- Guangdong Bioengineering Institute (Guangzhou Sugarcane Industry Research Institute), Guangdong Academy of Sciences, Guangzhou, China
Feathers are a major by-product of the poultry industry. They are mainly composed of keratins which have wide applications in different fields. Due to the increasing production of feathers from poultry industries, the untreated feathers could become pollutants because of their resistance to protease degradation. Feathers are rich in amino acids, which makes them a valuable source for fertilizer and animal feeds. Numerous bacteria and fungi exhibited capabilities to degrade chicken feathers by secreting enzymes such as keratinases, and accumulated evidence shows that feather-containing wastes can be converted into value-added products. This review summarizes recent progress in microbial degradation of feathers, structures of keratinases, feather application, and microorganisms that are able to secrete keratinase. In addition, the enzymes critical for keratin degradation and their mechanism of action are discussed. We also proposed the strategy that can be utilized for feather degradation. Based on the accumulated studies, microbial degradation of feathers has great potential to convert them into various products such as biofertilizer and animal feeds.
Introduction
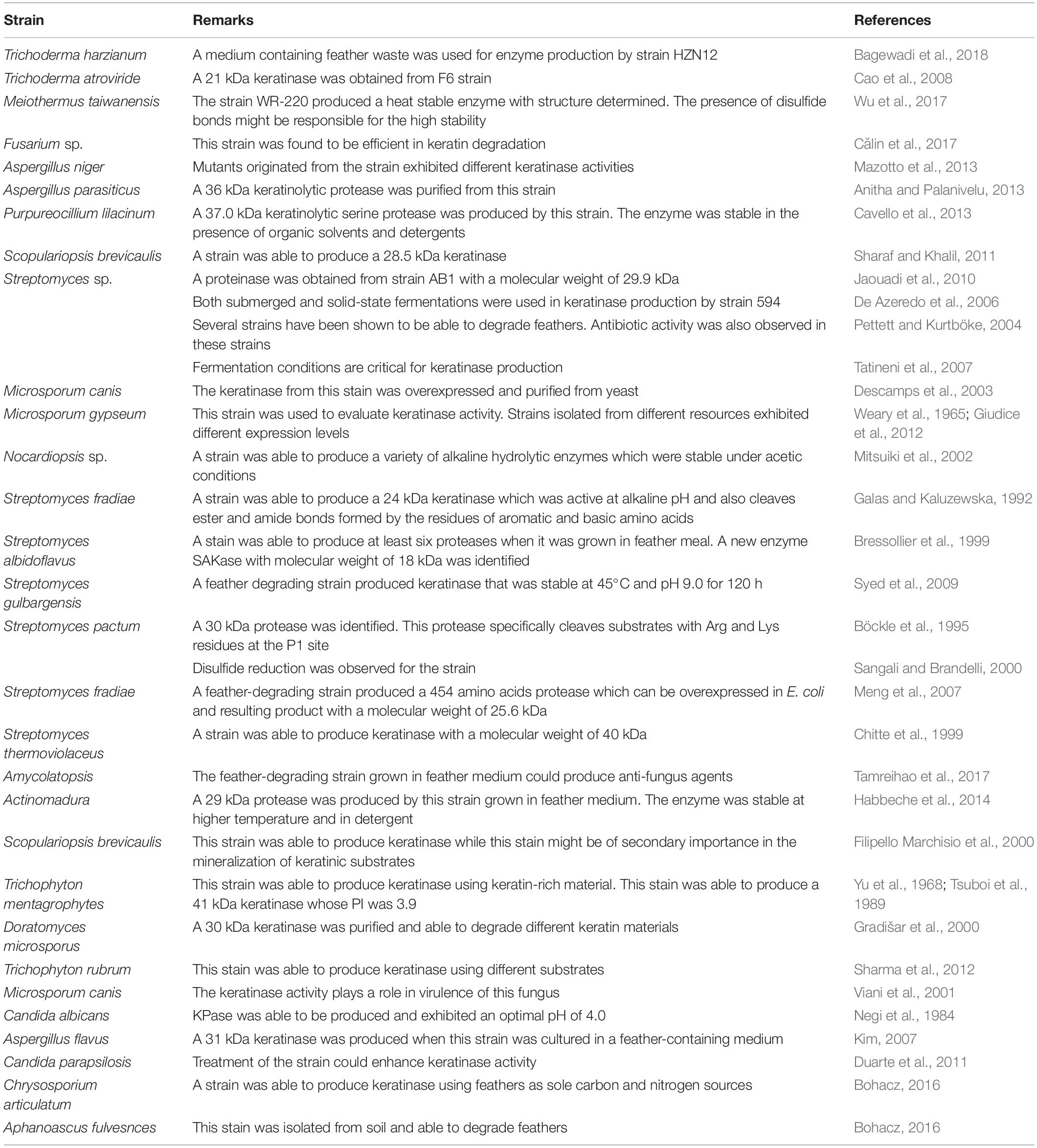
Feathers are an important by-product in poultry industry as they account for 5–7% of the body weight of chicken. It is estimated that approximately several million tons of feathers could be generated annually from poultry industry globally (Verma et al., 2017; da Silva, 2018). Feathers are usually collected and stored at certain areas before further treatment. As feathers might be mixed with blood, meat and grease, the storage conditions such as temperature and duration have to be carefully managed (Tesfaye et al., 2017). Feathers can be disposed by incineration, which is one effective method to destroy conceivable infection agents. Feathers can also be disposed through burial and controlled landfilling while special management is required to keep them from ground water (Tesfaye et al., 2017). Feathers have many applications. As described in a recent review (Tesfaye et al., 2017), feathers such as chicken feathers can be used for decorative applications, medical devices, fertilizer, dusters, bedding materials, and feedstocks (Figure 1) (Papadopoulos et al., 1986). The traditional feather processing methods such as chemical treatment and stem pressure cooking method could convert feathers into animal feeds, but the processes need a large amount of energy and some amino acids are destroyed during treatment (Papadopoulos, 1989; Latshaw et al., 1994; Wang and Parsons, 1997). Although feathers can be utilized as materials in different fields (Shanmugasundaram et al., 2018), a large amount of feathers are still released into the environment without proper treatment. Feathers have become one source of pollutant because of their recalcitrant nature (Brandelli et al., 2015). Untreated feather waste can sustain many pathogenic microorganisms and emit various pollutants such as nitrous oxide, ammonia, and hydrogen sulfide, which are a threat to the environment and people’s health (Tamreihao et al., 2019). Therefore, converting feathers into value-added products using economic methods is of great interest to many researchers (Kang et al., 2018). Accumulated studies have shown that feathers can be efficiently degraded by various microorganisms (Figure 1). Microbial conversion of feathers into value-added products such as biofertilizer and animal feeds should be used in poultry industries (Tamreihao et al., 2019).
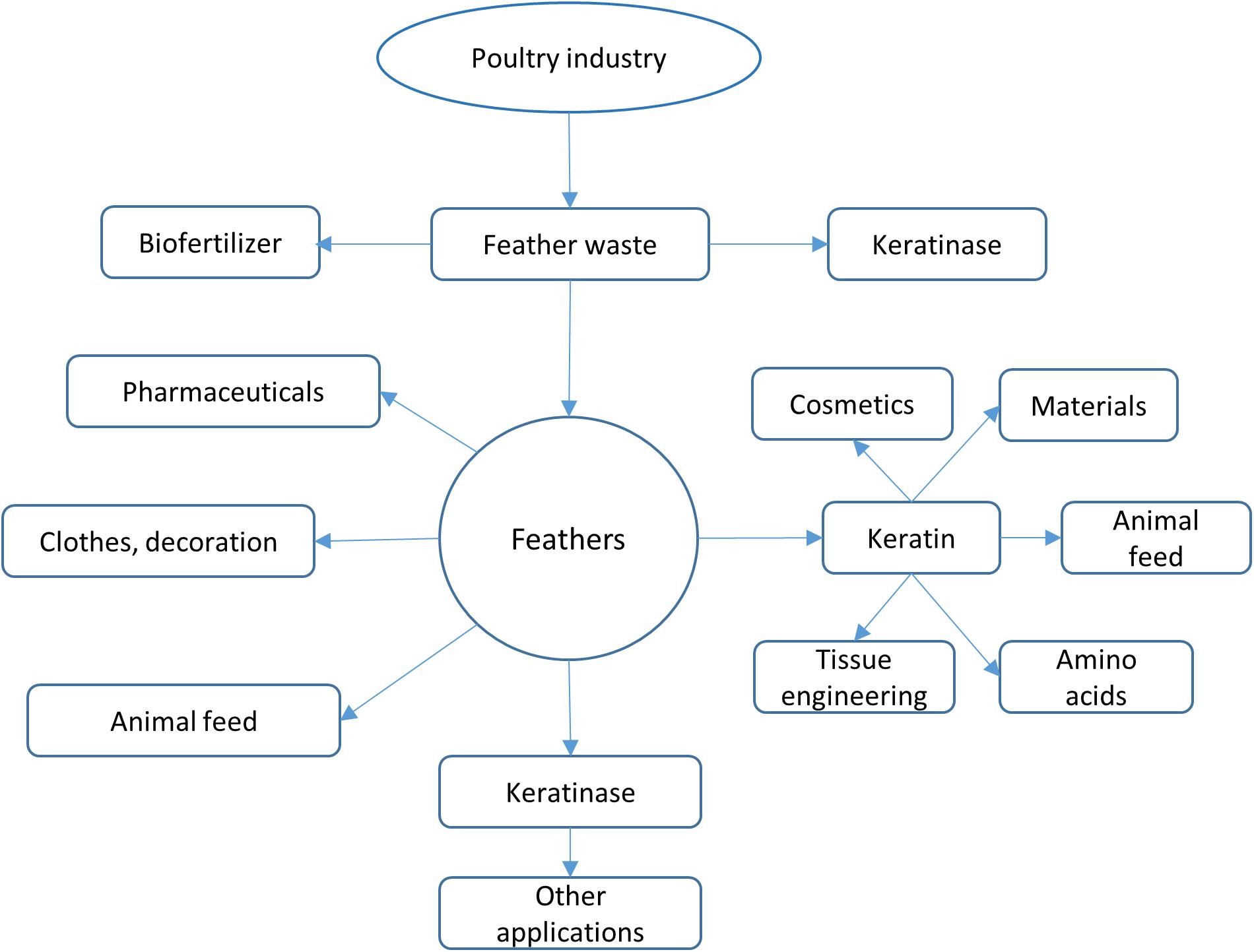
Figure 1. Feather applications. Feathers from poultry industry can be converted into various products. Feathers can be used directly or processed using different ways such as chemical treatment and microbial processing. The most attractive strategy to use feathers is to convert them into value-added products such as biofertilizer and high nutrient animal feeds.
In this review, progress in microbial degradation of feathers is described. Some bacteria and fungi used in feather degradation are summarized. As most studies used only a single strain in feather treatment, using microbial consortium in the treatment might be a more effective way in feather degradation. The enzymes that are secreted by microorganisms and their mechanism of action on protein degradation are discussed. Based on accumulated studies, feathers from poultry industry can be treated in an economic and environmentally friendly way and serve as a valuable source for other applications.
Keratin and Its Structures
The main component (over 90%) of feathers is keratin which is also a key structural element in other organs. Other components such as keratin-associated proteins are present in both feathers and other keratin-rich materials (Adav et al., 2018). Keratin forms fibrous structures and exists widely in nature. It is a recalcitrant structural protein and the third most abundant material after cellulose and chitin (Lange et al., 2016). Keratin can be divided into soft keratin whose cysteine content is less than 10% and hard keratin whose cysteine content is 10–14% (Jin et al., 2017). Hard keratin is normally present in hair, nails, wool, claws, and hooves while soft keratin is present in the epidermis of skin protecting cells from stress. Keratin forms recalcitrant polymers which are insoluble in water and organic solvent, resistant to degradation by the common proteolytic enzymes such as pepsin and trypsin. This is due to the fact that keratin forms high order fibrous structures through disulfide bonds, hydrogen bonds and hydrophobic interactions (Meyers et al., 2008).
Like normal proteins, keratins’ secondary structures contain both α-helix and β-sheet. Keratins can be classified as α-keratins and β-keratins, respectively. β-keratin contains a central domain which is rich in residues favoring to form β-sheet structures associated with the filament framework, an N- and a C-terminal domains which are associated with the matrix and form crosslinking via disulfide bonds (Fraser and Parry, 2011). The structures of β-pleated sheets are present in β-keratins and can form supramolecular fibril bundle. The α-helix coiled and coil structures are assembled in α-keratins and can form intermediate filaments through self-assembling (Lange et al., 2016). The contents of α- and β-keratins are different among various organs (Wu et al., 2015). For example, wool is normally composed of α-keratin (Fang et al., 2013) while feathers contain both keratins (Bodde et al., 2011). Based on analysis, the contents of α- and β-keratins at various position of feathers are different (Ng et al., 2014). The outer rachis of feathers consists mainly β-keratin. As β-keratins have a higher content of cysteine residues than α-keratins, a higher content of disulfide bonds is present (Tesfaye et al., 2017). The formation of covalent bonds is able to stabilize protein structure, which makes it difficult for feathers to be degraded by proteases (Wang et al., 2016).
Posttranslational modifications on keratins such as phosphorylation, sumoylation and glycosylation might also be important for their structure and stability (Snider and Omary, 2014). In some keratins, the uncoiled head structure contains both threonine and serine amino acids which could be the phosphorylation sites. Site-specific phosphorylation of the residues located at this domain could be a major facilitator of intermediate filament reorganization (Snider and Omary, 2014). As keratins can be classified into type I (acidic) and type II (basic) keratins based on the pH at which the proteins are neutral with total charges equal to zero (PI) (Bragulla and Homberger, 2009). Post-translational modification can affect the PI of keratins. Therefore, the conformation of keratins will be changed when the post-translational modification status is altered. If the modification site is mutated, lacking post-translational modification may lead to unsuccessful intermediate filament assembly (Bragulla and Homberger, 2009).
Feather Applications and Process
Keratin can form high-molecular weight complex without chemical modifications, making it attractive to be used as novel materials in many fields (Barone et al., 2005). Feathers from poultry have great potential to be used in several fields as they are one of the important source of keratin (Sharma and Gupta, 2016). As keratins can also be obtained from human resources, their corresponding products can be used as biomedical devices (Dias et al., 2010a). It was shown that keratin material derived from human hair clippings could be used as a new carrier/scaffold for carrying bone morphogenetic protein-2 in bone regeneration (de Guzman et al., 2013). Keratins were considered as a biodegradable material and have been used in wound healing applications (Wang et al., 2017). Keratins from different sources were proven to have biocompatibility with living systems, which makes them have potential applications as novel and valuable materials (Dias et al., 2010b; Ma et al., 2016). Keratins from feathers or other wastes can be converted into amino acids (Hill et al., 2010) which can be used as animal feeds (Onifade et al., 1998). Feathers from chickens were able to be converted into hydrogels for wound healing in a rat model (Wang et al., 2017). In addition, feathers also contain fat, water and minerals such as nitrogen, phosphorus, potassium, calcium, magnesium, iron, manganese, zinc, and copper, which makes them to serve as valuable energy and material sources (Tamreihao et al., 2019). Feathers of course can serve as an important source of amino acids as the keratin sequence consists of high contents of cysteine, glutamine, proline, and serine amino acids (Tesfaye et al., 2017). The contents of histidine, glycine, tryptophan, and glutamic acid are low. The feather hydrolysis products can serve as protein or amino feeds for animals or for microbial growth (Williams et al., 1991). Therefore, feathers can serve as important carbon and nitrogen sources for microbial culture.
Despite the potential applications of keratins derived from keratin-rich materials, feathers from poultry industries are not used efficiently. The feather by-products are normally mixed with other wastes such as blood, meat, skin, fat, and poultry dung (Tesfaye et al., 2017). Although feathers can be converted into feather meal by steam and chemical treatment (Kim and Patterson, 2000), it requires complicated steps in the poultry industry. Feather-containing wastes from poultry industry have to be stored properly before different treatment (Tesfaye et al., 2017). Storage of feather wastes is not a straightforward process as several parameters such as space, temperature and time need to be properly managed. Improper storage can cause growth of bacteria, yeast, fungi, and viruses or the formation of odors such as ammonia and hydrogen sulfide, which can threaten people’s health (Tesfaye et al., 2017). If feather wastes contain other impurities, separation is required before they are sent to process. Therefore, microbial processing of feathers from poultry industries is an economic and efficient way as feather wastes can be processed directly without pretreatment such as separation.
Microbial Degradation of Feathers
Although the nature of keratin-rich wastes such as feathers is resistant to degradation by common proteases, keratins are not accumulated in nature, suggesting that they are degraded by microorganisms (Williams and Shih, 1989). Studies have shown that many microorganisms are able to degrade such wastes by secreting keratinolytic and proteolytic enzymes-keratinases (Williams and Shih, 1989; Tamreihao et al., 2019). These microorganisms include bacteria, actinomycetes, and fungi (Cǎlin et al., 2017; Bohacz and Korniłłowicz-Kowalska, 2019). Numerous microorganisms have been isolated from different environments that are rich in keratin and have been applied to degrade keratin-containing wastes from different resources (Onifade et al., 1998; Chaturvedi et al., 2014).
Bacteria
Quite a few bacteria have been isolated from various environments especially the places rich in keratin-containing materials (Muthusamy et al., 2011). Soil samples from the poultry industry are a good source for isolating feather-degrading bacteria (Kim et al., 2001). The most frequently used method to identify screened strains is 16S rDNA sequencing (Khodayari and Kafilzadeh, 2018). The isolated bacteria were then used in degrading keratin-rich materials such as feathers and wools. Secreted proteases-keratinase by these organisms were responsible for cleaving the keratin proteins. Recent studies suggested that keratinases were functional synergistically with other enzymes. Bacteria have great potential to be used widely as they grow fast and their enzymes sustain the activity under different conditions. Some enzymes are still active at high temperature and at different pHs. Bacillus licheniformis is the most effective keratin-degrading bacterium in the genus (Manczinger et al., 2003). Other bacteria including Bacillus, Stenotrophomonas, Pseudomonas, Brevibacillus, Fusarium, Geobacillus, Chryseobacterium, Xanthomonas, Nesterenkonia, and Serratia are able to produce keratin-degrading enzymes (Table 1). The keratinases produced by bacteria exhibited a wide range of optimal temperature (28–90°C) and pH (5.8–11) (Tamreihao et al., 2019). The molecular weights of bacterial keratinase are different among different species. In addition, adding keratin or feathers in the cultural medium normally facilitates production of keratinase (Herzog et al., 2016), implying that the enzyme production might be an inducible process.
Extensive studies have been carried to isolate different types of keratin-degrading bacteria. Random mutagenesis using ethyl methanesulfonate was carried out to improve the activity of a keratin-degrading bacterium Bacillus subtilis LFB-FIOCRUZ 1266. The mutants exhibited higher feather degradation rate by 15% than the wild type strain. In addition, the mutants showed higher keratinolytic activity and sulfide yield than the wild type strain (de Paiva et al., 2018). Mutation using ultraviolet irradiation and N-methyl-N′-nitro-N-nitrosoguanidine treatment or N-methyl-N′-nitro-N-nitrosoguanidine treatment alone was carried out on B. subtilis. The resulting mutant exhibited higher keratinase activity (Cai et al., 2008) or higher feather degradation efficiency than the wild type. The authors also identified a 45 kDa protease playing important role in feather degradation (Wang et al., 2015). Apparently, random mutagenesis on selected strain is an efficient way to improve feather degradation efficiency, implying multiple enzymes might be involved in the degradation process. Site-directed mutagenesis to improve keratinase activity could also improve feather degradation efficiency (Liu et al., 2013).
Fungi and Actinomycetes
Degradation of keratin-rich material in nature is the result of cooperation of bacteria and other microorganisms (Lange et al., 2016). Fungi and actinomycetes have been found to be able to degrade keratin-rich materials such as feathers. Some fungi are pathogenic and present on the surface of human or animal skins. The keratinases secreted by these fungi are important for their invasion into the body (Gilardi, 1965). These fungi have special structures such as hyphae to facilitate keratin degradation (Korniłłowicz-Kowalska and Bohacz, 2011; Tridico et al., 2014). It was shown that high keratinase activity correlated with fast development of symptoms (Viani et al., 2001). The pathogenic fungi could secret keratinases while they must be avoided in applications due to the safety requirement. Some non-pathogenic fungi exhibited capability to degrade feathers and have potential for being approved for application in animal feed or biofertilizer (Bhange et al., 2016a). The amino acid sequences of keratinases produced by fungi and actinomycetes are different from those produced by bacteria. The keratinase producing fungi have been reviewed recently (Lange et al., 2016) and Table 2 also lists some fungi and actinomycetes that are able to produce keratinases and have potential to be used in feather degradation. In addition to feather degradation, some strains were able to produce antibiotics which inhibited bacterial growth, which has been demonstrated in several actinomycetes (Pettett and Kurtböke, 2004). This type of strain is very useful in feather treatment as feathers from poultry industries may carry different types of pathogens.
Keratinases
The term keratinase (EC3.4.21/24/99.11) is referring to a class of proteases which possess keratinolytic activities (Lange et al., 2016). The identified keratinases are serine or metallo protease with the capability to degrade keratinous proteins (Gupta and Ramnani, 2006; Sahni et al., 2015). The mechanism of action of these enzymes is still not completely understood as these enzymes alone could not degrade native keratins (Gupta and Ramnani, 2006). The keratinases produced by various bacteria and fungi exhibit different characteristics such as amino acid sequence, molecular weight, optimal pH and temperature toward keratins from different origins (Brandelli, 2008). As keratinases are able to cleave insoluble and recalcitrant keratins derived from keratin-rich wastes such as feathers, hair and wool, they have great potential of industrial applications. Accumulated studies have shown that keratinase can be used in several fields (Figure 2) including animal feed (Grazziotin et al., 2006), fertilizers, leather industries, biomedical fields, detergents, cosmetics and materials (Yue et al., 2011; Paul et al., 2016; Su et al., 2019). Keratinase produced by B. licheniformis PWD-1 was found to be able to degrade prions which are infectious agents and resistant to proteolytic and mild protein-destructive processes (Van de Wiel et al., 2003). This enzyme can be used to remove prions present in animal feed. Keratinase will play important roles in agricultural and environmental chemistry due to its ability to degrade keratins from various sources (da Silva, 2018).
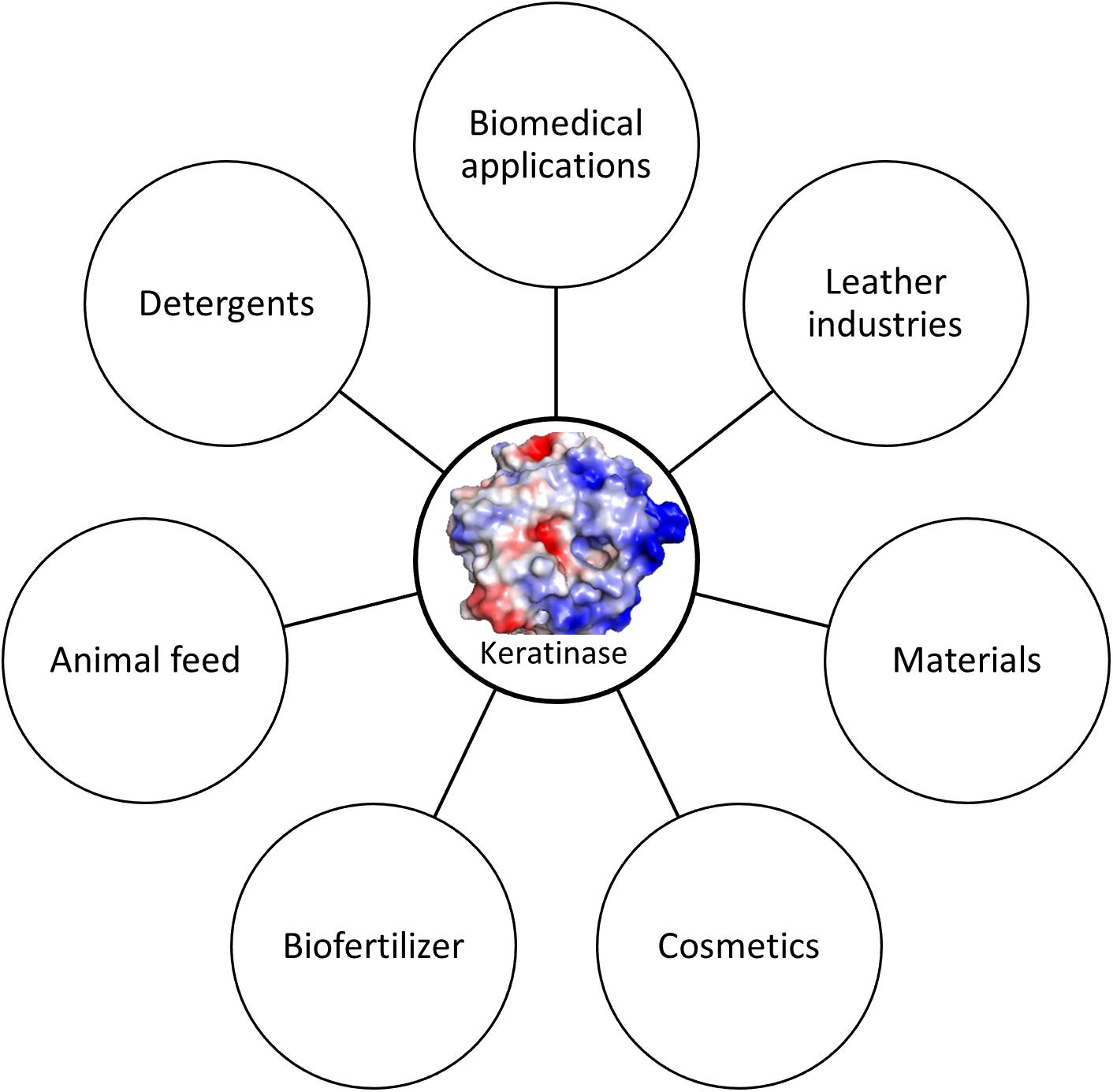
Figure 2. Keratinase applications. Keratinase can be used in different fields. The application can be enlarged when more stable enzymes are available.
Biochemical Analysis of Keratinases
Most researchers purified the enzyme from an organism and characterized the purified products using keratin-derived substrates such as azokeratin, keratin azure, human hair, cow horn, feather and keratin powder derived from different keratins (Gupta and Ramnani, 2006). The optimal enzymatic conditions including buffer pH, temperatures were obtained using these assays (Govarthanan et al., 2015). In addition, substrate specificity were analyzed by using substrates containing different amino acids (Brandelli et al., 2010). Keratinases from both bacteria and fungi exhibited activity under temperatures ranging from 28-90°C (Tamreihao et al., 2019) or even higher (Intagun and Kanoksilapatham, 2017). The enzymes could also sustain its activity at pH from 5 to 13 (Gupta and Ramnani, 2006; Brandelli et al., 2015). Studies also revealed that the enzymes from fungi, bacteria and other extremophiles exhibit higher optimal temperature, which gives rise to high efficiency in keratin degradation (Kanoksilapatham and Intagun, 2017). A heat-stable keratins from Meiothermus taiwanensis WR-220 was able to be over-expressed and purified by recombinant techniques (Wu et al., 2017). Biochemical and structural studies were able to be carried out to understand its mechanism of action. The presence of disulfide bonds in the protease contributes to the thermal stability (Wu et al., 2017). This information is very useful in enzyme engineering as some proteases do not contain cysteine residues. Introducing disulfide bonds will be a strategy to improve enzyme stability.
Recombinant Keratinase
Recombinant DNA technology has been used to clone the genes of keratinases from bacteria and fungi and overexpressed them in bacteria such as Escherichia coli and other cells (Descamps et al., 2003; Khardenavis et al., 2009). Although it is not an economic way to use recombinant keratinases to degrade wastes such as feathers, introducing a tag to facilitate protein purification makes it straightforward for enzyme production with high purity and high yield. This method is also very useful for characterizing enzymes using biochemical, biophysical and structural methods (Fang et al., 2014). In addition, when this method is used in protein engineering, the activity and yield of the mutants are easily evaluated (Fang et al., 2019). With optimized conditions, recombinant production of keratinase is possible to be used for industrial applications.
Structure and Substrate Reorganization Site of Keratinase
Structural studies have been carried out for several keratinases. One of the recent studies revealed the detailed structure of a keratinase (rMtaKer) from M. taiwanensis WR-220 (Wu et al., 2017), paving the way for understanding of mechanism of action of this class of enzymes. Intact rMtaKer consists of signal peptide, N-terminal pro-peptide (N-pro), and mature protease domain with a catalytic triad formed by Asp39, His72, and Ser224. The N-pro region was cleaved when rMtaKer was overexpressed in E. coli, resulting in the protease domain only. The purified rMtaKer exhibited active protease activity against several substrates such as feathers, milk, casein and elastin. The 1.5 Å crystal structure of the protease shows that keratinase is composed of seven-stranded parallel β-sheets with six α-helices and four β-sheets flanking around. The four β-sheets are made of two anti-parallel strands and possess the conserved catalytic triad formed by Asp39, His72, and Ser224.
The keratinase contains two calcium ions that are important for stabilizing the structures. The presence of the metal ions in keratinase explains the fact that many ions are important for affecting enzymatic activity. The first Ca2+ stabilizes the surface loop between α1 and β2, coordinated by oxygen atoms from Asp11, Asp14, Gln15, Ser21, and Thr23. The second Ca2+ exhibits close contacts with oxygen atoms of Val172, Gly175, Thr177, and two water molecules. These two metal ions are also present in other members of the subtilisin superfamily. Two disulfide bonds formed by residues Cys69-Cys101, and Cys165-Cys196 (Figure 3). As the disulfide bond is able to stabilize protein structure, the optimal temperature for rMtaKer is 65°C. The presence of the disulfide bonds might be one of the reasons for the thermal stability. Introducing disulfide bonds to other keratinase in protein engineering will be a strategy to improve protease thermal stability.
Figure 3. Structure and substrate binding of keratinases. (A) Crystal structure of rMtaKer. The structure of rMtaKer (PDB id 5WSL) is shown. The metal ions are shown as spheres. (B) Surface charge representation of rMtaKer. The substrate binding to the active site are shown as sticks. (C) The residues critical for disulfide bond formation are shown. (D) Crystals structure of other keratinase. The crystal structure of a bacterial keratinase (PDB id 1CSE) is shown. No cysteine residue is present in this enzyme.
The substrate specificity of keratinases is still not well known. A study showed that Nesterenkonia sp. AL20 was able to produce alkaline protease using chicken feather as the nutrient source. The protease was able to cleave tetra-peptide substrates with hydrophobic residues at the P1 site (Bakhtiar et al., 2005). In the structure of rMtaKer, self-cleavage was observed. Although rMtaKer is monomeric in solution, protein form oligomers in the crystals, which is due to crystal packing. The residues including Tyr278-Glu279-Asn280-Leu281-Tyr282 occupy the protease active site from the neighboring monomer (Figure 3). This structure reveals the residues that are critical for substrate binding. The protease cleavage sequence from P1 to P4 obtained in this study can be used for determining the optimal protease cleavage site.
Constructing More Active and Stable Enzyme
Keratinases from microorganisms are present in a pre-pro-form in which an inhibitory region suppresses the protease activity (Liu et al., 2014). This inhibitor region acts as an intermolecular chaperone critical for the folding of the mature protease domain by assisting its folding, affecting the structure of the protease domain and temporarily binding to the protease domain before activation through cis or trans modes (Liu et al., 2014). Mutations in the pro-peptide region can affect the folding rate of the protein, which leads to high enzyme production, conformational change of the mature enzyme and acceleration of enzyme maturation. Such studies have been carried out on proteases from several species (Takagi et al., 2001; Fang et al., 2010; Uehara et al., 2013; Su et al., 2019). Computational aided site-directed mutation was carried out for residues at different sites. Mutation of residues (Y94F) was able to improve substrate specificity. Some mutations such as A218S and A218G were able to improve the thermal stability of the enzymes. The mutants carrying mutations at this site exhibited highest activity at 70°C, which will broaden the application of keratinase (Fang et al., 2017). These mutagenesis studies are also helpful for understanding the dynamics and regulation of keratinase (Liu et al., 2013).
Mechanism of Keratin Degradation by Microorganisms
Structural studies on keratinases and feather degradation in vitro suggested that one keratinase is not enough to degrade keratin as keratinases do not have the activity to break the disulfide bonds. Several mechanisms have been proposed and it has been recognized that two steps may be involved in the keratinolytic process- sulfitolysis and proteolysis (Korniłłowicz-Kowalska and Bohacz, 2011). Sulfitolysis is required to cleave disulfide bonds and proteolysis is to cleave the protein (Yamamura et al., 2002b). Removal of the disulfide bonds by enzymes such as sulfide reductases or reducing agents such as sulfite secreted by the strain is critical for conformational changes of keratins, which makes more sites available for keratinase degradation (Lange et al., 2016). A study has shown that the crude enzyme exhibited higher keratin degrading rate than the purified enzyme. At least two enzymes are required for keratin degradation (Yamamura et al., 2002a). One enzyme is responsible for producing reduced keratin with cysteine residues in the reduced form so that its cleavage sites were exposed to protease. The following steps are also proposed in keratin degradation, namely sulfitolysis, proteolysis, and deamination (Yu et al., 1968). Bacteria and fungi exhibit different mechanisms for keratin degradation while their keratinases are able to cleave the polyproteins. In addition to sulfitolysis and proteolysis, mechanical destruction plays important roles in keratin degradation by fungi (Korniłłowicz-Kowalska and Bohacz, 2011). Despite such progress in understanding protease mechanism of action, more studies have to be carried out to identify the enzymes critical for keratin degradation.
Questions Need to Be Solved
Studies on keratinases produced by both bacteria and fungi have been carried out by researchers (Table 1) and reviewed (Gupta and Ramnani, 2006; Brandelli, 2008; Brandelli et al., 2010; Korniłłowicz-Kowalska and Bohacz, 2011; Gupta et al., 2013). Different keratinases have been purified and characterized from various stains (Table 1). Accumulated studies suggest that other enzymes such as disulfide reductase that can break disulfide bonds are important for keratinase degradation. Studies have shown that the whole-cell product has a higher keratin-degrading efficiency than the purified keratinase does. As proposed in a recent review, enzymatic degradation of keratins might be similar to decomposition of cellulose in which several enzymes are required in the process. Further studies identifying new components important for keratins degradation are important for understanding the mechanism of action of keratinases. It has been noted that keratins do not accumulated in the nature, suggesting that they might be degraded through the cooperation of microorganisms such as bacteria and fungi in the environment. Extensive studies have been working on screening strains that are able to degrade feathers or keratins while most studies were focused on a single strain in keratin treatment.
Perspective
Feathers from poultry industries can be a valuable resource instead of a waste or a threat to the environment. Based on accumulated studies, there is no doubt that feathers can be used in diverse fields. Microbial degradation of feathers is a feasible and economic way to make full use of the waste. Converting feathers into biofertilizer and animal feeds is easily applicable to various poultry industries. Despite the efforts spent in this direction, some works are still necessary to be carried out to build up more efficient systems that can be utilized in feather treatment.
Strategies in Strain Screening
Feather degrading pathogenic bacteria and fungi are useful for understanding the mechanism of action of keratinases, but these strains must not be used in feather treatment due to their pathogenic effect. These strains can be easily isolated as they exist widely in nature. Careful strain identification is needed in strain isolation and identification steps. In addition, the antifungal or antibacterial activity can be used as a parameter in the screening step. A study showed that feather-degrading B. subtilis S8 exhibited antifungal activity (Go et al., 2015) and other feather-degrading strains also exhibited antibiotic activity. When a strain is isolated and characterized, whole-cell mutagenesis is an efficient strategy to improve its feather degradation capabilities. All these studies are important to obtain potent microorganisms for feather degradation. It has been noted the fermentation conditions are critical to make the microorganisms to exhibit high yield of the target enzymes (Tatineni et al., 2007).
Microbial Consortium
Combination of several stains might be a strategy to degrade feathers efficiently. Indeed, a research team has screened a microbial consortium KMCG6 which is composed of Bacteroidetes and Proteobacteria. This consortium exhibited a high degradation level (Kang et al., 2018). Similar strategy can be utilized in screening suitable stains that are able to degrade feathers efficiently. It has been noted that microbial consortium or mixed microorganisms have different effects from a single microorganism in microbial degradation field. Careful experimental design is required in the steps of enrichment and screening. It might be possible that combining isolated stains to construct a microbial consortium is easy to obtain reproductive results in later applications.
Keratinase Identification
Many microorganisms are able to produce keratinases. With the development of genome sequencing techniques and accumulation of genome sequences in nucleotide databases, analyzing the information from the genome can provide a fast way to identify new enzymes that are critical for keratin degradation. Using this approach, the enzymes critical for feather degradation were analyzed for anaerobic bacterium Serratia marcescens EGD-HP20 (Fuke et al., 2018). This approach might not be sufficient to identify enzymes that are required for feather degradation as the enzymes secreted into the medium are affected by cultural conditions (Rajput and Gupta, 2013). In addition to optimizing fermentation conditions to achieve high feather-degrading efficiencies, identification of secreted enzymes critical for keratin degradation should be carried out using sensitive methods such as mass spectrometry (MS). Combination of DNA analysis with MS identification on secreted proteins will provide a reliable and fast way to identify necessary components in keratin degradation.
Keratinase Characterization
Several types of substrates have been used in enzymatic characterization while using keratin-driven substrates which may not be sensitive enough to characterize different enzymes due to the solubility. Developing an efficient enzymatic assay in which peptidic substrate is used will be useful for characterizing and comparing keratinases from different microorganisms. Based on recent crystal structure of a keratinase, the five-residue peptide Tyr278-Glu279-Asn280-Leu281-Tyr282 can serve as a keratinase substrate in protease activity assay. It has been noted that the optimization of the peptide sequence might be required for obtaining more sensitive substrate.
Enzyme Engineering
Protein engineering of keratinase has been successful in creating more stable enzymes under high temperatures and alkaline conditions. Bioinformation-guided point mutation is an efficient way to alter enzymatic characteristics (Fang et al., 2016, 2019). The structure of a thermal stable keratinase can be used to guide enzyme engineering. For example, disulfide bonds can be introduced into a keratinase to increase the enzyme stability (Wu et al., 2017). Replacing Ca2+-binding residues with other amino acids could possibly abolish mental binding activity of keratinases, which could reduce the reliance of metal ions in enzyme catalysis. Accumulated studies prove that it is feasible to construct more active and stable enzymes that are suitable for industrial applications.
Mixed Enzymes in Feather Degradation
Keratinases have great potential to be used in several fields. Accumulated studies have suggested that several enzymes are essential for keratin degradation. Mixed enzymes might be required for developing keratinase products which can have higher keratin degrading activity. More studies on enzymes such as disulfide reductase will be useful for designing keratinase products. In addition, adding some chemicals that can facilitate breakage of the disulfide bonds is also useful for increasing keratinase catalytic efficiency.
In summary, with the increase of global population, the requirement for poultry products such as broiler is growing annually. The by-product, feather is also an important source that can be converted from potential wastes to value added products. Microbial processing of feather is therefore a promising technique to generate versatile products such as biofertilizer, animal feed and keratinases. More studies are still required to understand mechanism action of feather degradation and set up an economic strategy for feather processing in large scales.
Author Contributions
QL drafted and revised the manuscript.
Funding
This research was supported by funding “Hundred-Talent Program” (Grant Numbers: 2020GDASYL-20200102010 and 2020GDASYL-20200102009), Guangdong Academy of Sciences, China.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author appreciates the support from Guangdong Bioengineering Institute (Guangzhou Sugarcane Industry Research Institute), Guangdong Academy of Sciences, Guangdong, China. The author also apologizes for the uncited references due to space limitation.
References
Abdel-Fattah, A. M., El-Gamal, M. S., Ismail, S. A., Emran, M. A., and Hashem, A. M. (2018). Biodegradation of feather waste by keratinase produced from newly isolated Bacillus licheniformis ALW1. J. Genet. Eng. Biotechnol. 16, 311–318. doi: 10.1016/j.jgeb.2018.05.005
Abdel-Naby, M. A., El-Refai, H. A., and Ibrahim, M. H. A. (2017). Structural characterization, catalytic, kinetic and thermodynamic properties of Keratinase from Bacillus pumilus FH9. Int. J. Biol. Macromol. 105, 973–980. doi: 10.1016/j.ijbiomac.2017.07.118
Adav, S. S., Subbaiaih, R. S., Kerk, S. K., Lee, A. Y., Lai, H. Y., Ng, K. W., et al. (2018). Studies on the proteome of human hair - identification of histones and deamidated keratins. Sci. Rep. 8:1599. doi: 10.1038/s41598-018-20041-9
Agrahari, S., and Wadhwa, N. (2012). Isolation and characterization of feather degrading enzymes from Bacillus megaterium SN1 isolated from Ghazipur poultry waste site. Appl. Biochem. Microbiol. 48, 175–181. doi: 10.1134/s0003683812020020
Anitha, T. S., and Palanivelu, P. (2013). Purification and characterization of an extracellular keratinolytic protease from a new isolate of Aspergillus parasiticus. Protein Express. Purific. 88, 214–220. doi: 10.1016/j.pep.2013.01.007
Arokiyaraj, S., Varghese, R., Ali Ahmed, B., Duraipandiyan, V., and Al-Dhabi, N. A. (2019). Optimizing the fermentation conditions and enhanced production of keratinase from Bacillus cereus isolated from halophilic environment. Saudi J. Biol. Sci. 26, 378–381. doi: 10.1016/j.sjbs.2018.10.011
Bach, E., Sant’Anna, V., Daroit, D. J., Corrêa, A. P. F., Segalin, J., and Brandelli, A. (2012). Production, one-step purification, and characterization of a keratinolytic protease from Serratia marcescens P3. Process Biochem. 47, 2455–2462. doi: 10.1016/j.procbio.2012.10.007
Bagewadi, Z. K., Mulla, S. I., and Ninnekar, H. Z. (2018). Response surface methodology based optimization of keratinase production from Trichoderma harzianum isolate HZN12 using chicken feather waste and its application in dehairing of hide. J. Environ. Chem. Eng. 6, 4828–4839. doi: 10.1016/j.jece.2018.07.007
Bakhtiar, S., Estiveira, R. J., and Hatti-Kaul, R. (2005). Substrate specificity of alkaline protease from alkaliphilic feather-degrading Nesterenkonia sp. AL20. Enzyme Microb. Technol. 37, 534–540. doi: 10.1016/j.enzmictec.2005.04.003
Barone, J. R., Schmidt, W. F., and Liebner, C. F. E. (2005). Thermally processed keratin films. J. Appl. Polym. Sci. 97, 1644–1651. doi: 10.1002/app.21901
Bhange, K., Chaturvedi, V., and Bhatt, R. (2016a). Ameliorating effects of chicken feathers in plant growth promotion activity by a keratinolytic strain of Bacillus subtilis PF1. Bioresour. Bioprocess. 3:13. doi: 10.1186/s40643-016-0091-y
Bhange, K., Chaturvedi, V., and Bhatt, R. (2016b). Simultaneous production of detergent stable keratinolytic protease, amylase and biosurfactant by Bacillus subtilis PF1 using agro industrial waste. Biotechnol. Rep. 10, 94–104. doi: 10.1016/j.btre.2016.03.007
Bhari, R., Kaur, M., Singh, R. S., Pandey, A., and Larroche, C. (2018). Bioconversion of chicken feathers by Bacillus aerius NSMk2: a potential approach in poultry waste management. Bioresour. Technol. Rep. 3, 224–230. doi: 10.1016/j.biteb.2018.07.015
Böckle, B., Galunsky, B., and Müller, R. (1995). Characterization of a keratinolytic serine proteinase from Streptomyces pactum DSM 40530. Appl. Environ. Microbiol. 61, 3705–3710.
Bockle, B., and Muller, R. (1997). Reduction of disulfide bonds by Streptomyces pactum during growth on chicken feathers. Appl. Environ. Microbiol. 63, 790–792.
Bodde, S. G., Meyers, M. A., and McKittrick, J. (2011). Correlation of the mechanical and structural properties of cortical rachis keratin of rectrices of the Toco Toucan (Ramphastos toco). J. Mechan. Behav. Biomed. Mater. 4, 723–732. doi: 10.1016/j.jmbbm.2011.01.010
Bohacz, J. (2016). Biodegradation of feather waste keratin by a keratinolytic soil fungus of the genus Chrysosporium and statistical optimization of feather mass loss. World J. Microbiol. Biotechnol. 33:13. doi: 10.1007/s11274-016-2177-2
Bohacz, J., and Korniłłowicz-Kowalska, T. (2019). Fungal diversity and keratinolytic activity of fungi from lignocellulosic composts with chicken feathers. Process Biochem. 80, 119–128. doi: 10.1016/j.procbio.2019.02.012
Bragulla, H. H., and Homberger, D. G. (2009). Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. J. Anat. 214, 516–559. doi: 10.1111/j.1469-7580.2009.01066.x
Brandelli, A. (2008). Bacterial keratinases: useful enzymes for bioprocessing agroindustrial wastes and beyond. Food Bioprocess Technol. 1, 105–116. doi: 10.1007/s11947-007-0025-y
Brandelli, A., Daroit, D. J., and Riffel, A. (2010). Biochemical features of microbial keratinases and their production and applications. Appl. Microbiol. Biotechnol. 85, 1735–1750. doi: 10.1007/s00253-009-2398-5
Brandelli, A., Sala, L., and Kalil, S. J. (2015). Microbial enzymes for bioconversion of poultry waste into added-value products. Food Res. Int. 73, 3–12. doi: 10.1016/j.foodres.2015.01.015
Bressollier, P., Letourneau, F., Urdaci, M., and Verneuil, B. (1999). Purification and characterization of a keratinolytic serine proteinase from Streptomyces albidoflavus. Appl. Environ. Microbiol. 65, 2570–2576.
Cai, C.-G., Lou, B.-G., and Zheng, X.-D. (2008). Keratinase production and keratin degradation by a mutant strain of Bacillus subtilis. J. Zhejiang University Sci. B 9, 60–67. doi: 10.1631/jzus.B061620
Cǎlin, M., Constantinescu-Aruxandei, D., Alexandrescu, E., Rǎut, I., Doni, M. B., Arsene, M.-L., et al. (2017). Degradation of keratin substrates by keratinolytic fungi. Electron. J. Biotechnol. 28, 101–112. doi: 10.1016/j.ejbt.2017.05.007
Callegaro, K., Welter, N., and Daroit, D. J. (2018). Feathers as bioresource: microbial conversion into bioactive protein hydrolysates. Process Biochem. 75, 1–9. doi: 10.1016/j.procbio.2018.09.002
Cao, L., Tan, H., Liu, Y., Xue, X., and Zhou, S. (2008). Characterization of a new keratinolytic Trichoderma atroviride strain F6 that completely degrades native chicken feather. Lett. Appl. Microbiol. 46, 389–394. doi: 10.1111/j.1472-765X.2008.02327.x
Cavello, I. A., Hours, R. A., Rojas, N. L., and Cavalitto, S. F. (2013). Purification and characterization of a keratinolytic serine protease from Purpureocillium lilacinum LPS # 876. Process Biochem. 48, 972–978. doi: 10.1016/j.procbio.2013.03.012
Chaturvedi, V., Bhange, K., Bhatt, R., and Verma, P. (2014). Production of kertinases using chicken feathers as substrate by a novel multifunctional strain of Pseudomonas stutzeri and its dehairing application. Biocatal. Agric. Biotechnol. 3, 167–174. doi: 10.1016/j.bcab.2013.08.005
Chitte, R. R., Nalawade, V. K., and Dey, S. (1999). Keratinolytic activity from the broth of a feather-degrading thermophilic Streptomyces thermoviolaceus strain SD8. Lett. Appl. Microbiol. 28, 131–136. doi: 10.1046/j.1365-2672.1999.00484.x
da Silva, R. R. (2018). Keratinases as an alternative method designed to solve keratin disposal on the environment: its relevance on agricultural and environmental chemistry. J. Agric. Food Chem. 66, 7219–7221. doi: 10.1021/acs.jafc.8b03152
De Azeredo, L. A. I., De Lima, M. B., Coelho, R. R. R., and Freire, D. M. G. (2006). Thermophilic protease production by Streptomyces sp. 594 in submerged and solid-state fermentations using feather meal. J. Appl. Microbiol. 100, 641–647. doi: 10.1111/j.1365-2672.2005.02791.x
de Guzman, R. C., Saul, J. M., Ellenburg, M. D., Merrill, M. R., Coan, H. B., Smith, T. L., et al. (2013). Bone regeneration with BMP-2 delivered from keratose scaffolds. Biomaterials 34, 1644–1656. doi: 10.1016/j.biomaterials.2012.11.002
de Paiva, D. P., de Oliveira, S. S. A., Mazotto, A. M., Vermelho, A. B., and de Oliveira, S. S. (2018). Keratinolytic activity of Bacillus subtilis LFB-FIOCRUZ 1266 enhanced by whole-cell mutagenesis. 3 Biotech 9:2. doi: 10.1007/s13205-018-1527-1
Descamps, F., Brouta, F., Vermout, S., Monod, M., Losson, B., and Mignon, B. (2003). Recombinant expression and antigenic properties of a 31.5-kDa keratinolytic subtilisin-like serine protease from Microsporum canis. FEMS Immunol. Med. Microbiol. 38, 29–34. doi: 10.1016/s0928-8244(03)00101-9
Dias, G. J., Mahoney, P., Swain, M., Kelly, R. J., Smith, R. A., and Ali, M. A. (2010a). Keratin–hydroxyapatite composites: biocompatibility, osseointegration, and physical properties in an ovine model. J. Biomed. Mater. Res. Part A 95A, 1084–1095. doi: 10.1002/jbm.a.32908
Dias, G. J., Peplow, P. V., McLaughlin, A., Teixeira, F., and Kelly, R. J. (2010b). Biocompatibility and osseointegration of reconstituted keratin in an ovine model. J. Biomed. Mater. Res. Part A 92A, 513–520. doi: 10.1002/jbm.a.32394
Duarte, T. R., Oliveira, S. S., Macrae, A., Cedrola, S. M. L., Mazotto, A. M., Souza, E. P., et al. (2011). Increased expression of keratinase and other peptidases by Candida parapsilosis mutants. Braz. J. Med. Biol. Res. 44, 212–216. doi: 10.1590/s0100-879x2011007500011
Fakhfakh, N., Ktari, N., Haddar, A., Mnif, I. H., Dahmen, I., and Nasri, M. (2011). Total solubilisation of the chicken feathers by fermentation with a keratinolytic bacterium, Bacillus pumilus A1, and the production of protein hydrolysate with high antioxidative activity. Process Biochem. 46, 1731–1737. doi: 10.1016/j.procbio.2011.05.023
Fang, N., Zhong, C.-Q., Liang, X., Tang, X.-F., and Tang, B. (2010). Improvement of extracellular production of a thermophilic subtilase expressed in Escherichia coli by random mutagenesis of its N-terminal propeptide. Appl. Microbiol. Biotechnol. 85, 1473–1481. doi: 10.1007/s00253-009-2183-5
Fang, Z., Sha, C., Peng, Z., Zhang, J., and Du, G. (2019). Protein engineering to enhance keratinolytic protease activity and excretion in Escherichia coli and its scale-up fermentation for high extracellular yield. Enzyme Microb. Technol. 121, 37–44. doi: 10.1016/j.enzmictec.2018.11.003
Fang, Z., Zhang, J., Du, G., and Chen, J. (2017). Rational protein engineering approaches to further improve the keratinolytic activity and thermostability of engineered keratinase KerSMD. Biochem. Eng. J. 127, 147–153. doi: 10.1016/j.bej.2017.08.010
Fang, Z., Zhang, J., Liu, B., Du, G., and Chen, J. (2013). Biodegradation of wool waste and keratinase production in scale-up fermenter with different strategies by Stenotrophomonas maltophilia BBE11-1. Bioresour. Technol. 140, 286–291. doi: 10.1016/j.biortech.2013.04.091
Fang, Z., Zhang, J., Liu, B., Du, G., and Chen, J. (2016). Enhancement of the catalytic efficiency and thermostability of Stenotrophomonas sp. keratinase KerSMD by domain exchange with KerSMF. Microb. Biotechnol. 9, 35–46. doi: 10.1111/1751-7915.12300
Fang, Z., Zhang, J., Liu, B., Jiang, L., Du, G., and Chen, J. (2014). Cloning, heterologous expression and characterization of two keratinases from Stenotrophomonas maltophilia BBE11-1. Process Biochem. 49, 647–654. doi: 10.1016/j.procbio.2014.01.009
Filipello Marchisio, V., Fusconi, A., and Querio, F. L. (2000). Scopulariopsis brevicaulis: a keratinophilic or a keratinolytic fungus? Mycoses 43, 281–292. doi: 10.1046/j.1439-0507.2000.00580.x
Fontoura, R., Daroit, D. J., Correa, A. P. F., Meira, S. M. M., Mosquera, M., and Brandelli, A. (2014). Production of feather hydrolysates with antioxidant, angiotensin-I converting enzyme- and dipeptidyl peptidase-IV-inhibitory activities. New Biotechnol. 31, 506–513. doi: 10.1016/j.nbt.2014.07.002
Fraser, R. D. B., and Parry, D. A. D. (2011). The structural basis of the filament-matrix texture in the avian/reptilian group of hard β-keratins. J. Struct. Biol. 173, 391–405. doi: 10.1016/j.jsb.2010.09.020
Fuke, P., Gujar, V. V., and Khardenavis, A. A. (2018). Genome annotation and validation of keratin-hydrolyzing proteolytic enzymes from Serratia marcescens EGD-HP20. Appl. Biochem. Biotechnol. 184, 970–986. doi: 10.1007/s12010-017-2595-0
Galas, E., and Kaluzewska, M. (1992). Proteinases of Streptomyces fradiae. III. Catalytic and some physico- chemical properties of keratinolytic proteinase. Acta Microbiol. Pol. 41, 169–177.
Gegeckas, A., Gudiukaitė, R., and Citavicius, D. (2014). Keratinolytic proteinase from Bacillus thuringiensis AD-12. Int. J. Biol. Macromol. 69, 46–51. doi: 10.1016/j.ijbiomac.2014.05.024
Gegeckas, A., Gudiukaitė, R., Debski, J., and Citavicius, D. (2015). Keratinous waste decomposition and peptide production by keratinase from Geobacillus stearothermophilus AD-11. Int. J. Biol. Macromol. 75, 158–165. doi: 10.1016/j.ijbiomac.2015.01.031
Gegeckas, A., Šimkutė, A., Gudiukaitė, R., and Čitavičius, D. J. (2018). Characterization and application of keratinolytic paptidases from Bacillus spp. Int. J. Biol. Macromol. 113, 1206–1213. doi: 10.1016/j.ijbiomac.2018.03.046
Gilardi, G. L. (1965). Nutrition of systemic and subcutaneous pathogenic fungi. Bacteriol. Rev. 29, 406–424.
Giudice, M. C., Reis-Menezes, A. A., Rittner, G. M. G., Mota, A. J., and Gambale, W. (2012). Isolation of Microsporum gypseum in soil samples from different geographical regions of Brazil, evaluation of the extracellular proteolytic enzymes activities (keratinase and elastase) and molecular sequencing of selected strains. Braz. J. Microbiol. 43, 895–902. doi: 10.1590/S1517-83822012000300007
Go, T.-H., Cho, K.-S., Lee, S.-M., Lee, O.-M., and Son, H.-J. (2015). Simultaneous production of antifungal and keratinolytic activities by feather-degrading Bacillus subtilis S8. Indian J. Microbiol. 55, 66–73. doi: 10.1007/s12088-014-0502-8
Govarthanan, M., Selvankumar, T., Selvam, K., Sudhakar, C., Aroulmoji, V., and Kamala-Kannan, S. (2015). Response surface methodology based optimization of keratinase production from alkali-treated feather waste and horn waste using Bacillus sp. MG-MASC-BT. J. Ind. Eng. Chem. 27, 25–30. doi: 10.1016/j.jiec.2014.12.022
Gradišar, H., Kern, S., and Friedrich, J. (2000). Keratinase of Doratomyces microsporus. Appl. Microbiol. Biotechnol. 53, 196–200. doi: 10.1007/s002530050008
Grazziotin, A., Pimentel, F. A., de Jong, E. V., and Brandelli, A. (2006). Nutritional improvement of feather protein by treatment with microbial keratinase. Anim. Feed Sci. Technol. 126, 135–144. doi: 10.1016/j.anifeedsci.2005.06.002
Grazziotin, A., Pimentel, F. A., Sangali, S., de Jong, E. V., and Brandelli, A. (2007). Production of feather protein hydrolysate by keratinolytic bacterium Vibrio sp. kr2. Bioresour. Technol. 98, 3172–3175. doi: 10.1016/j.biortech.2006.10.034
Gupta, R., and Ramnani, P. (2006). Microbial keratinases and their prospective applications: an overview. Appl. Microbiol. Biotechnol. 70:21. doi: 10.1007/s00253-005-0239-8
Gupta, R., Sharma, R., and Beg, Q. K. (2013). Revisiting microbial keratinases: next generation proteases for sustainable biotechnology. Critic. Rev. Biotechnol. 33, 216–228. doi: 10.3109/07388551.2012.685051
Habbeche, A., Saoudi, B., Jaouadi, B., Haberra, S., Kerouaz, B., Boudelaa, M., et al. (2014). Purification and biochemical characterization of a detergent-stable keratinase from a newly thermophilic actinomycete Actinomadura keratinilytica strain Cpt29 isolated from poultry compost. J. Biosci. Bioeng. 117, 413–421. doi: 10.1016/j.jbiosc.2013.09.006
Hamiche, S., Mechri, S., Khelouia, L., Annane, R., El Hattab, M., Badis, A., et al. (2019). Purification and biochemical characterization of two keratinases from Bacillus amyloliquefaciens S13 isolated from marine brown alga Zonaria tournefortii with potential keratin-biodegradation and hide-unhairing activities. Int. J. Biol. Macromol. 122, 758–769. doi: 10.1016/j.ijbiomac.2018.10.174
Han, M., Luo, W., Gu, Q., and Yu, X. (2012). Isolation and characterization of a keratinolytic protease from a feather-degrading bacterium Pseudomonas aeruginosa C11. Afr. J. Microbiol. Res. 6, 2211–2221. doi: 10.5897/AJMR11.921
Herzog, B., Overy, D. P., Haltli, B., and Kerr, R. G. (2016). Discovery of keratinases using bacteria isolated from marine environments. Syst. Appl. Microbiol. 39, 49–57. doi: 10.1016/j.syapm.2015.10.004
Hill, P., Brantley, H., and Van Dyke, M. (2010). Some properties of keratin biomaterials: kerateines. Biomaterials 31, 585–593. doi: 10.1016/j.biomaterials.2009.09.076
Intagun, W., and Kanoksilapatham, W. (2017). A review: biodegradation and applications of keratin degrading microorganisms and keratinolytic enzymes, focusing on thermophiles and thermostable serine proteases. Am. J. Appl. Sci. 14, 1016–1023. doi: 10.3844/ajassp.2017.1016.1023
Jaouadi, B., Abdelmalek, B., Fodil, D., Ferradji, F. Z., Rekik, H., Zaraî, N., et al. (2010). Purification and characterization of a thermostable keratinolytic serine alkaline proteinase from Streptomyces sp. strain AB1 with high stability in organic solvents. Bioresour. Technol. 101, 8361–8369. doi: 10.1016/j.biortech.2010.05.066
Jeevana Lakshmi, P., Kumari Chitturi, C. M., and Lakshmi, V. V. (2013). Efficient degradation of feather by keratinase producing Bacillus sp. Int. J. Microbiol. 2013:7. doi: 10.1155/2013/608321
Jeong, J.-H., Jeon, Y.-D., Lee, O.-M., Kim, J.-D., Lee, N.-R., Park, G.-T., et al. (2010a). Characterization of a multifunctional feather-degrading Bacillus subtilis isolated from forest soil. Biodegradation 21, 1029–1040. doi: 10.1007/s10532-010-9363-y
Jeong, J.-H., Lee, O. M., Jeon, Y.-D., Kim, J.-D., Lee, N.-R., Lee, C.-Y., et al. (2010b). Production of keratinolytic enzyme by a newly isolated feather-degrading Stenotrophomonas maltophilia that produces plant growth-promoting activity. Process Biochem. 45, 1738–1745. doi: 10.1016/j.procbio.2010.07.020
Jeong, J.-H., Park, K.-H., Oh, D.-J., Hwang, D.-Y., Kim, H.-S., Lee, C.-Y., et al. (2010c). Keratinolytic enzyme-mediated biodegradation of recalcitrant feather by a newly isolated Xanthomonas sp. P5. Polym. Degrad. Stabil. 95, 1969–1977. doi: 10.1016/j.polymdegradstab.2010.07.020
Jin, H.-S., Park, S. Y., Kim, K., Lee, Y.-J., Nam, G.-W., Kang, N. J., et al. (2017). Development of a keratinase activity assay using recombinant chicken feather keratin substrates. PLoS One 12:e0172712. doi: 10.1371/journal.pone.0172712
Kang, D., Herschend, J., Al-Soud, W. A., Mortensen, M. S., Gonzalo, M., Jacquiod, S., et al. (2018). Enrichment and characterization of an environmental microbial consortium displaying efficient keratinolytic activity. Bioresour. Technol. 270, 303–310. doi: 10.1016/j.biortech.2018.09.006
Kanoksilapatham, W., and Intagun, W. (2017). A review: biodegradation and applications of keratin degrading microorganisms and keratinolytic enzymes, focusing on thermophiles and thermostable serine proteases. Am. J. Appl. Sci. 14, 1016–1023. doi: 10.3844/ajassp.2017.1016.1023
Khardenavis, A. A., Kapley, A., and Purohit, H. J. (2009). Processing of poultry feathers by alkaline keratin hydrolyzing enzyme from Serratia sp. HPC 1383. Waste Manag. 29, 1409–1415. doi: 10.1016/j.wasman.2008.10.009
Khodayari, S., and Kafilzadeh, F. (2018). Separating keratinase producer bacteria from the soil of poultry farms and optimization of the conditions for maximum enzyme production. Eur. J. Exp. Biol. 8:35. doi: 10.21767/2248-9215.100076
Kim, J. D. (2007). Purification and characterization of a keratinase from a feather-degrading fungus, Aspergillus flavus strain K-03. Mycobiology 35, 219–225. doi: 10.4489/MYCO.2007.35.4.219
Kim, J. M., Choi, Y. M., and Suh, H. J. (2005). Preparation of feather digests as fertilizer with Bacillus pumilis KHS-1. J. Microbiol. Biotechnol. 15, 427–476.
Kim, J. M., Lim, W. J., and Suh, H. J. (2001). Feather-degrading Bacillus species from poultry waste. Process Biochem. 37, 287–291. doi: 10.1016/S0032-9592(01)00206-0
Kim, W. K., and Patterson, P. H. (2000). Nutritional value of enzyme- or sodium hydroxide-treated feathers from dead hens. Poult. Sci. 79, 528–534. doi: 10.1093/ps/79.4.528
Kojima, M., Kanai, M., Tominaga, M., Kitazume, S., Inoue, A., and Horikoshi, K. (2006). Isolation and characterization of a feather-degrading enzyme from Bacillus pseudofirmus FA30-01. Extremophiles 10, 229–235. doi: 10.1007/s00792-005-0491-y
Korniłłowicz-Kowalska, T., and Bohacz, J. (2011). Biodegradation of keratin waste: theory and practical aspects. Waste Manag. 31, 1689–1701. doi: 10.1016/j.wasman.2011.03.024
Kshetri, P., Roy, S. S., Sharma, S. K., Singh, T. S., Ansari, M. A., Prakash, N., et al. (2019). Transforming chicken feather waste into feather protein hydrolysate using a newly isolated multifaceted keratinolytic bacterium chryseobacterium sediminis RCM-SSR-7. Waste Biomass Valoriz. 10, 1–11. doi: 10.1007/s12649-017-0037-4
Laba, W., Choinska, A., Rodziewicz, A., and Piegza, M. (2015). Keratinolytic abilities of Micrococcus luteus from poultry waste. Braz. J. Microbiol. 46, 691–700. doi: 10.1590/S1517-838246320140098
Lange, L., Huang, Y., and Busk, P. K. (2016). Microbial decomposition of keratin in nature-a new hypothesis of industrial relevance. Appl. Microbiol. Biotechnol. 100, 2083–2096. doi: 10.1007/s00253-015-7262-1
Latshaw, J. D., Musharaf, N., and Retrum, R. (1994). Processing of feather meal to maximize its nutritional value for poultry. Anim. Feed Sci. Technol. 47, 179–188. doi: 10.1016/0377-8401(94)90122-8
Lee, H., Suh, D. B., Hwang, J. H., and Suh, H. J. (2002). Characterization of a kerationlytic metalloprotease from Bacillus sp. SCB-3. Appl. Biochem. Biotechnol. 97, 123–133. doi: 10.1385/abab
Lin, X., Inglis, G. D., Yanke, L. J., and Cheng, K.-J. (1999). Selection and characterization of feather-degrading bacteria from canola meal compost. J. Ind. Microbiol. Biotechnol. 23, 149–153. doi: 10.1038/sj.jim.2900706
Lin, X., Kelemen, D. W., Miller, E. S., and Shih, J. C. (1995). Nucleotide sequence and expression of kerA, the gene encoding a keratinolytic protease of Bacillus licheniformis PWD-1. Appl. Environ. Microbiol. 61, 1469–1474.
Lin, X., Lee, C.-G., Casale, E. S., and Shih, J. C. H. (1992). Purification and characterization of a keratinase from a feather-degrading Bacillus licheniformis Strain. Appl. Environ. Microbiol. 58, 3271–3275.
Liu, B., Zhang, J., Fang, Z., Du, G., Chen, J., and Liao, X. (2014). Functional analysis of the C-terminal propeptide of keratinase from Bacillus licheniformis BBE11-1 and its effect on the production of keratinase in Bacillus subtilis. Process Biochem. 49, 1538–1542. doi: 10.1016/j.procbio.2014.04.021
Liu, B., Zhang, J., Fang, Z., Gu, L., Liao, X., Du, G., et al. (2013). Enhanced thermostability of keratinase by computational design and empirical mutation. J. Ind. Microbiol. Biotechnol. 40, 697–704. doi: 10.1007/s10295-013-1268-4
Liu, Q., Long, K., Lu, F., and Chen, J. (2017). Biodegradation and antibacterial activity of a feather-degrading strain of bacterium. Biocatal. Agric. Biotechnol. 9, 195–200. doi: 10.1016/j.bcab.2017.01.002
Lo, W.-H., Too, J.-R., and Wu, J.-Y. (2012). Production of keratinolytic enzyme by an indigenous feather–degrading strain Bacillus cereus Wu2. J. Biosci. Bioeng. 114, 640–647. doi: 10.1016/j.jbiosc.2012.07.014
Ma, B., Qiao, X., Hou, X., and Yang, Y. (2016). Pure keratin membrane and fibers from chicken feather. Int. J. Biol. Macromol. 89, 614–621. doi: 10.1016/j.ijbiomac.2016.04.039
Manczinger, L., Rozs, M., Vágvölgyi, C., and Kevei, F. (2003). Isolation and characterization of a new keratinolytic Bacillus licheniformis strain. World J. Microbiol. Biotechnol. 19, 35–39. doi: 10.1023/a:1022576826372
Mazotto, A. M., Couri, S., Damaso, M. C. T., and Vermelho, A. B. (2013). Degradation of feather waste by Aspergillus niger keratinases: comparison of submerged and solid-state fermentation. Int. Biodeterior. Biodegrad. 85, 189–195. doi: 10.1016/j.ibiod.2013.07.003
Meng, K., Li, J., Cao, Y., Shi, P., Wu, B., Han, X., et al. (2007). Gene cloning and heterologous expression of a serine protease from Streptomyces fradiae var.k11. Can. J. Microbiol. 53, 186–195. doi: 10.1139/w06-122
Meyers, M. A., Chen, P.-Y., Lin, A. Y.-M., and Seki, Y. (2008). Biological materials: structure and mechanical properties. Prog. Mater. Sci. 53, 1–206. doi: 10.1016/j.pmatsci.2007.05.002
Mitsuiki, S., Sakai, M., Moriyama, Y., Goto, M., and Furukawa, K. (2002). Purification and some properties of a keratinolytic enzyme from an alkaliphilic Nocardiopsis sp. TOA-1. Biosci. Biotechnol. Biochem. 66, 164–167. doi: 10.1271/bbb.66.164
Muthusamy, G., Thangaswamy, S., and Arunprakash, S. (2011). Production of keratinolytic enzyme by a newly isolated feather degrading Bacillus sp. from chick feather waste. Int. J. Pharma Bio Sci. 2, 259–265.
Nam, G.-W., Lee, D.-W., Lee, H.-S., Lee, N.-J., Kim, B.-C., Choe, E.-A., et al. (2002). Native-feather degradation by Fervidobacterium islandicum AW-1, a newly isolated keratinase-producing thermophilic anaerobe. Arch. Microbiol. 178, 538–547. doi: 10.1007/s00203-002-0489-0
Negi, M., Tsuboi, R., Matsui, T., and Ogawa, H. (1984). Isolation and characterization of proteinase from Candida albicans: substrate specificity. J. Invest. Dermatol. 83, 32–36. doi: 10.1111/1523-1747.ep12261656
Ng, C. S., Chen, J.-J., Lu, M.-Y. J., Ho, M.-R., Wu, S.-M., Fan, W.-L., et al. (2014). Genomic organization, transcriptomic analysis, and functional characterization of Avian α- and β-Keratins in diverse feather forms. Genome Biol. Evol. 6, 2258–2273. doi: 10.1093/gbe/evu181
Onifade, A. A., Al-Sane, N. A., Al-Musallam, A. A., and Al-Zarban, S. (1998). A review: potentials for biotechnological applications of keratin-degrading microorganisms and their enzymes for nutritional improvement of feathers and other keratins as livestock feed resources. Bioresour. Technol. 66, 1–11. doi: 10.1016/S0960-8524(98)00033-9
Papadopoulos, M. C. (1989). Effect of processing on high-protein feedstuffs: a review. Biol. Wastes 29, 123–138. doi: 10.1016/0269-7483(89)90092-X
Papadopoulos, M. C., El Boushy, A. R., Roodbeen, A. E., and Ketelaars, E. H. (1986). Effects of processing time and moisture content on amino acid composition and nitrogen characteristics of feather meal. Anim. Feed Sci. Technol. 14, 279–290. doi: 10.1016/0377-8401(86)90100-8
Park, G.-T., and Son, H.-J. (2009). Keratinolytic activity of Bacillus megaterium F7-1, a feather-degrading mesophilic bacterium. Microbiol. Res. 164, 478–485. doi: 10.1016/j.micres.2007.02.004
Parrado, J., Rodriguez-Morgado, B., Tejada, M., Hernandez, T., and Garcia, C. (2014). Proteomic analysis of enzyme production by Bacillus licheniformis using different feather wastes as the sole fermentation media. Enzyme Microb. Technol. 57, 1–7. doi: 10.1016/j.enzmictec.2014.01.001
Paul, T., Halder, S. K., Das, A., Bera, S., Maity, C., Mandal, A., et al. (2013). Exploitation of chicken feather waste as a plant growth promoting agent using keratinase producing novel isolate Paenibacillus woosongensis TKB2. Biocatal. Agric. Biotechnol. 2, 50–57. doi: 10.1016/j.bcab.2012.10.001
Paul, T., Jana, A., Mandal, A. K., Mandal, A., Das Mohpatra, P. K., and Mondal, K. C. (2016). Bacterial keratinolytic protease, imminent starter for NextGen leather and detergent industries. Sustain. Chem. Pharm. 3, 8–22. doi: 10.1016/j.scp.2016.01.001
Pettett, L. M., and Kurtböke, D. I. (2004). Development of an environmentally friendly biofertilizer with keratin degrading and antibiotic producing actinomycetes. Actinomycetologica 18, 34–42. doi: 10.3209/saj.18_34
Poopathi, S., and Abidha, S. (2008). Biodegradation of poultry waste for the production of mosquitocidal toxins. Int. Biodeter. Biodegrad. 62, 479–482. doi: 10.1016/j.ibiod.2008.03.005
Rahayu, S., Syah, D., and Thenawidjaja Suhartono, M. (2012). Degradation of keratin by keratinase and disulfide reductase from Bacillus sp. MTS of Indonesian origin. Biocatal. Agric. Biotechnol. 1, 152–158. doi: 10.1016/j.bcab.2012.02.001
Rai, S. K., Konwarh, R., and Mukherjee, A. K. (2009). Purification, characterization and biotechnological application of an alkaline β-keratinase produced by Bacillus subtilis RM-01 in solid-state fermentation using chicken-feather as substrate. Biochem. Eng. J. 45, 218–225. doi: 10.1016/j.bej.2009.04.001
Rai, S. K., and Mukherjee, A. K. (2011). Optimization of production of an oxidant and detergent-stable alkaline β-keratinase from Brevibacillus sp. strain AS-S10-II: application of enzyme in laundry detergent formulations and in leather industry. Biochem. Eng. J. 54, 47–56. doi: 10.1016/j.bej.2011.01.007
Rajput, R., and Gupta, R. (2013). Thermostable keratinase from Bacillus pumilus KS12: production, chitin crosslinking and degradation of Sup35NM aggregates. Bioresour. Technol. 133, 118–126. doi: 10.1016/j.biortech.2013.01.091
Rajput, R., Sharma, R., and Gupta, R. (2010). Biochemical characterization of a thiol-activated, oxidation stable keratinase from Bacillus pumilus KS12. Enzyme Res. 2010:7. doi: 10.4061/2010/132148
Ramakrishna Reddy, M., Sathi Reddy, K., Ranjita Chouhan, Y., Bee, H., and Reddy, G. (2017). Effective feather degradation and keratinase production by Bacillus pumilus GRK for its application as bio-detergent additive. Bioresour. Technol. 243, 254–263. doi: 10.1016/j.biortech.2017.06.067
Riffel, A., Daroit, D. J., and Brandelli, A. (2011). Nutritional regulation of protease production by the feather-degrading bacterium Chryseobacterium sp. kr6. New Biotechnol. 28, 153–157. doi: 10.1016/j.nbt.2010.09.008
Rodríguez-Morgado, B., Gómez, I., Parrado, J., García, C., Hernández, T., and Tejada, M. (2015). Accelerated degradation of PAHs using edaphic biostimulants obtained from sewage sludge and chicken feathers. J. Hazard. Mater. 300, 235–242. doi: 10.1016/j.jhazmat.2015.05.045
Sahni, N., Sahota, P. P., and Gupta, P. U. (2015). Bacterial keratinases and their prospective applications: a review. Int. J. Curr. Microbiol. Appl. Sci. 4, 768–783.
Sangali, S., and Brandelli, A. (2000). Isolation and characterization of a novel feather-degrading bacterial strain. Appl. Biochem. Biotechnol. 87, 17–24. doi: 10.1385/abab:87:1:17
Sanghvi, G., Patel, H., Vaishnav, D., Oza, T., Dave, G., Kunjadia, P., et al. (2016). A novel alkaline keratinase from Bacillus subtilis DP1 with potential utility in cosmetic formulation. Int. J. Biol. Macromol. 87, 256–262. doi: 10.1016/j.ijbiomac.2016.02.067
Saravanan, K., and Dhurai, B. (2012). Exploration on the amino acid content and morphological structure in chicken feather fiber. J. Text. Apparel Technol. Manag. 7, 1–4.
Shanmugasundaram, O. L., Syed Zameer Ahmed, K., Sujatha, K., Ponnmurugan, P., Srivastava, A., Ramesh, R., et al. (2018). Fabrication and characterization of chicken feather keratin/polysaccharides blended polymer coated nonwoven dressing materials for wound healing applications. Mater. Sci. Eng. C 92, 26–33. doi: 10.1016/j.msec.2018.06.020
Sharaf, E. F., and Khalil, N. M. (2011). Keratinolytic activity of purified alkaline keratinase produced by Scopulariopsis brevicaulis (Sacc.) and its amino acids profile. Saudi J. Biol. Sci. 18, 117–121. doi: 10.1016/j.sjbs.2010.12.011
Sharma, A., Chandra, S., and Sharma, M. (2012). Difference in keratinase activity of dermatophytes at different environmental conditions is an attribute of adaptation to parasitism. Mycoses 55, 410–415. doi: 10.1111/j.1439-0507.2011.02133.x
Sharma, S., and Gupta, A. (2016). Sustainable management of keratin waste biomass: applications and future perspectives. Braz. Arch. Biol. Technol. 59:e16150684.
Snider, N. T., and Omary, M. B. (2014). Post-translational modifications of intermediate filament proteins: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 15:163. doi: 10.1038/nrm3753
Su, C., Gong, J.-S., Sun, Y.-X., Qin, J., Zhai, S., Li, H., et al. (2019). Combining pro-peptide engineering and multisite saturation mutagenesis to improve the catalytic potential of keratinase. ACS Synth. Biol. 8, 425–433. doi: 10.1021/acssynbio.8b00442
Swetlana, N., and Jain, P. C. (2010). Feather degradation by strains of Bacillus isolated from decomposing feathers. Braz. J. Microbiol. 41, 196–200. doi: 10.1590/S1517-838220100001000028
Syed, D. G., Lee, J. C., Li, W.-J., Kim, C.-J., and Agasar, D. (2009). Production, characterization and application of keratinase from Streptomyces gulbargensis. Bioresour. Technol. 100, 1868–1871. doi: 10.1016/j.biortech.2008.09.047
Takagi, H., Koga, M., Katsurada, S., Yabuta, Y., Shinde, U., Inouye, M., et al. (2001). Functional analysis of the propeptides of subtilisin E and aqualysin I as intramolecular chaperones. FEBS Lett. 508, 210–214. doi: 10.1016/s0014-5793(01)03053-8
Tamreihao, K., Devi, L. J., Khunjamayum, R., Mukherjee, S., Ashem, R. S., and Ningthoujam, D. S. (2017). Biofertilizing potential of feather hydrolysate produced by indigenous keratinolytic Amycolatopsis sp. MBRL 40 for rice cultivation under field conditions. Biocatal. Agric. Biotechnol. 10, 317–320. doi: 10.1016/j.bcab.2017.04.010
Tamreihao, K., Mukherjee, S., Khunjamayum, R., Devi, L. J., Asem, R. S., and Ningthoujam, D. S. (2019). Feather degradation by keratinolytic bacteria and biofertilizing potential for sustainable agricultural production. J. Basic Microbiol. 59, 4–13. doi: 10.1002/jobm.201800434
Tatineni, R., Doddapaneni, K. K., Potumarthi, R. C., and Mangamoori, L. N. (2007). Optimization of keratinase production and enzyme activity using response surface methodology with streptomyces sp7. Appl. Biochem. Biotechnol. 141:187. doi: 10.1007/bf02729061
Tesfaye, T., Sithole, B., and Ramjugernath, D. (2017). Valorisation of chicken feathers: a review on recycling and recovery route—current status and future prospects. Clean Technol. Environ. Policy 19, 2363–2378. doi: 10.1007/s10098-017-1443-9
Tiwary, E., and Gupta, R. (2010). Medium optimization for a novel 58kDa dimeric keratinase from Bacillus licheniformis ER-15: biochemical characterization and application in feather degradation and dehairing of hides. Bioresour. Technol. 101, 6103–6110. doi: 10.1016/j.biortech.2010.02.090
Tork, S., Aly, M. M., and Nawar, L. (2010). Biochemical and molecular characterization of a new local keratinase producing pseudomomanas sp., MS21. Asian J. Biotechnol. 2, 1–13. doi: 10.3923/ajbkr.2010.1.13
Tork, S. E., Shahein, Y. E., El-Hakim, A. E., Abdel-Aty, A. M., and Aly, M. M. (2013). Production and characterization of thermostable metallo-keratinase from newly isolated Bacillus subtilis NRC 3. Int. J. Biol. Macromol. 55, 169–175. doi: 10.1016/j.ijbiomac.2013.01.002
Tridico, S. R., Koch, S., Michaud, A., Thomson, G., Kirkbride, K. P., and Bunce, M. (2014). Interpreting biological degradative processes acting on mammalian hair in the living and the dead: which ones are taphonomic? Proc. R. Soc. B Biol. Sci. 281:20141755. doi: 10.1098/rspb.2014.1755
Tsuboi, R., Ko, I., Takamori, K., and Ogawa, H. (1989). Isolation of a keratinolytic proteinase from Trichophyton mentagrophytes with enzymatic activity at acidic pH. Infect. Immun. 57, 3479–3483.
Uehara, R., Ueda, Y., You, D.-J., Koga, Y., and Kanaya, S. (2013). Accelerated maturation of Tk-subtilisin by a Leu→Promutation at the C-terminus of the propeptide, which reduces the binding of the propeptide to Tk-subtilisin. FEBS J. 280, 994–1006. doi: 10.1111/febs.12091
Van de Wiel, D. F. M., Langeveld, J. P. M., Bossers, A., Garssen, G. J., Shih, G. C., Wang, J.-J., et al. (2003). Enzymatic degradation of prion protein in brain stem from infected cattle and sheep. J. Infect. Dis. 188, 1782–1789. doi: 10.1086/379664
Verma, A., Singh, H., Anwar, S., Chattopadhyay, A., Tiwari, K. K., Kaur, S., et al. (2017). Microbial keratinases: industrial enzymes with waste management potential. Crit. Rev. Biotechnol. 37, 476–491. doi: 10.1080/07388551.2016.1185388
Viani, F. C., Santos, J. I. D., Paula, C. R., Larson, C. E., and Gambale, W. (2001). Production of extracellular enzymes by Microsporum canis and their role in its virulence. Med. Mycol. 39, 463–468. doi: 10.1080/mmy.39.5.463.468
Wang, B., Yang, W., McKittrick, J., and Meyers, M. A. (2016). Keratin: structure, mechanical properties, occurrence in biological organisms, and efforts at bioinspiration. Prog. Mater. Sci. 76, 229–318. doi: 10.1016/j.pmatsci.2015.06.001
Wang, J., Hao, S., Luo, T., Cheng, Z., Li, W., Gao, F., et al. (2017). Feather keratin hydrogel for wound repair: preparation, healing effect and biocompatibility evaluation. Colloids Surf. B Biointerf. 149, 341–350. doi: 10.1016/j.colsurfb.2016.10.038
Wang, J.-J., and Shih, J. C. H. (1999). Fermentation production of keratinase from Bacillus licheniformis PWD-1 and a recombinant B. subtilis FDB-29. J. Ind. Microbiol. Biotechnol. 22, 608–616. doi: 10.1038/sj.jim.2900667
Wang, X., and Parsons, C. (1997). Effect of processing systems on protein quality of feather meals and hog hair meals. Poult. Sci. 76, 491–496. doi: 10.1093/ps/76.3.491
Wang, X. C., Zhao, H. Y., Liu, G., Cheng, X. J., and Feng, H. (2015). Improving production of extracellular proteases by random mutagenesis and biochemical characterization of a serine protease in Bacillus subtilis S1-4. Genet. Mol. Res. 15:gmr.15027831. doi: 10.4238/gmr.15027831
Weary, P. E., Canby, C. M., and Cawley, E. P. (1965). Keratinolytic activity of Microsporum canis and Microsporum gypseum. J. Invest. Dermatol. 44, 300–310. doi: 10.1038/jid.1965.54
Williams, C. M., Lee, C. G., Garlich, J. D., and Shih, J. C. H. (1991). Evaluation of a bacterial feather fermentation product, feather-lysate, as a feed protein1. Poult. Sci. 70, 85–94. doi: 10.3382/ps.0700085
Williams, C. M., and Shih, J. C. H. (1989). Enumeration of some microbial groups in thermophilic poultry waste digesters and enrichment of a feather-degrading culture. J. Appl. Bacteriol. 67, 25–35. doi: 10.1111/j.1365-2672.1989.tb04951.x
Wu, P., Ng, C. S., Yan, J., Lai, Y.-C., Chen, C.-K., Lai, Y.-T., et al. (2015). Topographical mapping of α- and β-keratins on developing chicken skin integuments: functional interaction and evolutionary perspectives. Proc. Natl. Acad. Sci. U.S.A. 112, E6770–E6779. doi: 10.1073/pnas.1520566112
Wu, W.-L., Chen, M.-Y., Tu, I. F., Lin, Y.-C., EswarKumar, N., Chen, M.-Y., et al. (2017). The discovery of novel heat-stable keratinases from Meiothermus taiwanensis WR-220 and other extremophiles. Sci. Rep. 7:4658. doi: 10.1038/s41598-017-04723-4
Yamamura, S., Morita, Y., Hasan, Q., Rao, S. R., Murakami, Y., Yokoyama, K., et al. (2002a). Characterization of a new keratin-degrading bacterium isolated from deer fur. J. Biosci. Bioeng. 93, 595–600. doi: 10.1016/S1389-1723(02)80243-2
Yamamura, S., Morita, Y., Hasan, Q., Yokoyama, K., and Tamiya, E. (2002b). Keratin degradation: a cooperative action of two enzymes from Stenotrophomonas sp. Biochem. Biophys. Res. Commun. 294, 1138–1143. doi: 10.1016/S0006-291X(02)00580-6
Yu, R. J., Harmon, S. R., and Blank, F. (1968). Isolation and purification of an extracellular keratinase of Trichophyton mentagrophytes. J. Bacteriol. 96, 1435–1436.
Yue, X. Y., Zhang, B., Jiang, D. D., Liu, Y. J., and Niu, T. G. (2011). Separation and purification of a keratinase as pesticide against root-knot nematodes. World J. Microbiol. Biotechnol. 27, 2147–2153. doi: 10.1007/s11274-011-0680-z
Zaraî Jaouadi, N., Rekik, H., Ben Elhoul, M., Zohra Rahem, F., Gorgi Hila, C., Slimene Ben Aicha, H., et al. (2015). A novel keratinase from Bacillus tequilensis strain Q7 with promising potential for the leather bating process. Int. J. Biol. Macromol. 79, 952–964. doi: 10.1016/j.ijbiomac.2015.05.038
Zhang, R.-X., Gong, J.-S., Su, C., Zhang, D.-D., Tian, H., Dou, W.-F., et al. (2016). Biochemical characterization of a novel surfactant-stable serine keratinase with no collagenase activity from Brevibacillus parabrevis CGMCC 10798. Int. J. Biol. Macromol. 93, 843–851. doi: 10.1016/j.ijbiomac.2016.09.063
Keywords: keratin, microorganism, protease, keratinase, bio fertilizer, fermentation
Citation: Li Q (2019) Progress in Microbial Degradation of Feather Waste. Front. Microbiol. 10:2717. doi: 10.3389/fmicb.2019.02717
Received: 30 August 2019; Accepted: 08 November 2019;
Published: 05 December 2019.
Edited by:
Kian Mau Goh, University of Technology, Malaysia, MalaysiaReviewed by:
Justyna Bohacz, University of Life Sciences in Lublin, PolandMuthusamy Govarthanan, University of Seoul, South Korea
Copyright © 2019 Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingxin Li, cWluZ3hpbl9saUBvdXRsb29rLmNvbQ==
 Qingxin Li
Qingxin Li