
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 November 2019
Sec. Microbial Physiology and Metabolism
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02559
Burkholderia pyrrocinia JK-SH007 is a plant growth-promoting bacteria (PGPB), that can promote the growth of poplar and other trees, and, production of the plant hormone indole-3-acetic acid (IAA) is one of the reasons for this effect. Therefore, the aims of this study were to evaluate the effect of the external environment on the synthesis of IAA by B. pyrrocinia JK-SH007 and to perform a functional analysis of its IAA synthesis pathway. In this study, IAA and its synthetic intermediates indole-3-acetamide (IAM), indole-3-pyruvic acid (IPyA), tryptamine (TAM), and indole-3-acetonitrile (IAN) were detected in B. pyrrocinia JK-SH007 fermentation broth by high-performance liquid chromatography and tandem mass spectrometry (HPLC-MS/MS), and these indolic compounds were also found in the cell-free extraction of B. pyrrocinia JK-SH007, but the genomic analysis of B. pyrrocinia JK-SH007 indicated that IAA biosynthesis was mainly through the IAM and TAM pathways. The effects of L-tryptophan (L-Trp), temperature and pH on the synthesis of IAA were investigated, and the results showed that L-Trp exerted a significant effect on IAA synthesis and that 37°C and pH 7 were the optimal conditions IAA production by B. pyrrocinia JK-SH007. In addition, the protein expression of tryptophan 2-monooxygenase and indoleacetamide hydrolase, which are the key enzymes of the indole acetamide-mediated IAA synthesis pathway, was analyzed, and their activity was verified by substrate feeding experiments. The results revealed the existence of an IAA synthesis pathway mediated by IAM and indicated that this pathway plays a role in B. pyrrocinia JK-SH007. This study lays the foundation for further exploration of the specific pathway and mechanism of IAA synthesis in B. pyrrocinia JK-SH007.
Plant growth-promoting bacteria (PGPB), which are ubiquitous in plants (Strobel et al., 2004), can help host plants absorb nutrients and improve their plant growth (Strobel et al., 2004; Santoyo et al., 2016). PGPB exert positive effects on plants via many mechanisms, including the production of indole-3-acetic acid (IAA), cytokinin, gibberellin, and ACC deaminase, nitrogen fixation, phosphorous solubilization and siderophore chelation of iron in soil (Glick, 2015; Olanrewaju et al., 2017; Ramakrishna et al., 2019). IAA is an auxin that plays an important role in plant growth and development (Benjamins and Scheres, 2008; Schaller et al., 2015) by stimulating cell elongation and rooting, increasing seed germination and seedling growth, and promoting plant growth at the shoot apex (Ahmad et al., 2005; Tsavkelova et al., 2007; Khamna et al., 2010; Zhang et al., 2017a, b). The ability to synthesize IAA is a common feature of PGPB (Olanrewaju et al., 2017), and although a large number of IAA-producing strains have been identified, studies on IAA biosynthesis-related genes or enzymes have generally been limited to a small number of model microorganisms, such as Azospirillum (Mandira and Srivastava, 2007), Pseudomonas (Patten and Glick, 2002a), and Rhizobium (Theunis et al., 2004). Various IAA biosynthetic pathways, including a tryptophan-dependent or tryptophan-independent pathways, have been proposed in plant-associated bacteria (Mcclerklin et al., 2018). The tryptophan-dependent pathways include the indole-3-acetamide (IAM), indole-3-pyruvic acid (IPyA), indole-3-acetonitrile (IAN), tryptamine (TAM), and tryptophan side-chain oxidase (TSO) pathways (Duca et al., 2014; Di et al., 2016), and the IAM pathway is one of the IAA synthesis pathways in which L-tryptophan (L-Trp) is a precursor. In this pathway, L-Trp is first converted into IAM by tryptophan 2-monooxygenase, which is encoded by the iaaM gene, and IAM is further catalyzed by indoleacetamide hydrolase, which is encoded by the iaaH gene, to synthesize IAA (Di et al., 2016). The IAM pathway is thought to exist in both phytopathogenic and beneficial bacteria (Gudrun et al., 1984; Theunis et al., 2004).
Indole-3-acetic acid production by bacteria is influenced by the culture conditions, growth stages and substrate availability, and the pressures exerted by different environmental conditions encountered by bacteria fine-tune the biosynthesis of this metabolite (Spaepen et al., 2007). The tryptophan concentration, pH, temperature, and carbon and nitrogen sources, among other variables, constitute the environmental conditions that are often encountered by bacteria, and these abiotic factors can change IAA biosynthesis (Ona et al., 2003, 2005; Malhotra and Srivastava, 2009; Molina et al., 2018). Therefore, the control of IAA biosynthesis requires a better understanding of mechanisms for optimizing these factors.
Burkholderia pyrrocinia JK-SH007 is an endophytic bacterium that was isolated from a branch of Populus × euramericana “Nanlin895” in Jiangsu Province, China, and belongs to the Burkholderia cepacia complex (Bcc) (Ren et al., 2011). This strain can significantly increase the enzymatic activity of poplar rhizosphere soil, which is conducive to the absorption of nutrients by plants and plays an important role in promoting poplar growth (Ren et al., 2011). In addition, this strain can colonize poplar, has endogenous characteristics, and can effectively inhibit the three pathogens causing poplar canker disease (Cytospora chrysosperma, Phomopsis macrospora, and Fusicoccum aesculi) (Ren et al., 2010, 2011, 2106). By increasing the ground diameter of pear trees, B. pyrrocinia JK-SH007 also promotes the growth of pear trees, improves the physiological indexes of leaves, increases the available phosphorus content in rhizosphere soil and improves fruit quality (Dou et al., 2017). Some species of Bcc have been widely reported as human opportunistic pathogens and are considered to be important pathogenic bacteria causing cystic fibrosis (CF) (Agodi et al., 2001; Campana et al., 2005; Mahenthiralingam et al., 2008). However, many Bcc strains are common biocontrol bacteria with many functions such as biological control, biodegradation, and plant growth (Bevivino et al., 2000; Sopheareth et al., 2013). Previous studies have shown that B. pyrrocinia JK-SH007 strain is not toxic to plant, animal or human (Ren, unpublished data), and it is in line with the safe application of Bcc. The strain does not contain the virulence genes BCESM and cblA (Ren et al., 2011), which is preliminarily proved to be biosafe. As a result, B. pyrrocinia JK-SH007 can be considered a potential PGPB (Ren et al., 2010, 2011, 2106).
However, the growth-promoting mechanism of B. pyrrocinia JK-SH007 remains unclear, although it is know that the synthesis of IAA plays a role in the growth-promoting effect of PGPB. A study of the characteristics and synthesis mechanism of the IAA produced by this strain can serve as a theoretical basis for understanding the growth-promoting effect of the strain on plants and thereby enhance its application in forests (Fan et al., 2018). Previous studies have shown that B. pyrrocinia JK-SH007 has the ability to synthesize IAA and that L-Trp exerts a significant effect on IAA synthesis. B. pyrrocinia JK-SH007 might harbor the IAM-mediated IAA synthesis pathway (Liu et al., 2019), but the conditions under which B. pyrrocinia JK-SH007 can synthesize IAA and the enzymes related to this pathway have not been elucidated. In this study, the ability of B. pyrrocinia JK-SH007 to synthesize IAA and the effects of different factors on its synthesis were analyzed, and the key rate-limiting enzymes in the IAA synthesis pathway of B. pyrrocinia JK-SH007 based on IAM, tryptophan 2-monooxygenase (iaaM) and indoleacetamide hydrolase (iaaH) were expressed and identified.
Burkholderia pyrrocinia JK-SH007 (strain number: CCTCC M209028), Escherichia coli BL21 (DE3) and the expression vector pET32a were provided by Jiangsu Key Laboratory for Prevention and Management of Invasive Species.
The Burkholderia pyrrocinia JK-SH007 strain was inoculated into tryptic soy broth (TSB) liquid medium at 1% and cultured with shaking at 30°C for 36 h to reach an OD600nm = 1.6). The fermentation broth was collected (filter with a 0.22 μm PES membrane filter unit) and sent to Nanjing Webiolotech Biotechnology Co., Ltd., for high-performance liquid chromatography and tandem mass spectrometry (HPLC-MS/MS) analysis.
Burkholderia pyrrocinia JK-SH007 cells cultured for 36 h (OD600nm = 1.6) were collected, and the bacteria washed three times with potassium phosphate buffer (pH 7.5) and suspended. The mixed bacterial solution was treated with an ultrasonic cell crushing apparatus under an ice bath for 15 min. The bacterial body was then destroyed, and the contents were released (Shokri and Emtiazi, 2010). The supernatant containing acellular extract was obtained by centrifugation at 10 000 × g for 10 min. The supernatant was combined with solution of 1.0 mg/mL L-Trp (Yuanye, Shanghai, China). After 30 min of incubation at 37°C, the reaction mixtures were sent to Nanjing Webiolotech Biotechnology Co., Ltd., for HPLC-MS/MS analysis.
The predicted and annotated gene sequences obtained from B. pyrrocinia JK-SH007 genome were analyzed by similarity analysis with KEGG (Kyoto Encyclopedia of Genes and Genomes) (Kanehisa et al., 2012) and NCBI (National Center for Biotechnology Information), and then each gene was allocated to the KEGG pathway maps of Trp metabolism and IAA biosynthesis. Based on this analysis, the whole IAA biochemical pathways was proposed and compared with the metabolite spectrum obtained by B. pyrrocinia JK-SH007 in vitro (Torres et al., 2018).
The Burkholderia pyrrocinia JK-SH007 strain was inoculated in TSB liquid medium at 1% and cultured at 30°C in the dark. The Microorganism IAA ELISA Kit (MLBIO, Shanghai, China) was used to determine the IAA content at 12, 16, 20, 24, 28, 32, 36, 40, 44, 48, 60, 72, 84, 96, 108, 120, 144, and 168 h, and the absorbances at OD600nm at these time points were determined.
An aseptic L-Trp solution (Yuanye, Shanghai, China) solution (obtained after removing bacteria using a 0.22 μm PES membrane filter unit) was added to TSB liquid medium and synthetic dropout (SD) liquid medium (18.22% D-Sorbitol, 0.67% yeast nitrogen base without amino acids, and 2% glucose, pH 5.8.) to final concentrations of 0.2, 0.5, 0.8, 1.0, 1.5, 2.0, 3.0, 4.0, 6.0, 8.0, and 10.0 mg/mL. TSB liquid medium or SD liquid medium without L-Trp solution was used as the control. B. pyrrocinia JK-SH007 was inoculated in the above mentioned media at 1%, and the IAA contents afterculture in these media for 48 h in the dark at 37°C were measured.
The aseptic L-Trp solution was added to TSB liquid culture medium to a final concentration of 0.5 mg/mL. The B. pyrrocinia JK-SH007 strain was inoculated into the culture medium at 1%, and the IAA contents and absorbances at OD600nm after culture with shaking at 15, 20, 25, 30, 37, and 42°C for 36 h in the dark were measured, 20 μL of B. pyrrocinia JK-SH007 was coated on TSB agar plates (containing 0.5 mg/mL L-Trp) and cultured at 15, 20, 25, 30, 37, and 42°C for 36 h as a control.
The pH value of TSB liquid culture medium was adjusted to 4, 5, 6, 7 and 8, and the aseptic L-Trp solution was then added to these culture media to a final concentration of 0.5 mg/mL. B. pyrrocinia JK-SH007 was inoculated into the culture media at 1%, and the IAA contents and OD600nm values after culture in these media for 36 h in the dark at 37°C were determined. Simultaneously, cultures of 20 μL of B. pyrrocinia JK-SH007 coated on TSB agar plates (containing 0.5 mg/mL L-Trp) with pH values of 4, 5, 6, 7, and 8, at 37°C for 36 h were used as a control.
Based on the whole genome sequencing analysis and annotation results of B. pyrrocinia JK-SH007 (unpublished data), and according to the sequence characteristics of the iaaM and iaaH genes and the recognition site of the restriction endonucleases of the expression vector pET32a, EcoRV, and EcoRI restriction sites (underlined portions) were added to the forward and reverse primers for the target gene (designed with Primer 5.0). The primer sequences were as follows:
iaaM, E-iaaM-F: 5′-CGATATCATGCCGACGCACCGCTC GGCA-3′ and E-iaaM-R: 5′-CGAATTCTCAGTCGCCG TACCGATAGCG-3′; and iaaH, E-iaaH-F: 5′-CGA TATCATGCGCGAGATAGCCGATGC-3′ and E-iaaH -R: 5′-CGAATTCTCAGGCGGCCGTCGCGTGCT-3′.
The primers were synthesized by Nanjing GenScript Biotechnology Co., Ltd. The iaaM and iaaH genes were amplified using the B. pyrrocinia JK-SH007 genome DNA as the template and the high-fidelity enzyme PrimeSTARHS® DNA Polymerase with GC Buffer (Takara, Dalian, China). The PCR system (50 μL) consisted of 25 μL of 2 × PrimeSTAR GC Buffer, 4 μL of dNTP mixture, 5 μL of Primer F/R, 2.5 μL of the DNA template, 0.5 μL of PrimeSTAR® HS DNA Polymerase and 8 μL of ddH2O. PCR amplification was performed using the following conditions: 10 min at 95°C, 40 cycles of 30 s at 95°C, 30 s at 64°C (iaaM)/68°C (iaaH) and 2 min at 72°C, and 72°C for 10 min. The correctness of the PCR product was observed by 1% agarose gel electrophoresis, and the PCR product was purified by gel-excision using the TaKaRa MiniBEST Agarose Gel DNA Extraction Kit Ver. 4.0 (Takara, Dalian, China).
The purified products of the iaaM and iaaH genes and the expression vector pET32a were double-digested with the QuickCut restriction enzymes EcoRV and EcoRI (Takara, Dalian, China) and incubated at 37°C for 2 h. After reaction at 37°C for 2 h, the products were recovered by 1% agarose gel electrophoresis. The digested product of pET32a was ligated with the iaaM and iaaH gene digestion products overnight at 16°C using T4 DNA Ligase (Takara, Dalian, China). The recombinant expression plasmid was transformed into E. coli BL21 (DE3), plated on Luria-Bertani (LB) solid medium containing ampicillin (Amp) antibiotic (100 μg/mL), and grown overnight at 37°C. A positive recombinant was selected for colony PCR verification and double-enzyme digestion verification, and the positive recombinant was then sent to Beijing Genomics Institute (BGI) for sequencing.
Escherichia coli BL21 (DE3) with the recombinant plasmid pET32a-iaaM/iaaH and E. coli BL21 (DE3) with the empty vector pET32a (used as a control) was inoculated into LB liquid medium (containing 100 μg/mL Amp) at 1%. Once the OD600nm value reached 0.6, Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM, and the culture was then induced overnight at 25°C. After centrifugation at 10 000 × g for 10 min, the supernatant was discarded, and the bacteria were washed twice with phosphate-buffered saline (PBS) buffer (10 mM, pH 7.2–7.4). The bacterial pellet was then resuspended, and for ultrasonic treatment, the resuspended bacterial liquid was crushed in an ice bath with a cell crushing apparatus until the optical density no longer increased and the bacterial suspension became clear. The supernatant and precipitate were collected for SDS-PAGE analysis.
Gene expression was induced overnight with 0.3 and 0.5 mM IPTG at 25°C under shaking conditions. The supernatant and precipitate after ultrasonic crushing were collected for SDS-PAGE analysis.
With an IPTG concentration of 0.5 mM, gene expression was induced after overnight incubation at temperatures of 15, 20, and 25°C. The supernatant and precipitate after ultrasonic crushing were collected for SDS-PAGE analysis.
The recombinant protein was subjected to SDS-PAGE electrophoresis, transferred to a PVDF membrane (0.45 μm), washed three times with PBST, blocked with a PBST solution containing 5% skimmed milk powder for 2 h, and incubated with 1:1 000 anti-His antibody (Tiangen Biotech, Beijing, China). The membrane was washed three times with PBST and added to anti-mouse IgG (H&L) AP conjugated (Promega, Madison, WI, United States) secondary antibody at a 1: 4 000 dilution. The membrane was incubated for 2 h at room temperature with gentle shaking and then washed three times with PBST, and color development was then induced by addition of the color development solution Western Blue® Stabilized Substrate for Alkaline phosphatase (Promega, Madison, WI, United States). The reaction was terminated by rinsing the membrane with distilled water immediately after the color development met the requirements.
The supernatant of the tryptophan 2-monooxygenase fusion protein and the inclusion bodies of the indoleacetamide hydrolase fusion protein were purified using a Ni-IDA-Sefinose column (Sang Biotech, Shanghai, China). The purified indoleacetamide hydrolase fusion protein inclusion bodies were placed in dialysis bag (8–14 kD), and then placed in renaturation buffer [0.3 mol/L NaCl, 100 mmol/L NaH2PO4, 2 mmol/L reduced glutathione, 0.2 mmol/L oxidized L- glutathione, 40 mmol/L glycine, and 10% glycerol (v/v), pH 8.0], with decreasing concentrations of urea (6, 4, 2, and 1 mol/L) for 8 h. Finally, the solution was dialyzed in PBS buffer (10 mM, pH 7.2–7.4) without urea for 12 h. The solution in the dialysis bag was removed and centrifuged at 10 000 × g for 30 min (Xia et al., 2013). The purified and renatured proteins were concentrated by Amicon® Ultra-15 centrifugal filtration (30 000 NMWL) and analyzed by SDS-PAGE. The protein concentration was determined with a BCA Protein Assay Kit (ComWin, Beijing, China).
One assay mixture (1 mL) consisted of 1 mg of L-Trp, PBS buffer (10 mM, pH 7.2–7.4), and 0.2 mg of tryptophan 2-monooxygenase and incubated at 37°C for 2 h, and the other assay mixture (1 mL) consisted of 1 mg of IAM (Yuanye, Shanghai, China), PBS buffer (10 mM, pH 7.2–7.4), and 0.2 mg of indoleacetamide hydrolase and incubated at 37°C for 2 h. A mixture of the two above mentioned reactions was sent to Nanjing Webiolotech Biotechnology Co., Ltd., for HPLC-MS/MS analysis.
The experimental data were analyzed by ANOVA, and Tukey’s post test using a significance level of P < 0.05. The values shown represent the standard deviations (SD) of the averages. All statistical analyses were performed using IBM SPSS Statistics 25.0 (IBM Inc., Armonk, NY, United States). Graphs were drawn using GraphPad Prism 6.0 (GraphPad Software, Inc., United States), and three biological replicates were included in each experiment.
Indole-3-acetic acid (Supplementary Figures S1A,C) and its synthetic intermediate, IAM (Supplementary Figures S1B,D), were detected in B. pyrrocinia JK-SH007 fermentation broth by HPLC-MS/MS analysis (a normal mass-to-charge ratio is obtained if the peak time of the sample is almost the same as or within a certain error from the peak time of the standard product, the mass-to-charge is normal), were detected in B. pyrrocinia JK-SH007 fermentation broth by HPLC-MS/MS analysis (Supplementary Figures S1C,D), which indicates that B. pyrrocinia JK-SH007 has the ability to synthesize IAA and that might harbor an IAM-mediated IAA synthesis pathway. In addition, the IAA precursors IPyA (Supplementary Figures S2A,B) IAN (Supplementary Figures S2C,D), and trace amounts of TAM (Supplementary Figures S2E,F) were detected (Table 1).
The cell-free extract of B. pyrrocinia JK-SH007 was incubated with L-Trp. IAA (Supplementary Figures S3A,C), IAM (Supplementary Figures S3B,D), IPyA (Supplementary Figures S4A,B), IAN (Supplementary Figures S4C,D) and TAM (Supplementary Figures S4E,F), in the reaction mixture were detected by HPLC-MS/MS (Table 2). These results showed the existence of enzymes related to IAA synthesis in B. pyrrocinia JK-SH007, that convert L-Trp into indole derivatives such as IAM and IAA. The existence of a tryptophan-dependent IAA synthesis pathway in B. pyrrocinia JK-SH007 was thus confirmed.
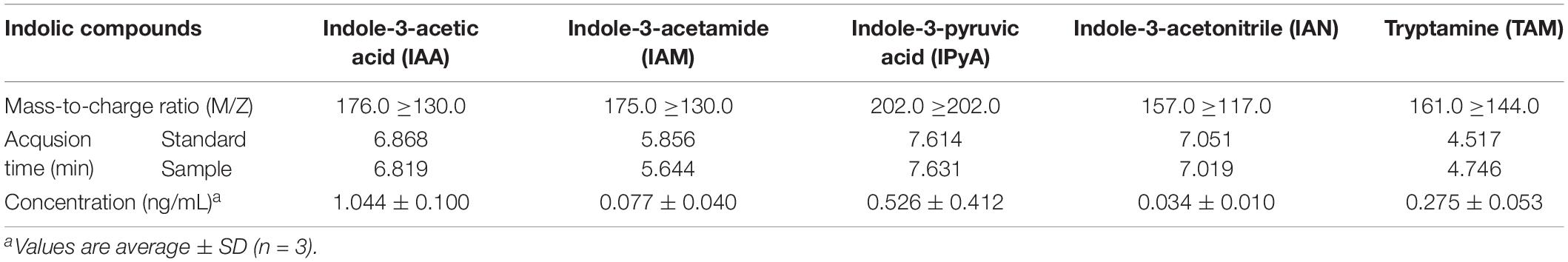
Table 2. HPLC-MS/MS analysis of indolic compounds produced in the cell-free extract of B. pyrrocinia JK-SH007.
The putative IAA biosynthetic pathways in the B. pyrrocinia JK-SH007 genome were identified based on the KEGG of Ttp metabolism and IAA biosynthesis. Mainly involved in the genes of the IAM, IPyA, IAN, and TAM pathways (Table 3 and Supplementary Figure S5). B. pyrrocinia JK-SH007 contains all the genes required for IAM and TAM pathways, but lacks the key genes for IPyA and IAN pathways. Explain that there is no complete IPyA or IAN pathway in B. pyrrocinia JK-SH007.
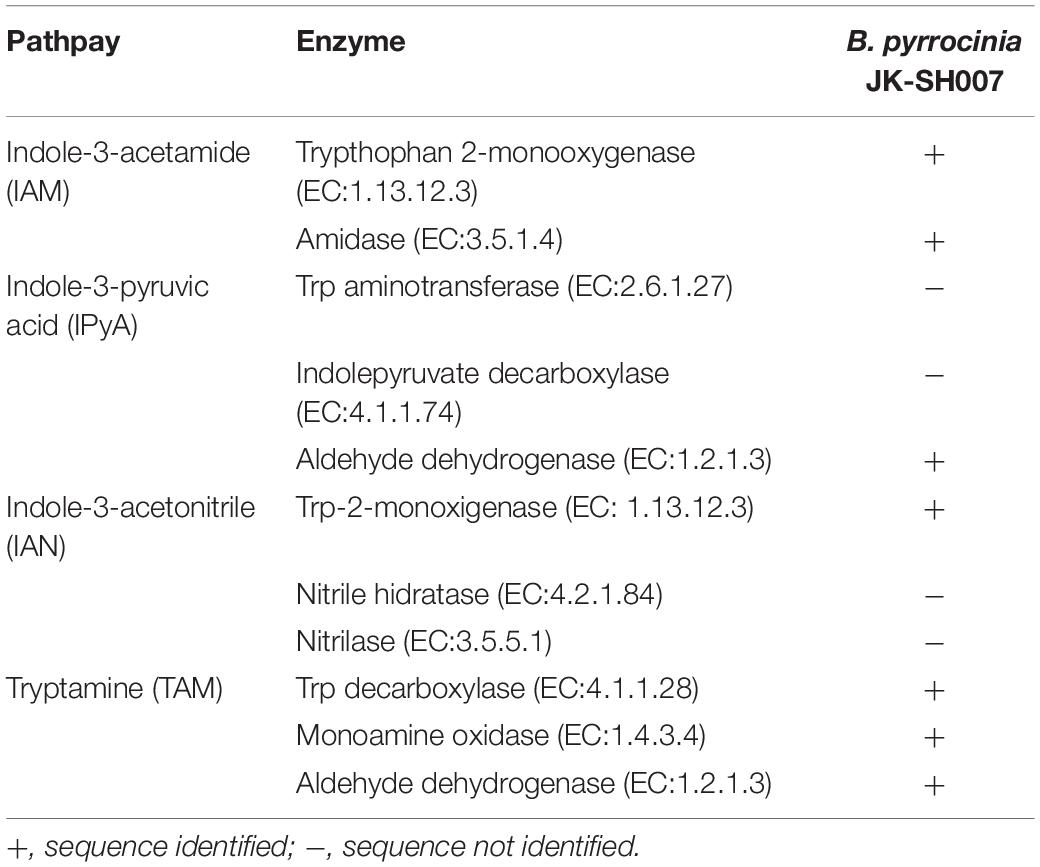
Table 3. Genomic analysis of indole-3-acetic acid (IAA) biosynthesis pathways in B. pyrrocinia JK-SH007.
The relationship between the IAA yield and the growth curve of B. pyrrocinia JK-SH007 was assessed (Figure 1A), and the increases in the number of bacteria at the early stage gradually increased the amount of IAA synthesized by B. pyrrocinia JK-SH007. During the culture of B. pyrrocinia JK-SH007, the amount of IAA synthesized at the stationary phase reached a maximum of 6.969 μg/mL at 36h and then decreased rapidly, which may be due to the degradation of IAA by B. pyrrocinia JK-SH007 itself.
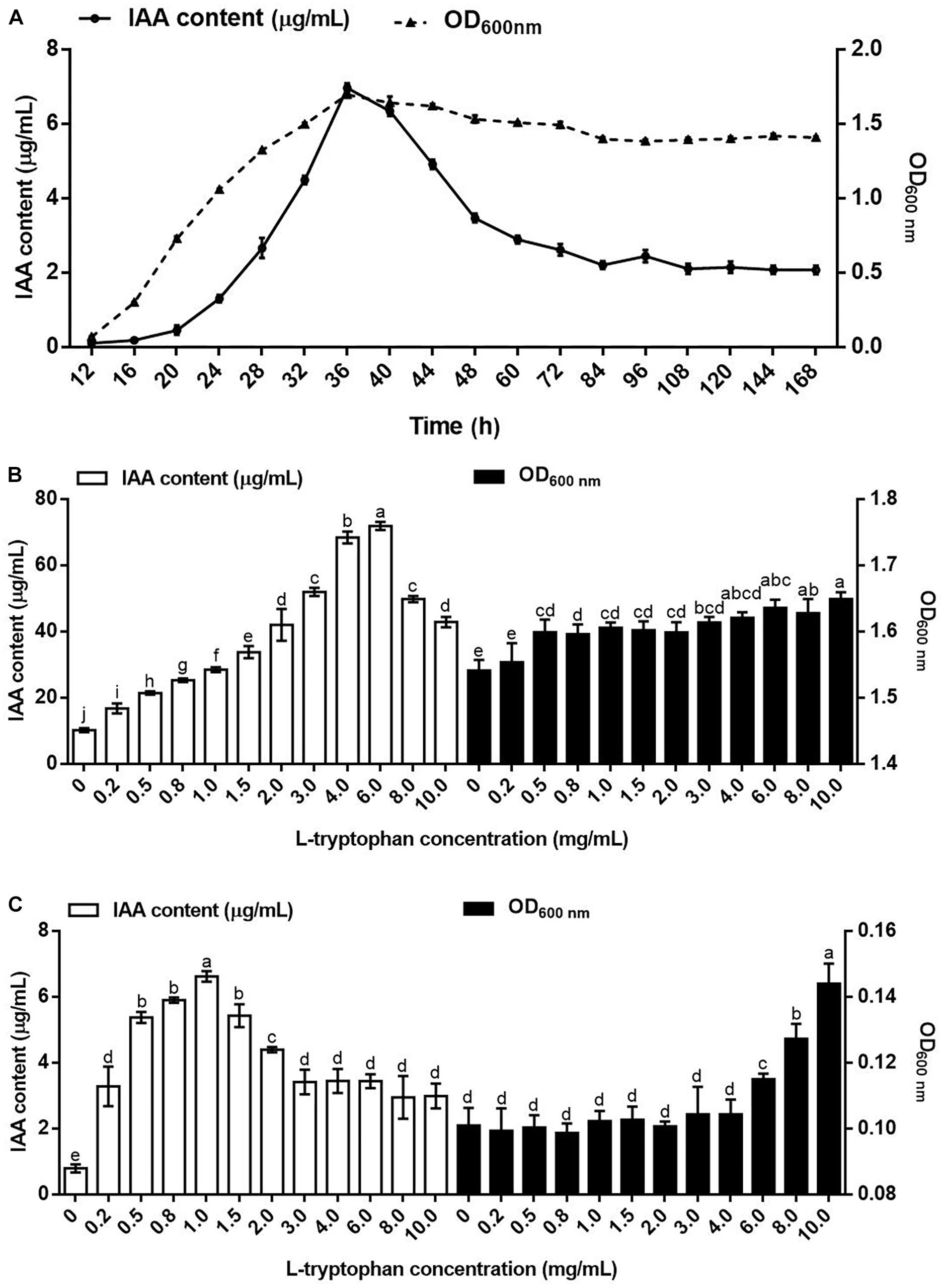
Figure 1. Indole-3-acetic acid in B. pyrrocinia JK-SH007. (A) Relationship between the growth of B. pyrrocinia JK-SH007 and IAA biosynthesis. (B) Effects of L-Trp on the synthesis of IAA by B. pyrrocinia JK-SH007 in TSB liquid medium. (C) Effects of L-Trp on the synthesis of IAA by B. pyrrocinia JK-SH007 in SD liquid medium. The error bars represent the SDs, and different letters indicate significant differences.
Within a specific range, with increasing concentrations of L-Trp, the ability of B. pyrrocinia JK-SH007 in TSB liquid medium to synthesize IAA continuously improved. Although the amount of IAA synthesized by B. pyrrocinia JK-SH007 decreased when the L-Trp concentration reached 8.0 and 10.0 mg/mL, it was still higher than that produced without L-Trp (Figure 1B).
In the absence of L-Trp in the SD liquid medium, B. pyrrocinia JK-SH007 produced very small amounts of IAA (0.795 μg/mL). Increases in the L-Trp concentration within a specific range improved the ability of B. pyrrocinia JK-SH007 to synthesize IAA, until the maximum IAA concentration of 6.621 μg/mL was obtained with an L-Trp concentration of 1.0 mg/mL. Although further increases in the L-Trp concentrations to levels above 1.5 mg/mL decreased the amount of IAA synthesized by B. pyrrocinia JK-SH007, this level of IAA produced remained higher than that produced in the absence of L-Trp (Figure 1C). The difference of IAA yield of B. pyrrocinia JK-SH007 between TSB medium and SD medium may be due to the lack of amino acids in SD medium, which leads to the fact that the growth state of B. pyrrocinia JK-SH007 is much lower than that of TSB medium, so it limits the production of B. pyrrocinia JK-SH007 IAA. However, in both media, L-Trp promoted the growth of B. pyrrocinia JK-SH007 to a certain extent. Overall, the results revealed that L-Trp exerted a very significant (P < 0.05) effect on the synthesis of IAA by B. pyrrocinia JK-SH007, which indicates that L-Trp is an important precursor in the synthesis of IAA by B. pyrrocinia JK-SH007. The results also showed that the amount of IAA synthesized by B. pyrrocinia JK-SH007 was related to its growth state.
Temperature substantially influenced the synthesis of IAA by and the growth of B. pyrrocinia JK-SH007. The results showed that B. pyrrocinia JK-SH007 could grow in the temperature range of 15–42°C, but temperatures that were too high or low were not conducive to the growth of B. pyrrocinia JK-SH007 or to IAA production. B. pyrrocinia JK-SH007 grew optimally at 25 and 30°C, but a temperature of 37°C resulted in an IAA yield of 18.608 μg/mL, which was significantly (P < 0.05) higher than that reached with the other temperatures (Figure 2A). The growth of B. pyrrocinia JK-SH007 on TSB agar plates was the same as that on TSB liquid medium.
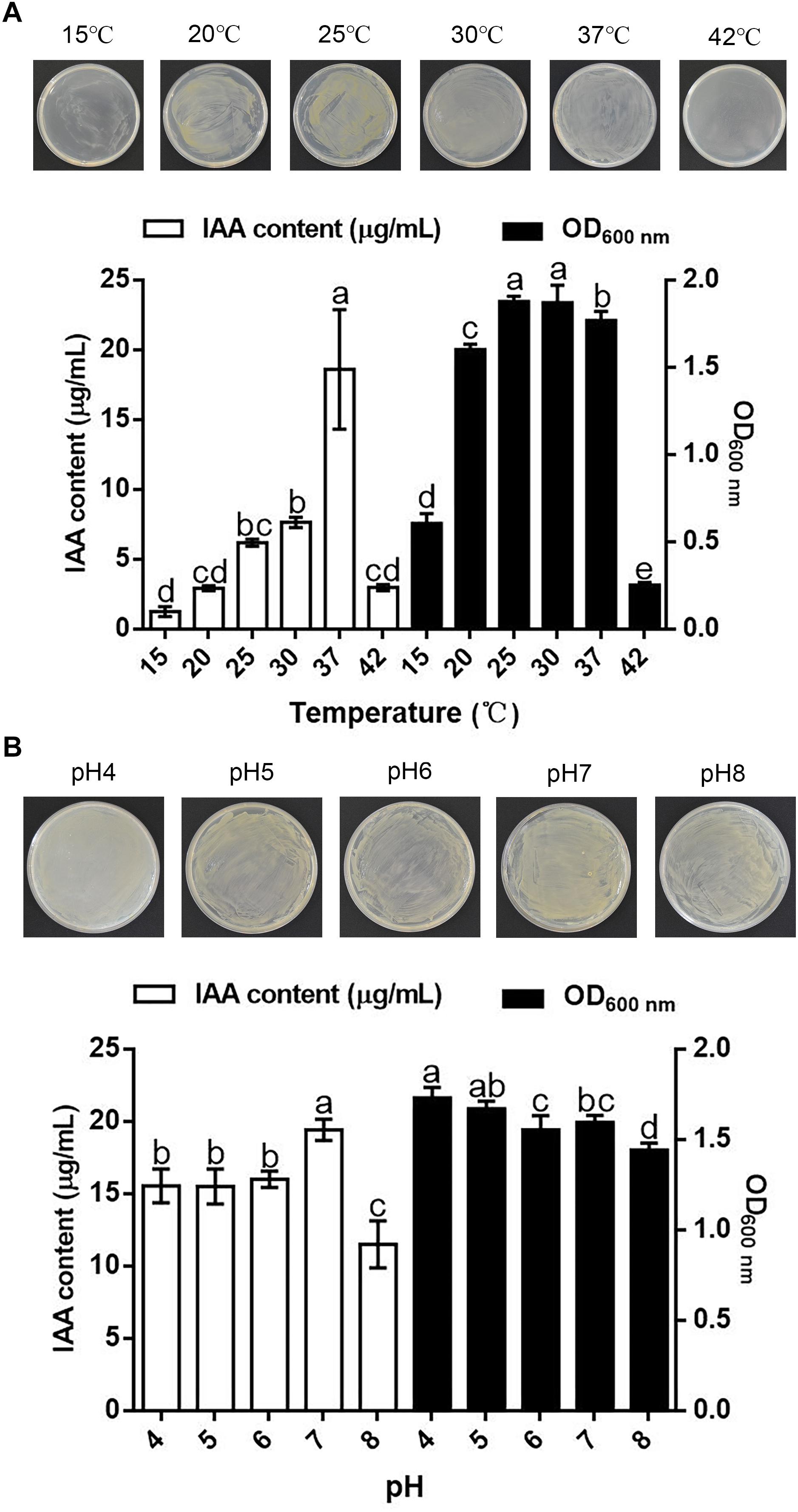
Figure 2. Effect of temperature and pH on the synthesis of IAA by B. pyrrocinia JK-SH007. (A) Effect of temperature on the synthesis of IAA by B. pyrrocinia JK-SH007. (B) Effect of pH on the synthesis of IAA by B. pyrrocinia JK-SH007. The error bars represent the SDs, and different letters indicate significant differences.
The results showed that pH exreted a certain effect on the synthesis of IAA by and the growth of B. pyrrocinia JK-SH007. The strain was able to grow over a broad pH range of 4–8, and the growth of B. pyrrocinia JK-SH007 on TSB agar plates was the same as that on TSB liquid medium. The highest IAA production yield was 19.432 μg/mL was reached with a pH value of 7, whereas a 40.77% lower yieldwas obtained at pH 8, and the fastest growth of B. pyrrocinia JK-SH007 was observed at pH 4 (Figure 2B).
The iaaM (GenBank accession MK238350) and iaaH genes (GenBank accession MK238351) were obtained by using B. pyrrocinia JK-SH007 genomic DNA as the template for the design of a primer with a restriction enzyme site (Supplementary Figures S6A,B). The PCR product obtained after restriction enzyme digestion was ligated with the digested expression vector pET32a and transformed into E. coli BL21 (DE3). After colony PCR verification (Supplementary Figures S6C,D), the recombinant plasmid was extracted and verified by double-enzyme digestion (Supplementary Figures S6E,F). The positive recombinant plasmid was also verified by sequencing. The correct pET32a-iaaM/iaaH recombinant plasmid (Supplementary Figures S6G,H) was therefore used in the subsequent experiments.
The expression of the tryptophan 2-monooxygenase and indoleacetamide hydrolase fusion proteins was analyzed by 6% SDS-PAGE. As shown in Figure 3A, bands consistent with the tryptophan 2-monooxygenase fusion protein (approximately 80 kD) can be observed in the supernatant and precipitate, whereas bands consistent with the indoleacetamide hydrolase fusion protein (approximately 60 kD) can be observed in the precipitate. The results indicated that the tryptophan 2-monooxygenase and indoleacetamide hydrolase fusion proteins were both expressed in E. coli BL21 (DE3), and the tryptophan 2-monooxygenase fusion protein was expressed in the supernatant and precipitate, mostly in the form of inclusion bodies, whereas the indoleacetamide hydrolase fusion protein was expressed mostly in inclusion bodies, but hardly in the supernatant (Figure 3A).
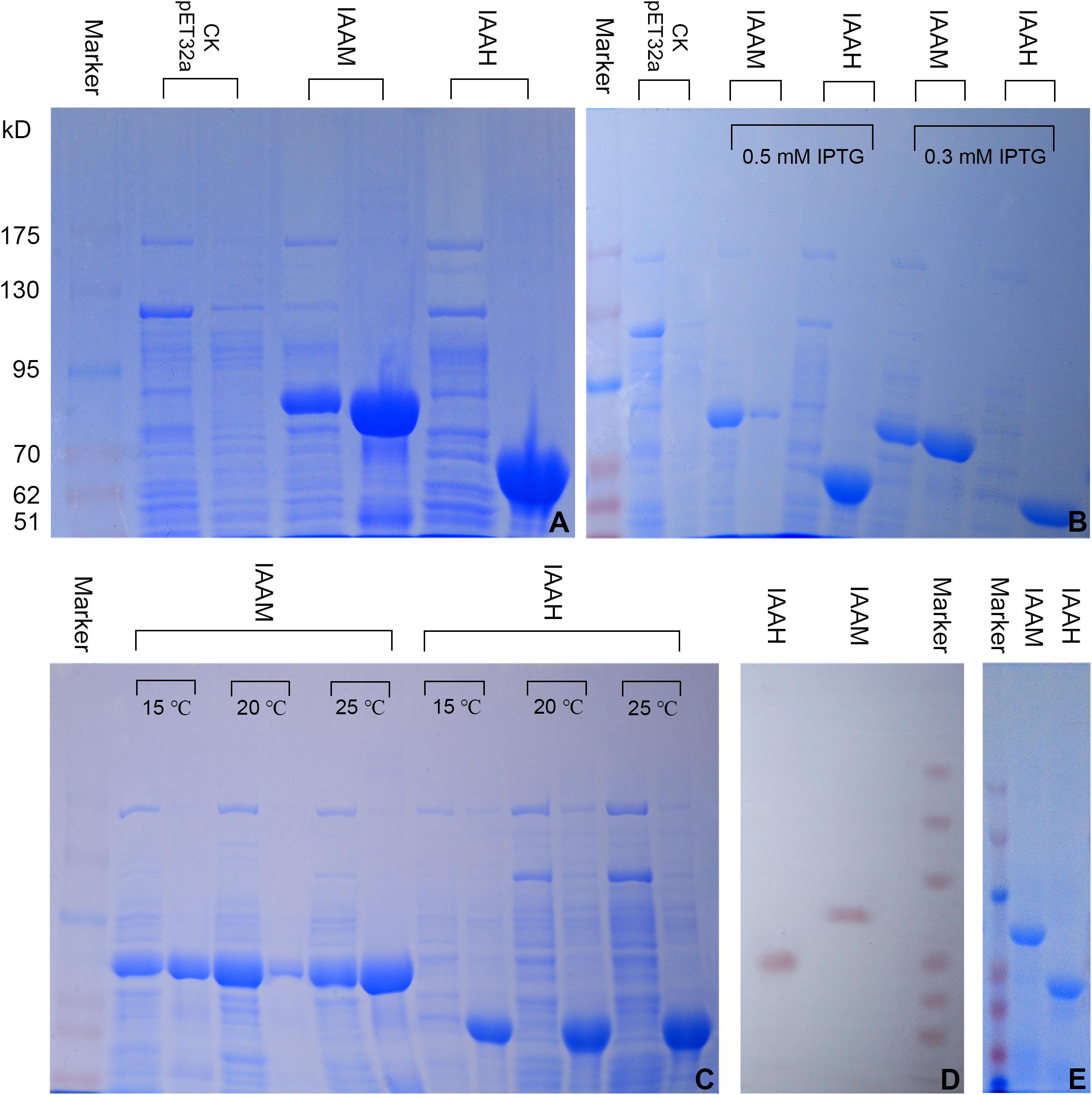
Figure 3. Prokaryotic expression, purification and Western blot identification of the tryptophan 2-monooxygenase (IAAM) and indoleacetamide hydrolase (IAAH) fusion proteins. (A) Expression of tryptophan 2-monooxygenase and indoleacetamide hydrolase (Opti-Protein Marker, the sizes from top to bottom are as follows: 175, 130, 95, 70, 62, and 51 kD; for each treatment, the left lane is the supernatant and the right is the precipitate; this information is also relevant to later figures). (B) Effect of the IPTG concentration on the expression of tryptophan 2-monooxygenase and indoleacetamide hydrolase. (C) Effect of the temperature on the expression of the tryptophan 2-monooxygenase and indoleacetamide hydrolase fusion proteins. (D) Western blot analysis of tryptophan 2-monooxygenase and indoleacetamide hydrolase. (E) Purified tryptophan 2-monooxygenase and indoleacetamide hydrolase.
The expression of the tryptophan 2-monooxygenase and indoleacetamide hydrolase fusion proteins was induced by IPTG at concentrations of 0.3 and 0.5 mM (Figure 3B). The expression of the tryptophan 2-monooxygenase fusion protein at an IPTG concentration of 0.5 mM was significantly higher than that obtained with an IPTG concentration of 0.3 mM. Although the expression of the indoleacetamide hydrolase fusion protein pbtained with an IPTG concentration of 0.5 mM was higher than that at an IPTG concentration of 0.3 mM, it was expressed solely in inclusion bodies.
The induced expression of the tryptophan 2-monooxygenase and indoleacetamide hydrolase fusion proteins at temperatures of 15, 20, and 25°C is shown in Figure 3C. The expression level of the tryptophan 2-monooxygenase fusion protein in the supernatant obtained with a temperature of 20°C than at 15 and 25°C. However, the indoleacetamide hydrolase fusion protein was expressed in large quantities at 15, 20, and 25°C but was present only in inclusion bodies. Therefore, expression of the tryptophan 2-monooxygenase and indoleacetamide hydrolase fusion proteins was induced by IPTG at a concentration of 0.5 mM and a temperature of 20°C to facilitate purification and subsequent experiments.
A Western blot analysis with His monoclonal antibody showed that the tryptophan 2-monooxygenase and indoleacetamide hydrolase (Figure 3D) fusion proteins induced by IPTG formed distinct protein imprinting bands at 80 kD and 60 kD, respectively, which confirmed the expression of the fusion proteins.
An IPTG concentration of 0.5 mM and a temperature of 20°C resulted in high expression of the tryptophan 2-monooxygenase and indoleacetamide hydrolase fusion proteins. The supernatant of the tryptophan 2-monooxygenase fusion protein and the inclusion bodies of the indoleacetamide hydrolase fusion protein were collected, and the indoleacetamide hydrolase fusion protein inclusion bodies were dissolved in a buffer containing 8 M urea. The tryptophan 2-monooxygenase and indoleacetamide hydrolase fusion proteins were purified on a Ni-IDA-Sefinose column. The purified proteins were analyzed by 6% SDS-PAGE. As shown in Figure 3E, single bands appeared in the two lanes, which indicated the successful purification of the tryptophan 2-monooxygenase and indoleacetamide hydrolase (Figure 3E) fusion proteins. The purified indoleacetamide hydrolase fusion protein inclusion bodies were renatured by dialysis refolding. The tryptophan 2-monooxygenase and indoleacetamide hydrolase fusion proteins were concentrated by ultrafiltration, and the concentration of each protein was determined using the BCA method. The concentration of the tryptophan 2-monooxygenase fusion protein was 635.616 μg/mL, and the concentration of the indoleacetamide hydrolase fusion protein was 772.325 μg/mL.
HPLC-MS/MS analysis (Table 4) showed that 2.015 ng/mL IAM (Supplementary Figure S7A) was generated in the product mixture obtained from the tryptophan 2-monooxygenase and L-Trp, and 25.645 ng/mL IAA (Supplementary Figure S7B) was generated from the reaction of indoleacetamide hydrolase and IAM. These findings indicate that the prokaryotic-expressed purified tryptophan 2-monooxygenase and indoleacetamide hydrolase enzymes were active and that the IAM-mediated IAA synthesis pathway plays a role in B. pyrrocinia JK-SH007.
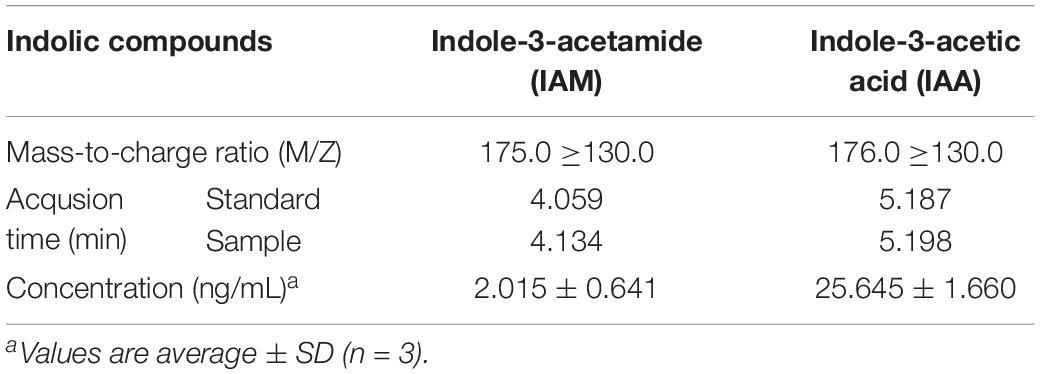
Table 4. HPLC-MS/MS analysis of indole-3-acetamide (IAM) and indole-3-acetic acid (IAA) in protein mixture.
The HPLC-MS/MS evaluation of the B. pyrrocinia JK-SH007 fermentation broth revealed that B. pyrrocinia JK-SH007 produces IAA, IAM, IPyA, IAN, and TAM, and in the experiment assessing the production of IAA by the B. pyrrocinia JK-SH007 cell-free extract, the total enzyme content, including the IAA biosynthetic enzymes, was obtained by breaking the bacterial cells through ultrasonic treatment, and the obtained cell-free extract reacted with the IAA synthesis precursor L-Trp to produce IAM, IPyA, IAN, TAM, and IAA (Shokri and Emtiazi, 2010), which further indicated that the tryptophan-dependent pathway plays an important role in the synthesis of IAA by B. pyrrocinia JK-SH007. However, the whole genome analysis and annotation of B. pyrrocinia JK-SH007 showed that it did not have the key genes of IPyA and IAN pathways, and contained the integrity of IAM and TAM pathways, indicating that B. pyrrocinia JK-SH007 might synthesize IAA through complete IAM and TAM pathways, rather than IPyA and IAN pathways.
The relationship between IAA synthesis by and the growth of B. pyrrocinia JK-SH007 was determined. The results showed that the amount of IAA synthesized by B. pyrrocinia JK-SH007 increased with increases in the number of B. pyrrocinia JK-SH007 bacteria at the early growth stage, potentially because IAA is a secondary metabolite. The highest amount of synthesized IAA was achieved once B. pyrrocinia JK-SH007 reached the stationary stage, which was also consistent with the results reported by Blinkov et al. (2014). The amount of IAA produced by B. pyrrocinia JK-SH007 decreased gradually after 36 h of cultivation, which might be due to the degradation of IAA by B. pyrrocinia JK-SH007 itself. Some studies have showed that some bacterial strains such as Paraburkholderia phytofirmans, Burkholderia phytofirmans use IAA as a sole carbon and energy source, and the degradation of IAA is important in plant growth promotion (Zúñiga et al., 2013; Donoso et al., 2017). A small amount of IAA could be synthesized by B. pyrrocinia JK-SH007 without the addition of exogenous L-Trp, and L-Trp has been detected as the main compound in some plant secretions (Kamilova et al., 2006). Within a specific concentration range, L-Trp significantly enhanced the ability of B. pyrrocinia JK-SH007 to synthesize IAA, and L-Trp is an important physiological precursor for the synthesis of IAA by B. pyrrocinia JK-SH007. This finding also proves that some microbes can produce a small amount of IAA in the absence of L-Trp but can generate a relatively large amount of IAA in the presence of IAA (Khalid et al., 2004; Zahir et al., 2010).
The optimal temperature and pH for IAA synthesis by B. pyrrocinia JK-SH007 were 37°C and 7, respectively. The activities of enzymes associated with the IAA biosynthesis pathway in B. pyrrocinia JK-SH007 were higher under these conditions than under the other tested conditions, which enhanced the production of IAA; however, the optimal growth conditions for B. pyrrocinia JK-SH007 were 25°C or 30°C and a pH of 4, which indicated that there was no direct relationship between the biosynthesis of IAA by B. pyrrocinia JK-SH007 and the optimal growth of its cells. The optimal temperature and pH for IAA production can reflect the ecological sites occupied by bacteria, at various temperatures and under the slightly acidic conditions between roots and the physiological conditions within plants (Duca et al., 2014). The above described experiments also showed that the yield of IAA was affected by the culture conditions, growth conditions, and substrate availability (Chaiharn and Lumyong, 2010).
Through the prokaryotic expression and characterization of tryptophan 2-monooxygenase and indoleacetamide hydrolase in B. pyrrocinia JK-SH007, this study proved that the IAM pathway is active in B. pyrrocinia JK-SH007, and the detection of IAM in the fermentation broth of B. pyrrocinia JK-SH007 confirmed this conclusion. However, other IAA synthesis intermediates, namely, IPyA, IAN and TAM were also detected in the fermentation broth of B. pyrrocinia JK-SH007. And the whole genome of B. pyrrocinia JK-SH007 does not have the key genes of IPyA and IAN pathways, but contains the integrity of IAM and TAM pathways, B. pyrrocinia JK-SH007 might also synthesize IAA through the TAM pathway, but further functional verification of these IAA synthesis pathways is needed. In fact, PGPB contain multiple IAA pathways (Valérie et al., 2007).
We demonstrated that the PGPB B. pyrrocinia JK-SH007 can produce the plant auxin IAA through the IAM pathway. This discovery not only extends the understanding of the IAM-mediated IAA synthesis pathway in Burkholderia cepacia but also indicates that the IAM-mediated IAA biosynthesis pathway has no clear boundary between plant pathogenic bacteria and PGPB (Patten and Glick, 2002a, b; Lin and Xu, 2013). The IAM-mediated IAA synthesis pathways have been observed in symbiotic bacteria such as Azospirillum spp., Rhizobium spp., and Bradyrhizobium spp. (Theunis et al., 2004; Malhotra and Srivastava, 2006; Spaepen and Vanderleyden, 2011).
In this study, the ability of B. pyrrocinia JK-SH007 to synthesize the auxin IAA was confirmed by HPLC-MS/MS, and the related IAA synthetic intermediates were also detected, IAA, and its synthetic intermediates were also found in the cell-free extraction of B. pyrrocinia JK-SH007, but the genomic analysis of B. pyrrocinia JK-SH007 indicated that IAA biosynthesis was mainly through the IAM and TAM pathways. The effects of L-Trp, temperature and pH on the production of IAA by B. pyrrocinia JK-SH007 were studied, and the results indicate that these factors might play a role in the interaction between plants and bacteria. Through the protein expression of tryptophan 2-monooxygenase and indoleacetamide hydrolase and the verification of their activities, this study proved that the IAM pathway exists in B. pyrrocinia JK-SH007 and plays an important role in the synthesis of IAA. These findings verify that an IAA biosynthesis pathway based on IAM is found in B. pyrrocinia JK-SH007 and that L-Trp is the precursor of this pathway. Future studies should further investigate the overall IAA biosynthesis pathway in B. pyrrocinia JK-SH007 and the interaction between IAA biosynthesis pathways. Obtaining an in-depth understanding of the growth-promoting mechanism of B. pyrrocinia JK-SH007 is necessary for the application of its growth-promoting function. This study also provides a reference for related research on endophytic bacteria that can synthesize the auxin IAA in plants to promote plant growth without causing host diseases.
The datasets generated for this study can be found in the GenBank accession MK238350 and GenBank accession MK238351.
W-HL, F-FC, J-RY, and C-EW performed the research. W-HL, H-HF, X-QF, and J-YS analyzed the data. All authors wrote the manuscript.
This work was supported by the Jiangsu Province Agricultural Independent Innovation Project [CX(16)1005].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank the Forest Pathology Laboratory of Nanjing Forestry University for their support and assistance, and Nanjing Webiolotech Biotechnology Co., Ltd., for technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02559/full#supplementary-material
Agodi, A., Mahenthiralingam, E., Barchitta, M., Giannino, V., Sciacca, A., and Stefani, S. (2001). Burkholderia cepacia complex infection in Italian patients with cystic fibrosis: prevalence, epidemiology, and genomovar status. J. Clin. Microbiol. 39, 2891–2896. doi: 10.1128/JCM.39.8.2891-2896.2001
Ahmad, I., Khan, M. S., and Ahmad, F. (2005). Indole acetic acid production by the indigenous isolates of Azotobacter and Fluorescent Pseudomonas in the presence and absence of tryptophan. Tur. J. Biol. 29, 29–34.
Benjamins, R., and Scheres, B. (2008). Auxin: the looping star in plant development[J]. Ann. Rev. Plant Biol. 59, 443–465. doi: 10.1146/annurev.arplant.58.032806.103805
Bevivino, A., Dalmastri, C., Tabacchioni, S., and Chiarini, L. (2000). Efficacy of Burkholderia cepacia MCI 7 in disease suppression and growth promotion of maize. Biol. Fertil. Soils 31, 225–231. doi: 10.1007/s003740050649
Blinkov, E. A., Tsavkelova, E. A., and Selitskaya, O. V. (2014). Auxin production by the Klebsiella planticola strain TSKhA-91 and its effect on development of cucumber(Cucumis sativus L.) Seeds. Microbiology 83, 531–538. doi: 10.1134/S0026261714050063
Campana, S., Taccetti, G., Ravenni, N., Favari, F., Cariani, L., Sciacca, A., et al. (2005). Transmission of Burkholderia cepacia complex: evidence for new epidemic clones infecting cystic fibrosis patients in Italy. J. Clin. Microbiol. 43, 5136–5142. doi: 10.1128/JCM.43.10.5136-5142.2005
Chaiharn, M., and Lumyong, S. (2010). Screening and optimization of indole-3-acetic acid production and phosphate solubilization from rhizobacteria aimed at improving plant growth. Curr. Microbiol. 62, 173–181. doi: 10.1007/s00284-010-9674-6
Di, D. W., Zhang, C., Luo, P., An, C. W., and Guo, G. Q. (2016). The biosynthesis of auxin: how many paths truly lead to IAA? Plant Growth Regul. 78, 275–285. doi: 10.1007/s10725-015-0103-5
Donoso, R., Leiva-Novoa, P., Zúñiga, A., Timmermann, T., Recabarren-Gajardo, G., and González, B. (2017). Biochemical and genetic basis of Indole-3-Acetic Acid (Auxin Phytohormone) degradation by the plant-growth-promoting Rhizobacterium Paraburkholderia phytofirmans PsJN. Appl. Environ. Microbiol. 83:e01991-16. doi: 10.1128/AEM.01991-16
Dou, C. Y., Wang, Y., Zhang, M. M., Jiang, J., and Ye, R. (2017). An application of three somatotrophic bacteria on pear in Shanghai. J. Nanjing Forest. Univ. 41, 186–190. doi: 10.3969/j.issn.1000-2006
Duca, D., Lorv, J., Patten, C. L., Rose, D., and Glick, B. R. (2014). Indole-3-acetic acid in plant–microbe interactions. Antonie van Leeuwenhoek 106, 85–125. doi: 10.1007/s10482-013-0095-y
Fan, B., Wang, C., Song, X. F., Ding, X. L., Wu, L. M., Wu, H. J., et al. (2018). Bacillus velezensis FZB42 in 2018: the gram-positive model strain for plant growth promotion and biocontrol. Front. Microbiol. 9:2491. doi: 10.3389/fmicb.2018.02491
Glick, B. R. (2015). “Introduction to plant growth-promoting bacteria,” in Beneficial Plant-Bacterial Interactions, (Cham: Springer International Publishing), doi: 10.1007/978-3-319-13921-0_1
Gudrun, S., Waffenschmidt, S., Weiler, E. W., and Joachim, S. (1984). The T-region of Ti plasmids codes for an enzyme synthesizing indole-3-acetic acid. Eur. J. Biochem. 138, 387–391. doi: 10.1111/j.1432-1033.1984.tb07927.x
Kamilova, F., Kravchenko, L. V., Shaposhnikov, A. I., Azarova, T., Makarova, N., and Lugtenberg, B. (2006). Organic acids, sugars, and L-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol. Plant Microbe Interact. 19, 250–256. doi: 10.1094/MPMI-19-0250
Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., and Tanabe, M. (2012). KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, D109–D114. doi: 10.1093/nar/gkr988
Khalid, A., Arshad, M., and Zahir, Z. A. (2004). Screening plant growth-promoting rhizobacteria for improving growth and yield of wheat. J. Appl. Microbiol. 96, 473–480. doi: 10.1046/j.1365-2672.2003.02161.x
Khamna, S., Yokota, A., Peberdy, J. F., and Lumyong, S. (2010). Indole-3-acetic acid production by Streptomyces Sp. isolated from some thai medicinal plant rhizosphere soils. Tur. J. Biol. 4, 23–32. doi: 10.5053/ejobios.2010.4.0.4
Lin, L., and Xu, X. (2013). Indole-3-acetic acid production by endophytic Streptomyces sp. En-1 isolated from medicinal plants. Curr. Microbiol. 67, 209–217. doi: 10.1007/s00284-013-0348-z
Liu, W. H., Chen, F. F., Ye, J. R., Wang, C. E., Kang, Y., and Fu, H. H. (2019). Identification of indole-3-acetamide IAA synthesis function and its dependent pathway in Burkholderia pyrrocinia JK-SH007. Scientia Silvae Sinicae 55, 121–129.
Mahenthiralingam, E., Baldwin, A., and Dowson, C. G. (2008). Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J. Appl. Microbiol. 104, 1539–1551. doi: 10.1111/j.1365-2672.2007.03706.x
Malhotra, M., and Srivastava, S. (2006). Targeted engineering of Azospirillum brasilense SM with indole acetamide pathway for indoleacetic acid over-expression. Can. J. Microbiol. 52, 1078–1084. doi: 10.1139/W06-071
Malhotra, M., and Srivastava, S. (2009). Stress-responsive indole-3-acetic acid biosynthesis by Azospirillum brasilense SM and its ability to modulate plant growth. Eur. J. Soil Biol. 45, 73–80. doi: 10.1016/j.ejsobi.2008.05.006
Mandira, M., and Srivastava, S. (2007). An ipdC gene knock-out of Azospirillum brasilense strain SM and its implications on indole-3-acetic acid biosynthesis and plant growth promotion. Antonie Van Leeuwenhoek 93, 425–433. doi: 10.1007/s10482-007-9207-x
Mcclerklin, S. A., Goo, L. S., Harper, C. P., Ron, N., Jez, J. M., and Kunkel, B. N. (2018). Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000. PLoS Pathog. 14:e1006811. doi: 10.1371/journal.ppat.1006811
Molina, R., Rivera, D., Mora, V., López, G., Rosas, S., Spaepen, S., et al. (2018). Regulation of IAA biosynthesis in Azospirillum brasilense under environmental stress conditions. Curr. Microbiol. 75, 1408–1418. doi: 10.1007/s00284-018-1537-6
Olanrewaju, O. S., Glick, B. R., and Babalola, O. O. (2017). Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 33:197. doi: 10.1007/s11274-017-2364-9
Ona, O., Smets, I., Gysegom, P., Bernaerts, K., Van Impe, J., Prinsen, E., et al. (2003). The effect of pH on indole-3-acetic acid (IAA) biosynthesis of Azospirillum brasilense Sp7. Symbiosis 35, 199–208. doi: 10.1111/j.1462-2920.2007.01529.x
Ona, O., Van Impe, J., Prinsen, E., and Vanderleyden, J. (2005). Growth and indole-3-acetic acid biosynthesis of Azospirillum brasilense Sp245 is environmentally controlled. FEMS Microbiol. Lett. 246, 125–132. doi: 10.1016/j.femsle.2005.03.048
Patten, C. L., and Glick, B. R. (2002a). Regulation of indoleacetic acid production in Pseudomonas putida GR12-2 by tryptophan and the stationary-phase sigma factor RpoS. Can. J. Microbiol. 48, 635–642. doi: 10.1139/w02-053
Patten, C. L., and Glick, B. R. (2002b). Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 68, 3795–3801. doi: 10.1128/AEM.68.8.3795-3801.2002
Ramakrishna, W., Yadav, R., and Li, K. F. (2019). Plant growth promoting bacteria in agriculture: two sides of a coin. Appl. Soil Ecol. 138, 10–18. doi: 10.1016/j.apsoil.2019.02.019
Ren, J. H., Ban, H. D., Ye, J. R., and Li, H. (2010). Fermentation conditions of antagonistic strain Burkholderia pyrrocinia JK-SH007 and its control effect on poplar canker disease. Chin. J. Biol. Control 26, 300–306. doi: 10.16409/j.cnki.2095-039x.2010.03.005
Ren, J. H., Li, H., Liu, H., Ye, J. R., and Wu, X. Q. (2106). Influence of Burkholderia pyrrocinia JK-SH007 on the microbial population and functional diversity of microbial communities in the rhizosphere soil of poplar. Scientia Silvae Sinicae 52, 126–133. doi: 10.11707/j.1001-7488.201605015
Ren, J. H., Ye, J. R., Liu, H., Xu, X. L., and Wu, X. Q. (2011). Isolation and characterization of a new Burkholderia pyrrocinia strain JK-SH007 as a potential biocontrol agent. World J. Microbiol. Biotechnol. 27, 2203–2215. doi: 10.1007/s11274-011-0686-6
Santoyo, G., Morenohagelsieb, G., Del, C. O. M., and Glick, B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99. doi: 10.1016/j.micres.2015.11.008
Schaller, G. E., Bishopp, A., and Kieber, J. J. (2015). The yin-yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell 27, 44–63. doi: 10.1105/tpc.114.133595
Shokri, D., and Emtiazi, G. (2010). Indole-3-acetic acid (IAA) production in symbiotic and non-symbiotic nitrogen-fixing bacteria and its optimization by Taguchi design. Curr. Microbiol. 61, 217–225. doi: 10.1007/s00284-010-9600-y
Sopheareth, M., Chan, S. R., Naing, K. W., Lee, Y. S., Hyun, H. N., Kim, Y. C., et al. (2013). Biocontrol of late llight (Phytophthora capsici) disease and growth promotion of pepper by Burkholderia cepacia MPC-7. Plant Pathol. J. 29, 67–76. doi: 10.5423/PPJ.OA.07.2012.0114
Spaepen, S., and Vanderleyden, J. (2011). Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 3:704. doi: 10.1101/cshperspect.a001438
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31, 425–448. doi: 10.1111/j.1574-6976.2007.00072.x
Strobel, G., Daisy, B., Castillo, U., and Harper, J. (2004). Natural products from endophytic microorganisms. J. Nat. Product. 67, 257–268. doi: 10.1021/np030397v
Theunis, M., Kobayashi, H., Broughton, W. J., and Prinsen, E. (2004). Flavonoids, NodD1, NodD2, and nod-box NB15 modulate expression of the y4wEFG locus that is required for indole-3-acetic acid synthesis in Rhizobium sp. strain NGR234. Mol. Plant Microbe Interact. 17, 1153–1161. doi: 10.1094/MPMI2004.17.10.1153
Torres, D., Benavidez, I., Donadio, Elias Mongiardini, Rosas, S., Spaepen, S., et al. (2018). New insights into auxin metabolism in, Bradyrhizobium japonicum. Res. Microbiol. 169, 313–323. doi: 10.1016/j.resmic.2018.04.002
Tsavkelova, E. A., Cherdyntseva, T. A., Botina, S. G., and Netrusov, A. I. (2007). Bacteria associated with orchid roots and microbial production of auxin. Microbiol. Res. 162, 69–76. doi: 10.1016/j.micres.2006.07.014
Valérie, G., Antoun, H., and Tweddell, R. J. (2007). Growth stimulation and fruit yield improvement of greenhouse tomato plants by inoculation with Pseudomonas putida or Trichoderma atroviride: possible role of indole acetic acid (IAA). Soil Biol. Biochem. 39, 1968–1977. doi: 10.1016/j.soilbio.2007.02.015
Xia, Q. Y., Gu, W. L., Lu, J., Zhang, X., Lu, X. H., and Wang, Y. G. (2013). The study on renaturation, expression and purification of chitinase from endophytic bacterium KKWB-5 against banana fusarium wilt in Escherichia coli. Chin. Agric. Sci. Bull. 29, 307–312. doi: 10.3969/j.issn.1000-6850.2013.33.054
Zahir, Z. A., Shah, M. K., Naveed, M., and Akhter, M. J. (2010). Substrate-dependent auxin production by Rhizobium phaseoli improves the growth and yield of Vigna radiata L. under salt stress conditions. J. Microbiol. Biotechnol. 20, 1288–1294. doi: 10.4014/jmb.1002.02010
Zhang, W. X., Fan, J. J., Tan, Q. Q., Zhao, M. M., and Cao, F. L. (2017a). Mechanisms underlying the regulation of root formation in Malus hupehensis stem cuttings by using exogenous hormones. J. Plant Growth Regul. 36, 174–185. doi: 10.1007/s00344-016-9628-8
Zhang, W. X., Fan, J. J., Tan, Q. Q., Zhao, M. M., Zhao, T., and Cao, F. L. (2017b). The effects of exogenous hormones on rooting process and the activities of key enzymes of Malus hupehensis stem cuttings. PLoS One 12:e0172320. doi: 10.1371/journal.pone.0172320
Zúñiga, A., Poupin, M. J., Donoso, R., Ledger, T., Guiliani, N., Gutierrez, R. A., et al. (2013). Quorum sensing and indole-3-acetic acid degradation play a role in colonization and plant growth promotion of Arabidopsis thaliana by Burkholderia phytofirmans PsJN. Mol. Plant Microbe Interact. 26, 546–553. doi: 10.1094/MPMI-10-12-0241-R
Keywords: Burkholderia pyrrocinia, indole-3-acetamide, tryptophan 2-monooxygenase, indoleacetamide hydrolase, protein expression
Citation: Liu W-H, Chen F-F, Wang C-E, Fu H-H, Fang X-Q, Ye J-R and Shi J-Y (2019) Indole-3-Acetic Acid in Burkholderia pyrrocinia JK-SH007: Enzymatic Identification of the Indole-3-Acetamide Synthesis Pathway. Front. Microbiol. 10:2559. doi: 10.3389/fmicb.2019.02559
Received: 14 July 2019; Accepted: 23 October 2019;
Published: 05 November 2019.
Edited by:
Biswarup Mukhopadhyay, Virginia Tech, United StatesReviewed by:
M. Oves, King Abdulaziz University, Saudi ArabiaCopyright © 2019 Liu, Chen, Wang, Fu, Fang, Ye and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Ren Ye, anJ5ZUBuamZ1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.