
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 09 April 2020
Sec. Fungi and Their Interactions
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00610
This article is part of the Research TopicFungal Primary and Secondary Metabolism and its Importance for Virulence and Biomedical ApplicationsView all 13 articles
 Joanna Tannous1†
Joanna Tannous1† Omer Barda2†
Omer Barda2† Dianiris Luciano-Rosario3
Dianiris Luciano-Rosario3 Dov B. Prusky2,4
Dov B. Prusky2,4 Edward Sionov2*
Edward Sionov2* Nancy P. Keller1,5,6*
Nancy P. Keller1,5,6*Penicillium expansum is one of the most harmful post-harvest pathogens of pomaceous fruits and the causal agent of blue rot disease. During infection, P. expansum produces the toxic secondary metabolites patulin and citrinin that can impact virulence and, further, render the fruit inedible. Several studies have shown that epigenetic machinery controls synthesis of secondary metabolites in fungi. In this regard, the epigenetic reader, SntB, has been reported to govern the production of multiple toxins in Aspergillus species, and impact virulence of plant pathogenic fungi. Here we show that deletion of sntB in P. expansum results in several phenotypic changes in the fungus including stunted vegetative growth, reduced conidiation, but enhanced germination rates as well as decreased virulence on Golden Delicious apples. In addition, a decrease in both patulin and citrinin biosynthesis in vitro and patulin in apples, was observed. SntB positively regulates expression of three global regulators of virulence and secondary metabolism (LaeA, CreA, and PacC) which may explain in part some of the phenotypic and virulence defects of the PeΔsntB strain. Lastly, results from this study revealed that the controlled environmental factors (low temperatures and high CO2 levels) to which P. expansum is commonly exposed during fruit storage, resulted in a significant reduction of sntB expression and consequent patulin and citrinin reduction. These data identify the epigenetic reader SntB as critical factor regulated in post-harvest pathogens under storage conditions and a potential target to control fungal colonization and decaying of stored fruit.
The ubiquitous fungus Penicillium expansum is the dominant post-harvest pathogen among fruits and vegetables, mainly pome fruits (Tannous et al., 2017a). Because of its ability to grow at low temperatures, P. expansum has also been associated with widespread spoilage of fruits during storage (Altunatmaz et al., 2012; Tannous et al., 2015). Besides the aesthetic aspect of its presence, contamination by P. expansum poses a health hazard due to the production of toxic secondary metabolites (mycotoxins) in contaminated fruit. Among the mycotoxins produced by P. expansum, the polyketide patulin is the most notable contaminant, given its long-established toxicity and prevalence during fruit infection (Puel et al., 2010; Tannous et al., 2017a). Citrinin is another polyketide mycotoxin reported to often co-occur with patulin on apples (Martins et al., 2002). This mycotoxin has been shown to be genotoxic and nephrotoxic (Flajs and Peraica, 2009). Due to the agricultural losses and serious health risks associated with the occurrence of these mycotoxins, a deeper understanding of what triggers their production is needed.
A few years ago, the complete genome sequence of P. expansum was unveiled by two research groups (Ballester et al., 2015; Li et al., 2015). Both papers provide new insights into secondary metabolism biosynthetic gene clusters, mainly those responsible for patulin and citrinin biosynthesis. Today, the biosynthetic pathways of patulin and citrinin synthesis are largely known (Artigot et al., 2009; Snini et al., 2014; Tannous et al., 2014, 2017b; He and Cox, 2016; Li et al., 2019). Recent studies were focused on the regulation of these two mycotoxins and other P. expansum secondary metabolites that could impact virulence and/or health hazards related to this pathogen. In this regard, the widely studied global regulator of secondary metabolism in fungi, LaeA, a member of the transcriptional Velvet Complex (Bayram et al., 2008), was shown to positively regulate patulin production (Kumar et al., 2016). Deletion of laeA in two P. expansum strains led to a significant decrease in patulin biosynthesis due to down-regulation of the expression of the patulin biosynthesis genes (Kumar et al., 2016). In contrast, citrinin biosynthesis was not subjected to laeA regulation in P. expansum (Kumar et al., 2016). Both patulin and citrinin synthesis were reduced as a result of deletion of veA, a second component of the Velvet Complex (El Hajj Assaf et al., 2018). Patulin was found to also be positively regulated by PacC, the pH regulatory transcription factor (Barad et al., 2014; Chen et al., 2018). Lastly, CreA, the carbon catabolite repressor has been reported to act as a positive regulator of both patulin and citrinin production (Tannous et al., 2018). In addition to their impact on secondary metabolism and some physiological traits, the loss of all these regulatory proteins resulted in attenuated virulence of P. expansum on apples (Kumar et al., 2016; El Hajj Assaf et al., 2018; Tannous et al., 2018).
In recent years, epigenetic and consequent histone post-translational modifications have been shown to play an important role in secondary metabolite production in fungi (Pfannenstiel and Keller, 2019), including regulation of several Aspergillus and Fusarium mycotoxins such as aflatoxin, fumonisin and trichothecenes (Visentin et al., 2012; Liu et al., 2015; Yang et al., 2016; Pfannenstiel et al., 2018). Most studies investigated the role of histone modifying enzymes, such as methylases or histone deacetylases in modulating secondary metabolite synthesis in fungi. However, more recently an additional histone modifying protein, the epigenetic reader SntB, was identified in Aspergillus species to regulate not only fungal development and secondary metabolism (Pfannenstiel et al., 2017) but also virulence in the aflatoxigenic fungus A. flavus (Pfannenstiel et al., 2018). SntB orthologs, called Snt2, had previously been described in the plant pathogens Fusarium oxysporum (Denisov et al., 2011) and Magnaporthe oryzae (He et al., 2018). These studies did not address the impact on secondary metabolism but found that the loss of snt2 resulted in reduced virulence of both pathogens on their respective hosts. Additionally, both studies reported defects in autophagy-dependent cell death pathways in the snt2 mutants.
To our knowledge, epigenetic control of mycotoxin production and virulence in P. expansum has not been addressed. Our work presents evidence that SntB regulates P. expansum development, patulin and citrinin production, and virulence on apples. SntB is a positive regulator of laeA, creA, and pacC expression, which may explain some of its impact on fungal biology. Moreover, this is the first report of epigenetic response(s) of a post-harvest pathogen to environmental stressors. Data from our study revealed a deregulation of sntB under low temperatures and high CO2 levels, two conditions widely applied during fruit storage to limit P. expansum growth. These results allowed us to propose epigenetic responses as a potential modulator of virulence of storage pathogens.
Penicillium expansum strain Pe-21 (also called Pe-d1) from Israel, was used to generate the Δku70 knock-out strain that was used as the parental control strain for all subsequent experiments. Derivative strains from P. expansum Pe-21 are listed in Supplementary Table S1. Strains were cultivated in glucose minimal medium (GMM) at 25°C for 5 days to collect fresh spores. Conidia were collected and adjusted using a hemocytometer to the indicated concentrations. Screening of fungal transformants was done on sorbitol minimal medium (SMM) agar supplemented with the appropriate antibiotic (hygromycin or phleomycin at the respective concentrations of 100 and 50 μg/ml). For physiological experiments, GMM agar and broth were used. To screen for secondary metabolite production in vitro, strains were cultivated on four different agar media chosen to represent a broad range of nutrient source, which are: Potato Dextrose Agar (PDA), Yeast Extract Sucrose Agar (YES), Czapek Yeast Extract Agar (CYA) and GMM, at 25°C for 10 days. Each medium was prepared as shown in Supplementary Table S2.
Competent cells of the yeast strain BJ5464 (MATalpha, ura3-52, trp1, leu2-Δ1, his3-Δ200, pep4:HIS3, prb1-Δ1.6R, can1, GAL) were used to facilitate appropriate plasmid construct assembly. The plasmids used in this study are listed in Supplementary Table S3.
In our recent work, a Δku70 strain of P. expansum Pe-21 was generated using hygromycin as a selectable marker (Tannous et al., 2018). Here a new P. expansum Δku70 was created to allow recycling of the hygromycin resistance gene that can be re-used as a selectable marker in future transformations. The new P. expansum Δku70, hygromycin sensitive (HygS) strain, termed TJT14.1, was constructed by employing the β-Rec/six site-specific recombination system described by Hartmann et al. (2010). The hygromycin recyclable marker used to construct the Δku70 strain is based on site-specific recombination that allows repetitive gene targeting. The elements of the self-excising β-rec/six blaster cassette are shown in Supplementary Figure S1A. The Δku70 knockout cassette was constructed using yeast recombination cloning (Wiemann et al., 2018). The three fragments (5′flank Ku70- hygromycin resistance cassette and 3′flank Ku70) were assembled into the pYHC-WA-pyrG plasmid. The plasmid was linearized by PCR using primers YS_F and YS_R (Supplementary Table S4A). After yeast recombinational cloning, the plasmid pJT1 was created and recovered from yeast DNA by transforming into E. coli. The nested primers Pe_KOKu70_NestedF and Pe_KOKu70_NestedR were used to amplify the entire deletion cassette from plasmid pJT1 using the Long Template Expand PCR System (Roche, Indianapolis, IN, United States). PCR reactions were performed according to the manufacturer’s instructions, and the construct was eluted with a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). This construct was used to transform protoplasts of the parental strain Pe-21 (Barad et al., 2016b).
The predicted sequence for the P. expansum sntB ortholog was obtained from GenBank (XM_016748327) by conducting a BLAST search with the A. flavus sntB gene sequence (AFLA_029990) against the P. expansum genome scaffolds. The generated HygS Δku70 strain, TJT14.1, was used to delete the sntB gene using the phleomycin resistance gene ble as a selectable marker (Silar, 1995). The resultant deletion strain was called TJT15.1. The schematic representation of the sntB gene replacement with the ble gene is depicted in Supplementary Figure S2A. The three amplified fragments (5′flank sntB, ble gene, and 3′flank sntB) were assembled using a double joint PCR protocol described by Lim et al. (2012). To confirm that the phenotype exhibited by the ΔsntB strain is caused by the deletion of this specific gene, TJT15.1 was complemented with a wild-type copy of the sntB gene using hygromycin as selectable marker to create TJT17.1. NotI and BsrgI restriction sites introduced respectively at the predicted promoter and terminator of the sntB gene, using primers Pe_sntBcomp_F and Pe_sntBcomp_R, were used to subclone sntB downstream of the hph gene into the pJT1 plasmid (Supplementary Figure S3A). The ligation of the digested insert into the recipient plasmid was performed using the T4 DNA ligase from New England Biolabs, according to the manufacturer’s instructions. The ligation reaction was later transformed into E. coli DH5alpha competent cells following the manufacturer’s directions (Thermo Fisher Scientific). Five bacterial colonies were picked and screened for successful ligations by conducting a diagnostic restriction digest with the same enzymes used for the cloning. The correct plasmids were then grown in 50 ml LB supplemented with ampicillin (100 μg/ml), and the plasmid DNA was isolated using Quantum Prep®Plasmid Midiprep Kit (Biorad) according to the manufacturers’ instructions. Ten micrograms of plasmid DNA was used for the transformation of the ΔsntB strain. The plasmid was linearized using swaI prior to fungal transformation.
To generate the different deletion and complement strains, the fungal transformation was performed following the polyethylene glycol-mediated protoplast transformation protocol described previously by Tannous et al. (2018). All transformants were screened by PCR and further subjected to southern blot to confirm the single insertion of the deletion cassette using [α32P] dCTP (PerkinElmer, United States) to label the DNA probes, following manufacturer’s instructions. In order to recycle the hph resistance cassette and gain back the sensitivity to hygromycin B in the Δku70 strain generated in this study, deletion strains were grown on minimal medium amended with 2% xylose to activate the ß-recombinase placed under the xylose-inducible promoter. The confirmation of the hygromycin sensitivity was done on GMM hygromycin plates and by PCR targeting the hph gene. Primers used for constructing the deletion cassettes, molecular cloning and mutants’ confirmation are listed in Supplementary Table S4A.
Both vegetative growth and conidial production were assessed on GMM agar. To measure the radial mycelial growth, agar plates were point-inoculated with 106 spores of each strain and incubated at 25°C. Radial growth was monitored by diameter measurements in the four cardinal directions. Measurements were recorded on alternate days for a 6 day period.
To quantify total conidia produced by the various strains, GMM top agar (0.7% (w/v) agar) containing 106 spores was overlaid on agar plates of the same media (20 ml) and incubated at 25°C in the dark. To accurately count conidia, 2 cm plugs from each plate were homogenized in 3 ml of 0.01% Tween 20 water, diluted and enumerated with a hemocytometer. Conidial production was quantified starting by the second day post-inoculation using three replicate plates per strain.
Germination assays were performed in sterile 12-well culture plates. The spore concentration of all strains was adjusted to 105 spores/ml in GMM broth, and one milliliter of each spore suspension was distributed into 3 replicate wells. Time-course microscopy was carried out over 24 h at 25°C using a Nikon Ti inverted microscope. Germination rate was monitored regularly by capturing images of each well hourly, beginning 4 h post-incubation. A number of spore germlings was counted for each strain and recorded. The percent of germinated spores was plotted against time, and the germination rates were determined.
Liquid cultures of P. expansum strains were grown in GMM supplemented with Yeast Extract (YE). The 25 ml cultures were inoculated using 108 spores and incubated for 14 h at 25°C under 250 rpm. After incubation, mycelia were harvested in 50 ml tubes and washed twice with Dulbecco’s Phosphate-Buffered Saline (DPBS) lacking calcium and magnesium (Life Technologies, Carlsbad, CA, United States). For the negative control, mycelia were incubated at 100°C for 45 min before proceeding with the protocol. Mycelia were later incubated at 37°C for 10 min in a 0.1% Evans Blue (Sigma Aldrich, St Louis, MO, United States) solution. After incubation, three wash steps were performed using DPBS again. Samples were mounted using Ibidi mounting Medium (Ibidi, Fitchburg, WI, United States).
Fluorescence microscopy was performed using Axio Imager A10 Microscope EC Plan-NEOFLUAR 40x/1.3 Oil DIC/∞/0.17 objective and a series 120 X-Cite R light source (EXFO) (Olympus Corporation, Shinjuku, TYO, Japan). Micrographs were acquired using the DIC (Differential Interphase Contrast) and TRITC (Tetramethylrhodamine) channels with uniform exposure settings for all samples: 6 ms for the DIC channel and 40 ms for the TRITC channel. The mean gray value area per hyphae of 10 micrographs per strain was calculated using Fiji (Schindelin et al., 2012).
Cultures intended for RNA isolation were grown on sterile 0.45 μm nitrocellulose membrane circles (Whatman, Kent, United Kingdom) placed on top of solid YES medium (20 g bacto yeast extract, 150 g sucrose, 15 g bacto agar per liter). A 106 fungal spores/ml solution (100 μl) was inoculated onto 55 mm petri dishes containing 10 ml of media. To test the impact of abiotic stressors, the plates were incubated at 28°C for 48 h in the dark and then transferred to the appropriate stress conditions for additional 48 h. For comparison between the WT, ΔsntB and complement strains, the plates were grown at 28°C for 4 days. After incubation, total RNAs were extracted from 100 mg of lyophilized mycelia using the Hybrid-R RNA isolation kit (GeneAll, Seoul, South Korea) according to the manufacturer protocol. The DNase and reverse-transcription reactions were performed on 1 μg of total RNA with the Maxima First-Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, United States) according to the manufacturer protocol. The cDNA samples were diluted 1:20 (v/v) with ultrapure water. Real time quantitative PCR was performed with the StepOnePlus system (Applied Biosystems, Waltham, MA, United States). PCR amplifications were performed with 3 μl of cDNA template in 10 μl of a reaction mixture containing 7 μl mix from the Fast SYBR green Master Mix (Applied Biosystems, Waltham, MA, United States) and 200 nM primers. PCRs were carried out with the following cycling program: 20 s at 95°C, followed by 40 cycles of 95°C for 3 s and 60°C for 20 s. The samples were normalized using β-tubulin gene (PEXP_025370) as endogenous control and the relative expression levels were measured using the 2(–ΔΔCt) analysis method (Livak and Schmittgen, 2001). Primers used for qRT-PCR analyses are listed in Supplementary Table S4B. Results were analyzed with StepOne software v2.3.
Agar plugs with fungal mycelia from gene expression experiments were used to evaluate mycotoxin production. For patulin analysis, 1 g agar was added to 2 ml of HPLC grade ethyl acetate (Bio-Lab, Jerusalem, Israel) and crushed to homogeneity. Patulin was extracted by shaking for 30 min at 150 RPM on an orbital shaking platform. The supernatant was transferred to a clean glass tube and was evaporated to dryness under a stream of gaseous nitrogen at 50°C. The residues were re-dissolved in 1 ml of the mobile phase (0.02 M ammonium acetate:acetonitrile 9:1), filtered through a 0.22 μm PTFE syringe filter (Agela Technologies, Tianjin, China) and kept at −20°C prior to HPLC analysis. For citrinin analysis, 1 g agar plug with fungal mycelia was added to 1 ml of methanol and crushed. The mycotoxin was extracted by shaking for 30 min at 150 RPM and centrifuged for 10 min at 6000 × g. The supernatant was filtered through a 0.22 μm PTFE syringe filter and kept at −20°C prior to HPLC analysis. The citrinin mobile phase consisted of acidified water (adjusted to pH 2.5 with acetic acid) and acetonitrile (50:50). Both mycotoxins were quantitatively analyzed by injection of 20 μl into a reverse phase UHPLC system (Waters ACQUITY Arc, FTN-R, Milford, MA, United States) using a Kinetex 3.5 μm XB-C18 (150 × 4.6 mm) column (Phenomenex, Torrance, CA, United States). The column temperature was maintained at 30°C and the flow rate was 1 ml/min. The patulin peak was detected with a photodiode array (PDA) detector at 280 nm; citrinin was detected with a fluorescence detector (331 nm excitation, 500 nm emission). Both mycotoxins were quantified by comparing with a calibration curve of the standard mycotoxins (Fermentek, Jerusalem, Israel).
SntB deletion and complement mutants confirmed by Southern blot analysis were tested on apples cv. Golden Delicious, obtained from a local orchard, to evaluate the role of sntB in the virulence process of P. expansum. As described previously by Tannous et al. (2018), apples were surface sterilized with 2% sodium hypochlorite solution and thoroughly rinsed with sterile distilled water. A single uniform 5 mm deep wound was made at the equator of each apple (put on its side) using sterile toothpicks. For each strain, 10 μl of a spore suspension at a concentration of 108 spores/ml were deposited into the wound. Apples were incubated at 25°C in the dark for 11 days. The diameter of the rotten spots were recorded every other day. Three technical replicates were performed for each strain, and the entire experiment was repeated twice for confirmation of the observed results. At the end of the incubation period, patulin was extracted from apples and quantified by HPLC as described earlier by Tannous et al. (2018). Moreover, to eliminate the effect of variability between apples and to compare the fruit colonization, the three strains (control, ΔsntB, and sntBC) were inoculated twice on the same apple. Six technical replicates were performed and the whole experiment was repeated three times for confirmation of the observed results. Representative images were taken 5 days post-inoculation.
For all experiments, values are stated as the mean ± standard error of the mean (SEM) of three independent replicates unless otherwise indicated. The statistical analysis of the data was performed using the one-way ANOVA. If one-way ANOVA reports a p value of < 0.05 further analyses were performed using Tukey’s single-step multiple comparison test to determine significant difference between the strains. Analyses were done using R Studio (R Studio Team, 2015, Boston, MA, United States) and GraphPad Prism software (GraphPad, San Diego, CA, United States).
The deduced amino acid sequence of SntB in P. expansum (XP_016595169) showed a high degree of identity with orthologous sequences from species of Aspergillus (ranging from 60 to 64%), whereas a lower percentage of identity was scored with the orthologous sequences from M. oryzae (around 46%) and F. oxysporum (around 42%). The sequence identity with the Snt2 of Saccharomyces cerevisiae was even lower (around 32%), and therefore it was excluded from the multisequence alignment. Alignment of deduced amino acid sequences of SntB revealed the presence of five highly conserved domains (boxes A–E) that are considered to be essential for the activity of this protein (Supplementary Figure S4). The first conserved domain (box A) consists of the Bromo adjacent homology (BAH) domain that may be involved in protein-protein interaction specialized in gene silencing (Callebaut et al., 1999). Box B carries the first plant homeodomain (PHD) finger which is a C4HC3 zinc-finger-like motif thought to be involved in epigenetics and chromatin-mediated transcriptional regulation (Aasland et al., 1995; Fair et al., 2001). The third domain (box C) contains the SANT (SWI3, ADA2, N-CoR, and TFIIIB) DNA-binding domains, belonging to a various set of proteins that share a common 3 alpha-helix bundle. Recent studies suggested that SANT domains might be a histone-tail-binding module (Boyer et al., 2004). In boxes D and E, are found two other plant homeodomain (PHD) fingers.
To identify the HygS Δku70:hph transformants, fifty colonies obtained using the pJT1 vector were found resistant to hygromycin. Monosporic transformants were further checked by direct specific PCR analysis targeting the ku70 gene using the primer set Pe_Ku70orf_F and Pe_Ku70orf_R. Amplification of the ORF was not observed in twenty three transformants, which were then analyzed by southern blotting. Only five out of twenty three transformants showed a single insertion of the cassette at the ku70 locus. To recycle the hph resistance cassette and gain back the sensitivity to hygromycin B, these five transformants were grown on minimal medium amended with 2% xylose. Four of these transformants were not able to grow on plates supplemented with hygromycin B after this treatment. The absence of hph gene was further confirmed by PCR using the primer set hph_F and hph_R (data not shown). A single Δku70, HygS strain labeled TJT14.1 was used for the following experiments as a control strain. Southern blot confirmation for the selected Δku70 is shown in Supplementary Figure S1B.
To determine the effects of sntB on growth, conidiation, and other cellular processes, sntB deletion (PeΔsntB) and complementation strains were generated. For sntB deletion, ten single transformants were initially selected on SMM agar supplemented with phleomycin (50 μg/ml). The phleomycin resistant strains were later confirmed by PCR for the absence of the ORF amplification (data not shown) and subjected to a final confirmation by southern blot analysis using probes specific for flanking regions of the sntB gene (Supplementary Figure S2B). Two DNA fragments of 1216 and 1189 kb corresponding respectively to the 5′ and 3′flanking regions of sntB were used to generate the radioactive probes utilized in this hybridization. Genomic DNA from both the control and PeΔsntB strains were digested separately with EcoRI and BanII. The presence of 5.4 and 2.5 kb bands in the Southern blot analysis confirmed the sntB deletion, while 4.2 and 1.6 kb bands corresponded to the control (Supplementary Figure S2B). One of the correct PeΔsntB deletion strains was used for the following experiments and was called TJT15.1.
With respect to the sntB complement strain, diagnostic PCR was performed to confirm the integration of the wild type allele in the ΔsntB strain using the same set of primers that amplify the sntB ORF (data not shown). The positive strains were later subjected to southern blot analysis using the sntB gene as a probe (Supplementary Figure S3B). Genomic DNA from both sntB deletion and complement strains were digested with NotI and BsrgI. As expected, the ΔsntB strain did not show any band, whereas the sntBc strain revealed the expected band of 6.3 kb (Supplementary Figure S3B). One of the correct sntBc strain was used for the following experiments and was labeled TJT17.1.
The impact of sntB deletion on P. expansum physiology was evaluated by virtue of four different determinants: pigmentation, radial growth, spore production and germination rate. Visual observation of growth plates showed distinct pigmentation patterns correlating with sntB loss on all media examined after 72 h of growth at 25°C (Figure 1A). Radial growth data collected on GMM agar after 6 days of culture at 25°C revealed a significant reduction of growth diameter when sntB is deleted in P. expansum (Figure 1B). While the ΔsntB strain (TJT15.1) reached an approximate growth diameter of 25 mm, the control strain (TJT14.1) grew to an area of 30 mm, 6 days post-inoculation. However, the sntB complement strain (TJT17.1) was 1 mm greater in growth diameter compared to the control strain at the end of the incubation period (Figure 1B). A similar pattern was observed on the three other tested media (Supplementary Figures S5A–C). Sporulation data gathered on the second and third days post-inoculation on GMM agar showed decreased spore production by the ΔsntB mutant compared to both the control and the sntB complement strains (Figure 1C). It remains unclear if the reduced spore count in the ΔsntB strain is due to growth impairment. However, the PeΔsntB mutant spores showed a faster germination rate compared to the control strains, starting 4 h post-inoculation. All strains reached equivalent germination rates 11 h post-inoculation (Figure 1D).
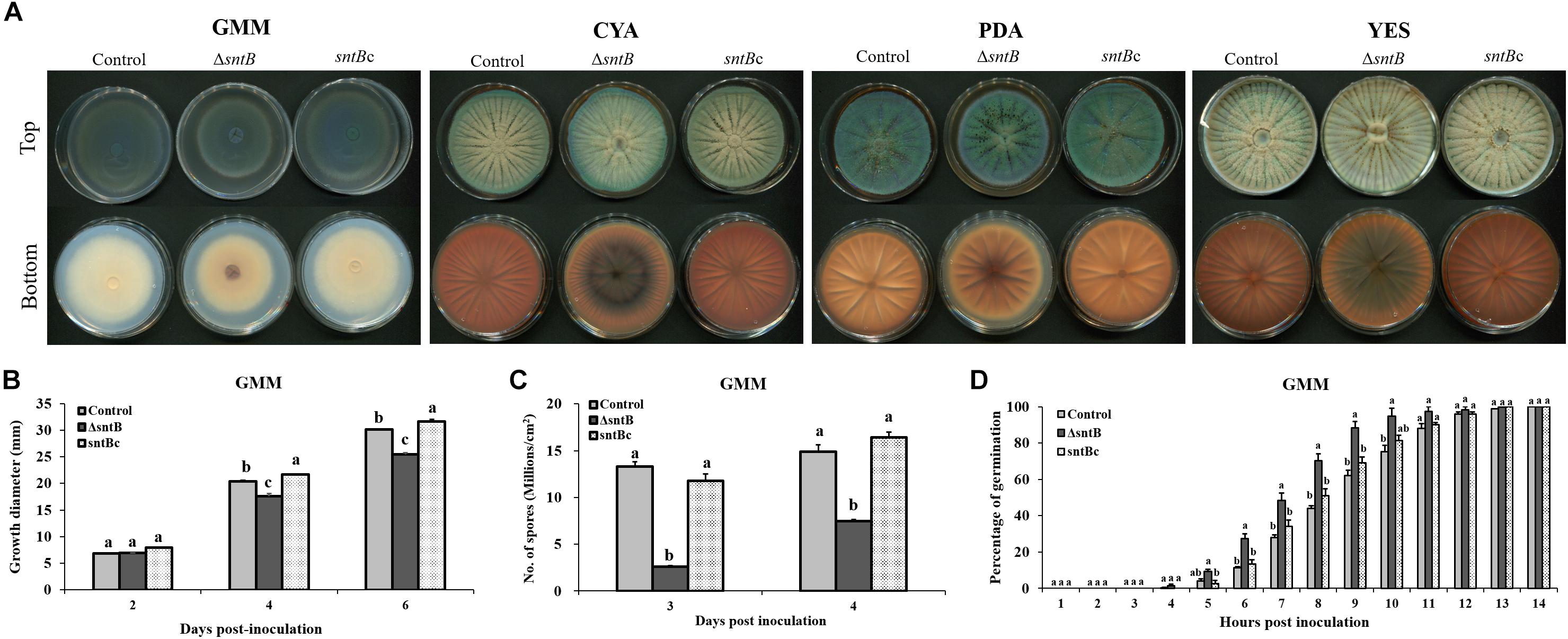
Figure 1. Physiological analysis of the control, sntB deletion and complement strains of P. expansum Pe-21. (A) Growth phenotype of P. expansum strains monitored on four different media (GMM, Glucose Minimal Medium; CYA, Czapek Yeast Extract Agar; PDA, Potato Dextrose Agar; YES, Yeast Extract Sucrose Agar) for 72 h at 25°C. (B) Radial growth, (C) conidia production were monitored on GMM agar, and (D) germination rate of P. expansum strains was measured on GMM broth. One-way ANOVA differences were considered significant when p < 0.05. Different letters above the columns indicate statistically significant differences between the strains as determined using Tukey’s single-step multiple comparison test.
Other studies have reported SntB homologs to be involved in autophagy mediated programed cell death (Denisov et al., 2011; He et al., 2018). To investigate if P. expansum SntB plays a role in cell death pathways, we first assessed in vitro cell viability of WT, ΔsntB, and sntB complement strains using Evans Blue Dye (Supplementary Figure S6A). Dead cells are permeable to this dye. Moreover, this staining fluoresces upon excitation with green light providing the opportunity to have quantitative data. After quantification and comparison of fluorescence using the average mean gray area for 10 micrographs per strain, we found no significant difference in cell viability between the assessed samples (Supplementary Figure S6B).
Loss of sntB resulted in significant reduction of patulin and citrinin levels in the mutant strain compared to the control strain when grown in YES media (Figures 2A,B). To determine if the changes in patulin and citrinin levels were correlated with transcript levels, the expression levels of selected biosynthetic genes in both patulin and citrinin gene clusters were analyzed and compared between the control, sntB deletion and complement strains. Comparison of gene expression between the control and ΔsntB mutant revealed that deletion of sntB resulted in a reduced expression level of the specific transcription factor of the patulin gene cluster, patL, and the polyketide synthase (PKS) gene, patK (Figure 2C). Similarly, sntB deletion resulted in a significant reduction of the expression level of the citrinin gene cluster transcription factor, ctnA, and citC, which encodes an oxidoreductase in citrinin biosynthetic pathway (Figure 2D). Production of both mycotoxins was recovered by the complement strain, although not to the level of the control strain, which was in agreement with gene expression analysis (Figures 2A–D). These results were not totally unexpected considering that previous publications have reported similar outcomes, where the complement strains partially restored the parental phenotype (Carvalho et al., 2010; Kwon et al., 2011; Pfannenstiel et al., 2017).

Figure 2. SntB-associated regulation of patulin and citrinin biosynthesis. The three Penicillium strains (control, ΔsntB, and sntBc) were grown on solidified YES media at 28°C. Patulin (A) and citrinin (B) levels, relative expressions of patulin (C) and citrinin cluster genes (D), and global transcription factors (E) were evaluated at 4 days post-inoculation. Error bars represent the standard error of the mean (SEM) across three technical replicates. One-way ANOVA differences were considered significant when p < 0.05. Different letters above the columns indicate statistically significant differences between the strains as determined using Tukey’s single-step multiple comparison test.
Moreover, we also asked if loss of sntB influenced expression of regulatory genes known to regulate both metabolites. Expression of creA encoding the transcription factor regulating carbon catabolite repression (Tannous et al., 2018), pacC encoding the pH regulatory transcription factor (Chen et al., 2018) and laeA encoding a methyltransferase involved in global regulation of fungal secondary metabolism (Kumar et al., 2016), were markedly downregulated in the knockout strain (>2-fold, p < 0.001) (Figure 2E). Overall, the complement strain expressed similar levels of the global regulatory genes compared to the control strain (Figure 2E).
Next, we analyzed the ability of the PeΔsntB mutant to infect apple cv. Golden Delicious. Apple fruits were inoculated with a conidial suspension of ΔsntB, sntBc, and the P. expansum control strains. Lesion development was monitored every 2 days, and the diameter was measured. As shown in Figures 3A,B, sntB disruption resulted in a significant reduction of decay development as the lesion diameter caused by ΔsntB was about 37% smaller than that of the control and sntB complement strains on day 11 post-inoculation. Similar to the results described in vitro, the production of patulin by the ΔsntB mutant was significantly reduced compared to the control and the sntB complement strain on apples (Figure 3C). As described in our previous work (Tannous et al., 2018), citrinin was not detectable on apples.
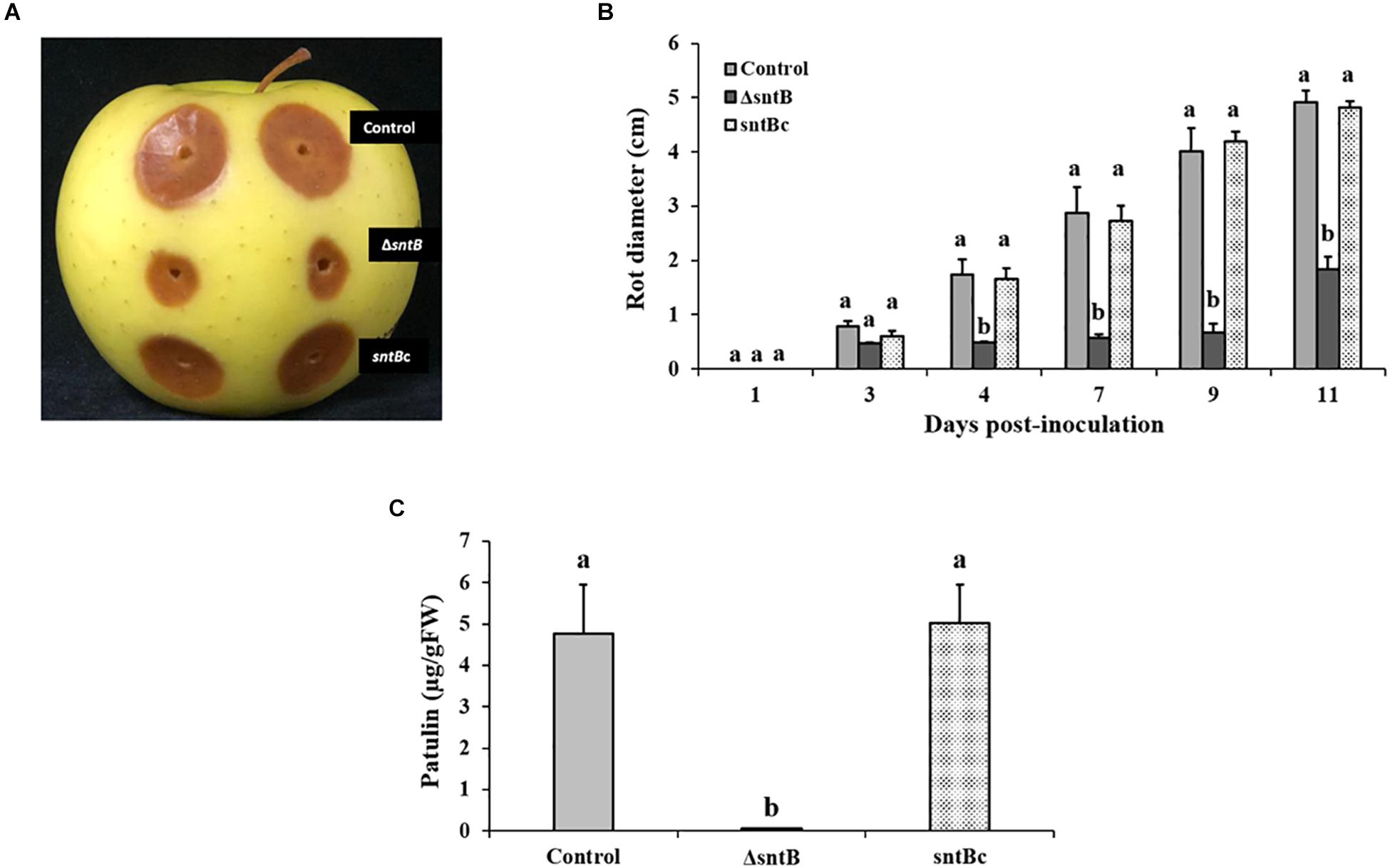
Figure 3. SntB is required for P. expansum pathogenicity and patulin production on apples. (A) Apples were inoculated with 106 spores and incubated at 25°C in the dark for 5 days. (B) Histogram showing the rot diameter on apples inoculated with the three Penicillium strains (control, ΔsntB, sntBc). (C) Histogram displaying patulin production on apples analyzed by HPLC. Each strain was inoculated with 3 reps. One-way ANOVA differences were considered significant when p < 0.05. Different letters above the columns indicate statistically significant differences between the strains as determined using Tukey’s single-step multiple comparison test.
To gain an understanding on how SntB itself may be regulated and possibly affect subsequent patulin and citrinin synthesis in conditions associated with fruit storage, we assessed the impact of temperature, light and CO2 levels on gene expression and mycotoxin synthesis. qRT-PCR analyses showed a significant downregulation of sntB when P. expansum was exposed to high CO2 levels and low temperatures (18 and 5°C) (Figure 4A). Metabolite analysis showed that patulin and citrinin levels were reduced after incubation at 5°C and high CO2, while their synthesis was significantly upregulated under continuous white light (Figures 4B,C). Interestingly, incubation of the control strain at 18°C resulted in a dramatic increase in patulin accumulation of more than 130 ppm compared to 1.2 ppm at 28°C (control condition). In contrast, citrinin production by the control strain was markedly inhibited at 18°C to a concentration of 0.014 ppm compared to 6.8 ppm under control condition (Figures 4B,C). The sntB deletion strain produced significantly lower amounts of both mycotoxins compared to the control strain, while their accumulation was recovered by the complement strain, under all tested conditions (Figures 4B,C). A similar pattern was observed for the relative expression of the two key genes in the patulin biosynthetic pathway, patK and patN (Figure 5A). However, the transcript level of the patulin specific transcription factor (patL) was significantly decreased at 18°C and under white light, in contrary to the aforementioned increase of the mycotoxin level under the same conditions. The expression levels of the citrinin transcription factor (ctnA) and another two keys genes involved in citrinin biosynthesis (citS and citC) correlated with the mycotoxin production under all tested conditions except white light, under which a downregulation of the tested genes was observed (Figure 5B).
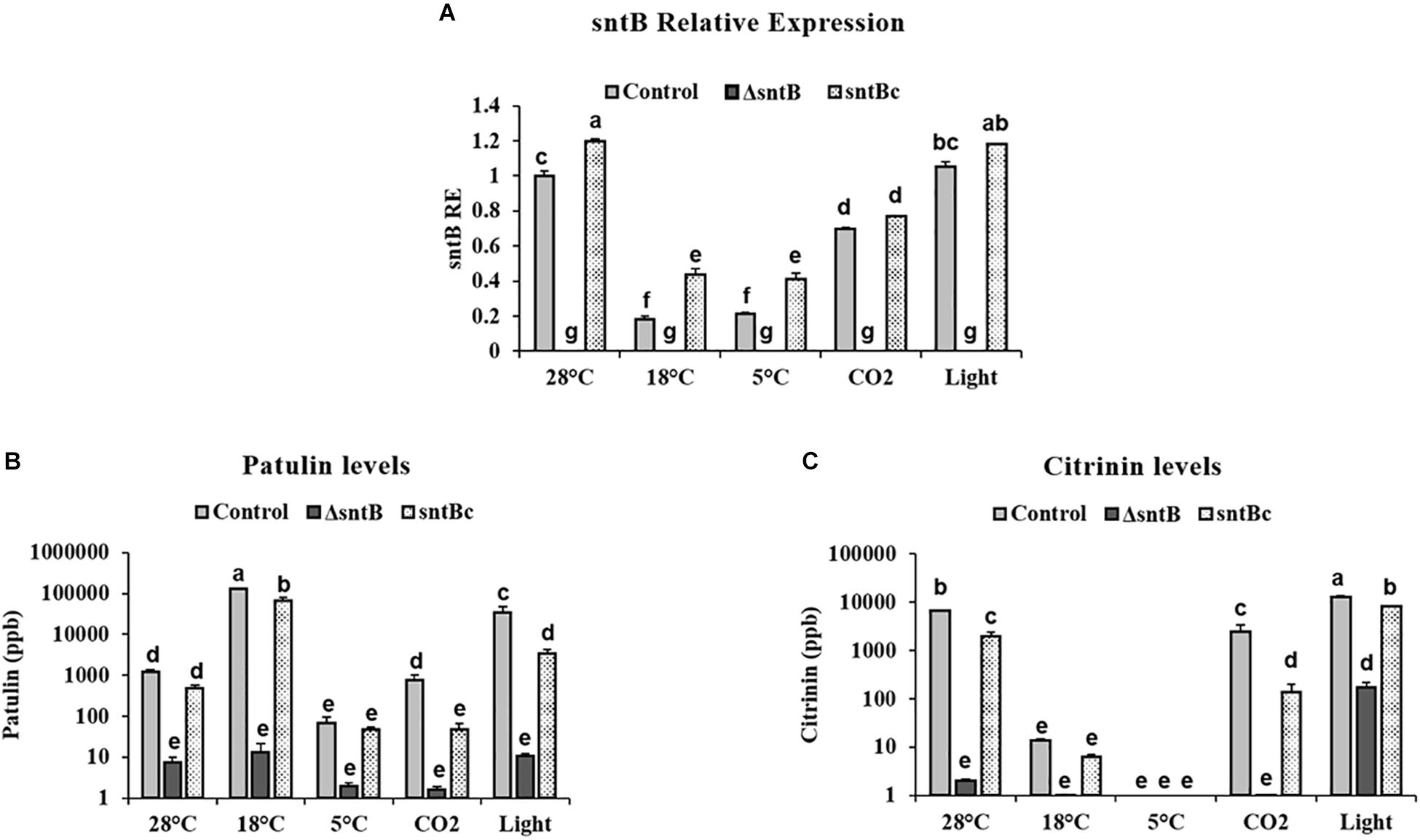
Figure 4. Effect of abiotic factors on sntB expression and mycotoxin accumulation. The three Penicillium strains (control, ΔsntB, and sntBc) were grown on YES agar media at 28°C for 48h in the dark and then subjected to different treatment conditions for additional 48 h. Control – 28°C incubation in the dark; 18–18°C incubation in the dark; 5–5°C incubation in the dark; light – 28°C incubation under continuous white fluorescent lighting; CO2 – 28°C incubation at 20% CO2 in the dark. (A) Total RNA was extracted and the relative expression of sntB was evaluated. (B) Patulin and (C) citrinin were extracted and subjected to HPLC analysis. Each strain was inoculated with 3 reps. One-way ANOVA differences were considered significant when p < 0.05. Different letters above the columns indicate statistically significant differences between the strains as determined using Tukey’s single-step multiple comparison test.
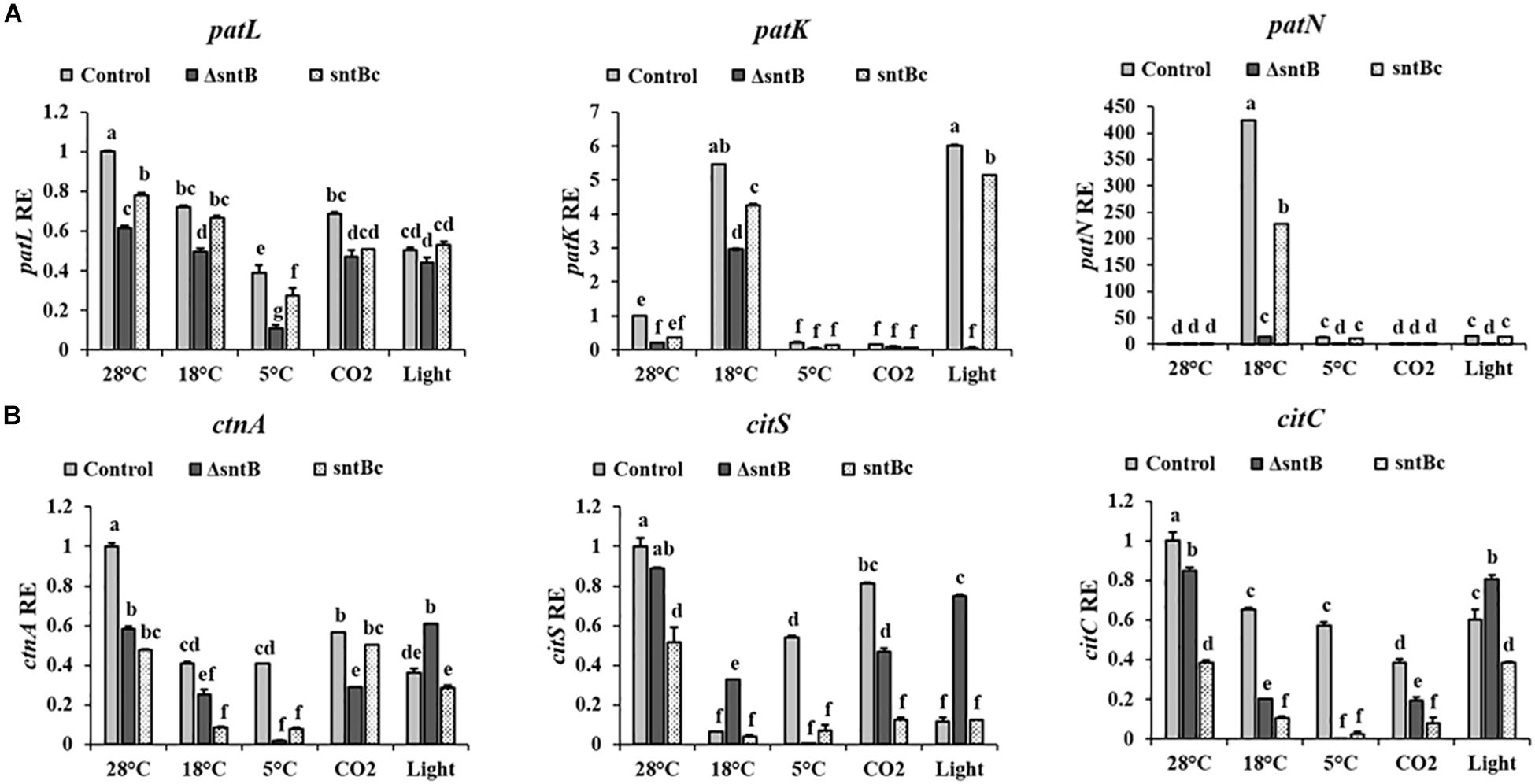
Figure 5. Effect of abiotic factors on relative expression of mycotoxin cluster genes. The three Penicillium strains (control, ΔsntB, and sntBc) were grown on YES agar media at 28°C for 48 h in the dark and then subjected to different treatment conditions as mentioned in Figure 4. RNA was extracted and the relative expression of patulin cluster genes (A), and citrinin cluster genes (B) was evaluated. Error bars represent the standard error of the mean (SEM) across three technical replicates. One-way ANOVA differences were considered significant when p < 0.05. Different letters above the columns indicate statistically significant differences between the strains as determined using Tukey’s single-step multiple comparison test.
Due to its cytotoxic, genotoxic and teratogenic properties, its wide presence in fruits and its virulence-associated function (Snini et al., 2015), patulin synthesis by the fruit pathogen P. expansum has been the topic of research of many studies resulting in an excellent understanding of the biosynthetic pathway of this mycotoxin (Li et al., 2019). Emphasis has also been recently focused on citrinin, another nephrotoxic mycotoxin of P. expansum, reported to often co-occur with patulin (Gimeno, 2005). The biosynthetic steps leading to citrinin and the gene cluster responsible for its production are currently known (He and Cox, 2016; Schmidt-Heydt et al., 2019). Yet, the regulatory pathways underlying the production of both mycotoxins are still not completely resolved, and concurrent research is dedicated to expanding current knowledge.
This paper reports on the identification of a new protein in P. expansum that adds to the short list of proteins recognized to play a role in the virulence of this pathogen (Barad et al., 2016a; Kumar et al., 2016; Chen et al., 2018; El Hajj Assaf et al., 2018; Tannous et al., 2018; Li et al., 2019). This current study is the first to explore a novel response of the post-harvest pathogen P. expansum to epigenetic perturbations through the deletion of the epigenetic reader SntB. In addition to its impact on the ability of the fungus to cause infection and proliferate within apple tissue, SntB also regulates secondary metabolism in P. expansum with negative effects on patulin and citrinin production in vitro (Figures 2A,B).
Penicillium expansum sntB encodes a protein with several histone interacting domains: a BAH domain, a SANT domain and three plant homeodomain (PHD) fingers. SntB is homologous to Snt2 in Saccharomyces cerevisiae and Schizosaccharomyces pombe (Roguev et al., 2004; Baker et al., 2013). In the latter, Snt2 was shown to be a member of a protein complex called Lid2C that includes the proteins Lid2, Ash2, Sdc1, and Jmj3 (Roguev et al., 2004). A different complex was found in S. cerevisiae, and this includes the subunits Ecm5 and Rpd3 (Shevchenko et al., 2008). Originally reported to be simply involved in the regulation of chromatin remodeling (Roguev et al., 2004), subsequent research has shown that the protein reader SntB is a putative virulence factor and a major regulator of fungal secondary metabolism (Denisov et al., 2011; Baker et al., 2013; He et al., 2018; Pfannenstiel et al., 2018). Loss of sntB in the mycotoxigenic seed pathogen A. flavus resulted in global misregulation of secondary metabolism, where some metabolites were up- and some down-regulated such as aflatoxin (Pfannenstiel et al., 2018; Greco et al., 2019). We see a similar global misregulation in the P. expansumΔsntB mutant.
The effect of SntB on various secondary metabolites may in part lay in its regulation of transcriptional complexes/transcription factors (Figure 2E). LaeA, a member of the Velvet Complex (VC) linking secondary metabolism to development (Bayram et al., 2008), regulates the expression of 12 putative backbone genes in P. expansum including patulin genes (Kumar et al., 2016). Along the same line, the carbon catabolite repressor, CreA, was proposed to post-transcriptionally regulate patulin and citrinin biosynthesis (Tannous et al., 2018). Similarly, deletion of sntB resulted in decreased biosynthesis of both mycotoxins (Figures 2A,B). Lastly, the reduced production of mycotoxins can also be potentially linked to the deregulation of the gene encoding the regulatory protein PacC (Figure 2E). Previous works have established that the pH-responsive zinc finger transcription factor PacC is essential for patulin biosynthesis in P. expansum. Barad et al. (2016a) provided the first evidence for the pivotal role of PacC in regulating patulin production in P. expansum. This paper showed that ammonia secretion by P. expansum during fruit colonization resulted in the activation of PacC and the subsequent increase of patulin accumulation (Barad et al., 2016a). These findings have later been confirmed by Chen et al. (2018) that showed a loss of patulin production by the ΔpacC mutant of P. expansum at pH values above 6.0 due to the significant down-regulation of all the genes in this cluster.
Whilst the deletion of sntB in P. expansum was not lethal, a large set of physiological characteristics were disturbed. This included decreased radial growth on all tested media, reduced conidial formation and accelerated spore germination (Figure 1). To the best of our knowledge, this is the first report to show the physiological effects of the epigenetic reader SntB in Penicillium. However, those findings were not unexpected as similar outcomes for sntB loss were observed in other ascomycetes. In A. flavus, the deletion of sntB has led to various morphological phenotypes, such as reduced radial growth and reduced sclerotia formation (Pfannenstiel et al., 2018). Along the same line, snt2 deletion mutants of Fusarium oxysporum and Neurospora crassa, displayed morphological aberrations, including a drop in conidia formation and biomass accumulation, delayed vegetative growth and recurrent hyphal septation (Denisov et al., 2011; Kronholm et al., 2016). Although there have been several studies involved with SntB in ascomycetes, further data are needed to establish a congruent storyline about the impact of SntB on fungal development. Several previous reports described the role of epigenetics in the regulation of phenotypic alterations in filamentous fungi. Epigenetic mechanisms, e.g., histone modifications and RNA interference pathway, were found to be involved in phenotypic plasticity (i.e., phenotypic changes in the colony morphology) of N. crassa across an array of controlled environments including temperature, pH, osmotic stress and sugar content (Kronholm et al., 2016). Comparable results were previously gathered in the dimorphic yeast Aureobasidium pullulans, where phenotypic switches from yeast to mycelial form are likely to be an epigenetic phenomenon (Slepecky and Starmer, 2009).
Apart from their impact on fungal morphology and secondary metabolism, a number of studies highlighted the crucial role of epigenetic modifications in the success of fungal plant pathogens. Virulence assays performed on apples showed that the deletion of the sntB gene had a profound effect on lesion development during infection as lesion diameter caused by ΔsntB was smaller than that of the control strain (Figure 3). Complementation of the deletion strain with the wild type copy of sntB restored the parental virulence phenotype (Figure 3). Our results were in agreement with previous reports on three other plant pathogens, A. flavus (Pfannenstiel et al., 2018), M. oryzae (He et al., 2018), and F. oxysporum (Denisov et al., 2011), where the deletion and/or disruption of sntB reduced virulence on the respective plant hosts.
How SntB regulates P. expansum virulence is still unknown. The studies of both M. oryzae and F. oxysporum suggested that decrease in virulence could be mediated by the autophagic programmed cell death. However, our results did not support a role for SntB in PCD in P. expansum (Supplementary Figure S6). One possible explanation for these results is the difference in protein architecture of P. expansum SntB compared to other reported species. In M. oryzae, MoSnt2 contains an ELM2 domain that P. expansum SntB lacks (He et al., 2018). In a similar manner, F. oxysporum f. sp. melonis Snt2 contains a GATA-Zn domain that is not present in P. expansum SntB (Denisov et al., 2011). The reported functional difference may also be explained by the distinctive lifestyles of plant pathogenic fungi. In M. oryzae, autophagy mediated cell death and MoSnt2 are required for appressoria formation, an essential penetration structure present in M. oryzae but not in P. expansum (Veneault-Fourrey et al., 2006; He et al., 2018).
Penicillium expansum sntB was found to be regulated by different environmental factors, such as temperature and CO2 levels, that are usually used to modulate the physiology of stored fruits (Hulme, 1971). Moreover our data revealed that the down regulation of sntB observed at low temperatures and high CO2 levels (Figure 4A) is accompanied by a significant decrease of patulin and citrinin production (Figures 4B,C). However, we found a negative correlation between patulin accumulation and patL expression in the WT under certain environmental conditions (incubation at 18°C and under light; Figure 5A). In addition to transcription factor of the patulin gene cluster (PatL), other regulatory proteins known to impact patulin biosynthesis in P. expansum, including transcription factors LaeA, CreA, and PacC associated with production of secondary metabolites and broad responses to carbon availability and pH, respectively (Kumar et al., 2016, 2018; Chen et al., 2018; Tannous et al., 2018). At the same time, patulin accumulation under all tested conditions is positively correlated with the transcript level of one of the key genes in the patulin biosynthetic pathway, patK (PKS). Those results are of great importance as they point out that the same environmental factors that regulate the host physiological response also regulate the expression of P. expansum sntB and mycotoxin production simultaneously. Therefore, the epigenetic reader SntB opens up new avenues of study in order to thoroughly understand P. expansum biology and virulence. This will allow better regulation of fungal decaying of stored fruit as well as mycotoxin contamination by P. expansum during fruit storage.
An increasing body of evidence points toward epigenetic mechanisms being responsible for a wide array of biological phenomena, including morphological development, virulence and secondary metabolism. This work represents the first step in the exploration of epigenetic regulation of development and pathogenesis in the post-harvest plant pathogen P. expansum. The present data shows that SntB is necessary for the biosynthesis of several secondary metabolites, including patulin and citrinin, and is required for full virulence on apples. sntB expression responds to environmental factors modulated during host storage and thus may provide clues to incorporate epigenetic control strategies against this storage disease.
All datasets generated for this study are included in the article/Supplementary Material.
NK, DP, ES, OB, and JT conceived the presented idea and designed the study. JT, OB, and DL-R carried out the experiments, collected, analyzed the data and designed the figures. JT wrote the manuscript with support from OB and DL-R on specific sections. NK, DP, and ES edited the manuscript. All authors discussed the results, commented on the manuscript, conceived, and planned the experiments.
This research was supported by Research Grant Award No. IS-5042-17C from BARD, the United States – Israel Binational Agricultural Research and Development Fund and in part by the Food Research Institute of the University of Wisconsin, Madison.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00610/full#supplementary-material
Aasland, R., Gibson, T. J., and Stewart, A. F. (1995). The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20, 56–59. doi: 10.1016/s0968-0004(00)88957-4
Altunatmaz, S. S., Issa, G., and Aydin, A. (2012). Detection of airborne psychrotrophic bacteria and fungi in food storage refrigerators. Braz. J. Microbiol. 43, 1436–1443. doi: 10.1590/s1517-83822012000400027
Artigot, M. P., Loiseau, N., Laffitte, J., Mas-Reguieg, L., Tadrist, S., Oswald, I. P., et al. (2009). Molecular cloning and functional characterization of two CYP619 cytochrome P450s involved in biosynthesis of patulin in Aspergillus clavatus. Microbiology 155, 1738–1747. doi: 10.1099/mic.0.024836-0
Baker, L. A., Ueberheide, B. M., Dewell, S., Chait, B. T., Zheng, D., and Allis, C. D. (2013). The yeast Snt2 protein coordinates the transcriptional response to hydrogen peroxide-mediated oxidative stress. Mol. Cell. Biol. 33, 3735–3748. doi: 10.1128/mcb.00025-13
Ballester, A.-R., Marcet-Houben, M., Levin, E., Sela, N., Selma-Lázaro, C., Carmona, L., et al. (2015). Genome, transcriptome, and functional analyses of Penicillium expansum provide new insights into secondary metabolism and pathogenicity. Mol. Plant Microbe Interact. 28, 232–248. doi: 10.1094/mpmi-09-14-0261-fi
Barad, S., Espeso, E. A., Sherman, A., and Prusky, D. (2016a). Ammonia activates pacC and patulin accumulation in an acidic environment during apple colonization by Penicillium expansum. Mol. Plant. Pathol. 17, 727–740. doi: 10.1111/mpp.12327
Barad, S., Sionov, E., and Prusky, D. (2016b). Role of patulin in post-harvest diseases. Fungal Biol. Rev. 30, 24–32. doi: 10.1016/j.fbr.2016.02.001
Barad, S., Horowitz, S. B., Kobiler, I., Sherman, A., and Prusky, D. (2014). Accumulation of the mycotoxin patulin in the presence of gluconic acid contributes to pathogenicity of Penicillium expansum. Mol. Plant Microbe Interact. 27, 66–77. doi: 10.1094/mpmi-05-13-0138-r
Bayram, Ö, Krappmann, S., Ni, M., Bok, J. W., Helmstaedt, K., Valerius, O., et al. (2008). VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320, 1504–1506. doi: 10.1126/science.1155888
Boyer, L. A., Latek, R. R., and Peterson, C. L. (2004). The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 5, 158–163. doi: 10.1038/nrm1314
Callebaut, I., Courvalin, J.-C., and Mornon, J.-P. (1999). The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 446, 189–193. doi: 10.1016/s0014-5793(99)00132-5
Carvalho, N. D., Arentshorst, M., Kwon, M. J., Meyer, V., and Ram, A. F. (2010). Expanding the ku70 toolbox for filamentous fungi: establishment of complementation vectors and recipient strains for advanced gene analyses. Appl. Microbiol. Biotechnol. 87, 1463–1473. doi: 10.1007/s00253-010-2588-1
Chen, Y., Li, B., Xu, X., Zhang, Z., and Tian, S. (2018). The pH-responsive PacC transcription factor plays pivotal roles in virulence and patulin biosynthesis in Penicillium expansum. Environ. Microbiol. 20, 4063–4078. doi: 10.1111/1462-2920.14453
Denisov, Y., Freeman, S., and Yarden, O. (2011). Inactivation of Snt2, a BAH/PHD-containing transcription factor, impairs pathogenicity and increases autophagosome abundance in Fusarium oxysporum. Mol. Plant Pathol. 12, 449–461. doi: 10.1111/j.1364-3703.2010.00683.x
El Hajj Assaf, C., Snini, S., Tadrist, S., Bailly, S., Naylies, C., et al. (2018). Impact of veA on the development, aggressiveness, dissemination and secondary metabolism of Penicillium expansum. Mol. Plant Pathol. 19, 1971–1983. doi: 10.1111/mpp.12673
Fair, K., Anderson, M., Bulanova, E., Mi, H., Tropschug, M., and Diaz, M. O. (2001). Protein interactions of the MLL PHD fingers modulate MLL target gene regulation in human cells. Mol. Cell. Biol. 21, 3589–3597. doi: 10.1128/mcb.21.10.3589-3597.2001
Flajs, D., and Peraica, M. (2009). Toxicological properties of citrinin. Arh. Hig Rada Toksikol. 60, 457–464.
Gimeno, A. (2005). Patulin and citrinin in Portuguese apples with rotten spots. J. Food Protect. 68, 1–7.
Greco, C., Pfannenstiel, B. T., Liu, J. C., and Keller, N. P. (2019). Depsipeptide Aspergillicins revealed by chromatin reader protein deletion. ACS Chem. Biol. 14, 1121–1128. doi: 10.1021/acschembio.9b00161
Hartmann, T., Dümig, M., Jaber, B. M., Szewczyk, E., Olbermann, P., Morschhäuser, J., et al. (2010). Validation of a self-excising marker in the human pathogen Aspergillus fumigatus by employing the β-rec/six site-specific recombination system. Appl. Environ. Microbiol. 76, 6313–6317. doi: 10.1128/aem.00882-10
He, M., Xu, Y., Chen, J., Luo, Y., Lv, Y., Su, J., et al. (2018). MoSnt2-dependent deacetylation of histone H3 mediates MoTor-dependent autophagy and plant infection by the rice blast fungus Magnaporthe oryzae. Autophagy 14, 1543–1561. doi: 10.1080/15548627.2018.1458171
He, Y., and Cox, R. J. (2016). The molecular steps of citrinin biosynthesis in fungi. Chem. Sci. 7, 2119–2127. doi: 10.1039/c5sc04027b
Kronholm, I., Johannesson, H., and Ketola, T. (2016). Epigenetic control of phenotypic plasticity in the filamentous fungus Neurospora crassa. G3 6, 4009–4022. doi: 10.1534/g3.116.033860
Kumar, D., Barad, S., Chen, Y., Luo, X., Tannous, J., Dubey, A., et al. (2016). LaeA regulation of secondary metabolism modulates virulence in Penicillium expansum and is mediated by sucrose. Mol. Plant Pathol. 18, 1150–1163. doi: 10.1111/mpp.12469
Kumar, D., Tannous, J., Sionov, E., Keller, N., and Prusky, D. (2018). Apple intrinsic factors modulating the global regulator, LaeA, the Patulin gene cluster and patulin accumulation during fruit colonization by Penicillium expansum. Front. Plant Sci. 9:1094. doi: 10.3389/fpls.2018.01094
Kwon, M. J., Arentshorst, M., Roos, E. D., van den Hondel, C. A., Meyer, V., and Ram, A. F. (2011). Functional characterization of Rho GTPases in Aspergillus niger uncovers conserved and diverged roles of Rho proteins within filamentous fungi. Mol. Microbiol. 79, 1151–1167. doi: 10.1111/j.1365-2958.2010.07524.x
Li, B., Chen, Y., Zong, Y., Shang, Y., Zhang, Z., Xu, X., et al. (2019). Dissection of patulin biosynthesis, spatial control and regulation mechanism in Penicillium expansum. Environ. Microbiol. 21, 1124–1139. doi: 10.1111/1462-2920.14542
Li, B., Zong, Y., Du, Z., Chen, Y., Zhang, Z., Qin, G., et al. (2015). Genomic characterization reveals insights into patulin biosynthesis and pathogenicity in Penicillium species. Mol. Plant Microbe Interact. 28, 635–647. doi: 10.1094/mpmi-12-14-0398-fi
Lim, F. Y., Sanchez, J. F., Wang, C. C., and Keller, N. P. (2012). Toward awakening cryptic secondary metabolite gene clusters in filamentous fungi. Methods Enzymol. 517:303. doi: 10.1016/b978-0-12-404634-4.00015-2
Liu, Y., Liu, N., Yin, Y., Chen, Y., Jiang, J., and Ma, Z. (2015). Histone H3K4 methylation regulates hyphal growth, secondary metabolism and multiple stress responses in Fusarium graminearum. Environ. Microbiol. 17, 4615–4630. doi: 10.1111/1462-2920.12993
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Martins, M. L., Gimeno, A., Martins, H. M., and Bernardo, F. (2002). Co-occurrence of patulin and citrinin in Portuguese apples with rotten spots. Food Addit. Contam. 19, 568–574. doi: 10.1080/02652030210121320
Pfannenstiel, B. T., Greco, C., Sukowaty, A. T., and Keller, N. P. (2018). The epigenetic reader SntB regulates secondary metabolism, development and global histone modifications in Aspergillus flavus. Fungal Genet. Biol. 120, 9–18. doi: 10.1016/j.fgb.2018.08.004
Pfannenstiel, B. T., and Keller, N. P. (2019). On top of biosynthetic gene clusters: how epigenetic machinery influences secondary metabolism in fungi. Biotechnol. Adv. 37:107345. doi: 10.1016/j.biotechadv.2019.02.001
Pfannenstiel, B. T., Zhao, X., Wortman, J., Wiemann, P., Throckmorton, K., Spraker, J. E., et al. (2017). Revitalization of a forward genetic screen identifies three new regulators of fungal secondary metabolism in the genus Aspergillus. MBio 8:e1246-17.
Puel, O., Galtier, P., and Oswald, I. P. (2010). Biosynthesis and toxicological effects of patulin. Toxins 2, 613–631. doi: 10.3390/toxins2040613
Roguev, A., Shevchenko, A., Schaft, D., Thomas, H., Stewart, A. F., and Shevchenko, A. (2004). A comparative analysis of an orthologous proteomic environment in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Mol. Cell. Proteom. 3, 125–132.
R Studio Team (2015). RStudio: Integrated Development for R. Boston, MA: RStudio, Inc. Available at: http://www.rstudio.com
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Schmidt-Heydt, M., Stoll, D., and Geisen, R. (2019). Whole-genome sequencing of the fungus Penicillium citrinum reveals the biosynthesis gene cluster for the mycotoxin citrinin. Microbiol. Resour. Announc. 8, e1419–e1418.
Shevchenko, A., Roguev, A., Schaft, D., Buchanan, L., Habermann, B., Sakalar, C., et al. (2008). Chromatin Central: towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome Biol. 9:R167.
Silar, P. (1995). Two new easy to use vectors for transformations. Fungal Genet. Rep. 42:73. doi: 10.4148/1941-4765.1353
Slepecky, R. A., and Starmer, W. T. (2009). Phenotypic plasticity in fungi: a review with observations on Aureobasidium pullulans. Mycologia 101, 823–832. doi: 10.3852/08-197
Snini, S. P., Tadrist, S., Laffitte, J., Jamin, E. L., Oswald, I. P., and Puel, O. (2014). The gene PatG involved in the biosynthesis pathway of patulin, a food-borne mycotoxin, encodes a 6-methylsalicylic acid decarboxylase. Int. J. Food Microbiol. 171, 77–83. doi: 10.1016/j.ijfoodmicro.2013.11.020
Snini, S. P., Tannous, J., Heuillard, P., Bailly, S., Lippi, Y., Zehraoui, E., et al. (2015). The patulin is a cultivar dependent aggressiveness factor favoring the colonization of apples by Penicillium expansum. Mol. Plant Pathol. 17, 920–930. doi: 10.1111/mpp.12338
Tannous, J., Atoui, A., El Khoury, A., Francis, Z., Oswald, I. P., Puel, O., et al. (2015). A study on the physicochemical parameters for Penicillium expansum growth and patulin production: effect of temperature, pH, and water activity. Food Sci. Nutr. 4, 611–622. doi: 10.1002/fsn3.324
Tannous, J., El Khoury, R., Snini, S. P., Lippi, Y., El Khoury, A., Atoui, A., et al. (2014). Sequencing, physical organization and kinetic expression of the patulin biosynthetic gene cluster from Penicillium expansum. Int. J. Food Microbiol. 189, 51–60. doi: 10.1016/j.ijfoodmicro.2014.07.028
Tannous, J., Keller, N. P., Atoui, A., El Khoury, A., Lteif, R., Oswald, I. P., et al. (2017a). Secondary metabolism in Penicillium expansum: emphasis on recent advances in patulin research. Crit. Rev. Food Sci. Nutr. 58, 2082–2098. doi: 10.1080/10408398.2017.1305945
Tannous, J., Snini, S. P., El Khoury, R., Canlet, C., Pinton, P., Lippi, Y., et al. (2017b). Patulin transformation products and last intermediates in its biosynthetic pathway, E-and Z-ascladiol, are not toxic to human cells. Arch. Toxicol. 91, 2455–2467. doi: 10.1007/s00204-016-1900-y
Tannous, J., Kumar, D., Sela, N., Sionov, E., Prusky, D., and Keller, N. P. (2018). Fungal attack and host defense pathways unveiled in near avirulent interactions of Penicillium expansum creA mutants on apples. Mol. Plant Pathol. 19, 2635–2650. doi: 10.1111/mpp.12734
Veneault-Fourrey, C., Barooah, M., Egan, M., Wakley, G., and Talbot, N. J. (2006). Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 312, 580–583. doi: 10.1126/science.1124550
Visentin, I., Montis, V., Döll, K., Alabouvette, C., Tamietti, G., Karlovsky, P., et al. (2012). Transcription of genes in the biosynthetic pathway for fumonisin mycotoxins is epigenetically and differentially regulated in the fungal maize pathogen Fusarium verticillioides. Eukaryot. Cell 11, 252–259. doi: 10.1128/ec.05159-11
Wiemann, P., Soukup, A. A., Folz, J. S., Wang, P.-M., Noack, A., and Keller, N. P. (2018). CoIN: co-inducible nitrate expression system for secondary metabolites in Aspergillus nidulans. Fungal Biol. Biotechnol. 5:6.
Keywords: Penicillium expansum, epigenetic regulation, SntB, epigenetic reader, virulence, secondary metabolism, patulin, citrinin
Citation: Tannous J, Barda O, Luciano-Rosario D, Prusky DB, Sionov E and Keller NP (2020) New Insight Into Pathogenicity and Secondary Metabolism of the Plant Pathogen Penicillium expansum Through Deletion of the Epigenetic Reader SntB. Front. Microbiol. 11:610. doi: 10.3389/fmicb.2020.00610
Received: 21 January 2020; Accepted: 19 March 2020;
Published: 09 April 2020.
Edited by:
Chengshu Wang, Institute of Plant Physiology and Ecology (SIBS-CAS), ChinaReviewed by:
Ana-Rosa Ballester, Institute of Agrochemistry and Food Technology (IATA), SpainCopyright © 2020 Tannous, Barda, Luciano-Rosario, Prusky, Sionov and Keller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edward Sionov, ZWR3YXJkc2lvQHZvbGNhbmkuYWdyaS5nb3YuaWw=; Nancy P. Keller, bnBrZWxsZXJAd2lzYy5lZHU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.