- Center for Soybean Research of the State Key Laboratory of Agrobiotechnology and School of Life Sciences, The Chinese University of Hong Kong, Shatin, Hong Kong
Sinorhizobium fredii is a dominant rhizobium on alkaline-saline land that can induce nitrogen-fixing symbiotic root nodules in soybean. Two S. fredii strains, CCBAU25509 and CCBAU45436, were used in this study to facilitate in-depth analyses of this species and its interactions with soybean. We have previously completed the full assembly of the genomes and detailed transcriptomic analyses for these two S. fredii strains, CCBAU25509 and CCBAU45436, that exhibit differential compatibility toward some soybean hosts. In this work, we performed high-throughput Orbitrap analyses of the whole proteomes and secretomes of CCBAU25509 and CCBAU45436 at different growth stages. Our proteomic data cover coding sequences in the chromosome, chromid, symbiotic plasmid, and other accessory plasmids. In general, we found higher levels of protein expression by genes in the chromosomal genome, whereas proteins encoded by the symbiotic plasmid were differentially accumulated in bacteroids. We identified secreted proteins from the extracellular medium, including seven and eight Nodulation Outer Proteins (Nops) encoded by the symbiotic plasmid of CCBAU25509 and CCBAU45436, respectively. Differential host restriction of CCBAU25509 and CCBAU45436 is regulated by the allelic type of the soybean Rj2(Rfg1) protein. Using sequencing data from this work and available in public databases, our analysis confirmed that the soybean Rj2(Rfg1) protein has three major allelic types (Rj2/rfg1, rj2/Rfg1, rj2/rfg1) that determine the host restriction of some Bradyrhizobium diazoefficiens and S. fredii strains. A mutant defective in the type 3 protein secretion system (T3SS) in CCBAU25509 allowed this strain to nodulate otherwise-incompatible soybeans carrying the rj2/Rfg1 allelic type, probably by disrupting Nops secretion. The allelic forms of NopP and NopI in S. fredii might be associated with the restriction imposed by Rfg1. By swapping the NopP between CCBAU25509 and CCBAU45436, we found that only the strains carrying NopP from CCBAU45436 could nodulate soybeans carrying the rj2/Rfg1 allelic type. However, no direct interaction between either forms of NopP and Rfg1 could be observed.
Introduction
Rhizobia are Gram-negative bacteria that form symbiotic root nodules in leguminous plants. They reduce atmospheric dinitrogen to ammonia which can then be efficiently used by host plants for growth and development, in return for carbon and other nutrients provided by their hosts. Thus, they constitute an important mechanism for sustainable agriculture by reducing the dependence on synthetic nitrogen fertilizers (Dixon and Kahn, 2004).
Most of the recognized legume-nodulating rhizobia belong to the genera Rhizobium, Mesorhizobium, Sinorhizobium (Ensifer), and Bradyrhizobium within the α-proteobacteria class (Sugawara et al., 2013). The genus Sinorhizobium can induce nodule formation on the roots of many host plants from the Leguminosae family. Sinorhizobium meliloti, and its close relative, Sinorhizobium medicae, can establish symbiosis with Medicago truncatula and Medicago sativa, while Sinorhizobium saheli and Sinorhizobium terangae form root and stem nodules with Sesbania or Acacia (woody legumes). Sinorhizobium fredii, on the other hand, has a very wide range of hosts, representing all three subfamilies of the Leguminosae family, including cultivated (Glycine max) and wild (G. soja) soybeans (Sugawara et al., 2013).
Bradyrhizobium (species japonicum and diazoefficiens) and Sinorhizobium (species fredii) are the two major genera that can form nodules with the important crop soybean. Soybean-nodulating B. japonicum and B. diazoefficiens are more prominent in acidic soils and exhibit high nitrogen fixation capacity (Man et al., 2008; Tian et al., 2012), and they have long been used as inoculants in the field to improve yield and reduce fertilizer use (Paau, 1989). On the other hand, S. fredii are often found in alkaline soils in Asia (Han et al., 2009) and could perform efficient nitrogen fixation with some Chinese soybean cultivars (Muñoz et al., 2016). Yet, the use of Sinorhizobium as inoculant is not very widespread in Asia.
High-quality reference genomes of S. fredii with good annotation have been released in recent years (Schmeisser et al., 2009; Schuldes et al., 2012; Weidner et al., 2012; Jiao et al., 2018; Dang et al., 2019). While the transcriptome of prokaryote RNA is polycistronic in nature, prediction of coding sequences (CDSs) by open reading frames (ORFs) in operons, RNA sequencing, and sequence homology searches are only indicative. Furthermore, since protein abundance and varieties are dependent on the rate of mRNA translation as well as protein degradation and post-translational modifications, transcriptomic analyses can only partially explain the proteome (Muers, 2011). High-throughput proteomics has therefore provided evidence to support the existence of potential coding sequences and provided quantitative measurement of proteins and their variants to better correlate with the biochemical functions under different conditions. Over the last 20 years, proteomics based on mass spectrometry (MS) has contributed significantly to protein studies by facilitating protein identification and quantification. MS supports both relative and absolute protein measurement at a larger scale without the need to generate antibodies (Liu et al., 2016).
In addition to the free-living forms, rhizobia also exist as bacteroids in the host cells inside root nodules. This unique cell stage undergoes massive changes in its transcriptome and proteome under the influence of the host plant. Study of the bacteroid proteome which is encapsulated by the host cell could be challenging (Marx et al., 2016), as it will require either the analysis of complex host-symbiont co-proteome or the isolation of bacteroids in order to study the bacteroid-specific proteome.
Furthermore, the initiation of the nodulation process requires signal exchange between the host and the bacteria, which involves nodulation factors (lipo-chitooligosaccharides), surface carbohydrates and extracellular proteins (Krishnan et al., 2003; Okazaki et al., 2013; Staehelin and Krishnan, 2015; Sugawara et al., 2018). Previous studies characterizing secreted proteins relied largely on forward genetic studies (Sugawara et al., 2018) and gel-based proteomics (Rodrigues et al., 2007) which limited the throughput and scope of the studies. Eight nodulation outer proteins (Nops) (NopA, NopB, NopC, NopD, NopI, NopP, NopL, and NopX) have been detected in S. fredii strain HH103 in the extracellular medium based on precipitated proteins in response to genistein induction (Rodrigues et al., 2007). In more recent years, several Nops have been functionally characterized in S. fredii for their involvement in nodulation processes and the findings were reviewed by Staehelin and Krishnan (2015). Thus far, little is known about the biochemical functions of the secretomes of rhizobia. Hence, an extensive catalog of secreted proteins and the effective comparison of secretomes between rhizobium strains would greatly help us understanding the roles of these proteins during host-rhizobium interactions.
Host-specific nodulation may rely on Nops, i.e., rhizobial effectors, translocated into the host cytoplasm through the type 3 protein secretion system (T3SS) (Krishnan and Pueppke, 1993). The T3SS is common to many bacterial pathogens targeting animals and plants (Staehelin and Krishnan, 2015). T3SS effector proteins have been well-characterized in various S. fredii strains, NGR234, USDA257, and HH103 (Krishnan et al., 2003; Marie et al., 2004; López-Baena et al., 2016). The effectors secreted by T3SS, upon entering the plant cell, may lead to effector-triggered immunity (ETI) mediated by resistance proteins (R proteins) in plants (Hurley et al., 2014). Rhizobial Nops can also help determine host specificity, such as in the cases of Sinorhizobium- and Bradyrhizobium-soybean interactions (Yang et al., 2010; Sugawara et al., 2018). Earlier, it was reported that allelic types of the soybean R protein Rj2(Rfg1) protein could determine host restriction of B. diazoefficiens and S. fredii (Yang et al., 2010). For example, soybean carrying the dominant type of the Rfg1 gene can restrict the nodulation by S. fredii USDA193 (Fan et al., 2017).
In response to flavonoids derived from host plants, various transcription factors (e.g., NodD1, NodD2, and ttsI) can trigger activation of genes (e.g., rhc, nol, and nop) responsible for T3SS (Wassem et al., 2008). In S. fredii, approximately 15 genes have been predicted to encode for the T3SS Nops (Kimbrel et al., 2013).
The mutants rhcJ and ttsI of B. japonicum USDA122 fail to secrete Nops and were able to nodulate otherwise non-compatible soybean plants, suggesting that effectors secreted via T3SS trigger incompatibility with soybean hosts (Tsukui et al., 2013). The determining factor in B. diazoefficiens strain USDA122 has been shown to be the NopP effector protein. Mutations in the nopP gene are tightly associated with host compatibility in soybean expressing the Rj2 R protein (Sugawara et al., 2018).
A negative effect on the early stages of soybean nodulation was observed upon the inactivation of the S. fredii HH103 T3SS, which suggests that T3SS is required to suppress plant-generated immune responses for successful nodulation in some cases (Jiménez-Guerrero et al., 2015). Random Tn5 insertion mutations in the T3SS gene cluster in an incompatible strain of S. fredii (CCBAU25509) led to nodule formation in G. max C08, suggesting a significant role of insertion sequences in the adaptive evolution of rhizobial compatibility (Zhao et al., 2017). However, mutations in a number of nop genes have been found affecting symbiosis (Staehelin and Krishnan, 2015; López-Baena et al., 2016). Recently, NopI of S. fredii HH103 was also found to determine nodulation numbers in soybean (Jiménez-Guerrero et al., 2017). In S. fredii HH103, NopI, and NopP show 48% amino acid sequence identity (Jiménez-Guerrero et al., 2017).
Here we make use of the high-throughput Orbitrap system to study the proteomes of two S. fredii strains, CCBAU45436 and CCBAU25509, during mid-log, stationary phase and in bacteroids using different soybean hosts. We have previously completed their genome assembly and transcriptome analyses (Jiao et al., 2018) and the data could serve as the basis for the proteomic analyses. The purpose is to build two important reference proteomes for these two strains of S. fredii that have been reported to exhibit differential host specificities toward soybean expressing the R protein, Rfg1 (Qi et al., 2014; Muñoz et al., 2016; Zhao et al., 2017). In addition, we also investigated the secretomes of both strains to get insights into the Nops that may be associated with host compatibility. The methods previously reported for Nops detection in the extracellular medium required specific antibodies that are labor-intensive and time-consuming to produce while our adapted method for detecting effector proteins is faster and more reliable, all without the need for generating antibodies. A randomly generated Tn5-inserted rhcN mutant mitigated the host incompatibility with G. max C08 by suppressing the secretion of T3SS-related Nops and disrupting the expressions of pilus assembly-associated genes. Furthermore, we have also identified NopP as a key component in determining genotype-specific nodulation of soybean.
Materials and Methods
Cultivation of Bacteria and Plants
Sinorhizobium fredii CCBAU25509 and CCBAU45436 were cultured at 28°C in yeast mannitol agar (YMA) medium with nalidixic acid (25 mg/L). Both wild type CCBAU25509 and CCAU45436 are resistant to nalidixic acid. Plant growth and inoculation was also carried out in accordance with the method described previously (Jiao et al., 2018). The surface-sterilized seeds of G. max (accession C08) and Glycine soja (accession W05) were germinated in vermiculite wetted with autoclaved water in the greenhouse. Following germination, 1 ml each of CCBAU25509 or CCBAU45436 suspension culture (OD600 = 0.8) per plant was inoculated onto the seedlings. Bacteroids were collected in 15 ml of extraction buffer [10 mM DTT, 300 mM sucrose, 10 mM phosphate buffer pH 7.0, 2 mM MgCl2 and 2% (w/v) PVP-40] by extensive grinding of 2–5 g nodules in a pre-cooled mortar and pestle and later purified on a Percoll gradient according to the method described previously (Li et al., 2013). The bacteroids were collected from the nodules, then washed with Milli-Q water and stored at −80°C until use.
Extraction of Total and Secreted Proteins
For the extraction of total proteins, S. fredii strains were grown in the YMA medium at 28°C until mid-log (∼15 h culture, OD600 = 0.7) and stationary phase (∼48 h culture, OD600 = 1.4), and the bacteria were collected by centrifugation at 4000 × g. The pellets obtained were washed twice in phosphate buffered saline (PBS) (pH 7.2) and subjected to lysis in a buffer composed of 2% SDS in 20 mM Tris-HCl (pH 8.8) at 95°C at 500 rpm in a Thermomixer for 30 min (Eppendorf, Germany) (Tanca et al., 2013). The protein-containing supernatant was extracted for 5 min at 12000 × g by centrifugation. Proteins were precipitated using chilled acetone at −20°C for 4 h and the resulting pellet was dissolved in 50 mM Tris-HCl buffer (pH 8.5) containing 8M urea. The same protocol was adopted for the extraction of proteins from bacteroids from C08 and W05 nodules. In total, three replicates were used to perform whole proteome analysis.
Extracellular proteins were induced from bacterial strains grown in 50 ml of RMS medium in a 500 ml flask (Broughton et al., 1986) with a final concentration of 10 μM genistein on an orbital shaker (180 rpm) for 28 h (about 109 bacteria ml–1) at 28°C (Jiménez-Guerrero et al., 2017). Two successive centrifugation steps at 4°C for 4000 × g and 8000 × g for 30 min were used to remove bacterial cells. The supernatants were mixed with 3 volumes of chilled acetone and kept for 24 h at −20°C. The mixtures were centrifuged at 22,000 × g at 4°C for 45 min and the resulting pellets were dried and resuspended in 300 μl of 8M urea in Tris-HCl buffer (pH 8). The same volume of extracted extracellular proteins were separated on NuPAGETM 4–12% bis-tris protein gels with MES running buffer. SilverXpressTM silver staining kit was used for visualizing the separated proteins (Thermo Scientific, United States). For secretome analysis, the experiment was repeated two times.
Digestion of proteins was performed using SMART digestTM trypsin kit (Thermo Scientific, United States) in solution. The reduction and alkylation of the tryptic peptides was achieved with the use of 10 mM dithiothreitol (DTT) at 56°C for 30 min and 25 mM iodoacetamide at room temperature for 25 min. The digested peptides were purified using PierceTM C-18 spin columns and finally dissolved in 0.1% formic acid (FA).
Orbitrap-Based LC-MS/MS Analysis
MS analysis was performed using an Orbitrap FusionTM LumosTM TribridTM Mass Spectrometer (Thermo Scientific, United States) interfaced with an LC UltiMate 3000 RSLCnano system. The peptide separation was carried out at 50°C with C-18 μ-precolumn (300−μm i.d. × 5 mm) followed by AcclaimTM PepMapTM RSLC, nanoViper C-18 column, 75 μm × 25 cm (Thermo Scientific, United States) at a flow rate of 0.3 μl/min, using mobile phase A [98% H2O, 1.9% acetonitrile (ACN) with 0.1% FA] and mobile phase B (98% ACN, 1.9% H2O with 0.1% FA). The following LC gradient was used to detect secretome proteins: 100% A for the initial 5 min, then 0% to 6% B for 3 min, then 6% B to 30% B for 42 min, then 30% B to 45% B for 10 min, then 45% B to 60% B for 10 min, then 60% B to 80% B for 5 min and an additional 5 min at 80% B followed by final re-equilibrium with 100% A for 10 min. We used the following LC gradient profile (100% A for the initial 5 min, then 0% B to 6% B for 3 min, then 6% B to 18% B for 40 min, then 18% B to 30% B for 10 min, then 30% B to 80% B for 2 min, then 80% B for an additional 5 min followed by re-equilibrium with 100% A for 10 min) for total protein detection.
The Orbitrap was set up in a data-dependent MS/MS mode under direct control of the Xcalibur software (version 4.1), where a full-scan spectrum (from 375 to 1500 m/z) was followed by tandem mass spectra (MS/MS). The instrument was operated in positive mode with a spray voltage of 2 kV, a capillary temperature of 300°C, and was calibrated before measurements. Full scans were performed in the Orbitrap with a resolution of 60,000 at 400 m/z, with a precursor ion selection (AGC > 4.0e5) and ion charge > 1. Higher energy collisional dissociation (HCD), performed at the far side of the C−trap, was chosen as the fragmentation method, by applying a 30% value for normalized collision energy, an isolation window of m/z 1.6, with a maximum injection time of 250 milli seconds (ms) and Orbitrap resolution of 15,000.
The Proteome Discoverer platform (version 2.3; Thermo Scientific, Germany), interfaced with an in−house Sequest server, was used for data parsing and protein identification, according to the following criteria: in-house database of CCBAU25509 and CCBAU45436, Enzyme Trypsin, Maximum Missed Cleavage Sites 2, Precursor Mass Tolerance 10 ppm, Fragment Mass Tolerance 0.2 Da, Cysteine Carbamidomethylation as Static modification, N−terminal Acetylation and oxidation as dynamic modifications. The Percolator algorithm was used for peptide validation [peptide confidence: q−value < 0.01, corresponding to false discovery rate (FDR) < 0.01], and only rank 1 peptides were considered. Peptide and protein grouping according to Proteome Discoverer’s algorithms were allowed, applying strict maximum parsimony principle. A Label-free quantification (LFQ) analysis was performed with three replicates for each treatment for whole proteome analysis.
Phylogenomic Analysis
In this study, 16S rRNA sequences were retrieved from 56 rhizobial genomes (Supplementary Table S1), including 10 S. fredii strains, 10 S. meliloti strains, 6 S. medicae strains, 9 B. japonicum strains, 5 Mesorhizobium sp. strains, 6 B. diazoefficiens strains, 4 B. elkanii strains, and 6 R. leguminosarum strains using the EzBioCloud database1 (Yoon et al., 2017). The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model (Tamura and Nei, 1993) in MEGA7 (Kumar et al., 2016).
Genome- and Transcriptome-Wide Identification of Nops in Selected Genomes
For deeper analyses of the nop genes in CCBAU25509 and CCBAU45436, we retrieved the predicted nop sequences from S. fredii NGR234, USDA257, and USDA207 (Kimbrel et al., 2013) for use in our search. To predict the upstream tts box sequences reported for nop gene regulation, we constructed HMMs (Hidden Markov Models) for tts box sequence homologs in CCBAU25509 and CCBAU45436 accordingly (Marie et al., 2004; Songwattana et al., 2017). Nops detected in the extracellular medium from S. fredii CCBAU25509 and CCBAU45436 were further analyzed for the presence or absence of the eight identified Nops in the 56 selected genomes using the BLAST tool available in the EzBioCloud database with a cutoff of e-value ≤ 1e–10 and protein sequence identity ≥ 40%. In complement of this, the proteins were search against NCBI database using BLASTP with the same cutoff. A heatmap was generated based on presence of a nop gene sequence in all 56 selected genomes. The protein sequences obtained were screened and aligned using the MEGA72 MUSCLE alignment tool. The information associated with the detected Nops, such as the CDS, the name of the rhizobial strain and genomic location, was also recorded. To check the growth stage-specific expression of all eight Nops during the mid-log and stationary phases and in the collected bacteroids, the RNA-Seq dataset was further analyzed using Fragments Per Kilobase of transcript, per Million mapped reads (FPKM) values collected from transcriptome datasets available (Jiao et al., 2018). The expression of the nopP and nopI genes was further confirmed by qPCR using One Step SYBR Prime ScriptTM RT-PCR Kit II (Takara Bio, Inc., Japan), according to the manufacturer’s instruction, and primers are listed in Supplementary Table S2.
Genotyping and Phenotyping of Soybean Accessions
Full length or a partial fragment of the Rj2(Rfg1) gene of 22 soybean accessions (Lam et al., 2010; Muñoz et al., 2016) were amplified from genomic DNA using iProof polymerase (BioRad) according to the manufacturer’s instruction. Primers for the amplification and sequencing are listed in Supplementary Table S2. For the nodulation test, surface-sterilized seeds were sown on wetted vermiculite followed by the addition of 1 ml overnight culture of CCBAU25509 or USDA122 per seed. Formation of effective nodules were confirmed 28 days post-inoculation. Re-sequencing data were retrieved from a previous publication (Zhou et al., 2015). Only the single-nucleotide polymorphisms (SNPs) at the seven previously reported polymorphic sites (E452K, I490R, Q731E, E736N, P743S, E756D, and R758S) distinguishing different alleles of Rj2(Rfg1) were analyzed (Yang et al., 2010). SNP on the 758th amino acid residue was missing in the dataset and was not included in the analyses. Accessions with missing data or heterozygous SNPs at the polymorphic sites of interest were discarded. Allelic forms were determined according to Yang et al. (2010).
Generation, Screening, and Plant Assays of the rhcN:Tn5 Mutant
Bi-parental mating was performed between the donor E. coli S17-1 λpir (Biomedal Life Science, Spain) harboring pUTmini-Tn5 km (Biomedal Life Science, Spain) and the recipient S. fredii CCBAU25509 by mixing their cultures in a ratio of 1:3 (Tsurumaru et al., 2008) on TY agar for conjugation and mutagenesis. Mutants were selected on a TY agar plate supplemented with nalidixic acid (50 mg/L) and kanamycin (50 mg/L). A total number of > 1.27 × 106 individual clones were inoculated into 2-day-old C08 seedlings. Nodules were collected 28 days after inoculation. The collected nodules were surface-sterilized with 5% household bleach and washed thoroughly with autoclaved Milli-Q water. Mutants were isolated aseptically by inserting autoclaved toothpicks into the core of the nodules and streaking on TY agar with nalidixic acid (50 mg/L) and kanamycin (50 mg/L) for selection.
The isolates grown on the selection medium were further subjected to PCR screening with Tn5 transposon-specific primers and one pair of CCBAU25509-specific primers (Supplementary Table S2) to eliminate possible contamination from other antibiotic-resistant strains. The PCR-confirmed mutants were re-inoculated into C08 to allow nodulation. Genomic DNA (gDNA) of the confirmed mutants were extracted using the CTAB buffer protocol. The Tn5 insertion site was determined by Y-linker PCR. In brief, gDNA was digested with NlaIII restriction endonuclease. Forty nanograms of the resulting gDNA were used to ligate with 1 μg of Y-linker in a 20 μl reaction at 16°C overnight (Kwon and Ricke, 2000). The ligated product was then amplified by PCR with Y-linker- and transposon-specific primers (Supplementary Table S2) for the selective amplification of the 5′-flanking region of the transposon and cloned into pMD20 T-vector (Takara Bio, Inc., Japan) and the successful cloning was confirmed by sequencing. The chlorophyll contents of soybean plants inoculated with wild type and mutant CCBAU25509 were measured in a portion of the leaf excised from the first trifoliate and calculated based on the recorded fresh weight, following a method described previously (Millard and Robinson, 1987).
Gene Expression Analysis
Wild type CCBAU25509 and the obtained rhcN:Tn5 mutant were grown in 50 ml TY medium and induced with 20 μM genistein or an equivalent volume of DMSO as control. Cultures were collected at OD600 = 0.4 and pelleted by centrifugation at 5000 × g. The RNA was extracted using TRIzol reagent (Invitrogen, United States) according to the manufacturer’s protocol with a modification in the lysis step in which ∼100 μl of glass powder was added per ml of TRIzol reagent. DNase I-treated RNA was diluted 10-fold for qPCR using One Step SYBR® Prime ScriptTM RT-PCR Kit II (Takara Bio, Inc., Japan) according to the manufacturer’s instruction, except that annealing and extension was performed at 55.7°C. Primer sequences for RT-qPCR can be found in Supplementary Table S2. The amplification efficiencies and specificities of the primers used for qPCR were validated with primer efficiency assays and melt curve analyses. GyrA was used as the housekeeping gene for normalization.
Generation of nopP-Exchanged Mutants of CCBAU25509 and CCBAU45436
The nopP coding sequences with ∼1 kb upstream/downstream were amplified from the genomic DNA of CCBAU25509 and CCBAU45436 and cloned between the XbaI and PstI sites of pK18mobsacB (ATCC® 87097TM). Swapping of the nopP genes was done using an established protocol (Schafer et al., 1994) with slight modifications. In brief, an 8,714 bp construct containing the 3,013 bp nopP fragment from CCBAU25509 (nopP25509) was transferred into CCBAU45436, and vice versa, by bi-parental mating using E. coli S17-1 λpir. TY agar supplemented with nalidixic acid (50 mg/L) and kanamycin (50 mg/L) was used for the selection of the first crossing-over event while TY agar supplemented with nalidixic acid (50 mg/L) and 10% sucrose was used to select for the second crossing over. Mutants with successfully exchanged nopPs were confirmed by sequencing. The resulting CCBAU25509nopP45436 and CCBAU45436nopP25509 were used to inoculate C08 for the assessment of nodulation phenotype. The identities of the bacteria within the nodules were confirmed using PCR to rule out possible cross-contamination. Numerical data were analyzed using one-way ANOVA followed by Tukey’s post hoc test using GraphPad Prism (ver. 8.1.1). Primers for generating and screening of the mutants can be found in Supplementary Table S2.
Yeast Two-Hybrid Assay
Full-length and selected domains of Rfg1 cDNA were amplified from C08 and W05 and were subcloned into pGBKT7 (Clontech, United States). nopP coding sequences were amplified from both CCBAU25509 and CCBAU45436 and were cloned into pGADT7-rec (Clontech, United States). Primers for the amplification of target sequences can be found in Supplementary Table S2. The pGBKT7 and pGADT7-rec constructs were transformed into Y2H gold and Y187 (Takara Bio, Inc., Japan), respectively, using lithium acetate/polyethylene glycol method (Gietz and Schiestl, 2007). Yeast mating and interaction analyses were done according to Yeast Protocols Handbook (Clontech, United States).
Results
Orbitrap-Based Total Proteomics of CCBAU25509 and CCBAU45436
We performed a comprehensive proteome profiling of CCBAU45436 and CCBAU25509 at mid-log and stationary phases, and in bacteroids with three biological replicates for each condition. The peptides obtained from the tryptic digestion of extracted proteomes were analyzed by Orbitrap FusionTM LumosTM TribridTM Mass Spectrometer coupled to a nano-liquid chromatography (LC).
On average, during the mid-log and stationary phases, we detected nearly 20,000 peptide spectrum matches (PSMs) corresponding to 11,000 peptides, or 2,000 proteins in each replicate with the false discovery rate (FDR) of q < 0.01 (Fisher’s exact test) (Figure 1A and Supplementary Tables S3, S4). Almost 600 proteins were detected from bacteroids collected from the nodules of G. max (C08) and G. soja (W05) plants inoculated with S. fredii CCBAU45436 and CCBAU25509, respectively, after 28 days (Figure 1A and Supplementary Tables S3, S4). Nodules could not be formed on C08 roots inoculated with CCBAU25509 due to the presence of the Rfg1 allele in C08, which rendered C08 incompatible with the rhizobium. The constructed circular proteomic and genomic maps of both strains provide a bird’s-eye view of the translating core and accessory genomes of S. fredii at various growth stages (Figure 1B).
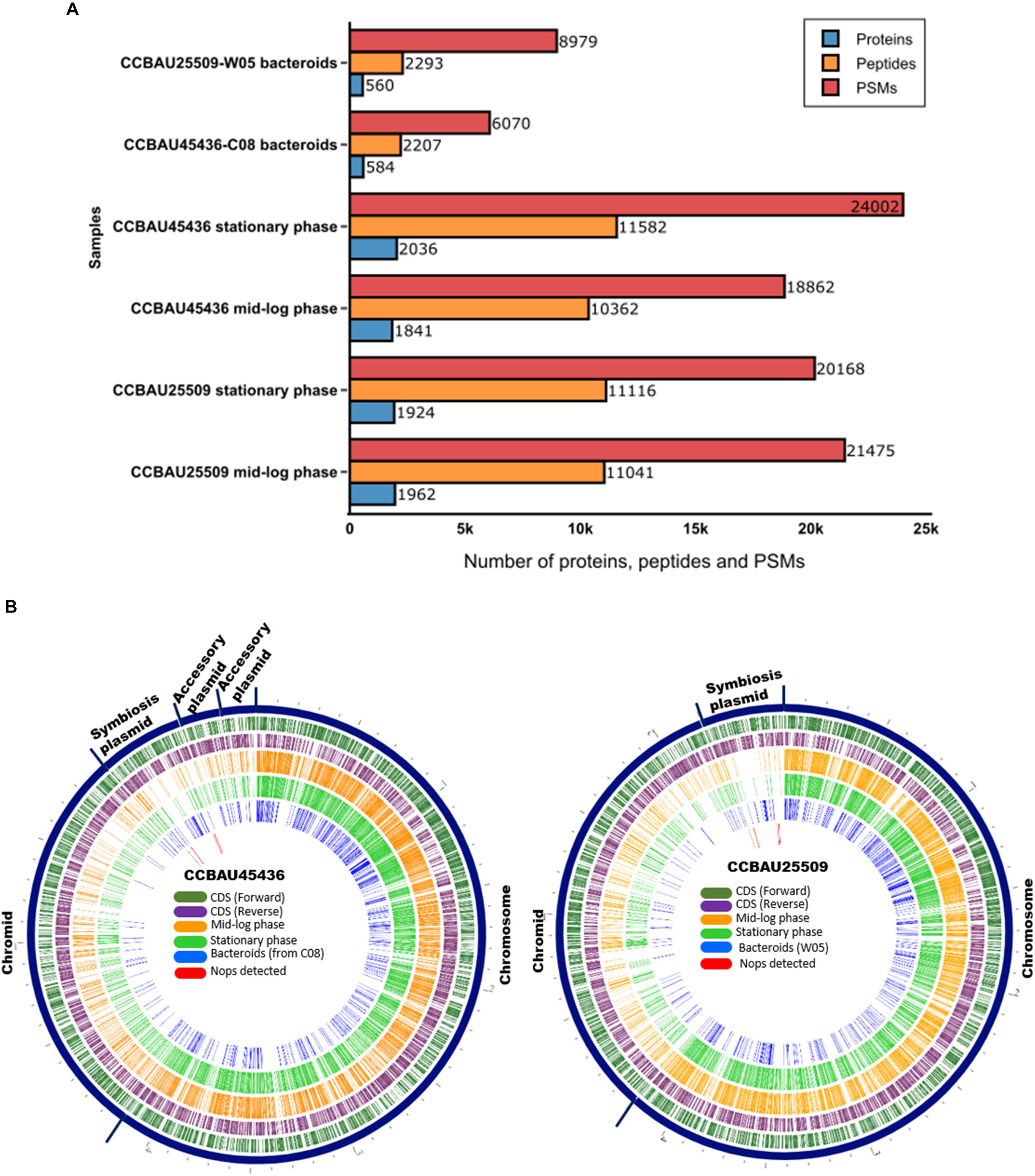
Figure 1. Proteomes of Sinorhizobium fredii CCBAU25509 and CCBAU45436. (A) Number of identified proteins, peptides, and peptide spectrum matches (PSMs) from CCBAU25509 and CCBAU45436 at various growth stages in one of the three replicates. Bacteroids of CCBAU25509 and CCBAU45436 were obtained by inoculating these strains into their compatible soybean hosts, W05 and C08, respectively. (B) Circular views of detected proteins overlaid on the genomic locations of the encoding genes.
During all three growth stages, a large proportion of the detected proteomes were encoded by the chromosome-borne CDSs of each strain (Figure 1B). This is consistent with the observation of a previous proteomic study of S. medicae strain (Berrabah et al., 2018). All proteome datasets had proteins spanning a range of molecular weights and isoelectric points (Supplementary Tables S3, S4). ABC transporter proteins, tricarboxylic acid (TCA) cycle proteins, cold shock proteins, DUF (domain of unknown function) proteins, ferredoxin family proteins, heat shock proteins, nitrogen fixation and regulatory proteins, phosphate transport proteins, outer membrane lipoproteins, and transcriptional regulator proteins made up the key families of proteins detected in the bacteroids collected from C08 and W05 nodules. The phasin family and the chaperonin proteins were abundantly found in all samples analyzed based on the number of peptides.
Gene ontology (GO) annotation analyses of all the identified proteins showed that the major portion of the proteome was found in the cytoplasm, integral component of membrane, periplasmic space, ribosome, plasma membrane, and cell membrane components of the cell at all growth stages (Supplementary Figure S1). At the molecular level, most of the proteome was involved in ATP binding, metal ion binding and transferase activity, oxidoreductase activity, as the structural constituent of ribosome, DNA-binding transcription factor activity, ATPase activity, hydrolase activity, rRNA binding, and pyridoxal phosphate binding, in both rhizobial strains at all three growth stages. A significant portion of the proteome was also involved in metabolic activities at all three growth stages, such as the regulation of transcription, carbohydrate metabolic processes, phosphorelay signal transduction system, transmembrane transport, cell redox homeostasis, TCA cycle, and nitrogen compound metabolic processes. On the other hand, a major portion of the proteins from the bacteroids were found to be involved in nitrogen fixation when classified according to biological processes.
The reproducibility among replicates was based on the calculated Log10 abundance of peptides using the available LFQ consensus and processing workflow in the Proteome Discoverer software (version 2.3). The LFQ peptides between each replicate had almost the same mean value, showing a high degree of reproducibility among the samples (Supplementary Figure S2). The abundantly expressed stage-specific peptides based on p-value < 0.05 are found in a comparison between mid-log and stationary phase LFQ datasets (Supplementary Figure S2).
By comparing the flagellar export proteins in the mid-log phase versus the stationary phase, carbonic anhydrase, DUF971 domain-containing protein, nitrogen regulatory protein P-II and protein-export chaperone, SecB, chaperonin, and the phasin family of proteins were once again found to be the most highly expressed proteins (Supplementary Figure S3 and Table S3). Some of the key quality parameters such as peptide and PSM confidence, q-value, and concatenated rank by server were found to be high (Supplementary Figure S4).
Correlation of mRNA and Protein Data
In the next step we compared the LFQ proteome data to RNA−sequencing data that have been recorded previously under the same growth stages by us (Jiao et al., 2018). The protein/mRNA−pairs from CCBAU25509 and CCBAU45436 during mid-log phase (CCBAU25509 vs. CCBAU45436: 1,869 vs. 1,863), stationary phase (1,665 vs. 1,871), and in bacteroids (527 vs. 509) were available to build correlations (Figure 2 and Supplementary Table S5). Among those pairs, proteins were quantified for all replicates. In mid-log phase, the correlation between LFQ protein abundance and mRNA expression levels was relatively high (r = 0.557 and 0.578 for CCBAU45436 and CCBAU25509, respectively), which is in the same range as the protein−mRNA correlation coefficient previously published for another Gram-negative bacterium (Kwon et al., 2014; Erdmann et al., 2018). A positive but weak correlation (r = 0.161 and 0.167) between LFQ protein and mRNA expression was observed during stationary phase. In bacteroids, the correlation between LFQ protein and mRNA expression is moderate (r = 0.386 and 0.442 for CCBAU25509 and CCBAU45436, respectively). These results are consistent with a general observation that the correlation between protein and mRNA levels is dependent on cell cycle and growth stage (Liu et al., 2016).
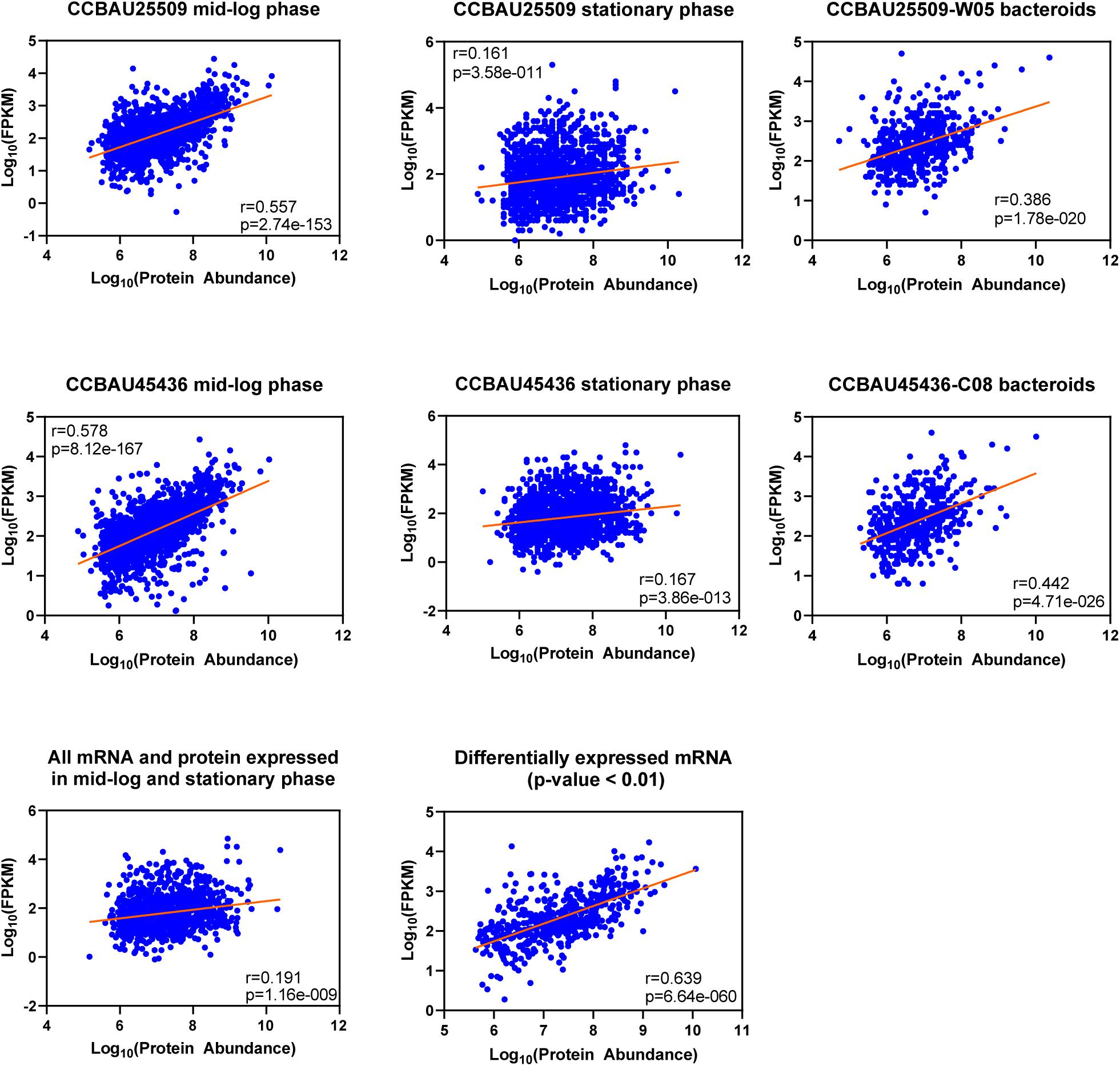
Figure 2. Overall correlation between the levels of label-free quantified (LFQ) proteins and mRNAs at different growth stages of S. fredii CCBAU25509 and CCBAU45436. The figures were drawn based on Log10 abundance values of proteome and transcriptome datasets. Averaged abundance of proteins of three biological replicates were used. W05: a G. soja accession; C08: a G. max accession. Pearson’s correlations were calculated using GraphPad Prism 8.2.1 at 95% confidence interval.
We also investigated the correlation between differentially expressed mRNA and their encoded proteins between mid-log phase and stationary phase in both strains combined. The correlation of differentially expressed mRNA/protein is much higher (r = 0.639) compared to the correlation of total expressed mRNA/protein (r = 0.191), indicating that gene expression with more dynamic changes may have higher mRNA-to-protein correlations (Erdmann et al., 2018). Since the number of expressed genes in bacteroids were much lower, we did not include them in this analysis.
Secretome-Wide Nops Detection in CCBAU25509 and CCBAU45436
To investigate host-specific differences between the CCBAU25509 and CCBAU45436 strains, we performed a secretome analysis to identify Nops. We used genistein to induce the production of Nops as suggested by previous researches (Jiménez-Guerrero et al., 2017). Seven out of eight known Nops (NopA, NopB, NopC, NopD, NopI, NopL, NopP) were detected from the extracellular medium of both CCBAU45436 and CCBAU25509 using the Orbitrap system (Table 1). On the other hand, NopX was missing from the CCBAU25509 secretome, despite that the intact nopX gene was present in the CCBAU25509 genome with the same coding sequence as that in CCBAU45436. We also identified some other proteins in the secretome such as a 10- and a 60-kDa chaperonin, ATP-binding cassette (ABC) transporters, translation initiation factors, 50S ribosomal proteins, 30S ribosomal proteins, ATP-binding proteins, succinyl-CoA synthetase, outer-membrane lipoproteins, periplasmic dipeptide transport proteins and TolB protein precursor (Supplementary Table S6). The presence of ribosomal proteins and some other cytoplasmic proteins in the secretome could be the result of a non-classical (leaderless) mode of secretion described previously (Bendtsen et al., 2005). All the identified Nops and other secreted proteins had q-values < 0.01 (Table 1 and Supplementary Table S6).
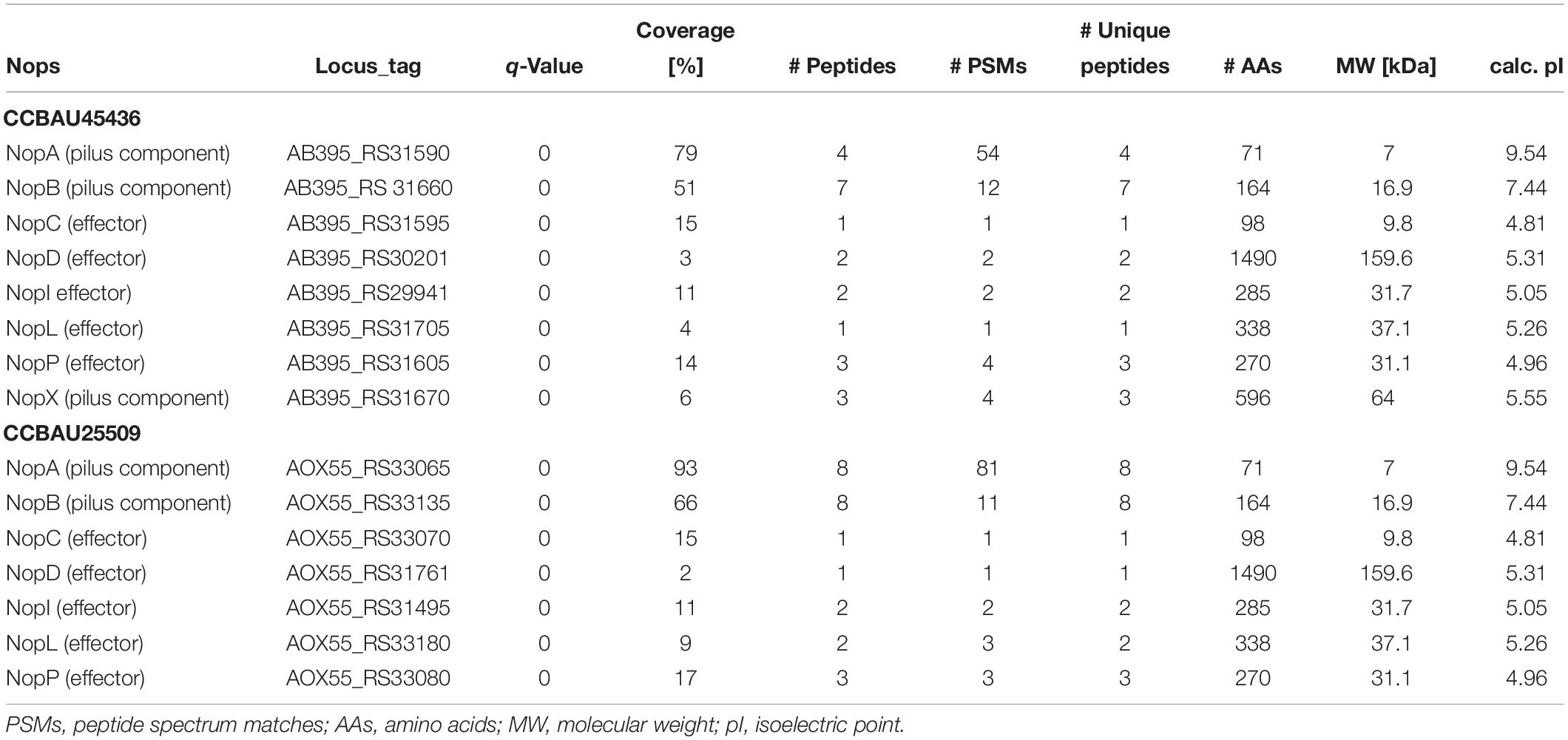
Table 1. Orbitrap-based detection of Nops in CCBAU45436 and CCBAU25509 in response to genistein induction.
In S. fredii CCBAU25509 and CCBAU45436, all eight nop genes are located on the symbiotic plasmid (Figure 1B), as reported in other S. fredii strains (Schmeisser et al., 2009; Schuldes et al., 2012; Weidner et al., 2012; Jiao et al., 2018; Dang et al., 2019). We collected the transcriptomic data of each nop gene from our previous work (Jiao et al., 2018) to evaluate their expression patterns during the mid-log and stationary phases without genistein induction, and in the nodules of G. max (C08) and G. soja (W05) (Supplementary Figure S5). Except nopX, all the other nop genes showed high expressions at the stationary phase. Higher expressions of all eight nop genes were found in the C08 nodules compared to the W05 nodules. Since the NopP and NopI proteins may play a role in host specificity (see below), the transcriptomic data of the nopP and nopI genes were confirmed with RT-qPCR (Supplementary Figure S6).
Genome-Wide nop Gene Distributions in Sinorhizobium and Closely Related Genera
Using the previously reported nop gene sequences from the strains, NGR234, USDA257 and USDA207, 18 known or putative nop genes (nopA, nopAA, nopB, nopBE, nopBF, nopBH, nopC, nopD, nopL, nopI, nopP, nopM1, nolT, nopU, nopV, nopX, and nopZ) were identified in CCBAU25509 and CCBAU45436 (Figure 3A and Supplementary Table S7). Six tts boxes were also identified in the upstream regions of six nop genes (nopAA, nopM1, nopC, nopB, nopP, and nopL) (Supplementary Table S7). In CCBAU25509, a membrane-anchored protein-coding gene and an Fnr-type transcriptional regulator were also found to be eventually regulated by tts boxes (Supplementary Table S7). In CCBAU45436, the flavin mononucleotide (FMN) reductase gene and a hypothetical protein-coding gene were found to have a tts box upstream of their respective coding sequences (Supplementary Table S7).
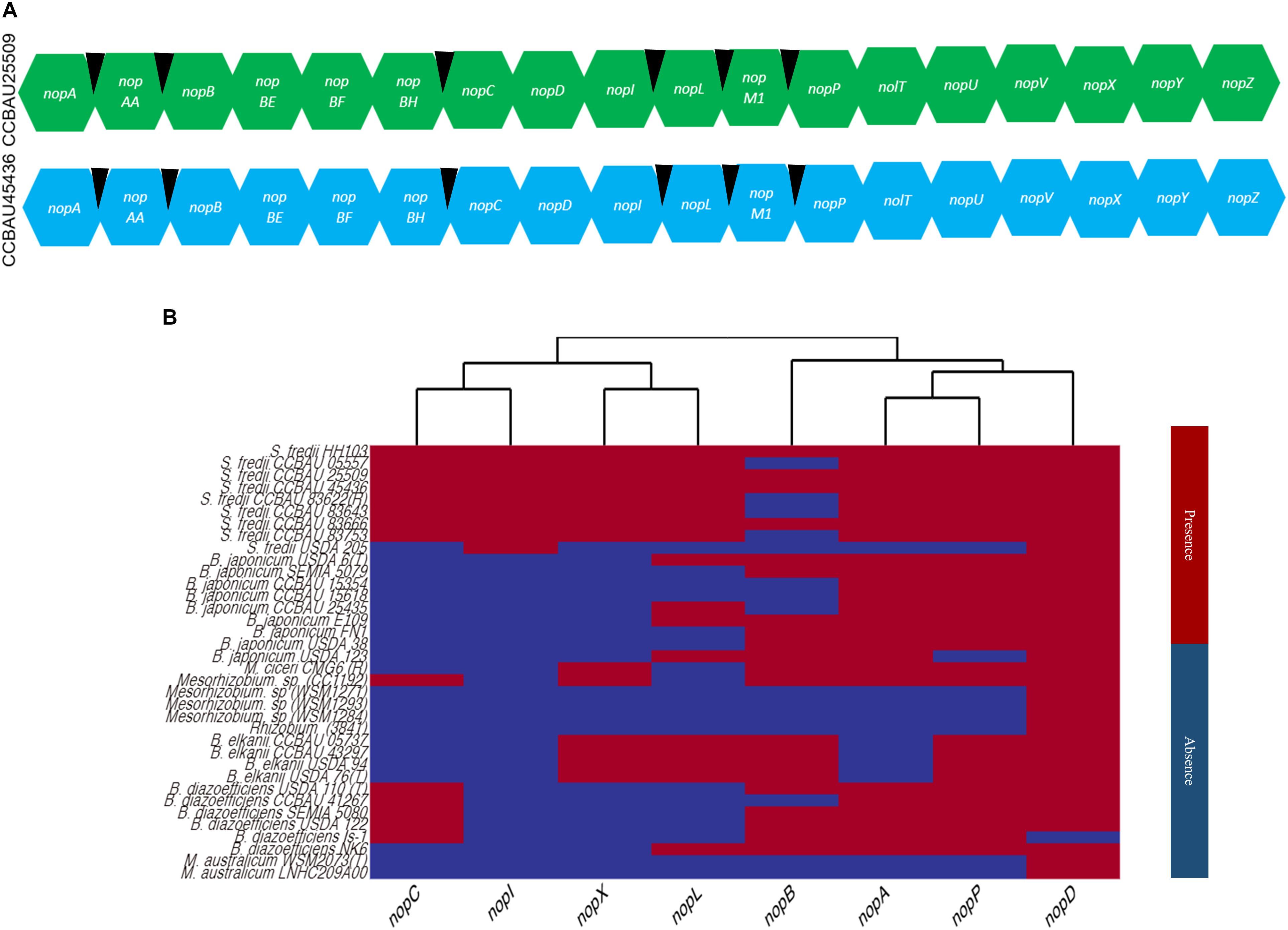
Figure 3. Known and putative nop genes identified in S. fredii CCBAU25509 and CCBAU45436 and the distribution of eight nop genes in 37 rhizobial strains. (A) Known and putative nop genes identified in the genomes of both S. fredii strains. Black wedges indicate the presence of tts boxes in the upstream regions of corresponding genes. (B) Cladogram showing the distribution of eight well-characterized nop genes in the genomes of 43 rhizobial strains belonging to four genera. Red and blue color indicate the presence or absence of a specific nop, respectively, in a particular genome.
To get a better idea of the evolution of the Nops we detected in the extracellular medium upon flavonoid induction, we next investigated the distribution of these nop genes in Sinorhizobium and other rhizobia.
We downloaded 56 annotated and assembled genomes of the genera Sinorhizobium, Mesorhizobium, Bradyrhizobium, and Rhizobium (Supplementary Table S1). The phylogenetic tree built based on the 16S rRNA sequences resulted in two major clades. Clade I was made up of Mesorhizobium, Sinorhizobium, and Rhizobium while clade II was made up of Bradyrhizobium. In general, the genus Sinorhizobium was closer to the genus Rhizobium than to the genus Mesorhizobium (Supplementary Figure S7).
We searched for the nop genes that encode for the eight Nops discussed above in all 56 genomes and generated a heat map based on the presence or absence of each nop gene in all four genera (Figure 3). Out of 56 selected genomes, only 36 genomes yielded position hits and their genomic locations and sequences are listed in Supplementary Table S7. We did not find any hit for a nop gene sequence from all selected genomes belonging S. medicae and S. meliloti strains. Overall, a lineage-specific pattern was observed among strains and species for the presence or absence of these nop genes (Figure 3). As expected, the observed amino acid sequence similarities of encoded Nops were found to be higher between species than between genera for a particular Nop (Supplementary Figure S8).
NopD, which is a putative T3SS effector protein with a SUMO protease domain, was found in 35 of the 36 genomes containing nop genes, and was only missing in B. diazoefficiens (Is-1) strain (Rodrigues et al., 2007; Sánchez et al., 2012) (Supplementary Figure S8).
NopA (a pilus subunit protein) and NopL (a T3SS effector protein), were found in 25 and 17 genomes, respectively (Figure 3 and Supplementary Table S7). The predicted protein sequence similarity is close to 50% among all orthologs found, showing a high degree of conservation (Supplementary Figure S8). NopA, was present only in S. fredii, B. japonicum, B. diazoefficiens and two strains of Mesorhizobium (Figure 3). Another T3SS pilus component protein, NopB, was observed only in S. fredii, B. elkanii, B. japonicum, and M. ciceri.
NopP (a T3SS effector protein), which determines host specificity in Bradyrhizobium (Sugawara et al., 2018), was only found in S. fredii, B. japonicum, B. elkanii, B. diazoefficiens, M. ciceri, and Mesorhizobium sp. (CC1192). The major translocator protein, NopX, was found only in B. elkanii, M. ciceri, Mesorhizobium sp. (CC1192) and S. fredii strains (Figure 3).
The identified sequences of NopX in B. elkanii and S. fredii strains showed a high sequence similarity > 75% (Supplementary Figure S8). A high sequence similarity (>75%) was also observed among the Nops within the genus Bradyrhizobium, but < 50% identity when these were compared to the NopP from S. fredii. The same pattern was observed for NopD (Supplementary Figure S8).
On the other hand, NopI (a T3SS effector protein) was particularly identified in S. fredii strains only and appeared to be species-specific (Figure 3). NopC (a T3SS effector protein) was only found in S. fredii, B. diazoefficiens, and Mesorhizobium sp. (CC1192).
Host Restriction of S. fredii Is Dependent on the Allelic Types of the Soybean Rj2/Rfg1 Protein
We selected 22 soybean accessions available in our laboratory (Lam et al., 2010) and performed PCR amplification and Sanger sequencing of the genomic regions encoding polymorphic amino acids (Yang et al., 2010) of the Rj2/Rfg1 protein. In addition to the three previously reported allelic types (Rj2/rfg1, rj2/rfg1, and rj2/Rfg1), a new allelic type with a previously unreported combination of amino acid polymorphism in the Rj2 region (E452, R490) was identified (Figure 4). All 22 soybean accessions were inoculated with the S. fredii strain CCBAU25509 and selected accessions were inoculated with the B. diazoefficiens strain USDA122 to characterize their nodulation phenotype (Figure 4). As expected, the Rj2/rfg1 and rj2/Rfg1 types exhibited restriction to B. japonicum strain USDA122 and S. fredii strain CCBAU25509 respectively, while the rj2/rfg1 allelic type imposed no restriction. The newly identified allelic type behaved like rj2. No Rj2/Rfg1 allelic type was found.
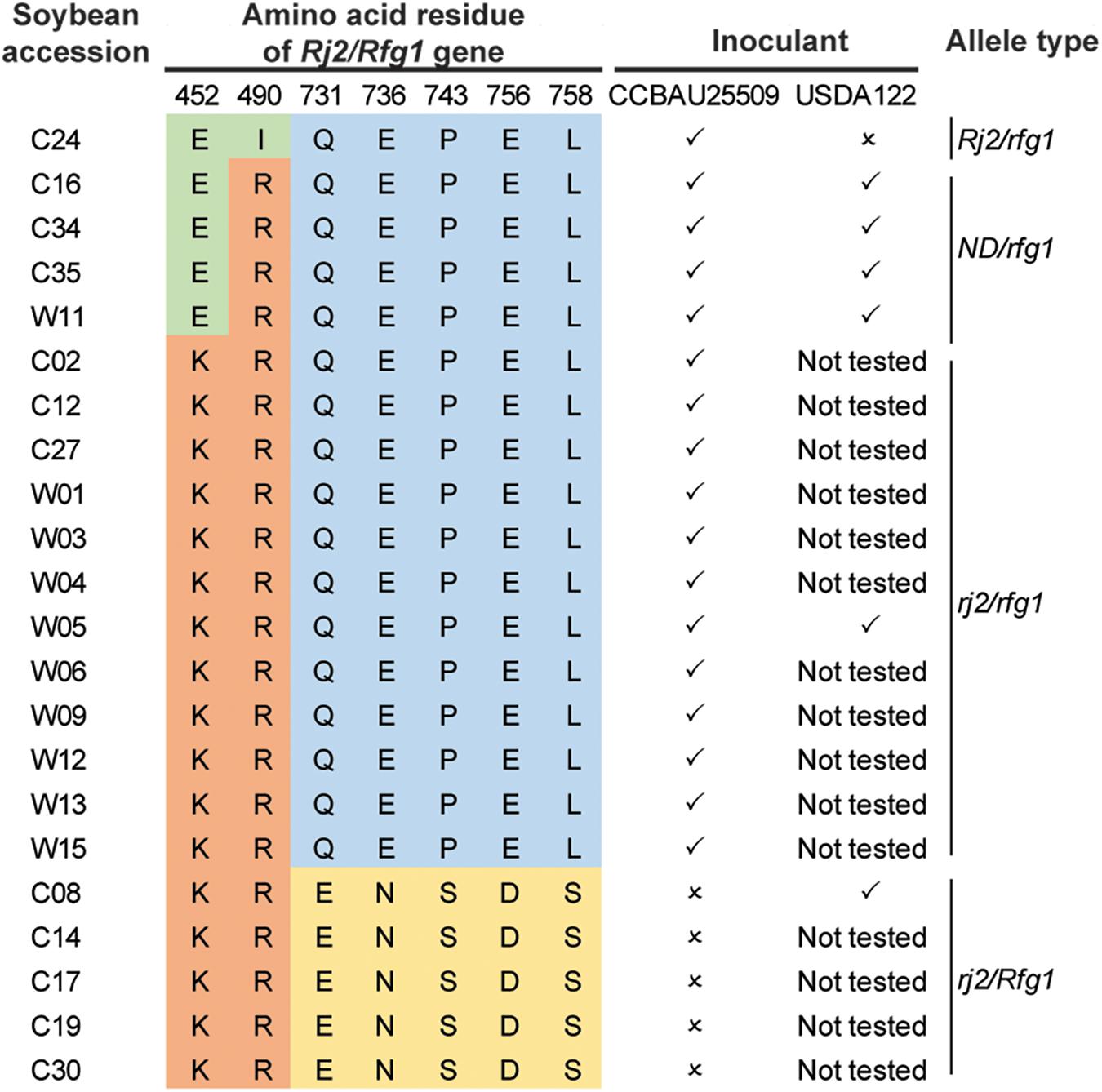
Figure 4. Genotypes and nodulation phenotypes of 22 soybean accessions. ✓: able to form nodules; ×: unable to form nodules. ND, not previously detected.
To further search for the allelic types of the Rj2(Rfg1) protein, we made use of the dataset from a large-scale re-sequencing project (Zhou et al., 2015). After removing accessions with ambiguous SNPs (either missing or heterozygous), 147 accessions showing clear allele types remained (Supplementary Table S8). Combining all available information, we found that the Rj2/rfg1 allele was the rarest while most of the accessions carried either the rj2/Rfg1 or the rj2/rfg1 allele.
T3SS in S. fredii Is Essential for Host Restriction
By taking advantage of the incompatibility of CCBAU25509 with the soybean host C08 (rj2/Rfg1), we performed Tn5 insertion mutagenesis in CCBAU25509 to select for mutants that could successfully nodulate C08. We generated a mixture of > 1.27 × 106 Tn5-insertion mutants by bi-parental mating of CCBAU25509 with E. coli S17-1 λpir harboring pUTmini-Tn5 km (Biomedal Life Science, Spain). The mixture of Tn5 mutants of CCBAU25509 was then used to inoculate 300 seedlings of C08. Four well-formed nodules resulted and were harvested at 28 days post-inoculation. Isolates from these four nodules were further confirmed by re-inoculating them into C08, of which three were able to form effective nodules after re-inoculation. The presence of the Tn5 insertion in these isolates were confirmed by PCR with strain-specific primers and Tn5-specific primers, and by Y-linker PCR and sequencing (Figure 5A). Results showed that these three isolates were likely the result of a single event having the mini-Tn5 transposon of 2,356 bp inserted in the middle of a coding region resembling an ortholog of rhcN (pSF25509a: 342,898.342,897 bp). RhcN was previously identified in the S. fredii strain NGR234 as a putative ATP synthase involved in the secretion of Nops through the lumen of the T3SS pilus (Staehelin and Krishnan, 2015).
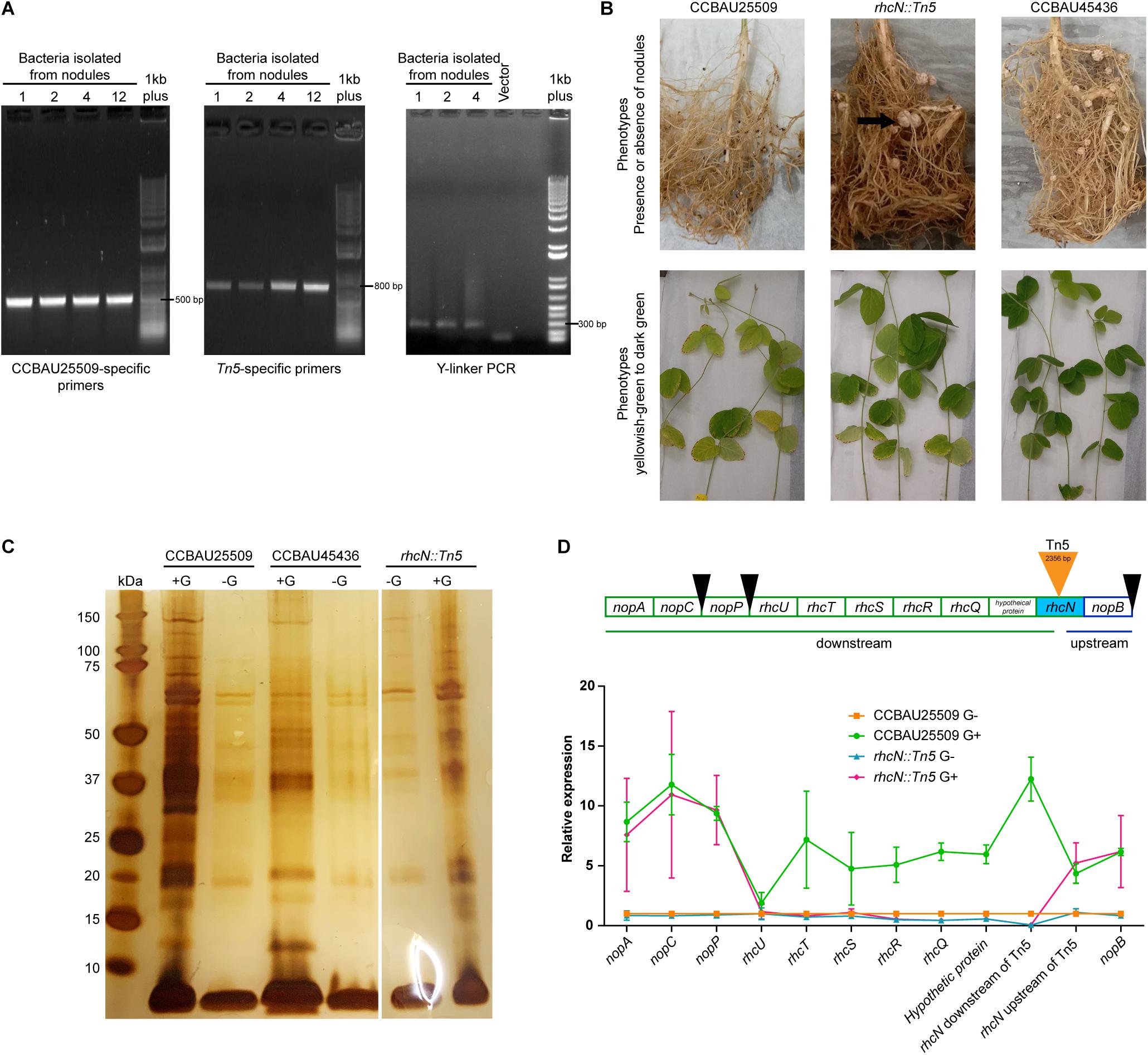
Figure 5. Characterization of the rhcN:Tn5 mutant. (A) PCR was performed to confirm that the rhcN:Tn5 mutant originated from CCBAU25509 (using CCBAU25509 primers) and contains a Tn5 insertion (using Tn5 primers and Y-linker PCR). Numbers 1, 2, 4, and 12 are rhizobia isolated from independent nodules. However, they were probably the same insertional event since the junction sequences were identical. Last lane on each gel is the 1 kb Plus DNA Ladder (Invitrogen). (B) Performance of the rhcN:Tn5 mutant inoculated onto the soybean accession C08 that carries the Rfg1 allele. Formation of root nodules (indicated by black arrows) and the leaf color (pale green or dark green) when the plants were grown under nitrogen-limiting conditions were recorded for the rhcN:Tn5 mutant, the non-compatible parent strain CCBAU25509, and the compatible strain CCBAU45436 28 days post-inoculation. (C) Silver staining visualization of the NuPAGE 4–12% bis-tris gel of the precipitated extracellular proteins from CCBAU25509, CCBAU45436 and the rhcN:Tn5 mutant with and without genistein (G) induction. (D) Upper panel: genes upstream and downstream of the rhcN operon. Black wedges indicate the upstream tts box sequences. Lower panel: relative expression patterns of these genes in CCBAU25509 and in the rhcN:Tn5 mutant with and without genistein (G). GyrA was used as the reference gene. Each data point represents the mean of three independent biological replicates. Error bar: standard deviation.
The rhcN:Tn5 insertion in CCBAU25509 removed host incompatibility with C08, which, when inoculated with this mutant, were successfully nodulated and exhibited a higher chlorophyll content (an indicator of nitrogen contents in planta) under nitrogen-limiting conditions (Figure 5B and Table 2).

Table 2. Performance of soybean accession C08 carrying the Rfg1 allele when inoculated with CCBAU25509, the rhcN:Tn5 mutant of CCBAU25509, or CCBAU45436 28 days post-inoculation.
To investigate the impact of the rhcN:Tn5 mutation on the secretion of Nops, we repeated the secretome experiment with the mutant strain. Secreted proteins that are not related to T3SS, such as the 60-kDa chaperonin, translation initiation factor IF-2, outer-membrane lipoproteins, etc., were still present in the secretome (Supplementary Table S6). On the other hand, no Nops, except NopA, were found in the secretome upon genistein induction (Figure 5C). Since the absence of Nops in the rhcN:Tn5 mutant improved the host compatibility, it could be inferred that Nops play a role in the incompatibility of CCBAU25509 with some soybean cultivars. Moreover, NopX, which was not detected in the secretome of the wild type CCBAU25509 (Table 1), was unlikely to have contributed to host incompatibility.
We then investigated the expression patterns of other genes in the same operon of rhcN, including the upstream nopB and other downstream genes: a hypothetical protein-coding gene, rhcQ, rhcR, rhcS, rhcT, rhcU, nopP, nopC, and nopA, in the presence or absence of genistein, using the gene expression patterns of the wild type strain CCBAU25509 as the control (Figure 5D). Without genistein, the expression levels of the above genes between CCBAU25509 and the rhcN:Tn5 mutant were comparable, except for rhcQ, rhcR and a hypothetical protein-coding gene, located immediately downstream of the Tn5 insertion, which had lower expressions in the rhcN mutant. In the presence of genistein, all the above-mentioned genes were up-regulated in the wild type strain of CCBAU25509. However, in the rhcN:Tn5 mutant, only nopB (located upstream of the Tn5 insertion site) and nopP, nopC, and nopA (far downstream of the insertion site) were induced by genistein. Their degree of induction by genistein in the rhcN:Tn5 mutant was comparable to that in the wild type CCBAU25509. In the rhcN:Tn5 mutant, the lower expression of hypothetical protein-coding gene, rhcQ, rhcR, rhcS, rhcT, rhcU genes compared to higher expression in CCBAU25509 in response to genistein suggests that the nodulation phenotype in the rhcN:Tn5 mutant may be due to the disruption of multiple components of the T3SS.
Conservation of NopP and NopI Protein Sequences in Host-Restricted S. fredii Strain
The Rj2/Rfg1 protein is the soybean determinant restricting nodulation by some B. japonicum, B. diazoefficiens, and S. fredii strains (Yang et al., 2010; Fan et al., 2017). Polymorphisms of seven amino acid residues (E452K, I490R, Q731E, E736N, P743S, E756D, and R758S) were used to define three allelic groups of Rj2/Rfg1 (Yang et al., 2010; Fan et al., 2017). Rj2/rfg1 restricts some B. japonicum and B. diazoefficiens strains; and rj2/Rfg1 restricts some S. fredii strains. On the other hand, rj2/rfg1 allows most B. japonicum, B. diazoefficiens, and S. fredii to form nodules.
We compared the sequences of Nops between Rfg1-compatible and Rfg1-incompatible S. fredii strains, such as CCBAU45436 and CCBAU25509, respectively. Protein sequences of most Nops were either completely identical between the two strains (NopA, NopC, and NopL) or showed no host-specific sequence conservation (NopB and NopX) (Supplementary Figure S9). The exceptions are the host-specific sequence conservation of NopP and NopI, which may be the culprits of host incompatibility. All amino acid substitutions in NopI and NopP were confirmed by PCR-based amplification, cloning and sequencing.
The NopP from Bradyrhizobium was reported to be a bacterial determinant of host incompatibility in soybean expressing the Rj2 protein, although no direct interaction between NopP and Rj2 has been found (Sugawara et al., 2018). In soybean expressing the Rfg1 allele, the NopP sequences in the restrictive S. fredii strains are invariable while there are some non-synonymous changes in the non-restrictive strains (Figure 6). We also observed two amino acid substitutions (K63E and R115Q) in NopP in the non-restrictive S. fredii CCBAU45436 compared to the restrictive CCBAU25509 (Figure 6A). One of the amino acids in the NopP of CCBAU25509 (K63) was observed at the same position as one previously reported in the NopP from B. diazoefficiens (USDA 122) (R60) and is proposed to be a key residue for Rj2-mediated incompatibility in soybean (Sugawara et al., 2018).
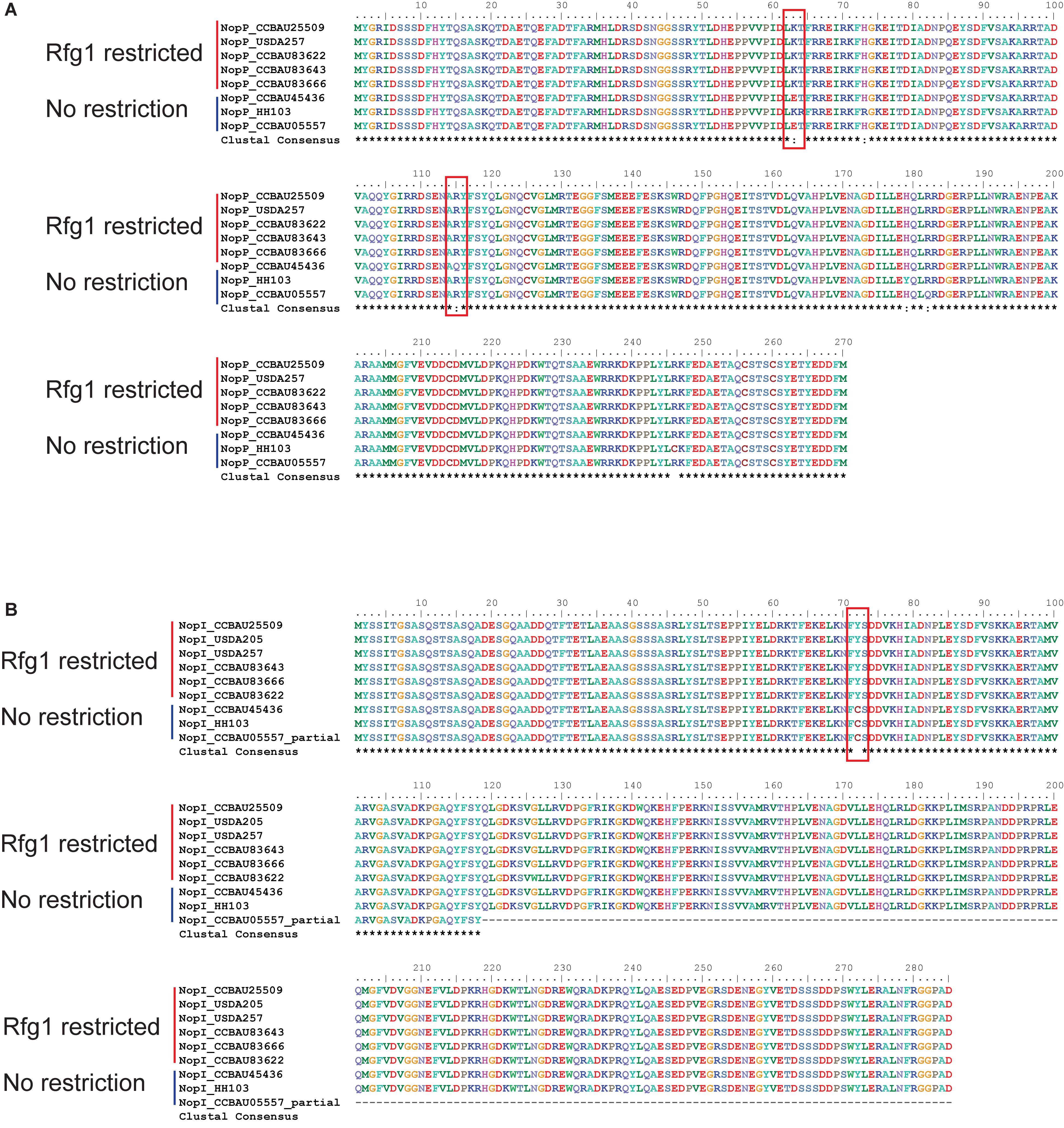
Figure 6. NopI and NopP protein sequence alignments between Rfg1-compatible and incompatible S. fredii strains. Shown host compatibility is based on observed phenotypes in previous publications (Yang et al., 2010; Jiménez-Guerrero et al., 2017; Jiao et al., 2018; Temprano-Vera et al., 2018). (A) NopP alignment showing K63E and R115Q amino acid substitutions. (B) NopI alignment showing a Y72C amino acid substitution.
NopI is a S. fredii-specific Nop which exhibits 48% sequence identity to NopP in the strain HH103 (Jiménez-Guerrero et al., 2017). The alignment of NopI showed an amino acid substitution (Y72C) in CCBAU45436 compared to CCBAU25509 (Figure 6B). We extracted the protein sequences of Nops from S. fredii strains with known host specificity for sequence alignments. NopI proteins are clearly divided into two groups among restrictive and non-restrictive S. fredii strains (Figure 6B).
NopP Played Essential Roles in Determining Host Compatibility
Here we compared the nodulation phenotypes among wild type CCBAU25509, wild type CCBAU45436, CCBAU25509 carrying only NopP from CCBAU45436 (25509nopP45436), and CCBAU45436 carrying only NopP from CCBAU25509 (45436nopP25509). The bacteria were inoculated into G. max C08 which carries the rj2/Rfg1 allele. Our results showed that only those strains carrying NopP from CCBAU45436 (wild type CCBAU45436 and 25509nopP45436) were able to nodulate C08 while those carrying NopP from CCBAU25509 (wild type CCBAU25509 and 45436nopP25509) could not (Figure 7 and Supplementary Figure S10).
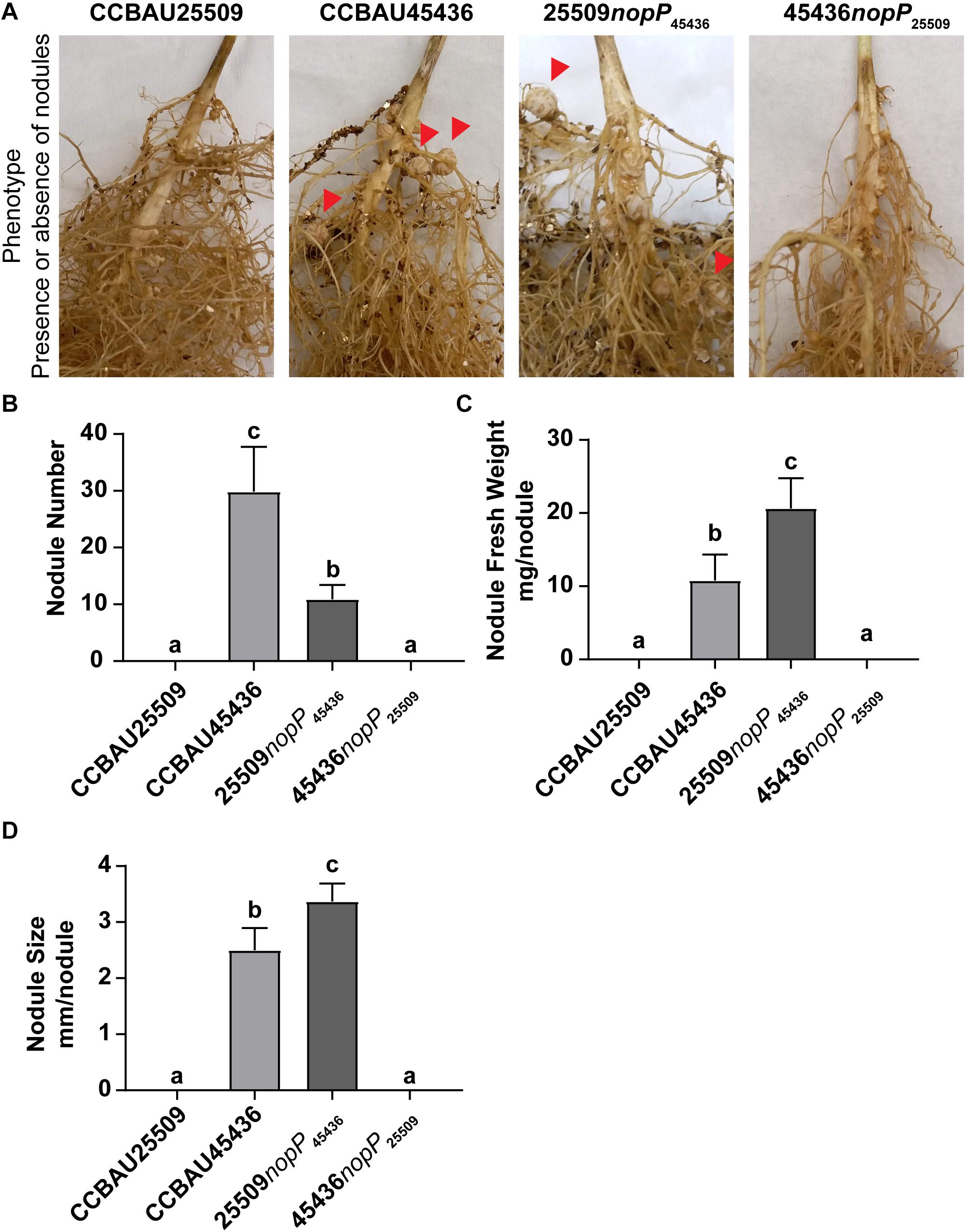
Figure 7. Nodulation phenotype of S. fredii strains carrying different NopP alleles. (A) Root of G. max C08 inoculated with wild type S. fredii CCBAU25509, wild type S. fredii CCBAU45436, mutant of CCBAU25509 carrying NopP from CCBAU45436 (25509nopP45436), and mutant of CCBAU45436 carrying NopP from CCBAU25509 (45436nopP25509). C08 carries the rj2/Rfg1 allele. Red arrows indicate some effective nodules formed on the roots. (B) Nodule number, (C) Nodule fresh weight and (D) Nodule size of the nodules harvested from the inoculated roots 28 days post-inoculation. N = 8. Error bar: standard deviation. Different letters above the bars indicate the results are statistically different at p < 0.05. The experiment has been repeated twice with consistent results (Supplementary Figure S10).
This finding led us to investigate if there is any direct interaction between Rfg1 and the two isoforms of sinorhizobial NopP using yeast two-hybrids (Supplementary Figure S11). The results did not demonstrate any direct interaction between Rfg1 and either forms of NopP. It is possible that NopP may bind to an unknown decoy in soybean which mediates the restriction functions of Rfg1.
Discussion
One important environmental value of leguminous plants is their ability to form root nodules which fix atmospheric dinitrogen molecules by converting them to ammonia through symbiotic relationships with soil rhizobia (Dixon and Kahn, 2004). Because of this unique property, cultivation of soybean, a leguminous crop, has become an integral part of sustainable agriculture. For example, a recent modeling study predicts that large-scale replacement of monocropping maize with intercropping maize-soybean could reduce the amount of nitrogen fertilizer used and subsequently alleviate air pollution problems caused by the emission of nitrogen oxides as a result of the excessive use of nitrogenous fertilizers (Fung et al., 2019).
To better characterize these rhizobial strains and their symbiotic relationships with soybean, we have previously completed the full genomes and conducted comprehensive transcriptomic analyses of two S. fredii strains, CCBAU25509 and CCBAU45436, which exhibit differential host compatibility toward soybean carrying the Rfg1 allele (Jiao et al., 2018). These two strains are descendants of a dominant sub-lineage in northeastern China (Temprano-Vera et al., 2018) and hence are representative strains.
To better characterize these two strains, we conducted high-throughput proteomic analyses to provide a unique view of proteins produced during mid-log phase, stationary phase, and in the bacteroid form. These data are a valuable resource for the study of S. fredii and symbiosis (Supplementary Tables S3, S4). The constructed circular proteome map with reference to its distribution within the genome suggests that the core genome has a higher priority in translation compared to plasmids (Figure 1B), which is consistent with previous research in Bradyrhizobium (Delmotte et al., 2014). These results are consistent with the findings in a previous proteome profiling experiment done in S. melioti (Sobrero et al., 2012; Berrabah et al., 2018). The most abundant protein species are chaperonins in all three growth stages, suggesting that they are the key proteins to sustain protein homeostasis (Wälti et al., 2017). The presence of phasin family proteins in all three growth stages of S. fredii indicates that they are multipurpose proteins which the cell always needs for regulating growth (Mezzina and Pettinari, 2016). The substantially expressed carbonic anhydrase protein in mid-log phase suggests that CO2 conversion to bicarbonate is necessary for growth in the free-living form (Merlin et al., 2003). The appearance of ABC transporter proteins, TCA cycle proteins, cold shock proteins, ferredoxin family proteins, nitrogen fixation and regulatory proteins, phosphate transport proteins, outer membrane lipoproteins, as well as transcriptional regulator proteins in bacteroids collected from nodules suggest that these proteins are important pillars of survival and nitrogen fixation in the symbiotic form.
In this study, we have established a new protocol to study the secretome by harnessing the power of Orbitrap. ABC transporters, ATP-binding protein, outer membrane lipoproteins, periplasmic dipeptide transport proteins, and TolB protein precursors were found in the extracellular medium of bacterial culture (Supplementary Table S6), suggesting possible roles in symbiosis (Broughton et al., 2000).
In addition to other secreted proteins, we also successfully detected eight previously reported Nops (NopA, NopB, NopC, NopD, NopI, NopL, NopP, and NopX) in the extracellular medium of bacterial culture upon genistein induction (Table 1). NopA, NopB, and NopX are components of the injectosome while the others, NopC, NopD, NopI, NopL and NopP, are possibly effector proteins secreted by T3SS into the host cytoplasm. These results are supported by previous studies performed on individual Nops in other Sinorhizobium strains (Krishnan et al., 2003; Marie et al., 2003; Lorio et al., 2004; Rodrigues et al., 2007; Zhang et al., 2011; Kim and Krishnan, 2014; Jiménez-Guerrero et al., 2015, 2017; Ge et al., 2016; Sugawara et al., 2018).
To analyze the genome-wide nop gene distribution in Sinorhizobium and other closely related genera, we downloaded 56 genomes covering Sinorhizobium, Mesorhizobium, Rhizobium, and Bradyrhizobium. Phylogenetic analyses using 16S rRNA differentiated Bradyrhizobium as a separate clade from the other three genera. Indeed, it was estimated that Bradyrhizobium had separated from the other clade before the emergence of legumes (Tian et al., 2012).
Despite the large evolutionary distance of Bradyrhizobium from the other three genera, on average there is a sequence similarity of > 50% among the eight Nops across all four genera (Supplementary Figure S8), suggesting a mutualistic co-evolution of T3SS effector proteins (Kimbrel et al., 2013).
Overall, the observed lineage-specific patterns of Nops (NopA, NopB, NopC, NopD, NopI, NopL, NopP, and NopX) distribution across all four genera indicate that they are distantly located in the symbiosis islands of their respective genomes, which cannot be explained solely by horizontal gene transfer or high genome-wide diversity (Kaneko et al., 2002; Kimbrel et al., 2013; Hungria et al., 2015) (Figure 3). In response to genistein induction, the observed expressions of Nops in the form of FPKM values are consistent with the previously published expression patterns of NopA, NopB, NopX, and NopP in S. fredii HH103 (Pérez-Montaño et al., 2016) (Supplementary Figure S5).
The presence of NopD in all four genera suggests that this effector protein in T3SS remains conserved during the adaptation to various hosts and geographic regions (Figure 3) (Kimbrel et al., 2013). NopI is absent in Bradyrhizobium (Figure 3) while some other Nops, such as NopE, NopH, pgl and NopF, are secreted by Bradyrhizobium but not by Sinorhizobium (Hempel et al., 2009). Although both Bradyrhizobium and Sinorhizobium strains can nodulate the same soybean host, they probably acquired divergent machineries for symbiosis that are functionally similar, independently of each other (Tian et al., 2012). The absence of NopX in B. diazoefficiens and B. japonicum suggests that, during divergence from B. elkanii, they might have lost this gene (Figure 3).
Between CCBAU25509 and CCBAU45436, there are seven common genistein-inducible Nops while NopX alone is undetectable in the extracellular medium of CCBAU25509 (Table 1). Although the nopX genes share an identical coding sequence in both CCBAU25509 and CCBAU45436, based on our transcriptome data (Supplementary Figure S5), it is likely that the expression of nopX gene in CCBAU25509 is turned off.
Bradyrhizobium diazoefficiens and S. fredii are restricted by some soybean hosts depending on the allelic type of the Rj2(Rfg1) protein. Extended analysis of soybean germplasm showed that there are three major allelic types: Rj2/rfg1, rj2/Rfg1, and rj2/rfg1, whereas Rj2/rfg1 seems to be a minor allelic type. CCBAU25509 is restricted by C08 which carries the Rfg1 allele while CCBAU45436 is not. It is largely unknown how Rfg1 recognizes the incompatible strain and restricts its nodulation. To investigate the role of T3SS in the host restriction of CCBAU25509, we constructed a mutant in CCBAU25509 that carries an insertional mutation in the rhcN gene. This insertion resulted in the deactivation of several T3SS components in the same operon (Figure 5D). As a result, the rhcN:Tn5 CCBAU25509 mutant could successfully nodulate the soybean accession that contains the Rfg1 allele.
Since the removal of T3SS allows CCBAU25509 to bypass the restriction imposed by Rfg1, the absence of NopX in the CCBAU25509 secretome is unlikely to be the cause of host incompatibility. While the sequence of NopA is conserved between CCBAU25509 and CCBAU45436, and could still be detected in the secretome of the rhcN:Tn5 CCBAU25509 mutant, the remaining six Nops were no longer detectable in the secretome of the mutant.
Interestingly, sequence alignments show that the protein sequences of NopP and NopI, but not the other Nops, are conserved in the Rfg1-restricted S. fredii strain (Figure 6 and Supplementary Figure S9). NopP and NopI are paralogs playing different roles in the nodulation of other rhizobial strains (Jiménez-Guerrero et al., 2017; Sugawara et al., 2018). The nopP of B. diazoefficiens was reported to genetically interact with Rj2 in determining the host specificity (Sugawara et al., 2018), while knocking out the nopI of S. fredii HH103 reduced the nodule numbers (Jiménez-Guerrero et al., 2017).
The highly conserved amino acid substitutions in NopI and NopP of CCBAU25509 could be potential amino acid candidates for studying the differential host compatibility between these two strains (Figure 6). In B. diazoefficiens (USDA122), a mutation in the same position (residue 63 of NopP from S. fredii and residue 60 of NopP from B. diazoefficiens) in NopP changes the compatibility toward soybeans carrying an Rj2 allele (Sugawara et al., 2018). However, B. diazoefficiens carrying the Rj2-incompatible NopP could still nodulate soybeans carrying the Rfg1 allele (Yang et al., 2010).
The effects of NopP in S. fredii on legume nodulation has been widely studied in the previous decades. However, specific studies on the effects of NopP in soybean nodulation are few. It is reported that disruption in what S. fredii HH103 could lead to an increase in nodule number on the roots of its Rfg1-carrying compatible soybean host (López-Baena et al., 2009).
In this study, by swapping the NopP between CCBUA25509 and CCBAU45436, it showed that only the strains carrying nopP45436 could successfully nodulate C08 (Figure 7 and Supplementary Figure S10). Although the nodule number generated by 25509nopP45436 was lower than that of wild type CCBAU45436, the restriction barrier between C08 and CCBAU25509 was overcome. On the other hand, unlike wild type CCBAU45436, CCBAU45436nopP25509 showed a restrictive nodulation phenotype similar to wild type CCBAU25509. This suggests that NopP may serve as a determinant in nodulation restriction by soybeans expressing Rfg1, based on its amino acid sequence, by either permitting nodulation (NopP45436 with E63 and Q115) or by restricting nodulation (NopP25509 with K63 and R115). It is speculated that the NopP25509 may serve as one of the effectors recognized by the Rfg1-expressing hosts and trigger the host defense mechanism while NopP45436 may serve as an effector that inhibits or escapes the host defense response. These results suggest that, similar to the NopP in Bradyrhizobium, the NopP in S. fredii also plays a role in host nodulation restrictions, and the amino acids at positions 63 and 115 may serve as restriction determinants between S. fredii and Rfg1-type soybean hosts.
Our yeast two-hybrid assay failed to detect any direct interaction between NopP and Rfg1 in vivo in the heterologous system (Supplementary Figure S11). However, it was reported that many effectors do not directly interact with the R proteins, but instead trigger defense responses by binding to decoys (Büttner, 2016). Of course, we cannot rule out the possibility that post-translational modifications in the native organism may be required for the interaction (Skorpil et al., 2005; Sugawara et al., 2018).
In summary, we provided comprehensive proteomic and secretomic information on two model strains of S. fredii to facilitate further in-depth analyses of S. fredii and its symbiotic relationships with different soybean accessions.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
H-ML coordinated this overall research strategy. HR and W-LC wrote the first draft of the manuscript. HR performed the Orbitrap-based proteomic and secretomic analyses. S-NT helped to optimize the LC gradient for secretome analysis. W-LC, W-TT, and L-YC identified and characterized the rhcN:Tn5 CCBAU25509 mutant. K-SW, C-YL, and F-LW characterized the nodulation genotype and phenotype of soybean accessions. MX performed soybean re-sequencing data analysis. C-YL and W-LC performed yeast two-hybrid experiments. W-LC created and characterized the NopP mutants. M-WL and H-ML edited and proofread the final manuscript.
Funding
This work was supported by the Hong Kong Research Grants Council General Research Fund (14108014), Area of Excellence Scheme (AoE/M-403/16), and the Lo Kwee-Seong Biomedical Research Fund awarded to H-ML, and the Shenzhen Virtual University Park Support Scheme to CUHK Shenzhen Research Institute (YFJGJS1.0).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Sinorhizobium fredii strains CCBAU25509 and CCBAU45436 were gifts from Prof. Changfu Tian of the China Agricultural University. B. diazoefficiens USDA122 was received from Prof. Kiwamu Minamisawa of Tohoku University. We thank Kwan-Pok Li for his assistance in mutant phenotyping. Qianwen Wang advised on sequence and transcriptome analyses. Prof. Kiwamu Minamisawa and Prof. Masayuki Sugawara provided critical comments. Jee Yan Chu copy-edited this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02569/full#supplementary-material
Footnotes
References
Bendtsen, J. D., Kiemer, L., Fausbøll, A., and Brunak, S. (2005). Non-classical protein secretion in bacteria. BMC Microbiol. 7:58. doi: 10.1186/1471-2180-5-58
Berrabah, F., Balliau, T., Aït-Salem, E. H., George, J., Zivy, M., Ratet, P., et al. (2018). Control of the ethylene signaling pathway prevents plant defenses during intracellular accommodation of the rhizobia. New Phytol. 219, 313–323. doi: 10.1111/nph.15142
Broughton, W. J., Jabbouri, S., and Perret, X. (2000). Keys to symbiotic harmony. J. Bacteriol. 182, 5641–5652. doi: 10.1128/jb.182.20.5641-5652.2000
Broughton, W. J., Wong, C. H., Lewin, A., Samrey, U., Myint, H., Meyer, H., et al. (1986). Identification of Rhizobium plasmid sequences involved in recognition of Psophocarpus, Vigna, and other legumes. J. Cell Biol. 102, 1173–1182. doi: 10.1083/jcb.102.4.1173
Büttner, D. (2016). Behind the lines–actions of bacterial type III effector proteins in plant cells. FEMS Microbiol. Rev. 40, 894–937. doi: 10.1093/femsre/fuw026
Dang, X., Xie, Z., Liu, W., Sun, Y., Liu, X., Zhu, Y., et al. (2019). The genome of ensifer alkalisoli YIC4027 provides insights for host specificity and environmental adaptations. BMC Genomics 20:643. doi: 10.1186/s12864-019-6004-7
Delmotte, N., Mondy, S., Alunni, B., Fardoux, J., Chaintreuil, C., Vorholt, J. A., et al. (2014). A proteomic approach of Bradyrhizobium/Aeschynomene root and stem symbioses reveals the importance of the fixA locus for symbiosis. Int. J. Mol. Sci. 15, 3660–3670. doi: 10.3390/ijms15033660
Dixon, R., and Kahn, D. (2004). Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2, 621–631. doi: 10.1038/nrmicro954
Erdmann, J., Preusse, M., Khaledi, A., Pich, A., and Häussler, S. (2018). Environment-driven changes of mRNA and protein levels in Pseudomonas aeruginosa. Environ. Microbiol. 20, 3952–3963. doi: 10.1111/1462-2920.14419
Fan, Y., Liu, J., Lyu, S., Wang, Q., Yang, S., and Lam, H.-M. (2017). The soybean Rfg1 gene restricts nodulation by Sinorhizobium fredii USDA193. Front. Plant Sci. 8:1548. doi: 10.3389/fpls.2017.01548
Fung, K. M., Tai, A. P. K., Yong, T., Liu, X., and Lam, H.-M. (2019). Co-benefits of intercropping as a sustainable farming method for safeguarding both food security and air quality. Environ. Res. Lett. 14:044011. doi: 10.1088/1748-9326/aafc8b
Ge, Y.-Y., Xiang, Q.-W., Wagner, C., Zhang, D., Xie, Z.-P., and Staehelin, C. (2016). The type 3 effector NopL of Sinorhizobium sp. strain NGR234 is a mitogen-activated protein kinase substrate. J. Exp. Bot. 67, 2483–2494. doi: 10.1093/jxb/erw065
Gietz, R. D., and Schiestl, R. H. (2007). Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 38–41. doi: 10.1038/nprot.2007.15
Han, L. L., Wang, E. T., Han, T. X., Liu, J., Sui, X. H., Chen, W. F., et al. (2009). Unique community structure and biogeography of soybean rhizobia in the saline-alkaline soils of Xinjiang. China. Plant Soil 324, 291–305. doi: 10.1007/s11104-009-9956-6
Hempel, J., Zehner, S., Göttfert, M., and Patschkowski, T. (2009). Analysis of the secretome of the soybean symbiont Bradyrhizobium japonicum. J. Biotechnol. 140, 51–58. doi: 10.1016/j.jbiotec.2008.11.002
Hungria, M., Menna, P., and Delamuta, J. R. (2015). “Bradyrhizobium, the ancestor of all rhizobia: phylogeny of housekeeping and nitrogen-fixation genes,” in Biological Nitrogen Fixation, ed. F. J. de Bruijn (Hoboken, NJ: Wiley).
Hurley, B., Subramaniam, R., Guttman, D. S., and Desveaux, D. (2014). Proteomics of effector-triggered immunity (ETI) in plants. Virulence 5, 752–760. doi: 10.4161/viru.36329
Jiao, J., Ni, M., Zhang, B., Zhang, Z., Young, J. P. W., Chan, T.-F., et al. (2018). Coordinated regulation of core and accessory genes in the multipartite genome of Sinorhizobium fredii. PLoS Gene. 14:e1007428. doi: 10.1371/journal.pgen.1007428
Jiménez-Guerrero, I., Pérez-Montaño, F., Medina, C., Ollero, F. J., and López-Baena, F. J. (2015). NopC is a Rhizobium-specific type 3 secretion system effector secreted by Sinorhizobium (Ensifer) fredii HH103. PLoS One 10:e0142866. doi: 10.1371/journal.pone.0142866
Jiménez-Guerrero, I., Pérez-Montaño, F., Medina, C., Ollero, F. J., and López-Baena, F. J. (2017). The Sinorhizobium (Ensifer) fredii HH103 nodulation outer protein NopI is a determinant for efficient nodulation of soybean and cowpea plants. Appl. Environ. Microbiol. 83:e2770-16. doi: 10.1128/AEM.02770-16
Kaneko, T., Nakamura, Y., Sato, S., Minamisawa, K., Uchiumi, T., Sasamoto, S., et al. (2002). Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 31, 189–197. doi: 10.1093/dnares/9.6.189
Kim, W.-S., and Krishnan, H. B. (2014). A nopA deletion mutant of Sinorhizobium fredii USDA257, a soybean symbiont, is impaired in nodulation. Curr. Microbiol. 68, 239–246. doi: 10.1007/s00284-013-0469-4
Kimbrel, J. A., Thomas, W. J., Jiang, Y., Creason, A. L., Thireault, C. A., Sachs, J. L., et al. (2013). Mutualistic co-evolution of type III effector genes in Sinorhizobium fredii and Bradyrhizobium japonicum. PLoS Pathog. 9:e1003204. doi: 10.1371/journal.ppat.1003204
Krishnan, H. B., Lorio, J., Kim, W. S., Jiang, G., Kim, K. Y., Deboer, M., et al. (2003). Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and exported by a type III protein secretion system in Sinorhizobium fredii USDA257. Mol. Plant Microbe Interact. 16, 617–625. doi: 10.1094/mpmi.2003.16.7.617
Krishnan, H. B., and Pueppke, S. G. (1993). Flavonoid inducers of nodulation genes stimulate Rhizobium fredii USDA257 to export proteins into the environment. Mol. Plant Microbe Interact. 6, 107–113.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kwon, T., Huse, H. K., Vogel, C., Whiteley, M., and Marcotte, E. M. (2014). Protein-to-mRNA ratios are conserved between Pseudomonas aeruginosa strains. J. Proteome Res. 13, 2370–2380. doi: 10.1021/pr4011684
Kwon, Y. M., and Ricke, S. C. (2000). Efficient amplification of multiple transposon-flanking sequences. J. Microbiol. Methods 41, 195–199. doi: 10.1016/s0167-7012(00)00159-7
Lam, H.-M., Xu, X., Liu, X., Chen, W., Yang, G., Wong, F.-L., et al. (2010). Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42, 1053–1059. doi: 10.1038/ng.715
Li, Y., Tian, C. F., Chen, W. F., Wang, L., Sui, X. H., and Chen, W. X. (2013). High-resolution transcriptomic analyses of Sinorhizobium sp. NGR234 bacteroids in determinate nodules of Vigna unguiculata and indeterminate nodules of Leucaena leucocephala. PLoS One 8:e70531. doi: 10.1371/journal.pone.0070531
Liu, Y., Beyer, A., and Aebersold, R. (2016). On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550. doi: 10.1016/j.cell.2016.03.014
López-Baena, F. J., Monreal, J. A., Pérez-Montaño, F., Guasch-Vidal, B., Bellogín, R. A., Vinardell, J. M., et al. (2009). The absence of nops secretion in Sinorhizobium fredii HH103 increases GmPR1 expression in williams soybean. Mol. Plant Microbe Interact. 22, 1445–1454. doi: 10.1094/MPMI-22-11-1445
López-Baena, F. J., Ruiz-Sainz, J. E., Rodriguez-Carvajal, M. A., and Vinardell, J. M. (2016). Bacterial molecular signals in the Sinorhizobium fredii-soybean symbiosis. Int. J. Mol. Sci. 17:E755. doi: 10.3390/ijms17050755
Lorio, J. C., Kim, W. S., and Krishnan, H. B. (2004). NopB, a soybean cultivar-specificity protein from Sinorhizobium fredii USDA257, is a type III secreted protein. Mol. Plant Microbe Interact. 17, 1259–1268. doi: 10.1094/mpmi.2004.17.11.1259
Man, C. X., Wang, H., Chen, W. F., Sui, X. H., Wang, E. T., and Chen, W. X. (2008). Diverse rhizobia associated with soybean grown in the subtropical and tropical regions of China. Plant Soil 310, 77–87. doi: 10.1007/s11104-008-9631-3
Marie, C., Deakin, W. J., Ojanen-Reuhs, T., Diallo, E., Reuhs, B., Broughton, W. J., et al. (2004). TtsI, a key regulator of rhizobium species NGR234 is required for type III-dependent protein secretion and synthesis of rhamnose-rich polysaccharides. Mol. Plant Microbe Interact. 17, 958–966. doi: 10.1094/mpmi.2004.17.9.958
Marie, C., Deakin, W. J., Viprey, V., Kopciñska, J., Golinowski, W., Krishnan, H. B., et al. (2003). Characterization of Nops, nodulation outer proteins, secreted via the type III secretion system of NGR234. Mol. Plant Microbe Interact. 16, 743–751. doi: 10.1094/mpmi.2003.16.9.743
Marx, H., Minogue, C. E., Jayaraman, D., Richards, A. L., Kwiecien, N. W., Siahpirani, A. F., et al. (2016). A proteomic atlas of the legume Medicago truncatula and its nitrogen-fixing endosymbiont Sinorhizobium meliloti. Nat. Biotech. 34, 1198–1205. doi: 10.1038/nbt.3681
Merlin, C., Masters, M., Mcateer, S., and Coulson, A. (2003). Why is carbonic anhydrase essential to Escherichia coli? J. Bacteriol. 185, 6415–6424. doi: 10.1128/jb.185.21.6415-6424.2003
Mezzina, M. P., and Pettinari, M. J. (2016). Phasins, multifaceted polyhydroxyalkanoate granule-associated proteins. Appl. Environ. Microbiol. 82, 5060–5067. doi: 10.1128/AEM.01161-16
Millard, P., and Robinson, D. (1987). Colorimetric determination of the total chlorophyll concentrations in potato leaves by liquid scintillation counting. Potato Res. 30, 491–494. doi: 10.1007/bf02361926
Muers, M. (2011). Transcriptome to proteome and back to genome. Nat. Rev. Genet. 12:518. doi: 10.1038/nrg3037
Muñoz, N., Qi, X., Li, M.-W., Xie, M., Gao, Y., Cheung, M.-Y., et al. (2016). Improvement in nitrogen fixation capacity could be part of the domestication process in soybean. Heredity 117, 84–93. doi: 10.1038/hdy.2016.27
Okazaki, S., Kaneko, T., Sato, S., and Saeki, K. (2013). Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc. Natl. Acad. Sci. U.S.A. 110, 17131–17136. doi: 10.1073/pnas.1302360110
Pérez-Montaño, F., Jiménez-Guerrero, I., Acosta-Jurado, S., Navarro-Gómez, P., Ollero, F. J., Ruiz-Sainz, J. E., et al. (2016). A transcriptomic analysis of the effect of genistein on Sinorhizobium fredii HH103 reveals novel rhizobial genes putatively involved in symbiosis. Sci. Rep. 6:31592. doi: 10.1038/srep31592
Qi, X., Li, M.-W., Xie, M., Liu, X., Ni, M., Shao, G., et al. (2014). Identification of novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Comm. 5:4340. doi: 10.1038/ncomms5340
Rodrigues, J. A., López-Baena, F. J., Ollero, F. J., Vinardell, J. M., Espuny, M. D. R., Bellogín, R. A., et al. (2007). NopM and NopD are rhizobial nodulation outer proteins: identification using LC-MALDI and LC-ESI with a monolithic capillary column. J. Proteome Res. 6, 1029–1037. doi: 10.1021/pr060519f
Sánchez, C., Mercante, V., Babuin, M. F., and Lepek, V. C. (2012). Dual effect of Mesorhizobium loti T3SS functionality on the symbiotic process. FEMS Microbiol. Lett. 330, 148–156. doi: 10.1111/j.1574-6968.2012.02545.x
Schafer, A., Tauch, A., Jager, W., Kalinowski, J., Thierbach, G., and Puhler, A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. doi: 10.1016/0378-1119(94)90324-7
Schmeisser, C., Liesegang, H., Krysciak, D., Bakkou, N., Le Quéré, A., Wollherr, A., et al. (2009). Rhizobium sp. strain NGR234 possesses a remarkable number of secretion systems. Appl. Environ. Microbiol. 75, 4035–4045. doi: 10.1128/AEM.00515-09
Schuldes, J., Orbegoso, M. R., Schmeisser, C., Krishnan, H. B., Daniel, R., and Streit, W. R. (2012). Complete genome sequence of the broad-host-range strain Sinorhizobium fredii USDA257. J. Bacteriol. 194:4483. doi: 10.1128/JB.00966-12
Skorpil, P., Saad, M. M., Boukli, N. M., Kobayashi, H., Ares-Orpel, F., Broughton, W. J., et al. (2005). NopP, a phosphorylated effector of rhizobium sp. strain NGR234, is a major determinant of nodulation of the tropical legumes Flemingia congesta and Tephrosia vogelii. Mol. Microbiol. 57, 1304–1317. doi: 10.1111/j.1365-2958.2005.04768.x
Sobrero, P., Schlüter, J.-P., Lanner, U., Schlosser, A., Becker, A., and Valverde, C. (2012). Quantitative proteomic analysis of the Hfq-regulon in Sinorhizobium meliloti 2011. PLoS One 7:e48494. doi: 10.1371/journal.pone.0048494
Songwattana, P., Noisangiam, R., Teamtisong, K., Prakamhang, J., Teulet, A., Tittabutr, P., et al. (2017). Type 3 secretion system (T3SS) of Bradyrhizobium sp. DOA9 and its roles in legume symbiosis and rice endophytic association. Front. Microbiol. 8:1810. doi: 10.3389/fmicb.2017.01810
Staehelin, C., and Krishnan, H. B. (2015). Nodulation outer proteins: double-edged swords of symbiotic rhizobia. Biochem. J. 470, 263–274. doi: 10.1042/BJ20150518
Sugawara, M., Epstein, B., Badgley, B. D., Unno, T., Xu, L., Reese, J., et al. (2013). Comparative genomics of the core and accessory genomes of 48 Sinorhizobium strains comprising five genospecies. Genome Biol. 14:R17. doi: 10.1186/gb-2013-14-2-r17
Sugawara, M., Takahashi, S., Umehara, Y., Iwano, H., Tsurumaru, H., Odake, H., et al. (2018). Variation in bradyrhizobial NopP effector determines symbiotic incompatibility with Rj2-soybeans via effector-triggered immunity. Nat. Commun. 9:3139. doi: 10.1038/s41467-018-05663
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526.
Tanca, A., Biosa, G., Pagnozzi, D., Addis, M. F., and Uzzau, S. (2013). Comparison of detergent-based sample preparation workflows for LTQ-Orbitrap analysis of the Escherichia coli proteome. Proteomics 13, 2597–2607. doi: 10.1002/pmic.201200478
Temprano-Vera, F., Rodríguez-Navarro, D. N., Acosta-Jurado, S., Perret, X., Fossou, R. K., Navarro-Gómez, P., et al. (2018). Sinorhizobium fredii strains HH103 and Ngr234 form nitrogen fixing nodules with diverse wild soybeans (Glycine soja) from central china but are ineffective on northern china accessions. Front. Microbiol. 9:2843. doi: 10.3389/fmicb.2018.02843
Tian, C. F., Zhou, Y. J., Zhang, Y. M., Li, Q. Q., Zhang, Y. Z., Li, D. F., et al. (2012). Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations. Proc. Natl. Acad. Sci. U.S.A. 109, 8629–8634. doi: 10.1073/pnas.1120436109
Tsukui, T., Eda, S., Kaneko, T., Sato, S., Okazaki, S., Kakizaki-Chiba, K., et al. (2013). The type III secretion system of Bradyrhizobium japonicum USDA122 mediates symbiotic incompatibility with Rj2 soybean plants. Appl. Environ. Microbiol. 79, 1048–1051. doi: 10.1128/AEM.03297-12
Tsurumaru, H., Yamakawa, T., Tanaka, M., and Sakai, M. (2008). Tn5 mutants of Bradyrhizobium japonicum is-1 with altered compatibility with Rj2-soybean cultivars. Soil Sci. Plant Nutr. 54, 197–203.
Wälti, M. A., Schmidt, T., Murray, D. T., Wang, H., Hinshaw, J. E., and Clore, G. M. (2017). Chaperonin GroEL accelerates protofibril formation and decorates fibrils of the het-s prion protein. Proc. Natl. Acad. Sci. U.S.A. 114, 9104–9109. doi: 10.1073/pnas.1711645114
Wassem, R., Kobayashi, H., Kambara, K., Le Quéré, A., Walker, G. C., Broughton, W. J., et al. (2008). TtsI regulates symbiotic genes in rhizobium species NGR234 by binding to tts boxes. Mol. Microbiol. 68, 736–748. doi: 10.1111/j.1365-2958.2008.06187.x
Weidner, S., Becker, A., Bonilla, I., Jaenicke, S., Lloret, J., Margaret, I., et al. (2012). Genome sequence of the soybean symbiont Sinorhizobium fredii HH103. J. Bacteriol. 194, 1617–1618. doi: 10.1128/JB.06729-11
Yang, S., Tang, F., Gao, M., Krishnan, H. B., and Zhu, H. (2010). R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. U.S.A. 107, 18735–18740. doi: 10.1073/pnas.1011957107
Yoon, S.-H., Ha, S.-M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing ezbiocloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Zhang, L., Chen, X.-J., Lu, H.-B., Xie, Z.-P., and Staehelin, C. (2011). Functional analysis of the type 3 effector nodulation outer protein L (NopL) from rhizobium sp. NGR234: symbiotic effects, phosphorylation, and interference with mitogen-activated protein kinase signaling. J. Biol. Chem. 286, 32178–32187. doi: 10.1074/jbc.M111.265942
Zhao, R., Liu, L. X., Zhang, Y. Z., Jiao, J., Cui, W. J., Zhang, B., et al. (2017). Adaptive evolution of rhizobial symbiotic compatibility mediated by co-evolved insertion sequences. ISME J. 12, 101–111. doi: 10.1038/ismej.2017.136
Keywords: Sinorhizobium fredii, proteome, secretome, nodulation outer proteins, soybean
Citation: Rehman HM, Cheung W-L, Wong K-S, Xie M, Luk C-Y, Wong F-L, Li M-W, Tsai S-N, To W-T, Chan L-Y and Lam H-M (2019) High-Throughput Mass Spectrometric Analysis of the Whole Proteome and Secretome From Sinorhizobium fredii Strains CCBAU25509 and CCBAU45436. Front. Microbiol. 10:2569. doi: 10.3389/fmicb.2019.02569
Received: 14 June 2019; Accepted: 23 October 2019;
Published: 12 November 2019.
Edited by:
Benjamin Gourion, UMR 2594 Laboratoire Interactions Plantes-Microorganismes (LIPM), FranceReviewed by:
Francisco Javier López-Baena, University of Seville, SpainChristian Staehelin, Sun Yat-sen University, China
Copyright © 2019 Rehman, Cheung, Wong, Xie, Luk, Wong, Li, Tsai, To, Chan and Lam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hon-Ming Lam, aG9ubWluZ0BjdWhrLmVkdS5oaw==
†These authors have contributed equally to this work
 Hafiz Mamoon Rehman
Hafiz Mamoon Rehman Wai-Lun Cheung
Wai-Lun Cheung Kwong-Sen Wong
Kwong-Sen Wong Min Xie
Min Xie Man-Wah Li
Man-Wah Li Hon-Ming Lam
Hon-Ming Lam