- 1Key Laboratory of Agro-ecological Processes in Subtropical Regions and Taoyuan Station of Agro-ecology Research, Institute of Subtropical Agriculture, Chinese Academy of Sciences, Changsha, China
- 2School of Minerals Processing and Bioengineering, Central South University, Changsha, China
- 3Key Laboratory of Biometallurgy, Ministry of Education, Central South University, Changsha, China
- 4Changsha Folianovo Biotechnology Co. Ltd., Changsha, China
The introduction of Acidithiobacillus thiooxidans A01 strengthens the positive interactions between physiologically distinct microorganisms and enhances the bioleaching ability of the consortium. However, the effect of introducing an exogenous strain, A. thiooxidans A01 on the structure and function of the adsorbed and planktonic microbial consortia during bioleaching of low-grade copper sulfide remains unclear. In this study, A. thiooxidans A01 was introduced into an indigenous leaching microbial community on the 0th (group B), 24th (group C), and 36th day (group D). Results revealed that the copper leaching efficiency was highest in group D, in which the Cu2+ concentration in the solution reached 251.5 mg/L on day 48, which was 18.5% higher than that of the control (group A, no addition of A. thiooxidans A01). Restriction fragment length polymorphism (RFLP) analysis of the microbial community in group D revealed the presence of Leptospirillum ferriphilum, Acidithiobacillus ferrooxidans, Acidithiobacillus caldus, Sulfobacillus sp., Acidiphilium spp., and Acidithiobacillus albertensis before introduction of A. thiooxidans A01 on the 36th or 48th day; however, A. albertensis was absent on day 48 in group A. Further, the proportion of dominant A. caldus, L. ferriphilum, and A. ferrooxidans became altered. The results of real-time PCR in group D showed that A. thiooxidans A01 was primarily adsorbed on the surface of the ore, with the adsorption reaching the maxima on day 42; while the free A. thiooxidans A01 in solution grew slowly, reaching its maximum concentration on day 45. Compared with that in the control group, the abundance of both free and attached A. caldus and Sulfobacillus sp. decreased following the introduction of A. thiooxidans A01, while that of L ferriphilum, A. ferrooxidans, and Acidiphilium sp. increased. Functional gene arrays data indicated that the abundance of genes involved in sulfide and iron oxidation in L. ferriphilum and A. ferrooxidans, as well as that of the metal (loid) resistance genes of A. ferrooxidans, L. ferriphilum, and Acidiphilium sp. increased, while the abundance of genes involved in sulfur metabolism in A. caldus and Sulfolobus spp. decreased. Taken together, these results provide useful information for application of bioleaching of copper sulfide in industry.
Introduction
Surface water and groundwater are often polluted in the process of mineral exploitation, primarily because sulfide dissolution leads to production of acidic water. These wastewaters often contain high concentration of elements such as copper, zinc, iron, sulfur, and arsenic, and thus called acid mine drainage (AMD). Many extremely eosinophilic microorganisms have been isolated from such acidic environments (Baker and Banfield, 2003; Akcil and Koldas, 2006; Cheng et al., 2009; Marchevsky et al., 2015). Among these microorganisms, different types of interactions such as competition, predation, mutualism, and synergy, have been described (Johnson, 1998). However, many studies have indicated that mixed cultures containing a variety of microorganisms tend to be more robust and more efficient in oxidizing sulfide minerals due to their extensive interactions (Akcil et al., 2007; Zhang et al., 2008; Deng et al., 2017). Further, the ecological function of the microbial community was often associated with its structure and diversity. On the one hand, the unique original microbial compositions may lead to distinct bioleaching efficiencies (Bacelar-Nicolau and Johnson, 1999; Zhang et al., 2007). However, on the other hand, many environmental factors, such as temperature and pH, may result in changes within the community structure and diversity during the bioleaching process (Davis-Belmar et al., 2012), subsequently resulting in varied dissolution rates at different stages (Mukherjee et al., 2004; Demergasso et al., 2005; Johnson, 2008).
In recent years, the continuous development of genome tools has promoted further analysis of microorganisms in complex environments. To study the structure and function of microbial community, functional gene arrays (FGAs), also termed the GeoChip, have been developed and proven to be a useful genomic technology for the thorough investigation of environmental and bioleaching processes (Mukherjea, 1971; Rodrìguez et al., 2003; Schadt et al., 2004; Yin et al., 2007; He et al., 2008, 2011, 2012). In our previous study (Liu et al., 2011b), we artificially constructed multiple co-culture systems of indigenous leaching microbial communities; introduced different types of leaching bacteria in different growth periods of indigenous communities; and studied the introduction mechanism at the gene, cell, and community levels. However, we encountered certain limitations. Firstly, the selected indigenous leaching microbial community was an artificial co-culture system. Generally, natural indigenous leaching microbial communities have a more stable structure and higher diversity (Demergasso et al., 2005; Huang et al., 2018). Secondly, the selected mineral was high purity pyrite not chalcopyrite. Chalcopyrite (CuFeS2) accounts for ∼70% of the Earth’s copper and is one of the most wide-spread copper sulfide minerals (Lu et al., 2016). However, the Cu extraction from chalcopyrite through acid leaching is an extremely time-consuming process with low efficiency (Ma et al., 2017). Therefore, it has greater practical significance and value to study introduction mechanisms in the leaching system of low-grade chalcopyrite with indigenous leaching microbial communities.
A leaching system comprises planktonic and adsorbed bacteria (Chen et al., 2017). However, studies on microbial ecology in leaching systems have primarily focused on planktonic microorganisms in leaching solutions, with little knowledge of the microbial ecology of mineral surface adsorption (Klocke et al., 2007; Nkulu et al., 2015). According to previous studies (Sand and Gehrke, 2006; Chen et al., 2008), the metal ion and organic matter content on the mineral surface during the bioleaching process is much higher than that in the leaching solution; indicating that the growth environment of microorganisms on the mineral surface may be quite different from that in the leaching solution. Moreover, the optimal introduction period can differ with the stage of the planktonic microbial community growth period. Therefore, it is of great significance to study the dynamic changes occurring within the community structure and functional genes in the two different growth environments, while also defining the optimal introduction period.
In this study, we selected natural microbial communities as indigenous microbial communities (Liu et al., 2011a). In the low-grade copper sulfide bioleaching system, A. thiooxidans A01 was introduced during the stable growth of planktonic and adsorbed microbial communities. Real-time polymerase chain reaction (RT-PCR), restriction fragment length polymorphism (RFLP), and FGAs were used to identify the indigenous populations before and after A. thiooxidans A01 introduction. The results underscore the complexity of the bioleaching processes and provide a novel approach to enhance the bioleaching efficiency of an indigenous consortium that may be useful in the bioleaching of low-grade copper sulfide.
Materials and Methods
Microorganism Preparation
The microbial consortium used herein has been described in our previous study (Liu et al., 2011b). In brief, the acclimation experiments consisted of an equal mixture of several AMD samples from different copper ores; these experiments were carried out in 0K basal medium containing 0.5% chalcopyrite. The indigenous consortium was obtained in this study after successive subculturing of five generations, with one generation developing every 60 days. A. thiooxidans A01 strain (FJ154526), an important member of genus Acidithiobacillus, was isolated from the AMD of the Dexing copper mine, China. This organism is an obligate sulfur oxidizer with strong sulfur-oxidizing properties; however, it is unable to utilize ferrous iron as an energy source (Yin et al., 2014). A. thiooxidans A01 was also acclimated for five generations in 0K medium containing 0.5% chalcopyrite to adapt the bioleaching system of chalcopyrite before commencing the bioleaching study. The strain was cultivated with elemental sulfur as the sole energy source at pH 2.0, 30°C, and 170 rpm, and subcultured every 2 weeks. The adapted A. thiooxidans A01 was used as the exogenous strain in this study. The 0K basal medium (g/L) is composed of (NH4)2SO4 (3), K2HPO4 (0.5), KCl (0.1), Ca(NO3)2 (0.01), and MgSO4⋅7H2O (0.5).
Mineral Components
The low-grade chalcopyrite used in this bioleaching experiment was obtained from Heilongjiang Province, China. The mineral sample was ground and sieved to obtain a particle size <75 μm. The chemical compositions of copper sulfide ore were analyzed by X-ray fluorescence (XRF), and the result indicated that the contents of Cu, Mo, CaO, and MgO were 0.51, 0.011, 4.63, and 1.29%, respectively. The contents of Ag and Au were 12 and 0.2 g/t, respectively. Mineral phases of copper in low-grade copper sulfide ore were determined by wet chemical analysis according to “DZG20.01-1991” (petromineralogy analysis) (Chun et al., 2016), and the results showed that this copper sulfide presented in the form of primary copper sulfide (74.5%), secondary copper sulfide (23.5%), free copper oxide (1.18%), and combined copper oxide (0.78%). The total content of metallic minerals was 5.71%, and the main metallic minerals included digenite and chalcopyrite, among which the total content of digenite was 0.78% and that of chalcopyrite was 4.85%. From the perspective of gangue minerals, it was mainly quartz, with a total content of 54.2%, including 36.1% alum and 12.5% other mineral elements.
Bioleaching Experiments
The bioleaching experiments were conducted in 250 mL shake flasks containing 100 mL of autoclaved 0K basal medium with 5% (w/v) ultraviolet-sterilized chalcopyrite sample at 170 rpm for 48 days. The experiment included two parts.
Part I sought to determine the most effective introduction period. The cultivated indigenous consortium and A. thiooxidans A01 were, respectively, centrifuged at 10,000 rpm/min for 5 min, and the cell pellet was inoculated into medium at a final cell density of 107 cell/mL. A. thiooxidans A01 was introduced into the indigenous consortium on days 0, 24, and 36, respectively (groups B, C, and D). Flasks without the introduction of A. thiooxidans A01 were used as a control (group A) and uninoculated flasks were used as abiotic controls. The introduction time was determined according to the 48 days of pre-experiment, in which the concentration of microorganisms adsorbed on the ore surface reached the maximum on the 24th day, while the concentration of microorganisms in solution reached the maximum on the 36th day (data not shown). During the bioleaching experiment, the temperature was adjusted every 8 h, and the circulating temperature was 25, 35, and 45°C, respectively. The circulating temperature was applied since the optimum growth temperature of microorganisms was diverse; hence, varied temperatures may allow the growth of a relatively rich microbial community. In the first 3 days of bioleaching, the pH was daily adjusted to 2.0 using hydrochloric acid and was no longer adjusted once a stable pH was maintained. Samples were withdrawn every 6 days to determine the concentration of copper and ferrous iron in the solution. The evaporated water and sampling loss were supplemented with sterilized distilled water periodically. All experiments were performed in triplicate.
Part II: Based on the experimental results from part I, introduction of A. thiooxidans A01 on the 36th day was identified as the most effective introduction period (group D). To analyze the cause for the improved bioleaching efficiency, we determined the microbial community structure, population dynamics, and community functional gene abundance before and after the introduction of A. thiooxidans A01 on the 36th day. A total of 39 flasks were inoculated with indigenous consortium (group A), 18 of which were introduced with A. thiooxidans A01 on day 36 (group D) as described in part I. The DNA from adsorbed and planktonic bacteria in groups A and D were extracted separately on days 38, 40, 42, 45, and 48. Concurrently, the total microbial DNA (including adsorbed and planktonic microorganisms) was extracted from group A on day 36 (before the introduction of A. thiooxidans A01) and from groups A and D on day 48 at the end of the bioleaching process. DNA samples from pure A. thiooxidans A01 were also extracted. All experiments were performed in triplicate with three flasks per sampling point. The pH and oxidation-redox potential (ORP) of group A and group D were also determined on days 6, 12, 18, 24, 30, 36, 38, 40, 42, 45, and 48.
Analytical Techniques
Planktonic bacteria were enumerated by direct counting using a Neubauer chamber hemocytometer. Copper and ferrous iron concentration in solution were determined by inductively coupled plasma-atomic emission spectrometry [ICP-AES: Baird Plasma Spetrovate PS-6 (N+1)] and o-phenanthroline spectrophotometric method, respectively. The pH and ORP value were determined using a pH meter (PHSJ-4A, Leici, Shanghai, China) and a platinum electrode with an Ag/AgCl reference electrode (218, Leici, Shanghai, China).
Suspended and adsorbed microorganisms were conducted as described previously (Zeng et al., 2010). The DNA samples from A. thiooxidans A01, groups A, B, C, and D were extracted using a TIANamp Bacteria DNA Kit (Tiangen Biotech, Co., Ltd., Beijing, China). The 16S rRNA gene amplification and cloning of consortium A and D on days 36 and 48 were conducted to analyze the microbial community following RFLP analysis. Based on the RFLP profile, a total of 16 representative clones were selected for sequencing. DNA sequence identification was executed using BLAST in GenBank1. Microarray hybridization was used to assess population dynamics and functional gene abundance of the microbial community. Real-time quantification polymerase chain reaction (RT-qPCR) was conducted to analyze the population dynamics of A. thiooxidans A01. All methods were detailed in our previous study (Liu et al., 2011a). The 16S rRNA sequences were submitted to GenBank, and the accession numbers assigned were JF313219, JF313232-313244, JF429148, and FJ154514.
Results
Bioleaching of Low-Grade Copper Sulfide With Consortium A, B, C, and D
Figure 1A depicted the copper concentration in the different groups during the bioleaching experiment. Copper concentration was highest in consortium D (251.5 mg/L) after 48 days, followed by consortium C (230.8 mg/L) and consortium B (215.1 mg/L). Copper content was lowest in group A (204.3 mg/L). Figure 1B showed the copper recovery (%) throughout the entire bioleaching period. The copper recovery was found to gradually increase from 20% on the 6th day to approximately 90% at the end of bioleaching. Following the introduction of A. thiooxidans A01 at different growth stages of the indigenous leaching microbial community, copper extraction was observed to improve to varying degrees compared with that in the control, most notably during the stable growth period (up to 98.6%). The copper recovery in groups B, C, and D was 4.3, 10.4, and 18.5% higher than that in group A. Figure 1C displayed the variation in ferrous iron concentration in solution during the bioleaching experiment. The ferrous content of the four groups increased from 629 mg/L on the 6th day to 1,340 mg/L on day 18, after which it began to decrease. The ferrous concentration of group B was slightly higher than other groups in the earlier period and was seen to decline slightly faster than group A in the later period. After introduction of A. thiooxidans A01 on day 24, a sudden descent of the ferrous concentration appeared in group C, and the ferrous concentration on day 48 reduced to 95 mg/L. The ferrous concentration in group D decreased after 36 days and finally trended toward zero.
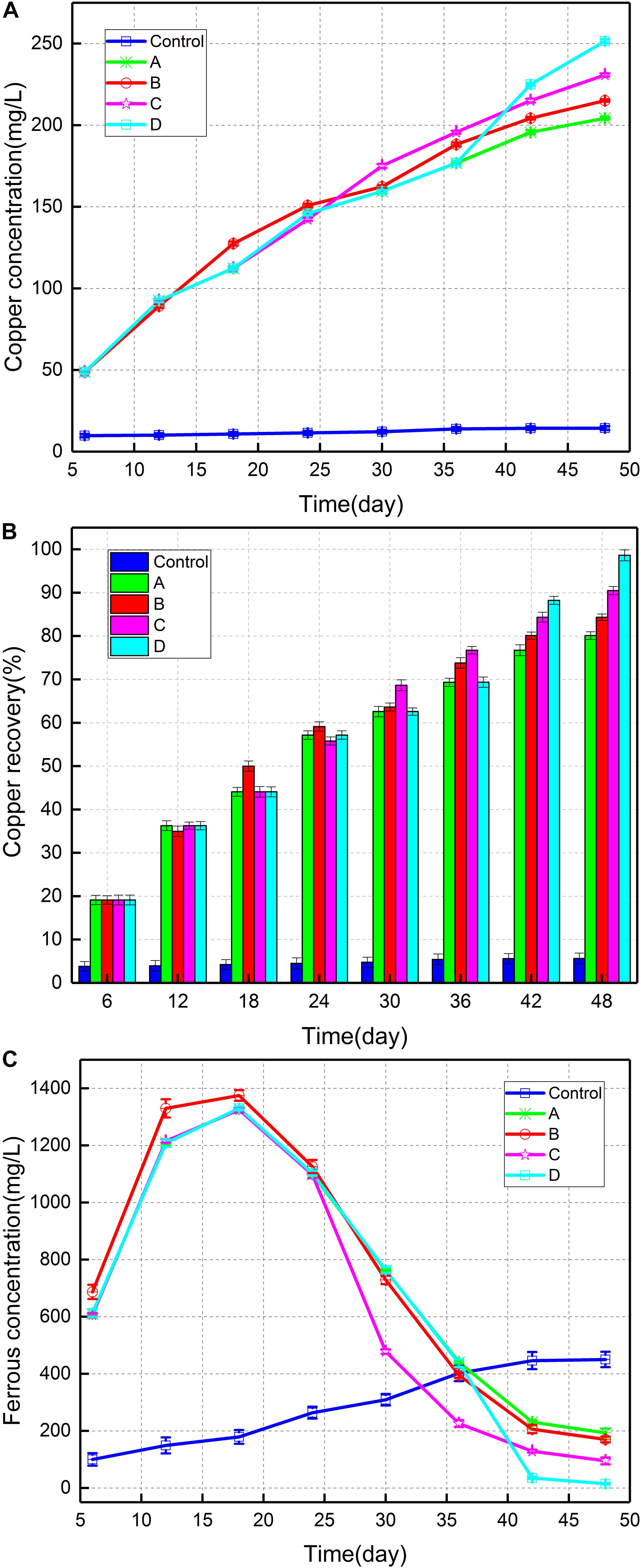
Figure 1. (A) Copper concentration of different consortia, (B) copper recovery (%), and (C) ferrous iron concentration.
Figure 2A depicted the change in potential of leaching solution of consortium A and D. In consortium A, the potential increased from 534 on day 38 to 575 mV on day 48. The potential of consortium D increased from 563 to 600 mV. Therefore, the introduction of A. thiooxidans A01 during the stable growth phase of planktonic bacteria led to a relative increase in potential. Figure 2B displayed the pH change in the leaching solution of consortium A and D. In consortium A, the pH decreased from 1.67 on day 38 to 1.45 on day 48. Similarly, the pH of consortium D decreased from 1.58 to 1.35. Thus, the introduction of A. thiooxidans A01 was associated with a relative decrease in pH value. The finding concerning consortium D indicated that the introduction during stable growth of the planktonic bacteria in the native indigenous bioleaching consortium maximally accelerated copper sulfide dissolution.
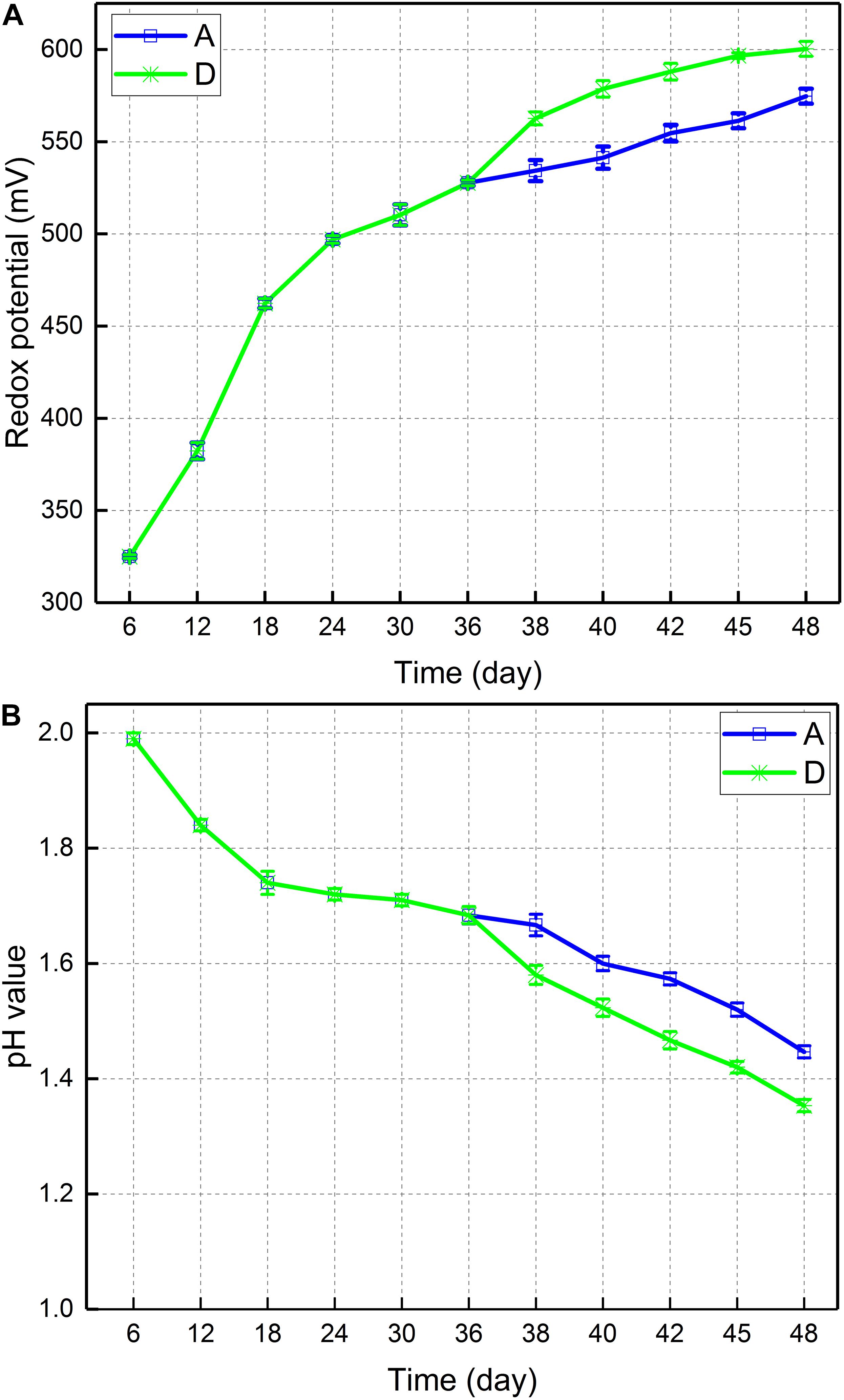
Figure 2. (A) Redox potential and (B) pH values during bioleaching of chalcopyrite by consortium A and D.
Community Structure Analysis
The finding that copper extraction of group D was the highest of the four consortia, prompted further analysis of the effect of introducing A. thiooxidans A01 during stable growth (on day 36) on the structure and function of the indigenous consortium, thereby determining why the ecological function of consortium D was improved.
Restriction fragment length polymorphism was applied to analyze the structure of the indigenous consortium A and D. Total DNA was extracted from consortium A on day 36 (before introduction of A. thiooxidans A01) and from consortia A and D on day 48 during the bioleaching process. Clones (n = 125) of each bacterial 16S rRNA clone library were analyzed by RFLP. A total of 14 operational taxonomic units (OTUs) termed L1 to L16, omitting L4 and L9, were identified (Table 1). The phylogenetic analysis based on all OTUs (including that from the original consortium) was performed to estimate the bacterial population. Six bacterial species including L. ferriphilum, A. ferrooxidans, A. caldus, Sulfobacillus sp., A. albertensis, and Acidiphilium sp. were detected in consortium A (Figure 3). Nearly all were widely common bacteria that were reported in previous studies. Among them, A. caldus, L. ferriphilum, and A. ferrooxidans were the dominant populations in the initial community, and their proportions in consortium A were 38.4, 30.4, and 17.6%, respectively (Table 1). Examination of consortium A on day 48 revealed the same population, except for the absence of A. albertensis. The primary difference was that the proportion of each population had changed, and the proportion of A. caldus, L. ferriphilum, and A. ferrooxidans was 43.2, 36, and 10.4%, respectively. Compared with consortium A, consortium D exhibited more diversity at day 48, with six species: L. ferriphilum (clones L7, L8, L11, and L14), A. caldus (clones L2, L3, and L15), A. ferrooxidans (clones L1 and L5), Sulfobacillus sp. (clone L10), A. albertensis (clone L12), and Acidiphilium sp. (clone L6). Alternatively, the proportion of each detected bacteria changed on day 48. The proportions of A. caldus, L. ferriphilum, and A. ferrooxidans were 32, 40, and 20.8%, respectively, in consortium D.
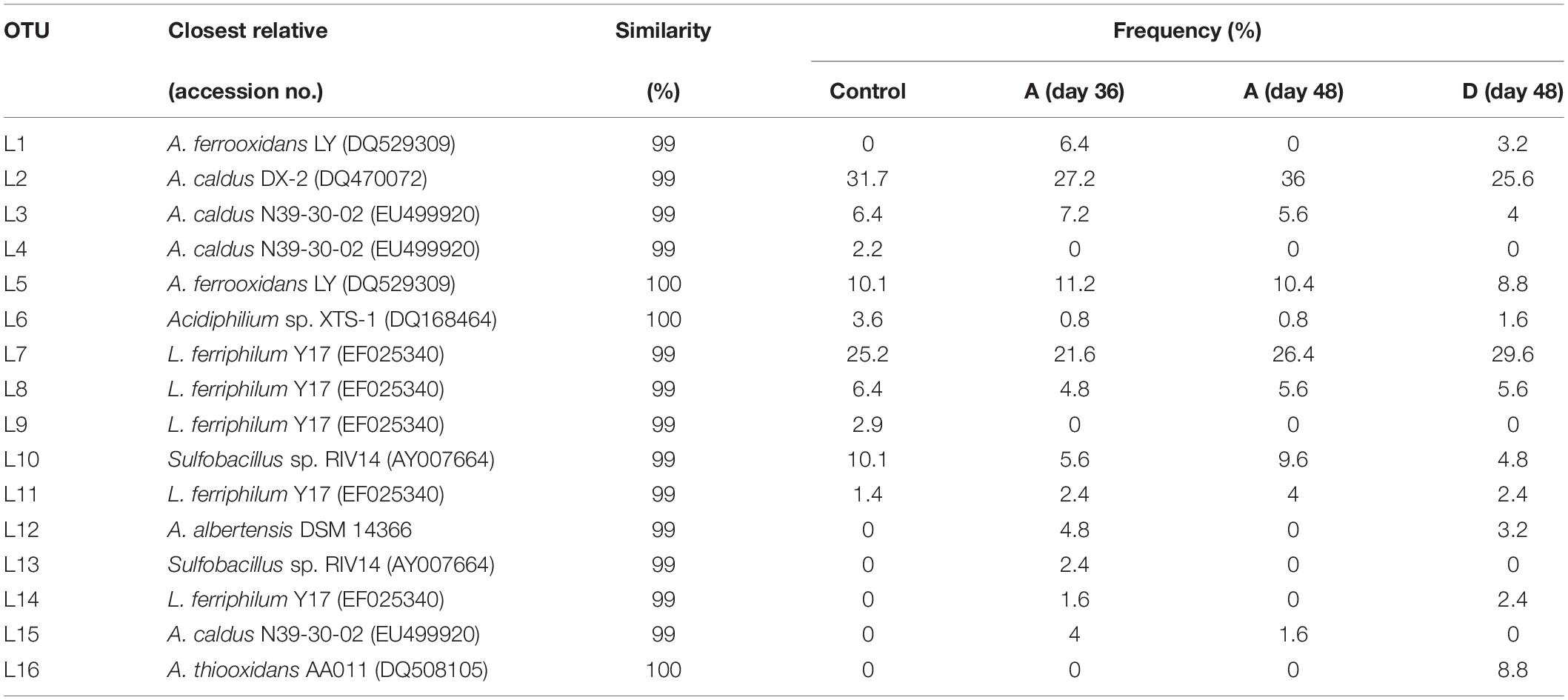
Table 1. Similarity of 16S rRNA gene sequences from clones arranged into groups according to RFLP patterns to sequences retrieved from databases.
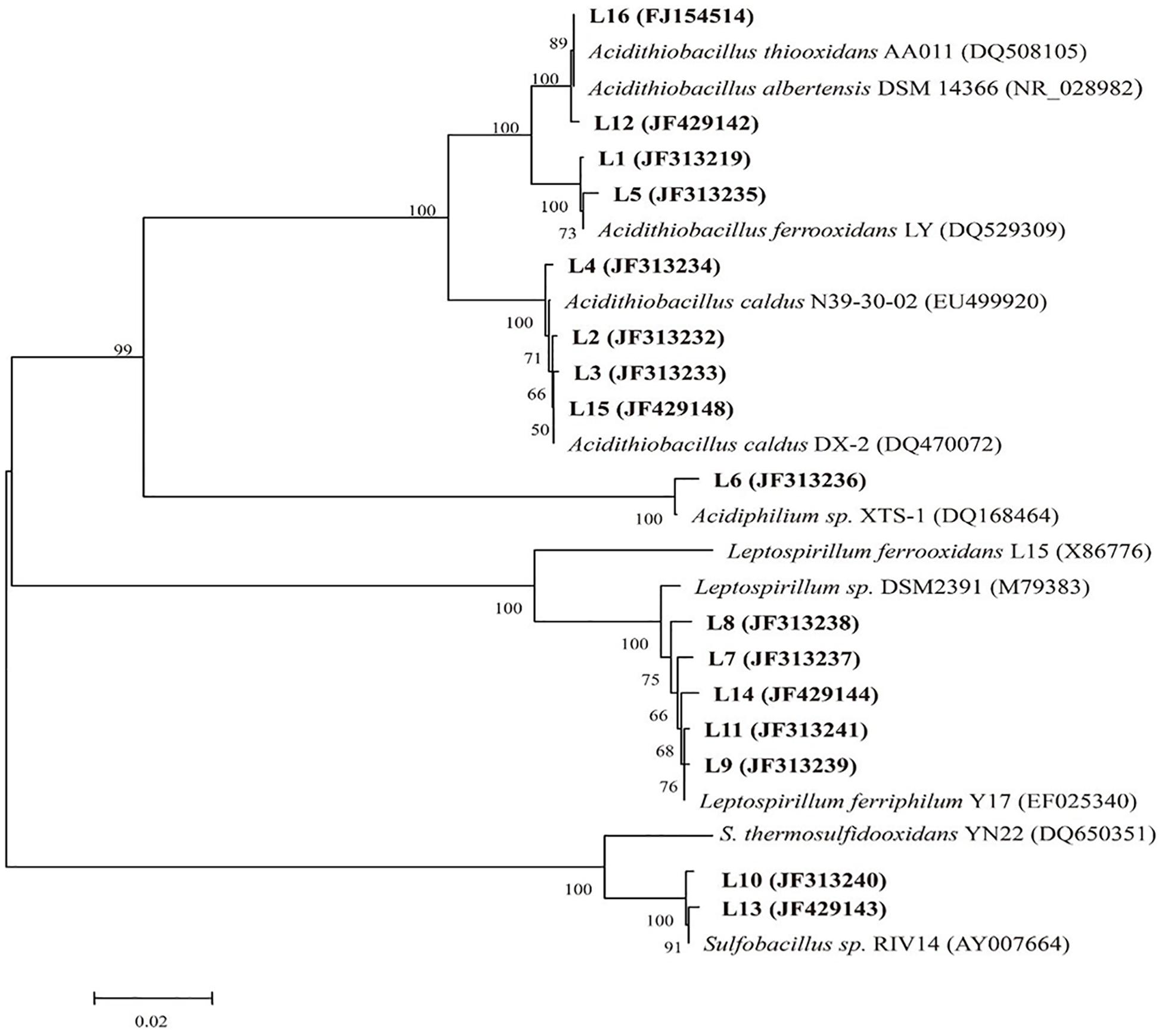
Figure 3. Phylogenetic tree according to the sequences of the partial 16S rRNA gene detected in this study (bold) and the reference sequences downloaded from the NCBI database.
A. thiooxidans A01 Dynamics Analysis
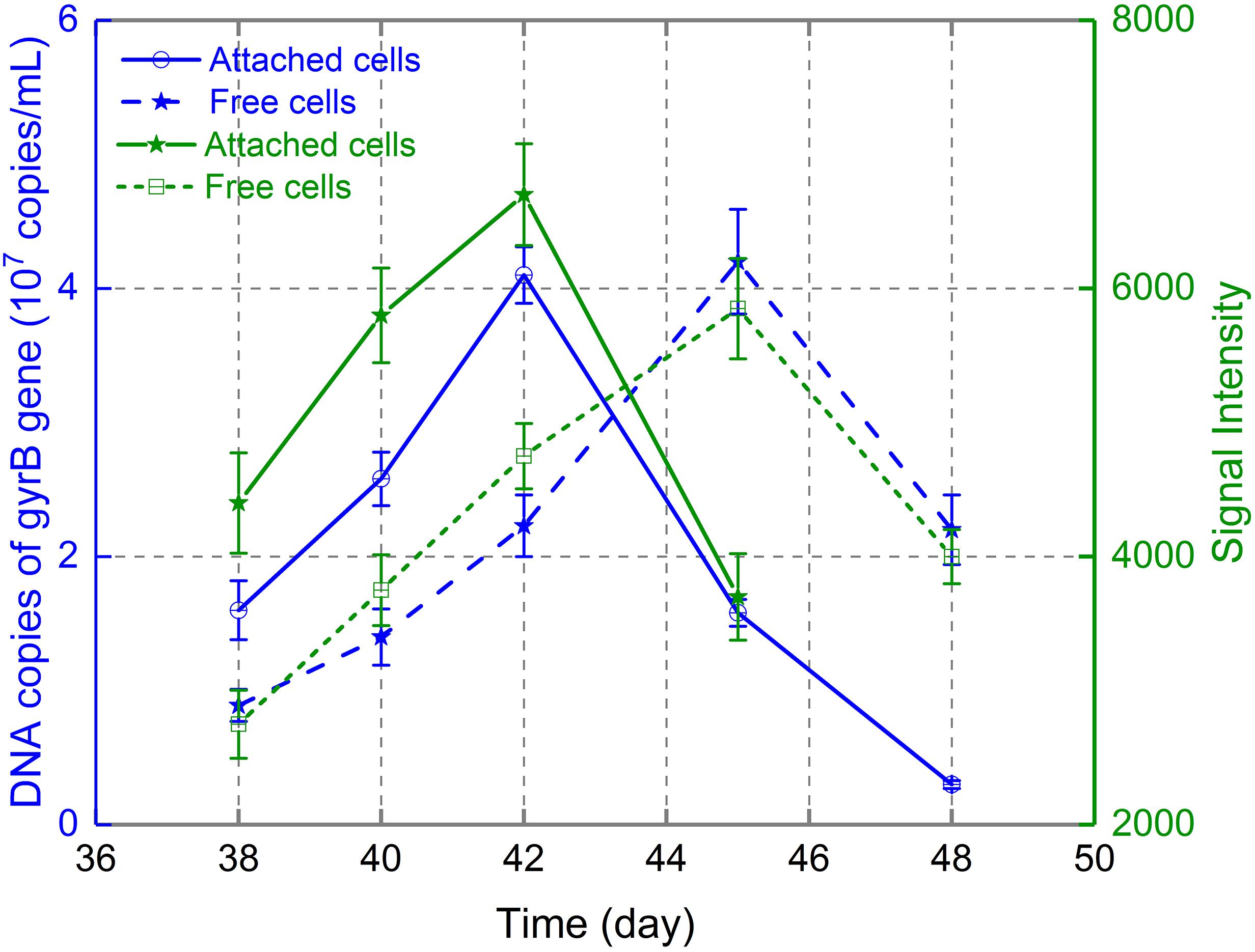
As the ratio of 16S rRNA copies to cell number is constant (Demergasso et al., 2005), the variation in 16S rRNA copy number in real-time PCR can reflect the variation in cell number during various bioleaching processes (Zhang X. et al., 2015; Deng et al., 2017). Therefore, real-time PCR was used to monitor the dynamic changes in free and attached A. thiooxidans A01 in this study. Extracted DNA from free and attached cells at days 38, 40, 42, 45, and 48 during the bioleaching of low-grade chalcopyrite was used for standard and RT-PCR. Figure 4 depicted the changes in 16S rRNA copies of A. thiooxidans A01 in leaching solution and on the mineral surface after its introduction to the indigenous consortium. The abundance of A. thiooxidans A01 attached to the mineral surface reached maximum density (4.08 × 107 copies/mL) at day 42 and decreased to 3.03 × 106 copies/mL by day 48. Planktonic A. thiooxidans A01 increased gradually from 7.82 × 106 copies/mL at day 38 to 4.32 × 107 copies/mL at day 45, and then decreased to 2.12 × 107 copies/mL at day 48. The microarray hybridization data showed a similar pattern.
Dominant Population Dynamics Analysis
The dominant population dynamics of the free and attached microorganisms was monitored using FGAs constructed as previously detailed and containing all acquired 16S rRNA sequences of acidophiles from GenBank (Yin et al., 2007; He et al., 2008). Thus, the array was suitable to analyze microbial structures and dynamics during the bioleaching processes. Figure 5 depicted the variation in the 16S rRNA hybridization signal ratios of the dominant population derived from the planktonic and attached bacteria in the consortium A and D. The abundance of the attached L. ferriphilum and A. ferrooxidans increased in consortium D compared with that in consortium A, while that of A. caldus and Sulfobacillus sp. was reduced (Figure 5A). The abundance of planktonic L. ferriphilum, A. ferrooxidans, and Acidiphilium spp. increased following introduction of A. thiooxidans A01, while the abundance of attached A. caldus and Sulfobacillus sp. decreased (Figure 5B).
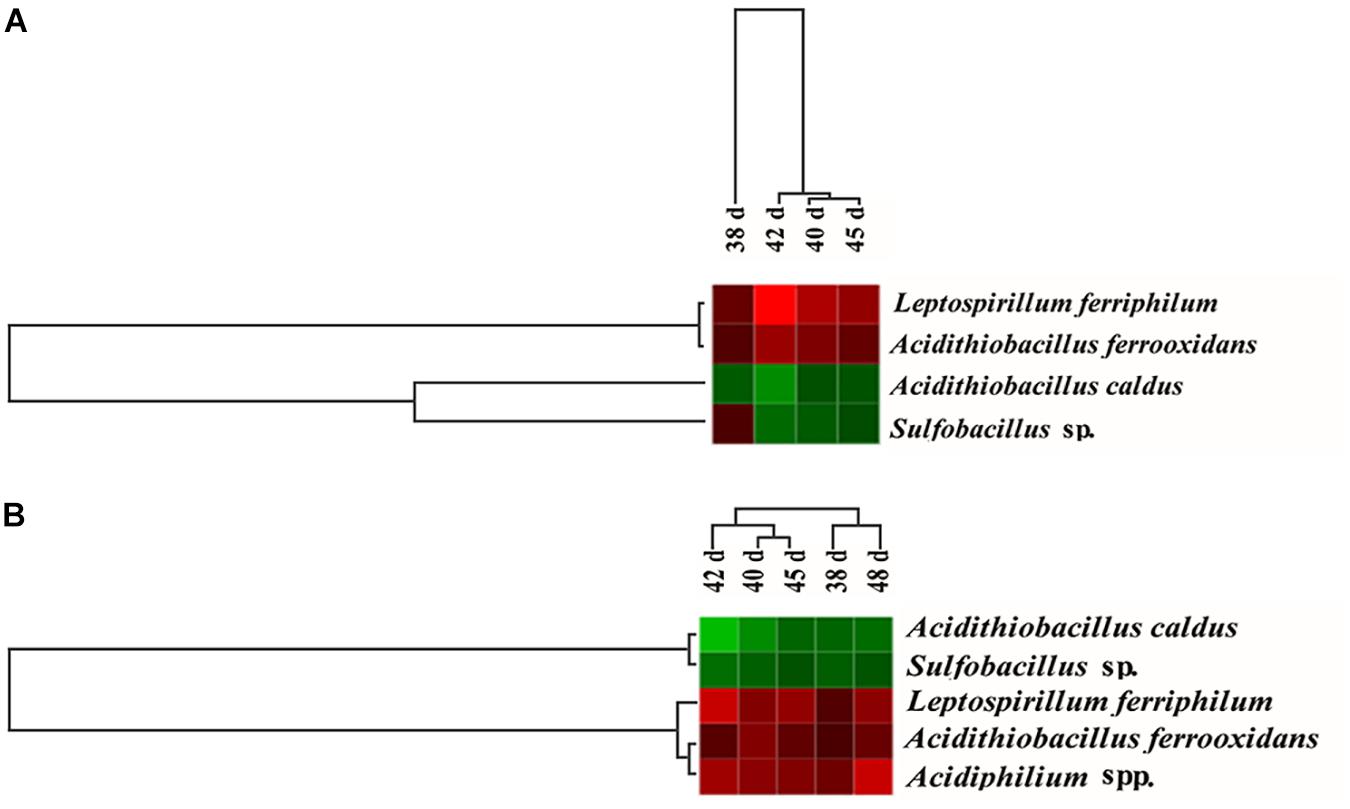
Figure 5. Hierarchical cluster analysis of the dynamics of the attached microorganisms (A) and the planktonic microorganisms (B) from 38 to 45 days according to the hybridization signal intensity ratios compared with the original consortium. Red means an obvious increase in hybridization signal, black means no significant change, and green means an obvious decrease in hybridization signal.
Changes in Microbial Functional Genes
To verify how the introduction of A. thiooxidans A01 influenced microbial ecological functions, the changes in functional genes of the planktonic and attached microorganisms were analyzed in detail. The consortium from group A was used as a control. Key genes involved in sulfur metabolism, iron oxidation, and metal resistance with the high hybridization signal intensities were clustered (Figure 6). With respect to functional genes related to iron metabolism, the abundance of genes encoding protoheme ferrolyase from L. ferriphilum increased significantly in both attached and planktonic samples. For functional genes related to sulfur metabolism, the abundance of the genes encoding sulfide-quinone reductase from A. ferrooxidans was obviously increased. However, the abundance of genes involved in sulfur metabolism from A. caldus and Sulfolobus spp. was decreased during the leaching process. For example, the abundance of the doxD gene related to the oxidation of inorganic sulfur complex from A. caldus decreased obviously in attached and planktonic samples. Furthermore, the abundance of some genes involved in metal resistance from A. ferrooxidans, L. ferriphilum, and Acidiphilium sp. was increased in consortium D.
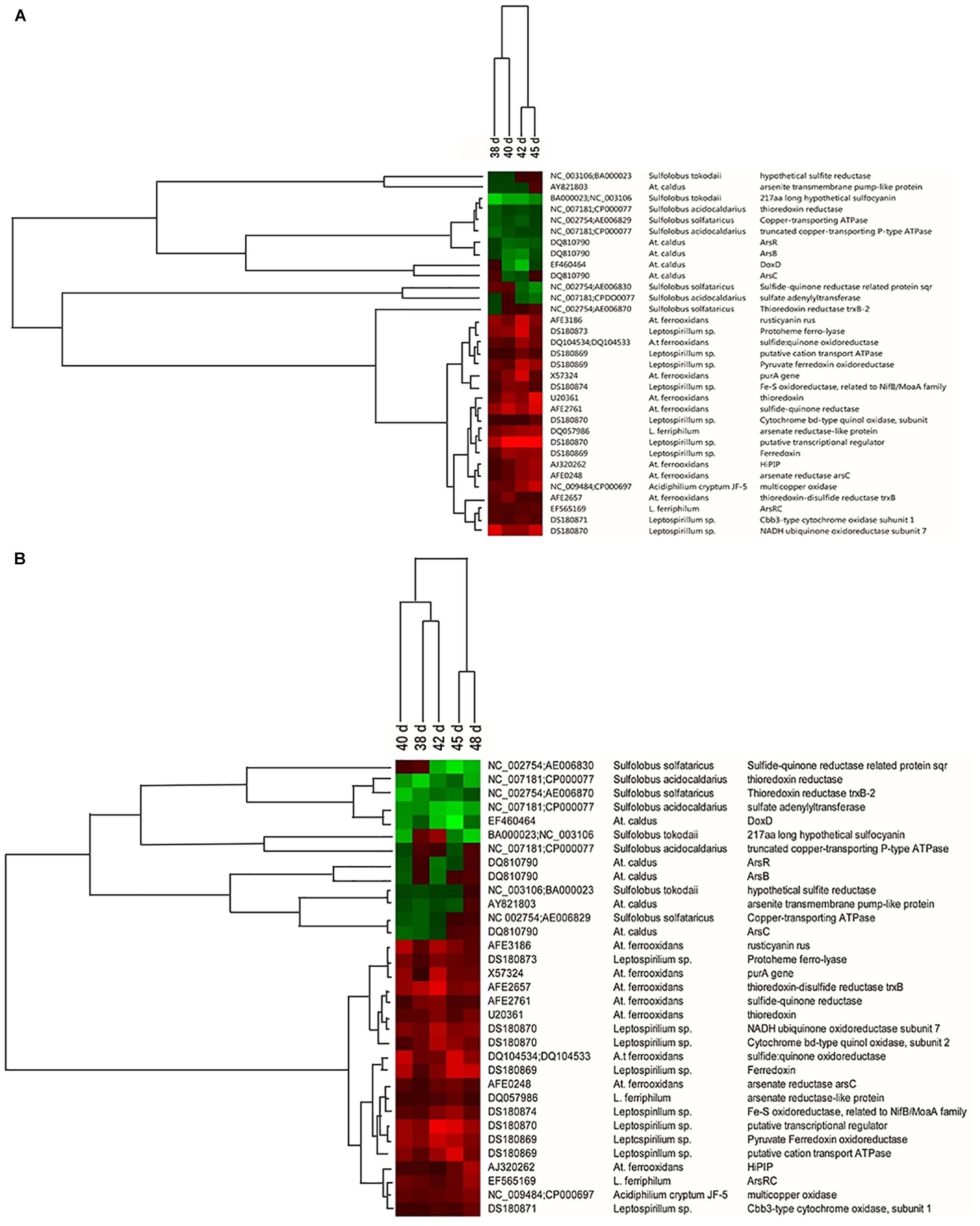
Figure 6. Hierarchical cluster analysis of the functional gene changes in ore surface (A) and leaching solution (B) after introduction of A. thiooxidans A01 from 38 to 45 days according to the hybridization signal intensity ratios compared with the control consortium. Red means an obvious increase in hybridization signal, black means no significant change, and green means an obvious decrease in hybridization signal.
Discussion
The introduction of A. thiooxidans A01 was advantageous for the dissolution of chalcopyrite (Figures 1A,B). Compared with that in the control group (group A), the redox potential increased immediately after A. thiooxidans A01 introduction (500–600 mv), which increased the oxidation of chalcopyrite and copper concentration. In addition, the pH of the solution decreased significantly with the subsequent rapid increase of redox potential (Figure 2A). This was consistent with the results of previous studies, wherein the redox potential of the solution was related to the ratio of Fe3+/Fe2+ (Zhang et al., 2014; Huang et al., 2018). The reason may be that the concentration of Fe3+ in the solution increased and the oxidation of sulfur produced a relatively large amount of sulfuric acid (Zhang X. et al., 2015). The results of the ferrous concentration in this study confirmed this finding (Figure 1C), as for which the ferrous concentration rapidly decreased following the introduction of A. thiooxidans A01, regardless of when it was introduced, and the rapid decrease of ferrous iron was attributed to the rapid oxidation of ferric iron. In accordance with the Nernst equation (Romero et al., 2003), the Fe3+/Fe2+ ratio is positively related to the redox potential. The pH value showed an overall downward trend during the entire bioleaching process. This explains the relatively high sulfur oxidation and precipitation of iron (jarosite) that have been previously described (Sandström and Petersson, 1997; Gómez et al., 1999; Zhang L. et al., 2015). These physicochemical parameter results were similar to those reported by Ma et al. (2017) in which they investigated the effect of an artificially designed proportion of microorganisms on chalcopyrite bioleaching, and found that the group with more sulfur-oxidizing bacteria induced higher copper extraction and redox potential as well as a lower pH and a faster oxidation of ferrous iron. Together, these results indicate that the introduction of sulfur-oxidizing bacteria can effectively promote the bioleaching of chalcopyrite.
Compared with the composition and structure of consortium A at day 48, the proportion of various microorganisms changed in consortium A and D at day 48 (Table 1). The results showed that the introduction of A. thiooxidans A01 during the stable period could promote the growth of L. ferriphilum and inhibit the growth of A. caldus. Similarly, the introduction of A. thiooxidans A01 during the stable period had differing effects on each population in consortium D (Figure 5). The reason for this observation may be that A. thiooxidans A01, as an obligate sulfur-oxidizing bacterium, was bound to join the competition of sulfur energy due to the need for growth after its introduction into the indigenous community. As a result of the intensified competition, the growth of identical sulfur-oxidizing bacteria, A. caldus and Sulfobacillus sp., were also inevitably affected, leading to their growth inhibition. For iron-oxidizing bacteria that use Fe2+ as an energy source, the introduction of A. thiooxidans A01 accelerated the oxidative decomposition of minerals (the direct mechanism), which led to the production of more Fe2+ (Liu et al., 2006). L. ferriphilum and A. ferrooxidans, thus, obtained increased energy to grow, thereby increasing their abundance in the community (Ma et al., 2018). Moreover, the introduction of exotic species could stimulate the enhancement of the metabolic activity of the community and thus provided more carbon and energy sources for the growth of heterotrophic bacteria, which may be one of the reasons for the increased abundance of heterotrophic Acidiphilium sp. in the community (Ullrich et al., 2015). In addition, in the latter stage, the abundance of organic matter accumulated from microbial biomass and metabolites provided more energy for the growth of heterotrophic bacteria. Acidiphilium sp., as a heterotrophic acidophile, may play an important role in the consumption of organics and, thus, reduce the inhibitory effect of organics on A. ferrooxidans and L. ferriphilum (Huang et al., 2018). For the planktonic microbial population in the leaching solution, the trend was consistent with that of the attached population following introduction. However, the degree of change varied for different timepoint or populations. According to previous studies (Harneit et al., 2006; Africa et al., 2012), microorganisms from mineral adapted cultures are capable of attaching to minerals more efficiently, and produce increased levels of EPS, which may concentrate the ferric iron to oxidatively attack chalcopyrite. When ferrous iron becomes trapped in the EPS layer, it gets released into the solution, thereby allowing the microorganisms in solution to grow rapidly (Zeng et al., 2010). Moreover, in the later stages of bioleaching, large amounts of jarosite and sulfur film form on the mineral surface, effectively blocking further oxidation. Thus, the introduction of A. thiooxidans A01 at this time period would attach to the mineral surface, thereby partially eliminating the inhibition of sulfur, which ultimately results in higher copper efficiency (Dong et al., 2013; Ma et al., 2017).
The changes in community function genes before and after introduction were also analyzed (Figure 6). With respect to functional genes related to sulfide oxidation, thioredoxin, sulfide: quinone oxidoreductase, sulfide–quinone reductase, thioredoxin–disulfide reductase trxB, and HiPIP were observed in A. ferrooxidans (Wakai et al., 2004). The abundance of these genes was significantly increased in both attached and planktonic samples, while the abundance of genes involved in sulfur metabolism from A. caldus and Sulfolobus spp. such as thioredoxin reductase, DoxD, and sulfide-quinone reductase-related protein sqr was decreased. However, in a previous study (Liu et al., 2011a), the abundance of genes involved in sulfur metabolism from A. caldus and Sulfolobus spp. was increased with increasing number of subculture, while genes involved in sulfur metabolism from A. ferrooxidans were decreased. These opposing results indicate that the introduction of A. thiooxidans A01 may exhibit synergistic effects with those of A. ferrooxidans and L. ferriphilum; however, it competes for energy with Sulfolobus spp. and A. caldus. Further, iron metabolism-related genes such as rusticyanin (rus) from A. ferrooxidans, ferredoxin and protoheme ferro-lyase from L. ferriphilum, and electron transfer chain-related genes were all increased following introduction of A. thiooxidans A01, thereby enhancing ferrous oxidation activity (Liu W. et al., 2011). Additionally, the abundance of arsenate reductase (arsC), arsenate reductase-like protein, and ArsRC genes involved in metal (loid) resistance in A. ferrooxidans, L. ferriphilum, and Acidiphilium sp. was increased in group D. Arsenic is often associated with sulfide minerals, and its toxicity becomes apparent after the dissolution of the mineral (Drewniak and Sklodowska, 2013). It has been reported that the arsC gene is responsible for reducing arsenate to arsenite, and that the arsB gene is involved in arsenite efflux, while arsR may encode a repressor of arsB (Butcher et al., 2000). The introduction of A. thiooxidans A01 promoted the further dissolution of chalcopyrite, resulting in more arsenic being released into the solution. These upregulated genes helped them to resist pressure and ensured survival (Luo et al., 2009).
Conclusion
The exogenous species A. thiooxidans A01 was introduced into an indigenous consortium on days 0, 24, and 36 (bioleaching cycle spanned 48 days). Copper concentration was 215.1, 230.8, and 251.5 mg/L, respectively, in the bioleaching system. After the introduction of A. thiooxidans A01, L. ferriphilum, A. ferrooxidans, A. caldus, Sulfobacillus sp., A. albertensis, and Acidiphilium were continuously detected, although their proportions changed during bioleaching. Only five microbial species (except for A. albertensis) were detected, and the proportions of A. caldus, L. ferriphilum, and A. ferrooxidans were 43.2, 36, and 10.45% in the control group. Real-time PCR and FGAs data suggested that A. thiooxidans A01 introduced at day 36 primarily attached to the mineral surface. The absorbance peaked at day 42 and then declined. In the supernatant, the cell number of A. thiooxidans A01 was initially low, peaked at day 45, and then decreased. The abundance of both attached and planktonic A. caldus and Sulfobacillus sp. decreased after the introduction of A. thiooxidans A01, while the abundance of L. ferriphilum, A. ferrooxidans, and Acidiphilium sp. all increased. FGAs data indicated that the abundance of genes involved in sulfide and iron oxidation from L. ferriphilum and A. ferrooxidans, and the metal (loid) resistance gene from A. ferrooxidans, L. ferriphilum, and Acidiphilium sp. increased, while the genes involved in sulfur metabolism from A. caldus and Sulfolobus spp. decreased. This research is of great significance to study the introduction mechanism of indigenous leaching microbial communities.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Author Contributions
YL designed and performed the experiments. YL and JW wrote the manuscript. HH and GC assisted in performing some experiments. XL, HL, and LS revised the manuscript. All authors read and approved the submitted version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (41771300, 41301274, and 51604308), the Youth Innovation Team Project of Institute of Subtropical Agriculture (2017QNCXTD_GTD), Chinese Academy of Sciences, and the National Key Research and Development Program of China (2017YFD0202000).
Conflict of Interest
GC was employed by company Changsha Folianovo Biotechnology Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Editage (www.editage.cn) for providing English language editing services.
Footnotes
References
Africa, C. J., Van Hille, R., and Harrison, S. (2012). Attachment of Acidithiobacillus ferrooxidans and Leptospirillum ferriphilum cultured under varying conditions to pyrite, chalcopyrite, low-grade ore and quartz in a packed column reactor. Appl. Microbiol. Biotechnol. 97, 1317–1324. doi: 10.1007/s00253-012-3939-x
Akcil, A., and Koldas, S. (2006). Acid mine drainage (AMD): causes, treatment and case studies. J. Clean. Product. 14, 1139–1145. doi: 10.1016/j.jclepro.2004.09.006
Akcil, A., Ciftci, H., and Deveci, H. (2007). Role and contribution of pure and mixed cultures of mesophiles in bioleaching of a pyritic chalcopyrite concentrate. Miner. Eng. 20, 310–318. doi: 10.1016/j.mineng.2006.10.016
Bacelar-Nicolau, P., and Johnson, D. (1999). Leaching of pyrite by acidophilic heterotrophic iron-oxidizing bacteria in pure and mixed cultures. Appl. Environ. Microbiol. 65, 585–590.
Baker, B. J., and Banfield, J. F. (2003). Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44, 139–152. doi: 10.1016/S0168-6496(03)00028-X
Butcher, B., Deane, S., and Rawlings, D. (2000). the chromosomal arsenic resistance genes of Thiobacillus ferrooxidans have an unusual arrangement and confer increased arsenic and antimony resistance to Escherichia coli. Appl. Environ. Microbiol. 66, 1826–1833. doi: 10.1128/AEM.66.5.1826-1833.2000
Chen, M., Zhang, L., Gu, G., Hu, Y., and Su, L. (2008). Effects of microorganisms on surface properties of chalcopyrite and bioleaching. Trans. Nonferrous Metals Soc. Chin. 18, 1421–1426. doi: 10.1016/S1003-6326(09)60019-60014
Chen, P., Xu, R., Yan, L., Wu, Z., Wei, Y., Zhao, W., et al. (2017). Properties of realgar bioleaching using an extremely acidophilic bacterium and its antitumor mechanism as an anticancer agent. Biol. Res. 50:17. doi: 10.1186/s40659-017-0122-y
Cheng, H., Hu, Y., Luo, J., Xu, B., and Zhao, J. (2009). Geochemical processes controlling fate and transport of arsenic in acid mine drainage (AMD) and natural systems. J. Hazard. Mater. 165, 13–26. doi: 10.1016/j.jhazmat.2008.10.070
Chun, T., Ning, C., Long, H., Li, J., and Yang, J. (2016). Mineralogical characterization of copper slag from tongling nonferrous metals group China. JOM 68, 2332–2340. doi: 10.1007/s11837-015-1752-1756
Davis-Belmar, C. S., Gallardo, I., Demergasso, C., and Rautenbach, G. (2012). Effect of organic extractant LIX 84IC, pH and temperature changes on bioleaching microorganisms during SX treatment. Hydrometallurgy 12, 135–139. doi: 10.1016/j.hydromet.2012.09.004
Demergasso, C. S., Galleguillos, P. A., Escudero, L. V., Zepeda, A. V. J., Castillo, D., and Casamayor, E. O. (2005). Molecular characterization of microbial populations in a low-grade copper ore bioleaching test heap. Hydrometallurgy 80, 241–253. doi: 10.1016/j.hydromet.2005.07.013
Deng, S., Gu, G., Wu, Z., and Xu, X. (2017). Bioleaching of arsenopyrite by mixed cultures of iron-oxidizing and sulfur-oxidizing microorganisms. Chemosphere 185, 403–411. doi: 10.1016/j.chemosphere.2017.07.037
Dong, Y., Lin, H., Xu, X., Zhang, Y., Gao, Y., and Zhou, S. (2013). Comparative study on the bioleaching, biosorption and passivation of copper sulfide minerals. Int. Biodeterior. Biodegradation 84, 29–34. doi: 10.1016/j.ibiod.2013.05.024
Drewniak, L., and Sklodowska, A. (2013). Arsenic-transforming microbes and their role in biomining processes. Environ. Sci. Pollut. Res. 20, 7728–7739. doi: 10.1007/s11356-012-1449-1440
Gómez, C., Blázquez, M. L., and Ballester, A. (1999). Bioleaching of a Spanish complex sulphide ore bulk concentrate. Miner. Eng. 12, 93–106. doi: 10.1016/S0892-6875(98)00122-128
Harneit, K., Göksel, A., Kock, D., Klock, J. H., Gehrke, T., and Sand, W. (2006). Adhesion to metal sulfide surfaces by cells of Acidithiobacillus ferrooxidans, Acidithiobacillus thiooxidans and Leptospirillum ferrooxidans. Hydrometallurgy 83, 245–254. doi: 10.1016/j.hydromet.2006.03.044
He, Z. L., Van Nostrand, J. D., Wu, L. Y., and Zhou, J. Z. (2008). Development and application of functional gene arrays for microbial community analysis. Trans. Nonferrous Metals Soc. Chin. 18, 1319–1327. doi: 10.1016/S1003-6326(09)60004-60002
He, Z., Deng, Y., and Zhou, J. (2012). Development of functional gene microarrays for microbial community analysis. Curr. Opin. Biotechnol. 23, 49–55. doi: 10.1016/j.copbio.2011.11.001
He, Z., Van Nostrand, J. D., Deng, Y., and Zhou, J. (2011). Development and applications of functional gene microarrays in the analysis of the functional diversity, composition, and structure of microbial communities. Front. Environ. Sci. Eng. China 5, 1–20. doi: 10.1007/s11783-011-0301-y
Huang, C., Qin, C., Feng, X., Liu, X., Yin, H., Jiang, L., et al. (2018). Chalcopyrite bioleaching of an in situ leaching system by introducing different functional oxidizers. RSC Adv. 8, 37040–37049. doi: 10.1039/C8RA07085G
Johnson, D. B. (1998). Biodiversity and ecology of acidophilic microorganisms. FEMS Microbiol. Ecol. 27, 307–317. doi: 10.1016/S0168-6496(98)00079-78
Johnson, D. B. (2008). Biodiversity and interactions of acidophiles: key to understanding and optimizing microbial processing of ores and concentrates. Trans. Nonferrous Metals Soc. China 18, 1367–1373. doi: 10.1016/S1003-6326(09)60010-60018
Klocke, M., Mähnert, P., Mundt, K., Souidi, K., and Linke, B. (2007). Microbial community analysis of a biogas-producing completely stirred tank reactor fed continuously with fodder beet silage as mono-substrate. Syst. Appl. Microbiol. 30, 139–151. doi: 10.1016/j.syapm.2006.03.007
Liu, J., Wang, Z., Geng, M., and Qiu, G. (2006). Progress in the study of polyphase interfacial interactions between microorganism and mineral in bio-hydrometallurgy. Min. Metall. Eng. 26, 40–44.
Liu, W., Lin, J., Pang, X., Cui, S., Mi, S., and Lin, J. (2011). Overexpression of rusticyanin in Acidithiobacillus ferrooxidans ATCC19859 increased Fe(II) oxidation activity. Curr. Microbiol. 62, 320–324. doi: 10.1007/s00284-010-9708-9700
Liu, Y., Yin, H., Liang, Y., Shen, L., Liu, Y., Fu, X., et al. (2011a). Changes in the composition of an acid mine drainage microbial community upon successive transfers in medium containing low-grade copper sulfide. Bioresour. Technol. 102, 9388–9394. doi: 10.1016/j.biortech.2011.05.095
Liu, Y., Yin, H., Zeng, W., Liang, Y., Liu, Y., Baba, N., et al. (2011b). The effect of the introduction of exogenous strain Acidithiobacillus thiooxidans A01 on functional gene expression, structure and function of indigenous consortium during pyrite bioleaching. Bioresour. Technol. 102, 8092–8098. doi: 10.1016/j.biortech.2011.06.012
Lu, D., Wang, W., Chang, Y., Xie, F., and Jiang, K. J. M. (2016). Thermodynamic analysis of possible chalcopyrite dissolution mechanism in sulfuric acidic aqueous solution. Metals 6:303. doi: 10.3390/met6120303
Luo, H., Shen, L., Yin, H., Li, Q., Chen, Q., Luo, Y., et al. (2009). Comparative genomic analysis of Acidithiobacillus ferrooxidans strains using the A. ferrooxidans ATCC 23270 whole-genome oligonucleotide microarray. Can. J. Microbiol. 55, 587–598. doi: 10.1139/W08-158
Ma, L., Wang, X., Feng, X., Liang, Y., Xiao, Y., Hao, X., et al. (2017). Co-culture microorganisms with different initial proportions reveal the mechanism of chalcopyrite bioleaching coupling with microbial community succession. Bioresour. Technol. 223, 121–130. doi: 10.1016/j.biortech.2016.10.056
Ma, L., Wang, X., Liu, X., Wang, S., and Wang, H. (2018). Intensified bioleaching of chalcopyrite by communities with enriched ferrous or sulfur oxidizers. Bioresour. Technol. 268, 415–423. doi: 10.1016/j.biortech.2018.08.019
Marchevsky, N., Urbieta, M. S., Bernardelli, C., Mas, M., and Donati, E. R. (2015). Zinc recovery during refractory ore biooxidation by an indigenous consortium. Int. J. Min. Process. 138, 30–37. doi: 10.1016/j.minpro.2015.03.009
Mukherjea, A. (1971). The convolution equation P=P ∗ Q of choqet and deny and relatively invariant measures on semi-groups. Ann. Institut. Fourier 21, 87–97. doi: 10.5802/aif.394
Mukherjee, A., Raichur, A. M., Modak, J. M., and Natarajan, K. A. (2004). Exploring process options to enhance metal dissolution in bioleaching of Indian Ocean nodules. J. Chem. Technol. Biotechnol. 79, 512–517. doi: 10.1002/jctb.1014
Nkulu, G., Gaydardzhiev, S., Mwema, E., and Compere, P. (2015). SEM and EDS observations of carrollite bioleaching with a mixed culture of acidophilic bacteria. Min. Eng. 75, 70–76. doi: 10.1016/j.mineng.2014.12.005
Rodrìguez, Y., Ballester, A., Blázquez, M. L., González, F., and Muñoz, J. A. (2003). New information on the chalcopyrite bioleaching mechanism at low and high temperature. Hydrometallurgy 71, 47–56. doi: 10.1016/S0304-386X(03)00173-177
Romero, R., Mazuelos, A., Palencia, I., and Carranza, F. (2003). Copper recovery from chalcopyrite concentrates by the BRISA process. Hydrometallurgy 70, 205–215. doi: 10.1016/S0304-386X(03)00081-81
Sand, W., and Gehrke, T. (2006). Extracellular polymeric substances mediate bioleaching/biocorrosion via interfacial processes involving iron(III) ions and acidophilic bacteria. Res. Microbiol. 157, 49–56. doi: 10.1016/j.resmic.2005.07.012
Sandström, Å, and Petersson, S. (1997). Bioleaching of a complex sulphide ore with moderate thermophilic and extreme thermophilic microorganisms. Hydrometallurgy 46, 181–190. doi: 10.1016/S0304-386X(97)00012-11
Schadt, C. W., Liebich, J., Chong, S. C., Gentry, T. J., He, Z., Pan, H., et al. (2004). Design and use of functional gene microarrays (FGAs) for the characterization of microbial communities. Methods Microbiol. 34, 331–368. doi: 10.1016/S0580-9517(04)34011-0
Ullrich, S. R., Poehlein, A., Voget, S., Hoppert, M., Daniel, R., Leimbach, A., et al. (2015). Permanent draft genome sequence of Acidiphilium sp. JA12-A1. Stand. Genom. Sci. 10:56. doi: 10.1186/s40793-015-0040-y
Wakai, S., Kikumoto, M., Kanao, T., and Kamimura, K. (2004). Involvement of Sulfide: quinone oxidoreductase in sulfur oxidation of an acidophilic iron-oxidizing bacterium, Acidithiobacillus ferrooxidans NASF-1. Biosci. Biotechnol. Biochem. 68, 2519–2528. doi: 10.1271/bbb.68.2519
Yin, H., Cao, L., Qiu, G., Wang, D., Kellogg, L., Zhou, J., et al. (2007). Development and evaluation of 50-mer oligonucleotide arrays for detecting microbial populations in acid mine drainages and bioleaching systems. J. Microbiol. Methods 70, 165–178. doi: 10.1016/j.mimet.2007.04.011
Yin, H., Zhang, X., Liang, Y., Xiao, Y., Niu, J., and Liu, X. (2014). Draft genome sequence of the Extremophile Acidithiobacillus thiooxidans A01, isolated from the wastewater of a coal dump. Genom. Announc. 2:e00222-14. doi: 10.1128/genomeA.00222-214
Zeng, W., Qiu, G., Zhou, H., Peng, J., Chen, M., Tan, S. N., et al. (2010). Community structure and dynamics of the free and attached microorganisms during moderately thermophilic bioleaching of chalcopyrite concentrate. Bioresour. Technol. 101, 7068–7075. doi: 10.1016/j.biortech.2010.04.003
Zhang, J., Zhang, X., Ni, Y., Yang, X., and Li, H. (2007). Bioleaching of arsenic from medicinal realgar by pure and mixed cultures. Process. Biochem. 42, 1265–1271. doi: 10.1016/j.procbio.2007.05.021
Zhang, L., Mao, F., Li, K., Wang, Y., Chen, X., and Zhou, H. (2015). Enhancement in copper extraction from chalcopyrite by re-inoculation of different acidophilic, moderately thermophilic microorganisms. Hydrometallurgy 156, 142–151. doi: 10.1016/j.hydromet.2015.06.009
Zhang, L., Wu, J., Wang, Y., Wan, L., Mao, F., Zhang, W., et al. (2014). Influence of bioaugmentation with Ferroplasma thermophilum on chalcopyrite bioleaching and microbial community structure. Hydrometallurgy 146, 15–23. doi: 10.1016/j.hydromet.2014.02.013
Zhang, R., Wei, M., Ji, H., Chen, X., Qiu, G., and Zhou, H. (2008). Application of real-time PCR to monitor population dynamics of defined mixed cultures of moderate thermophiles involved in bioleaching of chalcopyrite. Appl. Microbiol. Biotechnol. 81:1161. doi: 10.1007/s00253-008-1792-1798
Keywords: Acidithiobacillus thiooxidans A01, planktonic microbes, adsorbed microbes, functional gene arrays, bioleaching
Citation: Liu Y, Wang J, Hou H, Chen G, Liu H, Liu X and Shen L (2020) Effect of Introduction of Exogenous Strain Acidithiobacillus thiooxidans A01 on Structure and Function of Adsorbed and Planktonic Microbial Consortia During Bioleaching of Low-Grade Copper Sulfide. Front. Microbiol. 10:3034. doi: 10.3389/fmicb.2019.03034
Received: 07 September 2019; Accepted: 17 December 2019;
Published: 15 January 2020.
Edited by:
Jesus Angel Muñoz, Complutense University of Madrid, SpainReviewed by:
Decai Jin, Research Center for Eco-Environmental Sciences (CAS), ChinaLaura Castro, Complutense University of Madrid, Spain
Copyright © 2020 Liu, Wang, Hou, Chen, Liu, Liu and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Shen, bGlzaGVuQGNzdS5lZHUuY24=
†These authors have contributed equally to this work
 Yi Liu
Yi Liu Junjun Wang
Junjun Wang Haijun Hou1
Haijun Hou1 Gang Chen
Gang Chen Hongwei Liu
Hongwei Liu Xueduan Liu
Xueduan Liu Li Shen
Li Shen