- 1School of Life Sciences, University of Technology Sydney, Ultimo, NSW, Australia
- 2Climate Change Cluster, University of Technology Sydney, Ultimo, NSW, Australia
- 3Centre for Shellfish Research, Vancouver Island University, Nanaimo, BC, Canada
The Vibrio genus is notable for including several pathogens of marine animals and humans, yet characterization of Vibrio diversity using routine 16S rRNA sequencing methods is often constrained by poor resolution beyond the genus level. Here, a new high throughput sequencing approach targeting the heat shock protein (hsp60) as a phylogenetic marker was developed to more precisely discriminate members of the Vibrio genus in environmental samples. The utility of this new assay was tested using mock communities constructed from known dilutions of Vibrio isolates. Relative to standard and Vibrio-specific 16S rRNA sequencing assays, the hsp60 assay delivered high levels of fidelity with the mock community composition at the species level, including discrimination of species within the Vibrio harveyi clade. This assay was subsequently applied to characterize Vibrio community composition in seawater and delivered substantially improved taxonomic resolution of Vibrio species compared to 16S rRNA analysis. Finally, this assay was applied to examine patterns in the Vibrio community within oysters during a Pacific oyster mortality event. In these oysters, the hsp60 assay identified species-level Vibrio community shifts prior to disease onset, pinpointing V. harveyi as a putative pathogen. Given that shifts in the Vibrio community can precede, cause, and follow disease onset in numerous marine organisms, there is a need for an accurate high throughput assay for defining Vibrio community composition in natural samples. This Vibrio-centric hsp60 sequencing assay offers the potential for precise high throughput characterization of Vibrio diversity, providing an enhanced platform for dissecting Vibrio dynamics in the environment.
Introduction
The Vibrio genus is a group of Gram-negative marine bacteria that are ubiquitous in a number of different aquatic environments, including estuaries, the open ocean, and the deep-sea (Simidu and Tsukamoto, 1985; Thompson et al., 2004; Siboni et al., 2016). The Vibrio genus comprises high levels of metabolic diversity and can play important ecological roles in marine biogeochemical cycling (Urdaci et al., 1988; Svitil et al., 1997; Chimetto et al., 2008; Hunt et al., 2008; Grimes et al., 2009), but are most recognized for their often ecologically important relationships with a wide range of aquatic organisms including bivalves, cephalopods, polychaetes, fish, corals, and algae (Lee and Ruby, 1994; Grisez et al., 1997; Hood and Winter, 1997; Raguenes et al., 1997; Ben-Haim and Rosenberg, 2002; Nyholm and Nishiguchi, 2008; Miyashiro and Ruby, 2012; Lemire et al., 2015; Tout et al., 2015). Notably, while some of these relationships include mutualistic interactions (Huq et al., 1983; Thompson et al., 2006), many are pathogenic with diverse Vibrio species causing disease in aquatic animals (Goarant et al., 2000; Kushmaro et al., 2001; Ben-Haim and Rosenberg, 2002; Becker et al., 2004; Frans et al., 2011; Geng et al., 2014; Vezzulli et al., 2015) and within aquaculture settings, sometimes resulting in significant economic losses (Lafferty et al., 2015). Notably, some Vibrio species are also dangerous human pathogens (Daniels and Shafaie, 2000).
Given their ecological, economic, and human health significance, assessing Vibrio diversity and the presence of specific Vibrio species in the environment is an important objective and a wide suite of both culture-dependent (Donovan and Van Netten, 1995; Grisez et al., 1997) and -independent techniques have been applied (Lane et al., 1985; Dorsch et al., 1992). Among culture-independent techniques, 16S rRNA sequencing has been widely used to characterize Vibrio community diversity and composition, but typically delivers poor species-level resolution, particularly among highly genetically related Vibrio species (Nagpal et al., 1998; Gomez-Gil et al., 2004; Pascual et al., 2010; Cano-Gomez et al., 2011), thereby limiting examination of intra-genus heterogeneity (Poretsky et al., 2014). Previous attempts to employ Vibrio-specific 16S rRNA primers have incrementally improved the taxonomic resolution when characterizing Vibrio diversity (Yong et al., 2006) but the proportion of sequences that can be unambiguously assigned to Vibrio species often remains low (Siboni et al., 2016). Due to the inherent inadequacies of the 16S rRNA gene for correctly identifying Vibrio species, another gene target encoding the heat shock protein 60 (hsp60, also known as groEL or cpn60), a type I chaperonin protein that assists in protein folding (Hemmingsen et al., 1988; Farr et al., 2000; Brinker et al., 2001), has been proposed as a good candidate for Vibrio phylogenetic studies (Kwok et al., 2002). Previous studies utilizing hsp60 have primarily relied on Vibrio isolates for species identification (Preheim et al., 2011; Szabo et al., 2012; Silvester et al., 2017), but a recent study applied universal hsp60 primers to characterize Vibrio community diversity using high throughput amplicon sequencing (Jesser and Noble, 2018). While this approach delivered improved Vibrio species identification relative to 16S rRNA, it used universal, rather than Vibrio-centric, hsp60 primers and required bioinformatic filtering for Vibrio assigned species, as is often required for 16S rRNA sequencing.
One of the many areas where the development of a high precision assay for determining Vibrio community diversity would have great utility is within the aquaculture industry, where Vibrio infections cause substantial losses in stock and profits (Gay et al., 2004; Lafferty et al., 2015; Lemire et al., 2015; Bruto et al., 2017; Green et al., 2019), but the precise identity of the pathogen is often not well resolved or incorrectly assigned to the wrong species (Sawabe et al., 2013; Richards et al., 2014; Dubert et al., 2017; Green et al., 2019). Some Vibrio species, including Vibrio splendidus and Vibrio coralliilyticus, have negative impacts on oyster cultivation by causing mortality in hatcheries (Sugumar et al., 1998; Takahashi et al., 2000; Elston et al., 2008; Richards et al., 2015; King et al., 2019a). Outside of hatchery settings, a number of Vibrio species have been identified as oyster pathogens (Waechter et al., 2002; Saulnier et al., 2010; Duperthuy et al., 2011; Wendling et al., 2014; Bruto et al., 2017; Go et al., 2017; King et al., 2019a). Because of the complexity of oyster diseases, direct evidence for the involvement of Vibrio species often remains unproven; therefore, they are usually regarded as opportunistic pathogens (Garnier et al., 2007; Jenkins et al., 2013; Go et al., 2017; De Lorgeril et al., 2018). Culture-dependent studies have observed shifts in the Vibrio community preceding the onset of disease in oysters, whereby putatively pathogenic Vibrio bacteria replaced the benign Vibrio commensals (Lemire et al., 2015). To better understand the role of Vibrio species in disease events such as those afflicting oysters, a precise high throughput technique that characterizes this important bacterial group is required.
Here, a new Vibrio-centric hsp60 amplicon sequencing assay was developed, coupled with a customized Vibrio hsp60 sequence reference database, to precisely characterize Vibrio diversity in environmental samples using the Illumina sequencing platform. The utility of this new assay was first validated using mock Vibrio communities constructed from varying proportions of Vibrio isolates before moving to tests with natural seawater samples, and finally applying it to characterize patterns in Vibrio diversity during a Pacific oyster mortality event (Green et al., 2019).
Materials and Methods
Primer and Vibrio Reference Dataset Construction
In order to develop a reference dataset to aid the design of a new set of degenerate primers targeting the hsp60 gene, 100 Vibrio hsp60 coding sequences were collected from the NCBI repository and blasted against the NCBI nucleotide database (nt file). Sequences were extracted using extract_hitseqs_from_sequences.pl (Kahlke, 2019) and both accession numbers and their respective taxonomy were then extracted using list_basta_taxa.py provided by BASTA v1.3.2.3 (Kahlke and Ralph, 2019). The BLAST output was filtered to retain taxa assigned to the Vibrio genus using the filter_basta_fasta.py script also provided by BASTA (Kahlke and Ralph, 2019) and genes assigned as hsp60 and groEL were collected and added to our Vibrio-hsp60 dataset. Both of these genes were chosen because they have previously been assigned as the same gene, but are annotated differently (Silvester et al., 2017). The aforementioned steps generated 1926 Vibrio hsp60/groEL sequences. The Vibrio-hsp60 dataset was then aligned with MAFFT (Katoh et al., 2017) using the einse–reorder option and the dataset was cleaned (removal of short sequences and those sequences with “N” base pairs), yielding 1468 Vibrio hsp60/groEL sequences across more than 30 different Vibrio species. The aligned untrimmed hsp60 dataset was visualized using UGENE (Golosova et al., 2014) and highly conserved areas within the consensus sequence were chosen for primer construction. Primers were constructed using the Primer3Plus (Untergasser et al., 2007) software. The constructed degenerate primers were named Vib-hspF3-23 and Vib-hspR401-422 (Table 1), and their application resulted in the amplification of a 487 bp PCR product (Illumina adapters inclusive).
To ensure that the Vibrio reference dataset was constructed with accurately assigned Vibrio taxa and not partial hsp60 reads (which could possibly be assigned to the wrong taxa), we constructed a reference dataset using hsp60 sequences taken from whole genomes. First, all of the currently available complete Vibrio genomes were collected from the NCBI repository (185 genomes) and a BLAST database was constructed using these genomes. Vibrio hsp60 sequences were compared against this database and all hits at least 65% similar to the query hsp60 sequence and at least 400 base pairs long were extracted using extract_hitseqs_from_sequences.pl (Kahlke, 2019). BLAST hits were then visualized and trimmed to the primer locations in MEGA (version 7.0.26). To determine the coverage of Vibrio species in our Vibrio reference dataset, we compared the taxa in this trimmed dataset against the listed Vibrio species in the NCBI taxonomy database. Where possible, hsp60 sequences for missing Vibrio species were collected from incomplete whole genomes and added to the Vibrio reference dataset. This yielded a dataset comprising of 106 different Vibrio species incorporating 284 hsp60 sequences. In some instances, hsp60 was found in both Vibrio chromosomes. Where known, the second copy of the gene was named “group2.”
Mock Vibrio Community Preparation
Ten Vibrio species, spanning five clades (Sawabe et al., 2013; Turner et al., 2018) and incorporating species that are relevant to both human health (Daniels and Shafaie, 2000) and aquaculture diseases (Luna-González et al., 2002; Bruto et al., 2017; Go et al., 2017) were grown overnight in LB20 broth (per liter: 10 g tryptone, 5 g yeast extract, 20 g NaCl), with shaking at 28°C. Bacterial cells were enumerated using a Beckman CytoFLEX flow cytometer and cell counts were diluted to a standardized concentration across all strains. Three different mock Vibrio communities were prepared by mixing the 10 Vibrio species in different dilution ratios (Table 2). DNA was then extracted from mock assemblages using the Qiagen DNeasy UltraClean Microbial Kit (catalog: 12224-250) following the manufacturer’s instructions.
Mock Community PCR Conditions and Sequencing
DNA extracted from the mock bacterial communities was diluted to 10 ng μL–1 and used in a 50 μL PCR reaction volume as follows: 10 μL of 5× Hi-Fi Buffer (Bioline), 5 μL of 10 mM dNTPs, 2 μL of high-fidelity velocity polymerase (0.5 units μL–1; Bioline), 2.5 μL of 10 μM forward primer (VF169 or Vib-hspF3-23), 2.5 μL of 10 μM reverse primer (Vib-680R or Vib-hspR401-422), 2 μL of DNA template (10 ng μL–1), with the remaining volume made up with sterile water. The PCR mixture was then subjected to the following PCR conditions: one cycle of 98°C for 2 min, 30 cycles of 98°C for 30 s, 50°C for 30 s, and 72°C for 30 s, and a final extension time of 72°C for 10 min. PCR products were purified with a Bioline Isolate II PCR and Gel Kit (catalog: BIO-52059) using the manufacturer’s instructions. For 16S (341F and 805R) rRNA sequencing, extracted DNA was amplified by the Ramaciotti Centre for Genomics with the following PCR conditions: 95°C for 3 min, 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and a final extension at 72°C for 5 min.
Mock bacterial community amplicons were characterized on the Illumina MiSeq platform (Ramaciotti Centre for Genomics; Sydney, NSW, Australia) using the manufacturers guidelines, using three primer sets (Table 1): the universal 16S rRNA primers 341F and 805R (Herlemann et al., 2011); a previously published Vibrio-specific 16S rRNA primer pair, VF169 (Yong et al., 2006) and Vib-680R (Thompson et al., 2004; Siboni et al., 2016); and the Vibrio-centric hsp60 primer pair designed in this study, Vib-hspF3-23 and Vib-hspR401-422.
Mock Community Sequence Analysis
Bacterial 16S rRNA and hsp60 sequencing reads for the mock communities were processed as outlined in Kahlke (2018). Briefly, paired-end DNA sequences were joined using FLASH (Magoč and Salzberg, 2011) and subsequently trimmed using Mothur (Schloss et al., 2009) (PARAMETERS: universal 16S—maxhomop = 6, maxambig = 0, qaverage = 25, minlength = 491, maxlength = 501; Vibrio-specific 16S—maxhomop = 6, maxambig = 0, qaverage = 25, minlength = 533, maxlength = 534; hsp60—maxhomop = 6, maxambig = 0, qaverage = 25, minlength = 420, maxlength = 420). Trimmed sequence length was determined by the 2.5 and 97.5% tiles for the universal 16S and Vibrio-specific 16S assay, and 420 bp was chosen for the hsp60 assay as it closely corresponded to the amplicon size (excluding illumina adapters) and was the 75% tile. The resulting fragments were clustered at 97% into operational taxonomic units (OTUs) and chimeric sequences were identified and removed using vsearch (Rognes et al., 2016). To assign taxonomy, QIIME (Caporaso et al., 2010) was used with the RDP classifier against either the Silva v128 database (for 16S rRNA analyzed samples) or against our custom Vibrio-hsp60 reference dataset. Sequences were then rarefied to the same depth to remove the effect of sampling effort upon analysis.
Seawater Collection, 16S rRNA Sequencing, and Data Analysis
To test the newly designed Vibrio-centric hsp60 sequencing assay on seawater, water was collected from Sydney Harbour (33.839S, 151.254E) in the Austral summer. Seawater was filtered in triplicate through 0.22 μm membranes, before the filters were immediately snap frozen in liquid nitrogen. Microbial DNA was subsequently extracted from filters using a Qiagen DNeasy PowerWater kit (catalog: 14900-100-NF) and sent to the Ramaciotti Centre for Genomics (University of New South Wales, Sydney, NSW, Australia) for 16S rRNA (341F and 805R) sequencing on the Illumina MiSeq platform.
Raw 16S rRNA demultiplexed paired-end DNA sequences were joined using Flash (Magoč and Salzberg, 2011) and trimmed with Mothur (Schloss et al., 2009) (PARAMETERS: maxhomop = 6, maxambig = 0, qaverage = 25, minlength = 441, maxlength = 466); 96% of the sequences were successfully joined with FLASH and the trim lengths corresponded to the 25 and 97.5% tiles identified by Mothur. Fragments were then clustered into OTUs at 97% and chimeric sequences removed using vsearch (Rognes et al., 2016). QIIME (Caporaso et al., 2010) and the RDP classifier were then used to assign taxonomy against the Silva v128 database. Sequences were then rarefied.
DNA from seawater was also amplified using the Vib-hspF3-23 and Vib-hspR401-422 primer pair with the Illumina adapters added to the primers (Table 1). The 30 μL PCR reaction mixture was as follows: 6 μL of 5× Hi-Fi Buffer (Bioline), 3 μL of 10 mM dNTPs, 0.5 μL of high-fidelity velocity polymerase (2 units μL–1; Bioline), 1.5 μL of 20 μM forward primer, 1.5 μL of 20 μM reverse primer, 3.5 μL of template DNA, and the remainder (14 μL) made up with sterile water. The mixture was used with the following touchdown PCR conditions: one cycle of 98°C for 2 min, 5 cycles of 98°C for 30 s, 60°C for 30 s, and 72°C for 45 s, 21 cycles of 98°C for 30 s, 60°C for 30 s with a reduction of 0.5°C per cycle (60 to 50°C), and 72°C for 45 s, 16 cycles of 98°C for 30 s, 50°C for 30 s, and 72°C for 45 s, and a final extension time of 72°C for 10 min. Amplicons were then purified by the Ramaciotti Centre for Genomics, and characterized on the Illumina MiSeq platform using the manufacturer’s guidelines.
As the Vibrio-centric hsp60 primer pair in some scenarios was found to non-specifically amplify other (non-Vibrio) taxa, a further cleaning step was added to the data analysis. In the first instance, pair-ended sequences were joined using FLASH (Magoč and Salzberg, 2011) and trimmed using mothur (Schloss et al., 2009) (PARAMETERS: maxhomop = 5, maxambig = 0, qaverage = 25, minlength = 420, maxlength = 420). These fragments were then clustered at 97% into OTUs and chimeric sequences were removed using vsearch (Rognes et al., 2016). Non-Vibrio sequences were removed by BLASTing cleaned sequences against the Vibrio-hsp60 reference dataset and retaining those that were 90% similar to any sequence in that dataset. This fasta file was then used to assign taxonomy against the custom Vibrio-hsp60 reference dataset with the RDP classifier. Due to the large spread of sequences per sample, data were not rarefied, rather sequences were normalized to the number of sequences per sample to produce the relative abundance of each taxa for each sample (data analysis workflow is available at King et al., 2019b; https://doi.org/10.17605/OSF.IO/4798P).
Quantitative PCR (qPCR)
To provide an indication of Vibrio abundance, a quantitative PCR (qPCR) assay was used to quantify the number of Vibrio-specific 16S rRNA gene copies in each sample using the Vibrio-specific 16S rRNA gene primers Vib-567F and Vib2-r (Table 1) (Thompson et al., 2004). qPCR was performed using an epMotion 5075l Automated Liquid Handling System on a Bio-Rad CFX384 Touch Real-Time PCR Detection System with a seven-point calibration curve and a negative control. The calibration curve was created from 10-fold dilutions of a known quantity of amplicon DNA, measured by a Qubit fluorometer. All sample analyses were performed with three technical replicates, using the following reaction mixture: 2.5 μL iTaq Universal SYBR Green supermix, 0.4 mM of each forward and reverse primer, 1 μL of template DNA (50 ng μL-1), and the remainder made up with water. The qPCR cycling conditions were previously described (Siboni et al., 2016) and as follows: 95°C for 3 min followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. The resulting data were normalized to milliliters of collected water. A coefficient of variation (CV) was then calculated for the technical triplicates, and where necessary, samples with CV > 1% had a replicate removed from the analysis. A melting curve was added to the end of every run to confirm the presence of a single PCR product.
Laboratory-Induced Oyster Mortality Event
The newly designed Vibrio-centric hsp60 primer set was applied to examine patterns in Vibrio community diversity during a previously described laboratory-induced Pacific Oyster mortality event, where Vibrio species had previously been implicated as the cause of oyster mortality during a simulated marine heatwave (Green et al., 2019). Briefly, triploid Pacific oyster (Crassostrea gigas) spat were collected from Port Stephens, New South Wales, Australia, prior to a forecasted marine heat wave. Spat were held in two different temperature conditions (low 20 ± 1°C and high 25 ± 1°C) with and without antibiotics (100 units/mL of penicillin and 0.1 mg/mL of streptomycin) and monitored for 6 days. Spat were placed in sterilized glass tanks, and UV and 5 μm filter sterilized seawater were added and replaced daily. Triplicate oyster spat were sampled on days 0, 3, 4, 5, and 6, and dead oysters were removed and frozen at -80°C prior to processing. Cumulative mortality was 77.4 ± 10.7 and 3.4 ± 5.9% for the high and low temperature treatment, respectively, with antibiotics in the high temperature treatment reducing the cumulative mortality to 4.3 ± 3.7%, indicating a likely role of bacteria in the oyster mortality. Mortalities were greatest between days 3 and 5. For the purposes of this study, DNA extracted from spat exposed to the high and low temperature treatments from each sampling point were amplified with the Vibrio-centric hsp60 primer pair (Table 1). Vibrio dynamics were previously characterized within oyster tissues using a combination of culture-based approaches, qPCR and 16S rRNA amplicon sequencing (Green et al., 2019). Oyster DNA were subject to hsp60 PCR amplification, sequencing, and data analysis, and qPCR, as described above for the seawater samples, with the exception of DNA being diluted to 50 ng μL–1 for the PCR conditions.
Statistical Analyses
Comparisons of community compositions were performed using non-metric multidimensional scaling analysis (nMDS) with a Bray–Curtis dissimilarity index, applied to data normalized to the number of sequences per sample, with sequences less than 1% relative abundance removed, and then transformed (square root). Patterns observed in the nMDS analysis were statistically tested with a one-way PERMANOVA with 9999 permutations. To examine the similarity between each characterized community to the mock community, similarity indices were calculated with a Bray–Curtis dissimilarity index using data that were filtered, transformed, and with sequences assigned to the second chromosome (group 2) combined with their respective assigned species. To examine the contribution of individual Vibrio species to community dissimilarity, a SIMPER analysis with untransformed data was used with a Bray–Curtis dissimilarity index. To determine the relationship between Vibrio-specific 16S rRNA gene copies and the yield of cleaned hsp60 sequences from the QIIME analysis, an ordinary least squares linear regression was used. Spearman’s rank correlation was used to examine relationships between Vibrio species (summarized at the species level) and oyster mortality. All statistical comparisons were performed in the PAST statistical environment (Hammer et al., 2001).
Results and Discussion
Comparison of Vibrio Mock Community Characterization Using 16S rRNA and hsp60
Mock Vibrio communities consisting of 10 different Vibrio species were characterized using the 16S rRNA, Vibrio-specific 16S rRNA, and Vibrio-centric hsp60 primer pairs followed by Illumina MiSeq sequencing of the amplicons. When examined on an nMDS, the Vibrio community structure defined by the Vibrio-centric hsp60 assay clustered closer to the true mock community than the two 16S rRNA-bases assays, with the true mock community sitting within the 95% ellipses for each Vibrio-centric hsp60 characterized community (Figures 1A–C). Comparatively, the compositions of the 16S rRNA, Vibrio-specific 16S rRNA, and Vibrio-centric hsp60 characterized communities were on average 16, 25, and 77% similar to the true mock community, respectively (Figure 1D).
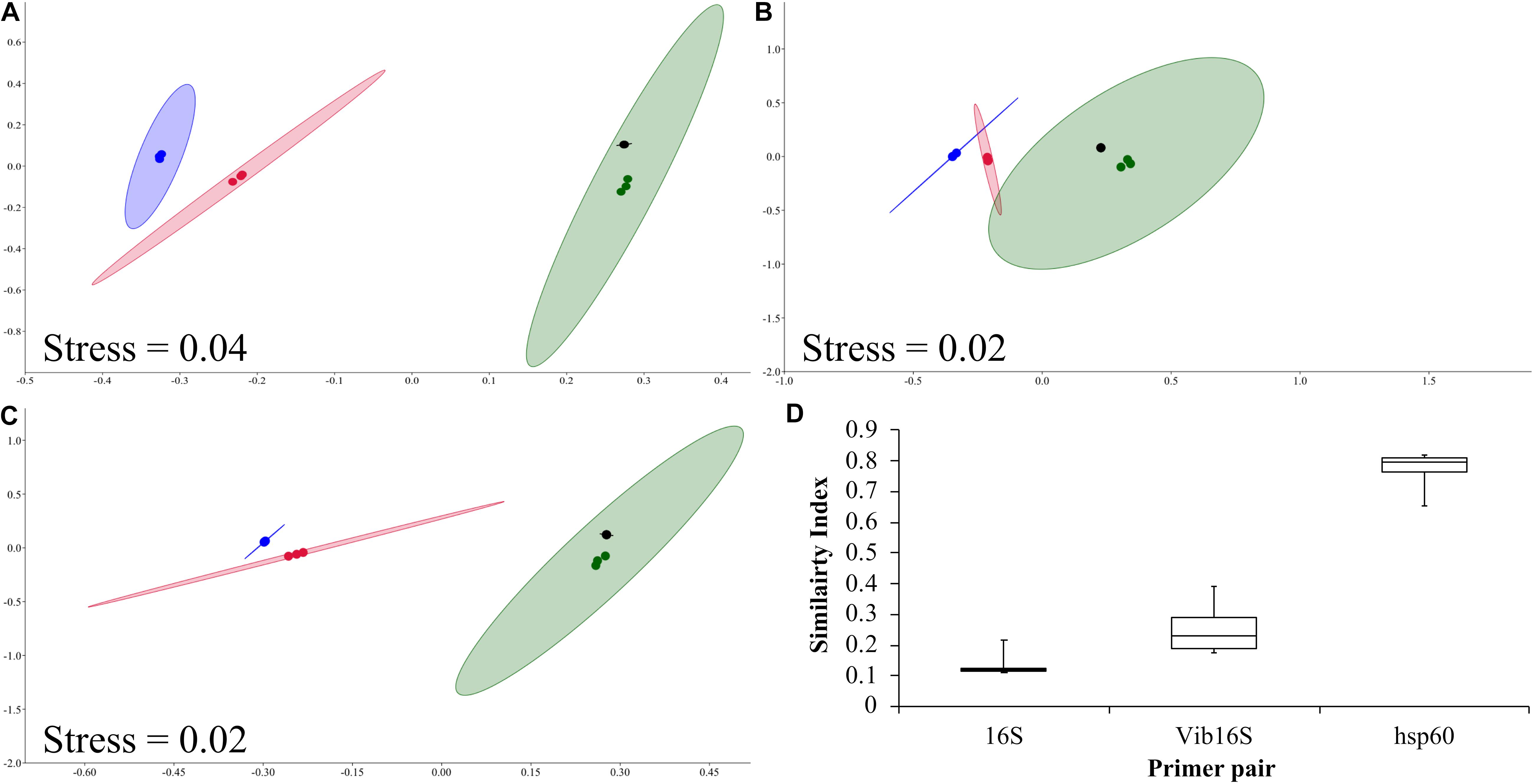
Figure 1. nMDS analysis of the 16S rRNA (blue dots), Vibrio-specific 16S rRNA (red dots), and Vibrio-centric hsp60 (green dots) characterized mock communities, and the true mock community (black dots). Mock communities 1, 2 and 3 are (A), (B) and (C) respectively. 95% ellipses are shown. (D) Box and whisker plot of Bray–Curtis similarity comparisons of community composition compared to the true mock communities. Data for all three mock communities are combined. For species assigned across two taxonomic assignments (e.g., group 2), they were combined with their respective species for D.
The Vibrio community data derived from the traditional V3–V4 16S rRNA primer pair was poorly characterized beyond the genus level, with only one Vibrio species in the mock community correctly identified (Figure 2). On average, sequences were not defined beyond the Vibrio genus level 90, 74, and 89% of the time in mock communities 1, 2, and 3, respectively. Of the sequences that were assigned beyond the Vibrio genus level, they were only assigned to Vibrio cholerae and Vibrio azureus. Notably, V. azureus was not part of the mock communities indicating not only imprecise, but incorrect taxonomic classification. V. azureus is within the Vibrio harveyi clade, for which it was probably incorrectly attributed (Yoshizawa et al., 2009). Sequences assigned to V. cholerae were correctly assigned but were marginally under-represented when compared to the real abundance within the mock community (6–23% for 16S rRNA; 7.5–30% for the mock communities).
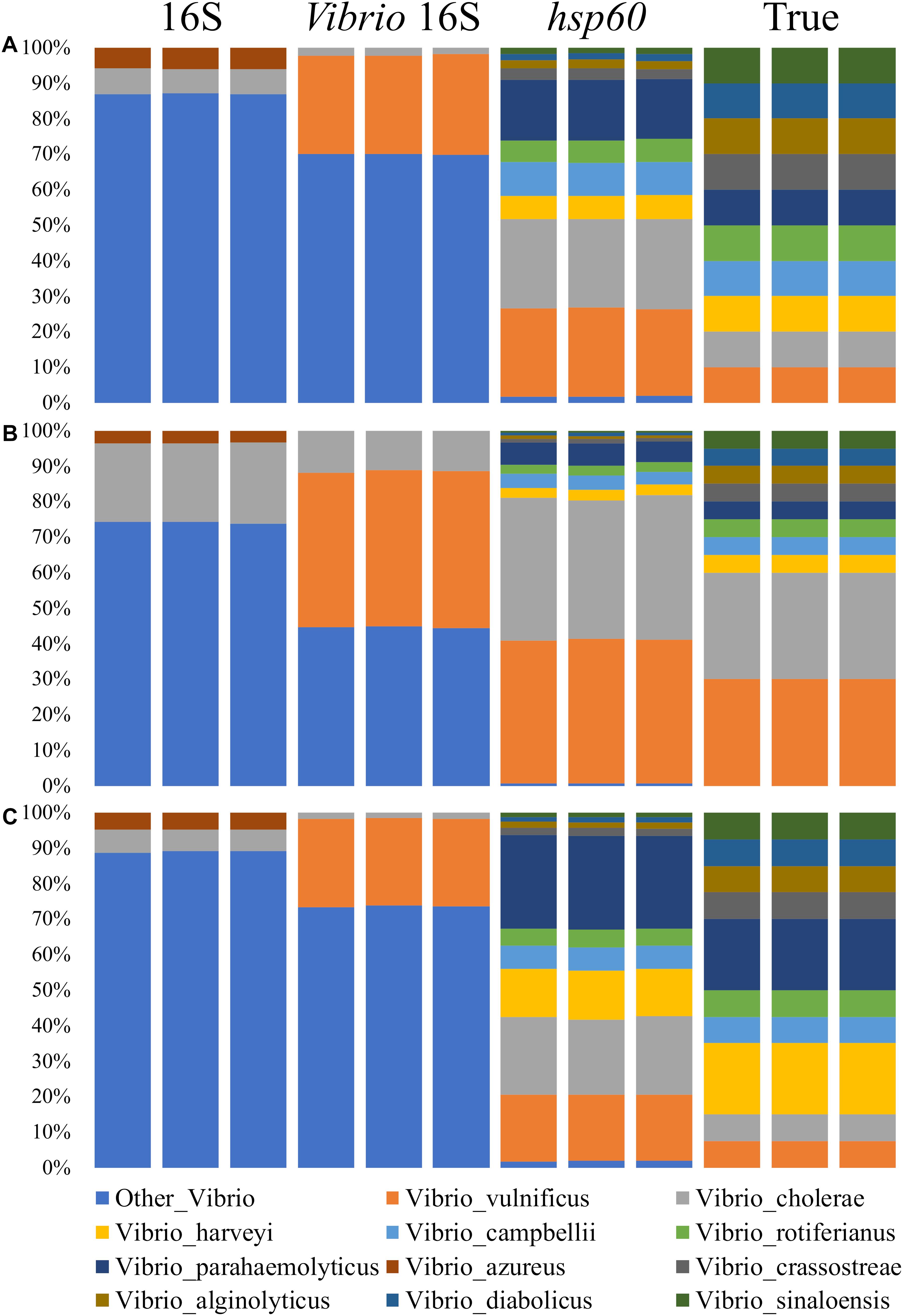
Figure 2. Comparison of amplicon sequenced phylogenetic markers for the Vibrio mock communities. Mock communities 1, 2 and 3 are (A), (B) and (C) respectively. Each bar represents the average across a biological triplicate for each sequencing assay. Communities were characterized using 16S rRNA V3-V4 (Herlemann et al., 2011), Vibrio-specific 16S rRNA (Thompson et al., 2004; Yong et al., 2006), and Vibrio-centric hsp60 primer pairs. The true mock community composition is also shown. Displayed data are relative abundance summarized at the species level. For sequences assigned to the second chromosome (group 2), they were combined with their respective species. Sequences representing less than 1% of the relative abundance were removed.
Similar to the 16S rRNA characterized community composition, the data derived from the Vibrio-specific 16S rRNA sequencing assay were only able to identify two Vibrio species in the mock community, with the majority of the sequences not resolved beyond the genus level. Although correctly identified as Vibrio, the majority of sequences could not be assigned to the species level 70, 45, and 74% of the time in mock communities 1, 2, and 3, respectively. For the remainder of the sequences assigned beyond the genus level, they were correctly assigned to Vibrio vulnificus and V. cholerae. Sequences assigned to V. vulnificus were over-represented when compared to the true mock community composition (25–44% for Vibrio-specific 16S rRNA; 7.5–30% for the mock communities), while sequences assigned to V. cholerae were under-represented (2–12% for Vibrio-specific 16S rRNA; 7.5–30% for the mock communities).
Relative to both of the 16S rRNA based sequencing assays, the Vibrio-centric hsp60 primer set identified the greatest number of species in the Vibrio community, with all of the species present in the mock community correctly identified by this assay. While all of the species were correctly identified, differences in the relative abundance of each species were observed when compared to the true mock community (Table 3). Vibrio campbellii was the best represented species with each Vibrio-centric hsp60 characterized mock community only showing a 1% difference to the true mock community, while Vibrio sinaloensis was the most under-represented species with differences of 4–8% for the Vibrio-centric hsp60 characterized communities. For V. vulnificus and V. cholerae, the only two correctly identified species in the 16S rRNA and Vibrio-specific 16S rRNA characterized communities, the Vibrio-centric hsp60 assay provided better representation for V. vulnificus (over-representation of 10–13% compared to the mock community) compared to the Vibrio-specific 16S rRNA assay (over-representation of 14–18%) and 16S rRNA assay (not identified). For V. cholerae, this species was under-represented in both the 16S rRNA (1–8%) and Vibrio-specific 16S rRNA assays (6–20%) compared to an over-representation in the Vibrio-centric hsp60 assay (9–14%). The exaggeration of V. vulnificus and V. cholerae could possibly be due to a greater hsp60 primer affinity to these species, or the presence of two copies of hsp60 in the genomes of these bacteria (one copy was identified in each chromosome for these two species).
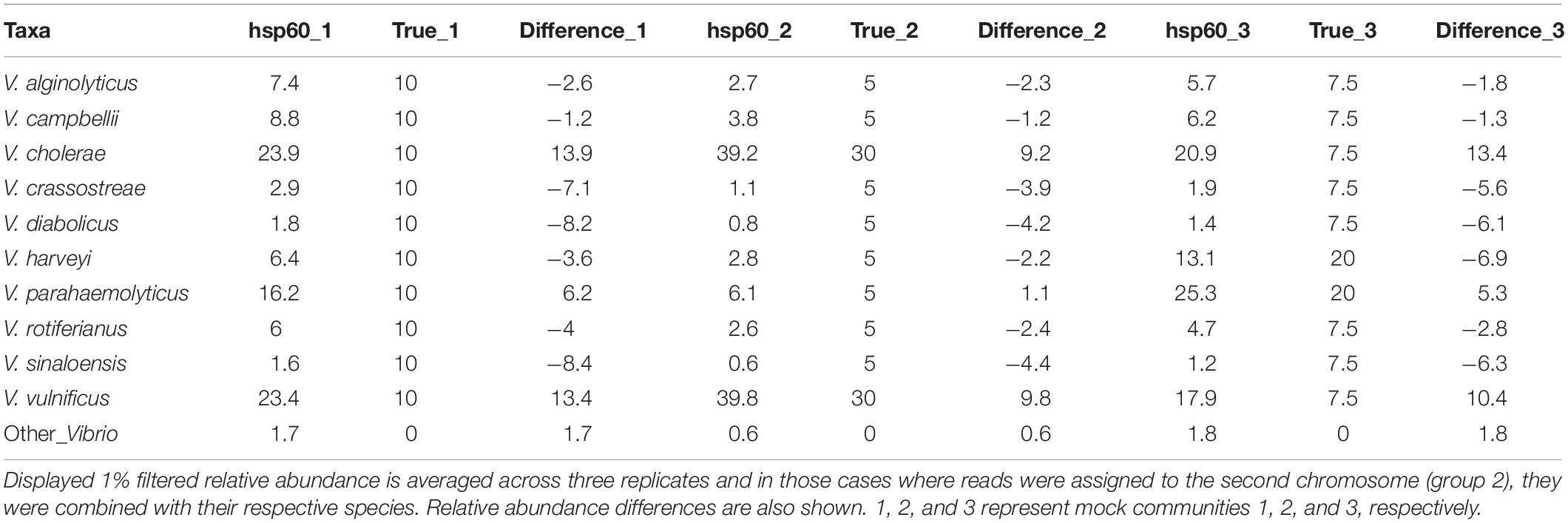
Table 3. Relative abundance comparisons between the Vibrio-centric hsp60 characterized mock communities and the true mock communities.
Notably, the Vibrio-centric hsp60 sequencing assay also distinguished members of the V. harveyi clade, a tight phylogenetic group within the Vibrio genus (Sawabe et al., 2013; Urbanczyk et al., 2013), which has previously had numerous incorrect taxonomic assignments to species within this clade caused by close 16S rRNA genetic similarity (Lin et al., 2010; Sawabe et al., 2013; Urbanczyk et al., 2013). This clade includes Vibrio parahaemolyticus, Vibrio alginolyticus, V. harveyi, V. campbellii, Vibrio diabolicus, and Vibrio rotiferianus (Sawabe et al., 2013; Turner et al., 2018), all of which were discriminated with the Vibrio-centric hsp60 sequencing assay. Many of these species are important pathogens (Daniels and Shafaie, 2000; Luna-González et al., 2002; Go et al., 2017) and therefore accurately identifying their presence in environmental samples is an important requisite of a Vibrio specific assay of this type.
Previous attempts to perform hsp60 amplicon sequencing have used universal hsp60 primers and filtered the data for the assigned Vibrio sequences (Jesser and Noble, 2018). However, only 0.5% of the total hsp60 data were assigned to Vibrio species in this previous study, and of these a significant proportion was unassigned to Vibrio species (Jesser and Noble, 2018), which was attributed to poor Vibrio species representation in the cpn60 database (Hill et al., 2004; Jesser and Noble, 2018). In contrast, our assay resulted in 21.1% of the data being retained, with 98.1–99.5% of this being successfully assigned at the species level. We believe that the higher specificity of our assay was provided in part by the boutique Vibrio hsp60 reference dataset that we developed, which encompasses 106 different Vibrio species compared to only 63 unique species in the cpn60 database (accessed August 2019).
Vibrio Diversity in Seawater
After confirming the utility of the Vibrio-centric hsp60 sequencing assay using mock communities, this assay was used to characterize Vibrio diversity in seawater samples collected from Sydney Harbour, with the measured community composition compared to that derived from traditional 16S rRNA sequencing (Figure 3). Sequences assigned to the Vibrio genus or Vibrio species only made up 0.13–0.17% of the total bacterial community using 16S rRNA sequencing, with the majority (59–77% relative abundance) of these sequences not resolved beyond the Vibrio genus level. In contrast, only 1.4–1.7% of the sequences when using the Vibrio-centric hsp60 sequencing assay were not assigned to a Vibrio species. Ten different Vibrio species were identified within the seawater samples using the Vibrio-centric hsp60 sequencing assay, most of which occurred in low abundance (1–4% relative abundance) except for V. azureus and Vibrio mediterranei, which comprised between 58–71 and 10–29% of the total Vibrio community, respectively. In contrast, only three Vibrio species were identified using the 16S rRNA assay. Both assays identified the presence of V. mediterranei, with similar levels of relative abundance (16S rRNA: 19–34%; hsp60: 10–29%). The capability of the Vibrio-centric hsp60 sequencing assay to resolve more sequences to the species level in seawater samples, relative to traditional 16S rRNA sequencing, highlights its utility when examining fine-scale patterns in Vibrio diversity.
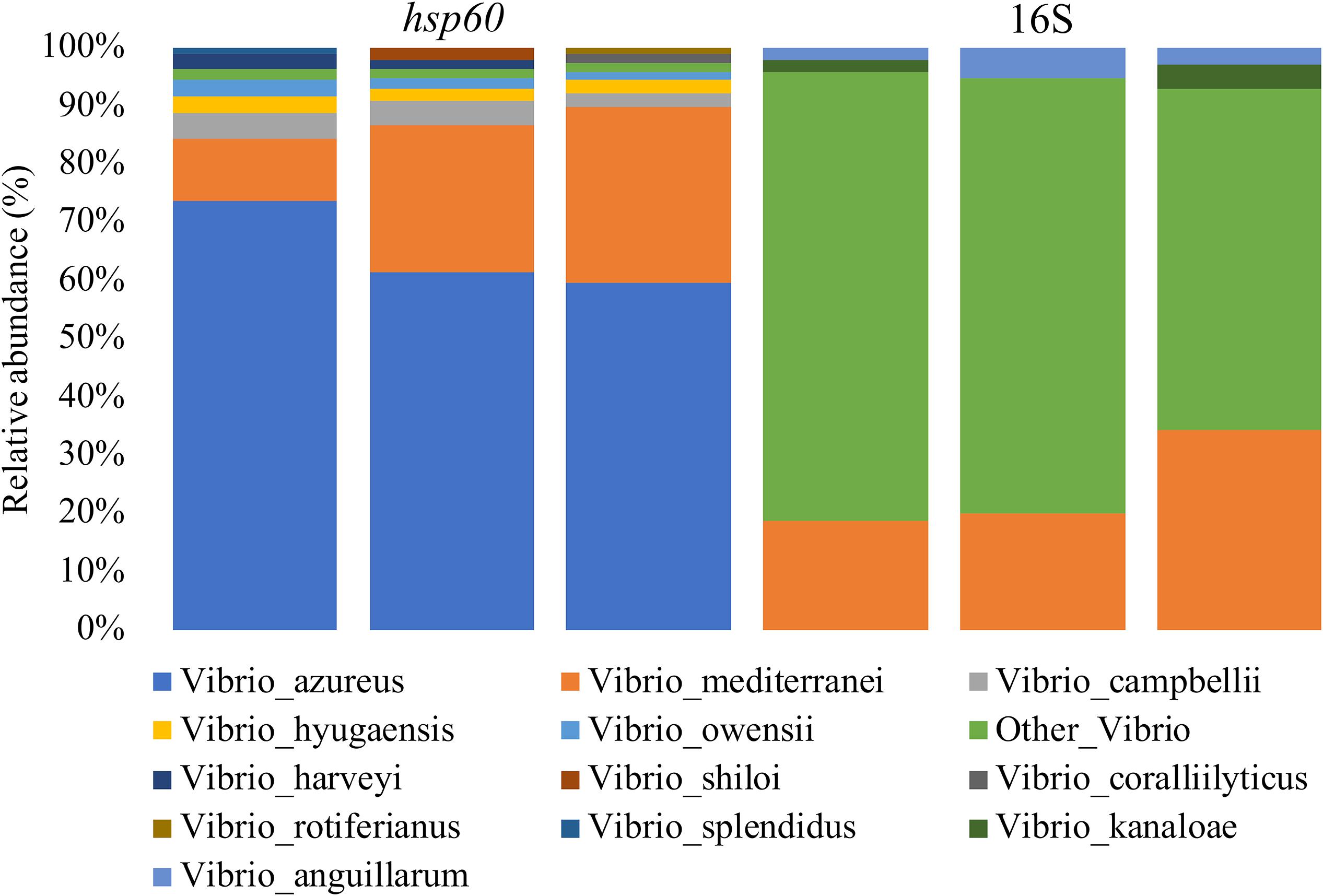
Figure 3. Vibrio diversity in seawater from Sydney Harbour. DNA was characterized with the Vibrio-centric hsp60 and 16S rRNA V3-V4 (Watermann et al., 2008) primer sets. Displayed data are relative abundance summarized at the species level.
Vibrio Abundance Determines Assay Efficacy
To determine whether Vibrio abundance influenced assay data yield, both seawater and oyster samples were used. As expected, samples with the greatest abundance of Vibrio, as determined using qPCR targeting Vibrio 16S rRNA gene copies, had the greatest number of hsp60 sequences (Supplementary Figure S1), with a significant relationship observed between Vibrio 16S rRNA gene copies and hsp60 sequences (R2 = 0.87; p = 0.0001) (Figure 4). It is possible that the low number of hsp60 sequences in samples with low Vibrio biomass was due to non-specific amplification of hsp60 sequences associated with other bacterial genera. However, the linear relationship between Vibrio abundance and hsp60 data yield indicates that when elevated levels of Vibrio are present within a sample, this assay delivers substantial capacity to probe the diversity of the community.
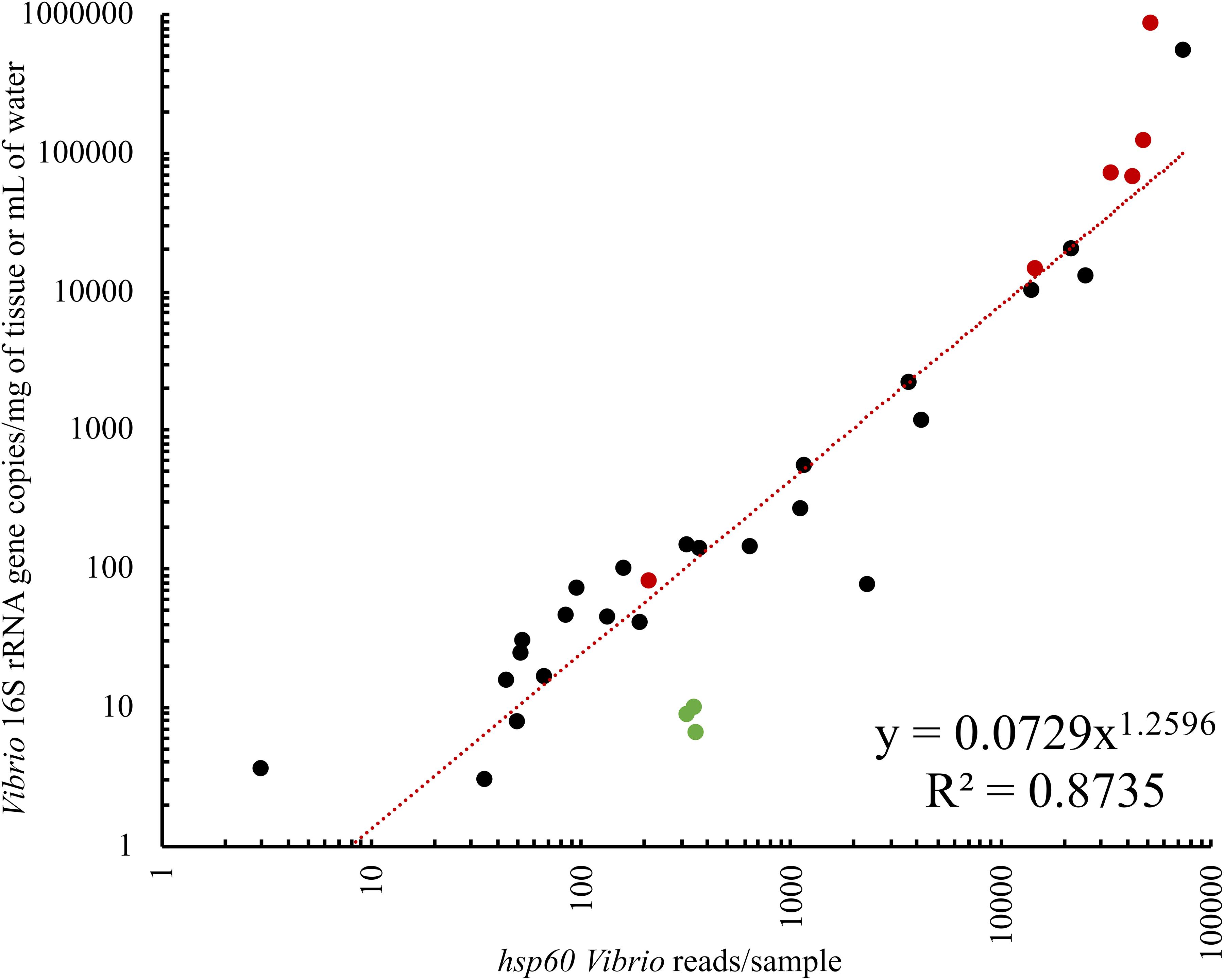
Figure 4. Ordinary least squares linear regression of Vibrio 16S rRNA gene copies and hsp60 sequences per sample. Vibrio 16S rRNA gene copies were determined with qPCR while hsp60 sequences refer to the analyzed Illumina sequencing data generated from the Vibrio-centric hsp60 assay. Black dots are oyster samples, red dots are oyster mortality samples, and green dots are seawater samples. Both axes are logarithmic in scale.
Vibrio Diversity During a Laboratory Induced Oyster Mortality Event
After confirming the utility of the Vibrio-centric hsp60 assay to track Vibrio community dynamics with high fidelity using a mock community and successfully applying it to characterize Vibrio diversity within natural seawater samples, it was next used to examine patterns in Vibrio diversity during a induced oyster mortality event. During a simulated heatwave event described in detail in Green et al. (2019), significant levels of oyster mortality were observed in oysters exposed to an increase in water temperature to 25°C (77.4 ± 10.7%), relative to oysters maintained at ambient temperature levels at 20°C (3.4 ± 5.9%). The Vibrio-centric hsp60 assay was applied on samples derived from this study, because previous analyses (culturing and 16S rRNA sequencing) indicated a potential involvement of Vibrio in the oyster mortality event (Green et al., 2019).
Using the Vibrio-centric hsp60 sequencing assay, the Vibrio community composition associated with Pacific oysters was significantly different between temperature treatments (F = 6.5, p = 0.0005; Supplementary Figure S2) and oyster mortality levels (F = 14.8, p = 0.0003 versus low temperature; F = 4.4, p = 0.013 versus high temperature). The “baseline” Vibrio community (Figure 5) on the first day of the experiment (day 0), 4 days prior to significant mortalities, was distributed across nine different species, with Vibrio brasiliensis, Vibrio chagasii, Vibrio fortis, and V. harveyi representing the dominant members of the Vibrio community with average relative abundances of 9, 20, 11, and 35%, respectively.
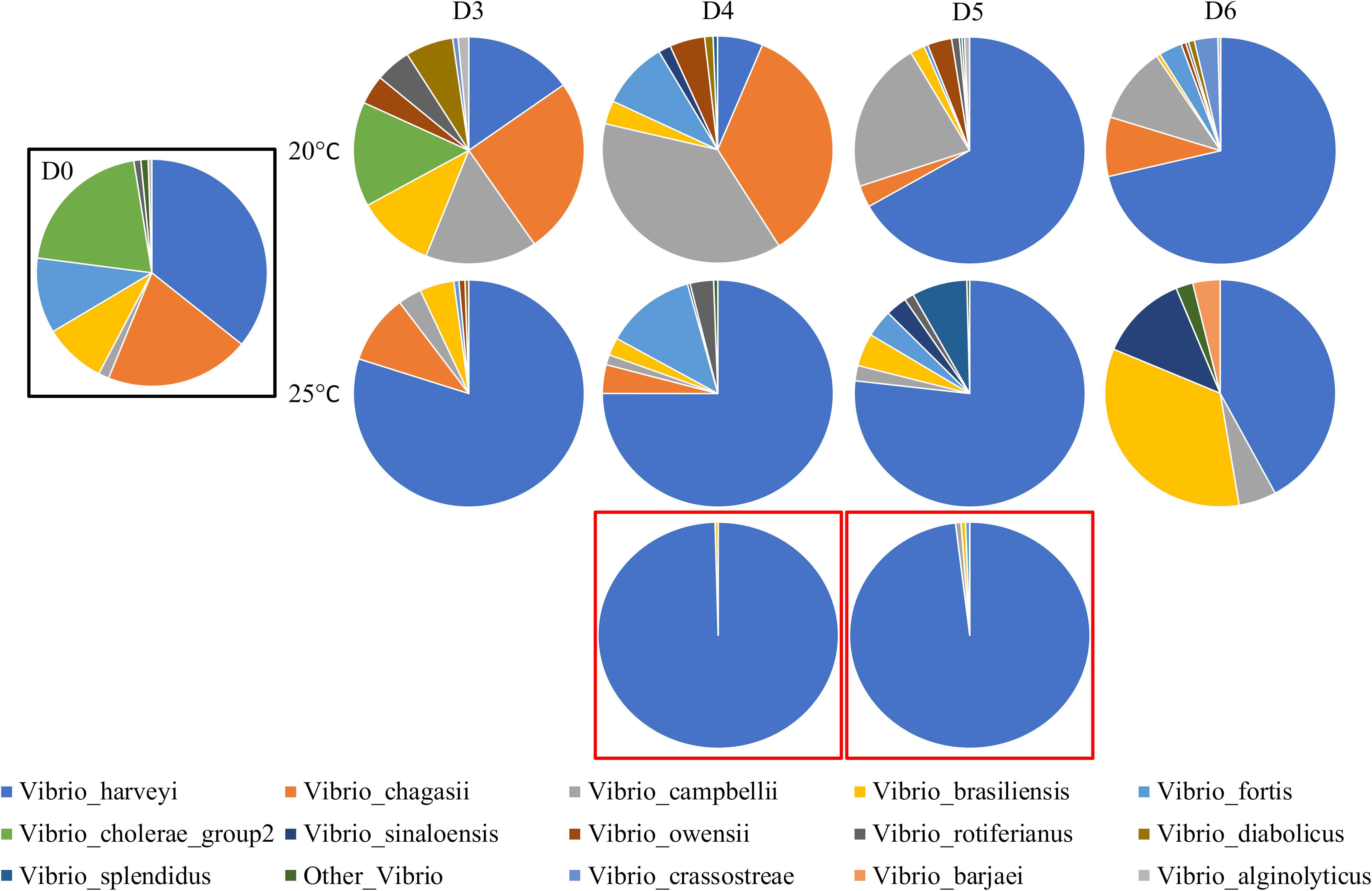
Figure 5. Vibrio community of C. gigas spat across 6 days and two temperature treatments. D0, D3, D4, D5, and D6 correspond to sampling days 0–6. Communities are averaged across three biological replicates and summarized at the species level. Communities in a black box are day 0. Communities in red boxes are dead C. gigas spat from the high (25°C) temperature treatment, taken on days 4 and 5, respectively. Sequences representing less than 1% of the relative abundance were removed.
When comparing the control and marine heatwave samples, the Vibrio communities were on average 56% dissimilar to each other. In the ambient temperature control, V. campbellii and V. chagasii were the most prominent members of the Vibrio community, contributing 18 and 15% to the community dissimilarity (21 and 17.6% average relative abundance, respectively; Supplementary Table S1), with the relative abundance of both species negatively correlated to temperature (rs = -0.4, p = 0.04; rs = -0.53, p = 0.008, respectively; Supplementary Table S2) and oyster mortality (rs = -0.45, p = 0.02; rs = -0.52, p = 0.007, respectively). On the other hand, V. harveyi dominated the Vibrio community in the high temperature marine heatwave treatments, whereby this species contributed 37% to the community dissimilarity between temperature treatments and was positively correlated to temperature (rs = 0.52, p = 0.011) and oyster mortality (rs = 0.55, p = 0.006). On days 3–5, V. harveyi increased substantially in relative abundance from 35% on day 0 to 73–75% of the whole community, followed by a decrease in relative abundance (41%) on day 6. This pattern is consistent with the results of a V. harveyi specific qPCR assay previously performed on these samples, where a significant increase in copies of the V. harveyi gyrase B gene was observed on days 3–5, followed by a decrease on day 6 (Green et al., 2019).
Notably, a sharp increase in the relative abundance of V. harveyi was also observed in the low temperature treatment on days 5 (6% on day 4 to 65%) and 6 (68%), which was again consistent with qPCR data (Green et al., 2019). Dead oyster samples collected on days 4 and 5 from the high temperature treatment were also completely dominated by V. harveyi, which represented 97 and 96% of the Vibrio community, respectively. Low levels of oyster mortality (2%) were observed in the low temperature treatment on day 6 (Green et al., 2019), which notably corresponded with an increase in the relative abundance of V. harveyi on the preceding day (6–65% from days 4 to 5), possibly indicating a time-delayed relationship for V. harveyi proliferation due to the lower temperature. V. harveyi was previously implicated as the causative agent behind this mortality event (Green et al., 2019) and a previous study implicated V. harveyi as a causative agent for an unknown mass mortality outbreak from the same region the oysters were sourced from (Port Stephens) (Go et al., 2017). The data derived from the Vibrio-centric hsp60 sequencing approach were able to unambiguously pinpoint the putative pathogen that increased in abundance prior to disease onset, as evidenced by previous culturing studies (Go et al., 2017; Green et al., 2019).
During the simulated marine heatwave, temperature was strongly correlated with oyster mortality (rs = 0.87, p = 0.0001) and may have provided a selective advantage for V. harveyi allowing for an increase in the relative abundance of this species, effectively replacing the putative commensal Vibrio species (Lemire et al., 2015) and/or temperature may have acted as an immunosuppressant in the oysters allowing for a shift in the Vibrio community preceding disease (Lokmer and Wegner, 2015). Interestingly, the oysters on day 6 in the high temperature treatment had a decreased number of sequences assigned to V. harveyi relative to the preceding days (75 to 45% from days 5 to 6). A possible explanation for this pattern is that a sub-population of surviving oysters exhibited higher tolerance to the elevated temperature conditions, avoided colonization by V. harveyi and survived.
These results indicate that temperature stressed Pacific oysters undergo a substantial shift in the composition of their Vibrio community, involving a dramatic increase in the relative abundance of V. harveyi, which precedes oyster mortality. Both the occurrence of elevated levels of the V. harveyi in oysters before and during mortality and the very high levels of V. harveyi in freshly deceased oysters further implicate this species in oyster mortality events, in agreement with previous studies (Saulnier et al., 2010; Segarra et al., 2010; Jenkins et al., 2013; Le Roux et al., 2016; Go et al., 2017).
Advantages and Disadvantages of the Vibrio-Centric hsp60 Assay
Due to the non-specific nature of our primer set, sometimes a significant proportion of hsp60 sequences was assigned to non-Vibrio taxa, including species assigned to the Labrenzia, Erythrobacter, Phaeobacter, and Pseudomonas genera (Supplementary Table S3), and were therefore removed. Despite this, the Vibrio-centric hsp60 assay delivered significantly enhanced species identification in all of the tested samples, relative to 16S rRNA, with community profiles displaying high levels of fidelity with the known composition of mock communities. Relative to 16S rRNA and previously published universal hsp60 primers (Jesser and Noble, 2018), our hsp60 assay also generated a greater yield of Vibrio-assigned data, with our hsp60 assays full potential attained in combination with our boutique Vibrio-database. In some instances, samples with a low Vibrio abundance will likely generate low numbers of cleaned hsp60 sequences. In this study, we removed sequences that were not at least 90% similar to any Vibrio hsp60 sequence in our boutique Vibrio database, which was derived from 106 different Vibrio species. The 90% threshold was chosen as lower thresholds (i.e., 85 or 80%) were found to include other taxa closely related to Vibrio (i.e., Photobacterium). While the number of sequences not assigned to a Vibrio species in our samples was low, it may be possible that some of these sequences could be unidentified Vibrio species or Vibrio species not in our Vibrio database. To determine the identity of these sequences assigned to the “Vibrio” genus, the representative sequence can be BLASTed against the NCBI database and where necessary, new hsp60 sequences can be added to the Vibrio database.
Conclusion
Most standard approaches for examining Vibrio diversity are constrained by poor taxonomic resolution beyond the genus level. This is often a significant limitation because Vibrio species are often implicated in disease events among both natural populations of marine organisms (Kushmaro et al., 2001; Austin and Zhang, 2006; Rubio-Portillo et al., 2014) and commercially important aquaculture species (Goarant et al., 2000; Becker et al., 2004; Frans et al., 2011; Geng et al., 2014; Vezzulli et al., 2015). Here, a Vibrio-centric hsp60 sequencing assay was created using primers tailored to Vibrio-centric hsp60 and used in combination with a custom-built Vibrio reference dataset including 106 Vibrio species. The sequencing assay was able to successfully identify every Vibrio species included within a mock community constructed with known dilutions of different Vibrio species. Despite an exaggeration in the relative abundance of some species, the Vibrio-centric hsp60 sequencing assay provided greatly superior taxonomic resolution when compared to conventional 16S rRNA sequencing methods. The Vibrio-centric hsp60 sequencing assay was subsequently successfully applied to seawater samples providing better discrimination of Vibrio diversity compared to 16S rRNA amplicon sequencing approaches, highlighting its utility in seawater. Next, the sequencing assay was able to unambiguously identify the Vibrio species that increased in abundance during an oyster mortality event, pinpointing a putative pathogen involved in the deaths of oysters following a simulated marine heatwave. This Vibrio-centric hsp60 sequencing assay offers the potential for high throughput characterization of Vibrio diversity while retaining a highly specific degree of taxonomic resolution in environmental samples, important for dissecting species level community dynamics and their relationship with the environment or disease.
Data Availability Statement
The raw fastq files generated for this study are available under BioProject number PRJNA580513. Data analysis pipeline is available at https://doi.org/10.17605/OSF.IO/4798P.
Author Contributions
WK and NS created the degenerate primers and optimized the PCR. WK, NS, and TK analyzed the data. TG performed the oyster mortality experiment and provided samples. ML and JS conceived and designed the study. WK, NS, ML, and JS wrote the manuscript.
Funding
This research was supported by an Australian Research Council Linkage Project (LP160101785) to JS and ML; a Cooperative Research Centre Project (CRC-P 2016-805; Future Oysters), led by the Australian Seafood Industry Pty Ltd in partnership with a number of Australian research organizations; and Ausgem, a research partnership initiated between the University of Technology Sydney and the New South Wales Department of Primary Industries.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02907/full#supplementary-material
References
Austin, B., and Zhang, X.-H. (2006). Vibrio harveyi: a significant pathogen of marine vertebrates and invertebrates. Lett. Appl. Microbiol. 43, 119–124. doi: 10.1111/j.1472-765X.2006.01989.x
Becker, P., Gillan, D., Lanterbecq, D., Jangoux, M., Rasolofonirina, R., Rakotovao, J., et al. (2004). The skin ulceration disease in cultivated juveniles of Holothuria scabra (Holothuroidea, Echinodermata). Aquaculture 242, 13–30. doi: 10.1016/j.aquaculture.2003.11.018
Ben-Haim, Y., and Rosenberg, E. (2002). A novel Vibrio sp. pathogen of the coral Pocillopora damicornis. Mar. Biol. 141, 47–55. doi: 10.1007/s00227-002-0797-6
Brinker, A., Pfeifer, G., Kerner, M. J., Naylor, D. J., Hartl, F. U., and Hayer-Hartl, M. (2001). Dual function of protein confinement in chaperonin-assisted protein folding. Cell 107, 223–233. doi: 10.1016/S0092-8674(01)00517-7
Bruto, M., James, A., Petton, B., Labreuche, Y., Chenivesse, S., Alunno-Bruscia, M., et al. (2017). Vibrio crassostreae, a benign oyster colonizer turned into a pathogen after plasmid acquisition. ISME J. 11, 1043–1052. doi: 10.1038/ismej.2016.162
Cano-Gomez, A., Høj, L., Owens, L., and Andreakis, N. (2011). Multilocus sequence analysis provides basis for fast and reliable identification of Vibrio harveyi-related species and reveals previous misidentification of important marine pathogens. Syst. Appl. Microbiol. 34, 561–565. doi: 10.1016/j.syapm.2011.09.001
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336.
Chimetto, L. A., Brocchi, M., Thompson, C. C., Martins, R. C., Ramos, H. R., and Thompson, F. L. (2008). Vibrios dominate as culturable nitrogen-fixing bacteria of the Brazilian coral Mussismilia hispida. Syst. Appl. Microbiol. 31, 312–319. doi: 10.1016/j.syapm.2008.06.001
Daniels, N. A., and Shafaie, A. (2000). A review of pathogenic Vibrio infections for clinicians. Infect. Med. 17, 665–685.
De Lorgeril, J., Lucasson, A., Petton, B., Toulza, E., Montagnani, C., Clerissi, C., et al. (2018). Immune-suppression by OsHV-1 viral infection causes fatal bacteraemia in Pacific oysters. Nat. Commun. 9:4215. doi: 10.1038/s41467-018-06659-3
Donovan, T. J., and Van Netten, P. (1995). Culture media for the isolation and enumeration of pathogenic Vibrio species in foods and environmental samples. Int. J. Food Microbiol. 26, 77–91. doi: 10.1016/0168-1605(95)00015-C
Dorsch, M., Lane, D., and Stackebrandt, E. (1992). Towards a phylogeny of the genus Vibrio based on 16S rRNA sequences. Int. J. Syst. Bacteriol. 42, 58–63. doi: 10.1099/00207713-42-1-58
Dubert, J., Barja, J. L., and Romalde, J. L. (2017). New insights into pathogenic vibrios affecting bivalves in hatcheries: present and future prospects. Front. Microbiol. 8:762–762. doi: 10.3389/fmicb.2017.00762
Duperthuy, M., Schmitt, P., Garzón, E., Caro, A., Rosa, R. D., Le Roux, F., et al. (2011). Use of OmpU porins for attachment and invasion of Crassostrea gigas immune cells by the oyster pathogen Vibrio splendidus. Proc. Natl. Acad. Sci. U.S.A. 108, 2993–2998. doi: 10.1073/pnas.1015326108
Elston, R. A., Hasegawa, H., Humphrey, K. L., Polyak, I. K., and Hase, C. C. (2008). Re-emergence of Vibrio tubiashii in bivalve shellfish aquaculture: severity, environmental drivers, geographic extent and management. Dis. Aqua. Organ. 82, 119–134. doi: 10.3354/dao01982
Farr, G. W., Furtak, K., Rowland, M. B., Ranson, N. A., Saibil, H. R., Kirchhausen, T., et al. (2000). Multivalent binding of nonnative substrate proteins by the chaperonin GroEL. Cell 100, 561–573. doi: 10.1016/S0092-8674(00)80692-3
Frans, I., Michiels, C. W., Bossier, P., Willems, K., Lievens, B., and Rediers, H. (2011). Vibrio anguillarum as a fish pathogen: virulence factors, diagnosis and prevention. J. Fish Dis. 34, 643–661. doi: 10.1111/j.1365-2761.2011.01279.x
Garnier, M., Labreuche, Y., Garcia, C., Robert, M., and Nicolas, J. L. (2007). Evidence for the involvement of pathogenic bacteria in summer mortalities of the pacific oyster Crassostrea gigas. Microb. Ecol. 53, 187–196. doi: 10.1007/s00248-006-9061-9
Gay, M., Renault, T., Pons, A. M., and Le Roux, F. (2004). Two Vibrio splendidus related strains collaborate to kill Crassostrea gigas: taxonomy and host alterations. Dis. Aqua. Organ. 62, 65–74. doi: 10.3354/dao062065
Geng, Y., Liu, D., Han, S., Zhou, Y., Wang, K. Y., Huang, X. L., et al. (2014). Outbreaks of vibriosis associated with Vibrio mimicus in freshwater catfish in China. Aquaculture 433, 82–84. doi: 10.1016/j.aquaculture.2014.05.053
Go, J., Deutscher, A., Spiers, Z., Dahle, K., Kirkland, P., and Jenkins, C. (2017). An investigation into mass mortalities of unknown aetiology in Pacific oysters, Crassostrea gigas, in Port Stephens, New South Wales, Australia. Dis. Aqua. Organ. 125, 227–242. doi: 10.3354/dao03146
Goarant, C., Herlin, J., Brizard, R., Marteau, A.-L., Martin, C., and Martin, B. (2000). Toxic factors of Vibrio strains pathogenic to shrimp. Dis. Aqua. Organ. 40, 101–107. doi: 10.3354/dao040101
Golosova, O., Henderson, R., Vaskin, Y., Gabrielian, A., Grekhov, G., Nagarajan, V., et al. (2014). Unipro UGENE NGS pipelines and components for variant calling, RNA-seq and ChIP-seq data analyses. PeerJ 2:e644. doi: 10.7717/peerj.644
Gomez-Gil, B., Soto-Rodriguez, S., Garcia-Gasca, A., Roque, A., Vazquez-Juarez, R., Thompson, F. L., et al. (2004). Molecular identification of Vibrio harveyi-related isolates associated with diseased aquatic organisms. Microbiology 150, 1769–1777. doi: 10.1099/mic.0.26797-0
Green, T. J., Siboni, N., King, W. L., Labbate, M., Seymour, J. R., and Raftos, D. (2019). Simulated marine heat wave alters abundance and structure of Vibrio populations associated with the pacific oyster resulting in a mass mortality event. Microb. Ecol. 77, 736–747. doi: 10.1007/s00248-018-1242-9
Grimes, D. J., Johnson, C. N., Dillon, K. S., Flowers, A. R., Noriea, N. F., and Berutti, T. (2009). What genomic sequence information has revealed about Vibrio Ecology in the ocean—a review. Microb. Ecol. 58, 447–460. doi: 10.1007/s00248-009-9578-9
Grisez, L., Reyniers, J., Verdonck, L., Swings, J., and Ollevier, F. (1997). Dominant intestinal microflora of sea bream and sea bass larvae, from two hatcheries, during larval development. Aquaculture 155, 387–399. doi: 10.1016/S0044-8486(97)00113-0
Hammer, Ø, Harper, D. A. T., and Ryan, P. D. (2001). Past: paleontological statistics software package for education and data analysis. Palaeontol. Electr. 4, 1–9.
Hemmingsen, S. M., Woolford, C., Van Der Vies, S. M., Tilly, K., Dennis, D. T., Georgopoulos, C. P., et al. (1988). Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature 333:330. doi: 10.1038/333330a0
Herlemann, D. P., Labrenz, M., Jurgens, K., Bertilsson, S., Waniek, J. J., and Andersson, A. F. (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–1579. doi: 10.1038/ismej.2011.41
Hill, J. E., Penny, S. L., Crowell, K. G., Goh, S. H., and Hemmingsen, S. M. (2004). cpnDB: a chaperonin sequence database. Genome Res. 14, 1669–1675. doi: 10.1101/gr.2649204
Hood, M. A., and Winter, P. A. (1997). Attachment of Vibrio cholerae under various environmental conditions and to selected substrates. FEMS Microbiol. Ecol. 22, 215–223. doi: 10.1111/j.1574-6941.1997.tb00373.x
Hunt, D. E., Gevers, D., Vahora, N. M., and Polz, M. F. (2008). Conservation of the chitin utilization pathway in the Vibrionaceae. Appl. Environ. Microbiol. 74, 44–51. doi: 10.1128/AEM.01412-07
Huq, A., Small, E. B., West, P. A., Huq, M. I., Rahman, R., and Colwell, R. R. (1983). Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 45, 275–283.
Jenkins, C., Hick, P., Gabor, M., Spiers, Z., Fell, S. A., Gu, X., et al. (2013). Identification and characterisation of an ostreid herpesvirus-1 microvariant (OsHV-1 micro-var) in Crassostrea gigas (Pacific oysters) in Australia. Dis. Aquat. Organ. 105, 109–126. doi: 10.3354/dao02623
Jesser, K. J., and Noble, R. T. (2018). Vibrio ecology in the Neuse River Estuary, North Carolina, characterized by Next-Generation Amplicon Sequencing of the Gene Encoding Heat Shock Protein 60 (hsp60). Appl. Environ. Microbiol. 84:e333-18. doi: 10.1128/aem.00333-18
Kahlke, T., and Ralph, P. J. (2019). BASTA – Taxonomic classification of sequences and sequence bins using last common ancestor estimations. Methods Ecol. Evol. 10, 100–103. doi: 10.1111/2041-210x.13095
Katoh, K., Rozewicki, J., and Yamada, K. D. (2017). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
King, W. L., Jenkins, C., Seymour, J. R., and Labbate, M. (2019a). Oyster disease in a changing environment: decrypting the link between pathogen, microbiome and environment. Mar. Environ. Res. 143, 124–140. doi: 10.1016/j.marenvres.2018.11.007
King, W. L., Siboni, N., and Kahlke, T. (2019b). A new high throughput sequencing assay for characterising the diversity of natural Vibrio communities and its application to a Pacific oyster mortality event - data analysis workflow. Open Sci. Framework. doi: 10.17605/OSF.IO/4798P (accessed November 4, 2019).
Kushmaro, A., Banin, E., Loya, Y., Stackebrandt, E., and Rosenberg, E. (2001). Vibrio shiloi sp. nov., the causative agent of bleaching of the coral Oculina patagonica. Int. J. Syst. Evol. Microbiol. 51, 1383–1388. doi: 10.1099/00207713-51-4-1383
Kwok, A. Y. C., Wilson, J. T., Coulthart, M., Ng, L. K., Mutharia, L., and Chow, A. W. (2002). Phylogenetic study and identification of human pathogenic Vibrio species based on partial hsp60 gene sequences. Can. J. Microbiol. 48, 903–910. doi: 10.1139/w02-089
Lafferty, K. D., Harvell, C. D., Conrad, J. M., Friedman, C. S., Kent, M. L., Kuris, A. M., et al. (2015). Infectious diseases affect marine fisheries and aquaculture economics. Annu. Rev. Mar. Sci. 7, 471–496. doi: 10.1146/annurev-marine-010814-015646
Lane, D. J., Pace, B., Olsen, G. J., Stahl, D. A., Sogin, M. L., and Pace, N. R. (1985). Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci.U.S.A. 82, 6955–6959. doi: 10.1073/pnas.82.20.6955
Le Roux, F., Wegner, K. M., and Polz, M. F. (2016). Oysters and vibrios as a model for disease dynamics in Wild Animals. Trends Microbiol. 24, 568–580. doi: 10.1016/j.tim.2016.03.006
Lee, K.-H., and Ruby, E. G. (1994). Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl. Environ. Microbiol. 60, 1565–1571.
Lemire, A., Goudenege, D., Versigny, T., Petton, B., Calteau, A., Labreuche, Y., et al. (2015). Populations, not clones, are the unit of vibrio pathogenesis in naturally infected oysters. ISME J. 9, 1523–1531. doi: 10.1038/ismej.2014.233
Lin, B., Wang, Z., Malanoski, A. P., O’grady, E. A., Wimpee, C. F., Vuddhakul, V., et al. (2010). Comparative genomic analyses identify the Vibrio harveyi genome sequenced strains BAA-1116 and HY01 as Vibrio campbellii. Environ. Microbiol. Rep. 2, 81–89. doi: 10.1111/j.1758-2229.2009.00100.x
Lokmer, A., and Wegner, K. M. (2015). Hemolymph microbiome of Pacific oysters in response to temperature, temperature stress and infection. ISME J. 9, 670–682. doi: 10.1038/ismej.2014.160
Luna-González, A., Maeda-Martínez, A. N., Sainz, J. C., and Ascencio-Valle, F. (2002). Comparative susceptibility of veliger larvae of four bivalve mollusks to a Vibrio alginolyticus strain. Dis. Aqua. Organ. 49, 221–226.
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Miyashiro, T., and Ruby, E. G. (2012). Shedding light on bioluminescence regulation in Vibrio fischeri. Mol. Microbiol. 84, 795–806. doi: 10.1111/j.1365-2958.2012.08065.x
Nagpal, M. L., Fox, K. F., and Fox, A. (1998). Utility of 16S–23S rRNA spacer region methodology: how similar are interspace regions within a genome and between strains for closely related organisms? J. Microbiol. Methods 33, 211–219. doi: 10.1016/S0167-7012(98)00054-2
Nyholm, S., and Nishiguchi, M. (2008). The evolutionary ecology of a sepiolid squid-Vibrio association: from cell to environment. Vie. Milieu 58:175.
Pascual, J., Macián, M. C., Arahal, D. R., Garay, E., and Pujalte, M. J. (2010). Multilocus sequence analysis of the central clade of the genus Vibrio by using the 16S rRNA, recA, pyrH, rpoD, gyrB, rctB and toxR genes. Int. J. Syst. Evol. Microbiol. 60, 154–165. doi: 10.1099/ijs.0.010702-0
Poretsky, R., Rodriguez-R, L. M., Luo, C., Tsementzi, D., and Konstantinidis, K. T. (2014). Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One 9:e93827. doi: 10.1371/journal.pone.0093827
Preheim, S. P., Boucher, Y., Wildschutte, H., David, L. A., Veneziano, D., Alm, E. J., et al. (2011). Metapopulation structure of Vibrionaceae among coastal marine invertebrates. Environ. Microbiol. 13, 265–275. doi: 10.1111/j.1462-2920.2010.02328.x
Raguenes, G., Christen, R., Guezennec, J., Pignet, P., and Barbier, G. (1997). Vibrio diabolicus sp. nov., a new polysaccharide-secreting organism isolated from a deep-sea hydrothermal vent polychaete annelid, Alvinella pompejana. Int. J. Syst. Bacteriol. 47, 989–995. doi: 10.1099/00207713-47-4-989
Richards, G. P., Bono, J. L., Watson, M. A., and Needleman, D. S. (2014). Complete genome sequence for the shellfish pathogen Vibrio coralliilyticus RE98 Isolated from a shellfish hatchery. Genome Announc. 2:e1253-14. doi: 10.1128/genomeA.01253-14
Richards, G. P., Watson, M. A., Needleman, D. S., Church, K. M., and Hase, C. C. (2015). Mortalities of Eastern and Pacific oyster Larvae caused by the pathogens Vibrio coralliilyticus and Vibrio tubiashii. Appl. Environ. Microbiol. 81, 292–297. doi: 10.1128/aem.02930-14
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Rubio-Portillo, E., Yarza, P., Peñalver, C., Ramos-Esplá, A. A., and Antón, J. (2014). New insights into Oculina patagonica coral diseases and their associated Vibrio spp. communities. ISME J. 8, 1794–1807. doi: 10.1038/ismej.2014.33
Saulnier, D., De Decker, S., Haffner, P., Cobret, L., Robert, M., and Garcia, C. (2010). A large-scale epidemiological study to identify bacteria pathogenic to Pacific Oyster Crassostrea gigas and correlation between virulence and metalloprotease-like activity. Microb. Ecol. 59, 787–798. doi: 10.1007/s00248-009-9620-y
Sawabe, T., Ogura, Y., Matsumura, Y., Gao, F., Amin, A., Mino, S., et al. (2013). Updating the Vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio tritonius sp. nov. Front. Microbiol. 4:414. doi: 10.3389/fmicb.2013.00414
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Segarra, A., Pepin, J. F., Arzul, I., Morga, B., Faury, N., and Renault, T. (2010). Detection and description of a particular Ostreid herpesvirus 1 genotype associated with massive mortality outbreaks of Pacific oysters, Crassostrea gigas, in France in 2008. Virus Res. 153, 92–99. doi: 10.1016/j.virusres.2010.07.011
Siboni, N., Balaraju, V., Carney, R., Labbate, M., and Seymour, J. R. (2016). Spatiotemporal dynamics of Vibrio spp. within the Sydney Harbour estuary. Front. Microbiol. 7:460. doi: 10.3389/fmicb.2016.00460
Silvester, R., Alexander, D., Antony, A. C., and Hatha, M. (2017). GroEL PCR- RFLP - An efficient tool to discriminate closely related pathogenic Vibrio species. Microb. Pathog. 105, 196–200. doi: 10.1016/j.micpath.2017.02.029
Simidu, U., and Tsukamoto, K. (1985). Habitat segregation and biochemical activities of marine members of the family vibrionaceae. Appl. Environ. Microbiol. 50, 781–790.
Sugumar, G., Nakai, T., Hirata, Y., Matsubara, D., and Muroga, K. (1998). Vibrio splendidus biovar II as the causative agent of bacillary necrosis of Japanese oyster Crassostrea gigas larvae. Dis. Aqua. Organ. 33, 111–118.
Svitil, A. L., Chadhain, S., Moore, J. A., and Kirchman, D. L. (1997). Chitin degradation proteins produced by the marine bacterium Vibrio harveyi growing on different forms of chitin. Appl. Environ. Microbiol. 63, 408–413.
Szabo, G., Preheim, S. P., Kauffman, K. M., David, L. A., Shapiro, J., Alm, E. J., et al. (2012). Reproducibility of Vibrionaceae population structure in coastal bacterioplankton. ISME J. 7:509. doi: 10.1038/ismej.2012.134
Takahashi, K. G., Nakamura, A., and Mori, K. (2000). Inhibitory effects of ovoglobulins on bacillary necrosis in larvae of the Pacific oyster, Crassostrea gigas. J. Invertebrate Pathol. 75, 212–217. doi: 10.1006/jipa.1999.4922
Thompson, F. L., Austin, B., and Swings, J. (eds). (2006). “The biology of vibrios,” in American Society for Microbiology. (Washington, DC: ASM Press)Google Scholar
Thompson, J. R., Randa, M. A., Marcelino, L. A., Tomita-Mitchell, A., Lim, E., and Polz, M. F. (2004). Diversity and dynamics of a North Atlantic coastal Vibrio community. Appl. Environ. Microbiol. 70, 4103–4110.
Tout, J., Siboni, N., Messer, L. F., Garren, M., Stocker, R., Webster, N. S., et al. (2015). Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis. Front. Microbiol. 6:432. doi: 10.3389/fmicb.2015.00432
Turner, J. W., Tallman, J. J., Macias, A., Pinnell, L. J., Elledge, N. C., Nasr Azadani, D., et al. (2018). Comparative genomic analysis of Vibrio diabolicus and six taxonomic synonyms: a first look at the distribution and diversity of the expanded Species. Front. Microbiol. 9:1893–1893. doi: 10.3389/fmicb.2018.01893
Untergasser, A., Nijveen, H., Rao, X., Bisseling, T., Geurts, R., and Leunissen, J. A. (2007). Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, W71–W74.
Urbanczyk, H., Ogura, Y., and Hayashi, T. (2013). Taxonomic revision of Harveyi clade bacteria (family Vibrionaceae) based on analysis of whole genome sequences. Int. J. Syst. Evol. Microbiol. 63, 2742–2751. doi: 10.1099/ijs.0.051110-0
Urdaci, M. C., Stal, L. J., and Marchand, M. (1988). Occurrence of nitrogen fixation among Vibrio spp. Arch. Microbiol. 150, 224–229. doi: 10.1007/BF00407784
Vezzulli, L., Pezzati, E., Stauder, M., Stagnaro, L., Venier, P., and Pruzzo, C. (2015). Aquatic ecology of the oyster pathogens Vibrio splendidus and Vibrio aestuarianus. Environ. Microbiol. 17, 1065–1080. doi: 10.1111/1462-2920.12484
Waechter, M., Le Roux, F., Nicolas, J. L., Marissal, E., and Berthe, F. (2002). Characterisation of Crassostrea gigas spat pathogenic bacteria. Comp. Rendus Biol. 325, 231–238. doi: 10.1016/S1631-0691(02)01428-2
Watermann, B. T., Herlyn, M., Daehne, B., Bergmann, S., Meemken, M., and Kolodzey, H. (2008). Pathology and mass mortality of Pacific oysters, Crassostrea gigas (Thunberg), in 2005 at the East Frisian coast, Germany. J. Fish Dis. 31, 621–630. doi: 10.1111/j.1365-2761.2008.00953.x
Wendling, C. C., Batista, F. M., and Wegner, K. M. (2014). Persistence, Seasonal Dynamics and Pathogenic Potential of Vibrio Communities from Pacific Oyster Hemolymph. PLoS One 9:e94256. doi: 10.1371/journal.pone.0094256
Yong, L., Guanpin, Y., Hualei, W., Jixiang, C., Xianming, S., Guiwei, Z., et al. (2006). Design of Vibrio 16S rRNA gene specific primers and their application in the analysis of seawater Vibrio community. J. Ocean Univ. China 5, 157–164. doi: 10.1007/BF02919216
Keywords: Vibrio, Vibrio communities, seawater, oyster (Crassostrea gigas), DNA sequencing, marine microbiology, hsp60
Citation: King WL, Siboni N, Kahlke T, Green TJ, Labbate M and Seymour JR (2019) A New High Throughput Sequencing Assay for Characterizing the Diversity of Natural Vibrio Communities and Its Application to a Pacific Oyster Mortality Event. Front. Microbiol. 10:2907. doi: 10.3389/fmicb.2019.02907
Received: 18 September 2019; Accepted: 03 December 2019;
Published: 20 December 2019.
Edited by:
Lasse Riemann, University of Copenhagen, DenmarkReviewed by:
Matthias Labrenz, Leibniz Institute for Baltic Sea Research (LG), GermanyJesus L. Romalde, University of Santiago de Compostela, Spain
Copyright © 2019 King, Siboni, Kahlke, Green, Labbate and Seymour. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maurizio Labbate, TWF1cml6aW8uTGFiYmF0ZUB1dHMuZWR1LmF1; Justin R. Seymour, SnVzdGluLlNleW1vdXJAdXRzLmVkdS5hdQ==
†These authors share first authorship
 William L. King
William L. King Nachshon Siboni
Nachshon Siboni Tim Kahlke
Tim Kahlke Timothy J. Green
Timothy J. Green Maurizio Labbate
Maurizio Labbate Justin R. Seymour
Justin R. Seymour