- 1Department of Plant Science and Landscape Architecture, University of Maryland, College Park, MD, United States
- 2Centre for Food Safety and Security Systems, University of Maryland, College Park, MD, United States
The enteric pathogen Salmonella enterica can interact with parts of the plant immune system despite not being a phytopathogen. Previous transcriptomic profiling of S. enterica associating with tomato suggested that Salmonella was responding to oxidative and nitrosative stress in the plant niche. We aimed to investigate whether Salmonella was eliciting generation of reactive oxygen species (ROS) and nitric oxide (NO), two components of the microbe-associated molecular pattern (MAMP)-triggered immunity (MTI) of plants. We also sought to determine whether this interaction had any measurable effects on Salmonella colonization of plants. Biochemical, gene expression and on-plant challenge assays of tomato vegetative and fruit organs were conducted to assess the elicitation of ROS and NO in response to Salmonella Newport association. The counter bacterial response and the effect of NO and ROS on Salmonella colonization was also investigated. We detected H2O2 in leaves and fruit following challenge with live S. Newport (p < 0.05). Conversely, NO was detected on leaves but not on fruit in response to S. Newport (p < 0.05). We found no evidence of plant defense attenuation by live S. Newport. Bacterial gene expression of S. Newport associating with leaves and fruit were indicative of adaptation to biotic stress in the plant niche. The nitrosative stress response genes hmpA and yoaG were significantly up-regulated in S. Newport on leaves and fruit tissue compared to tissue scavenged of NO or ROS (p < 0.05). Chemical modulation of these molecules in the plant had a restrictive effect on bacterial populations. Significantly higher S. Newport titers were retrieved from H2O2 scavenged leaves and fruit surfaces compared to controls (p < 0.05). Similarly, S. Newport counts recovered from NO-scavenged leaves, but not fruit, were higher compared to control (p < 0.05), and significantly lower on leaves pre-elicited to produce endogenous NO. We present evidence of Salmonella elicitation of ROS and NO in tomato, which appear to have a restricting effect on the pathogen. Moreover, bacterial recognition of ROS and NO stress was detected. This work shows that tomato has mechanisms to restrict Salmonella populations and ROS and NO detoxification may play an important role in Salmonella adaptation to the plant niche.
Introduction
Non-typhoidal Salmonella enterica is a leading cause of foodborne illness transmitted by fresh fruit and vegetables (Callejón et al., 2015). Traceback investigations of several salmonellosis outbreaks implicating fresh produce have pointed to contamination sources in crop production areas (Greene et al., 2008; Bennett et al., 2015). S. enterica is frequently isolated from water and soil in agricultural settings (Micallef et al., 2012; Bell et al., 2015; Callahan et al., 2019), suggesting that this enteric pathogen is able to cycle through various ecological niches and become established in the plant phyllosphere.
Salmonella enterica can survive and multiply on plants (Brandl and Mandrell, 2002), the success of which is influenced by multiple factors (Brandl et al., 2013), including plant genotype and organ (Barak et al., 2011; Han and Micallef, 2014), age (Brandl and Amundson, 2008; Zheng et al., 2013), surface metabolite profiles (Han and Micallef, 2016), as well as resident epiphytes (Poza-Carrion et al., 2013). Studies on S. enterica colonizing tomato fruit wounds, lettuce soft rot lesions and sprouts have identified specific sets of genes expressed under these conditions, including genes involved in amino acid biosynthesis, fatty acid metabolism, iron acquisition, attachment and stress response (Goudeau et al., 2013; Salazar et al., 2013; Tan et al., 2016; de Moraes et al., 2017, 2018). Research within our group investigating gene expression in S. enterica epiphytically colonizing tomato shoots and roots detected several genes involved in nitrosative and oxidative stress mitigation and multiple Salmonella pathogenicity island-2-encoded type III secretion system genes (T3SS-2) (Han et al., unpublished). These findings pointed to interplay between the enteric pathogen and the plant, as a result of tomato plant recognition and immune response.
Plants recognize potential microbial pathogens via MAMP interaction with pathogen recognition receptors (PRRs) (Jones and Dangl, 2006). This recognition initiates several strong yet transient signaling events to occur, beginning with an influx of calcium ions into the cell (Ranf et al., 2011) which induces a burst of reactive oxygen species (ROS) (Doke, 1983) and nitric oxide (NO) (Ma et al., 2008; Rasul et al., 2012). The generation of ROS and the more recently identified NO serve multiple purposes for the plant. The ROS burst can directly control the potential pathogen threat and, together with NO, may activate mitogen associated protein kinases (MAPKs) and signal the up-regulation of transcription factors that initiate transient defense responses. These include salicylic acid (Tsuda et al., 2008) and ethylene biosynthesis (Liu and Zhang, 2004) which in part comprise MAMP- triggered immunity (MTI) (Meng and Zhang, 2013). The non-plant pathogen S. Typhimurium and its flagellin 22 (flg22) have been shown to induce an ROS burst in tobacco and tomato leaf disks, respectively (Shirron and Yaron, 2011; Meng et al., 2013). Flagellin 22 from S. Typhimurium was recognized by tobacco and Arabidopsis thaliana through the FLS2 receptor, inducing MTI which was effective in restricting S. enterica and the plant pathogen Pseudomonas syringae pv. tomato (Meng et al., 2013; Garcia et al., 2014). The role of NO in plant defense is less understood but it was generated in Arabidopsis in response to lipopolysaccharide (LPS) challenge (Zeidler et al., 2004) and also required for abscisic acid-induced stomatal closure (Neill et al., 2002). Melotto et al. (2006) showed that Arabidopsis guard cells generated NO in response to flg22 and LPS, which was followed by stomatal closure. They also reported that S. enterica was able to trigger stomatal closure in Arabidopsis. Furthermore, flagellar mutants of S. Typhimurium were shown to better colonize wheat, alfalfa and Arabidopsis, suggesting that attenuation of MAMPs favored bacterial colonization (Iniguez et al., 2005). Finally, some studies have suggested that S. enterica may have the ability to suppress MTI in Arabidopsis and tobacco and the role of effector proteins has been invoked (Shirron and Yaron, 2011; Garcia et al., 2014; Neumann et al., 2014).
The current state of knowledge and our finding that S. enterica expresses oxidative and nitrosative stress genes, and T3SS-2 genes when colonizing tomato shoot and root surfaces (Han et al., unpublished), implied that S. enterica recognition by tomato induces the generation of both ROS and NO, and that S. enterica attempts to attenuate the plant immune response. We set out to test this hypothesis in leaves, but also more relevantly, on tomato fruit, in relation to both live and killed S. enterica to assess potential plant defense suppression. MTI induction in tomato leaves and fruit in response to S. enterica Newport (SeN) association was investigated by measuring NO and ROS generation. Moreover, we sought to describe the effect of this plant response on S. enterica populations. The reciprocal bacterial response and the effect of surface modulation of NO and ROS on S. enterica colonization of tomato leaves and fruit was also evaluated.
Materials and Methods
Cultivation of Plant Material
Tomato seeds cv. ‘Heinz-1706’ were obtained from the Tomato Genetics Resource Center (TGRC) from the University of California, Davis. After pre-treatment in 30% w/v polyethylene glycol solution at room temperature with shaking for 72 h, seeds were germinated in potting media (Sunshine LC1; Sungro Horticulture, Canada) at 25°C. Germinated seeds were transferred to fresh potting media supplemented with fertilizer (Osmocote controlled release fertilizer 18–6–12:nitrogen–phosphate–potash, The Scotts Company LLC., Marysville, OH, United States) and subjected to a 16 h-light/8 h-dark photoperiod and 26°C day temperature/18°C night temperature with 70% humidity (RH) at the University of Maryland Research Greenhouse. Plants were drip irrigated. Tomato seedlings were grown to five true leaves before experimentation, unless otherwise noted. Seedlings for experimentation were transported to a BSL-2 growth chamber (16 h-light/8 h-dark photoperiod and 23°C constant temperature with 70–80% RH) at least 5 days prior to inoculation and flood irrigated to a depth of 5 cm in trays every 4 days. Water was withheld 3 h before inoculating leaves and for the duration of experiments. For fruit, plants were either grown in the field (summer) at the Wye Research and Education Centre, Queenstown, MD, United States, or transplanted into 6 L pots to be grown in the greenhouse (winter) once they reached the 5-leaf stage. In the greenhouse, plants were fertilized once a week and treated with non-organophosphate containing pesticide once every 2 weeks for aphid and white fly management. Once plants reached maturity, red, ripe fruit was collected immediately prior to experimentation, rinsed with sterile water and air dried, unless otherwise stated.
Bacterial Strains
The Salmonella enterica Newport strain used was an environmental isolate collected from an irrigation pond that matched a recurring tomato outbreak strain (Greene et al., 2008). SeN had been previously adapted to rifampicin (rif) and was therefore maintained at −80°C in Brucella Broth (BD, Sparks, MD, United States) containing 15% glycerol and 50 μg/mL rifampicin (rif, Tokyo Chemical Industry, Portland, OR, United States). For each experiment, cultures of SeN were grown overnight on Trypticase Soy Agar (TSA; BD) + rif at 35°C. A single colony was selected, suspended in sterile water, and diluted to OD600 = 0.34 – approximately 8.5 log CFU/mL. Serial dilutions were made in sterile water for inoculum preparation and in 0.1% peptone water for bacterial quantification (BD Difco, Sparks, MD, United States). Cells were enumerated on TSArif. Pseudomonas syringae pv. maculicola ES4326 (Psm), a relative to P. syringae pv. tomato with similar virulence and in the same genospecies clade (genospecies III) (Preston, 2000) was grown in TSA at 25°C, and inoculum prepared as described above.
Detection of ROS in Leaves and Fruit
To detect the amount of H2O2 produced in leaves following SeN challenge, 3,3′-diaminobenzidine (DAB) staining was adapted from Bindschedler et al. (2006). Briefly, a third emerged leaflet from freshly watered, 5-leaved ‘Heinz-1706’ plants was syringe infiltrated into the abaxial surface with 500 uL of either SeN in sterile water at 8 log CFU/mL, heat-killed SeN, or sterile water (control) (N = 6 plants per treatment). Positive controls were conducted with Psm. Inoculated plants were incubated in a BSL-2 growth chamber. All experiments were performed in a complete randomized design (CRD) and repeated at least twice. At 0.1 and 24 hours post-inoculation (hpi), inoculated leaflets were excised and submerged in 5 mL DAB solution (1 mg/mL aqueous DAB (Alfa Aesar, Ward Hill, MA, United States), 200 mM Na2HPO4 (VWR, West Chester, PA, United States), 0.05% Tween 20 (Amresco, Solon, OH, United States) and 100 μL 3 N HCl). Samples were vacuum-infiltrated in a vacuum desiccator attached to the laboratory vacuum system for 4 min, then incubated in the dark at 23°C with shaking at 50 rpm for 4 h. At the end of staining, decolorizer solution was added (3:1:1 95% ethanol, glycerol, glacial acetic acid) and samples were incubated in a boiling water bath for 15 min. Decolorized leaflets were fixed to paper and imaged with an Epson V330 photo scanner. The stain, corresponding to H2O2 production, was analyzed for intensity via ImageJ2 FIJI package (Schindelin et al., 2012). Optical density in leaves was calculated using the formula
To detect a range of ROS produced from SeN challenge on fruit, staining with 6-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA; Invitrogen, Molecular Probes Inc., Eugene, OR, United States) was adapted from Shin and Schachtman (2004). Briefly, 3 mm × 3 mm sections of ripe tomato exocarp per fruit were excised with a sterile razor and placed in separate black 96-well plates to serve as technical replicates for one fruit (Corning, Nazareth, PA, United States). Aliquots of 150 μL deionized water were delivered to the sample wells and incubated overnight in the dark at 4°C to allow for dissipation of any ROS production due to injury. Immediately before experimentation, wells were washed with 100 μL sterile water. One hundred micro-liters of 8.0 log CFU/mL SeN, heat killed SeN (14 h only) or sterile water were delivered to sample wells (N = 4 fruit per treatment, with three technical replicates per fruit). Samples were vacuum-infiltrated in a vacuum desiccator for 5 min, then shaken at 100 rpm at 27°C for 3 or 14 hpi. At the time of sampling, 25 μM CM-H2DCFDA in water was added to each well. The fluorophore was allowed to react for 30 min at 23°C in the dark with shaking at 50 rpm before being imaged with a Synergy HTX Microplate reader (BioTek, Winooski, VT, United States) at 485 nm excitation, 520 nm emission with 50 gain.
Detection of NO in Leaves and Fruit
To measure amounts of NO release from tomato when challenged with SeN, 4,5-diaminofluorescein diacetate (DAF-2 DA; Fisher Scientific, Hampton, NH, United States) was used for its ability to complex intercellular NO as well as NO in solution (Rasul et al., 2012). For measurements on leaves, leaflets of mature ‘Heinz-1706’ plants grown in the research greenhouse were punched three times with a 3 mm hole puncher and cut tissue pieces were placed in separate wells in a black 96-well plate, to serve as technical replicates for each leaf (Corning, Nazareth, PA, United States) (N = 5 leaves per treatment, with three technical replicates per leaf). For measurements on fruit, 3 mm × 3 mm sections of ripe tomato exocarp were excised with a sterile razor (N = 5 fruit per treatment, with three technical replicates each). For both experiments, 150 μL deionized water was placed in the sample wells and plates were incubated overnight in the dark at 4°C to allow for the dissipation of injury related NO signal. All experiments were performed in a CRD and repeated at least twice. Prior to inoculation, tissues were washed twice with sterile water, then challenged with 100 μL of 8 log CFU/mL SeN, heat killed SeN, sterile water or 8 log CFU/mL Psm. Samples were vacuum-infiltrated for 5 min, then shaken at 100 rpm at 27°C. At 0.1, 1, and 3 h a final concentration of 15 μM DAF-2 DA in 50 mM Tris HCl pH 7.5 was delivered to the wells. Plates were incubated in the dark for 30 min at 27°C with shaking at 50 rpm and immediately read on the Synergy HTX (BioTek) at 485 nm excitation, 520 nm emission with 50 gain.
Targeted q-RT-PCR of SeN Genes Colonizing Leaf and Fruit Surfaces
To evaluate genes involved in nitrosative and oxidative stress responses in SeN colonizing the tomato phyllosphere, 3-leaf tomato seedlings cv. ‘Heinz-1706’ were pre-treated with water (native environment), 2-4-carboxyphenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO; NO-limiting environment) or CaCl2 (excess NO environment), then challenged with SeN. To achieve this, 48 plants grown in autoclaved LC-1 potting media (Sunshine LC1) were separated into three groups and aerosol-sprayed with either 1 mL 0.5% CaCl2, ddH2O or 0.2 mM cPTIO. The plants were allowed to air-dry for 30 min. The second emerged leaf was challenged with 7 log CFU/mL SeN, delivered as ten 2-μL spots onto the leaf surface. Plants were incubated in the BSL-2 growth chamber, as previously described. At 6 hpi, inoculated leaves of three plants were pooled to comprise one composite sample (N = 4 composite samples per treatment). Samples were immediately fixed in 2:1 RNAProtect Bacteria Reagent (Qiagen, Germantown, MD, United States):ddH2O. Samples were sonicated on a 8510 Branson Sonicator at full strength for 2 min to dislodge surface attached bacteria. The wash solution containing SeN was transferred to a fresh tube and processed as described below.
To evaluate the role of genes involved in colonization of tomato fruit, cv. ‘Heinz-1706’ mature red fruit were washed with 200 ppm sodium hypochlorite and triple rinsed with ddH2O. Fruit were then syringe-injected at the calyx with either 500 μL ddH2O or 0.25 mM ascorbic acid. Seven log CFU/mL SeN was delivered as five 20-μL spots on the fruit surface. Fruit were incubated in the BSL-2 growth chamber in identical conditions as seedlings. At 6 hpi, five fruit from each treatment were pooled to comprise one composite sample (N = 4 composite samples per treatment). Samples were placed in RNAlater Stabilization Solution (Invitrogen, Carlsbad, CA, United States). Fruit were vigorously vortexed for 3 min to dislodge attached cells. The wash solution containing SeN was transferred to a fresh tube and processed as described below.
In both experiments, 0.5 mL SeN inoculum in water was immediately fixed with RNAProtect Bacteria Reagent or RNAlater Stabilization Solution to serve as the baseline for gene expression. All samples were centrifuged at 5,000 × g for 30 min. Total RNA was extracted from the resultant pellet using the Qiagen RNeasy Mini kit (leaves) (Qiagen) or the Purelink RNA Isolation kit (fruit) (Invitrogen) with 45 min on-column DNA digestion (Invitrogen). Resulting total RNA was evaluated on the Nanodrop 1000 (Thermo Fisher) for quality. PCR of target genes was performed using 1 μL total RNA sample template to ensure depletion of gDNA. cDNA was synthesized with Verso cDNA kit (Thermo Scientific, Waltham, MA, United States) and 1 ng samples were subjected to q-PCR of genes using primers listed in Table 1. Primers were used at 100 nM concentration and verified to be 90–105% efficient. Plant material was verified to produce no off-target amplification before experimentation. In a series of experiments using TSB amended with treatment reagents, SeN gene expression was confirmed to be reflective of epiphytic habit on tomato surface and not an artifact of interaction with elicitor or scavenger (Supplementary Figure 1). Amplification was conducted on an ABI Step-One Plus (Applied Biosystems, Foster City, CA, United States) with SYBR as a reporter (PowerUPTM SYBR Green Master Mix, Thermo Fisher Scientific, Austin, TX, United States) using the following parameters: 50°C for 2 min, 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 59°C for 30 s. Melt curve analysis was included to ensure product specificity. The cutoff Ct was set to 36.5 cycles. Data were analyzed on the ABI Step One Plus instrument with the ΔΔCt method (Pfaffl, 2001) using RNA polymerase sigma factor rpoD as the endogenous control. Relative gene expression was compared to expression in SeN inoculum after internal normalization to rpoD expression.
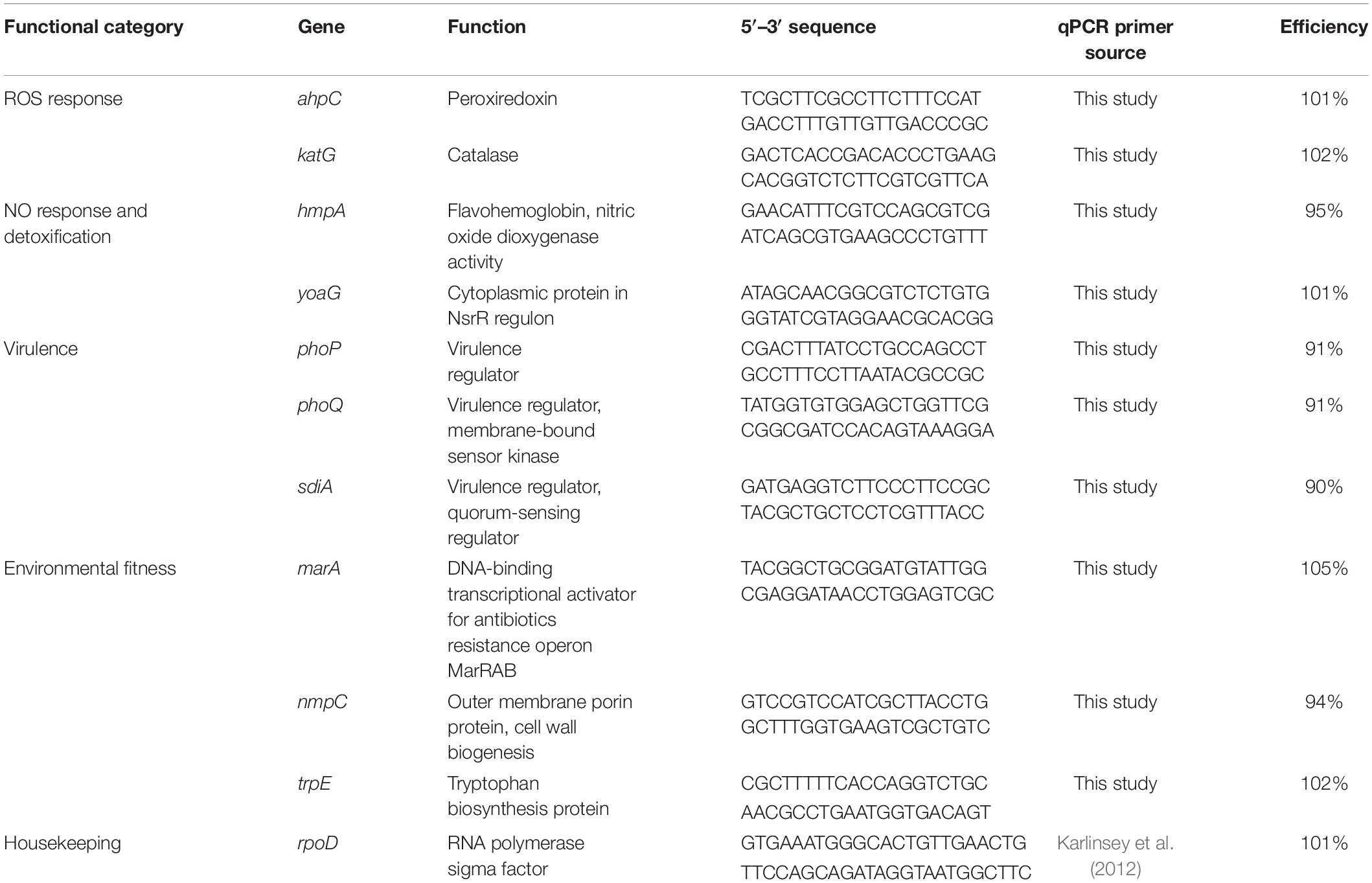
Table 1. List of genes, their qPCR primers and efficiencies used to examine S. Newport gene expression on tomato leaves and fruits.
Modulation of Endogenous Hydrogen Peroxide and Nitric Oxide Levels, and Plant Colonization Assays
To investigate the effect of plant derived ROS and NO on SeN survival on tomato surfaces, the third emerged leaf on 5-leaved ‘Heinz-1706’ seedlings or mature fruit were treated with reagents to either scavenge surface ROS (Bradley et al., 1992; Lee et al., 1999) or NO (Małolepsza and Różalska, 2005; Keshavarz-Tohid et al., 2016), or elicit production of NO (Chakraborty et al., 2016), then subsequently inoculated with SeN. The reagents employed, concentrations and application methods for leaves and fruit are detailed in Table 2. All experiments were performed in a CRD and repeated at least twice. After application of cPTIO and CaCl2, fruit (N = 10 for each treatment) and leaves (N = 18 for cPTIO experiments, N = 4 for CaCl2 experiments) were left to air-dry for 4 h at room temperature. Ascorbic acid-treated leaves (N = 3 per treatment) were left to dry for 2 h and fruit (N = 11 per treatment) were left to dry for 30 min prior to SeN inoculation. Following pretreatment, a suspension of 5.5 log CFU/mL SeN in water was applied to the surface of leaflets or fruit in ten 2-μL spots. Samples were incubated at 23°C at 75% RH for 12 h. To retrieve viable SeN, inoculated leaflets or fruit exocarp were cut and excised, respectively, as previously described, and diluted in 0.1% peptone water. Leaflets were hand-massaged and sonicated and fruit were hand-massaged and vortexed for 2 min before serially plating dilutions onto TSArif and incubating at 35°C for 20 h.

Table 2. Chemicals and application methodology for modulation of tomato leaf and fruit NO, ROS levels.
Statistical Analysis
Statistical analysis was conducted in JMP version 14.1.0. The degree of NO and ROS elicitation was analyzed for significance via Student’s t-test in pairwise comparisons, or ANOVA and orthogonal contrasts for a priori comparisons excluding the positive control (α = 0.05). Targeted gene expression data was analyzed for significance using ANOVA and post hoc via Dunnett’s test (inoculum control versus on-plant gene expression) or Tukey’s Honestly Significant Difference (cPTIO versus CaCl2 versus control on leaves), and Student’s t-test (ascorbic acid versus control on fruit) all at α = 0.05. For on-plant challenge assays, Student’s t-test was employed to compare treatment to water control (α = 0.05).
Results
Salmonella Newport Elicits H2O2 Production in Tomato Leaves and Fruit
A dark brown precipitate indicative of H2O2 production was detected immediately following inoculation of leaves with SeN (0.1 hpi) (Figures 1A,C). At 0.1 hpi, the brown precipitate deposited in SeN-challenged leaves (0.47 ± 0.10; p < 0.05) and heat-killed SeN-challenged leaves (0.45 ± 0.08; p < 0.05) was significantly darker than the negative control. Twenty-four hpi, heat-killed SeN (0.20 ± 0.05) and water control (0.23 ± 0.08) had comparable measurements, while leaves treated with live SeN exhibited darker staining (0.35 ± 0.18; p < 0.05) compared to control. The staining obtained with SeN was diffuse, as opposed to localized in spots as was observed for the virulent Psm pathogen (Figure 1C). Overall, we found no evidence of live SeN suppression of ROS generation.
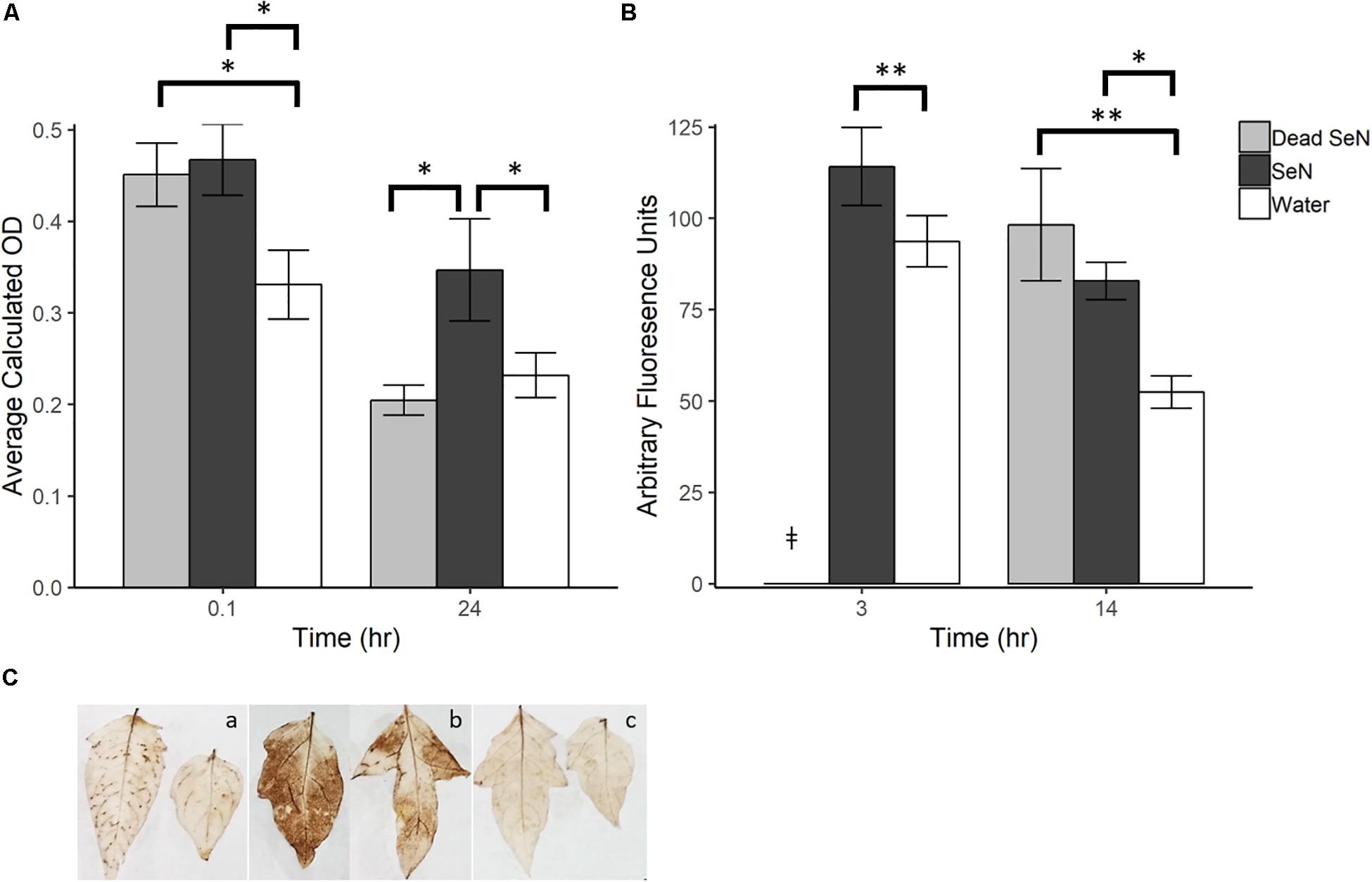
Figure 1. Plant derived ROS produced following challenge with sterile water (H2O), heat killed S. Newport (dead SeN) or live S. Newport. (A) Optical density showing degree of DAB stain in tomato leaves following SeN challenge via syringe infiltration. (B) CM-H2DCFDA fluorescence from tomato fruit peel challenged with SeN. (C) DAB staining of leaflets 24 h post-inoculation with (a) Pseudomonas syringae (Psm), (b) SeN, and (c) water. Error bars represent standard error of the mean and ∗∗ p < 0.01; ∗p ≤ 0.05. ‡denotes treatment was not included in that timepoint.
In fruit, at 3 h post-challenge, more ROS was detected in live SeN-treated exocarp than in water-treated exocarp (p < 0.01; Figure 1B). At 14 hpi, significantly more ROS was detected in exocarp samples treated with heat-killed (98.18 ± 51.0 Arbitrary fluoresence units, Au; p < 0.01) and live SeN (82.82 ± 17.0 Au; p < 0.05) compared to controls (51.41 ± 15.5 Au). Conversely to leaves, heat-killed SeN produced a similar signal to live SeN in fruit at the later timepoint. Taken together, these measurements suggest that live SeN can induce ROS generation in both tomato leaf and fruit exocarp tissue. Further, the differential ROS levels detected in tomato leaves inoculated with live or heat-killed SeN showed that ROS generation was more prolonged in response to live cells, potentially triggered by Salmonella activity in the leaf niche, and not solely from interaction with microbial surface cellular components.
NO Production Was Detected in Leaves but Not in Fruit Exocarp Challenged With SeN
Following leaf surface inoculation with SeN, NO was detected in all treatments (Figure 2). The tomato pathogen Psm-treated leaf sections served as a positive control and, as expected, induced significantly more NO than the water control at all sampling times (p < 0.001). At each timepoint, live SeN induced a stronger signal in leaves compared to water (87.67 ± 13.24, 100.6 ± 13.4 and 112.9 ± 18.8 Au, respectively; all p < 0.01; Figure 2A). Heat-killed SeN also induced NO at 3 hpi (p < 0.01). We did not observe any evidence of live SeN suppression of NO generation.
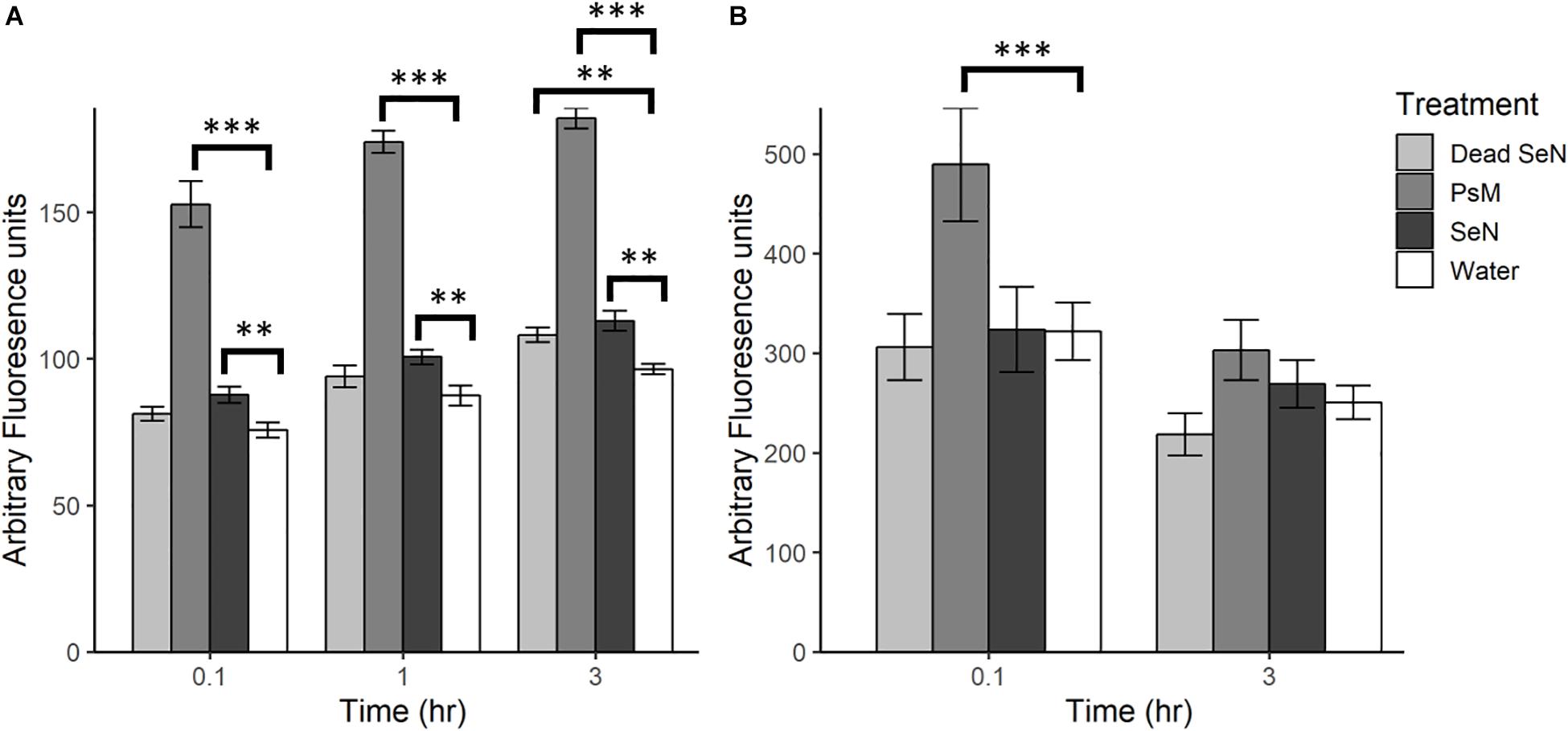
Figure 2. Plant derived NO produced following challenge with heat killed S. Newport (dead SeN), Pseudomonas syringae (Psm) or S. Newport (SeN) on (A) tomato leaves and (B) tomato fruit, measured with DAF-2 DA. Error bars represent standard error of the mean and ∗∗p < 0.01; ∗∗∗p ≤ 0.001.
Fruit tissue fluorescence for all treatments decreased over time by an average of 100.1 Au, with the smallest average decrease observed in live SeN-treated fruit tissue and the largest change in Psm treated leaf sections (Figure 2B). Psm produced significantly more NO than the water control at 0.1 hpi (p < 0.001) but not at 3 hpi. No significant exocarp production of NO was detected in SeN-treated fruit exocarp compared to the water control, either at 0.1 or 3 hpi.
Overall, fruit exocarp tissue produced a stronger NO signal than leaf tissue (p < 0.05) regardless of treatment. If NO was produced in relation to SeN, it was not strong enough to be detected with the method employed over the background NO being generated. Further, exocarp measurements exhibited a larger variation with a coefficient of variation (CV) of 0.52 compared to 0.36 for leaf sections. Taken together, these data suggest that live SeN can induce NO generation in tomato leaves, but not fruit.
Expression of ROS and NO Detoxification Genes Was Detected in SeN Colonizing Leaf Surfaces
To investigate specific bacterial responses to the observed elicited H2O2 and NO on tomato leaves, a targeted gene expression analysis was conducted on SeN cells inoculated onto native leaves, or leaves pre-treated to enhance or limit NO. We assayed genes responsible for NO detoxification, ROS mitigation and other environmental fitness factors (Table 1) that were previously found to be expressed in S. enterica Typhimurium colonizing tomato shoots and roots (Han et al., unpublished). On leaves, 78% of SeN samples displayed ≥|2| -fold change in gene expression compared to the inoculum. The flavohemoglobin hmpA, a main detoxifier of NO in oxygenated environments (Crawford and Goldberg, 1998) and yoaG, a cytoplasmic protein in the NsrR regulon (Lin et al., 2007), were shown to be differentially up-regulated in NO-excess and native leaves compared to the inoculum and to NO-limiting treatments (p < 0.05) (Figure 3). Compared to the inoculum, expression of hmpA increased threefold (p < 0.05) in SeN associating with native leaves and NO-excess leaves. Additionally, the yoaG gene showed an almost threefold increase on native leaves and a fourfold increase on NO-excess leaves (p < 0.05) compared to inoculum expression levels. The similar increases in gene expression in both NO-excess and native leaves, relative to the inoculum, suggest that SeN itself is serving as a strong NO elicitor on leaves. Finally, gene expression of both hmpA and yoaG in NO-limiting environments was comparable to expression levels in the inoculum. Taken together, these findings suggest that SeN may be countering NO and/or NO-regulated plant responses.
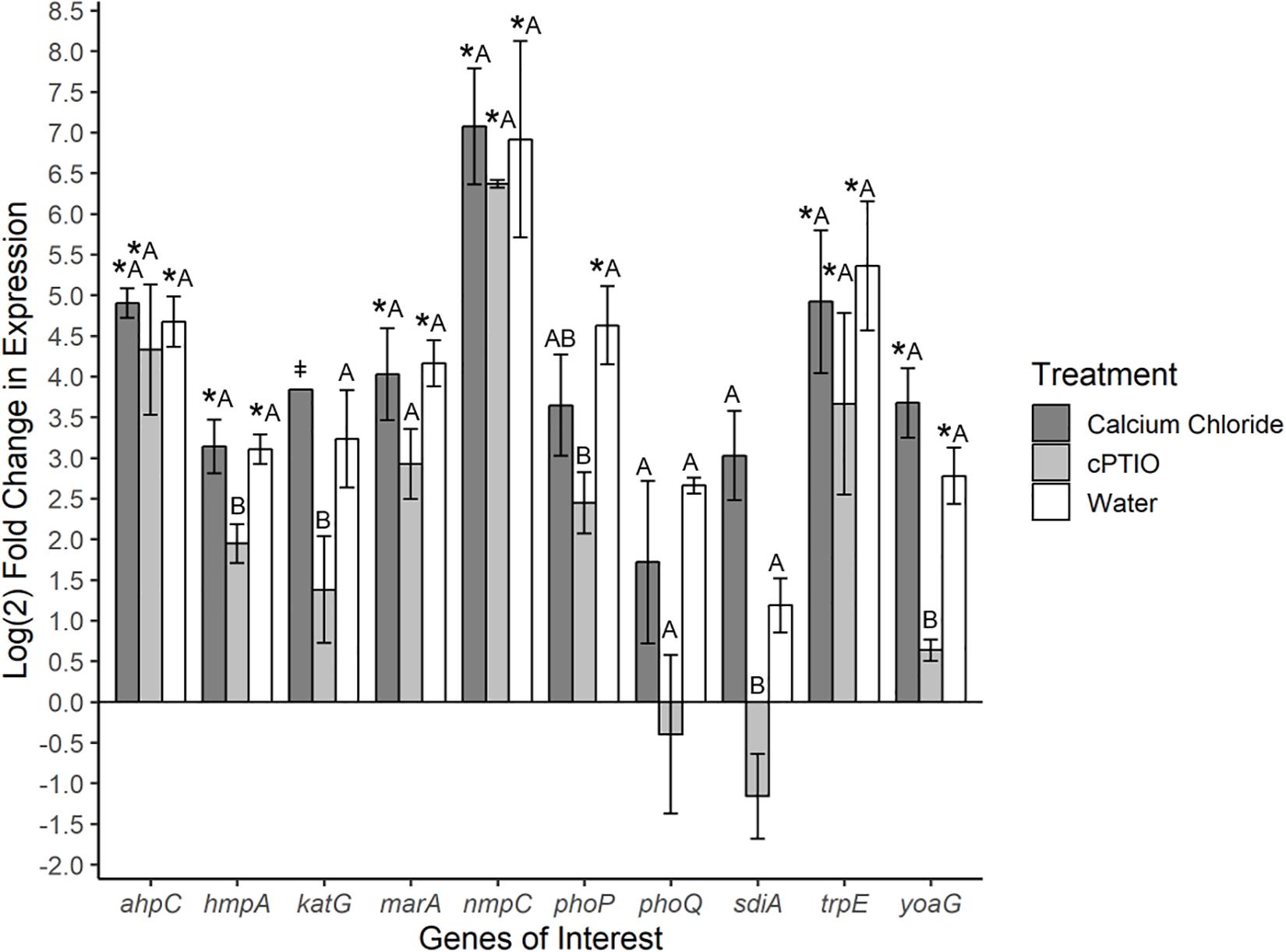
Figure 3. Log2-fold change in expression of S. Newport genes compared to expression in inoculum. ΔΔCt results of S. Newport colonizing 3-leaf tomato seedlings pretreated to reflect the native environment (H2O), NO-limiting (cPTIO) or NO-excess (CaCl2) environments normalized against expression of the sigma factor rpoD. N = 4 groups of three pooled plants per treatment. Error bars represent the standard error of the mean (SEM). ‡ denotes insufficient data to generate (SEM). Asterisks denote significance in gene transcription compared to inoculum according to Dunnett’s test (α = 0.05). Letters denote significant differences in gene transcription among treatments for each target gene using Tukey’s Honestly Significant Difference (α = 0.05).
The gene ahpC encodes an enzyme alkyl hydroperoxide reductase (Ahp) that protects cells from oxidative stress by catalyzing the reduction of hydrogen peroxide and organic peroxides (Seaver and Imlay, 2001). This gene was up-regulated in all treatments compared to inoculum (p < 0.05), but was not found to be differentially expressed among treatments. Expression of the catalase gene katG (Morgan et al., 1986) was not different from the inoculum, but differed between SeN on native versus cPTIO-treated leaves (p = 0.053). Overall, SeN appeared to be responding to oxidative stress on leaves.
The virulence factors phoQ two component system (Monsieurs et al., 2005) and the quorum sensing gene sdiA (Ahmer et al., 1998) both exhibited an increase in expression on native and NO-excess environments. The transcription of sdiA on CaCl2-treated leaves and of phoQ on cPTIO-treated leaves was higher and lower, respectively, than transcription in the inoculum but weakly statistically supported (p = 0.06). Compared to expression on NO-limiting environments, sdiA expression increased fourfold on NO-excess leaves and phoQ expression increasing threefold on native leaves (Figure 3). Transcription levels of sdiA on NO-excess and native environments were significantly different (p = 0.05). The multiple antibiotic resistance transcriptional regulator marA (Lee et al., 2015) was also significantly up-regulated in NO-excess and native plant environments compared to expression in the inoculum. This gene expression pattern suggests these genes may be important for leaf colonization. The outer membrane porin nmpC involved in H2O2 and other small molecule diffusion across cell membranes (Calderón et al., 2011) and trpE, a component of tryptophan biosynthesis, displayed uniform significant up-regulation in all treatments compared to inoculum (p < 0.05), indicating they may not be directly affected by plant derived NO stress.
Expression of NO and ROS Detoxification Genes Was Detected in SeN Colonizing Fruit Surface
Gene expression was also investigated in SeN associating with the surface of tomato fruit. Ascorbic acid was employed as an ROS scavenger (ROS-limiting environment) before challenging fruit with SeN and comparing expression profiles to SeN on water treated tomato fruit (native environment). In total, 72% of SeN on fruit samples displayed a ≥|2| -fold change in gene expression compared to the inoculum (Figure 4). SeN on native fruit exhibited significant up-regulation of hmpA and yoaG compared to the inoculum (p < 0.05). These genes were also up-regulated in the native fruit environment compared to the ROS-limiting environment (p < 0.05). Transcription of the oxidative stress gene ahpC was several log fold-change higher on native versus ascorbic-acid treated fruit (p < 0.05). The findings suggest SeN on fruit was countering plant derived NO stress and provides evidence that NO induction by SeN may be affected by ROS activity.
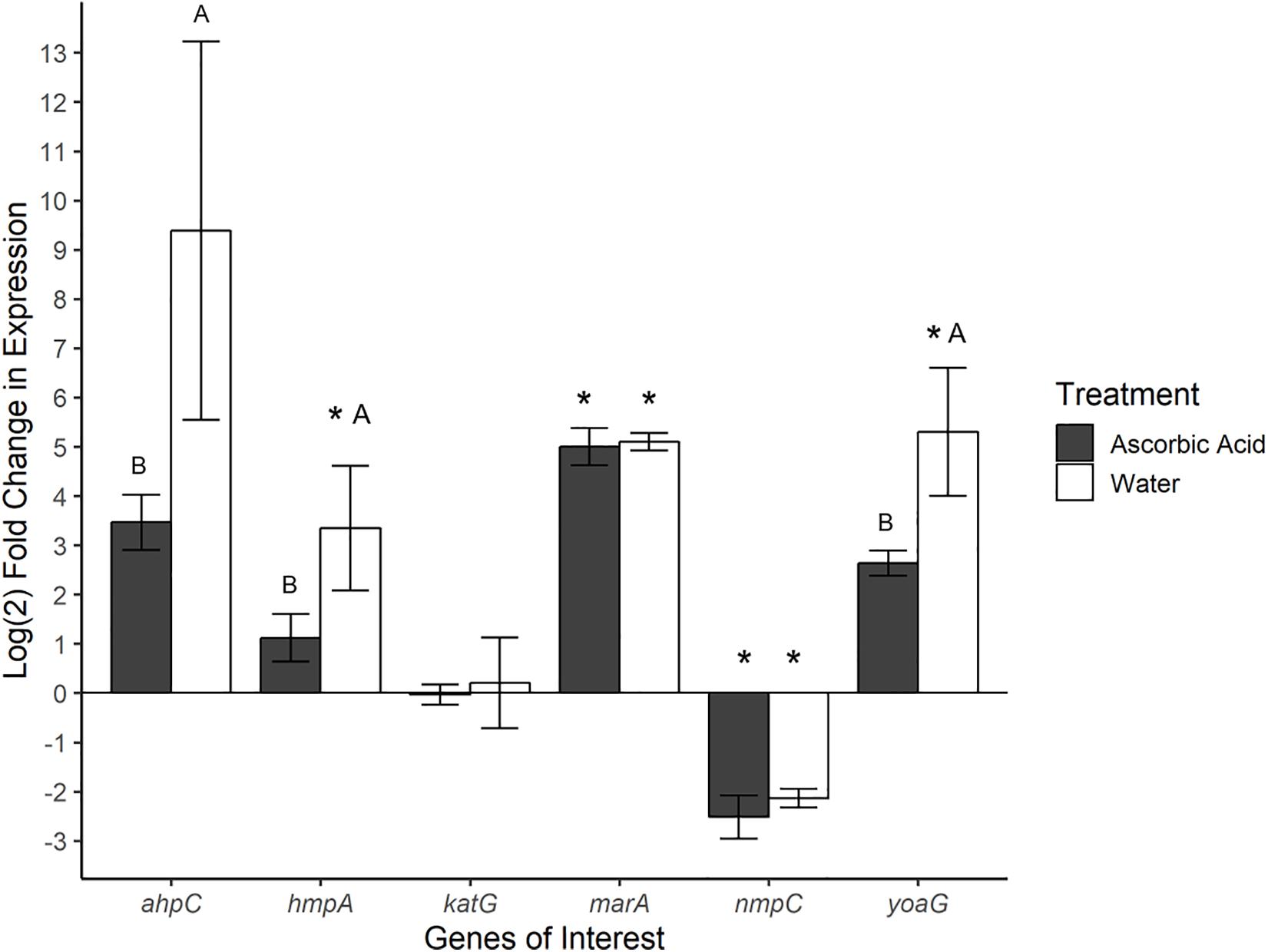
Figure 4. Log2-fold change in gene expression of S. Newport genes compared to expression in inoculum. ΔΔCt results of S. Newport colonizing tomato fruit pretreated to reflect the native fruit environment (H2O) or ROS-limiting (AscA) environments normalized against expression of sigma factor rpoD. Error bars represent the standard error of the mean. Asterisks denote significance in gene transcription compared to inoculum via Dunnett’s test (α = 0.05). Letters denote significant differences in expression between treatments for each target gene (Student’s t-test, α = 0.05).
As seen in SeN on leaves, transcription levels of marA were higher in SeN on native and ROS-scavenged fruit (p < 0.05) compared to inoculum and appeared unaffected by modulation of ROS. Down-regulation of nmpC was detected in SeN on both fruit treatments (p < 0.05), compared to inoculum.
Modulating Tomato Surface NO Levels Significantly Affected SeN Colonization of Leaves, but Not Fruit
To evaluate whether SeN colonization of tomato surfaces was significantly affected by modulated levels of plant-derived H2O2 and NO, a series of 12 h on-plant SeN challenge assays were conducted on leaves and fruit. SeN counts recovered from leaves pre-elicited to produce endogenous NO were almost 2 log lower at 12 hpi (p < 0.001), measured at 3.32 ± 0.2 log CFU/leaflet, compared to 5.15 ± 0.3 log CFU/leaflet recovered from mock-treated leaves (Figure 5A). Conversely, SeN counts recovered from NO-scavenged leaves were higher (4.85 ± 0.5 log CFU/leaflet) compared to sterile water treated leaves (p < 0.05; Figure 5B). This effect was not observed on fruit. Regardless of pre-treatment, SeN was retrieved at higher titers with smaller coefficients of variation (CV), on leaf compared to fruit samples, in both NO scavenged tissue (CVfruit = 0.39 and CVleaves = 0.10) and NO elicited tissue (CVfruit = 0.42 and CVleaves = 0.24).
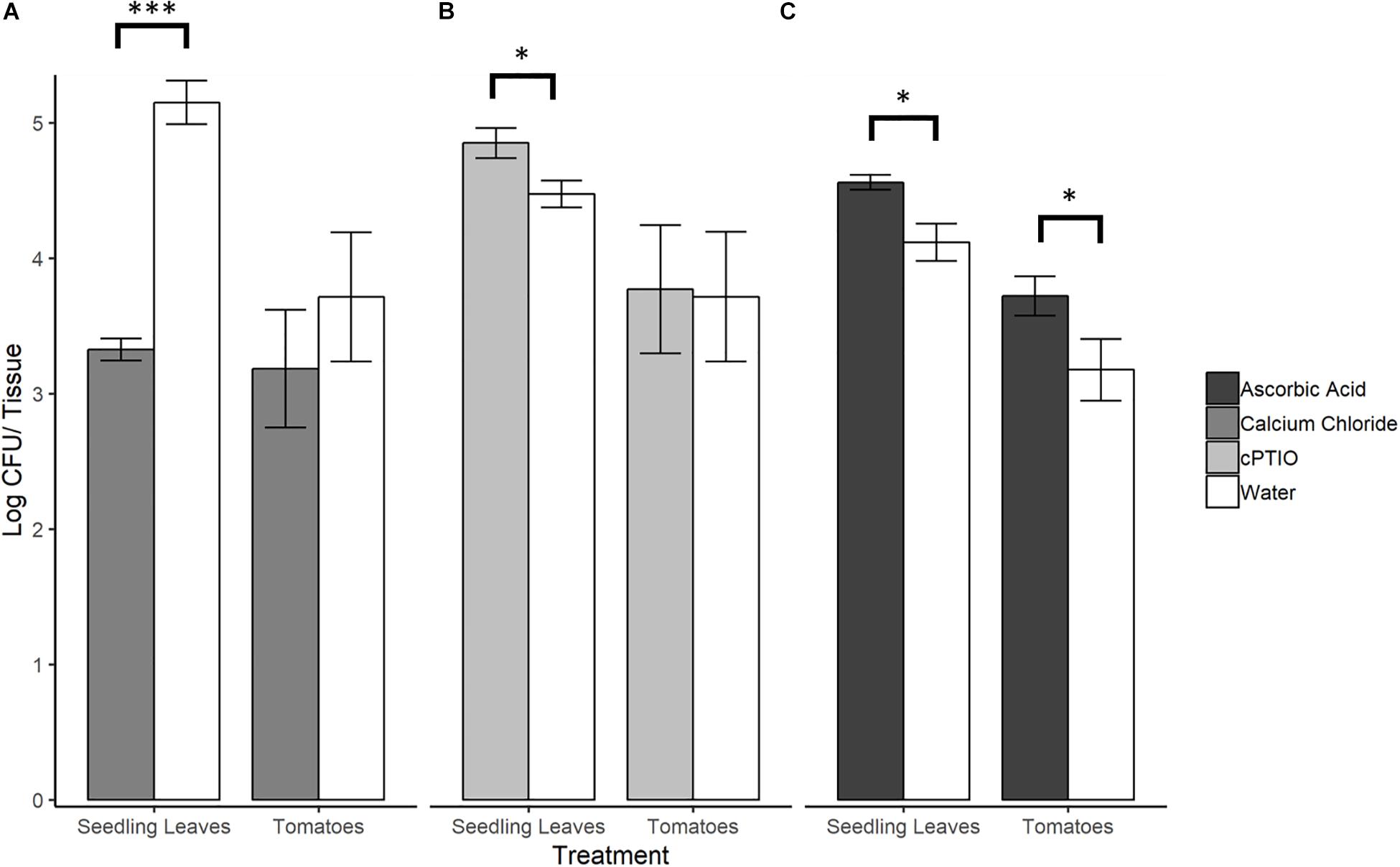
Figure 5. S. Newport counts retrieved from tomato seedling leaves and fruit after pre-treatment of plant tissue with (A) NO elicitor, (B) NO scavenger, or (C) H2O2 scavenger. Error bars represent standard error of the mean. Asterisks denote statistically significant difference by Student’s t-test between treatments and water control (α = 0.05) with ∗∗∗p < 0.001 and ∗p ≤ 0.05.
Scavenging H2O2 on Tomato Leaves and Fruit Favors SeN
When ROS was scavenged from plant leaf and fruit samples with ascorbic acid, significantly higher SeN counts were retrieved from scavenged leaves (4.56 ± 0.10 log CFU/leaflet) and fruit surfaces (3.27 ± 0.47 log CFU/fruit) compared to control (p < 0.05; Figure 5C). On fruit, regardless of treatment, SeN retrieval was ∼0.8 log CFU/unit lower than on leaves, with a larger coefficient of variation (CV) in retrieval from fruit compared to leaves (CVfruit = 0.20 and CVleaves = 0.07).
Discussion
In the present study, biochemical, gene expression and on-plant colonization assays of both vegetative tissue and ripe mature fruit provide evidence to support tomato plant recognition and response to Salmonella Newport. Tomato leaves and fruit generated ROS and leaves also generated NO in response to the enteric pathogen. In turn, Salmonella interpreted the mounted plant response as a stress, evidenced by differential ROS and NO detoxification gene up-regulation in Salmonella colonizing plant surfaces. Salmonella, therefore, may need to respond to plant-derived stimuli to ensure successful epiphytic colonization. Importantly, this study shows that tomato plants possess mechanisms capable of restricting Salmonella populations on leaf and fruit surfaces. ROS was detected in both leaf and fruit samples, and the reciprocal bacterial response was consistent with ROS negatively impacting Salmonella colonization, restricting bacterial counts. NO induction and an adverse effect of NO on Salmonella colonization was also detected on leaves, but not fruit. The ROS and NO bursts induced Salmonella to express genes needed for ROS and NO detoxification on leaves and NO detoxification on fruit. Despite no detected NO in fruit tissue subsequent to Salmonella challenge, and no effect of NO on bacterial restriction on fruit, bacterial gene expression results suggested Salmonella perception of NO on fruit. This work provides evidence that plant-derived NO is generated in response to S. enterica recognition.
Nitric oxide is required for PAMP-induced stomatal closure (Melotto et al., 2006). The production of NO in tomato leaves in response to Salmonella could therefore be signaling this innate immune response. This may also explain why NO was detected on leaves but not fruit, as tomato fruit lack stomata (Rančić et al., 2010). Low levels of NO, however, may have been present even on fruit, as transcription of NsR regulon genes was detected in Salmonella on both leaves and fruit. Up-regulation of NO detoxification gene hmpA during tomato surface colonization indicated that S. Newport perceived plant-derived NO as a stressor, corroborated by lower bacterial counts of S. Newport on NO-elicited seedling leaves. The NsrR regulon, controlled by the nitric oxide sensing transcriptional repressor NsrR, plays an important role in nitrosative stress resistance during infection. Within this regulon, the flavohemoglobin HmpA is identified as the main protein responsible for NO detoxification activities in the presence of an oxygenated environment (Karlinsey et al., 2012). A transcriptomic study of S. Typhimurium on tomato leaves and roots identified multiple up-regulated NsrR regulon genes, ygbA, ytfE, yoaG STM1808 and yfhH (Han et al., unpublished), which in E. coli is known to offer an NsrR binding site (Partridge et al., 2009). Supporting this, we assayed two genes in the NsrR regulon, hmpA and yoaG, both of which were differentially up-regulated compared to all native fruit and leaf environments. Han et al. (unpublished) measured gene expression in cells that had been colonizing plants for multiple days and did not detect hmpA. In our study, gene expression was assayed following 6 h of plant association, to capture SeN responses to early MTI. In fact, hmpA expression implies an early release of NO by the plant, and the need for S. Newport to mitigate its immediate effects. The NO detoxification-associated genes hmpA, nfrA and ygbA were also found to be up-regulated in soft rot macerated cilantro and lettuce leaf tissue caused by the plant pathogen Dickeya dadantii (Goudeau et al., 2013). In the present study, Salmonella NO detoxification was reported in the absence of a plant pathogen or tissue injury. Tomato plants produced NO upon perception of Salmonella which, in turn, led the bacteria to switch on reactive nitrogen species (RNS) detoxification machinery. This action may be necessary in order for Salmonella to successfully persist on some plant surfaces.
Evidence of ROS elicitation in tomato colonized with S. Newport was also strong, consistent with other reports of MTI induction in plants associating with this enteropathogen (Schikora et al., 2008; Shirron and Yaron, 2011; Meng et al., 2013; Garcia et al., 2014). The response we detected on leaves was diffuse across the leaf tissue and similar to staining reported for avirulent pathogens, as opposed to virulent pathogens (Großkinsky et al., 2012). Although Salmonella is not known to enter the apoplastic space (Potnis et al., 2014), the assay we used required infiltration, such that the response may have been more pronounced than what occurs when Salmonella is associating with the leaf epiphytically or residing in sub-stomatal chambers. In any case, ROS had a restrictive effect on Salmonella populations on the surface of both leaves and fruit and evidence of bacterial detoxification of ROS stress while colonizing leaf surfaces was detected via up-regulation of S. enterica ahpC. By contrast, we did not detect expression of the catalase katG. AhpC and the catalase encoded by katG are both known to scavenge H2O2 and organic hydroperoxides, but we only detected consistent expression of ahpC in Salmonella associated with leaves. In macrophages, ahpC expression is stronger than katG expression (Hébrard et al., 2009). Ahp was reported to be a more efficient scavenger at low concentrations of H2O2 (below 20 μM) than catalase, which becomes the primary active enzyme at higher H2O2 concentrations (Seaver and Imlay, 2001). From our findings, therefore, we infer that levels of H2O2 reaching Salmonella on the tomato plant surface were in concentrations insufficient to induce catalase activity.
Modulation of NO on leaves did not impact ahpC expression, but the use of an ROS scavenger on fruit reduced transcription of ahpC. Transcription of the NsrR-regulated hmpA and yoaG genes in Salmonella on fruit was also higher when ROS was not attenuated, suggesting that higher levels of ROS may be related to NO levels. A large degree of interconnectivity exists between NO and ROS signaling in plant tissue (Romero-Puertas and Sandalio, 2016). On native and ascorbic acid-treated fruit tissue, Salmonella would have been responding to NO levels induced by the enteropathogen itself. However, NO has been shown to regulate ascorbate peroxidase (APX) (Yang et al., 2015), the enzyme that uses ascorbate as an electron donor to reduce H2O2 to H2O. On ascorbic acid-treated fruit, therefore, the depleted H2O2 environment could have signaled the attenuation of NO, such that Salmonella would have been responding to lower NO levels reflected in reduced hmpA and yoaG transcription.
Other than NO and ROS stress, SeN gene expression on leaves and fruit was indicative of adaptation to a novel environment. In leaves, trpE, a gene in the tryptophan biosynthesis pathway which has been associated with biofilm development (Hamilton et al., 2009), was up-regulated in all environments. Biofilm formation is known to enhance the capacity of pathogenic bacteria to survive stresses in the environment and during host infection. Thus, the present work provides more evidence to the growing body of work which defines attachment as paramount to survival in the phyllosphere (Barak et al., 2005, 2007). Also up-regulated in all leaf and fruit environments was the gene marA. In addition to their importance as regulators of xenobiotic efflux, marRAB may have indirect effects on expression of iron metabolism genes, membrane composition genes, and the stress related sigma factor rpoS (Lee et al., 2015). In S. enterica transcriptomic surveys (Wang et al., 2010; Brankatschk et al., 2014), the use of such machinery was found to be pertinent to survival in stress-inducing environments. In our study, nmpC (ompD, STM1572) displayed differential expression among plant tissue types, up-regulated compared to inoculum in Salmonella on leaf surfaces but down-regulated on fruit surfaces. NmpC is one of the most abundant outer membrane porins of S. enterica, used to transport molecules, including H2O2, into the cell and toxins out of the cell (Calderón et al., 2011). Expression of nmpC was down-regulated in S. Typhimurium exposed to H2O2 (Calderón et al., 2011). Additionally, NmpC may be needed for adherence and recognition of S. Typhimurium to human macrophages and epithelial cells during the initial stages of infection (Hara-Kaonga and Pistole, 2004) and is also involved in host cell recognition in mammalian models (Ipinza et al., 2014; van der Heijden et al., 2016). In our study, we observed up-regulation of both nmpC and ROS detoxification gene ahpC on leaves. However, nmpC was down-regulated in SeN on native fruit where ahpC expression was more variable. These findings suggest an additional function of this porin, perhaps engaging in efflux activity of other xenobiotics. Taken together, targeted gene expression on both leaves and fruit provide evidence that S. enterica attaches to and recognizes various stressors on plant surfaces.
Working with NO modulation on fruit proved challenging. While up-regulation of bacterial NO detoxification genes was detected on fruit, we could not easily measure NO released from fruit tissue challenge with various biotic treatments. Salmonella produced a fluorescent response in fruit exocarp greater than that of leaves but was not consistent. It is possible that exocarp excision generated high levels of NO, even in the negative control, and masking the weaker signal elicited by the pathogen. This could also explain why DAF-2 DA fluorescence diminished over time, even in the positive control. Further, ascorbic acid significantly affected S. enterica counts on fruit whereas NO modulation did not. This could be attributed to NO and ROS endogenous levels in mature red fruit at the time of study. Ripe red fruit are known to have lower concentrations of nitric oxide compared to mature green fruit, as NO is involved in regulating ethylene production and thus facilitating the ripening process (Ya’acov et al., 1998). Most NO modulation studies in fruit responding to plant pathogens are routinely conducted with mature green fruit (Lai et al., 2011; Zheng et al., 2011a, b; Zhu and Tian, 2012). The transition from green to red fruit is marked by accruement of high ROS concentrations (Kumar et al., 2016). Petrasch et al. (2019) investigated ROS detoxification by fungal pathogens in red ripe fruit. Thus, the ascorbic acid injected in our plant colonization assays could have been targeting ripening-related ROS, hence providing a more hospitable environment for colonizing S. enterica. Mature ripe fruit tissue also has lower levels of pathogen recognition response capacity compared to vegetative tissue, perhaps due in part to the breakdown of cellular wall components during ripening (Cantu et al., 2009). S. enterica studies on ripe and unripe tomato fruit have found the organism proliferates more readily in ripe red compared to mature green tomato fruit (Barak et al., 2011). Taken together, these data imply mechanisms of S. enterica restriction are plant tissue-specific and may be facilitated or confounded by the fruit ripening process. Regardless, more research is needed to evaluate the interconnectivity between ripening and pathogen defense, both in the contexts of plant and human pathogens.
Overall, higher titers of S. Newport were consistently retrieved from leaves compared to fruit, an observation which has been reported elsewhere (Barak et al., 2011; Han and Micallef, 2014; Gu et al., 2018). Supporting evidence can be found in Salmonella field sampling studies. For example in a multi-year field study, sampling tomato leaves and fruit for wild S. enterica colonization found only leaves returned positive S. enterica result, never fruit (Gu et al., 2018). This tissue-specific variability in Salmonella carrying capacity could be due to relative abundance and composition of nutrients on the surface of the different plant organs, higher relative humidity on leaves as a result of transpiration, and a higher and more rugged surface area or attachment on leaves compared to fruit. Higher proportions of fatty acids have been seen in fruit washes of tomato cv. ‘Heinz-1706’ compared to seedling shoot or mature leaf washes, and this correlated negatively with S. enterica growth (Han and Micallef, 2016). While the leaves of the tomato are not eaten, in the field leaves and tomato fruit are in constant contact with one another, serving as a contamination source for fruit. Vegetative matter is commonly picked up during harvest of tomato fruit and could lead to widespread contamination if appropriate Good Agricultural Practices are not followed. Cross-contamination of fruit from contaminated vegetative matter is a potential risk when using recirculating water to wash tomatoes without the appropriate concentration of sanitizer (Bolten et al., 2020).
We noted that SeN retrieval from leaves was less variable than fruit, suggesting a potential heterogeneity of response to unique stressors present on the latter plant organ. Diversification of stress response or “bet hedging” has been documented in intercellular S. enterica interaction with ROS and other stress agents (Helaine and Holden, 2013; Burton et al., 2014; Helaine et al., 2014). Tomato fruit may be a uniquely harsh plant niche for S. enterica for which “bet hedging” may be a significant strategy to ensure long term survival. Understanding tissue specific enteropathogen–plant interactions therefore can help devise strategies to minimize fruit contamination. Yet, enteropathogen–plant interaction data relevant to agricultural situations, which relate directly to salmonellosis outbreak-causing S. enterica strains, remain limited.
Salmonella enterica mitigation of host-derived NO and ROS is crucial for successful invasion in animal host models. These processes have been well documented (Vazquez-Torres and Fang, 2001; Zheng et al., 2011a; Spector and Kenyon, 2012; van der Heijden et al., 2015). Presence of RNS in the mammalian gut increased overall colonization fitness of S. enterica, possibly because it can outcompete some resident microflora (Stecher et al., 2007). This compounds the importance of investigating the presence of analogous interactions in S. enterica–plant associations. Perception and response to NO and ROS, which can be short-term restricting agents, together with other important stress and host adaptation responses, may lead to long-term persistence in the field. As ROS and RNS may be present in multiple scenarios in the agricultural setting (Diaz and Plummer, 2018), mitigating these compounds may be a key factor for S. enterica persistence in between entry into an animal host. Future work should investigate the ability of Salmonella to mitigate these stresses on plants and whether they are shared by all serovars, or specific to serovars that are regularly implicated in produce outbreaks. This is essential to continue to elucidate Salmonella adaptation to non-animal host environments.
Deciphering the highly nuanced and complex plant-associated lifestyle of this enteric pathogen is imperative to inform strategies to minimize successful Salmonella contamination and persistence in an agricultural setting and help in identifying plant traits or cultivars that are unfavorable for Salmonella colonization. Furthermore, how non-plant pathogenic microorganisms, including enteric pathogens, interact with the plant immune system to colonize the phyllosphere is poorly understood. The MAMPs recognized by plants that result in MTI are highly conserved among microbes. It has recently been suggested that plants cannot differentiate between pathogens and commensals, and micro-organisms must evade or attenuate plant immunity to colonize plants (Teixeira et al., 2019). Untangling the role that plant immunity plays in the establishment of human pathogens on plants would add to knowledge about microbiome assembly, while also elucidating mechanisms that can lead to enhanced food safety.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
SM and AF conceptualized the study and designed the experiments. AF conducted the experiments. AF, SB, and BS designed and conducted q-RT-PCR experiments. AF and SM analyzed and interpreted the data and wrote the manuscript. SB and BS reviewed and edited the manuscript. SM administered the project.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Shunyuan Xiao for generously providing Pseudomonas syringae pv. maculicola ES4326. This project was completed using UMD start-up funds to SM. We also thank Megan Holbert and Sydney Wallace at the University of Maryland research greenhouse, and Michael Newell at the Central Maryland Wye Research and Education Centre, Queenstown Maryland, for assistance in tomato cultivation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00391/full#supplementary-material
References
Ahmer, B. M. M., Van Reeuwijk, J., Timmers, C. D., Valentine, P. J., and Heffron, F. (1998). Salmonella Typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J. Bacteriol. 180, 1185–1193. doi: 10.1128/jb.180.5.1185-1193.1998
Barak, J. D., Gorski, L., Naraghi-Arani, P., and Charkowski, A. O. (2005). Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl. Environ. Microbiol. 71, 5685–5691. doi: 10.1128/aem.71.10.5685-5691.2005
Barak, J. D., Jahn, C. E., Gibson, D. L., and Charkowski, A. O. (2007). The role of cellulose and O-antigen capsule in the colonization of plants by Salmonella enterica. Mol. Plant Microbe Interact. 20, 1083–1091. doi: 10.1094/mpmi-20-9-1083
Barak, J. D., Kramer, L. C., and Hao, L. (2011). Colonization of tomato plants by Salmonella enterica Is cultivar dependent, and type 1 trichomes are preferred colonization sites. Appl. Environ. Microbiol. 77, 498–504. doi: 10.1128/AEM.01661-10
Bell, R. L., Zheng, J., Burrows, E., Allard, S., Wang, C. Y., Keys, C. E., et al. (2015). Ecological prevalence, genetic diversity, and epidemiological aspects of Salmonella isolated from tomato agricultural regions of the Virginia Eastern Shore. Front. Microbiol. 6:415. doi: 10.3389/fmicb.2015.00415
Bennett, S. D., Littrell, K. W., Hill, T. A., Mahovic, M., and Behravesh, C. B. (2015). Multistate foodborne disease outbreaks associated with raw tomatoes, United States, 1990–2010: a recurring public health problem. Epidemiol. Infect. 143, 1352–1359. doi: 10.1017/s0950268814002167
Bindschedler, L. V., Dewdney, J., Blee, K. A., Stone, J. M., Asai, T., Plotnikov, J., et al. (2006). Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J. 47, 851–863. doi: 10.1111/j.1365-313X.2006.02837.x
Bolten, S., Gu, G., Luo, Y., Van Haute, S., Zhou, B., Millner, P., et al. (2020). Salmonella inactivation and cross-contamination on cherry and grape tomatoes under simulated wash conditions. Food Microbiol. 87:103359. doi: 10.1016/j.fm.2019.103359
Bradley, D. J., Kjellbom, P., and Lamb, C. J. (1992). Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell 70, 21–30. doi: 10.1016/0092-8674(92)90530-P
Brandl, M. T., and Amundson, R. (2008). Leaf age as a risk factor in contamination of lettuce with Escherichia coli O157: H7 and Salmonella enterica. Appl. Environ. Microbiol. 74, 2298–2306. doi: 10.1128/aem.02459-07
Brandl, M. T., Cox, C. E., and Teplitski, M. (2013). Salmonella interactions with plants and their associated microbiota. Phytopathology 103, 316–325. doi: 10.1094/phyto-11-12-0295-rvw
Brandl, M. T., and Mandrell, R. E. (2002). Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. Appl. Environ. Microbiol. 68, 3614–3621. doi: 10.1128/aem.68.7.3614-3621.2002
Brankatschk, K., Kamber, T., Pothier, J. F., Duffy, B., and Smits, T. H. M. (2014). Transcriptional profile of Salmonella enterica subsp enterica serovar Weltevreden during alfalfa sprout colonization. Microb. Biotechnol. 7, 528–544. doi: 10.1111/1751-7915.12104
Burton, N. A., Schürmann, N., Casse, O., Steeb, A. K., Claudi, B., Zankl, J., et al. (2014). Disparate impact of oxidative host defenses determines the fate of Salmonella during systemic infection in mice. Cell Host Microbe 15, 72–83. doi: 10.1016/j.chom.2013.12.006
Calderón, I. L., Morales, E., Caro, N. J., Chahúan, C. A., Collao, B., Gil, F., et al. (2011). Response regulator ArcA of Salmonella enterica serovar Typhimurium downregulates expression of OmpD, a porin facilitating uptake of hydrogen peroxide. Res. Microbiol. 162, 214–222. doi: 10.1016/j.resmic.2010.11.001
Callahan, M. T., Van Kessel, J. A., and Micallef, S. A. (2019). Salmonella enterica recovery from river waters of the Maryland Eastern Shore reveals high serotype diversity and some multidrug resistance. Environ. Res. 168, 7–13. doi: 10.1016/j.envres.2018.09.012
Callejón, R. M., Rodríguez-Naranjo, M. I., Ubeda, C., Hornedo-Ortega, R., Garcia-Parrilla, M. C., and Troncoso, A. M. (2015). Reported foodborne outbreaks due to fresh produce in the United States and European Union: trends and causes. Foodborne Pathog. Dis. 12, 32–38. doi: 10.1089/fpd.2014.1821
Cantu, D., Blanco-Ulate, B., Yang, L., Labavitch, J. M., Bennett, A. B., and Powell, A. L. T. (2009). Ripening-regulated susceptibility of tomato fruit to Botrytis cinerea requires NOR but Not RIN or ethylene. Plant Physiol. 150, 1434–1449. doi: 10.1104/pp.109.138701
Chakraborty, N., Ghosh, S., Chandra, S., Sengupta, S., and Acharya, K. (2016). Abiotic elicitors mediated elicitation of innate immunity in tomato: an ex vivo comparison. Physiol. Mol. Biol. Plants 22, 307–320. doi: 10.1007/s12298-016-0373-z
Crawford, M. J., and Goldberg, D. E. (1998). Regulation of the Salmonella Typhimurium flavohemoglobin gene - A new pathway for bacterial gene expression in response to nitric oxide. J. Biol. Chem. 273, 34028–34032. doi: 10.1074/jbc.273.51.34028
de Moraes, M. H., Soto, E. B., Salas González, I., Desai, P., Chu, W., Porwollik, S., et al. (2018). Genome-wide comparative functional analyses reveal adaptations of Salmonella sv. Newport to a Plant Colonization Lifestyle Front. Microbiol. 9:877. doi: 10.3389/fmicb.2018.00877
de Moraes, M. H., Desai, P., Porwollik, S., Canals, R., Perez, D. R., Chu, W., et al. (2017). Salmonella persistence in tomatoes requires a distinct set of metabolic functions identified by transposon insertion sequencing. Appl. Environ. Microbiol. 83:0e3028-e16. doi: 10.1128/AEM.03028-16
Diaz, J. M., and Plummer, S. (2018). Production of extracellular reactive oxygen species by phytoplankton: past and future directions. J. Plankton Res. 40, 655–666. doi: 10.1093/plankt/fby039
Doke, N. (1983). Involvement of superoxide anion generation in the hypersensitive response of potato tuber tissues to infection with an incompatible race of Phytophthora infestans and to the hyphal wall components. Physiol. Plant Pathol. 23, 345–357. doi: 10.1016/0048-4059(83)90019-X
Garcia, A. V., Charrier, A., Schikora, A., Bigeard, J., Pateyron, S., de Tauzia-Moreau, M.-L., et al. (2014). Salmonella enterica flagellin is recognized via FLS2 and activates PAMP-Triggered Immunity in Arabidopsis thaliana. Mol. Plant 7, 657–674. doi: 10.1093/mp/sst145
Goudeau, D. M., Parker, C. T., Zhou, Y. G., Sela, S., Kroupitski, Y., and Brandl, M. T. (2013). The Salmonella transcriptome in lettuce and cilantro soft rot reveals a niche overlap with the animal host intestine. Appl. Environ. Microbiol. 79, 250–262. doi: 10.1128/aem.02290-12
Greene, S. K., Daly, E. R., Talbot, E. A., Demma, L. J., Holzbauer, S., Patel, N. J., et al. (2008). Recurrent multistate outbreak of Salmonella Newport associated with tomatoes from contaminated fields, 2005. Epidemiol. Infect. 136, 157–165. doi: 10.1017/s095026880700859x
Großkinsky, D. K., Koffler, B. E., Roitsch, T., Maier, R., and Zechmann, B. (2012). Compartment-specific antioxidative defense in Arabidopsis against virulent and avirulent Pseudomonas syringae. Phytopathol 102, 662–673. doi: 10.1094/PHYTO-02-12-0022-R
Gu, G., Strawn, L., Oryang, D., Zheng, J., Reed, E., Ottesen, A., et al. (2018). Agricultural practices influence Salmonella contamination and survival in pre-harvest tomato production. Front. Microbiol. 9:2451. doi: 10.3389/fmicb.2018.02451
Hamilton, S., Bongaerts, R. J. M., Mulholland, F., Cochrane, B., Porter, J., Lucchini, S., et al. (2009). The transcriptional programme of Salmonella enterica serovar Typhimurium reveals a key role for tryptophan metabolism in biofilms. BMC Genomics 10:599. doi: 10.1186/1471-2164-10-599
Han, S., and Micallef, S. A. (2014). Salmonella Newport and Typhimurium colonization of fruit differs from leaves in various tomato cultivars. J. Food Prot. 77, 1844–1850. doi: 10.4315/0362-028x.jfp-13-562
Han, S., and Micallef, S. A. (2016). Environmental Metabolomics of the Tomato Plant Surface Provides Insights on Salmonella enterica Colonization. Appl. Environ. Microbiol. 82, 3131–3142. doi: 10.1128/aem.00435-16
Hara-Kaonga, B., and Pistole, T. G. (2004). OmpD but not OmpC is involved in adherence of Salmonella enterica serovar Typhimurium to human cells. Can. J. Microbiol. 50, 719–727. doi: 10.1139/W04-056
Hébrard, M., Viala, J. P. M., Méresse, S., Barras, F., and Aussel, L. (2009). Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance. J. Bacteriol. 191, 4605–4614. doi: 10.1128/JB.00144-09
Helaine, S., Cheverton, A. M., Watson, K. G., Faure, L. M., Matthews, S. A., and Holden, D. W. (2014). Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343, 204–208. doi: 10.1126/science.1244705
Helaine, S., and Holden, D. W. (2013). Heterogeneity of intracellular replication of bacterial pathogens. Curr. Opin. Microbiol. 16, 184–191. doi: 10.1016/j.mib.2012.12.004
Iniguez, A. L., Dong, Y. M., Carter, H. D., Ahmer, B. M. M., Stone, J. M., and Triplett, E. W. (2005). Regulation of enteric endophytic bacterial colonization by plant defenses. Mol. Plant Microb. Interact. 18, 169–178. doi: 10.1094/MPMI-18-0169
Ipinza, F., Collao, B., Monsalva, D., Bustamante, V. H., Luraschi, R., Alegría-Arcos, M., et al. (2014). Participation of the Salmonella OmpD Porin in the Infection of RAW264.7 Macrophages and BALB/c Mice. PLoS One 9:e111062. doi: 10.1371/journal.pone.0111062
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Karlinsey, J. E., Bang, I.-S., Becker, L. A., Frawley, E. R., Porwollik, S., Robbins, H. F., et al. (2012). The NsrR regulon in nitrosative stress resistance of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 85, 1179–1193. doi: 10.1111/j.1365-2958.2012.08167.x
Keshavarz-Tohid, V., Taheri, P., Taghavi, S. M., and Tarighi, S. (2016). The role of nitric oxide in basal and induced resistance in relation with hydrogen peroxide and antioxidant enzymes. J. Plant Physiol. 199, 29–38. doi: 10.1016/j.jplph.2016.05.005
Kumar, V., Irfan, M., Ghosh, S., Chakraborty, N., Chakraborty, S., and Datta, A. (2016). Fruit ripening mutants reveal cell metabolism and redox state during ripening. Protoplasma 253, 581–594. doi: 10.1007/s00709-015-0836-z
Lai, T., Wang, Y., Li, B., Qin, G., and Tian, S. (2011). Defense responses of tomato fruit to exogenous nitric oxide during postharvest storage. Postharvest Biol. Technol. 62, 127–132. doi: 10.1016/j.postharvbio.2011.05.011
Lee, J.-J., Hsuan, S.-L., Kuo, C.-J., Wu, Y.-C., and Chen, T.-H. (2015). MarA and RamA regulate virulence in Salmonella enterica serovar Choleraesuis. Vet. Microbiol. 181, 323–327. doi: 10.1016/j.vetmic.2015.09.006
Lee, S., Choi, H., Suh, S., Doo, I.-S., Oh, K.-Y., Jeong Choi, E., et al. (1999). Oligogalacturonic acid and chitosan reduce stomatal aperture by inducing the evolution of reactive oxygen species from guard cells of Tomato and Commelina communis. Plant Physiol. 121, 147–152. doi: 10.1104/pp.121.1.147
Lin, H. Y., Bledsoe, P. J., and Stewart, V. (2007). Activation of yeaR-yoaG operon transcription by the nitrate-responsive regulator NarL is independent of oxygen-responsive regulator Fnr in Escherichia coli K-12. J. Bacteriol. 189, 7539–7548. doi: 10.1128/JB.00953-07
Liu, Y., and Zhang, S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. 16, 3386–3399. Plant Cell doi: 10.1105/tpc.104.026609
Ma, W., Smigel, A., Tsai, Y. C., Braam, J., and Berkowitz, G. A. (2008). Innate immunity signaling: cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiol. 48, 818–828. doi: 10.1104/pp.108.125104
Małolepsza, U., and Różalska, S. (2005). Nitric oxide and hydrogen peroxide in tomato resistance: nitric oxide modulates hydrogen peroxide level in o-hydroxyethylorutin-induced resistance to Botrytis cinerea in tomato. Plant Physiol. Biochem. 43, 623–635. doi: 10.1016/j.plaphy.2005.04.002
Melotto, M., Underwood, W., Koczan, J., Nomura, K., and He, S. Y. (2006). Plant Stomata Function in Innate Immunity against Bacterial Invasion. Cell 126, 969–980. doi: 10.1016/j.cell.2006.06.054
Meng, F., Altier, C., and Martin, G. B. (2013). Salmonella colonization activates the plant immune system and benefits from association with plant pathogenic bacteria. Environ. Microbiol. 15, 2418–2430. doi: 10.1111/1462-2920.12113
Meng, X., and Zhang, S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266. doi: 10.1146/annurev-phyto-082712-102314
Micallef, S. A., Rosenberg Goldstein, R. E., George, A., Kleinfelter, L., Boyer, M. S., McLaughlin, C. R., et al. (2012). Occurrence and antibiotic resistance of multiple Salmonella serotypes recovered from water, sediment and soil on mid-Atlantic tomato farms. Environ. Res. 114, 31–39. doi: 10.1016/j.envres.2012.02.005
Monsieurs, P., De Keersmaecker, S., Navarre, W. W., Bader, M. W., De Smet, F., McClelland, M., et al. (2005). Comparison of the PhoPQ regulon in Escherichia coli and Salmonella Typhimurium. J. Mol. Evol 60, 462–474. doi: 10.1007/s00239-004-0212-7
Morgan, R. W., Christman, M. F., Jacobson, F. S., Storz, G., and Ames, B. N. (1986). Hydrogen peroxide-inducible proteins in Salmonella Typhimurium overlap with heat shock and other stress proteins. Proc. Natl. Acad. Sci. U.S.A. 83, 8059–8063. doi: 10.1073/pnas.83.21.8059
Neill, S. J., Desikan, R., Clarke, A., and Hancock, J. T. (2002). Nitric oxide is a novel component of abscisic acid signaling in stomatal guard cells. Plant Physiol. 128, 13–16. doi: 10.1104/pp.010707
Neumann, C., Fraiture, M., Hernàndez-Reyes, C., Akum, F. N., Virlogeux-Payant, I., Chen, Y., et al. (2014). The Salmonella effector protein SpvC, a phosphothreonine lyase is functional in plant cells. Front. Microbiol. 5:548. doi: 10.3389/fmicb.2014.00548
Partridge, J. D., Bodenmiller, D. M., Humphrys, M. S., and Spiro, S. (2009). NsrR targets in the Escherichia coli genome: new insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility. Mo. Microbiol. 73, 680–694. doi: 10.1111/j.1365-2958.2009.06799.x
Petrasch, S., Silva, C. J., Mesquida-Pesci, S. D., Gallegos, K., van den Abeele, C., Papin, V., et al. (2019). Infection strategies deployed by Botrytis cinerea, Fusarium acuminatum, and Rhizopus stolonifer as a function of tomato fruit ripening stage. Front. Plant Sci. 10:223. doi: 10.3389/fpls.2019.00223
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Potnis, N., Soto-Arias, J. P., Cowles, K. N., van Bruggen, A. H. C., Jones, J. B., and Barak, J. D. (2014). Xanthomonas perforans colonization influences Salmonella enterica in the tomato phyllosphere. Appl. Environ. Microbiol. 80, 3173–3180. doi: 10.1128/AEM.00345-14
Poza-Carrion, C., Suslow, T., and Lindow, S. (2013). Resident Bacteria on Leaves Enhance Survival of Immigrant Cells of Salmonella enterica. Phytopathol 103, 341–351. doi: 10.1094/PHYTO-09-12-0221-FI
Preston, G. M. (2000). Pseudomonas syringae pv. tomato: the right pathogen, of the right plant, at the right time. Mol. Plant Pathol. 1, 263–275. doi: 10.1046/j.1364-3703.2000.00036.x
Rančić, D., Quarrie, S. P., and Pećinar, I. (2010). “Anatomy of tomato fruit and fruit pedicel during fruit development,” in Microscopy: Science, Technology, Applications and Education, eds A. Méndez-Vilas and J. Díaz, (Badajoz: Formatex), 851–861.
Ranf, S., Eschen-Lippold, L., Pecher, P., Lee, J., and Scheel, D. (2011). Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J. 68, 100–113. doi: 10.1111/j.1365-313X.2011.04671.x
Rasul, S., Dubreuil-Maurizi, C., Lamotte, O., Koen, E., Poinssot, B., Alcaraz, G., et al. (2012). Nitric oxide production mediates oligogalacturonide-triggered immunity and resistance to Botrytis cinerea in Arabidopsis thaliana. Plant Cell Environ. 35, 1483–1499. doi: 10.1111/j.1365-3040.2012.02505.x
Romero-Puertas, M. C., and Sandalio, L. M. (2016). Nitric oxide level is self-regulating and also regulates its ROS partners. Front. Plant Sci. 7:316. doi: 10.3389/fpls.2016.00316
Salazar, J. K., Deng, K. P., Tortorello, M. L., Brandl, M. T., Wang, H., and Zhang, W. (2013). Genes ycfR, sirA and yigG contribute to the surface attachment of Salmonella enterica Typhimurium and Saintpaul to Fresh Produce. PLoS One 8:e57272. doi: 10.1371/journal.pone.0057272
Schikora, A., Carreri, A., Charpentier, E., and Hirt, H. (2008). The dark side of the salad: Salmonella typhimurium overcomes the innate immune response of Arabidopsis thaliana and shows an endopathogenic lifestyle. PLoS One. 3:e2279. doi: 10.1371/journal.pone.0002279
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji - an Open Source platform for biological image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Seaver, L. C., and Imlay, J. A. (2001). Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183, 7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001
Shin, R., and Schachtman, D. P. (2004). Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc. Natl. Acad. Sci. U.S.A. 101, 8827–8832. doi: 10.1073/pnas.0401707101
Shirron, N., and Yaron, S. (2011). Active suppression of early immune response in tobacco by the human pathogen Salmonella Typhimurium. PLoS One 6:e18855. doi: 10.1371/journal.pone.0018855
Spector, M. P., and Kenyon, W. J. (2012). Resistance and survival strategies of Salmonella enterica to environmental stresses. Food Res. Int. 45, 455–481. doi: 10.1016/j.foodres.2011.06.056
Stecher, B., Robbiani, R., Walker, A. W., Westendorf, A. M., Barthel, M., Kremer, M., et al. (2007). Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5:e244. doi: 10.1371/journal.pbio.0050244
Tan, M. S. F., White, A. P., Rahman, S., and Dykes, G. A. (2016). Role of Fimbriae, Flagella and Cellulose on the Attachment of Salmonella Typhimurium ATCC 14028 to Plant Cell Wall Models. PLoS One 11:e0158311. doi: 10.1371/journal.pone.0158311
Teixeira, P. J. P. L., Colaianni, N. R., Fitzpatrick, C. R., and Dangl, J. L. (2019). Beyond pathogens: microbiota interactions with the plant immune system. Curr. Opinion Microbiol. 49, 7–17. doi: 10.1016/j.mib.2019.08.003
Tsuda, K., Sato, M., Glazebrook, J., Cohen, J. D., and Katagiri, F. (2008). Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 53, 763–775. doi: 10.1111/j.1365-313X.2007.03369.x
van der Heijden, J., Bosman, E. S., Reynolds, L. A., and Finlay, B. B. (2015). Direct measurement of oxidative and nitrosative stress dynamics in Salmonella inside macrophages. Proc. Natl. Acad. Sci. U.S.A. 112, 560–565. doi: 10.1073/pnas.1414569112
van der Heijden, J., Reynolds, L. A., Deng, W., Mills, A., Scholz, R., Imami, K., et al. (2016). Salmonella rapidly regulates membrane permeability to survive oxidative stress. mBio 7:e01238-16. doi: 10.1128/mBio.01238-16
Vazquez-Torres, A., and Fang, F. C. (2001). Oxygen-dependent anti-Salmonella activity of macrophages. Trends Microbiol. 9, 29–33. doi: 10.1016/s0966-842x(00)01897-7
Wang, S., Phillippy, A. M., Deng, K., Rui, X., Li, Z., Tortorello, M. L., et al. (2010). Transcriptomic Responses of Salmonella enterica serovars enteritidis and typhimurium to chlorine-based oxidative stress. Appl. Environ. Microbiol. 76, 5013–5024. doi: 10.1128/AEM.00823-10
Ya’acov, Y. L., Wills, R. B. H., and Ku, V. V.-V. (1998). Evidence for the function of the free radical gas—nitric oxide (NO)—as an endogenous maturation and senescence regulating factor in higher plants. Plant Physiol. Biochem. 36, 825–833. doi: 10.1016/s0981-9428(99)80020-5
Yang, H., Mu, J., Chen, L., Feng, J., Hu, J., Li, L., et al. (2015). S-nitrosylation positively regulates ascorbate peroxidase activity during plant stress responses. Plant Physiol. 167, 1604–1615. doi: 10.1104/pp.114.255216
Zeidler, D., Zähringer, U., Gerber, I., Dubery, I., Hartung, T., Bors, W., et al. (2004). Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc. Natl. Acad. Sci. U.S.A. 101, 15811–15816. doi: 10.1073/pnas.0404536101
Zheng, J., Allard, S., Reynolds, S., Millner, P., Arce, G., Blodgett, R. J., et al. (2013). Colonization and Internalization of Salmonella enterica in Tomato Plants. Appl. Environ. Microbiol. 79, 2494–2502. doi: 10.1128/AEM.03704-12
Zheng, Y., Shen, L., Yu, M., Fan, B., Zhao, D., Liu, L., et al. (2011a). Nitric oxide synthase as a postharvest response in pathogen resistance of tomato fruit. Postharvest Biol. Technol. 60, 38–46. doi: 10.1016/j.postharvbio.2010.12.003
Zheng, Y., Sheng, J., Zhao, R., Zhang, J., Lv, S., Liu, L., et al. (2011b). Preharvest l-Arginine treatment induced postharvest disease resistance to Botrysis cinerea in Tomato Fruits. J. Agric. Food Chem. 59, 6543–6549. doi: 10.1021/jf2000053
Keywords: human pathogens on plants, nitrosative stress, oxidative stress, food safety, Salmonella–tomato interaction, nitric oxide, ROS
Citation: Ferelli AMC, Bolten S, Szczesny B and Micallef SA (2020) Salmonella enterica Elicits and Is Restricted by Nitric Oxide and Reactive Oxygen Species on Tomato. Front. Microbiol. 11:391. doi: 10.3389/fmicb.2020.00391
Received: 25 November 2019; Accepted: 25 February 2020;
Published: 13 March 2020.
Edited by:
Max Teplitski, University of Florida, United StatesReviewed by:
Juan Manuel Cevallos-Cevallos, ESPOL Polytechnic University, EcuadorNeha Potnis, Auburn University, United States
Andree George, Western Regional Research Center (USDA-ARS), United States
Copyright © 2020 Ferelli, Bolten, Szczesny and Micallef. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shirley A. Micallef, c21pY2FsbEB1bWQuZWR1
†Present address: Samantha Bolten, Environmental Microbial and Food Safety Laboratory, United States Department of Agriculture – Agricultural Research Service, Beltsville, MD, United States
 Angela Marie C. Ferelli
Angela Marie C. Ferelli Samantha Bolten
Samantha Bolten Brooke Szczesny
Brooke Szczesny Shirley A. Micallef
Shirley A. Micallef