
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 13 March 2020
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00425
This article is part of the Research TopicAntimicrobial Resistance As a Global Public Health Problem: How Can We Address It?View all 50 articles
 Mingju Hao1†
Mingju Hao1† Xiaohong Shi1†
Xiaohong Shi1† Jingnan Lv2†
Jingnan Lv2† Siqiang Niu3
Siqiang Niu3 Shiqing Cheng4
Shiqing Cheng4 Hong Du2
Hong Du2 Fangyou Yu5
Fangyou Yu5 Yi-Wei Tang6
Yi-Wei Tang6 Barry N. Kreiswirth7
Barry N. Kreiswirth7 Haifang Zhang2*
Haifang Zhang2* Liang Chen7
Liang Chen7Objective: The emergence of carbapenem-resistant and hypervirulent Klebsiella pneumoniae (CR-hvKp) strains poses a significant public threat, and effective antimicrobial therapy is urgently needed. Recent studies indicated that apramycin is a potent antibiotic with good activity against a range of multi-drug resistant pathogens. In this study, we evaluated the in vitro activity of apramycin against clinical CR-hvKp along with carbapenem-resistant non-hvKp (CR-non-hvKp) isolates.
Methods: Broth microdilution method was used to evaluate the in vitro activities of apramycin, gentamicin, amikacin, imipenem, meropenem, doripenem, ertapenem and other comparator “last-resort” antimicrobial agents, including ceftazidime-avibactam, colistin and tigecycline, against eighty-four CR-hvKp and forty CR-non-hvKp isolates collected from three Chinese hospitals. Multilocus Sequence typing (MLST), molecular capsule typing (wzi sequencing) and antimicrobial resistance genes were examined by PCR and Sanger sequencing. Pulsed-field gel electrophoresis and next generation sequencing were conducted on selected isolates.
Results: Among the 84 CR-hvKp isolates, 97.6, 100, 97.6, and 100% were resistant to imipenem, meropenem, doripenem and ertapenem, respectively. Apramycin demonstrated an MIC50/MIC90 of 4/8 μg/mL against the CR-hvKp isolates. In contrast, the MIC50/MIC90 for amikacin and gentamicin were >64/>64 μg/mL. All CR-hvKp isolates were susceptible to ceftazidime-avibactam, colistin and tigecycline with the MIC50/MIC90 values of 0.5/1, 0.25/0.5, 1/1, respectively. For CR-non-hvKp, The MIC50/90 values for apramycin, gentamicin and amikacin were 2/8, >64/>64, and >64/>64 μg/mL, respectively. There were no statistical significance in the resistance rates of antimicrobial agents between CR-hvKp and CR-non-hvKp groups (p > 0.05). Genetic analysis revealed that all CR-hvKp isolates harbored blaKPC–2, and 94% (n = 79) belong to the ST11 high-risk clone. 93.6% (44/47) of amikacin or gentamicin resistant strains carried 16S rRNA methyltransferases gene rmtB.
Conclusion: Apramycin demonstrated potent in vitro activity against CR-hvKp isolates, including those were resistant to amikacin or gentamicin. Further studies are needed to evaluate the applicability of apramycin to be used as a therapeutic antibiotic against CR-hvKp infections.
Klebsiella pneumoniae is a clinically important pathogen, causing a wide range of diseases including pneumonia, urinary tract infections (UTIs), bloodstream infections, in hospital settings among neonates, elderly and immunocompromised individuals (Paczosa and Mecsas, 2016; Bengoechea and Sa Pessoa, 2019). Hypervirulent K. pneumoniae (hvKp) is an increasing recognized pathotype that is more virulent than classical K. pneumoniae (cKp), which showed the propensity to cause serious, life-threatening infections in otherwise healthy individuals in the community (Shon et al., 2013; Gu et al., 2018).
Historically, hvKp isolates are susceptible to most antibiotics, however, multidrug-resistant hvKp, especially carbapenem-resistant strains, are increasingly reported in the past few years (Cejas et al., 2014; Li et al., 2014; Siu et al., 2014). The emergence of carbapenem-resistant and hypervirulent K. pneumoniae (CR-hvKp) was either due to the acquisition of a pLVPK-like virulence plasmid by classic carbapenem-resistant K. pneumoniae strains (Gu et al., 2018) or through acquisition of a carbapenemase-encoding plasmid by hvKp isolates (Shon et al., 2013). The rmpA, rmpA2, iutA, iucABCD and iroBCDN genes harbored by pLVPK-like virulence plasmid is responsible for the virulence phenotype (Yang et al., 2019). A recent study from China reported a fatal outbreak of ventilator associated pneumonia caused by ST11 CR-hvKp strains (Gu et al., 2018). More recently, one hypermucoviscos isolate with KPC-3 carbapenemase-encoding plasmid was firstly described in the United States, which also exhibited colistin heteroresistance (Wozniak et al., 2019). The rapid dissemination of CR-hvKp strains in different global regions underscores the urgent need of appropriate antibiotic therapy against these life-threating pathogens.
Aminoglycosides including amikacin, gentamicin and tobramycin, which belong to the 4,6-disubstituted deoxystreptamine (DOS) subclass, are among the few drugs that retain certain in vitro activity against CRE (Livermore et al., 2011). However, the acquisition of 16S rRNA methylases confer high level resistance to the 4,6-disubstituted DOS aminoglycosides, which imposes serious challenge for clinical management (Hu et al., 2017). Apramycin is of the 4-monosubstituted DOS subclass, markedly different in its chemical structure from other clinically used aminoglycoside antibiotics. Its unique structure makes apramycin molecules inherently resilient to almost all resistance determinants commonly found in multi-drug resistant strains (Juhas et al., 2019). Indeed, recent studies indicated that apramycin is a potent antibiotic with good activity against a range of clinical pathogens, including multidrug-resistant Mycobacterium tuberculosis, carbapenem-resistant Enterobacteriaceae, spectinomycin-resistant Neisseria gonorrhoeae and Staphylococcus aureus (Smith and Kirby, 2016; Hu et al., 2017; Truelson et al., 2018; Galani et al., 2019; Riedel et al., 2019). In order to evaluate whether apramycin can be served as an alternative antibiotic to treat the CR-hvKp infections, we examined the in vitro activity of apramycin against a collection of CR-hvKp isolates collected from three different hospitals in China. Its in vitro activity was compared with the results from other aminoglycosides (amikacin and gentamicin) and some “last resort” antibiotics, including colistin, tigecycline and ceftazidime-avibactam.
Eighty-four unique (one isolate per patient) CR-hvKp isolates, including 65 isolates from our previous study (Yu et al., 2018a), were included. They were collected from three hospitals across different regions in China (Shandong, Jiangsu and Shanghai) between 2014 and 2019. These isolates were obtained from sputum (53.6%, 45/84), urine (14.3%, 12/84), blood (14.3%, 12/84), pus (4.8%, 4/84), ascites (3.6%, 3/84) and other sources (9.5%, 8/84). In this study, we defined an hvKp strain based upon demonstration of a positive string test (hypermucoviscosity) and co-harboring the genes rmpA (rmpA or rmpA2) and iutA as previously (Yu et al., 2018a). All 84 isolates were hypermucoviscous (string test >5 mm) and were positive for the rmpA/rmpA2 and iutA by PCR analysis (Yu et al., 2018b). In addition, 40 unique strains of carbapenem resistant non-hypervirulent K. pneumoniae (CR-non-hvKp) isolates, with negative hypermucoviscous phenotype and negative rmpA/rmpA2 and iutA PCR results, were collected from the same three hospitals, and were included to compare with the susceptibility results of CR-hvKp.
The antimicrobial susceptibility was assessed via a minimum inhibitory concentration (MIC) broth microdilution method. Results were interpreted using Clinical Laboratory Standards Institute (CLSI) breakpoints (Clinical and Laboratory Standards Institute, 2019) with the exception of tigecycline and polymyxin B, which were interpreted using EUCAST breakpoints (The European Committee on Antimicrobial Susceptibility Testing, 2019). National Antimicrobial Resistance Monitoring System (NARMS) breakpoints for enteric bacteria were used for the interpretation of apramycin susceptibility, in which strains were classified as susceptible (MIC ≤ 8 μg/mL), intermediate (MIC = 16∼32 μg/mL), or resistant (MIC ≥ 64 μg/mL) (National Antibiotic Resistance Monitoring System [NARMS], 2002). MICs were performed in duplicate on two separate days. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as the quality control strains in each experiment. K. pneumoniae ATCC 700603 was used as quality control strain for ceftazidime-avibactam which was tested against ceftazidime-avibactam and ceftazidime alone to confirm the activity of avibactam in the combination.
All CR-Kp isolates including CR-hvKp and CR-non-hvKp were genotyped by multilocus Sequence Typing (MLST) and wzi sequencing (Brisse et al., 2013). Pulsed-field gel electrophoresis (PFGE) of Xbal-digested genomic DNA samples of 15 selected CR-hvKp isolates was performed with a CHEF MAPPER XA apparatus (Bio-Rad, United States). PFGE patterns were analyzed using GelJ software 2.0v (Heras et al., 2015).
A multiplex PCR was used to detect KPC, NDM, VIM, IMP, and OXA-48-like carbapenemase genes, followed by Sanger sequencing (Chen et al., 2011; Chavda et al., 2016; Evans et al., 2016). Common aminoglycoside modifying enzyme (AME) and RMT-encoding genes were determined by PCR and Sanger sequencing. AME coding genes, including aac(3′)-IIa, aac(3′)-IId, aac(3′)-IV, aac(6′)-IIa, aph(3′)-Ia, aph(3′)-Ib, ant(2′)-Ia and ant(3′)-Ia, and ARM/RMT methyltransferases coding genes, including armA, rmtA, rmtB, rmtC, rmtD and npmA (Wu et al., 2009), were examined. The primers used in this study were listed in Supplementary Table S1.
The apramycin resistant isolate, KpApr172, was subject to next generation sequencing using the Illumina HiSeq system (Illumina, San Diego, CA, United States). Genomic DNA was isolated using a Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, United States). Sequencing reads were de novo assembled using Spades 3.12.0 (Bankevich et al., 2012). In silico multi-locus sequence typing was performed using MLST 2.0 (Larsen et al., 2012) while the acquired antimicrobial resistance genes were identified using ResFinder 3.0 (Zankari et al., 2012). The plasmid replicons in the sequenced isolates were identified using PlasmidFinder 2.0 (Carattoli et al., 2014). The whole-genome sequence of KpApr172 was deposited in the GenBank database under accession number WUJI00000000.
Chi-square and Fisher’s exact analysis were employed to compare resistance rate of antimicrobial agents between CR-hvKp and CR-non-hvKp groups. Statistical analyses were performed using GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, United States). P value of <0.05 was considered to indicate statistically significant differences.
The in vitro activities of apramycin and other antimicrobials against 84 clinical isolates CR-hvKp and 40 CR-non-hvKp strains were summarized in Table 1. The CR-hvKp isolates showed high-level resistance to four tested carbapenems, with MIC50/90 values 128/>256, 128/> 256, 64/128 and 128/>256 μg/mL for meropenem, imipenem, doripenem, and ertapenem, respectively, 97.6, 100, 97.6, and 100% of the isolates were resistant to imipenem, meropenem, doripenem and ertapenem. All isolates were resistant to ceftazidime and 97.6% (n = 82) isolates were resistant to aztreonam. Apramycin exhibited significantly better antimicrobial activity than the clinical standard-of-care aminoglycosides gentamicin and amikacin, with MIC50/90 values 4/8 μg/mL). The MIC ranged from 1 to 16 μg/mL (Figure 1). By contrast, the MIC50/90 values for gentamicin and amikacin were both >64/>64 μg/mL.
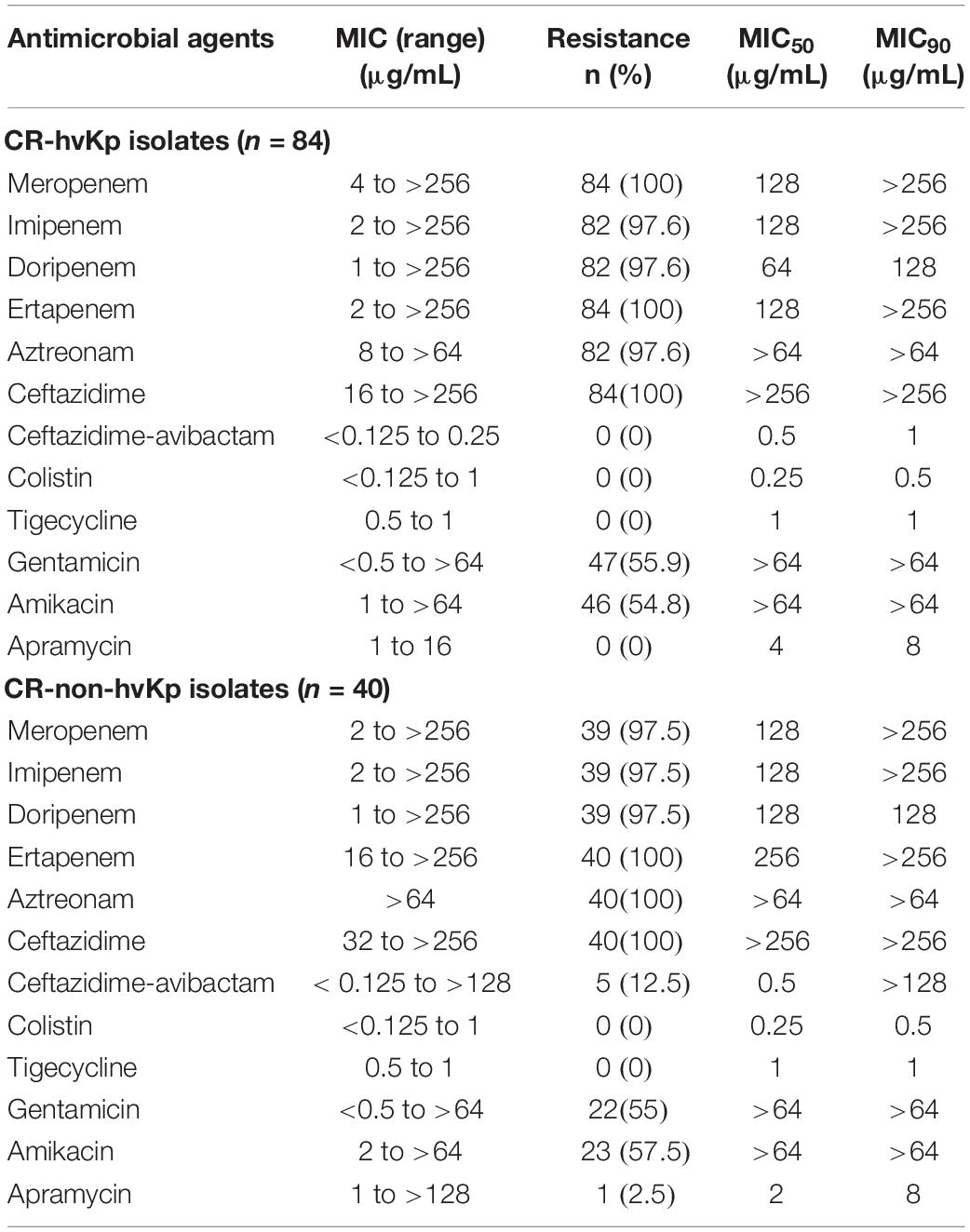
Table 1. In vitro susceptibility of different antibiotics against 84 CR-hvKp and 40 CR-non-hvKp isolates.
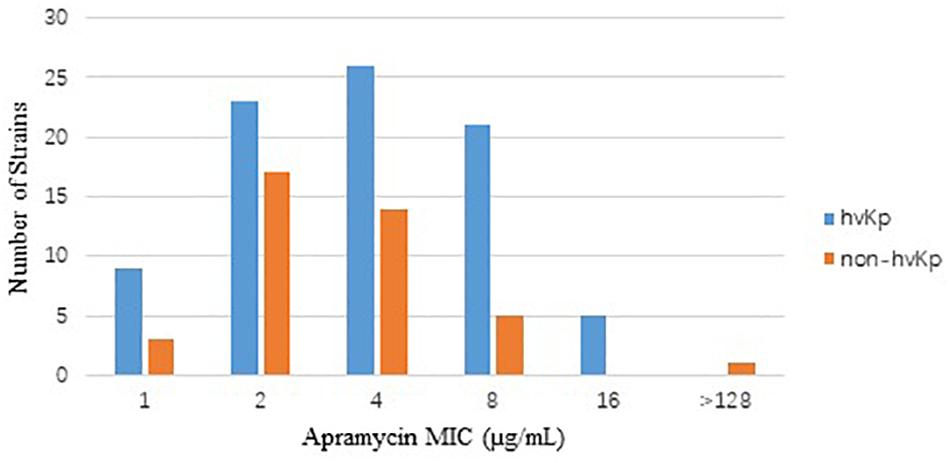
Figure 1. Distribution of apramycin MIC in CR-hvKp (n = 84) and CR-non-hvKp (n = 40) isolates. hvKp: hypervirulent Klebsiella pneumonia, non-hvKp: non-hypervirulent Klebsiella pneumonia.
For CR-non-hvKp, 2.5, 55, and 57.5% of the isolates were resistant to apramycin, gentamicin and amikacin, respectively. The MIC50/90 values for apramycin, gentamicin and amikacin were 2/8, >64/>64, and >64/> 4 μg/mL, respectively. One isolate was resistant to all three aminoglycosides including apramycin (MIC > 128 μg/mL). Five NDM producing CR-non-hvKp isolates were resistant to ceftazidime-avibactam (MIC > 128 μg/mL), but they remained susceptible to colistin and tigecycline. There were no statistical significance in the resistance rates of antimicrobial agents between CR-hvKp and CR-non-hvKp groups (p > 0.05).
Multilocus sequence typing showed that most clinical CR-hvKp strains (94%, 79/84) belong to ST11, the most prevalent CR-Kp ST in China (Gu et al., 2018), while the remaining five isolates were from ST268 (n = 2), ST65 (n = 1), ST412 (n = 1), and ST595 (n = 1). Among the 79 ST11 isolates, 44 have the capsular polysaccharide wzi allele 64 (wzi64) (corresponding with capsular type KL64), and the other 35 isolates carry wzi209 (corresponding with capsular KL47). For CR-non-hvKp strains, 70% (28/40) of the isolates were identified as ST11, carrying wzi64 (n = 18) and wzi209 (n = 10), respectively. The other 12 CR-non-hvKp isolates were from ST2407 (n = 3), ST1308 (n = 3), ST25 (n = 2), ST307 (n = 1), ST1308 (n = 1), ST412 (n = 1), and ST48 (n = 1). To further explore the relatedness of individual CR-hvKp strains, we randomly selected 15 ST11 CR-hvKp strains from the three hospitals for PFGE analysis (Supplementary Figure S1). The results showed these isolates generally clustered by hospitals and capsular KL types, but displayed various PFGE profiles. Our results suggested that apramycin is actively against CR-hvKp and CR-non-hvKp isolates most prevalent STs from different geographical regions (Jiangsu, Shanghai, and Shandong) in China.
The distribution of carbapenemase and aminoglycoside resistance genes were summarized in Table 2. All CR-hvKp isolates and 72.5% (29/40) of CR-non-hvKp isolates harbored blaKPC–2. Five CR-non-hvKp strains harbored blaNDM–1 (n = 4) and blaNDM–5 (n = 1). 93.6% (44/47) of the CR-hvKp and 95.6% (22/23) of the CR-non-hvKp isolates that were resistant to amikacin or gentamicin carried the RMT gene rmtB.
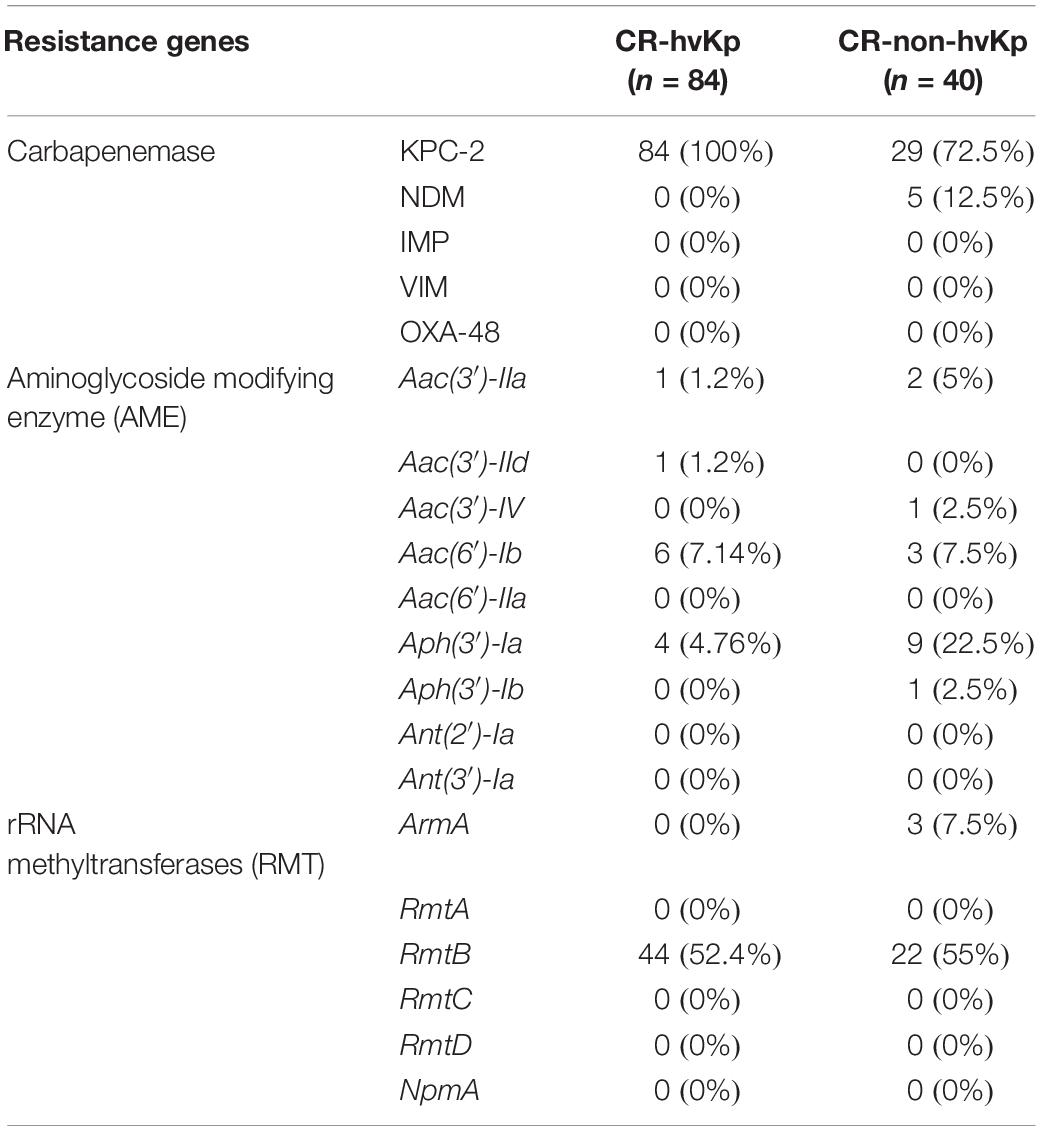
Table 2. Prevalence of carbapenemase and aminoglycoside resistance genes in CR-hvKp and CR-non-hvKp isolates.
Notably, one CR-non-hvKp isolate (KpApr172) was resistant to apramycin. PCR showed that it contains the aminoglycoside acetyltransferase 3-IV gene aac(3)-IV, which encodes resistance to apramycin, gentamicin, netilmicin and tobramycin. Further next generation sequencing revealed that KpApr172 was a strain ST11 and harbored the KL47 capsular type. Resistance gene mining indicated that KpApr172 harbored 17 antimicrobial resistance genes encoding resistance to β-lactams (blaKPC–2, blaNDM–5, blaCTX–M–3, and blaSHV–12), aminoglycosides [armA, aadA2, aac(3)-IId, aac(3)-IV and aph(4)-Ia], macrolid [msr(E) and mph(E)], bleomycin (ble), florfenicol (floR), quaternary ammonium compound (qacEΔ1) and sulfonamide-trimethoprim (sul1, sul2 and dfrA12). The quinolone resistance-determining region (QRDR) genes gyrA, gyrB, parC and parE were further examined, and we observed mutations encoding amino acid substitutions at Ser83-Ile and Asp87-Gly within the QRDR regions of GyrA, and Ser80-Ile in ParC. Examination of the outer membrane protein OmpK35 and OmpK36 genes revealed a premature stop codon at AA63 in OmpK35, due to an adenine (A) deletion mutation at nt 82, and the glycine and aspartic acid insertion at AA134. In silico plasmid replicon typing identified five plasmid groups in KpApr172, belonging to incompatibility groups X3, R, FII and ColE-like (n = 2), respectively.
In order to further characterize the transferability of apramycin resistance gene aac(3)-IV, we attempted to transfer them via conjugation to a recipient E. coli J53 AzR strain (data not shown), however, we were not able to transfer the aac(3)-IV by conjugation, suggesting the gene may be harbored by a non-conjugative plasmid or on the chromosome. Examination of the aac(3)-IV-containing contig from the genome assemblies revealed that aac(3)-IV was located on a 13,397 bp segment, along with the resistance genes, floR, sul2 and aph(4)-Ia (Figure 2). BLASTn analysis showed that the sequences of this contig were identical (100% query coverage and 100% identities) to the corresponding regions in some plasmids, including pHNAH67 (KX246266, from E. coli of chicken), pCFSA122-1 (CP033224, from Salmonella enterica of pork), pSH16G4466 (MK477617, from Salmonella enterica in human), pXGE1mcr (KY990887, from E. coli of cow feces), pHNSHP45-2 (KU341381, from E coli of pig feces), and chromosome of E. coli YSP8-1 (CP037910, from pig feces) from animal or human sources in China.
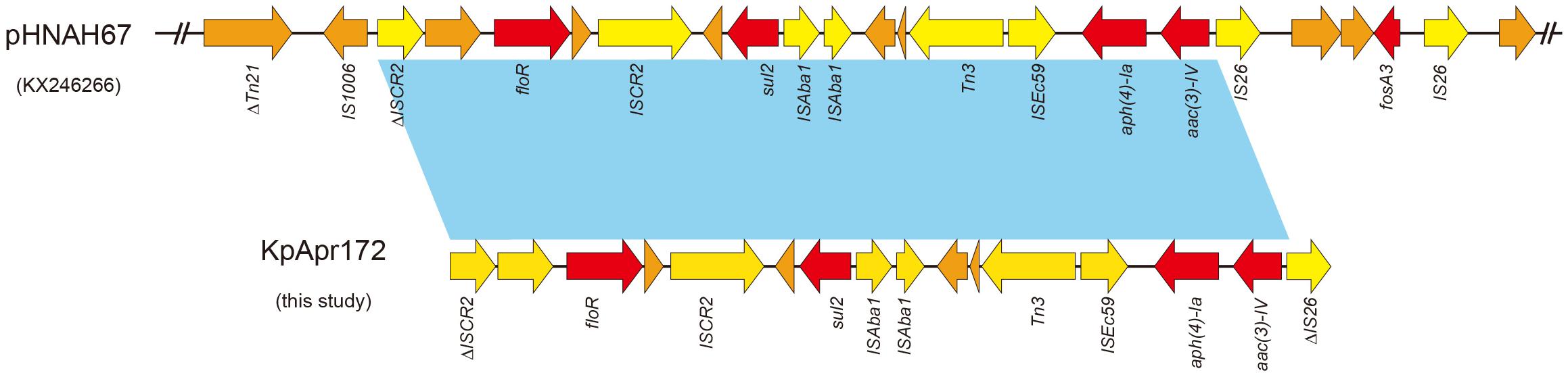
Figure 2. Sequence structure of aac(3)-IV-containing element in pHNAH67 (KX246266) and KpApr172 (this study). Colored arrows indicate open reading frames, with red, yellow and orange arrows representing resistance genes, mobile elements and additional genes, respectively. Light blue shading denotes regions of shared 100% homology between different elements.
Aminoglycosides resistance is mostly due to the chemical modification by aminoglycoside-modifying enzymes (AMEs), including acetyltransferases (AACs), phosphotransferases (APHs) and nucleotidyltransferases (ANTs) (Ramirez and Tolmasky, 2010). Some studies had shown AAC(6′)-Ib is one of the most prevalent and clinically relevant AMEs, which confers the resistance to amikacin and other aminoglycosides (Ramirez et al., 2013). A Greek study showed more than 70% of carbapenemases-producing Enterobacteriaceae strains carry aac(6′)-Ib gene (Galani et al., 2019). However, only 7.14% of CR-hvKp and 7.5% of CR-non-hvKp isolates were found to carry this gene in our study. By contrast, we found a high frequency of ARM/RMT methyltransferases gene in our study. ARM/RMT methyltransferases were found to confer high levels of resistance to most aminoglycosides other than apramycin (Galimand et al., 2003). ARM/RMT genes armA and rmt had been frequently described in clinical isolates throughout America, Europe and India (Fritsche et al., 2008). Our study showed 93.6% (44/47) CR-hvKp and 95.6% (22/23) CR-non-hvKp amikacin or gentamycin resistant strains contained the 16S rRNA methylase gene rmtB. These results suggested that gentamycin and amikacin have limited activities against clinical CRKp isolates, including CR-hvKp, in Chinese hospitals.
In comparison, our results showed that all CR-hvKp isolates were susceptible to apramycin, whereas ∼ 50% of them were resistant to amikacin or gentamycin. Similarly, almost all CR-non-hvKp strains (39/40) were susceptible to apramycin irrespective of the presence of KPC-2 or NDM carbapenemase genes. In addition, our results were in accordance with previous studies that apramycin is not be inactivated by general AMEs and methylases (Livermore et al., 2011), making it an potential antibiotic against clinical amikacin or gentamycin resistant strains.
Nevertheless, apramycin resistance has also been described in clinical isolates worldwide (Threlfall et al., 1986; Chaslus-Dancla et al., 1989, 1991; Hunter et al., 1993; Johnson et al., 1995; Mathew et al., 2003; Zhang et al., 2009). Production of the acetylase aac(3′)-IV was the major mechanism underlying apramycin resistance in clinical isolates. The aac(3′)-IV gene encoding apramycin resistance was usually harbored by a mobile plasmid (Chaslus-Dancla et al., 1989; Zhang et al., 2009). In this study, we identified one aac(3′)-IV-harboring apramycin resistant isolate KpApr172 in CR-non-hvKp with a high MIC value of >128 μg/ml. Genomic analysis showed that KpApr172 belonged to the epidemic CRKp ST11 clone, while the aac(3′)-IV-harboring contigs showed highly identity to plasmid or chromosome sequences of isolates from animal sources. Since apramycin is only licensed for use in farm animals in China, we suspected that horizontal transfer of apramycin resistant genes from animals to human might be responsible for the dissemination of apramycin resistance (Yates et al., 2004). Therefore, aac(3′)-IV gene should be preferably screened for apramycin resistant isolates in clinical settings.
In this study no CR-hvKp isolates were found to be resistant to the “last resort” antimicrobial agents, including ceftazidime-avibactam, colistin and tigecycline, however, the treatment arsenal for CR-hvKp could be easily compromised by the spread of mobile resistant genes. Plasmid-mediated colistin and tigecycline resistance mechanisms, MCR-1 and Tet(x), have been described in China (Liu et al., 2016; He et al., 2019; Sun et al., 2019). In addition, the FDA approved cefetazidime-avibactam is ineffective against metallo-β-lactamase (e.g., NDM) producers. Furthermore, the original KPC-2 ST11 CR-hvKp outbreak study showed that despite being in vitro susceptible to tigecycline and colistin, long-term treatment with colistin or tigecycline (alone or in combination with several other antibiotics) was not able to eradicate KPC-2-producing ST11 CR-hvKp isolates, resulting in fatal outcomes (Gu et al., 2018). This convergence of increased resistance and hypervirulence of CR-hvKp underscores the need for the improvement of the antimicrobial treatment choices.
Our study showed apramycin had a potent in vitro activity to CR-hvKp irrespective of most commonly occurring carbapenemases and aminoglycoside modifying enzymes. In addition, recent studies had showed that apramycin was associated with fewer ototoxic and nephrotoxic side effects in comparison with other human use aminoglycosides antibiotics (Perzynski et al., 1979; Matt et al., 2012; Ishikawa et al., 2019). These findings suggested apramycin and apramycin-derivative compounds could be a promising antibiotic for the treatment of CR-hvKp infections.
To our knowledge, this is the first study to examine the activities of apramycin against CR-hvKp clinical isolates. Our results indicate that apramycin remain highly active against CR-hvKp. These data support a possible role for apramycin in the treatment of infections due to this hypervirulent and multidrug-resistant strains. A further clinical evaluation of apramycin treatment efficacy is warranted.
The whole-genome sequence of KpApr172 was deposited in the GenBank database under accession number WUJI00000000.
MH, XS, and JL contributed to work, data analysis, and manuscript preparation. SN, SC, HD, and FY analyzed the data. Y-WT and BK prepared the manuscript. HZ and LC contributed to study design, data analysis, and manuscript preparation.
This study was supported by the Jiangsu Overseas Visiting Scholar Program for University Prominent Young and Middle-aged Teachers and Presidents, the Natural Science Foundation of Jiangsu Province (BK20181173), the grant from the Medicine and Health Science Technology Development Project of Shandong Province, China (2017WS007), Key Research and Development Project of Jiangsu Provincial Science and Technology Department (BE2017654), Gusu Health Youth Talent of Suzhou, Jiangsu Youth Medical Talents Program (QN-866 and 867), and the Cultivate Fund from Shandong Provincial Qianfoshan Hospital (No. QYPY2019NSFC0623). This work was in part supported by the grants from the National Institutes of Health (R01AI090155 to BK).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00425/full#supplementary-material
FIGURE S1 | Dendrogram of PFGE patterns of 15 CR-hvKp isolates from hospitals in Shanghai (a), Suzhou (b), and Jinan (c), respectively. PFGE typing revealed most of the CR-hvKp strains not belong to the same clone.
TABLE S1 | Primers used in this study.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bengoechea, J. A., and Sa Pessoa, J. (2019). Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol. Rev. 43, 123–144. doi: 10.1093/femsre/fuy043
Brisse, S., Passet, V., Haugaard, A. B., Babosan, A., Kassis-Chikhani, N., Struve, C., et al. (2013). wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J. Clin. Microbiol. 51, 4073–4078. doi: 10.1128/JCM.01924-13
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Cejas, D., Fernandez Canigia, L., Rincon Cruz, G., Elena, A. X., Maldonado, I., Gutkind, G. O., et al. (2014). First isolate of KPC-2-producing Klebsiella pneumonaie sequence type 23 from the Americas. J. Clin. Microbiol. 52, 3483–3485. doi: 10.1128/JCM.00726-14
Chaslus-Dancla, E., Glupcznski, Y., Gerbaud, G., Lagorce, M., Lafont, J. P., and Courvalin, P. (1989). Detection of apramycin resistant Enterobacteriaceae in hospital isolates. FEMS Microbiol. Lett. 52, 261–265. doi: 10.1016/0378-1097(89)90208-5
Chaslus-Dancla, E., Pohl, P., Meurisse, M., Marin, M., and Lafont, J. P. (1991). High genetic homology between plasmids of human and animal origins conferring resistance to the aminoglycosides gentamicin and apramycin. Antimicrob. Agents Chemother. 35, 590–593. doi: 10.1128/aac.35.3.590
Chavda, K. D., Satlin, M. J., Chen, L., Manca, C., Jenkins, S. G., Walsh, T. J., et al. (2016). Evaluation of a Multiplex PCR assay to rapidly detect Enterobacteriaceae with a broad range of beta-lactamases directly from perianal swabs. Antimicrob. Agents Chemother. 60, 6957–6961. doi: 10.1128/AAC.01458-16
Chen, L., Mediavilla, J. R., Endimiani, A., Rosenthal, M. E., Zhao, Y., Bonomo, R. A., et al. (2011). Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (bla KPC) variants. J. Clin. Microbiol. 49, 579–585. doi: 10.1128/JCM.01588-10
Clinical and Laboratory Standards Institute (2019). Performance Standards for Antimicrobial Susceptibility Testing. M100 standard, 29th ed.
Evans, S. R., Hujer, A. M., Jiang, H., Hujer, K. M., Hall, T., Marzan, C., et al. (2016). Rapid molecular diagnostics, antibiotic treatment decisions, and developing approaches to inform empiric therapy: PRIMERS I and II. Clin. Infect. Dis. 62, 181–189. doi: 10.1093/cid/civ837
Fritsche, T. R., Castanheira, M., Miller, G. H., Jones, R. N., and Armstrong, E. S. (2008). Detection of methyltransferases conferring high-level resistance to aminoglycosides in Enterobacteriaceae from Europe, North America, and Latin America. Antimicrob. Agents Chemother. 52, 1843–1845. doi: 10.1128/AAC.01477-07
Galani, I., Nafplioti, K., Adamou, P., Karaiskos, I., Giamarellou, H., Souli, M., et al. (2019). Nationwide epidemiology of carbapenem resistant Klebsiella pneumoniae isolates from Greek hospitals, with regards to plazomicin and aminoglycoside resistance. BMC Infect. Dis. 19:167. doi: 10.1186/s12879-019-3801-1
Galimand, M., Courvalin, P., and Lambert, T. (2003). Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 47, 2565–2571. doi: 10.1128/aac.47.8.2565-2571.2003
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2018). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4, 1450–1456. doi: 10.1038/s41564-019-0445-2
Heras, J., Dominguez, C., Mata, E., Pascual, V., Lozano, C., Torres, C., et al. (2015). GelJ–a tool for analyzing DNA fingerprint gel images. BMC Bioinformatics 16:270. doi: 10.1186/s12859-015-0703-0
Hu, Y., Liu, L., Zhang, X., Feng, Y., and Zong, Z. (2017). In vitro activity of neomycin, streptomycin, paromomycin and apramycin against carbapenem-resistant Enterobacteriaceae clinical strains. Front. Microbiol. 8:2275. doi: 10.3389/fmicb.2017.02275
Hunter, J. E., Hart, C. A., Shelley, J. C., Walton, J. R., and Bennett, M. (1993). Human isolates of apramycin-resistant Escherichia coli which contain the genes for the AAC(3)IV enzyme. Epidemiol. Infect. 110, 253–259. doi: 10.1017/s0950268800068175
Ishikawa, M., Garcia-Mateo, N., Cusak, A., Lopez-Hernandez, I., Fernandez-Martinez, M., Muller, M., et al. (2019). Lower ototoxicity and absence of hidden hearing loss point to gentamicin C1a and apramycin as promising antibiotics for clinical use. Sci. Rep. 9:2410. doi: 10.1038/s41598-019-38634-3
Johnson, A. P., Malde, M., Woodford, N., Cunney, R. J., and Smyth, E. G. (1995). Urinary isolates of apramycin-resistant Escherichia coli and Klebsiella pneumoniae from Dublin. Epidemiol. Infect. 114, 105–112. doi: 10.1017/s0950268800051955
Juhas, M., Widlake, E., Teo, J., Huseby, D. L., Tyrrell, J. M., Polikanov, Y. S., et al. (2019). In vitro activity of apramycin against multidrug-, carbapenem- and aminoglycoside-resistant Enterobacteriaceae and Acinetobacter baumannii. J Antimicrob Chemother. 74, 944–952. doi: 10.1093/jac/dky546
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Li, W., Sun, G., Yu, Y., Li, N., Chen, M., Jin, R., et al. (2014). Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin. Infect. Dis. 58, 225–232. doi: 10.1093/cid/cit675
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Livermore, D. M., Mushtaq, S., Warner, M., Zhang, J. C., Maharjan, S., Doumith, M., et al. (2011). Activity of aminoglycosides, including ACHN-490, against carbapenem-resistant Enterobacteriaceae isolates. J. Antimicrob. Chemother. 66, 48–53. doi: 10.1093/jac/dkq408
Mathew, A. G., Arnett, D. B., Cullen, P., and Ebner, P. D. (2003). Characterization of resistance patterns and detection of apramycin resistance genes in Escherichia coli isolated from swine exposed to various environmental conditions. Int. J. Food Microbiol. 89, 11–20. doi: 10.1016/s0168-1605(03)00124-7
Matt, T., Ng, C. L., Lang, K., Sha, S. H., Akbergenov, R., Shcherbakov, D., et al. (2012). Dissociation of antibacterial activity and aminoglycoside ototoxicity in the 4-monosubstituted 2-deoxystreptamine apramycin. Proc. Natl. Acad. Sci. U.S.A. 109, 10984–10989. doi: 10.1073/pnas.1204073109
National Antibiotic Resistance Monitoring System [NARMS] (2002). National Antibiotic Resistance Monitoring System (NARMS) Working Group. Annual Report. Available at: https://www.cdc.gov/narms/annual/2002/2002ANNUALREPORTFINAL.pdf (accessed May 2019).
Paczosa, M. K., and Mecsas, J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661. doi: 10.1128/MMBR.00078-15
Perzynski, S., Cannon, M., Cundliffe, E., Chahwala, S. B., and Davies, J. (1979). Effects of apramycin, a novel aminoglycoside antibiotic on bacterial protein synthesis. Eur. J. Biochem. 99, 623–628. doi: 10.1111/j.1432-1033.1979.tb13295.x
Ramirez, M. S., Nikolaidis, N., and Tolmasky, M. E. (2013). Rise and dissemination of aminoglycoside resistance: the aac(6’)-Ib paradigm. Front. Microbiol. 4:121. doi: 10.3389/fmicb.2013.00121
Ramirez, M. S., and Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat 13, 151–171. doi: 10.1016/j.drup.2010.08.003
Riedel, S., Vijayakumar, D., Berg, G., Kang, A. D., Smith, K. P., and Kirby, J. E. (2019). Evaluation of apramycin against spectinomycin-resistant and -susceptible strains of Neisseria gonorrhoeae. J. Antimicrob. Chemother. 74, 1311–1316. doi: 10.1093/jac/dkz012
Shon, A. S., Bajwa, R. P., and Russo, T. A. (2013). Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4, 107–118. doi: 10.4161/viru.22718
Siu, L. K., Huang, D. B., and Chiang, T. (2014). Plasmid transferability of KPC into a virulent K2 serotype Klebsiella pneumoniae. BMC Infect. Dis. 14:176. doi: 10.1186/1471-2334-14-176
Smith, K. P., and Kirby, J. E. (2016). Evaluation of apramycin activity against carbapenem-resistant and -susceptible strains of Enterobacteriaceae. Diagn. Microbiol. Infect. Dis 86, 439–441. doi: 10.1016/j.diagmicrobio.2016.09.002
Sun, J., Chen, C., Cui, C. Y., Zhang, Y., Liu, X., Cui, Z. H., et al. (2019). Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 4, 1457–1464. doi: 10.1038/s41564-019-0496-4
The European Committee on Antimicrobial Susceptibility Testing (2019). Breakpoint Tables for Interpretation of MICs and Zone Diameters, version 9.0.
Threlfall, E. J., Rowe, B., Ferguson, J. L., and Ward, L. R. (1986). Characterization of plasmids conferring resistance to gentamicin and apramycin in strains of Salmonella typhimurium phage type 204c isolated in Britain. J. Hyg. 97, 419–426. doi: 10.1017/s0022172400063609
Truelson, K. A., Brennan-Krohn, T., Smith, K. P., and Kirby, J. E. (2018). Evaluation of apramycin activity against methicillin-resistant, methicillin-sensitive, and vancomycin-intermediate Staphylococcus aureus clinical isolates. Diagn. Microbiol. Infect. Dis. 92, 168–171. doi: 10.1016/j.diagmicrobio.2018.05.018
Wozniak, J. E., Band, V. I., Conley, A. B., Rishishwar, L., Burd, E. M., Satola, S. W., et al. (2019). A nationwide screen of carbapenem-resistant Klebsiella pneumoniae reveals an isolate with enhanced virulence and clinically undetected colistin heteroresistance. Antimicrob. Agents Chemother. 63, 107–119. doi: 10.1128/AAC.00107-19
Wu, Q., Zhang, Y., Han, L., Sun, J., and Ni, Y. (2009). Plasmid-mediated 16S rRNA methylases in aminoglycoside-resistant Enterobacteriaceae isolates in Shanghai China. Antimicrob. Agents Chemother. 53, 271–272. doi: 10.1128/AAC.00748-08
Yang, X., Wai-Chi Chan, E., Zhang, R., and Chen, S. (2019). A conjugative plasmid that augments virulence in Klebsiella pneumoniae. Nat. Microbiol. 4, 2039–2043. doi: 10.1038/s41564-019-0566-7
Yates, C. M., Pearce, M. C., Woolhouse, M. E., and Amyes, S. G. (2004). High frequency transfer and horizontal spread of apramycin resistance in calf faecal Escherichia coli. J. Antimicrob. Chemother. 54, 534–537. doi: 10.1093/jac/dkh353
Yu, F., Lv, J., Niu, S., Du, H., Tang, Y. W., Bonomo, R. A., et al. (2018a). In vitro activity of Ceftazidime-Avibactam against carbapenem-resistant and Hypervirulent Klebsiella pneumoniae Isolates. Antimicrob. Agents Chemother. 62, 1031–1018. doi: 10.1128/AAC.01031-18
Yu, F., Lv, J., Niu, S., Du, H., Tang, Y. W., Pitout, J. D. D., et al. (2018b). Multiplex PCR analysis for rapid detection of Klebsiella pneumoniae carbapenem-resistant (Sequence Type 258 [ST258] and ST11) and Hypervirulent (ST23, ST65, ST86, and ST375) strains. J. Clin. Microbiol. 56:JCM.00731-18. doi: 10.1128/JCM.00731-18
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: apramycin, susceptibility, carbapenem resistance, hypervirulent Klebsiella pneumoniae, aminoglycoside
Citation: Hao M, Shi X, Lv J, Niu S, Cheng S, Du H, Yu F, Tang Y-W, Kreiswirth BN, Zhang H and Chen L (2020) In vitro Activity of Apramycin Against Carbapenem-Resistant and Hypervirulent Klebsiella pneumoniae Isolates. Front. Microbiol. 11:425. doi: 10.3389/fmicb.2020.00425
Received: 14 September 2019; Accepted: 27 February 2020;
Published: 13 March 2020.
Edited by:
Luciene Andrade Da Rocha Minarini, Federal University of São Paulo, BrazilReviewed by:
Qi Wang, Peking University People’s Hospital, ChinaCopyright © 2020 Hao, Shi, Lv, Niu, Cheng, Du, Yu, Tang, Kreiswirth, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haifang Zhang, aGFpZmFuZ3poYW5nQHNpbmEuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.