
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 26 March 2020
Sec. Food Microbiology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00512
 Huizhen Li1,2†
Huizhen Li1,2† Fei Liu1†
Fei Liu1† Jingjing Lu1,2
Jingjing Lu1,2 Jialu Shi1,2
Jialu Shi1,2 Jiaqi Guan1,2
Jiaqi Guan1,2 Fenfen Yan1,2
Fenfen Yan1,2 Bailiang Li1,2*
Bailiang Li1,2* Guicheng Huo1,2*
Guicheng Huo1,2*The global prevalence of obesity is rising year by year, which has become a public health problem worldwide. In recent years, animal studies and clinical studies have shown that some lactic acid bacteria possess an anti-obesity effect. In our previous study, mixed lactobacilli (Lactobacillus plantarum KLDS1.0344 and Lactobacillus plantarum KLDS1.0386) exhibited anti-obesity effects in vivo by significantly reducing body weight gain, Lee’s index and body fat rate; however, its underlying mechanisms of action remain unclear. Therefore, the present study aims to explore the possible mechanisms for the inhibitory effect of mixed lactobacilli on obesity. C57BL/6J mice were randomly divided into three groups including control group (Control), high fat diet group (HFD) and mixed lactobacilli group (MX), and fed daily for eight consecutive weeks. The results showed that mixed lactobacilli supplementation significantly improved blood lipid levels and liver function, and alleviated liver oxidative stress. Moreover, the mixed lactobacilli supplementation significantly inhibited lipid accumulation in the liver and regulated lipid metabolism in epididymal fat pads. Notably, the mixed lactobacilli treatment modulated the gut microbiota, resulting in a significant increase in acetic acid and butyric acid. Additionally, Spearman’s correlation analysis found that several specific genera were significantly correlated with obesity-related indicators. These results indicated that the mixed lactobacilli supplementation could manipulate the gut microbiota and its metabolites (acetic acid and butyric acid), resulting in reduced liver lipid accumulation and improved lipid metabolism of adipose tissue, which inhibited obesity.
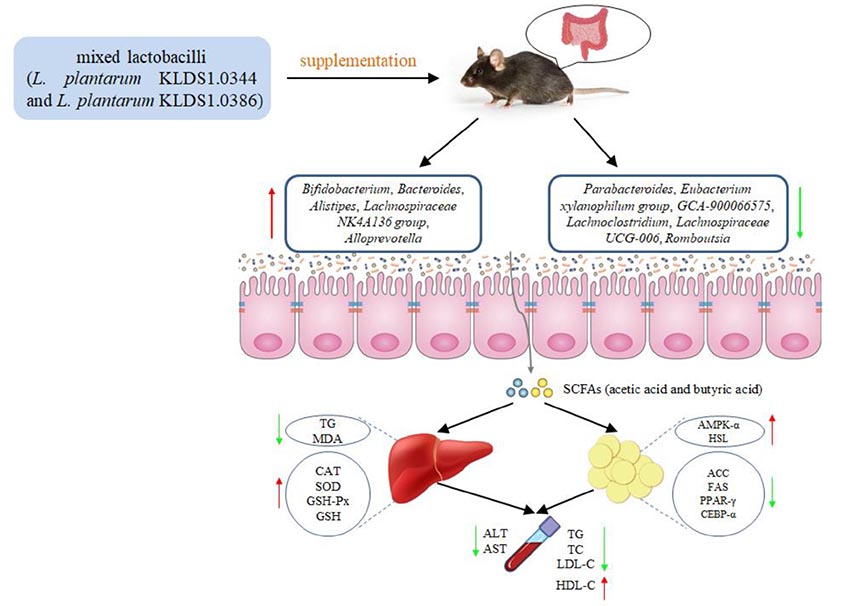
Graphical Abstract. The mixed lactobacilli (Lactobacillus plantarum KLDS1.0344 and Lactobacillus plantarum KLDS1.0386) prevented high fat diet-induced obesity via regulating gut microbiota and lipid metabolism of the adipose tissue and inhibiting liver lipid accumulation in mice.
Obesity has become a worldwide epidemic disease and has a serious impact on the healthy development of the human body, which is a major hidden danger to public health (Risk and Factor Collaboration, 2016; Lu J. et al., 2016; Solas et al., 2017). The researchers collected data from 195 countries and found that the prevalence of obesity worldwide has increased dramatically since 1980 (Reuter and Mrowka, 2019). In 2015, approximately 603 million adults and 107 million children suffered from obesity, with an overall prevalence of 12% for adults and 5% for children (GBD 2015 Obesity Collaborators, 2017). Obesity is defined by the World Health Organization as an excessive accumulation of fat that may be harmful to health and is diagnosed when body mass index (BMI) ≥30 kg/m2 (Prospective Studies and Collaboration, 2009). In general, the intake of a large amount of high-sugar and high-fat food, sedentary work style and less physical exercise cause the energy intake to be larger than the energy consumption of the body, which will lead to the accumulation of excessive triglyceride in liver, kidney week and adipose tissue, thus causing obesity (Hurt et al., 2010; Heymsfield and Wadden, 2017; Bluher, 2019). Obesity can increase the risk of various diseases such as cardiovascular disease (Oikonomou and Antoniades, 2019), type 2 diabetes (Dietz, 2017), osteoarthritis (Schott et al., 2018), Alzheimer’s disease (Solas et al., 2017), anxiety (Ogrodnik et al., 2019), depression (Tyrrell et al., 2018), and certain cancers [for example breast cancer (Picon-Ruiz et al., 2017; Hao et al., 2018), colorectal cancer (Wunderlich et al., 2018), pancreatic cancer (Zaytouni et al., 2017), stomach cancer (Murphy et al., 2018) and liver cancer (Shin et al., 2013)].
At present, the medications approved by the US Food and Drug Administration (FDA) for long-term weight management mainly reduce energy or food intake by causing fat malabsorption, promoting satiety, delaying gastric emptying, decreasing appetite, or acting on the central nervous system pathway, but they all have certain side effects such as nausea, diarrhea, vomiting, dry mouth, constipation, dizziness and fecal incontinence (Davidson et al., 1999; Smith et al., 2010; Gadde et al., 2011; Apovian et al., 2013; Xavier et al., 2015). For some patients with obesity to a certain extent, gastric banding, Roux-en-Y gastric bypass and vertical-sleeve gastrectomy, three types of surgery for obesity, are common treatments (Neylan et al., 2016; Schauer et al., 2016). Although more effective than drug intervention, these procedures are more risky (American College of Cardiology/American Heart Association Task Force on Practice Guidelines Obesity Expert Panel, 2013, 2014) and may cause some complications (Almalki et al., 2017). Therefore, safer and healthful non-drug therapies have been proposed, including the use of probiotics. Lactobacillus rhamnosus Lb102 and Bifidobacterium animalis ssp. lactis Bf141 isolated by Le Barz et al. from fermented milk products and human feces, respectively, were used to interfere with mice fed with high-fat diet and it was found that they could effectively alleviate the onset of obesity and reduce the content of liver fat in mice (Le Barz et al., 2019).
Lactobacillus plantarum KLDS1.0344 and L. plantarum KLDS1.0386 were isolated from traditional fermented dairy products in Inner Mongolia, China, and preserved in our laboratory. In addition, our previous studies have demonstrated that L. plantarum KLDS1.0344 and L. plantarum KLDS1.0386 have strong acid and bile salt resistance, high cell adhesion activities and lipid metabolism regulation properties (Tang et al., 2016; Jin et al., 2017; Lu et al., 2018; Yan et al., 2019; Yue et al., 2019). Hence, we investigated whether a mixture of L. plantarum KLDS1.0344 and L. plantarum KLDS1.0386 could prevent obesity. The results showed that the combined treatment of L. plantarum KLDS1.0344 and L. plantarum KLDS1.0386 could significantly improve some obesity-related indicators in high fat diet-fed mice, including body weight gain, Lee’ s index, and body fat rate, etc. which established its effect in inhibiting obesity (Lu J. et al., 2019). However, the mechanism of the combined treatment of L. plantarum KLDS1.0344 and L. plantarum KLDS1.0386 to inhibit obesity was still unclear. Therefore, the aim of this study was to further explore whether the combined intervention of L. plantarum KLDS1.0344 and L. plantarum KLDS1.0386 prevented obesity by reducing liver lipid accumulation and regulating lipid metabolism and gut microbiota.
Triglyceride (TG), total cholesterol (TC), low density lipoprotein cholesterol (LDL-C), high density lipoprotein cholesterol (HDL-C), antioxidant enzymes, glutathione (GSH), malondialdehyde (MDA), alanine aminotransferase (ALT) and aspartate aminotransferase (AST) assay kits were obtained from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). PrimeScriptTM RT reagent Kit with gDNA Eraser was obtained from Takara Biomedical Technology (Beijing) Co. Ltd. (Beijing, China).
Lactobacillus plantarum KLDS1.0344 and L. plantarum KLDS1.0386 were inoculated in De Man Rogosa Sharpe (MRS) broth, respectively, according to 2% inoculation amount, cultured at 37°C, subcultured every 24 h, cultured in the third generation to 18 h, then centrifuged at 2500 g at 4°C for 10 min to harvest bacterial cell, and washed with a sterile phosphate buffered saline (PBS) for three times. The washed L. plantarum KLDS1.0344 and L. plantarum KLDS1.0386 were separately resuspended with sterile PBS to reach a concentration of 5 × 108 CFU/mL, and then the two were mixed at a ratio of 1: 1. Bacteria were freshly prepared daily during the 8-week experiment.
Male Specific Pathogen-Free (SPF) C57BL/6J mice, aged 21–28 days, were provided by Beijing Vital River Laboratory Animal Technology Co. Ltd. (Beijing, China) (Approval No. SCXK (JING) 2012-0001). These mice were housed in plastic cages under environmentally controlled conditions (temperature, 20–22°C; relative humidity, 50 ± 10%; and lighting, 12 h light/12 h dark cycle) and given free access to standard diet and water to acclimate to the environment for a week before the start of the experiment. The animal experiment protocol was approved by the Institutional Animal Care and Use Committee of the Northeast Agricultural University under the approved protocol number Specific pathogen free rodent management (SRM)-06.
After one week of acclimatization, the mice were randomly assigned to the following three groups: control group (Control), high fat diet group (HFD) and mixed lactobacilli group (MX), with 8 mice in each group. Mice in the control group were fed D12450B control diet, while the others were fed D12492 high fat diet. The feed was manufactured and supplied by Beijing Keao Xieli Feed Co. Ltd. (Beijing, China), and its formula is shown in Supplementary Table S1. From 9: 00 to 11: 00 am every day, the control group and the high fat diet group mice were gavaged with 0.2 mL of sterile PBS solution, and for the mixed lactobacilli group, the mice was administered with 0.2 mL of the mixed lactobacilli suspension (108 CFU). During the whole experiment, the padding and water were changed twice a week, and the high-fat diet was changed every day to prevent the oxidation of fat to produce odor which affected the mice to eat. The entire experiment lasted for 8 weeks. Eight weeks later, all mice were removed from diet for 12 h and then were anesthetized and sacrificed. The serum samples were obtained via centrifugation at 1500 g for 10 min at 4°C, and stored at −80°C for further analysis. Livers and epididymal fat pads were stored at −80°C. The cecal contents of the mice were collected into sterile tubes and stored at −80°C for later use.
The concentration of TC, TG, LDL-C and HDL-C in serum of each group of mice were measured with the commercial assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s protocols. The final data of TC, TG, LDL-C and HDL-C are reported as mmol/L.
The activity of ALT and AST in serum of each group of mice were measured with the commercial assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s protocols. The levels of ALT and AST are reported as U/L.
Liver homogenates were centrifuged at 1150 g for 10 min at 4°C and the protein concentration was measured by bicinchoninic acid (BCA) method using a commercial kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Then, the levels of TG, catalase (CAT), superoxide dismutase (SOD), glutathione peroxidase (GSH-Px), GSH and MDA in the mouse livers were measured using commercially available kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) following the manufacturer’s instructions.
To determine the expression of key genes for lipid metabolism in epididymal fat pads, mRNA levels of adenosine 5′-monophosphate-activated protein kinase-α (AMPK-α), hormone-sensitive lipase (HSL), peroxisome proliferators-activated receptor-γ (PPAR-γ), CAATT/enhancer binding proteins α (C/EBPα), fatty acid synthetase (FAS) and acetyl CoA carboxylase (ACC) were assessed by quantitative real-time polymerase chain reaction (qPCR). Total RNA of the tissues was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. The mRNA was reverse transcribed into cDNA using the PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara, Otsu, Japan). Quantitative real-time PCR was performed using reverse-transcribed cDNA as a template, and ABI7500 fluorescence quantitative PCR instrument (Applied Biosystems, Woolston, Warrington, United Kingdom) and SYBR Green PCR Master Mix (Applied Biosystems, Woolston, Warrington, United Kingdom) were used according to the manufacturer’s protocols. Specific forward and reverse primer sequences for quantitative real-time PCR are listed in Supplementary Table S2. All reactions were performed in triplicate. Relative quantification of gene expression were analyzed with the 2–ΔΔ Ct method. The target gene levels were calculated relative to β-actin and the data were shown as fold changes.
Total bacterial DNA of the cecal contents in each group (n = 3) was extracted using the E.Z.N.A.® Stool DNA Kit (Omega Bio-Tek, Norcross, GA, United States) according to the manufacturer’s instructions. The extracted DNA was checked by agarose gel electrophoresis and quantified using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, United States). The V3-V4 hypervariable region of the bacterial 16S rDNA was amplified by PCR using forward primer 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The products of PCR were purified with AxyPrep DNA Gel Extraction Kit (Axygen Bioscience, Union City, CA, United States), quantified using Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, United States), and pooled in equimolar ratios. PCR amplicons sequencing was performed on Illumina Miseq platform (Illumina Inc. San Diego, CA, United States) following the standard protocols. The resulting raw reads were merged with FLASH software (V1.2.11) (Magoč and Salzberg, 2011) and quality-filtered using QIIME software (V1.7.0) (Caporaso et al., 2010). High-quality clean tags were obtained by using the UCHIME algorithm to identify and remove the chimeric sequences (Edgar et al., 2011). The tags with nucleotide 97% sequence identity were clustered into the same operational taxonomic units (OTUs) using Uparse software (V7.0.1001) (Edgar, 2013). These OTUs were subjected to analysis using the Greengenes database by PyNAST software (Version 1.2) and were annotated to taxonomic information (Yilmaz et al., 2013). The species abundance of microorganisms in the three groups at phylum and genus levels were compared. Linear Discriminant Analysis Effect Size (LEfSe) was used to identify potential microbial biomarkers associated with different treatments with an effect size threshold of 2 (Segata et al., 2011).
In order to determine the level of SCFAs in the intestine, 50 mg the cecal contents were mixed in 0.3 mL pure water, treated with a ball mill at 45 Hz for 4 min, ultrasonically treated in ice water bath for 5 min, and centrifuged at 5000 g for 20 min at 4°C. The supernatant (0.2 mL) and 0.3 mL pure water were mixed evenly, treated with a ball mill at 45 Hz for 4 min, ultrasonically treated in ice water bath for 5 min, and centrifuged at 5000 g for 20 min at 4°C. After that, 0.3 mL the supernatant was uniformly mixed with the original 0.2 mL the supernatant, and 0.1 mL 50% H2SO4 and 0.4 mL the internal standard solution (2-Methylvaleric acid dissolved in diethyl ether to 50 μg/mL) was added. Next, the sample was centrifuged at 12000 g for 15 min at 4°C and allowed to stand at −20°C for 30 min. The supernatant was transferred to a sample vial and detected by gas chromatography-mass spectrometry (GC-MS). GC-MS detection was performed using an Agilent 7890 gas chromatography mass spectrometer equipped with an Agilent HP-FFAP capillary column (30 m × 250 μm × 0.25 μm, J&W Scientific, Folsom, CA, United States). Specific chromatographic conditions were used with reference to the method previously described (Zheng et al., 2013). Acetic acid, butyric acid, propionic acid, valeric acid (Sigma, St. Louis, MO, United States) were used as the standards. The concentration of each SCFA was determined according to a standard curve obtained from seven different concentrations of standards.
All experiments were performed with at least three replicates and all experimental data were displayed as mean ± standard deviation (SD). Analysis of the data was carried out using SPSS 20.0 software (SPSS Inc. Chicago, IL, United States). Statistical differences among groups were determined using one-way analysis of variance (ANOVA), followed by Duncan’s multiple range test. The Spearman’s rank correlation coefficients between the relative abundance of mixed lactobacilli-manipulated gut microbiome and obesity-related indicators were determined using R software 3.4.1 for correlational statistical analysis. P-values < 0.05 were considered to be statistically significant.
The blood lipid levels of the three groups of mice are shown in Figure 1. Compared with the control group, mice in the HFD group showed a dramatical increase in the serum levels of TG, TC, and LDL-C and significant decrease in the serum level of HDL-C (p < 0.01), indicating that abnormal blood lipid metabolism was occurred in the HFD group. Conversely, oral administration of mixed lactobacilli markedly inhibited these changes of lipid parameters in the serum (p < 0.01).
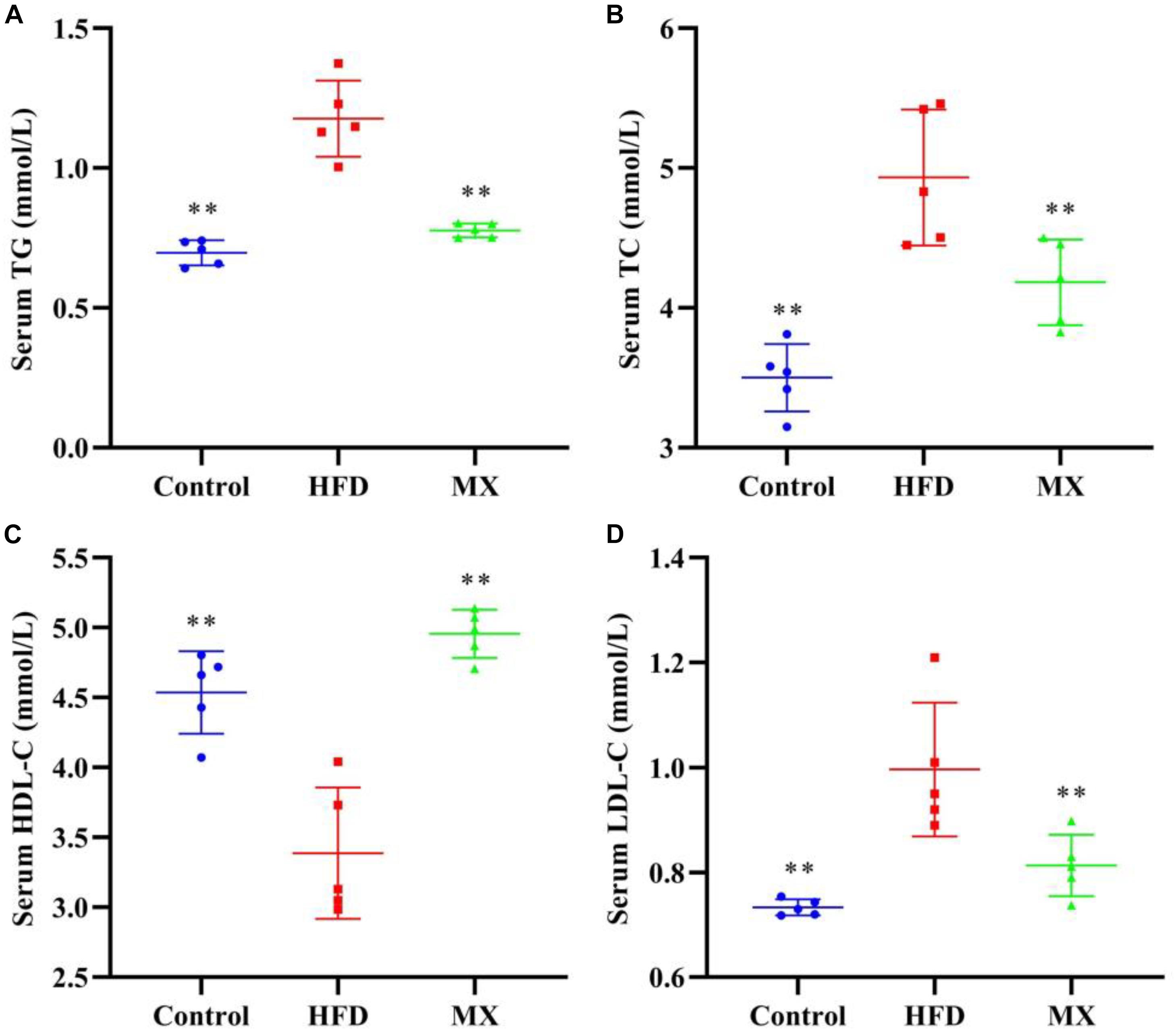
Figure 1. Effects of mixed lactobacilli on blood lipids. Control, control group; HFD, high fat diet group; and MX, mixed lactobacilli group. (A) Serum triglyceride (TG) level; (B) Serum total cholesterol (TC) level; (C) Serum high density lipoprotein cholesterol (HDL-C) level; and (D) Serum low density lipoprotein cholesterol (LDL-C) level. All data are represented as mean ± SD. **p < 0.01: significant difference compared with mice in the HFD group.
Serum ALT and AST levels, which are commonly used as indicators for evaluating liver function, were measured. As shown in Figures 2A,B, in a comparison between the control and HFD groups, the HFD group showed significantly higher levels of ALT and AST in serum than that of the control group (p < 0.01), indicating liver damage in these mice. Notably, treatment with mixed lactobacilli significantly reduced serum ALT and AST levels induced by high fat diet (p < 0.01).
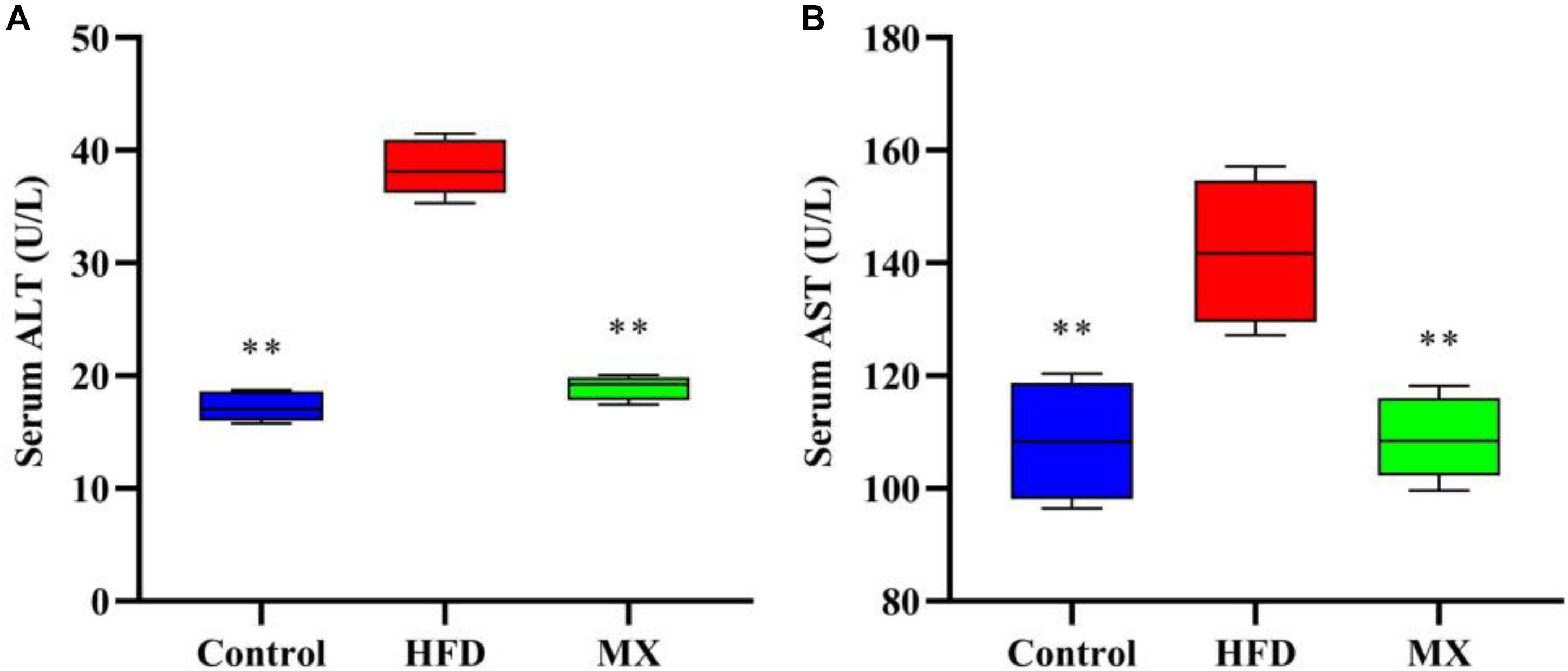
Figure 2. Effects of mixed lactobacilli on liver function. Control, control group; HFD, high fat diet group; and MX, mixed lactobacilli group. (A) Serum alanine aminotransferase (ALT) level; and (B) Serum aspartate aminotransferase (AST) level. All data are represented as mean ± SD. All data are represented as mean ± SD. **p < 0.01: significant difference compared with mice in the HFD group.
To determine lipid accumulation in the liver of mice, we examined the levels of TG in the liver of the three groups of mice. As shown in Figure 3, the liver TG concentration was pronouncedly higher in the HFD group when compared with the control group (p < 0.01). However, the mixed lactobacilli administration markedly decreased the high levels of TG induced by high fat diet (p < 0.01).
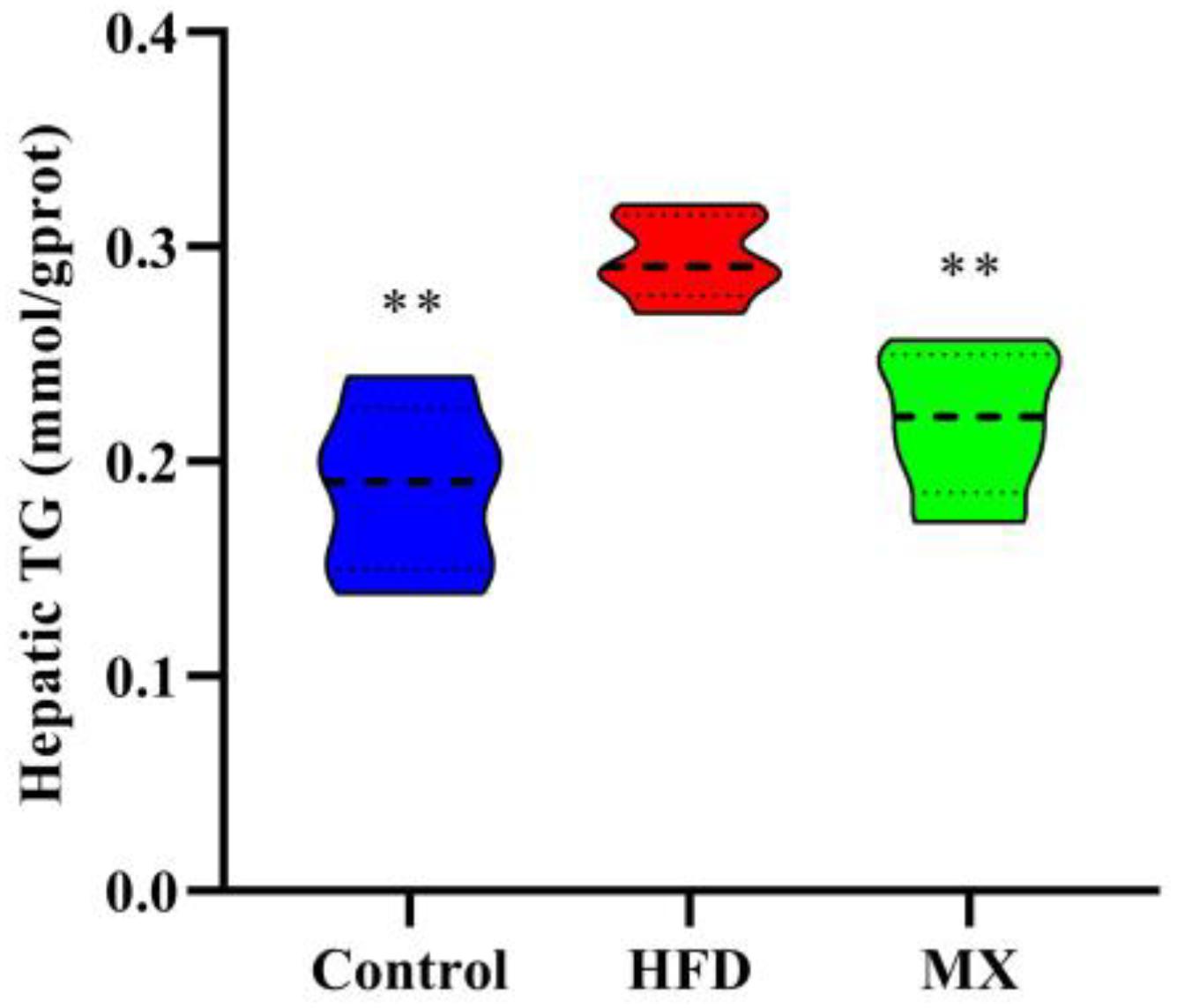
Figure 3. Effect of mixed lactobacilli on hepatic triglyceride (TG). Control, control group; HFD, high fat diet group; and MX, mixed lactobacilli group. All data are represented as mean ± SD. **p < 0.01: significant difference compared with mice in the HFD group.
The levels of antioxidant enzymes, GSH and MDA in the liver of the mice were measured. As depicted in Figures 4A–D, the levels of CAT, SOD, GSH-Px and GSH in the liver of the mice of the HFD group were significantly decreased as compared with that of the control group (p < 0.01). However, these reduced levels were significantly elevated by mixed lactobacilli supplementation (p < 0.01). Furthermore, mixed lactobacilli treatment significantly reduced liver MDA levels induced by high fat diet (Figure 4E, p < 0.01). These findings suggested that mixed lactobacilli administration could enhance the antioxidant capacity of the mice liver.
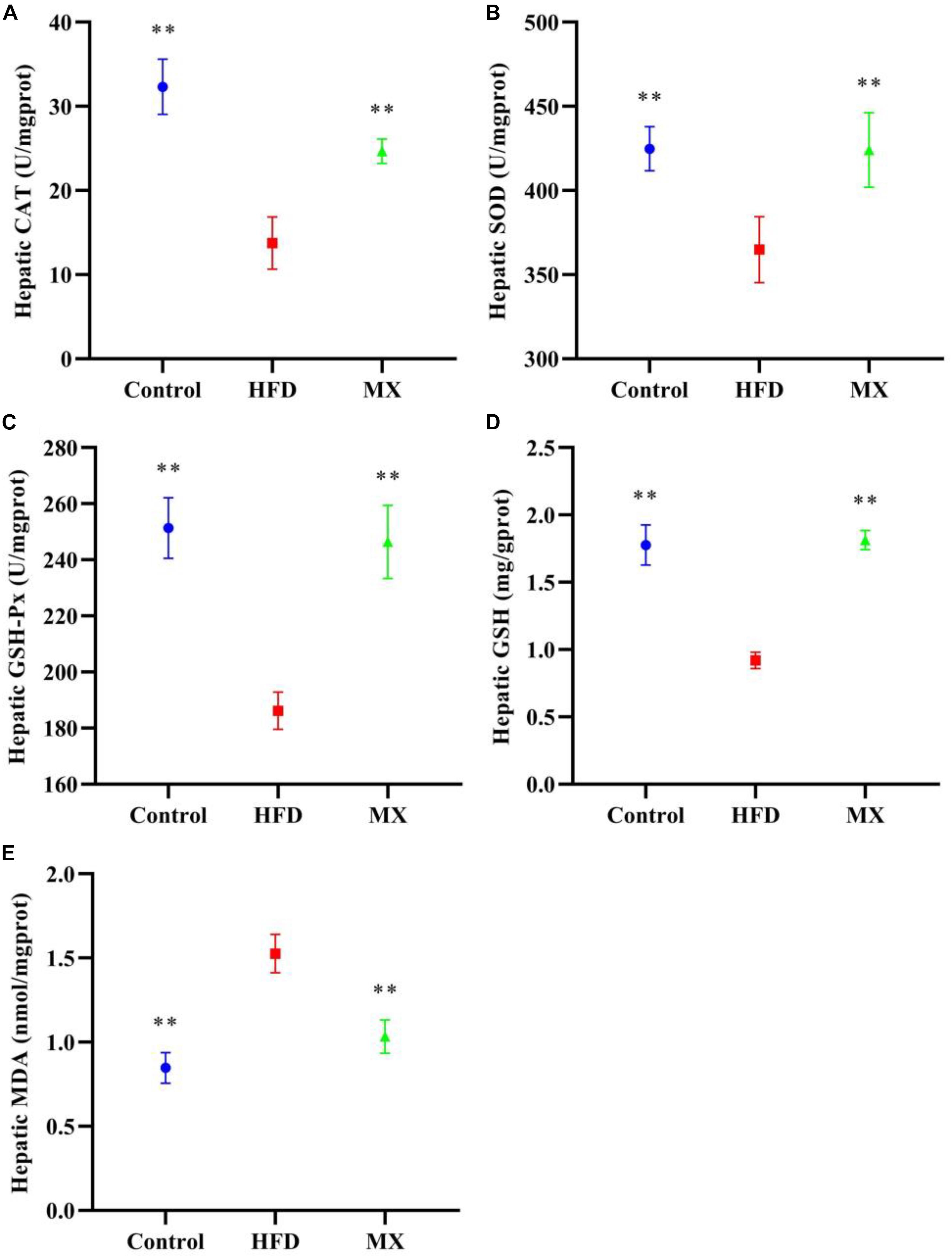
Figure 4. Effect of mixed lactobacilli on hepatic antioxidant enzymes, glutathione (GSH) and malondialdehyde (MDA). Control, control group; HFD, high fat diet group; and MX, mixed lactobacilli group. (A) Catalase (CAT); (B) Superoxide dismutase (SOD); (C) Glutathione peroxidase (GSH-Px); (D) GSH; and (E) MDA. All data are represented as mean ± SD. **p < 0.01: significant difference compared with mice in the HFD group.
The results and comparisons of mRNA expression of key genes for lipid metabolism in the epididymal fat pad are illustrated in Figure 5. As shown in Figures 5A,B, high fat diet feeding caused a significant down-regulation of mRNA levels of AMPK-α and HSL in the epididymal fat pad as compared to the control group (p < 0.01), which was normalized by the mixed lactobacilli supplementation. In addition, as shown in Figures 5C–F, the mRNA expression levels of ACC, FAS, PPAR-γ, and C/EBP-α in the epididymal fat pad of the mice in the HFD group were significantly higher than those in the control group (p < 0.01 or p < 0.05). However, after treatment with the mixed lactobacilli, the mRNA expression levels of ACC, FAS, and PPAR-γ were significantly decreased (p < 0.01), and the mRNA expression level of C/EBP-α was also decreased but not significant (p > 0.05).
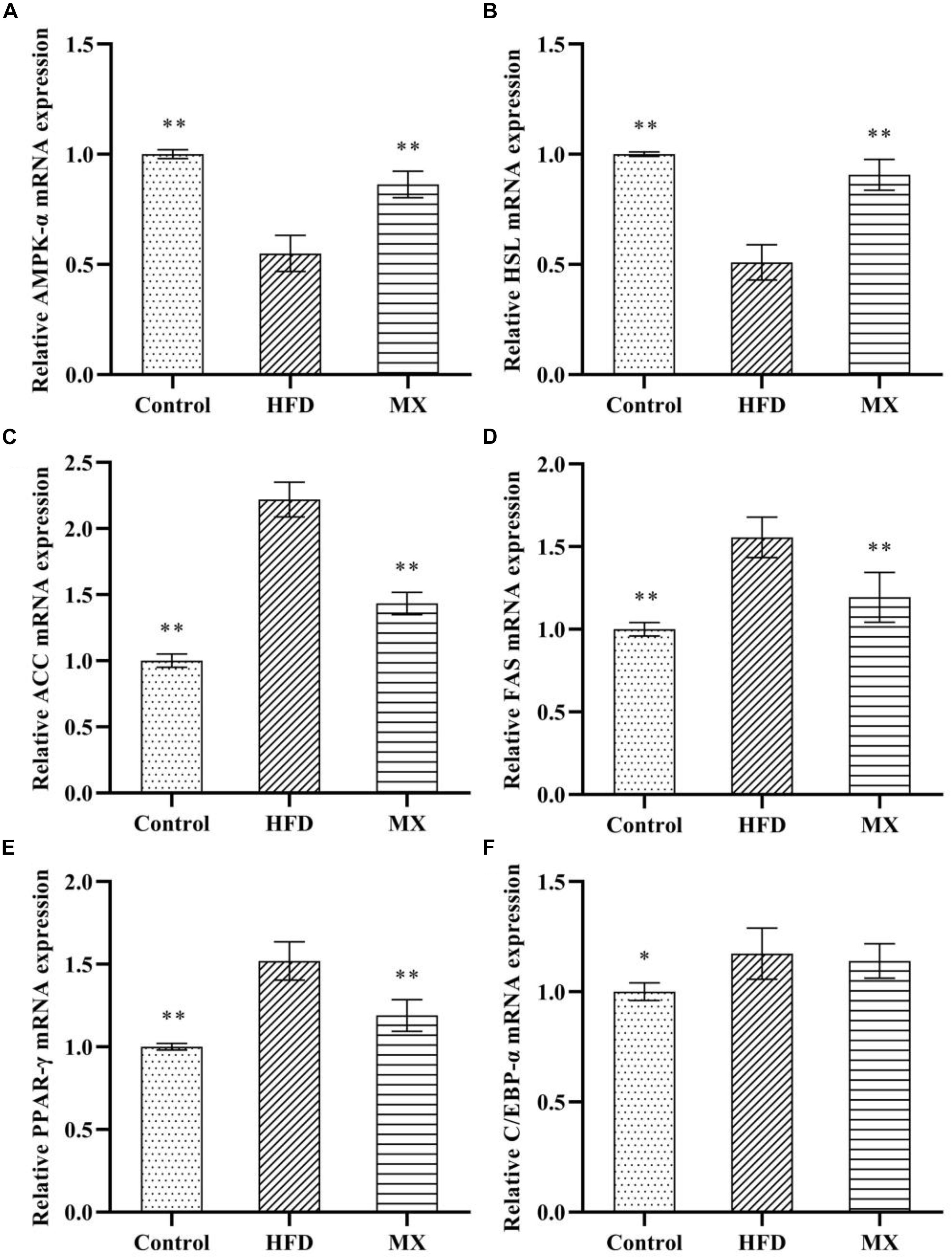
Figure 5. Effect of mixed lactobacilli on lipid metabolism regulating genes in epididymal fat pads. Control, control group; HFD, high fat diet group; and MX, mixed lactobacilli group. (A) Adenosine 5′-monophosphate-activated protein kinase-α (AMPK-α); (B) Hormone-sensitive lipase (HSL); (C) Acetyl CoA carboxylase (ACC); (D) Fatty acid synthetase (FAS); (E) Peroxisome proliferators-activated receptor-γ (PPAR-γ); and (F) CAATT/enhancer binding proteins α (C/EBPα). All data are represented as mean ± SD. ∗p < 0.05 and ∗∗p < 0.01: significant difference compared with mice in the HFD group.
To investigate whether mixed lactobacilli have an important role in the bacterial communities of high fat diet-fed mice, the cecal gut microbiota of the mice was analyzed by sequencing the 16S rDNA variable region V3-V4. At the phylum level, the dominant components in all groups were Firmicutes and Bacteroidetes, with a ratio of more than 85% (Figure 6A). Compared to the control group, the HFD group exhibited a higher relative abundance of Firmicutes and a lower relative abundance of Bacteroides, representing 79.67 and 9.43%, respectively. Consistently, a significant increase in the ratio of Firmicutes to Bacteroides in the HFD group was observed compared to the control group (p < 0.01, Table 1). However, mixed lactobacilli treatment attenuated the increase in Firmicutes, the decrease in Bacteroidetes and the increase in Firmicutes-to-Bacteroidetes ratio induced by high fat diet.
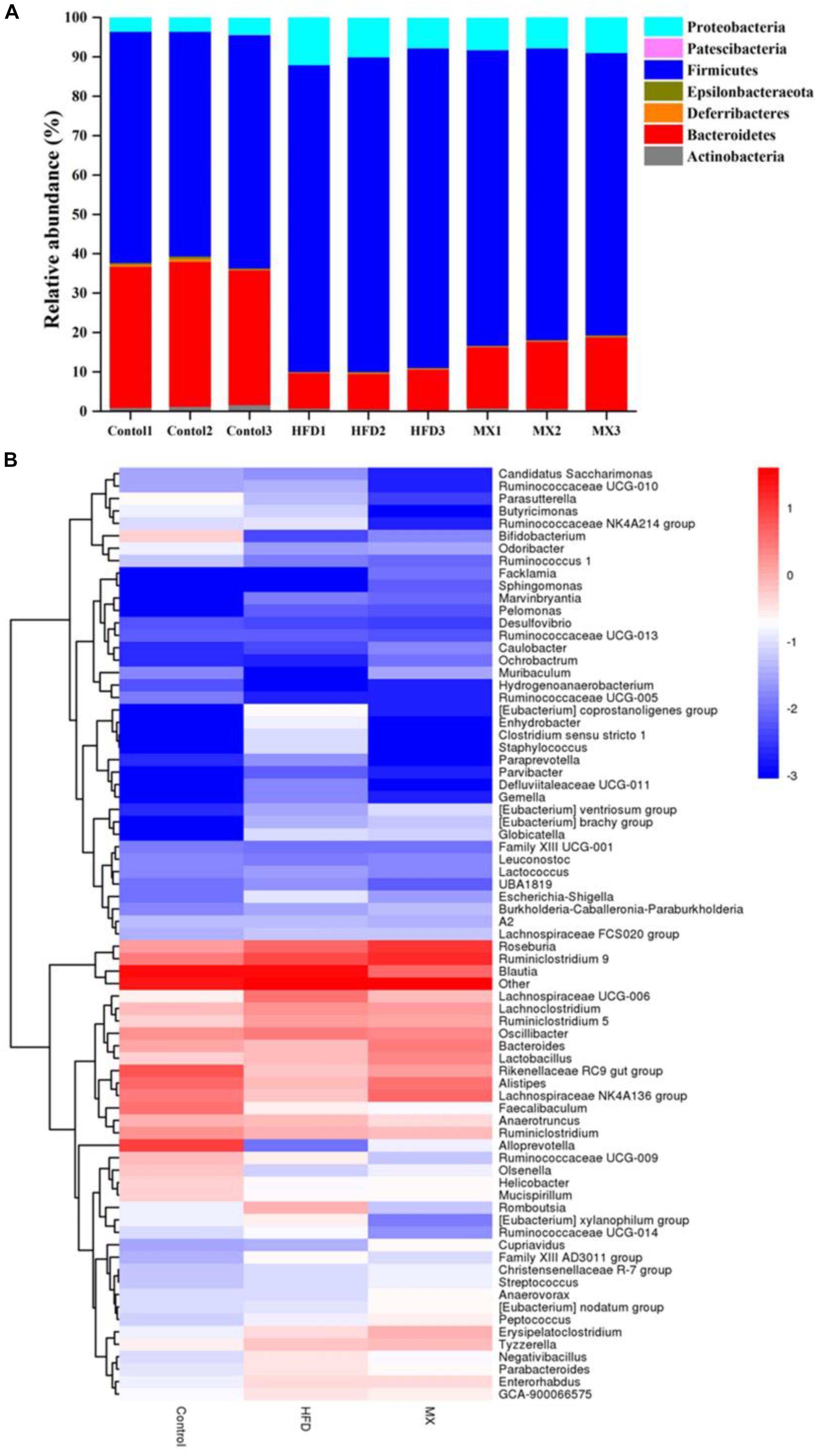
Figure 6. Changes of the gut microbial population at the phylum level following mixed lactobacilli administration. Control, control group; HFD, high fat diet group; and MX, mixed lactobacilli group. (A) Stacked bar plot of gut microbiota composition at the phylum level; (B) Heatmap of gut microbiota composition at the genus level. n = 3 per group.
The distribution of gut microbiota at the genus level in different groups was shown by the genera abundance heatmap (Figure 6B). Compared with the control group, the relative abundances of Bifidobacterium, Bacteroides, Alistipes, Lachnospiraceae NK4A136 group and Alloprevotella were decreased in the HFD group but the relative abundances of Parabacteroides, Eubacterium xylanophilum group, GCA-900066575, Lachnoclostridium, Lachnospiraceae UCG-006 and Romboutsia were increased, all of which were inhibited by mixed lactobacilli supplementation (Figure 6B). Collectively, these results implied that mixed lactobacilli consumption clearly modulated the taxonomic composition of the intestinal flora of mice fed with high fat diet.
To identify predominant microbiota in each group, LEfSe analysis was performed. The resulting cladogram (Figure 7) disclosed that Bacteroidetes, Alloprevotella and Alistipes were more dominant in the control group than the other two groups. The HFD group was enriched with Firmicutes, Lachnospiraceae UCG-006, Lachnoclostridium, Romboutsia, Parabacteroides, GCA-900066575 and Eubacterium xylanophilum group, while the MX group was enriched with Lachnospiraceae NK4A136 group and Bacteroides. The histogram of the Latent Dirichlet Allocation (LDA) scores (Figure 8) further revealed a clear difference between the control, HFD and MX groups in terms of the composition of biological clades, which was in agreement with the aforementioned results.
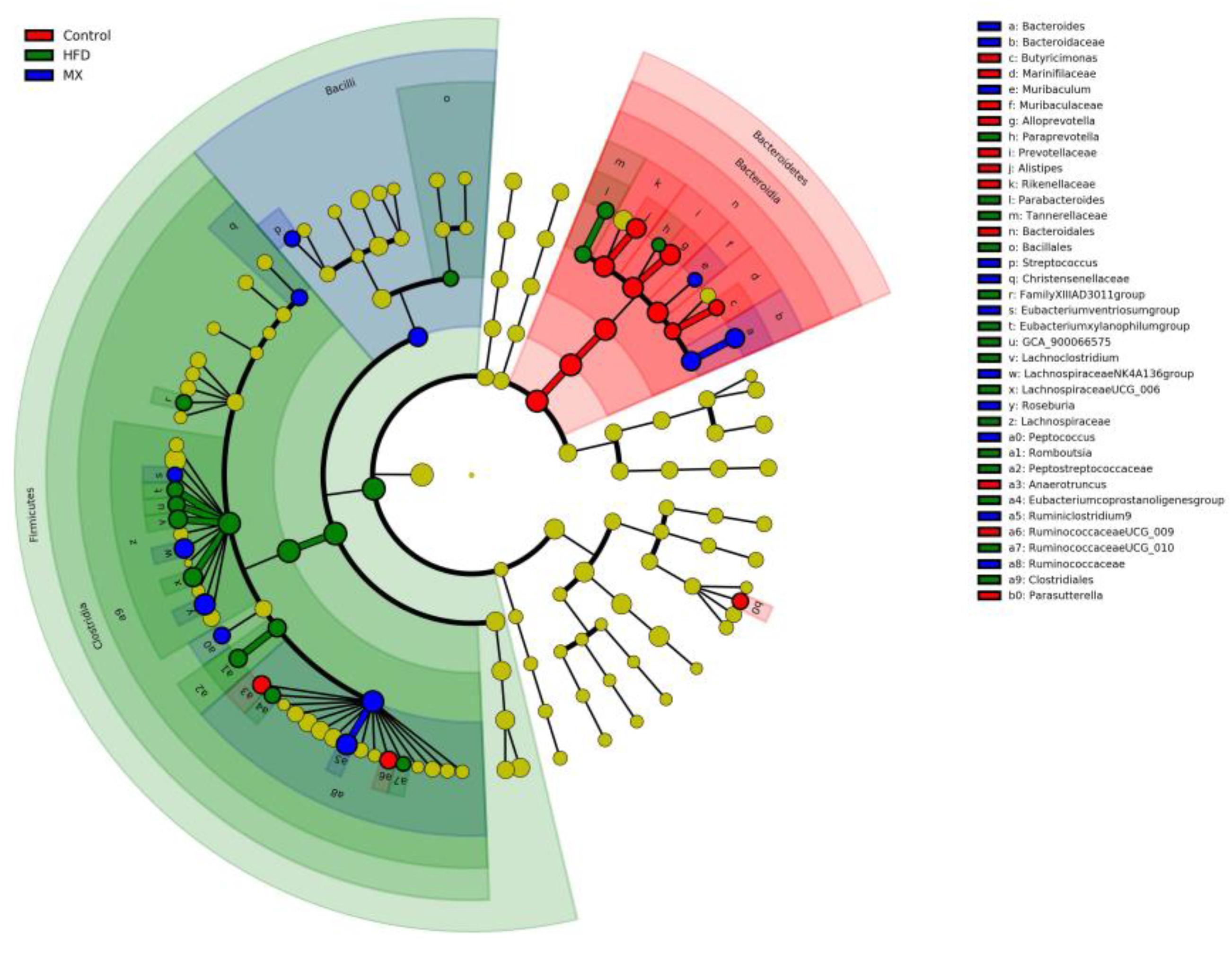
Figure 7. Linear Discriminant Analysis Effect Size (LEfSe) comparison of gut microbiota between the Control, HFD and MX groups. Control, control group; HFD, high fat diet group; and MX, mixed lactobacilli group. n = 3 per group.
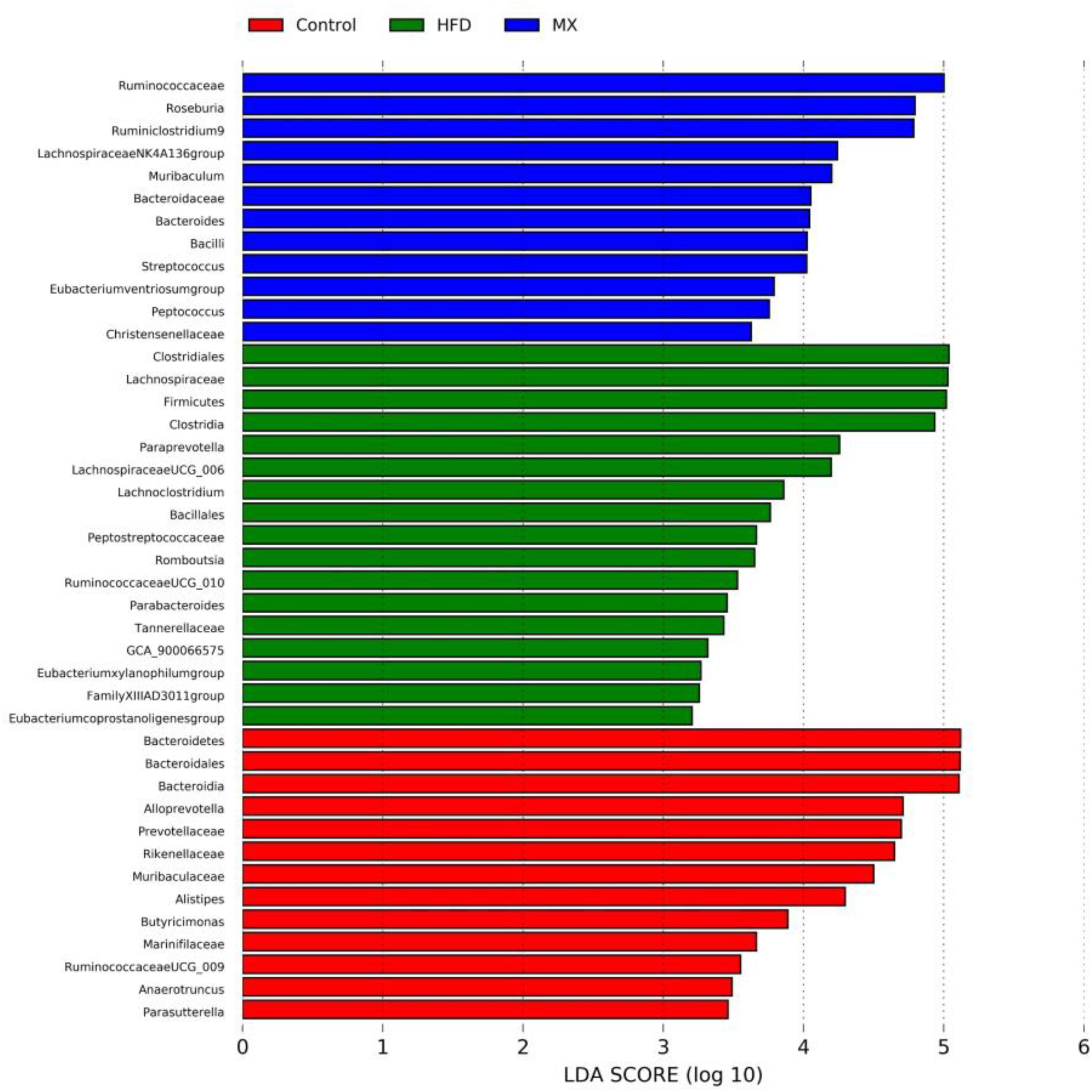
Figure 8. The Latent Dirichlet Allocation (LDA) scores indicate the effect size and ranking of each differentially abundant taxon between the Control, HFD and MX groups. Control, control group; HFD, high fat diet group; and MX, mixed lactobacilli group. n = 3 per group.
To explore changes in SCFAs metabolism in the intestine, the levels of acetic acid, butyric acid, propionic acid and valeric acid in the cecal contents of different groups of mice were determined by GC-MS (Supplementary Figure S1). As shown in Table 2, the levels of acetic acid and butyric acid were strikingly decreased in the HFD group compared with the control group (p < 0.01), whereas supplement with mixed lactobacilli significantly increased the levels of the two SCFAs (p < 0.05 and p < 0.01). However, after 8 weeks of mixed lactobacilli treatment, the levels of propionic acid and valeric acid did not change significantly.
The correlations between the relative abundances of the dominant gut microbial community at the genus level (the top 35 genera according to the relative abundance) and obesity-related indicators were determined by Spearman’s correlation analysis (Figure 9). Obesity-related indicators included short chain fatty acids (acetic acid, butyric acid, propionic acid and valeric acid), genes of epididymal adipose tissue (AMPK-α, HSL, ACC, FAS, PPAR-γ, and C/EBP-α), hepatic parameters (TG, CAT, SOD, GSH-Px, GSH, MDA, ALT, and AST) and blood lipids (TG, TC, HDL-C, and LDL-C). The relative abundances of Bifidobacterium, Bacteroides, Alistipes, Lachnospiraceae NK4A136 group and Alloprevotella were positively correlated with the levels of acetic acid and butyric acid, while the relative abundances of Parabacteroides, Eubacterium xylanophilum group, GCA-900066575, Lachnoclostridium, Lachnospiraceae UCG-006 and Romboutsia were negatively correlated with the levels of acetic acid and butyric acid. Moreover, the relative abundances of Bifidobacterium, Alistipes and Alloprevotella showed significant positive correlations with the mRNA expression levels of AMPK-α and HSL (p < 0.01 or p < 0.05) and significant negative correlations with the mRNA expression levels of ACC, FAS and PPAR-γ (p < 0.01 or p < 0.05), but the relative abundances of Parabacteroides, GCA-900066575, Lachnoclostridium and Lachnospiraceae UCG-006 showed the opposite trend (p < 0.01 or p < 0.05). In addition, Bifidobacterium, Bacteroides, Alistipes, Lachnospiraceae NK4A136 group and Alloprevotella were negatively correlated with TG and MDA in the liver and TG, TC, LDL-C, ALT and AST in the serum, whereas they were positively correlated with CAT, SOD, GSH-Px and GSH in the liver and HDL-C in the serum, but Parabacteroides, Eubacterium xylanophilum group, GCA-900066575, Lachnoclostridium, Lachnospiraceae UCG-006 and Romboutsia were opposite.
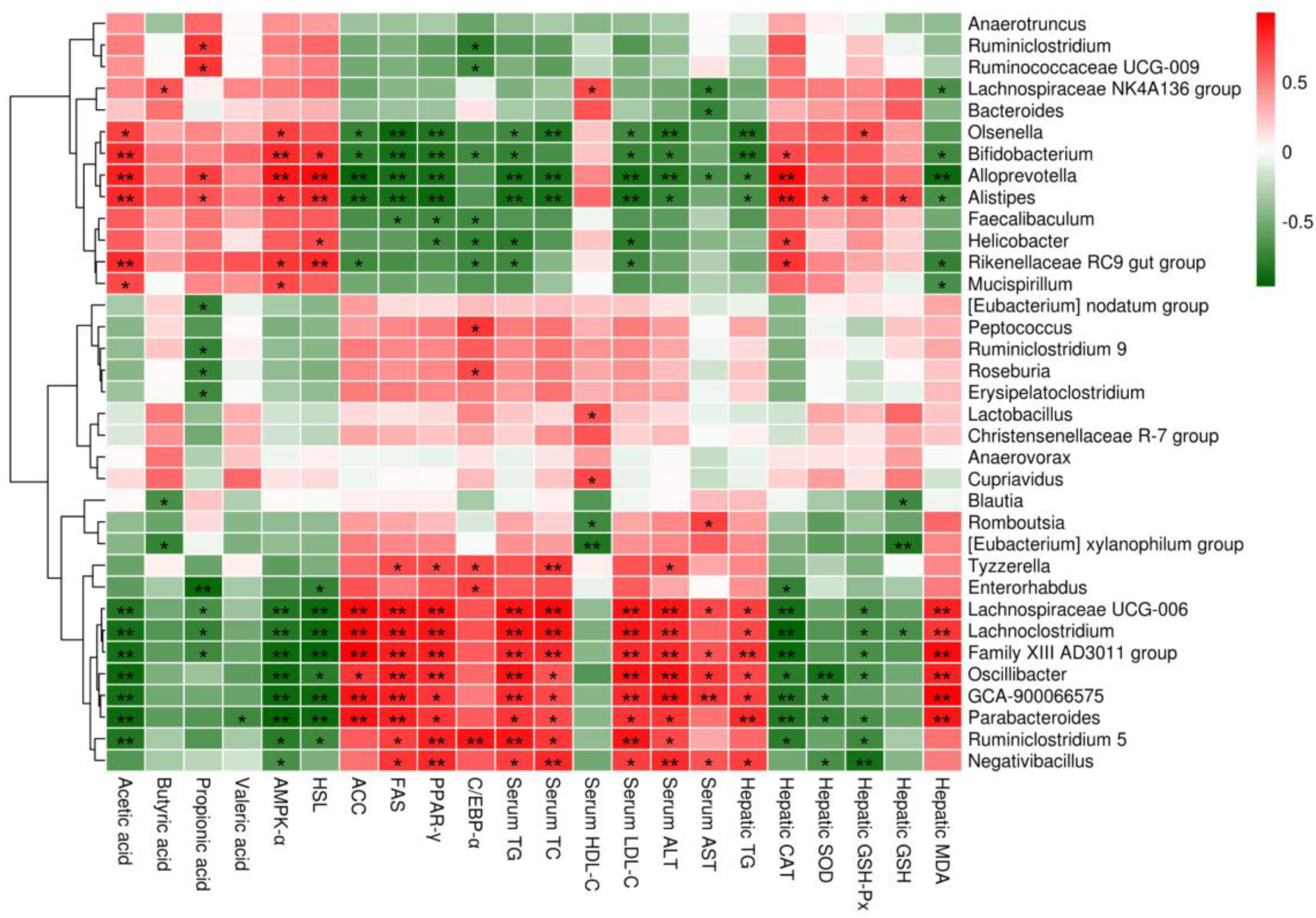
Figure 9. Heatmap of Spearman’s correlation between the relative abundance of gut microbiome at the genus level and obesity-related indicators. Short chain fatty acids: acetic acid, butyric acid, propionic acid and valeric acid; genes of epididymal adipose tissue: adenosine 5′-monophosphate-activated protein kinase-α (AMPK-α), hormone-sensitive lipase (HSL), acetyl CoA carboxylase (ACC), fatty acid synthetase (FAS), peroxisome proliferators-activated receptor-γ (PPAR-γ) and CAATT/enhancer binding proteins α (C/EBPα); hepatic parameters: triglyceride (TG), catalase (CAT), superoxide dismutase (SOD), glutathione peroxidase (GSH-Px), glutathione (GSH), malondialdehyde (MDA), alanine aminotransferase (ALT) and aspartate aminotransferase (AST); blood lipids: TG, total cholesterol (TC), high density lipoprotein cholesterol (HDL-C) and low density lipoprotein cholesterol (LDL-C). Significant correlations are indicated by *p < 0.05 and **p < 0.01.
Obesity, a metabolic disease, is becoming more common and is prone to other metabolic complications (such as cardiovascular disease and type 2 diabetes) (Sonnenburg and Backhed, 2016; Dahiya et al., 2017), pharmacotherapy causes adverse effects (Srivastava and Apovian, 2018; Rosa-Gonçalves and Majerowicz, 2019), so it is imperative to seek natural and non-toxic anti-obesity substances. Lactic acid bacteria have been used in fermented dairy products for more than 100 years (Aryana and Olson, 2017) and are generally regarded as safe (GRAS) (Özogul and Hamed, 2017). Moreover, to date, a large amount of evidence has shown that some lactic acid bacteria have an effective anti-obesity effect in animal research and clinical research (Dahiya et al., 2017). Previous researches in our laboratory have demonstrated that L. plantarum KLDS1.0344 and L. plantarum KLDS1.0386 have strong acid and bile salt resistance, high cell adhesion activities and lipid metabolism regulation properties (Tang et al., 2016; Jin et al., 2017; Lu et al., 2018; Yan et al., 2019; Yue et al., 2019). Therefore, further researches were carried out and it was found that mixed lactobacilli (L. plantarum KLDS1.0344 and L. plantarum KLDS1.0386) could prevent the formation of obesity in high fat diet-fed mice (Lu J. et al., 2019), which confirmed the probiotic properties of the mixed lactobacilli. However, the underlying mechanisms were unclear. Thus, in this study, the possible mechanisms by which the same lactobacilli strains could prevent obesity were mined.
The anti-obesity effect of mixed lactobacilli in vivo was studied using the D12492 high fat diet-induced obesity model, which is a widely used model for obesity research. Convincing evidence has demonstrated that obesity is often accompanied by dyslipidemia, such as elevated levels of TC, TG, and LDL-C, as well as decreased HDL-C levels, which are risk factors for cardiovascular disease (Hunter and Hegele, 2017; Kotsis et al., 2017). Thus, after 8 weeks of animal feeding, the levels of TC, TG, LDL-C and HDL-C in the serum of the three groups of mice were measured to evaluate their blood lipid metabolism. Our data indicated that compared with the control group, the serum levels of TC, TG and LDL-C were significantly increased, and the serum levels of HDL-C were significantly decreased in the HFD group, as expected. However, such changes in mice fed a high fat diet were reversed by the mixed lactobacilli treatment, implying an improvement in metabolic dysfunction. The findings were in agreement with previous research that a probiotic L. plantarum strain isolated from the homemade kumiss could effectively inhibit serum TC, TG, LDL-C and HDL-C changes from feeding with high fat diet (Wang et al., 2012).
Obesity is frequently characterized by the development of non-alcoholic fatty liver disease (NAFLD) (Michelotti et al., 2013; Canfora et al., 2019). The liver, an important site of lipid metabolism in the body, maintains the balance of lipid synthesis and decomposition under normal conditions, whereas a high-fat diet breaks this balance, causing excessive lipid accumulation (that is, hepatic steatosis) and oxidative stress in the liver, indicating the occurrence of liver injury (Rotman and Sanyal, 2017; Friedman et al., 2018). On this account, we aimed to determine the preventive effect of mixed lactobacilli intervention on NAFLD in high fat diet-fed mice. First, hepatic lipid accumulation was evaluated by measuring TG concentration. A significant elevated TG concentration in the liver was observed in the HFD group, which was consistent with the large number of lipid droplets in liver histopathological sections of the HFD group observed in our previous studies (Lu J. et al., 2019), implying that hepatic steatosis occurred. These results were in agreement with the earlier reports in which, obese mice suffered from non-alcoholic hepatic steatosis (Liou et al., 2019). However, it is noteworthy that mixed lactobacilli treatment substantially attenuated hepatic steatosis. Second, we assessed liver oxidative stress by analyzing antioxidant indices and lipid peroxidation biomarkers, including SOD, CAT, GSH-Px, GSH, and MDA. SOD, a critical antioxidant, can convert superoxide radical anions (O2–, incompletely reduced forms of oxygen) into hydrogen peroxide (H2O2), which in turn is catalyzed into water by CAT and GSH-Px (Turrens, 2003; Borrelli et al., 2018). GSH as a non-enzymatic antioxidant can directly scavenge reactive oxygen species (ROS) by binding with them (Anu et al., 2014). MDA is the end product of free radical-mediated lipid peroxidation and is currently considered a reliable biomarker related to oxidative stress (Wang et al., 2013). Our results demonstrated that oral administration of mixed lactobacilli significantly increased levels of SOD, CAT, GSH-Px, and GSH while significantly reducing MDA levels in high fat diet-fed mice, suggesting amelioration of liver oxidative stress. Finally, serum ALT and AST levels, often used to determine the extent of liver function damage (Zhao et al., 2017a), were measured. We found that serum levels of ALT and AST were significantly decreased in the MX group. Similarly, it has been previously stated that the treatment of the probiotic mixture (6 Lactobacillus and 3 Bifidobacterium) reduced serum ALT and AST levels in high fat diet-fed rats (Liang et al., 2018). Taken together, the mixed lactobacilli could inhibit liver lipid accumulation, enhance liver antioxidant capacity and improve liver function.
To further explore the potential mechanisms by which mixed lactobacilli inhibited obesity induced by high fat diet in mice, we examined the expression of lipid metabolism-related genes in epididymal adipose tissue. Adipose tissue, as one of the main sites for storing triglycerides, is an important organ regulating lipid metabolism (Schneeberger et al., 2015; Park et al., 2017). Extensive amounts of reports have shown that PPAR-γ and C/EBP-α are key transcription factors for adipocyte differentiation in adipose tissue (Prestwich and Macdougald, 2007). Park et al. (2014) demonstrated that L. plantarum LG42 supplementation strikingly decreased mRNA expression of PPAR-γ and C/EBP-α in high fat diet-fed mice. In this study, compared with HFD group, mixed lactobacilli treatment significantly down-regulated PPAR-γ mRNA levels, and also lowered the mRNA level of C/EBP-α but not significant. The AMPK pathway is a classical pathway that regulates lipid metabolism. AMPK is a known cellular energy sensor that shuts down anabolic pathways such as fatty acid synthesis (Hardie and Pan, 2002). ACC and FAS are key enzymes in fatty acid synthesis (Liou et al., 2019). Specifically, activation of AMPK-α stimulates ACC phosphorylation, which blocks the expression of FAS (Liou et al., 2019). HSL is the rate-limiting enzyme in the breakdown of triglycerides in adipose tissue (Liou et al., 2019). In the present study, the mRNA levels of AMPK-α and HSL were remarkably reduced in the HFD group compared with the control group, while the mRNA levels of ACC and FAS were significantly elevated, and these changes were completely eliminated by the treatment of mixed lactobacilli, similar to the study by Qiao et al. (Qiao et al., 2015).
Accumulating evidence suggests that the gut microbiota is a key environmental factor in the development of obesity (Walker and Julian, 2013). For instance, a previous study showed that germ-free, lean mice transplanted with intestinal microbiota from obese mice became obese, while those transplanted with intestinal flora from lean mice remained lean (Turnbaugh et al., 2006). Accordingly, gut microbiota is considered as a new target for the prevention and treatment of obesity. Subsequently, to investigate whether the mixed lactobacilli exerted its anti-obesity effects also through regulating the gut microbiota, we determined the intestinal bacteria composition of the three groups of mice. At the phylum level, we observed that the relative abundance of Firmicutes increased and the relative abundance of Bacteroidetes decreased in the HFD group compared with that in the control group, resulting in an significant increase in the ratio of Firmicutes/Bacteroidetes, which is consistent with many previous reports (Ilseung et al., 2012; Chang et al., 2015; Zhang et al., 2018). The researchers found that Firmicutes could produce more harvestable energy than Bacteroidetes, so in the case of an increase in the relative abundance of Firmicutes and a decrease in the relative abundance of Bacteroidetes, the absorption of calories increased and then promoted obesity (Turnbaugh et al., 2006; Komaroff, 2017). Noteworthily, the mixed lactobacilli supplementation in high fat diet-fed mice attenuated the increase in Firmicutes, the decrease in Bacteroidetes and the increase in the ratio of Firmicutes/Bacteroidetes. At the genus level, our results reflected that the relative abundances of Bifidobacterium, Bacteroides, Alistipes, Lachnospiraceae NK4A136 group and Alloprevotella decreased while that of Parabacteroides, Eubacterium xylanophilum group, GCA-900066575, Lachnoclostridium, Lachnospiraceae UCG-006 and Romboutsia increased in the HFD group compared with the control group. In line with our results, earlier studies found that the abundances of Bifidobacterium (Zhu et al., 2018), Bacteroides (Zhang et al., 2015; Martinez et al., 2017), Alistipes (Zhu et al., 2018), Lachnospiraceae NK4A136 group (Hu et al., 2019), and Alloprevotella (Wang et al., 2019) were negatively correlated with obesity, and the abundances of Parabacteroides (Lieber et al., 2018; Zhao et al., 2018), Eubacterium xylanophilum group (Zhu J. et al., 2019), GCA-900066575 (Zhu J. et al., 2019), Lachnoclostridium (Zhao et al., 2017b), Lachnospiraceae UCG-006 (Kang et al., 2019), and Romboutsia (Zhao et al., 2018) were positively correlated with obesity. Bifidobacterium, a beneficial microbial species, producing lactic acid and acetic acid, can reduce intestinal pH and inhibit the growth of various detrimental bacteria to maintain intestinal health (Wang R. et al., 2018). Bacteroides, Alistipes, Lachnospiraceae NK4A136 group and Alloprevotella are also capable of producing SCFAs such as acetic acid and butyric acid (Borton et al., 2017; Gotoh et al., 2017; Jiang et al., 2018; Wang J. et al., 2018; Yin et al., 2018). It was reported that the abundance of Bacteroides was reduced in individuals with atherosclerotic cardiovascular disease (Jie et al., 2017) and post-inflammatory irritable bowel syndrome (Chen et al., 2017). Alistipes can effectively inhibit inflammation via preventing LPS-induced TNF-α release at higher concentrations (Canfora et al., 2015) and has been found at lower levels in the gut of patients with hepatocellular carcinoma (Ren et al., 2019), colitis (Jiang et al., 2018) and non-alcoholic fatty liver disease (Tang et al., 2018). Previous studies have indicated that Lachnospiraceae NK4A136 group may have an anti-inflammatory effect (Li et al., 2019) and its relative abundance was reduced in mice with immune dysfunction caused by cyclophosphamide (Zhu G. et al., 2019). Alloprevotella has been shown to be significantly reduced in mice with metabolic syndrome (Shang et al., 2017), a disease easily caused by obesity. According to previous studies, Parabacteroides was enriched in individuals with type 2 diabetes (Qin et al., 2012) and Behcet’s disease (Ye et al., 2018). Eubacterium xylanophilum group, GCA-900066575, Lachnoclostridium, Lachnospiraceae UCG-006 and Romboutsia belong to Lachnospiraceae, which may suppress the growth of SCFA-producing bacteria (Duncan et al., 2002) and were found to be associated with metabolic disorders and colon cancer (Keishi and Kikuji, 2014; Meehan and Beiko, 2014). Importantly, the mixed lactobacilli treatment reversed the changes in several of the above genus.
Changes in the gut microbiota cause changes in the SCFAs levels, which are negatively correlated with obesity (Coelho et al., 2018). Lu Y. et al. (2016) have shown that supplementation of SCFAs in the diet dramatically inhibited the body weight gain in high fat diet-fed mice. SCFAs produced by bacteria may induce the release of gut-derived satiety hormones, such as peptide YY (PYY) and glucagon-like peptide-1 (GLP-1), which suppress food intake and increase satiety (Canfora et al., 2015). Furthermore, studies have indicated that SCFAs can reach the liver through the portal vein, thereby activating the nuclear erythroid 2-related factor 2 (Nrf-2) pathway to alleviate oxidative stress (Li et al., 2018) and activating the AMPK pathway to inhibit lipid accumulation (Canfora et al., 2015). In addition, SCFAs have been reported to regulate lipid metabolism in adipose tissue by modulating related signaling pathways (Gijs et al., 2015). Therefore, we determined the levels of acetic acid, butyric acid, propionic acid and valeric acid in each group by GC-MS. Intriguingly, compared to the HFD group, mixed lactobacilli supplementation significantly increased the concentrations of acetic acid and butyric acid, which was in agreement with changes in the intestinal microbiota. Based on the above results, we speculated that the mixed lactobacilli treatment might attenuate liver oxidative stress, reduce liver lipid accumulation and improve lipid metabolism of adipose tissue by regulating intestinal microbiota and metabolites. In line with our finding, Yin et al. (2018) concluded that one of the potential mechanisms of melatonin inhibiting obesity might related to the increased levels of acetic acid and butyric acid.
To further confirm the role of gut microbiota in anti-obesity, we also analyzed the correlation between the relative abundance of mixed lactobacilli-manipulated gut microbiota and obesity-related indicators through Spearman’s correlation analysis. In general, Bifidobacterium, Bacteroides, Alistipes, Lachnospiraceae NK4A136 group and Alloprevotella were positively correlated with acetic acid and butyric acid in intestine, AMPK-α and HSL in epididymal adipose tissue, CAT, SOD, GSH-Px and GSH in liver, and HDL-C in serum, while they were negatively correlated with ACC, FAS and PPAR-γ in epididymal adipose tissue, TG and MDA in liver, and TG, TC, LDL-C, ALT and AST in serum, but Parabacteroides, Eubacterium xylanophilum group, GCA-900066575, Lachnoclostridium, Lachnospiraceae UCG-006 and Romboutsia displayed the opposite trend. Therefore, the results also indicated that mixed lactobacilli administration could effectively modulate the gut microbiota induced by high fat diet, and then improve obesity-related indicators.
The mixed lactobacilli intervention could alleviate the changes induced by high fat diet including disordered blood lipids, liver oxidative stress and liver injury. Further, the mixed lactobacilli intervention modulated the gut microbiota of high fat diet-fed mice, resulting in increased SCFAs (acetic acid and butyric acid), which regulated lipid metabolism in adipose tissue and reduced liver lipid accumulation, thereby preventing obesity. Hence, our results offer significant insight into the oral administration of mixed lactobacilli to suppress obesity in high fat diet-fed mice.
All datasets generated for this study are included in the article/Supplementary Material.
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of the Northeast Agricultural University.
HL did the experiments and wrote the manuscript. FL and JL did the experiments. JS, JG, and FY processed the data. BL and GH corrected the manuscript.
This study was financially supported by the National Key Research and Development Program of China (No. 2017YFD0400303).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00512/full#supplementary-material
Almalki, O. M., Lee, W. J., Chen, J. C., Ser, K. H., Lee, Y. C., and Chen, S. C. (2017). Revisional gastric bypass for failed restrictive procedures: comparison of Single-Anastomosis (Mini-) and Roux-en-Y Gastric Bypass. Obes. Surg. 28, 970–975. doi: 10.1007/s11695-017-2991-0
American College of Cardiology/American Heart Association Task Force on Practice Guidelines Obesity Expert Panel, 2013 (2014). Expert Panel Report: guidelines (2013) for the management of overweight and obesity in adults. Obesity 22, S41–S410.
Anu, R., Amit, K., Vivek, S., Brijesh, Y., Ruchi, T., Sandip, C., et al. (2014). Oxidative stress, prooxidants, and antioxidants: the interplay. Biomed Res. Int. 2014:761264.
Apovian, C. M., Aronne, L., Rubino, D., Still, C., Wyatt, H., Burns, C., et al. (2013). A randomized, phase 3 trial of naltrexone SR/bupropion SR on weight and obesity-related risk factors (COR-II). Obesity 21, 935–943. doi: 10.1002/oby.20309
Aryana, K. J., and Olson, D. W. (2017). A 100-year review: yogurt and other cultured dairy products. J. Dairy Sci. 100, 9987–10013. doi: 10.3168/jds.2017-12981
Bluher, M. (2019). Obesity: global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 15, 288–298. doi: 10.1038/s41574-019-0176-8
Borrelli, A., Bonelli, P., Tuccillo, F. M., Goldfine, I. D., Evans, J. L., Buonaguro, F. M., et al. (2018). Role of gut microbiota and oxidative stress in the progression of non-alcoholic fatty liver disease to hepatocarcinoma: current and innovative therapeutic approaches. Redox Biol. 15, 467–479. doi: 10.1016/j.redox.2018.01.009
Borton, M. A., Sabag-Daigle, A., Wu, J., Solden, L. M., O’Banion, B. S., Daly, R. A., et al. (2017). Chemical and pathogen-induced inflammation disrupt the murine intestinal microbiome. Microbiome 5:47. doi: 10.1186/s40168-017-0264-8
Canfora, E. E., Jocken, J. W., and Blaak, E. E. (2015). Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 11, 577–591. doi: 10.1038/nrendo.2015.128
Canfora, E. E., Meex, R. C., Venema, K., and Blaak, E. E. (2019). Gut microbial metabolites in obesity. NAFLD and T2DM. Nat. Rev. Endocrinol. 15, 261–273. doi: 10.1038/s41574-019-0156-z
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336.
Chang, C.-J., Lin, C.-S., Lu, C.-C., Martel, J., Ko, Y.-F., Ojcius, D. M., et al. (2015). Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat. Commun. 6:7489. doi: 10.1038/ncomms8489
Chen, Y., Xiao, S., Gong, Z., Zhu, X., Yang, Q., Li, Y., et al. (2017). Wuji wan formula ameliorates diarrhea and disordered colonic motility in post-inflammation irritable bowel syndrome rats by modulating the gut microbiota. Front. Microbiol. 8:2307. doi: 10.3389/fmicb.2017.02307
Coelho, O. G. L., Cândido, F. G., and Alfenas, R. C. G. (2018). Dietary fat and gut microbiota: mechanisms involved in obesity control. Crit. Rev. Food Sci. Nutr. 59, 3045–3053. doi: 10.1080/10408398.2018.1481821
Dahiya, D. K., Puniya, M., Shandilya, U. K., Dhewa, T., Kumar, N., Kumar, S., et al. (2017). Gut microbiota modulation and its relationship with obesity using prebiotic fibers and probiotics: a review. Front. Microbiol. 8:563. doi: 10.3389/fmicb.2017.00563
Davidson, M. H., Hauptman, J., Digirolamo, M., Foreyt, J. P., et al. (1999). Weight control and risk factor reduction in obese subjects treated for 2 years with orlistat: a randomized controlled trial. JAMA 281, 235–242.
Dietz, W. H. (2017). Obesity and excessive weight gain in young adults: new targets for prevention. JAMA 318, 241–242.
Duncan, S. H., Adela, B., Stewart, C. S., Pryde, S. E., and Flint, H. J. (2002). Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl. Environ. Microbiol. 68, 5186–5190. doi: 10.1128/aem.68.10.5186-5190.2002
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Friedman, S. L., Neuschwander-Tetri, B. A., Rinella, M., and Sanyal, A. J. (2018). Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 24, 908–922. doi: 10.1038/s41591-018-0104-9
Gadde, K. M., Allison, D. B., Ryan, D. H., Peterson, C. A., Troupin, B., Schwiers, M. L., et al. (2011). Effects of low-dose, controlled-release, phentermine plus topiramate combination on weight and associated comorbidities in overweight and obese adults (CONQUER): a randomised, placebo-controlled, phase 3 trial. Lancet 377, 1341–1352. doi: 10.1016/S0140-6736(11)60205-5
GBD 2015 Obesity Collaborators, Afshin, A., Forouzanfar, M. H., Reitsma, M. B., Sur, P., Estep, K., et al. (2017). Health effects of overweight and obesity in 195 countries over 25 years. N. Engl. J. Med. 377, 13–27.
Gijs, D. B., Aycha, B., Albert, G., Karen, V. E., Rick, H., Dijk, T. H., et al. (2015). Short-chain fatty acids protect against high-fat diet-induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes Metab. Res. Rev. 64, 2398–2408.
Gotoh, A., Nara, M., Sugiyama, Y., Sakanaka, M., Yachi, H., Kitakata, A., et al. (2017). Use of Gifu Anaerobic Medium for culturing 32 dominant species of human gut microbes and its evaluation based on short-chain fatty acids fermentation profiles. Biosci. Biotechnol. Biochem. 81, 2009–2017. doi: 10.1080/09168451.2017.1359486
Hao, J., Zhang, Y., Yan, X., Yan, F., Sun, Y., Zeng, J., et al. (2018). Circulating adipose fatty acid binding protein is a new link underlying obesity-associated breast/mammary tumor development. Cell Metab. 28, 689–705.e5. doi: 10.1016/j.cmet.2018.07.006
Hardie, D. G., and Pan, D. A. (2002). Regulation of fatty acid synthesis and oxidation by the AMP-activated protein kinase. Biochem. Soc. Trans. 30, 1064–1070.
Heymsfield, S. B., and Wadden, T. A. (2017). Mechanisms, pathophysiology, and management of obesity. N. Engl. J. Med. 376, 254–266.
Hu, S., Wang, J., Xu, Y., Yang, H., Wang, J., Xue, C., et al. (2019). Anti-inflammation effects of fucosylated chondroitin sulphate from Acaudina molpadioides by altering gut microbiota in obese mice. Food Funct. 10, 1736–1746. doi: 10.1039/c8fo02364f
Hunter, P. M., and Hegele, R. A. (2017). Functional foods and dietary supplements for the management of dyslipidaemia. Nat. Rev. Endocrinol. 13, 278–288.
Hurt, R. T., Kulisek, C., Buchanan, L. A., and Mcclave, S. A. (2010). The obesity epidemic: challenges, health initiatives, and implications for gastroenterologists. Gastroenterol. Hepatol. 6, 780–792.
Ilseung, C., Shingo, Y., Laura, C., Methé, B. A., Jiri, Z., Kelvin, L., et al. (2012). Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488, 621–626. doi: 10.1038/nature11400
Jiang, L., Lv, J., Liu, J., Hao, X., Ren, F., and Guo, H. (2018). Donkey milk lysozyme ameliorates dextran sulfate sodium-induced colitis by improving intestinal barrier function and gut microbiota composition. J. Funct. Foods 48, 144–152.
Jie, Z., Xia, H., Zhong, S. L., Feng, Q., Li, S., Liang, S., et al. (2017). The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 8:845. doi: 10.1038/s41467-017-00900-1
Jin, D., Yu, S., Li, B., Li, N., Yan, F., and Huo, G. (2017). Effect of Lactobacillus plantarum KLDS1.0386 on the cholesterol metabolism in C57BL/6J mice. Sci. Technol. Food Ind. 38, 275–279. doi: 10.13386/j.issn1002-0306.2017.23.050
Kang, Y., Li, Y., Du, Y., Guo, L., Chen, M., Huang, X., et al. (2019). Konjaku flour reduces obesity in mice by modulating the composition of the gut microbiota. Int. J. Obes. 43, 1631–1643. doi: 10.1038/s41366-018-0187-x
Keishi, K., and Kikuji, I. (2014). Intestinal colonization by a Lachnospiraceae bacterium contributes to the development of diabetes in obese mice. Microbes Environ. 29, 427–430. doi: 10.1264/jsme2.ME14054
Komaroff, A. L. (2017). The microbiome and risk for obesity and diabetes. JAMA 317, 355–356. doi: 10.1001/jama.2016.20099
Kotsis, V., Antza, C., Doundoulakis, G., and Stabouli, S. (2017). Obesity, Hypertension, and Dyslipidemia. Obesity 11, 227–241.
Le Barz, M., Daniel, N., Varin, T. V., Naimi, S., Demers-Mathieu, V., Pilon, G., et al. (2019). In vivo screening of multiple bacterial strains identifies Lactobacillus rhamnosus Lb102 and Bifidobacterium animalis ssp. lactis Bf141 as probiotics that improve metabolic disorders in a mouse model of obesity. FASEB J. 33, 4921–4935. doi: 10.1096/fj.201801672R
Li, B., Evivie, S. E., Lu, J., Jiao, Y., Wang, C., Li, Z., et al. (2018). Lactobacillus helveticus KLDS1. 8701 alleviates D-galactose-induced aging by regulating Nrf-2 and gut microbiota in mice. Food Funct. 9, 6586–6598. doi: 10.1039/c8fo01768a
Li, J., Wu, T., Li, N., Wang, X., Chen, G., and Lyu, X. (2019). Bilberry anthocyanin extract promotes intestinal barrier function and inhibits digestive enzyme activity by regulating the gut microbiota in aging rats. Food Funct. 10, 333–343. doi: 10.1039/c8fo01962b
Liang, Y., Lin, C., Zhang, Y., Deng, Y., Liu, C., and Yang, Q. (2018). Probiotic mixture of Lactobacillus and Bifidobacterium alleviates systemic adiposity and inflammation in non-alcoholic fatty liver disease rats through Gpr109a and the commensal metabolite butyrate. Inflammopharmacology 26, 1051–1055. doi: 10.1007/s10787-018-0479-8
Lieber, A. D., Beier, U. H., Xiao, H., Wilkins, B. J., Jiao, J., Li, X. S., et al. (2018). Loss of HDAC6 alters gut microbiota and worsens obesity. FASEB J. 33, 1098–1109. doi: 10.1096/fj.201701586R
Liou, C. J., Lee, Y. K., Ting, N. C., Chen, Y. L., Shen, S. C., Wu, S. J., et al. (2019). Protective Effects of Licochalcone A Ameliorates Obesity and Non-Alcoholic Fatty Liver Disease Via Promotion of the Sirt-1/AMPK Pathway in Mice Fed a High-Fat Diet. Cells 8:447. doi: 10.3390/cells8050447
Lu, J., Bi, Y., and Guang, N. (2016). Curbing the obesity epidemic in China. Lancet Diabetes Endocrinol. 4, 470–471.
Lu, J., Li, B., Li, Z., Li, H., and Huo, G. (2018). Antioxidant properties and inhibition of fat deposition of Lactobacillus plantarum KLDS1.0344 in vitro. Food Sci. Tecnol. 43, 1–7. doi: 10.13684/j.cnki.spkj.2018.10.001
Lu, J., Song, Y., Yue, Y., and Huo, G. (2019). Two Lactobacillus plantarum combined to Inhibit the formation of obesity Induced by high fat in mice. Food Sci. Technol. 40, 286–290. doi: 10.13386/j.issn1002-0306.2019.19.049
Lu, Y., Fan, C., Li, P., Lu, Y., Chang, X., and Qi, K. (2016). Short chain fatty acids prevent high-fat-diet-induced obesity in mice by regulating G protein-coupled receptors and gut microbiota. Sci. Rep. 6:37589. doi: 10.1038/srep37589
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Martinez, K., Devlin, J. C., Lacher, C. R., Yin, Y., Cai, Y., Wang, J., et al. (2017). Increased weight gain by C-section: functional significance of the primordial microbiome. Sci. Adv. 3:eaao1874. doi: 10.1126/sciadv.aao1874
Meehan, C. J., and Beiko, R. G. (2014). A phylogenomic view of ecological specialization in the lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol. Evol. 6, 703–713. doi: 10.1093/gbe/evu050
Michelotti, G. A., Machado, M. V., and Diehl, A. M. (2013). NAFLD. NASH and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 10, 656–665.
Murphy, N., Jenab, M., and Gunter, M. J. (2018). Adiposity and gastrointestinal cancers: epidemiology, mechanisms and future directions. Nat. Rev. Gastroenterol. Hepatol. 15, 659–670. doi: 10.1038/s41575-018-0038-1
Neylan, C. J., Kannan, U., Dempsey, D. T., Williams, N. N., and Dumon, K. R. (2016). The surgical management of obesity. Gastroenterol. Clin. North Am. 45, 689–703.
Ogrodnik, M., Zhu, Y., Langhi, L. G. P., Tchkonia, T., Kruger, P., Fielder, E., et al. (2019). Obesity-induced cellular senescence drives anxiety and impairs neurogenesis. Cell Metab. 29, 1061–1077.e8.
Oikonomou, E. K., and Antoniades, C. (2019). The role of adipose tissue in cardiovascular health and disease. Nat. Rev. Cardiol. 16, 83–99.
Özogul, F., and Hamed, I. (2017). The importance of lactic acid bacteria for the prevention of bacterial growth and their biogenic amines formation: a review. Crit. Rev. Food Sci. Nutr. 58, 1660–1670. doi: 10.1080/10408398.2016.1277972
Park, J. E., Oh, S. H., and Cha, Y. S. (2014). Lactobacillus plantarum LG42 isolated from gajami sik-hae decreases body and fat pad weights in diet-induced obese mice. J. Appl. Microbiol. 116, 145–156. doi: 10.1111/jam.12354
Park, S., Ji, Y., Jung, H. Y., Park, H., Kang, J., Choi, S. H., et al. (2017). Lactobacillus plantarum HAC01 regulates gut microbiota and adipose tissue accumulation in a diet-induced obesity murine model. Appl. Microbiol. Biotechnol. 101, 1605–1614. doi: 10.1007/s00253-016-7953-2
Picon-Ruiz, M., Morata-Tarifa, C., Valle-Goffin, J. J., Friedman, E. R., and Slingerland, J. M. (2017). Obesity and adverse breast cancer risk and outcome: Mechanistic insights and strategies for intervention. CA Cancer J. Clin. 67, 378–397. doi: 10.3322/caac.21405
Prestwich, T. C., and Macdougald, O. A. (2007). Wnt/β-catenin signaling in adipogenesis and metabolism. Curr. Opin. Cell Biol. 19, 612–617.
Prospective Studies, and Collaboration. (2009). Body-mass index and cause-specific mortality in 900000 adults: collaborative analyses of 57 prospective studies. Lancet 373, 1083–1096.
Qiao, Y., Sun, J., Xia, S., Li, L., Li, Y., Wang, P., et al. (2015). Effects of different Lactobacillus reuteri on inflammatory and fat storage in high-fat diet-induced obesity mice model. J. Funct. Foods 14, 424–434.
Qin, J., Li, Y., Cai, Z., Li, S., Zhu, J., Zhang, F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. doi: 10.1038/nature11450
Ren, Z., Li, A., Jiang, J., Zhou, L., Yu, Z., Lu, H., et al. (2019). Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 68, 1014–1023. doi: 10.1136/gutjnl-2017-315084
Reuter, S., and Mrowka, R. (2019). Obesity, adipocytes and insulin resistance-Friends for life? Acta Physiol. 225:e13258.
Risk, N. C. D., and Factor Collaboration. (2016). Trends in adult body-mass index in 200 countries from 1975 to 2014: a pooled analysis of 1698 population-based measurement studies with 19⋅2 million participants. Lancet 387, 1377–1396. doi: 10.1016/S0140-6736(16)30054-X
Rosa-Gonçalves, P., and Majerowicz, D. (2019). Pharmacotherapy of obesity: limits and perspectives. Am. J. Cardiovasc. Drugs 19, 349–364.
Rotman, Y., and Sanyal, A. J. (2017). Current and upcoming pharmacotherapy for non-alcoholic fatty liver disease. Gut 66, 180–190. doi: 10.1136/gutjnl-2016-312431
Schauer, P. R., Mingrone, G., Ikramuddin, S., and Wolfe, B. (2016). Clinical outcomes of metabolic surgery: efficacy of glycemic control, weight loss, and remission of diabetes. Diabetes Care 39, 902–911. doi: 10.2337/dc16-0382
Schneeberger, M., Everard, A., Gómez-Valadés, A. G., Matamoros, S., Ramírez, S., Delzenne, N. M., et al. (2015). Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci. Rep. 5:16643. doi: 10.1038/srep16643
Schott, E. M., Farnsworth, C. W., Grier, A., Lillis, J. A., Soniwala, S., Dadourian, G. H., et al. (2018). Targeting the gut microbiome to treat the osteoarthritis of obesity. JCI Insight 3:e95997.
Segata, N., Izard, J., Waldron, L., and Gevers, D. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60.
Shang, Q., Song, G., Zhang, M., Shi, J., Xu, C., Hao, J., et al. (2017). Dietary fucoidan improves metabolic syndrome in association with increased Akkermansia population in the gut microbiota of high-fat diet-fed mice. J. Funct. Foods 28, 138–146.
Shin, Y., Tze Mun, L., Koji, A., Hiroaki, K., Seidai, S., Seiichi, O., et al. (2013). Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101. doi: 10.1038/nature12347
Smith, S. R., Weissman, N. J., Anderson, C. M., Matilde, S., Emil, C., Scott, S., et al. (2010). Multicenter, placebo-controlled trial of lorcaserin for weight management. N. Engl. J. Med. 363, 245–256.
Solas, M., Milagro, F. I., Ramírez, M. J., and Martínez, J. A. (2017). Inflammation and gut-brain axis link obesity to cognitive dysfunction: plausible pharmacological interventions. Curr. Opin. Pharmacol. 37, 87–92. doi: 10.1016/j.coph.2017.10.005
Sonnenburg, J. L., and Backhed, F. (2016). Diet-microbiota interactions as moderators of human metabolism. Nature 535, 56–64. doi: 10.1038/nature18846
Srivastava, G., and Apovian, C. M. (2018). Current pharmacotherapy for obesity. Nat. Rev. Endocrinol. 14, 12–24.
Tang, W., Yao, X., Xia, F., Yang, M., Chen, Z., Zhou, B., et al. (2018). Modulation of the gut microbiota in rats by hugan qingzhi tablets during the treatment of high-fat-diet-induced nonalcoholic fatty liver disease. Oxid. Med. Cell. Longev. 2018:7261619. doi: 10.1155/2018/7261619
Tang, Y., Yu, S., Guo, L., Wang, N., and Huo, G. (2016). Screening and study on probiotic characteristics of a cholesterol-lowering Lactobacillus. Sci. Technol. Food Ind. 37, 142–144. doi: 10.13386/j.issn1002-0306.2016.01.020
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Vincent, M., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031.
Turrens, J. F. (2003). Mitochondrial formation of reactive oxygen species. J. Physiol. 552, 335–344.
Tyrrell, J., Mulugeta, A., Wood, A. R., Zhou, A., Beaumont, R. N., Tuke, M. A., et al. (2018). Using genetics to understand the causal influence of higher BMI on depression. Int. J. Epidemiol. 48, 834–848.
Walker, A. W., and Julian, P. (2013). Microbiology. Fighting obesity with bacteria. Science 341, 1069–1070.
Wang, J., Qin, C., He, T., Kai, Q., Sun, W., Xin, Z., et al. (2018). Alfalfa-containing diets alter luminal microbiota structure and short chain fatty acid sensing in the caecal mucosa of pigs. J. Anim. Sci. Biotechnol. 9:11. doi: 10.1186/s40104-017-0216-y
Wang, J., Wang, P., Li, D., Hu, X., and Chen, F. (2019). Beneficial effects of ginger on prevention of obesity through modulation of gut microbiota in mice. Eur. J. Nutr. 59, 699–718.
Wang, J., Zhang, H., Chen, X., Chen, Y., Menghebilige, and Bao, Q. (2012). Selection of potential probiotic lactobacilli for cholesterol-lowering properties and their effect on cholesterol metabolism in rats fed a high-lipid diet. J. Dairy Sci. 95, 1645–1654. doi: 10.3168/jds.2011-4768
Wang, R., Li, H., Yang, X., Xue, X., Deng, L., Shen, J., et al. (2018). Genetically obese human gut microbiota induces liver steatosis in germ-free mice fed on normal diet. Front. Microbiol. 9:1602. doi: 10.3389/fmicb.2018.01602
Wang, Y., Li, Y., Xie, J., Zhang, Y., Wang, J., Sun, X., et al. (2013). Protective effects of probiotic Lactobacillus casei Zhang against endotoxin-and d-galactosamine-induced liver injury in rats via anti-oxidative and anti-inflammatory capacities. Int. Immunopharmacol. 15, 30–37. doi: 10.1016/j.intimp.2012.10.026
Wunderlich, C. M., Ackermann, P. J., Ostermann, A. L., Adams-Quack, P., Vogt, M. C., Tran, M. L., et al. (2018). Obesity exacerbates colitis-associated cancer via IL-6-regulated macrophage polarisation and CCL-20/CCR-6-mediated lymphocyte recruitment. Nat. Commun. 9:1646.
Xavier, P. S., Arne, A., Ken, F., Frank, G., Alfredo, H., Michel, K., et al. (2015). A Randomized, Controlled Trial of 3.0 mg of Liraglutide in Weight Management. N. Engl. J. Med. 373, 11–22.
Yan, F., Li, N., Yue, Y., Wang, C., Zhao, L., Evivie, S. E., et al. (2019). Screening for potential novel probiotics with dipeptidyl peptidase IV-inhibiting activity for type 2 diabetes attenuation in vitro and in vivo. Front. Microbiol. 10:2855. doi: 10.3389/fmicb.2019.02855
Ye, Z., Zhang, N., Wu, C., Zhang, X., Wang, Q., Huang, X., et al. (2018). A metagenomic study of the gut microbiome in Behcet’s disease. Microbiome 6:135. doi: 10.1186/s40168-018-0520-6
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2013). The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648.
Yin, J., Li, Y., Han, H., Chen, S., Gao, J., Liu, G., et al. (2018). Melatonin reprogramming of gut microbiota improves lipid dysmetabolism in high-fat diet-fed mice. J. Pineal Res. 65:e12524. doi: 10.1111/jpi.12524
Yue, Y., Yan, F., Li, B., Song, Y., Shi, J., Guan, J., et al. (2019). Screening of Lactobacillus with inhibitory of HT-29 cell proliferation and probiotic function. Food Sci. 1–13.
Zaytouni, T., Tsai, P. Y., Hitchcock, D. S., DuBois, C. D., Freinkman, E., Lin, L., et al. (2017). Critical role for arginase 2 in obesity-associated pancreatic cancer. Nat. Commun. 8:242. doi: 10.1038/s41467-017-00331-y
Zhang, X., Zhang, M., Ho, C.-T., Guo, X., Wu, Z., Weng, P., et al. (2018). Metagenomics analysis of gut microbiota modulatory effect of green tea polyphenols by high fat diet-induced obesity mice model. J. Funct. Foods 46, 268–277. doi: 10.1039/c7fo01570d
Zhang, X., Zhao, Y., Xu, J., Xue, Z., Zhang, M., Pang, X., et al. (2015). Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci. Rep. 5:14405. doi: 10.1038/srep14405
Zhao, L., Chen, Y., Xia, F., Abudukerimu, B., Zhang, W., Guo, Y., et al. (2018). A Glucagon-Like peptide-1 receptor agonist lowers weight by modulating the structure of gut microbiota. Front. Endocrinol. 9:233. doi: 10.3389/fendo.2018.00233
Zhao, L., Jiang, Y., Ni, Y., Zhang, T., Duan, C., Huang, C., et al. (2017a). Protective effects of Lactobacillus plantarum C88 on chronic ethanol-induced liver injury in mice. J. Funct. Foods 35, 97–104.
Zhao, L., Zhang, Q., Ma, W., Tian, F., Shen, H., and Zhou, M. (2017b). A combination of quercetin and resveratrol reduces obesity in high-fat diet-fed rats by modulation of gut microbiota. Food Funct. 8, 4644–4656. doi: 10.1039/c7fo01383c
Zheng, X., Qiu, Y., Zhong, W., Baxter, S., Su, M., Li, Q., et al. (2013). A targeted metabolomic protocol for short-chain fatty acids and branched-chain amino acids. Metabolomics 9, 818–827.
Zhu, G., Jiang, Y., Yao, Y., Wu, N., Luo, J., Hu, M., et al. (2019). Ovotransferrin ameliorates the dysbiosis of immunomodulatory function and intestinal microbiota induced by cyclophosphamide. Food Funct. 10, 1109–1122. doi: 10.1039/c8fo02312c
Zhu, J., Kong, Y., Yu, J., Shao, S., Mao, M., Zhao, M., et al. (2019). Consumption of drinking water N-Nitrosamines mixture alters gut microbiome and increases the obesity risk in young male rats. Environ. Pollut. 248, 388–396. doi: 10.1016/j.envpol.2019.02.012
Keywords: Lactobacillus plantarum, obesity, lipid accumulation, lipid metabolisim, gut microbiota, short chain fatty acids
Citation: Li H, Liu F, Lu J, Shi J, Guan J, Yan F, Li B and Huo G (2020) Probiotic Mixture of Lactobacillus plantarum Strains Improves Lipid Metabolism and Gut Microbiota Structure in High Fat Diet-Fed Mice. Front. Microbiol. 11:512. doi: 10.3389/fmicb.2020.00512
Received: 03 January 2020; Accepted: 10 March 2020;
Published: 26 March 2020.
Edited by:
Rosanna Tofalo, University of Teramo, ItalyReviewed by:
Stefano Raimondi, University of Modena and Reggio Emilia, ItalyCopyright © 2020 Li, Liu, Lu, Shi, Guan, Yan, Li and Huo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bailiang Li, MTU4NDYwOTIzNjJAMTYzLmNvbQ==; Guicheng Huo, Z3VpY2hlbmdodW9AMTI2LmNvbQ==
†
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.