferring the analytes onto the column. The GC temperature program started with an initial temperature of 50°C, held for 1 min, then heated to 100°C at 10°C min–1, then to 250°C at 4°C min–1 and finally to 300°C at 20°C min–1. The final temperature was maintained for 5 min. Compounds were identified by their mass spectra, retention times and elution order in comparison to a commercial reference standard (Sigma Aldrich C4-C24 FAME mix, inclusive of unsaturated compounds). Individual FAMEs were quantified by their peak areas relative to the peak area of an internal standard (methyl tetracosanoate, Sigma-Aldrich) and corrected for variable levels of ionization by a response factor that was calculated from analyses of the reference standard mix at the start of each analytical sequence. In addition, correlations between different temperature and fatty acids were also assessed in this study using GraphPad Prism 5.00.
In the current study, 42 different bacterial isolates representing 23 different genera were identified by 16S rRNA sequencing (Tables 1, 2). Pseudomonas was the most dominant genus followed by Arthrobacter, Brevundimonas, and Massilia. In addition, cell membrane fatty acids analyses revealed that in 24 out of 42 bacterial strains straight-chain monounsaturated fatty acids (MUFAs) were the major group of cell membrane (CM) fatty acid (FA), whereas 18 species were found to have branched fatty acids, both saturated and monounsaturated, as the dominant FAs in their cell membranes. The most important membrane fatty acids found in the 42 cold-tolerant bacterial species are listed in Tables 1, 2. The main individual n-MUFAs detected were n-C15:1(cis–10), n-C16:1(cis–9), and n-C18:1(tr–9), whilst the dominant br-FAs are i-C15:0, a-C15:0, and a-C17:0. Most of the bacteria isolates were found to possess a-C15:0 (15 bacteria isolates) followed by n-C16:1(cis–9) (12 bacteria isolates) and n-C18:1(tr–9) (10 bacteria isolates) in their cell membranes. Only 1 species was found to produce i-C15:0, whereas 2 of each isolate produced ai-C17:0 and n-C15:1(cis–10) as the primary cell membrane fatty acid moieties. Very few of the bacterial species studied produce polyunsaturated fatty acids (Table 3).
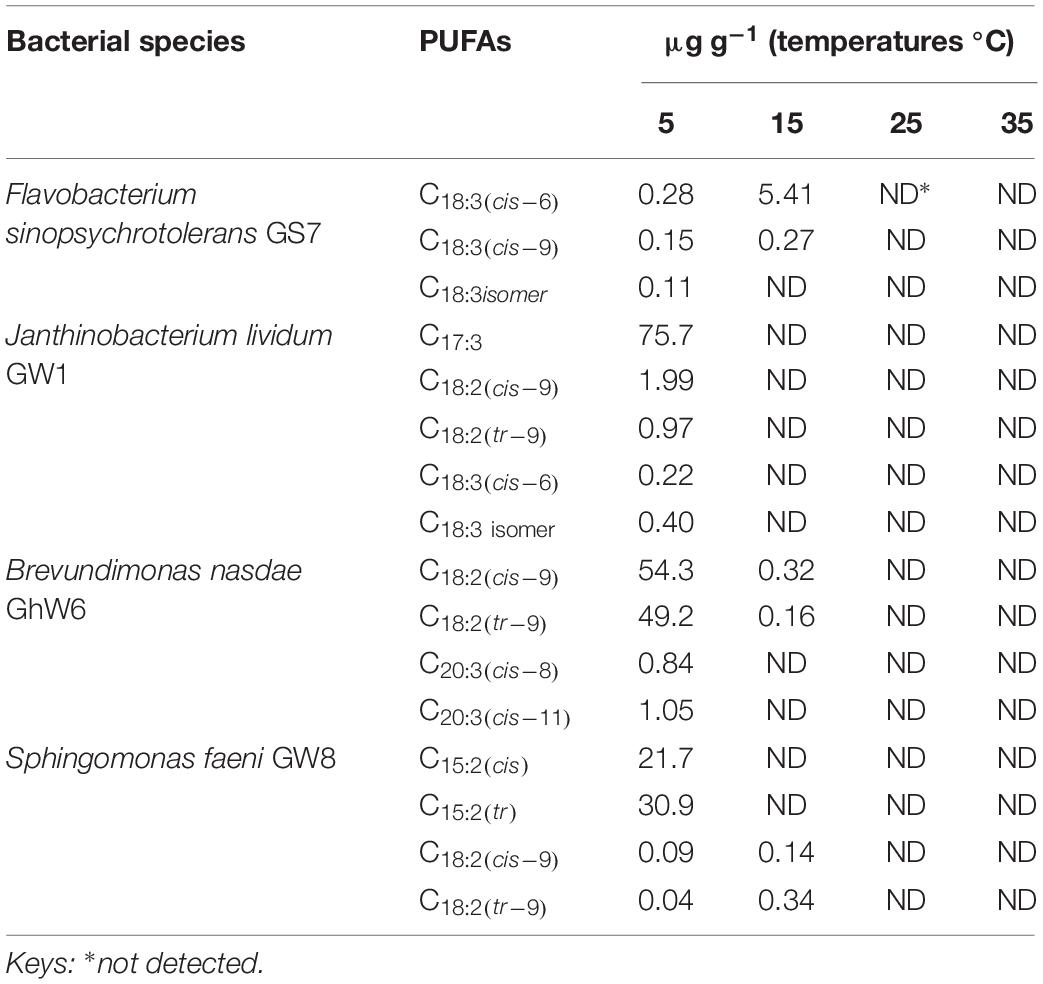
Table 3. Distribution of polyunsaturated fatty acids (PUFAs) in bacteria cell membranes at different temperatures.
In 32 out of 42 bacterial species, either br-FAs or n-MUFAs accounted for more than 70% of all quantified cell membrane FAs. In Paracoccus hibiscisoli GS9, n-MUFAs account for 94% of the determined FAs, followed by Arthrobacter nitroguajacolicus GS13 (89.6%), Flavobacterium sinopsychrotolerans GS7 (89.0%), Massilia aurea HI1 (84.3%) and Brevundimonas intermedia GS21 (83.7%), with 11 species showing relative amounts of n-MUFAs of more than 70%. The proportion of br-FAs reaches 98.5% in Arthrobacter sulfureus GhS9 followed by Acidovorax radices GW9 (98.3%), Arthrobacter agilis GS1 (97.9%), Sphingomonas faeni GW8 (96.7%), Rhizobium herbae GS14 (96.6%), Enterobacter mori GhS12 (95.2%), Bacillus simplex HS7 (93.8%), Arthrobacter psychrolactophilus HS1 (93.7%), Pseudarthrobacter scleromae HS2 (91.1%) and Staphylococcus equorum GhS5 (90.2%). The remaining 7 strains contain 70% br-FA in their cell membranes. Some strains show a dominance of a single compound such as n-C18:1(tr–9) in Paracoccus hibiscisoli GS9 and Rhizobium giardinii HI5, with 93.3 and 61.7% of the quantified FAs, respectively.
In the current study, the effect of different temperatures on the distribution of fatty acids in the cell membranes of 10 bacterial species have been comprehensively determined (Table 4). The selected bacteria belong to Gram-negative (e.g., Flavobacterium sinopsychrotolerans GS7, Paracoccus hibiscisoli GS9, Janthinobacterium lividum GW1, Sphingomonas faeni GW8, Brevundimonas nasdae-GhW6, Rhizobium giardinii HI5 and Pseudomonas extremaustralis-HS8) and Gram-positive groups (e.g., Sporosarcina psychrophila GS15, Staphylococcus equorum GhS5 and Arthrobacter psychrolactophilus HS1). Correlations were used to represent association of temperatures with cell membrane fatty acids (Tables 5, 6). The Gram-negative strains accumulated straight-chain monounsaturated fatty acids (n-MUFAs) as the major group of cell membrane fatty acids except Sphingomonas faeni GW8 (Table 4). The highest quantity of n-MUFAs were produced at lower temperature, i.e., 5 and 15°C, and lower levels at higher temperature, i.e., 25 and 35°C, but n-MUFAs still formed the major group of cell membrane fatty acids at all temperatures, except for Janthinobacterium lividum GW1, which generated relative levels of straight-chain saturated fatty acids (n-SFA) of up to 58% at 35°C as compared to 36% of n-MUFAs. In addition, Gram-positive bacteria revealed predominantly branched-chain fatty acids, both saturated and monounsaturated, as the major groups among the analyzed cell membrane FAs (Table 4). However, at all temperatures, bacterial strains tend to incorporate higher levels of saturated branched-chain fatty acids into their cell membranes compared to branched-chain monounsaturated fatty acids (br-MUFAs). Saturated fatty acids occur only at very low levels (between 1.8 and 4%) in Gram-positive bacteria at all temperatures. In addition, Gram-positive bacteria exhibit the highest levels of saturated branched-chain fatty acids (br-SFAs) in their cell membranes at higher temperature, e.g., 25 and 35°C, and gradually decreasing levels of br-SFAs with decreased temperature, i.e., 5 and 15°C. Similarly, the highest levels of branched chain monounsaturated fatty acids were seen at low temperature (5 and 15°C) as compared to high temperature (25 and 35°C).
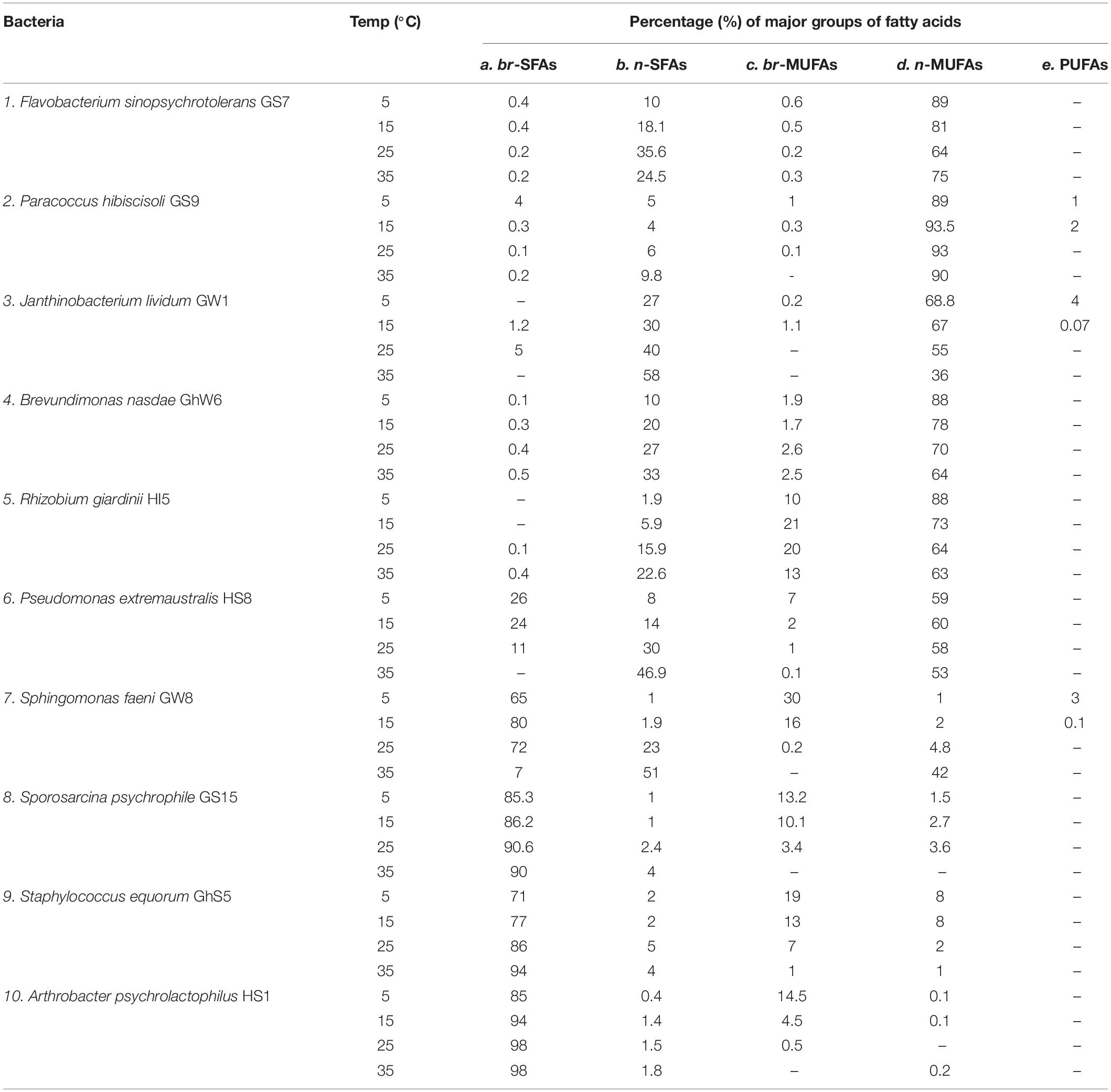
Table 4. Temperature derived distributions of the major groups of fatty acids in cell membranes of bacteria species.
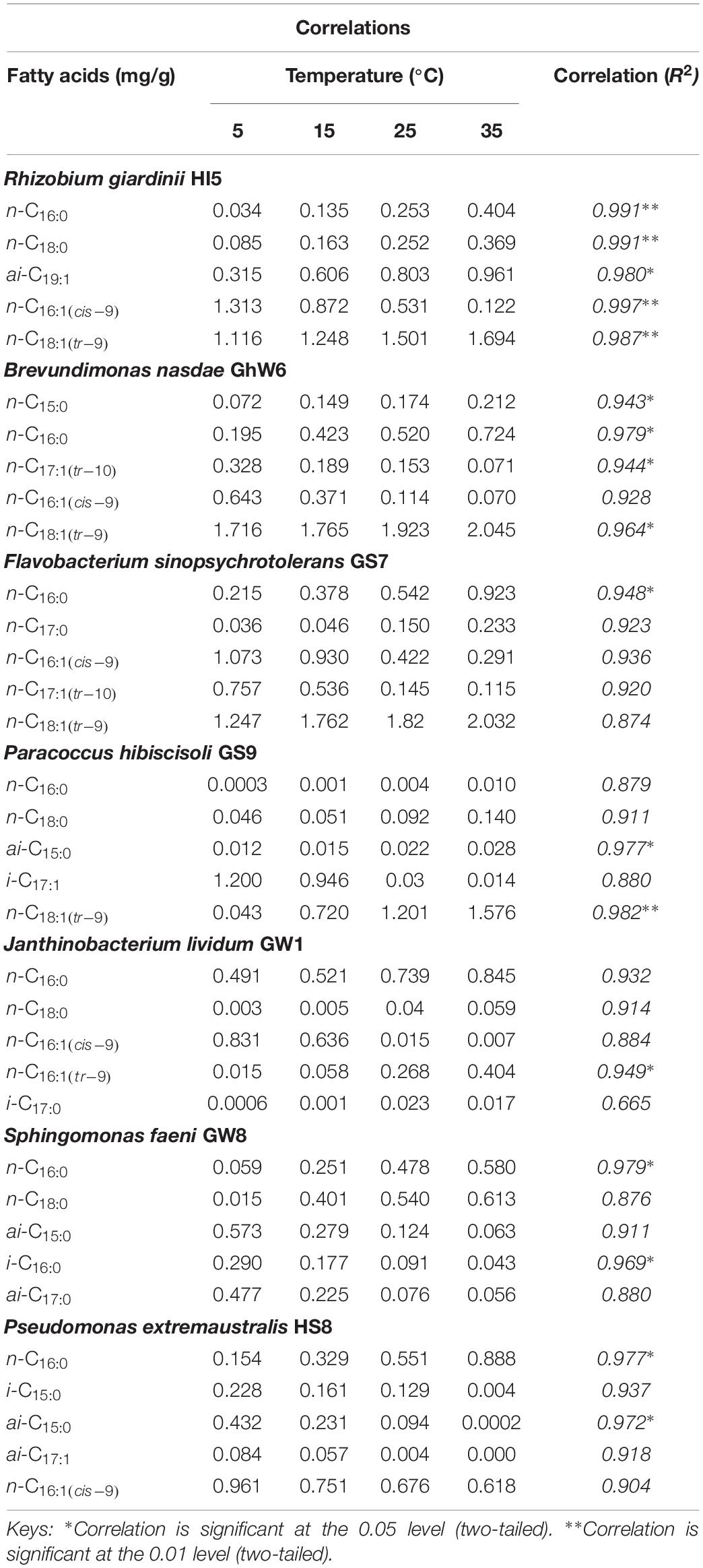
Table 5. Correlations of cell membrane fatty acids produced by Gram-negative bacteria species at different temperature.
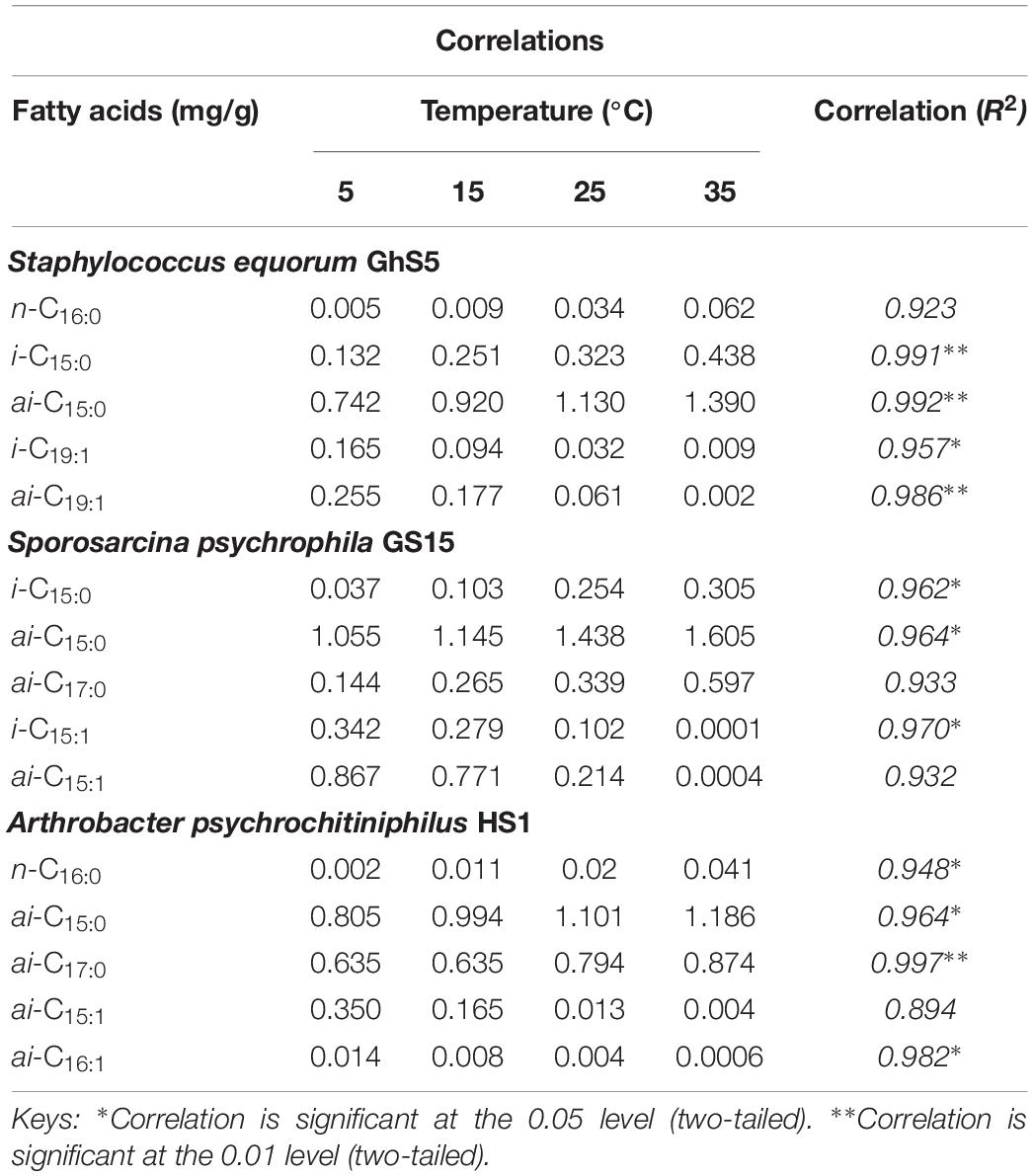
Table 6. Correlations of cell membrane fatty acids produced by Gram-positive bacteria species at different temperature.
Temperature greatly influences the distribution of individual fatty acids in cell membranes of all bacterial strains (Table 5). The straight-chain fatty acids such as C15:0, C16:0, C17:0, and C18:0 occur at higher levels at higher temperatures such as 25 and 35°C but at lower levels under lower temperatures, i.e., 5 and 15°C. Similarly, Gram-negative bacteria were observed to contain high levels of n-C16:1(cis–9) and n-C18:1(tr–9) with increasing temperature, although their production was also high at lower temperatures as well. Monounsaturated fatty acids such as n-C16:1(tr–9) and n-C17:1(tr–10) were also among those fatty acids whose production was enhanced at higher temperatures but decreased at low temperatures. Likewise, high levels of the branched saturated fatty acid, i-C17:0, were observed at 35°C but with lower levels at 5°C. Sphingomonas faeni GW8 produces i-C16:0 at higher levels at lower temperatures but at lower levels at 25 and 35°C. In addition, Gram positive bacterial strains were observed to generate the branched chain saturated component, a-C15:0, at high levels in their cell membranes. The levels of br-SFAs such as i-C15:0 and a-C15:0 gradually elevated with increasing temperature but with no sufficient difference. On the other hand, branched chain monounsaturated fatty acids such as i-C15:1, a-C15:1, i-C19:1 and a-C19:1 were produced by bacteria at lower levels at 25 and 35°C but found at higher levels at 5 and 15°C. Moreover, the straight chain fatty acids such as C16:0 and C18:0 were observed at high levels at increased temperature such as 25 and 35°C but in at low levels at lower temperature, i.e., at 5 and 15°C.
The present work aimed to study the distribution of saturated and unsaturated, normal and branched phospholipid fatty acids (PLFAs) in cell membranes of cold-tolerant bacterial strains belonging to both Gram-negative and Gram-positive groups. The comprehensive PLFA profiles of bacteria from Pakistan glaciers obtained in this study could be used to identify and quantify bacteria in cold habitats as each bacterial species displays a unique cell membrane fatty acid profile. Most importantly, the saturated C16:0 fatty acid is the most common FA, observed at high levels in all types of bacteria. Using PLFAs as bio-markers for bacterial identification and quantification continues to increase in popularity in the scientific community (Willers et al., 2015). PLFAs are the primary component of bacterial cell membranes; they degrade very rapidly upon cell death (White et al., 2009). Therefore, they are considered to be more representative of living bacterial biomass and more accurate than DNA-based approaches as PLFAs are more readily transformed following cell−death compared to DNA (Feinstein et al., 2009). Several researchers have used PLFA profiles for the quantification of bacteria in a given environment in order to understand function of genes involved in metabolic activities, for the screening of pathogenic bacteria, or the determination of community structures and bacterial diversity (Kellogg et al., 2001; Jungblut et al., 2009; Frostegard et al., 2011; Buhring et al., 2012; Naeher et al., 2012). Here we suggest that the comparison of PLFA distributions of bacteria isolated from cold environments with bacteria inhabiting warmer habitats, for identification purposes, would be misguided as fatty acid composition and quantity alter greatly alongside changes in the surrounding environment (Willers et al., 2015). However, PLFAs may be useful in association with the study of physiological changes in bacteria.
Most of the Gram-negative bacteria exhibit n-MUFAs as the major group of cell membrane FAs while those of Gram-positive species are dominated by br-SFAs, a characteristic pattern previously reported by Frostegard and Baath (1996). The a-C15:0, n-C16:0 and n-C18:1 as the dominant cell membrane FAs in Gram-positive species in the current study corroborates with the literature (Gillan et al., 1983; Breulmann et al., 2014; Dong et al., 2014; Fichtner et al., 2014; Reinsch et al., 2014). More specifically, n-C16:1 and the monounsaturated fatty acid n-C18:1 were the major types of fatty acid in cell membranes of the studied bacterial species summarized Table 1. Previous research has shown n-C16:1 and n-C18:1 as the most predominant PLFA and signature biomarkers of Gram-negative bacteria (Buckeridge et al., 2013; Djukic et al., 2013; Tavi et al., 2013; Zheng et al., 2013; Banks et al., 2014; Lange et al., 2014; Reinsch et al., 2014; Zhang et al., 2014).
Apart from general interspecies PLFA distributions we evaluated the effect of growth temperature on the relative distributions and levels of major FAs in cell membranes of Gram-negative and Gram-positive bacteria. Notably, increasing temperature caused a substantial decline in the relative amount of br-SFAs and br-MUFAs, in particular, in Gram-positive species (Sporosarcina psychrophila GS15, Sphingomonas faeni GW8, Staphylococcus equorum GhS5, Arthrobacter psychrolactophilus HS1) from about 90% at 5 and 15°C to less than 1% at 35°C while the amount of saturated normal FAs increased from an initial range of 0.4–2% to 1.8–51%. These observations corroborate the role of br-FAs in the adaptability of these bacteria toward changes in temperature. It has been reported that br-FAs maintain the normal liquid-crystalline (fluid) state of bacterial PLFA by lowering the melting point and, thus, enabling membranes to perform normal cellular function at low temperature (Suutari and Laakso, 1994; Bowles et al., 1996; Sun et al., 2012). An increased production of br-FAs by psychrophilic bacteria in response to lower temperatures has been observed previously, e.g., for the food pathogen Listeria monocytogenes (Annous et al., 1997). However, to the best of our knowledge, this is the first time that such temperature adaption is reported for bacteria species Sporosarcina psychrophila, Sphingomonas faeni, Staphylococcus equorum, and Arthrobacter psychrolactophilus. Other Gram-positive bacteria for which equivalent changes in PLFA composition at lower temperature has been observed include Staphylococci, incorporating increasing amounts of br-MUFAs into their cell membranes (Onyango and Alreshidi, 2018). Suutari and Laakso (1992) reported increased production of lower-melting point a-FAs at low temperature for Bacillus subtilis and Bacillus megaterium but decreased i-FA production. Similarly, Annous et al. (1997) reported an increase in levels of short-chain and a-branched-chain FAs in the cell membrane of Listeria monocytogenes with the temperature decreasing from 45 to 5°C.
Changes in FA distributions from different growth temperatures were similarly pronounced for the Gram-negative bacteria investigated, Flavobacterium sinopsychrotolerans GS7, Paracoccus hibiscisoli GS9, Janthinobacterium lividum GW1, Brevundimonas nasdae GhW6, Rhizobium giardinii HI5 and Pseudomonas extremaustralis HS8. In particular, the relative levels of saturated fatty acids such as C15:0, C16:0, C17:0, and C18:0 vary significantly. High levels of saturated FAs lower the membrane fluidity, thus, helping bacteria to survive at higher temperatures (Knothe and Dunn, 2009). Although n-MUFAs were the main group of membrane FAs of the investigated Gram-negative bacterial strains at all temperatures, the relative level of n-C16:1(cis–9) decreased considerably with increasing temperature. The accumulation of MUFAs especially with cis-conformation has been reported as a means of lowering the FA melting point and improving cell membrane fluidity at low temperatures (Russell, 1989; Mangelsdorf et al., 2009). For example, saturated n-C16:0 has a melting point of 63°C whereas the introduction of one double bond, i.e., converting n-C16:0 to n-C16:1, lowers the melting point significantly to −1°C (Knothe and Dunn, 2009).
As far as we know, membrane FA adaption to growth temperature has not yet been reported for the above-mentioned Gram-negative species. Similar patterns were observed by Theberge et al. (1996) for the FA composition of Rhizobium leguminosarum at temperatures of 10, 15, 22, and 30°C, with higher levels of MUFAs and C18:1, in particular, at lower temperatures. Bajerski et al. (2017) evaluated the FA profile of Chryseobacterium frigidisoli and found monounsaturated i-C17:1 at high levels at lower temperature (10°C), confirming the role of unsaturated membrane FAs with a lower melting point in cold adaptation (Russell, 1984, 1989). In contrast, Pseudomonas syringae, which was isolated from ice samples of Antarctica, contains high levels of saturated and trans-monounsaturated FAs although fatty acids with the cis-isomers were also detected (Kiran et al., 2005).
Overall, the observed shifts in the distributions of saturated and unsaturated, normal and branched FAs with growth temperature appear consistent across the investigated 10 strains of Gram-positive and Gram-negative bacteria. This points toward the potential for the development of temperature-sensitive molecular ratios for (non-polar) glacial settings with high proportions of bacterial biomass such as glacial ice, melted water and sediments. Such approaches remain to be tested in situ.
The analysis of cell membrane fatty acids of 42 cold-tolerant bacteria isolated from various glaciers of the Karakoram Mountain Range has revealed straight-chain monounsaturated fatty acids and branched-chain fatty acids as the dominant compounds among cell membrane fatty acids in Gram-positive and Gram-negative species, respectively. In addition, fatty acid distributions in bacterial cell membranes are significantly affected by growth temperature. In particular, a pronounced shift from enhanced proportions of monounsaturated to saturated branched and normal fatty acids with increasing temperature has been observed in Gram-negative bacteria species, whereas a notable increase in branched and normal saturated fatty acids with increased temperatures has witnessed in Gram-positive bacteria. Our study demonstrates the role of fatty acids for the maintenance of cell membrane fluidity in bacteria living in non-polar glacial habitats. The observed temperature-dependent shifts in fatty acid distributions may furthermore contribute to the development of temperature-sensitive molecular proxies in order to track temperature changes in (sub-) glacial environmental archives with high proportions of bacterial biomass. Finally, the capability of psychrophilic bacteria to produce biodesirable fatty acids, e.g., polyunsaturated fatty acids, represents an alternative source of such fatty acids for heterotrophic organisms in glacial environments.
All datasets generated for this study are included in the article.
NH, FH, and AA designed the research work. NH, MR, AH, and FH collected the samples from glaciers. AS, NH, and MR isolated and identified bacteria species. IB, JH, and NH carried out the GS/MS and afterward data analysis. FH and AA supervised the whole research work. FH and NH wrote the manuscript. All authors read and approved the final manuscript.
Funds for the research work were provided by Commonwealth Scholarship Commission, United Kingdom. In addition, NERC Life Sciences Mass Spectrometry Facility, School of Chemistry, University of Bristol, also supported this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are thankful to Dr. Simmon Cobb, Senior Teaching Laboratory Technician and Mr. James Williams, the Analytical Research Technician in LOWTEX for providing technical help during research work.
Annous, B. A., Becker, L. A., Bayles, D. O., Labeda, D. P., and Wilkinson, B. J. (1997). Critical role of anteiso-C15: 0 fatty acid in the growth of Listeria monocytogenes at low temperatures. Appl. Environ. Microbiol. 63, 3887– 3894.
Bajerski, F., and Wagner, D. (2013). Bacterial succession in Antarctic soils of two glacier forefields on Larsemann Hills, East Antarctica. FEMS. Microbiol. Ecol. 85, 128–142. doi: 10.1111/1574-6941.12105
Bajerski, F., Wagner, D., and Mangelsdorf, K. (2017). Cell membrane fatty acid composition of Chryseobacterium frigidisoli PB4T, isolated from Antarctic glacier forefield soils, in response to changing temperature and pH conditions. Front. Microbiol. 8:677. doi: 10.3389/fmicb.2017.00677
Banks, M. L., Kennedy, A. C., Kremer, R. J., and Eivazi, F. (2014). Soil microbial community response to surfactants and herbicides in two soils. Appl. Soil Ecol. 74, 12–20.
Bowles, B. L., Foglia, T. A., and Juneja, V. K. (1996). Temperature induced shifts in the fatty acid profile of Staphylococcus aureus WRRC B124 1. J. Rap. Methods Aut. Microbiol. 4, 235–245.
Breulmann, M., Masyutenko, N. P., Kogut, B. M., Schroll, R., Dörfler, U., Buscot, F., et al. (2014). Short-term bioavailability of carbon in soil organic matter fractions of different particle sizes and densities in grassland ecosystems. Sci. Total Environ. 497, 29–37. doi: 10.1016/j.scitotenv.2014.07.080
Buckeridge, K. M., Banerjee, S., Siciliano, S. D., and Grogan, P. (2013). The seasonal pattern of soil microbial community structure in mesic low arctic tundra. Soil Biol. Biochem. 65, 338–347.
Buhring, S. I., Schubotz, F., Harms, C., Lipp, J. S., Amils, R., and Hinrichs, K. U. (2012). Lipid signatures of acidophilic microbial communities in an extreme acidic environment–Río Tinto, Spain. Org. Geochem. 47, 66–77.
Cossins, A. R., and MacDonald, A. G. (1984). Homeoviscous theory under pressure: II. The molecular order of membranes from deep-sea fish. BBA Biomembranes 776, 144–150.
Crowe, J. H., Hoekstra, F. A., and Crowe, L. M. (1992). Anhydrobiosis. Annu. Rev. Physiol. 54, 579–599.
DeLong, E. F., and Yayanos, A. A. (1986). Biochemical function and ecological significance of novel bacterial lipids in deep-sea procaryotes. Appl. Environ. Microbiol. 51, 730–737.
Denich, T. J., Beaudette, L. A., Lee, H., and Trevors, J. T. (2003). Effect of selected environmental and physico-chemical factors on bacterial cytoplasmic membranes. J. Microbiol. Methods 52, 149–182.
Djukic, I., Zehetner, F., Watzinger, A., Horacek, M., and Gerzabek, M. H. (2013). In situ carbon turnover dynamics and the role of soil microorganisms therein: a climate warming study in an Alpine ecosystem. FEMS Microbiol. Ecol. 83, 112–124. doi: 10.1111/j.1574-6941.2012.01449.x
Dong, H. Y., Kong, C. H., Wang, P., and Huang, Q. L. (2014). Temporal variation of soil friedelin and microbial community under different land uses in a long-term agroecosystem. Soil Biol. Biochem. 69, 275–281.
El Razak, A. A., Ward, A. C., and Glassey, J. (2014). Screening of marine bacterial producers of polyunsaturated fatty acids and optimisation of production. Microbial. Ecol. 67, 454–464. doi: 10.1007/s00248-013-0332-y
Feinstein, L. M., Sul, W. J., and Blackwood, C. B. (2009). Assessment of bias associated with incomplete extraction of microbial DNA from soil. Appl. Environ. Microbiol. 75, 5428–5433. doi: 10.1128/AEM.00120-09
Fichtner, A., Von Oheimb, G., Härdtle, W., Wilken, C., and Gutknecht, J. L. M. (2014). Effects of anthropogenic disturbances on soil microbial communities in oak forests persist for more than 100 years. Soil Biol. Biochem. 70, 79–87.
Frostegard, A., and Baath, E. (1996). The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol. Fert. Soils 22, 59–65.
Frostegard, A., Tunlid, A., and Baah, E. (2011). Use and misuse of PLFA measurements in soils. Soil Biol. Biochem. 43, 1621–1625.
Ganzert, L., Lipski, A., Hubberten, H. W., and Wagner, D. (2011). The impact of different soil parameters on the community structure of dominant bacteria from nine different soils located on Livingston Island, South Shetland Archipelago, Antarctica. FEMS Microbiol. Ecol. 76, 476–491. doi: 10.1111/j.1574-6941.2011.01068.x
Georlette, D., Blaise, V., Collins, T., D’Amico, S., Gratia, E., Hoyoux, A., et al. (2004). Some like it cold: biocatalysis at low temperatures. FEMS Microbiol. Rev. 28, 25–42.
Gillan, F. T., Johns, R. B., Verheyen, T. V., Nichols, P. D., Esdaile, R. J., and Bavor, H. J. (1983). Monounsaturated fatty acids as specific bacterial markers in marine sediments. Adv. Org. Geochem. 1, 198–206. doi: 10.1016/j.scitotenv.2011.10.015
Hassan, N., Rafiq, M., Hayat, M., Shah, A. A., and Hasan, F. (2016). Psychrophilic and psychrotrophic fungi: a comprehensive review. Rev. Environ. Sci. Biotechnol. 15, 147–172.
Hazel, J. R., and Williams, E. E. (1990). The role of alterations in membrane lipid composition in enabling physiological adaptation of organisms to their physical environment. Prog. Lipid Res. 29, 167–227.
Hoshino, T., and Matsumoto, N. (2012). Cryophilic fungi to denote fungi in the cryosphere. Fungal Biol. Rev. 26, 102–105.
Jungblut, A. D., Allen, M. A., Burns, B. P., and Neilan, B. A. (2009). Lipid biomarker analysis of cyanobacteria-dominated microbial mats in meltwater ponds on the McMurdo Ice Shelf, Antarctica. Org. Geochem. 40, 258–269.
Kellogg, J. A., Bankert, D. A., Withers, G. S., Sweimler, W., Kiehn, T. E., and Pfyffer, G. E. (2001). Application of the Sherlock Mycobacteria Identification System using high-performance liquid chromatography in a clinical laboratory. J. Clin. Microbiol. 39, 964–970.
Kiran, M. D., Annapoorni, S., Suzuki, I., Murata, N., and Shivaji, S. (2005). Cis–trans isomerase gene in psychrophilic Pseudomonas syringae is constitutively expressed during growth and under conditions of temperature and solvent stress. Extremophiles 9, 117–125.
Knothe, G., and Dunn, R. O. (2009). A comprehensive evaluation of the melting points of fatty acids and esters determined by differential scanning calorimetry. J. Am. Oil Chem. Soc. 86, 843–856.
Lange, M., Habekost, M., Eisenhauer, N., Roscher, C., Bessler, H., Engels, C., et al. (2014). Biotic and abiotic properties mediating plant diversity effects on soil microbial communities in an experimental grassland. PLoS One 9:e96182. doi: 10.1371/journal.pone.0096182
Mangelsdorf, K., Finsel, E., Liebner, S., and Wagner, D. (2009). Temperature adaptation of microbial communities in different horizons of Siberian permafrost-affected soils from the Lena Delta. Chem. D Erde Geochem. 69, 169–182.
Mansilla, M. C., Cybulski, L. E., Albanesi, D., and de Mendoza, D. (2004). Control of membrane lipid fluidity by molecular thermosensors. J. Bacteriol. 186, 6681–6688.
Margesin, R., Fonteyne, P. A., Schinner, F., and Sampaio, J. P. (2007). Rhodotorula psychrophila sp. nov., Rhodotorula psychrophenolica sp. nov. and Rhodotorula glacialis sp. nov., novel psychrophilic basidiomycetous yeast species isolated from alpine environments. Int. J. Syst. Evol. Microbiol. 57, 2179–2184.
Margesin, R., and Miteva, V. (2011). Diversity and ecology of psychrophilic microorganisms. Res. Microbiol. 162, 346–361. doi: 10.1016/j.resmic.2010.12.004
Montiel, P. O. (2000). Soluble carbohydrates (trehalose in particular) and cryoprotection in polar biota. Cryo Lett. 21, 83–90.
Naeher, S., Smittenberg, R. H., Gilli, A., Kirilova, E. P., Lotter, A. F., and Schubert, C. J. (2012). Impact of recent lake eutrophication on microbial community changes as revealed by high resolution lipid biomarkers in Rotsee (Switzerland). Org. Geochem. 49, 86–95.
Nichols, D. S., Brown, J. L., Nichols, P. D., and McMeekin, T. A. (1997). Production of eicosapentaenoic and arachidonic acids by an Antarctic bacterium: response to growth temperature. FEMS Microbiol. Lett. 152, 349–354.
Onyango, L. A., and Alreshidi, M. M. (2018). Adaptive metabolism in staphylococci: survival and persistence in environmental and clinical settings. J. Pathog. 2018:1092632. doi: 10.1155/2018/1092632
Reinsch, S., Michelsen, A., Sárossy, Z., Egsgaard, H., Schmidt, I. K., Jakobsen, I., et al. (2014). Short-term utilization of carbon by the soil microbial community under future climatic conditions in a temperate heathland. Soil Biol. Biochem. 68, 9–19.
Russell, N. J. (1984). Mechanisms of thermal adaptation in bacteria: blueprints for survival. Trends Biochem. Sci. 9, 108–112.
Russell, N. J. (1989). Functions of lipids: structural roles and membrane functions. Microbial. Lipid 2, 279–365.
Russell, N. J. (1990). Cold adaptation of microorganisms. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 326, 595–611.
Sinensky, M. (1974). Homeoviscous adaptation—a homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc. Natl. Acad. Sci.U.S.A. 71, 522–525.
Singh, S. M., Puja, G., and Bhat, D. J. (2006). Psychrophilic fungi from Schirmacher oasis, East Antarctica. Curr. Sci. 90, 1388–1392.
Sun, Y., Wilkinson, B. J., Standiford, T. J., Akinbi, H. T., and O’Riordan, M. X. (2012). Fatty acids regulate stress resistance and virulence factor production for Listeria monocytogenes. J. Bacteriol. 194, 5274–5284. doi: 10.1128/JB.00045-12
Suutari, M., and Laakso, S. (1992). Unsaturated and branched chain-fatty acids in temperature adaptation of Bacillus subtilis and Bacillus megaterium. BBA Lipids Lipid Metab. 1126, 119–124.
Suutari, M., and Laakso, S. (1994). Microbial fatty acids and thermal adaptation. Crit. Rev. Microbiol. 20, 285–328.
Tavi, N. M., Martikainen, P. J., Lokko, K., Kontro, M., Wild, B., Richter, A., et al. (2013). Linking microbial community structure and allocation of plant-derived carbon in an organic agricultural soil using 13CO2 pulse-chase labelling combined with 13C-PLFA profiling. Soil Biol. Biochem. 58, 207–215.
Theberge, M. C., Prevost, D., and Chalifour, F. P. (1996). The effect of different temperatures on the fatty acid composition of Rhizobium leguminosarum bv. viciae in the faba bean symbiosis. New Phytol. 134, 657–664.
White, P. M., Potter, T. L., and Strickland, T. C. (2009). Pressurized liquid extraction of soil microbial phospholipid and neutral lipid fatty acids. J. Agric. Food Chem. 57, 7171–7177. doi: 10.1021/jf901257n
Willers, C., Jansen van Rensburg, P. J., and Claassens, S. (2015). Microbial signature lipid biomarker analysis–an approach that is still preferred, even amid various method modifications. J. Appl. Microbiol. 118, 1251–1263.
Wirsen, C. O., Jannasch, H. W., Wakeham, S. G., and Canuel, E. A. (1986). Membrane lipids of a psychrophilic and barophilic deep-sea bacterium. Curr. Microbiol. 14, 319–322.
Wynn-Williams, D. D. (1990). “Ecological aspects of Antarctic microbiology,” in Advances in Microbial Ecology, ed. K. C. Marshall (Boston, MA: Springer), 71–146.
Zhang, H., Ding, W., He, X., Yu, H., Fan, J., and Liu, D. (2014). Influence of 20–year organic and inorganic fertilization on organic carbon accumulation and microbial community structure of aggregates in an intensively cultivated sandy loam soil. PLoS One 9:e92733. doi: 10.1371/journal.pone.0092733
Keywords: cell membrane, cold adaptation, fatty acids, non-polar glaciers, psychrophilic bacteria
Citation: Hassan N, Anesio AM, Rafiq M, Holtvoeth J, Bull I, Haleem A, Shah AA and Hasan F (2020) Temperature Driven Membrane Lipid Adaptation in Glacial Psychrophilic Bacteria. Front. Microbiol. 11:824. doi: 10.3389/fmicb.2020.00824
Received: 06 February 2020; Accepted: 07 April 2020;
Published: 14 May 2020.
Edited by:
Pierre Amato, UMR 6296 Institut de Chimie de Clermont-Ferrand (ICCF), FranceReviewed by:
B. J. Wilkinson, Illinois State University, United StatesCopyright © 2020 Hassan, Anesio, Rafiq, Holtvoeth, Bull, Haleem, Shah and Hasan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fariha Hasan, ZmFyaWhhaGFzYW5AeWFob28uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.