- 1Graduate Institute of Biomedical Sciences, College of Medicine, Chang Gung University, Taoyuan, Taiwan
- 2Department of Medical Biotechnology and Laboratory Science, College of Medicine, Chang Gung University, Taoyuan, Taiwan
- 3Department of Laboratory Medicine, Chang Gung Memorial Hospital, Taoyuan, Taiwan
Staphylococcus aureus spreads rapidly on the surface of soft agar medium. The spreading depends on the synthesis of biosurfactants, i.e., phenol soluble modulins (PSMs), which facilitate colony spreading of S. aureus. Our earlier study demonstrated that water accumulates in a colony is important to modulate colony spreading of S. aureus. The current study screened a transposon-based mutant library of S. aureus HG001 and obtained four non-spreading mutants with mutations in hemY and ctaA, which are involved in heme synthesis. The spreading ability of these mutants was restored when the mutants are transformed with a plasmid encoding hemY or ctaA, respectively. HemY mutants, which do not synthesize heme B, were able to spread on agar medium supplemented with hemin, a heme B derivative. By contrast, hemin supplementation did not rescue the spreading of the ctaA mutant, which lacks heme B and heme A, indicating that heme A is also critical for colony spreading. Moreover, mutations in hemY and ctaA had little effect on PSMs production but affect ATP production and water accumulation in the colony. In conclusion, this study sheds light on the role of heme synthesis and energy production in the regulation of S. aureus colony spreading, which is important for understanding the movement mechanisms of bacteria lacking a motor apparatus.
Introduction
Bacteria use different types of motility, including swarming, swimming, twitching, gliding, and sliding, to move across a surface or through a liquid to avoid dangers and acquire nutrients (Olsen et al., 2013; Vater et al., 2014). Many pathogenic members of the genera Proteus, Serratia, Vibrio, Bacillus, Clostridium, Escherichia, and Salmonella swarm, which is critical to the establishment of infection by these bacteria (Allison et al., 1992; Jones et al., 2004; Kim and Surette, 2005; Lai et al., 2005; Callegan et al., 2006; Jaques and McCarter, 2006; Armbruster et al., 2013). However, Staphylococcus aureus, which does not have pili or flagella, uses a different strategy, called colony spreading, to move across the surface of soft agar and animal tissues (Kaito and Sekimizu, 2007).
Many factors, including wall teichoic acid (WTA) and extracellular DNA, are involved in colony spreading of S. aureus (Kaito and Sekimizu, 2007; Kaito et al., 2011a). Furthermore, cell wall associated factors such as fibronectin-binding protein A/B (FnbpA/B) and clumping factor A/B (ClfA/B), both of which promote biofilm formation (O’Neill et al., 2008), antagonize colony spreading (Tsompanidou et al., 2012). Recent studies have demonstrated that host factors, such as albumin and high-density lipoproteins in serum, stimulate S. aureus colony spreading (Omae et al., 2014). Additionally, spreading motility depends on the activation of the accessory gene regulator (Agr) quorum-sensing system, which is responsible for the expression of the biosurfactant phenol-soluble modulins (PSMs) (Tsompanidou et al., 2013). S. aureus expresses a variety of PSMs, including PSMα1–4, PSMβ1–2, and PSMγ, with PSMγ more commonly known as δ-hemolysin (Queck et al., 2008; Aoyagi et al., 2014). Among the PSMs produced by S. aureus, PSMα1–4 are the most important for colony spreading (Tsompanidou et al., 2013). By contrast, colony spreading of S. aureus is inhibited by PSMγ (Omae et al., 2012) and the newly identified psm-mec gene; psm-mec is located in the fudoh locus in the type II and III staphylococcal chromosomal cassettes mec (SCCmec) (Kaito et al., 2008), which also contains the mecA gene responsible for methicillin resistance (Kaito et al., 2011b).
A recent study on Bacillus subtilis swarming revealed that the organism extracts water from agar and produces surfactin, which reduces the surface tension of water (Ke et al., 2015), ultimately allowing the colony to form flowing water-filled channels that facilitate the swarming of bacteria, resulting in rapid expansion of the colony (Ke et al., 2015). Extraction of water from agar to facilitate swarming is not limited to B. subtilis. Water is also found in swarms formed by Escherichia coli and Salmonella enterica (Chen et al., 2007; Ping et al., 2014). Our earlier study found that water accumulates in spreading colonies of S. aureus (Lin et al., 2016). In E. coli, it has been suggested that lipopolysaccharide (LPS) serves as an osmolyte to extract water from agar (Ping et al., 2014). However, B. subtilis and S. aureus do not contain LPS, and whether these organisms use an osmolyte to extract water remains unknown. To further investigate the mechanisms of colony spreading, this study screened a transposon-based mutant library of S. aureus HG001 and obtained non-spreading mutants with mutations in genes involved in the synthesis of heme, an iron-containing porphyrin compound that participates in aerobic respiration and energy production (Hederstedt, 2012; Dailey et al., 2017). The results demonstrate that heme deficiency has little effect on PSMs expression but greatly impacts ATP production. Moreover, the spreading colonies of heme-deficient mutants accumulate less water, indicating that heme plays a role in energy generation and water extraction during colony spreading.
Materials and Methods
Strains, Culture Conditions, and Chemicals
Staphylococcus aureus HG001, a derivative of S. aureus NCTC8325 (Herbert et al., 2010), was used in a spreading assay and for the generation of a transposon mutant library. M47, M99, D19, and M60 mutants with defects in colony spreading were selected from the transposon mutant library using a spreading assay (Table 1). E. coli EPI300 (Epicenter Technologies, Madison, WI, United States) was used as a host for cloning. S. aureus RN4220, which produces β-hemolysin (Nair et al., 2011), were used for assessment of δ-hemolytic activity. S. aureus SA113 (ATCC35556) and its isogenic mutant SA113ΔtagO, which contains a deletion in tagO and does not produce WTA (Weidenmaier et al., 2004) were used in a spreading assay, tiled plate assay, and ATP assay. S. aureus NCTC8325-4, a derivative of S. aureus NCTC8325 and its isogenic mutant NCTC8325-4ΔhemB, which contains a deletion in hemB and does not produce hemes (von Eiff et al., 1997), were used in a spreading assay, tiled plate assay, ATP assay, and enzyme activity assay. Bacteria were cultured in tryptic soy broth (TSB) and tryptic soy agar (TSA) (Oxoid, Basingstoke, United Kingdom). Antibiotic-resistant colonies were selected on media that contained ampicillin (100 μg/ml), spectinomycin (100 μg/ml), erythromycin (5 μg/ml), and chloramphenicol (10 μg/ml). Hemin, tunicamycin and N,N′-dicyclohexylcarbodiimide (DCCD) purchased from Sigma–Aldrich, Inc. (St. Louis, MO, United States) were added to TSA-0.25 and TSA-0.4.
Spreading and Gravity-Flow Experiments
For spreading assays, TSA containing 0.25 (TSA-0.25) or 0.4% (TSA-0.4) agarose (Seakem, Lonza Rockland, Inc.) was used. After autoclaving, medium was placed in a water bath at 55°C for 30 min. Plates containing 25 ml TSA-0.25 or TSA-0.4 were prepared and were then incubated at room temperature for 20 min before inoculation. Each plate was inoculated with 2 μl (1 × 107 CFU) of an overnight culture and was then allowed to dry for 15 min in a laminar flow hood. The plates were incubated at 37°C for 24 h to allow bacteria to spread. To determine the presence of water within colonies, the TSA-0.4 plates were tilted at a 30° angle during incubation according to the method described previously (Lin et al., 2016).
Transposon Mutagenesis
A mutant library was generated using the bursa aurealis transposon-based insertional mutagenesis method (Bae et al., 2004). Briefly, S. aureus HG001 was sequentially transformed with pBursa and pFA545, which contains genes encoding mariner transposase and confers resistance to tetracycline and ampicillin (Bae et al., 2004). The resulting transformants were spread on TSA containing erythromycin (TSAerm) and were incubated at 30°C overnight. After induction of transposase expression and curing the plasmid by incubating the transformants at 43°C for 2 days, cells containing transposon insertions in the chromosome were selected on TSAerm. Approximately 9000 mutants were collected and screened for their inability to spread on TSA-0.25 plates. Mutants that were defective in spreading were identified using a spreading assay (Lin et al., 2016). The sites of transposon insertion in the selected mutants were analyzed by sequencing as previously described (Bae et al., 2004). Briefly, total DNA of S. aureus was isolated and digested with the restriction enzyme AciI. The digested DNA fragments were self-ligated and amplified by inverse PCR using primers complementary to the border sequences of the transposon (Bae et al., 2004). The PCR products were then sequenced to identify the location of transposon insertion using the genome sequence of S. aureus NCTC8325 (accession number: NC_007795.1) (Herbert et al., 2010), which is the parental strain of S. aureus HG001.
Plasmids
The plasmid pPSM-gfp (Lin et al., 2016), which contains the gfp gene transcribed from the psmα promoter, was used to determine quorum-sensing activation in spreading colonies. The hemY gene was amplified by PCR using the primers hemY-F (5′-CGGTCTAGAGGTGGTTTATCACCATTAGC) and hemY-R (5′-TTACCCGGGCTTACAACTCTGCGATTACT); ctaA was amplified using the primers ctaA-F (5′-CGGTCTAGA GCGTGCCATTAAAATTACGG) and ctaA-R (5′-TTACCCGGG ATGCGCCACCCATAATTAAA) and tagO was amplified using the primers tagO-F (5′-CGGTCTAGATAGCACTTGTT ACTGCAGCA) and tagO-R (5′-TTACCCGGGATCCCATA CAGCTATGCTTT). The amplified DNA fragments were digested with XbaI and SmaI and were then inserted into the XbaI–SmaI sites in pHY300PLK (TaKaRa, Japan) to generate pHY-hemY, pHY-ctaA, and pHY-tagO.
PSMs Extraction and Detection
Phenol soluble modulins were extracted according to a method described previously (Lin et al., 2018). Briefly, an overnight culture was harvested and adjusted with TSB to an OD578 of 7.0. Following centrifugation at 2000 × g for 10 min, the supernatants were separated into two parts. One part of supernatant was used to extract PSMs using a butanol method (Lin et al., 2018). The other part of supernatant was used to isolate the exoproteins using Amicon Ultra-4 centrifugal filters (Millipore, Billerica, MA, United States). Both PSMs and exoproteins were analyzed using SDS-PAGE, followed by Coomassie blue staining (0.1% Coomassie brilliant blue R250, 10% acetic acid, 50% methanol). Synthetic PSMα3 (Kelowna International Scientific Inc., Taiwan) was used as a size marker to indicate the location of PSMs on the SDS-PAGE gel. The total amounts of exoproteins in the culture medium were used as an internal control. The intensity of protein bands in each lane was quantified by using a densitometer. The amount of PSMs was normalized to the amount of exoproteins.
Assessment of δ-Hemolytic Activity
Expression of δ-hemolysin was evaluated by cross-streaking test strains perpendicularly to S. aureus RN4220, which produces β-hemolysin, according to the method described previously (Herbert et al., 2010). β-Hemolysin enhances δ-hemolysin-mediated lysis of sheep red blood cells (Herbert et al., 2010). Therefore, δ-hemolytic activity can be visualized as a zone of enhanced hemolysis at the intersection of S. aureus RN4220 and the streaked test strain.
Determination of Agr Expression in Spreading Colonies
Activation of the agr operon was determined according to a method described previously (Lin et al., 2016). In brief, S. aureus HG001(pPSM-gfp) and S. aureus M60(pPSM-gfp) were inoculated on TSA-0.4 agar and cultured at 37°C for 5 h. Colonies that formed on TSA-0.4 were observed under an upright confocal laser-scanning microscope (CLSM) (Leica, TCS-SP2). Green fluorescence expression by the colony indicated quorum-sensing activation.
ATP Assay
Tryptic soy agar-0.4 plates were inoculated with 2 μl overnight bacterial culture and incubated at 37°C for 3 h. The cells on the surface of the TSA-0.4 plates were harvested, washed, suspended in PBS, and enumerated spectrophotometrically at OD578. The amount of intracellular ATP was measured by using a BacTiter-Glo Microbial Cell Viability Assay Kit (Promega, Madison, WI, United States). A 100-μl aliquot of cells was mixed with an equal volume of the assay reagent. The mixture was incubated at room temperature with shaking for 10 min. After incubation, the luminescence signal, which is proportional to the amount of ATP, was detected using a luminometer (Promega, GloMax Explorer GM-3510). ATP concentration was calculated based on an ATP standard curve that was prepared using adenosine 5′-triphosphate. The ATP level in the cells was normalized to the absorbance of the bacterial culture at 578 nm (A578).
Measurement of Cytochrome Oxidase Activity
The enzyme activity was determined using cytochrome c oxidase assay kit (CYTOCOX1, Sigma–Aldrich, Inc., St. Louis, MO, United States) according to the manufacturer’s instructions. A 2-μl overnight bacterial culture was inoculated on TSA-0.4 plates. After incubation at 37°C for 3 h, the cells on the surface of the plates were harvested, washed, and enumerated spectrophotometrically at OD578. Bacterial suspensions were mixed with assay buffer and ferrocytochrome c substrate solution. The decrease in absorbance at 550 nm of ferrocytochrome c caused by its oxidation to ferricytochrome c was measured. Enzyme activity was then calculated and presented as Units/ml. Definition of one unit is to oxidize 1.0 μ mole of ferrocytochrome c per min at pH 7.0 and 25°C. The value of each sample was normalized to the absorbance of the bacterial culture at 578 nm (A578).
RNA Isolation and Real-Time RT-PCR
Bacterial cells were suspended in 0.5 mg/ml lysostaphin (Sigma–Aldrich, Inc., St. Louis, MO, United States) and incubated at 37°C for 15 min. RNA was then isolated and purified by using TRIzol reagent (Invitrogen, Waltham, MA, United States) according to the manufacturer’s instructions. The transcripts of qoxA and atpA were reverse transcribed and amplified using the primers: qoxA-F (5′-CATTTGTATCCTGCACTTACTG) and qoxA-R (5′-CGTTGCTGCTTTAGCTATTC); atpA-F (5′-TGGTGTTCGACTGGTAATG) and atpA-R (5′-TTC GGTTCAGACCTTGAT), respectively. The gyrB mRNA, which was used as an internal control, was amplified using primers F1 (5′-ACGGATAACGGACGTGGTATCCCA) and R1 (5′-GCCACCGCCGAATTTACCACCA). RNA transcripts were quantified by real-time reverse transcription-PCR (RT-PCR; Bio-Rad). To monitor the specificity of RT-PCR, the PCR products were analyzed by melting-curve analysis. Experiment was performed at least three times and each sample was prepared in triplicate.
Statistical Analysis
Data were analyzed by two-tailed Student’s t-test using GraphPad Prism software version 5.0 (La Jolla, CA, United States). A p-value of <0.05 was considered statistically significant. Data are presented as the mean ± SD.
Results
Involvement of the Heme Synthesis System in Colony Spreading
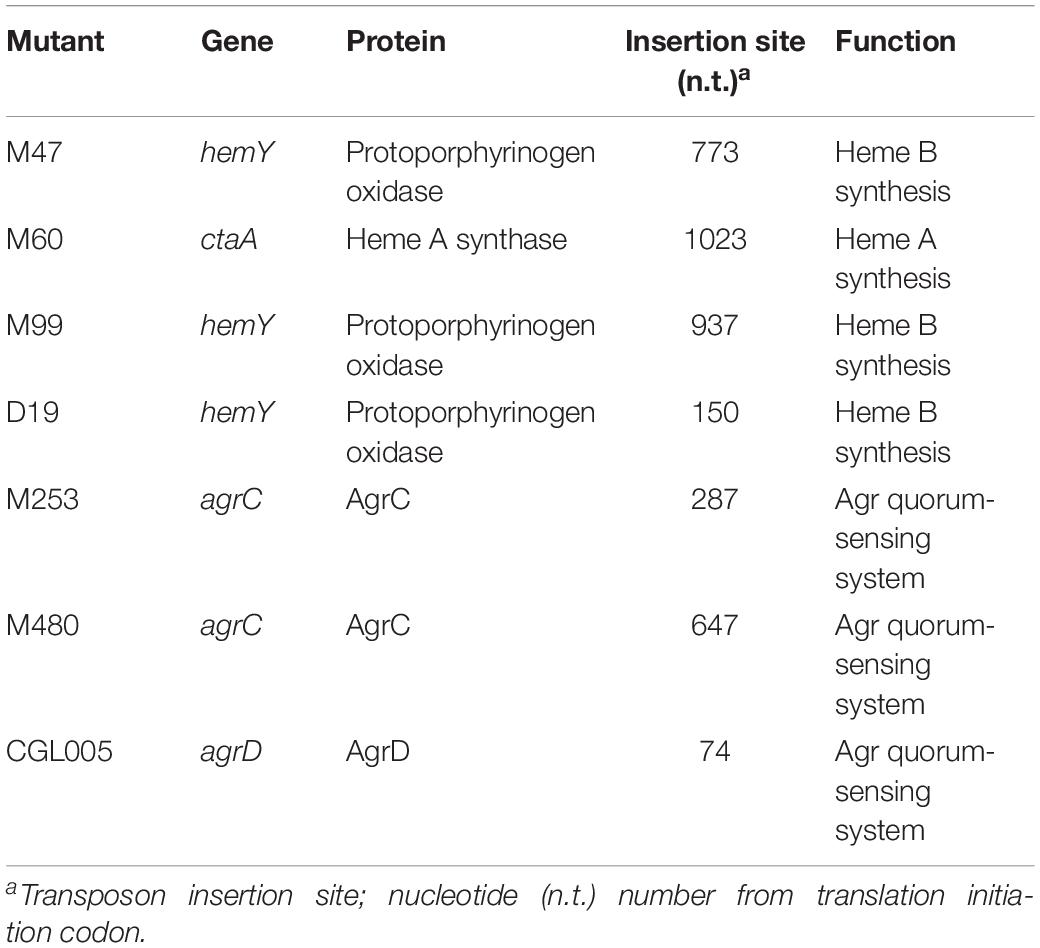
To identify the genes that are involved in colony spreading, S. aureus HG001 was mutagenized with a mariner-based transposon, bursa aurealis (Bae et al., 2004). Approximately 9000 colonies harboring transposon insertions were obtained. These colonies were subsequently screened for their ability to spread on TSA-0.25. Among the mutants that were defective in colony spreading, M253, M480, and CGL005 contained transposon insertions in agrC and agrD (Table 1), verifying the importance of the Agr quorum-sensing system in S. aureus spreading (Tsompanidou et al., 2011). Mutants M47, M99, and D19 had a transposon insertion in different locations in hemY (Figure 1B). HemY encodes protoporphyrinogen oxidase, which converts protoporphyrinogen IX into protoporphrin IX, a precursor of protoheam (heme B) (Panek and O’Brian, 2002; Figure 1A). Mutant M60 contained an insertion in ctaA, which encodes heme A synthase (Hederstedt, 2012). When these mutants were genetically complemented with a plasmid carrying hemY or ctaA, the colony spreading activities of the complemented mutants were restored (Figures 2Ag–j), indicating that hemY and ctaA are crucial to colony spreading.
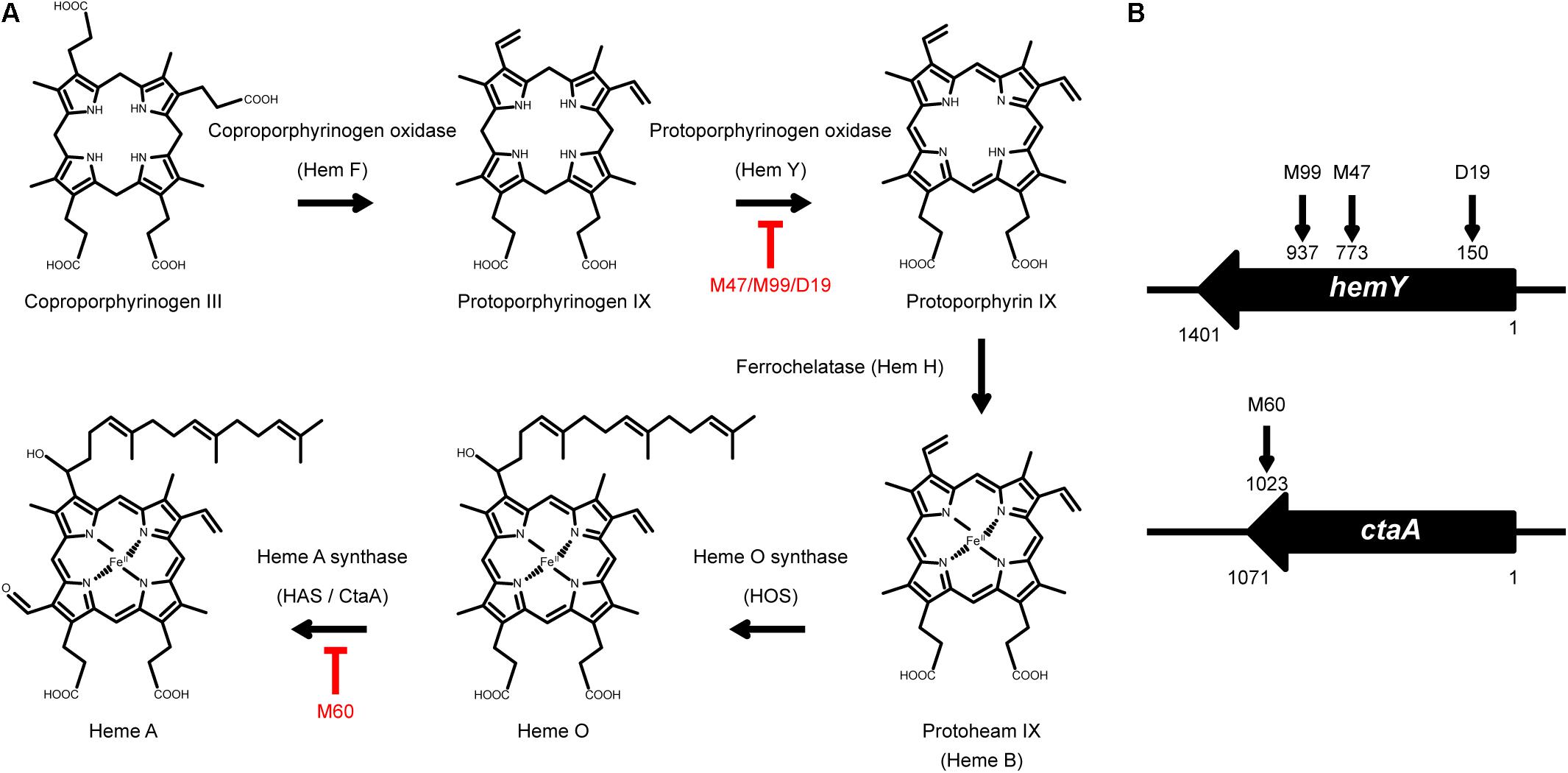
Figure 1. The heme synthesis pathway and transposon insertion mutations in non-spreading mutants. (A) Schematic illustration of the heme synthesis pathway and mutants that are defective in heme synthesis. (B) The vertical arrows on hemY and ctaA indicate the location of transposon insertions in M99, M47, D19, and M60.
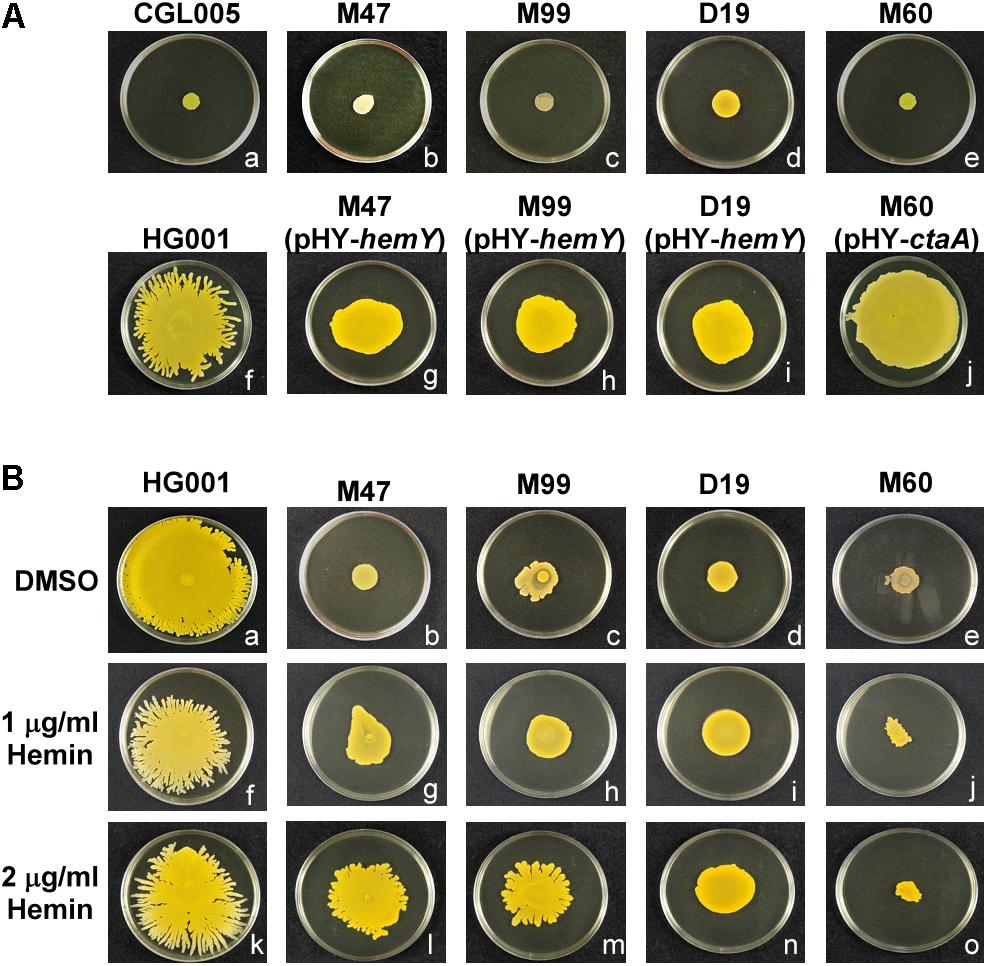
Figure 2. Morphology of spreading colonies formed by heme-deficient mutants. (A) S. aureus strains CGL005 (a), M47 (b), M99 (c), D19 (d), M60 (e), HG001 (f), M47 (pHY-hemY) (g), M99 (pHY-hemY) (h), D19 (pHY-hemY) (i), and M60 (pHY-ctaA) (j) were inoculated on TSA-0.25 plates. After 24 h of incubation at 37°C, the morphology of spreading colonies was observed. Strain CGL005 is a non-spreading mutant. (B) TSA-0.25 plates containing 1 μg/ml (f–j) or 2 μg/ml (k–o) hemin were inoculated with S. aureus strains HG001 (a,f,k), M47 (b,g,l), M99 (c,h,m), D19 (d,i,n), and M60 (e,j,o). After incubation for 24 h, morphology of spreading colonies was observed. Plates containing DMSO (a–e) were used as a solvent control. The pictures shown are representative of at least three independent experiments.
We also incorporated hemin, a heme B derivative, into the TSA-0.25 plates to determine whether hemin addition rescued the spreading defect of the heme-deficient mutants. The results showed that adding 1 or 2 μg/ml hemin into the TSA-0.25 plates restored the spreading ability of the M47, M99, and D19 mutants (Figures 2Bf–i,k–n). However, hemin could not restore the spreading ability of M60, indicating that heme A, which is located downstream of the heme synthesis pathway, is also crucial for S. aureus colony spreading (Figures 1A, 2Bj,o).
Mutations Affecting Heme Synthesis Impair Water Accumulation in Spreading Colonies
To determine whether heme deficiency affected water accumulation in spreading colonies, the plates were tilted at a 30° angle during culturing to observe the flow of accumulated water in the colonies with gravity. The tilted plate assay showed that after incubation for 3 h, water flowed out of the S. aureus HG001 colony toward gravity (Figure 3Af). However, only relatively small amount of water flowed out from M47, M60, M99, and D19 colonies (Figures 3Ab–e), indicating that mutations in heme synthesis genes affected water accumulation in the spreading colonies. Furthermore, water accumulation was restored in the spreading colonies of the complemented mutants (Figures 3Ag–j). Adding hemin to the medium also rescued the water accumulation phenotypes of the heme-deficient mutants M47, M99, and D19 (Figures 3Bb–d) but not that of M60 (Figure 3Be). These results verified that hemY and ctaA are critical not only for colony spreading but also for water accumulation.
Figure 3. Water accumulation in spreading colonies. (A) TSA-0.4 plates were inoculated with overnight cultures of S. aureus strains CGL005 (a), M47 (b), M99 (c), D19 (d), M60 (e), HG001 (f), M47 (pHY-hemY) (g), M99 (pHY-hemY) (h), D19 (pHY-hemY) (i), and M60 (pHY-ctaA) (j). (B) TSA-0.4 plates containing 2 μg/ml hemin were inoculated with overnight cultures of S. aureus strains HG001 (a), M47 (b), M99 (c), D19 (d), and M60 (e). Plates were tilted 30° and incubated for 3 h to observe the flow of water out of the colonies. Strain CGL005 is a non-spreading mutant. The pictures shown are representative of at least three independent experiments.
Activation of Quorum Sensing in Spreading Colonies of Heme-Deficient Mutants
It is well known that, in S. aureus, the cell density-dependent Agr quorum-sensing system regulates hemolysins and PSMs synthesis (Queck et al., 2008) and is required for colony spreading (Tsompanidou et al., 2011). Our previous study showed that an increased bacterial density within a spreading colony triggers the Agr quorum-sensing response to transcribe psm genes and produce PSMs, which facilitate colony spreading (Lin et al., 2016). To examine whether defects in heme synthesis affected the activity of the Agr system, resulting in the reduction of spreading, we analyzed Agr functions based on the production of δ-hemolysin and PSMs. Since δ-hemolysin-mediated lysis of sheep red blood cells is enhanced by β-hemolysin, which is produced by S. aureus RN4220 (Herbert et al., 2010), therefore, δ-hemolytic activity can be visualized as a zone of enhanced hemolysis at the intersection of S. aureus RN4220 and the streaked test strains. δ-Hemolysin assay showed an enhanced hemolytic area at the intersection of streaks of the heme-deficient mutants M47, M99, D19, and M60 and strain RN4220 after 24 h of incubation, indicating that the mutants produced δ-hemolysin (Figure 4A). To further evaluate Agr functions, we examined PSMs production by extracting PSMs from the media of S. aureus HG001, M60, and M60 (pHY-ctaA) cultures. PSMs are small peptides that are secreted into the culture medium. As antibodies monitoring exoproteins are unavailable, we used the total amount of exoproteins as an internal control to correct the relative amount of PSMs in each sample. The results demonstrated that there was no significant difference in the amount of PSMs produced by the three strains (Figure 4B), indicating that heme deficiency does not influence PSMs expression. Our previous study revealed that quorum-sensing occurs in spreading colonies of S. aureus HG001 after 3 h of culturing (Lin et al., 2016). The current study found that the cell density of the colony formed by the non-spreading mutant M60 was similar to that of S. aureus HG001 during the first 5 h of culture (Figure 4C). Furthermore, a reporter plasmid, pPSM-gfp, in which the promoter of the psmα operon that is controlled by Agr quorum-sensing, activates the transcription of a gfp reporter gene, was transformed into S. aureus HG001 and the M60 mutant. We found that under a CLSM, the cells in the colony of S. aureus HG001 did not exhibit much green fluorescence before 3 h culturing (Figures 4Da,b). However, strong green fluorescent signals were detected after 5 h incubation (Figure 4Dc). The similar results were observed in the colony of M60 mutant (Figures 4Dd–f). The CLSM analysis revealed that a quorum-sensing response occurred in the spreading colony of the S. aureus HG001 and M60 mutant (Figure 4D). Overall, these results clarified that the inability of the heme-deficient mutants to spread is not due to Agr dysfunction.
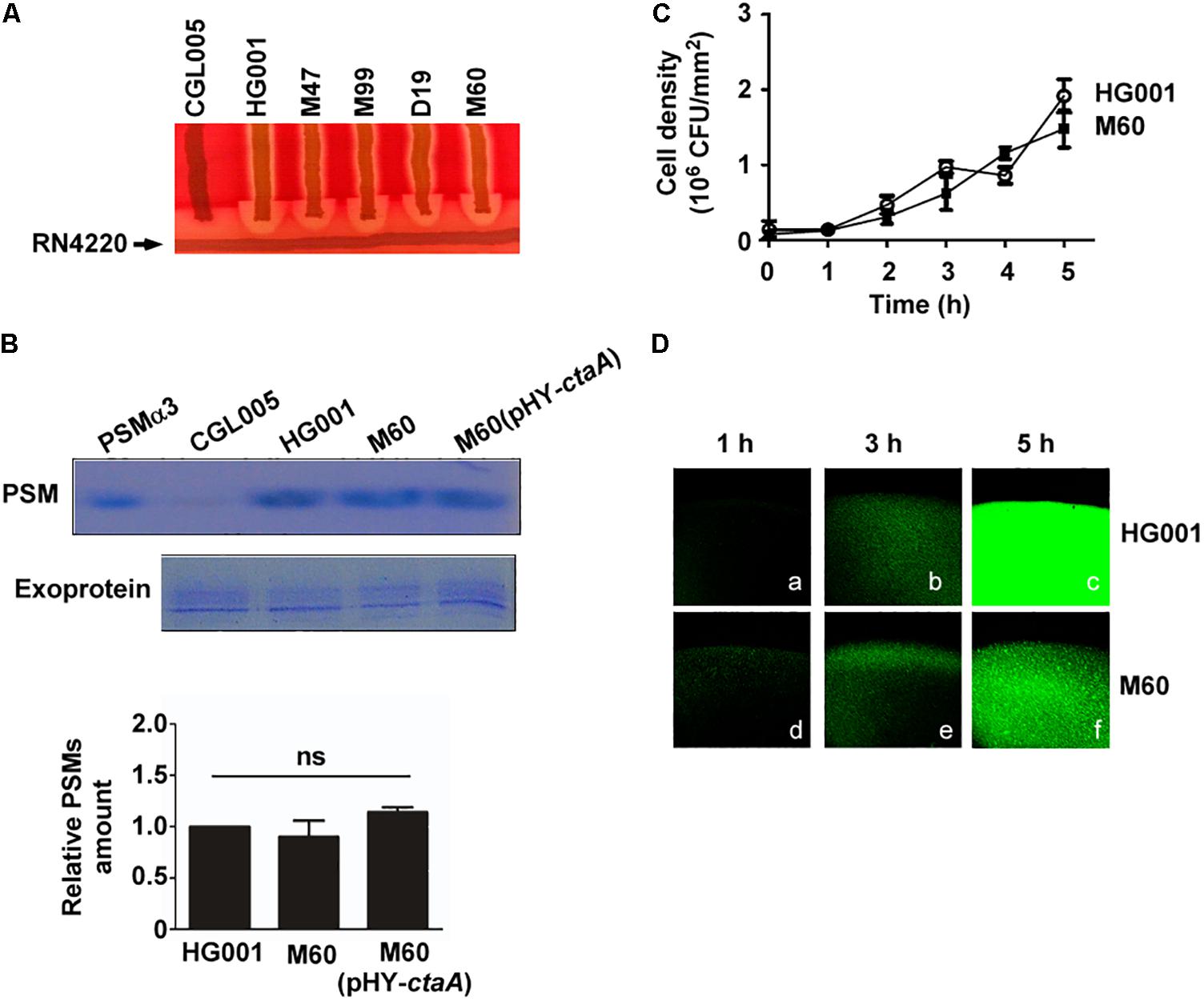
Figure 4. Agr activity in heme-deficient mutants. (A) δ-Hemolysin activities of S. aureus strains CGL005, HG001, M47, M99, D19, and M60 were determined by cross-streaking the strains perpendicularly to S. aureus RN4220 on blood agar plates. The picture shown is representative of at least three independent experiments. (B) PSMs and exoproteins were isolated from the media of S. aureus HG001, M60, and M60 (pHY-ctaA) cultures. PSMs were extracted with butanol. The exoproteins in the culture medium were isolated using Amicon Ultra-4 centrifugal filters. Both PSMs and exoproteins were analyzed by SDS-PAGE followed by Coomassie blue staining (Top). Synthetic PSMα3 was used as a size marker to indicate the location of PSMs on the SDS-PAGE gel. Strain CGL005, which is an Agr deficiency mutant and does not produce PSMs, was used as a control. Exoproteins in culture medium were used as loading control. The intensity of protein bands in each lane was quantified by using a densitometer. The amount of PSMs was normalized to the amount of exoproteins. The relative amount of PSMs from S. aureus HG001 was set at 1 to calculate the relative amount of PSMs from strains M60 and M60 (pHY-ctaA) (Bottom). The gel shown is representative of three independent experiments. Data are presented as the mean of three independent experiments. Error bars denote standard deviations. ns indicates no significant difference. (C) TSA-0.4 plates were inoculated with S. aureus HG001 and M60 and incubated for 5 h. The cell number in a spreading colony was determined by CFU enumeration. Cell density (CFU/mm2) was then calculated. Data are presented as the mean of three independent experiments. Error bars denote standard deviations. (D) After 1, 3, or 5 h of incubation on TSA-0.4 plates, images of spreading colonies formed by S. aureus HG001 (pPSM-gfp) (a–c) and M60 (pPSM-gfp) (d–f) were acquired under a CLSM.
Heme Deficiency Impairs Energy Production and Colony Spreading
Staphylococcus aureus can utilize aerobic respiration to generate energy. Aerobic respiration depends on enzymes such as heme-containing cytochrome oxidases to catalyze a series of electron transport chain reactions, which drive proton translocation across the cytoplasmic membrane and produce ATP via the F0F1-ATPase (Mobius et al., 2010; Zeden et al., 2018). Therefore, we proposed that mutations in the heme synthesis pathway may impair cytochrome oxidase activity and decrease energy production. To test this hypothesis, cytochrome oxidase activity assays and ATP assays were conducted. The results showed that the activity of cytochrome oxidases in S. aureus HG001 was 0.65 unit/ml (Figure 5A). In the heme-deficient mutant M60, the activity of cytochrome oxidases decreased to 0.2 unit/ml (Figure 5A). However, the activity of cytochrome oxidases increased to 0.5 unit/ml when pHY-ctaA was introduced into M60 (Figure 5A). Similar results were obtained for detection of ATP production (Figure 5B). The results revealed that lack of heme A diminished the activity of cytochrome oxidases and led to decrease ATP production. Furthermore, heme A deficiency did not affect the transcription of qoxA and atpA, which encode cytochrome oxidase subunit and ATPase subunit, respectively (Figures 5C,D). In addition, an inhibitor of ATPase, DCCD (Joung et al., 2016), was used to clarify whether the requirement of energy is crucial to water accumulation and colony spreading. The results showed that adding DCCD into culture medium not only reduced ATP production (Figure 5F) but also decreased water accumulation (Figures 5Ed–f) and colony spreading (Figures 5Ea–c). These results revealed that heme deficiency impaired ATP production and resulted in decreased water accumulation and colony spreading.
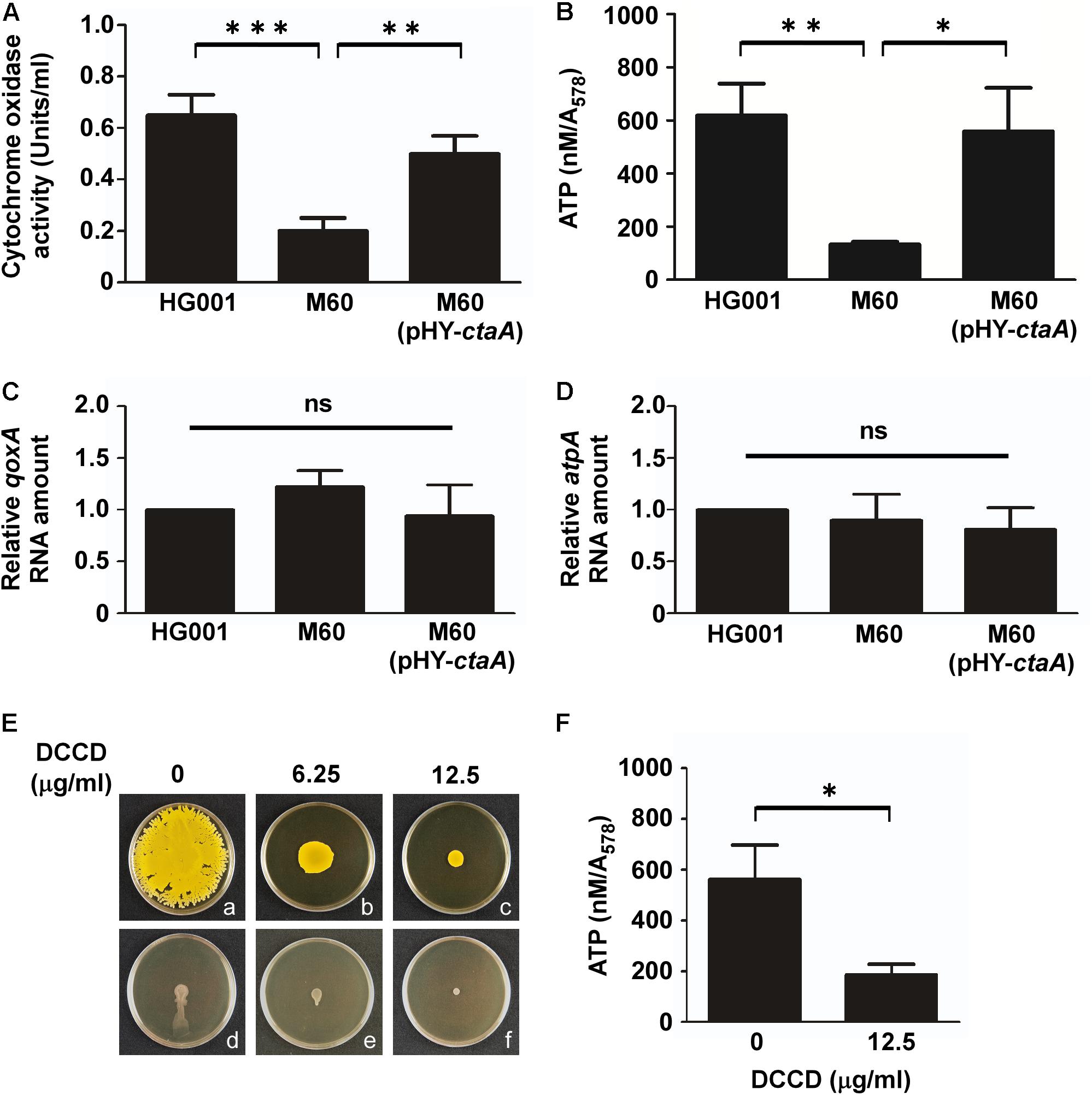
Figure 5. Mutation of ctaA and inhibition of ATPase decrease ATP production and colony spreading. After incubation on plates for 3 h, the cells in colonies of S. aureus HG001, M60, and M60 (pHY-ctaA) were harvested. The activity of cytochrome oxidase was determined using an enzyme assay kit (A). The concentrations of intracellular ATP were determined and normalized to the absorbance of the bacterial cultures at 578 nm (A578) (B). Total RNA was isolated from S. aureus HG001, M60, and M60 (pHY-ctaA). Transcriptions of qoxA (C) and atpA (D) were determined by quantitative RT-PCR. The amount of qoxA and atpA transcripts was normalized to the amount of gyrB RNA. The RNA amounts of qoxA and atpA in S. aureus HG001 were set at 1 to calculate the relative amounts of qoxA and atpA in strains M60 and M60 (pHY-ctaA). (E) S. aureus HG001 was inoculated on DCCD-containing TSA-0.25 (a–c) or TSA-0.4 (d–f) plates. After 24 h of incubation at 37°C, the morphology of spreading colonies on TSA-0.25 plates was observed (a–c). TSA-0.4 plates containing DCCD were tilted 30° and incubated for 3 h to observe the flow of water out of the colonies (d–f). (F) The cells on DCCD-containing TSA-0.4 plates were harvested for determination of the ATP concentrations. The pictures shown in part (E) are representative of at least three independent experiments. Data are presented as the mean of three independent experiments. Error bars denote standard deviations. Significant differences are denoted as ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.005. ns indicates no significant difference.
Discussion
Heme is the prosthetic group of cytochrome oxidases, which are important for electron transport and energy production (Mobius et al., 2010; Hederstedt, 2012). Previous studies have shown that electron-transport-defective mutants have characteristics similar to those of small cell variants (SCVs), which exhibit reduced colony size and slow growth (von Eiff et al., 1997; Hammer et al., 2013). Despite that our heme-deficient mutants exhibited phenotypes typical of SCVs, such as slow growth and decreased colony size, the cell densities of the colonies formed by the mutants reached the threshold required to trigger the quorum-sensing response to initiate colony spreading (Figures 4C,D). Although both strains HG001 and M60 reached a comparable cell density to trigger the quorum-sensing response (Figures 4C,D), the total number of cells in the colony of the M60 mutant is less than that of the strains HG001 because of the decreased colony size of the M60 mutant. Therefore, under a CLSM, the fluorescence intensity of the strains HG001 is stronger than that of the M60 mutant (Figure 4D). Furthermore, defects in heme synthesis had little effect on the expression of PSMs (Figure 4B). These results indicate that the non-spreading phenotype of the heme-deficient mutants is not attributable to slow growth or Agr dysfunction. However, the colony spreading assay and tiled plate assay showed that mutations in heme synthesis genes affected water accumulation and colony spreading (Figures 2, 3). We also used another heme deficiency mutant S. aureus NCTC8325-4ΔhemB, which also exhibited typical phenotypes of SCVs, to clarify the importance of heme synthesis to water accumulation and colony spreading (Supplementary Figure S2).
Staphylococcus aureus is able to spread on a moist semisolid surface (Kaito and Sekimizu, 2007). Our earlier study also showed that increasing the agarose concentration decreases colony spreading (Lin et al., 2016). However, increasing the volume of culture medium in the plate can rescue S. aureus spreading on plates containing a high concentration of agarose (Lin et al., 2016). These results indicate that the amount of water in the medium influences the ability of S. aureus to spread. Additionally, there is overwhelming evidence supporting the idea that water is recruited from the culture medium by a bacterial swarm (Wu and Berg, 2012; Ping et al., 2014; Ke et al., 2015; Yang et al., 2017). Work by Wu and Berg (2012) showed that a water reservoir traveling with a swarm fuels the spreading of E. coli. B. subtilis swarm in water, and the water surface tension modulates the swarming mechanics (Ke et al., 2015). A recent study on the swarming motility of P. aeruginosa demonstrated that their ability to draw water from the culture medium is the essential factor determining whether this bacteria can spread over the agar surface (Yang et al., 2017). Similarly, our previous study showed that water accumulation in spreading colonies to elicit water surface tension and is crucial for facilitating colony spreading of S. aureus (Lin et al., 2016). However, the underlying mechanism by which S. aureus extracts water from the medium is still unknown.
Based on the screening of a transposon mutant library, we found that mutations in genes involved in heme synthesis impaired the spreading of S. aureus (Table 1). When the heme-deficient mutants were cultured on tilted plates, the colonies formed by the mutants had less water flowing out (Figure 3A), suggesting that heme synthesis affects water accumulation during colony spreading of S. aureus. Since hemes are involved in energy production, we hypothesize that S. aureus utilizes an energy-dependent mechanism to extract water from the culture medium. Water accumulation in a colony increases the surface tension to restrict colony expansion. The cells grow, and the cell density in the colony increases, which triggers the quorum-sensing response to activate expression of PSMs. As water is continuously extracted from the medium, PSMs weaken the water surface tension, allowing water to flow. S. aureus is then carried by the flowing water across the plate surface.
Although many studies strongly suggest that water recruitment is crucial for bacterial swarming and colony spreading, the exact molecule to extract water is still unknown. In E. coli, it has been suggested that LPS may serve as the osmolyte to extract water from agar and facilitates its swarming (Ping et al., 2014). In S. aureus, mutations in the tagO gene, which encodes the first enzyme TagO of the WTAs synthesis pathway, impair the spreading ability of S. aureus (Kaito and Sekimizu, 2007), indicating WTAs are required for colony spreading of S. aureus. Both LPS and WTAs are glycopolymers that attached to the cell surface of bacteria. Therefore, we posit that WTAs are likely as an osmolyte and involved in water extraction during colony spreading of S. aureus. To test the hypothesis, S. aureus HG001 was cultured on plates that contained tunicamycin, an inhibitor of TagO (Campbell et al., 2011). The results showed that tunicamycin inhibited colony spreading of S. aureus in a dose dependent manner (Supplementary Figure S1A). Furthermore, inhibition of TagO by tunicamycin (Supplementary Figure S1A) or deletion of tagO in S. aureus strain SA113 (SA113ΔtagO) (Supplementary Figure S1B) decreased water accumulation in spreading colony, indicating that WTAs are required for water extraction during colony spreading of S. aureus. WTAs are synthesized intracellularly and exported to the cell surface by an energy-dependent ABC (ATP-binding cassette) transporter (Winstel et al., 2014), indicating that energy is required to facilitate translocation of WTAs across membrane. Accordingly, lack of heme in heme deficiency mutants results in reduction of ATP production and may decrease the amount of WTAs attached to cell surface, by which diminish the water extraction and disable colony spreading. However, the mechanisms need to be further investigated.
Although the ability of S. aureus to move across a soft solid surface is well known, colony spreading of S. aureus is currently defined as a form of passive motility (Pollitt and Diggle, 2017). A recent study showed that S. aureus forms comet structures at the tips of spreading colony dendrites that facilitate colonies extension on soft agar (Pollitt et al., 2015). This comet-type movement exhibits the characteristics of active motility (Pollitt et al., 2015). The current study finds that heme is required for both energy production and colony spreading. Additionally, the results indicate that energy consumption is involved in colony spreading and imply that colony spreading is a form of energy-dependent active motility. Furthermore, both colony spreading and comet motility of S. aureus require Agr-induced expression of PSM-type biosurfactants (Pollitt et al., 2015; Kizaki et al., 2016). Biosurfactants are also required for the swarming motility of B. subtilis, Serratia marcescens, and Pseudomonas aeruginosa (Kearns, 2010), suggesting that S. aureus shares a common mechanism with these actively motile bacteria to move on agar surfaces.
In nature, the ability to move rapidly gives bacteria advantages to obtain nutrient and food. Moreover, the significance of motility in bacterial pathogenesis is well documented (Jones et al., 2004). It is known that the motility of P. mirabilis, E. coli, Vibrio cholera, and S. marcescens is closely associated with their pathogenesis (Jones et al., 2004; Syed et al., 2009; Shanks et al., 2013; Garcia-Heredia et al., 2016). An earlier study showed that S. aureus is capable of spreading on fresh pork meat, showing that this organism is able to spread on soft tissues (Tsompanidou et al., 2013). Previous studies also demonstrated that most invasive S. aureus clinical strains are able to spread, indicating that spreading is a common characteristic shared by strains that cause invasive infections in human (Kaito et al., 2008). The results of this study provide new insights into the movement mechanisms of bacteria that lack a motor apparatus and elucidate how S. aureus spreads, which is important for understanding how this pathogen moves in human tissues to cause disease.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
M-HL and C-CL conceived and designed the study, performed the experiments, analyzed the data, and wrote the manuscript.
Funding
This work was supported by grants from the Chang Gung Memorial Hospital (CGMH), Taiwan (CMRPD1F0061-3 and CMRPD1J0271-3), and the Ministry of Science and Technology (MOST), Taiwan (MOST 108-2320-B-182-028 and 105-2320-B-182-027-MY3).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank M-LH and the Microscopy Core Laboratory at the Linkou Chang Gung Memorial Hospital (CGMH), Taiwan, for microscopy support. The authors would also like to thank Shih-Tung Liu for his critiques.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00170/full#supplementary-material
References
Allison, C., Lai, H. C., and Hughes, C. (1992). Co-ordinate expression of virulence genes during swarm-cell differentiation and population migration of Proteus mirabilis. Mol. Microbiol. 6, 1583–1591. doi: 10.1111/j.1365-2958.1992.tb00883.x
Aoyagi, T., Kaito, C., Sekimizu, K., Omae, Y., Saito, Y., Mao, H., et al. (2014). Impact of psm-mec in the mobile genetic element on the clinical characteristics and outcome of SCCmec-II methicillin-resistant Staphylococcus aureus bacteraemia in Japan. Clin. Microbiol. Infect 20, 912–919. doi: 10.1111/1469-0691.12575
Armbruster, C. E., Hodges, S. A., and Mobley, H. L. (2013). Initiation of swarming motility by Proteus mirabilis occurs in response to specific cues present in urine and requires excess L-glutamine. J. Bacteriol. 195, 1305–1319. doi: 10.1128/JB.02136-12
Bae, T., Banger, A. K., Wallace, A., Glass, E. M., Aslund, F., Schneewind, O., et al. (2004). Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. U.S.A 101, 12312–12317. doi: 10.1073/pnas.0404728101
Callegan, M. C., Novosad, B. D., Ramirez, R., Ghelardi, E., and Senesi, S. (2006). Role of swarming migration in the pathogenesis of bacillus endophthalmitis. Invest Ophthalmol. Vis. Sci. 47, 4461–4467. doi: 10.1167/iovs.06-0301
Campbell, J., Singh, A. K., Santa Maria JP Jr, Kim, Y., Brown, S., Swoboda, J. G., et al. (2011). Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 6, 106–116. doi: 10.1021/cb100269f
Chen, B. G., Turner, L., and Berg, H. C. (2007). The wetting agent required for swarming in Salmonella enterica serovar typhimurium is not a surfactant. J. Bacteriol. 189, 8750–8753. doi: 10.1128/JB.01109-07
Dailey, H. A., Dailey, T. A., Gerdes, S., Jahn, D., Jahn, M., O’brian, M. R., et al. (2017). Prokaryotic heme biosynthesis: multiple pathways to a common essential product. Microbiol. Mol. Biol. Rev. 81:e00048-16. doi: 10.1128/MMBR.00048-16
Garcia-Heredia, A., Garcia, S., Merino-Mascorro, J. A., Feng, P., and Heredia, N. (2016). Natural plant products inhibits growth and alters the swarming motility, biofilm formation, and expression of virulence genes in enteroaggregative and enterohemorrhagic Escherichia coli. Food Microbiol. 59, 124–132. doi: 10.1016/j.fm.2016.06.001
Hammer, N. D., Reniere, M. L., Cassat, J. E., Zhang, Y., Hirsch, A. O., Indriati Hood, M., et al. (2013). Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. MBio 4 4:.00241-13. doi: 10.1128/mBio.00241-13
Hederstedt, L. (2012). Heme A biosynthesis. Biochim. Biophys. Acta 1817, 920–927. doi: 10.1016/j.bbabio.2012.03.025
Herbert, S., Ziebandt, A. K., Ohlsen, K., Schafer, T., Hecker, M., Albrecht, D., et al. (2010). Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect Immun. 78, 2877–2889. doi: 10.1128/IAI.00088-10
Jaques, S., and McCarter, L. L. (2006). Three new regulators of swarming in Vibrio parahaemolyticus. J. Bacteriol. 188, 2625–2635. doi: 10.1128/JB.188.7.2625-2635.2006
Jones, B. V., Young, R., Mahenthiralingam, E., and Stickler, D. J. (2004). Ultrastructure of Proteus mirabilis swarmer cell rafts and role of swarming in catheter-associated urinary tract infection. Infect Immun. 72, 3941–3950. doi: 10.1128/IAI.72.7.3941-3950.2004
Joung, D. K., Mun, S. H., Choi, S. H., Kang, O. H., Kim, S. B., Lee, Y. S., et al. (2016). Antibacterial activity of oxyresveratrol against methicillin-resistant Staphylococcus aureus and its mechanism. Exp. Ther. Med. 12, 1579–1584. doi: 10.3892/etm.2016.3486
Kaito, C., Hirano, T., Omae, Y., and Sekimizu, K. (2011a). Digestion of extracellular DNA is required for giant colony formation of Staphylococcus aureus. Microb. Pathog 51, 142–148. doi: 10.1016/j.micpath.2011.04.007
Kaito, C., Omae, Y., Matsumoto, Y., Nagata, M., Yamaguchi, H., Aoto, T., et al. (2008). A novel gene, fudoh, in the SCCmec region suppresses the colony spreading ability and virulence of Staphylococcus aureus. PLoS One 3:e3921. doi: 10.1371/journal.pone.0003921
Kaito, C., Saito, Y., Nagano, G., Ikuo, M., Omae, Y., Hanada, Y., et al. (2011b). Transcription and translation products of the cytolysin gene psm-mec on the mobile genetic element SCCmec regulate Staphylococcus aureus virulence. PLoS Pathog 7:e1001267. doi: 10.1371/journal.ppat.1001267
Kaito, C., and Sekimizu, K. (2007). Colony spreading in Staphylococcus aureus. J. Bacteriol. 189, 2553–2557. doi: 10.1128/JB.01635-06
Ke, W. J., Hsueh, Y. H., Cheng, Y. C., Wu, C. C., and Liu, S. T. (2015). Water surface tension modulates the swarming mechanics of Bacillus subtilis. Front. Microbiol. 6:1017. doi: 10.3389/fmicb.2015.01017
Kearns, D. B. (2010). A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8, 634–644. doi: 10.1038/nrmicro2405
Kim, W., and Surette, M. G. (2005). Prevalence of surface swarming behavior in Salmonella. J. Bacteriol. 187, 6580–6583. doi: 10.1128/JB.187.18.6580-6583.2005
Kizaki, H., Omae, Y., Tabuchi, F., Saito, Y., Sekimizu, K., and Kaito, C. (2016). Cell-surface phenol soluble modulins regulate Staphylococcus aureus Colony Spreading. PLoS One 11:e0164523. doi: 10.1371/journal.pone.0164523
Lai, H. C., Soo, P. C., Wei, J. R., Yi, W. C., Liaw, S. J., Horng, Y. T., et al. (2005). The RssAB two-component signal transduction system in Serratia marcescens regulates swarming motility and cell envelope architecture in response to exogenous saturated fatty acids. J. Bacteriol. 187, 3407–3414. doi: 10.1128/JB.187.10.3407-3414.2005
Lin, M. H., Ke, W. J., Liu, C. C., and Yang, M. W. (2016). Modulation of Staphylococcus aureus spreading by water. Sci. Rep. 6:25233. doi: 10.1038/srep25233
Lin, M. H., Li, C. C., Shu, J. C., Chu, H. W., Liu, C. C., and Wu, C. C. (2018). exoproteome profiling reveals the involvement of the foldase PrsA in the cell surface properties and pathogenesis of Staphylococcus aureus. Proteomics 18:e1700195. doi: 10.1002/pmic.201700195
Mobius, K., Arias-Cartin, R., Breckau, D., Hannig, A. L., Riedmann, K., Biedendieck, R., et al. (2010). Heme biosynthesis is coupled to electron transport chains for energy generation. Proc. Natl. Acad. Sci. U.S. A. 107, 10436–10441. doi: 10.1073/pnas.1000956107
Nair, D., Memmi, G., Hernandez, D., Bard, J., Beaume, M., Gill, S., et al. (2011). Whole-genome sequencing of Staphylococcus aureus strain RN4220, a key laboratory strain used in virulence research, identifies mutations that affect not only virulence factors but also the fitness of the strain. J. Bacteriol. 193, 2332–2335. doi: 10.1128/JB.00027-11
Olsen, J. E., Hoegh-Andersen, K. H., Casadesus, J., Rosenkranzt, J., Chadfield, M. S., and Thomsen, L. E. (2013). The role of flagella and chemotaxis genes in host pathogen interaction of the host adapted Salmonella enterica serovar Dublin compared to the broad host range serovar S. Typhimurium. BMC Microbiol. 13:67. doi: 10.1186/1471-2180-13-67
Omae, Y., Sekimizu, K., and Kaito, C. (2012). Inhibition of colony-spreading activity of Staphylococcus aureus by secretion of delta-hemolysin. J. Biol. Chem. 287, 15570–15579. doi: 10.1074/jbc.M112.357848
Omae, Y., Sekimizu, K., and Kaito, C. (2014). Identification of Staphylococcus aureus colony-spreading stimulatory factors from mammalian serum. PLoS One 9:e97670. doi: 10.1371/journal.pone.0097670
O’Neill, E., Pozzi, C., Houston, P., Humphreys, H., Robinson, D. A., Loughman, A., et al. (2008). A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins. FnBPA and FnBPB. J. Bacteriol. 190, 3835–3850. doi: 10.1128/JB.00167-08
Panek, H., and O’Brian, M. R. (2002). A whole genome view of prokaryotic haem biosynthesis. Microbiology 148, 2273–2282. doi: 10.1099/00221287-148-8-2273
Ping, L., Wu, Y., Hosu, B. G., Tang, J. X., and Berg, H. C. (2014). Osmotic pressure in a bacterial swarm. Biophys. J. 107, 871–878. doi: 10.1016/j.bpj.2014.05.052
Pollitt, E. J., Crusz, S. A., and Diggle, S. P. (2015). Staphylococcus aureus forms spreading dendrites that have characteristics of active motility. Sci. Rep. 5:17698. doi: 10.1038/srep17698
Pollitt, E. J. G., and Diggle, S. P. (2017). Defining motility in the Staphylococci. Cell Mol. Life Sci. 74, 2943–2958. doi: 10.1007/s00018-017-2507-z
Queck, S. Y., Jameson-Lee, M., Villaruz, A. E., Bach, T. H., Khan, B. A., Sturdevant, D. E., et al. (2008). RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32, 150–158. doi: 10.1016/j.molcel.2008.08.005
Shanks, R. M., Lahr, R. M., Stella, N. A., Arena, K. E., Brothers, K. M., Kwak, D. H., et al. (2013). A Serratia marcescens PigP homolog controls prodigiosin biosynthesis, swarming motility and hemolysis and is regulated by cAMP-CRP and HexS. PLoS One 8:e57634. doi: 10.1371/journal.pone.0057634
Syed, K. A., Beyhan, S., Correa, N., Queen, J., Liu, J., Peng, F., et al. (2009). The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J. Bacteriol. 191, 6555–6570. doi: 10.1128/JB.00949-09
Tsompanidou, E., Denham, E. L., Becher, D., De Jong, A., Buist, G., Van Oosten, M., et al. (2013). Distinct roles of phenol-soluble modulins in spreading of Staphylococcus aureus on wet surfaces. Appl. Environ. Microbiol. 79, 886–895. doi: 10.1128/AEM.03157-12
Tsompanidou, E., Denham, E. L., Sibbald, M. J., Yang, X. M., Seinen, J., Friedrich, A. W., et al. (2012). The sortase A substrates FnbpA, FnbpB, ClfA and ClfB antagonize colony spreading of Staphylococcus aureus. PLoS One 7:e44646. doi: 10.1371/journal.pone.0044646
Tsompanidou, E., Sibbald, M. J., Chlebowicz, M. A., Dreisbach, A., Back, J. W., Van Dijl, J. M., et al. (2011). Requirement of the agr locus for colony spreading of Staphylococcus aureus. J. Bacteriol. 193, 1267–1272. doi: 10.1128/JB.01276-10
Vater, S. M., Weisse, S., Maleschlijski, S., Lotz, C., Koschitzki, F., Schwartz, T., et al. (2014). Swimming behavior of Pseudomonas aeruginosa studied by holographic 3D tracking. PLoS One 9:e87765. doi: 10.1371/journal.pone.0087765
von Eiff, C., Heilmann, C., Proctor, R. A., Woltz, C., Peters, G., and Gotz, F. (1997). A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 179, 4706–4712. doi: 10.1128/jb.179.15.4706-4712.1997
Weidenmaier, C., Kokai-Kun, J. F., Kristian, S. A., Chanturiya, T., Kalbacher, H., Gross, M., et al. (2004). Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10, 243–245. doi: 10.1038/nm991
Winstel, V., Xia, G., and Peschel, A. (2014). Pathways and roles of wall teichoic acid glycosylation in Staphylococcus aureus. Int. J. Med. Microbiol. 304, 215–221. doi: 10.1016/j.ijmm.2013.10.009
Wu, Y., and Berg, H. C. (2012). Water reservoir maintained by cell growth fuels the spreading of a bacterial swarm. Proc. Natl. Acad. Sci. U.S.A. 109, 4128–4133. doi: 10.1073/pnas.1118238109
Yang, A., Tang, W. S., Si, T., and Tang, J. X. (2017). Influence of physical effects on the swarming motility of Pseudomonas aeruginosa. Biophys. J. 112, 1462–1471. doi: 10.1016/j.bpj.2017.02.019
Zeden, M. S., Schuster, C. F., Bowman, L., Zhong, Q., Williams, H. D., and Grundling, A. (2018). Cyclic di-adenosine monophosphate (c-di-AMP) is required for osmotic regulation in Staphylococcus aureus but dispensable for viability in anaerobic conditions. J. Biol. Chem. 293, 3180–3200. doi: 10.1074/jbc.M117.818716
Keywords: Staphylococcus aureus, colony spreading, water accumulation, heme, ATP
Citation: Liu C-C and Lin M-H (2020) Involvement of Heme in Colony Spreading of Staphylococcus aureus. Front. Microbiol. 11:170. doi: 10.3389/fmicb.2020.00170
Received: 09 October 2019; Accepted: 24 January 2020;
Published: 11 February 2020.
Edited by:
Thomas Dandekar, Julius Maximilian University of Würzburg, GermanyReviewed by:
Lígia M. Saraiva, New University of Lisbon, PortugalAngela Wilks, University of Maryland, Baltimore, United States
Copyright © 2020 Liu and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mei-Hui Lin, dGhlYUBtYWlsLmNndS5lZHUudHc=
 Chao-Chin Liu
Chao-Chin Liu Mei-Hui Lin
Mei-Hui Lin