- 1Key Laboratory of Agro-Ecological Processes in Subtropical Region, National Engineering Laboratory for Pollution Control and Waste Utilization in Livestock and Poultry Production, Hunan Provincial Engineering Research Center for Healthy Livestock and Poultry Production, Scientific Observing and Experimental Station of Animal Nutrition and Feed Science in South-Central, Ministry of Agriculture, Institute of Subtropical Agriculture, Chinese Academy of Sciences, Changsha, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Laboratory of Basic Biology, Hunan First Normal University, Changsha, China
- 4Hunan International Joint Laboratory of Animal Intestinal Ecology and Health, Laboratory of Animal Nutrition and Human Health, College of Life Sciences, Hunan Normal University, Changsha, China
Maternal gut microflora changes dramatically during perinatal period and plays a vital role in animal health and reproductive performance. However, little is known about the microbial differences between sows with different productive capacities during perinatal period. Hence, this study explored fecal microbial diversity, composition, metabolic functions, and phenotypes differences between high productive capacity (HPC, litter size ≥ 15) and low productive capacity (LPC, litter size ≤ 7) sows during late pregnancy (LP, the third day before due date) and early stage after parturition (EAP, the third day after parturition) as well as serum biochemical indices differences after parturition. Results showed that HPC sows had lower microbial richness at LP stage and higher microbial diversity at EAP stage than LPC sows. Several genera belonging to the Prevotellaceae family exhibited higher abundance, while some genera belonging to the Ruminococcaceae family exhibited lower abundance in HPC sows compared to LPC sows at LP stage. Moreover, the relative abundance of Eubacterium_coprostanoligenes_group and Ruminococcaceae_UCG-014 in HPC sows was significantly higher than that in LPC sows at EAP stage. The predicted metabolic functions related to Lipopolysaccharide biosynthesis were significantly higher in HPC sows at LP stage. Further, HPC sows had significantly higher blood urea nitrogen (BUN) and high-density lipoprotein cholesterol (HDL-C) levels after parturition, and there were strong correlations between BUN level and the relative abundance of genera belonging to the Ruminococcaceae families. These results indicated that the HPC sows may experience greater inflammation than LPC sows at LP stage. Inflammation environment might impact health but promote parturition. The microbial differences at EAP stage might be beneficial to hemostasis and anti-inflammation, which might contribute to postpartum recovery in HPC sow.
Introduction
Pregnant mothers undergo various changes in immunity, metabolism, steroid hormone production and behaviors (Linzer and Fisher, 1999). Uterine contraction, pain and the increasing level of plasma cortisol contribute to physiological stresses, which might lead to dysregulated secretion of oxytocin, oxidative stress, liver damage and impaired immunity (Lawrence et al., 1992; van de Ligt et al., 2002; Verheyen et al., 2007; Jarvis et al., 2010; Marek et al., 2013). Further, some females develop the metabolic syndrome such as decreased insulin sensitivity in late pregnancy (Barbour et al., 2007). Reduced insulin sensitivity may lead to decreased feed intake in sows during lactation (Père and Etienne, 2007). The perinatal period (late pregnancy, LP and early stage after parturition, EAP) is a critical stage for sows, during which most of piglets died under unreasonable management (Bäckström, 1973). However, supervision and nutritional feed supplements can improve sow health and reproductive performance (Holyoake et al., 1995; Kim et al., 2007). Thus, changes that occur in sows during perinatal period need to be further explored, which will guide human interventions to improve sow reproductivity.
The microbiome is of significant importance to a host, which affects the host’s immune system, metabolism, emotion and cognition (Chu and Mazmanian, 2013; Rothschild et al., 2018; Sarkar et al., 2018). Gestation changes the microbiota structure, while intestinal microbes remain stable during lactation (Liu et al., 2019). It was reported that intestinal microflora changed dramatically throughout pregnancy (Koren et al., 2012). In particular, stools from the third trimester contained lower microbial diversity and a higher abundance of Proteobacteria and Actinobacteria, which contributed to increased adiposity and insulin insensitivity (Koren et al., 2012). Regarding the host’s immune system, SCFAs produced by microbiota are critical for epithelial barrier function, tumor suppression, antioxidation, cytokine production and anti-inflammatory effects (Maslowski and Mackay, 2011; Maynard et al., 2012). Moreover, microflora plays a vital role in reproductive performance. Probiotics feed supplements have been shown to improve sow reproductive capacity by influencing gut microbiota (Alexopoulos et al., 2004; Tsukahara et al., 2018; Cao et al., 2019).
The gut microbiota is a center regulator of host metabolism (Schoeler and Caesar, 2019). The gut-liver axis enables the host to control and shape the gut microbiota and affect animal’s feeding behavior and energy metabolism (Ringseis et al., 2019). Gut microbiota undergo a significant shift during pregnancy (Santacruz et al., 2010; Huang et al., 2019). Santacruz et al. (2010) showed that gut microbiota is associated with body weight, weight gain and biochemical parameters in pregnant women. Huang et al. (2019) showed that host-microbiota interactions during the perinatal period impact host metabolism of sows. However, gut microbial differences between sows with different productive capacities and its relationship with serum biochemical indices remain elusive.
Therefore, this study investigated differences in gut microbiota between low productive capacity (LPC, litter size ≤ 7) and high productive capacity (HPC, litter size ≥ 15) sows during perinatal period. Further, health status between sows with different reproductivity after parturition was compared using serum biochemical indices to determine its correlations with microbiota.
Materials and Methods
Experimental Design and Ethics Statement
This experiment followed guidelines for animal research approved by the Animal Welfare Committee of the Institute of Subtropical Agriculture, Chinese Academy of Sciences. The feeding experiments were performed at the pig breeding farm in Hunan province, which had no history of serious bacterial or viral infections. Eighty multiparous hybrid pregnant sows (Landrace × Yorkshire) were used in the experiments. They had similar expected dates of confinement, backfat thicknesses and no medical history. They were in 3–6 birth order and the same physical condition. After parturition, 6 HPC sows (litter size ≥ 15) and 6 LPC sows (litter size ≤ 7) were chosen according to their litter sizes (Supplementary Table S1) and they all received the amoxicillin treatment to diminish postpartum inflammation for 3 days. All sows were provided with the same commercial formula feed once at LP stage and twice after parturition every day. Sows were raised individually in a piggery with hard plastic slatted flooring and they had free access to water through nipple drinkers.
Sample Collection
At the third day before due date (late stage of pregnancy), feces of all the 80 sows were collected. And feces of 6 HPC as well as 6 LPC sows were collected at the third day after parturition (early stage after parturition). Sterile centrifugal tubes were used to collect fresh feces, which were immediately frozen in liquid nitrogen, and stored at −80°C. Samples were grouped as follows: A, C: sows with high productive capacity (HPC) at LP and EAP stage separately; B, D: sows with low productive capacity (LPC) at LP and EAP stage separately. About 5mL of blood was collected the day after parturition via the auricular vein with vacuum tubes and centrifuged at 1500 g for 15 min. Subsequently, the supernatant was stored at −20°C before further determination.
Microbiota Analysis Based on 16S RNA High-Throughput Sequencing
Microbiota analysis was conducted using 6 fecal samples in each group from different sows and about 0.25 g of each was used to extract bacterial DNA using CTAB/SDA method. The composition and diversity of microflora were analyzed by 16S rRNA high-throughput sequencing. 16S rRNA genes of V4 regions were amplified using 515F: 5′-GTGCCAGCMGCCGCGGTAA-3′ and 806R: 5′-GGACTACHVGGGTWTCTAAT-3′ primers with barcodes. Illumina HiSeq 2500 platform was used to conduct paired-end sequencing. Raw tags were assembled and filtered under specific conditions (Bokulich et al., 2013) to obtain clean data using the QIIME (V1.7.0) (Caporaso et al., 2010) and FLASH (V1.2.7) (Tanja and Salzberg, 2011). Sequences were analyzed and operational taxonomic units (OUT) were determined using UPARSE (v7.0.1001) (Edgar, 2013). Sequences were assigned to the same OTUs at a 97% similarity level. The GreenGene Database1 (Desantis et al., 2006) based on RDP classifier (V 2.2)2 (Qiong et al., 2007) was used to assigned sequences to a taxonomic level. The assembled HiSeq sequences obtained in the present study were submitted to NCBI Sequence Read Archive (SRA, No. PRJNA565644).
Metagenome Prediction, Functional Metabolic Pathways and Metabolic Phenotypes Analysis
Functional metagenomes were predicted using PICRUSt (V1.1.3) (Langille et al., 2013). OTUs were verified using the Genome Prediction Tutorial for PICRUSt. Normalized 16S rRNA data were analyzed to predict metagenomes using the database of Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology3. Functional differences were explored using STAMP (V2.1.3) (Parks et al., 2014). Additionally, microbial phenotypes were predicted using Bugbase4 based on 16S RNA data and mapping files following provided instructions (Ward et al., 2017b).
Determination of Serum Biochemical Indices and Their Correlations With Microbial Abundance
Serum biochemical parameters including blood urea nitrogen (BUN), creatinine (CREA), triglyceride (TG), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), glucose (GLU), AST, ALT, total protein (TP), albumin (ALB), globulin (GLO), ration of albumin/globulin (A/G), and cholesterol (CHO) were determined by TBA-120FR biochemistry analyzer (Toshiba Medical Systems Corporation, Tokyo, Japan). IgM, IgA and IgG were analyzed using an enzyme-linked immunosorbent assay kit (Cusabio Biotech Co., Hubei, China) following provided instructions. The correlations between microbial abundance at the genus level and serum biochemical indices were evaluated by Spearman’s correlation analysis and visualized diagrams were created using R (V2.15.3).
Statistical Analysis
Statistical analyses were performed using SPASS 22 (SPSS Inc.). Alpha and beta community diversity were calculated with QIIME (V1.7.0). R (V2.15.3) and GraphPad Prism (V8.0.2) were used to create visualized diagrams. For PICRUSt results, differences between groups were analyzed by one-way ANOVA and the Tukey–Kramer multiple comparisons test. Paired T-test (LP vs. EAP) or independent T-test (HPC vs. LPC) was used after accessing normality with Shapiro–Wilk W-test to analyze microbial alpha diversity and serum biochemical indices. Variability in the data was expressed as means ± SD, and level of P < 0.05 was considered significant. Wilcoxon signed-rank test (LP vs. EAP) or Mann–Whitney U-test (HPC vs. LPC) was applied to analyze gut microbial phenotypes differences.
Results
Diversity Changes in Gut Microbiota
In total, 24 fecal samples were used to perform 16S rRNA high-throughput sequencing using the Illumina HiSeq 2500 platform. On average, 62136 tags were verified, and 1183 OTUs per sample with 97% sequence similarity were obtained. Diversity differences between groups were accessed. Based on Chao1 and ACE, LPC sows contained significantly more observed species and higher microbial richness than HPC sows at LP stage (ACE, P < 0.001; Chao1, P < 0.01). At EAP stage, the microbial diversity (represented by Shannon and Simpson) of HPC sows was significantly higher than LPC sows (Shannon, P < 0.01; Simpson, P < 0.05) (Figure 1).
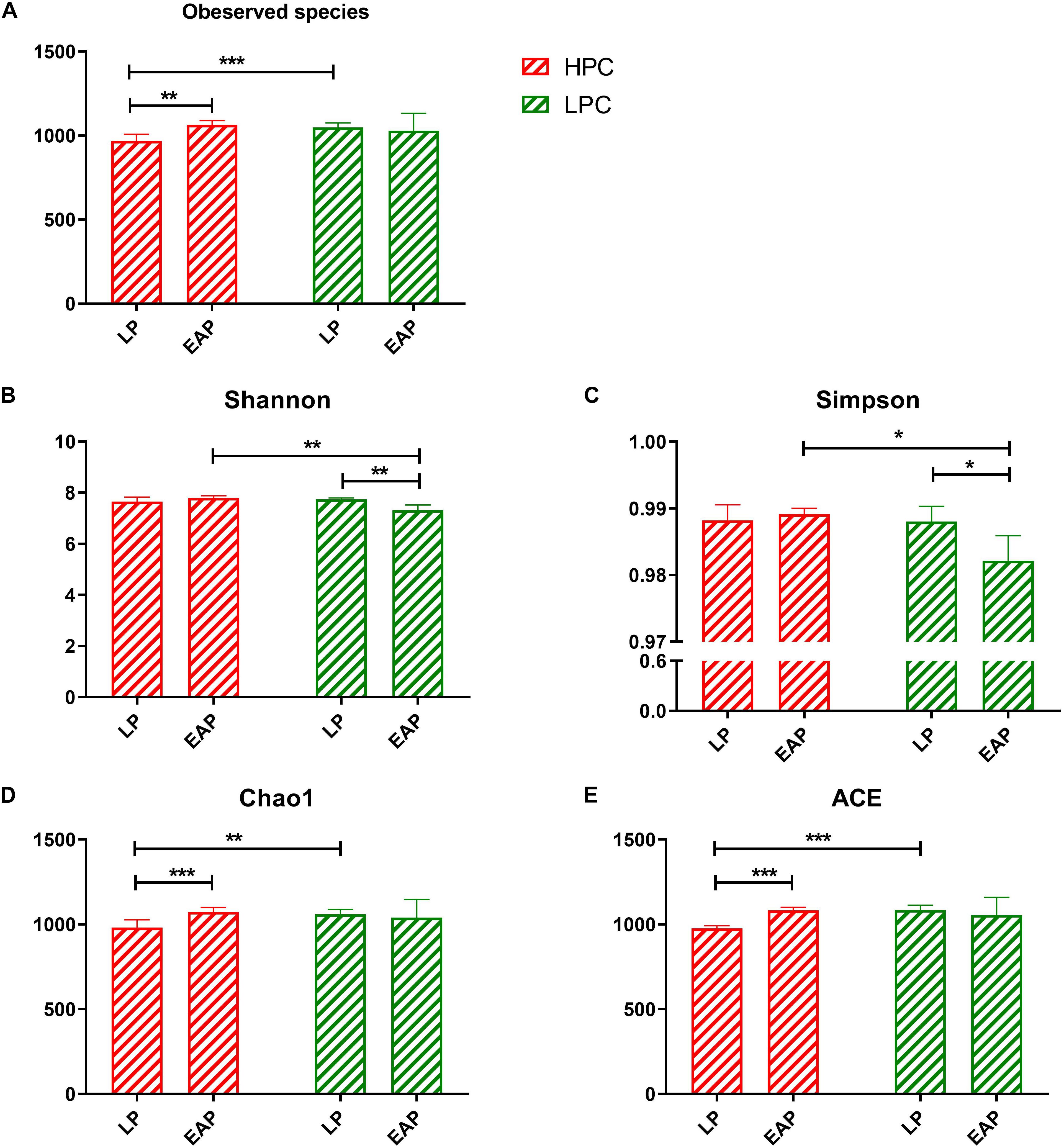
Figure 1. Alpha diversity and richness of gut microbiota from high productive capacity (HPC) and low productive capacity (LPC) sows at late pregnancy (LP) and early stage after parturition (EAP) stage. (A–E) Indices for alpha diversity and richness. Paired T-test (LP vs. EAP) or independent T-test (HPC vs. LPC) was used after accessing normality with Shapiro–Wilk W-test. Data are presented as mean ± SD (n = 6), ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Composition Changes of Gut Microbiota
To evaluate the fecal microbial differences caused by reproductive capacities, we compared β-diversity and composition of the four groups. Results of the PCoA analysis based on Bray–Curtis dissimilarity and non-metric multidimensional scaling (NMDS) showed distinct separation patterns of group A and group B as well as group C and group D (Figures 2A,B), which suggested distinct microbial differences between LPC and HPC sows. Unweighted pair-group method with arithmetic mean (UPGMA) results disclosed that Firmicutes, Bacteroidetes, Proteobacteria, and Spirochaetes were the predominant floras (Figure 2C). The relative abundance of Firmicutes was accounted for at least 50% followed by Bacteroidetes, Proteobacteria and Spirochactes. Heatmap tree was used to show genera differences among groups and their phylogenic relationships (Figure 2D). For HPC sows, gut microbiota was mainly enriched in genera belonging to Prevotellaceae at LP stage and genera belonging to Ruminococcaceae at EAP stage. For LPC sows, genera belonging to Firmicutes (Lactobacillus, Family_XIII_AD3011_group, Streptococcus, Oscillospira) were the predominant microflora at LP stage, while these four genera were decreased at EAP stage.
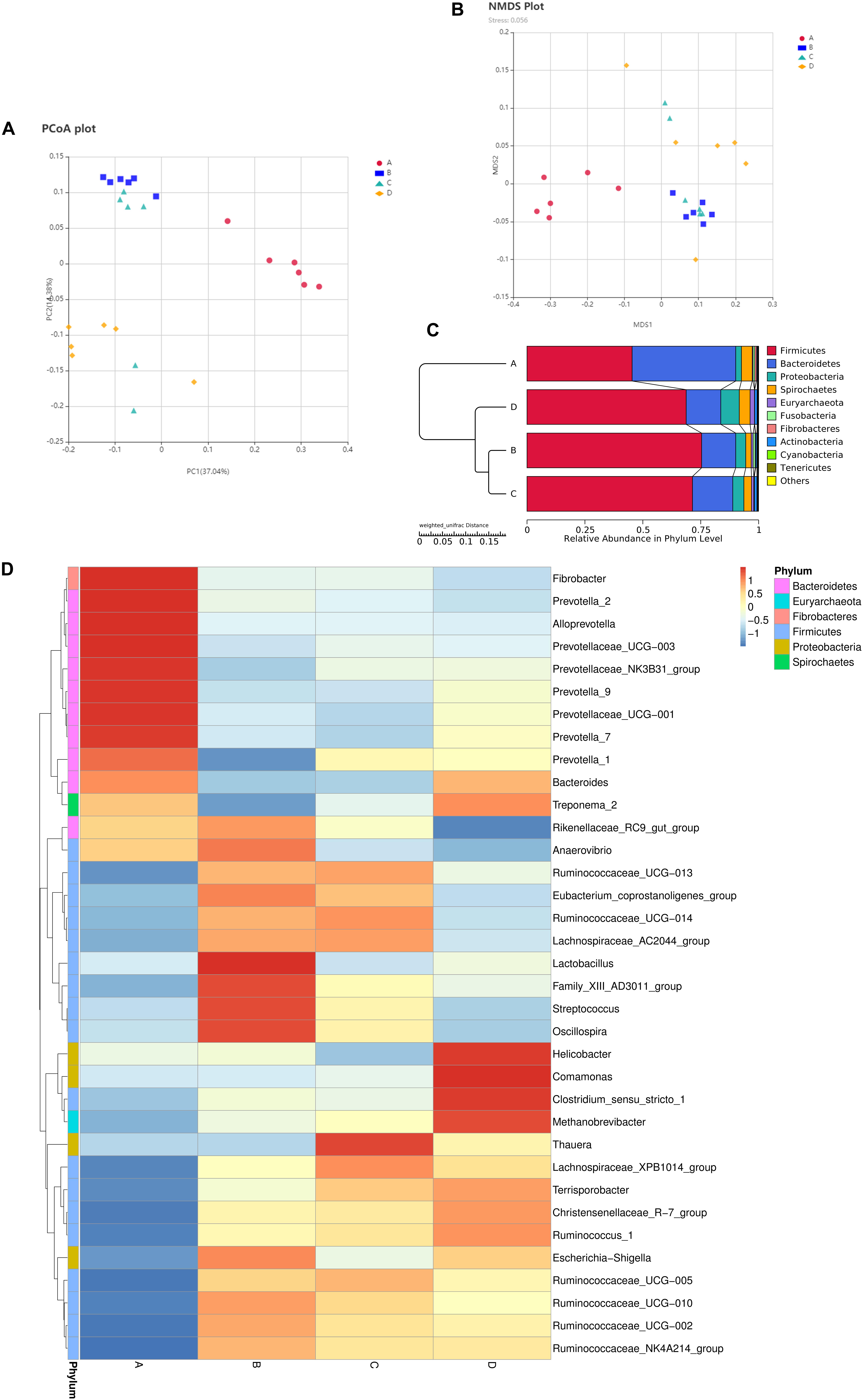
Figure 2. Composition differences of gut microbiota in sows. (A,B) Principal coordinate analysis (PCoA) and non-metric multidimensional scaling (NMDS) analyses. (C) Unweighted pair-group method with arithmetic mean (UPGMA) analysis. (D) Heatmap showing significantly different genera among groups. n = 6. A,C: sows with high productive capacity (HPC) at late pregnancy (LP) and early stage after parturition (EAP) separately; B,D: sows with low productive capacity (LPC) at late pregnancy (LP) and early stage after parturition (EAP) separately.
Differences in gut microflora between groups at the genus level were explored using T-test bar plots. At LP stage, the relative abundance of Prevotellaceae_NK3B31_group, Alloprevotella and Prevotella-2 in HPC sows was significantly higher while the relative abundance of Ruminococcaceae_UCG-005, Ruminococcaceae_UCG-002 and Ruminococcaceae_NK4A214_group was significantly lower than LPC sows (Figure 3A). Further, the relative abundance of Eubacterium_coprostanoligenes_group, Ruminococcaceae_UCG-014, and Phascolarctobacterium in HPC sows was significantly higher than LPC sows at EAP stage (Figure 3B). In addition, gut microbial compositions were analyzed using the linear discriminant analysis (LDA) effect size (LEfSe) method (Figure 4), the results of which coincided with the T-test results, suggesting significant gut microbial differences between sows with different reproductive capacities during different stages of perinatal period.
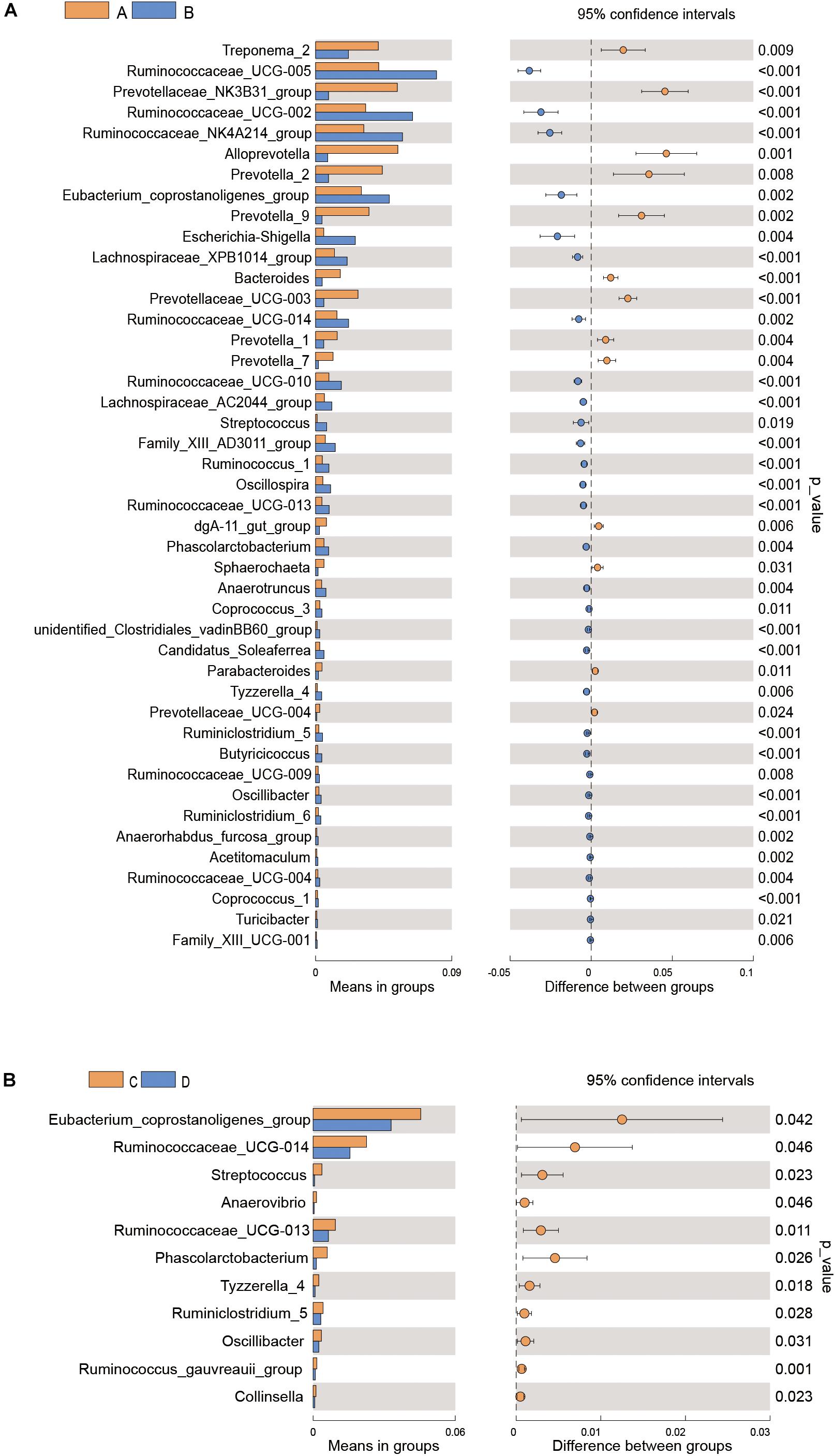
Figure 3. T-test bar plot of significantly different gut microbial species in sows at genus level (A,B). n = 6. A, C: sows with high productive capacity (HPC) at late pregnancy (LP) and early stage after parturition (EAP) separately; B, D: sows with low productive capacity (LPC) at late pregnancy (LP) and early stage after parturition (EAP) separately.
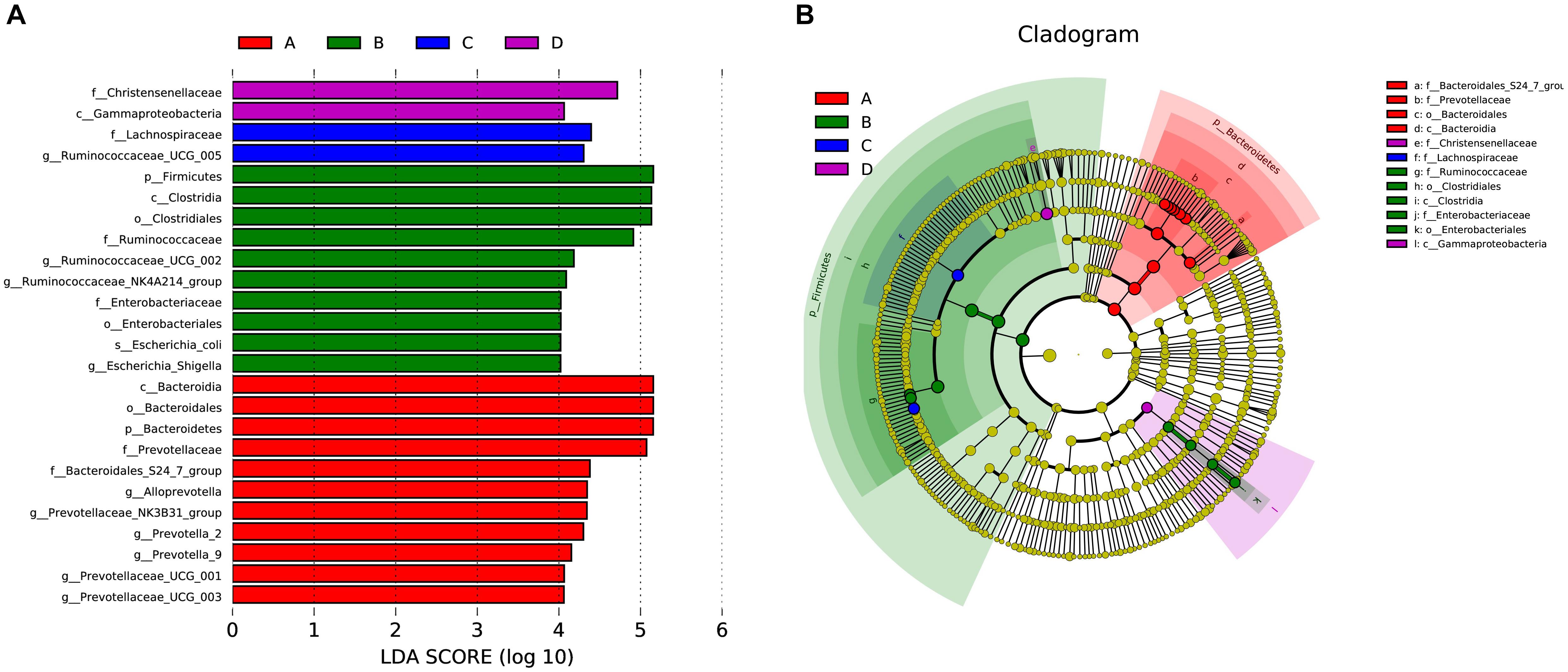
Figure 4. LEfSe analysis of gut microbial composition in sows with different productive capacities during perinatal period; (A) Histogram of the LDA scores, showing the biomarker taxa (LDA score > 4). (B) Cladogram obtained from LEfSe analysis, indicating the phylogenetic distribution of microbiota. n = 6. A, C: sows with high productive capacity (HPC) at late pregnancy (LP) and early stage after parturition (EAP) separately; B, D: sows with low productive capacity (LPC) at late pregnancy (LP) and early stage after parturition (EAP) separately.
Metabolic Functional Changes of Gut Microbiota
Based on the significant differences in bacteria composition, we analyzed metabolic functional changes. PICRUSt was applied to produce metagenome based on 16S rRNA sequencing results at KEGG taxonomy level 3. PCA analysis based on KEGG annotation demonstrated clear clustering between group A and group B as well as group C and group D (Figure 5B). In addition, the heatmap showed the distributions of significantly different functional pathways among groups (Figure 5A), of which differential pathways related to microbial metabolism were selected.
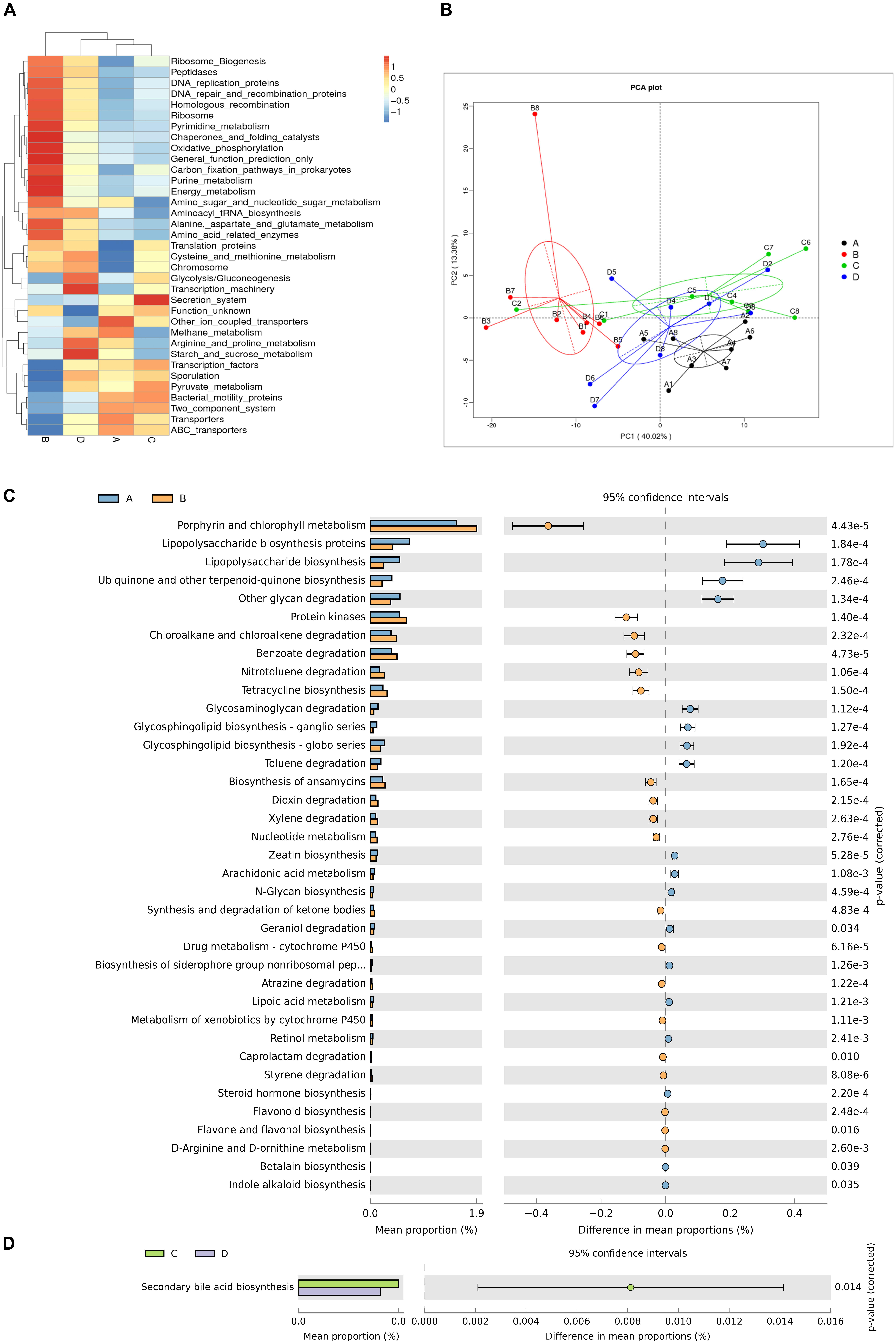
Figure 5. Differences in metabolic functions of gut microbiota. (A) Heatmap showing significantly different functional pathways. (B) Principal components analysis (PCA) plot of functional profiles among groups. (C,D) T-test bar plot of significantly differed metabolic pathways. n = 6. A, C: sows with high productive capacity (HPC) at late pregnancy (LP) and early stage after parturition (EAP) separately; B, D: sows with low productive capacity (LPC) at late pregnancy (LP) and early stage after parturition (EAP) separately.
Microbial gene functions related to metabolic pathways such as Lipopolysaccharide biosynthesis were significantly higher while pathways such as Porphyrin and chlorophyll metabolism and Protein kinases were significantly lower in HPC sows compared to LPC sows at LP stage (Figure 5C). Microbial gene functions related to secondary bile acid biosynthesis were also higher in HPC sows than LPC sows at EAP stage (Figure 5D).
Metabolic Phenotypic Changes in Gut Microbiota
To explore differences in bacterial metabolic phenotypes between HPC and LPC sows, BugBase was used and the results are shown in Figure 6. For gram stain, the relative abundance of gram-negative and gram-positive bacteria was significantly higher and lower (P < 0.01), respectively, in HPC sows than LPC sows at LP stage. Moreover, biofilm formation and potentially pathogenic capacity of HPC sow gut microflora were significantly higher (P < 0.01) than LPC sows at LP stage.
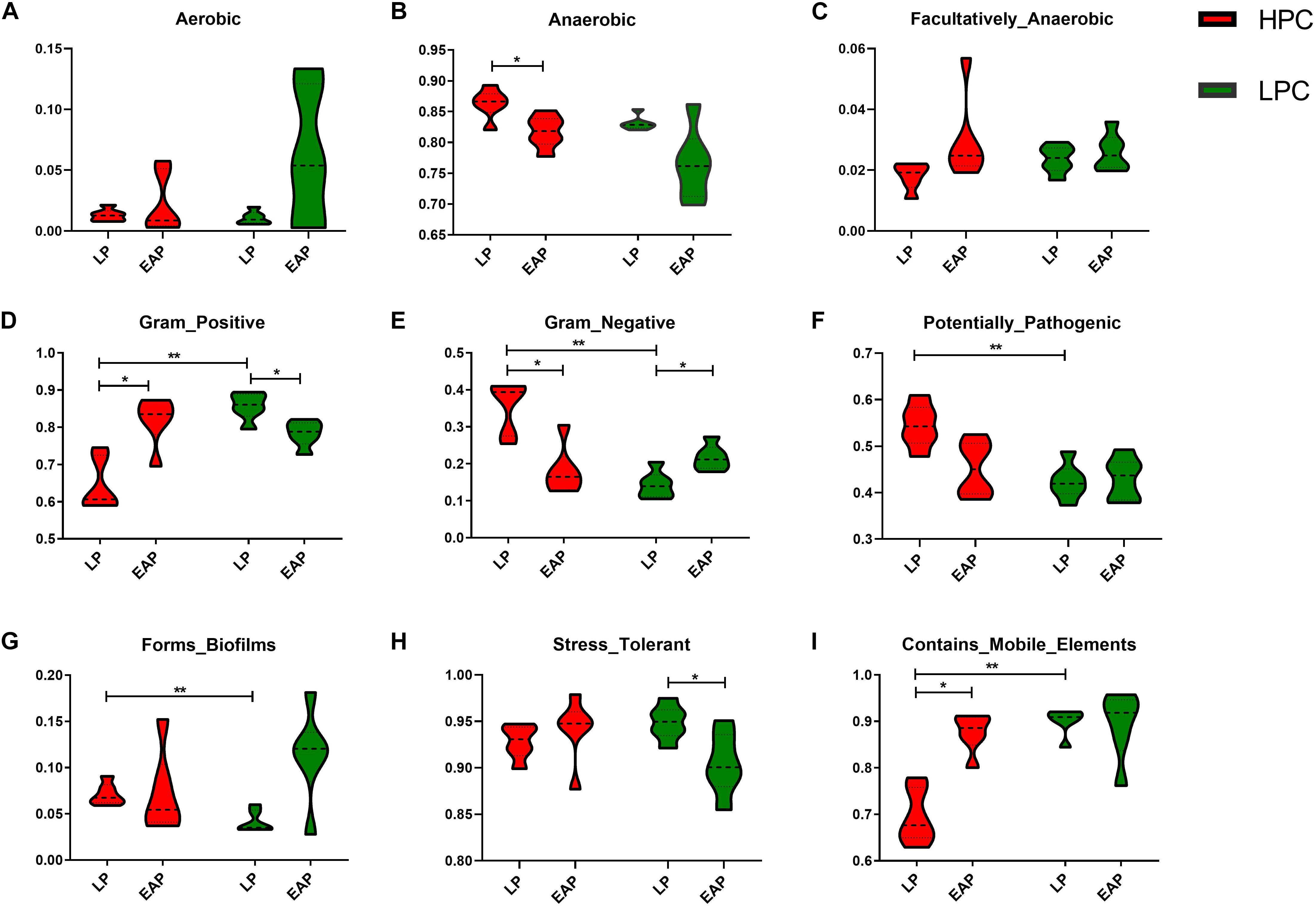
Figure 6. Metabolic phenotypes of gut microbiota from high productive capacity (HPC) and low productive capacity (LPC) sows at late pregnancy (LP) and early stage after parturition (EAP) stage. (A–I) Different indices included in microbial phenotypes. Wilcoxon signed-rank test (LP vs. EAP) or Mann–Whitney U-test (HPC vs. LPC) was applied. ∗P < 0.05, ∗∗P < 0.01 (n = 6).
Serum Biochemical Indices and Correlation With Gut Microbial Abundance in Sows
Gut microbiota participates in the regulation of host’s immunity. Thus, we analyzed serum biochemical indices and their correlations with fecal microbial abundance. R software was used to perform Spearman correlation analysis of gut microbiota at the genus level at EAP stage (Figure 7C). BUN and HDL-C levels of HPC sows were significantly lower than LPC sows after parturition (Figures 7A,B). Further, the BUN level showed negative correlations with some genera belonging to Ruminococcaceae (Ruminococcaceae_UCG-013, r = −0.66, P < 0.05; Ruminococcaceae_UCG-005, r = −0.72, P < 0.01).
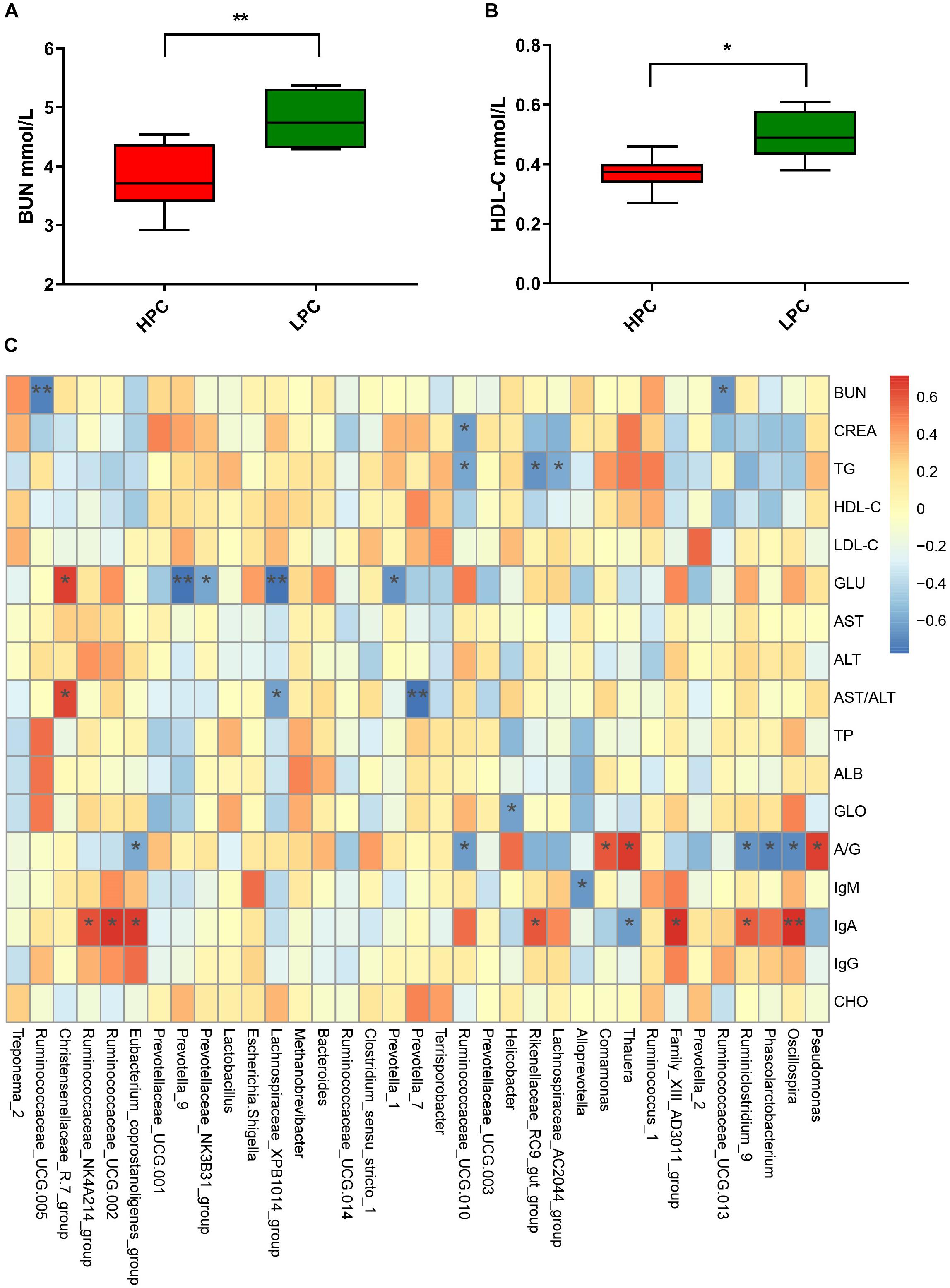
Figure 7. Serum biochemical indices differences in sows and correlations with gut microbial abundance during early stage after parturition. (A) Blood urea nitrogen (BUN, n = 6). (B) Serum high-density lipoprotein cholesterol (HDL-C, n = 6). (C) Heatmap of correlations between gut microbial abundance and serum biochemical indices at genus level. Paired T-test (LP vs. EAP) or independent T-test (HPC vs. LPC) was used after accessing normality with Shapiro–Wilk W-test to analyze serum biochemical indices differences. The correlations between microbial abundance at the genus level and serum biochemical indices were evaluated by Spearman’s correlation analysis. ∗P < 0.05, ∗∗P < 0.01. HPC, high productive capacity; LPC, low productive capacity; BUN, blood urea nitrogen; CREA, creatinine; TG, triglyceride; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; GLU, glucose; AST, aspartate aminotransferase; ALT, alanine aminotransferase; TP, total protein; ALB, albumin; GLO, globulin; A/G, ration of albumin/globulin; CHO, cholesterol.
Discussion
Composition, activity, and coevolution of gut microbiota with hosts are of great importance to animal health (Elson and Alexander, 2015; Woo and Alenghat, 2017). Remarkable changes occur in sow gut microbiota during pregnancy and lactation (Cheng et al., 2018; Liu et al., 2019). However, little is known about whether sows with different reproductive capacities have the same gut microbial performance during perinatal period. Therefore, we first investigated the large gut microbial variances between HPC sows (litter size ≥ 15) and LPC sows (litter size ≤ 7) as well as the correlation with serum biochemical indices during late pregnancy and early stage after parturition.
Bacteroidetes, Firmicutes, Proteobacteria, and Actinobacteria were the most abundant phyla in most mammals (Ley et al., 2008), consistent with our study that Firmicutes and Bacteroidetes were the most abundant phyla in all groups (Figure 2C). In this study, the microbial diversity of HPC sows was significantly higher than LPC sows at EAP stage. Previous study reported that higher diversity of gut microbiota represented greater plasticity in response to perturbations (Bäckhed et al., 2005; Kitano and Oda, 2006) and has been used as a health indicator (Shanahan, 2010). Thus, the significant gut microbial diversity of HPC sows may contribute to effective management of various disturbances during lactation. However, the gut microbiota richness of HPC sows was significantly lower than that in LPC sows at LP stage. Lower microflora richness has been linked to insulin resistance, dyslipidemia, and inflammation (Le Chatelier et al., 2013) and is also a significant marker of gut health (Lozupone et al., 2012). The results indicated that HPC sows might experience greater inflammation at LP stage. However, a pro-inflammatory environment is favorable for the contraction of the uterus, expulsion of the baby, and rejection of the placenta during the late pregnancy (Mor and Cardenas, 2010). Thus, the α-diversity differences might lead to greater plasticity to perturbations and a favorable environment for parturition in HPC sows.
We found that Bacteroidetes and Firmicutes were the most abundant phyla in all groups. The relative abundance of Firmicutes and Bacteroidetes is associated with energy metabolism of their hosts (Ley et al., 2005, 2006; Komaroff, 2017). These two bacteria might enhance intestinal function to meet energy needs for parturition. Compared to LPC sows, HPC sows had greater relative abundance of Bacteroidetes at LP stage and Firmicutes and Bacteroidetes at EAP stage. The differences in microflora might potentially contribute to distinct reproductive capacity due to their functions in energy metabolism. At the genus level, the relative abundance of Alloprevotella, Prevotella-2, and Prevotellaceae_NK3B31_group bacteria in HPC sows was higher while the relative abundance of Ruminococcaceae_UCG-005, Ruminococcaceae_UCG-002 and Ruminococcaceae_NK4A214_group was significantly lower than LPC sows at LP stage. Prevotellaceae is correlated with amino acids, energy and vitamins metabolism (Zhang et al., 2018). Ruminococcaceae is negatively associated with Lipopolysaccharide (LPS) biosynthesis and lower Ruminococcaceae might contribute to inflammatory environment. However, inflammation during LP stage might contribute to the initiation of pregnancy (Mor and Cardenas, 2010; Kang et al., 2017). Differences of Prevotellaceae and Ruminococcaceae might provide better condition for farrowing and promote fetal growth at LP stage. The relative abundance of Eubacterium_coprostanoligenes_group and Ruminococcaceae_UCG-014 in HPC sows was significantly higher than LPC sows at EAP stage (Figure 3B). Eubacterium can produce SCFAs from amino acids and SCFAs possess anti-inflammatory effects (Kanauchi et al., 1999; Tedelind et al., 2007). Scarpa et al. (2011) reported that Eubacteriaceae was negatively related to polymorphonuclear cell and monocyte infiltration. Further, butyrate-producing Ruminococcaceae could reduce LPS biosynthesis (Kang et al., 2017), suggesting that HPC sows might suffer less inflammation at EAP stage, which might contribute to postpartum recovery.
PICRUSt was used to analyze metabolic functional changes. Microbial gene functions related to metabolic pathways such as LPS biosynthesis were significantly higher in HPC sows compared to LPC sows at LP stage. LPS is a component of the cell walls of gram-negative bacteria, which could lead to severe inflammation by upregulating the expression of interleukin-1 and tumor necrosis factor in the lung (Ulich et al., 1991). PICRUSt results indicated that HPC sows suffered greater inflammation than LPC sows. Conversely, LPS plays a role in the adhesion of microflora to gut mucosa (Nevola et al., 1985), and it is important for the activation of immune responses (Takeda and Akira, 2005). Moreover, LPS contributes to a pro-inflammatory environment, which promotes the initiation of parturition (Norman et al., 2007). These results indicated that gut microbiota in HPC sows might lead to a more favorable physiological status for parturition than LPC sows at LP stage. Microbial gene functions related to metabolic pathways such as secondary bile acid biosynthesis in HPC sows were significantly higher compared to LPC sows at EAP stage. Secondary bile acids generated by gut microbial enzymes are important signaling molecules and metabolic regulators to host’s pathways (Valdes et al., 2018). Low concentrations of secondary acids exhibit anti-inflammatory effect by reducing pro-inflammatory cytokines while high concentrations can cause DNA damage, oxidative stress and apoptosis (Ajouz et al., 2014; Ward et al., 2017a). The underlying cause of the secondary bile acids variances between LPC and HPC sows remains to be further elucidated.
There were significant differences in gut microbial metabolic phenotypes between sows with different reproductivity during different periods. The relative abundance of gram-positive and anaerobic bacteria in HPC sows was significantly higher than LPC sows at LP stage, which might be a marker for sows with different reproductivity at LP stage. Further, stress-tolerant bacteria were more abundant in HPC sows than LPC sows at EAP stage, indicating that HPC sows could better handle various stresses after parturition. Further, it has been reported that bacterial genes coding cell surface proteins (including lipopolysaccharide biosynthesis proteins) play a role in biofilm formation (Theunissen et al., 2010). The relative abundance of biofilm forming bacteria in HPC sows was significantly higher than LPC sows at LP stage (Figure 6), which was consistent with more metabolic pathways related to LPS biosynthesis in HPC sows (Figure 5C). Correspondingly, the biofilm formation of bacteria is associated with drug resistance (Stewart and Costerton, 2001). However, the mechanism of biofilm formation differences remains to be further studied. Further, there were higher abundances of potentially pathogenic bacteria in HPC sows than LPC sows, which might lead to a pro-inflammatory environment at LP stage.
Blood urea nitrogen is the final catabolism product of proteins, which could reflect the amino acids balance (Coma et al., 1995). HPC sows had lower BUN levels, indicating that amino acids in HPC sows were more balanced than LPC sows. Further, BUN level showed a negative correlation with Ruminococcaceae. Ruminococcaceae is related to amino acids metabolism (Zhang et al., 2018). Thus, the BUN differences might be caused by Ruminococcaceae. High-density lipoprotein cholesterol is negatively related to cardiovascular disease (Gordon et al., 1989), and it promotes prostaglandin I2 synthetase activity (Beitz and Förster, 1980). HDL-C level decreases after parturition, which may be affected by hormonal, body composition or life-style changes (Lewis et al., 1996). The mechanism underlying the difference in HDL-C levels between groups needs to be further explored.
Conclusion
We found tremendous microbial diversity, composition, metabolic functions, phenotypes and serum indices differences between HPC and LPC sows during perinatal period, especially at the LP stage. Microbial richness was significantly lower at LP stage, while microbial diversity was significantly higher at EAP stage in HPC sows. Additionally, there were also significant differences in BUN and HDL-C levels after parturition. This study discloses great microbial differences between HPC and LPC sows during perinatal period, which might lead to an inflammatory environment at LP stage and an anti-inflammatory environment at EAP stage, and these differences might promote high productive capacity. However, further studies are needed to explain causes of the microbial differences and their relationships with productive capacity.
Data Availability Statement
The datasets generated for this study can be found in the NCBI Sequence Read Archive (No. PRJNA565644).
Ethics Statement
The animal study was reviewed and approved by the Animal Welfare Committee of the Institute of Subtropical Agriculture, Chinese Academy of Sciences.
Author Contributions
JZ, XX, LZ, XK, BT, and YY designed the study. JZ and XX carried out the animal trials and sample analysis. YS, JZ, and XX wrote and revised the manuscript.
Funding
This project was funded by the National Program on Key Basic Research Project (2017YFD0500503), Natural Science Foundation of Hunan Province (2018JJ1028), and Research Foundation of Education Bureau of Hunan Province, China (18B476).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
YS specially thanks XX for her support and guidance. YS acknowledges JZ and Ming Qi for their help in various aspects.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.03047/full#supplementary-material
Footnotes
- ^ http://greengenes.lbl.gov/Download/
- ^ http://sourceforge.net/projects/rdp-classifier/
- ^ https://www.genome.jp/kegg/ko.html
- ^ https://bugbase.cs.umn.edu/
References
Ajouz, H., Mukherji, D., and Shamseddine, A. (2014). Secondary bile acids: an underrecognized cause of colon cancer. World J. Surg. Oncol 12:164. doi: 10.1186/1477-7819-12-164
Alexopoulos, C., Georgoulakis, I., Tzivara, A., Kritas, S., Siochu, A., and Kyriakis, S. (2004). Field evaluation of the efficacy of a probiotic containing Bacillus licheniformis and Bacillus subtilis spores, on the health status and performance of sows and their litters. J. Anim. Physiol. Anim. Nutr. 88, 381–392. doi: 10.1111/j.1439-0396.2004.00492.x
Bäckhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., and Gordon, J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307, 1915–1920. doi: 10.1126/science.1104816
Bäckström, L. (1973). Environment and animal health in piglet production. a field study of incidences and correlations. Acta Vet. Scand. Suppl. 41, 1–240.
Barbour, L. A., McCurdy, C. E., Hernandez, T. L., Kirwan, J. P., Catalano, P. M., and Friedman, J. E. (2007). Cellular mechanisms for insulin resistance in normal pregnancy and gestational diabetes. Diabetes Care (Suppl. 2):S112-9. doi: 10.2337/dc07-s202
Beitz, J., and Förster, W. (1980). Influence of human low density and high density lipoprotein cholesterol on the in vitro prostaglandin I2 synthetase activity. Biochim. Biophys. Acta 620, 352–355. doi: 10.1016/0005-2760(80)90126-5
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Cao, M., Li, Y., Wu, Q. J. J., Zhang, P., Li, W. T. T., Mao, Z. Y. Y., et al. (2019). Effects of dietary Clostridium butyricum addition to sows in late gestation and lactation on reproductive performance and intestinal microbiota. J. Anim. Sci. 97, 3426–3439. doi: 10.1093/jas/skz186
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cheng, C., Wei, H., Yu, H., Xu, C., Jiang, S., and Peng, J. (2018). Metabolic syndrome during perinatal period in sows and the link with gut microbiota and metabolites. Front. Microbiol. 9:1989. doi: 10.3389/fmicb.2018.01989
Chu, H., and Mazmanian, S. K. (2013). Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat. Immunol. 14, 668–675. doi: 10.1038/ni.2635
Coma, J., Carrion, D., and Zimmerman, D. R. (1995). Use of plasma urea nitrogen as a rapid response criterion to determine the lysine requirement of pigs. J. Anim. Sci. 73, 472–481. doi: 10.2527/1995.732472x
Desantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/aem.03006-05
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Elson, C. O., and Alexander, K. L. (2015). Host-microbiota interactions in the intestine. Dig. Dis. 33, 131–136. doi: 10.1159/000369534
Gordon, D. J., Probstfield, J. L., Garrison, R. J., Neaton, J. D., Castelli, W. P., Knoke, J. D., et al. (1989). High-density lipoprotein cholesterol and cardiovascular disease. four prospective American studies. Circulation 79, 8–15. doi: 10.1161/01.cir.79.1.8
Holyoake, P. K., Dial, G. D., Trigg, T., and King, V. L. (1995). Reducing pig mortality through supervision during the perinatal period. J. Anim. Sci. 73, 3543–3551.
Huang, X., Gao, J., Zhao, Y., He, M., Ke, S., Wu, J., et al. (2019). Dramatic remodeling of the gut microbiome around parturition and its relationship with host serum metabolic changes in sows. Front. Microbiol. 10:2123. doi: 10.3389/fmicb.2019.02123
Jarvis, S., Lawrence, A. B., Mclean, K. A., Deans, L. A., Chirnside, J., Calvert, S. K., et al. (2010). The effect of piglet expulsion in the sow on plasma cortisol, adrenocorticotropic hormone and β-endorphin. Zuchthygiene 34, 89–94. doi: 10.1046/j.1439-0531.1999.00174.x
Kanauchi, O., Fujiyama, Y., Mitsuyama, K., Araki, Y., and Bamba, T. (1999). Increased growth of Bifidobacterium and Eubacterium by germinated barley foodstuff, accompanied by enhanced butyrate production in healthy volunteers. Int. J. Mol. Med. 3, 175–179.
Kang, C., Wang, B., Kaliannan, K., Wang, X., Lang, H., Hui, S., et al. (2017). Gut microbiota mediates the protective effects of dietary capsaicin against chronic low-grade inflammation and associated obesity induced by high-fat diet. mBio 8:e470-17. doi: 10.1128/mBio.00470-17
Kim, S. W., Mateo, R. D., Yin, Y.-L., and Wu, G. (2007). Functional amino acids and fatty acids for enhancing production performance of sows and piglets. Asian-Australas J. Anim. Sci. 20, 295–306. doi: 10.5713/ajas.2007.295
Kitano, H., and Oda, K. (2006). Robustness trade-offs and host-microbial symbiosis in the immune system. Mol. Syst. Biol. 2:2006.0022. doi: 10.1038/msb4100039
Koren, O., Goodrich, J. K., Cullender, T. C., Spor, A., Laitinen, K., Backhed, H. K., et al. (2012). Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 150, 470–480. doi: 10.1016/j.cell.2012.07.008
Langille, M. G. I., Jesse, Z., Gregory, J. C., Daniel, M. D., Dan, K., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Lawrence, A. B., Petherick, J. C., Mclean, K., Gilbert, C. L., Chapman, C., and Russell, J. A. (1992). Naloxone prevents interruption of parturition and increases plasma oxytocin following environmental disturbance in parturient sows. Physiol. Behav. 52, 917–923. doi: 10.1016/0031-9384(92)90371-8
Le Chatelier, E., Nielsen, T., Qin, J., Prifti, E., Hildebrand, F., Falony, G., et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. doi: 10.1038/nature12506
Lewis, C. E., Funkhouser, E., Raczynski, J. M., Sidney, S., Bild, D. E., and Howard, B. V. (1996). Adverse effect of pregnancy on High Density Lipoprotein (HDL) cholesterol in young adult women: the CARDIA study. Am. J. Epidemiol. 144, 247–254. doi: 10.1093/oxfordjournals.aje.a008919
Ley, R. E., Bäckhed, F., Turnbaugh, P., Lozupone, C. A., Knight, R. D., and Gordon, J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102, 11070–11075. doi: 10.1073/pnas.0504978102
Ley, R. E., Micah, H., Catherine, L., Turnbaugh, P. J., Rob Roy, R., Stephen, B. J., et al. (2008). Evolution of mammals and their gut microbes. Science 320, 1647–1651. doi: 10.1126/science.1155725
Ley, R. E., Turnbaugh, P. J., Klein, S., and Gordon, J. I. (2006). Human gut microbes associated with obesity. Nature 444, 1022–1023. doi: 10.1038/4441022a
Linzer, D. I., and Fisher, S. J. (1999). The placenta and the prolactin family of hormones: regulation of the physiology of pregnancy. Mol. Endocrinol. 13, 837–840. doi: 10.1210/mend.13.6.0286
Liu, H., Hou, C., Li, N., Zhang, X., Zhang, G., Yang, F., et al. (2019). Microbial and metabolic alterations in gut microbiota of sows during pregnancy and lactation. FASEB J. 33, 4490–4501. doi: 10.1096/fj.201801221RR
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., and Knight, R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. doi: 10.1038/nature11550
Marek, S., Roman, D. B., Mariola, B., and Marcin, K. (2013). The influence of the duration of the expulsive stage of parturition on the occurrence of postpartum oxidative stress in sows with uncomplicated, spontaneous farrowings. Theriogenology 80, 706–711. doi: 10.1016/j.theriogenology.2013.05.015
Maslowski, K. M., and Mackay, C. R. (2011). Diet, gut microbiota and immune responses. Nat. Immunol. 12, 5–9. doi: 10.1038/ni0111-5
Maynard, C. L., Elson, C. O., Hatton, R. D., and Weaver, C. T. (2012). Reciprocal interactions of the intestinal microbiota and immune system. Nature 489, 231–241. doi: 10.1038/nature11551
Mor, G., and Cardenas, I. (2010). REVIEW ARTICLE: the immune system in pregnancy: a unique complexity. Am. J. Reprod. Immunol. 63, 425–433. doi: 10.1111/j.1600-0897.2010.00836.x
Nevola, J. J., Stocker, B. A., Laux, D. C., and Cohen, P. S. (1985). Colonization of the mouse intestine by an avirulent Salmonella typhimurium strain and its lipopolysaccharide-defective mutants. Infect. Immun. 50, 152–159.
Norman, J. E., Bollapragada, S., Yuan, M., and Nelson, S. M. (2007). Inflammatory pathways in the mechanism of parturition. BMC Pregnancy Childbirth 7:S7. doi: 10.1186/1471-2393-7-S1-S7
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Père, M.-C., and Etienne, M. (2007). Insulin sensitivity during pregnancy, lactation, and postweaning in primiparous gilts. J. Anim. Sci. 85, 101–110. doi: 10.2527/jas.2006-130
Qiong, W., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/aem.00062-07
Ringseis, R., Gessner, D. K., and Eder, K. (2019). The gut-liver axis in the control of energy metabolism and food intake in animals. Annu. Rev. Anim. Biosci. doi: 10.1146/annurev-animal-021419-083852 [Epub ahead of print].
Rothschild, D., Weissbrod, O., Barkan, E., Kurilshikov, A., Korem, T., Zeevi, D., et al. (2018). Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215. doi: 10.1038/nature25973
Santacruz, A., Collado, M. C., Garcia-Valdes, L., Segura, M. T., Martin-Lagos, J. A., Anjos, T., et al. (2010). Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br. J. Nutr. 104, 83–92. doi: 10.1017/S0007114510000176
Sarkar, A., Harty, S., Lehto, S. M., Moeller, A. H., Dinan, T. G., Dunbar, R. I. M., et al. (2018). The microbiome in psychology and cognitive neuroscience. Trends Cogn. Sci. 22, 611–636. doi: 10.1016/j.tics.2018.04.006
Scarpa, M., Grillo, A., Faggian, D., Ruffolo, C., Bonello, E., D’Incà, R., et al. (2011). Relationship between mucosa-associated microbiota and inflammatory parameters in the ileal pouch after restorative proctocolectomy for ulcerative colitis. Surgery 150, 56–67. doi: 10.1016/j.surg.2011.02.009
Schoeler, M., and Caesar, R. (2019). Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Dis. doi: 10.1007/s11154-019-09512-0 [Epub ahead of print].
Shanahan, F. (2010). Probiotics in perspective. Gastroenterology 139, 1808–1812. doi: 10.1053/j.gastro.2010.10.025
Stewart, P. S., and Costerton, J. W. (2001). Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138. doi: 10.1016/s0140-6736(01)05321-1
Takeda, K., and Akira, S. (2005). Toll-like receptors in innate immunity. Int. Immunol. 17, 1–14. doi: 10.1093/intimm/dxh186
Tanja, M., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Tedelind, S., Westberg, F., Kjerrulf, M., and Vidal, A. (2007). Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: a study with relevance to inflammatory bowel disease. World J. Gastroenterol. 13, 2826–2832. doi: 10.3748/wjg.v13.i20.2826
Theunissen, S., De Smet, L., Dansercoer, A., Motte, B., Coenye, T., Van Beeumen, J. J., et al. (2010). The 285 kDa Bap/RTX hybrid cell surface protein (SO4317) of Shewanella oneidensis MR-1 is a key mediator of biofilm formation. Res. Microbiol. 161, 144–152. doi: 10.1016/j.resmic.2009.12.002
Tsukahara, T., Inatomi, T., Otomaru, K., Amatatsu, M., Romero-Pérez, G. A., and Inoue, R. (2018). Probiotic supplementation improves reproductive performance of unvaccinated farmed sows infected with porcine epidemic diarrhea virus. Anim. Sci. J. 89, 1144–1151. doi: 10.1111/asj.13040
Ulich, T. R., Watson, L. R., Yin, S. M., Guo, K. Z., Wang, P., Thang, H., et al. (1991). The intratracheal administration of endotoxin and cytokines.1. Characterization of LPS-induced IL-1 and TNF messenger-RNA expression and the LPS-induced, IL-1-induced, and TNF-induced inflammatory infiltrate. Am. J. Pathol. 138, 1485–1496.
Valdes, A. M., Walter, J., Segal, E., and Spector, T. D. (2018). Role of the gut microbiota in nutrition and health. BMJ 361:k2179. doi: 10.1136/bmj.k2179
van de Ligt, J., Lindemann, M., Harmon, R., Monegue, H. J., and Cromwell, L. G. (2002). Effect of chromium tripicolinate supplementation on porcine immune response during the postweaning period. J. Anim. Sci. 80, 449–455. doi: 10.2527/2002.802449x
Verheyen, A. J. M., Maes, D. G. D., Mateusen, B., Deprez, P., Janssens, G. P. J., Lange, L. D., et al. (2007). Serum biochemical reference values for gestating and lactating sows. Vet. J. 174, 92–98. doi: 10.1016/j.tvjl.2006.04.001
Ward, J. B. J., Lajczak, N. K., Kelly, O. B., O’Dwyer, A. M., Giddam, A. K., Ni Gabhann, J., et al. (2017a). Ursodeoxycholic acid and lithocholic acid exert anti-inflammatory actions in the colon. Am. J. Physiol. Gastrointest. Liver Physiol. 312, G550–G558. doi: 10.1152/ajpgi.00256.2016
Ward, T., Larson, J., Meulemans, J., Hillmann, B., Lynch, J., Sidiropoulos, D., et al. (2017b). BugBase predicts organism level microbiome phenotypes. BioRxiv [preprint]. doi: 10.1101/133462
Woo, V., and Alenghat, T. (2017). Host–microbiota interactions: epigenomic regulation. Curr. Opin. Immunol. 44, 52–60. doi: 10.1016/j.coi.2016.12.001
Keywords: gut microbiota, productive capacity, perinatal period, sows, serum immunity
Citation: Shao Y, Zhou J, Xiong X, Zou L, Kong X, Tan B and Yin Y (2020) Differences in Gut Microbial and Serum Biochemical Indices Between Sows With Different Productive Capacities During Perinatal Period. Front. Microbiol. 10:3047. doi: 10.3389/fmicb.2019.03047
Received: 19 September 2019; Accepted: 18 December 2019;
Published: 17 January 2020.
Edited by:
Hua Xiang, Institute of Microbiology (CAS), ChinaReviewed by:
Xiangfang Zeng, China Agricultural University (CAU), ChinaYulan Liu, Wuhan Polytechnic University, China
Copyright © 2020 Shao, Zhou, Xiong, Zou, Kong, Tan and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xia Xiong, eHhAaXNhLmFjLmNu; Xiangfeng Kong, bm5reGZAaXNhLmFjLmNu
†These authors have contributed equally to this work
 Yirui Shao
Yirui Shao Jian Zhou
Jian Zhou Xia Xiong
Xia Xiong Lijun Zou
Lijun Zou Xiangfeng Kong
Xiangfeng Kong Bie Tan
Bie Tan Yulong Yin
Yulong Yin