
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 28 April 2020
Sec. Infectious Agents and Disease
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00776
Lysine malonylation (Kmal) is a new post-translational modification (PTM), which has been reported in several prokaryotic and eukaryotic species. Although Kmal can regulate many and diverse biological processes in various organisms, knowledge about this important PTM in the apicomplexan parasite Toxoplasma gondii is limited. In this study, we performed the first global profiling of malonylated proteins in T. gondii tachyzoites using affinity enrichment and Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Three experiments performed in tandem revealed 294, 345, 352 Kmal sites on 203, 236, 230 malonylated proteins, respectively. Computational analysis showed the identified malonylated proteins to be localized in various subcellular compartments and involved in many cellular functions, particularly mitochondrial function. Additionally, one conserved Kmal motif with a strong bias for cysteine was detected. Taken together, these findings provide the first report of Kmal profile in T. gondii and should be an important resource for studying the physiological roles of Kmal in this parasite.
Toxoplasmosis, caused by the protozoan parasite Toxoplasma gondii, is estimated to affect approximately one-third of the world population (Montoya and Liesenfeld, 2004). This parasite has the ability to infect almost all mammalian and avian species (Webster, 2010; Robert-Gangneux and Darde, 2012; Liu et al., 2015). The life cycle of T. gondii involves definitive host (members of the cat family; Felidae) and intermediate (mammals, including humans) host. During its development, the parasite progresses through three main morphological stages, including one replicative stage (tachyzoite), which is associated with the acute phase of infection, the dormant stage (bradyzoites-containing tissue cyst), which is associated with latent form of infection, and the environmentally resistant oocyst stage. The life cycle of T. gondii includes asexual reproduction, which involves the formation of tachyzoites and bradyzoites-containing cysts in the intermediate host and sexual reproduction which involves the formation of oocysts in the feline intestinal epithelium. In order to adapt to different environments and survive inside various tissues within different hosts, the parasite tightly regulates its metabolic and protein functions at the post-translational level (Xiao et al., 2010; Dubey et al., 2017).
Lysine post-translational modifications (PTMs), such as acetylation (Choudhary et al., 2009), methylation (Peng et al., 2011), succinylation (Lin et al., 2012; Hirschey and Zhao, 2015), and ubiquitination (Hershko and Ciechanover, 1998), play key roles in broadening the functional diversity of proteins and impact significantly on the regulation of protein functions in prokaryotic and eukaryotic organisms (Lin et al., 2012; Hirschey and Zhao, 2015). Lysine succinylation has been investigated in T. gondii, where it was found to be involved in a broad range of cellular functions (Li et al., 2014). Also, lysine acetylation has been studied in three T. gondii strains belonging to three different genotypes and the level of acetylation was found to correlate with the parasite strain virulence and has been found widespread on proteins of diverse functions in T. gondii (Jeffers and Sullivan, 2012; Wang et al., 2019).
Post-translational modification of proteins via lysine malonylation (Kmal) has been reported across many metabolic pathways, such as fatty acid synthesis and oxidation (Hirschey and Zhao, 2015), mitochondrial respiration (Xie et al., 2012), glycolysis (Peng et al., 2011; Lin et al., 2012), and modification of histones (Hershko and Ciechanover, 1998). Lysine malonylation was firstly observed in mammalian cells and bacterial cells (Lin et al., 2012). Since then, there have been growing interests in exploring the regulatory roles of Kmal in various microbial species, such as Escherichia coli (Qian et al., 2016), Cyanobacteria (Ma et al., 2017), and Saccharopolyspora erythraea (Xu et al., 2016). Although Kmal can regulate many crucial and diverse cellular processes (He et al., 2012), its existence and function in T. gondii remain unknown.
In the present study, we characterized malonylated proteins in T. gondii tachyzoites using Liquid chromatography-tandem mass spectrometry (LC-MS/MS) coupled with sensitive immune-affinity purification. Three parallel experiments were performed, which identified 294, 345, 352 Kmal sites on 203, 236, 230 proteins, respectively. Functional analyses showed predominant presence of malonylated proteins in metabolic processes, such as glycolysis/gluconeogenesis, aminoacyl-tRNA biosynthesis, pentose phosphate pathway, and fatty acid biosynthesis. To our knowledge, this study is the first to characterize protein malonylation in T. gondii. Our data lay the foundation for future investigations into the biological functions of malonylated proteins in T. gondii and in the context of host–parasite interaction.
Tachyzoites of T. gondii RH strain were maintained by serial passage in human foreskin fibroblast (HFF) monolayers, which were grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, United States) supplemented with 10% fetal calf serum (FBS, Gibco, United States), 100 U/ml antibiotics (penicillin–streptomycin solution). T. gondii tachyzoites and HFF monolayers were cultivated in a 5% CO2 humidified incubator at 37°C. The tachyzoites and cell debris were harvested when the infected HFF monolayer was lysed. The mixture was washed several times with phosphate buffered saline (PBS) and passed through a 25 gauge needle. Then, the parasites were filtrated using a 3 μm membrane filters (Millipore) in order to remove the cell debris, and stored at −80°C until use.
The parasite pellets were sonicated with 12 short bursts of 10 s, followed by intervals of 10 s on ice for three times in lysis buffer [8 M urea, 1% Protease inhibitor cocktail and for PTM experiments, deacetylase inhibitors were also added to the lysis buffer, e.g., 3 μM trichostatin A (TSA) and 50 mM nicotinamide (NAM)], and the remaining debris was removed by centrifugation at 12,000 g for 10 min at 4°C. Finally, the pellet was discarded, and the protein concentration was examined with BCA kit.
Proteins (20 μg) of tachyzoites were electrophoresed on 12% SDS-PAGE and transferred to PVDF membrane. TBST buffer (25 mM Tris–HCl, 150 mM NaCl, 0.1% Tween20, pH 8.0) with 5% BSA was used to block the membrane for 60 min. Then, the membrane was incubated with anti-malonyllysine antibody (1:500, catalog no. PTM-901; PTM Biolabs, Hangzhou, China) in TBST with 2.5% BSA overnight at 4°C. The membrane was washed three times with TBST, followed by incubation with horseradish-peroxidase-conjugated Goat anti-Rabbit IgG (1:5000; Thermo) for 60 min at room temperature. After washing the membrane three times, an ECL kit was used for protein visualization.
The protein solution was reduced with 5 mM dithiothreitol (DTT) for 30 min at 56°C and then alkylated with 11 mM iodoacetamide (IAA) for 15 min at 25°C away from light. The proteins were diluted by adding 100 mM NH4CO3 until the urea concentration became <2 M. Finally, trypsin was added at 1:50 trypsin-to-protein mass ratio for the first digestion overnight and 1:100 trypsin-to-protein mass ratio for a second 4 h-digestion.
The tryptic peptides were fractionated by high pH reverse-phase HPLC using Thermo Betasil C18 column (5 μm particles, 10 mm ID, 250 mm length). The peptides were separated using a gradient of 8–32% acetonitrile (pH 9.0) over 60 min into 60 fractions. Thereafter, the separated peptides were combined into four fractions as described previously (Liu et al., 2016).
The tryptic peptides dissolved in NETN buffer (100 mM NaCl, 1 mM EDTA, 50 mM Tris–HCl, 0.5% NP-40, pH 8.0) were incubated with pre-washed anti-malonyl-K antibody beads (PTM Biolabs, Hangzhou, China) at 4°C overnight with gentle shaking (Zhang et al., 2016). Then, the beads were washed four times with NETN buffer and twice with H2O. The bound peptides were eluted from the beads with 0.1% trifluoroacetic acid (TFA; Sigma-Aldrich, Saint Louis, United States) and were cleaned using C18 ZipTips (Merck Millipore, Billerica, United States).
The tryptic peptides were dissolved in 0.1% formic acid (solvent A), directly loaded onto a reverse-phase analytical column (15-cm length, 75 μm i.d.) at a constant flow rate of 700 nL/min on an EASY-nLC 1000 UPLC system. The gradient included an increase from 6–23% solvent B (0.1% formic acid in 98% acetonitrile) over 26 min, followed by 8 min from 23–35%, and reaching to 80% in 3 min and holding at 80% for the last 3 min. The peptides were subjected to a nanospray ionization (NSI) source followed by tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo) coupled online to an ultra-performance liquid chromatography (UPLC). The m/z scan range was 350–1800 for full scan, and intact peptides were detected in the Orbitrap at a resolution of 70,000. A data-dependent procedure was performed, selecting the 20 most intense ions for MS/MS with 30 s dynamic exclusion at a resolution of 17,500.
The MS/MS data were processed using MaxQuant search engine (v.1.5.2.8). Tandem mass spectra were searched against Uniprot T. gondii database (ToxoDB 46, 8322 sequences downloaded on March 18, 2020) concatenated with reverse decoy database. Trypsin/P was specified as cleavage enzyme allowing up to 4 missing cleavages. The mass tolerance for fragment ions was set as 0.02 Da. Cysteine carbamidomethylation was specified as fixed modification. Methionine oxidation, and Kmal were specified as variable modifications. False discovery rate (FDR) was adjusted to <1% and minimum score for modified peptides was set >40. All other parameters in MaxQuant were set to default values. The site localization probability was set as >0.75. Label-free quantification was performed using the MaxQuant label free quantification (LFQ) algorithm (Cox et al., 2014), by comparing the abundance of the same peptides across runs, with both ion intensities and spectral counts used for this purpose.
Subcellular localization of all malonylated proteins identified in three separate experiments was predicted using WOLF PSORT1. For Motif analysis, the sequence models contained amino acids at specific positions of the malonyl-21-mers (10 amino acids upstream and downstream of the Kmal sites) in all protein sequences were predicted by MoMo software (motif-x algorithm). The T. gondii protein sequences from the UniProt database was used as background database parameter, and other parameters were set as default.
GO annotation was performed against UniProt-GOA database2. Firstly, the identified protein identifiers (IDs) were converted to UniProt IDs and then mapped to GO IDs. If the identified malonylated proteins were not annotated by UniProt-GOA database, the InterProScan software were used to annotate protein’s GO function based on protein sequence alignment method. Also, an online service tool, KEGG Automatic Annotation Server (KAAS)3, was used to annotate protein’s KEGG database description. Then, the annotation result was mapped to the KEGG pathway database using KEGG mapper. The protein domains were annotated by using InterProScan software and the InterPro domain database4 based on protein sequence alignment. A two-tailed Fisher’s exact test was employed for the GO/KEGG/Domain enrichment analysis of the differentially malonylated proteins against all identified proteins. The terms with P values <0.05 were considered significant.
All differentially abundant malonylated proteins were searched against the Search Tool for Recurring Instances of Neighboring Genes (STRING) database5 for Protein–Protein interaction (PPI). The interaction score was set with high confidence (≧0.7), and all other parameters were set as default. The PPI network was visualized by Cytoscape (version 3.5.0).
Our study provides the first characterization of Kmal, a newly discovered type of lysine acylation, in T. gondii. We have identified multiple malonylated proteins with a wide range of functions, such as metabolism and immune response, as discussed below.
To detect the abundance of malonylated proteins in T. gondii tachyzoite extracts, western blotting was performed prior to the proteomic experiments (Figure 1A). The global malonylome analysis of T. gondii was performed using affinity enrichment followed by high-resolution LC-MS/MS. Three parallel experiments were performed, which identified 294 Kmal sites on 203 proteins in Exp.1 (Supplementary Table S1), 345 Kmal sites on 236 proteins in Exp.2 (Supplementary Table S2), and 352 Kmal sites on 230 proteins in Exp.3 (Supplementary Table S3). A total of 160 proteins were overlapped between Exp.1 and Exp.2, 151 proteins were overlapped between Exp.1 and Exp.3, 170 proteins were overlapped between Exp.2 and Exp.3, and 138 proteins were overlapped among these three experiments. The low number of overlapped proteins may be attributed to the low incidence of this PTM and the resolution of the methods used, resulting in less repetitions. Overall, 506 Kmal sites across 326 proteins were detected. The number of Kmal proteins in T. gondii was higher than that reported in common wheat, S. erythraea (Xu et al., 2016; Liu et al., 2018), but was lower than that detected in E. coli (Qian et al., 2016), rhizobacterium Bacillus amyloliquefaciens FZB42 (Fan et al., 2017) and mammals (Colak et al., 2015), however was close to the number of malonylated proteins identified in Cyanobacteria (Ma et al., 2017). As shown in Figure 1B, a total of 326 proteins were modified/malonylated at more than two lysine residues. For example about 73% (239 out of 326) of the identified malonylated proteins were modified at one lysine residue, 14% (46 out of 326) were modified at two lysine residues, and 13% (41 out of 326) were modified at ≥ two lysine residues. The finding that 27% of the malonylated proteins were modified at multiple lysine residues/sites is in agreement with the results detected in wheat and S. erythraea (Xu et al., 2016; Liu et al., 2018), indicating some conservation in the level of protein malonylation between T. gondii and other organisms, such as bacteria and plants, however the level and site of malonylation may vary among different organisms.
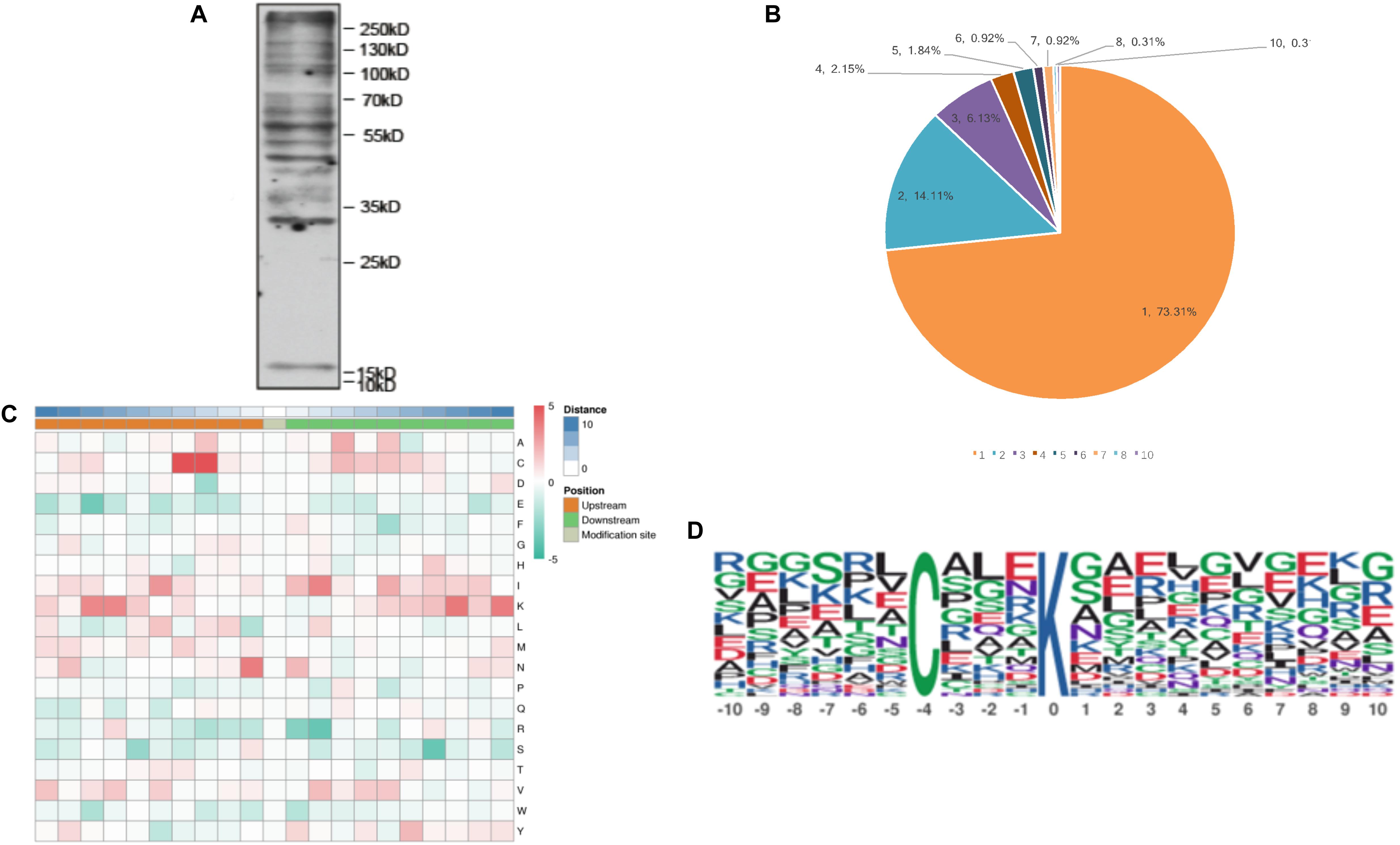
Figure 1. The properties of the lysine malonylation peptides in Toxoplasma gondii. (A) Western blot analysis of tachyzoite lysate probed with anti-malonylation antibodies. (B) The number of Kmal sites for the malonylated protein detected in T. gondii. (C) Heat map showing different types of amino acids at positions −10 to +10 from the malonylated lysine residue. Red and green color denotes high and low frequency, respectively. (D) Sequence motif logos showing the Kmal sites detected in proteins and the position-specific amino acids’ composition surrounding the Kmal sites.
Interestingly, T. gondii Elongation Factor 1-Alpha (TgEF-1α) was the largest malonylated protein with 10 malonylated sites, which seems to have functions related to cell motility, protein turnover, cell growth, and signal transduction (Ridgley et al., 1996), DNA replication and repair protein networks (Toueille et al., 2007), suggesting an important, conserved role for Kmal in the activities of this protein. The second highly malonylated protein was the eukaryotic mitochondrial porins which plays a role in transporting tRNAs and apoptosis (Flinner et al., 2013). T. gondii 14-3-3 protein had six Kmal sites, this protein is found in the parasitophorous vacuole and the excreted/secreted parasite antigens. T. gondii 14-3-3 protein has been suggested to play a role in the modulation of the migratory properties of host cells (Weidner et al., 2016) and host immunity (Assossou et al., 2004). In the present study, five Kmal sites were identified in heat shock protein 70 (Hsp70). A previous study showed that the heat shock protein was the most widely succinylated protein, which can be succinylated on up to 17 independent lysine residues (Li et al., 2014), and the heat shock protein can also be ubiquitinated (Salmon de Monerri et al., 2015). The heat shock protein was shown to play a critical role in modulating host immune response during Plasmodium falciparum infection (Pooe et al., 2017) and was also associated with protective immunity against T. gondii (Kikumura et al., 2010). These results suggest that Kmal participates in the regulation of various biological processes of different cellular components and influences several biological functions in T. gondii.
To further examine the properties of Kmal sites in T. gondii, the flanking sequences from ten amino acids upstream to ten amino acids downstream of the modified site were studied using the MoMo software and hierarchical cluster analysis. As shown in Figure 1C, the frequency of cysteine (C) residue at the position −4 to −3 was highest, the positively charged lysine (K) residue was enriched at the positions −10 to −6 and +10 to +5, whereas the negatively charged residues glutamate (E) and phenylalanine (F) were not observed, this result agree with Kmal reported in common wheat (Liu et al., 2018). Previous study showed that G residues and E residues are highly enriched surrounding the acetylated lysine, while G is overrepresented in the positions on the N-terminal side (Jeffers and Sullivan, 2012). Therefore, the additional characteristics of the surrounding amino acids are not obvious from the motifs, except in some specific positions.
One motif, C (X3) Kmal (Kmal indicates the lysine malonylation site, and X represents a random amino acid residue), was obtained (Figure 1D). Except C (X3) Kmal, K (X6) Kmal, Kmal (X6) K, and K (X5) Kmal were also detected in S. erythraea (Xu et al., 2016). However, the number of Kmal motifs was much lower than lysine acetylation motifs (Li et al., 2016; Lv et al., 2016; Zhang et al., 2016) and succinylation motifs (Pan et al., 2015; Jin and Wu, 2016). The sequence logos show a strong bias for cysteine (C), which was observed at the −3 and −4 positions (Figure 1C), similar to the Kmal bias for arginine (R) detected in the common wheat (Liu et al., 2018).
To understand the primary function of malonylated proteins, enriched GO terms at 2nd level in the three GO categories: biological process (BP), cellular component (CC) and molecular function (MF) were identified (Supplementary Table S4). In terms of BP, 57, 56, 56 and 56 malonylated proteins were enriched in cellular metabolic process, organic substance metabolic process, nitrogen compound metabolic process and primary metabolic process, accounting for 11% of all identified malonylated proteins in T. gondii, respectively (Figure 2A). Regarding the MF category, 16, 12, and 12% of the malonylated proteins were related to protein binding, organic cyclic compound binding and heterocyclic compound binding, respectively (Figure 2B). Also, we used WoLF PSORT to further predict the subcellular locations of the identified proteins (Horton et al., 2007). As shown in Figure 2C, most of the malonylated proteins in T. gondii were located in the cytoplasm (30%) and nucleus (28%), the rest of malonylated proteins were predicted to be located in plasma membrane (15%) extracellular (11%) and mitochondria (10%) (Figure 2C).
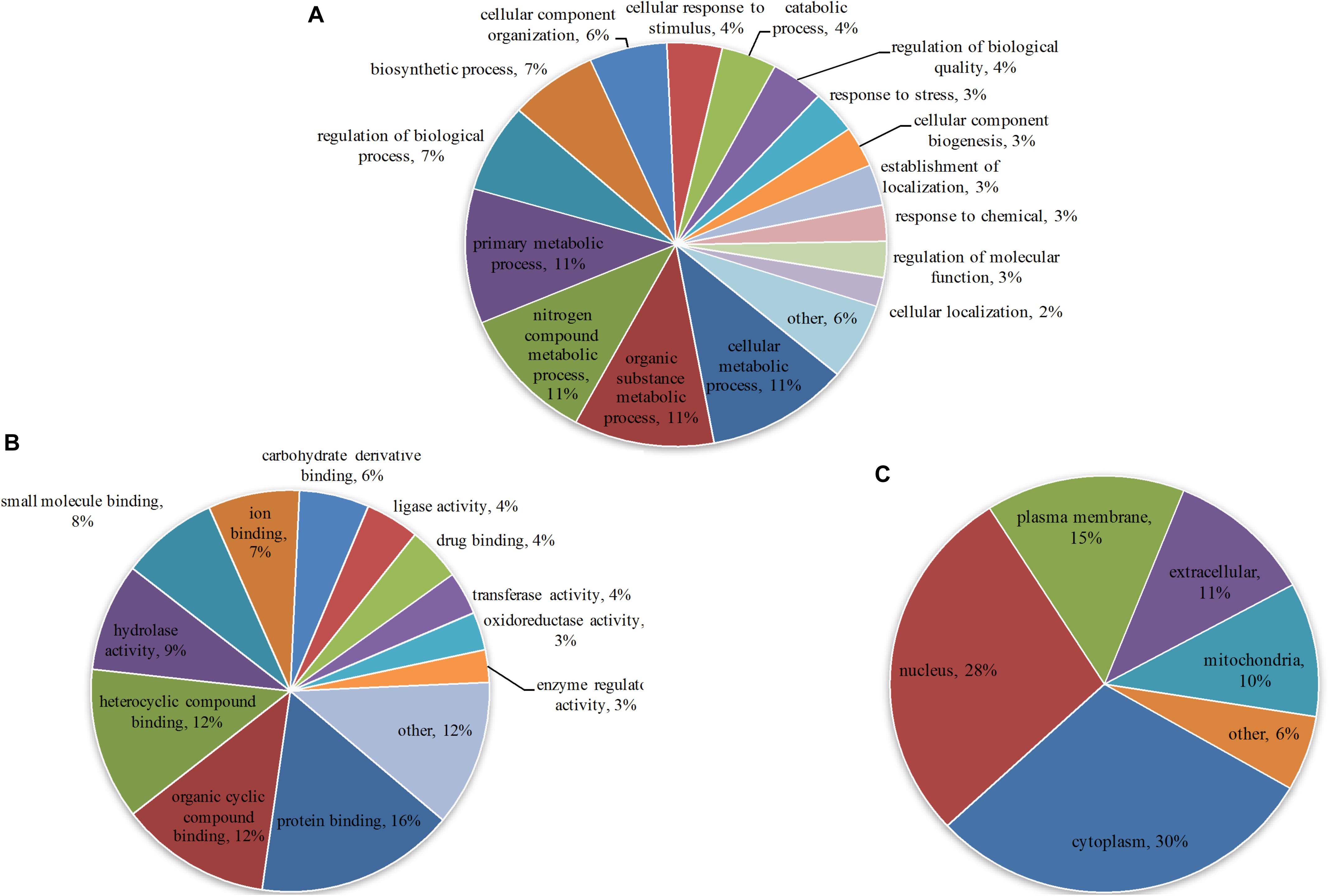
Figure 2. Gene Ontology (GO) functional annotation of the identified malonylated proteins in Toxoplasma gondii, including categories: (A) Biological process, (B) molecular function and (C) the GO annotation of the subcellular locations of the malonylated proteins, including the percentages of malonylated proteins involved in specific GO terms in relation to the total malonylated proteins.
The GO enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed in T. gondii, as shown in Supplementary Table S4. Based on the GO enrichment results of the CC category, malonylated proteins were significantly located in the cell body (Figure 3A). In terms of MF, the most significantly enriched malonylated proteins were related to ligase activity and small molecule binding (Figure 3B). Regarding BP, malonylated proteins were mainly implicated in purine metabolism and monocarboxylic acid biosynthetic processes processes (Figure 3C). Interestingly, seven malonylated proteins in T. gondii belonged to the pentose phosphate pathway (Figure 4A). A highly proliferative organism such as T. gondii is an avid auxotrophic for many metabolites and must consume these, and carbon sources (e.g., glucose) from the host cell to sustain its growth and proliferation. The pentose phosphate pathway along with glycolysis and mitochondrial respiration are used by the intracellular tachyzoites of T. gondii to generate energy and essential anabolic precursors from glucose (MacRae et al., 2012). In addition, eight malonylated proteins in T. gondii belonged to the fatty acid biosynthesis (Figure 4B). These are involved in the growth, development, and reproduction of T. gondii (Fu et al., 2019). Malonyl-CoA can serve as a substrate of fatty acid synthesis and as an inhibitor of fatty acid oxidation (Foster, 2012). Thus, malonylation might indirectly regulate these important processes in T. gondii. KEGG pathway analysis showed that most malonylated proteins are enriched in glycolysis/gluconeogenesis, aminoacyl-tRNA biosynthesis, fatty acid biosynthesis (Figure 5A). Furthermore, enriched protein domain analysis showed that malonylated proteins are significantly enriched in ribosomal protein L7Ae/L30e/S12e/Gadd45 family, glutamate/Leucine/Phenylalanine/Valine dehydrogenase, phosphoglycerate kinase and pyridine nucleotide-disulfide oxidoreductase were significantly enriched in the malonylated proteins (Figure 5B).
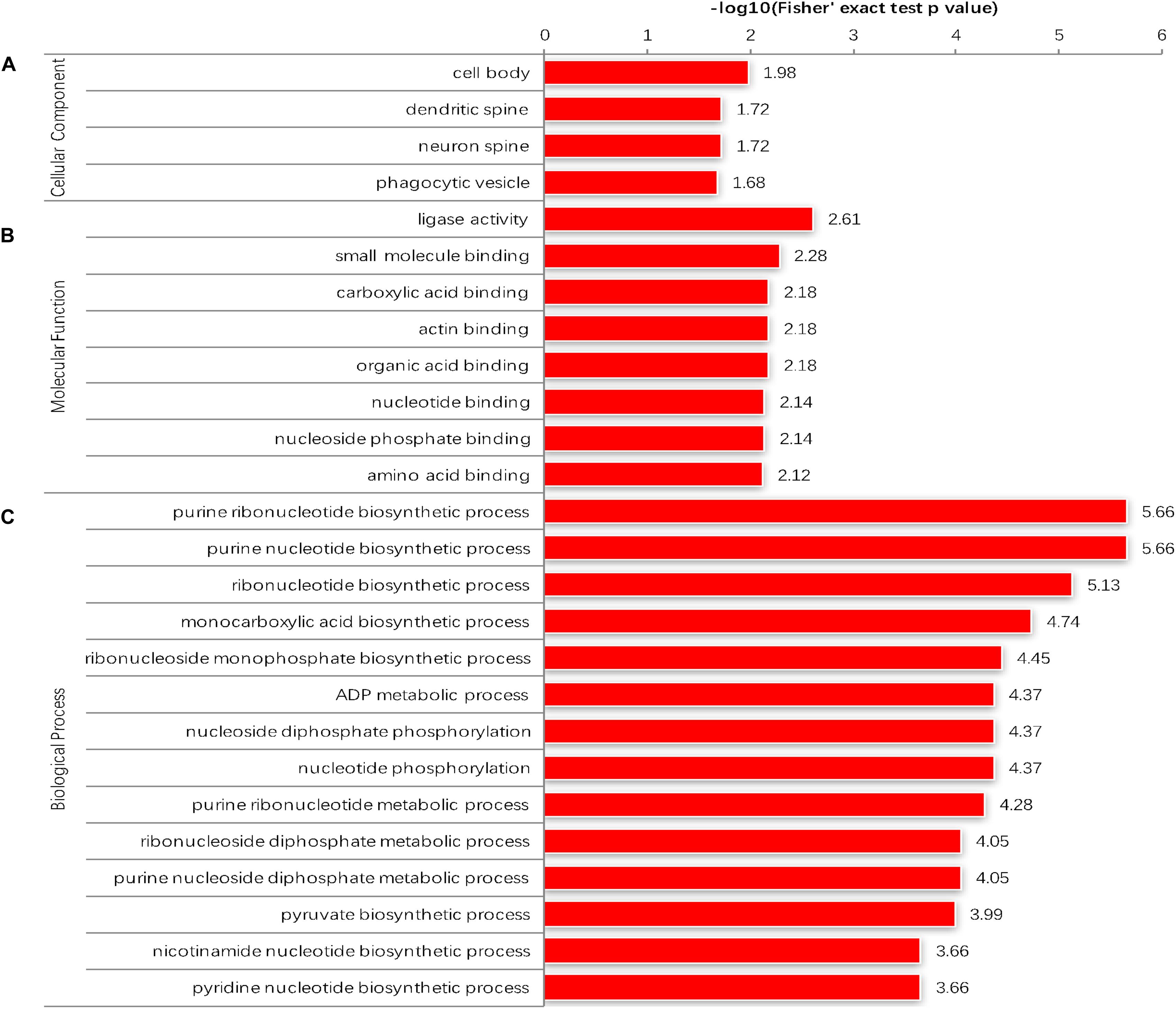
Figure 3. Enrichment analysis of the malonylated proteins in Toxoplasma gondii according to the categories of (A) cellular component. (B) Molecular function. (C) Biological process.
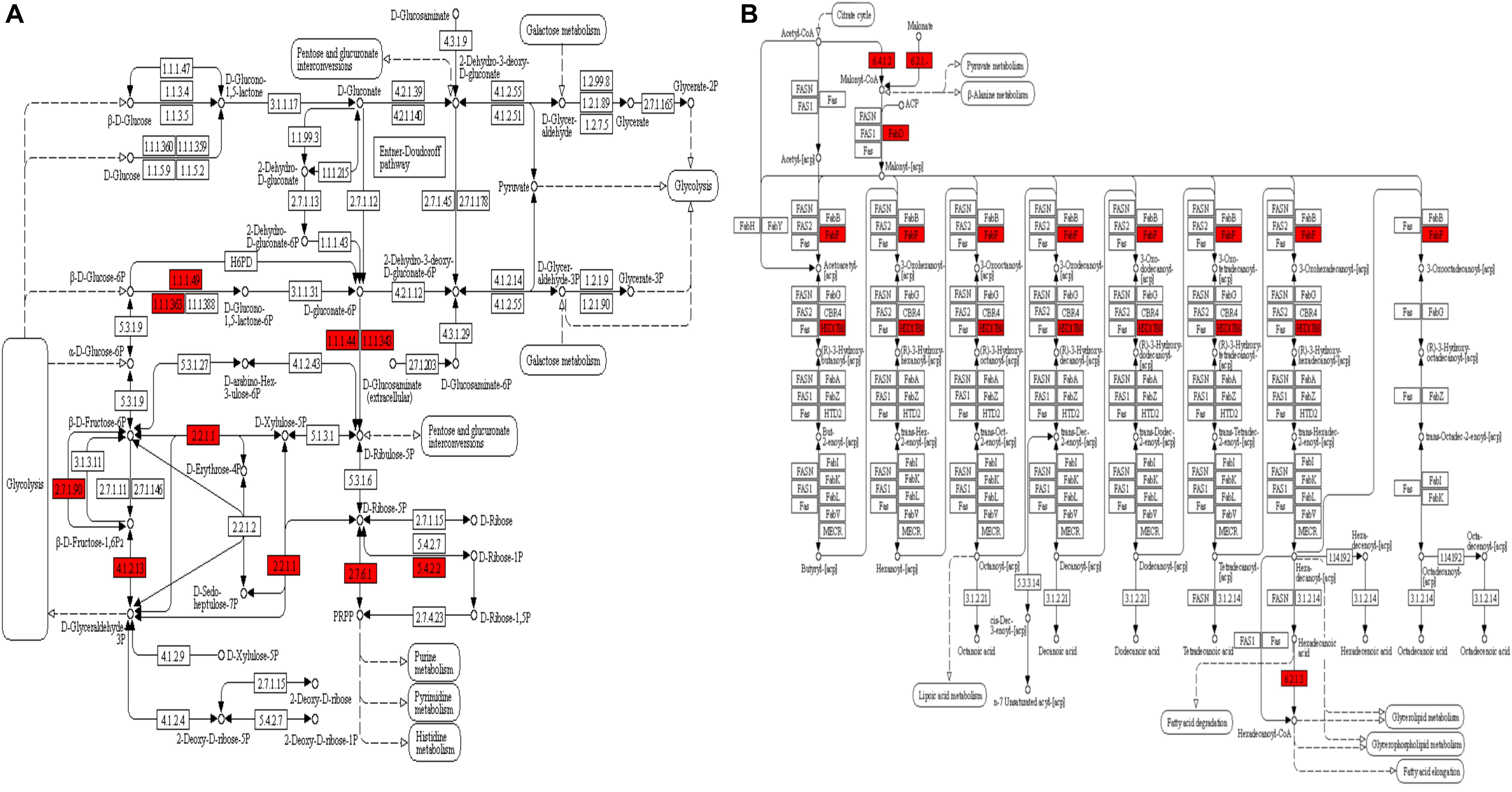
Figure 4. KEGG pathway enrichment analysis of the malonylated proteins in Toxoplasma gondii. (A) Alterations in the pentose phosphate pathway with significantly enriched modified proteins showing inside red boxes. (B) Alterations in the fatty acid biosynthesis pathway with significantly enriched modified proteins are shown inside red boxes.
The malonylated protein fructose-bisphosphate aldolase, which plays a central role in glycolysis and gluconeogenesis pathways, has been considered as a potential target for drug development against pathogenic bacteria (Ziveri et al., 2017). Another malonylated protein (leucyl aminopeptidase; LAP) is a functional aminopeptidase in the cytoplasm of T. gondii (Jia et al., 2010). Lactate dehydrogenase (LDH1) was also identified as a malonylated protein, and has been previously shown to catalyze the conversion of pyruvate and lactate in anaerobic growth conditions and used for energy supply (Abdelbaset et al., 2017). T. gondii expresses two different lactate dehydrogenase enzyme genes, LDH1 is only expressed in the tachyzoite stage whereas LDH2 is preferentially expressed in the bradyzoite stage. Given the important role of this enzyme in the parasite virulence, phenotypic differentiation, and latent infection, the potential of using T. gondii LDH mutant strains as vaccine candidates has been suggested (Abdelbaset et al., 2017). Five Kmal sites were identified in glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in the present study, this protein is also succinylated and ubiquitinated in T. gondii, and the glyceraldehyde-3-phosphate dehydrogenase I (GAPDH I) was found in the parasite cytoplasm, succinylated on two lysines (Li et al., 2014; Salmon de Monerri et al., 2015). Previous reports showed that phosphorylation and glycation of GAPDH are involved in the regulation of (PPI) and intracellular localization of the enzyme, and that the aggregation of GAPDH can be affected by all types of PTMs (Muronetz et al., 2016). Lysine 213 of GAPDH can be malonylated in macrophages following lipopolysaccharide (LPS) stimulation, which causes its dissociation from several inflammatory mRNAs, promoting translation (Galván-Peña et al., 2019), implying that Kmal has important roles in these processes. A total of 83 malonylated proteins identified in the present study had also been identified as acetylated proteins in a previous study (Jeffers and Sullivan, 2012). KEGG pathway analysis showed that these 83 proteins were mainly enriched in glycolysis/gluconeogenesis, carbon fixation in photosynthetic organisms, aminoacyl-tRNA biosynthesis and biosynthesis of antibiotics (Figure 6), which indicate that malonylation and acetylation may have similarities or synergistic effects on some functions and mechanisms. Further studies are warranted to elucidate the relationship between malonylation and acetylation in T. gondii.
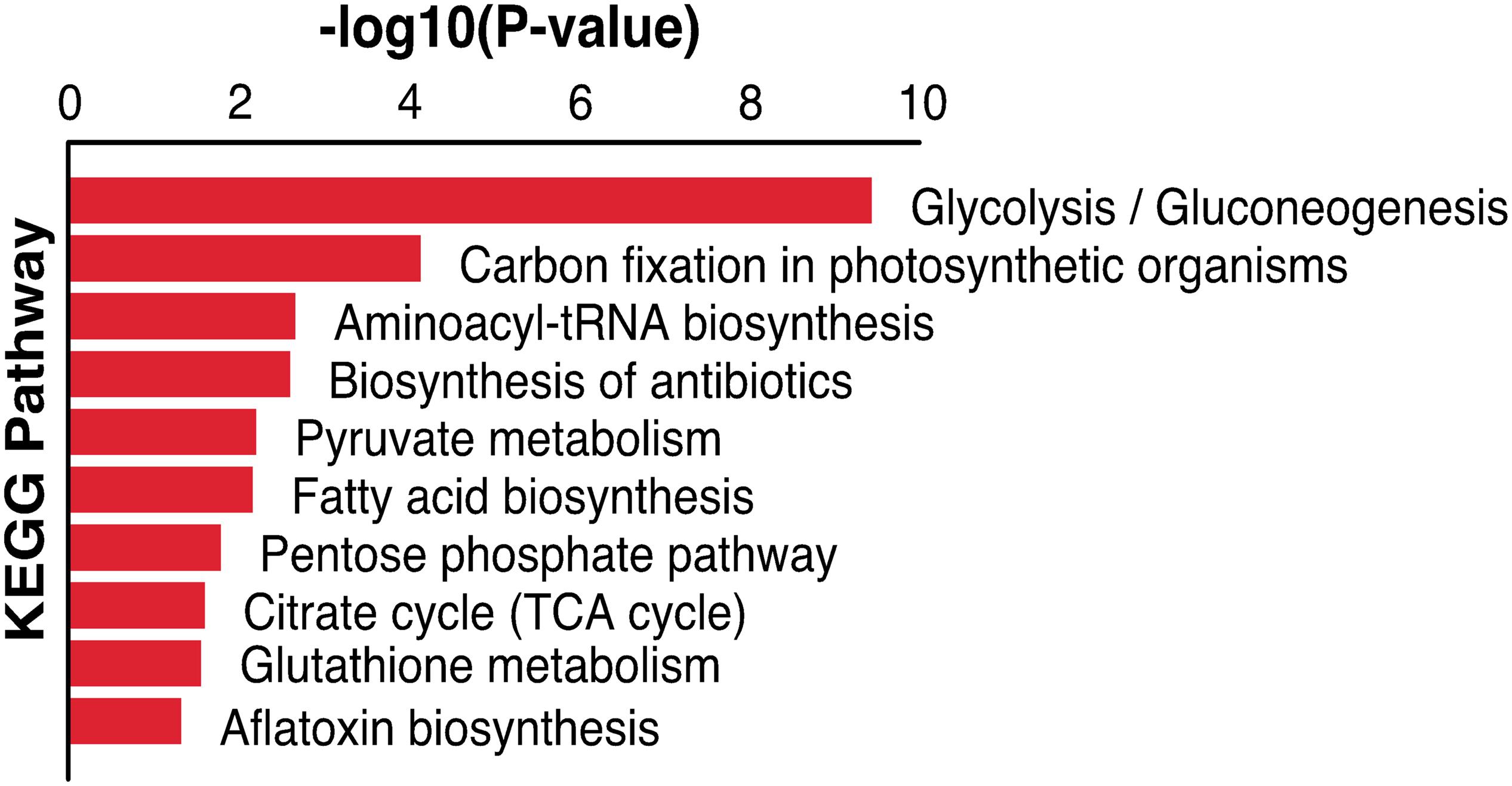
Figure 6. KEGG pathway enrichment analysis of both malonylated and acetylated proteins in Toxoplasma gondii.
The PPI analysis was performed by searching the STRING database and PPI networks were visualized using Cytoscape software in order to further identify the major biological processed affected by Kmal in T. gondii (Shannon et al., 2003; Figure 7, Supplementary Table S5). Using MCODE (Minimal Common Oncology Data Elements), a number of highly associated subnetworks of Kmal proteins were identified, including glycolysis/gluconeogenesis, ribosome, proteasome and aminoacyl-tRNA biosynthesis processes, which are in agreement with the KEGG pathway enrichment analysis, suggesting that these processes play important roles in shaping the proteomic landscape of T. gondii.
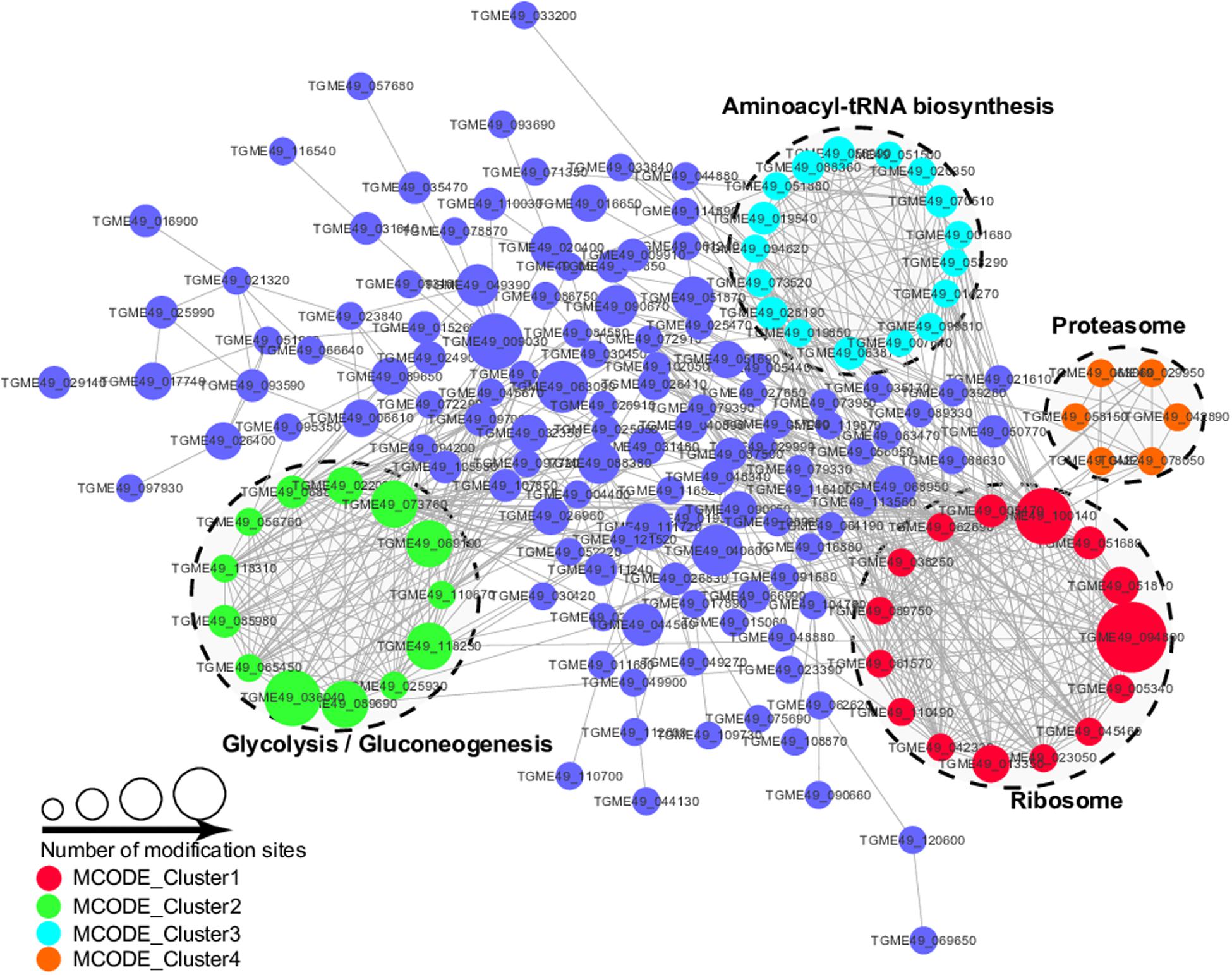
Figure 7. Protein–Protein interaction networks of malonylated proteins in Toxoplasma gondii. Nodes represent the malonylated proteins and edges represent interactors between malonylated proteins. The color of the edges denotes the combined score of interactors.
In this study, we described the first malonylome profile in T. gondii using mass spectrometry in combination with immune-affinity purification. Over 300 proteins were found to be malonylated and were localized in different cellular compartments. These malonylated proteins were associated with diverse biological and metabolic processes, suggesting that Kmal is involved in the regulation of T. gondii physiology. We also found that some important proteins related to T. gondii invasion, emergence and virulence were malonylated, which suggest that Kmal plays an important role in the regulation of functions of these critical proteins of T. gondii. Our findings will serve as a useful resource for further investigation of the functions of Kmal, especially in the context of parasite interaction with the host cells. Further studies should focus on the experimental verification of the functions of these malonylated proteins to reveal the exact roles of malonylation in T. gondii. Analysis of the extent of differences and similarities in Kmal between different developmental stages (bradyzoites, tachyzoites and oocysts) and between distinct genotypes of T. gondii is warranted, which in turn will facilitate the elucidation of mechanisms underlying the phenotypic transformation and varying virulence of T. gondii.
All the mass spectrometry data have been submitted to the ProteomeXchange Consortium with the identifier PXD015809.
RD, F-CL, and X-QZ conceived and designed the research. L-BN performed the research, analyzed the data and drafted the manuscript. Q-LL, HE, and N-JH contributed materials, reagents, and analysis tools. HE, F-CL, RD, and X-QZ critically revised the manuscript. All authors reviewed and approved the final version of the manuscript.
Project support was provided by the National Natural Science Foundation of China (Grant No. 31802182), National Key Research and Development Program of China (Grant No. 2017YFD0500403), and the International Science and Technology Cooperation Project of Gansu Provincial Key Research and Development Program (Grant No. 17JR7WA031).
N-JH was employed by Jingjie PTM Biolabs (Hangzhou) Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Jingjie PTM Biolabs (Hangzhou, China) for assistance in LC-MS/MS analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00776/full#supplementary-material
TABLE S1 | The identified Kmal sites in T. gondii in Exp.1.
TABLE S2 | The identified Kmal sites in T. gondii in Exp.2.
TABLE S3 | The identified Kmal sites in T. gondii in Exp.3.
TABLE S4 | Annotations of malonylated proteins including information on Gene Ontology (GO), protein domains, KEGG pathways, and subcellular localization.
TABLE S5 | Protein interaction network of the identified malonylated proteins.
Abdelbaset, A. E., Fox, B. A., Karram, M. H., Abd Ellah, M. R., Bzik, D. J., and Igarashi, M. (2017). Lactate dehydrogenase in Toxoplasma gondii controls virulence, bradyzoite differentiation, and chronic infection. PLoS One 12:e0173745. doi: 10.1371/journal.pone.0173745
Assossou, O., Besson, F., Rouault, J. P., Persat, F., Ferrandiz, J., Martine, M., et al. (2004). Characterization of an excreted/secreted antigen form of 14-3-3 protein in Toxoplasma gondii tachyzoites. FEMS Microbiol. Lett. 234:25.
Choudhary, C., Kumar, C., Gnad, F., Nielsen, M. L., Rehman, M., and Walther, T. C. (2009). Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840. doi: 10.1126/science
Colak, G., Pougovkina, O., Dai, L., Tan, M., Brinke, H., Huang, H., et al. (2015). Proteomic and biochemical studies of lysine malonylation suggest its malonic acidaria-associated regulatory role in mitochondrial function and fatty acid oxidation. Mol. Cell. Proteomics 14, 3056–3071. doi: 10.1074/mcp.M115.048850
Cox, J., Hein, M. Y., Luber, C. A., Paron, I., Nagaraj, N., and Mann, M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526. doi: 10.1074/mcp.M113.031591
Dubey, R., Staker, B. L., Foe, I. T., Bogyo, M., Myler, P. J., and Ngô, H. M. (2017). Gubbels M.J. Membrane skeletal association and post-translational allosteric regulation of Toxoplasma gondii GAPDH1. Mol. Microbiol. 103, 618–634. doi: 10.1111/mmi
Fan, B., Li, Y. L., Li, L., Peng, X. J., Bu, C., Wu, X. Q., et al. (2017). Malonylome analysis of rhizobacterium Bacillus amyloliquefaciens FZB42 reveals involvement of lysine malonylation in polyketide synthesis and plant-bacteria interactions. J. Proteome 154, 1–12. doi: 10.1016/j.jprot.2016.11.022
Flinner, N., Ellenrieder, L., Stiller, S. B., Becker, T., Schleiff, E., and Mirus, O. (2013). Mdm10 is an ancient eukaryotic porin co-occurring with the ERMES complex. Biochim. Biophys. Acta 1833, 3314–3325. doi: 10.1016/j.bbamcr.2013.10.006
Foster, D. W. (2012). Malonyl-CoA: the regulator of fatty acid synthesis and oxidation. J. Clin. Invest. 122, 1958–1959. doi: 10.1172/jci63967
Fu, Y., Cui, X., Liu, J., Zhang, X., Zhang, H., Yang, C., et al. (2019). Synergistic roles of acyl-CoA binding protein (ACBP1) and sterol carrier protein 2 (SCP2) in Toxoplasma lipid metabolism. Cell. Microbiol. 21:e12970. doi: 10.1111/cmi.12970
Galván-Peña, S., Carroll, R. G., Newman, C., Hinchy, E. C., Palsson-McDermott, E., and Robinson, E. K. (2019). Malonylation of GAPDH is an inflammatory signal in macrophages. Nat. Commun. 10:338. doi: 10.1038/s41467-018-08187-6
He, W., Newman, J. C., Wang, M. Z., Ho, L., and Verdin, E. (2012). Mitochondrial sirtuins: regulators of protein acylation and metabolism. Trends Endocrin. Met. 23, 467–476. doi: 10.1016/j.tem.2012.07.004
Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. doi: 10.1146/annurev.biochem.67.1.425
Hirschey, M. D., and Zhao, Y. (2015). Metabolic regulation by lysine malonylation, succinylation and glutarylation. Mol. Cell. Proteomics 14, 2308–2315. doi: 10.1074/mcp.R114.046664
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Jeffers, V., and Sullivan, W. J. (2012). Lysine acetylation is widespread on proteins of diverse function and localization in the protozoan parasite Toxoplasma gondii. Eukaryot. Cell 11, 735–742. doi: 10.1128/EC.00088-12
Jia, H., Nishikawa, Y., Luo, Y., Yamagishi, J., Sugimoto, C., and Xuan, X. (2010). Characterization of a leucine aminopeptidase from Toxoplasma gondii. Mol. Biochem. Parasitol. 170, 1–6. doi: 10.1016/j.molbiopara.2009.11.005
Jin, W., and Wu, F. (2016). Proteome-wide identification of lysine succinylation in the proteins of tomato (Solanum lycopersicum). PLoS One 11:e0147586. doi: 10.1371/journal.pone.0147586
Kikumura, A., Fang, H., Mun, H. S., Uemura, N., Makino, M., Sayama, Y., et al. (2010). Protective immunity against lethal anaphylactic reaction in Toxoplasma gondii-infected mice by DNA vaccination with T. gondii-derived heat shock protein 70 gene. Parasitol. Int. 59, 105–111. doi: 10.1016/j.parint.2010.03.006
Li, D., Lv, B., Tan, L., Yang, Q., and Liang, W. (2016). Acetylome analysis reveals the involvement of lysine acetylation in diverse biological processes in Phytophthora sojae. Sci. Rep. 6:29897. doi: 10.1038/srep29897
Li, X., Hu, X., Wan, Y., Xie, G., Li, X., Chen, D., et al. (2014). Systematic identification of the lysine succinylation in the protozoan parasite Toxoplasma gondii. J. Proteome Res. 13, 6087–6095. doi: 10.1021/pr500992r
Lin, H., Su, X., and He, B. (2012). Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem. Biol. 7, 947–960. doi: 10.1021/cb3001793
Liu, J., Wang, G., Lin, Q., Liang, W., Gao, Z., Mu, P., et al. (2018). Systematic analysis of the lysine malonylome in common wheat. BMC Genomics 19:209. doi: 10.1186/s12864-018-4535-y
Liu, L., Wang, G., Song, L., Lv, B., and Liang, W. (2016). Acetylome analysis reveals the involvement of lysine acetylation in biosynthesis of antibiotics in Bacillus amyloliquefaciens. Sci. Rep. 6:20108. doi: 10.1038/srep20108
Liu, Q., Wang, Z. D., Huang, S. Y., and Zhu, X. Q. (2015). Diagnosis of toxoplasmosis and typing of Toxoplasma gondii. Parasit. Vectors 8:292. doi: 10.1186/s13071-015-0902-6
Lv, B., Yang, Q., Li, D., Liang, W., and Song, L. (2016). Proteome-wide analysis of lysine acetylation in the plant pathogen Botrytis cinerea. Sci. Rep. 6:29313. doi: 10.1038/srep29313
Ma, Y., Yang, M., Lin, X., Liu, X., Huang, H., and Ge, F. (2017). Malonylome analysis reveals the involvement of lysine malonylation in metabolism and photosynthesis in cyanobacteria. J. Proteome Res. 16, 2030–2043. doi: 10.1021/acs.jproteome.7b00017
MacRae, J. I., Sheiner, L., Nahid, A., Tonkin, C., Striepen, B., and McConville, M. J. (2012). Mitochondrial metabolism of glucose and glutamine is required for intracellular growth of Toxoplasma gondii. Cell Host Microbe 12, 682–692. doi: 10.1016/j.chom.2012.09.013
Montoya, J. G., and Liesenfeld, O. (2004). Toxoplasmosis. Lancet 363, 1965–1976. doi: 10.1016/S0140-6736(04)16412-X
Muronetz, V. I., Barinova, K. V., Stroylova, Y. Y., Semenyuk, P. I., and Schmalhausen, E. V. (2016). Glyceraldehyde-3-phosphate dehydrogenase: aggregation mechanisms and impact on amyloid neurodegenerative diseases. Int. J. Biol. Macromol. 100, 55–66. doi: 10.1016/j.ijbiomac.2016.05.066
Pan, J., Chen, R., Li, C., Li, W., and Ye, Z. (2015). Global analysis of protein lysine succinylation profiles and their overlap with lysine acetylation in the marine bacterium Vibrio parahemolyticus. J. Proteome Res. 14, 4309–4318. doi: 10.1021/acs.jproteome.5b00485
Peng, C., Lu, Z., Xie, Z., Cheng, Z., Chen, Y., Tan, M., et al. (2011). The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteomics 10, M111.012658. doi: 10.1074/mcp.M111.012658
Pooe, A., Gabriele, K., Heine, H., and Shonhai, A. (2017). Plasmodium falciparum heat shock protein 70 lacks immune modulatory activity. Protein Peptide Lett. 24, 503–510. doi: 10.2174/0929866524666170214141909
Qian, L., Nie, L., Chen, M., Liu, P., Zhu, J., Zhai, L., et al. (2016). Global profiling of protein lysine malonylation in Escherichia coli reveals its role in energy metabolism. J. Proteome Res. 15, 2060–2071. doi: 10.1021/acs.jproteome.6b00264
Ridgley, E. L., Xiong, Z. H., Kaur, K. J., and Ruben, L. (1996). Genomic organization and expression of elongation factor-1 alpha genes in Trypanosoma brucei. Mol. Biochem. Parasitol. 79, 119–123. doi: 10.1016/0166-6851(96)02639-4
Robert-Gangneux, F., and Darde, M. L. (2012). Epidemiology of and diagnostic strategies for toxoplasmosis. Clin. Microbiol. Rev. 25, 264–296. doi: 10.1128/CMR.05013-11
Salmon de Monerri, N. C., Yakubu, R. R., Chen, A. L., Bradley, P. J., Nieves, E., Weiss, L. M., et al. (2015). The ubiquitin proteome of Toxoplasma gondii reveals roles for protein ubiquitination in cell-cycle transitions. Cell Host Microb. 18, 621–633. doi: 10.1016/j.chom.2015.10.014
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Toueille, M., Saint-Jean, B., Castroviejo, M., and Benedetto, J. P. (2007). The elongation factor 1A: a novel regulator in the DNA replication/repair protein network in wheat cells? Plant Physiol. Biochem. 45, 113–118. doi: 10.1016/j.plaphy.2007.01.006
Wang, Z. X., Hu, R. S., Zhou, C. X., He, J. J., Elsheikha, H., and Zhu, X. Q. (2019). Label-free quantitative acetylome analysis reveals Toxoplasma gondii genotype-specific acetylomic signatures. Microorganisms 30, 7. doi: 10.3390/microorganisms7110510
Webster, J. P. (2010). Dubey, j. p. toxoplasmosis of animals and humans. Parasit. Vectors 3:112. doi: 10.1186/1756-3305-3-112
Weidner, J. M., Kanatani, S., Uchtenhagen, H., Varas-Godoy, M., Schulte, T., Engelberg, K., et al. (2016). Migratory activation of parasitized dendritic cells by the protozoan Toxoplasma gondii 14-3-3 protein. Cell. Microbiol. 18, 1537–1550. doi: 10.1111/cmi.12595
Xiao, H., El Bissati, K., Verdier-Pinard, P., Burd, B., Zhang, H., Kim, K., et al. (2010). Post-translational modifications to Toxoplasma gondii alpha- and beta-tubulins include novel C-terminal methylation. J. Proteome Res. 9, 359–372. doi: 10.1021/pr900699a
Xie, Z., Dai, J., Dai, L., Tan, M., Cheng, Z., Wu, Y., et al. (2012). Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics 11, 100–107. doi: 10.1074/mcp.m111.015875
Xu, J. Y., Xu, Z., Zhou, Y., and Ye, B. C. (2016). Lysine malonylome may affect the central metabolism and erythromycin biosynthesis pathway in Saccharopolyspora erythraea. J. Proteome Res. 15:1685. doi: 10.1021/acs.jproteome.6b00131
Zhang, Y., Song, L., Liang, W., Mu, P., Wang, S., and Lin, Q. (2016). Comprehensive profiling of lysine acetylproteome analysis reveals diverse functions of lysine acetylation in common wheat. Sci. Rep. 6:21069. doi: 10.1038/srep21069
Keywords: Toxoplasma gondii, toxoplasmosis, post-translational modifications, lysine malonylation, malonylome
Citation: Nie L-B, Liang Q-L, Du R, Elsheikha HM, Han N-J, Li F-C and Zhu X-Q (2020) Global Proteomic Analysis of Lysine Malonylation in Toxoplasma gondii. Front. Microbiol. 11:776. doi: 10.3389/fmicb.2020.00776
Received: 08 January 2020; Accepted: 31 March 2020;
Published: 28 April 2020.
Edited by:
Lihua Xiao, South China Agricultural University, ChinaReviewed by:
Si-Yang Huang, Yangzhou University, ChinaCopyright © 2020 Nie, Liang, Du, Elsheikha, Han, Li and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Du, ZHVydWkxOTcxMDdAMTI2LmNvbQ==; Fa-Cai Li, bGk3ODU2MTI3MEAxNjMuY29t; Xing-Quan Zhu, eGluZ3F1YW56aHUxQGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.