- 1Aix Marseille Univ, IRD, MEPHI, Faculté de Médecine et de Pharmacie, Marseille, France
- 2IHU Méditerranée Infection, Marseille, France
- 3Assistance Publique des Hôpitaux de Marseille, Marseille, France
- 4Institut des Sciences de l’Évolution, CNRS-IRD-UM2, CC065, Université Montpellier 2, Montpellier, France
Colistin is considered as a last resort antibiotic. The re-use of this antibiotic highlighted the emergence of colistin resistance mediated by chromosomal and plasmidic resistance mechanisms. Five colistin-resistant Klebsiella pneumoniae strains from Laos and Thailand were analyzed by Next Generation Sequencing (NGS) approaches to determine their colistin resistance mechanisms. Antimicrobial susceptibility testing, conjugation and transformation were performed on these strains. Moreover, whole genome sequencing (WGS) combining Illumina (MiSeq) and Oxford Nanopore technologies (MinION) was realized to obtain closed genomes and plasmids. Resistome analyses as well as location of mcr genes and its genetic environments were done in silico. All five strains had colistin MIC of 32 mg/L and were positive for mcr-3 variants including additionally positive for a mcr-8 variant gene. The novel variants were named mcr-3.21, mcr-3.26, mcr-3.28, and mcr-8.3 genes. The mcr-3 variants genes were located on plasmids IncP1, IncFII, and IncI1 type, while mcr-8.3 gene was found on an IncFII type plasmid. The genetic environment of mcr-3.21 and mcr-3.26 genes were composed of a composite transposon ISKpn40- mcr-3-dgkA- ISKpn40. Concerning mcr-8.3 gene, a similar genetic environment of mcr-8.1 gene surrounded by ISIX2 and IS903B was observed. To the best of our knowledge, this is the first description of the novel variants mcr-3.21, mcr-3.26, mcr-3.28 and mcr-8.3 genes as well as the first study on co-occurrence of mcr-3 and mcr-8 genes. Spread and evolution of mcr genes should be monitored.
Introduction
Antibiotic resistance is a major problem worldwide. This phenomenon has required adjustments in therapeutic approaches, including the use of powerful and broad-spectrum antibiotics, such as carbapenems against multi-drug resistant Gram-negative bacteria. The emergence of carbapenem resistant Enterobacteriaceae has led to the re-use of an old antibiotic: colistin (Biswas et al., 2012). This new therapeutic strategy promoted the apparition of colistin resistance mediated by chromosomal and plasmidic resistance mechanisms (Baron et al., 2016). In Klebsiella pneumoniae, mgrB inactivation appears to be the predominant mechanism of chromosomal resistance to colistin. Changes in amino acids in the two-component control systems PmrAB, PhoPQ, and CrrAB are also known to decrease colistin sensitivity by adding 4-amino-4-deoxy-l-arabinose and/or phosphoethanolamine to lipopolysaccharide (Baron et al., 2016). The mcr-1 gene encoding a phosphoethanolamine transferase is the most widespread mechanism of colistin resistance mediated by a plasmid reported in the literature. To date, nine different plasmid-mediated colistin resistance genes have been described, noted from mcr-1 to mcr-9, which are isolated from bacteria in human, animal and environmental samples (Liu et al., 2016; Yin et al., 2017; Wang et al., 2018; Carroll et al., 2019). Although mcr-1 has been mainly reported in Escherichia coli from animal origins, K. pneumoniae can also harbor mcr genes (Yin et al., 2017). In a previous study, we isolated 32 colistin-resistant K. pneumoniae strains from human and animal samples collected in Laos, Thailand and France (Olaitan et al., 2014). 20 of these isolates had an inactivation of the mgrB gene but 12 strains had no colistin resistance mechanisms identified. In this work, we re-analyzed five of these strains by a Next-Generation Sequencing (NGS) approach to finally identify the mechanism of colistin resistance of these isolates.
Materials and Methods
Bacterial Strains and Antimicrobial Susceptibility Testing
Five colistin-resistant K. pneumoniae strains isolated in 2012 from stool samples of healthy humans from Laksip, Laos (LH102-A, LH94, LH375) and from Thailand (TH114, TH164) were reanalyzed by NGS approach to determine their colistin resistance mechanisms. Approval was given by the Ministry of Health Council of Medical Sciences, National Ethics Committee for Health Research (NHCHR), Lao PDR [no.51/NECHR] and by the Ethical Committee of Mahidol University, Bangkok, Thailand [no.0517.1116/661]. Each participant was informed and signed a consent form. Consents are kept at the Rodolphe Mérieux Laboratory in Laos, Vientiane, for Laos and at the Department of Helminthology, Faculty of Tropical Medicine, Mahidol University, Bangkok, for Thailand.
All strains had intact mgrB gene (Olaitan et al., 2014). Antimicrobial susceptibility testing (AST) was determined by minimum inhibitory concentrations (MICs) using microdilution with UMIC test (Biocentric, Bandol, France) for colistin and using the E-test method (bioMérieux, Marcy l’Etoile, France) for others antibiotics in accordance with EUCAST recommendations.
Plasmid Transferability
To confirm mcr genes location biologically, conjugation was performed with an azide-resistant E. coli J53 strain. Selection of transconjuguants was done on MacConkey agar (Beckton Dickinson, Le Pont de Claix, France) supplemented with 120 mg/L sodium azide and 2 mg/L colistin. In case of unsuccessful conjugation, transformation of pure plasmid by electroporation method with Top10 electrocompetents E. coli (Thermo Fisher Scientific, Waltham, MA, United States) was performed and transformants were selected on LB agar (Beckton Dickinson, France) with 2 mg/L colistin. Plasmid curing was performed at high temperature on K. pneumoniae LH94 strain (Letchumanan et al., 2015). Then, the presence of mcr genes and plasmid types were controlled by standard PCR (Carattoli et al., 2005; García-Fernández et al., 2009; Rebelo et al., 2018; Wang et al., 2018).
Genomic Analysis
All strains were sequenced using both MiSeq (Illumina Inc., San Diego, CA, United States) and MinION (Oxford Nanopore Technologies Inc., United Kingdom) technologies. Spades genome assembler was used to assemble long read sequencing data generated by Nanopore and short read data generated by Illumina (Bankevich et al., 2012). Genomes were annotated with RAST (Aziz et al., 2008), resistance gene using both ARG-ANNOT (Gupta et al., 2014) and blastN alignment, and the presence of plasmid was screened using PlasmidFinder software (Carattoli et al., 2014). oriTfinder was used to detect the presence of transfer-related modules of the sequenced plasmids: oriT region, relaxase, T4SS and T4CP (Li et al., 2018). The sequence type (ST) of K. pneumoniae strains was performed in silico by multilocus sequence typing (MLST) analysis using the MLST database (Larsen et al., 2012). The presence of known mutations conferring colistin resistance on the mgrB, pmrA, pmrB, phoP, phoQ, crrA and crrB genes has been also investigated in silico using both nucleic and amino acids alignments. Mutations of mcr genes have been verified by Sanger sequencing. The genetic environment of mcr genes has been reconstituted by comparing the sequence of genes surrounding the mcr-like genes against the NCBI database, using blastX parameter. These environments were also compared to those of mcr-3.1 (KY924928), mcr-3.11 (MG552133) and mcr-8.1 (MG736312) genes. The plasmid pKP91 (MG736312) carrying the first mcr-8.1 gene described in the literature was compared with the plasmid pLH94-8 (CP035204) of the strain LH94 that harbor a mcr-8.3 gene using a pairwise comparison of their Average Nucleotide Identity based on Blast (ANIb) on Jspecies software (Richter et al., 2016). CGViewServer was used to represent the complete sequences of plasmids carrying mcr genes (Stothard et al., 2017).
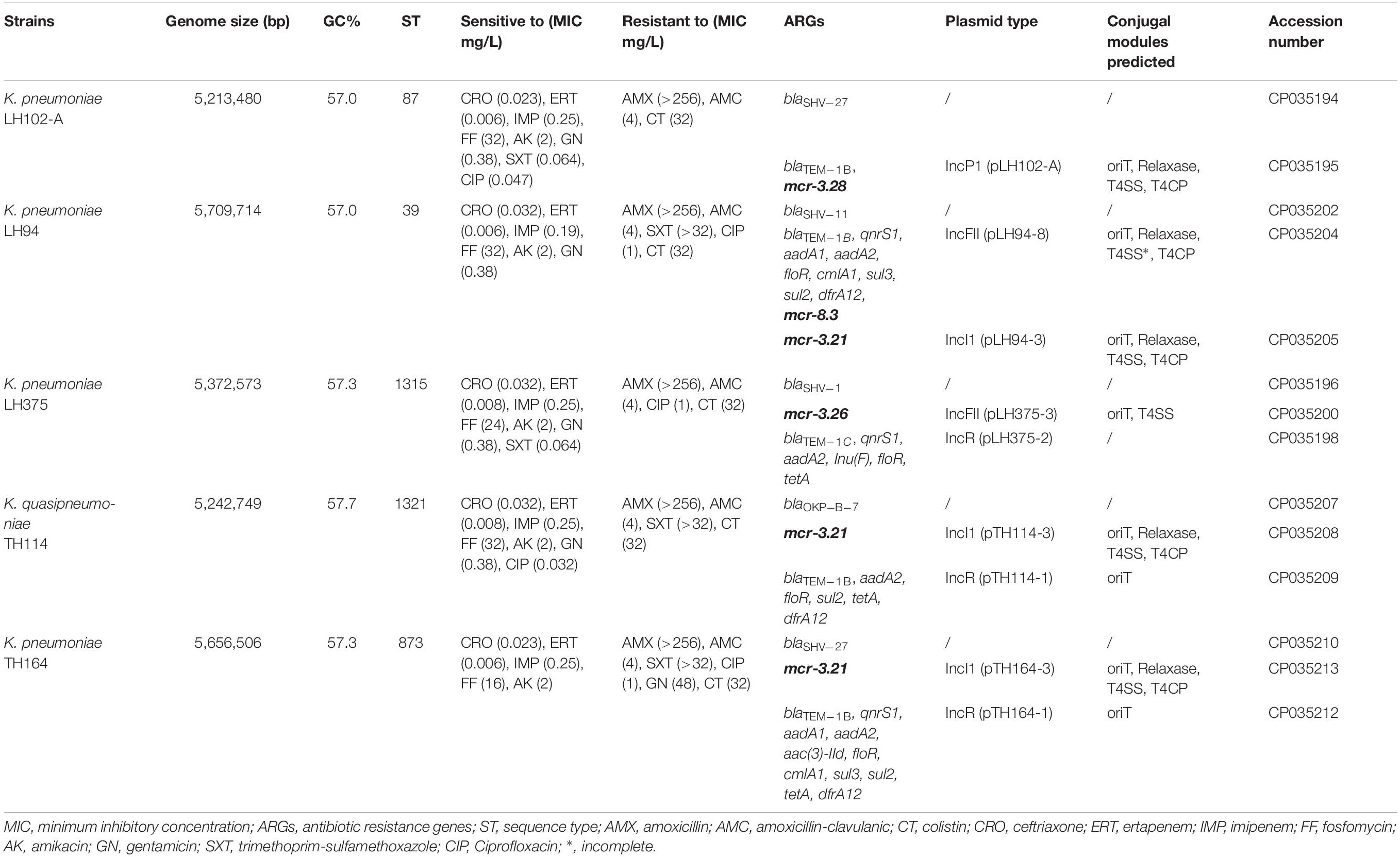
Genomes of LH102-A, LH94, LH375, TH114, and TH164 strains have been submitted to GenBank under accession numbers CP035194-CP035195, CP035202-CP035206, CP035196-CP035201, CP035207-CP035209, and CP035210-CP035213, respectively (Table 1). Novel variants of mcr-3 and mcr-8 genes were detected and assigned by NCBI (Partridge et al., 2018) as mcr-3.21 (AWY10763), mcr-3.26 (AWY10762), mcr-3.28 (AXI82466) and mcr-8.3 (AXI82467) (Figures 1A,B).
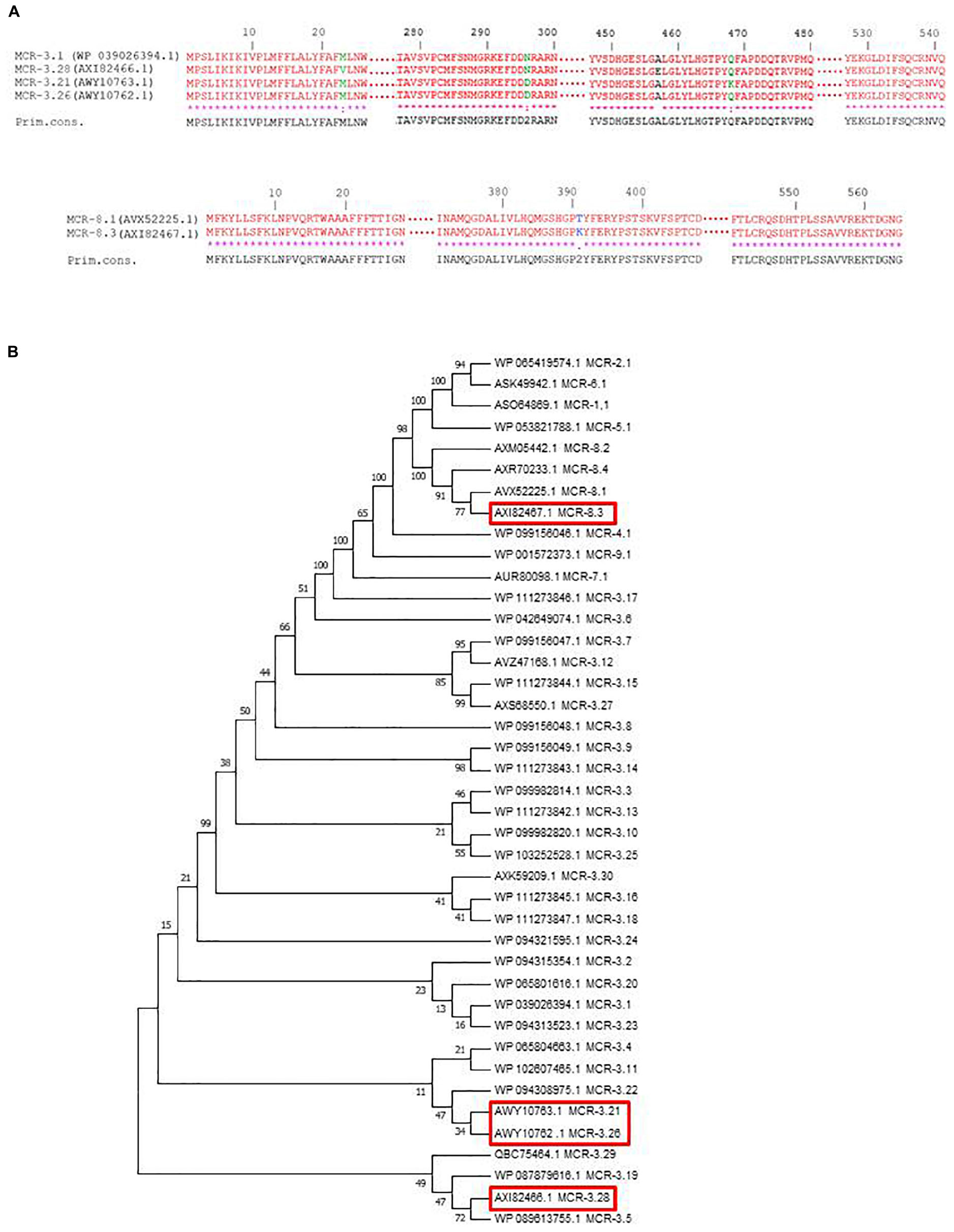
Figure 1. (A) Amino acid sequence alignments of the new MCR-3 and MCR-8 variants using Clustal W on NPS@ website. (B) Phylogenetic tree of MCR variants, including MCR-3 and MCR-8 variants. Sequences were aligned using MUSCLE and phylogenetic interferences were obtained using the neighbor-joining method within the MEGA 7 software. Numbers at the nodes are percentages of bootstrap values obtained by repeating the analysis 500 times to generate a majority consensus tree. Evolutionary distances were computed using the Poisson correction method and are expressed in units of the number of amino acid substitutions per site.
Results
All K. pneumoniae strains had a colistin MIC at 32 mg/L (Table 1). They remain susceptible to ceftriaxone, ertapenem, imipenem, fosfomycin and amikacin but were resistant to amoxicillin and amoxicillin-clavulanic acid. Resistance to trimethoprim-sulfamethoxazole, ciprofloxacin and gentamicin was inconstantly observed (Table 1). The resistome analysis of these strains indicated the presence of genes encoding resistance for beta-lactams (blaSHV, blaOKP–B, blaTEM), aminoglycosides (aadA) sulphonamides (sul), fluoroquinolones (qnrS1), fosfomycin, phenicol (floR), tetracycline (tetA), trimethoprim (dfrA), and colistin (mcr) (Table 1).
Genome size of the five Klebsiella spp. strains ranged from 5,213,480 to 5,709,714 bp with chromosomes and plasmids included for each strain. Blast analysis of RpoB protein and the specific presence of the blaOKP–B–7 gene instead of blaSHV gene allow to reclassify TH114 isolate as a Klebsiella quasipneumoniae strain (Table 1). According to the MLST analysis, each strain belonged to different sequence types (ST39, ST97, ST873, ST1315, and ST1321).
We observed no disruption or known mutations in the mgrB gene or in the different two-component systems pmrAB, phoPQ, and crrAB of these five genomes. However, K. pneumoniae LH102-A and LH375 carried a mcr-3.28 and a mcr-3.26 gene, respectively. The strains LH94, TH164 and TH114 harbored a mcr-3.21 gene, but LH94 isolate also carried mcr-8.3 gene (Table 1). Compared to mcr-3.1 gene, mcr-3.21 displayed two amino acids substitution (N296D and Q468K), while mcr-3.26 had only one amino acid change (N296D). For the mcr-3.28 gene, two mutations (M23V and A457E) were identified. The mcr-8.3 gene is distinguished from mcr-8.1 by an amino acid substitution T391K (Figures 1A,B). In each strain, the mcr-3 gene was present on one plasmid but lastly only conjugate in LH102-A, LH375 and TH114 strains. The transconjuguants and transformants isolates had a colistin MIC of 2 mg/L and were susceptible to antibiotics previously tested except for amoxicillin in a transconjugant LH102-A strain which was consistent with the presence of a blaTEM–1B one the plasmid harboring the mcr gene (Table 1). The mcr-8 and mcr-3 genes of the LH94 strain were not found in the original strain after curing plasmid. In addition, the mcr-8 gene was not detected in transformants of the LH94 strain carrying the mcr-3 gene, which confirms its presence on another plasmid. The presence of potential transfer-related modules, including the origin of transfer (oriT) region, the relaxase gene, T4CP gene and the type IV secretion system gene cluster (T4SS) were detected in some strains (Table 1).
The reconstitution of plasmids was achieved using reads from both Miseq and Minion sequencer. The three mcr-3.21 genes were carried by an IncI1 plasmid type (LH94, TH114 and TH164) whereas the mcr-3.28 gene was harbored by an IncP1 (LH102-A) and the mcr3.26 by an IncFII (LH375). The mcr-8.3 gene was also found on an IncFII plasmid type (LH94) (Table 1). Plasmid structure of the five plasmids carrying mcr-3 genes and the plasmid containing the mcr-8 gene was represented in Figure 2. Plasmids LH94, TH114, and TH164 belonging to IncI1 plasmid type were very similar, except for TH164 that carried more transposases genes around mcr gene. Although the close genetic environment of mcr genes was very similar between the five plasmids, the LH375 and LH102A ones belong to other plasmid type and so were different from the IncI1 plasmids. The genetic environments of mcr-3.21 and mcr-3.26 genes in strains TH114, LH94, TH164, and LH375 were similar and composed of a composite transposon ISKpn40- mcr-3.21-dgkA- ISKpn40 such as mcr-3.11 gene (Figure 3A; Xiang et al., 2018). Concerning the mcr-3.28 gene, it was framed by two mobile elements as follow: TnAs2- mcr-3.28- dgkA-IS26. For mcr-8.3 gene, a similar genetic environment of mcr-8.1 gene was observed (Figure 3B). It was interesting to note the presence of two genes annotated as a response regulator transcription factor CopR for the first one (EQH49_28300) and as a HAMP domain-containing histidine kinase BaeS for the second one (EQH49_28305). The combination of these two protein families generally constitutes a two-component system. These systems are often involved in colistin resistance in Enterobacteriaceae (pmrAB, crrAB or phoPQ). The presence of a two-component system on a plasmid carrying the mcr-8 gene requires further analysis to understand its role in colistin resistance. The plasmids pLH94-8 (CP035204) and pKP91 (MG736312) harboring mcr-8 gene shared 95.42% of identity.
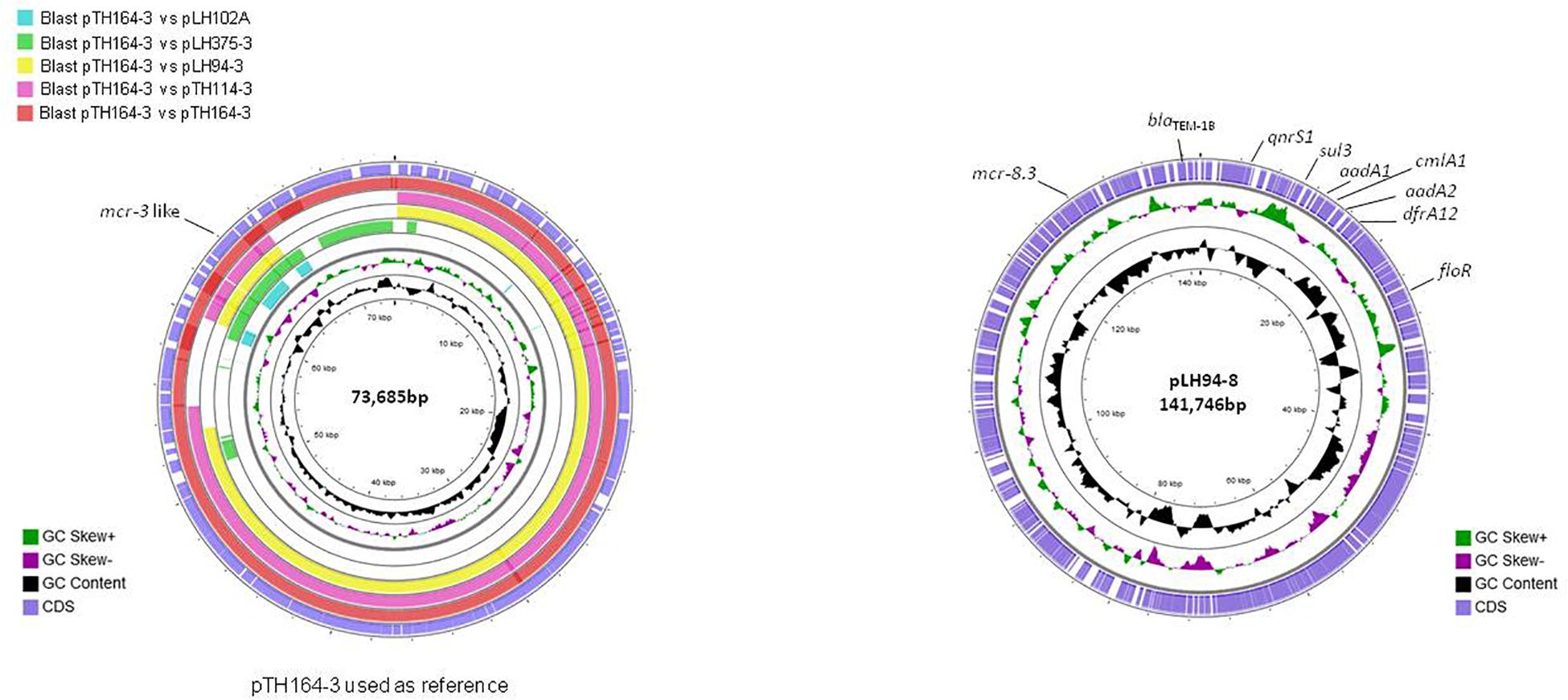
Figure 2. Circular representation of the five complete mcr-3 plasmids (left) and the mcr-8 plasmid (right). Figure constructed using CGView software.
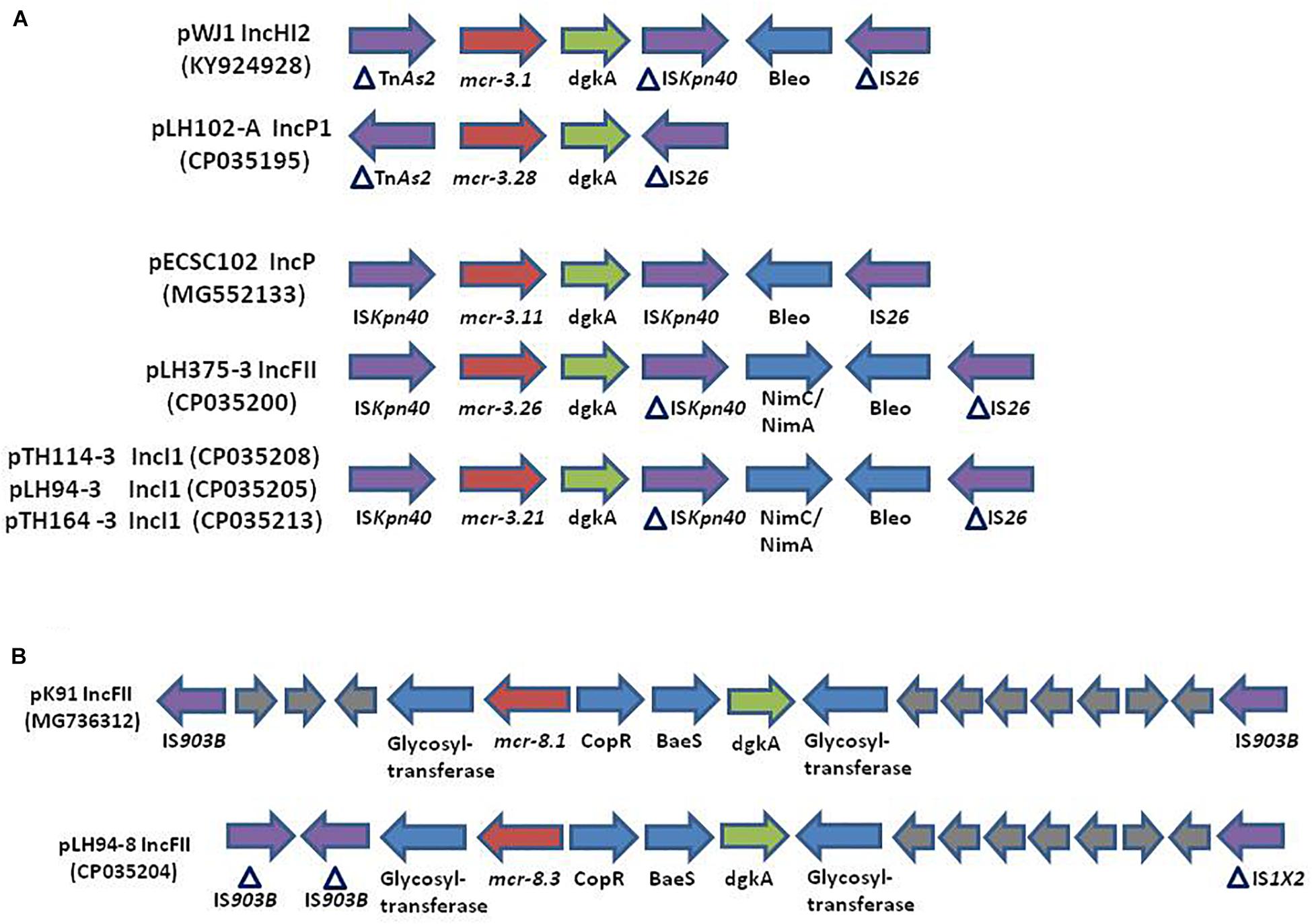
Figure 3. (Not to scale) (A) Comparison of the genetic environment of the mcr-3 gene in plasmids: pWJ1 (KY924928), pLH102-A (CP035195), pECSC102 (MG552133), pLH375-3 (CP035200), pTH114-3 (CP035208), pLH94-3 (CP035205), and pTH164-3 (CP035213). (B) Comparison of the genetic environment of the mcr-8 gene in plasmid pK91 (MG736312) and in the plasmid of strain LH94 carrying mcr-8.3 gene: pLH94-8 (CP035204). HP, hypothetical protein, IS, insertion sequence, Tn, transposon.
Discussion
Following plasmid-mediated colistin resistance gene mcr-1, the mcr-3 gene is the most widely described variants found in the literature, while mcr-2, and mcr-4 to mcr-8 remain uncommon. The MCR-3 protein shares only 32.5% amino acid with MCR-1 which makes impossible its detection with previously described mcr-1 primers (Yin et al., 2017). Since its first description in 2017, thirty variants of mcr-3 gene have been deposit in NCBI and this gene has been observed in different countries and in different strains such as E. coli, Aeromonas spp., and K. pneumoniae (Figure 1B; Yin et al., 2017; Fukuda et al., 2018; Shen et al., 2018).
The presence of K. pneumoniae strains carrying the mcr-3 gene in samples from humans in Thailand has already been described (Yin et al., 2017) but never in Laos. Similarly to the transposon Tn6330 (ISApl1-mcr-1-ORF-ISApl1), a composite transposon ISKpn40- mcr-3-dgkA- ISKpn40 has been reported in several type of plasmids (Xiang et al., 2018). It is known the integration of this type of transposon into a variable region is mediated by insertion sequences (IS), that are then lost to stabilize the integration of the transposon (Snesrud et al., 2018). The genetic environment of our mcr-8.3 gene was similar to that of the mcr-8.1 gene in the reference plasmid pK91 which was isolated from a swine fecal sample in China (Wang et al., 2018). However, unlike plasmid pK91, the pLH94-8 plasmid also harbors other antibiotic resistance genes. The co-occurrence of different antibiotic resistance genes could explain the presence of two mcr genes in a same isolate (Bi et al., 2015). Indeed, this mcr-8 gene could have been selected because it was harbored by a plasmid that also conferred resistance to beta-lactam, aminoglycoside, sulphonamide, fluoroquinolone, phenicol and trimethoprim.
The first description of the mcr-8 gene was done in K. pneumoniae strains isolated from animals and human in 2016 in China. This gene was found on a conjugative IncFII type plasmid (Wang et al., 2018). To date, the mcr-8 gene and its variants have only been described in China in Stenotrophomonas spp., Raoultella ornithinolytica and K. quasipneumoniae from animal and environmental origins (Li et al., 2019; Wang et al., 2019; Yang et al., 2019). Since MCR-8 had only 30.26 to 39.96% identity with other MCR, it is possible that the prevalence of mcr-8 gene is underestimated. Reanalysis of previous works will probably respond to this question. In our study, the detection of a mcr-8.3 gene in a K. pneumoniae strain isolated from a 2012 sample from Laos raises the question of the spread of this gene. Moreover, the emergence of novel variants for mcr-3 and mcr-8 gene indicates that this gene has evolved over time. Furthermore, co-location in the same strain on different plasmids encoding different mcr genes and others antibiotic resistance genes increases the risk of spread of colistin resistance (Long et al., 2019). Antibiotic resistance genes and their transferability are a major risk for human health. The “One health” approach promoting systemic coordination of three sectors: environment, animals and humans could be beneficial for the effective control of global antibiotic resistance problems (Ryu et al., 2017).
To the best of our knowledge, this is the first study describing the novels variants mcr-3.21, mcr-3.26, mcr-3.28, mcr-8.3 genes and demonstrating the co-existence of MCR-3 and MCR-8 in a K. pneumoniae isolate from Laos.
Data Availability Statement
The datasets analyzed in this manuscript are publicly available on GenBank. Requests to access the datasets should be directed tobGluZGEuaGFkamFkakB1bmktYW11LmZy.
Author Contributions
LH wrote the manuscript, performed the experiments, and analyzed the data. SB helped for bioinformatics analysis and to draft the manuscript. AO isolated the strains and helped to draft the manuscript. SM was responsible for the collection of samples and helped to draft the manuscript. J-MR conceived the study, participated in designing and coordination, and helped to draft the manuscript. All authors read and approved the final draft of the manuscript.
Funding
This work was supported by the ANR SEA-EU-NET FarmResist (IZJFZ3-177614) and the French ANR FutureHealthSEA (ANR-17-CE35-0003-02). It was also supported by the French Government under the “Investissements d’avenir” (Investments for the Future) program managed by the Agence Nationale de la Recherche (ANR, fr: National Agency for Research) (reference: Méditerranée Infection 10-IAHU-03), Région Provence-Alpes-Côte d’Azur, and European funding FEDER PRIMI.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank CookieTrad for proofreading the text.
References
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Baron, S., Hadjadj, L., Rolain, J. M., and Olaitan, A. O. (2016). Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int. J. Antimicrob. Agents. 48, 583–591. doi: 10.1016/j.ijantimicag.2016.06.023
Bi, D., Jiang, X., Sheng, Z.-K., Ngmenterebo, D., Tai, C., Wang, M., et al. (2015). Mapping the resistance-associated mobilome of a carbapenem-resistant Klebsiella pneumoniae strain reveals insights into factors shaping these regions and facilitates generation of a ‘resistance-disarmed’ model organism. J. Antimicrob. Chemother. 70, 2770–2774. doi: 10.1093/jac/dkv204
Biswas, S., Brunel, J.-M., Dubus, J.-C., Reynaud-Gaubert, M., and Rolain, J.-M. (2012). Colistin: an update on the antibiotic of the 21st century. Expert Rev. Anti. Infect. Ther. 10, 917–934. doi: 10.1586/eri.12.78
Carattoli, A., Bertini, A., Villa, L., Falbo, V., Hopkins, K. L., and Threlfall, E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., and Wiedmann, M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 10:e852-19. doi: 10.1128/mBio.00853-19
Fukuda, A., Usui, M., Okubo, T., Tagaki, C., Sukpanyatham, N., and Tamura, Y. (2018). Co-harboring of cephalosporin (bla)/colistin (mcr) resistance genes among Enterobacteriaceae from flies in Thailand. FEMS Microbiol. Lett. 365:fny178. doi: 10.1093/femsle/fny178
García-Fernández, A., Fortini, D., Veldman, K., Mevius, D., and Carattoli, A. (2009). Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J. Antimicrob. Chemother. 63, 274–281. doi: 10.1093/jac/dkn470
Gupta, S. K., Padmanabhan, B. R., Diene, S. M., Lopez-Rojas, R., Kempf, M., Landraud, L., et al. (2014). ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58, 212–220. doi: 10.1128/AAC.01310-13
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Letchumanan, V., Chan, K.-G., and Lee, L.-H. (2015). An insight of traditional plasmid curing in Vibrio species. Front. Microbiol. 6:735. doi: 10.3389/fmicb.2015.00735
Li, J., Liu, S., Fu, J., Yin, J., Zhao, J., Zhong, C., et al. (2019). Co-occurrence of colistin and meropenem resistance determinants in a Stenotrophomonas strain isolated from sewage water. Microb. Drug Resist. 25, 317–325. doi: 10.1089/mdr.2018.0418
Li, X., Xie, Y., Liu, M., Tai, C., Sun, J., Deng, Z., et al. (2018). oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 46, W229–W234. doi: 10.1093/nar/gky352
Liu, Y.-Y., Wang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Long, H., Feng, Y., Ma, K., Liu, L., McNally, A., and Zong, Z. (2019). The co-transfer of plasmid-borne colistin-resistant genes mcr-1 and mcr-3.5, the carbapenemase gene blaNDM-5 and the 16S methylase gene rmtB from Escherichia coli. Sci. Rep. 9:696. doi: 10.1038/s41598-018-37125-1
Olaitan, A. O., Diene, S. M., Kempf, M., Berrazeg, M., Bakour, S., Gupta, S. K., et al. (2014). Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int. J. Antimicrob. Agents 44, 500–507. doi: 10.1016/j.ijantimicag.2014.07.020
Partridge, S. R., Di Pilato, V., Doi, Y., Feldgarden, M., Haft, D. H., Klimke, W., et al. (2018). Proposal for assignment of allele numbers for mobile colistin resistance (mcr) genes. J. Antimicrob. Chemother. 73, 2625–2630. doi: 10.1093/jac/dky262
Rebelo, A. R., Bortolaia, V., Kjeldgaard, J. S., Pedersen, S. K., Leekitcharoenphon, P., Hansen, I. M., et al. (2018). Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro. Surveill. 23:17-00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672
Richter, M., Rosselló-Móra, R., Oliver Glöckner, F., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Ryu, S., Kim, B. I., Lim, J.-S., Tan, C. S., and Chun, B. C. (2017). One health perspectives on emerging public health threats. J. Prev. Med. Public Heal. 50, 411–414. doi: 10.3961/jpmph.17.097
Shen, Y., Xu, C., Sun, Q., Schwarz, S., Ou, Y., Yang, L., et al. (2018). Prevalence and genetic analysis of mcr-3 -positive aeromonas species from humans, retail meat, and environmental water samples. Antimicrob. Agents Chemother. 62:e404-18. doi: 10.1128/AAC.00404-18
Snesrud, E., McGann, P., and Chandler, M. (2018). The birth and demise of the ISApl1-mcr-1-ISApl1 composite transposon: the vehicle for transferable colistin resistance. mBio 9:e02381-17. doi: 10.1128/mBio.02381-17
Stothard, P., Grant, J. R., and Van Domselaar, G. (2017). Visualizing and comparing circular genomes using the CGView family of tools. Brief. Bioinform. 20, 1576–1582. doi: 10.1093/bib/bbx081
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 7:122. doi: 10.1038/s41426-018-0124-z
Wang, X., Wang, Y., Zhou, Y., Wang, Z., Wang, Y., Zhang, S., et al. (2019). Emergence of colistin resistance gene mcr-8 and its variant in Raoultella ornithinolytica. Front. Microbiol. 10:228. doi: 10.3389/fmicb.2019.00228
Xiang, R., Liu, B.-H., Zhang, A.-Y., Lei, C.-W., Ye, X.-L., Yang, Y.-X., et al. (2018). Colocation of the polymyxin resistance gene mcr-1 and a variant of mcr-3 on a plasmid in an Escherichia coli isolate from a chicken farm. Antimicrob. Agents Chemother. 62:e00501-18. doi: 10.1128/AAC.00501-18
Yang, X., Liu, L., Wang, Z., Bai, L., and Li, R. (2019). Emergence of mcr-8.2-bearing Klebsiella quasipneumoniae of animal origin. J. Antimicrob. Chemother. 74, 2814–2817. doi: 10.1093/jac/dkz213
Keywords: colistin, mcr, whole genome sequencing, resistance, epidemiology – descriptive
Citation: Hadjadj L, Baron SA, Olaitan AO, Morand S and Rolain J-M (2019) Co-occurrence of Variants of mcr-3 and mcr-8 Genes in a Klebsiella pneumoniae Isolate From Laos. Front. Microbiol. 10:2720. doi: 10.3389/fmicb.2019.02720
Received: 18 July 2019; Accepted: 08 November 2019;
Published: 26 November 2019.
Edited by:
Laura Villa, National Institutes of Health (ISS), ItalyReviewed by:
Jian-Hua Liu, South China Agricultural University, ChinaHong-Yu OU, Shanghai Jiao Tong University, China
Copyright © 2019 Hadjadj, Baron, Olaitan, Morand and Rolain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jean-Marc Rolain, amVhbi1tYXJjLnJvbGFpbkB1bml2LWFtdS5mcg==
 Linda Hadjadj1,2
Linda Hadjadj1,2 Abiola Olumuyiwa Olaitan
Abiola Olumuyiwa Olaitan Serge Morand
Serge Morand Jean-Marc Rolain
Jean-Marc Rolain