- 1Department of Biochemistry, Genetics and Microbiology, University of Pretoria, Pretoria, South Africa
- 2Forestry, Agriculture and Biotechnology Institute, University of Pretoria, Pretoria, South Africa
The complexity of plant microbial communities provides a rich model for investigating biochemical and regulatory strategies involved in interbacterial competition. Within these niches, the soft rot Enterobacteriaceae (SRE) represents an emerging group of plant–pathogens causing soft rot/blackleg diseases resulting in economic losses worldwide in a variety of crops. A preliminary screening using next-generation sequencing of 16S rRNA comparatively analyzing healthy and diseased potato tubers, identified several taxa from Proteobacteria to Firmicutes as potential potato endophytes/plant pathogens. Subsequent to this, a range of molecular and computational techniques were used to determine the contribution of antimicrobial factors such as bacteriocins, carbapenem and type VI secretion system (T6SS), found in an aggressive SRE (Pectobacterium carotovorum subsp. brasiliense strain PBR1692 – Pcb1692) against these endophytes/plant pathogens. The results showed growth inhibition of several Proteobacteria by Pcb1692 depends either on carbapenem or pyocin production. Whereas for targeted Firmicutes, only the Pcb1692 pyocin seems to play a role in growth inhibition. Furthermore, production of carbapenem by Pcb1692 was observably dependent on the presence of environmental iron and oxygen. Additionally, upon deletion of fur, slyA and expI regulators, carbapenem production ceased, implying a complex regulatory mechanism involving these three genes. Finally, the results demonstrated that although T6SS confers no relevant advantage during in vitro competition, a significant attenuation in competition by the mutant strain lacking a functional T6SS was observed in planta.
IMPORTANCE
Soft rot Enterobacteriaceae (SRE) represents important phytopathogens causing soft rot/blackleg diseases in a variety of crops leading to huge economic losses worldwide. These pathogens have been isolated alongside other bacteria from different environments such as potato tubers, stems, roots and from the soil. In these environments, SREs coexist with other bacteria where they have to compete for scarce nutrients and other resources. In this report, we show that Pectobacterium carotovorum subsp. brasiliense strain PBR1692 – Pcb1692, which represents one of the SREs, inhibits growth of several different bacteria by producing different antimicrobial compounds. These antimicrobial compounds can be secreted inside or outside the plant host, allowing Pcb1692 to effectively colonize different types of ecological niches. By analyzing the genome sequences of several SREs, we show that other SREs likely deploy similar antimicrobials to target other bacteria.
Introduction
For phytopathogenic bacteria, more is known about interactions with their hosts and virulence factors recruited to enforce successful colonization while relatively less is known about microbe–microbe interactions and how such interactions impact on niche colonizations (Bhunia, 2018; Glazebrook and Roby, 2018; Rodriguez-Moreno et al., 2018; Xin et al., 2018). Studies on the mammalian gut microbiome, the advent of new sequencing technologies, availability of many ‘omics’ data sets such as metagenomics, transcriptomics and proteomics are some of the factors that have brought microbial interactions to the fore (Knief et al., 2012; Yu and Zhang, 2012; Kõiv et al., 2015; Bost et al., 2018; Clayton et al., 2018). Thus, the past few decades have seen a rapid growth in the body of literature on microbial interactions.
Bacteria exist in complex multispecies communities which are mostly characterized by competition and to a lesser extent, cooperation (Stubbendieck and Straight, 2015; Bauer et al., 2018; García-Bayona and Comstock, 2018). In these interactions, survival depends on the ability to compete for resources in a given niche. Mechanisms of competition can be classified as exploitative or interference (Hibbing et al., 2010; Cornforth and Foster, 2013; Martin et al., 2017; Tyc et al., 2017; García-Bayona and Comstock, 2018). Exploitative competition is competition whereby microbes compete for scarce resources and the winner typically limits nutrient availability from competitors. Interference competition is characterized by production of specialized metabolites (antibacterial compounds). Microbes have a large arsenal of antimicrobial weapons that mediate competition and these include contact-dependent inhibition (CDI), such as the type VI secretion system (T6SS), antibiotic and/or bacteriocin production (Coulthurst et al., 2005; Aoki et al., 2010; Desriac et al., 2010; Russell et al., 2011, 2014; Grinter et al., 2012; Costa et al., 2015; Shyntum et al., 2015; Hachani et al., 2016; Wall, 2016; Verster et al., 2017; Bernal et al., 2018; Trunk et al., 2018). Over the years, many studies have interrogated mechanisms of the different antimicrobials ‘singularly’ while fewer studies have systematically investigated these factors in concert. For example, a growing number of studies have shown the role, mechanism of action and targets of T6SS, T5SS, and bacteriocins in bacterial competitions (Aoki et al., 2010; Bernal et al., 2018). There is therefore a need to study these systems collectively and under similar experimental conditions in order to gain better insight into their relative contribution to bacterial competition.
The T6SS is typically made up of 15–23 different proteins amongst which 13 proteins (TssA-M) are highly conserved in a wide range of bacteria and encode structural components of the T6SS (Mougous et al., 2006; Pukatzki et al., 2006). This secretion system has been extensively studied in several plant and animal pathogens and shown to secrete bactericidal effectors which inhibit growth of targeted bacteria lacking cognate immunity proteins (Poole et al., 2011; Bernal et al., 2018). The T6SS is regulated by quorum sensing, the ferric uptake regulator (Fur), histone-like nucleoid structuring protein (H-NS), RNA binding proteins (Hfq and RmsA) and several two-component systems (Silverman et al., 2012; Miyata et al., 2013; Salomon et al., 2014). In addition to the T6SS, Pectobacterium spp., upon induction with DNA damaging agents, secrete low molecular weight bacteriocins such as carocin (S1K, S2, and D), pectocin (P, M1, and M2) and the phage-like bacteriocin, carotovoricin (Ctv) (Chuang et al., 2007; Chan et al., 2009, 2011; Roh et al., 2010; Grinter et al., 2012). Ctv is encoded by the ctv gene cluster, containing 18 to 23 different genes encoding the lysis cassette, tail sheath and tail fiber (Yamada et al., 2006). Similar to T6SS effectors, bacteriocins are bactericidal effectors which play a role in both inter and intraspecies competition. In addition to bacteriocins, some Pectobacterium, Dickeya, and Serratia spp. produce the β-lactam antibiotic, 1-carbapen-2-em-3-carboxylic acid (carbapenem), which is targeted against different bacteria species including strains closely related to the carbapenem-producing strain. Carbapenem is encoded by carA/B/C/D/E/F/G/H genes, with carF and carG encoding immunity proteins (CarF/G) (McGowan et al., 1995, 1997, 2005). The car gene cluster was also shown to be regulated by the quorum sensing regulator, CarR, the stress response regulator SlyA (Hor) and Hfq (McGowan et al., 1995; Wang et al., 2018).
Soft Rot Enterobacteriaceae (SRE), comprising of Dickeya and Pectobacterium spp. is a group of pathogens causing huge economic losses on potato production worldwide (Adeolu et al., 2016; Charkowski, 2018). In addition to potato, SREs also cause disease in several monocots and dicots including pepper, avocado, onion, sugar beet, cacti, chicory, wasabi, rice, cabbage, marigold, burdock, cucumber, cassava, lettuce, pumpkin, and many other plants (Ma et al., 2007). They cause blackleg and soft rot diseases in the field or during post-harvest storage, respectively. Decaying potato tubers are characterized by multispecies bacterial communities that are both Gram positive and negative (Kõiv et al., 2015; Charkowski, 2018). Furthermore, within the SRE, it is quite common to isolate bacteria from Pectobacterium spp. as well as Dickeya spp., the two primary genera within Enterobacteriaceae from a single infected plant (Kõiv et al., 2015). Of these, Pectobacterium carotovorum subsp. brasiliense is an emerging SRE with global distribution (van der Merwe et al., 2010; Panda et al., 2012; Onkendi and Moleleki, 2014; Moraes et al., 2017; Ma et al., 2018; Voronina et al., 2019). Previous studies have shown that Pcb strain PBR1692 (referred to henceforth as Pcb1692) is able to inhibit growth of Pectobacterium carotovorum subsp. carotovorum WPP14 (Pcc) and P. atrosepticum SCRI1043 suggesting that many more bacteria can be inhibited by Pcb1692 thus giving it a fitness advantage, in a given ecological niche (Marquez-Villavicencio et al., 2011; Durrant, 2016). In this study, we use a combination of metagenomics, in silico analyses and various in vitro and in planta competition assays to identify and characterize bacterial targets, and the distinctive roles played by the different antimicrobial produced by Pcb1692 in bacterial competition, their regulation and possible contribution toward adaptation to diverse ecological niches.
Materials and Methods
Strains and Growth Conditions
All bacterial strains used in this study are listed in Supplementary Table S1. Bacterial strains were cultured in either Luria-Bertani (LB) medium or M9 minimum medium (Miller, 1972) supplemented with 0.4% sugar (glucose, sucrose, or glycerol) and grown aerobically at 37 or 28°C with or without agitation, as required by the given experimental conditions. Growth medium was supplemented with either 100 μg/ml ampicillin, 50 μg/ml kanamycin, 15 μg/ml gentamycin, 15 μg/ml tetracycline, or 50 μg/ml chloramphenicol. Antibiotics were purchased from (Sigma-Aldrich).
DNA Isolation, Miseq Illumina Sequencing, and Data Analysis
Six diseased (soft rot disease) and six healthy potato tuber were collected from Gauteng, North West and Limpopo Province in South Africa. Genomic DNA (gDNA) was extracted from 1 g of fresh tissue using the ZYMO RESEARCH Quick-gDNA MiniPrep kit, according to the manufacturer’s instruction. DNA from either healthy or diseased tubers was pooled and aliquots used for 16S rRNA PCR amplification and Illumina Miseq sequencing. The 16S rRNA PCR was performed to amplify v3–v4 variable regions using primers 419F (ACTCCTACGGGAGGCAGCAG) and 806R (GGACTACHVGGGTWTCTAAT) (Fadrosh et al., 2014). The PCR reaction mixture consisted of 2.5 mM dNTPs, 10 × DreamTaq buffer (supplemented with 20 mM MgCl2), 0.5 U DreamTaq Polymerase (Fermentas), 10 μM each forward and reverse primer, 100 ng DNA template and nuclease free water up to the final reaction volume of 25 μl. PCR amplification conditions were as follows: denaturation at 95°C for 3 min, followed by 35 cycles at 95°C 30 s, 58°C 45 s, 72°C 2 min and the final extension at 72°C for 5 min. The purified PCR products (260/280 ratio ≥ 1.8) were sent to the Agricultural Research Council (ARC) Biotechnology platform, Pretoria, South Africa for 16S metagenomics library preparation, modified with Illumina specific adapters (Klindworth et al., 2013). Thereafter, the pooled samples were subjected to Miseq Illumina sequencing. Paired-end reads were trimmed using Trimmomatic v0.38 (Bolger et al., 2014), allowing minimum nucleotide quality average of 30 per 5 bp window in the sequences. Illumina adapters were removed using Cutadapt v1.18 (Martin, 2011). Chloroplast sequences were filtered out based on sequence identity assessed through Blastn (Camacho et al., 2009) with 16S rRNA acquired from Solanum tuberosum. High-quality reads were then submitted to SILVAngs rRNA-based data analysis pipeline under default settings (Quast et al., 2012). A total of 327,032 and 475,616 paired-end reads from diseased and healthy potato, respectively, were processed. After the initial systematic quality trimming, the 63,841 and 68,015 remaining reads from diseased and healthy samples, respectively, were filtered for chloroplast sequences. A total of 12,379 and 35,526 chloroplast sequences were removed from diseased and healthy samples respectively, and the remaining high-quality reads were analyzed using the SILVAngs pipeline (Quast et al., 2012). Subsequently, a total of 35,177 and 805 reads respectively from diseased and healthy samples were successfully classified into bacterial taxa. The Krona package (Ondov et al., 2011) was used to generate radial visualization of the resulting taxonomic profiles.
Plant Extracts
Potato tuber extracts were prepared as previously described, with slight modifications (Mattinen et al., 2007). In summary, five grams of surface sterilized potato tubers (flesh and peel) were ground in 100 ml of distilled water using a blender. Plant tissue was removed by repeated centrifugation at 5000 rpm for 30 min at 4°C. The clarified plant extract was filter sterilized twice using a 0.2 μm filters, and aliquots stored at −16°C. Iron was selectively removed from potato tuber extracts by adding 200 μM of 2,2′-dipyridyl (Sigma Aldrich) dissolved in chloroform in a 1:1 ratio. The resulting mixture was vortexed for 2 min and iron free supernatant collected by centrifugation at 5000 rpm for 10 min at 4°C. The M9 agar plates were then supplemented with a 1:10 dilution of either plant extracts or plant extracts pre-treated with 2,2′-dipyridyl.
Orthology Predictions and Binding Sites Analyses
Orthologs of bacteriocins and car genes were predicted by using OrthoMCL (Li et al., 2003) pipeline to analyze complete protein datasets from 100 Pectobacterium and Dickeya strains obtained from the RefSeq database as previously described (Bellieny-Rabelo et al., 2019b). Upstream regions of expR, slyA, carR and carA from Pectobacterium betavasculorum strain NCPPB2795, Pectobacterium atrosepticum strain ICMP19972, Pectobacterium carotovorum subsp. carotovorum strain NCPPB312 and Pcb1692 genomes were retrieved from the NCBI online database1. Then the respective sequences of those regions were scanned using FIMO software from MEME package (Bailey et al., 2006) for the presence of putative Fur binding sites. This analysis was conducted using all known Fur binding sites found in MEME databases for prokaryotes: Prodoric (release 8.9) (Munch et al., 2003), RegTransBase v4 (Kazakov et al., 2006), and CollectTF (Kilic et al., 2014).
In vitro Bacterial Competition Assays
Inter and intra species bacterial competition assays were performed as previously described (Shyntum et al., 2015). The complete list of bacteria used in this assay is provided in Supplementary Table S1. In summary, targeted bacteria were transformed with plasmid pMP7605 conferring gentamycin resistance (Lagendijk et al., 2010). Overnight cultures of Pcb1692 and targeted bacteria were normalized to an OD600 = 0.1, mixed in a 1:1 ratio and 20 μl spotted on LB or M9 agar containing no antibiotics. When required, M9 was supplemented with 0.01, 1, 10, or 50 μM of ferric sulfate (FeSO4.7H2O), magnesium sulfate (MgSO4.7H2O), manganese chloride (MnCl2.4H2O), zinc sulfate (ZnSO4.7H2O), nickel sulfate (NiSO4), cobalt chloride (CoCl2.H2O) or copper chloride (CuCl.H2O). Experiments were performed in triplicates and repeated three independent times.
Bacterial spots were briefly air dried and allowed to grow for 16 h at 28°C. When required, bacterial competition assays were performed anaerobically by placing agar plated in an anaerobic jar (Merck) containing moistened Anaerocult A (Merck) which absorbs oxygen and a moistened blue Anaerotest strip (Merck) which turns white in the absence of oxygen. Anaerobic jars were then tightly sealed and incubated overnight at 4°C. Thereafter, overnight spots were scraped off, serially diluted and plated on LB agar supplemented with gentamycin (15 μg/ml). The effect of iron on bacterial competition was determined using the following agar plates: (1) LB agar, (2) M9 minimal agar, (3) M9 supplemented with FeSO4.7H2O (10 μM), (4) M9 supplemented with potato tuber extracts, (5) M9 supplemented with FeSO4.7H2O (10 μM) and potato tuber extracts, (6) M9 supplemented with potato tuber extracts pre-treated with the iron chelator 2,2′-dipyridyl (200 μM) and (7) complementation of plate 6 listed above by the addition FeSO4.7H2O (10 μM) to potato tubers pre-treated with the 2,2′-dipyridyl (200 μM). All assays were performed in triplicates and three independent times. The results are presented as CFU/ml of target bacteria.
Generation of Pectobacterium carotovorum subsp. brasiliense Mutant Strains
The different Pcb1692 mutant strains were generated by site-directed mutagenesis using the lambda red recombination technique (Datsenko and Wanner, 2000). In summary, overlap extension polymerase chain reaction (PCR) was used to generate a gene knockout cassette by fusing the upstream and downstream regions flanking the targeted gene to the kanamycin resistance genes (Shyntum et al., 2015; Tanui et al., 2017). The fused PCR product was then electroporated into Pcb1692 harboring pKD20 and transformants were selected on nutrient agar supplemented with either 50 μg/ml kanamycin or 50 μg/ml chloramphenicol. The list of primers used in this study is provided in Supplementary Table S2. The integrity of each Pcb1692 mutant strain was confirmed by PCR analyses, nucleotide sequencing and Southern blot analysis (results not shown). The list of mutant strains generated in this study is provided in Supplementary Table S3.
Complementation of the Pcb1692 Mutant Strains
The list of primers used in this study is provided in Supplementary Table S2. The Pcb1692 fur, expI, PCBA_RS02805 (pyocin, designated pyo), PCBA_RS02805:PCBA_RS02810 (pyocin and immunity genes, designated pyoI) and the T6SS genes tssA/B including their putative promoter sequences were PCR amplified and individually cloned into CloneJet 1.3 to generate pfur, pexpI, ppyo, ppyoI, and pT6, respectively. The tetracycline resistance gene (tetr) with its downstream promoter sequence was amplified from plasmid pME6031 (Heeb et al., 2000) with primers tetF/tetR. The ampicillin resistance gene located in plasmids pfur, pexpI, ppyo, ppyoI, pT6, and pJET-slyA (Bellieny-Rabelo et al., 2019a) was removed by inverse PCR, using primers IrF/IrR. The linearized vectors were then ligated to tetr to generate pJET3-fur, pJET3-slyA and pJET3-expI, pJET3-pyo, pJET3-pyoI, and pJET3-T6. The Pcb1692 fur promoter sequence was amplified with FurF/R and ligated to cloneJet 1.3 to generate pfur-1. The ampr gene in pfur-1 was replaced with tetr, as previously described, generating pJET4. The promoterless Pcb1692 carC and carFG genes were amplified by PCR, followed by restriction digested with SpeI and cloned into pJET4 previously digested with SpeI, to generate pJET4-carC and pJET4-carFG, respectively. Plasmids pJET3-fur, pJET3-slyA, pJET3-expI, pJET4-carC, pJET3-pyo, pJET3-pyoI, and pJET3-T6 were transformed into their corresponding mutant strains to generate the complemented mutant strains, Pcb1692Δfurpfur, Pcb1692ΔslypslyA, Pcb1692ΔexpIpexpI, Pcb1692ΔcarCpcarC, Pcb1692Δpyoppyo, Pcb1692ΔpyoIppyoI, and Pcb1692ΔT6pT6. Plasmid pJET4-carFG was transformed into D. dadantii in order to experimentally determine if the CarF and CarG proteins confer immunity to the carbapenem produced by Pcb1692. All transformants were maintained in LB supplemented with 15 μg/ml tetracycline.
In planta Competition Assay
In planta competition assays were performed as previously described (Marquez-Villavicencio et al., 2011) with slight modifications. Targeted bacteria (D. chrysanthemi, D. dadantii, Pcc, P. atrosepticum and PcbG4P5 were transformed with plasmid pMP7605 conferring gentamycin resistance (Lagendijk et al., 2010), as previously described. Wild-type Pcb1692, Pcb1692ΔT6, and Pcb1692ΔT6pT6 strains were grown overnight in LB supplemented with appropriate antibiotics. The optical density of overnight bacterial cultures was adjusted to OD600 = 1, resuspended in 1 X PBS, mixed in a 1:1 ratio and inoculated into surface sterilized potato tubers (cv. Mondial). Inoculated tubers were placed in moist plastic containers, sealed, and incubated for 72 h at 25°C. Macerated tuber tissue was scooped out and the CFU/ml of surviving targeted bacteria determined by serial dilutions on LB supplemented with gentamycin (15 μg/ml). The assays were performed three independent times each consisting of three technical repeats.
Detection of Carbapenem and Bacteriocin Production by Pcb1692
Carbapenem production was determined by spot-on-lawn assays. Carbapenem production was determined for the following strains: wild-type Pcb1692, Pcb1692 mutant strains, HPI01 and laboratory stocks of Pcb strains whose genome sequences have not been determined (strains CC1, CC2, XT3, XT10, 358, G4P5, and G4P7). In summary, a 10 μl culture of Pcb strains, at OD600 = 0.1 was spotted on a lawn of freshly prepared targeted bacteria and a clear zone around Pcb was indicative of carbapenem production. To determine the role of iron in carbapenem production, lawns of targeted bacteria were prepared in M9 or M9 supplemented with iron (10 μM). Cultures of Pcb strains, OD600 = 0.1 were then spotted onto the M9 and production of a clear zone determined 24 h post-inoculation at 28°C. Bacteriocin production was determined by spot-on-lawn overlay method, as previously described (Roh et al., 2009). In summary, 10 μl of overnight cultures of Pcb strains were spotted on LB or M9, air dried and incubated overnight. Thereafter, bacteriocin production was induced by either UV irradiation or mitomycin C treatment. For mitomycin C induction, 10 μl of mitomycin C (50 μg/ml) was inoculated on top of Pcb strains, air dried and incubated in the dark for 5 h. For UV irradiation, cultures of Pcb strains were exposed to UV light for 10 s and incubated in the dark for 5 h. Thereafter, bacteriocin-induced Pcb cultures were killed with chloroform fumes for 10 min and then air dried for 30 min. Twenty microliters of overnight cultures of targeted bacteria was inoculated into 25 ml of cooled 0.7% agar and overlayed on bacteriocin induced Pcb1692 strains. Clear zones around Pcb strains were indicative of bacteriocin production. In control experiments, Pcb strains were not exposed to UV or mitomycin C. All experiments were repeated three times. To determine the ability of Pcb1692 strains to produce carbapenem and bacteriocins under anaerobic conditions, the experimental procedures were the same as above except that bacteria were grown anaerobically in anaerobic jars (Merck) containing moistened Anaerocult A (Merck) which absorbs oxygen and moistened blue Anaerotest strips (Merck) which turns white in the absence of oxygen. Anaerobic jars were then tightly sealed and incubated overnight.
Statistical Analysis
All experiments were performed in triplicates and three independent times. Where applicable, an unpaired, two-tailed Student’s t-test was performed to determine statistical significance and a p-value less than 0.05 (p < 0.05) was considered to be a statistically significant difference.
Results
The Microbiome of Potato Tubers
To understand typical microbial flora on potato tubers and thus infer potential competitors, we performed 16S rRNA metagenomics of diseased and healthy potato tuber tissues. We found overall a high representation of bacteria from the phylum Proteobacteria in both diseased (51%) and healthy potato tubers (75%). On the other hand, Firmicutes made up 2 and 28% of bacteria in healthy and diseased potato tubers, respectively, implying an increasing population of Firmicutes as tubers progress from a healthy to a diseased state. In both diseased and healthy tissues, the dominant orders within the Gammaproteobacteria were Enterobacteriales (56 and 31%, respectively) and Pseudomonadales (25 and 59%, respectively) (Figure 1). Within Enterobacteriales, the analyses identified the following genera: Pantoea, Enterobacter, Serratia together with genera such as Yersinia, Klebsiella, Salmonella, Escherichia, Citrobacter, and Shigella in both healthy and diseased potato (Figure 1). As can be expected, soft rot bacteria within the Pectobacterium and Dickeya genera were well-represented in diseased potato (Figure 1).
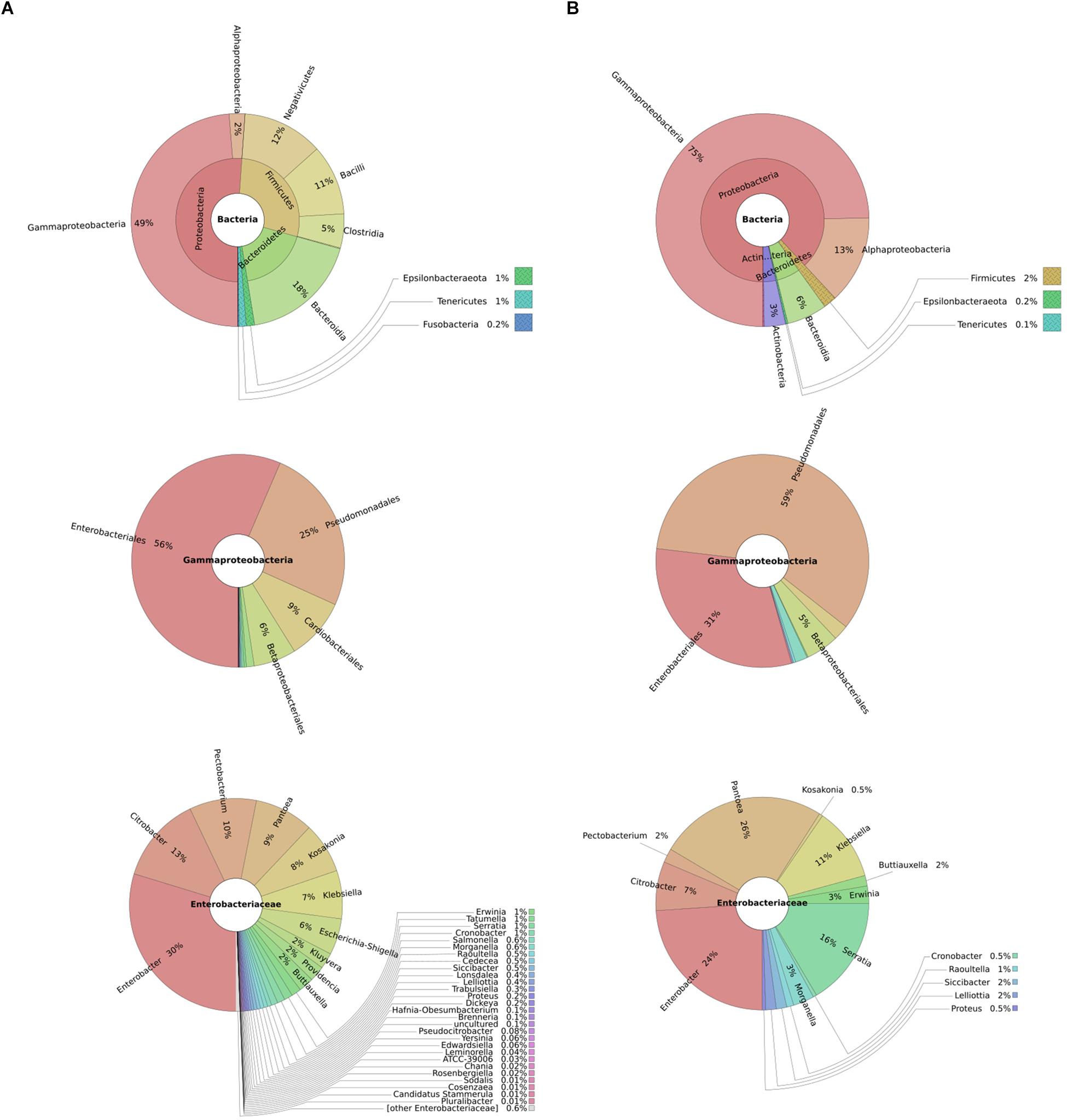
Figure 1. Taxonomic profile of diseased potatoes assessed through shotgun sequencing: metagenomics analyses represented by composite pie-charts depict the taxonomic representation obtained from sequenced dataset. For each graph, outer radiuses represent the relative proportion of reads assigned to different taxa of the inner circle taxon. Charts under column (A) represents the relative proportion of reads found in diseased potato tubers while (B) represents reads from healthy potato tubers.
Bacterial Growth Inhibition by Pcb1692 Is Associated With Inter and Intra Species Competition
Pcb1692 has previously been shown to inhibit growth of some closely related SREs such as Pcc and D. dadantii (Marquez-Villavicencio et al., 2011; Durrant, 2016). In this study, we wanted to determine the spectrum of bacteria killed by Pcb1692 in competition assays. We hypothesized that Pcb strains will likely compete with bacteria that they typically share the same niche. We used the metagenomics data to identify bacteria (at genus level) generally found in both healthy and diseased potato tubers. We combined the metagenomics data analysis with literature searches to identify bacteria (genus and species) previously isolated from diseased potato tubers (Hollis, 1951; Bulgarelli et al., 2012; Gupta et al., 2014; Schreiter et al., 2014; Donn et al., 2015; Hong et al., 2015; Kõiv et al., 2015). With this combined information, we selected a number of different bacteria for subsequent competition assays. We chose to use type strains as they have either draft or full genome sequences publicly available. This choice was necessitated by the need to perform in silico analyses in order to understand the bases of susceptibility or resistance of target strains, to killing by Pcb1692. Target cells were co-cultured in water or with Pcb1692. As has been shown by others, the results show that Pcb1692 can inhibit growth of closely related Pectobacterium and Dickeya species when co-cultured in LB agar (Figure 2). The magnitude of this inhibition is strain specific. For example, up to a 6-log reduction in CFU/ml (p < 0.01) was observed when Pcb1692 was co-cultured with some members of SRE within the phylum Proteobacteria, such as P. atrosepticum, Pcc, Pectobacterium carotovorum subsp. odoriferum (Pco), and D. dadantii while a 1–3 log reduction (p < 0.05) was observed for P. paradisiaca, P. cipripedii, P. cacticida and P. betavasculorum. The study also demonstrates that within Proteobacteria, Pcb1692’s competitive ability is not only restricted to SREs. Our results show that Pcb1692 can inhibit other members of Enterobacteriaceae such as Enterobacter cowanii, Salmonella typhimurium, Escherichia coli and Serratia marcescens and one Pseudomonad (Pseudomonas aeruginosa) (Figure 2). The ability of Pcb to inhibit P. aeruginosa, S. typhimurium, E. coli, E. cowanii and S. marcescens, which have previously been isolated from potato tubers (Malek et al., 2013; Kõiv et al., 2015) likely gives Pcb a fitness advantage in potato tubers, however, this needs to be experimentally verified. Interestingly, the results also showed that Pcb1692 was able to significantly inhibit growth of other closely related Pcb strains such as G4P5, G4P7, XT3, and XT10 (5 log reduction in CFU/ml p < 0.01) but not strains HPI01, 358, CCI, and CC2 (Figure 2 and Supplementary Table S4). The intra and interspecies inhibition observed could either be due to nutrient competition, phage induction, small metabolite production, or secretion of effectors by Pcb1692 for which closely related strains lack the cognate immunity factor, a premise that is supported by our subsequent in silico analysis. In addition, the data also showed that Pcb1692 was unable to inhibit in vitro growth of other Proteobacteria such as D. chrysanthemi, P. wasabiae, S. ficaria, Pantoea ananatis, Pantoea stewartii subsp. indologenes, and Firmicutes such as Bacillus subtilis and B. cereus in rich LB medium. Together, these findings demonstrate that Pcb1692 readily inhibits growth of both (1) most SREs analyzed in this study, including closely related Pcb strains, as well as (2) non-SREs.
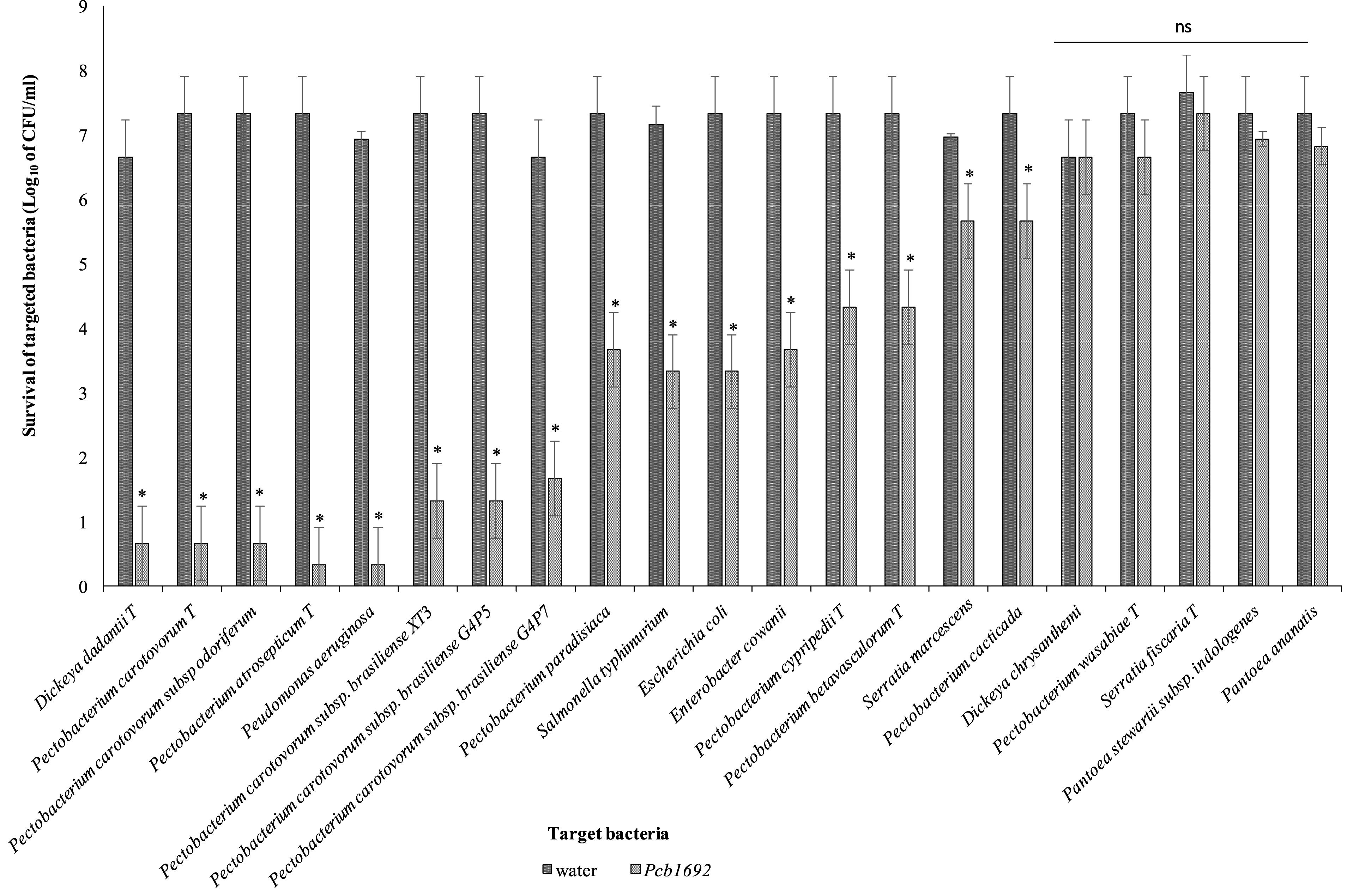
Figure 2. Interbacterial competition between Pectobacterium carotovorum subsp. brasiliense (Pcb1692) and targeted bacteria: Pcb1692 and targeted bacteria were co-cultured in a 1:1 ratio on LB and the log10 CFU/ml of surviving targeted bacteria determined 24 h post-incubation. Experiments were performed in triplicates and repeated three independent times. Bars represent mean values, error bars represent one standard deviation and asterisk represent p-values for the differences in CFU/ml of recovered target bacteria when co-cultured with Pcb1692 relative to water control as determined by the two-tailed Student’s t-test. p < 0.05 was considered to be statistically significant. P, Pectobacterium.
Iron in Potato Tuber Extracts Plays an Essential Role in Bacterial Competition
Potato tubers contain several metals such as iron, potassium, zinc, copper, manganese, lead, cadmium; however, these metals are sequestered in different plant compartments and not readily available to bacteria in the intracellular spaces (Luis et al., 2011; Tack, 2014). Some of these metals play a role in virulence and bacterial competition, by either acting as enzyme co-factors or co-activator/-repressors of some transcriptional regulators (Fones and Preston, 2013). We therefore investigated the effect of some of these metals on the observed ability of Pcb1692 to kill targeted bacteria in vitro. For this part of the study, Pcb1692 was co-cultured with Pcc and D. dadantii in M9 minimal media supplemented with different metals at concentrations 0.01 to 50 μM, representing physiological levels typically found in potato tubers (Luis et al., 2011; Tack, 2014) (see section “Materials and Methods”). The results showed that while low concentrations of copper and cobalt (0.01 and 1 μM) had no effect on competition, higher concentrations of both metals (10 and 50 μM) inhibited growth of both Pcb1692 and Pcc (Supplementary Data Sheet S1). In addition, different concentrations of magnesium, manganese, zinc, nickel (0.01, 1, 10, and 50 μM) had no effect on bacterial competition (Supplementary Data Sheet S1). On the other hand, the addition of ferric iron (10 or 50 μM) to M9 minimal medium enhanced the ability of Pcb1692 to kill either Pcc, Pco, or D. dadantii while lower concentrations (0.1 and 1 μM) had no effect on bacteria competition (Figure 3 and Supplementary Data Sheet S2). Therefore, 10 μM ferric iron was used to supplement M9 medium in all downstream assays associated with iron. For competition assays, target bacteria were co-cultured in water (controls) or with Pcb1692 (Figure 3). Figure 3 shows that when Pco was co-cultured with Pcb1692 in LB (rich) medium, fewer cells of target bacteria were recovered compared to controls (p < 0.05). However, a 1–2 log increase in the number of target bacteria cells recovered after co-culture in nutrient-poor M9 medium (relative to rich medium) was observed. This indicated a statistically significant reduction (p < 0.05) in the ability of Pcb1692 to kill targeted bacteria in nutrient-poor medium compared to LB rich medium. The magnitude of inhibition by Pcb1692 significantly increased by 2 log when M9 agar was supplemented with potato tuber extracts. Thus, adding tuber extracts to minimal medium enriched the medium and hence a similar outcome on competition as with rich LB medium was observed. When M9 was supplemented with iron, fewer competitor/target cells were recovered (relative to rich medium), indicating a statistically significant (p < 0.05) increase in the Pcb1692 killing ability. Addition of the iron chelator 2,2′-dipyridyl to M9 tuber-supplemented medium eliminated ability of Pcb1692 to kill competing bacteria, while addition of 10 μM of ferric iron to M9 media restored competition in the absence of plant extracts. Together these findings demonstrate that the presence of iron increased the ability of Pcb1692 to inhibit targeted bacteria. In the next sections, we report the approaches used to establish whether or not the effect of iron is associated with the expression of T6SS, carbapenem or bacteriocin production in Pcb1692.
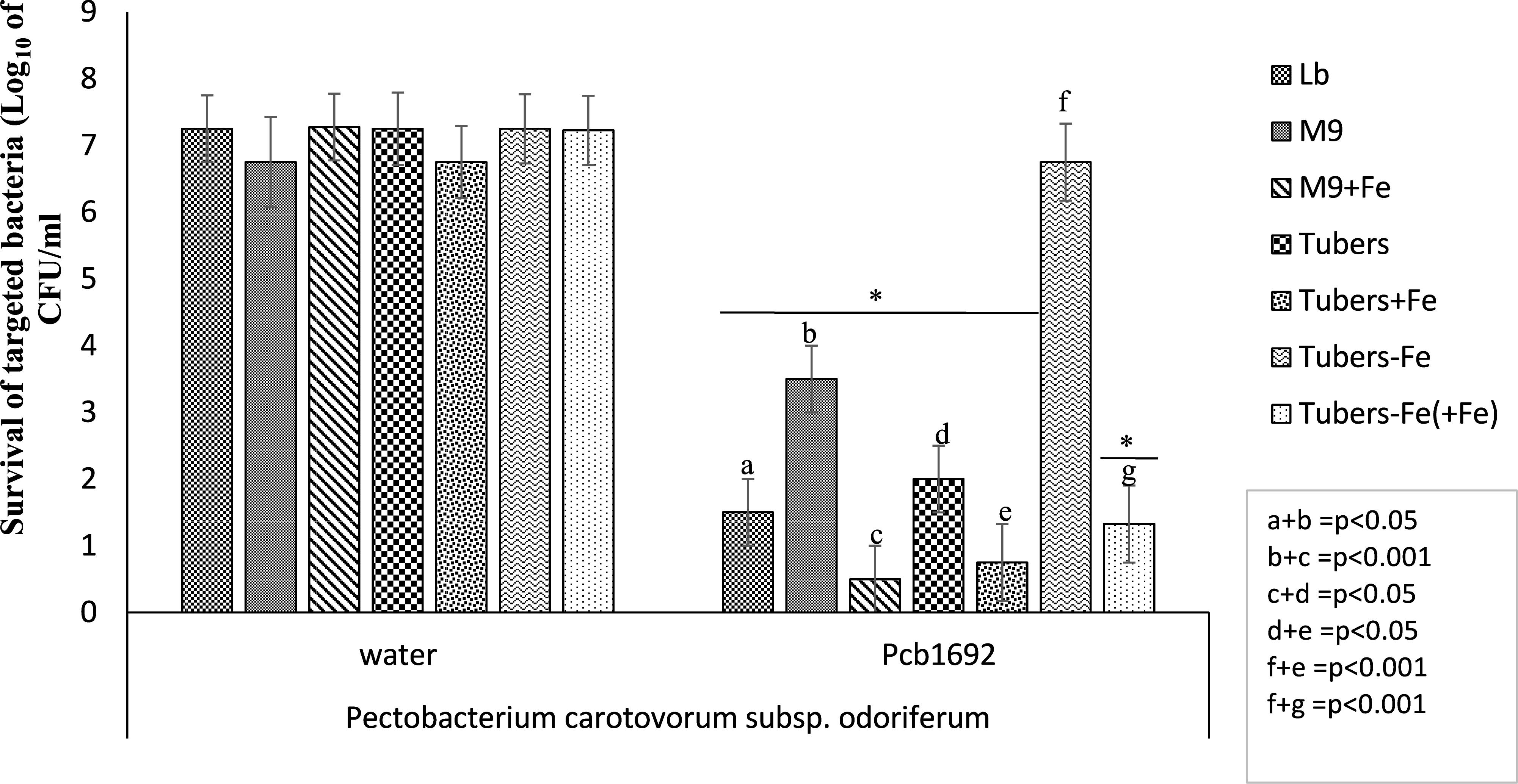
Figure 3. Effect of iron in interbacterial competition: Pectobacterium carotovorum subsp. brasiliense 1692 and targeted bacteria were co-cultured in a 1:1 ratio in either M9, M9 supplemented with either potato tuber extracts or 10 μM ferric iron. The iron in plant extracts was removed using 2,2′-dipyridyl. Results are presented as the CFU/ml of recovered targeted bacteria following 24 h co-culture. Asterisk indicates statistically significant differences p < 0.05 in the recovery of targeted bacteria co-cultured with Pcb1692 relative to water control as determined by two-tailed Student’s t-test. Pco, Pectobacterium carotovorum subsp. odoriferum. Experiments were performed in triplicates and repeated three independent times. “a–g” represent the different competition conditions used in this study and corresponds to the pattern fill shown in the key LB, M9, M9+Fe, Tuber, Tuber+Fe, Tuber-Fe, and tuber-Fe(+Fe).
In silico Identification of Genes Associated With Antibacterial Activity
A transcriptome profiling experiment performed previously in our laboratory, showed that Pcb1692 genes encoding the T6SS and bacteriocins (carotovoricin and pyocin) were up-regulated during in planta (potato tuber) infection (Bellieny-Rabelo et al., 2019b). The same study also showed that gene homologs of the Pcb1692 T6SS were found to be present in the genome sequences of all publicly available genome sequences of Dickeya and Pectobacterium (Bellieny-Rabelo et al., 2019b). Similarly, while homologous sequences of Pcb1692 carotovoricin gene cluster were present in all publicly available genome sequences of Pectobacterium spp. it is missing from the genome sequences of all Dickeya spp. except D. dianthicola RNS049 and D. zeae strains CSLRW192, EC1, and ZJU1202. The above analysis was extended to include orthology relationship between the Pcb1692 protein sequences associated with the carbapenem and the S-type pyocin in order to determine their presence or absence in other Pectobacterium and Dickeya spp.
The Pcb1692 car gene cluster was found to contain all six conserved genes required for synthesis of carbapenem (carA/B/C/D/E/H) including genes encoding the carbapenem immunity proteins (CarF/G). Homologous car gene clusters were also identified in P. betavasculorum ATCC 43762 (T), Pcc ATCC 15713(T), D. chrysanthemi NCPPB 3533, D. zeae CSL RW192, D. zeae Ech1591, and several strains of Pcb (Figure 4). This gene cluster was found to be missing from all publicly available genome sequences of P. wasabiae, P. atrosepticum, P. parmentieri, D. solani, and D. dadantii (exempting D. dadantii subsp. dieffenbachiae NCPPB2976) (Sheet 1 in Supplementary Table S5). Similarly, the car gene cluster was found to be missing from the genome sequence of Pcb strains (BC1, YCD21, YCD49, YCD62, and YCD65). Interestingly, although lacking the car gene cluster, P. wasabiae strains (CFBP3304, NCPPB3701, and NCPPB3702), P. parmentieri strains (CFIA1002 and RNS08421a) and Pcb strains (BC1, YCD21, YCD49, YCD62, and YCD65) all retained the carbapenem immunity gene carF/carG, suggesting that they may be resistant to the carbapenem produced by some Pectobacterium and Dickeya spp.
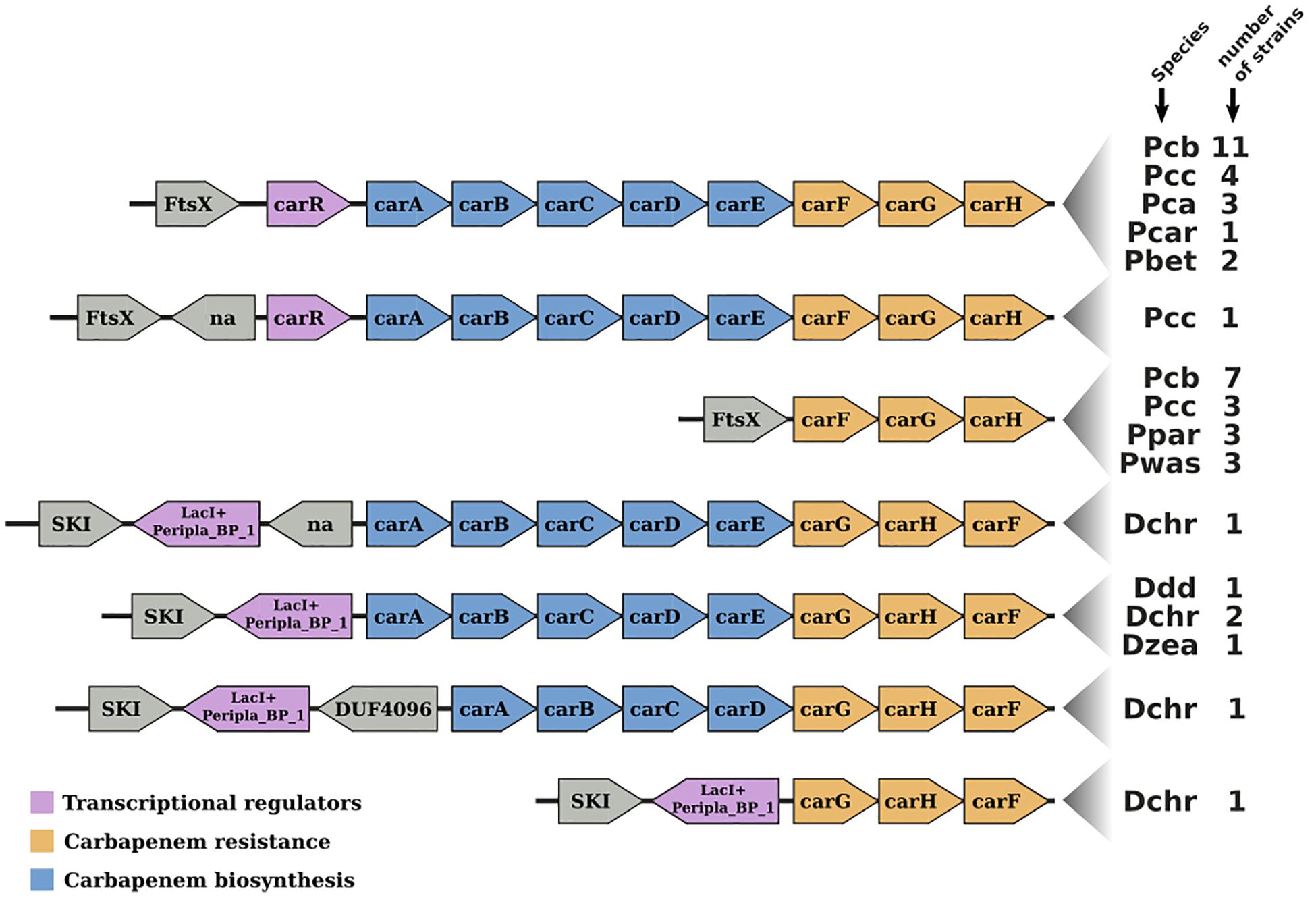
Figure 4. Gene-neighborhood screening of the carbapenem gene cluster (car) in Soft-Rott-Enterobacteriaceae (SRE): 100 SRE genomes were analyzed and all bacteria strains that conserve segments of the car cluster were screened. All known car genes are labeled in the figure. Genes unrelated with the car cluster are labeled using functional domains detected in the respective encoded proteins when applicable, otherwise these are labeled with “na.”
Furthermore, we identified a putative S-type pyocin encoded by the Pcb1692 PCBA_RS02805. This protein (794 amino acids) was predicted to contain a colicin-like bacteriocin tRNase domain (Pfam: PF03515) (Bateman et al., 2004), suggesting it might kill other bacteria by degrading their tRNA. Homologs of the Pcb1692 pyocin were identified in the genome sequence of Pcb strains (LMG21371, HPI01, YCD29, ICMP19477, BC1), Pcc strains (PCC21, NCPPB3395, and YCD57) and P. betavasculorum NCPPB2795 but missing from all publicly available genome sequences of P. atrosepticum, P. wasabiae, and Dickeya (Sheet 2 in Supplementary Table S5). Downstream of the Pcb1692 PCBA_RS02805 gene is PCBA_RS02810 encoding the cognate S-pyocin immunity factor (∼90 amino acids), with homologs present in all SREs encoding S-pyocin. Interestingly, although the genome sequences of Pco strains (S6, S6-2, Q106, Q32, and Q3) and Pcc strains (Y57 and 3F-3) do not have genes encoding the homologous Pcb1692 S-pyocin, they encode S-pyocin immunity factors 100% identical to the one found in Pcb1692 (PCBA_RS02810). Overall, this analysis demonstrates that the Pcb1692 toxin/antitoxin modules analyzed in this study are not conserved in all bacteria including some strains of Pcb and may therefore be a determining factor in the outcome of inter or intra species competition among SRE.
The Pcb1692 Carbapenem Gene Cluster Plays a Role in Bacterial Competition in vitro
In order to investigate the relative contribution of the Pcb1692 T6SS, carotovoricin, carbapenem, and the putative S-type pyocin in competition, we generated Pcb1692 mutants with deletions in genes associated with the production of S-pyocin (Pcb1692Δpyo), carbapenem (Pcb1692ΔcarC), carotovoricin (Pcb1692Δlyt and Pcb1692Δfer) and biosynthesis of a functional T6SS (Pcb1692ΔT6). Targeted bacteria (D. dadantii or Pcc), earlier shown to be inhibited by wild-type Pcb1692, were co-cultured in vitro with either water or the different Pcb1692 mutant strains. The results show that the CFU/ml of recovered targeted bacteria following co-culture with the PcbΔT6, Δlyt, Δfer, and Δpyo mutant strains was similar to the CFU/ml of recovered targeted bacteria following co-culture with the wild-type strain. However, the Pcb1692ΔcarC mutant strain lost the ability to inhibit growth of D. dadantii and Pcc (Figures 5A,C). Trans-expression of the carC gene in Pcb1692ΔcarC restored the ability of the complemented strain to inhibit targeted bacteria, similar to wild-type Pcb1692 (Figures 5A,C). Together, these findings suggest that carbapenem production by Pcb1692 is associated with inter and intrabacterial competition in vitro. Trans-expression of the Pcb1692 carF/G genes in D. dadantii prevented growth inhibition of this strain when co-cultured with wild-type Pcb1692 confirming that growth inhibition of these bacteria is associated with Pcb1692 carbapenem and that the Pcb1692 carF/G genes confer immunity to the Pcb1692 carbapenem (Figure 5B). In addition, our results showed that while Pcb strains CC1, CC2, HPI01, and 358 can produce carbapenem, which inhibit growth of D. dadantii (Figure 6), Pcb strains XT3, XT10, G4P5, and G4P7 do not produce carbapenem under the same experimental conditions (Supplementary Table S4). Similarly, the carbapenem produced by Pcb strains 1692, CC1, CC2, HPI01, and 358 was found to inhibit growth of Pcb strains XT3, XT10, G4P5 and G4P7, suggesting that the latter group of strains lack the cognate carbapenem immunity factor. Together, in vitro and in silico analyses uncover the contrasting conservation patterns of the carbapenem biosynthesis cluster in SREs, highlighting its critical role in interference competition among these organisms. With this information, we proceed toward assessing possible regulatory processes involved in the control of carbapenem biosynthesis.
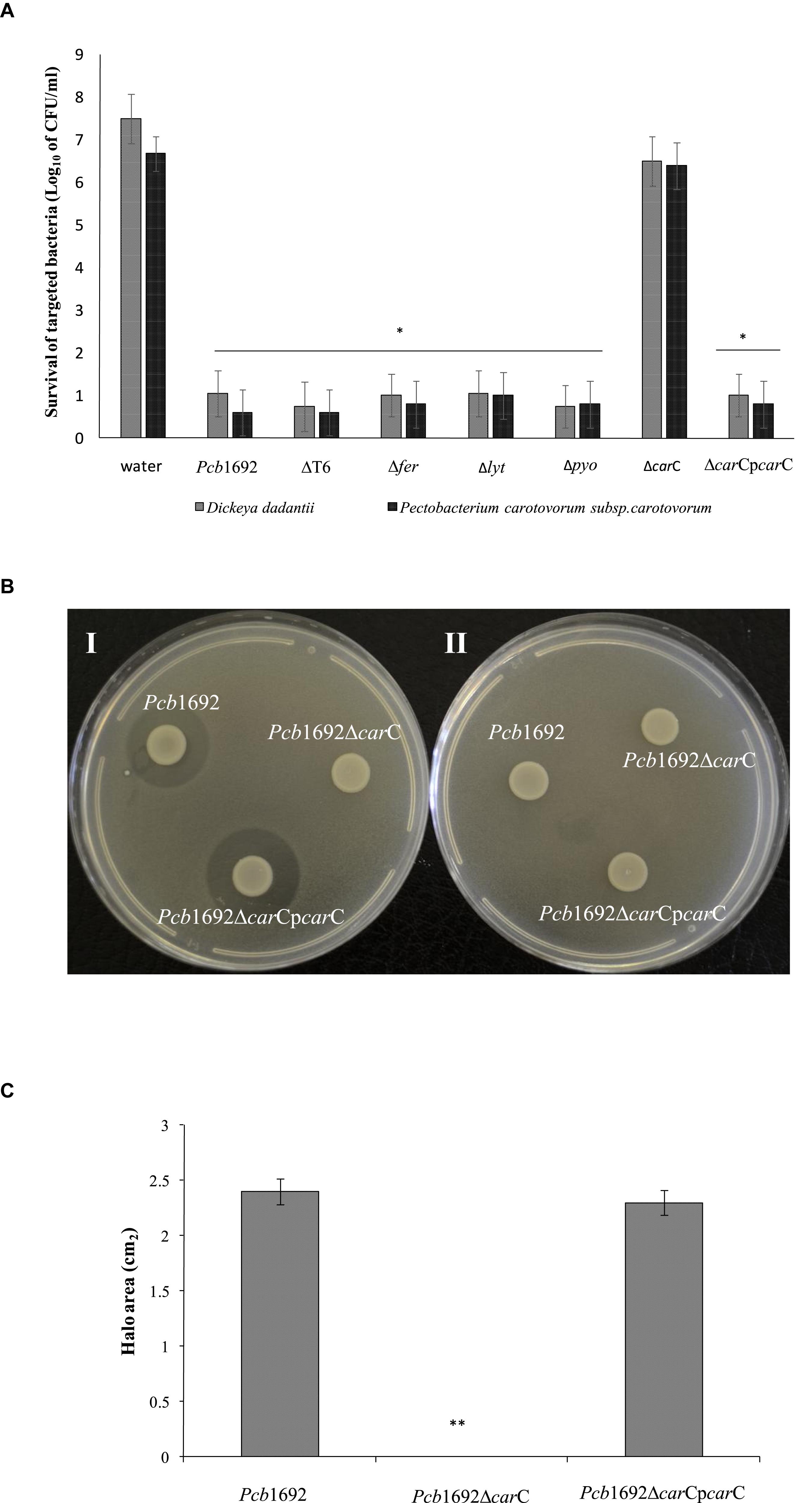
Figure 5. Interbacterial competition with Pectobacterium carotovorum subsp. Brasiliense 1692 mutant strains. (A) Wild-type Pcb1692 and its isogenic mutant (T6, fer, lyt, and carC) were co-cultured with D. dadantii and Pcc for 24 h. The data shows the log10 CFU/ml of recovered D. dadantii and Pcc following co-culture. The horizontal line with an asterisk represents the range of samples with statistically significant differences (p < 0.05) relative to water controls. (B) (i) Clearing zone due to carbapenem produced by strains of Pcb1692 spotted on a lawn of D. dadantii with circularize plasmid PJET4, (ii) No clearing zones when of Pcb1692 is spotted on a lawn of D. dadantii expressing carF and G resistance genes from plasmid pJET4-carFG (C) Halo area was measured using ImageJ and data presented as the mean, error bars represent one standard deviation and asterisk represent statistically significant difference (p < 0.05) as determined by the two-tailed Student’s t-test. Experiments were performed in triplicates and repeated three independent times.
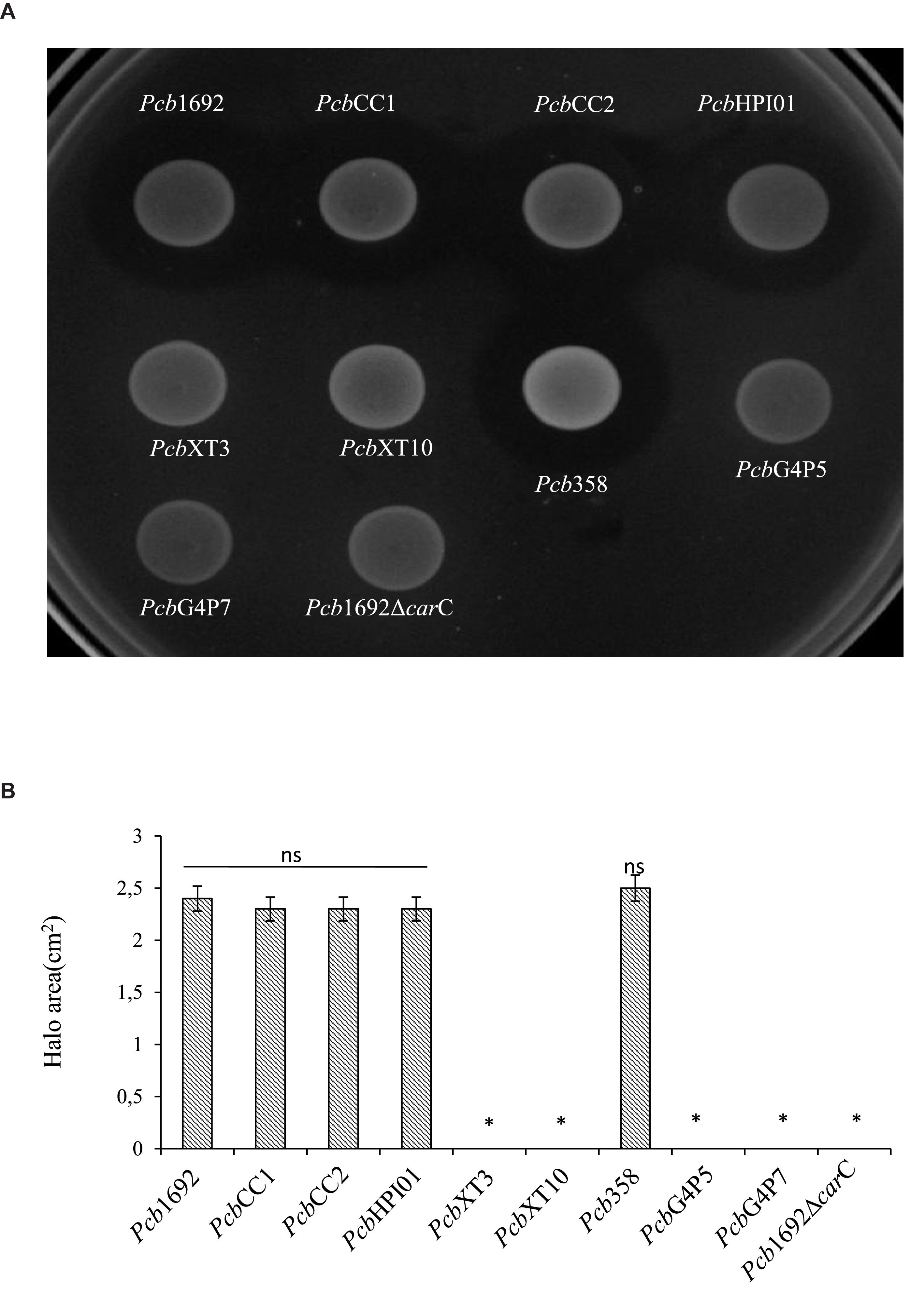
Figure 6. Carbapenem production by environmental strains of Pectobacterium carotovorum subsp. brasiliense. The ability of laboratory strains of Pcb isolated from South Africa to produce carbapenem when spotted on a lawn of Dickeya dadantii was determined by the spot-on-lawn assay. (A) The presence of a clear zone around the spotted Pcb was indicative of carbapenem production. Pcb1692 was used as a control given that this strain produces carbapenem. (B) Halo area was measured using ImageJ and data presented as the mean, error bars represent one standard deviation and asterisk represent statistically significant difference (p < 0.05) as determined by the two-tailed Student’s t-test. Experiments were performed in triplicates and repeated three independent times.
Iron Is Required for Carbapenem Production by Pcb1692 in Minimal Media and the Car Gene Cluster Is Regulated by Fur, SlyA, and ExpI
Having demonstrated that the Pcb1692 carbapenem plays a role in in vitro interbacterial competition, we next investigated how this gene cluster is regulated and under which conditions Pcb1692 produces carbapenem. To this end, we included Pcb1692 transcriptional regulator mutant strains including the ferric uptake regulator (Tanui et al., 2017), N-acyl homoserine lactone synthase (ExpI) (Moleleki et al., 2017) and the stress response regulator SlyA. The results showed that while wild-type Pcb1692 produced a clear zone of inhibition when cultured on a lawn of P. atrosepticum, the Pcb mutant strains (ΔcarC, ΔslyA, ΔexpI, and Δfur) did not produce this clear zone associated with carbapenem production (Figures 7A,C). Loss of carbapenem production could be complemented by trans-expression of corresponding genes in the Pcb1692 mutant strains (Figures 7A,C). In addition, while wild-type Pcb1692 and the complemented mutant strains produced carbapenem on LB, they all lost the ability to produce carbapenem on M9 supplemented with 0.4% of glucose (Figure 7B). Interestingly, carbapenem production was restored when M9 was supplemented with 10 μM of ferric iron (Supplementary Data Sheet S3). These findings implicate iron and the iron homeostasis protein Fur in the regulation of carbapenem production in Pcb1692. Aiming to predict whether Fur is able to regulate carbapenem production in Pcb1692 directly, or indirectly, we performed in silico analysis using the MEME package (Bailey et al., 2006) to search for fur binding sites in the promoter regions of carR, slyA, expI, and expR. Interestingly, the analysis identified fur binding sites in the promoter sequences of carR, slyA, and expR but not in the promoter sequences of expI and carA (the first gene in the car gene cluster), however, this still needs to be experimentally verified (Supplementary Table S6). Given that CarR, and not ExpR, is required for carbapenem production (Rivet, 1998; Coulthurst et al., 2005; Hamed et al., 2013), the Pcb1692 Fur protein may be able to indirectly control the transcription of carbapenem production via carR, slyA, or both.
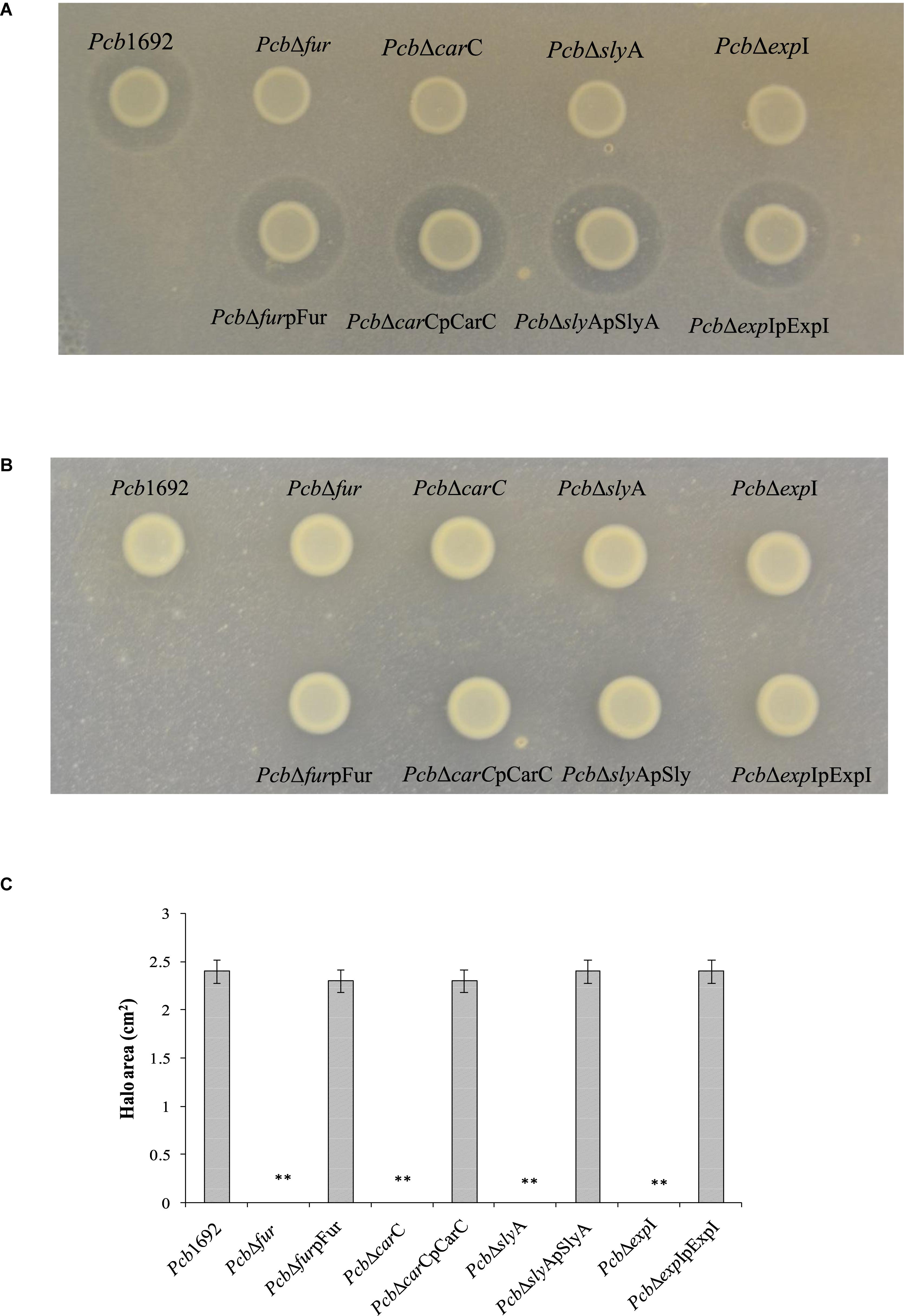
Figure 7. Iron is required for production of carbapenem by Pectobacterium carotovorum subsp. brasiliense 1692. Pcb1692 wild-type mutant and complemented mutant strains were spotted on a lawn of P. atrosepticum and clear zone around the spotted Pcb strains was indicative of carbapenem production. (A) Carbapenem production on LB agar by Pcb strains. (B) No detectable levels of carbapenem were observed on M9. (C) Halo area was measured using ImageJ and data presented as the mean, error bars represent one standard deviation and asterisk represent statistical significant difference (p < 0.05) as determined by the two-tailed Student’s t-test. Experiments were performed in triplicates and repeated three independent times.
The Pcb1692 T6SS Plays a Role in Interspecies but Not Intraspecies Competition in planta
Given the Pcb1692 T6SS did not appear to play a role in bacterial competition in vitro, but was upregulated in potato tubers, we reasoned that Pcb1692 T6SS could play a role in competition in planta. To determine the contribution of the Pcb1692 T6SS in bacterial competition, in planta, standardized cultures of Pcb1692, Pcb1692ΔT6, or Pcb1692ΔT6-pT6 were independently co-inoculated with targeted bacteria (D. chrysanthemi, D. dadantii, Pcc, or PcbG4P5) into surface sterilized potato tubers, and CFU/ml of targeted bacteria enumerated 3 days post-inoculation. The results show no reduction in the CFU/ml of PcbG4P5 when co-inoculated in potato tubers with either wild-type Pcb1692, Pcb1692ΔT6, or Pcb1692ΔT6-pT6 suggesting that T6SS-related effectors secreted by Pcb1692 cannot inhibit growth of this particular strain (Figure 8). However, a statistically significant (p < 0.05) 2–3 log reduction in the CFU/ml was observed when D. dadantii, Pcc, or D. chrysanthemi were co-inoculated with the wild-type Pcb1692 and Pcb1692ΔT6-pT6, this inhibition was completely lost when targeted strains were co-inoculated with Pcb1692ΔT6 (Figure 8). These results demonstrate the involvement of the Pcb1692 T6SS in interbacterial competition in potato tubers.
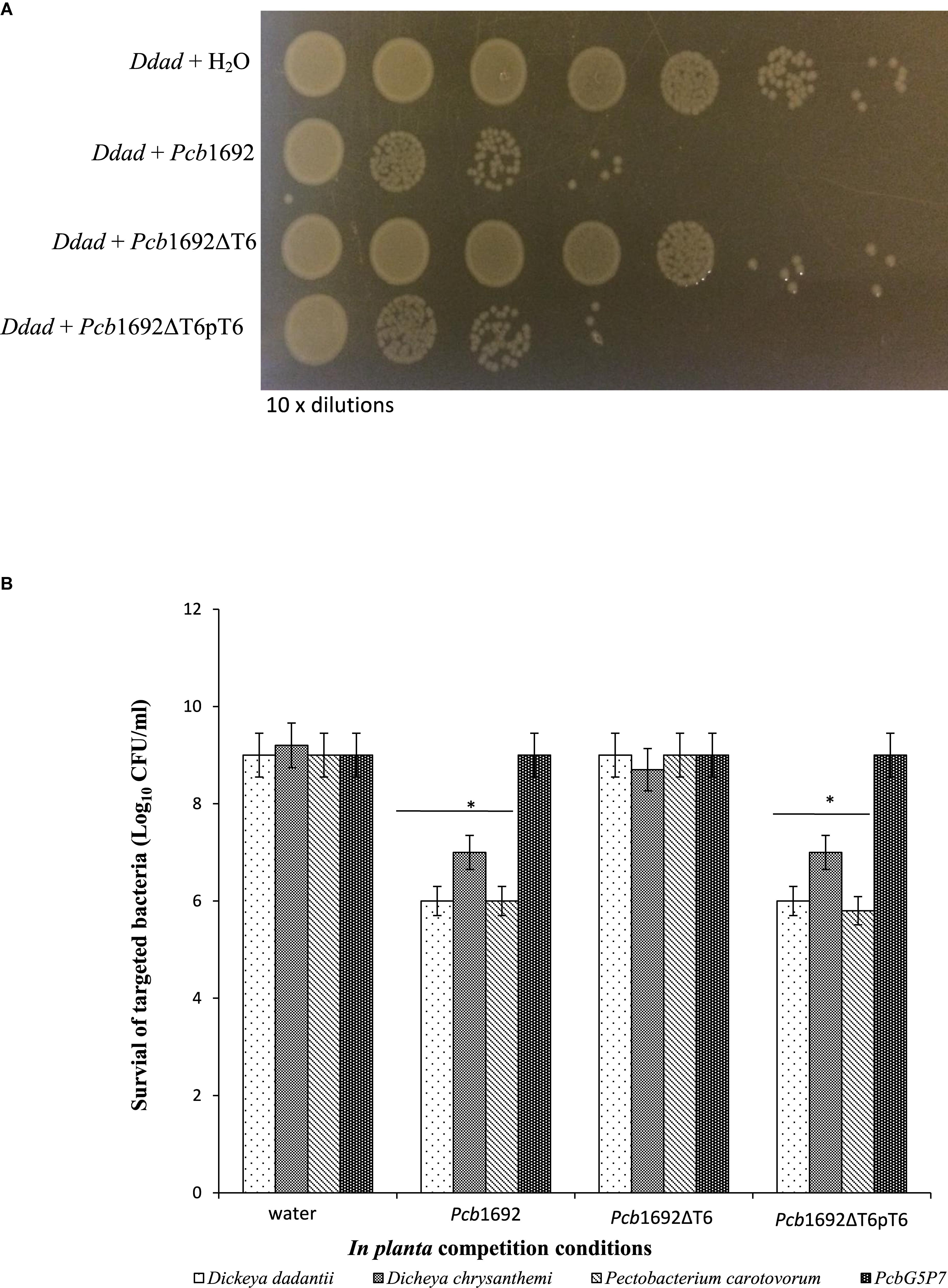
Figure 8. Effect of the T6SS in Pcb1692 anti-bacterial competition in planta. Gentamycin resistant targeted bacteria (Dickeya dadantii, D. chrysanthemi, and Pcc) were mixed in a 1:1 ratio with either water, wild-type Pcb1692, Pcb1692ΔT6 or Pcb1692ΔT6pT6, inoculated into potato tubers and incubated for 24 h and (A) survival of D. dadantii enumerated by 10-times serial dilution on gentamycin (15 μg/ml) agar. (B) Quantification of survival of targeted bacteria presented as log10CFU/ml. Asterisks indicated statistically significant differences (p < 0.05) relative to water control, as determined by the two-tailed Student’s t-test. Experiments were performed in triplicates and repeated three independent times.
We earlier demonstrated that although the in vitro production of carbapenem by Pcb1692 plays a role in both intra and interspecies competition, it does not inhibit growth of D. chrysanthemi. The D. chrysanthemi strain used in this study was found to be resistant to ampicillin, suggesting that this strain might produce a β-lactamase which degrades carbapenem. Conversely, we found that the T6SS of Pcb1692 inhibits growth of D. chrysanthemi in planta but not in vitro. These findings together with our in vitro competition assays suggest that depending on the ecological niche, Pcb1692 can use either the T6SS or carbapenem to inhibit growth of several bacteria species. This is further supported by the fact that the Pcb1692 car gene cluster, unlike the T6SS, is not differentially expressed in planta and may thus be recruited by Pcb1692 to eliminate competitors while residing on the rhizosphere or plant surfaces (Bellieny-Rabelo et al., 2019b). However, following ingress into plant tissue Pcb1692 may recruit mainly the T6SS for bacterial competition.
Pcb1692 Produces Bacteriocin(S) Following Mitomycin C Induction
Despite the fact that the genome sequence of Pcb1692 has genes encoding pyocin and carotovoricin, no role was determined for these bacteriocins in the assays we conducted thus far. Bacteriocin production is usually induced in response to DNA damaging agents which trigger SOS response (Chuang et al., 2007; Chan et al., 2011). Thus, we investigated the ability of Pcb1692 to produce these bacteriocins following induction with either mitomycin C or UV irradiation, using the spot-on-lawn overlay method (see section “Materials and Methods”). The results showed that wild-type Pcb1692, Pcb1692ΔcarC, Pcb1692Δfur, Pcb1692ΔslyA, and Pcb1692ΔexpI all produced bacteriocins following mitomycin C treatment and no bacteriocins were produced in control experiments (Figure 9). Considering that the above mentioned Pcb1692 mutant strains do not produce carbapenem, this indicated that the clear zones around the spotted bacteria may either represent Pcb1692 S-type pyocin, carotovoricin, a combination of both bacteriocins or a yet to be identified bacteriocin. In order to determine whether these clear zones were due to secretion of the putative S-type pyocin by Pcb1692, we generated a Pcb1692ΔpyoI double mutant strain lacking both the pyocin and putative immunity genes but with intact genes associated with carbapenem and carotovoricin production. Interestingly, when Pcb1692ΔpyoI was overlaid on mitomycin C-induced Pcb1692 strains, we observed clear zones of inhibition around wild-type Pcb1692 and all mutant strains used in this study except Pcb1692Δpyo, which lacks the S-type pyocin but has the immunity gene (Figure 10). In addition, the complemented pyocin mutant strain Pcb1692Δpyoppyo was restored in its ability to inhibit growth of Pcb1692ΔpyoI similar to wild-type Pcb1692 while no Pcb1692 strain could kill the complemented pyoI mutant Pcb1692ΔpyoIppyoI (Figure 10). Together these findings demonstrate that following mitomycin C induction, Pcb1692 secretes the S-type pyocin, which inhibits growth of bacteria lacking the immunity factor. To evaluate the host range of killing by the Pcb1692 S-type pyocin, the overlay assay was repeated with different bacteria and the results show that bacteriocins produced by Pcb1692 can inhibit growth of P. atrosepticum, Pcc, Pco, D. dadantii, S. typhimurium, and E. coli but not the type strains of S. marcescens and D. chrysanthemi including some environmental strains of Pcc isolated from South Africa (Supplementary Table S4). The results also showed that the Pcb1692 carotovoricin isogenic mutant strains, Pcb1692Δfer and Pcb1692Δlyt produced bacteriocins similar to the wild-type. This could be because the Pcb1692 carotovoricin mutant strains still produced S-type pyocin or because the ferredoxin gene located within the ctv gene cluster of Pcb1692 may not be essential for carotovoricin production given that it is not conserved in the genome sequence of several SREs that have this gene cluster (Bellieny-Rabelo et al., 2019b). Similarly, the carotovoricin lytic cassette has three lytic genes associated with cell lysis and release of carotovoricin, thus presenting the possibility of functional complementation following deletion of any one the lytic genes, in our case, Pcb1692 Δlyt.
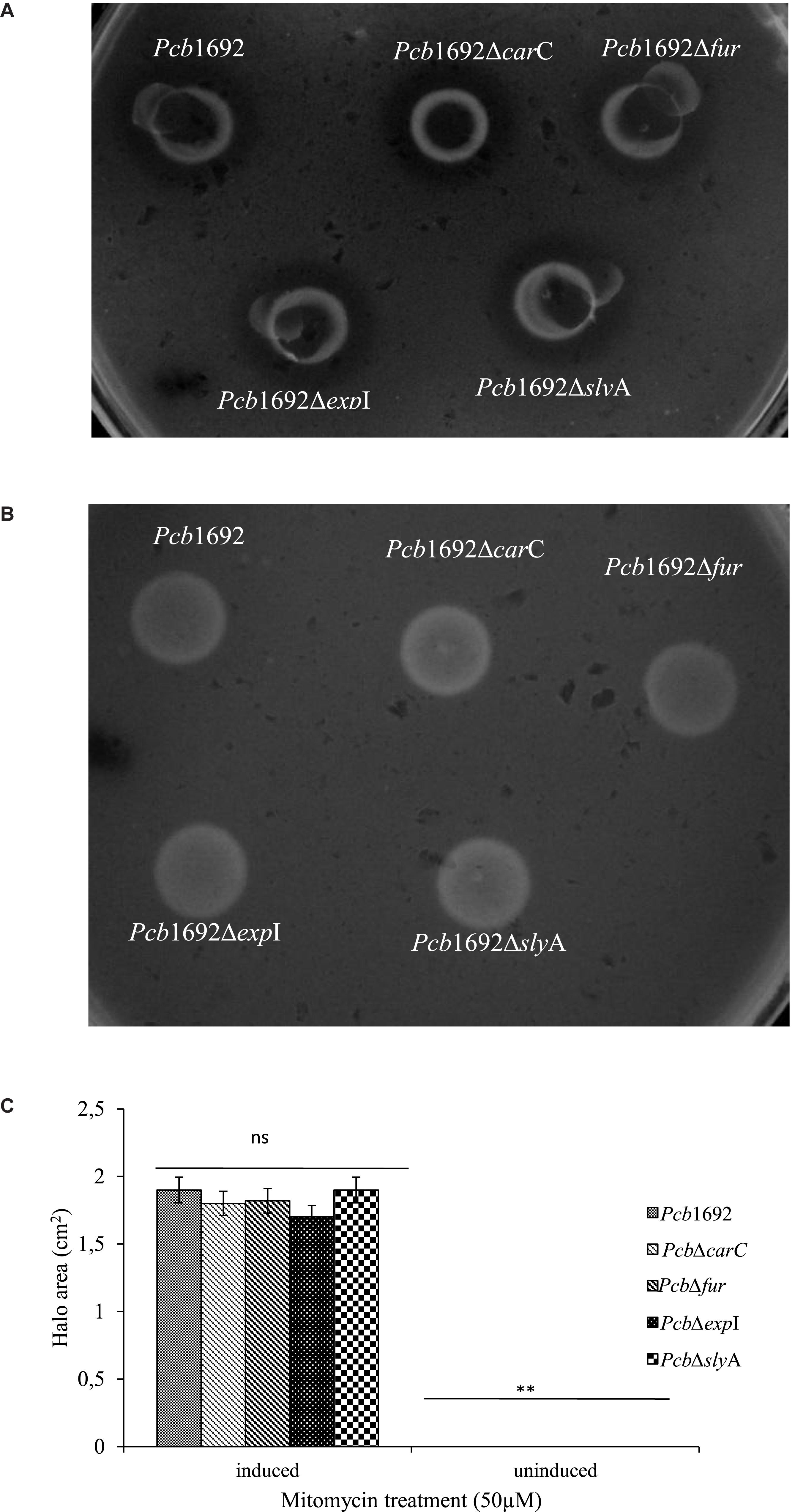
Figure 9. Bacteriocin production by strains of Pectobacterium carotovorum subsp. brasiliense 1692. Pcb1692 and its isogenic mutant strains were spotted on LB and grown overnight. (A) Bacteriocin production was induced with mitomycin C (B) control plate not induced. Dickeya dadantii was used as the indicator strain. Clear zones are indicative of bacteriocin production by Pcb1692. (C) Halo area was measured and ImageJ and data presented as the mean, error bars represent one standard deviation and asterisk represent statistical significant difference (p < 0.05) as determined by the two-tailed Student’s t-test. Experiments were performed in triplicates and repeated three independent times. Ns, not statistically significant.
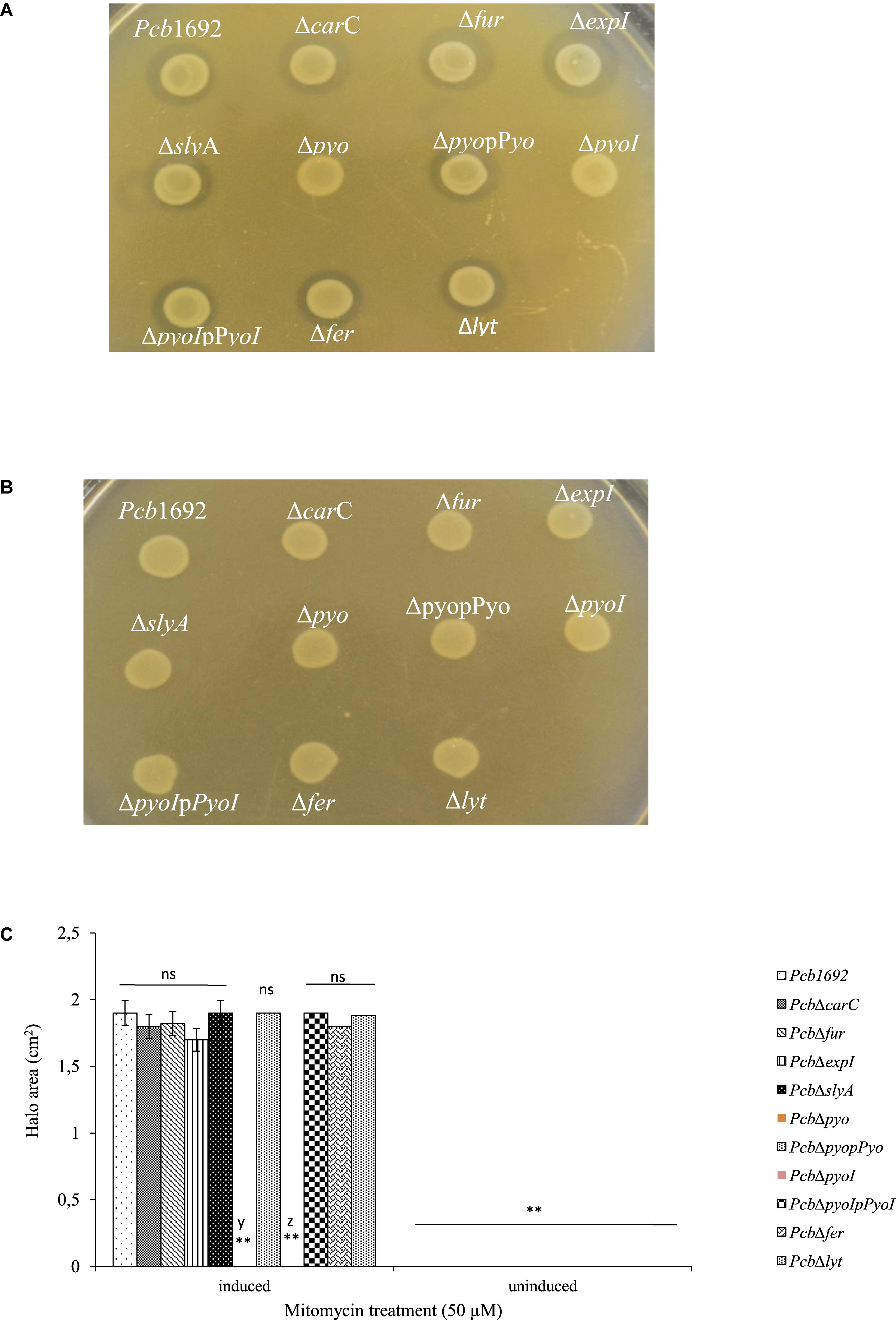
Figure 10. Production and detection of the S-type pyocin of Pectobacterium carotovorum subsp. brasiliense 1692. Pcb1692 and its isogenic mutant strains including Pcb1692Δpyo, Pcb1692ΔpyoI and their respective complemented strains were spotted on LB and grown overnight. Bacteriocin production was induced with mitomycin C for 5 h. Induced cells were killed with chloroform vapor and overlaid with (A) a lawn of either Pcb1692ΔpyoI or (B) Pcb1692ΔpyoIppyoI. Clear zones are indicative of bacteriocin production. (C) Halo area was measured and ImageJ and data presented as the mean, error bars represent one standard deviation and asterisk represent statistical significant difference (p < 0.01) as determined by the two-tailed Student’s t-test. Experiments were performed in triplicates and repeated three independent times. Ns, not statistically significant; y, Pcb1692Δpyo and z, Pcb1692ΔpyoI.
Pcb1692 Produces Bacteriocins but Not Carbapenem Under Anaerobic Conditions
Previous studies have shown that oxygen is required for carbapenem production (Axelrood et al., 1988; Coulthurst et al., 2005). Therefore, to better understand the role played by oxygen in bacterial competition and the biological significance of carbapenem and bacteriocin production in the context of potato tubers, we assayed the ability of Pcb1692 to produce these antimicrobial compounds under aerobic or anaerobic conditions. The results demonstrated that Pcb1692 produces carbapenem under aerobic but not anaerobic conditions (Figure 11A). In addition, following mitomycin C treatment Pcb1692 and its isogenic mutant strains Pcb1692ΔcarC, Pcb1692ΔT6, Pcb1692Δpyo, Pcb1692Δfer, and Pcb1692Δlyt all produced bacteriocins when overlaid with either Pcc, Pco, P. atrosepticum, D. dadantii or Bacillus cereus under both aerobic and anaerobic conditions (Figure 11B and Supplementary Table S4). These findings clearly demonstrate that although bacteriocins are generally considered to have a narrow host range of killing (Riley and Gordon, 1999), bacteriocins produced by Pcb1692 may have a wide host range of killing which includes the Firmicutes and B. cereus. We further investigated the role played by the Pcb1692 T6SS in competition under anaerobic conditions, which mimics one of the conditions in planta where there is limited oxygen. Under aerobic conditions, no targeted bacteria were recovered when co-cultured with either Pcb1692 wild-type, mutant or complemented mutant strains. This could be ascribed to previously demonstrated production of Pcb1692 carbapenem under aerobic conditions (Figure 11A). Pcb1692 killing ability on target bacteria under anaerobic conditions was only slightly reversed in the Pcb1692ΔT6 mutant strain. Thus, we could conclude that the presence or absence of oxygen did not affect T6SS dependent in vitro bacterial competition (Figure 11C). Together these findings suggest that Pcb1692 might secrete bacteriocins but not carbapenem following ingress into potato tubers, where conditions are mostly anaerobic. Furthermore, the presence or absence of oxygen did not affect the magnitude of growth inhibition of targeted bacteria by Pcb1692ΔT6 relative to wild-type. This therefore supports our previous findings that the T6SS of Pcb1692 may only be functionally active in planta.
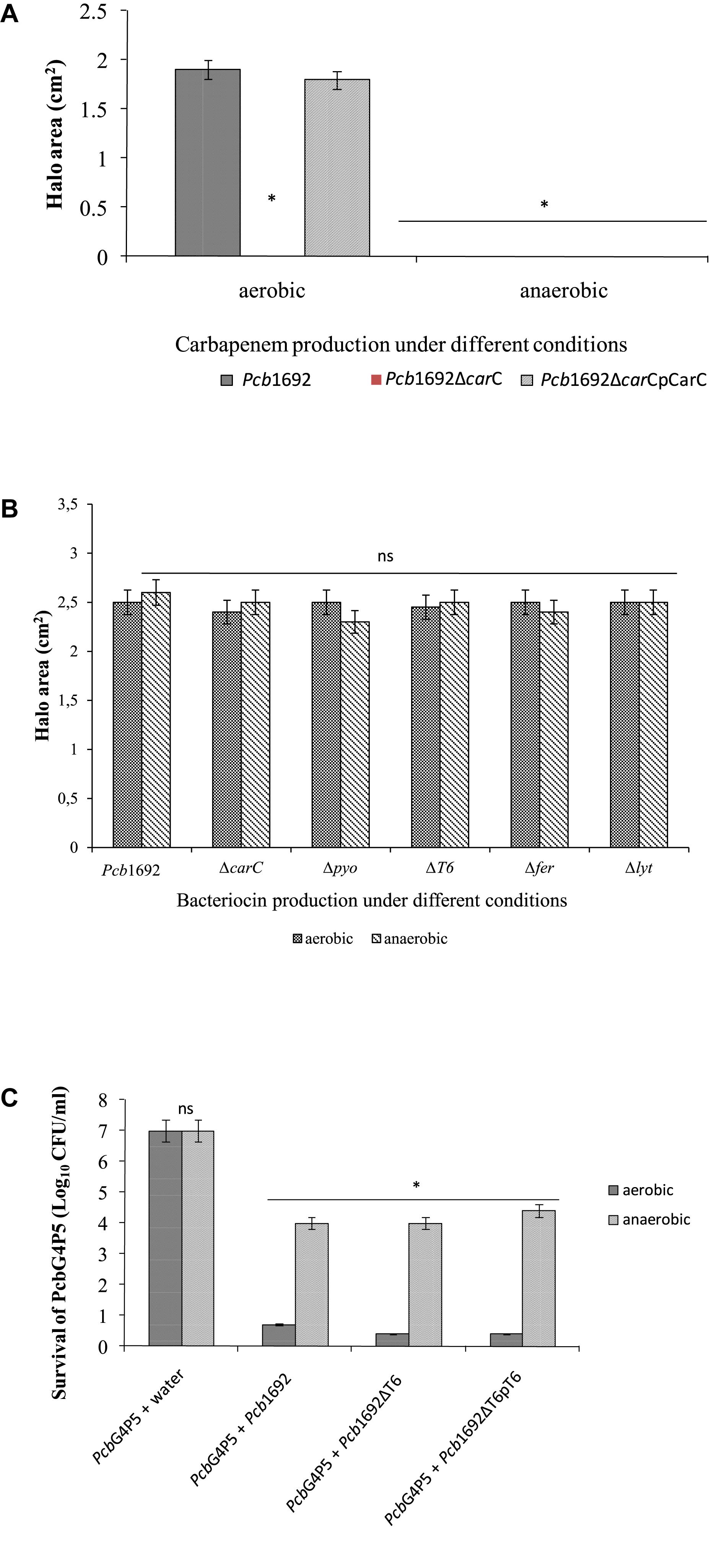
Figure 11. Effect of oxygen in competition, antibiotic and bacteriocin production by Pectobacterium carotovorum subsp. brasiliense 1692. (A) Carbapenem production by Pcb1692 strains spotted on a lawn of PcbG4P5 and incubated aerobically and anaerobically. (B) Bacteriocin production by Pcb1692 strains induced with mitomycin C and overlaid with Bacillus cereus. (C) Interbacterial competition assays between Pcb1692 and the targeted strain PcbG4P5, data is presented as CFU/ml of recovered targeted bacteria following 24 h incubation aerobically and anaerobically. Halo area was measured using ImageJ and data presented as the mean, error bars represent one standard deviation and asterisk represent statistical significant difference (p < 0.05) as determined by the two-tailed Student’s t-test. Experiments were performed in triplicates and repeated three independent times.
Discussion
Inducing Conditions Associated With Antimicrobial Production by Pcb1692
Bacteria exist as complex multispecies communities in any given niche (Kõiv et al., 2015; García-Bayona and Comstock, 2018). The interactions within these communities can be characterized by cooperation between species or more commonly, by competition (Stubbendieck and Straight, 2016; Chassaing and Cascales, 2018). Interference competition is mediated by production of an arsenal of antimicrobial weapons. In this study, we set out to explore microbe–microbe interactions that shape population structures of soft rot Enterobacteriaceae with a specific focus on interference competition. We used Pcb1692, an emerging soft rot pathogen of potatoes, as a model to help us understand factors shaping dynamics of microbial communities which can be found in decaying potato tubers. We were intrigued by the vast number of different antibacterial compounds encoded in the genome of Pcb1692 and other SREs, some of which were upregulated during Pcb1692 infection of potato tubers (Bellieny-Rabelo et al., 2019b). We hypothesized that these different systems likely target different types of bacteria and are likely deployed to enable Pcb1692 to colonize different environments. To test this hypothesis, we first performed a 16S metagenomics analysis of healthy and diseased potato tubers in order to determine the bacteria genus and strains (previously isolated from potato tubers in other studies) that can be used in competition against Pcb1692. The selected taxa used in this study such as Serratia, Escherichia, Salmonella, Bacillus, Pseudomonas, Enterobacter, Pectobacterium, and Dickeya represent bacteria that Pcb will likely encounter either in potato tubers, rhizosphere, or phyllosphere. Thereafter, we used a combination of in silico analyses, systematic mutagenesis of different antimicrobial systems and in vitro as well as in planta competition assays. To this end, we examined the role of different antimicrobial compounds produced by Pcb1692, including the T6SS, carbapenem, carotovoricin and bacteriocins, in bacteria competition. Our results identified some of the conditions associated with production of antimicrobial compounds by Pcb1692 such as (1) constitutive production of carbapenem in vitro under aerobic conditions, (2) iron-mediated secretion of carbapenem, under nutrient-limiting conditions, (3) SOS-mediated bacteriocin production, and (4) the role T6SS of Pcb1692 in bacteria competition when inoculated in potato tubers. This study sets an important milestone in interdisciplinary studies addressing the battery of compounds involved in both inter- and intra-specific bacterial competition.
The aforementioned inducing conditions mimic conditions likely encountered byPcb1692 whether residing outside (both in the rhizo- and phyllo-sphere) or when colonizing intercellular spaces within potato tubers. For example, in the phyllosphere, availability of oxygen likely allows Pcb to produce carbapenem which inhibits growth of several Proteobacteria within the families Enterobacteria and Pseudomonadaceae, including E. coli, Salmonella typhimurium, P. aeruginosa and Pcb strains isolated from different diseased plant materials. While some of these bacteria represent enteric pathogens, members of the genera Salmonella, Pseudomonas, and Escherichia have previously been isolated from the rhizosphere and endosphere of several different plants including potato tubers, suggesting that Pcb and other SREs will compete with these bacteria (Hollis, 1951; Bulgarelli et al., 2012; Gupta et al., 2014; Schreiter et al., 2014; Donn et al., 2015; Hong et al., 2015; Kõiv et al., 2015). Similarly, the possibility of Pcb to encounter SOS-inducing conditions in the phyllosphere such as UV irradiation, nutrient limitation and production of antimicrobial compounds by other bacteria suggest that Pcb can potentially produce bacteriocins while residing in this niche. Once in the rhizosphere, Pcb encounters conditions of limited oxygen and SOS inducing conditions such as antimicrobial compounds produced by other bacteria (Erill et al., 2007; Goyal and Mattoo, 2014; Fang et al., 2017; Thorgersen et al., 2017; Cooper et al., 2018; García-Bayona and Comstock, 2018; Leaden et al., 2018), suggesting that Pcb1692 may be able to produce bacteriocins but not carbapenem while residing in the rhizosphere, however, this needs to experimentally verified. Finally, once inside potato tubers Pcb encounters several stresses which can trigger bacteria SOS such as nutrient starvation, iron deficiency, chromate stress, osmotic stress, oxidative and acidic stress, and β-lactam antibiotics produced by either bacteria endophytes or the host plant, leading to phage induction and bacteriocin production (Erill et al., 2007; Goyal and Mattoo, 2014; Fang et al., 2017; Thorgersen et al., 2017; Cooper et al., 2018; García-Bayona and Comstock, 2018; Leaden et al., 2018).
Role of Carbapenem and Bacteriocin in Bacteria Competition, Resistance Mechanisms
Our results show that the different antimicrobials produced by Pcb1692 target different genera. For example, Pcb1692 deploys carbapenem and bacteriocins to inhibit growth of all the aforementioned targeted bacteria, except S. marcescens, D. chrysanthemi, and P. wasabiae. Our in silico analysis showed that some of the SREs analyzed in this study such as D. dadantii, Pco, and P. atrosepticum do not have genes associated with carbapenem production or resistance, which could account for the observed 5-log reduction in the CFU/ml of these strains when co-cultured with Pcb1692 (Figure 2). Similarly, the genome sequences of several closely related strains of Pcb do not have homologs of car genes which could in part explain the growth inhibition observed when Pcb1692 was co-cultured with Pcb (XT3, XT10, G4P5, and G4P7). In this regard, PCR amplification failed to amplify the carF and carG genes in Pcb (XT3, XT10, G4P5, and G4P7), however, because of the possibility of sequence variation and genetic heterogeneity of Pcb strains and SREs in general (Lee et al., 2014; Adeolu et al., 2016), one cannot rule out the presence or absence of a gene in a given bacterial genome solely based of PCR. The inability of the Pcb1692 carbapenem to inhibit growth of D. chrysanthemi and P. wasabiae and S. marcescens may be explained as follows: (1) The in silico analysis showed that although the genome sequences of D. chrysanthemi and P. wasabiae do not contain gene homologs associated with carbapenem biosynthesis genes they do contain homologs of carF and carG associated with carbapenem resistance, (2) although in silico analysis data we presented did not include analysis of the genome sequence of S. marcescens ATCC 13880T (Daligault et al., 2014), our analysis indicated that the genome sequence of this strain has homologs of carA (GSMA_00116) and carB (GSMA_00117) but missing the remaining car genes. Additionally, carbapenem resistance has been observed in several Proteobacteria including Klebsiella and Serratia and this resistance has been associated with either production of β-lactamases, the constitutive expression of efflux pumps, reduced membrane permeability, or modification of penicillin-binding protein targets (Yang et al., 2012; Lutgring and Limbago, 2016). The ability of any bacteria to deploy either a single or a combination of the aforementioned carbapenem resistance mechanisms may in part explain the differences observed in the magnitude of inhibition shown in Figure 2. Similarly, following induction with mitomycin C, Pcb1692 produces bacteriocins which inhibit growth of several Proteobacteria and the Firmicutes and B. cereus. Our in silico analysis showed that homologs of the Pcb1692 pyocin and immunity factor are not conserved in genome sequences of most SREs analyzed including some closely related strains of Pcb. While this finding could in part be explained the observed killing (halo formation) of these strain, it does not explain resistance of the S. marcescens, P. wasabiae, and D. chrysanthemi to these bacteriocins. Like carbapenem resistance, it is possible that these bacteria may have developed different mechanisms associated with bacteriocin resistance such as phage superinfection, modification of membrane, DNA or RNA target sites or rapid efflux or neutralization of bacteriocins (Jarvis, 1967; Ming and Daeschel, 1993; Mazzotta et al., 1997; Hyman and Abedon, 2010).
The Role Played by the Pcb1692 T6SS in Bacteria Competition
Previous studies have shown that while the T6SS of some bacteria is constitutively expressed and actively secretes antimicrobial compounds under laboratory conditions, the same system needs to be induced or activated in other bacteria by different cues such as temperature, osmolarity, oxygen, pH, plant extracts, and several divalent metals including iron (Miyata et al., 2013). For example, some T6SS genes of Pectobacterium atrosepticum are upregulated in the presence of plant extracts while the T6SS of Pcb1692 and Dickeya is massively upregulated when inoculated into potato tubers (Mattinen et al., 2007; Bellieny-Rabelo et al., 2019b; Raoul des Essarts et al., 2019). However, for SREs and most other plant pathogens, the exact role this secretion system plays in bacterial competition remains largely unknown (Bernal et al., 2018). Our results demonstrate that the Pcb1692ΔT6SS was attenuated in its ability to inhibit D. chrysanthemi, D. dadantii and Pcc, relative to wild-type when co-inoculated into potato tubers. These findings clearly demonstrate that Pcb1692 uses the T6SS to inhibit growth of D. chrysanthemi in planta but not in vitro. Therefore, it is possible that this system is recruited to inhibit growth of some endophytes that likely share the ‘potato tuber niche’ with Pcb. On the other hand, the inability of the T6SS of Pcb1692 to inhibit growth of the closely related PcbG5P7 may be due to the presence of overlapping T6-related toxin/immunity modules in their genome. This hypothesis is supported by the reported conservation of one of the T6-islands encoding several predicted toxin/antitoxin modules in different strains of Pcb (Bellieny-Rabelo et al., 2019b). Overall, our analysis suggests that once inside potato tubers, the T6SS of Pcb1692 is activated and alongside its arsenal of bacteriocins, inhibits growth of targeted bacteria.
The Role Played by Iron, Oxygen, and Fur on Carbapenem Production
Iron is an essential element which acts as a cofactor of several enzymes, a constituent of heme and is required for biosynthesis of iron–sulfur clusters (Fe–S) found in several proteins (Andrews et al., 2003; Seo et al., 2014). Because of its importance, bacteria and their host plants are continuously engaged in an arms race for iron. During these interactions, the host plant limits iron availability to invading microbes by sequestering iron in proteins such as ferritin while the bacteria by producing siderophore, which are high-affinity iron uptake systems capable of chelating iron from the plant (Boughammoura et al., 2007; Skaar, 2010). Despite its involvement in several biological processes, both excess and limited iron is detrimental to bacteria. Furthermore, intracellular levels of iron are regulated by the ferric uptake regulator (Fur) (Chandrangsu et al., 2017). In the presence of iron, Fur binds to iron and the FurFe2+ complex represses expression of several genes including iron uptake gene while concomitantly activating expression of genes associated with iron efflux, virulence, stress response and several metabolic processes (Foster, 1991; Andrews et al., 2003; Delany et al., 2004; Hassan and Troxell, 2013; Seo et al., 2014; Porcheron and Dozois, 2015). Interestingly, our results showed that carbapenem production by Pcb1692 is dependent on iron and may be regulated directly or indirectly by Fur. This is based on the fact that Pcb1692Δfur mutant strain lost the ability to produce carbapenem compared to the wild-type strain. Similarly, the results showed that wild-type Pcb1692 only produced carbapenem when M9 was supplemented with iron. Identification of putative Fur binding sites in slyA and carR promoter regions in Pcb1692, Pcc, Pco, and Pca further suggests that Fur might indirectly regulate expression of the carbapenem genes by binding to the promoter sequences of slyA and carR. In addition to iron and Fur, our results demonstrate that Pcb1692 produces detectable levels of carbapenem and bacteriocins when spotted on a lawn of targeted bacteria and incubated aerobically, however, when the above assay was repeated under anaerobic conditions, Pcb1692 lost the ability to produce carbapenem but not bacteriocins.
The requirement for iron and Fur in the biosynthesis of carbapenem is consistent with the finding that CarE is a putative ferredoxin with 2 Fe–S clusters while the carbapenem synthase CarC protein is an oxygenase which requires iron and oxygen and 2-oxoglutarate as co-factors (Hamed et al., 2013; Dunham et al., 2018). The requirement for oxygen by CarC is also supported by our findings which showed that that Pcb1692 does not produce carbapenem under anaerobic conditions. Therefore, the effect of iron and oxygen may have a direct effect on functionality of CarC and CarE and hence carbapenem production in the apoplast where there is limited iron and oxygen. It is important to note that the ability of Pcb and SREs in general to produce carbapenem and bacteriocins in planta has not been determined in this study including previous studies partly because of the difficulty associated with identification and quantification of antibiotics and bacteriocins produced by bacteria in planta. Therefore, we can only cautiously speculate on the physiological roles these compounds play during in planta bacteria competition based on the assay conditions which mimic in planta conditions conducted in the current study.
Conclusion
Our results demonstrate that Pcb deploys an arsenal of antimicrobial compounds targeted at different bacteria and recruited under different environmental conditions to give Pcb1692 a competitive advantage over different endophytes likely encountered during various stages of potato tuber invasion. This competitive advantage spans phyllosphere which is typically aerobic, rhizosphere and intercellular spaces inside potato tubers which are typically characterized by anaerobiosis and iron-limiting conditions. For example, the Pcb1692 T6SS is recruited in planta following perception of unknown plant signals and inhibits members of the Enterobacteriaceae. Furthermore, Pcb1692 produces carbapenem and bacteriocins, which inhibit several gram-negative and gram-positive bacteria including B. cereus. While Pcb1692 can produce bacteriocins aerobically and anaerobically, carbapenem production was found to be dependent on the availability of oxygen and iron. Finally, the results showed that carbapenem production by Pcb1692 is regulated by the concerted action of Fur, SlyA and ExpI.
Data Availability Statement
The 16S metagenomics raw data has been deposited in the Sequence Read 567 Archive database on NCBI under the Accession No. PRJNA509544.
Author Contributions
DS and LM conceived the study and designed the experiments. NS performed the metagenomic study. DB-R analyzed metagenomics data and performed the in silico analysis. DS, NN, and AG performed the experiments. DS, DB-R, and LM analyzed the data and provided technical and scientific discussion. DS wrote the manuscript. NN, AG, DB-R, and LM revised the manuscript. All authors read and approved the final manuscript.
Funding
This research study was funded by the National Research Foundation (NRF), South Africa through Competitive Funding for Rated Researchers (CFRR) 98993, Bioinformatics and Functional Genomics (BFG 93685), Foundational Biodiversity Information Programmes (FBIP 86951), and The Technology Transfer Fund (RTF) 98654. DS was supported by NRF BFG Post-Doctoral Fellowship. DB-R was supported by University of Pretoria Post-Doctoral Fellowship. NN, AG, and NS were jointly funded by Potato South Africa Transformation Bursary and the NRF Grant Holder Linked Bursary.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This manuscript has been released as a Pre-Print at bioRxiv, 497016 (Shyntum et al., 2018).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02379/full#supplementary-material
Footnotes
References
Adeolu, M., Alnajar, S., Naushad, S., and Gupta, R. S. (2016). Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 66, 5575–5599. doi: 10.1099/ijsem.0.001485
Andrews, S. C., Robinson, A. K., and Rodríguez-Quiñones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. doi: 10.1016/s0168-6445(03)00055-x
Aoki, S. K., Diner, E. J., de Roodenbeke, C. T. K., Burgess, B. R., Poole, S. J., Braaten, B. A., et al. (2010). A widespread family of polymorphic contact-dependent toxin delivery systems in Bacteria. Nature 468, 439–442. doi: 10.1038/nature09490
Axelrood, P. E., Rella, M., and Schroth, M. N. (1988). Role of antibiosis in competition of Erwinia strains in potato infection courts. Appl. Environ. Microbiol. 54, 1222–1229.
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34(Suppl. 2), W369–W373.
Bateman, A., Coin, L., Durbin, R., Finn, R. D., Hollich, V., Griffiths-Jones, S., et al. (2004). The Pfam protein families database. Nucleic Acids Res. 32(Suppl. 1), D138–D141.
Bauer, M. A., Kainz, K., Carmona-Gutierrez, D., and Madeo, F. (2018). Microbial wars: competition in ecological niches and within the microbiome. Microbial. Cell 5, 215–219. doi: 10.15698/mic2018.05.628
Bellieny-Rabelo, D., Nkomo, N. P., Shyntum, D. Y., and Moleleki, L. N. (2019a). Horizontally Acquired Quorum Sensing Regulators Recruited by the PhoP Regulatory Network Expand Host-Adaptation Repertoire in The Phytopathogen Pectobacterium Carotovorum. Biorxiv. [preprint]. doi: 10.1101/776476
Bellieny-Rabelo, D., Tanui, C. K., Miguel, N., Kwenda, S., Shyntum, D. Y., and Moleleki, L. N. (2019b). Transcriptome and comparative genomics analyses reveal new functional insights on key determinants of pathogenesis and interbacterial competition in Pectobacterium and Dickeya spp. Appl. Environ. Microbiol. 85:e02050-18.. doi: 10.1128/AEM.02050-18
Bernal, P., Llamas, M. A., and Filloux, A. (2018). Type VI secretion systems in plant-associated Bacteria. Environ. Microbiol. 20, 1–15. doi: 10.1111/1462-2920.13956
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bost, A., Martinson, V. G., Franzenburg, S., Adair, K. L., Albasi, A., Wells, M. T., et al. (2018). Functional variation in the gut microbiome of wild Drosophila populations. Mol. Ecol. 27, 2834–2845. doi: 10.1111/mec.14728
Boughammoura, A., Franza, T., Dellagi, A., Roux, C., Matzanke-Markstein, B., and Expert, D. (2007). Ferritins, bacterial virulence and plant defence. Biometals 20, 347–353. doi: 10.1007/s10534-006-9069-0
Bulgarelli, D., Rott, M., Schlaeppi, K., van Themaat, E. V. L., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Chan, Y.-C., Wu, H.-P., and Chuang, D.-Y. (2009). Extracellular secretion of Carocin S1 in Pectobacterium carotovorum subsp. Carotovorum occurs via the type III secretion system integral to the bacterial flagellum. BMC Microbiol. 9:181. doi: 10.1186/1471-2180-9-181
Chan, Y.-C., Wu, J.-L., Wu, H.-P., Tzeng, K.-C., and Chuang, D.-Y. (2011). Cloning, purification, and functional characterization of Carocin S2, a ribonuclease bacteriocin produced by Pectobacterium carotovorum. BMC Microbiol. 11:99. doi: 10.1186/1471-2180-11-99
Chandrangsu, P., Rensing, C., and Helmann, J. D. (2017). Metal homeostasis and resistance in Bacteria. Nat. Rev. Microbiol. 15, 338–350. doi: 10.1038/nrmicro.2017.15
Charkowski, A. O. (2018). The changing face of bacterial soft-rot diseases. Annu. Rev. Phytopathol. 56, 269–288. doi: 10.1146/annurev-phyto-080417-045906
Chassaing, B., and Cascales, E. (2018). Antibacterial weapons: targeted destruction in the microbiota. Trends Microbiol. 26, 329–338. doi: 10.1016/j.tim.2018.01.006
Chuang, D.-Y., Chien, Y.-C., and Wu, H.-P. (2007). Cloning and expression of the Erwinia carotovora subsp. carotovora gene encoding the low-molecular-weight bacteriocin carocin S1. J. Bacteriol. 189, 620–626. doi: 10.1128/jb.01090-06
Clayton, J. B., Al-Ghalith, G. A., Long, H. T., Van Tuan, B., Cabana, F., Huang, H., et al. (2018). Associations between nutrition, gut microbiome, and health in a novel nonhuman primate Model. Sci. Rep. 8:11159. doi: 10.1038/s41598-018-29277-x
Cooper, B., Islam, N., Xu, Y., Beard, H. S., Garrett, W. M., Gu, G., et al. (2018). Quantitative proteomic analysis of staphylococcus aureus treated with punicalagin, a natural antibiotic from pomegranate that disrupts iron homeostasis and induces SOS. Proteomics 18:e1700461. doi: 10.1002/pmic.201700461
Cornforth, D. M., and Foster, K. R. (2013). Competition sensing: the social side of bacterial stress responses. Nat. Rev. Microbiol. 11, 285–293. doi: 10.1038/nrmicro2977
Costa, T. R., Felisberto-Rodrigues, C., Meir, A., Prevost, M. S., Redzej, A., Trokter, M., et al. (2015). Secretion systems in gram-negative Bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 13, 343–359. doi: 10.1038/nrmicro3456
Coulthurst, S. J., Barnard, A. M., and Salmond, G. P. (2005). Regulation and biosynthesis of carbapenem antibiotics in Bacteria. Nat. Rev. Microbiol. 3, 295-306.
Daligault, H., Davenport, K., Minogue, T., Broomall, S., Bruce, D., Chain, P., et al. (2014). Genome assembly of Serratia marcescens type strain ATCC 13880. Genome Announc. 2:e00967-14. doi: 10.1128/genomeA.00967-14
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Delany, I., Rappuoli, R., and Scarlato, V. (2004). Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol. Microbiol. 52, 1081–1090. doi: 10.1111/j.1365-2958.2004.04030.x
Desriac, F., Defer, D., Bourgougnon, N., Brillet, B., Le Chevalier, P., and Fleury, Y. (2010). Bacteriocin as weapons in the marine animal-associated Bacteria warfare: inventory and potential applications as an aquaculture probiotic. Mar. Drugs 8, 1153–1177. doi: 10.3390/md8041153
Donn, S., Kirkegaard, J. A., Perera, G., Richardson, A. E., and Watt, M. (2015). Evolution of bacterial communities in the wheat crop rhizosphere. Environ. Microbiol. 17, 610–621. doi: 10.1111/1462-2920.12452
Dunham, N. P., Chang, W.-C., Mitchell, A. J., Martinie, R. J., Zhang, B., Bergman, J. A., et al. (2018). Two distinct mechanisms for C–C desaturation by Iron (II)-and 2-(Oxo) glutarate-Dependent oxygenases: importance of α-heteroatom Assistance. J. Am. Chem. Soc. 140, 7116–7126. doi: 10.1021/jacs.8b01933
Durrant, A. (2016). Antimicrobial Production by Pectobacterium Carotovorum Subspecies Brasiliensis and its Role in Competitive Fitness of the Potato Pathogen. Philadlphia, PA: Lincoln University.
Erill, I., Campoy, S., and Barbé, J. (2007). Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol. Rev. 31, 637–656. doi: 10.1111/j.1574-6976.2007.00082.x
Fadrosh, D. W., Ma, B., Gajer, P., Sengamalay, N., Ott, S., Brotman, R. M., et al. (2014). An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2:6. doi: 10.1186/2049-2618-2-6
Fang, Y., Mercer, R. G., McMullen, L. M., and Gänzle, M. G. (2017). Induction of Shiga toxin prophage by abiotic environmental stress in food. Appl. Environ. Microbiol. 83, e1378–e1317. doi: 10.1128/AEM.01378-17
Fones, H., and Preston, G. M. (2013). The impact of transition metals on bacterial plant disease. FEMS Microbiol. Rev. 37, 495–519. doi: 10.1111/1574-6976.12004
Foster, J. W. (1991). Salmonella acid shock proteins are required for the adaptive acid tolerance response. J. Bacteriol. 173, 6896–6902. doi: 10.1128/jb.173.21.6896-6902.1991
García-Bayona, L., and Comstock, L. E. (2018). Bacterial antagonism in host-associated microbial communities. Science 361:eaat2456. doi: 10.1126/science.aat2456
Glazebrook, J., and Roby, D. (2018). Plant biotic interactions: from conflict to collaboration. Plant J. 93, 589–591. doi: 10.1111/tpj.13812
Goyal, R. K., and Mattoo, A. K. (2014). Multitasking antimicrobial peptides in plant development and host defense against biotic/abiotic stress. Plant Sci. 228, 135–149. doi: 10.1016/j.plantsci.2014.05.012
Grinter, R., Milner, J., and Walker, D. (2012). Ferredoxin containing bacteriocins suggest a novel mechanism of iron uptake in Pectobacterium spp. PloS One 7:e33033. doi: 10.1371/journal.pone.0033033
Gupta, A., Gopal, M., Thomas, G. V., Manikandan, V., Gajewski, J., Thomas, G., et al. (2014). Whole genome sequencing and analysis of plant growth promoting Bacteria isolated from the rhizosphere of plantation crops coconut, cocoa and arecanut. PLoS One 9:e104259. doi: 10.1371/journal.pone.0104259
Hachani, A., Wood, T. E., and Filloux, A. (2016). Type VI secretion and anti-host effectors. Curr. Opin. Microbiol. 29, 81–93. doi: 10.1016/j.mib.2015.11.006
Hamed, R. B., Gomez-Castellanos, J. R., Henry, L., Ducho, C., McDonough, M. A., and Schofield, C. J. (2013). The enzymes of β-lactam biosynthesis. Nat. Prod. Rep. 30, 21–107. doi: 10.1039/C2NP20065A
Hassan, H. M., and Troxell, B. (2013). Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic Bacteria. Front. Cell. Infect. Microbiol. 3:59. doi: 10.3389/fcimb.2013.00059
Heeb, S., Itoh, Y., Nishijyo, T., Schnider, U., Keel, C., Wade, J., et al. (2000). Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated Bacteria. Mol. Plant Microbe Int. 13, 232–237. doi: 10.1094/mpmi.2000.13.2.232
Hibbing, M. E., Fuqua, C., Parsek, M. R., and Peterson, S. B. (2010). Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25. doi: 10.1038/nrmicro2259
Hong, Y., Liao, D., Hu, A., Wang, H., Chen, J., Khan, S., et al. (2015). Diversity of endophytic and rhizoplane bacterial communities associated with exotic Spartina alterniflora and native mangrove using Illumina amplicon sequencing. Can. J. Microbiol. 61, 723–733. doi: 10.1139/cjm-2015-0079
Hyman, P., and Abedon, S. T. (2010). Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 70, 217–248. doi: 10.1016/S0065-2164(10)70007-1
Jarvis, B. (1967). Resistance to nisin and production of nisin-inactivating enzymes by several Bacillus species. Microbiology 47, 33–48. doi: 10.1099/00221287-47-1-33
Kazakov, A. E., Cipriano, M. J., Novichkov, P. S., Minovitsky, S., Vinogradov, D. V., Arkin, A., et al. (2006). RegTransBase-a database of regulatory sequences and interactions in a wide range of prokaryotic genomes. Nucleic Acids Res. 35(Suppl. 1), D407–D412.
Kilic, S., White, E. R., Sagitova, D. M., Cornish, J. P., and Erill, I. (2014). CollecTF: a database of experimentally validated transcription factor-binding sites in Bacteria. Nucleic Acids Res. 42, D156–D160. doi: 10.1093/nar/gkt1123
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1–e1. doi: 10.1093/nar/gks808
Knief, C., Delmotte, N., Chaffron, S., Stark, M., Innerebner, G., Wassmann, R., et al. (2012). Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 6, 1378–1390. doi: 10.1038/ismej.2011.192
Kõiv, V., Roosaare, M., Vedler, E., Kivistik, P. A., Toppi, K., Schryer, D. W., et al. (2015). Microbial population dynamics in response to Pectobacterium atrosepticum infection in potato tubers. Sci. Rep. 5:11606. doi: 10.1038/srep11606
Lagendijk, E. L., Validov, S., Lamers, G. E., De Weert, S., and Bloemberg, G. V. (2010). Genetic tools for tagging gram-negative Bacteria with mCherry for visualization in vitro and in natural habitats, biofilm and pathogenicity studies. FEMS Microbiol. Lett. 305, 81–90. doi: 10.1111/j.1574-6968.2010.01916.x
Leaden, L., Silva, L. G., Ribeiro, R. A., dos Santos, N. M., Lorenzetti, A. P., Alegria, T. G., et al. (2018). Iron deficiency generates oxidative stress and activation of the SOS response in Caulobacter crescentus. Front. Microbiol. 9:2014. doi: 10.3389/fmicb.2018.02014
Lee, D. H., Kim, J.-B., Lim, J.-A., Han, S.-W., and Heu, S. (2014). Genetic diversity of Pectobacterium carotovorum subsp. Brasiliensis isolated in Korea. Plant pathol. J. 30, 117–124. doi: 10.5423/PPJ.OA.12.2013.0117
Li, L., Stoeckert, C. J. Jr., and Roos, D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. doi: 10.1101/gr.1224503
Luis, G., Rubio, C., González-Weller, D., Gutiérrez, A. J., Revert, C., and Hardisson, A. (2011). Comparative study of the mineral composition of several varieties of potatoes (Solanum tuberosum L.) from different countries cultivated in Canary Islands (Spain.). Int. J. Food Sci. Technol. 46, 774–780. doi: 10.1111/j.1365-2621.2011.02556.x
Lutgring, J. D., and Limbago, B. M. (2016). The problem of carbapenemase producing carbapenem-resistant Enterobacteriaceae detection. J. Clin. Microbiol. 54, 529–534. doi: 10.1128/JCM.02771-15
Ma, B., Hibbing, M. E., Kim, H.-S., Reedy, R. M., Yedidia, I., Breuer, J., et al. (2007). Host range and molecular phylogenies of the soft rot enterobacterial genera Pectobacterium and Dickeya. Phytopathology 97, 1150–1163. doi: 10.1094/PHYTO-97-9-1150
Ma, X., Schloop, A., Swingle, B., and Perry, K. L. (2018). Pectobacterium and Dickeya responsible for potato blackleg disease in New York State in 2016. Plant Dis. 102, 1834–1840. doi: 10.1094/PDIS-10-17-1595-RE
Malek, M., Acharjee, M., and Rahman, T. (2013). Microbiological profile of potato samples collected from bangladesh agricultural research institute (BARI) and notification of anti-bacterial traits. Stamford J. Microbiol. 3, 21–25. doi: 10.3329/sjm.v3i1.22747
Marquez-Villavicencio, M. D. P., Groves, R. L., and Charkowski, A. O. (2011). Soft rot disease severity is affected by potato physiology and Pectobacterium taxa. Plant Dis. 95, 232–241. doi: 10.1094/PDIS-07-10-0526
Martin, M., Dragoš, A., Hölscher, T., Maróti, G., Bálint, B., Westermann, M., et al. (2017). De novo evolved interference competition promotes the spread of biofilm defectors. Nat. Commun. 8:15127. doi: 10.1038/ncomms15127
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12.
Mattinen, L., Nissinen, R., Riipi, T., Kalkkinen, N., and Pirhonen, M. (2007). Host-extract induced changes in the secretome of the plant pathogenic bacterium Pectobacterium atrosepticum. Proteomics 7, 3527–3537. doi: 10.1002/pmic.200600759
Mazzotta, A. S., Crandall, A. D., and Montville, T. J. (1997). Nisin resistance in Clostridium botulinum spores and vegetative cells. Appl. Environ. Microbiol. 63, 2654–2659.
McGowan, S., Sebaihia, M., Jones, S., Yu, B., Bainton, N., Chan, P., et al. (1995). Carbapenem antibiotic production in Erwinia carotovora is regulated by CarR, a homologue of the LuxR transcriptional activator. Microbiology 141, 541–550. doi: 10.1099/13500872-141-3-541
McGowan, S., Sebaihia, M., O’leary, S., Hardie, K., Williams, P., Stewart, G., et al. (1997). Analysis of the carbapenem gene cluster of Erwinia carotovora: definition of the antibiotic biosynthetic genes and evidence for a novel β-lactam resistance mechanism. Mol. Microbiol. 26, 545–556. doi: 10.1046/j.1365-2958.1997.6001974.x
McGowan, S. J., Barnard, A. M., Bosgelmez, G., Sebaihia, M., Simpson, N. J., Thomson, N. R., et al. (2005). Carbapenem antibiotic biosynthesis in Erwinia carotovora is regulated by physiological and genetic factors modulating the quorum sensing-dependent control pathway. Mol. Microbiol. 55, 526–545. doi: 10.1111/j.1365-2958.2004.04397.x
Miller, J. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Ming, X., and Daeschel, M. A. (1993). Nisin resistance of foodborne Bacteria and the specific resistance responses of Listeria monocytogenes Scott A. J. Food Prot. 56, 944–948. doi: 10.4315/0362-028X-56.11.944
Miyata, S. T., Bachmann, V., and Pukatzki, S. (2013). Type VI secretion system regulation as a consequence of evolutionary pressure. J. Med. Microbiol. 62, 663–676. doi: 10.1099/jmm.0.053983-0
Moleleki, L. N., Pretorius, R. G., Tanui, C. K., Mosina, G., and Theron, J. (2017). A quorum sensing-defective mutant of Pectobacterium carotovorum ssp. brasiliense 1692 is attenuated in virulence and unable to occlude xylem tissue of susceptible potato plant stems. Mol. Plant Pathol. 18, 32–44. doi: 10.1111/mpp.12372
Moraes, A., Souza, E., Mariano, R., Silva, A., Lima, N., Peixoto, A., et al. (2017). First report of Pectobacterium aroidearum and Pectobacterium carotovorum subsp. brasiliensis causing soft rot of cucurbita pepo in brazil. Plant Dis. 101:379. doi: 10.1094/pdis-08-16-1168-pdn
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. doi: 10.1126/science.1128393
Munch, R., Hiller, K., Barg, H., Heldt, D., Linz, S., Wingender, E., et al. (2003). PRODORIC: prokaryotic database of gene regulation. Nucleic Acids Res. 31, 266–269. doi: 10.1093/nar/gkg037
Ondov, B. D., Bergman, N. H., and Phillippy, A. M. (2011). Interactive metagenomic visualization in a Web browser. BMC Bioinformat. 12:385. doi: 10.1186/1471-2105-12-385
Onkendi, E. M., and Moleleki, L. N. (2014). Characterization of Pectobacterium carotovorum subsp. carotovorum and brasiliense from diseased potatoes in Kenya. Eur. J. Plant pathol. 139, 557–566. doi: 10.1007/s10658-014-0411-z
Panda, P., Fiers, M., Armstrong, K., and Pitman, A. (2012). First report of blackleg and soft rot of potato caused by Pectobacterium carotovorum subsp. brasiliensis in New Zealand. New Dis. Rep. 26, 2044–2588.
Poole, S. J., Diner, E. J., Aoki, S. K., Braaten, B. A., de Roodenbeke, C. T. K., Low, D. A., et al. (2011). Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genetics 7:e1002217. doi: 10.1371/journal.pgen.1002217
Porcheron, G., and Dozois, C. M. (2015). Interplay between iron homeostasis and virulence: fur and RyhB as major regulators of bacterial pathogenicity. Vet. Microbiol. 179, 2–14. doi: 10.1016/j.vetmic.2015.03.024
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. 103, 1528–1533. doi: 10.1073/pnas.0510322103
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Raoul des Essarts, Y., Pédron, J., Blin, P., Van Dijk, E., Faure, D., and Van Gijsegem, F. (2019). Common and distinctive adaptive traits expressed in Dickeya dianthicola and Dickeya solani pathogens when exploiting potato plant host. Environ. Microbiol. 21, 1004–1018. doi: 10.1111/1462-2920.14519
Riley, M. A., and Gordon, D. M. (1999). The ecological role of bacteriocins in bacterial competition. Trends Microbiol. 7, 129–133. doi: 10.1016/s0966-842x(99)01459-6
Rivet, M. M. (1998). Investigation into the Regulation of Exoenzyme Production in Erwinia Carotovora Subspecies Carotovora. Coventry: University of Warwick.
Rodriguez-Moreno, L., Ebert, M. K., Bolton, M. D., and Thomma, B. P. (2018). Tools of the crook-infection strategies of fungal plant pathogens. Plant J. 93, 664–674. doi: 10.1111/tpj.13810
Roh, E., Lee, S., Lee, Y., Ra, D., Choi, J., Moon, E., et al. (2009). Diverse antibacterial activity of Pectobacterium carotovorum subsp. carotovorum isolated in Korea. J. Microbiol. Biotechnol. 19, 42–50.
Roh, E., Park, T.-H., Kim, M.-I., Lee, S., Ryu, S., Oh, C.-S., et al. (2010). Characterization of a new bacteriocin, Carocin D, from Pectobacterium carotovorum subsp. carotovorum Pcc21. Appl. Environ. Microbiol. 76, 7541–7549. doi: 10.1128/AEM.03103-09
Russell, A. B., Hood, R. D., Bui, N. K., LeRoux, M., Vollmer, W., and Mougous, J. D. (2011). Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347. doi: 10.1038/nature10244
Russell, A. B., Peterson, S. B., and Mougous, J. D. (2014). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148. doi: 10.1038/nrmicro3185
Salomon, D., Klimko, J. A., and Orth, K. (2014). H-NS regulates the Vibrio parahaemolyticus type VI secretion system 1. Microbiology 160(Pt 9), 1867–1873. doi: 10.1099/mic.0.080028-0
Schreiter, S., Ding, G.-C., Heuer, H., Neumann, G., Sandmann, M., Grosch, R., et al. (2014). Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front. Microbiol. 5:144. doi: 10.3389/fmicb.2014.00144
Seo, S. W., Kim, D., Latif, H., O’Brien, E. J., Szubin, R., and Palsson, B. O. (2014). Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat. Commun. 5:4910. doi: 10.1038/ncomms5910
Shyntum, D. Y., Nkomo, N., Gricia, A. R., Shigange, N. L., Bellieny-Rabelo, D., and Moleleki, L. N. (2018). The Impact of Type VI Secretion System, Bacteriocins and Antibiotics on Competition Amongst Soft-Rot Enterobacteriaceae: Regulation of Carbapenem Biosynthesis by Iron and the Transcriptional regulator Fur. bioRxiv. [preprint]. doi: 10.1101/497016
Shyntum, D. Y., Theron, J., Venter, S. N., Moleleki, L. N., Toth, I. K., and Coutinho, T. A. (2015). Pantoea ananatis utilizes a type VI secretion system for pathogenesis and bacterial competition. Mol. Plant Microbe Int. 28, 420–431. doi: 10.1094/MPMI-07-14-0219-R
Silverman, J. M., Brunet, Y. R., Cascales, E., and Mougous, J. D. (2012). Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol. 66, 453–472. doi: 10.1146/annurev-micro-121809-151619
Skaar, E. P. (2010). The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathogens 6:e1000949. doi: 10.1371/journal.ppat.1000949
Stubbendieck, R. M., and Straight, P. D. (2015). Escape from lethal bacterial competition through coupled activation of antibiotic resistance and a mobilized subpopulation. PLoS Genetics 11:e1005722. doi: 10.1371/journal.pgen.1005722
Stubbendieck, R. M., and Straight, P. D. (2016). Multifaceted interfaces of bacterial competition. J. Bacteriol. 198, 2145–2155. doi: 10.1128/JB.00275-16
Tack, F. M. (2014). Trace elements in potato. Potato Res. 57, 311–325. doi: 10.1007/s11540-014-9268-y
Tanui, C. K., Shyntum, D. Y., Priem, S. L., Theron, J., and Moleleki, L. N. (2017). Influence of the ferric uptake regulator (Fur) protein on pathogenicity in Pectobacterium carotovorum subsp. brasiliense. PloS One 12:e0177647. doi: 10.1371/journal.pone.0177647
Thorgersen, M. P., Lancaster, W. A., Ge, X., Zane, G. M., Wetmore, K. M., Vaccaro, B. J., et al. (2017). Mechanisms of chromium and uranium toxicity in Pseudomonas stutzeri RCH2 Grown under anaerobic nitrate-reducing conditions. Front. Microbiol. 8:1529. doi: 10.3389/fmicb.2017.01529
Trunk, K., Peltier, J., Liu, Y.-C., Dill, B. D., Walker, L., Gow, N. A., et al. (2018). The type VI secretion system deploys antifungal effectors against microbial competitors. Nat. Microbiol. 3, 920–931. doi: 10.1038/s41564-018-0191-x
Tyc, O., Song, C., Dickschat, J. S., Vos, M., and Garbeva, P. (2017). The ecological role of volatile and soluble secondary metabolites produced by soil bacteria. Trends Microbiol. 25, 280–292. doi: 10.1016/j.tim.2016.12.002
van der Merwe, J. J., Coutinho, T. A., Korsten, L., and van der Waals, J. E. (2010). Pectobacterium carotovorum subsp. brasiliensis causing blackleg on potatoes in South Africa. Eur. J. Plant Pathol. 126, 175–185. doi: 10.1007/s10658-009-9531-2
Verster, A. J., Ross, B. D., Radey, M. C., Bao, Y., Goodman, A. L., Mougous, J. D., et al. (2017). The landscape of type VI secretion across human gut microbiomes reveals its role in community composition. Cell Host Microbe 22, 411.e4–419.e4. doi: 10.1016/j.chom.2017.08.010
Voronina, M. V., Kabanova, A. P., Shneider, M. M., Korzhenkov, A. A., Toschakov, S. V., Miroshnikov, K. K., et al. (2019). First report of pectobacterium carotovorum subsp. brasiliense causing blackleg and stem rot disease of potato in russia. Plant Dis. 103:364.
Wall, D. (2016). Kin recognition in Bacteria. Annu. Rev. Microbiol. 70, 143–160. doi: 10.1146/annurev-micro-102215-095325
Wang, C., Pu, T., Lou, W., Wang, Y., Gao, Z., Hu, B., et al. (2018). Hfq, a RNA chaperone, contributes to virulence by regulating plant cell wall–degrading enzyme production, type VI secretion system expression, bacterial competition, and suppressing host defense response in Pectobacterium carotovorum. Mol. Plant Microbe Int. 31, 1166–1178. doi: 10.1094/mpmi-12-17-0303-r
Xin, X.-F., Kvitko, B., and He, S. Y. (2018). Pseudomonas syringae: what it takes to be a pathogen. Nat. Rev. Microbiol. 16, 316–328. doi: 10.1038/nrmicro.2018.17
Yamada, K., Hirota, M., Niimi, Y., Nguyen, H. A., Takahara, Y., Kamio, Y., et al. (2006). Nucleotide sequences and organization of the genes for carotovoricin (Ctv) from Erwinia carotovora indicate that Ctv evolved from the same ancestor as Salmonella typhi prophage. Biosci. Biotechnol. Biochem. 70, 2236–2247. doi: 10.1271/bbb.60177
Yang, H., Cheng, J., Hu, L., Zhu, Y., and Li, J. (2012). Mechanisms of antimicrobial resistance in Serratia marcescens. Afr. J. Microbiol. Res. 6, 4427–4437.
Keywords: Enterobacteriaceae, type VI secretion system, Fur, carbapenem, Pectobacterium brasiliense, bacterial competition, potato microbiome, microbial interactions
Citation: Shyntum DY, Nkomo NP, Shingange NL, Gricia AR, Bellieny-Rabelo D and Moleleki LN (2019) The Impact of Type VI Secretion System, Bacteriocins and Antibiotics on Bacterial Competition of Pectobacterium carotovorum subsp. brasiliense and the Regulation of Carbapenem Biosynthesis by Iron and the Ferric-Uptake Regulator. Front. Microbiol. 10:2379. doi: 10.3389/fmicb.2019.02379
Received: 05 August 2019; Accepted: 30 September 2019;
Published: 18 October 2019.
Edited by:
Jayachandran N. Kizhakkedathu, The University of British Columbia, CanadaReviewed by:
Sébastien Bontemps-Gallo, Institut Pasteur de Lille, FranceYong-Qiang He, Guangxi University, China
Copyright © 2019 Shyntum, Nkomo, Shingange, Gricia, Bellieny-Rabelo and Moleleki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lucy Novungayo Moleleki, bHVjeS5tb2xlbGVraUB1cC5hYy56YQ==
 Divine Yufetar Shyntum
Divine Yufetar Shyntum Ntombikayise Precious Nkomo
Ntombikayise Precious Nkomo Ntwanano Luann Shingange
Ntwanano Luann Shingange Alessandro Rino Gricia
Alessandro Rino Gricia Daniel Bellieny-Rabelo
Daniel Bellieny-Rabelo Lucy Novungayo Moleleki
Lucy Novungayo Moleleki