- Department of Microbiology, Faculty of Medicine, Kuwait University, Kuwait City, Kuwait
Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) has been reported to colonize and cause infections in animals as well as in humans. LA-MRSA isolates have only recently been identified in patients admitted to Kuwait hospitals. This study was conducted to characterize LA-MRSA isolates obtained from patients admitted to Kuwait hospitals. A total of 202 (7.1%) of 2,823 MRSA isolates obtained from clinical samples in 2016 and 2017 in 11 public Kuwait hospitals were assigned to lineages previously known to be associated with livestock. They were characterized using antibiogram, spa typing, and DNA microarray for the assignment of clonal complexes (CCs) and detection of antibiotic resistance and virulence determinants. Identification as putative LA-MRSA clones was based on the molecular definition inferred from DNA microarray. The LA-MRSA isolates consisted of CC96 (N = 31), CC97 (N = 169), and CC398 (N = 2). Isolates belonging to CC96 and CC398 were resistant to erythromycin and clindamycin mediated by erm(A) and erm(C). CC97 isolates were multiresistant to gentamicin, kanamycin, erythromycin, clindamycin, tetracycline, chloramphenicol, fusidic acid, trimethoprim, and ciprofloxacin and harbored aacA-aphD, erm(A), erm(C), msr(A), tet(K), cat, fusC, and dfrS1. In total, 35 spa types were identified among the isolates. CC398 isolates consisted of t899 and t034. Ten spa types were identified among CC96 with t11822 (N = 13) as the most prevalent. CC97 consisted of 26 spa types with most belonging to t267 (N = 73) followed by t359 (N = 39). CC398 was composed of CC398-MRSA-IV and CC398-MRSA-V (PVL+). CC96 belonged to CC96-MRSA-IV and CC96-MRSA-IV (PVL+) Central Asian caMRSA/WA MRSA-119. CC97 consisted of six strains including CC97-MRSA-V (fusC+), CC97-MRSA-IV WA MRSA-54/63, CC97-MRSA-V, CC97-MRSA-(V+fus), CC97-MRSA-(mec VI+fus), and CC97-MRSA (mecV/VT+fus+ccrAB2). Whereas CC96 and CC97 isolates were identified in 2016 and 2017, CC398 isolates were detected only in 2016. This study identified four LA-MRSA clones among MRSA isolated from patients in Kuwait hospitals in 2016–2017 with CC97-MRSA-V (fusC+) as the dominant clone. The presence of LA-MRSA with different genetic backgrounds suggests its independent acquisition from different sources.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is an opportunistic pathogen that colonizes humans as well as animals (Kluytmans et al., 1997). In humans, MRSA causes a wide range of infections including skin and soft tissue infections (SSTIs), and invasive infections such as pneumonia and endocarditis (Gordon and Lowy, 2008). Since it was first reported in the United Kingdom (Jevons, 1961), MRSA has evolved into three types, including health-care-associated MRSA (HA-MRSA), which was isolated from patients in health-care settings (Campanile et al., 2010); community-acquired MRSA (CA-MRSA), which was initially isolated from healthy individuals with no previous exposure to health-care facilities (Udo et al., 1993); and the new strains that were associated with livestock in the early 2000s (Armand-Lefevre et al., 2005; Voss et al., 2005), which were designated as livestock-associated MRSA (LA-MRSA). Initially, livestock-associated S. aureus isolates causes major problems in agriculture and are the leading cause of bovine mastitis (Fluit, 2012). In addition, MRSA isolates associated with livestock (called LA-MRSA) belonging to specific lineages such as ST398 have been reported to cause infections in animals and animal handlers (Armand-Lefevre et al., 2005; Voss et al., 2005). In recent years, these LA-MRSA lineages were able to break the species barrier to colonize and cause infections in humans with or without contact with livestock (Fitzgerald, 2012; Hetem et al., 2013).
Several molecular typing methods including staphylococcal protein A (spa) typing, multilocus sequence typing (MLST), pulsed-field gel electrophoresis, staphylococcal cassette chromosome mec (SCCmec) typing, DNA microarray, and whole genome sequencing (WGS) have been used to characterize and identify S. aureus lineages including LA-MRSA. Several LA-MRSA lineages have been identified including CC398 and CC9, which are predominant in Europe and Asia, respectively (Lo et al., 2012; Chuanga and Huang, 2015). Other clones associated with livestock include ST72, ST97, ST5, ST1, and ST433 (Lo et al., 2012; Chuanga and Huang, 2015).
The SCCmec genetic element is a mobile genetic element that confers methicillin resistance and resistance to other beta-lactam antibiotics to susceptible strains following its acquisition. The SCCmec element is variable in structural organization and the carriage of additional genetic structures such as transposons and insertion sequence elements. The high diversity in its structural organization and composition has formed the basis of SCCmec typing of MRSA strains (Hiramatsu et al., 2013). HA-MRSA isolates carry SCCmec types I, II, and III; CA-MRSA isolates carry SCCmec types IV, V, and VI; and LA-MRSA can carry any of the SCCmec types associated with CA-MRSA or HA-MRSA. For example, CC9 isolated from pigs in Asia were reported to harbor SCCmec III, SCCmec V, or SCCmec IX element (Cui et al., 2009; Neela et al., 2009). Similarly, CC398 isolates have been reported to carry different SCCmec types including SCCmec IV, SCCmec V, and SCCmec IX (van Loo et al., 2007).
Staphylococcus aureus is endowed with multiple virulence factors, such as toxins, enzymes, hemolysins, and leukocidins including Panton–Valentine leukocidin (PVL), that enhance the capacity of the bacterium to cause disease in humans. Genomic studies have revealed that LA-MRSA clones such as CC398 lack or rarely carry specific virulence factors including PVL and toxic shock syndrome toxin (TSST), which are considered major contributors in S. aureus infections (Jamrozy et al., 2012; Price et al., 2012). PVL is a pore-forming cytotoxin that plays a major role in S. aureus infections by targeting leukocytes (Maltezou and Giamarellou, 2006).
Although HA-MRSA and CA-MRSA have been widely reported in patients attending Kuwait hospitals (Boswihi et al., 2016), LA-MRSA has only recently been detected among MRSA isolates obtained from patients admitted to Kuwait hospitals. This paper reports the molecular characterization of LA-MRSA isolates obtained from patients in Kuwait hospitals in 2016–2017. LA-MRSA isolates selected for this study were identified based on their clonal complex (CC), which was determined by DNA microarray.
Materials and Methods
Ethical Approval
The study did not require ethical approval, because all the MRSA isolates were obtained as part of routine diagnostic microbiology investigations.
Methicillin-Resistant Staphylococcus aureus Strains
A total of 4726 MRSA isolates were obtained from clinical samples in 2016 (N = 2305) and 2017 (N = 2421) in 11 public Kuwait hospitals. MRSA isolates were obtained from clinical samples submitted to the clinical microbiology diagnostic laboratory in the 11 hospitals. The isolates were identified using biochemical tests and tube coagulase at the diagnostic microbiology laboratory. Once it was identified as MRSA in the diagnostic laboratories, the isolates were sent to MRSA Reference Laboratory located in the Department of Microbiology, Faculty of Medicine, Kuwait University, for molecular typing. The information accompanying the submitted MRSA isolates was sample ID, date of isolation, patient location, patient ID, and clinical source. The isolates were subcultured twice on brain–heart infusion agar (BHIA) plates to obtain pure colonies and incubated at 35°C for 18 h. Pure cultures were preserved in beads and stored at −20 and −80°C. They were recovered on BHIA and incubated at 35°C prior to testing.
Antibiotic Susceptibility Testing
Antibiotic susceptibility testing was performed using the disc diffusion method according to the Clinical Laboratory Standards Institute (CLSI) (Clinical and Laboratory Standard Institute, 2015). The susceptibility testing was performed with 13 antibiotics including benzyl penicillin (10 U), cefoxitin (30 μg), kanamycin (30 μg), mupirocin (200 and 5 μg), gentamicin (10 μg), erythromycin (15 μg), clindamycin (2 μg), chloramphenicol (30 μg), tetracycline (10 μg), trimethoprim (2.5 μg), fusidic acid (10 μg), rifampicin (5 μg), and ciprofloxacin (5 μg). Minimum inhibitory concentration (MIC) for cefoxitin, vancomycin, and teicoplanin were determined with Etest strips (bioMérieux, Marcy l’Etoile, France) according to the manufacturer’s instructions. S. aureus strains ATCC25923 and ATCC29213 were used as quality control strains for the disc diffusion and MIC determination, respectively. The D-test was used to test for inducible resistance to clindamycin. Susceptibility to fusidic acid was interpreted according to the British Society to Antimicrobial Chemotherapy (BSAC) (British Society to Antimicrobial Chemotherapy [BSAC], 2013).
DNA Isolation for Amplification
Three to five identical colonies of an overnight culture were picked using a sterile loop and suspended in a microfuge tube containing 50 μl of lysostaphin (150 μg/ml) and 10 μl of RNase (10 μg/ml) solution. The tube was incubated at 37°C in the heating block (ThermoMixer, Eppendorf, Hamburg, Germany) for 20 min. To each sample, 50 μl of proteinase K (20 mg/ml) and 150 μl of Tris buffer (0.1 M) were added and mixed by pipetting. The tube was then incubated at 60°C in the water bath (VWR Scientific Co., Shellware Lab, United States) for 10 min. The tube was transferred to a heating block at 95°C for 10 min in order to inactivate proteinase K activity. Finally, the tube was centrifuged, and the extracted DNA was stored at 4°C till used for PCR.
Spa Typing
Amplification of spa gene was performed using synthetic primers published previously (Harmsen et al., 2003) in a total volume of 25 μl. The PCR protocol consisted of an initial denaturation at 94°C for 4 min, followed by 25 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 1 min, and extension for 3 min at 72°C, and a final cycle with a single extension for 5 min at 72°C. Five microliters of the PCR product was analyzed by 1.5% agarose gel electrophoresis to confirm amplification. The PCR product was purified using MicroElute Cycle-Pure Spin kit (Omega Bio-Tek Inc., United States) according to the manufacturer’s protocol. The purified DNA was used for sequencing PCR. A total of 10 μl of the sequencing reaction mixture containing 2 μl of big dye terminator mix, 2 μl of 5× sequencing buffer, 3 μl of nuclease-free water, 1 μl of 3.2 pM primer (forward and reverse), and 2 μl of purified DNA were prepared. The sequencing PCR protocol consisted of initial denaturation for 1 min at 94°C, followed by 25 cycles of denaturation for 10 s at 96°C, annealing at 55°C for 5 s, and extension for 4 min at 66°C. Ultra-Sep Dye Terminator Removal kit (Omega Bio-Tek Inc., United States) was used to purify DNA. Purified DNA was sequenced in an automated 3130 × 1 genetic analyzer (Applied Biosystems, United States) in accordance with the manufacturer’s protocol. The sequence of spa gene was analyzed using the Ridom Staph Type software (Ridom GmbH, Wurzburg, Germany). The software detected the spa repeats and assigned the spa type for each isolate.
DNA Microarray
The S. aureus Genotyping kit 2.0 (Alere GmbH, Germany) was used for clonal assignment and the detection of genes encoding antibiotic resistance and virulence factors for MRSA isolates representing each spa type identified using the protocol provided by the manufacturer (Monecke et al., 2008).
Results
Detection of Livestock-Associated Methicillin-Resistant Staphylococcus aureus Isolates
A total of 4,726 MRSA isolates collected from patients in 11 hospitals in 2016–2017 were investigated by spa typing. From this, 2,823 isolates selected on the basis of spa types were analyzed by DNA microarray to assign the isolates into CCs. The 2,823 isolates included isolates from all clinical samples in all hospitals with the same spa types. The DNA microarray results identified 202 (7.1%) of the 2,823 isolates as LA-MRSA isolates. The isolates were defined as LA-MRSA solely on the basis of molecular rather than epidemiological definition. The LA-MRSA isolates that belonged to CC96 (31 isolates), CC97 (169 isolates), and CC398 (2 isolates) were identified as LA-MRSA. The CC96 isolates were obtained in 2016 (N = 21) and 2017 (N = 10). Eighty-three and 86 CC97 isolates were isolated in 2016 and 2017, respectively. CC398 isolates were only detected in 2016. Of the non-LA-MRSA isolates, the dominant CCs were CC5 (N = 796), CC22 (N = 397), CC8 (N = 304), CC1 (N = 239), CC6 (N = 223), CC30 (N = 179), CC80 (N = 178), and CC88 (N = 88). This report focuses on the characteristics of the LA-MRSA isolates.
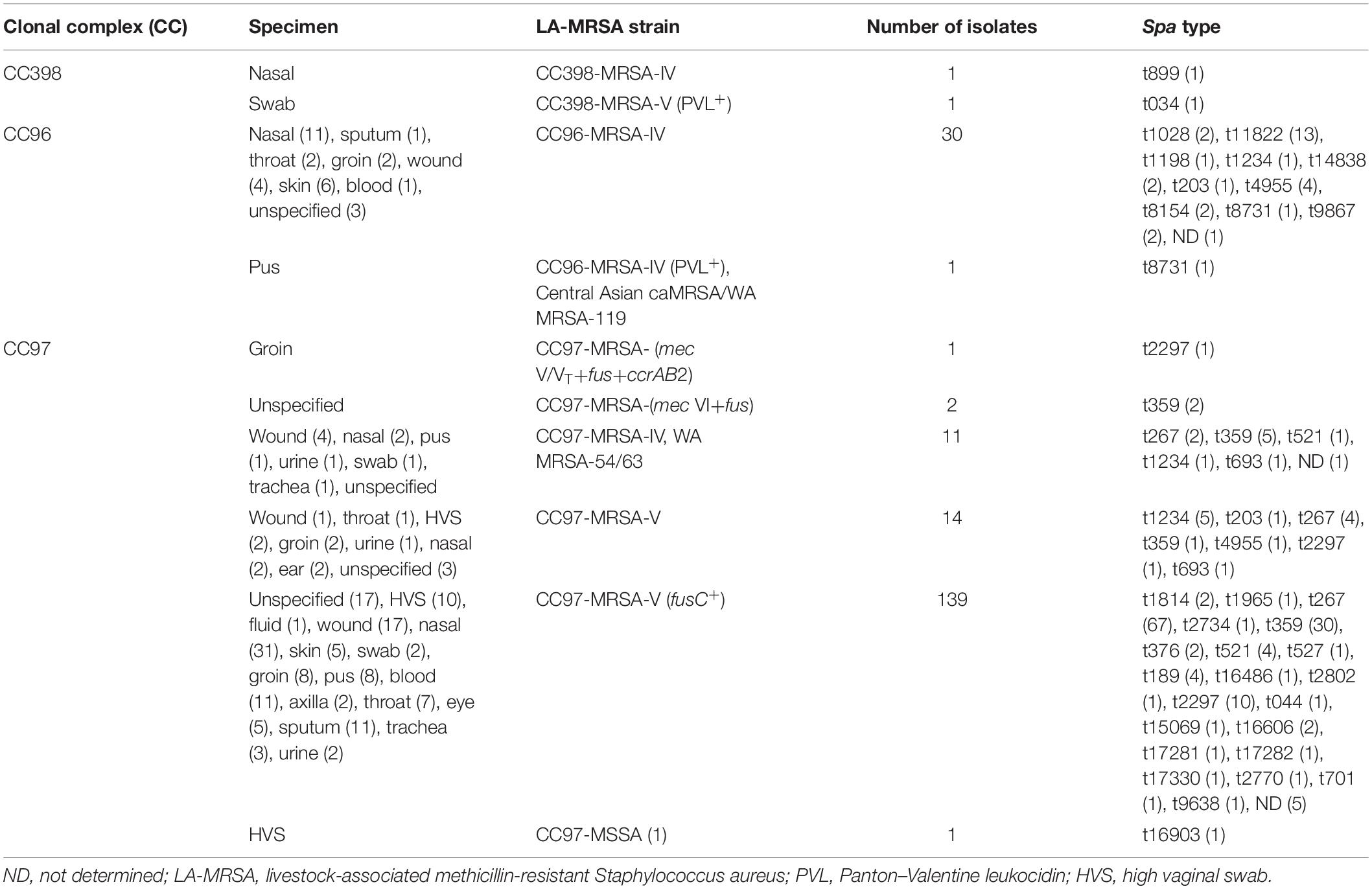
The characteristics of the LA-MRSA are summarized in Table 1. The results for each isolate is presented in the Supplementary Table S1. The LA-MRSA isolates were obtained from clinical samples of patients treated in Kuwait hospitals. The samples were collected from 137 inpatients and 50 outpatients. The locations of 17 patients were not specified. The isolates were obtained from nasal swab (47; 23.2%), skin and soft tissue (47; 23.2%), groin (13; 6.4%), high vaginal swab (HVS) (13; 6.4%), blood (12; 5.9%), sputum (12; 5.9%), throat (10; 4.9%), eye (5; 2.4%), trachea (4; 1.9%), urine (4; 1.9%), axilla (2; 0.9%), ear (2; 0.9%), and fluid (1; 0.5%). The source of 30 (14.8%) isolates was unspecified.
The CC398 belonged to two genotypes including PVL-negative CC398-MRSA-IV/t899 and PVL-positive CC398-MRSA-V/t034.
Thirty CC96 isolates belonged to CC96-MRSA-IV, and one isolate belonged to CC96-MRSA-IV (PVL+), also known as the Central Asian caMRSA/WA MRSA-119.
Seven different subtypes were identified among the CC97 isolates. These included CC97-MRSA-V (fusC+) (139 isolates), CC97-MRSA-V (14 isolates), CC97-MRSA-IV WA MRSA-54/63 (11 isolates), CC97-MRSA-(mec VI+fus) (2 isolates), CC97-MRSA-(mec V/VT+fus+ccrAB2) (1 isolate), CC97-MRSA-(V+fus) (1 isolate), and CC97-MSSA (1 isolate).
The LA-MRSA isolates belonged to 35 spa types. CC398 isolates consisted of two spa types, t899 and t034. Ten spa types were identified among CC96 isolates including t11822 (N = 13), t4955 (N = 4), t1028 (N = 2), t8154 (N = 2), t8731 (N = 2), t9867 (N = 2), t14838 (N = 2), t1234 (N = 1), t1198 (N = 1), and t203 (N = 1). Spa type was not determined (ND) for one isolate.
Twenty-six spa types were identified among CC97 isolates, with t267 (N = 67), t359 (N = 39), and t2297 (N = 12) as the common spa types in this lineage. The other spa types were detected less frequently. Spa types t203, t1234, and t4955 were identified in both CC96 and CC97 isolates (Table 1).
Antibiotic Susceptibility Testing and Antibiotic Resistance Genes
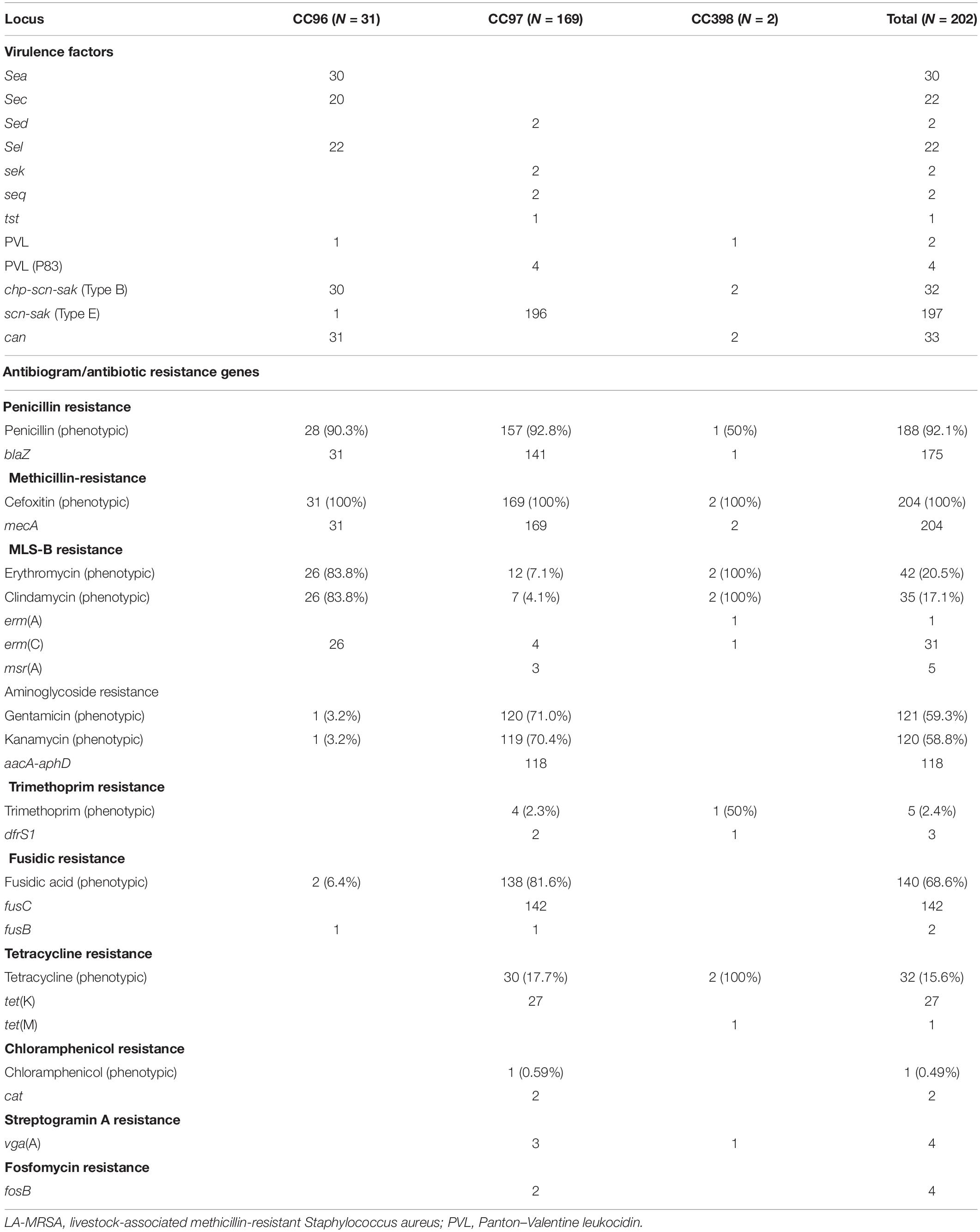
All LA-MRSA isolates were tested for susceptibility to antimicrobial agents. The results for the disk susceptibility testing and the genetic resistance determinants are summarized in Table 2. Antibiotic susceptibility testing showed that all LA-MRSA isolates were resistant to cefoxitin and were positive for mecA. A total of 187 (92.5%) isolates were resistant to penicillin mediated by blaZ. Sixteen CC97 isolates were phenotypically resistant to penicillin by disc diffusion method but lacked blaZ (Haveri et al., 2005).
Gentamicin resistance and kanamycin resistance were detected in 120 CC97 isolates, with only 118 isolates positive for aacA–aphD. One CC96 and two CC97 isolates were phenotypically resistant to gentamicin and kanamycin but lacked any of the aminoglycoside resistance genes in the DNA microarray panel (Table 2).
Macrolide–lincosamide–streptogramin-B (MLS-B) resistance was detected in 20.7% of the LA-MRSA. All CC96 isolates resistant to erythromycin and clindamycin carried erm(C). MLS-B resistance in the CC398 isolates was mediated by erm(A) and erm(C). Four CC97 erythromycin- and clindamycin-resistant isolates carried erm(C), three isolates were resistant only to erythromycin carried msr(A), and five isolates phenotypically resistant to erythromycin and clindamycin lacked any of the MLS-B resistance genes in the microarray panels.
Fusidic acid resistance genes fusB and fusC were identified in CC96 and CC97 isolates. Two CC96 isolates were phenotypically resistant to fusidic acid, but only one isolate carried fusB. fusB was also detected in one isolate belonging to CC97, whereas 142 isolates carried fusC (Table 2).
Tetracycline resistance was detected in 30 CC97 isolates with 27 isolates carrying tet(K). Tetracycline resistance gene, tet(M), was found in one CC398 isolate (Table 2). Trimethoprim resistance gene dfrS1 was detected in one CC398 isolate and in two of the four phenotypically resistant CC97 isolates. Chloramphenicol resistance mediated by cat was detected in a single CC97 isolate (Table 2).
vga(A), which confers resistance to streptogramin A compound (not tested phenotypically in this study), was found in three CC97 isolates and one isolate belonging to CC398. Fosfomycin was not tested in this study. However, two isolates belonging to CC97 carried fosB, which confers fosfomycin resistance (Table 2).
Virulence Encoding Genes
DNA microarray demonstrated that all LA-MRSA isolates carried genes for virulence factors including genes for adhesions, accessory gene regulators (agr), capsular polysaccharides (cap), and enzymes but varied in their carriage of genes for exotoxins (Table 2). All isolates were positive for agr and cap but differed in the types of agr and cap alleles. CC398 and CC97 carried agr 1 and cap 5, whereas CC96 isolates carried agr III and cap 8.
Of the 204 LA-MRSA isolates, 80 (39.6%) isolates carried enterotoxins. Two CC97 isolates carried sed, sek, and seq, whereas CC96 isolates variably carried sec, sea, and sel (Table 2). No enterotoxins were detected among CC398 isolates (Table 2).
Gene for TSST-1, tst, was detected in one CC97 isolate. PVL was only found in two CC96 isolates.
All isolates carried microbial surface components recognizing adhesive matrix molecules (MSCRAMMs). However, they varied in the carriage of collagen adhesion (cna). The cna gene was detected in CC96 and CC398 isolates but not in CC97 isolates (Table 2).
The immune evasion cluster (IEC) genes (scn, chp, and sak) were identified in all LA-MRSA isolates. CC398 carried scn, chp, and sak encoding genes (IEC type B). All CC97 and one CC96 isolates lacked chp and carried scn and sak (IEC type E).
Discussion
Livestock-associated methicillin-resistant S. aureus was initially isolated from livestock and later in isolates from humans who were in contact with livestock (Kinross et al., 2017). Subsequently, LA-MRSA isolated from individuals with no contact with livestock was reported in different places including Italy (Pan et al., 2009), Spain (Lozano et al., 2011), Australia (Monecke et al., 2011), and Saudi Arabia (Monecke et al., 2012). In this study, we characterized LA-MRSA obtained from human patients in Kuwait hospitals. The result of the study revealed that LA-MRSA constituted 7.1% of MRSA isolated from patients in hospitals in Kuwait in 2016–2017. The low prevalence of LA-MRSA reported in this study is similar to results reported in patients in Europe including the United Kingdom (Harrison et al., 2017); Luxembourg, Poland, and Norway (Kinross et al., 2017); and Asian countries such as China, Taiwan, Japan, and Malaysia (Chuanga and Huang, 2015). Human colonization with LA-MRSA is more common in areas with high density of livestock (Cuny et al., 2013; Kinross et al., 2017). Nevertheless, colonization with LA-MRSA isolates was also reported in people with no contact with livestock. A study published in 2013 in the United States reported CC398 MSSA as the dominant strain among detainees in the Dallas County Jail (David et al., 2013), which showed the transmission of these isolates in the absence of an animal source.
The LA-MRSA isolates belonged to three CCs, including CC96, CC97, and CC398, in this study. CC97 was found in 169 isolates, making CC97 the dominant LA-MRSA clone in Kuwait hospitals in 2016–2017. Prior to this report, CC97 isolates were reported to cause an outbreak in a neonatal intensive care unit (ICU) in a Kuwait hospital in 2007 (Udo et al., 2011; Udo and Al-Sweih, 2017) and was detected in four isolates in another hospital in 2010 (Boswihi et al., 2016). The increase in the prevalence of CC97 in recent years in Kuwait may suggest an increased transmission among patients in hospitals. CC97 has also been sporadically reported in patients in Saudi Arabia (Monecke et al., 2012), Spain (Lozano et al., 2011; Reynaga et al., 2018), and Australia (Monecke et al., 2011). Although CC97 is rarely reported in humans (Grundmann et al., 2010; Schaumburg et al., 2012), it is commonly isolated from cattle, and it is considered one of the most common causes of bovine mastitis (Smith et al., 2005; Aires-de-Sousa et al., 2007; Sung et al., 2008) and one of the most common in the Italian pig industry (Battisti et al., 2010; Feltrin et al., 2016). The increase in the prevalence of CC97 among human patients observed in this study is of concern, as it highlights the increasing transmission of LA-MRSA among human patients. The CC97 isolates consisted of 26 spa types and six genotypes, revealing the diversity of the isolates. CC97-MRSA-V (fusC+)-t267 was the dominant strain carrying few enterotoxin genes (sed, sek, and seq) and bovine PVL (P83), which is similar to the bovine CC97 isolates reported in Italy (Feltrin et al., 2016). Spa type t267 was also reported in isolates obtained from bovine in Portugal (Conceição et al., 2017), Switzerland (Boss et al., 2016), and Brazil (Rabello et al., 2007). The other spa types associated with CC97 isolates in this study (t359, t521, t2297) have also been reported in patients as well as animals in other studies (Albrecht et al., 2015; Feltrin et al., 2016), making them successful zoonotic subtypes.
Few reports have described the virulence profiles of CC97 isolates. A study in South Africa (Schmidt et al., 2017) showed that CC97 isolated from bovine and humans carried few enterotoxins genes including sec and sel. Similarly, the CC97 isolates in this study harbored sec and sel, suggesting that sec and sel may be common features of CC97 MRSA isolates.
CC97 isolates in this study were multiresistant to antibiotics, including resistance to gentamicin, kanamycin, erythromycin, clindamycin, and tetracycline. Similarly, multiresistant strains of CC97 were isolated from human patients in Saudi Arabia (Monecke et al., 2012) and in bovine in Spain (Gómez-Sanz et al., 2010). In contrast, non-multiresistant isolates of CC97 were obtained from dairy milk in China (Wang et al., 2018). We observed differences between the antibiotic susceptibility patterns and the carriage of antibiotic resistance genes in CC97 isolates obtained in this study. Penicillin resistance was detected in 157 of the CC97 isolates, but only 141 carried blaZ, which could be due to a lack of signal to blaZ, blaI, and blaR. A similar observation was reported by Williamson et al. (2014) in two CC398-t034 isolates in which blaZ could not be detected by DNA microarray, although they were resistant to penicillin and the resistance was confirmed by the detection of penicillinase with nitrocefin. A similar pattern was also observed in isolates resistant to erythromycin, gentamicin, kanamycin, trimethoprim, and tetracycline, in which the corresponding resistance genes could not be detected by the arrays. This could be due to the presence of other resistance mechanisms that are not in the DNA microarray panel or due to intrinsic resistance in these isolates. Intrinsic resistance was documented in S. aureus with penicillin-binding protein 2a (PBP2a), which renders the effectiveness of the beta-lactam antibiotics.
CC96 was the second most common LA-MRSA clone detected in this study. Isolates from this lineage were first detected in Kuwait in 2016 (21 isolates) and then in 2017 (10 isolates). CC96 MRSA isolates are rare in humans with only single isolates reported previously from Russia (Mendes et al., 2012) and Saudi Arabia (Senok et al., 2016; Mat Azis et al., 2017). However, ST96-MSSA is a common pathogen of rabbits, where it causes different infections (Mendes et al., 2012; Viana et al., 2015; Merz et al., 2016; Moreno-Grúa et al., 2018). An isolate of ST96-MRSA belonging to spa type t1190 was isolated from a rabbit meat sample that could not be characterized as either CA-MRSA or HA-MRSA (European Food Safety Authority, 2017), suggesting that a previously ST96-MSSA/t1190 had acquired the mecA determinant. Although the ST96-MRSA in this study belongs to different spa types, we argue that they are probably related to the rabbit ST96 lineage. Furthermore, the CC96-MRSA in this study belonged to agr III and cap8 and were resistant to erythromycin mediated by ermC similar to ST96-MSSA isolates isolated from rabbits (Merz et al., 2016). The lack of information on the epidemiology and the genetic characteristics of ST96-MRSA in the literature warrants further studies to describe the origin, prevalence, and molecular characteristics of the emerging CC96-MRSA.
Since its discovery in the Netherlands in the early 2000s, CC398 has become the most common LA-MRSA clone circulating in Europe (Butaye et al., 2016). This is the first report of CC398 in human patients in Kuwait hospitals and as far as we know in the Gulf Cooperation Council (GCC) countries. CC398 is the most prevalent lineage in pigs (Butaye et al., 2016; Feltrin et al., 2016), but it has also been reported in horses, poultry, cattle, and companion animals (Butaye et al., 2016). The CC398-MRSA was classified into two strains carrying SCCmec IV and V each and belonged to spa types t899 and t034, respectively. The CC398-IV-t899 isolate was PVL negative, similar to isolates obtained from animals in United Kingdom (Bortolami et al., 2017) and human patients in Spain (Lozano et al., 2012), Italy (Pan et al., 2009), and Denmark (Larsen et al., 2016). MRSA belonging to spa type t899 has been described as a hybrid LA-MRSA of CC9 and CC398 (Larsen et al., 2016; Sharma et al., 2018).
Apparently, CC398-MRSA of animal origin usually lacks enterotoxins and PVL (Feltrin et al., 2016), whereas the early-branching East Asian strain is positive for PVL (Yu et al., 2008). In this study, one CC398-MRSA-V isolate was positive for PVL, as has also been reported in human patients in China (Yu et al., 2008), Sweden (Welinder-Olsson et al., 2008), Finland (Salmenlinna et al., 2010), and New Zealand (Williamson et al., 2014), suggesting that our strain may belong to the human branch of CC398. The lack of enterotoxins in these two CC398-MRSA isolates is consistent with previous results suggesting that absence of enterotoxins may be a characteristic feature of isolates from this lineage (Lozano et al., 2012; Feltrin et al., 2016).
CC398 strains of animal origin usually lack the IEC genes that facilitate the colonization and invasion of MRSA in human hosts (Price et al., 2012). The two CC398 isolates detected in this study carried the IEC genes scn, chp, and sak (IEC type B). Similarly, recent studies by Cuny et al. (2015) and Pérez-Moreno et al. (2017) also reported CC398 isolates of human origin carrying the IEC genes. The presence of IEC genes in these isolates may explain the ability of these strains to jump from livestock and successfully adapt and colonize human beings. One of the CC398 isolates belonged to spa type t899.
The major limitation of this study is the lack of information on the patients’ travel history or contact with animals. Therefore, it is difficult to determine if these LA-MRSA strains were acquired by contact with livestock or by household members who are in contact with livestock. Nevertheless, the detection of these clones in human patients is significant because it shows their expansion beyond the usual livestock hosts, which may pose new problems for infection control.
Conclusion
In conclusion, this study described the characteristics of LA-MRSA strain belonging to CC96, CC97, and CC398 in patients in Kuwait hospitals. Genotyping showed that our isolates are diverse and belonged to different lineages including CC398, which are prevalent in Europe. This is the first report of CC398 LA-MRSA in Kuwait. The study also revealed that most of the isolates belonging to CC97 expressed resistance to multiple classes of antibiotics. The CC97 and CC398 MRSA isolates shared characteristics similar to those obtained from bovine and human patients in Europe. These observations suggest that LA-MRSA isolates were introduced to Kuwait via different routes. Further surveillance studies are required to monitor future transmission patterns of these isolates. Although LA-MRSA may have the same virulence potential and causes similar infections as S. aureus, identifying these isolates can inform on their origin, which will help in controlling the spread of MRSA in the clinical settings.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
SB, BM, BN, TV, and ST carried out the laboratory work. SB performed the data analysis. EU performed the experimental design. SB and EU carried out the manuscript writing and editing. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer SM declared a past co-authorship, with several of the authors, SB and EU, to the handling editor.
Acknowledgments
The authors are grateful to the technical staff in the MRS Reference Laboratory located in Microbiology Department, Faculty of Medicine, for their technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02912/full#supplementary-material
References
Aires-de-Sousa, M., Parente, C. E., Vieira-da-Motta, O., Bonna, I. C., Silva, D. A., and de Lencastre, H. (2007). Characterization of Staphylococcus aureus isolates from buffalo, bovine, ovine, and caprine milk samples collected in Rio de Janeiro State, Brazil. Appl. Environ. Microbiol. 73, 3845–3849. doi: 10.1128/aem.00019-07
Albrecht, V. S., Limbago, B. M., Moran, G. J., Krishnadasan, A., Gorwitz, R. J., McDougal, L. K., et al. (2015). Staphylococcus aureus colonization and strain type at various body sites among patients with a closed abscess and uninfected controls at U.S. emergency departments. J. Clin. Microbiol. 53, 3478–3484. doi: 10.1128/JCM.01371-15
Armand-Lefevre, L., Ruimy, R., and Andremont, A. (2005). Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg. Infect. Dis. 11, 711–714. doi: 10.3201/eid1105.040866
Battisti, A., Franco, A., Merialdi, G., Hasman, H., Iurescia, M., Lorenzetti, R., et al. (2010). Heterogeneity among methicillin-resistant Staphylococcus aureus from Italian pig finishing holdings. Vet. Microbiol. 142, 361–366. doi: 10.1016/j.vetmic.2009.10.008
Bortolami, A., Verin, R., Chantrey, J., Corrò, M., Ashpole, I., Lopez, J., et al. (2017). Characterization of livestock-associated methicillin-resistant Staphylococcus aureus CC398 and mecC-positive CC130 from Zoo Animals in the United Kingdom. Microb. Drug Resist. 23, 908–914. doi: 10.1089/mdr.2017.0161
Boss, R., Cosandey, A., Luini, M., Artursson, K., Bardiau, M., Breitenwieser, F., et al. (2016). Bovine Staphylococcus aureus: subtyping, evolution, and zoonotic transfer. J. Dairy Sci. 99, 515–528. doi: 10.3168/jds.2015-9589
Boswihi, S. S., Udo, E. E., and Al-Sweih, N. (2016). Shifts in the clonal distribution of methicillin-resistant Staphylococcus aureus in Kuwait Hospitals: 1992-2010. PLoS One 11:e0162744. doi: 10.1371/journal.pone.0162744
British Society to Antimicrobial Chemotherapy [BSAC], (2013). Available at: http://bsac.org.uk/susceptibility (accessed February 21, 2019).
Butaye, P., Argudín, M. A., and Smith, T. C. (2016). Livestock-associated MRSA and its current evolution. Curr. Clin. Microbiol. Rep. 3, 19–31. doi: 10.1007/s40588-016-0031-9
Campanile, F., Bongiorno, D., Borbone, S., and Stefani, S. (2010). Methicillin-resistant Staphylococcus aureus evolution- the multiple facets of an old pathogen. Eur. Infect. Dis. 4, 70–76.
Chuanga, Y. Y., and Huang, Y. C. (2015). Livestock-associated methicillin-resistant Staphylococcus aureus in Asia: an emerging issue? Int. J. Antimicrob. Agents 45, 334–340. doi: 10.1016/j.ijantimicag.2014.12.007
Clinical and Laboratory Standard Institute, (2015). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Second Informational Supplement M100-S25. Wayne, PA: CLSI.
Conceição, T., de Lencastre, H., and Aires-de-Sousa, M. (2017). Healthy bovines as reservoirs of major pathogenic lineages of Staphylococcus aureus in Portugal. Microb. Drug Resist. 23, 845–851. doi: 10.1089/mdr.2017.0074
Cui, S., Li, J., Hu, C., Jin, S., Li, F., Guo, L., et al. (2009). Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. J. Antimicrob. Chemother. 64, 680–683. doi: 10.1093/jac/dkp275
Cuny, C., Abdelbary, M., Layer, F., Werner, G., and Witte, W. (2015). Prevalence of the immune evasion gene cluster in Staphylococcus aureus CC398. Vet. Microbiol. 177, 219–223. doi: 10.1016/j.vetmic.2015.02.031
Cuny, C., Köck, R., and Witte, W. (2013). Livestock associated MRSA (LAMRSA) and its relevance for humans in Germany. Int. J. Med. Microbiol. 303, 331–337. doi: 10.1016/j.ijmm.2013.02.010
David, M. Z., Siegel, J., Lowy, F. D., Zychowski, D., Taylor, A., Lee, C. J., et al. (2013). Asymptomatic carriage of sequence type 398, type t571 methicillin-susceptible Staphylococcus aureus in an urban jail: a newly emerging, transmissible pathogenic strain. J. Clin. Microbiol. 51, 2443–2447. doi: 10.1128/JCM.01057-13
European Food Safety Authority, (2017). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 15:5077.
Feltrin, F., Alba, P., Kraushaar, B., Ianzano, A., Argudín, M. A., Di Matteo, P., et al. (2016). A livestock-associated, multidrug-resistant, methicillin-resistant Staphylococcus aureus clonal complex 97 lineage spreading in dairy cattle and pigs in Italy. Appl. Environ. Microbiol. 82, 816–821. doi: 10.1128/AEM.02854-15
Fitzgerald, J. R. (2012). Human origin for livestock-associated methicillin-resistant Staphylococcus aureus. mBio 3:e00082-12. doi: 10.1128/mBio.00082-12
Fluit, A. C. (2012). Livestock-associated Staphylococcus aureus. Clin. Microbiol. Infect. 18, 735–744. doi: 10.1111/j.1469-0691.2012.03846.x
Gómez-Sanz, E., Torres, C., Lozano, C., Fernández-Pérez, R., Aspiroz, C., Ruiz-Larrea, F., et al. (2010). Detection, molecular characterization, and clonal diversity of methicillin-resistant Staphylococcus aureus CC398 and CC97 in Spanish slaughter pigs of different age groups. Foodborne Pathog. Dis. 7, 1269–1277. doi: 10.1089/fpd.2010.0610
Gordon, R. J., and Lowy, F. D. (2008). Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46, S350–S359. doi: 10.1086/533591
Grundmann, H., Aanensen, D. M., van den Wijngaard, C. C., Spratt, B. G., Harmsen, D., Friedrich, A. W., et al. (2010). Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 7:e1000215. doi: 10.1371/journal.pmed.1000215
Harmsen, D., Claus, H., Witte, W., Rothgänger, J., Claus, H., Turnwald, D., et al. (2003). Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41, 5442–5448. doi: 10.1128/jcm.41.12.5442-5448.2003
Harrison, E. M., Coll, F., Toleman, M. S., Blane, B., Brown, N. M., Török, M. E., et al. (2017). Genomic surveillance reveals low prevalence of livestock-associated methicillin-resistant Staphylococcus aureus in the East of England. Sci. Rep. 7:7406. doi: 10.1038/s41598-017-07662-2
Haveri, M., Suominen, S., Rantala, L., Honkanen-Buzalski, T., and Pyörälä, S. (2005). Comparison of phenotypic and genotypic detection of penicillin G resistance of Staphylococcus aureus isolated from bovine intramammary infection. Vet. Microbiol. 106, 97–102. doi: 10.1016/j.vetmic.2004.12.015
Hetem, D. J., Bootsma, M. C., Troelstra, A., and Bonten, M. J. (2013). Transmissibility of livestock-associated methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 19, 1797–1802. doi: 10.3201/eid1911.121085
Hiramatsu, K., Ito, T., Tsubakishhita, S., Sasaki, T., Takeuchi, F., Morimoto, Y., et al. (2013). Genomic basis for methicillin resistance in Staphylococcus aureus. Infect. Chemother. 45, 117–136. doi: 10.3947/ic.2013.45.2.117
Jamrozy, D. M., Fielder, M. D., Butaye, P., and Coldham, N. G. (2012). Comparative genotypic and phenotypic characterisation of methicillin-resistant Staphylococcus aureus ST398 isolated from animals and humans. PLoS One 7:e40458. doi: 10.1371/journal.pone.0040458
Jevons, M. (1961). Celbenin-resistant staphylococci. Br. Med. J. 1, 124–125. doi: 10.1136/bmj.1.5219.124-a
Kinross, P., Petersen, A., Skov, R., Van Hauwermeiren, E., Pantosti, A., Laurent, F., et al. (2017). Livestock-associated meticillin-resistant Staphylococcus aureus (MRSA) among human MRSA isolates, European Union/European Economic Area countries, 2013. Euro Surveill. 22:16-00696. doi: 10.2807/1560-7917.ES.2017.22.44.16-00696
Kluytmans, J., van Belkum, A., and Verbrugh, H. (1997). Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10, 505–520. doi: 10.1128/cmr.10.3.505
Larsen, J., Stegger, M., Andersen, P. S., Petersen, A., Larsen, A. R., Westh, H., et al. (2016). Evidence for human adaptation and foodborne transmission of livestock-associated methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 63, 1349–1352.
Lo, Y. P., Wan, M. T., Chen, M. M., Su, H. Y., Lauderdale, T. L., and Chou, C. C. (2012). Molecular characterization and clonal genetic diversity of methicillin- resistant Staphylococcus aureus of pig origin in Taiwan. Comp. Immunol. Microbiol. Infect. 35, 513–521. doi: 10.1016/j.cimid.2012.05.001
Lozano, C., Gómez-Sanz, E., Benito, D., Aspiroz, C., Zarazaga, M., and Torres, C. (2011). Staphylococcus aureus nasal carriage, virulence traits, antibiotic resistance mechanisms, and genetic lineages in healthy humans in Spain, with detection of CC398 and CC97 strains. Int. J. Med. Microbiol. 301, 500–505. doi: 10.1016/j.ijmm.2011.02.004
Lozano, C., Rezusta, A., Gómez, P., Gómez-Sanz, E., Báez, N., Martin-Saco, G., et al. (2012). High prevalence of spa types associated with the clonal lineage CC398 among tetracycline-resistant methicillin-resistant Staphylococcus aureus strains in a Spanish hospital. J. Antimicrob. Chemother. 67, 330–334. doi: 10.1093/jac/dkr497
Maltezou, H. C., and Giamarellou, H. (2006). Community acquired methicillin resistant Staphylococcus aureus infections. Int. J. Antimicrob. Agents 27, 87–96.
Mat Azis, N., Pung, H. P., Abdul Rachman, A. R., Amin Nordin, S., Sarchio, S. N. E., Suhaili, Z., et al. (2017). A persistent antimicrobial resistance pattern and limited methicillin-resistance-associated genotype in a short-term Staphylococcus aureus carriage isolated from a student population. J. Infect. Public Health 10, 156–164. doi: 10.1016/j.jiph.2016.02.013
Mendes, R. E., Deshpande, L. M., Smyth, D. S., Shopsin, B., Farrell, D. J., and Jones, R. N. (2012). Characterization of methicillin-resistant Staphylococcus aureus strains recovered from a phase IV clinical trial for linezolid versus vancomycin for treatment of nosocomial pneumonia. J. Clin. Microbiol. 50, 3694–3702. doi: 10.1128/JCM.02024-12
Merz, A., Stephan, R., and Johler, S. (2016). Genotyping and DNA microarray based characterization of Staphylococcus aureus isolates from rabbit carcasses. Meat Sci. 112, 86–89. doi: 10.1016/j.meatsci.2015.11.002
Monecke, S., Coombs, G., Shore, A. C., Coleman, D. C., Akpaka, P., Borg, M., et al. (2011). A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936. doi: 10.1371/journal.pone.0017936
Monecke, S., Jatzwauk, L., Weber, S., Slickers, P., and Ehricht, R. (2008). DNA microarray-based genotyping of methicillin-resistant Staphylococcus aureus strains from Eastern Saxony. Clin. Microbiol. Infect. 14, 534–545. doi: 10.1111/j.1469-0691.2008.01986.x
Monecke, S., Skakni, L., Hasan, R., Ruppelt, A., Ghazal, S. S., Hakawi, A., et al. (2012). Characterisation of MRSA strains isolated from patients in a hospital in Riyadh, Kingdom of Saudi Arabia. BMC Microbiol. 12:146. doi: 10.1186/1471-2180-12-146
Moreno-Grúa, E., Pérez-Fuentes, S., Muñoz-Silvestre, A., Viana, D., Fernández-Ros, A. B., Sanz-Tejero, C., et al. (2018). Characterization of livestock-associated methicillin-resistant Staphylococcus aureus isolates obtained from commercial Rabbitries located in the Iberian Peninsula. Front. Microbiol. 9:1812. doi: 10.3389/fmicb.2018.01812
Neela, V., Arif, M. Z., Shamsudin, M. N., van Belkum, A., Khoon, L. Y., and Ghaznavi, E. (2009). Prevalence of ST9 methicillin-resistant Staphylococcus aureus among pigs and pig handlers in Malaysia. J. Clin. Microbiol. 47, 4138–4140. doi: 10.1128/JCM.01363-09
Pan, A., Battisti, A., Zoncada, A., Bernieri, F., Boldini, M., Franco, A., et al. (2009). Community-acquired methicillin-resistant Staphylococcus aureus ST398 infection, Italy. Emerg. Infect. Dis. 15, 845–847.
Pérez-Moreno, M. O., Centelles-Serrano, M. J., Nogales-López, J., Domenech-Spanedda, M. F., Lozano, C., and Torres, C. (2017). Unusual presence of the immune evasion gene cluster in livestock-associated MRSA of lineage CC398 causing peridural and psoas abscesses in a poultry farmer. Enferm. Infecc. Microbiol. Clin. 35, 651–654. doi: 10.1016/j.eimc.2016.07.008
Price, L. B., Stegger, M., Hasman, H., Aziz, M., Larsen, J., Andersen, P. S., et al. (2012). Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3:e00305-11. doi: 10.1128/mBio.00305-11
Rabello, R. F., Moreira, B. M., Lopes, R. M., Teixeira, L. M., Riley, L. W., and Castro, A. C. (2007). Multilocus sequence typing of Staphylococcus aureus isolates recovered from cows with mastitis in Brazilian dairy herds. J. Med. Microbiol. 56, 1505–1511. doi: 10.1099/jmm.0.47357-0
Reynaga, E., Torres, C., Garcia-Nuñez, M., Navarro, M., Vilamala, A., Puigoriol, E., et al. (2018). Prevalence of MRSA ST398 carriage in nursing home residents in an area of Spain with a high density of pig farming. Infect. Control Hosp. Epidemiol. 39, 90–93. doi: 10.1017/ice.2017.244
Salmenlinna, S., Lyytikainen, O., Vainio, A., Myllyniemi, A. L., Raulo, S., Kanerva, M., et al. (2010). Human cases of methicillin-resistant Staphylococcus aureus CC398, Finland. Emerg. Infect. Dis. 16, 1626–1629. doi: 10.3201/eid1610.091571
Schaumburg, F., Köck, R., Mellmann, A., Richter, L., Hasenberg, F., Kriegeskorte, A., et al. (2012). Population dynamics among methicillin resistant Staphylococcus aureus in Germany during a 6-year period. J. Clin. Microbiol. 50, 3186–3192. doi: 10.1128/JCM.01174-12
Schmidt, T., Kock, M. M., and Ehlers, M. M. (2017). Molecular characterization of Staphylococcus aureus isolated from bovine mastitis and close human contacts in South African dairy herds: genetic diversity and inter-species host transmission. Front. Microbiol. 8:511. doi: 10.3389/fmicb.2017.00511
Senok, A., Ehricht, R., Monecke, S., Al-Saedan, R., and Somily, A. (2016). Molecular characterization of methicillin-resistant Staphylococcus aureus in nosocomial infections in a tertiary-care facility: emergence of new clonal complexes in Saudi Arabia. New Microbes New Infect. 14, 13–18. doi: 10.1016/j.nmni.2016.07.009
Sharma, M., AbuOun, M., Nunez-Garcia, J., Rogers, J., Welchman, D., Teale, C., et al. (2018). MRSA spa type t899 from food animals in the UK. Vet. Rec. 182, 697–698. doi: 10.1136/vr.k2576
Smith, E. M., Green, L. E., Medley, G. F., Bird, H. E., Fox, L. K., Schukken, Y. H., et al. (2005). Multilocus sequence typing of intercontinental bovine Staphylococcus aureus isolates. J. Clin. Microbiol. 43, 4737–4743. doi: 10.1128/jcm.43.9.4737-4743.2005
Sung, J. M., Lloyd, D. H., and Lindsay, J. A. (2008). Staphylococcus aureus host specificity: comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology 154, 1949–1959. doi: 10.1099/mic.0.2007/015289-0
Udo, E. E., and Al-Sweih, N. (2017). Dominance of community-associated methicillin-resistant Staphylococcus aureus clones in a maternity hospital. PLoS One 12:e0179563. doi: 10.1371/journal.pone.0179563
Udo, E. E., Aly, N. Y., Sarkhoo, E., Al-Sawan, R., and Al-Asar, A. S. (2011). Detection and characterization of an ST97-SCCmec-V community-associated methicillin-resistant Staphylococcus aureus clone in a neonatal intensive care unit and special care baby unit. J. Med. Microbiol. 60, 600–604. doi: 10.1099/jmm.0.028381-0
Udo, E. E., Pearman, J. W., and Grubb, W. B. (1993). Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J. Hosp. Infect. 25, 97–108. doi: 10.1016/0195-6701(93)90100-e
van Loo, I., Huijsdens, X., Tiemersma, E., de Neeling, A. J., van de Sande-Bruinsma, N., Beaujean, D., et al. (2007). Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg. Infect. Dis. 13, 1834–1839. doi: 10.3201/eid1312.070384
Viana, D., Selva, L., Penadés, M., and Corpa, J. M. (2015). Screening of virulence genes in Staphylococcus aureus isolates from rabbits. World Rabbit Sci. 23, 185–195.
Voss, A., Loeffen, F., Bakker, J., Klaassen, C., and Wulf, M. (2005). Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11, 1965–1966.
Wang, W., Lin, X., Jiang, T., Peng, Z., Xu, J., Yi, L., et al. (2018). Prevalence and characterization of Staphylococcus aureus cultured from raw milk taken from dairy cows with mastitis in Beijing, China. Front. Microbiol. 9:1123. doi: 10.3389/fmicb.2018.01123
Welinder-Olsson, C., Florén-Johansson, K., Larsson, L., Oberg, S., Karlsson, L., and Ahrén, C. (2008). Infection with Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus t034. Emerg. Infect. Dis. 14, 1271–1272. doi: 10.3201/eid1408.071427
Williamson, D. A., Bakker, S., Coombs, G. W., Tan, H. L., Monecke, S., and Heffernan, H. (2014). Emergence and molecular characterization of clonal complex 398 (CC398) methicillin-resistant Staphylococcus aureus (MRSA) in New Zealand. J. Antimicrob. Chemother. 69, 1428–1430. doi: 10.1093/jac/dkt499
Keywords: livestock-associated methicillin-resistant Staphylococcus aureus, molecular typing, DNA microarray, antibiotic resistance genes, virulence factors
Citation: Boswihi SS, Udo EE, Mathew B, Noronha B, Verghese T and Tappa SB (2020) Livestock-Associated Methicillin-Resistant Staphylococcus aureus in Patients Admitted to Kuwait Hospitals in 2016–2017. Front. Microbiol. 10:2912. doi: 10.3389/fmicb.2019.02912
Received: 05 March 2019; Accepted: 03 December 2019;
Published: 08 January 2020.
Edited by:
Ziad Daoud, University of Balamand, LebanonReviewed by:
Phil Giffard, Charles Darwin University, AustraliaStefan Monecke, Leibniz Institute of Photonic Technology (IPHT), Germany
Copyright © 2020 Boswihi, Udo, Mathew, Noronha, Verghese and Tappa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edet E. Udo, ZWRldEBoc2MuZWR1Lmt3
 Samar S. Boswihi
Samar S. Boswihi Edet E. Udo
Edet E. Udo Bindu Mathew
Bindu Mathew