- Department of Molecular Microbiology, Institute of Microbiology, Faculty of Biology, University of Warsaw, Warsaw, Poland
Yersinia enterocolitica exhibits a dual lifestyle, existing as both a saprophyte and a pathogen colonizing different niches within a host organism. OmpR has been recognized as a regulator that controls the expression of genes involved in many different cellular processes and the virulence of pathogenic bacteria. Here, we have examined the influence of OmpR and varying temperature (26°C vs. 37°C) on the cytoplasmic proteome of Y. enterocolitica Ye9N (bio-serotype 2/O:9, low pathogenicity). Differential label-free quantitative proteomic analysis indicated that OmpR affects the cellular abundance of a number of proteins including subunits of urease, an enzyme that plays a significant role in acid tolerance and the pathogenicity of Y. enterocolitica. The impact of OmpR on the expression of urease under different growth conditions was studied in more detail by comparing urease activity and the transcription of ure genes in Y. enterocolitica strains Ye9N and Ye8N (highly pathogenic bio-serotype 1B/O:8). Urease expression was higher in strain Ye9N than in Ye8N and in cells grown at 26°C compared to 37°C. However, low pH, high osmolarity and the presence of urea did not have a clear effect on urease expression in either strain. Further analysis showed that OmpR participates in the positive regulation of three transcriptional units encoding the multi-subunit urease (ureABC, ureEF, and ureGD) in strain Ye9N, but this was not the case in strain Ye8N. Binding of OmpR to the ureABC and ureEF promoter regions was confirmed using an electrophoretic mobility shift assay, suggesting that this factor plays a direct role in regulating the transcription of these operons. In addition, we determined that OmpR modulates the expression of a ureR-like gene encoding a putative regulator of the ure gene cluster, but in the opposite manner, i.e., positively in Ye9N and negatively in Ye8N. These findings provide some novel insights into the function of OmpR in adaptation strategies of Y. enterocolitica.
Introduction
Yersinia enterocolitica, a Gram-negative bacterium, exhibits a dual lifestyle, existing as both a saprophyte widely distributed in nature and a pathogen colonizing many different niches within host organisms. Y. enterocolitica has been isolated from different sources, including food, clinical material and certain warm- and cold-blooded animals (Fuchs et al., 2011; Fàbrega and Vila, 2012). In humans, Y. enterocolitica causes yersiniosis – an acute or chronic foodborne disease manifested by a variety of clinical symptoms, especially gastrointestinal (Bottone, 1997; Carniel et al., 2006). Cases of yersiniosis occur worldwide, and in Europe it is the third most common food-borne gastroenteritis after campylobacteriosis and salmonellosis (European Food Safety Authority and European Centre for Disease Prevention, and Control [ECDC], 2017; Chlebicz and Śliżewska, 2018).
Yersinia enterocolitica is a member of the genus Yersinia (family Yersiniaceae, previously Enterobacteriaceae; Adeolu et al., 2016), which includes two other human pathogens: Y. pseudotuberculosis and the plague bacillus Y. pestis, and at least 18 other species considered harmless for humans (Bottone, 2015). Y. enterocolitica is a heterogeneous species comprising many bio-serotypes differing in pathogenicity (Howard et al., 2006). Of six biotypes (1A, 1B, 2, 3, 4 and 5) separated according to their biochemical properties, biotype 1A is considered non-pathogenic, biotype 1B is highly pathogenic to humans and lethal in a mouse model of infection, while biotypes 2 to 5 exhibit low virulence (unable to kill mice) (Bottone, 1997). Strains of biotypes 1A and 2−5 belong to the Y. enterocolitica subsp. palearctica (Neubauer et al., 2000) and are generally found in Europe and Japan (termed “Old World strains”). The most virulent biotype 1B strains belong to the Y. enterocolitica subsp. enterocolitica (Neubauer et al., 2000) and are dominant in North America (“New World strains”). Y. enterocolitica can be divided into approximately 70 serotypes, of which only eleven have been associated with disease in humans, with the majority of cases involving just four virulent serotypes: O:8 (biotype 1B), O:3 (biotype 4), O:9 (biotype 2), and O:5,27 (biotypes 2 and 3) (Bottone, 1997; Fàbrega and Vila, 2012).
Yersinia enterocolitica synthesizes numerous virulence factors that are active at different stages of an infection and whose expression is altered according to the diverse growth conditions experienced by these bacteria in mammalian hosts (Straley and Perry, 1995; Marceau, 2005; Erhardt and Dersch, 2015; Chen et al., 2016). It was previously demonstrated that a functional urease is essential for the survival of Y. enterocolitica O:9 during passage through the stomach (De Koning-Ward and Robins-Browne, 1995; Young et al., 1996), and is also necessary for full virulence in oral infections in the rat model (Gripenberg-Lerche et al., 2000). In addition, a non-enzymatic biological effect of urease may be to trigger the inflammatory process that is thought to underly the arthritogenicity of Y. enterocolitica (Probst et al., 1993). Ureases appear to play a vital role in the survival of Y. enterocolitica cells in the natural environment by degrading urea in the soil and water, which is utilized by this saprophyte as the sole nitrogen source (Young et al., 1996).
Ureases are highly conserved enzymes widely distributed among bacterial species (Mobley et al., 1995; Konieczna et al., 2012; Kappaun et al., 2018). They play an important role in acid resistance and the virulence of several human pathogens, including Proteus mirabilis, Helicobacter pylori and Staphylococcus aureus (Stingl et al., 2002; Rutherford, 2014; Zhou et al., 2019). Bacterial ureases are multi-subunit nickel-dependent enzymes which catalyze the hydrolysis of urea into ammonia (urea amidohydrolase, EC 3.5.1.5). They are typically comprised of three distinct subunits (UreA, subunit γ; UreB, subunit β; and UreC, subunit α) encoded by genes organized into an operon (Mobley et al., 1995; Konieczna et al., 2012). For assembly of the holoenzyme, four accessory/regulatory proteins are required, which are encoded by genes located in the same ure locus (Farrugia et al., 2013; Kappaun et al., 2018).
Different environmental conditions influence the production/activity of the urease complex in bacteria. Some bacterial ureases are regulated by nutrient conditions, pH, urea concentration or nitrogen limitation, whereas others are unaffected by environmental signals and are synthesized constitutively (Mobley et al., 1995; Patra and Aschenbach, 2018). Several regulators of ure gene expression have been identified in ureolytic bacteria. In Klebsiella species, the regulator NtrC activates transcription of the ure promoters in response to nitrogen deprivation (Collins et al., 1993; Magasanik, 1993). Urease expression in Y. pseudotuberculosis is positively regulated by the regulator OmpR and negatively by the regulators RovM and CsrA (Hu et al., 2009; Dai et al., 2018). In many ureolytic pathogenic bacteria, e.g., P. mirabilis, Providencia stuartii, Escherichia coli and Salmonella species, the regulator UreR positively influences expression of the ure genes in the presence of urea (Mobley et al., 1985; Mulrooney et al., 1988; D’Orazio and Collins, 1993; D’Orazio and Collins, 1995; Poore et al., 2001). UreR is a member of the AraC/XylS family of transcriptional regulators and is encoded by the ureR gene that can be located on the chromosome or a plasmid (Nicholson et al., 1993; Island and Mobley, 1995; Poore and Mobley, 2003).
The two-component regulatory system EnvZ/OmpR, consisting of the histidine kinase EnvZ and response regulator OmpR, is involved in the control of various cellular processes, and functions in a number of bacteria (Stock et al., 1989; Hoch and Silhavy, 1995). In response to changes in the environment (osmolarity and pH) the kinase EnvZ phosphorylates OmpR. Conformational changes in this phosphorylated regulator allow it to bind to DNA and modulate gene expression (Kenney, 2002). There is now a considerable body of evidence indicating that OmpR is involved in the regulation of fundamental physiological properties in Yersinia species (Gao et al., 2011; Gueguen et al., 2013; Reboul et al., 2014; Bontemps-Gallo et al., 2019). Our research on Y. enterocolitica strain Ye9N (bio-serotype 2/O:9) suggests that OmpR performs a variety of functions, some of which differ from those described in other bacteria (Brzostek et al., 2012). We have shown that OmpR plays a regulatory function in modulating the abundance of outer membrane proteins, including virulence factors like invasin Inv and adhesin YadA (Brzostek et al., 2007; Skorek et al., 2013; Nieckarz et al., 2016). In contrast to E. coli, we found that the OmpR of Y. enterocolitica is involved in the positive control of flagellum synthesis and motility (Raczkowska et al., 2011). OmpR-dependent regulation of the components of the Y. enterocolitica multidrug efflux pump AcrAB-TolC was also demonstrated (Raczkowska et al., 2015). Another study showed that an ompR mutant of Y. enterocolitica strain Ye9N exhibits a reduced ability to survive under acidic conditions in vitro and within macrophages, which indicates that OmpR is involved in the adaptation of this bacterium to low pH (Brzostek et al., 2003).
In this study, the impact of OmpR and varying temperature (26°C vs. 37°C) on the cytoplasmic proteome of strain Ye9N (2/O:9) was investigated by shotgun label-free quantitative proteomic analysis (LC-MS/MS). Our results indicated that the loss of OmpR in this strain affects the production of a number of proteins, including subunits of urease. In spite of significant advances in our understanding of urease function in Y. enterocolitica, the molecular mechanisms controlling ure expression remain unclear. Therefore, to verify the proteomic data and to gain a greater understanding of the role of OmpR in regulating urease production in Y. enterocolitica, urease activity and ure gene transcription were further examined in strain Ye9N (low pathogenic bio-serotype 2/O:9) and strain Ye8N (highly pathogenic bio-serotype 1B/O:8). Our investigations revealed that the regulator OmpR plays a somewhat different role in controlling urease expression in these two pathogenic Y. enterocolitica strains. These findings provide some novel insights into the function of OmpR in the adaptation strategy of Y. enterocolitica.
Materials and Methods
Bacterial Strains and Growth Conditions
The strains and plasmids used in this study are described in Supplementary Table S1. Y. enterocolitica strains were cultured at 26°C or 37°C in LB medium (10 g/l tryptone, 5 g/l yeast extract, 5 g/l NaCl). E. coli strains were grown at 37°C in LB medium. Antibiotics were used at the following concentrations: nalidixic acid (Nal) – 30 μg/ml, chloramphenicol (Cm) – 25 μg/ml, kanamycin (Km) – 50 μg/ml, gentamicin (Gm) – 40 μg/ml, tetracycline (Tet) – 12.5 μg/ml, trimethoprim (Tp) – 50 μg/ml.
Isolation of Cytoplasmic Proteins for Shotgun Label-Free Quantitative Proteomic Analysis
Triplicate overnight cultures of Y. enterocolitica strains Ye9N (the wild-type strain) and AR4 (OmpR-deficient mutant) were grown in LB, pH 7.0 at 26°C or 37°C with shaking (150 rpm) to an OD600 of 3.0. Samples of 25 ml were centrifuged (8000 × g, 20 min, 4°C), and the cell pellets were flash frozen in liquid nitrogen and stored at −80°C prior to fractionation. Each of the bacterial pellets was resuspended in 12.5 ml of lysis buffer (200 mM Tris−HCl pH 8.0, 0.5 M sucrose, 250 μg/ml lysozyme, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride) and incubated at 4°C for 1 h. The cell suspensions were then sonicated on ice for 18 cycles of 30 s, separated by 30 s rest intervals, using a Sonics Vibra-Cell VCX 130 (Sonics & Materials, Newtown, CT, United States). The cell lysates were centrifuged for 10 min at 8000 × g at 4°C to remove unbroken cells and debris, then the supernatants were centrifuged at high speed (35,000 × g, 1.5 h, 4°C) to pellet total membranes. The protein concentrations in the supernatants (cytoplasmic protein extracts) were measured using the RC-DC protein assay (Bio-Rad, Hercules, United States) and equalized by dilution with lysis buffer. The protein extracts were precipitated by adding 4 volumes of cold acetone and holding at −20°C overnight. The samples were then centrifuged for 20 min at 14,000 × g at 4°C. The resultant protein pellets were rinsed with cold acetone twice, each time resuspending and centrifuging for 20 min at 14,000 × g at 4°C. After carefully removing the supernatants, the protein precipitates were briefly air-dried and resuspended in 40 μl of 5× Invitrosol LC/MS Protein Solubilizer (Invitrogen Life Science Technologies). To each sample, 4 μl of 20% (w/v) urea stock solution was added to facilitate dissolution of the pellet. The samples were then vortexed for 3 min, incubated at 60°C for 25 min and diluted with 160 μl of 25 mM ammonium bicarbonate to produce a final Invitrosol concentration of 1×.
Sample preparation for mass spectrometry analyses were performed as described previously (Nieckarz et al., 2016). The cytoplasmic proteomes of strains Ye9N and AR4 were compared to produce a differential proteome list. Proteins whose abundance differed significantly (q-value ≤ 0.05) between the wild-type strain Ye9N and OmpR-deficient mutant AR4, according to MS analysis, were defined by the ratios of ≤0.67 (protein more abundant in AR4) or ≥1.5 (protein less abundant in AR4).
Acid-Sensitivity Assay
The acid-sensitivity assay was performed essentially as described by Kakoschke et al. (2014). To measure survival in acid, overnight cultures grown in LB at 26°C or 37°C were diluted to 107 CFU/ml in PBS (pH 7.0). Then, 0.5 ml of each bacterial cell suspension was mixed with an equal volume of PBS acidified with acetic acid to pH 4.0 (acid stress), or PBS at pH 7.0 (control). The cell suspensions were incubated at 26°C or 37°C for 90 min and then dilutions were plated on LB plates to determine the number of viable cells as CFU/ml. The percentage survival value was defined as the CFU/ml after acid treatment × 100/CFU/ml of the control.
Molecular Biology Techniques
DNA manipulations, polymerase chain reactions (PCRs), restriction digests, ligations and DNA electrophoresis, were performed according to standard protocols (Sambrook and Russell, 2001). Plasmid and genomic DNA were isolated using a Plasmid Miniprep DNA purification Kit and GeneMATRIX Bacterial & Yeast Genomic DNA Purification Kit (EurX), respectively. When amplified fragments were used for cloning, the PCR was performed using DreamTaq DNA polymerase or Phusion High-Fidelity DNA polymerase (Thermo Scientific). Oligonucleotide primers for PCR and sequencing were purchased from Sigma Aldrich and are listed in Supplementary Table S2. DNA fragments amplified by PCR were purified using a PCR/DNA Clean Up kit (EurX). The plasmids used in this study are described in Supplementary Table S1. DNA sequencing was performed by Genomed S.A. (Warsaw, Poland).
Construction of ureABC:lacZ, ureEF:lacZ, ureGD:lacZ, and ureR:lacZ Transcriptional Fusion Plasmids
To obtain ureABC:lacZ, ureEF:lacZ, ureGD:lacZ and ureR:lacZ transcriptional fusions, DNA fragments containing the promoters of the ureABC, ureEF and ureGD operons, and the ureR-like gene were amplified from Y. enterocolitica chromosomal DNA by PCR using the primer pairs NureABCKpnIL/NureABCKpnIP, LureEFEcoRI/PureEFKpnI, LureGDEcoRI/PureGDKpnI, and EcoRIReg1/KpnIReg2, respectively (Supplementary Table S2). The amplified fragments were digested with KpnI (in the case of the ureABC operon promoter) or EcoRI/KpnI and cloned into the corresponding sites of reporter vector pCM132Gm [derivative of plasmid pCM132 (Marx and Lidstrom, 2001) containing a gentamicin resistance cassette], upstream of a promoterless lacZ gene. The resulting constructs were verified by PCR using the primer pair pCM132GmSPR1/pCM132GmSPR2 (flanking the EcoRI and KpnI recognition sequences) followed by sequencing of the amplicons. The correct orientation of the cloned ureABC promoter was verified by PCR using the primer pair pCM132GmSPR1/lacZSprP and by sequencing. The constructs pCM132Gm-ureABC:lacZ, pCM132Gm-ureEF:lacZ, pCM132Gm-ureGD:lacZ and pCM132Gm-ureR:lacZ were introduced into E. coli CC118 λpir and transferred by conjugation into Y. enterocolitica Ye9N, the ompR mutant AR4, Ye8N and the ompB mutant KJ4, selecting transconjugants on LB plates containing Gm and Nal. The presence of these constructs in the Y. enterocolitica strains was confirmed by plasmid isolation and PCR with the primer pair pCM132GmSPR1/pCM132GmSPR2.
β-Galactosidase Assays
β-galactosidase assays were performed essentially as described by Thibodeau et al. (2004), using 96-well flat-bottomed plates (Nest Sc. Biotech.) and a Sunrise plate reader (Tecan). Briefly, cultures grown to stationary phase at 26°C or 37°C were diluted with LB medium to an OD600 of 0.4–0.5 and 80 μl of each cell suspension were then mixed with 20 μl of POPCulture Reagent (EMD Millipore Corp) and incubated for 15 min to cause cell lysis. In the wells of a microtiter plate, 20 μl of each cell lysate were mixed with 130 μl of Z Buffer and 30 μl of ONPG (4 mg/ml). For kinetic assays, the absorbance at 415 nm (relative to a blank) was measured at time intervals of 10 s, with 2 s of shaking before each reading. The assays were performed at 26°C and monitored for up to 20 min. The β-galactosidase activity was expressed in Miller units calculated as described previously (Thibodeau et al., 2004). Each assay was performed at least in triplicate. To test the effect of acid pH, high osmolarity and urea on reporter gene expression, cultures grown to stationary phase at 26°C were diluted to an OD600 of ∼ 0.4 in LB medium. Then 1 ml of each culture was centrifuged (4000 × g, 5 min, room temperature) and the cell pellets were resuspended in 1 ml of LB (86 mM NaCl, pH 7.0), LB adjusted to pH 4.5, LB supplemented with NaCl to a final concentration of 350 mM or LB containing 100 mM urea. These cultures were incubated for 2 h at 26°C with shaking (150 rpm). After two washes in LB medium by centrifugation (4000 × g, 5 min, room temperature) and gentle re-suspension, β-galactosidase activity in the final cell suspensions was measured.
Electrophoretic Mobility Shift Assays (EMSAs)
The in vitro interaction between phosphorylated OmpR (OmpR-P) and the promoters of selected genes was examined using EMSAs, as described previously (Jaworska et al., 2018), with some modifications. The primers listed in Supplementary Table S2 were used in PCRs with Y. enterocolitica Ye9N genomic DNA to amplify DNA fragments comprising the regulatory regions of the operons ureABC (−305 to + 7 relative to the A of the ATG start codon of ureA), ureEF (−352 to + 33 relative to the A of the ATG start codon of ureE), ureGD (−272 to + 33 relative to the A of the ATG start codon of ureG), and gene ureR-like (−310 to + 52 relative to the A of the ATG start codon of ureR-like). Increasing amounts of recombinant OmpR-His6 were phosphorylated in vitro by incubation for 30 min at room temperature in reaction buffer (50 mM Tris-HCl pH 8.0, 100 mM KCl, 1 mM EDTA, 1 mM DTT, 20 mM MgCl2, 12% glycerol, 100 μg/ml BSA, 0.1% Triton X-100) containing 20 mM acetyl phosphate (lithium potassium acetyl phosphate; Sigma-Aldrich). After phosphorylation, 0.05 pmoles of the respective test fragments were added to form the separate EMSA binding reactions. As a negative control, a 211-bp fragment of the Y. enterocolitica 16S rRNA gene amplified by PCR (Supplementary Table S2) was also included in each reaction. The binding reactions were analyzed by electrophoresis (∼3 h at 110V) on 4.2% native polyacrylamide gels (19:1 acrylamide/bis-acrylamide, 0.2 × TBE, 2% glycerol). SYBRgreen 1× solution (Invitrogen) was used to stain the DNA bands in the gels, which were visualized with a GE Healthcare AI600 imager.
Semi-Quantitative Urease Enzyme Assay
Overnight cultures of Y. enterocolitica strains grown in LB, pH 7.0 at 26°C or 37°C were adjusted to an OD600 of 1.0 (approximately 109 bacterial cells per ml). Then 1 ml of each cell suspension was centrifuged (4000 × g, 3 min, 25°C). The cell pellets were resuspended in 0.1 ml of LB and 10 μl lots were spotted onto the surface of urease enzyme assay plates [Christensen’s urea agar – 1 g/L peptone, 1 g/L D(+)-glucose, 2 g/L potassium dihydrogen phosphate, 5 g/L sodium chloride, 0.012 g/L phenol red, 12 g/L agar, pH 6.8 with or without 2% urea]. Following incubation at 26°C or 37°C for 24 h, urease activity was observed as pink zones around the bacterial growth. Urease catalyzes the hydrolysis of urea to ammonia, which causes an increase in pH indicated by the pH indicator phenol red (Young et al., 1996).
Quantitative Urease Enzyme Assay
The quantitative urease activity assay was based on the measurement of ammonia formed by enzymatic hydrolysis of urea, using Nessler’s reagent. Briefly, triplicate cultures of the Y. enterocolitica strains were grown overnight at 26°C in LB medium with shaking (150 rpm). Then, 1 ml of each culture was centrifuged (4000 × g, 3 min, 25°C) and the cell pellets washed once in Christensen’s urea medium [1 g/L peptone, 1 g/L D(+)-glucose, 2 g/L potassium dihydrogen phosphate, 5 g/L sodium chloride pH 6.8, with 2% urea] and resuspended in the same. The cell suspensions were diluted to an OD600 of 0.1 in Christensen’s urea medium and incubated for 2 h at 26°C with shaking (150 rpm). The OD600 of the cultures was measured to confirm no change, then 90 μl aliquots were mixed with 180 μl of 10% Nessler’s reagent (Sigma Aldrich) in the wells of a 96-well flat-bottomed plate (Nest Sc. Biotech.). Controls containing only Christensen’s urea medium were also included. The absorbance at 420 nm (relative to a blank) was measured 5 min after mixing the cells with Nessler’s reagent, using a TECAN Infinite M200PRO microplate reader. The levels of ammonia produced were determined from a standard curve constructed using defined concentrations of NH4Cl prepared fresh in the assay buffer (Young et al., 1996).
Construction of an ompB (ompRenvZ) Deletion Mutant
The ompB:Tp deletion mutant of Y. enterocolitica Ye8N (replacement with Tp resistance gene cassette) was obtained by homologous recombination using suicide vector pDS132 (Philippe et al., 2004), carrying the mutagenic DNA fragment constructed by overlap extension PCR using primers listed in Supplementary Table S2. Briefly, three DNA fragments were PCR-amplified using strain Ye8N genomic DNA (for flanking regions) or plasmid p34E-Tp (for the Tp cassette) as the templates. A mixture of these three amplicons was then used as the template with flanking primers in a PCR. The mutagenic fragment was cloned into pDS132. The resulting construct pDSompB was transferred from E. coli CC118 λpir to Y. enterocolitica strain Ye8N by triparental mating, with E. coli strain DH5α carrying plasmid pRK2013 as a helper. To select for the second recombination, the transconjugant strains were plated on LB agar containing Tp and 10% (w/v) sucrose. Sucrose-resistant colonies were screened for the loss of the plasmid (Cm resistance). The correct allelic exchange was verified by PCR using primers listed in Supplementary Table S2 and by sequencing.
Construction of Plasmid pompB for Complementation
To complement the ompB mutation, the coding sequence of the ompB (ompRenvZ) operon with the native ribosome binding site was amplified by PCR using Ye8N chromosomal DNA as the template with primers OmpB1 and OmpB2 (Supplementary Table S2). The amplified fragment was digested with EcoRI/BamHI and cloned into the corresponding sites of reporter vector pBBR1MCS-2, containing a kanamycin resistance cassette (Kovach et al., 1995). The resulting construct pompB was verified by sequencing and used to transform E. coli S17 λpir. This plasmid was then introduced into the ompB mutant strain KJ4 by biparental conjugation. Exconjugants were selected on LB agar plates containing trimethoprim and kanamycin.
Bioinformatic Analyses
Bioinformatic analyses of the mass spectrometric data were performed as described previously (Nieckarz et al., 2016). In silico analyses of the ure gene cluster and ureR-like gene in Y. enterocolitica strains were performed on a shotgun genome sequence of Y. enterocolitica subsp. palearctica Ye9N (bio-serotype 2/O:9; NCBI/GenBank: JAALCX000000000) and Y. enterocolitica subsp. enterocolitica 8081 (bio-serotype 1B/O:8; NCBI/GenBank: AM286415). The Needleman-Wunsch algorithm was used for sequence alignment (Altschul et al., 1997). Promoter prediction was performed using the web-based software BPROM in the Softberry package (Solovyev and Salamov, 2011). Manual homology searches of predicted urease sub-units were conducted by BLAST analysis1 to describe a urease protein complex. The MOTIF bioinformatics tool provided by GenomeNet, Japan, was used to search for sequence motifs2. The identified proteins were described according to the UniProt databases or GenBank, or homologous sequences obtained using BLAST. Statistical analyses were performed using Prism 5 software (v. 5.01, GraphPad). Student’s t-test was used to determine statistically significant differences.
Results
Effect of Temperature on the Cytoplasmic Proteome of Y. enterocolitica Strain Ye9N
To study the effect of temperature on the cytoplasmic proteome of Y. enterocolitica, cells of wild-type strain Ye9N, grown in standard lysogeny broth (LB) medium at 26°C or 37°C, were fractionated using ultracentrifugation and the cytoplasmic fraction was analyzed by shotgun label-free quantitative LC-MS/MS. This analysis revealed 23 differentially expressed proteins (q-value ≤ 0.05, at least two peptides per protein, minimal acceptable fold change 1.5, Table 1). The more abundant proteins at the higher growth temperature included molecular chaperones such as GroEL (Hsp60; ∼4-fold) together with its co-chaperone GroES (∼7-fold), DnaK (Hsp70, ∼1.7-fold), ClpB (∼6-fold) and HtpG, a prokaryotic Hsp90 homolog (∼4-fold). In contrast, the level of trigger factor (TF), a ribosome-associated peptidyl cis/trans isomerase, which represents the only ribosome-associated chaperone known in bacteria (Hoffmann et al., 2010), was decreased at 37°C (∼2.7-fold). The level of YopE, the secreted effector of the Yersinia Ysc-Yop T3SS, which disrupts the actin cytoskeleton of eukaryotic cells (Viboud and Bliska, 2005; Aepfelbacher et al., 2007; Trosky et al., 2008), was higher at 37°C compared to 26°C (∼9-fold), confirming previous reports (Lambert de Rouvroit et al., 1992; Akopyan et al., 2011). Upregulation of YerA (∼6.7-fold), a chaperone required for YopE secretion at 37°C, was also noted (Parsot et al., 2003). Proteins more abundant at the lower temperature (26°C) included the periplasmic maltose-binding protein MalE (∼14.39-fold), amino acid metabolic enzymes histidine ammonia-lyase HutH (∼7.57-fold) and succinylornithine transaminase AstC (∼7.99-fold), as well as bacterioferritin Bfr (∼7.43-fold). Interestingly, this proteomic analysis revealed downregulation at 37°C of a structural subunit of urease, UreC (∼2.3-fold), and the urease accessory proteins UreE (∼2.8-fold) and UreG (∼3.2-fold).
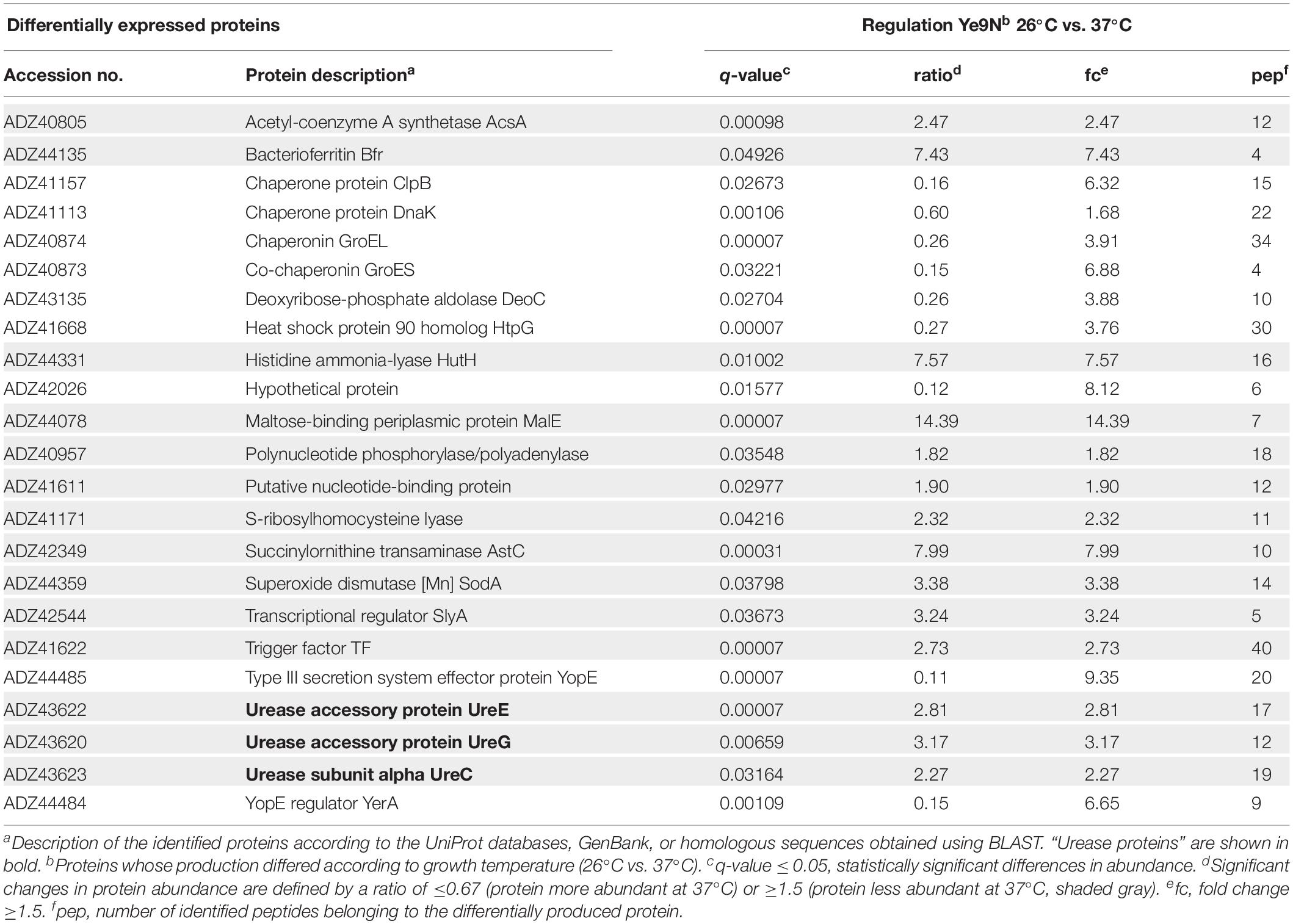
Table 1. Comparison of the cytoplasmic proteins produced by Y. enterocolitica wild-type strain Ye9N grown in LB medium at 26°C and 37°C.
Differences in Cytoplasmic Protein Abundance Between the Parental Y. enterocolitica Strain and an ompR Mutant
Shotgun Proteomic analysis was applied to identify cytoplasmic proteins subject to regulation by OmpR. We analyzed cytoplasmic fractions of the wild-type strain Ye9N and the isogenic ompR null mutant AR4, cultured at 26°C or 37°C in LB medium, by LC-MS/MS analysis. The mutant AR4 (ΔompR:Km) was constructed previously by a reverse genetics PCR-based strategy (Brzostek et al., 2003). The proteomic analysis revealed 28 differentially expressed proteins (q-value ≤ 0.05, at least two peptides per protein, minimal acceptable fold change 1.5, Table 2). The major difference in cytoplasmic protein abundance between the strains at 26°C, was a more than 130-fold decrease in the level of HU-alpha, a subunit of histone-like bacterial protein important in maintenance of the nucleoid structure (Boubrik et al., 1991; Zhou et al., 2005; Kamashev et al., 2017), in the ompR mutant. This indicated that OmpR positively affects hupA gene expression. It was previously demonstrated that the Y. pestis PhoP protein, which belongs to the OmpR-family of two-component system response regulators, positively influences the production of HU-alpha (Zhou et al., 2005). Differential quantitative LC-MS/MS analysis of cytoplasmic samples revealed that urease subunit gamma UreA was less abundant (∼3-fold) in the ompR-negative strain AR4 compared with the parental strain at 26°C, suggesting positive OmpR-dependent regulation.
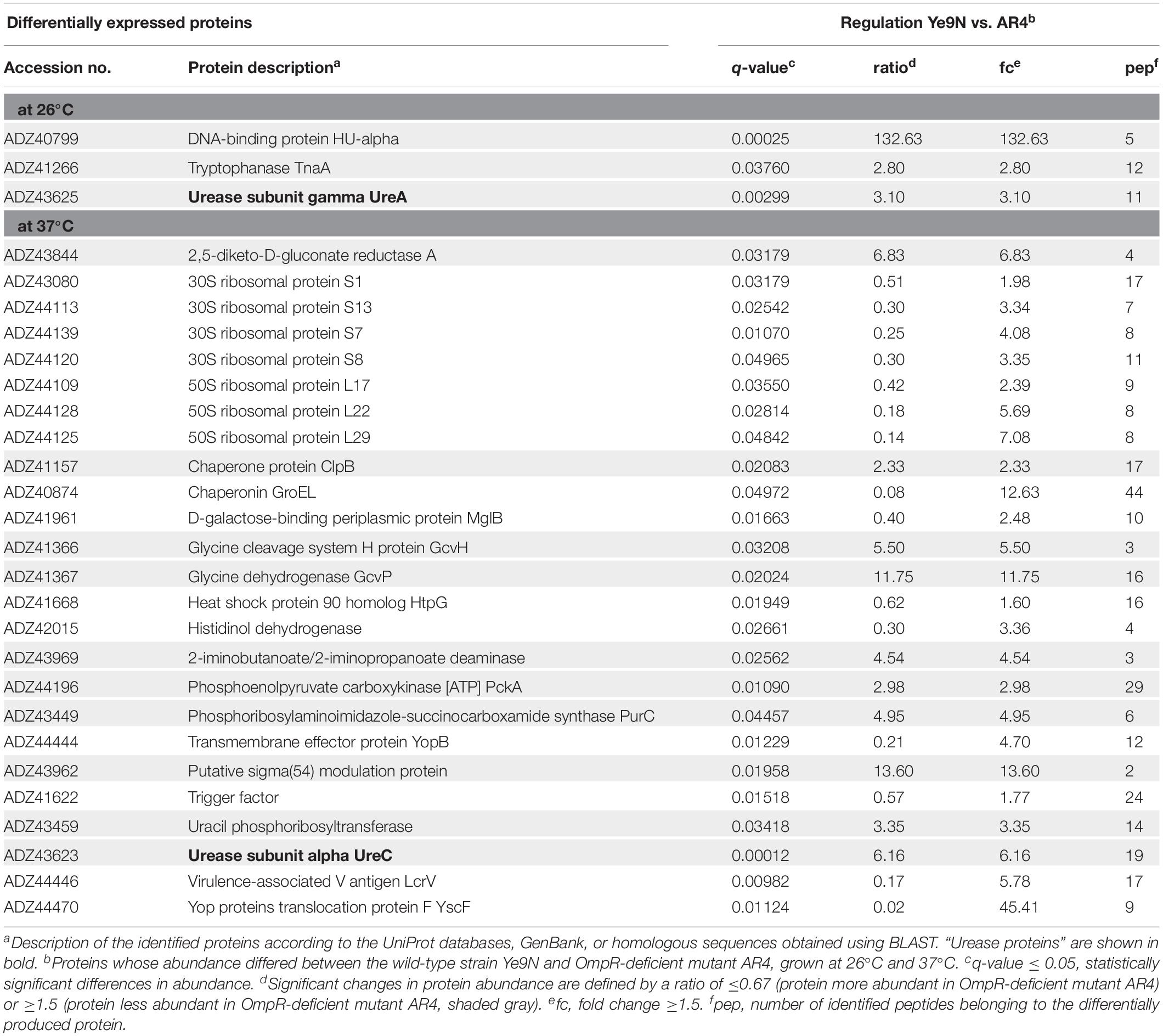
Table 2. Comparison of the cytoplasmic proteins produced by Y. enterocolitica Ye9N and isogenic ompR mutant AR4 at 26°C and 37°C.
The proteomic analysis of cytoplasmic fractions from cells grown at 37°C suggested that OmpR exerts a positive influence on the expression of the structural subunit of urease, UreC, since levels of this protein were reduced in the ompR mutant at this temperature (∼6-fold). In addition, we observed a two-fold decrease in the abundance of the chaperone ClpB in the ompR mutant. Proteins displaying reduced levels in the ompR mutant at 37°C included the putative sigma(54) modulation protein (∼13.6-fold) and the housekeeping enzymes 2,5-diketo-D-gluconate reductase A (∼6.8-fold) and glycine dehydrogenase GcvP (∼11.8-fold). In agreement with our previous finding, the absence of the OmpR regulator also caused an increase in the proteins of the Ysc-Yop T3SS (Nieckarz et al., 2016). These subunits of the Yersinia injectisome, which translocates the effector Yop proteins into host cells, include the translocation pore protein YopB (∼4.7-fold), forming a pore in the host cell membrane, the needle tip protein LcrV (∼5.8-fold) and the protein YscF, which polymerizes into a needle-like structure (45-fold) (Lee et al., 2000; Nordfelth and Wolf-Watz, 2001; Dewoody et al., 2013). Our proteomic data also revealed an increased amount of ribosomal proteins in the ompR mutant strain AR4 compared to the parent strain Ye9N at 37°C (∼ 2- to 7-fold), as well as higher levels of the chaperones GroEL (∼12.6-fold) and HtpG (∼1.6-fold).
The results of the proteomic analysis indicated that OmpR affects (both positively and negatively) the production of a number of proteins that serve a variety of functions. Notably, two of the three structural subunits of urease (UreA and UreC) were identified among the positively regulated OmpR-dependent targets.
Temperature and OmpR Influence the Production of Urease in Y. enterocolitica
To investigate the influence of OmpR on the production of urease by Y. enterocolitica we examined the activity of this enzyme in the wild-type strains Ye9N (a low-level pathogenic strain of bio-serotype 2/O:9) and Ye8N (a high-level pathogenic strain of bio-serotype 1B/O:8), and their respective mutants AR4 (ΔompR:Km) and KJ4. The mutant KJ4, lacking the ompB operon (ΔompRenvZ:Tp), was constructed in the present study by targeted deletion via homologous recombination (see MATERIALS AND METHODS).
Ureolytic activity was estimated in a semi-quantitative manner by plating Y. enterocolitica strains on Christensen’s urea agar and monitoring the change in color after 24-h incubation at 26°C or 37°C (Figure 1A). Higher urease activity was observed in strain Ye9N than in Ye8N at both temperatures. Moreover, the ureolytic activity of Ye9N was higher at 26°C than 37°C. The ompR mutant (AR4) exhibited a decreased level of urease production compared to Ye9N at both temperatures. In contrast, no visible difference was observed between Ye8N and KJ4 (ompB), irrespective of the growth temperature. To confirm that OmpR promotes urease production in Ye9N, plasmid pompR carrying the wild-type ompR allele was used to complement the mutation in strain AR4. Complementation caused a significant increase in urease activity in this ompR mutant at 26°C but not at 37°C. However, expression of the ompB operon in trans (plasmid pompB carrying both the ompR and envZ genes) in the ompB mutant KJ4, did not affect urease activity at either of the tested temperatures.
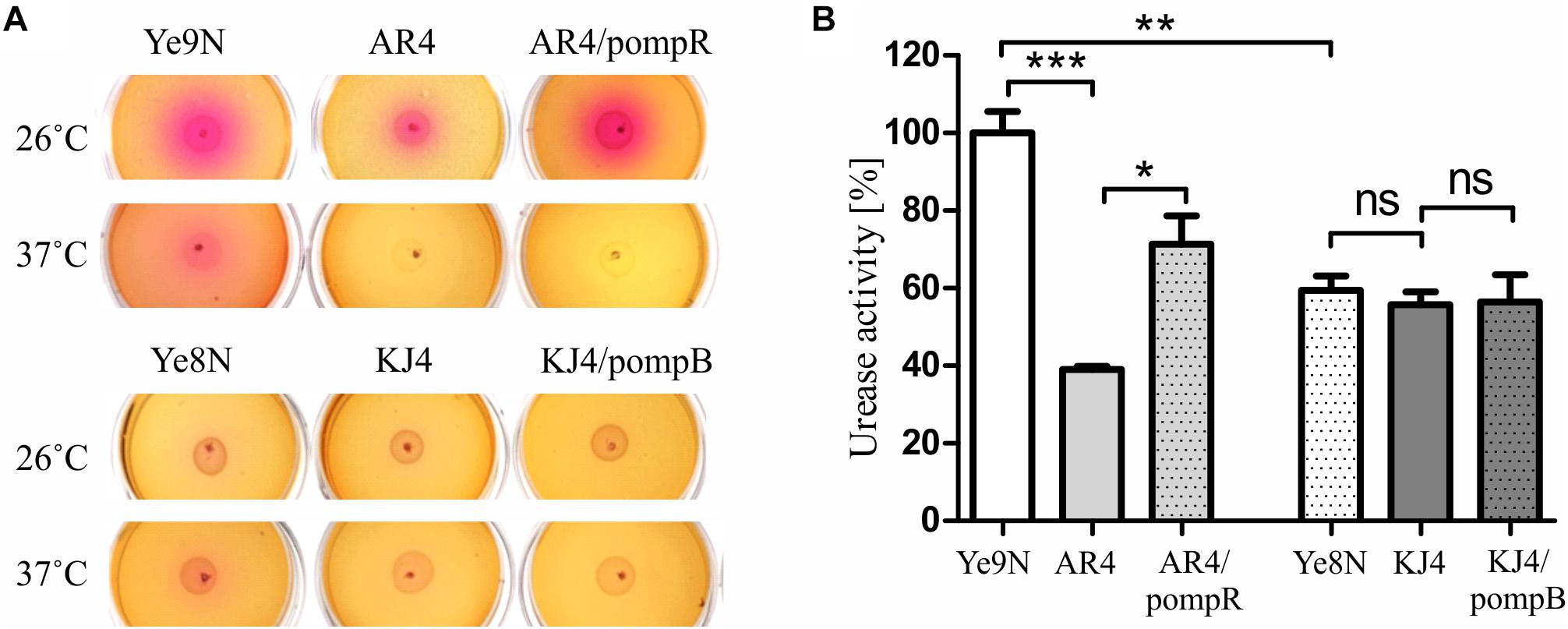
Figure 1. Changes in urease activity in Y. enterocolitica strains differing in their OmpR content. Ureolytic activity was compared in the strains Ye9N (wt), AR4 (ompR), complemented AR4 (ompR/pompR), Ye8N (wt), KJ4 (ompB), complemented KJ4 (ompB/pompB) (A) Results of a semi-quantitative urease activity assay based on a color change in Christensen’s urea agar after 24-h growth at 26°C or 37°C. (B) Quantitative urease activity determined using an assay based on the measurement of ammonia formed by enzymatic hydrolysis of urea in cultures grown at 26°C in Christensen’s medium supplemented with 2% of urea, using Nessler’s reagent. The concentration of ammonium ions is presented as a percentage, taking the value for the wild-type strain Ye9N as 100%. Significance was calculated using Student’s unpaired t-test (***P ≤ 0.001; **P ≤ 0.01; and *P ≤ 0.05; ns, not significant, P > 0.05). Data are from three independent experiments.
To further examine the impact of OmpR on the production of urease we employed a quantitative assay using Nessler’s reagent. The results presented in Figure 1B confirmed the Christensen’s urea agar data. Strain Ye9N displayed the highest urease activity, and that detected in the ompR-deficient strain AR4 was significantly lower (2.6-fold). Moreover, complementation of the ompR mutation in strain AR4 caused a clearly visible increase in urease activity (1.8-fold). There was no difference in urease activity between strains Ye8N, mutant KJ4 and KJ4/pompB. These findings showed that the level of ureolytic activity is dependent on the Y. enterocolitica strain, the presence of OmpR protein, as well as the temperature.
OmpR Enhances the Survival of Y. enterocolitica Strains Ye9N and Ye8N at Acidic pH, but to Different Degrees
To examine whether there are other phenotypic differences between the Y. enterocolitica strains Ye9N and Ye8N, their survival abilities at acidic pH was analyzed. It was previously demonstrated that the regulator OmpR is necessary for the survival of Y. enterocolitica strains of bio-serotypes 1B/O:8 and 2/O:9 in acidic conditions in vitro (Dorrell et al., 1998; Brzostek et al., 2003). However, in these separate studies, the acid stress conditions and temperatures applied to these strains were different. Thus, it was thought necessary to examine the effect of OmpR on the susceptibility to acid stress of strains Ye9N and Ye8N by studying them in parallel. The acid survival abilities of these strains and their OmpR-deficient mutants were analyzed by exposing cells grown at 26°C or 37°C to pH 4.0 for 90 min, and then determining the viable cell number. This was expressed as a percentage of the number of viable cells following exposure to pH 7.0 for the same length of time (Figure 2). Compared to wild-type strain Ye9N, its ompR mutant AR4 displayed decreases in% survival in acid conditions of 46% at 26°C and 26% at 37°C (Figure 2A). Similarly, the OmpR-deficient mutant KJ4, exhibited decreased survival in acid conditions compared to wild-type parent strain Ye8N: by 23% at 26°C and 61% at 37°C (Figure 2B). To confirm that OmpR promotes acid survival in Y. enterocolitica, plasmids pompR and pompB carrying the wild-type ompR alleles were used to complement the mutations in strain AR4 and KJ4, respectively. Complementation caused a significant increase in survival at pH 4.0, except for strain AR4 grown at 37°C. Together, these findings showed that both Y. enterocolitica strains are able to survive acid shock at the two tested temperatures, but strain Ye9N is more acid sensitive than Ye8N at 26°C. Moreover, OmpR promotes resistance to acidic stress. Strong OmpR-dependent acid survival was observed for strain Ye9N at 26°C and for strain Ye8N at 37°C. Thus, the impact of this regulator on acid survival is both strain specific and affected by temperature.
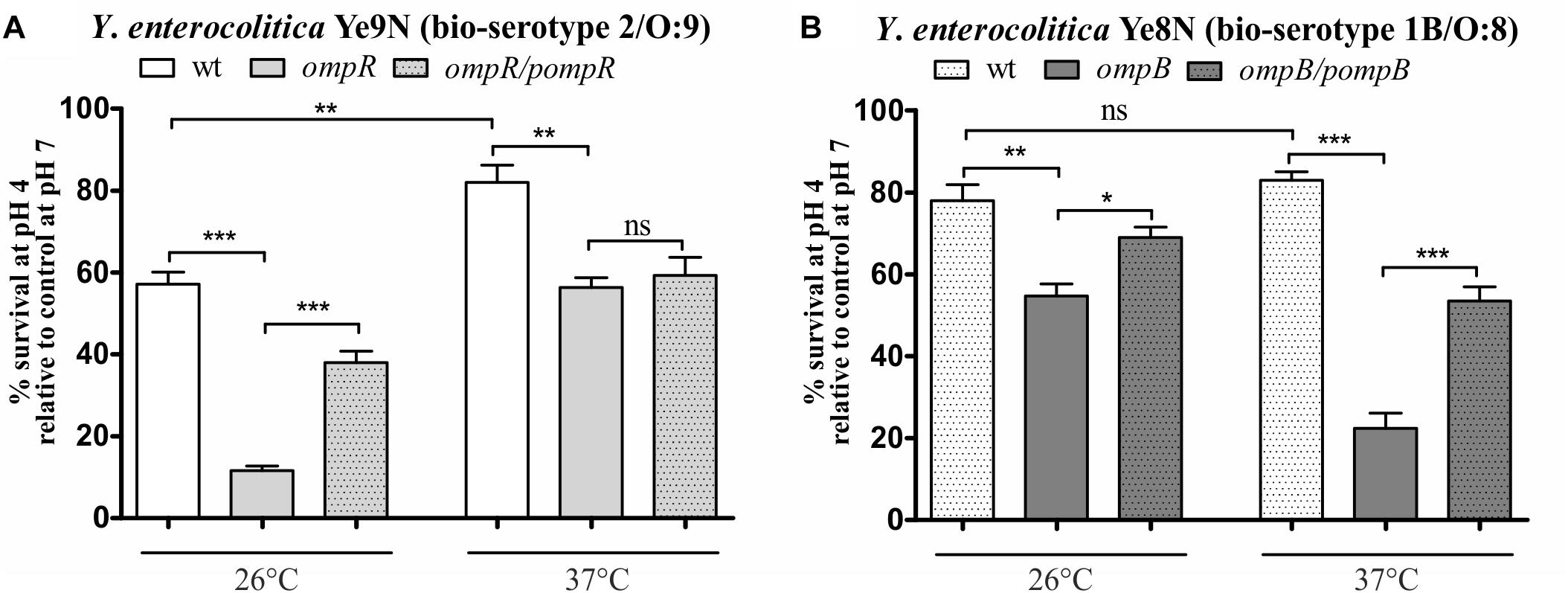
Figure 2. OmpR-dependent survival of Y. enterocolitica strains of bio-serotypes 2/O:9 (A) and 1B/O:8 (B) upon exposure to acid stress at 26°C or 37°C. The low pH sensitivity of the following strains was compared: Ye9N (2/O:9, wt), AR4 (ompR mutant), complemented AR4 (ompR/pompR), Ye8N (1B/O:8, wt), KJ4 (ompB mutant), complemented KJ4 (ompB/pompB). Bacterial viability following 90-min exposure to pH 4.0 is expressed relative to that of cells incubated in PBS at pH 7.0 (control). The data represent mean values with standard deviations, obtained in at least three independent experiments. Significance was calculated using Student’s unpaired t-test (***P ≤ 0.001; **P ≤ 0.01; and *P ≤ 0.05; ns, not significant, P > 0.05).
Genomic Organization of the ure Locus of Y. enterocolitica and Sequence Comparisons
A shotgun genome sequence has recently been generated for Y. enterocolitica strain Ye9N (2/O:9). The availability of raw sequence data files of the Ye9N genome (NCBI/GenBank: JAALCX000000000) allowed us to examine the ure locus (contig 39) and compare it to that of strain 8081 (1B/O:8, NCBI/GenBank: AM286415,Thomson et al., 2006), whose derivative is strain Ye8N. Bioinformatic analysis revealed the presence of chromosomally located multigenic ure loci in both strains (Figure 3 and Supplementary Figure S1A). Within each locus, the ure genes are all in the same transcriptional orientation. In these strains, the ure locus is flanked by conserved genes: upstream is a gene encoding a hypothetical protein, while downstream there is the yut gene encoding a urea transporter (Sebbane et al., 2002). Using BPROM software, three putative promoters (−10 and −35 motifs) located upstream of ureA, ureE and ureG were identified within the urease cluster of both strains (Figure 3 and Supplementary Figure S1A), suggesting its organization in three operons, ureABC, ureEF and ureGD, encoding the structural (UreABC) and accessory/regulatory proteins (UreEF and UreGD). Alignment of the nucleotide sequences of ureABC, ureEF and ureGD of strain Ye9N with whose of strain 8081 showed 97%, 98% and 99% sequence identity, respectively. Nucleotide sequence alignments of the intergenic region upstream of ureABC of the analyzed strains revealed 98% sequence identity. Analogous alignments of the intergenic regions upstream of ureEF showed 99% identity, while sequences upstream of ureGD exhibited 99% identity.
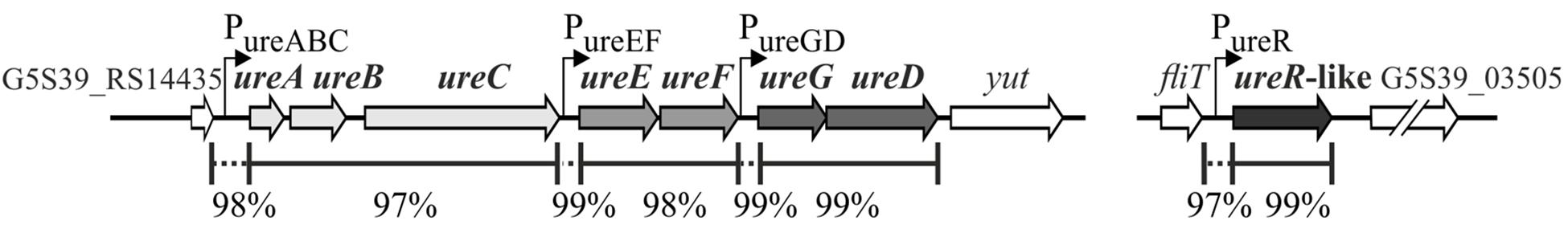
Figure 3. The genomic organization of the ure gene cluster and ureR-like gene in Y. enterocolitica strain Ye9N (2/O:9) (NCBI/GenBank: JAALCX010000039 and NCBI/GenBank: JAALCX010000005, respectively). Percentage identity of the DNA sequences of the Ye9N strain with the equivalent sequences of Ye8N (1B/O:8) (derivative of 8081, NCBI/GenBank: AM286415.1) is shown. Intergenic regions between the operons/genes are marked by dotted lines. The ure gene cluster is flanked upstream by the gene encoding a hypothetical protein (G5S39_RS14435) and downstream by the yut gene. The ureR-like gene is localized between the fliT gene and the gene encoding a metal-dependent phosphohydrolase (G5S39_03505). The positions of putative promoters are indicated by arrows. The locus tags in the Ye9N strain are indicated.
The sequence conservation of polypeptides comprising the urease system of both strains was examined further. The urease structural proteins UreA, UreB and UreC, as well as the accessory/regulatory proteins of the Y. enterocolitica strain Ye9N exhibit a high degree of amino acid sequence identity to those encoded by the corresponding open reading frames (ORFs) of Y. enterocolitica 8081 (98−100%) (data not shown).
Finally, using BLASTp searches to analyze the genomes of both Y. enterocolitica strains we identified a putative transcriptional regulator of the AraC family (289 aa) with homology to the regulator UreR identified in other ureolytic bacteria. Amino acid sequence alignment of this protein (strain 8081, GenBank: CAL12570) revealed 71% similarity (56% identity) to the urease operon transcriptional activator of Serratia ficaria (GenBank: SNW02772.1) and 43% similarity (22% identity) to UreR of P. mirabilis (GenBank: Z18752.1). Therefore, for consistency, this AraC-like protein of Y. enterocolitica will be termed UreR-like from here on. In silico analysis of this protein sequence using the MOTIF online tool (GenomeNet, Japan), identified helix-turn-helix motifs and an AraC-like binding domain.
The UreR-like protein of both strain 8081 (1B/O:8) (NCBI/GenBank: AM286415.1) and strain Ye9N (contig 5) is encoded by a gene located within a cluster of flagellar structural and assembly genes (downstream of the fliT gene and upstream of the gene encoding metal-dependent phosphohydrolase), and not in the vicinity of the ure cluster (Figure 3 and Supplementary Figure S1B). The nucleotide sequence identity of ureR-like genes of strains Ye9N and 8081 is 99% (and Supplementary Figure S1B), while that of the intergenic regions upstream of the ureR-like genes is 97%.
OmpR Positively Affects the Expression of ureABC, ureEF, and ureGD in Y. enterocolitica Strain Ye9N
To examine the impact of regulator OmpR on the activity of urease operon promoters in different Y. enterocolitica strains, we constructed transcriptional fusions of the putative promoter sequences with the lacZ gene in plasmid pCM132Gm. The PureABC, PureEF, and PureGD promoters located upstream of the corresponding operons were identified using the bacterial promoter prediction program BPROM (Solovyev and Salamov, 2011). The three reporter gene constructs were introduced into the wild-type Y. enterocolitica strains Ye9N and Ye8N, and their respective OmpR-deficient derivatives AR4 (ompR mutant) and KJ4 (ompB mutant). The expression of the transcriptional fusions was quantified by measuring β-galactosidase activity in cells grown to stationary phase at 26°C or 37°C in LB medium (Figure 4). The activity of the ureABC promoter in the ompR mutant (AR4 strain) was lower than in the wild-type strain Ye9N at 26°C and 37°C (19 and 33% lower, respectively) (Figure 4A). To confirm positive OmpR-dependent regulation of this urease operon in strain Ye9N, a plasmid carrying a wild-type copy of the ompR gene (pompR in Figure 4) was used to complement the ompR mutation in strain AR4. This caused a statistically significant increase in the level of β-galactosidase activity from ureABC:lacZ at both tested temperatures, indicating that OmpR acts as a positive regulator of the operon encoding the three structural subunits of urease in this strain. The positive regulatory effect of OmpR was also observed for fusions carrying the promoter regions of the ureEF (Figure 4B) and ureGD (Figure 4C) operons. Reduced activity of ureEF:lacZ (18% and 24% decrease in activity) and ureGD:lacZ (38% and 17% decrease in activity) fusions was observed in the ompR mutant strain AR4 compared to the wild-type Ye9N at 26°C and 37°C, respectively. However, an effect of complementation with plasmid pompR was only detected for the reporter fusion ureGD:lacZ at 37°C. Interestingly, the OmpR-dependent regulation of urease operon transcription was not observed in Y. enterocolitica Ye8N, at either 26°C or 37°C. However, the β-galactosidase activity of ureABC:lacZ was increased in complemented strain KJ4/pompB at both temperatures. We concluded that OmpR positively and specifically influences ure expression in Y. enterocolitica strain Ye9N, but its role is less clear in strain Ye8N.
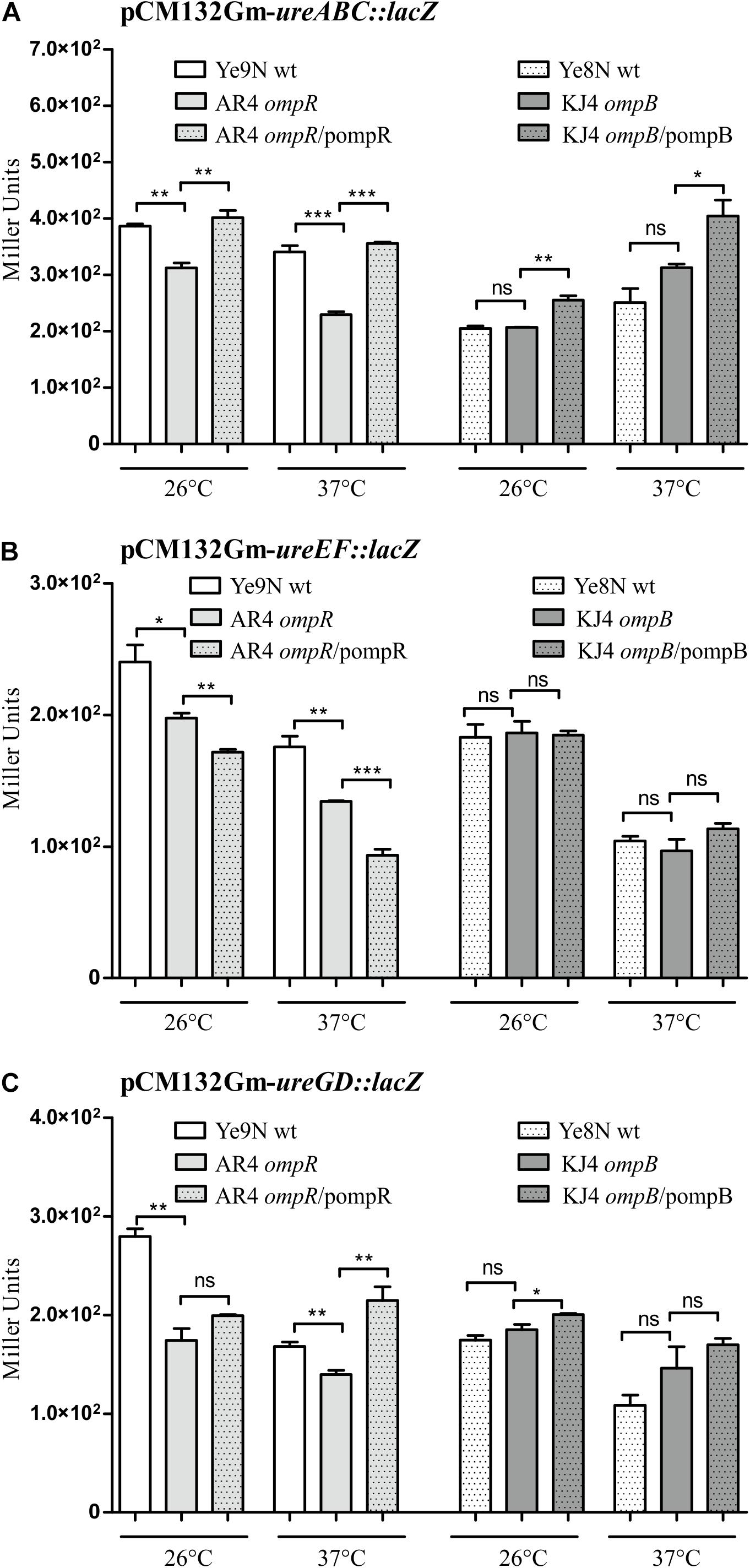
Figure 4. OmpR-dependent regulation of ureABC, ureEF and ureGD operon expression in Y. enterocolitica. β-galactosidase activity of strains carrying transcriptional fusions with lacZ expressed from plasmids pCM132Gm-ureABC:lacZ (A), pCM132Gm-ureEF:lacZ (B) and pCM132Gm-ureGD:lacZ (C), with or without OmpR, i.e., Ye9N (wt), AR4 (ompR mutant), AR4/pompR (complemented ompR mutant, ompR/pompR), Ye8N (wt), KJ4 (ompB mutant), KJ4/pompB (complemented ompB mutant, ompB/pompB). Strains were grown at 26°C or 37°C to stationary phase in LB medium. The data represent mean β-galactosidase activity values (Miller units) with the standard deviation from at least two independent experiments, each performed using at least triplicate cultures of each strain. Significance was calculated using Student’s unpaired t-test (***P ≤ 0.001; **P ≤ 0.01; and *P ≤ 0.05; ns, not significant, P > 0.05).
With regard to the influence of temperature on urease promoter activity, β-galactosidase production by each urease fusion was higher at 26°C compared to 37°C in Y. enterocolitica strain Ye9N: by 12% for ureABC:lacZ, 27% for ureEF:lacZ and 40% for ureGD:lacZ. In the case of strain Ye8N, temperature had no significant effect on expression of the ureABC reporter fusion. However, for ureEF:lacZ and ureGD:lacZ, lower β-galactosidase activity was observed at 37°C compared to 26°C. Furthermore, we found higher ure operon expression in strain Ye9N than in Ye8N at both tested temperatures. These data indicated OmpR- and temperature-dependent modulation of the Y. enterocolitica Ye9N ure operons that parallels urease activity (see Figure 1).
The expression of many OmpR regulon genes in E. coli and Salmonella is osmoregulated and/or modulated in response to pH conditions (Chakraborty and Kenney, 2018). Next, we tested whether the expression of urease transcriptional units changes in response to high osmolarity and/or acidic conditions. Moreover, we were curious to see if urea could influence ure cluster gene expression. The effect of 350 mM NaCl, pH 4.5 and 100 mM urea on the β-galactosidase activity produced by ure operon:lacZ transcriptional fusions was tested in stationary phase cells of the wild-type Y. enterocolitica strains Ye9N and Ye8N grown at 26°C (Figure 5). No effect of high osmolarity (350 mM NaCl) or low pH (pH 4.5) was observed for the ureABC:lacZ fusion in strains Ye9N and Ye8N (Figure 5A). Only the addition of 100 mM urea caused a slight induction of ureABC expression in both tested strains. Measurements of β-galactosidase activity demonstrated that ureEF:lacZ expression was lower in strain Ye9N grown under acidic and high osmolarity conditions (Figure 5B). For strain Ye8N, a decrease in the expression of ureEF:lacZ was only observed in the presence of 350 mM NaCl. The addition of 100 mM urea significantly upregulated ureEF:lacZ expression only in strain Ye8N. The ureEF pattern of expression in response to high osmolarity, low pH and urea was also observed for the ureGD:lacZ fusion in strain Ye9N (Figure 5C). In strain Ye8N, significant changes in ureGD:lacZ expression were recorded in cells grown at pH 4.5 (downregulation) and in the presence of urea (upregulation).
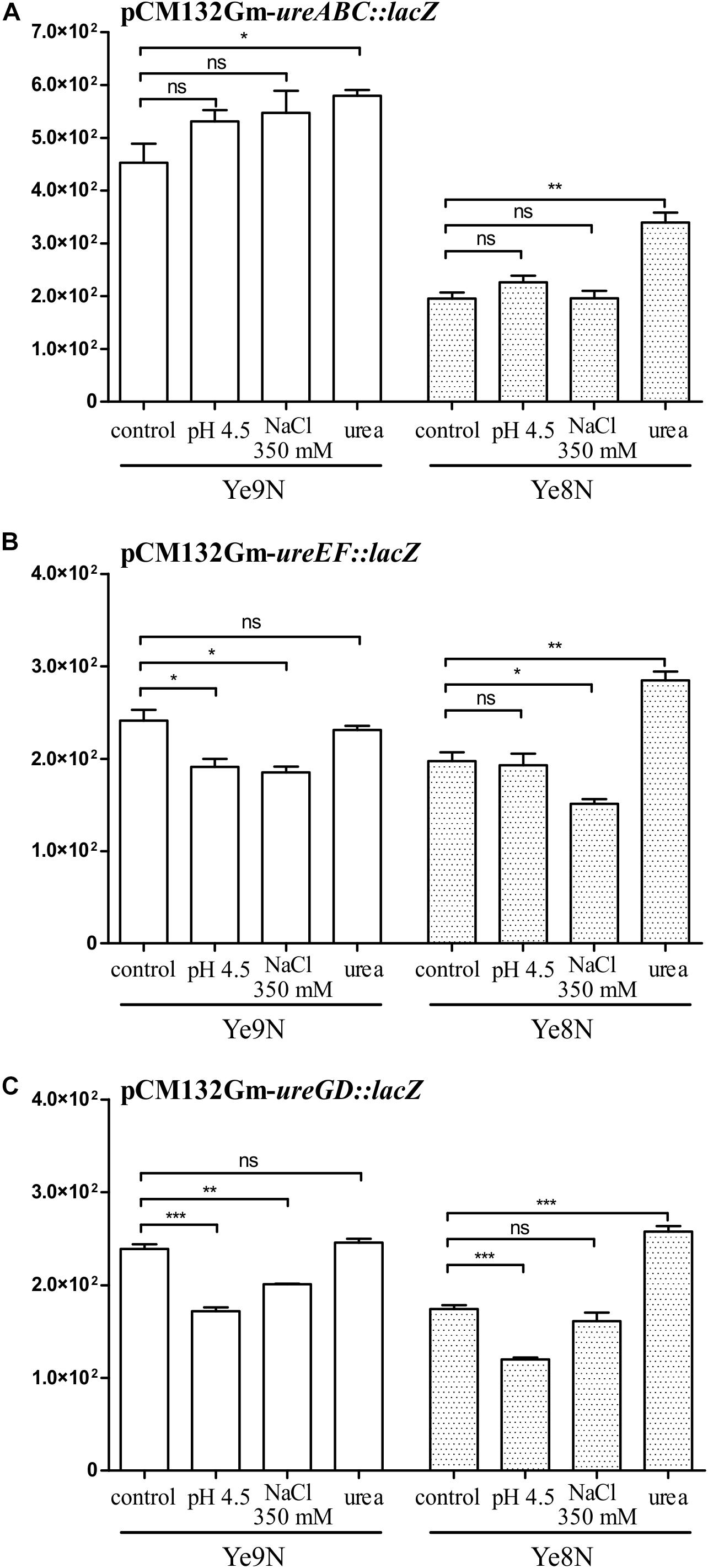
Figure 5. The effect of acid pH, high osmolarity and urea on the expression of the ureABC, ureEF and ureGD operons of Y. enterocolitica. β-galactosidase activity of strains Ye9N and Ye8N carrying transcriptional fusions with lacZ expressed from plasmids pCM132Gm-ureABC:lacZ (A), pCM132Gm-ureEF:lacZ (B), and pCM132Gm-ureGD:lacZ (C) was analyzed after incubating stationary phase cells at 26°C for 2 h in LB medium (86 mM NaCl, pH 7.0), LB adjusted to pH 4.5, LB supplemented with NaCl to a final concentration of 350 mM or LB containing 100 mM urea. The data represent mean β-galactosidase activity values (Miller units) with the standard deviation from at least two independent experiments, each performed using at least triplicate cultures of each strain. Significance was calculated using Student’s unpaired t-test (***P ≤ 0.001; **P ≤ 0.01; and *P ≤ 0.05; ns, not significant, P > 0.05).
OmpR Regulates the Expression of a Gene Encoding a UreR-Like Putative Transcriptional Regulator
To examine whether Y. enterocolitica ureR-like expression responds to the presence of 100 mM urea, acidic conditions (pH 4.5) or high osmolarity (350 mM NaCl), and to test the influence of the regulator OmpR, we constructed a transcriptional fusion of the putative ureR-like promoter sequence identified by BPROM (Solovyev and Salamov, 2011) with the lacZ gene in vector pCM132Gm. This construct pCM132Gm- ureR:lacZ was introduced into the strains used to test the urease operon fusions, i.e., Ye9N and Ye8N and their OmpR-deficient derivatives, AR4 (ompR mutant) and KJ4 (ompB mutant).
As shown in Figure 6A, high osmolarity had no effect on ureR:lacZ expression in either of the strains Ye9N or Ye8N. A 2-h exposure to an acidic environment resulted in decreased ureR-like expression in the strain Ye9N. A slight stimulatory effect of urea (1.2-fold) was observed, but only in the strain Ye8N, which parallels the induction of ure gene expression by urea seen in this strain.
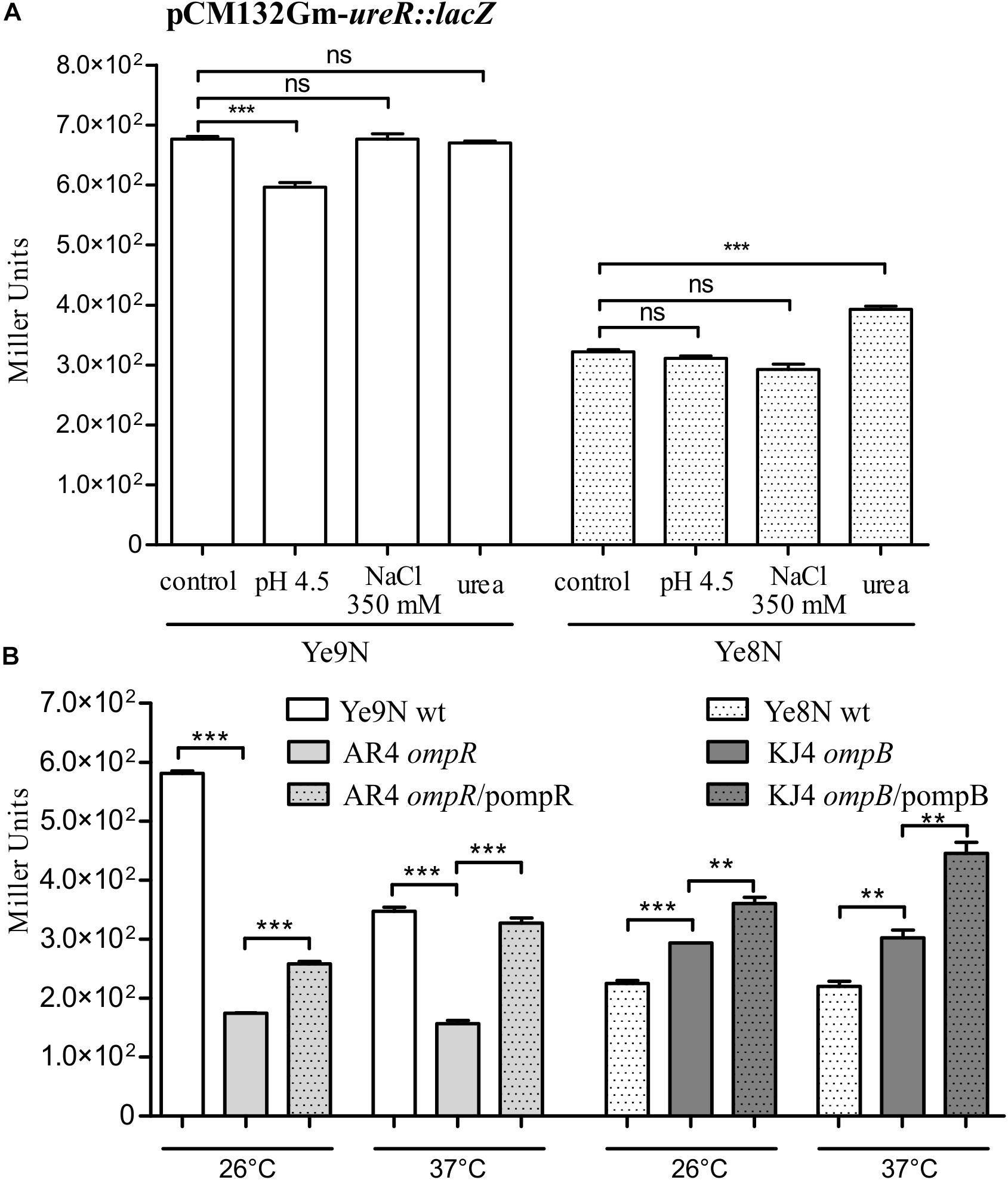
Figure 6. Analysis of ureR-like gene expression in Y. enterocolitica by measuring the β-galactosidase activity of strains carrying the plasmid pCM132Gm-ureR:lacZ. (A) The effect of acid pH, high osmolarity and urea on the ureR-like expression in Y. enterocolitica strains Ye9N and Ye8N was analyzed after incubating stationary phase cells at 26°C for 2 h in LB (86 mM NaCl, pH 7.0; control), LB adjusted to pH 4.5, LB supplemented with NaCl to a final concentration of 350 mM or LB containing 100 mM urea. (B) OmpR-dependent regulation of ureR-like gene expression in Ye9N (wt), AR4 (ompR mutant), AR4/pompR (complemented ompR mutant, ompR/pompR), Ye8N (wt), KJ4 (ompB mutant) and KJ4/pompB (complemented ompB mutant, ompB/pompB). Strains were grown at 26°C or 37°C to stationary phase in LB medium. The data represent mean β-galactosidase activity values (Miller units) with the standard deviation from at least two independent experiments, each performed using at least triplicate cultures of each strain. Significance was calculated using Student’s unpaired t-test (***P ≤ 0.001 and **P ≤ 0.01; ns, not significant, P > 0.05).
The ureR:lacZβ-galactosidase activity detected in the ompR mutant (strain AR4) was lower than that measured in the wild-type strain Ye9N at 26°C or 37°C (3.3- and 2.2-fold lower, respectively), and this effect was complemented by the wild-type ompR allele provided in trans (Figure 6B). These data suggested that OmpR is a positive regulator of the ureR-like gene in strain Ye9N. Interestingly, we found that the absence of the ompB in strain KJ4 resulted in the upregulation of ureR-like gene expression at both tested temperatures (by 1.3-fold at 26°C and 1.4-fold at 37°C). However, the addition of plasmid-encoded ompB to strain KJ4 did not complement the ompB deletion and even upregulated the ureR-like gene expression. These findings indicated that OmpR has the opposite regulatory effect on ureR-like gene expression in the two tested Y. enterocolitica strains, i.e., positive in Ye9N and negative in Ye8N.
OmpR Binds Directly to the ureABC, ureEF and ureR-Like Regulatory Regions
We next tested whether OmpR may influence ure operon expression directly. In silico analysis of the regulatory sequence of ureABC (Figure 7A left panel) identified two 20-bp putative OmpR-binding sites (OBS), with 45% and 40% homology to the E. coli OmpR consensus sequence [TTTTACTTTTTG(A/T)AACATAT] (Maeda et al., 1991; Harlocker et al., 1995). No potential OBS were recognized in either the ureEF or the ureGD regulatory regions.
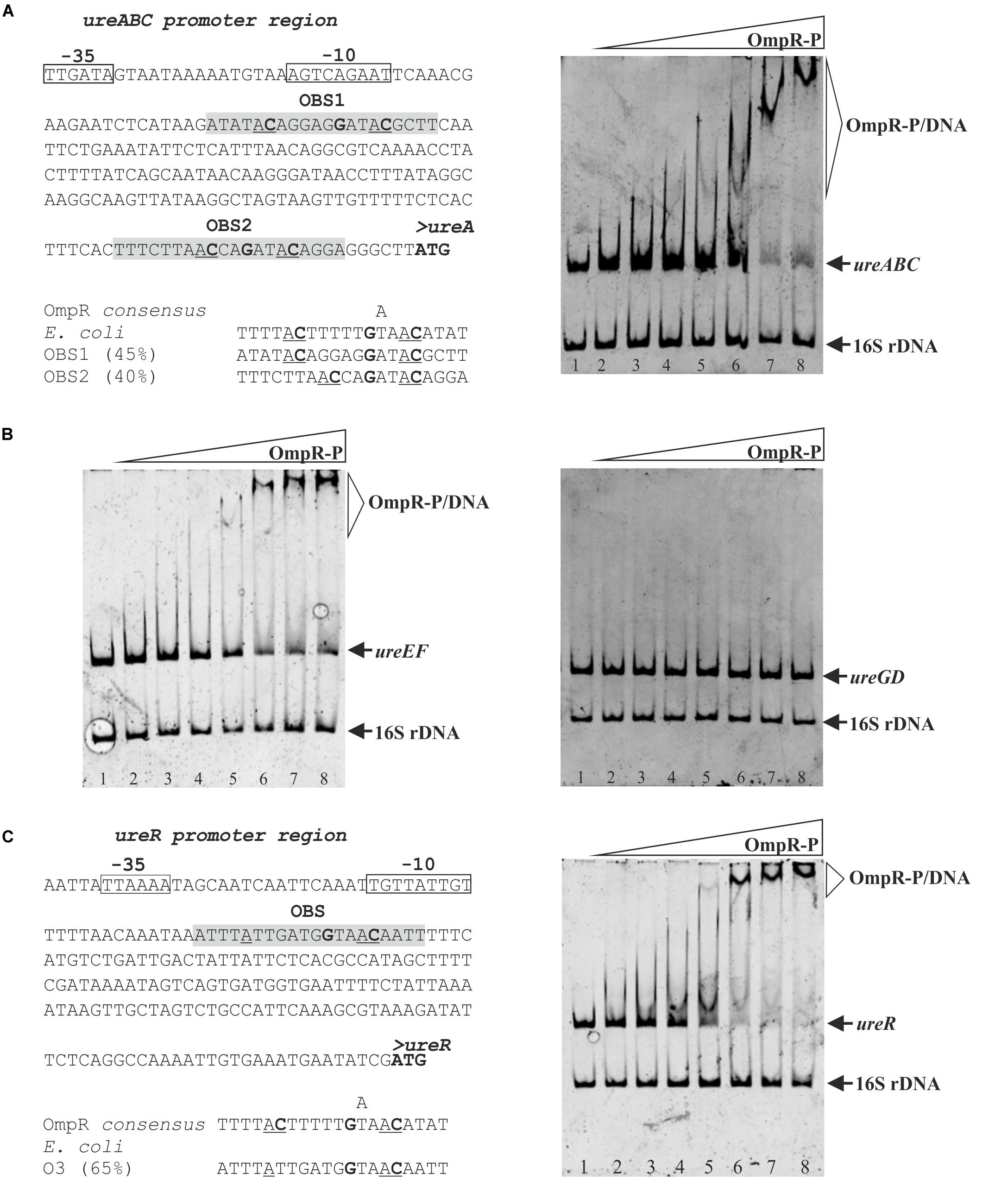
Figure 7. Interaction of OmpR with the ureABC, ureEF, ureGD and ureR-like regulatory regions of Y. enterocolitica. (A, left panel) Putative OmpR-binding sites (OBS) in the promoter region of the ureABC operon of Y. enterocolitica identified based on similarity to the E. coli consensus sequence (% identity values are shown). (A, right panel) Binding of phosphorylated OmpR to the ureABC regulatory region examined using an EMSA. (B) Binding of phosphorylated OmpR to the ureEF and ureGD regulatory regions examined using EMSAs. (C, left panel) Putative OmpR-binding site in the promoter of the ureR-like gene of Y. enterocolitica (% identity to the E. coli consensus sequence is shown) (C, right panel) interaction of OmpR with the ureR-like regulatory region examined using an EMSA. In the in silico analysis (A,C, left panels) the start codons are shown in bold. The likely –10 and –35 promoter elements are marked. The potential OBS in the promoter regions of ureABC and the ureR-like gene are shaded gray with the central GXXAC motifs marked. In the EMSAs, different concentrations of OmpR-P were incubated with DNA fragments representing the ureABC (312 bp), ureEF (385 bp), ureGD (375 bp), and ureR-like (363 bp) regulatory regions. A 16S rDNA fragment (211 bp) was included in each reaction as a non-specific binding control. The reaction mixtures contained 0.05 pmol of each fragment and increasing amounts of OmpR-P: for the ure operon fragments (A,B), lanes 1 – no protein, lanes 2 – 0.93 μM, lanes 3 – 1.24 μM, lanes 4 – 1.85 μM, lanes 5 – 2.47 μM, lanes 6 – 3.71 μM, lanes 7 – 4.33 μM, and lanes 8 – 4.95 μM; for the ureR-like fragment (C), lane 1 – no protein, lane 2 – 0.31 μM, lane 3 – 0.46 μM, lane 4 – 0.62 μM, lane 5 – 0.77 μM, lane 6 – 0.93 μM, lane 7 – 1.85 μM, and lane 8 – 2.78 μM. The identities of the bands resolved by electrophoresis on native 4.2% polyacrylamide gels are indicated.
To determine whether OmpR could modulate ureABC, ureEF and ureGD expression by directly binding to the regulatory regions of these operons, we employed electrophoretic mobility shift assays (EMSAs) (Figure 7A right panel and 7B). DNA fragments containing the regulatory regions of the three urease operons were amplified by PCR and used in EMSAs with increasing concentrations of phosphorylated OmpR-His6 (OmpR-P). A specific shifted DNA-protein complex was formed by the interaction between the ureABC regulatory region fragment and phosphorylated OmpR (Figure 7A right panel). We observed a stepwise increase in shifted bands of decreasing mobility as the OmpR concentration was increased from 2.47 μM up to 4.95 μM, suggesting that more than one binding site is present. Although no potential OBS were identified in the ureEF promoter sequence, phosphorylated OmpR also interacted with this regulatory region and a slower migrating DNA-protein band appeared at an OmpR-P concentration of 2.47 μM (Figure 7B left panel). As shown in Figure 7B right panel, OmpR-P did not interact with the regulatory region of ureGD, which is consistent with the apparent absence of OBS. OmpR was unable to bind the negative control 16S rDNA fragment included in each binding reaction.
Following the demonstration that OmpR regulates ureR-like gene expression in Y. enterocolitica, we hypothesized that this regulator may do so directly. One potential OBS was identified in the ureR-like gene regulatory region by in silico analysis (Figure 7C left panel). The predicted OBS exhibits high sequence identity (65%) to the consensus OmpR-binding sequence of E. coli. An EMSA was then used to examine the ability of the phosphorylated OmpR regulator to bind to the ureR-like regulatory region in vitro (Figure 7C right panel). This analysis revealed the appearance of specific slower migrating OmpR-DNA complexes concomitantly with the disappearance of the ureR-like regulatory region fragment band. In comparison with the EMSAs performed using the ureABC and ureEF regulatory region fragments (Figures 7A,B), the OmpR-P/ureR-like complexes were formed at a lower OmpR concentration (i.e., at 0.77 μM rather than 2.47 μM) suggesting higher binding affinity of OmpR for the ureR-like target.
Discussion
This study started by examining the impact of OmpR, the response regulator of the two-component signal transduction system EnvZ/OmpR, on the cytoplasmic proteome of Y. enterocolitica Ye9N (bio-serotype 2/O:9) at 26°C (optimum temperature for growth) and 37°C (the body temperature of mammalian hosts). The results of the proteomic analysis indicated that temperature and OmpR affect (both positively and negatively) the production of a number of proteins that serve a variety of functions. OmpR was found to be involved in the expression of some regulatory proteins, including HU-alpha, sigma 54 and TF (Dillon and Dorman, 2010; Hoffmann et al., 2010; Danson et al., 2019), as well as effector proteins of the Type III Secretion System, the major virulence factors of pathogenic Y. enterocolitica (Cornelis, 2002). Thus, OmpR appears to influence the adaptive and virulence properties of Y. enterocolitica.
Interestingly, differential analysis of the cytoplasmic proteome at 26°C and 37°C identified components of the urease system among the set of temperature-regulated proteins. The levels of UreC, UreE and UreG were reduced at 37°C compared to 26°C, which is in line with the findings of a study carried out on the low pathogenic Y. enterocolitica strain W227 serotype O:9 (De Koning-Ward and Robins-Browne, 1997). It was suggested previously that higher expression of urease at moderate temperature, might be important for the adaptive ability of Y. enterocolitica during the earliest stage of pathogenesis and assists passage through the acidic environment of the stomach (Young et al., 1996; De Koning-Ward and Robins-Browne, 1997). It has also been proposed that low-temperature-induced expression of the ure genes observed in strain W22703 (bio-serotype 2/O:9) may play a role in the adaptive abilities of low-pathogenic Y. enterocolitica strains during bacteria-insect interactions (Heermann and Fuchs, 2008; Fuchs et al., 2011).
A significant finding of our quantitative proteomic analysis is that the regulator OmpR positively affects the abundance of components of the Y. enterocolitica urease system at both tested temperatures (26°C and 37°C). The impact of the OmpR regulator on urease expression was examined in more detail in Y. enterocolitica strain Ye9N of bio-serotype 2/O:9 and strain Ye8N of bio-serotype 1B/O:8 (derivative of clinical isolate 8081). The high-level pathogenic strain 8081, isolated in the United States, is the most studied Y. enterocolitica strain with respect to its virulence factors and their contribution to pathogenesis in mammals (Portnoy et al., 1981). The low-level pathogenic strain Ye9N (bio-serotype 2/O:9), isolated in Poland, has been the research object of many studies, especially concerning the role of OmpR in regulating the virulence properties and adaptive abilities of Y. enterocolitica (Brzostek et al., 2003, 2007; Raczkowska et al., 2011, 2015; Skorek et al., 2013; Nieckarz et al., 2016, 2017).
Upon analyzing these two strains in parallel, higher levels of urease activity were observed in Ye9N than in Ye8N, and in cells grown at 26°C compared to 37°C. More interestingly, OmpR exerted a positive impact on urease activity in the strain Ye9N grown at 26°C. At 37°C this effect was less obvious since a wild-type copy of ompR expressed from a plasmid did not complement the decrease in urease activity caused by the ompR mutation. This suggests that a more complex regulatory mechanism influences the production/activity of urease at 37°C in the presence of OmpR supplied in trans. No positive impact of OmpR on urease activity was observed in strain Ye8N, irrespective of the growth temperature. Thus, OmpR-dependent regulation of urease activity seems to be characteristic of particular Y. enterocolitica strains. Phenotypic variations among closely related strains of Y. enterocolitica, even those belonging to the same bio-serotype, have been highlighted previously (Garzetti et al., 2012).
Other phenotypic studies of Y. enterocolitica strains Ye9N and Ye8N have demonstrated that OmpR increases, to a varying degree, the survival of both strains in acidic conditions. Since these acid survival tests were not performed in the presence of urea, the observed differences were not related to urease activity and this suggested distinct roles for OmpR in other acid-tolerance mechanisms operating in these bacteria at 26°C and 37°C. Intra-species differences in acid survival are not specific to Y. enterocolitica strains, since variability in acid resistance has also been reported among isolates of Salmonella Typhimurium (Berk et al., 2005). It may be concluded that the role of OmpR in acid survival of Y. enterocolitica strains is complex and involves urease-dependent and -independent mechanisms.
In the course of this study we verified the hypothesis that OmpR directly regulates expression of the ure operons in Y. enterocolitica. Firstly, promoter-lacZ reporter gene fusion assays revealed that ure operon expression is higher in strain Ye9N than in Ye8N, and higher at 26°C than at 37°C, which is consistent with the proteomic data and urease activity tests. Interestingly, in the case of strain Ye8N, temperature significantly affected expression of ureEF:lacZ and ureGD:lacZ reporter fusions but not ureABC:lacZ. Thus, the overall thermoregulation of urease activity depends on the expression of the structural genes. Secondly, we demonstrated that OmpR acts as a positive regulator of the ureABC, ureEF and ureGD operons in strain Ye9N, both at 26°C and 37°C. The OmpR-dependent regulation of urease cluster transcription was not observed in strain Ye8N, even though the ure loci of both strains exhibit high nucleotide sequence identity (98%). It has been relatively well established that although Y. enterocolitica strains possess the same virulence factors, their expression profiles can differ significantly (Schmühl et al., 2019). This may be the consequence of the finding that even small alterations in the content and organization of the genetic information may alter the expression pattern of virulence genes and their regulators in response to environmental factors (Uliczka et al., 2011; Erhardt and Dersch, 2015).
The generally accepted mechanism of OmpR activation in bacteria is phosphorylation of this regulator by its partner kinase EnvZ in response to high osmolarity and low pH (Russo and Silhavy, 1990; Hoch and Silhavy, 1995; Schwan et al., 2002; Rentschler et al., 2013). This study has revealed that OmpR positively regulates expression of the ureABC, ureEF and ureGD operons in Y. enterocolitica strain Ye9N. However, no effect of high osmolarity (350 mM NaCl) or low pH (pH 4.5) was observed in the case of the ureABC operon encoding the urease subunits. We presume that a combination of environmental signals or some other stimuli activate OmpR to regulate ureABC expression. It was previously suggested that the activation of Y. enterocolitica urease by low pH is not due to increased ure gene expression (Young et al., 1996). Interestingly, the acid-induction of urease cluster transcriptional units has been demonstrated in Y. pseudotuberculosis strain YPIII, and OmpR was identified as playing a key role in this regulation (Hu et al., 2009). However, Y. pseudotuberculosis differs considerably from Y. enterocolitica in genomic content and the degree of pathogenicity (Galindo et al., 2011) and it is distantly related from an evolutionary point of view (McNally et al., 2016; Tan et al., 2016). Moreover, in contrast to Y. enterocolitica O:9, urease does not seem to be involved in the virulence of Y. pseudotuberculosis (Riot et al., 1997).
Despite the failure to detect induction by acidic conditions, slightly higher expression of ureABC in strain Ye9N and of all ure operons in strain Ye8N were observed in the presence of urea. The induction of urease structural genes by urea has been reported in Proteus and Providentia species, with levels 5- to 25-fold greater than those in uninduced cultures (Mobley et al., 1995). In these species, the urease induction is mediated by the transcriptional regulator UreR, which is encoded by a gene located upstream of the ureDABCEFG cluster (Mobley et al., 1995). No such ureR-like gene is present in the vicinity of the Y. enterocolitica ure cluster. Through comparative bioinformatic analysis we identified an AraC-like protein with 71% similarity (56% identity) to the urease operon transcriptional activator of Serratia ficaria. However, amino acid sequence alignment with UreR of P. mirabilis revealed only 43% similarity (22% identity). In this study, we employed transcriptional fusion analysis and an EMSA to show that OmpR directly regulates the expression of this ureR-like gene in Y. enterocolitica strains Ye9N and Ye8N. Interestingly this gene is regulated in the opposite manner in these strains (positively in Ye9N and negatively in Ye8N), but the nature of this reciprocal regulation is unknown. Possibly, the strain-specific differences in OmpR-dependent regulation of the urease cluster that we observed are due to its opposite effect on the expression of the UreR-like regulator. The function of this putative Y. enterocolitica UreR regulator requires further investigation.
Finally, in the course of this study we verified the hypothesis that OmpR directly regulates the expression of the ure operons in Y. enterocolitica strain Ye9N. Putative OBS were identified in the regulatory regions of the studied transcriptional units based on similarity to the consensus sequence described for OmpR of E. coli (Maeda et al., 1991; Harlocker et al., 1995). The results of EMSAs demonstrated that phosphorylated OmpR can specifically bind to the ureABC and ureEF promoter regions, which suggests direct regulation. In contrast, OmpR-dependent regulation of ureGD expression seems to be indirect.
Conclusion
In summary, this study has revealed different roles for OmpR in the regulation of urease expression in two Y. enterocolitica strains representing different bio-serotypes with distinct pathogenic abilities. Thus, it has increased our understanding of the function of OmpR in the survival of Y. enterocolitica under conditions where the activity of urease is crucial.
At this time it cannot be concluded that these differences in the role of OmpR are a common characteristic of other Y. enterocolitica strains of these bio-serotypes. Further comparative analysis of multiple representatives of particular bio-serotypes is required to determine whether OmpR-dependent urease expression is a strain- or bio-serotype-specific adaptation to increase survival in the natural environment, or within certain hosts or particular host niches.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
MN designed the study and prepared the figures. MN, PK, and KJ performed the experiments. MN, AR, and KB analyzed the data. MN and KB wrote the manuscript and provided the financial support.
Funding
This work was supported by grants from the National Science Center, Poland (PRELUDIUM grant UMO-2015/19/N/NZ6/00923 and OPUS grant UMO-2011/01/B/NZ6/01845). Publication co-financed from the resources of the University of Warsaw.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Michał Dadlez’s group from the Mass Spectrometry Laboratory, IBB PAS, Warsaw, Poland, for their excellent technical support in the MS analyses.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00607/full#supplementary-material
FIGURE S1 | Characterization of the ure gene cluster and ureR-like gene in Y. enterocolitica strains: Ye9N (2/O:9), (contig 39 – NCBI/GenBank: JAALCX010000039 and contig 5 – NCBI/GenBank: JAALCX010000005) and 8081 (1B/O:8), (NCBI/GenBank: AM286415). Nucleotide sequence alignments of the ure gene cluster composed of the seven ORFs encoding the structural (UreABC) and accessory (UreEFGD) proteins of the urease system, and the intergenic regions (A) and the ureR-like gene encoding the putative UreR-like transcriptional regulator (B). The Needleman-Wunsch algorithm was used to draw the alignment. Percentage identity of the DNA sequences in the alignments is shown. The likely −10 and −35 promoter elements, indicated by BPROM software, are underlined. The start codons are marked in green and the stop codons are marked in gray. The ure gene cluster is flanked upstream by the gene encoding a hypothetical protein and downstream by the yut gene. The ureR- like gene is localized between the fliT gene and the gene encoding a metal-dependent phosphohydrolase. The locus tags in the Ye9N strain are indicated.
TABLE S1 | Strains and plasmids used in this study.
TABLE S2 | Oligonucleotide primers used in this study. The table shows the sequences of primers used for cloning, the construction of transcriptional fusions, and to generate regulatory region fragments for EMSAs.
Footnotes
References
Adeolu, M., Seema, A., Naushad, S., and Gupta, R. S. (2016). Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morgane. Int. J. Syst. Evol. Microbiol. 66, 5575–5599. doi: 10.1099/ijsem.0.001485
Aepfelbacher, M., Trasak, C., and Ruckdeschel, K. (2007). Effector functions of pathogenic Yersinia species. Thromb. Haemost. 98, 521–529. doi: 10.1160/th07-03-0173
Akopyan, K., Edgren, T., Wang-Edgren, H., Rosqvist, R., Fahlgren, A., Wolf-Watz, H., et al. (2011). Translocation of surface-localized effectors in type III secretion. Proc. Natl. Acad. Sci. U.S.A. 108, 1639–1644. doi: 10.1073/pnas.1013888108
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Berk, P. A., Jonge, R., Zwietering, M. H., Abee, T., and Kieboom, J. (2005). Acid resistance variability among isolates of Salmonella enterica serovar Typhimurium DT104. J. Appl. Microbiol. 99, 859–866. doi: 10.1111/j.1365-2672.2005.02658.x
Bontemps-Gallo, S., Fernandez, M., Dewitte, A., Raphaël, E., Gherardini, F. C., Elizabeth, P., et al. (2019). Nutrient depletion may trigger the Yersinia pestis OmpR-EnvZ regulatory system to promote flea-borne plague transmission. Mol. Microbiol. 112, 1471–1482. doi: 10.1111/mmi.14372
Bottone, E. J. (1997). Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10, 257–276. doi: 10.1128/cmr.10.2.257-276.1997
Bottone, E. J. (2015). Yersinia enterocolitica: revisitation of an enduring human pathogen. Clin. Microbial Newslett. 37, 1–8. doi: 10.1016/j.clinmicnews.2014.12.003
Boubrik, F., Bonnefoy, E., and Rouviere-Yaniv, J. (1991). HU and IHF: similarities and differences. In Escherichia coli, the lack of HU is not compensated for by IHF. Res. Microbiol. 142, 239–247. doi: 10.1016/0923-2508(91)90036-a
Brzostek, K., Brzóstkowska, M., Bukowska, I., Karwicka, E., and Raczkowska, A. (2007). OmpR negatively regulates expression of invasin in Yersinia enterocolitica. Microbiology 153, 2416–2425. doi: 10.1099/mic.0.2006/003202-0
Brzostek, K., Raczkowska, A., and Zasada, A. (2003). The osmotic regulator OmpR is involved in the response of Yersinia enterocolitica O:9 to environmental stresses and survival within macrophages. FEMS Microbiol. Lett. 228, 265–271. doi: 10.1016/s0378-1097(03)00779-1
Brzostek, K., Skorek, K., and Raczkowska, A. (2012). OmpR, a central integrator of several cellular responses in Yersinia enterocolitica. Adv. Exp. Med. Biol. 954, 325–334. doi: 10.1007/978-1-4614-3561-7_40
Carniel, E., Autenrieth, I., Cornelis, G., Fukushima, H., Guinet, F., Isberg, R., et al. (2006). “Y. enterocolitica and Y. pseudotuberculosis,” in The Prokaryotes, eds M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (New York, NY: Springer), 270–398.
Chakraborty, S., and Kenney, L. J. (2018). A new role of OmpR in acid and osmotic stress in Salmonella and E. coli. Front. Microbiol. 9:2656. doi: 10.3389/fmicb.2018.02656
Chen, S., Thompson, K. M., and Francis, M. S. (2016). Environmental regulation of Yersinia pathophysiology. Front. Cell. Infect. Microbiol. 6:25. doi: 10.3389/fcimb.2016.00025
Chlebicz, A., and Śliżewska, K. (2018). Campylobacteriosis Salmonellosis, Yersiniosis, and Listeriosis as zoonotic foodborne diseases: a review. Int. J. Environ. Res. Public Health 15:863. doi: 10.3390/ijerph15050863
Collins, C. M., Gutman, D. M., and Laman, H. (1993). Identification of a nitrogen-regulated promoter controlling expression of Klebsiella pneumoniae urease genes. Mol. Microbiol. 8, 187–198. doi: 10.1111/j.1365-2958.1993.tb01215.x
Cornelis, G. R. (2002). The yersinia ysc-yop ‘type III’ weaponry. Nat. Rev. Mol. Cell Biol. 3, 742–752.
Dai, Q., Xu, L., Xiao, L., Zhu, K., Song, Y., Li, C., et al. (2018). RovM and CsrA negatively regulate urease expression in Yersinia pseudotuberculosis. Front. Microbiol. 9:348. doi: 10.3389/fmicb.2018.00348
Danson, A. E., Jovanovic, M., Buck, M., and Zhang, X. (2019). Mechanisms of σ54-dependent transcription initiation and regulation. J. Mol. Biol. 431, 3960–3974. doi: 10.1016/j.jmb.2019.04.022
De Koning-Ward, T. F., and Robins-Browne, R. M. (1995). Contribution of urease to acid tolerance in Yersinia enterocolitica. Infect. Immun. 63, 3790–3795. doi: 10.1128/iai.63.10.3790-3795.1995
De Koning-Ward, T. F., and Robins-Browne, R. M. (1997). A novel mechanism of urease regulation in Yersinia enterocolitica. FEMS Microbiol. Lett. 147, 221–226. doi: 10.1111/j.1574-6968.1997.tb10245.x
Dewoody, R. S., Merritt, P. M., and Marketon, M. M. (2013). Regulation of the Yersinia type III secretion system: traffic control. Front. Cell. Infect. Microbiol. 3:4. doi: 10.3389/fcimb.2013.00004
Dillon, S. C., and Dorman, C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195. doi: 10.1038/nrmicro2261
D’Orazio, S. E. F., and Collins, C. M. (1993). The plasmid-encoded urease gene cluster of the family Enterobacteriaceae is positively regulated by UreR, a member of the AraC family of transcriptional activators. J. Bacteriol. 175, 3459–3467. doi: 10.1128/jb.175.11.3459-3467.1993
D’Orazio, S. E. F., and Collins, C. M. (1995). UreR activates transcription at multiple promoters within the plasmid-encoded urease locus of the Enterobacteriaceae. Mol. Microbiol. 16, 145–155. doi: 10.1111/j.1365-2958.1995.tb02399.x
Dorrell, N., Li, S. R., Everest, P. H., Dougan, G., and Wren, B. W. (1998). Construction and characterisation of a Yersinia enterocolitica O:8 ompR mutant. FEMS Microbiol. Lett. 165, 145–151.
Erhardt, M., and Dersch, P. (2015). Regulatory principles governing Salmonella and Yersinia virulence. Front. Microbiol. 6:949. doi: 10.3389/fmicb.2015.00949
European Food Safety Authority and European Centre for Disease Prevention, and Control [ECDC] (2017). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 15:e5077. doi: 10.2903/j.efsa.2017.5077
Fàbrega, A., and Vila, J. (2012). Yersinia enterocolitica: pathogenesis, virulence and antimicrobial resistance. Enferm. Infecc. Microbiol. Clin. 30, 24–32. doi: 10.1016/j.eimc.2011.07.017
Farrugia, M. A., Macomber, L., and Hausinger, R. P. (2013). Biosynthesis of the urease metallocenter. J. Biol. Chem. 288, 13178–13185. doi: 10.1074/jbc.R112.446526
Fuchs, T. M., Brandt, K., Starke, M., and Rattei, T. (2011). Shotgun sequencing of Yersinia enterocolitica strain W22703 (biotype 2, serotype O:9): genomic evidence for oscillation between invertebrates and mammals. BMC Genomics 12:168. doi: 10.1186/1471-2164-12-168
Galindo, C. L., Rosenzweig, J. A., Kirtley, M. L., and Chopra, A. K. (2011). Pathogenesis of Y. enterocolitica and Y. pseudotuberculosis in Human Yersiniosis. J. Pathog. 2011:182051. doi: 10.4061/2011/182051
Gao, H., Zhang, Y., Han, Y., Yang, L., Liu, X., Guo, Z., et al. (2011). Phenotypic and transcriptional analysis of the osmotic regulator OmpR in Yersinia pestis. BMC Microbiol. 11:39. doi: 10.1186/1471-2180-11-39
Garzetti, D., Bouabe, H., Heesemann, J., and Rakin, A. (2012). Tracing genomic variations in two highly virulent Yersinia enterocolitica strains with unequal ability to compete for host colonization. BMC Genomics 13:467. doi: 10.1186/1471-2164-13-467
Gripenberg-Lerche, C., Zhang, L., Ahtonen, P., Toivanen, P., and Skurnik, M. (2000). Construction of urease-negative mutants of Yersinia enterocolitica serotypes O:3 and O:8: role of urease in virulence and arthritogenicity. Infect. Immun. 68, 942–947. doi: 10.1128/IAI.68.2.942-947.2000
Gueguen, E., Durand, E., Zhang, X. Y., d’Amalric, Q., Journet, L., and Cascales, E. (2013). Expression of a Yersinia pseudotuberculosis type VI secretion system is responsive to envelope stresses through the OmpR transcriptional activator. PLoS One 8:e66615. doi: 10.1371/journal.pone.0066615
Harlocker, S. L., Bergstrom, L., and Inouye, M. (1995). Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J. Biol. Chem. 270, 26849–26856. doi: 10.1074/jbc.270.45.26849
Heermann, R., and Fuchs, T. M. (2008). Comparative analysis of the Photorhabdus luminescens and the Yersinia enterocolitica genomes: uncovering candidate genes involved in insect pathogenicity. BMC Genomics 9:40. doi: 10.1186/1471-2164-9-40
Hoch, J. A., and Silhavy, T. J. (1995). Two-component signal transduction. Washington, DC: American Society of Microbiology Press.
Hoffmann, A., Bukau, B., and Kramer, G. (2010). Structure and function of the molecular chaperone Trigger Factor. Biochim. Biophys. Acta 1803, 650–661. doi: 10.1016/j.bbamcr.2010.01.017
Howard, S. L., Gaunt, M. W., Hinds, J., Witney, A. A., Stabler, R., and Wren, B. W. (2006). Application of comparative phylogenomics to study the evolution of Yersinia enterocolitica and to identify genetic differences relating to pathogenicity. J. Bacteriol. 188, 3645–3653. doi: 10.1128/JB.188.10.3645-3653.2006
Hu, Y., Lu, P., Wang, Y., Ding, L., Atkinson, S., and Chen, S. (2009). OmpR positively regulates urease expression to enhance acid survival of Yersinia pseudotuberculosis. Microbiology 155, 2522–2531. doi: 10.1099/mic.0.028381-0
Island, M. D., and Mobley, H. L. T. (1995). Proteus mirabilis urease: operon fusion and linker insertion analysis of ure gene organization, regulation, and function. J. Bacteriol. 177, 5653–5660. doi: 10.1128/jb.177.19.5653-5660.1995
Jaworska, K., Nieckarz, M., Ludwiczak, M., Raczkowska, A., and Brzostek, K. (2018). OmpR-mediated transcriptional regulation and function of two heme receptor proteins of Yersinia enterocolitica bio-serotype 2/O:9. Front. Cell. Infect. Microbiol. 8:333. doi: 10.3389/fcimb.2018.00333
Kakoschke, T., Kakoschke, S., Magistro, G., Schubert, S., Borath, M., Heesemann, J., et al. (2014). The RNA chaperone Hfq impacts growth, metabolism and production of virulence factors in Yersinia enterocolitica. PLoS One 9:e86113. doi: 10.1371/journal.pone.0086113
Kamashev, D., Agapova, Y., Rastorguev, S., Talyzina, A. A., Boyko, K. M., Korzhenevskiy, D. A., et al. (2017). Comparison of histone-like HU protein DNA-binding properties and HU/IHF protein sequence alignment. PLoS One 12:e0188037. doi: 10.1371/journal.pone.0188037
Kappaun, K., Piovesan, A. R., Carlini, C. R., and Ligabue-Braun, R. (2018). Ureases: historical aspects, catalytic and non-catalytic properties – a review. J. Adv. Res. 13, 3–17. doi: 10.1016/j.jare.2018.05.010
Kenney, L. J. (2002). Structure/function relationships in OmpR and other winged-helix transcription factors. Curr. Opin. Microbiol. 5, 135–141. doi: 10.1016/s1369-5274(02)00310-7
Konieczna, I., Zarnowiec, P., Kwinkowski, M., Kolesinska, B., Fraczyk, J., Kaminski, Z., et al. (2012). Bacterial urease and its role in long-lasting human diseases. Curr. Protein Pept. Sci. 13, 789–806. doi: 10.2174/138920312804871094
Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M., et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176. doi: 10.1016/0378-1119(95)00584-1
Lambert de Rouvroit, C., Sluiters, C., and Cornelis, G. R. (1992). Role of the transcriptional activator, VirF, and temperature in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol. Microbiol. 6, 395–409. doi: 10.1111/j.1365-2958.1992.tb01483.x
Lee, V. T., Tam, C., and Schneewind, O. (2000). LcrV, a substrate for Yersinia enterocolitica type III secretion, is required for toxin targeting into the cytosol of HeLa cells. J. Biol. Chem. 275, 36869–36875.
Maeda, S., Takayanagi, K., Nishimura, Y., Maruyama, T., Sato, K., and Mizuno, T. (1991). Activation of the osmoregulated ompC gene by the OmpR protein in Escherichia coli: a study involving synthetic OmpR-binding sequences. J. Biochem. 110, 324–327.
Magasanik, B. (1993). The regulation of nitrogen utilization in enteric bacteria. J. Cell. Biochem. 51, 34–40.
Marceau, M. (2005). Transcriptional regulation in Yersinia: an update. Curr. Issues Mol. Biol. 7, 151–177.
Marx, C. J., and Lidstrom, M. E. (2001). Development of improved versatile broadhost-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147, 2065–2075. doi: 10.1099/00221287-147-8-2065
McNally, A., Thomson, N. R., Reuter, S., and Wren, B. W. (2016). ‘Add, stir andreduce’: Yersinia spp. as model bacteria for pathogen evolution. Nat. Rev. Microbiol. 14, 177–190. doi: 10.1038/nrmicro.2015.29
Mobley, H. L., Chippendale, G. R., Fraiman, M. H., Tenney, J. H., and Warren, J. W. (1985). Variable phenotype of Providencia stuartii due to plasmid-encode traits. J. Clin. Microbiol. 22, 851–853.
Mobley, H. L., Island, M. D., and Hausinger, R. P. (1995). Molecular biology of microbial ureases. Microbiol. Rev. 59, 451–480.
Mulrooney, S. B., Lynch, M. J., Mobley, H. L., and Hausinger, R. P. (1988). Purification, characterization, and genetic organization of recombinant Providencia stuartii urease expressed by Escherichia coli. J. Bacteriol. 170, 2202–2207.
Neubauer, H., Aleksic, S., Hensel, A., Finke, E. J., and Meyer, H. (2000). Yersinia enterocolitica 16S rRNA gene types belong to the same genospecies but form three homology groups. Int. J. Med. Microbiol. 290, 61–64.
Nicholson, E. B., Concaugh, E. A., Foxall, P. A., Island, M. D., and Mobley, H. L. (1993). Proteus mirabilis urease: transcriptional regulation by UreR. J. Bacteriol. 175, 465–473.
Nieckarz, M., Raczkowska, A., Debski, J., Kistowski, M., Dadlez, M., Heesemann, J., et al. (2016). Impact of OmpR on the membrane proteome of Yersinia enterocolitica in different environments: repression of major adhesin YadA and heme receptor HemR. Environ. Microbiol. 18, 997–1021. doi: 10.1111/1462-2920.13165
Nieckarz, M., Raczkowska, A., Jaworska, K., Stefańska, E., Skorek, K., Stosio, D., et al. (2017). The role of OmpR in the expression of genes of the KdgR regulon involved in the uptake and depolymerization of oligogalacturonides in Yersinia enterocolitica. Front. Cell. Infect. Microbiol. 7:366. doi: 10.3389/fcimb.2017.00366
Nordfelth, R., and Wolf-Watz, H. (2001). YopB of Yersinia enterocolitica is essential for YopE translocation. Infect. Immun. 69, 3516–3518.
Parsot, C., Hamiaux, C., and Page, A. L. (2003). The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6, 7–14.
Patra, A. K., and Aschenbach, J. R. (2018). Ureases in the gastrointestinal tracts of ruminant and monogastric animals and their implication in urea-N/ammonia metabolism. J. Adv. Res. 13, 39–50. doi: 10.1016/j.jare.2018.02.005
Philippe, N., Alcaraz, J. P., Counsage, E., Geiselmann, J., and Schneider, D. (2004). Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51, 246–255. doi: 10.1016/j.plasmid.2004.02.003
Poore, C. A., Coker, C., Dattelbaum, J. D., and Mobley, H. L. T. (2001). Identification of the domains of UreR, an AraC-like transcriptional regulator of the urease gene cluster in Proteus mirabilis. J. Bacteriol. 183, 4526–4535.
Poore, C. A., and Mobley, H. L. T. (2003). Differential regulation of the Proteus mirabilis urease gene cluster by UreR and H-NS. Microbiology 149, 3383–3394. doi: 10.1099/mic.0.26624-0
Portnoy, D. A., Moseley, S. L., and Falkow, S. (1981). Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect. Immun. 31, 775–782.
Probst, P., Hermann, E., Meyer zum Buschenfelde, K. H., and Fleischer, B. (1993). Identification of the Yersinia enterocolitica urease beta subunit as a target antigen for human synovial T lymphocytes in reactive arthritis. Infect. Immun. 61, 4507–4509.
Raczkowska, A., Skorek, K., Bielecki, J., and Brzostek, K. (2011). OmpR controls Yersinia enterocolitica motility by positive regulation of flhDC expression. Antonie Van Leeuweanhoek 99, 381–394. doi: 10.1007/s10482-010-9503-8
Raczkowska, A., Trzos, J., Lewandowska, O., Nieckarz, M., and Brzostek, K. (2015). Expression of the AcrAB components of the AcrAB-TolC multidrug efflux pump of Yersinia enterocolitica is subject to dual regulation by OmpR. PLoS One 10:e0124248. doi: 10.1371/journal.pone.0124248
Reboul, A., Lemaitre, N., Titecat, M., Merchez, M., Deloison, G., Ricard, I., et al. (2014). Yersinia pestis requires the 2-component regulatory system OmpR-EnvZ to resist innate immunity during the early and late stages of plague. J. Infect. Dis. 210, 1367–1375. doi: 10.1093/infdis/jiu274
Rentschler, A. E., Lovrich, S. D., Fitton, R., Enos-Berlage, J., and Schwan, W. R. (2013). OmpR regulation of the uropathogenic Escherichia coli fimB gene in an acidic/high osmolality environment. Microbiology 159, 316–327. doi: 10.1099/mic.0.059386-0
Riot, B., Berche, P., and Simonet, M. (1997). Urease is not involved in the virulence of Yersinia pseudotuberculosis in mice. Infect. Immun. 65, 1985–1990.
Russo, F. D., and Silhavy, T. J. (1990). EnvZ controls the concentration of phosphoryled OmpR to mediate osmoregulation of the porin genes. J. Mol. Biol. 222, 567–580.
Rutherford, J. C. (2014). The emerging role of urease as a general microbial virulence Factor. PLoS Pathog. 10:e1004062. doi: 10.1371/journal.ppat.1004062
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Schmühl, C., Beckstette, M., Heroven, A. K., Bunk, B., Spröer, C., McNally, A., et al. (2019). Comparative transcriptomic profiling of Yersinia enterocolitica O:3 and O:8 reveals major expression differences of fitness- and virulence-relevant genes indicating ecological separation. mSystems 4:e00239-18. doi: 10.1128/mSystems.00239-18
Schwan, W. R., Lee, J. L., Lenard, F. A., Matthews, B. T., and Beck, M. T. (2002). Osmolarity and pH growth conditions regulate fim gene transcription and type 1 pilus expression in uropathogenic Escherichia coli. Infect. Immun. 70, 1391–1402.
Sebbane, F., Bury-Mone, S., Cailliau, K., Browaeys-Poly, E., De Reuse, H., and Simonet, M. (2002). The Yersinia pseudotuberculosis Yut protein, a new type of urea transporter homologous to eukaryotic channels and functionally interchangeable in vitro with the Helicobacter pylori UreI protein. Mol. Microbiol. 45, 1165–1174. doi: 10.1046/j.1365-2958.2002.03096.x
Skorek, K., Raczkowska, A., Dudek, B., Miêtka, K., Guz-Regner, K., Pawlak, A., et al. (2013). Regulatory protein OmpR influences the serum resistance of Yersinia enterocolitica O:9 by modifying the structure of the outer membrane. PLoS One 8:e79525. doi: 10.1371/journal.pone.0079525
Solovyev, V., and Salamov, A. (2011). “Automatic annotation of microbial genomes and metagenomic sequences,” in Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies, ed. R. W. Li (Hauppauge: NY: Nova Science Publishers), 61–78.
Stingl, K., Altendorf, K., and Bakker, E. P. (2002). Acid survival of Helicobacter pylori: how does urease activity trigger cytoplasmic pH homeostasis? Trends Microbiol. 10, 70–74.
Stock, J. B., Ninfa, A. J., and Stock, A. M. (1989). Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53, 450–490.
Straley, S. C., and Perry, R. D. (1995). Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 3, 310–317. doi: 10.1016/S0966-842X(00)88960-X
Tan, S. Y., Tan, I. K. P., Tan, M. F., Dutta, A., and Choob, S. W. (2016). Evolutionary study of Yersinia genomes deciphers emergence of human pathogenic species. Sci Rep. 6:36116. doi: 10.1038/srep36116
Thibodeau, S. A., Fang, R., and Joung, J. K. (2004). High-throughput betagalactosidase assay for bacterial cell-based reporter systems. Biotechniques 36, 410–415. doi: 10.2144/04363BM07
Thomson, N. R., Howard, S., Wren, B. W., Holden, M. T., Crossman, L., Challis, G. L., et al. (2006). The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2:e206.
Trosky, J. E., Liverman, A. D., and Orth, K. (2008). Yersinia outer proteins: Yops. Cell. Microbiol. 10, 557–565.
Uliczka, F., Pisano, F., Schaake, J., Stolz, T., Rohde, M., Fruth, A., et al. (2011). Unique cell adhesion and invasion properties of Yersinia enterocolitica O:3, the most frequent cause of human Yersiniosis. PLoS Pathog. 7:e1002117. doi: 10.1371/journal.ppat.1002117
Viboud, G. I., and Bliska, J. B. (2005). Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu. Rev. Microbiol. 59, 69–89.
Young, G. M., Amid, D., and Miller, V. L. (1996). A bifunctional urease enhances survival of pathogenic Yersinia enterocolitica and Morganella morganii at low pH. J. Bacteriol. 178, 6487–6495. doi: 10.1128/jb.178.22.6487-6495.1996
Zhou, C., Bhinderwala, F., Lehman, M. K., Thomas, V. C., Chaudhari, S. S., Yamada, K. J., et al. (2019). Urease is an essential component of the acid response network of Staphylococcus aureus and is required for a persistent murine kidney infection. PLoS Pathog. 15:e1007538. doi: 10.1371/journal.ppat.1007538
Keywords: Yersinia enterocolitica, OmpR regulator, urease activity, urease-subunits genes, proteome, strain-specific differences
Citation: Nieckarz M, Kaczor P, Jaworska K, Raczkowska A and Brzostek K (2020) Urease Expression in Pathogenic Yersinia enterocolitica Strains of Bio-Serotypes 2/O:9 and 1B/O:8 Is Differentially Regulated by the OmpR Regulator. Front. Microbiol. 11:607. doi: 10.3389/fmicb.2020.00607
Received: 03 January 2020; Accepted: 19 March 2020;
Published: 08 April 2020.
Edited by:
Ulrike Kappler, The University of Queensland, AustraliaReviewed by:
Thilo Fuchs, Friedrich Loeffler Institute, GermanyHuaiqi Jing, Chinese Center for Disease Control and Prevention, China
Copyright © 2020 Nieckarz, Kaczor, Jaworska, Raczkowska and Brzostek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marta Nieckarz, bmllY2thcnptYXJ0YUBnbWFpbC5jb20=; Katarzyna Brzostek, a2Jyem9zdGVrQGJpb2wudXcuZWR1LnBs
†Present address: Marta Nieckarz, Laboratory for Molecular Infection Medicine Sweden, Department of Molecular Biology, Umeå University, Umeå, Sweden
 Marta Nieckarz
Marta Nieckarz Patrycja Kaczor
Patrycja Kaczor Karolina Jaworska
Karolina Jaworska Adrianna Raczkowska
Adrianna Raczkowska Katarzyna Brzostek
Katarzyna Brzostek