
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 17 April 2020
Sec. Microbiotechnology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00634
This article is part of the Research Topic Exploring the Growing Role of Cyanobacteria in Industrial Biotechnology and Sustainability View all 10 articles
Sustainable production of biofuels and biochemicals has been broadly accepted as a solution to lower carbon dioxide emissions. Besides being used as lubricants or detergents, oleochemicals are also attractive biofuels as they are compatible with existing transport infrastructures. Cyanobacteria are autotrophic prokaryotes possessing photosynthetic abilities with mature genetic manipulation systems. Through the introduction of exogenous or the modification of intrinsic metabolic pathways, cyanobacteria have been engineered to produce various bio-chemicals and biofuels over the past decade. In this review, we specifically summarize recent progress on photosynthetic production of fatty acids, fatty alcohols, fatty alk(a/e)nes, and fatty acid esters by genetically engineered cyanobacteria. We also summarize recent reports on fatty acid and lipid metabolisms of cyanobacteria and provide perspectives for economic cyanobacterial oleochemical production in the future.
Since the Industrial Revolution, the level of global carbon dioxide together with other greenhouse gases (GHGs) significantly increased due to human activities (Ainsworth et al., 2020). Cumulative anthropogenic emissions of CO2 have been considered as the main driver of global warming (Ainsworth et al., 2020). Transportation is a major contributor to the global CO2 emission, representing 65% of the world oil consumption and 24% of global CO2 emissions due to the direct combustion of fuels (Li et al., 2019; Solaymani, 2019). With the worldwide concerns about global warming, biofuels have been embraced as promising alternatives to fossil fuels, because they are renewable and generally can lower carbon emissions (Demirbas, 2009; Gaurav et al., 2017).
Oleochemicals are a large group of fatty acid derivatives, including fatty acids, fatty alcohols, fatty alk (a/e)nes, and fatty acid methyl/ethyl esters and waxes (Pfleger et al., 2015). They can be used as biodiesels, lubricants, and surfactants, and others (Yu et al., 2014; Pfleger et al., 2015; Marella et al., 2018). Compared with ethanol, which is another popular biofuel molecule, lipid-derived biodiesels have been considered to be better biofuel molecules due to their high energy density and compatibility with the existing liquid fuel infrastructure (i.e., fuel engines, refinery equipment, and transportation pipelines) (Lu, 2010).
Traditionally, crop oils and animal fats (Figure 1) were used as feedstocks for the production of oleochemicals by chemical or enzymatic processes (Pfleger et al., 2015). However, this traditional route for oleochemical production will compete with crops for arable land, decrease food production, and raise serious concerns about food security (Graham-Rowe, 2011). Microalgae have been considered as promising feedstocks for oleochemicals (Figure 1) because of their higher lipid productivities per ground area than oleaginous agricultural crops, as well as the lack of competition they would provide for agricultural land (Mata et al., 2010; Wijffels and Barbosa, 2010). Besides, abundant lignocellulosic biomass has become another ideal feedstock for the production of oleochemicals (Figure 1), in the context of large-scale metabolic engineering efforts in microbial systems (Lee et al., 2008; Alper and Stephanopoulos, 2009; Peralta-Yahya and Keasling, 2010; Keasling, 2012). Some heterotrophic model microorganisms, such as Escherichia coli and Saccharomyces cerevisiae, have been genetically modified to produce many kinds of biofuels and bio-chemicals including oleochemicals from lignocellulosic sugars (Atsumi et al., 2008; Steen et al., 2010; Buijs et al., 2013). Some recent review articles have summarized biosynthesis pathways, metabolic engineering strategies, and challenges for the production of oleochemicals by heterotrophic microbes (Janssen and Steinbuchel, 2014; Pfleger et al., 2015; Marella et al., 2018).
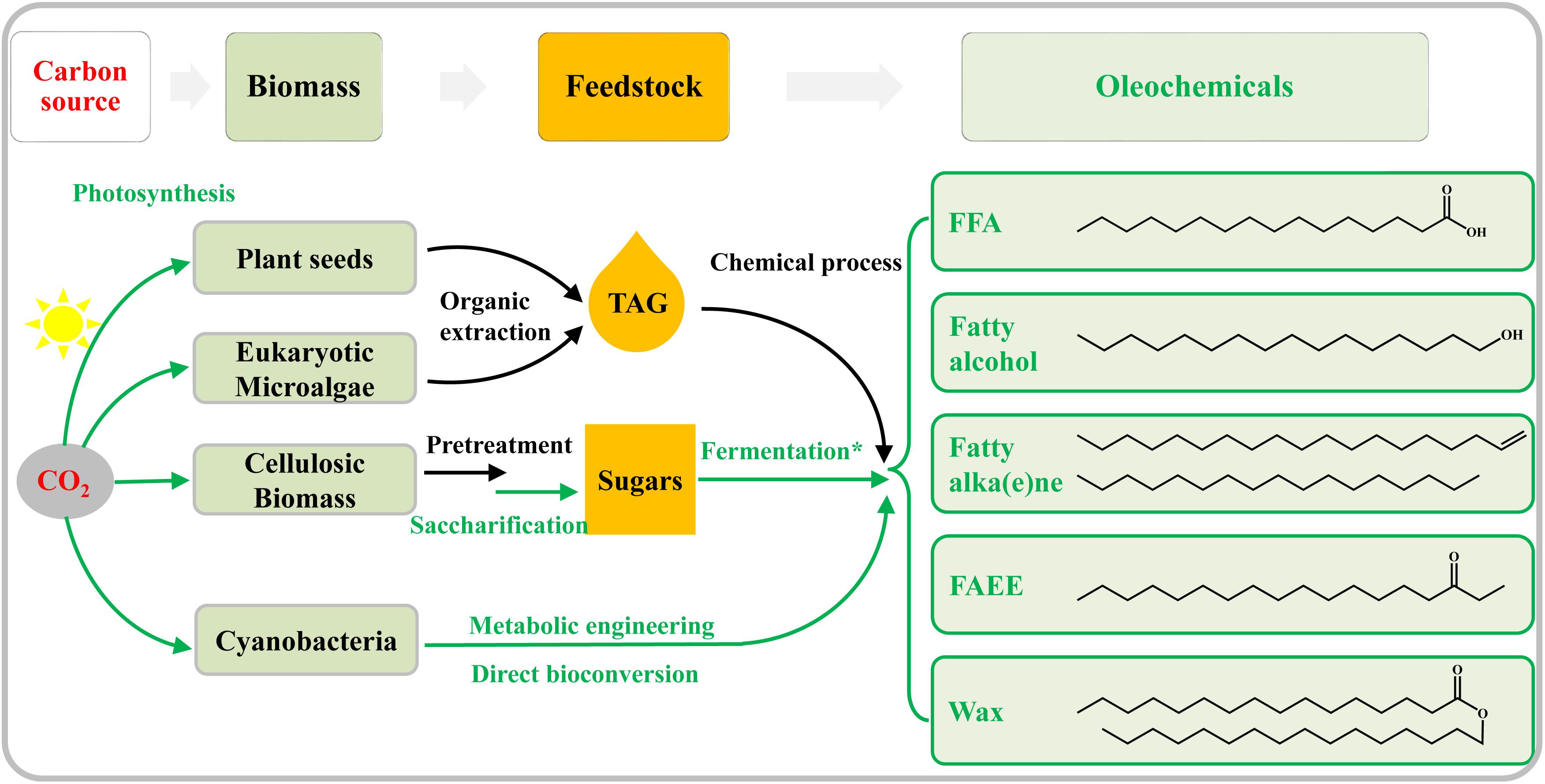
Figure 1. Overview of both traditional and emerging technologies for the bio-production of oleochemicals. The green lines show enzymatic or biological conversion, whereas physical or chemical processes are in black. Vegetable oil seeds are traditionally utilized to produce oleochemicals. Oleaginous eukaryotic microalgae are attractive alternatives to plant oil. Abundant cellulosic biomass is first hydrolyzed to sugars, and the latter is then fermented to produce oleochemicals by the engineered heterotrophic microbes (indicated by the asterisk). Harboring photosynthesis ability, cyanobacteria can be genetically modified to direct convert CO2 to oleochemicals.
Cyanobacteria are the only prokaryotes capable of performing oxygen-evolving photosynthesis (Hagemann and Hess, 2018), and have been the genetic models for photosynthesis research for decades (Wang et al., 2018). They were initially not considered to be useful to the Aquatic Species Program for biofuel production, because most of them do not naturally accumulate storage lipids in the form of triacylglycerol (TAG) as some oleaginous eukaryotic microalgae do (Sheehan et al., 1998). However, cyanobacteria have emerged as novel chassis strains for the production of biofuels and bio-chemicals since 2009, owing to their photosynthetic abilities and reliable genetic systems (Angermayr et al., 2009; Atsumi et al., 2009; Dexter and Fu, 2009; Lindberg et al., 2009). Engineered cyanobacteria are able to produce various compounds directly from CO2, bypassing the need for fermentable sugars and arable land (Lai and Lan, 2015). In the past decade, photosynthetic production of various compounds, including oleochemicals (Figure 1), has been achieved in several model cyanobacteria through metabolic engineering (Zhou and Li, 2010; Angermayr et al., 2015; Savakis and Hellingwerf, 2015; Oliver et al., 2016; Xiong et al., 2017). This review summarizes current knowledge on the metabolism of fatty acids and membrane lipids in cyanobacteria, provides the current status of metabolic engineering strategies for producing oleochemicals, and discusses key challenges and possible solutions in the field.
The biosynthesis of membrane lipids in cyanobacteria has been investigated since the 1980s (Naoki and Norio, 1982) and was followed by systematical works by Murata and co-workers in the 1990s (Wada and Murata, 1990; Wada and Murata, 1998) and 2000s (Sato and Wada, 2009). Unlike heterotrophic prokaryotes, the vast majority of cyanobacteria have thylakoid membranes in their cytoplasm where photosynthesis takes place (Rexroth et al., 2011). Both cytoplasmic (plasma) and thylakoid membranes of cyanobacteria include four major polar glycerolipids: monogalactosyl diacylglycerol (MGDG), digalactosyl diacylglycerol (DGDG), sulfoquinovosyldiacylglycerol (SQDG), and phosphatidylglycerol (PG) (Los and Mironov, 2015). Despite a report indicating the occurrence of neutral lipid droplets including triacylglycerol (TAG) in the cyanobacterium Nostoc punctiforme PCC73102 (hereafter Npu73102) (Peramuna and Summers, 2014), it is noteworthy that the above four polar lipids still serve as the dominant sink for fatty acids in cyanobacteria.
Same as the widely studied fatty acid biosynthesis pathway in E. coli, cyanobacterial fatty acid biosynthesis pathways are composed of reactions catalyzed by two protein complexes, namely, acetyl-CoA carboxylase (ACCase) and type II fatty acid synthase (FAS) encoded by fab genes (Sato and Wada, 2009). In brief, acetyl-CoA is firstly converted to malonyl-CoA by acetyl-CoA carboxylase, and then to malonyl-ACP by malonyl-CoA:ACP transacylase (FabD) (Figure 2). Subsequently, butyryl-ACP is generated by sequential reactions catalyzed by β-ketoacyl-ACP synthase III (FabH), β-ketoacyl-ACP reductase (FabG), β-hydroxyacyl-ACP dehydrase (FabZ), and enoyl-ACP reductase (FabI). The fatty acid chain is then elongated with an acetyl unit from malonyl-ACP for each condensation-reduction-dehydration-reduction cycle (Figure 2) (Sato and Wada, 2009). For most cyanobacteria, palmitoyl-ACP (C16) and stearoyl-ACP (C18) are used as precursors for the biosynthesis of membrane lipids. Contrary to previous findings in E. coli (Yu et al., 2011), it was proved that FabH, which condenses malonyl-ACP with acetyl-CoA to form acetoacetyl-ACP, is the sole rate-limiting enzyme of FAS in Synechococcus sp. PCC7002 (hereafter Syn7002) (Kuo and Khosla, 2014).
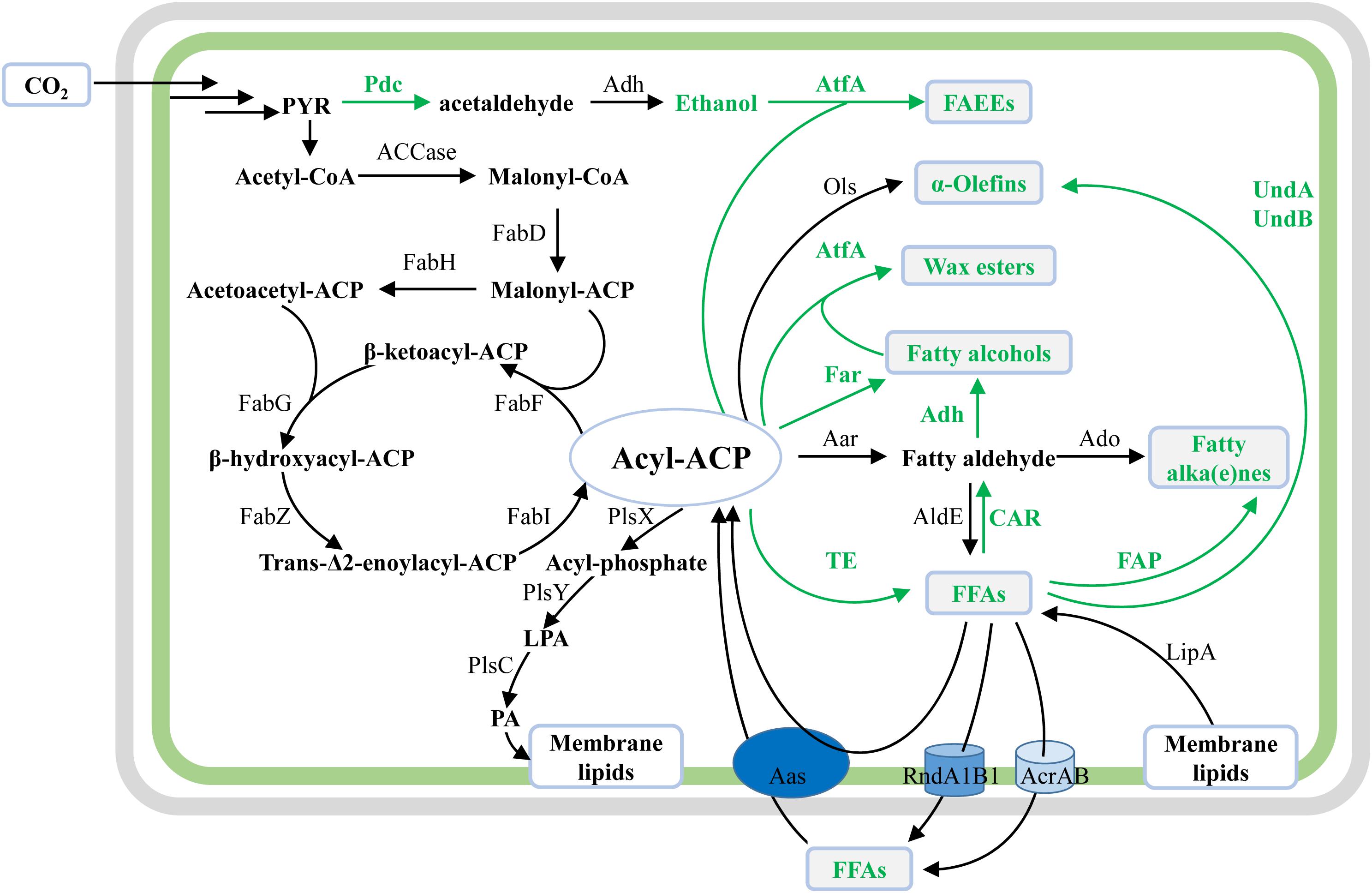
Figure 2. Schematic overview of native and synthetic pathways for biosynthesis of membrane lipids and oleochemicals in cyanobacteria. The illustration shows the native biosynthesis pathways for membrane lipids, together with synthetic pathways for cyanobacterial oleochemical production. For synthetic pathways, enzymes and reaction direction are shown in green. For native cyanobacterial pathways, enzymes and reaction direction are in black. Oleochemicals discussed in this work are shown in green texts and in gray boxes.
For the biosynthesis of the four polar glycerolipids mentioned above, phosphatidic acid (PA) is synthesized as the common precursor by the acylation of both sn-1 and 2 positions of glycerol-3-phosphate (G3P) with the long-chain fatty acyl-A (C16 or C18) by different acyltransferases (Sato and Wada, 2009). Specifically, the fatty acyl-ACPs are first activated by an inorganic phosphate group by phosphate acyltransferase (PlsX), and subsequently transferred to the sn-1 position of G3P by acylglycerol-phosphate acyltransferase (AGPAT or PlsY), resulting in lysophosphatidic acid (LPA) (Figure 2). Secondly, lysophosphatidic acid acyltransferase (LPAAT or PlsC) catalyzes the transfer of fatty acid chains to the sn-2 position of LPA in the PA biosynthesis, resulting in PA (Figure 2). Although the over-expression of the putative PlsX enhanced lipid production in Synechocystis sp. PCC 6803 (hereafter Syn6803) (Towijit et al., 2018), the detailed enzymatic characteristics of both PlsX and PlsY are still unknown. Sll1848 was identified as the primary LPAAT with a high specificity for 16:0-ACP (Weier et al., 2005), whereas Sll1752 was characterized as the secondary LPAAT that prefers stearoyl and oleoyl substrates (C18) in Syn6803 (Okazaki et al., 2006) (Figure 2).
As summarized by previous reviews (Wada and Murata, 1998; Sato and Wada, 2009), different polar head groups are further transferred to the sn-3 position of PA to synthesize four major polar glycerolipids in cyanobacteria. Finally, MGDG, DGDG, SQDG, and PG have a head group of 1β-galactose, digalactose, 6-deoxy-6-sulfo-α1-glucose, and sn-glycerol 1-phosphate at their sn-3 position of the glycerol moiety, respectively, besides two acyl groups esterified at the sn-1 and sn -2 positions (Sato and Wada, 2009).
As reviewed previously, cyanobacterial desaturases were classified as acyl-lipid desaturases rather than acyl-CoA or acyl-ACP desaturases, which means the fatty acid chain would be desaturated only when fatty acids are bound to membrane lipids (Murata and Wada, 1995; Sato and Wada, 2009; Los and Mironov, 2015). In response to the cold stress, the fatty acid chains can be stepwise desaturated at the Δ9, Δ12, ω3, and Δ6 positions by four specific desaturases, namely, DesC, DesA, DesB, and DesD, respectively. The fatty acid chain length of cyanobacteria varies from C14 to C18, whereas the number of double bonds in the fatty acid chains may vary from 0 to 4, which is controlled by the activities of the above desaturases (Los and Mironov, 2015). The fatty acid composition determined by the chain length and the numbers of double bonds can be used for the classification of cyanobacterial strains (Wada and Murata, 1998).
As the physical barrier of cells and sites of photosynthesis and respiration, the cytoplasmic and thylakoid membranes of cyanobacteria are sensitive to various environmental stimuli. On the one hand, glycerolipid and fatty acid compositions were observed to change with alterations of growth temperature, light illumination intensity, carbon dioxide, and pH (Wada and Murata, 1990; Sakamoto and Bryant, 2002; Cuellar-Bermudez et al., 2015). On the other hand, membrane lipids play an active role in the acclimation of cyanobacteria to different environmental conditions, including high temperature (Nanjo et al., 2010) and low temperature (Murata and Wada, 1995; Sakamoto and Bryant, 2002). Despite these observations on the physiological roles of lipids, little is known about the regulation of fatty acid and lipid metabolisms in cyanobacteria.
Hik33 has been identified as the sensory histidine kinase of a two-component system which perceives the low-temperature signal and controls the expression of desB gene (Suzuki et al., 2000; Mikami et al., 2002; Murata and Los, 2006). Additionally, the global regulator LexA was found to repress the expression of some fab genes (Kizawa et al., 2017), while a transcriptional regulator, CyAbrB2, was found to inhibit the FFA production in Syn6803 (Kawahara et al., 2016). The global nitrogen regulator PII protein was shown to negatively regulate cyanobacterial fatty acid biosynthesis by transcriptional control (Verma et al., 2018) or by interacting with biotin carboxyl carrier protein (BCCP) which is a subunit of ACCase (Hauf et al., 2016).
A decade ago, it was shown that the elimination of fatty acid β-oxidation by disrupting the fadD or fadE gene (Figure 2), over-expression of a thioesterase (TE) gene to release FFAs, and over-expression of ACCase have been demonstrated to be effective approaches for the overproduction of free fatty acids (FFAs) in some heterotrophic microbes like E. coli (Lu et al., 2008). Similar strategies were also adopted soon afterward for cyanobacterial FFA production (Table 1).
There is no complete fatty acid β-oxidation pathway for FFA degradation in cyanobacteria, based on bioinformatics analysis. However, acyl–acyl carrier protein synthetases (Aas) were proven to be able to re-activate FFAs to acyl-ACPs (Figure 2), and the latter can be incorporated into the membrane lipids through the above-mentioned acyltransferases in cyanobacteria (Kaczmarzyk and Fulda, 2010). The disruption of Aas led to the FFA accumulation and secretion in both Syn6803 and Synechococcus elongatus PCC7942 (hereafter Syn7942) (Kaczmarzyk and Fulda, 2010). This strategy has been adopted by most research efforts on cyanobacterial FFA production (Liu et al., 2011a; Ruffing and Jones, 2012; Ruffing, 2014) (Table 1).
Hydrolysis of acyl-ACP to FFA by thioesterases can release the feedback inhibition of acyl-ACP to some enzymes of FAS-II and has in turn been confirmed to be an effective strategy to enhance FFA production in E. coli (Heath and Rock, 1996a, b; Lu et al., 2008). However, there is no gene encoding a thioesterase in cyanobacteria. Engineering efforts for improving FFA production in cyanobacteria began nearly a decade ago (Liu et al., 2011a). In this work, a truncated E. coli TE gene ’tesA and three plant TE genes were heterologously expressed in the aas mutant of Syn6803 to achieve the production and secretion of FFAs (Liu et al., 2011a). Other thioesterases from Arabidopsis thaliana (FatB) (Hu et al., 2013), Chlamydomonas reinhardtii (Fat1) (Ruffing, 2013a), Acinetobacter baylyi (’AcTesA) (Afrin et al., 2018), and Arachis hypogaea L. (AhFatA, AhFatB) (Chen et al., 2017) were also functionally expressed in cyanobacteria for FFA production (Table 1), yielding long-chain (C16–C18) FFAs in most cases.
Medium-chain fatty acids (MCFAs, C4–C12) are valuable precursors to gasoline, but are not typical products of microbial fatty acid synthesis (Torella et al., 2013). Different from the above-mentioned thioesterases, thioesterases from Cinnamomum camphorum (CcFatB1), Umbellularia californica (UcFatB1), Cuphea hookeriana (ChFatB1), and Anaerococcus tetradius (Tes3) prefer medium chain length acyl-ACP substrates. When producing MCFAs, they were expressed in cyanobacteria to control the chain lengths of the FFA products (Murata and Wada, 1995; Work et al., 2015; Yunus and Jones, 2018). It is noteworthy that these short or medium chain length specific thioesterases should be expressed in the aas mutant to avoid the reactivation and the elongation of FFAs (Table 1). In addition, the replacement of the native FabH with a Chaetoceros ketoacyl-ACP synthase III in the lauric acid-secreting strain of Syn7002 increased MCFA synthesis up to five-fold (Gu et al., 2016).
Besides the activity of acyl–acyl carrier protein synthetase, cyanobacterial Aas was also identified as a FFA importer (von Berlepsch et al., 2012) (Figure 2). And the inactivation of Aas resulted in the FFA secretion in some cyanobacterial strains, such as Syn6803, Syn7942 (Kaczmarzyk and Fulda, 2010), and Syn7002 (Ruffing, 2014), indicating that FFAs can be exported out of cyanobacterial cells by active or passive transport. It was proved that weakening cell walls by the deletion of the possible surface protein (Sll1951) and the peptidoglycan assembly protein (PBP2) as well as by ampicillin treatment led to the decrease of intracellular FFA amounts and the increase of overall FFA production in Syn6803 (Liu et al., 2011a).
Moreover, a RND-type FFA efflux system (RndA1B1) was identified by genomic analysis of a spontaneous mutant of the FFA-producing strain of Syn6803 (Kato et al., 2015) (Figure 2). Furthermore, the highest FFA yield (0.36 g/g dry cell weight) up to now has been achieved in the RndA1B over-expressing strain of Syn7942 through in situ removal of the FFA product from the culture medium by an isopropyl myristate (IM) overlay (Kato et al., 2017) (Figure 3). Recently, Sll0180 and Slr2131, homologs to AcrA and AcrB of E. coli respectively, were identified to be another FFA efflux system (Bellefleur et al., 2019) (Figure 2). Replacing the native slr2131 with the E. coli acrB gene significantly increased the extracellular FFA concentration of Syn6803 (Bellefleur et al., 2019).
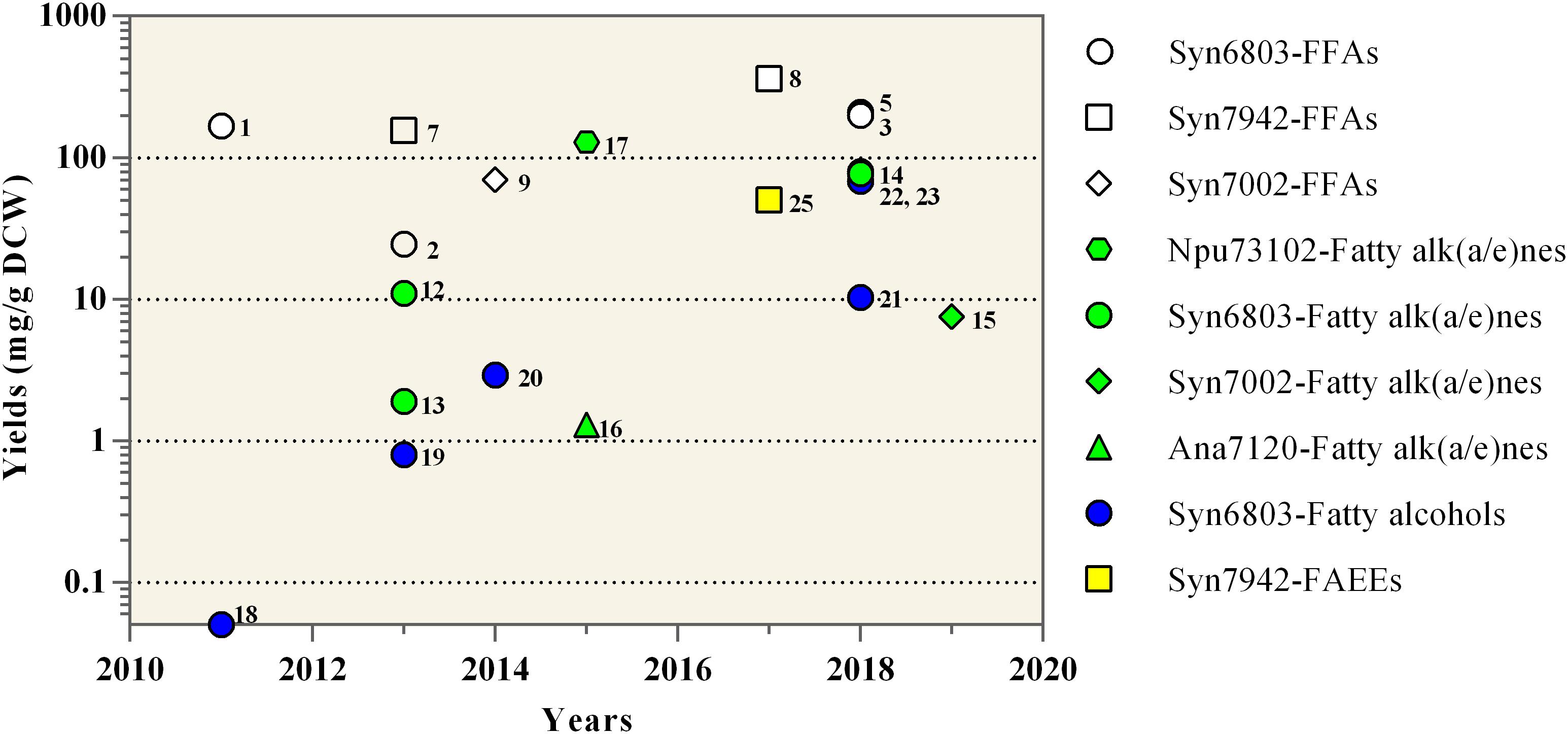
Figure 3. Yields of oleochemicals reported from engineered cyanobacteria. Maximum oleochemical yields in each study are shown at the years in which the study has been published. The detailed information for each data point including the reference is listed in Table 1 and can be retrieved using the adjacent number. The reported works on Syn6803, Syn7942, Syn7002, Npu73102, and Ana7120 are shown as circles, squares, diamonds, hexagons, and triangles, respectively. The work on the production of FFAs, Fatty alk(a/e)nes, Fatty alcohols, and FAEEs are shown as white, green, blue, and yellow colors, respectively.
In fact, little FFAs was found in the cells of cyanobacteria grown under the normal culture condition (Kaczmarzyk and Fulda, 2010). Isotope labeling experiments indicated that they are released from membrane lipids (Kaczmarzyk and Fulda, 2010). Lipases were considered to be responsible for the releasing of FFAs from membrane lipids, and heterologous expression of the foreign lipase resulted in the increase of FFAs in Syn6803 (Liu et al., 2011b). sll1969 is the only candidate lipase gene in the genome of Syn6803. The deletion of this gene decreased the FFA production, but did not completely block the FFA production (Gao et al., 2012a), suggesting it is not the only pathway for endogenous FFAs biosynthesis in Syn6803.
Besides the lipase, two cyanobacterial aldehyde dehydrogenases (AldE), namely Synpcc7942_0489 from Syn7942 (Kaiser et al., 2013) and Slr0091 from Syn6803 (Trautmann et al., 2013), were proven sufficient to oxidize fatty aldehyde precursors into fatty acids (Figure 2). It is noteworthy that these two aldehyde dehydrogenases are also able to utilize aldehyde substrates with shorter chain lengths (C8 to 12) (Kaiser et al., 2013) or apocarotenals (Trautmann et al., 2013), besides long-chain fatty aldehydes. For the purpose of FFA over-production in cyanobacteria, overexpression of acyl-ACP reductase (Aar) in the presence of AldE was proven to be a successful strategy (Kaiser et al., 2013) (Table 1).
Syn7942 encodes only one Δ9 dedaturase gene (desC) in its genome and has only saturated and monounsaturated (Δ9) fatty acid chains in its membranes. Heterologous expression of the Δ12 desaturase gene (desA) from Syn6803 led to the conversion of endogenous monounsaturated fatty acids into dienoic fatty acids (Δ9, 12) and in turn changed the fatty acid compositions of Syn7942 (Wada et al., 1990). This modification further enhanced host tolerance to chilling (Wada et al., 1990) and strong light illumination (Gombos et al., 1997). In a recent study, the heterogeneous expression of two desaturases (DesA and DesB) from Syn7002 conferred an ability of producing alpha-linolenic acid (ALA; Δ9, 12, 15) to Syn7942 (Santos-Merino et al., 2018). Further, the ALA content of the desaturases-expressing mutant was improved to levels as high as 22.6% of the total lipids, by two metabolic engineering approaches designated as the fabF overexpression and the fadD disruption (Santos-Merino et al., 2018).
Since 1960s, cyanobacteria were known to be able to naturally produce linear and branched fatty alkanes and alkenes with carbon chain lengths ranging from 15 to 19, besides membrane lipids (Han et al., 1968; Winters et al., 1969). However, the cyanobacterial biosynthesis pathways of fatty alk(a/e)nes were not identified until 2010 (Schirmer et al., 2010).
A two-enzyme pathway, consisting of acyl-ACP reductase (Aar) and aldehyde-deformylating oxygenase (Ado), was first identified by both comparative genomic and enzymatic analysis (Schirmer et al., 2010). In the Aar-Ado pathway, Aar catalyzes the conversion of acyl-ACP to fatty aldehydes, and Ado oxidizes and deformylates aldehydes to alk(a/e)nes (Figure 2), including pentadecane, heptadecane, 8-heptadecene, or 7-methylheptadecane.
A year later, the second native cyanobacterial alkene biosynthesis pathway (olefin synthase, Ols) was characterized in Syn7002 (Mendez-Perez et al., 2011). Harnessing a modular type I polyketide synthase (Ols), fatty acyl-ACP precursors are elongated and decarboxylated to synthesize terminal 1-alkenes (Figure 2), including 1-heptadecene, 1-non-adecene, or 1,14-nanadecadiene. It is worth noting that all alk(a/e)nes-producing cyanobacteria harbor only one of the two above-mentioned pathways but never both in nature (Coates et al., 2014; Klahn et al., 2014). However, it was proved that both Ols and Aar-Ado pathways can co-exist in one engineered marine cyanobacterium (Yoshino et al., 2015; Knoot and Pakrasi, 2019). And the Aar-Ado pathway, as well as two non-cyanobacterial alkane biosynthesis genes, can complement an Ols knockout strain of Syn7002 (Knoot and Pakrasi, 2019).
Despite the fact that alkane biosynthesis pathways were characterized, little is known about the physiological roles of alk(a/e)nes in cyanobacteria. Alkanes were shown to accumulate in thylakoid and cytoplasmic membranes of Syn6803 (Lea-Smith et al., 2016) as well as in lipid droplets of Npu73102 (Peramuna and Summers, 2014; Peramuna et al., 2015). Through reverse genetic approaches, it was found that cyanobacterial alkanes might play roles in regulating redox balance and reductant partitioning in photosynthesis (Berla et al., 2015), and modulating membrane flexibility, which is required for optimal cell division, size, and growth (Lea-Smith et al., 2016). In addition, alkanes were proven to be required for cyanobacterial tolerance to abiotic stresses including cold (Berla et al., 2015) and salt (Yamamori et al., 2018).
With the carbon chain lengths ranging from C15 to 19, cyanobacterial alk(a/e)nes could be directly used in diesel and jet engines, and have attracted great attention from academics. On the one hand, chemical structures and profiles of cyanobacterial alk(a/e)nes were examined across a wide range of cyanobacterial species (Liu et al., 2013; Coates et al., 2014; Zhu et al., 2018). On the other hand, several model cyanobacterial species were metabolically engineered for improving their alk(a/e)nes production (Mendez-Perez et al., 2011; Hu et al., 2013; Wang et al., 2013; Kageyama et al., 2015; Peramuna et al., 2015) (Table 1). Among these engineering approaches, the over-expression of endogenous or heterogeneous alk(a/e)ne biosynthesis genes was widely used and proven to be successful (Xie et al., 2017). In addition, the cyanobacterial alk(a/e)ne production can be further improved by increasing the copy numbers of these genes through inserting them into different genomic loci (Wang et al., 2013).
As expected, the over-expression of the multi-subunit acetyl-CoA carboxylase, which catalyzes the first step of fatty acid biosynthesis, was proven to be an effective way to enhance the cyanobacterial alkane production (Tan et al., 2011; Wang et al., 2013). The cyanobacterial alkane production can also be improved by blocking the competing pathway, like the poly-β-hydroxybutyrate (PHB) pathway (Wang et al., 2013). Different from cyanobacterial FFA production, Aas is beneficial for cyanobacterial alkane production in the Aar-Ado pathway (Gao et al., 2012b). The over-expression of Aas promoted cyanobacterial alkane production, because the acyl-ACP precursors of the Aar-Ado pathway are mainly from the Aas-mediated reactivation of FFAs, which are from the hydrolysis of membrane lipids rather than the de novo fatty acid biosynthesis pathway (Gao et al., 2012b). Similarly, the over-expression of lipolytic enzymes, which release the FFA by hydrolyzing membrane lipids, was also beneficial for alkane production (Wang et al., 2013; Peramuna et al., 2015). For example, the heptadecane production in Npu73102 was significantly improved by over-expression of Aar, Ado, and a lipase candidate Npun_F5141, together with the high light illumination, reaching 12.9% of dry cell weight (DCW) (Peramuna et al., 2015) (Figure 3).
In addition to the above engineering approaches on cyanobacterial native alkane biosynthesis pathways, several synthetic metabolic pathways were recently constructed and evaluated for alkane production in cyanobacteria (Yunus et al., 2018; Knoot and Pakrasi, 2019). In brief, three newly identified fatty acid decarboxylases were heterologously expressed in cyanobacteria successfully for converting FFA precursors to Cn–1 alk(a/e)ne end-products (Figure 2), including UndA (Rui et al., 2014) and UndB (Rui et al., 2015) from Pseudomonas fluorescens Pf-5, together with fatty acid photodecarboxylase (FAP) from Chlorella variabilis (Sorigue et al., 2017). Although catalyzing similar reactions with another two 1-alkenes producing enzymes, namely Ols from Synechococcus (Mendez-Perez et al., 2011) and OleTJE from Jeotgalicoccus (Rude et al., 2011), both UndA and UndB prefer FFA substrates with medium-chain lengths (C10–C16) rather than long-chain substrates (Rui et al., 2014, 2015). Recently, a phenylalanine 239 to alanine mutation of UndA (UndA-F239A) increased its enzymatic activities toward long chain fatty acids and improved its compatibility with cyanobacterial fatty acid compositions (Knoot and Pakrasi, 2019). FAP from eukaryotic algae mediates the light-driven conversion of fatty acid substrates to alkanes, with a wide range of substrate chain lengths (C12 to C18) and a higher substrate specificity to hexadecanoic acid (Sorigue et al., 2017). When expressing in the aas mutant of Syn6803 harboring a truncated E. coli thioesterase ’TesA, FAP can markedly improve alkane production (Yunus et al., 2018). And the total alkane yield was further increased to 77.1 mg/g DCW through the removal of the chloroplast transit peptide of FAP (’FAP) and the increase of light illumination (Yunus et al., 2018) (Table 1 and Figure 3).
Fatty alcohols can be used in the manufacture of cosmetics, detergents, lubricants, and potentially as biofuels (d’Espaux et al., 2017). Similar to several reports on microbial production of fatty alcohols (Steen et al., 2010; d’Espaux et al., 2017), cyanobacterial fatty alcohol production (Table 1) was mainly realized by heterologous expression of fatty acid reductases (Far) which utilize fatty acyl-ACP or acyl-CoA as substrates and NADH or NADPH as cofactors (Tan et al., 2011; Yao et al., 2014; Kaczmarzyk et al., 2018). All the Fars that worked well in cyanobacteria are from plants (Tan et al., 2011) and bacterium (Yao et al., 2014), whereas cyanobacteria expressing the Fars from mice failed to produce any fatty alcohols (Tan et al., 2011). The engineered strain of Syn6803 harboring the Far from jojoba (Simmondsia chinensis) produced only 0.05 mg fatty alcohols per gram DCW (Tan et al., 2011). Then, the cyanobacterial fatty alcohol yield was dramatically improved to 0.76 mg/g DCW by increasing the copy numbers of the plant Far-expressing cassettes and blocking both the PHB and glycogen biosynthesis pathways (Qi et al., 2013) (Table 1).
Compared to the plant Fars, a bacterial Far (Maqu_2220 from Marinobacter aquaeolei VT8) showed better substrate preferences to long-chain fatty acyl CoA/ACP (C16–C18) (Hofvander et al., 2011) and better performance in the engineered strain of Syn6803 for fatty alcohol production (Yao et al., 2014). For further improving fatty alcohol production of this Maqu_2220-expressing strain, the inactivation of the fatty alkane biosynthesis pathway which competes with Far for acyl-ACP precursors was shown to be effective, resulting in 2.9 mg/g DCW fatty alcohols (Yao et al., 2014). Recently, phosphate acyltransferase PlsX was identified as another key node in C18 fatty acyl-ACP consumption, and the fatty alcohol yield of Syn6803 was increased to 10.4 mg/g DCW by transcriptional inhibition of plsX using CRISPR-interference (CRISPRi) technique (Kaczmarzyk et al., 2018) (Table 1).
Besides Fars, carboxylic acid reductase (CAR) from Mycobacterium marinum, which can effectively convert FFAs into corresponding fatty aldehydes, was also used for cyanobacterial fatty alcohol production (Yunus and Jones, 2018; Yunus et al., 2018). In the presence of Ado, heterologous expression of CAR in the FFA-producing strain of Syn6803 unexpectedly led to the conversion of most of the FFA pool into corresponding fatty alcohols rather than fatty alk(a/e)nes, resulting in ∼68 mg/g DCW fatty alcohols (Yunus et al., 2018). It was speculated that the Ado failed in competition with native aldehyde reductases (or alcohol dehydrogenases, Adh) (Yunus et al., 2018). For producing medium chain-length fatty alcohols, Tes3 from A. tetradius, rather than E. coli ’TesA was co-expressed in the aas mutant of Syn6803 together with CAR and its maturation protein Sfp. Through the optimization of promoters and ribosomal binding sites and in situ product extraction with isopropyl myristate, the titers of 1-octanol and 1-decanol of the above mutant were increased to more than 100 mg/L, which is the highest titer of cyanobacterial fatty alcohols to date (Yunus and Jones, 2018) (Table 1). However, it should be noted that this 1-octanol producing strain displayed genetic instability and reduced 1-octanol production during continuous sub-culturing (Yunus and Jones, 2018).
Industrially, fatty acid esters are produced by transesterification of vegetable oils or animal fats with an alcohol in the presence of a suitable catalyst. For microbial production of fatty acid esters, the transesterification process is performed enzymatically by a multi-functional wax ester synthase/acyl-CoA:diacylglycerol acetyl transferase (WS/DGAT) (Janssen and Steinbuchel, 2014). Due to its wide substrate specificities to alcohols with various carbon lengths, the WS/DGAT (AtfA) from Acinetobacter baylyi ADP1 was normally used in cyanobacteria to mediate the combination of the activated fatty acids with ethanol (Lee et al., 2017) or fatty alcohols (Kaiser et al., 2013) (Table 1). To achieve wax ester production in Syn7942, AtfA was co-expressed with the Aar as well as a long-chain alcohol dehydrogenase from Syn6803 (Adh, Slr1192) or A. bayli (ACIAD3612) (Kaiser et al., 2013). Given the fact that FFA accumulates in the wax-producing strain as the byproduct (Kaiser et al., 2013), the endogenous aldE gene might be a candidate target for improving wax production by metabolic engineering.
For fatty acid ethyl esters (FAEEs) production in Syn7942, an ethanol biosynthesis pathway was firstly constructed by introducing both pyruvate decarboxylase and alcohol dehydrogenase from Zymomonas mobilis (Lee et al., 2017). The further expression of the AtfA in the ethanol-producing strain resulted in 40% ethanol reduction and the detection of trace concentrations of palmitic acid ethyl ester (Lee et al., 2017). A synthetic phosphoketolase pathway containing a phosphoketolase from A. nidulans (XpkA) and a phosphotransacetylase from B. subtilis (Pta) was then introduced to increase the acetyl-CoA pool and the FAEE production (Lee et al., 2017). The FAEE production was finally increased further to 50.0 mg/g DCW by culture optimization (Lee et al., 2017) (Figure 3). However, the low substrate specificities of the WS/DGAT to ethanol (Stoveken et al., 2005) could be a bottleneck for the FAEE production, considering the appearance of ethanol byproducts.
Increasing the substrate supply and blocking the competitive pathways are routine strategies for improving the production of target products. Different from heterotrophic microbes, autotrophic cyanobacteria utilize CO2 rather than sugars as carbon sources, using the Calvin-Bassham-Benson (CBB) pathway. Thus, the RuBisCO, which is the key enzyme in the CBB cycle, has been considered as an ideal target for improving cyanobacterial carbon fixation. Over-expression of the RuBisCO from Syn7942 in Syn7002 led to a more than three-fold increase in FFA production (Ruffing, 2014). However, the same strategy did not work to improve the FFA production of Syn7942 (Ruffing, 2013a).
For directing the fixed carbon flux to de novo fatty acid biosynthesis pathway, a heterologous phosphoketolase pathway, which was discovered to be efficient for increasing the supply of acetyl-CoA precursor, was introduced into the FAEE-producing strain of Syn7942, and greatly enhanced the FAEE production (Lee et al., 2017). In addition, over-expression of the ACCase has been considered to direct the carbon flux into the fatty acid pathway, and was confirmed effective for FFA over-production in E. coli (Lu et al., 2008). However, it did not always work for improving FFA production of cyanobacteria. For example, the over-expression of an ACCase from C. reinhardtii led to a 20% increase of the specific FFA production in the FFA-producing strain of Syn7942 (Ruffing, 2013a) and a 56% increase of total fatty alk(a/e)nes in Syn6803 (Tan et al., 2011) (Table 1), but there was no significant change of FFA yield in Syn6803 using a similar approach (Liu et al., 2011a).
As a major carbon sink, glycogen can account for more than 50% of the DCW in some cyanobacteria treated by the stressed conditions (Song et al., 2016), which indicates that glycogen biosynthesis competes with the fatty acid biosynthesis pathway for carbon flux. However, the complete disruption of the glycogen biosynthesis always resulted in an increase of cyanobacterial susceptibility to stress conditions (Luan et al., 2019), and the reconfiguration of electron flow in photosynthesis (Work et al., 2015). Thus, the deletion of glgC, the key gene for cyanobacterial glycogen biosynthesis, showed a slight increase of fatty alcohol production in Syn6803 (Qi et al., 2013) and no increase of lauric acid production in Syn7002 (Work et al., 2015).
As hydrophobic compounds, oleochemicals inevitably interact with cell membranes, which are sites of photosynthesis and respiration, and will result in a series of physiological effects, including reduced photosynthetic yields, chlorophyll-a degradation, changes in the cellular localization of the light-harvesting pigments (Ruffing and Jones, 2012), increased reactive oxygen species (ROS), cell membrane permeability (Ruffing, 2013b), and impaired cell growth (Kamarainen et al., 2012; Ruffing and Jones, 2012).
A dozen candidate genes were identified by comparative transcriptome analyses with potentials to mitigate FFA toxicity. The disruption of two porins and the overexpression of ROS-degrading proteins were confirmed to be effective in reducing the toxic effects of FFA production and recovering cell growth (Ruffing, 2013b). Furthermore, transporters specific to oleochemicals are promising candidates to secrete oleochemicals out of cells. The inactivation of Aas, which also functions as a FFA uptake transporter, was found to be able to alleviate FFA toxicity of cyanobacteria (von Berlepsch et al., 2012). Recently, it was found that the over-expression of a RND-type FFA exporter (RndA1B1) or the native or foreign (AcrAB) efflux systems for FFAs enhanced FFA secretion and cell growth (Kato et al., 2015; Bellefleur et al., 2019). Besides transporters, in situ removal of oleochemicals from the culture medium by some organic solutes was demonstrated to be able to significantly increase cyanobacterial cell tolerance to fatty alcohol, FFA, or FAEE (Kato et al., 2017; Lee et al., 2017; Yunus and Jones, 2018).
In the past decade, cyanobacteria were successively engineered to produce oleochemicals directly from CO2, inspired by successful strategies on oleochemical production by E. coli. Although these efforts proved the concept of engineering cyanobacteria for oleochemical production, the engineered strains are far from being used for commercial applications mainly due to their poor production ability. To improve cyanobacterial oleochemical production, more intensive efforts are needed in the future.
As shown in Figure 2, all fatty acyl chains of oleochemicals are ultimately from the de novo fatty acid biosynthesis pathway in cyanobacteria. The fundamental works on the regulatory mechanisms of cyanobacterial fatty acid metabolism are required to identify potential targets for unlocking and boosting fatty acid biosynthesis. Excretion of oleochemicals out of cells by transporters can alleviate the toxicity of FFA and was shown to be an effective strategy for improving cyanobacterial FFA production (Kato et al., 2015, 2017). However, native or foreign transporters for other oleochemicals still need to be discovered and evaluated in cyanobacteria. In addition, it should be helpful for improving cyanobacterial production of oleochemicals by introducing a heterologous reductant regenerating system to balance the reductant generation and utilization, considering the fact that reductant is needed in in oleochemical biosynthesis pathways.
Moreover, genetic instability of engineered cyanobacteria (Jones, 2014) is another potential issue for future commercial application. Despite limited observation up to now (Takahama et al., 2003; Jones, 2014), genetic instability can randomly result in some mutations on the genes associated with the production traits. And these mutations will be enriched and finally lead to the failure of cyanobacterial production of some toxic compounds like oleochemicals, if they are beneficial for fitness. For reliable production of oleochemicals in engineered cyanobacteria, key genes associated with genetic fidelity should be identified though systematic genetic analysis and then modified in cyanobacteria to construct the chassis with a higher genetic stability. On the other hand, some inducible promoters or novel genome editing tools should be used to drive the oleochemical biosynthesis only when cyanobacterial cells stop to propagate, reducing the chance of spreading the mutation through the population in long-term culturing.
XT and SY conceived the outline and revised the manuscript. LW, LC, and XT drafted the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (31871303, 21978071, and U1932141), the National Key Research and Development Program of China (2018YFC1801200), Key Laboratory of Biofuel, Chinese Academy of Sciences (CASKLB201802), and State Key Laboratory of Freshwater Ecology and Biotechnology (2018FB09).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Guodong Luan for proofreading and valuable suggestions on this manuscript.
Enzymes: Aar, acyl-ACP reductase; Aas, acyl–acyl carrier protein synthetases; ACCase, acetyl-CoA carboxylase; ACP, acyl carrier protein; Adh, alcohol dehydrogenase; Ado, aldehyde deformylating oxygenase; AldE, aldehyde dehydrogenase; AtfA, wax ester synthase/acyl-CoA:diacylglycerol acetyl transferase from Acinetobacter baylyi ADP1; CAR, carboxylic acid reductase; FabD, malonyl-CoA:ACP transacylase; FabF, b-ketoacyl -ACP synthase II; FabG, β-ketoacyl -ACP reductase; FabH, β-ketoacyl -ACP synthase III; FabI, enoyl-ACP reductase; FabZ, β-hydroxyacyl -ACP dehydrase; FAP, fatty acid photodecarboxylase (FAP); Far, fatty acid reductase; LipA, lipase; Ols, olefin synthase; Pdc, pyruvate decarboxylase; PlsC, lysophosphatidic acid acyltransferase; PlsX, phosphate acyltransferase; PlsY, acylglycerol-phosphate acyltransferase; RndA1B1, a cyanobacterial RND-type efflux system; RuBisCO, ribulose 1, 5-bisphosphate carboxylase/oxygenase; Sfp, phosphopantetheinyl transferase; TE, thioesterase; UndA, UndB, fatty acid decarboxylases from Pseudomonas sp. Compounds: FAEEs, fatty acid ethyl esters; FFAs, free fatty acids; IM, isopropyl myristate; LPA, lysophosphatidic acid; PA, phosphatidic acid; PHB, poly-β-hydroxybutyrate; PYR, pyruvate.
Afrin, S., Khan, M. R. I., Zhang, W., Wang, Y., Zhang, W., He, L., et al. (2018). Membrane-located expression of thioesterase from Acinetobacter baylyi enhances free fatty acid production with decreased toxicity in Synechocystis sp, PCC6803. Front. Microbiol. 9, 2842. doi: 10.3389/fmicb.2018.02842
Ainsworth, E. A., Lemonnier, P., and Wedow, J. M. (2020). The influence of rising tropospheric carbon dioxide and ozone on plant productivity. Plant Biol. 22(Suppl. 1), 5–11. doi: 10.1111/plb.12973
Alper, H., and Stephanopoulos, G. (2009). Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat. Rev. Microbiol. 7, 715–723. doi: 10.1038/nrmicro2186
Angermayr, S. A., Hellingwerf, K. J., Lindblad, P., and Teixeira De Mattos, M. J. (2009). Energy biotechnology with cyanobacteria. Curr. Opin. Biotechnol. 20, 257–263. doi: 10.1016/j.copbio.2009.05.011
Angermayr, S. A., Rovira, A. G., and Hellingwerf, K. J. (2015). Metabolic engineering of cyanobacteria for the synthesis of commodity products. Trends Biotechnol. 33, 352–361. doi: 10.1016/j.tibtech.2015.03.009
Atsumi, S., Hanai, T., and Liao, J. C. (2008). Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89. doi: 10.1038/nature06450
Atsumi, S., Higashide, W., and Liao, J. C. (2009). Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 27, 1177–1180. doi: 10.1038/nbt.1586
Bellefleur, M. P. A., Wanda, S. Y., and Curtiss, R. III (2019). Characterizing active transportation mechanisms for free fatty acids and antibiotics in Synechocystis sp. PCC 6803. BMC Biotechnol. 19:5. doi: 10.1186/s12896-019-0500-3
Berla, B. M., Saha, R., Maranas, C. D., and Pakrasi, H. B. (2015). Cyanobacterial alkanes modulate photosynthetic cyclic electron flow to assist growth under cold stress. Sci. Rep. 5:14894. doi: 10.1038/srep14894
Buijs, N. A., Siewers, V., and Nielsen, J. (2013). Advanced biofuel production by the yeast Saccharomyces cerevisiae. Curr. Opin. Chem. Biol. 17, 480–488. doi: 10.1016/j.cbpa.2013.03.036
Chen, G., Chen, J., He, Q. F., Zhang, Y., Peng, Z. Y., Fan, Z. X., et al. (2017). Functional expression of the Arachis hypogaea L. Acyl-ACP thioesterases AhFatA and AhFatB enhances fatty acid production in Synechocystis sp PCC6803. Energies 10:2093. doi: 10.3390/en10122093
Coates, R. C., Podell, S., Korobeynikov, A., Lapidus, A., Pevzner, P., Sherman, D. H., et al. (2014). Characterization of cyanobacterial hydrocarbon composition and distribution of biosynthetic pathways. PLoS One 9:e85140. doi: 10.1371/journal.pone.0085140
Cuellar-Bermudez, S. P., Romero-Ogawa, M. A., Vannela, R., Lai, Y. S., Rittmann, B. E., and Parra-Saldivar, R. (2015). Effects of light intensity and carbon dioxide on lipids and fatty acids produced by Synechocystis sp. PCC6803 during continuous flow. Algal Res. 12, 10–16. doi: 10.1016/j.algal.2015.07.018
Demirbas, A. (2009). Political, economic and environmental impacts of biofuels: a review. Appl. Energy 86, S108–S117.
d’Espaux, L., Ghosh, A., Runguphan, W., Wehrs, M., Xu, F., Konzock, O., et al. (2017). Engineering high-level production of fatty alcohols by Saccharomyces cerevisiae from lignocellulosic feedstocks. Metab. Eng. 42, 115–125. doi: 10.1016/j.ymben.2017.06.004
Dexter, J., and Fu, P. (2009). Metabolic engineering of cyanobacteria for ethanol production. Energ. Environ. Sci. 2, 857–864.
Gao, Q., Tan, X., and Lu, X. (2012a). [Characterization of a key gene in membrane lipid cycle in Synechocystis sp. PCC6803]. Sheng Wu Gong Cheng Xue Bao 28, 1473–1481.
Gao, Q., Wang, W., Zhao, H., and Lu, X. (2012b). Effects of fatty acid activation on photosynthetic production of fatty acid-based biofuels in Synechocystis sp. PCC6803. Biotechnol. Biofuels 5:17. doi: 10.1186/1754-6834-5-17
Gaurav, N., Sivasankari, S., Kiran, G. S., Ninawe, A., and Selvin, J. (2017). Utilization of bioresources for sustainable biofuels: a review. Renew. Sustain. Energy Rev. 73, 205–214. doi: 10.1016/j.rser.2017.01.070
Gombos, Z., Kanervo, E., Tsvetkova, N., Sakamoto, T., Aro, E. M., and Murata, N. (1997). Genetic enhancement of the ability to tolerate photoinhibition by introduction of unsaturated bonds into membrane glycerolipids. Plant Physiol. 115, 551–559. doi: 10.1104/pp.115.2.551
Gu, H., Jinkerson, R. E., Davies, F. K., Sisson, L. A., Schneider, P. E., and Posewitz, M. C. (2016). Modulation of medium-chain fatty acid synthesis in Synechococcus sp. PCC 7002 by replacing FabH with a chaetoceros ketoacyl-acp synthase. Front. Plant Sci. 7:690. doi: 10.3389/fpls.2016.00690
Hagemann, M., and Hess, W. R. (2018). Systems and synthetic biology for the biotechnological application of cyanobacteria. Curr. Opin. Biotechnol. 49, 94–99. doi: 10.1016/j.copbio.2017.07.008
Han, J., Mccarthy, E. D., Hoeven, W. V., Calvin, M., and Bradley, W. H. (1968). Organic geochemical studies, ii. A preliminary report on the distribution of aliphatic hydrocarbons in algae, in bacteria, and in a recent lake sediment. Proc. Natl. Acad. Sci. U.S.A. 59, 29–33. doi: 10.1073/pnas.59.1.29
Hauf, W., Schmid, K., Gerhardt, E. C. M., Huergo, L. F., and Forchhammer, K. (2016). Interaction of the nitrogen regulatory protein GlnB (PII) with biotin carboxyl carrier protein (BCCP) controls acetyl-coa levels in the cyanobacterium Synechocystis sp, PCC 6803. Front. Microbiol. 7:1700. doi: 10.3389/fmicb.2016.01700
Heath, R. J., and Rock, C. O. (1996a). Inhibition of beta-ketoacyl-acyl carrier protein synthase III (FabH) by acyl-acyl carrier protein in Escherichia coli. J. Biol. Chem. 271, 10996–11000. doi: 10.1074/jbc.271.18.10996
Heath, R. J., and Rock, C. O. (1996b). Regulation of fatty acid elongation and initiation by acyl-acyl carrier protein in Escherichia coli. J. Biol. Chem. 271, 1833–1836. doi: 10.1074/jbc.271.4.1833
Hofvander, P., Doan, T. T., and Hamberg, M. (2011). A prokaryotic acyl-CoA reductase performing reduction of fatty acyl-CoA to fatty alcohol. FEBS Lett. 585, 3538–3543. doi: 10.1016/j.febslet.2011.10.016
Hu, P., Borglin, S., Kamennaya, N. A., Chen, L., Park, H., Mahoney, L., et al. (2013). Metabolic phenotyping of the cyanobacterium Synechocystis 6803 engineered for production of alkanes and free fatty acids. Appl. Energy 102, 850–859. doi: 10.1016/j.apenergy.2012.08.047
Janssen, H. J., and Steinbuchel, A. (2014). Fatty acid synthesis in Escherichia coli and its applications towards the production of fatty acid based biofuels. Biotechnol. Biofuels 7:7. doi: 10.1186/1754-6834-7-7
Jones, P. R. (2014). Genetic instability in cyanobacteria - an elephant in the room? Front. Bioeng. Biotechnol. 2:12. doi: 10.3389/fbioe.2014.00012
Kaczmarzyk, D., Cengic, I., Yao, L., and Hudson, E. P. (2018). Diversion of the long-chain acyl-ACP pool in Synechocystis to fatty alcohols through CRISPRi repression of the essential phosphate acyltransferase PlsX. Metab. Eng. 45, 59–66. doi: 10.1016/j.ymben.2017.11.014
Kaczmarzyk, D., and Fulda, M. (2010). Fatty acid activation in cyanobacteria mediated by acyl-acyl carrier protein synthetase enables fatty acid recycling. Plant Physiol. 152, 1598–1610. doi: 10.1104/pp.109.148007
Kageyama, H., Waditee-Sirisattha, R., Sirisattha, S., Tanaka, Y., Mahakhant, A., and Takabe, T. (2015). Improved alkane production in nitrogen-fixing and halotolerant cyanobacteria via abiotic stresses and genetic manipulation of alkane synthetic genes. Curr. Microbiol. 71, 115–120. doi: 10.1007/s00284-015-0833-7
Kaiser, B. K., Carleton, M., Hickman, J. W., Miller, C., Lawson, D., Budde, M., et al. (2013). Fatty aldehydes in cyanobacteria are a metabolically flexible precursor for a diversity of biofuel products. PLoS One 8:e58307. doi: 10.1371/journal.pone.0058307
Kamarainen, J., Knoop, H., Stanford, N. J., Guerrero, F., Akhtar, M. K., Aro, E. M., et al. (2012). Physiological tolerance and stoichiometric potential of cyanobacteria for hydrocarbon fuel production. J. Biotechnol. 162, 67–74. doi: 10.1016/j.jbiotec.2012.07.193
Kato, A., Takatani, N., Ikeda, K., Maeda, S. I., and Omata, T. (2017). Removal of the product from the culture medium strongly enhances free fatty acid production by genetically engineered Synechococcus elongatus. Biotechnol. Biofuels 10:141. doi: 10.1186/s13068-017-0831-z
Kato, A., Takatani, N., Use, K., Uesaka, K., Ikeda, K., Chang, Y., et al. (2015). Identification of a cyanobacterial RND-type efflux system involved in export of free fatty acids. Plant Cell Physiol. 56, 2467–2477. doi: 10.1093/pcp/pcv150
Kawahara, A., Sato, Y., Saito, Y., Kaneko, Y., Takimura, Y., Hagihara, H., et al. (2016). Free fatty acid production in the cyanobacterium Synechocystis sp. PCC 6803 is enhanced by deletion of the cyAbrB2 transcriptional regulator. J. Biotechnol. 220, 1–11. doi: 10.1016/j.jbiotec.2015.12.035
Keasling, J. D. (2012). Synthetic biology and the development of tools for metabolic engineering. Metab. Eng. 14, 189–195. doi: 10.1016/j.ymben.2012.01.004
Kizawa, A., Kawahara, A., Takashima, K., Takimura, Y., Nishiyama, Y., and Hihara, Y. (2017). The LexA transcription factor regulates fatty acid biosynthetic genes in the cyanobacterium Synechocystis sp. PCC 6803. Plant J. 92, 189–198. doi: 10.1111/tpj.13644
Klahn, S., Baumgartner, D., Pfreundt, U., Voigt, K., Schon, V., Steglich, C., et al. (2014). Alkane biosynthesis genes in cyanobacteria and their transcriptional organization. Front. Bioeng. Biotechnol. 2:24. doi: 10.3389/fbioe.2014.00024
Knoot, C. J., and Pakrasi, H. B. (2019). Diverse hydrocarbon biosynthetic enzymes can substitute for olefin synthase in the cyanobacterium Synechococcus sp. PCC 7002. Sci. Rep. 9:1360. doi: 10.1038/s41598-018-38124-y
Kuo, J., and Khosla, C. (2014). The initiation ketosynthase (FabH) is the sole rate-limiting enzyme of the fatty acid synthase of Synechococcus sp. PCC 7002. Metab. Eng. 22, 53–59. doi: 10.1016/j.ymben.2013.12.008
Lai, M. C., and Lan, E. I. (2015). Advances in metabolic engineering of cyanobacteria for photosynthetic biochemical production. Metabolites 5, 636–658. doi: 10.3390/metabo5040636
Lea-Smith, D. J., Ortiz-Suarez, M. L., Lenn, T., Nürnberg, D. J., Baers, L. L., Davey, M. P., et al. (2016). Hydrocarbons are essential for optimal cell size, division, and growth of cyanobacteria. Plant Physiol. 172, 1928–1940. doi: 10.1104/pp.16.01205
Lee, H. J., Choi, J., Lee, S.-M., Um, Y., Sim, S. J., Kim, Y., et al. (2017). Photosynthetic CO2 conversion to fatty acid ethyl esters (FAEEs) using engineered cyanobacteria. J. Agric. Food Chem. 65, 1087–1092. doi: 10.1021/acs.jafc.7b00002
Lee, S. K., Chou, H., Ham, T. S., Lee, T. S., and Keasling, J. D. (2008). Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 19, 556–563. doi: 10.1016/j.copbio.2008.10.014
Li, F., Cai, B., Ye, Z., Wang, Z., Zhang, W., Zhou, P., et al. (2019). Changing patterns and determinants of transportation carbon emissions in Chinese cities. Energy 174, 562–575. doi: 10.1016/j.energy.2019.02.179
Lindberg, P., Park, S., and Melis, A. (2009). Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab. Eng. 12, 70–79. doi: 10.1016/j.ymben.2009.10.001
Liu, A., Zhu, T., Lu, X., and Song, L. (2013). Hydrocarbon profiles and phylogenetic analyses of diversified cyanobacterial species. Appl. Energy 111, 383–393. doi: 10.1016/j.apenergy.2013.05.008
Liu, X., Sheng, J., and Curtiss Iii, R. (2011a). Fatty acid production in genetically modified cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 108, 6899–6904. doi: 10.1073/pnas.1103014108
Liu, X., Fallon, S., Sheng, J., and Curtiss, R. (2011b). CO2-limitation-inducible green recovery of fatty acids from cyanobacterial biomass. Proc. Natl. Acad. Sci. U.S.A. 108, 6905–6908. doi: 10.1073/pnas.1103016108
Los, D. A., and Mironov, K. S. (2015). Modes of fatty acid desaturation in cyanobacteria: an update. Life 5, 554–567. doi: 10.3390/life5010554
Lu, X. (2010). A perspective: Photosynthetic production of fatty acid-based biofuels in genetically engineered cyanobacteria. Biotechnol. Adv. 28, 742–746. doi: 10.1016/j.biotechadv.2010.05.021
Lu, X., Vora, H., and Khosla, C. (2008). Overproduction of free fatty acids in E. coli: implications for biodiesel production. Metab. Eng. 10, 333–339. doi: 10.1016/j.ymben.2008.08.006
Luan, G., Zhang, S., Wang, M., and Lu, X. (2019). Progress and perspective on cyanobacterial glycogen metabolism engineering. Biotechnol. Adv. 37, 771–786. doi: 10.1016/j.biotechadv.2019.04.005
Marella, E. R., Holkenbrink, C., Siewers, V., and Borodina, I. (2018). Engineering microbial fatty acid metabolism for biofuels and biochemicals. Curr. Opin. Biotechnol. 50, 39–46. doi: 10.1016/j.copbio.2017.10.002
Mata, T. M., Martins, A. A., and Caetano, N. S. (2010). Microalgae for biodiesel production and other applications: a review. Renew. Sustain. Energy Rev. 14, 217–232. doi: 10.1016/j.rser.2009.07.020
Mendez-Perez, D., Begemann, M. B., and Pfleger, B. F. (2011). Modular synthase-encoding gene involved in alpha-olefin biosynthesis in Synechococcus sp Strain PCC 7002. Appl. Environ. Microbiol. 77, 4264–4267. doi: 10.1128/AEM.00467-11
Mikami, K., Kanesaki, Y., Suzuki, I., and Murata, N. (2002). The histidine kinase Hik33 perceives osmotic stress and cold stress in Synechocystis sp PCC 6803. Mol. Microbiol. 46, 905–915. doi: 10.1046/j.1365-2958.2002.03202.x
Murata, N., and Los, D. A. (2006). Histidine kinase Hik33 is an important participant in cold-signal transduction in cyanobacteria. Physiol. Plant. 126, 17–27. doi: 10.1111/j.1399-3054.2006.00608.x
Murata, N., and Wada, H. (1995). Acyl-lipid desaturases and their importance in the tolerance and acclimatization to cold of cyanobacteria. Biochem. J. 308(Pt 1), 1–8. doi: 10.1042/bj3080001
Nanjo, Y., Mizusawa, N., Wada, H., Slabas, A. R., Hayashi, H., and Nishiyama, Y. (2010). Synthesis of fatty acids de novo is required for photosynthetic acclimation of Synechocystis sp. PCC 6803 to high temperature. Biochim. Biophys. Acta 1797, 1483–1490. doi: 10.1016/j.bbabio.2010.03.014
Naoki, S., and Norio, M. (1982). Lipid biosynthesis in the blue-green alga, Anabaena variabilis: I. Lipid classes. Biochim. Biophys. Acta 710, 271–278. doi: 10.1016/0005-2760(82)90109-6
Okazaki, K., Sato, N., Tsuji, N., Tsuzuki, M., and Nishida, I. (2006). The significance of C16 fatty acids in the sn-2 positions of glycerolipids in the photosynthetic growth of Synechocystis sp. PCC6803. Plant Physiol. 141, 546–556. doi: 10.1104/pp.105.075796
Oliver, N. J., Rabinovitch-Deere, C. A., Carroll, A. L., Nozzi, N. E., Case, A. E., and Atsumi, S. (2016). Cyanobacterial metabolic engineering for biofuel and chemical production. Curr. Opin. Chem. Biol. 35, 43–50. doi: 10.1016/j.cbpa.2016.08.023
Peralta-Yahya, P. P., and Keasling, J. D. (2010). Advanced biofuel production in microbes. Biotechnol. J. 5, 147–162. doi: 10.1002/biot.200900220
Peramuna, A., Morton, R., and Summers, M. L. (2015). Enhancing alkane production in cyanobacterial lipid droplets: a model platform for industrially relevant compound production. Life 5, 1111–1126. doi: 10.3390/life5021111
Peramuna, A., and Summers, M. L. (2014). Composition and occurrence of lipid droplets in the cyanobacterium Nostoc punctiforme. Arch. Microbiol. 196, 881–890. doi: 10.1007/s00203-014-1027-6
Pfleger, B. F., Gossing, M., and Nielsen, J. (2015). Metabolic engineering strategies for microbial synthesis of oleochemicals. Metab. Eng. 29, 1–11. doi: 10.1016/j.ymben.2015.01.009
Qi, F., Yao, L., Tan, X., and Lu, X. (2013). Construction, characterization and application of molecular tools for metabolic engineering of Synechocystis sp. Biotechnol. Lett. 35, 1655–1661. doi: 10.1007/s10529-013-1252-0
Rexroth, S., Mullineaux, C. W., Ellinger, D., Sendtko, E., Rögner, M., and Koenig, F. (2011). The plasma membrane of the cyanobacterium Gloeobacter violaceus contains segregated bioenergetic domains. Plant Cell 23:2379. doi: 10.1105/tpc.111.085779
Rude, M. A., Baron, T. S., Brubaker, S., Alibhai, M., Del Cardayre, S. B., and Schirmer, A. (2011). Terminal olefin (1-Alkene) biosynthesis by a novel P450 fatty acid decarboxylase from Jeotgalicoccus species. Appl. Environ. Microbiol. 77, 1718–1727. doi: 10.1128/AEM.02580-10
Ruffing, A. M. (2013a). Borrowing genes from Chlamydomonas reinhardtii for free fatty acid production in engineered cyanobacteria. J. Appl. Phycol. 25, 1495–1507. doi: 10.1007/s10811-013-9993-7
Ruffing, A. M. (2013b). RNA-Seq analysis and targeted mutagenesis for improved free fatty acid production in an engineered cyanobacterium. Biotechnol. Biofuels 6:15. doi: 10.1186/1754-6834-6-113
Ruffing, A. M. (2014). Improved free fatty acid production in cyanobacteria with Synechococcus sp. PCC 7002 as host. Front. Bioeng. Biotechnol. 2:17. doi: 10.3389/fbioe.2014.00017
Ruffing, A. M., and Jones, H. D. (2012). Physiological effects of free fatty acid production in genetically engineered Synechococcus elongatus PCC 7942. Biotechnol. Bioeng. 109, 2190–2199. doi: 10.1002/bit.24509
Rui, Z., Harris, N. C., Zhu, X., Huang, W., and Zhang, W. (2015). Discovery of a family of desaturase-like enzymes for 1-Alkene biosynthesis. ACS Catal. 5, 7091–7094. doi: 10.1021/acscatal.5b01842
Rui, Z., Li, X., Zhu, X., Liu, J., Domigan, B., Barr, I., et al. (2014). Microbial biosynthesis of medium-chain 1-alkenes by a nonheme iron oxidase. Proc. Natl. Acad. Sci. U.S.A. 111, 18237–18242. doi: 10.1073/pnas.1419701112
Sakamoto, T., and Bryant, D. A. (2002). Synergistic effect of high-light and low temperature on cell growth of the Delta12 fatty acid desaturase mutant in Synechococcus sp. PCC 7002. Photosynth. Res. 72, 231–242.
Santos-Merino, M., Garcillán-Barcia, M. P., and De La Cruz, F. (2018). Engineering the fatty acid synthesis pathway in Synechococcus elongatus PCC 7942 improves omega-3 fatty acid production. Biotechnol. Biofuels 11:239. doi: 10.1186/s13068-018-1243-4
Sato, N., and Wada, H. (2009). “Lipid biosynthesis and its regulation in cyanobacteria,” in Lipids in Photosynthesis: Essential and Regulatory Functions, eds H. Wada, and N. Murata, (Dordrecht: Springer), 157–177. doi: 10.1007/978-90-481-2863-1_8
Savakis, P., and Hellingwerf, K. J. (2015). Engineering cyanobacteria for direct biofuel production from CO2. Curr. Opin. Biotechnol. 33, 8–14. doi: 10.1016/j.copbio.2014.09.007
Schirmer, A., Rude, M. A., Li, X., Popova, E., and Del Cardayre, S. B. (2010). Microbial biosynthesis of alkanes. Science 329, 559–562. doi: 10.1126/science.1187936
Sheehan, J., Dunahay, T., Benemann, J., and Roessler, P. (1998). A look back at the U.S. Department of Energy’s Aquatic Species Program: Biodiesel from Algae. Close-Out Report. Golden: National Renewable Energy Lab.
Solaymani, S. (2019). CO2 emissions patterns in 7 top carbon emitter economies: the case of transport sector. Energy 168, 989–1001. doi: 10.1016/j.energy.2018.11.145
Song, K., Tan, X., Liang, Y., and Lu, X. (2016). The potential of Synechococcus elongatus UTEX 2973 for sugar feedstock production. Appl. Microbiol. Biotechnol. 100, 7865–7875. doi: 10.1007/s00253-016-7510-z
Sorigue, D., Legeret, B., Cuine, S., Blangy, S., Moulin, S., Billon, E., et al. (2017). An algal photoenzyme converts fatty acids to hydrocarbons. Science 357, 903–907. doi: 10.1126/science.aan6349
Steen, E. J., Kang, Y., Bokinsky, G., Hu, Z., Schirmer, A., Mcclure, A., et al. (2010). Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463, 559–562. doi: 10.1038/nature08721
Stoveken, T., Kalscheuer, R., Malkus, U., Reichelt, R., and Steinbuchel, A. (2005). The wax ester synthase/acyl coenzyme a:diacylglycerol acyltransferase from Acinetobacter sp. Strain ADP1: characterization of a novel type of acyltransferase. J. Bacteriol. 187, 1369–1376. doi: 10.1128/jb.187.4.1369-1376.2005
Suzuki, I., Los, D. A., Kanesaki, Y., Mikami, K., and Murata, N. (2000). The pathway for perception and transduction of low-temperature signals in Synechocystis. EMBO J. 19, 1327–1334. doi: 10.1093/emboj/19.6.1327
Takahama, K., Matsuoka, M., Nagahama, K., and Ogawa, T. (2003). Construction and analysis of a recombinant cyanobacterium expressing a chromosomally inserted gene for an ethylene-forming enzyme at the psbAI locus. J. Biosci. Bioeng. 95, 302–305. doi: 10.1016/s1389-1723(03)80034-8
Tan, X., Yao, L., Gao, Q., Wang, W., Qi, F., and Lu, X. (2011). Photosynthesis driven conversion of carbon dioxide to fatty alcohols and hydrocarbons in cyanobacteria. Metab. Eng. 13, 169–176. doi: 10.1016/j.ymben.2011.01.001
Torella, J. P., Ford, T. J., Kim, S. N., Chen, A. M., Way, J. C., and Silver, P. A. (2013). Tailored fatty acid synthesis via dynamic control of fatty acid elongation. Proc. Natl. Acad. Sci. U.S.A. 110, 11290–11295. doi: 10.1073/pnas.1307129110
Towijit, U., Songruk, N., Lindblad, P., Incharoensakdi, A., and Jantaro, S. (2018). Co-overexpression of native phospholipid-biosynthetic genes plsX and plsC enhances lipid production in Synechocystis sp. PCC 6803. Sci. Rep. 8:13510. doi: 10.1038/s41598-018-31789-5
Trautmann, D., Beyer, P., and Al-Babili, S. (2013). The ORF slr0091 of Synechocystis sp. PCC6803 encodes a high-light induced aldehyde dehydrogenase converting apocarotenals and alkanals. FEBS J. 280, 3685–3696. doi: 10.1111/febs.12361
Verma, E., Chakraborty, S., Tiwari, B., and Mishra, A. K. (2018). Transcriptional regulation of acetyl CoA and lipid synthesis by PII protein in Synechococcus PCC 7942. J. Basic Microbiol. 58, 187–197. doi: 10.1002/jobm.201700467
von Berlepsch, S., Kunz, H.-H., Brodesser, S., Fink, P., Marin, K., Flügge, U.-I., et al. (2012). The acyl-acyl carrier protein synthetase from Synechocystis sp. PCC 6803 mediates fatty acid import. Plant Physiol. 159, 606–617. doi: 10.1104/pp.112.195263
Wada, H., Gombos, Z., and Murata, N. (1990). Enhancement of chilling tolerance of a cyanobacterium by genetic manipulation of fatty acid desaturation. Nature 347, 200–203. doi: 10.1038/347200a0
Wada, H., and Murata, N. (1990). Temperature-induced changes in the fatty acid composition of the cyanobacterium, Synechocystis PCC6803. Plant Physiol. 92, 1062–1069. doi: 10.1104/pp.92.4.1062
Wada, H., and Murata, N. (1998). “Membrane lipids in cyanobacteria,” in Lipids in Photosynthesis: Structure, Function and Genetics, eds S. Paul-André and M. Norio, (Dordrecht: Springer), 65–81. doi: 10.1007/0-306-48087-5_4
Wang, B., Eckert, C., Maness, P. C., and Yu, J. (2018). A genetic toolbox for modulating the expression of heterologous genes in the cyanobacterium Synechocystis sp. PCC 6803. ACS Synth. Biol. 7, 276–286. doi: 10.1021/acssynbio.7b00297
Wang, W., Liu, X., and Lu, X. (2013). Engineering cyanobacteria to improve photosynthetic production of alka(e)nes. Biotechnol. Biofuels 6:69. doi: 10.1186/1754-6834-6-69
Weier, D., Muller, C., Gaspers, C., and Frentzen, M. (2005). Characterisation of acyltransferases from Synechocystis sp. PCC6803. Biochem. Biophys. Res. Commun. 334, 1127–1134. doi: 10.1016/j.bbrc.2005.06.197
Wijffels, R. H., and Barbosa, M. J. (2010). An outlook on microalgal biofuels. Science 329, 796–799. doi: 10.1126/science.1189003
Winters, K., Parker, P. L., and Van Baalen, C. (1969). Hydrocarbons of blue-green algae: Geochemical signfficance. Science 163, 467–468. doi: 10.1126/science.163.3866.467
Work, V. H., Melnicki, M. R., Hill, E. A., Davies, F. K., Kucek, L. A., Beliaev, A. S., et al. (2015). Lauric acid production in a glycogen-less strain of Synechococcus sp. PCC 7002. Front. Bioeng. Biotechnol. 3:48. doi: 10.3389/fbioe.2015.00048
Xie, M., Wang, W., Zhang, W., Chen, L., and Lu, X. (2017). Versatility of hydrocarbon production in cyanobacteria. Appl. Microbiol. Biotechnol. 101, 905–919. doi: 10.1007/s00253-016-8064-9
Xiong, W., Cano, M., Wang, B., Douchi, D., and Yu, J. (2017). The plasticity of cyanobacterial carbon metabolism. Curr. Opin. Chem. Biol. 41, 12–19. doi: 10.1016/j.cbpa.2017.09.004
Yamamori, T., Kageyama, H., Tanaka, Y., and Takabe, T. (2018). Requirement of alkanes for salt tolerance of cyanobacteria: Characterization of alkane synthesis genes from salt-sensitive Synechococcus elongatus PCC7942 and salt-tolerant Aphanothece halophytica. Lett. Appl. Microbiol. 67, 299–305. doi: 10.1111/lam.13038
Yao, L., Qi, F., Tan, X., and Lu, X. (2014). Improved production of fatty alcohols in cyanobacteria by metabolic engineering. Biotechnol. Biofuels 7:94. doi: 10.1186/1754-6834-7-94
Yoshino, T., Liang, Y., Arai, D., Maeda, Y., Honda, T., Muto, M., et al. (2015). Alkane production by the marine cyanobacterium Synechococcus sp. NKBG15041c possessing the alpha-olefin biosynthesis pathway. Appl. Microbiol. Biotechnol. 99, 1521–1529. doi: 10.1007/s00253-014-6286-2
Yu, A. Q., Pratomo Juwono, N. K., Leong, S. S., and Chang, M. W. (2014). Production of fatty acid-derived valuable chemicals in synthetic microbes. Front. Bioeng. Biotechnol. 2:78. doi: 10.3389/fbioe.2014.00078
Yu, X., Liu, T., Zhu, F., and Khosla, C. (2011). In vitro reconstitution and steady-state analysis of the fatty acid synthase from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 108, 18643–18648. doi: 10.1073/pnas.1110852108
Yunus, I. S., and Jones, P. R. (2018). Photosynthesis-dependent biosynthesis of medium chain-length fatty acids and alcohols. Metab. Eng. 49, 59–68. doi: 10.1016/j.ymben.2018.07.015
Yunus, I. S., Wichmann, J., Wordenweber, R., Lauersen, K. J., Kruse, O., and Jones, P. R. (2018). Synthetic metabolic pathways for photobiological conversion of CO2 into hydrocarbon fuel. Metab. Eng. 49, 201–211. doi: 10.1016/j.ymben.2018.08.008
Zhou, J., and Li, Y. (2010). Engineering cyanobacteria for fuels and chemicals production. Protein Cell 1, 207–210. doi: 10.1007/s13238-010-0043-9
Keywords: cyanobacterium, oleochemicals, metabolic engineering, carbon dioxide conversion, lipid metabolism
Citation: Wang L, Chen L, Yang S and Tan X (2020) Photosynthetic Conversion of Carbon Dioxide to Oleochemicals by Cyanobacteria: Recent Advances and Future Perspectives. Front. Microbiol. 11:634. doi: 10.3389/fmicb.2020.00634
Received: 12 January 2020; Accepted: 20 March 2020;
Published: 17 April 2020.
Edited by:
Daniel Ducat, Michigan State University, United StatesReviewed by:
Anne M. Ruffing, Sandia National Laboratories (SNL), United StatesCopyright © 2020 Wang, Chen, Yang and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shihui Yang, U2hpaHVpLllhbmdAaHVidS5lZHUuY24=; Xiaoming Tan, eGlhb21pbmcudGFuQGh1YnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.