
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 13 March 2020
Sec. Microbial Symbioses
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00300
This article is part of the Research TopicDesigning Bio-Formulations Based On Organic Amendments, Beneficial Microbes And Their MetabolitesView all 20 articles
 Xiao Xu1,2†
Xiao Xu1,2† Mingwei Shao1,2†
Mingwei Shao1,2† Caiping Yin1
Caiping Yin1 Zhenchuan Mao3*
Zhenchuan Mao3* Jingjing Shi2
Jingjing Shi2 Xinyuan Yu2
Xinyuan Yu2 Ying Wang2
Ying Wang2 Feifei Sun2
Feifei Sun2 Yinglao Zhang1,2*
Yinglao Zhang1,2*The phylogenetic diversity of fungi isolated from the Odontotermes formosanus was investigated by dilution-plate method, combined with morphological characteristics and 5.8S rDNA sequencing. Thirty-nine fungi were isolated and purified from O. formosanus, which were belonging to two phyla and four classes (Sordariomycetes, Dothideomycetes, Eurotiomycetes, Agaricomycetes). Furthermore, nine bacterial 16S rRNA sequences were obtained from total fungal genomic DNA. All bacterial symbionts were segmented into four genera: Bacillus, Methylobacterium, Paenibacillus, and Trabulsiella. The antimicrobial activities of all endophytic fungi extracts were tested by using the filter paper method against Escherichia coli (ATCC 8739), Bacillus subtilis (ATCC 6633), Staphylococcus aureus (ATCC 6538), and Canidia albicans (ATCC 10231). The results exhibited that 25 extracts (64%) exhibited antibacterial activity against at least one of the tested bacterial strains. Furthermore, the secondary metabolites 1 [5-hydroxyramulosin (1a):biatriosporin M (1b) = 2:1] from the Pleosporales sp. BYCDW4 exhibited potent antimicrobial activities against E. coli, C. albicans, B. subtilis, and S. aureus with the inhibition zone diameter (IZD) of 13.67, 14.33, 12.17, and 11.33 mm, respectively, which were comparable with those of the positive control. 1-(2,5-Dihydroxyphenyl)-3-hydroxybutan-1-one (2) from the Microdiplodia sp. BYCDW8 showed medium inhibitory activities against B. subtilis and S. aureus, with the IZD range of 8.32–9.13 mm. In conclusion, the study showed the diversity of insect symbionts could be expected to develop the resource of new species and antibiotics.
The symbiotic relationships, kinds of organisms including microorganisms, animals, and plants can be seen everywhere in the world (Blackwell and Vega, 2018; Grigorescu et al., 2018). For example, the first direct visual evidence of a not-yet-cultured CFB bacterium is detected in a mycorrhizal fungus (Barbieri et al., 2000). Many others with well-studied examples including the sponge and symbiotic strains, nitrogen-fixing bacteria, and legumes live in symbiotic association (Trainer and Charles, 2006; Webster et al., 2008). Compared with other interactions, the insect symbiont interactions still represent a largely unknown field (Park et al., 2018).
There are few researches reported that symbioses of insects are an important unexploited microbial resource for human beings. For example, insect gut microbes, especially fungi in suppression of unwanted microorganisms (parasites, microorganic pathogens, etc.), protecting the hosts and maintaining the ordinary growth and behavior, are of interest to insects (Lu et al., 2016; Beemelmanns et al., 2017; Van Arnam et al., 2017). Based on this symbiosis, several new immunosuppressive polyketides were obtained from a fungus attached to mantis, Daldinia eschscholzii IFB-TL01 (Zhang et al., 2008, 2011). In addition, insect symbiosis is also an important source of new microorganisms. For instance, 57 entomopathogenic fungi were analyzed, including one species which was firstly reported (Nishi and Sato, 2017). Many new species were also isolated from aquatic insects, among which new genera were found (White and Lichtwardt, 2004; White et al., 2018). Nonetheless, extensive, methodical, and biologically correlative researches about the insect microorganisms were restricted, especially when the complexity of insect symbiotic system was taken into consideration.
Termites, belonging to the class Insecta and the order Isoptera in the phylum Arthropoda, are a kind of eusocial and hemimetabolous insects (Mathew et al., 2012). Multitudinous and particular microbial floras reside in the termites, most of which can help the host to digest and utilize their food (Zhu et al., 2012; Li et al., 2017). Odontotermes formosanus as a termite species is mainly distributed in south areas of the line between Yellow River and Yangtze River in China. As far as we know, research concentrated on O. formosanus fungi derived from China have been rather rare (Chou et al., 2007; Shinzato et al., 2007). Underexploited O. formosanus symbionts can offer a pathway to finding unreported biological resources, which will solve the dilemma that needs to be dealt with regarding new drugs, especially antibiotics, which we are in sore need of due to increasing bacterial resistance to existing antibiotics (Procópio et al., 2012; Bi et al., 2013; Elahwany et al., 2013). Herein, the study was intended to explore the phylogenetic diversity, bacterial symbionts, antimicrobial potency, and compounds of the fungi associated with O. formosanus.
A total of 50 termite samples were collected from the rotten wood in a grove in Zhejiang Normal University (29°00′17.37″N, 119°29′54.84″E, Jinhua city, China) during early spring in March 2016. The 50 collected samples were starved for 12 h and surface-sterilized in 75% ethanol for 2 min, followed by rinsing in sterilized water three times (30 s each). Sterile forceps were used to dissect samples to get the guts. The guts and degutted bodies of termites were fully homogenized separately in 0.5 ml sterile water. Then, the homogenates were diluted in a 10-fold series (i.e., 10–1, 10–2, 10–3), and aliquots of 200 μl from each dilution were spread onto 10 isolation media (Supplementary Table S1). Pure colonies of fungi from the appropriate dilution were transferred into new MEA medium. Isolated strains were preserved on MEA slants at 4°C until use. Fungal species were grouped via molecular sequence data. The fungi were stored at our institute.
DNA sequencing was performed according to the methods detailed previously (Hoffman and Arnold, 2010; Shao et al., 2015). Symbiotic fungi were transferred into ME medium (20 g malt extract, 20 g sucrose, 1 g peptone in 1 L of distilled H2O) and cultured at 28 ± 0.5°C on rotary shakers for 5–6 days. The growing fungal mycelia were used to afford samples for genomic DNA extraction. Fungal total DNA was amplified using the Fast DNA Extraction Kit (BioTeke, Beijing, China) as claimed by the manufacturer’s specification. The genomic DNA was stored at 4°C until use. The primers ITS1/ITS4 and 27F/1492R were used to amplify 5.8S rDNA and 16S rRNA based on the fungal genomic DNA. Qualified PCR samples were used for sequencing (Sangon Biotech Co., Ltd., Shanghai, China).
As described previously (Shao et al., 2015), all resulting sequences’ affiliation which were returned from Sangon Biotech Company were recognized by the available data in BLAST from the National Center for Biotechnology Information (NCBI) database. Sequence alignment and neighbor-joining phylogenetic analysis were carried out using MEGA software version 5.1. Bootstrap analysis of tree construction on the strength of the sequences was accustomed to estimate the neighbor-joining information based on 1,000 replicates (Felsenstein, 1985). The 5.8S rDNA sequences that had been obtained were placed in NCBI with the accession numbers MG820065–MG820103. Besides, endosymbiotic bacterial 16S rRNA sequences were deposited in NCBI with the accession numbers MG825089–MG825096.
Every fungus was grown on MEA medium at 28 ± 0.5°C for 3–4 days. Then, pieces of fresh mycelium were inoculated into 1-L Erlenmeyer flasks each containing 150 ml of ME liquid medium, after 2–3 days of incubation at 28 ± 0.5°C in a shaker rotating at 180 rpm. A 20-ml suspension of the fungus was transferred as seed into 1-L Erlenmeyer flasks each containing 400 ml of ME liquid medium. The flask cultures were incubated at 28 ± 0.5°C for 1 week.
A total of 26 L of fermentation broth of BYCDW4 was filtered and extracted with EtOAc (3 × 26 L) at room temperature. The solvent was then evaporated in vacuo to give a black-brown crude extract (13.5 g). The obtained extract was separated by column chromatography (CC) using silica gel (SiO2: 200–300 mesh, 15 g) and eluting with a stepwise gradient of CH2Cl2/MeOH (100:0–100:16, v/v) to afford six primary fractions (A to F). Then the white needle crystal (compound 1, 5.84 g) were recrystallized by CH2Cl2/MeOH continuously up to no impurity.
The dark-yellow mixture of BYCDW8 (1.5 g) was obtained with the method as above. Compound 2 (14.5 mg) was isolated and purified from subfraction B-2, which was given from further chromatography over silica gel in fraction B (0.4 g, CH2Cl2/MeOH, 100:1).
The structures of compounds 1 and 2 were primarily analyzed by mass and 1H/13C-nuclear magnetic resonance (NMR) spectroscopies. The electrospray ionization mass spectrometry of the purified metabolites was recorded on a TripeTOF 4600 instrument (Bruker, Billerica, MA, United States). 1H/13C NMR spectra were measured with a Bruker AVANCE-600 (Bruker, Switzerland) spectrometer, and chemical shifts were reported as parts per million (δ) by referring to tetramethylsilane as internal standards. The structure of compound 1a was further determined by single crystal X-ray.
The antimicrobial activities of all the 39 fungal crude extracts and the secondary metabolites were evaluated using filter-paper method as described previously (Li et al., 2014). Three tested pathogens (Escherichia coli ATCC 8739, Bacillus subtilis ATCC 6633, Staphylococcus aureus ATCC 6538) were cultivated on NA medium (NaCl 3 g, peptone 10 g, beef extract 3 g, agar 18–20 g, distilled H2O 1,000 ml) in 37°C, whereas the pathogenic Candida albicans (ATCC 10231) was inoculated on PDA medium in 28°C. Then 200 μl of the overnight fermentation broth of the pathogens was inoculated on the corresponding plate. Next, 5 μl of the tested objects including crude extracts, metabolites, and positive control drugs which were dissolved completely by acetone with the concentration of 6 mg/ml was pipetted onto a sterile filter disk with a diameter of 5 mm. Gentamicin sulfate and amphotericin B were used as the positive control of pathogenic bacteria and yeast, respectively. Then, all the processed filter papers were placed on the above pre-prepared medium. Each test was set up three repeats. Plates were cultivated at constant temperature incubator and the inhibition zone was measured at 24–36 h.
In this study, a total of 39 symbiotic fungi were isolated from O. formosanus (Table 1 and Figure 1). The ITS1-5.8S-ITS2 region of all strains were sequenced and compared with available GenBank reference sequences. Sequences analysis showed that all fungi were attached to the phyla Ascomycota and Basidiomycota, 35 strains of which were grouped into three classes [Sordariomycetes (23.1%), Dothideomycetes (53.8%), and Eurotiomycetes (15.4%)] within Ascomycota. The other three strains (8%) were distributed in the Agaricomycetes within the phylum Basidiomycota (Figure 1).
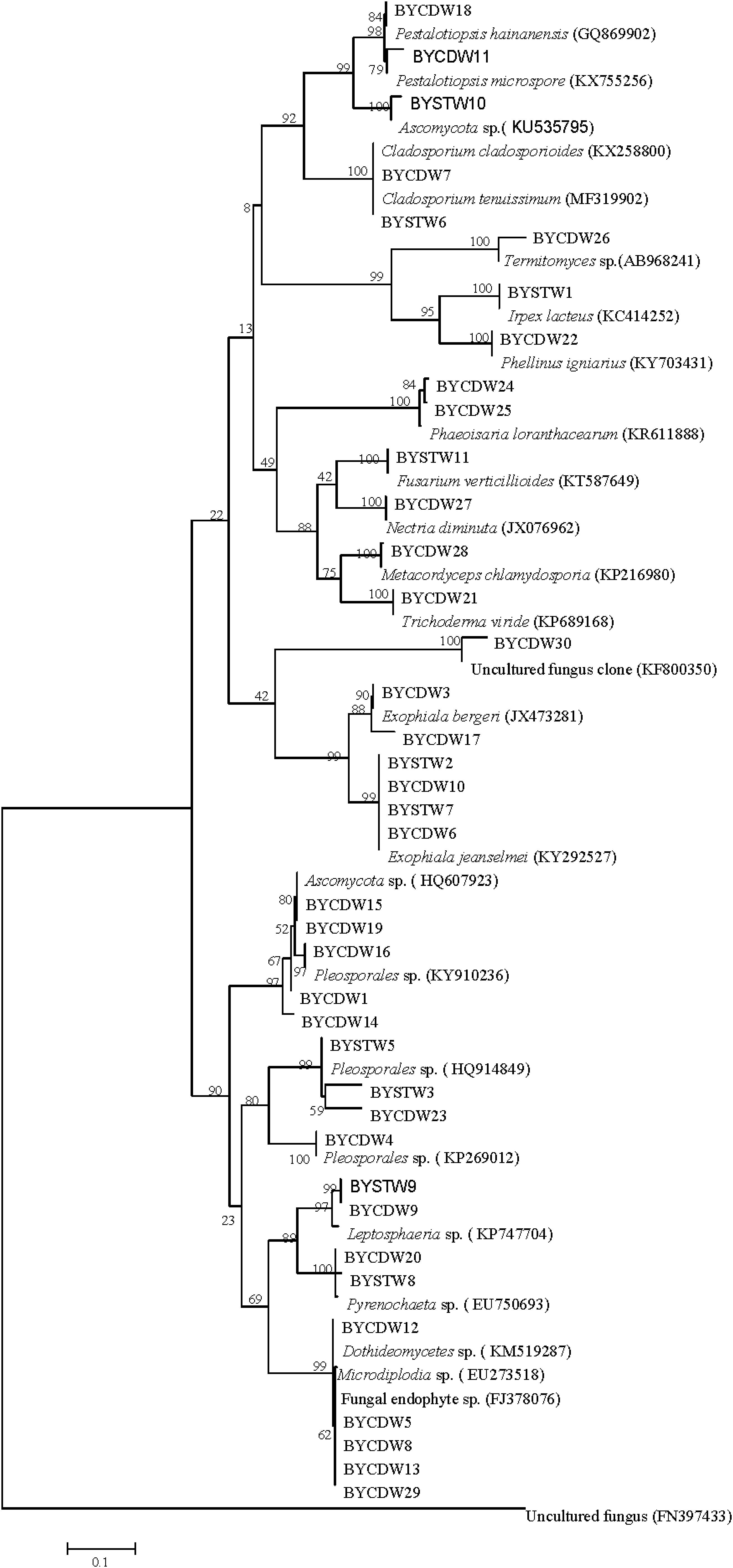
Figure 1. Neighbor-joining phylogenetic tree of ITS rDNA sequences of O. formosanus fungal isolates. Bootstrap values were calculated using 1,000 replications.
The fungi of Dothideomycetes were dominant species in phylogenetic diversity of cultivable fungi from O. formosanus. The largest number (21) of isolates was distributed in three orders, including the Capnodiales, Venturiales, and Pleosporales. Among of them, two strains belonging to Capnodiales showed highly similar to Cladosporium cladosporioides with identity of 99%. The strain BYCDW30 showed only 92% similarity to O. constricta, which indicated a potential new species. The most frequent order identified was the Pleosporales, which included six genera based on phylogenetic tree analysis (Figure 1).
The strains of Sordariomycetes (9) were mainly included in the three classes (Xylariales, Pleurotheciales, and Hypocreales). Four of them were grouped into the family Hyporcreaceae and were identified as Fusarium verticillioides, Metacordyceps chlamydosporia, Nectria diminuta, and Trichoderma viride, respectively (Figure 1). Two strains showed similar to Phaeoisaria loranthacearum with a low identity of 96%. Two other strains belonging to Xylariales were identified as Pestalotiopsis hainanensis and P. microspora, respectively.
Six isolates of the class Eurotiomycetes were mainly concentrated in the genus Exophiala grouped into the order Chaetothyriales. Four strains showed highly similar to Exophiala jeanselmei with an identity of more than 99%. Another two strains were similar to E. bergeri with sequence match of 94% and 99%, respectively. What was worth mentioning was that the BYCDW17 showed a sequence match of 94% with E. bergeri, which indicated that it might not have been deposited in the GenBank database or represent a potential new species (Landeweert et al., 2003).
The fungi of class Agaricomycetes belonging to the phylum Basidiomycota were mainly distributed in the orders Agaricales and Polyporales. The strain BYCDW26 of Agaricales was a Termitomyces fungus, which had the low similar sequence with an identity of 88.10%. Another two strains belonging to Polyporales were identified as Irpex lacteus and Phellinus igniarius, respectively.
A total of nine bacterial symbionts 16S rRNA was obtained from the fungi associated with O. formosanus in this study (Supplementary Table S2). These bacterial symbionts mainly pertained to the class Dothideomycetes and identified as Bacillales (Bacillus, Paenibacillus), Rhizobiales (Methylobacterium), and Enterobacterium (Trabulsiella). Furthermore, six bacteria were putatively divided into genus Bacillus.
The antimicrobial results are shown in Table 2. Of the all isolates, 25 extracts (64%) exhibited antibacterial activity against at least one of the tested bacterial strains. Especially, BYCDW8, BYCDW4 (Supplementary Figure S1), BYCDW13, and BYCDW19 exhibited remarkable inhibitory activities against S. aureus (ATCC 6538) with the inhibition zone diameter (IZD) of more than 20 mm. These four fungi were all attached to Pleosporales in the class Dothideomycetes. Furthermore, the strain BYCDW4 possessed notable inhibitory effect against E. coli (ATCC 8739) with the IZD of 21.07 mm, which was stronger than the positive gentamicin sulfate with the IZD of 17.67 mm. However, other strains did not express remarkable activity against E. coli. The crude extracts of BYCDW3, BYCDW4, and BYCDW25 exhibited great inhibition effect on C. albicans (ATCC 10231), which were equivalent to that of amphotericin with the IZD of 19.17 mm. And among them, the crude extract of BYCDW25 was the most prominent. Further analysis showed that the inhibition activities of the fungal extracts against gram-positive bacteria was much better than the gram-negative bacteria. In addition, the potential new isolates BYCDW24 and BYCDW25 (in the order Pleurotheciales) presented moderate antimicrobial activities against several pathogens.
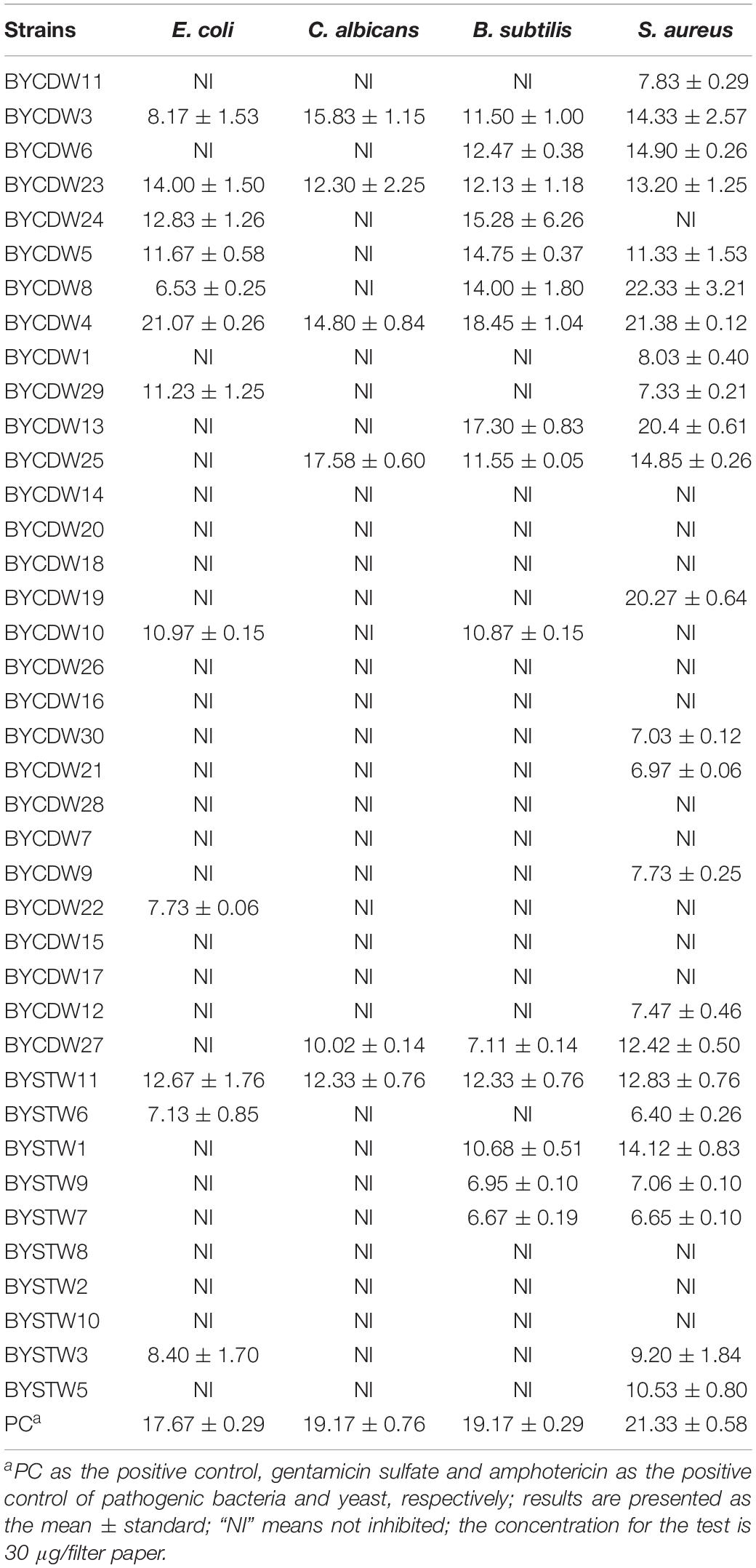
Table 2. Antimicrobial activities of 39 fungal crude extracts against pathogenic bacteria and yeast (mm).
Secondary metabolites were identified by spectroscopic analyses, including HR–ESI-MS, NMR, and comparisons with the data described in the previous literatures. The constituents were identified as compound 1 [5-hydroxyramulosin (1a): biatriosporin M (1b) = 2:1] from BYCDW4 and 1-(2,5-dihydroxyphenyl)-3-hydroxybutan-1-one (2) (Figure 2) from BYCDW8 based on the following data.
Compound 1 was obtained as a white needle crystal, which is a mixture (5-hydroxyramulosin (Osterhage et al., 2002) and biatriosporin M (Zhou et al., 2016)) with the proportion of 2:1 from the data of 1H NMR. 5-Hydroxyramulosin (1a): HR–ESI-MS: [M + H]+ m/z 199.0964 (calcd for 199.0970), [M + Na]+ m/z 221.0785 (calcd for 221.0790); 1H NMR (CDCl3) δ: 13.30 (s, 1H), 4.48 (m, 1H), 4.05 (m, 1H), 2.68 (m, 1H), 2.59 (m, 1H), 2.38 (m, 1H), 2.05 (m, 1H), 1.72 (m, 1H), 1.96 (m, 1H), 1.84 (m, 1H), 1.42 (d, 6.3, 3H); 13C NMR (CDCl3) δ: 174.3, 172.4, 92.2, 76.2, 66.0, 37.4, 32.7, 27.9, 24.3, 21.8. Biatriosporin M (1b): HR–ESI-MS: [M + H]+ m/z 197.0807 (calcd for 197.0814), [M + Na]+ m/z 219.0625 (calcd for 219.0633); 1H NMR (CDCl3) δ: 12.70 (s, 1H), 6.66 (dd, 9.9, 5.4, 1H), 6.25 (d, 9.9, 1H), 4.50 (m, 1H), 3.99 (m, 1H), 2.81 (m, 1H), 1.88 (m, 1H), 1.80 (m, 1H), 1.47 (d, 6.3, 3H); 13C NMR (CDCl3) δ: 172.5, 166.0, 138.7, 125.8, 90.9, 75.9, 63.2, 36.0, 30.3, 21.6. Compound 1a was further identified based on single-crystal X-ray diffraction data (Supplementary Table S3 and Supplementary Figure S2). Compound 1b was a dehydrogenated derivative of 1a. Owing to their similar polarity, we were not able to isolate them by conventional separation methods. To the best of our knowledge, this was the first report that compound 1 was isolated from the fungus with high yield of 43.3% (compound/crude extract = 5.84 g:13.50 g). Worth mentioning was that compound 1a (5-hydroxyramulosin) was reported with good antifungal activity against the fungal pathogen Aspergillus niger (IC50 = 2.10 μg/ml) and remarkable cytotoxicity to murine leukemia cells (IC50 = 2.10 μg/ml) (Santiago et al., 2012).
1-(2,5-Dihydroxyphenyl)-3-hydroxybutan-1-one (2) (Hong et al., 2018): light green-orange powder. HR-ESI-MS: [M + H]+ m/z 197.0804 (calcd for 197.0814).1H NMR (acetone-d6) δ: 11.77 (br s, 1H), 8.19 (br s, 1H), 7.34 (d, 2.94, 1H), 7.09 (dd, 2.94, 8.88, 1H), 6.80 (d, 8.88, 1H), 4.37 (m, 1H), 3.92 (br s, 1H), 3.19 (dd, 8.04, 17.52, 1H), 3.03 (dd, 4.62, 17.52, 1H), 1.26 (d, 6.20, 3H). 13C NMR (acetone-d6) δ: 206.6, 156.6, 150.4, 125.6, 120.3, 119.3, 115.9, 64.8, 48.2, 23.7.
The antimicrobial activities of the compounds isolated from the strain BYCDW4 and BYCDW8 is shown in Table 3. Under the test concentration of 30 μg/filter paper, compound 1 [5-hydroxyramulosin (1a):biatriosporin M (1b) = 2:1] exhibited some inhibitory activities against all the test pathogens. More specifically, the inhibitory effect on E. coli was the most significant with the IZD of 13.7 mm, which was slightly weaker than that of positive gentamicin sulfate with IZD of 17.7 mm. Compound 1 also showed good antimicrobial activities against C. albicans, B. subtilis, and S. aureus with the IZD of 14.33, 12.17, and 11.33 mm, respectively, which were comparable with those of the positive control. Compound 2 exhibited medium inhibitory activity against B. subtilis and S. aureus, with the IZD range of 8.32–9.13 mm, while it had no inhibition effect on C. albicans.
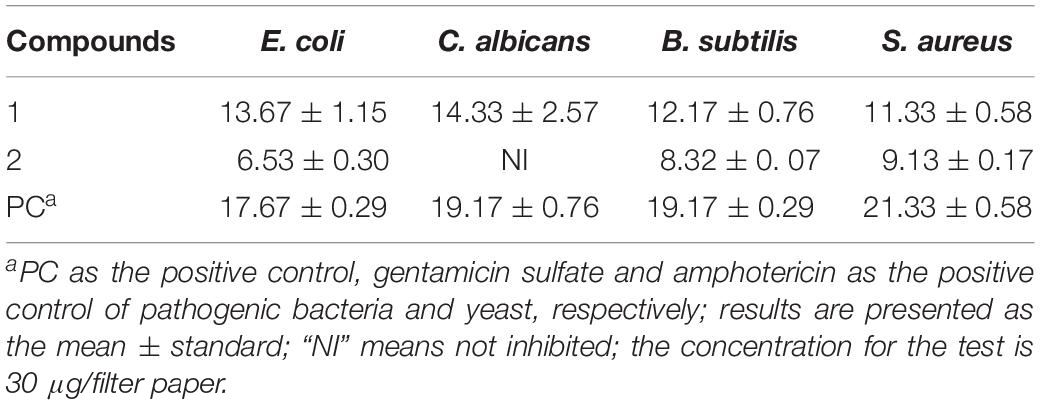
Table 3. Antimicrobial activities of compounds isolated from BYCDW4 and BYCDW8 against the test pathogens (mm).
Microorganisms isolated from the bioprospecting underexploited environments with rich microbial biodiversity has been an important source of active natural products and new microorganism resources. A great variety of cultivable microbial flora was reported to exist in insects (Douglas, 2015; Shao et al., 2017; Blackwell and Vega, 2018). These important microbial resources were increasingly reported to be important sources of new natural products (Carr et al., 2012; Adnani et al., 2017; Xiao et al., 2017). In this study, the diversity of symbiotic fungi associated with O. formosanus and endophytic bacteria were studied. Thirty-nine fungi, including some potential new species, were isolated by culture-dependent method and molecular biological identification. To our knowledge, this is the first time that the phylogenetic diversity of cultivable and biological activity screening of secondary metabolites of fungi isolated from O. formosanus was studied, which will provide an important microbial resource for the discovery of new species and natural products.
Today, multidrug resistance of pathogens represents one of the major challenges to treat infectious diseases in community- and hospital-acquired infection diseases. There is also an urgent need for new antibiotics against gram-negative bacteria which have already been resistant to existing carbapenems and third-generation cephalosporins (Luther et al., 2019). In our study, the crude extract of potential new fungus BYCDW4 showed especially notable inhibitory effect against gram-negative E. coli (ATCC 8739) with the IZD of 21.07 mm, which was stronger than that of the positive gentamicin sulfate. The inhibition effect of crude extract against E. coli was better than purified compound 1 with the IZD of 13.6 mm. This might be the result of the synergistic action of multiple compounds, or the more active compounds have not been separated from the crude extract. Further research on other metabolites of BYCDW4 will be expected to develop new antibiotics for the treatment of gram-negative bacteria.
The potential new isolates BYCDW24 and BYCDW25 (in the order Pleurotheciales) presented selectively antimicrobial activities against the pathogens. BYCDW25 had good inhibitory activity against C. albicans and nearly no inhibition to E. coli, while BYCDW24 showed certain inhibitory effect on E. coli but no effect on C. albicans. As we reported, the BYCDW24 and BYCDW25 shared the same evolutionary relationship except the BYCDW25 harboring one endophytic bacterium. Similar to endobacteria of insects, the endobacteria of fungi showed a range of behaviors from mutualism to antagonism (Bonfante and Desiro, 2017). It has been reported that endobacterium affects the activity of host fungus (Partida-Martinez and Hertweck, 2005; Lackner and Hertweck, 2011). Whether the endobacterium from BYCDW25 played an important role in the biological activity should be further explored.
It is reported that the majority of symbiotic bacteria was identified as Bacillus from the fungus-growing termite (Mathew et al., 2012; Zhu et al., 2012). In our experiments, 66.7% (6/9) symbiotic bacteria from the cultivable fungi were identified as Bacillus, which proved that these dominant bacterial symbionts can also inhabit fungi isolated from the gut of O. formosanus.
In this study, 39 fungi were isolated and identified from O. formosanus, which were belonging to two phyla and four classes. Furthermore, nine bacterial 16S rRNA sequences were obtained from total fungal genomic DNA. All bacterial symbionts belonged to four genera: Bacillus, Methylobacterium, Paenibacillus, and Trabulsiella. The antimicrobial bioassay showed that 25 fungal extracts (64%) exhibited antibacterial activities against at least one of the tested bacterial strains. Furthermore, the secondary metabolites 1 [5-hydroxyramulosin (1a):biatriosporin M (1b) = 2:1] from the Pleosporales sp. BYCDW4 exhibited potent antimicrobial activities against all tested pathogens with the IZD of more than 11.3 mm, which were comparable with those of the positive control. 1-(2,5-Dihydroxyphenyl)-3-hydroxybutan-1-one (2) from the Microdiplodia sp. BYCDW8 showed medium antibacterial activities against B. subtilis and S. aureus. In conclusion, the report showed the diversity of insect symbionts had a potential to develop the resource of new species and antibiotics.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
YZ designed the research and supervised the study. XX, MS, CY, XY, FS, and ZM performed the experiments and analyzed the data. XX and MS wrote the manuscript. All authors revised the manuscript and approved the final version for submission.
This work was supported by the National Natural Science Foundation of China (NSFC) (31770007) and National Key R&D Program (2017YFD0200600).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00300/full#supplementary-material
Adnani, N., Rajski, S. R., and Bugni, T. S. (2017). Symbiosis-inspired approaches to antibiotic discovery. Nat. Prod. Rep. 34, 784–814. doi: 10.1039/c7np00009j
Barbieri, E., Potenza, L., Rossi, I., Sisti, D., Giomaro, G., Rossetti, S., et al. (2000). Phylogenetic characterization and in situ detection of a Cytophaga–Flexibacter–Bacteroides phylogroup bacterium in Tuber borchii Vittad, Ectomycorrhizal mycelium. Appl. Environ. Microbiol. 66, 5035–5042. doi: 10.1128/AEM.66.11.5035-5042.2000
Beemelmanns, C., Ramadhar, T. R., Kim, K. H., Klassen, J. L., Cao, S., Wyche, T. P., et al. (2017). Macrotermycins A-D, glycosylated macrolactams from a termite-associated Amycolatopsis sp. M39. Org. Lett. 19, 1000–1003. doi: 10.1021/acs.orglett.6b03831
Bi, S. F., Guo, Z. K., Jiang, N., Jiao, R. H., Ge, H. M., and Tan, R. X. (2013). New alkaloid from Streptomyces koyangensis residing in Odontotermes formosanus. J. Asian Nat. Prod. Res. 15, 422–425. doi: 10.1080/10286020.2013.76724
Blackwell, M., and Vega, F. E. (2018). Lives within lives: hidden fungal biodiversity and the importance of conservation. Fungal Ecol. 35, 127–134. doi: 10.1016/j.funeco.2018.05.011
Bonfante, P., and Desiro, A. (2017). Who lives in a fungus? The diversity, origins and functions of fungal endobacteria living in mucoromycota. ISME J. 11, 1727–1735. doi: 10.1038/ismej.2017.21
Carr, G., Poulsen, M., Klassen, J. L., Hou, Y., Wyche, T. P., Bugni, T. S., et al. (2012). Microtermolides A and B from termite-associated Streptomyces sp. and structural revision of vinylamycin. Org. Lett. 14, 2822–2825. doi: 10.1021/ol301043p
Chou, J. H., Sheu, S. Y., Lin, K. Y., Chen, W. M., Arun, A. B., and Young, C. C. (2007). Comamonas odontotermitis sp. nov., isolated from the gut of the termite Odontotermes formosanus. Int. J. Syst. Evol. Microbiol. 57, 887–891. doi: 10.1099/ijs.0.64632-0
Douglas, A. E. (2015). Multiorganismal insects: diversity and function of resident microorganisms. Annu. Rev. Entomol. 60, 17–34. doi: 10.1146/annurev-ento-010814-020822
Elahwany, A. M., Ghozlan, H. A., Elsharif, H. A., and Sabry, S. A. (2013). Phylogenetic diversity and antimicrobial activity of marine bacteria associated with the soft coral Sarcophyton glaucum. J. Basic Microb. 55, 2–10. doi: 10.1002/jobm.201300195
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Lackner, G., and Hertweck, C. (2011). Impact of endofungal bacteria on infection biology, food safety, and drug development. PLoS Pathog. 7:e1002096. doi: 10.1371/journal.ppat.1002096
Grigorescu, A. S., Renoz, R., Sabri, A., Foray, V., Hance, T., and Thonart, P. (2018). Accessing the hidden microbial diversity of aphids: an illustration of how culture-dependent methods can be used to decipher the insect microbiota. Microb. Ecol. 75, 1035–1048. doi: 10.1007/s00248-017-1092-x
Hoffman, M. T., and Arnold, A. E. (2010). Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl. Environ. Microbiol. 76:4063. doi: 10.1128/AEM.02928-09
Hong, Q. T., Cheol, K. D., Phan, V. K., Chau, V. M., Nguyen, X. N., Bui, H. T., et al. (2018). Macrolide and phenolic metabolites from the marine-derived fungus Paraconiothyrium sp. VK-13 with anti-inflammatory activity. J. Antibiot. 71, 826–830. doi: 10.1038/s41429-018-0073-8
Landeweert, R., Leeflang, P., Kuyper, T. W., Hoffland, E., Rosling, A., Wernars, K., et al. (2003). Molecular identification of ectomycorrhizal mycelium in soil horizons. Appl. Environ. Microbiol. 69, 327–333. doi: 10.1128/AEM.69.1.327-333.2003
Li, H., Yelle, D. J., Li, C., Yang, M., Ke, J., Zhang, R., et al. (2017). Lignocellulose pretreatment in a fungus-cultivating termite. Proc. Natl. Acad. Sci. U.S.A. 114, 4709–4714. doi: 10.1073/pnas.1618360114
Li, S., Shao, M. W., Lu, Y. H., Kong, L. C., Jiang, D. H., and Zhang, Y. L. (2014). Phytotoxic and antibacterial metabolites from Fusarium proliferatum ZS07 Isolated from the gut of long-horned grasshoppers. J. Agric. Food Chem. 62, 8997–9001. doi: 10.1021/jf502484n
Lu, Y. H., Jin, L. P., Kong, L. C., and Zhang, Y. L. (2016). Phytotoxic, antifungal and immunosuppressive metabolites from Aspergillus terreus QT122 isolated from the gut of dragonfly. Curr. Microbiol. 74, 1–6. doi: 10.1007/s00284-016-1157-y
Luther, A., Urfer, M., Zahn, M., Mueller, M., Wang, S. Y., Mondal, M., et al. (2019). Chimeric peptidomimetic antibiotics against Gram-negative bacteria. Nature 576, 452–458. doi: 10.1038/s41586-019-1665-6
Mathew, G. M., Ju, Y. M., Lai, C. Y., Mathew, D. C., and Huang, C. C. (2012). Microbial community analysis in the termite gut and fungus comb of Odontotermes formosanus: the implication of Bacillus as mutualists. FEMS Microbiol. Ecol. 79, 504–517. doi: 10.1111/j.1574-6941.2011.01232.x
Nishi, O., and Sato, H. (2017). Species diversity of the entomopathogenic fungi Metarhizium anisopliae and M. flavoviride species complexes isolated from insects in Japan. Mycoscience 58, 472–479. doi: 10.1016/j.myc.2017.06.008
Osterhage, C., König, G. M., Jones, P. G., and Wright, A. D. (2002). 5-Hydroxyramulosin, a new natural product produced by Phoma tropica, a marine-derived fungus isolated from the alga Fucus spiralis. Planta Med. 68, 1052–1054. doi: 10.1055/s-2002-35670
Park, J. M., You, Y. H., Back, C. G., Kim, H. H., Ghim, S. Y., and Park, J. H. (2018). Fungal load in Bradysia agrestis, a phytopathogen-transmitting insect vector. Symbiosis 74, 145–158. doi: 10.1007/s13199-017-0494-3
Partida-Martinez, L. P., and Hertweck, C. (2005). Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437, 884–888. doi: 10.1038/nature03997
Procópio, R. E. D. L., Silva, I. R. D., Martins, M. K., Azevedo, J. L. D., and Araújo, J. M. D. (2012). Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 16, 466–471. doi: 10.1016/j.bjid.2012.08.014
Santiago, C., Fitchett, C., Munro, M. H., Jalil, J., and Santhanam, J. (2012). Cytotoxic and antifungal activities of 5-hydroxyramulosin, a compound produced by an endophytic fungus isolated from Cinnamomum mollisimum. Evid Based Compl. Alt. 2012:689310. doi: 10.1155/2012/689310
Shao, M. W., Lu, Y. H., Miao, S., Zhang, Y., Chen, T. T., and Zhang, Y. L. (2015). Diversity, bacterial symbionts and antibacterial potential of gut-associated fungi isolated from the Pantala flavescens Larvae in China. PLoS One 10:e0134542. doi: 10.1371/journal.pone.0134542
Shao, Y., Chen, B., Sun, C., Ishida, K., Hertweck, C., and Boland, W. (2017). Symbiont-derived antimicrobials contribute to the control of the Lepidopteran gut microbiota. Cell Chem. Biol. 24, 66–75. doi: 10.1016/j.chembiol.2016.11.015
Shinzato, N., Muramatsu, M., Matsui, T., and Watanabe, Y. (2007). Phylogenetic analysis of the gut bacterial microflora of the fungus-growing termite Odontotermes formosanus. Biosci. Biotechnol. Biochem. 71, 906–915. doi: 10.1271/bbb.60540
Trainer, M. A., and Charles, T. C. (2006). The role of PHB metabolism in the symbiosis of rhizobia with legumes. Appl. Microbiol. Biotechol. 71, 377–386. doi: 10.1007/s00253-006-0354-1
Van Arnam, E. B., Currie, C. R., and Clardy, J. (2017). Defense contracts: molecular protection in insect–microbe symbioses. Chem. Soc. Rev. 47, 1638–1651. doi: 10.1039/C7CS00340D
Webster, N. S., Cobb, R. E., and Negri, A. P. (2008). Temperature thresholds for bacterial symbiosis with a sponge. ISME J. 2, 830–842. doi: 10.1038/ismej.2008.42
White, M. M., and Lichtwardt, R. W. (2004). Fungal symbionts (Harpellales) in Norwegian aquatic insect larvae. Mycologia 96, 891–910. doi: 10.2307/3762122
White, M. M., Valle, L. G., Lichtwardt, R. W., Siri, A., Strongman, D. B., William, R. T., et al. (2018). New species and emendations of Orphella: taxonomic and phylogenetic reassessment of the genus to establish the orphellales, for stonefly gut fungi with a twist. Mycologia 110, 147–178. doi: 10.1080/00275514.2018.1448198
Xiao, Y. S., Zhang, B., Zhang, M., Guo, Z. K., Deng, X. Z., Shi, J., et al. (2017). Rifamorpholines A–E, potential antibiotics from locust-associated actinobacteria Amycolatopsis sp. Hca4. Org. Biom. Chem. 15, 3909–3916. doi: 10.1039/C7OB00614D
Zhang, Y. L., Ge, H. M., Zhao, W., Dong, H., Xu, Q., Li, S. H., et al. (2008). Unprecedented immunosuppressive polyketides from Daldinia eschscholzii, a mantis-associated fungus. Angew Chem. Int. Ed. 120, 5823–5826. doi: 10.1002/ange.200801284
Zhang, Y. L., Zhang, J., Jiang, N., Lu, Y. H., Wang, L., Xu, S. H., et al. (2011). Immunosuppressive polyketides from mantis-associated Daldinia eschscholzii. J. Am. Chem. Soc. 133, 5931–5940. doi: 10.1021/ja110932p
Zhou, Y. H., Zhang, M., Zhu, R. X., Zhang, J. Z., Xie, F., Li, X. B., et al. (2016). Heptaketides from an endolichenic fungus Biatriospora sp. and their antifungal activity. J. Nat. Prod. 79, 2149–2158. doi: 10.1021/acs.jnatprod.5b00998
Keywords: Odontotermes formosanus, fungal diversity, bacterial symbionts, antimicrobial activities, secondary metabolites
Citation: Xu X, Shao M, Yin C, Mao Z, Shi J, Yu X, Wang Y, Sun F and Zhang Y (2020) Diversity, Bacterial Symbionts, and Antimicrobial Potential of Termite-Associated Fungi. Front. Microbiol. 11:300. doi: 10.3389/fmicb.2020.00300
Received: 29 October 2019; Accepted: 10 February 2020;
Published: 13 March 2020.
Edited by:
Santiago Gutierrez, Universidad de León, SpainReviewed by:
M. Oves, King Abdulaziz University, Saudi ArabiaCopyright © 2020 Xu, Shao, Yin, Mao, Shi, Yu, Wang, Sun and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenchuan Mao, bWFvemhlbmNodWFuQGNhYXMuY24=; Yinglao Zhang, emhhbmd5bEBhaGF1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.