- 1College of Plant Sciences, Jilin University, Changchun, China
- 2School of Life Sciences, Henan University, Kaifeng, China
Fungal sexual development requires the involvement of a large number of functional genes. Fungal genes encoding sexual differentiation process proteins (SDPs), isps, have been known for decades. isp4/SDP and its homologs function as oligopeptide transporters (OPTs), yet their roles in reproduction are unknown. Here, we genetically analyzed all four isp4/SDP homologs in the sexual species Chaetomium thermophilum and asexual species Thermomyces lanuginosus. Using single gene deletion mutants, we found that T. lanuginosus SDP (TlSDP) participated in asexual sporulation, whereas the other homologs participated in sexual morphogenesis. In complementary tests, C. thermophilum SDPs (CtSDP1-3) restored sporulation defects in TlSDP deletion strains (ΔTlSDP), and their translated proteins, which were localized onto the cytomembrane, possessed OPT activity. Interestingly, CtSDP2 accumulated at the top of the hyphae played a distinct role in determining the sexual cycle, glutathione transport, and lifespan shortening. A unique 72nt-insertion fragment (72INS) was discovered in CtSDP2. Biological analysis of the 72INS deletion and DsRED-tagged fusion strains implied the involvement of 72INS in fungal growth and development. In contrast to TlSDP, which only contributes to conidial production, the three CtSDPs play important roles in sexual and asexual reproduction, and CtSDP2 harbors a unique functional 72INS that initiates sexual morphogenesis.
Introduction
Ascomycota, the largest fungal phylum, is mostly defined according to sexual morphological features, particularly the sexual structures in which ascospores are formed (Cavalier-Smith, 2010). Sexual and asexual reproduction are common in ascomycota to produce offspring, but they have completely different features. During asexual reproduction, conidia are generally created from the tips of conidiophores, which are morphologically similar to vegetative hyphae. On the contrary, sexual reproduction of fungi is quite complex. Aggregation of vegetative mycelia indicates the initiation of sexual reproduction. Related hyphal cells differentiate into sterile and fertile hyphal cells, and finally the fruiting mycelium mature hyphae into the ascocarp. The ascogonia and antheridia are two key structures during sexual sporogenesis Hook-shaped structures known as croziers determine the formation of ascospores in the dikaryotic stage of filamentous fungi (Sautter and Hock, 1982). Interestingly, sexual reproduction can be triggered in supposedly asexual fungi, such as Candida albicans and Trichoderma reesei (Taylor et al., 1999; Magee and Magee, 2004; Seidl et al., 2009; Dyer and O’Gorman, 2011; Dyer and O’Gorman, 2012). Penicillium chrysogenum, which has been considered asexual for more than 100 years, has been shown to reproduce sexually based on MAT gene organization (Böhm et al., 2013).
Some genes associated with sexual differentiation, including lsdA, nsdD, and veA, have been successfully characterized (Lee et al., 2001; Kim et al., 2002; Han et al., 2010; Palmer et al., 2013). Sexual differentiation process proteins gene (SDPs) have been studied extensively in the fission yeast Schizosaccharomyces pombe (Sato et al., 1994). In S. pombe, five isp genes (isp3-isp7) were identified and they were preferentially expressed during sporogensis. Further study revealed that they were involved in diverse biological/biochemical functions (Yanagida, 2009; Shimanuki et al., 2010; Laor et al., 2014). Isp4 has been found up-regulated by nitrogen starvation-induced meiosis and found to encode an oligopeptide transporter (OPT). Subsequently, isp4 homologs have been extensively characterized in yeast (Lubkowitz et al., 1998; Bourbouloux et al., 2000; Hauser et al., 2000; Gonzalez-Lopez et al., 2002; Lubkowitz et al., 2010; Oliver and Joachim, 2010), but only a few OPTs have been reported in filamentous fungi. In Schizophyllum commune, the mtd1 gene is regulated during the mating reaction, and the results of complementation of gene disruption are in accordance with an OPT (Lengeler and Kothe, 1999). In Colletotrichum gloeosporioides, CgOPT1 disruption resulted in fewer spores, reduced pigmentation, and less severe pathogenic effects (Chague et al., 2009). In Aspergillus fumigatus, the octuple OPT gene deletion mutant that expressed no OPT genes displayed normal growth on various substrates (Thomas et al., 2011). Recently, members of the OPT gene family in Phanerochaete chrysosporium and Ganoderma lucidum have been analyzed in yeast systems (Xiang et al., 2013, 2017). Although the biochemical transport functions of the OPT genes described above have been confirmed, little is known about their biological roles, particularly in sexual differentiation.
The thermophilic fungi Thermomyces lanuginosus and Chaetomium thermophilum (McGrath and Varshavsky, 1989) coexist in the same environments, which have relatively high temperatures and abundant lignocellulosic compost. However, the two thermophilic fungi seem to have different reproduction patterns. T. lanuginosus reproduces asexually, while C. thermophilum reproduces sexually. By analyzing genes up-regulated during sporogenesis, three homologs of yeast isp4 (Sato et al., 1994) were identified in C. thermophilum (termed CtSDP1–CtSDP3), and single was identified in T. lanuginosus (termed TlSDP). We hypothesized that the identified genes were involved in regulating sexual or asexual reproduction in filamentous fungi. To verify this hypothesis and obtain direct insight into the biological roles of the four identified SDP homologs, several single gene deletion strains and homologous gene complementation strains were created, and phenotypes related to stages of growth, development, and sporogenesis were analyzed. All SDP genes, as expected, were found to be important for sporogenesis of their strains, but only CtSDP2 was required for sexual morphogenesis. CtSDP2 appears to determine the formation of sexual structures. We also found a 72nt-insertion fragment in CtSDP2 and investigated its role in sexual morphogenesis.
Materials and Methods
Fungal Strains
Thermomyces lanuginosus strain 9W and Chaetomium thermophilum strain S4 were isolated from a sample of cow dung compost (CDC) in Jilin province, which is located in northeastern China. Wild-type and mutant strains were cultured at 50°C on potato dextrose agar media (PDA; 200 g/L peeled potato, 20 g/L glucose, and 16 g/L agar) or complete minimal media (CM; 1 g/L yeast extract, 0.5 g/L casein enzymatic hydrolyzate, 0.5 g/L casamino acids hydrolyzate, 10 g/L glucose, 1 g/L Ca(NO3)2⋅4H2O, 0.2 g/L KH2PO4, 0.25 g/L MgSO4⋅7H2O, 0.15 g/L NaCl, and 16 g/L agar).
Vectors and Construction Strategies
Serial plasmids were employed for genetic transformation. Based on the PXEH vector, a vector containing the upstream and downstream flanking regions of the target gene was reconstructed to accomplish gene-specific deletions. This vector also conferred kanamycin resistance in Agrobacterium tumefaciens and Escherichia coli and harbored the hygromycin B phosphotransferase (hyg) gene as a selection marker for fungal deletion mutants. Briefly, the flanking sequences of the TlSDP gene were PCR-amplified from T. lanuginosus 9W DNA genome. The left (988 bp) and right (1095 bp) border flanking sequences were amplified and validated. PCR primers were designed to introduce the required enzyme cutting sites; the left flanking sequence was XhoI and SacI, and the right flanking sequence was BamHI and HindIII. Both flanking sequences were digested, purified and ligated into the PXEH vector. The TlSDP-PXEH vector was confirmed by PCR, enzyme digestion and sequencing validation, after which it was used for further research. To create single gene deletion mutants of C. thermophilum for CtSDP1, CtSDP2, and CtSDP3, the corresponding flanking sequences were amplified from C. thermophilum S4 DNA genome. Deletion vectors CtSDP1-PXEH, CtSDP2-PXEH, and CtSDP3-PXEH were reconstructed according to the approach described above. ΔCtSDP1, ΔCtSDP2, and ΔCtSDP3 were created by gene replacement (Supplementary Figure S1 and Supplementary Tables S1, S2).
To ensure that the obtained mutant phenotype could be attributed to the desired deletion and to investigate sub-localization of the protein, several single-gene and polygenic complementary vectors were constructed based on vectors such as pKD7-RED, pKAICR80, and pKAFCR100. pKD7-RED contains the DsRED gene as a subcellular localization tag and the G418-resistance gene as a selection marker. Plasmids pKAICR80 and pKAFCR100 contain the DsGFP gene as a tag and neomycin phosphotransferase II as a selection marker. For the construction of single-gene complementary vectors, the RFP in the vector was removed. The main distinction between the complementary vector and the corresponding sub-localization vector (which contains both the complemented gene and the fluorescent protein tag genes) is the presence or absence of the DsRED tag gene. The same construction strategies and primers were used for both types of vectors.
The pKD7-RED vector was used for single-gene and sub-localization tag gene expression. The TlSDP coding sequence was amplified from 9W cDNA sequences, whereas the coding sequences of CtSDP1, CtSDP2, and CtSDP3 were amplified from S4 cDNA sequences. The PCR-amplified fragments were digested with restriction enzymes and ligated into vectors digested with the same enzymes. For pKD7-RED-TlSDP, pKD7-RED-CtSDP1 and pKD7-RED-CtSDP2, XbaI and SalI were used, whereas BamHI and SmaI were used for pKD7-RED-CtSDP3. Each of the successfully constructed vectors was transformed into the ΔTlSDP strain to generate four complemented strains (ΔTlSDP/TlSDP, ΔTlSDP/CtSDP1, ΔTlSDP/CtSDP2, and ΔTlSDP/CtSDP3). The 72nt-insertion fragment (72INS) was deleted from pKD7-RED-CtSDP2 using La-TAQ PCR amplification, resulting in the reconstructed vector ΔTlSDP:[CtSDP2Δ72INS-RED] (Supplementary Table S2).
The pKAICR80 vector and the revamped pKAFCR100 vector were used for polygenic expression. The coding sequences of CtSDP1, CtSDP2, and CtSDP3 were amplified as described above. Each of the digested fragments was ligated into the same double-digested pKAFCR80 vector, resulting in three transition plasmids: pKAFCR80-CtSDP1, pKAFCR80-CtSDP2, and pKAFCR80-CtSDP3. For pKAFCR80-CtSDP1, XbalI, and KpnI enzyme cutting sites were added to both sides of the CtSDP1 sequence; for pKAFCR80-CtSDP2 and pKAFCR80-CtSDP3, SmaI/SacI and XbaI/XhoI were added. The transition plasmids were double digested with Claim and PmacI, respectively. The recovered 35S-P-CtSDP1-NOS-T fragment was ligated to the revamped pKAFCR100 plasmid and digested with the same enzymes, after which we obtained the pKAFCR100-CtSDP1 vector. Next, the 35S-P-CtSDP2-NOS-T fragment was ligated into the pKAFCR100-CtSDP1 vector and double-digested with Claim and HapI to obtain the pKAFCR100-CtSDP1-CtSDP2 vector. Double-gene expression vectors pKAFCR100-CtSDP1-CtSDP3 and pKAFCR100-CtSDP2-CtSDP3 were constructed in the same manner. Finally, the 35S-P-CtSDP3-NOS-T fragment was ligated into the pKAFCR100-CtSDP1-CtSDP2 vector and double-digested with ClaI and HapI, resulting in the three-gene expression vector pKAFCR100-CtSDP1-CtSDP2-CtSDP3. Using the same strategy, double and triple gene expression vectors were constructed, after which the ΔTlSDP/CtSDP12, ΔTlSDP/CtSDP13, ΔTlSDP/CtSDP23, and ΔTlSDP/CtSDP123 mutant strains were obtained.
The primer pairs used for a gene cloning and vector construction are listed in Supplementary Table S1. All primers were designed using DNAMAN. PCR cloning and analyses were performed using high fidelity fusion polymerase (Fermentas). Restriction enzymes, T4 DNA ligase, SYBR Premix Ex Taq and other DNA-modifying enzymes were used as recommended by the suppliers (TaKaRa, Dalian).
ATMT Transformation
Agrobacterium tumefaciens strain AGL-1 was used for transformation of T. lanuginosus strain 9W and C. thermophilum strain S4. The transformation process was modified based on a prior study (Khan et al., 2014). AGL-1 carrying a deletion or expression vector was cultured overnight in a rotatory shaker (180 rpm) at 28°C in 10 mL Luria-Bertani (LB) medium with 50 μg/mL rifampicin and 50 μg/mL kanamycin. Subsequently, 2.0 mL of the bacteria solution was centrifuged at 2,400 × g for 10 min, after which the precipitate was resuspended in inducible medium supplemented with 200 μM acetosyringone (AS) to achieve an optical density at 600 nm (OD600) of between 0.2 and 0.4, as assessed using a microplate reader (Molecular Devices, Sunnyvale, CA, United States). The medium was cultured with agitation at 180 rpm for 8–10 h at 28°C until an OD600 value of 0.8 was reached. T. lanuginosus 9W was cultured on PDA at 50°C for 4–5 days. The conidia were washed from the clones using induction liquid medium (IM) and adjusted to a final concentration of 1 × 105 conidia/mL. C. thermophilum S4 was cultured on CM at 50°C for nearly 7 days. The cleistocarp was harvested by scraping the agar surface, allowing the release of ascospores by shock or compression. The ascospores were filtered through four layers of sterile gauze, washed in sterilized distilled water, and adjusted to a final concentration of 1 × 105 spores/mL.
Sterile Hybond N membranes (Amersham Biosciences, Piscataway, NJ, United States) were placed on solid IM + AS plates. The corresponding concentration of T. lanuginosus conidia was mixed with the AGL-1 culture in equal volumes, after which 100 μL of the resulting mixture was pipetted onto the plate, spread evenly with a sterilized SS-Spreader, and air-dried. The plates were co-cultured in the dark at 25°C for 2 days. The filters were transferred onto selective PDA medium containing 80 μg/mL hygromycin B at 50°C in the dark until colonies appeared. The expression transformants were selected with 50 μg/mL G418 rather than hygromycin B. The control group was treated with distilled water.
Bioinformatic Analysis
Nucleotide and protein sequences were downloaded from the Fungal Genomes Database1 and analyzed using PubMed online tools and the SWISS-MODEL database for protein modeling2. Multiple sequence alignment was performed with the sequences of SDPs using DNAMAN. A phylogenetic tree was established via the neighbor-joining tree available in MEGA7.0.9, and the schematic diagram of the carrier was constructed using IBS 1.0.1 software.
Oligopeptide Transporter Activity Assay
Minimal medium base [1:50 dilution of the stock solution (26 g L–1 KCl, 26 g L–1 MgSO4, 76 g L–1 KH2PO4, 50 mL L–1 trace elements solution, pH 6.5)] was supplemented with glucose to a final concentration of 1%. Growth assays were carried out in Petri dishes (40 mm × 8 mm) filled with 10 mL solid growth medium. Each plate contained protease inhibitors [2 mM AEBSF (Sigma), 5 mM Pepstatin A (Sigma) and 1 mM EDTA (Sigma)] and 2–3 mg of a defined oligopeptide [KLLG, GGFL, or LWMR (Qiangyao BioTech, Shanghai)] in 10 mL culture medium as the only available nitrogen source.
Microscopic Observation
In order to observe fungal development structures, such as hook-like structures, coil–coil structures, or related structures, the corresponding strains were cultured in a PDA plate in which coverslips were obliquely inserted into the culture medium. The coverslips were removed from the plate and observed under a microscope (Nikon 80i, Japan) after the mycelia covered all of each coverslip.
For the subcellular localization, the corresponding strains were cultured in a PDA plate in which coverslips were obliquely inserted into the culture medium for 5 days and observed via fluorescence microscopy after the mycelia covered all of each coverslip. The coverslips were removed from the PDA plate, washed twice gently with double distilled water, and incubated with 100 μL DiO perchlorate (Yeasen Biology, Shanghai, China) for 2–20 min. The cell membrane dye DiO was used to stain the cell membrane. Green fluorescence from DiO and red fluorescence from DsRED2 were observed under an Olympus Xa21 microscope (Olympus, Tokyo, Japan).
Growth Rate and Lifespan Measurements
Lifespan was assayed according to the method of Geydan et al. (2012) with slight modification. In brief, lifespan was measured in time (days) and in length (cm) of continuous growth using Petri dishes (150 mm × 25 mm) filled with 65 mL of CM medium, culture in 50°C. Each measurement was performed at least in duplicate for each medium. The Petri dishes were incubated under an angle of 30–45°. Growth was scored three times every 5 days. Explants from the rims of over-grown Petri dishes were transferred to new Petri dishes with fresh medium to allow continued growth, and this process was repeated until the growth rate declined significantly. Strains were classified as non-senescent when the growth rate did not decline and there were no morphological changes. According to Geydan et al. (2012), fast senescing cultures showed a decline in growth rate and morphological changes, which were often followed by complete cessation of growth.
Measurement of Conidial Germination and Germ Tube Elongation
We investigated whether conidial growth was defective based on conidial production, the conidial germination rate, and bud tube elongation. First, conidial production was assessed by growing 5-mm mycelial plugs of wild-type and mutant strains separately on PDA plates at 50°C. After 5 days of culture, the conidia of the strains were washed with distilled water and counted. To measure the germination rate, 100 μL of the conidial suspension (104 conidia/mL) was mixed with 100 μL sterilized distilled water, added to each well of a 96-well plate, and incubated at 50°C. Once per hour, the appropriate experimental group was placed on a microscope slide and observed using a Nikon YS100 microscope (Nikon, Japan). Germ tube elongation was measured using photography software. Images of conidia germination were collected in the 5th hour. Each assay was independently repeated three times for each strain.
Measurement and Qualitative Detection of Endogenous H2O2
The hydrogen peroxide content was determined as previously described for plants (Cui et al., 2017). Hydrogen peroxide was extracted by homogenizing 3 g of mycelia from wild-type and mutant strains in 6 mL of cold acetone. The homogenate was centrifuged at 3,500 × g for 5 min at room temperature, and the resulting supernatant was designated as the sample extract. Thereafter, 0.1 mL of titanium reagent [5% (w/v) titanic sulfate in concentrated H2SO4] was added to 1 mL of the sample extract, followed by the addition of 0.2 mL of strong aqua ammoniac to precipitate the peroxide-titanium complex. The precipitated sample was centrifuged at 3,000 × g for 10 min at room temperature, the supernatant was discarded, and the precipitate was solubilized in 5 mL of 2M H2SO4. The absorbance of the samples was determined at 415 nm against a 2M H2SO4 blank. The hydrogen peroxide concentration in the samples was determined by comparing the absorbance against a standard curve of 0–5 μM titanium-H2O2 complex. This experiment was performed in triplicate and independently repeated three times for each strain.
Lucigenin (N,N′-dimethyl-9,9′-bisacridinium dinitrate) (Invitrogen, Cat#: L 6868) is a chemiluminescence probe that emits blue-green fluorescence when it reacts with a variety of reducing agents. The lucigenin signal was excited by an argon laser at a wavelength of 457 nm. The fluorescence was used to qualitatively measure changes in the H2O2 concentration. Hyphae used in this experiment were treated with 0.01M PBS for 2 h before incubation with 10 μM lucigenin. Confocal images were collected using an Olympus Xa21 microscope (Olympus, Tokyo, Japan).
Nitrogen Source Utilization Test
Nitrogen source utilization tests were conducted to determine the functions of TlSDP, CtSDP1, CtSDP2, and CtSDP3. Peptone, glutamic acid, reduced glutathione (GSH), tripeptide (KLG), and tetrapeptide (GGFL) were added separately to minimal medium [2 g (NH4)2SO4, 0.2 g MgSO4, 0.01 g CaCl2, 0.005 g FeSO4, 0.002 g MnCl2, 10.5 g K2HPO4, and 4.5 g KH2PO4 in 1000 mL distilled water, pH 7.5] replace the original nitrogen source ((NH4)2SO4) in the MM medium. Next, 5-mm mycelial plugs of the wild-type and mutant strains were cultured on the medium described above at 50°C for 5 days. The colonies were observed every day, and the colony diameter was measured after 5 days. Each sample was tested in three technical replicates. The experiment was repeated three times.
Quantitative Real-Time PCR (qRT-PCR)
The conidia suspension was incubated in CM medium at 50°C for 4 days in a rotatory shaker (180 rpm) or left stationary. Mycelia were collected by filtration, and TRIzol (TaKaRa, Dalian) was used to extract total RNA from the samples. First-strand cDNA was synthesized using the EasyScript First-Stand cDNA Synthesis SuperMix kit (Trans). Subsequently, qRT-PCR was performed using the PCRmax Eco system. Relative quantification of each DNA sample was performed using a DNA binding dye (TB Green remix Ex Taq II, TaKaRa). The thermal program was as follows: 50°C for 60 s and 95°C for 90 s, followed by 40 cycles of 95°C for 5 s, 60°C for 34 s, and 72°C for 30 s, with a final step at 95°C for 15 s, 60°C for 60 s, and 95°C for 15 s. Each sample was tested in three replicates in each experiment, and the actin gene was used as an internal standard. The relative quantification of each sample was calculated as follows: 2–ΔCq (Cq = Cqgene-Cqactin). For the figures, the expression quantity of the wild-type group was defined as 1, and the relative values of the other samples were calculated accordingly. The primer sequences used for qRT-PCR are listed in the Supplementary Information.
Statistical Analysis
All experiments were repeated at least three times. The mean ± standard deviation of the colony diameter, germination rate, and relative expression were determined using GraphPad Prism 7.00 software. Data were analyzed using InStat3. The threshold for statistical significance was p < 0.05.
Results
T. lanuginosus and C. thermophilum Inhabiting the Same Environment Reproduce in Distinct Patterns
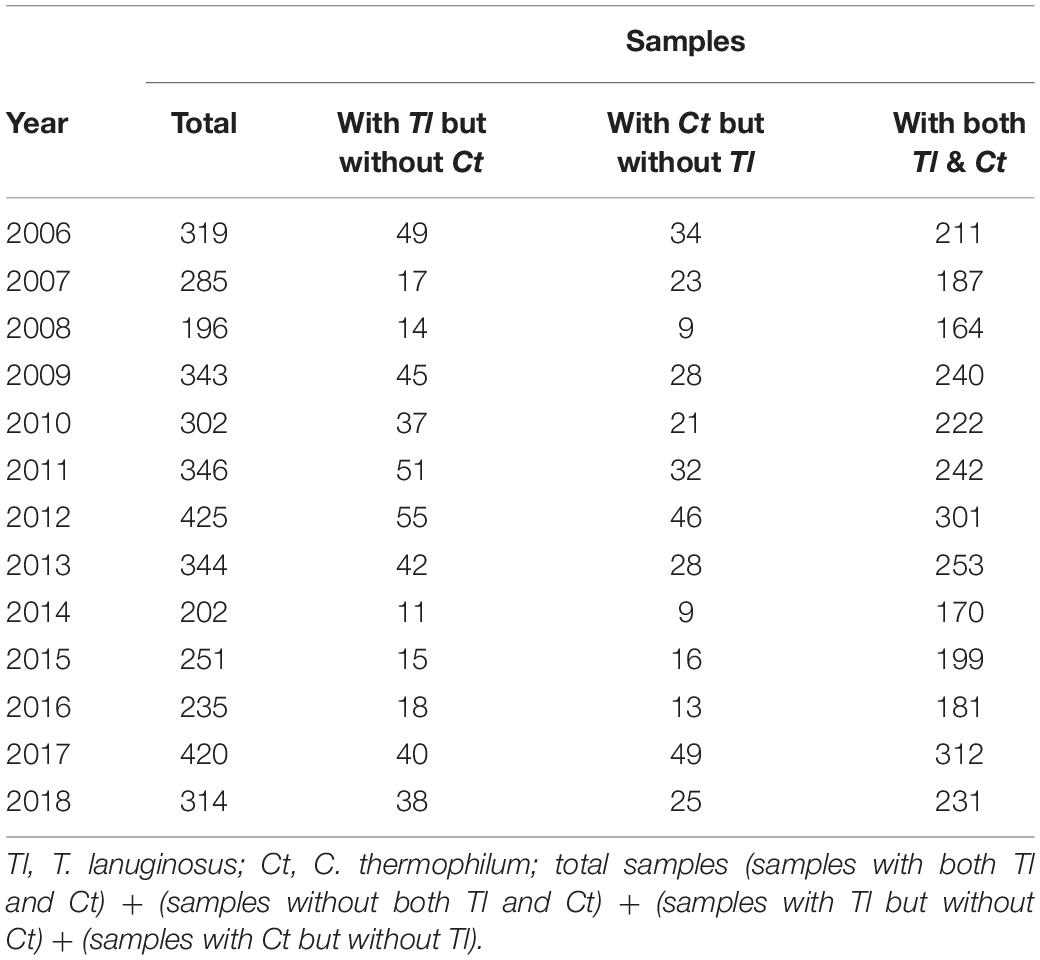
Thermophilic molds are distributed ubiquitously and thrive in a variety of natural habitats, including soils, composts, wood chip piles, nesting materials of birds and other animals, and municipal refuse (Singh et al., 2016). In our laboratory, a large number of thermophilic fungi have been isolated and identified from cow dung, straw or compost piles in northeastern China since 2006. During our investigations, we found that sexual reproduction in C. thermophilum and asexual reproduction in T. lanuginosus were often found in the same samples (Figure 1A and Table 1). T. lanuginosus is classified as a Deuteromycete, which is defined as unicellular or septate with asexual reproduction by the formation of aleurioconidia (Hauser et al., 2000). Initially the colonies appear white and grow rapidly, reaching 9 cm in diameter at 50°C within 5 days, after which turn gray or greenish-gray beginning from the center of the colony. C. thermophilum is a thermotolerant, floccose fungus that produces typical concentric rings of growth. The ascomata of C. thermophilum are dark brown, superficial, globose, subglobose, or ovate. The peridium is brown, of textura angularis or irregularis. The sexual ascospores (Figure 1A) appear globose or subglobose.
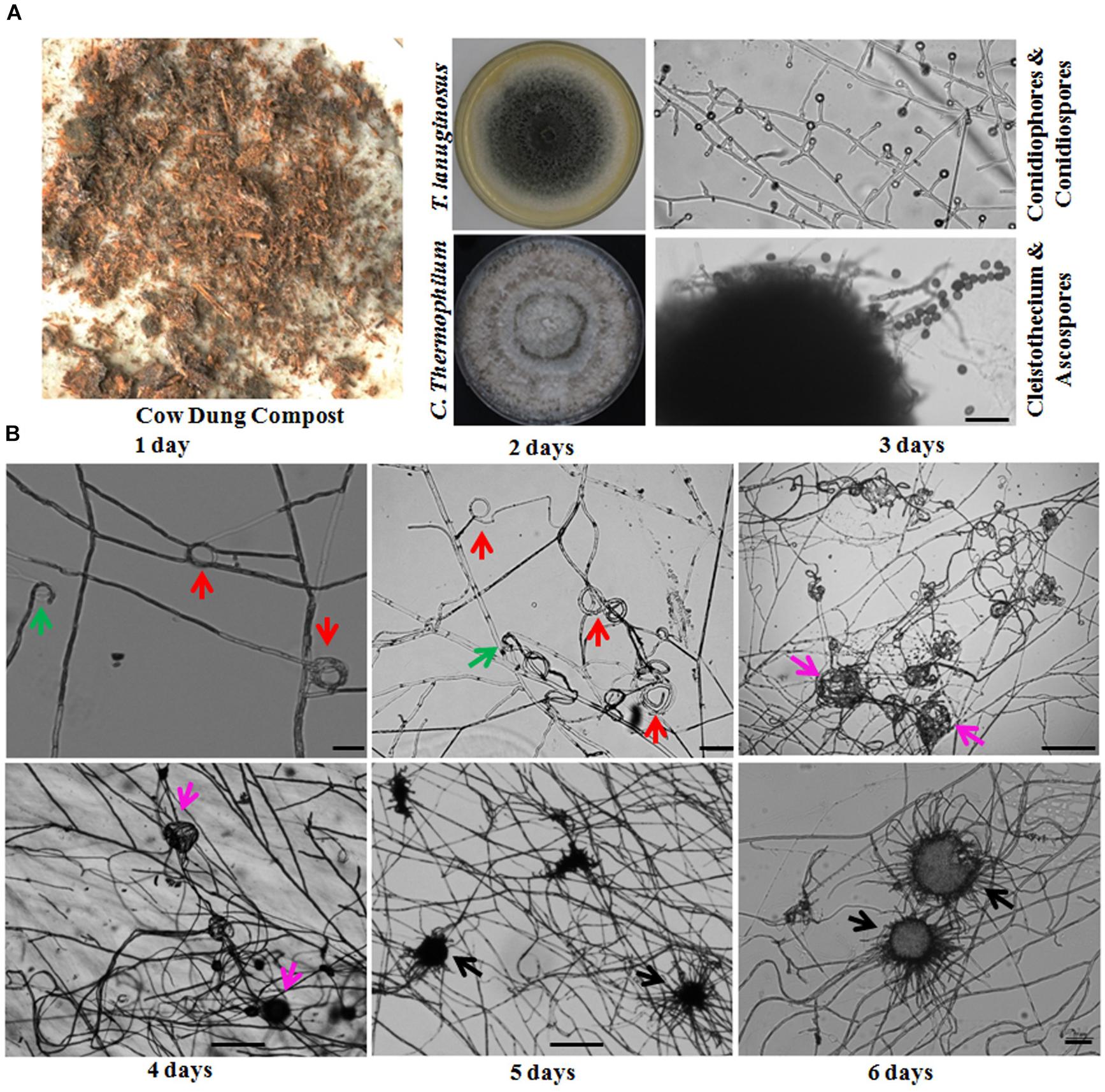
Figure 1. Thermomyces lanuginosus and Chaetomium thermophilum isolated from the same environment display different reproductive patterns. (A) A representative sample and the isolated T. lanuginosus and C. thermophilum strains. The cow dung sample was collected from Changchun, and the isolated strains were cultivated on CM plates under normal culture conditions for certain period; the T. lanuginosus conidiophores and conidiospores formed after 5 days, and the C. thermophilum cleistothecium and ascospore formed after 7 days. Bar = 20 μm. (B) Sexual lifecycle of C. thermophilum. The germinated ascospores were inoculated on CM media. The hook-like (green arrows), coil coil-like (red arrows), sclerotia-like (pink arrows), and immature cleistothecium structures (black arrows) formed at the apex of the hyphae on the different day. Bar = 50 μm.
The life cycle of the thermophilic fungus T. lanuginosus is comprised of a vegetative mycelium growth stage and a conidial reproductive stage. When T. lanuginosus conidia are inoculated on CM plates under normal culture conditions (pH 7.0, 50–55°C, in darkness), the conidia immediately germinate and enter the mycelia growth stage. After 2 days of vegetative growth, T. lanuginosus enter the reproductive stage and subsequently produce conidia. On the fourth or fifth day, a large number of mature conidia are formed, which can germinate and enter the next life cycle (Supplementary Figure S2). In contrast to T. lanuginosus, the life cycle of C. thermophilum is extremely complex. It must go through a stage of morphogenesis and then produce sexual ascospores. As the starting point of development, germinated ascospores were prepared in advance and inoculated on CM media. After 1 day of growth and development, hook-shaped or coil–coil structures formed at the apex of the hyphae, which represented the initiation of sexual reproduction. Over the next 5 days, the vegetative mycelia gradually aggregated into spherical structures. Next, the related hyphal cells differentiated into sterile and fertile hyphal cells. Finally, the fruiting mycelium, containing both sterile and fertile hyphae, matured into the ascocarp (Figure 1B). As the ascospores mature and are released, a sexual reproductive cycle is completed.
Four isp4/SDP Homologs and Their Expression Profiles
Based on the isp4 protein sequence reported by Sato et al. (1994), we performed a Blast-p search of the genomes of T. lanuginosus and C. thermophilum1. The four genes identified by the Blast-p search were designated TlSDP, CtSDP1, CtSDP2, and CtSDP3.
To measure the expression levels of the identified genes, samples for mRNA extraction were collected at different stages according to the time course of growth and development (1, 2, 3, 4, and 5 days). By using qRT-PCR analysis, TlSDP expression was found to increase from the vegetative growth stage to the conidial formation and maturation stage (Figure 2A). Tlmat, a mating type determination gene that is essential for sexual reproduction in sexual fungi, appeared to be up-regulated during fungal development (Figure 2A), suggesting the involvement of TlSDP and Tlmat in the growth and development of T. lanuginosus.
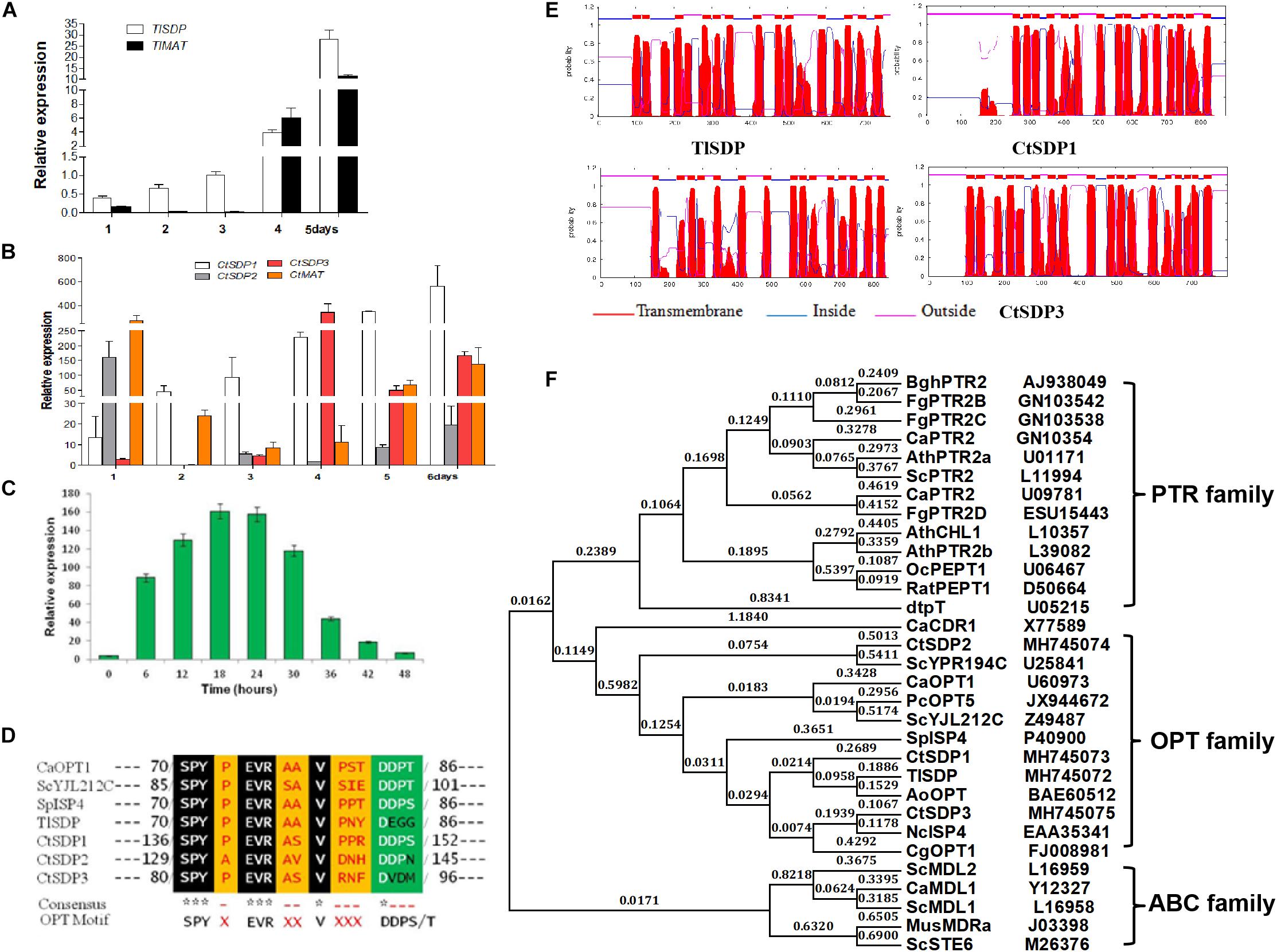
Figure 2. The isp4/SDP homologs in T. lanuginosus and C. thermophilum. (A) The expression pattern of TlSDP and TlMAT over 5 days. (B) The expression patterns of CtSDP1, CtSDP2, and CtSDP3 over 6 days. (C) The expression pattern of CtSDP2 over 48 h. (D) The single isp4 homolog in T. lanuginosus (TlSDP) and the three isp4 homologs in C. thermophilum (CtSDP1, CtSDP2, CtSDP3) contain the consensus sequence SPYxEVRxxVxxxDDP typical of OPTs. (E) TMHMM posterior probabilities for WEBSEQUENCE (on-line analysis). The analysis predicted 13–18 transmembrane-spanning domains, represented as red peaks. (F) Phylogenetic tree of oligopeptide transporter protein (OPT) sequences from various species. The full protein sequences from different eukaryotic organisms were analyzed, which revealed three major groups. The four isp4/SDPs homologs in this study grouped into the OPT clade.
Like TlSDP, CtSDP1 expression was continuously elevated from the vegetative growth stage to the ascospores formation and maturation stage (Figure 2B). However, CtSDP3 expression appeared to remain at a low level for the first 3 days but reached a much higher level on the fourth day, after which it decreased, but remained relatively high (Figure 2B). Surprisingly, CtSDP2 expression was highest on the first day. On the second day, CtSDP2 expression suddenly dropped from 160% to less than 5%; over the next 5 days, the relative expression level of CtSDP2 remained at a level of less than 30%, although its expression level increased slightly beginning on the third day (Figure 2B). Considering the complexity of sexual morphogenesis in C. thermophilum, the distinct expression patterns of the three CtSDPs may reflect their different roles in sexual reproduction.
In order to obtain a more detailed expression profile of CtSDP2, fungal samples were collected every 6 h during the initial 48-h period. In ascospores that had just germinated, the expression level of CtSDP2 was very low. CtSDP2 expression gradually increased with germ tube elongation and hyphal growth. The expression level was highest from 18 to 24 h, after which it gradually decreased and reached its lowest level at 48 h (Figure 2C). Given the large number of hook-like structures at the top of the hyphae formed at this stage, CtSDP2 is likely to be a relatively important factor during sexual morphogenesis.
Next, the structures of the proteins predicted to be encoded by TlSDP, CtSDP1, CtSDP2, and CtSDP3 were analyzed using online tools. The corresponding predicted proteins (TlSDP and CtSDP1–CtSDP3) contain the consensus sequence SPYxEVRxxVxxxDDP (Figure 2D), which has been found in all previously studied OPTs (Lubkowitz et al., 1997, 1998). All four proteins contained 13–18 transmembrane-spanning domains3 and were most likely to be localized in the plasma membrane (Figure 2E and Supplementary Figure S3A). The phylogenetic analysis of SDP proteins was performed by the neighbor joining method (Rogaeva et al., 1999) with proteins from other fungi and plants (Supplementary Table S3B). The phylogenetic tree showed two major clades among the selected proteins. One clade contained members of the ABC family, whereas the other clade was further divided into two sub-branches containing the PTR and OPT families (Figure 2F). In the OPT family, CtSDP1 and CtSDP3 possessed close evolutionary relationships to TlSDP, but not to CtSDP2 (Supplementary Figure S3B). These analyses clearly demonstrate that the four SDPs belong to the OPT family and that CtSDP2 differs from the other three proteins.
CtSDP1–3 Play Different Roles in TlSDP Deletion Strain (ΔTlSDP)
First, we constructed single gene deletion mutants (ΔTlSDP, ΔCtSDP1, ΔCtSDP2, and ΔCtSDP3) to verify that these genes function in their own strain (Supplementary Figures S1A, S6). In comparison with the T. lanuginosus wild-type strain, ΔTlSDP exhibited defective conidia, increased growth rate, and bud tube elongation (Figures 3B–E). The phenotype was more complex in the C. thermophilum single gene deletion mutants. In CM media, there was no significant change in colony growth among the deletion mutant strains, but significant alteration occurred at sexual morphogenesis (Figure 3A). ΔCtSDP2 loss resulted in no sexual structures and no formation of the cleistothecium or ascospores (Figure 3A), confirming the requirement of CtSDP2 for sexual structure initiation. In the ΔCtSDP1 and ΔCtSDP3 mutants, the cleistothecium and ascospores were produced, but the ΔCtSDP1 mutant appeared to show distorted and retarded cleistothecium formation. The ΔCtSDP3 mutant produced a smaller number of cleistothecia in comparison with the wild-type strain (Figure 3A). Interestingly, in the three mutant strains, loss of a single gene did not affect the expression of the other two genes (Supplementary Figure S4A), implying no mutual regulation among them.
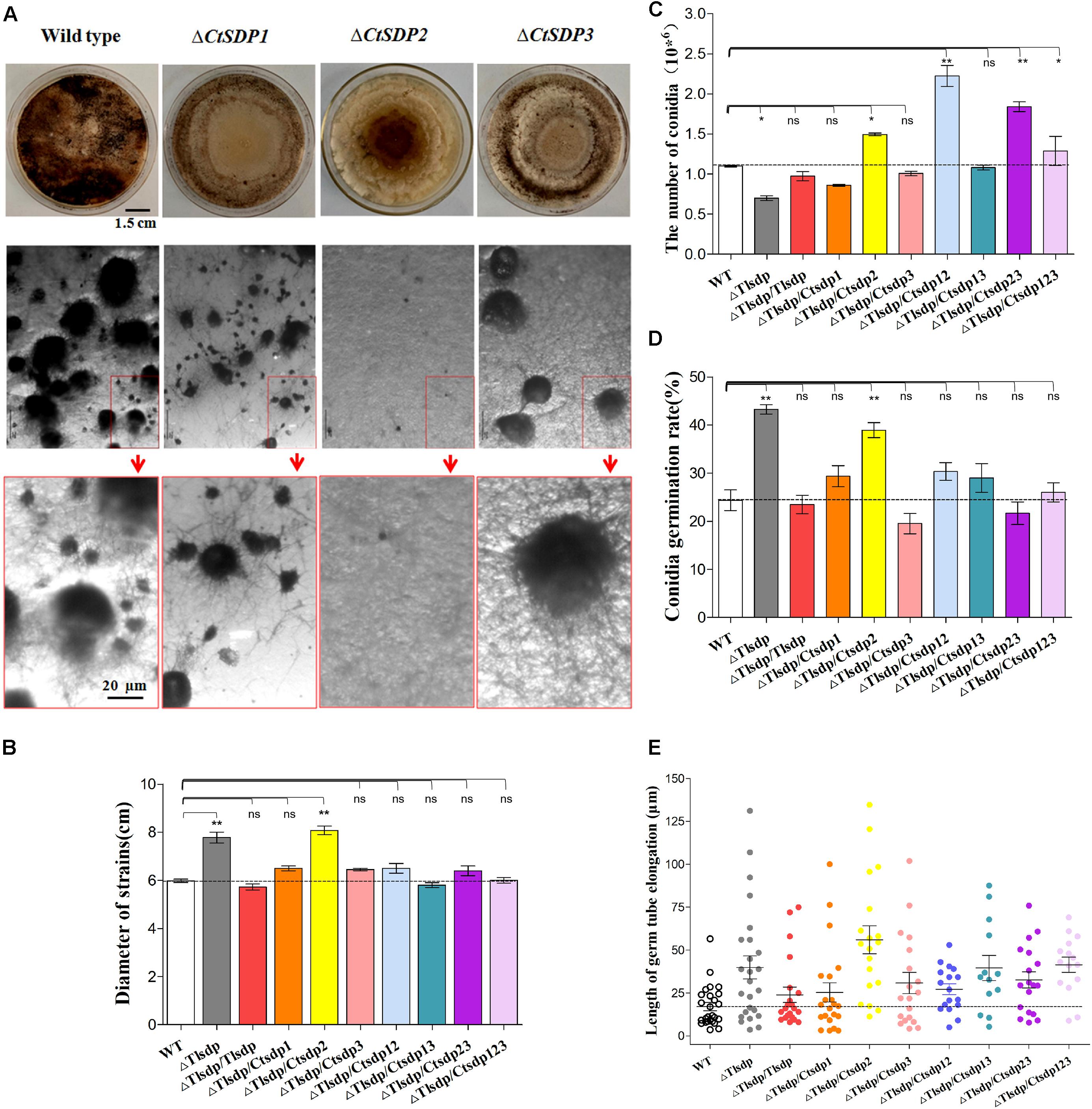
Figure 3. Biological roles of isp4/SDP genes in T. lanuginosus and C. thermophilum. (A) Colony morphologies of single-gene deletion mutants (ΔCtSDP1, ΔCtSDP2, and ΔCtSDP3) and the wild-type strains grown on CM plates at 50°C for 7 days (the first row); the representative microphotographic positions were selected randomly from the colonies of single-gene deletion mutants. The black spheroids represent sexual structures (the second row); the enlarged microphotographic pictures of colonies are shown in red rectangle boxes (the third row). Comparisons of (B) colony growth, (C) conidial production, (D) conidial germination rate, and (E) germ tube elongation between the ΔTlSDP, wild-type, and complemented strains (single, double, or triple expression of the three CtSDPs, based on the ΔTlSDP strains), grown on CM media at 50°C for 5 h. Error bars represent standard deviation. Significant differences between the wild-type strains and the two mutant strains are indicated as: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; or ns, not significant.
To further verify the functional relationship of the four SDP genes, three CtSDP genes were constructed and transformed into the ΔTlSDP mutant strain (single, double, and triple transformation) (Supplementary Figures S1B–D). These strains were subjected to assessments of colony growth, conidial production and germination, and germ tube elongation. In comparison with the wild-type (T. lanuginosus) and ΔTlSDP/TlSDP strains, all complemented strains, with the exception of ΔTlSDP/CtSDP2, produced colonies of a similar size (Figure 3B and Supplementary Figure S4B). Interestingly, only ΔTlSDP/CtSDP2 was not able to restore the growth phenotype of ΔTlSDP (Figure 3B), implying that CtSDP1 and CtSDP3, but not CtSDP2, play the same role as TlSDP in growth.
With regard to sporulation, all SDP genes restored the defective conidia production of the ΔTlSDP strain (Figure 3C). Furthermore, sporulation of ΔTlSDP/CtSDP2, ΔTlSDP/CtSDP12, ΔTlSDP/CtSDP23, and ΔTlSDP/CtSDP123 was higher or significantly higher than that of the wild-type strain during growth on PDA or CM media plates for 4 days (Figure 3C), further indicating the importance of CtSDP2 in sporogenesis.
Conidial vitality was investigated by measuring the conidial germination rate and assessing germ tube elongation. The germination rate of the ΔTlSDP/CtSDP2 strains was significantly greater than that of the wild-type strain (Figure 3D). In addition, assessment of germ tube elongation showed that the germinated spores continued to grow and develop (Figure 3E). The biological activities of the complemented strains suggested that the four tested genes are involved in growth, as well as the formation and development of conidia. Moreover, CtSDPs, especially CtSDP2, may play dual roles in sexual and asexual reproduction.
Subcellular Location and Oligopeptide Transporter Activity of Four SDPs Genes
In order to verify whether the four SDPs has OPT activity, experiments assessing protein subcellular localization and nitrogen source utilization were performed. First, four DsRED-tagged SDP complementation strains based on ΔTlSDP were constructed. In these strains, every SDP gene was fused with DsRED, which was mainly used to determine the subcellular location of the fused protein (Supplementary Figure S1B). After 60 h of cultivation, DsRED red fluorescence co-localized with DiO green fluorescence on the hyphal cell membrane, septa and conidial surface in all four complementation samples, confirming that the four tested genes are localized on the cell membrane (Figure 4A). In addition, in the CtSDP2/DsRED fusion strains, the merged fluorescence signal was markedly accumulated at the top, branch, and septum of the growth hyphae (Figure 4B), implying that CtSDP2 is associated with fungal growth and development.
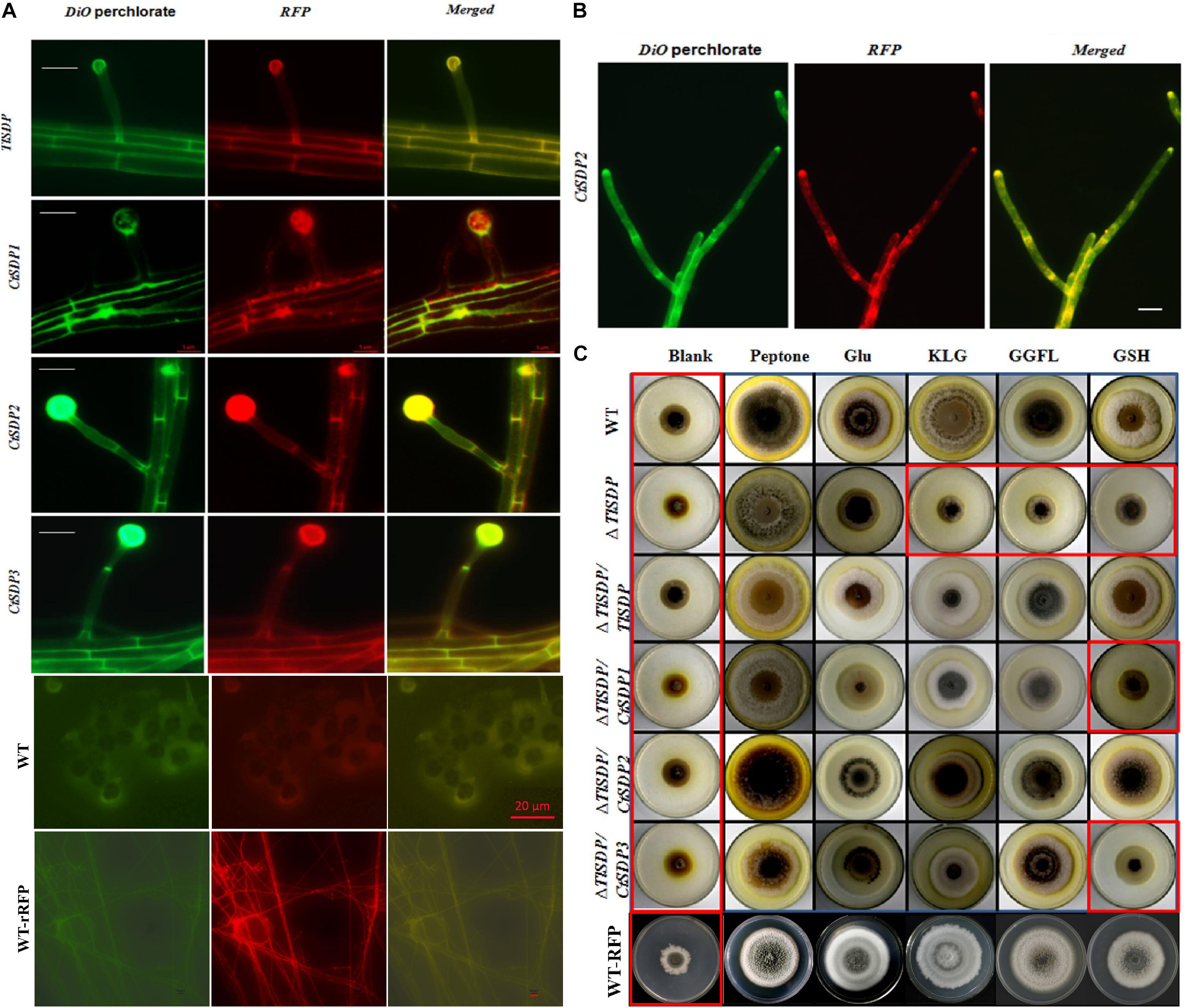
Figure 4. The OPT activity and subcellular locations of the four isp4/SDP proteins. (A) Intracellular localization of four SDP proteins. In the merged image, DiO perchlorate (green fluorescence) colocalized with DsRED (red fluorescence), appearing as yellow areas. Merged, green fluorescence overlapped with red fluorescence of DsRED. Images were acquired after 36 h of growth in CM medium. Bar = 10 μm. (B) CtSDP2/DsRED accumulated at the top, branch, and septum of the growth hyphae. Bar = 10 μm. (C) Biological analysis of OPT activity in a series of complemented strains expressing the four SDP genes. Growth experiments were performed on MM media lacking a nitrogen source (blank) and MM media supplemented with peptone, glutamic acid, GSH, tripeptide (KLG), or tetrapeptide (GGFL) as the sole nitrogen resource with all the tested strains. Strains in red rectangular boxes are inhibited in growth (thus, no OPT activity).
The coding proteins of Isp4 and its homologs are OPTs (Sato et al., 1994), and the four SDP proteins identified in this study also contain OPT family special sequence SPYxEVRxxVxxxDDP. To verify the transporter ability of the SDPs, growth experiments on MM media were carried out with different nitrogen sources. T. lanuginosus wild-type, ΔTlSDP/TlSDP and ΔTlSDP strains grew poorly on MM media lacking a nitrogen source, but the wild-type and ΔTlSDP/TlSDP strains grew well when synthesized tripeptides (KLG) or tetrapeptides (GGFL) were added as the sole nitrogen resource (Figure 4C). This experiment confirmed that TlSDP is an OPT, at least for the oligopeptide used in this research.
The growth of ΔTlSDP/CtSDP1, ΔTlSDP/CtSDP2, and ΔTlSDP/CtSDP3 was assessed on nitrogen-deficient media or media supplemented with a nitrogen source such as tetrapeptides. Additionally, protease inhibitors were added to limit degradation of oligopeptides by secreted proteases. Among several oligopeptides tested, the tetrapeptide GGFL revealed the most obvious growth phenotype. On nitrogen-deficient media supplemented with GGFL, ΔTlSDP growth was extremely retarded; however, all the three complemented strains, as well as ΔTlSDP/TlSDP, grew normally (Figure 4C). Unexpectedly, when GSH was used as the sole nitrogen source, only TlSDP and CtSDP2 were able to restore the defective growth of ΔTlSDP (Figure 4C), suggesting that TlSDP and CtSDP2 are involved in glutathione transportation.
Life Span Is Extended in ΔCtSDP2 and Shortened in ΔTlSDP/CtSDP2
During incubation and storage with PDA or CM at room temperature or 50°C, almost all isolated C. thermophilum strains in our lab were susceptible to aging and death in fewer than 60 days (Supplementary Table S4). Life span analysis of the C. thermophilum wild-type S4 strain and the three ΔCtSDP strains was carried out according to Geydan et al. (2012) and Cui et al. (2017). In the third culture cycle, growth of the S4, ΔCtSDP1, and ΔCtSDP3 strains stopped on the 15th–18th day, meaning the life span of these fungi was less than 58 days (Figure 5A). However, the ΔCtSDP2 strain has an extended lifespan of more than 65 days (Figure 5A). Thus, we speculate that CtSDP2 has a negative effect on fungal lifespan.
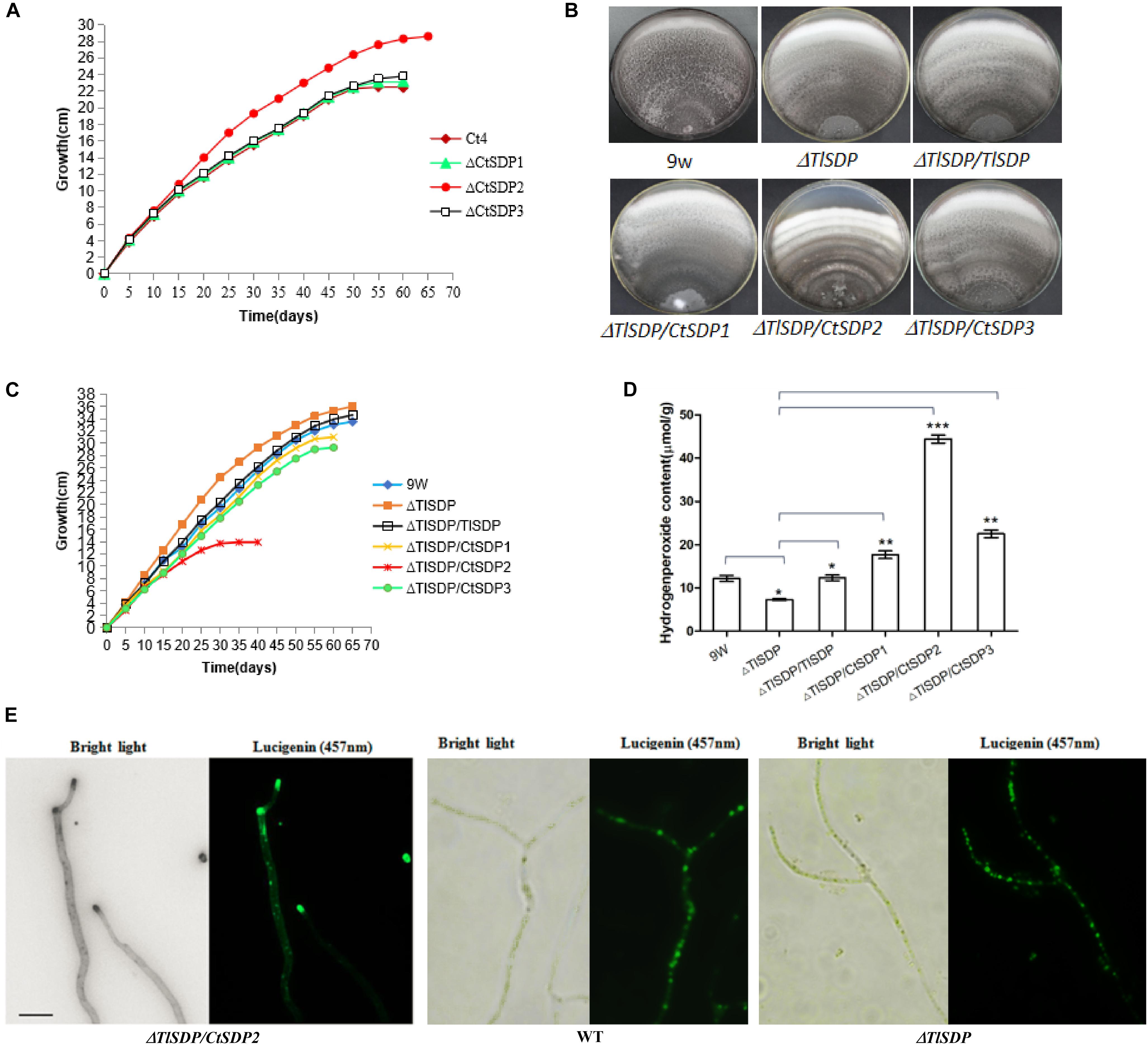
Figure 5. Lifespan is extended in ΔCtSDP2 and shortened in ΔTlSDP/CtSDP2. (A) ΔCtSDP2 exhibited constant growth for greater than 65 days. When compared to the ΔCtSDP2 strain, the ΔCtSDP1, ΔCtSDP3, and S4 wild-type strains exhibited slowed growth phases, followed by a halt in growth at approximately less than 58 days. Growth curves were established from 30 subcultures of six independent spores for each genotype (S4, ΔCtSDP1, ΔCtSDP2, and ΔCtSDP3). (B) The mycelial phenotype of ΔTlSDP, ΔTlSDP/TlSDP, ΔTlSDP/CtSDP1, ΔTlSDP/CtSDP2, and ΔTlSDP/CtSDP3 inoculated on CM medium at 20 days. (C) The growth curves of ΔTlSDP, ΔTlSDP/TlSDP, and ΔTlSDP/CtSDP1-3 inoculated on CM medium. The ΔTlSDP/CtSDP2 strains displayed a slow growth phase, followed by a halt in the growth phase at approximately less than 35 days. Other tested strains exhibited constant growth for more than 60–65 days. Growth curves were established from 30 subcultures of six independent spores for each genotype. (D) Hydrogen peroxide (H2O2) content of the ΔTlSDP, ΔTlSDP/TlSDP, ΔTlSDP/CtSDP1, ΔTlSDP/CtSDP2, and ΔTlSDP/CtSDP3 strains. Significant differences between the wild-type strains and the two mutant strains are indicated as: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; or ns, not significant. Error bars represent standard deviation. (E) The ΔTlSDP/CtSDP2 strains accumulate more H2O2 at the top of their hyphae. The light (left) and fluorescence confocal (right) microscope observations of a ΔTlSDP/CtSDP2 hypha. Bar = 10 μm.
Next, we performed lifespan measurement for the T. lanuginosus wild-type strain and five mutants (ΔTlSDP/TlSDP, ΔTlSDP, ΔTlSDP/CtSDP1, ΔTlSDP/CtSDP2, and ΔTlSDP/CtSDP3). In the first culture cycle, the ΔTlSDP/CtSDP2 strains showed obvious mycelial growth reduction and concentric undulate hyphae (Figure 5B). In the second 20-day culture cycle, ΔTlSDP/CtSDP2 growth stopped on the 15th day, indicating that the maximum lifespan of the ΔTlSDP/CtSDP2 strain was approximately 35 days (Figure 5C). The growth of the ΔTlSDP strains on CM was linear at 65 days, indicating that its lifespan was greater than 65 days. The lifespans of the ΔTlSDP/TlSDP, ΔTlSDP/CtSDP1, and ΔTlSDP/CtSDP3 strains ranged from 60 to 65 days (Figure 5C). These results show that CtSDP2, which has a negative effect on the lifespan of C. thermophilum, plays a similar role in heterologous species.
Reactive oxygen species (ROS) are associated with reduced longevity (Bianchi and Falcioni, 2016; Nita and Grzybowski, 2016), and ROS production has been regarded as the primary cause of biological aging (Harman, 1972; Bianchi and Falcioni, 2016; Nita and Grzybowski, 2016). Thus, we compared the hydrogen peroxide content of the ΔTlSDP and ΔTlSDP/CtSDP2 strains. The hydrogen peroxide content of the ΔTlSDP/CtSDP2 strain was significantly higher than that of the ΔTlSDP strain (Figure 5D). In addition, accumulation of hydrogen peroxide at the tops of hyphae was observed in the ΔTlSDP/CtSDP2 strain (Figure 5E), but not in the ΔTlSDP strain, partially explaining the relationship between longevity and CtSDP2 expression.
The 72-nt Insertion Fragment (72INS) of CtSDP2 and Its Role in Sexual Development
To determine why the role of CtSDP2 differs from the four SDPs, their protein sequences were aligned (Supplementary Figure S5). An extra oligopeptide fragment consisting of 24 amino acid residues from Arg518 to Leu541 (Figure 6A) was found in CtSDP2. This 24-amino acid oligopeptide fragment is encoded by 72 nucleotides (nt) that appear to be inserted into the original sequence of CtSDP2 (Supplementary Figure S7), and it was designated 72INS. A website Blast search indicated that 72INS might be associated with surface proteins and leucine rich repeat proteins (Figure 6B).
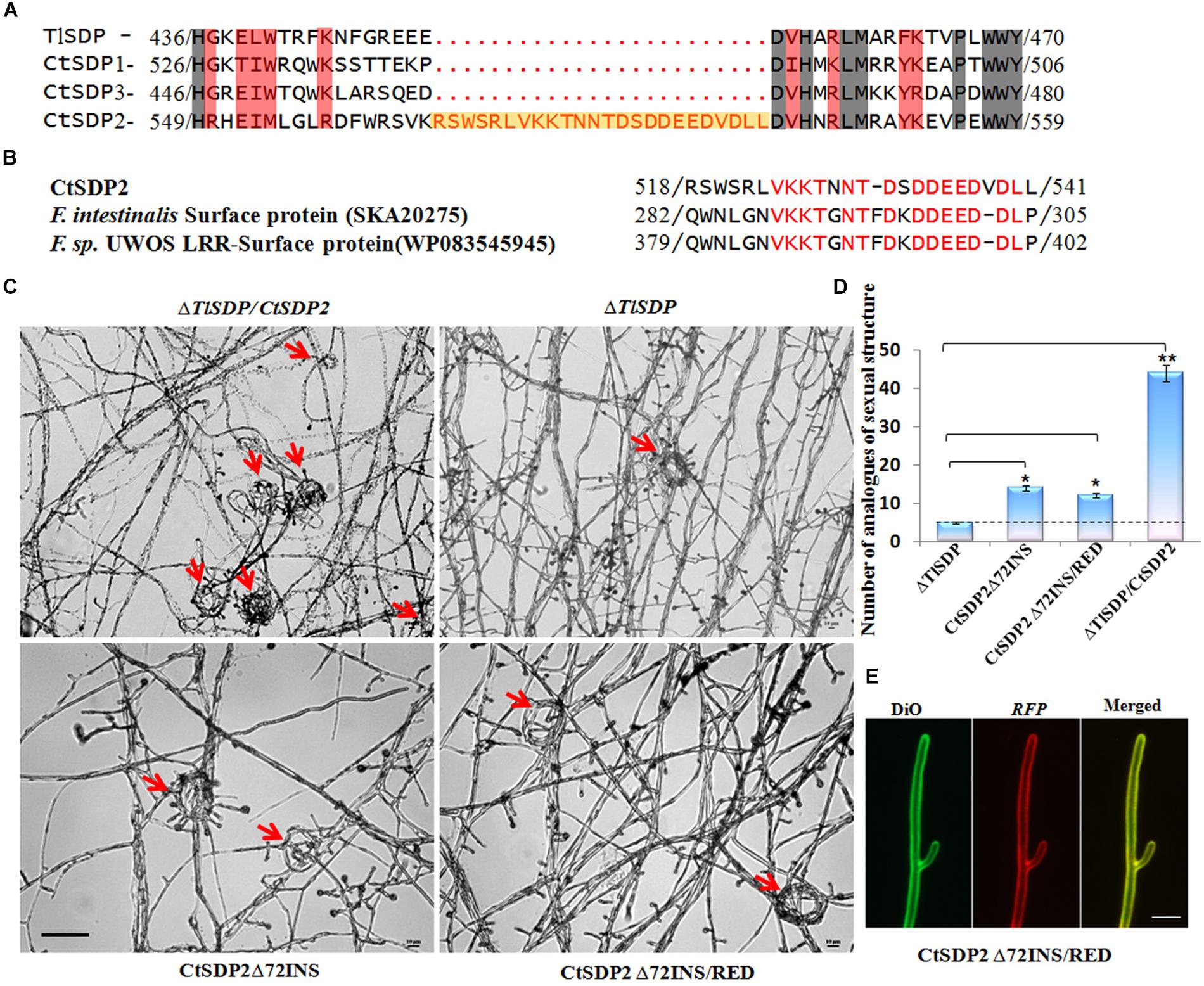
Figure 6. The 72-nt insertion fragment (72INS) of CtSDP2 and its role in sexual development. (A) Alignments of the predicted amino acid sequences of TlSDP, CtSDP1, CtSDP2, and CtSDP3. An extra oligopeptide fragment (72INS) consisting of 24 amino acid residues between 518 (Arg) and 541 (Leu) is highlighted in the CtSDP2 protein. (B) Amino acid sequences similar to the 72INS (24 amino acid residues) of CtSDP2. Identical amino acid residues are indicated by red letters in CtSDP2, F. intestinalis surface protein (SKA20275), and F. sp. UWOS LRR surface protein (WP083545945). (C) Phenotypic comparison of the ΔTlSDP/CtSDP2, ΔTlSDP, CtSDP2Δ72INS, and CtSDP2Δ72INS/RED strains. Phenotypes of the tested strains grown on solid CM for 4 days were observed. The analogs of the sexual structure, such as coil coil-like structures (red arrows), were observed. Bar = 50 μm. (D) The average number of analogs of sexual structures of the tested strains observed in different microscope vision fields. Significant differences between the relative strains are indicated as: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; or ns, not significant. (E) Intracellular localization of the CtSDP2Δ72INS/RED protein. In the merged image, DiO perchlorate (green fluorescence) colocalized with DsRED (red fluorescence), appearing as yellow areas. CtSDP2Δ72INS/RED proteins no longer accumulated on the top of the hyphae, and the hyphal septa at the apex of the hyphae or near the hyphal branches were barely observed. Images were acquired after 36 h of growth in CM medium. Bar = 10 μm.
In order to explore the function of 72INS, we constructed two 72INS-deletion vectors based on the pKD7-RED-CtSDP2 vectors (Supplementary Figures S1B,C), in which 72INS was deleted using La-TAQ PCR amplification (Supplementary Table S1). The transformant containing CtSDP2Δ72INS (CtSDP2 lacking 72INS) was obtained and compared with strains containing the corresponding full-length sequences in the ΔTlSDP strain. No significant differences were found between the tested strains in terms of colony phenotype, growth rate, sporogenesis, or OPT absorption. However, the number of hook-shaped or coil–coil structures was sharply reduced in the 72INS deletion strains (Figures 6C,D). Therefore, 72INS may contribute to the initiation of sexual morphogenesis, given that CtSDP2 is required for sexual structure formation in C. thermophilum (Figure 3A) and the ΔTlSDP/CtSDP2 strain formed more analogs of sexual structures (hook-like or coil–coil structures) than did the ΔTlSDP/CtSDP2Δ72INS strain (Figure 6C). The DsRED-tagged proteins did not accumulate at the tops of the hyphae. In addition, hyphal septa at the hyphal apex or near the hyphal branches were hardly observed (Figure 6E). These results reflect the involvement of 72INS in CtSDP2 accumulation at the tops of hyphae and septum formation.
Discussion
Peptide transport across membranes is accomplished by diverse organisms. At present, three families of transporters have been intensively studied: the ABC (ATP binding cassette) family, the PTR (peptide transport) family, and the OPT family (Stacey et al., 2002). The ABC superfamily transports a large variety of substances (not restricted to peptides) (Payne and Smith, 1994). The PTR family transports amino acids, dipeptides, tripeptides, and nitrate (Stacey et al., 2002). In contrast to the ABC and PTR families, OPTs predominantly transport tetrapeptides and pentapeptides (Lubkowitz et al., 1997, 1998). In this study, the unique TlSDP gene of T. lanuginosus was cloned and biologically analyzed through creating the TlSDP gene deletion strains and complementation strains. As to the three SDPs in C. thermophilum, we successfully obtained three single gene deletion mutants (ΔCtSDP1-3), and at least three independent transformation colonies for each CtSDP mutants were isolated and checked for the defects. Unfortunately, we did not get the corresponding complementary strains. (We have tried many times, and each time we screened nearly 1000 candidate transformants, but they were identified as not having complementary genes.) Therefore, based on the ΔTlSDP strains, we transformed each of CtSDP1-3 in the T. lanuginosus system, which we have successfully used in previous study. The four SDPs showed tetrapeptide transport activity according to the results of complementary growth experiments under nitrogen deficiency conditions (Figure 4C), confirming their assignment to the OPT family (Figure 2F). In addition, TlSDP and CtSDP2, in contrast with the other two SDPs, possessed the ability to transport glutathione, suggesting that TlSDP and CtSDP2 are glutathione transporter like Hgt1p (Opt1p) (Bourbouloux et al., 2000) and HGT1 (Dworeck et al., 2009).
Peptide transport systems are involved in multiple cellular processes, such as yeast mating and fungal pathogenicity (Kuchler et al., 1989; McGrath and Varshavsky, 1989; Stacey et al., 2002; Chague et al., 2009). The Blumeria graminis PTR2 gene was reported to be expressed in infection structures (Droce et al., 2015). In addition, the Fusarium graminearum PTR2 gene is associated with perithecium development and conidial production (Droce et al., 2017). Interestingly, our SDPs seemed to share some similarities with PTR2 regarding sexual development-related biological functions.
The TlSDP is the only isp4 homolog in T. lanuginosus. Along with the three CtSDP genes, TlSDP was shown to be an OPT member based on growth experiments using complementary transformants (Figure 4C), The presence of fewer OPTs implied that an alternative system is required for nitrogen source absorption in T. lanuginosus. In the growth assay for OPT identification, differences in colony growth between the SDP deletion strains and the complementary strains were observed only when protease inhibitors were added to nitrogen-deficient media supplemented with polypeptides (data not shown), indicating that a powerful protease-dependent secretory degradation system exists in T. lanuginosus. Surely, other peptide transport systems, including members of the PTR families, may be involved in the utilization of nitrogen resources, because multiple sequences similar to those of PTR members were found in the genomes of T. lanuginosus and C. thermophilum4,5. With regard to nitrogen uptake, the important, but inessential, OPT activity of CtSDPs does not fully explain their biological functions in sexual development, especially the initial role of CtSDP2 in sexual reproduction. Thus, the molecular mechanism of SDPs in sexual reproduction must be further studied.
In this study, we have created a series of mutants and complementary transformant strains, which provide rich materials for functional analyses of different SDPs. Each of the three deletion mutants of CtSDP1–CtSDP3 contains the other two genes; therefore, the phenotype of the single-gene deletion strains did not entirely reflect the effect of the deleted gene. However, the requirement of CtSDP2 for sexual morphogenesis was confirmed, because no sexual structures were observed in the ΔCtSDP2 strain. Although we failed to obtain double- or triple-deletion CtSDP mutants, the ΔTlSDP strain served as an important genetic tool that enabled us to study CtSDPs. In addition, the reduced conidial production and defective asexual reproduction of the ΔTlSDP strain made it suitable for experiments assessing sporulation and sexual reproduction. Biological analyses of the functions of CtSDP1, CtSDP2, and CtSDP3 were performed using the ΔTlSDP system. The observed enhancement of conidial production in the ΔTlSDP/CtSDP strain indicates the dual role of CtSDPs.
CtSDP2 expression tended to reduce the longevity of fungus (Figures 5A,B). This effect was probably associated with energy metabolism, given the enormous energy requirements for growth and development (Cui et al., 2017). H2O2 and other ROS induce aging, and the production of free ROS, such as H2O2, in cells is the primary cause for biological aging (Harman, 1972; Falcioni and Bianchi, 2016; Nita and Grzybowski, 2016). In comparison with the ΔTlSDP strains, ΔTlSDP/CtSDP2 accumulated more H2O2 (Figure 5D), and the lifespan of the ΔTlSDP/CtSDP2 strain was much shorter than those of the ΔTlSDP strains (Figure 5C).
Insertion and deletion events cannot be accurately identified if ancestral sequence information is deficient. Human genetic evidence suggests that insertion and deletion are major sources of gene defects (Smith et al., 1997; Rogaeva et al., 1999; Mcgovern et al., 2005; Volfovsky et al., 2009). In Bacillus subtilis, Escherichia coli, and Saccharomyces cerevisiae, insertions and deletions are not randomly distributed and are likely to occur more often in essential proteins and those that are highly connected, indicating a possible role of sequence insertions and deletions in the regulation and modification of protein–protein interactions (Chan et al., 2007). In our research, a 24-amino acid oligopeptide fragment (encoded by 72 nt DNA sequence) was only found in CtSDP2, suggesting that a 72-nt DNA fragment was inserted into the original sequence of CtSDP2, if the other three SDPs are ancestral sequence. Sequences similar to 72INS are contained in some surface proteins and are likely to play important roles in initial adherence to mucosal tissue, as well as in long-term survival of the pathogen on mucosal surfaces (de Miguel et al., 2010). Thus, CtSDP2 gained extra functions in comparison with the other three SDPs (Figure 6A).
Insertion and deletion are evolutionary changes in the sequence length of DNA and protein molecules (Britten et al., 2003; Tao et al., 2007). To our knowledge, the functions of insertions or deletions are not well-understood. An in-depth study of 72INS will provide a valuable reference for us to carry out functional insertion sequence research. This study is the first to link the expression of SDP/isp4 genes to their sexual differentiation functions in C. thermophilum and T. lanuginosus. In particularly, we confirmed that CtSDP2 harbors a unique functional 72INS that is required for sexual morphogenesis.
Data Availability Statement
The datasets generated for this study can be found in NCBI, accession numbers MH745073, MH745074, MH745075, and MH745072.
Author Contributions
S-HZ, X-LX, and YW conceptualized the study. X-LX, YW, Y-YS, G-MP, and S-HZ worked on the data curation. X-LX, YW, Y-YS, G-MP, L-NC, and S-HZ did the formal analysis. S-HZ and YW were responsible for the funding acquisition and the project administration. X-LX, YW, Y-YS, G-MP, L-NC, GW, and S-HZ carried out the investigation. X-LX, YW, Y-YS, and S-HZ worked on the methodology. S-HZ supervised the study and X-LX wrote, reviewed and edited the original draft.
Funding
This research was financially supported by the National Natural Science Foundation of China (Grant Nos. 31671972 and 31670141) and a project of the Ministry of Science and Technology of China (Grant No. 2016YFD0300703).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02994/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/assembly/GCA_000315935.1/
- ^ https://www.swissmodel.expasy.org/
- ^ http://www.cbs.dtu.dk/services/TMHMM/
- ^ http://fungi.ensembl.org
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
References
Bianchi, V. E., and Falcioni, G. (2016). Reactive oxygen species, health and longevity. AIMS Mol. Sci. 4, 479–504.
Böhm, J., Hoff, B., O’Gorman, C. M., Wolfers, S., Klix, V., Binger, D., et al. (2013). Sexual reproduction and mating-type–mediated strain development in the penicillin-producing fungus Penicillium chrysogenum. Proc. Natl. Acad. Sci. U.S.A. 110, 1476–1481. doi: 10.1073/pnas.1217943110
Bourbouloux, A., Shahi, P., Chakladar, A., Delrot, S., and Bachhawat, A. K. (2000). Hgt1p, a high affinity glutathione transporter from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 275, 13259–13265. doi: 10.1074/jbc.275.18.13259
Britten, R. J., Rowen, L., Williams, J., and Cameron, R. A. (2003). Majority of divergence between closely related DNA samples is due to indels. Proc. Natl. Acad. Sci. U.S.A. 100, 4661–4665. doi: 10.1073/pnas.0330964100
Cavalier-Smith, T. (2010). A revised six-kingdom system of life. Biol. Rev. Camb. Philos. Soc. 73, 203–266. doi: 10.1111/j.1469-185x.1998.tb00030.x
Chague, V., Maor, R., and Sharon, A. (2009). CgOpt1, a putative oligopeptide transporter from Colletotrichum gloeosporioides that is involved in responses to auxin and pathogenicity. BMC Microbiol. 9:173. doi: 10.1186/1471-2180-9-173
Chan, S. K., Hsing, M., Hormozdiari, F., and Cherkasov, A. (2007). Relationship between insertion/deletion (indel) frequency of proteins and essentiality. BMC Bioinformatics 8:227. doi: 10.1186/1471-2105-8-227
Cui, X., Wei, Y., Xie, X. L., Chen, L. N., and Zhang, S. H. (2017). Mitochondrial and peroxisomal Lon proteases play opposing roles in reproduction and growth but co-function in the normal development, stress resistance and longevity of Thermomyces lanuginosus. Fungal Genet. Biol. 103, 42–54. doi: 10.1016/j.fgb.2017.04.002
de Miguel, N., Lustig, G., Twu, O., Chattopadhyay, A., Wohlschlegel, J. A., and Johnson, P. J. (2010). Proteome analysis of the surface of Trichomonas vaginalis reveals novel proteins and strain-dependent differential expression. Mol. Cell. Proteomics 9, 1554–1566. doi: 10.1074/mcp.M000022-MCP201
Droce, A., Holm, K. B., Olsson, S., Frandsen, R. J., Sondergaard, T. E., Sørensen, J. L., et al. (2015). Expression profiling and functional analyses of BghPTR2, a peptide transporter from Blumeria graminis f. sp. hordei. Fungal Biol. 119, 551–559. doi: 10.1016/j.funbio.2015.02.007
Droce, A., Sørensen, J. L., Sondergaard, T. E., Rasmussen, J. J., Lysøe, E., and Giese, H. (2017). PTR2 peptide transporters in Fusarium graminearum influence secondary metabolite production and sexual development. Fungal Biol. 121, 515–527. doi: 10.1016/j.funbio.2017.02.003
Dworeck, T., Wolf, K., and Zimmermann, M. (2009). SpOPT1, a member of the oligopeptide family (OPT) of the fission yeast Schizosaccharomyces pombe, is involved in the transport of glutathione through the outer membrane of the cell. Yeast 26, 67–73. doi: 10.1002/yea.1652
Dyer, P. S., and O’Gorman, C. M. (2011). A fungal sexual revolution: Aspergillus and Penicillium show the way. Curr. Opin. Microbiol. 14, 649–654. doi: 10.1016/j.mib.2011.10.001
Dyer, P. S., and O’Gorman, C. M. (2012). Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol. Rev. 36, 165–192. doi: 10.1111/j.1574-6976.2011.00308.x
Falcioni, G., and Bianchi, V. E. (2016). Reactive oxygen species, health and longevity. AIMS Mol. Sci. 3, 479–504.
Geydan, T. D., Debets, A. J., Verkley, G. J., and van Diepeningen, A. D. (2012). Correlated evolution of senescence and ephemeral substrate use in the Sordariomycetes. Mol. Ecol. 21, 2816–2828. doi: 10.1111/j.1365-294X.2012.05569.x
Gonzalez-Lopez, C. I., Roman, S., Sylvie, B. R., and Claude, G. (2002). Genetic control of extracellular protease synthesis in the yeast Yarrowia lipolytica. Genetics 160, 417–427.
Han, K., Han, K., Yu, J. H., Chae, K., Jahng, K., and Han, D. (2010). The nsdD gene encodes a putative GATA-type transcription factor necessary for sexual development of Aspergillus nidulans. Mol. Microbiol. 41, 299–309. doi: 10.1046/j.1365-2958.2001.02472.x
Harman, D. (1972). Free radical theory of aging: dietary implications. Am. J. Clin. Nutr. 25, 839–843. doi: 10.1093/ajcn/25.8.839
Hauser, M., Donhardt, A. M., Barnes, D., Naider, F., and Becker, J. M. (2000). Enkephalins are transported by a novel eukaryotic peptide uptake system. J. Biol. Chem. 275, 3037–3041. doi: 10.1074/jbc.275.5.3037
Khan, I. A., Wang, Y., Li, H.-J., Lu, J.-P., Liu, X.-H., and Lin, F.-C. (2014). Disruption and molecular characterization of calpains-related (MoCAPN1, MoCAPN3 and MoCAPN4) genes in Magnaporthe oryzae. Microbiol. Res. 169, 844–854. doi: 10.1016/j.micres.2014.03.003
Kim, H. S., Han, K. Y., Kim, K. J., Han, D. M., Jahng, K. Y., and Chae, K.-S. (2002). The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet. Biol. 37, 72–80. doi: 10.1016/s1087-1845(02)00029-4
Kuchler, K., Sterne, R. E., and Thorner, J. (1989). Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J. 8, 3973–3984. doi: 10.1002/j.1460-2075.1989.tb08580.x
Laor, D., Cohen, A., Pasmanik-Chor, M., Oron-Karni, V., Kupiec, M., and Weisman, R. (2014). Isp7 is a novel regulator of amino acid uptake in the TOR signaling pathway. Mol. Cell. Biol. 34, 794–806. doi: 10.1128/MCB.01473-13
Lee, D. W., Kim, S., Kim, S. J., Han, D. M., Jahng, K.-Y., and Chae, K.-S. (2001). The lsdA gene is necessary for sexual development inhibition by a salt in Aspergillus nidulans. Curr. Genet. 39, 237–243. doi: 10.1007/s002940100206
Lengeler, K., and Kothe, E. (1999). Mated: a putative peptide transporter of Schizophyllum commune expressed in dikaryons. Curr. Genet. 36, 159–164. doi: 10.1007/s002940050486
Lubkowitz, M. A., Barnes, D., Breslav, M., Burchfield, A., Naider, F., and Becker, J. M. (1998). Schizosaccharomyces pombe isp4 encodes a transporter representing a novel family of oligopeptide transporters. Mol. Microbiol. 28, 729–741. doi: 10.1046/j.1365-2958.1998.00827.x
Lubkowitz, M. A., Barnes, D., Breslav, M., Burchfield, A., Naider, F., and Becker, J. M. (2010). Schizosaccharomyces pombe isp4 encodes a transporter representing a novel family of oligopeptide transporters. Mol. Microbiol. 28, 729–741. doi: 10.1046/j.1365-2958.1998.00827.x
Lubkowitz, M. A., Hauser, L., Breslav, M., Naider, F., and Becker, J. M. (1997). An oligopeptide transport gene from Candida albicans. Microbiology 143, 387–396. doi: 10.1099/00221287-143-2-387
Magee, P. T., and Magee, B. B. (2004). Through a glass opaquely: the biological significance of mating in Candida albicans. Curr. Opin. Microbiol. 7, 661–665. doi: 10.1016/j.mib.2004.10.003
Mcgovern, D. P. B., Pirro, H., Tariq, A., van Heel, D. A., Moffatt, M. F., Alisoun, C., et al. (2005). Association between a complex insertion/deletion polymorphism in NOD1 (CARD4) and susceptibility to inflammatory bowel disease. Hum. Mol. Genet. 14, 1245–1250. doi: 10.1093/hmg/ddi135
McGrath, J. P., and Varshavsky, A. (1989). The yeast STE6 gene encodes a homologue of the mammalian multidrug resistance P-glycoprotein. Nature 340, 400–444.
Nita, M., and Grzybowski, A. (2016). The role of the reactive oxygen species and oxidative stress in the pathomechanism of the age-related ocular diseases and other pathologies of the anterior and posterior eye segments in adults. Oxid. Med. Cell. Longev. 2016, 1–23. doi: 10.1155/2016/3164734
Oliver, R., and Joachim, M. U. (2010). A family of oligopeptide transporters is required for growth of Candida albicans on proteins. Mol. Microbiol. 60, 795–812. doi: 10.1111/j.1365-2958.2006.05136.x
Palmer, J. M., Theisen, J. M., Duran, R. M., Grayburn, W. S., Calvo, A. M., and Keller, N. P. (2013). Secondary metabolism and development is mediated by LlmF control of VeA subcellular localization in Aspergillus nidulans. PLoS Genet. 9:e1003193. doi: 10.1371/journal.pgen.1003193
Payne, J. W., and Smith, M. W. (1994). Peptide transport by Micro-organisms. Adv. Microb. Physiol. 36, 1–80. doi: 10.1016/s0065-2911(08)60176-9
Rogaeva, E. A., Premkumar, S., Grubber, J., Serneels, L., Scott, W. K., Kawarai, T., et al. (1999). An α-2-macroglobulin insertion-deletion polymorphism in Alzheimer disease. Nat. Genet. 22, 21–22. doi: 10.1038/8732
Sato, S., Suzuki, H., Widyastuti, U., Hotta, Y., and Tabata, S. (1994). Identification and characterization of genes induced during sexual differentiation in Schizosaccharomyces pombe. Curr. Genet. 26, 31–37. doi: 10.1007/bf00326301
Sautter, C., and Hock, B. (1982). Crozier formation in Byssochlamys nivea. Mycologia 74, 256–264. doi: 10.1080/00275514.1982.12021498
Seidl, V., Seibel, C., Kubicek, C. P., and Schmoll, M. (2009). Sexual development in the industrial workhorse Trichoderma reesei. Proc. Natl. Acad. Sci. U.S.A. 106, 13909–13914. doi: 10.1073/pnas.0904936106
Shimanuki, M., Chung, S. Y., Kawasaki, Y., Uehara, L., Tsutsumi, C., Hatanaka, M., et al. (2010). Two-step, extensive alterations in the transcriptome from G0 arrest to cell division in Schizosaccharomyces pombe. Genes Cells 12, 677–692. doi: 10.1111/j.1365-2443.2007.01079.x
Singh, B., Poçasfonseca, M. J., Johri, B. N., and Satyanarayana, T. (2016). Thermophilic molds: biology and applications. Crit. Rev. Microbiol. 42, 985–1006. doi: 10.3109/1040841X.2015.1122572
Smith, M. W., Dean, M., Carrington, M., Winkler, C., Huttley, G. A., Lomb, D. A., et al. (1997). Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort. Science 277, 959–965. doi: 10.1126/science.277.5328.959
Stacey, M. G., Koh, S., Becker, J., and Stacey, G. (2002). AtOPT3, a member of the oligopeptide transporter family, is essential for embryo development in Arabidopsis. Plant Cell 14, 2799–2811. doi: 10.1105/tpc.005629
Tao, S., Fan, Y., Wang, W., Ma, G., Liang, L., and Shi, Q. (2007). Patterns of insertion and deletion in mammalian genomes. Curr. Genomics 8, 370–378. doi: 10.2174/138920207783406479
Taylor, J. W., Jacobson, D. J., and Fisher, M. C. (1999). THE EVOLUTION OF ASEXUAL FUNGI: reproduction, speciation and classification. Immunology 121, 383–391.
Thomas, H., Cairns, T. C., Patrick, O., Joachim, M. U., Bignell, E. M., and Sven, K. (2011). Oligopeptide transport and regulation of extracellular proteolysis are required for growth of Aspergillus fumigatus on complex substrates but not for virulence. Mol. Microbiol. 82, 917–935. doi: 10.1111/j.1365-2958.2011.07868.x
Volfovsky, N., Oleksyk, T. K., Cruz, K. C., Truelove, A. L., Stephens, R. M., and Smith, M. W. (2009). Genome and gene alterations by insertions and deletions in the evolution of human and chimpanzee chromosome 22. BMC Genomics 10:51. doi: 10.1186/1471-2164-10-51
Xiang, Q., Shen, K., Yu, X., Zhao, K., Gu, Y., Zhang, X., et al. (2017). Analysis of the oligopeptide transporter gene family in Ganoderma lucidum: structure, phylogeny, and expression patterns. Genome 60, 293–302. doi: 10.1139/gen-2016-0120
Xiang, Q., Wang, Z., Zhang, Y., and Wang, H. (2013). An oligopeptide transporter gene family in Phanerochaete chrysosporium. Gene 522, 133–141. doi: 10.1016/j.gene.2013.03.069
Keywords: isp4/SDP (sexual differentiation process protein), OPT (oligopeptide transporter), sexual reproduction, 72nt-insertion fragment (72INS), C. thermophilum, T. lanuginosus
Citation: Xie X-L, Wei Y, Song Y-Y, Pan G-M, Chen L-N, Wang G and Zhang S-H (2020) Genetic Analysis of Four Sexual Differentiation Process Proteins (isp4/SDPs) in Chaetomium thermophilum and Thermomyces lanuginosus Reveals Their Distinct Roles in Development. Front. Microbiol. 10:2994. doi: 10.3389/fmicb.2019.02994
Received: 22 October 2019; Accepted: 10 December 2019;
Published: 06 January 2020.
Edited by:
Jae-Hyuk Yu, University of Wisconsin–Madison, United StatesReviewed by:
Hokyoung Son, Seoul National University, South KoreaHee-Soo Park, Kyungpook National University, South Korea
Copyright © 2020 Xie, Wei, Song, Pan, Chen, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shi-Hong Zhang, emhhbmdfc2hAamx1LmVkdS5jbg==
 Xiang-Li Xie
Xiang-Li Xie Yi Wei1
Yi Wei1 Gang Wang
Gang Wang Shi-Hong Zhang
Shi-Hong Zhang