
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 November 2019
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02468
This article is part of the Research Topic Characterization Of Mobile Genetic Elements Associated With Acquired Resistance Mechanisms View all 15 articles
 Zhe Yin1†
Zhe Yin1† Lingfei Hu1†
Lingfei Hu1† Qiaoxiang Cheng2
Qiaoxiang Cheng2 Xiaoyuan Jiang1
Xiaoyuan Jiang1 Yanan Xu2
Yanan Xu2 Wenhui Yang1
Wenhui Yang1 Huiying Yang1
Huiying Yang1 Yuee Zhao1
Yuee Zhao1 Bo Gao1
Bo Gao1 Jinglin Wang1
Jinglin Wang1 Erhei Dai2*
Erhei Dai2* Dongsheng Zhou1*
Dongsheng Zhou1*Three different MDR plasmids p16005813A, p16005813B, and p16005813C, which carried a total of 18 non-redundant resistance genes or gene loci, were identified in a single clinical isolate of Leclercia adecarboxylata. The p16005813A backbone showed very low levels of identity to all DNA sequences available in public databases and carried a repA gene that could not assigned into any of known incompatibility groups. The IncFII-family p16005813B and pECAZ161_KPC had essentially identical backbones. p16005813C belonged to an IncR single-replicon plasmid. p16005813A, p16005813B, and p16005813C harbored three different novel MDR regions as their sole accessory modules. The MDR region of p16005813B manifested as Tn6505, which was generated from insertion of blaIMP–8-carrying In655 instead of In4 into the Tn1696 backbone. Other key antibiotic resistance elements included Tn2, IS26–mph(A)–mrx–mphR(A)–IS6100 unit, chrA region, In27, and aacC2–tmrB region in the MDR region of p16005813A, and ΔTn9 carrying catA1, In609, and IS26–tetA(C)–tetR(C)–IS26 unit in the MDR region of p16005813C. This was the first report of coexistence of three different MDR plasmids, and that of occurrence of IMP-encoding plasmid and blaIMP–8 gene in L. adecarboxylata.
Leclercia adecarboxylata, a Gram-negative rod of the Enterobacteriaceae family, exists widely in nature and shares many biochemical features with Escherichia coli (Anuradha, 2014; Spiegelhauer et al., 2018). As an extremely rare human pathogen, L. adecarboxylata causes monomicrobial infection in immune suppressed patients and ones with underlying medical conditions, and it is also found as part of a causative agent of polymicrobial infections in immunocompetent subjects, requiring other coinfecting bacteria to establish infection (Anuradha, 2014; Spiegelhauer et al., 2018).
L. adecarboxylata is generally susceptible to cephalosporins, carbapenems, tetracyclines, aminoglycosides, quinolones, and chloramphenicol (Stock et al., 2004). Nevertheless, L. adecarboxylata isolates have evolved to acquire foreign antibiotic resistance genes, which encode extended-spectrum β-lactamases (ESBLs) SHV-12 (Mazzariol et al., 2003) and CTX-M-3 (Shin et al., 2012), and carbapenemases IMP-1 (GenBank accession number KJ531212), IMP-4 (Betteridge et al., 2013; Leung et al., 2013), KPC-2 (Geffen et al., 2013; Weingarten et al., 2018), NDM-1 (Sun et al., 2015; Hoyos-Mallecot et al., 2017; Riazzo et al., 2017) and VIM-1 (Papagiannitsis et al., 2013; Papousek et al., 2017); from these strains, one NDM-encoding plasmid pP10164-NDM (accession number KP900016) (Sun et al., 2015), one VIM-encoding pLec-476cz (accession number KY320277) (Papousek et al., 2017), and several KPC-encoding plasmids (Weingarten et al., 2018) have been fully sequenced.
Only a small amount of MDR L. adecarboxylata strains, each harboring multiple acquired resistance genes, have been reported (Shin et al., 2012; Garcia-Fulgueiras et al., 2014; Sun et al., 2016). Notably, complete genome sequencing has been applied to only one of those MDR strains designated P10164, which harbors pP10164-NDM that is not a MDR plasmid (Sun et al., 2015), and two MDR plasmids pP10164-2 and pP10164-3 (accession numbers KX710093 and KX710094, respectively), as shown in our previous study (Sun et al., 2016). This follow-up study disclosed co-occurrence three different multi-drug resistant (MDR) plasmids, p16005813A (unknown incompatibility group), p16005813B (IncFII), and p16005813C (IncR), containing a total of 18 non-redundant resistance genes or gene loci, in a single clinical L. adecarboxylata isolate.
L. adecarboxylata 16005813 was isolated in 2016 from a sputum specimen of an infant with pneumonia in a public hospital in Ningbo, China.
Genomic DNA was isolated from the 16005813 isolate using an DNeasy UltraClean Microbial Kit or a Large Construct Kit (Qiagen, NW, Germany), and then sequenced from a sheared DNA library with average size of 15 kb (ranged from 10 to 20 kb) on a PacBio RSII sequencer (Pacific Biosciences, CA, United States), as well as a paired-end library with an average insert size of 400 bp (ranged from 150 to 600 kb) on a HiSeq sequencer (Illumina, CA, United States). The paired-end short Illumina reads were used to correct the long PacBio reads utilizing proovread (Hackl et al., 2014), and then the corrected PacBio reads were assembled de novo utilizing SMARTdenovo1. Open reading frames (ORFs) and pseudogenes were predicted using RAST 2.0 (Brettin et al., 2015) with default parameters, combined with BLASTP/BLASTN (Boratyn et al., 2013) searches against the UniProtKB/Swiss-Prot database (Boutet et al., 2016) and the RefSeq database (O’leary et al., 2016). Annotation of resistance genes, mobile elements, and other features was carried out using the online databases including CARD (Jia et al., 2017), ResFinder (Zankari et al., 2012), ISfinder (Siguier et al., 2006), INTEGRALL (Moura et al., 2009), and Tn Number Registry (Roberts et al., 2008). Multiple and pairwise sequence comparisons were performed using MUSCLE 3.8.31 (Edgar, 2004) and BLASTN, respectively. Gene organization diagrams were drawn in Inkscape 0.48.12.
Plasmid conjugal transfer experiments were carried out with the rifampin-resistant E. coli EC600 being used as a recipient and the 16005813 isolate as a donor, as described previously (Jiang et al., 2017). Bacteria were spotted on Muller-Hinton agar (BD Biosciences, NJ, United States) plates containing 1000 μg/ml rifampin together with 10 μg/ml azithromycin, 2 μg/ml meropenem or 20 μg/ml tetracycline, for selecting an E. coli transconjugant that carried mph (A) (p16005813A), blaIMP (B) (p16005813B), or tet (C) (p16005813C), respectively. Plasmid electroporation experiments were carried out as described previously (Ouyang et al., 2018). Bacteria were spotted on Super Optimal Broth agar plates containing 10 μg/ml azithromycin, 2 μg/ml meropenem or 20 μg/ml tetracycline, for selecting an E. coli electroporant that carried p16005813A, p16005813B, or p16005813C, respectively. Each transconjugant or electroporant was further confirmed by PCR detection of rep (replication initiation) and/or blaIMP–8 genes carried by relevant plasmids, followed by sequencing of PCR amplicons using the primers for PCR amplification (Supplementary Table S1).
Activity of carbapenemases in bacterial cell extracts was determined by a modified Carba NP test (Feng et al., 2016). Bacterial antimicrobial susceptibility was tested by bioMérieux VITEK 2 and interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.
The complete sequence of plasmids p16005813A, p16005813B, and p16005813C, and that of the 16005813 chromosome, were submitted to GenBank under accession numbers MK036891, MK036884, MK036885, and CP036199, respectively.
The 16005813 isolate carried three plasmids p16005813A, p16005813B, and p16005813C, which exhibited circularly closed sequences 138,399 bp, 45,490 bp, and 61,463 bp in length and contained 149, 56, and 76 ORFs, respectively (Table 1). Each plasmid contained the backbone (responsible for plasmid replication and maintenance, and/or conjugal transfer), with insertion of a single accessory module (acquired DNA region associated and bordered with mobile elements) (Table 1 and Figure 1). A total of 18 non-redundant genes or gene loci, which were involved in resistance to β-lactams including carbapenems (blaIMP–8, blaCTX–M–9 and blaTEM–1), aminoglycosides (aacC2, aacA4, and aadA2), tetracyclines [tet (C)], chloramphenicol (catA1 and catB8), macrolides [mph (A)], trimethoprim (dfrA12), sulfonamide (sul1), quaternary ammonium compounds (qacED1), chromate (chrA), tunicamycin (tmrB), copper (cop), mercury (mer) and arsenic (Zankari et al., 2012), were found in these three plasmids (Table 2). The 16005813 chromosome carried several putative intrinsic resistance genes (including a fosfomycin resistance gene, and several gene loci encoding putative MDR proteins), but harbored none of accessory resistance regions (Supplementary Table S2).
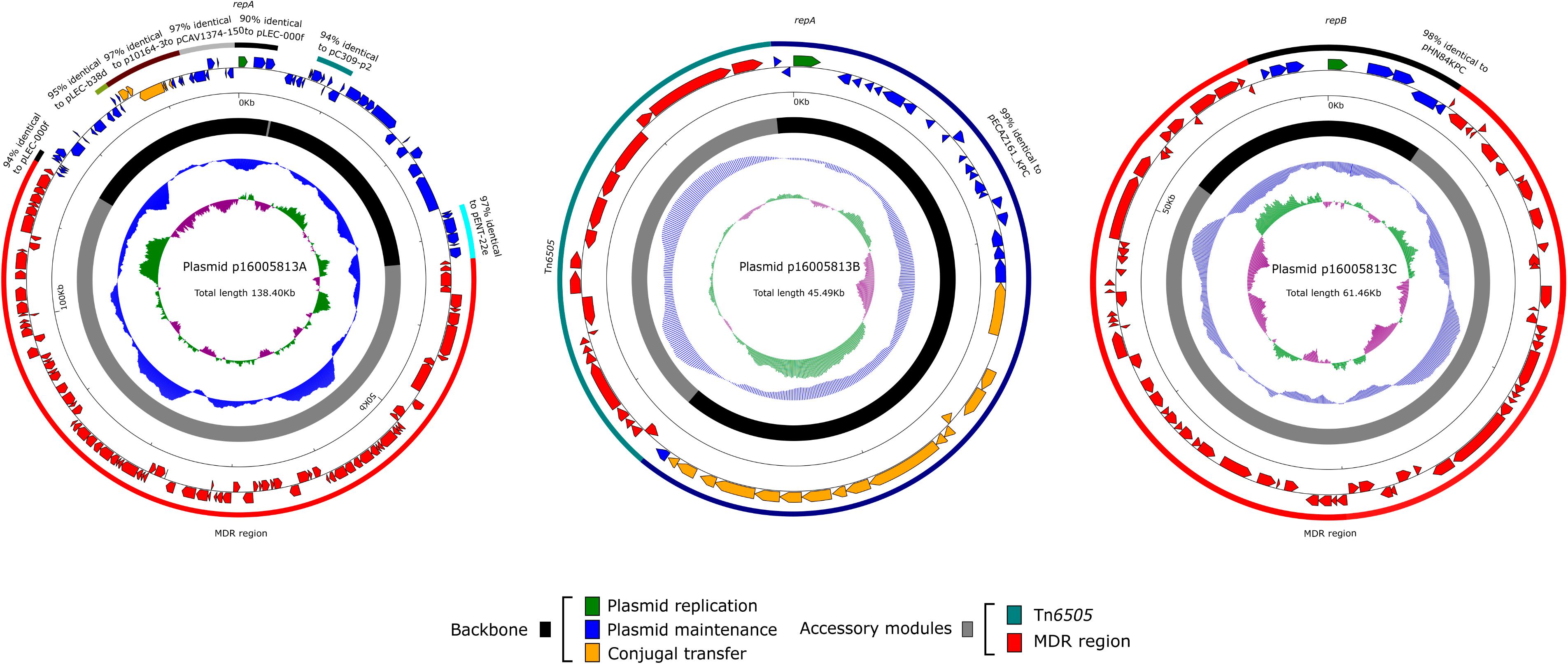
Figure 1. Plasmid schematic maps. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and color, respectively. The innermost circle presents GC-skew [(G–C)/(G + C)], with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle presents GC content.
p16005813A, p16005813B, and p16005813C could be transferred from the 16005813 isolate into TOP10 through electroporation, generating the electroporations p16005813A-TOP10, p16005813B-TOP10, and p16005813C-TOP10 (Table 3), respectively. Successful conjugation transfer of p16005813B, but not p16005813A and p16005813C, into EC600 generated the transconjugant p16005813B-EC600, which was consistent with the fact that only p16005813B carried a complete set of conjugal transfer genes. Strains 16005813, p16005813B-TOP10, and p16005813B-EC600 had metallo-β-lactamase activity and were resistant to ceftazidime and imipenem (Table 3), which was resulted from encoding of IMP enzyme by p16005813B.
The p16005813A backbone showed very low levels of identity to all DNA sequences available in public databases. The replication initiation gene repA of p16005813A showed 100% BLAST coverage and 94% nucleotide identity to the counterparts in pLEC-5e18 (accession number CP026390) and pLEC-000f (accession number CP026170) from L. adecarboxylata; all these RepA proteins could not assigned into any of known incompatibility groups. A 121-bp region containing six imperfect direct repeats of GTGtGTcataacATG was located 89-bp upstream of repA of p16005813A, and might function as RepA-binding iterons. Key determinants for plasmid maintenance include a type Ia partitioning system ParABC, two distinct type II toxin-antitoxin systems VapBC and ParDE involved in post-segregational killing. Residual conjugal transfer determinants TrbA, MobC and mutated NikAB were found, which was consistent with the p16005813A’s nature of not self-transmissible.
p16005813B and pECAZ161_KPC (accession number CP019010) had essentially identical backbones and each of them carried a single accessory module, but the two accessory modules were different with respective to resistance genes and mobile elements harbored, and inserted at two different sites of the backbone. Key backbone genes of p16005813B include a novel replication initiation gene repA belonging to the IncFII family, ccdBA encoding a type II toxin-antitoxin system, and a set of P-type type IV secretion system genes (tviF3/12/13/15/17) involved in conjugal transfer.
The p16005813C backbone was composed of repB (replication initiation) and its iterons, parAB (type Ia partitioning), umuCD (SOS mutagenesis), and resD (multimer resolvase), but lacked retA (group IIB intron-encoding reverse transcriptase), and vagCD (toxin-antitoxin) compared to the IncR single-replicon reference plasmid pHN84KPC (accession number KY296104) with the most complete IncR backbone. IncR single-replicon plasmids including p16005813 lacked conjugal transfer genes and thus were not self-transmissible (Chen et al., 2014; Compain et al., 2014).
Each of p16005813A, p16005813B and p16005813C harbored a MDR region as the sole accessory module. The MDR region (83.1 kb in length, Figure 2) of p16005813A was generated from insertion of a 61-kb region into a truncated (losing of tnsABCD) version of the sil–cop region, which was originally found in the IncHI2 reference plasmid R478 (accession number BX664015) (Gilmour et al., 2004) and composed of a Tn7-like core transposition module tnsABCD together with the silver (sil) and copper (cop) resistance loci. Various derivatives of the sil–cop region were presented in IncHI2 plasmids (Sun et al., 2016; Liang et al., 2017). The 61-kb region comprised Tn2, and two 28-kb repeat regions, each of which harbored a mer-containing Tn21 remnant + IS26–mph(A)–mrx–mphR(A)–IS6100 unit + chrA region as found in pP10159-5 (Ouyang et al., 2018), In27, and aacC2–tmrB region as found in pA708-IMP (accession number MF344567).
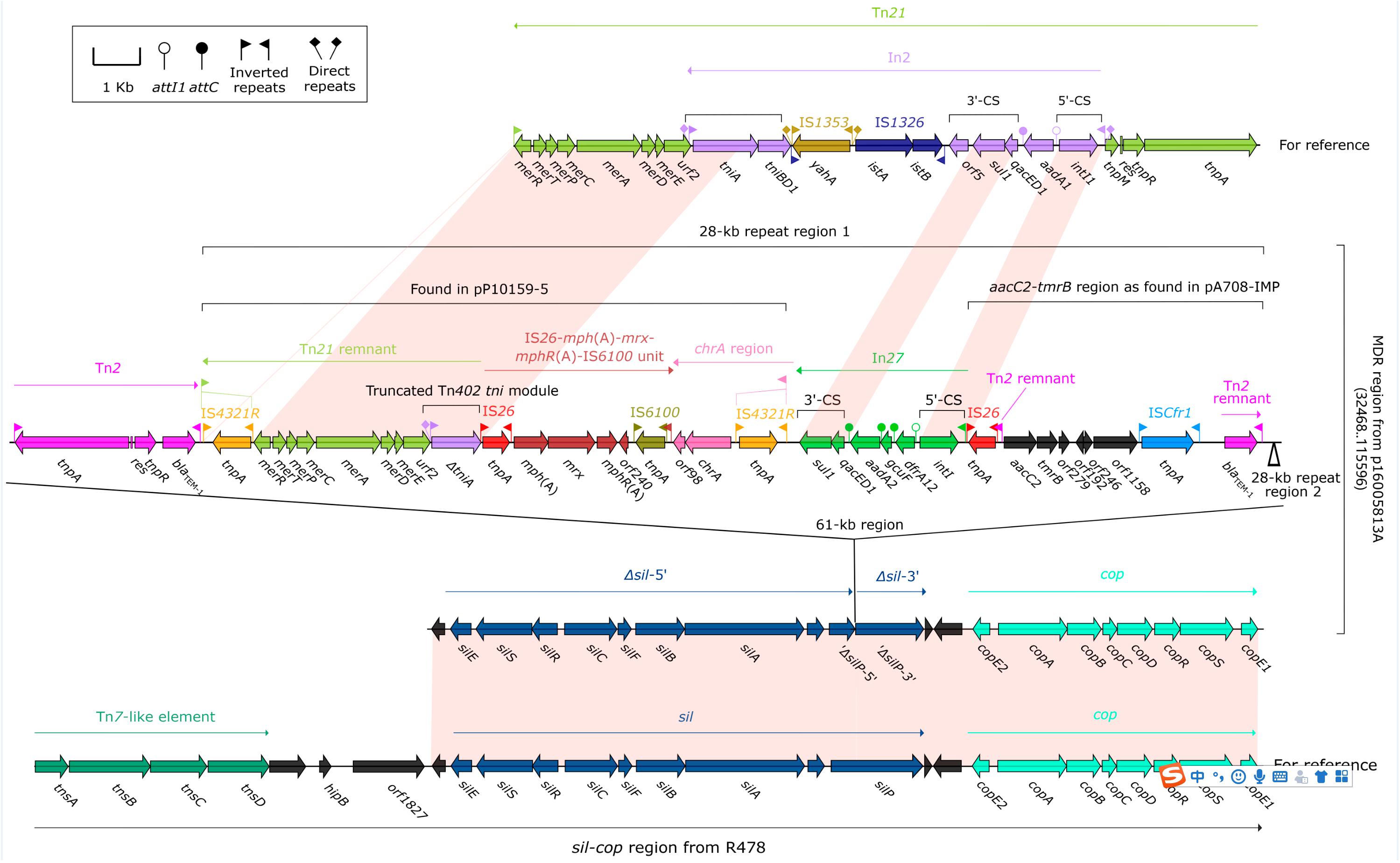
Figure 2. Organization of the MDR region of p16005813A and comparison with related regions. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity). Numbers in brackets indicate e nucleotide positions within corresponding plasmids. The accession numbers of the sil–cop region from R478, Tn21, and Tn2 for reference are BX664015, AF071413, and HM749967, respectively.
Tn6505 (16.9 kb in length, Figure 3) from p16005813B was a novel derivative of Tn1696 (Partridge et al., 2001) belonged to the Tn21 subgroup of Tn3 transposon family, and flanked by 5 bp direct repeats (DRs; target site duplication signals for transposition). Tn6505 differed from Tn1696 by (i) insertion of blaIMP–8-carrying In655 instead of In4 at the same position within the resolution (res) site, and (ii) interruption of IRLTn6505 (inverted repeat right of Tn6505) by IS5075 that was a hunter of terminal IRs of Tn21 subgroup transposons (Partridge and Hall, 2003).
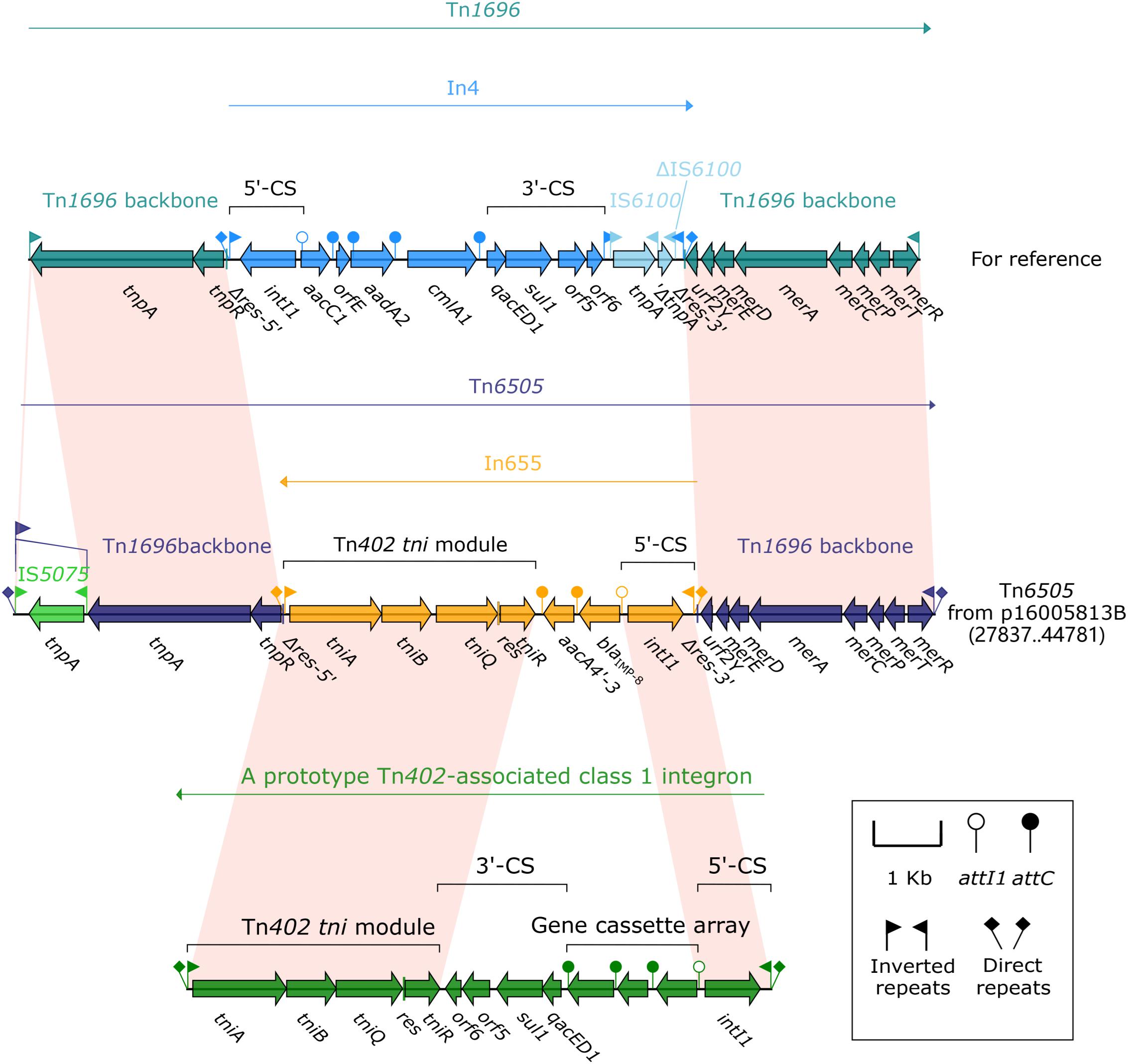
Figure 3. Organization of the MDR region (Tn6505) of p16005813B and comparison with related regions. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity). Numbers in brackets indicate e nucleotide positions within corresponding plasmids. The accession number of Tn1696 for reference is U12338.
The MDR region (52.2 kb in length, Figure 4) from p16005813C carried at least four resistance modules, namely Tn6322 (Sun et al., 2016), ΔTn9 carrying catA1 (Alton and Vapnek, 1979), In609, IS26–tetA(C)–tetR(C)–IS26 unit (Sun et al., 2016). Tn6322 was composed of a Tn21 core transposition module tnpAR–res together with a mer locus, and its IRL was interrupted by IS4321R (a homolog of IS5075). In609 was a complex class 1 integron carried variable region 1 (VR1: gene cassette gcuD2–aacA4′-17–gcuE14–catB8) and VR2 (blaCTX–M–9).
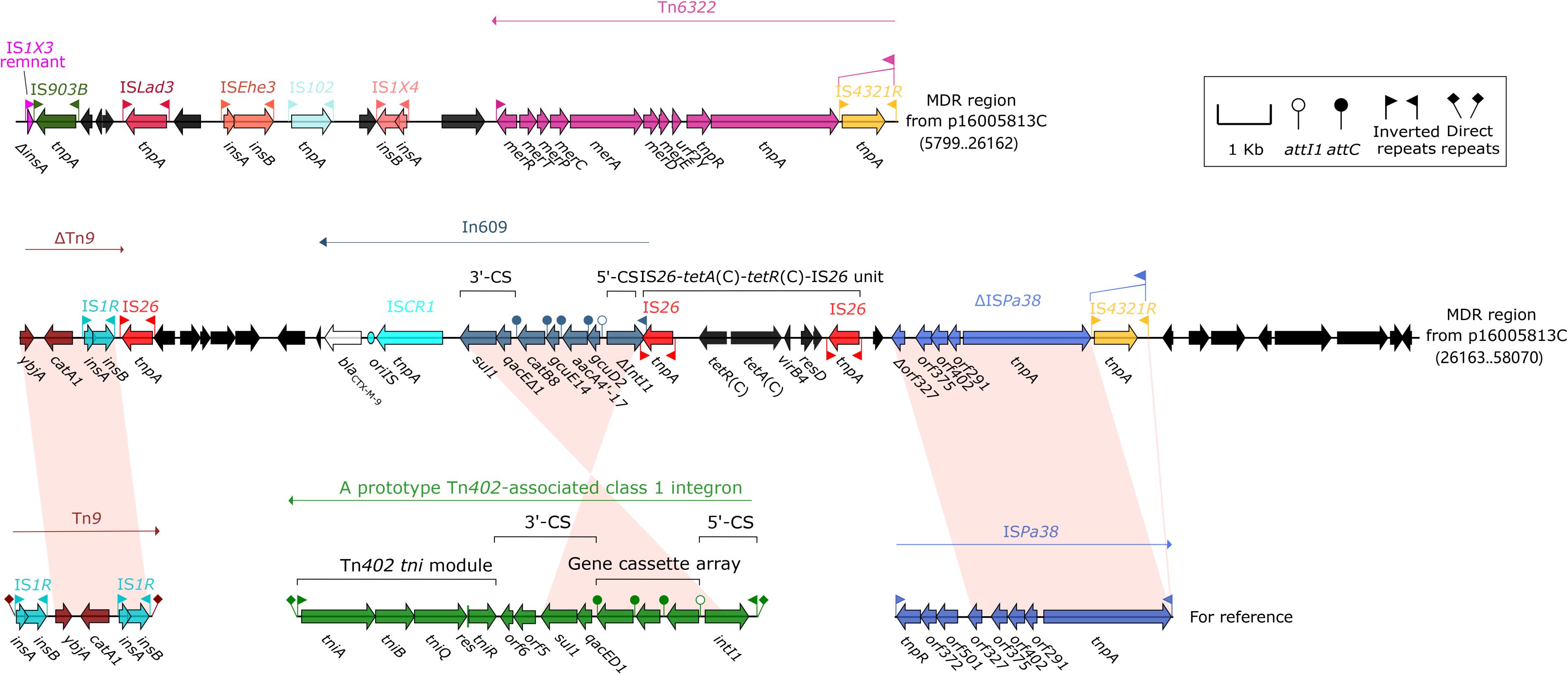
Figure 4. Organization of the MDR region of p16005813C and comparison with related regions. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity). Numbers in brackets indicate e nucleotide positions within corresponding plasmids. The accession numbers of Tn9 and ISPa38 for reference are LN830952, and MH909331, respectively.
This is the first report of coexistence of three different MDR plasmids, and that of occurrence of IMP-encoding plasmid and blaIMP–8 gene in L. adecarboxylata. blaIMP–8 was located in In655, representing an ancestral Tn402-associated integron (containing a complete tniTn402 core transposition module) at stage I of evolution as defined previously (Jiang et al., 2017). The three co-existent MDR plasmids carried a large amount of resistance genes, making the relevant L. adecarboxylata isolate tend to be extensively drug resistant. Epidemiological investigation of multi-drug resistant L. adecarboxylata needed to be carried out in China.
The datasets generated for this study can be found in the complete sequence of plasmids p16005813A, p16005813B, and p16005813C, and that of the 16005813 chromosome, were submitted to GenBank under accession numbers MK036891, MK036884, MK036885, and CP036199, respectively.
This study uses the clinical bacterial isolate obtained from a public hospital in Ningbo, China. The study needs not to be reviewed or approved by the ethics committee of the hospital, because the bacterial isolate involved in this study was part of the routine hospital laboratory procedure. The research involving biohazards and all related procedures were approved by the Biosafety Committee of the Beijing Institute of Microbiology and Epidemiology.
DZ and ED conceived the study and designed experimental procedures. ZY, LH, QC, XJ, and YX performed the experiments. ZY, LH, DZ, and ED analyzed the data. WY, HY, YZ, BG, and JW contributed to reagents and materials. DZ, ZY, LH, and ED wrote the manuscript.
This work was supported by the National Science and Technology Major Project (2018ZX10733402) of China and the Foundation of State Key Laboratory of Pathogen and Biosecurity (SKLPBS1810) of China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02468/full#supplementary-material
Alton, N. K., and Vapnek, D. (1979). Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature 282, 864–869. doi: 10.1038/282864a0
Anuradha, M. (2014). Leclercia adecarboxylata isolation: case reports and review. J. Clin. Diagn. Res. 8, DD03–DD04.
Betteridge, T., Merlino, J., Natoli, J., Cheong, E. Y., Gottlieb, T., and Stokes, H. W. (2013). Plasmids and bacterial strains mediating multidrug-resistant hospital-acquired infections are coresidents of the hospital environment. Microb. Drug Resist. 19, 104–109. doi: 10.1089/mdr.2012.0104
Boratyn, G. M., Christiam, C., Cooper, P. S., George, C., Amelia, F., Ning, M., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, 29–33. doi: 10.1093/nar/gkt282
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/swiss-prot, the manually annotated section of the uniprot knowledgebase: how to use the entry view. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Chen, Y. T., Lin, J. C., Fung, C. P., Lu, P. L., Chuang, Y. C., Wu, T. L., et al. (2014). KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J. Antimicrob. Chemother. 69, 628–631. doi: 10.1093/jac/dkt409
Compain, F., Frangeul, L., Drieux, L., Verdet, C., Brisse, S., Arlet, G., et al. (2014). Complete nucleotide sequence of two multidrug-resistant IncR plasmids from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 58, 4207–4210. doi: 10.1128/AAC.02773-13
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Feng, W., Zhou, D., Wang, Q., Luo, W., Zhang, D., Sun, Q., et al. (2016). Dissemination of IMP-4-encoding pIMP-HZ1-related plasmids among Klebsiella pneumoniae and Pseudomonas aeruginosa in a Chinese teaching hospital. Sci. Rep. 6:33419. doi: 10.1038/srep33419
Garcia-Fulgueiras, V., Seija, V., Aguerrebere, P., Cordeiro, N., and Vignoli, R. (2014). First report of a clinical isolate of Leclercia adecarboxylata harbouring multiple resistance genes in Uruguay and review of the literature. J. Glob. Antimicrob. Resist. 2, 77–81. doi: 10.1016/j.jgar.2014.01.002
Geffen, Y., Adler, A., Paikin, S., Khabra, E., Gorenshtein, S., Aronov, R., et al. (2013). Detection of the plasmid-mediated KPC-2 carbapenem-hydrolysing enzyme in three unusual species of the Enterobacteriaceae family in Israel. J. Antimicrob. Chemother. 68, 719–720. doi: 10.1093/jac/dks443
Gilmour, M. W., Thomson, N. R., Sanders, M., Parkhill, J., and Taylor, D. E. (2004). The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid 52, 182–202. doi: 10.1016/j.plasmid.2004.06.006
Hackl, T., Hedrich, R., Schultz, J., and Forster, F. (2014). proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 30, 3004–3011. doi: 10.1093/bioinformatics/btu392
Hoyos-Mallecot, Y., Rojo-Martin, M. D., Bonnin, R. A., Creton, E., Navarro Mari, J. M., and Naas, T. (2017). Draft genome sequence of NDM-1-Producing Leclercia adecarboxylata. Genome Announc. 5:e00135-17. doi: 10.1128/genomeA.00135-17
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jiang, X., Yin, Z., Yin, X., Fang, H., Sun, Q., Tong, Y., et al. (2017). Sequencing of blaIMP-carrying IncN2 plasmids, and comparative genomics of IncN2 plasmids harboring class 1 integrons. Front. Cell Infect. Microbiol. 7:102. doi: 10.3389/fcimb.2017.00102
Leung, G. H., Gray, T. J., Cheong, E. Y., Haertsch, P., and Gottlieb, T. (2013). Persistence of related blaIMP–4 metallo-beta-lactamase producing Enterobacteriaceae from clinical and environmental specimens within a burns unit in Australia - a six-year retrospective study. Antimicrob. Resist. Infect. Control 2:35. doi: 10.1186/2047-2994-2-35
Liang, Q., Yin, Z., Zhao, Y., Liang, L., Feng, J., Zhan, Z., et al. (2017). Sequencing and comparative genomics analysis of the IncHI2 plasmids pT5282-mphA and p112298-catA and the IncHI5 plasmid pYNKP001-dfrA. Int. J. Antimicrob. Agents 49, 709–718. doi: 10.1016/j.ijantimicag.2017.01.021
Mazzariol, A., Zuliani, J., Fontana, R., and Cornaglia, G. (2003). Isolation from blood culture of a Leclercia adecarboxylata strain producing an SHV-12 extended-spectrum beta-lactamase. J. Clin. Microbiol. 41, 1738–1739. doi: 10.1128/jcm.41.4.1738-1739.2003
Moura, A., Soares, M., Pereira, C., Leitão, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
O’leary, N. A., Wright, M. W., Rodney, B. J., Stacy, C., Diana, H., Rich, M. V., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Ouyang, J., Sun, F., Zhou, D., Feng, J., Zhan, Z., Xiong, Z., et al. (2018). Comparative genomics of five different resistance plasmids coexisting in a clinical multi-drug resistant Citrobacter freundii isolate. Infect. Drug Resist. 11, 1447–1460. doi: 10.2147/IDR.S165818
Papagiannitsis, C. C., Studentova, V., Hrabak, J., Kubele, J., Jindrak, V., and Zemlickova, H. (2013). Isolation from a nonclinical sample of Leclercia adecarboxylata producing a VIM-1 metallo-beta-lactamase. Antimicrob. Agents Chemother. 57, 2896–2897. doi: 10.1128/aac.00052-13
Papousek, I., Papagiannitsis, C. C., Medvecky, M., Hrabak, J., and Dolejska, M. (2017). Complete nucleotide sequences of two VIM-1-encoding plasmids from Klebsiella pneumoniae and Leclercia adecarboxylata isolates of czech origin. Antimicrob. Agents Chemother. 61:e02648-16. doi: 10.1128/AAC.02648-16
Partridge, S. R., Brown, H. J., Stokes, H. W., and Hall, R. M. (2001). Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45, 1263–1270. doi: 10.1128/aac.45.4.1263-1270.2001
Partridge, S. R., and Hall, R. M. (2003). The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J. Bacteriol. 185, 6371–6384. doi: 10.1128/jb.185.21.6371-6384.2003
Riazzo, C., Lopez-Cerero, L., Rojo-Martin, M. D., Hoyos-Mallecot, Y., Fernandez-Cuenca, F., Martin-Ruiz, J. L., et al. (2017). First report of NDM-1-producing clinical isolate of Leclercia adecarboxylata in Spain. Diagn. Microbiol. Infect. Dis. 88, 268–270. doi: 10.1016/j.diagmicrobio.2017.04.013
Roberts, A. P., Chandler, M., Courvalin, P., Guedon, G., Mullany, P., Pembroke, T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60, 167–173. doi: 10.1016/j.plasmid.2008.08.001
Shin, G. W., You, M. J., Lee, H. S., and Lee, C. S. (2012). Catheter-related bacteremia caused by multidrug-resistant Leclercia adecarboxylata in a patient with breast cancer. J. Clin. Microbiol. 50, 3129–3132. doi: 10.1128/JCM.00948-12
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, 32–36.
Spiegelhauer, M. R., Andersen, P. F., Frandsen, T. H., Nordestgaard, R. L. M., and Andersen, L. P. (2018). Leclercia adecarboxylata: a case report and literature review of 74 cases demonstrating its pathogenicity in immunocompromised patients. Infect. Dis. 51, 179–188. doi: 10.1080/23744235.2018.1536830
Stock, I., Burak, S., and Wiedemann, B. (2004). Natural antimicrobial susceptibility patterns and biochemical profiles of Leclercia adecarboxylata strains. Clin. Microbiol. Infect. 10, 724–733. doi: 10.1111/j.1469-0691.2004.00892.x
Sun, F., Yin, Z., Feng, J., Qiu, Y., Zhang, D., Luo, W., et al. (2015). Production of plasmid-encoding NDM-1 in clinical Raoultella ornithinolytica and Leclercia adecarboxylata from China. Front. Microbiol. 6:458. doi: 10.3389/fmicb.2015.00458
Sun, F., Zhou, D., Sun, Q., Luo, W., Tong, Y., Zhang, D., et al. (2016). Genetic characterization of two fully sequenced multi-drug resistant plasmids pP10164-2 and pP10164-3 from Leclercia adecarboxylata. Sci. Rep. 6:33982. doi: 10.1038/srep33982
Weingarten, R. A., Johnson, R. C., Conlan, S., Ramsburg, A. M., Dekker, J. P., Lau, A. F., et al. (2018). Genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring carbapenem resistance. mBio 9:e02011-17. doi: 10.1128/mBio.02011-17
Keywords: Leclercia adecarboxylata, multidrug resistance, plasmid, mobile elements, blaIMP–8
Citation: Yin Z, Hu L, Cheng Q, Jiang X, Xu Y, Yang W, Yang H, Zhao Y, Gao B, Wang J, Dai E and Zhou D (2019) First Report of Coexistence of Three Different MDR Plasmids, and That of Occurrence of IMP-Encoding Plasmid in Leclercia adecarboxylata. Front. Microbiol. 10:2468. doi: 10.3389/fmicb.2019.02468
Received: 16 June 2019; Accepted: 15 October 2019;
Published: 05 November 2019.
Edited by:
Costas C. Papagiannitsis, University of Thessaly, GreeceReviewed by:
Ibrahim Bitar, Charles University, CzechiaCopyright © 2019 Yin, Hu, Cheng, Jiang, Xu, Yang, Yang, Zhao, Gao, Wang, Dai and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erhei Dai, ZGFpZWgyMDA4QDEyNi5jb20=; Dongsheng Zhou, ZG9uZ3NoZW5nemhvdTE5NzdAZ21haWwuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.