
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 08 November 2019
Sec. Evolutionary and Genomic Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02557
This article is part of the Research Topic Plasmid Transfer: Mechanisms, Ecology, Evolution, and Applications View all 34 articles
Plasmids are key vehicles of horizontal gene transfer and contribute greatly to bacterial genome plasticity. In this work, we studied a group of plasmids from enterobacteria that encode phylogenetically related mobilization functions that populate the previously non-described MOBQ4 relaxase family. These plasmids encode two transfer genes: mobA coding for the MOBQ4 relaxase; and mobC, which is non-essential but enhances the plasmid mobilization frequency. The origin of transfer is located between these two divergently transcribed mob genes. We found that MPFI conjugative plasmids were the most efficient helpers for MOBQ4 conjugative dissemination among clinically relevant enterobacteria. While highly similar in their mobilization module, two sub-groups with unrelated replicons (Rep_3 and ColE2) can be distinguished in this plasmid family. These subgroups can stably coexist (are compatible) and transfer independently, despite origin-of-transfer cross-recognition by their relaxases. Specific discrimination among their highly similar oriT sequences is guaranteed by the preferential cis activity of the MOBQ4 relaxases. Such a strategy would be biologically relevant in a scenario of co-residence of non-divergent elements to favor self-dissemination.
Mobilizable plasmids are small genetic elements transmissible by conjugation with the assistance of a helper conjugative plasmid. They encode a relaxase, and usually a relaxase accessory protein (RAP), which are in charge of the conjugative DNA processing at a specific site of the origin of transfer (oriT) called nic. Mobilizable plasmids lack the transfer genes required for establishing a conjugative bridge (mating pair formation system, MPF) to the recipient cell, as well as the type IV coupling protein (T4CP) that puts in contact relaxosome and MPF and thus depend on conjugative plasmids to be transferred (Garcillán-Barcia and de la Cruz, 2013).
According to their relaxase, transmissible plasmids were phylogenetically classified into MOB families (Francia et al., 2004; Garcillán-Barcia et al., 2009). Currently, nine relaxase MOB classes are defined, and five of them (MOBP, MOBF, MOBQ, MOBH, and MOBC) are prevalent in transmissible plasmids hosted in γ-Proteobacteria. Plasmids gathered in a relaxase MOB family share similar genomic traits. Relaxase MOB classification has thus shown to be a good predictor of the plasmid backbone (Garcillán-Barcia and de la Cruz, 2013; Fernandez-Lopez et al., 2017). Mobilizable plasmids resident in γ-Proteobacteria form phylogenetically related clusters mainly within two relaxase MOB classes: MOBP and MOBQ (Garcillán-Barcia et al., 2009). Relevant examples are ColE1-like plasmids, grouped in family MOBP5; IncQ1 plasmids, such as RSF1010/R1162, gathered in MOBQ11; and IncQ2 plasmids, such as pTC-F14, in family MOBP14 (Garcillán-Barcia et al., 2009; Garcillán-Barcia and de la Cruz, 2013). An additional clade of small plasmids encoding MOBQ relaxases, previously classified as MOBQu, and here redefined as MOBQ4, was observed in a phylogenetic reconstruction of this relaxase family (Garcillán-Barcia et al., 2009).
A pair of degenerate primers specific for MOBQ4 plasmids was implemented in the Degenerate PCR MOB Typing (DPMT) approach developed by Alvarado et al. (2012) to detect and classify transmissible plasmids. This method revealed the abundance of MOBQ4 plasmids in clinical isolates of enterobacteria (Alvarado et al., 2012; Garcillán-Barcia et al., 2015), previously unnoticed by other plasmid typing methods. Whole-genome sequencing of clinical E. coli isolates also uncovered the presence of this kind of plasmids (Brolund et al., 2013; de Toro et al., 2014; Lanza et al., 2014). Prototype plasmids pIGWZ12 and ColE9-J (ColE2-like) cluster within the MOBQ4 clade. They are stable, theta-replicating, high copy-number, narrow host-range plasmids, whose replication systems have been extensively studied (Yasueda et al., 1989, 1994; Yagura et al., 2006; Zaleski et al., 2006, 2015). Here, we uncovered the diversity of MOBQ4 plasmids, determined the helper conjugative plasmids responsible for their dissemination, and established their behavior in terms of stability and transfer.
MOBQ4 plasmid derivatives were constructed by isothermal assembly of linear DNA fragments from PCR reactions, following the Gibson method (Gibson et al., 2009, 2015). The MOBQ41 backbone (replication and mobilization regions), based on the complete sequence of the pE2022_4 plasmid [GenBank Acc. No. KT693143 (Lanza et al., 2014)], was linked to a kanamycin-resistance gene [coordinates 272 to 1216 of pSEVA211, GenBank Acc. No. JX560326 (Silva-Rocha et al., 2013)] and a cerulean fluorescent protein gene [coordinates 41 to 1091 of pNS2-φVL (Dunlop et al., 2008)], generating plasmid pRC1. The MOBQ42 backbone (replication and mobilization regions) was obtained by PCR amplification from the E. coli isolate HUMV 04/979 (Garcillán-Barcia et al., 2015), which contains a ColE9-J-like plasmid (coordinates 5102 to 7577, GenBank Acc. No. NC_011977.1). It was joined to a chloramphenicol resistance gene (coordinates 272–1072 of pSEVA311, GenBank Acc. No. JX560331 (Silva-Rocha et al., 2013)] and mCherry fluorescent protein gene (cfp, coordinates 1092–2117 of pNS2-φVL (Dunlop et al., 2008)], generating plasmid pRC2. MOBQ4 plasmids lacking the mobC ORF (from start to stop codon) were constructed by self-ligation of a single PCR fragment from either pRC1 or pRC2, producing plasmids pRC3 and pRC4, respectively.
Additional plasmids were constructed to delimit the oriT region. A schematic representation of the fragments included in each construction is depicted in Figure 1. Such fragments were individually assembled to coordinates 1–1030 and 1360–3001 of vector pSEVA631 (GenBank Acc. No. JX560348). Plasmids pRC5 and pRC6 contained a fragment including the mobC gene, the 178bp intergenic region between mobC and mobA and the first 400 nucleotides of the mobA gene from pRC1 and pRC2, respectively. Plasmids pRC7 and pRC8 included only the 178bp intergenic fragment (Supplementary Figure S1), located between genes mobA and mobC of pRC1 and pRC2, respectively. Plasmids pRC14 and pRC15 contain the oriT regions of pRC7 and pRC8 but cloned in the inverse orientation. Plasmids pRC11 and pRC9, respectively included portions 1–70 and 71–178 of the intergenic fragment between genes mobA and mobC of pRC1, while the same portions from pRC2 were included in pRC12 and pRC10, respectively. A pSEVA631 fragment containing coordinates 1–1030 and 1360–3001 was self-ligated, generating the non-mobilizable vector pRC13, which was used as a control in the mating experiments.
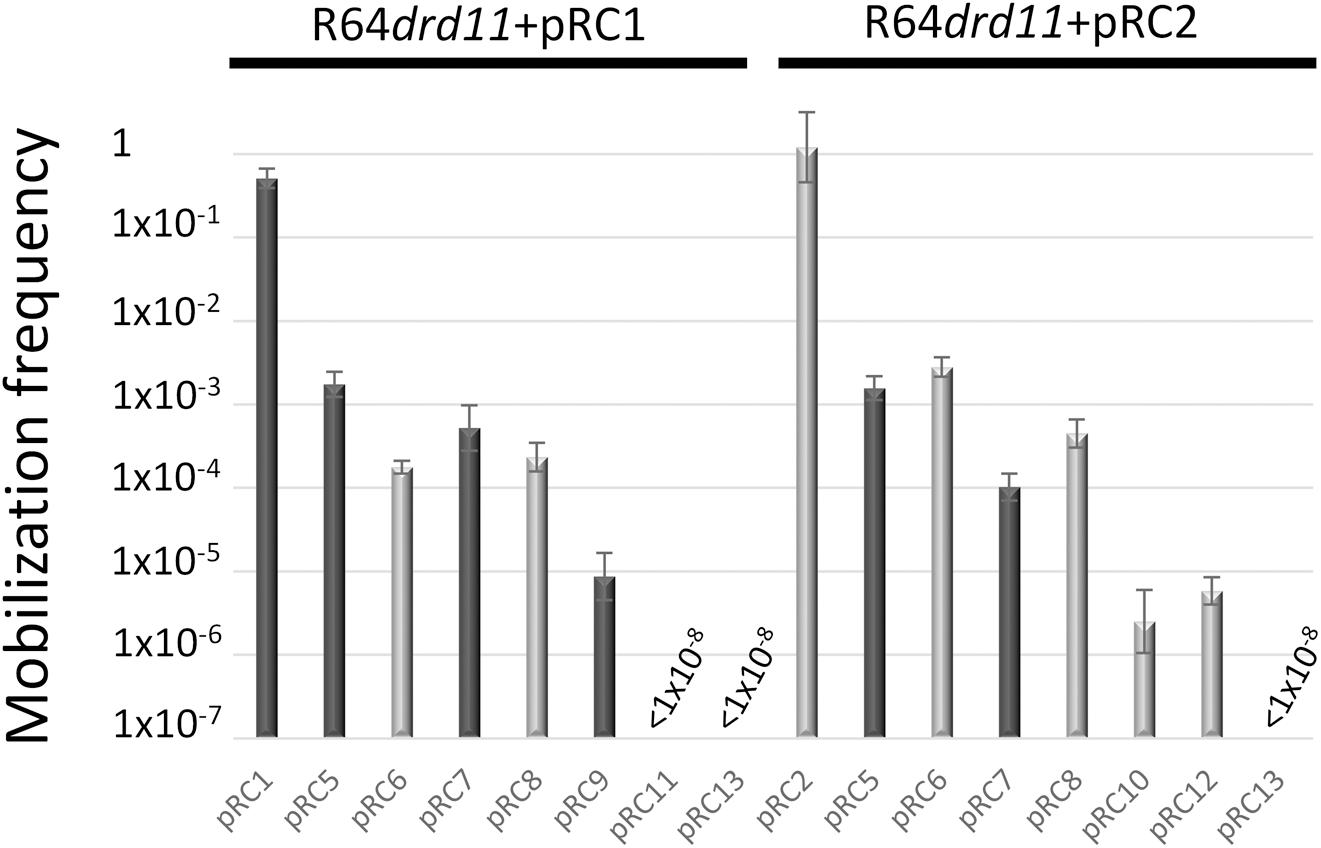
Figure 1. Schematic representation of the MOBQ4 DNA segments included in a series of recombinant plasmids. The mobilization region of MOBQ4 plasmids includes mobC and mobA genes, represented by large, horizontal gray arrows. The extent of the mobilization region included in each construction is represented by a gray bar. The plasmid names for the MOBQ41–based constructions are listed in the left column, while those for MOBQ42-based constructions are in the right column. Plasmids pRC1, pRC2, pRC3, and pRC4 also include the replication module of MOBQ41 or MOBQ42 plasmids.
Plasmids pRC1 and pRC2 were introduced in the recA+ and recA– isogenic strains UB1636 (F– lys his trp rpsL) (Achtman et al., 1971) and UB1637 (F– lys his trp rpsL recA56) (de la Cruz and Grinsted, 1982), either independently to check for their stability or both together to check for their compatibility. Single colonies were inoculated in Lysogeny-Broth (LB) supplemented with kanamycin at 50 μg/ml (for pRC1-containing strains) or chloramphenicol at 25 μg/ml (for pRC2-containing strains) and grown to saturation at 37°C with agitation (150 rpm). A volume of 9.7 μl was transferred from saturated cultures to 10 mL of fresh LB media without antibiotics and grown to saturation in the same conditions. Rounds of transfer and growth were repeated up to 80 generations. The proportion of plasmid-bearing cells in the population was monitored by replica-plating 100 colonies in LB-agar supplemented with the appropriate antibiotics every 10 generations. A larger number of cells was inspected by fluorescence microscopy and, in the case of pRC1-containing cells, also by flow cytometry. Live cells were visualized using a Leica AF6500 microscope at 63x magnification. CFP and mCherry signals were monitored using BP filters (Excitation 434/17 – Emission 479/40 for CFP, Excitation 562/40 – Emission 641/75 for mCherry). Images were obtained using an iXon885 EM CCD Camera (Andor) and up to 1000 cells were analyzed in each case. Fluorescence emission was measured by flow cytometry using a FACS Canto II flow cytometer (Becton Dickinson) equipped with a 488 nm solid state laser for excitation. The cyan fluorescence of 20,000 events was detected using a 525/20 filter.
Conjugative plasmids used in this work are listed in Supplementary Table S1. They were tested as helpers of the MOBQ4 plasmids in surface mating experiments, following the procedure described by del Campo et al. (2012). E. coli strain DH5α (F– endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG purB20 φ80dlacZΔM15 Δ(lacZYA-argF)U169, hsdR17(rK–mK+(), λ–) (Grant et al., 1990) containing different plasmid combinations was used as donor and BW25113 (lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78), BW25993 (lacIq hsdR514 ΔaraBADAH33 ΔrhaBADLD78) (Datsenko and Wanner, 2000) as recipient. Donor and recipient strains were mixed in a 1:1 ratio, deposited onto an LB-agar surface and incubated for 1 h at 37°C (except when drR27 was used as a helper, in which case matings were carried out at 25°C). Then, the mixture was resuspended in LB and plated in the presence of appropriate antibiotics. Conjugation frequencies were expressed as the number of transconjugants per donor cell.
The 300 N-terminal residues of the MobA relaxase of plasmid ColE9-J were used as a query in a BLASTP search (Altschul et al., 1997) (e-value: 1xE-3). The homologous sequences were aligned using MUSCLE (Edgar, 2004). TrimAl v1.4 was used to calculate the average identity between sequences in the alignment (Capella-Gutiérrez et al., 2009). ProtTest 3 was used to estimate the best model of protein evolution for our set (Guindon and Gascuel, 2003; Darriba et al., 2011). RAxML version 7.2.7 (Stamatakis, 2006) was used for phylogenetic reconstruction. Using the JTTGAMMA model 10 maximum likelihood (ML) searches trees were inferred and support values were assigned to each node of the best tree from 1000 bootstrap searches. Relaxase of the pXF5847 plasmid (GenBank Acc. no. YP_009076807.1) was used as outgroup.
Phyre2 was used to predict the 3D structure of the MobA relaxase domains of plasmids pE2022_4 and ColE9-J (Kelley et al., 2015), which were visualized using PyMOL (Schrödinger, 2015).
MOBQ is a broad relaxase class that encompasses several families, each of which includes related plasmid backbones: MOBQ1 comprises relaxases of mobilizable broad host-range IncQ1-like plasmids; MOBQ2, conjugative relaxases of pTi and many rhizobial plasmids; MOBQ3, conjugative broad host-range plasmids resident in gram-positive, such as pIP501 (Garcillán-Barcia et al., 2009). In this previous study, many MOBQ plasmids were not ascribed to a specific subclassification due to either low resolution of the clades or lack of information on the plasmid members. Here, we focused on one of these poorly defined clades, now named MOBQ4, prompted by the fact that these relaxases have been recurrently detected in enterobacterial clinical isolates (Alvarado et al., 2012; Brolund et al., 2013; de Toro et al., 2014; Lanza et al., 2014; Garcillán-Barcia et al., 2015).
The phylogenetic reconstruction, based on the first N-terminal 300 residues of MOBQ4 relaxases produced two clusters, MOBQ41 and MOBQ42 (Figure 2A and Supplementary Table S2). This relaxase domain contains the three relaxase motifs (Figure 2B) and share 84% average amino acid identity (97 and 90% for individual MOBQ41 and MOBQ42 groups, respectively). The 3D structure prediction of the relaxase domain of MOBQ41 and MOBQ42 plasmids rendered MOBQ relaxases NES [plasmid pLW1043, PDB Acc. No. 4HT4 (Edwards et al., 2013)] and MobA [plasmid R1162/RSF1010, PDB Acc. No. 2NS6, (Monzingo et al., 2007)] as best hits (100% confidence). The superimposed structures pointed to MOBQ4 amino acids Y25 (motif I), E87 and E89 (motif II), and H125, H133 and H135 (motif III) as homologs of the MobA_R1162 catalytic residues Y25, E74 and E76, and H112, H120 and H122, respectively (Figure 2C). Contrary to the high conservation of the N-terminal domain among members of both MOBQ4 subgroups, the amino acid identity of the C-terminal part of the MOBQ4 relaxases dropped to 35%. This C-terminal domain exhibited low homology to SogL primases of IncI1 plasmids.
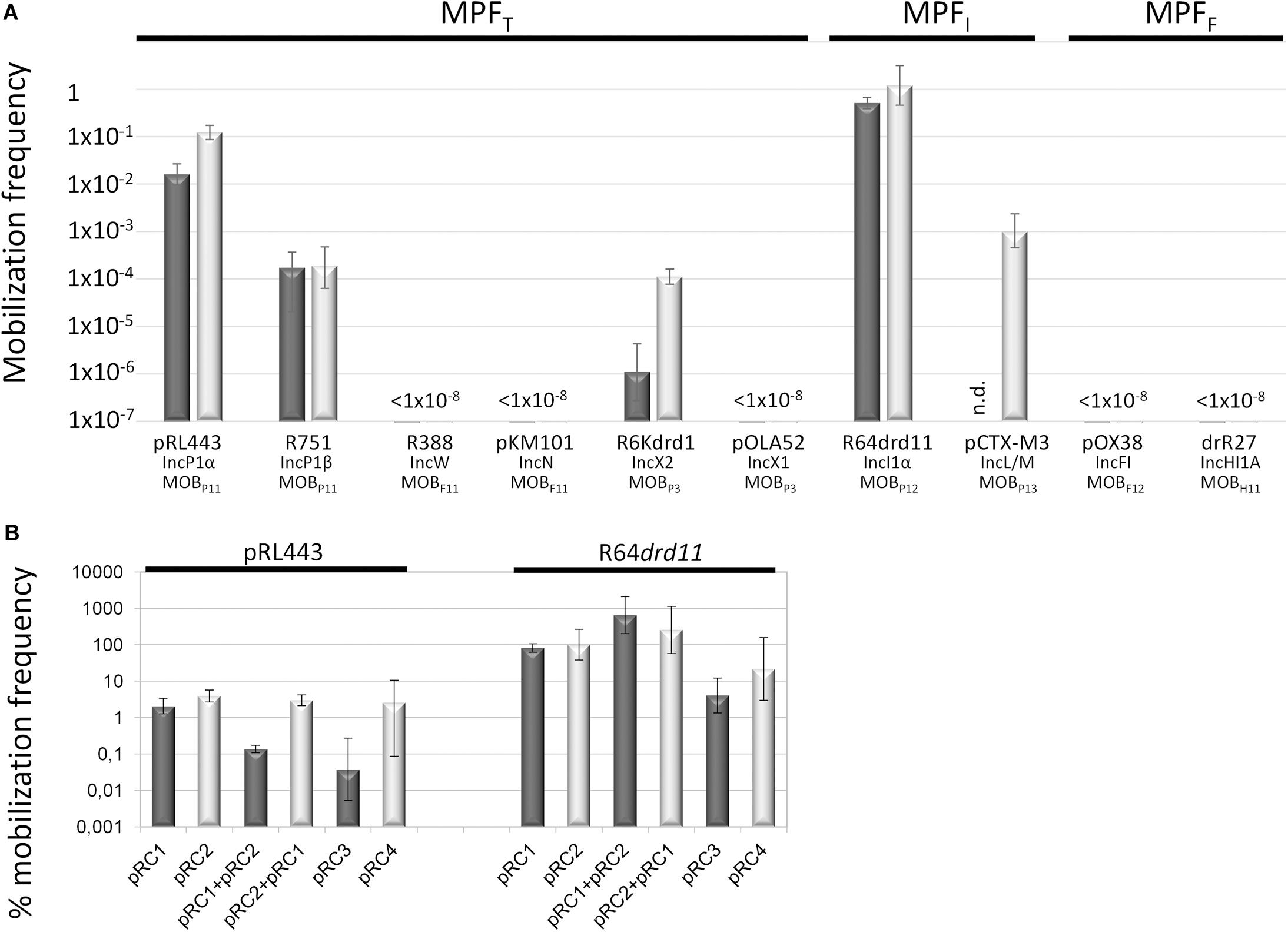
Figure 2. The MOBQ4 relaxase family. (A) Maximum-likelihood phylogenetic reconstruction of the N-terminal domain of MOBQ4 relaxases is shown. MobA relaxase of plasmid pXF5843 was used as outgroup. Bootstrap values of relevant nodes are indicated. Families Q41 and Q42 are shadowed in dark and light gray, respectively. A prototype backbone of each MOBQ4 group is represented to the right of the corresponding clade. Genes of the mobilization module are represented in the same gray color pattern. The elements of the replication module are dotted (for MOBQ41) or striped (for MOBQ42). Genes of the colicin operon and the hypothetical proteins are depicted in a white background. (B) Multiple alignment of the three conserved MOBQ relaxase motifs (Garcillán-Barcia et al., 2009). (1) MOBQ41 relaxases, with the exception of pMG828-2; (2) pMG828-2; (3) MOBQ42 relaxases. Putative catalytic residues are indicated by black triangles over the amino acid sequences. (C) Protein 3D-structure superposition of MOBQ41, MOBQ42 and MOBQ11 relaxases. In gray, the minimal relaxase domain of MobA of plasmid R1162/RSF1010 used as a model (PDB 2NS6); in blue and magenta the predicted structures of the homologous domains of the MOBQ41 (MobA of pE2022_4) and MOBQ42 (MobA of ColE9-J) relaxases, respectively. Key residues of MOBQ4 relaxases are highlighted as sticks.
Each MOBQ4 subclade groups highly related backbones (Figure 2A). MOBQ41 are cryptic, small-size plasmids (Supplementary Table S2). Their backbone contains only four genes encoding a replication initiation protein (Rep), a relaxase (MobA), a putative relaxase accessory protein (MobC) and a hypothetical protein. The genes for the last two are generally not annotated. Besides the above-mentioned replication and mobilization genes, MOBQ42 plasmids also contain a colicin operon, including colicin, immunity and lysis genes, following the synteny of Group A nuclease colicins (Cascales et al., 2007). Plasmids ColE9-J and pO111_4 contain a second, partial colicin operon.
The MOBQ4 subdivision in two relaxase groups matches with the presence of two different replicons (Supplementary Table S2) and this family thus encompasses at least two plasmid species as defined by Fernandez-Lopez et al. (2017). MOBQ41 plasmids encode a replication initiation protein that belongs to the Rep_3 superfamily [PF01051 in the Pfam classification (Finn et al., 2016)], with no defined group in the PlasmidFinder classification (Carattoli et al., 2014). MOBQ42 plasmids encode ColE2-like initiators (Pfam PF03090 + PF08708), classified as Col156 by PlasmidFinder. Plasmids pIGWZ12 and ColE9-J exemplify each cluster. They are stable, theta-replicating, high copy number plasmids (15 and 10 copies per chromosome molecule, respectively (Takechi et al., 1994; Zaleski et al., 2012). The origin of replication of plasmid pIGWZ12 was located upstream the rep gene. It contains iterons, an A+T rich region and four DnaA boxes (Zaleski et al., 2006, 2015). The iterons were found to be the incompatibility determinants (Zaleski et al., 2015). ColE2-like plasmids, such as ColE9-J, form a group of closely related elements that share an identical priming mechanism, mediated by the plasmid-encoded Rep protein (Horii and Itoh, 1988; Itoh and Horii, 1989; Yasueda et al., 1989; Hiraga et al., 1994). The origin of replication consists of 32 bp located downstream of the rep gene, containing two directly repeated sequences (Kido et al., 1991; Nomura et al., 1991; Yagura and Itoh, 2006; Yagura et al., 2006). In ColE2-like plasmids, the rep gene expression is post-transcriptionally controlled by a plasmid-encoded RNA (RNAI), which binds the untranslated 5′ region of the rep mRNA, preventing its translation (Sugiyama and Itoh, 1993; Takechi et al., 1994; Yasueda et al., 1994). MOBQ42 plasmids contain a cer-like site (Hiraga et al., 1994), an indication that they use a host site-specific recombination system for resolving multimers to monomers as ColE1-like plasmids do (Summers and Sherratt, 1984, 1988; Summers, 1998).
All completely sequenced MOBQ4 plasmids come from hosts of the Enterobacteriaceae family (Supplementary Table S2). They were isolated from different backgrounds: Salmonella enterica isolated from pork meat (pSD4.0) (Bleicher et al., 2013), pork feces (p4_TW-Stm6) (Dyall-Smith et al., 2017) and human systemic infection (pYU39_5.1) (Calva et al., 2015), multidrug-resistant environmental E. coli (pSMS35_4) (Fricke et al., 2008), commensal E. coli (pSE11-6) (Oshima et al., 2008), enterohemorrhagic E. coli strains of the O26 and O111 serogroups (pO26-S4 and pO111_4) (Ogura et al., 2009; Fratamico et al., 2011), extended-spectrum beta-lactamase producing E. coli clinical isolates (pE2022_4, pFV9873_1, pEC147-3 and pEC08-6) (Brolund et al., 2013; Lanza et al., 2014), E. coli isolated from human urinary tract (pVR50F) (Beatson et al., 2015) and bloodstream infections (pSF-468-4) (Stephens et al., 2015), as well as porcine extraintestinal pathogenic E. coli strain (p2PCN033) (Liu et al., 2015), among others (Supplementary Table S2). None of these plasmids contain antibiotic-resistance genes. There is still no clue on the selective advantage provided by the cryptic MOBQ41 plasmids. In the case of MOBQ42 plasmids, the fact that all carry colicin operons, a priori an advantageous trait for the bacterial host, could explain the abundance of this type of plasmids. For example, the MOBQ42 plasmid pDPT1 was stably acquired by a Vietnamese Shigella sonnei strain in the mid-1990s, and became fixed in the evolving bacterial population (Holt et al., 2013). The colicin E5 produced by pDPT1 was highly bactericidal against non-immune Shigella and E. coli strains. The acquisition of the pDPT1 colicin plasmid, coinciding with the high increase of dysentery produced by this strain, suggests that pDPT1 conferred a beneficial function to its host (Holt et al., 2013).
To study the MOBQ4 plasmids, two derivatives were constructed, pRC1 and pRC2. They included the replication and mobilization modules of the MOBQ41 and MOBQ42 backbones, respectively. Antibiotic-resistance and fluorescent protein genes were also included as reporters. Plasmid stability and compatibility were assayed in recA+ and recA– E. coli strains by propagating the plasmids either alone or in combination during 80 generations. Despite the cargoes loaded in plasmids pRC1 and pRC2, the percentage of plasmid retention in the bacterial population was 100%, suggesting that the MOBQ4 backbone confers a minimized fitness cost to its enterobacterial host (San Millan and MacLean, 2017). Besides stability in E. coli, both MOBQ4 plasmid species also exhibited full compatibility (100% retention of both after 100 generations), as could be expected due to their different replicons (Novick, 1987), and ruling out other plasmid-encoded traits out of the replication module that could interfere with the stable vertical inheritance of each other.
Since mobilizable plasmids do not encode the mating pair formation system neither the T4CP, their transfer relies on auto-transmissible plasmids. We wondered which conjugative plasmids could be responsible for the dissemination of the MOBQ4 plasmids. Not all conjugative plasmids are equally efficient at supplying these functions to a specific mobilizable plasmid (Cabezón et al., 1994, 1997). The contacts established between the relaxosome of the mobilizable plasmid and the T4CP-MPF of the helper plasmid are crucial in the transfer process. ColE1-like MOBP5 plasmids are efficiently mobilized by IncF-MOBF12 (e.g., F) and IncI1-MOBP12 (e.g., R64drd11) plasmids (Cabezón et al., 1997). IncQ1-MOBQ1 plasmids, such as RSF1010, are transferred by IncP1-MOBP11 helper plasmids (e.g., RP4) (Cabezón et al., 1997; Meyer, 2009). pMV158-like plasmids (MOBV1) are mobilized by IncP1-MOBP11 and Inc18-MOBQ3 (e.g., pIP501) plasmids (Lorenzo-Díaz et al., 2014).
We looked for reports providing indirect evidence on MOBQ4 plasmid mobilization through conjugation. In a survey for the presence of transmissible plasmids in a multidrug E. coli collection, MOBQ4 transconjugants were obtained from seven out of the eight MOBQ4 containing clinical isolates (Garcillán-Barcia et al., 2015). In all cases, a MOBP12-MPFI plasmid, presumptively the helper, was also present in both, donor and transconjugant cells. Similarly, the MOBQ41 plasmid pSD4.0 and the IncI1 plasmid pSD107 were found in E. coli transconjugants arisen from a mating with Salmonella enterica (Bleicher et al., 2013).
Three conjugative MPF types (MPFT, MPFF, and MPFI) are prevalent in Enterobacteriaceae (Smillie et al., 2010; Guglielmini et al., 2014), the taxonomic family where MOBQ4 plasmids have been found. In this study, a set of conjugative plasmids representative of these MPF families were tested as helpers for the mobilization of MOBQ4 plasmids (Supplementary Table S1). Not all of them were equally efficient (Figures 3A,B and Supplementary Table S3). R64drd11, the prototype of IncI1α-MOBP12 plasmids, which encodes a MPFI conjugative apparatus, was the most efficient helper. Another MPFI plasmid, pCTX-M3 (IncL/M-MOBP13), was also an efficient helper. Co-residence with MPFI plasmids has been reported for the MOBQ4 plasmids pSE11-6 (Oshima et al., 2008), pSD4.0 (Bleicher et al., 2013), pEC147-4 (Brolund et al., 2013), pO26-S4 (Fratamico et al., 2011), pDPT1 (Holt et al., 2013), and pE2022_4 (Lanza et al., 2014).
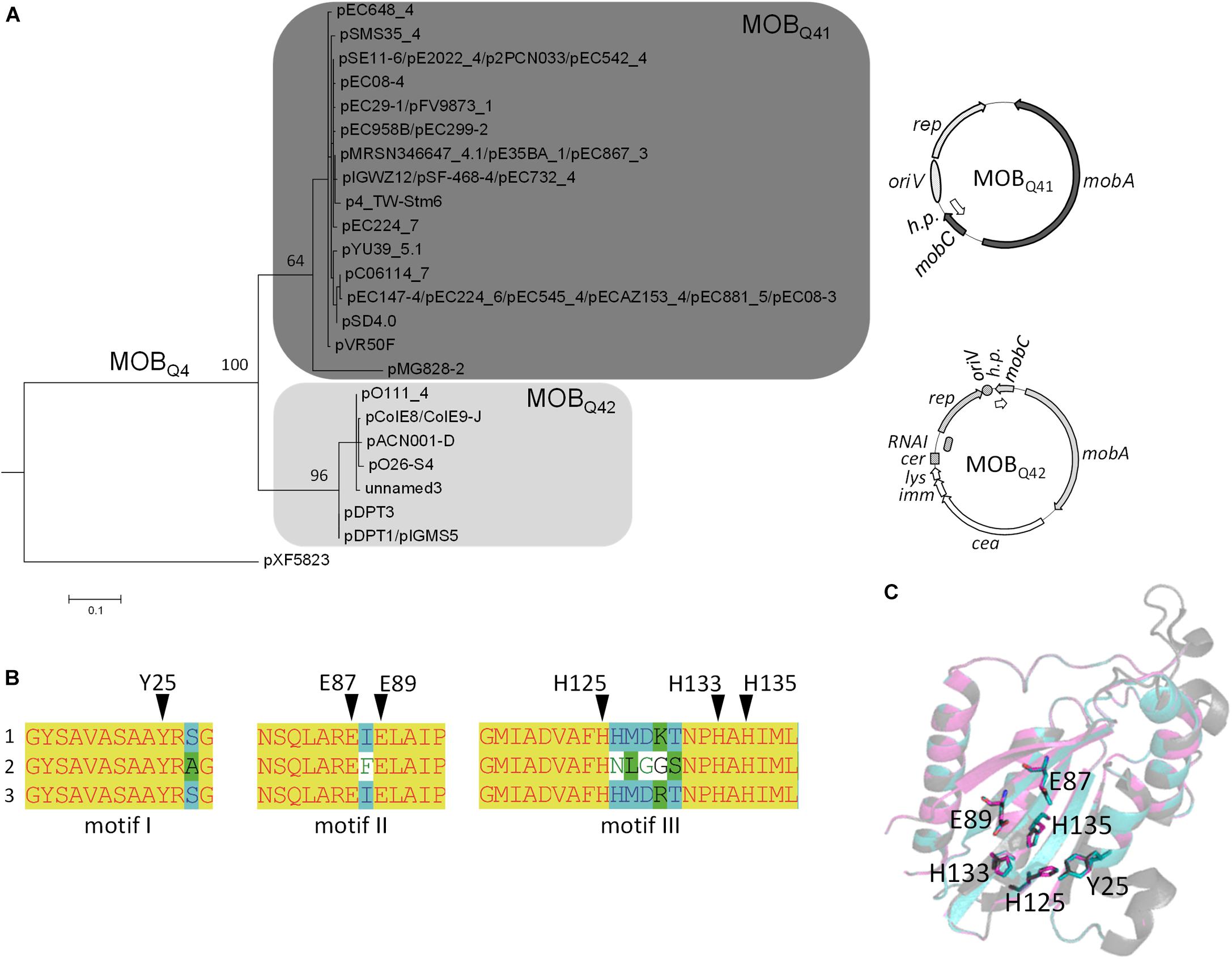
Figure 3. Mobilization frequencies of MOBQ4 plasmids by a series of helper plasmids. The mobilization frequency was calculated as the number of transconjugants containing the MOBQ4 plasmid per donor cell. Figures are the average of at least six independent experiments. (A) Dark- and light-gray bars indicate the mobilization frequencies of pRC1 (MOBQ41) and pRC2 (MOBQ42) plasmids, respectively. Below the bars the helper plasmid used in each case, as well as its corresponding Inc and MOB groups, are indicated. The MPF types of the helper plasmids are indicated in the upper part of the figure. (B) The mobilization efficiencies of the MOBQ4 plasmids are relativized to the helper plasmid transfer rates (100%). The mobilization efficiencies of MOBQ41 and MOBQ42-based constructions are represented by dark- and light-gray bars, respectively. pRC1 + pRC2 indicates the mobilization frequency of pRC1 when coresident with pRC2. pRC2 + pRC1 indicates the mobilization frequency of pRC2 when coresident with pRC1.
On the other hand, MPFF-type plasmids [e.g., IncF-MOBF12 (F) or IncHI1-MOBH11 (R27) plasmids], which show high prevalence in enterobacteria, were not appropriate for MOBQ4 mobilization. MPFT plasmids behaved unevenly as MOBQ4 mobilizers. IncP1-MOBP11 (RP4 and R751) and IncX2-MOBP3 (R6Kdrd1) plasmids rendered MOBQ4 transconjugants, while IncW-MOBF11 (R388), IncN-MOBF11 (pKM101) or IncX1-MOBP3 (pOLA52) did not. Contrary to IncP, IncW and IncN plasmids, most IncF, IncI1, IncH, and IncX plasmids are naturally repressed for conjugation. In this study, we used derepressed variants of IncF (pOX38 and R100-1), IncI1α (R64drd11), IncHI1 (drR27), and IncX2 (R6Kdrd1) plasmids, but not a derepressed IncX1. IncX1 and IncX2 plasmids are highly similar in their conjugation genes. Taking into account that the IncX2 derepressed plasmid R6Kdrd1 was not efficient at mobilizing MOBQ4 plasmids (Figure 3A and Supplementary Table S3), and that the IncX1 plasmid pOLA52 self-transfers at low frequency (around 10–4 per donor) (Sørensen et al., 2003), the lack of mobilization of the MOBQ4 plasmids pRC1 and pRC2 by pOLA52 is not surprising. The widely different mobilization efficiencies displayed by the two IncP1-MOBP11 helpers used is more curious. RP4 and R751 are prototypes of the α and β divisions of the IncP1 backbones, respectively. Despite the high conservation of their transfer genes, the kanamycin-sensitive RP4 derivative, pRL443, was 100–1000 times more efficient than R751 as a MOBQ4 helper. Noticeable differences were also observed for these two conjugative plasmids at transferring IncQ2-MOBP14 mobilizable plasmids pTC-F14 and pTF-FC2 (van Zyl et al., 2003). The common characteristic of the MOBQ4 mobilizers was their belonging to the MOBP relaxase class. This could indicate that the MOBQ4 relaxosomes interact more efficiently with the T4 encoded by these MOBP plasmids.
Bacterial co-infection with multiple plasmids is common in nature (San Millan et al., 2014). Co-residence of compatible plasmids may lead to intracellular interactions that negatively or positively affect plasmid transfer rates (Gama et al., 2017a,b,c; Getino et al., 2017). Among them, plasmid-encoded fertility inhibition systems that block transmission of unrelated plasmids from the same donor cell have been intensively studied (Maindola et al., 2014; Gama et al., 2018; Getino and de la Cruz, 2018). Besides, competition of two relaxosomes for the same T4CP-MPF can result in the preponderance of one them (Cascales et al., 2005), a fact relevant for any mobilizable plasmid. Cohabitation of two or more mobilizable plasmids that use the same mating apparatus could affect each other’s transfer. To test whether the mobilization of the MOBQ41 plasmid was affected by co-residence with a MOBQ42 plasmid and vice versa, pRC1 and pRC2 were introduced conjointly with the helper plasmid (either pRL443 or R64drd11) in the same cell (Figure 3B). Curiously, presence of pRC1 did not produce a significant variation in pRC2 transfer. In turn, pRC2 produced one-log decrease in pRC1 transfer by pRL443. However, this moderate negative effect was not exhibited when using R64drd11 as a helper: on the contrary, pRC2 presence resulted in one-log increase in pRC1 transfer. Testing different combinations of MOBQ41, MOBQ42 and helpers would be necessary to deeper assess the impact of residing together in MOBQ4 horizontal propagation.
Many conjugative and mobilizable plasmids encode RAPs that recognize and bind their cognate oriT sequence probably favoring a single-stranded state around the nic site (de la Cruz et al., 2010). Deletion of RAP genes trwA of R388 (Moncalián et al., 1997), nikA of R64 (Furuya et al., 1991), mobB and mobC of plasmids pTC-F14 and pTF-FC2 (van Zyl et al., 2003), traJ and traK of RP4 (Guiney et al., 1989), mobC of R1162/RSF1010 (Brasch and Meyer, 1986), and mbeC of ColE1 (Varsaki et al., 2009) resulted in drastic decrease of plasmid transfer. All MOBQ4 plasmids encode a gene, called mobC, which is located adjacent to oriT and transcribed opposite to the mobA relaxase gene (Figure 1). Most of the mobC genes are not annotated, so we updated their annotation, as listed in Supplementary Table S2. The MobC proteins of MOBQ4 plasmids are small (less than 100 amino acids) and showed no homology to other RAPs (by using PSI-Blast). To check whether MobC plays a role in the MOBQ4 plasmid mobilization, mobC deletion mutants were constructed from pRC1 and pRC2, respectively producing pRC3 and pRC4 (Figure 1). A moderate decrease in mobilization was observed in the mobC– variants: 1.5-log reduction for pRC3 and 0.6-log for pRC4, when using R64drd11 as a helper (Figure 3B). MobC is thus not absolutely essential for MOBQ4 plasmid mobilization. This is an interesting difference to other plasmid groups, which should be further investigated. It is conceivable that some MOBQ4 plasmids can be found, the mobilization of which is independent of RAPs.
The 178 bp intergenic region comprised between the mobC and mobA genes of MOBQ4 plasmids was assembled with an oriT-lacking fragment of vector pSEVA631. The resulting constructions, pRC7 (for MOBQ41) and pRC8 (for MOBQ42) (Figure 1), were introduced in donor strains to check for their mobilization. The transfer proteins were supplied in trans: the corresponding mobilizable plasmid (pRC1 or pRC2) provided the relaxosomal proteins, while the conjugative plasmid (R64drd11) supplied the T4CP and MPF. Plasmids pRC7 and pRC8 were transferred to the recipient population, but 1000-fold less efficiently than their corresponding mobA+mobC+ partners (pRC1 and pRC2) (Figure 4). This result was confirmed by using plasmids pRC14 and pRC15, instead of pRC7 and pRC8, in the mobilization experiments. Plasmids pRC14 and pRC15 contained the same oriT region present in pRC7 and pRC8, but cloned in the inverse orientation. Besides, to avoid losing any oriT-related function, larger segments including also the mobC gene and the first 431 bp of the mobA gene [pRC5 and pRC6 (Figure 1)], were analyzed. Here again relaxase, T4CP and MPF components were provided in trans. Plasmids pRC5 and pRC6 behave similarly to pRC7 and pRC8, and were mobilized at least 500-fold less than pRC1 and pRC2 (Figure 4).
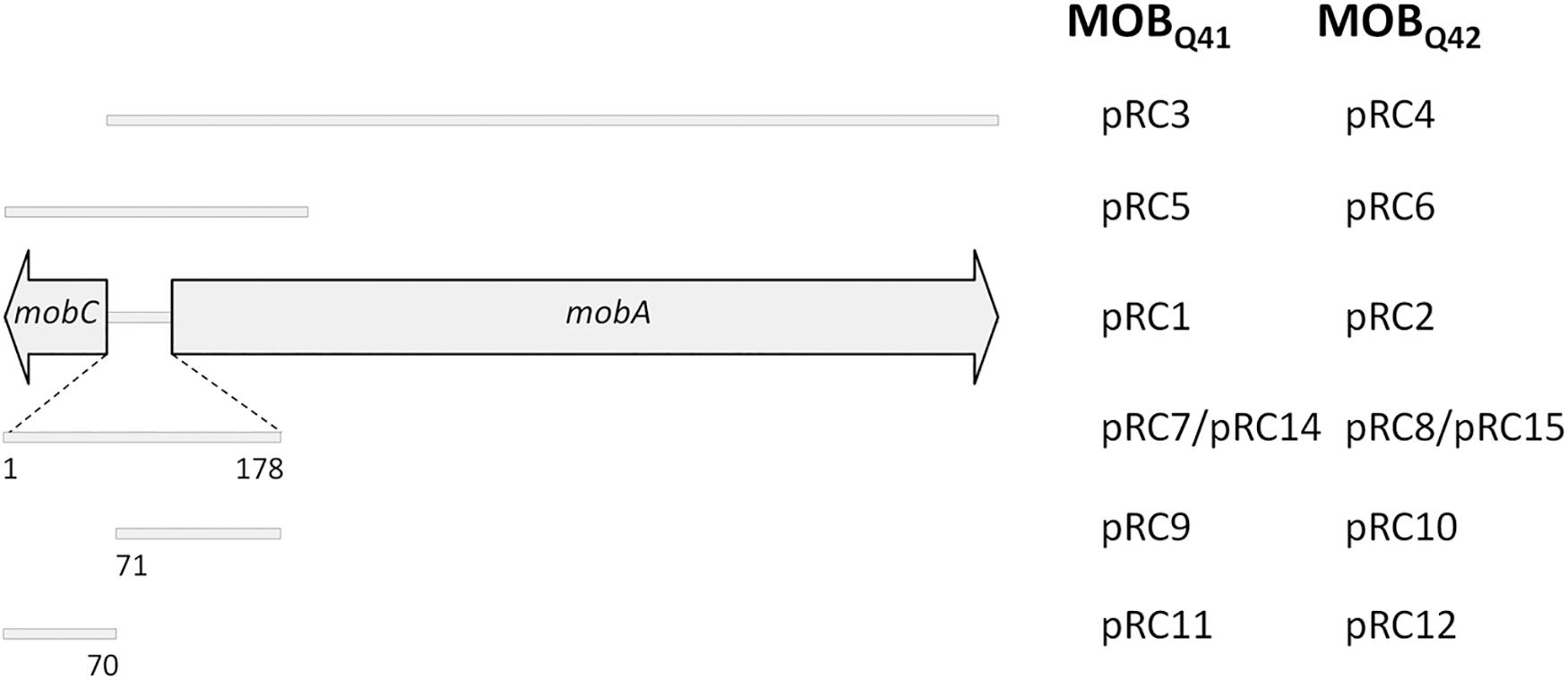
Figure 4. In trans mobilization of oriT fragments. R64drd11-mediated mobilization frequencies of pRC1 and plasmids containing fragments of its oriT are represented by dark-gray bars, while those of pRC2 and its derivatives are in light-gray bars. The bars represent the average of at least six experiments.
MOBQ4 relaxases showed thus a cis-acting preference for their oriTs, performing at least 500-fold better on a cis than on a trans oriT substrate. The cis-acting preference is a characteristic exhibited by some DNA-binding proteins, such as the TnpA transposases of Tn10, Tn5 and Tn903 (Morisato et al., 1983; Derbyshire et al., 1990; DeLong and Syvanen, 1991). Relaxases generally lack a cis preference for their oriTs. There are only a few examples of relaxases that show preference for a cis-encoded substrate. The MOBP relaxase of transposon Tn1549 was found to be cis-acting (Tsvetkova et al., 2010). Notably, all plasmid-encoded cis-acting relaxases have been reported in members of the MOBQ class: TraA of plasmid pRetCFN42d (MOBQ2) (Pérez-Mendoza et al., 2006) and TraA of plasmid pIP501 (MOBQ3) (Arends et al., 2012). Nevertheless, other MOBQ relaxases, such as Nes_pSK41 (Pollet et al., 2016), as well as MobA of plasmids R1162/RSF1010 and pSC101 (Brasch and Meyer, 1986; Derbyshire and Willetts, 1987; Meyer, 2000) worked efficiently in trans.
The MOBQ4 relaxases were also tested for their specificity to act on a non-cognate MOBQ4 oriT. The oriTs of MOBQ41 and MOBQ42 plasmids differ in 10 nucleotides along their 178bp sequence (Supplementary Figure S1). Mobilization frequencies of oriT_MOBQ42 plasmids pRC6 or pRC8 by the MOBQ41 plasmid pRC1 + R64drd11, as well as oriT_MOBQ41 plasmids pRC5 or pRC7 by the MOBQ42 plasmid pRC2 + R64drd11, were similar to that obtained for the cognate systems, varying no more than one log (Figure 4).
To further delimit the oriT of MOBQ4 plasmids, the 178bp oriT fragments cloned in pRC7 and pRC8 (see Supplementary Figure S1) were subdivided in two portions, one containing oriT nucleotides 1–70 (pRC11 and pRC12) and the other containing oriT nucleotides 71–178 (pRC9 and pRC10) (Figure 1 and Supplementary Figure S1). Disruption of the 178bp oriT region resulted in a drastic loss of conjugation efficiency of the oriT-containing plasmid (Figure 4), as previously reported for pIGWZ12 (Zaleski et al., 2015).
The cis-acting preference of the MOBQ4 relaxases shown here is an example of biological orthogonality (de Lorenzo, 2011), that is, a mechanism to avoid interference. It implies that when two MOBQ4 plasmids are present in the same cell, the contribution of oriT cross-recognition by the heterologous MOBQ4 relaxase to plasmid transfer is not substantial. This feature could be essential to guarantee their efficient transfer, given the fact that both types of MOBQ4 plasmids use the same repertoire of conjugative helpers and share the same hosts.
MOBQ41 and MOBQ42 plasmids are able to coexist and spread in the E. coli population without affecting each other largely. They disseminate through bacterial conjugation, aided specially by MPFI conjugative plasmids, but neither of the MOBQ4 plasmids dominates the horizontal transfer process. Co-residence of MOBQ41 and MOBQ42 plasmids in the same host neither hindered nor boosted considerably their respective mobilization frequencies. Since both plasmids (MOBQ41 and MOBQ42) have a narrow host-range (they circulate among enterobacteria), their coexistence in natural environments is likely. In such ecological setting, specific discrimination among their highly similar oriT sequences would be guaranteed by the preferential cis activity of the MOBQ4 relaxase. Such strategy would be biologically relevant in a scenario of co-residence of non-divergent elements to favor self-dissemination.
All datasets generated for this study are included in the article/Supplementary Material.
MG-B and FC conceived the study and designed the experiments. RC-L, AC, and MG-B performed the experiments. RC-L, AC, MG-B, and FC interpreted the data. MG-B and FC wrote the manuscript.
This work was supported by the Spanish Ministry of Economy and Competitiveness (BFU2017-86378-P, AEI/FEDER, UE, to FC) and Consejo Superior de Investigaciones Científicas (201820I143 to MG-B). We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors want to thank María Aramburu and Raúl Fernández-López for their technical assistance with the flow cytometer and the fluorescence microscopy, respectively. This manuscript has been released as a Pre-Print at bioRxiv (Garcillán-Barcia et al., 2019).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02557/full#supplementary-material
Achtman, M., Willetts, N., and Clark, A. J. (1971). Beginning a genetic analysis of conjugational transfer determined by the F factor in Escherichia coli by isolation and characterization of transfer-deficient mutants. J. Bacteriol. 106, 529–538.
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Alvarado, A., Garcillán-Barcia, M. P., and de la Cruz, F. (2012). A degenerate primer MOB typing (DPMT) method to classify gamma-proteobacterial plasmids in clinical and environmental settings. PLoS One 7:e40438. doi: 10.1371/journal.pone.0040438
Arends, K., Schiwon, K., Sakinc, T., Hübner, J., and Grohmann, E. (2012). Green fluorescent protein-labeled monitoring tool to quantify conjugative plasmid transfer between gram-positive and gram-negative bacteria. Appl. Environ. Microbiol. 78, 895–899. doi: 10.1128/AEM.05578-5511
Beatson, S. A., Ben Zakour, N. L., Totsika, M., Forde, B. M., Watts, R. E., Mabbett, A. N., et al. (2015). Molecular analysis of asymptomatic bacteriuria Escherichia coli strain VR50 reveals adaptation to the urinary tract by gene acquisition. Infect. Immun. 83, 1749–1764. doi: 10.1128/IAI.02810-2814
Bleicher, A., Schöfl, G., Rodicio, M. D. R., and Saluz, H. P. (2013). The plasmidome of a Salmonella enterica serovar derby isolated from pork meat. Plasmid 69, 202–210. doi: 10.1016/j.plasmid.2013.01.001
Brasch, M. A., and Meyer, R. J. (1986). Genetic organization of plasmid R1162 DNA involved in conjugative mobilization. J. Bacteriol. 167, 703–710. doi: 10.1128/jb.167.2.703-710.1986
Brolund, A., Franzén, O., Melefors, O., Tegmark-Wisell, K., and Sandegren, L. (2013). Plasmidome-analysis of ESBL-producing escherichia coli using conventional typing and high-throughput sequencing. PLoS One 8:e65793. doi: 10.1371/journal.pone.0065793
Cabezón, E., Lanka, E., and de la Cruz, F. (1994). Requirements for mobilization of plasmids RSF1010 and ColE1 by the IncW plasmid R388: trwB and RP4 traG are interchangeable. J. Bacteriol. 176, 4455–4458. doi: 10.1128/jb.176.14.4455-4458.1994
Cabezón, E., Sastre, J. I., and de la Cruz, F. (1997). Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 254, 400–406. doi: 10.1007/s004380050432
Calva, E., Silva, C., Zaidi, M. B., Sanchez-Flores, A., Estrada, K., Silva, G. G. Z., et al. (2015). Complete genome sequencing of a multidrug-resistant and human-invasive Salmonella enterica serovar typhimurium strain of the emerging sequence type 213 genotype. Genome Announc. 3:e663-15. doi: 10.1128/genomeA.00663-615
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Cascales, E., Atmakuri, K., Liu, Z., Binns, A. N., and Christie, P. J. (2005). Agrobacterium tumefaciens oncogenic suppressors inhibit T-DNA and VirE2 protein substrate binding to the VirD4 coupling protein. Mol. Microbiol. 58, 565–579. doi: 10.1111/j.1365-2958.2005.04852.x
Cascales, E., Buchanan, S. K., Duché, D., Kleanthous, C., Lloubès, R., Postle, K., et al. (2007). Colicin biology. Microbiol. Mol. Biol. Rev. 71, 158–229. doi: 10.1128/MMBR.00036-36
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. doi: 10.1093/bioinformatics/btr088
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
de la Cruz, F., Frost, L. S., Meyer, R. J., and Zechner, E. L. (2010). Conjugative DNA metabolism in gram-negative bacteria. FEMS Microbiol. Rev. 34, 18–40. doi: 10.1111/j.1574-6976.2009.00195.x
de la Cruz, F., and Grinsted, J. (1982). Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J. Bacteriol. 151, 222–228.
de Lorenzo, V. (2011). Beware of metaphors: chasses and orthogonality in synthetic biology. Bioeng. Bugs 2, 3–7. doi: 10.4161/bbug.2.1.13388
de Toro, M., Garcillán-Barcia, M. P., and De La Cruz, F. (2014). Plasmid diversity and adaptation analyzed by massive sequencing of Escherichia coli plasmids. Microbiol. Spectr. 2, 219–235. doi: 10.1128/microbiolspec.PLAS-0031-2014
del Campo, I., Ruiz, R., Cuevas, A., Revilla, C., Vielva, L., and de la Cruz, F. (2012). Determination of conjugation rates on solid surfaces. Plasmid 67, 174–182. doi: 10.1016/j.plasmid.2012.01.008
DeLong, A., and Syvanen, M. (1991). Trans-acting transposase mutant from Tn5. Proc. Natl. Acad. Sci. U.S.A. 88, 6072–6076. doi: 10.1073/pnas.88.14.6072
Derbyshire, K. M., Kramer, M., and Grindley, N. D. (1990). Role of instability in the cis action of the insertion sequence IS903 transposase. Proc. Natl. Acad. Sci. U.S.A. 87, 4048–4052. doi: 10.1073/pnas.87.11.4048
Derbyshire, K. M., and Willetts, N. S. (1987). Mobilization of the non-conjugative plasmid RSF1010: a genetic analysis of its origin of transfer. Mol. Gen. Genet. 206, 154–160. doi: 10.1007/bf00326551
Dunlop, M. J., Cox, R. S., Levine, J. H., Murray, R. M., and Elowitz, M. B. (2008). Regulatory activity revealed by dynamic correlations in gene expression noise. Nat. Genet. 40, 1493–1498. doi: 10.1038/ng.281
Dyall-Smith, M. L., Liu, Y., and Billman-Jacobe, H. (2017). Genome sequence of an australian monophasic Salmonella enterica subsp. enterica typhimurium Isolate (TW-Stm6) carrying a large plasmid with multiple antimicrobial resistance genes. Genome Announc. 5, e793–17. doi: 10.1128/genomeA.00793-717
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edwards, J. S., Betts, L., Frazier, M. L., Pollet, R. M., Kwong, S. M., Walton, W. G., et al. (2013). Molecular basis of antibiotic multiresistance transfer in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 110, 2804–2809. doi: 10.1073/pnas.1219701110
Fernandez-Lopez, R., Redondo, S., Garcillan-Barcia, M. P., and de la Cruz, F. (2017). Towards a taxonomy of conjugative plasmids. Curr. Opin. Microbiol. 38, 106–113. doi: 10.1016/j.mib.2017.05.005
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Francia, M. V., Varsaki, A., Garcillán-Barcia, M. P., Latorre, A., Drainas, C., and de la Cruz, F. (2004). A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28, 79–100. doi: 10.1016/j.femsre.2003.09.001
Fratamico, P. M., Yan, X., Caprioli, A., Esposito, G., Needleman, D. S., Pepe, T., et al. (2011). The complete DNA sequence and analysis of the virulence plasmid and of five additional plasmids carried by shiga toxin-producing Escherichia coli O26:H11 strain H30. Int. J. Med. Microbiol. 301, 192–203. doi: 10.1016/j.ijmm.2010.09.002
Fricke, W. F., Wright, M. S., Lindell, A. H., Harkins, D. M., Baker-Austin, C., Ravel, J., et al. (2008). Insights into the environmental resistance gene pool from the genome sequence of the multidrug-resistant environmental isolate Escherichia coli SMS-3-5. J. Bacteriol. 190, 6779–6794. doi: 10.1128/JB.00661-668
Furuya, N., Nisioka, T., and Komano, T. (1991). Nucleotide sequence and functions of the oriT operon in IncI1 plasmid R64. J. Bacteriol. 173, 2231–2237. doi: 10.1128/jb.173.7.2231-2237.1991
Gama, J. A., Zilhão, R., and Dionisio, F. (2017a). Co-resident plasmids travel together. Plasmid 93, 24–29. doi: 10.1016/j.plasmid.2017.08.004
Gama, J. A., Zilhão, R., and Dionisio, F. (2017b). Conjugation efficiency depends on intra and intercellular interactions between distinct plasmids: plasmids promote the immigration of other plasmids but repress co-colonizing plasmids. Plasmid 93, 6–16. doi: 10.1016/j.plasmid.2017.08.003
Gama, J. A., Zilhão, R., and Dionisio, F. (2017c). Multiple plasmid interference - pledging allegiance to my enemy’s enemy. Plasmid 93, 17–23. doi: 10.1016/j.plasmid.2017.08.002
Gama, J. A., Zilhão, R., and Dionisio, F. (2018). Impact of plasmid interactions with the chromosome and other plasmids on the spread of antibiotic resistance. Plasmid 99, 82–88. doi: 10.1016/j.plasmid.2018.09.009
Garcillán-Barcia, M. P., Cuartas Lanza, R., Cuevas, A., and de la Cruz, F. (2019). Comparative analysis of MOBQ4 plasmids demonstrates that MOBQ is a cis-acting enriched relaxase protein family. bioRxiv doi: org/10.1101/726927 [Preprint]
Garcillán-Barcia, M. P., and de la Cruz, F. (2013). Ordering the bestiary of genetic elements transmissible by conjugation. Mob. Genet. Elements 3:e24263. doi: 10.4161/mge.24263
Garcillán-Barcia, M. P., Francia, M. V., and de la Cruz, F. (2009). The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33, 657–687. doi: 10.1111/j.1574-6976.2009.00168.x
Garcillán-Barcia, M. P., Ruiz del Castillo, B., Alvarado, A., de la Cruz, F., and Martínez-Martínez, L. (2015). Degenerate primer MOB typing of multiresistant clinical isolates of E. coli uncovers new plasmid backbones. Plasmid 77, 17–27. doi: 10.1016/j.plasmid.2014.11.003
Getino, M., and de la Cruz, F. (2018). Natural and artificial strategies to control the conjugative transmission of plasmids. Microbiol. Spectr 6, 1–25. doi: 10.1128/microbiolspec.MTBP-0015-2016
Getino, M., Palencia-Gándara, C., Garcillán-Barcia, M. P., and de la Cruz, F. (2017). PifC and osa, plasmid weapons against rival conjugative coupling proteins. Front. Microbiol. 8:2260. doi: 10.3389/fmicb.2017.02260
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., and Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Gibson, M. K., Forsberg, K. J., and Dantas, G. (2015). Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 9, 207–216. doi: 10.1038/ismej.2014.106
Grant, S. G., Jessee, J., Bloom, F. R., and Hanahan, D. (1990). Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. U.S.A. 87, 4645–4649. doi: 10.1073/pnas.87.12.4645
Guglielmini, J., Néron, B., Abby, S. S., Garcillán-Barcia, M. P., de la Cruz, F., and Rocha, E. P. C. (2014). Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 42, 5715–5727. doi: 10.1093/nar/gku194
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Guiney, D. G., Deiss, C., Simnad, V., Yee, L., Pansegrau, W., and Lanka, E. (1989). Mutagenesis of the Tra1 core region of RK2 by using Tn5: identification of plasmid-specific transfer genes. J. Bacteriol. 171, 4100–4103. doi: 10.1128/jb.171.7.4100-4103.1989
Hiraga, S., Sugiyama, T., and Itoh, T. (1994). Comparative analysis of the replicon regions of eleven ColE2-related plasmids. J. Bacteriol. 176, 7233–7243. doi: 10.1128/jb.176.23.7233-7243.1994
Holt, K. E., Thieu Nga, T. V., Thanh, D. P., Vinh, H., Kim, D. W., Vu Tra, M. P., et al. (2013). Tracking the establishment of local endemic populations of an emergent enteric pathogen. Proc. Natl. Acad. Sci. U.S.A. 110, 17522–17527. doi: 10.1073/pnas.1308632110
Horii, T., and Itoh, T. (1988). Replication of ColE2 and ColE3 plasmids: the regions sufficient for autonomous replication. Mol. Gen. Genet. 212, 225–231. doi: 10.1007/bf00334689
Itoh, T., and Horii, T. (1989). Replication of ColE2 and ColE3 plasmids: in vitro replication dependent on plasmid-coded proteins. Mol. Gen. Genet. 219, 249–255. doi: 10.1007/bf00261184
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kido, M., Yasueda, H., and Itoh, T. (1991). Identification of a plasmid-coded protein required for initiation of ColE2 DNA replication. Nucleic Acids Res. 19, 2875–2880. doi: 10.1093/nar/19.11.2875
Lanza, V. F., de Toro, M., Garcillán-Barcia, M. P., Mora, A., Blanco, J., Coque, T. M., et al. (2014). Plasmid flux in Escherichia coli ST131 sublineages, analyzed by plasmid constellation network (PLACNET), a new method for plasmid reconstruction from whole genome sequences. PLoS Genet. 10:e1004766. doi: 10.1371/journal.pgen.1004766
Liu, C., Zheng, H., Yang, M., Xu, Z., Wang, X., Wei, L., et al. (2015). Genome analysis and in vivo virulence of porcine extraintestinal pathogenic Escherichia coli strain PCN033. BMC Genomics 16:717. doi: 10.1186/s12864-015-1890-1899
Lorenzo-Díaz, F., Fernández-López, C., Garcillán-Barcia, M. P., and Espinosa, M. (2014). Bringing them together: plasmid pMV158 rolling circle replication and conjugation under an evolutionary perspective. Plasmid 74, 15–31. doi: 10.1016/j.plasmid.2014.05.004
Maindola, P., Raina, R., Goyal, P., Atmakuri, K., Ojha, A., Gupta, S., et al. (2014). Multiple enzymatic activities of ParB/Srx superfamily mediate sexual conflict among conjugative plasmids. Nat. Commun. 5:5322. doi: 10.1038/ncomms6322
Meyer, R. (2000). Identification of the mob genes of plasmid pSC101 and characterization of a hybrid pSC101-R1162 system for conjugal mobilization. J. Bacteriol. 182, 4875–4881. doi: 10.1128/jb.182.17.4875-4881.2000
Meyer, R. (2009). Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid 62, 57–70. doi: 10.1016/j.plasmid.2009.05.001
Moncalián, G., Grandoso, G., Llosa, M., and de la Cruz, F. (1997). oriT-processing and regulatory roles of TrwA protein in plasmid R388 conjugation. J. Mol. Biol. 270, 188–200. doi: 10.1006/jmbi.1997.1082
Monzingo, A. F., Ozburn, A., Xia, S., Meyer, R. J., and Robertus, J. D. (2007). The structure of the minimal relaxase domain of MobA at 2.1 a resolution. J. Mol. Biol. 366, 165–178. doi: 10.1016/j.jmb.2006.11.031
Morisato, D., Way, J. C., Kim, H. J., and Kleckner, N. (1983). Tn10 transposase acts preferentially on nearby transposon ends in vivo. Cell 32, 799–807. doi: 10.1016/0092-8674(83)90066-90061
Nomura, N., Masai, H., Inuzuka, M., Miyazaki, C., Ohtsubo, E., Itoh, T., et al. (1991). Identification of eleven single-strand initiation sequences (ssi) for priming of DNA replication in the F, R6K, R100 and ColE2 plasmids. Gene 108, 15–22. doi: 10.1016/0378-1119(91)90482-q
Ogura, Y., Ooka, T., Iguchi, A., Toh, H., Asadulghani, M., Oshima, K., et al. (2009). Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 106, 17939–17944. doi: 10.1073/pnas.0903585106
Oshima, K., Toh, H., Ogura, Y., Sasamoto, H., Morita, H., Park, S.-H., et al. (2008). Complete genome sequence and comparative analysis of the wild-type commensal Escherichia coli strain SE11 isolated from a healthy adult. DNA Res. 15, 375–386. doi: 10.1093/dnares/dsn026
Pérez-Mendoza, D., Lucas, M., Muñoz, S., Herrera-Cervera, J. A., Olivares, J., de la Cruz, F., et al. (2006). The relaxase of the rhizobium etli symbiotic plasmid shows nic site cis-acting preference. J. Bacteriol. 188, 7488–7499. doi: 10.1128/JB.00701-706
Pollet, R. M., Ingle, J. D., Hymes, J. P., Eakes, T. C., Eto, K. Y., Kwong, S. M., et al. (2016). Processing of nonconjugative resistance plasmids by conjugation nicking enzyme of staphylococci. J. Bacteriol. 198, 888–897. doi: 10.1128/JB.00832-815
San Millan, A., Heilbron, K., and MacLean, R. C. (2014). Positive epistasis between co-infecting plasmids promotes plasmid survival in bacterial populations. ISME J. 8, 601–612. doi: 10.1038/ismej.2013.182
San Millan, A., and MacLean, R. C. (2017). Fitness costs of plasmids: a limit to plasmid transmission. Microbiol. Spectr 5, 601–612. doi: 10.1128/microbiolspec.MTBP-0016-2017
Silva-Rocha, R., Martínez-García, E., Calles, B., Chavarría, M., Arce-Rodríguez, A., de Las Heras, A., et al. (2013). The standard european vector architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 41, D666–D675. doi: 10.1093/nar/gks1119
Smillie, C., Garcillán-Barcia, M. P., Francia, M. V., Rocha, E. P. C., and de la Cruz, F. (2010). Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74, 434–452. doi: 10.1128/MMBR.00020-10
Sørensen, A. H., Hansen, L. H., Johannesen, E., and Sørensen, S. J. (2003). Conjugative plasmid conferring resistance to olaquindox. Antimicrob. Agents Chemother. 47, 798–799. doi: 10.1128/aac.47.2.798-799.2003
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Stephens, C. M., Skerker, J. M., Sekhon, M. S., Arkin, A. P., and Riley, L. W. (2015). Complete genome sequences of four Escherichia coli ST95 isolates from bloodstream infections. Genome Announc 3:e1241-15. doi: 10.1128/genomeA.01241-1215
Sugiyama, T., and Itoh, T. (1993). Control of ColE2 DNA replication: in vitro binding of the antisense RNA to the Rep mRNA. Nucleic Acids Res. 21, 5972–5977. doi: 10.1093/nar/21.25.5972
Summers, D. (1998). Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Mol. Microbiol. 29, 1137–1145. doi: 10.1046/j.1365-2958.1998.01012.x
Summers, D. K., and Sherratt, D. J. (1984). Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell 36, 1097–1103. doi: 10.1016/0092-8674(84)90060-90066
Summers, D. K., and Sherratt, D. J. (1988). Resolution of ColE1 dimers requires a DNA sequence implicated in the three-dimensional organization of the cer site. EMBO J. 7, 851–858. doi: 10.1002/j.1460-2075.1988.tb02884.x
Takechi, S., Yasueda, H., and Itoh, T. (1994). Control of ColE2 plasmid replication: regulation of rep expression by a plasmid-coded antisense RNA. Mol. Gen. Genet. 244, 49–56. doi: 10.1007/bf00280186
Tsvetkova, K., Marvaud, J.-C., and Lambert, T. (2010). Analysis of the mobilization functions of the vancomycin resistance transposon Tn1549, a member of a new family of conjugative elements. J. Bacteriol. 192, 702–713. doi: 10.1128/JB.00680-689
van Zyl, L. J., Deane, S. M., and Rawlings, D. E. (2003). Analysis of the mobilization region of the broad-host-range IncQ-like plasmid pTC-F14 and its ability to interact with a related plasmid, pTF-FC2. J. Bacteriol. 185, 6104–6111. doi: 10.1128/jb.185.20.6104-6111.2003
Varsaki, A., Moncalián, G., Garcillán-Barcia, M., del, P., Drainas, C., and de la Cruz, F. (2009). Analysis of ColE1 MbeC unveils an extended ribbon-helix-helix family of nicking accessory proteins. J. Bacteriol. 191, 1446–1455. doi: 10.1128/JB.01342-1348
Yagura, M., and Itoh, T. (2006). The rep protein binding elements of the plasmid ColE2-P9 replication origin. Biochem. Biophys. Res. Commun. 345, 872–877. doi: 10.1016/j.bbrc.2006.04.168
Yagura, M., Nishio, S.-Y., Kurozumi, H., Wang, C.-F., and Itoh, T. (2006). Anatomy of the replication origin of plasmid ColE2-P9. J. Bacteriol. 188, 999–1010. doi: 10.1128/JB.188.3.999-1010.2006
Yasueda, H., Horii, T., and Itoh, T. (1989). Structural and functional organization of ColE2 and ColE3 replicons. Mol. Gen. Genet. 215, 209–216. doi: 10.1007/bf00339719
Yasueda, H., Takechi, S., Sugiyama, T., and Itoh, T. (1994). Control of ColE2 plasmid replication: negative regulation of the expression of the plasmid-specified initiator protein, Rep, at a posttranscriptional step. Mol. Gen. Genet. 244, 41–48. doi: 10.1007/bf00280185
Zaleski, P., Wawrzyniak, P., Sobolewska, A., Łukasiewicz, N., Baran, P., Romańczuk, K., et al. (2015). pIGWZ12–A cryptic plasmid with a modular structure. Plasmid 79, 37–47. doi: 10.1016/j.plasmid.2015.04.001
Zaleski, P., Wawrzyniak, P., Sobolewska, A., Mikiewicz, D., Wojtowicz-Krawiec, A., Chojnacka-Puchta, L., et al. (2012). New cloning and expression vector derived from Escherichia coli plasmid pIGWZ12; a potential vector for a two-plasmid expression system. Plasmid 67, 264–271. doi: 10.1016/j.plasmid.2011.12.011
Keywords: mobilizable plasmids, horizontal gene transfer, MOBQ relaxase, cis-acting relaxase, plasmid coexistence, bacterial conjugation
Citation: Garcillán-Barcia MP, Cuartas-Lanza R, Cuevas A and de la Cruz F (2019) Cis-Acting Relaxases Guarantee Independent Mobilization of MOBQ4 Plasmids. Front. Microbiol. 10:2557. doi: 10.3389/fmicb.2019.02557
Received: 02 September 2019; Accepted: 23 October 2019;
Published: 08 November 2019.
Edited by:
Eva M. Top, University of Idaho, United StatesReviewed by:
Ana P. Tedim, Institute of Health Sciences Studies of Castilla y León (IECSCYL), SpainCopyright © 2019 Garcillán-Barcia, Cuartas-Lanza, Cuevas and de la Cruz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. Pilar Garcillán-Barcia, Z2FyY2lsbXBAdW5pY2FuLmVz; Fernando de la Cruz, ZGVsYWNydXpAdW5pY2FuLmVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.