- 1Department of Biochemical Engineering and Biotechnology, Indian Institute of Technology Delhi, New Delhi, India
- 2Agroécologie, AgroSup Dijon, Institut National de la Recherche Agronomique, Université Bourgogne – Franche-Comté, Dijon, France
Due to rhizodeposits and various microbial interactions, the rhizosphere is an extremely dynamic system, which provides a conductive niche not only for bacteria beneficial to plants but also for those that might pose a potential threat to humans. The importance of bioinoculants as biocontrol agents to combat phytopathogens has been widely recognized. However, little information exists with respect to their role in inhibiting human pathogens in the rhizosphere. The present study is an attempt to understand the impact of an established bacterial consortium, Azotobacter chroococcum, Bacillus megaterium, and Pseudomonas fluorescens, on the survivability of Listeria monocytogenes in the rhizosphere of Cajanus cajan and Festuca arundinacea. An experiment conducted in Hoagland’s medium in the presence of C. cajan demonstrated that the presence of bioinoculants impaired growth of L. monocytogenes compared to that observed in their absence. On the other hand, in the presence of F. arundinacea, no significant differences were observed in the population dynamics of L. monocytogenes in the presence or absence of the bioinoculants. Agar plate assay through cross streak method revealed the inhibition of L. monocytogenes by bioinoculants. Potential bioactive compounds were identified by ultra-performance liquid chromatography-mass spectrometry (UPLC-MS). These results suggest that agricultural amendments can act as protective agents against human pathogens while enforcing plant growth promotion.
Introduction
As an extremely dynamic living system, soil contains a rich and diverse microbiota (Vig et al., 2008). The rhizosphere is the area surrounding the roots where processes mediated by microorganisms are influenced by the root system. In this habitat, rhizodeposits and microbial interactions shape structurally and functionally diverse niches (Hinsinger and Marschner, 2006; Pierret et al., 2007; Jones and Hinsinger, 2008; Hinsinger et al., 2009; Raaijmakers et al., 2009). This rich microbial diversity is important for plant’s health, and for the maintenance of soil fertility (Schreiter et al., 2014). It offers a favorable environment not only to beneficial bacteria but also to those that can be a potential threat to humans (Berg et al., 2005; Tyler and Triplett, 2008). These groups include foodborne human pathogens such as Listeria monocytogenes, Salmonella enterica, and Escherichia coli O157:H7 (Mendes et al., 2013).
Listeria monocytogenes is a well-known human pathogen and a microorganism that can survive for long duration in soil and on plant material (Vivant et al., 2013a, b). Its transmission from soil to plants and vegetables is of significance as consumption of raw vegetable produce may be a source of human contamination (Franz et al., 2010; Ajayeoba et al., 2015; Tang et al., 2017; Oyinloye et al., 2018). Death incidences from this food-borne pathogen have been reported all around the world (Bialvaei et al., 2018; Salama et al., 2018). Moreover, the fate of L. monocytogenes in soil depends on the characteristics of the biotic environment (Vivant et al., 2013a, b). Hence, it becomes important to assess the fate of L. monocytogenes during growth of plants inoculated with plant growth promoting consortia.
Bioinoculants are live microorganisms selected to promote plant health and protect against phytopathogens. The use of bioinoculants as biocontrol agents to combat plant pathogens has been studied extensively (Yin et al., 2013; Basu et al., 2017; Igiehon and Babalola, 2017; Liu et al., 2018; Keote et al., 2019). The efficiency of a consortium composed of A. chroococcum A-41, B. megaterium MTCC 453, and P. fluorescens MTCC 9768 (called ABP hereafter) as bioinoculant promoting growth of C. cajan has been established (Sharma et al., 2017). Azotobacter chroococcum is a free-living diazotrophic microorganism. B. megaterium is a key organism for the biocontrol of plant diseases. This species has also been found to solubilize inorganic phosphates by producing organic acids like 2- ketogluconic-, glycolic-, oxalic-, malonic-, and succinic- acid. P. fluorescens strains exhibit biocontrol properties by producing enzymes like chitinase and β-1, 3-glucanase (Kumar et al., 2007), protecting the plant roots against parasitic fungi such as Fusarium or Pythium, as well as some phytophagous nematodes. They also secrete various secondary metabolites, viz. siderophores, indole acetic acid (IAA) and 2, 4-diacetyl phloroglucinol, which aid in plant growth promotion (Kumar et al., 2007). In the present study, it was hypothesized that these agricultural amendments could exert adverse effects on L. monocytogenes, thus preventing invasion and persistence of the pathogen in the rhizosphere. Two model crops were implemented in the present study. Cajanus cajan (pigeonpea, a dicot plant) is an important legume crop and is widely cultivated in Asia, Africa, and the Caribbean Islands. It is an ideal source of food, fodder, and firewood in agroforestry systems. India leads in terms of the production and consumption of C. cajan, with about 90% of global production (Kumar et al., 2014). Owing to the deep tap root system, C. cajan UPAS-120, which is an early maturing variety, enhances soil fertility, prevents soil erosion, and is able to tolerate drought. F. arundinacea (tall fescue, a monocot plant) is an important forage grass throughout its native Europe, as well as a phytoremediation plant. The reason for choosing two different crops was to assess antagonistic activity of bioinoculants under two different soil settings (dicotyledonous versus monocotyledonous plants).
Introduction of large number of bioinoculants in soil, in excess to their natural population, can lead to disturbance and unbalance of resident communities. These effects on resident soil microbial communities other than the target organism are called ‘non-target effects’ (Winding et al., 2004). Such non-target effect of the above mentioned ABP consortium has been demonstrated where the consortium exerted a negative impact on Gram-negative enteric bacteria during cultivation of C. cajan (Sharma et al., 2017), but their role in combating human pathogens dwelling in soil has not been studied in depth. Hence, in the present study, the impact of this agricultural amendment was studied on the survivability of the human pathogen L. monocytogenes, as little information is available regarding non-target antagonistic effects on human pathogenic bacteria (Sheikh, 2010).
The objectives of the study were (a) to assess the impact of bioinoculants on the survivability of L. monocytogenes, and (b) to identify putative bioactive compounds against L. monocytogenes. The study demonstrates antagonistic activities of the bioinoculants against the foodborne pathogen L. monocytogenes highlighting their new potential.
Materials and Methods
Plant Hosts
The model crops for the study were C. cajan (cultivar UPAS-120) and F. arundinacea (cultivar Méandre). Seeds of C. cajan were procured from National Seeds Corporation Ltd. (NSC), New Delhi, India. F. arundinacea is commonly known as tall fescue. It is an important forage grass throughout Europe. The seeds were procured from Carneau, France.
Microbial Strains and Media
The bioinoculants (ABP) used in this study was a consortium composed of A. chroococcum A-41, B. megaterium MTCC 453, and P. fluorescens MTCC 9768. A. chroococcum A-41 was obtained from the Division of Microbiology, Indian Agricultural Research Institute (IARI), New Delhi, India. B. megaterium MTCC 453 and P. fluorescens MTCC 9768 were procured from the Institute of Microbial Technology, Chandigarh, India. L. monocytogenes L9, a rifampicin-resistant derivative of L. monocytogenes EGD-e was used (Lemunier et al., 2005). Glycerol stocks of all the strains were maintained at −80°C in Tryptone Soy Broth (TSB, Conda, Spain). Bacteria were cultured in TSB. Solid medium (tryptone soy agar: TSA) was prepared by adding 15 g/L of agar in TSB.
Generation and Selection of Spontaneous Antibiotic-Resistant Mutants
Antibiotic-resistant mutants were used to avail of specific selective agar media for each of the three bioinoculant species and L. monocytogenes, thereby aiding in the specific enumeration of each bioinoculant species and pathogenic bacterium. Spontaneous antibiotic resistant mutants of bioinoculants were generated by exposure to UV (Miller, 1992). The exposure resulted in the killing of cells and a kill curve was generated. The time of exposure was optimized for 99% killing. Antibiotic-resistant mutants were screened by assessing the ability of the strains to grow on their respective antibiotic supplemented TSA (streptomycin for A. chroococcum, kanamycin for B. megaterium, and ampicillin for P. fluorescens) containing 100 μg/mL antibiotic (Nautiyal, 1996). Comparison of growth of mutants and their parental strains is considered a suitable assay for competitiveness (Nautiyal, 1996; Fischer et al., 2010). Therefore, the mutants of bioinoculants were assessed in terms of growth rate with respect to their parental strains. Bioscreen assays (EL800, BioTek, United States) were performed for 48 h, wherein the growth rate and growth profile were assessed by simultaneous inoculation of all the strains in triplicates in a 96 well plate (OD 600 nm, 27°C). The growth rate was monitored real-time.
In addition to growth rates and profiles, the ecological fitness of different mutants of the three bioinoculants was compared in terms of their plant growth promoting (PGP) properties [production of ammonium, siderophores, IAA, protease, catalase, and phosphate solubilization]. The mutant possessing the maximum closeness to the parental type in terms of both growth rate and PGP properties was selected for further studies.
Assay for NH3 Production
The mutants were tested for the production of ammonia in peptone water containing (per liter) 10 g peptone, 5 g NaCl, 5 g yeast extract, and pH adjusted to 7.6 (Demutskaya and Kalinichenko, 2010). Freshly grown cultures (OD = 0.1) were inoculated in 10 mL peptone water in each tube and incubated for 48 h at 28°C ± 2°C. After incubation, the broth was centrifuged. Nessler’s reagent (0.5 mL) was added to the supernatant obtained after centrifugation of the broth. The development of brown to yellow color was considered a positive test for ammonia production.
Production of Siderophores
The isolates (OD = 1) were inoculated separately (1% v/v inoculum) in sterile succinate medium (SM) containing (per liter) 6 g K2HPO4, 3 g KH2PO4, 0.2 g MgSO4.7H2O, 1 g (NH4)2SO4, 4 g succinic acid, pH adjusted to 7.0 (Meyer and Abdallah, 1978) and incubated on a rotary shaker (Eppendorf, 5804R, United States) at 120 rpm and 28°C ± 2°C. Supernatants of 24–36 h grown cultures were tested for siderophore production (Schwyn and Neilands, 1987).
IAA Production
Indole acetic acid production was carried out in Luria broth supplemented with a filter-sterilized solution of 10 g/L L-tryptophan. The liquid medium was inoculated with 0.1 mL bacterial cultures adjusted to an optical density of 0.5 measured at 660 nm by a spectrophotometer. Inoculated media were incubated at 28°C ± 2°C for 48 h. After incubation, the cells were separated from the culture broth by centrifugation at 5000 rpm for 15 min. For the estimation of IAA, the supernatant (2 mL) was mixed with two drops of ortho-phosphoric acid and 4 mL of Salkowaski’s reagent (50 mL, 35% perchloric acid, 1 mL 0.5 N FeCl3 solution). The development of pink color indicated IAA production (Brick et al., 1991).
Production of Protease and Catalase
The isolates were checked for the production of protease (Sokol et al., 1979) and catalase (MacFaddin, 2000). Protease production was determined using a skim milk agar medium, which contained (per liter) 5 g pancreatic digest of casein, 2.5 g yeast extract, 1 g glucose, 7% skim milk solution, and 15 g of agar. An appropriate dilution of a mid-log-phase broth culture was spread on the plate. After 24 h of incubation, the development of a halo zone surrounding the colony was considered a positive result. Catalase producing isolates were screened by the addition of 6% H2O2 (v/v) on the colonies grown on Luria Agar plates and by observing the formation of bubbles (evolution of oxygen).
Phosphate Solubilization
To detect the ability of bacteria to solubilize phosphate, mutants were spot inoculated onto Pikovskaya’s agar containing (per liter) 2.5 g tricalcium phosphate, 13 g glucose, 0.5 g (NH4)SO4, 0.2 g NaCl, 0.1 g MgSO4.7H2O, 0.2 g KCl, 0.5 g yeast extract, trace MnSO4, trace FeSO4.7H2O, 15 g agar and pH adjusted to 7.2. Plates were incubated for 3 days at 28°C ± 2°C. The strains demonstrating a halo zone around the colonies were considered positive (Nautiyal, 1999).
In vitro Plant Growth Experiment on Hoagland Mineral Agar Upon Inoculation With Bioinoculants and/or L. monocytogenes
A modified Hoagland medium [500 μM KH2PO4, 5 mM KNO3, 1 mM MgSO4, 2.5 mM Ca(NO3)2, 50 μM H3BO3, 50 nM CoCl2, 50 nM CuSO4, 15 μM ZnSO4, 2.5 μM KI, 50 μM MnSO4, 3 μM Na2Mo4 2H2O, 50 μM Fe EDTA] was used for growth experiment with F. arundinacea and C. cajan in vitro (Arnon and Hoagland, 1940). When necessary, Vitro agar (Conda Laboratories, Torrejón de Ardoz Madrid, Spain) 8 g/L was added. The seeds of C. cajan and F. arundinacea were rinsed with water and then disinfected by keeping them in a solution of sodium hypochlorite (2.6%) for 60 min at 53°C. The seeds were then washed eight times with sterile water. Disinfected seeds were placed onto sterile soft agar and incubated at 25°C for germination. After germination, the seedlings were transferred onto sterile Hoagland plates (five seedlings sown at a regular distance per plate) and further incubated for 5 days in a growth chamber (18°C, 8 h night, 24°C, 16 h day).
Bacterial inocula were prepared by incubating cultures in TSB at 30°C for 24 h. Five mL of culture of L. monocytogenes L9 or bioinoculants were collected by centrifugation (5 min, 10,000 × g) and washed two times in 5 mL of Hoagland medium. The pellet was suspended in 10 mL of Hoagland medium. After adequate dilution in Hoagland medium, root systems were inoculated with 200 μL/plant of the bacterial suspension (2 × 106 cfu/root). Three different treatments were used; T1: L. monocytogenes alone, T2: co-inoculation of the ABP bioinoculant (consortium of A. chroococcum, B. megaterium, and P. fluorescens) with L. monocytogenes (LM + ABP), and T3: bioinoculant ABP alone. After the sampling at the start of the experiment (time point 0) and on 1st day, sampling was done every alternate day for 15 days. Three Hoagland plates were taken per treatment per sampling point via destructive sampling (n = 15). At each sampling, the plant roots were cut from the shoot and the enumeration was assessed on the roots.
Cross Streak Assay of Bioinoculants With L. monocytogenes
To assess the inhibition of L. monocytogenes by the bioinoculants, cross streak assay was performed (Montano and Henderson, 2013). This assay was done on TSA plates with three different strengths (full strength TSA, 1/10th TSA, and 1/100th TSA). The three bioinoculant strains, and L. monocytogenes (all strains in their log phase with 1 × 105 cfu/mL) were streaked onto the plates and incubated for 24 h at 30°C, after which observations were made at the intersects of the streaks.
Enumeration of Bioinoculants and L. monocytogenes
For the enumeration of bioinoculant species and L. monocytogenes, TSA medium supplemented with respective antibiotics at the concentration of 100 μg/mL was used. Streptomycin, kanamycin, ampicillin, and rifampicin supplemented TSA was used for the enumeration of A. chroococcum, B. megaterium, P. fluorescens, and L. monocytogenes, respectively. Sampling was performed by uprooting the plants with sterile forceps. Five plants were taken per replicate and three biological replicates were used. The roots of five plants were then cut from their respective shoots and placed in 50 mL sterile plastic tubes along with 0.7 g sterile glass beads (<106 μm, acid-washed glass beads, Sigma) and 10 mL of tryptone salt solution (1 g tryptone, 8.5 g NaCl per liter). It was then vortexed for 2 min at maximum speed before serial dilution and spot inoculation on respective selective media. The plates were then incubated for 24 h at 30°C before enumeration. Sterile conditions were maintained throughout the experiment.
Detection of Potential Inhibitors of the Pathogen
To assess the production of molecules responsible for the inhibition of L. monocytogenes, an agar plate diffusion assay was developed (Bhattacharjee et al., 2006). To accomplish this, supernatants of cultures (A. chroococcum; B. megaterium; P. fluorescens; A. chroococcum + L. monocytogenes; B. megaterium + L. monocytogenes, and P. fluorescens + L. monocytogenes) inoculated with similar initial numbers (1.0 × 105 cfu of each bacteria per mL of TSB) and grown overnight at 30°C were extracted in several solvents with different polarities. The solvents used for the extractions were hexane (polarity- 0.1), petroleum ether (polarity- 0.1), ethyl ether (polarity- 2.8), chloroform (polarity- 4.1), ethyl acetate (polarity- 4.4), and acetone (polarity- 5.1) (Jangir et al., 2018). As a negative control, the uninoculated medium was extracted with dimethyl sulfoxide (DMSO) and all the above-mentioned solvents. Uninoculated medium supplemented with kanamycin and ampicillin served as positive controls. L. monocytogenes (1.0 × 106 cfu/mL of TSA) was inoculated in molten TSA before pouring the agar on plates (pour plate method). After solidification of the plates, wells were made in the plates, and the supernatants previously extracted from different cultures were poured into the wells. The plates were incubated overnight at 30°C and then checked for inhibition zones. The next step was to run TLC plates for direct bioautography assay. To accomplish this, it was necessary to select the mobile phase that could resolve the extracts obtained from chloroform to generate maximum number of bands.
Once the mobile phase was selected, fresh TLC plate with chloroform-extracted samples was run again for direct bioautography assay. TLC was run for 40 min, followed by band visualization under UV light. TLC plates were then subjected to direct bioautography test (Fisher and Lautner, 1961; Nicolaus et al., 1961) to check which fraction of the extract resulted in inhibition. The test is a colorimetric assay and is used to assess metabolically active cells. It is based on the principle of conversion of reducing tetrazolium salt of 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) dye to its insoluble form of formazan (purple color) by live cells. TLC plates were dipped for 10 s into the culture broth containing L. monocytogenes at a very early log phase (1 × 104 cfu/mL). After 10 min at room temperature, the plates were then sprayed with the aqueous solution of MTT dye (2.5 mg/mL). The TLC plates were then incubated inside the laminar hood for 4-5 h. Bands of interest exhibiting inhibition zones were extracted in DMSO for further analysis by UPLC-MS on Waters Acquity UPLC system (Waters Corp., Milford, MA, United States) and Applied Biosystems AP14000 tandem quadrupole mass spectrometer (Foster City, CA, United States) coordinated by Analyst software. The column used was Acquity UPLC® BEH C18 (1.7 μ, Waters Corp., United States). The mobile phase consisted of 0.1% formic acid (solvent A) and acetonitrile (solvent B). Bioactive components were then identified using the metabolite search tool of METLIN1 metabolomics database. Ten ppm was set as the tolerance on the mass of precursor ion. The structures of the bioactive compounds were procured from PubChem2.
Statistical Analysis
All experiments were performed at least in triplicate. The standard deviation was calculated for each treatment. Analysis of variance (ANOVA) was performed on the data using SPSS Statistics 16.0 for Windows® (SPSS Inc., Chicago, IL, United States). Two-way ANOVA was performed with treatments and time points as independent variables and cfu as the dependent variable. Tukey’s HSD post hoc test was employed to compare means. Significantly different values (p < 0.05) between different sampling time points and between different treatments for the same time point are marked by lower case letters.
Results
Generation of Antibiotic-Resistant Mutants
Antibiotic-resistant mutants were generated for specific tracking of the three bioinoculant bacterial species (A. chroococcum, B. megaterium, and P. fluorescens). Streptomycin-resistant mutant M3 of A. chroococcum (as all mutants in this case exhibited growth profile similar to that of the respective parental strain, M3 was selected on the basis of its maximum closeness to the parental strain with respect to amount of IAA production), kanamycin-resistant mutant M2 of B. megaterium, and ampicillin-resistant mutant M7 of P. fluorescens were selected. These mutants possessed the maximum closeness to their parental type in terms of growth rate (generation time of A. chroococcum = 1.3 h, B. megaterium = 1.5 h, and P. fluorescens = 51 min) and PGP properties, such as phosphate solubilization and production of IAA, protease, catalase, siderophores, and ammonia (Supplementary Table S1).
Impact of Bionoculants on the Population Dynamics of Root-Associated L. monocytogenes in C. cajan and F. arundinacea Plant Models Grown in vitro in Hoagland’s Medium
The experiment was carried out in vitro in Hoagland’s medium to assess the effect of bioinoculants on the dynamics of the population of L. monocytogenes and vice-versa in the presence of C. cajan and F. arundinacea.
In C. cajan plant model, in the absence of bioinoculants, the population of L. monocytogenes increased over time on the roots during the 1st week of the experiment (Figure 1A). During the 2nd week, the population of L. monocytogenes on the roots was in the range of 1.6 × 107–4.4 × 107 cfu per root. When the roots were colonized by ABP, no growth was observed. The population of L. monocytogenes was significantly lower than in the control experiment after 1 week of incubation and until the end of the experiment (Figure 1A). The population was one to two log lower than in the control experiment (8.0 × 105–3.3 × 106 cfu per root) during the 2nd week of the experiment. Each bioinoculant species maintained its numbers in the order of 105–108 cfu per root over a period of 2 weeks (Figures 1B–D). L. monocytogenes did not significantly affect the growth of A. chroococcum throughout the experiment resulting in a population of A. chroococcum in the range of 1.1 × 107–3.6 × 108 cfu per root during co-cultivation with L. monocytogenes (Figure 1B). The population of B. megaterium on the roots was found to be in the range of 1.1 × 107–8.5 × 108 cfu per root in the presence of L. monocytogenes and 6.3 × 107–7.5 × 108 cfu per root in the absence of L. monocytogenes. At the start of the experiment, there was an increase in the number of B. megaterium in the treatment containing L. monocytogenes and consortium (Figure 1C). However, towards the end of the experiment, the population of B. megaterium reduced slightly, and thereafter maintained an almost steady-state (1.6 × 107–6.2 × 107 cfu per root). In the case of P. fluorescens, lower abundance was observed in the presence of L. monocytogenes as the experiment progressed (Figure 1D). During the 2nd week of the experiment, the population of P. fluorescens was in the range of 1.0 × 106–1.5 × 106 cfu per root in both treatments.
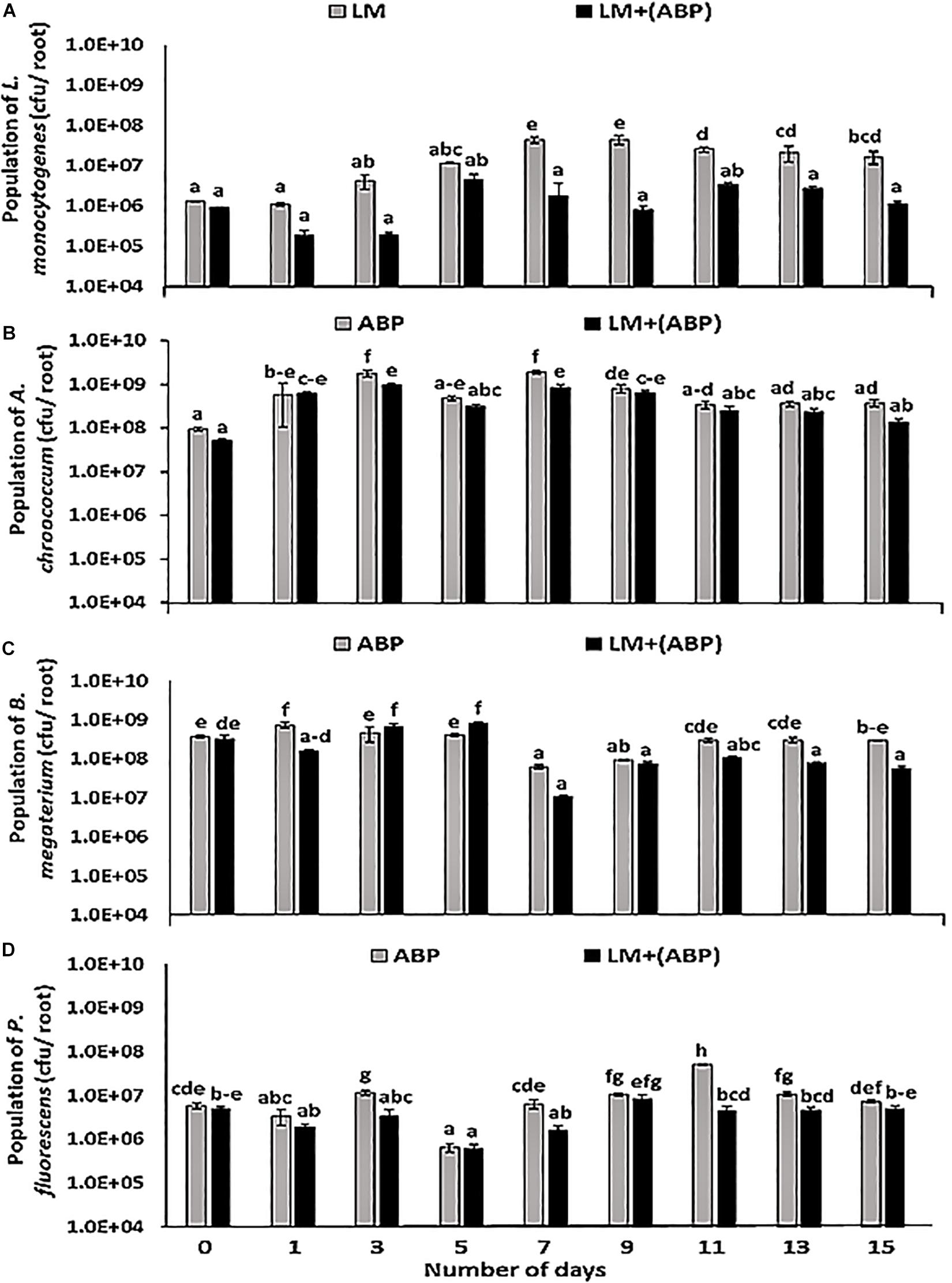
Figure 1. Population dynamics of (A) Listeria monocytogenes, (B) Azotobacter chroococcum, (C) Bacillus megaterium, and (D) Pseudomonas fluorescens during incubation in the Cajanus cajan plant model for 15 days, assayed by plating samples on TSA containing rifampicin, streptomycin, kanamycin, and ampicillin, respectively. Significantly different values (p < 0.05) between different treatments and time points are marked by lower case letters. Standard deviation (n = 3) is represented by error bars.
In F. arundinacea plant model, the population of L. monocytogenes increased with time in control experiments and on plants inoculated with ABP. No significant differences were observed between the two treatments (Figure 2A). In the absence of bioinoculants, the population of L. monocytogenes on the roots was in the range of 7.6 × 105–4.4 × 107 cfu per root, compared with 7.0 × 105–3.8 × 107 cfu per root in the presence of ABP (Figure 2A). Each of the bioinoculant maintained its numbers in the order of 105–108 cfu per root throughout the experiment. In the case of A. chroococcum, L. monocytogenes did not significantly affect its population. Initially, the population of A. chroococcum was in the range of 5.4 × 108 cfu per root in both the treatments, after which a dip (66.7%) in the population of A. chroococcum was observed. Thereafter, the population remained stable in the range of 1.0 × 107–2.2 × 108 cfu per root (Figure 2B). No significant differences were observed in the population of B. megaterium in the presence and the absence of L. monocytogenes throughout the experiment, resulting in a population of B. megaterium in the range of 3.2 × 106–7.8 × 108 cfu per root (Figure 2C). In the case of P. fluorescens, the presence of L. monocytogenes resulted in a significant reduction in the numbers of P. fluorescens towards the end of the 1st week of the experiment (Figure 2D). However, during the 2nd week of the experiment, the population of P. fluorescens was significantly higher in the treatment containing L. monocytogenes, in contrast with the treatment without L. monocytogenes where the population was in the range of 1.0 × 107–1.5 × 107 cfu per root.
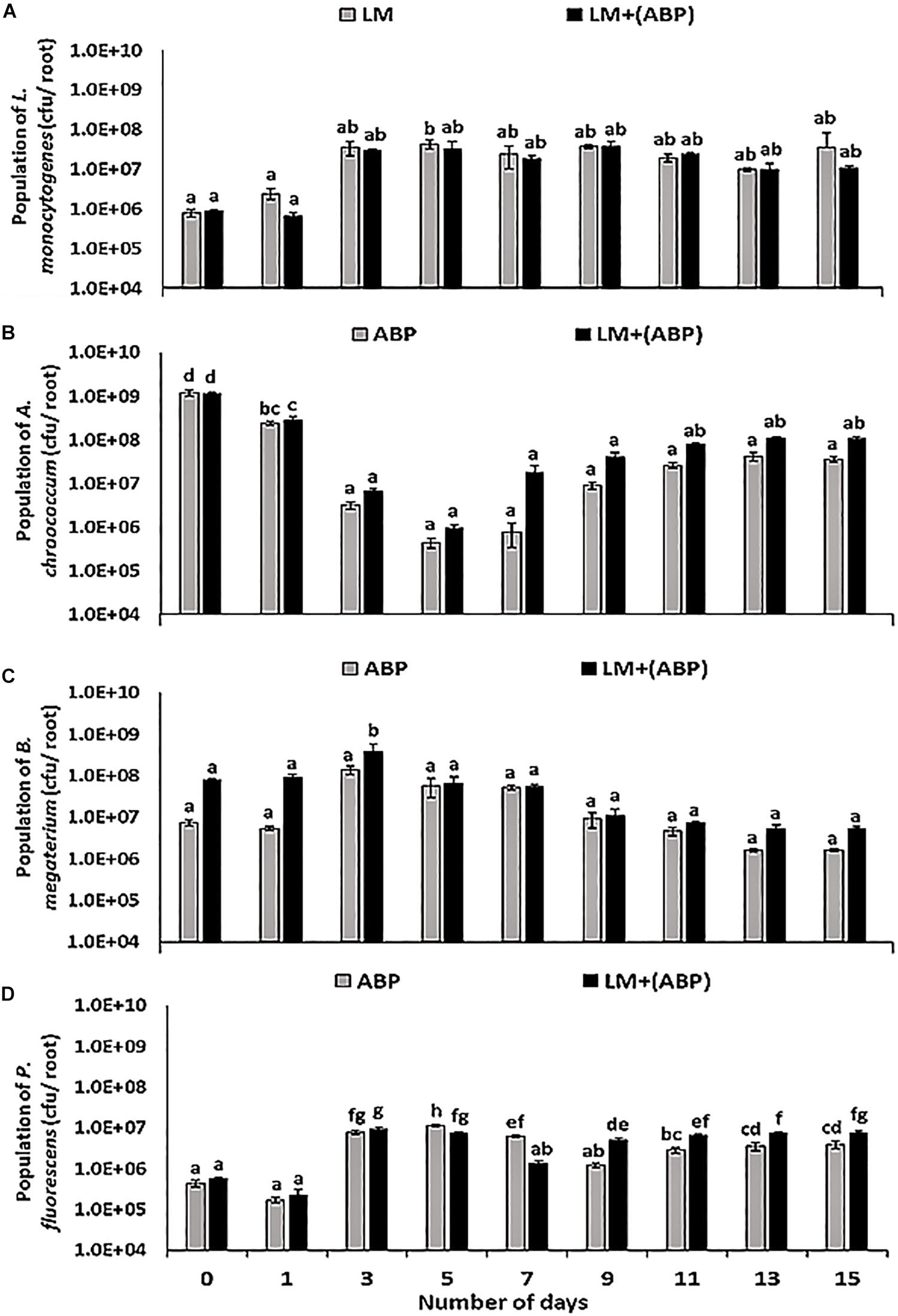
Figure 2. Population dynamics of (A) L. monocytogenes, (B) A. chroococcum, (C) B. megaterium, and (D) P. fluorescens during incubation in the F. arundinacea plant model for 15 days, assayed by plating samples on TSA containing rifampicin, streptomycin, kanamycin and ampicillin, respectively. Significantly different values (p < 0.05) between different treatments and time points are marked by lower case letters. Standard deviation (n = 3) is represented by error bars.
Evidence of Antagonistic Activity and Detection of Bioactive Component(s)
After observing a significant impact of the bioinoculants on the reduction of L. monocytogenes populations on the roots of C. cajan grown in Hoagland’s medium, the next step was to investigate possible antagonistic activities of A. chroococcum, B. megaterium, and P. fluorescens linked to the production of inhibitory compounds. Initially cross streak assay was performed on solid medium (TSA plates). It was observed that the growth of L. monocytogenes was inhibited by A. chroococcum on 1/100th TSA and B. megaterium on both 1/10th TSA and 1/100th TSA (Supplementary Figure S1). No zone of inhibition could be observed with P. fluorescens.
To further investigate production of antagonistic molecules, an inhibition assay was set up after extraction of supernatants in a range of solvents. No inhibition zone was observed with the negative controls, whereas prominent zones of inhibition were observed with uninoculated medium supplemented with kanamycin and ampicillin used as positive controls (Figure 3A). Inhibition zones were observed in chloroform extracts of all bioinoculant species monocultures and co-cultures with L. monocytogenes (Figure 3B).

Figure 3. Evidence of inhibitory activity of bioinoculant species’ supernatant extracts in an agar plate diffusion assay. (A) negative controls (uninoculated medium extracted with DMSO, chloroform and ethyl acetate) and positive controls (kanamycin and ampicillin); (B) chloroform extracts from culture of A. chroococcum alone (A), co-culture of A. chroococcum and L. monocytogenes (A2), culture of B. megaterium alone (B), co-culture of B. megaterium and L. monocytogenes (B2), culture of P. fluorescens alone (P), and co-culture of P. fluorescens and L. monocytogenes (P2).
The presence of an inhibition zone in all the samples extracted with chloroform led to the selection of chloroform as extracting solvent for further processing. The next step was to select a suitable mobile phase to run TLC (based on maximum resolution with respect to number of bands generated). Among all solvents tested, chloroform resulted in the highest number of bands as observed on the chromatogram under UV light (Supplementary Figure S2), hence, it was selected as a mobile phase for direct bioautography test. A final TLC plate was run taking supernatant extracted in chloroform as samples and chloroform as mobile phase (Figure 4A).
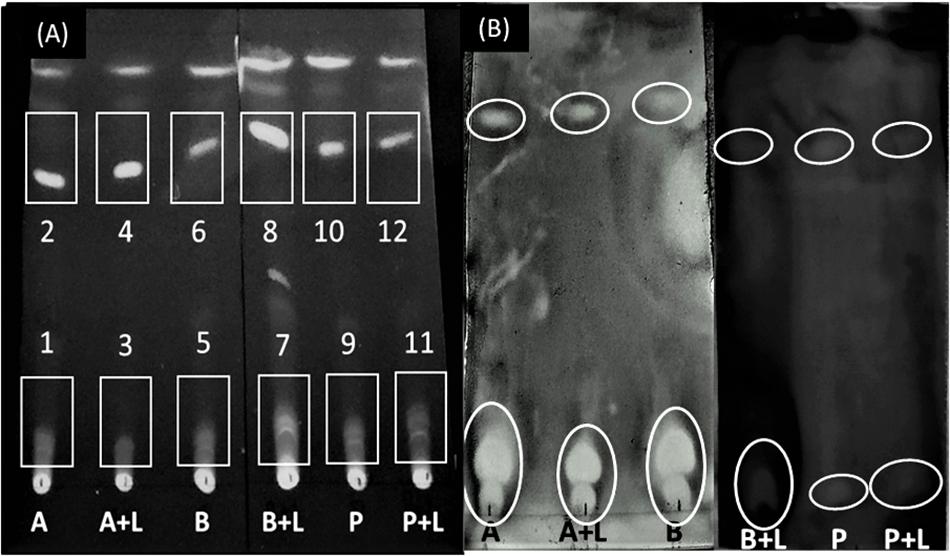
Figure 4. Separation of bioactive compounds against L. monocytogenes. (A) Thin Layer Chromatogram of chloroform-extracted samples using chloroform as a mobile phase. The numbers identify the spots where inhibition of L. monocytogenes was observed. (B) Direct bioautography test using 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide (MTT) dye. A, A. chroococcum; B, B. megaterium; P, P. fluorescens; L, L. monocytogenes. Inhibition zones have been encircled.
Direct bioautography was performed to confirm the bands showing inhibition against L. monocytogenes. Inhibition zones were evidenced with all ABP samples (Figure 4B). Bands of interest were extracted in DMSO and were subjected to UP-LCMS for further analysis. Specific peaks were observed in the chromatograms (Supplementary Figure S3) suggesting A, B, and P produced different bioactive compounds. With the help of the METLIN database, these compounds were identified (Table 1). None of the compounds identified in co-cultures were detected in supernatants of pure cultures of L. monocytogenes (data not shown). Moreover, no inhibition of A. chroococcum, B. megaterium, and P. fluorescens could be observed by L. monocytogenes L9 supernatants under our experimental conditions (data not shown).
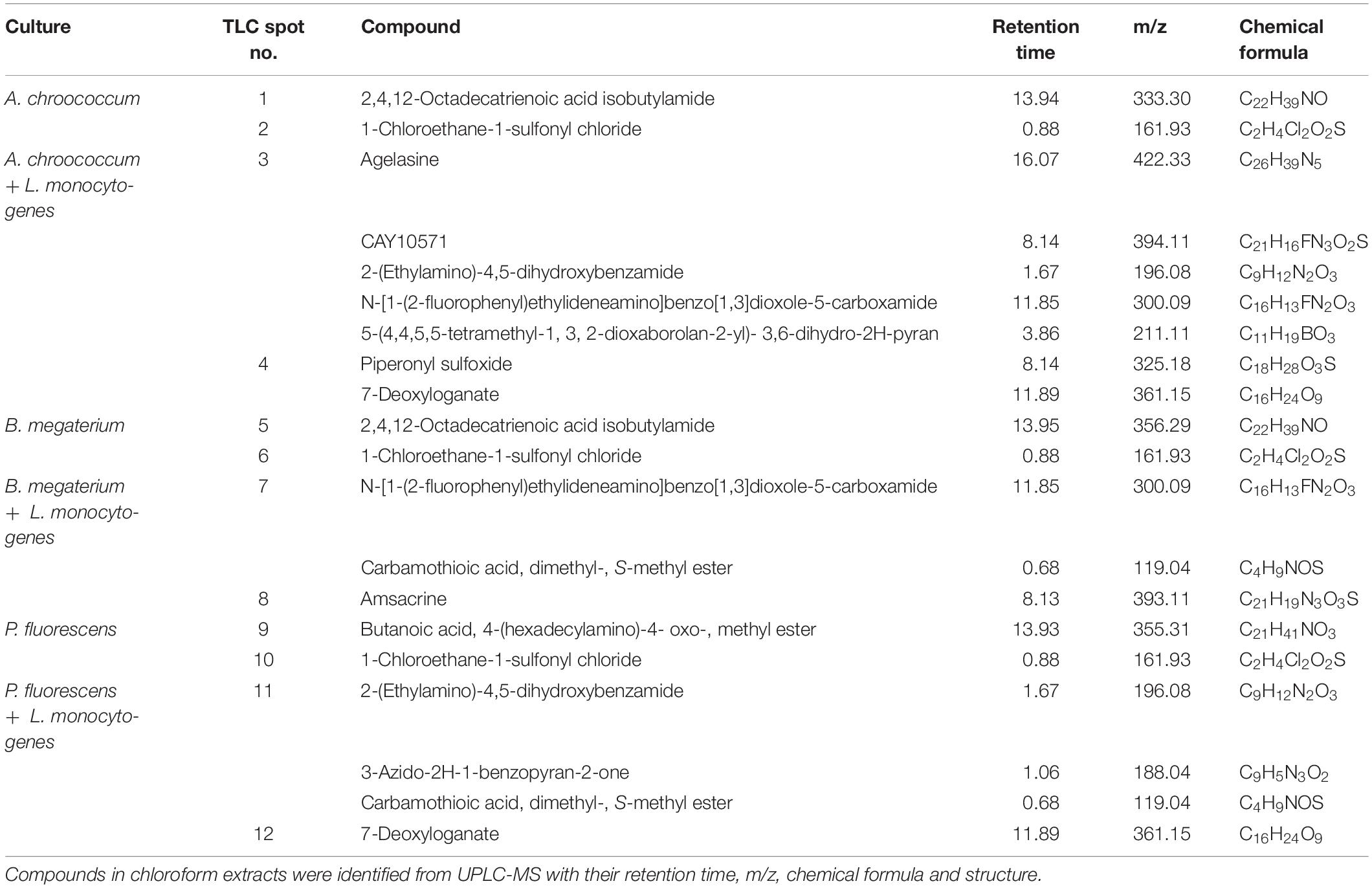
Table 1. Major non-volatile compounds released by Azotobacter chroococcum, Bacillus megaterium, and Pseudomonas fluorescens in pure cultures, and during co-cultivation with Listeria monocytogenes.
Discussion
Listeria monocytogenes is a ubiquitous human foodborne pathogen dwelling in many habitats such as soil, decaying vegetation, water, sediments, wastewater treatment plants, food processing environments and foodstuffs (McLauchlin, 1987; Ly and Müller, 1990; Aureli et al., 1992). It causes listeriosis, a food-borne infection that affects children, immune-compromised individuals, and pregnant women (Cossart and Toledo-Arana, 2008). In case of ruminants, systemic listeriosis occurs by oral ingestion of L. monocytogenes (Lecuit, 2007). Death incidences caused by this pathogen have been reported all over the world (Centers for Disease Control and Prevention- United States, 2013 EFSA Panel on Biological Hazards (BIOHAZ) et al., 2018) (Bialvaei et al., 2018; Salama et al., 2018). The use of bioinoculants to combat plant pathogens has been studied extensively (Yin et al., 2013; Basu et al., 2017; Igiehon and Babalola, 2017; Liu et al., 2018; Keote et al., 2019), but their role in combating human pathogens dwelling in the soil is still in its infancy.
A bacterial consortium consisting of A. chroococcum A-41, B. megaterium MTCC 453, and P. fluorescens MTCC 9768 was shown to exert a negative impact on Gram-negative enteric bacteria in our previous field study (Sharma et al., 2017). The present study was conducted to assess their impact specifically on the fate of L. monocytogenes during root colonization of C. cajan and F. arundinacea. The bioinoculants were previously designed to protect C. cajan against phytopathogens. The reason for choosing C. cajan was establishment of non-target effects of the bioinoculants in the rhizosphere of C. cajan in our previous study (Sharma et al., 2017). F. arundinacea was chosen because it is widely used in pastures as a grazing grass for ruminants. L. monocytogenes, is a zoonotic agent that causes lethal infections and mastitis in ruminants (Nightingale et al., 2004; Papić et al., 2020). Moreover, incidence of L. monocytogenes is higher in grassland than in other outdoor settings (Wang et al., 2017). The rhizosphere of this type of plant may be a preferential habitat, and it may be important in the routes of transfer of L. monocytogenes. In addition to this, we wanted to examine the response of a dicot as well as a monocot plant on overall study.
An experiment performed in Hoagland’s medium with C. cajan showed a significant reduction (93%) in the population of L. monocytogenes when roots were inoculated with ABP consortium, suggesting strong effect of the bioinoculants leading to impaired growth of L. monocytogenes. It was previously demonstrated that the biotic environment and especially microbial diversity is the main driver of the inhibition of L. monocytogenes in complex habitats (Vivant et al., 2013a). However, when experiments were performed in the F. arundinacea model, the population of L. monocytogenes was not affected by the presence of the bioinoculant species. As one plant is monocot and the other plant is dicot, it can be hypothesized that environmental conditions surrounding the roots of the two plant models are different. For example, the quantity and composition of root exudates may differ according to the plant model, resulting in differences in the physiology and fitness of the bioinoculants and/or of L. monocytogenes. Study conducted by Yishay et al. (2008) showed that the plant pathogen Pectobacterium carotovorum ssp. carotovorum originated from monocots exhibited higher virulence towards its monocot host, as compared to isolates from dicot plant. Another study conducted by Liu et al. (2013) suggested differences in microbial diversity when comparing the rhizospheres of monocot species with dicot species. Higher fungal diversity was found in the rhizosphere of monocot plants, which could be due to enhanced below-ground plant biomass. In case of rhizospheres of dicot species, there was abundance of both Gram-negative and Gram-positive bacteria. This could be attributed to the fact that deeper roots of dicot plants resulted in higher number of attachment sites for bacteria to flourish. Indeed, the populations of A. chroococcum and B. megaterium were lower in the experiments conducted in the F. arundinacea model compared to the C. cajan model. Many mechanisms underlie dynamics of complex microbial communities where niche exclusion, metabolic competition for resources and antibiosis can shape interactions between coexisting microbial species (Ghoul and Mitri, 2016). In our model system, several mechanisms could affect growth of L. monocytogenes. In order to investigate antibiosis and inhibitory effects of bioinoculants on L. monocytogenes, cross streak assay was performed in vitro on plates containing TSA of different strengths. Inhibition could be observed, and the extent of this inhibition was dependent on nutrient concentration. It confirmed that the impact of ABP on the populations of L. monocytogenes varied according to the conditions of its environment. The reason for visible inhibition at 1/10th and 1/100th strength of TSA may be attributed to competitive exclusion, i.e., increased competition for the available nutrients in the medium (Moran et al., 2016).
In a study conducted by Buchanan and Bagi (1999), it was observed that P. fluorescens was able to inhibit L. monocytogenes, and this inhibition was dependent on temperature, pH and sodium chloride levels in brain-heart infusion broth. To the best of our knowledge, this is the only report mentioning inhibition of L. monocytogenes by P. fluorescens. On the other hand, no reports are available with respect to inhibition of L. monocytogenes by either A. chroococcum or B. megaterium.
It was previously found that supercritical fluid extraction (SFE-CO2) extracts from C. cajan were effective in the inhibition of certain human pathogens (Zu et al., 2010). Apart from displaying various plant growth promoting properties, antagonistic activities of Azotobacter, Bacillus, and Pseudomonas species against phytopathogens have been documented previously (Xu et al., 2014; Mora et al., 2015; Munjal et al., 2016; Viscardi et al., 2016; Fernandez et al., 2017; Hennessy et al., 2017). Tailocins, pyrazine and siderophores were among these inhibitory compounds. So far, there has not been any report wherein the inhibitory activities of these classes of molecules have been found against L. monocytogenes. This prompted us to investigate whether or not inhibitory molecules could be produced by the bioinoculant species during incubation with L. monocytogenes in our plant model. In the present study, chloroform extraction of supernatants of each bioinoculant species resulted in the separation and identification of several compounds. Interestingly, major differences were observed between pure and co-cultures. This might suggest that the presence of L. monocytogenes could trigger release of specific compounds. Alternatively, some of these compounds could be produced by L. monocytogenes in the co-cultures.
Some of the compounds identified are known biologically active molecules. Agelasines are known to possess antimicrobial and cytotoxic properties (Nakamura et al., 1984; Vik et al., 2006). Biological activity of several compounds containing a piperonyl ring has been reported against viruses (Day et al., 2002), amoebae (Wani et al., 2012), cancerous cells (de Olivetra et al., 2010; Mathew et al., 2011), and they possess anti-proliferative and anticonvulsant properties (Prasanthi et al., 2013). Another study demonstrated high antibacterial activities of piperonyl triazoles against Bacillus subtilis, E. coli, Klebsiella pneumoniae, and Micrococcus luteus (Srinivas and Sunitha, 2016). 7-Deoxyloganate is a terpenoid and this class is known to possess antimicrobial/antibacterial activities (Knobloch et al., 1989; Dorman and Deans, 2000; Tada et al., 2002). According to their chain length, fatty acids develop antibacterial activities. It was shown that Mycobacterium bovis and M. tuberculosis had maximum susceptibility to tetradecanoic acid, linolenic (cis, cis, cis-9, 12, 15-octadecatrienoic acid) and arachidonic acids (Kondo and Kanai, 1977). Antimicrobial activity of medium and long chain free fatty acids against L. monocytogenes has been reported (Lillebaek et al., 2017). Amsacrine is a small molecule that inhibits DltB (D-alanylation of lipoteichoic acids) (Neuhaus and Baddiley, 2003). This membrane-embedded enzyme is needed for D-alanylation of lipoteichoic acids. This modification has been found to be responsible for virulence in Staphylococcus aureus (Pasquina et al., 2016). Several compounds containing pyran rings have been tested for their microbiocide properties (Rajan et al., 2017) and derivatives of 2H-pyran (thiourea and benzene sulfonylurea) possess antimicrobial activities (Faidallah et al., 2011). Carboxamides are not only being used as site-specific fungicides (Jangir et al., 2018) but have also been shown to possess antibacterial properties (Jeong and Moloney, 2013). Hence, it suggests that at least part of the inhibitory effects caused by these bioinoculants could be attributed to antibiosis triggered by the release of bioactive compounds during root colonization.
The present study focused on only one strain of L. monocytogenes under in vitro conditions. Assessing the susceptibility of various strains of L. monocytogenes to these bioinoculants, in more complex environments such as soil, forms the future perspective of the work. Additionally, employing the purified form of the identified compounds for inhibition assay is promising for future study.
Conclusion
Inhibition of L. monocytogenes was observed during co-culture with bioinoculants, but this effect depended on the conditions of the environment. It was observed in C. cajan plant model but not in F. arundinacea model. Similarly, inhibition could be observed in complex medium but only under limiting nutrient availability. Antagonistic activity was recorded and bioactive compounds could be separated by chromatography. The results of this study demonstrate that bioinoculation of plants can contribute to the control of human pathogens and document positive non-target effects of bioinoculants. As L. monocytogenes is the causative agent of listeriosis, a life-threatening condition in at-risk people (adults aged 65 or older, pregnant women and their newborns and people with weakened immune systems), its survival in soil, and especially on vegetables, generates health hazards. The inhibition of this human pathogen by the selected bioinoculants introduces an entirely novel aspect of the potential implementation of this agricultural amendment for the benefit of not only plant growth, but also human health.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
PP and SS contributed to conception and design of the study. RS, LG, and DG performed experiments. RS performed statistical analyses. RS, PP, SS, LG, and DG wrote the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
Funding
This work was supported by the Department of Biotechnology, Government of India (Grant No. BT/PR5499/AGR/21/355/2012). RS thanks Council of Scientific and Industrial Research, India, for providing Fellowship, and Raman-Charpak Student Exchange Program between India and France for specifically supporting this work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors sincerely thank Dr. Lucinda Elizabeth Doyle for diligent language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00350/full#supplementary-material
Footnotes
References
Ajayeoba, T. A., Atanga, O. O., Obadina, A. O., Bankole, M. O., and Adelowo, O. O. (2015). The incidence and distribution of Listeria monocytogenes in ready-to-eat vegetables in South-Western Nigeria. Food Sci. Nutr. 4, 59–66. doi: 10.1002/fsn3.263
Arnon, D. I., and Hoagland, D. R. (1940). Crop production in artificial culture solutions and in soil with reference to factors influencing yield and absorption of inorganic nutrients. Soil Sci. 50, 363–384.
Aureli, P., Costantini, A., and Zolea, S. (1992). Antimicrobial activity of some plant essential oils against Listeria monocytogenes. J. Food Prot. 55, 344–348. doi: 10.4315/0362-028x-55.5.344
Basu, S., Rabara, R., and Negi, S. (2017). Towards a better greener future – an alternative strategy using biofertilizers. I: plant growth promoting bacteria. Plant Gene 12, 43–49. doi: 10.1016/j.plgene.2017.07.004
Berg, G., Zachow, C., Lottmann, J., Götz, M., Costa, R., and Smalla, K. (2005). Impact of plant species and site on rhizosphere-associated fungi antagonistic to Verticillium dahlia kleb. Appl. Environ. Microbiol. 71, 4203–4213. doi: 10.1128/aem.71.8.4203-4213.2005
Bhattacharjee, I., Chatterjee, S. K., Chatterjee, S., and Chandra, G. (2006). Antibacterial potentiality of Argemone mexicana solvent extracts against some pathogenic bacteria. Mem. Inst. Oswaldo Cruz 101, 645–648. doi: 10.1590/s0074-02762006000600011
Bialvaei, A. Z., Sheikhalizadeh, V., Mojtahedi, A., and Irajian, G. (2018). Epidemiological burden of Listeria monocytogenes in Iran. Iran J. Basic Med. Sci. 21, 770–780.
Brick, J. M., Bostock, R. M., and Silversone, S. E. (1991). Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microbiol. 57, 535–538. doi: 10.1128/aem.57.2.535-538.1991
Buchanan, R. L., and Bagi, L. K. (1999). Microbial competition: effect of Pseudomonas fluorescens on the growth of Listeria monocytogenes. Food Microbiol. 16, 523–529. doi: 10.1006/fmic.1998.0264
Cossart, P., and Toledo-Arana, A. (2008). Listeria monocytogenes, a unique model in infection biology: an overview. Microbes Infect. 10, 1041–1050. doi: 10.1016/j.micinf.2008.07.043
Day, S. H., Lin, Y. C., Tsai, M. L., Tsao, L., Ko, H., Chung, M., et al. (2002). Potent cytotoxic lignans from Justicia procumbens and their effects on nitric oxide and tumor necrosis factor-α production in mouse macrophages. J. Nat. Prod. 65, 379–381. doi: 10.1021/np0101651
de Olivetra, A. N., Bocca, C. C., Carvalho, J. E., Ruiz, A. L., Silva, T. P., Rittner, R., et al. (2010). New substituted 4-arylaminoquinazolines as potent inhibitors of breast tumor cell lines: in vitro and docking experiments. Eur. J. Med. Chem. 45, 4339–4342. doi: 10.1016/j.ejmech.2010.04.034
Demutskaya, L. N., and Kalinichenko, I. E. (2010). Photometric determination of ammonium nitrogen with the Nessler reagent in drinking water after its chlorination. J. Water Chem. Technol. 32, 90–94. doi: 10.3103/s1063455x10020049
Dorman, H. J., and Deans, S. G. (2000). Antimicrobial agents from plants: antibacterial activity of plant volatile oils. J. Appl. Microbiol. 88, 308–316. doi: 10.1046/j.1365-2672.2000.00969.x
EFSA Panel on Biological Hazards (BIOHAZ), Ricci, A., Allende, A., Bolton, D., Chemaly, M., Davies, R., et al. (2018). Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 16:e05134. doi: 10.2903/j.efsa.2018.5134
Faidallah, H. M., Khan, K. A., and Asiri, A. M. (2011). Synthesis and characterization of a novel series of benzenesulfonylurea and thiourea derivatives of 2H-pyran and 2H-pyridine-2-ones as antibacterial, antimycobacterial and antifungal agents. Chem. Eur. J. 2, 243–250. doi: 10.5155/eurjchem.2.2.243-250.257
Fernandez, M., Godino, A., Príncipe, A., Morales, G. M., and Fischer, S. (2017). Effect of a Pseudomonas fluorescens tailocin against phytopathogenic Xanthomonas observed by atomic force microscopy. J. Biotechnol. 256, 13–20. doi: 10.1016/j.jbiotec.2017.07.002
Fischer, S. E., Jofré, E. C., Cordero, P. V., Gutiérrez, M. F. J., and Mori, G. B. (2010). Survival of native Pseudomonas in soil and wheat rhizosphere and antagonist activity against plant pathogenic fungi. Antonie Leeuwenhoek 97, 241–251. doi: 10.1007/s10482-009-9405-9
Fisher, R., and Lautner, H. (1961). Zum papier chromatographischen nachweis von Penicillin präparaten. Arch. Der. Pharm. 294, 1–7.
Franz, E., Tromp, S. O., Rijgersberg, H., and van der Fels-Klerx, H. J. (2010). Quantitative microbial risk assessment for Escherichia coli O157:H7, Salmonella, and Listeria monocytogenes in leafy green vegetables consumed at salad bars. J. Food Prot. 73, 274–285. doi: 10.4315/0362-028x-73.2.274
Ghoul, M., and Mitri, S. (2016). The ecology and evolution of microbial competition. Trends Microbiol. 24, 833–845. doi: 10.1016/j.tim.2016.06.011
Hennessy, R. C., Glaring, M. A., Olsson, S., and Stougaard, P. (2017). Transcriptomic profiling of microbe-microbe interactions reveals the specific response of the biocontrol strain P.fluorescens In5 to the phytopathogen Rhizoctonia solani. BMC Res. 10:376. doi: 10.1186/s13104-017-2704-8
Hinsinger, P., Bengough, A. G., Vetterlein, D., and Young, I. M. (2009). Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant Soil 321, 117–152. doi: 10.1007/s11104-008-9885-9
Hinsinger, P., and Marschner, P. (2006). Rhizosphere – perspective and challenges – a tribute to Lorenz Hiltner. Plant Soil 283, 1–333.
Igiehon, N. O., and Babalola, O. O. (2017). Biofertilizers and sustainable agriculture: exploring arbuscular mycorrhizal fungi. Appl. Microbiol. Biotechnol. 101, 4871–4881. doi: 10.1007/s00253-017-8344-z
Jangir, M., Pathak, R., Sharma, S., and Sharma, S. (2018). Biocontrol mechanisms of Bacillus sp., isolated from tomato rhizosphere, against Fusarium oxysporum f. sp. lycopersici. Biocontrol 123, 60–70. doi: 10.1016/j.biocontrol.2018.04.018
Jeong, Y., and Moloney, M. G. (2013). Synthesis and antibacterial activity of monocyclic 3-carboxamide tetramic acids. Beilstein J. Org. Chem. 9, 1899–1906. doi: 10.3762/bjoc.9.224
Keote, G. A., Reddy, M. S. P., Kumar, A., and Wasnikar, A. R. (2019). Bio-inoculants against chick pea wilt incited by Fusarium oxysporum f. sp. ciceri. J. Pharmacogn. Phytochem. 8, 1590–1594.
Knobloch, K., Pauli, A., and Iberl, B. (1989). Antibacterial and antifungal properties of essential oils components. J. Essent. Oil Res. 1, 119–128. doi: 10.1080/10412905.1989.9697767
Kondo, E., and Kanai, K. (1977). The relationship between the chemical structure of fatty acids and their mycobactericidal activity. Jpn. J. Med. Sci. Biol. 30, 171–178. doi: 10.7883/yoken1952.30.171
Kumar, C. V., Sameer Mula, M. G., Singh, I. P., Saxena, R. K., Rao, N. V. P. R. G., and Varshney, R. K. (2014). Pigeonpea Perspective in India. 1st Philippine Pigeonpea Congress. Batac: Mariano Marcos State University.
Kumar, V., Kumar, A., Verma, V. C., Gond, S. K., and Kharwar, R. N. (2007). Induction of defense enzymes in Pseudomonas fluorescens treated chickpea roots against Macrophomina phaseolina. Indian Phytopath. 60, 289–295.
Lecuit, M. (2007). Human listeriosis and animal models. Microbes Infect. 9, 1216–1225. doi: 10.1016/j.micinf.2007.05.009
Lemunier, M., Francou, C., Rousseaux, S., Houot, S., Dantigny, P., Piveteau, P., et al. (2005). Long-term survival of pathogenic and sanitation indicator bacteria in experimental biowaste composts. Appl. Environ. Microbiol. 71, 5779–5786. doi: 10.1128/aem.71.10.5779-5786.2005
Lillebaek, E. M. S., Nielsen, S. L., Thomasen, R. S., Faergeman, N. J., and Kallipolitis, B. H. (2017). Antimicrobial medium- and long-chain free fatty acids prevent PrfA-dependent activation of virulence genes in Listeria monocytogenes. Res. Microbiol. 168, 547–557. doi: 10.1016/j.resmic.2017.03.002
Liu, K., Mclnroy, J. A., Hu, C. H., and Kloepper, J. W. (2018). Mixtures of plant-growth-promoting rhizobacteria enhance biological control of multiple plant diseases and plant-growth promotion in the presence of pathogens. Plant Dis. 102, 67–72. doi: 10.1094/pdis-04-17-0478-re
Liu, W. L., Pan, X. C., Zhang, C. B., and Wang, J. (2013). Characterization of substrate microbial communities in vertical flow mesocosms as impacted by both planting pattern and species richness. Res. Microbiol. 164, 941–948. doi: 10.1016/j.resmic.2013.08.001
Ly, T. M. C., and Müller, H. E. (1990). Ingested Listeria monocytogenes survive and multiply in protozoa. J. Med. Microbiol. 33, 51–54. doi: 10.1099/00222615-33-1-51
MacFaddin, J. F. (2000). Biochemical Tests for Identification of Medical Bacteria, 3rd Edn. Philadelphia, PA: Lippincott Williams and Wilkins.
Mathew, A., Mary Sheeja, T. L., Kumar, A., and Radha, K. (2011). Design, synthesis and biological evaluation of pyrazole analogues of natural piperine. J. Med. Chem. 3, 48–56.
McLauchlin, J. (1987). Listeria monocytogenes, recent advances in the taxonomy and epidemiology of listeriosis in humans. J. Appl. Microbiol. 63, 1–11. doi: 10.1111/j.1365-2672.1987.tb02411.x
Mendes, A., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Meyer, J. M., and Abdallah, M. A. (1978). The florescent pigment of Pseudomonas fluorescens biosynthesis, purification and physical-chemical properties. J. Gen. Microbiol. 107, 319–328. doi: 10.1099/00221287-107-2-319
Miller, K. G. (1992). “Middle eocene to oligocene stable isotopes, climate, and deep-water history: the terminal eocene event,” in Eocene-Oligocene Climatic and Biotic Evolution, eds D. R. Prothero, and W. A. Berggren, (Princeton: Princeton University Press), 160–177. doi: 10.1515/9781400862924.160
Montano, E. T., and Henderson, L. O. (2013). “Studies of antibiotic production by cave bacteria,” in Cave Microbiomes: A Novel Resource for Drug Discovery, ed. N. Cheeptham, (New York, NY: Springer), 109–130. doi: 10.1007/978-1-4614-5206-5_6
Mora, I., Cabrefiga, J., and Montesinos, E. (2015). Cyclic lipopeptide biosynthetic genes and products, and inhibitory activity of plant-associated Bacillus against phytopathogenic bacteria. PLoS One 10:e0127738. doi: 10.1371/journal.pone.0127738
Moran, J. C., Crank, E. L., Ghabban, H. A., and Horsburgh, M. J. (2016). Deferred growth inhibition assay to quantify the effect of bacteria-derived antimicrobials on competition. J. Vis. Exp. 115, 2989–2992.
Munjal, V., Nadakkakath, A. V., Sheoran, N., Kundu, A., Venugopal, V., Subaharan, K., et al. (2016). Genotyping and identification of broad spectrum antimicrobial volatiles in black pepper root endophytic biocontrol agent, Bacillus megaterium BP17. Biol. Control 92, 66–76. doi: 10.1016/j.biocontrol.2015.09.005
Nakamura, H., Wu, H., Ohizumi, Y., and Hirata, Y. (1984). Agelasine-A, -B, -C and -D, novel bicyclic diterpenoids with a 9-methyladeninium unit possessing inhibitory effects on Na, K-ATPase from the okinawa sea sponge Agelas sp. Tetrahedron Lett. 25, 2989–2992. doi: 10.1016/s0040-4039(01)81345-9
Nautiyal, C. S. (1996). A method for selection and characterization of rhizosphere- competent bacteria of chickpea. Curr. Microbiol. 34, 12–17. doi: 10.1007/s002849900136
Nautiyal, C. S. (1999). An efficient microbiological growth medium for screening phosphorus solubilizing microorganisms. FEMS Microbiol. Lett. 170, 2017–2021.
Neuhaus, F. C., and Baddiley, J. A. (2003). Continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in Gram-positive bacteria. Microbiol. Mol. Biol. Rev 67, 686–723. doi: 10.1128/mmbr.67.4.686-723.2003
Nicolaus, B. J. R., Coronelli, C., and Binaghi, A. (1961). Microbiological determination of antibiotics by thin layer chromatography. Farmaco. Ed. Prat. 16, 349–370.
Nightingale, K. K., Schukken, Y. H., Nightingale, C. R., Fortes, E. D., Ho, A. J., Her, Z., et al. (2004). Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 70, 4458–4467. doi: 10.1128/aem.70.8.4458-4467.2004
Oyinloye, M. A., David, O. M., Muhammad, A., and Famurewa, O. (2018). Occurrence and molecular characterisation of Listeria species in some fresh-cut vegetables and environmental samples. J. Adv. Microbiol. 12, 1–12. doi: 10.9734/jamb/2018/43046
Papić, B., Kušar, D., Zdovc, I., Golob, M., and Pate, M. (2020). Retrospective investigation of listeriosis outbreaks in small ruminants using different analytical approaches for whole genome sequencing-based typing of Listeria monocytogenes. Infect. Genet. Evol. 77, 1–9.
Pasquina, L., Maria, J. P. S., Wood, B. M., Moussa, S. H., Matano, L., Santiago, M., et al. (2016). A synthetic lethal approach for compound and target identification in Staphylococcus aureus. Nat. Chem. Biol. 12, 40–45. doi: 10.1038/nchembio.1967
Pierret, A., Doussan, C., Capowiez, Y., Bastardie, F., and Pagès, L. (2007). Root functional architecture: a framework for modeling the interplay between roots and soil. Vadose Zone J. 6, 269–281. doi: 10.2136/vzj2006.0067
Prasanthi, G., Prasad, K. V., and Bharathi, K. (2013). Design, synthesis and evaluation of dialkyl 4-(benzo[d][1,3]dioxol-6-yl)-1,4-dihydro-2,6-dimethyl-1-substituted pyridine-3,5-dicarboxylates as potential anticonvulsants and their molecular properties prediction. Eur. J. Med. Chem. 66, 516–525. doi: 10.1016/j.ejmech.2013.06.006
Raaijmakers, J. M., Paulitz, T. C., Steinberg, C., Alabouvette, C., and Moënne-Loccoz, Y. (2009). The rhizosphere: a playground and battlefield for soil borne pathogens and beneficial microorganisms. Plant Soil 321, 341–361. doi: 10.1007/s11104-008-9568-6
Rajan, R., Renold, P., Mahajan, A., Stierli, D., and Beaudegnies, R. (2017). United States Patent No. US 20170000133A1. Available at: https://patents.google.com/patent/US20170000133A1/en (accessed March 2, 2020).
Salama, P. J., Embarek, P. K. B., Bagaria, J., and Fall, I. S. (2018). Learning from listeria: safer food for all. Lancet 391, 2305–2306. doi: 10.1016/s0140-6736(18)31206-6
Schreiter, S., Ding, G., Heuer, H., Neumann, G., Sandmann, M., Grosch, R., et al. (2014). Effect of the soil-type on the microbiome in the rhizosphere of field-grown lettuce. Front. Microbiol. 5:144. doi: 10.3389/fmicb.2014.00144
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Sharma, R., Paliwal, J. S., Chopra, P., Dogra, D., Pooniya, V., Bisaria, V. S., et al. (2017). Survival, efficacy and rhizospheric effects of bacterial inoculants on Cajanus cajan. Agric. Ecosyst. Environ. 240, 244–252. doi: 10.1016/j.agee.2017.02.018
Sheikh, H. M. A. (2010). Antimicrobial activity of certain bacteria and fungi isolated from soil mixed with human saliva against pathogenic microbes causing dermatological diseases. Saudi J. Biol. Sci. 17, 331–339. doi: 10.1016/j.sjbs.2010.06.003
Sokol, P. A., Ohman, D. E., and Iglewski, B. H. (1979). A more sensitive plate assay for detection of protease production by Pseudomonas aeruginosa. J. Clin. Microbiol. 9, 538–540.
Srinivas, A., and Sunitha, M. (2016). Synthesis of piperonyltriazoles as anti-microbial agents. Indian J. Chem. 55, 102–109.
Tada, Y., Shikishima, Y., Takaishi, Y., Shibata, H., Higuti, T., Honda, G., et al. (2002). Coumarins and γ-pyronederivatives from Prangos pabularia: antibacterial activity and inhibition of cytokine release. Phytochemistry 59, 649–654.
Tang, J. Y. H., Razali, N. A. S., Jalil, L. A., Mat-Sa’ad, S. H., Nakaguchi, Y., Nishibuchi, M., et al. (2017). Detection of Listeria spp. and Listeria monocytogenes in vegetables by loop-mediated isothermal amplification (LAMP) and multiplex polymerase chain reaction (PCR). J. Fundam. Appl. Sci. 9, 698–714.
Tyler, H. L., and Triplett, E. W. (2008). Plants as a habitat for beneficial and/or human pathogenic bacteria. Annu. Rev. Phytopathol. 46, 53–73. doi: 10.1146/annurev.phyto.011708.103102
Vig, K., Singh, D. K., Agarwal, H. C., Dhawan, A. K., and Dureja, P. (2008). Soil microorganisms in cotton fields sequentially treated with insecticides. Ecotoxicol. Environ. Saf. 69, 263–276. doi: 10.1016/j.ecoenv.2006.12.008
Vik, A., Hedner, E., Charnock, C., Samuelsen, Ø, Larsson, R., Gundersen, L., et al. (2006). (+)-Agelasine D: improved synthesis and evaluation of antibacterial and cytotoxic activities. J. Nat. Prod. 69, 381–386. doi: 10.1021/np050424c
Viscardi, S., Ventorino, V., Duran, P., Maggio, A., Pascale, S. D., Mora, M. L., et al. (2016). Assessment of plant growth promoting activities and abiotic stress tolerance of Azotobacter chroococcum strains for a potential use in sustainable agriculture. J. Soil Sci. Plant Nutr. 16, 848–863.
Vivant, A., Garmyn, D., Maron, P., Nowak, V., and Piveteau, P. (2013a). Microbial diversity and structure are drivers of the biological barrier effect against Listeria monocytogenes in soil. PLoS One 8:e76991. doi: 10.1371/journal.pone.0076991
Vivant, A. L., Garmyn, D., and Piveteau, P. (2013b). Listeria monocytogenes, a down-to-earth pathogen. Front. Cell Infect.Microbiol. 3:87. doi: 10.3389/fcimb.2013.00087
Wang, Y., Lu, L., Lan, R., Salazar, J. K., Liu, J., Xu, J., et al. (2017). Isolation and characterization of Listeria species from rodents in natural environments in China. Emerg. Microbes Infect. 6, 1–15.
Wani, M. Y., Bhat, A. R., Azam, A., Choi, I., and Athar, F. (2012). Probing the antiamoebic and cytotoxicity potency of novel tetrazole and triazine derivatives. Eur. PMC 48, 313–320. doi: 10.1016/j.ejmech.2011.12.033
Winding, A., Binnerup, S. J., and Pritchard, H. (2004). Non-target effects of bacterial biological control agents suppressing root pathogenic fungi. FEMS Microbiol. Ecol. 47, 129–141. doi: 10.1016/s0168-6496(03)00261-7
Xu, M., Sheng, J., Chen, L., Men, Y., Gan, L., Guo, S., et al. (2014). Bacterial community compositions of tomato (Lycopersicum esculentum Mill.) seeds and plant growth promoting activity of ACC deaminase producing Bacillus subtilis (HYT-12-1) on tomato seedlings. World J. Microbiol. Biotechnol. 30, 835–845. doi: 10.1007/s11274-013-1486-y
Yin, D., Wang, N., Xia, F., Li, Q., and Wang, W. (2013). Impact of biocontrol agents Pseudomonas fluorescens 2P24 and CPF10 on the bacterial community in the cucumber rhizosphere. Eur. J. Soil Biol. 59, 36–42. doi: 10.1016/j.ejsobi.2013.09.001
Yishay, M., Burdman, S., Valverde, A., Luzzatto, T., Ophir, R., and Yedidia, I. (2008). Differential pathogenicity and genetic diversity among Pectobacterium carotovorum ssp. carotovorum isolates from monocot and dicot hosts support early genomic divergence within this taxon. Environ. Microbiol. 10, 2746–2759. doi: 10.1111/j.1462-2920.2008.01694.x
Keywords: Azotobacter chroococcum, Bacillus megaterium, Pseudomonas fluorescens, Cajanus cajan, Festuca arundinacea, inhibition, UPLC-MS, bioactive compounds
Citation: Sharma R, Gal L, Garmyn D, Bisaria VS, Sharma S and Piveteau P (2020) Evidence of Biocontrol Activity of Bioinoculants Against a Human Pathogen, Listeria monocytogenes. Front. Microbiol. 11:350. doi: 10.3389/fmicb.2020.00350
Received: 05 December 2019; Accepted: 17 February 2020;
Published: 11 March 2020.
Edited by:
Camille Eichelberger Granada, Universidade do Vale do Taquari - Univates, BrazilReviewed by:
Daiane Heidrich, Universidade do Vale do Taquari - Univates, BrazilCameron Parsons, North Carolina State University, United States
Houhui Song, Zhejiang Agriculture & Forestry University, China
Vânia Borges Ferreira, Universidade Católica Portuguesa, Portugal
Copyright © 2020 Sharma, Gal, Garmyn, Bisaria, Sharma and Piveteau. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shilpi Sharma, c2hpbHBpQGRiZWIuaWl0ZC5hYy5pbg==; Pascal Piveteau, UGFzY2FsLnBpdmV0ZWF1QGlucmFlLmZy
†Present address: Pascal Piveteau, INRAE, Rennes, France
 Richa Sharma1
Richa Sharma1 Laurent Gal
Laurent Gal Shilpi Sharma
Shilpi Sharma Pascal Piveteau
Pascal Piveteau