
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 November 2019
Sec. Evolutionary and Genomic Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02556
 Charlotte Peeters1
Charlotte Peeters1 Evelien De Canck1
Evelien De Canck1 Margo Cnockaert1
Margo Cnockaert1 Evie De Brandt1
Evie De Brandt1 Cindy Snauwaert2
Cindy Snauwaert2 Bart Verheyde1
Bart Verheyde1 Eliza Depoorter1
Eliza Depoorter1 Theodore Spilker3
Theodore Spilker3 John J. LiPuma3
John J. LiPuma3 Peter Vandamme1,2*
Peter Vandamme1,2*Comparative analysis of partial gyrB, recA, and gltB gene sequences of 84 Pandoraea reference strains and field isolates revealed several clusters that included no taxonomic reference strains. The gyrB, recA, and gltB phylogenetic trees were used to select 27 strains for whole-genome sequence analysis and for a comparative genomics study that also included 41 publicly available Pandoraea genome sequences. The phylogenomic analyses included a Genome BLAST Distance Phylogeny approach to calculate pairwise digital DNA–DNA hybridization values and their confidence intervals, average nucleotide identity analyses using the OrthoANIu algorithm, and a whole-genome phylogeny reconstruction based on 107 single-copy core genes using bcgTree. These analyses, along with subsequent chemotaxonomic and traditional phenotypic analyses, revealed the presence of 17 novel Pandoraea species among the strains analyzed, and allowed the identification of several unclassified Pandoraea strains reported in the literature. The genus Pandoraea has an open pan genome that includes many orthogroups in the ‘Xenobiotics biodegradation and metabolism’ KEGG pathway, which likely explains the enrichment of these species in polluted soils and participation in the biodegradation of complex organic substances. We propose to formally classify the 17 novel Pandoraea species as P. anapnoica sp. nov. (type strain LMG 31117T = CCUG 73385T), P. anhela sp. nov. (type strain LMG 31108T = CCUG 73386T), P. aquatica sp. nov. (type strain LMG 31011T = CCUG 73384T), P. bronchicola sp. nov. (type strain LMG 20603T = ATCC BAA-110T), P. capi sp. nov. (type strain LMG 20602T = ATCC BAA-109T), P. captiosa sp. nov. (type strain LMG 31118T = CCUG 73387T), P. cepalis sp. nov. (type strain LMG 31106T = CCUG 39680T), P. commovens sp. nov. (type strain LMG 31010T = CCUG 73378T), P. communis sp. nov. (type strain LMG 31110T = CCUG 73383T), P. eparura sp. nov. (type strain LMG 31012T = CCUG 73380T), P. horticolens sp. nov. (type strain LMG 31112T = CCUG 73379T), P. iniqua sp. nov. (type strain LMG 31009T = CCUG 73377T), P. morbifera sp. nov. (type strain LMG 31116T = CCUG 73389T), P. nosoerga sp. nov. (type strain LMG 31109T = CCUG 73390T), P. pneumonica sp. nov. (type strain LMG 31114T = CCUG 73388T), P. soli sp. nov. (type strain LMG 31014T = CCUG 73382T), and P. terrigena sp. nov. (type strain LMG 31013T = CCUG 73381T).
Members of the genus Pandoraea have emerged as rare opportunistic pathogens in persons with cystic fibrosis (Jørgensen et al., 2003; Johnson et al., 2004; Pimentel and MacLeod, 2008; Kokcha et al., 2013; Ambrose et al., 2016; Martina et al., 2017; See-Too et al., 2019) and several cases of chronic colonization and patient-to-patient transfer in this patient group have been reported (Jørgensen et al., 2003; Atkinson et al., 2006; Degand et al., 2015; Pugès et al., 2015; Ambrose et al., 2016; Dupont et al., 2017; Greninger et al., 2017). In addition to causing infection in cystic fibrosis patients, Pandoraea isolates have been recovered from blood and from samples from patients with chronic obstructive pulmonary disease or chronic granulomatous disease (Coenye et al., 2000; Daneshvar et al., 2001). Although the small number of patients involved and underlying diseases make it difficult to identify these bacteria as the cause of clinical deterioration (Martina et al., 2017; Green and Jones, 2018), one report described sepsis, multiple organ failure and death in a non-cystic fibrosis patient who underwent lung transplantation for sarcoidosis (Stryjewski et al., 2003).
Of the 11 validly named Pandoraea species, six (i.e., Pandoraea apista, Pandoraea norimbergensis, Pandoraea pulmonicola, Pandoraea pnomenusa, Pandoraea sputorum, and Pandoraea fibrosis) have been recovered from human clinical specimens (Coenye et al., 2000; See-Too et al., 2019), while Pandoraea faecigallinarum, Pandoraea oxalativorans, Pandoraea terrae, Pandoraea thiooxydans, and Pandoraea vervacti have been isolated from environmental samples (Anandham et al., 2010; Sahin et al., 2011; Jeong et al., 2016). An uncultivated endosymbiont of the trypanosomatid Novymonas esmeraldas represents an additional Pandoraea species which was provisionally named Candidatus Pandoraea novymonadis (Kostygov et al., 2017).
A growing number of reports demonstrate that soil and water represent the natural habitats of Pandoraea bacteria where they can be part of rhizosphere communities (Anandham et al., 2010; Jurelevicius et al., 2010; Peeters et al., 2016; Dong et al., 2018) and be involved in oxalate degradation (Jin et al., 2007; Sahin et al., 2011). The latter suggests they may be important contributors to soil formation, soil fertility and retention, and cycling of elements necessary for plant growth (Sahin, 2003). These free-living Pandoraea bacteria are often enriched in polluted soils and participate in the biodegradation of complex organic substances including lignin (Shi et al., 2013; Kumar et al., 2018b; Liu et al., 2019), biodiesel and petroleum by-products (de Paula et al., 2017; Sarkar et al., 2017; Tirado-Torres et al., 2017), p-xylene (Wang et al., 2015), δ-hexachlorocyclohexane (Pushiri et al., 2013), di-n-butyl phthalate (Yang et al., 2018), biphenyl, benzoate and naphthalene (Uhlik et al., 2012), and tetracycline (Wu et al., 2019) and β-lactam antibiotics (Crofts et al., 2017). A particularly well-documented Pandoraea strain, i.e., JB1T (LMG 31106T), was isolated in the 1980s from garden soil (Parsons et al., 1988) and was able to use biphenyl, 2-, 3- and 4-chloro-biphenyl, m-toluate, p-toluate naphthalene, m-hydroxybenzoate and diphenylmethane (Springael et al., 1996). Although this strain also represented a separate novel Pandoraea species, it was not formally classified (Coenye et al., 2000) pending the availability of more than one strain representing the same novel species, a taxonomic practice that has been largely abandoned today.
The genome sequences of several strains with bioremediation potential have been reported, but a growing number of studies fail to provide species level identification of such strains (Pushiri et al., 2013; Chan et al., 2015; Kumar M. et al., 2016; Crofts et al., 2017; Liu et al., 2018; Wu et al., 2019). In addition, in our studies on the diversity and epidemiology of opportunistic pathogens in persons with cystic fibrosis, we isolated a considerable number of Pandoraea strains that represent novel species (unpublished data). The present study aimed to clarify the taxonomy and formally name these novel Pandoraea species, and to make reference cultures and whole-genome sequences of each of these versatile bacteria publicly available.
Isolates representing novel Pandoraea species are listed in Table 1, along with their isolation source details. These strains were initially assigned to the genus Pandoraea on the basis of sequence analysis of 16S rRNA, gyrB or recA genes (data not shown). Well-characterized reference strains and recent field isolates identified in the present study as established Pandoraea species are listed in Supplementary Table S1. Strains were grown aerobically on Tryptone Soya Agar (Oxoid) and incubated at 28°C. Cultures were preserved in MicroBankTM vials at −80°C.
DNA was extracted using an automated Maxwell® DNA preparation instrument (Promega, United States). The final extract was treated with RNAse (2 mg/ml, 5 μL per 100 μL extract) and incubated at 37°C for 1 h. DNA quality was checked using 1% agarose gel electrophoresis and DNA quantification was performed using the QuantiFluor ONE dsDNA system and the Quantus fluorometer (Promega, United States). DNA was stored at −20°C prior to further analysis.
Nearly complete 16S rRNA sequences were obtained as described previously (Peeters et al., 2013).
Partial recA gene sequences (663 bp) were amplified by PCR using forward primer 5′-AGG ACG ATT CAT GGA AGA WAG C-3′ and reverse primer 5′-GAC GCA CYG AYG MRT AGA ACT T-3′ (Spilker et al., 2009). Each 25 μl PCR reaction consisted of 1x PCR buffer (Qiagen), 1 U of Taq polymerase (Qiagen), 250 μM of each dNTP (Applied Biosystems), 1 × Q-solution (Qiagen), 1 μM of each primer and 2 μl of DNA (Peeters et al., 2013). PCR was performed using a Veriti 96 Well Thermal Cycler (Applied Biosystems). Initial denaturation for 2 min at 94°C was followed by 30 cycles of 30 s at 94°C, 45 s at 58°C and 1 min at 72°C, and a final elongation for 10 min at 72°C. Amplicons were purified using a NucleoFast 96 PCR clean-up kit (Macherey-Nagel). Sequencing primers (one per sequencing reaction) were the same as the amplification primers. Sequence analysis was performed with an Applied Biosystems 3130xl Genetic Analyzer and protocols of the manufacturer using the BigDye Terminator Cycle Sequencing Ready kit. Sequence assembly was performed using BioNumerics v7.6 (Applied Maths, Belgium).
Partial gyrB sequences (573 bp) were amplified by PCR using forward primer 5′-GAC AAY GGB CGY GGV RTB CC-3′ (this study) and reverse primer 5′-YTC GTT GWA RCT GTC GTT CCA CTG C-3′ (Spilker et al., 2009). The PCR protocol was the same as for recA, except that 2 μM of primer was used and an annealing temperature of 60°C. Sequencing primers (one per sequencing reaction) were 5′-ACG ACA AGC ACG ARC CSA AGC G-3′ (this study) and the same reverse primer as for amplification. Sequence analysis and assembly were performed as described above for the recA gene.
Partial gltB sequences were amplified by PCR using forward primer 5′-CTG CAT CAT GAT GCG CAA GTG-3′ (Spilker et al., 2009) and reverse primer 5′-GTT GCC ACG GAA RTC GTT GG-3′ (this study). The PCR protocol was the same as for recA, except that 0.4 μM of primer was used. Sequencing primers (one per sequencing reaction) were the same as the amplification primers. Sequence analysis and assembly were performed as described above for the recA gene.
Gene sequences of recA, gyrB, and gltB were aligned based on their amino acid sequences using Muscle (Edgar, 2004) in MEGA7 (Kumar S. et al., 2016). Phylogenetic trees were constructed using RAxML v8.2.11 (Stamatakis, 2014) with the GTRCAT substitution model and 1000 bootstrap analyses. Visualization and annotation of the phylogenetic trees was performed using iTOL (Letunic and Bork, 2016).
The genome sequences of 27 strains (Table 2 and Supplementary Table S2) were determined using the Illumina HiSeq4000 platform (PE150) at the Oxford Genomics Centre. Quality reports were created by FastQC. Reads were trimmed using Trimmomatic (Bolger et al., 2014) with the MAXINFO:50:0.8 and MINLEN:50 options. Genome size was estimated using kmc (Kokot et al., 2017) and reads were subsampled with seqtk1 to 80x coverage depth for assembly. Assembly was performed using SPAdes v3.12.0 (Bankevich et al., 2012) with error correction, default k-mer sizes (21, 33, 55, 77) and mismatch correction. Contigs were filtered on length (minimum 500 bp) and coverage (minimum 0.5x and maximum 8x overall coverage). Raw reads were mapped against the assemblies using bwa mem (Li, 2013) and contigs were polished using Pilon 1.22 (Walker et al., 2014) with default parameters. Quast (Gurevich et al., 2013) was used to create quality reports of the resulting assemblies. Annotation was performed using Prokka 1.12 (Seemann, 2014) with a genus-specific database based on publicly available genomes.
All 41 publicly available (January 29, 2019) whole-genome sequences of Pandoraea bacteria were downloaded from the NCBI database (Table 2). Burkholderia cenocepacia J2315T was used as an outgroup in the phylogenomic analyses. For strains B-6 (Liu et al., 2018), E26 (Chan et al., 2015), PE-S2R-1 and PE-S2T-3 (Crofts et al., 2017) no annotation was available and therefore annotation was performed using Prokka as described above.
The GBDP approach was used to calculate pairwise digital DNA–DNA hybridization (dDDH) values and their confidence intervals (formula 2) using the Genome-to-Genome Distance Calculator (GGDC 2.12) under recommended settings (Meier-Kolthoff et al., 2013). ANI values were calculated with the OrthoANIu algorithm (Yoon et al., 2017). Whole-genome phylogeny was assessed based on 107 single-copy core genes found in a majority of bacteria (Dupont et al., 2012) using bcgTree (Ankenbrand and Keller, 2016). Visualization and annotation of the phylogenetic tree was performed using iTOL (Letunic and Bork, 2016).
To enable a comparative genomic study, each protein-coding gene (CDS) in the 68 Pandoraea genomes (n = 331,123) was functionally classified using the COG (Galperin et al., 2015) and KEGG orthologies (Kanehisa and Goto, 2000; Kanehisa et al., 2017). COGs were assigned by a reversed position-specific BLAST (RPSBLAST v2.6.0+) with an e-value cut-off of 1E-3 against the NCBI conserved domain database (CDD v3.16) (Tange, 2011). KEGG orthology was inferred using the KEGG automated annotation server (KAAS) (Moriya et al., 2007). Based on COG and K numbers, each CDS was assigned to the respective COG category and KEGG hierarchy. In case multiple COG categories were defined for the same COG, the first category was considered as the primary category. Protein orthologous groups (orthogroups) were inferred using OrthoFinder v2.2.7 (Emms and Kelly, 2015) with default parameters. For each orthogroup, we mapped the genomes and species in which it was present, the specificity (core, multiple species, single species or single isolate), and COG and KEGG functional classification.
Data mapping, visualization and statistical analyses were performed using RStudio with R v3.5.2. Pearson’s chi-square analyses were used to test the association between different sets of categorical variables. When a significant relationship was found between two variables, we further examined the standardized Pearson residuals. Standardized Pearson residuals with high absolute values indicate a lack of fit of the null hypothesis of independence in each cell (Agresti, 2002) and thus indicate observed cell frequencies in the contingency table that are significantly higher or lower than expected based on coincidence.
The G + C content of all strains was calculated from their genome sequences using Quast (Gurevich et al., 2013).
Biochemical characterization was performed as described previously (Draghi et al., 2014).
After a 24 h incubation period at 28°C on Tryptone Soya Agar (BD), a loopful of well-grown cells was harvested and fatty acid methyl esters were prepared, separated and identified using the Microbial Identification System (Microbial ID) as described previously (Vandamme et al., 1992).
The 16S rRNA gene sequences determined in the present study are publicly available through the GenBank/EMBL/DDBJ accession numbers listed in the species descriptions. Because the 16S rRNA sequences of Pandoraea species show high levels of similarity (Coenye et al., 2000; Daneshvar et al., 2001), gyrB gene sequence analysis has been introduced for species level identification of Pandoraea isolates (Coenye and LiPuma, 2002). To provide more robust phylogenetic analysis, partial sequences of the gyrB gene, and also of the recA and gltB genes were generated for a total of 84 Pandoraea reference strains and field isolates, and were used to construct phylogenetic trees (Figure 1 and Supplementary Figures S1, S2). The gltB, gyrB and gltB sequences determined in the present study are publicly available through the GenBank/EMBL/DDBJ accession numbers listed in Figure 1 and Supplementary Figures S1, S2 and in the species descriptions.
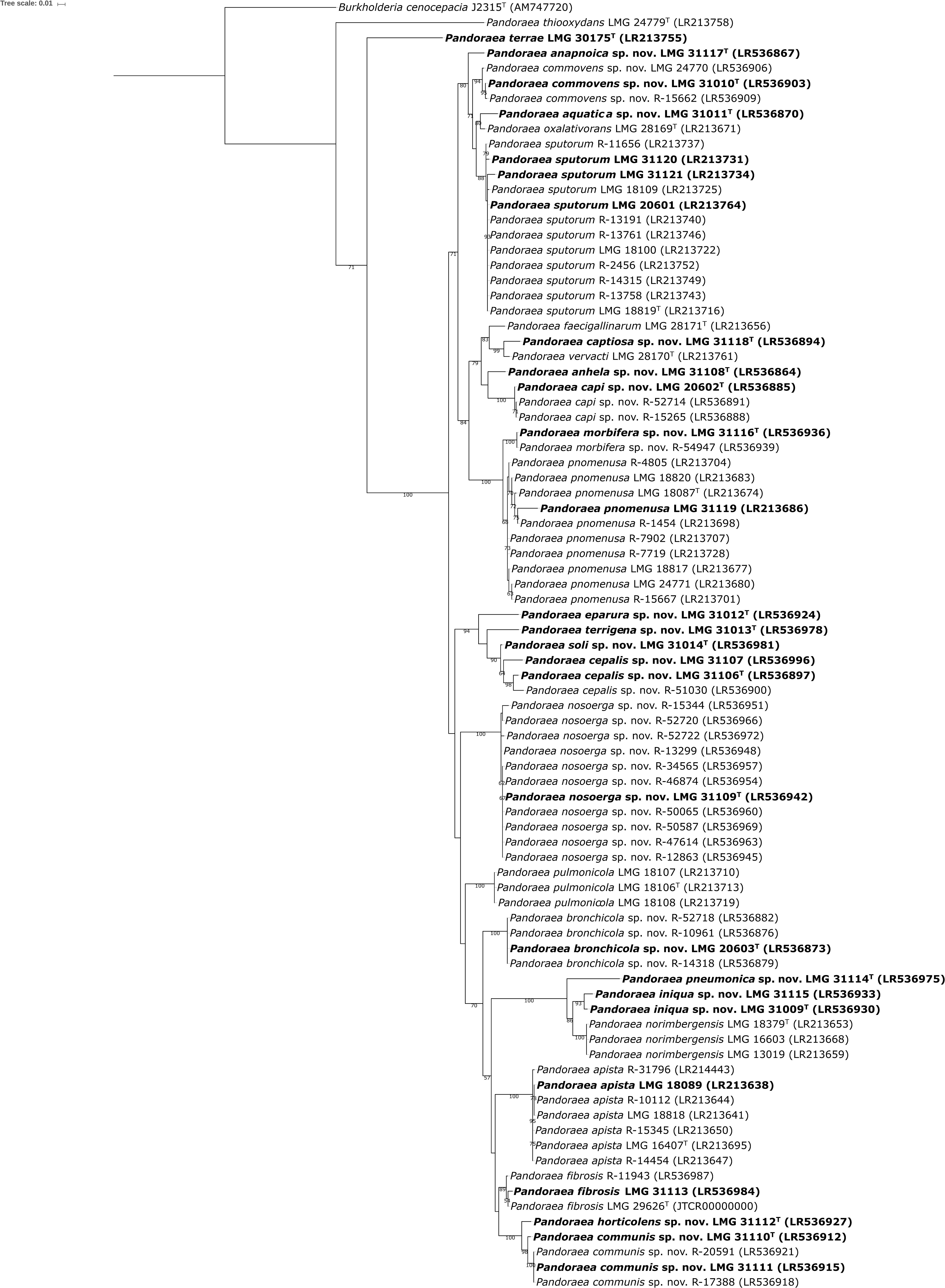
Figure 1. Phylogenetic tree based on partial gyrB sequences of all Pandoraea strains examined. Sequences (495–573 bp) were aligned based on their amino acid sequences and phylogeny was inferred using the Maximum Likelihood method and GTRCAT substitution model in RAxML. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches if greater than 50%. Burkholderia cenocepacia J2315T was used as outgroup. The scale bar indicates the number of substitutions per site. Isolates selected for whole-genome sequencing are shown in bold character type.
Overall, the three phylogenetic trees had comparable topologies, but while taxonomic reference strains of established Pandoraea species (Supplementary Table S1) and several groups of field isolates formed well-delineated clusters, others did not (Figure 1 and Supplementary Figures S1, S2). Each of these phylogenetic trees was therefore used to select a total of 27 isolates (shown in bold character type in Figure 1 and Supplementary Figures S1, S2) for whole-genome sequence analysis. These included 6 isolates that were tentatively assigned to established Pandoraea species using single locus sequence analyses, 20 isolates that clustered separately or whose assignment was equivocal, and P. terrae LMG 30175T, the sole Pandoraea type strain for which there was no publicly available whole-genome sequence at the time of writing.
The assembly of the Illumina HiSeq 150 bp paired end reads resulted in assemblies with 12–113 contigs and a total of 4.86–6.45 Mbp (Table 2 and Supplementary Table S2). The number of predicted CDS in the newly sequenced genomes ranged from 4,266 to 5,652 (Table 2). No clustered regularly interspaced short palindromic repeats (CRISPRs) were identified. The annotated assemblies of these 27 genomes were submitted to the European Nucleotide Archive and are publicly available through the GenBank/EMBL/DDBJ accession numbers listed in Table 2 and in the species descriptions. The G + C content of the newly sequenced strains, as calculated from their genome sequences, ranged from 62.3 to 66.1 mol% (Table 2). These values are similar those of other Pandoraea genomes, except for Ca. Pandoraea novymonadis that has a G + C content of 43.8% (Kostygov et al., 2017).
The 27 genomes from the present study were compared to all 41 publicly available Pandoraea genomes (GenBank database, January 29, 2019), which included 6 unclassified Pandoraea strains (Pushiri et al., 2013; Chan et al., 2015; Crofts et al., 2017; Kumar et al., 2018a; Liu et al., 2018). Pairwise dDDH and ANI values among the 68 genome sequences were calculated and are listed in Supplementary Tables S3, S4, respectively. Species delineation based on the 70% dDDH (Meier-Kolthoff et al., 2013) and 95–96% ANI thresholds (Yoon et al., 2017) yielded 30 species, which included the 11 validly named species, Ca. Pandoraea novymonadis, a total of 17 novel species for which we propose the names shown in Table 1, and a novel species represented by strains PE-S2R-1 and PE-S2T-3 (Crofts et al., 2017) (see below). One of these novel species, i.e., Pandoraea cepalis, corresponds with Pandoraea genomospecies 1, which we reported earlier (Coenye et al., 2000). Two novel species, i.e., Pandoraea capi and Pandoraea bronchicola, correspond with Pandoraea genomospecies 3 and 4, respectively, reported by Daneshvar et al. (2001). Finally, the phylogenomic data (Figure 2 and Supplementary Tables S3, S4), but also each of the single locus sequence analyses, showed that Pandoraea genomospecies 2 LMG 20602 should be classified as P. sputorum, which contradicts earlier wet-lab DNA-DNA hybridization results (Daneshvar et al., 2001).
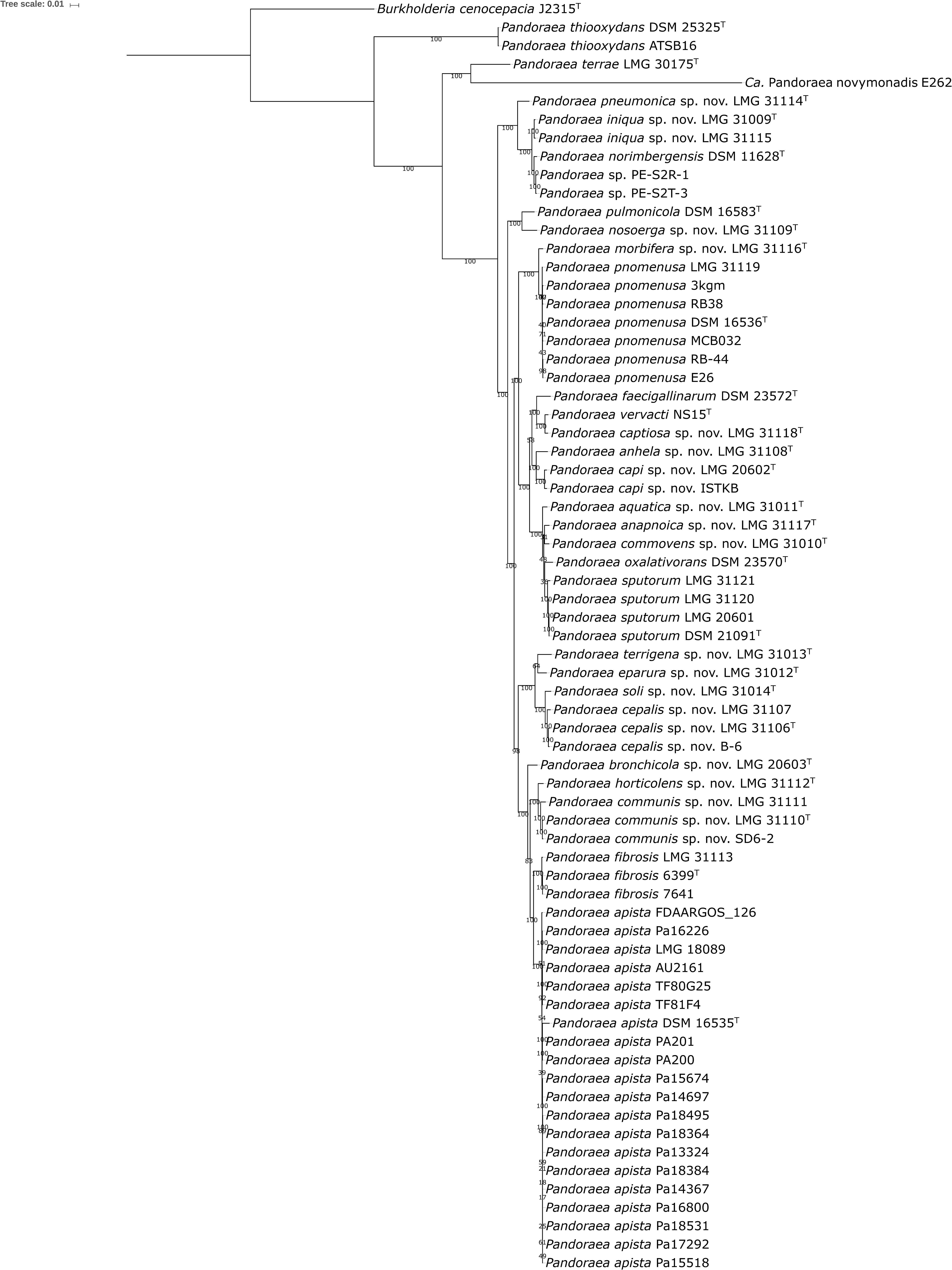
Figure 2. Phylogenetic tree based on 107 single-copy core genes. BcgTree was used to extract the nucleotide sequence of 107 single-copy core genes and to construct their phylogeny by partitioned maximum-likelihood analysis. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. Burkholderia cenocepacia J2315T was used as outgroup. Bar, 0.01 changes per nucleotide position.
The use of dDDH and ANI threshold levels was generally straightforward, yet some pairs of strains showed values close to the generally applied taxonomic threshold levels (Supplementary Tables S3, S4) (Meier-Kolthoff et al., 2013; Yoon et al., 2017). The two strains classified as P. capi showed 96.4% ANI and 69.6% dDDH with a dDDH confidence interval of 66.6–72.5%, and these strains were therefore classified as the same species. Similarly, the three strains classified as P. cepalis showed 96.2–98.4% ANI, 68.4–86.0% dDDH, and the 70% dDDH threshold level was in the confidence interval; these strains were therefore classified as one species. P. soli LMG 31014T showed 95.0–95.8% ANI and 60.7–65.0% dDDH toward P. cepalis strains, and the 70% dDDH threshold level was not part of the confidence interval, so this strain was classified as a separate species. Similarly, P. horticolens LMG 31112T showed 95.0-95.3% ANI and 60.0-62.2% dDDH toward P. communis, and the 70% dDDH threshold level was not part of the confidence interval so this strain was also classified as a separate species.
The phylogenomic analyses also allowed us to identify 4 out of 6 unclassified Pandoraea strains for which genome sequences are publicly available: strain ISTKB (Kumar M. et al., 2016) was assigned to P. capi, strain B-6 (Liu et al., 2018) to P. cepalis, strain SD6-2 (Pushiri et al., 2013) to P. communis, and strain E26 (Chan et al., 2015) to P. pnomenusa (Figure 2 and Table 2). Finally, strains PE-S2R-1 and PE-S2T-3 (Crofts et al., 2017) formed a separate cluster, which represented yet another novel Pandoraea species that remains to be formally classified (Supplementary Tables S3, S4).
The phylogenomic tree based on 107 single-copy marker genes was well resolved and the clusters delineated by dDDH and ANI formed monophyletic groups with a high bootstrap support (Figure 2). The clades in the phylogenomic tree of the present study showed a branching order similar to a previously published tree based on 119 conserved proteins (Kostygov et al., 2017). The results of the phylogenomic analyses along with the clustering in the individual recA, gyrB, and gltB single locus sequence analyses (Figure 1 and Supplementary Figures S1, S2) were used to identify each of the 84 isolates included in the present study. P. sputorum strain LMG 31121 clustered with the remaining P. sputorum strains in the gyrB and gltB trees but grouped aberrantly in the recA tree. In addition, P. cepalis proved particularly difficult to identify through single locus sequence analysis as it exhibited more variation in each of the sequences examined (Figure 1 and Supplementary Figures S1, S2) than any other Pandoraea species.
The type strains of each of 11 established Pandoraea species and of 17 novel Pandoraea species reported in the present study were included in an extensive phenotypic characterization. Among Pandoraea species, P. thiooxydans not only occupies a separate phylogenetic position (Figures 1, 2 and Supplementary Figures S1, S2) but also has a distinctive phenotype (Table 3). While all other Pandoraea species show normal growth on general microbiological growth media (i.e., they generate colonies of 1–4 mm in diameter after 2 days of incubation at 37°C), P. thiooxydans LMG 24779T requires prolonged incubation up to 7 days before the same colony size was obtained.
The following biochemical characteristics were shared by all Pandoraea strains investigated: growth at 15, 28, and 37°C, but not at 4°C; growth in the presence of 0–4% NaCl, but not in the presence of 6–10% NaCl; growth at pH 6, 7, and 8, but not at pH 4, 5, or 9. No anaerobic growth. Oxidase activity is present. No hydrolysis of starch or casein. No DNase activity. No denitrification. Assimilation of L-malate, but not L-arabinose, D-mannose, D-mannitol, N-acetylglucosamine, maltose or adipate. No fermentation of glucose. No indole production, esculin hydrolysis, arginine dihydrolase, urease or PNP-β-galactosidase activity, or liquefaction of gelatin. Leucine arylamidase activity is present, but no C8-ester-lipase, C14-lipase, valine or cystine arylamidase, trypsin, chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase or α-fucosidase activity.
An overview of biochemical characteristics useful for distinguishing the type strains of Pandoraea species is shown in Table 3.
The fatty acid profiles of all type strains are shown in Table 4. Both quantitative and qualitative differences were present. The predominant fatty acids in all strains investigated were C16:0, C17:0 cyclo, C16:0 3-OH, C18:1 ω7c, C19:0 cyclo ω8c, summed feature 2 (comprising C14:0 3-OH, C16:1 iso I, an unidentified fatty acid with equivalent chain length of 10.928, or C12:0 ALDE, or any combination of these fatty acids), or summed feature 3 (comprising C16:1 ω7c or C15:0 iso 2-OH or both).
The 68 Pandoraea genomes in the present study comprised 331,123 CDS, of which 273,692 (83%) and 128,054 (39%) could be assigned to the COG and KEGG orthologies, respectively (Supplementary Table S5). Orthologous genes were identified to determine the conserved genome content of the genus Pandoraea. Ortholog analysis revealed 10,783 orthogroups (325,879 CDS) in total, of which 738 (51,633 CDS) were present in all genomes, 897 (62,692 CDS) were present in all genomes except Ca. Pandoraea novymonadis, 8,003 (207,937 CDS) were present in multiple species, 1,130 (3,581 CDS) were species-specific and 15 (36 CDS) were isolate-specific (Figure 3). For further analyses, the core orthogroups were defined as those present in all genomes or all genomes except Ca. Pandoraea novymonadis (n = 1,635). COG and KEGG could be assigned to 7,243 (67%) and 3,655 (34%) of a total of 10,783 orthogroups (Supplementary Table S6). A previous pan genome analysis of 36 Pandoraea genomes by Wu et al. (2019) revealed a core genome of 1,903 CDS. As shown by these authors, the pan genome of Pandoraea is open (Wu et al., 2019) and the number of core genes decreases with an increasing number of genomes analyzed.
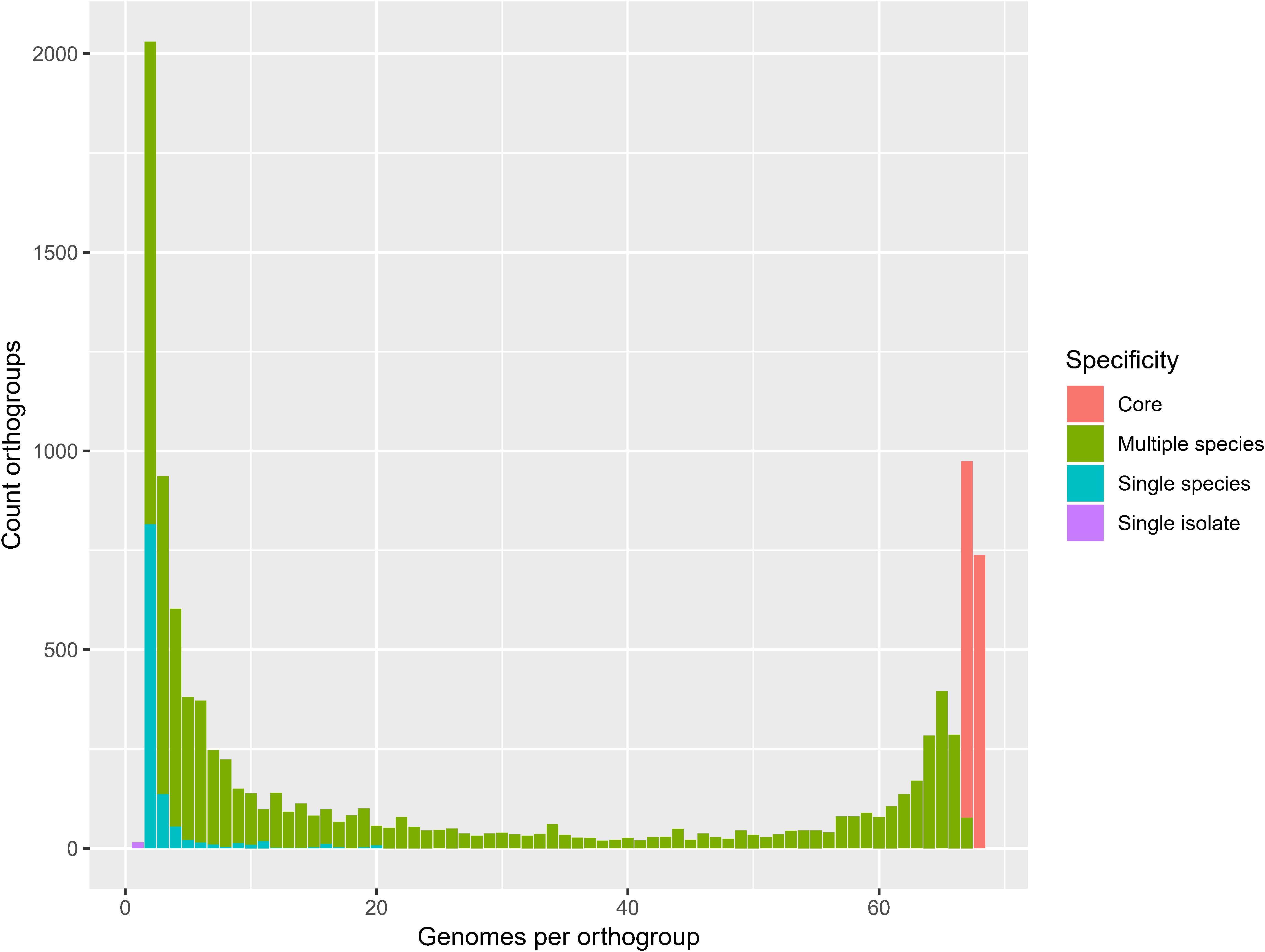
Figure 3. Genomes per orthogroup. Core, present in all genomes or all genomes except Ca. Pandoraea novymonadis.
The frequency of orthologous versus non-orthologous CDS varied significantly per isolate [X2(67) = 7423, p < 0.001] and species [X2(29) = 5863, p < 0.001]. The number of non-orthologous CDS per genome ranged from 0 to 632, with P. terrae LMG 30175T showing the highest percentage of non-orthologous CDS (Figure 4 and Supplementary Table S7). To identify biological functions that were over- or underrepresented in the core genome, we looked at the COG and KEGG functional classification of the orthogroups versus their specificity (core, multiple species, single species or single isolate). The specificity of the orthogroups varied significantly among the COG categories [X2(66) = 522, p < 0.001] and highest levels of the KEGG pathways [X2(10) = 130, p < 0.001]. The core orthogroups were significantly enriched in the COG categories Translation, ribosomal structure and biogenesis (J), Posttranslational modification, protein turnover, chaperones (O), Nucleotide transport and metabolism (F) and Coenzyme transport and metabolism (H) (Figure 5 and Supplementary Table S8) and in the KEGG pathway Genetic Information Processing (09120) (Figure 6 and Supplementary Table S9).
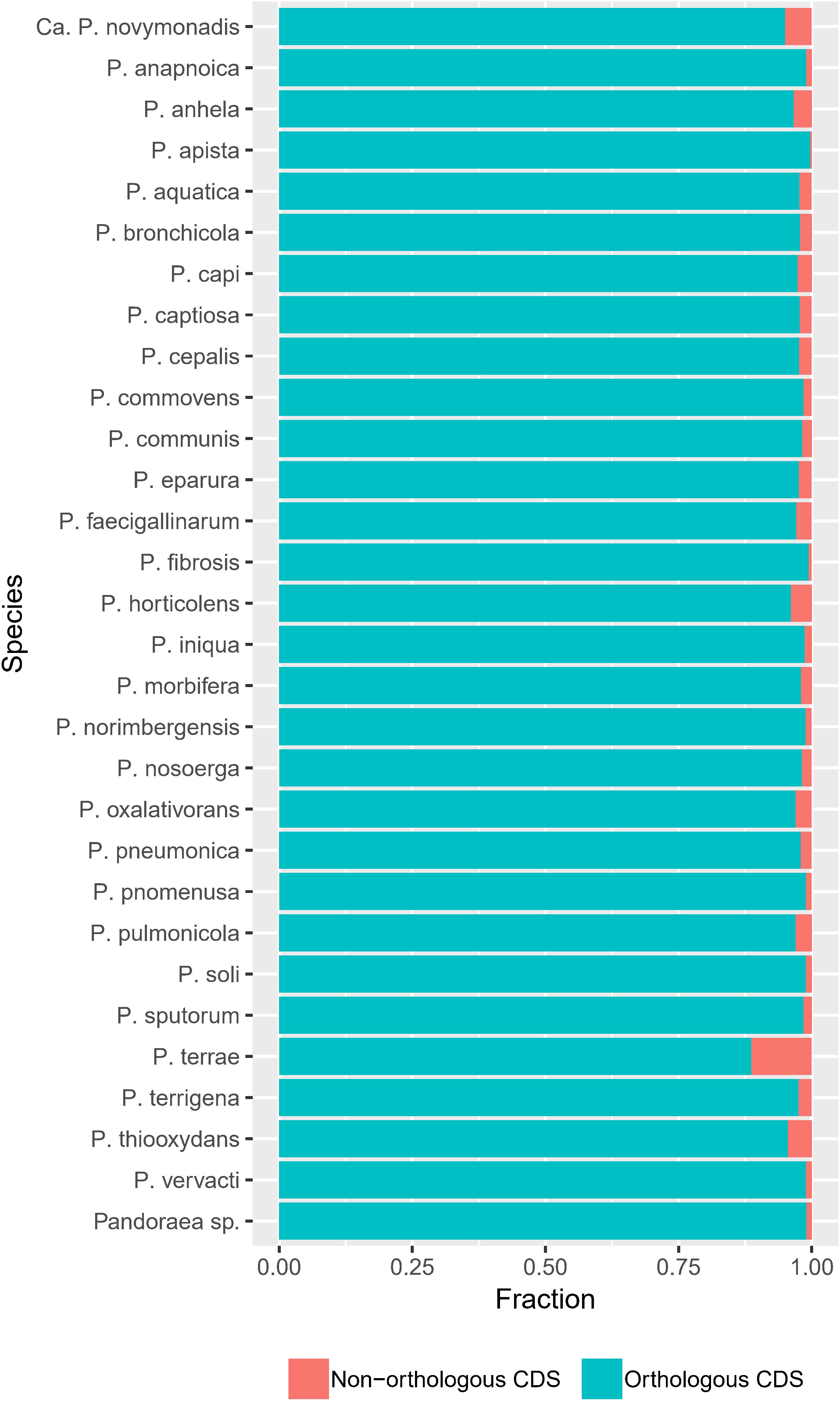
Figure 4. The frequency of orthologous versus non-orthologous CDS varies among species. Bar plots show the number of orthologous and non-orthologous CDS per species [X2(29) = 5863, p < 0.001].
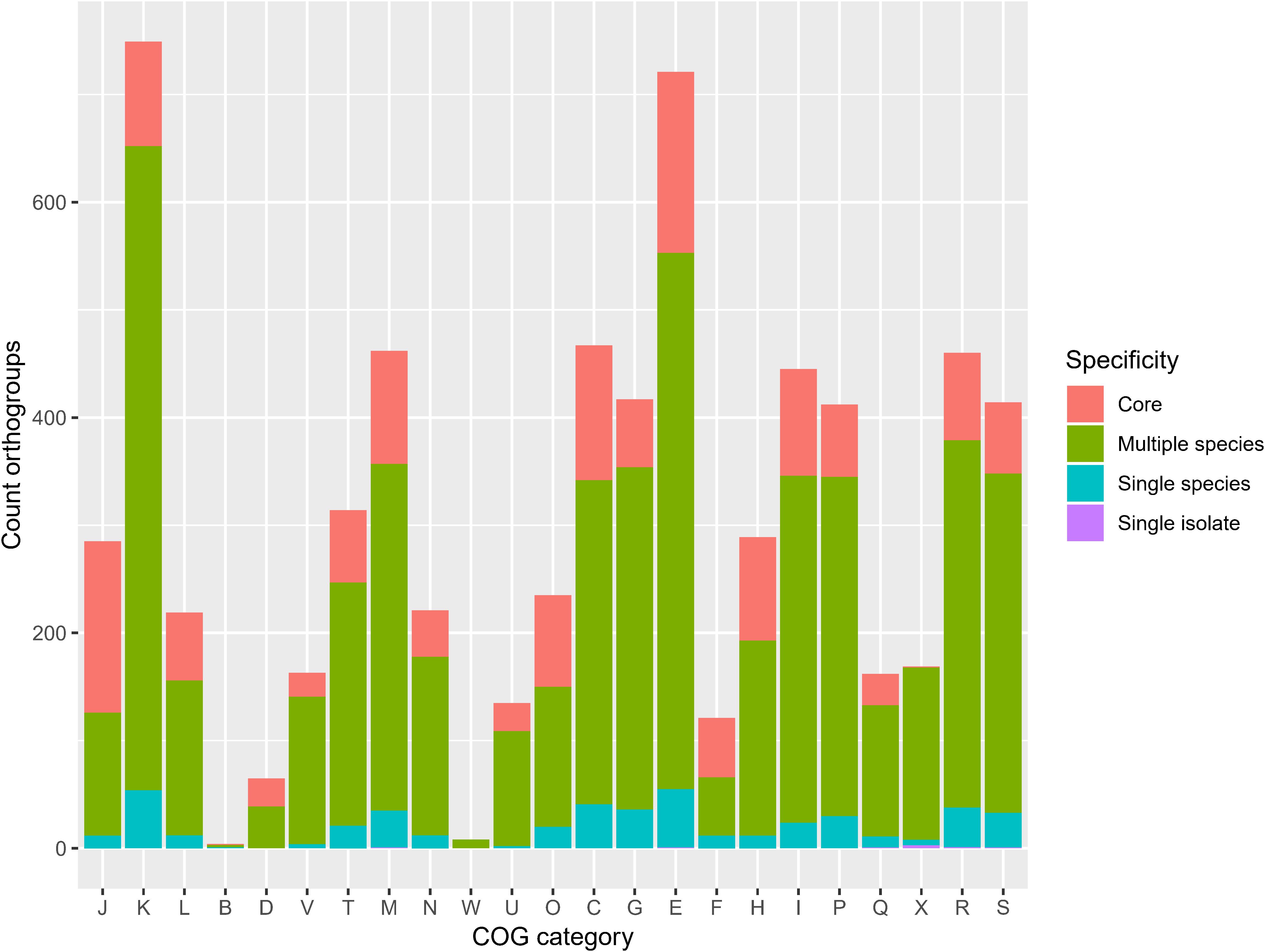
Figure 5. Orthogroup specificity varies among COG categories. Bar plot shows the number of orthogroups and their specificity per COG category [X2(66) = 522, p < 0.001]. J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; B, chromatin structure and dynamics; D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; W, extracellular structures; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, chaperones; X, mobilome: prophages, transposons; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown.
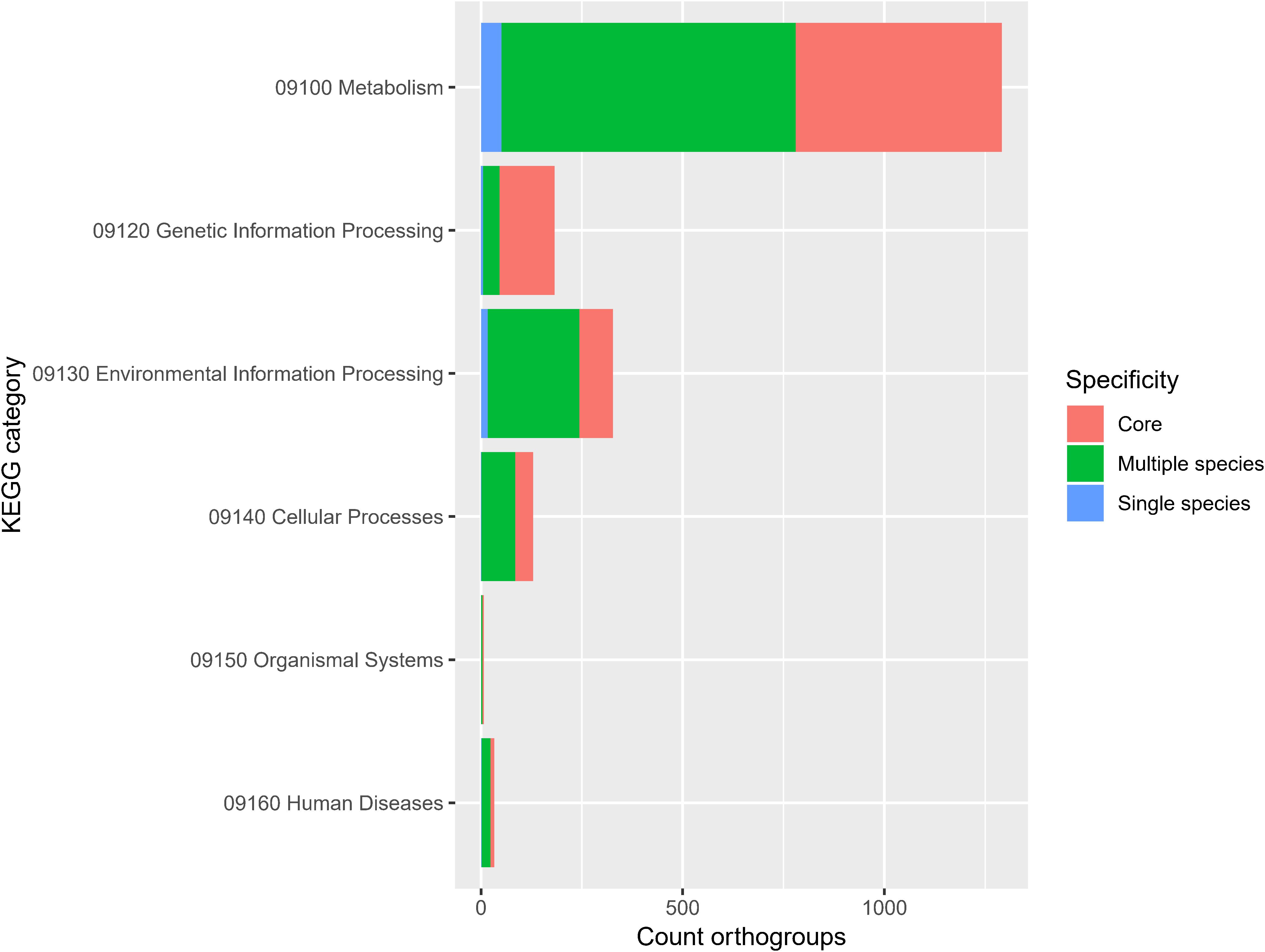
Figure 6. Orthogroup specificity varies among KEGG categories. Bar plot shows the number of orthogroups and their specificity per KEGG category [X2(10) = 130, p < 0.001].
Because many Pandoraea strains participate in the biodegradation of recalcitrant xenobiotics (Uhlik et al., 2012; Pushiri et al., 2013; Shi et al., 2013; Wang et al., 2015; Crofts et al., 2017; de Paula et al., 2017; Sarkar et al., 2017; Tirado-Torres et al., 2017; Kumar et al., 2018b; Yang et al., 2018; Liu et al., 2019; Wu et al., 2019), we specifically looked at the orthogroups in the KEGG pathway Xenobiotics biodegradation and metabolism (Figure 7). Most orthogroups in this pathway were present in multiple species (n = 28) and some were even present in the core Pandoraea genome (n = 6). This confirmed the potential of Pandoraea for degrading xenobiotics. In particular, the widespread capacity to utilize benzoate derivatives (Figure 7, pathways 362, 364, 627, and 633) explains why several strains have the potential to degrade lignin (Shi et al., 2013; Kumar et al., 2018a; Liu et al., 2019) and other aromatic compounds (Springael et al., 1996; Uhlik et al., 2012; Wang et al., 2015). Finally, P. fibrosis and P. thiooxydans showed a unique capacity to degrade specific compounds (Figure 7). P. fibrosis was only recently described and named after its origin from a cystic fibrosis patient (See-Too et al., 2019) but its unique capacity to degrade nitrotoluene derivatives is yet another example of the versatility in one Pandoraea species.
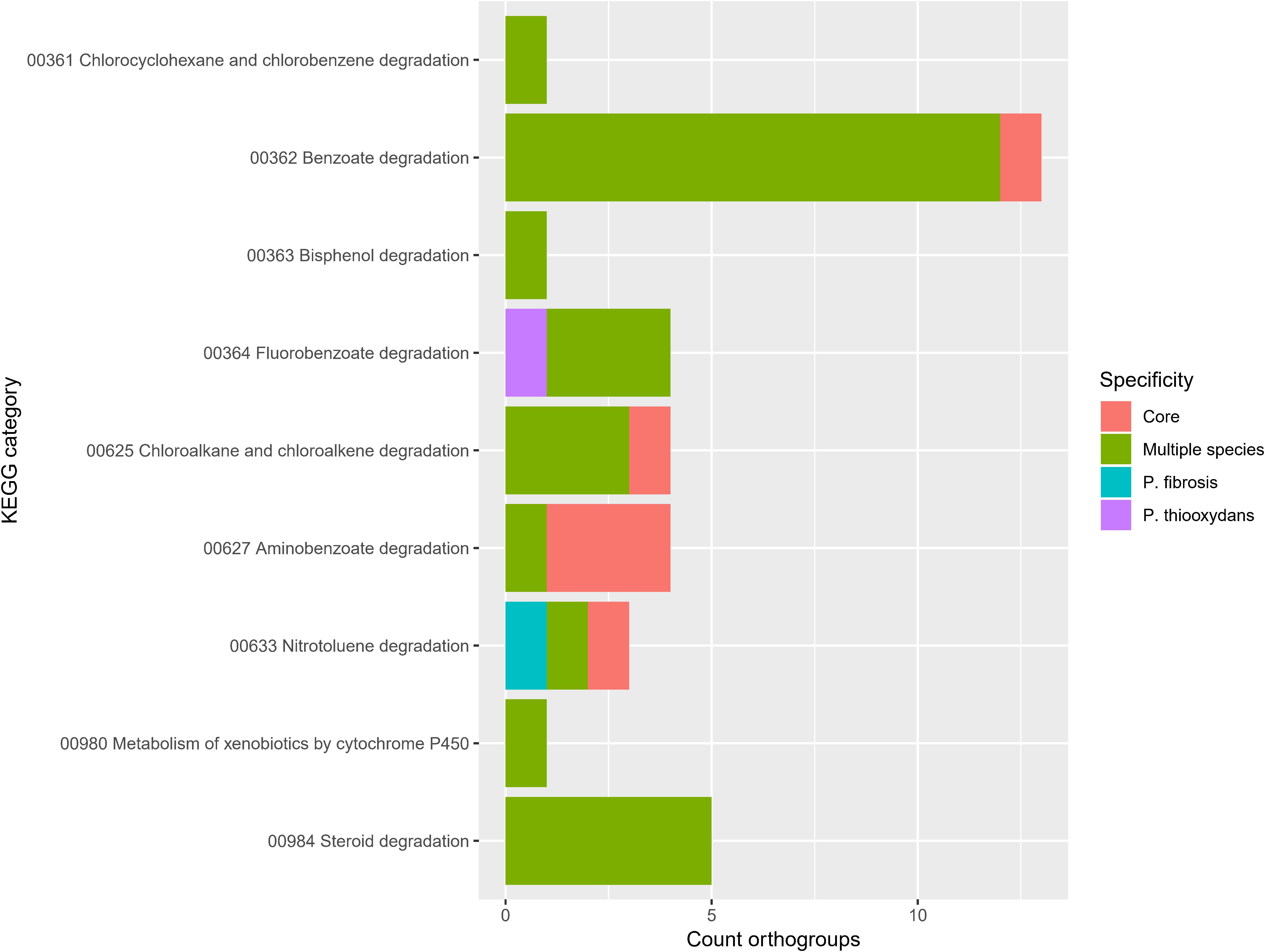
Figure 7. Orthogroup specificity in KEGG pathway Xenobiotics biodegradation and metabolism. Bar plot shows the number of orthogroups and their specificity.
The present study extends the number of formally named Pandoraea species considerably and makes reference cultures and their whole-genome sequences publicly available. The genus Pandoraea further emerges as a group of environmental bacteria with strong biodegradation capacities and as opportunistic human pathogens, especially in persons with cystic fibrosis. Within this genus, P. thiooxydans and P. terrae and Candidatus P. novymonadis cluster outside the main Pandoraea lineage. The aberrant phylogenomic position of the former is further supported by a distinctive phenotype. The classification of these bacteria within this monophyletic genus could therefore be questioned.
Taking into account the source and identification of strains ISTKB (a rhizospheric soil isolate, Kumar M. et al., 2016) and B-6 (an eroded bamboo slip isolate, Liu et al., 2018), and, to be as comprehensive as possible, also some additional unpublished own data (JL and PV), the novel species P. aquatica, P. capi, P. cepalis, P. commovens, P. communis, and P. iniqua, but also the established species P. faecigallinarum, P. norimbergensis, P. pnomenusa, and P. fibrosis, have all been isolated from both human clinical and environmental sources. Thus far, the novel species P. anapnoica, P. anhela, P. bronchicola, P. captiosa, P. morbifera, P. nosoerga, and P. pneumonica, but also the established species P. apista, P. pulmonicola, and P. sputorum, have all been isolated from human clinical sources only; while the novel species P. eparura, P. horticolens, P. soli and P. terrigena, and the established species P. oxalativorans, P. terrae, P. thiooxydans, and P. vervacti have thus far been isolated from environmental samples only.
The present study provides genomic, chemotaxonomic and phenotypic data that enable a formal proposal of 17 novel Pandoraea species as outlined below. By making reference cultures and whole-genome sequences of each of these versatile bacteria publicly available, we aim to contribute to future knowledge about the metabolic versatility and pathogenicity of these organisms.
Pandoraea anapnoica sp. nov. (a.na.pnoi’ca. Gr. masc. adj. anapnoikos, affecting respiration; N.L. fem. adj. anapnoica affecting respiration).
The phenotypic description is as presented above and in Table 3.
Isolated from human clinical samples in the United States.
The type strain is LMG 31117T (=CCUG 73385T) and was isolated from a cystic fibrosis specimen in the United States in 1999. Its G + C content is 62.4 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 31117T are publicly available through the accession numbers LR536847, LR536866–LR536868, and CABPSP010000000, respectively.
Pandoraea anhela sp. nov. (an.he’la. L. fem. adj. anhela breath-taking).
The phenotypic description is as presented above and in Table 3.
Isolated from human clinical samples in the United States.
The type strain is LMG 31108T (=CCUG 73386T) and was isolated from a cystic fibrosis specimen in the United States in 2006. Its G + C content is 63.4 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 31108T are publicly available through the accession numbers LR536848, LR536863-LR536865 and CABPSB010000000, respectively.
Pandoraea aquatica sp. nov. (a.qua’ti.ca. L. fem. adj. aquatica aquatic).
The phenotypic description is as presented above and in Table 3.
Isolated from human clinical samples in the United States and from pond water in Belgium.
The type strain is LMG 31011T (=CCUG 73384T) and was isolated from pond water in a greenhouse in Belgium in 2013. Its G + C content is 62.9 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 31011T are publicly available through the accession numbers LR536849, LR536869–LR536871, and CABPSN010000000, respectively.
Pandoraea bronchicola sp. nov. (bron.chi’co.la. L. neut. pl. n. bronchia, the bronchial tubes; L. suff. -cola [from L. n. incola] a dweller, inhabitant; N.L. fem. n. bronchicola a dweller of bronchi, coming from the bronchi).
The phenotypic description is as presented above and in Table 3.
Isolated from human clinical samples in the United States.
The type strain is LMG 20603T (= ATCC BAA-110T = CDC H652T) and was isolated from cystic fibrosis sputum in the United States in 1998. Its G + C content is 63.0 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 20603T are publicly available through the accession numbers LR536994, LR536872–LR536874, and CABPST010000000, respectively.
Pandoraea capi sp. nov. (ca’pi. Gr. masc. n. kapos, breath; N.L. gen. n. capi, referring to the lung as niche of these bacteria).
The phenotypic description is as presented above and in Table 3.
Isolated from human clinical samples in the United States and from rhizospheric soil in India.
The type strain is LMG 20602T (=ATCC BAA-109T = CDC G9805T) and was isolated from sputum of a non-cystic fibrosis patient in the United States in 1996. Its G + C content is 63.4 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 20602T are publicly available through the accession numbers LR536850, LR536884–LR536886, and CABPRV010000000, respectively.
Pandoraea captiosa sp. nov. (cap.ti.o’sa. L. fem. adj. captiosa, harmful, disadvantageous).
The phenotypic description is as presented above and in Table 3.
Isolated from human clinical samples in the United States.
The type strain is LMG 31118T (=CCUG 73387T) and was isolated from a cystic fibrosis specimen in the United States in 2008. Its G + C content is 63.3 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 31118T are publicly available through the accession numbers LR536851, LR536893–LR536895, and CABPSQ010000000, respectively.
Pandoraea cepalis sp. nov. [ce.pa’lis. Gr. n. kepos, garden; -alis L. adjective forming suffix, pertaining to; N.L. fem. adj. cepalis pertaining to garden (soil)].
The phenotypic description is as presented above and in Table 3.
Isolated from soil and water samples in Belgium and the Netherlands, from human clinical samples in the United States, and from historical bamboo slips in China.
The type strain is LMG 31106T (=CCUG 39680T) and was isolated from garden soil in The Netherlands. Its G + C content is 63.7 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 31106T are publicly available through the accession numbers LR536852, LR536896–LR536898, and CABPSL010000000, respectively.
Pandoraea commovens sp. nov. (com.mo’vens. L. v. commovere, to trouble, upset; L. pres. part. commovens troubling).
The phenotypic description is as presented above and in Table 3.
Isolated from human clinical samples in Belgium and the United States, from soil samples in Belgium, and from plant roots in India.
The type strain is LMG 31010T (=CCUG 73378T) and was isolated from sputum of a cystic fibrosis patient in Belgium in 2002. Its G + C content is 62.6 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 31010T are publicly available through the accession numbers LR536853, LR536902–LR536904, and CABPSA010000000, respectively.
Pandoraea communis sp. nov. (com.mu’nis. L. fem. adj. communis common, widespread).
The phenotypic description is as presented above and in Table 3.
Isolated from human clinical, soil and water samples in Belgium, and from soil in Australia.
The type strain is LMG 31110T (=CCUG 73383T) and was isolated from sputum of a cystic fibrosis patient in Belgium in 2012. Its G + C content is 62.6 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 31110T are publicly available through the accession numbers LR536854, LR536911-LR536913 and CABPSJ010000000, respectively.
Pandoraea eparura sp. nov. (ep.a.ru’ra. Gr. masc. adj. eparouros, attached to the soil; N.L. fem. adj. eparura attached to the soil).
The phenotypic description is as presented above and in Table 3.
The type (and thus far only) strain is LMG 31012T (=CCUG 73380T) and was isolated from soil of a house plant in Belgium in 2003. Its G + C content is 63.7 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 31012T are publicly available through the accession numbers LR536855, LR536923–LR536925, and CABPSH010000000, respectively.
Pandoraea horticolens sp. nov. (hor.ti’co.lens. L. n. hortus garden; L. v. colere to dwell; L. pres. part. colens dwelling; N.L. part. adj. horticolens because the type strain was isolated from garden [soil]).
The phenotypic description is as presented above and in Table 3.
The type (and thus far only) strain is LMG 31112T (=CCUG 73379T) and was isolated from garden soil in Belgium in 2003. Its G + C content is 62.3 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 31112T are publicly available through the accession numbers LR536857, LR536926-LR536928 and CABPSM010000000, respectively.
Pandoraea iniqua sp. nov. (in.i’qua. L. fem. adj. iniqua disadvantageous, hostile).
The phenotypic description is as presented above and in Table 3.
Isolated from soil samples in Belgium and human clinical samples in the United States.
The type strain is LMG 31009T (=CCUG 73377T) and was isolated from maize rhizosphere soil in Belgium in 2002. Its G + C content is 63.1 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 31009T are publicly available through the accession numbers LR536856, LR536929–LR536931, and CABPSF010000000, respectively.
Pandoraea morbifera sp. nov. (mor.bi’fe.ra, L. fem. adj. morbifera that brings disease).
The phenotypic description is as presented above and in Table 3.
Isolated from human clinical samples in the United States.
The type strain is LMG 31116T (=CCUG 73389T) and was isolated from a cystic fibrosis specimen in the United States in 2006. Its G + C content is 64.7 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 31116T are publicly available through the accession numbers LR536858, LR536935–LR536937, and CABPSD010000000, respectively.
Pandoraea nosoerga sp. nov. (no.so.er’ga, Gr. masc. adj. nosoergos, causing sickness; N.L. fem. adj. nosoerga).
The phenotypic description is as presented above and in Table 3.
Isolated from human clinical samples in Australia, Belgium, Germany, United Kingdom and the United States.
The type strain is LMG 31109T (=CCUG 73390T) and was isolated from a cystic fibrosis specimen in the United States in 2008. Its G + C content is 66.1 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 31109T are publicly available through the accession numbers LR536859, LR536941-LR536943 and CABPSC010000000, respectively.
Pandoraea pneumonica sp. nov. (pneu.mo’ni.ca, Gr. masc. adj. pneumonikos, of the lungs; N.L. fem. adj. pneumonica).
The phenotypic description is as presented above and in Table 3.
The type (and thus far only) strain is LMG 31114T (=CCUG 73388T) and was isolated from a cystic fibrosis specimen in the United States in 2009. Its G + C content is 62.5 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 31114T are publicly available through the accession numbers LR536861, LR536974–LR536976, and CABPSK010000000, respectively.
Pandoraea soli sp. nov. (so’li. L. gen. n. soli of soil, the source of the type strain).
The phenotypic description is as presented above and in Table 3.
The type (and thus far only) strain is LMG 31014T (=CCUG 73382T) and was isolated from soil of a house plant in Belgium in 2003. Its G + C content is 63.6 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 31014T are publicly available through the accession numbers LR536860, LR536980–LR536982, and CABPSG010000000, respectively.
Pandoraea terrigena sp. nov. (ter.ri’ge.na. L. fem. n. terra soil; L. v. gignere to bear; L. fem. n. terrigena [nominative in apposition] born of, or from, soil, soil-born).
The phenotypic description is as presented above and in Table 3.
The type (and thus far only) strain is LMG 31013T (=CCUG 73381T) and was isolated from soil of a house plant in Belgium in 2003. Its G + C content is 63.5 mol% (calculated based on its genome sequence). The 16S rRNA, gltB, gyrB, recA and whole-genome sequence of LMG 31013T are publicly available through the accession numbers LR536862, LR536977–LR536979, and CABPRU010000000, respectively.
The datasets generated for this study can be found in the European Nucleotide Archive PRJEB30806, PRJEB30685, PRJEB30686, PRJEB30687, PRJEB30688, PRJEB30689, PRJEB30690, PRJEB30807, PRJEB30745, PRJEB30746, PRJEB30808, PRJEB30691, PRJEB30692, PRJEB30809, PRJEB30810, PRJEB30693, PRJEB30694, PRJEB30695, PRJEB30696, PRJEB30697, PRJEB30699, PRJEB30700, PRJEB30701, PRJEB30698, PRJEB30811, PRJEB30702, PRJEB30703, PRJEB30812, PRJEB30704, PRJEB30705, PRJEB30706, PRJEB30707, PRJEB30708, PRJEB30714, PRJEB30709, PRJEB30710, PRJEB30711, PRJEB30712, PRJEB30713, PRJEB30813, PRJEB30814, PRJEB30815, PRJEB30755, PRJEB30724, PRJEB30756, PRJEB30725, PRJEB30726, PRJEB30727, PRJEB30728, PRJEB30721, PRJEB30722, PRJEB30723, PRJEB30757, PRJEB30715, PRJEB30716, PRJEB30717, PRJEB30753, PRJEB30752, PRJEB30754, PRJEB30740, PRJEB30741, PRJEB30742, PRJEB30743, PRJEB30718, PRJEB30744, PRJEB30748, PRJEB30749, PRJEB30750, PRJEB30751, PRJEB30729, PRJEB30730, PRJEB30731, PRJEB30732, PRJEB30733, PRJEB30734, PRJEB30735, PRJEB30736, PRJEB30737, PRJEB30738, PRJEB30739, PRJEB30747, PRJEB30720, and PRJEB30719.
PV, JL, and CP conceived the study. PV and CP wrote the manuscript. EDB, EDC, TS, and CP performed single locus sequence analyses. CP performed phylogenetic analyses. CP, ED, and BV carried out the genomic data analyses. EDC, MC, EDB, and CS carried out wet-lab phenotypical analyses. PV and JL generated the required funding. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Part of this work was performed in the framework of the Belgian National Reference Centre for Burkholderia, supported by the Ministry of Social Affairs through a fund within the National Health Insurance System. This funding agency had no role in study design, data collection and interpretation, or the decision to submit the work for publication. JL and TS receive support from the Cystic Fibrosis Foundation (United States).
We thank the Oxford Genomics Centre at the Welcome Centre for Human Genetics (funded by Wellcome Trust grant reference 203141/Z/16/Z) for the generation and initial processing of the sequencing data. We thank colleagues of the following referring laboratories for depositing some of the strains listed in Table 1: D. Piérard (Department of Microbiology and Infection Control, Universitair Ziekenhuis Brussel, Vrije Universiteit Brussel, Brussels, Belgium), G. Ieven (Laboratory of Medical Microbiology, Universitair Ziekenhuis Antwerpen, Antwerp, Belgium), H. Franckx (Revalidation Centre Zeepreventorium, De Haan, Belgium) and K. De Boeck (Department of Pediatric Pulmonology and Infectious Diseases, Universitair Ziekenhuis Leuven, Leuven, Belgium).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02556/full#supplementary-material
ANI, average nucleotide identity; COG, cluster of orthologous groups; dDDH, digital DNA–DNA hybridization; GBDP, genome blast distance phylogeny; GGDC, genome-to-genome distance calculator; KEGG, kyoto encyclopedia of genes and genomes.
Agresti, A. (2002). “Inference for contingency tables,” in Categorical Data Analysis, ed. A. Agresti, (Hoboken, NJ: John Wiley & Sons, Inc), 70–114. doi: 10.1002/0471249688.ch3
Ambrose, M., Malley, R. C., Warren, S. J. C., Beggs, S. A., Swallow, O. F. E., McEwan, B., et al. (2016). Pandoraea pnomenusa isolated from an Australian patient with cystic fibrosis. Front. Microbiol. 7:692. doi: 10.3389/fmicb.2016.00692
Anandham, R., Indiragandhi, P., Kwon, S. W., Sa, T. M., Jeon, C. O., Kim, Y. K., et al. (2010). Pandoraea thiooxydans sp. nov., a facultatively chemolithotrophic, thiosulfate-oxidizing bacterium isolated from rhizosphere soils of sesame (Sesamum indicum L.). Int. J. Syst. Evol. Microbiol. 60, 21–26. doi: 10.1099/ijs.0.012823-0
Ankenbrand, M. J., and Keller, A. (2016). bcgTree: automatized phylogenetic tree building from bacterial core genomes. Genome 59, 783–791. doi: 10.1139/gen-2015-0175
Atkinson, R. M., LiPuma, J. J., Rosenbluth, D. B., and Dunne, W. M. (2006). Chronic colonization with Pandoraea apista in cystic fibrosis patients determined by repetitive-element-sequence PCR. J. Clin. Microbiol. 44, 833–836. doi: 10.1128/JCM.44.3.833-836.2006
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Chan, K., Yin, W.-F., Tee, K. K., Chang, C., and Priya, K. (2015). Pandoraea sp. strain E26: discovery of its quorum-sensing properties via whole-genome sequence analysis. Genome Announc. 3, 26–27. doi: 10.1128/genomeA.00565-15
Coenye, T., Falsen, E., Hoste, B., Ohlén, M., Goris, J., Govan, J. R. W., et al. (2000). Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pulmonicola sp. nov., Pandoraea pnomenusa sp. nov., Pandoraea sputorum sp. nov. and Pandoraea norimbergensis comb. nov. Int. J. Syst. Evol. Microbiol. 50, 887–899. doi: 10.1099/00207713-50-2-887
Coenye, T., and LiPuma, J. J. (2002). Use of the gyrB gene for the identification of Pandoraea species. FEMS Microbiol. Lett. 208, 15–19. doi: 10.1111/j.1574-6968.2002.tb11053.x
Crofts, T. S., Wang, B., Spivak, A., Gianoulis, T. A., Forsberg, K. J., Gibson, M. K., et al. (2017). Draft genome sequences of three β-lactam-catabolizing soil Proteobacteria. Genome Announc. 5, 10–12. doi: 10.1128/genomeA.00653-17
Daneshvar, M. I., Hollis, D. G., Steigerwalt, A. G., Whitney, A. M., Spangler, L., Douglas, M. P., et al. (2001). Assignment of CDC weak oxidizer group 2 (WO-2) to the genus Pandoraea and characterization of three new Pandoraea genomo species. J. Clin. Microbiol. 39, 1819–1826. doi: 10.1128/JCM.39.5.1819-1826.2001
de Paula, F. C., de Paula, C. B. C., Gomez, J. G. C., Steinbüchel, A., and Contiero, J. (2017). Poly(3-hydroxybutyrate-co-3-hydroxyvalerate) production from biodiesel by-product and propionic acid by mutant strains of Pandoraea sp. Biotechnol. Prog. 33, 1077–1084. doi: 10.1002/btpr.2481
Degand, N., Lotte, R., Deconde Le Butor, C., Segonds, C., Thouverez, M., Ferroni, A., et al. (2015). Epidemic spread of Pandoraea pulmonicola in a cystic fibrosis center. BMC Infect. Dis. 15:583. doi: 10.1186/s12879-015-1327-8
Dong, L., Xu, J., Zhang, L., Cheng, R., Wei, G., Su, H., et al. (2018). Rhizospheric microbial communities are driven by Panax ginseng at different growth stages and biocontrol bacteria alleviates replanting mortality. Acta Pharm. Sin. B 8, 272–282. doi: 10.1016/j.apsb.2017.12.011
Draghi, W. O., Peeters, C., Cnockaert, M., Snauwaert, C., Wall, L. G., Zorreguieta, A., et al. (2014). Burkholderia cordobensis sp. nov., from agricultural soils. Int. J. Syst. Evol. Microbiol. 64, 2003–2008. doi: 10.1099/ijs.0.059667-0
Dupont, C., Aujoulat, F., Chiron, R., Condom, P., Jumas-Bilak, E., and Marchandin, H. (2017). Highly diversified Pandoraea pulmonicola population during chronic colonization in cystic fibrosis. Front. Microbiol. 8:1892. doi: 10.3389/fmicb.2017.01892
Dupont, C. L., Rusch, D. B., Yooseph, S., Lombardo, M.-J., Alexander Richter, R., Valas, R., et al. (2012). Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 6, 1186–1199. doi: 10.1038/ismej.2011.189
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Ee, R., Ambrose, M., Lazenby, J., Williams, P., Chan, K.-G., and Roddam, L. (2015). Genome sequences of two Pandoraea pnomenusa isolates recovered 11 months apart from a cystic fibrosis patient. Genome Announc. 3:e1389-14. doi: 10.1128/genomeA.01389-14
Emms, D. M., and Kelly, S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. doi: 10.1186/s13059-015-0721-2
Galperin, M. Y., Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2015). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269. doi: 10.1093/nar/gku1223
Green, H., and Jones, A. M. (2018). Emerging Gram-negative bacteria: pathogenic or innocent bystanders. Curr. Opin. Pulm. Med. 24, 592–598. doi: 10.1097/MCP.0000000000000517
Greninger, A. L., Streithorst, J., Golden, J. A., Chiu, C. Y., and Miller, S. (2017). Complete genome sequence of sequential Pandoraea apista isolates from the same cystic fibrosis patient supports a model of chronic colonization with in vivo strain evolution over time. Diagn. Microbiol. Infect. Dis. 87, 1–6. doi: 10.1016/j.diagmicrobio.2016.10.013
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Jeong, S. E., Lee, H. J., Jia, B., and Jeon, C. O. (2016). Pandoraea terrae sp. nov., isolated from forest soil, and emended description of the genus Pandoraea Coenye et al., 2000. Int. J. Syst. Evol. Microbiol. 66, 3524–3530. doi: 10.1099/ijsem.0.001229
Jin, Z.-X., Wang, C., Dong, W., and Li, X. (2007). Isolation and some properties of newly isolated oxalate-degrading Pandoraea sp. OXJ-11 from soil. J. Appl. Microbiol. 103, 1066–1073. doi: 10.1111/j.1365-2672.2007.03363.x
Johnson, L. N., Han, J.-Y., Moskowitz, S. M., Burns, J. L., Qin, X., and Englund, J. A. (2004). Pandoraea bacteremia in a cystic fibrosis patient with associated systemic illness. Pediatr. Infect. Dis. J. 23, 881–882. doi: 10.1097/01.inf.0000136857.74561.3c
Jørgensen, I. M., Johansen, H. K., Frederiksen, B., Pressler, T., Hansen, A., Vandamme, P., et al. (2003). Epidemic spread of Pandoraea apista, a new pathogen causing severe lung disease in cystic fibrosis patients. Pediatr. Pulmonol. 36, 439–446. doi: 10.1002/ppul.10383
Jurelevicius, D., Korenblum, E., Casella, R., Vital, R. L., and Seldin, L. (2010). Polyphasic analysis of the bacterial community in the rhizosphere and roots of Cyperus rotundus L. grown in a petroleum-contaminated soil. J. Microbiol. Biotechnol. 20, 862–870. doi: 10.4014/jmb.0910.10012
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y., and Morishima, K. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361. doi: 10.1093/nar/gkw1092
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kokcha, S., Bittar, F., Reynaud-Gaubert, M., Mely, L., Gomez, C., Gaubert, J.-Y., et al. (2013). Pandoraea pulmonicola chronic colonization in a cystic fibrosis patient, France. New Microbes New Infect. 1, 27–29. doi: 10.1002/2052-2975.16
Kokot, M., Dlugosz, M., and Deorowicz, S. (2017). KMC 3: counting and manipulating k-mer statistics. Bioinformatics 33, 2759–2761. doi: 10.1093/bioinformatics/btx304
Kostygov, A. Y., Butenko, A., Nenarokova, A., Tashyreva, D., Flegontov, P., Lukes, J., et al. (2017). Genome of Ca. Pandoraea novymonadis, an endosymbiotic bacterium of the trypanosomatid Novymonas esmeraldas. Front. Microbiol. 8:1940. doi: 10.3389/fmicb.2017.01940
Kumar, M., Gazara, R. K., Verma, S., Kumar, M., Verma, P. K., and Thakur, I. S. (2016). Genome sequence of Pandoraea sp. ISTKB, a lignin-degrading Betaproteobacterium, isolated from Rhizospheric soil. Genome Announc. 4, 275–287. doi: 10.1128/genomeA.01240-16
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kumar, M., Mishra, A., Singh, S. S., Srivastava, S., and Thakur, I. S. (2018a). Expression and characterization of novel laccase gene from Pandoraea sp. ISTKB and its application. Int. J. Biol. Macromol. 115, 308–316. doi: 10.1016/j.ijbiomac.2018.04.079
Kumar, M., Verma, S., Gazara, R. K., Kumar, M., Pandey, A., Verma, P. K., et al. (2018b). Genomic and proteomic analysis of lignin degrading and polyhydroxyalkanoate accumulating β-proteobacterium Pandoraea sp. ISTKB. Biotechnol. Biofuels 11, 1–23. doi: 10.1186/s13068-018-1148-2
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [preprint]. Available at: http://arxiv.org/abs/1303.3997. (accessed June 8, 2015).
Liu, D., Yan, X., Si, M., Deng, X., Min, X., Shi, Y., et al. (2019). Bioconversion of lignin into bioplastics by Pandoraea sp. B-6: molecular mechanism. Environ. Sci. Pollut. Res. 26, 2761–2770. doi: 10.1007/s11356-018-3785-1
Liu, D., Yan, X., Zhuo, S., Si, M., Liu, M., Wang, S., et al. (2018). Pandoraea sp. B-6 assists the deep eutectic solvent pretreatment of rice straw via promoting lignin depolymerization. Bioresour. Technol. 257, 62–68. doi: 10.1016/j.biortech.2018.02.029
Martina, P. F., Martínez, M., Frada, G., Alvarez, F., Leguizamón, L., Prieto, C., et al. (2017). First time identification of Pandoraea sputorum from a patient with cystic fibrosis in Argentina: a case report. BMC Pulm. Med. 17:33. doi: 10.1186/s12890-017-0373-y
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H.-P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. doi: 10.1093/nar/gkm321
Parsons, J. R., Sijm, D. T. H. M., van Laar, A., and Hutzinger, O. (1988). Biodegradation of chlorinated biphenyls and benzoic acids by a Pseudomonas strain. Appl. Microbiol. Biotechnol. 29, 81–84. doi: 10.1007/BF00258355
Peeters, C., Depoorter, E., Praet, J., and Vandamme, P. (2016). Extensive cultivation of soil and water samples yields various pathogens in patients with cystic fibrosis but not Burkholderia multivorans. J. Cyst. Fibros. 15, 769–775. doi: 10.1016/j.jcf.2016.02.014
Peeters, C., Zlosnik, J. E. A., Spilker, T., Hird, T. J., LiPuma, J. J., and Vandamme, P. (2013). Burkholderia pseudomultivorans sp. nov., a novel Burkholderia cepacia complex species from human respiratory samples and the Rhizosphere. Syst. Appl. Microbiol. 36, 483–489. doi: 10.1016/j.syapm.2013.06.003
Pimentel, J. D., and MacLeod, C. (2008). Misidentification of Pandoraea sputorum isolated from sputum of a patient with cystic fibrosis and review of Pandoraea species infections in transplant patients. J. Clin. Microbiol. 46, 3165–3168. doi: 10.1128/JCM.00855-08
Pugès, M., Debelleix, S., Fayon, M., Mégraud, F., and Lehours, P. (2015). Persistent infection because of Pandoraea sputorum in a young cystic fibrosis patient resistant to antimicrobial treatment. Pediatr. Infect. Dis. J. 34, 1135–1137. doi: 10.1097/INF.0000000000000843
Pushiri, H., Pearce, S. L., Oakeshott, J. G., Russell, R. J., and Pandey, G. (2013). Draft genome sequence of Pandoraea sp. strain SD6-2, isolated from lindane-contaminated Australian soil. Genome Announc. 1:e00415-13. doi: 10.1128/genomeA.00415-13
Sahin, N. (2003). Oxalotrophic bacteria. Res. Microbiol. 154, 399–407. doi: 10.1016/S0923-2508(03)00112-8
Sahin, N., Tani, A., Kotan, R., Sedlacek, I., Kimbara, K., and Tamer, A. U. (2011). Pandoraea oxalativorans sp. nov., Pandoraea faecigallinarum sp. nov. and Pandoraea vervacti sp. nov., isolated from oxalate-enriched culture. Int. J. Syst. Evol. Microbiol. 61, 2247–2253. doi: 10.1099/ijs.0.026138-0
Sarkar, P., Roy, A., Pal, S., Mohapatra, B., Kazy, S. K., Maiti, M. K., et al. (2017). Enrichment and characterization of hydrocarbon-degrading bacteria from petroleum refinery waste as potent bioaugmentation agent for in situ bioremediation. Bioresour. Technol. 242, 15–27. doi: 10.1016/j.biortech.2017.05.010
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
See-Too, W. S., Ambrose, M., Malley, R., Ee, R., Mulcahy, E., Manche, E., et al. (2019). Pandoraea fibrosis sp. nov., a novel Pandoraea species isolated from clinical respiratory samples. Int. J. Syst. Evol. Microbiol. 69, 645–651. doi: 10.1099/ijsem.0.003147
Shi, Y., Chai, L., Tang, C., Yang, Z., Zheng, Y., Chen, Y., et al. (2013). Biochemical investigation of kraft lignin degradation by Pandoraea sp. B-6 isolated from bamboo slips. Bioprocess Biosyst. Eng. 36, 1957–1965. doi: 10.1007/s00449-013-0972-9
Spilker, T., Baldwin, A., Bumford, A., Dowson, C. G., Mahenthiralingam, E., and LiPuma, J. J. (2009). Expanded multilocus sequence typing for Burkholderia species. J. Clin. Microbiol. 47, 2607–2610. doi: 10.1128/JCM.00770-09
Springael, D., van Thor, J., Goorissen, H., Ryngaert, A., De Baere, R., Van Hauwe, P., et al. (1996). RP4:Mu3A-mediated in vivo cloning and transfer of a chlorobiphenyl catabolic pathway. Microbiology 142, 3283–3293. doi: 10.1099/13500872-142-11-3283
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stryjewski, M. E., LiPuma, J. J., Messier, R. H., Reller, L. B., and Alexander, B. D. (2003). Sepsis, multiple organ failure, and death due to Pandoraea pnomenusa infection after lung transplantation. J. Clin. Microbiol. 41, 2255–2257. doi: 10.1128/JCM.41.5.2255-2257.2003
Tirado-Torres, D., Acevedo-Sandoval, O., Rodriguez-Pastrana, B. R., and Gayosso-Canales, M. (2017). Phylogeny and polycyclic aromatic hydrocarbons degradation potential of bacteria isolated from crude oil-contaminated site. J. Environ. Sci. Heal. Part A 52, 897–904. doi: 10.1080/10934529.2017.1316170
Uhlik, O., Wald, J., Strejcek, M., Musilova, L., Ridl, J., Hroudova, M., et al. (2012). Identification of bacteria utilizing biphenyl, benzoate, and naphthalene in long-term contaminated soil. PLoS One 7:e40653. doi: 10.1371/journal.pone.0040653
Vandamme, P., Vancanneyt, M., Pot, B., Mels, L., Hoste, B., Dewettinck, D., et al. (1992). Polyphasic taxonomic study of the emended genus Arcobacter with Arcobacter butzleri comb. nov. and Arcobacter skirrowii sp. nov., an Aerotolerant bacterium isolated from veterinary specimens. Int. J. Syst. Bacteriol. 42, 344–356. doi: 10.1099/00207713-42-3-344
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wang, X., Wang, Q., Li, S., and Li, W. (2015). Degradation pathway and kinetic analysis for p-xylene removal by a novel Pandoraea sp. strain WL1 and its application in a biotrickling filter. J. Hazard. Mater. 288, 17–24. doi: 10.1016/j.jhazmat.2015.02.019
Wu, X., Wu, X., Shen, L., Li, J., Yu, R., Liu, Y., et al. (2019). Whole genome sequencing and comparative genomics analyses of Pandoraea sp. XY-2, a new species capable of biodegrade tetracycline. Front. Microbiol. 10:33. doi: 10.3389/fmicb.2019.00033
Yang, J., Guo, C., Liu, S., Liu, W., Wang, H., Dang, Z., et al. (2018). Characterization of a di-n-butyl phthalate-degrading bacterial consortium and its application in contaminated soil. Environ. Sci. Pollut. Res. 25, 17645–17653. doi: 10.1007/s11356-018-1862-0
Keywords: Pandoraea, novel species, cystic fibrosis microbiology, comparative genomics, xenobiotics, biodegradation, opportunistic pathogens
Citation: Peeters C, De Canck E, Cnockaert M, De Brandt E, Snauwaert C, Verheyde B, Depoorter E, Spilker T, LiPuma JJ and Vandamme P (2019) Comparative Genomics of Pandoraea, a Genus Enriched in Xenobiotic Biodegradation and Metabolism. Front. Microbiol. 10:2556. doi: 10.3389/fmicb.2019.02556
Received: 07 September 2019; Accepted: 23 October 2019;
Published: 06 November 2019.
Edited by:
Iain Sutcliffe, Northumbria University, United KingdomReviewed by:
Martin W. Hahn, University of Innsbruck, AustriaCopyright © 2019 Peeters, De Canck, Cnockaert, De Brandt, Snauwaert, Verheyde, Depoorter, Spilker, LiPuma and Vandamme. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter Vandamme, UGV0ZXIuVmFuZGFtbWVAdWdlbnQuYmU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.