
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 February 2020
Sec. Infectious Agents and Disease
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.00284
This article is part of the Research Topic Pathogenomics of the genus Brucella and beyond View all 31 articles
The genus Brucella comprises major pathogenic species causing disease in livestock and humans, e.g. B. melitensis. In the past few years, the genus has been significantly expanded by the discovery of phylogenetically more distant lineages comprising strains from diverse wildlife animal species, including amphibians and fish. The strains represent several potential new species, with B. inopinata as solely named representative. Being genetically more distant between each other, relative to the “classical” Brucella species, they present distinct atypical phenotypes and surface antigens. Among surface protein antigens, the Omp2a and Omp2b porins display the highest diversity in the classical Brucella species. The genes coding for these proteins are closely linked in the Brucella genome and oriented in opposite directions. They share between 85 and 100% sequence identity depending on the Brucella species, biovar, or genotype. Only the omp2b gene copy has been shown to be expressed and genetic variation is extensively generated by gene conversion between the two copies. In this study, we analyzed the omp2 loci of the non-classical Brucella spp. Starting from two distinct ancestral genes, represented by Australian rodent strains and B. inopinata, a stepwise nucleotide reduction was observed in the omp2b gene copy. It consisted of a first reduction affecting the region encoding the surface L5 loop of the porin, previously shown to be critical in sugar permeability, followed by a nucleotide reduction in the surface L8 loop-encoding region. It resulted in a final omp2b gene size shared between two distinct clades of non-classical Brucella spp. (African bullfrog isolates) and the group of classical Brucella species. Further evolution led to complete homogenization of both omp2 gene copies in some Brucella species such as B. vulpis or B. papionis. The stepwise omp2b deletions seemed to be generated through recombination with the respective omp2a gene copy, presenting a conserved size among Brucella spp., and may involve short direct DNA repeats. Successive Omp2b porin alteration correlated with increasing porin permeability in the course of evolution of Brucella spp. They possibly have adapted their porin to survive environmental conditions encountered and to reach their final status as intracellular pathogen.
Members of the genus Brucella are Gram-negative, facultative, intracellular bacteria that can infect many species of animals and man. Until the 1990s, six species were classically recognized within the genus Brucella: B. abortus, B. melitensis, B. suis, B. ovis, B. canis, and B. neotomae (Corbel and Brinley Morgan, 1984; Moreno et al., 2002; Godfroid et al., 2011). This classification was mainly based on differences in pathogenicity, host preference, and phenotypic characteristics (Alton et al., 1988). Since then, with the help of modern molecular typing methods [e.g. Multiple Loci Sequence Analysis (MLSA) and Multiple Loci VNTR Analysis (MLVA)] and whole genome sequencing (WGS), a number of new species representing mostly wildlife isolates and showing very different phenotypes have been validly published. In chronological order it concerns the species (i) B. ceti and B. pinnipedialis isolated from marine mammals, with cetaceans (dolphin, porpoise, and whale species) and pinnipeds (various seal species) as preferred hosts, respectively (Foster et al., 2007); (ii) B. microti isolated initially from the common vole but found later also in red foxes, in soil, and most recently in marsh frogs (Scholz et al., 2008a, b, 2009; Jaÿ et al., 2018); (iii) B. inopinata isolated from human (Scholz et al., 2010); (iv) B. papionis isolated from baboons (Whatmore et al., 2014); and (v) the latest B. vulpis species isolated from red foxes (Scholz et al., 2016a). Novel Brucella strains representing potentially novel species have also been isolated from Australian rodents (Tiller et al., 2010a), a wide variety of frog species (Eisenberg et al., 2012; Fischer et al., 2012; Scholz et al., 2016b; Soler-Lloréns et al., 2016; Al Dahouk et al., 2017; Kimura et al., 2017; Mühldorfer et al., 2017), and surprisingly also from fish namely from a bluespotted ribbontail ray (Taeniura lymma) (Eisenberg et al., 2017). The genus Brucella nowadays is thus not restricted to mammal species. The most recent potential new species reported is isolated from a dog in Costa Rica in the early 1980s (Guzmán-Verri et al., 2019).
Brucella spp. consist nowadays of two major groups. The first represents the species termed “classical” and consists of the six initially recognized species together with the more recent species B. ceti, B. pinnipedialis, B. microti, and B. papionis. The further subdivision in MLSA or MLVA genotypes proved to be helpful for subtyping of some species. For example B. ceti genotype ST23 is currently mainly found in several cetacean species (porpoise, dolphin, whale), whereas B. ceti genotype ST26 seems more restricted to dolphin species (Maquart et al., 2009; Whatmore et al., 2016; Vergnaud et al., 2018). The first group has been characterized and investigated in detail because of its importance in causing disease in animals and humans. The second major group is represented by lineages phylogenetically more distant from the classical species and still poorly characterized. This group comprises several distinct subgroups consisting of isolates from diverse wildlife animal species cited above, i.e. Red foxes, Australian rodents, several frog species, the isolate from a ray fish, and interestingly also two isolates from human cases. These subgroups represent several potential new species, with B. inopinata, isolated from a human case, as solely named representative. Being genetically more distant between each other, relative to the classical Brucella species, they present distinct atypical phenotypes and surface antigens. For example several amphibian subgroups and the human isolate Brucella sp. BO2 present distinct unidentified O antigens, that are not typable by polyclonal or monoclonal antibodies used for serotyping of the classical species (Tiller et al., 2010b; Zygmunt et al., 2012; Al Dahouk et al., 2017).
Among outer membrane proteins, the Omp2 porins have been shown to display the highest diversity within the classical Brucella species, which allowed to differentiate them at the species, biovar, or genotype level (Ficht et al., 1990, 1996; Cloeckaert et al., 1995, 2001). More precisely, the porins are encoded by the omp2 locus, consisting of two closely related genes omp2a and omp2b, separated by approximately 830 bp and oriented in opposite directions (Ficht et al., 1989; Marquis and Ficht, 1993). Gene diversity is extensively generated by recombination between both copies called also gene conversion (Santoyo and Romero, 2005), thus resulting in omp2a and omp2b gene copies sharing from 82 to 100% nucleotide identity depending on the species, biovar or genotype. The highest divergence between the gene copies in classical species was found in B. microti and B. melitensis (82.4 and 83.4% identity, respectively, see Supplementary Figure S1). In contrast, complete homogenization of the copies (100% identity) was observed for B. ceti ST26 or B. papionis (Supplementary Figure S1). Several other situations may be observed and in former studies Brucella strains were sometimes described in a simple way as carrying either (i) both the omp2a and omp2b gene copies (e.g. B. melitensis), (ii) two omp2a gene copies (e.g. B. ovis), or (iii) two omp2b gene copies (e.g. B. ceti) (Ficht et al., 1990, 1996; Cloeckaert et al., 2001). The release of additional sequences showed that the situation is more complex because of the existence of numerous intermediate situations, including also B. ovis whose gene copies are actually not 100% omp2a identical. Only the omp2b gene copy has been shown to be expressed (Marquis and Ficht, 1993), and the genetic exchanges that may occur with its omp2a counterpart may affect the sugar permeability of the expressed porin as previously shown for naturally chimeric Omp2a and Omp2b porin variants (Paquet et al., 2001). The evaluated Omp2a variant showed a more efficient pore in sugar diffusion than the Omp2b variant. Differences in their respective L5 surface-exposed loop were suggested to play a major role in permeability, the L5 loop of Omp2a being shorter and containing less negatively charged residues than that of Omp2b.
Porins being important first line players to resist and adapt to environmental conditions, in the present study we analyzed the omp2 loci and their encoded porins from non-classical Brucella spp. to identify possible evolutionary paths that may have contributed to the adaptation to their final status as intracellular mammal or human pathogens.
Brucella strains analyzed were from previous studies (Scholz et al., 2010, 2016a; Tiller et al., 2010a, b; Zygmunt et al., 2012; Al Dahouk et al., 2017; Eisenberg et al., 2017), and are listed in the Supplementary Figure S1. Bacterial cultures and DNA extraction were performed as described previously (Cloeckaert et al., 1995). For the African bullfrog isolates of this study, DNA was extracted from killed bacterial cells provided by Dr. S. Al Dahouk (BfR, Berlin, Germany). PCR amplification of the omp2a and omp2b genes was performed as described previously (Cloeckaert et al., 1995, 2001). The PCR products were sequenced at Genome Express (Meylan, France). Nucleotide sequence analysis and alignments were done using Clustal Omega at EBI1. GenBank nucleotide accession numbers are indicated in Supplementary Figure S1. omp2 sequences were aligned using BioNumerics version 7.6.3 (Applied Maths, Belgium) with default parameters and the multiple alignment was used to produce a Maximum Parsimony tree. Comparative Omp2 protein analyses were done using a previously published Omp2a and Omp2b predicted and functionally validated topology model (Paquet et al., 2001).
Whole genome SNP (wgSNP) analysis was done as previously described (Vergnaud et al., 2018). Full genome sequences and assemblies were converted into artificial reads. SNPs were identified by mapping sequencing reads on a reference genome within BioNumerics. Assembly GCF_000007125 (B. melitensis 16 M) was used as reference. The Bio-Neighbor joining algorithm with bootstrap analysis embedded in BioNumerics was used for clustering analysis (Gascuel, 1997).
As shown in Figures 1, 2 and Supplementary Figure S1, omp2 sequence diversity analysis allowed to separate the non-classical Brucella spp. of this study in nine distinct groups (A to I), that correlated perfectly well with other previously published molecular methods such as MLSA, MLVA, IS711 profiling, or whole genome phylogeny (Figure 3). A second observation was a relative lower nucleotide sequence identity between their respective omp2a and omp2b gene copy ranging from 78.9 to 83.5%, in comparison to those of classical Brucella species ranging from 82.4 to 100% (Figure 2 and Supplementary Figure S1). Considering all omp2 sequences from both classical and non-classical Brucella spp., two separate clusters were observed in the clustering analysis. The “omp2a” cluster appeared less diverse than the “omp2b” cluster (Figure 1). All omp2a representatives shared the same size (1104 bp). In contrast, the size of omp2b representatives varied from 1089 to 1128 bp. To analyze in more detail the molecular basis behind this diversity, in particular recombination events, we have used the two most divergent omp2 gene copies, namely those of Brucella spp. isolated from Australian rodents, to designate them as either reference omp2a and reference omp2b gene copy. It actually provided a more comprehensive evolutionary picture, establishing a clearer link between classical and non-classical strains, than if the most divergent gene copies observed in the classical species only had been used as in previous studies (eg. B. melitensis or B. microti) (Cloeckaert et al., 2001; Al Dahouk et al., 2012). It also allowed to explain omp2b gene size variation, through stepwise genetic reduction, from the size observed for Australian rodent strains to that of the classical Brucella species. Starting from these designated reference omp2a and omp2b gene copies, the effect of genetic recombination events was observed in all groups of strains. Recombination events are schematized in Figure 1 by boxes containing either uncolored segments (omp2b) or yellow-colored segments (containing omp2a-specific nucleotides) along the groups of strains studied, and these genetic changes are more detailed in Figure 2. The events observed consisted mostly of converting some specific omp2b regions into omp2a, but the opposite situation (converting omp2a regions to omp2b) also occurred for some genotypes, resulting in almost complete (e.g. B. vulpis or B. ceti ST23) or complete (e.g. B. ceti ST26 or B. papionis) homogenization of both gene copies. Of note is that homogenization did not involve the same omp2 regions for the examples cited above (see Figure 2), resulting in the placement of the homogenized copies in either the omp2a or the omp2b cluster of the omp2 clustering analysis, for, respectively, B. vulpis and the other species/genotypes (Figure 1).
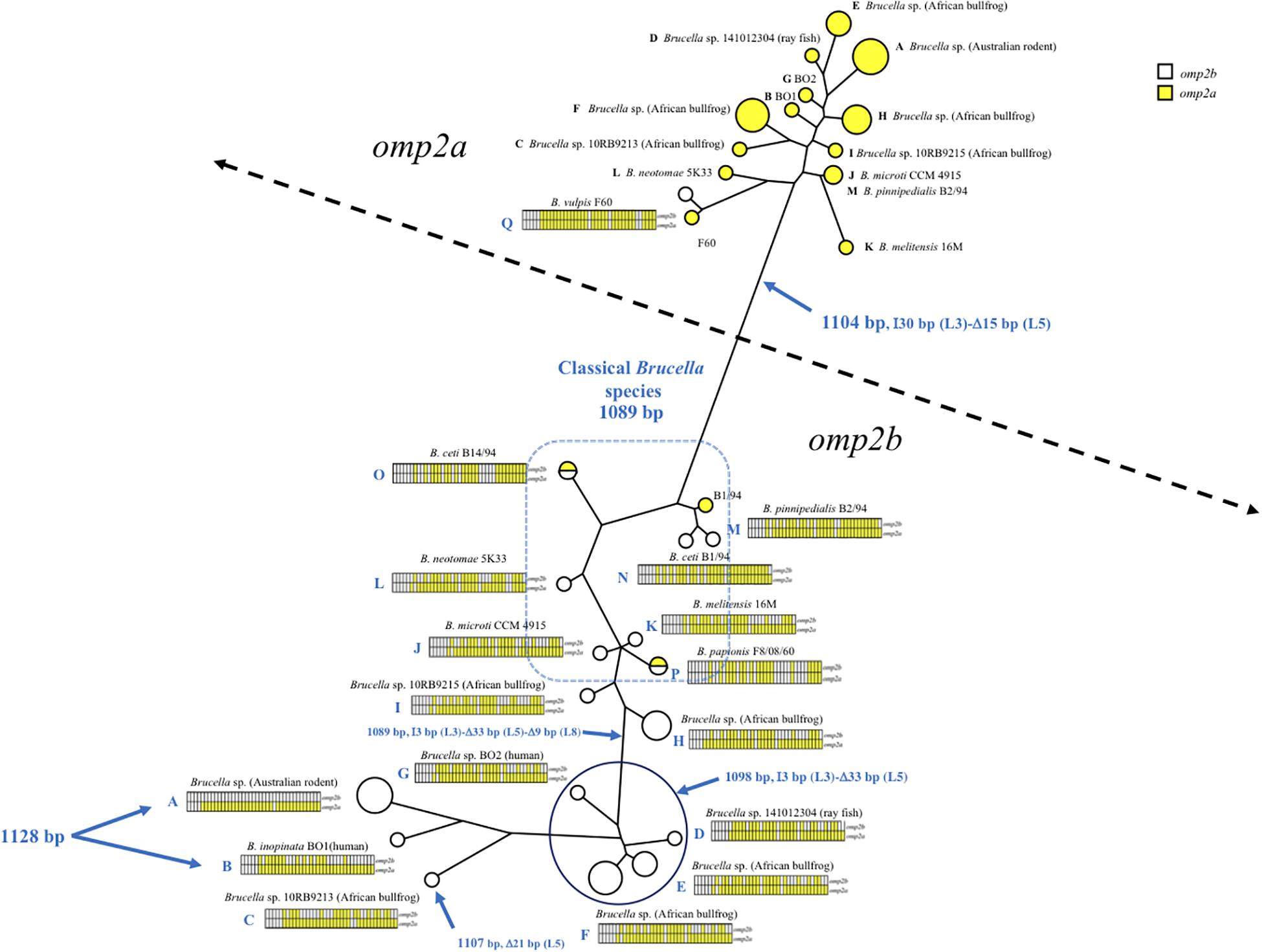
Figure 1. Brucella spp. omp2 clustering analysis, constructed using the omp2a and omp2b sequences of the non-classical and classical Brucella species of this study (GenBank accession numbers are in Supplementary Figure S1). The strains are represented by circles, increasing in size for strains with identical sequences. Empty circles represent the omp2b sequences while yellow-colored circles represent the omp2a sequences of the respective strains. Two half-colored circles are present in the tree for strains carrying identical omp2a and omp2b sequences, i.e. B. papionis F8/08/60 and B. ceti B14/94. The gene conversion events for each strain or groups of strains are schematized by sliced boxes representing omp2b (upper part) and omp2a (lower part), colored in yellow when omp2a-specific sequences are present. Relevant omp2 size variations are indicated in blue, with the Greek I and D letters meaning insertion and deletion, respectively. The size in bp of each event, as well as the encoded segment concerned (L3, L5, and L8) are indicated.
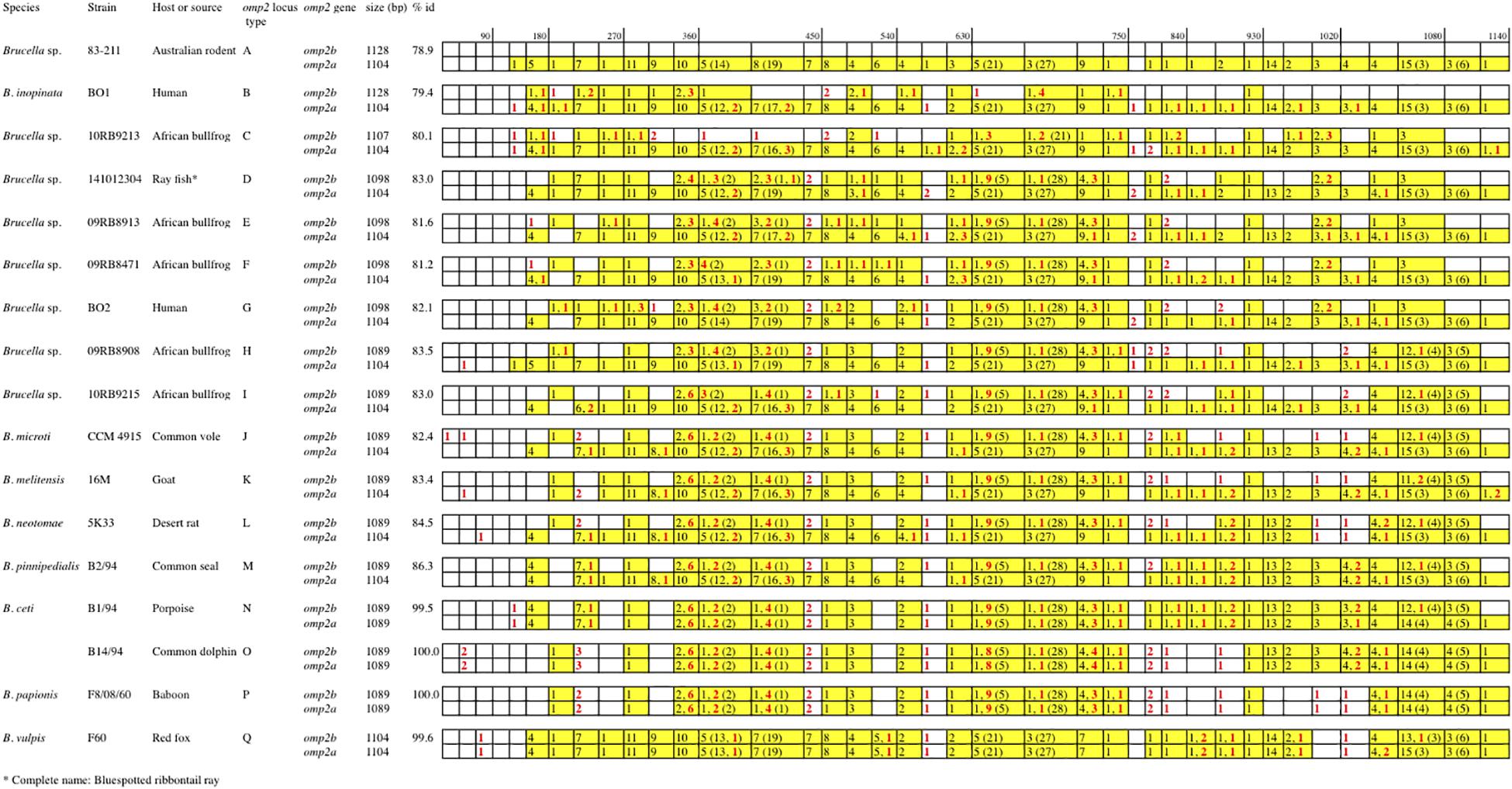
Figure 2. Schematic multiple nucleotide sequence alignment of the omp2a and omp2b genes of Brucella strains. This schematic representation was done according to the detailed alignment show in the Supplementary Figure S1. Nucleotide sequences are represented by rectangles divided into boxes of 30 nucleotides. The Brucella sp. 83–211 omp2b gene sequence was used as a reference (white boxes). The boxes containing omp2a-specific nucleotides are colored in yellow. The numbers in the corresponding boxes indicate the number of omp2a-specific nucleotides present in the sequence considered. Numbers in parentheses represent insertions and deletions. Numbers in red indicate nucleotide differences that are not due to gene conversion.
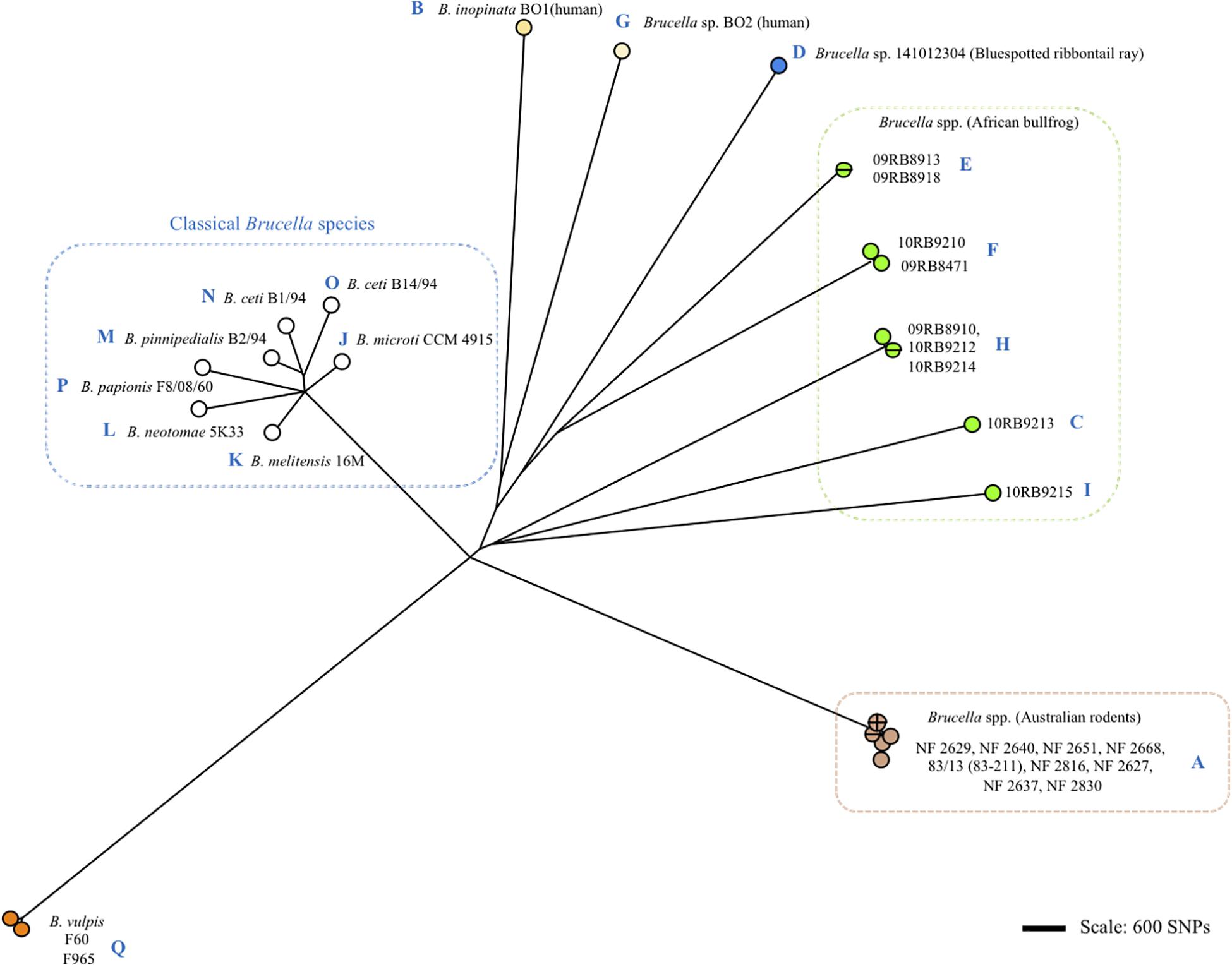
Figure 3. Clustering of Brucella spp. and representative classical strains of this study by whole genome SNP calling and Bio-NJ analysis. The color code reflects animal or human host assignment. Strain names are indicated (strain 83/13 is an alias of 83–211). Blue letters A to Q indicate the corresponding omp2 locus type according to Figure 2 and Supplementary Figure S1.
Regarding omp2b gene size variation, departing from a 1128 bp-sized omp2b gene as represented in Australian rodent Brucella spp. (group A) and the human B. inopinata BO1 isolate (group B), a number of consecutive indel events were observed including: (i) a deletion of 21 bp in the surface-encoding loop L5 in Brucella sp. 10RB9213 isolated from an African bullfrog; (ii) a deletion of 33 bp in the same region in four groups of isolates representing two groups from African bullfrogs (groups E and F), one isolate from ray fish (strain 141012304, group D) and the human isolate BO2 (group G); and (iii) and a 9 bp deletion in the region encoding the L8 surface-exposed loop was detected in two groups of African bullfrog strains (H and I), to reach the omp2b size of classical species (1089 bp). Those latter African bullfrog Brucella spp. thus present the same omp2b size as the classical species represented in Figure 1 (from B. microti to the marine mammal species B. ceti and B. pinnipedialis).
Relative to omp2b of classical species (1089 bp), the 1104 bp-sized omp2a differs by an insertion of 30 bp in the region encoding the L3 surface loop and a deletion of 15 bp in the region encoding the L5 surface loop (Figure 1). The conserved size of the omp2a gene copy in both the non-classical and classical species suggests that, as previously observed for classical species (Ficht et al., 1996; Cloeckaert et al., 2002), this gene copy may have been used to convert its omp2b counterpart and thus generate the successive omp2b gene deletions in the course of evolution of the non-classical species.
Although the Omp2b diversity generated through gene conversion appeared sequential, it did not correlate with the global phylogeny of Brucella spp. as shown in Figure 3. More precisely, we did not observe a clear sequential evolutionary link among the non-classical species which appeared genetically far more distant between each other than is reflected by the diversity of their omp2 locus. It may be explained by the essential functional nature of the Omp2b porin (Laloux et al., 2010), that actually would allow only variation depending on the external or intracellular environmental pressure and favor convergent evolution. In addition, although some SNPs were observed between omp2 sequences (indicated in Figures and Supplementary Figures S1–S3), Omp2b variation relied mainly on gene conversion and not on SNP variation as for the whole phylogeny of Brucella spp.
As described above omp2a or omp2b insertions or deletions were predominantly located in regions encoding three predicted surface exposed loops, namely L3, L5, and L8 (Paquet et al., 2001). Figure 4 shows a nucleotide sequence alignment of each region of representative strains of this study, complete sequence alignment is shown in Supplementary Figure S2. The L3 encoding region is characterized by either a 33 bp or 30 bp deletion in omp2b relative to omp2a in omp2 groups A, B, and C Brucella strains and the other groups, respectively. This 3 bp difference is difficult to explain according to the sequence alignment, but nevertheless in the shorter deletion, omp2a-specific sequence fragments remain suggesting that the deletion in omp2b resulted from an intra-molecular recombination event between omp2a and omp2b for this specific region. The L3 loop has been shown to play a major role as constriction loop in most bacterial porins (Jap and Walian, 1996). However, in Brucella spp. this loop did not appear critical for sugar permeability according to a previous study (Paquet et al., 2001). According to the same study the L5 loop appeared to be a critical determinant in Brucella porin sugar permeability and could participate in the formation of the pore “external mouth,” which serves to prescreen the solute in porins of known structure (Paquet et al., 2001). Consistent with this, two deletions, a first of 21 bp and the second of 33 bp, in the L5 encoding region of the Omp2b porin were identified in the present study, starting from the omp2 groups A and B strains to reach a minimal L5 size in the other groups of strains, with J to O strains representing the classical Brucella species (Figures 1, 4). In addition, it must be noted that omp2b from B. vulpis (group P in Figures 1, 2) represents a separate situation as its omp2b copy has converted to omp2a in the L5 region with a concomitant additional 15 bp deletion relative to other omp2b sequences (Figure 4), thus making it the shortest Omp2b encoding L5 region of strains of this study. A similar situation has been reported previously for B. abortus strain 45/20 for which the same omp2b to omp2a sequence conversion was observed in this segment (Paquet et al., 2001). The different L5 situations are detailed in the nucleotide sequence alignment of Figure 4. In addition to segmental gene conversion-mediated exchanges between the respective omp2a and omp2b gene copy, short 6 bp direct DNA repeats may have contributed to generate the different deletions observed in the L5-encoding segment. At the amino acid sequence level it resulted in the loss of either 7 or 11 amino acids including one or two negatively charged residues, respectively (Figure 5), that may possibly be important on ion conductance and ion selectivity of the L5 loop (Paquet et al., 2001). According to previous data, the sequential loss of amino acids observed correlates with increased sugar permeability to reach the highest permeation rate for porin variants containing the shortest Omp2a-type L5 loop (Figure 5; Paquet et al., 2001).
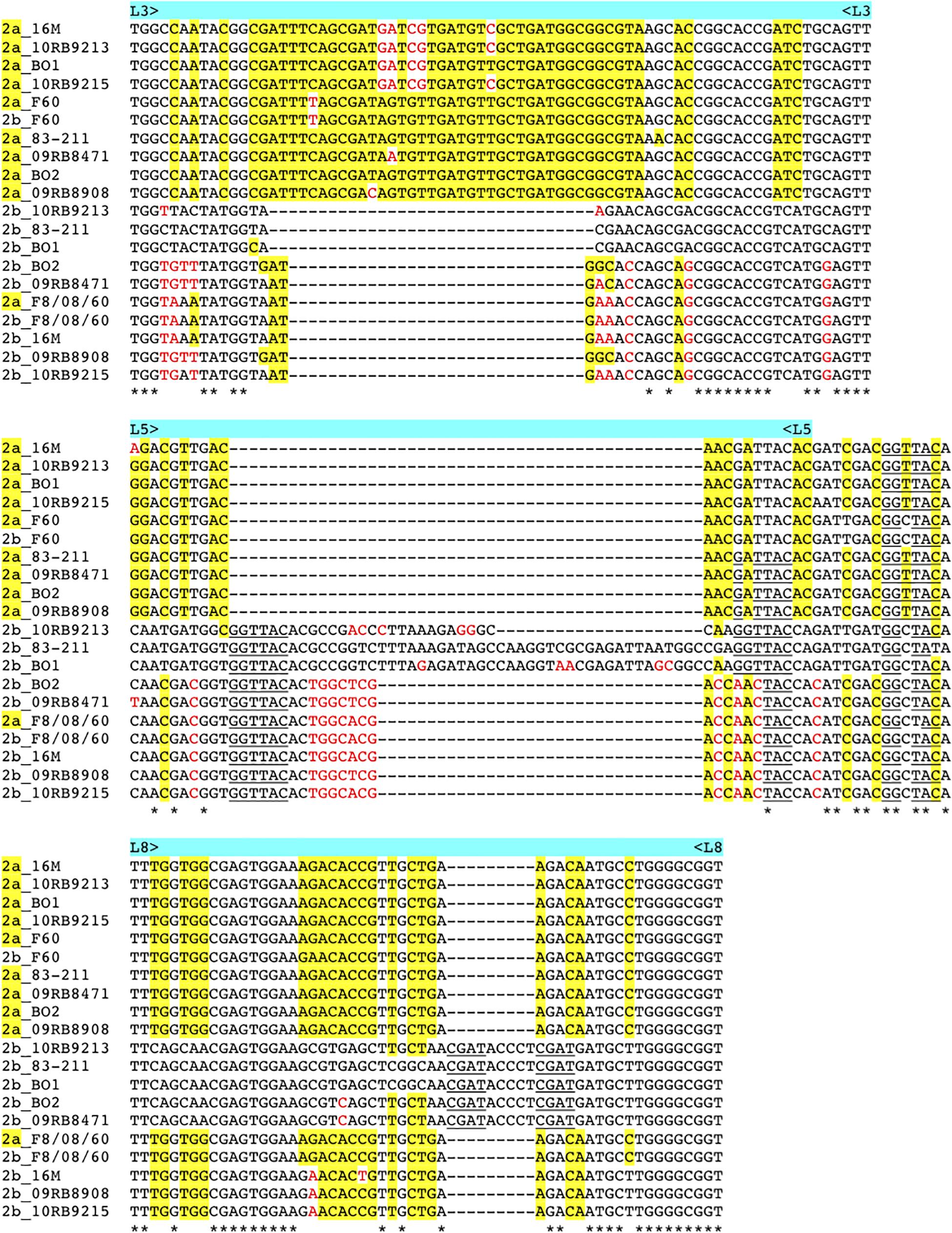
Figure 4. Multiple nucleotide sequence alignment of the omp2 segments encoding the L3, L5, and L8 loops (upper, middle, and lower panel, respectively), from representative strains containing insertions or deletions. Strains and omp2 genes used are indicated on the left of each segment, omp2a genes are indicated as 2a in yellow. omp2a-specific nucleotides are highlighted in yellow, according to the omp2a and omp2b reference sequences used from Brucella sp. 83–211. Nucleotides colored in red indicate differences that are not due to gene conversion, according to the same reference sequences. Direct DNA repeats are underlined. *Indicates identical nucleotides.

Figure 5. Amino acid sequence alignment of the Omp2b and Omp2a L5 loops from representative strains of this study, showing the progressive loss of amino acids in accordance with the respective nucleotide sequence data shown in Figures 1–3. The strains are indicated on the left, when Omp2a is used the corresponding strain number is highlighted in yellow. The predicted L5 loop, according to Paquet et al. (2001), is highlighted in blue. Underlined B9 and B10 sequences indicate the predicted flanking transmembrane β-strands (Paquet et al., 2001). Omp2a-specific amino acids are highlighted in yellow, in accordance with the Omp2a and Omp2b reference sequence used from Brucella sp. 83–211. Omp2 amino acids colored in red are independent from gene conversion according to the same reference sequences. The middle panel of the figure highlights the negatively charged amino acids of the segment. The right panel indicate the available sugar permeability data from previously published strains (Paquet et al., 2001). *Indicates identical amino acids.
The last genetic reduction observed in the sequential course of Omp2b porin evolution shown in Figure 1, consisted of a 9 bp deletion in the region encoding the surface L8 loop, and it finally ended up to the size of omp2b and the respective Omp2b porin protein observed for the classical species. As shown in Figure 5 this short deletion, as observed for the L5 segment, may be the result of gene conversion using the respective omp2a copy as template, and two short 4 bp direct repeats may also have contributed to create this deletion. In addition it created an omp2a-like segment at the 3′ end of the respective omp2b sequence, which did not appear as clearly when using as references omp2a and omp2b genes from classical species (e.g. B. melitensis 16 M) (Cloeckaert et al., 2001). As a consequence group H and I non-classical Brucella spp. (African bullfrog isolates), possess Omp2b porins with an Omp2a-like L8 surface loop at the amino acid sequence level, as seen in all classical Brucella species (Supplementary Figure S3). The L8 surface loop was previously shown to be an important target site recognized by monoclonal antibodies directed against conformational epitopes of the Omp2b porin (Paquet et al., 2001). We thus predict that the genetic loss in the L8 encoding region has created antigenic variability of the corresponding protein in the course of evolution of Brucella spp., and possibly may have affected antibody induction and recognition in some animal species. In several bacterial species gene conversion appears to have a prime importance in the generation of antigenic variation, an interesting mechanism whereby some bacterial pathogens are able to avoid the host immune system (Santoyo and Romero, 2005). For example, in Anaplasma marginale the major surface protein Msp2 is encoded by a single expression site, and diversity is achieved by gene conversion of chromosomally encoded msp2 pseudogenes (Graça et al., 2015). A model was proposed where msp2, through gene conversion, progressively incorporates changes to produce an msp2 repertoire capable of generating sufficient antigenic variants to escape immunity and establish persistent infection (Graça et al., 2019). We may see a similar situation from the present omp2 study, with the progressive changes of the Omp2b porin cited above, altering the porin both functionally and antigenically.
This study provided evidence for a progressive genetic loss in the omp2b gene encoding the major outer membrane porin, from non-classical Brucella spp. to the classical pathogenic Brucella species. This genetic loss appears mainly mediated by segmental gene conversion events with its silent omp2a gene copy, and concerns especially the regions encoding the L5 surface loop for porin function and the L8 loop for antigenic variability. The progressive loss in the L5 loop correlates with increasing sugar permeability of the porin, and could be related to environmental adaptation to survive conditions from a possible extracellular aquatic environment (e.g. amphibian or fish) to the intracellular macrophagic environment of the classical pathogenic species causing disease in livestock and humans.
The new sequence data of this study were deposited in Genbank. The accession numbers are listed in Supplementary Figure S1.
AC conceived and designed the study. AC, GV, and MZ analyzed the data, and drafted the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Nelly Bernardet for expert technical assistance and Dr. S. Al Dahouk (BfR, Berlin, Germany) for providing killed bacterial cells used to extract DNA analyzed in this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00284/full#supplementary-material
Al Dahouk, S., Hofer, E., Tomaso, H., Vergnaud, G., Le Flèche, P., Cloeckaert, A., et al. (2012). Intraspecies biodiversity of the genetically homologous species Brucella microti. Appl. Environ. Microbiol. 78, 1534–1543. doi: 10.1128/AEM.06351-11
Al Dahouk, S., Köhler, S., Occhialini, A., Jiménez de Bagüés, M. P., Hammerl, J. A., Eisenberg, T., et al. (2017). Brucella spp. of amphibians comprise genomically diverse motile strains competent for replication in macrophages and survival in mammalian hosts. Sci. Rep. 7:44420. doi: 10.1038/srep44420
Alton, G. G., Jones, L. M., Angus, R. D., and Verger, J. M. (1988). Techniques for the Brucellosis Laboratory. Paris: INRA.
Cloeckaert, A., Verger, J. M., Grayon, M., and Grépinet, O. (1995). Restriction site polymorphism of the genes encoding the major 25 kDa and 36 kDa outer-membrane proteins of Brucella. Microbiology 141, 2111–2121. doi: 10.1099/13500872-141-9-2111
Cloeckaert, A., Verger, J. M., Grayon, M., Paquet, J. Y., Garin-Bastuji, B., Foster, G., et al. (2001). Classification of Brucella spp. isolated from marine mammals by DNA polymorphism at the omp2 locus. Microbes Infect. 3, 729–738. doi: 10.1016/s1286-4579(01)01427-7
Cloeckaert, A., Vizcaíno, N., Paquet, J. Y., Bowden, R. A., and Elzer, P. H. (2002). Major outer membrane proteins of Brucella spp.: past, present and future. Vet. Microbiol. 90, 229–247. doi: 10.1016/s0378-1135(02)00211-0
Corbel, M. J., and Brinley Morgan, W. J. (1984). “Genus Brucella meyer and shaw 1920, 173AL,” in Bergey’s Manual of Systematic Bacteriology, eds N. R. Krieg, and J. G. Holt, (Baltimore, MD: Williams and Wilkins), 377–390.
Eisenberg, T., Hamann, H. P., Kaim, U., Schlez, K., Seeger, H., and Schauerte, N. (2012). Isolation of potentially novel Brucella spp. from frogs. Appl. Environ. Microbiol. 78, 3753–3755. doi: 10.1128/AEM.07509-11
Eisenberg, T., Riße, K., Schauerte, N., Geiger, C., Blom, J., and Scholz, H. C. (2017). Isolation of a novel ‘atypical’ Brucella strain from a bluespotted ribbontail ray (Taeniura lymma). Antonie Van Leeuwenhoek 110, 221–234. doi: 10.1007/s10482-016-0792-4
Ficht, T. A., Bearden, S. W., Sowa, B. A., and Adams, L. G. (1989). DNA sequence and expression of the 36-kilodalton outer membrane protein gene of Brucella abortus. Infect. Immun. 57, 3281–3291. doi: 10.1128/iai.57.11.3281-3291.1989
Ficht, T. A., Bearden, S. W., Sowa, B. A., and Marquis, H. (1990). Genetic variation at the omp2 porin locus of the brucellae: species-specific markers. Mol. Microbiol. 4, 1135–1142. doi: 10.1111/j.1365-2958.1990.tb00688.x
Ficht, T. A., Husseinen, H. S., Derr, J., and Bearden, S. W. (1996). Species-specific sequences at the omp2 locus of Brucella type strains. Int. J. Syst. Bacteriol. 46, 329–331. doi: 10.1099/00207713-46-1-329
Fischer, D., Lorenz, N., Heuser, W., Kämpfer, P., Scholz, H. C., and Lierz, M. (2012). Abscesses associated with a Brucella inopinata-like bacterium in a big-eyed tree frog (Leptopelis vermiculatus). J. Zoo Wildl. Med. 43, 625–628. doi: 10.1638/2011-0005r2.1
Foster, G., Osterman, B. S., Godfroid, J., Jacques, I., and Cloeckaert, A. (2007). Brucella ceti sp. nov. and Brucella pinnipedialis sp. nov. for Brucella strains with cetaceans and seals as their preferred hosts. Int. J. Syst. Evol. Microbiol. 57, 2688–2693. doi: 10.1099/ijs.0.65269-0
Gascuel, O. (1997). BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14, 685–695. doi: 10.1093/oxfordjournals.molbev.a025808
Godfroid, J., Scholz, H. C., Barbier, T., Nicolas, C., Wattiau, P., Fretin, D., et al. (2011). Brucellosis at the animal/ecosystem/human interface at the beginning of the 21st century. Prev. Vet. Med. 102, 118–131. doi: 10.1016/j.prevetmed.2011.04.007
Graça, T., Ku, P. S., Silva, M. G., Turse, J. E., Hammac, G. K., and Brown, W. C. (2019). Segmental variation in a duplicated msp2 pseudogene generates Anaplasma marginale antigenic variants. Infect. Immun. 87:e00727-18.
Graça, T., Paradiso, L., Broschat, S. L., Noh, S. M., and Palmer, G. H. (2015). Primary structural variation in Anaplasma marginale Msp2 efficiently generates immune escape variants. Infect. Immun. 83, 4178–4184. doi: 10.1128/IAI.00851-815
Guzmán-Verri, C., Suárez-Esquivel, M., Ruíz-Villalobos, N., Zygmunt, M. S., Gonnet, M., Campos, E., et al. (2019). Genetic and phenotypic characterization of the etiological agent of canine orchiepididymitis smooth Brucella sp. BCCN84.3. Front. Vet. Sci. 6:175. doi: 10.3389/fvets.2019.00175
Jap, B. K., and Walian, P. J. (1996). Structure and functional mechanism of porins. Physiol. Rev. 76, 1073–1088. doi: 10.1152/physrev.1996.76.4.1073
Jaÿ, M., Girault, G., Perrot, L., Taunay, B., Vuilmet, B., Rossignol, F., et al. (2018). Phenotypic and molecular characterization of Brucella microti-like bacteria from a domestic marsh frog (Pelophylax ridibundus). Front. Vet. Sci. 5:283. doi: 10.3389/fvets.2018.00283
Kimura, M., Une, Y., Suzuki, M., Park, E. S., Imaoka, K., and Morikawa, S. (2017). Isolation of Brucella inopinata-like bacteria from White’s and Denny’s tree frogs. Vector Borne Zoonotic Dis. 17, 297–302. doi: 10.1089/vbz.2016.2027
Laloux, G., Deghelt, M., de Barsy, M., Letesson, J. J., and De Bolle, X. (2010). Identification of the essential Brucella melitensis porin Omp2b as a suppressor of Bax-induced cell death in yeast in a genome-wide screening. PLoS One 5:e13274. doi: 10.1371/journal.pone.0013274
Maquart, M., Le Flèche, P., Foster, G., Tryland, M., Ramisse, F., Djønne, B., et al. (2009). MLVA16 typing of 295 marine mammal Brucella isolates from different animal and geographic origins identifies 7 major groups within Brucella ceti and Brucella pinnipedialis. BMC Microbiol. 9:145. doi: 10.1186/1471-2180-9-145
Marquis, H., and Ficht, T. A. (1993). The omp2 gene locus of Brucella abortus encodes two homologous outer membrane proteins with properties characteristic of bacterial porins. Infect. Immun. 61, 3785–3790. doi: 10.1128/iai.61.9.3785-3790.1993
Moreno, E., Cloeckaert, A., and Moriyón, I. (2002). Brucella evolution and taxonomy. Vet. Microbiol. 90, 209–227. doi: 10.1016/s0378-1135(02)00210-9
Mühldorfer, K., Wibbelt, G., Szentiks, C. A., Fischer, D., Scholz, H. C., Zschöck, M., et al. (2017). The role of ‘atypical’ Brucella in amphibians: are we facing novel emerging pathogens? J. Appl. Microbiol. 122, 40–53. doi: 10.1111/jam.13326
Paquet, J. Y., Diaz, M. A., Genevrois, S., Grayon, M., Verger, J. M., de Bolle, X., et al. (2001). Molecular, antigenic, and functional analyses of Omp2b porin size variants of Brucella spp. J. Bacteriol. 183, 4839–4847. doi: 10.1128/jb.183.16.4839-4847.2001
Santoyo, G., and Romero, D. (2005). Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol. Rev. 29, 169–183. doi: 10.1016/j.femsre.2004.10.004
Scholz, H. C., Hofer, E., Vergnaud, G., Le Flèche, P., Whatmore, A. M., Al Dahouk, S., et al. (2009). Isolation of Brucella microti from mandibular lymph nodes of red foxes, Vulpes vulpes, in lower Austria. Vector Borne Zoonotic Dis. 9, 153–156. doi: 10.1089/vbz.2008.0036
Scholz, H. C., Hubalek, Z., Nesvadbova, J., Tomaso, H., Vergnaud, G., Le Flèche, P., et al. (2008b). Isolation of Brucella microti from soil. Emerg. Infect. Dis. 14, 1316–1317. doi: 10.3201/eid1408.080286
Scholz, H. C., Hubalek, Z., Sedlácek, I., Vergnaud, G., Tomaso, H., Al Dahouk, S., et al. (2008a). Brucella microti sp. nov., isolated from the common vole Microtus arvalis. Int. J. Syst. Evol. Microbiol. 58, 375–382. doi: 10.1099/ijs.0.65356-65350
Scholz, H. C., Mühldorfer, K., Shilton, C., Benedict, S., Whatmore, A. M., Blom, J., et al. (2016b). The change of a medically important genus: worldwide occurrence of genetically diverse novel Brucella species in exotic frogs. PLoS One 11:e0168872. doi: 10.1371/journal.pone.0168872
Scholz, H. C., Nöckler, K., Göllner, C., Bahn, P., Vergnaud, G., Tomaso, H., et al. (2010). Brucella inopinata sp. nov., isolated from a breast implant infection. Int. J. Syst. Evol. Microbiol. 60, 801–808. doi: 10.1099/ijs.0.011148-11140
Scholz, H. C., Revilla-Fernández, S., Al Dahouk, S., Hammerl, J. A., Zygmunt, M. S., Cloeckaert, A., et al. (2016a). Brucella vulpis sp. nov., isolated from mandibular lymph nodes of red foxes (Vulpes vulpes). Int. J. Syst. Evol. Microbiol. 66, 2090–2098. doi: 10.1099/ijsem.0.000998
Soler-Lloréns, P. F., Quance, C. R., Lawhon, S. D., Stuber, T. P., Edwards, J. F., Ficht, T. A., et al. (2016). A Brucella spp. isolate from a Pac-Man frog (Ceratophrys ornata) reveals characteristics departing from classical Brucellae. Front. Cell. Infect. Microbiol. 6:116. doi: 10.3389/fcimb.2016.00116
Tiller, R. V., Gee, J. E., Frace, M. A., Taylor, T. K., Setubal, J. C., Hoffmaster, A. R., et al. (2010a). Characterization of novel Brucella strains originating from wild native rodent species in North Queensland, Australia. Appl. Environ. Microbiol. 76, 5837–5845. doi: 10.1128/AEM.00620-610
Tiller, R. V., Gee, J. E., Lonsway, D. R., Gribble, S., Bell, S. C., Jennison, A. V., et al. (2010b). Identification of an unusual Brucella strain (BO2) from a lung biopsy in a 52 year-old patient with chronic destructive pneumonia. BMC Microbiol. 10:23. doi: 10.1186/1471-2180-10-23
Vergnaud, G., Hauck, Y., Christiany, D., Daoud, B., Pourcel, C., Jacques, I., et al. (2018). Genotypic expansion within the population structure of classical Brucella species revealed by MLVA16 typing of 1404 Brucella isolates from different animal and geographic origins, 1974-2006. Front. Microbiol 9:1545. doi: 10.3389/fmicb.2018.01545
Whatmore, A. M., Davison, N., Cloeckaert, A., Al Dahouk, S., Zygmunt, M. S., Brew, S. D., et al. (2014). Brucella papionis sp. nov., isolated from baboons (Papio spp.). Int. J. Syst. Evol. Microbiol. 64, 4120–4128. doi: 10.1099/ijs.0.065482-65480
Whatmore, A. M., Koylass, M. S., Muchowski, J., Edwards-Smallbone, J., Gopaul, K. K., and Perrett, L. L. (2016). Extended multilocus sequence analysis to describe the global population structure of the genus Brucella: phylogeography and relationship to biovars. Front. Microbiol. 7:2049. doi: 10.3389/fmicb.2016.02049
Keywords: Brucella, Omp2 porin, loop, diversity, gene conversion, evolution
Citation: Cloeckaert A, Vergnaud G and Zygmunt MS (2020) Omp2b Porin Alteration in the Course of Evolution of Brucella spp. Front. Microbiol. 11:284. doi: 10.3389/fmicb.2020.00284
Received: 09 December 2019; Accepted: 07 February 2020;
Published: 24 February 2020.
Edited by:
Miklos Fuzi, Semmelweis University, HungaryReviewed by:
Zeliang Chen, Shenyang Agricultural University, ChinaCopyright © 2020 Cloeckaert, Vergnaud and Zygmunt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Axel Cloeckaert, QXhlbC5DbG9lY2thZXJ0QGlucmFlLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.