
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 24 October 2019
Sec. Food Microbiology
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02443
This article is part of the Research TopicDevelopments in Campylobacter, Helicobacter & Related Organisms Research – CHRO 2019View all 18 articles
 Than Linh Quyen1
Than Linh Quyen1 Steen Nordentoft2†
Steen Nordentoft2† Aaydha Chidambara Vinayaka2
Aaydha Chidambara Vinayaka2 Tien Anh Ngo2†
Tien Anh Ngo2† Pia Engelsmenn2
Pia Engelsmenn2 Yi Sun3
Yi Sun3 Mogens Madsen1
Mogens Madsen1 Dang Duong Bang2*
Dang Duong Bang2* Anders Wolff1*
Anders Wolff1*Campylobacteriosis is one of the most common foodborne diseases worldwide. Two Campylobacter species – C. jejuni and C. coli in poultry and poultry products are considered to be the main source of human campylobacteriosis. Therefore, studying Campylobacter status in poultry flocks is needed to prevent transmission of disease and reduce human risk, health cost, and economic losses. In this study, we adapted and used a Loop-Mediated Isothermal Amplification (LAMP) assay for specific, sensitive, simple and cost-effective rapid detection of C. jejuni and C. coli in the poultry production chain. Amplified LAMP products were detected using a small, low-cost portable commercial blue LED transilluminator and a direct visual detection strategy was demonstrated. By using optimized conditions for amplification a limit of detection (LOD) of 50 CFU/ml was achieved for testing of C. jejuni and C. coli in spiked chicken feces without enrichment. The method took 60–70 min from receiving the samples to the final results (including 30 min for amplification). The optimized LAMP showed a relative accuracy of 98.4%, a specificity of 97.9%, and a sensitivity of 100% in comparison to real-time PCR method. Cohen’s kappa index also showed an excellent agreement (0.94) between the two methods. The results showed that the method is specific, sensitive and is suitable to develop for rapid detection of Campylobacter spp. at poultry production.
Campylobacteriosis is one of the leading causes of bacterial diarrhea worldwide (CDC, 2017; Helwigh et al., 2017). Two Campylobacter species, C. jejuni and C. coli account for the majority of human campylobacteriosis (Lawson et al., 1999; Tresse et al., 2017). Poultry and poultry products are considered to be the main sources for disease transmission (Wingstrand et al., 2006; Powell et al., 2012). The prevalence of Campylobacter in broiler flocks remains high (Sibanda et al., 2018). Data obtained from an electronically distributed survey in Denmark reported that 63% of the broiler farms tested positive for Campylobacter (Sandberg et al., 2015). Thus, there is an urgent need for a fast and simple method suitable for the detection of Campylobacter within poultry production chains.
Identification of C. jejuni and C. coli using conventional bacterial cultures in combination with biochemical-based assay are time-consuming (requiring more than 4 days) and laborious (Biesta-Peters et al., 2019). Therefore, several alternate methods have been developed and reported for the detection of Campylobacter spp. (Supplementary Table S1). Although ELISA (enzyme-linked immunosorbent assay) (RIDASCREEN® Campylobacter, R-Biopharm AG, Darmstadt, Germany) and real-time polymerase chain reaction (real-time PCR) (Alves et al., 2016) could detect Campylobacter in much shorter time (within 2 h) from fecal materials, the limit of detection remains high (Supplementary Table S1). Moreover, real-time PCR require sophisticated and expensive equipment to amplify and detect the presence of Campylobacter. Therefore, both PCR and real-time PCR are not suitable for rapid detection of the pathogens in the production chains. LAMP has been used to overcome the drawbacks of the PCR. LAMP is faster than PCR (de Paz et al., 2014; Sabike and Yamazaki, 2019) and can be performed under constant temperature in a range of 60–65°C, thus eliminating the need for sophisticated thermal control as in PCR (Mori et al., 2013; Sun et al., 2015). LAMP has several advantages such as fast reaction, simple operation, low cost, high sensitivity and specificity (Njiru, 2012; Velders et al., 2018). Moreover, the LAMP reaction is more tolerant to inhibitors in comparison to PCR and real-time PCR assays (Stedtfeld et al., 2014; Kosti et al., 2015). The LAMP reaction produces large amounts of amplified products (dsDNA). It can therefore even be detected by naked eyes when using appropriate DNA staining techniques (Xie et al., 2014). With these advantages, the LAMP may be suitable for rapid detection of pathogens in the poultry production chains.
LAMP was developed for the detection of Campylobacter spp. in poultry samples such as meat, carcass swabs, and fecal samples (Yamazaki et al., 2008, 2009b; Sabike et al., 2016; Romero and Cook, 2018; Sabike and Yamazaki, 2019). To study the epidemiology of Campylobacter spp. and to prevent transmission in the production chain, the time taken for detection of Campylobacter is crucial. However, to detect the presence of the Campylobacter spp. in cloacal swabs, ceca, meat and environmental cleaning samples, an enrichment step of 22 to 24 h is needed (Yamazaki et al., 2009b; Sabike et al., 2016; Sabike and Yamazaki, 2019). Consequently, it takes a total of at least 24–26 h for sample enrichment, preparation, amplification, and detection. Moreover, Campylobacter grows slowly and requires specific microaerobic conditions to grow, which makes it difficult to apply this method for the detection of Campylobacter spp. in the production chains. In contrast, stool specimens may not require an enrichment step since Campylobacter infected chicken feces may contain up to 109 Campylobacter per gram (Corry and Atabay, 2001; Hermans et al., 2010; Addis and Sisay, 2015). However, the content of inhibitors in feces is frequently high, which could inhibit the LAMP reaction (Schrader et al., 2012). In one study, to detect Campylobacter spp. directly from poultry feces with a limit of detection (LOD) of 1.2–1.4 CFU per test, the stool samples had to be diluted 1:4000 to reduce the inhibition effects (Yamazaki et al., 2008). Therefore, the method could not detect the fecal samples containing less than 1.2 × 106 CFU of C. coli or 1.4 × 106 CFU of C. jejuni per gram of stool specimen.
In this study, to provide a simple, rapid, cost-effective and sensitive method suitable for rapid detection of Campylobacter spp. within poultry production chains, we have developed an optimized LAMP assay with smaller reaction volume, a shorter reaction time, and higher sensitivity using a commercial LAMP kit. We also evaluated the use of a commercial mini UV transilluminator, which is small, simple, low cost and portable for LAMP detection. Moreover, for evaluation of the performance of the optimized LAMP method, a conventional real-time PCR was used in parallel to study the epidemiology of Campylobacter infection in a broiler farm.
Chromosomal DNA from all bacteria strains used in this study (listed in Table 1) was isolated using DNeasy Blood and Tissue kit (Qiagen, Germany). The DNA concentration was determined by NanoDrop (Thermo Scientific, United States).
Preparation of DNA from fecal sock samples: Each pair of boot socks was placed in a stomacher bag containing 200 ml of saline (0.9% NaCl). Fecal materials were released by gentle manipulation of the socks for 2 min. 1 ml of the suspension was collected and centrifuged at 5000 × g for 5 min. After discarding the supernatant, the pellet was used for DNA extraction by an automate KingFisherTM Purification system (Thermo Fisher, Copenhagen, Denmark) or stored at minus 20°C for later use. Further, the pellet was treated with 200 μl of lysis buffer consisting of 190 μl of magnetic lysis buffer and 10 μl of Proteinase K (20 mg/ml) followed by incubation for 10 min at room temperature. 100 μl of the treated sample was used to purify DNA by an automate KingFisherTM Purification system using the Magnesil KF Genomic DNA kit (Promega, Denmark) described previously (Lund et al., 2003). Finally, the supernatant was used as the template for real-time PCR and the LAMP assay.
A real-time PCR targeting the 16S rRNA gene of thermophilic Campylobacter species (C. jejuni, C. coli) was used as a reference method to evaluate the performance of the LAMP (Josefsen et al., 2010). Primer sequences used for real-time PCR were listed in Supplementary Table S2. Each reaction contained 25 μl of master mixture consisting of 1X buffer, 2.5 mM of MgCl2, 2.5 U of Tth enzyme (Roche, Denmark), 6.96% Glycerol (Merck, Germany), 0.6 mM of dNTPs, 0.01 mg/ml of BSA, 0.5 μM of forward primer OT-1559, 0.5 μM of reverse primer 18-1, 0.076 μM of Campylobacter LNA probe, 0.06 μM of IAC probe, 0.24 × 10–9 μM of IAC (primers used for construction of the internal amplification control) (Supplementary Table S2) (Life Technology Europe, Roskilde, Denmark), 10 μl of DNA template and 2.22 μl of PCR grade water (Sigma-Aldrich, Denmark). The real-time PCR conditions were 95°C for 3 min, followed by 40 cycles of 95°C for 15 s, 60°C for 60 s and 72°C for 30 s. The real-time PCR was performed on a MxPro-Mx3005P (Agilent Technologies ApS Glostrup, Denmark).
The LAMP assay was carried out using a Loopamp Campylobacter detection kit (Eiken Chemical Co., Ltd., Tokyo, Japan). The kit consists of a set of specific primers to recognize the oxidoreductase gene from C. jejuni and aspartate kinase gene from C. coli. The kit does not specifically identify C. jejuni or C. coli but can detect both species. LAMP reaction contained 12.5 μl of the master mixture consisting of 6.25 μl of the reaction mixture, 1.25 μl of primer mixture Campylobacter, 0.5 μl of Bst DNA polymerase, 2.5 μl of distilled water and 2 μl of the template. In the LAMP reactions, PCR grade water was used as negative control, and purified genomic DNA was used as positive control. The LAMP reaction was performed on a Dri-Block® DB-2TC (TECHNE, Staffordshire, United Kingdom) at a constant temperature of 65°C for 30 or 60 min. The reactions were terminated by heating up to 80°C for 2 min.
Initially, fecal sock samples were pre-confirmed for the absence of C. jejuni and C. coli by both conventional culture and real-time PCR. Different dilutions of Campylobacter cells for spiking in the fecal sock samples were prepared from serial 10-fold dilutions in saline water from stock cultures (OD600 = 0.3, spectrophotometer UV-1600PC) of a C. jejuni CCUG 11284 and a C. coli CCUG 11283 separately. 100 μl of each dilution was used to spike in 1 ml suspension of the Campylobacter negative fecal sock samples. The mixtures were centrifuged at 5000g for 5 min and the pellets were then used for DNA extraction as described above. The samples were analyzed by both LAMP and real-time PCR methods. Further, 100 μl of each dilution from 10–1 to 10–8 were spread on blood agar plates for determining colony forming unit (CFU) and the plates were incubated at 41.5°C in the microaerobic atmosphere. The CFU was determined by colonies counting after 48 h of incubation.
Evaluation of assays precision between real-time PCR and LAMP was calculated based on relative accuracy, relative specificity, relative sensitivity and Cohen’s kappa index as described previously (Chin et al., 2016; Vinayaka et al., 2019) using following formulas:
Where:
PA: the positive agreement between the real-time PCR and LAMP methods;
NA: the negative agreement between the real-time PCR and LAMP methods;
N: total number of samples (NA + PA + PD + ND);
PD: false positives in the LAMP method;
ND: false negatives in the LAMP method;
N−: total number of negative results (NA + PD);
N+: total number of positive results (PA + PD);
P(o): (PA + NA)/N; and
P(e): {(positive recovery in real-time PCR/total number of tested samples (N)) × (negative recovery in real-time PCR/total number of tested samples (N))} + {(negative recovery in LAMP/total number of tested samples (N)) × (negative recovery in real-time PCR/total number of tested samples (N))}.
LAMP products were visually detected by two different methods: direct visual detection under UV light by staining DNA using SYBR® Safe DNA intercalating dye and agarose gel electrophoresis.
Direct visual detection using SYBR® Safe staining DNA: The SYBR® Safe (10 000X concentrate in DMSO, Life Technology, Denmark) was diluted 1:10, and 1 μl of diluted dye was added to each tube after LAMP reaction. The tubes were observed under UV light from a portable DR22 blue LED transilluminator (Supplementary Figure S1) (Clare Chemical Research, Inc., United States).
Gel electrophoresis detection: After LAMP reactions, 5 μl of each amplified LAMP product were loaded on 2% agarose gel containing 1X of SYBR® Safe DNA Gel Stain (Invitrogen, Life Technologies, United States). Gel electrophoresis was carried out at 100 volts for 60 min and the gel electrophoresis patterns were observed under Bio-Rad Gel Doc 2000 UV transilluminator (Bio-Rad Life Science, Denmark).
To evaluate the ability of the method for rapid detection of Campylobacter, an evaluation trial was conducted in a large broiler farm located in Jutland, Denmark. The farm comprises 16 houses that are placed in blocks of four. All the four blocks have the same design with an entrance in the middle leading to a corridor from where there are doors to access to the 4 houses (Figure 1). During the poultry production period, fecal samples were collected from 16 houses, separately. Sampling was done on a weekly basis for four weeks by the boot sock sampling method as described previously (Food Standard Agency annual report, 2004/05). Briefly, elastic textile bands (Qualicum Scientific Ltd./Solar Biological Inc, United Kingdom) moistured with tryptone buffer (SteriSox, SODIBOX Nevez, France) were placed on clean boots. Fecal samples were collected from the floor of each poultry house by a farmer walking around the house. The socks were then placed in zipper bags and sent to the laboratory. The samples were processed in the lab as described above.
Different volumes of the LAMP reaction ranging from 3 to 25 μl were selected and tested using 2 ng of C. jejuni DNA as a template. It is estimated that 2 ng DNA C. jejuli corresponds to 1.13 × 106 genome equivalents (Chin et al., 2017). We observed LAMP products in all the reaction volumes tested such as 25, 12.5, and 6 μl (Figure 2A). Even small reaction volumes of 3 and 5 μL were sufficient enough to generate good LAMP signals (result not shown). The results indicated that the reaction volume had no influence on the LAMP efficiency and assay principle.
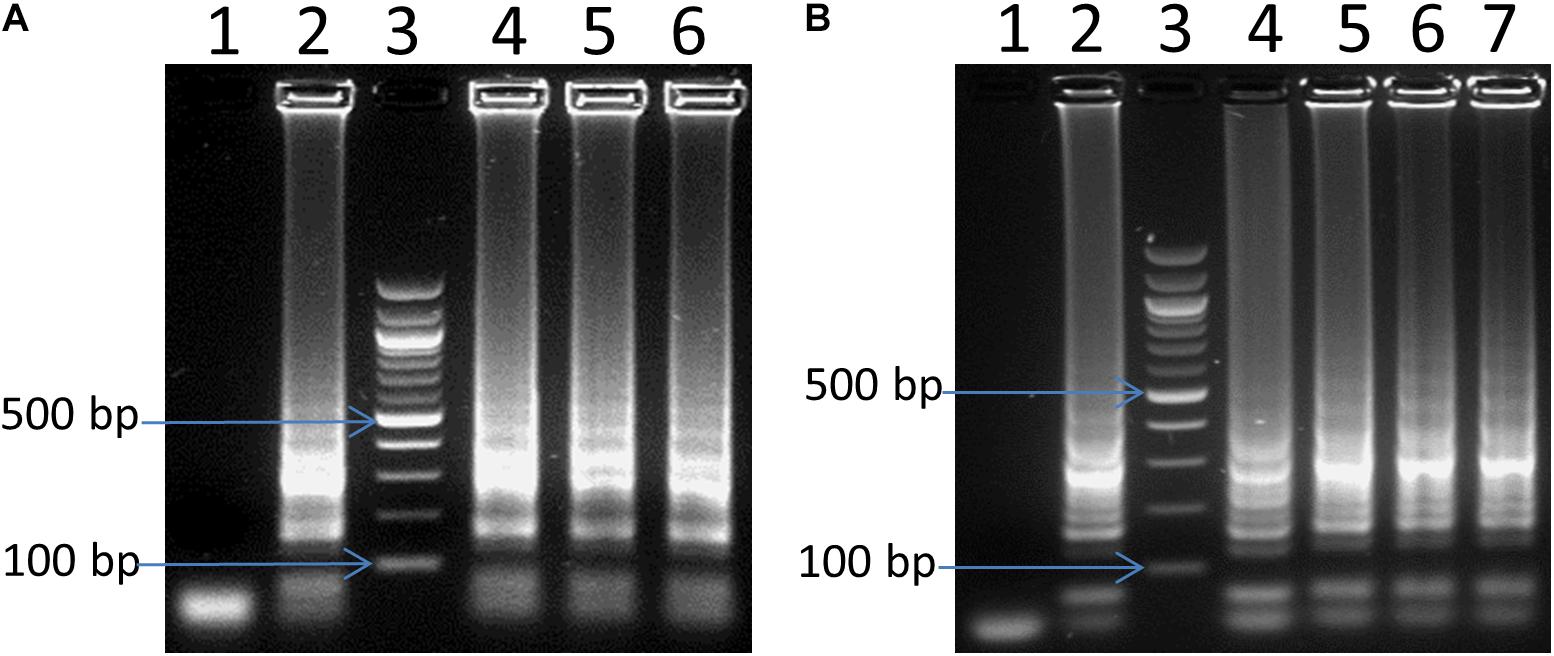
Figure 2. Optimization of LAMP for reaction volume (A) and amplification time (B). In panel (A), lane 4, 5 and 6: 6, 12.5, and 25 μL reaction volume, respectively. In panel (B), lane 4, 5, 6 and 7: 15 min, 30 min, 45 and 60 min, respectively. In both panels (A,B): lane 1: negative control, lane 2: positive control, and lane 3: 100 bp ladder.
Different reaction times of 5, 10, 15, 30, 45, and 60 min using 2 ng of Campylobacter DNA as a template were also tested to define a suitable reaction time for the LAMP assay. There were no visible LAMP amplifications for 5 and 10 min reactions, but LAMP products were visible after 15 min (Figure 2B). More amplified LAMP products were obtained after 30 min, whereas the intensity remained the same as the reaction went on for 45 and 60 min (Figure 2B). Hence, 30 min of incubation has been selected for further experiments.
It is intended to adopt this technique into the poultry production chains wherein, the cost of each test is a primary concern. Many studies of LAMP assay were reported using different volumes (12.5 μl, 25 μl, even 50 μl) for LAMP reactions (Kato et al., 2005; Wang et al., 2008; Yamazaki et al., 2008, 2009b; Peng et al., 2011; Zhang et al., 2013). However, until now, no study has addressed appropriate reaction volume for LAMP assay. The cost of one Campylobacter Detection Kit with 48 reactions (25 μl per reaction as recommended by manufacture) from Eiken Ltd., Japan was 4456,50 DKK. As a consequence, each reaction (25 μl as recommended by the manufacturer) cost ∼93 DKK which was a rather high cost for screening pathogens at farms. Therefore, the result of this study showing reaction volume did not influence the efficiency of the assay will help reduction of the cost when using LAMP for the detection of pathogens at farms. Further, KingFisherTM Purification system used in this study concentrates the samples and provides a good quality of DNA template. The cost of DNA extraction and purification is ∼23 DKK per sample and it is possible to process 24 samples at once in approximately 20 min.
Diagnosis time of foodborne disease screening is considered to be vital for preventing transmission and decreasing economic losses in broiler production. Rapid detection strategies with high specificity and sensitivity are of much relevance for broiler production. In this study, after 30 min of the LAMP reaction we were able to detect the presence of C. coli/C. jejuni in fecal samples as low as 50 CFU/mL without enrichment. The reaction time in this study was shorter in comparison to previous reports (35–60 min) (Iwamoto et al., 2003; Kato et al., 2005; Wang et al., 2008; Yamazaki et al., 2009a; Yang et al., 2010; Zhang et al., 2012, 2013). Zhuang et al. (2014) reported a LAMP reaction that can detect Salmonella in chicken feces within 25 min with a detection limit of 200 CFU per reaction that was 200 times higher than this study. Total time from sample preparation to the final result in this study was 60–70 min, and it is much shorter than previous reports (Kato et al., 2005; Okamura et al., 2008; Yamazaki et al., 2008, 2009b; Techathuvanan et al., 2010).
To compare the sensitivity of the optimized LAMP assay, reaction time of 30 min was compared with 60 min of amplification. A serial dilution of C. jejuni DNA ranging from 0.1 pg to 2 ng was prepared and used as a template for the LAMP reaction. Figure 3 shows that there were no LAMP amplified products when 0.1 pg of DNA template was used in both reaction times. In contrast, LAMP amplified products were observed with all other template concentrations tested. This result confirmed that, the optimized LAMP has similar sensitivity at 30 and 60 min of amplification.
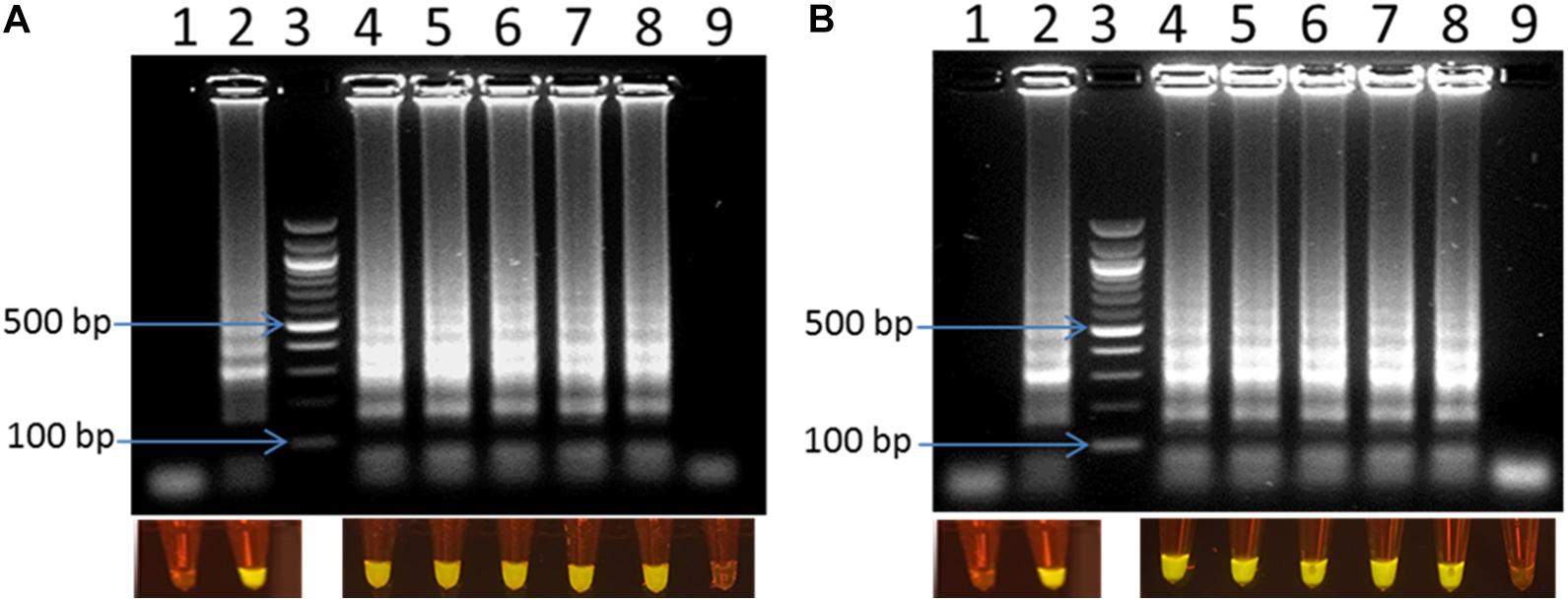
Figure 3. Visual detection of LAMP products on 2% agarose gel electrophoresis and using SYBR® Safe staining DNA for sensitivity of the LAMP using pure DNA of C. jejuni in 30 min (A) and 60 min (B). In both panels: lane 1: negative control, lane 2: positive control, lane 3: 100 bp ladder, and lane 4–9: 2 ng, 0.2 ng, 20 pg, 2 pg, 1 pg, 0.1 pg DNA of C. jejuni, respectively.
The optimized LAMP assay was also tested with the whole cell of C. jejuni and C. coli spiked into chicken fecal samples as described above. LODs of 1 CFU/reaction (corresponding to 50 CFU/ml) were observed for both C. jejuni and C. coli within 30 min of amplification (Figure 4). The optimized LAMP assay, in this study, showed better performance with lower LOD and shorter reaction time than previous reports (1.2–1.4 CFU and 5 CFU per reaction within 60 min) (Yamazaki et al., 2008; Romero and Cook, 2018).
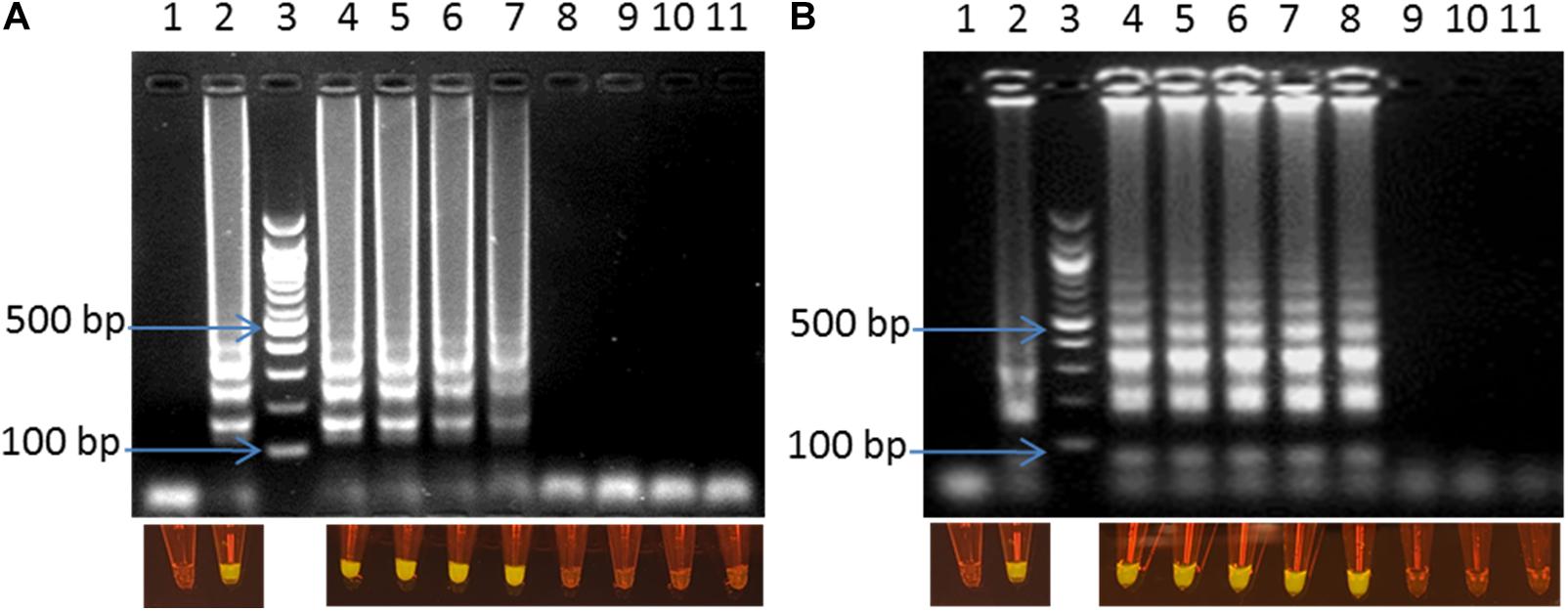
Figure 4. Visual detection of LAMP products on 2% agarose gel electrophoresis and using SYBR® Safe staining DNA for the sensitivity of the fecal spiked sample with C. jejuni (A) and C. coli (B). In both panels: lane 1: negative control, lane 2: positive control, lane 3: 100bp ladder, and lane 4–11: 10–1–10–8 dilution.
Efficiency of the optimized LAMP assay was also tested with real poultry fecal samples. A total of 64 boot-sock samples were collected and tested for the presence of C. jejuni and C. coli. Results showed that out of 64 samples tested, 17 samples were positive and 47 samples were negative for C. jejuni and C. coli (Table 2). There was 100% coincidence between the gel electrophoresis-based detection method and the visual observation using blue LED transilluminator (Figure 5). On the other hand, in the real-time PCR method, 18 samples were found positive and 46 samples were found negative with C. jejuni/C. coli. Comparing the results of the two methods showed a relative accuracy, specificity, and sensitivity of 98.43, 97.87, and 100%, respectively for the LAMP method. Cohen’s kappa index showed an excellent agreement between the real-time PCR and the LAMP (Cohen’s kappa = 0.94) (Table 2). One sample tested positive with the real-time PCR but negative with LAMP assay was further confirmed by conventional PCR using a different primer set developed in our lab. Out of four tests performed with this sample, the results were negative for C. jejuni/C. coli in 2 attempts and positive only for C. jejuni in another 2 attempts (data not shown). This difference in the results between the two methods may be attributed to a low number of target concentration in the tested sample.
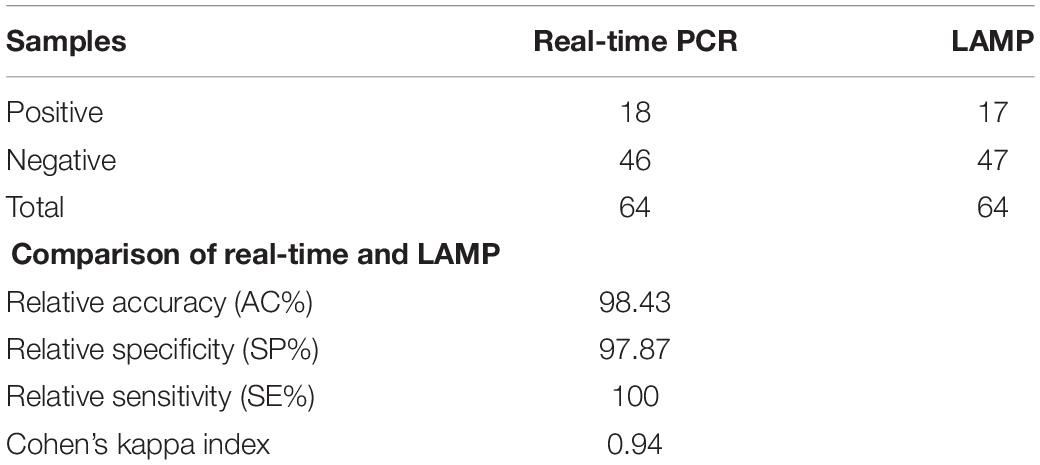
Table 2. Comparison of real-time PCR and LAMP assay for detection of chicken feces from sock samples.
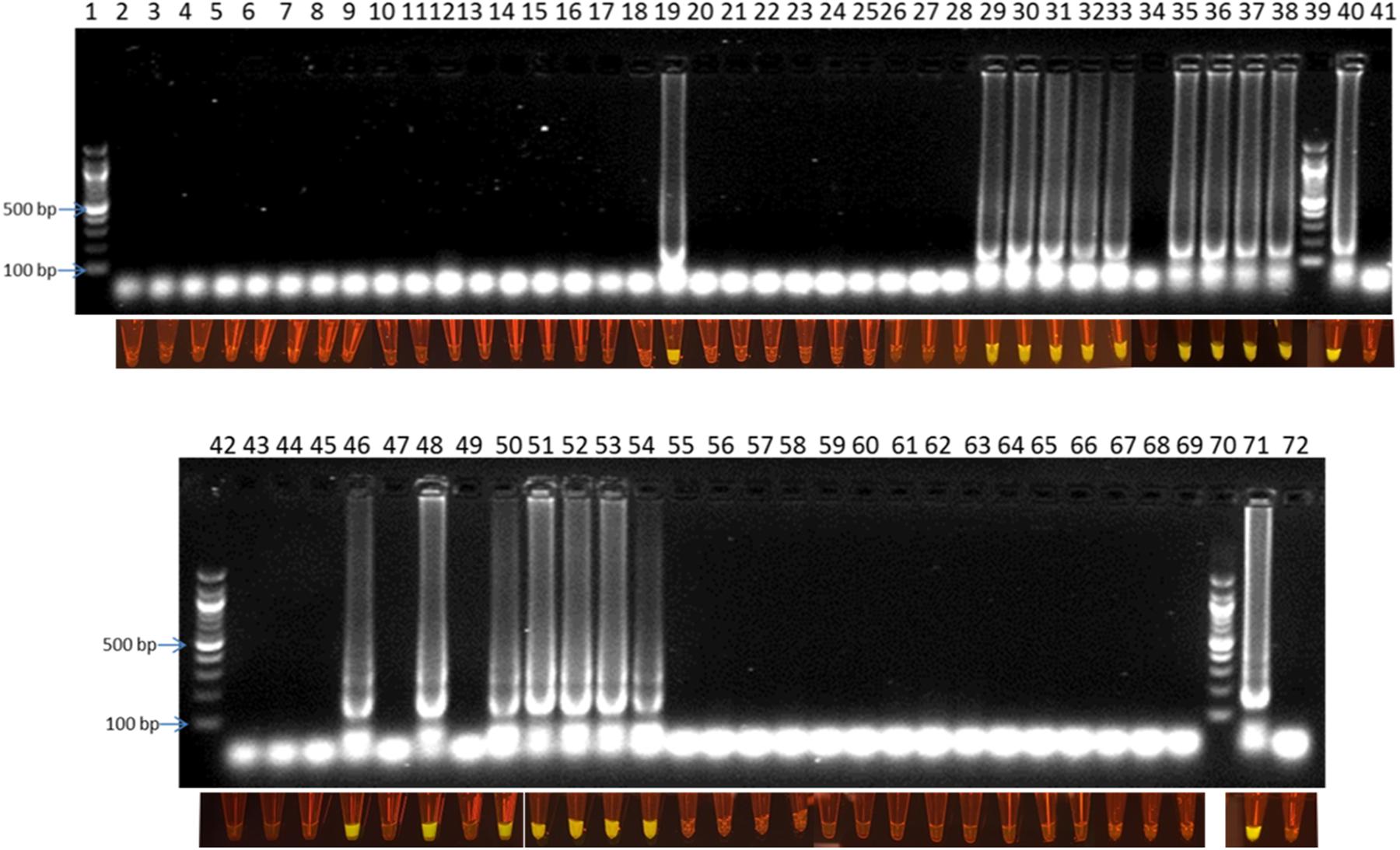
Figure 5. Visual detection of LAMP products on 2% agarose gel electrophoresis and using SYBR® Safe staining DNA for chicken fecal samples: lane 1, 39, 42, and 70: 100 bp ladder, lane 40 and 71: positive control, lane 41 and 72: negative control, and lane 2–38 and 43–69: samples from rotation 1, 2 and 3.
LAMP products could be detected by gel electrophoresis after amplification. Figure 3 showed that positive amplification was observed in positive control and with DNA target from 1 pg to 2 ng. On the other hand, the products could also be observed directly by adding 1 μl of 1:10 SYBR® Safe. Under a commercial portable DR22 blue LED transilluminator, both positive control and the reactions with DNA target concentrations ranging from 1 pg to 2 ng of DNA changed from transparent to yellow, while no color change was observed in the negative control and with 0.1 pg of DNA template (Figure 3). This result showed that there was no difference in the detection limit between the gel electrophoresis and color-change observation by the low cost commercial DR22 blue LED transilluminator. The coincidence of results of gel electrophoresis and direct visual detection were also observed in Figures 4, 5.
Twenty six bacterial strains as listed in Table 1 were tested for the specificity of the assay. As one can see in Supplementary Figure S2, LAMP positive reactions were observed only with C. jejuni and C. coli DNA templates, while no LAMP amplification was detected from the reactions using DNA from the other 24 bacterial reference strains that included 10 other Campylobacter species. The results showed that the LAMP assay was highly specific for C. jejuni and C. coli.
For this study, we have collected fecal samples weekly from sixteen broiler houses for testing Campylobacter through three rotations of poultry production (Table 3). After each rotation, the houses were cleaned and disinfected before starting a new rotation. The first rotation lasted 4 weeks. In the first and second week, Campylobacter was not detected in any of the sixteen houses. In the third week, Campylobacter was found in house number 2 in block 1 and house 16 in block 4. Only a week later, Campylobacter was detected in all the houses of block 1 and block 4. The second rotation lasted only 2 weeks. In the first week, Campylobacter was detected in house number 14 and 16 of block 4. One week later, Campylobacter was found in all the houses in block 4 and also house number 12 in block 3. The third rotation in this study lasted four weeks and no Campylobacter was found in the first and second week. In the third week, Campylobacter was found in house number 12 in block 3. In the fourth week, all houses in block 3 were positive for Campylobacter. The results showed clearly that, once Campylobacter was introduced into the broiler houses, it was transmitted easily and quickly to other broilers houses and other broilers blocks.
Campylobacter is abundant in cloaca, cecum and large intestine of poultry, up to 109 CFU/g of feces (Corry and Atabay, 2001; Hermans et al., 2010). After excretion, Campylobacter survives from a minimum of 2 days up to 14 days in the feces (Smith et al., 2016). Therefore, the feces may play a key role in the transmission of Campylobacter. Wild animals such as crawling insects, arthropods and flies have been shown to be vectors that can transfer Campylobacter from outside into broiler houses as well as from fecal materials inside broiler houses to other broiler houses (Sheppard, 2014). A study carried out in Denmark showed that there were 8.2 and 70.2% of flies caught outside a broiler house positive with Campylobacter as confirmed with conventional culture and PCR method, respectively (Hald et al., 2004). This study suggests that flies may play an important role in Campylobacter infection of broiler flocks during summer. Besides, air and dust are also considered as a source of the transmission (Berrang et al., 2004; Wilson, 2004). Bull et al. (2006) reported that approximately 6% of air samples were positive for Campylobacter once the broiler flocks were positive with Campylobacter (Bull et al., 2006). Furthermore, in the early stage of the colonization, Campylobacter can spread into the air of the broiler house (Olsen et al., 2009).
In addition, water may also contribute as a source of Campylobacter transmission in broilers houses. Bull et al. (2006) reported that approximately 31% of water samples from drinkers were positive with Campylobacter once flocks were positive with Campylobacter (Bull et al., 2006). Moreover, Campylobacter can persist for long periods in well-water and can also survive under various conditions for days (Buswell et al., 1998), weeks (Korhonen and Martikalnon, 1991), and even months (Rollins and Colwell, 1986; Thomas et al., 2002) in different aqueous environment (Schallenberg et al., 2005).
Although humidity in the broiler house is not a direct source of the transmission, it can influence the transmission of Campylobacter. Line (2006) showed that, at low relative humidity (30% ± 10%), the colonization of Campylobacter was delayed compared with high relative humidity (80% ± 10%) since it has been shown that water can enhance the survival of Campylobacter in broiler flocks (Trigui et al., 2015).
In summary, the present study describes a simple and rapid LAMP assay for the detection of C. jejuni and C. coli in chicken feces. The assay conditions were optimized for low reaction volume and shorter time of reaction. With the optimized conditions, it was possible to detect C. jejuni and C. coli in spiked chicken feces as low as 50 CFU/ml within 60–70 min in total. The LAMP assay was compared with an in house real-time PCR. Cohen’s kappa index showed excellent agreement between the two methods. The optimized LAMP method was used to study the transmission of Campylobacter at a Danish poultry farm. The results confirmed the capability of the LAMP technique as a rapid screening method for the detection of Campylobacter spp. at poultry production.
All datasets generated for this study are included in the manuscript/Supplementary Files.
DB, AW, AV, SN, MM, and YS designed the work. TQ and PE performed the experiments. DB, TQ, AW, AV, SN, TN, and YS wrote the manuscript.
SN is currently employed by the company Novo Nordisk A/S, Bagsværd, Denmark. TN is currently employed by Vinmec HealthCare System, Hanoi, Vietnam.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was supported by the Department of Biotechnology and Biomedicine, Technical University of Denmark, Denmark; a national GUDP-funded project through the ISOAMKIT Project No. 34009-15-1038 (www.isoamkit.com/en/); two EU H2020-funded projects, SMARTDIAGNOS, grant no. 687697 (www.smartdiagnos.eu), and VIVALDI, grant no. 773422 (www.vivaldi-ia.eu); and the EMIDA project, grant no. 3405-11-0433.
We thank Kirsten Michaëlis from DTU Food for technical assistant.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02443/full#supplementary-material
Addis, M., and Sisay, D. (2015). A review on major food-borne bacterial illnesses. J. Trop. Dis. 3:176. doi: 10.4176/2329-891X.1000176
Alves, J., Hirooka, E. Y., and de Oliveira, T. C. R. M. (2016). Development of a multiplex real-time PCR assay with an internal amplification control for the detection of Campylobacter spp. and Salmonella spp. in chicken meat. LWT Food Sci. Technol. 72, 175–181. doi: 10.1016/j.lwt.2016.04.051
Berrang, M. E., Northcutt, J. K., and Dickens, J. A. (2004). The contribution of airborne contamination to Campylobacter counts on defeathered broiler carcasses. J. Appl. Poult. Res. 13, 1–4. doi: 10.1093/japr/13.1.1
Biesta-Peters, E. G., Jongenburger, I., de Boer, E., and Jacobs-Reitsma, W. F. (2019). Validation by interlaboratory trials of EN ISO 10272 - microbiology of the food chain - Horizontal method for detection and enumeration of Campylobacter spp. - part 1: detection method. Int. J. Food Microbiol. 288, 39–46. doi: 10.1016/j.ijfoodmicro.2018.05.007
Bull, S. A., Allen, V. M., Domingue, G., Jørgensen, F., Frost, J. A., Ure, R., et al. (2006). Sources of Campylobacter spp. colonizing housed broiler flocks during rearing. Appl. Environ. Microbiol. 72, 645–652. doi: 10.1128/AEM.72.1.645
Buswell, C. M., Herlihy, Y. M., Lawrence, L. M., McGuiggan, J. T. M., Marsh, P. D., Keevil, C. W., et al. (1998). Extended survival and persistence of Campylobacter spp. water and aquatic biofilms and their detection by immunofluorescent-antibody and -rRNA staining. Appl. Environ. Microbiol. 64, 733–741.
CDC (2017). Foodborne Diseases Active Surveillance Network (FoodNet): FoodNet 2015 Surveillance Report (Final Data). Atlanta: U.S. Department of Health and Human Services.
Chin, W. H., Sun, Y., Høgberg, J., Hung, T. Q., Wolff, A., and Bang, D. D. (2017). Solid-phase PCR for rapid multiplex detection of Salmonella spp. at the subspecies level, with amplification efficiency comparable to conventional PCR. Anal. Bioanal. Chem. 409, 2715–2726. doi: 10.1007/s00216-017-0216-y
Chin, W. H., Sun, Y., Høgberg, J., Quyen, T. L., Engelsmann, P., Wolff, A., et al. (2016). Direct PCR – A rapid method for multiplexed detection of different serotypes of Salmonella in enriched pork meat samples. Mol. Cell. Probes 32, 24–32. doi: 10.1016/j.mcp.2016.11.004
Corry, J. E. L., and Atabay, H. I. (2001). Poultry as a source of Campylobacter and related organisms. J. Appl. Microbiol. 90, 96S–114S. doi: 10.1046/j.1365-2672.2001.01358.x
de Paz, H. D., Brotons, P., and Muñoz-Almagro, C. (2014). Molecular isothermal techniques for combating infectious diseases: towards low-cost point-of-care diagnostics. Expert Rev. Mol. Diagn. 14, 827–843. doi: 10.1586/14737159.2014.940319
Hald, B., Bang, D. D., Pedersen, K., Dybdahl, J., Madsen, M., and Study, T. (2004). Flies and Campylobacter broiler flocks. Emerg. Infect. Dis. 10, 1490–1492.
Helwigh, B., Julia, C., and Luise, M. (2017). Anonymous, 2016. Annual Report on Zoonoses in Denmark 2017. Søborg: National Food Institute.
Hermans, D., Martel, A., van Deun, K., Verlinden, M., van Immerseel, F., Garmyn, A., et al. (2010). Intestinal mucus protects Campylobacter jejuni in the ceca of colonized broiler chickens against the bactericidal effects of medium-chain fatty acids. Poult. Sci. 89, 1144–1155. doi: 10.3382/ps.2010-00717
Iwamoto, T., Sonobe, T., and Hayashi, K. (2003). Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J. Clin. Microbiol. 41, 2616–2622. doi: 10.1128/JCM.41.6.2616-2622.2003
Josefsen, M. H., Lofstrom, C., Hansen, T. B., Christensen, L. S., Olsen, J. E., and Hoorfar, J. (2010). Rapid quantification of viable Campylobacter bacteria on chicken carcasses, using real-time PCR and propidium monoazide treatment, as a tool for quantitative risk assessment. Appl. Environ. Microbiol. 76, 5097–5104. doi: 10.1128/AEM.00411-10
Kato, H., Yokoyama, T., Kato, H., and Arakawa, Y. (2005). Rapid and simple method for detecting the toxin B gene of Clostridium difficile in stool specimens by loop-mediated isothermal amplification. J. Clin. Microbiol. 43, 6108–6112. doi: 10.1128/JCM.43.12.6108-6112.2005
Korhonen, L. K., and Martikalnon, P. J. (1991). Survival of Escherichia coli and Campylobacter jejuni in untreated and filtered lake water. J. Appl. Bacteriol. 71, 379–382. doi: 10.1111/j.1365-2672.1991.tb03804.x
Kosti, T., Ellis, M., Williams, M. R., Stedtfeld, T. M., Kaneene, J. B., Stedtfeld, R. D., et al. (2015). Thirty-minute screening of antibiotic resistance genes in bacterial isolates with minimal sample preparation in static self-dispensing 64 and 384 assay cards. Appl. Microbiol. Biotechnol. 99, 7711–7722. doi: 10.1007/s00253-015-6774-z
Lawson, A. J., Logan, J. M., O’neill, G. L., Desai, M., and Stanley, J. (1999). Large-scale survey of Campylobacter species in human gastroenteritis by PCR and PCR-enzyme-linked immunosorbent assay. J. Clin. Microbiol. 37, 3860–3864.
Line, J. E. (2006). Influence of relative humidity on transmission of Campylobacter jejuni in broiler chickens. Poult. Sci. 85, 1145–1150. doi: 10.1093/ps/85.7.1145
Lund, M., Wedderkopp, A., Waino, M., Nordentoft, S., Bang, D. D., Pedersen, K., et al. (2003). Evaluation of PCR for detection of Campylobacter in a national broiler surveillance programme in Denmark. J. Appl. Microbiol. 94, 929–935. doi: 10.1046/j.1365-2672.2003.01934.x
Mori, Y., Kanda, H., and Notomi, T. (2013). Loop-mediated isothermal amplification (LAMP): recent progress in research and development. J. Infect. Chemother. 19, 404–411. doi: 10.1007/s10156-013-0590-0
Njiru, Z. K. (2012). Loop-mediated isothermal amplification technology: towards point of care diagnostics. PLoS Negl. Trop. Dis. 6:e1572. doi: 10.1371/journal.pntd.0001572
Okamura, M., Ohba, Y., Kikuchi, S., Suzuki, A., Tachizaki, H., Takehara, K., et al. (2008). Loop-mediated isothermal amplification for the rapid, sensitive, and specific detection of the O9 group of Salmonella in chickens. Vet. Microbiol. 132, 197–204. doi: 10.1016/j.vetmic.2008.04.029
Olsen, K. N., Lund, M., Skov, J., Christensen, L. S., and Hoorfar, J. (2009). Detection of Campylobacter bacteria in air samples for continuous real-time monitoring of Campylobacter colonization in broiler flocks. Appl. Environ. Microbiol. 75, 2074–2078. doi: 10.1128/AEM.02182-08
Peng, Y., Xie, Z., Liu, J., Pang, Y., Deng, X., Xie, Z., et al. (2011). Visual detection of H3 subtype avian influenza viruses by reverse transcription loop-mediated isothermal amplification assay. Virol. J. 8:337. doi: 10.1186/1743-422X-8-337
Powell, L. F., Lawes, J. R., Clifton-Hadley, F. A., Rodgers, J., Harris, K., Evans, S. J., et al. (2012). The prevalence of Campylobacter spp. in broiler flocks and on broiler carcases, and the risks associated with highly contaminated carcases. Epidemiol. Infect. 140, 2233–2246. doi: 10.1017/S0950268812000040
Rollins, D. M., and Colwell, R. R. (1986). Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl. Environ. Microbiol. 52, 531–538.
Romero, M. R., and Cook, N. (2018). A rapid LAMP-based method for screening poultry samples for Campylobacter without enrichment. Front. Microbiol. 9:2401. doi: 10.3389/fmicb.2018.02401
Sabike, I. I., Uemura, R., Kirino, Y., Mekata, H., Sekiguchi, S., Okabayashi, T., et al. (2016). Use of direct LAMP screening of broiler fecal samples for Campylobacter jejuni and Campylobacter coli in the positive flock identification strategy. Front. Microbiol. 7:1582. doi: 10.3389/fmicb.2016.01582
Sabike, I. I., and Yamazaki, W. (2019). Improving the detection accuracy and time for Campylobacter jejuni and Campylobacter coli in naturally infected live and slaughtered chicken broilers using a real-time fluorescent loop-mediated isothermal amplification approach. J. Food Prot. 82, 189–193. doi: 10.4315/0362-028X.JFP-18-179
Sandberg, M., Sørensen, L. L., Steenberg, B., Chowdhury, S., Ersbøll, A. K., and Alban, L. (2015). Risk factors for Campylobacter colonization in Danish broiler flocks, 2010 to 2011. Poult. Sci. 94, 447–453. doi: 10.3382/ps/peu065
Schallenberg, M., Bremer, P. J., Henkel, S., Launhardt, A., and Burns, C. W. (2005). Survival of Campylobacter jejuni in water: effect of grazing by the freshwater crustacean Daphnia cannata (Cladocera). Appl. Environ. Microbiol. 71, 5085–5088. doi: 10.1128/AEM.71.9.5085-5088.2005
Schrader, C., Schielke, A., Ellerbroek, L., and Johne, R. (2012). PCR inhibitors - occurrence, properties and removal. J. Appl. Microbiol. 113, 1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x
Sheppard, S. K. (2014). Campylobacter Ecology and Evolution | Book. Norfolk: Caister Academic Press.
Sibanda, N., McKenna, A., Richmond, A., Ricke, S. C., Callaway, T., Stratakos, A. C., et al. (2018). A review of the effect of management practices on Campylobacter prevalence in poultry farms. Front. Microbiol. 9:2002. doi: 10.3389/fmicb.2018.02002
Smith, S., Meade, J., Gibbons, J., McGill, K., Bolton, D., and Whyte, P. (2016). The impact of environmental conditions on Campylobacter jejuni survival in broiler faeces and litter. Infect. Ecol. Epidemiol. 6:31685. doi: 10.3402/iee.v6.31685
Stedtfeld, R. D., Stedtfeld, M., Kronlein, M., Seyrig, G., Ste, R. J., Cupples, A. M., et al. (2014). DNA extraction-free quantification of Dehalococcoides spp. in groundwater using a hand-held device. Environ. Sci. Technol. 48, 13855–13863. doi: 10.1021/es503472h
Sun, Y., Linh, Q. T., Hung, T. Q., Chin, W. H., Wolff, A., and Dang Duong, B. (2015). A lab-on-a-chip system with integrated sample preparation and loop-mediated isothermal amplification for rapid and quantitative detection of Salmonella spp. in food samples. Lab Chip 15, 1898–1904. doi: 10.1039/C4LC01459F
Techathuvanan, C., Draughon, F. A., and D’Souza, D. H. (2010). Loop-mediated isothermal amplification (LAMP) for the rapid and sensitive detection of Salmonella Typhimurium from pork. J. Food Sci. 75, M165–M172. doi: 10.1111/j.1750-3841.2010.01554.x
Thomas, C., Hill, D., and Mabey, M. (2002). Culturability, injury and morphological dynamics of thermophilic Campylobacter spp. within a laboratory-based aquatic model system. J. Appl. Microbiol. 92, 433–442. doi: 10.1046/j.1365-2672.2002.01550.x
Tresse, O., Alvarez-Ordóñez, A., and Connerton, I. F. (2017). Editorial: about the foodborne pathogen Campylobacter. Front. Microbiol. 8:1908. doi: 10.3389/fmicb.2017.01908
Trigui, H., Thibodeau, A., Fravalo, P., Letellier, A. P., and Faucher, S. (2015). Survival in water of Campylobacter jejuni strains isolated from the slaughterhouse. Springerplus 4:799. doi: 10.1186/s40064-015-1595-1
Velders, A. H., Schoen, C., and Saggiomo, V. (2018). Loop-mediated isothermal amplification (LAMP) shield for Arduino DNA detection. BMC Res. Notes 11:93. doi: 10.1186/s13104-018-3197-9
Vinayaka, A. C., Ngo, T. A., Kant, K., Engelsmann, P., Dave, V. P., Shahbazi, M.-A., et al. (2019). Rapid detection of Salmonella enterica in food samples by a novel approach with combination of sample concentration and direct PCR. Biosens. Bioelectron. 129, 224–230. doi: 10.1016/j.bios.2018.09.078
Wang, L., Shi, L., Alam, M. J., Geng, Y., and Li, L. (2008). Specific and rapid detection of foodborne Salmonella by loop-mediated isothermal amplification method. Food Res. Int. 41, 69–74. doi: 10.1016/j.foodres.2007.09.005
Wilson, I. G. (2004). Airborne Campylobacter infection in a poultry worker: case report and review of the literature. Commun. Dis. Public Health 7, 349–353.
Wingstrand, A., Neimann, J., Engberg, J., Nielsen, E. M., Gerner-Smidt, P., Wegener, H. C., et al. (2006). Fresh chicken as main risk factor for campylobacteriosis. Denmark. Emerg. Infect. Dis. 12, 280–284. doi: 10.3201/eid1202.050936
Xie, L., Xie, Z. Z. Z., Zhao, G., Liu, J., Pang, Y., Deng, X., et al. (2014). A loop-mediated isothermal amplification assay for the visual detection of duck circovirus. Virol. J. 11, 76. doi: 10.1186/1743-422X-11-76
Yamazaki, W., Taguchi, M., Ishibashi, M., Kitazato, M., Nukina, M., Misawa, N., et al. (2008). Development and evaluation of a loop-mediated isothermal amplification assay for rapid and simple detection of Campylobacter jejuni and Campylobacter coli. J. Med. Microbiol. 57, 444–451. doi: 10.1099/jmm.0.47688-0
Yamazaki, W., Taguchi, M., Ishibashi, M., Nukina, M., Misawa, N., and Inoue, K. (2009a). Development of a loop-mediated isothermal amplification assay for sensitive and rapid detection of Campylobacter fetus. Vet. Microbiol. 136, 393–396. doi: 10.1016/j.vetmic.2008.11.018
Yamazaki, W., Taguchi, M., Kawai, T., Kawatsu, K., Sakata, J., Inoue, K., et al. (2009b). Comparison of loop-mediated isothermal amplification assay and conventional culture methods for detection of Campylobacter jejuni and Campylobacter coli in naturally contaminated chicken meat samples. Appl. Environ. Microbiol. 75, 1597–1603. doi: 10.1128/AEM.02004-08
Yang, J., Yang, R., Cheng, A., Wang, M., Fu, L., Yang, S., et al. (2010). A simple and rapid method for detection of Goose Parvovirus in the field by loop-mediated isothermal amplification. Virol. J. 7, 14. doi: 10.1186/1743-422X-7-14
Zhang, J., Zhu, J., Ren, H., Zhu, S., Zhao, P., Zhang, F., et al. (2013). Rapid visual detection of highly pathogenic Streptococcus suis serotype 2 isolates by use of loop-mediated isothermal amplification. J. Clin. Microbiol. 51, 3250–3256. doi: 10.1128/JCM.01183-13
Zhang, Y., Shan, X., Shi, L., Lu, X., Tang, S., Wang, Y., et al. (2012). Development of a fimY-based loop-mediated isothermal amplification assay for detection of Salmonella in food. Food Res. Int. 45, 1011–1015. doi: 10.1016/j.foodres.2011.02.015
Keywords: campylobacteriosis, Campylobacter spp., loop mediated isothermal amplification, broiler fecal sample, broiler chicken production, rapid detection
Citation: Quyen TL, Nordentoft S, Vinayaka AC, Ngo TA, Engelsmenn P, Sun Y, Madsen M, Bang DD and Wolff A (2019) A Sensitive, Specific and Simple Loop Mediated Isothermal Amplification Method for Rapid Detection of Campylobacter spp. in Broiler Production. Front. Microbiol. 10:2443. doi: 10.3389/fmicb.2019.02443
Received: 20 June 2019; Accepted: 10 October 2019;
Published: 24 October 2019.
Edited by:
Nicolae Corcionivoschi, Agri-Food and Biosciences Institute (AFBI), United KingdomReviewed by:
Xiaonan Lu, University of British Columbia, CanadaCopyright © 2019 Quyen, Nordentoft, Vinayaka, Ngo, Engelsmenn, Sun, Madsen, Bang and Wolff. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dang Duong Bang, ZGRiYUBmb29kLmR0dS5kaw==; Anders Wolff, YXdvbEBkdHUuZGs=
†Present address: Steen Nordentoft, Novo Nordisk A/S, Bagsværd, Denmark Tien Anh Ngo, Vinmec Biobank, Vinmec HealthCare System, Hanoi, Vietnam
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.