- 1Stream Biofilm and Ecosystem Research Laboratory, School of Architecture, Civil and Environmental Engineering, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland
- 2Groupe de recherche interuniversitaire en limnologie, Département des sciences biologiques, Université du Québec à Montréal, Montréal, QC, Canada
Despite the recognition of streams and rivers as sources of methane (CH4) to the atmosphere, the role of CH4 oxidation (MOX) in these ecosystems remains poorly understood to date. Here, we measured the kinetics of MOX in stream sediments of 14 sites to resolve the ecophysiology of CH4 oxidizing bacteria (MOB) communities. The streams cover a gradient of land cover and associated physicochemical parameter and differed in stream- and porewater CH4 concentrations. Michealis–Menten kinetic parameter of MOX, maximum reaction velocity (Vmax), and CH4 concentration at half Vmax (KS) increased with CH4 supply. KS values in the micromolar range matched the CH4 concentrations measured in shallow stream sediments and indicate that MOX is mostly driven by low-affinity MOB. 16S rRNA gene sequencing identified MOB classified as Methylococcaceae and particularly Crenothrix. Their relative abundance correlated with pmoA gene counts and MOX rates, underscoring their pivotal role as CH4 oxidizers in stream sediments. Building on the concept of enterotypes, we identify two distinct groups of co-occurring MOB. While there was no taxonomic difference among the members of each cluster, one cluster contained abundant and common MOB, whereas the other cluster contained rare operational taxonomic units (OTUs) specific to a subset of streams. These integrated analyses of changes in MOB community structure, gene abundance, and the corresponding ecosystem process contribute to a better understanding of the distal controls on MOX in streams.
Introduction
In streambeds, microorganisms organized in surface-attached and matrix-enclosed biofilms drive biogeochemical processes that regulate the outgassing of carbon dioxide (CO2), methane (CH4), and nitrous oxide (N2O) (Battin et al., 2016). While the study of stream CO2 biogeochemistry has received considerable attention over the last decade, the biogeochemistry of CH4 (Crawford et al., 2014; Trimmer et al., 2015; Stanley et al., 2016) and specifically the microbial ecology of aerobic CH4 oxidizing bacteria (MOB) in streams remains poorly understood (Trimmer et al., 2015, 2010; Stanley et al., 2016). Streams outgas substantial amounts of CH4 originating from groundwater and methanogenesis within the streambed (Chaudhary et al., 2014; Stanley et al., 2016). Globally, streams and rivers may emit 26.8 Tg CH4 per year, which is similar to the combined contribution of other natural CH4 sources such as wildfires, termites, CH4 hydrates, and permafrost (Stanley et al., 2016). The presence of wetlands and human activity in stream catchments are positively related to streamwater CH4 concentrations and CH4 evasion fluxes (Chaudhary et al., 2014; Stanley et al., 2016). Land cover is linked to streamwater CH4 concentration, as the excess delivery of organic matter and nutrients leads to oxygen depletion and thus stimulates anaerobic methanogenesis.
MOB constitute the main microbial processors of CH4 at oxic/anoxic interfaces in stream sediments (Trimmer et al., 2010, 2015; Buriánková et al., 2012; Shelley et al., 2014, 2015) where MOB can account for up to 30% of prokaryotic cell numbers (Buriánková et al., 2012). CH4 oxidation (MOX) can be an important ecosystem process, equivalent to up to 46% of net photosynthetic production in chalk streams (Trimmer et al., 2010; Shelley et al., 2014) and reducing CH4 evasion fluxes by 70% (Zaiss et al., 1981; de Angelis and Scranton, 1993). Among MOB, low-affinity CH4 oxidizers with substrate affinity constants KS [i.e., the substrate concentration for which the MOX rate is half the maximum reaction velocity (Vmax)] in the micromolar range are adapted to elevated CH4 concentrations typical for hot spots of methanogenesis. In contrast, high-affinity oxidizers with KS values in the nanomolar range are adapted to atmospheric CH4 concentrations (Bender and Conrad, 1992). Independent of their kinetic affinity, known MOB are distributed across Methylocystaceae, Beijerinckiaceae (both α-Proteobacteria, or type II MOB), Methylococcaceae, Methylothermaceae (both γ-Proteobacteria, or type I MOB), and Methylacidiphilaceae (Verrucomicrobia) (Knief, 2015). Proteobacterial MOB dominate methanotrophic communities in most environments and were previously associated with two life strategies (Ho et al., 2013): γ-proteobacterial MOB are generally responsive to changes in CH4 concentrations by rapidly inducing pmoA gene expression (coding for a subunit of the particulate CH4 monooxygenase). Although numerically not abundant in many environments, they functionally dominate MOX when CH4 concentrations are high. Recent studies revealed the importance of Crenothrix, a filamentous type I MOB in lakes (Oswald et al., 2017) and caves (Karwautz et al., 2018). In contrast, α-proteobacterial MOB typically feature more stable populations with less responsiveness to fluctuating CH4 concentrations (Ho et al., 2013).
The aim of this study was to link MOX kinetics to MOB community structure and genetic potential of stream sedimentary communities. Experimenting with stream sediments sampled across a large environmental gradient (including alpine, natural, urban, and agriculture-dominated catchments), we hypothesized that the kinetics of MOX depend on CH4 supply, which we expected to predictably vary with land use. By characterizing changes in the MOB community (membership and overall structure) and measuring both the gene and corresponding ecosystem processes across environmental gradients, we aimed to better understand the distal controls on MOX in streams. Elucidating the changes in MOX and MOB communities is critical, given the apparent relationship between land use, nutrient, and CH4 dynamics in streams (Stanley et al., 2016) and recent predictions that eutrophication will continue to increase during the 21st century (Sinha et al., 2017).
Materials and Methods
Study Sites and Sampling
We sampled 14 sites (located between 378 and 1,189 m above sea level) of nine streams in Switzerland between March and July 2016. Using QGIS, we derived the percent land cover for each of the catchments using European Environment Agency’s 2018 Corine Land Cover (CLC, Version 20) database. Dominant land cover types included non-irrigated arable land, mixed forest, coniferous forest, natural grasslands, bare rock, discontinuous urban fabric, and land principally occupied by agriculture with significant areas of natural vegetation and transitional woodland-shrub (Supplementary Table S1).
At each site, we collected triplicate benthic sediment samples (down to 5 cm depth) from which we retained the sandy fraction (<2 mm). Sediment subsamples for DNA extraction were immediately transferred to LifeGuard Soil Preservation Solution (Mobio) and stored at −80°C until processing. Subsamples for flow cytometric cell counting were immediately fixed with 2.5% formaldehyde (final concentration) and kept cool (4°C). Sediments were transported cooled to the laboratory for kinetic analyses (see below). Porewater was sampled using a 1.5-cm diameter piezometer connected to a syringe. For the determination of dissolved CH4 and CO2, we collected 30 ml stream- and porewater into 60 ml syringes flushed with N2 and containing 0.05 g of NaN3 to stop biological activity. Stream- and porewater dissolved oxygen (DO) was directly measured with an LDO probe (Hach) equipped with a flow-through cell connected to the piezometer. Another set of stream- and porewater samples were filtered [0.2 μm polytetrafluoroethylene (PTFE) filters] and treated with 1 M HCl for the measurement of Fe(II) or 5% (w/v) zinc acetate for S(-II) measurements. Stream- and porewater samples for the determination of dissolved organic carbon (DOC) concentration were filtered through pre-combusted (450°C for 4 h) GF/F filters (Whatman).
Geochemical Analyses
Anions and cations were measured by ion chromatography (DX-3000, Dionex) with an IonPac AS11-HC column (anions) and an IonPac CS16 Cation-Exchange column (cations). S(-II) and Fe(II) were measured photometrically (Cline, 1969; Stookey, 1970). DOC concentration was measured using a Sievers M5310C TOC Analyzer. CO2 and CH4 concentrations in the streamwater and porewater were measured with the headspace equilibration method and using cavity ring-down spectroscopy (Picarro G2201-I). First, we added ultrapure N2 (30 ml) to the syringes containing the sample, which we shook (1 min) to allow gases to equilibrate with the headspace. The gases were then transferred to a gas-tight glass syringe (N2-flushed) and diluted to a final volume of 50 ml with N2. Partial pressures of CO2 and CH4 in the headspace samples were converted to gas concentration in the water samples as described previously (Bagnoud et al., 2016). The surface area of dry sediments was measured using the Brunauer–Emmett–Teller method (Gemini 2375 V4.01; Micromeritics). All rates and microbial parameters were normalized to sediment surface area.
Methane Oxidation Kinetics
We derived MOX kinetics from laboratory incubations in round, 11 cm in diameter and 8 cm in height Plexiglas chambers (volume: 760 cm3). The chambers featured in- and outlet valves at the lid and were sealed airtight using screws and a rubber seal. Prior to the experiments, the respective streamwater was equilibrated with air amended with 1% CH4 (Segers, 1998). For each site, triplicated incubations were set up with approximately 25 g of sediment (wet weight), which covered the bottom of the chambers with a thin layer. The chambers were then filled headspace-free using CH4-amended streamwater. To avoid diffusion limitation, magnetic stirring bars in the center of the incubation chambers ensured continuous mixing of the water within the chambers (sediment was initially pushed toward the sides of the chambers but then remained undisturbed). Triplicated incubations without sediment for each site were used to estimate the contribution of streamwater microbial communities to MOX in the incubations. The incubations were performed in the dark and at 20°C ± 2°C. Concentrations of dissolved O2 (DO), CH4, and CO2 were monitored twice per day in each chamber over 3–5 days, depending on MOX rates. DO was measured using optical sensors (SP-PSt3-NAU, Presens, Germany) mounted to the inside of the chambers. The optical DO reading was obtained through the Plexiglass. For the measurement of CH4 and CO2, 30 ml of water was withdrawn from the chambers and measured using the headspace equilibration method and cavity ring-down spectroscopy as described above. The removed water was simultaneously replaced with unamended streamwater (equilibrated with air), and we accounted for dilution effects by measuring CH4, CO2, and O2 concentrations therein.
Kinetic parameters of MOX were modeled based on Michaelis–Menten:
where V is the MOX velocity (or rate), Vmax the maximum reaction velocity, S the CH4 concentration, and Ks the concentration for which V = Vmax/2 (half-saturation constant).
16S rDNA Amplicon Sequencing and Bioinformatic Analyses
DNA was extracted from sediments collected in the field using the Fast DNA spin kit (MP Bio). 16S rRNA genes were amplified using the 515F/806R primers (Caporaso et al., 2011) overhanged with adapter sequences according to the MiSeq manufacturer’s instructions, using an annealing temperature of 50°C. Library preparation and sequencing on an Illumina MiSeq platform (250 bp paired-end) was performed by the LGTF sequencing facility (University of Lausanne).
Reads were analyzed using the UPARSE pipeline (Edgar, 2013), including the removal of unique reads, reads with an expected error >1, and chimeric sequences. Reads were clustered to operational taxonomic units (OTUs) at 97% sequence identity. The RDP Classifier v.1.12 was used to assign taxonomy to OTUs with a confidence threshold of 0.8 (Wang et al., 2007). Raw and representative OTU sequences were deposited to NCBI SRA and NCBI Genbank, respectively. Both datasets can be accessed under BioSample numbers SAMN07174972-SAMN07174985.
A reference tree for Methylococcacea was constructed using published sequences (Bowman, 2015) using IQ-TREE v.1.5.3 (Nguyen et al., 2015). References were aligned with MAFFT v.7.309 (Katoh and Standley, 2013). Sequences of Methylococcaceae OTUs were added to the reference alignment using MAFFT. RAxML v.8.2.9 (Stamatakis, 2014) was used to compute the evolutionary placements of the OTUs on the tree.
Quantitative PCR
We used the primer pair A189 F/Mb661 R (Holmes et al., 1995; Costello and Lidstrom, 1999) for the quantification of the pmoA gene. Reactions were performed in duplicate on a MIC instrument (Bio Molecular Systems) using the SensiFAST SYBR No-ROX kit (Bioline) containing 1 × PCR buffer, 500 nM of each primer, 0.1 ng μl−1 bovine serum albumin (BSA; Invitrogen) and 2.5 μl template DNA. Various five-fold dilutions of the DNA extracts (up to 1:125 or 1:625 in water) were examined to assess the efficiency of the amplification assay for each sample. For every sample, the dilution resulting in a ΔCq closer to the theoretical value for five-fold dilutions (2.3 cycles) was used for further analysis. After an initial denaturation at 94°C for 5 min, 45 cycles were performed at 95°C for 15 s, 60°C for 20 s, and 72°C for 30 s. Data acquisition was performed at 82°C for 5 s to avoid fluorescent signal from primer dimers. Melt curve analysis was performed from 72 to 95°C in 0.3°C s−1 increments. Agarose gel electrophoresis was used in addition to verify the formation of expected PCR product. Reactions containing all reagents except template DNA were included as negative controls. Amplification efficiency of standard curves ranged from 91 to 95% (R2 > 0.99), while corresponding amplification efficiencies of samples showed only minor inhibition of the amplification reaction in a few of them (amplification efficiency 84 to 110% and R2 > 0.99). A minimum sensitivity of 10 target molecules per reaction was achieved.
Bacterial Abundance
Bacterial cells associated with sediments were enumerated using flow cytometry (Novocyte, Acea) (Besemer et al., 2009). Cells were detached and separated from the sediment by shaking (1 h) and sonication (1 min; 1-Hz pulses; 14% amplitude) in pyrophosphate solution (0.025 mM). Samples were diluted (1:10) with MilliQ-water, stained with SYTO13 (Molecular Probes) (25 μM) and identified in plots of fluorescence at 520 nm and side scatter of the 488-nm laser. Volumetric counts, corrected for dilution with fixative and dye, were normalized to sediment surface.
Statistical Analyses
We used principal component analysis (PCA) to characterize the gradient of streams sampled. For this, we used the normalized concentrations of streamwater NO3–, SO42–, Cl–, and Ca2+ and DOC, pH, temperature, and altitude of each site as well as the percent coverage of the major land cover categories (Supplementary Table S1). Next, we fitted stream- and porewater CH4 concentrations onto the PCA using the function envfit implemented in the R package vegan (Oksanen et al., 2017). Pearson’s correlation analyses, linear models, and non-metric multidimensional scaling (nMDS) ordinations were also computed using R. A generalized additive model (function ordisurf) was used to fit pmoA copy numbers to an nMDS based on Bray–Curtis dissimilarities using the R package vegan. In analogy to the identification of community types (Gonze et al., 2017) for which samples are grouped by community composition and form distinct clusters (i.e., enterotypes in the gut microbiome) (Costea et al., 2018), we investigated the contribution of MOB to the microbial composition landscape. For this, we excluded the most oligotrophic sample Adn-1, which contained only a single methanotrophic OTU. Next, we visualized the result of the nMDS ordination as the density landscape of OTUs scores. We then highlighted the location of methanotrophic OTUs in this community composition landscape and identified two distinct clusters of MOB OTUs using k-mean clustering. Finally, we extracted non-MOB OTUs that co-cluster with MOB clusters in the community composition landscape and assessed whether taxonomic groups were relatively overrepresented. We interpret the relative overrepresentation of a taxonomic group associated with the MOB clusters as signs of a positive association of these co-occurring groups.
Results
Geochemical Characterization of Streams
The 14 sites covered a wide gradient of environmental conditions (Supplementary Table S2 and Figure 1A). The PCA revealed three classes of sites: catchments of Vev-1 and Adn-1 were characterized by natural vegetation, mainly grasslands and bare rocks. These sites were located at high altitude; streamwater temperature and the concentration of ions and DOC were low. Vey-1, Vey-2, Ven-1, and Ven-2 formed another group of samples, with catchments covered to approximately 30% by mixed and coniferous forests and elevated concentrations of ions and DOC in the streamwater. Finally, Geb-1, Sen-1, Sor-1, Sor-2, Sor-3, Com-1, Com-2, and Tal-1 formed another group of samples with arable land and urban regions dominating the catchments. They featured elevated ion and DOC concentrations and higher temperatures as compared to the sites at high altitude. However, compared to the sites dominated by forests, these sites were characterized by elevated pH. Fitting stream and shallow porewater CH4 concentrations to the ordination showed that sites at elevated altitude were generally characterized by low CH4 concentrations in both stream- and porewater. The group of samples with substantial areas covered by forests had elevated CH4 concentration in the streamwater, whereas the sites dominated by agriculture and urban areas were characterized by high porewater CH4 concentrations.
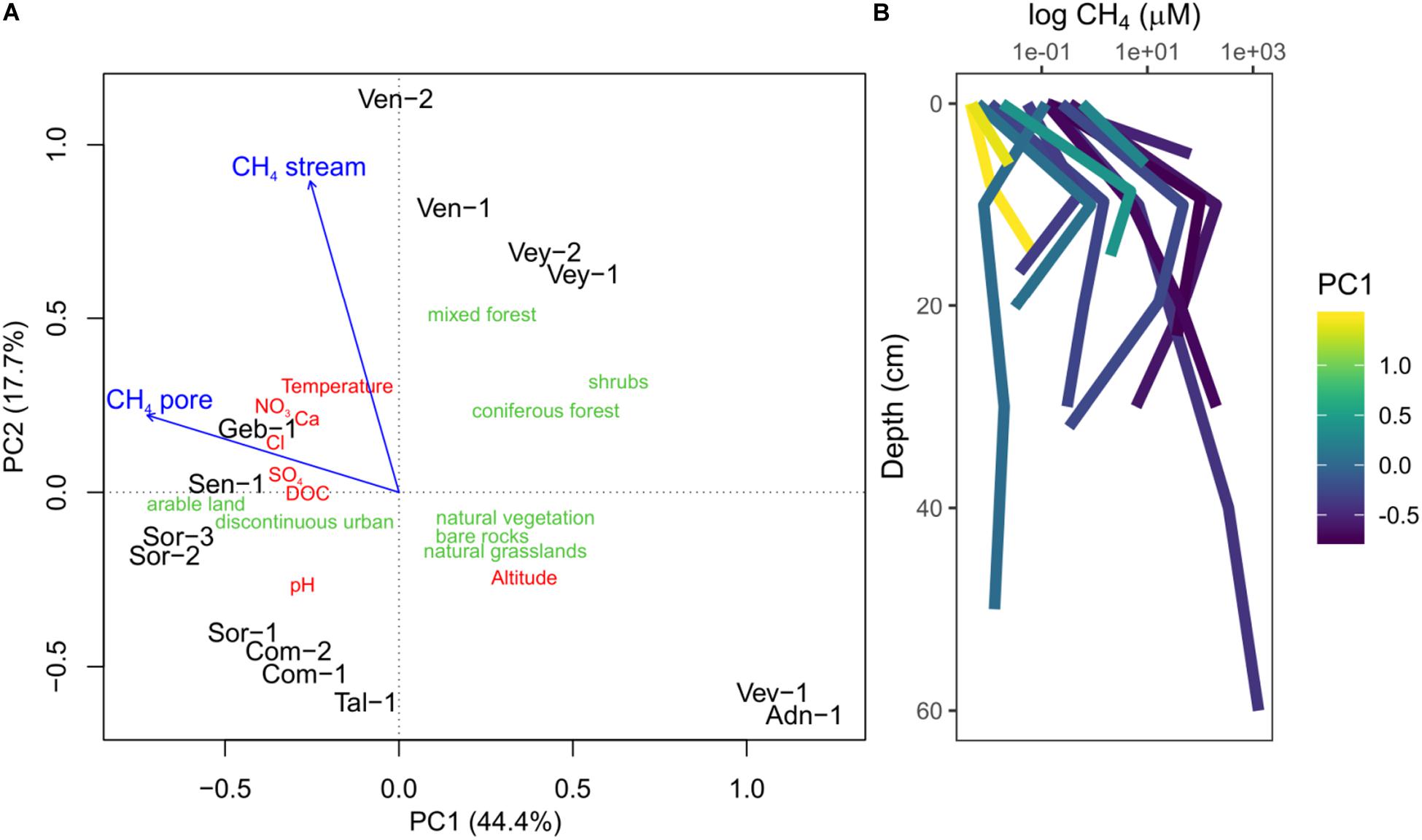
Figure 1. Principal component analysis (PCA) using stream environmental characteristics (red) and land cover type (green) to cluster sampling sites (A). The sampling sites group into three categories: sites at high altitude dominated by bare rock and natural vegetation (Vev-1, And-1), sites with substantial areas of the catchment covered by forests (Ven-2, Ven-1, Vey-2, Vey-1), and sites dominated by arable land and urban areas (Geb-1, Sen-1, Sor-1, Sor-2, Sor-3, Com-1, Com-2, Tal-1). Stream- and porewater methane (CH4) concentrations were fitted onto the ordination (blue arrows) and were elevated in sites with large areas of the catchment dominated by arable land and forests. Concentration profiles of CH4 measured in streambeds (B) color coded according to the site scores on the first PC (PC1). Measurements at 0 cm represent streamwater. Shown are averages of replicated measurements at each depth. Note the peak in CH4 concentration at approximately 10 cm depth in most sites.
In these 14 streams, CH4 concentrations were higher in the porewater (average ± standard deviation; 77.75 ± 234.75 μM) than in the streamwater (0.134 ± 0.159 μM) (Figure 1B), except for stream Ven-1. Porewater CH4 concentrations often peaked around a depth of 10 cm (Figure 1B and Supplementary Figure S1). Concomitantly with CH4, CO2 concentration increased on average from 90 ± 78 μM in the streamwater to 541 ± 445 μM in the porewater, while porewater DO concentration decreased from 322 ± 44 to 95 ± 104 μM with depth. Porewater in some streams (Com-2, Sen-1, Sor-3, Ven-2) became anoxic below 10-cm depth (Supplementary Figure S1). Porewater Fe(II) and S(-II) concentrations ranged from 0 to 27 μM and 0 to 4 μM, respectively, and were only above detection limit (0.3 μM) in streams with elevated nitrate concentrations (e.g., Geb-1, Sen-1, Sor-1, Sor-2, Sor-3; Supplementary Figure S1). Concentrations of Fe(II) and NH4+ (ranging from 0 to 278 μM) followed patterns similar to CH4 in the streambeds. Porewater concentrations of SO42– and NO3– ranged from 70 to 458 μM and from <1 to 402 μM, respectively, and typically decreased with streambed depth (Supplementary Figure S1). Streamwater CH4 concentration and the ratio of CH4:CO2, a proxy for in situ CH4 production (Stanley et al., 2016), were higher in streams with elevated NO3– concentrations (Supplementary Figure S2). Unlike the other streams, Vey-2 drains a wetland with low-redox environments releasing high amounts of CH4 but low amounts of nitrate (Supplementary Table S1). Phosphate concentrations were below the detection limit (5 μM) for all measured samples.
Kinetics of Methane Oxidation in Sedimentary Biofilms
Contrasting incubations with and without sediments revealed that MOX was primarily associated with sediments (Supplementary Figures S3–S5). The dynamics of MOX of incubations with sediments were well captured by Michaelis–Menten kinetics (Supplementary Figures S6). Overall, Vmax ranged between 0 and 53.9 nmol m–2 h−1, and KS ranged between 0 and 10.6 μM. MOX was undetectable in the two alpine sites (Vmax = 0 and KS = 0). Average Vmax and KS in sites dominated by forests reached 20.1 ± 22.7 nmol m–2 h−1 (average ± standard deviation) and 5.3 ± 3.8 μM. Highest Vmax and KS were detected in Vey-2, the site draining a wetland. Excluding this site reduced average Vmax to 8.8 ± 3.2 nmol m–2 h−1 and KS to 3.6 ± 1.9 μM for this group of samples. Streams dominated by agriculture and urban areas reached an average Vmax of 11.7 ± 8.4 nmol m–2 h−1 and KS of 2.7 ± 2.4. Both Vmax and KS reflected the CH4 supply of the respective streams (Figure 2). Excluding Vey-2, Vmax was positively related to streamwater (R2 = 0.92, p < 0.01) and porewater (R2 = 0.69, p < 0.01) CH4 concentrations. Similarly, KS was also positively related to stream- and porewater CH4 concentrations (R2 = 0.84, p < 0.01 and R2 = 0.45, p = 0.01, respectively) when excluding Vey-2. Excluding site Ven-1, which had a very low porewater CH4 concentration, KS values reached on average 88.1% of the measured porewater CH4 concentrations, however, with considerable variation (range: 4–533%). Given that CH4 concentrations experienced by MOB are in the range of the KS values, putative in situ MOX rates estimated by the Michealis–Menten model (Eq. 1) ranged between 0 and 27 nmol CH4 m–2 h−1 (Supplementary Table S3). Assuming that 50% of the oxidized CH4 was used for biomass production (Trimmer et al., 2015), these putative in situ MOX rates translate into carbon fixation rates ranging from 0 and 13.5 nmol C m–2 h−1 (Supplementary Table S3).
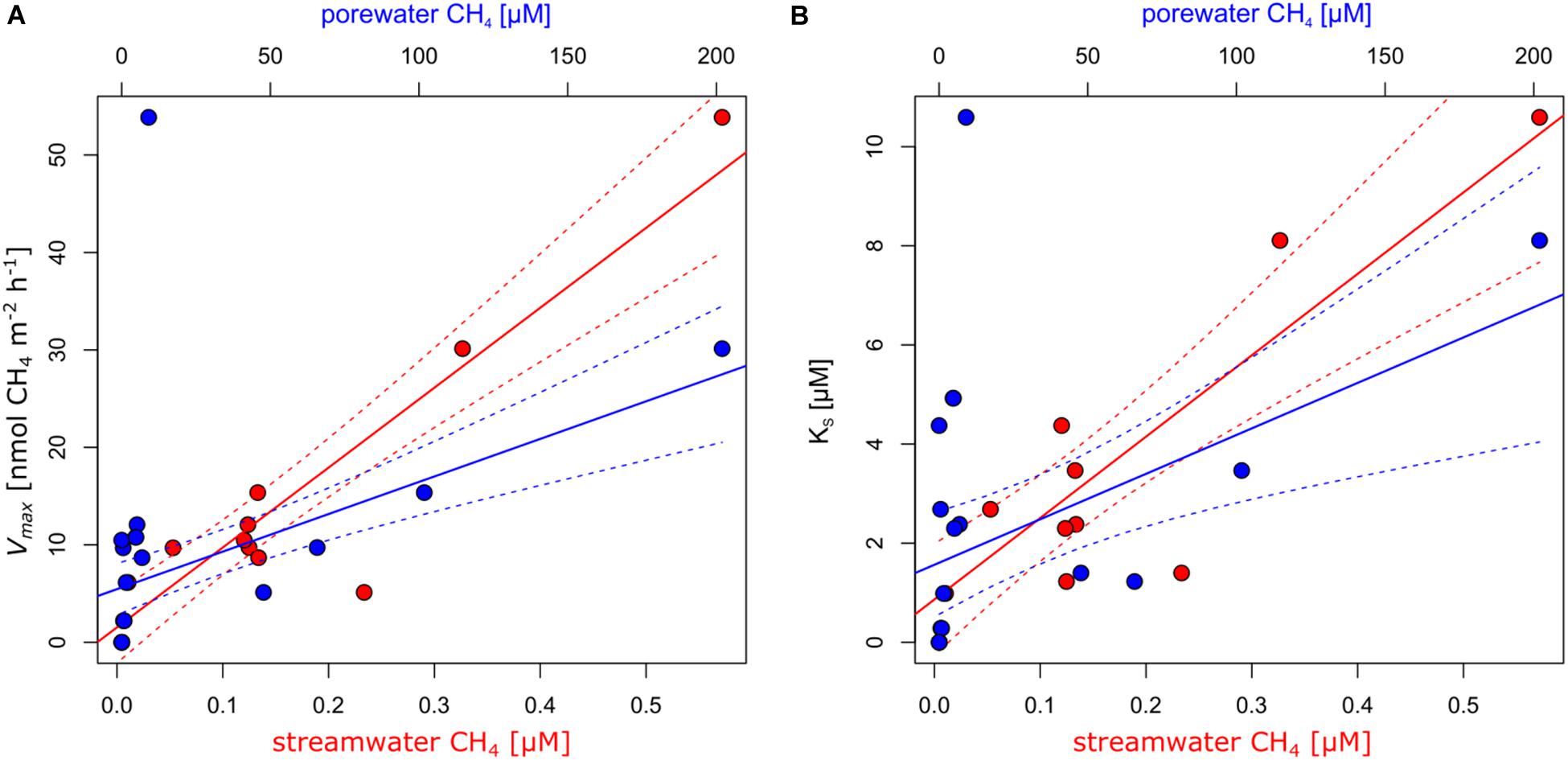
Figure 2. Methane (CH4) oxidation (MOX) kinetics constants maximum reaction velocity [Vmax; (A)] and concentration at half Vmax [KS; (B)] were significantly related to streamwater (red) and porewater (blue) CH4 concentrations. The lines represent linear regression models, and the dashed lines represent the respective 95% confidence interval.
Relating Methane Oxidizing Bacteria to Biofilm Community Structure
We measured pmoA gene counts to quantify the abundance of MOB and their contribution to total bacterial abundance and MOX. Total cell abundance ranged on average from 4.6 × 106 to 3.6 × 108 cells m–2 (Supplementary Table S3). qPCR revealed that the number of cells harboring a pmoA coding gene ranged from below detection limit to 3.8 × 106 cells m–2, with an average of 8.7 ± 9.89 × 105 cells m–2 (Supplementary Table S3). Thus, pmoA-harboring cells contributed on average 0.7 ± 0.69% to total cell abundance, with a maximum contribution of 2.65% in the sediment of Sen-1.
16S rRNA gene amplicon sequencing resulted in 3.22 million reads that clustered into 16,364 OTUs (Supplementary Table S4 and Figure S7) with β-Proteobacteria (22 ± 6%), α-Proteobacteria (20 ± 4%), γ-Proteobacteria (8 ± 2%), and Sphingobacteria (7 ± 2%) dominating the communities. In general, we found relatively little taxa turnover among the various streams, with 97% of the most abundant OTUs (i.e., >1% relative abundance, n = 4,900) being present in all 14 sites. More specifically, the 86 most abundant OTUs, accounting for 45.6% of all reads, were detected in all samples. These most abundant OTUs included members of the genera Rhodoferax (5 ± 2%), Novosphingobium (2 ± 1%), and Nitrospira (1 ± 0.4%). Across all streams, we identified 36 obligate methanotrophic OTUs annotated as Methylococcaceae (γ-Proteobacteria; i.e., low-affinity, type-I MOB). The relative abundance of methanotrophic OTUs ranged between 0.001% in the alpine stream Adn-1 and 2% in the lowland stream Vey-2 with an average of 0.56 ± 0.58% (Figure 3). Refining their taxonomic assignment revealed that on average 60% of MOB were affiliated with Crenothrix spp. across all sites (Figure 4). The most abundant Crenothrix-related OTU, OTU222, was consistently detected in all 14 sites and numerically dominated the methanotrophic community in 13 samples. Seven out of the 36 methanotrophic OTUs were detected in 13 out of 14 sites. They include four Crenothrix OTUs, OTU222 (relative abundance of 40.5 ± 18.2% of the methanotrophic community fraction), OTU155 (20.2 ± 10.1%), OTU1184 (2.4 ± 2.3%), and OTU2012 (3.5 ± 3.9%); one OTU classified as Methylobacter (OTU4584, 6.8 ± 5.1%); and one OTU classified as Methylomonas (OTU621, 4.2 ± 2.8%). Across all streams, the relative abundance of Methylococcaceae was positively correlated with streamwater CH4 concentration (R2 = 0.85, p < 0.01). In turn, Vmax was positively related to the relative abundance of Methylococcaceae (R2 = 0.89, p < 0.01) as well as to the number of pmoA-harboring cells (R2 = 0.99, p < 0.01).
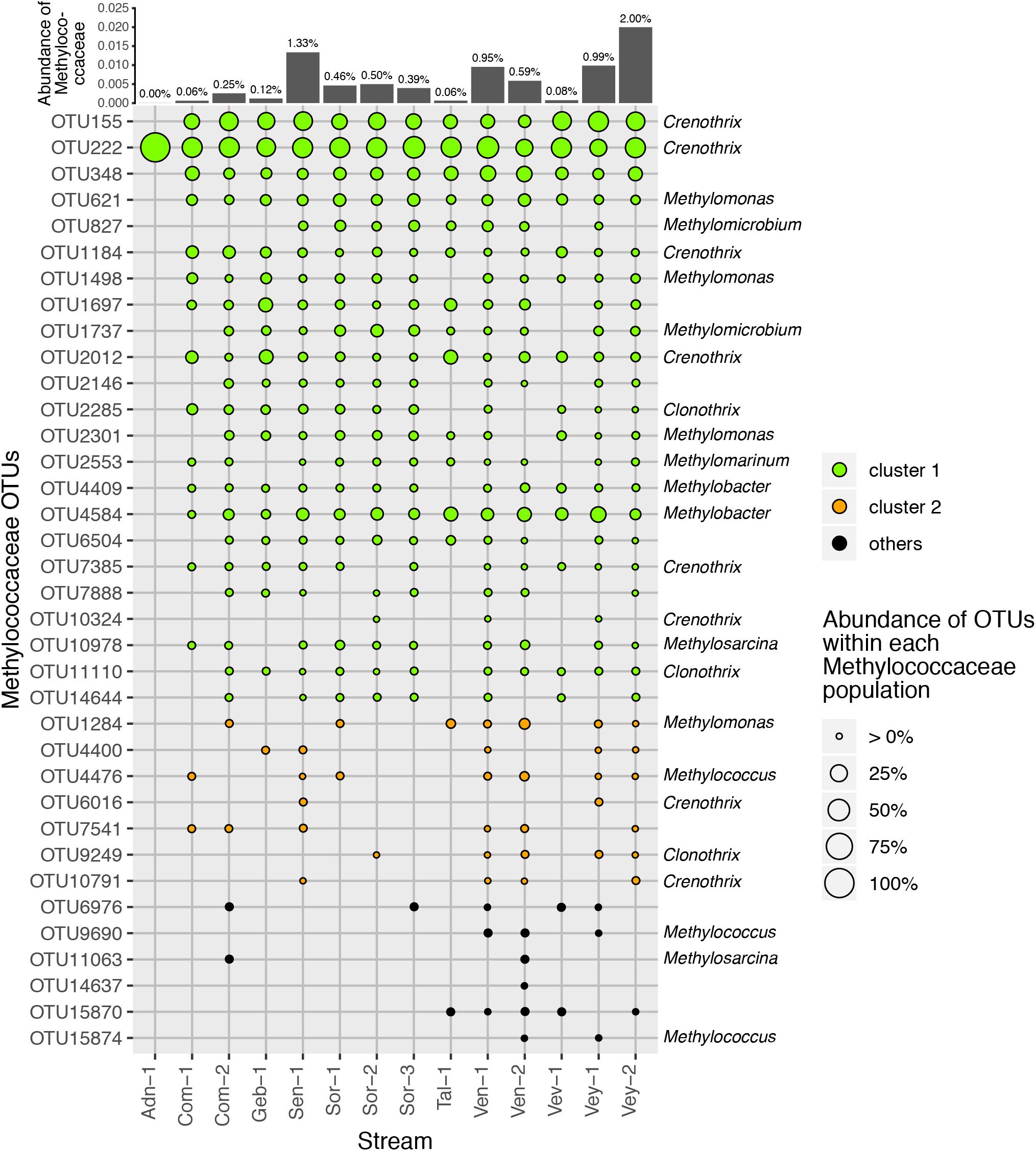
Figure 3. Operational taxonomic units (OTUs) classified as Methylococcaceae and particularly Crenothrix were present in all samples. The relative abundance of each OTU is represented by the symbol size, the color of each symbol reflects cluster affiliation (see Figure 5). The relative abundance of Methylococcaceae to the total microbial community is indicated for each sample at the top. The genus affiliation (if classified) for each OTU is indicated to the right side of the plot.
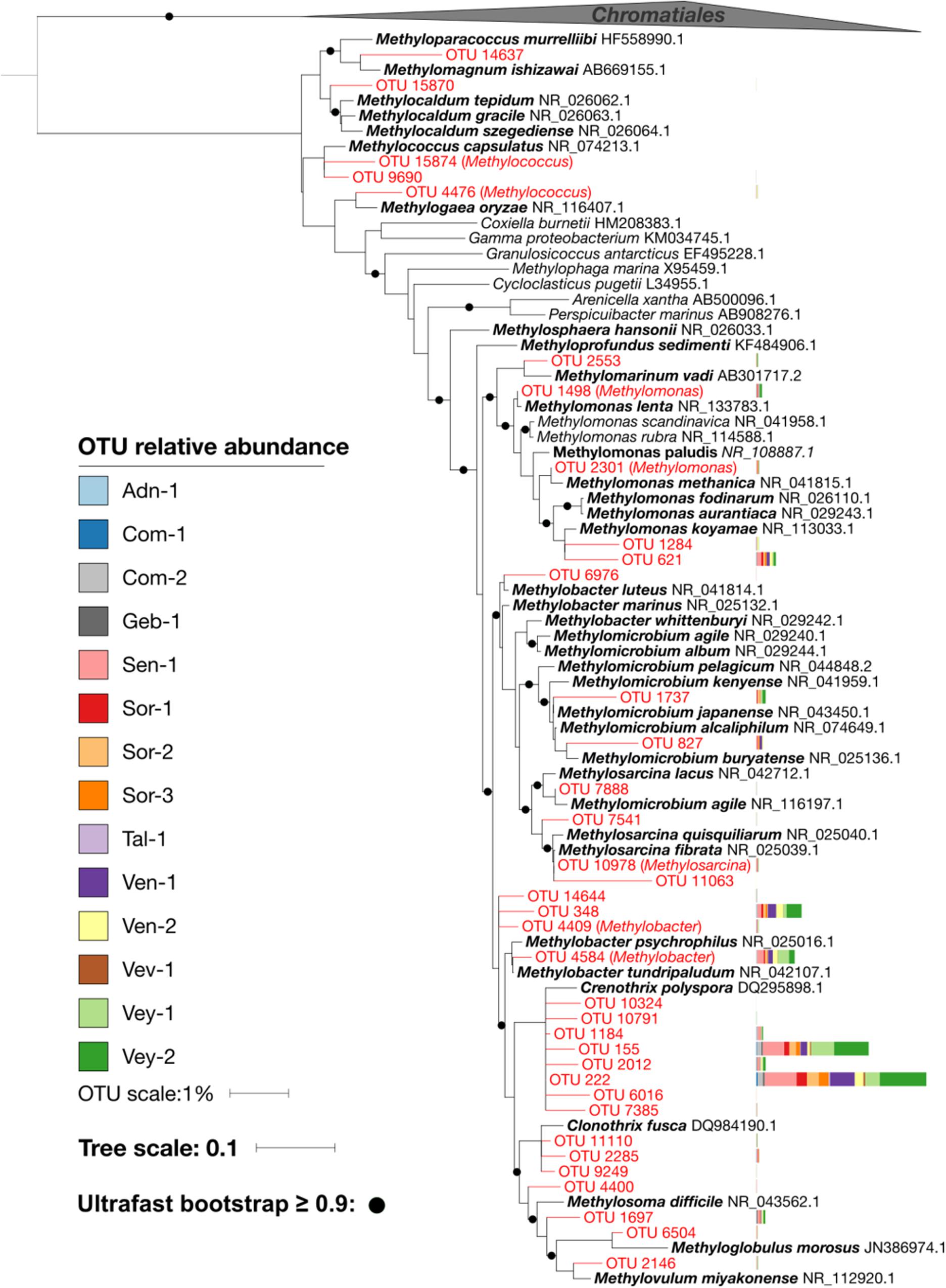
Figure 4. Placement of Methylococcaceae operational taxonomic units (OTUs) (red) in a reference phylogenetic tree. Two Chromatiales sequences were used as outgroup. Bootstrap values greater than 0.9 are highlighted with black circles. The relative abundance of OTUs in each sample is represented as stacked bars. The majority of and the most abundant Methylococcaceae OTUs cluster with Crenothrix polyspora.
Community composition shifted along the environmental gradient, and this pattern co-varied significantly with pmoA gene counts (log-transformed; R2 = 0.80, p < 0.05) and Vmax (R2 = 0.69, p < 0.05; Figure 5A). The microbial composition landscape featured 13 distinct density peaks at a density cutoff of 0.5 (estimated kernel densities ranged from 0 to 6.53). Each of these peaks in the community landscape reflects a group of co-varying OTUs (Figure 5B). Methanotrophic OTUs formed two distinct clusters containing 23 (cluster 1) and seven (cluster 2) OTUs. Six rather low-abundant methanotrophic OTUs were not comprised within these two clusters. Cluster 1 was dominated by the most abundant methanotrophic OTUs Crenothrix, Methylobacter, and Methylomonas. Members of this cluster were found in streams with high CH4 concentration. Cluster 2 was dominated by OTUs affiliated with Methylomonas, Methylogaea, and Methylosarcina but also contained low-abundant OTUs classified as Crenothrix and Clonothrix. Members of cluster 2 were only detected in a subset of streams (Sen-1, Ven-1, Ven-2, Vey-1, and Vey-2), all of which, except Sen-1, were dominated by forests in their catchments (Figure 1A). A convex hull around cluster 1 contained 1,189 non-MOB OTUs, whereas MOB in cluster 2 co-varied with 348 non-methanotrophic OTUs. Both clusters were dominated by unclassified bacterial OTUs (accounting for 21.9 and 67.5% of reads in cluster 1 and cluster 2, respectively), but OTUs classified as non-methanotrophic γ-Proteobacteria (21.2 and 3.2%), δ-Proteobacteria (11.6 and 2.7%), and Acidobacteria (5.5 and 9.4%) also co-varied with the two methanotrophic clusters. In contrast, abundant taxonomic groups such as β-Proteobacteria, α-Proteobacteria, and Bacteroidetes were underrepresented in both methanotrophic clusters (Supplementary Figure S8).
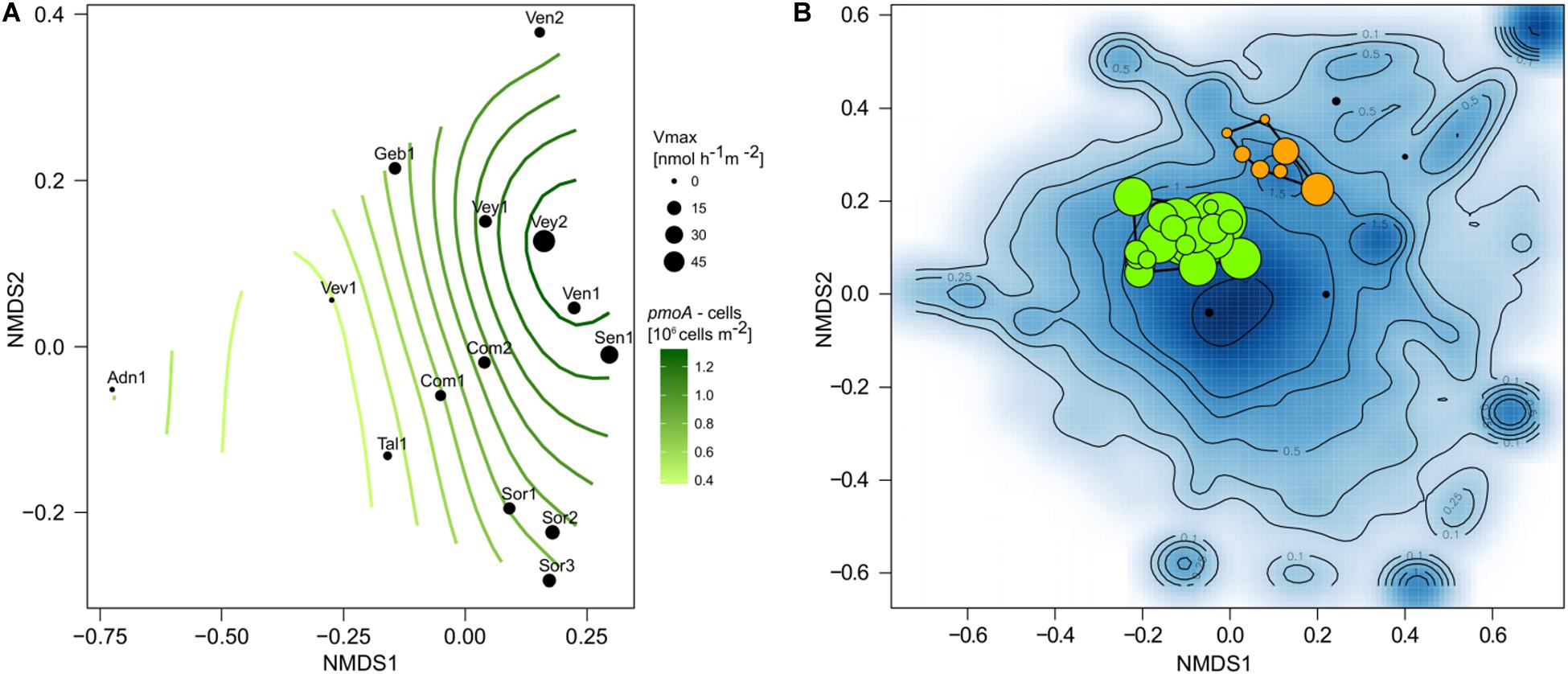
Figure 5. Non-metric multidimensional scaling ordination (nMDS) based on Bray–Curtis similarities of stream sediment communities (A). Maximum reaction velocity (Vmax) measured at each site is indicated by symbol size, and a GAM model (surface isoclines) represents the distribution of pmoA gene counts among communities. Microbial community landscape featuring distinct density peaks of co-occurring operational taxonomic units (OTUs) (B). Shown is the density distribution of species scores (blue background). Isoclines highlight distinct groups of OTUs with similar distribution among the samples. Methanotrophic OTUs (colored circles) mainly fall within two regions and can be grouped in cluster 1 (green) and cluster 2 (orange). Symbol size represents the relative abundance of methanotrophic OTUs.
Discussion
Our study provides insights into the ecophysiology of stream sediment MOB by studying MOX kinetics, MOB community composition, and co-occurrence patterns with non-MOB across a gradient of land cover and associated physicochemical characteristics. Taken together, the results underpin the coupling between CH4 supply, MOX kinetics, and MOB structure in streams. Specifically, we found that Vmax and KS increased in streams with elevated CH4 supply. KS values in the μM range matched those measured in shallow streambeds and suggest that low-affinity MOB adapted to high CH4 concentrations drive MOX in stream sediments. In line with this, MOB communities were dominated by Crenothrix, a type-I MOB belonging to the Methylococcaceae family. Our work thus suggests that the attenuation of CH4 by MOX scales with CH4 availability in streams. CH4 availability across these streams was linked to land cover and particularly NO3–, an indicator of human impact. The ratio of streamwater CH4:CO2, a proxy for the contribution of methanogenesis to ecosystem metabolism (Stanley et al., 2016) was also elevated in streams with high NO3– concentrations. This reflects the potential for in situ CH4 production linked to oxygen-poor microhabitats in the streambed of streams with elevated NO3– concentration and draining catchments dominated by agriculture. As reported earlier (Trimmer et al., 2009, 2010), the ecological niche of stream MOX appears to be restricted to benthic sediments, which agrees with the absence of MOX in streamwater incubations. This is relevant, given the estimated CH4 emission of 26.8 Tg CH4 per year from streams and rivers (Stanley et al., 2016) and the role of MOX as a sink of CH4 at the interface between supersaturated groundwater and streamwater, from which CH4 evades to the atmosphere.
The relationships between the relative abundance of Methylococcaceae, pmoA-harboring cells, and both Vmax and KS point to the functional relevance of these MOB in streams. Our finding that the potential for MOX is related to pmoA copy numbers is consistent with the prevalence of aerobic MOB for the membrane-bound particulate form of CH4 monooxygenase (pMMO, one subunit being encoded by pmoA) as compared to the soluble form (sMMO) (Ho et al., 2013; Knief, 2015). Both the relative abundance of Methylococcaceae and pmoA-harboring cell numbers were related to Vmax, highlighting the link between MOB abundance and MOX activity (at non-limiting CH4 concentrations). In contrast, KS is independent of the abundance of MOB but depends on the overall affinity of CH4 monooxygenases. As substrate affinity exerts a selective force, KS values are expected to match in situ substrate concentrations (Tilman, 1977; Martens-Habbena et al., 2009; Kits et al., 2017). KS values reported here were on average approximately 50 times greater than streamwater CH4 concentrations but in the range of shallow porewater CH4 concentrations. This suggests that MOB in shallow stream sediments thrive on elevated CH4 concentrations. CH4 concentrations increased with sediment depth, a pattern that was noted earlier (Sanders et al., 2007; Rulík et al., 2013; Bednařík et al., 2015; Mach et al., 2015). The interface between oxygen-rich streamwater and CH4-enriched porewater may therefore represent a hot spot for MOX. However, given the dominance of Methylococcaceae and particularly Crenothrix across all sites, the variability of KS may reflect ecophysiological adaptations of these key players to CH4 supply. Since streambed CH4 concentrations vary in space and time (Stanley et al., 2016; Crawford et al., 2017; Natchimuthu et al., 2017), flush-feeding on transient CH4 pulses may be one of the strategies to maintain methanotrophic activity in stream sediments (Stanley et al., 2016). A comparable phenomenon has been reported for rice paddy soils, where methanotrophs can transiently uphold high-affinity MOX activity (Cai et al., 2016). The fact that MOB communities were dominated by type I MOB which are responsive to changes in CH4 concentrations may support this notion. Kinetic information of MOX in streams is scarce, but reanalyzing published data (Shelley et al., 2015; Trimmer et al., 2015) (Supplementary Material), we found our MOX kinetic estimates match those reported from English chalk streams and a lowland stream in the Czech Republic (Figure 6).
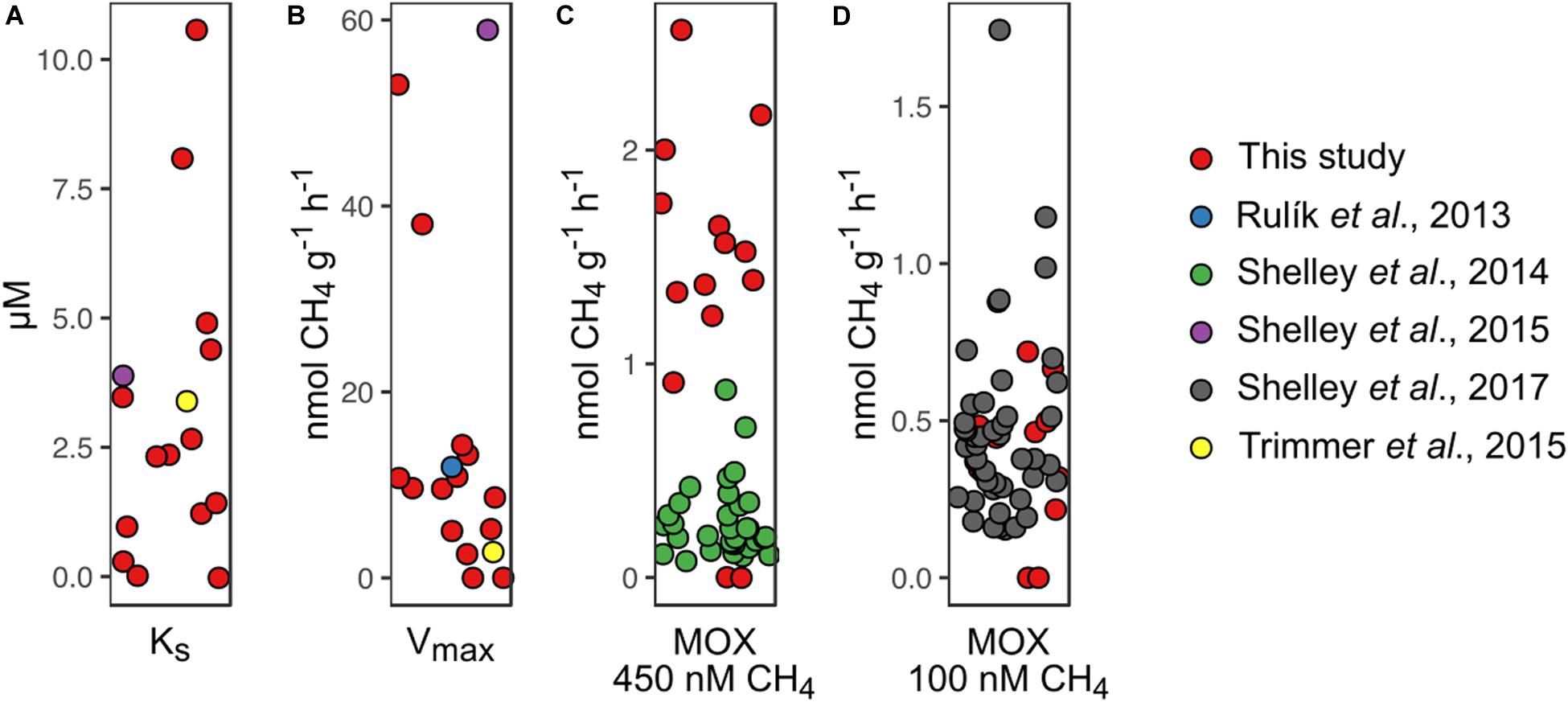
Figure 6. Comparison of methane (CH4) oxidation (MOX) rates measured in this study with MOX rates obtained by reanalyzing data from other studies. (A) Concentration at half Vmax (KS), (B) maximum reaction velocity (Vmax), (C) MOX rates at an initial CH4 concentration of 450 nM, and (D) MOX rates at an initial CH4 concentration of 100 nM are shown.
Type I pmoA (i.e., γ-proteobacterial) is typically expressed by low-affinity MOB in environments with elevated and fluctuating CH4 concentrations (Ho et al., 2013). Methylococcaceae are such type I MOB (Bowman, 2015), and KS values in the micromolar range corroborate the low substrate affinity of these MOB. Our results contrast those from chalk streams, where pmoA gene sequencing revealed equitable distributions of type I and II MOB (Trimmer et al., 2015). While the presence of type II MOB within the sediments of chalk streams may be attributable to comparatively low CH4 concentration, we did not find type II MOB in streams with low CH4 supply. However, methanotrophs appear to be rare (i.e., <2% of reads in our 16S rRNA gene survey) and the MOB inventory might not have been fully captured by the 16S rRNA gene sequencing approach. Sequencing of the genes encoding pMMO (pmoCAB) might be a more sensitive approach, and future investigations may consider this strategy. Yet, the microbial community landscape analysis revealed two clusters of MOB (Figure 5B)—one containing most of the abundant MOB OTUs and reflecting a shared core community and another cluster of rare MOB, shared only among a subset of streams with forested catchments. There was no consistent taxonomic separation of these clusters. Therefore, whereas a dichotomy between type I and II dominated MOB communities may reflect MOB lifestyle strategies, a more nuanced continuum of trait and taxa distributions appears more likely to underlie the adaptations of MOB to the wide range and variable CH4 concentrations within and among streams. For instance, a whole suite of pmoA-encoded enzymes may cover a large gradient of CH4 affinities (Baani and Liesack, 2008).
Strikingly, we found across all streams only MOB belonging to the Methylococcaceae family. Most abundant among the Methylococcaceae was Crenothrix, accounting for 60% of the reads. Crenothrix is a large filamentous γ-Proteobacteria that was originally identified as an aerobic CH4 oxidizer in drinking water distribution networks (Völker et al., 1977; Stoecker et al., 2006). Crenothrix has recently been reported as a critical component of the methanotrophic communities in temperate lakes (Oswald et al., 2017), a cave ecosystem (Karwautz et al., 2018), and in a eutrophic littoral wetland (Siljanen et al., 2011). The ecological success of Crenothrix might be related to its ability to use NO3– besides oxygen as an electron acceptor (Oswald et al., 2017). In the microbial community landscape analysis, Nitrospira, a nitrite-oxidizing bacterium, is among the most abundant non-methanotrophic OTUs falling within the methanotrophic cluster 1. This co-occurrence may indicate a syntrophic relationship between Crenothrix and Nitrospira in stream sediments—a hypothesis that requires further investigation. Nevertheless, our study highlights the key role of Crenothrix in CH4 cycling in freshwater ecosystems.
Methanotrophic carbon fixation can account for up to 46% of benthic phototrophic production (Shelley et al., 2014) and putative carbon fixation rates of up to 13.5 nmol C m–2 h−1 estimated here highlight the functional relevance of MOB at the ecosystem scale. Mediated by diverse interactions with methylotrophs and heterotrophs, MOB may thus significantly contribute to the functioning of stream benthic communities. For instance, MOB were previously shown to sustain diverse communities of non-methanotrophic bacteria in biofilms in a mineral spring cave (Karwautz et al., 2018) and in marine hydrocarbon seeps (Paul et al., 2017). In our study, microbial community structure co-varied with Vmax and pmoA-harboring cell numbers. Such analyses cannot resolve to which extent this may be due to the role of interactions among microbes or due to shared environmental preferences along the environmental gradient. However, MOB can produce copious amounts of extracellular polymeric substances (EPSs) (Strong et al., 2015; Karwautz et al., 2018), which is understood as an energy-spilling mechanism to prevent the accumulation of toxic formaldehyde. Given the importance of biofilm formation for benthic stream communities (Battin et al., 2016), EPS production may be yet another important role MOB fulfill in stream ecosystems.
In conclusion, this work contributes to our understanding of the ecology of MOB in streams by identifying the coupling between CH4 supply and the kinetics of methanotrophic activity. Substrate affinities of a phylogenetically homogeneous MOB population dominated by Crenothrix, particularly in lowland streams impacted by agriculture, suggest that the attenuation of CH4 by MOX might scale with CH4 availability in streams. This is important with respect to the greenhouse gas potential of CH4.
Data Availability Statement
The datasets generated for this study can be found in the NCBI SRA SAMN07174972–SAMN07174985.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Nicolas Escoffier, Amber Ulseth, Sabine Flury McGinnis, Janine Rüegg, David Scheidweiler, Mauricio Morales, and Robert Niederdorfer for help during field work. Rizlan Bernier-Latmani is thanked for the CH4 used for the kinetic experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00771/full#supplementary-material
References
Baani, M., and Liesack, W. (2008). Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SC2. Proc. Natl. Acad. Sci. U.S.A. 105, 10203–10208. doi: 10.1073/pnas.0702643105
Bagnoud, A., Chourey, K., Hettich, R. L., de Bruijn, I., Andersson, A. F., Leupin, O. X., et al. (2016). Reconstructing a hydrogen-driven microbial metabolic network in Opalinus Clay rock. Nat. Commun. 7:12770. doi: 10.1038/ncomms12770
Battin, T. J., Besemer, K., Bengtsson, M. M., Romani, A. M., and Packmann, A. I. (2016). The ecology and biogeochemistry of stream biofilms. Nat. Rev. Microbiol. 14, 251–263. doi: 10.1038/nrmicro.2016.15
Bednařík, A., Čáp, L., Maier, V., and Rulík, M. (2015). Contribution of methane benthic and atmospheric fluxes of an experimental area (sitka stream). CLEAN Soil Air Water 43, 1136–1142. doi: 10.1002/clen.201300982
Bender, M., and Conrad, R. (1992). Kinetics of CH4 oxidation in oxic soils exposed to ambient air or high CH4 mixing ratios. FEMS Microbiol. Lett. 101, 261–269. doi: 10.1016/0378-1097(92)90823-7
Besemer, K., Singer, G., Hödl, I., and Battin, T. J. (2009). Bacterial community composition of stream biofilms in spatially variable-flow environments. Appl. Environ. Microbiol. 75, 7189–7195. doi: 10.1128/AEM.01284-09
Bowman, J. P. (2015). Methylococcaceae, in: Bergey’s Manual of Systematics of Archaea and Bacteria. Hoboken, NJ: John Wiley & Sons, Ltd.
Buriánková, I., Brablcová, L., Mach, V., Hýblová, A., Badurová, P., Cupalová, J., et al. (2012). Methanogens and methanotrophs distribution in the hyporheic sediments of a small lowland stream. Fundam. Appl. Limnol. Arch. Fr Hydrobiol. 181, 87–102. doi: 10.1127/1863-9135/2012/0283
Cai, Y., Zheng, Y., Bodelier, P. L. E., Conrad, R., and Jia, Z. (2016). Conventional methanotrophs are responsible for atmospheric methane oxidation in paddy soils. Nat. Commun. 7:11728. doi: 10.1038/ncomms11728
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chaudhary, P. P., Wright, A.-D. G., Brablcová, L., Buriánková, I., Bednařík, A., and Rulík, M. (2014). Dominance of methanosarcinales phylotypes and depth-wise distribution of methanogenic community in fresh water sediments of sitka stream from czech republic. Curr. Microbiol. 69, 809–816. doi: 10.1007/s00284-014-0659-8
Cline, J. D. (1969). Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14, 454–458. doi: 10.4319/lo.1969.14.3.0454
Costea, P. I., Hildebrand, F., Arumugam, M., Bäckhed, F., Blaser, M. J., Bushman, F. D., et al. (2018). Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 3, 8–16. doi: 10.1038/s41564-017-0072-8
Costello, A. M., and Lidstrom, M. E. (1999). Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 65, 5066–5074.
Crawford, J. T., Loken, L. C., West, W. E., Crary, B., Spawn, S. A., Gubbins, N., et al. (2017). Spatial heterogeneity of within-stream methane concentrations. J. Geophys. Res. Biogeosci. 122, 1036–1048. doi: 10.1002/2016JG003698
Crawford, J. T., Lottig, N. R., Stanley, E. H., Walker, J. F., Hanson, P. C., Finlay, J. C., et al. (2014). CO2 and CH4 emissions from streams in a lake-rich landscape: patterns, controls, and regional significance. Glob. Biogeochem. Cycles 28:2013GB004661. doi: 10.1002/2013GB004661
de Angelis, M. A., and Scranton, M. I. (1993). Fate of methane in the Hudson River and Estuary. Glob. Biogeochem. Cycles 7, 509–523. doi: 10.1029/93GB01636
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Gonze, D., Lahti, L., Raes, J., and Faust, K. (2017). Multi-stability and the origin of microbial community types. ISME J. 11, 2159–2166. doi: 10.1038/ismej.2017.60
Ho, A., Kerckhof, F.-M., Luke, C., Reim, A., Krause, S., Boon, N., et al. (2013). Conceptualizing functional traits and ecological characteristics of methane-oxidizing bacteria as life strategies. Environ. Microbiol. Rep. 5, 335–345. doi: 10.1111/j.1758-2229.2012.00370.x
Holmes, A. J., Costello, A., Lidstrom, M. E., and Murrell, J. C. (1995). Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132, 203–208. doi: 10.1016/0378-1097(95)00311-r
Karwautz, C., Kus, G., Stöckl, M., Neu, T. R., and Lueders, T. (2018). Microbial megacities fueled by methane oxidation in a mineral spring cave. ISME J. 12, 87–100. doi: 10.1038/ismej.2017.146
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kits, K. D., Sedlacek, C. J., Lebedeva, E. V., Han, P., Bulaev, A., Pjevac, P., et al. (2017). Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature 549, 269–272. doi: 10.1038/nature23679
Knief, C. (2015). Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front. Microbiol. 6:1346. doi: 10.3389/fmicb.2015.01346
Mach, V., Blaser, M. B., Claus, P., Chaudhary, P. P., and Rulík, M. (2015). Methane production potentials, pathways, and communities of methanogens in vertical sediment profiles of river Sitka. Terr. Microbiol. 6:506. doi: 10.3389/fmicb.2015.00506
Martens-Habbena, W., Berube, P. M., Urakawa, H., de la Torre, J. R., and Stahl, D. A. (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461, 976–979. doi: 10.1038/nature08465
Natchimuthu, S., Wallin, M. B., Klemedtsson, L., and Bastviken, D. (2017). Spatio-temporal patterns of stream methane and carbon dioxide emissions in a hemiboreal catchment in Southwest Sweden. Sci. Rep. 7:39729. doi: 10.1038/srep39729
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2017). vegan: Community Ecology Package. R Package Version 24–22.
Oswald, K., Graf, J. S., Littmann, S., Tienken, D., Brand, A., Wehrli, B., et al. (2017). Crenothrix are major methane consumers in stratified lakes. ISME J. 11, 2124–2140. doi: 10.1038/ismej.2017.77
Paul, B. G., Ding, H., Bagby, S. C., Kellermann, M. Y., Redmond, M. C., Andersen, G. L., et al. (2017). Methane-oxidizing bacteria shunt carbon to microbial mats at a marine hydrocarbon seep. Front. Microbiol. 8:186. doi: 10.3389/fmicb.2017.00186
Rulík, M., Mach, V., Brablcová, L., Buriánková, I., Badurová, P., and Gratzová, K. (2013). Methanogenic System of a Small Lowland Stream Sitka, Czech Republic, in: Biomass Now-Cultivation Utilization. London: IntechOpen, 395–426.
Sanders, I. A., Heppell, C. M., Cotton, J. A., Wharton, G., Hildrew, A. G., Flowers, E. J., et al. (2007). Emission of methane from chalk streams has potential implications for agricultural practices. Freshw. Biol. 52, 1176–1186. doi: 10.1111/j.1365-2427.2007.01745.x
Segers, R. (1998). Methane production and methane consumption: a review of processes underlying wetland methane fluxes. Biogeochemistry 41, 23–51. doi: 10.1023/A:1005929032764
Shelley, F., Abdullahi, F., Grey, J., and Trimmer, M. (2015). Microbial methane cycling in the bed of a chalk river: oxidation has the potential to match methanogenesis enhanced by warming. Freshw. Biol. 60, 150–160. doi: 10.1111/fwb.12480
Shelley, F., Grey, J., and Trimmer, M. (2014). Widespread methanotrophic primary production in lowland chalk rivers. Proc. R. Soc. Lond. B Biol. Sci. 281:20132854. doi: 10.1098/rspb.2013.2854
Siljanen, H. M. P., Saari, A., Krause, S., Lensu, A., Abell, G. C. J., Bodrossy, L., et al. (2011). Hydrology is reflected in the functioning and community composition of methanotrophs in the littoral wetland of a boreal lake. FEMS Microbiol. Ecol. 75, 430–445. doi: 10.1111/j.1574-6941.2010.01015.x
Sinha, E., Michalak, A. M., and Balaji, V. (2017). Eutrophication will increase during the 21st century as a result of precipitation changes. Science 357, 405–408. doi: 10.1126/science.aan2409
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stanley, E. H., Casson, N. J., Christel, S. T., Crawford, J. T., Loken, L. C., and Oliver, S. K. (2016). The ecology of methane in streams and rivers: patterns, controls, and global significance. Ecol. Monogr. 86, 146–171. doi: 10.1890/15-1027.1
Stoecker, K., Bendinger, B., Schöning, B., Nielsen, P. H., Nielsen, J. L., Baranyi, C., et al. (2006). Cohn’s Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc. Natl. Acad. Sci. U. S. A. 103, 2363–2367. doi: 10.1073/pnas.0506361103
Stookey, L. L. (1970). Ferrozine - a new spectrophotometric reagent for iron. Anal. Chem. 42, 779–781. doi: 10.1021/ac60289a016
Strong, P. J., Xie, S., and Clarke, W. P. (2015). Methane as a resource: can the methanotrophs add value? Environ. Sci. Technol. 49, 4001–4018. doi: 10.1021/es504242n
Tilman, D. (1977). Resource competition between plankton algae: an experimental and theoretical approach. Ecology 58, 338–348. doi: 10.2307/1935608
Trimmer, M., Hildrew, A. G., Jackson, M. C., Pretty, J. L., and Grey, J. (2009). Evidence for the role of methane-derived carbon in a free-flowing, lowland river food web. Limnol. Oceanogr. 54, 1541–1547. doi: 10.4319/lo.2009.54.5.1541
Trimmer, M., Maanoja, S., Hildrew, A. G., Pretty, J. L., and Grey, J. (2010). Potential carbon fixation via methane oxidation in well-oxygenated river bed gravels. Limnol. Oceanogr. 55, 560–568. doi: 10.4319/lo.2010.55.2.0560
Trimmer, M., Shelley, F. C., Purdy, K. J., Maanoja, S. T., Chronopoulou, P.-M., and Grey, J. (2015). Riverbed methanotrophy sustained by high carbon conversion efficiency. ISME J. 9, 2304–2314. doi: 10.1038/ismej.2015.98
Völker, H., Schweisfurth, R., and Hirsch, P. (1977). Morphology and ultrastructure of crenothrix polyspora cohn. J. Bacteriol. 131, 306–313.
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Keywords: Methylococcaceae, Crenothrix, Michaelis–Menten, enterotypes, pmoA
Citation: Bagnoud A, Pramateftaki P, Bogard MJ, Battin TJ and Peter H (2020) Microbial Ecology of Methanotrophy in Streams Along a Gradient of CH4 Availability. Front. Microbiol. 11:771. doi: 10.3389/fmicb.2020.00771
Received: 17 January 2020; Accepted: 31 March 2020;
Published: 14 May 2020.
Edited by:
Jonathan P. Zehr, University of California, Santa Cruz, United StatesReviewed by:
Adrian Ho, Leibniz University Hannover, GermanyRonald Oremland, United States Geological Survey (USGS), United States
Copyright © 2020 Bagnoud, Pramateftaki, Bogard, Battin and Peter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hannes Peter, aGFubmVzLnBldGVyQGVwZmwuY2g=
†Present address: Alexandre Bagnoud, Haute École d’Ingénierie et de Gestion du Canton de Vaud, Institut de Génie Thermique, Yverdon-les-Bains, Switzerland Matthew J. Bogard, Department of Biological Sciences, University of Lethbridge, Lethbridge, AB, Canada
 Alexandre Bagnoud
Alexandre Bagnoud Paraskevi Pramateftaki
Paraskevi Pramateftaki Matthew J. Bogard2†
Matthew J. Bogard2† Hannes Peter
Hannes Peter