- 1Department of Earth and Environmental Sciences, The University of Manchester, Manchester, United Kingdom
- 2Department of Soil and Crop Sciences, College of Agricultural Sciences, Colorado State University, Fort Collins, CO, United States
- 3Rawwater Engineering Company Limited, Culcheth, United Kingdom
Produced waters from hydraulically fractured shale formations give insight into the microbial ecology and biogeochemical conditions down-well. This study explores the potential for sulfide production by persistent microorganisms recovered from produced water samples collected from the Marcellus shale formation. Hydrogen sulfide is highly toxic and corrosive, and can lead to the formation of “sour gas” which is costly to refine. Furthermore, microbial colonization of hydraulically fractured shale could result in formation plugging and a reduction in well productivity. It is vital to assess the potential for sulfide production in persistent microbial taxa, especially when considering the trend of reusing produced waters as input fluids, potentially enriching for problematic microorganisms. Using most probable number (MPN) counts and 16S rRNA gene sequencing, multiple viable strains of bacteria were identified from stored produced waters, mostly belonging to the Genus Halanaerobium, that were capable of growth via fermentation, and produced sulfide when supplied with thiosulfate. No sulfate-reducing bacteria (SRB) were detected through culturing, despite the detection of relatively low numbers of sulfate-reducing lineages by high-throughput 16S rRNA gene sequencing. These results demonstrate that sulfidogenic produced water populations remain viable for years post production and, if left unchecked, have the potential to lead to natural gas souring during shale gas extraction.
Introduction
Shale gas is an unconventional hydrocarbon being utilized increasingly as a cleaner alternative to coal (Jenner and Lamadrid, 2013). Natural gas is recovered from shale formations by means of hydraulic fracturing; the high pressure injection of water-based fluids to force open fractures in the rock, allowing gas to flow to the surface (Lee et al., 2011; Mouser et al., 2016). Input fluids can then interact with the shale formation leading to changes in the geochemistry of the produced waters (Schramm, 2011; Murali Mohan et al., 2013a; Akob et al., 2015). Studies on produced waters from the Marcellus shale show an inverse correlation between dissolved organic carbon and salinity (Cluff et al., 2014), with salinity increasing with time (Hayes, 2009; Schramm, 2011; Barbot et al., 2013; Murali Mohan et al., 2013a; Cluff et al., 2014; Akob et al., 2015; Rowan et al., 2015). Fracturing fluids typically include sand and a number of organic and inorganic additives, to hold open newly formed fractures and allow operations to run as efficiently as possible (Stringfellow et al., 2014; Elsner and Hoelzer, 2016). Included among common additives are biocides, used to prevent microbial activity from interfering with gas extraction (Elsner and Hoelzer, 2016). Despite the addition of biocides and the extreme conditions encountered downhole including high salinity, pressure, reducing conditions, and moderately high temperatures (Fichter et al., 2012; Akob et al., 2015; Daly et al., 2016; Nixon et al., 2019) prior studies have identified microbial communities in produced waters from fractured shales in the Antrim, Barnett, Burkett, Utica, Bakken, and Marcellus formations in the United States (Davis et al., 2012; Struchtemeyer and Elshahed, 2012; Murali Mohan et al., 2013a, b; Strong et al., 2013; Wuchter et al., 2013; Cluff et al., 2014; Akob et al., 2015; Daly et al., 2016; Liang et al., 2016; Lipus et al., 2017; Evans et al., 2018) as well as the Sichuan Basin in China (Zhang et al., 2017). A common finding in many of these studies, is a transition from high diversity microbial communities in injected fluids, resembling the freshwater environment the water was sourced from, to an enriched and lower diversity community of mostly halotolerant and halophilic facultative and strict anaerobes, such as Halanaerobium (Davis et al., 2012; Struchtemeyer and Elshahed, 2012; Murali Mohan et al., 2013a; Strong et al., 2013; Cluff et al., 2014; Akob et al., 2015; Daly et al., 2016; Mouser et al., 2016; Vikram et al., 2016; Booker et al., 2017; Lipus et al., 2017; Borton et al., 2018; Evans et al., 2019).
Some halotolerant microorganisms common to fractured shales can impact negatively on shale gas extraction. Microbial colonization of fractured shale may lead to sour gas production, microbially induced corrosion and bioclogging, with implications for both human safety and well productivity. Metabolic by-products such as organic acids and to a greater extent hydrogen sulfide can corrode steel infrastructure leading to environmental hazards and lost revenue (Eden et al., 1993). Hydrogen sulfide is also highly flammable, toxic to human health (Guidotti, 2010) and can result in “sour gas” which is costly to refine (Eden et al., 1993; Fichter et al., 2012). Sulfide may be produced through the reduction of sulfur species (such as sulfate and thiosulfate) coupled to the direct oxidation of organics or hydrogen by sulfate-reducing (Grein et al., 2013) and thiosulfate-reducing bacteria (TRB) (Ravot et al., 1997). Alternatively biogenic sulfide can also result from the transfer of reducing equivalents onto thiosulfate during fermentation reactions (Booker et al., 2017). Additionally, the formation of microbial biofilms may impede gas flow and result in fracture clogging (Bottero et al., 2010). Evidence suggests that organic chemicals used during hydraulic fracturing may stimulate these deleterious processes, for example via organic acid production from fermentation reactions, and sulfide production from sulfate and thiosulfate reduction pathways (Struchtemeyer et al., 2011; Liang et al., 2016; Booker et al., 2017, 2019; Nixon et al., 2017; Evans et al., 2019). Sulfate-reducing bacteria (SRB) have been identified previously in production fluids through next generation sequencing but their in situ role in sulfide production is undetermined (Davis et al., 2012; Struchtemeyer and Elshahed, 2012; Liang et al., 2016), whereas TRB have been isolated from these environments and represent an alternative route to sulfide production (Ravot et al., 1997, 2005; Liang et al., 2016; Booker et al., 2017; Lipus et al., 2017).
Microorganisms in production fluids could prove especially problematic when considering the trend for their reuse in future fracturing operations. This practice reduces the demand on freshwater supplied and circumvents challenges related to disposal (Gregory et al., 2011; Murali Mohan et al., 2013b; Cluff et al., 2014). Produced waters are often stored above ground in impoundments or sealed tanks prior to reuse or disposal, undergoing aeration or biocide treatment prior to reuse, with variable success in regard to microbial control (Murali Mohan et al., 2013b). It is therefore essential to understand the potential for stored produced waters to cause further microbial problems in shale gas extraction. However, relatively little is known about the metabolism of viable microorganisms in production waters (Daly et al., 2016; Vikram et al., 2016; Booker et al., 2017). Given the potential for deleterious microbial processes to affect shale gas extraction, there remains a critical need to assess the metabolic capabilities and resilience of persistent taxa recovered from hydraulically fractured shale systems. Here, we assess the persistence of problematic microorganisms in produced waters collected from a shale gas well in the Marcellus formation. Samples were stored for several 100 days between sampling and the analysis reported here, giving a good view of the potential role of persistent microbes present in souring reactions. Culture based approaches were used to investigate the presence of sulfate-reducing, thiosulfate-reducing and fermentative bacteria (FB), and the composition of most probable number (MPN) enrichment cultures for these organisms was characterized and compared to the microbial composition of the produced fluids soon after collection, to identify the most persistent taxa. Collectively, these data confirm the numerical dominance and long-term persistence of fermentative TRB, alongside a far lower number of SRB, highlighting the importance of developing appropriate diagnostic tools to help control microbial sulfide production.
Materials and Methods
Sampling
Produced water samples were collected over several 100 days as part of a Marcellus Shale Energy and Environment Laboratory project (MSEEL, 2015) in order to characterize the changes to production fluid geochemistry and microbiology, and full details of sample collection have been published previously (Evans et al., 2018; Daly et al., 2019). Prior research has shown that over this period of time, produced fluids become more saline and the community less diverse, with halotolerant bacteria dominating later stage fluids (Daly et al., 2016). Produced water samples from the gas-water-separator of the MIP-3H shale gas well in West Virginia (herein known as Marcellus-4) were collected daily. The authors had access to stored samples collected at 29, 79, 142, and 303 days after hydraulic fracturing. These samples were collected between December 2015 and September 2016 and selected for use in this study representing a spread from early flowback to late production fluids. Samples from the same well have been reported on in prior studies (Evans et al., 2018; Daly et al., 2019) Prior to use in this study, samples were stored at room temperature under anoxic conditions for varying periods of time. The first set of experiments were conducted on samples stored for 808, 758, 695, and 534 days, respectively. Produced water samples were stored for an additional 291 days prior to the second set of MPN experiments conducted using TRB l medium with the addition of yeast extract.
Most Probable Number (MPN) Counts
Most probable number counts were used to estimate the number of viable heterotrophic, sulfate-reducing and thiosulfate-reducing microorganisms in the four stored produced water samples, all dilution series were run in triplicate and incubated at 40°C, comparable to previously reported temperatures (Akob et al., 2015) for up to 112 days; the most positive endpoints then underwent 16S rRNA sequencing. Strict anaerobic techniques were used throughout (Miller and Wolin, 1974). A starting dilution of 1 ml inoculum into 9 ml media was diluted ten-fold in serial dilutions to a final concentration of 1:107this was repeated for each sample in triplicate. The salinities of the selective media were chosen to reflect the high salinities previously measured in produced fluid samples (Daly et al., 2016). Growth was identified visually based on increased turbidity (fermentative bacteria MPNs) or the presence of a black precipitate (iron sulfide formation in sulfate-reducing and thiosulfate-reducing MPNs). MPN counts were then calculated from the vials scored as positive (Oblinger and Koburger, 1975). When estimating MPN counts for sulfate-reducing and thiosulfate-reducing cultures a pitch-black vial (compared to the colorless control) was deemed a positive result. Sulfate-reducing and thiosulfate-reducing media were amended with iron (II) chloride, which reacts with sulfide to form a black FeS precipitate in the presence of sulfide, an anticipated by-product of microbial thiosulfate- and sulfate-reduction. Addition of sodium sulfide as a reducing agent would lead to false positive results, and therefore we opted to add sodium thioglycolate to SRB media instead [as in Postgate media (Tanner, 1989)]. The fermentative medium lacked iron(II) chloride and hence sodium sulfide was used as a reducing agent. For the FB a positive score was based on visible turbidity, compared to the control. Since this medium contained resazurin (a redox-sensitive indicator dye), optical density readings could not be conducted aerobically. MPN enrichment media are described below.
Fermentative Bacteria (FB) Medium
This medium was adapted from Spring et al. (2015) to target viable FB in the produced water samples (Spring et al., 2015). The medium contained (in grams per liter deionized water unless otherwise stated): NaCl (80.0), MgCl2.6H2O (6.0), NH4Cl2 (1.0), CaCl.2H2O (0.4), K2HPO4 (0.4), KCl (1.5), yeast extract (0.2), Na2CO3 (0.75), L-Cysteine- HCL- H2O (0.3), D-glucose (2.0), and 10 ml vitamin solution. The vitamin solution contained (in mg per liter deionized water) biotin (2.0), folic acid (2.0), pyridoxine-HCl (10.0), riboflavin (5.0), thiamine (5.0), nicotinic acid (5.0), pantothenic acid (5.0), vitamin B-12 (0.1), p-aminobenzoic acid (5.0), and thioctic acid (5.0). The medium was dispensed into 10 ml glass serum vials, purged with an 80:20 mix of N2:CO2 for 10 min, and autoclaved at 121°C for 20 min. Prior to inoculation, all FB aliquots were amended with 1.25 mM sodium sulfide from a sterile anoxic stock and the pH adjusted to 7.3–7.5.
Sulfate-Reducing Bacteria (SRB) Medium
This medium was modified from the Postgate B recipe (Tanner, 1989) and contained (in grams per liter deionized water): KH2PO4 (0.5), CaSO4.2H2O (1.26), NH4Cl (1.00), MgSO4.7H2O (2.00), Yeast extract (1.00), FeSO4.7H2O, ascorbic acid (0.10), sodium thioglycolate (0.10), sodium acetate (2.56), sodium lactate (3.5), and NaCl (100.00). The pH was adjusted to 7.1, before being dispensed, purged with N2:CO2 and autoclaved as above.
Thiosulfate-Reducing Bacteria (TRB) Medium
This bespoke medium contained (in grams per liter deionized water): NaHCO3 (2.5), NH4Cl (0.25), NaH2PO4H2O (0.06), KCl (0.1), sodium acetate (2.56), sodium lactate (3.5) NaCl (100), Na2S2O3.5H2O (4.27), 10 ml vitamin solution (as above), and 10 ml mineral solution. The mineral solution contained (in mg per liter deionized water) nitrilotriacetic acid (1.5), MgSO4 (3.0), MnSO4.H2O (0.5), NaCl (1.0), FeSO4.7H2O (0.1), CaCl2.2H2O (0.1), CoCl2.6H2O (0.1), ZnCl2 (0.13), CuSO4.5H2O (0.01), AlK (SO4)2.12H2O (0.01), H3BO3 (0.01), NaMoO4 (0.025), NiCl2.6H2O (0.024), and NaWO4.2H2O (0.025). The medium was dispensed, purged with N2:CO2 and autoclaved as above. Filter-sterilized iron (II) chloride was then added from anoxic stock to a final concentration of 1.8 mM (as an indicator for sulfide production)and the pH adjusted to 6.8–7.0. Subsequent TRB MPNs were repeated with the addition of 0.2 g/L yeast extract to the medium prior to autoclave, which was otherwise prepared as above.
Ion Chromatography
Acetate, lactate and thiosulfate concentrations in MPN cultures were measured using high performance ion chromatography (IC) (Dionex ICS5000 dual channel, Hemel Hempstead, United Kingdom) as described previously (Nixon et al., 2018). Mean changes in concentration of anions in MPN tests were assessed for significance using a Student t test (2-tailed, type 2, critical value of 0.05).
DNA Extraction and 16S rRNA Gene Amplification and Sequencing
The standard protocol describing 16S rRNA gene sequencing carried out on samples shortly after sample collection is precisely detailed in Borton et al. (2017).
At the time of inoculation of the MPN series, 1 ml samples of inocula were collected and frozen at −20°C. Further 1 ml samples were collected from positive end points at the conclusion of MPN experiments and stored at −20°C. DNA was extracted from 0.2 ml aliquots of these samples using the DNeasy PowerLyzerTM PowerSoil Kit (Qiagen, Manchester, United Kingdom). Positive (Escherichia coli) and negative (ultrapure water) controls were also run. Sequencing of PCR amplicons of 16S rRNA was conducted with the Illumina® MiSeqTM platform (Illumina, San Diego, CA, United States) targeting the V4 hyper variable region (forward primer, 515F, 5′-GTGYCAGCMGCCGCGGTAA-3′; reverse primer, 806R, 5′-GGACTACHVGGGTWTCTAAT-3′) for two 250-bp paired-end sequencing (Caporaso et al., 2011, 2012). PCR amplification was performed using Roche FastStartTM High Fidelity PCR System (Roche Diagnostics Ltd., Burgess Hill, United Kingdom) in 50 μl reactions under the following conditions: initial denaturation at 95°C for 2 min, followed by 36 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 1 min, and a final extension step of 5 min at 72°C. The PCR products were purified and normalized to ∼20 ng each using the SequalPrep Normalization Kit (Fisher Scientific, Loughborough, United Kingdom). The PCR amplicons from all samples were pooled in equimolar ratios. The run was performed using a 4 pM sample library spiked with 4 pM PhiX to a final concentration of 10% in accordance with Kozich et al., 2013 (Kozich et al., 2013). Raw sequences were divided into samples by barcodes (up to one mismatch was permitted) using a sequencing pipeline as follows. Quality control and trimming was performed using Cutadapt (Compeau et al., 2013), FastQC (Andrews, 2010), and Sickle (Joshi and Fass, 2011). MiSeqTM error correction was performed using SPAdes (Nurk et al., 2013). Forward and reverse reads were incorporated into full-length sequences with Pandaseq (Masella et al., 2012). Chimeras were removed using ChimeraSlayer (Haas et al., 2011), and OTU’s were generated with UPARSE (Edgar, 2013). OTUs were classified by Usearch (Edgar, 2010) at the 97% similarity level, and singletons were removed. Rarefaction analysis was conducted using the original detected OTUs in Qiime (Caporaso et al., 2010). Taxonomic assignment was performed by the RDP naïve bayesian classifier version 2.2 (Wang et al., 2007), used in combination with the Silva SSU 132 ribosomal RNA gene database (Quast et al., 2013). The OTU tables were rarefied to the sample containing the lowest number of sequences, all samples having less than 5,000 sequences were removed from analyses prior to the rarefaction step. The step size used was 2000 and 10 iterations were performed at each step. Both sequencing reaction mix controls and extraction controls were sequenced alongside the samples and any shared OTU’s found to be present in these were manually deleted from the Sample OTU table prior to final analysis.
Accession Numbers
Raw sequencing data obtained in this study are available at the NCBI Sequencing Read Archive1 under the accession numbers PRJNA578032, SAMN13315659, SAMN13315660, SAMN13315661, and SAMN13315662.
Results
Most Probable Number Counts
To quantify the numbers of persistent culturable sulfide-producing bacteria in produced waters from the Marcellus shale gas well studied, a series of MPN enrichments were set up targeting sulfate-reducing and TRB (alongside FB) using samples that had been collected 29, 79, 142, and 303 days after hydraulic fracturing, and then stored for 534–808 days at room temperature as shown in Table 1 prior to cultivation. The term “persistent” is used here to describe cells that survive storage conditions and can be cultured under the conditions provided. The formation of black precipitate (iron sulfide) in thiosulfate and sulfate-reducing media was scored as positive and indicative of sulfide production. An increase in turbidity was scored as positive growth in the FB medium. All MPN vials were incubated at 40°C for 112 days prior to scoring.
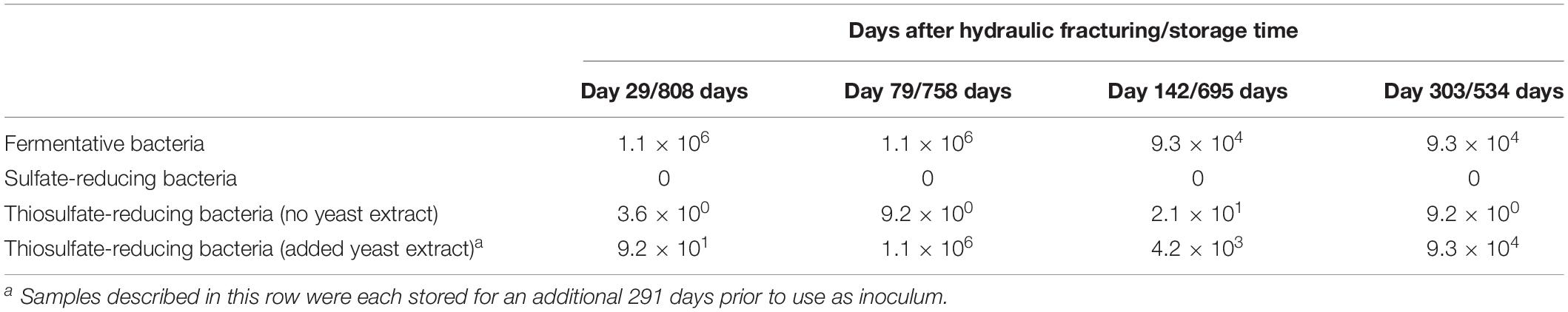
Table 1. Most probable number (MPN) counts (cells/ml) after 112 days incubation and the average cell counts from time of sample collection.
Most probable number counts (Table 1) showed that across all the samples analyzed, FB were more numerous than dissimilatory sulfate- or TRB. In initial MPN tests targeting dissimilatory TRB, using a fully defined selective medium, only low numbers of TRB were cultured in samples (following storage for 534–808 days), with a maximum MPN count of just 21 cells/ml when inoculated with the sample from day 142 post hydraulic fracturing. After an additional 291 days storage, the inoculum was used to repeat the TRB l MPNs with the addition of yeast extract, and MPN counts increased considerably, peaking at 1.1 × 106 cells/ml, despite their prolonged storage. Indeed, MPNs conducted in TRB medium amended with yeast extract were up to several orders of magnitude greater than those conducted in the absence of yeast extract, suggesting that yeast extract has a role in stimulating the growth of TRB.
While the estimated number of persistent FB was initially high (approximately 1.1 × 106) in MPNs inoculated with produced waters collected 29 and 79 days after hydraulic fracturing (and stored for 808 and 758 days respectively), MPN counts were approximately two orders of magnitude lower in the MPNs inoculated with later produced waters (collected 142 and 303 days after hydraulic fracturing and stored for 695 and 534 days, respectively). The MPN counts for TRB in tests with added yeast extract, inoculated with the produced waters collected 79 and 303 days after hydraulic fracturing (and stored for 1049 and 825 days, respectively), approached the MPN counts obtained from FB. The number of persistent TRB estimated in the presence of yeast extract increased from 9.2 × 101 to 1.1 × 106 between produced waters from 29 to 79 days after hydraulic fracturing, and MPN counts were maintained in the range of 4.2 × 103 and 9.3 × 104 for the later samples. The equivalent number of thiosulfate-reducing and FB estimated from MPN tests on samples collected 79 and 303 days post hydraulic fracturing, after hundreds of days storage suggests that the majority of FB detected were also capable of thiosulfate reduction. No culturable SRB were detected in the SRB MPNs across any of the four stored produced waters tested (Table 1).
Ion Chromatography to Support Thiosulfate Reduction in Relevant MPN Counts
After 112 days incubation the initial TRB MPNs (without yeast extract) showed a statistically significant (p < 0.05) decrease in the average concentration of lactate, acetate and thiosulfate relative to abiotic controls. Positive MPN end-points from TRB l medium with the addition of yeast extract also showed a statistically significant (p < 0.05) decrease in thiosulfate concentrations after the same incubation period. In addition, positive end points from the thiosulfate-reducing MPNs, with the addition of yeast extract, did not show a significant decrease in the concentration of lactate or acetate after a total of 112 days incubation, despite the significant reduction of thiosulfate, suggesting that here thiosulfate-reduction may instead be coupled to the fermentation of compounds present in yeast extract rather than acetate or lactate metabolism.
Microbial Community Composition Based on 16S rRNA Gene Sequencing
Microbial community composition in enrichment cultures from the positive MPN tubes was assessed using 16S rRNA gene sequencing with the illumina® MiSeqTMplatform. The aims were to compare how community composition changed between sample collection and their use as inocula in MPN tests reported here (Figure 1), and to assess which taxa became enriched in MPN tests targeting fermentative (Figure 2), sulfate-reducing and thiosulfate-reducing (Figures 3, 4) microorganisms. 16S rRNA gene sequencing was conducted on all positive MPN end-points. As such, data are presented for fermentative and thiosulfate-reducing enrichments only, as no SRB l MPNs were scored as positive. In the figures the Shannon H diversity index is displayed above each bar as a measure of change in diversity between samples.
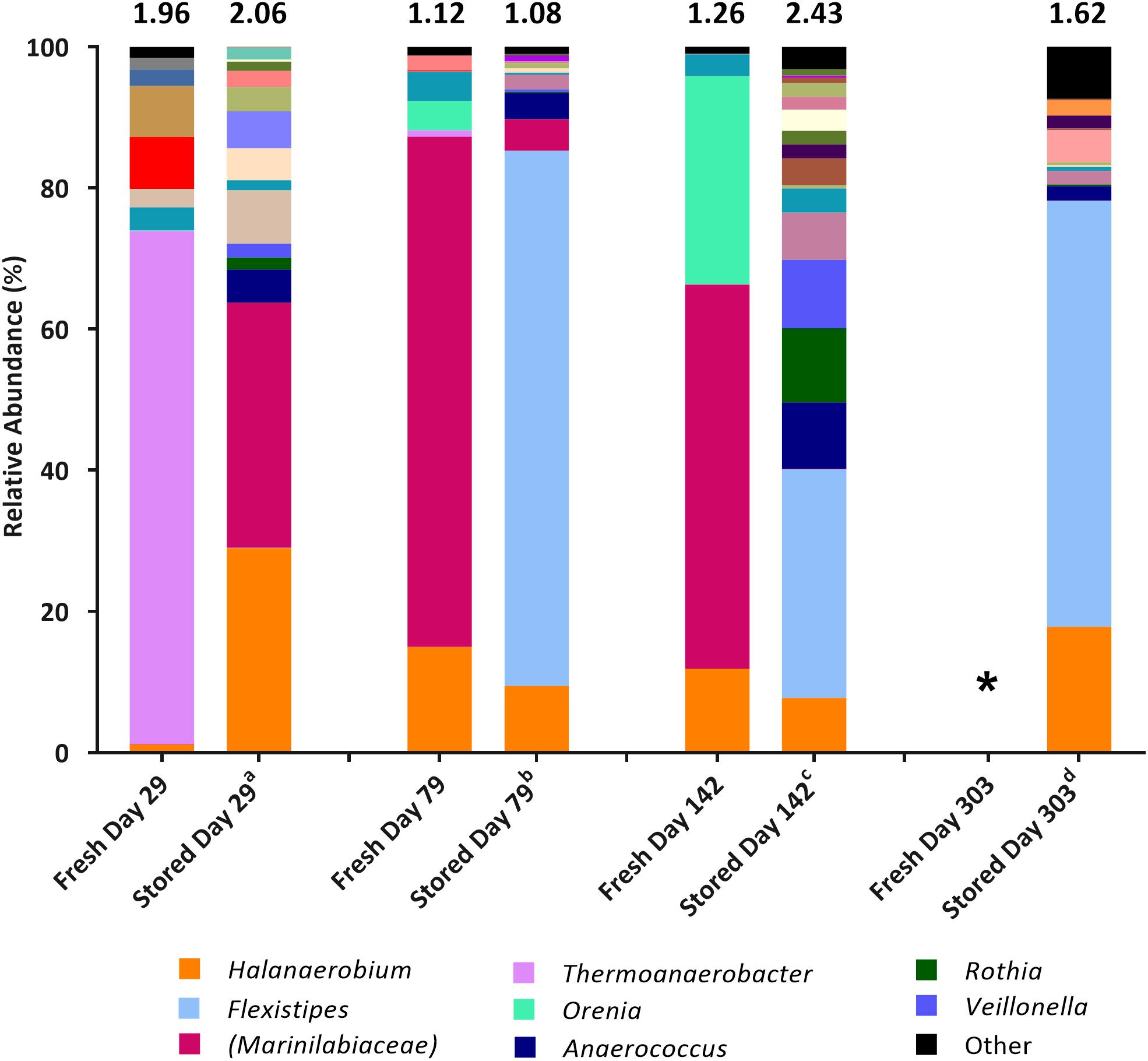
Figure 1. Genus-level microbial community comparison based on 16S rRNA gene sequencing from fresh samples (DNA extracted shortly after collection) compared with stored samples (DNA extracted at the time of initial MPN inoculation). DNA was extracted from fresh samples shortly after sample collection, DNA was extracted from stored samples at the time of initial MPN inoculation. Where genus level could not be resolved the last matched taxonomic level was assigned in brackets. “Other” is comprised of taxa that represent less than 1% relative abundance combined across all samples. The Shannon diversity index calculated from operational taxonomic units is displayed above each bar. a 808 days storage, b 758 days storage, c 695 days in storage, and d 534 days in storage. * denotes 16S rRNA gene data unavailable. Key taxa are highlighted in legend.
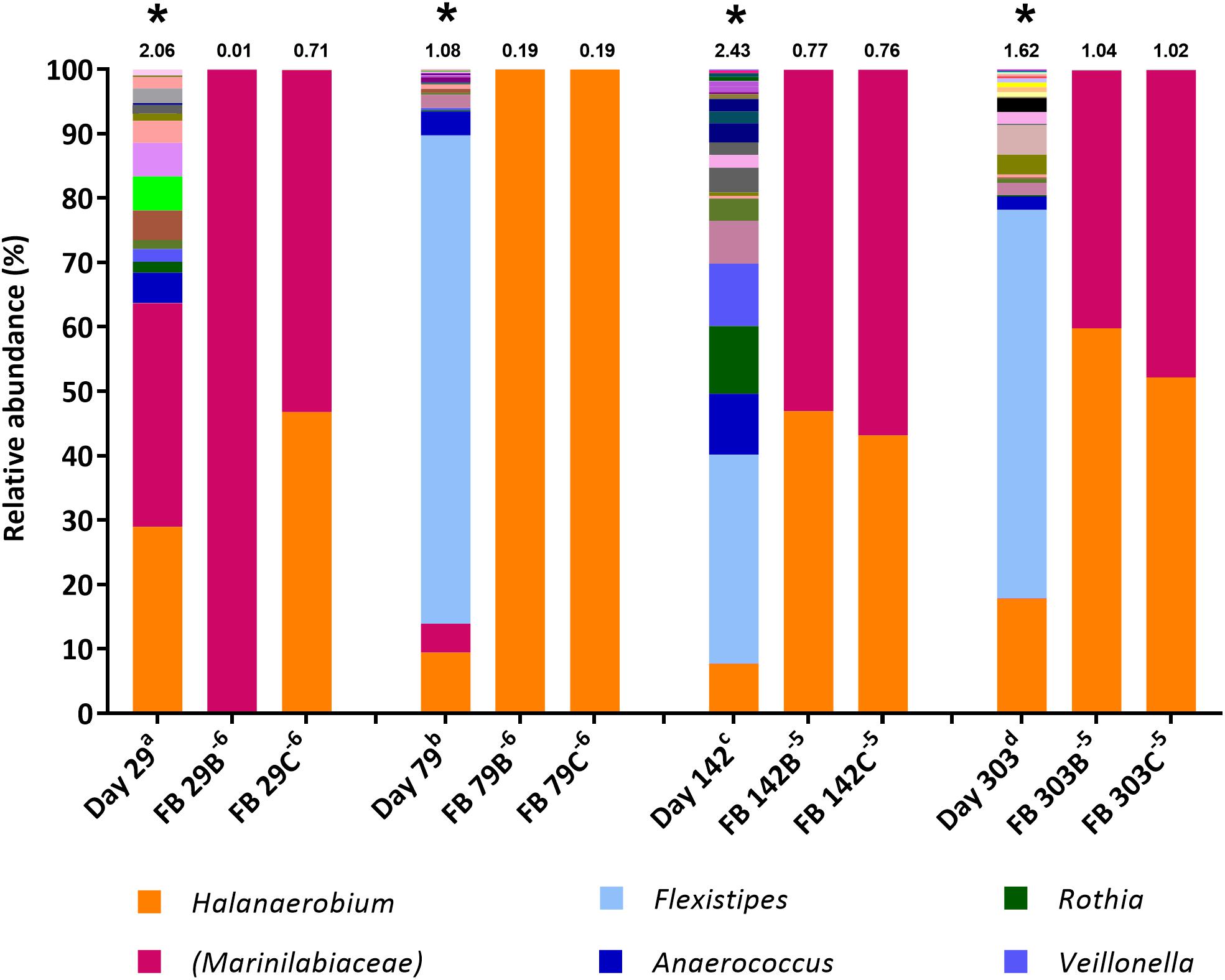
Figure 2. Genus-level microbial community composition of fermentative bacteria (FB) MPN enrichments based on 16S rRNA gene sequencing. Enrichments were conducted at 8% salinity. Where genus level could not be resolved the last matched taxonomic level was assigned in brackets. The Shannon diversity index calculated from operational taxonomic units is displayed above each bar. a 808 days storage, b 758 days storage, c 695 days in storage, and d 534 days storage. * indicates inocula. Key taxa are highlighted in legend. Enrichments were conducted in triplicate where A, B, C refers to specific replica and superscript power refers to dilution factor of the positive end-points sequenced.
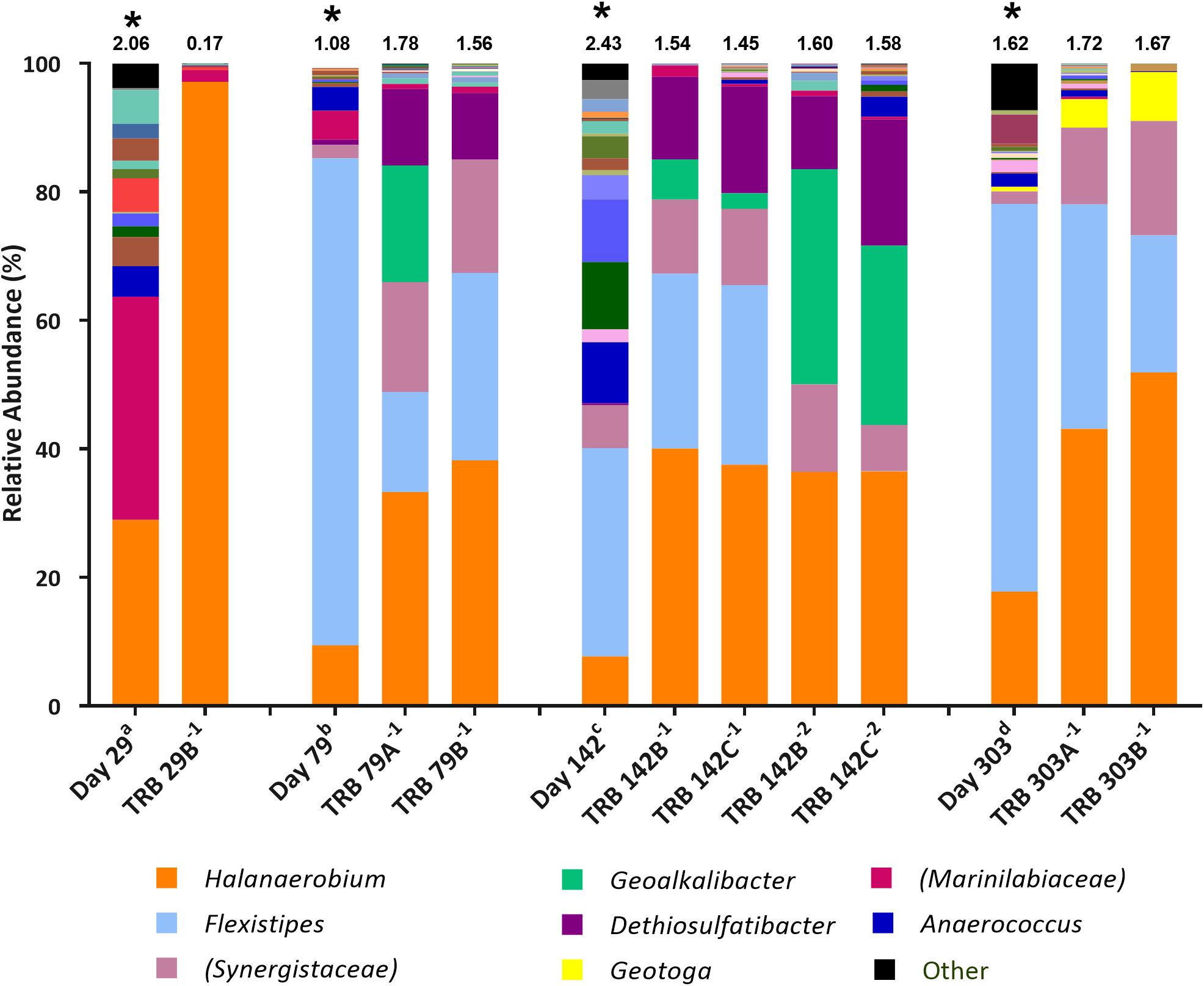
Figure 3. Genus-level microbial community composition of MPN enrichments for TRB based on 16S rRNA gene sequencing. Enrichments were conducted at 10% salinity. Genera that constitute less than 0.1% combined abundance were grouped as “other.” Where genus level could not be resolved the highest resolved taxonomic level is given in brackets. The Shannon diversity index calculated from operational taxonomic units is displayed above each bar. a 808 days storage, b 758 days storage, c 695 days in storage, and d 534 days storage, * indicates inocula. Key taxa are highlighted in legend. Enrichments were conducted in triplicate where A, B, C refers to specific replica and superscript power refers to dilution factor of the positive end-points sequenced.
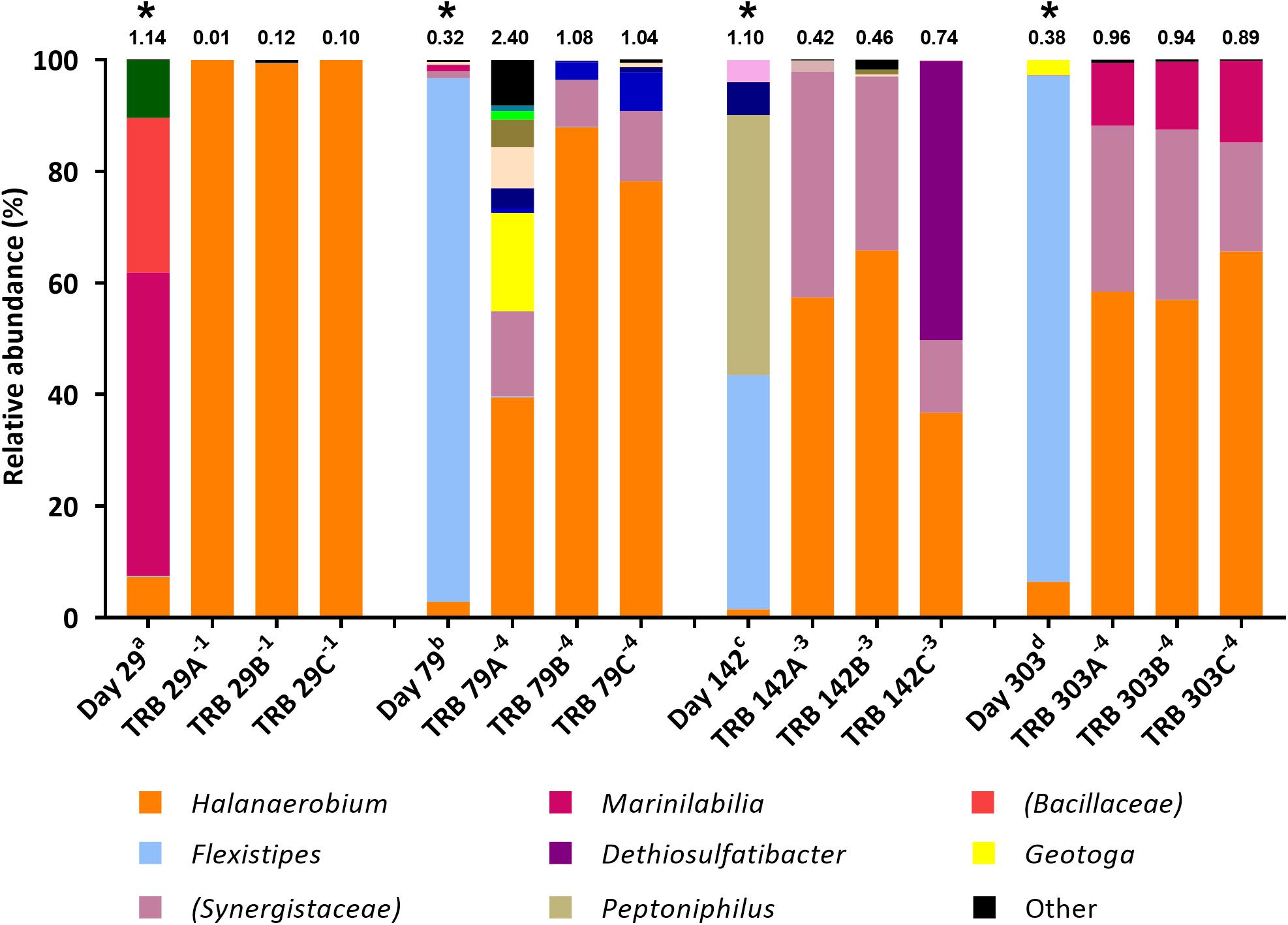
Figure 4. Genus-level microbial community composition of TRB MPN enrichments amended with yeast extract, based on 16S rRNA gene sequencing. Enrichments were conducted at 10% salinity. Where identity at genus level could not be resolved the highest resolved taxonomic level is given in brackets. The Shannon diversity index calculated from operational taxonomic units is displayed above each bar. a 808 days storage, b 758 days storage, c 695 days in storage, and d 534 days storage, * indicates inocula. Key taxa are highlighted in legend. Enrichments were conducted in triplicate where A, B, C refers to specific replica and superscript power refers to dilution factor of the positive end-points sequenced.
Persistent Community Members in Aged Produced Water Samples
Community composition analysis showed few similarities at the time of sample collection, compared to when the stored samples were sequenced at the time of initial MPN inoculation, some 534–808 days after sample collection (Figure 1). Generally, species diversity increased during sample storage, likely a result of previously dominant taxa diminishing in numbers over time so that lower abundance (previously undetected) taxa represented a larger proportion of the remaining community. Sequences closely associated to the prevailing taxa Halanaerobium and Marinilabiaceae were present at the time of inoculation, but generally decreased in relative abundance over time (comparing community compositions in freshly sampled waters, and those of the stored samples used as inocula) with the exception of the Day 29 sample. Taxa not detected at the time of collection, or that were present only at low relative abundance (such as Anaerococcus and Flexistipes), represented a higher proportion of the microbial population present at time of inoculation (Figure 1). SRB affiliated with the genera Desulfotomaculum and Desulfovibrio, identified at the time of sample collection (combined relative abundance of <3.43% across all samples), were not detected in the inoculum.
Community Composition of Positive Fermentative MPNs
The FB medium (Spring et al., 2015) used in the MPN cultures for fermentative microorganisms is a non-restrictive sugar-containing medium containing both the fermentable carbon substrate glucose and yeast extract as a source of additional nutrients. MPNs in FB medium exhibited rapid and extensive growth (observed as turbidity) compared with MPNs initially set up using the thiosulfate-reducing and sulfate-reducing media. Figure compares the microbial community compositions of produced water inocula to the highest dilution FB MPN enrichments that were scored positive.
The diversity of 16S rRNA genes was lower in positive enrichments, compared to the inocula, with a Shannon H index as low as 0.01 in one enrichment (Figure 2), dominated almost entirely by OTUs belonging to the family Marinilabiaceae (99.91% relative abundance). Organisms most closely affiliated with Marinilabiaceae were detected across the inocula at relative abundances ranging from 0.02% in the stored day 303 sample to 34.7% in the stored day 29 sample (stored for 534 and 808 days, respectively, before use as MPN inocula). Although stored produced water collected 142 and 303 days post hydraulic fracturing contained less than 1% Marinilabiaceae, this increased to 53–56% and 40–47%, respectively, when transferred to FB medium. Organisms most closely related with the family Marinilabiaceae or genus Halanaerobium appear to be similarly dominant, with a combined abundance of over 99.8% in all enrichments. MPN enrichments for FB inoculated with the stored produced water collected 79 days post hydraulic fracturing were dominated by members of the genus Halanaerobium, which comprised over 99.9% relative abundance.
Community Composition of Positive Thiosulfate-Reducing MPNs
Halanaerobium species were found to increase in relative abundance across thiosulfate-reducing enrichments compared with inocula (Figure 3). The highest enrichment of Halanaerobium species was observed in MPNs inoculated with the day 29 produced water (30.0% relative abundance), with a final relative abundance of 97.2% in the MPN enrichment, thus highlighting Halanaerobium as a key player in sulfide production. Compared to the inocula, the relative abundance of Dethiosulfatibacter species increased ten-fold in all enrichments. Similarly Geoalkalibacter species, not detected in the inocula, were observed in positive enrichments inoculated with day 79 and day 142 produced waters. Organisms most closely affiliated with Geotoga species increased in relative abundance in TRB MPNs inoculated with the day 303 sample from less than 0.1% to 7.63%. Flexistipes species declined in relative abundance in TRB MPNs, but still remained the second most abundant genus after Halanaerobium at a dilution of 10–1 in all enrichments. Marinilabiaceae was detected in all starting inocula but their relative abundance declined across all enrichments to below 2%.
Subsequent TRB MPN enrichments amended with yeast extract were inoculated 9 months after the initial TRB MPNs. As such, community composition of these older inocula were assessed with 16S rRNA gene sequencing. Compared with the inocula used in initial TRB MPNs (Figure 3), Shannon diversity H index had decreased by the time of their use in yeast extract-amended TRB MPNs (Figure 4). Generally the relative abundance of Halanaerobium species in the MPN inocula had decreased over time while the relative abundance of Flexistipes species had increased over time.
Addition of yeast extract to TRB medium led to comparable community composition in positive enrichments (Figure 4) to that of FB enrichments (Figure 2). In all yeast extract-amended TRB positive enrichments, the relative abundance of Halanaerobium species increased, becoming the dominant lineage in most cases (Figure 4). Similarly, members of the family Synergistaceae increased in relative abundance from below 2.0% in the inocula to between 8.4% and 40.5% relative abundance in positive thiosulfate-reducing MPNs inoculated with day 142 and 303 produced waters, exhibiting up to a 6-fold increase in relative abundance (Figure 4).
In contrast to TRB positive MPNs without added yeast extract, the relative abundance of organisms most closely affiliated with Flexistipes dropped to below 0.2%. Dethiosulfatibacter was undetected in inocula yet became the dominant taxon in one positive enrichment inoculated with day 142 sample (50.0%).
Discussion
Hydraulic fracturing as a means of recovering natural gas from shale is known to create new microbial ecosystems in the deep subsurface (Daly et al., 2016). Some microbial processes in fractured shales are known to negatively impact on natural gas extraction, for instance through hydrocarbon souring and microbial-induced corrosion (Johnson et al., 2008). Here, we assessed the presence of fermentative and sulfidogenic bacteria in shale gas production fluids with a focus on persistent species surviving several hundred days in storage. Using a targeted MPN enrichment approach coupled to DNA-based microbial composition analysis, we demonstrated that dominant halotolerant taxa in fractured shales are persistent and have the potential to sour gas, contribute to corrosion and proliferate in the presence of fermentative substrates, potentially reducing permeability and thus natural gas recovery.
Microbial community composition of the production fluids shows a marked shift following storage, with communities changing between sample collection and initial testing (Figure 1), and further changes occurring between initial MPNs (Figures 2, 3) and follow up MPNs (Figure 4). After the long periods since their collection, produced water samples used in MPN enrichments still contained relatively high abundances of prominent taxa previously reported in fractured shale ecosystems, including species of Halanaerobium and Marinilabiliaceae (Wuchter et al., 2013; Fichter et al., 2017). Persistent microorganisms identified here were able to withstand nutrient-limiting storage conditions and grow in targeted MPN enrichments.
Fermentative and thiosulfate-reducing microorganisms were observed in MPN enrichments inoculated with all production fluids. In contrast, although 16S rRNA gene sequencing detected sequences affiliated with sulfate-reducing microorganisms at the time of sample collection, none were detected through MPN culturing (Table 1), in line with previous studies (Struchtemeyer and Elshahed, 2012). Fermentative enrichments were dominated by members of the Halanaerobium genus and Marinilabiliaceae family (Figure 1). This is unsurprising given that FB medium was the only selective growth medium containing glucose as members of both taxa can utilize glucose (Denger et al., 2002; Liang et al., 2016). Initial thiosulfate-reducing MPN enrichments led to increased abundances of taxa affiliated with Geoalkalibacter, Geotoga, Dethiosulfatibacter, and Synergistaceae (Figure 3), although subsequent enrichments amended with yeast extract were dominated by – Halanaerobium (Figure 4). Collectively, these findings demonstrate that a diverse array of fermentative and sulfidogenic microorganisms remain viable in production fluids for long periods of time. Although not all taxa putatively identified through 16S rRNA gene sequencing were culturable within the selective media used, many were amenable to laboratory culturing and quantification. The authors acknowledge that the metabolic activity witnessed under laboratory conditions may not reflect metabolic activity in situ.
Persistent Sulfidogenic Taxa
No SRB were observed in MPN tests (Table 1), despite their detection in produced waters shortly after collection. Sequences affiliated with SRB have been detected in drilling muds and produced water impoundments (Struchtemeyer et al., 2011; Murali Mohan et al., 2013b) and transcriptomic evidence suggests that the sulfate reduction pathway is active in produced waters (Vikram et al., 2016). The lack of viable SRB cultured from the production fluids following storage, suggests they are not persistent taxa and therefore pose limited threat long-term when considering the recycling of produced waters after extended holding periods. However, we acknowledge that the culturing conditions used may not have been conducive to the growth of all SRB.
Although sulfate-reducing MPNs were scored negative, a number of sulfidogenic taxa were observed in thiosulfate-reducing enrichments, including representatives of the genera Halanaerobium, Dethiosulfatibacter, Geoalkalibacter, and Geotoga (Figures 3, 4). It is well known that Halanaerobium species are able to colonize hydraulically fractured shale ecosystems, regardless of geography (Mouser et al., 2016; Lipus et al., 2017) and are ubiquitous across shale gas wells in the United States (Struchtemeyer and Elshahed, 2012; Murali Mohan et al., 2013a; Strong et al., 2013; Cluff et al., 2014; Akob et al., 2015; Daly et al., 2016; Mouser et al., 2016; Booker et al., 2017; Lipus et al., 2017; Borton et al., 2018). The moderate halophile Halanaerobium congolense has also been shown to reduce both thiosulfate and elemental sulfur to sulfide, but lacks the ability to reduce sulfate to sulfide (Ravot et al., 2005). Metabolic pathway analyses have revealed that Halanaerobium species have the potential for thiosulfate reduction, acid production and biofilm formation (Lipus et al., 2017), potentially making this a highly problematic microorganism during shale gas extraction. Indeed this ability has led to an emerging interest in the role that Halanaerobium species may have in the biogenesis of sulfide and corrosive organic acids, such as acetate (Booker et al., 2017, 2019). It is therefore unsurprising that Halanaerobium spp. were detected across all produced water samples studied here, and showed increased relative abundance across all targeted enrichments, even after prolonged periods in storage.
Dethiosulfatibacter, another known thiosulfate-reducing bacterium (Takii et al., 2007; Qian et al., 2019), was also enriched in positive TRB MPNs, with and without yeast extract, despite low abundance in the inocula (Figures 3, 4). Members of this genus were also detected in production fluids from a 4.1 km shale gas well in the Sichuan Basin, China, and relative abundance was found to increase between produced water production and storage (Zhang et al., 2017). Together these findings indicate Dethiosulfatibacter may contribute to corrosion and souring during shale gas extraction.
Also enriched in thiosulfate-reducing enrichments was Geoalkalibacter. Although not detected in produced water samples, its increased abundance in enrichments from day 79 to day 142 samples suggest that it is capable of thiosulfate reduction. Geoalkalibacter was not detected in any enrichments that used yeast extract containing media, despite reports that yeast extract is required as a source of growth factors (Zavarzina et al., 2006), perhaps due to being outcompeted by dominant Halanaerobium strains (Figure 4). Isolates of this genus have not been shown to mediate thiosulfate reduction, although G. ferrihydriticaus and G. subterraneus can produce sulfide from the reduction of elemental sulfur (Zavarzina et al., 2006; Greene et al., 2009). Geoalkalibacter species have been identified previously in produced waters collected from shale gas wells in the Antrim shale formation (Wuchter et al., 2013). When taken together, these findings suggest members of this genus may in fact be capable of thiosulfate reduction, and could play a role in souring and microbially induced corrosion during shale gas extraction.
Members of the genus Geotoga are also capable of reducing elemental sulfur to form hydrogen sulfide (Davey et al., 1993), but to the best of our knowledge is not known to be capable of thiosulfate reduction. Geotoga species were detected at low levels (<1%) in the initial day 79 and 303 produced water samples, but showed a ten-fold increase in relative abundance when cultured in thiosulfate-reducing media (Figure 3). Geotoga species were observed in similar sulfidogenic enrichments using produced waters from the Marcellus shale, although the added electron acceptor in that study was sulfate in the form of barite (Wuchter et al., 2013). Laboratory reactor experiments established with Utica produced waters and amended with glycine betaine also gave rise to an enrichment in Geotoga (Borton et al., 2018). Proteomic evidence for carbon-based metabolism in Geotoga in these experiments highlighted its importance in persistent shale metabolic networks (Borton et al., 2018). Our findings also demonstrate it is a persistent member of fractured shale communities, and may also contribute to biogenic sulfide production.
Evidently, many sulfide producing taxa have the potential to remain persistant and metabolically active for many years post fracturing, as illustrated through the community composition analysis of positive thiosulfate-reducing enrichments (Figures 3, 4). Although SRB have been identified previously in production fluids through next generation sequencing (Davis et al., 2012; Struchtemeyer and Elshahed, 2012; Liang et al., 2016) no SRB were shown to be viable in these experiments, in agreement with Struchtemeyer and Elshahed (2012). Multiple Halanaerobium species have been isolated from shale environments and exhibit thiosulfate-reducing capabilities (Ravot et al., 1997, 2005; Fichter et al., 2012; Liang et al., 2014, 2016; Booker et al., 2017). It appears more likely that thiosulfate-reduction is the more prominent sulfidogenic pathway in fractured shale communities, as evidenced by the widespread presence of the thiosulfate-reducing bacterium Halanaerobium (Davis et al., 2012; Struchtemeyer and Elshahed, 2012; Murali Mohan et al., 2013a; Strong et al., 2013; Cluff et al., 2014; Akob et al., 2015; Daly et al., 2016; Mouser et al., 2016; Vikram et al., 2016; Booker et al., 2017; Lipus et al., 2017; Borton et al., 2018; Evans et al., 2019).
Biogenic sulfide production in conventional oil reservoirs has long been attributed to the activity of SRB, and targeted culture-based MPN enrichments for these organisms have been effective in diagnosing a potential biogenic souring problem. Molecular analyses targeting genes in the sulfate reduction pathway have also been described (Muyzer and Stams, 2008). However, in line with our own findings, these approaches will not detect the diverse array of taxa in fractured shale communities capable of sulfide production via thiosulfate reduction. The long-term persistence of sulfidogenic lineages demonstrated in this study further highlights the need for effective diagnostic tools and control strategies targeted to thiosulfate-reducing microorganisms.
Influence of Yeast Extract
Yeast extract was added as a source of trace nutrients to the second round of thiosulfate-reducing MPNs. However, given the strong enrichment for Halanaerobium in yeast extract-amended thiosulfate-reducing enrichments, the yeast extract may have served directly as a growth substrate for Halanaerobium (Figure 4). MPN counts were several orders of magnitude lower in the initial TRB medium compared with fermentative MPNs cultured in FB medium and thiosulfate-reducing MPNs supplemented with yeast extract (Table 1), confirming that yeast extract enhances growth in these cultures.
Most probable number enrichments conducted in TRB medium with the addition of yeast extract visually formed iron sulfide within days of inoculation, as opposed to months in thiosulfate-reducing MPNs devoid of yeast extract. In contrast to initial thiosulfate-reducing MPN enrichments, no significant decrease in acetate or lactate was observed in yeast-amended thiosulfate-reducing enrichments, suggesting components of yeast extract were serving as alternative electron donors coupled to thiosulfate reduction. Indeed, prior work suggests that Halanaerobium uses thiosulfate as a sink for reducing equivalents during fermentation reactions, although this process was not coupled directly to growth (Booker et al., 2017). Yeast extract has also been linked previously to increased sulfide production via thiosulfate reduction, and it has been suggested that yeast extract may itself serve as an electron donor (Liang et al., 2014). One plausible explanation for this is that amino acids within yeast extract could be being fermented by Halanaerobium. Amino acids such as lysine and alanine have been suggested as fermentable substrates for Halanaerobium based on metagenomic data and these amino acids are likely present within yeast extract (Daly et al., 2016; Borton et al., 2018). Yeast extract may also provide additional growth factors or trace nutrients that are otherwise in limited supply and further accelerate growth rate.
Other Noteworthy Taxa
In addition to the taxa discussed above, which are likely to catalyze thiosulfate reduction, other microbes that require further discussion include members of the genus Flexistipes and family Marinilabiaceae. Following storage, members of the genus Flexistipes increased in relative abundance, becoming the most abundant taxa at 79, 142, and 303 days post hydraulic fracturing, prior to inoculation in the MPN series (Figure 1). The genus Flexistipes has been previously identified in deep marine environments and are strict anaerobes with a requirement for NaCl and a short generation time of just 8.5 h under optimum conditions, so it is unsurprising that they are able to dominate the stored samples (Fiala et al., 1990; Lapidus et al., 2011). However, their relative abundance decreased dramatically when transferred into MPN cultures; for example, produced waters collected 79 and 303 days after hydraulic fracturing and stored for 1049 and 825 days, respectively, prior to use as MPN inocula consisted of over 90% Flexistipes, this dropped to below, 0.2% when transferred into thiosulfate-reducing medium containing yeast extract (Figure 4). Based on these observations it is unlikely that Flexistipes play a direct role in sulfide production as they were out compete by Halanaerobium despite their initial high abundance. However, this drop in relative abundance may also be attributed to unsuitable growth media. FB enrichments were dominated by the members of the family Marinilabiaceae and/or genus Halanaerobium, able to proliferate despite the high salinity (8% NaCl), moderate temperature (40°C) and the availability of electron donors/acceptors within the FB medium. However, when transferred to thiosulfate-reducing MPN cultures, the relative abundance of Marinilabiaceae declined dramiatically across all enrichments to below 2%, suggesting that they are unlikley to play a role in sulfide production, this conclusion is supported by studies showing strains of Marinilabiaceae being incapable of sulfide production via thiosulfate reduction (Yang et al., 2014). Such taxa could still have a role in biofouling through fermentation of organic substrates or fracture clogging, for example.
Conclusion
In summary, this work has shown that many metabolically active sulfide-producing taxa can persist for long periods of time post fracturing, including Halanaerobium and Dethiosulfatibacter species. Such microorganisms may produce hydrogen sulfide, a chemical which is hazardous to human health (Guidotti, 2010) and can cause souring reactions resulting in sour gas and the corrosion of steel infastructure (Eden et al., 1993; Fichter et al., 2012). Although classical SRB were not abundent in the aged produced waters, a key finding in this study was the identification of the TRB Halanaerobium, which was highlighted as a potentially problematic and persistent microorganism within aged produced water communities. This persistence could have significant implications in regard to both sulfide production and biofouling when considering the storage or recycling of produced waters, and needs to be studied in more depth using samples stored appropriately on site. Although produced water is treated prior to reuse with biocides and/or aeration these problematic microorganisms could still go on to seed new wells and some studies suggest that this could lead to a rise in biocide resistance (Russell, 1995; Struchtemeyer and Elshahed, 2012; Murali Mohan et al., 2013b) Clearly new detection and control methods must be installed to combat the problems posed by such persistent and problematic microorganisms. Such studies are now ongoing using cultures from a range of hydraulically fractured shale systems, facilitated by the relative ease of laboratory cultivation of appropriate model organisms such as Halanaerobium using selective media, as demonstrated in this study.
Data Availability Statement
The datasets generated for this study can be found in NCBI SRA accession PRJNA578032 refers to the microcosm experiments and SAMN13315659, SAMN13315660, SAMN13315661, and SAMN13315662 refer to samples “Fresh Day” 29, 79, 142, and 303, respectively.
Author Contributions
LC, SN, and JL contributed to conception and design of the experiments. RD collected initial samples. MW and KW provided the samples for experiments conducted by LC. DNA was extracted from experiments by LC and 16S rRNA gene sequencing carried out by CB. LC carried out the data analysis, prepared figures and LC and SN wrote the manuscript with contributions and revisions from all authors.
Funding
LC acknowledges support from the Natural Environment Research Council CASE studentship and CASE partner Rawwater Engineering Company Limited along with technical support staff CB and Alastair Bewsher. SN acknowledges support from a Natural Environment Research Council Research Fellowship (NE/R013462/1). JL acknowledges financial support from the Royal Society.
Conflict of Interest
BE was employed by Rawwater Engineering Company Limited. Authors declare that this study received funding from Rawwater Engineering Company Limited as CASE sponsors for LC’s NERC-funded Ph.D., providing background industry context to the work and comments on the manuscript.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Akob, D. M., Cozzarelli, I. M., Dunlap, D. S., Rowan, E. L., and Lorah, M. M. (2015). Organic and inorganic composition and microbiology of produced waters from Pennsylvania shale gas wells. Appl. Geochem. 60, 116–125. doi: 10.1016/j.apgeochem.2015.04.011
Andrews, S. (2010). FastQC: A Quality Control Tool For High Throughput Sequence Data. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed September 16, 2019).
Barbot, E., Vidic, N. S., Gregory, K. B., and Vidic, R. D. (2013). Spatial and temporal correlation of water quality parameters of produced waters from Devonian-age shale following hydraulic fracturing. Environ. Sci. Technol. 47, 2562–2569. doi: 10.1021/es304638h
Booker, A. E., Borton, M. A., Daly, R. A., Welch, S. A., Nicora, C. D., Hoyt, D. W., et al. (2017). Sulfide generation by dominant Halanaerobium microorganisms in hydraulically fractured shales. mSphere 2:e0257-17. doi: 10.1128/mSphereDirect.00257-217
Booker, A. E., Hoyt, D. W., Meulia, T., Eder, E., Nicora, C. D., Purvine, S. O., et al. (2019). Deep-subsurface pressure stimulates metabolic plasticity in shale-colonizing Halanaerobium spp. Appl. Environ. Microbiol. 85, 1–16. doi: 10.1128/AEM.00018-19
Borton, M. A., Hoyt, D. W., Roux, S., Daly, R. A., Welch, S. A., Nicora, C. D., et al. (2018). Coupled laboratory and field investigations resolve microbial interactions that underpin persistence in hydraulically fractured shales. PNAS 115, E6585–E6594. doi: 10.1073/pnas.1800155115
Borton, M. A., Sabag-Daigle, A., Wu, J., Solden, L. M., O’Banion, B. S., Daly, R. A., et al. (2017). Chemical and pathogen-induced inflammation disrupt the murine intestinal microbiome. Microbiome 5, 1–15. doi: 10.1186/s40168-017-0264-268
Bottero, S., Picioreanu, C., Delft, T., Enzien, M., Control, D. M., van Loosdrecht, M. C. M., et al. (2010). “Formation damage and impact on gas flow caused by biofilms growing within proppant packing used in hydraulic fracturing,” in Proceedings - SPE International Symposium on Formation Damage Control, Piscataway, NJ.
Caporaso, J., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F., Costello, E., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl.), 4516–4522. doi: 10.1073/pnas.1000080107
Cluff, M. A., Hartsock, A., MacRae, J. D., Carter, K., and Mouser, P. J. (2014). Temporal changes in microbial ecology and geochemistry in produced water from hydraulically fractured Marcellus shale gas wells. Environ. Sci. Technol. 48, 6508–6517. doi: 10.1021/es501173p
Compeau, P. E. C., Pevzner, P. A., Tesler, G., Papoutsoglou, G., Roscito, J. G., Dahl, A., et al. (2013). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi: 10.14806/ej.17.1.200
Daly, R. A., Borton, M. A., Wilkins, M. J., Hoyt, D. W., Kountz, D. J., Wolfe, R. A., et al. (2016). Microbial metabolisms in a 2.5-km-deep ecosystem created by hydraulic fracturing in shales. Nat. Microbiol. 1, 1–9. doi: 10.1038/nmicrobiol.2016.146
Daly, R. A., Roux, S., Borton, M. A., Morgan, D. M., Johnston, M. D., Booker, A. E., et al. (2019). Viruses control dominant bacteria colonizing the terrestrial deep biosphere after hydraulic fracturing. Nat. Microbiol. 4, 352–361. doi: 10.1038/s41564-018-0312-316
Davey, M. E., Wood, W. A., Key, R., Nakamura, K., and Stahl, D. A. (1993). Isolation of three species of geotoga and petrotoga: two new genera, representing a new lineage in the bacterial line of descent distantly related to the “thermotogales.”. Syst. Appl. Microbiol. 16, 191–200. doi: 10.1016/S0723-2020(11)80467-80464
Davis, J. P., Struchtemeyer, C. G., and Elshahed, M. S. (2012). Bacterial communities associated with production facilities of two newly drilled thermogenic natural gas wells in the barnett shale (Texas. USA). Microb. Ecol. 64, 942–954. doi: 10.1007/s00248-012-0073-73
Denger, K., Warthmann, R., Ludwig, W., and Schink, B. (2002). Anaerophaga thermohalophilia gen. nov. sp., A Moderately Thermohalophilic, strictly anaerobic fermentative bacterium. Int. J. Syst. Evol. Microbiol. 52, 173–178. doi: 10.1099/00207713-52-1-173
Eden, B., Laycock, P. J., and Fielder, M. (1993). Oilfield Resevoir Souring. HSE Books. Available at: http://www.hse.gov.uk/research/othpdf/200-399/oth385.pdf (accessed September 16, 2019).
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Elsner, M., and Hoelzer, K. (2016). Quantitative survey and structural classification of hydraulic fracturing chemicals reported in unconventional gas production. Environ. Sci. Technol. 50, 3290–3314. doi: 10.1021/acs.est.5b02818
Evans, M. V., Getzinger, G., Luek, J. L., Hanson, A. J., McLaughlin, M. C., Blotevogel, J., et al. (2019). In situ transformation of ethoxylate and glycol surfactants by shale-colonizing microorganisms during hydraulic fracturing. ISME J. 13, 2690–2700. doi: 10.1038/s41396-019-0466-0
Evans, M. V., Panescu, J., Hanson, A. J., Welch, S. A., Sheets, J. M., Nastasi, N., et al. (2018). Members of marinobacterand arcobacterinfluence system biogeochemistry during early production of hydraulically fractured natural gas wells in the appalachian basin. Front. Microbiol. 9:2646. doi: 10.3389/fmicb.2018.02646
Fiala, G., Woese, C. R., Langworthy, T. A., and Stetter, K. O. (1990). Flexistipes sinusarabici, a novel genus and species of eubacteria occurring in the Atlantis II deep brines of the Red Sea. Arch. Microbiol. 154, 120–126. doi: 10.1007/bf00423320
Fichter, J., Janes, C., Summer, E. J., Mills, A., and Summer, E. J. (2017). Use of A Methodological Panel to Identify the Source of Problematic Microbial Contamination in an Oil Shale Field. Houston, TX: NACE.
Fichter, J., Moore, R., and Summer, E. (2012). How hot is too hot for bacteria? A technical study assessing bacterial establishment in downhole drilling, fracturing and stimulation operation. NACE Int. 3, 2402–2422.
Greene, A. C., Patel, B. K. C., and Yacob, S. (2009). Geoalkalibacter subterraneus sp. nov., an anaerobic Fe(III)- and Mn(IV)-reducing bacterium from a petroleum reservoir, and emended descriptions of the family Desulfuromonadaceae and the genus Geoalkalibacter. Int. J. Syst. Evol. Microbiol. 59, 781–785. doi: 10.1099/ijs.0.001537-1530
Gregory, K. B., Vidic, R. D., and Dzombak, D. A. (2011). Water management challenges associated with the production of shale gas by hydraulic fracturing. Elements 7, 181–186. doi: 10.2113/gselements.7.3.181
Grein, F., Ramos, A. R., Venceslau, S. S., and Pereira, I. A. C. (2013). Unifying concepts in anaerobic respiration: insights from dissimilatory sulfur metabolism. Biochim. Biophys. Acta Bioenerg. 1827, 145–160. doi: 10.1016/j.bbabio.2012.09.001
Guidotti, T. L. (2010). Hydrogen sulfide: advances in understanding human toxicity. Int. J. Toxicol. 29, 569–581. doi: 10.1177/1091581810384882
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Hayes, T. (2009). Sampling and Analysis of Water Streams Associated with the Development of Marcellus Shale Gas. Final Report Prepared for Marcellus Shale Coalition. Des Plaines, IL: Gas Technology Institute.
Jenner, S., and Lamadrid, A. J. (2013). Shale gas vs. coal: policy implications from environmental impact comparisons of shale gas, conventional gas, and coal on air, water, and land in the United States. Energy Policy 53, 442–453. doi: 10.1016/j.enpol.2012.11.010
Johnson, K., French, K., Fichter, J. K., and Oden, R. (2008). Use of Microbiocides in Barnett Shale Gas Well Fracturing Fluids to Control Bacteria Related Problems. Houston, TX: NACE International.
Joshi, N., and Fass, J. (2011). Sickle: A sliding-window, Adaptive, Quality-Based Trimming Tool for FastQ Files (Version 1.33).
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the Miseq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/aem.01043-1013
Lapidus, A., Chertkov, O., Nolan, M., Lucas, S., Hammon, N., Deshpande, S., et al. (2011). Genome sequence of the moderately Thermophilic halophile Flexistipes sinusarabici strain (MAS10t). Stand. Genomic Sci. 5, 86–96. doi: 10.4056/sigs.2235024
Lee, D. S., Herman, J. D., Elsworth, D., Kim, H. T., and Lee, H. S. (2011). A critical evaluation of unconventional gas recovery from the marcellus shale, northeastern United States. KSCE J. Civ. Eng. 15, 679–687. doi: 10.1007/s12205-011-0008-4
Liang, R., Davidova, I. A., Marks, C. R., Stamps, B. W., Harriman, B. H., Stevenson, B. S., et al. (2016). Metabolic capability of a predominant Halanaerobium sp. in hydraulically fractured gas wells and its implication in pipeline corrosion. Front. Microbiol. 7:988. doi: 10.3389/fmicb.2016.00988
Liang, R., Grizzle, R. S., Duncan, K. E., McInerney, M. J., and Suflita, J. M. (2014). Roles of thermophilic thiosulfate-reducing bacteria and methanogenic archaea in the biocorrosion of oil pipelines. Front. Microbiol. 5:89. doi: 10.3389/fmicb.2014.00089
Lipus, D., Vikram, A., Ross, D., Bain, D., Gulliver, D., Hammack, R., et al. (2017). Predominance and metabolic potential of Halanaerobium spp. in produced water from hydraulically fractured Marcellus shale wells. Am. Soc. Microbiol. 83, 1–15. doi: 10.1128/AEM.02659-16
Masella, A. P., Bartram, A. K., Truszkowski, J. M., Brown, D. G., and Neufeld, J. D. (2012). PANDAseq: paired-end assembler for illumina sequences. BMC Bioinform. 13:31. doi: 10.1186/1471-2105-13-31
Miller, T. L., and Wolin, M. J. (1974). A serum bottle modification of the hungate technique for cultivating obligate anaerobes. Appl. Microbiol. 27, 985–987. doi: 10.1128/aem.27.5.985-987.1974
Mouser, P. J., Borton, M., Darrah, T. H., Hartsock, A., and Wrighton, K. C. (2016). Hydraulic fracturing offers view of microbial life in the deep terrestrial subsurface. FEMS Microbiol. Ecol. 92, 1–18. doi: 10.1093/femsec/iw166
MSEEL, (2015). Project Overview. Available at: www.mseel.org/(accessed December 16, 2019).
Murali Mohan, A., Hartsock, A., Bibby, K. J., Hammack, R. W., Vidic, R. D., and Gregory, K. B. (2013a). Microbial community changes in hydraulic fracturing fluids and produced water from shale gas extraction. Environ. Sci. Technol. 47, 13141–13150. doi: 10.1021/es402928b
Murali Mohan, A., Hartsock, A., Hammack, R. W., Vidic, R. D., and Gregory, K. B. (2013b). Microbial communities in flowback water impoundments from hydraulic fracturing for recovery of shale gas. FEMS Microbiol. Ecol. 86, 567–580. doi: 10.1111/1574-6941.12183
Muyzer, G., and Stams, A. J. M. (2008). The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 6, 441–454. doi: 10.1038/nrmicro1892
Nixon, S. L., Daly, R. A., Borton, M. A., Solden, L. M., Welch, S. A., Cole, D. R., et al. (2019). Genome-resolved metagenomics extends the environmental distribution of the verrucomicrobia phylum to the deep terrestrial subsurface. Am. Soc. Microbiol. 4, 1–18. doi: 10.1128/mSphere.00613-619
Nixon, S. L., van Dongen, B. E., Boothman, C., Small, J. S., and Lloyd, J. R. (2018). Additives in plasticised polyvinyl chloride fuel microbial nitrate reduction at high pH: implications for nuclear waste disposal. Front. Environ. Sci. 6:97. doi: 10.3389/fenvs.2018.00097
Nixon, S. L., Walker, L., Streets, M. D. T., Eden, B., Boothman, C., Taylor, K. G., et al. (2017). Guar gum stimulates biogenic sulfide production at elevated pressures: Implications for shale gas extraction. Front. Microbiol. 8:679. doi: 10.3389/fmicb.2017.00679
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A. A., Korobeynikov, A., Lapidus, A., et al. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20, 714–737. doi: 10.1089/cmb.2013.0084
Oblinger, J. L., and Koburger, J. A. (1975). Understanding and teaching the most probable number technique. J. Milk Food Technol. 38:6. doi: 10.4315/0022-2747-38.9.540
Qian, J., Zhang, M., Jing, R., Bai, L., Zhou, B., Zhao, M., et al. (2019). Thiosulfate as the electron acceptor in sulfur bioconversion-associated process (SBAP) for sewage treatment. Water Res. 163, 114850. doi: 10.1016/j.watres.2019.07.017
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
Ravot, G., Casalot, L., Ollivier, B., Loison, G., and Magot, M. (2005). RdlA, a new gene encoding a rhodanese-like protein in Halanaerobium congolense and other thiosulfate-reducing anaerobes. Res. Microbiol. 156, 1031–1038. doi: 10.1016/j.resmic.2005.05.009
Ravot, G., Magot, M., Ollivier, B., Patel, B. K., Ageron, E., Grimont, P. A., et al. (1997). Haloanaerobium congolense sp. nov., an anaerobic, moderately halophilic, thiosulfate- and sulfur-reducing bacterium from an African oil field. FEMS Microbiol. Lett. 147, 81–88. doi: 10.1111/j.1574-6968.1997.tb10224.x
Rowan, E. L., Engle, M. A., Kraemer, T. F., Schroeder, K. T., Hammack, R. W., and Doughten, M. W. (2015). Geochemical and isotopic evolution of water produced from Middle devonian marcellus shale gas wells, appalachian basin, pennsylvania. Am. Assoc. Pet. Geol. Bull. 99, 181–206. doi: 10.1306/07071413146
Russell, A. D. (1995). Mechanisms of bacterial resistance to biocides. Int. Biodeterior. Biodegrad. 36, 247–265. doi: 10.1016/0964-8305(95)00056-59
Schramm, E. (2011). What is Flowback, and How Does It Differ From Produced Water? Inst. Energy Environ. Res. Northeast. Pennsylvania Clear. Available at: https://termine.com/archives/494 (accessed July 5, 2019).
Spring, S., Brinkmann, N., Murrja, M., Spröer, C., Reitner, J., and Klenk, H. P. (2015). High diversity of culturable prokaryotes in a lithifying hypersaline microbial mat. Geomicrobiol. J. 32, 332–346. doi: 10.1080/01490451.2014.913095
Stringfellow, W. T., Domen, J. K., Camarillo, M. K., Sandelin, W. L., and Borglin, S. (2014). Physical, chemical, and biological characteristics of compounds used in hydraulic fracturing. J. Hazard. Mater. 275, 37–54. doi: 10.1016/j.jhazmat.2014.04.040
Strong, L. C., Gould, T., Kasinkas, L., Sadowsky, M. J., Aksan, A., and Wackett, L. P. (2013). Biodegradation in waters from hydraulic fracturing: chemistry, microbiology, and engineering. J. Environ. Eng. 140:B4013001. doi: 10.1061/(ASCE)EE.1943-7870.0000792
Struchtemeyer, C. G., Davis, J. P., and Elshahed, M. S. (2011). Influence of the drilling mud formulation process on the bacterial communities in thermogenic natural gas wells of the Barnett Shale. Appl. Environ. Microbiol. 77, 4744–4753. doi: 10.1128/AEM.00233-211
Struchtemeyer, C. G., and Elshahed, M. S. (2012). Bacterial communities associated with hydraulic fracturing fluids in thermogenic natural gas wells in North Central Texas, USA. FEMS Microbiol. Ecol. 81, 13–25. doi: 10.1111/j.1574-6941.2011.01196.x
Takii, S., Hanada, S., Tamaki, H., Ueno, Y., Sekiguchi, Y., Ibe, A., et al. (2007). Dethiosulfatibacter aminovorans gen. nov., sp. nov., a novel thiosulfate-reducing bacterium isolated from coastal marine sediment via sulfate-reducing enrichment with Casamino acids. Int. J. Syst. Evol. Microbiol. 57, 2320–2326. doi: 10.1099/ijs.0.64882-64880
Tanner, R. S. (1989). Monitoring sulfate-reducing bacteria: comparison of enumeration media. J. Microbiol. Methods 10, 83–90. doi: 10.1016/0167-7012(89)90004-90003
Vikram, A., Lipus, D., and Bibby, K. (2016). Metatranscriptome analysis of active microbial communities in produced water samples from the Marcellus Shale. Microb. Ecol. 72, 571–581. doi: 10.1007/s00248-016-0811-z
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-67
Wuchter, C., Banning, E., Mincer, T. J., Drenzek, N. J., and Coolen, M. J. L. (2013). Microbial diversity and methanogenic activity of antrim shale formation waters from recently fractured wells. Front. Microbiol. 4:367. doi: 10.3389/fmicb.2013.00367
Yang, S. H., Seo, H. S., Woo, J. H., Oh, H. M., Jang, H., Lee, J. H., et al. (2014). Carboxylicivirga gen. nov. in the family Marinilabiliaceae with two novel species, Carboxylicivirga mesophila sp. nov. and Carboxylicivirga taeanensis sp. nov., and reclassification of Cytophaga fermentans as Saccharicrinis fermentans gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 64, 1351–1358. doi: 10.1099/ijs.0.053462-53460
Zavarzina, D. G., Kolganova, T. V., Boulygina, E. S., Kostrikina, N. A., Tourova, T. P., and Zavarzin, G. A. (2006). Geoalkalibacter ferrihydriticus gen. nov. sp. nov., the first alkaliphilic representative of the family Geobacteraceae, isolated from a soda lake. Microbiology 75, 673–682. doi: 10.1134/s0026261706060099
Keywords: thiosulfate-reducing bacteria, biogenic sulfide, Halanaerobium, non-conventional gas, microbiology
Citation: Cliffe L, Nixon SL, Daly RA, Eden B, Taylor KG, Boothman C, Wilkins MJ, Wrighton KC and Lloyd JR (2020) Identification of Persistent Sulfidogenic Bacteria in Shale Gas Produced Waters. Front. Microbiol. 11:286. doi: 10.3389/fmicb.2020.00286
Received: 04 October 2019; Accepted: 07 February 2020;
Published: 21 February 2020.
Edited by:
Terry C. Hazen, The University of Tennessee, Knoxville, United StatesReviewed by:
Lisa Gieg, University of Calgary, CanadaRegina Lamendella, Juniata College, United States
Copyright © 2020 Cliffe, Nixon, Daly, Eden, Taylor, Boothman, Wilkins, Wrighton and Lloyd. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lisa Cliffe, TGlzYS5DbGlmZmVAbWFuY2hlc3Rlci5hYy51aw==
 Lisa Cliffe
Lisa Cliffe Sophie L. Nixon
Sophie L. Nixon Rebecca A. Daly
Rebecca A. Daly Bob Eden
Bob Eden Kevin G. Taylor
Kevin G. Taylor Christopher Boothman
Christopher Boothman Michael J. Wilkins
Michael J. Wilkins Kelly C. Wrighton
Kelly C. Wrighton Jonathan R. Lloyd
Jonathan R. Lloyd