
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 12 February 2020
Sec. Fungi and Their Interactions
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02921
This article is part of the Research Topic Aspergillus-Derived Mycotoxins In The Feed And Food Chain View all 22 articles
Species of the highly diverse fungal genus Aspergillus are well-known agricultural pests, and, most importantly, producers of various mycotoxins threatening food safety worldwide. Mycotoxins are studied predominantly from the perspectives of human and livestock health. Meanwhile, their roles are far less known in nature. However, to understand the factors behind mycotoxin production, the roles of the toxins of Aspergilli must be understood from a complex ecological perspective, taking mold-plant, mold-microbe, and mold-animal interactions into account. The Aspergilli may switch between saprophytic and pathogenic lifestyles, and the production of secondary metabolites, such as mycotoxins, may vary according to these fungal ways of life. Recent studies highlighted the complex ecological network of soil microbiotas determining the niches that Aspergilli can fill in. Interactions with the soil microbiota and soil macro-organisms determine the role of secondary metabolite production to a great extent. While, upon infection of plants, metabolic communication including fungal secondary metabolites like aflatoxins, gliotoxin, patulin, cyclopiazonic acid, and ochratoxin, influences the fate of both the invader and the host. In this review, the role of mycotoxin producing Aspergillus species and their interactions in the ecosystem are discussed. We intend to highlight the complexity of the roles of the main toxic secondary metabolites as well as their fate in natural environments and agriculture, a field that still has important knowledge gaps.
The lifestyles of Aspergillus species associated with plants range from saprophytes and symptomless endophytes to weak and opportunistic phytopathogens. The shift between these lifestyles is the result of global transcriptome changes, primarily affecting secondary metabolite (SM) production (e.g., Reverberi et al., 2013). The principal and well-known mycotoxins produced by the Aspergilli are ochratoxin A (OTA) and aflatoxins (AFs), as well as less-prominent toxins like patulin (Keller et al., 2005). These toxins are found in different agricultural commodities (Varga et al., 2004), and are tightly regulated with different threshold limits depending on the matrix (Cano et al., 2016).
Due to the importance of SMs in plant pathogenesis and animal toxicoses, understanding their regulation and biosynthesis is crucial but still hindered by notable knowledge gaps. The species A. flavus, for example, has been predicted to possess 56 SM biosynthesis gene clusters (Keller et al., 2005), but only some secondary metabolites, e.g., AFs (Yu et al., 2004), aflatrem (Nicholson et al., 2009), piperazine (Forseth et al., 2013), asparasone (Malysheva et al., 2014), cyclopiazonic acid (CPA) (Chang et al., 2009), and kojic acid (Terabayashi et al., 2010) have been assigned to a particular gene cluster (Ehrlich and Mack, 2014). A. flavus thus might produce metabolites besides well-known mycotoxins that could be underrated contributors to its toxicity to humans and animals.
Initially, it was hypothesized that mycotoxin production helps fungi to compete with other organisms for nutrient sources like fruits or seeds (Janzen, 1977). Mycotoxins are now also known to act as chemical signals between representatives of different kingdoms, e.g., as inhibitors of quorum sensing (QS), virulence factors in pathogens, or as protectors of sclerotia from insect predation (Ciegler, 1983; Wicklow et al., 1994; Desjardins and Hohn, 1997; Rasmussen et al., 2005; Rohlfs et al., 2010).
Due to their economic and public health importance, the research on mycotoxins has so far mostly been focused on animal husbandry, the food chain, and human aspects. However, for a comprehensive understanding of toxigenic molds’ ecology and of the evolutionary pressures shaping mycotoxin production, interactions with the micro- and macroflora and fauna in different habitats need to be considered and investigated. The study of the overall role of microbial SMs in natural habitats is a previously mostly neglected, but an emerging field (O’Brien and Wright, 2011).
AFs are produced by as much as 16 species (Frisvad et al., 2019), most notably by A. flavus and A. parasiticus. A wide range of Aspergillus spp. produces the AF precursor sterigmatocystin (ST), which is also a carcinogenic compound. The ST/AF polyketide biosynthetic pathways are perhaps the most thoroughly studied ones in fungi (Cleveland et al., 2009; Khaldi et al., 2010).
The most common AF-producing species and the most common member of section Flavi is A. flavus, which possesses two distinct morphotypes, namely the “L-type” with big sclerotia (with average diameter of >400 μm), and the “S-type” that produces small sclerotia (under 400 μm) (Gilbert et al., 2018). However, several additional and often newly delimited species (A. aflatoxiformans, A. arachidicola, A. austwickii, A. cerealis, A. minisclerotigenes, A. mottae, A. pipericola, and A. texensis) have been characterized by S-type sclerotia. Earlier reports on S-type A. flavus may have referred to any of these species, including those that produce both aflatoxin B1 (AFB1) and aflatoxin G1 (AFG1) (so-called SBG strains) (Singh et al., 2018; Frisvad et al., 2019).
While the ecological role of ST is not known in detail, it is presumably antagonistic to organisms competing for resources with ST producers. Both AFs and ST have been reported to be phytotoxic (Stoessl, 1981; McLean et al., 1995). AFs inhibit plant photosynthesis by hindering chlorophyll and carotenoid synthesis (Anjorin and Inje, 2014), leading to virescence or albinism in the contaminated plants (Reiss, 1978). However, in plant pathogenesis, the role of these mycotoxins needs to be investigated as non-aflatoxigenic strains also have the potential to colonize plant hosts, e.g., on cotton bolls (Cotty, 2007), and these types of strains are isolated frequently.
Soil is the natural habitat for A. flavus, and AF production is considered to give a fitness advantage in that environment (Drott et al., 2017). Selective forces that maintain the polymorphism of non-aflatoxigenic and aflatoxigenic colonies are mainly unknown. Resource competition among the closely related strains is modulated by factors such as chemical composition and pH of the soil or nutrient and water availability (Ehrlich, 2014). Moreover, competition between aflatoxigenic and non-aflatoxigenic strains is strain-dependent, and it must be noted that non-aflatoxigenic strains are not necessarily atoxigenic, as they may produce toxins other than AFs. Under high fungal density, non-aflatoxigenic strains can outcompete both toxigenic and other non-aflatoxigenic populations (Cotty, 2006). Aflatoxigenic isolates were shown to have lower fitness than non-aflatoxigenic isolates in wide temperature ranges (25–42°C) (Drott et al., 2019). This may explain the success of the latter in competition. The metabolic cost of AF production seems to explain the low fitness as AFB1 itself does not affect the growth of A. flavus at concentrations as high as 500 ng g−1 (Drott et al., 2019), orders of magnitude higher than what can be measured in soils (0.6–5.5 ng·g−1) (Accinelli et al., 2008). Inoculation of soil with non-aflatoxigenic strains also protects crops from AF contamination during storage (Dorner and Cole, 2002; Bandyopadhyay et al., 2016).
AFB1 is transient in soils with a half-life of approximately 5 days at 28°C; however, it is produced continuously as long as there is a substrate, e.g., corn residues (Accinelli et al., 2008). High A. flavus levels (log10 3.1–4.5 cfu·g−1), AFB1 production, and expression of the AF biosynthetic genes (aflG, aflD, aflP, aflR, and aflS; Ehrlich et al., 2005) have been reported in the former study.
Studies on AFB1 transformation in soil or purified mineral systems have identified AFs B2 (AFB2) and G2 (AFG2) as the primary transformation products using thin-layer chromatography. However, the more sophisticated HPLC-MS technique did not detect these molecules in spiked soils. In an aqueous-soil environment, a new structure, B2a (AFB2a), was detected as the single primary transformation product. AFB2a is a hydrolytic product of AFB1 and the soil acting as an acid catalyst (Starr et al., 2017) (Figure 1).
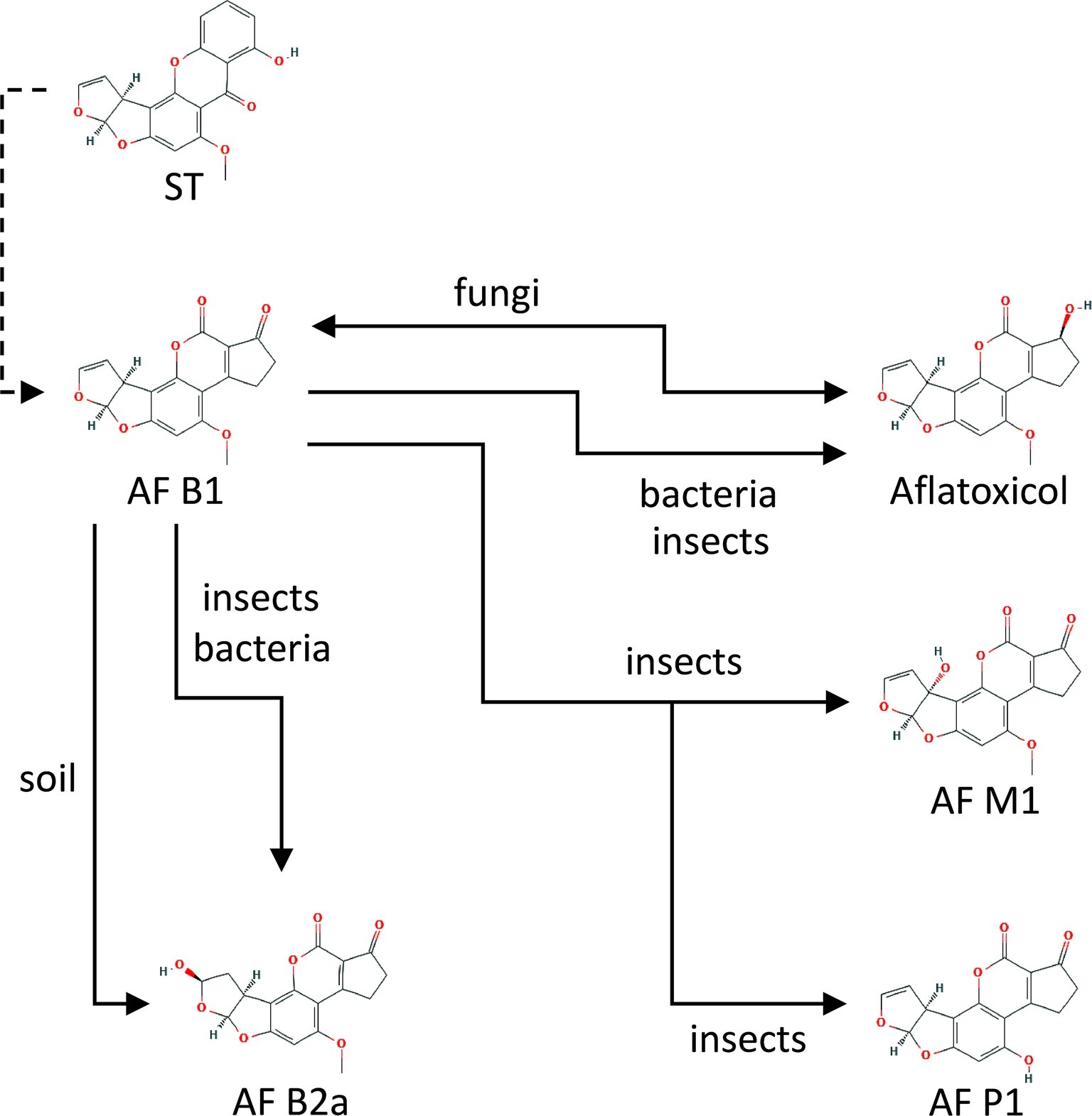
Figure 1. Main chemical conversions of aflatoxin B1 (AFB1) under interaction with different organisms and soil. Sterigmatocystin (ST) is a chemical precursor of aflatoxin B1 (AFB1) in aflatoxigenic fungi. The further conversion processes are explained in details in the text. Source: National Center for Biotechnology Information. PubChem Compound Database (accessed June 6, 2019) (Bolton et al., 2008).
AFs taken up through plant roots can be accumulated, transported to other tissues (e.g., in groundnut seedlings; Hariprasad et al., 2015; Snigdha et al., 2015), degraded, metabolized, or masked, or can be diffused back to the medium (e.g., in maize; Mertz et al., 1980).
Various fungi can inhibit AF accumulation. In an in vitro soil environment, Fusarium oxysporum was able to inhibit AF production at different temperatures (25 and 30°C) and fumonisins accumulated instead of AFB1 (Falade et al., 2016). On the contrary, inhibitory effect by A. flavus on Fusarium oxysporum f. sp. niveum and Fusarium solani f. sp. cucurbitae has also been described with an inhibition rate exceeding 50 % in in vitro and greenhouse experiments. Hyperparasitism of A. niger, A. flavus, and A. terreus on F. oxysporum f. sp. melonis was also demonstrated (Boughalleb-M’Hamdi et al., 2018).
Gliotoxin (an epipolythiodioxopiperazine) has internal disulfide bridges that conjugate proteins (Spikes et al., 2008). Gliotoxin biosynthesis and regulation are reviewed by Dolan et al. (2015). The compound is implicated in the formation of reactive oxygen species (ROS) by redox cycling and is generally broadly cytotoxic (Gardiner et al., 2005). Therefore, its detoxification is only possible by its biosynthetic enzymes (Scharf et al., 2018). One of the significant gliotoxin producers besides biocontrol Trichoderma ssp. is A. fumigatus, a saprophyte and an opportunistic animal pathogen. Gliotoxin produced by this fungus acts as a virulence factor mediating systemic mycosis in susceptible vertebrates (Latgé, 2001; Scharf et al., 2016) and presumably in insects (Reeves et al., 2004). A. fumigatus possesses a self-protecting system against gliotoxin (Schrettl et al., 2010; O’Keeffe et al., 2014). RNA-seq revealed 164 differentially expressed genes (DEGs) in A. fumigatus treated with external gliotoxin, and besides gliotoxin biosynthesis genes, helvolic acid biosynthesis genes, siderophore-iron transport genes showed altered expression (O’Keeffe et al., 2014). High temperature and humidity during crop maturation may favor A. fumigatus presence and toxin production. Gliotoxin enters the food chain and reaches the most sensitive farm animals, like horses and poultry (Pena et al., 2010). However, there is no threshold limit for this molecule.
In composted mineral soil with a natural microbiota, the toxin may function as an antibiotic, effectively controlling the damping-off disease of Zinnia elegans (zinnia) seedlings caused by the fungus Rhizoctonia solani and the water mold Pythium ultimum (Lumsden et al., 1992). A strong correlation between the presence of bacterial peptidoglycan, lipopolysaccharide, or lipoteichoic acid in soil and the gliotoxin secretion of A. fumigatus was described by Svahn et al. (2014). This finding was potentially relevant for drug discovery research, and parallelism was found with the increased virulence of A. fumigatus in case of bacterial co-infection.
Several Aspergilli in sections Circumdati (such as A. steynii and A. westerdijkiae), Flavi, and Nigri (e.g., A. carbonarius and A. niger; Palencia et al., 2010) are well-known producers of OTA, a mycotoxin teratogenic, carcinogenic, immunosuppressive, and nephrotoxic in animals (Samson et al., 2014). All studied OTA-producing fungi have a consensus OTA biosynthetic pathway with four highly conserved biosynthetic genes in a cluster and a bZIP transcription factor (Wang et al., 2018).
OTA induced necrotic lesions on Arabidopsis thaliana leaves via induction of an oxidative burst by elevated ROS (hydrogen peroxide and superoxide anion) levels (Peng et al., 2010). Meanwhile, the downregulation of the antioxidant defense enzymes in host plants and up-regulation of lipid peroxidation were detected, along with root growth inhibition of seedlings (Peng et al., 2010). Infiltration of 4-week-old A. thaliana leaves with 2 mM and 1 mM OTA solutions in vitro resulted in macroscopic lesions (Wang et al., 2012), and the growth of A. thaliana was repressed, while cell death was detected with characteristic hypersensitive response-type lesions on the excised leaves. Cell death did not only result in a manifestation of oxidative burst but the deposition of phenols and callose (Peng et al., 2010) as well. McLean (1996) investigated the effect of the toxin on germinating Zea mays embryos. Interestingly, there was no linear relationship between the inhibitory effect and the OTA concentrations as 10 μg·ml−1 OTA was inhibitory, while 5 or 25 μg·ml−1 OTA was stimulatory for root and shoot growth.
Soil type, in connection with microbial activity, affects OTA half-life. In soils with higher microbial activity, like planted soils, faster degradation could be measured (Mortensen et al., 2006) caused by the microbial biomass (e.g., Barberis et al., 2014). Regulation of OTA biosynthesis can be modulated by volatile organic carbons (VOCs) as observed for A. carbonarius and fruit ketones, C-8 alcohols, and trans-nerolidol (Zhang et al., 2017).
Patulin is a polyketide mycotoxin produced by Penicillium spp. and to a lesser extent, various Aspergilli (Zhang et al., 2008). It is frequently found in fresh fruits or fruit juices and jams contaminated with blue mold rot (Logrieco et al., 2003). Like clavatol, patulin inhibits numerous plant pathogenic fungi and water molds in vitro, i.e., Fusarium oxysporum f. sp. cucumerinum, Botrytis cinerea, Didymella bryoniae, Rhizoctonia solani, and Pythium ultimum (Zhang et al., 2008). Patulin and clavatol produced by Aspergillus clavatonanicus endophyte of Taxus mairei have been shown to antagonize plant pathogens (Zhang et al., 2008). Interestingly, Botha et al. (2018) reported that A. clavatus produced higher concentration of tremorgenic mycotoxins (i.e., tryptoquivaline A, deoxytryptoquivaline A, and deoxynortryptoquivaline) than concomitant patulin and cytochalasin E. Patulin, similarly to penicillic acid has the potential to interfere with bacterial QS communication in soil (Rasmussen et al., 2005), hinting at its potentially manifold ecological roles in microbial communities.
The neurotoxic CPA is an indole-tetramic acid produced by 13 species in section Flavi (Frisvad et al., 2019). It inhibits endoplasmic reticulum calcium ATPases at nanomolar concentrations, and therefore, it is an inducer of cell death in plants (Chang et al., 2009). Usually, CPA and AFs are concomitant mycotoxins. Most A. flavus strains synthesize AFs B1 and B2 besides CPA, although some strains also synthesize AFs G1 and G2 (Geiser et al., 2000; Cardwell and Cotty, 2002). In contrast, A. parasiticus strains produce all four AFs without CPA biosynthesis (Dorner et al., 1984). Moreover, a “sleeping” CPA cluster was activated by the overexpression of a general secondary metabolism regulator gene (laeA) in A. fumisynnematus (Hong et al., 2015).
CPA was proposed to modify calcium homeostasis, mitochondria, and cytoplasm membranes based on animal studies (Riley and Goeger, 1992). This mycotoxin serves as a critical pathogenicity factor that enables the saprophytic lifestyle of A. flavus (Chalivendra et al., 2017), presumably, through its good iron-chelating characteristics (Riley and Goeger, 1992).
It is well-known that multiple mechanisms are involved in host plant defense systems in response to A. flavus infection and AF accumulation. Peanut was found to have evolved complex defense mechanisms to resist pathogens, such as blocking the invasion and activating a range of defense responses (Holbrook and Stalker, 2003). Eight hundred forty-two candidate genes were recognized for A. flavus resistance in post-harvest seeds (Wang et al., 2016a). Genes involved in defensive responses to A. flavus and AF biosynthesis were stimulated in resistant genotype (Wang et al., 2016b).
The plant cell wall, the first line of defense against microbial pathogens, is primarily made up of polysaccharides cellulose, hemicellulose, and pectin. While opportunistic fungi usually infect plants through wounds (e.g., mechanical or pest damages), pathogenic ones actively penetrate cell walls, often through the secretion of a range of polysaccharide-degrading enzymes such as pectinesterase, arabinofuranosidase, mannosidase, and galacturonidase along with amylases or proteases (Whitehead et al., 1995; Bellincampi et al., 2014; Wang et al., 2016b). In peanuts resistant to A. flavus infection, feruloyl esterase, pectinesterase, arabinofuranosidase, mannosidase, polygalacturonase, and galacturonidase fungal activities were significantly downregulated compared to the sensitive plants (Wang et al., 2016a). Resistance to A. flavus infection is naturally the most critical factor in avoiding AF exposure to consumers. Pod infection, seed invasion, and AF production in the cotyledon are the crucial steps to be considered (Nigam et al., 2009). The first interaction between the plant and the mold is at the pod shell, where the pathogen resistance depends on the shell structure. The second barrier is the undamaged seed coat. Upon a successful invasion, A. flavus colonizes the seed cotyledon and produces AFs. In a proteomic study, a total of 29 seed proteins showed differential expression between the resistant and susceptible peanut cultivars under drought stress in response to A. flavus (Wang et al., 2010). Under drought stress, AF production was consistent in peanut pods even if roots of those plants were well watered. Meanwhile, AF was not produced in well-watered peanuts pods, while roots were under drought stress (Sanders et al., 1993).
The data suggest that drought stress is the most critical factor in the interaction of the plant and the fungal agent. Therefore, watering of the fields is crucial along with the improvement of the plant’s resistance by genetic modification or selection.
Pathogenesis in maize depends on environmental factors (e.g., Payne and Widstrom, 1992; Kebede et al., 2012; Fountain et al., 2014), metabolic state of the kernels (Chen et al., 2010; Jiang et al., 2011), physiological state of the fungus (Jayashree and Subramanyam, 2000), and time elapsed following infection (Scott and Zummo, 1994; Betrán and Isakeit, 2004). Vitreous compared to softer dent type endosperm was positively correlated with AF contamination and resistance to ear rot (Betrán and Isakeit, 2004; Llorente et al., 2004).
Since maize is a favorable host for the Aspergilli, especially for A. flavus, and the plant’s resistance is genetically determined, much effort was invested worldwide to develop resistant maize genotypes. Recent breeding investigations focused on quantitative trait loci (QTL) for AF resistance (Kelley et al., 2012; Fountain et al., 2015), and the studies demonstrated that the resistance to A. flavus is highly quantitative and is not conferred by a single gene. Any given QTL was found to account for a rather low level of phenotypic variance explained regarding AF resistance. Resistance thus has a polygenic nature with a combination of multiple traits being involved in the resistant phenotype (Fountain et al., 2014; Yin et al., 2014). Maize inbred lines were found also to vary in their tolerance to CPA (Chalivendra et al., 2017). Moreover, CPA tolerance of the root was in a significant correlation to silk resistance under fungal colonization (Mideros et al., 2012).
During infection, mycelia were detected inside the scutellum, exhibiting a biofilm-like formation at the endosperm-scutellum interface (Dolezal et al., 2013). This biofilm-like structure bears resemblance to the biofilm of A. fumigatus in the human lung (Loussert et al., 2010). In situ hybridization of RNA showed the expression of the pathogenesis-related protein gene in the aleurone and scutellum of maize seed (PRms) during A. flavus infection (Shu et al., 2015). Transcripts of the maize sucrose synthase-encoding gene (shrunken-1; Sh1) were detected in the embryo in non-infected kernels, but the gene was up-regulated in the aleurone and scutellum under A. flavus infection. Moreover, the transcripts of PRms and Sh1 showed accumulation in the seeds before infection (Shu et al., 2015).
A recent study was conducted on expression profiling of 267 unigenes (mostly genes of metabolism, stress response and disease resistance) in a mapping population derived from a cross between susceptible and resistant parent plants (Dhakal et al., 2017). It revealed that many genes involved in the synthesis and hydrolysis of starch and sugar mobilization and others related to energy production and/or precursors of lignin and phytoalexins used in the defense response were highly expressed (Dolezal et al., 2014; Shu et al., 2015; Dhakal et al., 2017).
Apart from Fusarium infection (Mesterházy, 2008), A. flavus causes the most economic loss on cornfields. However, co-infection by these genera is not investigated in detail, and only some aspects are known like the inhibitory effect on AFB1 production by Fusarium (Falade et al., 2016), and inhibitory and hyper-parasitic effect of A. flavus on Fusaria (Boughalleb-M’Hamdi et al., 2018). Moreover, the physiological effects of the co-produced mycotoxins like CPA and AFs or the effect of the co-infection on mycotoxin productions is rarely investigated (e.g., Marín et al., 2001; Giorni et al., 2016).
Cottonseed can be contaminated pre-and postharvest by Aspergilli. A comparative transcriptome analysis was performed investigating the genes expressed differentially in corn, peanut, and cotton under aflatoxigenic A. flavus infection (Bedre et al., 2015). Only 26 common genes were identified as candidate A. flavus resistance genes in all the three plants. Six of these genes coded for Fe(II)-dependent oxygenase superfamily proteins and 2-oxoglutarate. In response to both non-aflatoxigenic and aflatoxigenic strains, genes encoding alcohol dehydrogenase, UDP glycosylation transferase, and helix loop helix protein were induced (Bedre et al., 2015). Upregulation of primary metabolism modulated signal transduction cascades that were essential to plant defense responses (Rojas et al., 2014). In the pericarp, sucrose and starch metabolism besides glycerolipid metabolism were upregulated under infection with non-aflatoxigenic A. flavus. The metabolic pathways activated by the presence of non-aflatoxigenic A. flavus in the plant pericarp and seeds compared to aflatoxigenic A. flavus activated pathways can lead to possible target genes to develop fungal stress tolerance and resistance in cotton (Bedre et al., 2015).
Phytohormones are well-known mediators of fungus-plant interactions with different roles. The abscisic acid (ABA) (Hauser et al., 2011; Xin et al., 2012), salicylic acid (SA) (Janda and Ruelland, 2014), and ethylene (ET) (Bleecker and Kende, 2002; Ton et al., 2002) phytohormonal pathways in plants can act against A. flavus and AF production by mediating and channeling many stress-response genes (Bari and Jones, 2009). Transcriptomic analysis revealed DEGs of phytohormone production and signaling in response to AF production in peanut (Wang et al., 2016a). Moreover, DEGs concerning ABA production and signaling showed higher expression in a sensitive peanut genotype than in the resistant plants (Wang et al., 2016b).
Determining the roles of ET is challenging as disease symptoms seem to be either reduced or enhanced or not affected depending on the pathogen-host interaction (Bleecker and Kende, 2002). It inhibits AF biosynthesis in A. flavus through alleviation of oxidative stress (Huang et al., 2009). However, DEGs involved in ET production were downregulated in response to AF production, and most of them were also repressed in the resistant genotype. Wang et al. (2016b) concluded that ET might suppress resistance to AF production, and later Wang et al. (2017) found that ET emitted by infected seed facilitated the colonization by A. flavus but not AF production in maize, potentially opening up biotechnological applications.
Contrary, SA is suppressive for some fungi (Seyfferth and Tsuda, 2014). SA inhibited mycelial growth and mycotoxin formation of A. flavus in vitro, and the in vivo evaluation resulted in more significant inhibitory effects for the intact treated pistachio fruit as for injured ones (Panahirad et al., 2014).
Jasmonates are lipid-derived signals compounds in plant growth and development in response to stresses like pathogen attack or drought (Wasternack, 2014). Jasmonic acid (JA) and its metabolites, members of the oxylipin family, are synthesized in the alpha-linolenic acid pathway. Many of them modify gene expression in a regulatory network with synergistic and antagonistic effects concerning other plant hormones such as SA, auxin, ET, and ABA (Wasternack, 2007). Metabolism of alpha-linolenic acid was upregulated in pericarp under both non-aflatoxigenic and toxigenic A. flavus infection in comparison to seeds. Similarly, the alkaloid biosynthetic pathway was more intensively upregulated in the pericarp under both non-aflatoxigenic and toxigenic A. flavus infection than in the seed. In tobacco host plants, the alkaloid biosynthesis was increased in response to insect foraging and application of JA (Todd et al., 2010). Therefore, it was suggested that the JA-regulated defense response is also stimulated as an answer to A. flavus infection (Bedre et al., 2015).
Furthermore, in the case of the aflatoxigenic A. flavus infection, upregulation of arachidonic acid (AA) metabolism was detected in seeds, exceeding that under non-aflatoxigenic infection in the pericarp. AA has a role in plants as a signaling compound, and it stimulates plant defense responses through fatty acids. Meanwhile, pathogen AA triggers plant innate immunity resulting in defense responses and programmed plant cell death (Savchenko et al., 2010).
PR proteins are disease resistance proteins induced in the host plant in response to pathogen infection (Bravo et al., 2003; Luo et al., 2011). Identification and characterization of such plant genes have importance in reducing fungal pathogenicity. In maize, PR-protein genes included PR-1, PR-4, PR-5, PR-10, and chitinase (Dhakal et al., 2017).
The plant hydrolytic enzymes like β-1,3-glucanases and chitinases show antifungal activity owing to the degradation of fungal cell wall components (Cordero et al., 1994; Dolezal et al., 2014). Plant chitinases also have lysozyme activity and are active in preventing mycelial development (Collinge and Slusarenko, 1987; Collinge et al., 1993). The gene expression of chitinase 2 and PR-10 was reported to be upregulated in maize seeds during fungal infection (Cordero et al., 1994). In vitro PR-10 protein possessed antifungal activity against A. flavus, and its production was upregulated upon A. flavus infection in a resistant maize hybrid but not in a susceptible one (Chen et al., 2006). RNAi gene silencing driven repression of PR-10 resulted in an increased susceptibility to A. flavus and AF production (Chen et al., 2010). Moreover, overexpression of chitinase genes (Cletus et al., 2013) resulted in resistance against fungal infection in rice (Baisakh et al., 2001) and peanut (Rohini and Sankara Rao, 2001; Prasad et al., 2013).
Besides chitinases (Singh et al., 2015), lectins are also involved in the plant defense mechanisms (Dang and Van Damme, 2015) and probably play an essential role in inhibiting AF production (Hawkins et al., 2015). In resistant and sensitive plant genotypes, chitinase showed different expression levels (Wang et al., 2016a). Eleven chitinase encoding transcripts were expressed differentially in pericarp and seed during infection by both aflatoxigenic and non-aflatoxigenic strains in cotton (Bedre et al., 2015), while in maize seven chitinase genes were associated with the increased in vivo resistance to A. flavus infection and AF accumulation (Hawkins et al., 2015).
Production of the PR maize seed protein, ZmPRms, was recently shown to be involved in resistance to A. flavus and other pathogens in a seed-specific RNA interference study (Majumdar et al., 2017). A. flavus infection increased significantly on corn kernels with downregulated ZmPRms with a concomitant 4.5–7.5-fold higher accumulation of AFs, presenting the protein’s role in evading infection and toxin accumulation (Majumdar et al., 2017).
Plants also produce cell wall polygalacturonase-inhibiting proteins to counteract the activity of fungal polygalacturonases (Kalunke et al., 2015), enzymes that catalyze the hydrolysis of the α-(1–4) linkages between the D-galacturonic acid units in homogalacturonan resulting in cell separation in the plant tissues. The interaction between polygalacturonases and inhibiting proteins promoted the formation of oligogalacturonides, which evoked further defense responses (Federici et al., 2006). In peanut, Wang et al. (2016b) showed that all six DEGs of polygalacturonase-inhibiting proteins were upregulated to a much higher level in a resistant genotype than in a sensitive one.
Plant’s linoleic acid and 9- and 13-hydroperoxy fatty acids (9S- and 13S-HPODE oxylipin products) have a substantial effect on the differentiation processes of Aspergillus spp. Both 9S- and 13S-HPODE alter secondary metabolism in A. parasiticus and A. nidulans (Gardner, 1995; Burow et al., 1997). They also increase the production of the conidiospores in A. nidulans and A. flavus, and, in A. nidulans, elevate cAMP levels (Calvo et al., 1999; Affeldt et al., 2012). Additionally, A. flavus infection of peanut seeds promoted linoleate 9-LOX expression and 9S-HPODE accumulation. 13S-HPODE producing lipoxygenase alleles (PnLOX2 and PnLOX3) were highly expressed in mature seed, but these genes were repressed between 5-fold and 250-fold during A. flavus infection. The outcomes of these investigations proposed that 9S-HPODE is a susceptibility, while 13S-HPODE is a resistance factor during Aspergillus spp. infection (Tsitsigiannis et al., 2005). Similarly, linoleic acid host-derived oxylipins were also suggested to drive mycotoxin synthesis (Burow et al., 1997; Brodhagen et al., 2008; Reverberi et al., 2010). 13S-HPODE repressed expression of ST and AF biosynthetic pathway genes at concentrations of 10 and 100 μM and, in this way, significantly reduced ST and AF production in both A. nidulans (ST producer) and A. parasiticus (AF producer) in vitro (Burow et al., 1997). The maize ZmLOX3-mediated pathway acted as a root-specific suppressor of all three major defense signaling pathways (Gao et al., 2008a,b).
The oxylipin-driven processes are complicated further by fungal oxylipin production. A. flavus single lipoxygenase produced oxylipins influence host responses. Reverberi et al. (2010) found that a lox-like gene mutant A. ochraceus was not only failed to produce 13S-HPODE, but a sharp decrease was detected in its OTA production. The conidium formation was also delayed, and the sclerotium production was increased in the cultures. Moreover, seeds infected with the A. ochraceus mutant could not produce normal 9S-HPODE levels or induce the defensive PR1, suggesting the importance of the fungal 13S-HPODE in the regulation of host defense response. The oxylipin profile of the maize kernels inoculated with wild type and lox mutant A. flavus strains showed elevated levels of HPODE and diHODES, also suggesting that the fungal Lox produces compounds that suppress plant oxylipin production. The ΔAflox1 mutant strain was able to produce AF only on kernels, but not in axenic culture (Scarpari et al., 2014), revealing the complexity of the metabolic interactions.
PSIB α oxylipins derived from linoleic acid in A. nidulans were also reminiscent of those produced from seed fatty acids, and the infected seeds were able to influence the fungal development imitating and interfering with signals controlling conidiogenesis (Prost et al., 2005).
Oxidative stress is a critical factor that can stimulate the synthesis of AF and other SMs (Reverberi et al., 2010, 2013). H2O2 and other oxidative agents (Fanelli et al., 1985; Jayashree and Subramanyam, 2000; Narasaiah et al., 2006) activate AF biosynthesis in Aspergillus sect. Flavi (Reverberi et al., 2008). At the plant-pathogen boundary, ROS production is an essential feature that contributed to Aspergillus virulence besides SM production (Reverberi et al., 2013). In seeds contaminated with Aspergilli, a burst of H2O2 was detectable within a few hours of infection (Lamb and Dixon, 1997; Kachroo et al., 2003; Reverberi et al., 2008; Peng et al., 2010). For A. flavus, it appeared that lowering H2O2 levels in the corn embryo helps to prevent A. flavus infection and AF accumulation (Magbanua et al., 2007).
Among the stress-related transcripts, the presence of fungal superoxide dismutase in the dent samples indicated oxidative stress, known to be coupled to the production of AFs (Jayashree and Subramanyam, 2000; Fountain et al., 2015, 2016). It is arising that oxidative stress in fungi plays an essential role not only in SM biosynthesis but also in plant-fungal interactions. Within plant tissues, environmental stresses, e.g., drought and heat stress, may also result in the accumulation of ROS and play an essential role in communication between plants and the Aspergilli (Fountain et al., 2014).
In various plant seeds (e.g., maize, sunflower), the processes of lipoperoxidation induce a change in the ratio of oxidants and antioxidants, in favor of ROS accumulation in fungal cells and stimulating synthesis of AFs in A. flavus and A. parasiticus (Fabbri et al., 1983; Burow et al., 1997; Reverberi et al., 2008; Gao and Kolomiets, 2009). The SM production may be considered as the result of fungal cell response to incomplete scavenging of ROS (Reverberi et al., 2008; Hong et al., 2013).
At the plant’s side, DEGs and antioxidant transcripts of glutathione S-transferase, ferredoxin, copper amine oxidase, ascorbate peroxidase, and peroxidase involved in ROS processing and scavenging showed amplified activity during infection with both non-aflatoxigenic and toxigenic A. flavus (Bedre et al., 2015). Plant peroxidases also contributed to the response to AF production. DEG peroxidases showed a significantly higher expression in an A. flavus resistant peanut genotype than in a sensitive one, indicating better management of ROS in the former during fungal infection (Wang et al., 2016a).
Genes of the phenylpropanoid biosynthetic pathway that produce antimicrobial phytoalexins, phenolic substances, and lignin in plants (Collinge and Slusarenko, 1987; Lawton and Lamb, 1987) were found to show higher expression and more rapid activation in an A. flavus resistant maize genotype than in a sensitive one. Moreover, biosynthesis genes of phenylpropanoids, flavonoids, stilbenoids, diarylheptanoids, and gingerol were enriched only in the resistant maize genotype (Wang et al., 2016a). DEGs analysis in cotton inoculated with aflatoxigenic and non-aflatoxigenic A. flavus also revealed some significant variances in the expression rates of the genes taking part in the defense mechanisms. For instance, in the pericarp, the phenylpropanoid pathway was enriched at a higher level under aflatoxigenic strain infection than under non-aflatoxigenic infection (Bedre et al., 2015).
The flavonoid pathway is essential in the production of several antifungal compounds and, therefore, it is related to defense reactions (Treutter, 2005). In seeds, the flavonoid biosynthesis pathway was the utmost upregulated under non-aflatoxigenic A. flavus infection exceeding the pericarp (Bedre et al., 2015). Numerous studies illustrated the potential impact that flavonoids could exert on SM production. Rutin (quercetin-3-rutinoside) was demonstrated as an effective inhibitor of AFB1 production (Chitarrini et al., 2014). Naringin (flavanone-7-O-glycoside), hesperidin (3′,5,7-trihydroxy 4′-methoxy flavanones 7-rutinoside), and some plant glucosides were characterized for their capacity to restrain mycotoxin production (e.g., patulin by Penicillium expansum, A. terreus, and Byssochlamys fulva; Salas et al., 2012). Similarly, the growth of A. parasiticus and its AFB1 production were repressed by methanolic extracts of Ephedra major roots (Bagheri-Gavkosh et al., 2009). The inhibition of the growth and AFB1 production of A. parasiticus was attributed to quercetin and p-coumaric acid flavonoid compounds. In peanut, some stilbenoids (arachidin-1, arachidin-3, and chiricanine A) caused changes in growth rate, mycelial morphology, and spore germination of A. flavus (Sobolev et al., 2018). Moreover, a significant decrease or almost complete suppression of AF production was revealed in A. parasiticus, A. flavus and A. nomius (Sobolev et al., 2018). Similarly, plants with high concentrations of other antioxidants like β-carotene, β-cryptoxanthin, and total provitamin A also had a reduced amount of AF contamination than hybrids with low carotenoid contents (Suwarno et al., 2019). The relative ease of plant breeding for increased provitamin A as compared to breeding directly for AF resistance suggested novel approaches to suppress AF contamination.
Plants metabolize xenobiotic compounds such as mycotoxins as part of their defense mechanisms. In plants, similar to animals, phase I metabolism (enzymatic transformation such as oxidation, reduction, or hydrolysis), phase II process (sulfatation, glucosidation, glucuronidation) (Coleman et al., 1997; Berthiller et al., 2009), and phase III detoxification (sequestration of compounds conjugated to glucose or glutathione into a vacuole or their permanent attachment to the plant cell wall) (Berthiller et al., 2013) can be differentiated. The chemical transformations in phase I are typical for lipophilic compounds, and most of the hydrophilic compounds are not affected by this phase. In phase I, oxidations are catalyzed by the cytochrome P-450 system, while the hydrolysis is catalyzed by esterases and amidases (Coleman et al., 1997).
Plant-metabolized mycotoxins have been identified mostly for Fusarium toxins (HT-2 toxin, T-2 toxin, nivalenol, fusarenon-X, deoxynivalenol, zearalenone, fusaric acid; Berthiller et al., 2013) or insecticidal destruxins from Metarhizium anisopliae (Pal et al., 2007). The metabolism of some Alternaria toxin derivatives and Aspergillus mycotoxins was studied using plant cell cultures (Ruhland et al., 1996) and germinating cereals and vegetables (Ruhland et al., 1997). The same OTA derivatives were isolated from all the tested plant species, and the conversion was nearly complete (Berthiller et al., 2013). However, the quantitative distribution strongly depended on the plant species. In addition to ochratoxin α, the main derivatives were (4R)- and (4S)-4-hydroxy-ochratoxin A and β-glucosides of both isomers were detected. Ochratoxin α is considered as a non-toxic molecule, whereas hydroxy-ochratoxin A is as potent immunosuppressant as OTA (Berthiller et al., 2013).
The lack of current studies on plant-modified and masked Aspergillus mycotoxins calls for attention to a considerable gap in the understanding of mycotoxins’ fate and ecological roles, especially in the case of toxins produced by plant pathogens, such as A. flavus.
The possible interactions of fungi in the genus Aspergillus with the micro- and macrobiota of the soil can be very diverse ranging from direct physical contact, through non-contact biochemical/enzymatic interactions (e.g., via biotransformation), up to volatile organic compounds (VOCs) exerting their effects without physical contact between competing organisms.
Actinomycetes (e.g., Verheecke et al., 2014), Lactobacilli (e.g., Romanens et al., 2019), Bifidobacteria (e.g., Ghazvini et al., 2016), and Bacilli (Siahmoshteh et al., 2017) are the best-studied groups from these aspects. Several studies have conducted screening on microbial collections to find potential biocontrol isolates that inhibit mold growth, testing (1) bacteria ranging from endophytes and rhizosphere species (Wang et al., 2013); (2) traditional fermentation products (Ahlberg et al., 2017); (3) various other samples where natural interactions with toxigenic molds are far less plausible, as in halophilic soils (Jafari et al., 2018) or fish intestines (Veras et al., 2016). The effects on toxin production and the underlying mechanisms of growth and toxigenic nature are, similarly to yeasts, less understood and often not attempted to uncover. OTA biodetoxification was reviewed by Chen et al. (2018) in detail. Microbes can affect OTA concentration by degradation or absorption and at gene regulation level. OTA biosynthesis genes (acpks, acOTApks, and acOTAnrps) and the general SM regulator veA of A. carbonarius were downregulated upon co-culturing with Streptomyces isolates, with a concomitant decrease in OTA production (El Khoury et al., 2017). While acOTAnrps and acOTApks, along with laeA, a general regulator of fungal secondary metabolism, were found to be downregulated by Lactobacillus plantarum (Lappa et al., 2018).
Close physical interaction between bacteria and fungi induced otherwise silent biosynthesis genes in A. nidulans (Schroeckh et al., 2009). These are from a wide range of gene clusters known as silent or non-expressed ones of merely predicted SMs (Keller et al., 2005). For example, the direct physical interaction between A. nidulans and actinomycetes resulted in orsellinic acid and lecanoric acid production via chromatin remodeling (Netzker et al., 2015) of the fungal culture (Schroeckh et al., 2009). Intimate interaction was also described for plant root-Bacillus subtilis-A. niger interactions, where B. subtilis attached on the surface of the plant root and onto fungal mycelia. Transcriptomic data revealed that both the fungus and the bacterium modified their metabolism during the interaction. The antifungal and antibacterial defense mechanisms of both B. subtilis and A. niger were reduced upon attachment of bacteria to the mycelia (Benoit et al., 2015). Furthermore, bacterial-fungal interaction can also affect plants negatively, for example, Salmonella enterica subsp. enterica serovar. Typhimurium established biofilm on A.niger hyphae, where the bacterial growth was promoted, while the bacterial biofilm protected the fungus in a mutualistic relationship (Balbontín et al., 2014). Regarding the maize plant, the co-colonization has more adverse consequences on plant growth than colonization by either microbe individually.
Mycotoxins in soil are subjects of microbial biotransformation, detoxification, or degradation. A wide variety of microorganisms can biotransform mycotoxins (reviewed by Verheecke et al., 2016). Most studies were conducted with AFB1 due to its high toxicity and carcinogenicity. Several bacteria and fungi, including Rhizopus sp. (Cole et al., 1972), Hypomyces rosellus (Dactylium dendroides), and Corynebacterium rubrum (Mann and Rehm, 1976) convert AFB1 to aflatoxicol (Figure 1) reducing its C-3 keto on the cyclopentanone ring. AFB1 degradation of Nocardia corynebacteroides (Flavobacterium aurantiacum) was reported first by Ciegler et al. (1966). However, AFB1 was only metabolized partially and mostly adsorbed to N. corynebacteroides cells (Line and Brackett, 1995).
Bacteria can reduce the amount of AFB1 by forming AFB2 with lower toxicity, and by making other compounds (AFG2, aflatoxicol) undetectable. Myxococcus fulvus reduced AFB1 by 80.7% (Guan et al., 2010). Teniola et al. (2005) studied Rhodococcus erythropolis, and a remarkable reduction (70%) of AFB1 was observed with cell-free extracts, and an almost total (over 90%) degradation was detected within 4 h. Nocardia asteroides was also able to transform AFB1 to another fluorescent product (Arai et al., 1967).
Among fungi, Rhizopus species, such as R. arrhizus (Cole et al., 1972), R. oryzae (Knol et al., 1990; Faraj et al., 1993; Varga et al., 2005) and R. oligosporus (Kusumaningtyas et al., 2006) have been described as being able to degrade AFB1, whereas several other Rhizopus species (Cole et al., 1972) also have been shown to remove AFG1. Non-aflatoxigenic A. flavus isolates, Rhizopus sp., A. niger, and A. glaucus (Eurotium herbariorum) converted AFB1 to aflatoxicol (Figure 1) and vice versa (Nakazato et al., 1990). Alternaria sp., Phoma sp., Trichoderma sp., and Sporotrichum sp. have been found to lower AFB1 to 65–99% of the original concentrations (Shantha, 1999). Other fungi, such as Hypomyces rosellus (Dactylium dendroides) (Detroy and Hesseltine, 1968), Mucor ambiguous, Trichoderma viride (Mann and Rehm, 1976), Armillaria tabescens (Liu et al., 1998), Phoma sp. (Shantha, 1999), Pleurotus ostreatus (Motomura et al., 2003), and Trametes versicolor (Zjalic et al., 2006) have also been described to lower AFB1 concentrations. OTA degradation was demonstrated when applying Bacillus licheniformis (Petchkongkaew et al., 2008), Brevibacterium species (B. linens, B. iodinum, B. epidermidis, B. casei) (Rodriguez et al., 2011), Acinetobacter calcoaceticus (Hwang and Draughon, 1994), and Phenylobacterium immobile (Wegst and Lingens, 1983). Cell-free supernatants of Pseudomonas putida reduced OTA concentration by 8.45–25.70% (Rodriguez et al., 2011). The dimorphic fungus Apiotrichum mycotoxinivorans (Trichosporon mycotoxinivorans) also degraded OTA (Molnar et al., 2004). Aspergillus species such as A. niger, A. fumigatus, A. japonicus, and section Nigri species were also able to remove OTA from liquid media (Varga et al., 2000; Abrunhosa et al., 2002, 2014; Bejaoui et al., 2006). Patulin degradation was rarely demonstrated. However, for example, the yeast Rhodosporidium kratochvilovae was shown to decrease patulin concentration, whereas the concentration of desoxypatulinic acid increased with time (Castoria et al., 2011). Another possible detoxification mechanism is done by PGUG enzyme from yeast Meyerozyma guilliermondii (Chen et al., 2017) or by oxidoreductase from bacteria Gluconobacter oxydans (Ricelli et al., 2007). Besides the antagonistic effects of yeasts on mycotoxin production, the cytotoxic and inhibitory effects of the toxins on yeasts (summarized in Figure 2) have also been investigated in some cases (reviewed by Pfliegler et al., 2015). In these studies, the well-known model organisms Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe have been studied. The toxic effects of AF and OTA, among other mycotoxins, negatively affected the yield of maize mash fermentation processes (Kłosowski et al., 2010), suggesting considerable toxicity. The mechanism of the AF toxic action was shown to be a DNA replication block (Fasullo et al., 2010). Mutagenic effects were detected after ST exposure (Kuczuk et al., 1978). Furthermore, patulin was found to induce oxidative stress and DNA damage both in fission and budding yeasts (Horváth et al., 2012; Papp et al., 2012; Ianiri et al., 2013), with an additional effect of fluidization of the cytoplasm membrane in S. pombe (Horváth et al., 2010).
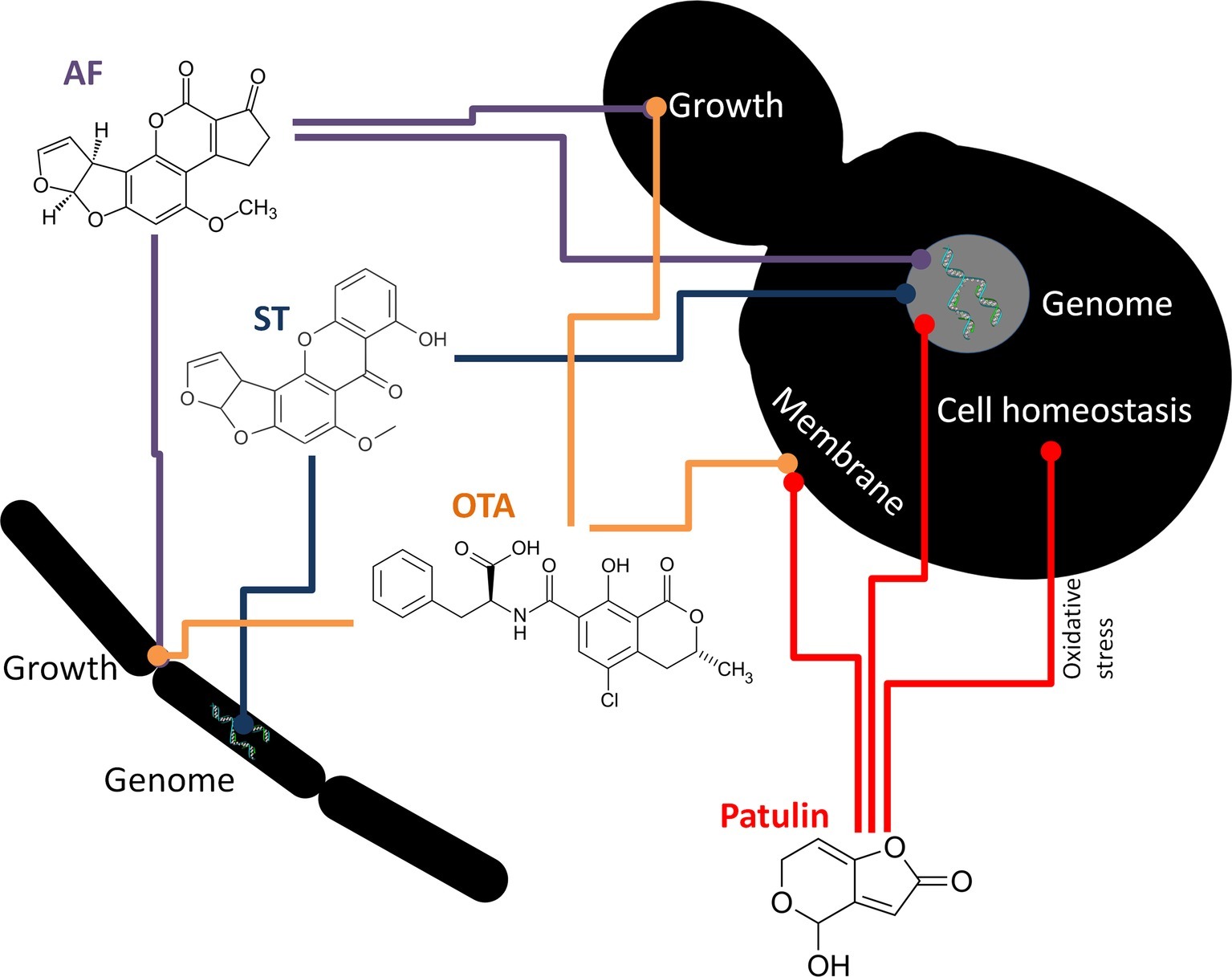
Figure 2. Mechanisms of action of some Aspergillus mycotoxins on bacteria (left) and yeasts (right). Colored lines represent antagonistic/damaging effects. AF, aflatoxin; ST, sterigmatocystin; OTA, ochratoxin A.
Yeasts utilize general and oxidative stress response pathways along with potential degradation mechanisms to resist mycotoxin exposure (Iwahashi et al., 2006; Ianiri et al., 2013); thus, variation in sensitivity to mycotoxins is not a surprise. Indeed, Hanseniaspora uvarum, S. cerevisiae, and Kluyveromyces marxianus were all found to be resistant to AF and OTA (Angioni et al., 2007). Aspergillus mycotoxin toxicity to bacteria is far less understood. Madhyastha et al. (1994) found Bacillus and Brevibacillus spp. to be highly susceptible to AFB1, but mostly resistant to OTA (except for B. brevis and B. cereus). Tested strains of Pseudomonas, Salmonella, Listeria, and Escherichia were usually unaffected by mycotoxins. Additionally, Kuczuk et al. (1978) demonstrated the mutagenic effects of ST on S. Typhimurium.
Biodegradation techniques with higher effectiveness may be developed based on existing data and novel research, by further identifying microorganisms capable of biodegrading mycotoxins, by confirming non-toxicity of degradation compounds, by improving both their toxin tolerance and their degradation abilities, and by testing various modes of application.
Fungi interact with plants through VOCs. This phenomenon could play an essential role in fungal pathogenesis. VOCs released by pathogenic fungi could influence plants before any physical interaction between the two organisms. Some VOCs (fatty acid derivatives, terpenoids, phenylpropanoids) are lipophilic; they are small (less than 300 Da) and have high vapor pressure (0.01 kPa or higher at 20°C) and are well known as signal molecules among various organisms. Some of the VOCs (e.g., C15H24) were found to be unique to aflatoxigenic A. flavus (Zeringue et al., 1993). Different fungal-bacterial interaction leads to the specific initiation of fungal SM genes. The two-way volatile interaction between A. flavus and Ralstonia solanacearum, a similarly widespread and economically crucial soil-borne pathogenic bacterium of peanut, was studied by Spraker et al. (2014). R. solanacearum decreased the production of its major virulence factor extracellular polysaccharide in response to A. flavus VOCs, while A. flavus responded to the bacterial VOCs by reducing conidiospore production and by increasing AF production on peanut. Arbuscular mycorrhizae are also affected by the Aspergilli. Funneliformis mosseae (Glomus mosseae) decreased the saprobic A. niger population through its effect on the plant, whereas A. niger inhibited F. mosseae in its extramatrical stage through the production of soluble substances or VOCs (McAllister et al., 1995).
Application of some special yeasts may cause a direct inhibition of mycotoxin production of filamentous fungi, independently of their growth suppressing effect (Petersson et al., 1998; Hua et al., 2014). However, the effect on toxin production is rarely separated from the growth-inhibiting effect due to methodological constraints. Wickerhamomyces anomalus (Pichia anomala) is the best-characterized yeast species from this aspect. Hua et al. (2014) recognized 2-phenyl ethanol (2-PE), a volatile compound produced by W. anomalus as both growth and AF biosynthesis inhibitor in A. flavus. AF biosynthesis genes aflR (a positive regulator), aflC (polyketide synthase, an early gene in the AF pathway), aflS (transcription enhancer), aflK (versicolorin B synthase), and aflO (O-methyltransferase B) were downregulated more than 10,000-fold following 2-PE treatment. Altered expression patterns were also observed for chromatin-modifying genes (MYST1, MYST2, MYST3, hdaA, gcn5, rpdA), influencing mold growth negatively (Hua et al., 2014). On the contrary, a subsequent characterization of the temporal transcriptome response of A. flavus to smaller, subinhibitory 2-PE concentration revealed inhibition of CPA and AF biosynthesis genes that can be attributed to stimulating active growth of the mold, a condition that does not favor SM production (Chang et al., 2015). These results highlighted the complexity of fungus-fungus interactions depending on the metabolic state and VOC concentration as delicately controlled as the production of mycotoxins (Figure 3).
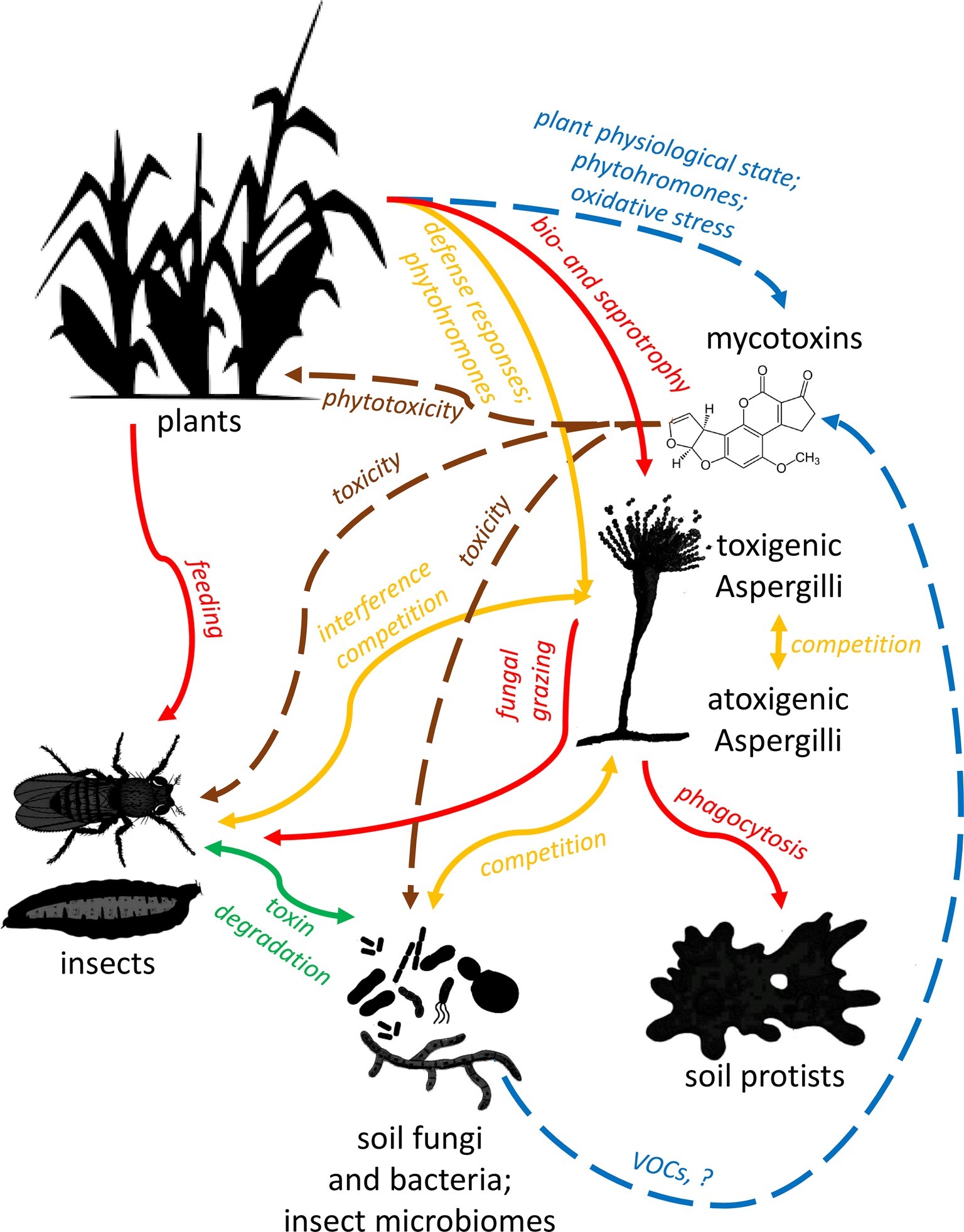
Figure 3. Schematic summary of ecological interactions of plants, fungi, insects, microbes, and Aspergilli. Red lines represent trophic relationships, with arrows pointing towards predators and herbivores. Orange lines represent competitive relationships, while green ones show mutualistic relations. Brown lines signal toxic effects of mycotoxins on various organisms, and blue lines show modulating effects of plants and microbes on toxin production. Note that trophic interactions and pathogenicity of soil microbiota are only considered in relation to aflatoxigenic Aspergilli and their toxins in this review and figure.
Streptomyces isolates decreased AF levels when co-cultured with A. flavus, and this effect was also linked to suppressing AF regulator gene expression (Verheecke et al., 2015). Subsequently, S. alboflavus VOCs (mainly dimethyl trisulfide and benzenamine) were shown to play a critical role in this effect, downregulating genes involved in AF biosynthesis in addition to growth inhibition (Yang et al., 2019). Along with W. anomalus, Hanseniaspora uvarum and Pichia kluyveri yeasts were also found to produce VOCs (most notably 2-PE) that hindered the growth and OTA production of A. ochraceus (Masoud et al., 2005; Masoud and Kaltoft, 2006). A follow-up study showed that 2-PE inhibition of OTA production byin A. carbonarius and A. ochraceus isolates was also inhibited by 2-PE, though was caused by the downregulation of their non-ribosomal peptide synthase, polyketide synthase, and monooxygenase genes (Farbo et al., 2018) and the regulatory veA and laeA genes (Amaike and Keller, 2009).
Another VOC, ethylacetate, was involved in the biocontrol effects of Saccharomyces, Metschnikowia, and W. anomalus yeasts against various molds, including A. carbonarius (Oro et al., 2018). VOCs were also responsible for the biocontrol effect of Candida friedrichii, Candida intermedia, Lachancea thermotolerans, and Cyberlindnera jadinii (Fiori et al., 2014). However, this effect was species-specific. Only C. friedrichii reduced mold growth significantly, while the others only inhibited the fungal sporulation.
Finally, it should be noted that yeast-mold, and bacteria-mold interactions through VOCs and other factors, including growth inhibition mechanisms and the mechanisms of gene expression alterations in mycotoxin gene clusters, mostly have been tested in solid and liquid co-cultures, i.e., isolated from the plant host. Studies based on results of the last decades thus should focus on disentangling the interplay among microbes in vivo, both to understand the microbial ecology of mycotoxin production in crops and to evaluate the utilization strategies.
Secretion of mycotoxins and escape from phagocytosis are strategies evolved in molds to counter predation in the natural environment. A. fumigatus and free-living amoebal species are both abundant soil organisms with antagonistic relationships. Mechanisms of A. fumigatus to avoid ingestion by amoebae were modeled with Acanthamoeba castellanii (Van Waeyenberghe et al., 2013). Intra-amoebal passage left a fraction of the consumed conidia viable. These spores were able to escape the food vacuoles after phagocytosis and germinated intra-cytoplasmatically, resulting in amoebal death. Interactions with mammalian and avian macrophages and A. fumigatus have been compared to these processes, leading to the hypothesis that the ability of the fungus to kill and escape macrophages is a pre-adoptive trait developed in their original ecological niche, namely the soil (Van Waeyenberghe et al., 2013).
Similarly, the slime mold Dictyostelium discoideum efficiently consumed fungal spores upon contact with A. fumigatus, but the ingestion was more intensive when conidia contained lower amounts of the green spore pigment dihydroxy naphthalene (DHN) melanin (Hillmann et al., 2015). Conidia could survive phagocytosis, and the intracellular germination began only after some hours of co-incubation, which leads to a fatal disruption of the predatory cell. Furthermore, both organisms secreted cross-inhibitory factors that could block fungal growth or induce amoebal aggregation (caused by fungal gliotoxin) with subsequent cell lysis, respectively (Figure 3). A. fumigatus and related ascomycetes produced the above mentioned DHN melanin in their spores. However, A. terreus is a DHN-melanin synthesis deficient fungus and, instead, had a tyrosinase (TyrP), and an unusual NRPS-like enzyme (MelA) expressed under conidiation. MelA produced aspulvinone E, which is stimulated for polymerization by TyrP. The new pigment, Asp-melanin, in addition to its usual function conferring resistance against UV radiation, hindered phagocytosis by soil amoeba. Contrary to DHN melanin, Asp-melanin did not prevent acidification of phagolysosomes. Therefore, it is probable that it contributes to the endurance of A. terreus conidia in an acidic environment (Geib et al., 2016).
Furthermore, the antibiotic compound fumagillin produced by A. fumigatus is active against microsporidia and several amoebae but is also poisonous when administered to mammals (Stevanovic et al., 2008). However, this substance was widely used in apiculture against amoebal disease (Bailey, 1955).
Recently, roles of fungal SMs in the ecosystem have been demonstrated by toxicological, behavioral, and experimental evolutionary setups with a still limited number of arthropod species. Using fruit fly larvae (Drosophila), the role of AF in protection from fungivores is linked to its role in interference competition (Drott et al., 2017), supporting Janzen’s (1977) old and not universally accepted hypothesis (Sherratt et al., 2006). Janzen postulated a fitness advantage of AF production in the presence of soil microbes, vertebrates, or arthropods with which the fungus engages in interference competition. Recent experiments have shown that deterring arthropods indeed confers a fitness advantage to the fungus colonizing nutrient-rich sources (e.g., decaying fruits, seeds, dung, and carrion) (Drott et al., 2017), in addition to the more straightforward and previously described (Caballero Ortiz et al., 2013; Doll et al., 2013) deterring effect on fungal grazers. Mycotoxin production by colonizing fungi may create an adverse environment for arthropods competing for these nutrition sources (Rohlfs and Churchill, 2011). The fact that arthropods, especially insects, are not only competitors of the Aspergilli, but their feeding may predispose the plant or the harvested plant product upon which it feeds to Aspergillus infection (Beti et al., 1995; Niu et al., 2008; Ni et al., 2011) further illustrates the complicated tripartite ecological interactions of these molds with plants and arthropods (summarized in Figure 3).
Naturally, the production of AFs may exert selective pressure on exposed arthropods to evolve resistance or tolerance mechanisms that can manifest in detoxification mechanisms or active antagonism towards the fungus. Arthropods are very diverse in their interactions with toxigenic molds, ranging from high susceptibility to remarkable tolerance, presumably, resulting from the variable nature of this evolutionary pressure across habitats. Variation in susceptibility to AF and other mycotoxins has been detected by various studies focusing on mycophagous mites (Racovitza, 2009), Drosophila species (Rohlfs and Obmann, 2009), soldier fly larvae (Hermetia illucens) (Bosch et al., 2017; Camenzuli et al., 2018), the maize weevil (Sitophilus zeamais) (Drott et al., 2017), the yellow and lesser mealworms (Tenebrio molitor and Alphitobius diaperinus) (Bosch et al., 2017; Camenzuli et al., 2018), the navel orangeworm (Amyelois transitella) (Niu et al., 2009), the cabbage looper (Trichoplusia ni) (Zeng et al., 2013), or the corn earworm (Helicoverpa zea) (Zeng et al., 2006; Niu et al., 2008, 2009). It is plausible that species feeding on highly contaminated food sources are selected towards higher tolerance. Maize weevils are remarkable from this aspect: no mortality increase was observed among these pests even when their food sources contained up to 30,000 μg kg−1 AFB1 (Drott et al., 2017).
Additionally, using Drosophila melanogaster as a model organism, within-species variation in tolerating mycotoxins has also been observed (Rohlfs, 2006). This intraspecific variation may enable populations to adapt to increased fungal competition and mycotoxin exposure, as demonstrated with the same fly species and A. nidulans in an experimental evolutionary setup (Trienens and Rohlfs, 2011). The authors concluded that evolved lineages were more tolerant both to fungal and to purified ST exposure without increased resistance, i.e., without increased ability to impair fungal growth. At the same time, grazing by D. melanogaster larvae induced resistance in A. nidulans. Grazing activated the expression of many putative resistance genes of the fungus, along with laeA, the key SM regulator gene (Amaike and Keller, 2011). The reaction to the fungivores co-occurred with gene expression changes in signal transduction, epigenetic regulation, and SM biosynthesis. Reciprocal insect-fungus interactions may select the Aspergilli for inducible resistance resulting in higher fitness in habitats with a high abundance of fungivores (Caballero Ortiz et al., 2013).
Feeding by D. melanogaster larvae induced synthesis of methyl farnesoate and juvenile hormone-III in A. nidulans upon expressing a heterologous regulatory protein (Nielsen et al., 2013). It indicates the probable importance of juvenile hormone biosynthesis in fungal-insect antagonistic relationships while also raising possibilities in insecticidal strategies, given the developmental and metabolic importance of juvenile hormones in arthropods (Nielsen et al., 2013). Vice versa, insects may also develop behavioral adaptations to respond to toxic fungal competitors. For example, Drosophila larvae have been shown to aggregate around aflatoxigenic A. nidulans colonies suppressing fungal growth, improving the chance of larval survival to the adult stage in natural habitats (Rohlfs, 2005; Trienens et al., 2017).
Another fungal-bacterial-insect interaction was described with the connection of an endophytic herbivore, Dendroctonus rufipennis (spruce beetle), which is accompanied by an invasion of its galleries by several fungal species (e.g., A. fumigatus, A. nomius, Leptographium abietinum, Trichoderma harzianum) (Cardoza et al., 2006). Trichoderma and Aspergilli significantly decreased the survival and reproduction of spruce beetle in controlled circumstances. Adult spruce beetle insects exuded an oral secretion, which inhibited the growth of tested fungi except for A. nomius or disrupted the fungal morphology in a dose-dependent way. Oral secretions on microbiological media revealed presence of bacteria responsible for the antifungal activity. The isolated bacteria belonged to the Actinobacteria, Firmicutes, Betaproteobacteria, and Gammaproteobacteria taxa that showed species-specific inhibitory activities (Cardoza et al., 2006).
Tolerance requires effective detoxification of food-derived AFs, mechanisms of which have recently been uncovered, but so far only in a few species. H. zea has been shown to predispose the plant upon which it feeds to Aspergillus infection and concomitant AF contamination, and this pest insect was shown to be able to efficiently metabolize AFB1 into the less toxic AFP1 (Figure 1) using cytochrome P450 monooxygenases (Niu et al., 2008). However, the action of these monooxygenase enzymes is not yet fully understood, as some results indicate that bioactivation, not detoxification may also result from their activity in insects (Zeng et al., 2006, 2013). Larvae of A. transitella, a significant pest of almonds and pistachios have been shown to metabolize AFB1 into three biotransformation products, mainly aflatoxicol, and to negligible amounts of AFM1 and AFB2a (Figure 1). The relatively high production of aflatoxicol may reflect a detoxifying adaptation arising from the often mold-infected habitats of the A. transitella (Lee and Campbell, 2000). The codling moth Cydia pomonella, a pest infecting walnuts and pome fruits, produced none to low levels of AFB1 biotransformation products, suggesting a lower level of detoxification capability (Lee and Campbell, 2000).
A further aspect of insect mycotoxin tolerance and indirect mold-microbiome interactions may also be relevant: the effects of insect symbionts during mycotoxin exposure (Figure 3). Insect microbial symbionts are ubiquitous, incredibly diverse, and their interactions with their hosts are far from being wholly understood (e.g., Dowd and Vega, 2004). At least one symbiotic yeast-like species, Symbiotaphrina kochii, can enzymatically detoxify and utilize mycotoxins as carbon sources (along with plant allochemicals and insecticides, even as sole carbon sources) (Shen and Dowd, 1991). More recently, Rohlfs and Kürschner (2010) reported that increased diversity of dietary yeast species benefited Drosophila larvae competing with, and exposed to the toxins of A. nidulans, by apparently ameliorating the effects of the toxins. These works call attention to the highly under-researched interactions of invertebrate gut microbiotas and toxins. It is plausible that the microbiome of insects and other arthropods, especially of those that are fungal grazers or face interference competition from molds, is an essential factor contributing to the observed variation in resistance to AF and other mycotoxins, and hence the ability of certain arthropods to compete with highly toxigenic molds.
Finally, the application of entomopathogenic fungi is a capable alternative to chemical control of insects, e.g., mosquitoes. Aspergillus clavatus from Oedaleus senegalensis (Senegalese locust) was highly pathogenic against Culex quinquefasciatus, Aedes aegypti, and Anopheles gambiae mosquito larvae. Application of A. clavatus using spore concentrations ranging between 4.3 and 21 × 107 ml−1 resulted in 11–68% mortality against C. quinquefasciatus, and 37–100% against A. aegypti (Seye et al., 2010). Moreover, also in pheromone production, a possible biotechnological application is hiding. The VOC spiroketal (E)-conophthorin (7-methyl-1,6-dioxaspiro[4.5]decane) (Beck and Higbee, 2015) and the isomeric chalcogran are recognized as semiochemicals of some scolytid beetles. Conophthorin is produced by both insects and plants and widely known as a non-host plant VOC from the bark of angiosperm species. Interestingly, VOC production was tested as a response to primary fatty acids of the host plants by non-aflatoxigenic and aflatoxigenic A. flavus, as well as A. niger, A. parasiticus, Penicillium glabrum, and Rhizopus stolonifera. On linoleic acid, these fungi formed both spiroketals, while those on linolenic acid emitted only chalcogran. Conversely, no production was detected on palmitic and oleic acid, which also adds a new level of insect-plant-Aspergillus VOC interaction (Beck et al., 2012).
Non-aflatoxigenic knockout and low toxin-producing strains of Aspergillus are less capable of antagonizing insect populations (Regulin and Kempken, 2018). In addition to balancing selection on mycotoxin production, it must be noted that insect adaptation to mold competition seems to favor tolerance instead of resistance (Trienens and Rohlfs, 2011). Thus, selective pressure on fungi competing with insects is less likely to fuel co-evolutionary arms races or Red Queen dynamics (Rabajante et al., 2015) that would clearly favor more toxigenic strains.
Because of their economic and public health importance, research on fungal SM mycotoxins has mostly been focused on animal husbandry, the food chain, and human aspects. However, genome data analyses of numerous fungi and the analytical measurements revealed that most of the predicted SM-associated clusters are silent, demonstrating that fungi continue to be a yet undiscovered resource of biologically active molecules. It was also concluded that A. flavus might produce metabolites besides well-known mycotoxins that could be underrated contributors to the toxicity to humans and animals. By changing the culture conditions or the genetic regulation to activate silent clusters, new molecules may be discovered that later can be available for medicine or selective biocontrol of fungi or higher eukaryotes.
For a comprehensive understanding of toxigenic molds’ ecology and the evolutionary pressures shaping mycotoxin production, interactions with the micro- and macroflora and fauna in different habitats need to be considered and investigated. The study of the overall role of microbial SMs in natural habitats is now an emerging field. However, the lack of current studies on plant-modified and masked Aspergillus mycotoxins calls for attention to a considerable gap in our understanding of mycotoxins’ fate and ecological roles.
Some interaction research revealed new levels of regulations of SM gene expressions through chemical interactions even without direct physical contact. Metabolomic studies at the level of VOCs can boost our knowledge to solve the puzzle of the interactions.
Microbial symbionts of insects are ubiquitous and incredibly diverse; however, their interactions with their hosts are far from being wholly understood. The review also calls attention to the highly under-researched interactions of invertebrate gut microbiotas and mycotoxins. The microbiome of insects and other arthropods is an essential factor contributing to the observed variation in resistance to AF and other mycotoxins, and, hence, in the ability of certain arthropods to compete with highly toxigenic molds.
Recently developed and applied plant protection or soil fertilization agents also should be studied focusing on their effects on interkingdom interactions in soil, or on plants and in plant tissues. In connection with this, the recently approved non-aflatoxigenic A. flavus strains and fungal preparations are also a subject for further research on interactions of the soil macro- and microbiota. Studying metabolic pathways in pericarp and seeds that are activated differentially by non-aflatoxigenic and aflatoxigenic A. flavus may help to identify possible target genes to increase plant tolerance and resistance and to fight AF contamination. Mycotoxin biodegradation techniques with higher effectiveness may also be developed based on the existing data and novel research by identifying further microorganisms capable of biodegrading mycotoxins, by improving both their toxin tolerance and their degradation abilities, and by modification of the application.
This article also wanted to attract attention to the fact that most of the direct and indirect yeast-mold and bacteria-mold interactions have been tested only in in vitro conditions. Such studies targeted fungal growth inhibition mechanisms and the gene expression alterations in SM gene clusters. Therefore, studies initiated by the results of the last decades should focus on disentangling the interplay in vivo, both to understand the microbial ecology of mycotoxin production in crops and to evaluate the utilization strategies. Therefore, greenhouse or microplot experiments should be applied for the extended data collection.
IP encouraged TP and WP to investigate the literature on interaction. IP and ZG supervised the writing of this work. TP took the lead in writing the manuscript. WP prepared the figures and wrote sections about yeast-fungal, insect-fungal interactions. TP prepared the sections considering microbial, plant, and soil interactions. ZG prepared the section about masked mycotoxins. All authors discussed the review and contributed to the final manuscript.
Project no. 2018-1.2.1-NKP-2018-00002 has been implemented with the support provided by the National Research, Development and Innovation Fund of Hungary, financed under the 2018-1.2.1-NKP funding scheme. This project was also financed by the National Research, Development and Innovation Fund project K119494 and by the Higher Education Institutional Excellence Programme. This study was supported by grants from the Ministry of Innovation and Technology in Hungary (NKFIH-1150-6/2019), within the framework of the Biotechnology thematic program of the University of Debrecen, Hungary.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abrunhosa, L., Inês, A., Rodrigues, A. I., Guimarães, A., Pereira, V. L., Parpot, P., et al. (2014). Biodegradation of ochratoxin A by Pediococcus parvulus isolated from Douro wines. Int. J. Food Microbiol. 188, 45–52. doi: 10.1016/j.ijfoodmicro.2014.07.019
Abrunhosa, L., Serra, R., and Venancio, A. (2002). Biodegradation of ochratoxin A by fungi isolated from grapes. J. Agric. Food Chem. 50, 7493–7496. doi: 10.1021/jf025747i
Accinelli, C., Abbas, H. K., Zablotowicz, R. M., and Wilkinson, J. R. (2008). Aspergillus flavus aflatoxin occurrence and expression of aflatoxin biosynthesis genes in soil. Can. J. Microbiol. 54, 371–379. doi: 10.1139/W08-018
Affeldt, K. J., Brodhagen, M., and Keller, N. P. (2012). Aspergillus oxylipin signaling and quorum sensing pathways depend on G protein-coupled receptors. Toxins 4, 695–717. doi: 10.3390/toxins4090695
Ahlberg, S., Joutsjoki, V., Laurikkala, S., Varmanen, P., and Korhonen, H. (2017). Aspergillus flavus growth inhibition by Lactobacillus strains isolated from traditional fermented Kenyan milk and maize products. Arch. Microbiol. 199, 457–464. doi: 10.1007/s00203-016-1316-3
Amaike, S., and Keller, N. P. (2009). Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus. Eukaryot. Cell 8, 1051–1060. doi: 10.1128/EC.00088-09
Amaike, S., and Keller, N. P. (2011). Aspergillus flavus. Annu. Rev. Phytopathol. 49, 107–133. doi: 10.1146/annurev-phyto-072910-095221
Angioni, A., Caboni, P., Garau, A., Farris, A., Orro, D., Budroni, M., et al. (2007). In vitro interaction between ochratoxin A and different strains of Saccharomyces cerevisiae and Kloeckera apiculata. J. Agric. Food Chem. 55, 2043–2048. doi: 10.1021/jf062768u
Anjorin, A. T., and Inje, T. (2014). Effect of total aflatoxin on the growth characteristics and chlorophyll level of sesame (Sesamum indicum L.). New York Sci. J. 7, 8–13. http://www.sciencepub.net/newyork/ny0704/002_23713ny070414_8_13.pdf
Arai, T., Tatsuya, I., and Koyama, Y. (1967). Antimicrobial activity of aflatoxins. J. Bacteriol. 93, 59–64. doi: 10.1128/JB.93.1.59-64.1967
Bagheri-Gavkosh, S., Bigdeli, M., Shams-Ghahfarokhi, M., and Razzaghi-Abyaneh, M. (2009). Inhibitory effects of Ephedra major host on Aspergillus parasiticus growth and aflatoxin production. Mycopathologia 168, 249–255. doi: 10.1007/s11046-009-9220-x
Bailey, L. (1955). Control of amoeba disease by the fumigation of combs and by fumagillin. Bee World 36, 162–163. doi: 10.1080/0005772X.1955.11094896
Baisakh, N., Datta, K., Oliva, N., Ona, I., Rao, G. J. N., Mew, T. W., et al. (2001). Rapid development of homozygous transgenic rice using anther culture harboring rice chitinase gene for enhanced sheath blight resistance. Plant Biotechnol. 18, 101–108. doi: 10.5511/plantbiotechnology.18.101
Balbontín, R., Vlamakis, H., and Kolter, R. (2014). Mutualistic interaction between Salmonella enterica and Aspergillus niger and its effects on Zea mays colonization. Microbial Biotech. 7, 589–600. doi: 10.1111/1751-7915.12182
Bandyopadhyay, R., Ortega-Beltran, A., Akande, A., Mutegi, C., Atehnkeng, J., Kaptoge, L., et al. (2016). Biological control of aflatoxins in Africa: current status and potential challenges in the face of climate change. World Mycotoxin J. 9, 771–789. doi: 10.3920/WMJ2016.2130
Barberis, C. L., Pena, G., Carranza, C., and Magnoli, C. E. (2014). Effect of indigenous mycobiota on ochratoxin A production by Aspergillus carbonarius isolated from soil. Mycotoxin Res. 30, 1–8. doi: 10.1007/s12550-013-0181-z
Bari, R., and Jones, J. D. G. (2009). Role of plant hormones in plant defense responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Beck, J. J., and Higbee, B. S. (2015). Plant- or fungal-produced conophthorin as an important component of host plant volatile-based attractants for agricultural lepidopteran insect pests. ACS Symposium Series: Discovery and Synthesis of Crop Protection Products. Washington DC, USA: American Chemical Society, 111–127.
Beck, J. J., Mahoney, N. E., Cook, D., and Gee, W. S. (2012). Generation of the volatile spiroketals conophthorin and chalcogran by fungal sporeson polyunsaturated fatty acids common to almonds and pistachios. J. Agric. Food Chem. 60, 11869–11876. doi: 10.1021/jf304157q
Bedre, R., Rajasekaran, K., Mangu, V. R., Sanchez Timm, L. E., Bhatnagar, D., and Baisakh, N. (2015). Genome-wide transcriptome analysis of cotton (Gossypium hirsutum L.) identifies candidate gene signatures in response to aflatoxin producing fungus Aspergillus flavus. PLoS One 10:e0138025. doi: 10.1371/journal.pone.0138025
Bejaoui, H., Mathieu, F., Taillandier, P., and Lebrihi, A. (2006). Biodegradation of ochratoxin A by Aspergillus section Nigri species isolated from French grapes: a potential means of ochratoxin A decontamination in grape juices and musts. FEMS Microbiol. 255, 203–208. doi: 10.1111/j.1574-6968.2005.00073.x
Bellincampi, D., Cervone, F., and Lionetti, V. (2014). Plant cell wall dynamics and wall-related susceptibility in plant-pathogen interactions. Front. Plant Sci. 5:228. doi: 10.3389/fpls.2014.00228
Benoit, I., van den Esker, M. H., Patyshakuliyeva, A., Mattern, D. J., Blei, F., Zhou, M., et al. (2015). Bacillus subtilis attachment to Aspergillus niger hyphae results in mutually altered metabolism. Environ. Microbiol. 17, 2099–2113. doi: 10.1111/1462-2920.12564
Berthiller, F., Crews, C., Dall’Asta, C., De Saeger, S., Haesaert, G., Karlovsky, P., et al. (2013). Masked mycotoxins: a review. Mol. Nutr. Food Res. 57, 165–186. doi: 10.1002/mnfr.201100764
Berthiller, F., Schuhmacher, R., Adam, G., and Krska, R. (2009). Formation, determination and significance of masked and other conjugated mycotoxins. Anal. Bioanal. Chem. 395, 1243–1252. doi: 10.1007/s00216-009-2874-x
Beti, J. A., Phillips, T. W., and Smalley, E. B. (1995). Effects of maize weevils (Coleoptera: Curculionidae) on production of aflatoxin B1 by Aspergillus flavus in stored corn. J. Econ. Entomol. 88, 1776–1782. doi: 10.1093/jee/88.6.1776
Betrán, F. J., and Isakeit, T. (2004). Aflatoxin accumulation in maize hybrids of different maturities. Agron. J. 96, 565–570. doi: 10.2134/agronj2004.5650
Bleecker, A. B., and Kende, H. (2002). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Bolton, E. E., Wang, Y., Thiessen, P. A., and Bryant, S. H. (2008). “PubChem: integrated platform of small molecules and biological activities” in Annual Reports in Computational Chemistry. eds. R. A. Wheeler and D. C. Spellmeyer (Oxford: Elsevier), 217–241. doi: 10.1016/S1574-1400(08)00012-1
Bosch, G., Van Der Fels-Klerx, H. J., De Rijk, T. C., and Oonincx, D. G. A. B. (2017). Aflatoxin B1 tolerance and accumulation in black soldier fly larvae (Hermetia illucens) and yellow mealworms (Tenebrio molitor). Toxins 9:185. doi: 10.3390/toxins9060185
Botha, C. J., Truter, M., and Sulyok, M. (2018). Multimycotoxin analysis of South African Aspergillus clavatus isolates. Mycotoxin Res. 34, 91–97. doi: 10.1007/s12550-017-0303-0
Boughalleb-M’Hamdi, N., Salem, I. B., and M’Hamdi, M. (2018). Evaluation of the efficiency of Trichoderma, Penicillium, and Aspergillus species as biological control agents against four soil-borne fungi of melon and watermelon. Egypt. J. Biol. Pest Control 28:25. doi: 10.1186/s41938-017-0010-3
Bravo, J. M., Campo, S., Murillo, I., Coca, M., and Segund, B. S. (2003). Fungus and wound-induced accumulation of mRNA containing a class II chitinase of the pathogenesis-related protein 4 (PR-4) family of maize. Plant Mol. Biol. 52, 745–759. doi: 10.1023/A:1025016416951
Brodhagen, M., Tsitsigiannis, D. I., Hornung, E., Goebel, C., Feussner, I., and Keller, N. P. (2008). Reciprocal oxylipin-mediated cross-talk in the Aspergillus-seed pathosystem. Mol. Microbiol. 67, 378–391. doi: 10.1111/j.1365-2958.2007.06045.x
Burow, G. B., Nesbitt, T. C., Dunlap, J., and Keller, N. P. (1997). Seed lipoxygenase products modulate Aspergillus mycotoxin biosynthesis. Mol. Plant-Microbe Interact. 10, 380–387. doi: 10.1094/MPMI.1997.10.3.380
Caballero Ortiz, S., Trienens, M., and Rohlfs, M. (2013). Induced fungal resistance to insect grazing: reciprocal fitness consequences and fungal gene expression in the Drosophila-Aspergillus model system. PLoS One 8:e74951. doi: 10.1371/journal.pone.0074951
Calvo, A. M., Hinze, L. L., Gardner, H. W., and Keller, N. P. (1999). Sporogenic effect of polyunsaturated fatty acids on development of Aspergillus spp. Appl. Environ. Microbiol. 65, 3668–3673. doi: 10.1128/AEM.65.8.3668-3673.1999
Camenzuli, L., van Dam, R., de Rijk, T., Andriessen, R., van Schelt, J., and van der Fels-Klerx, H. J. I. (2018). Tolerance and excretion of the mycotoxins aflatoxin B1, zearalenone, deoxynivalenol, and ochratoxin A by Alphitobius diaperinus and Hermetia illucens from contaminated substrates. Toxins 10:91. doi: 10.3390/toxins10020091
Cano, P., Puel, O., and Oswald, I. P. (2016). “Mycotoxins: fungal secondary metabolites with toxic properties” in Fungi: applications and management strategies. eds. S. K. Deshmukh, J. K. Misra, J. P. Tewari, and T. Papp (Boca Raton: CRC Press), 318–371.
Cardoza, Y. J., Klepzig, K. D., and Raffa, K. F. (2006). Bacteria in oral secretions of an endophytic insect inhabitant agonistic fungi. Ecol. Entomol. 31, 636–645. doi: 10.1111/j.1365-2311.2006.00829.x
Cardwell, K. F., and Cotty, P. J. (2002). Distribution of Aspergillus section Flavi among field soils from the four agroecological zones of the republic of Bénin, West Africa. Plant Dis. 86, 434–439. doi: 10.1094/PDIS.2002.86.4.434
Castoria, R., Mannina, L., Durán-Patrόn, R., Maffei, F., Sobolev, A. P., De Felice, D. V., et al. (2011). Conversion of the mycotoxin patulin to the less toxic desoxypatulinic acid by the biocontrol yeast Rhodosporidium kratochvilovae strain LS11. J. Agric. Food Chem. 59, 11571–11578. doi: 10.1021/jf203098v
Chalivendra, S. C., DeRobertis, C., Chang, P.-K., and Damann, K. E. (2017). Cyclopiazonic acid is a pathogenicity factor for Aspergillus flavus and a promising target for screening germplasm for ear rot resistance. Mol. Plant-Microbe Interact. 30, 361–373. doi: 10.1094/MPMI-02-17-0026-R
Chang, P.-K., Ehrlich, K., and Fujii, I. (2009). Cyclopiazonic acid biosynthesis of Aspergillus flavus and Aspergillus oryzae. Toxins 1, 74–99. doi: 10.3390/toxins1020074
Chang, P.-K., Hua, S., Sarreal, S., and Li, R. (2015). Suppression of aflatoxin biosynthesis in Aspergillus flavus by 2-phenylethanol is associated with stimulated growth and decreased degradation of branched-chain amino acids. Toxins 7, 3887–3902. doi: 10.3390/toxins7103887
Chen, Z.-Y., Brown, R. L., Damann, K. E., and Cleveland, T. E. (2010). PR10 expression in maize and its effect on host resistance against Aspergillus flavus infection and aflatoxin production. Mol. Plant Pathol. 11, 69–81. doi: 10.1111/j.1364-3703.2009.00574.x
Chen, Z.-Y., Brown, R. L., Rajasekaran, K., Damann, K. E., and Cleveland, T. E. (2006). Identification of a maize kernel pathogenesis-related protein and evidence for its involvement in resistance to Aspergillus flavus infection and aflatoxin production. Phytopathology 96, 87–95. doi: 10.1094/PHYTO-96-0087
Chen, W., Li, C., Zhang, B., Zhou, Z., Shen, Y., Liao, X., et al. (2018). Advances in biodetoxification of ochratoxin A – a review of the past five decades. Front. Microbiol. 9:1386. doi: 10.3389/fmicb.2018.01386
Chen, Y., Peng, H. M., Wang, X., Li, B. Q., Long, M. Y., and Tian, S. P. (2017). Biodegradation mechanisms of patulin in Candida guilliermondii: an iTRAQ-based proteomic analysis. Toxins 9:48. doi: 10.3390/toxins9020048
Chitarrini, G., Nobili, C., Pinzari, F., Antonini, A., De Rossi, P., Del Fiore, A., et al. (2014). Buckwheat achenes antioxidant profile modulates Aspergillus flavus growth and aflatoxin production. Int. J. Food Microbiol. 189, 1–10. doi: 10.1016/j.ijfoodmicro.2014.07.029
Ciegler, A., Lillehoj, E. B., Peterson, R. E., and Hall, H. H. (1966). Microbial detoxification of aflatoxin. Appl. Microbiol. 14, 934–939. doi: 10.1128/aem.14.6.934-939.1966
Ciegler, A. (1983). Do mycotoxins function in ecological processes? J. Food Saf. 5, 23–30. doi: 10.1111/j.1745-4565.1983.tb00452.x
Cletus, J., Balasubramanian, V., Vashisht, D., and Sakthivel, N. (2013). Transgenic expression of plant chitinases to enhance disease resistance. Biotechnol. Lett. 35, 1719–1732. doi: 10.1007/s10529-013-1269-4
Cleveland, T. E., Yu, J., Fedorova, N., Bhatnagar, D., Payne, G. A., Nierman, W. C., et al. (2009). Potential of Aspergillus flavus genomics for applications in biotechnology. Trends Biotechnol. 27, 151–157. doi: 10.1016/j.tibtech.2008.11.008
Cole, R. J., Kirksey, J. W., and Blankenship, B. R. (1972). Conversion of aflatoxin B1 to isomeric hydroxy compounds by Rhizopus spp. J. Agric. Food Chem. 20, 1100–1102. doi: 10.1021/jf60184a040
Coleman, J., Blake-Kalff, M., and Davies, E. (1997). Detoxification of xenobiotics by plants; chemical modification and vacuolar compartmentation. Trends Plant Sci. 2, 144–151. doi: 10.1016/S1360-1385(97)01019-4
Collinge, D. B., Kragh, K. M., Mikkelsen, J. D., Nielsen, K. K., Rasmussen, U., and Vad, K. (1993). Plant chitinases. Plant J. 3, 31–40. doi: 10.1046/j.1365-313X.1993.t01-1-00999.x
Collinge, D. B., and Slusarenko, A. J. (1987). Plant gene-expression in response to pathogens. Plant Mol. Biol. 9, 389–410. doi: 10.1007/BF00014913
Cordero, M. J., Raventos, D., and San Segundo, B. (1994). Differential expression and induction of chitinases and beta-glucanases in response to fungal infection during germination of maize seeds. Mol. Plant-Microbe Interact. 7, 23–31. doi: 10.1094/MPMI-7-0023
Cotty, P. J. (2006). “Biocompetitive exclusion of toxigenic fungi” in The mycotoxin factbook. eds. D. Barug, D. Bhatnagar, H. P. van Egdmond, J. W. van der Kamp, W. A. van Osenbruggen, and A. Visconti (The Netherlands: Wageningen Academic Publishers), 179–197.
Cotty, P. J. (2007). Influence of field application of an atoxigenic strain of Aspergillus flavus on the populations of A. flavus infecting cotton bolls and on the aflatoxin content of cottonseed. Phytopathology 84:1270. doi: 10.1094/phyto-84-1270
Dang, L., and Van Damme, E. J. M. (2015). Toxic proteins in plants. Phytochemistry 117, 51–64. doi: 10.1016/j.phytochem.2015.05.020
Desjardins, A. E., and Hohn, T. M. (1997). Mycotoxins in plant pathogenesis. Mol. Plant-Microbe Interact. 10, 147–152. doi: 10.1094/MPMI.1997.10.2.147
Detroy, R. W., and Hesseltine, C. W. (1968). Isolation and biological activity of a microbial conversion product of aflatoxin B1. Nature 219:967. doi: 10.1038/219967a0
Dhakal, R., Chai, C., Karan, R., Windham, G. L., Williams, W. P., and Subudhi, P. K. (2017). Expression profiling coupled with in-silico mapping identifies candidate genes for reducing aflatoxin accumulation in maize. Front. Plant Sci. 8:503. doi: 10.3389/fpls.2017.00503
Dolan, S. K., O’Keeffe, G., Jones, G. W., and Doyle, S. (2015). Resistance is not futile: gliotoxin biosynthesis, functionality and utility. Trends Microbiol. 23, 419–428. doi: 10.1016/j.tim.2015.02.005
Dolezal, A. L., Obrian, G. R., Nielsen, D. M., Woloshuk, C. P., Boston, R. S., and Payne, G. A. (2013). Localization, morphology and transcriptional profile of Aspergillus flavus during seed colonization. Mol. Plant Pathol. 14, 898–909. doi: 10.1111/mpp.12056
Dolezal, A. L., Shu, X., OBrian, G. R., Nielsen, D. M., Woloshuk, C. P., Boston, R. S., et al. (2014). Aspergillus flavus infection induces transcriptional and physical changes in developing maize kernels. Front. Microbiol. 5:384. doi: 10.3389/fmicb.2014.00384
Doll, K., Chatterjee, S., Scheu, S., Karlovsky, P., and Rohlfs, M. (2013). Fungal metabolic plasticity and sexual development mediate induced resistance to arthropod fungivory. Proc. R. Soc. B Biol. Sci. 280:20131219. doi: 10.1098/rspb.2013.1219
Dorner, J. W., and Cole, R. J. (2002). Effect of application of nontoxigenic strains of Aspergillus flavus and A. parasiticus on subsequent aflatoxin contamination of peanuts in storage. J. Stored Prod. Res. 38, 329–339. doi: 10.1016/S0022-474X(01)00035-2
Dorner, J. W., Cole, R. J., and Diener, U. L. (1984). The relationship of Aspergillus flavus and Aspergillus parasiticus with reference to production of aflatoxins and cyclopiazonic acid. Mycopathologia 87, 13–15.
Dowd, P. F., and Vega, F. (2004). “The role of yeasts as insect endosymbionts” in Insect-fungal associations: Ecology and evolution. eds. F. E. Vega and M. Blackwell (New York: Oxford University Press), 211–244.
Drott, M. T., Debenport, T., Higgins, S. A., Buckley, D. H., and Milgroom, M. G. (2019). Fitness cost of aflatoxin production in Aspergillus flavus when competing with soil microbes could maintain balancing selection. MBio 10:e02782-18. doi: 10.1128/mBio.02782-18
Drott, M. T., Lazzaro, B. P., Brown, D. L., Carbone, I., and Milgroom, M. G. (2017). Balancing selection for aflatoxin in Aspergillus flavus is maintained through interference competition with, and fungivory by insects. Proc. R. Soc. B Biol. Sci. 284:20172408. doi: 10.1098/rspb.2017.2408
Ehrlich, K. C. (2014). Non-aflatoxigenic Aspergillus flavus to prevent aflatoxin contamination in crops: advantages and limitations. Front. Microbiol. 5:50. doi: 10.3389/fmicb.2014.00050
Ehrlich, K. C., and Mack, B. M. (2014). Comparison of expression of secondary metabolite biosynthesis cluster genes in Aspergillus flavus, A. parasiticus, and A. oryzae. Toxins 6, 1916–1928. doi: 10.3390/toxins6061916
Ehrlich, K. C., Yu, J., and Cotty, P. J. (2005). Aflatoxin biosynthesis gene clusters and flanking regions. J. Appl. Microbiol. 99, 518–527. doi: 10.1111/j.1365-2672.2005.02637.x
El Khoury, R., Mathieu, F., Atoui, A., Kawtharani, H., El Khoury, A., Afif, C., et al. (2017). Ability of soil isolated actinobacterial strains to prevent, bind and biodegrade ochratoxin A. Toxins 9:222. doi: 10.3390/toxins9070222
Fabbri, A. A., Fanelli, C., Panfili, G., Passi, S., and Fasella, P. (1983). Lipoperoxidation and aflatoxin biosynthesis by Aspergillus parasiticus and A. flavus. Microbiol. 129, 3447–3452. doi: 10.1099/00221287-129-11-3447
Falade, T. D. O., Syed Mohdhamdan, S. H., Sultanbawa, Y., Fletcher, M. T., Harvey, J. J. W., Chaliha, M., et al. (2016). In vitro experimental environments lacking or containing soil disparately affect competition experiments of Aspergillus flavus and co-occurring fungi in maize grains. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 33, 1241–1253. doi: 10.1080/19440049.2016.1198048
Fanelli, C., Fabbri, A. A., Pieretti, S., Finotti, E., and Passi, S. (1985). Effect of different antioxidants and free radical scavengers on aflatoxin production. Mycotoxin Res. 1, 65–69. doi: 10.1007/BF03192005
Faraj, M. K., Smith, J. E., and Harran, G. (1993). Aflatoxin biodegradation: effects of temperature and microbes. Mycol. Res. 97, 1388–1392. doi: 10.1016/S0953-7562(09)80174-3
Farbo, M. G., Urgeghe, P. P., Fiori, S., Marcello, A., Oggiano, S., Balmas, V., et al. (2018). Effect of yeast volatile organic compounds on ochratoxin A-producing Aspergillus carbonarius and A. ochraceus. Int. J. Food Microbiol. 284, 1–10. doi: 10.1016/j.ijfoodmicro.2018.06.023
Fasullo, M., Chen, Y., Bortcosh, W., Sun, M., and Egner, P. A. (2010). Aflatoxin B1-associated DNA adducts stall S phase and stimulate Rad51 foci in Saccharomyces cerevisiae. J. Nucleic Acids 2010:456487. doi: 10.4061/2010/456487
Federici, L., Di Matteo, A., Fernandez-Recio, J., Tsernoglou, D., and Cervone, F. (2006). Polygalacturonase inhibiting proteins: players in plant innate immunity? Trends Plant Sci. 11, 65–70. doi: 10.1016/j.tplants.2005.12.005
Fiori, S., Urgeghe, P. P., Hammami, W., Razzu, S., Jaoua, S., and Migheli, Q. (2014). Biocontrol activity of four non- and low-fermenting yeast strains against Aspergillus carbonarius and their ability to remove ochratoxin A from grape juice. Int. J. Food Microbiol. 189, 45–50. doi: 10.1016/j.ijfoodmicro.2014.07.020
Forseth, R. R., Amaike, S., Schwenk, D., Affeldt, K. J., Hoffmeister, D., Schroeder, F. C., et al. (2013). Homologous NRPS-like gene clusters mediate redundant small-molecule biosynthesis in Aspergillus flavus. Angew. Chem. Int. Ed. Eng. 52, 1590–1594. doi: 10.1002/anie.201207456
Fountain, J. C., Bajaj, P., Nayak, S. N., Yang, L., Pandey, M. K., Kumar, V., et al. (2016). Responses of Aspergillus flavus to oxidative stress are related to fungal development regulator, antioxidant enzyme, and secondary metabolite biosynthetic gene expression. Front. Microbiol. 7:2048. doi: 10.3389/fmicb.2016.02048
Fountain, J. C., Khera, P., Yang, L., Nayak, S. N., Scully, B. T., Lee, R. D., et al. (2015). Resistance to Aspergillus flavus in maize and peanut: molecular biology, breeding, environmental stress, and future perspectives. Crop J. 3, 229–237. doi: 10.1016/j.cj.2015.02.003
Fountain, J. C., Scully, B. T., Ni, X., Kemerait, R. C., Lee, R. D., Chen, Z.-Y., et al. (2014). Environmental influences on maize-Aspergillus flavus interactions and aflatoxin production. Front. Microbiol. 5:40. doi: 10.3389/fmicb.2014.00040
Frisvad, J. C., Hubka, V., Ezekiel, C. N., Hong, S.-B., Nováková, A., Chen, A. J., et al. (2019). Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 93, 1–63. doi: 10.1016/j.simyco.2018.06.001
Gao, X., and Kolomiets, M. V. (2009). Host-derived lipids and oxylipins are crucial signals in modulating mycotoxin production by fungi. Toxin Rev. 28, 79–88. doi: 10.1080/15569540802420584
Gao, X., Starr, J., Göbel, C., Engelberth, J., Feussner, I., Tumlinson, J., et al. (2008a). Maize 9-lipoxygenase ZmLOX3 controls development, root-specific expression of defense genes, and resistance to root-knot nematodes. Mol. Plant-Microbe Interact. 21, 98–109. doi: 10.1094/MPMI-21-1-0098
Gao, X., Starr, J., Göbel, C., Engelberth, J., Feussner, I., Tumlinson, J., et al. (2008b). Maize 9-lipoxygenase ZmLOX3 controls development, root-specific expression of defense genes, and resistance to root-knot nematodes. Mol. Plant-Microbe Interact. 21, 98–109. doi: 10.1094/MPMI-21-1-0098
Gardiner, D. M., Waring, P., and Howlett, B. J. (2005). The epipolythiodioxopiperazine (ETP) class of fungal toxins: distribution, mode of action, functions and biosynthesis. Microbiology 151, 1021–1032. doi: 10.1099/mic.0.27847-0
Gardner, H. W. (1995). Biological roles and biochemistry of the lipoxygenase pathway. Hort Sci. 30, 197–205. doi: 10.21273/HORTSCI.30.4.197
Geib, E., Gressler, M., Viediernikova, I., Hillmann, F., Jacobsen, I. D., Nietzsche, S., et al. (2016). A non-canonical melanin biosynthesis pathway protects Aspergillus terreus conidia from environmental stress. Cell Chem. Biol. 23, 587–597. doi: 10.1016/j.chembiol.2016.03.014
Geiser, D. M., Dorner, J. W., Horn, B. W., and Taylor, J. W. (2000). The phylogenetics of mycotoxin and sclerotium production in Aspergillus flavus and Aspergillus oryzae. Fungal Genet. Biol. 31, 169–179. doi: 10.1006/fgbi.2000.1215
Ghazvini, R. D., Kouhsari, E., Zibafar, E., Hashemi, S. J., Amini, A., and Niknejad, F. (2016). Antifungal activity and aflatoxin degradation of Bifidobacterium bifidum and Lactobacillus fermentum against toxigenic Aspergillus parasiticus. Open Microbiol. J. 10, 197–201. doi: 10.2174/1874285801610010197
Gilbert, M. K., Mack, B. M., Moore, G. G., Downey, D. L., Lebar, M. D., Joardar, V., et al. (2018). Whole genome comparison of Aspergillus flavus L-morphotype strain NRRL 3357 (type) and S-morphotype strain AF70. PLoS One 13:e0199169. doi: 10.1371/journal.pone.0199169
Giorni, P., Bertuzzi, T., and Battilani, P. (2016). AF in maize, a multifaceted answer of Aspergillus flavus governed by weather, host-plant and competitor fungi. J. Cereal Sci. 70, 256–262. doi: 10.1016/j.jcs.2016.07.004
Guan, S., Zhao, L., Ma, Q., Zhou, T., Wang, N., Hu, X., et al. (2010). In vitro efficacy of Myxococcus fulvus ANSM068 to biotransform aflatoxin B1. Int. J. Mol. Sci. 11, 4063–4079. doi: 10.3390/ijms11104063
Hariprasad, P., Vipin, A. V., Karuna, S., Raksha, R. K., and Venkateswaran, G. (2015). Natural aflatoxin uptake by sugarcane (i L.) and its persistence in jaggery. Environ. Sci. Pollut. Res. 22, 6246–6253. doi: 10.1007/s11356-014-3851-2
Hauser, F., Waadt, R., and Schroeder, J. I. (2011). Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21, R346–R355. doi: 10.1016/j.cub.2011.03.015
Hawkins, L. K., Mylroie, J. E., Oliveira, D. A., Smith, J. S., Ozkan, S., Windham, G. L., et al. (2015). Characterization of the maize chitinase genes and their effect on Aspergillus flavus and aflatoxin accumulation resistance. PLoS One 10:e0126185. doi: 10.1371/journal.pone.0126185
Hillmann, F., Novohradská, S., Mattern, D. J., Forberger, T., Heinekamp, T., Westermann, M., et al. (2015). Virulence determinants of the human pathogenic fungus Aspergillus fumigatus protect against soil amoeba predation. Environ. Microbiol. 17, 2858–2869. doi: 10.1111/1462-2920.12808
Holbrook, C. C., and Stalker, H. T. (2003). “Peanut breeding and genetic resources” in Plant breeding reviews. Vol. 22. ed. J. Janick (Wiley), 297–356.
Hong, E. J., Kim, N. K., Lee, D., Kim, W. G., and Lee, I. (2015). Overexpression of the laeA gene leads to increased production of cyclopiazonic acid in Aspergillus fumisynnematus. Fungal Biol. 119, 973–983. doi: 10.1016/j.funbio.2015.06.006
Hong, S.-Y., Roze, L. V., Wee, J., and Linz, J. E. (2013). Evidence that a transcription factor regulatory network coordinates oxidative stress response and secondary metabolism in Aspergilli. Microbiol. Open 2, 144–160. doi: 10.1002/mbo3.63
Horváth, E., Nagy, G., Turáni, M., Balogh, E., Papp, G., Pollak, E., et al. (2012). Effect of the fungal mycotoxin patulin on the chromatin structure of fission yeast Schizosaccharomyces pombe. J. Basic Microbiol. 52, 642–652. doi: 10.1002/jobm.201100515
Horváth, E., Papp, G., Belágyi, J., Gazdag, Z., Vágvölgyi, C., and Pesti, M. (2010). In vivo direct patulin-induced fluidization of the plasma membrane of fission yeast Schizosaccharomyces pombe. Food Chem. Toxicol. 48, 1898–1904. doi: 10.1016/j.fct.2010.04.031
Hua, S. S. T., Beck, J. J., Sarreal, S. B. L., and Gee, W. (2014). The major volatile compound 2-phenylethanol from the biocontrol yeast, Pichia anomala, inhibits growth and expression of aflatoxin biosynthetic genes of Aspergillus flavus. Mycotoxin Res. 30, 71–78. doi: 10.1007/s12550-014-0189-z
Huang, J.-Q., Jiang, H.-F., Zhou, Y.-Q., Lei, Y., Wang, S.-Y., and Liao, B.-S. (2009). Ethylene inhibited aflatoxin biosynthesis is due to oxidative stress alleviation and related to glutathione redox state changes in Aspergillus flavus. Int. J. Food Microbiol. 130, 17–21. doi: 10.1016/j.ijfoodmicro.2008.12.027
Hwang, C. A., and Draughon, F. A. (1994). Degradation of ochratoxin A by Acinetobacter calcoaceticus. J. Food Prot. 57, 410–414. doi: 10.4315/0362-028X-57.5.410
Ianiri, G., Idnurm, A., Wright, S. A. I., Durán-Patrón, R., Mannina, L., Ferracane, R., et al. (2013). Searching for genes responsible for patulin degradation in a biocontrol yeast provides insight into the basis for resistance to this mycotoxin. Appl. Environ. Microbiol. 79, 3101–3115. doi: 10.1128/AEM.03851-12
Iwahashi, Y., Hosoda, H., Park, J.-H., Lee, J.-H., Suzuki, Y., Kitagawa, E., et al. (2006). Mechanisms of patulin toxicity under conditions that inhibit yeast growth. J. Agric. Food Chem. 54, 1936–1942. doi: 10.1021/jf052264g
Jafari, S., Aghaei, S.-S., Afifi-Sabet, H., Shams-Ghahfarokhi, M., Jahanshiri, Z., Gholami-Shabani, M., et al. (2018). Exploration, antifungal and antiaflatoxigenic activity of halophilic bacteria communities from saline soils of Howze-Soltan playa in Iran. Extremophiles 22, 87–98. doi: 10.1007/s00792-017-0979-2
Janda, M., and Ruelland, E. (2014). Magical mystery tour: salicylic acid signalling. Environ. Exp. Bot. 114, 117–128. doi: 10.1016/j.envexpbot.2014.07.003
Janzen, D. H. (1977). Why fruits rot, seeds mold, and meat spoils. Am. Nat. 111, 691–713. doi: 10.1086/283200
Jayashree, T., and Subramanyam, C. (2000). Oxidative stress as a prerequisite for aflatoxin production by Aspergillus parasiticus. Free Radic. Biol. Med. 29, 981–985. doi: 10.1016/S0891-5849(00)00398-1
Jiang, T., Zhou, B., Luo, M., Abbas, H. K., Kemerait, R., Lee, R. D., et al. (2011). Expression analysis of stress-related genes in kernels of different maize (Zea mays L.) inbred lines with different resistance to aflatoxin contamination. Toxins 3, 538–550. doi: 10.3390/toxins3060538
Kachroo, A., He, Z., Patkar, R., Zhu, Q., Zhong, J., Li, D., et al. (2003). Induction of H2O2 in transgenic rice leads to cell death and enhanced resistance to both bacterial and fungal pathogens. Transgenic Res. 12, 577–586. doi: 10.1023/A:1025896513472
Kalunke, R. M., Tundo, S., Benedetti, M., Cervone, F., De Lorenzo, G., and D’Ovidio, R. (2015). An update on polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein that protects crop plants against pathogens. Front. Plant Sci. 6:146. doi: 10.3389/fpls.2015.00146
Kebede, H., Abbas, H., Fisher, D., and Bellaloui, N. (2012). Relationship between aflatoxin contamination and physiological responses of corn plants under drought and heat stress. Toxins 4, 1385–1403. doi: 10.3390/toxins4111385
Keller, N. P., Turner, G., and Bennett, J. W. (2005). Fungal secondary metabolism—from biochemistry to genomics. Nat. Rev. Microbiol. 3, 937–947. doi: 10.1038/nrmicro1286
Kelley, R. Y., Williams, W. P., Mylroie, J. E., Boykin, D. L., Harper, J. W., Windham, G. L., et al. (2012). Identification of maize genes associated with host plant resistance or susceptibility to Aspergillus flavus infection and aflatoxin accumulation. PLoS One 7:e36892. doi: 10.1371/journal.pone.0036892
Khaldi, N., Seifuddin, F. T., Turner, G., Haft, D., Nierman, W. C., Wolfe, K. H., et al. (2010). SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 47, 736–741. doi: 10.1016/j.fgb.2010.06.003
Kłosowski, G., Mikulski, D., Grajewski, J., and Błajet-Kosicka, A. (2010). The influence of raw material contamination with mycotoxins on alcoholic fermentation indicators. Bioresour. Technol. 101, 3147–3152. doi: 10.1016/j.biortech.2009.12.040
Knol, W., Bol, J., and Huis in’t Veld, J. H. J. (1990). “Detoxification of aflatoxin B1 in feeds by Rhizopus oryzae in solid state” in Food biotechnology. Vol. 2. eds. P. Zeuthen, J. C. Cheftel, C. Eriksson, T. R. Gormley, P. Linko, and K. Paulus (London: Elsevier Applied Science), 132–136.
Kuczuk, M. H., Benson, P. M., Heath, H., and Hayes, A. W. (1978). Evaluation of the mutagenic potential of mycotoxins using Salmonella typhimurium and Saccharomyces cerevisiae. Mutat. Res. 53, 11–20.
Kusumaningtyas, E., Widiastuti, R., and Maryam, R. (2006). Reduction of aflatoxin B1 in chicken feed by using Saccharomyces cerevisiae, Rhizopus oligosporus, and their combination. Mycopathologia 162, 307–311. doi: 10.1007/s11046-006-0047-4
Lamb, C., and Dixon, R. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 251–275. doi: 10.1146/annurev.arplant.48.1.251
Lappa, I. K., Mparampouti, S., Lanza, B., and Panagou, E. Z. (2018). Control of Aspergillus carbonarius in grape berries by Lactobacillus plantarum: a phenotypic and gene transcription study. Int. J. Food Microbiol. 275, 56–65. doi: 10.1016/j.ijfoodmicro.2018.04.001
Latgé, J.-P. (2001). The pathobiology of Aspergillus fumigatus. Trends Microbiol. 9, 382–389. doi: 10.1016/S0966-842X(01)02104-7
Lawton, M. A., and Lamb, C. J. (1987). Transcriptional activation of plant defense genes by fungal elicitor, wounding, and infection. Mol. Cell. Biol. 7, 335–341. doi: 10.1128/MCB.7.1.335
Lee, S. E., and Campbell, B. C. (2000). In vitro metabolism of aflatoxin B 1 by larvae of navel orangeworm, Amyelois transitella (Walker) (Insecta, Lepidoptera, Pyralidae) and codling moth, Cydia pomonella (L.) (Insecta, Lepidoptera, Tortricidae). Arch. Insect Biochem. Physiol. 45, 166–174. doi: 10.1002/1520-6327(200012)45:4<166::AID-ARCH4>3.0.CO;2-8
Line, J. E., and Brackett, R. E. (1995). Role of toxin concentration and second carbon source in microbial transformation of aflatoxin B1 by Flavobacterium aurantiacum. J. Food Prot. 58, 1042–1044. doi: 10.4315/0362-028X-58.9.1042
Liu, D.-L., Yao, D.-S., Liang, R., Ma, L., Cheng, W.-Q., and Gu, L.-Q. (1998). Detoxification of aflatoxin B1 by enzymes isolated from Armillariella tabescens. Food Chem. Toxicol. 36, 563–574. doi: 10.1016/S0278-6915(98)00017-9
Llorente, C., Betrán, F., Bockholt, A., and Fojt, F. (2004). Registration of Tx772 maize. Crop Sci. 44, 1036–1037. doi: 10.2135/cropsci2004.1036a
Logrieco, A., Bottalico, A., Mulé, G., Moretti, A., and Perrone, G. (2003). Epidemiology of toxigenic fungi and their associated mycotoxins for some Mediterranean crops. Eur. J. Plant Pathol. 109, 645–667. doi: 10.1023/A:1026033021542
Loussert, C., Schmitt, C., Prevost, M. C., Balloy, V., Fadel, E., Philippe, B., et al. (2010). In vivo biofilm composition of Aspergillus fumigatus. Cell. Microbiol. 12, 405–410. doi: 10.1111/j.1462-5822.2009.01409.x
Lumsden, R. D., Locke, J. C., Adkins, S. T., Walter, J. F., and Ridout, C. J. (1992). Isolation and localization of the antibiotic gliotoxin produced by Gliocladium virens from alginate prill in soil and soilless media. Phytopathology 82:230. doi: 10.1094/Phyto-82-230
Luo, M., Brown, R. L., Chen, Z. Y., Menkir, A., Yu, J., and Bhatnagar, D. (2011). Transcriptional profiles uncover Aspergillus flavus-induced resistance in maize kernels. Toxins 3, 766–786. doi: 10.3390/toxins3070766
Madhyastha, M. S., Marquardt, R. R., Masi, A., Borsa, J., and Frohlich, A. (1994). Comparison of toxicity of different mycotoxins to several species of bacteria and yeasts: use of Bacillus brevis in a disc diffusion assay. J. Food Prot. 57, 48–53. doi: 10.4315/0362-028X-57.1.48
Magbanua, Z. V., De Moraes, C. M., Brooks, T. D., Williams, W. P., and Luthe, D. S. (2007). Is catalase activity one of the factors associated with maize resistance to Aspergillus flavus? Mol. Plant-Microbe Interact. 20, 697–706. doi: 10.1094/MPMI-20-6-0697
Majumdar, R., Rajasekaran, K., Sickler, C., Lebar, M., Musungu, B. M., Fakhoury, A. M., et al. (2017). The pathogenesis-related maize seed (PRms) gene plays a role in resistance to Aspergillus flavus infection and aflatoxin contamination. Front. Plant Sci. 8:1758. doi: 10.3389/fpls.2017.01758
Malysheva, S., Arroyo-Manzanaresb, N., Cary, J., Ehrlich, K., van den Bussched, J., Vanhaecked, L., et al. (2014). Identification of novel metabolites from Aspergillus flavus by high resolution and multiple stage mass spectrometry. Food Addit. Contam. Part A 31, 111–120. doi: 10.1080/19440049.2013.859743
Mann, R., and Rehm, H. J. (1976). Degradation products from aflatoxin B1 by Corynebacterium rubrum, Aspergillus niger, Trichoderma viride and Mucor ambiguus. Eur. J. Appl. Microbiol. 2, 297–306. doi: 10.1007/BF01278613
Marín, S., Albareda, X., Ramos, A. J., and Sanchis, V. (2001). Impact of environment and interactions of Fusarium verticillioides and Fusarium proliferatum with Aspergillus parasiticus on fumonisin B 1 and aflatoxins on maize grain. J. Sci. Food Agric. 81, 1060–1068. doi: 10.1002/jsfa.894
Masoud, W., and Kaltoft, C. H. (2006). The effects of yeasts involved in the fermentation of Coffea arabica in East Africa on growth and ochratoxin A (OTA) production by Aspergillus ochraceus. Int. J. Food Microbiol. 106, 229–234. doi: 10.1016/j.ijfoodmicro.2005.06.015
Masoud, W., Poll, L., and Jakobsen, M. (2005). Influence of volatile compounds produced by yeasts predominant during processing of Coffea arabica in East Africa on growth and ochratoxin A (OTA) production by Aspergillus ochraceus. Yeast 22, 1133–1142. doi: 10.1002/yea.1304
McAllister, C. B., Garcia-Romera, I., Martin, J., Godeas, A., and Ocampo, J. A. (1995). Interaction between Aspergillus niger Van Tiegh. and Glomus mosseae (Nicol. & Gerd.) Gerd. & Trappe. New Phytol. 129, 309–316. doi: 10.1111/j.1469-8137.1995.tb04301.x
McLean, M. (1996). The phytotoxicity of selected mycotoxins on mature, germinating Zea mays embryos. Mycopathologia 132, 173–183. doi: 10.1007/BF01103984
McLean, M., Watt, M. P., Berjak, P., and Dutton, M. F. (1995). Aflatoxin B1 - its effects on an in vitro plant system. Food Addit. Contam. 12, 435–443. doi: 10.1080/02652039509374327
Mertz, D., Lee, D., Zuber, M., and Lillehoj, E. (1980). Uptake and metabolism of aflatoxin by Zea mays. J. Agric. Food Chem. 28, 963–966. doi: 10.1021/jf60231a003
Mesterházy, A. (2008). Resistance of corn to Fusarium ear rot and its relation to seedling resistance. J. Phytopathol. 103, 218–231. doi: 10.1111/j.1439-0434.1982.tb01746.x
Mideros, S. X., Windham, G. L., Williams, W. P., and Nelson, R. J. (2012). Tissue-specific components of resistance to Aspergillus ear rot of maize. Phytopathology 102, 787–793. doi: 10.1094/PHYTO-12-11-0355
Molnar, O., Schatzmayr, G., Fuchs, E., and Prillinger, H. (2004). Trichosporon mycotoxinivorans sp. nov., a new yeast species useful in biological detoxification of various mycotoxins. Syst. Appl. Microbiol. 27, 661–671. doi: 10.1078/0723202042369947
Mortensen, G. K., Strobel, B. W., and Hansen, H. C. B. (2006). Degradation of zearalenone and ochratoxin A in three Danish agricultural soils. Chemosphere 62, 1673–1680. doi: 10.1016/j.chemosphere.2005.06.037
Motomura, M., Toyomasu, T., Mizuno, K., and Shinozawa, T. (2003). Purification and characterization of an aflatoxin degradation enzyme from Pleurotus ostreatus. Microbiol. Res. 158, 237–242. doi: 10.1078/0944-5013-00199
Nakazato, M., Morozumi, S., Saito, K., Fujinuma, K., Nishima, T., and Kasai, N. (1990). Interconversion of aflatoxin B1 and aflatoxicol by several fungi. Appl. Environ. Microbiol. 56, 1465–1470. doi: 10.1128/AEM.56.5.1465-1470.1990
Narasaiah, K. V., Sashidhar, R. B., and Subramanyam, C. (2006). Biochemical analysis of oxidative stress in the production of aflatoxin and its precursor intermediates. Mycopathologia 162, 179–189. doi: 10.1007/s11046-006-0052-7
Netzker, T., Fischer, J., Weber, J., Mattern, D. J., König, C. C., Valiante, V., et al. (2015). Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front. Microbiol. 6:299. doi: 10.3389/fmicb.2015.00299
Ni, X., Wilson, J. P., David Buntin, G., Guo, B., Krakowsky, M. D., Dewey Lee, R., et al. (2011). Spatial patterns of aflatoxin levels in relation to ear-feeding insect damage in pre-harvest corn. Toxins 3, 920–931. doi: 10.3390/toxins3070920
Nicholson, M. J., Koulman, A., Monahan, B. J., Pritchard, B. L., Payne, G. A., and Scott, B. (2009). Identification of two aflatrem biosynthesis gene loci in Aspergillus flavus and metabolic engineering of Penicillium paxilli to elucidate their function. Appl. Environ. Microbiol. 75, 7469–7481. doi: 10.1128/AEM.02146-08
Nielsen, M. T., Klejnstrup, M. L., Rohlfs, M., Anyaogu, D. C., Nielsen, J. B., Gotfredsen, C. H., et al. (2013). Aspergillus nidulans synthesize insect juvenile hormones upon expression of a heterologous regulatory protein and in response to grazing by Drosophila melanogaster larvae. PLoS One 8:e73369. doi: 10.1371/journal.pone.0073369
Nigam, S. N., Waliyar, F., Aruna, R., Reddy, S. V., Kumar, P. L., Craufurd, P. Q., et al. (2009). Breeding peanut for resistance to aflatoxin contamination at ICRISAT. Peanut Sci. 36, 42–49. doi: 10.3146/AT07-008.1
Niu, G., Siegel, J., Schuler, M. A., and Berenbaum, M. R. (2009). Comparative toxicity of mycotoxins to navel orangeworm (Amyelois transitella) and corn earworm (Helicoverpa zea). J. Chem. Ecol. 35, 951–957. doi: 10.1007/s10886-009-9675-8
Niu, G., Wen, Z., Rupasinghe, S. G., Ren, S. Z., Berenbaum, M. R., and Schuler, M. A. (2008). Aflatoxin B1 detoxification by CYP321A1 in Helicoverpa zea. Arch. Insect Biochem. Physiol. 69, 32–45. doi: 10.1002/arch.20256
O’Brien, J., and Wright, G. D. (2011). An ecological perspective of microbial secondary metabolism. Curr. Opin. Biotechnol. 22, 552–558. doi: 10.1016/j.copbio.2011.03.010
O’Keeffe, G., Hammel, S., Owens, R. A., Keane, T. M., Fitzpatrick, D. A., Jones, G. W., et al. (2014). RNA-seq reveals the pan-transcriptomic impact of attenuating the gliotoxin self-protection mechanism in Aspergillus fumigatus. BMC Genomics 15:894. doi: 10.1186/1471-2164-15-894
Oro, L., Feliziani, E., Ciani, M., Romanazzi, G., and Comitini, F. (2018). Volatile organic compounds from Wickerhamomyces anomalus, Metschnikowia pulcherrima and Saccharomyces cerevisiae inhibit growth of decay causing fungi and control postharvest diseases of strawberries. Int. J. Food Microbiol. 265, 18–22. doi: 10.1016/j.ijfoodmicro.2017.10.027
Pal, S., St. Leger, R. J., and Wu, L. P. (2007). Fungal peptide destruxin A plays a specific role in suppressing the innate immune response in Drosophila melanogaster. J. Biol. Chem. 282, 8969–8977. doi: 10.1074/jbc.M605927200
Palencia, E. R., Hinton, D. M., and Bacon, C. W. (2010). The black Aspergillus species of maize and peanuts and their potential for mycotoxin production. Toxins 2, 399–416. doi: 10.3390/toxins2040399
Panahirad, S., Zaare-Nahandi, F., Mohammadi, N., Alizadeh-Salteh, S., and Safaie, N. (2014). Effects of salicylic acid on Aspergillus flavus infection and aflatoxin B1 accumulation in pistachio (Pistacia vera L.) fruit. J. Sci. Food Agric. 94, 1758–1763. doi: 10.1002/jsfa.6488
Papp, G., Horváth, E., Mike, N., Gazdag, Z., Belágyi, J., Gyöngyi, Z., et al. (2012). Regulation of patulin-induced oxidative stress processes in the fission yeast Schizosaccharomyces pombe. Food Chem. Toxicol. 50, 3792–3798. doi: 10.1016/j.fct.2012.07.001
Payne, G. A., and Widstrom, N. W. (1992). Aflatoxin in maize. Crit. Rev. Plant Sci. 10, 423–440. doi: 10.1080/07352689209382320
Pena, G. A., Pereyra, C. M., Armando, M. R., Chiacchiera, S. M., Magnoli, C. E., Orlando, J. L., et al. (2010). Aspergillus fumigatus toxicity and gliotoxin levels in feedstuff for domestic animals and pets in Argentina. Lett. Appl. Microbiol. 50, 77–81. doi: 10.1111/j.1472-765X.2009.02756.x
Peng, X.-L., Xu, W.-T., Wang, Y., Huang, K.-L., Liang, Z., Zhao, W., et al. (2010). Mycotoxin ochratoxin A-induced cell death and changes in oxidative metabolism of Arabidopsis thaliana. Plant Cell Rep. 29, 153–161. doi: 10.1007/s00299-009-0808-x
Petchkongkaew, A., Taillandier, P., Gasaluck, P., and Lebrihi, A. (2008). Isolation of Bacillus spp. from Thai fermented soybean (Thua-nao): screening for aflatoxin B1 and ochratoxin A detoxification. J. Appl. Microbiol. 104, 1495–1502. doi: 10.1111/j.1365-2672.2007.03700.x
Petersson, S., Hansen, M. W., Axberg, K., Hult, K., and Schnürer, J. (1998). Ochratoxin A accumulation in cultures of Penicillium verrucosum with the antagonistic yeast Pichia anomala and Saccharomyces cerevisiae. Mycol. Res. 102, 1003–1008. doi: 10.1017/S0953756297006047
Pfliegler, W. P., Pusztahelyi, T., and Pócsi, I. (2015). Mycotoxins - prevention and decontamination by yeasts. J. Basic Microbiol. 55, 805–818. doi: 10.1002/jobm.201400833
Prasad, K., Bhatnagar-Mathur, P., Waliyar, F., and Sharma, K. K. (2013). Overexpression of a chitinase gene in transgenic peanut confers enhanced resistance to major soil borne and foliar fungal pathogens. J. Plant Biochem. Biotechnol. 22, 222–233. doi: 10.1007/s13562-012-0155-9
Prost, I., Dhondt, S., Rothe, G., Vicente, J., Rodriguez, M. J., Kift, N., et al. (2005). Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol. 139, 1902–1913. doi: 10.1104/pp.105.066274
Rabajante, J. F., Tubay, J. M., Uehara, T., Morita, S., Ebert, D., and Yoshimura, J. (2015). Red queen dynamics in multi-host and multi-parasite interaction system. Sci. Report. 5:10004. doi: 10.1038/srep10004
Racovitza, A. (2009). The influence of various moulds on the multiplication of some mycophagous mites. J. Gen. Microbiol. 57, 379–381. doi: 10.1099/00221287-57-3-379
Rasmussen, T. B., Skindersoe, M. E., Bjarnsholt, T., Phipps, R. K., Christensen, K. B., Jensen, P. O., et al. (2005). Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology 151, 1325–1340. doi: 10.1099/mic.0.27715-0
Reeves, E. P., Messina, C. G. M., Doyle, S., and Kavanagh, K. (2004). Correlation between gliotoxin production and virulence of Aspergillus fumigatus in Galleria mellonella. Mycopathologia 158, 73–79. doi: 10.1023/B:MYCO.0000038434.55764.16
Regulin, A., and Kempken, F. (2018). Fungal genotype determines survival of Drosophila melanogaster when competing with Aspergillus nidulans. PLoS One 13:e0190543. doi: 10.1371/journal.pone.0190543
Reiss, J. (1978). “Effects of mycotoxins on higher plants, algae, fungi and bacteria” in Mycotoxic fungi, mycotoxins, mycotoxicoses: An encyclopaedic handbook volume 3. Mycotoxicoses of man and plants: Mycotoxin control and regulatory practices. eds. T. D. Wyllie and L. G. Morehouse (New York, NY, USA: Marcel Dekker Inc.), 119–143.
Reverberi, M., Punelli, M., Scala, V., Scarpari, M., Uva, P., Mentzen, W. I., et al. (2013). Genotypic and phenotypic versatility of Aspergillus flavus during maize exploitation. PLoS One 8:e68735. doi: 10.1371/journal.pone.0068735
Reverberi, M., Punelli, F., Scarpari, M., Camera, E., Zjalic, S., Ricelli, A., et al. (2010). Lipoperoxidation affects ochratoxin A biosynthesis in Aspergillus ochraceus and its interaction with wheat seeds. Appl. Microbiol. Biotechnol. 85, 1935–1946. doi: 10.1007/s00253-009-2220-4
Reverberi, M., Zjalic, S., Ricelli, A., Punelli, F., Camera, E., Fabbri, C., et al. (2008). Modulation of antioxidant defense in Aspergillus parasiticus is involved in aflatoxin biosynthesis: a role for the Ap yapA gene. Eukaryot. Cell 7, 988–1000. doi: 10.1128/EC.00228-07
Ricelli, A., Baruzzi, F., Solfrizzo, M., Morea, M., and Fanizzi, F. P. (2007). Biotransformation of patulin by Gluconobacter oxydans. Appl. Environ. Microbiol. 73, 785–792. doi: 10.1128/AEM.02032-06
Riley, R. T., and Goeger, D. E. (1992). “Cylopiazonic acid: speculation on its function in fungi” in Handbook of applied mycology. Vol. 5. Mycotoxins in ecological systems. eds. D. Bhatnagar, E. B. Lillehoj, and D. K. Arora (New York: Marcel Dekker), 385–402.
Rodriguez, H., Reveron, I., Doria, F., Costantini, A., De Las, R. B., Muňoz, R., et al. (2011). Degradation of ochratoxin A by Brevibacterium species. J. Agric. Food Chem. 59, 10755–10760. doi: 10.1021/jf203061p
Rohini, V. K., and Sankara Rao, K. (2001). Transformation of peanut (Arachis hypogaea L.) with tobacco chitinase gene: variable response of transformants to leaf spot disease. Plant Sci. 160, 889–898. doi: 10.1016/S0168-9452(00)00462-3
Rohlfs, M. (2005). Clash of kingdoms or why Drosophila larvae positively respond to fungal competitors. Front. Zool. 2:2. doi: 10.1186/1742-9994-2-2
Rohlfs, M. (2006). Genetic variation and the role of insect life history traits in the ability of Drosophila larvae to develop in the presence of a competing filamentous fungus. Evol. Ecol. 20, 271–289. doi: 10.1007/s10682-006-0002-3
Rohlfs, M., Albert, M., Keller, N. P., and Kempken, F. (2010). Secondary chemicals protect mould from fungivory. Biol. Lett. 3, 523–525. doi: 10.1098/rsbl.2007.0338
Rohlfs, M., and Churchill, A. C. L. (2011). Fungal secondary metabolites as modulators of interactions with insects and other arthropods. Fungal Genet. Biol. 48, 23–34. doi: 10.1016/j.fgb.2010.08.008
Rohlfs, M., and Kürschner, L. (2010). Saprophagous insect larvae, Drosophila melanogaster, profit from increased species richness in beneficial microbes. J. Appl. Entomol. 134, 667–671. doi: 10.1111/j.1439-0418.2009.01458.x
Rohlfs, M., and Obmann, B. (2009). Species-specific responses of dew fly larvae to mycotoxins. Mycotoxin Res. 25, 103–112. doi: 10.1007/s12550-009-0015-1
Rojas, C. M., Senthil-Kumar, M., Tzin, V., and Mysore, K. S. (2014). Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 5:17. doi: 10.3389/fpls.2014.00017
Romanens, E., Leischtfeld, S. F., Volland, A., Stevens, M. J. A., Krähenmann, U., Isele, D., et al. (2019). Screening of lactic acid bacteria and yeast strains to select adapted anti-fungal co-cultures for cocoa bean fermentation. Int. J. Food Microbiol. 290, 262–272. doi: 10.1016/j.ijfoodmicro.2018.10.001
Ruhland, M., Engelhardt, G., and Wallnoefer, P. R. (1996). Transformation of the mycotoxin ochratoxin A in plants. 2. Time course and rates of degradation and metabolite production in cell-suspension cultures of different crop plants. Mycopathologia 134, 97–102. doi: 10.1007/BF00436871
Ruhland, M., Engelhardt, G., and Wallnoefer, P. R. (1997). Transformation of the mycotoxin ochratoxin A in artificially contaminated vegetables and cereals. Mycotox. Res. 13, 54–60. doi: 10.1007/BF02945066
Salas, M. P., Reynoso, C. M., Céliz, G., Daz, M., and Resnik, S. L. (2012). Efficacy of flavanones obtained from citrus residues to prevent patulin contamination. Food Res. Int. 48, 930–934. doi: 10.1016/j.foodres.2012.02.003
Samson, R. A., Visagie, C. M., Houbraken, J., Hong, S. B., Hubka, V., Klaassen, C. H., et al. (2014). Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 78, 141–173. doi: 10.1016/j.simyco.2014.07.004
Sanders, T. H., Cole, R. J., Blankenship, P. D., and Dorner, J. W. (1993). Aflatoxin contamination of peanuts from plants drought stressed in pod or root zones 1. Peanut Sci. 20, 5–8. doi: 10.3146/i0095-3679-20-1-2
Savchenko, T., Walley, J. W., Chehab, E. W., Xiao, Y., Kaspi, R., Pye, M. F., et al. (2010). Arachidonic acid: an evolutionarily conserved signaling molecule modulates plant stress signaling networks. Plant Cell 22, 3193–3205. doi: 10.1105/tpc.110.073858
Scarpari, M., Punelli, M., Scala, V., Zaccaria, M., Nobili, C., Ludovici, M., et al. (2014). Lipids in Aspergillus flavus-maize interaction. Front. Microbiol. 5:74. doi: 10.3389/fmicb.2014.00074
Scharf, D. H., Brakhage, A. A., and Mukherjee, P. K. (2016). Gliotoxin – bane or boon? Environ. Microbiol. 18, 1096–1109. doi: 10.1111/1462-2920.13080
Scharf, D. H., Dworschak, J. D., Chankhamjon, P., Scherlach, K., Heinekamp, T., Brakhage, A. A., et al. (2018). Reconstitution of enzymatic carbon-sulfur bond formation reveals detoxification-like strategy in fungal toxin biosynthesis. ACS Chem. Biol. 13, 2508–2512. doi: 10.1021/acschembio.8b00413
Schrettl, M., Carberry, S., Kavanagh, K., Haas, H., Jones, G. W., O’Brien, J., et al. (2010). Self-protection against gliotoxin – a component of the gliotoxin biosynthetic cluster, GliT, completely protects Aspergillus fumigatus against exogenous gliotoxin. PLoS Pathog. 6:e1000952. doi: 10.1371/journal.ppat.1000952
Schroeckh, V., Scherlach, K., Nutzmann, H.-W., Shelest, E., Schmidt-Heck, W., Schuemann, J., et al. (2009). Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 106, 14558–14563. doi: 10.1073/pnas.0901870106
Scott, G. E., and Zummo, E. (1994). Kernel infection and aflatoxin production in maize by Aspergillus flavus relative to inoculation and harvest dates. Plant Dis. 78:123. doi: 10.1094/PD-78-0123
Seye, F., Faye, O., Ndiaye, M., Njie, E., and Marie Afoutou, J. (2010). Pathogenicity of the fungus, Aspergillus clavatus, isolated from the locust, Oedaleus senegalensis, against larvae of the mosquitoes Aedes aegypti, Anopheles gambiae and Culex quinquefasciatus. J. Insect Sci. 9, 1–7. doi: 10.1673/031.009.5301
Seyfferth, C., and Tsuda, K. (2014). Salicylic acid signal transduction: the initiation of biosynthesis, perception and transcriptional reprogramming. Front. Plant Sci. 5:697. doi: 10.3389/fpls.2014.00697
Shantha, T. (1999). Fungal degradation of aflatoxin B1. Nat. Toxins 7, 175–178. doi: 10.1002/1522-7189(200009/10)7:5<175::AID-NT63>3.0.CO;2-M
Shen, S. K., and Dowd, P. F. (1991). Detoxification spectrum of the cigarette beetle symbiont Symbiotaphrina kochii in culture. Entomol. Exp. Appl. 60, 51–59. doi: 10.1111/j.1570-7458.1991.tb01522.x
Sherratt, T. N., Wilkinson, D. M., and Bain, R. S. (2006). Why fruits rot, seeds mold and meat spoils: a reappraisal. Ecol. Model. 192, 618–626. doi: 10.1016/j.ecolmodel.2005.07.030
Shu, X., Livingston, D. P., Franks, R. G., Boston, R. S., Woloshuk, C. P., and Payne, G. A. (2015). Tissue-specific gene expression in maize seeds during colonization by Aspergillus flavus and Fusarium verticillioides. Mol. Plant Pathol. 16, 662–674. doi: 10.1111/mpp.12224
Siahmoshteh, F., Siciliano, I., Banani, H., Hamidi-Esfahani, Z., Razzaghi-Abyaneh, M., Gullino, M. L., et al. (2017). Efficacy of Bacillus subtilis and Bacillus amyloliquefaciens in the control of Aspergillus parasiticus growth and aflatoxins production on pistachio. Int. J. Food Microbiol. 254, 47–53. doi: 10.1016/j.ijfoodmicro.2017.05.011
Singh, H. R., Deka, M., and Das, S. (2015). Enhanced resistance to blister blight in transgenic tea (Camellia sinensis) by overexpression of class I chitinase gene from potato (Solanum tuberosum). Funct. Integr. Genom. 15, 461–480. doi: 10.1007/s10142-015-0436-1
Singh, P., Orbach, M. J., and Cotty, P. J. (2018). Aspergillus texensis: a novel aflatoxin producer with S morphology from the United States. Toxins 10:E513. doi: 10.3390/toxins10120513
Snigdha, M., Hariprasad, P., and Venkateswaran, G. (2015). Transport via xylem and accumulation of aflatoxin in seeds of groundnut plant. Chemosphere 119, 524–529. doi: 10.1016/j.chemosphere.2014.07.033
Sobolev, V., Arias, R., Goodman, K., Walk, T., Orner, V., Faustinelli, P., et al. (2018). Suppression of aflatoxin production in Aspergillus species by selected peanut (Arachis hypogaea) stilbenoids. J. Agric. Food Chem. 66, 118–126. doi: 10.1021/acs.jafc.7b04542
Spikes, S., Xu, R., Nguyen, C. K., Chamilos, G., Kontoyiannis, D. P., Jacobson, R. H., et al. (2008). Gliotoxin production in Aspergillus fumigatus contributes to host-specific differences in virulence. J. Infect. Dis. 197, 479–486. doi: 10.1086/525044
Spraker, J. E., Jewell, K., Roze, L. V., Scherf, J., Ndagano, D., Beaudry, R., et al. (2014). A volatile relationship: profiling an inter-kingdom dialogue between two plant pathogens, Ralstonia solanacearum and Aspergillus flavus. J. Chem. Ecol. 40, 502–513. doi: 10.1007/s10886-014-0432-2
Starr, J. M., Rushing, B. R., and Selim, M. I. (2017). Solvent-dependent transformation of aflatoxin B1 in soil. Mycotoxin Res. 33, 197–205. doi: 10.1007/s12550-017-0278-x
Stevanovic, J., Stanimirovic, Z., Radakovic, M., and Stojic, V. (2008). In vitro evaluation of the clastogenicity of fumagillin. Environ. Mol. Mutagen. 49, 594–601. doi: 10.1002/em.20409
Stoessl, A. (1981). “Structure and biogenetic relations: fungal nonhost-specific” in Toxins in plant disease. ed. R. D. Durbin (New York: Academic Press), 109–219.
Suwarno, W. B., Hannok, P., Palacios-Rojas, N., Windham, G., Crossa, J., and Pixley, K. V. (2019). Provitamin A carotenoids in grain reduce aflatoxin contamination of maize while combating vitamin A deficiency. Front. Plant Sci. 10:30. doi: 10.3389/fpls.2019.00030
Svahn, K. S., Göransson, U., Chryssanthou, E., Olsen, B., Sjölin, J., and Strömstedt, A. A. (2014). Induction of gliotoxin secretion in Aspergillus fumigatus by bacteria-associated molecules. PLoS One 9:e93685. doi: 10.1371/journal.pone.0093685
Teniola, O. D., Addo, P. A., Brost, I. M., Färber, P., Jany, K.-D., Alberts, J. F., et al. (2005). Degradation of aflatoxin B1 by cell-free extracts of Rhodococcus erythropolis and Mycobacterium fluoranthenivorans sp. nov. Int. J. Food Microbiol. 105, 111–117. doi: 10.1016/j.ijfoodmicro.2005.05.004
Terabayashi, Y., Sano, M., Yamane, N., Marui, J., Tamano, K., Sagara, J., et al. (2010). Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal Genet. Biol. 47, 953–961. doi: 10.1016/j.fgb.2010.08.014
Todd, A. T., Liu, E., Polvi, S. L., Pammett, R. T., and Page, J. E. (2010). A functional genomics screen identifies diverse transcription factors that regulate alkaloid biosynthesis in Nicotiana benthamiana. Plant J. 62, 589–600. doi: 10.1111/j.1365-313X.2010.04186.x
Ton, J., Van Pelt, J. A., Van Loon, L. C., and Pieterse, C. M. J. (2002). Differential effectiveness of salicylate-dependent and jasmonate/ethylene-dependent induced resistance in Arabidopsis. Mol. Plant-Microbe Interact. 15, 27–34. doi: 10.1094/MPMI.2002.15.1.27
Treutter, D. (2005). Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol. 7, 581–591. doi: 10.1055/s-2005-873009
Trienens, M., Kraaijeveld, K., and Wertheim, B. (2017). Defensive repertoire of Drosophila larvae in response to toxic fungi. Mol. Ecol. 26, 5043–5057. doi: 10.1111/mec.14254
Trienens, M., and Rohlfs, M. (2011). Experimental evolution of defense against a competitive mold confers reduced sensitivity to fungal toxins but no increased resistance in Drosophila larvae. BMC Evol. Biol. 11:206. doi: 10.1186/1471-2148-11-206
Tsitsigiannis, D. I., Kunze, S., Willis, D. K., Feussner, I., and Keller, N. P. (2005). Aspergillus infection inhibits the expression of peanut 13 S-HPODE-forming seed lipoxygenases. Mol. Plant-Microbe Interact. 18, 1081–1089. doi: 10.1094/mpmi-18-1081
Van Waeyenberghe, L., Baré, J., Pasmans, F., Claeys, M., Bert, W., Haesebrouck, F., et al. (2013). Interaction of Aspergillus fumigatus conidia with Acanthamoeba castellanii parallels macrophage-fungus interactions. Environ. Microbiol. Rep. 5, 819–824. doi: 10.1111/1758-2229.12082
Varga, J., Péteri, Z., Tábori, K., Téren, J., and Vágvölgyi, C. (2005). Degradation of ochratoxin A and other mycotoxins by Rhizopus isolates. Int. J. Food Microbiol. 99, 321–328. doi: 10.1016/j.ijfoodmicro.2004.10.034
Varga, J., Rigo, K., and Teren, J. (2000). Degradation of ochratoxin A by Aspergillus species. Int. J. Food Microbiol. 59, 1–7. doi: 10.1016/S0168-1605(00)00230-0
Varga, J., Tóth, B., Mesterházy, Á., Téren, J., and Fazekas, B. (2004). “Mycotoxigenic Fungi and Mycotoxins in foods and feeds in Hungary” in An overview on toxigenic fungi and mycotoxins in Europe. eds. A. Logrieco and A. Visconti (Dordrecht: Springer).
Veras, F. F., Correa, A. P. F., Welke, J. E., and Brandelli, A. (2016). Inhibition of mycotoxin-producing fungi by Bacillus strains isolated from fish intestines. Int. J. Food Microbiol. 238, 23–32. doi: 10.1016/j.ijfoodmicro.2016.08.035
Verheecke, C., Liboz, T., Anson, P., Zhu, Y., and Mathieu, F. (2015). Streptomyces – Aspergillus flavus interactions: impact on aflatoxin B accumulation. Food Add. Cont. Part A 32, 572–576. doi: 10.1080/19440049.2014.1003336
Verheecke, C., Liboz, T., Darriet, M., Sabaou, N., and Mathieu, F. (2014). In vitro interaction of actinomycetes isolates with Aspergillus flavus: impact on aflatoxins B1 and B2 production. Lett. Appl. Microbiol. 58, 597–603. doi: 10.1111/lam.12233
Verheecke, C., Liboz, T., and Mathieu, F. (2016). Microbial degradation of aflatoxin B1: current status and future advances. Int. J. Food Microbiol. 237, 1–9. doi: 10.1016/j.ijfoodmicro.2016.07.028
Wang, H., Lei, Y., Wan, L., Yan, L., Lv, J., Dai, X., et al. (2016a). Comparative transcript profiling of resistant and susceptible peanut post-harvest seeds in response to aflatoxin production by Aspergillus flavus. BMC Plant Biol. 16:54. doi: 10.1186/s12870-016-0738-z
Wang, H., Lei, Y., Yan, L., Wan, L., Ren, X., Chen, S., et al. (2016b). Functional genomic analysis of Aspergillus flavus interacting with resistant and susceptible peanut. Toxins 8:46. doi: 10.3390/toxins8020046
Wang, S., Park, Y.-S., Yang, Y., Borrego, E. J., Isakeit, T., Gao, X., et al. (2017). Seed-derived ethylene facilitates colonization but not aflatoxin production by Aspergillus flavus in maize. Front. Plant Sci. 8:415. doi: 10.3389/fpls.2017.00415
Wang, Y., Peng, X., Xu, W., Luo, Y., Zhao, W., Hao, J., et al. (2012). Transcript and protein profiling analysis of OTA-induced cell death reveals the regulation of the toxicity response process in Arabidopsis thaliana. J. Exp. Bot. 63, 2171–2187. doi: 10.1093/jxb/err447
Wang, Y., Wang, L., Wu, F., Liu, F., Wang, Q., Zhang, X., et al. (2018). A consensus ochratoxin A biosynthetic pathway: insights from the genome sequence of Aspergillus ochraceus and a comparative genomic analysis. Appl. Environ. Microbiol. 84:e01009-18. doi: 10.1128/AEM.01009-18
Wang, K., Yan, P., Ding, Q., Wu, Q., Wang, Z., and Peng, J. (2013). Diversity of culturable root-associated/endophytic bacteria and their chitinolytic and aflatoxin inhibition activity of peanut plant in China. World J. Microbiol. Biotechnol. 29, 1–10. doi: 10.1007/s11274-012-1135-x
Wang, T., Zhang, E., Chen, X., Li, L., and Liang, X. (2010). Identification of seed proteins associated with resistance to pre-harvested aflatoxin contamination in peanut (Arachis hypogaea L). BMC Plant Biol. 10:267. doi: 10.1186/1471-2229-10-267
Wasternack, C. (2007). Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100, 681–697. doi: 10.1093/aob/mcm079
Wasternack, C. (2014). Action of jasmonates in plant stress responses and development – applied aspects. Biotechnol. Adv. 32, 31–39. doi: 10.1016/j.biotechadv.2013.09.009
Wegst, W., and Lingens, F. (1983). Bacterial degradation of ochratoxin A. FEMS Microbiol. Lett. 17, 341–344. doi: 10.1111/j.1574-6968.1983.tb00433.x
Whitehead, M. P., Shieh, M. T., Cleveland, T. E., Cary, J. W., and Dean, R. A. (1995). Isolation and characterization of polygalacturonase genes (pecA and pecB) from Aspergillus flavus. Appl. Environ. Microbiol. 61, 3316–3322. doi: 10.1128/AEM.61.9.3316-3322.1995
Wicklow, D. T., Dowd, P. F., and Gloer, J. B. (1994). “Antiinsectan effects of Aspergillus metabolites” in The genus Aspergillus. Federation of European Microbiological Societies Symposium Series. Vol. 69. eds. K. A. Powell, A. Renwick, and J. F. Peberdy (Boston, MA: Springer).
Xin, M., Wang, X., Peng, H., Yao, Y., Xie, C., Han, Y., et al. (2012). Transcriptome comparison of susceptible and resistant wheat in response to powdery mildew infection. Genom. Proteom. Bioinforma 10, 94–106. doi: 10.1016/j.gpb.2012.05.002
Yang, M., Lu, L., Pang, J., Hu, Y., Guo, Q., Li, Z., et al. (2019). Biocontrol activity of volatile organic compounds from Streptomyces alboflavus TD-1 against Aspergillus flavus growth and aflatoxin production. J. Microbiol. 57, 396–404. doi: 10.1007/s12275-019-8517-9
Yin, Z., Wang, Y., Wu, F., Gu, X., Bian, Y., Wang, Y., et al. (2014). Quantitative trait locus mapping of resistance to Aspergillus flavus infection using a recombinant inbred line population in maize. Mol. Breed. 33, 39–49. doi: 10.1007/s11032-013-9932-y
Yu, J., Chang, P.-K., Ehrlich, K. C., Cary, J. W., Bhatnagar, D., Cleveland, T. E., et al. (2004). Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 70, 1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004
Zeng, R. S. L., Niu, G., Wen, Z., Schuler, M. A., and Berenbaum, M. R. (2006). Toxicity of aflatoxin B1 to Helicoverpa zea and bioactivation by cytochrome P450 monooxygenases. J. Chem. Ecol. 32, 1459–1471. doi: 10.1007/s10886-006-9062-7
Zeng, R. S., Wen, Z., Niu, G., and Berenbaum, M. R. (2013). Aflatoxin B1: toxicity, bioactivation and detoxification in the polyphagous caterpillar Trichoplusia ni. Insect Sci. 20, 318–328. doi: 10.1111/1744-7917.12007
Zeringue, H. J. Jr., Bhatnagar, D., and Cleveland, T. E. (1993). C15H24 volatile compounds unique to aflatoxigenic strains of Aspergillus flavus. Appl. Environ. Microbiol. 59, 2264–2270. doi: 10.1128/AEM.59.7.2264-2270.1993
Zhang, X., Cheng, Z., Ma, L., and Li, J. (2017). A study on accumulation of volatile organic compounds during ochratoxin A biosynthesis and characterization of the correlation in Aspergillus carbonarius isolated from grape and dried vine fruit. Food Chem. 277, 55–63. doi: 10.1016/j.foodchem.2016.12.061
Zhang, C. L., Zheng, B. Q., Lao, J. P., Mao, L. J., Chen, S. Y., Kubicek, C. P., et al. (2008). Clavatol and patulin formation as the antagonistic principle of Aspergillus clavatonanicus, an endophytic fungus of Taxus mairei. Appl. Microbiol. Biotechnol. 78, 833–840. doi: 10.1007/s00253-008-1371-z
Keywords: Aspergillus, aflatoxin, mycotoxin, plant, insect, microbe, soil, interaction
Citation: Pfliegler WP, Pócsi I, Győri Z and Pusztahelyi T (2020) The Aspergilli and Their Mycotoxins: Metabolic Interactions With Plants and the Soil Biota. Front. Microbiol. 10:2921. doi: 10.3389/fmicb.2019.02921
Received: 31 July 2019; Accepted: 04 December 2019;
Published: 12 February 2020.
Edited by:
Anindya Chanda, University of South Carolina, United StatesReviewed by:
Jeffrey William Cary, Agricultural Research Service (USDA), United StatesCopyright © 2020 Pfliegler, Pócsi, Győri and Pusztahelyi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tünde Pusztahelyi, cHVzenRhaGVseWlAYWdyLnVuaWRlYi5odQ==; cHVzenRhaGVseWlAeWFob28uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.