
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 February 2020
Sec. Infectious Agents and Disease
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00288
This article is part of the Research Topic Recent Advances in the Controversial Human Pathogens Pneumocystis, Microsporidia and Blastocystis View all 12 articles
Blastocystis is a genetically diverse microbial eukaryote thriving in the gut of humans and other animals. While Blastocystis has been linked with gastrointestinal disorders, its pathogenicity remains controversial. Previous reports have suggested that one out of six humans could be carrying Blastocystis in their gut, while the numbers could be even higher in animals. Most studies on Blastocystis are either exclusively targeting the organism itself and/or the associated prokaryotic microbiome, while co-occurrence of other microbial eukaryotes has been mainly ignored. Herein, we aimed to explore presence and genetic diversity of Blastocystis along with the commonly occurring eukaryotes Cryptosporidium, Eimeria, Entamoeba and Giardia in the gut of asymptomatic animals from two conservation parks in the United Kingdom. Building upon a previous study, a total of 231 fecal samples were collected from 38 vertebrates, which included 12 carnivorous and 26 non-carnivorous species. None of the animals examined herein showed gastrointestinal symptoms. The barcoding region of the small subunit ribosomal RNA was used for subtyping of Blastocystis. Overall, 47% of animal species were positive for Blastocystis. Twenty six percent of samples carried more than one subtypes, including the newly identified hosts Scottish wildcat, bongo and lynx. Fifty three percent of samples carried at least another microbial eukaryote. Herewith, we discuss potential implications of these findings and the increasingly blurred definition of microbial parasites.
The gut microbiome comprises the collective genomes of microbial symbionts and is composed of bacteria, fungi, viruses and protists within the gastrointestinal (GI) tract of a host (Blaser, 2014). Though literature associated with bacterial microbiota is increasing, studies on the rest of the microbiome components are just beginning to surface. Historically, presence of protists in the gut has been considered as parasitism, thus these microbial eukaryotes have been subject to rigorous elimination in both humans and other animals (Parfrey et al., 2011). Despite this, current data demonstrates that some protists are more common than previously thought, raising the possibility of commensalistic or even mutualistic roles in the gut ecosystem (Lukes et al., 2015; Chudnovskiy et al., 2016). In this regard, no other protist has been studied more extensively than the anaerobic stramenopile Blastocystis. Its prevalence in humans has been estimated to a staggering one billion (Stensvold, 2012). Though a similar estimation for animals is not available, data from numerous animal studies covering broad range of hosts strongly suggest that colonization rate in animals is likely higher than in humans.
Blastocystis is extremely heterogeneous genetically (Gentekaki et al., 2017). Based on the SSU rRNA gene, Blastocystis from avian and mammalian hosts is divided into 17 subtypes, which are considered separate species (Stensvold and Clark, 2016b). Nonetheless, there are many sequences originating from ectothermic hosts that do not belong to any of the designated subtypes (Yoshikawa et al., 2016). The various subtypes of Blastocystis do not seem to be host-specific. For example, ST1 to ST9 have been identified in humans, but also in other hosts (Stensvold and Clark, 2016b). The exception seems to be ST9, which has yet to be identified in a non-human host (Stensvold and Clark, 2016a). ST10 to ST17 have been found only in animals so far, with the exception of ST12, which has also been identified in humans (Ramirez et al., 2016).
Though Blastocystis has been found in individuals with gastrointestinal symptoms, asymptomatic carriage is also common (Scanlan et al., 2014; AbuOdeh et al., 2016; Nieves-Ramirez et al., 2018; Yowang et al., 2018; Mardani Kataki et al., 2019). In vitro experiments using cell lines have shown the invasion potential of some strains/subtypes of Blastocystis (Puthia et al., 2008; Wawrzyniak et al., 2012), with no evidence to date that this also occurs in vivo (Clark et al., 2013). Experimental infections in mouse models have been achieved only after an inoculum of considerable size (up to 4 × 107) is administered (Moe et al., 1997; Elwakil and Hewedi, 2010). Recent studies on animals have shown that Blastocystis exists asymptomatically in a broad array of hosts (Betts et al., 2018; Wang et al., 2018b). Collectively, these findings highlight the uncertainty surrounding pathogenicity status of Blastocystis in both humans and other animals.
Presence of multiple Blastocystis subtypes in humans is not often reported (Whipps et al., 2010; Scanlan and Stensvold, 2013). To our knowledge, only a few reports have demonstrated mixed colonization in animals (AbuOdeh et al., 2016; Cian et al., 2017; Betts et al., 2018). In our previous work, Betts et al. (2018) examined Blastocystis distribution in a wildlife park in the United Kingdom, and identified various genetic isolates in a number of different animals across the park. Importantly, we also demonstrated presence of up to four subtypes in healthy captive animals (Betts et al., 2018). At that time, while microscopically screening the fecal samples, we noted presence of other protists as well. Most previous studies have been focused on identifying single target protist species, but only a few have focused on co-occurrence of multiple microbial eukaryotes in the gut. Herein, we have expanded the study area to include an additional wildlife park. We aimed to further characterize presence of Blastocystis isolates along with additional microbial eukaryotes across a broad range of taxa in the two parks.
Two zoos situated in the Southeast, United Kingdom were sampled in this study: 1) Wildwood Conservation Park, Herne Bay, Kent, United Kingdom (51°19′54.1″N 1°07′10.1″E). This is a small conservation park housing native vertebrate and invertebrate species from the United Kingdom and mainland Europe with the exception of the red-necked wallaby (Macropus rufogriseus). The park is actively involved in breeding and re-introduction programs for native animals including the European water vole (Arvicola amphibious) and Scottish wildcat (Felis silvestris silvestris), and 2) Howletts Wild Animal park, Canterbury, Kent, United Kingdom (51°16′11.8″N 1°09′25.0″E). This is a large zoo with over 400 animals from 50 vertebrate and invertebrate species from across the globe. The zoo has a large primate collection, including one of the largest family groups of western lowland gorilla (Gorilla gorilla gorilla) in the world. The zoo is involved in a number of re-introductory schemes, mainly into national parks. Both zoos closely monitor animal health, through licensed veterinarians once a month. To our knowledge none of the animals in this study presented symptomatic gastrointestinal diseases or diarrhea.
A total of 231 fresh fecal samples have been collected from 38 vertebrate species between July 2016 and March 2019 (Supplementary Table S1). One hundred and eighteen samples were from a previous collection (Accession numbers of Blastocystis positive samples: MF186640-MF186709; Betts et al., 2018) and the rest were newly collected. Sixty-seven of these samples were from nine vertebrate species collected from Howletts Zoo between November 2017 and February 2019 and the remaining samples were collected form 31 species at Wildwood. Samples from gray wolf (Canis lupus) and European bison (Bison bonasus) were collected from both zoos. Sampling covered a total of 33 mammalian species, four bird species and one reptile (Supplementary Table S1). In both zoos, a minimum of one fecal sample was collected from each enclosure. In enclosures where more than one animal resided, between two and five samples were collected, each of which was considered as individual sample. For some water voles (Arvicola amphibious), a number of repeat collections were carried out over the course of 12 months (Supplementary Table S1). Fresh fecal samples were collected in the morning either before or shortly after enclosures were cleaned. For some animals, including avian species and the reptile; where age of fecal sample is difficult to determine, multiple samples were collected. Zookeepers supervised all collections.
Once collected, fecal samples were stored at 4°C in sterile falcon tubes within 1 h of collection until DNA extraction. In some instances, heat fixed slides were prepared. Within an hour of sampling, a small amount of fecal sample from the water voles and other randomly selected animals were separately inoculated in four sterile falcon tubes containing the following media: two tubes containing modified LYSGM [16⋅07 mM potassium phosphate dibasic, 2⋅94 mM potassium phosphate monobasic, 128⋅34 mM sodium chloride, 2⋅5 g L–1 yeast extract, 0⋅5 g L–1 liver extract, 5% adult bovine (Sigma)/horse serum (Gibco); modified TYSGM-9, without mucin (Diamond, 1982)1 ], two tubes of TYM (22⋅2 g L–1 trypticase peptone, 11⋅1 g L–1 yeast extract, 16⋅23 mM maltose, 9⋅17 mML-cysteine, 1⋅26 mM L-ascorbic acid, 5⋅1 mM potassium phosphate dibasic, 6⋅53 mM potassium phosphate monobasic) (Diamond, 1957, 1983) enriched with 5% fetal bovine serum (FBS; Sigma) and 2 tubes with 0⋅5% Liver Digest (LD) medium (0⋅5 g L–1 Oxoid liver extract). The tubes were incubated at 35°C. samples were examined for Blastocystis under the microscope every 3–5 days. After initially leaving the cultures for 2 weeks, they were subcultured every 10 days.
Genomic DNA was extracted directly from a minimum of 250 mg of fresh fecal sample or culture pellet using the Microbiome DNA Purification Kit Purelink (Fisher, United Kingdom) to the manufacturer’s instructions. DNA was eluted in 100 μl elution buffer and aliquotted. The working stock was stored at −20°C, while the rest was placed at −80°C for long-term storage. Extracted DNA was used for the polymerase chain reaction (PCR) with specific primers targeting regions of interest (Supplementary Table S2). PCR was carried out using the 2X PCRBIO Taq DNA Polymerase (PCRBIOSYSTEMS). Reagents per 25 μl reaction were as follows: PCRBIO Taq mix, 0.4 μM forward primer, 0.4 μM reverse primer, 19 μl nuclease free water and 2 μl DNA (ranging in concentration 10–50 ng/μl). Details of amplification conditions for all species in this study are provided in Table 1.
Fragments amplified to the correct size were excised and extracted using the Thermo Scientific GeneJET Gel Extraction Kit (following manufacturer’s instructions) purified gel extracts were eluted in 30–50 μl of elution buffer. If PCR reactions were left for 7 days before ligation, a polyadenylation reaction was carried out with the following protocol: per reaction 0.25 μl GoTaq DNA Polymerase (Promega), 7 μl Gel extraction, 2 μl 5X GoTaq Buffer (Promega), 0.5 mM MgCl2, 2.5 mM dATP (Promega) and 0.3 μl nuclease water at 72°C for 30 min. 1.5 μl of polyadenylation product or gel extract was cloned using the pGEM-T easy vector system I (Promega) following manufacturer’s protocol. Between 3 and 10 colonies per transformation were grown in 5 ml overnight cultures. Plasmid DNA was extracted using the GeneJET Plasmid Miniprep Kit (following manufacturer’s instructions). Before sequencing, a restriction digest using EcoRI (Promega) was carried out to confirm fragment insertion, per 10 μl reaction, 0.25 μl EcoRI, 5 μl miniprep elution, 1 μl 10X buffer H and 3.75 μl dH20 was incubated at 37°C for 2 h and visualized on a 1.5% agarose gel. Positive samples were sequenced using both the T7 or SP6 universal primers by Eurofins, United Kingdom.
Raw reads were trimmed to remove remaining vector fragments and unambiguous bases at the ends of the reads. BLAST search using the newly obtained sequences against the non-redundant (nr) database was used to identify sequence positive clones. A dataset was assembled including all new sequences in addition to reference sequences encompassing the breadth of diversity of Blastocystis and an alignment was carried out using MAFFT v.7 (Katoh and Toh, 2010). Alignment contained four outgroup taxa for a total of 171 taxa. After aligning with MAFFT, ambiguous positions were masked with trimAl (Capella-Gutierrez et al., 2009). Following trimming, the alignment contained 1326 positions. A maximum likelihood tree was constructed using the RAxML software version 8 (Stamatakis, 2014, 2015) on the online platform CIPRES (Miller et al., 2010).2 For each dataset bootstrap support was calculated from 1000 replicates.
Blastocystis was cultured in tubes containing both types of media. Isolates from fox, lynx, wallaby, elk and otter grew at 35°C, while the ones from water voles grew at room temperature (Supplementary Figure S1). We were unable to establish cultures from other hosts.
Building upon sampling from a previous study, a total of 231 fecal samples from 38 vertebrate species were examined. It should be noted that the percent positive percentages herein are the minimum since PCR amplification rather than qPCR was used. Blastocystis was detected in 18/38 species (47%). A total of 255 clones were sequence positive for Blastocystis; 184 of these clones were from the current study. Of the 12-carnivorous species only three (pine marten, lynx and Scottish wild cat) were sequence positive for Blastocystis (25%, Table 2). There were no sequence positives for badger, European brown bear, otter, polecat, red and arctic foxes, stoat, gray and Iberian wolves, despite having multiple samples from different time points from these species. For non-carnivorous species, 15/26 (58%) were sequence positive, while barnacle and pink footed geese, four lined snake, hedgehog, water shrew, raven, red billed chough, black and brown rats, pied tamarind and black rhinoceros were negative (Table 2). Blastocystis was found in all artiodactyl species examined, but not all fecal samples were sequence positive. Sequence positive results for samples were as follows: Carnivora 3/50 (6%); Artiodactyla 20/36 (56%); Anseriformes 0/2 (0%); Squamata 0/1 (0%); Eulopotyphia 0/0 (0%); Passeriformes 0/4 (0%); Rodentia 29/81 (36%); Diprotodontia 2/5 (40%); Primates 27/43 (63%); Perissodactyla 0/0 (0%).
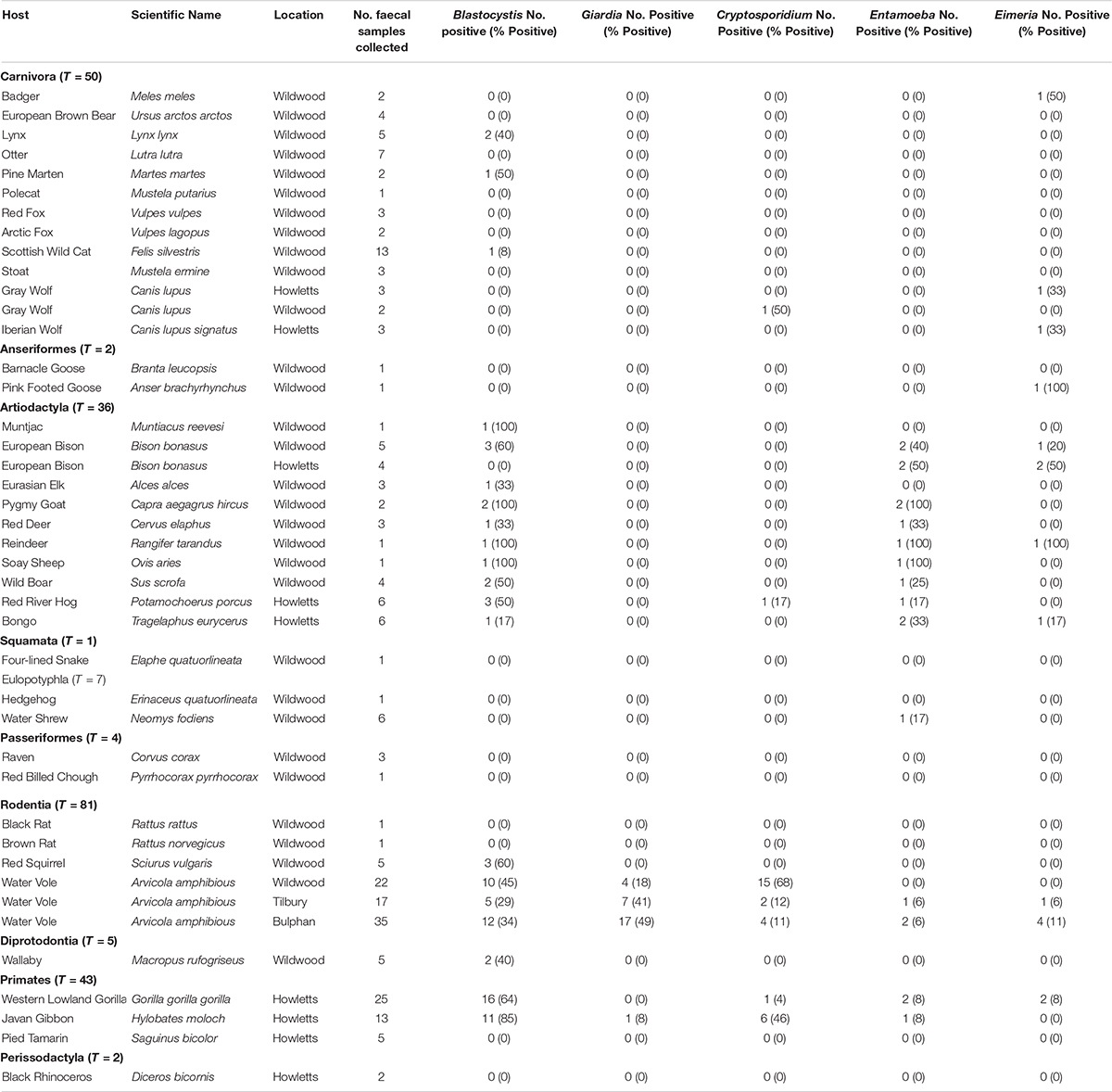
Table 2. Prevalence of Blastocystis, Giardia, Cryptosporidium, Entamoeba, and Eimeria in study animals.
Regarding subtypes from cultures, we only looked at water voles as their cultures were numerous. We found only ST1 and ST4, while the rest of the STs found in the faces were not recovered.
In total, 10 known subtypes were detected: ST1, ST2, ST3, ST4, ST5, ST8, ST10, ST13, ST14, and ST15 (Table 3). Of those, ST2, ST3, ST8 and ST15 were not found in our previous collection. Subtype 4 was the most commonly isolated, found in 83/255 (33%) clones across 11 species. This was followed by ST2, isolated from 80/255 samples (31%); ST10 27/255 (11%); ST1 26/255 (10%); ST14 17/255 (7%); ST5 13/255 (5%); ST3 and ST15 4/255 (2%); ST13 1/255 (0.4%). Three sequences grouped with the B. lapemi clade.
All artiodactyls, except for the European Bison (Bison bonasus) housed at Howletts, had at least one positive ST identification. The subtypes found in this group coincided with published data with most isolates belonging to ST5, ST10 and ST14. ST5 was present in 6/36 (17%) samples; ST10 in 10/36 (28%) samples; ST14 in 7/36 (19%); ST4 in 2/36 (6%) samples; ST1 and ST13 both 1/36 (3%). 5/36 samples exhibited co-occurrence with two or more STs. The bongo calf (Tragelaphus eurycerus) –shared the same STs (10 and 14) with its mother as opposed to the father, who is housed separately and in whom we only detected ST14.
Eighty-one samples from four species belonging to the order Rodentia are presented in this study. Brown rat (Rattus norvegicus) and black rat (Rattus rattus) yielded no Blastocystis positive isolates. ST2 and ST4 were detected in three samples were from Red squirrel (Sciurus vulgaris). Water vole (Arvicola amphibious) samples accounted for a total of 26/81 (32%) positive Blastocystis samples and 88 positive clones. a total of 74 water vole samples have been taken to date, 26/74 (35%) are sequence positive for one or more STs. The large sample number is due to the sizable cohort in the study, which included repeat sampling over an extended period of time. Three groups of water vole were sampled: captive voles from Wildwood (22 samples) and wild caught voles from two areas in Essex, United Kingdom; Tilbury (17 samples) and Bulphan (35 samples). The wild caught voles were routinely screened over the course of 10–12 months. Amongst sequence positive samples the captive voles had a total of 30 positive clones obtained from 9/22 (41%) positive samples; Tilbury voles had 28 positive clones from 5/17 (29%) samples, while Bulphan voles yielded 29 clones from 11/35 (31%) positive samples. ST4 was the most commonly identified across both captive and wild voles, representing 76/88 (86%) of the clones and 23/26 (88%) samples. ST1, ST15 and a subtype placing with B. lapemi were all identified in two samples, ST1 and B. lapemi clade ST were isolated in captive voles, whereas ST15 was found in one wild vole across repeat sample time points. ST10 and ST14 were identified in one sample each from captive voles. Co-occurrence of two or more STs was identified in four voles, all of which were captive. ST4 was present in all of these co-occurrence instances along with ST1, ST10, ST14, and B. lapemi clade ST.
A total of 43 non-human primate (NHP) samples were collected from Howletts zoo as follows: 25 gorillas (Gorilla gorilla gorilla) samples from four family groups ranging in size (G1, G3 G4 and G5) and one individual were collected across two collection times, 13 Javan gibbon (Hylobates moloch) samples from individuals across seven groups ranging in size (A-G) and five pied tamarin (Saguinus bicolor) samples from group enclosures. Of these samples, 16/25 gorillas (64%); 11/13 (85%) Javan gibbons were sequence positive for at least one ST, while no Blastocystis was detected in any of the pied tamarins (Table 4). In terms of clones, for the gorillas, 64 positive clones were sequenced, of which 45/64 (70%) were ST2; 9/64 (14%) ST1; 8/64 (13%) ST3; and 2/64 (3%) were ST5. There were no notable differences observed among family groups. Specifically, all family groups had a relatively high incidence of ST2, while ST5 was only reported from family group 5. Co-colonization with two STs was seen in four of the gorilla samples (Table 4). The Javan gibbons represent one of the highest proportions of sequence positive clones for Blastocystis STs, from the 11 positive samples, 45 clones were sequenced. ST1 represented 18/45 (40%) of these clones; ST2 17/45 (38%); ST3 and ST5 both 4/45 (9%); ST8 1/45 (2%); ST15 1/45 (2%). Of the gibbon groups, Group F was the only one to not have any sequence positive data across two sample collections. Of all the groups, Group G was only sampled from once as its members were released to the wild between collections. Differences were observed among groups between the sample collections. For example, ST5 and ST15 were detected in Group B upon first collection, yet in the second ST1 and ST2 were found.
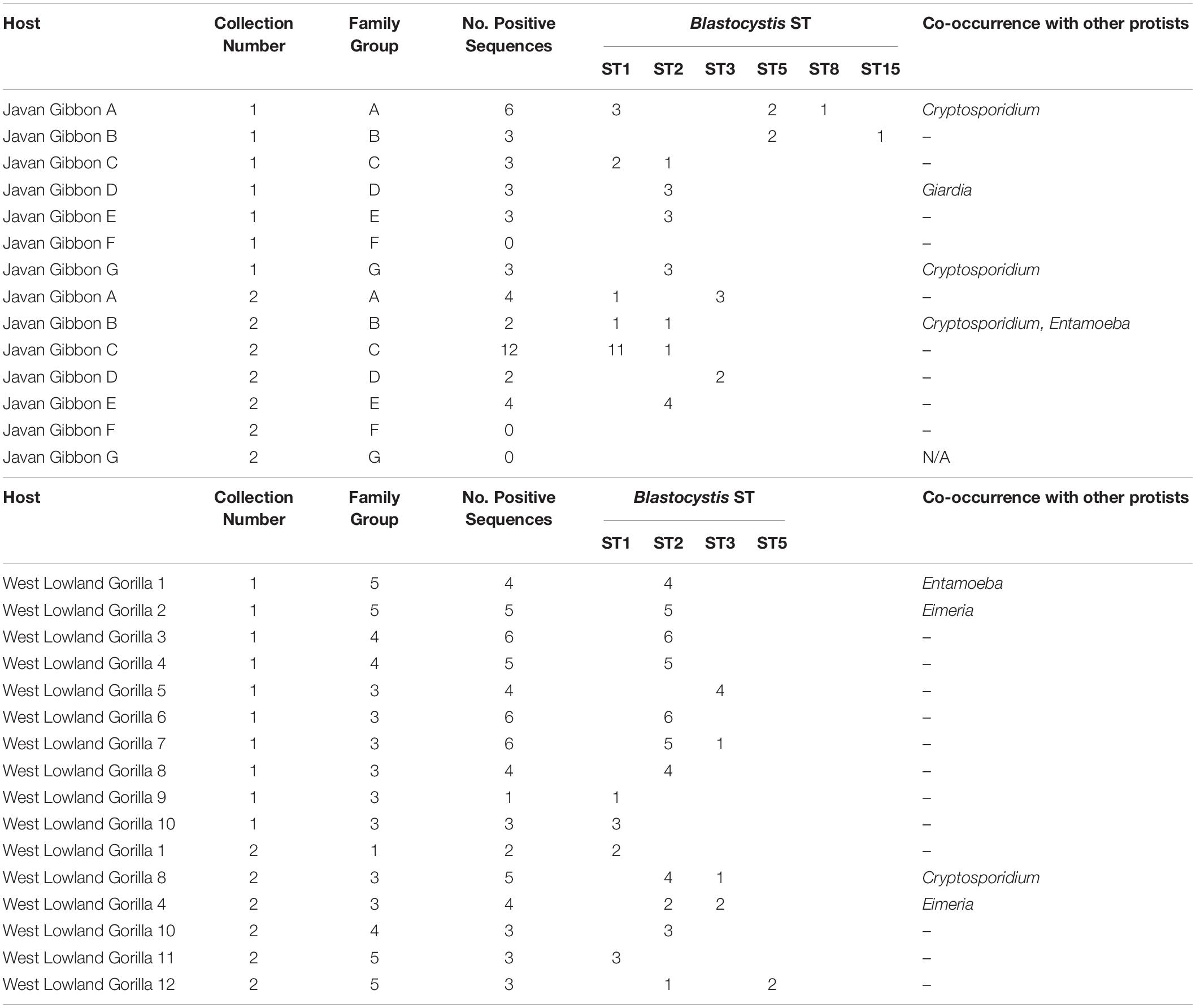
Table 4. Blastocystis subtyping in captive Javan gibbons (Hylobates moloch) and West Lowland gorillas (Gorilla gorilla gorilla) from two sample collections with co-occurrence of other protists within sampled groups.
In general, differences in ST distribution and prevalence are seen between the two zoos, the most obvious attribute to this is the differences in sampled taxa. Samples from Wildwood were comprised largely of members from the orders Rodentia, Artiodactyla and Carnivora, with Water voles and Scottish wild cats being sampled several times. Samples from Howletts were mainly from NHPs and other members of the Artiodactyla. The European bison and gray wolf were the only species sampled across both parks. Notably, Blastocystis was not isolated from any wolf or bison samples from Howletts, even though the bison housed at Wildwood and Howletts are related. The differences in ST distribution among the parks reflect the taxa housed within. Wildwood comprises largely of ST4 and ST10, STs commonly associated with rodents and hooved animals, whereas ST2, ST1 and ST5 are isolated on Howletts and are commonly associated with NHPs.
In total, 25 of the Blastocystis positive samples harbored more than one subtype; specifically, two subtypes were detected in 22 samples, three subtypes in two samples, while one sample contained four subtypes.
Newly generated sequences have been submitted to GenBank (MN526748- MN526930).
Fecal samples were screened for Cryptosporidium, Eimeria, Entamoeba, Giardia and Isospora. Of the 81 Blastocystis positive samples, 43 (53%) harbored at least one of the above-mentioned protists in addition to Blastocystis (Table 5). Of those, 35 samples had one additional protist as follows: 14 cases from samples of Rodentia (all water voles), 13 from Artiodactyla (three from European bisons, three from bongos, two from Red river hogs, two from pygmy goats, one from wild boar, one from soay sheep, and one from red deer) and eight from NHPs (four from gorillas and three from Javan gibbons). Seven samples carried Blastocystis and two other protists: four Rodentia (all from water voles), two NHPs (both from Javan gibbons), and one from Artiodactyla (reindeer). A single sample from water vole was found with three other protists. The widest range of host species where co-occurrence was noted in the Artiodactyla. Cryptosporidium was detected in 31 (13%) samples and co-occurred with Blastocystis in 19 cases (61%); 22 (9%) samples were positive Entamoeba, 14 of which (64%) were found with Blastocystis; 29 (12%) samples harbored Giardia which co-occurred with Blastocystis in 10 cases (35%); 17 (7%) samples were positive for Eimeria, while nine were found with Blastocystis. Of the three (1%) Isospora positive samples, none co-occurred with Blastocystis.
All Blastocystis sequences grouped together with maximum support (100BS) (Figure 1). Newly acquired sequences belong to ST1, ST2, ST3, ST4, ST5, ST8, ST10, ST14, ST15, and the B. lapemi clade. In agreement with previous studies, ST15, ST16 and ST17 along with sequences originating from ectotherms placed in the most basal positions (Alfellani et al., 2013; Yowang et al., 2018). Subtypes 3, 4, 8, and 10 grouped together, while subtypes 7, 9 and 6 formed a clade. Two of the water vole sequences grouped within the clade formed by B. lapemi and B. pythoni. Subtypes 1, 2 and 11 grouped together and sister to the clade formed by subtypes 5, 12, 13, and 14.
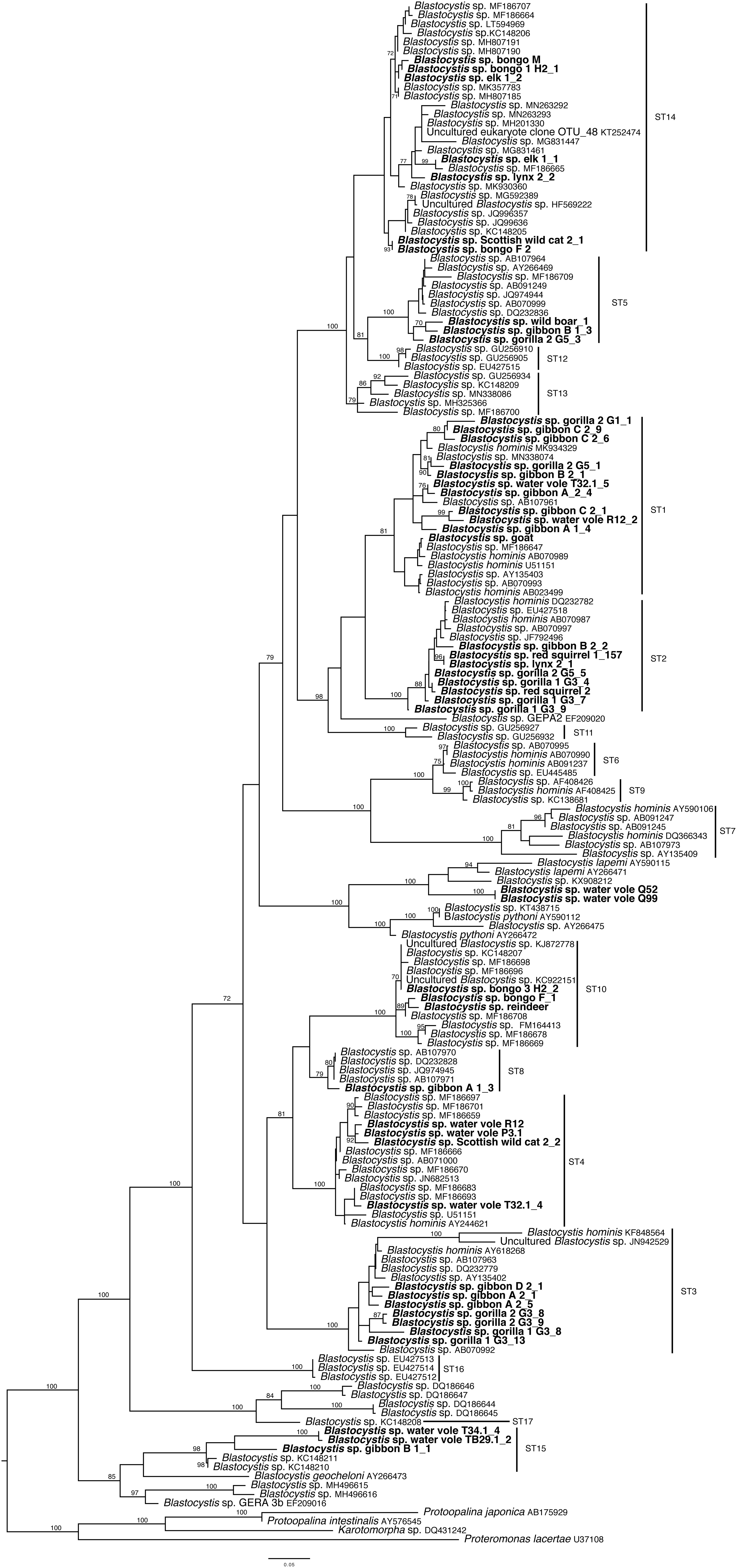
Figure 1. Maximum likelihood phylogenetic tree inferred from 171 sequences and 1326 sites using RAxML v. 8. New sequences are in bold lettering. Numerical values indicate bootstrap support values and only those of over 70 are shown.
Animals from 38 species from two animal parks in the United Kingdom were sampled over a period of 3 years. Eighty-two samples from 47% of all animal species were sequence positive for Blastocystis. Of those 82, (21/82) 26% were found to harbor more than one ST, while 53% also harbored other protists. Blastocystis was present in animals from both parks. As expected, ST4 was dominant in rodents, whereas ST10 and ST14 dominated in artiodactyls. In primates, ST1 and ST2 were dominant. We reported Blastocystis presence in the Lynx and the Scottish wild cat for the first time. Both of these animals are carnivorous. Our study confirms previous findings on reduced presence and often absence of Blastocystis in carnivores and high prevalence in artiodactyls (Alfellani et al., 2013; Cian et al., 2017, Zhao et al., 2017). It is well known that dietary, behavioral and environmental factors shape bacterial communities, though this has yet to be shown for microbial eukaryotes. In that vein, a possible explanation for the above observation could be that captive carnivores consume a diet consisting of almost exclusively refrigerated meat, which is devoid of other eukaryotes. This considerably reduces contamination. Nonetheless, a recent study on free-living carnivorous animals confirmed presence of Blastocystis in only 1.6% of hosts (Calero-Bernal et al., 2019), suggesting that additional factors might account for the low prevalence. Artiodactyls are herbivorous animals that consume exclusively fiber, while carnivores consume only animal protein. Thus the two also differ considerably in the overall structure and physiology of their respective gastrointestinal tracts. Both diet and physiology likely contribute to microbiota composition, and as a result, the microbial communities of artiodactyls and carnivores differ considerably (Sanders et al., 2015; Nishida and Ochman, 2018). In general, herbivores, to which artiodactyls belong, harbor high microbial diversity, while carnivores encompass the least diverse microbial communities amongst mammals (Nishida and Ochman, 2018). High microbial diversity and specific microbial profiles are linked to presence of Blastocystis in human studies though a causative link has yet to be established (Andersen et al., 2015; Audebert et al., 2016; Iebba et al., 2016; O’Brien Andersen et al., 2016; Beghini et al., 2017; Forsell et al., 2017; Nieves-Ramirez et al., 2018; Tito et al., 2019). A similar result has also been obtained from a study focusing on wild chimpanzees (Renelies-Hamilton et al., 2019). Given the high prevalence of Blastocystis in artiodactyls it would be interesting to explore whether such specific profiles exist in these animals as well.
As in our previous study (Betts et al., 2018), we identified multiple subtypes of Blastocystis in the same host. In addition to the elk, pygmy goat, red deer and water vole hosts bearing multiple subtypes, we add the Scottish wildcat (ST4 and ST14), bongo (ST10, ST14), and lynx (ST2, ST14). Previous reports also noticed presence of multiple STs in animals (Fayer et al., 2012; Badparva et al., 2015; AbuOdeh et al., 2016). Cian et al., documented several instances of mixed colonization of subtypes (11%), especially in primates and artiodactyls (Cian et al., 2017), while Wang et al., reported mixed colonization in 58% of a pig population (Wang et al., 2014). Collectively these data strengthen previously raised hypotheses that occurrence of multiple subtypes in animals is not unusual, but rather common (Fayer et al., 2012; Betts et al., 2018). Thus, a logical extension of this study would be to disentangle whether co-occurring subtypes occupy distinct functional niches in the complex gut ecosystem, a direction that has also been suggested by Beghini et al. (2017).
Co-occurrence of Blastocystis with Entamoeba, Giardia, Cryptosporidium and Eimeria in multiple animal species across the two parks was also examined. Most previous studies have either looked for multiple parasites from single animal species or have targeted one microbial eukaryote in various hosts (Fayer et al., 2012; Parsons et al., 2015; Enriquez et al., 2016, 2019; Jacob et al., 2016). Herein, Blastocystis did not co-occur with other protists in any of the carnivores, even though we did observe co-occurrence of Cryptosporidium and Eimeria in gray and Iberian wolves. The case of artiodactyls is particularly notable. Eight out of ten artiodactyls that were Blastocystis positives co-occurred with an Entamoeba species. Out of these, three co-occurred with Blastocystis, Entamoeba and Eimeria (European bison, reindeer, bongo), while one animal had Blastocystis, Entamoeba and Cryptosporidium (red river hog). Significantly, none of the animals exhibited diarrheal episodes or other obvious gastrointestinal symptoms as confirmed by zookeepers and licensed veterinarians. Typically, microbial eukaryotes in animals are identified and reported upon onset of gastrointestinal symptoms. Herein, we sampled and detected gut protists before presentation of symptoms, though the possibility that some of the animals might have had symptoms before they were brought into the parks cannot be excluded. Asymptomatic carriage of a single or multiple protists in animals is not uncommon and the concern of zoonotic transmission has often been articulated (Fayer et al., 2012; Cian et al., 2017; Desoubeaux et al., 2018; Udonsom et al., 2018; Wang et al., 2018a; Enriquez et al., 2019). In case of zoonosis, detecting the reservoir is difficult as there is no reason to check the original host for presence of pathogens. The level and type of interaction among Blastocystis, other microbial eukaryotes (including fungi) and the rest of the host microbiome is unclear. Future animal studies should focus on exploring the eukaryotic component of the gut microbiome rather than targeting individual microbial species, in order to shed light on the role of eukaryome as a whole in the gut ecosystem. Combination of in vitro and in vivo targeted metagenomics and metabolomics approaches along with network analysis will greatly increase our understanding of these issues.
The case of Blastocystis is of interest. In the past, co-occurrence of Blastocystis with pathogens in stool samples of humans with gastrointestinal symptoms was likely one of the reasons for its controversial pathogenicity. Since adaptation of the subtyping system, the argument has been framed around specific subtypes or strains being pathogenic. Nonetheless, in a rather anthropocentric approach, assessment of the pathogenic potential of Blastocystis has focused primarily on humans and the “human” subtypes ST1 to ST9, while non-human metazoans and the rest of the subtypes have been largely overlooked. Moreover, the health status of animal subjects in many studies is not reported. When animals happen to have diarrhea the subtype present in these animals is often not mentioned, rather percent overall occurrence of individual subtypes is emphasized. Consequently, Blastocystis pathogenicity in animals is not well understood. It would be interesting to see whether any of the animals sampled herein will present any symptoms in the future. To that end, we have communicated with the zoo stuff to inform us in case symptoms develop in any of these animals.
To determine to which subtype the new sequences belonged, phylogenetic analysis was performed. Two of the newly generated sequences, both of which come from water voles, did not group with any of the known subtypes, but as sister to Blastocystis lapemi. There are two sequences designated as B. lapemi in the database, both of which originated from sea snakes (Yoshikawa et al., 2004; Noel et al., 2005). A third sequence that also groups within the clade and is genetically distinct comes from a monitor lizard. Therefore, either B. lapemi is not limited to sea snakes or all these sequences represent different species. In the absence of a culture and a full SSU rRNA sequence we designate those three sequences as Blastocystis sp. Four sequences – one coming from gibbon and three from water voles – group with ST15. Water vole is a newly reported host for ST15. Previously, Betts et al. (2018) had reported a potentially novel subtype, but had refrained from establishing it as such since the whole sequence was not available. Since then, several studies focusing on animals have contributed significantly toward populating previously isolate-sparse subtypes. As a result, the phylogenetic landscape of Blastocystis is changing. Expanded taxon sampling including several additional ST14 isolates from the database and from the current study has shown that ST14 is now divided into three distinct subclades, with new isolates populating all three. The previously suspected novel sequence (Betts et al., 2018) groups in one of the three. Thus, either ST14 has high intra-subtype divergence or it must be separated to at least two maybe even three subtypes. Nonetheless several subtypes harbor a high degree of genetic diversity except for ST4, which is the least genetically diverse (Stensvold and Clark, 2016b; Beghini et al., 2017). Given the variable degree of intra-subtype diversity, caution should be taken when establishing new subtypes. Genetic diversity within subtypes should be properly assessed. Commonly, closely related sequences from specific subtypes are included in the analysis, while more divergent representatives are not, leading to establishment of erroneous STs. Finally, the whole SSU rRNA region should be sequenced and phylogenies should include the breadth of Blastocystis diversity. Consistent approaches to subtyping Blastocystis will further elucidate the variety of subtypes that exist and their associations with specific hosts (El Safadi et al., 2016; Betts et al., 2018; Robertson et al., 2019).
In the current study, we employed cloning and demonstrated the presence of multiple subtypes within a single host and also presence of multiple eukaryotes within a host. We would like to emphasize that DNA was mainly extracted directly from fresh fecal samples without culturing in Jones media. Even though we still cannot guarantee that all subtypes present in the stool samples were amplified, selective pressures and constraints that culturing imposes were circumvented. In working with fecal samples other issues came to light. One of them is primer specificity. Eukaryotic microbe primers amplify the microbe of interest provided it is there. Our screening showed that all pairs of specific primers and most especially those of Blastocystis and Entamoeba also amplified several other eukaryotes. For example, approximately ∼40% of the sequenced clones did not correspond to Blastocystis specific sequences. Development of new Blastocystis-specific primers that will amplify a large fragment of the SSU rRNA gene are urgently needed, since this will reduce the costs of cloning and sequencing.
Herein we have identified asymptomatic carriage of multiple microbial eukaryotes in a number of animal species. This is defined as presence of multiple Blastocystis subtypes in single hosts and in many cases these co-occur with up to three other microbial eukaryotes. Given the higher prevalence of overlap of microbial eukaryotes in animals and especially in artiodactyls, the latter might provide a model not only for studying the spectrum of parasitism (Rueckert et al., 2019), but also the associated microbial communities and how those relate with the different parts of this spectrum.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
EB carried out the collections, culturing, collected and analyzed all the data, and wrote a first draft of the manuscript. AT and EG directed research, planned experiments, analyzed data, and wrote the manuscript.
This research was supported by BBSRC research grant (BB/M009971/1) to Dr. AT. Dr. EG is supported by The Thailand Research Fund (RSA6080048). We would like to thank the University of Kent for sponsoring ADT’s trip to Thailand under a GCRF grant. EB was supported by a Ph.D. studentship from the School of Biosciences at the University of Kent.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank members of the Dr. Tsaousis laboratory, Adele Thomasz, Vicki Breakell, Angus I. Carpenter and Hazel Ryan from the Wildwood Trust and Adrian Harland from the Howllets Zoo and the zookeepers from both zoos for assisting with sample collection and accommodating us during our visitations.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00288/full#supplementary-material
AbuOdeh, R., Ezzedine, S., Samie, A., Stensvold, C. R., and ElBakri, A. (2016). Prevalence and subtype distribution of Blastocystis in healthy individuals in Sharjah, United Arab Emirates. Infect. Genet. Evol. 37, 158–162. doi: 10.1016/j.meegid.2015.11.021
Alfellani, M. A., Taner-Mulla, D., Jacob, A. S., Imeede, C. A., Yoshikawa, H., Stensvold, C. R., et al. (2013). Genetic diversity of Blastocystis in livestock and zoo animals. Protist 164, 497–509. doi: 10.1016/j.protis.2013.05.003
Andersen, L. O., Bonde, I., Nielsen, H. B., and Stensvold, C. R. (2015). A retrospective metagenomics approach to studying Blastocystis. FEMS Microbiol. Ecol. 91:fiv072. doi: 10.1093/femsec/fiv072
Audebert, C., Even, G., Cian, A., Blastocystis Investigation Group, Loywick, A., Merlin, S., et al. (2016). Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota. Sci. Rep. 6:25255. doi: 10.1038/srep25255
Badparva, E., Sadraee, J., and Kheirandish, F. (2015). Genetic diversity of Blastocystis isolated from cattle in khorramabad, iran. Jundishapur J. Microbiol. 8:e14810. doi: 10.5812/jjm.14810
Beghini, F., Pasolli, E., Truong, T. D., Putignani, L., Caccio, S. M., and Segata, N. (2017). Large-scale comparative metagenomics of Blastocystis, a common member of the human gut microbiome. ISME J. 11, 2848–2863. doi: 10.1038/ismej.2017.139
Betts, E. L., Gentekaki, E., Thomasz, A., Breakell, V., Carpenter, A. I., and Tsaousis, A. D. (2018). Genetic diversity of Blastocystis in non-primate animals. Parasitology 145, 1228–1234. doi: 10.1017/S0031182017002347
Blaser, M. J. (2014). The microbiome revolution. J. Clin. Invest. 124, 4162–4165. doi: 10.1172/JCI78366
Calero-Bernal, R., Santín, M., Maloney, J. G., Martín-Pérez, M., Habela, M. A., Fernández-García, J. L., et al. (2019). Blastocystis sp. subtype diversity in wild carnivore species from Spain. J. Eukaryot. Microbiol. doi: 10.1111/jeu.12772 [Epub ahead of print].
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chudnovskiy, A., Mortha, A., Kana, V., Kennard, A., Ramirez, J. D., Rahman, A., et al. (2016). Host-protozoan interactions protect from mucosal infections through activation of the Inflammasome. Cell 167, 444–456.e14. doi: 10.1016/j.cell.2016.08.076
Cian, A., El Safadi, D., Osman, M., Moriniere, R., Gantois, N., Benamrouz-Vanneste, S., et al. (2017). Molecular epidemiology of Blastocystis sp. in various animal groups from two french zoos and evaluation of potential zoonotic risk. PLoS One 12:e0169659. doi: 10.1371/journal.pone.0169659
Clark, C. G., van der Giezen, M., Alfellani, M. A., and Stensvold, C. R. (2013). Recent developments in Blastocystis research. Adv. Parasitol. 82, 1–32. doi: 10.1016/B978-0-12-407706-5.00001-0
Desoubeaux, G., Peschke, R., Le-Bert, C., Fravel, V., Soto, J., Jensen, E. D., et al. (2018). Seroprevalence survey for Microsporidia in common bottlenose Dolphin (Tursiops truncatus): example of a quantitative approach based on immunoblotting. J. Wildl. Dis. 54, 870–873. doi: 10.7589/2017-11-287
Diamond, L. S. (1957). The establishment of various trichomonads of animals and man in axenic cultures. J. Parasitol. 43, 488–490.
Diamond, L. S. (1982). A new liquid medium for xenic cultivation of Entamoeba histolytica and other lumen-dwelling protozoa. J. Parasitol. 68, 958–959.
Diamond, L. S. (1983). “Lumen dwelling protozoa: Entamoeba, trichomonads, and Giardia,” in In Vitro Cultivation of Protozoan Parasites, ed. J. B. Jensen (Boca Raton, FL: CRC Press), 67–109.
El Safadi, D., Cian, A., Nourrisson, C., Pereira, B., Morelle, C., Bastien, P., et al. (2016). Prevalence, risk factors for infection and subtype distribution of the intestinal parasite Blastocystis sp. from a large-scale multi-center study in France. BMC Infect. Dis. 16:451. doi: 10.1186/s12879-016-1776-8
Elwakil, H. S., and Hewedi, I. H. (2010). Pathogenic potential of Blastocystis hominis in laboratory mice. Parasitol. Res. 107, 685–689. doi: 10.1007/s00436-010-1922-y
Enriquez, G. F., Garbossa, G., Macchiaverna, N. P., Argibay, H. D., Bua, J., Gürtler, R. E., et al. (2016). Is the infectiousness of dogs naturally infected with Trypanosoma cruzi associated with poly-parasitism? Vet. Parasitol. 223, 186–194. doi: 10.1016/j.vetpar.2016.04.042
Enriquez, G. F., Macchiaverna, N. P., Argibay, H. D., López Arias, L., Farber, M., Gürtler, R. E., et al. (2019). Polyparasitism and zoonotic parasites in dogs from a rural area of the Argentine Chaco. Vet. Parasitol. Reg. Stud. Rep. 16:100287. doi: 10.1016/j.vprsr.2019.100287
Fayer, R., Santin, M., and Macarisin, D. (2012). Detection of concurrent infection of dairy cattle with Blastocystis, Cryptosporidium, Giardia, and Enterocytozoon by molecular and microscopic methods. Parasitol. Res. 111, 1349–1355. doi: 10.1007/s00436-012-2971-1
Forsell, J., Bengtsson-Palme, J., Angelin, M., Johansson, A., Evengard, B., and Granlund, M. (2017). The relation between Blastocystis and the intestinal microbiota in Swedish travellers. BMC Microbiol. 17:231. doi: 10.1186/s12866-017-1139-7
Gentekaki, E., Curtis, B. A., Stairs, C. W., Klimes, V., Elias, M., Salas-Leiva, D. E., et al. (2017). Extreme genome diversity in the hyper-prevalent parasitic eukaryote Blastocystis. PLoS Biol. 15:e2003769. doi: 10.1371/journal.pbio.2003769
Iebba, V., Santangelo, F., Totino, V., Pantanella, F., Monsia, A., Di Cristanziano, V., et al. (2016). Gut microbiota related to Giardia duodenalis, Entamoeba spp. and Blastocystis hominis infections in humans from Cote d’Ivoire. J. Infect. Dev. Ctries. 10, 1035–1041. doi: 10.3855/jidc.8179
Jacob, A. S., Busby, E. J., Levy, A. D., Komm, N., and Clark, C. G. (2016). Expanding the Entamoeba universe: new hosts yield novel ribosomal lineages. J. Eukaryot. Microbiol. 63, 69–78. doi: 10.1111/jeu.12249
Katoh, K., and Toh, H. (2010). Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26, 1899–1900. doi: 10.1093/bioinformatics/btq224
Lukes, J., Stensvold, C. R., Jirka Pomajbaková, K., and Wegener Parfrey, L. (2015). Are human intestinal eukaryotes beneficial or commensals? PLoS Pathog. 11:e1005039. doi: 10.1371/journal.ppat.1005039
Mardani Kataki, M., Tavalla, M., and Beiromvand, M. (2019). Higher prevalence of Blastocystis hominis in healthy individuals than patients with gastrointestinal symptoms from Ahvaz, southwestern Iran. Comp. Immunol. Microbiol. Infect. Dis. 65, 160–164. doi: 10.1016/j.cimid.2019.05.018
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES science gateway for inference of large phylogenetic trees,” in Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, 1–8.
Moe, K. T., Singh, M., Howe, J., Ho, L. C., Tan, S. W., Chen, X. Q., et al. (1997). Experimental Blastocystis hominis infection in laboratory mice. Parasitol. Res. 83, 319–325. doi: 10.1007/s004360050256
Nieves-Ramirez, M. E., Partida-Rodriguez, O., Laforest-Lapointe, I., Reynolds, L. A., Brown, E. M., Valdez-Salazar, A., et al. (2018). Asymptomatic intestinal colonization with Protist Blastocystis is strongly associated with distinct microbiome ecological patterns. mSystems 3:e00007-18. doi: 10.1128/mSystems.00007-18
Nishida, A. H., and Ochman, H. (2018). Rates of gut microbiome divergence in mammals. Mol. Ecol. 27, 1884–1897. doi: 10.1111/mec.14473
Noel, C., Dufernez, F., Gerbod, D., Edgcomb, V. P., Delgado-Viscogliosi, P., Ho, L. C., et al. (2005). Molecular phylogenies of Blastocystis isolates from different hosts: implications for genetic diversity, identification of species, and zoonosis. J. Clin. Microbiol. 43, 348–355. doi: 10.1128/jcm.43.1.348-355.2005
O’Brien Andersen, L., Karim, A. B., Roager, H. M., Vigsnæs, L. K., Krogfelt, K. A., Licht, T. R., et al. (2016). Associations between common intestinal parasites and bacteria in humans as revealed by qPCR. Eur. J. Clin. Microbiol. Infect. Dis. 35, 1427–1431. doi: 10.1007/s10096-016-2680-2
Parfrey, L. W., Walters, W. A., and Knight, R. (2011). Microbial eukaryotes in the human microbiome: ecology, evolution, and future directions. Front. Microbiol. 2:153. doi: 10.3389/fmicb.2011.00153
Parsons, M. B., Travis, D., Lonsdorf, E. V., Lipende, I., Roellig, D. M. A., Kamenya, S., et al. (2015). Epidemiology and molecular characterization of Cryptosporidium spp. in humans, wild primates, and domesticated animals in the greater gombe ecosystem, tanzania. PLoS Negl. Trop. Dis. 9:e0003529. doi: 10.1371/journal.pntd.0003529
Puthia, M. K., Lu, J., and Tan, K. S. (2008). Blastocystis ratti contains cysteine proteases that mediate interleukin-8 response from human intestinal epithelial cells in an NF-kappaB-dependent manner. Eukaryot. Cell 7, 435–443. doi: 10.1128/ec.00371-07
Ramirez, J. D., Sanchez, A., Hernandez, C., Florez, C., Bernal, M. C., Giraldo, J. C., et al. (2016). Geographic distribution of human Blastocystis subtypes in South America. Infect. Genet. Evol. 41, 32–35. doi: 10.1016/j.meegid.2016.03.017
Renelies-Hamilton, J., Noguera-Julian, M., Parera, M., Paredes, R., Pacheco, L., Dacal, E., et al. (2019). Exploring interactions between Blastocystis sp., Strongyloides spp. and the gut microbiomes of wild chimpanzees in Senegal. Infect. Genet. Evol. 74:104010. doi: 10.1016/j.meegid.2019.104010
Robertson, L. J., Clark, C. G., Debenham, J. J., Dubey, J. P., Kváč, M., Li, J., et al. (2019). Are molecular tools clarifying or confusing our understanding of the public health threat from zoonotic enteric protozoa in wildlife? Int. J. Parasitol. Parasites Wildl. 9, 323–341. doi: 10.1016/j.ijppaw.2019.01.010
Rueckert, S., Betts, E. L., and Tsaousis, A. D. (2019). The symbiotic spectrum: Where do the gregarines fit? Trends Parasitol. 35, 687–694. doi: 10.1016/j.pt.2019.06.013
Sanders, J. G., Beichman, A. C., Roman, J., Scott, J. J., Emerson, D., McCarthy, J. J., et al. (2015). Baleen whales host a unique gut microbiome with similarities to both carnivores and herbivores. Nat. Commun. 6:8285. doi: 10.1038/ncomms9285
Scanlan, P. D., and Stensvold, C. R. (2013). Blastocystis: getting to grips with our guileful guest. Trends Parasitol. 29, 523–529. doi: 10.1016/j.pt.2013.08.006
Scanlan, P. D., Stensvold, C. R., Rajilic-Stojanovic, M., Heilig, H. G., De Vos, W. M., O’Toole, P. W., et al. (2014). The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol. Ecol. 90, 326–330. doi: 10.1111/1574-6941.12396
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stamatakis, A. (2015). Using RAxML to infer phylogenies. Curr. Protoc. Bioinformatics 51, 6.14.1–14.14. doi: 10.1002/0471250953.bi0614s51
Stensvold, C. R. (2012). Thinking Blastocystis out of the box. Trends Parasitol. 28:305. doi: 10.1016/j.pt.2012.05.004
Stensvold, C. R., and Clark, C. G. (2016a). Current status of Blastocystis: a personal view. Parasitol. Int. 65, 763–771. doi: 10.1016/j.parint.2016.05.015
Stensvold, C. R., and Clark, C. G. (2016b). Molecular identification and subtype analysis of Blastocystis. Curr. Protoc. Microbiol. 43, 20A.2.1–20A.2.10. doi: 10.1002/cpmc.17
Tito, R. Y., Chaffron, S., Caenepeel, C., Lima-Mendez, G., Wang, J., Vieira-Silva, S., et al. (2019). Population-level analysis of Blastocystis subtype prevalence and variation in the human gut microbiota. Gut 68, 1180–1189. doi: 10.1136/gutjnl-2018-316106
Udonsom, R., Prasertbun, R., Mahittikorn, A., Mori, H., Changbunjong, T., Komalamisra, C., et al. (2018). Blastocystis infection and subtype distribution in humans, cattle, goats, and pigs in central and western Thailand. Infect. Genet. Evol. 65, 107–111. doi: 10.1016/j.meegid.2018.07.007
Wang, J., Gong, B., Liu, X., Zhao, W., Bu, T., Zhang, W., et al. (2018a). Distribution and genetic diversity of Blastocystis subtypes in various mammal and bird species in northeastern China. Parasit. Vectors 11:522. doi: 10.1186/s13071-018-3106-z
Wang, J., Gong, B., Yang, F., Zhang, W., Zheng, Y., and Liu, A. (2018b). Subtype distribution and genetic characterizations of Blastocystis in pigs, cattle, sheep and goats in northeastern China’s Heilongjiang Province. Infect. Genet. Evol. 57, 171–176. doi: 10.1016/j.meegid.2017.11.026
Wang, W., Bielefeldt-Ohmann, H., Traub, R. J., Cuttell, L., and Owen, H. (2014). Location and pathogenic potential of Blastocystis in the porcine intestine. PLoS One 9:e103962. doi: 10.1371/journal.pone.0103962
Wawrzyniak, I., Texier, C., Poirier, P., Viscogliosi, E., Tan, K. S., Delbac, F., et al. (2012). Characterization of two cysteine proteases secreted by Blastocystis ST7, a human intestinal parasite. Parasitol. Int. 61, 437–442. doi: 10.1016/j.parint.2012.02.007
Whipps, C. M., Boorom, K., Bermudez, L. E., and Kent, M. L. (2010). Molecular characterization of Blastocystis species in Oregon identifies multiple subtypes. Parasitol. Res. 106, 827–832. doi: 10.1007/s00436-010-1739-8
Yoshikawa, H., Abe, N., and Wu, Z. (2004). PCR-based identification of zoonotic isolates of Blastocystis from mammals and birds. Microbiology 150, 1147–1151. doi: 10.1099/mic.0.26899-0
Yoshikawa, H., Koyama, Y., Tsuchiya, E., and Takami, K. (2016). Blastocystis phylogeny among various isolates from humans to insects. Parasitol. Int. 65, 750–759. doi: 10.1016/j.parint.2016.04.004
Yowang, A., Tsaousis, A. D., Chumphonsuk, T., Thongsin, N., Kullawong, N., Popluechai, S., et al. (2018). High diversity of Blastocystis subtypes isolated from asymptomatic adults living in Chiang Rai, Thailand. Infect. Genet. Evol. 65, 270–275. doi: 10.1016/j.meegid.2018.08.010
Keywords: Blastocystis, genetic diversity, subtyping, co-occurrence, phylogeny, micro-eukaryome
Citation: Betts EL, Gentekaki E and Tsaousis AD (2020) Exploring Micro-Eukaryotic Diversity in the Gut: Co-occurrence of Blastocystis Subtypes and Other Protists in Zoo Animals. Front. Microbiol. 11:288. doi: 10.3389/fmicb.2020.00288
Received: 02 October 2019; Accepted: 10 February 2020;
Published: 25 February 2020.
Edited by:
Christen Rune Stensvold, Statens Serum Institut (SSI), DenmarkReviewed by:
Hamed Mirjalali, Shahid Beheshti University of Medical Sciences, IranCopyright © 2020 Betts, Gentekaki and Tsaousis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eleni Gentekaki, Z2VudGVrYWtpLmVsZUBtZnUuYWMudGg=; Anastasios D. Tsaousis, QS5Uc2FvdXNpc0BrZW50LmFjLnVr; dHNhb3VzaXMuYW5hc3Rhc2lvc0BnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.