- College of Life Sciences, Shanghai Normal University, Shanghai, China
Iron–sulfur (Fe–S) clusters are ancient and ubiquitous cofactors and are involved in many important biological processes. Unlike the non-photosynthetic bacteria, cyanobacteria have developed the sulfur utilization factor (SUF) mechanism as their main assembly pathway for Fe–S clusters, supplemented by the iron–sulfur cluster and nitrogen-fixing mechanisms. The SUF system consists of cysteine desulfurase SufS, SufE that can enhance SufS activity, SufBC2D scaffold complex, carrier protein SufA, and regulatory repressor SufR. The S source for the Fe–S cluster assembly mainly originates from L-cysteine, but the Fe donor remains elusive. This minireview mainly focuses on the biogenesis pathway of the Fe–S clusters in cyanobacteria and its relationship with iron homeostasis. Future challenges of studying Fe–S clusters in cyanobacteria are also discussed.
Introduction
As cofactors of proteins, iron–sulfur (Fe–S) clusters participate in many important physiological processes, including respiration, photosynthesis, nitrogen fixation, amino acid and purine metabolism, RNA modification, and DNA replication, as well as repair and regulation of gene expression (Beinert et al., 1997; Johnson et al., 2005; Lill, 2009; Balk and Pilon, 2011; Maio and Rouault, 2015). Owing to their photosynthetic autotrophic lifestyle, cyanobacteria are particularly rich in Fe–S clusters. During evolution, cyanobacteria have developed many membrane-embedded photosynthetic protein complexes and electron carriers that contain Fe–S clusters (Table 1). As a consequence, the demand for iron (Fe) in cyanobacteria far exceeds that in other, non-photosynthetic organisms. For example, the Fe quota of oxygenic photosynthetic cyanobacterium Synechocystis species strain PCC 6803 (hereafter Synechocystis 6803) cells is one order of magnitude higher than that of non-photosynthetic bacterium Escherichia coli (Finney and O’Halloran, 2003; Keren et al., 2004).
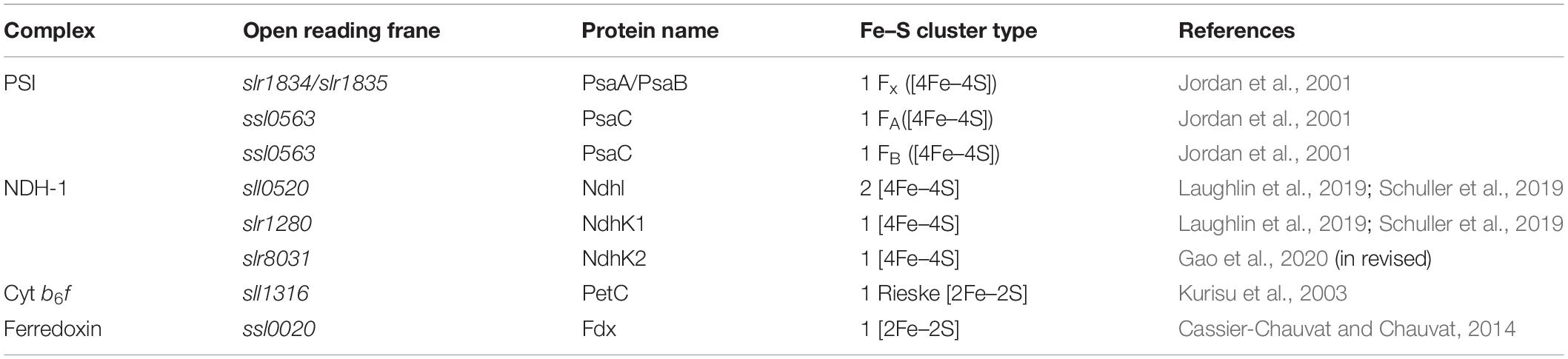
Table 1. Fe–S cluster proteins of photosynthetic complexes in the cyanobacterium Synechocystis 6803.
The Fe–S clusters mainly exist as [2Fe–2S], [4Fe–4S], and [3Fe–4S] types, and their assemblages of Fe ions (+ 2 or + 3 formal oxidation states) and inorganic sulfide (S2–) are coordinated to proteins typically by cysteine ligations at each Fe of the Fe–S cluster (Peters and Broderick, 2012) (for reviews, see Beinert, 2000; Lill, 2009; Balk and Pilon, 2011). However, His, Arg, and Glu residues can also be involved in Fe–S cluster coordination (Berkovitch et al., 2004; Meyer, 2008).
The early earth richly contained reducing Fe and S (Wächtershäuser, 1992), and consequently, Fe–S clusters are believed to spontaneously assemble into primitive biological macromolecules by using suitable ligands (Meyer, 2008). The atmosphere started to become oxidized by oxygenic photosynthesis after the proliferation of cyanobacteria between 3.2 and 2.4 billion years ago (Brocks et al., 1999) and severely limited the assembly of Fe–S clusters (Chapman and Schopf, 1983).
Moreover, reactive oxygen species (ROS), as by-product of oxygen metabolism, also damaged Fe–S clusters (Sutton et al., 2004; Wallace et al., 2004). As a consequence, free Fe could produce ROS through a Fenton reaction to damage cells further (Latifi et al., 2009). Under aerobic conditions, a number of dedicated proteins for Fe–S clusters biogenesis are adapted in cyanobacteria. Therefore, an effective balance between Fe acquisition and protection against oxidative stress is critical for cyanobacteria to survive in their habitat. Many researchers have reviewed the assembly of Fe–S clusters in bacteria and plants (Lill, 2009; Balk and Pilon, 2011; Mettert and Kiley, 2015; Lu, 2018). This minireview will focus on the Fe–S cluster biogenesis and its relationship with Fe homeostasis in cyanobacteria. The challenges of studying Fe–S clusters in cyanobacteria are also discussed.
Fe–S Clusters Biogensis
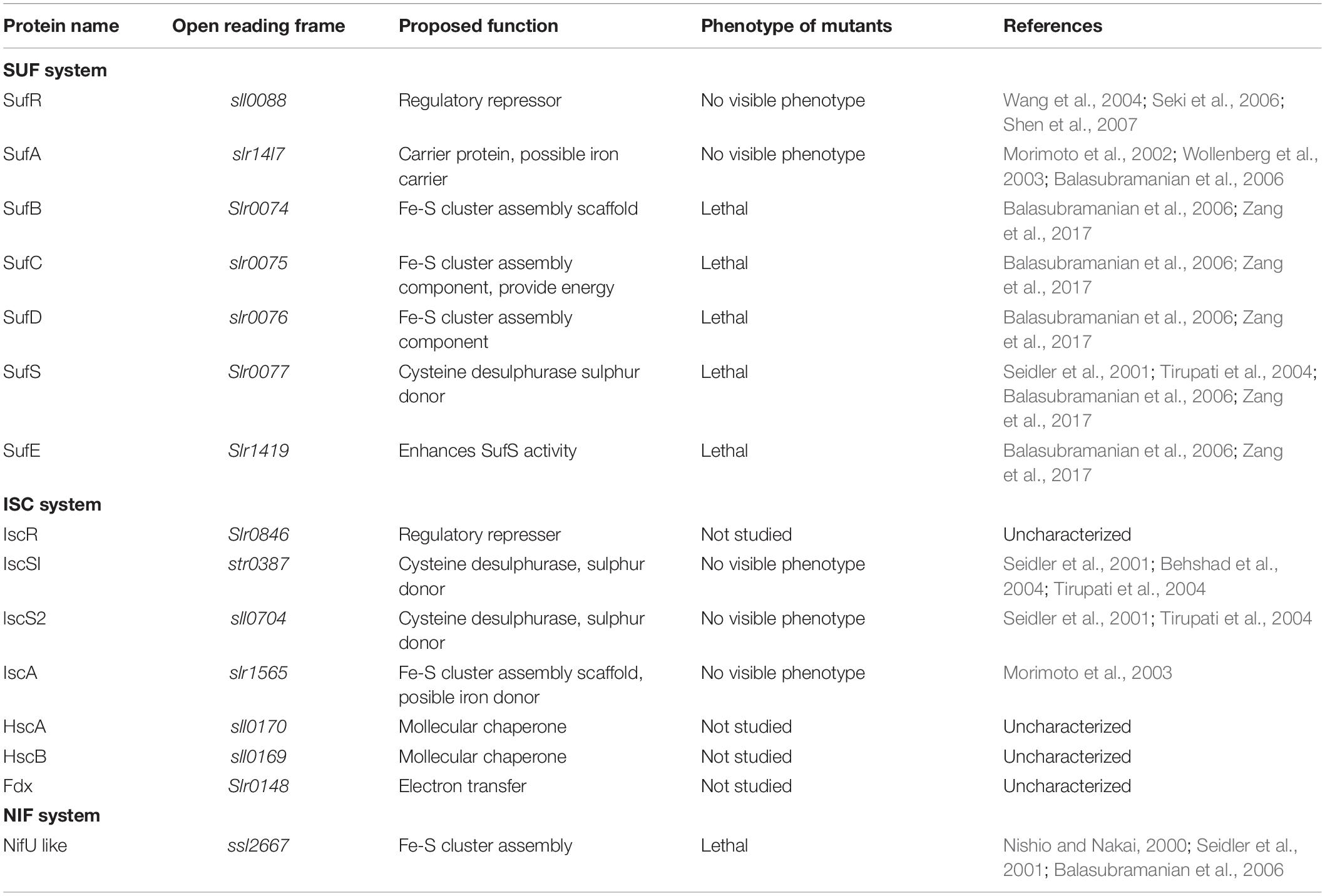
So far, three major mechanisms have been identified for the assembly of Fe–S clusters, including the nitrogen-fixing (NIF), iron–sulfur cluster (ISC), and S utilization factor (SUF) (Johnson et al., 2005; Lill, 2009). The NIF system is the first discovery of Fe–S cluster biosynthesis pathway in Azotobacter vinelandii, and its function is specific to the assembly of Fe–S clusters for the nitrogenase in NIF organisms (Jacobson et al., 1989a, b). Meanwhile, the isc gene region was identified in A. vinelandii using a biochemical approach, and its products are suggested to participate in Fe–S cluster assembly as housekeeping role and are distributed across almost all domains of life, from some archaea and gram-negative bacteria to yeasts, plants, animals, and humans (Zheng et al., 1998; Lill, 2009; Rouault, 2012). The SUF system is the third discovery of Fe–S cluster biosynthesis pathway (Takahashi and Tokumoto, 2002). Compared with ISC system, SUF system is less widespread and is found only in archaea, most gram-positive bacteria, the chloroplasts of plants, and green algae (Takahashi et al., 1986, 1991; Takahashi and Tokumoto, 2002; Albrecht et al., 2010; Santos et al., 2014; Selbach et al., 2014). In E. coli, the SUF system is activated only in response to conditions of oxidative stress or Fe starvation (Outten et al., 2004; Outten, 2015). During evolution, cyanobacteria choose SUF as their major system for Fe–S cluster biosynthesis (Balasubramanian et al., 2006; Ayala-Castro et al., 2008; Outten, 2015), and all core suf genes cannot be knocked out completely in the cyanobacterium Synechocystis 6803 (Tirupati et al., 2004; Balasubramanian et al., 2006; Zang et al., 2017). In higher plants, the importance of SUF system has been verified by analyzing its mutants (Xu and Møller, 2006; Hu et al., 2017). The phylogenetic distribution of the SUF system indicates a coevolutionary relationship with photosynthetic energy storing pathways (Zang et al., 2017). This may be a reason why cyanobacteria chose SUF system as their major synthesis pathway for Fe–S clusters.
These three different mechanisms follow a common biosynthetic rule. The overall biogenesis process can be divided two main steps: (1) de novo assembly of Fe–S cluster on the scaffold protein by recruiting Fe and S and (2) transferring the Fe–S cluster from the scaffold protein to target apo-proteins (Apo) (Figure 1; Lill, 2009; Balk and Pilon, 2011). As shown in Table 2, the main components involved in Fe–S cluster biosynthesis are identified in cyanobacteria using sequence alignment, reverse genetics, physiology, and biochemistry approaches.
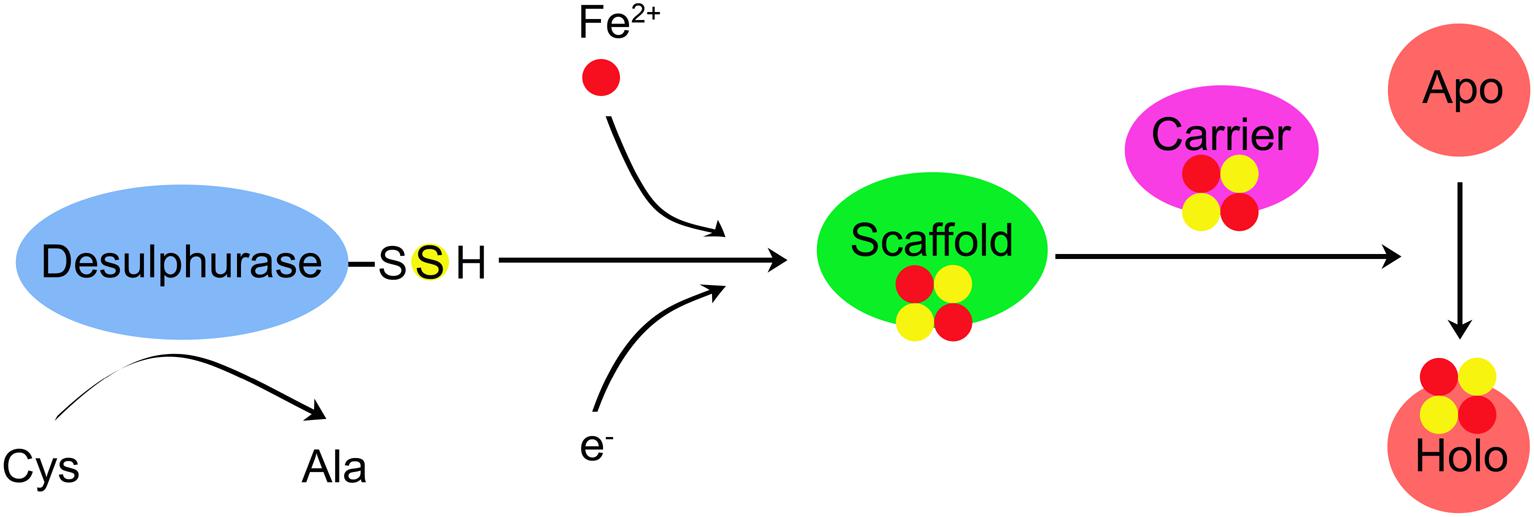
Figure 1. A proposed principle for the Fe–S cluster biogenesis. Three Fe–S cluster systems have been identified in cyanobacteria, including the nitrogen-fixing (NIF), iron–sulfur (Fe–S) cluster (ISC), and S utilization factor (SUF). Three different machines may follow a common biosynthetic rule. The overall biogenesis process can be divided two main steps: (1) de novo assembly of Fe–S cluster on the scaffold protein by recruiting Fe and S; (2) transferring Fe–S cluster from the scaffold protein to target apo-proteins (apo-protein) and then are assembled into the polypeptide chain. Cysteine (Cys) is converted to alanine (Ala) by the Cys desulfurase. Electrons are needed for the reduction of S0 (Cys) to S2– (Fe–S cluster). The source of Fe is not yet known. De novo assembly of Fe–S cluster is performed on the scaffold. The newly assembled Fe–S cluster is transferred to the carrier protein, which delivers the Fe–S cluster to recipient Apo and converts recipient Apo into holo-protein (Holo).
SUF Mechanism
In archaea, the components of SUF system are relatively simple, and its minimal functional core consists only of SufBC (Anbar et al., 2007). During evolution from archaea to bacteria, many components of this system are added, including SufA-SufE and SufS (Zheng et al., 2001; Lee et al., 2004; Outten et al., 2004). In oxygenic photosynthetic organisms, cyanobacteria and higher plants retain the components of SUF system in E. coli and choose this system as their major Fe–S cluster assembly pathways (Outten, 2015). This appears to be an evolutionary choice in response to the rise of oxygen (Boyd et al., 2014).
The SufABCDSE proteins are well characterized in E. coli. SufA is a scaffold protein that can transfer the [2Fe–2S] cluster into Apo (Ollagnier-de-Choudens et al., 2004; Vinella et al., 2009). SufB forms a stable complex with SufC and SufD with a 1:2:1 stoichiometry, and subsequently, the SufBC2D complex functions as a new type of scaffold for the formation of Fe–S clusters (Chahal et al., 2009; Wollers et al., 2010). SufS, a pyridoxal 5′-phosphate-dependent cysteine desulfurase, possesses a low catalytic activity (Mihara et al., 1999, 2000) and can be fully activated upon binding with SufE to form SufSE complex, which can transfer S atoms into SufB (Loiseau et al., 2003; Outten et al., 2003; Layer et al., 2007).
In the cyanobacterial genome, the sufB, sufC, sufD, and sufS (sufBCDS operon) are arranged with the same transcriptional direction; sufA is not included in the sufBCDS operon, and sufR is located at upstream of sufB with an opposite transcriptional direction (Wang et al., 2004; Seki et al., 2006; Shen et al., 2007; Bai et al., 2018). Cyanobacterial SufR can coordinate two [4Fe–4S]2+, 1+ clusters and functions as a transcriptional repressor of the sufBCDS operon and an autoregulator itself (Shen et al., 2007). The dual functions of SufR depend on the redox state of [4Fe–4S]2+, 1+ clusters (Shen et al., 2007). The transcription level of SufR is also regulated by light, oxidative stress, and Fe deficiency (Wang et al., 2004; Seki et al., 2006; Vuorijoki et al., 2017). Specifically, SufR represses the promoter of sufBCDS operon (P1, not P2; two promoters P1 and P2 for sufBCDS operon) under moderate light conditions, and P1 activation results from the derepression by the high light shift (Seki et al., 2006). Under the conditions of oxidative stress and Fe deficiency, expression levels of the sufBCDS genes were elevated in ΔsufR (Wang et al., 2004; Vuorijoki et al., 2017). Therefore, sufR is also a transcriptional repressor of the suf operon under Fe-limiting conditions. Similar to bacterial and plastid SufA, little is known regarding whether cyanobacterial SufA functions as assembly scaffold or carrier with Fe or Fe–S cluster. In the cyanobacterium Synechocystis 6803, in vitro purified SufA appears to only bind Fe (Morimoto et al., 2002). However, the recombinant protein exists as a dimer that can bind a [2Fe–2S] cluster and then transfer into Apo of [2Fe–2S] and [4Fe–4S] clusters (Wollenberg et al., 2003). As a consequence, deletion of sufA exhibited a chlorosis compared with the wild type under Fe-deficient conditions, regardless of a similar growth phenotype under standard growth conditions (Balasubramanian et al., 2006). Similarly, in vitro purified plastid SufA can bind a [2Fe–2S] cluster (Abdel-Ghany et al., 2005; Yabe and Nakai, 2006) and transfer the Fe–S cluster into apo-ferredoxin (apo-Fdx) (Abdel-Ghany et al., 2005). However, the phenotype of mutant was the same with wild type even under Fe-deficient conditions in Arabidopsis (Yabe and Nakai, 2006). Collectively, it suggested that SufA may be an Fe–S cluster carrier protein and not assembly scaffold in oxygenic photosynthetic organisms. In cyanobacteria, SufBC2D is proposed to be a major scaffold complex of Fe–S cluster assembly, although the experimental evidence is absent.
It was previously reported that knockout of each of sufBCDS and sufE genes was lethal in cyanobacteria (Tirupati et al., 2004; Balasubramanian et al., 2006; Zang et al., 2017) and in higher plants (Xu and Møller, 2006; Hu et al., 2017). This indicates that the SUF system is essential to carry out oxygenic photosynthesis. In cyanobacteria and higher plants, the SUF system was reported to be involved in the biogenesis of the Fe–S clusters for photosystem I (PSI) (Wang et al., 2004; Shen and Golbeck, 2006). In chloroplasts, High Chlorophyll Fluorescence 101 (HCF101) was reported to function as a scaffold protein for assembly of the [4Fe–4S] cluster (Schwenkert et al., 2010). In the Δhcf101 mutant, the levels of [4Fe–4S] proteins of PSI were severely reduced in chloroplasts (Lezhneva et al., 2004; Stöckel and Oelmüller, 2004), suggesting that HCF101 may be required for biosynthesis of Fe–S clusters in PSI. Slr0067, a counterpart of HCF101 in Synechocystis 6803 (Lezhneva et al., 2004), and interacts with NdhI, a subunit of NDH-1 complex, as deduced from the results of yeast two-hybrid assay (Dai et al., 2013). NdhI contains two [4Fe–4S] clusters (Laughlin et al., 2019; Schuller et al., 2019). Thus, Slr0067 may be involved in formation of [4Fe–4S] clusters of NDH-1 in cyanobacteria. Furthermore, NDH-1 interacts PSI to form a supercomplex NDH-1-PSI (Peng et al., 2008; Gao et al., 2016), but the interrelationship between Slr0067/HCF101 and the supercomplex needs to be further investigated.
HCF101/Slr0067 is a conserved protein and also exists in non-photosynthetic organisms. The counterpart in Salmonella enterica is ApbC that is required for the maturation of the Fe–S clusters proteins in thiamine biosynthetic pathway (Skovran and Downs, 2003; Boyd et al., 2008, 2009). In addition, HCF101/Slr0067 with homology to NBP35 is a P-loop NTPase in cytosolic Fe–S cluster protein assembly (CIA) machinery (Bych et al., 2008; Balk and Schaedler, 2014). NBP35 can interact with Cfd1 (cytosolic Fe–S cluster deficient) to form a heterotetrameric complex as a scaffold in Fe–S cluster protein maturation in yeast and mammals (Netz et al., 2007, 2012; Balk and Schaedler, 2014). However, Cdf1, a counterpart of NBP35, lacks the N-terminal Fe–S cluster-binding domain (Roy et al., 2003; Hausmann et al., 2005) and is also not identified in green lineage (Bych et al., 2008; Kohbushi et al., 2009). Collectively, NBP35 is considered to function as a homodimer and can assemble [2Fe–2S] and [4Fe–4S] clusters on C- and N-terminal domains, respectively, in green lineage (Bych et al., 2008; Kohbushi et al., 2009).
Based on the above analyses, a model of cyanobacterial SUF system for Fe–S cluster synthesis is schematically represented in Figure 2. Fe–S cluster biogenesis is initiated by SufS, which converts L-cysteine (Cys) to L-alanine (Ala). Sulfane (S0) is transferred from SufS to SufE (S transferase) and then to SufB of SufBC2D scaffold complex and bound as a persulfide (S2–). Putative Fe and electron (for reduction of S0 to S2–) donors are still unknown. SufC has an ATPase activity, thus coupling ATP hydrolysis with the formation of Fe–S clusters. Subsequently, the newly assembled Fe–S cluster is transferred to the carrier protein, which delivers the Fe–S cluster to Apo to form holo-protein (Holo).
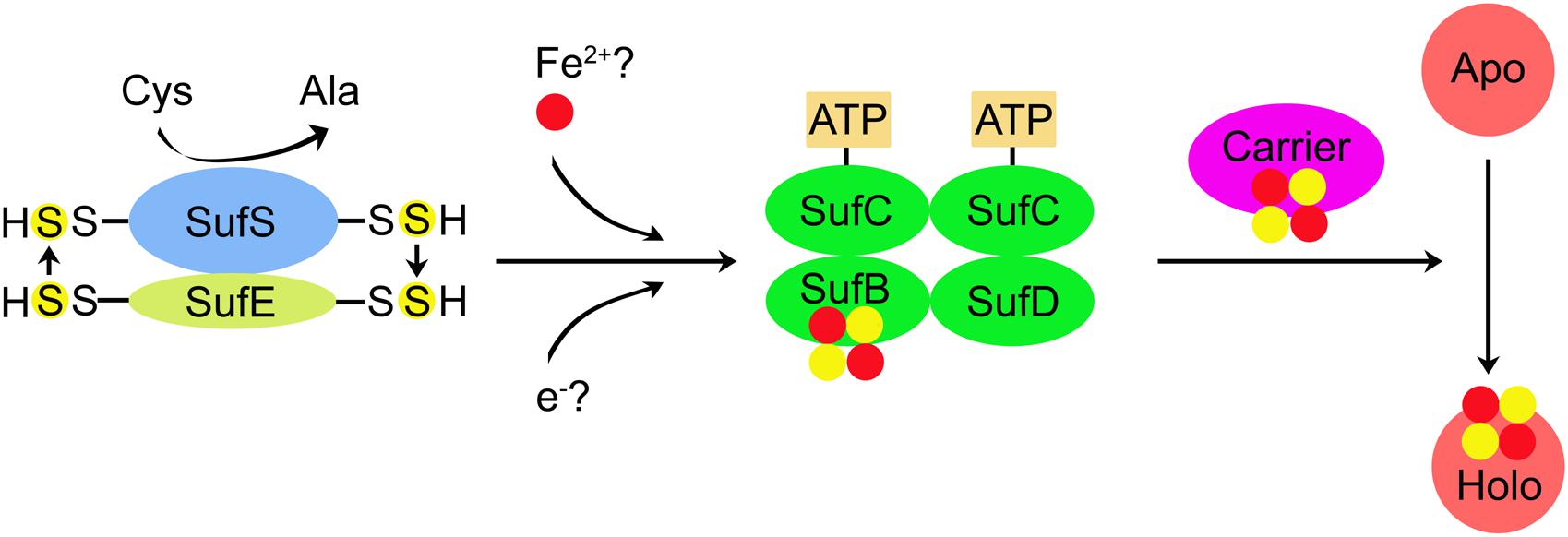
Figure 2. A proposed model for the assembly of Fe–S clusters by SUF system in cyanobacteria. Fe–S cluster biogenesis is initiated by SufS (cysteine desulfurase), which converts cysteine (Cys) to alanine (Ala). Sulfane (S0) is transferred from SufS to SufE (sulfur transferase) and then to SufB of SufBC2D scaffold complex and bound as a persulfide (S2–). Putative Fe and electron (for reduction S0 to S2–) donors are still unknown. SufC has an ATPase activity, thus coupling ATP hydrolysis with the formation of Fe–S clusters. Subsequently, the newly assembled Fe–S cluster is transferred to the carrier protein, which delivers the Fe–S cluster to apo-protein (Apo) and further converts Apo to holo-protein (Holo). SufA and Slr0067 (Synechocystis 6803) may function as the carrier proteins.
ISC Mechanism
Cyanobacterial genome contains almost all homologs of ISC system from E. coli (Table 2), although this system is less important in cyanobacteria. The ISC assembly system encoded by iscR-iscSUA-hscBA-fdx has been well studied in E. coli. Among them, IscR suppresses the expression of gene cluster isc (Fleischhacker et al., 2012) as a global regulator for Fe–S cluster biogenesis (Schwartz et al., 2001). IscS is pyridoxal 5′-phosphate–dependent cysteine desulfurase (Flint, 1996), and it is also a major cysteine desulfurase that can catalyze the reaction of L-cysteine to L-alanine and lead to release the S element required for Fe–S cluster formation (Schwartz et al., 2000). Two cysteine desulfurases (IscS1 and IscS2) were previously identified in cyanobacteria, but their absence did not affect the growth of cells under normal growth conditions (Seidler et al., 2001; Behshad et al., 2004; Tirupati et al., 2004). Although IscS1 and IscS2 were absent, SufS may supply S for the ISC system. SufS is essential for the growth and thus plays a dominant role in cysteine desulfurization for Fe–S cluster biogenesis in cyanobacteria. In contrast, the function of IscS1 and IscS2 on cysteine desulfurization is relatively minor. Consequently, deletion of iscS1 and iscS2 did not affect the growth of cyanobacterial cells. Two heat shock cognate proteins, HscB and HscA, specifically interact with IscU (Silberg et al., 2004; Tapley and Vickery, 2004) and promote an ATP-dependent reaction that the assembled Fe–S clusters are transferred from IscU into Apo (Chandramouli and Johnson, 2006; Bonomi et al., 2008; Alderson et al., 2014). It is worthy of note that a typical IscU is missing in some non–nitrogen-fixation cyanobacteria, for example, Synechocystis 6803 (Kaneko et al., 1996; Seidler et al., 2001). This may be that IscU mainly functions for the assembly of Fe–S clusters proteins related to nitrogenase in NIF organisms.
IscA is a scaffold for Fe–S cluster assembly (Ding and Clark, 2004) that can transfer [2Fe–2S] cluster into Apo (Ollagnier-de-Choudens et al., 2004). Ferredoxin may provide electrons for the Fe–S cluster assembly (Chandramouli et al., 2007; Shi et al., 2012). In Synechocystis 6803, IscA can also bind a [2Fe–2S] cluster, but the presence of IaiH (IscA-interacting Heat-repeats–containing protein) is required for their stable binding (Morimoto et al., 2003). Only three cysteine residues are conserved in IscA (Morimoto et al., 2002), and IaiH may be required to provide another cysteine to further stabilize the [2Fe–2S] cluster. Although IscA is able to bind [2Fe–2S] cluster in vitro in the absence of IaiH (Morimoto et al., 2002, 2003), it was shown that nearly all cellular IscA and IaiH exist as a complex (Morimoto et al., 2003). This suggests that IscA interacts with IaiH to form a complex that may perform physiological functions in vivo. The functions of other members of cyanobacterial ISC system need to be further investigated in the future.
NIF Mechanism
In the cyanobacterial NIF system, only one scaffold protein NfuA is involved in Fe–S cluster assembly. Nfus are U-type proteins and contain a typical Nfu domain that shares a high sequence identity with the C-terminal domain of NifU (Angelini et al., 2008). The binding forms of NfuA with Fe–S cluster in cyanobacteria are different. In the cyanobacterium Synechocystis 6803, in vitro purified NfuA can transfer a labile [2Fe–2S] cluster into apo-Fdx (Nishio and Nakai, 2000). By contrast, in the cyanobacterium Synechococcus species PCC 7002, NufA can transfer the [4Fe–4S] cluster into PsaC, a subunit of PSI complex, via their interaction (Jin et al., 2008). Furthermore, complete segregation of ΔnfuA mutant was not obtained, indicating that NfuA is indispensable for cell growth and supporting that NfuA functions as the scaffold protein in the NIF system (Seidler et al., 2001; Balasubramanian et al., 2006).
In order to perform the Fe–S cluster assembly of nitrogenase in A. vinelandii, a series of genes (nifUSVWZM) are necessary. They gradually lose the function of biological nitrogen fixation in cyanobacteria, possibly because of the purpose of carrying out photosynthesis. As a consequence, their encoding products retain only one scaffold protein to involve in Fe–S cluster assembly. Higher plants have completely lost the NIF mechanism during evolution.
Iron Homeostasis
Iron and S meet at the scaffold protein, leading to the biosynthesis of Fe–S clusters. Release of an excessive free Fe damages cyanobacterial cells, regardless of the fact that Fe is important for Fe–S cluster synthesis. As a consequence, it is very important to maintain Fe homeostasis in cyanobacterial cells. It has been proposed that Fe donor or carrier and Fe storage proteins play an important role in Fe homeostasis.
Iron Donor
It is well known that S for the Fe–S cluster assembly comes from L-cysteine catalyzed by desulfurase SufS or IscS. However, Fe donor remains elusive. Frataxin is an important mitochondrial protein and its decrease causes Friedreich’s ataxia (FRDA), a lethal neurodegenerative disease (Campuzano et al., 1996). This protein has been proposed as a possible Fe donor for the Fe–S cluster biogenesis (Yoon and Cowan, 2003; Layer et al., 2006). Frataxin was further found to interact with the S donor IscS and the scaffold protein IscU for Fe–S cluster biogenesis (Layer et al., 2006; Adinolfi et al., 2009; Shi et al., 2010). BLAST searches unveiled that frataxin is highly conserved from bacteria to human (Babcock et al., 1997) but is absent in the genome of cyanobacteria1. Based on previous studies, we speculate that there are several reasons for the absence of frataxin in cyanobacteria: (1) regardless of a phylogenetic co-occurrence of frataxin with proteins of the Isc operon in cells (Huynen et al., 2001), ISC system is not a main Fe–S assembly machine in cyanobacteria; (2) frataxin and its homologs have a weak Fe-binding activity (Ding et al., 2007; Lu et al., 2010; Stemmler et al., 2010), inconsistent with the high-Fe demand in cyanobacteria. To cope with the high-Fe demand, it is logical to hypothesize that cyanobacteria lose frataxin with low-Fe affinity. During evolution, it appears plausible that cyanobacteria might have chosen a protein with high-Fe affinity as their Fe donor, although we do not know who this protein is.
Alternative Fe donor proteins are suggested to be IscA and SufA because they have a high affinity for Fe-binding activity in E. coli and cyanobacteria (Wollenberg et al., 2003; Ding et al., 2004; Lu et al., 2008; Landry et al., 2013). Unfortunately, these studies are carried out in vitro, and Fe donors proposed have not been shown to interact with cysteine desulfurases or scaffold proteins (Py and Barras, 2010).
Moreover, a phenotype analysis under standard growth conditions has failed to provide any strong evidence that supports a role for IscA/SufA in cellular Fe homeostasis (Seidler et al., 2001; Djaman et al., 2004; Balasubramanian et al., 2006). Therefore, IscA/SufA may only be used for transferring Fe or Fe–S cluster into Apo as carrier protein. However, there is a notable and interesting question that there are subtle regulatory mechanism defects in IscA/SufA. Absence of IscA will result in mistakenly sensing Fe limitation in cyanobacterial cells as deduced from the increased Fe stress-induced protein A (IsiA) protein, regardless of the fact that cells are under the Fe-sufficient conditions (Balasubramanian et al., 2006). IsiA is chlorophyll a-binding protein that forms around PSI under Fe limitation and thus is usually selected as a marker for Fe deficiency in cyanobacteria (Melkozernov et al., 2003; Ryan-Keogh et al., 2012). Nevertheless, the inappropriate Fe limitation response in ΔiscA is ameliorated by additionally inactivating the suf gene (Balasubramanian et al., 2006). Thus, IscA plays an important role in sensing to Fe levels in cyanobacterial cells.
Iron Storage Protein
Iron storage proteins are considered to be important ways for regulating Fe homeostasis in cyanobacteria. Two types of Fe storage proteins are present in cyanobacteria: bacterioferritin (BFR) and DNA-binding proteins from starved cells (DPS) (Keren et al., 2004; Castruita et al., 2006; Shcolnick et al., 2007). These storage proteins are involved in the storage, release, and transfer of Fe. As a consequence, they play an important role in Fe homeostasis.
In cyanobacteria, multiple bfr genes are present in genome (Keren et al., 2004). Bfr proteins have heme or di-Fe binding site in response to different physiological functions. In E. coli, it has been reported that hemeless Bfr accumulates four times more Fe than a Bfr that binds heme, in vitro (Andrews et al., 1995). This suggests that while the di-Fe center is needed for Fe acquisition, the heme may be needed for Fe extraction from the Bfr structure. Bfrs store Fe in a cavity at the center of their 24-mer ultrastructure. Iron enters the Bfr complex as Fe2+ and is oxidized on its way to the central cavity (Carrondo, 2003; Lewin et al., 2005). In cyanobacterium Synechocystis 6803, there are two bfr genes, bfrA and bfrB. Targeted mutagenesis of each of them resulted in poor growth under Fe-deprived conditions (Keren et al., 2004), however, inactivation of both genes did not cause a more severe phenotype (Keren et al., 2004). This result suggests the possible presence of a heteromultimeric structure of cyanobacterial BFR, in which one subunit ligates a di-Fe center, whereas the other accommodates heme binding.
DNA-binding proteins from starved cells proteins are a subgroup of the ferritin family that lack the fifth helix found in other ferritins (Andrews et al., 2003). During evolution, DPS divided into different functions. It functions as Fe storage proteins, DNA-binding proteins protecting against oxidative stress, cold shock proteins, neutrophile activators, and pili components (Andrews et al., 2003). In cyanobacterium Synechococcus species PCC 7942, DpsA binds a heme (Peña and Bullerjahn, 1995), and inactivation of DpsA results in slow growth rates on the Fe-depleted media (Sen et al., 2000). However, a Dps family protein MrgA in cyanobacterium Synechocystis 6803 cells appears to have a specific role in intracellular Fe trafficking, rather than in Fe storage (Shcolnick et al., 2007). MrgA can catalyze similar reactions as BFR, oxidizing Fe2+ to Fe3+ using hydrogen peroxide (H2O2) (Lewin et al., 2005). However, MrgA may be located downstream of BFR and may not affect the total Fe storage. It coordinates the dynamic balance of Fe in vivo mainly through BFR (Li et al., 2004; Shcolnick et al., 2007, 2009). Therefore, Fe storage proteins are an important strategy for cyanobacteria to regulate Fe balance and protect cells.
Differentiation of Fe–S Cluster Pathways Between Cyanobacteria and Bacteria
Although cyanobacteria inherit the biosynthetic pathways of Fe–S clusters, changes have taken place in the process of using these pathways to synthesize Fe–S clusters. Cyanobacteria choose SUF mechanism, which has higher tolerance to oxidative stress in bacteria as the main Fe–S cluster assembly pathway, supplemented by ISC and NIF mechanisms.
In bacteria, ISC is the housekeeping Fe–S cluster assembly system (Lill, 2009; Ding, 2016), whereas SUF is induced when bacteria encounter Fe-limited or oxidative stress (Outten et al., 2004; Outten, 2015). However, cyanobacteria adopt a different Fe–S cluster assembly strategy from bacteria. Sulfur utilization factor is a dominating Fe–S cluster assembly mechanism, whereas ISC mechanism is auxiliary in cyanobacteria. It is possible that the Fe–S cluster synthesis system in cyanobacteria is distinct from other prokaryotes for several reasons: (1) cyanobacteria are prokaryotes with photosynthetic characteristics, in which abundant Fe–S cluster proteins participate in photosynthetic electron transport in thylakoid membrane (Table 1). For example, consistent with Arabidopsis thaliana, SufA in the cyanobacterium Synechocystis 6803 contains five Cys residues, however, IscA contains only three Cys residues in non-photosynthetic organisms (Balasubramanian et al., 2006). Because SUF system may be involved in Fe–S cluster assembly of PSI (Yu et al., 2003; Wang et al., 2004), the components specific to the assembly of the Fe–S clusters in photosynthetic complexes were formed during evolution. (2) The reduced bioavailability of Fe and S by oxygenic photosynthesis drives the production of additional components of SUF system in response to the oxidative stress. Under conditions of anaerobic or very low concentration of oxygen, the core SufBC scaffold complex is sufficient to assemble Fe–S clusters protein because of presence of the majority of soluble Fe2+ and S2– (Boyd et al., 2014). With the increase in oxygen levels, SufD, SufS, and SufE are added into the SUF system in order to adapt an environment of decreased bioavailability of Fe and S (Boyd et al., 2014). Undoubtedly, cyanobacteria choose the SUF system as a dominant Fe–S cluster biosynthetic mechanism. (3) Reactive oxygen species produced by oxygenic metabolism from photosynthetic electron transport and other oxygenic metabolism pathways will damage the Fe–S clusters in proteins. Excessive electron accumulation in photosystem II (PSII) and PSI, especially under high light stress conditions, will combine with oxygen to produce ROS directly damaging Fe–S clusters. Sulfur utilization factor system is activated by high light and promotes Fe–S cluster biogenesis to compensate for the high light stress (Seki et al., 2006). Furthermore, free Fe could produce more deleterious ROS through a Fenton reaction to damage cyanobacterial cells. Collectively, in order to cope with the side effects of photosynthesis, cyanobacteria primarily select the SUF system to assemble Fe–S clusters and optimize this system to adapt to their inhabit environment.
Perspectives
Fe–S cluster proteins are essential for many biological processes. During evolution, three assembly pathways for Fe–S clusters, SUF, ISC, and NIF, are formed in cyanobacteria. Over several decades, despite many progresses in biosynthesis of Fe–S clusters, thorough basis structure, detailed biochemical characteristics, and functional molecular mechanism are yet unknown. Some key components specific to the Fe and electron donors of SUF machinery for Fe–S cluster biogenesis need to be further characterized. Additionally, cyanobacteria inherited an SUF system from bacteria, but this system in cyanobacteria has a higher tolerance to oxidative stress in comparison with that in bacteria because of high oxidative stress raised by oxygenic photosynthesis. However, the functional mechanism is not yet uncovered. It has been proposed that the SUF system may be associated with the biosynthesis of Fe–S clusters in photosynthetic membrane protein complexes, including PSI and NDH-1 (Lezhneva et al., 2004; Stöckel and Oelmüller, 2004; Dai et al., 2013). With the exception of Slr0067, however, no other Fe–S assembly proteins of the SUF system have been identified to interact with the photosynthetic membrane protein complexes in cyanobacteria.
The functional roles of many components of Fe–S cluster assembly systems identified in cyanobacteria were proposed based on their counterparts in bacteria and higher plants. To unravel the specific roles of these components and the regulatory network of Fe–S cluster assembly and transfer pathways, further studies are required in cyanobacteria in the future.
Author Contributions
FG wrote the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant number 31700205).
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Abdel-Ghany, S. E., Ye, H., Garifullina, G. F., Zhang, L., Pilon-Smits, E. A., and Pilon, M. (2005). Iron-sulfur cluster biogenesis in chloroplasts. Involvement of the scaffold protein CpIscA. Plant Physiol. 138, 161–172. doi: 10.1104/pp.104.058602
Adinolfi, S., Iannuzzi, C., Prischi, F., Pastore, C., Iametti, S., Martin, S. R., et al. (2009). Bacterial frataxin CyaY is the gatekeeper of iron-sulfur cluster formation catalyzed by IscS. Nat. Struct. Mol. Biol. 16, 390–396. doi: 10.1038/nsmb.1579
Albrecht, A. G., Netz, D. J., Miethke, M., Pierik, A. J., Burghaus, O., Peuckert, F., et al. (2010). SufU is an essential iron-sulfur cluster scaffold protein in Bacillus subtilis. J. Bacteriol. 192, 1643–1651. doi: 10.1128/JB.01536-09
Alderson, T. R., Kim, J. H., Cai, K., Frederick, R. O., Tonelli, M., and Markley, J. L. (2014). The specialized Hsp70 (HscA) interdomain linker binds to its nucleotide-binding domain and stimulates ATP hydrolysis in both cis and trans configurations. Biochemistry 53, 7148–7159. doi: 10.1021/bi5010552
Anbar, A. D., Duan, Y., Lyons, T. W., Arnold, G. L., Kendall, B., Creaser, R. A., et al. (2007). A whiff of oxygen before the great oxidation event? Science 317, 1903–1906. doi: 10.1126/science.1140325
Andrews, S. C., Le Brun, N. E., Barynin, V., Thomson, A. J., Moore, G. R., Guest, J. R., et al. (1995). Site-directed replacement of the coaxial heme ligands of bacterioferritin generates heme-free variants. J. Biol. Chem. 270, 23268–23274. doi: 10.1074/jbc.270.40.23268
Andrews, S. C., Robinson, A. K., and Rodríguez-Quiñones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. doi: 10.1016/S0168-6445(03)00055-X
Angelini, S., Gerez, C., Ollagnier-de Choudens, S., Sanakis, Y., Fontecave, M., and Barras, F. (2008). NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J. Biol. Chem. 283, 14084–14091. doi: 10.1074/jbc.M709405200
Ayala-Castro, C., Saini, A., and Outten, F. W. (2008). Fe-S cluster assembly pathways in bacteria. Microbiol. Mol. Biol. Rev. 72, 110–125. doi: 10.1128/MMBR.00034-07
Babcock, M., de Silva, D., Oaks, R., Davis-Kaplan, S., Jiralerspong, S., Montermini, L., et al. (1997). Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 276, 1709–1712. doi: 10.1126/science.276.5319.1709
Bai, Y., Chen, T., Happe, T., Lu, Y., and Sawyer, A. (2018). Iron–sulphur cluster biogenesis via the SUF pathway. Metallomics 10, 1038–1052. doi: 10.1039/c8mt00150b
Balasubramanian, R., Shen, G., Bryant, D. A., and Golbeck, J. H. (2006). Regulatory roles for IscA and SufA in iron homeostasis and redox stress responses in the cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 188, 3182–3191. doi: 10.1128/JB.188.9.3182-3191.2006
Balk, J., and Pilon, M. (2011). Ancient and essential: the assembly of iron–sulfur clusters in plants. Trends Plant Sci. 16, 218–226. doi: 10.1016/j.tplants.2010.12.006
Balk, J., and Schaedler, T. A. (2014). Iron cofactor assembly in plants. Annu. Rev. Plant Biol. 65, 125–153. doi: 10.1146/annurev-arplant-050213-035759
Behshad, E., Parkin, S. E., and Bollinger, J. M. (2004). Mechanism of cysteine desulfurase Slr0387 from Synechocystis sp. PCC 6803: kinetic analysis of cleavage of the persulfide intermediate by chemical reductants. Biochemistry 43, 12220–12226. doi: 10.1021/bi049143e
Beinert, H. (2000). Iron-sulfur proteins: ancient structures, still full of surprises. J. Biol. Inorg. Chem. 5, 2–15. doi: 10.1007/s007750050002
Beinert, H., Holm, R. H., and Munck, E. (1997). Iron-sulfur clusters: nature’smodular, multipurpose structures. Science 277, 653–659. doi: 10.1126/science.277.5326.653
Berkovitch, F., Nicolet, Y., Wan, J. T., Jarrett, J. T., and Drennan, C. L. (2004). Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 303, 76–79. doi: 10.1126/science.1088493
Bonomi, F., Iametti, S., Morleo, A., Ta, D., and Vickery, L. E. (2008). Studies on the mechanism of catalysis of iron-sulfur cluster transfer from IscU [2Fe2S] by HscA/HscB chaperones. Biochemistry 47, 12795–12801. doi: 10.1021/bi801565j
Boyd, E. S., Thomas, K. M., Dai, Y., Boyd, J. M., and Outten, F. W. (2014). Interplay between oxygen and Fe–S cluster biogenesis: insights from the Suf pathway. Biochemistry 53, 5834–5847. doi: 10.1021/bi500488r
Boyd, J. M., Pierik, A. J., Netz, D. J., Lill, R., and Downs, D. M. (2008). Bacterial ApbC can bind and effectively transfer iron-sulfur clusters. Biochemistry 47, 8195–8202. doi: 10.1021/bi800551y
Boyd, J. M., Sondelski, J. L., and Downs, D. M. (2009). Bacterial ApbC protein has two biochemical activities that are required for in vivo function. J. Biol. Chem. 284, 110–118. doi: 10.1074/jbc.M807003200
Brocks, J. J., Logan, G. A., Buick, R., and Summons, R. E. (1999). Archean molecular fossils and the early rise of eukaryotes. Science 285, 1033–1036. doi: 10.1126/science.285.5430.1033
Bych, K., Netz, D. J., Vigani, G., Bill, E., Lill, R., Pierik, A. J., et al. (2008). The essential cytosolic iron-sulfur protein Nbp35 acts without Cfd1 partner in the green lineage. J. Biol. Chem. 283, 35797–35804. doi: 10.1074/jbc.M807303200
Campuzano, V., Montermini, L., Molto, M. D., Pianese, L., Cossee, M., Cavalcanti, F., et al. (1996). Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423–1427. doi: 10.1126/science.271.5254.1423
Carrondo, M. A. (2003). Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBO J. 22, 1959–1968. doi: 10.1093/emboj/cdg215
Cassier-Chauvat, C., and Chauvat, F. (2014). Function and regulation of ferredoxins in the cyanobacterium, Synechocystis PCC6803: recent advances. Life 4, 666–680. doi: 10.3390/life4040666
Castruita, M., Saito, M., Schottel, P. C., Elmegreen, L. A., Myneni, S., Stiefel, E. I., et al. (2006). Overexpression and characterization of an iron storage and DNA-binding Dps protein from Trichodesmium erythraeum. Appl. Environ. Microbiol. 72, 2918–2924. doi: 10.1128/AEM.72.4.2918-2924.2006
Chahal, H. K., Dai, Y., Saini, A., Ayala-Castro, C., and Outten, F. W. (2009). The SufBCD Fe-S scaffold complex interacts with SufA for Fe-S cluster transfer. Biochemistry 48, 10644–10653. doi: 10.1021/bi901518y
Chandramouli, K., and Johnson, M. K. (2006). HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry 45, 11087–11095. doi: 10.1021/bi061237w
Chandramouli, K., Unciuleac, M. C., Naik, S., Dean, D. R., Huynh, B. H., and Johnson, M. K. (2007). Formation and properties of [4Fe–4S] clusters on the IscU scaffold protein. Biochemistry 46, 6804–6811. doi: 10.1021/bi6026659
Chapman, D. J., and Schopf, J. W. (1983). “Biological and biochemical effects of the development of an aerobic environment,” in Earth’s earliest biosphere: Its origin and evolution (A 84-43051 21-51), ed. J. W. Schopf (Princeton, NJ: Princeton University Press), 302–320.
Dai, H., Zhang, L., Zhang, J., Mi, H., Ogawa, T., and Ma, W. (2013). Identification of a cyanobacterial CRR6 protein, Slr1097, required for efficient assembly of NDH-1 complexes in Synechocystis sp. PCC 6803. Plant J. 75, 858–866. doi: 10.1111/tpj.12251
Ding, H. (2016). “Iron homeostasis and iron–sulfur cluster assembly in Escherichia coli,” in Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria, ed. F.J. de Bruijn, (Hoboken, NJ: Wiley), 203–214. doi: 10.1002/9781119004813.ch17
Ding, H., and Clark, R. J. (2004). Characterization of iron binding in IscA, an ancient iron-sulphur cluster assembly protein. Biochem. J. 379, 433–440. doi: 10.1042/BJ20031702
Ding, H., Clark, R. J., and Ding, B. (2004). IscA mediates iron delivery for assembly of iron-sulfur clusters in IscU under the limited accessible free iron conditions. J. Biol. Chem. 279, 37499–37504. doi: 10.1074/jbc.M404533200
Ding, H., Yang, J., Coleman, L. C., and Yeung, S. (2007). Distinct iron binding property of two putative iron donors for the iron-sulfur cluster assembly: IscA and the bacterial frataxin ortholog CyaY under physiological and oxidative stress conditions. J. Biol. Chem. 282, 7997–8004. doi: 10.1074/jbc.M609665200
Djaman, O., Outten, F. W., and Imlay, J. A. (2004). Repair of oxidized iron-sulfur clusters in Escherichia coli. J. Biol. Chem. 279, 44590–44599. doi: 10.1074/jbc.M406487200
Finney, L. A., and O’Halloran, T. V. (2003). Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300, 931–936. doi: 10.1126/science.1085049
Fleischhacker, A. S., Stubna, A., Hsueh, K. L., Guo, Y., Teter, S. J., Rose, J. C., et al. (2012). Characterization of the [2Fe-2S] cluster of Escherichia coli transcription factor IscR. Biochemistry 51, 4453–4462. doi: 10.1021/bi3003204
Flint, D. H. (1996). Escherichia coli contains a protein that is homologous in function and N-terminal sequence to the protein encoded by the nifS gene of Azotobacter vinelandii and that can participate in the synthesis of the Fe-S cluster of dihydroxy-acid dehydratase. J. Biol. Chem. 271, 16068–16074.
Gao, F., Zhao, J., Chen, L., Battchikova, N., Ran, Z., Aro, E. M., et al. (2016). The NDH-1L-PSI supercomplex is important for efficient cyclic electron transport in cyanobacteria. Plant Physiol. 172, 1451–1464. doi: 10.1104/pp.16.00585
Gao, F., Zhao, J., Zhang, N., Jiang, J., Yang, X., Wei, L., et al. (2020). A vast majority of NdhM id redundant under growth temperature but is important under high temperature for accumulation of a large complex of NDH-1 in cyanobacteria. Plant cell Physiol. (in revised)Google Scholar
Hausmann, A., Netz, D. J., Balk, J., Pierik, A. J., Muhlenhoff, U., and Lill, R. (2005). The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery. Proc. Natl. Acad. Sci. U.S.A. 102, 3266–3271. doi: 10.1073/pnas.0406447102
Hu, X., Kato, Y., Sumida, A., Tanaka, A., and Tanaka, R. (2017). The SUFBC2D complex is required for the biogenesis of all major classes of plastid Fe-S proteins. Plant J. 90, 235–248. doi: 10.1111/tpj.13483
Huynen, M. A., Snel, B., Bork, P., and Gibson, T. J. (2001). The phylogenetic distribution of frataxin indicates a role in iron-sulfur cluster protein assembly. Hum. Mol. Genet. 10, 2463–2468. doi: 10.1093/hmg/10.21.2463
Jacobson, M. R., Brigle, K. E., Bennett, L. T., Setterquist, R. A., Wilson, M. S., Cash, V. L., et al. (1989a). Physical and genetic map of the major nif gene cluster from Azotobacter vinelandii. J. Bacteriol. 171, 1017–1027. doi: 10.1128/jb.171.2.1017-1027.1989
Jacobson, M. R., Cash, V. L., Weiss, M. C., Laird, N. F., Newton, W. E., and Dean, D. R. (1989b). Biochemical and genetic analysis of the nifUSVWZM cluster from Azotobacter vinelandii. Mol. Gen. Genet. 219, 49–57. doi: 10.1007/bf00261156
Jin, Z., Heinnickel, M., Krebs, C., Shen, G., Golbeck, J. H., and Bryant, D. A. (2008). Biogenesis of iron-sulfur clusters in photosystem I: holo-NfuA from the cyanobacterium Synechococcus SP. PCC 7002 rapidly and efficiently transfers [4Fe-4S] clusters to apo-PsaC in vitro. J. Biol. Chem. 283, 28426–28435. doi: 10.1074/jbc.M803395200
Johnson, D. C., Dean, D. R., Smith, A. D., and Johnson, M. K. (2005). Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281. doi: 10.1146/annurev.biochem.74.082803.133518
Jordan, P., Fromme, P., Witt, H. T., Klukas, O., Saenger, W., and Krauss, N. (2001). Threedimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411, 909–917. doi: 10.1038/35082000
Kaneko, T., Sato, S., Kotani, H., Tanaka, A., Asamizu, E., Nakamura, Y., et al. (1996). Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803.II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3, 109–136. doi: 10.1093/dnares/3.3.109
Keren, N., Aurora, R., and Pakrasi, H. B. (2004). Critical roles of bacterioferritins in iron storage and proliferation of cyanobacteria. Plant Physiol. 135, 1666–1673. doi: 10.1104/pp.104.042770
Kohbushi, H., Nakai, Y., Kikuchi, S., Yabe, T., Hori, H., and Nakai, M. (2009). Arabidopsis cytosolic Nbp35 homodimer can assemble both [2Fe-2S] and [4Fe–4S] clusters in two distinct domains. Biochem. Biophys. Res. Commun. 378, 810–815. doi: 10.1016/j.bbrc.2008.11.138
Kurisu, G., Zhang, H., Smith, J. L., and Cramer, W. A. (2003). Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science 302, 1009–1014. doi: 10.1126/science.1090165
Landry, A. P., Cheng, Z., and Ding, H. (2013). Iron binding activity is essential for the function of IscA in iron-sulphur cluster biogenesis. Dalton T 42, 3100–3106. doi: 10.1039/c2dt32000b
Latifi, A., Ruiz, M., and Zhang, C. C. (2009). Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 33, 258–278. doi: 10.1111/j.1574-6976.2008.00134.x
Laughlin, T. G., Bayne, A. N., Trempe, J. F., Savage, D. F., and Davies, K. M. (2019). Structure of the complex I-like molecule NDH of oxygenic photosynthesis. Nature 566, 411–414. doi: 10.1038/s41586-019-0921-0
Layer, G., Gaddam, S. A., Ayala-Castro, C. N., Ollagnier-de Choudens, S., Lascoux, D., Fontecave, M., et al. (2007). SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J. Biol. Chem. 282, 13342–13350. doi: 10.1074/jbc.M608555200
Layer, G., Ollagnier-de Choudens, S., Sanakis, Y., and Fontecave, M. (2006). Iron sulfur cluster biosynthesis: characterization of Escherichia coli CyaY as an iron donor for the assembly of [2Fe-2S] clusters in the scaffold IscU. J. Biol. Chem. 281, 16256–16263. doi: 10.1074/jbc.M513569200
Lee, J. H., Yeo, W. S., and Roe, J. H. (2004). Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor. Mol. Microbiol. 51, 1745–1755. doi: 10.1111/j.1365-2958.2003.03946.x
Lewin, A., Moore, G. R., and Le Brun, N. E. (2005). Formation of protein-coated iron minerals. Dalton Trans. 22, 3597–3610. doi: 10.1039/b506071k
Lezhneva, L., Amann, K., and Meurer, J. (2004). The universally conserved HCF101 protein is involved in assembly of [4Fe–4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J. 37, 174–185. doi: 10.1046/j.1365-313X.2003.01952.x
Li, H., Singh, A. K., McIntyre, L. M., and Sherman, L. A. (2004). Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp. strain PCC 6803. J. Bacteriol. 186, 3331–3345. doi: 10.1128/JB.186.11.3331-3345.2004
Lill, R. (2009). Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838. doi: 10.1038/nature08301
Loiseau, L., Ollagnier-de-Choudens, S., Nachin, L., Fontecave, M., and Barras, F. (2003). Biogenesis of Fe-S cluster by the bacterial Suf system: SufS and SufE form a new type of cysteine desulfurase. J. Biol. Chem. 278, 38352–38359. doi: 10.1074/jbc.M305953200
Lu, J., Bitoun, J. P., Tan, G., Wang, W., Min, W., and Ding, H. (2010). Iron-binding activity of human iron–sulfur cluster assembly protein hIscA1. Biochem. J. 428, 125–131. doi: 10.1042/BJ20100122
Lu, J., Yang, J., Tan, G., and Ding, H. (2008). Complementary roles of SufA and IscA in the biogenesis of iron–sulfur clusters in Escherichia coli. Biochem. J. 409, 535–543. doi: 10.1042/BJ20071166
Lu, Y. (2018). Assembly and transfer of Iron–Sulfur clusters in the plastid. Front. Plant Sci. 9:336. doi: 10.3389/fpls.2018.00336
Maio, N., and Rouault, T. A. (2015). Iron–sulfur cluster biogenesis in mammalian cells: new insights into the molecular mechanisms of cluster delivery. Biochim. Biophys. Acta-Mol. Cell. Res. 1853, 1493–1512. doi: 10.1016/j.bbamcr.2014.09.009
Melkozernov, A. N., Bibby, T. S., Lin, S., Barber, J., and Blankenship, R. E. (2003). Time-resolved absorption and emission show that the CP43′ antenna ring of iron-stressed Synechocystis sp. PCC6803 is efficiently coupled to the photosystem I reaction center core. Biochemistry 42, 3893–3903. doi: 10.1021/bi026987u
Mettert, E. L., and Kiley, P. J. (2015). How is Fe-S cluster formation regulated? Annu. Rev. Microbiol. 69, 505–526. doi: 10.1146/annurev-micro-091014-104457
Meyer, J. (2008). Iron-sulfur protein folds, iron-sulfur chemistry, and evolution. J. Biol. Inorg. Chem. 13, 157–170. doi: 10.1007/s00775-007-0318-7
Mihara, H., Kurihara, T., Yoshimura, T., and Esaki, N. (2000). Kinetic and mutational studies of three NifS homologs from Escherichia coli: mechanistic difference between L-cysteine desulfurase and L-selenocysteine lyase reactions. J. Biochem. 127, 559–567. doi: 10.1093/oxfordjournals.jbchem.a022641
Mihara, H., Maeda, M., Fujii, T., Kurihara, T., Hata, Y., and Esaki, N. (1999). A nifS-like gene, csdB, Encodes an Escherichia coli counterpart of mammalian selenocysteine lyase gene cloning, purification, characterization and preliminary x-ray crystallographic studies. J. Biol. Chem. 274, 14768–14772. doi: 10.1074/jbc.274.21.14768
Morimoto, K., Nishio, K., and Nakai, M. (2002). Identification of a novel prokaryotic HEAT-repeats-containing protein which interacts with a cyanobacterial IscA homolog. FEBS Lett. 519, 123–127. doi: 10.1016/s0014-5793(02)02736-9
Morimoto, K., Sato, S., Tabata, S., and Nakai, M. (2003). A HEAT-repeats containing protein, IaiH, stabilizes the iron-sulfur cluster bound to the cyanobacterial IscA homologue, IscA2. J. Biochem. 134, 211–217. doi: 10.1093/jb/mvg131
Netz, D. J., Pierik, A. J., Stümpfig, M., Bill, E., Sharma, A. K., Pallesen, L. J., et al. (2012). A bridging [4Fe–4S] cluster and nucleotide binding are essential for function of the Cfd1-Nbp35 complex as a scaffold in iron-sulfur protein maturation. J. Biol. Chem. 287, 12365–12378. doi: 10.1074/jbc.M111.328914
Netz, D. J., Pierik, A. J., Stümpfig, M., Mühlenhoff, U., and Lill, R. (2007). The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol. Nat. Chem. Biol. 3, 278–286. doi: 10.1038/nchembio872
Nishio, K., and Nakai, M. (2000). Transfer of iron–sulfur cluster from NifU to apoferredoxin. J. Biol. Chem. 275, 22615–22618. doi: 10.1074/jbc.C000279200
Ollagnier-de-Choudens, S., Sanakis, Y., and Fontecave, M. (2004). SufA/IscA: reactivity studies of a class of scaffold proteins involved in [Fe-S] cluster assembly. J. Biol. Inorg. Chem. 9, 828–838. doi: 10.1007/s00775-004-0581-9
Outten, F. W. (2015). Recent advances in the Suf Fe-S cluster biogenesis pathway: beyond the Proteobacteria. Biochim. Biophys. Acta 1853, 1464–1469. doi: 10.1016/j.bbamcr.2014.11.001
Outten, F. W., Djaman, O., and Storz, G. (2004). A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 52, 861–872. doi: 10.1111/j.1365-2958.2004.04025.x
Outten, F. W., Wood, M. J., Munoz, F. M., and Storz, G. (2003). The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J Biol Chem. 278, 45713–45719. doi: 10.1074/jbc.M308004200
Peña, M. M. O., and Bullerjahn, G. S. (1995). The DpsA protein of Synechococcus sp. strain PCC7942 is a DNA-binding hemoprotein linkage of the Dps and bacterioferritin protein families. J. Biol. Chem. 270, 22478–22482. doi: 10.1074/jbc.270.38.22478
Peng, L., Shimizu, H., and Shikanai, T. (2008). The chloroplast NAD(P)H dehydrogenase complex interacts with photosystem I in Arabidopsis. J. Biol. Chem. 283, 34873–34879. doi: 10.1074/jbc.M803207200
Peters, J. W., and Broderick, J. B. (2012). Emerging paradigms for complex iron-sulfur cofactor assembly and insertion. Annu. Rev. Biochem. 81, 429–450. doi: 10.1146/annurev-biochem-052610-094911
Py, B., and Barras, F. (2010). Building Fe–S proteins: bacterial strategies. Nat. Rev. Microbiol. 8, 436–446. doi: 10.1038/nrmicro2356
Rouault, T. A. (2012). Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease. Dis. Model Mech. 5, 155–164. doi: 10.1242/dmm.009019
Roy, A., Solodovnikova, N., Nicholson, T., Antholine, W., and Walden, W. E. (2003). A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J. 22, 4826–4835. doi: 10.1038/sj.emboj.7600518
Ryan-Keogh, T. J., Macey, A. I., Cockshutt, A. M., Moore, C. M., and Bibby, T. S. (2012). The cyanobacterial chlorophyll-binding-protein IsiA acts to increase the vivo effective absorption cross-section of PSI under iron limitation. J. Phycol. 48, 145–154. doi: 10.1111/j.1529-8817.2011.01092.x
Santos, J. A., Alonso-García, N., Macedo-Ribeiro, S., and Pereira, P. J. B. (2014). The unique regulation of iron-sulfur cluster biogenesis in a Gram-positive bacterium. Proc. Natl. Acad. Sci. U.S.A. 111, E2251–E2260. doi: 10.1073/pnas.1322728111
Schuller, J. M., Birrell, J. A., Tanaka, H., Konuma, T., Wulfhorst, H., Cox, N., et al. (2019). Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer. Science 363, 257–260. doi: 10.1126/science.aau3613
Schwartz, C. J., Djaman, O., Imlay, J. A., and Kiley, P. J. (2000). The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 97, 9009–9014. doi: 10.1073/pnas.160261497
Schwartz, C. J., Giel, J. L., Patschkowski, T., Luther, C., Ruzicka, F. J., Beinert, H., et al. (2001). IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 14895–14900. doi: 10.1073/pnas.251550898
Schwenkert, S., Netz, D. J., Frazzon, J., Pierik, A. J., Bill, E., Gross, J., et al. (2010). Chloroplast HCF101 is a scaffold protein for [4Fe–4S] cluster assembly. Biochem. J. 425, 207–214. doi: 10.1042/BJ20091290
Seidler, A., Jaschkowitz, K., and Wollenberg, M. (2001). Incorporation of iron–sulphur clusters in membrane-bound proteins. Biochem. Soc. Trans. 29, 418–421. doi: 10.1042/bst0290418
Seki, A., Nakano, T., Takahashi, H., Matsumoto, K., Ikeuchi, M., and Tanaka, K. (2006). Light-responsive transcriptional regulation of the suf promoters involved in cyanobacterium Synechocystis sp. PCC 6803 Fe-S cluster biogenesis. FEBS Lett. 580, 5044–5048. doi: 10.1016/j.febslet.2006.08.027
Selbach, B. P., Chung, A. H., Scott, A. D., George, S. J., Cramer, S. P., and Dos Santos, P. C. (2014). Fe–S cluster biogenesis in Gram-positive bacteria: SufU is a zinc-dependent sulfur transfer protein. Biochemistry 53, 152–160. doi: 10.1021/bi4011978
Sen, A., Dwivedi, K., Rice, K. A., and Bullerjahn, G. S. (2000). Growth phase and metal-dependent regulation of the dpsA gene in Synechococcus sp. strain PCC 7942. Arch. Microbiol. 173, 352–357. doi: 10.1007/s002030000153
Shcolnick, S., Shaked, Y., and Keren, N. (2007). A role for mrgA, a DPS family protein, in the internal transport of Fe in the cyanobacterium Synechocystis sp. PCC6803. Biochim. Biophys. Acta 1767, 814–819. doi: 10.1016/j.bbabio.2006.11.015
Shcolnick, S., Summerfield, T. C., Reytman, L., Sherman, L. A., and Keren, N. (2009). The mechanism of iron homeostasis in the unicellular cyanobacterium Synechocystis sp. PCC 6803 and its relationship to oxidative stress. Plant Physiol. 150, 2045–2056. doi: 10.1104/pp.109.141853
Shen, G., Balasubramanian, R., Wang, T., Wu, Y., Hoffart, L. M., Krebs, C., et al. (2007). SufR coordinates two [4Fe–4S]2+, 1+ clusters and functions as a transcriptional repressor of the sufBCDS operon and an autoregulator of sufR in cyanobacteria. J. Biol. Chem. 282, 31909–31919. doi: 10.1074/jbc.M705554200
Shen, G., and Golbeck, J. H. (2006). Assembly of the bound iron–sulfur clusters in photosystem I. Photosystem I. Dordrecht: Springer, 529–548. doi: 10.1007/978-1-4020-4256-0_31
Shi, R., Proteau, A., Villarroya, M., Moukadiri, I., Zhang, L., Trempe, J. F., et al. (2010). Structural basis for Fe–S cluster assembly and tRNA thiolation mediated by IscS protein–protein interactions. PLoS Biol. 8:e1000354. doi: 10.1371/journal.pbio.1000354
Shi, Y., Ghosh, M., Kovtunovych, G., Crooks, D. R., and Rouault, T. A. (2012). Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis. Biochim. Biophys. Acta 1823, 484–492. doi: 10.1016/j.bbamcr.2011.11.002
Silberg, J. J., Tapley, T. L., Hoff, K. G., and Vickery, L. E. (2004). Regulation of the HscA ATPase reaction cycle by the co-chaperone HscB and the iron sulfur cluster assembly protein IscU. J. Biol. Chem. 279, 53924–53931. doi: 10.1074/jbc.M410117200
Skovran, E., and Downs, D. M. (2003). Lack of the ApbC or ApbE protein results in a defect in [Fe-S] cluster metabolism in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185, 98–106. doi: 10.1128/jb.185.1.98-106.2003
Stemmler, T. L., Lesuisse, E., Pain, D., and Dancis, A. (2010). Frataxin and mitochondrial FeS cluster biogenesis. J. Biol. Chem. 285, 26737–26743. doi: 10.1074/jbc.R110.118679
Stöckel, J., and Oelmüller, R. (2004). A novel protein for Photosystem I biogenesis. J. Biol. Chem. 279, 10243–10251. doi: 10.1074/jbc.M309246200
Sutton, V. R., Stubna, A., Patschkowski, T., Münck, E., Beinert, H., and Kiley, P. J. (2004). Superoxide destroys the [2Fe-2S]2+ cluster of FNR from Escherichia coli. Biochemistry 43, 791–798. doi: 10.1021/bi0357053
Takahashi, Y., Mitsui, A., Hase, T., and Matsubara, H. (1986). Formation of the iron–sulfur cluster of ferredoxin in isolated chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 83, 2434–2437. doi: 10.1073/pnas.83.8.2434
Takahashi, Y., Mitsui, A., and Matsubara, H. (1991). Formation of the Fe–S cluster of ferredoxin in lysed spinach chloroplasts. Plant Physiol. 95, 97–103. doi: 10.1104/pp.95.1.97
Takahashi, Y., and Tokumoto, U. (2002). A third bacterial system for the assembly of iron–sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 277, 28380–28383. doi: 10.1074/jbc.C200365200
Tapley, T. L., and Vickery, L. E. (2004). Preferential substrate binding orientation by the molecular chaperone HscA. J. Biol. Chem. 279, 28435–28442. doi: 10.1074/jbc.M400803200
Tirupati, B., Vey, J. L., Drennan, C. L., and Bollinger, J. M. (2004). Kinetic and structural characterization of Slr0077/SufS, the essential cysteine desulfurase from Synechocystis sp. PCC 6803 Biochemistry 43, 12210–12219. doi: 10.1021/bi0491447
Vinella, D., Brochier-Armanet, C., Loiseau, L., Talla, E., and Barras, F. (2009). Iron-sulfur (Fe/S) protein biogenesis: phylogenomic and genetic studies of A-type carriers. PLoS Genet. 5:e1000497. doi: 10.1371/journal.pgen.1000497
Vuorijoki, L., Tiwari, A., Kallio, P., and Aro, E. M. (2017). Inactivation of iron–sulfur cluster biogenesis regulator SufR in Synechocystis sp. PCC 6803 induces unique iron-dependent proteinlevel responses. Biochim. Biophys. Acta 1861, 1085–1098. doi: 10.1016/j.bbagen.2017.02.020
Wächtershäuser, G. (1992). Groundworks for an evolutionary biochemistry: the iron-sulphur world. Prog. Biophys. Mol. Biol. 58, 85–201. doi: 10.1016/0079-61079290022-x
Wallace, M. A., Liou, L. L., Martins, J., Clement, M. H., Bailey, S., Longo, V. D., et al. (2004). Superoxide inhibits 4Fe–4S cluster enzymes involved in amino acid biosynthesis cross-compartment protection by CuZn-superoxide dismutase. J. Biol. Chem. 279, 32055–32062. doi: 10.1074/jbc.M403590200
Wang, T., Shen, G., Balasubramanian, R., McIntosh, L., Bryant, D. A., and Golbeck, J. H. (2004). The sufR gene (sll0088 in Synechocystis sp. strain PCC 6803) functions as a repressor of the sufBCDS operon in iron–sulfur cluster biogenesis in cyanobacteria. J. Bacteriol. 186, 956–967. doi: 10.1128/jb.186.4.956-967.2004
Wollenberg, M., Berndt, C., Bill, E., Schwenn, J. D., and Seidler, A. (2003). A dimer of the FeS cluster biosynthesis protein IscA from cyanobacteria binds a [2Fe–2S] cluster between two protomers and transfers it to [2Fe–2S] and [4Fe–4S] apo proteins. Eur. J. Biochem. 270, 1662–1671. doi: 10.1014/j.1432-1033.2003.03522.x
Wollers, S., Layer, G., Garcia-Serres, R., Signor, L., Clemancey, M., Latour, J. M., et al. (2010). Iron-sulfur (Fe-S) cluster assembly: the SufBCD complex is a new type of Fe-S scaffold with a flavin redox cofactor. J. Biol. Chem. 285, 23331–23341. doi: 10.1074/jbc.M110.127449
Xu, X. M., and Møller, S. G. (2006). AtSufE is an essential activator of plastidic and mitochondrial desulfurases in Arabidopsis. EMBO J. 25, 900–909. doi: 10.1038/sj.emboj.7600968
Yabe, T., and Nakai, M. (2006). Arabidopsis AtIscA-I is affected by deficiency of Fe-S cluster biosynthetic scaffold AtCnfU-V. Biochem. Biophs. Res. Commun. 340, 1047–1052. doi: 10.1016/j.bbrc.2005.12.104
Yoon, T., and Cowan, J. A. (2003). Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J. Am. Chem. Soc. 125, 6078–6084. doi: 10.1021/ja027967i
Yu, J., Shen, G., Wang, T., Bryant, D. A., Golbeck, J. H., and McIntosh, L. (2003). Suppressor mutations in the study of photosystem I biogenesis: sll0088 is a previously unidentified gene involved in reaction center accumulation in Synechocystis sp. strain PCC 6803. J. Bacteriol. 185, 3878–3887. doi: 10.1128/JB.185.13.3878-3887.2003
Zang, S. S., Jiang, H. B., Song, W. Y., Chen, M., and Qiu, B. S. (2017). Characterization of the sulfur-formation (suf) genes in Synechocystis sp. PCC 6803 under photoautotrophic and heterotrophic growth conditions. Planta 246, 927–938. doi: 10.1007/s00425-017-2738-0
Zheng, L., Cash, V. L., Flint, D. H., and Dean, D. R. (1998). Assembly of iron–sulfur clusters: identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273, 13264–13272. doi: 10.1074/jbc.273.21.13264
Keywords: Fe–S clusters, SUF mechanism, ISC mechanism, iron homeostasis, cyanobacteria
Citation: Gao F (2020) Iron–Sulfur Cluster Biogenesis and Iron Homeostasis in Cyanobacteria. Front. Microbiol. 11:165. doi: 10.3389/fmicb.2020.00165
Received: 26 October 2019; Accepted: 23 January 2020;
Published: 28 February 2020.
Edited by:
Patricia Coutinho Dos Santos, Wake Forest University, United StatesReviewed by:
Franklin Wayne Outten, University of South Carolina, United StatesHuangen Ding, Louisiana State University, United States
Copyright © 2020 Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fudan Gao, Z2FvZmRAc2hudS5lZHUuY24=
 Fudan Gao
Fudan Gao