- 1School of Biological Sciences, University of Southampton, Southampton, United Kingdom
- 2National Infection Service, Public Health England, Salisbury, United Kingdom
In its native environment of rotting vegetation, the soil nematode Caenorhabditis elegans encounters a range of bacteria. This includes species from the ESKAPE group of pathogens that pose a clinical problem in acquired hospital infections. Here, we investigated three Gram-negative members of the ESKAPE group, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Acinetobacter baumannii. Pathogenicity profiles as measured by time to kill adult C. elegans showed that P. aeruginosa was the most pathogenic, followed by K. pneumoniae, while C. elegans cultured on A. baumannii exhibited the same survival as those on the standard laboratory food source for C. elegans, Escherichia coli OP50. The pathogenicity was paralleled by a reduction in time that C. elegans resided on the bacterial lawn with the most pathogenic strains triggering an increase in the frequency of food-leaving. Previous reports indicate that gut colonization is a feature of pathogenicity, but we found that the most pathogenic strains were not associated with the highest level of colonization. Indeed, clearance of P. aeruginosa strains from the C. elegans gut was independent of bacterial pathogenicity. We show that this clearance is regulated by neuromodulation as C. elegans mutants in unc-31 and egl-3 have enhanced clearance of P. aeruginosa. Intriguingly this is also not linked to their pathogenicity. It is likely that there is a dynamic balance occurring in the C. elegans intestinal environment between maintaining a healthy, beneficial microbiota and removal of pathogenic bacteria.
Introduction
The nematode worm Caenorhabditis elegans has been used to investigate several fundamental biological processes and has emerged as a useful model for bacterial infection (Brenner, 2009; Khan et al., 2018). C. elegans is a bacteriophagous free living nematode which resides in the soil and on rotting fruit or vegetation (Schulenburg and Félix, 2017). Ingestion of microorganisms in these habitats is reflected in the recent observation that environmental bacteria can colonize the worm gut generating a complex microbiota (Zhang et al., 2017). Pathogenic bacteria also exist in these environments and nematodes have evolved mechanisms to respond to bacterial pathogens (Cohen and Troemel, 2015). C. elegans can be infected by and reacts to bacterial pathogens including Pseudomonas aeruginosa, Staphylococcus aureus, Salmonella enterica serovar Typhimurium, and Serratia marcescens, as well as several non-human bacterial pathogens (Tan et al., 1999a; Aballay et al., 2000, 2003; Garsin et al., 2001; Kurz et al., 2003). Bacterial pathogenicity in C. elegans can be investigated using several approaches including fast and slow killing (Darby et al., 1999; Mahajan-Miklos et al., 1999; Tan et al., 1999a). There is also increasing evidence that measures of C. elegans behavior in response to bacteria can be a useful surrogate of the underlying pathogenicity that ultimately progresses to death (Meisel and Kim, 2014). C. elegans can remain on bacterial lawns for extended periods of time which is known as dwelling behavior (Ben Arous et al., 2009). On lawns that contain potentially pathogenic bacteria or other aversive perturbations, C. elegans exhibit increased motility, food leaving events, and overall increased time off the lawn (Melo and Ruvkun, 2012). This is described as enhancement of food leaving or food aversion behavior (Melo and Ruvkun, 2012). These behaviors are also similar to other learnt behaviors of C. elegans in relation to aversive or repellent odorants or other chemicals (Meisel and Kim, 2014). This indicates that neuromodulatory pathways in C. elegans can be activated as part of the host response to distinct bacterial pathogens to enable the worms survival (Mills et al., 2012).
In this study, C. elegans was utilized to compare a selection of Gram-negative opportunistic pathogens. We provide insight into the relative pathogenicity of five strains each of Klebsiella pneumoniae, Acinetobacter baumannii, and P. aeruginosa (members of the ESKAPE group of pathogens). These pathogens are becoming more problematic in a clinical environment, especially in infections with immunocompromised individuals (Brenner, 1974) and therefore understanding their virulence is of fundamental importance. The well-defined pathogenic strain in C. elegans P. aeruginosa PA14 was used as a known comparison. We relate pathogen induced killing, nematode food aversion, and bacterial gut colonization. Results shown in this study reinforce the emergent view for complex host–pathogen interactions by implying several interacting determinants are controlling bacterial pathogenicity in C. elegans. Furthermore, through analysis of specific mutants that are defective in neuromodulation, we show that the nervous system is actively engaged in regulating the gut microbiome.
Materials and Methods
C. elegans and Bacterial Strain Maintenance
Caenorhabditis elegans strains used in this study were Bristol N2, DA509 unc-31 (e928) IV, XA3741 egl-3 (ok979). All strains were maintained on 5 cm Nematode growth media (NGM) plates with Escherichia coli OP50 as a food source according to standard methods (Brenner, 1974). Infection with Galleria mellonella was performed according to previous protocols (Wand et al., 2012). The bacterial strains used in this study are clinical strains with a variety of drug resistance markers that have previously been studied and are listed in Table 1 (Wand et al., 2012, 2013; Benthall et al., 2015). The experiments used numerical coding (P1-5, K1-5, and A1-5) to define the different bacterial strains and were therefore performed blind as to the identity of the bacteria with the code only being revealed at the end of the experiments conducted in this paper; the original strain names are used throughout the manuscript. Bacterial MLST typing was based on the Pasteur MLST typing. All strains were grown in LB broth with aeration or on LB agar plates at 37°C from glycerol stocks kept at −80°C unless otherwise stated.
Preparation of Assay Plates
Bacterial cultures were prepared from cultures grown overnight at 37°C in 5 mL of LB broth. The following day a 100 μL sample of the overnight culture was sub-cultured into 10 mL of LB at 37°C and grown until an OD600 of 0.8 was reached; 50 μL of this fresh culture was pipetted onto the center of 5 cm NGM plates and left to grow overnight at 20°C to form a lawn for C. elegans behavioral experiments.
C. elegans Killing Assay
15-20 L4 plus 1 C. elegans were transferred onto plates prepared as described above. Worms were observed each day and recorded as dead when they did not respond to being touched with a platinum wire. Animals that had crawled off the side of the plate were censured and removed from the assay. Individual worm populations were transferred to a new NGM plate with a fresh lawn of the same bacteria every other day to ensure the original adult population was the only one being observed and were devoid of any progeny produced during the experiment. The life span of C. elegans cultivated on the distinct bacterial strains was measured to assess the relative killing time. The data were plotted in a Kaplan Meier plot with the mean and 95% killing time being measured.
C. elegans Behavioral Assays
Caenorhabditis elegans populations were developmentally synchronized by picking L4 larval stage defined by the presence of a developing vulva. L4 worms were then maintained overnight on OP50 to become L4 plus 1 (young adults). At the start of the assay, seven young L4 plus 1 C. elegans were transferred onto the middle of the bacterial lawn and observed at 2, 6, 24, and 48 h after this initial transfer (Supplementary Figure S1). Food aversion was measured at these time points by the proportion of the original seven worms that were off the bacterial lawn. For egg laying and progeny production, previous published methods were utilized (Scott et al., 2017).
Determination of C. elegans Bacterial Colonization
Determining the number of bacterial cells present in the C. elegans intestine was based on previous protocols (Begun et al., 2007; Kawli and Tan, 2008; Portal-Celhay et al., 2012). Following an indicated time of exposure to the bacteria, approximately 10 worms were paralyzed by being picked into a 5 μL drop of 25 mM levamisole in M9 buffer on an unseeded 5 cm NGM plate. These paralyzed worms with inhibited pharyngeal pumping were washed three times with 1 mM sodium azide. The worms were then further washed twice with 5 μL drops of M9. For homogenization, C. elegans were picked into an Eppendorf containing sterile PBS with 1% Triton X-100 and lyzed using a glass pestle. The exact number of worms that lyzed was analyzed by visual inspection. The worm lysates were then serially diluted in PBS and plated on LB agar. Following overnight growth at 37°C, the number of colony forming units (CFUs) per animal was calculated. For experiments where P. aeruginosa colonization was measured after being transferred to OP50, the diluted worm lysates were plated on cetrimide agar for specific selection of P. aeruginosa. The pharyngeal pumping rate of C. elegans was measured by observing worms under 60x magnification while the worms were feeding on bacterial lawns. One contraction and relaxation of the grinder portion of the terminal bulb was considered one pharyngeal pumping event. This was measured continuously for 60 s.
Measure of Clearance of Bacteria From the C. elegans Intestine
Animals were fed on a lawn of fresh P. aeruginosa for 24 h and then analyzed for bacterial colonization as described above to measure the number of bacteria colonizing C. elegans. In addition, C. elegans was also exposed to P. aeruginosa for 24 h and then transferred to a lawn of OP50 for an additional 24 h. Animals were analyzed for P. aeruginosa CFUs as above by being plated on cetrimide agar. To calculate the efficacy with which P. aeruginosa cells are removed from C. elegans, the fold clearance was calculated by dividing the number of P. aeruginosa CFUs at 24 h by the number of P. aeruginosa measured 24 h post-OP50 transfer.
Statistical Analysis
All statistical analysis was done in Graphpad Prism and the individual tests used as indicated in figure legends). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ****P < 0.0001.
Results
The initial aim was to identify the pathogenicity of the 15 clinical strains of P. aeruginosa, K. pneumoniae, and A. baumannii relative to P. aeruginosa strain PA14. These strains were selected because they had previously shown a range of lethality in another invertebrate model G. mellonella (Supplementary Figure S2) (Wand et al., 2012, 2013; Benthall et al., 2015). For P. aeruginosa, which is naturally highly virulent in Galleria with an infectious dose of less than 100 CFUs, only one strain (GH97) showed reduced virulence in Galleria. To understand which strains were more pathogenic in C. elegans, worms were exposed to each bacterial strain at 20°C until death was observed (Figure 1). These killing experiments were designed to mimic the conditions on which a standard lifespan of C. elegans would be measured with the animals feeding on the laboratory standard food source of E. coli OP50 alone.
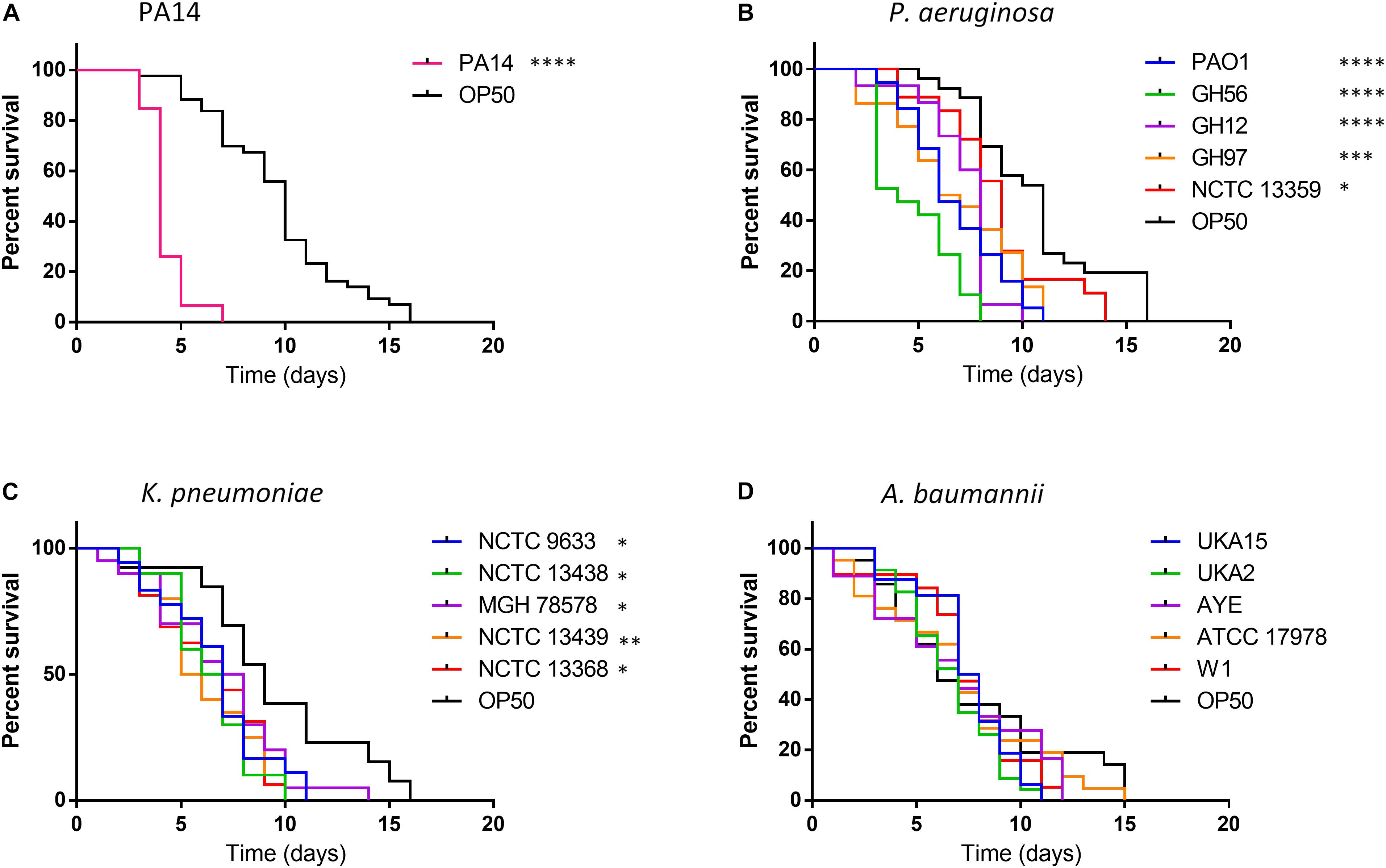
Figure 1. Survival of C. elegans exposed to strains of P. aeruginosa, K. pneumoniae, and A. baumannii relative to E. coli OP50. C. elegans survival after challenge with P. aeruginosa PA14 (A), other P. aeruginosa strains (B), K. pneumoniae (C) and A. baumannii (D) was recorded every day until 100% mortality was observed in the E. coli OP50 control. A log-rank (Mantel–Cox) test was used to compare the mortality of all strains relative to OP50 and significance is indicated. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ****P < 0.0001.
Exposure to OP50 led to total mortality after 16 days, conversely killing of C. elegans with the hypervirulent P. aeruginosa PA14 was more rapid with all worms’ dead after 7 days exposure (Figure 1A). For other P. aeruginosa strains tested killing was enhanced relative to OP50 with total mortality occurring between 8–11 days for all strains except NCTC 13359 (14 days) with strain GH56 showing a C. elegans mortality profile similar to PA14 (Figure 1B). This strain showed enhanced virulence relative to the other P. aeruginosa strains (Supplementary Table S1). All K. pneumoniae strains tested also showed significantly enhanced killing relative to OP50 with total C. elegans mortality occurring 10–14 days for all strains after initial challenge (Figure 1C). For A. baumannii, there appeared to be no significant time difference in mortality between any of the strains and OP50 (Figure 1D). Unlike P. aeruginosa, K. pneumoniae and A. baumannii showed little strain to strain variation in virulence in C. elegans (Supplementary Table S1). Due to the variation in virulence with P. aeruginosa strains, the whole genome sequences were analyzed for the presence of potential virulence factors. GacA and ToxA have been shown to be important virulence factors for slow killing of C. elegans by P. aeruginosa (Tan et al., 1999b). Analysis of the amino acid sequences for these proteins showed slight sequence variation but this and the presence/absence of other specific virulence factors previously described (Gallagher and Manoil, 2001; Feinbaum et al., 2012) cannot readily explain the enhanced virulence of P. aeruginosa strains PA14 and GH56 (Supplementary Table S2).
C. elegans Food Aversion Behavior to the Range of Bacterial Pathogens
Caenorhabditis elegans has been shown to exhibit food aversion when exposed to various pathogenic bacteria. Therefore, we sought to understand whether measurements of food aversion are indicative of pathogenicity and worm killing. The number of worms off the bacterial lawn was measured at given time-points (2, 6, 24, and 48 h after initial exposure). Exposing adult wild-type C. elegans to lawns of OP50 showed that as time increased, the number of worms existing off the food source increased. In comparison, there was a greater tendency for C. elegans to remain off the food source if that food source was P. aeruginosa PA14 (Figure 2A). This was also true for two other P. aeruginosa strains (GH56 and GH97) (Figure 2B). For K. pneumoniae (Figure 2C) and A. baumannii (Figure 2D), there was little difference in enhanced food aversion with only challenge with K. pneumoniae strain NCTC 13368 and A. baumannii strain UKA2 leading to an increased number of worms off the food at the 48 h time-point. Indeed, certain A. baumannii strains as a food source appeared to show reduced food aversion relative to OP50 at the later time points (strains ATCC 17978 and W1). This could be caused by visual depletion of the OP50 lawn relative to the bacterial lawns from the ESKAPE pathogens. Visual inspection of the lawns from all A. baumannii, K. pneumoniae, and P. aeruginosa strains did show that there was no noticeable depletion of the bacterial lawns throughout the 48 h assay with the lawns of some strains, specifically certain K. pneumoniae strains, becoming thicker. This is reflected in their mucoid phenotype which is linked to the presence of the rmpA gene (Hsu et al., 2011). The screen for food aversion suggested that exposure of C. elegans to K. pneumoniae NCTC 13368, A. baumannii UKA2, P. aeruginosa GH56, and P. aeruginosa GH97 generated an aversive response as measured by an increased propensity to be off the bacterial lawn. Those strains that resulted in enhanced levels of food aversion were also pathogenic indicating that enhanced food aversion can be a predictor of bacterial pathogenicity in C. elegans. Egg laying and hatching, and the ability to develop larval progeny are measures of C. elegans health. Worms which are maintained in environments containing harmful chemicals or varying quality of food might be expected, given their reduced time present on food, to produce fewer eggs. When egg laying and progeny production were investigated, there was found to be no significant difference between the number of eggs present on the bacterial lawn at 24 h between any of the strains and OP50 (Supplementary Figure S3). However, the number of progeny at 48 h was significantly reduced for all strains relative to OP50 except for strains K. pneumoniae NCTC 9633 and A. baumannii W1 (Figure 3).
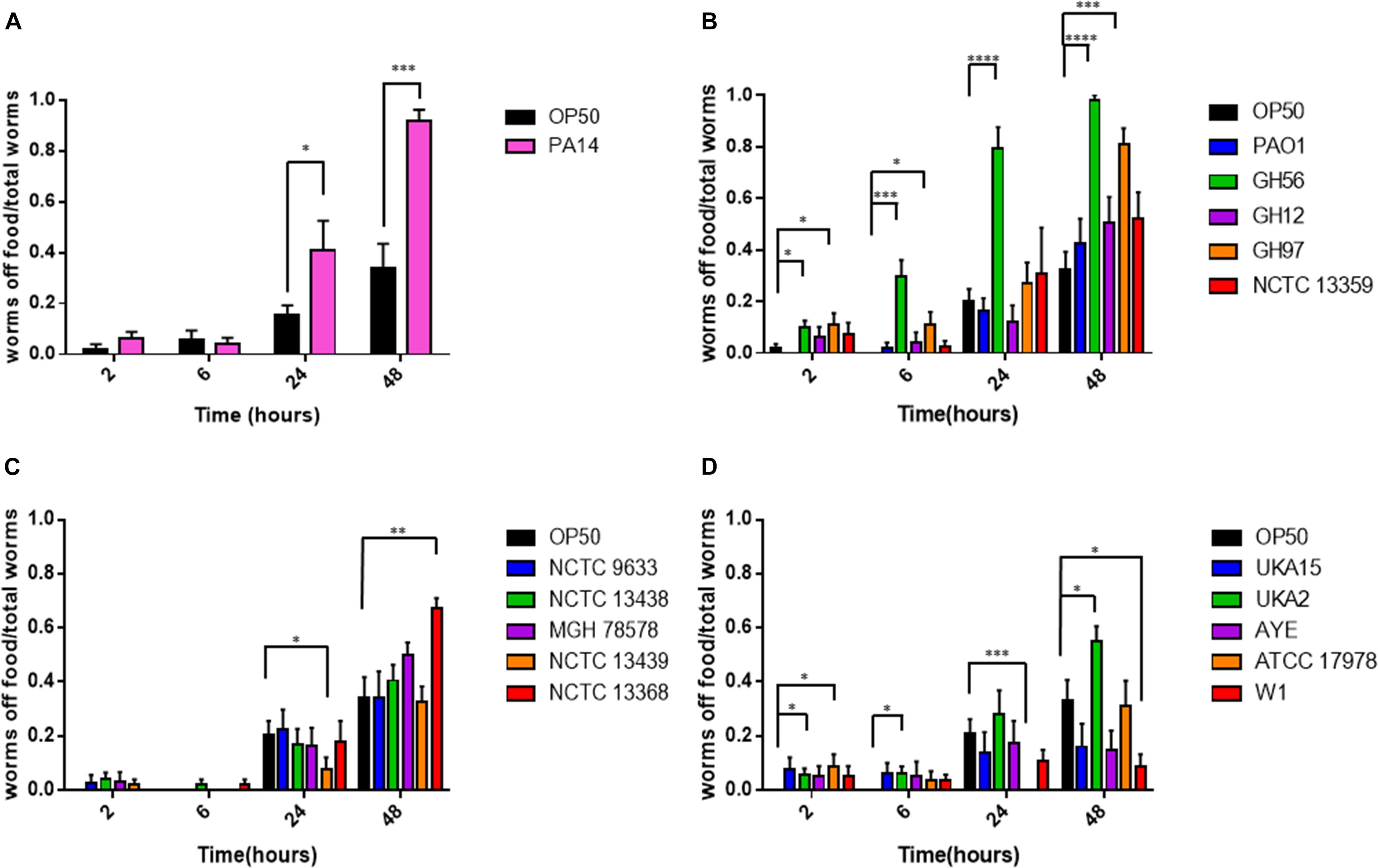
Figure 2. Food aversion for different bacterial pathogens relative to E. coli OP50. The proportion of worms off food for P. aeruginosa PA14 (A), other P. aeruginosa strains (B), K. pneumoniae (C), and A. baumannii (D) was measured at various time-points after exposure to the bacterial lawn as shown. These experiments were performed in at least triplicate for all strains. Error bars represent ± SEM. Analysis is by one-way ANOVA with Tukey’s multiple comparison. Significance relative to OP50 at the relevant time-point is shown. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ****P < 0.0001.
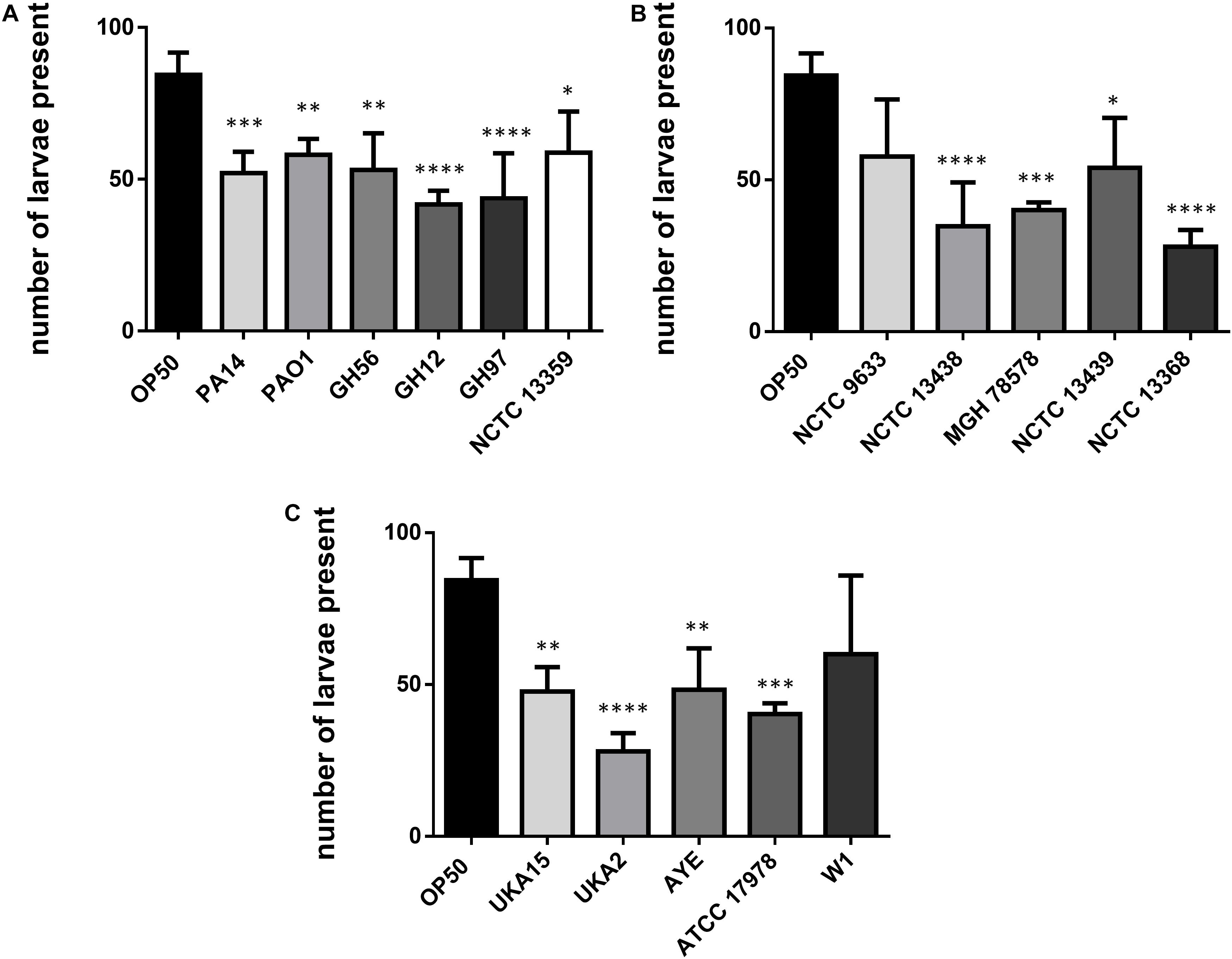
Figure 3. Number of C. elegans progeny after 48 h exposure to bacterial pathogens. Results are shown for P. aeruginosa (A), K. pneumoniae (B), and A. baumannii (C) strains and were compared to E. coli OP50. These experiments were performed in at least triplicate for all strains. Error bars represent ± SEM. Analysis is by one-way ANOVA with Tukey’s multiple comparison. Significance relative to OP50 is shown. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ****P < 0.0001.
Colonization Levels of Bacteria in C. elegans
To investigate possible determinants for the spectrum of virulence exhibited in the range of strains we examined the ability of all strains to colonize the gut of C. elegans after ingestion. Worms were exposed to individual bacterial strains for 48 h (the last time-point in the food aversion assay) after which the C. elegans were homogenized and the number of CFUs per worm calculated (Figure 4A). There were relatively few strains which showed a significant change in the bacterial number isolated after 48 h relative to OP50. Statistical analysis revealed that there was no significant variation between the colonization levels of any two bacterial strains when compared intra-species. However, when the average colonization levels for each species of bacteria was compared with the others, it was shown that K. pneumoniae had significantly reduced colonization numbers relative to P. aeruginosa and A. baumannii (Figure 4B). To understand whether colonization levels were affected by the rate of C. elegans ingestion of bacteria, the pharyngeal pumping rate was measured at 48 h relative to OP50 while the worms were feeding. For all strains tested, the pumping rate per minute was approximately 240. There was no significant change between OP50 and the other strains tested with the exception of PA14 exposed animals which had reduced pharyngeal pumping with a rate of 223 pumps per minute.
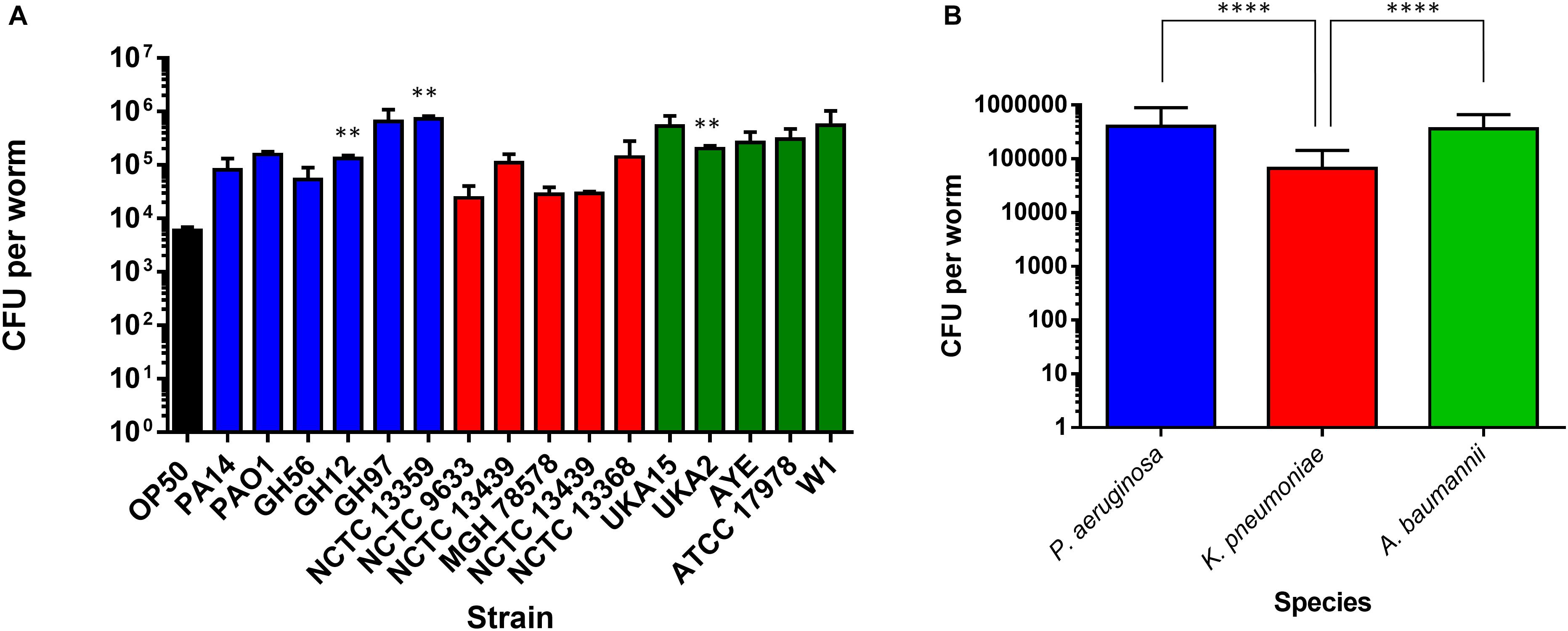
Figure 4. Colonization of the C. elegans intestine by bacterial pathogens. The ability of strains of P. aeruginosa, K. pneumoniae, and A. baumannii to colonize the C. elegans intestine was measured after 48 h exposure (A) and compared to E. coli OP50. These experiments were performed in at least triplicate for all strains. Error bars represent ± SEM. Analysis is by one-way ANOVA with Tukey’s multiple comparison. Significance relative to OP50 is shown. Comparison of mean CFU per species of bacteria in colonization of C. elegans after 48 h exposure (B). Error bars represent ± SEM. Analysis is by student’s unpaired t-test and significance relative to the other bacterial species is shown. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ****P < 0.0001.
Previous studies investigating PA14 had proposed that colonization was an important determinant of pathogenicity. To extend our comparative observations which suggest that this is not the case, we considered the time taken to cause death in 75% of worms. When P. aeruginosa strains were compared with OP50, there appeared to be no significant difference between colonization levels at 48 h and the longer time-points, and no significant differences between the P. aeruginosa strains and OP50 (Supplementary Figure S4).
Since the levels of colonization at 48 h did not appear to correlate with pathogenicity, we proceeded to examine the ability of the colonizing bacteria to be cleared from the C. elegans intestine once the worms were transferred to an E. coli OP50 food source (Figure 5). We focused on P. aeruginosa strains since these exhibited the greatest variation in killing (Figure 1B) and for comparison with PA14. Results showed that after exposure to individual P. aeruginosa strains for 24 h strains PA14, PAO1, and GH56 were cleared more rapidly following additional 24 h exposure to OP50 (Figure 6). Apart from PAO1 having enhanced clearance relative to strains GH12, GH97, and NCTC 13359, there was no other significant difference in the clearance of any P. aeruginosa strains relative to another. We did notice increased variation in clearance rates in PA14 and GH56 which may account for this lack of significance in clearance rates, although these results do indicate that C. elegans is able to clear more pathogenic strains (PA14, GH56) with the same efficiency as those which are less pathogenic (NCTC 13359).
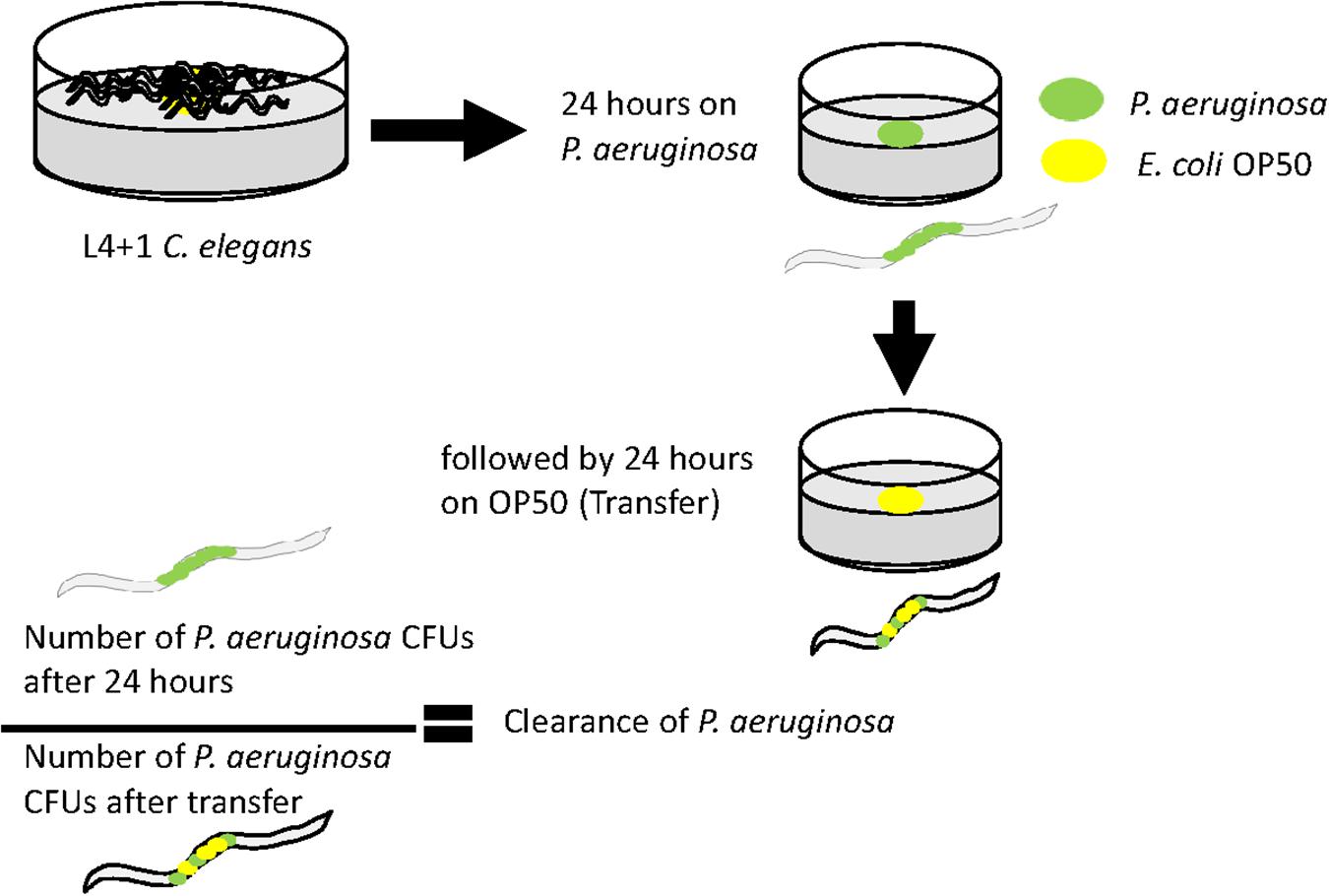
Figure 5. Methods for measuring clearance of P. aeruginosa from the C. elegans intestine after transfer to lawns of E. coli OP50. Age-synchronized 1 day old adult hermaphrodites were grown on a lawn of each P. aeruginosa strain before transfer to a lawn of E. coli OP50 for a further 24 h. Worms were analyzed for P. aeruginosa content immediately before and after transfer to E. coli OP50. The ratio of the CFU’s at each of these time points is an indicator of the clearance of P. aeruginosa from the intestine during exposure to E. coli.
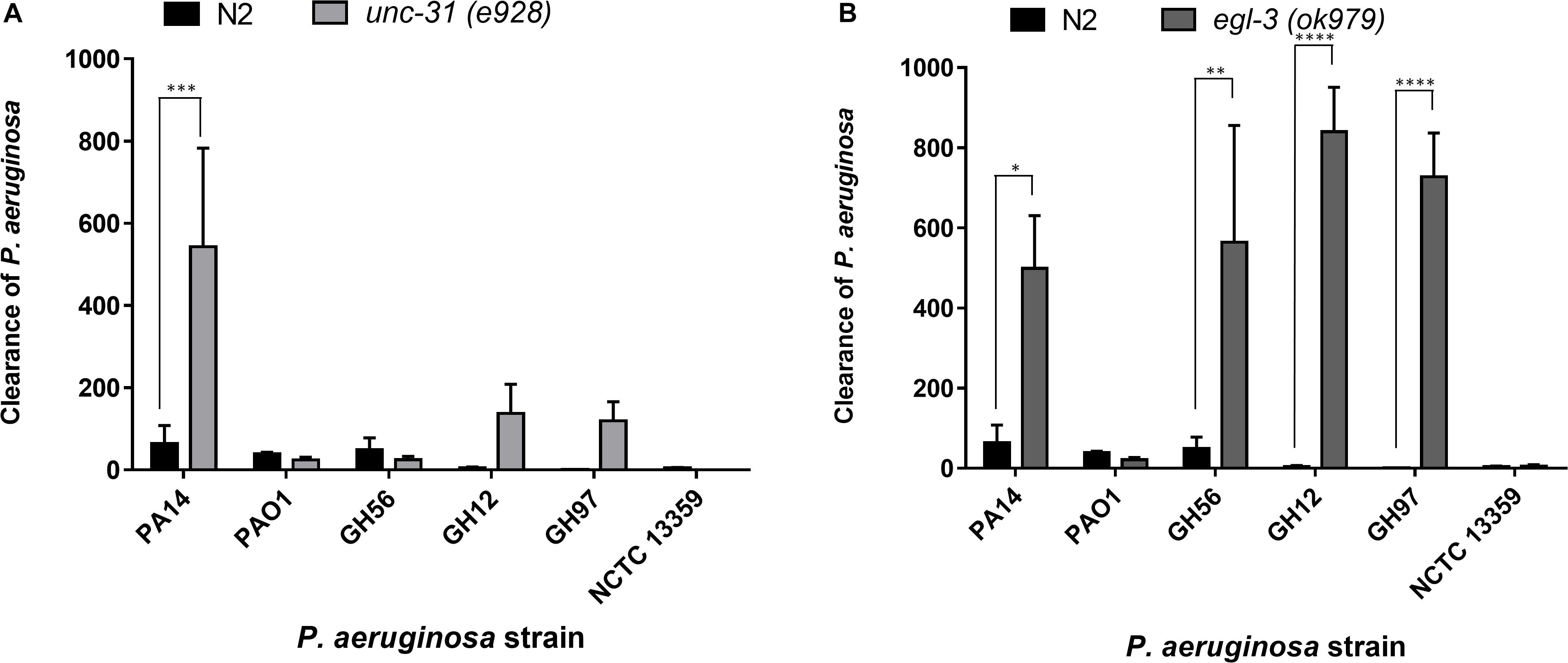
Figure 6. Clearance of P. aeruginosa strains from the C. elegans intestine is highly dependent on intact neuromodulation. The clearance of different strains of P. aeruginosa from the C. elegans intestine in unc-31 (e928) (A) and egl-3 (ok979) (B) mutant worms was measured using methods outlined in Figure 5. These experiments were performed in at least triplicate for all strains. Error bars represent ± SEM. Analysis is by one-way ANOVA with Sidaks multiple comparison. Significance relative to N2 worms is shown. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ****P < 0.0001.
Previous research has highlighted the importance of the neural network in pathogen avoidance (Cao et al., 2017). It has also indicated that C. elegans mutants with deficiencies in neuronal signaling show enhanced clearance rates of pathogenic bacteria after transfer to a non-pathogenic food source (Kawli and Tan, 2008). These previous experiments concluded a critical role for neuropeptide mediated modulation of neural signaling in the processes that control gut clearance. This was based on deficiencies in mutants that play a role in vesicle mediated exocytosis that initiates such neural signaling. Here we extended this analysis and investigated clearance from distinct mutants that have identified roles in peptide function but have not been investigated in the context of gut clearance. Clearance experiments using the mutant unc-31(e928) nematode, which is deficient in several aspects of neuronal function including neuropeptide signaling, revealed that PA14 is cleared more effectively in mutant unc-31(e928) worms than N2 worms (Figure 6A), but that clearance for other P. aeruginosa strains appeared to not be significantly enhanced. This suggested that the comparative approach provided an intersectional insight to further probe the relationship between neuromodulation and a generic role in bacterial pathogenicity. We also examined clearance rates in egl-3(ok979) worms, which are deficient in neuropeptide processing and lacks > 100 important neuropeptide transmitters including the previously implicated insulin related peptide INS-7 (Kawli and Tan, 2008). There was enhanced clearance of four of the P. aeruginosa strains (PA14, GH56, GH12, GH97) in these mutants relative to N2 worms but not for two others (PAO1 and NCTC 13359) (Figure 6B). This observation suggests a broader impact of Egl-3 and peptide processing (i.e., the ability to generate bioactive peptides) over determinants like Unc-31 and Unc-64 (Kawli and Tan, 2008) that mediate peptide secretion.
Discussion
Robust models of bacterial pathogenesis are required to address the challenges presented by antimicrobial resistance. C. elegans has emerged as a promising model for investigating bacterial infection and virulence (Marsh and May, 2012). However, previous studies have only investigated a very limited number of strains. In this study, we systemically screened several K. pneumoniae, A. baumannii, and P. aeruginosa isolates to determine their pathogenicity against C. elegans. We related this to their aversive influence on C. elegans behavior and provide insight into the differential response of the nematode to infections with varying pathogenicity.
The ability of C. elegans to regulate lawn occupancy is governed by food abundance and quality (Shtonda and Avery, 2006; Milward et al., 2011). C. elegans will move off lawns of more pathogenic bacteria (Anderson and McMullan, 2018) and results here show that those strains which are more pathogenic (P. aeruginosa GH56 and PA14), as measured by time to kill C. elegans, also resulted in an increased food aversion. This aversion is not linked to food depletion since visual inspection of the lawns indicated that lawns from all ESKAPE bacteria were intact and this is likely reflected in the different nutrient requirements for these bacteria relative to OP50. Behavior linked to food aversion is governed by components of the sensory neurons such as tax-2 and tax-4 (Milward et al., 2011). Experiments on food aversion using mutants in these genes suggest that this aversion is an exteroceptive response to bacteria after ingestion (Milward et al., 2011). The residence time of C. elegans on bacterial lawns is also regulated by population density and the presence of larvae; as the population increases, worms are more likely to leave the bacterial lawn (Milward et al., 2011; Scott et al., 2017). It is important to note that all the pathogenic strains of bacteria tested here reduced the fecundity of C. elegans resulting in lower numbers of larvae appearing over 24 h compared to worms cultured on E. coli OP50. Therefore, the increased food aversion observed with the pathogenic strains is not an indirect consequence of an effect on population density. The number of C. elegans progeny produced is not directly related to the number of eggs on the bacterial lawn indicating that while the presence of these pathogenic ESKAPE pathogens has no significant effect on egg production, it does imply an effect on egg hatching.
Bacteria can avoid lysis by the grinder section of the C. elegans pharynx (Avery and Shtonda, 2003) and colonize the post pharyngeal intestine in C. elegans. The ability of different bacteria to colonize the C. elegans intestine is an important determinant of virulence and pathogenicity (Kawli and Tan, 2008). In addition, previous studies using P. aeruginosa have implicated the nematode gut colonization in driving facets of the pathogenic responses (Schulenburg and Ewbank, 2004). However, our results show that all strains from the same species colonized C. elegans to a similar degree which was not dependent upon their relative pathogenicity. Longer term colonization with P. aeruginosa did also not appear to differentiate between strains with different pathogenicity profiles. This observation that colonization is not the primary determinant of pathogenicity is consistent with a previous study which showed that slow killing of C. elegans by strains of Burkholderia pseudomallei occurs in the absence of gut colonization (Ooi et al., 2012). In our analysis, colonization appeared to be species rather than strain dependent with higher numbers of bacteria isolated from the C. elegans gut colonized with A. baumannii or P. aeruginosa than K. pneumoniae strains. Therefore, C. elegans gut conditions may be more amenable to A. baumannii and P. aeruginosa colonization or these bacteria are better equipped to bypass the pharyngeal system of C. elegans (Avery and Shtonda, 2003).
Bacterial colonization of the intestine can be reversed by the response of the nematode to the infection. We found that wild-type (N2) C. elegans showed variation in the clearance levels for different P. aeruginosa strains but that overall bacterial pathogenicity was not a simple predictor of the clearance response. Thus, this more extensive comparison of pathogenicity for a range of bacterial isolates reveals that the determinants which drive interactions between the nematode host and the bacterial pathogen are more complex than previously appreciated. The enhanced clearance of specific P. aeruginosa strains from the C. elegans intestine using egl-3 (Kass et al., 2001) and unc-31 mutants (Avery et al., 1993), which are disrupted in aspects of neuropeptide maturation and release, respectively, showed the importance of these proteins in post bacterial colonization clearance. Indeed, there is evidence that unc-31 mutants are more resistant to infection with P. aeruginosa PA14 due to the expression of increased levels of immune effectors (Evans et al., 2008). However, the absence of these genes has no effect on the clearance of P. aeruginosa strains PAO1 and NCTC 13359. This suggests that while having a fully functioning range of neuropeptides reduces the number of bacteria that are cleared from C. elegans, there are other unknown aspects which are equally important in bacterial clearance (Montalvo-Katz et al., 2013).
The existence of a complex C. elegans gut microbiota which aids in protection against infection by pathogens has been reported (Kissoyan et al., 2019). The C. elegans gut microbiota was found to include Pseudomonas species and Enterobacteriaceae including Klebsiella species (Dirksen et al., 2016). It was observed that the four strains which are more highly cleared by egl-3 mutants included P. aeruginosa PA14 and GH56, the most pathogenic strains based on previous killing experiments, suggesting that this enhanced clearance in the mutant may be more selective toward more harmful strains. In addition, the more general effect of Egl-3 which is essential in proteolytic processing that converts pro-proteins to their bioactive peptides indicates that this biological function, rather than peptide secretion, is more important in defining the landscape of the bacterial host interaction. In this regard, it should be noted that Egl-3 although primarily associated in the processing of prohormones is also responsible for generating processed peptides that encode extracellular adhesion or bactericidal activities that could clearly impact host–pathogen interactions (Thacker and Rose, 2000; Dierking et al., 2016). This further suggests that colonizing bacteria elicit a differential response from the worm depending on the nature of the interaction occurring in the intestinal tissue and may indicate that specific Klebsiella and Pseudomonas strains are treated as commensals rather than pathogenic. Studies on the natural microbiome of C. elegans have shown that its physiology is affected differently by isolates from the same genera. This suggests that genetic differences at the strain level are important in pathogenicity (Samuel et al., 2016). Our results are consistent with this. While there was a clear trend suggesting that P. aeruginosa strains were more pathogenic than K. pneumoniae which in turn was more pathogenic than A. baumannii, there was strain to strain variation not just in terms of ability to kill C. elegans but also in terms of food aversion and clearance. All A. baumannii strains had similar virulence and food aversion profiles compared to E. coli OP50, which was observed in previous studies (Vallejo et al., 2015), but appeared to colonize in greater numbers.
This study has shown evidence of a multi-faceted host response due to bacterial colonization and that the nematode responses are driven by the properties of the bacterial strain or species. The evidence presented here indicates that neuropeptides and/or neuromodulation are important in regulating this balance but are not the only factor. It is likely that there is a dynamic balance occurring in the C. elegans intestinal environment between maintaining a healthy beneficial microbiota and removal of pathogenic bacteria. This would be especially true in the natural environment of C. elegans when animals are constantly moving between different bacterial food sources. The comparative approach we have used here highlights the rich biology of bacterial host interactions in C. elegans (Zhang et al., 2017) and should inform on the use of model organisms utilizing both environmental and clinical relevant strains for studying bacterial pathogenicity in C. elegans.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
ES and MW performed in vitro experiments. All the authors analyzed the data, designed the study, wrote the manuscript, read, and approved the final version of the manuscript.
Funding
ES was funded by a Doctoral Training Program Grant from the Biotechnology and Biological Sciences Research Council (BBSRC) and Public Health England (PHE). MW was funded by PHE Grant in Aid No. 109506.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.03113/full#supplementary-material
References
Aballay, A., Drenkard, E., Hilbun, L. R., and Ausubel, F. M. (2003). Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr. Biol. 13, 47–52. doi: 10.1016/s0960-9822(02)01396-9
Aballay, A., Yorgey, P., and Ausubel, F. M. (2000). Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 10, 1543–1545.
Anderson, A., and McMullan, R. (2018). Neuronal and non-neuronal signals regulate Caenorhabditis elegans avoidance of contaminated food. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373:20170255. doi: 10.1098/rstb.2017.0255
Avery, L., Bargmann, C. I., and Horvitz, H. R. (1993). The Caenorhabditis elegans unc-31 gene affects multiple nervouse system controlled functions. Genetics 134, 455–464.
Avery, L., and Shtonda, B. B. (2003). Food transport in the C. elegans pharynx. J. Exp. Biol. 206, 2441–2457. doi: 10.1242/jeb.00433
Begun, J., Gaiani, J. M., Rohde, H., Mack, D., Calderwood, S. B., Ausubel, F. M., et al. (2007). Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLos Pathog. 3:e57. doi: 10.1371/journal.ppat.0030057
Ben Arous, J., Laffont, S., and Chatenay, D. (2009). Molecular and sensory basis of a food related two-state behavior in C. elegans. PLoS One 4:e7584. doi: 10.1371/journal.pone.0007584
Benthall, G., Touzel, R. E., Hind, C. K., Titball, R. W., Sutton, J. M., Thomas, R. J., et al. (2015). Evaluation of antibiotic efficacy against infections caused by planktonic or biofilm cultures of Pseudomonas aeruginosa and Klebsiella pneumoniae in Galleria mellonella. Int. J. Antimicrob. Agents 46, 538–545. doi: 10.1016/j.ijantimicag.2015.07.014
Brenner, S. (2009). In the beginning was the worm. Genetics 182, 413–415. doi: 10.1534/genetics.109.104976
Cao, X., Kajino-Sakamoto, R., Doss, A., and Aballay, A. (2017). Distinct roles of sensory neurons in mediating pathogen avoidance and neuropeptide-dependent immune regulation. Cell Rep. 21, 1442–1451. doi: 10.1016/j.celrep.2017.10.050
Cohen, L. B., and Troemel, E. R. (2015). Microbial pathogenesis and host defense in the nematode C. elegans. Curr. Opin. Microbiol. 23, 94–101. doi: 10.1016/j.mib.2014.11.009
Darby, C., Cosma, C. L., Thomas, J. H., and Manoil, C. (1999). Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 96, 15202–15207.
Dierking, K., Yang, W., and Schulenburg, H. (2016). Antimicrobial effectors in the nematode Caernorhabditis elegans: an outgroup to the Arthropoda. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371:20150299. doi: 10.1098/rstb.2015.0299
Dirksen, P., Marsh, S. A., Braker, I., Heitland, N., Wagner, S., Nakad, R., et al. (2016). The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol. 14:38. doi: 10.1186/s12915-016-0258-1
Evans, E. A., Kawli, T., and Tan, M. W. (2008). Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signalling pathway in Caenorhabditis elegans. PLoS Pathog. 4:e1000175. doi: 10.1371/journal.ppat.1000175
Feinbaum, R. L., Urbach, J. M., Liberati, N. T., Djonovic, S., Adonizio, A., Carvunis, A.-R., et al. (2012). Genome-wide identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model. PLoS Pathog. 8:e1002813. doi: 10.1371/journal.ppat.1002813
Gallagher, L. A., and Manoil, C. (2001). Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183, 6207–6214. doi: 10.1128/jb.183.21.6207-6214.2001
Garsin, D. A., Sifri, C. D., Mylonakis, E., Qin, X., Singh, K. V., Murray, B. E., et al. (2001). A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. U.S.A. 98, 10892–10897. doi: 10.1073/pnas.191378698
Hsu, C. R., Lin, T. L., Chen, Y. C., Chou, H. C., and Wang, J. T. (2011). The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiol 157, 3446–3457. doi: 10.1099/mic.0.050336-0
Kass, J., Jacob, T. C., Kim, P., and Kaplan, J. M. (2001). The EGL-3 proprotein convertase regulates mechanosensory responses of Caenorhabditis elegans. J. Neurosci. 21, 9265–9272. doi: 10.1523/jneurosci.21-23-09265.2001
Kawli, T., and Tan, M. W. (2008). Neuroendocrine signals modulate the innate immunity of Caenorhabditis elegans through insulin signalling. Nat. Immunol. 9, 1415–1424. doi: 10.1038/ni.1672
Khan, F., Jain, S., and Oloketuyi, S. F. (2018). Bacteria and bacterial products: Foe and friends to Caenorhabditis elegans. Microbiol. Res. 215, 102–113. doi: 10.1016/j.micres.2018.06.012
Kissoyan, K. A. B., Drechsler, M., Stange, E. L., Zimmermann, J., Kaleta, C., Bode, H. B., et al. (2019). Natural C. elegans microbiota protects against infection via production of a cyclic lipopeptide of the viscosin group. Curr. Biol. 29, 1030.e5–1037.e5. doi: 10.1016/j.cub.2019.01.050
Kurz, C. L., Chauvet, S., Andrès, E., Aurouze, M., Vallet, I., Michel, G. P., et al. (2003). Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J. 22, 1451–1460. doi: 10.1093/emboj/cdg159
Mahajan-Miklos, S., Tan, M. W., Rahme, L. G., and Ausubel, F. M. (1999). Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa Caenorhabditis elegans pathogenesis model. Cell 96, 47–56. doi: 10.1016/s0092-8674(00)80958-7
Marsh, E. K., and May, R. C. (2012). Caenorhabditis elegans, a model organism for investigating immunity. Appl. Environ. Microbiol. 78, 2075–2081. doi: 10.1128/AEM.07486-11
Meisel, J. D., and Kim, D. H. (2014). Behavioural avoidance of pathogenic bacteria by Caenorhabditis elegans. Trends Immunol. 35, 465–470. doi: 10.1016/j.it.2014.08.008
Melo, J. A., and Ruvkun, G. (2012). Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defences. Cell 149, 452–466. doi: 10.1016/j.cell.2012.02.050
Mills, H., Wragg, R., Hapiak, V., Castelletto, M., Zahratka, J., Harris, G., et al. (2012). Monoamines and neuropeptides interact to inhibit aversive behaviour in Caenorhabditis elegans. EMBO J. 31, 667–678. doi: 10.1038/emboj.2011.422
Milward, K., Busch, K. E., Murphy, R. J., de Bono, M., and Olofsson, B. (2011). Neuronal and molecular substrates for optimal foraging in Caenorhabditis elegans. Proc. Natl. Acad. Aci. U.S.A. 108, 20672–20677. doi: 10.1073/pnas.1106134109
Montalvo-Katz, S., Huang, H., Appel, M. D., Berg, M., and Shapira, M. (2013). Association with soil bacteria enhances p38-dependent infection resistance in Caenorhabditis elegans. Infect. Immun. 81, 514–520. doi: 10.1128/IAI.00653-12
Ooi, S. K., Lim, T. Y., Lee, S. H., and Nathan, S. (2012). Burkholderia pseudomallei kills Caenorhabditis elegans through virulence mechanisms distinct from intestinal lumen colonization. Virulence 3, 485–496. doi: 10.4161/viru.21808
Portal-Celhay, C., Bradley, E. R., and Blaser, M. J. (2012). Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans. BMC Microbiol. 12:17. doi: 10.1186/1471-2180-12-49
Samuel, B. S., Rowedder, H., Braendle, C., Félix, M. A., and Ruvkun, G. (2016). Caenorhabditis elegans responses to bacteria from its natural habitats. Proc. Natl. Acad. Aci. U.S.A. 113, E3941–E3949. doi: 10.1073/pnas.1607183113
Schulenburg, H., and Ewbank, J. J. (2004). Diversity and specificity in the interaction between Caenorhabditis elegans and the pathogen Serratia marcescens. BMC Evol. Biol. 4:49.
Schulenburg, H., and Félix, M. A. (2017). The natural biotic environment of Caenorhabditis elegans. Genetics 206, 55–86. doi: 10.1534/genetics.116.195511
Scott, E., Hudson, A., Feist, E., Calahorro, F., Dillon, J., de Freitas, R., et al. (2017). An oxytocin-dependent social interaction between larvae and adult C. elegans. Sci. Rep. 7:10122. doi: 10.1038/s41598-017-09350-7
Shtonda, B. B., and Avery, L. (2006). Dietary choice behaviour in Caenorhabditis elegans. J. Exp. Biol. 209, 89–102. doi: 10.1242/jeb.01955
Tan, M. W., Mahajan-Miklos, S., and Ausubel, F. A. (1999a). Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 96, 715–720. doi: 10.1073/pnas.96.2.715
Tan, M. W., Rahme, L. G., Sternberg, J. A., Tompkins, R. G., and Ausubel, F. M. (1999b). Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. U.S.A. 96, 2408–2413.
Thacker, C., and Rose, A. M. (2000). A look at the Caenorhabditis elegans Kex2/Subtilisin-like proprotein convertase family. Bioessays 22, 545–553. doi: 10.1002/(sici)1521-1878(200006)22:6<545::aid-bies7>3.0.co;2-f
Vallejo, J. A., Beceiro, A., Rumbo-Feal, S., Rodríguez-Palero, M. J., Russo, T. A., and Bou, G. (2015). Optimisation of Caenorhabditis elegans model for studying the pathogenesis of opportunistic Acinetobacter baumannii. Int. J. Antimicrob. Agents [Epub ahead of print].
Wand, M. E., Bock, L. J., Turton, J. F., Nugent, P. G., and Sutton, J. M. (2012). Acinetobacter baumannii virulence is enhanced in Galleria mellonella following biofilm adaptation. J. Med. Microbiol. 61, 470–477. doi: 10.1099/jmm.0.037523-0
Wand, M. E., McCowen, J. W., Nugent, P. G., and Sutton, J. M. (2013). Complex interactions of Klebsiella pneumoniae with the host immune system in a Galleria mellonella infection model. J. Med. Microbiol. 62, 1790–1798. doi: 10.1099/jmm.0.063032-0
Keywords: Caenorhabditis elegans, Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, food aversion, pathogenicity, colonization, neuropeptide
Citation: Scott E, Holden-Dye L, O’Connor V and Wand ME (2020) Intra Strain Variation of the Effects of Gram-Negative ESKAPE Pathogens on Intestinal Colonization, Host Viability, and Host Response in the Model Organism Caenorhabditis elegans. Front. Microbiol. 10:3113. doi: 10.3389/fmicb.2019.03113
Received: 06 September 2019; Accepted: 24 December 2019;
Published: 21 January 2020.
Edited by:
Laurence Rahme, Harvard Medical School, United StatesReviewed by:
Yiorgos Apidianakis, University of Cyprus, CyprusNatalia V. Kirienko, Rice University, United States
Copyright © 2020 Scott, Holden-Dye, O’Connor and Wand. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthew E. Wand, bWF0dGhldy53YW5kQHBoZS5nb3YudWs=
 Euan Scott1
Euan Scott1 Lindy Holden-Dye
Lindy Holden-Dye Vincent O’Connor
Vincent O’Connor Matthew E. Wand
Matthew E. Wand