
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 17 January 2020
Sec. Microbial Physiology and Metabolism
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.03026
This article is part of the Research Topic What Is known and What Remains To Be Discovered About Bacterial Outer Membrane Vesicles View all 18 articles
Membrane vesicles (MVs) are nanoparticles composed of lipid membranes that are produced by both Gram-negative and Gram-positive bacteria. MVs have been assigned diverse biological functions, and they show great potential for applications in various fields. However, the mechanisms underlying their functions and biogenesis are not completely understood. Accumulating evidence shows that MVs are heterogenous, and different types of MVs with different compositions are released from the same species. To understand the origin and function of these MVs, determining the biochemical properties of MVs is important. In this review, we will discuss recent progress in understanding the biochemical composition and properties of MVs.
Membrane vesicles (MVs), which are produced by most bacteria, have diverse biological functions. These functions are not only interesting from a biological perspective but also for their great potential for broad applications in immunology and biotechnology.
Membrane vesicles consist of various types of lipids derived from cellular membranes along with numerous other biomolecules, such as membrane, periplasmic, and cytoplasmic proteins; DNA; RNA; and low molecular mass organic compounds that confer various biological functions (Brown et al., 2015; Schwechheimer and Kuehn, 2015; Dauros-Singorenko et al., 2018; Toyofuku et al., 2019). Recent studies have shown that there are different pathways of MV biogenesis, which produce different types of MVs (Toyofuku et al., 2019). Thus, the mechanism of MV formation determines its biochemical composition. Knowledge of the composition of MVs is important for understanding both the mechanisms of biogenesis and their biological functions. A number of studies have examined the biochemical properties of MVs, and in this review, we will summarize the recent progress in understanding the basic properties of MVs.
In the following sections, we summarize biochemical analyses of bacterial MVs that provided insights into their biogenesis.
Classical MVs are often referred as outer membrane vesicles (OMVs), which are generated through blebbing of the outer membrane in Gram-negative bacteria (Schwechheimer and Kuehn, 2015; Jan, 2017). Hoekstra et al. (1976) reported that MVs from <volume>Escherichia coli are derived from the outer membrane. In this study, they demonstrated that the lipid composition, SDS-PAGE protein profile, and specific activities of several membrane enzymes in these MVs are similar to those in the outer membrane (Hoekstra et al., 1976). This pioneering study showed that some proteins, including lipoproteins, were less abundant in OMVs than in the cellular outer membrane, suggesting that OMVs may originate from specific outer membrane regions (Hoekstra et al., 1976). Lpp is the most abundant lipoprotein in E. coli, and its lipid moiety is anchored to the outer membrane (Schwechheimer et al., 2013). Lpp exists in both “free” and “bound” forms, which are respectively outer membrane-anchored or covalently cross-linked to peptidoglycan through a linkage between the outer membrane and peptidoglycan layer (Schwechheimer et al., 2013). The cross-link between Lpp and peptidoglycan is formed at the C-terminal lysine by L,D-transpeptidases in E. coli (Magnet et al., 2007). Wensink and Witholt (1981) reported that E. coli OMVs contained only 35% “free” lipoprotein and almost no “bound” lipoprotein. Several studies have provided supporting results to these observations, suggesting that membrane-peptidoglycan cross-linking plays a role in OMV formation in Gram-negative bacteria (Figure 1). For example, in E. coli, a lack of either Lpp or the L,D-transpeptidases YcfS, YbiS, and ErfK resulted in increased MV production compared with that in a wild type strain (Schwechheimer et al., 2013). In Salmonella typhimurium, a lack of Lpp or a mutation in the C-terminal lysine of Lpp also increased MV production (Deatherage et al., 2009). In Neisseria meningitidis, several proteins anchored to the outer or inner membrane through peptidoglycan, such as the pilus pore PilQ, peptidoglycan-binding protein RmpM, and the multidrug efflux pump channel protein MtrE, were less abundant in OMVs than in the outer membrane (Lappann et al., 2013).
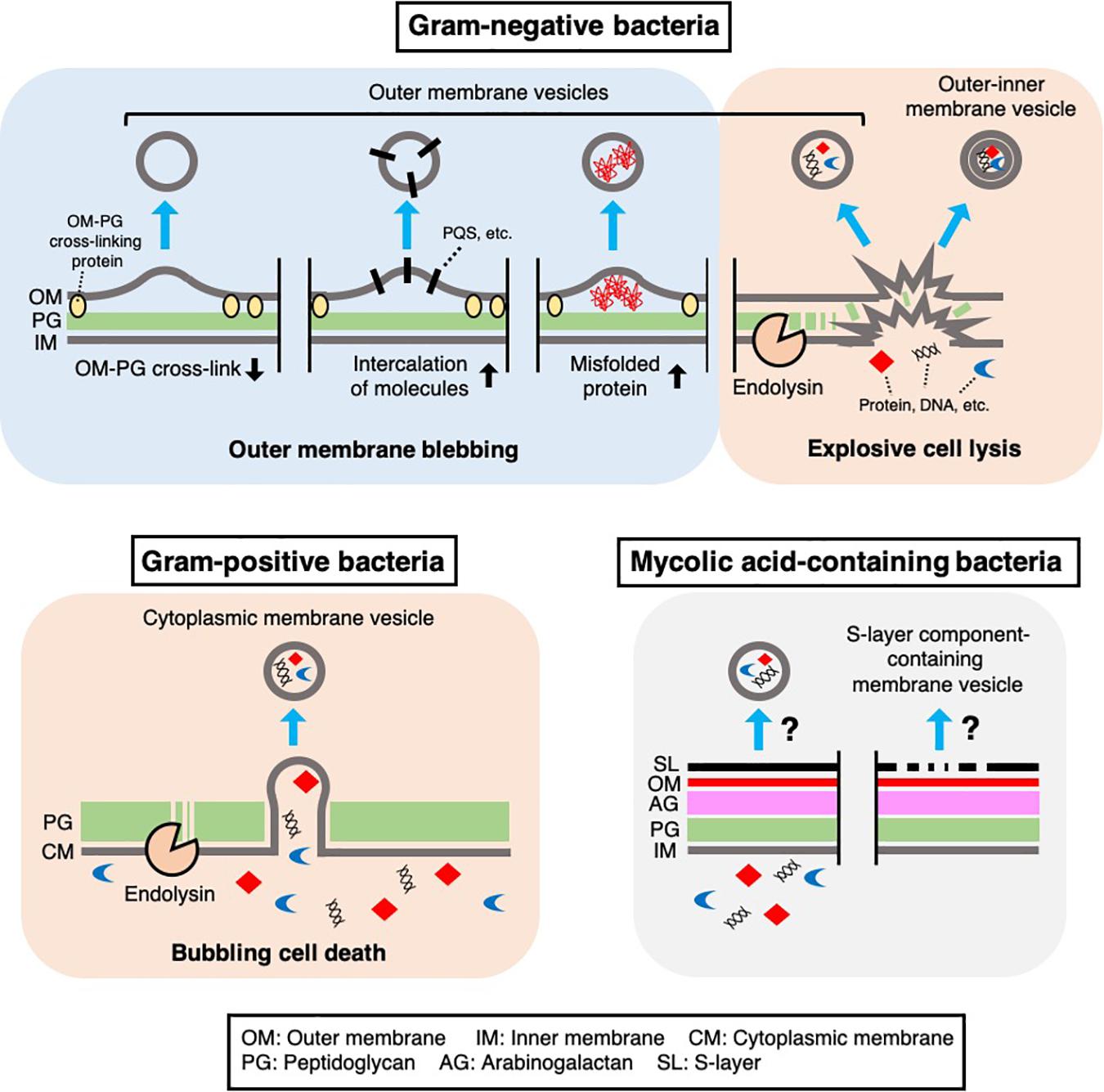
Figure 1. Mechanisms of MV biogenesis. Several mechanisms underlying MV formation in bacteria have been proposed. In Gram-negative bacteria, outer membrane vesicles and outer-inner membrane vesicles are produced through outer membrane blebbing or explosive cell lysis. Outer membrane blebbing is induced by structural changes in the cell envelope, such as a decrease in outer membrane-peptidoglycan cross-linking proteins, intercalation of molecules [such as Pseudomonas Quinolone Signal (PQS)] in the membrane, or the accumulation of misfolded proteins at specific regions of the cell envelope. Explosive cell lysis is triggered by phage-derived endolysin, which degrades the cell wall. In Gram-positive bacteria, cytoplasmic membrane vesicles are produced through bubbling cell death, in which phage-derived endolysin degrades the cell wall and the cytoplasmic membrane protrudes through the resulting holes in the peptidoglycan. In mycolic acid-containing bacteria, the mechanism of MV formation remains unknown, although there is evidence that these bacteria produce MVs containing inner membrane lipids or cell envelope associated-proteins such as S-layer component proteins.
In addition to the above proteins, OmpA is thought to be anchored to peptidoglycan through a non-covalent interaction with diaminopimelic acid, which cross-links two peptide stems to the peptidoglycan of Gram-negative bacteria (Smith et al., 2007; Park et al., 2012). Notably, a lack of OmpA also leads to increased OMV production in various Gram-negative bacteria (Sonntag et al., 1978; Song et al., 2008; Deatherage et al., 2009). These observations provide evidence for the following model of OMV formation: depletion of certain cell envelope-associated proteins, such as Lpp and OmpA, at a specific site in the outer membrane weakens outer membrane-peptidoglycan cross-linking and promotes blebbing of the outer membrane and subsequent OMV formation (Figure 1). Lpp and OmpA can be downregulated by activation of σE, suggesting that Gram-negative bacteria may modulate OMV production in response to the accumulation of misfolded outer membrane proteins in the cell envelope (Schwechheimer et al., 2013).
Further studies showed that the accumulation of misfolded proteins in the periplasm is involved in OMV formation. In E. coli, the periplasmic sensor protease DegS binds to the exposed peptides of misfolded outer membrane proteins (OMPs) and initiates signal transduction through the σE pathway upon cell envelope stress. The dual function protease/chaperone DegP is regulated by σE and prevents the accumulation of misfolded proteins in the periplasm. A lack of DegP leads to increased OMV production in a temperature-dependent manner (McBroom and Kuehn, 2007). In addition, when an OMP sequence-fused cytochrome was expressed that is misfolded and accumulates in the periplasm, the chimeric protein was enriched in OMVs compared to a control periplasm protein (McBroom and Kuehn, 2007). Based on this finding and those of related studies (McBroom et al., 2006; Schwechheimer and Kuehn, 2013), it has been proposed that the accumulation of misfolded proteins, such as OMPs, expands the physical distance between the outer membrane and peptidoglycan, leading to OMV formation, which releases these toxic components into the extracellular space (Figure 1).
Several proteomic analyses of MVs that were regarded as OMVs showed that inner membrane proteins and cytoplasmic proteins are also abundant in MVs. These results were often thought to be contaminating cell fragments or debris, such as protein aggregates. Still, a large proportion of cytoplasmic proteins are frequently detected in carefully purified MV fractions (Berleman et al., 2014; Kulkami et al., 2014; Oliver et al., 2017). To explain this, Kadurugamuwa and Beveridge suggested that localized and transient breakage in the peptidoglycan, catalyzed by autolysin, leads to the formation of OMVs containing inner and outer membrane components and cytoplasmic materials in Pseudomonas aeruginosa (Kadurugamuwa and Beveridge, 1995; Clarke, 2018). Eighteen years later, another group showed clear images of double-bilayer OMVs from Shewanella vesiculosa, which were called outer-inner membrane vesicles (O-IMVs) (Pérez-Cruz et al., 2013). Although the O-IMVs were estimated to represent only 0.1% of the total MVs in this bacterium under the condition used, proteomic analyses identified some cytoplasmic proteins in the purified MV fraction (Pérez-Cruz et al., 2013). A more recent study showed that expression of an endolysin encoded by a prophage triggers MV formation in P. aeruginosa (Turnbull et al., 2016; Toyofuku et al., 2019). In the proposed mechanism, the cell wall is degraded by endolysin, which triggers explosive cell lysis, causing the fragmentated membrane to round up and form MVs (Figure 1). During this process, nearby intracellular components, such as DNA, become trapped in the MVs (Turnbull et al., 2016; Toyofuku et al., 2019). Explosive cell lysis triggered by DNA damage can lead to MV formation in biofilms and under anoxic conditions (Toyofuku et al., 2014; Florez et al., 2017; Cooke et al., 2019). In addition, a similar route of MV formation has been suggested in Stenotrophomonas maltophilia, as production of O-IMVs and phages was detected in response to ciprofloxacin stress (Devos et al., 2017). The involvement of cell lysis in MV formation would explain why inner membrane and cytoplasmic components are detected in many proteomic analyses of OMV fractions (Kulkami et al., 2014; Olaya-Abril et al., 2014; Oliver et al., 2017; Avila-Calderón et al., 2018; Taheri et al., 2018).
Lipidomics has recently attracted much interest as it provides fundamental information about the biochemical properties and the structural bases of MVs (Lynch and Alegado, 2017). Lipid analyses are performed using various chromatography and mass spectrometry techniques, focusing on the structures of polar head groups, chain length, and saturation of fatty acid moieties. Typically, phosphoglycerolipids (PLs) are the most abundant lipids in MVs, and numerous other lipids, including glycerolipids and lipopolysaccharides (LPS), have also been detected in MVs by lipidomic analyses. As described above, Hoekstra et al. (1976) performed lipid analyses of E. coli OMVs and found that their lipid composition and phospholipid/protein ratio were similar to those of the outer membrane. However, the ratio of unsaturated fatty acids to cyclopropane fatty acids was significantly higher in OMVs (1.56) than in the outer membrane (0.19). Since unsaturated fatty acids are substituted by cyclopropane fatty acids when cells enter stationary phase, the authors suggested that these OMVs were released from cells during exponential phase (Hoekstra et al., 1976).
The structures of the fatty acid moieties are often suggested to be important factors in the biogenesis of MVs because these fatty acids affect the rigidity and fluidity of the lipid membrane. Several studies have analyzed the saturation of fatty acids in MVs and the outer membrane and discussed its significance in MV biogenesis. For example, the ratio of saturated fatty acids to unsaturated and/or branched fatty acids in OMVs was higher than that in the outer membrane of P. aeruginosa (Tashiro et al., 2011) and Prochlorococcus MED4 (Biller et al., 2014), while opposite results were obtained in Pseudomonas syringae (Kulkami et al., 2014). In some cases, fatty acid saturation was not significantly different between OMVs and the outer membrane (Fulsunder et al., 2014; Resch et al., 2016). More direct evidence was shown in Shewanella livingstonensis, where depletion of a branched fatty acid, eicosapentaenoic acid (EPA), increased MV production (Yokoyama et al., 2017). EPA was also involved in the folding of the outer membrane proteins in this bacterium (Dai et al., 2012), and may influence MV formation through the accumulation of misfolded proteins.
In addition to fatty acid moieties, lipid polar head groups may play a role in MV formation, as these head groups influence lipid conformation. Phosphatidylethanolamine (PE) is a typical conical lipid that can cause membrane curvature by clustering or sequestration (Agrawal and Ramachandran, 2019). In Haemophilus influenzae, the PE content of OMVs from PL transporter mutants (hypervesiculation mutants), which transport PLs from the outer membrane to the inner membrane, was two-fold higher than that of wild type (Roier et al., 2016). Differences in PE content between MVs and the outer membrane have also been reported in P. aeruginosa (Tashiro et al., 2011). Local and asymmetrical PE accumulation or depletion in the membrane leaflet may cause structural changes in the lipid membrane that ultimately lead to blebbing of the membrane to form MVs. Interestingly, another cone-shaped lipid molecule, deacylated LPS, has been shown to induce MV production when accumulated in the membrane (Elhenawy et al., 2016).
Compared to Gram-negative bacteria, little is known about the biogenesis and composition of MVs in Gram-positive bacteria (Brown et al., 2015; Liu et al., 2018). Given that Gram-positive bacteria lack an outer membrane in their cell envelope, an important question in MV biogenesis is how the vesicles pass through the thick peptidoglycan layer.
There are several hypotheses regarding the biogenesis of MVs in Gram-positive bacteria (reviewed in Brown et al., 2015; Toyofuku et al., 2019). Notably, cell wall modification is thought to be a key process in MV formation in these bacteria. For example, endolysin is involved in MV formation by Bacillus subtilis (Toyofuku et al., 2017a, 2019). In contrast to the explosive cell lysis induced by the action of endolysin in P. aeruginosa cells (Turnbull et al., 2016), the cytoplasmic membrane of B. subtilis cells protrudes through the holes in the peptidoglycan that are formed by the endolysin while the cell morphology remain intact (Toyofuku et al., 2017a, 2019) (Figure 1). Endolysin-triggered MV formation has also been demonstrated in another Gram-positive bacterium, Staphylococcus aureus (Andreoni et al., 2019). Another peptidoglycan-hydrolyzing enzyme, autolysin, has been suggested to induce MV formation in S. aureus further indicating that cell wall damage is a key step in MV formation in Gram-positive bacteria (Wang et al., 2018). In this study, proteomic analysis showed that autolysins were present in MVs. Gene deletion experiments further showed that autolysins, such as Sle1, facilitate MV release by hydrolyzing peptidoglycan, particularly at sites of active cell division (Wang et al., 2018).
Several studies have focused on the lipid compositions of MVs from Gram-positive bacteria. Resch et al. (2016) reported the accumulation of phosphatidylglycerol (PG) and the depletion of cardiolipin (CL) in MVs isolated from Streptococcus pyogenes culture. It can be assumed that the accumulation of cylindrical lipids, such as PG, and the depletion of conical lipids, such as CL, lead to MV formation in this bacterium; however, how these MVs pass through the cell wall is unclear. In addition, MVs from Propionibacterium acnes possessed a remarkedly reduced amount of triacylglycerol (TG) compared to the cell membrane (Jeon et al., 2018), suggesting that the biochemical and physical properties of these MVs may be largely different from those of the cell membrane.
In addition to typical Gram-positive bacteria, there is evidence that mycolic acid-containing bacteria also produce MVs (Marsollier et al., 2007; Prados-Rosales et al., 2011; Theresia et al., 2018; Chiplunkar et al., 2019) (Figure 1). These bacteria include clinically and industrially important Mycobacterium and Corynebacterium species and are characterized by unique cell envelope structures that contain mycolic acid-containing outer membrane. Prados-Rosales et al. (2011) reported seven Mycobacterium species that produce MVs. In the study, they performed proteomic analyses of MVs from Mycobacterium bovis Bacillus Calmette-Guérin (BCG), Mycobacterium tuberculosis H37Rv, and Mycobacterium smegmatis. These analyses revealed that MVs from BCG and M. tuberculosis H37Rv were enriched in lipoproteins, including well-known TLR2 ligands, whereas no lipoproteins were detected in M. smegmatis MVs (Prados-Rosales et al., 2011). In addition, the total extractable lipids in BCG MVs predominantly consisted of polar lipids, such as PE and diacylated phosphatidylinositol dimannoside (Ac2PIM2), while mycolic acid esters were not detected in the extracted lipids (Prados-Rosales et al., 2011). Given that mycolic acid esters are major lipids in the outer membrane of Mycobacteria, it is possible that these MVs may originate from the inner membrane (Prados-Rosales et al., 2011) (Figure 1). Prados-Rosales and colleagues also reported that the composition of MVs from M. tuberculosis is influenced by iron availability. Under iron-deficient conditions, acylated glycerides and PE were enriched in the MVs, whereas acyl trehalose, an important mycobacterial cell wall component, was more abundant in MVs produced under iron-sufficient conditions (Prados-Rosales et al., 2014; Rodriguez and Prados-Rosales, 2016). Interestingly, Rath et al. (2013) reported that a cytosolic membrane-associated protein, VirR, controls MV production and cargo selection in M. tuberculosis (Rodriguez and Prados-Rosales, 2016). VirR contains a disordered domain, suggestive of a binding partner, in its N-terminus and was found to be associated with several proteins, including lipoproteins (Rath et al., 2013). Notably, the VirR C-terminus has a LytR family transcriptional regulator domain that plays important roles in the formation and maintenance of the cell envelope (Brown et al., 2015).
Corynebacterium, another genus of mycolic acid-containing bacteria, also produces MVs. Theresia et al. (2018) reported that EGTA, a calcium chelator, induced MV production by Corynebacterium glutamicum. CspB, a major protein component of the para-crystalline surface layer (S-layer) of the bacterium, was predominantly detected in these MVs. The authors proposed that depletion of calcium ions altered the integrity of the S-layer and subsequently triggered the release of CspB-containing MVs (Theresia et al., 2018) (Figure 1). Other proteins detected in these MVs included CspA, CmytC, and CmytB, which are cell envelope-associated proteins (Theresia et al., 2018).
Although the mechanisms underlying MV formation in Mycobacteria and Corynebacteria are still unclear, identification of additional regulatory proteins and vesiculation-inducing factors should provide clues as to how MVs are formed and released beyond the complex cell envelope of these mycolic acid-containing bacteria.
Many biochemical analyses have revealed that certain proteins and lipids are selectively accumulated in MVs through their biogenesis. Although how these molecules are selected is largely unknown, here we describe recent observations concerning the mechanisms underlying their selection.
As described above, the accumulation of misfolded proteins in OMVs has been reported in Gram-negative bacteria, suggesting that, in some cases, OMV cargo selection is a consequence of the cell envelope stress response (McBroom and Kuehn, 2007; Olofsson et al., 2010; Schwechheimer and Kuehn, 2013). Additionally, Bonnington and Kuehn (2016) reported that the LPS composition of OMVs from Salmonella enterica changed in response to various stresses, such as low pH. The authors hypothesized that these cells may use OMV formation as a way to selectively remove environmentally disadvantageous LPS species from the outer membrane under certain conditions. Another study showed that the size and lipid composition of Klebsiella pneumoniae OMVs was altered after polymyxin treatment, suggesting that the lipid composition of OMVs reflects the outer membrane remodeling associated with cell envelope stress induced by polymyxin (Jasim et al., 2018). In contrast, several studies have suggested a mechanism in which certain proteins determine the MV composition. In Vibrio cholerae, proteomic analysis revealed that DegP was present in the MVs (Altindis et al., 2014). Interestingly, this study demonstrated the importance of DegP in the incorporation of at least nine proteins into OMVs (Altindis et al., 2014). The authors suggested that DegP can control the protein composition of OMVs by acting as a chaperone for certain proteins. Although the determinants of protein composition in bacterial MVs are still largely unknown, it is possible that the Bam complex, which catalyzes the assembly of outer membrane proteins, may, in part, determine protein cargo selection (Bonnington and Kuehn, 2014; Hussain and Bernstein, 2018).
Haurat et al. (2011) reported that anionic LPS (A-LPS) plays a critical role in OMV protein cargo selection in Porphyromonas gingivalis. In this study, gingipains and TonB-dependent outer membrane proteins were excluded from OMVs in the absence of A-LPS. Yokoyama et al. (2017) reported another example of lipid-dependent cargo selection, showing that a lack of EPA altered the protein composition of OMVs. In addition, the VacJ/Yrb lipid transporter system has been suggested to be involved in phospholipid accumulation in the outer leaflet of the outer membrane and the consequent OMV formation in H. influenzae (Roier et al., 2016).
Although the mechanism underlying MV cargo selection in Bacteroides fragilis is still unknown, it is noteworthy that most of the OMV-exclusive proteins were acidic hydrolases, whereas alkaline proteins were mainly found in the outer membrane (Elhenawy et al., 2014). The authors also demonstrated that an acidic hydrolase from another Bacteroides bacterium was heterologously expressed and selectively packed in B. fragilis OMVs (Elhenawy et al., 2014). Therefore, at least in some Bacteroides species, there may be an interesting mechanism in which the cargo proteins are selectively packed into OMVs based on their function or biochemical properties, such as pI.
Another interesting observation is a correlation between OMV size and protein contents (Turner et al., 2018). The authors found less protein content and diversity in small Helicobacter pylori OMVs (20–100 nm) than in larger OMVs (90–450 nm). These OMV may have originated from different formation routes leading to different protein contents.
Membrane vesicles play important roles in bacteria-host interactions. Bacterial MVs often have immunomodulating activities in host animals due to the presence of numerous molecules with microorganism-associated molecular patterns (MAMPs), including DNA, RNA, lipoproteins, LPS, and peptidoglycan (Kaparakis-Liaskos and Ferrero, 2015; Tan et al., 2018; Wang et al., 2019). For this reason, MVs have been intensively investigated for their potential in the development as novel vaccine platforms. Diverse hydrolytic enzymes are contained in the MVs of pathogens, suggesting that they may act in infection processes, such as the invasion of epithelial barriers (Olofsson et al., 2010; Lappann et al., 2013; Liu et al., 2018; Zarzecka et al., 2019). In addition, proteins involved in biofilm formation have been detected in MVs, suggest their potential role in biofilm formation and pathogen colonization in host animals (Altindis et al., 2014; Wagner et al., 2018).
In MV-dependent immunomodulation, the attachment and uptake of MVs by host cells has been proposed as an initial step (Kaparakis-Liaskos and Ferrero, 2015; Tan et al., 2018; Wang et al., 2019). So far, it is proposed that bacterial MVs are taken up by mammalian host cells through similar routes as other extracellular vesicles (EVs) (Figure 2). In mammalian cells, EVs (originating from mammalian cells) are taken up by recipient cells through phagocytosis or clathrin, caveolin-mediated endocytosis (Mulcahy et al., 2014). There is also evidence showing that lipid rafts are involved in EV uptake (Mulcahy et al., 2014). In addition, EVs may also fuse with the plasma membrane or be internalized via macropinocytosis, during which the EVs are enclosed into the lumen of macropinosomes or caught in membrane raffles before entry (Mulcahy et al., 2014).
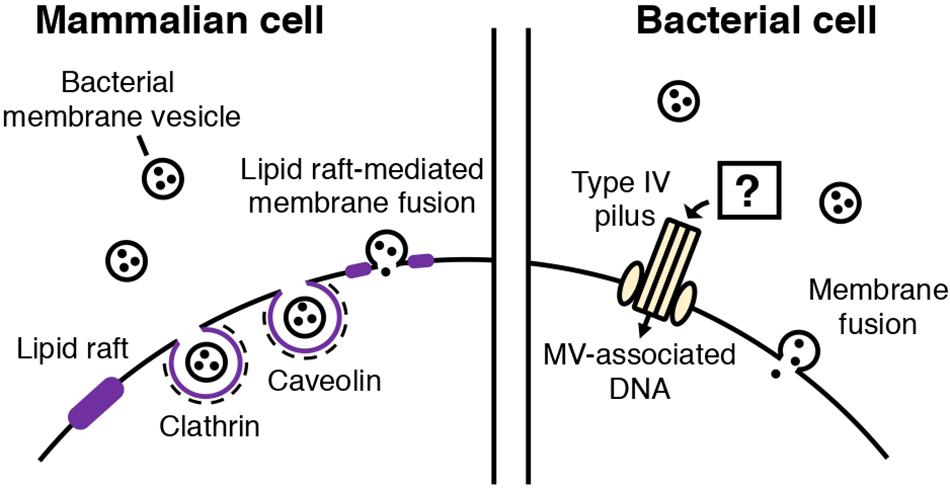
Figure 2. Routes of MV entry into mammalian and bacterial cells. In mammalian cells, bacterial MVs are thought to be internalized through several routes. Cholesterol-rich lipid rafts in the plasma membrane of the mammalian cell mediates MV entry through caveolin-mediated endocytosis or fusion of the lipid raft and bacterial MV. Clathrin-mediated endocytosis is also involved in MV internalization. In bacterial cells, two types of MV entry have been proposed: MV components, such as DNA, may be internalized into bacterial cells through type IV pili, or MVs may also fuse with the cellular membrane of the bacterial cell, depending on the membrane composition.
Schaar et al. (2011) reported that Moraxella catarrhalis OMVs enter host cells by binding to lipid rafts associated with caveolin, after interaction with Toll-like receptor 2 (TLR2). The authors also identified the adhesins and virulence factors responsible for triggering the immune response (Schaar et al., 2011). Several studies have also shown that specific, potent inhibitors of endocytosis decrease MV entry into host cells (Kesty et al., 2004; Bomberger et al., 2009; Furuta et al., 2009; Parker et al., 2010; Schaar et al., 2011; Crowley et al., 2013; O’Donoghue et al., 2017). MV fusion with the host cell membranes has been shown (Kesty et al., 2004). It was also suggested that the route of cellular entry may vary according to the MV composition and size. Parker et al. (2010) showed that H. pylori OMVs are internalized via clathrin-mediated endocytosis in the absence of VacA toxin, while VacA+ OMVs can be taken up through several routes. Turner et al. (2018) reported that smaller H. pylori OMVs (20–100 nm) preferentially entered host cells via caveolin-mediated endocytosis instead of clathrin-mediated endocytosis, while larger OMVs (90–450 nm) entered via several pathways including endocytosis. Based on the result that OMV size appears to be correlated with their protein composition, the authors proposed that the difference in the mechanisms of OMV entry may reflect the variation in OMV size and protein composition. O’Donoghue et al. (2017) showed that the LPS composition of an OMV determines major route and kinetics of host cell entry. OMVs lacking O-antigen are internalized via clathrin-mediated endocytosis, while in the presence of O-antigen, OMVs are likely to enter via lipid raft-dependent and receptor-independent routes (O’Donoghue et al., 2017).
Compared to MV entry into mammalian cells, MV uptake by bacterial cells is poorly understood. Although the entry of the membranous components or cargo into bacterial cells is an important step for MV function (Klieve et al., 2005; Mashburn and Whiteley, 2005; Domingues and Nielsen, 2017; Tashiro et al., 2017; Toyofuku et al., 2017b), the underlying mechanisms are unclear.
Fulsunder et al. (2014) showed that MVs mediate horizontal gene transfer between E. coli and Acinetobacter baylyi. In their experiments, a plasmid harboring an antibiotic resistance gene was transferred from one bacterium to the other via OMVs. They also demonstrated that OMVs were attached to or internalized by the recipient cells via transmission electron microscopy using immunogold-labeled OMVs. Importantly, the authors showed that the competence proteins of A. baylyi, such as ComA and ComB, play a role in the uptake of DNA delivered by OMVs. Therefore, it has been suggested that OMVs are lysed upon contact with the outer membrane of the bacterium, followed by type IV pilus-mediated transport of DNA (Fulsunder et al., 2014).
It has also been proposed that MVs transport their contents into bacterial cells through membranous fusion (Kadurugamuwa and Beveridge, 1996; Kim et al., 2016; Tashiro et al., 2017). For the membranes to fuse, they need to come into contact. Reducing intermembrane hydration repulsion, decreasing bilayer surface density or polarity, and increasing the hydrophobicity of the intermembrane hydrophilic region are known to bring two membranes in close contact (Ohki, 1982; Cevc et al., 1985; Rand and Parsegian, 1989; Burgess et al., 1992; Mondal Roy and Sarker, 2011). Kadurugamuwa suggested that two divalent cations, Mg2+ and Ca2+, which form salt bridges between MVs and the outer membrane, initiate membrane fusion and deliver the autolysin cargo to the cell (Kadurugamuwa and Beveridge, 1996; Wang et al., 2016; Oshima and Sumitomo, 2017). Tashiro and colleagues showed that Buttiauxella agrestis MVs selectively interact with Buttiauxella species. Based on Derjaguin-Landau-Verwey-Overbeek (DLVO) theory and physicochemical analyses, they suggested that van der Waal’s forces and electric repulsion energy are involved in the selective interaction of the bacteria with MVs (Tashiro et al., 2017).
Given that bacterial MVs can fuse with the lipid rafts in eukaryotic cells, microdomains in bacterial cells may also function as the contact sites for MVs. Functional microdomains in the membrane that contain certain lipids, such as PE, CL, diacylglycerols, cholesterols, or polyisoprenoids, have been suggested in various bacteria, including both Gram-negative and Gram-positive species (Matsumoto et al., 2006; LaRocca et al., 2010, 2013; López and Kolter, 2010; Toledo et al., 2018) although their compositions and structural bases remain largely unknown. PE and CL, which are inverted hexagonal phase-forming lipids, are major components of bacterial MVs, and the transition from the lamellar bilayer phase to the inverted hexagonal phase could facilitate the merging of lipids required for membrane fusion, once MVs come in contact with the cell (Powell and Marsh, 1985; Lewis and McElhaney, 1993; Kinnunen, 1996; Mondal Roy and Sarker, 2011). Some proteins, such as dynamin-like protein DynA, that mediate lipid mixing (Guo and Bramkamp, 2019), may also be involved in vesicle fusion, and this requires further investigations.
As we have reviewed here, biochemical approaches are powerful tools for elucidating the composition, biogenesis, and function of MVs. Proteomic and lipidomic analyses indicate that the composition of MVs depends on the growth conditions (Choi et al., 2014; Kim et al., 2016; Wagner et al., 2018; Taboada et al., 2019), suggesting that the MVs are heterogenous and their functions differ depending on the environment. On the other hand, recent studies have shown different pathways of MV formation, which lead to different MV compositions. Given this heterogeneity, we need to develop methods that enable separation and analysis of different types of MVs in cell culture as well as natural settings and body fluids. Currently, most MVs are isolated and purified by density-gradient ultracentrifugation or gel-filtration chromatography in which the particles are separated depending on their density or size in liquid solutions. Although these procedures can exclude the major contaminants (such as flagella and protein aggregates) from the purified MV solution, other techniques separating and sorting MVs depending on different properties, such surface charges, are required to investigate more precisely the biochemical properties of each MVs produced via different routes. To fully understand the true functions of each MV particles, we also need to understand the molecular mechanisms of how each types of MVs deliver their cargos to the target cells. Integrating biochemical information with imaging techniques and molecular biological approaches, may help us tackle such challenges.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
MT was supported by a Grant-in-Aid for Scientific Research (19H02866) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT). TN was supported by Grant-in-Aid for Early-Career Scientists (19K15726) from MEXT. NN was supported by the Japan Science and Technology Agency (JST) (ERATO project JPMJER 1502).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Agrawal, A., and Ramachandran, R. (2019). Exploring the links between lipid geometry and mitochondrial fission: emerging concepts. Mitochondrion 49, 305–313. doi: 10.1016/j.mito.2019.07.010
Altindis, E., Fu, Y., and Mekalanos, J. J. (2014). Proteomic analysis of Vibrio cholerae outer membrane vesicles. Proc. Natl. Acad. Sci. U.S.A. 111, E1548–E1556. doi: 10.1073/pnas.1403683111
Andreoni, F., Toyofuku, M., Menzi, C., Kalawong, R., Mairpady Shambat, S., François, P., et al. (2019). Antibiotics stimulate formation of vesicles in Staphylococcus aureus in both phage-dependent and -independent fashions and via different routes. Antimicrob. Agents Chemother. 63:e1439-18. doi: 10.1128/AAC.01439-18
Avila-Calderón, E. D., Otero-Olarra, J. E., Flores-Romo, L., Peralta, H., Aguilera-Arreola, M. G., Morales-García, M. R., et al. (2018). The outer membrane vesicles of Aeromonas hydrophila ATCC7966TM: a proteomic analysis and effect on host cells. Front. Microbiol. 9:2765. doi: 10.3389/fmicb.2018.02765
Berleman, J. E., Allen, S., Danielewicz, M. A., Remis, J. P., Gorur, A., Cunha, J., et al. (2014). The lethal cargo of Myxococcus xanthus outer membrane vesicles. Front. Microbiol. 5:474. doi: 10.3389/fmicb.2014.00474
Biller, S. J., Schubotz, F., Roggensack, S. E., Thompson, A. W., Summons, R. E., and Chisholm, S. W. (2014). Bacterial vesicles in marine ecosystems. Science 343, 183–186. doi: 10.1126/science.1243457
Bomberger, J. M., MacEachran, D. P., Coutermarsh, B. A., Ye, S., O’Toole, G. A., and Stanton, B. A. (2009). Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 5:e1000382. doi: 10.1371/journal.ppat.1000382
Bonnington, K. E., and Kuehn, M. J. (2014). Protein selection and export via outer membrane vesicles. Biochim. Biophys. Acta 1843, 1612–1619. doi: 10.1016/j.bbamcr.2013.12.011
Bonnington, K. E., and Kuehn, M. J. (2016). Outer membrane vesicle production facilitates LPS remodeling and outer membrane maintenance in Salmonella during environmental transitions. mBio 7:e001532-16. doi: 10.1128/mBio.01532-16
Brown, L., Wolf, J. M., Prados-Rosales, R., and Casadevall, A. (2015). Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 13, 620–630. doi: 10.1038/nrmicro3480
Burgess, S. W., McIntosh, T. J., and Lentz, B. R. (1992). Modulation of poly(ethylene glycol)-induced fusion by membrane hydration: importance of interbilayer separation. Biochemistry 31, 2653–2661. doi: 10.1021/bi00125a004
Cevc, G., Seddon, J. M., and Marsh, D. (1985). Thermodynamic and structural properties of phosphatidylserine bilayer membranes in the presence of lithium ions and protons. Biochim. Biophys. Acta 814, 141–150. doi: 10.1016/0005-2736(85)90429-8
Chiplunkar, S. S., Silva, C. A., Bermudez, L. E., and Danelishvili, L. (2019). Characterization of membrane vesicles released by Mycobacterium avium in response to environment mimicking the macrophage phagosome. Future Microbiol. 14, 293–313. doi: 10.2217/fmb-2018-0249
Choi, C. W., Park, E. C., Yun, S. H., Lee, S. Y., Lee, Y. G., Hong, Y., et al. (2014). Proteomic characterization of the outer membrane vesicle of Pseudomonas putida KT2440. J. Proteome Res. 13, 4298–4309. doi: 10.1021/pr500411d
Clarke, A. J. (2018). The “hole” story of predatory outer-membrane vesicles. Can. J. Microbiol. 64, 589–599. doi: 10.1139/cjm-2017-0466
Cooke, A. C., Nello, A. V., Ernst, R. K., and Schertzer, J. W. (2019). Analysis of Pseudomonas aeruginosa biofilm membrane vesicles supports multiple mechanisms of biogenesis. PLoS One 14:e0212275. doi: 10.1371/journal.pone.0212275
Crowley, J. T., Toledo, A. M., LaRocca, T. J., Coleman, J. L., London, E., and Benach, J. L. (2013). Lipid exchange between Borrelia burgdorferi and host cells. PLoS Pathog. 9:e1003109. doi: 10.1371/journal.ppat.1003109
Dai, X. Z., Kawamoto, J., Sato, S. B., Esaki, N., and Kurihara, T. (2012). Eicosapentaenoic acid facilitates the folding of an outer membrane protein of the psychrotrophic bacterium, Shewanella livingstonensis Ac10. Biochem. Biophys. Res. Commun. 425, 363–367. doi: 10.1016/j.bbrc.2012.07.097
Dauros-Singorenko, P., Blenkiron, C., Phillips, A., and Swift, S. (2018). The functional RNA cargo of bacterial membrane vesicles. FEMS Microbiol. Lett. 365:5. doi: 10.1093/femsle/fny023
Deatherage, B. L., Lara, J. C., Bergsbaken, T., Rassoulian Barrett, S. L., Lara, S., and Cookson, B. T. (2009). Biogenesis of bacterial membrane vesicles. Mol. Microbiol. 72, 1395–1407. doi: 10.1111/j.1365-2958.2009.06731.x
Devos, S., Van Putte, W., Vitse, J., Van Driessche, G., Stremersch, S., Van Den Broek, W., et al. (2017). Membrane vesicle secretion and prophage induction in multidrug-resistant Stenotrophomonas maltophilia in response to ciprofloxacin stress. Environ. Microbiol. 19, 3930–3937. doi: 10.1111/1462-2920.13793
Domingues, S., and Nielsen, K. M. (2017). Membrane vesicles and horizontal gene transfer in prokaryotes. Curr. Opin. Microbiol. 38, 16–21. doi: 10.1016/j.mib.2017.03.012
Elhenawy, W., Bording-Jorgensen, M., Valguarnera, E., Haurat, M. F., Wine, E., and Feldman, M. F. (2016). LPS remodeling triggers formation of outer membrane vesicles in Salmonella. mBio 7:e00940-16. doi: 10.1128/mBio.00940-16
Elhenawy, W., Debelyy, M. O., and Feldman, M. F. (2014). Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio 5:e00909-14. doi: 10.1128/mBio.00909-14
Florez, C., Raab, J. E., Cooke, A. C., and Schertzer, J. W. (2017). Membrane distribution of the Pseudomonas quinolone signal modulates outer membrane vesicle production in Pseudomonas aeruginosa. mBio 8:e001034-17. doi: 10.1128/mBio.01034-17
Fulsunder, S., Harms, K., Flaten, G. E., Johnsen, P. J., Chopade, B. A., and Nielsen, K. M. (2014). Gene transfer potential of outer membrane vesicles of Acinetobacter baylyi and effects of stress on vesiculation. Appl. Environ. Microbiol. 80, 3469–3483. doi: 10.1128/AEM.04248-13
Furuta, N., Tsuda, K., Omori, H., Yoshinori, T., Yoshimura, F., and Amano, A. (2009). Porphyromonas gingivalis outer membrane vesicles enter human epithelial cells via an endocytic pathway and are sorted to lysosomal compartments. Infect. Immun. 77, 4187–4196. doi: 10.1128/IAI.00009-09
Guo, L., and Bramkamp, M. (2019). Bacterial dynamin-like protein DynA mediates lipid and content mixing. FASEB J. 33, 11746–11757. doi: 10.1096/fj.201900844RR
Haurat, M. F., Aduse-Opoku, J., Rangarajan, M., Dorobantu, L., Gray, M. R., Curtis, M. A., et al. (2011). Selective sorting of cargo proteins into bacterial membrane vesicles. J. Biol. Chem. 286, 1269–1276. doi: 10.1074/jbc.M110.185744
Hoekstra, D., van der Laan, J. W., de Leij, L., and Witholt, B. (1976). Release of outer membrane fragments from normally growing Escherichia coli. Biochim. Biophys. Acta 455, 889–899. doi: 10.1016/0005-2736(76)90058-4
Hussain, S., and Bernstein, H. D. (2018). The Bam complex catalyzes efficient insertion of bacterial outer membrane proteins into membrane vesicles of variable lipid composition. J. Biol. Chem. 293, 2959–2973. doi: 10.1074/jbc.RA117.000349
Jan, A. T. (2017). Outer membrane vesicles (OMVs) of Gram-negative bacteria: a perspective update. Front. Microbiol. 8:1053. doi: 10.3389/fmicb.2017.01053
Jasim, R., Han, M. L., Zhu, Y., Hu, X., Hussein, M. H., Lin, Y. W., et al. (2018). Lipidomic analysis of the outer membrane vesicles from paired polymyxin-susceptible and -resistant Klebsiella pneumoniae clinical isolates. Int. J. Mol. Sci. 19:E2356. doi: 10.3390/ijms19082356
Jeon, J., Park, S. C., Her, J., Lee, J. W., Han, J. K., Kim, Y. K., et al. (2018). Comparative lipidomic profiling of the human commensal bacterium Propionibacterium acnes and its extracellular vesicles. RSC Adv. 8, 15241–15247. doi: 10.1039/C7RA13769A
Kadurugamuwa, J. L., and Beveridge, T. J. (1995). Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 177, 3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995
Kadurugamuwa, J. L., and Beveridge, T. J. (1996). Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J. Bacteriol. 178, 2767–2774. doi: 10.1128/jb.178.10.2767-2774.1996
Kaparakis-Liaskos, M., and Ferrero, R. L. (2015). Immune modulation by bacterial outer membrane vesicles. Nat. Rev. Immunol. 15, 375–387. doi: 10.1038/nri3837
Kesty, N. C., Mason, K. M., Reedy, M., Miller, S. E., and Kuehn, M. J. (2004). Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 23, 4538–4549. doi: 10.1038/sj.emboj.7600471
Kim, Y., Edwards, N., and Fenselau, C. (2016). Extracellular vesicle proteomes reflect developmental phases of Bacillus subtilis. Clin. Proteomics 13:6. doi: 10.1186/s12014-016-9107-z
Kinnunen, P. K. J. (1996). On the molecular-level mechanisms of peripheral protein-membrane interactions induced by lipids forming inverted non-lamellar phases. Chem. Phys. Lipids 8, 151–166. doi: 10.1016/0009-3084(96)02579-0
Klieve, A. V., Yokoyama, M. T., Forster, R. J., Ouwerkerk, D., Bain, P. A., and Mawhinney, E. L. (2005). Naturally occurring DNA transfer system associated with membrane vesicles in cellulolytic Ruminococcus spp. of ruminal origin. Appl. Environ. Microbiol. 71, 4248–4253. doi: 10.1128/AEM.71.8.4248-4253.2005
Kulkami, H. M., Swamy, ChV, and Jagannadham, M. V. (2014). Molecular characterization and functional analysis of outer membrane vesicles from the Antarctic bacterium Pseudomonas syringae suggest a possible response to environmental conditions. J. Proteome Res. 13, 1345–1358. doi: 10.1021/pr4009223
Lappann, M., Otto, A., Becher, D., and Vogel, U. (2013). Comparative proteome analysis of spontaneous outer membrane vesicles and purified outer membranes of Neisseria meningitidis. J. Bacteriol. 195, 4425–4435. doi: 10.1128/JB.00625-13
LaRocca, T. J., Crowley, J. T., Cusack, B. J., Pathak, P., Benach, J., London, E., et al. (2010). Cholesterol lipids of Borrelia burgdorferi form lipid rafts and are required for the bactericidal activity of a complement-independent antibody. Cell Host Microbe 8, 331–342. doi: 10.1016/j.chom.2010.09.001
LaRocca, T. J., Pathak, P., Chiantia, S., Toledo, A., Silvius, J. R., Benach, J. L., et al. (2013). Proving lipid rafts exist: membrane domains in the prokaryote Borrelia burgdorferi have the same properties as eukaryotic lipid rafts. PLoS Pathog. 9:e103353. doi: 10.1371/journal.ppat.1003353
Lewis, R. N., and McElhaney, R. N. (1993). Calorimetric and spectroscopic studies of the polymorphic phase behavior of a homologous series of n-saturated 1,2-diacyl phosphatidylethanolamine. Biophys. J. 64, 1081–1096. doi: 10.1016/S0006-3495(93)81474-7
Liu, Y., Defourny, K. A. Y., Smid, E. J., and Abee, T. (2018). Gram-positive bacterial extracellular vesicles and their impact on health and disease. Front. Microbiol. 9:1502. doi: 10.3389/fmicb.2018.01502
López, D., and Kolter, R. (2010). Functional microdomains in bacterial membranes. Genes Dev. 24, 1893–1902. doi: 10.1101/gad.1945010
Lynch, J. B., and Alegado, R. A. (2017). Spheres of hope, packets of doom: the good and bad of outer membrane vesicles in interspecies and ecological dynamics. J. Bacteriol. 199:e00012-e17. doi: 10.1128/JB.00012-17
Magnet, S., Bellais, S., Dubost, L., Fourgeaud, M., Mainardi, J. L., Petit-Frère, S., et al. (2007). Identification of the L, D-transpeptidases responsible for attachment of the Braun lipoprotein to Escherichia coli peptidoglycan. J. Bacteriol. 189, 3927–3931. doi: 10.1128/JB.00084-07
Marsollier, L., Brodin, P., Jackson, M., Korduláková, J., Tafelmeyer, P., Carbonnelle, E., et al. (2007). Impact of Mycobacterium ulcerans biofilm on transmissibility to ecological niches and Buruli ulcer pathogenesis. PLoS Pathog. 3:e62. doi: 10.1371/journal.ppat.0030062
Mashburn, L. M., and Whiteley, M. (2005). Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437, 422–425. doi: 10.1038/nature03925
Matsumoto, K., Kusaka, J., Nishibori, A., and Hara, H. (2006). Lipid domains in bacterial membranes. Mol. Microbiol. 61, 1110–1117. doi: 10.1111/j.1365-2958.2006.05317.x
McBroom, A. J., Johnson, A. P., Vemulapalli, S., and Kuehn, M. J. (2006). Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 188, 5385–5392. doi: 10.1128/JB.00498-06
McBroom, A. J., and Kuehn, M. J. (2007). Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63, 545–558. doi: 10.1111/j.1365-2958.2006.05522.x
Mondal Roy, S., and Sarker, M. (2011). Membrane fusion induced by small molecules and ions. J. Lipids 2011, 528784. doi: 10.1155/2011/528784
Mulcahy, L. A., Pink, R. C., and Carter, D. R. (2014). Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 3:24641. doi: 10.3402/jev.v3.24641
O’Donoghue, E. J., Sirisaengtaksin, N., Browning, D. F., Bielska, E., Hadis, M., Fernandez-Trillo, F., et al. (2017). Lipopolysaccharide structure impacts the entry kinetics of bacterial outer membrane vesicles into host cells. PLoS Pathog. 13:e1006760. doi: 10.1371/journal.ppat.1006760
Ohki, S. (1982). A mechanism of divalent ion-induced phosphatidylserine membrane fusion. Biochim. Biophys. Acta 689, 1–11. doi: 10.1016/0005-2736(82)90182-1
Olaya-Abril, A., Prados-Rosales, R., McConnell, M. J., Martín-Peña, R., González-Reyes, J. A., Jiménez-Munguía, I., et al. (2014). Characterization of protective extracellular membrane-derived vesicles produced by Streptococcus pneumoniae. J. Proteomics. 106, 46–60. doi: 10.1016/j.jprot.2014.04.023
Oliver, C., Hernández, M. A., Tandberg, J. I., Valenzuela, K. N., Lagos, L. X., Haro, R. E., et al. (2017). The proteome of biologically active membrane vesicles from Piscirickettsia salmonis LF-89 type strain identifies plasmid-encoded putative toxins. Front. Cell. Infect. Microbiol. 7:420. doi: 10.3389/fcimb.2017.00420
Olofsson, A., Vallström, A., Petzold, K., Tegtmeyer, N., Schleucher, J., Carlsson, S., et al. (2010). Biochemical and functional characterization of Helicobacter pylori vesicles. Mol. Microbiol. 77, 1539–1555. doi: 10.1111/j.1365-2958.2010.07307.x
Oshima, A., and Sumitomo, K. (2017). Vesicle fusion with bilayer lipid membrane controlled by electrostatic interaction. Biochem. Biophys. Rep. 11, 58–63. doi: 10.1016/j.bbrep.2017.06.003
Park, J. S., Lee, W. C., Yeo, K. J., Ryu, K. S., Kumarasiri, M., Hesek, D., et al. (2012). Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB J. 26, 219–228. doi: 10.1096/fj.11-188425
Parker, H., Chitcholtan, K., Hampton, M. B., and Keenan, J. I. (2010). Uptake of Helicobacter pylori outer membrane vesicles by gastric epithelial cells. Infect. Immun. 78, 5054–5061. doi: 10.1128/IAI.00299-10
Pérez-Cruz, C., Carrión, O., Delgado, L., Martinez, G., López-Iglesias, C., and Mercade, E. (2013). New type of outer membrane vesicle produced by the Gram-negative bacterium Shewanella vesiculosa M7T: implications for DNA content. Appl. Environ. Microbiol. 79, 1874–1881. doi: 10.1128/AEM.03657-12
Powell, G. L., and Marsh, D. (1985). Polymorphic phase behavior of cardiolipin derivatives studied by 31P NMR and X-ray diffraction. Biochemistry 24, 2902–2908. doi: 10.1021/bi00333a013
Prados-Rosales, R., Baena, A., Martinez, L. R., Luque-Garcia, J., Kalscheuer, R., Veeraraghavan, U., et al. (2011). Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J. Clin. Invest. 121, 1471–1483. doi: 10.1172/JCI44261
Prados-Rosales, R., Weinrick, B. C., Piqué, D. G., Jacobs, W. R. Jr., Casadevall, A., and Rodriguez, G. M. (2014). Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. J. Bacteriol. 196, 1250–1256. doi: 10.1128/JB.01090-13
Rand, R. P., and Parsegian, V. A. (1989). Hydration forces between phospholipids bilayers. Biochim. Biophys. Acta 988, 351–376. doi: 10.1016/0304-4157(89)90010-5
Rath, P., Huang, C., Wang, T., Wang, T., Li, H., Prados-Rosales, R., et al. (2013). Genetic regulation of vesiculogenesis and immunomodulation in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 110, E4790–E4797. doi: 10.1073/pnas.1320118110
Resch, U., Tsatsaronis, J. A., Le Rhun, A., Stübiger, G., Rohde, M., Kasvandik, S., et al. (2016). A two-component regulatory system impacts extracellular membrane-derived vesicle production in Group A Streptococcus. mBio 7:e00207-16. doi: 10.1128/mBio.00207-16
Rodriguez, G. M., and Prados-Rosales, R. (2016). Functions and importance of mycobacterial extracellular vesicles. Appl. Microbiol. Biotechnol. 100, 3887–3892. doi: 10.1007/s00253-016-7484-x
Roier, S., Zingl, F. G., Cakar, F., Durakovic, S., Kohl, P., Eichmann, T. O., et al. (2016). A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun. 7, 10515. doi: 10.1038/ncomms10515
Schaar, V., de Vries, S. P., Perez Vidakovics, M. L., Bootsma, H. J., Larsson, L., Hermans, P. W., et al. (2011). Multicomponent Morexella catarrhalis outer membrane vesicles induce an inflammatory response and are internalized by human epithelial cells. Cell. Microbiol. 13, 432–449. doi: 10.1111/j.1462-5822.2010.01546.x
Schwechheimer, C., and Kuehn, M. J. (2013). Synthetic effect between envelope stress and lack of outer membrane vesicle production in Escherichia coli. J. Bacteriol. 195, 4161–4173. doi: 10.1128/JB.02192-12
Schwechheimer, C., and Kuehn, M. J. (2015). Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol. 13, 605–619. doi: 10.1038/nrmicro3525
Schwechheimer, C., Sullivan, C. J., and Kuehn, M. J. (2013). Envelope control of outer membrane vesicle production in Gram-negative bacteria. Biochemistry 52, 3031–3040. doi: 10.1021/bi400164t
Smith, S. G., Mahon, V., Lambert, M. A., and Fagan, R. P. (2007). A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol. Lett. 273, 1–11. doi: 10.1111/j.1574-6968.2007.00778.x
Song, T., Mika, F., Lindmark, B., Liu, Z., Schild, S., Bishop, A., et al. (2008). A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol. Microbiol. 70, 100–111. doi: 10.1111/j.1365-2958.2008.06392.x
Sonntag, I., Schwarz, H., Hirota, Y., and Henning, U. (1978). Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major membrane proteins. J. Bacteriol. 136, 280–285.
Taboada, H., Meneses, N., Dunn, M. F., Vargas-Lagunas, C., Buchs, N., Castro-Mondragón, J. A., et al. (2019). Proteins in the periplasmic space and outer membrane vesicles of Rhizobium etli CE3 grown in minimal medium are largely distinct and change with growth phase. Microbiology 165, 638–650. doi: 10.1099/mic.0.000720
Taheri, N., Mahmud, A. K. M. F., Sandblad, L., Fällman, M., Wai, S. N., and Fahlgren, A. (2018). Campylobacter jejuni bile exposure influences outer membrane vesicles protein content and bacterial interaction with epithelial cells. Sci. Rep. 8:16996. doi: 10.1038/s41598-018-35409-0
Tan, K., Li, R., Huang, X., and Liu, Q. (2018). Outer membrane vesicles: current status and future direction of these novel vaccine adjuvants. Front. Microbiol. 9:783. doi: 10.3389/fmicb.2018.00783
Tashiro, Y., Hasegawa, Y., Shintani, M., Takaki, K., Ohkuma, M., Kimbara, K., et al. (2017). Interaction of bacterial membrane vesicles with specific species and their potential for delivery to target cells. Front. Microbiol. 8:571. doi: 10.3389/fmicb.2017.00571
Tashiro, Y., Inagaki, A., Shimizu, M., Ichikawa, S., Takaya, N., Nakajima-Kambe, T., et al. (2011). Characterization of phospholipids in membrane vesicles derived from Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 75, 605–607. doi: 10.1271/bbb.100754
Theresia, N. M., Aida, K., Takada, A., Iwai, N., and Wachi, M. (2018). Effects of EGTA on cell surface structures of Corynebacterium glutamicum. Arch. Microbiol. 200, 281–289. doi: 10.1007/s00203-017-1445-3
Toledo, A., Huang, Z., Coleman, J. L., London, E., and Benach, J. L. (2018). Lipid rafts can form in the inner and outer membranes of Borrelia burgdorferi and have different properties and associated proteins. Mol. Microbiol. 108, 63–76. doi: 10.1111/mmi.13914
Toyofuku, M., Cárcamo-Oyarce, G., Yamamoto, T., Eisenstein, F., Hsiao, C. C., Kurosawa, M., et al. (2017a). Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat. Commun. 8:481. doi: 10.1038/s41467-017-00492-w
Toyofuku, M., Morinaga, K., Hashimoto, Y., Uhl, J., Shimamura, H., Inaba, H., et al. (2017b). Membrane vesicle-mediated bacterial communication. ISME J. 11, 1504–1509. doi: 10.1038/ismej.2017.13
Toyofuku, M., Nomura, N., and Eberl, L. (2019). Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 17, 13–24. doi: 10.1038/s41579-018-0112-2
Toyofuku, M., Zhou, S., Sawada, I., Takaya, N., Uchiyama, H., and Nomura, N. (2014). Membrane vesicle formation is associated with pyocin production under denitrifying conditions in Pseudomonas aeruginosa PAO1. Environ. Microbiol. 16, 2927–2938. doi: 10.1111/1462-2920.12260
Turnbull, L., Toyofuku, M., Hynen, A. L., Kurosawa, M., Pessi, G., Petty, N. K., et al. (2016). Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 7:11220. doi: 10.1038/ncomms11220
Turner, L., Bitto, N. J., Steer, D. L., Lo, C., D’Costa, K., Ramm, G., et al. (2018). Helicobacter pylori outer membrane vesicle size determines their mechanisms of host cell entry and protein content. Front. Immunol. 9:1466. doi: 10.3389/fimmu.2018.01466
Wagner, T., Joshi, B., Janice, J., Askarian, F., Škalko-Basnet, N., Hagestad, O. C., et al. (2018). Enterococcus faecium produces membrane vesicles containing virulence factors and antimicrobial resistance related proteins. J. Proteomics 187, 28–38. doi: 10.1016/j.jprot.2018.05.017
Wang, S., Gao, J., and Wang, Z. (2019). Outer membrane vesicles for vaccination and targeted drug delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 11:e1523. doi: 10.1002/wnan.1523
Wang, X., Thompson, C. D., Weidenmaier, C., and Lee, J. C. (2018). Release of Staphylococcus aureus extracellular vesicles and their application as a vaccine platform. Nat. Commun. 9:1379. doi: 10.1038/s41467-018-03847-z
Wang, Z., Ma, Y., Khalil, H., Wang, R., Lu, T., Zhao, W., et al. (2016). Fusion between fluid liposomes and intact bacteria: study of driving parameters and in vitro bactericidal efficacy. Int. J. Nanomedicine 11, 4025–4036. doi: 10.2147/IJN.S55807
Wensink, J., and Witholt, B. (1981). Outer-membrane vesicles released by normally growing Escherichia coli contain very little lipoprotein. Eur. J. Biochem. 116, 331–335. doi: 10.1111/j.1432-1033.1981.tb05338.x
Yokoyama, F., Kawamoto, J., Imai, T., and Kurihara, T. (2017). Characterization of extracellular membrane vesicles of an Antarctic bacterium, Shewanella livingstonensis Ac10, and their enhanced production by alteration of phospholipid composition. Extremophiles 21, 723–731. doi: 10.1007/s00792-017-0937-z
Keywords: membrane vesicles, proteome, lipidome, cargo selection, endocytosis, membrane fusion
Citation: Nagakubo T, Nomura N and Toyofuku M (2020) Cracking Open Bacterial Membrane Vesicles. Front. Microbiol. 10:3026. doi: 10.3389/fmicb.2019.03026
Received: 01 November 2019; Accepted: 17 December 2019;
Published: 17 January 2020.
Edited by:
Alejandro J. Yañez, Austral University of Chile, ChileCopyright © 2020 Nagakubo, Nomura and Toyofuku. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masanori Toyofuku, dG95b2Z1a3UubWFzYW5vcmkuZ2ZAdS50c3VrdWJhLmFjLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.