
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 April 2020
Sec. Microbial Physiology and Metabolism
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00691
The tertiary branched short-chain 2-hydroxyisobutyric acid (2-HIBA) has been associated with several metabolic diseases and lysine 2-hydroxyisobutyrylation seems to be a common eukaryotic as well as prokaryotic post-translational modification in proteins. In contrast, the underlying 2-HIBA metabolism has thus far only been detected in a few microorganisms, such as the betaproteobacterium Aquincola tertiaricarbonis L108 and the Bacillus group bacterium Kyrpidia tusciae DSM 2912. In these strains, 2-HIBA can be specifically activated to the corresponding CoA thioester by the 2-HIBA-CoA ligase (HCL) and is then isomerized to 3-hydroxybutyryl-CoA in a reversible and B12-dependent mutase reaction. Here, we demonstrate that the actinobacterial strain Actinomycetospora chiangmaiensis DSM 45062 degrades 2-HIBA and also its precursor 2-methylpropane-1,2-diol via acetone and formic acid by employing a thiamine pyrophosphate-dependent lyase. The corresponding gene is located directly upstream of hcl, which has previously been found only in operonic association with the 2-hydroxyisobutyryl-CoA mutase genes in other bacteria. Heterologous expression of the lyase gene from DSM 45062 in E. coli established a 2-hydroxyisobutyryl-CoA lyase activity in the latter. In line with this, analysis of the DSM 45062 proteome reveals a strong induction of the lyase-HCL gene cluster on 2-HIBA. Acetone is likely degraded via hydroxylation to acetol catalyzed by a MimABCD-related binuclear iron monooxygenase and formic acid appears to be oxidized to CO2 by selenium-dependent dehydrogenases. The presence of the lyase-HCL gene cluster in isoprene-degrading Rhodococcus strains and Pseudonocardia associated with tropical leafcutter ant species points to a role in degradation of biogenic short-chain ketones and highly branched organic compounds.
2-Hydroxyisobutyric acid (2-HIBA) is a short-chain carboxylic acid bearing a tertiary hydroxyl group (Figure 1A). It has been viewed at as a mainly xenobiotic compound associated with the degradation of the structurally related tert-butyl moieties of the fuel oxygenates methyl and ethyl tert-butyl ether (Rohwerder et al., 2006). Additionally, 2-HIBA and its methyl ester can be formed as intermediates in industrial processes for the production of acrylic glass (Rohwerder and Müller, 2010), i.e., poly(methyl methacrylate). In the last years, however, 2-HIBA has been proposed as biomarker for several metabolic diseases, such as diabetes mellitus and adiposity (Li et al., 2009; Elliott et al., 2015), and increased titers are also associated with alcohol abuse (Irwin et al., 2018). Furthermore, it has recently been discovered that lysine 2-hydroxyisobutyrylation is an important post-translational modification of histones and other proteins (Dai et al., 2014; Huang et al., 2017). All this indicates now that 2-HIBA and its metabolism is much more widespread in both prokaryotes and eukaryotes than previously assumed. Nevertheless, enzymes specifically catalyzing the synthesis and degradation of 2-HIBA and its CoA thioester 2-hydroxyisobutyryl-CoA have thus far only been characterized in a few bacterial strains.
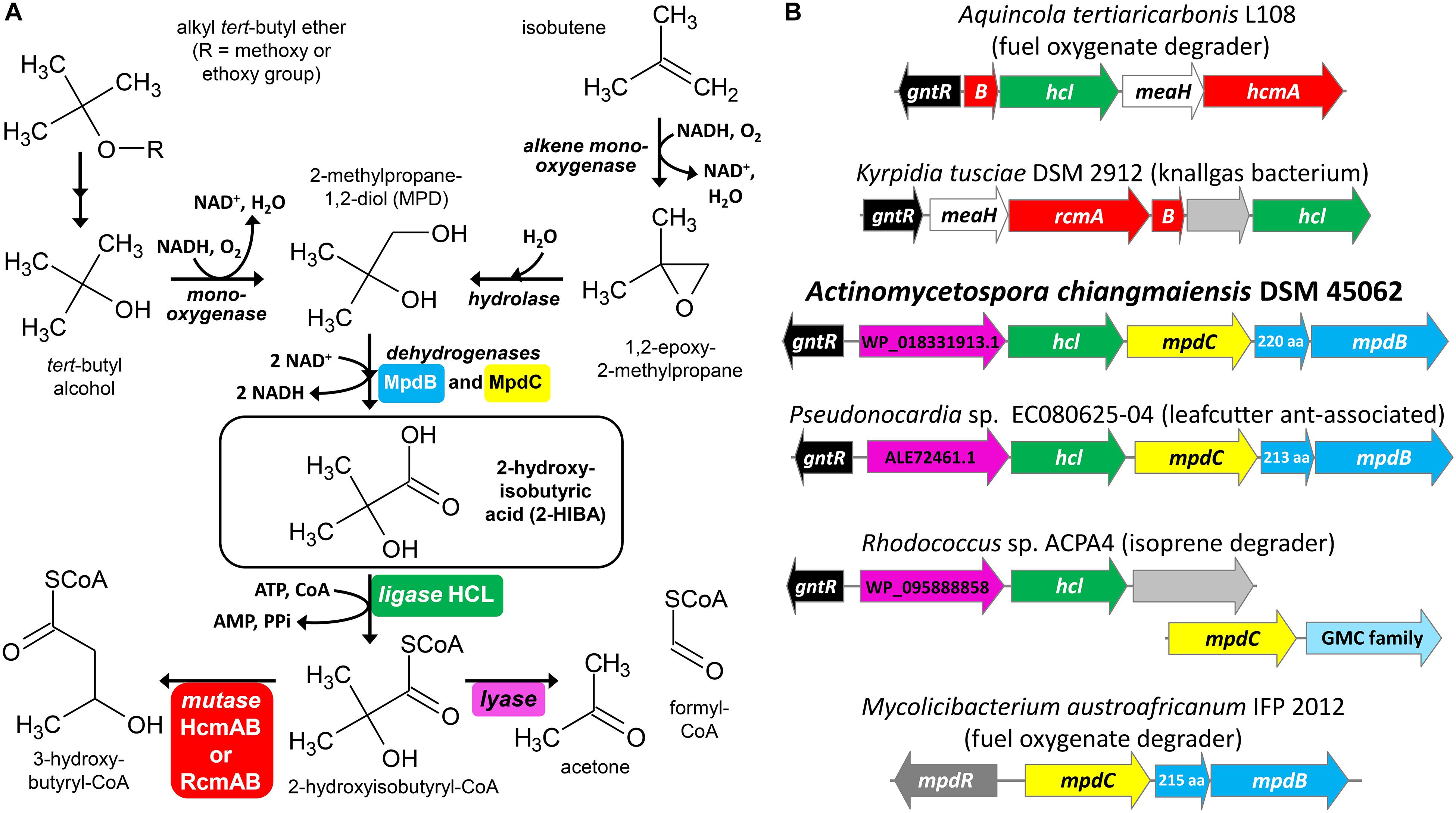
Figure 1. Bacterial conversion of tert-butyl compounds. (A) Proposed pathways for the degradation of fuel oxygenate ethers and isobutene via MPD and 2-HIBA. (B) Examples for gene clusters with mpdBC and/or hcl genes encoding the enzymes for the two-step oxidation of MPD to 2-HIBA and its activation to 2-hydroxyisobutyryl-CoA, respectively. The magenta colored genes from strain DSM 45062 and in the clusters of strains EC080625-04 and ACPA4 correspond to proteins WP_018331913.1, ALE72461.1 and WP_095888858.1, respectively, which likely function as TPP-dependent 2-hydroxyacyl-CoA lyases, for example, for the cleavage of 2-hydroxyisobutyryl-CoA to acetone and formyl-CoA. GMC family, glucose-methanol-choline oxidoreductase family.
Under oxic conditions, alkyl tert-butyl ethers are degraded via tert-butyl alcohol which is further oxidized to 2-methylpropane-1,2-diol (MPD) and 2-HIBA (Figure 1A) (Zahn et al., 2019). Accordingly, the MPD and 2-HIBA pathways have been studied first in aerobic fuel oxygenate degraders, such as the betaproteobacterium Aquincola tertiaricarbonis L108 (Schäfer et al., 2011, 2012; Schuster et al., 2012) and the actinobacterium Mycolicibacterium austroafricanum IFP 2012 (Francois et al., 2002, 2003; Lopes Ferreira et al., 2006a). In the latter, the dehydrogenases MpdB and MpdC catalyze the oxidation of MPD and 2-hydroxyisobutyraldehyde, respectively, to 2-HIBA. The subsequent steps might proceed via isopropanol and acetone, but have not yet been elucidated in strain IFP 2012 or other actinobacteria (Lopes Ferreira et al., 2006b). In contrast, strain A. tertiaricarbonis L108 CoA activates 2-HIBA employing the specific 2-HIBA-CoA ligase (HCL) (Zahn et al., 2019). The CoA thioester is then isomerized by the B12-dependent acyl-CoA mutase HcmAB to (S)-3-hydroxybutyryl-CoA (Yaneva et al., 2012; Kurteva-Yaneva et al., 2015). The mutase route has also been found in the Gram-positive bacterium Kyrpidia tusciae DSM 2912 (Weichler et al., 2015). Interestingly, here the mutase RcmAB, which is only distantly related to HcmAB, catalyzes the isomerization reaction, preferentially forming (R)-3-hydroxybutyryl-CoA. And Bacillus massiliosenegalensis JC6 even possesses both 2-hydroxyisobutyryl-CoA mutase forms (Rohde et al., 2017). The latter and also other bacterial strains bearing the mutase-HCL gene cluster (Figure 1B) have not been associated with the degradation of fuel oxygenates or other xenobiotics, but mostly seem to be isolated from pristine environments, such as geothermal solfatara ponds, marine dinoflagellates, agricultural soil and root nodules from Fabaceae species (Weichler et al., 2015; Zahn et al., 2019). This might be indicative of a natural source of 2-HIBA or a related carboxylic acid not yet identified. In this connection, it has been shown that isobutene degradation in strain Mycobacterium sp. ELW1 proceeds via the corresponding epoxide, MPD and 2-HIBA (Kottegoda et al., 2015), likely employing a plasmid-borne (pELW1-1, NZ_CP032156.1) alkene monooxygenase, an epoxide hydrolase and MpdBC-related dehydrogenase steps (Figure 1A).
As mentioned above, 2-HIBA might not be degraded exclusively via the mutase pathway and it has been speculated whether an alternative route could lead to isopropanol or acetone via a cleavage reaction, for example, a decarboxylation of the free carboxylic acid in the actinobacterial strains IFP 2012 and Mycolicibacterium vaccae JOB5 (Lopes Ferreira et al., 2006b). However, for these reactions neither the relevant metabolites nor the enzyme activities have been unambiguously demonstrated. Here, we investigate MPD and 2-HIBA metabolism in the actinobacterial strain Actinomycetospora chiangmaiensis DSM 45062, which has previously been isolated from tropical rainforest soil, Chiang Mai, Thailand (Jiang et al., 2008). Inspection of the genome sequence (NZ_ARBI00000000.1) revealed the absence of genes encoding 2-hydroxyisobutyryl-CoA mutases. However, hcl is present in a gene cluster comprising mpdB and mpdC as well as a gene predicted to encode a thiamine pyrophosphate (TPP)-dependent enzyme. This suggests a metabolic sequence from MPD via 2-HIBA to 2-hydroxyisobutyryl-CoA which might then be cleaved to acetone and formyl-CoA employing a TPP-dependent lyase reaction (Figure 1A). The postulated pathway is corroborated by detection of the corresponding metabolites and by proteome analysis showing a strong induction of the lyase-HCL gene cluster in strain A. chiangmaiensis DSM 45062 when grown on 2-HIBA. In addition, enzymatic acetone formation from 2-hydroxyisobutyryl-CoA was demonstrated in the wild-type strain and after heterologous expression of the lyase gene in E. coli.
In the fuel oxygenate-degrading A. tertiaricarbonis L108 and M. petroleiphilum PM1 as well as in many other bacterial strains (Yaneva et al., 2012; Weichler et al., 2015), the gene encoding the specific ligase for the CoA-activation of 2-HIBA, hcl, is in operon-like organization with the 2-hydroxyisobutyryl-CoA mutase genes, hcmAB or rcmAB, and meaH encoding a mutase-associated G-protein chaperone. In addition, upstream on the same or the complement strand, a gene for a putative transcriptional regulator of the GntR family is usually present (Figure 1B). However, a BLASTP search (NCBI non-redundant protein sequences) revealed that hcl also occurs in gene clusters lacking the mutase genes. In strain A. chiangmaiensis DSM 45062, for example, hcl is associated with two genes annotated as mpdB and mpdC (Figure 1B). The corresponding proteins MpdB and MpdC are closely related to the alcohol and aldehyde dehydrogenases, respectively, from M. austroafricanum IFP 2012 (76 and 72% sequence identity, respectively, at 99% query coverage) which have previously been demonstrated to be involved in the two-step oxidation of the methyl tert-butyl ether metabolite MPD to 2-HIBA via 2-hydroxyisobutyraldehyde (Lopes Ferreira et al., 2006a). This indicates that strain A. chiangmaiensis DSM 45062 might be capable of converting MPD to 2-hydroxyisobutyryl-CoA. Further degradation, however, does obviously not employ a B12-dependent mutase, but seems to proceed via an alternative route. Interestingly, the first ORF in the hcl gene cluster, NCBI RefSeq WP_018331913.1, is predicted to encode a protein of 590 aa length belonging to the TPP-requiring decarboxylase superfamily (Vogel and Pleiss, 2014) and might function as a 2-hydroxyacyl-CoA lyase. The human peroxisomal 2-hydroxyphytanoyl-CoA lyase HACL1 (NP_036392.2), for example, catalyzes the decomposition of 2-hydroxyphytanoyl-CoA (2-hydroxy-3,7,11,15-tetramethylhexadecanoyl-CoA) to the aldehyde pristanal (2,6,10,14-tetramethylpentadecanal) and formyl-CoA (Foulon et al., 1999, 2005). In addition, it has recently been demonstrated that HACL1 and related prokaryotic TPP-dependent enzymes can even catalyze the reversible acyloin condensation of formyl-CoA with short- and medium-chain carbonyl compounds (Chou et al., 2019; Burgener et al., 2020). Although these 2-hydroxyacyl-CoA lyases and the putative lyase from DSM 45062 are only distantly related (<30% sequence identity at about 90% query coverage), this provisional, only sequence-based functional assignment of WP_018331913.1 let us speculate about a mutase-independent pathway for the degradation of MPD and 2-HIBA proceeding via the decomposition of 2-hydroxyisobutyryl-CoA to acetone and formyl-CoA (Figure 1A).
In order to elucidate the 2-HIBA pathway in strain DSM 45062, cultures were incubated in a mineral salt medium supplemented either with 5.5 mM MPD or 5.5 mM 2-HIBA as main carbon sources. On both substrates, growth was poor and biomass was produced exclusively as small cell aggregates (up to about 3 mm, Supplementary Figure S1). Based on dry weight increase, roughly 0.2 g biomass were formed per g of MPD and 2-HIBA consumed. Degradation of MPD was accompanied by temporary formation of up to 3.3 and 2.4 mM 2-HIBA and acetone, respectively (Figure 2A). Besides, low amounts of formic acid (<0.2 mM) could be detected. Likewise, 2-HIBA was slowly degraded via acetone and formic acid (Figure 2B), confirming the postulated cleavage of the C4 carboxylic acid into C3 and C1 compounds (Figure 1A).
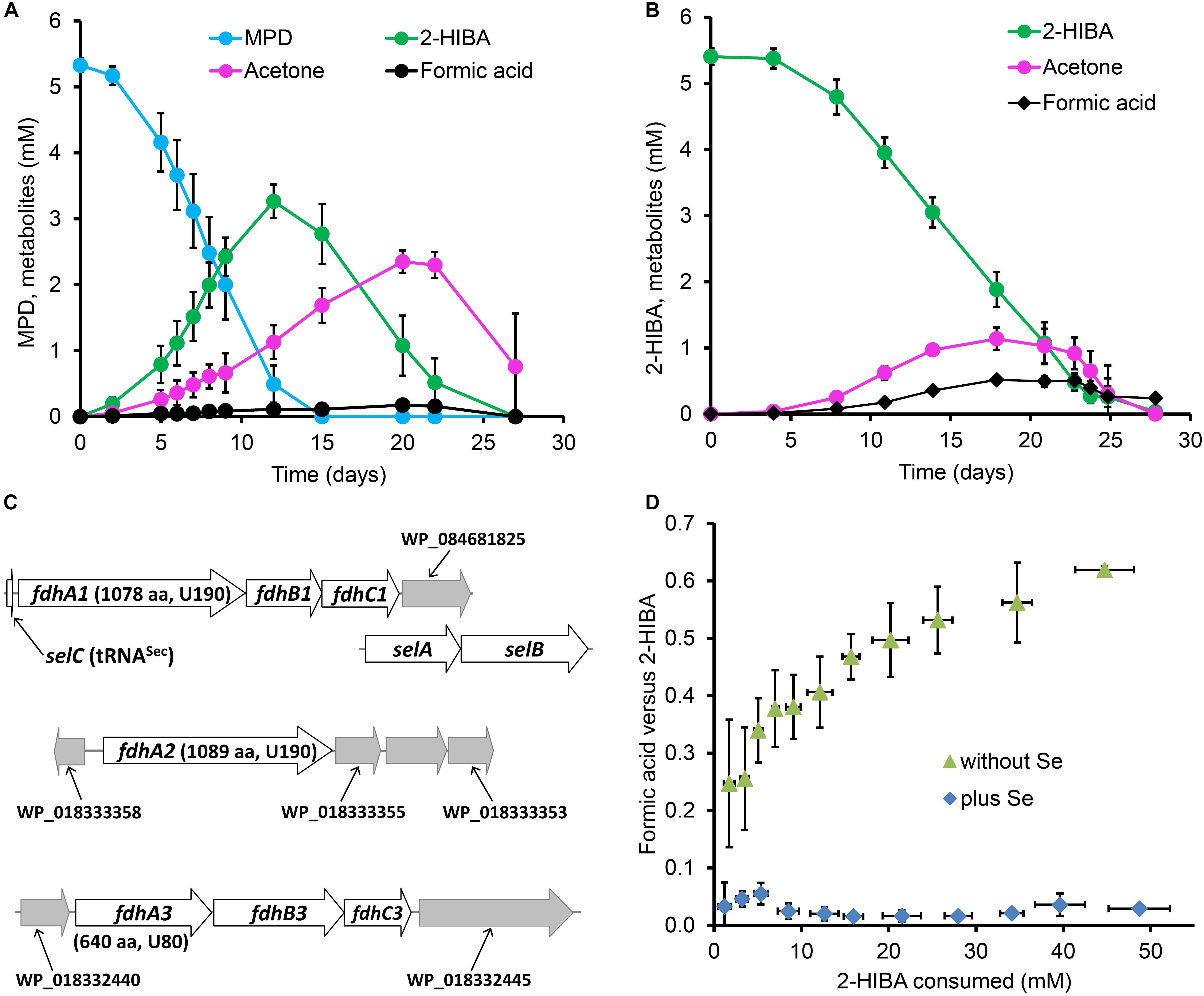
Figure 2. Metabolites of tert-butyl compound conversion and selenium dependence of formic acid removal in strain A. chiangmaiensis DSM 45062. Cultures degrading (A) MPD and (B) 2-HIBA as main carbon source when incubated in mineral salt medium not supplemented with selenium. (C) Gene clusters encoding selenocysteine-containing FDHs and associated genes. Predicted position of the selenocysteine in the corresponding FdhA subunits and the protein length are indicated. (D) Molar ratio of formic acid formation versus 2-HIBA consumption in fed-batch cultures with 2-HIBA as main carbon source incubated in mineral salt medium with or without selenium supplementation.
From the DSM 45062 genome, three multicomponent formate dehydrogenase (FDH) systems with alpha subunits bearing a selenocysteine residue within the conserved domain for binding the molybdenum/tungsten of the metallopterin cofactor (Hartmann et al., 2015) can be deduced (Figure 2C, Supplementary Figures S2–S4), whereas a selenium-independent formic acid oxidation is not encoded. Accordingly, substantial accumulation of formic acid was observed in prolonged fed-batch experiments when enabling degradation of up to 50 mM 2-HIBA in medium not supplemented with selenite (Figure 2D). However, this accumulation was not stoichiometric but only reached a molar ratio of about 0.6. Nevertheless, when supplementing the medium with selenite (0.11 μM), formic acid formation was almost negligible (Figure 2D) which is clearly corroborative of a selenocysteine-containing FDH step involved in degradation of the C1 cleavage product from 2-HIBA. Supplementation of the medium with tungsten (0.12 μM as sodium tungstate) did not show any effect on formic acid production from 2-HIBA, indicating that the active FDHs in strain DSM 45062 are binding the molybdenum-containing cofactor (Gladyshev et al., 1994). In contrast to the 2-HIBA cultures, fed-batch growth on acetone as main carbon source did not require supplementation with selenium, in line with a degradation pathway not proceeding via formic acid formation.
For identifying the key enzyme(s) catalyzing the 2-HIBA cleavage reaction, the proteome of DSM 45062 cells from fed-batch acetone and 2-HIBA cultures (both supplemented with selenite) was compared. Interestingly, expression of the complete lyase-HCL cluster is strongly induced on 2-HIBA resulting in an abundance of the corresponding gene products between 1.2 and 2.7% relative abundance of the whole proteome (Figure 3A and Table 1). In stark contrast, these proteins are almost absent in cells grown on acetone. Cells from both culture variants, however, show strong induction of a gene cluster encoding components of a binuclear iron monooxygenase (Figure 3B and Table 1) closely related to the MimABCD system involved in acetone hydroxylation in strains Mycolicibacterium goodii 12523 and Mycolicibacterium smegmatis mc2155 (Furuya et al., 2011, 2015). Hence, also in strain DSM 45062 acetone is likely hydroxylated to acetol (1-hydroxy-2-propanone) and might be subsequently oxidized to methylglyoxal and pyruvate (Figure 4). Enzyme candidates for catalyzing these alcohol and aldehyde oxidations are the putative isopropanol dehydrogenase of the Mim cluster (Adh1, WP_018330678.1) and a 507-aa aldehyde dehydrogenase (WP_018333199.1), respectively. The latter is only induced on acetone (abundance 5.2 ± 1.1%) and might be replaced by MpdC in 2-HIBA-grown cells. Besides, several other putative alcohol and aldehyde dehydrogenases are substantially induced on acetone, 2-HIBA or both substrates and might play a role in acetone degradation as well, for example, the Adh1-related WP_018331648.1 (Table 1). Among these enzymes are also alcohol dehydrogenases (WP_018332348.1 mainly induced on acetone at an abundance of about 4% and WP_018334013.1 on both substrates at values between 0.4 and 0.8%) that are predicted to use 4-nitroso-N,N-dimethylaniline as artificial and mycofactocin as endogenous cofactor, as has recently been elucidated for a related dehydrogenase involved in ethanol oxidation (Krishnamoorthy et al., 2019). Remarkably, among the most abundant proteins is also a putative catalase/peroxidase (WP_033414714.1 with an abundance between 1.6 and 4.2%) which is indicative of high stress by reactive oxygen species, possibly produced in the course of uncoupled MimABCD activity. As expected from the cultivation experiments, FDH gene clusters as well as the machinery for selenocysteine synthesis and its incorporation into proteins (SelD, SelA, and SelB) were expressed on 2-HIBA. However, the corresponding proteins were only detected at low abundance, with values between 0.001 and 0.02% (Table 1). Compared to acetone-grown cells, the fold-change in FdhA1, FdhB1 and the three FDH3 subunits was significant, albeit showing only maximal up-regulation of about threefold.
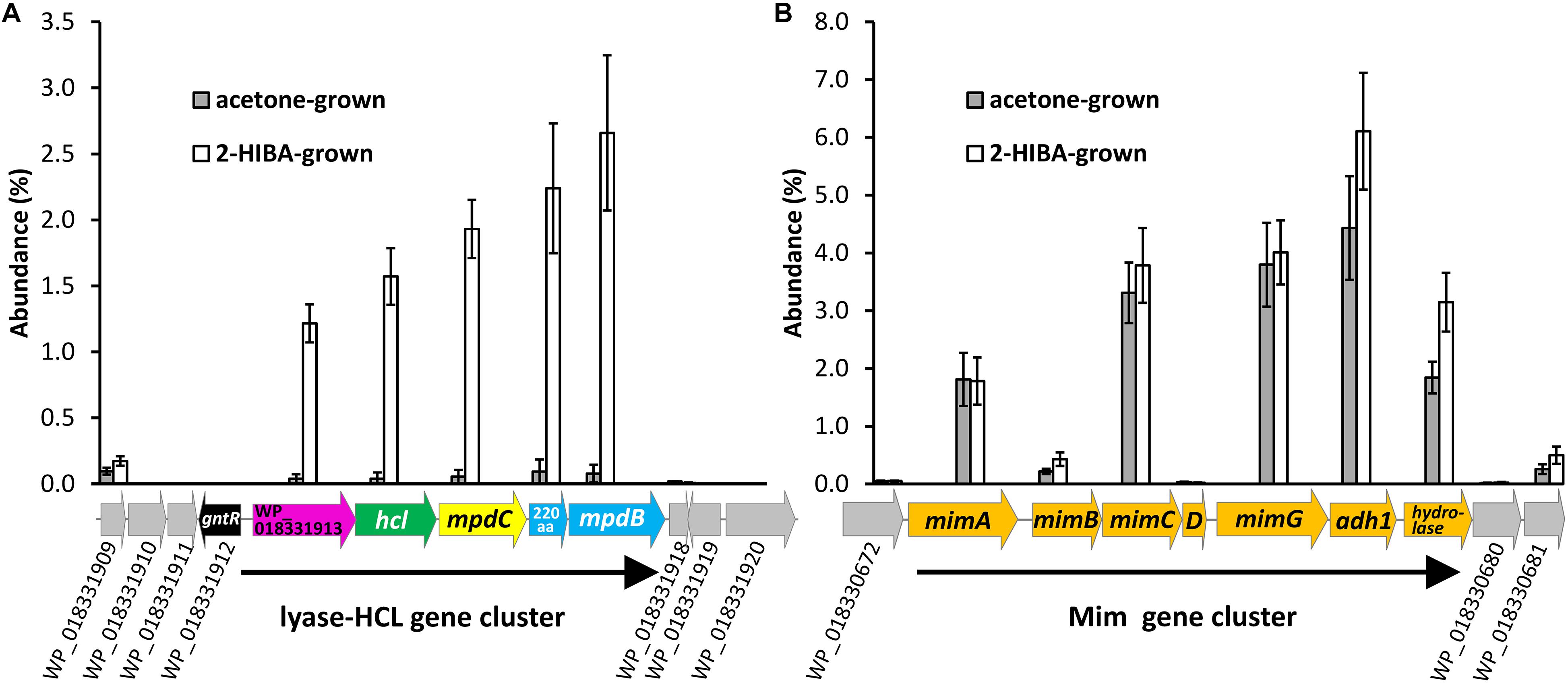
Figure 3. Induction of (A) the lyase-HCL and (B) the Mim gene clusters in fed-batch cultures of A. chiangmaiensis DSM 45062 grown on 2-HIBA or acetone in mineral salt medium supplemented with selenium. For comparison, abundance of proteins encoded by genes directly up- and downstream of the clusters is shown as well.
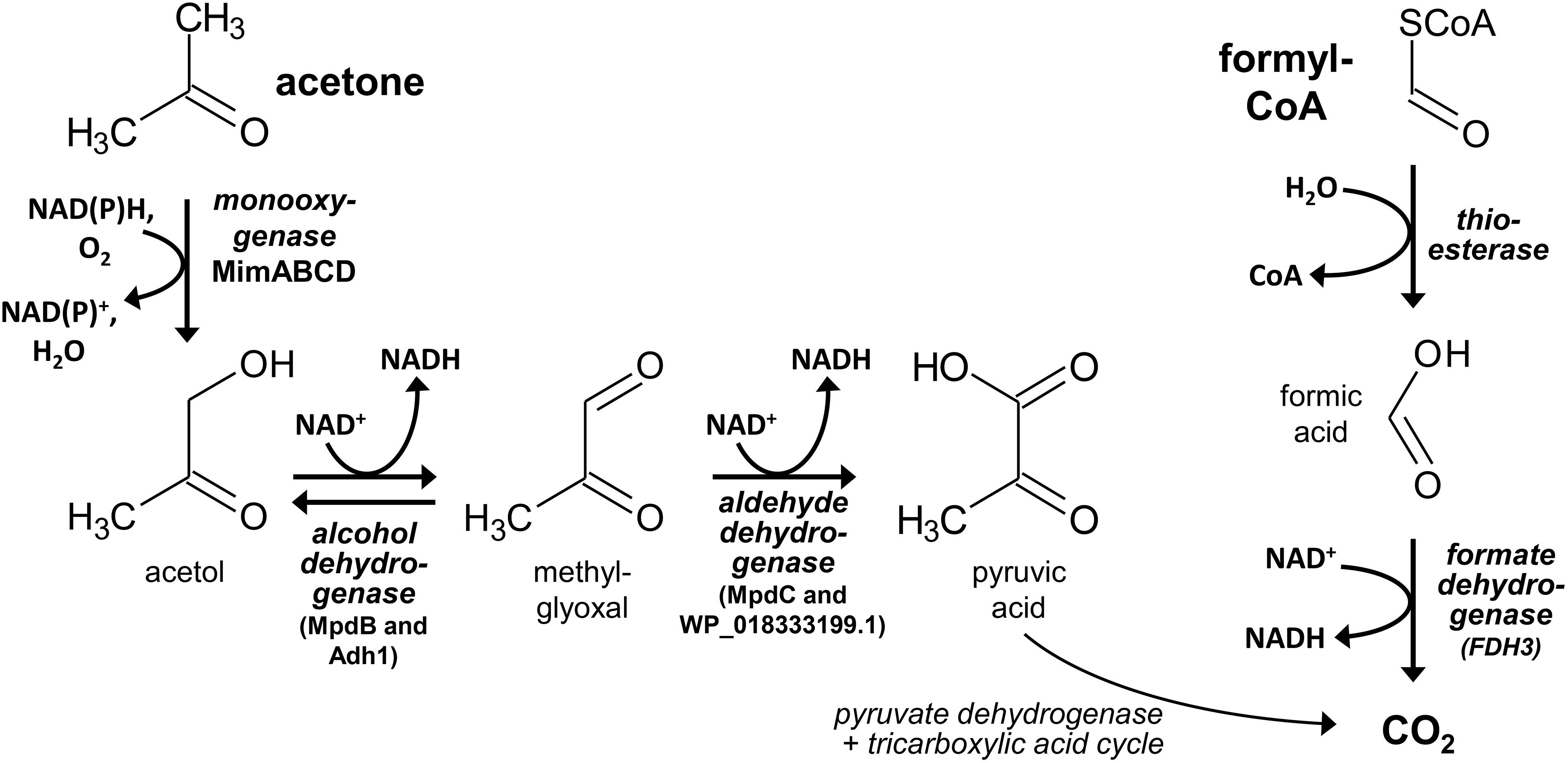
Figure 4. Proposed reaction steps for the degradation of the 2-hydroxyisobutyryl-CoA cleavage products acetone and formyl-CoA in strain A. chiangmaiensis DSM 45062. Likely, hydroxylation of acetone and the subsequent acetol oxidation to methylglyoxal are catalyzed by the Mim cluster diiron monooxygenase and alcohol dehydrogenase Adh1 (WP_018330678.1), respectively. Only in 2-HIBA-grown cells, alcohol oxidation may also involve MpdB and methylglyoxal could be oxidized by MpdC. By contrast, the aldehyde oxidation is likely taken over by dehydrogenase WP_018333199.1 when cells are incubated on acetone. Formyl-CoA may be hydrolyzed to formic acid, which can be oxidized by 2-HIBA-induced cytoplasmic NAD+-dependent formate dehydrogenase FDH3.
In line with the proteome results demonstrating significant expression of the predicted lyase gene, corresponding to WP_018331913.1, only in 2-HIBA-grown cells, crude extracts from the latter clearly show acetone formation at a rate of about 3 nmol min–1 mg protein–1 when incubated with 2-hydroxyisobutyryl-CoA, while this activity was not detected in cell-free preparations derived from acetone and glucose cultures (Figure 5A). Moreover, the 2-hydroxyisobutyryl-CoA lyase activity is stimulated by supplementation with TPP (optimal concentration at about 200 μM), in line with a cleavage mechanism dependent on this cofactor (Figure 5B). Similar activity of about 6 nmol min–1 mg–1 was established in crude extracts from E. coli cells heterologously expressing the lyase gene, confirming the role of WP_018331913.1 in cleaving 2-hydroxyisobutyryl-CoA to acetone and formyl-CoA. Interestingly, these crude extracts also show butanone formation from 2-hydroxy-2-methylbutyryl-CoA at nearly the same rate (about 5 nmol min–1 mg–1), whereas a corresponding cleavage of 2-hydroxy-2-ethylbutyryl-CoA to 3-pentanone was not detected.
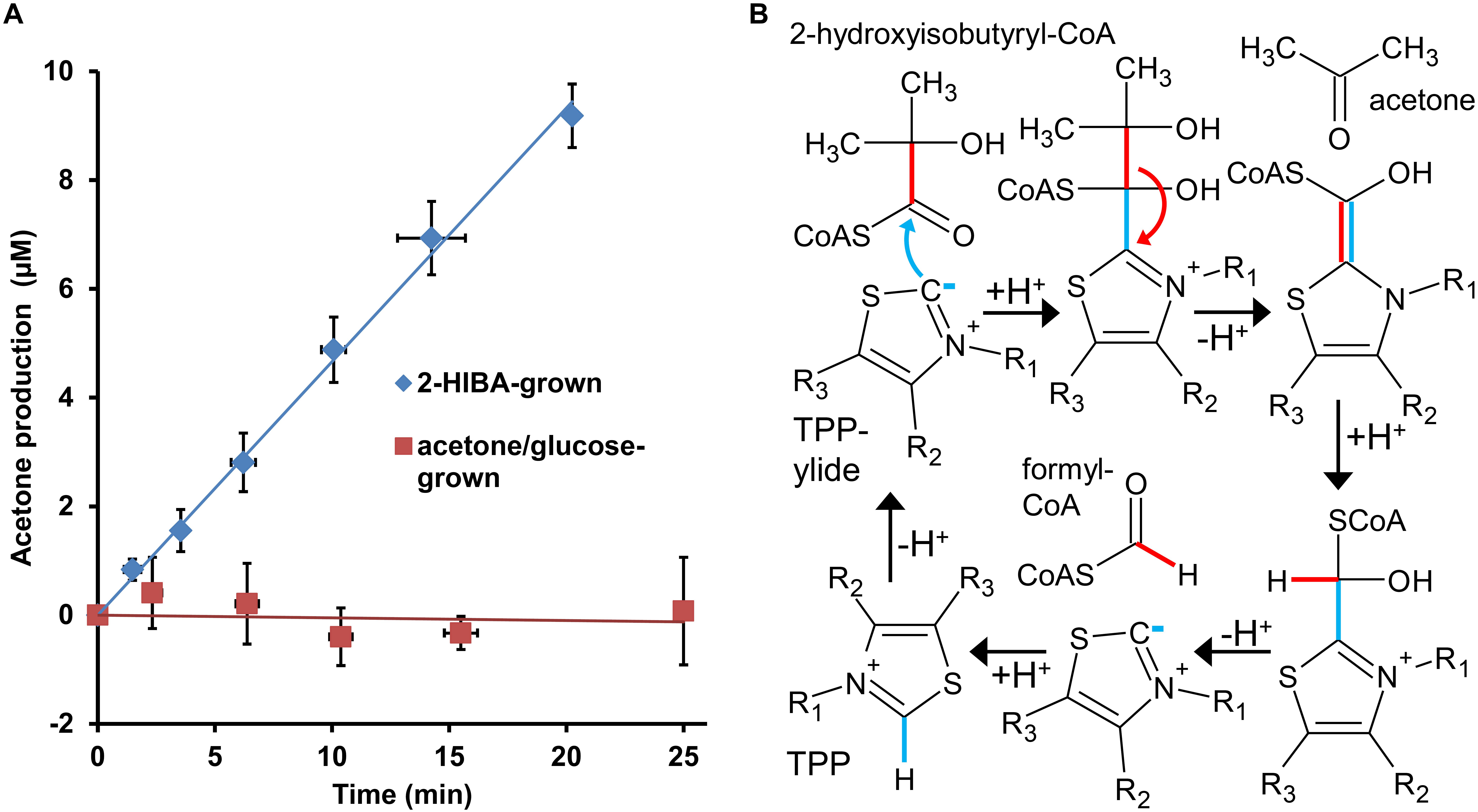
Figure 5. 2-Hydroxyisobutyryl-CoA lyase activity in strain A. chiangmaiensis DSM 45062. (A) TPP-dependent acetone formation from 2-hydroxyisobutyryl-CoA in crude extracts of cells grown on main carbon sources as indicated. The lyase activity obtained from 2-HIBA-grown cells amounts to 2.9 ± 0.1 nmol min–1 mg protein–1, while it is absent when the strain was incubated on acetone or glucose. (B) Mechanism for the reverse acyloin condensation of 2-hydroxyisobutyryl-CoA proposed to be catalyzed by the lyase from strain DSM 45062. The TPP-dependent reaction starts by deprotonation of the cofactor to yield the active ylide form. For simplicity, only the thiazole ring of TPP is shown.
In the search for a 2-hydroxyisobutyryl-CoA mutase-independent degradation pathway for short-chain tertiary branched 2-hydroxyacids, we found that the actinobacterial strain A. chiangmaiensis DSM 45062 is capable of oxidizing MPD to 2-HIBA and subsequent cleavage to acetone and formic acid. Likely, this cleavage reaction involves the corresponding CoA thioester and proceeds via formyl-CoA formation catalyzed by a novel TPP-dependent 2-hydroxyacyl-CoA lyase.
The strong induction of the lyase-HCL and Mim gene clusters in 2-HIBA-grown cells of strain DSM 45062 clearly indicates that 2-HIBA is initially activated to 2-hydroxyisobutyryl-CoA and that the cleavage product acetone is oxidized via formation of acetol and methylglyoxal, which is a highly toxic ketoaldehyde (Subedi et al., 2008). Efficient removal of the latter is, therefore, of outmost importance and seems to be guaranteed by activities of the aldehyde dehydrogenases MdpC and the acetone-induced WP_18333199.1. Besides, acetol degradation might also involve reduction to propane-1,2-diol and further oxidation to lactaldehyde and lactic acid, as previously suggested for E. coli (Subedi et al., 2008). In addition to the Mim cluster-associated alcohol dehydrogenase Adh1 and the two aldehyde dehydrogenases mentioned above, various other dehydrogenases are substantially induced and might be involved in acetol, methylglyoxal and lactaldehyde degradation as well. In contrast to acetone metabolism, no specific enzyme for the hydrolysis of formyl-CoA, the postulated second product of the 2-hydroxyisobutyryl-CoA lyase reaction, could be identified. Likely, it is hydrolyzed to CoA and formic acid by constitutive thioesterases with broad substrate specificity (Figure 4). Finally, the oxidation of formic acid turned out to be clearly dependent on selenium and highly likely involves the selenocysteine-containing membrane-bound FDH1 and cytoplasmic FDH3 identified in strain DSM 45062, although the detected abundance of the corresponding proteins was > 50-fold lower than for the other proteins of the lyase-dependent pathway, for example, the lyase, HCL and the large acetone monooxygenase subunit MimA. This result was equally obtained by two different proteome preparation methods and, therefore, seems not to be caused by inefficient protein extraction. More likely, the observed abundance is already sufficient to remove the formic acid produced during 2-HIBA cleavage, as selenocysteine-containing FDHs can show a > 100-fold increased catalytic efficiency compared to their cysteine counterparts (Axley et al., 1991). The general catalytic superiority of selenium over sulfur and its higher resistance to permanent oxidation (Reich and Hondal, 2016; Maroney and Hondal, 2018) are likely the drivers for the widespread occurrence of selenocysteine-containing enzymes in anaerobic as well as aerobic microorganisms (Peng et al., 2016), despite the requirement for selenium-sequestration and incorporation mechanisms.
During prolonged cultivation of strain DSM 45062 on 2-HIBA under selenium-limiting conditions, we observed a molar ratio of formic acid accumulation versus 2-HIBA consumption of only 0.6. This slightly understoichiometric accumulation might be resulting from partial assimilation of formic acid, possibly via reduction to formaldehyde and lyase-catalyzed acyloin condensation with formyl-CoA to glycolyl-CoA, as has been demonstrated for other 2-hydroxyacyl-CoA lyases (Chou et al., 2019). However, the novel lyase of strain DSM 45062 has not yet been biochemically characterized, due to the low activities obtained in crude extracts of wild type cells and after heterologous expression under the assay conditions applied. Considering the abundance of the enzyme in 2-HIBA-grown cells (Table 1), a specific activity of about 250 nmol min–1 mg–1 (corresponding to a kcat of 0.26 s–1) can be deduced, indicating that enzyme preparation and assay protocols still need to be optimized. In addition, a significant overexpression in E. coli could not be established, likely due to insufficient soluble expression as previously observed for human HACL1 and other 2-hydroxyacyl-CoA lyases from prokaryotic origin (Chou et al., 2019; Burgener et al., 2020). In order to purify the enzyme from strain DSM 45062 for kinetic and structural characterization, production of the recombinant protein has to be improved, for example, by employing an actinobacterial expression system such as M. smegmatis (Bashiri and Baker, 2015). Likewise, suitable purification protocols have to be established, as our preliminary attempts to remove elution buffer and to concentrate the lyase after standard immobilized metal ion affinity chromatography resulted in precipitation of the heterologous protein. The thus far characterized 2-hydroxyacyl-CoA lyase from Rhodospirillales bacterium URHD0017 (Chou et al., 2019) shows a relatively low Km value with formyl-CoA (Km = 200 ± 50 μM) for the synthase reaction with short-chain aldehydes and ketones. However, the latter compounds are poor substrates which leads to low catalytic efficiency values for the production of 2-hydroxyacyl-CoAs. In particular, the Km value for acetone exceeds 1 M, rating the enzyme rather unsuitable for developing an alternative biotechnological route to the important building block chemical methyl methacrylate via 2-HIBA (Rohwerder and Müller, 2010). Attempts to improve the enzyme by mutating various active site residues, however, did not yet result in enhanced synthesis efficiencies of the Rhodospirillales enzyme (Chou et al., 2019). Due to the involvement in a dissimilatory pathway for the degradation of 2-HIBA, we expect that the lyase from DSM 45062 is more specific for 2-hydroxyisobutyryl-CoA. Consequently, it might be a better candidate for the biotechnological production of the methyl methacrylate precursor from acetone and C1 compounds. In addition, 2-hydroxy-2-methylbutyryl-CoA is converted at equal rates, while the C6 acyl moiety of 2-hydroxy-2-ethylbutyryl-CoA seems already too bulky for the active site of the enzyme.
Interestingly, another 2-hydroxyacyl-CoA lyase candidate (IMG locus tag DebiaDRAFT_04574) belonging to the TPP-requiring enzyme superfamily has recently been identified in the deltaproteobacterium Desulfococcus biacutus KMRActS (Frey et al., 2018). The 685-aa protein is induced in acetone-grown cells (Gutierrez Acosta et al., 2014) and has been proposed to be involved in the anaerobic degradation of the carbonyl compound via 2-hydroxyisobutyryl-CoA formation (Frey et al., 2018). In strain KMRActS, the latter is isomerized by an RcmAB-like mutase to the common metabolite 3-hydroxybutyryl-CoA. Hence, for this energy efficient pathway, both the lyase and the mutase activities seem to be combined in one strain. Although not closely related to the lyase from DSM 45062 (22% identical residues at 84% query cover) and the other thus far identified 2-hydroxyacyl-CoA lyases (Chou et al., 2019), it is reasonable to assume that the deltaproteobacterial enzyme is likewise using formyl-CoA for the condensation reaction, possibly provided by a CoA-acylating formaldehyde dehydrogenase (Chou et al., 2019) or formyl-CoA transferase reaction (Berthold et al., 2008). However, as this pathway is for acetone and not for 2-HIBA degradation, strain KMRActS does not possess HCL for specifically activating the branched carboxylic acid. In conclusion, bacterial 2-hydroxyacyl-CoA lyases have evolved several times and are involved in pathways for the degradation of short-chain ketones and highly branched compounds, such as acetone and isobutene. In this connection, it is worth mentioning that a conserved lyase-HCL gene cluster is also found in Pseudonocardia strains (Figure 1B) isolated from the exoskeleton of fungus-farming leafcutter ants (Sit et al., 2015). In this case, 2-HIBA may be formed in the ants’ nest as a metabolite from isobutene produced in the course of fungal degradation of leucine-rich plant material, as the branched amino acid is usually processed via 3-hydroxyisovaleric acid which is known to be readily converted to isobutene and CO2 by eukaryotic mevalonate pyrophosphate decarboxylase (Gogerty and Bobik, 2010). Surprisingly, a lyase-HCL gene cluster variant is also present in isoprene-degrading Rhodococcus strains. In the latter, the MPD dehydrogenase MpdB is replaced by an only distantly related alcohol dehydrogenase of the glucose-methanol-choline oxidoreductase family (Figure 1B). Nevertheless, this may be indicative of a second bacterial isoprene oxidation pathway, besides the well-established route via a glutathione adduct (van Hylckama Vlieg et al., 1999; McGenity et al., 2018). In the proposed lyase-dependent route, isoprene would be oxidized as shown for isobutene in Figure 1A via the corresponding 1,2-epoxide and 1,2-diol. Finally, the potential lyase substrate 2-hydroxy-2-methylbut-3-enoyl-CoA could be formed which is likely cleaved to methyl vinyl ketone and formyl-CoA. Another substrate for the lyase may be formed by isoprene reduction to isoamylene (Kronen et al., 2019). The latter alkene could be processed analogous to isobutene and isoprene oxidation to 2-hydroxy-2-methylbutyryl-CoA, which is readily used as substrate by the lyase from strain DSM 45062. Considering that natural isoprene emissions on earth are about equivalent to methane emissions and amount to 500 and 600 Mt year–1 (Guenther et al., 2006, 2012), this volatile organic compound may be indeed the main driver for the evolution of actinobacterial 2-hydroxyacyl-CoA lyase.
Chemicals were purchased from Th. Geyer (Renningen, Germany) at the highest purity available. The lyase 2-hydroxyacyl-CoA substrates tested in this study were synthesized from the free carboxylic acids and CoA via thiophenyl esters (Padmakumar et al., 1993). 2-Hydroxy-2-methylbutyric acid was used as racemic mixture of the (R)- and (S)-enantiomers. The CoA thioesters were characterized and quantified by HPLC-based methods as previously described (Yaneva et al., 2012). MPD was from Taros Chemicals (Dortmund, Germany). Cultures of A. chiangmaiensis DSM 45062 (Jiang et al., 2008) were purchased from the German Collection of Microorganisms and Cell Cultures GmbH (Braunschweig, Germany) and grown at 30°C in a mineral salt medium containing the following (in milligrams L–1): NH4Cl, 760; KH2PO4, 680; K2HPO4, 970; CaCl2 × 6 H2O, 27; MgSO4 × 7 H2O, 71.2; FeSO4 × 7 H2O, 14.94; CuSO4 × 5 H2O, 0.785; MnSO4 × 4 H2O, 0.81; ZnSO4 × 7 H2O, 0.44; Na2MoO4 × 2 H2O, 0.25; CoCl2 × 6 H2O, 0.4; biotin, 0.02; folic acid, 0.02; pyridoxine-HCl, 0.1; thiamine-HCl, 0.05; riboflavin, 0.05; nicotinic acid, 0.05; DL-Ca-pantothenate, 0.05; p-aminobenzoic acid, 0.05; lipoic acid, 0.05, and cobalamin, 0.05; initial pH was 7.5. The medium was supplemented with 200 mg L–1 yeast extract and either glucose, 2-HIBA or acetone as main carbon source (500 to 1000 mg L–1). The trace elements selenium (Na2SeO3 × 5 H2O at 0.03 mg L–1) and tungsten (Na2WO4 × 2 H2O at 0.04 mg L–1) were added as indicated. In fed-batch experiments on 2-HIBA or acetone, substrate concentrations were maintained between 200 and 1500 mg L–1 (Supplementary Figure S5). After consumption of about 1000 mg L–1 of carbon source, the culture was supplemented with another quantum of 100 mg L–1 yeast extract. The pH was adjusted manually to values between 6.5 and 7.5 with 10% aqueous solutions of NaOH and sulfuric acid. Data shown on substrate consumption and metabolite formation in DSM 45062 cultures represent mean values and standard deviations from at least five independent experiments. Cultures of E. coli BL21 (DE3) (Invitrogen) were grown in lysogeny broth at 30°C.
Cells from fed-batch cultures growing on either acetone or 2-HIBA as main carbon source in mineral salt medium supplemented with selenium were harvested by centrifugation (10 min, 4°C, 5000 × g), washed twice in mineral salt medium and stored at −30°C. Proteome analyses were performed as described previously (Haange et al., 2019) with five replicates for each treatment. Mass spectrometric analysis of eluted peptides was performed on a Q Exactive HF mass spectrometer (Thermo Fisher Scientific, Waltham, MA, United States) coupled with a TriVersa NanoMate (Advion, Ltd., Harlow, United Kingdom) source in LC chip coupling mode. MS data processing was performed using Proteome Discoverer (v.2.2, Thermo Fischer Scientific, Waltham, MA, United States) against the protein-coding sequence of A. chiangmaiensis DSM 45062 (from NCBI March 2019) supplemented with protein-coding entries of FdhA1 and FdhA2 (see Figure 2C), in total 5,504 sequence entries. Search settings for Sequest HT search engine were set to trypsin (Full), max. missed cleavage sites: 2, precursor mass tolerance: 10 ppm, fragment mass tolerance: 0.05 Da. Carbamidomethylation of cysteines was specified as a fixed modification and the oxidation of methionine and N-acetylation of the protein N-terminus as a variable modification. Proteins were considered as identified when at least one unique peptide passed the false discovery rate below 0.01. In total, we were able to identify 2,848 protein groups comprised of 20,561 peptides, so the coverage of the A. chiangmaiensis DSM 45062 proteome was approximately 52%.
The 2-hydroxyacyl-CoA lyase gene (WP_018331913.1) was cloned into expression vector pASG-IBA43 according to the manufacturer’s protocol (IBA Lifesciences) after amplification using the Q5 High-Fidelity DNA Polymerase Master Mix (New England Biolabs), genomic DNA from strain DSM 45062, forward primer 5′-AGC GGC TCT TCA ATG GCG GAC CGG CAG GAC-3′ and reverse primer 5′-AGC GGC TCT TCT CCC GAT CCC TTC CTG ACG GCG-3′. The PCR conditions were initial denaturation of 3 min at 94°C, 25 cycles of 30 s 94°C, 45 s 60°C and 2 min 72°C, and a final elongation of 5 min at 72°C. Chemically competent cells of strain E. coli BL21 (DE3) were transformed with the constructed lyase-bearing vector via heat shock at 42°C. Heterologous expression of the lyase in lysogeny broth supplemented with 100 mg L–1 ampicillin was induced at 30°C for 5 to 7 hours by 200 μg L–1 anhydrotetracycline after an initial growth at 30°C from an optical density at 550 nm of 0.1 to a value of 0.6. Cells were harvested by centrifugation and mechanically disrupted with glass beads (Yaneva et al., 2012) in phosphate buffer (50 mM potassium phosphate, pH 7.2).
Rates of enzymatic 2-hydroxyacyl-CoA cleavage were determined in a discontinuous GC-based assay. In the case of crude extracts obtained from DSM 45062 cultures via mechanical disruption (see above), 1 mM MgCl2, 200 μM TPP, 312 μM 2-hydroxyisobutyryl-CoA and 0.16 g L–1 protein were mixed in phosphate buffer (50 mM potassium phosphate, pH. 7.2) at 30°C and 200-μL samples were taken at several time points within a total incubation of up to 25 min (Figure 5A). The samples were immediately transferred into 10-mL headspace GC vials, closed and incubated for 5 min at 70°C to quench the reaction. Then, ketone formation was analyzed by GC methods as described below. The data shown represent mean values and standard deviations from at least five independent experiments. Likewise, lyase activity in E. coli crude extracts obtained after heterologous expression was quantified at 30°C mixing 1 mM MgCl2, 200 μM TPP and 0.31 g L–1 protein with 430 μM of 2-hydroxyisobutyryl-CoA, racemic 2-hydroxy-2-methylbutyryl-CoA or 2-hydroxy-2-ethylbutyryl-CoA. As control, extracts from non-induced cultures as well as from the strain without the lyase expression plasmid were tested for lyase activity against 2-hydroxyisobutyryl-CoA. The detection limit for the lyase assay amounts to an enzymatic ketone (acetone, 2-butanone, or 3-pentanone) formation of 0.1 nmol min–1 mg–1.
In DSM 45062 cultures, consumption of substrates and formation of metabolites were routinely monitored by HPLC (Shimadzu Corporation) equipped with a Hi-Plex H column (300 mm × 7.7 mm; Agilent Technologies) and refractive index detector (Becher et al., 2018). In addition, acetone was quantified by headspace GC (Agilent Technologies) employing an Optima Delta-3 column (60 m × 0.32 mm × 0.25 μm; Macherey-Nagel) and flame ionization detection (Becher et al., 2018). Formation of ketones in bacterial cultures and in the lyase assays was verified by a GC system (Agilent Technologies) equipped with an Optima Delta-3 column (30 m × 0.25 mm × 0.25 μm; Macherey-Nagel) and mass spectrometer (Becher et al., 2018). The resulting mass spectra were compared with spectra from pure standard chemicals as well as with most-probable matches by the National Institute of Standards and Technology library database. Proteins were quantified using the Bradford reagent from Merck (Darmstadt, Germany).
Similarity of proteins was determined by BLAST (Altschul et al., 1997) and sequences were aligned by Clustal Omega (Sievers and Higgins, 2018). For identifying the selenocysteine insertion sequence (SECIS) element accompanying the selenocysteine UGA codon in FdhA subunit genes (Supplementary Figure S2), the bSECISearch tool1 was used (Zhang and Gladyshev, 2005). Secondary RNA structures were drawn with Forna2 (Kerpedjiev et al., 2015). Sequences of FDH subunits and other predicted proteins with ambiguous annotation were searched for functional sites with the NCBI Conserved Domain tool (Marchler-Bauer et al., 2017) and by InterPro3 (Finn et al., 2017).
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcaino et al., 2014; Perez-Riverol et al., 2019) with the dataset identifier PXD017800.
TR, M-TR, and JP conceived and directed the project. M-TR, NJ, and JP performed the experiments and all the authors were involved in data analysis as well as method design. TR wrote the manuscript with substantial input from all the authors.
The work was funded by the Program Topic “Chemicals in the Environment” within the Research Program “Terrestrial Environment” of the Helmholtz Association. JP has received funding from the Peoples Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme under the REA grant agreement number 624857.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank C. Dilßner and M. Neytschev (UFZ) for excellent technical assistance with CoA thioester synthesis, strain cultivation and HPLC analyses. In addition, we thank Birgit Würz (UFZ) for invaluable analytical advice and help with GC mass spectrometry. We are also indebted to L. von Wintzingerode, A. Grunwald, and J. Grabengießer (UFZ) for assistance in the cultivation and enzyme assay experiments. Many thanks to K. Eismann (UFZ) as well, for help with the proteome analysis and fruitful discussions regarding different protein extraction methods.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00691/full#supplementary-material
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Axley, M. J., Böck, A., and Stadtman, T. C. (1991). Catalytic properties of an Escherichia coli formate dehydrogenase mutant in which sulfur replaces selenium. Proc Natl Acad Sci U S A 88, 8450–8454. doi: 10.1073/pnas.88.19.8450
Bashiri, G., and Baker, E. N. (2015). Production of recombinant proteins in Mycobacterium smegmatis for structural and functional studies. Protein Sci 24, 1–10. doi: 10.1002/pro.2584
Becher, E., Heese, A., Claussen, L., Eisen, S., Jehmlich, N., Rohwerder, T., et al. (2018). Active site alanine preceding catalytic cysteine determines unique substrate specificity in bacterial CoA-acylating prenal dehydrogenase. FEBS Lett 592, 1150–1160. doi: 10.1002/1873-3468.13019
Berthold, C. L., Toyota, C. G., Richards, N. G., and Lindqvist, Y. (2008). Reinvestigation of the catalytic mechanism of formyl-CoA transferase, a class III CoA-transferase. J Biol Chem 283, 6519–6529. doi: 10.1074/jbc.M709353200
Burgener, S., Socorro Cortina, N., and Erb, T. J. (2020). Oxalyl-CoA Decarboxylase Enables Nucleophilic One-Carbon Extension of Aldehydes to Chiral alpha-Hydroxy Acids. Angew Chem Int Ed Engl 59, 5526–5530. doi: 10.1002/anie.201915155
Chou, A., Clomburg, J. M., Qian, S., and Gonzalez, R. (2019). 2-Hydroxyacyl-CoA lyase catalyzes acyloin condensation for one-carbon bioconversion. Nat Chem Biol 15, 900–906. doi: 10.1038/s41589-019-0328-0
Dai, L., Peng, C., Montellier, E., Lu, Z., Chen, Y., Ishii, H., et al. (2014). Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat Chem Biol 10, 365–370. doi: 10.1038/nchembio.1497
Elliott, P., Posma, J. M., Chan, Q., Garcia-Perez, I., Wijeyesekera, A., Bictash, M., et al. (2015). Urinary metabolic signatures of human adiposity. Sci Transl Med 7285ra262. doi: 10.1126/scitranslmed.aaa5680
Finn, R. D., Attwood, T. K., Babbitt, P. C., Bateman, A., Bork, P., Bridge, A. J., et al. (2017). InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res 45, D190–D199. doi: 10.1093/nar/gkw1107
Foulon, V., Antonenkov, V. D., Croes, K., Waelkens, E., Mannaerts, G. P., Van Veldhoven, P. P., et al. (1999). Purification, molecular cloning, and expression of 2-hydroxyphytanoyl-CoA lyase, a peroxisomal thiamine pyrophosphate-dependent enzyme that catalyzes the carbon-carbon bond cleavage during alpha-oxidation of 3-methyl-branched fatty acids. Proc Natl Acad Sci U S A 96, 10039–10044. doi: 10.1073/pnas.96.18.10039
Foulon, V., Sniekers, M., Huysmans, E., Asselberghs, S., Mahieu, V., Mannaerts, G. P., et al. (2005). Breakdown of 2-hydroxylated straight chain fatty acids via peroxisomal 2-hydroxyphytanoyl-CoA lyase: a revised pathway for the alpha-oxidation of straight chain fatty acids. J Biol Chem 280, 9802–9812. doi: 10.1074/jbc.M413362200
Francois, A., Garnier, L., Mathis, H., Fayolle, F., and Monot, F. (2003). Roles of tert-butyl formate, tert-butyl alcohol and acetone in the regulation of methyl tert-butyl ether degradation by Mycobacterium austroafricanum IFP 2012. Appl Microbiol Biotechnol 62, 256–262. doi: 10.1007/s00253-003-1268-9
Francois, A., Mathis, H., Godefroy, D., Piveteau, P., Fayolle, F., and Monot, F. (2002). Biodegradation of methyl tert-butyl ether and other fuel oxygenates by a new strain, Mycobacterium austroafricanum IFP 2012. Appl Environ Microbiol 68, 2754–2762. doi: 10.1128/aem.68.6.2754-2762.2002
Frey, J., Schneider, F., Huhn, T., Spiteller, D., Schink, B., and Schleheck, D. (2018). Two enzymes of the acetone degradation pathway of Desulfococcus biacutus: coenzyme B12 -dependent 2-hydroxyisobutyryl-CoA mutase and 3-hydroxybutyryl-CoA dehydrogenase. Environ Microbiol Rep 10, 283–292. doi: 10.1111/1758-2229.12637
Furuya, T., Hirose, S., Osanai, H., Semba, H., and Kino, K. (2011). Identification of the monooxygenase gene clusters responsible for the regioselective oxidation of phenol to hydroquinone in mycobacteria. Appl Environ Microbiol 77, 1214–1220. doi: 10.1128/aem.02316-10
Furuya, T., Nakao, T., and Kino, K. (2015). Catalytic function of the mycobacterial binuclear iron monooxygenase in acetone metabolism. FEMS Microbiol Lett 362, fnv136. doi: 10.1093/femsle/fnv136
Gladyshev, V. N., Khangulov, S. V., Axley, M. J., and Stadtman, T. C. (1994). Coordination of selenium to molybdenum in formate dehydrogenase H from Escherichia coli. Proc Natl Acad Sci U S A 91, 7708–7711. doi: 10.1073/pnas.91.16.7708
Gogerty, D. S., and Bobik, T. A. (2010). Formation of isobutene from 3-hydroxy-3-methylbutyrate by diphosphomevalonate decarboxylase. Appl Environ Microbiol 76, 8004–8010. doi: 10.1128/aem.01917-10
Guenther, A., Karl, T., Harley, P., Wiedinmyer, C., Palmer, P. I., and Geron, C. (2006). Estimates of global terrestrial isoprene emissions using MEGAN (Model of Emissions of Gases and Aerosols from Nature). Atmospheric Chemistry and Physics 6, 3181–3210. doi: 10.5194/acp-6-3181-2006
Guenther, A. B., Jiang, X., Heald, C. L., Sakulyanontvittaya, T., Duhl, T., Emmons, L. K., et al. (2012). The Model of Emissions of Gases and Aerosols from Nature version 2.1 (MEGAN2.1): an extended and updated framework for modeling biogenic emissions. Geoscientific Model Development 5, 1471–1492. doi: 10.5194/gmd-5-1471-2012
Gutierrez Acosta, O. B., Schleheck, D., and Schink, B. (2014). Acetone utilization by sulfate-reducing bacteria: draft genome sequence of Desulfococcus biacutus and a proteomic survey of acetone-inducible proteins. BMC Genomics 15:584. doi: 10.1186/1471-2164-15-584
Haange, S. B., Jehmlich, N., Hoffmann, M., Weber, K., Lehmann, J., von Bergen, M., et al. (2019). Disease Development Is Accompanied by Changes in Bacterial Protein Abundance and Functions in a Refined Model of Dextran Sulfate Sodium (DSS)-Induced Colitis. J Proteome Res 18, 1774–1786. doi: 10.1021/acs.jproteome.8b00974
Hartmann, T., Schwanhold, N., and Leimkuhler, S. (2015). Assembly and catalysis of molybdenum or tungsten-containing formate dehydrogenases from bacteria. Biochim Biophys Acta 1854, 1090–1100. doi: 10.1016/j.bbapap.2014.12.006
Huang, J., Luo, Z., Ying, W., Cao, Q., Huang, H., Dong, J., et al. (2017). 2-Hydroxyisobutyrylation on histone H4K8 is regulated by glucose homeostasis in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 114, 8782–8787. doi: 10.1073/pnas.1700796114
Irwin, C., Mienie, L. J., Wevers, R. A., Mason, S., Westerhuis, J. A., van Reenen, M., et al. (2018). GC-MS-based urinary organic acid profiling reveals multiple dysregulated metabolic pathways following experimental acute alcohol consumption. Sci Rep 8, 5775. doi: 10.1038/s41598-018-24128-1
Jiang, Y., Wiese, J., Tang, S. K., Xu, L. H., Imhoff, J. F., and Jiang, C. L. (2008). Actinomycetospora chiangmaiensis gen. nov., sp. nov., a new member of the family Pseudonocardiaceae. Int J Syst Evol Microbiol 58(Pt 2), 408–413. doi: 10.1099/ijs.0.64976-0
Kerpedjiev, P., Hammer, S., and Hofacker, I. L. (2015). Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 31, 3377–3379. doi: 10.1093/bioinformatics/btv372
Kottegoda, S., Waligora, E., and Hyman, M. (2015). Metabolism of 2-methylpropene (isobutylene) by the aerobic bacterium Mycobacterium sp. strain ELW1. Appl Environ Microbiol 81, 1966–1976. doi: 10.1128/aem.03103-14
Krishnamoorthy, G., Kaiser, P., Lozza, L., Hahnke, K., Mollenkopf, H. J., and Kaufmann, S. H. E. (2019). Mycofactocin Is Associated with Ethanol Metabolism in Mycobacteria. mBio 10, doi: 10.1128/mBio.00190-19
CrossRef Full Texte00190-19,
Kronen, M., Lee, M., Jones, Z. L., and Manefield, M. J. (2019). Reductive metabolism of the important atmospheric gas isoprene by homoacetogens. Isme j 13, 1168–1182. doi: 10.1038/s41396-018-0338-z
Kurteva-Yaneva, N., Zahn, M., Weichler, M. T., Starke, R., Harms, H., Müller, R. H., et al. (2015). Structural basis of the stereospecificity of bacterial B12-dependent 2-hydroxyisobutyryl-CoA mutase. J Biol Chem 290, 9727–9737. doi: 10.1074/jbc.M115.645689
Li, X., Xu, Z., Lu, X., Yang, X., Yin, P., Kong, H., et al. (2009). Comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry for metabonomics: Biomarker discovery for diabetes mellitus. Anal Chim Acta 633, 257–262. doi: 10.1016/j.aca.2008.11.058
Lopes Ferreira, N., Labbe, D., Monot, F., Fayolle-Guichard, F., and Greer, C. W. (2006a). Genes involved in the methyl tert-butyl ether (MTBE) metabolic pathway of Mycobacterium austroafricanum IFP 2012. Microbiology 152(Pt 5), 1361–1374. doi: 10.1099/mic.0.28585-0
Lopes Ferreira, N., Malandain, C., and Fayolle-Guichard, F. (2006b). Enzymes and genes involved in the aerobic biodegradation of methyl tert-butyl ether (MTBE). Appl Microbiol Biotechnol 72, 252–262. doi: 10.1007/s00253-006-0494-3
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45, D200–D203. doi: 10.1093/nar/gkw1129
Maroney, M. J., and Hondal, R. J. (2018). Selenium versus sulfur: Reversibility of chemical reactions and resistance to permanent oxidation in proteins and nucleic acids. Free Radic Biol Med 127, 228–237. doi: 10.1016/j.freeradbiomed.2018.03.035
McGenity, T. J., Crombie, A. T., and Murrell, J. C. (2018). Microbial cycling of isoprene, the most abundantly produced biological volatile organic compound on Earth. Isme j 12, 931–941. doi: 10.1038/s41396-018-0072-6
Padmakumar, R., Gantla, S., and Banerjee, R. (1993). A rapid method for the synthesis of methylmalonyl-coenzyme A and other CoA-esters. Anal Biochem 214, 318–320. doi: 10.1006/abio.1993.1494
Peng, T., Lin, J., Xu, Y. Z., and Zhang, Y. (2016). Comparative genomics reveals new evolutionary and ecological patterns of selenium utilization in bacteria. Isme j 10, 2048–2059. doi: 10.1038/ismej.2015.246
Perez-Riverol, Y., Csordas, A., Bai, J., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J., et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47, D442–D450. doi: 10.1093/nar/gky1106
Reich, H. J., and Hondal, R. J. (2016). Why Nature Chose Selenium. ACS Chem Biol 11, 821–841. doi: 10.1021/acschembio.6b00031
Rohde, M. T., Tischer, S., Harms, H., and Rohwerder, T. (2017). Production of 2-Hydroxyisobutyric Acid from Methanol by Methylobacterium extorquens AM1 Expressing (R)-3-Hydroxybutyryl Coenzyme A-Isomerizing Enzymes. Appl Environ Microbiol 83, doi: 10.1128/aem.02622-16
CrossRef Full Text e02622-16,
Rohwerder, T., Breuer, U., Benndorf, D., Lechner, U., and Müller, R. H. (2006). The alkyl tert-butyl ether intermediate 2-hydroxyisobutyrate is degraded via a novel cobalamin-dependent mutase pathway. Appl Environ Microbiol 72, 4128–4135. doi: 10.1128/aem.00080-06
Rohwerder, T., and Müller, R. H. (2010). Biosynthesis of 2-hydroxyisobutyric acid (2-HIBA) from renewable carbon. Microb Cell Fact 9, 13. doi: 10.1186/1475-2859-9-13
Schäfer, F., Muzica, L., Schuster, J., Treuter, N., Rosell, M., Harms, H., et al. (2011). Formation of alkenes via degradation of tert-alkyl ethers and alcohols by Aquincola tertiaricarbonis L108 and Methylibium spp. Appl Environ Microbiol 77, 5981–5987. doi: 10.1128/aem.00093-11
Schäfer, F., Schuster, J., Wurz, B., Hartig, C., Harms, H., Müller, R. H., et al. (2012). Synthesis of short-chain diols and unsaturated alcohols from secondary alcohol substrates by the Rieske nonheme mononuclear iron oxygenase MdpJ. Appl Environ Microbiol 78, 6280–6284. doi: 10.1128/aem.01434-12
Schuster, J., Schäfer, F., Hubler, N., Brandt, A., Rosell, M., Hartig, C., et al. (2012). Bacterial degradation of tert-amyl alcohol proceeds via hemiterpene 2-methyl-3-buten-2-ol by employing the tertiary alcohol desaturase function of the Rieske nonheme mononuclear iron oxygenase MdpJ. J Bacteriol 194, 972–981. doi: 10.1128/jb.06384-11
Sievers, F., and Higgins, D. G. (2018). Clustal Omega for making accurate alignments of many protein sequences. Protein Sci 27, 135–145. doi: 10.1002/pro.3290
Sit, C. S., Ruzzini, A. C., Van Arnam, E. B., Ramadhar, T. R., Currie, C. R., and Clardy, J. (2015). Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants. Proc Natl Acad Sci U S A 112, 13150–13154. doi: 10.1073/pnas.1515348112
Subedi, K. P., Kim, I., Kim, J., Min, B., and Park, C. (2008). Role of GldA in dihydroxyacetone and methylglyoxal metabolism of Escherichia coli K12. FEMS Microbiol Lett 279, 180–187. doi: 10.1111/j.1574-6968.2007.01032.x
van Hylckama Vlieg, J. E., Kingma, J., Kruizinga, W., and Janssen, D. B. (1999). Purification of a glutathione S-transferase and a glutathione conjugate-specific dehydrogenase involved in isoprene metabolism in Rhodococcus sp. strain AD45. J Bacteriol 181, 2094–2101.
Vizcaino, J. A., Deutsch, E. W., Wang, R., Csordas, A., Reisinger, F., Rios, D., et al. (2014). ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 32, 223–226. doi: 10.1038/nbt.2839
Vogel, C., and Pleiss, J. (2014). The modular structure of ThDP-dependent enzymes. Proteins 82, 2523–2537. doi: 10.1002/prot.24615
Weichler, M. T., Kurteva-Yaneva, N., Przybylski, D., Schuster, J., Müller, R. H., Harms, H., et al. (2015). Thermophilic Coenzyme B12-Dependent Acyl Coenzyme A (CoA) Mutase from Kyrpidia tusciae DSM 2912 Preferentially Catalyzes Isomerization of (R)-3-Hydroxybutyryl-CoA and 2-Hydroxyisobutyryl-CoA. Appl Environ Microbiol 81, 4564–4572. doi: 10.1128/aem.00716-15
Yaneva, N., Schuster, J., Schäfer, F., Lede, V., Przybylski, D., Paproth, T., et al. (2012). Bacterial acyl-CoA mutase specifically catalyzes coenzyme B12-dependent isomerization of 2-hydroxyisobutyryl-CoA and (S)-3-hydroxybutyryl-CoA. J Biol Chem 287, 15502–15511. doi: 10.1074/jbc.M111.314690
Zahn, M., Kurteva-Yaneva, N., Schuster, J., Krug, U., Georgi, T., Müller, R. H., et al. (2019). Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle. J Mol Biol 431, 2747–2761. doi: 10.1016/j.jmb.2019.05.027
Keywords: degradation pathway, isobutene, tert-butyl alcohol, fuel oxygenate, Mycolicibacterium, 2-hydroxyacyl-CoA lyase, acyloin condensation
Citation: Rohwerder T, Rohde M-T, Jehmlich N and Purswani J (2020) Actinobacterial Degradation of 2-Hydroxyisobutyric Acid Proceeds via Acetone and Formyl-CoA by Employing a Thiamine-Dependent Lyase Reaction. Front. Microbiol. 11:691. doi: 10.3389/fmicb.2020.00691
Received: 10 February 2020; Accepted: 25 March 2020;
Published: 15 April 2020.
Edited by:
Ivan A. Berg, University of Münster, GermanyReviewed by:
Wolfgang Buckel, University of Marburg, GermanyCopyright © 2020 Rohwerder, Rohde, Jehmlich and Purswani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thore Rohwerder, dGhvcmUucm9od2VyZGVyQHVmei5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.