- 1National Key Program of Microbiology and Department of Microbiology, College of Life Sciences, Nankai University, Tianjin, China
- 2Beijing Key Laboratory of Genetic Engineering Drug and Biotechnology, Institute of Biochemistry and Molecular Biology, School of Life Sciences, Beijing Normal University, Beijing, China
Autophagy is mainly a catabolic process, which is used to cope with nutrient deficiency and various stress conditions. Human environment often imposes various stresses on Cryptococcus neoformans, a major fungal pathogen of immunocompromised individuals; therefore, autophagic response of C. neoformans to these stresses often determines its survival in the host. However, a systematic study on how autophagy related (ATG) genes influence on autophagic flux, virulence, stress response and pathogenicity of C. neoformans is lacking. In this study, 22 ATG-deficient strains were constructed to investigate their roles in virulence, pathogenesis, stress response, starvation tolerance and autophagic flux in C. neoformans. Our results showed that Atg6 and Atg14-03 significantly affect the growth of C. neoformans at 37°C and laccase production. Additionally, atg2Δ and atg6Δ strains were sensitive to oxidative stress caused by hydrogen peroxide. Approximately half of the atgΔ strains displayed higher sensitivity to 1.5 M NaCl and remarkably lower virulence in the Galleria mellonella model than the wild type. Autophagic flux in C. neoformans was dependent on the Atg1-Atg13, Atg5-Atg12-Atg16, and Atg2-Atg18 complexes and Atg11. Cleavage of the green fluorescent protein (GFP) from Atg8 was difficult to detect in these autophagy defective mutants; however, it was detected in the atg3Δ, atg4Δ, atg6Δ and atg14Δ strains. Additionally, no homologs of Saccharomyces cerevisiae ATG10 were detected in C. neoformans. Our results indicate that these ATG genes contribute differentially to carbon and nitrogen starvation tolerance in C. neoformans compared with S. cerevisiae. Overall, this study advances our knowledge of the specific roles of ATG genes in C. neoformans.
Introduction
Autophagy is mainly a catabolic process, which is induced by nutrient deficiency and various stress conditions (Yang and Klionsky, 2010). Saccharomyces cerevisiae exhibits two major forms of autophagy: macroautophagy and microautophagy (Reggiori and Klionsky, 2013). Macroautophagy (hereafter referred to as autophagy) is a fundamental function of eukaryotic cells and is well conserved from S. cerevisiae to humans (Nakatogawa et al., 2009). The most important and characteristic morphological feature of autophagy is the de novo formation of double-membrane autophagosome, which contains either bulk cytoplasm or selected cargos, depending on the inducing conditions (Inoue and Klionsky, 2010). The formation, expansion, fusion with vacuole and degradation of autophagosome in the vacuole constitute major steps in autophagy that depend on a series of autophagy related (ATG) genes.
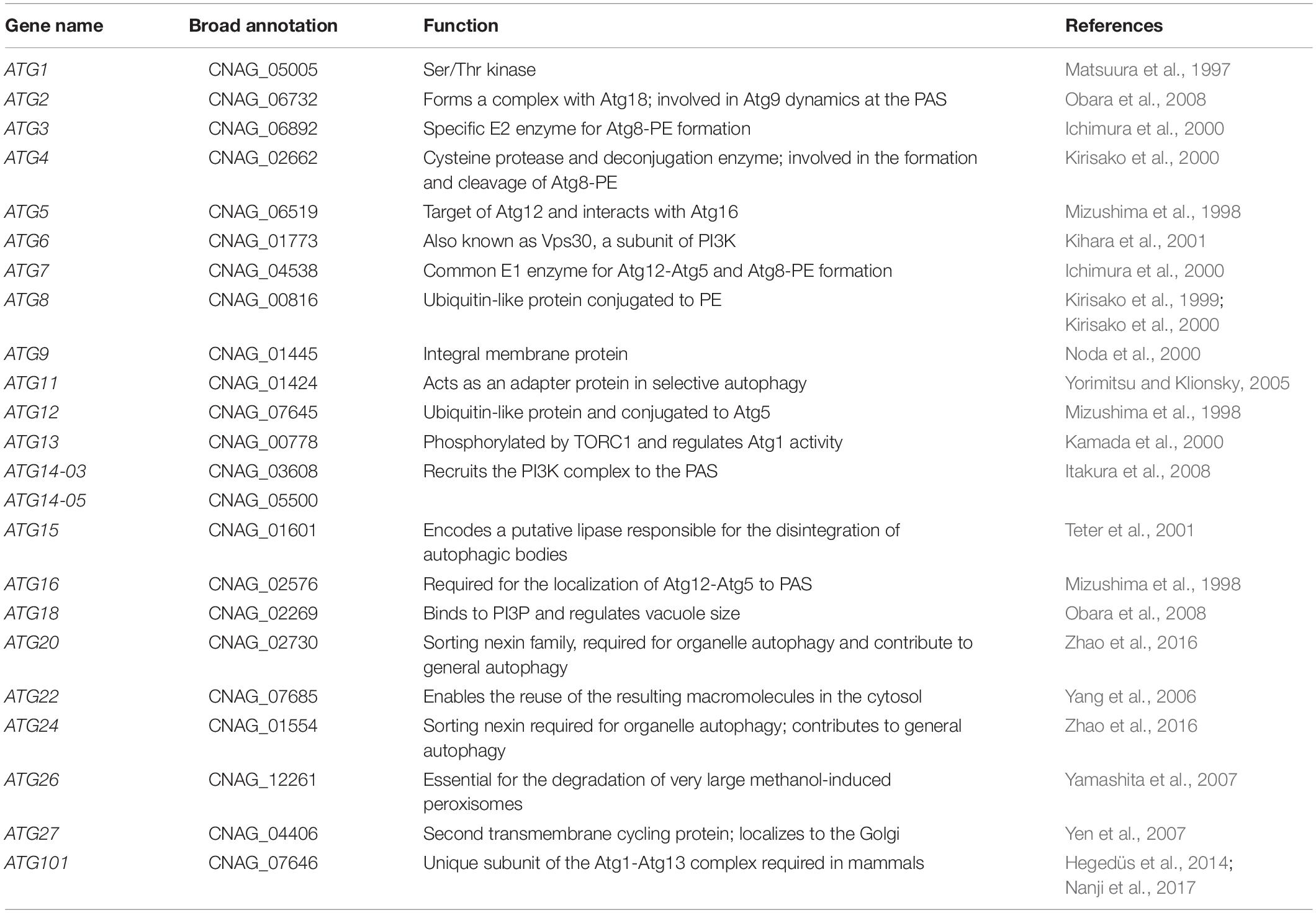
At present, more than 40 ATG genes have been identified in fungi, and the proteins encoded by most of these genes form several complexes during the autophagic process (Lin et al., 2019). ATG1, the first identified ATG gene, encodes a Ser/Thr kinase (Matsuura et al., 1997). Atg13, which binds to Atg1, is highly phosphorylated by TORC1 (Target of Rapamycin Complex) kinase under nutrient proficient conditions. However, Atg13 is rapidly dephosphorylated upon starvation or rapamycin treatment (Kamada et al., 2000). The Atg1-Atg13 complex comprises the most upstream Atg subfamily in the hierarchy of localization of Atg proteins to the pre-autophagosomal structure (PAS) (Kawamata et al., 2008). Phosphatidylinositol-3-phosphate (PI3P) is essential for autophagosome formation and resides in the autophagosomal membrane. Formation of PI3P depends on the PI3K complex I, which contains Vps34, Vps15, Vps30/Atg6, Atg14 and Atg38 (Petiot et al., 2000; Kihara et al., 2001). Atg8 is a small hydrophilic ubiquitin-like protein, whose accumulation is upregulated upon starvation. Atg8 is immediately cleaved by cysteine protease Atg4 to expose the C-terminal glycine (Gly) residue, which is essential for subsequent conjugation reactions (Kirisako et al., 1999, 2000). In conjugation with phosphatidylethanolamine (PE), Atg8 is involved in the elongation of the isolation membrane; this is mediated by Atg7 and Atg3, which are E1 and E2 enzymes in the ubiquitylation reaction, respectively (Ichimura et al., 2000). Atg4 is also a deconjugation enzyme that liberates Atg8 from the Atg8-PE complex in the outer membrane of autophagosome after fusion with the vacuole (Kirisako et al., 2000). Similar to the Atg8-PE system, Atg12 is catalyzed by Atg7 (E1 enzyme) and Atg10 (E2 enzyme) and then conjugated with Atg5 via an isomerized peptide bond with a lysine residue at the middle of Atg5 (Mizushima et al., 1998). the Atg12-Atg5 complex has an E3-like function in the lipidation of Atg8 (Hanada et al., 2007). Atg16 is required for the localization of Atg12-Atg5 to PAS in a PI3P-dependent manner (Suzuki et al., 2007). Atg9 is the only known integral membrane protein involved in the regulation of autophagosome formation by interacting with the Atg18-Atg2 complex in a PI3P-dependent manner (Noda et al., 2000; Obara et al., 2008) (Figure 1).
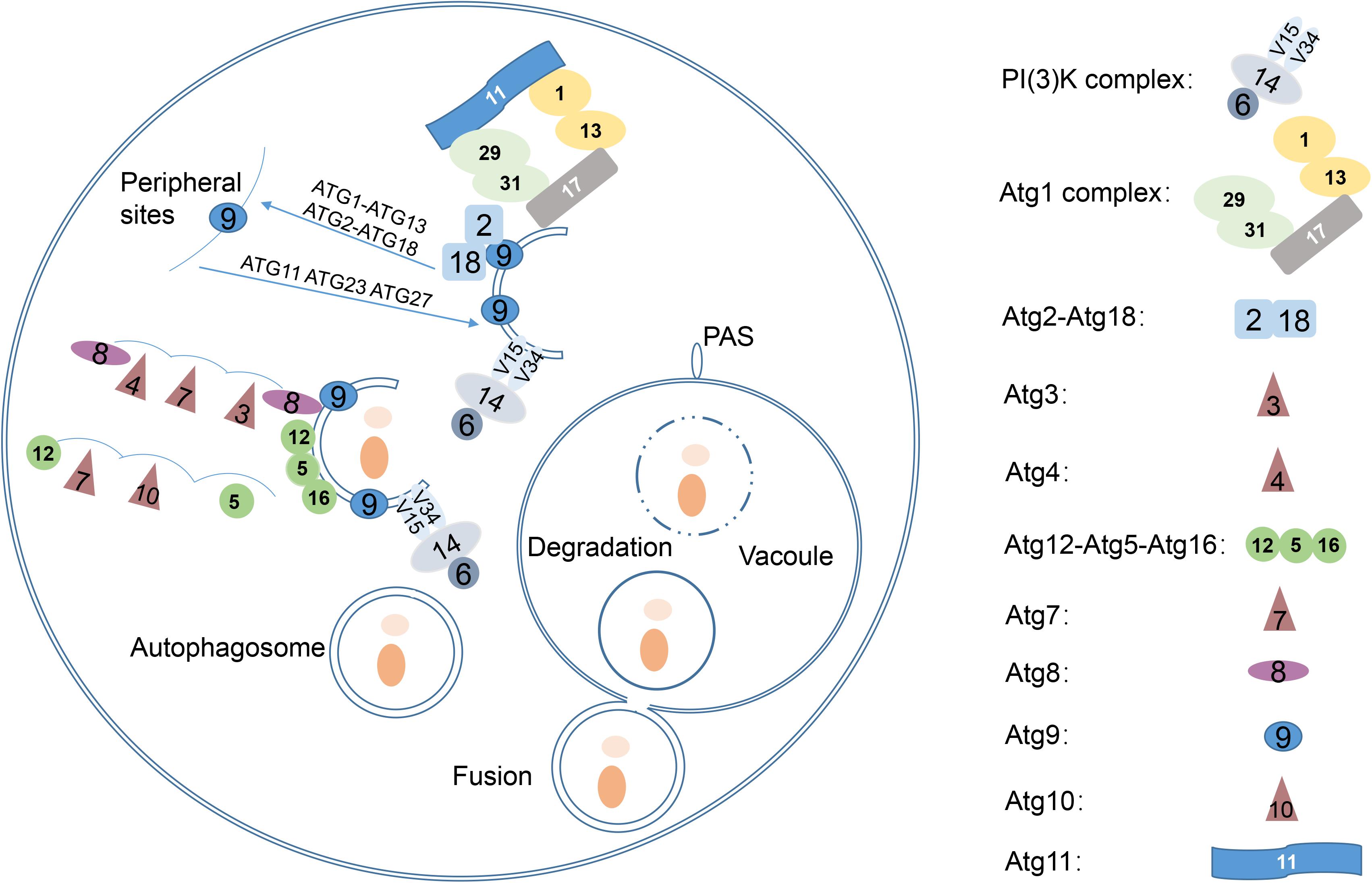
Figure 1. Overview of the autophagy pathway in yeast. Five key stages of autophagy are as follows: (1) Phagophore nucleates at the PAS and sequesters vesicle formation in yeast; (2) phagophore expands to sequester the cargo; (3) phagophore closes the double-membrane autophagosomes; (4) autophagosome fuses with the vacuole, releasing the inner vesicle, known as the autophagic body; (5) autophagic bodies are degraded by vacuolar hydrolases, and the products are released into the cytosol by various permeases. Revised from Nakatogawa et al., 2009.
C. neoformans is a major fungal pathogen of immunocompromised hosts, such as individuals who have human immunodeficiency virus (HIV)/AIDS or are subjects of organ transplant and long-term corticosteroid treatment (Park et al., 2009). C. neoformans infection is generally caused by the inhalation of yeast cells or spores that colonize the lung, resulting in cause pulmonary infection, and then disseminate through blood vessels, causing cryptococcal meningitis (Rajasingham et al., 2017). Recent studies have shown that several ATG genes are involved in the virulence of C. neoformans. Deletion of VPS34 leads to defective formation of Atg8-labeled vesicles and dramatic attenuation of virulence in mouse models of infection (Hu et al., 2008). Knockdown of the ATG8 gene led to higher sensitivity to starvation stress and attenuated virulence in both intravenous and intranasal murine models (Hu et al., 2008). Deletion of ATG7 led to increased cellular size and decreased survival rate in the lungs of infected mice (Oliveira et al., 2016). A recently published study suggests that different ATG genes are also involved in non-autophagic functions and contribute to virulence beyond their core functions in autophagy (Ding et al., 2018).
Despite these studies, a systematic investigation of how these ATG genes affect stress response, virulent factors, pathogenicity and autophagic flux under carbon and nitrogen starvation conditions is lacking. In this study, we generated 22 ATG mutants, and investigated the impacts of these genes on autophagic flux, virulence, stress response and pathogenicity of C. neoformans.
Materials and Methods
Strains, Plasmids and Culture Conditions
C. neoformans var. grubii strain H99 was used as the wild type (WT) in this study. The CRISPR-Cas9 system was used to knock out 22 ATG genes in C. neoformans. All vectors were amplified in Escherichia coli DH5α using ampicillin as the selection marker. Hygromycin B-resistance or Geneticin 418-resistance was used as the selection marker for transformed yeast cells. Yeast strains were cultured at 30°C with shaking at 180 rpm. The complete YPD medium (2% glucose, 2% Bacto peptone and 1% yeast extract [pH 6.0]) supplemented with 200 μg/mL hygromycin B or 40 μg/mL G418 were used for routine growth. Nitrogen starvation medium (SD-N; 0.17% yeast nitrogen base [without amino acids and ammonium sulfate] and 2% glucose), carbon starvation medium (SD-C; 0.17% yeast nitrogen base [without amino acids and ammonium sulfate], 0.5% ammonium sulfate, 0.5% casamino acids, 0.002% tryptophan, 0.002% adenine and 0.002% uracil) (Adachi et al., 2017), Nitrogen and carbon starvation medium (PBS; 0.8% NaCl, 0.02% KCl, 0.15% Na2HPO4 and 0.024% KH2PO4 [pH 7.4]) were used separately for nitrogen, carbon, nitrogen and carbon starvation treatments, respectively.
Construction of ATG Deletion (atgΔ) Mutants and Green Fluorescent Protein (GFP)-Atg8 Expression Strains
Each of the 22 ATG genes was deleted using the CRISPR-Cas9 system described previously (Wang et al., 2016). The plasmid contained double BspQI restriction sites between the U6 promoter and guide RNA. The target sequences (guiding RNA) were inserted into BspQI sites by annealing two reverse complementary sequences (TTG-N19/AAC-N19), followed by ligation using T4 ligase. Two homologous fragments were ligated to flank the hygromycin resistance (HygR) gene by double digestion with ClaI/HindIII and BamHI/XbaI, followed by ligation with T4 DNA ligase or In-Fusion HD Cloning kits (Takara). The pBS-Hyg-Atg-Cas9-gDNA plasmid was linearized using either KpnI or XbaI, and 1.5 μg of linearized fragments were transformed into the strain H99 by electroporation (Wang et al., 2016).
To construct the EGFP-Atg8 fusion protein expression plasmid, the ATG8 promoter (1,000 bp upstream of the start codon) and EGFP open reading frame (ORF) were ligated by overlapping PCR, and the resultant fragment was inserted into the KpnI site in pBS-G418 by In-Fusion HD Cloning to produce an intermediate plasmid. The ORF and terminator of ATG8 were then amplified from the genome of C. neoformans and ligated into the ClaI/HindIII sites of the intermediate plasmid to generate final plasmid pBS-G418-EGFP-Atg8. The final plasmid was linearized using KpnI, and 3 μg of the linearized product was transformed into the strain H99 and atgΔ mutants by electroporation. Yeast strains used in this study were listed in the Supplementary Table S1. Primers used in this study were listed in the Supplementary Table S2.
In vitro Virulent Factors Assays
Overnight (16-h) cultures in YPD liquid medium were harvested by centrifugation and washed three times with sterile PBS. Ten-fold gradient dilutions of WT and mutant strains were spotted on YPD agar plates and incubated at 37°C to investigate the thermotolerance of mutants. Laccase activity was assayed on Asn agar (0.1% glucose, 100 mg/L norepinephrine [TCI, Tokyo, Japan], 0.05% MgSO4-7H2O) by incubation in the dark for 3 days. Capsule formation was induced by incubating the WT and mutant cells in maltose medium (2% maltose, 0.1% Bacto peptone and 0.2% yeast extract) at 30°C for 4 days. Cells were stained with India ink and observed under a Nikon Eclipse 80i fluorescence microscope (Nikon, Tokyo, Japan).
To quantify laccase activity, overnight cultures were transferred to 0.1% Asn liquid medium for 5 h and diluted to a concentration of 1 × 108 cells/mL. Then, 100 μL of each yeast culture was placed in a 48-well clear flat-bottom plate. Then, 80 μL of 20 mg/mL 2, 2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) and 820 μL of 0.1 M Sodium acetate (pH 6.5) was added to each culture. Sterile 0.1% Asn liquid medium without any culture was used as a negative control. The plate was incubated at 30°C for 3 h. The absorbance of each culture was measured at 420 nm. The final EU of each strain was calculated using the following equation, modified from Alvarado-Ramírez et al., 2008:
Stress Response and Drug Sensitivity Assays
Ten-fold gradient dilutions of WT and atgΔ mutant cultures were spotted on YPD agar medium containing 0.01% SDS, 1.5 M NaCl, 2 M KCl, 1% Congo red, 10 mM H2O2, 1 M sorbitol and various antifungal drugs including Itraconazole (0.01 μg/mL), Fluconazole (6 μg/mL), Amphotericin B (0.1 μg/mL) and incubated at 30°C for 3 days.
Total RNA Isolation and Quantitative Real-Time PCR (qRT-PCR) Analysis
Total RNA was isolated using the RNAprep pure Plant Kit (Tiangen). Approximately 500 ng of total RNA was reverse-transcribed with oligo dT18 primer. Then, qRT-PCR was performed using 10 μL of SYBR green master mix (Roche), 0.5 μL of each primer, 8 μL of DNase/RNase-free water and 1 μL of cDNA or distilled water (negative control). Gene expression levels were determined using the ΔΔCT method, and the Actin1 (ACT1) gene was used for data normalization.
Pathogenicity of atgΔ Mutants in Galleria mellonella
The pathogenicity of atgΔ mutants in G. mellonella was assessed as described previously (Mylonakis et al., 2005). Larvae (0.2–0.3 g) without any dark spots were incubated at 28°C until the day before infection. Yeast cells were grown overnight in YPD liquid medium, washed three times with PBS and resuspended in PBS containing kanamycin (50 μg/mL) to a final concentration of 1 × 108 cells/mL. Then, 20 larvae injected into the last right proleg with 10 μL of yeast suspension. Another group of 20 larvae injected with 10 μL of PBS containing kanamycin served as a control. Larvae were incubated at 28°C, and their survival rate was monitored daily. Survival curves were drawn using the GraphPad Prism 5 software (GraphPad, San Diego, CA, United States).
Starvation Survival Assays
Yeasts were grown to mid-log phase in YPD liquid medium, harvested, washed three times with sterile PBS, and resuspended in 5 mL of SD-N, SD-C or PBS to a final concentration of 1 × 108 cells/mL. The cultures were incubated at 30°C with shaking at 180 rpm and starved for different durations. Subsequently, serially diluted cell suspensions at a starting concentration of 1 × 108 cells/mL were spotted on YPD agar plates. The plates were incubated at 30°C for 3 days to determine survival under starvation conditions.
Fluorescence Microscopy and Western Blotting
Yeast cells expressing EGFP-Atg8 were grown to mid-log phase in YPD medium, starved in SD-N or SD-C medium for 4 h and observed by both fluorescence microscopy and differential interference contrast microscopy (DIC) using laser scanning confocal microscope LSM710 (Zeiss, Germany).
Total protein was extracted by resuspending the cell pellet in 100 μL RIPA lysis buffer (1% PMSF, 1% PIC mix) containing an equal volume acid-washed glass bead. The mixture was vortexed for 1 min, incubated on ice for 1 min, and repeated five times. The lysate was obtained by centrifugation at 12,000 rpm for 10 min at 4°C. The concentration of yeast lysate was determined with using the BCA kit. Then, equal amount of total protein was mixed with 6 × SDS loading buffer (300 mM Tris-HCl [pH 6.8], 12% SDS, 60% glycerin, 0.6% bromophenol blue and 0.6% β-mercaptoethanol) and boiled for 5 min. Western blotting was performed as described previously (Zhu et al., 2001). Briefly, samples were separated by 12% SDS-PAGE at a constant voltage of 100 mV for approximately 2 h. Then, the separated yeast proteins were transferred to a PVDF membrane using Trans Blot Turbo (BioRad). The membrane was blocked by incubation in 5% (w/v) skim milk at 25°C for 2 h. To detect the GFP-Atg8 and internal control, membrane was incubated with anti-GFP (1:3,000; Santa Cruz Biotechnology) or anti-β-actin (1:3,000; GenScript) primary antibody, followed by anti-mouse IgG HRP-linked secondary antibody (1:5,000; Cell Signal). The ratio of cleaved to non-cleaved GFP protein was measured to monitor the autophagy flux.
Results
Characterization of ATG Genes in C. neoformans
Amino acid sequences of S. cerevisiae Atg proteins were used to identify Atg orthologs in C. neoformans var. grubii H99 strain1. A total of 23 Atg proteins were identified, based on the presence of conserved domains (Table 1). However, the genome of C. neoformans strain H99 did not contain orthologs of Atg10, an important and specific E2 enzyme required for Atg12-Atg5 conjugation (Shintani et al., 1999). Although Tongbao Liu’s group has claimed the identification of Atg10 in C. neoformans, it showed extremely low similarity to known homologs, implying that the role of this protein in Atg12-Atg5 conjugation needs further verification (personal communication). Orthologs of Atg17, 19, 21, 23, 29 and 31 were also not found in C. neoformans, possibly because of the very low level of similarity or the absence of these proteins in C. neoformans. Interestingly, a homolog of mammalian Atg101 (CNAG_07646) was identified in C. neoformans; this represents one of the major difference between S. cerevisiae and higher eukaryotes, as Atg29 and Atg31 in S. cerevisiae have been replaced by Atg101 in mammals (Hegedüs et al., 2014; Nanji et al., 2017). This suggests that C. neoformans is closer to mammals than to S. cerevisiae, which has important implications given that C. neoformans is a human pathogen. Two Atg14 homologous proteins (CNAG_03608 and CNAG_05500, later denoted as Atg14-03 and Atg14-05, respectively) were also identified in C. neoformans; Atg14 is a specific subunit of one of the PtdIns 3-kinase complexes responsible for targeting the complex to the probable site of autophagosome formation (Figure 2).
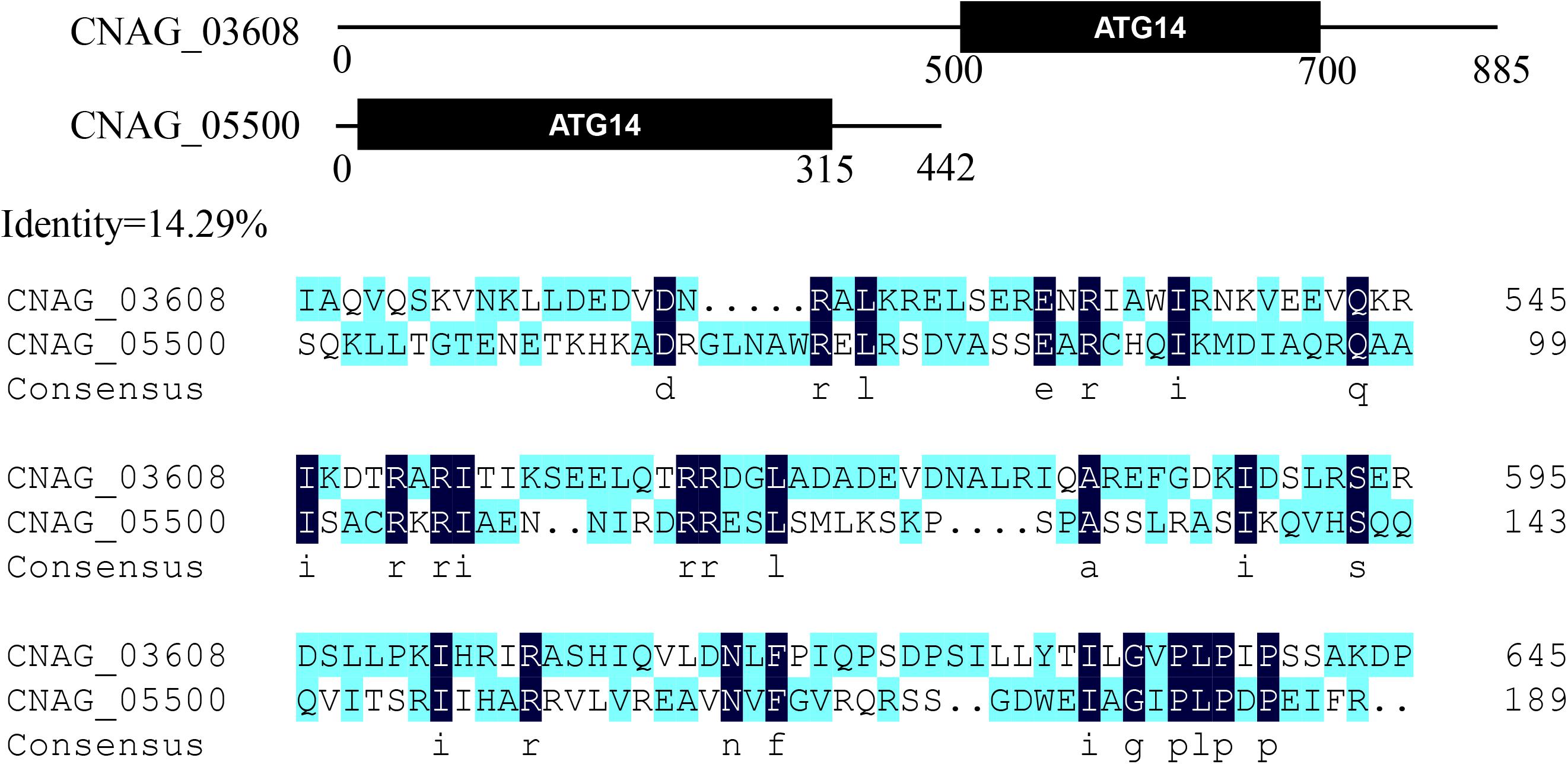
Figure 2. Two Atg14 homologous proteins in Cryptococcus neoformans. Putative conserved domains of Atg14-03 (CNAG_03608) and Atg14-05 (CNAG_05500), and amino acid sequence alignments of two Atg14 homologs are shown.
Confirmation of the Deletion of the 22 ATG Genes
To investigate the function of ATG genes, we replaced the genomic copy of each ATG gene with the HygR gene using the CRISPR-Cas9 suicide cassette system (Supplementary Figure S1A). To confirm whether the replacement cassette was correctly inserted, the putative atgΔ transformants were screened by PCR with three primer pairs, Atg-LF/Hyg-R, Hyg-F/Atg-LR, and Atg-LF/Atg-LR, in the last electropherogram, which produced 1,212-bp, 1561-bp and 4,532-bp PCR products, respectively, in the true transformants, whereas the WT produced a 2,426-bp PCR product using the Atg-LF/Atg-LR primer pair (Supplementary Figure S1B). The atgΔ strains produced a band that was 2,106 bp larger than H99 because of the insertion of hygromycin cassette. No PCR product could be amplified from the H99 genome using the primer pairs Atg-LF/Hyg-R and Hyg-F/Atg-LR (data not shown).
Contribution of ATG Genes to Autophagic Flux Under Nitrogen and Carbon Starvation Conditions
Studies have shown that ATG genes regulate autophagic flux in S. cerevisiae; however, whether these genes play similar roles in C. neoformans needs further investigation. One method to monitor autophagic flux is to follow the release of free GFP moiety from the GFP-Atg8 fusion protein by western blotting after phagosomes fuse with the vacuole (Cheong and Klionsky, 2008). To confirm that ATG genes are functionally associated with autophagy in C. neoformans, we constructed 15 strains expressing GFP-Atg8 fusion protein, and analyzed the release of GFP-Atg8 in these mutants.
We first analyzed autophagic flux induced under nitrogen starvation conditions. The results showed that the deletion of ATG1, ATG13, ATG11, ATG7, ATG5 and ATG12 led to significantly lower autophagic flux, as no GFP release was detected in the mutant strains after nitrogen starvation for 6 h, whereas significant GFP release was detected in the WT, indicating that these genes are essential for boosting autophagic flux under nitrogen starvation conditions (Figure 3A). We also found that autophagic flux decreased in atg2Δ, atg9Δ, atg18Δ and atg16Δ mutants under nitrogen starvation conditions, indicating that autophagic flux is also dependent on these proteins to some extent. However, deletion of Atg6 and Atg14, two subunits of the PI3K complex I, did not affect the cleavage of GFP-Atg8. Autophagic bodies were easily observed in the atg6Δ and atg14Δ mutants under nitrogen starvation, indicating the ATG6 and ATG14 are not essential for the formation of autophagic bodies (Supplementary Figure S2). Deletion of two components of Atg8 ubiquitin-like conjugation system, Atg3 and Atg4, also did not affect the cleavage of GFP-Atg8. These results suggest that these genes are not directly be involved in the regulation of autophagic flux in C. neoformans.
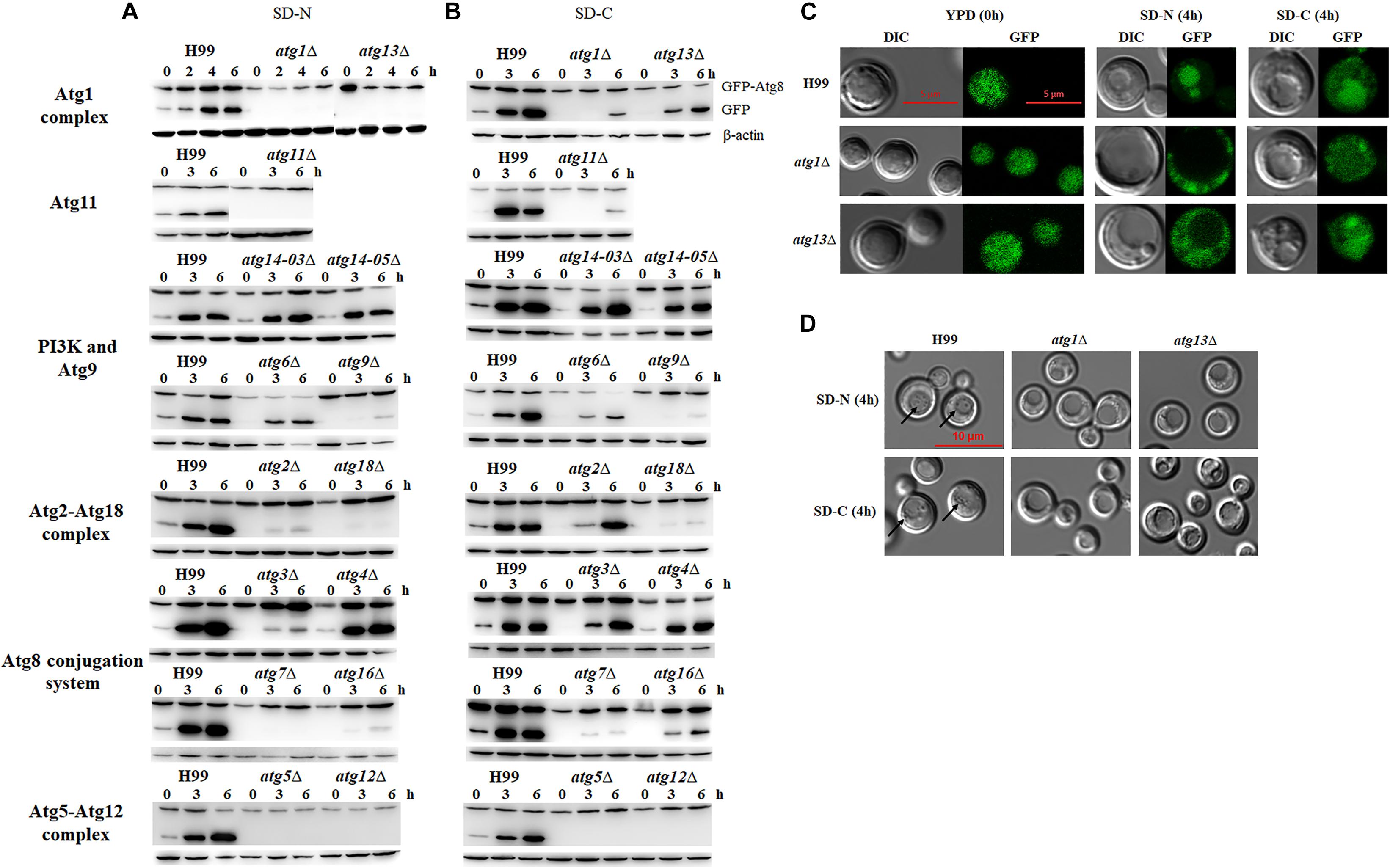
Figure 3. Nitrogen or carbon starvation induces autophagy in C. neoformans. (A,B) Analysis of GFP-Atg8 processing by western blotting. H99 and atgΔ strains transformed with a plasmid expressing the EGFP-Atg8 fusion protein were collected after shifting to nitrogen starvation medium (SD-N) (A) or carbon starvation medium (SD-C) (B) at the indicated times. Total proteins were extracted and subjected to immunoblot analysis; β-actin was used as the loading control. (C) Localization of EGFP-Atg8 upon nutrient starvation. H99, atg1Δ and atg13Δ cells in log phase (starved; 0 h) or after shifting to SD-N or SD-C medium containing 10 μg/mL nocodazole for 4 h were observed by both fluorescence microscopy and differential interferential contrast (DIC) with a laser scanning confocal microscope. (D) Autophagic body formation. H99 and atgΔ strains were incubated in SD-N or SD-C medium containing 1 mM PMSF and 10 μg/mL nocodazole for 4 h and subjected to DIC microscopy. Arrows indicate autophagic bodies within vacuoles.
Carbon is essential for all living organisms, and the energy produced by the metabolism of carbon sources can support almost all vital activities. Here, we tested whether deletion of the ATG genes affects autophagic flux under carbon starvation conditions, i.e., in the SD-C medium. When the WT cells were transferred from YPD to SD-C medium 3h, the release of GFP from the GFP-Atg8 fusion protein was significantly increased, indicating the boosting effect of glucose starvation on autophagic flux. In mutant cells, only the deletion of ATG5 and ATG12 led to the total loss of autophagic flux after starvation for 6 h; however, the deletion of ATG1, ATG7, ATG9, ATG11, ATG16 and ATG18 resulted in significantly reduced but not total loss of GFP release, indicating their involvement in maintaining autophagic flux upon carbon starvation (Figure 3B).
In comparison with nitrogen starvation, carbon starvation was more efficient in the induction of autophagic flux in C. neoformans, as the release of GFP was still visible even after the ATG genes were deleted (Figures 3A,B). In other words, these genes play less important roles in controlling autophagic flux under glucose starvation conditions than under nitrogen starvation conditions. Although the deletion of ATG1 and ATG11 led to decreased autophagic flux after starvation for 3 h, autophagic flux was enhanced again when mutant cells were incubated for 6 h under glucose starvation conditions. Similar results were obtained for other ATG mutants, except atg5Δ and atg12Δ, indicating that long-term starvation-induced GFP release might not depend on these genes.
To further confirm the roles of these ATG genes in the regulation of autophagic flux, localization of GFP fluorescence and formation of autophagic body in the vacuole were observed in the atg1Δ and atg13Δ mutant cells grown in SD-N or SD-C medium for 4 h by laser scanning confocal microscopy. Fluorescence was detected in the entire cells of the WT and atg1Δ and atg13Δ mutants under nutrient proficient conditions (Figure 3C). When the cells were incubated in the SD-N medium for 4 h, fluorescence relocated to vacuole in the WT but remained in the cytosol in the atg1Δ and atg13Δ mutants, confirming the important roles of ATG1 and ATG13 in regulating autophagic flux under nitrogen starvation conditions. However, when the cells were shifted from YPD to SD-C for 4 h, partial fluorescence in cytosol of the WT and atg13Δ mutant relocated to the vacuole; no relocation was detected in the atg1Δ mutant. These results indicated that the ATG genes perform non-essential roles in regulating autophagic flux under glucose starvation conditions.
To further confirm this result, autophagic body formation in the vacuole was observed using DIC microscopy under different starvation conditions. Autophagic bodies were observed in vacuole of WT cells under nitrogen starvation conditions but not in the vacuoles of atg1Δ and atg13Δ mutants, confirming the essential roles of ATG1 and ATG13 in autophagic flux. Under glucose starvation conditions, autophagic bodies was observed in the WT but not in the atg1Δ and atg13Δ mutants (Figure 3D). These results indicated that ATG genes play different roles in enhancing autophagic flux under nitrogen and carbon starvation conditions.
Contribution of ATG Genes to Traditional Virulent Factors
C. neoformans is a model system for studying fungal pathogenesis and exhibits three well-known pathogenesis-associated virulent factors, including the ability to grow at 37°C, antioxidant melanin and polysaccharide capsule (Williamson, 1997; Zhu and Williamson, 2004). Therefore, we first analyzed these three virulent factors in each of the 22 atgΔ mutants. The atg6Δ and atg24Δ mutants almost completely lost their abilities to grow at 37°C (Figure 4A), indicating that ATG6 and ATG24 are essential for thermotolerance. The atg7Δ, atg8Δ, atg12Δ, atg14-03Δ and atg20Δ mutants also exhibited decreased growth rate at 37°C but continued to propagate slowly, indicating their possible involvement in thermotolerance. Deletion of other ATG genes did not affect thermotolerance. Therefore, the contribution of these genes to thermotolerance showed the following order: ATG6 > ATG24 > ATG14-03 > ATG7, 8, 12, 20 > other ATG genes.
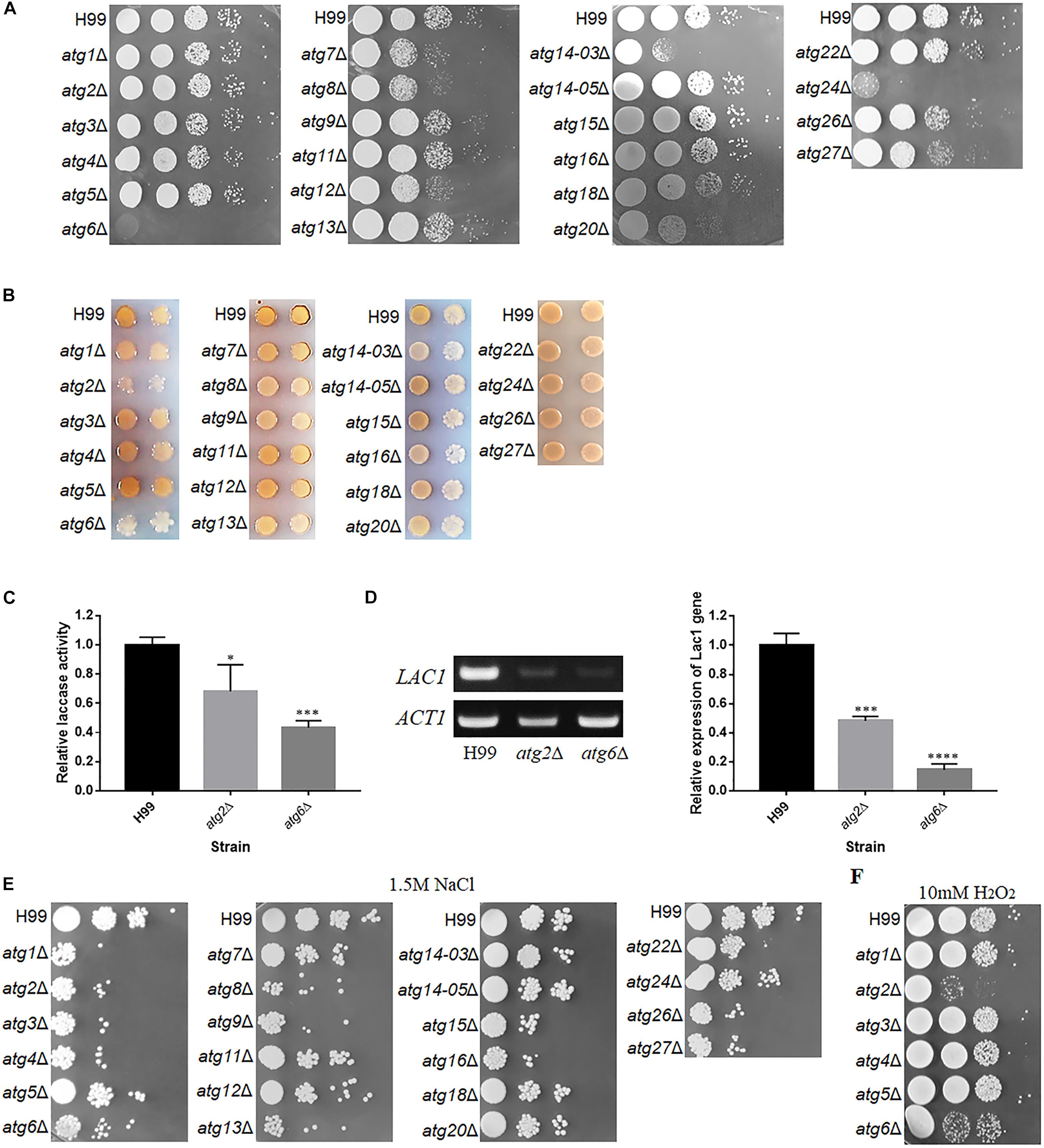
Figure 4. Phenotypes of atgΔ mutants of C. neoformans. (A) Thermotolerance test. Serial 10-fold dilutions of H99 and atgΔ strains were spotted onto YPD medium and incubated at 37°C. (B) Laccase activity assay to determine melanin production. Each strain was spotted and cultured on Asn medium containing 0.1% glucose and 100 mg/L norepinephrine at 30°C for 3 days. (C) Relative laccase activity of H99, atg2Δ and atg6Δ strains using ABTS as a substrate. The assay was performed three times. Asterisks indicate significant differences between the wild-type (H99) and mutant strains (∗P < 0.05; ∗∗∗P < 0.001; Student’s t-test). (D) Qualitative and quantitative analysis of LAC1 expression in atg2Δ and atg6Δ by reverse transcription PCR (RT-PCR) and quantitative real-time (qRT-PCR) upon induction by 0.1% glucose for 5 h. The assay was performed three times. Asterisks indicate significant differences (∗∗∗P < 0.001; ****P < 0.0001; Student’s t-test). (E,F) Sensitivity of atgΔ to NaCl and H2O2. Serial 10-fold dilutions of H99 and atgΔ strains were spotted on agar plates containing 1.5 M NaCl or 10 mM H2O2 and incubated at 30°C.
The production of melanin was significantly lower in the atg2Δ, atg6Δ, atg14-03Δ and atg16Δ mutants compared with the WT (Figure 4B). However, atg2Δ and atg6Δ mutants showed poor growth on 0.1% Asn, possibly because of inadequate nutrition. Therefore, the impact of the deletion of ATG2 and ATG6 on laccase activity was further determined in liquid culture using ABTS as the substrate. The laccase activity of atg2Δ and atg6Δ decreased by 30 and 60%, respectively, compared with the WT (Figure 4C). Consistent with this observation, the expression of LAC1, a laccase gene, was significantly down-regulated to 14.5% in the atg6Δ mutant, compared with the WT, as determined by qRT-PCR (Figure 4D). These results confirmed that Atg2 and Atg6 regulate laccase activity and consequently melanin production. However, deletion of other ATG genes did not affect the production of melanin.
The size of the capsule induced on maltose medium was also determined for these mutants. We found that the disruption of 20 ATG genes led to a smaller capsule than the WT; however, all mutants retained the ability to produce the capsule, implying that these ATG genes play insignificant roles in capsule synthesis (Supplementary Figure S3), and most of these genes positively control capsule formation. These results indicate that although ATG genes contribute differentially to the three traditional virulent factors, deletion of these genes dramatically affect thermotolerance at physiological temperature.
To confirm these phenotypes, we selected four defective strains (atg1Δ,atg7Δ,atg8Δ, and atg9Δ), and its complementation strains were constructed by co-transforming the ATG cassettes and the pBS-G418 plasmid. First, we verified the complementation strains by PCR. WT-sized bands were amplified in the correct transformants, but a larger band was amplified in the defective strains (Supplementary Figure S4A). Then, we measured the high temperature tolerance of these strains as well as capsule induction in low-iron medium and melanin production on L-DOPA agar plates (Ding et al., 2018). The atg7Δ and atg8Δ strains showed slight sensitivity to high temperatures; however, normal phenotype was restored in the respective complementation strains (Supplementary Figure S4B). The size of capsule in atg1Δ, atg7Δ, atg8Δ, and atg9Δ strains was smaller than the WT; this phenotype was also restored in the respective complementation strains (Supplementary Figure S4C). Deletion of none of the four genes affected the production of melanin compared with the WT (Supplementary Figure S4D).
Contribution of ATG Genes to Stress Tolerance and Antifungal Drugs Resistance
In addition to traditional virulent factors, stress tolerance and antifungal resistance of fungal pathogens are also related to pathogenesis. Therefore, WT and atgΔ strains were serially diluted and spotted on YPD agar medium containing 1.5 M NaCl, 2 M KCl or H2O2. Interestingly, atg1Δ, atg2Δ, atg3Δ, atg4Δ, atg6Δ, atg8Δ, atg9Δ, atg13Δ, atg15Δ, atg16Δ, atg26Δ and atg27Δ displayed higher sensitivity to 1.5 M NaCl (Figure 4E) and 2 M KCl (data not shown) than the WT, indicating their contributions to osmotic stress resistance. The atg2Δ and atg6Δ strains also displayed higher sensitivity to oxidative stress caused by H2O2 than the WT (Figure 4F), implying these two gene play important antioxidative roles.
The same method was used to determine the sensitivity of these mutants to various antifungal drugs. However, none of the mutants showed high sensitivity to antifungal drugs (Supplementary Figure S5), indicating that either these genes are not involved in antifungal activity to the specific drugs tested in this study or the concentration of drugs used in this investigation was not high enough to elicit a response.
Contribution of ATG Genes to Starvation Tolerance
Because autophagy is important for eukaryotic organisms to withstand starvation tolerance, and survival under starvation condition has been clearly linked to pathogenicity of several pathogens (Hu et al., 2008), starvation tolerance was further analyzed for these C. neoformans mutants under nitrogen starvation (SD-N) or carbon starvation (SD-C) or both carbon and nitrogen starvation (PBS) conditions.
In the 12-day nitrogen starvation treatment, atg1Δ, atg2Δ, atg3Δ, atg5Δ, atg6Δ, atg7Δ, atg8Δ, atg9Δ, atg12Δ, atg14-03Δ, atg14-05Δ, atg15Δ and atg18Δ mutants showed lower survival rate than the WT; the survival rate of atg2Δ, atg6Δ, atg8Δ, atg9Δ, atg12Δ, and atg18Δ strains was significantly lower than the WT in SD-N medium (Figure 5A).
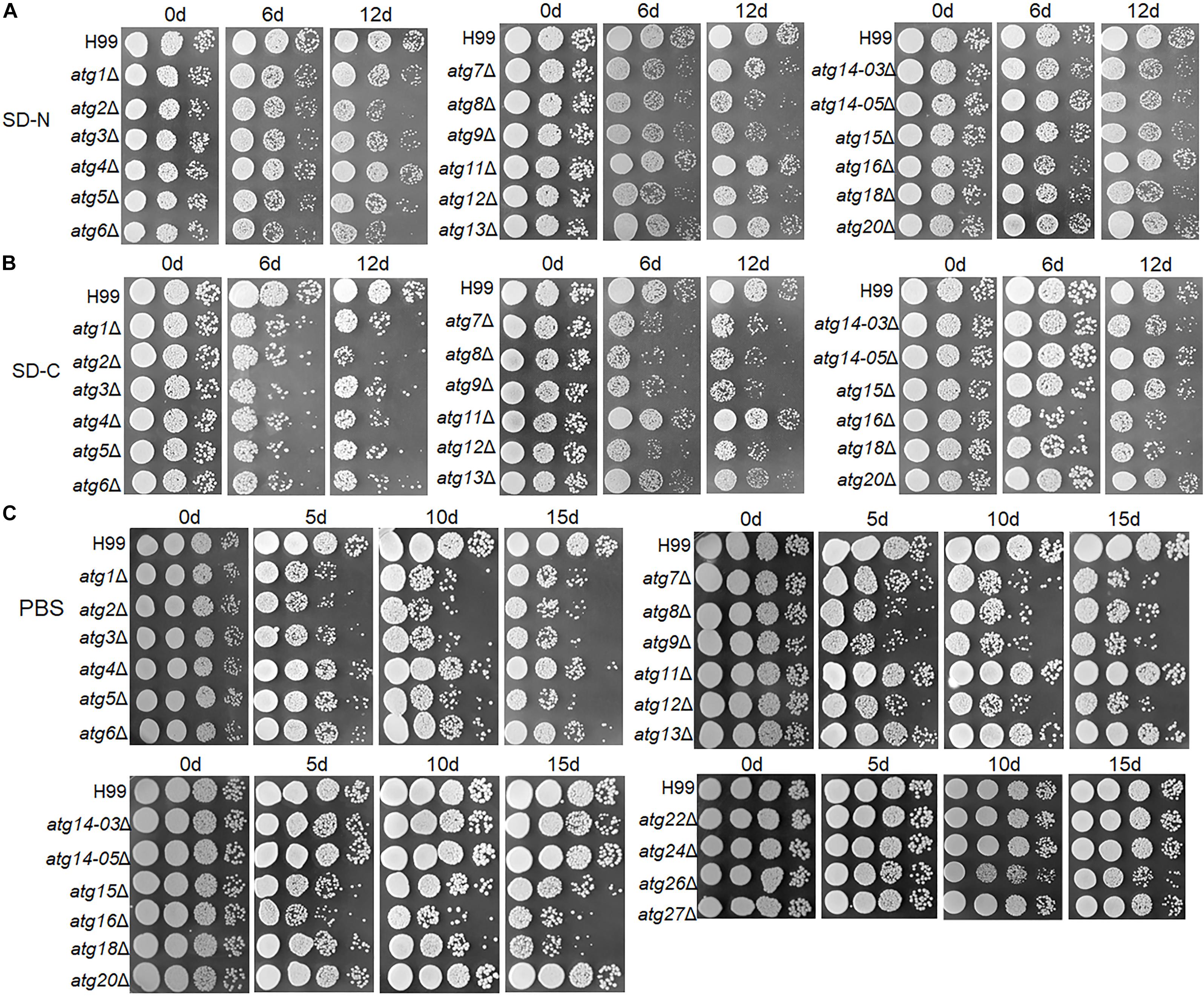
Figure 5. Tolerance of atgΔ strains to nitrogen and/or carbon starvation. (A–C) H99 and atgΔ strains were grown in liquid YPD for 16 h and then grown on SD-N medium (A), SD-C medium (B) or PBS (C) for the indicated durations. Dilutions were grown on YPD plates for 2 days.
In the 6-day glucose starvation treatment, all mutants (except atg11Δ, atg13Δ, atg14-03Δ, atg14-05Δ, atg15Δ and atg20Δ) showed significantly lower survival rate than the WT (Figure 5B).
Under both glucose and nitrogen starvation conditions (i.e., in the PBS buffer), atg1Δ, atg2Δ, atg3Δ, atg4Δ, atg5Δ, atg6Δ, atg7Δ, atg8Δ, atg9Δ, atg12Δ, atg15Δ, atg16Δ and atg18Δ mutants showed significantly lower survival rate than the WT. The remaining mutants did not show a drastic reduction in survival rate even after starvation for 15 days (Figure 5C).
These results demonstrate that the ATG genes contribute differentially to starvation tolerance, and most these genes play more important roles in tolerance to carbon-only or carbon and nitrogen starvation than in tolerance to nitrogen-only starvation.
Contribution of ATG Genes to the Pathogenicity of C. neoformans in the G. mellonella Model
Our results showed differential roles of ATG genes in the regulation of pathogenesis-related virulence. Next, we investigated whether the inference of these virulent factors would eventually lead to a change in pathogenesis. To perform this experiment, we used the G. mellonella model, which has been widely used in evaluating pathogenesis of C. neoformans (Mylonakis et al., 2005). We found that atg5Δ, atg6Δ, atg7Δ, atg8Δ, atg9Δ, atg11Δ, atg12Δ, atg14-03Δ, atg15Δ, atg16Δ, atg20Δ and atg24Δ strains showed remarkably attenuated pathogenesis in this invertebrate model, whereas atg6Δ, atg11Δ and atg14-03Δ were almost entirely avirulent (Figure 6). The other mutant strains did not show attenuated pathogenesis. Our results further confirmed that PI3K complex I signaling is a virulence-associated trait during infection with C. neoformans.
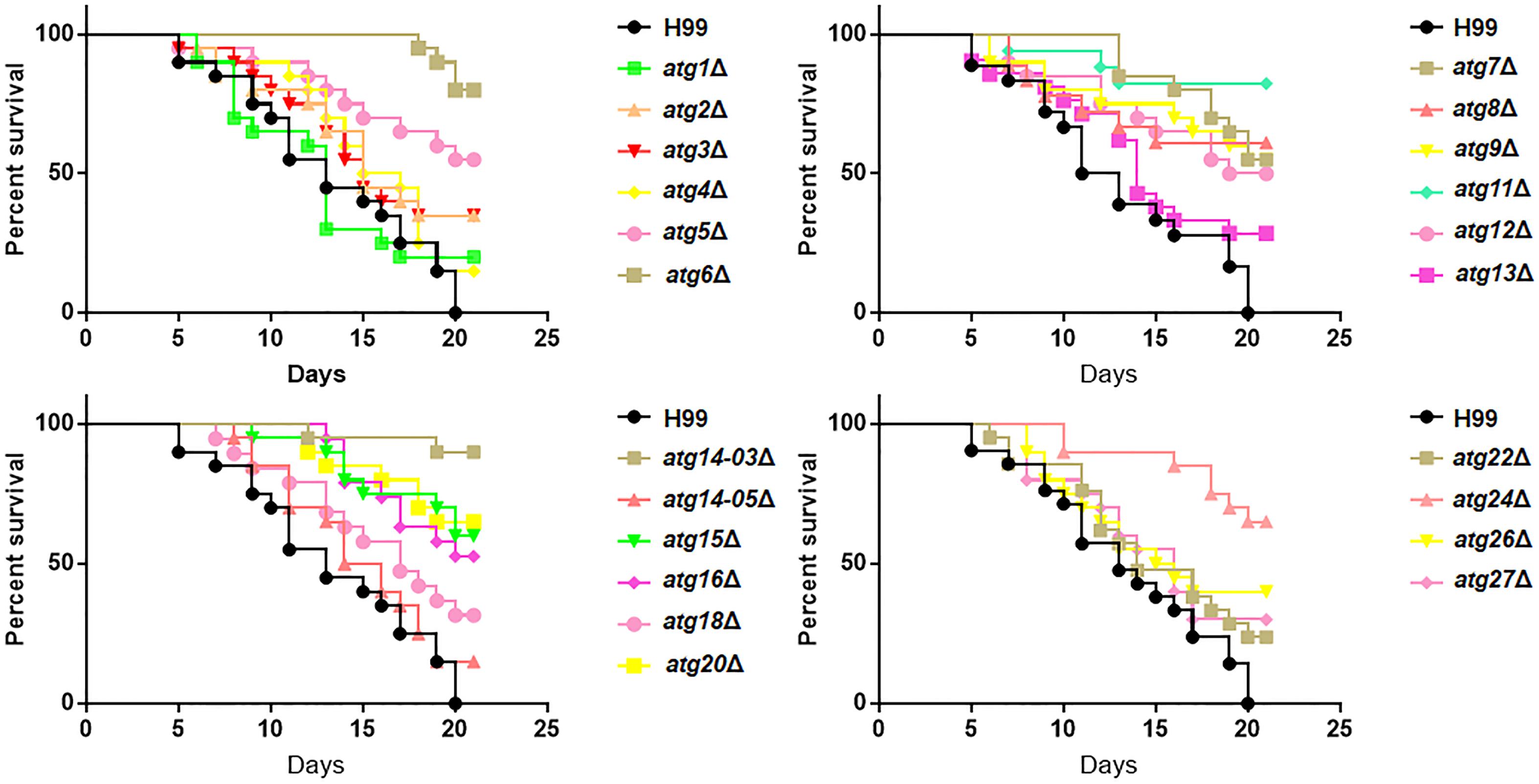
Figure 6. Role of ATG genes in the virulence of C. neoformans. Survival of Galleria mellonella larvae infected with 106 cells of H99 or atgΔ strains. The larvae were incubated at 30°C, and their survival was monitored daily.
Discussion
Autophagy is a highly conserved process essential for sustaining cellular integrity, homeostasis and survival (Mizushima et al., 2008). Although approximately 18 core genes have been confirmed to be essential for the critical autophagic steps (Farré and Subramani, 2016), their specific roles need to be investigated in different organisms, considering the influence of evolutionary forces on autophagic processes. In this study, 23 Atg-related homologs of S. cerevisiae were identified through BLAST searches of the C. neoformans protein database (Table 1), suggesting that the autophagy machinery is relatively well conserved between S. cerevisiae and C. neoformans.
Interestingly, several S. cerevisiae ATG gene homologs were not identified in C. neoformans including ATG10, 17, 19, 21, 23, 29 and 31. The lack of E2-like enzyme Atg10 is an interesting finding, as Atg10 is responsible for catalyzing the conjugation of Atg12 to Atg5 and is essential for autophagosome formation in S. cerevisiae (Shintani et al., 1999). However, the appearance of a simple non-covalent system in apicomplexan parasites, Plasmodium and Toxoplasma, and yeast species, Komagataella phaffii, might partially explain the absence of Atg10 in C. neoformans. This non-covalent Atg12-Atg5 complex retains the ability to facilitate Atg8-PE conjugation (Pang et al., 2019). Therefore, whether C. neoformans also adopts a similar non-covalent interaction system is worth investigating. Atg17 is reportedly conserved among yeast and filamentous fungal species. Although a possible homolog of yeast Atg17 has been found in C. neoformans (CNAG_01538) (Palmer et al., 2008), it was actually found to have very low homology to ScAtg17 in the structural maintenance of chromosomes (SMC) domain. Atg19 functions as a receptor for the cytoplasm-to-vacuole targeting (Cvt) cargo and mediate selective autophagy in S. cerevisiae (Chang and Huang, 2007). The absence of this homolog in C. neoformans indicates that the Cvt process might proceed with a different receptor. Atg21 is also essential for selective autophagy in the Cvt pathway or mitophagy and has been proposed to be required for the recruitment of Atg8 to PAS, especially in the Cvt pathway (Meiling et al., 2004a). Atg23 is required for the maturation proaminopeptidase I but not for autophagy (Meiling et al., 2004b). Thus, our data suggest that either the Cvt pathway is absent in C. neoformans or utilizes a different set of enzymes. Atg29 and Atg31 together with Atg17 were found to constitutively form the Atg17-Atg29-Atg31 ternary complex, which acts as an essential scaffold for the organization of PAS in S. cerevisiae (Kabeya et al., 2009). The absence of this complex suggests that autophagy induction and PAS organization are completely different in C. neoformans. A putative mammalian homolog, Atg101, was unexpectedly found in C. neoformans. In mammalian cells, Atg101 is critical for autophagy together with ULK, Atg13 and FIP200 (Jung et al., 2009); however, it is not conserved in budding yeast. Therefore, whether Atg101 in C. neoformans plays a similar role as the mammalian Atg101 is an interesting question. Unfortunately, we could not obtain atg101Δ mutant cells for unknown reasons.
Only a few ATG genes have been reported to be involved in regulating classical virulent factors. Deletion of Vps34 leads to significantly reduced melanin production (Hu et al., 2008). In our study, only putative ATG genes were analyzed. However, because Vps34, Atg14 and Atg6 form a complex to mediate the formation of autophagosome, deletion of ATG6 and ATG14 should provide some clues about the function of the PI3K complex. Our results showed that the deletion of ATG6 and ATG14-03 affected laccase production, indicating that the PI3K complex does regulate melanin production in C. neoformans (Figure 4B). However, deletion of another homolog, Atg14-05, did not result in significantly reduced melanin production, which raises a question regarding the role of Atg14-05 in autophagy in C. neoformans. Interestingly, in a previous study, the atg1Δ, atg8Δ, and atg9Δ mutants are not different from the WT in the growth at 30°C and 37°C or in the production of melanin. The atg7Δ mutant cause slight growth defect at both 30°C and 37°C and produce less melanin than the WT strain after 48-h incubation. These atgΔ mutants are able to produce a polysaccharide capsule similar in size to WT (Ding et al., 2018). However, there are some differences between our study and that of Ding et al. The atg7Δ and atg8Δ mutants resulted in slight sensitivity to high temperatures, and these four atgΔ mutants led to smaller capsules (Figure 4 and Supplementary Figure S4). As studies have pointed out that heat stress induces autophagy to insure protein homeostasis at high temperatures (Zhao et al., 2009), deletion of the heat shock transcription factor 1 (HSF1) increases basal autophagy levels (Dokladny et al., 2013), moreover, the transcriptions of ATG genes in C. neoformans are significant increase when transferred from 30 to 37°C in YPD medium (Gontijo et al., 2017), we speculate that autophagy can also be induced by high temperature in C. neoformans. Atg7 and Atg8 are the core proteins in autophagy process, and they caused slight growth defects at high temperatures which was more significant at 38°C (data not show). Capsules are induced with maltose medium (low nitrogen) and LIM medium (low iron). An important way for responding to low nitrogen and low iron conditions is the Pka1-cyclicAMP (cAMP) pathway (O’Meara and Alspaugh, 2012). Autophagic bodies was also observed in the vacuoles of the WT induced in maltose medium (data not show). However, we cannot explain how the deletion of the ATG gene reduces the size of the capsule based on our existing results. Except for the studies on the abovementioned ATG genes, no reports are available on the influence of ATG genes on virulent factors.
Atg20 and Atg24, the sorting nexins required for selective autophagy including the Cvt pathway, pexophagy and mitophagy (Zhao et al., 2016), were also required for the growth of C. neoformans at 37°C, indicating that selective autophagy might also be involved in regulating this virulent factor. A number of ATG transcripts have been reported to be induced during macrophage internalization. Suppression of ATG8 transcripts with RNAi resulted in severely attenuated fungal virulence, and reduction in the virulence of the vps34Δ mutant is caused by a defect in starvation tolerance and loss of the ability to form autophagic bodies during starvation and after phagocytosis by macrophages (Hu et al., 2008). By contrast, autophagy is dispensable for virulence in other pathogenic fungi including Candida albicans and Aspergillus fumigatus (Palmer et al., 2007; Richie et al., 2007). In our study, 12 atgΔ strains showed remarkably attenuated virulence in the invertebrate model of cryptococcosis (Figure 6), demonstrating a close relationship between autophagy and microbial virulence in C. neoformans.
Previous research has shown that the high osmolality glycerol (HOG) pathway controls the resistance of cells to osmotic stress in C. neoformans (Bahn et al., 2005). As most of atgΔ mutants showed reduced growth under high osmotic pressure, whether these ATG genes interact with HOG1 (MAPK) pathway to cope with high salt conditions deserves our attention. The atg2Δ and atg6Δ mutants were also sensitive to 10 mM H2O2 treatment, which often leads to the production of reactive oxygen species (ROS; Figure 4F). The deletion of ATG1 in Candida glabrata results in higher intracellular ROS than the WT, especially upon induction by H2O2 (Shimamura et al., 2019). Ethanol stress can also induce autophagy, which is regulated by H2O2 and superoxide anion (O2–) derived from the mitochondrial electron transport chain (mtETC), and autophagy in turn contributes to the elimination of H2O2 and O2– (Jing et al., 2018). In C. neoformans, whether this relationship exists between ROS elimination and autophagy is worth exploring.
Autophagy is a conserved cellular mechanism essential for survival upon starvation. In this study, most atgΔ mutants were sensitive to nitrogen starvation or carbon starvation, especially in the absence of glucose (Figures 5A,B). Under nitrogen starvation conditions, atg1Δ, atg13Δ, atg11Δ, atg7Δ, atg5Δ and atg12Δ mutants completely lost the ability to process GFP-Atg8 (Figure 3A) indicating significantly reduced autophagic flux. The Atg1-Atg13 complex acts as a scaffold for the recruitment of other Atg proteins essential for autophagy, and deletion of ATG1 and ATG13 blocks autophagic flux in S. cerevisiae and S. pombe (Kawamata et al., 2008; Mukaiyama et al., 2009). Atg11 acts as a scaffold for recruiting core Atg proteins and target proteins for selective degradation (Yorimitsu and Klionsky, 2005). Our analyses revealed that Atg11 was necessary for bulk autophagy. Atg7 is a unique E1 enzyme that activates two ubiquitin-like molecules, Atg8 and Atg12, and transfers them to different E2 enzymes (Yamaguchi et al., 2012). The formation of the Atg8-PE complex depends on the activity of the Atg12-Atg5 complex, which functions as an E3-like enzyme to enhance Atg8 lipidation (Hanada et al., 2007). The importance of Atg7, Atg5 and Atg12 in the formation of the Atg8-PE complex is also reflected in our results, and the activity of the E3-like enzyme is not completely dependent on Atg16, which is dispensable for the E3-like function of Atg12-Atg5 in vitro but it is required for Atg8-PE formation in vivo (Suzuki et al., 2001). The atg2Δ, atg18Δ and atg9Δ mutants only processed a small proportion of GFP-Atg8 in this study. Both these proteins are involved in the formation of autophagosomal membranes (Noda et al., 2000; Obara et al., 2008). Unlike previous studies, residual autophagy activity was detected in the deletion mutants of ATG6 and ATG14, both of which encode subunits of the PI3K complex (Figure 3A and Supplementary Figure S2). Considering that Atg14 has two homologous proteins in C. neoformans, single knock does not affect autophagy, which is worthy of further study whether it is due to functional redundancy. In the absence of a functional PI3K complex, Atg8 can be expressed and conjugated to PE, although autophagy will not take place (Itakura et al., 2008; Fogel et al., 2013). VPS34, a core subunit of PI3K, has been confirmed to be involved in the formation of autophagosomes in C. neoformans (Hu et al., 2008). Atg3 (E2 enzyme) and Atg4 (novel cysteine protease) were not important for the autophagy process in C. neoformans.
AMP-activated kinases (AMPKs) activate autophagy, whereas protein kinase A (PKA) and TORC1 inhibit autophagy (Stephan and Herman, 2006). During glucose starvation, AMPK is activated, whereas PKA is inhibited (Thevelein and Winde, 1999; Kamada et al., 2000; Aoh et al., 2013). Autophagy is induced in S. cerevisiae in response to abrupt carbon starvation, when cells are grown in the presence of glycerol, but not glucose, as the carbon source (Adachi et al., 2017). Consistent with these effects, autophagy was also induced under carbon starvation in C. neoformans (Figure 3B). The cleavage of GFP from the GFP-Atg8 fusion protein was hardly detected in atg7Δ, atg5Δ and atg12Δ, which are components of the ATG conjugation system and required for GFP cleavage, suggesting that lipidation of Atg8 is essential for its delivery to the vacuole. However, the cleavage of GFP from GFP-Atg8 was significantly enhanced compared to SD-N medium in other defective ATG strains. Carbon starvation also inactivates TORC1, which induced microautophagy resembling diauxic shift (carbon starvation)-induced microautophagy (Iwama and Ohsumi, 2019). The molecular mechanism of microautophagy does not require core Atg proteins but is regulated by the TORC1-Nem1/Spo7-Pah1 axis in budding yeast (Rahman et al., 2018). Therefore, we propose that the processing of GFP-Atg8 after long-term glucose starvation might be caused by microautophagy.
Conclusion
In this study, we investigated the contribution of traditional core ATG genes to cryptococcal virulent factors, stress response, starvation tolerance, pathogenesis and autophagic flux in C. neoformans. The atgΔ strains displayed higher sensitivity to 1.5 M NaCl, lower thermotolerance and remarkably lower pathogenesis. Although autophagic flux depends on a few core complexes, it also displayed specific features under nitrogen and glucose starvation conditions (Supplementary Table S3). Overall, this study provides very useful information on the functions of ATG genes in C. neoformans.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
XDZ, DW, and XZ conceived and designed the study. XZ, WF, XYZ, and CL performed the experiments. XM and XL provided assistance during in the experiments. XZ analyzed the data and wrote the manuscript. DW and XDZ reviewed and edited the manuscript. All authors read and approved the manuscript.
Funding
This work was continuously supported by the National Natural Science Foundation of China (Grants #81871629, #31470251, and #31872388).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02690/full#supplementary-material
Footnotes
References
Adachi, A., Koizumi, M., and Ohsumi, Y. (2017). Autophagy induction under carbon starvation conditions is negatively regulated by carbon catabolite repression. J. Biol. Chem. 292, 19905–19918. doi: 10.1074/jbc.m117.817510
Alvarado-Ramírez, E., Torres-Rodríguez, J. M., Sellart, M., and Vidotto, V. (2008). Laccase activity in Cryptococcus gattii strains isolated from goats. Rev. Iberoam. Micol. 25, 150–153. doi: 10.1002/yea.1629
Aoh, Q. L., Hung, C. W., and Duncan, M. C. (2013). Energy metabolism regulates clathrin adaptors at the trans-golgi network and endosomes. Mol. Biol. Cell. 24, 832–847. doi: 10.1091/mbc.e12-10-0750
Bahn, Y. S., Kojima, K., Cox, G. M., and Heitman, J. (2005). Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell. 16, 2285–2300. doi: 10.1091/mbc.e04-11-0987
Chang, C. Y., and Huang, W. P. (2007). Atg19 mediates a dual interaction cargo sorting mechanism in selective autophagy. Mol. Biol. Cell. 18, 919–929. doi: 10.1091/mbc.e06-08-0683
Cheong, H., and Klionsky, D. J. (2008). Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 451, 1–26. doi: 10.1016/S0076-6879(08)03201-1
Ding, H., Caza, M., Dong, Y., Arif, A. A., Horianopoulos, L. C., Hu, G., et al. (2018). ATG genes influence the virulence of Cryptococcus neoformans through contributions beyond core autophagy functions. Infect. Immun. 86, 1–24. doi: 10.1128/iai.00069-18
Dokladny, K., Zuhl, M. N., Mandell, M., Bhattacharya, D., Schneider, S., Deretic, V., et al. (2013). Regulatory coordination between two major intracellular homeostatic systems: heat shock response and autophagy. J. Biol. Chem. 288, 14959–14972. doi: 10.1074/jbc.M113.462408
Farré, J. C., and Subramani, S. (2016). Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat. Rev. Mol. Cell. Biol. 17, 537–552. doi: 10.1038/nrm.2016.74
Fogel, A. I., Dlouhy, B. J., Wang, C., Ryu, S. W., Neutzner, A., Hasson, S. A., et al. (2013). Role of membrane association and Atg14-dependent phosphorylation in beclin1-mediated autophagy. Mol. Cell. Biol. 33, 3675–3688. doi: 10.1128/MCB.00079-13
Gontijo, F. D. A., Melo, A. T. D., Pascon, R. C., Fernandes, L., Paes, H. C., Alspaugh, J. A., et al. (2017). The role of aspartyl aminopeptidase (Ape4) in Cryptococcus neoformans virulence and authophagy. Plos One. 12:e0177461. doi: 10.1371/journal.pone.0177461
Hanada, T., Noda, N. N., Satomi, Y., Ichimura, Y., Fujioka, Y., Takao, T., et al. (2007). The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282, 37298–37302. doi: 10.1074/jbc.c700195200
Hegedüs, K., Nagy, P., Gáspári, Z., and Juhász, G. (2014). The putative HORMA domain protein Atg101 dimerizes and is required for starvation-induced and selective autophagy in Drosophila. Biomed. Res. Int. 2014, 1–13. doi: 10.1155/2014/470482
Hu, G., Hacham, M., Waterman, S. R., Panepinto, J., Shin, S., Liu, X., et al. (2008). PI3K signaling of autophagy is required for starvation tolerance and virulenceof Cryptococcus neoformans. J. Clin. Invest. 118, 1186–1197. doi: 10.1172/jci32053
Ichimura, Y., Kirisako, T., Takao, T., Satomi, Y., Shimonishi, Y., Ishihara, N., et al. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492. doi: 10.1038/35044114
Inoue, Y., and Klionsky, D. J. (2010). Regulation of macroautophagy in Saccharomyces cerevisiae. Semin. Cell. Dev. Biol. 21, 664–670. doi: 10.1016/j.semcdb.2010.03.009
Itakura, E., Kishi, C., Inoue, K., and Mizushima, N. (2008). Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 19, 5360–5372. doi: 10.1091/mbc.E08-01-0080
Iwama, R., and Ohsumi, Y. (2019). Analysis of autophagy activated during changes in carbon source availability in yeast cells. J. Biol. Chem. 294, 5590–5603. doi: 10.1074/jbc.RA118.005698
Jing, H., Liu, H., Zhang, L., Gao, J., Song, H., and Tan, X. (2018). Ethanol induces autophagy regulated by mitochondrial ROS in Saccharomyces cerevisiae. J. Microbiol. Biotechnol. 28, 1982–1991. doi: 10.4014/jmb.1806.06014
Jung, C. H., Jun, C. B., Ro, S. H., Kim, Y. M., Otto, N. M., Cao, J., et al. (2009). ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 20, 1992–2003. doi: 10.1091/mbc.E08-12-1249
Kabeya, Y., Noda, N. N., Fujioka, Y., Suzuki, K., Inagaki, F., and Ohsumi, Y. (2009). Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 389, 612–615. doi: 10.1016/j.bbrc.2009.09.034
Kamada, Y., Funakoshi, T., Shintani, T., Nagano, K., Ohsumi, M., and Ohsumi, Y. (2000). Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell. Biol. 150, 1507–1513. doi: 10.1083/jcb.150.6.1507
Kawamata, T., Kamada, Y., Kabeya, Y., Sekito, T., and Ohsumi, Y. (2008). Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell. 19, 2039–2050. doi: 10.1091/mbc.E07-10-1048
Kihara, A., Noda, T., Ishihara, N., and Ohsumi, Y. (2001). Two distinct Vps34 phosphatidylinositol 3–kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell. Biol. 152, 519–530. doi: 10.1083/jcb.152.3.519
Kirisako, T., Baba, M., Ishihara, N., Miyazawa, K., Ohsumi, M., Yoshimori, T., et al. (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell. Biol. 147, 435–446. doi: 10.1083/jcb.147.2.435
Kirisako, T., Ichimura, Y., Okada, H., Kabeya, Y., Mizushima, N., Yoshimori, T., et al. (2000). The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell. Biol. 151, 263–276. doi: 10.1083/jcb.151.2.263
Lin, H. Y., Wang, J. J., Feng, M. G., and Ying, S. H. (2019). Autophagy-related gene ATG7 participates in the asexual development, stress response and virulence of filamentous insect pathogenic fungus Beauveria bassiana. Curr. Genet. 65, 1015–1024. doi: 10.1007/s00294-019-00955-1
Matsuura, A., Tsukada, M., Wada, Y., and Ohsumi, Y. (1997). Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192, 245–250. doi: 10.1016/s0378-1119(97)00084-x
Meiling, W. K., Barth, H., Voss, C., Eskelinen, E. L., Epple, U. D., and Thumm, M. (2004a). Atg21 is required for effective recruitment of Atg8 to the preautophagosomal structure during the Cvt pathway. J. Biol. Chem. 279, 37741–37750. doi: 10.1074/jbc.M401066200
Meiling, W. K., Bratsika, F., and Thumm, M. (2004b). ATG23, a novel gene required for maturation of proaminopeptidase I, but not for autophagy. FEMS Yeast Res. 4, 459–465. doi: 10.1016/S1567-1356(03)00207-1
Mizushima, N., Levine, B., Cuervo, A. M., and Klionsky, D. J. (2008). Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075. doi: 10.1038/nature06639
Mizushima, N., Noda, T., Yoshimori, T., Tanaka, Y., Ishii, T., George, M. D., et al. (1998). A protein conjugation system essential for autophagy. Nature 395, 395–398. doi: 10.1038/26506
Mukaiyama, H., Kajiwara, S., Hosomi, A., Giga-Hama, Y., Tanaka, N., Nakamura, T., et al. (2009). Autophagy-deficient Schizosaccharomyces pombe mutants undergo partial sporulation during nitrogen starvation. Microbiology 155, 3816–3826. doi: 10.1099/mic.0.034389-0
Mylonakis, E., Moreno, R., Khoury, J. B. E., Idnurm, A., Heitman, J., Calderwood, S. B., et al. (2005). Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect. Immun. 73, 3842–3850. doi: 10.1128/iai.73.7.3842-3850.2005
Nakatogawa, H., Suzuki, K., Kamada, Y., and Ohsumi, Y. (2009). Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell. Biol. 10, 458–467. doi: 10.1038/nrm2708
Nanji, T., Liu, X., Chew, L. H., Li, F. K., Biswas, M., Yu, Z. Q., et al. (2017). Conserved and unique features of the fission yeast core Atg1 complex. Autophagy. 13, 2018–2027. doi: 10.1080/15548627.2017.1382782
Noda, T., Kim, J., Huang, W. P., Baba, M., Tokunaga, C., Ohsumi, Y., et al. (2000). Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell. Biol. 148, 465–480. doi: 10.1083/jcb.148.3.465
Obara, K., Sekito, T., Niimi, K., and Ohsumi, Y. (2008). The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J. Biol. Chem. 283, 23972–23980. doi: 10.1074/jbc.m803180200
Oliveira, D. L., Fonseca, F. L., Zamith-Miranda, D., Nimrichter, L., Rodrigues, J., Pereira, M. D., et al. (2016). The putative autophagy regulator Atg7 affects the physiology and pathogenic mechanisms of Cryptococcus neoformans. Future Microbiol. 11, 1405–1419. doi: 10.2217/fmb-2016-0090
O’Meara, T. R., and Alspaugh, J. A. (2012). The Cryptococcus neoformans capsule: a sword and a shield. Clin. Microbiol. Rev. 25, 387–408. doi: 10.1128/cmr.00001-12
Palmer, G. E., Askew, D. S., and Williamson, P. R. (2008). The diverse roles of autophagy in medically important fungi. Autophagy 4, 982–988. doi: 10.4161/auto.7075
Palmer, G. E., Kelly, M. N., and Sturtevant, J. E. (2007). Autophagy in the pathogen Candida albicans. Microbiology 153, 51–58. doi: 10.1099/mic.0.2006/001610-0
Pang, Y., Yamamoto, H., Sakamoto, H., Oku, M., Mutungi, J. K., Sahani, M. H., et al. (2019). Evolution from covalent conjugation to non-covalent interaction in the ubiquitin-like ATG12 system. Nat. Struct. Mol. Biol. 26, 289–296. doi: 10.1038/s41594-019-0204-3
Park, B. J., Wannemuehler, K. A., Marston, B. J., Govender, N., Pappas, P. G., and Chiller, T. M. (2009). Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. Aids 23, 525–530. doi: 10.1097/qad.0b013e328322ffac
Petiot, A., Ogier-Denis, E., Blommaart, E. F., Meijer, A. J., and Codogno, P. (2000). Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 275, 992–998. doi: 10.1074/jbc.275.2.992
Rahman, M. A., Terasawa, M., Mostofa, M. G., and Ushimaru, T. (2018). The TORC1–Nem1/Spo7–Pah1/lipin axis regulates microautophagy induction in budding yeast. Biophys. Res. Commun. 504, 505–512. doi: 10.1016/j.bbrc.2018.09.011
Rajasingham, R., Smith, R. M., Park, B. J., Jarvis, J. N., Govender, N. P., Chiller, T. M., et al. (2017). Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet. Infect. Dis. 17, 873–881. doi: 10.1016/S1473-3099(17)30243-8
Reggiori, F., and Klionsky, D. J. (2013). Autophagic processes in yeast: mechanism, machinery and regulation. Genetics 194, 341–361. doi: 10.1534/genetics.112.149013
Richie, D. L., Fuller, K. K., Fortwendel, J., Miley, M. D., McCarthy, J. W., Feldmesser, M., et al. (2007). Unexpected link between metal ion deficiency and autophagy in Aspergillus fumigatus. Eukaryot. Cell. 6, 2437–2447. doi: 10.1128/ec.00224-07
Shimamura, S., Miyazaki, T., Tashiro, M., Takazono, T., Saijo, T., Yamamoto, K., et al. (2019). Autophagy-inducing factor Atg1 is required for virulence in the pathogenic fungus Candida glabrata. Front. Microbiol. 10:27. doi: 10.3389/fmicb.2019.00027
Shintani, T., Mizushima, N., Ogawa, Y., Matsuura, A., Noda, T., Ohsumi, Y., et al. (1999). Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 18, 5234–5241. doi: 10.1093/emboj/18.19.5234
Stephan, J. S., and Herman, P. K. (2006). The regulation of autophagy in eukaryotic cells: do all roads pass through Atg1? Autophagy 2, 146–148. doi: 10.4161/auto.2.2.2485
Suzuki, K., Kirisako, T., Kamada, Y., Mizushima, N., Noda, T., and Ohsumi, Y. (2001). The pre−autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971–5981. doi: 10.1093/emboj/20.21.5971
Suzuki, K., Kubota, Y., Sekito, T., and Ohsumi, Y. (2007). Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 12, 209–218. doi: 10.1111/j.1365-2443.2007.01050.x
Teter, S. A., Eggerton, K. P., Scott, S. V., Kim, J., Fischer, A. M., and Klionsky, D. J. (2001). Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J. Biol. Chem. 276, 2083–2087. doi: 10.1074/jbc.C000739200
Thevelein, J. M., and Winde, J. H. D. (1999). Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33, 904–918. doi: 10.1046/j.1365-2958.1999.01538.x
Wang, Y., Wei, D., Zhu, X., Pan, J., Zhang, P., Huo, L., et al. (2016). A ‘suicide’ CRISPR-Cas9 system to promote gene deletion and restoration by electroporation in Cryptococcus neoformans. Sci. Rep. 6:31145. doi: 10.1038/srep31145
Williamson, P. R. (1997). Laccase and melanin in the pathogenesis of Cryptococcus neoformans. Front. Biosci. 2:e99–e107. doi: 10.2741/a231
Yamaguchi, M., Matoba, K., Sawada, R., Fujioka, Y., Nakatogawa, H., Yamamoto, H., et al. (2012). Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7. Nat. Struct. Mol. Biol. 19:1250. doi: 10.1038/nsmb.2451
Yamashita, S. I., Oku, M., and Sakai, Y. (2007). Functions of PI4P and sterol glucoside necessary for the synthesis of a nascent membrane structure during pexophagy. Autophagy 3, 35–37. doi: 10.4161/auto.3311
Yang, Z., Huang, J., Geng, J., Nair, U., and Klionsky, D. J. (2006). Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell. 17, 5094–5104. doi: 10.1091/mbc.e06-06-0479
Yang, Z., and Klionsky, D. J. (2010). Eaten alive: a history of macroautophagy. Nat. Cell. Biol. 12, 814–822. doi: 10.1038/ncb0910-814
Yen, W. L., Legakis, J. E., Nair, U., and Klionsky, D. J. (2007). Atg27 is required for autophagy-dependent cycling of Atg9. Mol. Biol. Cell. 18, 581–593. doi: 10.1091/mbc.e06-07-0612
Yorimitsu, T., and Klionsky, D. J. (2005). Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell. 16, 1593–1605. doi: 10.1091/mbc.e04-11-1035
Zhao, D., Liu, X. M., Yu, Z. Q., Sun, L. L., Xiong, X., Dong, M. Q., et al. (2016). Atg20- and Atg24-family proteins promote organelle autophagy in fission yeast. J. Cell. Sci. 129, 4289–4304. doi: 10.1242/jcs.194373
Zhao, Y., Gong, S., Shunmei, E., and Zou, J. (2009). Induction of macroautophagy by heat. Mol. Biol. Rep. 36:2323. doi: 10.1007/s11033-009-9451-4
Zhu, X., Gibbons, J., Garcia-Rivera, J., Casadevall, A., and Williamson, P. R. (2001). Laccase of Cryptococcus neoformans is a cell wall-associated virulence factor. Infect. Immun. 69, 5589–5596. doi: 10.1128/iai.69.9.5589-5596.2001
Keywords: Cryptococcus neoformans, ATG genes, autophagy, Atg8, starvation, virulence, stress tolerance
Citation: Zhao X, Feng W, Zhu X, Li C, Ma X, Li X, Zhu X and Wei D (2019) Conserved Autophagy Pathway Contributes to Stress Tolerance and Virulence and Differentially Controls Autophagic Flux Upon Nutrient Starvation in Cryptococcus neoformans. Front. Microbiol. 10:2690. doi: 10.3389/fmicb.2019.02690
Received: 08 July 2019; Accepted: 06 November 2019;
Published: 26 November 2019.
Edited by:
Chengshu Wang, Shanghai Institutes for Biological Sciences (CAS), ChinaReviewed by:
Changbin Chen, Institut Pasteur of Shanghai (CAS), ChinaLinqi Wang, Institute of Microbiology (CAS), China
Copyright © 2019 Zhao, Feng, Zhu, Li, Ma, Li, Zhu and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xudong Zhu, emh1MTExODdAYm51LmVkdS5jbg==; Dongsheng Wei, d2VpZG9uZ3NoZW5nQG5hbmthaS5lZHUuY24=
 Xueru Zhao
Xueru Zhao Weijia Feng1
Weijia Feng1 Chenxi Li
Chenxi Li Xudong Zhu
Xudong Zhu Dongsheng Wei
Dongsheng Wei