
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 05 December 2019
Sec. Evolutionary and Genomic Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02826
This article is part of the Research TopicWhat can long-read technology and comparative microbial genomics reveal about evolution and spreading of pathogens under the ONE Health perspective?View all 10 articles
Fluoroquinolones are used to treat serious bacterial infections, including those caused by Escherichia coli and Salmonella enterica. The emergence of plasmid-mediated quinolone resistance (PMQR) represent a new challenge to the successful treatment of Gram-negative infections. As part of a long-term strategy to generate a reference database of closed plasmids from antimicrobial resistant foodborne bacteria, we performed long-read sequencing of 11 E. coli isolates from retail meats that were non-susceptible to ciprofloxacin. Each of the isolates had PMQR genes, including qnrA1, qnrS1, and qnrB19. The four qnrB19 genes were carried on two distinct ColE-type plasmids among isolates from pork chop and ground turkey and were identical to plasmids previously identified in Salmonella. Seven other plasmids differed from any other sequences in GenBank and comprised IncF and IncR plasmids that ranged in size from 48 to 180 kb. These plasmids also contained different combinations of resistance genes, including those conferring resistance to beta-lactams, macrolides, sulfonamides, tetracycline, and heavy metals. Although relatively few isolates have PMQR genes, the identification of diverse plasmids in multiple retail meat sources suggests the potential for further spread of fluoroquinolone resistance, including through co-selection. These results highlight the value of long-read sequencing in characterizing antimicrobial resistance genes of public health concern.
Fluoroquinolones such as ciprofloxacin are critically important antimicrobials in human medicine. They are used to treat serious bacterial infections caused by Gram-negative and Gram-positive organisms (Parry and Threlfall, 2008; Camins et al., 2011). Thus, fluoroquinolone resistance is a public health issue that can lead to treatment failures and the use of alternative agents with greater side effects.
Fluoroquinolones are bactericidal antimicrobials that disrupt DNA replication in bacterial cells. Fluoroquinolone resistance is often mediated by mutations in the DNA gyrase and topoisomerase genes, with gyrA mutations being the most common mechanism in Gram-negative bacteria (Reyna et al., 1995). In recent years, plasmid-mediated quinolone resistance (PMQR) has become more frequent and can spread resistance through horizontal gene transmission. Known genes include qnr, aac(6′)-Ib-cr, qepA, and oqxAB (Ruiz et al., 2012). There are several types of qnr genes, including qnrA, qnrB, qnrC, qnrD, qnrE, qnrS, and qnrVC, which together have over one hundred named alleles (Ruiz, 2019). The presence of these genes differs by bacterial species, with qnrA, qnrB, and qnrS being most common among Salmonella enterica and Escherichia coli in the United States (Tyson et al., 2017b).
Although PMQR often results in only low-level fluoroquinolone resistance, this can then result in continued selection for gyrA mutants with even higher levels of resistance (Kim et al., 2009). Transmissible fluoroquinolone resistance is significant since it can lead to the rapid spread of resistance in bacterial species of public health importance. Cross-species and cross-genus transfer of resistance determinants is also possible. For example, qnrB genes are thought to have originated in Citrobacter spp. (Ribeiro et al., 2015), but have now been found in many pathogens, including E. coli, S. enterica, and Klebsiella pneumoniae, among others (Gay et al., 2006; Jacoby et al., 2006; Robicsek et al., 2006).
The National Antimicrobial Resistance Monitoring System (NARMS) is a One Health surveillance system in the United States that monitors antimicrobial resistance of foodborne pathogens from food animals, retail meats, and humans. Previous NARMS surveillance has found that the prevalence of PMQR genes in the food supply in the United States is low (McDermott et al., 2016), although ColE plasmids carrying qnrB19 have been found in Salmonella from swine sources (Tyson et al., 2017a).
Plasmids containing multiple resistance genes can also co-select for resistance, as the use of one antimicrobial can select for resistance to additional drug classes (Vien le et al., 2012). Thus, it is important to identify and characterize resistance plasmids, particularly those conferring fluoroquinolone resistance.
In this paper, we report the use of long-read sequencing to characterize PMQR plasmids among E. coli isolated from retail meats in the United States, showing the diversity of mechanisms and potential co-selection.
Escherichia coli strains were isolated from routine NARMS retail meat sampling from 13 states in 2015–2017 (NARMS, 2017). Antimicrobial susceptibility testing was performed per CLSI methods, with ciprofloxacin non-susceptibility defined per CLSI M100-S29 as MICs ≥ 0.125 μg/mL (CLSI, 2019).
Genomic DNA was extracted with DNeasy Blood and Tissue Kits (Qiagen, Valencia, CA, United States) per the manufacturer’s instructions. Whole-genome sequencing was performed on the Pacific Biosciences (PacBio) Sequel Sequencer, as previously described (Tyson et al., 2019). Continuous long reads were assembled by the PacBio Hierarchical Genome Assembly Process (HGAP4.8) program. Assembly of the qnrB19 plasmids was done using CLC Genomics Workbench version 10.0.
The closed genomes were annotated by the Prokaryotic Genome Annotation Pipeline (PGAP) version 4.8 (Tatusova et al., 2016). Resistance genes were identified by the NCBI Pathogen Detection Pipeline by AMRFinder (Feldgarden et al., 2019). BLASTn was also used to compare plasmids identified with those in GenBank. Plasmid typing was determined by PlasmidFinder, comparing with the database to known plasmid types with 95% sequence identity and 60% sequence length (Carattoli et al., 2014). Multi-locus sequence typing (MLST) was done with assembled sequences using MLST 2.0 (Larsen et al., 2012), using the E. coli scheme previously described (Wirth et al., 2006). Sequences were submitted to GenBank, with BioSamples in Table 1 and plasmid nucleotide accession numbers listed in Table 2.
From 2015 to 2017, NARMS recovered 3,267 E. coli isolates from retail meat sampling. We performed short-read sequencing on approximately 1,500 of these isolates to identify resistance mechanisms. Most isolates with fluoroquinolone resistance mechanisms had gyrA mutations, comprising 42 isolates. Another 11 isolates lacked gyrase mutations but carried PMQR genes, including qnrA1, qnrS1, and qnrB19. Isolates with these genes were from a variety of sources, including retail chicken, turkey, beef, and pork (Table 1). There were no isolates with both PMQR genes and gyrA mutations.
To further characterize the isolates with PMQR genes, we performed long-read sequencing using Pacific Biosciences technology. From this sequencing, we obtained closed, circular chromosomes and plasmids from each isolate.
Four of the isolates possessed qnrB19 genes, which were found on small plasmids as expected. Long-read sequencing is not optimal for plasmids under 10 kb, so plasmids were closed using short-read sequencing data. We identified two distinct ColE-type plasmids of approximately 3 kb (Table 2) containing these genes. Interestingly, one of the isolates was from ground turkey, and had a different plasmid from the other three isolates, which were from pork chop. The four isolates were genetically distinct, comprising three different E. coli sequence types (STs) (Table 1). The two isolates that were ST10 were from different sources and not within 50 single-nucleotide polymorphisms (SNPs) of each other in the NCBI Isolates Browser (Feldgarden et al., 2019). The two plasmids we found were identical to those identified in swine sources of Salmonella in the United States (Tyson et al., 2017a) and have also been found in E. coli and Salmonella in South America (Pallecchi et al., 2010).
Three isolates had plasmids containing qnrA genes, with two isolates from ground turkey and one from ground beef. The ground turkey isolates were both identified as ST540 from in silico MLST (Larsen et al., 2012), and were 15 SNPs away from each other according to the NCBI Isolates Browser (Feldgarden et al., 2019). These two isolates had nearly identical PMQR plasmids of approximately 126 kb each, indicating likely clonal spread of this strain and its plasmid. These IncF plasmids had limited homology to known plasmids, and contained genes conferring resistance to beta-lactams, aminoglycosides, phenicols, sulfonamides, and tetracycline (Table 2). A graphical representation of one of these plasmids is shown in Figure 1. Interestingly, this plasmid also contains the iroN, iroBCDE, and sitABCD genes, which are involved in iron uptake and may contribute to virulence (Russo et al., 2002; Sabri et al., 2008). Furthermore, the plasmid contains the copB gene, which exports copper and confers copper resistance (Vollmecke et al., 2012), as well as the mer operon, which encodes mercury resistance (Hamlett et al., 1992). Thus, the presence of copper or mercury could co-select for fluoroquinolone resistance.
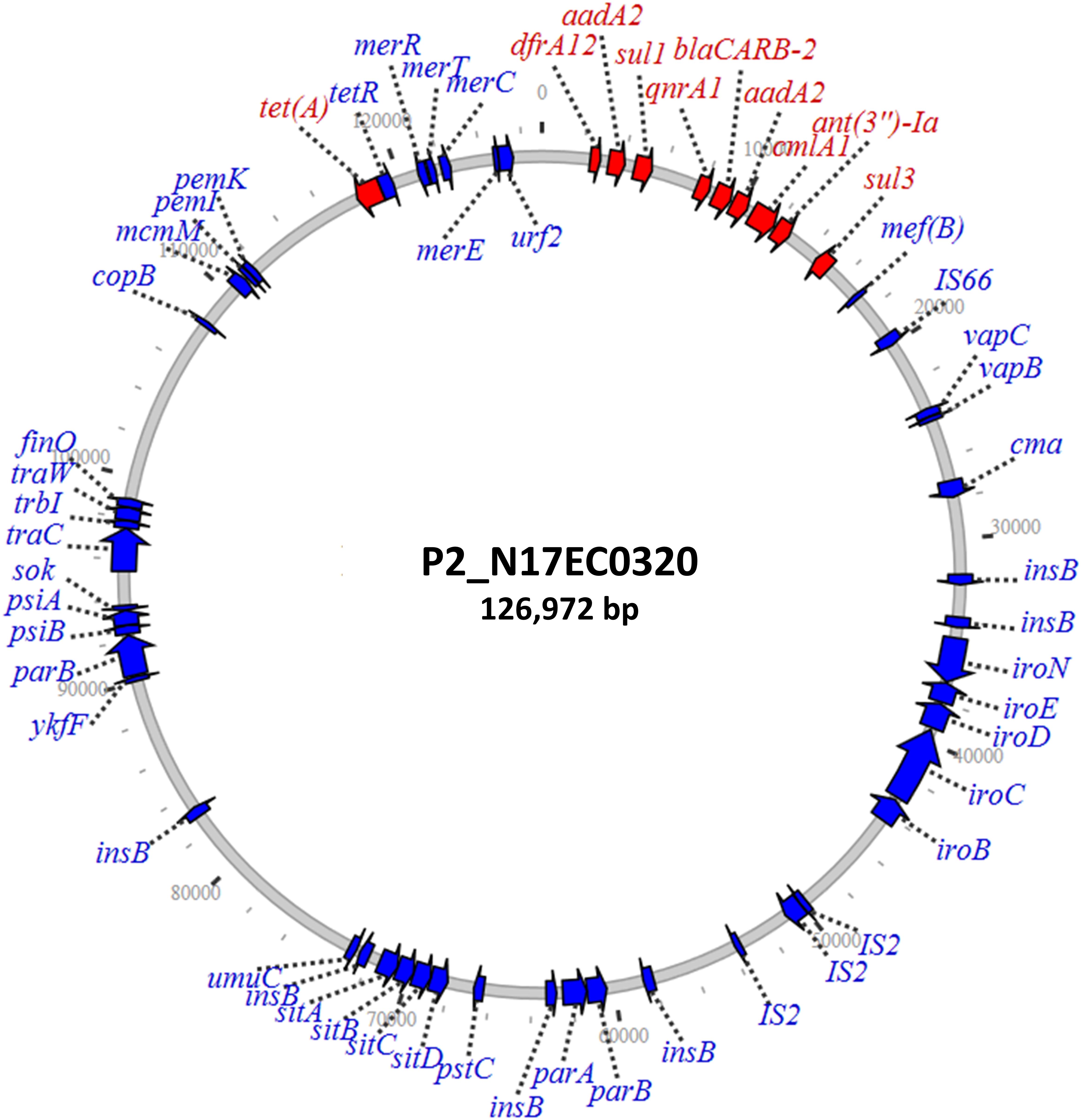
Figure 1. IncF plasmid containing qnrA1 and other resistance genes. Resistance genes are depicted in red, with additional annotated genes depicted in blue.
The qnrA plasmid from the ground beef isolate comprised a 48-kb IncR replicon (Figure 2) with limited similarity to known plasmids. This plasmid had additional genes conferring resistance to beta-lactams, macrolides, phenicols, and sulfonamides. Since fluoroquinolones, beta-lactams, and macrolides are some of the most important antimicrobials used to treat serious Gram-negative infections, potential transfer of this plasmid to other pathogens could compromise the effectiveness of multiple potential therapies. The plasmid also contained the vapBC toxin–antitoxin system, which plays a role in greater plasmid stability (Winther and Gerdes, 2012) that may help with plasmid persistence. This toxin–antitoxin system was also present in the IncF plasmids containing qnrA.
Four isolates had qnrS on unique IncF and IncR plasmids, with sizes from 81 to 180 kb (Table 2). These four isolates included those from retail pork, chicken, and turkey. One of these was an IncF plasmid from N16EC0879 that also had blaCTX–M–55, tetA, and aac(3)-IId (Figure 3). The presence of blaCTX–M–55 is noteworthy, as this is an extended-spectrum beta-lactamase gene that confers resistance to cephalosporins. Since cephalosporins such as ceftriaxone are used in human medicine, the combined transfer of both blaCTX–M–55 and qnrS1 on one plasmid could compromise therapy to multiple drug classes. While all four resistance genes were within a 15-kb fragment on the plasmid, only qnrS1 and aac(3)-IId were on an insertion sequence together, an IS2 element. This plasmid also contained the iron uptake genes sitABCD and iucAC (Gordon et al., 2017), in addition to macAB, which may contribute to virulence (Yamanaka et al., 2008; Turlin et al., 2014). This plasmid also had the copB copper resistance gene, as described in the IncF plasmids with qnrA. The two isolates with qnrS on IncR plasmids also had the sil operon genes, which confer silver resistance (Table 2; Andrade et al., 2018).
Only a subset of the PMQR plasmids had identifiable conjugal transfer genes, such as those in the tra locus (Ilangovan et al., 2015). In fact, only the IncF plasmids had known conjugal transfer genes, indicating that the other plasmids may not be self-transmissible, or may require helper plasmids for successful spread through conjugation.
We report detailed sequence data on fluoroquinolone non-susceptible E. coli from retail meats. This work shows the value of long-read sequencing in de novo characterization of AMR plasmids. Using only short-read sequencing data makes it difficult to accurately identify plasmids or fully characterize them (George et al., 2017). Using short-read sequencing data alone we have accurately identified resistance genes in E. coli (Tyson et al., 2015), but not which are co-located on plasmids. In addition, assemblies from short-read data have difficulty identifying multiple copies of the same gene. For instance, some plasmids in this study had multiple copies of aadA and sul genes, but short-read data assemblies typically only identify single copies of resistance genes (Xia et al., 2017; Su et al., 2019). Understanding the full plasmid structure also helps uncover potential co-selection, including to heavy metals and other biocides. These details are important in assessing the nature of resistant microbial hazards in food and other sources. Fluoroquinolone use is relatively low in food animal production in the United States, but most PMQR plasmids also had genes conferring resistance to tetracycline, which is the highest selling antimicrobial for food animals in the United States (FDA, 2018). This means that tetracycline use in food animals could result in continued selection for fluoroquinolone resistance in E. coli.
Interestingly, E. coli from all retail meat food types had PMQR genes. This contrasts with prior findings of swine as the major contributor to PMQR in retail meat Salmonella (Tyson et al., 2017a). Most plasmids in this study had minimal homology to known plasmids. This demonstrates the importance of increased sequencing of plasmids even in well-studied bacteria such as E. coli, since completely new plasmids are still being discovered.
One interesting finding was that some E. coli had the same qnrB19 plasmids as those in Salmonella from swine and retail pork from NARMS sampling (Tyson et al., 2017a). This reflects a likely transmission of plasmids between E. coli and Salmonella, including in non-swine sources. As observed in Salmonella, these isolates were diverse, indicating little serotype-specific barriers to transmission. These plasmids are small and likely not self-transmissible due to the lack of conjugation genes; each isolate had at least one additional plasmid, indicating a potential method for qnrB19 plasmid transmission.
Bacteria from this study were generic E. coli unlikely to cause foodborne disease in humans, so the direct risk of these bacteria impacting human health or treatment is low. Furthermore, none of the isolates were within 50 SNPs of any human isolates in the NCBI Isolates Browser. Nevertheless, as a source of resistance of human health concern, these bacteria could transfer resistance plasmids to pathogenic E. coli or to other genera. The tracking and reporting of PMQR in these bacteria is essential for a One Health strategy to identify emerging public health threats, and is enhanced by long-read sequencing for de novo characterization of novel plasmids.
The datasets generated for this study can be found in the NCBI BioSamples SAMN12587179, SAMN12587180, SAMN12587181, SAMN12587182, SAMN12587183, SAMN12587184, SAMN102 21061, SAMN10221115, SAMN10221118, SAMN10221255, and SAMN10221523.
GT conceived and coordinated the study. GT and PM wrote the manuscript. CL performed the sequencing and did the sequencing analysis. C-HH did the sequencing analysis and made the figures. SB-J performed the antimicrobial susceptibility testing. All authors contributed to finalizing the manuscript.
This work was funded by the U.S. Food and Drug Administration as part of routine work.
The views expressed in this article are those of the authors and do not necessarily reflect the official policy of the Department of Health and Human Services, the U.S. Food and Drug Administration, or the U.S. Government. Reference to any commercial materials, equipment, or process does not in any way constitute approval, endorsement, or recommendation by the U.S. Food and Drug Administration.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Andrade, L. N., Siqueira, T. E. S., Martinez, R., and Darini, A. L. C. (2018). Multidrug-resistant CTX-M-(15, 9, 2)- and KPC-2-producing Enterobacter hormaechei and Enterobacter asburiae isolates possessed a set of acquired heavy metal tolerance genes including a chromosomal sil Operon (for acquired silver resistance). Front. Microbiol. 9:539. doi: 10.3389/fmicb.2018.00539
Camins, B. C., Marschall, J., DeVader, S. R., Maker, D. E., Hoffman, M. W., and Fraser, V. J. (2011). The clinical impact of fluoroquinolone resistance in patients with E coli bacteremia. J. Hosp. Med. 6, 344–349. doi: 10.1002/jhm.877
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
CLSI, (2019). Performance Standards for Antimicrobial Susceptibility Testing, 29th Edn, Pennsylvania: CLSI.
FDA, (2018). 2017 Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals. Maryland: FDA.
Feldgarden, M., Brover, V., Haft, D. H., Prasad, A. B., Slotta, D. J., Tolstoy, I., et al. (2019). Validating the NCBI AMRFinder tool and resistance gene database using antimicrobial resistance genotype-phenotype correlations in a collection of NARMS isolates. Antimicrob. Agents Chemother. 63:e00483-19. doi: 10.1128/AAC.00483-419
Gay, K., Robicsek, A., Strahilevitz, J., Park, C. H., Jacoby, G., Barrett, T. J., et al. (2006). Plasmid-mediated quinolone resistance in non-Typhi serotypes of Salmonella enterica. Clin. Infect. Dis. 43, 297–304. doi: 10.1086/505397
George, S., Pankhurst, L., Hubbard, A., Votintseva, A., Stoesser, N., Sheppard, A. E., et al. (2017). Resolving plasmid structures in Enterobacteriaceae using the MinION nanopore sequencer: assessment of MinION and MinION/Illumina hybrid data assembly approaches. Microb. Genom. 3:e000118. doi: 10.1099/mgen.0.000118
Gordon, D. M., Geyik, S., Clermont, O., O’Brien, C. L., Huang, S., Abayasekara, C., et al. (2017). Fine-scale structure analysis shows epidemic patterns of clonal complex 95, a cosmopolitan Escherichia coli lineage responsible for extraintestinal infection. mSphere 2:e00168-17. doi: 10.1128/mSphere.00168-117
Hamlett, N. V., Landale, E. C., Davis, B. H., and Summers, A. O. (1992). Roles of the Tn21 merT, merP, and merC gene products in mercury resistance and mercury binding. J. Bacteriol. 174, 6377–6385. doi: 10.1128/jb.174.20.6377-6385.1992
Ilangovan, A., Connery, S., and Waksman, G. (2015). Structural biology of the Gram-negative bacterial conjugation systems. Trends Microbiol. 23, 301–310. doi: 10.1016/j.tim.2015.02.012
Jacoby, G. A., Walsh, K. E., Mills, D. M., Walker, V. J., Oh, H., Robicsek, A., et al. (2006). qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50, 1178–1182. doi: 10.1128/AAC.50.4.1178-1182.2006
Kim, H. B., Park, C. H., Kim, C. J., Kim, E. C., Jacoby, G. A., and Hooper, D. C. (2009). Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrob. Agents Chemother. 53, 639–645. doi: 10.1128/aac.01051-1058
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-6011
McDermott, P. F., Tyson, G. H., Kabera, C., Chen, Y., Li, C., Folster, J. P., et al. (2016). Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal salmonella. Antimicrob. Agents Chemother. 60, 5515–5520. doi: 10.1128/AAC.01030-1016
NARMS, (2017). NARMS Integrated Report: 2015. Laurel, MD: National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS).
Pallecchi, L., Riccobono, E., Sennati, S., Mantella, A., Bartalesi, F., Trigoso, C., et al. (2010). Characterization of small ColE-like plasmids mediating widespread dissemination of the qnrB19 gene in commensal Enterobacteria. Antimicrob. Agents Chemother. 54, 678–682. doi: 10.1128/aac.01160-1169
Parry, C. M., and Threlfall, E. J. (2008). Antimicrobial resistance in typhoidal and nontyphoidal salmonellae. Curr. Opin. Infect. Dis. 21, 531–538. doi: 10.1097/QCO.0b013e32830f453a
Reyna, F., Huesca, M., Gonzalez, V., and Fuchs, L. Y. (1995). Salmonella typhimurium gyrA mutations associated with fluoroquinolone resistance. Antimicrob. Agents Chemother. 39, 1621–1623. doi: 10.1128/aac.39.7.1621
Ribeiro, T. G., Novais, A., Branquinho, R., Machado, E., and Peixe, L. (2015). Phylogeny and comparative genomics unveil independent diversification trajectories of qnrB and genetic platforms within particular Citrobacter species. Antimicrob. Agents Chemother. 59, 5951–5958. doi: 10.1128/AAC.00027-15
Robicsek, A., Strahilevitz, J., Sahm, D. F., Jacoby, G. A., and Hooper, D. C. (2006). qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 50, 2872–2874. doi: 10.1128/AAC.01647-1645
Ruiz, E., Saenz, Y., Zarazaga, M., Rocha-Gracia, R., Martinez-Martinez, L., Arlet, G., et al. (2012). qnr, aac(6′)-Ib-cr and qepA genes in Escherichia coli and klebsiella spp.: genetic environments and plasmid and chromosomal location. J. Antimicrob. Chemother. 67, 886–897. doi: 10.1093/jac/dkr548
Ruiz, J. (2019). Transferable mechanisms of quinolone resistance from 1998 onward. Clin. Microbiol. Rev. 32:e00007-19. doi: 10.1128/cmr.00007-19
Russo, T. A., McFadden, C. D., Carlino-MacDonald, U. B., Beanan, J. M., Barnard, T. J., and Johnson, J. R. (2002). IroN functions as a siderophore receptor and is a urovirulence factor in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 70, 7156–7160. doi: 10.1128/iai.70.12.7156-7160.2002
Sabri, M., Caza, M., Proulx, J., Lymberopoulos, M. H., Bree, A., Moulin-Schouleur, M., et al. (2008). Contribution of the SitABCD, MntH, and FeoB metal transporters to the virulence of avian pathogenic Escherichia coli O78 strain chi7122. Infect. Immun. 76, 601–611. doi: 10.1128/IAI.00789-787
Su, M., Satola, S. W., and Read, T. D. (2019). Genome-based prediction of bacterial antibiotic resistance. J. Clin. Microbiol. 57, 281–285. doi: 10.1128/JCM.01405-1418
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Turlin, E., Heuck, G., Simoes Brandao, M. I., Szili, N., Mellin, J. R., Lange, N., et al. (2014). Protoporphyrin (PPIX) efflux by the MacAB-TolC pump in Escherichia coli. Microbiol. Open 3, 849–859. doi: 10.1002/mbo3.203
Tyson, G. H., Li, C., Ceric, O., Reimschuessel, R., Cole, S., Peak, L., et al. (2019). Complete genome sequence of a carbapenem-resistant Escherichia coli isolate with bla NDM-5 from a dog in the United States. Microbiol. Resour. Announc. 8:e00872-19. doi: 10.1128/mra.00872-819
Tyson, G. H., McDermott, P. F., Li, C., Chen, Y., Tadesse, D. A., Mukherjee, S., et al. (2015). WGS accurately predicts antimicrobial resistance in Escherichia coli. J. Antimicrob. Chemother. 70, 2763–2769. doi: 10.1093/jac/dkv186
Tyson, G. H., Tate, H. P., Zhao, S., Li, C., Dessai, U., Simmons, M., et al. (2017a). Identification of plasmid-mediated quinolone resistance in Salmonella isolated from swine ceca and retail pork chops in the United States. Antimicrob. Agents Chemother. 61:e001318-17. doi: 10.1128/aac.01318-1317
Tyson, G. H., Zhao, S., Li, C., Ayers, S., Sabo, J. L., Lam, C., et al. (2017b). Establishing genotypic cutoff values to measure antimicrobial resistance in Salmonella. Antimicrob. Agents Chemother. 61:e02140-16. doi: 10.1128/aac.02140-2116
Vien le, T. M., Minh, N. N., Thuong, T. C., Khuong, H. D., Nga, T. V., Thompson, C., et al. (2012). The co-selection of fluoroquinolone resistance genes in the gut flora of vietnamese children. PLoS One 7:e42919. doi: 10.1371/journal.pone.0042919
Vollmecke, C., Drees, S. L., Reimann, J., Albers, S. V., and Lubben, M. (2012). The ATPases CopA and CopB both contribute to copper resistance of the thermoacidophilic archaeon Sulfolobus solfataricus. Microbiology 158(Pt 6), 1622–1633. doi: 10.1099/mic.0.055905-55900
Winther, K. S., and Gerdes, K. (2012). Regulation of enteric vapBC transcription: induction by VapC toxin dimer-breaking. Nucleic Acids Res. 40, 4347–4357. doi: 10.1093/nar/gks029
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x
Xia, Y., Li, A. D., Deng, Y., Jiang, X. T., Li, L. G., and Zhang, T. (2017). MinION nanopore sequencing enables correlation between resistome phenotype and genotype of coliform bacteria in municipal sewage. Front. Microbiol. 8:2105. doi: 10.3389/fmicb.2017.02105
Keywords: PacBio, antimicrobial resistance, Escherichia coli, plasmids, fluoroquinolones
Citation: Tyson GH, Li C, Hsu C-H, Bodeis-Jones S and McDermott PF (2019) Diverse Fluoroquinolone Resistance Plasmids From Retail Meat E. coli in the United States. Front. Microbiol. 10:2826. doi: 10.3389/fmicb.2019.02826
Received: 04 September 2019; Accepted: 21 November 2019;
Published: 05 December 2019.
Edited by:
Ludmila Chistoserdova, University of Washington, United StatesReviewed by:
Liang Li, The Lundquist Institute, United StatesCopyright © 2019 Tyson, Li, Hsu, Bodeis-Jones and McDermott. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gregory H. Tyson, R3JlZ29yeS5UeXNvbkBmZGEuaGhzLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.